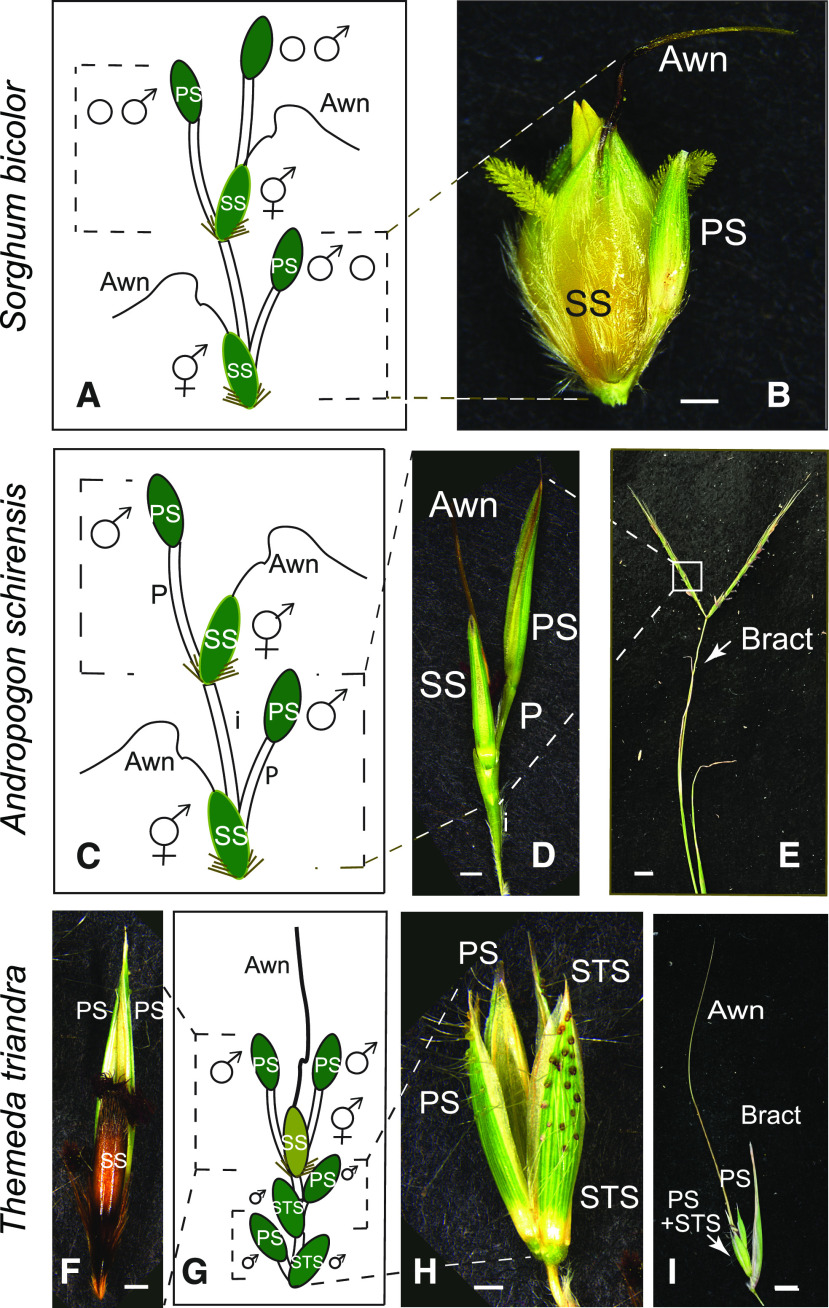
Apparently nonfunctional floral structures (spikelets) provide carbon for the developing grain in sorghum and two related wild species, thereby contributing to yield (fitness).
Abstract
Sorghum (Sorghum bicolor) and its relatives in the grass tribe Andropogoneae bear their flowers in pairs of spikelets in which one spikelet (seed-bearing or sessile spikelet [SS]) of the pair produces a seed and the other is sterile or male (staminate). This division of function does not occur in other major cereals such as wheat (Triticum aestivum) or rice (Oryza sativa). Additionally, one bract of the SS spikelet often produces a long extension, the awn, that is in the same position as, but independently derived from, that of wheat and rice. The function of the sterile spikelet is unknown and that of the awn has not been tested in Andropogoneae. We used radioactive and stable isotopes of carbon, RNA sequencing of metabolically important enzymes, and immunolocalization of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) to show that the sterile spikelet assimilates carbon, which is translocated to the largely heterotrophic SS. The awn shows no evidence of photosynthesis. These results apply to distantly related species of Andropogoneae. Removal of sterile spikelets in sorghum significantly decreases seed weight (yield) by ∼9%. Thus, the sterile spikelet, but not the awn, affects yield in the cultivated species and fitness in the wild species.
INTRODUCTION
The grain and bioenergy crop sorghum (Sorghum bicolor), along with most of the 1200 species in the grass tribe Andropogoneae, produces more sterile than fertile flowers in the inflorescence (Clayton and Renvoize, 1986; Schneider and Vegettti, 1992; Watson and Dallwitz, 1992 onward; McKone et al., 1998; Le Roux and Kellogg, 1999; Vegetti, 1999; Hodge and Kellogg, 2014; Gladman et al., 2019). This pattern is counterintuitive in that the plant appears to be sacrificing potential reproductive output by taking many of its floral structures “off line.” The sterile flowers must therefore have another function maintained through evolution to compensate for the loss of reproductive potential.

The seed of sorghum, like that of all grasses, is enclosed in floral bracts (glumes and lemmas) that together form a terminal unit known as a spikelet (a little spike; Clayton and Renvoize, 1986; Watson and Dallwitz, 1992 onward; Kellogg, 2015). The seed-bearing or sessile spikelet (SS) sits directly on the inflorescence axis and is paired with a pedicellate (stalked) spikelet (PS; Figures 1A and 1B). Unlike the seed-bearing SS, which is bisexual, the PS is generally sterile in sorghum, although in some lines the PS produces stamens (Burow et al., 2014; Jiao et al., 2018; Gladman et al., 2019). Presence of a PS, and its shape, size, and often sex expression, appears to be genetically fixed among plants of any given accession (E.A.K., personal observation).
Figure 1.
Spikelet Pair Structures.
(A) to (I) S. bicolor (see [A] and [B]), A. schirensis (see [C] to [E]), and T. triandra (see [F] to [I]). (A), (C), and (G) Spikelet pairs, marked by dashed lines. Sex expression of each spikelet is indicated. The SS includes a bisexual flower and bears the seed and also bears a twisted awn from the lemma (floral bract).
(A) Terminal spikelet (S. bicolor) is morphologically identical to a PS. PS may be either sterile (most commonly) or staminate.
(B) Spikelet pair of S. bicolor accession SAP-15 (PI 656014). Bar = 1 mm.
(C) PS is staminate.
(D) Spikelet pair of A. schirensis. Bar = 1 mm.
(E) Inflorescence of A. schirensis, showing two branches, each bearing 9 to 10 spikelet pairs. The uppermost leaf bears a reduced blade (arrow). Bar = 1 cm.
(F) Spikelet pair of T. triandra, showing the dark indurate SS, with two greenish PSs behind. Bar = 1 mm.
(G) Inflorescence structure in T. triandra, with three spikelet pairs and a terminal spikelet that is morphologically identical to the PS. Spikelets in the proximal two pairs (both pedicellate and sessile) are all staminate. STS, staminate sessile spikelet.
(H) Proximal spikelet pairs of T. triandra, here called PS and STS since all four are staminate. Bar = 1 mm. STS, staminate sessile spikelet.
(I) Inflorescence branch of T. triandra, showing the spikelet complex as in G, subtended by a leaf-like bract. Bar = 5 mm. STS, staminate sessile spikelet.
The function of the PS is unknown in sorghum or in any other member of Andropogoneae in most of which the PS is also sterile or staminate (Clayton and Renvoize, 1986; Watson and Dallwitz, 1992 onward; Vegetti, 1999; Kellogg, 2000, 2015; Arthan et al., 2017). Because grasses are wind pollinated (Niklas, 1987; Friedman and Harder, 2004; Cresswell et al., 2010), the PS cannot function in pollinator attraction as sterile flowers do in some other angiosperms (Stuessy et al., 1986; Krannitz and Maun, 1991; Nielsen et al., 2002; Jin et al., 2010; Morales et al., 2013). In some wild species, the PS may function in seed dispersal (Davidse, 1987; Doust et al., 2014; Linder et al., 2018) and/or in controlling pollen-to-ovule ratio (Connor, 1981, 1987). However, the dispersal function is not universal nor is it relevant in cultivated sorghum in which the grains are not shed, and pollen production in the PS only occurs in a few accessions.
The spikelet pair in sorghum may also include an awn on the SS (Figures 1A and 1B; Sieglinger et al., 1934). The awn, when present, is a slender extension of the lemma (floral bract) of the upper flower and may be twisted and geniculate. Like the PS, the presence, shape, and size of the awn appear to be genetically fixed within an accession (Jowett, 1968), and its function has not been demonstrated (“the awn is not known to have any protective or physiologic value to the sorghum plant”; Sieglinger et al., 1934). In some wild Andropogoneae, the awn is hygroscopic and can help orient the spikelet in the soil to enhance germination (Peart, 1979, 1981, 1984; Peart and Clifford, 1987). Awns in wheat (Triticum aestivum) and rice (Oryza sativa) are vascularized and assimilate carbon that is then transferred to the grain (Grundbacher, 1963; Motzo and Giunta, 2002; Li et al., 2006; Simkin et al., 2020), often contributing appreciably to yield; however, wheat and rice have solitary (unpaired) spikelets and the awns may have evolved independently of those in Andropogoneae (Teisher et al., 2017). Carbon assimilation has not been assessed in awns or more generally in any floral structures of Andropogoneae.
The PS, the awn, or both could produce photosynthate that contributes to grain filling. The PS is green, which suggests that it could be photosynthetically active, but its tiny size (3 to 6 mm long) makes it unclear whether it could assimilate and export enough carbon to contribute materially to the carbon economy of other floral organs. The SS is slightly longer (∼4 to 9 mm) and more broadly ovate, but it does not stay green as long as the PS. By contrast, the awn turns brown soon after heading, suggesting limited capacity for photosynthesis. If so, its function must differ from that of the wheat or rice awn. The awn is also small (4 to 18 mm) and in most sorghum lines is missing entirely.
Here we test the hypothesis that the PS and awn contribute photosynthate to the developing seed in sorghum and other members of the tribe Andropogoneae. We report a combination of radioactive and stable carbon isotopic labeling and metabolite analyses, RNA sequencing (RNA-seq) experiments, immunolocalization, morphological observations with scanning electron microscopy, and spikelet removal experiments. Our goal was to assess photosynthetic capacity in these organs and their potential contributions to yield and fitness. Given that Andropogoneae, including sorghum, are C4 NADP-dependent malic enzyme (NADP-ME) subtype (Hattersley and Watson, 1975, 1992), we also considered whether the anatomy, gene expression, and ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) localization in the PS were consistent with a two-cell C4 shuttle, as suggested for inflorescence tissues in a few other grasses (Pengelly et al., 2011; Rangan et al., 2016a, 2016b; Henry et al., 2017).
RESULTS
SS and Awn Are Carbon Sinks; PS Is a Source
For an initial assessment of carbon assimilation, we used a pulse-chase experiment to assess whether sorghum spikelets or the awn could take up 14C. Inflorescence branches were exposed to 14C for 1 h. Measurements were standardized by weight (dpm/mg) and are presented as a percentage of the total counts in spikelets and awns (Figure 2A; Supplemental Table 1). The PS accounted for a significantly greater percentage of 14CO2 taken up than the SS or awn (P < 0.0000001), with the latter being scarcely detectable. This was true whether the inflorescences were intact (attached) or whether the spikelets and awns were detached and lying on the bottom of the flask, suggesting that relative carbon uptake by the individual structures was consistent regardless of the effects of transpiration (difference nonsignificant, P > 0.997).
Figure 2.
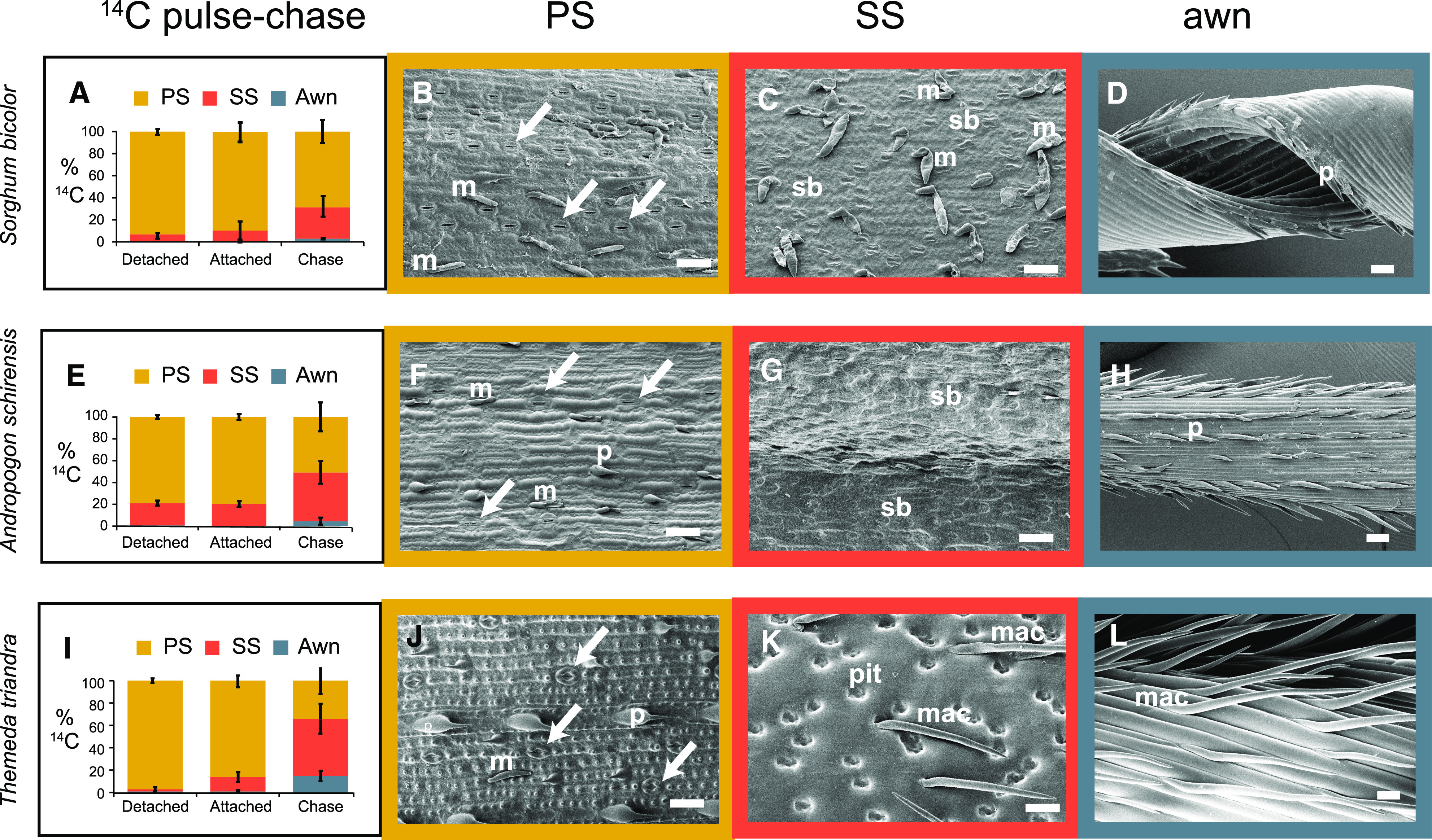
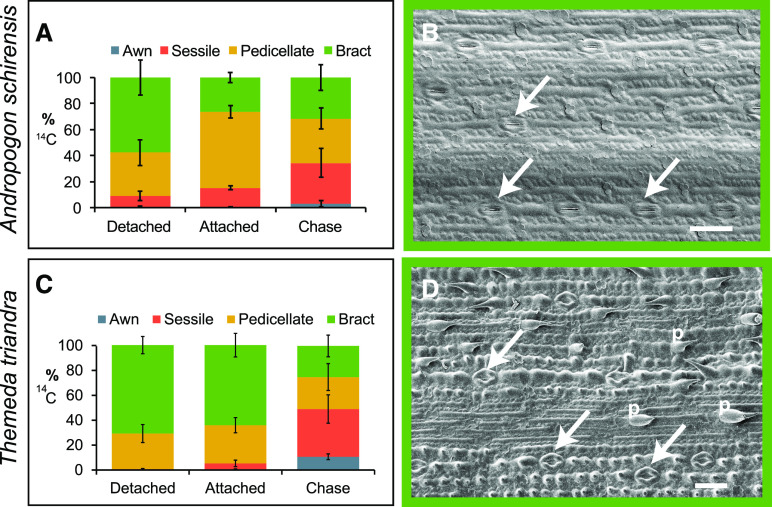
Results of 14C Pulse-Chase Experiments and Distribution of Stomata.
(A) to (L) S. bicolor (see [A] to [D]), A. schirensis (see [E] to [H]), and T. triandra (see [I] to [L]).
(A), (E), and (I) Percent dpm/mg for each organ after 1-h exposure to 14C with organs removed from the axis (detached), inflorescence intact (attached), or after 24-h chase (chase). Plot includes mean percentages and sds (n = 3).
(B), (F), and (J) Abaxial epidermis of PSs showing rows of stomata (arrows) and bicellular microhairs (M) and prickles (P).
(C), (G), and (K) Abaxial epidermis of SS showing no stomata, but bicellular microhairs (M) and silica bodies (sb) as well as large pits (pit) and macrohairs (mac) in T. triandra.
(D), (H), and (L) Awn showing no stomata or other epidermal structures, except for prickles in sorghum (D) and A. schirensis (H) and long microhairs in T. triandra (L).
Bar = 50 µm; note that (B) and (C) are more highly magnified than (D).
Intact inflorescences were also subjected to a pulse chase with 1-h pulse labeling followed by 24-h chase period in air (i.e., no additional labeled carbon provided). Over 24 h, the fraction of counts in the PS decreased and that of the SS increased significantly (0.01 < P < 0.5), indicating translocation of carbon from one structure to the other and other parts of the plant (Figure 2A; Supplemental Table 1). The fraction in the awn also increased but was still small relative to the spikelets and not significantly different from the 1-h labeling experiment.
To determine whether our result was specific to sorghum or might apply generally to Andropogoneae, we repeated the experiment with the distantly related species Themeda triandra and Andropogon schirensis that, like sorghum, have an SS and PS, with an awn on the SS (Figures 1C to 1I). These two species represent distinct clades of Andropogoneae and diverged ∼15 million years ago (Estep et al., 2014). The results were similar to those for sorghum: most 14C appeared in the PS after 1 h of labeling, whether attached to the inflorescence or not (difference nonsignificant, 0.5 < P < 1.0), but after a 24–h chase the proportion of label in the PS had decreased and that in the SS had increased significantly (0.0003 < P < 0.0022 [A. schirensis] and 1 × 10−5 < P < 4 × 10−5 [T. triandra]; Figures 2E and 2I; Supplemental Tables 2 and 3). Awns were scarcely labeled.
The surface morphology of each organ is consistent with what would be expected from the 14C results. The outer bracts (glumes) of the PS have obvious stomata in all three species (Figures 2B, 2F, and 2J) and are similar in this respect to leaves. By contrast, stomata are absent from the surface of the glumes of the SS (Figures 2C, 2G, and 2K), except near the apex in sorghum and on the sides in A. schirensis. No stomata were found on awns (Figures 2D, 2H, and 2L), unlike wheat (Li et al., 2006). Scanning electron microscopy data combined with the 14C data suggest that the PS may contribute photosynthate to the SS, whereas the awn may not.
PS Produces Isotopically Labeled Intermediates of Carbon Assimilation; SS and Awns Do Not
If the PS is indeed a source of carbon for the grain, it should produce metabolites characteristic of photosynthesis. To test this hypothesis, intact inflorescences were exposed to 13CO2 for 30, 60, or 300 s and then key metabolites were assessed using liquid chromatography–tandem mass spectrometry (LC-MS/MS). Knowing that inflorescence tissues often lack a fully developed C4 cycle, even in C4 plants such as sorghum or maize (Zea mays; Pengelly et al., 2011), we looked for evidence of photosynthetic metabolites, whether from C4 (e.g., malate), C3 (Calvin–Benson cycle intermediates such as phosphoglyceric acid [PGA] and fructose-6-phosphate), Suc (UDP-glucose [UDPG]), or starch (ADP-glucose [ADPG]) biosynthetic pathways. Conversely, we expected that SS and awn should be metabolically distinct, likely heterotrophic. Note that for this analysis, we were not attempting to determine whether the tissue should be classified as C3 or C4 but rather to determine which of several pathways might be active.
As in the 14C experiments, more carbon was labeled in the PS than in either the SS or awn. In a principal component analysis (PCA) of the metabolite results, the first principal component (PC1) accounted for more than 87% of the variance, distinguishing between values for unlabeled metabolites (loading just below 0) and those for labeled metabolites (loading positively; Figure 3A). By 5 min, metabolite labeling was greatest in the PS, whether considering the weighted average of all isotopologues for each metabolite (i.e., one data point per metabolite per organ and time point; Figure 3A) or individual isotopologues (i.e., three or four data points per metabolite per organ and time point; Figure 3B). Overall effects of organ, time, and organ with time were all significant (generally P ≤ 0.0001; Supplemental Tables 4A and 5A). Values for the PS were significantly different from those of either of the other two organs (generally P < 0.0001; Supplemental Tables 4B, 4C, 5B, and 5C), but the SS and awn were not significantly different from each other. In addition, the 5-min time point was significantly different from the 30- or 60-s pulses for most metabolites (P < 0.0001), with malate being an important exception. Malate was labeled between 30 and 60 s, possibly signifying NADP-ME C4 activity, but not between 60 and 300 s, indicating that initial labeling had stopped by 60 s (Supplemental Tables 4B and 4C).
Figure 3.
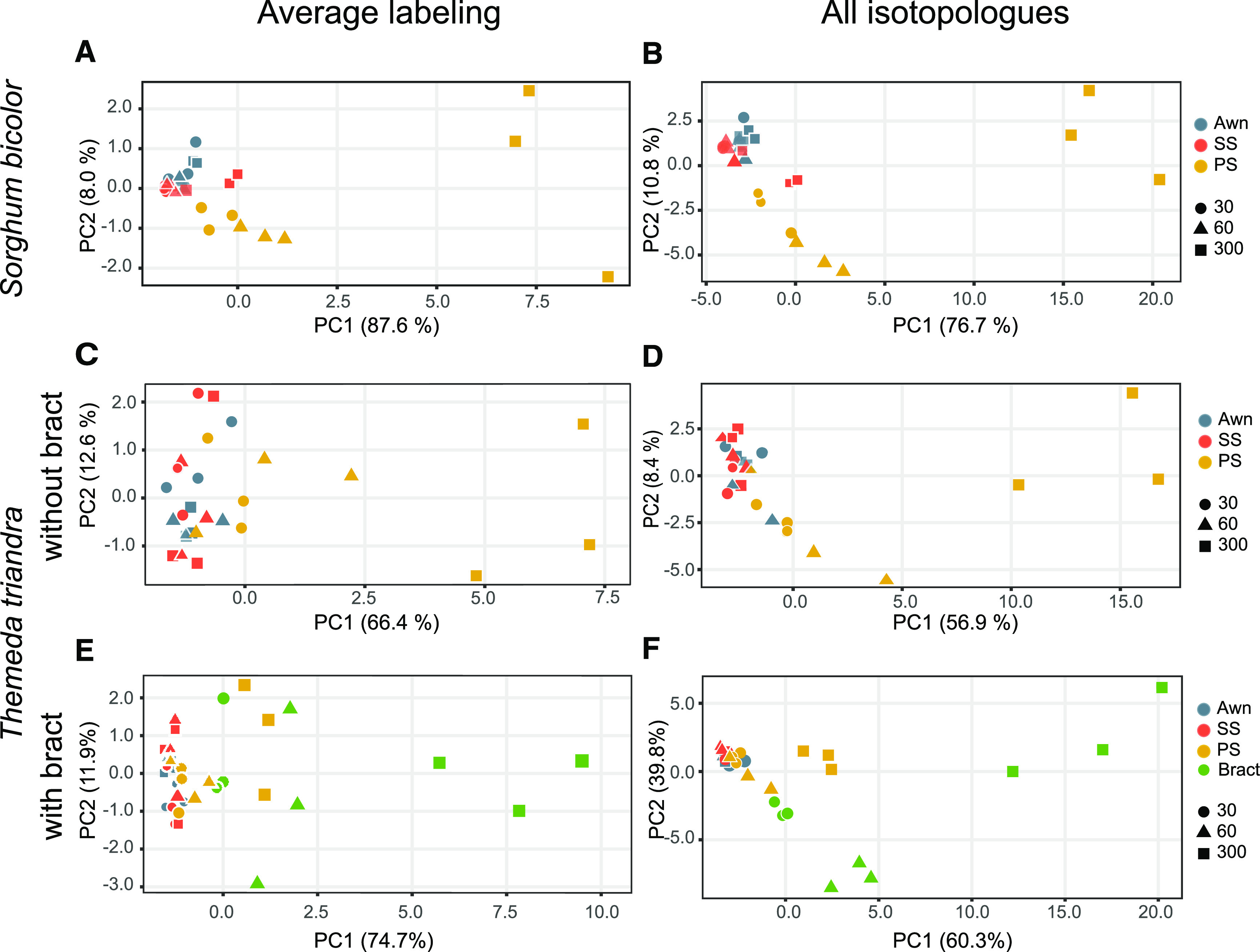
PCA of 13C-Labeled Metabolites.
(A) to (F) S. bicolor (see [A] and [B]) and T. triandra (see [C] to [F]). Values for awn and SS are not significantly different in either species for any time point nor for most individual metabolites. (A), (C), and (E) Average labeling. Each point is the weighted average of all labeled isotopologues for a given metabolite, organ, and time point. (B), (D), and (F) All isotopologues.
(A) Values for PS are significantly different from the other organs. See Supplemental Table 4.
(B) Awn and SS are significantly different for P5P and UDPG, but not otherwise; PS is significantly different from the other two organs except for PYR. Values at 30 and 60 s not significantly different for most metabolites, but the 300-s time point is distinct. See Supplemental Table 5.
(C) Values for PS are significantly different for six of nine metabolites, with the greatest difference in labeling at 300 s. See Supplemental Table 6.
(D) Awn and SS are significantly different for P5P, but not otherwise; PS is significantly different from the other two organs except for P5P, PGA, and PYR. Values at 30 and 60 s are similar for most metabolites, but the 300-s time point is distinct. See Supplemental Table 7.
(E) and (F) Values including bract; data for awn, SS and PS are the same as those in (C). (E) Bract is significantly different from all other organs for six out of eight metabolites. Values for PS are significantly different from awn and SS for only aspartate and PYR. The 300-s time point is significantly different from others. See Supplemental Table 11.
(F) Results are similar to those for average labeling, with bract being significantly different from all other organs for six out of eight metabolites and isotoplogues, and the greatest difference in labeling occurring at 300 s. See Supplemental Table 12.
Organs are distinguished by color, and time points are distinguished by shape. 30, values at 30 s of labeling; 60, values at 60 s of labeling; 300, values at 300 s of labeling.
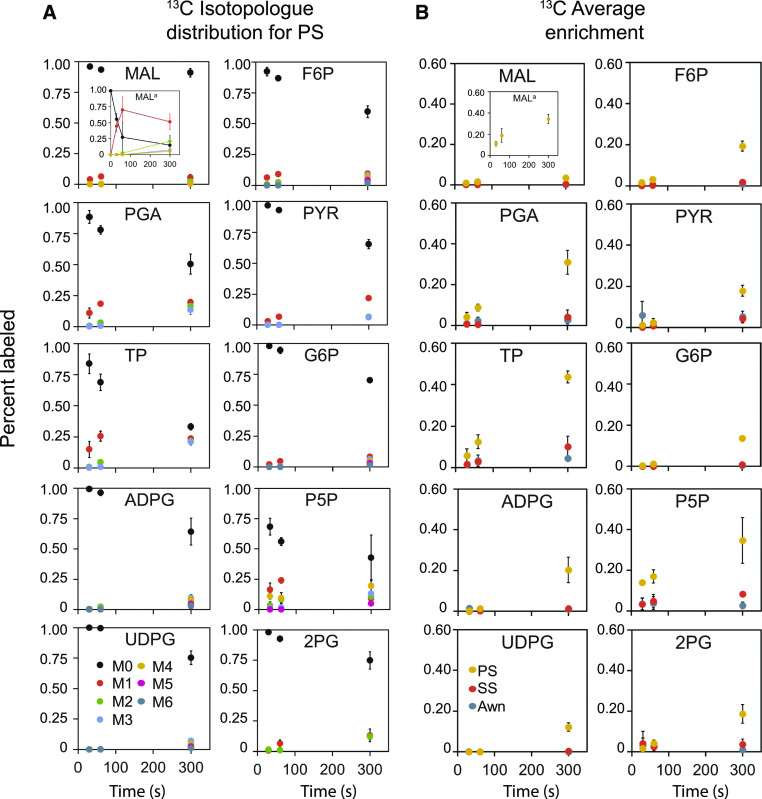
At the onset of a 13CO2 labeling experiment, the drop in the unlabeled isotopologue fraction for central metabolites indicates carbon assimilation. After correction for natural abundance, we see such a drop in metabolites in the PS (Figure 4A) along with increasing average fractional enrichment (Figure 4B), showing that primary metabolites were significantly labeled by 300 s. To further account for differing biomass compositions among organs that can confound metabolite pool sizes, we calculated the 13C atom equivalents formed (Supplemental Figure 1; inspired by Arrivault et al., 2017), which further confirmed that the PS is the only organ with significant 13C labeling and thus carbon assimilation.
Figure 4.
13C Labeling for Individual Metabolites at Three Time Points.
(A) 13C isotopologue distribution.
(B) Average labeling.
(A) and (B) Points are mean fractions for isotopologue distributions and average labeling; bars are sds, n = 3. Colors distinguish the three organs. Most label accumulation occurs in the PS and can be seen at 300 s. Insets represent the calculation of the active pool for malate. 2PG, 2-phosphoglycolate; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; MAL, malate; P5P, pentose 5 phosphates; PYR, pyruvate; TP, triose phosphates.
Labeling in malate appeared low, with the unlabeled isotopologue fraction, M0, scarcely decreasing between the 30- and 300-s time points (P = 0.0132; Figure 4A; Supplemental Table 5). However, most plant cells have a large inactive pool of malate (Arrivault et al., 2017), and we hypothesized that this pool might obscure changes in the dynamically labeled portion. By calculating the active malate pool in 13C, we found rapid incorporation at the 30- and 60-s time points in the PS (insets in Figures 4A and 4B). The rapid initial rise in the single-labeled active 13C-malate pool would be expected if there is a functional two-cell C4 shuttle using NADP-ME.
PGA, the immediate product of Rubisco-based carbon assimilation, and triose phosphate were rapidly labeled, as shown by the relative drop in M0 and increase in M1 (black and orange dots in Figure 4A), with the remaining unlabeled fraction in triose phosphate constituting <40% of the total pool in the PS after 5 min. Fructose-6-phosphate, which is also tied to the Calvin–Benson cycle, showed a similar but weaker labeling pattern. Pyruvate (PYR), a product of glycolysis and several steps removed from the Calvin–Benson cycle, was labeled (P ≤ 0.0001) linearly, as were other hexose and nucleotide hexose phosphates (glucose-6-phosphate, UDPG, and ADPG; all P ≤ 0.0001; Figures 4A and 4B; Supplemental Figure 1) used in Suc and starch biosynthesis, respectively. The linear slope of labeling in PYR and others within similar time frames indicates the delay due to C4-based CO2 delivery to the Calvin–Benson cycle and has been observed in other C4 studies (Weissmann et al., 2016; Arrivault et al., 2017), but not in C3 studies that result in hyperbolic labeling curves with less delay for three carbon intermediates during the same time frame (Ma et al., 2014). We also looked for 2-phosphoglycolate, glyoxylate, and glycolate, intermediates associated with photorespiration that could indicate C3 metabolism, although low levels of photorespiration may also occur in C4 plants (Zelitch et al., 2009). Typically, only a subset of these particular metabolites is reliably detected or reported (Weissmann et al., 2016; Arrivault et al., 2017). 2-Phosphoglycolate was labeled in the PS by 300 s, but not in the other organs (Figures 4A and 4B).
To test the generality of our results, we repeated the 13CO2 experiments with T. triandra. As in sorghum, the PS was clearly distinct from the SS and awn in a PCA. PC1 explained 66% of the variance and the overall pattern of the PCA was similar to that in sorghum (average labeling: Figure 3C; Supplemental Table 6; all isotopologues: Figure 3D; Supplemental Table 7), suggesting that the results may apply to many other members of Andropogoneae. Labeling of individual metabolites was also similar between the two species, except that aspartate labeling was significant (P < 0.001) and ADPG, PYR, and P5P labeling were nonsignificant (Supplemental Tables 6 and 7).
Photosynthetic Genes Are Expressed in Leaf and PS, but Not SS or Awn
To determine whether the metabolite data corresponded to underlying patterns of gene expression in the three organ types, we compared transcriptomes for spikelets and awns to those for leaves, focusing on a hand-curated set of 1441 genes encoding enzymes of central carbon metabolism, including photorespiration and also major transporters required for C4 photosynthesis (hereafter called metabolic genes; Supplemental Data Set). A PCA of the transcript data separated the leaf and SS along PC1 (Supplemental Figure 2A), whereas values for awn and PS separated most clearly along PC2, a pattern nearly identical to that of a PCA for all transcripts (Supplemental Figure 2B). The PCA indicates distinct differences in carbon metabolism among the four organ types. Of the 1441 metabolic genes, 922 were differentially expressed (DE) in at least one pairwise comparison of organs. A heatmap of these 922 transcripts (Supplemental Figure 3) found that genes upregulated in leaves were downregulated in the SS and vice versa. Expression in awns and PSs was clearly different from that of the other two organs. A distinct set of genes was upregulated in awns, but these appeared unrelated to either photosynthesis or sugar metabolism.
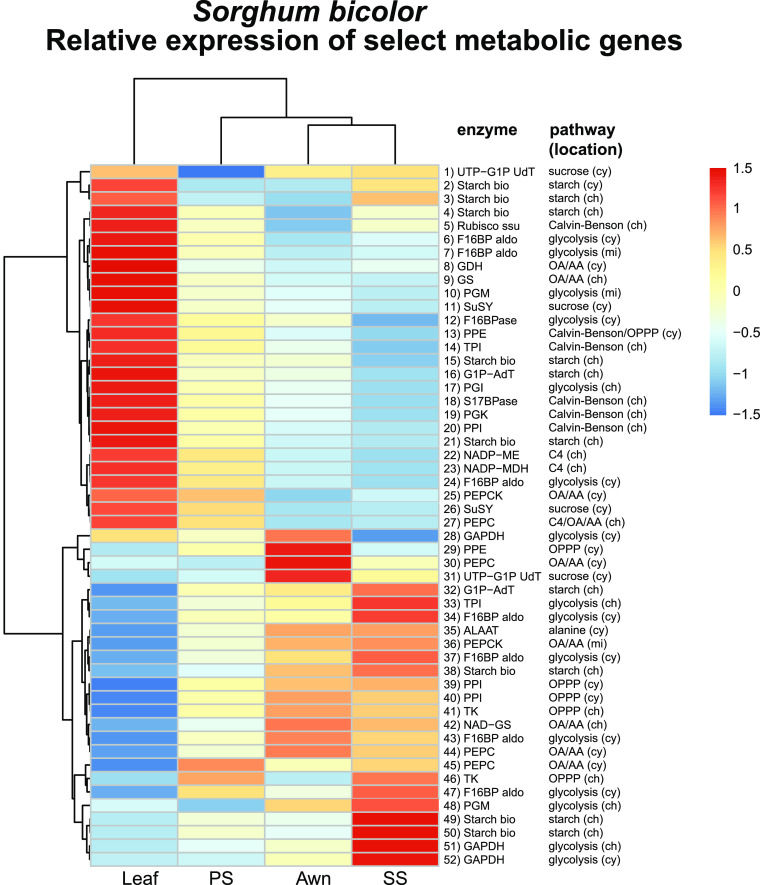
We examined expression of genes encoding the enzymes that directly produced the metabolites measured with 13C and other related central intermediates, a 52-gene subset of the 922 DE genes. Genes were assigned a provisional subcellular location by TargetP (Emanuelsson et al., 2007), although this should be regarded as a preliminary hypothesis. A heatmap of this subset (Figure 5) produced a pattern similar to that for the full set of metabolic genes (Supplemental Figure 3). Genes specific to C4 metabolism, including those encoding the copy of phosphoenolpyruvate carboxylase located on chromosome 10 (no. 27 in the figure), NADP-dependent malate dehydrogenase, and NADP-ME were highly expressed in the leaf, followed by the PS. The gene for the non-C4 phosphoenolpyruvate carboxykinase (PEPCK) located on chromosome 4 (no. 36 in the figure) was expressed highly in the leaf and PS (Figure 5), whereas the C4-based PEPCK gene located on chromosome 1 was not detected (and thus is not shown in Figure 5), indicating that the sorghum PS, like sorghum leaves, lacks an active PEPCK pathway (Wang et al., 2014; Döring et al., 2016). The small subunit of RuBisCO (transcript no. 5) was strongly expressed in the leaf, moderately in the spikelets, and scarcely at all in the awn.
Figure 5.
S. bicolor.
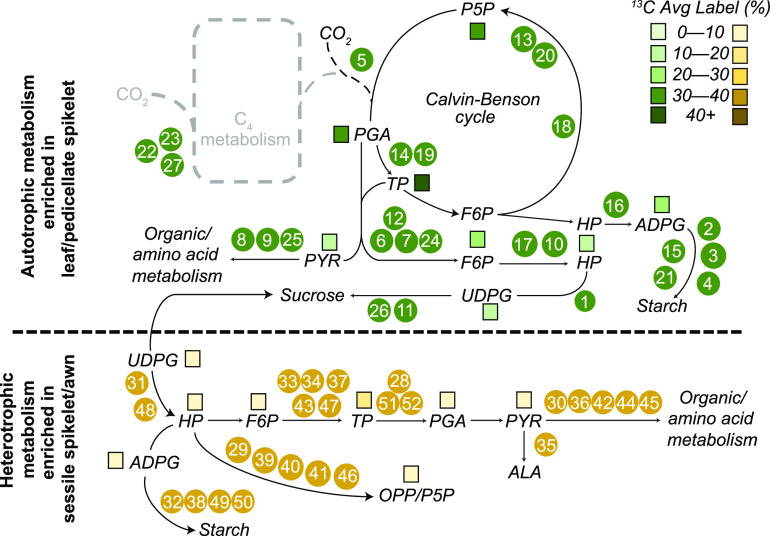
Relative expression of genes encoding biosynthetic enzymes immediately responsible for producing the metabolites labeled with 13C, a subset extracted from full set of 922 DE metabolic genes in Supplemental Figure 3. Colors reflect scaled z-scores of log2-normalized expression values. Labels of genes indicate enzyme name, biochemical process, and subcellular localization: 1) UTP-G1P UdT, UTP-glucose-1-phosphate-uridyltransferase, Sobic.002G291200.1; 2) starch synthase, Sobic.001G239500.2; 3) starch synthase, Sobic.010G047700.1; 4) starch synthase, Sobic.002G116000.1; 5) ribulose-1,5-bisphosphate carboxylase/oxygenase, small subunit, Sobic.005G042000.1; 6) F16BP aldo, fructose-1,6-bis-phosphate aldolase, Sobic.005G056400.1; 7) F16BP aldo, fructose-1,6-bis-phosphate aldolase, Sobic.008G053200.1; 8) GDH, glutamate dehydrogenase, Sobic.003G188400.1; 9) GS, glutamine synthetase, Sobic.006G249400.1; 10) PGM, phosphoglucomutase, Sobic.003G222500.1; 11) SuSY, sucrose phosphate synthase, Sobic.004G068400.1; 12) F16BPase, fructose-1,6-bisphosphatase, Sobic.003G367500.1; 13) PPE, phosphopentose epimerase, Sobic.001G491000.1; 14) TPI, triose phosphate isomerase, Sobic.002G277100.1; 15) starch synthase, Sobic.006G221000.1; 16) G1P-AdT, glucose-1-phosphate adenyltransferase, Sobic.007G101500.1; 17) PGI, phosphoglucoisomerase, Sobic.002G230600.1; 18) S17BPase, sedoheptulose 1,7-bisphosphatase, Sobic.003G359100.1; 19) PGK, phosphoglycerate kinase, Sobic.009G183700.1; 20) PPI, phosphopentose isomerase, Sobic.001G069000.1; 21) starch synthase, Sobic.004G238600.1; 22) NADP-ME, NADP-malic enzyme, Sobic.003G036200.1; 23) NADP-MDH, NADP-malate dehydrogenase, Sobic.007G166300.1; 24) F16BPase, fructose-1,6-bisphosphatase, Sobic.010G188300.1; 25) PEPCK, phosphoenol pyruvate carboxykinase, Sobic.004G338000.1; 26) SuSY, sucrose phosphate synthase, Sobic.003G403300.1; 27) PEPC, phosophoenol pyruvate carboxylase, Sobic.010G160700.1; 28) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.005G159000.1; 29) PPE, phosphopentose epimerase, Sobic.002G257300.1; 30) PEPC, phosphoenol pyruvate carboxylase, Sobic.004G106900.1; 31) UTP-G1P UdT, Sobic.006G213100.1; 32) G1P-AdT, glucose-1-phosphate adenyltransferase, Sobic.002G160400.1; 33) TPI, triose phosphate isomerase, Sobic.003G072300.2; 34) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.004G146000.1; 35) ALAAT, alanine amino transferase, Sobic.001G260701.1; 36) PEPCK, phosphoenol pyruvate carboxykinase, Sobic.006G198400.2; 37) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.003G393900.1; 38) starch synthase, Sobic.007G068200.1; 39) PPI, phosphopentose isomerase, Sobic.003G182400.1; 40) PPI, phosphopentose isomerase, Sobic.008G135701.1; 41) TK, transketolase, Sobic.010G024000.2; 42) NAD-GS, Sobic.003G258800.1; 43) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.003G096000.2; 44) PEPC, phosphoenol pyruvate carboxylase, Sobic.003G301800.1; 45) PEPC, phosphoenol pyruvate carboxylase, Sobic.002G167000.1; 46) TK, transketolase_Sobic.009G062800.1; 47) F16BP aldo, fructose-1,6-bisphosphate aldolase, Sobic.009G242700.1; 48) PGM, phosphoglucomutase, Sobic.001G116500.1; 49) starch synthase, Sobic.010G022600.1; 50) starch synthase, Sobic.010G093400.1; 51) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.004G056400.1; 52) GAPDH, glyceraldehyde-3-phosphate dehydrogenase, Sobic.004G205100.1. cy, cytosolic localized; ch, chloroplast localized; mi, mitochondrial localized; OA/AA, organic acid/amino acid metabolism.
Analogous data for T. triandra produced similar, but not identical, findings. Because T. triandra spikelets are subtended by a large leaf-like bract (see below), expression of the 769 metabolic genes in the spikelets was compared to that of the bract rather than to a foliage leaf. PCA of these transcripts placed the awns and PS at opposite sides of PC1 (32.9% of variance); the three replicate bract samples were distinguished from the other organs on PC2 (Supplemental Figure 4). Of the 769 genes, 322 were DE between at least one pair of organs. Relative expression of these genes showed a different pattern for each of the four organs (Supplemental Figure 5). Because the variability among samples of T. triandra was higher than that in sorghum, fewer genes were significantly DE, but overall trends were similar. Awns in T. triandra had a distinct set of upregulated genes that were not reflected in photosynthetic metabolism. Of the genes encoding enzymes that could produce the 13C metabolites, only 24 were DE (Supplemental Figure 6). (Note that T. triandra lacks a reference genome, so the genes are labeled with the names of their putative sorghum orthologs.) Genes related to photosynthesis are upregulated in the leaf-like bract and downregulated in the SS, whereas the situation is opposite for genes involved in starch and Suc metabolism. The PS is similar to both bracts and SSs, whereas awns have a unique expression profile. Unlike sorghum, genes for aspartate metabolism are DE, and the C4 isoform of PEPCK (no. 19 in Supplemental Figure 6) is moderately expressed in the SS, PS, and bract, suggesting that this species may have an active PCK type of C4 photosynthesis and use aspartate to transport carbon between the bundle sheath and mesophyll.
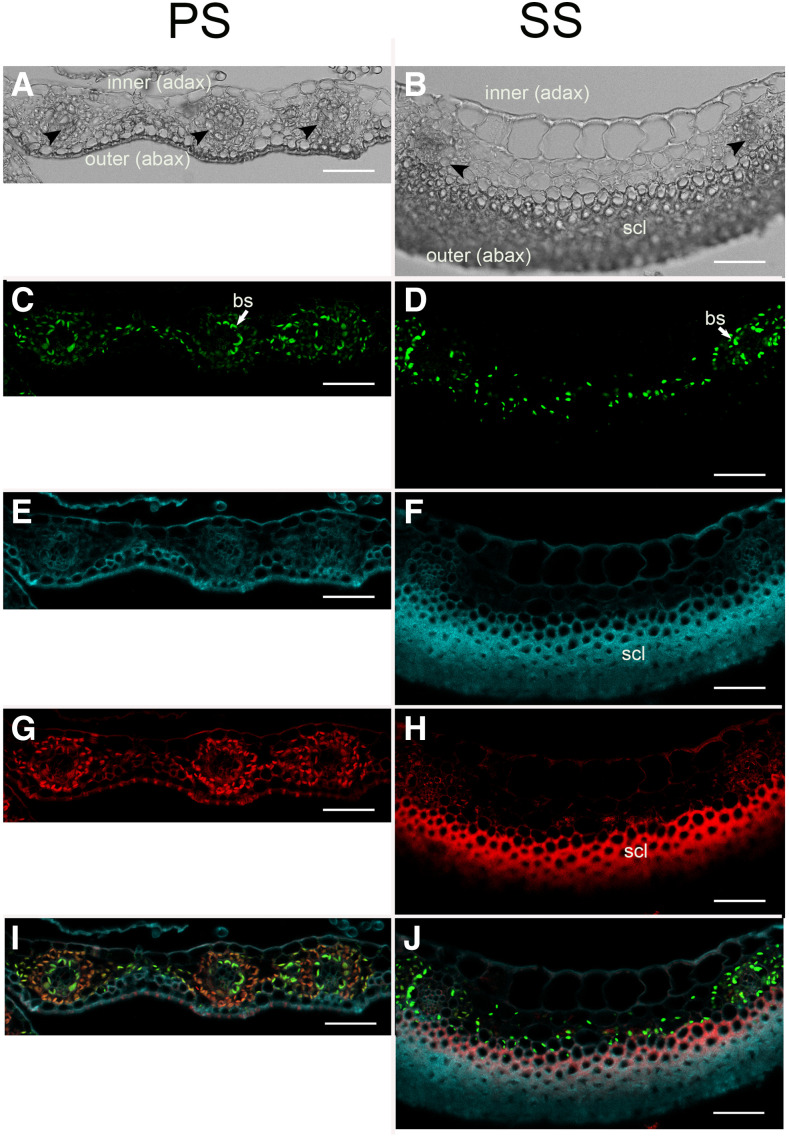
Rubisco Is Expressed in Both Bundle Sheath and Mesophyll Cells
If the PS and SS were similar to C4 leaves, we would expect that Rubisco would be restricted to cells of the bundle sheath and absent in the inter-vascular regions (Dengler and Nelson, 1999). In addition, veins would be separated by no more than two mesophyll cells. Immunolocalization showed abundant Rubisco in bundle sheath cells of both the PS and SS and also in most of the intervening cells (Figure 6; controls in Supplemental Figure 7). Veins are separated by more than two mesophyll cells, suggesting that a C4 shuttle might be operating around the veins but that other mesophyll cells may be relying on C3-like metabolism. In the PS, this interpretation is consistent with the 13C and gene expression data that show some labeling of the C4 metabolite malate as well as labeling of C3 intermediates of the Calvin–Benson cycle plus 2-phosphogylcolate, indicating photorespiration, and expression of C4- and C3-related enzymes.
Figure 6.
Immunofluorescence of Rubisco Large Subunit in Sorghum Spikelets.
(A), (C), (E), (G), and (I) Cross section of glume of PS.
(B), (D), (F), (H), and (J) Cross section of glume of SS.
(A) and (B) Bright field.
(C) and (D) Anti-Rubisco large subunit with Alexa 488 as the secondary antibody; emission window 509 to 585 nm.
(E) and (F) Lignin autofluorescence; emission window 415 to 485 nm.
(G) and (H) Chlorophyll autofluorescence; emission window 661 to 779 nm. Autofluorescence in sclerenchyma in (H) is nonspecific.
(I) and (J) Merge of Rubisco, chlorophyll, and lignin channels. Black arrowheads, vascular bundles; inner (adax), facing the inside of the spikelet; outer (abax), facing the outside of the spikelet; bs and white arrows, bundle sheath; scl, sclerenchyma. Bars = 50 µm.
In the SS, Rubisco-expressing cells are restricted to the adaxial (inner) side of the glume. This tissue is not green, and the plastids do not fluoresce in the red confocal channel (compare Figures 6D with 6F, and 6G with 6H; red color in Figures 6H and 6J appears to be nonspecific fluorescence of thick-walled cells). By contrast, the abaxial (outer) side is made up of several layers of sclerenchymatous cells. The thickness of the outer layers of the SS glume, the lack of stomata (Figure 2C) and chlorophyll, and the lack of labeled 13C intermediates in the metabolite experiments (Figure 3) together suggest that the SS is assimilating relatively little carbon from the surrounding air (except possibly near the very tip where it is similar to the PS; data not shown). We speculate that the Rubisco-expressing chloroplasts in the inner cells of the glume instead could be refixing respired CO2, as is common in floral and fruiting structures (Simkin et al., 2020), although validating this would require a different set of experiments than those presented here.
PS Contributes Significantly to Seed Weight (Yield)
To test the influence of PS function on plant grain yield, individual PSs were removed from alternating panicle branches at anthesis. Four genotypes differing in inflorescence architecture and relative size of the PS and SS (Figures 7A to 7D) were tested, with 20 treated plants (spikelets removed) and 20 untreated plants per genotype, grown in a randomized complete block design. In 78 of the 80 treated plants, seed size was smaller on branches from which PSs had been removed; one plant failed to set seed and in the other seed size was larger on the branches with the PS removed. The overall effect of treatment was significant (P = 0.000263), with seeds from which the adjacent PS was removed being significantly smaller than those on unmanipulated plants (Figures 7E and 7F; Supplemental Table 8). Differences among genotypes accounted for the greatest fraction (41%) of the overall variance, which was expected given substantial differences in seed size, panicle architecture, and time to maturity (Supplemental Table 8). Within any given genotype, however, treatment effects were highly significant. Overall, the PS contributed 8.82% to seed weight, with values of 8.08, 9.72, and 13.61 for each individual genotype (Supplemental Table 8). The effect in accession 597971, 7.80%, was nonsignificant as a result of highly variable seed filling in all plants and particularly high variance in the control plants; however, the result was highly significant (P = 8.73e–08) if only the manipulated plants were considered (Figure 7F).
Figure 7.
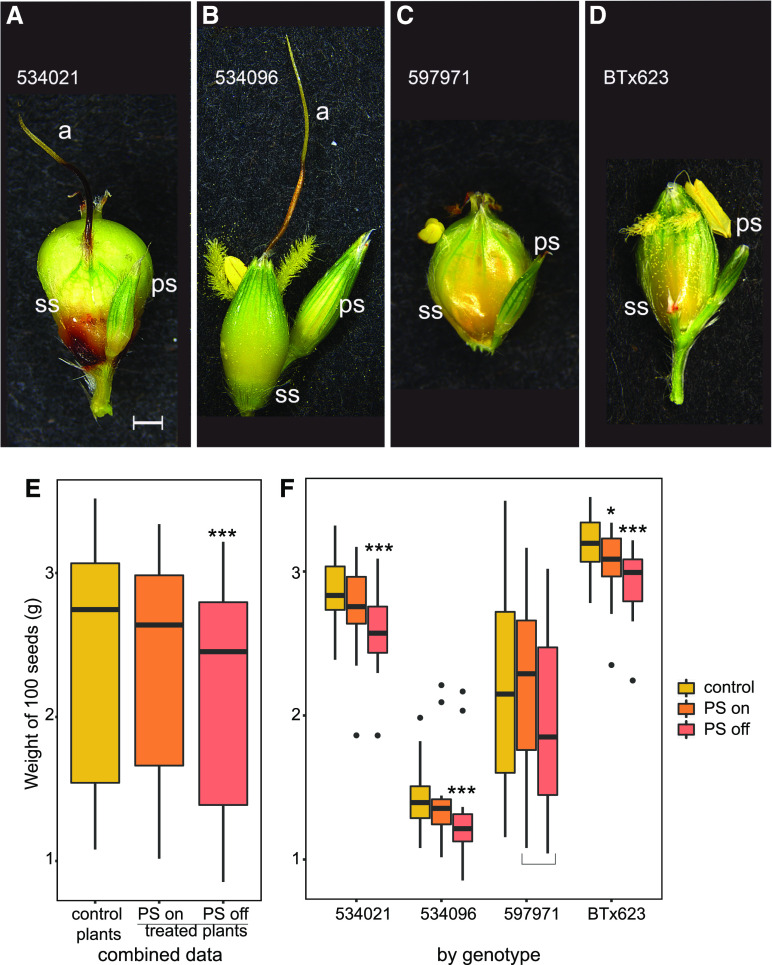
Spikelet Removal Experiments.
(A) to (D) Representative spikelet pair for each accession. (A) Jola Nandyal (534021). (B) SO85 (PI 534096). (C) SAP-170 (PI 597971). (D) BT×623 (PI 564163). Bar = 1 mm. All spikelets to the same scale.
(E) and (F) Results of removal experiments. 100-seed weight (g) from control (unmanipulated) plants and manipulated plants. In the latter plants, PSs from some branches were untouched (on) and those from other branches were removed (off). (E) Results from all genotypes combined, showing an 8.82% reduction in average weight with spikelet removal. (F) Average seed weight for each genotype analyzed separately.
(A) to (F) Effect sizes for 534021, 534096, and BT×623 were 8.08, 13.61, and 9.72%, respectively. Effect size for 597971, 7.80%, was nonsignificant because of high variance in the control plants; however, a comparison of seeds from only the manipulated plants (bracketed) was highly significant. ***P < 0.001; *P < 0.05. Box plot center line, median; upper and lower limits of boxes, first and third quartiles; whiskers, up to (down to) 1.5× the interquartile range; points, outliers. See also Supplemental Table 8. a, awn; ps, pedicellate spikelet; ss, seed-bearing spikelet.
Bracts Enhance Carbon Assimilation in Wild Species
The contribution and position of photosynthetic organs to seed production was also considered by analyzing the bracts in T. triandra and A. schirensis, which are much closer to the spikelets than the flag leaves of sorghum. Our 14C pulse-chase experiments tested whether these played a role in carbon assimilation.
In A. schirensis, the upper modified leaf (“bract”) is reduced largely to a sheath, with only a short blade (Figure 1E). As expected, this modified leaf assimilated 14C, although the difference in percent counts between the pulse and chase for the attached bract was nonsignificant (P = 0.9987; Figure 8A; Supplemental Table 9). The leaf sheath bears stomata throughout (Figure 8B), as expected. Stem segments including the flag leaf node and the lowermost inflorescence node contained some 14C, but there was little signal from the internode in between, suggesting that little carbon was being transported. The leaf is thus not a major source of photosynthate for the inflorescence within the time frame and conditions of this experiment (anthesis), and the increased 14C in SSs during the chase was largely accounted for by the drop in PS values.
Figure 8.
Results of 14C Pulse-Chase Experiments and Distribution of Stomata in Bracts.
(A) to (D) A. schirensis (see [A] and [B]) and T. triandra (see [C] and [D]). (A) and (C) 14C results including bract. Percent dpm for each organ after 1-h exposure to 14C with organs removed from the axis (detached), inflorescence intact (attached), or after 24-h chase. Plot includes mean percentages and sds; n = 3.
(A) Values for bracts and PS are significantly lower and higher, respectively, when attached to the axis rather than detached, whereas values for SS did not differ significantly. Percent counts in the bract were not significantly different after the 24-h chase as compared with attached, but were significant relative to detached. Percentages in the PS significantly decreased relative to attached, but not relative to detached. See also Supplemental Table 9. After 1 h, most counts are in the bract when organs are detached from the stem but in the PS when they are attached.
(B) and (D) Abaxial epidermis of bract showing rows of stomata (arrows) and prickles (P). Bar = 50 µm.
(C) Values for detached and attached organs after 1 h are not significantly different. Values for bract and SS significantly decrease and increase, respectively, after the 24-h chase. Values for PS are not significantly different. See also Supplemental Table 10.
By contrast, the bract of T. triandra is closely connected to the spikelets, separated by an internode of only 2 to 4.5 mm (Figure 1I). The bract accounted for 64 to 71% of the fixed 14C in the 1-h pulse (Supplemental Table 10) and that percentage dropped to 25% in the chase (P < 0.0001), with a corresponding increase in the amount in the SS (P < 0.01; Figure 8C; Supplemental Table 10). The epidermal pattern of the bract is leaf like, with extensive stomata (Figure 8D). Results of 13C assimilation were qualitatively similar to those for 14C (Figures 3E and 3F) and also similar to those for A. schirensis, with the bract being significantly different from the other three organs (Supplemental Tables 11 and 12). Thus, adding a leaf-like structure close to the spikelets can enhance carbon assimilation in the form of PS or bract and relative contribution is dependent on architecture and location.
DISCUSSION
Our data suggest that carbon capture in sterile spikelets may help compensate for the ∼50% reduction in meristems caused by having non-seed-bearing spikelets paired with seed-bearing spikelets, as summarized in Figure 9. It is unexpected that one set of floral structures would contribute carbon to the metabolism of other flowers; previously, flowers and inflorescences were reported only to contribute fixed carbon to their own metabolism (Bazzaz and Carlson, 1979; Vemmos and Goldwin, 1994; Antlfinger and Wendel, 1997; Aschan and Pfanz, 2003; Earley et al., 2009). Reproductive tissues of oilseeds are also often green and may assimilate or re-assimilate carbon (King et al., 1998; Furbank et al., 2004; Ruuska et al., 2004; Schwender et al., 2004; Goffman et al., 2005; Allen et al., 2009) and use photosynthetic light energy for ATP or reductant production (Browse and Slack, 1985; Asokanthan et al., 1997; Ruuska et al., 2004; Rolletschek et al., 2005). A number of these studies focused on the contribution of sunlight to metabolically demanding processes such as fatty acid biosynthesis that generate large amounts of CO2.
Figure 9.
Summary of Biochemical Pathways Assessed by 13C and RNA-Seq Data.
Numbered dots correspond to enzymes whose expression is shown in Figure 5. Colored squares reflect percent average label in 13C assays of metabolites. Top part of figure shows autotrophic metabolism and is enriched in the leaf and PS; bottom part of figure shows heterotrophic metabolism, enriched in the SS.
The PS exhibits autotrophic metabolism with labeled metabolites and transcript accumulation qualitatively similar to, but less significant than, a leaf. It can assimilate CO2 whether attached or detached from other structures, consistent with the leaf-like epidermal morphology of the glumes, with stomata distributed over most of the surface (Figures 2B, 2F, and 2J). The PS contributes 8 to 13% to yield (Figures 7E and 7F), an estimate consistent with earlier reports that photosynthesis from the entire sorghum inflorescence may account for 6 to 18% of yield (Fischer and Wilson, 1971; Evans and Wardlaw, 1976). We can now attribute that contribution to yield to a particular component of the inflorescence.
By contrast, the SS in sorghum appears to be largely heterotrophic at anthesis. Carbon assimilation is lower than in the PS but still easily detected, and stomata are much less common on the surface of the glumes (Figure 2C). Rubisco is strongly expressed in both bundle sheath and mesophyll cells but only on the adaxial (internal) side of the glumes (Figure 6). Expression of transcripts encoding proteins involved in photosynthesis was low in the SS, whereas genes involved in starch synthesis were relatively high (Figure 5; Supplemental Figure 3). Decarboxylating enzymes, found in other experiments to be upregulated in tobacco (Nicotiana tabacum) stems (Hibberd and Quick, 2002), were not upregulated in the SS, providing no evidence for decarboxylation of transported four carbon compounds.
We hypothesized initially that awns in sorghum should be similar to those in wheat, barley (Hordeum vulgare), and rice (Bort et al., 1996; Li et al., 2006; Maydup et al., 2010; Sanchez-Bragado et al., 2014; Hu et al., 2019, and references therein), which are large, green, and provide photosynthate to the developing grain. However, no evidence of CO2 assimilation was found in awns whether isolated or intact (Figure 2A), none of the metabolites were labeled (Figures 3 and 4), and no stomata were present (Figure 2D). While the awns exhibit distinct gene expression patterns (Figure 5; Supplemental Figures 2 and 3), they are generally more similar to the SS than to either the PS or the leaf or bract. Awns in Andropogoneae are derived independently from those in wheat, barley, or rice (Teisher et al., 2017), and, as shown here, differ in structure and physiology.
The results for sorghum apply to other members of the tribe Andropogoneae, as shown by data on the unrelated species A. schirensis and T. triandra. In both species, the SS is bisexual as in sorghum, has a firm outer glume, and bears an awn (Figure 1). The PS in these species is staminate (male). PSs in these species are photosynthetic and export carbon to the heterotrophic SS at anthesis (Figures 2E and 2F). The awns do not assimilate carbon and lack any evidence of carbon assimilation (Figures 2E, 2F, and 3C to 3F), although, as in sorghum, in T. triandra they produce awn-specific gene transcripts (Supplemental Figures 4, 5, and 6).
Because the structure of the spikelet pair is conserved among most of ∼1200 species of Andropogoneae (maize being an exception; Clayton and Renvoize, 1986; Watson and Dallwitz, 1992 onward; Vegetti, 1999; Kellogg, 2000, 2015; Arthan et al., 2017), we can infer that the function of the PS as a nurse tissue has been fixed and maintained by natural selection over ∼15 million years of evolution. The proximity of PSs to seeds may enable delivery of photoassimilates during stress such as drought-induced reduction in transpiration, as described for awns in wheat (Hu et al., 2019; Simkin et al., 2020).
The small photosynthetic PS boosts fitness or yield, but placing a leaf-like structure in the inflorescence closer than the flag leaf appears to be even more favorable. The bract in T. triandra assimilates considerably more carbon than the PS and transfers that carbon to the SS (Figure 8C). Metabolically and anatomically it is leaf like. The bract in A. schirensis is also photosynthetically active but appears to transfer little carbon to the developing inflorescence (Figure 8A). This may reflect the distance of translocation within the short time frames of these experiments. While the distance from the flag leaf to the panicle in sorghum is generally at least 30 cm and often twice that, the corresponding distance in A. schirensis is 14 to 21 cm and that in T. triandra is 2 to 4.5 mm (Figures 1E and 1I).
Although the PS is undoubtedly photosynthetic, it appears to be neither wholly C3 nor C4. A number of photosynthetic intermediates were significantly labeled in the PS, and the relative expression of photosynthetic enzymes is higher in the PS than the SS or awns but lower than in leaves or bracts, respectively (Figure 5; Supplemental Figures 3, 5, and 6), although these data do not distinguish the mode of carbon assimilation (Hibberd and Furbank, 2016). Rubisco is present in the bundle sheath (as in C4 tissue) but also throughout the mesophyll (as in C3 tissue; Figure 6). Veins in the PS and SS are also separated by more than two cells (Figure 6), suggesting the possibility of a limited C4 shuttle, as demonstrated in other inflorescence-related structures (Langdale et al., 1988; Pengelly et al., 2011). We also found labeled 2-phosphoglycolate in PSs and some unlabeled 2-phosphoglycolate in other organs, indicating limited photorespiration, consistent with either C3 or C4 Rubisco-based assimilation, although 2-phosphoglycolate phosphatase, an enzyme specific to the photorespiratory pathway, was not DE.
In summary, despite its small size, the PS in sorghum and Andropogoneae can make a significant contribution to yield by fixing carbon and translocating it to the SS, which holds the developing seed. Awns, in contrast, are carbon sinks, with limited metabolic activity. These results reflect millions of years of evolution, over which the spikelet pair has been selected and conserved. The PS may thus have contributed to fitness in natural populations and could also be a useful target for sorghum improvement in agricultural settings.
METHODS
Plant Material
Sorghum bicolor accessions BT×623 (PI 564163), SO85 (PI 534096), Jola Nandyal (PI 534021), and SAP-170 (PI 597971) were obtained via the USDA Germplasm Resource Information Network. All except SO85 are members of the Sorghum Association Panel (Casa et al., 2008). BT×623 is the line from which the reference genome sequence was obtained (Paterson et al., 2009; McCormick et al., 2018). Additional experiments were conducted on Themeda triandra (PI 208197) and Andropogon schirensis (Pasquet s.n.).
The three species represent different major clades of Andropogoneae (Estep et al., 2014). In addition, they differ in inflorescence structure. In Sorghum bicolor, the PSs are usually sterile, whereas in the other two species, the PSs are staminate (Figures 1A, 1C, and 1G). In Themeda triandra, the SS is associated with two staminate PSs and is subtended by two additional pairs of staminate spikelets, for a total of six staminate spikelets per sessile bisexual spikelet (Figures 1F, 1G, and 1H). For the purposes of this experiment, all six were considered similar and were treated together. In addition, the set of spikelets in T. triandra is closely subtended and partially enclosed by a large bract (Figure 1I). In A. schirensis, the uppermost leaf, which we refer to here as a bract, is reduced to a sheath and minimal blade (Figure 1E). The distance between the node of the leaf and the inflorescence node is 14 to 21 cm. S. bicolor has no bracts or inflorescence-associated leaves. In all three species, only the seed-bearing SS bears an awn.
Pulse-Chase Experiments to Determine 14C Assimilation
Labeling
To determine which structures assimilated and fixed CO2, we traced the localization of 14C in S. bicolor SO85, T. triandra, and A. schirensis. Plants were collected at anthesis in the greenhouse between 10 am and 11 am. Culms were cut with a razor blade and placed directly into tap water before being transferred to 250-mL Erlenmeyer flasks containing filter paper and 1 mL of water, with two flasks per species. One flask (A) contained one (T. triandra and S. bicolor) or two (A. schirensis) intact inflorescences and was used for the 24-h pulse-chase experiments. The second flask (B) for each species held one or two intact inflorescences and was used for the 1-h pulse experiment. Flask B also contained an additional inflorescence dissected into SSs, PSs, awns, and bracts or leaves to determine 14C assimilation without connection to the rest of the inflorescence. Each experiment used ∼40 to 80 mg of tissue, equating to ∼15 awns, 22 bracts, 16 to 20 SSs, and 40 staminate spikelets for T. triandra. For A. schirensis, a similar amount of biomass required ∼25 awns, 22 SSs, 30 staminate spikelets, or 1 to 2 leaves. For S. bicolor, we used 2.5 cm of one leaf, ∼6 to 8 SSs, ∼30 staminate spikelets, and all available awns from one inflorescence.
A plastic tube containing 12.5 µCi of 14C sodium bicarbonate was placed in each flask and maintained in an upright position by attachment to a plastic rod. Flasks were capped with airtight septa closures, and 1 mL of 6 N H2SO4 was added directly to each plastic tube using a syringe, releasing a pulse of 14CO2 into the flask. All samples were incubated for 1 h in a growth chamber at ∼350 µE.
Processing
After 1 h, all flasks were purged with air (30 to 60 s) to remove the 14CO2. The radioactive gas stream was captured in a reservoir containing 2 liters of 2 N KOH. For flask A for each species, the airtight septum and closure were replaced with a sponge top prior to incubation in the growth chamber under continuous illumination for the 24-h chase period. The contents of flask B were analyzed immediately. Detached awns, SSs and PSs, and leaves were separated, weighed, and transferred to tubes containing cold methanol:chloroform (7:3, v:v) and steel beads. Simultaneously, the intact inflorescence was dissected, and individual components were treated identically to the detached samples. Tissues were homogenized at 30 cycles/s in a bead homogenizer for two 5-min intervals and were stored at –20°C for 48 h. After 24 h, samples from flask A were dissected, weighed, and processed in the same way and stored at –20°C for 24 h.
All samples were extracted sequentially. The first extraction was based on 1.5 mL of 7:3 (v:v) methanol:chloroform that was used to homogenize and store tissues after labeling. Next, 200 µL from the methanol:chloroform extract was combined with 5 mL of scintillation fluid (HIONIC FLUOR; PerkinElmer). The remaining extract was removed from the residual biomass, and water (2 mL) was added. The biomass was bead homogenized as described above, centrifuged at low-to-medium speed (∼5000 rpm) for several minutes at room temperature, and then 200 µL was combined with 5 mL of scintillation fluid. Residual biomass was then treated with tissue solubilizer (ScintiGest), incubated overnight at 60°C, and prepared for scintillation identically to prior extracts. Scintillation counting in dpm was performed on a Beckman Coulter LS-6000TA scintillation counter and included recording 14C photon emissions for 5 min. After background subtraction from a blank that contained identical amounts of solvent and scintillation cocktail, total radioactivity per amount of tissue was calculated by accounting for differences in volume and mass.
Analysis of 14C Data
The total 14C assimilation (dpm/mg) for each organ was calculated by summing the counts from the three serial extractions and accounting for the total volumes of each extraction. Next, the percentage of label within awns, PSs, and SSs was determined by calculating the fractional 14C assimilation in each organ. Experiments were repeated three times over a 10-month span using individual plants harvested from a greenhouse. Percent label for each organ was averaged across experiments, and means and standard deviations were plotted in Excel (Microsoft). Significance was assessed using analysis of variance (type 1 ANOVA, balanced design).
13C Labeling in Planta
Labeling
The 13C isotopic labeling studies were performed on inflorescences of T. triandra and S. bicolor accession SO85. Themeda and Sorghum plants were grown in the greenhouse until anthesis. The intact inflorescence was placed in a deflated plastic bag (inflated volume ∼2 to 3 liters). A 10-mL serological pipette was fed into the bag with its tip near the apex of the inflorescence. The other end of the pipette was connected by hose to a tank of synthetic air comprising 13CO2:N2:O2 at a ratio of 0.033:78:21.967. For each timed treatment, the bag was rapidly inflated (∼15 L/min) and then the flow of gas was decreased to ∼2 L/min. Structures were labeled for 30, 60 or 300 s. Three replicate experiments were done for each time point for each species. For sorghum, each replicate was a single inflorescence from a single plant, making a total of nine inflorescences (3 replicates × 3 time points). For Themeda, each replicate included all flowering structures on a single tiller from a single very large plant, making a total of nine tillers (3 replicates × 3 time points). All experiments were conducted in the morning between 10 am and 11 am to minimize circadian effects. The synthetic air pumped into the bag flowed from the release of the pipette at the tip along the inflorescence structures and exited the bag at the point where the bag opening was grasped around the culm. At the end of the labeling period, the inflorescence was cut immediately and dropped in a large pool of liquid nitrogen in a Styrofoam box (22 × 33 × 15 cm) to quench metabolism. During labeling and quenching, plant tissues were exposed to greenhouse light levels between 250 and 400 µE m–2 s–1 from a combination of metal halide and high-pressure sodium lights.
Processing
Frozen tissue was dissected in liquid nitrogen to separate SSs and PSs, awns, and bracts. Each sample was then ground in liquid nitrogen with a mortar and pestle and extracted with methanol:chloroform (7:3, v:v) and then through addition of water to segregate polar and nonpolar phases, as described previously by Ma et al. (2014, 2017). Pipes (12 nmol) was added to each sample as an internal standard.
LC-MS/MS
A QTRAP 6500 tandem mass spectrometer linked to two Shimadzu LC-20AD pumps working in coordination with a SIL-20AC/HT autosampler was used to assess and quantify the isotopic labeling in intermediates of primary metabolism. Extract from ∼5% of the total harvested sample was injected. Standards for metabolites were run separately to establish retention time and, in some cases, confirm identification of isomers. Separation on the liquid chromatograph involved an ion pair method (Ma et al., 2017) with a flowrate of 300 µL/min and a binary gradient buffer combination with buffer A (11 mM acetic acid with 10 mM tributylamine as the ion pair) and buffer B (100% methanol). The method differed from prior work as the ramp profile was shortened with 3-min equilibration at 5% B; 10-min ramp to 35% B; 2-min ramp to 95% B, hold for 3 min, return to 5% B within 2 min; and equilibration there for 11 min, resulting in a total run time of 31 min. The source inlet temperature (550°C), curtain gas (35 psi), and auxiliary gases (both set to 60 psi) were chosen based on optimal peak response. Declustering potential, collision energy, and collision exit potential for individual mass transition pairs of multiple reaction monitoring were based on prior work by Ma et al. (2014, 2017).
Analysis of Mass Spectrometry Data
Peak intensities for individual mass traces that represent a precursor–product ion combination were measured using Analyst 1.6 (AB Sciex). The relative percent combination of isotopologues was calculated as well as 13C average labeling that is defined as (), with n equal to the number of carbons and mi defining the relative isotopologue abundance for each of the i isotopologues measured. Estimates were corrected for natural abundance, following Fernandez et al. (1996), and using the isotopomer network compartmental analysis computational platform (Young, 2014). Data were additionally compared through plots of the isotopologue distributions and 13C atom equivalents formed. All isotope labeling plots included standard deviations. Average labeling data were initially analyzed in raw, uncorrected form with PCA to assess reproducibility of results. Statistical significance of results was assessed with ANOVA (type 1, balanced design). Both PCA and ANOVA used standard programs in R, as presented in the text and figures.
Estimate of 13C Atom Equivalents Formed
The 13C average labeling and 13C isotopologue distributions reflect the dynamics of isotope incorporation into different organs and capture more information than static measurements of metabolite levels alone; however, relative labeling comparisons cannot fully account for the variation in biomass composition and differences in active and inactive pools between organs that dilute the rate of 13C labeling. Full comparisons of one metabolite to another in the strictest sense require a computational flux model that would be technically challenging and well beyond the scope of this study, given the small organs investigated here. In Supplemental Figure 1, the 13C atom equivalent plots provide a comparison of individual metabolites across the three organs compared in this study. The 13C isotopologues were normalized by the ratio of total metabolite peak signal to the median peak signal of all metabolites examined within that run and then weighted for the number of carbons per isotopologue and summed to get atom equivalents.
Malate Active Pool Estimates
The unlabeled fraction in malate leveled off by 60 s and did not vary significantly to 300 s, indicative of an inactive pool (Ma et al., 2014; Arrivault et al., 2017; Allen and Young, 2020). The active pool of malate was calculated by subtracting the unlabeled fraction that remains at long time points from all unlabeled fractions during the pulse and rescaling the unlabeled and labeled isotopologues.
Rubisco Immunolocalization
Sorghum spikelets at anthesis were fixed as for scanning electron microscopy (below) and sectioned with standard methods used for histology (Ruzin, 1999). Immunofluorescence assays were adapted from Lucas et al. (1995). Specifically, sections were de-paraffinized with Histo-Clear for 10 min twice and rehydrated with 100, 100, 90, 70, 50, and 30% (v/v) ethanol. After brief washing in water and PBS, the slides were treated with 0.1 mg/mL Proteinase K solution in PBS buffer for 10 min at room temperature, washed in PBS three times (5 min each), and incubated in blocking reagent (2% dry milk in PBS) for 2 h at room temperature. The slides were then probed with anti-Rubisco large subunit (rabbit, AS03 037; Agrisera) with 1:250 (v:v) dilution for 4 h at room temperature, followed by two washes with blocking reagents (the second wash was overnight at 4°C). Samples were then incubated with goat anti-rabbit IgG secondary antibody Alexa Fluor 488 (Thermo Fisher Scientific), with 1:100 (v:v) dilution for 2 h at room temperature, followed by two washes in blocking reagent for 15 min each, and stored in PBS buffer.
Fluorescence was obtained using a 20× HC PL APO (numerical aperture 0.7) objective lens on an inverted TCS SP8-X confocal microscope (Leica Microsystems) using the 405, 499, and 649 nm laser excitation lines. Alexa Fluor 488–labeled Rubisco signal was detected by PMT3 detector using an emission window of 509 to 585 nm. Lignin and chlorophyll autofluorescence were detected in parallel by hybrid detector HyD2 using emission windows of 415 to 485 and 661 to 779 nm, respectively. Bright-field images were obtained through the transmitted light detector. Images from individual channels were merged using Leica Application Suite software.
Sorghum Spikelet Removal Experiments
Removal of PSs
The impact on grain weight of removing PSs was tested in four accessions (genotypes) of S. bicolor: SAP-170 (PI 597971), BT×623 (PI 564163), Jola Nandyal (534021), and SO85 (PI 534096), referred to in this article by their PI numbers except for BT×623, which is the line used for the reference genome sequence. Forty seedlings of each genotype, total 160 plants, were grown in individual pots in a greenhouse at the Donald Danforth Plant Science Center in 2019, under metal halide and high-pressure sodium lights set to 14-h days, 28°C daytime and 22°C nighttime temperatures, and relative humidity 40% days and 50% nights. For each accession, 20 plants were left unmanipulated (designated “on,” or control plants). The other 20 plants were manipulated (designated “off”). When the plants reached anthesis, PSs were removed with forceps from alternating branches of the treated plant, which thus had a mix of treated and untreated branches (designated “off-off” and “off-on,” respectively). By removing PSs from alternating branches, variation along the inflorescence was averaged out. The untreated branches of the manipulated plants (off-on) thus served as an internal control. Treated branches were marked. Plants were grown to maturity, the inflorescence removed and dried at 40°C for 4 to 5 d, and spikelets harvested. Caryopses (“seeds”) were removed from the glumes and floral bracts.
Data Analysis
Seeds for genotypes BT×623 and 534021 were counted with a Seedburo 800 Count-a-Pak seed counter at the USDA in Columbia, Missouri. Seeds for genotypes 597971 and 534096 were too small for reliable counts on the seed counter, so they were spread on a flatbed scanner (Epson Perfection V550 or Epson WorkForce DS-50000, 300 dpi resolution, grayscale), imaged, and weighed. The resulting JPG files were converted to binary images using Fiji (Abramoff et al., 2004), and the Analyze Particles tool was used to count the seeds in each image. Total seed weight was divided by the number of seeds to estimate the weight of 100 seeds. The statistical effect of PS removal on seed weight was assessed using a mixed effect linear model (lmer function in R packages lme4 [Bates et al., 2014] and lmerTest [Kuznetsova et al., 2015]), where block, row, and column were specified as fixed effects and genotype was treated as a random effect. All data were visualized using ggplot2 (Wickham, 2016).
Removal of Awns
Preliminary experiments found that the effect of awn removal was appreciably less that than of spikelet removal, with a nonsignificant effect of 1% or less depending on genotype. Limited resources prevented us from carrying out an awn removal experiment large enough to demonstrate whether an effect that small actually exists.
Scanning Electron Microscopy
Plant material was fixed in formalin:acetic acid:50% alcohol (10:5:85 by volume) for a minimum of 24 h and then transferred to 50% (v/v) ethanol. Material was then taken through a standard ethanol dehydration series (70, 80, 90, 95, 100, 100, and 100% [v/v] ethanol) with at least 24 h per stage. Samples were dried in a SamDri-780 critical point drier at the Washington University Center for Cellular Imaging, coated with gold palladium, and imaged on a Merlin Field Emission scanning electron microscope at 5.0 kV (Zeiss). Photographs were adjusted for brightness, contrast, and input levels using Adobe Photoshop.
RNA-Seq
To complement 13C mass spectrometric measurements of metabolites, expression levels for genes controlling photosynthesis and sugar and starch metabolism were estimated with RNA-seq. As with the 13C experiments, material of S. bicolor and T. triandra was harvested at anthesis and immediately frozen in liquid nitrogen and dissected while frozen. Material was harvested between 10 am and 11 am to minimize circadian effects. For sorghum, mature leaves and inflorescences were harvested from three plants and used as individual biological replicates. Inflorescences were hand dissected into PS, SS, and awn samples. For T. triandra, inflorescences from three tillers from one individual were dissected into bract, PS, SS, and awn, with each tiller representing a biological replicate. Each of the 24 samples (2 species × 4 organs × 3 biological replicates) was individually ground to a fine powder using liquid nitrogen. To extract total RNA, ∼100 mg of tissue was added to 1 mL of TRIzol reagent (15596026; Thermo Fisher Scientific) and the samples were vortexed. Next, 0.2 mL of chloroform was added and the samples were shaken for 15 s and incubated at room temperature for 3 min, followed by centrifugation at 13,000g for 15 min at 4°C. The upper aqueous phase was removed and added to 0.5 mL of isopropanol. The sample was incubated on ice for 10 min and centrifuged as described above to obtain a nucleic acid pellet. The pellet was washed in 70% (v/v) ethanol, air dried briefly, and dissolved in 100 μL of diethyl pyrocarbonate–treated water. Total RNA was cleaned and concentrated using an RNeasy kit (74104; Qiagen) including on-column DNase treatment (79254; Qiagen). Total RNA was quantified with a Qubit RNA HS kit (Q32852; Thermo Fisher Scientific). At least one sample for each organ was run on an RNA Pico Bioanalyzer microfluidics chip (Agilent) to calculate RNA integrity numbers. All RNA integrity numbers were in the range 7.8 to 9.5.
The 24 RNA-seq libraries were prepared using SENSE mRNA library prep kit V2 for Illumina (001.24; Lexogen) with 500-ng input for all samples except for the three T. triandra awn samples (160, 420, and 350 ng, respectively). The protocol was adjusted to produce a mean insert size of ∼413 bp. Each library was indexed uniquely and amplified by 12 cycles of PCR for all samples except for the three T. triandra awn samples, which were amplified using 14 cycles. Final libraries were quantified using a Qubit DNA HS kit (Q32854; Thermo Fisher Scientific). Select libraries were also run on a High Sensitivity DNA Bioanalyzer microfluidic chip (Agilent) to confirm library size in the desired range (∼400 bp). Libraries were pooled by organism (3 biological replicates × 4 organs, each), resulting in 2 pools of 12 libraries at ∼0.8 nM per library (10 nM final pool concentration). These pools were sequenced using Illumina HiSeq4000 paired end (2 × 150 bp) technology at Michigan State University Genomics facility (https://rtsf.natsci.msu.edu/genomics/sequencing-services/).
Data Analysis
See Supplemental Table 13 for numbers of transcripts at each step.
Read-mapping in sorghum
Reads for S. bicolor were trimmed using Trimmomatic (Bolger et al., 2014) and mapped to the sorghum reference genome (Phytozome, Sbicolor_454_v3.1.1) using HISAT (Kim et al., 2015). To create expression level analysis, htseq-count was used to count the number of reads per gene for each sample (Anders et al., 2015).
Read-mapping and identification of putative orthologs in Themeda
Reads for T. triandra were downloaded and submitted to Trinity (Grabherr et al., 2011) for automated trimming, quality filtering, assembly, and expression quantification. Coding sequences and peptides were extracted using transdecoder (Haas et al., 2013). Putative orthologs of the metabolic genes in sorghum were identified using OrthoFinder (Emms and Kelly, 2015) on the longest isoform of peptide sequences from the sorghum and maize genomes and the predicted T. triandra peptides. If an orthogroup contained a sorghum metabolism gene as identified in the sorghum transcriptome analysis, the T. triandra peptides in that group were blasted against the sorghum peptides. If the T. triandra gene’s best match was a sorghum metabolism gene, it was considered that gene’s putative ortholog. The list of Themeda metabolism genes generated from this approach was then used for downstream analyses. Read counts per gene were generated with htseq-count (Anders et al., 2015).
Normalization for library size and log2
All normalization was performed in R (R Core Team, 2013). Raw transcript counts for each sample were normalized for library size across all samples for S. bicolor and T. triandra using the calcNormFactors function in the edgeR package (Robinson et al., 2010) and filtered to remove transcripts with fewer than one normalized read count in at least three samples, effectively discarding unexpressed transcripts. Next, library size-normalized read counts were log2 transformed. This step led to 17,232 unique transcripts in sorghum and 20,826 in T. triandra.
Construction of the metabolic gene set
We developed a custom list of genes involved in carbon metabolism to enable comparison between the RNA-seq data and the carbon isotope data. Manually annotated pathway descriptions are based on the Kyoto Encyclopedia of Genes and Genome (KEGG) pathway (Kanehisa, 2019; Kanehisa and Goto, 2000; Kanehisa et al., 2019) and biochemical textbook descriptions (Buchanan et al., 2002) supported by comparative organ expression profile data. Enzyme Commission numbers of enzymes involved in carbon metabolism were retrieved from the KEGG pathway database (https://www.genome.jp/kegg/kegg3a.html) for glycolysis/gluconeogenesis, the tricarboxylic acid cycle, pentose phosphate pathway, pentose glucuronate interconversions, Fru/Man interconversions, Gal metabolism, starch Suc metabolism, sugar nucleotide metabolism, PYR metabolism, glyoxylate/dicarboxylate metabolism, oxidative phosphorylation, photosynthesis, Calvin Benson cycle, etc. In instances where an enzyme could not be unambiguously assigned to a pathway, TargetP (Emanuelsson et al., 2007), a protein localization prediction tool, was used to determine possible organellar targeting and to guide descriptions. Genes not obviously targeted to chloroplasts or mitochondria were assumed to be cytoplasmic. A subset of C4 genes was identified based on phylogenetic literature and comparative genomic analysis (Wang et al., 2009; Huang et al., 2017). Expression levels of genes encoding these enzymes for each species and tissue were then extracted from the full transcriptomes using custom Python scripts (https://github.com/ekellogg-lab/pedicellate-spikelet-carbon), giving us a total of 3505 loci related to carbon metabolism. The intersection of this list with the log2-normalized filtered transcripts left us with 1441 transcripts in sorghum and 769 in T. triandra. We refer to this list as our set of metabolic genes.
Identification of DE genes
Differential expression analysis was performed in R (R Core Team, 2013). Tukey tests were performed on the expression values of the metabolic gene set using the function TukeyHSD in conjunction with ANOVA to generate all possible pairwise organ comparisons for each transcript in our metabolically-relevant gene list. The P-values generated by this approach were additionally corrected for multiple testing using p.adjust with method set to Benjamini–Hochberg. We identified 922 DE genes in sorghum with at least one significant pairwise organ comparison Benjamini–Hochberg-corrected P-value < 0.05), and 322 DE genes were identified in T. triandra.
Data display
All normalized transcripts and the normalized set of metabolic genes were analyzed by PCA using the function prcomp with log2-transformed expression values and visualized with functions in the ggplot2 package (Wickham, 2016). DE genes were displayed as expression heatmaps and were plotted using pheatmap (Kolde, 2015) with the ward.D2 clustering option and row scaling.
Genes to compare with 13C data
To connect the RNA-seq and 13C data directly, we generated a list of 36 enzymes that could produce the metabolites we assayed with 13C; some of these are encoded by more than one gene. We then used this list of enzymes to generate a small focused subset of the DE metabolic genes, giving us 52 and 24 DE genes in S. bicolor and T. triandra, respectively.
Data Availability
Raw reads from the RNA-seq experiment have been deposited at the Sequence Read Archive of the National Center for Biotechnology Information (accession numbers SAMN15618964 to SAMN15618987; BioProject ID: PRJNA648104). Data matrices for downstream analyses have been deposited at Dryad (https://datadryad.org).
Accession Numbers
See the Supplemental Data Set for the accession numbers referenced in this article.
Supplemental Data
Supplemental Figure 1. 13C atom equivalents produced over time.
Supplemental Figure 2. Sorghum bicolor, PCA of gene expression data.
Supplemental Figure 3. Sorghum bicolor, heat map of the 922 differentially expressed metabolic genes, by organ.
Supplemental Figure 4. Themeda triandra, PCA of gene expression data for the 769 select metabolic genes.
Supplemental Figure 5. Themeda triandra, heat map of 322 differentially expressed metabolic genes, by organ.
Supplemental Figure 6. Themeda triandra, relative expression of 24 genes encoding biosynthetic enzymes immediately responsible for producing the metabolites labeled with 13C.
Supplemental Figure 7. Immunolocalization, negative control with Alexa 488 secondary antibody only and no Rubisco antibody.
Supplemental Table 1. Sorghum bicolor, 14C data and analyses for pulse-chase experiments.
Supplemental Table 2. Andropogon schirensis, 14C data and analyses for pulse-chase experiments.
Supplemental Table 3. Themeda triandra, 14C data and analyses for pulse-chase experiments.
Supplemental Table 4. Sorghum bicolor, 13C data, p values for ANOVA (Type 1, two way), average labeling.
Supplemental Table 5. Sorghum bicolor, 13C data, p values for ANOVA (Type 1, two way), isotopomers of each metabolite analyzed separately.
Supplemental Table 6. Themeda triandra, 13C data, p values for ANOVA (Type 1, two way), average labeling.
Supplemental Table 7. Themeda triandra, 13C data, p values for ANOVA (Type 1, two way), isotopomers of each metabolite analyzed separately.
Supplemental Table 8. General linear model for effect of removal of pedicellate spikelet.
Supplemental Table 9. Andropogon schirensis, 14C data and analyses for pulse-chase experiments, including bract.
Supplemental Table 10. Themeda triandra, 14C data and analyses for pulse-chase experiments, including bract.
Supplemental Table 11. Themeda triandra, including the bract; 13C data, p values for ANOVA (Type 1, two way), average labeling.
Supplemental Table 12. Themeda triandra, including bract, 13C data, p values for ANOVA (Type 1, two way), isotopomers of each metabolite analyzed separately.
Supplemental Table 13. Numbers of unique transcripts after successive filters and related figures.
Supplemental Data Set. Expression values of genes.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
spikelet AmiGo: PO:0009051 AmiGO2
Acknowledgments
We thank Kirk Czymmek (Advanced Bioimaging Laboratory at the Danforth Plant Science Center [DDPSC]) for imaging support, Sherry Flint-Garcia (U.S. Department of Agriculture-Agricultural Research Service) for access to the seed counter, Sally Fabbri (Plant Growth Facility, DDPSC) for plant care, Brad Evans (Proteomics and Mass Spectrometry Core Facility, DDPSC) for technical assistance, James Fitzpatrick and Matthew Joens (Washington University Center for Cellular Imaging) for help with scanning electron microscopy, and the editors and three anonymous reviewers for comments that greatly improved the article. This work was supported in part by National Science Foundation (NSF grant DEB-1457748 to E.A.K.), and through U.S. Department of Agriculture-Agricultural Research Service (support to D.K.A.). The confocal Leica SPX-8 was acquired through an NSF Major Research Instrumentation Program (grant DBI-1337680), and mass spectrometry data was obtained on an instrument funded by NSF Major Research Instrumentation Program (grant DBI-1427621).
AUTHOR CONTRIBUTIONS
T.A-E., V.C., D.K.A., and E.A.K conceived and visualized the research, created the methodology, and wrote the original draft of the article. All authors performed the experiments and produced the figures. D.G., V.C., D.K.A., and E.A.K performed the formal analysis. All authors reviewed and edited the article. E.A.K. and D.K.A. supervised the project. E.A.K. and D.K.A. were project administrators and acquired funding. T.A-E. and E.A.K. took the photos for Figure 1. T.A-E. ran the assays, D.K.A. and E.A.K. did the data analysis and created the figures for Figure 2. T.A-E., D.K.A., and E.A.K. did the experiments for Figures 2A, 2E, and 2I. E.A.K. (with assistance of Matthew Joens) took the images for Figures 2B to 2C, 2F to 2H, and 2J to 2L (SEM). T.A-E., D.K.A., and E.A.K. did the experiments for Figure 3, T.A-E., D.K.A. and V.C. generated the LC-MS/MS data, D.K.A. and V.C. did the data analyses, and V.C. and E.A.K. created the figures. T.A-E., D.K.A., and E.A.K. did the experiments for Figure 4 and Supplemental Figure 1, T.A-E., D.K.A. and V.C. generated the LC-MS/MS data, D.K.A. did the data analyses, and D.K.A. and E.A.K. created the figures. T.A-E., D.K.A., and E.A.K. collected the samples for Figure 5 and Supplemental Figures 2 to 6, T.A-E. prepared RNA for sequencing, T.A-E., D.M.G. and V.C. analyzed the data, V.C. did the statistical analyses, and V.C. and E.A.K. created the figures. Sectioning, immunolocalization, and photomicroscopy for Figure 6 and Supplemental Figure 7 were done by Y.Y. Images in Figures 7A to 7D were created by T.A-E., the experiment was conducted by T.A-E., L.M.J., Y.Y., D.K.A., and E.A.K., data analysis by V.C., and box plots were made by V.C. and E.A.K. Contributions for Figure 8 are the same as for Figure 2. Figure 9 was created by D.K.A.
Footnotes
Articles can be viewed without a subscription.
References
- Abramoff M.D., Magalhaes P.J., Ram S.J.(2004). Image processing with ImageJ. Biophoton. Int. 11: 36–42. [Google Scholar]
- Allen D.K., Ohlrogge J.B., Shachar-Hill Y.(2009). The role of light in soybean seed filling metabolism. Plant J. 58: 220–234. [DOI] [PubMed] [Google Scholar]
- Allen D.K., Young J.D.(2020). Tracing metabolic flux through time and space with isotope labeling experiments. Curr. Opin. Biotechnol. 64: 92–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P.T., Huber W.(2015). HTSeq--A Python framework to work with high-throughput sequencing data. Bioinformatics 31: 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antlfinger A., Wendel L.(1997). Reproductive effort and floral photosynthesis in Spiranthes cernua (Orchidaceae). Am. J. Bot. 84: 769–780. [PubMed] [Google Scholar]
- Arrivault S., Obata T., Szecówka M., Mengin V., Guenther M., Hoehne M., Fernie A.R., Stitt M.(2017). Metabolite pools and carbon flow during C4 photosynthesis in maize: 13CO2 labeling kinetics and cell type fractionation. J. Exp. Bot. 68: 283–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthan W., McKain M.R., Traiperm P., Welker C.A.D., Teisher J.K., Kellogg E.A.(2017). Phylogenomics of Andropogoneae (Panicoideae: Poaceae) of mainland Southeast Asia. Syst. Bot. 42: 418–431. [Google Scholar]
- Aschan G., Pfanz H.(2003). Non-foliar photosynthesis - A strategy of additional carbon acquistion. Flora 198: 81–97. [Google Scholar]
- Asokanthan P.S., Johnson R.W., Griffith M., Krol M.(1997). The photosynthetic potential of canola embryos. Physiol. Plant 101: 353–360. [Google Scholar]
- Bates D., Maechler M., Bolker B., Walker S.(2014). lme4: Linear mixed-effects models using Eigen and S4., Volume 6 (R package version 1.0; ), pp. 1–23. [Google Scholar]
- Bazzaz F.A., Carlson R.W.(1979). Contribution to reproductive effort by photosynthesis of flowers and fruits. Nature 279: 554–555. [Google Scholar]
- Bolger A.M., Lohse M., Usadel B.(2014). Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 30: 2114–2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bort J., Brown R.H., Araus J.L.(1996). Refixation of respiratory CO2 in the ears of C3 cereals. J. Exp. Bot. 47: 1567–1575. [Google Scholar]
- Browse J., Slack C.R.(1985). Fatty-acid synthesis in plastids from maturing safflower and linseed cotyledons. Planta 166: 74–80. [DOI] [PubMed] [Google Scholar]
- Buchanan B., Gruissem W., Jones R.L.(2002). Biochemistry and Molecular Biology of Plants.. (Rockville, MD: American Society of Plant Physiologists; ). [Google Scholar]
- Burow G., Xin Z., Hayes C., Burke J.(2014). Characterization of a multiseeded (msd1) mutant of sorghum for increasing grain yield. Crop Sci. 54: 2030–2037. [Google Scholar]
- Casa A.M., Pressoir G., Brown P.J., Mitchell S.E., Rooney W.L., Tuinstra M.R., Franks C.D., Kresovich S.(2008). Community resources and strategies for association mapping in sorghum. Crop Sci. 48: 30–40. [Google Scholar]
- Clayton W.D., Renvoize S.A.(1986). Genera Graminum: Grasses of the World.. (London: Her Majesty’s Stationery Office; ). [Google Scholar]
- Connor H.E.(1981). Evolution of reproductive systems in the Gramineae. Ann. Mo. Bot. Gard. 68: 48–74. [Google Scholar]
- Connor H.E.(1987). Reproductive biology in the grasses In Grass Systematics and Evolution, Soderstrom T.R., Hilu K.W., Campbell C.S., and Barkworth M.E., eds (Washington, DC: Smithsonian Institution; ), pp. 117–132. [Google Scholar]
- Cresswell J.E., Krick J., Patrick M.A., Lahoubi M.(2010). The aerodynamics and efficiency of wind pollination in grasses. Funct. Ecol. 24: 706–713. [Google Scholar]
- Davidse G.(1987). Fruit dispersal in the Poaceae In Grass Systematics and Evolution, Soderstrom T.R., Hilu K.W., Campbell C.S., and Barkworth M.E., eds (Washington, DC: Smithsonian Institution; ), pp. 143–155. [Google Scholar]
- Dengler N.G., Nelson T.(1999). Leaf structure and development in C4 plants In C4 Plant Biology, Sage R.F., and Monson R.K., eds (San Diego: Academic Press; ), pp. 133–172. [Google Scholar]
- Döring F., Streubel M., Bräutigam A., Gowik U.(2016). Most photorespiratory genes are preferentially expressed in the bundle sheath cells of the C4 grass Sorghum bicolor. J. Exp. Bot. 67: 3053–3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doust A.N., Mauro-Herrera M., Francis A., Shand L.(2014). Morphological diversity and genetic regulation of seed dispersal in grasses. Am. J. Bot. 101: 1759–1769. [DOI] [PubMed] [Google Scholar]
- Earley E.J., Ingland B., Winkler J., Tonsor S.J.(2009). Inflorescences contribute more than rosettes to lifetime carbon gain in Arabidopsis thaliana (Brassicaceae). Am. J. Bot. 96: 786–792. [DOI] [PubMed] [Google Scholar]
- Emanuelsson O., Brunak S., von Heijne G., Nielsen H.(2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2: 953–971. [DOI] [PubMed] [Google Scholar]
- Emms D.M., Kelly S.(2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16: 157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estep M.C., McKain M.R., Vela Diaz D., Zhong J., Hodge J.G., Hodkinson T.R., Layton D.J., Malcomber S.T., Pasquet R., Kellogg E.A.(2014). Allopolyploidy, diversification, and the Miocene grassland expansion. Proc. Natl. Acad. Sci. USA 111: 15149–15154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans L.T., Wardlaw I.F.(1976). Aspects of the comparative physiology of grain yield in cereals. Adv. Agron. 28: 301–359. [Google Scholar]
- Fernandez C.A., Des Rosiers C., Previs S.F., David F., Brunengraber H.(1996). Correction of 13C mass isotopomer distributions for natural stable isotope abundance. J. Mass Spectrom. 31: 255–262. [DOI] [PubMed] [Google Scholar]
- Fischer K.S., Wilson G.L.(1971). Studies of grain production in Sorghum vulgare. II. Sites responsible for grain dry matter production during the post-anthesis period. Aust. J. Agric. Res. 22: 39–47. [Google Scholar]
- Friedman J., Harder L.D.(2004). Inflorescence architecture and wind pollination in six grass species. Funct. Ecol. 18: 851–860. [Google Scholar]
- Furbank R.T., White R., Palta J.A., Turner N.C.(2004). Internal recycling of respiratory CO2 in pods of chickpea (Cicer arietinum L.): the role of pod wall, seed coat, and embryo. J. Exp. Bot. 55: 1687–1696. [DOI] [PubMed] [Google Scholar]
- Gladman N., et al. (2019). Fertility of pedicellate spikelets in sorghum is controlled by a jasmonic acid regulatory module. Int. J. Mol. Sci. 20: 4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goffman F.D., Alonso A.P., Schwender J., Shachar-Hill Y., Ohlrogge J.B.(2005). Light enables a very high efficiency of carbon storage in developing embryos of rapeseed. Plant Physiol. 138: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 29: 644–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundbacher F.J.(1963). The physiological function of the cereal awn. Bot. Rev. 29: 366–381. [Google Scholar]
- Haas B.J., et al. (2013). De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8: 1494–1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattersley P.W., Watson L.(1975). Anatomical parameters for predicting photosynthetic pathways of grass leaves: The “maximum lateral cell count” and the “maximum cells distant count”. Phytomorphology 25: 325–333. [Google Scholar]
- Hattersley P.W., Watson L.(1992). Diversification of photosynthesis In Grass Evolution and Domestication, Chapman G.P., ed (Cambridge: Cambridge University Press; ), pp. 38–116. [Google Scholar]
- Henry R.J., Rangan P., Furtado A., Busch F.A., Farquhar G.D.(2017). Does C4 photosynthesis occur in wheat seeds? Plant Physiol. 174: 1992–1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibberd J.M., Furbank R.T.(2016). Wheat genomics: Seeds of C4 photosynthesis. Nat. Plants 2: 16172. [DOI] [PubMed] [Google Scholar]
- Hibberd J.M., Quick W.P.(2002). Characteristics of C4 photosynthesis in stems and petioles of C3 flowering plants. Nature 415: 451–454. [DOI] [PubMed] [Google Scholar]
- Hodge J.G., Kellogg E.A.(2014). Patterns of inflorescence development of three prairie grasses (Andropogoneae, Poaceae). Int. J. Plant Sci. 175: 963–974. [Google Scholar]
- Hu L., Zhang Y., Xia H., Fan S., Song J., Lv X., Kong L.(2019). Photosynthetic characteristics of non-foliar organs in main C3 cereals. Physiol. Plant. 166: 226–239. [DOI] [PubMed] [Google Scholar]
- Huang P., Studer A.J., Schnable J.C., Kellogg E.A., Brutnell T.P.(2017). Cross species selection scans identify components of C4 photosynthesis in the grasses. J. Exp. Bot. 68: 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Lee Y.K., Gladman N., Chopra R., Christensen S.A., Regulski M., Burow G., Hayes C., Burke J., Ware D., Xin Z.(2018). MSD1 regulates pedicellate spikelet fertility in sorghum through the jasmonic acid pathway. Nat. Commun. 9: 822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin B., Wang L., Wang J., Teng N.J., He X.D., Mu X.J., Wang Y.L.(2010). The structure and roles of sterile flowers in Viburnum macrocephalum f. keteleeri (Adoxaceae). Plant Biol. (Stuttg) 12: 853–862. [DOI] [PubMed] [Google Scholar]
- Jowett D.(1968). Inheritance of glume size and awn length in sorghum. Crop Sci. 8: 342–345. [Google Scholar]
- Kanehisa M.(2019). Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28: 1947–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Goto S.(2000). KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28: 27–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M., Sato Y., Furumichi M., Morishima K., Tanabe M.(2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 47 (D1): D590–D595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellogg E.A.(2000). Molecular and morphological evolution in Andropogoneae In Grasses: Systematics and Evolution, Jacobs S.W.L., and Everett J.E., eds (Melbourne: CSIRO; ), pp. 149–158. [Google Scholar]
- Kellogg E.A.(2015). Poaceae In Families and Genera of Vascular Plants, Kubitzki K., ed (Berlin: Springer; ), pp. 1–416. [Google Scholar]
- Kim D., Langmead B., Salzberg S.L.(2015). HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 12: 357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King S.P., Badger M.R., Furbank R.T.(1998). CO2 refixation characteristics of developing canola seeds and silique wall. Aust. J. Plant Physiol. 25: 377–386. [Google Scholar]
- Kolde R. (2015). pheatmap: Pretty Heamaps. R package version 1.0.2. [Google Scholar]
- Krannitz P.G., Maun M.A.(1991). An experimental study of flora display size and reproductive success in Viburnum opulus: Importance of grouping. Can. J. Bot. 69: 394–399. [Google Scholar]
- Kuznetsova A., Christensen R.H., Bavay C., Brockhoff P.B.(2015). Automated mixed ANOVA modeling of sensory and consumer data. Food Qual. Prefer. 40: 31–38. [Google Scholar]
- Langdale J.A., Zelitch I., Miller E., Nelson T.(1988). Cell position and light influence C4 versus C3 patterns of photosynthetic gene expression in maize. EMBO J. 7: 3643–3651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roux L.G., Kellogg E.A.(1999). Floral development and the formation of unisexual spikelets in the Andropogoneae (Poaceae). Am. J. Bot. 86: 354–366. [PubMed] [Google Scholar]
- Li X., Wang H., Li H., Zhang L., Teng N., Lin Q., Wang J., Kuang T., Li Z., Li B., Zhang A., Lin J.(2006). Awns play a dominant role in carbohydrate production during the grain-filling stages in wheat (Triticum aestivum). Physiol. Plant 127: 701–709. [Google Scholar]
- Linder H.P., Lehmann C.E.R., Archibald S., Osborne C.P., Richardson D.M.(2018). Global grass (Poaceae) success underpinned by traits facilitating colonization, persistence and habitat transformation. Biol. Rev. Camb. Philos. Soc. 93: 1125–1144. [DOI] [PubMed] [Google Scholar]
- Lucas W.J., Bouché-Pillon S., Jackson D.P., Nguyen L., Baker L., Ding B., Hake S.(1995). Selective trafficking of KNOTTED1 homeodomain protein and its mRNA through plasmodesmata. Science 270: 1980–1983. [DOI] [PubMed] [Google Scholar]
- Ma F., Jazmin L.J., Young J.D., Allen D.K.(2014). Isotopically nonstationary 13C flux analysis of changes in Arabidopsis thaliana leaf metabolism due to high light acclimation. Proc. Natl. Acad. Sci. USA 111: 16967–16972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma F., Jazmin L.J., Young J.D., Allen D.K.(2017). Isotopically nonstationary metabolic flux analysis (INST-MFA) of photosynthesis and photorespiration in plants. Methods Mol. Biol. 1653: 167–194. [DOI] [PubMed] [Google Scholar]
- Maydup M.L., Antonietta M., Guiamet J.J., Graciano C., López J.R., Tambussi E.A.(2010). The contribution of ear photosynthesis to grain filling in bread wheat (Triticum aestivum L.). Field Crops Res. 119: 48–58. [Google Scholar]
- McCormick R.F., et al. (2018). The Sorghum bicolor reference genome: Improved assembly, gene annotations, a transcriptome atlas, and signatures of genome organization. Plant J. 93: 338–354. [DOI] [PubMed] [Google Scholar]
- McKone M., Lund C., O’Brien J.(1998). Reproductive biology of two dominant prairie grasses (Andropogon gerardii and Sorghastrum nutans, Poaceae): Male-biased sex allocation in wind-pollinated plants? Am. J. Bot. 85: 776–783. [PubMed] [Google Scholar]
- Morales C.L., Traveset A., Harder L.D.(2013). Sterile flowers increase pollinator attraction and promote female success in the Mediterranean herb Leopoldia comosa. Ann. Bot. 111: 103–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motzo R., Giunta F.(2002). Awnedness affects grain yield and kernel weight in near-isogenic lines of durum wheat. Aust. J. Agric. Res. 53: 1285–1293. [Google Scholar]
- Nielsen L.R., Philipp M., Siegismund H.R.(2002). Selective advantage of ray florets in Scalesia affinis and S. pedunculata (Asteraceae), two endemic species from the Galápagos. Evol. Ecol. 16: 139–153. [Google Scholar]
- Niklas K.J.(1987). Pollen capture and wind-induced movement of compact and diffuse grass panicles - Implications for pollination efficiency. Am. J. Bot. 74: 74–89. [Google Scholar]
- Paterson A.H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Peart M.H.(1979). Experiments on the biological significance of the morphology of seed-dispersal units in grasses. J. Ecol. 67: 843–863. [Google Scholar]
- Peart M.H.(1981). Further experiments on the biological significance of the morphology of seed-dispersal units in grasses. J. Ecol. 69: 425–436. [Google Scholar]
- Peart M.H.(1984). The effects of morphology, orientation and position of grass diaspores on seedling survival. J. Ecol. 72: 437–453. [Google Scholar]
- Peart M.H., Clifford H.T.(1987). The influence of diaspore morphology and soil-surface properties on the distribution of grasses. J. Ecol. 75: 569–576. [Google Scholar]
- Pengelly J.J., Kwasny S., Bala S., Evans J.R., Voznesenskaya E.V., Koteyeva N.K., Edwards G.E., Furbank R.T., von Caemmerer S.(2011). Functional analysis of corn husk photosynthesis. Plant Physiol. 156: 503–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P., Furtado A., Henry R.J.(2016a). Commentary: New evidence for grain specific C4 photosynthesis in wheat. Front. Plant Sci. 7: 1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangan P., Furtado A., Henry R.J.(2016b). New evidence for grain specific C4 photosynthesis in wheat. Sci. Rep. 6: 31721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2013). R: A language and environment for statistical computing.. (Vienna: R Foundation for Statistical Computing; ). [Google Scholar]
- Robinson M.D., McCarthy D.J., Smyth G.K.(2010). edgeR: A bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolletschek H., Radchuk R., Klukas C., Schreiber F., Wobus U., Borisjuk L.(2005). Evidence of a key role for photosynthetic oxygen release in oil storage in developing soybean seeds. New Phytol. 167: 777–786. [DOI] [PubMed] [Google Scholar]
- Ruuska S.A., Schwender J., Ohlrogge J.B.(2004). The capacity of green oilseeds to utilize photosynthesis to drive biosynthetic processes. Plant Physiol. 136: 2700–2709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzin S.E.(1999). Plant Microtechnique and Microscopy. (New York: Oxford University Press; ). [Google Scholar]
- Sanchez-Bragado R., Molero G., Reynolds M.P., Araus J.L.(2014). Relative contribution of shoot and ear photosynthesis to grain filling in wheat under good agronomical conditions assessed by differential organ δ13C. J. Exp. Bot. 65: 5401–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider M.P., Vegettti A.C.(1992). The synflorescence in species of Sorghinae (Andropogoneae-Poaceae). Beitr. Biol. Pflanzen 67: 225–239. [Google Scholar]
- Schwender J., Goffman F., Ohlrogge J.B., Shachar-Hill Y.(2004). Rubisco without the Calvin cycle improves the carbon efficiency of developing green seeds. Nature 432: 779–782. [DOI] [PubMed] [Google Scholar]
- Sieglinger J.B., Swanson A.F., Martin J.H.(1934). Inheritance of awn development in sorghums. J. Agric. Res. 49: 663–668. [Google Scholar]
- Simkin A.J., Faralli M., Ramamoorthy S., Lawson T.(2020). Photosynthesis in non-foliar tissues: Implications for yield. Plant J. 101: 1001–1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuessy T.F., Spooner D.M., Evans K.A.(1986). Adaptive significance of ray corollas in Helianthus grosseserratus (Compositae). Am. Midl. Nat. 115: 191–197. [Google Scholar]
- Teisher J.K., McKain M.R., Schaal B.A., Kellogg E.A.(2017). Polyphyly of Arundinoideae (Poaceae) and evolution of the twisted geniculate lemma awn. Ann. Bot. 120: 725–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vegetti A.C.(1999). Typology of the synflorescence of Andropogoneae (Poaceae), additional comments. Feddes Repert. 110: 111–126. [Google Scholar]
- Vemmos S.N., Goldwin G.K.(1994). The photosynthetic activity of Cox’s Orange Pippin apple in relation to fruit setting. Ann. Bot. 73: 385–391. [Google Scholar]
- Wang X., Gowik U., Tang H., Bowers J.E., Westhoff P., Paterson A.H.(2009). Comparative genomic analysis of C4 photosynthetic pathway evolution in grasses. Genome Biol. 10: R68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Bräutigam A., Weber A.P., Zhu X.G.(2014). Three distinct biochemical subtypes of C4 photosynthesis? A modelling analysis. J. Exp. Bot. 65: 3567–3578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson L., Dallwitz M.J. (1992. onward) The grass genera of the world: descriptions, illustrations, identification, and information retrieval; including synonyms, morphology, anatomy, physiology, phytochemistry, cytology, classification, pathogens, world and local distribution, and references. Version 15. delta-intkey.com.
- Weissmann S., Ma F., Furuyama K., Gierse J., Berg H., Shao Y., Taniguchi M., Allen D.K., Brutnell T.P.(2016). Interactions of C4 subtype metabolic activities and transport in maize are revealed through the characterization of DCT2 mutants. Plant Cell 28: 466–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickham H.(2016). ggplot2: Elegant Graphics for Data Analysis.. (New York: Springer-Verlag; ). [Google Scholar]
- Young J.D.(2014). INCA: A computational platform for isotopically non-stationary metabolic flux analysis. Bioinformatics 30: 1333–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zelitch I., Schultes N.P., Peterson R.B., Brown P., Brutnell T.P.(2009). High glycolate oxidase activity is required for survival of maize in normal air. Plant Physiol. 149: 195–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Raw reads from the RNA-seq experiment have been deposited at the Sequence Read Archive of the National Center for Biotechnology Information (accession numbers SAMN15618964 to SAMN15618987; BioProject ID: PRJNA648104). Data matrices for downstream analyses have been deposited at Dryad (https://datadryad.org).