Physical and functional interactions between pectin and cellulose are required for normal wall assembly and plant growth, as revealed in analyses of mutants for a pectin methyltransferase.
Abstract
Pectins are abundant in the cell walls of dicotyledonous plants, but how they interact with other wall polymers and influence wall integrity and cell growth has remained mysterious. Here, we verified that QUASIMODO2 (QUA2) is a pectin methyltransferase and determined that QUA2 is required for normal pectin biosynthesis. To gain further insight into how pectin affects wall assembly and integrity maintenance, we investigated cellulose biosynthesis, cellulose organization, cortical microtubules, and wall integrity signaling in two mutant alleles of Arabidopsis (Arabidopsis thaliana) QUA2, qua2 and tsd2. In both mutants, crystalline cellulose content is reduced, cellulose synthase particles move more slowly, and cellulose organization is aberrant. NMR analysis shows higher mobility of cellulose and matrix polysaccharides in the mutants. Microtubules in mutant hypocotyls have aberrant organization and depolymerize more readily upon treatment with oryzalin or external force. The expression of genes related to wall integrity, wall biosynthesis, and microtubule stability is dysregulated in both mutants. These data provide insights into how homogalacturonan is methylesterified upon its synthesis, the mechanisms by which pectin functionally interacts with cellulose, and how these interactions are translated into intracellular regulation to maintain the structural integrity of the cell wall during plant growth and development.
INTRODUCTION
The primary cell walls of plants are mainly composed of cellulose, hemicelluloses, and pectins, which interact to form structural networks and tune wall mechanical strength, thus regulating cell shape determination and organ morphogenesis (Cosgrove, 2005). Cellulose is synthesized by cellulose synthase complexes (CSCs) at the plasma membrane (Paredez et al., 2006), and its β-1,4-glucan chains are extruded into the apoplast and coalesce into microfibrils, the patterned deposition of which can be guided by cortical microtubules (Baskin et al., 2004). Xyloglucan, the major hemicellulose in the primary walls in eudicots, is thought to interact with cellulose microfibrils at “biomechanical hotspots” that control wall expansibility (Park and Cosgrove, 2012) and to bind to the surface of cellulose (Zheng et al., 2018). Pectins are complex acidic polysaccharides that are rich in galacturonic acid (GalA) and include homogalacturonan (HG), rhamnogalacturonan I (RG-I), and RG-II domains as well as xylogalacturonan and apiogalacturonan in some species (Atmodjo et al., 2013). HG is an unbranched homopolymer of α-1,4-linked GalA (Atmodjo et al., 2013). After synthesis in the Golgi, HG is delivered to the apoplast in a highly methylesterified form and is demethylesterified in the wall by pectin methylesterases (PMEs), releasing protons and methanol (Driouich et al., 2012). The patterns of this demethylesterification determine whether HG will form Ca2+-mediated cross-links and/or become susceptible to degradation by pectinases. Pectins function to control wall thickness, cell adhesion, and tissue integrity during plant development (Caffall and Mohnen, 2009). Pectin-modifying genes can thus influence wall and tissue mechanics; for example, Arabidopsis (Arabidopsis thaliana) plants overexpressing the pectin-degrading gene POLYGALACTURONASE INVOLVED IN EXPANSION2 or lacking PME35 have stems with altered mechanical robustness, supporting a function for pectins in regulating tissue mechanics during development (Hongo et al., 2012; Xiao et al., 2017).

In Arabidopsis, multiple glycosyltransferases, methyltransferases, and acetyltransferases are required for pectin biosynthesis (Atmodjo et al., 2013). Several proteins involved in HG biosynthesis have been identified and characterized. GLUCURONATE 4-EPIMERASE1 (GAE1) functions in pectin biosynthesis by converting UDP-d-glucuronic acid to UDP-d-galacturonic acid and also influences cell wall integrity maintenance (Bethke et al., 2016). Arabidopsis GALACTURONOSYLTRANSFERASE1 (GAUT1) displays HG:GalA transferase activity and can form a complex with the homologous GAUT7 to synthesize high-molecular-weight polymeric HG via a “two-phase” mechanism (Sterling et al., 2006; Atmodjo et al., 2011; Amos et al., 2018). In grasses and woody dicot plants, GAUT4 possesses HG:GalA transferase activity, and knockdown of GAUT4 expression promotes plant growth and enhances biomass degradability (Biswal et al., 2018). GAUT11, another confirmed HG α-1,4 GalA transferase, catalyzes HG elongation and likely functions in RG-I production in seed mucilage (Voiniciuc et al., 2018). QUASIMODO1 (QUA1/GAUT8) encodes a putative glycosyltransferase and is likely required for HG biosynthesis, since qua1 mutant plants have reduced uronic acid content in their walls and display cell adhesion defects (Bouton et al., 2002; Orfila et al., 2005; Durand et al., 2009; Verger et al., 2018). Either during or soon after polymerization, GalA residues in HG can be acetylated at O2 or O3 positions, and the carboxyl groups of most GalA residues in HG are methylesterified by methyltransferases (Atmodjo et al., 2013), but the proteins that perform these modifications have not yet been positively identified.
Several candidate HG methyltransferase genes have been identified in Arabidopsis. Arabidopsis QUA2, also named TUMOROUS SHOOT DEVELOPMENT2 (TSD2), encodes an annotated pectin methyltransferase, although methyltransferase activity has not previously been biochemically demonstrated for its gene product (Krupková et al., 2007; Mouille et al., 2007). HG-derived GalA from extracted pectin in the qua2 mutant is reduced by 50%, and both qua2 and tsd2 mutant seedlings have shorter hypocotyls with cell-cell adhesion defects in hypocotyl epidermal cells (Krupková et al., 2007; Mouille et al., 2007). Mutants for another allele of QUA2, osu1, are more sensitive to imbalanced carbon/nitrogen conditions, suggesting that QUA2 is also a key modulator of carbon and nitrogen nutrient balance during plant development (Gao et al., 2008). In addition, mutants of a rice (Oryza sativa) putative ortholog of AtQUA2 show altered wall composition and developmental defects (Qu et al., 2016; Xu et al., 2017). OsQUA2 shares high sequence similarity with AtQUA2, and mutation of OsQUA2 results in reduced HG methylesterification in culm-sieve element and root cell walls, affecting Suc transport and root development, respectively (Qu et al., 2016; Xu et al., 2017). QUA3, another putative HG methyltransferase, influences pectin methylesterification and regulates cell wall biosynthesis in Arabidopsis suspension-cultured cells (Miao et al., 2011). Two more Arabidopsis genes, COTTON GOLGI RELATED2 (CGR2) and CGR3, encode putative HG methyltransferases, and cgr2 cgr3 double mutants display cell wall, photosynthesis, and growth defects (Kim et al., 2015; Weraduwage et al., 2016). Although cgr2 cgr3 double mutants show altered patterns of HG methylesterification in immunolabeling experiments and overexpression lines for either gene show increased general methyltransferase activity, these proteins were not shown to have specific HG methyltransferase activity (Kim et al., 2015).
HG has long been implicated in cell-cell adhesion in eudicots. For example, the cell adhesion defect in the Arabidopsis friable1 (frb1) mutant, in which a putative O-fucosyltransferase is mutated, has been attributed to alterations in pectin structure (Neumetzler et al., 2012). However, a simple model in which HG “glues” plant cells together is likely incorrect: a suppressor mutation of qua2, named esmeralda1, targets another putative O-fucosyltransferase and does not rescue the pectin content defect of the qua2 mutation (Verger et al., 2016). In addition, loss of FRB1 function results in expression changes in wall integrity-related genes, indicating that FRB1 is required for normal cell wall integrity (Neumetzler et al., 2012). Thus, wall integrity signaling that includes the sensing of pectin levels, configurations, interactions, or modifications is likely to influence cell-cell adhesion (Verger et al., 2016, 2018).
The above studies on the structure and function of pectins, plus many other analyses of wall polymer biosynthesis and function, lay a foundation to explore the complex mechanisms and implications of interactions among the different components of plant cell walls. Studies focusing on interactions between cellulose microfibrils and xyloglucan have led to a revision of the cellulose-xyloglucan load-bearing model, called the “biomechanical hotspot” model (Park and Cosgrove, 2012, 2015; Wang et al., 2013). An absence of detectable xyloglucan in the Arabidopsis xxt1 xxt2 mutant disrupts cellulose biosynthesis and cellulose microfibril patterning, supporting the importance of cellulose-xyloglucan interactions in determining wall structure (Xiao et al., 2016). Pectins are thought to interpenetrate into the cellulose-xyloglucan network (Somerville et al., 2004). Recent evidence demonstrating that pectins interact closely with cellulose microfibrils supports a role for pectin-cellulose interactions in maintaining cell wall integrity. 2D and 3D magic-angle-spinning (MAS) solid-state NMR experiments with Arabidopsis have detected extensive close associations between pectins and cellulose in native primary walls (Dick-Pérez et al., 2011; Wang et al., 2015), and experiments in which bacterial cellulose hydrogels were synthesized in pectin solutions revealed intimate interactions between subsets of pectins and cellulose microfibrils in vitro (Lopez-Sanchez et al., 2016). Chemical genetic screening has uncovered a drug, cobtorin, that disrupts cellulose-microtubule coalignment and can be counteracted by overexpressing pectin-modifying genes or pectinase treatment, implying that pectins might be involved in the deposition of cellulose microfibrils in alignment with cortical microtubules (Yoneda et al., 2010). Aside from pectin interactions with cellulose, acidic pectins can also interact with structural glycoproteins such as the extensins AtEXT3 and AtEXT4 to form pectate-extensin complexes, which might template further orderly deposition of nascent cell walls during cytokinesis (Cannon et al., 2008). Pectins have also been posited to cross-link covalently with hemicelluloses (Popper and Fry, 2008; Cornuault et al., 2014). Despite these advances, the exploration of how pectins affect cellulose biosynthesis and organization during cell wall assembly is still in its early stages.
In this study, we determined the molecular function of QUA2, which encodes an HG methyltransferase that is critical for normal HG biosynthesis. Then, we used two allelic Arabidopsis mutants with defects in pectin biosynthesis, qua2 and tsd2, to investigate the molecular mechanisms that functionally link pectin and cellulose in the cell wall and the effects of pectin deficiency on cellulose, the cytoskeleton, and wall and tissue integrity. The resulting data reveal functional interactions between pectin and cellulose during cell wall assembly and provide new evidence at the genetic, molecular, and cellular levels of the links between pectins, cellulose, the microtubule cytoskeleton, and wall integrity sensing during plant cell growth and organ morphogenesis.
RESULTS
Purified QUA2 Shows Methyltransferase Activity on Polygalacturonic Acid In Vitro
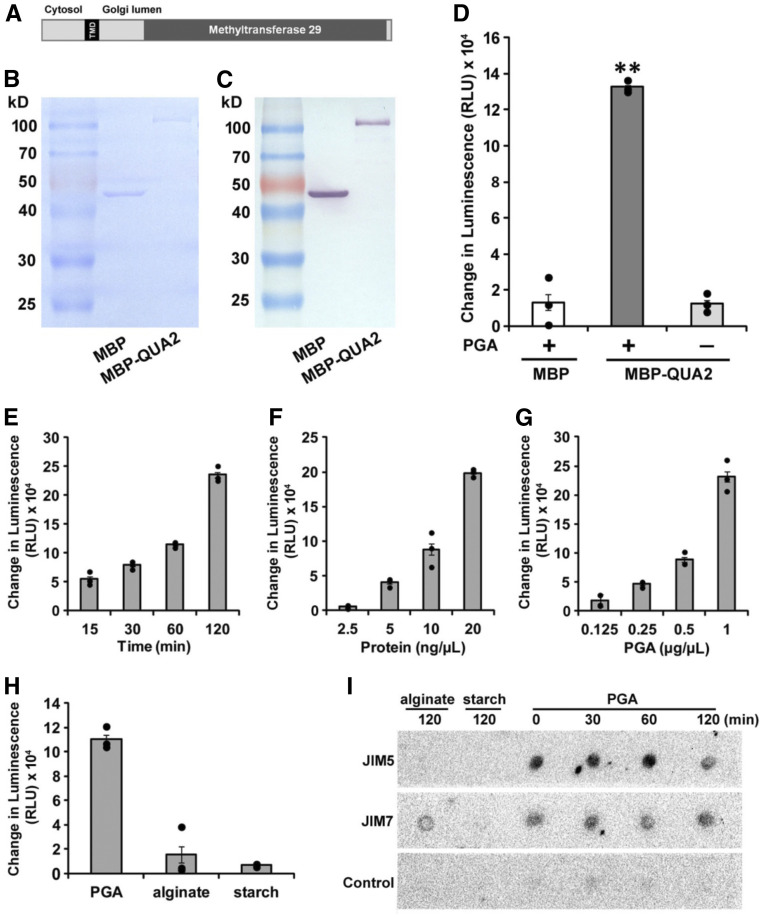
Although HG methyltransferase activity has long been detected in crude membrane fractions from plants, no purified protein has been demonstrated to possess this enzymatic activity in vitro (Goubet and Mohnen, 1999; Ibar and Orellana, 2007; Miao et al., 2011; Kim et al., 2015). The QUA2 protein is predicted to contain 684 amino acids and to be anchored in the Golgi membrane by a single transmembrane domain near its N terminus. Its C terminus is predicted to reside in the Golgi lumen and harbors a large catalytic region, which contains a putative S-adenosylmethionine (SAM)-dependent methyltransferase domain (Figure 1A). To determine the enzymatic activity of QUA2, a sequence including the methyltransferase domain of QUA2 without the transmembrane domain (amino acids 150 to 684) was cloned and expressed in Escherichia coli. After amylose affinity chromatography and size exclusion chromatography, a truncated QUA2 protein fused with maltose binding protein (MBP) and a His tag (MBP-QUA2) was purified to homogeneity as verified by SDS-PAGE (Figure 1B) and immunodetection with an anti-His antibody (Figure 1C).
Figure 1.
QUA2 Displays Methyltransferase Activity on PGA In Vitro.
(A) Schematic of predicted protein structure of QUA2. The black box represents the transmembrane domain (TMD). The dark gray box indicates the catalytic domain, which is in the SAM-dependent methyltransferase_29 superfamily. The N and C termini of QUA2 putatively face the cytosol and Golgi lumen, respectively.
(B) SDS-PAGE and Coomassie blue staining of purified MBP and MBP-fused QUA2 proteins. MBP and MBP-QUA2 are predicted to migrate at ∼40 and ∼100 kD, respectively.
(C) Immunoblot of purified MBP and MBP-fused QUA2 proteins using an anti-polyhistidine antibody.
(D) Methyltransferase assay with purified proteins. For each 50-μL reaction, 1 µg of purified MBP or MBP-QUA2 protein was incubated with 50 μg of PGA in the presence of 10 μM SAM, 2 mM Mg2+, and 0.25 M Suc. Enzymatic activity is proportional to the accumulated reaction product SAH, which is converted into ADP, then to ATP, and finally monitored by luciferase activity. The change in luminescence (in relative light units [RLU]) is the net luminescence over background, which is calculated as the raw luminescence minus the background luminescence (no-enzyme control).
(E) to (G) Methyltransferase activity of purified MBP-QUA2 is time- and concentration-dependent. Reactions were set up as described in (D), except for the indicated protein concentrations (F) or PGA concentrations (G). After incubation at 30°C for 2 h except in (E), the reactions were stopped by the addition of 0.5% trifluoroacetic acid and analyzed for SAH content.
(H) Substrate specificity test of QUA2. Methyltransferase assays were performed when supplied with 50 μg of the indicated polysaccharide. In the presence of PGA, the SAH generated is more than sevenfold and 16-fold higher than when alginate and starch were supplied, respectively.
(I) Immuno dot blot of the product of the methyltransferase reaction of MBP-QUA2 using monoclonal antibodies JIM5 (recognizing low-methylesterified HG) and JIM7 (recognizing methylesterified HG). Products reacted in the presence of the indicated substrate for the indicated time were adsorbed to a nitrocellulose membrane for immunoblotting. Control dots were processed in the absence of primary antibody.
Each column in (D) to (H) represents the average of three replicates, and error bars show se. **, P < 0.001, Student’s t test.
In general, SAM-dependent methyltransferases use SAM as a methyl donor and DNA, proteins, or other molecules as methyl acceptors to generate S-adenosylhomocysteine (SAH) and methylated products (Fontecave et al., 2004). In HG methylation, methyl groups from donor SAM molecules are thought to be transferred to C-6 carboxyls of GalA residues, resulting in the accumulation of SAH. Therefore, the concentration of SAH produced will be proportional to methyltransferase activity. Purified MBP or MBP-QUA2 was incubated with SAM and polygalacturonic acid (PGA; the fully demethylated form of HG) for 2 h. After the completion of the reactions, the generated SAH was converted into ADP, which was subsequently converted into ATP and measured by luciferase activity (Hsiao et al., 2016). In comparison with the MBP-only control, the reaction including both MBP-QUA2 and PGA yielded significant methyltransferase activity (Figure 1D). When PGA was omitted, accumulated SAH remained at background levels (Figure 1D). The methyltransferase assay was also performed for varying durations or supplied with different concentrations of MBP-QUA2 or PGA. The generated SAH increased with increasing incubation time or concentration of reaction components (Figures 1E to 1G), indicating that the methyltransferase activity of QUA2 is time- and concentration-dependent. The substrate specificity of MBP-QUA2 was investigated using the polysaccharides PGA, alginate, and starch. As shown in Figure 1H, PGA supported significantly higher methyltransferase activity than the other two polysaccharides. To further investigate whether PGA acts as the methyl acceptor during the reaction, reaction products using different polysaccharides as substrates were recovered after varying incubation times and used for immunoblotting with JIM5 (recognizing low-methylesterified HG) and JIM7 (recognizing methylesterified HG) antibodies. The products using PGA as substrate showed binding with JIM5 and JIM7, although binding intensity with JIM7 was weaker than with JIM5. JIM5 binding to the PGA reaction products diminished slightly with increasing reaction time (Figure 1I). Taken together, these results demonstrate that QUA2 is a functional HG methyltransferase in vitro.
Shorter Hypocotyls and Cell-Cell Adhesion Defects Are Evident in qua2 and tsd2 Mutants
To investigate the biological function of QUA2, we examined the phenotypes of two QUA2 mutants. qua2 and tsd2 mutants contain point mutations in the QUA2 (At1g78240) gene that result in stop codons (Supplemental Figure 1). Both mutants have independently been shown to have shorter hypocotyls and cell adhesion defects in epidermal cells of hypocotyls (Krupková et al., 2007; Mouille et al., 2007). To confirm these hypocotyl phenotypes simultaneously, seeds of the Arabidopsis wild-type Columbia-0 (Col) ecotype controls and qua2 and tsd2 mutants were sown on Murashige and Skoog (MS) plates without Suc and grown in the dark. qua2 and tsd2 etiolated seedlings had shorter hypocotyls than Col controls from 2 to 6 d of growth, and tsd2 hypocotyls were slightly shorter than qua2 hypocotyls (Figures 2A and 2B). Six-day-old etiolated Col seedlings had an average hypocotyl length of 1.59 ± 0.17 (sd) cm, whereas 6-d-old etiolated qua2 and tsd2 mutants had average hypocotyl lengths of 0.59 ± 0.12 and 0.51 ± 0.09 cm, respectively (n ≥ 30 seedlings per genotype; Figures 2A and 2B). Similar to previous reports (Krupková et al., 2007; Mouille et al., 2007), epidermal cells in 3-d-old etiolated qua2 and tsd2 hypocotyls were often detached from other cells (Figure 2C); in addition, pairs of guard cells in 6-d-old light-grown cotyledons were also sometimes separated from one another as observed after propidium iodide staining (Figure 2D), although these defects were less evident in true leaves. Abnormal cell separation was not observed in Col controls (Figures 2C and 2D). These data indicate that cell adhesion defects occur in at least two tissues of qua2 and tsd2 mutant plants.
Figure 2.
qua2 and tsd2 Mutants Have Shorter Hypocotyls and Show Cell Adhesion Defects.
(A) Six-day-old etiolated Arabidopsis Col, qua2, and tsd2 seedlings. Bar = 1 cm.
(B) Quantification of hypocotyl length in dark-grown seedlings 2 to 6 d after sowing (DAS; n ≥ 30 seedlings per genotype per time point). qua2 and tsd2 hypocotyls were much shorter than Col controls, and tsd2 hypocotyls were shorter than qua2 hypocotyls. Error bars represent sd. *, P < 0.05 and **, P < 0.001, Student’s t test.
(C) Hypocotyls from 3-d-old dark-grown Col, qua2, and tsd2 seedlings. Bar = 100 µm.
(D) Images of stomata present on the abaxial side of cotyledons of 6-d-old light-grown Col, qua2, and tsd2 seedlings. Cells were labeled with 100 µg/mL propidium iodide and imaged by spinning-disk confocal microscopy. Bar = 10 µm.
Mutations in QUA2 Limit Adult Plant Growth
After investigating seedling phenotypes, we next observed the phenotypes of adult plants. Three-week-old qua2 and tsd2 plants had smaller rosettes and shorter petioles compared with Col controls when grown in a light regime of 16 h of light/8 h of dark: average rosette diameter in qua2 and tsd2 plants was 3.74 ± 0.56 and 3.07 ± 0.96 cm, respectively, whereas it was 5.41 ± 0.66 cm in Col (n = 16 rosettes per genotype; Supplemental Figures 2A and 2B), and petiole length and leaf area in both mutants were smaller than in the Col wild type (Supplemental Figures 2C and 2D). Mature qua2 and tsd2 plants grown for 8 weeks had shorter stems with respective average heights of 28.12 ± 2.64 and 23.94 ± 2.54 cm, whereas average stem height was 40.00 ± 3.06 cm in Col controls (n = 16 plants per genotype; Supplemental Figures 2E and 2F). Both rosette diameter and stem length were significantly smaller in tsd2 plants than in qua2 plants (Supplemental Figures 2B and 2F). These data indicate that pectin deficiency impairs growth in the mutant plants and that the tsd2 mutation, which disrupts the first exon of QUA2/TSD2, causes more severe effects on plant growth than the qua2 mutation, which occurs in the seventh exon (Supplemental Figure 1). Together, these results indicate the involvement of QUA2 in cell elongation to maintain plant growth, even though cell adhesion defects were not as evident during adult growth in the mutants.
QUA2 Is Expressed in Multiple Tissues
Microarray data indicate that QUA2 is highly expressed in most tissues except mature pollen (Winter et al., 2007). To provide more detailed information about the spatiotemporal patterning of QUA2 expression, we generated multiple transformant lines of Col plants expressing a construct in which the GUS gene was fused to 2.5 kb of the promoter region of QUA2 and performed GUS staining experiments across multiple developmental stages and tissues. In young seedlings grown either in light or in dark, GUS staining was evident in cotyledons, roots, and hypocotyls (Supplemental Figures 3A to 3C). In adult plants, GUS staining was detected in expanding rosette leaves (Supplemental Figure 3D), distal and proximal ends of developing siliques (Supplemental Figure 3E), flowers (Supplemental Figures 3F and 3G), and xylem, phloem, and parenchyma cells of stems (Supplemental Figure 3H). Together, these data indicate that QUA2 is expressed across a wide range of tissues and might function in the growth of multiple organs during plant development.
Uronic Acid Levels Are Reduced, but Degree of HG Methylesterification Is Not, in qua2 and tsd2 Mutants
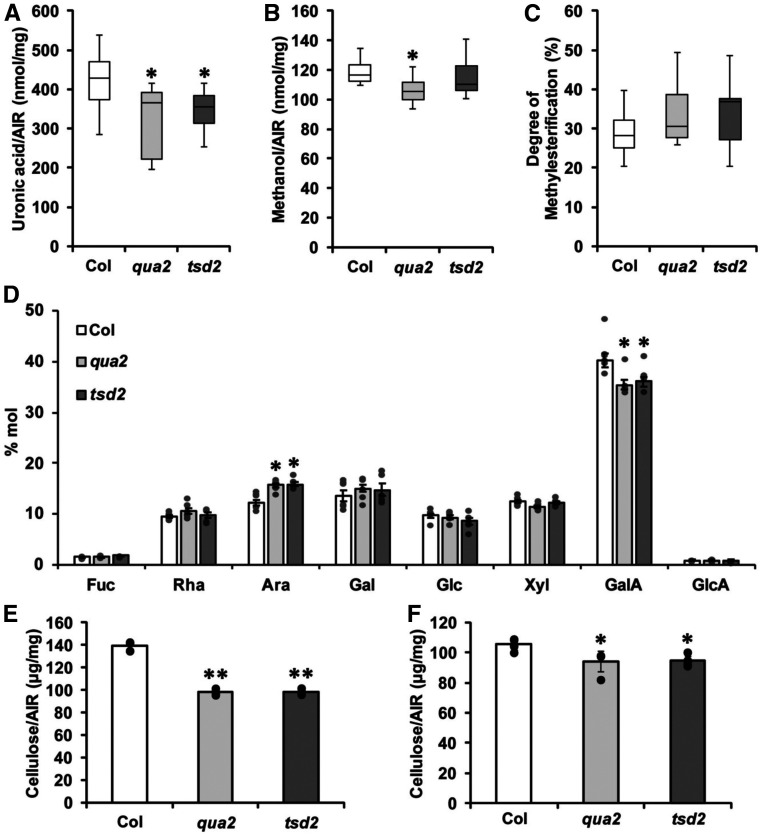
As the above results show, QUA2 is an HG methyltransferase that is expected to function during the biosynthesis of pectic HG. However, previous studies reported an ∼50% reduction in extractable HG, rather than total HG, in the qua2 mutant (Mouille et al., 2007) and no significant reduction in total uronic acid in the tsd2 mutant (Krupková et al., 2007), with no significant reduction in the degree of HG methylesterification in either mutant. To simultaneously compare both qua2 and tsd2 mutants with Col controls in the same set of experiments, we measured total uronic acid content and monosaccharide composition in 6-d-old etiolated seedlings. Total uronic acid content in qua2 and tsd2 mutants was significantly lower than in Col controls on a mass fractional basis (Figure 3A). To estimate the degree of pectin methylesterification, we measured methanol release upon saponification from cell walls of 6-d-old dark-grown seedlings. The amounts of released methanol did not differ significantly in qua2 or tsd2 samples from that released from Col samples (Figure 3B). Normalizing by the amount of HG in the mutants, the degree of HG methylesterification did not differ significantly from that in Col controls (Figure 3C). The lower uronic acid content in qua2 and tsd2 plants is consistent with results from sugar monosaccharide composition analysis, in which cell wall material from both mutants displayed significantly less GalA on a mol% basis, with arabinose content being significantly higher in the mutants (Figure 3D). Together, these results suggest that mutations in QUA2 might result in the synthesis of an unstable or incomplete form of HG that is rapidly degraded, resulting in lower HG levels in the wall, but that QUA2 is not solely responsible for HG methylesterification.
Figure 3.
qua2 and tsd2 Mutants Have Reduced Pectin and Cellulose Content.
(A) Total uronic acid content from 6-d-old etiolated Col, qua2, and tsd2 seedlings (n = 5 technical replicates from three biological replicates per genotype).
(B) Methanol released from cell walls of 6-d-old etiolated Col, qua2, and tsd2 seedlings (n = 5 technical replicates from three biological replicates per genotype).
(C) Calculated degree of pectin methylesterification from 6-d-old etiolated Col, qua2, and tsd2 seedlings (n = 5 technical replicates from three biological replicates per genotype).
(A) to (C) The bottom whisker is from minimum to quartile 1, the bottom box is from quartile 1 to median, the top box is from median to quartile 3, and the top whisker is from quartile 3 to maximum.
(D) Monosaccharide composition from cell walls of 6-d-old etiolated Col, qua2, and tsd2 seedlings (n ≥ 5 technical replicates per genotype).
(E) and (F) Crystalline cellulose content from 6-d-old etiolated seedlings (E) and the leaves (F) of 4-week-old Col, qua2, and tsd2 plants (n = 5 technical replicates per genotype).
In (D) to (F), error bars represent sd. *, P < 0.05 and **, P < 0.001, Student’s t test.
Cellulose Biosynthesis Is Inhibited in qua2 and tsd2 Mutants
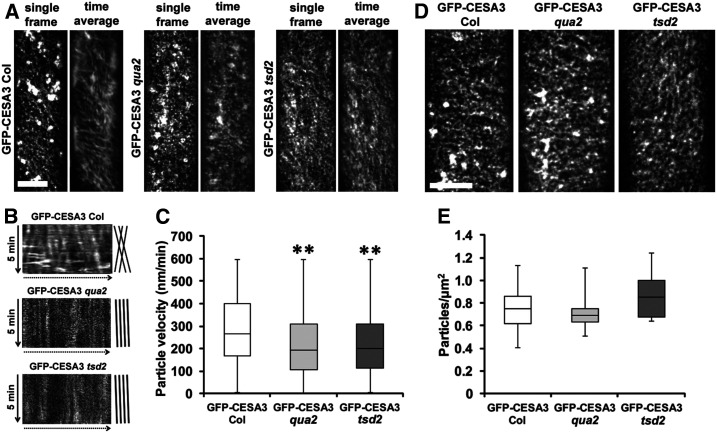
To investigate whether pectin deficiency in qua2 and tsd2 mutants affects cellulose biosynthesis, we first measured crystalline cellulose content according to the Updegraff method (Updegraff, 1969) from alcohol-insoluble residue (AIR) of 6-d-old etiolated seedlings. Average cellulose content was 139 ± 4 µg/mg AIR in Col, whereas it was roughly 100 µg/mg AIR in both qua2 and tsd2 mutants, representing a reduction in cellulose content of ∼30% (Figure 3E). Cellulose content was also decreased by ∼10% in rosettes leaves of 4-week-old qua2 and tsd2 plants relative to Col controls (Figure 3F). Based on the finding of reduced cellulose content in qua2 and tsd2 mutants, we next investigated whether cellulose biosynthesis was affected in the mutants. Transgenic lines expressing GFP-CESA3, a marker for primary wall CSCs (Desprez et al., 2007), were produced in the qua2 and tsd2 mutant backgrounds and in the Col background as a control (Xiao et al., 2016). To monitor CSC dynamics in mutants, time-lapse images of GFP-CESA3 over 5 min at 5-s intervals were recorded in epidermal cells at the tops of 3-d-old etiolated hypocotyls and GFP-CESA3 particle movement was analyzed and quantified. Fewer linear tracks of CSCs were present in time-averaged projections in both qua2 and tsd2 mutants compared with the well-organized CSC arrays evident in Col cells, and the average speed of GFP-CESA3 particles was 223.8 ± 0.4 (se) nm/min (n = 965 particles, 11 cells from 10 seedlings) in qua2 hypocotyls and 224.4 ± 0.3 nm/min (n = 1441 particles, 17 cells from 9 seedlings) in tsd2 hypocotyls, which were significantly slower than the average speed of 284.9 ± 0.4 nm/min (n = 1016 particles, 14 cells from 7 seedlings) in Col controls (Figure 4A; Supplemental Movie). The slower GFP-CESA3 particles in both mutants were also evident in kymographs, which showed steeper (slower) slopes and more unidirectional movement from CSC tracks as opposed to shallower (faster) slopes and bidirectional movement in Col controls (Figure 4B). This finding was confirmed when particle-tracking software was used to calculate particle speeds for many hundreds of GFP-CESA3 particles for each genotype (Figure 4C). GFP-CESA3 particle density from z-series images in qua2 and tsd2 mutants was 0.82 ± 0.11 and 0.90 ± 0.12 particles/µm2, respectively, which were not significantly different from a density of 0.88 ± 0.11 particles/µm2 in Col controls (Figures 4D and 4E). Therefore, disruption of pectin results in both reduced GFP-CESA3 particle speed and reduced cellulose content in the qua2 and tsd2 mutants, strongly indicating that cellulose synthesis is defective in the mutants.
Figure 4.
CSC Motility Is Reduced in qua2 and tsd2 Etiolated Hypocotyls.
(A) Single frames and time averages of time-lapse data sets with 61 frames (5-min duration, 5-s interval) from 3-d-old etiolated GFP-CESA3 Col, GFP-CESA3 qua2, and GFP-CESA3 tsd2 hypocotyls showing GFP-CESA3 particles and trajectories. Bar = 10 μm.
(B) Kymographs of linear tracks showing movement directions of GFP-CESA3 particles in (A) from Col, qua2, and tsd2.
(C) Box plot of GFP-CESA3 particle velocity (n ≥ 965 particles from at least 11 cells per genotype). **, P < 0.001, Student’s t test.
(D) Maximum projections from z-series of 3-d-old etiolated seedlings expressing GFP-CESA3. Bar = 10 μm.
(E) Box plot of average particle density (n ≥ 9873 particles from at least 13 cells per genotype) from 3-d-old etiolated GFP-CESA3 Col, GFP-CESA3 qua2, and GFP-CESA3 tsd2 hypocotyls.
In (C) and (E), the bottom whisker is from minimum to quartile 1, the bottom box is from quartile 1 to median, the top box is from median to quartile 3, and the top whisker is from quartile 3 to maximum.
Cellulose Organization and Morphology Are Disrupted in qua2 and tsd2 Hypocotyl Cells
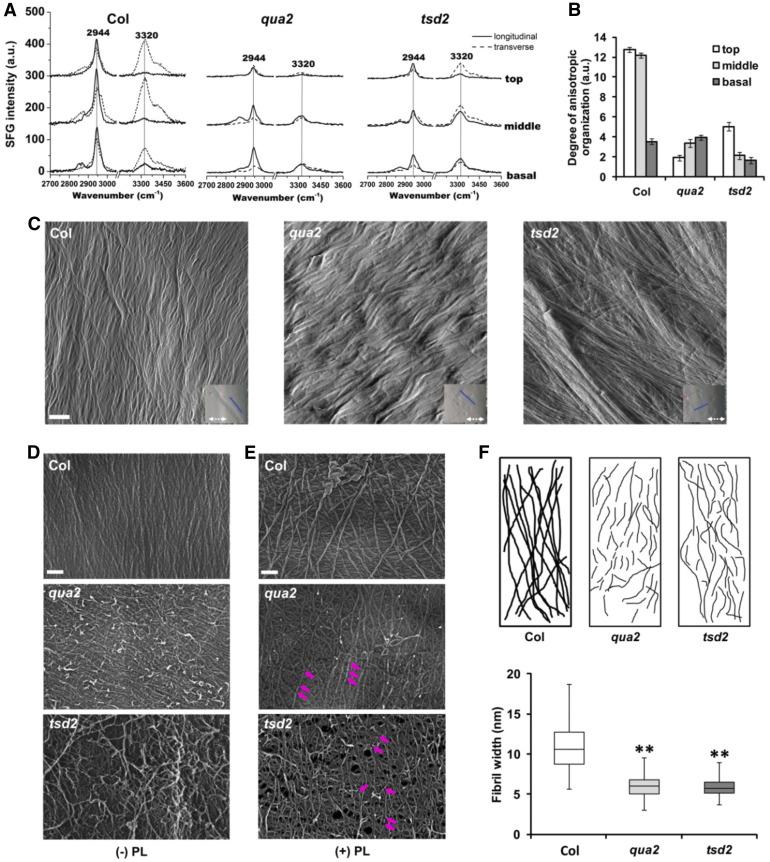
The change in cellulose biosynthesis we detected in the qua2 and tsd2 mutants prompted us to further explore how HG deficiency might affect cellulose microfibril arrangement in mutant walls. As a nondestructive method, sum frequency generation (SFG) can selectively detect the coherence of crystalline cellulose without interference from an amorphous polymer matrix (Makarem et al., 2019) and is sensitive to the mesoscale ordering of cellulose microfibrils over depths of hundreds of nanometers in plant cell walls (Lee et al., 2014; Makarem et al., 2017). Given that 6-d-old etiolated seedlings contain most of the deposited cellulose that is produced in etiolated seedlings, we observed SFG spectra in 6-d-old etiolated mutant and Col hypocotyls. The data displayed similar peak features along the lengths of the hypocotyls (Figure 5A), with sharp CH peaks centered at 2944 cm–1 and broad OH peaks centered at 3320 cm–1, as well as shoulder peaks at 3450 cm–1. Peak intensity at 2944 cm–1 can be used to estimate cellulose content (Barnette et al., 2012; Makarem et al., 2017). Compared with Col controls, SFG spectra from qua2 and tsd2 mutants had reduced CH peak intensities (Figure 5A), indicating lower cellulose content in the mutants, consistent with the above results from Updegraff assays. SFG intensity at 2944 cm–1 from middle and basal hypocotyls in all three genotypes was slightly higher than that from top regions (Figure 5A), suggesting that more cellulose had been deposited in lower regions, which is consistent with reported measures of wall synthesis and thicknesses in etiolated hypocotyls of Arabidopsis (Refrégier et al., 2004; Derbyshire et al., 2007). The relative orientation of cellulose microfibrils can be quantified by comparing OH:CH peak area ratio in the experimental geometry, in which the laser incidence plane is aligned either parallel (longitudinal) or transverse to the hypocotyl elongation axis (Kafle et al., 2014a, 2014b; Chen et al., 2017). The lower ratio of (OH:CHtransverse ratio) to (OH:CHlongitudinal ratio) in qua2 and tsd2 mutants (Figure 5B) indicated that cellulose microfibrils are aligned in a more random manner in the mutant cell walls than in Col controls.
Figure 5.
Cellulose Is Disorganized in qua2 and tsd2 Etiolated Hypocotyls.
(A) SFG signal intensity from top, middle, and basal hypocotyl regions of 6-d-old etiolated Col, qua2, and tsd2 seedlings. a.u., Arbitrary units.
(B) Degree of anisotropic organization in fibrils of Col, qua2, and tsd2 genotypes (n = 6 location points from a bundle of 10 seedlings). Error bars represent se. a.u.,Arbitrary units.
(C) AFM images from top regions of 3-d-old etiolated Col, qua2, and tsd2 hypocotyls. Blue lines in inset images indicate cell long axis; white dashed arrows indicate scan direction. Bar = 200 nm.
(D) and (E) FESEM images from top regions of 3-d-old etiolated Col, qua2, and tsd2 hypocotyls without (D) and with (E) pectate lyase (PL) treatment. Purple arrows display the positions where fibrils were fragmented. Bars = 100 nm.
(F) Diagram of fibril patterning from FESEM images in (E).
(G) Quantification of fibril width from FESEM images (n ≥ 100 cellulose microfibrils from at least three images). The bottom whisker is from minimum to quartile 1, the bottom box is from quartile 1 to median, the top box is from median to quartile 3, and the top whisker is from quartile 3 to maximum. Error bars represent sd. **, P < 0.001, Student’s t test.
To further examine cell wall organization in qua2 and tsd2 mutants, we observed wall structure in the innermost (newly deposited) layers of the wall in epidermal cells from the hypocotyls of 3-d-old etiolated seedlings by atomic force microscopy (AFM), which detects stiffer wall elements such as cellulose microfibrils and cross-linked matrix components, as well as by field emission scanning electron microscopy (FESEM) without or with treatment by pectate lyase to remove some matrix polysaccharides from the innermost face of the wall (Zhang et al., 2016). Compared with a relatively uniform distribution of fibrils evident in AFM images of Col controls, AFM images of qua2 mutants showed thicker, kinked fibrils, whereas fibrils in tsd2 cells showed large bundles of fibrils; in both mutant alleles, fibrils ran almost perpendicular to the longitudinal axis of the cell (Figure 5C). In FESEM images of Col controls, walls and fibrils also displayed uniform patterning, whereas walls were more randomly organized with less dense, crooked fibrils in both mutants, with the differences being more obvious in tsd2 than in qua2 mutants (Figures 5D and 5E). Bright fragments on the wall surface in FESEM images in mutants without pectate lyase treatment (Figure 5D) were reminiscent of structures of Ca2+ cross-linked matrix polysaccharides in onion (Allium cepa) epidermal walls (Zhang et al., 2016), implying that these structures might represent fragments of demethylesterified HG that are overrepresented in qua2 and tsd2 mutants and are not integrated tightly into the wall matrix upon delivery to the apoplast. Long fibrils that might represent cellulose microfibrils appeared to be more fragmented in both mutants after pectate lyase treatment (Figure 5E), as depicted in the diagram in Figure 5F. Moreover, fibrils in FESEM micrographs from mutants were thinner than those in Col controls after treatment with pectate lyase (Figure 5G). In addition, we observed porous features in tsd2 walls (Figure 5E). The above data highlight the function of HG in supporting normal cellulose synthesis and orientation, as well as the assembly of properly integrated primary walls, in Arabidopsis.
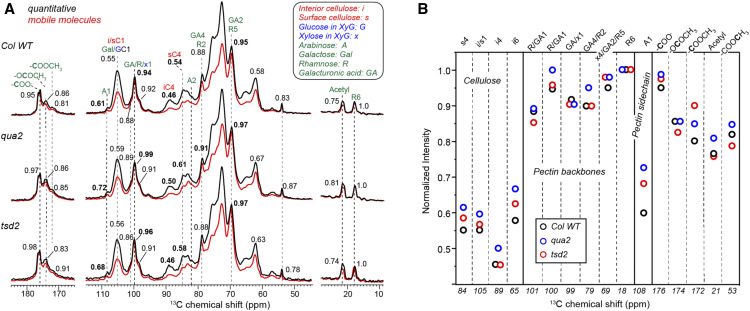
Reduced Pectin Deposition in Mutants Promotes the Increased Molecular Mobility of Pectin and Cellulose
We also detected a substantial change in the molecular mobility of cell wall components in qua2 and tsd2 mutants using solid-state NMR experiments. Mobility is site-specifically monitored by comparing the relative intensity of a spectrum that only selects mobile molecules with a quantitative spectrum (Figure 6A; Phyo et al., 2017). For each peak, the intensity ratio between the two spectra is quantified, which represents the relative mobility of each carbon site (Figure 6B). Compared with the wild-type sample, both mutants showed higher mobility for pectin backbones (GalA C1 at 100 ppm and GalA C4/Rha C1 at 69 ppm), pectin side chains (Ara C1 at 108 ppm), surface cellulose (sC4 at 84 ppm), and interior cellulose (iC4 at 89 ppm). A few exceptions were evident at carbon sites with special properties; for example, the Rha C6 resonating at 18 ppm remained highly mobile in all three samples, as it is a CH3 group that rotates freely. The qua2 mutant generally had the highest intensity ratios, indicating that this mutant has the most dynamic molecules among the three samples. Enhanced pectin mobility should originate from a perturbed backbone and side chain structure and domain organization, which shortens 13C relaxation and enhances pectin intensity in the 13C direct polarization spectrum measured with short recycle delays, but it is unclear why cellulose also showed an increase of 5 to 10% in mobility in the qua2 mutant. Since cellulose is typically considered to be rigid, the change in cellulose intensity might be attributed to the enhanced spin exchange caused by its stronger interactions with more mobile pectin in the qua2 mutant. The tsd2 mutant displayed mobilities between the wild-type and qua2 samples.
Figure 6.
qua2 and tsd2 Mutants Have Increased Mobility for Both Cellulose and Pectin as Detected by NMR.
(A) 13C direct polarization (DP) spectra measured with short recycle delays of 2 s for probing mobile molecules (red) and 35 s for quantitative detection (black) at 293K. The relative intensities between these two spectra are indicated for resolved cellulose and pectin peaks, with the higher relative intensity representing higher mobility.
(B) Relative intensity ratios between the 2-s DP that selects the mobile molecules and quantitative DP spectra for wild-type (black), qua2 (blue), and tsd2 (red) cell walls.
Cortical Microtubule Orientation Is Disrupted in the Absence of Normal Wall Structure in qua2 and tsd2 Hypocotyl Cells
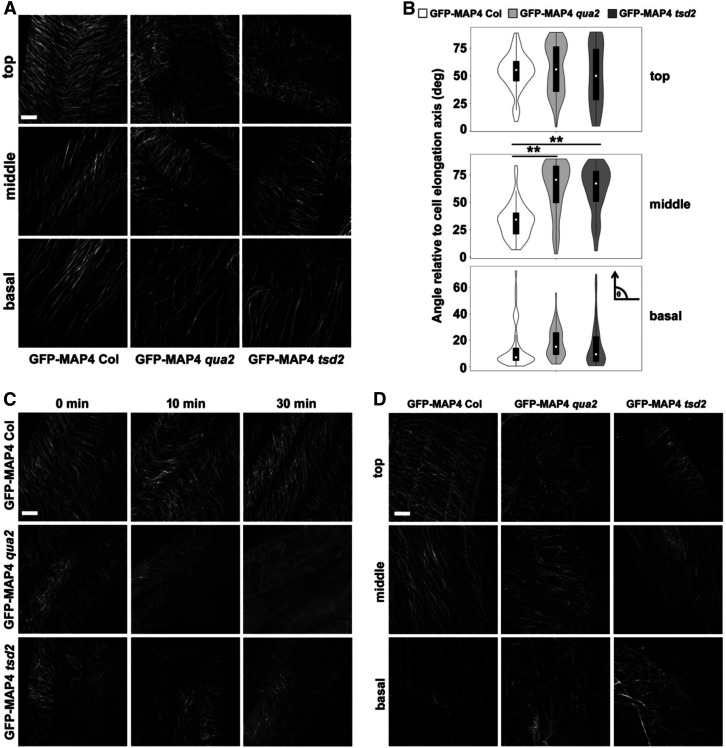
Given the functions of cortical microtubules in tissue mechanics and cellulose deposition, and the alteration of wall structure in qua2 and tsd2 mutants, we next investigated whether the disruption of wall biosynthesis in these mutants affects the patterning and dynamics of cortical microtubules. Transgenic lines expressing GFP-MAP4 (Marc et al., 1998) were generated in the Col, qua2, and tsd2 backgrounds. To observe the patterning of microtubules, z-stack images were collected from epidermal cells of top, middle, and basal hypocotyls in 3-d-old etiolated seedlings. Relative to the cell elongation axis, microtubules in the tops of Col hypocotyls were angled mostly from 40 to 70°, whereas microtubules in qua2 and tsd2 hypocotyl tops were more randomly distributed (Figures 7A and 7B). In comparison with the oblique microtubule angles in the middle regions of hypocotyls of Col controls that centered around 30° relative to the cell elongation axis, microtubules in the same regions of qua2 and tsd2 hypocotyls tended to be more transverse, with angles primarily between 50 and 90° (Figures 7A and 7B). In Col basal hypocotyls, most microtubules were longitudinal, whereas in qua2 and tsd2 mutants, some microtubules were oblique. We noticed that some microtubules in the qua2 background were wavier than in Col controls (Figure 7A). The aberrant orientation of microtubules in qua2 and tsd2 mutants implies that disruption of wall biosynthesis and cell adhesion defects affect cytoskeletal patterning in these genotypes, and/or vice versa.
Figure 7.
Cortical Microtubule Patterning Is Altered, and Microtubules Are More Sensitive to Oryzalin Treatment and Mechanical Stress, in qua2 and tsd2 Alleles.
(A) Maximum projections of z-stack images of microtubules from top, middle, and basal hypocotyls of 3-d-old etiolated GFP-MAP4 Col, GFP-MAP4 qua2, and GFP-MAP4 tsd2 seedlings. Bar = 10 μm.
(B) Violin plots of microtubule angles relative to the cell elongation axis in epidermal cells from top, middle, and basal hypocotyls (n ≥ 50 microtubules from at least 12 cells from three seedlings per genotype). The arrow in the small inset indicates the cell elongation axis. **, P < 0.001, Student’s t test.
(C) Cortical microtubule patterning in top hypocotyl regions of 3-d-old etiolated seedlings of GFP-MAP4 Col, GFP-MAP4 qua2, and GFP-MAP4 tsd2 seedlings treated with 10 mM oryzalin over time. Diffuse labeling in the bottom right-hand panels likely represents soluble GFP-MAP4. Bar = 10 μm.
(D) Projections of z-stack images of microtubules from top, middle, and basal hypocotyl regions of 3-d-old etiolated seedlings of GFP-MAP4 Col, GFP-MAP4 qua2, and GFP-MAP4 tsd2 genotypes subjected to a stress of 0.49 N. Bar = 10 μm.
Microtubules in qua2 and tsd2 Mutants Are Hypersensitive to Oryzalin and Mechanical Stress
To test whether microtubule stability is affected in pectin-deficient mutants, we grew Col, qua2, and tsd2 seedlings on MS medium containing oryzalin, a microtubule-depolymerizing drug. In treatments with different oryzalin concentrations, hypocotyls in 6-d-old etiolated qua2 and tsd2 seedlings were significantly shorter than Col controls (Supplemental Figures 4A and 4B) and were more significantly inhibited in elongation as a percentage of hypocotyl length without drug treatment than Col seedlings (Supplemental Figures 4C and 4D). Conversely, qua2 and tsd2 seedlings had wider hypocotyls, both in absolute (Supplemental Figures 4E and 4F) and relative (Supplemental Figures 4G and 4H) terms. Growth inhibition and more severe cell adhesion defects were observed in 3-d-old etiolated seedlings of the mutant genotypes when grown in the presence of 200 nM oryzalin (Supplemental Figure 5).
To further test the sensitivity of microtubules to oryzalin in the two mutants, 3-d-old etiolated seedlings expressing GFP-MAP4 in Col, qua2, and tsd2 backgrounds were incubated with 10 μM oryzalin in the dark for different time intervals, and z-series images were recorded from epidermal cells of top regions of hypocotyls. After treatment for 10 min, GFP-MAP4-labeled microtubules began to depolymerize in qua2 and tsd2 hypocotyls, and were nearly absent after 240 min of oryzalin treatment, whereas they remained relatively intact in Col controls (Figure 7C; Supplemental Figure 6).
Cortical microtubules respond to mechanical stresses to maintain plant organ morphology (Hamant et al., 2008), and application of external mechanical pressure can result in reorientation of microtubules in growing Arabidopsis leaves and hypocotyls (Jacques et al., 2013; Robinson and Kuhlemeier, 2018). To monitor the responses of microtubules to external pressure, we mounted 3-d-old etiolated seedlings of GFP-MAP4-expressing Col, qua2, and tsd2 genotypes in a chamber with a large cover glass at the bottom, two seedlings spaced 10 mm apart in 40 μL of water, and a smaller cover glass on top. A balance weight was gently placed on the top cover slip to apply external pressure to the seedlings for 10 s before imaging. Z-series images from top, middle, and basal hypocotyl cells of 3-d-old etiolated seedlings were analyzed. With an applied force of 0.49 N, cells showed fragmented microtubules with a frequency of 61.8 and 60.7% in the top of hypocotyls and 52.8 and 71.4% in the middle of hypocotyls of qua2 and tsd2 seedlings, respectively, and microtubules were fragmented in all basal hypocotyl cells in both mutants, whereas microtubules remained relatively intact in the top and middle hypocotyl cells, with fewer cells exhibiting microtubule fragmentation in basal hypocotyls of Col seedlings (Figure 7D; Supplemental Figure 7A). When a force of 0.98 N was applied, almost all cells in all three regions of qua2 and tsd2 mutants exhibited fragmented microtubules, whereas fewer cells had fragmented microtubules in Col hypocotyls (Supplemental Figure 7B). Together, these data imply that mutations in QUA2 result in microtubules that are more sensitive to oryzalin treatment and mechanical stress.
Mutations in QUA2 Result in Altered Expression of Wall Synthesis and Integrity-Sensing Genes
To examine the effects of disruption of cell wall biosynthesis and organization in qua2 and tsd2 mutants at the transcriptional level, we measured transcript levels of genes related to pectin and cellulose biosynthesis by qPCR (Table 1). The expression of some pectin biosynthesis genes, including GAE1, GAUT1, GAUT7, and GAUT12, was significantly downregulated, although GAUT8 and GAUT15 expression did not change, whereas genes encoding two pectin-interacting extensins, EXT3 and EXT4 (Cannon et al., 2008), were upregulated. FAD-LINKED OXIDOREDUCTASE (FADLOX), a gene involved in stress response and Ca2+ signaling (Verger et al., 2016), had higher transcript abundance in both mutants. The pectin modification gene PME35 (Hongo et al., 2012) had decreased expression. All three major CESA genes that function in primary wall biosynthesis, CESA1, CESA3, and CESA6, along with COBRA and CELLULOSE SYNTHASE INTERACTIVE1 (CSI1), were significantly downregulated. The effects of mutation of QUA2 on transcriptional regulation of microtubule stability and wall integrity signaling were also investigated using qPCR (Table 1). Several microtubule-related genes, including MICROTUBULE-ASSOCIATED PROTEIN70-1 (MAP70-1), MAP70-5, CLASP, FRAGILE FIBER1 (FRA1), and KATANIN1 (KTN1; Korolev et al., 2005, 2007; Stoppin-Mellet et al., 2006; Ambrose et al., 2007), were significantly downregulated in both mutants, and MAP20 had lower expression in qua2 but not tsd2. However, the expression levels of MAP65-1 did not differ significantly among all three genotypes. Receptor kinases, including FERONIA (FER), HERKULES1 (HERK1), THESEUS (THE1), KINESIN13A (KIN-13A), FEI1, and FEI2 have been termed cell wall sensors (Hématy and Höfte, 2008; Xu et al., 2008; Guo et al., 2009). All of the above genes were significantly downregulated in both mutants, whereas all five WALL ASSOCIATED KINASE (WAK) genes (Kohorn and Kohorn, 2012) except WAK3 were downregulated in the tsd2 mutant but not in the qua2 mutant (Table 1). These data imply that cells respond to the disruptions in wall structure induced by mutations in QUA2 by reducing the synthesis of both pectin and cellulose, increasing the expression of wall-related structural proteins and dampening wall integrity signaling.
Table 1. Expression Levels of Wall and Microtubule-Related Genes.
| Gene Name | Relative Expression | ||
|---|---|---|---|
| qua2 | tsd2 | ||
| Cellulose synthesis-related genes | |||
| CESA1 | 0.58 ± 0.05** | 0.48 ± 0.01** | |
| CESA3 | 0.69 ± 0.03* | 0.51 ± 0.06* | |
| CESA6 | 0.50 ± 0.03* | 0.43 ± 0.02** | |
| KOR1 | 0.42 ± 0.02** | 0.33 ± 0.03** | |
| COBRA | 0.72 ± 0.04* | 0.70 ± 0.02* | |
| CSI1 | 0.54 ± 0.02** | 0.42 ± 0.01** | |
| Microtubule-related genes | |||
| MAP20 | 0.59 ± 0.09* | 0.86 ± 0.12 | |
| MAP65-1 | 0.91 ± 0.02 | 0.87 ± 0.03 | |
| MAP70-1 | 0.62 ± 0.04** | 0.58 ± 0.01** | |
| MAP70-5 | 0.22 ± 0.01** | 0.26 ± 0.02** | |
| CLASP | 0.57 ± 0.05** | 0.41 ± 0.04** | |
| FRA1 | 0.46 ± 0.01** | 0.31 ± 0.04** | |
| KTN1 | 0.69 ± 0.04** | 0.62 ± 0.04** | |
| Cell wall integrity genes | |||
| FER | 0.65 ± 0.01* | 0.56 ± 0.03* | |
| HERK1 | 0.36 ± 0.01** | 0.31 ± 0.02** | |
| FEI1 | 0.68 ± 0.01** | 0.50 ± 0.07** | |
| FEI2 | 0.70 ± 0.05* | 0.59 ± 0.03** | |
| KIN13A | 0.48 ± 0.02** | 0.40 ± 0.01** | |
| THE1 | 0.41 ± 0.06* | 0.25 ± 0.02** | |
| WAK1 | 0.94 ± 0.01 | 0.18 ± 0.03* | |
| WAK2 | 1.07 ± 0.31 | 0.42 ± 0.13* | |
| WAK3 | 1.68 ± 0.24 | 0.94 ± 0.23 | |
| WAK4 | 0.99 ± 0.20 | 0.20 ± 0.06* | |
| WAK5 | 0.77 ± 0.29 | 0.15 ± 0.06** | |
| Pectin-related genes | |||
| GAE1 | 0.71 ± 0.10* | 0.53 ± 0.04* | |
| GAUT1 | 0.68 ± 0.06* | 0.65 ± 0.07* | |
| GAUT7 | 0.68 ± 0.09* | 0.63 ± 0.06** | |
| GAUT8 | 1.08 ± 0.06 | 1.08 ± 0.03 | |
| GAUT12 | 0.47 ± 0.07** | 0.19 ± 0.02** | |
| GAUT15 | 1.04 ± 0.20 | 0.76 ± 0.03 | |
| EXT3 | 1.66 ± 0.09* | 2.16 ± 0.07** | |
| EXT4 | 5.79 ± 0.57** | 5.25 ± 0.49** | |
| PME35 | 0.0065 ± 0.001** | 0.0036 ± 0.003** | |
Detection of mRNA expression levels is shown from 3-d-old etiolated Col, qua2, and tsd2 seedlings by qPCR. EF1-a was amplified as an internal control. Expression levels in Col were normalized to 1, and the expression levels of target genes in qua2 and tsd2 seedlings were calculated relative to these values. Data present averages and sd (n = 3 technical replicates). Asterisks indicate significant differences (*, P < 0.05 and **, P < 0.001, Student’s t test). See also the Supplemental Table.
DISCUSSION
Previous studies have determined that pectic HG is methylated in the Golgi apparatus before being secreted into the cell wall (Goubet and Mohnen, 1999). Methyltransferase activity has been detected in Golgi-enriched fractions and isolated microsomes, and the catalytic domains of putative methyltransferases face the Golgi lumen (Goubet and Mohnen, 1999; Ibar and Orellana, 2007; Miao et al., 2011; Kim et al., 2015). A series of putative methyltransferases have been identified, including QUA2/TSD2, QUA3, CGR2, and CGR3 in Arabidopsis and their homologs in other species, but none of them has yet been characterized for enzymatic activity in vitro. In this study, we heterologously expressed QUA2 in E. coli and purified it to homogeneity. The in vitro methyltransferase activity of QUA2 was confirmed using demethylesterified HG (PGA) as a substrate and a sensitive and robust method to test for methyltransferase activity (Hsiao et al., 2016). These data reveal the molecular function of QUA2 as a functional HG methyltransferase in vitro (Figure 1).
We also analyzed two mutant alleles of QUA2, qua2 and tsd2, which result from mutations at different positions in the QUA2 gene, and found that tsd2 mutants display more severe phenotypes such as shorter hypocotyls and smaller plants (Figure 2; Supplemental Figure 2). The tsd2 mutation disrupts the first exon of QUA2, possibly leading to more severe effects on protein function than the qua2 lesion, which resides in the seventh exon (Supplemental Figure 1). Previously, it was found that extractable HG-derived GalA from cell walls of qua2 leaves was 50% lower than in wild-type controls (Mouille et al., 2007). Our data confirmed lower GalA levels in monosaccharide composition analyses and lower total uronic acid content in both mutant alleles, although the estimated degree of pectin methylesterification did not show significant changes in etiolated seedlings of qua2 or tsd2 mutants (Figure 3). These data suggest at least three possible scenarios: first, mutations in QUA2 might inhibit HG biosynthesis without altering HG methylesterification; second, HG in qua2 and tsd2 mutants might be secreted with a lower degree of methylesterification and therefore be subject to faster degradation in the apoplast; and third, given that GAUTs and putative pectin methyltrasferases (PMTs) can coexist in protein complexes, presumably to coordinate the synthesis of highly methylesterified HG (Atmodjo et al., 2013), the loss of active QUA2 from HG-biosynthetic complexes that normally contain this protein might result in loss of function for those complexes, leading to reduced overall HG synthesis but an unchanged degree of methylesterification in the remaining HG synthesized by other complexes.
In the plant cell wall, different structural components interact with each other to form a unified, multi-network architecture. Hence, it is likely that a defect in one constituent will affect the structure and assembly of other wall components. The extensive interaction of HG with cellulose detected by solid-state NMR highlights the potential for this interaction to help define the structure and function of plant cell walls (Dick-Pérez et al., 2011), although pectins do not competitively bind to cellulose with as much affinity as xyloglucan in vitro (Zykwinska et al., 2008). Pectic RG-I has also been shown to interact with cellulose (Zykwinska et al., 2005; Broxterman and Schols, 2018). Both pectin and cellulose play important roles in the assembly of cell walls during cell expansion, but it has remained unclear whether and how pectic polysaccharides might influence cellulose biosynthesis and microfibril deposition. Here, we used the pectin-deficient mutants qua2 and tsd2 to explore the effects of pectin deficiency on cellulose synthesis and wall organization using different microscopy techniques, including spinning-disk confocal microscopy, SFG, AFM, and FESEM. We found that cellulose synthesis was inhibited with reduced crystalline cellulose content, lower speed of CESA particles in the plasma membrane, and altered fibril organization in the walls in qua2 and tsd2 mutants compared with Col controls (Figures 4 and 5; Supplemental Movie). These observations provide evidence to support the idea that pectins influence the deposition of cellulose in the wall, which is also supported by the fact that CESA-overexpressing plants contain higher cellulose and pectin levels (Hu et al., 2018). Studies of physical interactions between wall components have largely focused on hemicellulose-cellulose interactions, while pectin-cellulose interactions have long been understudied. To help fill in this gap, we investigated wall dynamics using NMR in qua2 and tsd2 mutants and wild-type controls (Figure 6). The results showed that the partial reduction in pectin might facilitate stronger interactions between pectic polysaccharides and cellulose, since an increased molecular mobility was detected for both classes of wall polymers in both mutants. Future experiments investigating the detailed molecular interactions between pectin subtypes and cellulose in wild-type and qua2/tsd2 mutant walls should provide even more information about how pectin influences cellulose mobility in the wall, and vice versa.
By constituting a shape-constraining and anisotropic extracellular matrix, the cell wall regulates cellular and tissue mechanics during plant growth and development. Cellulose has been hypothesized to be the predominant mechanical actor in the cell wall, but matrix polysaccharides are emerging as important mechanical regulators as well. As the major hemicellulose in eudicot cell walls, xyloglucan has been demonstrated to influence cell wall mechanics and plant growth and to functionally interact with cellulose (Miedes et al., 2013; Xiao et al., 2016). Pectins have also been shown to influence plant mechanics during growth due to their unique physical and chemical properties (Peaucelle et al., 2011, 2015; Bou Daher et al., 2018). Compared with wild-type controls or the highly aligned and bundled fibrils in top hypocotyl regions of xxt1 xxt2 mutants (Xiao et al., 2016), the apparent structure of the cell wall in qua2 and tsd2 hypocotyls observed here was very different: fibrils in untreated qua2 walls seem thicker, possibly representing bundled cellulose or cellulose coated with remaining pectins (Figures 5C and 5D), and both qua2 and tsd2 mutants displayed thinner, more crooked, and more randomly distributed fibrils after removal of pectin by pectate lyase treatment (Figures 5E to 5G). We also observed fragmented fibrils after pectate lyase treatment in qua2 and tsd2 mutants (Figure 5E), suggesting that cellulose microfibrils in the mutants might be more fragile and more sensitive to pectin removal. An alternative possibility is that normal cellulose-pectin interactions in the wild type reduce cellulose mobility (Figure 6), and this stabilizing effect is diminished when HG structure is disrupted in the mutant. Evidence for pectin stabilization of cellulose can be seen in the increases of cellulose microfibril mobility after treatments that soften HG (Zhang et al., 2019). Porous features in the walls of tsd2 hypocotyls after pectate lyase application (Figure 5E) are in keeping with a posited function of HG in determining the extent of wall porosity (Baron-Epel et al., 1988). Defective HG in both qua2 and tsd2 also apparently enhances molecular mobility in the wall and promotes interactions of the remaining pectin with other wall components such as cellulose in order to maintain wall structural integrity (Figures 5 and 6). As the results show, tsd2 displays more severe phenotypes in general than qua2, including reduced hypocotyl length, dwarf adult plants, and adhesion defects. However, the NMR data in the qua2 mutant show the highest molecular mobility, implying strong interactions between pectin and cellulose in this mutant. Given that cellulose exists as stiff fibrils in the wall, the enhanced mobility of cellulose in the mutants might result from its increased interaction with dynamic pectin molecules. It is evident that mutation of QUA2 leads to reduced HG and cellulose synthesis and remodeled polymer interactions. The above evidence underscores the functional interactions between pectin and cellulose and the effects of pectin defects on cellulose deposition and wall assembly.
Previous studies have shown that the defects in cellulose synthesis affect the alignment of cortical microtubules (Paredez et al., 2008). Additionally, microtubule patterning and stability are aberrant in xxt1 xxt2 hypocotyls that lack detectable xyloglucan (Xiao et al., 2016). Pectin modification might trigger mechanical and growth symmetry breaking, subsequently resulting in cortical microtubule reorientation (Peaucelle et al., 2015). On the other hand, tubulin perturbation also affects pectin deposition during cell wall formation (Swamy et al., 2015). Additionally, cortical microtubules respond to mechanical signals concomitant with the alteration of microtubule orientation (Hamant et al., 2008). These findings imply close coordination between cell wall synthesis, wall mechanics, and microtubule behavior. Given the reduced pectin and cellulose contents in qua2 and tsd2 plants, we sought to investigate microtubule organization and dynamics in these mutants. In different regions of qua2 and tsd2 hypocotyls, compared with wild-type controls, microtubules display altered orientation (Figures 7A and 7B). Compared with the more longitudinal microtubules in middle regions in xxt1 xxt2 hypocotyls (Xiao and Anderson, 2016), the microtubules in qua2 and tsd2 hypocotyls were more transverse relative to the cell elongation axis, which might be due to cell swelling caused by defects in cell adhesion. Microtubules in qua2 and tsd2 hypocotyl cells were more sensitive to oryzalin treatment (Figure 7C). This suggests either that the drug gets to the microtubules more quickly in the qua2 and tsd2 hypocotyl cells or that the microtubule polymerization process itself is perturbed in qua2 and tsd2 mutants (Rajangam et al., 2008). Microtubules in qua2 and tsd2 hypocotyl cells were also more sensitive to mechanical stress (Figure 7D), underscoring the role of microtubules in mechanical responsiveness (Hamant et al., 2019). Accordingly, these data indicate a potential role for QUA2 in microtubule formation. A recent study reported an alteration of tension stress patterns due to mechanical discontinuity of the epidermis in a mutant of QUA1, which encodes a putative pectin synthetic protein, leading to the disruption of microtubule alignment with the direction of maximum tensile stress (Verger et al., 2018). It is possible that the alteration of microtubule alignment we observed in qua2 and tsd2 mutants is due to epidermal discontinuity in these mutants, which disrupts the transduction of tensile stress across the epidermis. However, we cannot rule out other causes of microtubule disruption triggered by pectin and cellulose deficiency in the mutants.
Plants respond to internal cues such as turgor pressure and growth factors, and external environmental cues such as light, temperature, and pathogen infection, through cell wall remodeling under the control of cell wall integrity signaling pathways. Pectin is involved in maintaining cell wall integrity (Bouton et al., 2002; Krupková et al., 2007; Mouille et al., 2007; Neumetzler et al., 2012; Liu et al., 2014). However, it has not yet been clearly defined how pectin might interface with wall integrity-signaling pathways. To begin to answer the question of whether wall deficiency in the absence of QUA2/TSD2 function perturbs wall integrity signaling, we investigated the expression of cell wall integrity-related genes by qPCR: FER, HERK1, FEI1, FEI2, KIN13A, and THE1 had lower expression levels in qua2 and tsd2 mutants (Table 1). FER is thought to sense cell wall integrity and mechanical stress on the plasma membrane (Cheung and Wu, 2011; Lindner et al., 2012). The extracellular domain of FER has been shown to interact with pectin in vitro, suggesting that pectin might function as a ligand of FER (Feng et al., 2018). Although functional interaction between FER and pectin in vivo remains to be determined, FER might perceive wall integrity signals through its interaction with pectin. Arabidopsis WAK receptors can bind to pectin in cell walls and are required for cell expansion during plant development (Kohorn and Kohorn, 2012). In tsd2, the expression levels of several WAK genes (Table 1) were reduced, although there was no significant change in WAK expression in the qua2 mutant. A previous study showed that mechanical stimuli can induce extensin gene expression (Tiré et al., 1994). In our data, the expression of EXT3 and EXT4 was increased in qua2 and tsd2 mutants, implying a potential alteration of extensin-pectin interactions (Cannon et al., 2008). Taken together, many genes related to wall integrity sensing are downregulated in qua2 and tsd2 mutants, indicating that stimuli resulting from changes in wall structure and mechanics might activate signaling involved in cell wall integrity maintenance, resulting in feedback-mediated downregulation of the cognate genes.
It has remained elusive why reduction in pectin content sometimes causes cell adhesion defects. In qua2 and tsd2 mutants, cell adhesion defects might be directly attributable to lower HG levels, whereas another mutant with cell adhesion defects, frb1, does not display a significant decrease in pectin content, although mutation of FRB1 affects pectin methylesterification (Neumetzler et al., 2012). However, a reduced degree of pectin methylesterification has not been reported to cause cell adhesion phenotypes in Arabidopsis double mutants for the putative methyltransferases CGR2 and CGR3 (Kim et al., 2015). The suppressor mutation esmeralda1 rescues adhesion defects in qua1, qua2, and frb1 but does not restore pectin content, suggesting that cell adhesion defects might result from altered pectin-related feedback signaling from the state of the pectin in the cell wall (Verger et al., 2016). This implies that pectin modulates cell adhesion via a complicated mechanism potentially involving the physical adhesion between pectin molecules, pectin interactions with other wall components, pectin metabolism, and/or related signaling pathways. In addition to pectins, other wall components such as hemicelluloses might also function in cell adhesion (Ordaz-Ortiz et al., 2009).
Considering different cellular signaling pathways, it is worthwhile to ask whether there is a change in pectin structure and/or cross-linking in qua2 and tsd2 mutants that triggers calcium-mediated signal transduction. Ca2+ can cross-link demethylesterified HG molecules into macromolecular networks, increasing wall stiffness (Vincken et al., 2003), which implies that changes in Ca2+ concentration might disrupt the integrity of pectin networks to further influence wall mechanical properties. Ca2+ ions can also act as second messengers, functioning in the response of plants to environmental cues (Riveras et al., 2015; Wilkins et al., 2016). In qua1 mutants, the concentration of Ca2+ was dramatically increased in the cytosol, indicating the role of QUA1 in regulating cytoplasmic Ca2+ signaling (Zheng et al., 2017). FADLOX expression has been shown to respond to Ca2+-dependent protein kinases, which are Ca2+ sensors (Boudsocq et al., 2010). In addition, Ca2+ is involved in the interactions of pectins with cellulose (Lopez-Sanchez et al., 2017). In qua2 and tsd2 mutants, wall synthesis was inhibited and FADLOX expression was induced, suggesting that alteration in wall structure might activate Ca2+ signaling in response to disruption of wall integrity, although this idea needs to be explored further.
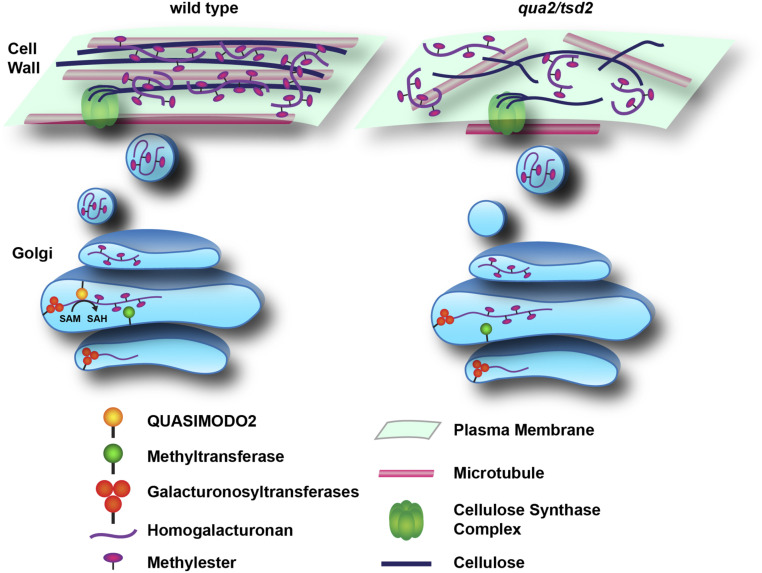
In summary, we propose a schematic for how pectin deficiency in qua2 and tsd2 mutants might affect pectin HG biosynthesis, cellulose biosynthesis, wall integrity, and microtubule stability (Figure 8). In qua2 and tsd2 mutants, the absence of a pectin methyltransferase, QUA2, hampers the biosynthesis of pectic HG in the Golgi and decreases its accumulation in the wall, and the consequent alteration of wall structure influences the activity of plasma membrane-localized CSCs. Loss of wall integrity destabilizes cortical microtubules, further decreasing the aligned deposition of cellulose in the wall. As pectin is reduced in mutants, the orientation of cellulose microfibrils that interact with pectins is changed, concomitant with increased molecular mobility of both cellulose and pectin. The experimental application of pectate lyase, which removes matrix polysaccharides, reveals thinner, more easily fragmented fibrils in the mutants, suggesting that cellulose structure might be fundamentally altered, as supported by our SFG data. Finally, the disruption of wall structure likely triggers changes in wall integrity signaling to respond to the deficiency of pectin and/or cellulose biosynthesis during plant developmental processes.
Figure 8.
Schematic of How Loss of the Pectin Methyltransferase QUA2 Affects HG Biosynthesis, Cellulose Biosynthesis, and Wall Architecture.
In wild-type plants (left), QUA2 functions as a methyltransferase to help synthesize highly methylesterified HG. qua2 and tsd2 mutants (right) produce lower amounts of HG and accumulate less HG in the cell wall, which influences the activity of plasma membrane-localized CSCs guided by cortical microtubules, decreasing cellulose deposition in the wall. The reduced HG content in the wall also alters cellulose microfibril patterning. The disruption of wall structure due to lower pectin and/or cellulose alters microtubule patterning and enhances the molecular mobility of pectin and cellulose, possibly due to increased interactions between them.
METHODS
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col as well as qua2 (qua2-1) and tsd2 (tsd2-1) mutants, a gift from Tanya Falbel (University of Wisconsin), were used in this study. Transgenic plants expressing GFP-CESA3 (Desprez et al., 2007), GFP-MAP4 (Marc et al., 1998), and QUA2pro-GUS, respectively, were generated using the Agrobacterium tumefaciens-based floral dipping method (Clough and Bent, 1998) with A. tumefaciens strain GV3101. Positive transformants were screened on MS plates containing 2.2 g/L MS salts (Caisson Laboratories), 0.6 g/L MES (Research Organics), 1% (w/v) Suc, 0.8% (w/v) agar-agar (Research Organics), and 25 mg/mL hygromycin (Omega Scientific), pH 5.6. Seedlings were transplanted from MS plates to soil for growth under a photoperiod of 16 h of white fluorescent light (4100K, 150 to 200 μmol m−2 s−1)/8 h of dark in a 22°C growth chamber.
Protein Expression and Purification
The catalytic domain of QUA2 (amino acids 150 to 684) was subcloned into pMBP-parallel2 by PCR. The construct containing the QUA2 catalytic domain with an N-terminal 6xHis-MBP tag was transformed and expressed in Escherichia coli strain BL21 (DE3) cells. After induction with 0.2 mM isopropyl β-d-thiogalactopyranoside overnight at 16°C, the cells were pelleted at 4000g for 10 min at 4°C. The pellets were lysed in a buffer containing 25 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM Tris (2-carboxyethyl)phosphine-HCl, 250 mM Suc, and 1 mM phenylmethylsulfonyl fluoride by French press. The lysate was then centrifuged at 25,000g for 30 min at 4°C. The supernatants were loaded onto amylose resin using gravity columns at 4°C, which was subsequently washed three times with a buffer containing 25 mM HEPES, pH 7.5, 150 mM NaCl, 0.5 mM Tris (2-carboxyethyl)phosphine-HCl, and 250 mM Suc. Then 10 mM maltose was added to the washing buffer and used to elute target proteins. The target proteins were further purified on a Superdex200 10/30 (GE Healthcare) column equilibrated with 25 mM Tris-HCl, pH 8.0, 150 mM NaCl, 250 mM Suc, and 2 mM DTT. The peak fractions were pooled and snap-frozen in liquid nitrogen for storage.
The purified MBP (negative control) and MBP-QUA2 proteins were resolved on 12% SDS-PAGE gels and then stained with Coomassie blue or immunolabeled with anti-polyhistidine. For immunoblot analysis, separated proteins were transferred onto nitrocellulose membranes (GE Healthcare) in a Bio-Rad Mini-Transfer Cell at 100 V for 60 min. After blocking in Tris-buffered saline (TBS) with Tween 20 (25 mM Tris base, 150 mM NaCl, and 0.1% [v/v] Tween 20, pH 7.5) containing 5% (w/v) nonfat milk for 30 min, the membrane was incubated overnight with a monoclonal anti-polyhistidine primary antibody (Sigma-Aldrich, catalog No. H1029, 1:5000). Then the membrane was washed three times in TBS with Tween 20 and incubated with alkaline phosphatase-conjugated goat anti-mouse secondary antibody (Sigma-Aldrich, catalog no. A9316, 1:5000) for 30 min. After thorough washing, the membrane was developed in 5-bromo-4-chloro-3′-indolyl phosphate p-toluidine salt/nitroblue tetrazolium chloride substrate solution (Amresco).
Enzymatic Activity Assay
The methyltransferase assay was performed by incubating the purified MBP-QUA2 catalytic domain or MBP alone (negative control) in the buffer containing 50 mM HEPES, pH 7.5, 2 mM MgCl2, 0.25 M Suc, 10 μM SAM (Promega), and 50 μg of PGA (Sigma-Aldrich, catalog no. 81325) at 30°C for 2 h. The PGA substrate was dissolved in 37.5 mM sodium acetate and the pH was adjusted to 7.5. After completion of the reaction, the reaction product, SAH, was monitored using the MTase-Glo Methyltransferase Assay kit (Promega) according to the manufacturer’s directions. Briefly, 20 μL of reaction products was transferred onto a Corning 96-well white plate and mixed with 5 μL of 5× MTase-Glo reagent, which converts SAH into ADP. After the addition of 25 μL of MTase-Glo Detection Solution, the generated ADP was converted into ATP, which was monitored by a luminescent reaction. Luminescence was recorded using a plate-reading luminometer (Synergy H1 microplate reader, BioTek). The change in luminescence represents background-subtracted luminescence. For time- and concentration-dependent enzyme assays, the indicated reaction times, protein, or PGA concentration were applied. Upon reaction completion, 5 μL of 0.5% (v/v) trifluoroacetic acid (Sigma-Aldrich, catalog no. T6608) was mixed with 20 μL of reaction product to stop the reaction. To test the substrate specificity of QUA2, alginate (Sigma-Aldrich, catalog No. A1112) and starch (Sigma-Aldrich, catalog No. S9765), two other polysaccharides, were individually added to reactions.
Immuno Dot Blot Analysis
Immuno dot blotting was performed as previously described with minor modifications (Pérez-Pérez et al., 2019). Aliquots of 3 μL of reaction products were spotted onto nitrocellulose membranes (GE Healthcare) and allowed to air-dry for 1 h. The membrane was blocked with TBS containing 1.5% (w/v) nonfat milk. Subsequently, the membrane was incubated with a primary antibody (Agrisera [JIM5, catalog no. AS184194; JIM7, catalog no. AS184195], 1:100) in blocking solution for 2 h at room temperature. After washing three times in TBS, the membrane was incubated with peroxidase-conjugated goat anti-rat secondary antibody (SeraCare, catalog no. 5220-0364, 1:1000) for 1 h. After washing three times in TBS, the blot was developed using Western ECL Substrate Solution (Bio-Rad, catalog no. 170-5060). The experiment was replicated three times.
Measurements of Hypocotyl Length and Width, Rosette Diameter, and Plant Height
Seeds were surface-sterilized in 30% (v/v) bleach + 0.1% (w/v) SDS, washed four times with sterile water, stored at 4°C for 3 d, and sown on MS plates without Suc. After exposure to light for 2 to 4 h to stimulate seed germination, seedlings were grown in the dark for 2 to 6 d at 22°C. Plates with etiolated seedlings were scanned using an HP Scanjet 8300 scanner at 600 dots per inch, and hypocotyl length was measured using ImageJ. Hypocotyl width was recorded on a Zeiss Observer SD spinning-disk confocal microscope with a 20× objective and quantified in ImageJ. Adult plants were imaged using a D5100 DSLR digital camera (Nikon). Rosette diameters, petiole length, and leaf area were measured in ImageJ, and plant height was measured using a ruler.
Preparation of AIR
Seedlings growing in the dark for 6 d were harvested and ground into a fine powder in liquid nitrogen with a mortar and pestle. The powder was suspended in 30 mL of 1:1 chloroform:methanol (v/v) with shaking for 1 h at room temperature. After precipitation by centrifugation at 4500g for 10 min at room temperature, the residue was washed four times in 70% (v/v) ethanol by resuspension and centrifugation at 4500g for 10 min at room temperature. The final AIR was suspended and centrifuged at 4500g for 10 min at room temperature in 100% acetone and air-dried in a chemical fume hood at room temperature (Fry, 1988).
Measurements of Uronic Acids and Methanol Release
For measurements of total uronic acids (Filisetti-Cozzi and Carpita, 1991), AIR was extracted from 6-d-old dark-grown seedlings. Samples with 1 mg of AIR were suspended in 141 μL of water in a glass tube. Fourteen microliters of 4 M sulfamic acid (Thermo Fisher Scientific)-potassium sulfamate (pH 1.6) was added and mixed thoroughly. Then 845 μL of concentrated H2SO4 containing 75 mM sodium tetraborate (Sigma-Aldrich) was added, and the solution was mixed vigorously by vortexing for 10 s. The solution was heated in a boiling water bath at 100°C for 5 min. After cooling down samples in a room temperature water bath, an aliquot of each sample was diluted 50-fold by adding 20 μL of sample to 980 μL of 85% (v/v) H2SO4. After thorough mixing, initial A525 was recorded in a plastic cuvette using a NanoDrop 2000C spectrophotometer. An aliquot of 28 μL of 0.15% (w/v) m-hydroxydiphenyl (Sigma-Aldrich) in 0.5% (w/v) NaOH was added to the diluted sample and mixed by inversion, and the mix was allowed to incubate at room temperature for 5 min before A525 was measured. GalA (Sigma-Aldrich) was used as a standard to calculate concentrations.
To measure released methylesters (Müller et al., 2013), ∼1 mg of AIR was weighed out for each experiment. After washing in 1 mL of water and centrifugation at 21,130g for 10 min at room temperature, 400 μL of 0.5 M NaOH was added to samples to incubate at room temperature for 1 h to release methylesters, and then 200 μL of 1 M HCl was added and mixed by vortexing for neutralization. Samples were centrifuged at 2400g for 10 min at room temperature, and 125 μL of supernatant was added to 125 μL of 20 mM HEPES buffer (pH 7.5). Samples were oxidized in 250 μL of HEPES buffer containing 0.03 units of alcohol oxidase (Sigma-Aldrich) with shaking for 15 min at room temperature. An aliquot of 250 μL of assay buffer (20 mM acetyl acetone, 50 mM acetic acid, and 2 M ammonium acetate) was then added to each sample and incubated at 60°C for 15 min. After the solution was cooled in a room temperature water bath, A412 was measured using a NanoDrop 2000C spectrophotometer. Methanol was used for a standard curve to calculate total methylester content. Assuming that methanol was released primarily from HG, the degree of HG methylesterification was estimated by dividing nmol of released methanol by nmol of uronic acids in the same amount of AIR. Note that this is an indirect estimate of the true degree of HG methylesterification and depends on two independent assays, one for uronic acid content and the other for methylester release.
Monosaccharide Composition Analysis
The monosaccharide analysis protocol was derived from De Ruiter et al. (1992). For each experiment, 1 mg of ground AIR sample and 500 μL of 3 N methanolic hydrochloric acid (Supelco, catalog No. 33051) were mixed and incubated at 80°C for 16 h, cooled on ice, and dried with filtered air at room temperature. An aliquot of 200 μL of 2 M trifluoroacetic acid was added and mixed thoroughly with dried wall residue for hydrolysis, incubated at 121°C for 2 h, and cooled on ice. The samples were centrifuged at 14,000g for 5 min at room temperature and air-dried. The samples were then resuspended with double distilled water and filtered with 0.2 μm of Millex-LG syringe-driven filter units (catalog No. SLLGR04NL) and analyzed by high-performance anion exchange chromatography with pulsed amperometric detection on a CarboPac PA20 column at a flow rate of 0.5 mL/min (Dionex ICS-5000 Capillary Reagent-Free IC System, Thermo Fisher Scientific). The column was first eluted with 10 mM sodium hydroxide for 15 min and then ramped to an equal mixture of 100 mM sodium hydroxide and 100 mM sodium acetate over 25 min. Sugar monosaccharide composition was quantified based on peaks in the samples using standards (Fuc, Rha, Ara, Gal, Glc, and Xyl at 100 mg/mL; GalA and glucuronic acid at 1 mg/mL; Sigma-Aldrich).
Cellulose Content Measurements
Tissue from 6-d-old dark-grown seedlings and 4-week-old rosette leaves from Arabidopsis Col, qua2, and tsd2 genotypes were used to measure crystalline cellulose content following a procedure described by Xiao et al. (2016) based on the Updegraff method (Updegraff, 1969).
SFG Imaging
Etiolated 6-d-old seedlings of Arabidopsis Col, qua2, and tsd2 genotypes were harvested and immersed in deuterium oxide overnight in the dark for water exchange before SFG analysis. Ten to 15 seedlings were grouped and aligned closely into a bundle on a glass cover slip. After adding deuterium oxide, another glass cover slip was added on the top to seal the sample and keep the seedlings wet. Three regions from the top, middle, and base of each set of hypocotyls were analyzed, and two SFG spectra at each location were collected in longitudinal and transverse directions relative to the hypocotyl growth axis. Due to spatial variability across the samples, SFG spectra from nine locations of each hypocotyl region were collected and averaged for each region. The hydroxyl:alkyl (OH:CH) area ratio was calculated by dividing the OH peak area (3150 to 3700 cm–1) with the CH peak area (2700 to 3050 cm–1) for each SFG spectrum.
In the SFG system, two high-intensity femtosecond laser pulses with wavelengths at 800 nm and tunable IR region (2.5 to 10 µm) were synchronized and overlapped on the samples. The 85-fs, 800-nm output pulses were generated at 2 kHz from a chirped pulse amplification system (Coherent, Libra) with a set of Ti:sapphire oscillator/amplifiers using an 80-kHz laser running at 20 fs and pumped by a frequency-doubled Nd:YLF diode-pumped laser producing 250-ns, 527-nm pulses at 2 kHz. By splitting the 800-nm output, 75% of the output was used to generate a tunable infrared (IR) line through an OPG/OPA system (Coherent, Opera Solo) equipped with a nonlinear difference frequency generation (NDFG) stage, and the remaining 25% of the pulses passed through a pair of angle-tuned Fabry-Pérot etalons (TecOptics) for spectral narrowing. Both the p-polarized IR and the s-polarized visible laser were overlapped at the sample surface at an incidence angle of 45° with respect to the surface plane. The s-polarized SFG signal was selected and detected in the transmission geometry. The SFG signal was filtered with a 775-nm short-pass filter, dispersed with a volume-phase holographic grating (Andor, Holospec), and then detected with a back-illuminated deep-depletion charge-coupled device DU420A-BEX2-DD camera (Andor). The probe size of the SFG system was estimated to be ∼120 µm along the laser incidence plane direction and ∼80 µm perpendicular to the incidence plane. The probe depth was measured to be ∼20 to 25 µm from the top external surface of the sample. SFG spectra were obtained by stitching multiple NDFG spectra together because each NDFG setting only provides an IR profile with 150 to 200 cm–1 bandwidth. SFG spectra for cellulose need to be collected from 2700 to 3700 cm–1. After stitching, each SFG spectrum was normalized by the incident IR profile with the same frequency region (Lee et al., 2016).
AFM
AFM sample preparation and imaging followed Xiao et al. (2016) with minor modifications. Seedlings were grown in the dark at 22°C for 3 d before harvesting hypocotyls. Tissues were ground in liquid nitrogen and rinsed in 20 mM HEPES (pH 7.0) with 0.1% (v/v) Tween 20 at least three times with spinning in between rinses until the supernatant was clear. A volume of fresh buffer equaling 150 μL was added to the pellet and vortexed. The tubes were spun down, supernatant was removed, and 100 μL of fresh buffer was added. Tubes were put on a rocker for 30 min to wash and disperse the pellet thoroughly. A final spin was done to remove the supernatant, and 50 μL of 20 mM HEPES, pH 7.5, buffer with no detergent was added to the pellet and mixed. Drops of 5 μL of suspension were placed on glass slides and allowed to dry almost completely to aid in sticking the cuticle sides of cells to the glass. Slides were gently rinsed with 20 mM HEPES buffer to remove unbound material.
AFM imaging was performed on a Dimension Icon system with a NanoScope V controller (Bruker) using the PeakForce QNM program under fluid conditions. A ScanAsyst Fluid+ tip was calibrated using software to determine deflection sensitivity and spring constant. The AFM system is contained inside a vibration isolation hood (Technical Manufacturing) to reduce noise interference and allow for high-resolution images of biological materials. PeakForce QNM scanning parameters were controlled by the ScanAsyst software for most settings to achieve the best resolution for both topography and peak force error images, except for the scan rate, which was set to 0.8 Hz, and the peak force, which was set to 800 pN. Samples were imaged at a size of 2 × 2 µm with 512 × 512 pixels. Images were processed in Nanoscope Analysis 1.8v software (Bruker) and exported as TIFs.
FESEM
FESEM experiments were performed in apical hypocotyls from 3-d-old dark-grown seedlings without or with 10 µg/mL pectate lyase treatment (Megazyme E-PLYCJ, lot 130901a) following the protocol described by Xiao et al. (2016), except that the critical point dryer used in this experiment was a manually operated BAL-TEC CPD030.
13C Labeling and Solid-State NMR Analysis
Never-dried primary cell walls were prepared as described (Wang et al., 2015). Arabidopsis seedlings were grown in the dark for 14 d using a liquid medium with 13C-labeled glucose as the sole carbon source. Seedlings were harvested, frozen, and ground in liquid nitrogen. The resulting materials were then washed with 1.5% (w/v) SDS for 3 h, treated with amylase, and washed with water to remove starch, cell membranes, and proteins and deactivate enzymes. To inhibit microbial contamination, 0.02% (w/v) NaN3 was used in all solutions. Bulk water was removed by centrifugation at 4500g for 1 h at room temperature (White et al., 2014). For each sample, ∼80 mg of hydrated seedlings was packed into a 4-mm MAS NMR rotor for measurements. Solid-state NMR experiments were conducted on a 400-MHz Bruker AVANCE spectrometer at 9.4 Tesla using a 4-mm MAS probe. Typical radiofrequency field strengths were 62.5 kHz for 13C and 62.5 to 83 kHz for 1H. 13C chemical shifts were externally referenced to the adamantane CH2 peak at 38.48 ppm on the tetramethylsilane scale. All spectra were recorded at 298K under 10-kHz MAS frequency. 1D 13C spectra were measured using 13C direct polarization with a long recycle delay of 35 s for quantitatively detecting all polysaccharides, whose 13C-T1 relaxation times are well below 6 s in uniformly 13C-labeled plant samples (Wang et al., 2015), and a short recycle delay of 2 s for selecting mobile molecules with short relaxation times. The intensity ratios between these two spectra represent the relative mobility of cell wall molecules.
Confocal Microscopy
Imaging experiments were performed according to Xiao et al. (2016). Seedlings were placed in 40 μL of water in a chamber composed of a microscope slide and cover slip separated by two pieces of double-stick tape. For pressure experiments, an additional chamber was designed with a smaller cover glass on the top with a large cover glass at the bottom. Images were taken on an Observer SD spinning-disk confocal microscope with a 100× 1.40 NA oil-immersion objective (Zeiss). Propidium iodide fluorescence was detected with a 561-nm excitation laser and 617/73-nm emission filter. GFP fluorescence was detected using a 488-nm excitation laser and 525/50-nm emission filter. GFP-CESA3 and GFP-MAP4 images were recorded in hypocotyl cells of dark-grown seedlings. Time-lapse and z-series images were analyzed using ImageJ and Imaris (Chen et al., 2010).
Drug Treatments
To analyze hypocotyl elongation after growth on oryzalin (Chem Service, N-12729), oryzalin was added onto MS plates at different concentrations, seeds were sown on the plates, and hypocotyl length and width from 6-d-old dark-grown seedlings were imaged and measured in ImageJ. Drug treatments for live-cell imaging were identical to those described previously (Xiao et al., 2016). Three-day-old dark-grown seedlings were submerged in 1 mL of liquid half-strength MS medium containing 10 μM oryzalin dissolved in DMSO (Sigma-Aldrich) and incubated in darkness for 10 to 240 min before imaging with spinning-disk confocal microscopy.
Gene Expression Detection by qPCR
Tissue from 3-d-old dark-grown Col, qua2, and tsd2 seedlings was used for total RNA extraction with a Plant RNA Kit (Omega Bio-Tek). Genomic DNA in samples was removed with RNase-free DNase I (NEB). RNA concentration was measured by spectrophotometer (NanoDrop 2000C), and first-strand cDNA was synthesized using qScript cDNA SuperMix (Quanta Biosciences) from DNase I-treated RNA. qPCR was performed using SYBR Green FastMix (Quanta Biosciences) with cDNA and gene-specific primers on a StepOne Plus Real-Time PCR machine (Applied Biosystems). The transcript levels of target genes relative to EF1 as a reference gene were calculated using the ΔΔCT method. Primer sequences are listed in the Supplemental Table.
GUS Activity Analysis
In addition to measurement of mRNA expression in different tissues by qPCR, the spatial expression pattern of QUA2 was examined by histochemical analysis. Twenty independent transgenic lines expressing QUA2pro:GUS were obtained (primer sequences are listed in the Supplemental Table), and T2 plants were used for GUS staining analysis. Tissues were immersed in buffer containing 50 mM sodium phosphate, pH 7.2, 0.2% (v/v) Triton X-100, and 2 mM 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid and cyclohexylammonium salt and incubated at 37°C in the dark for 4 to 16 h, which was dependent on the tissue age. Young dark-grown and light-grown seedlings were stained for 4 h, and flowers, siliques, and stems of 5-week-old QUA2pro:GUS adult plants were stained for 16 h. Pigment was removed from tissue with 70% (v/v) ethanol, and images of GUS staining were recorded using a Zeiss Discovery V12 fluorescence dissecting microscope.
Statistical Analyses
All experiments were repeated at least twice. Data are represented as averages ± sd or se, as specified. Analyses were performed using Student’s t test (two-tailed) or one-way ANOVA with Tukey’s test to evaluate the significant difference, and the P value thresholds are shown as P < 0.05 or P < 0.001. Statistical results are available in Supplemental File 1.
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: CESA1 (At4g32410), CESA3 (At5g05170), CESA6 (At5g64740), CLASP (At2g20190), COBRA (At5g60920), CSI1 (At2g22125), EF1-a (At5g60390), EXT3 (At1g21310), EXT4 (At1g76930), FEI1 (At1g31420), FEI2 (At2g35620), FER (At3g51550), FADLOX (At1g26380), FRA1 (At5g47820), GAE1 (At4g30440), GAUT1 (At3g61130), GAUT7 (At2g38650), GAUT8 (At3g25140), GAUT12 (At5g54690), GAUT15 (At3g58790), HERK1 (At3g46290), KIN13A (At3g16630), KOR1 (At5g49720), KTN1 (At1g80350), MAP20 (At5g37478), MAP65-1 (At5g55230), MAP70-1 (At1g68060), MAP70-5 (At4g17220), PME35 (At3g59010), QUA2/TSD2 (At1g78240), THE1 (At5g54380), WAK1 (At1g21250), WAK2 (At1g21270), WAK3 (At1g21240), WAK4 (At1g21210), and WAK5 (At1g21230).
Supplemental Data
Supplemental Figure 1. Related to Figure 2. Schematic gene structure of QUASIMODO2.
Supplemental Figure 2. Related to Figure 2. qua2 and tsd2 plants are smaller and shorter than wild type.
Supplemental Figure 3. Related to Figure 2. QUA2pro:GUS expression patterns in different plant tissues.
Supplemental Figure 4. Related to Figure 7. Hypocotyls and microtubules of qua2 and tsd2 mutants are more sensitive to oryzalin treatment.
Supplemental Figure 5. Related to Figure 7. Tissue integrity in hypocotyls of qua2 and tsd2 mutants is sensitive to oryzalin treatment.
Supplemental Figure 6. Related to Figure 7. Cortical microtubules in qua2 and tsd2 mutants are more sensitive to 10 mM oryzalin treatment for 30 min to 240 min.
Supplemental Figure 7. Related to Figure 7. Cortical microtubules of qua2 and tsd2 are more sensitive to mechanical stress.
Supplemental Table. Primers used in this study.
Supplemental File. Detailed results of statistical analyses.
Supplemental Movie. GFP-CESA3 particle motility in 3-d-old dark grown Col, qua2, and tsd2 seedlings.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
S-adenosyl- Gramene: S-Adenosyl-L-methionine (SAMe)
S-adenosyl- Araport: S-Adenosyl-L-methionine (SAMe)
Acknowledgments
We thank Liza Wilson and Sydney Duncombe for technical assistance with AFM experiments, Tanya Falbel for providing seeds of qua2 and tsd2 mutants, Richard Cyr for providing the GFP-MAP4 expression construct, and Nicole R. Brown for helpful comments on the article. With the exceptions below, this work was supported as part of The Center for Lignocellulose Structure and Formation, an Energy Frontier Research Center funded by the U.S. Department of Energy, Office of Science, Basic Energy Sciences (grant DE-SC0001090 to S.K., D.J.C., and C.T.A.). Analysis of QUA2 protein activity was supported by Fundamental Research Funds for the Central Universities (grants YJ20173 and SCU2019D013 to C.X.), and the China Postdoctoral Science Foundation (grant 2018M643465 to J.D.).
AUTHOR CONTRIBUTIONS
J.D., C.X., S.Q., S.H.K., T.W., D.J.C., and C.T.A. designed the research; J.D., C.X., A.K., L.W., S.H., W.J.B., S.N.K., Y.Z., Y.R., and M.R. performed research; J.D., C.X., S.H.K., T.W., D.J.C., and C.T.A. analyzed data; J.D., C.X., T.W., and C.T.A. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Ambrose J.C., Shoji T., Kotzer A.M., Pighin J.A., Wasteneys G.O.(2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19: 2763–2775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos R.A., Pattathil S., Yang J.Y., Atmodjo M.A., Urbanowicz B.R., Moremen K.W., Mohnen D.(2018). A two-phase model for the non-processive biosynthesis of homogalacturonan polysaccharides by the GAUT1:GAUT7 complex. J. Biol. Chem. 293: 19047–19063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atmodjo M.A., Hao Z., Mohnen D.(2013). Evolving views of pectin biosynthesis. Annu. Rev. Plant Biol. 64: 747–779. [DOI] [PubMed] [Google Scholar]
- Atmodjo M.A., Sakuragi Y., Zhu X., Burrell A.J., Mohanty S.S., Atwood J.A. III, Orlando R., Scheller H.V., Mohnen D.(2011). Galacturonosyltransferase (GAUT)1 and GAUT7 are the core of a plant cell wall pectin biosynthetic homogalacturonan:galacturonosyltransferase complex. Proc. Natl. Acad. Sci. USA 108: 20225–20230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnette A.L., Lee C., Bradley L.C., Schreiner E.P., Park Y.B., Shin H., Cosgrove D.J., Park S., Kim S.H.(2012). Quantification of crystalline cellulose in lignocellulosic biomass using sum frequency generation (SFG) vibration spectroscopy and comparison with other analytical methods. Carbohydr. Polym. 89: 802–809. [DOI] [PubMed] [Google Scholar]
- Baron-Epel O., Gharyal P.K., Schindler M.(1988). Pectins as mediators of wall porosity in soybean cells. Planta 175: 389–395. [DOI] [PubMed] [Google Scholar]
- Baskin T.I., Beemster G.T., Judy-March J.E., Marga F.(2004). Disorganization of cortical microtubules stimulates tangential expansion and reduces the uniformity of cellulose microfibril alignment among cells in the root of Arabidopsis. Plant Physiol. 135: 2279–2290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bethke G., Thao A., Xiong G., Li B., Soltis N.E., Hatsugai N., Hillmer R.A., Katagiri F., Kliebenstein D.J., Pauly M., Glazebrook J.(2016). Pectin biosynthesis is critical for cell wall integrity and immunity in Arabidopsis thaliana. Plant Cell 28: 537–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswal A.K., et al. (2018). Sugar release and growth of biofuel crops are improved by downregulation of pectin biosynthesis. Nat. Biotechnol. 36: 249–257. [DOI] [PubMed] [Google Scholar]
- Bou Daher F., Chen Y., Bozorg B., Clough J., Jönsson H., Braybrook S.A.(2018). Anisotropic growth is achieved through the additive mechanical effect of material anisotropy and elastic asymmetry. eLife 7: e38161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudsocq M., Willmann M.R., McCormack M., Lee H., Shan L., He P., Bush J., Cheng S.H., Sheen J.(2010). Differential innate immune signalling via Ca2+ sensor protein kinases. Nature 464: 418–422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouton S., Leboeuf E., Mouille G., Leydecker M.T., Talbotec J., Granier F., Lahaye M., Höfte H., Truong H.N.(2002). QUASIMODO1 encodes a putative membrane-bound glycosyltransferase required for normal pectin synthesis and cell adhesion in Arabidopsis. Plant Cell 14: 2577–2590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broxterman S.E., Schols H.A.(2018). Interactions between pectin and cellulose in primary plant cell walls. Carbohydr. Polym. 192: 263–272. [DOI] [PubMed] [Google Scholar]
- Caffall K.H., Mohnen D.(2009). The structure, function, and biosynthesis of plant cell wall pectic polysaccharides. Carbohydr. Res. 344: 1879–1900. [DOI] [PubMed] [Google Scholar]
- Cannon M.C., Terneus K., Hall Q., Tan L., Wang Y., Wegenhart B.L., Chen L., Lamport D.T., Chen Y., Kieliszewski M.J.(2008). Self-assembly of the plant cell wall requires an extensin scaffold. Proc. Natl. Acad. Sci. USA 105: 2226–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Ehrhardt D.W., Somerville C.R.(2010). Mutations of cellulose synthase (CESA1) phosphorylation sites modulate anisotropic cell expansion and bidirectional mobility of cellulose synthase. Proc. Natl. Acad. Sci. USA 107: 17188–17193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Lee C.M., Wang H.-F., Jensen L., Kim S.H.(2017). Experimental and theoretical study of azimuth angle and polarization dependences of sum-frequency-generation vibrational spectral features of uniaxially aligned cellulose crystals. J. Phys. Chem. C 121: 18876–18886. [Google Scholar]
- Cheung A.Y., Wu H.M.(2011). THESEUS 1, FERONIA and relatives: A family of cell wall-sensing receptor kinases? Curr. Opin. Plant Biol. 14: 632–641. [DOI] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F.(1998). Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743. [DOI] [PubMed] [Google Scholar]
- Cornuault V., Manfield I.W., Ralet M.C., Knox J.P.(2014). Epitope detection chromatography: A method to dissect the structural heterogeneity and inter-connections of plant cell-wall matrix glycans. Plant J. 78: 715–722. [DOI] [PubMed] [Google Scholar]
- Cosgrove D.J.(2005). Growth of the plant cell wall. Nat. Rev. Mol. Cell Biol. 6: 850–861. [DOI] [PubMed] [Google Scholar]
- Derbyshire P., Findlay K., McCann M.C., Roberts K.(2007). Cell elongation in Arabidopsis hypocotyls involves dynamic changes in cell wall thickness. J. Exp. Bot. 58: 2079–2089. [DOI] [PubMed] [Google Scholar]
- De Ruiter G.A., Schols H.A., Voragen A.G., Rombouts F.M.(1992). Carbohydrate analysis of water-soluble uronic acid-containing polysaccharides with high-performance anion-exchange chromatography using methanolysis combined with TFA hydrolysis is superior to four other methods. Anal. Biochem. 207: 176–185. [DOI] [PubMed] [Google Scholar]
- Desprez T., Juraniec M., Crowell E.F., Jouy H., Pochylova Z., Parcy F., Höfte H., Gonneau M., Vernhettes S.(2007). Organization of cellulose synthase complexes involved in primary cell wall synthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 104: 15572–15577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dick-Pérez M., Zhang Y., Hayes J., Salazar A., Zabotina O.A., Hong M.(2011). Structure and interactions of plant cell-wall polysaccharides by two- and three-dimensional magic-angle-spinning solid-state NMR. Biochemistry 50: 989–1000. [DOI] [PubMed] [Google Scholar]
- Driouich A., Follet-Gueye M.L., Bernard S., Kousar S., Chevalier L., Vicré-Gibouin M., Lerouxel O.(2012). Golgi-mediated synthesis and secretion of matrix polysaccharides of the primary cell wall of higher plants. Front. Plant Sci. 3: 79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand C., Vicré-Gibouin M., Follet-Gueye M.L., Duponchel L., Moreau M., Lerouge P., Driouich A.(2009). The organization pattern of root border-like cells of Arabidopsis is dependent on cell wall homogalacturonan. Plant Physiol. 150: 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng W., et al. (2018). The FERONIA receptor kinase maintains cell-wall integrity during salt stress through Ca2+ signaling. Curr. Biol. 28: 666–675.e665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filisetti-Cozzi T.M., Carpita N.C.(1991). Measurement of uronic acids without interference from neutral sugars. Anal. Biochem. 197: 157–162. [DOI] [PubMed] [Google Scholar]
- Fontecave M., Atta M., Mulliez E.(2004). S-Adenosylmethionine: Nothing goes to waste. Trends Biochem. Sci. 29: 243–249. [DOI] [PubMed] [Google Scholar]
- Fry S.C.(1988). The Growing Plant Cell Wall: Chemical and Metabolic Analysis.. (New York: Longman Scientific & Technical; ). [Google Scholar]
- Gao P., Xin Z., Zheng Z.L.(2008). The OSU1/QUA2/TSD2-encoded putative methyltransferase is a critical modulator of carbon and nitrogen nutrient balance response in Arabidopsis. PLoS One 3: e1387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goubet F., Mohnen D.(1999). Subcellular localization and topology of homogalacturonan methyltransferase in suspension-cultured Nicotiana tabacum cells. Planta 209: 112–117. [DOI] [PubMed] [Google Scholar]
- Guo H., Li L., Ye H., Yu X., Algreen A., Yin Y.(2009). Three related receptor-like kinases are required for optimal cell elongation in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 106: 7648–7653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamant O., Heisler M.G., Jönsson H., Krupinski P., Uyttewaal M., Bokov P., Corson F., Sahlin P., Boudaoud A., Meyerowitz E.M., Couder Y., Traas J.(2008). Developmental patterning by mechanical signals in Arabidopsis. Science 322: 1650–1655. [DOI] [PubMed] [Google Scholar]
- Hamant O., Inoue D., Bouchez D., Dumais J., Mjolsness E.(2019). Are microtubules tension sensors? Nat. Commun. 10: 2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hématy K., Höfte H.(2008). Novel receptor kinases involved in growth regulation. Curr. Opin. Plant Biol. 11: 321–328. [DOI] [PubMed] [Google Scholar]
- Hongo S., Sato K., Yokoyama R., Nishitani K.(2012). Demethylesterification of the primary wall by PECTIN METHYLESTERASE35 provides mechanical support to the Arabidopsis stem. Plant Cell 24: 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao K., Zegzouti H., Goueli S.A.(2016). Methyltransferase-Glo: A universal, bioluminescent and homogenous assay for monitoring all classes of methyltransferases. Epigenomics 8: 321–339. [DOI] [PubMed] [Google Scholar]
- Hu H., et al. (2018). Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol. J. 16: 976–988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibar C., Orellana A.(2007). The import of S-adenosylmethionine into the Golgi apparatus is required for the methylation of homogalacturonan. Plant Physiol. 145: 504–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacques E., Verbelen J.P., Vissenberg K.(2013). Mechanical stress in Arabidopsis leaves orients microtubules in a ‘continuous’ supracellular pattern. BMC Plant Biol. 13: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kafle K., Shi R., Lee C.M., Mittal A., Park Y.B., Sun Y.-H., Park S., Chiang V., Kim S.H.J.C.(2014a). Vibrational sum-frequency-generation (SFG) spectroscopy study of the structural assembly of cellulose microfibrils in reaction woods. Cellulose 21: 2219–2231. [Google Scholar]
- Kafle K., Xi X., Lee C.M., Tittmann B.R., Cosgrove D.J., Park Y.B., Kim S.H.J.C.(2014b). Cellulose microfibril orientation in onion (Allium cepa L.) epidermis studied by atomic force microscopy (AFM) and vibrational sum frequency generation (SFG) spectroscopy. Cellulose 21: 1075–1086. [Google Scholar]
- Kim S.J., Held M.A., Zemelis S., Wilkerson C., Brandizzi F.(2015). CGR2 and CGR3 have critical overlapping roles in pectin methylesterification and plant growth in Arabidopsis thaliana. Plant J. 82: 208–220. [DOI] [PubMed] [Google Scholar]
- Kohorn B.D., Kohorn S.L.(2012). The cell wall-associated kinases, WAKs, as pectin receptors. Front. Plant Sci. 3: 88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolev A.V., Buschmann H., Doonan J.H., Lloyd C.W.(2007). AtMAP70-5, a divergent member of the MAP70 family of microtubule-associated proteins, is required for anisotropic cell growth in Arabidopsis. J. Cell Sci. 120: 2241–2247. [DOI] [PubMed] [Google Scholar]
- Korolev A.V., Chan J., Naldrett M.J., Doonan J.H., Lloyd C.W.(2005). Identification of a novel family of 70 kDa microtubule-associated proteins in Arabidopsis cells. Plant J. 42: 547–555. [DOI] [PubMed] [Google Scholar]
- Krupková E., Immerzeel P., Pauly M., Schmülling T.(2007). The TUMOROUS SHOOT DEVELOPMENT2 gene of Arabidopsis encoding a putative methyltransferase is required for cell adhesion and co-ordinated plant development. Plant J. 50: 735–750. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Kafle K., Huang S., Kim S.H.(2016). Multimodal broadband vibrational sum frequency generation (MM-BB-V-SFG) spectrometer and microscope. J. Phys. Chem. B 120: 102–116. [DOI] [PubMed] [Google Scholar]
- Lee C.M., Kafle K., Park Y.B., Kim S.H.(2014). Probing crystal structure and mesoscale assembly of cellulose microfibrils in plant cell walls, tunicate tests, and bacterial films using vibrational sum frequency generation (SFG) spectroscopy. Phys. Chem. Chem. Phys. 16: 10844–10853. [DOI] [PubMed] [Google Scholar]
- Lindner H., Müller L.M., Boisson-Dernier A., Grossniklaus U.(2012). CrRLK1L receptor-like kinases: Not just another brick in the wall. Curr. Opin. Plant Biol. 15: 659–669. [DOI] [PubMed] [Google Scholar]
- Liu H., Ma Y., Chen N., Guo S., Liu H., Guo X., Chong K., Xu Y.(2014). Overexpression of stress-inducible OsBURP16, the β subunit of polygalacturonase 1, decreases pectin content and cell adhesion and increases abiotic stress sensitivity in rice. Plant Cell Environ. 37: 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Sanchez P., Martinez-Sanz M., Bonilla M.R., Wang D., Gilbert E.P., Stokes J.R., Gidley M.J.(2017). Cellulose-pectin composite hydrogels: Intermolecular interactions and material properties depend on order of assembly. Carbohydr. Polym. 162: 71–81. [DOI] [PubMed] [Google Scholar]
- Lopez-Sanchez P., Martinez-Sanz M., Bonilla M.R., Wang D., Walsh C.T., Gilbert E.P., Stokes J.R., Gidley M.J.(2016). Pectin impacts cellulose fibre architecture and hydrogel mechanics in the absence of calcium. Carbohydr. Polym. 153: 236–245. [DOI] [PubMed] [Google Scholar]
- Makarem M., Lee C.M., Kafle K., Huang S., Chae I., Yang H., Kubicki J.D., Kim S.H.J.C.(2019). Probing cellulose structures with vibrational spectroscopy. Cellulose 26: 35–79. [Google Scholar]
- Makarem M., Sawada D., O’Neill H.M., Lee C.M., Kafle K., Park Y.B., Mittal A., Kim S.H.(2017). Dependence of sum frequency generation (SFG) spectral features on the mesoscale arrangement of SFG-active crystalline domains interspersed in SFG-inactive matrix: A case study with cellulose in uniaxially aligned control samples and alkali-treated secondary cell walls of plants. J. Phys. Chem. C 121: 10249–10257. [Google Scholar]
- Marc J., Granger C.L., Brincat J., Fisher D.D., Kao T., McCubbin A.G., Cyr R.J.(1998). A GFP-MAP4 reporter gene for visualizing cortical microtubule rearrangements in living epidermal cells. Plant Cell 10: 1927–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao Y., Li H.Y., Shen J., Wang J., Jiang L.(2011). QUASIMODO 3 (QUA3) is a putative homogalacturonan methyltransferase regulating cell wall biosynthesis in Arabidopsis suspension-cultured cells. J. Exp. Bot. 62: 5063–5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miedes E., Suslov D., Vandenbussche F., Kenobi K., Ivakov A., Van Der Straeten D., Lorences E.P., Mellerowicz E.J., Verbelen J.P., Vissenberg K.(2013). Xyloglucan endotransglucosylase/hydrolase (XTH) overexpression affects growth and cell wall mechanics in etiolated Arabidopsis hypocotyls. J. Exp. Bot. 64: 2481–2497. [DOI] [PubMed] [Google Scholar]
- Mouille G., Ralet M.C., Cavelier C., Eland C., Effroy D., Hématy K., McCartney L., Truong H.N., Gaudon V., Thibault J.F., Marchant A., Höfte H.(2007). Homogalacturonan synthesis in Arabidopsis thaliana requires a Golgi-localized protein with a putative methyltransferase domain. Plant J. 50: 605–614. [DOI] [PubMed] [Google Scholar]
- Müller K., Levesque-Tremblay G., Bartels S., Weitbrecht K., Wormit A., Usadel B., Haughn G., Kermode A.R.(2013). Demethylesterification of cell wall pectins in Arabidopsis plays a role in seed germination. Plant Physiol. 161: 305–316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumetzler L., Humphrey T., Lumba S., Snyder S., Yeats T.H., Usadel B., Vasilevski A., Patel J., Rose J.K., Persson S., Bonetta D.(2012). The FRIABLE1 gene product affects cell adhesion in Arabidopsis. PLoS One 7: e42914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ordaz-Ortiz J.J., Marcus S.E., Knox J.P.(2009). Cell wall microstructure analysis implicates hemicellulose polysaccharides in cell adhesion in tomato fruit pericarp parenchyma. Mol. Plant 2: 910–921. [DOI] [PubMed] [Google Scholar]
- Orfila C., Sørensen S.O., Harholt J., Geshi N., Crombie H., Truong H.N., Reid J.S., Knox J.P., Scheller H.V.(2005). QUASIMODO1 is expressed in vascular tissue of Arabidopsis thaliana inflorescence stems, and affects homogalacturonan and xylan biosynthesis. Planta 222: 613–622. [DOI] [PubMed] [Google Scholar]
- Paredez A.R., Persson S., Ehrhardt D.W., Somerville C.R.(2008). Genetic evidence that cellulose synthase activity influences microtubule cortical array organization. Plant Physiol. 147: 1723–1734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredez A.R., Somerville C.R., Ehrhardt D.W.(2006). Visualization of cellulose synthase demonstrates functional association with microtubules. Science 312: 1491–1495. [DOI] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J.(2012). A revised architecture of primary cell walls based on biomechanical changes induced by substrate-specific endoglucanases. Plant Physiol. 158: 1933–1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park Y.B., Cosgrove D.J.(2015). Xyloglucan and its interactions with other components of the growing cell wall. Plant Cell Physiol. 56: 180–194. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Braybrook S.A., Le Guillou L., Bron E., Kuhlemeier C., Höfte H.(2011). Pectin-induced changes in cell wall mechanics underlie organ initiation in Arabidopsis. Curr. Biol. 21: 1720–1726. [DOI] [PubMed] [Google Scholar]
- Peaucelle A., Wightman R., Höfte H.(2015). The control of growth symmetry breaking in the Arabidopsis hypocotyl. Curr. Biol. 25: 1746–1752. [DOI] [PubMed] [Google Scholar]
- Pérez-Pérez Y., Carneros E., Berenguer E., Solís M.T., Bárány I., Pintos B., Gómez-Garay A., Risueño M.C., Testillano P.S.(2019). Pectin de-methylesterification and AGP increase promote cell wall remodeling and are required during somatic embryogenesis of Quercus suber. Front. Plant Sci. 9: 1915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phyo P., Wang T., Xiao C., Anderson C.T., Hong M.(2017). Effects of pectin molecular weight changes on the structure, dynamics, and polysaccharide interactions of primary cell walls of Arabidopsis thaliana: Insights from solid-state NMR. Biomacromolecules 18: 2937–2950. [DOI] [PubMed] [Google Scholar]
- Popper Z.A., Fry S.C.(2008). Xyloglucan-pectin linkages are formed intra-protoplasmically, contribute to wall-assembly, and remain stable in the cell wall. Planta 227: 781–794. [DOI] [PubMed] [Google Scholar]
- Qu L., Wu C., Zhang F., Wu Y., Fang C., Jin C., Liu X., Luo J.(2016). Rice putative methyltransferase gene OsTSD2 is required for root development involving pectin modification. J. Exp. Bot. 67: 5349–5362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajangam A.S., et al. (2008). MAP20, a microtubule-associated protein in the secondary cell walls of hybrid aspen, is a target of the cellulose synthesis inhibitor 2,6-dichlorobenzonitrile. Plant Physiol. 148: 1283–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Refrégier G., Pelletier S., Jaillard D., Höfte H.(2004). Interaction between wall deposition and cell elongation in dark-grown hypocotyl cells in Arabidopsis. Plant Physiol. 135: 959–968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riveras E., Alvarez J.M., Vidal E.A., Oses C., Vega A., Gutiérrez R.A.(2015). The calcium ion is a second messenger in the nitrate signaling pathway of Arabidopsis. Plant Physiol. 169: 1397–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S., Kuhlemeier C.(2018). Global compression reorients cortical microtubules in Arabidopsis hypocotyl epidermis and promotes growth. Curr. Biol. 28: 1794–1802.e1792. [DOI] [PubMed] [Google Scholar]
- Somerville C., Bauer S., Brininstool G., Facette M., Hamann T., Milne J., Osborne E., Paredez A., Persson S., Raab T., Vorwerk S., Youngs H.(2004). Toward a systems approach to understanding plant cell walls. Science 306: 2206–2211. [DOI] [PubMed] [Google Scholar]
- Sterling J.D., Atmodjo M.A., Inwood S.E., Kumar Kolli V.S., Quigley H.F., Hahn M.G., Mohnen D.(2006). Functional identification of an Arabidopsis pectin biosynthetic homogalacturonan galacturonosyltransferase. Proc. Natl. Acad. Sci. USA 103: 5236–5241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoppin-Mellet V., Gaillard J., Vantard M.(2006). Katanin’s severing activity favors bundling of cortical microtubules in plants. Plant J. 46: 1009–1017. [DOI] [PubMed] [Google Scholar]
- Swamy P.S., et al. (2015). Tubulin perturbation leads to unexpected cell wall modifications and affects stomatal behaviour in Populus. J. Exp. Bot. 66: 6507–6518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiré C., De Rycke R., De Loose M., Inzé D., Van Montagu M., Engler G.(1994). Extensin gene expression is induced by mechanical stimuli leading to local cell wall strengthening in Nicotiana plumbaginifolia. Planta 195: 175–181. [DOI] [PubMed] [Google Scholar]
- Updegraff D.M.(1969). Semimicro determination of cellulose in biological materials. Anal. Biochem. 32: 420–424. [DOI] [PubMed] [Google Scholar]
- Verger S., Chabout S., Gineau E., Mouille G.(2016). Cell adhesion in plants is under the control of putative O-fucosyltransferases. Development 143: 2536–2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger S., Long Y., Boudaoud A., Hamant O.(2018). A tension-adhesion feedback loop in plant epidermis. eLife 7: e34460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vincken J.P., Schols H.A., Oomen R.J., McCann M.C., Ulvskov P., Voragen A.G., Visser R.G.(2003). If homogalacturonan were a side chain of rhamnogalacturonan I: Implications for cell wall architecture. Plant Physiol. 132: 1781–1789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voiniciuc C., Engle K.A., Günl M., Dieluweit S., Schmidt M.H., Yang J.Y., Moremen K.W., Mohnen D., Usadel B.(2018). Identification of key enzymes for pectin synthesis in seed mucilage. Plant Physiol. 178: 1045–1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Park Y.B., Caporini M.A., Rosay M., Zhong L., Cosgrove D.J., Hong M.(2013). Sensitivity-enhanced solid-state NMR detection of expansin’s target in plant cell walls. Proc. Natl. Acad. Sci. USA 110: 16444–16449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Park Y.B., Cosgrove D.J., Hong M.(2015). Cellulose-pectin spatial contacts are inherent to never-dried Arabidopsis primary cell walls: Evidence from solid-state nuclear magnetic resonance. Plant Physiol. 168: 871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weraduwage S.M., Kim S.J., Renna L., Anozie F.C., Sharkey T.D., Brandizzi F.(2016). Pectin methylesterification impacts the relationship between photosynthesis and plant growth. Plant Physiol. 171: 833–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White P.B., Wang T., Park Y.B., Cosgrove D.J., Hong M.(2014). Water-polysaccharide interactions in the primary cell wall of Arabidopsis thaliana from polarization transfer solid-state NMR. J. Am. Chem. Soc. 136: 10399–10409. [DOI] [PubMed] [Google Scholar]
- Wilkins K.A., Matthus E., Swarbreck S.M., Davies J.M.(2016). Calcium-mediated abiotic stress signaling in roots. Front. Plant Sci. 7: 1296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J.(2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS One 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Anderson C.T.(2016). Interconnections between cell wall polymers, wall mechanics, and cortical microtubules: Teasing out causes and consequences. Plant Signal. Behav. 11: e1215396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao C., Barnes W.J., Zamil M.S., Yi H., Puri V.M., Anderson C.T.(2017). Activation tagging of Arabidopsis POLYGALACTURONASE INVOLVED IN EXPANSION2 promotes hypocotyl elongation, leaf expansion, stem lignification, mechanical stiffening, and lodging. Plant J. 89: 1159–1173. [DOI] [PubMed] [Google Scholar]
- Xiao C., Zhang T., Zheng Y., Cosgrove D.J., Anderson C.T.(2016). Xyloglucan deficiency disrupts microtubule stability and cellulose biosynthesis in Arabidopsis, altering cell growth and morphogenesis. Plant Physiol. 170: 234–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S.L., Rahman A., Baskin T.I., Kieber J.J.(2008). Two leucine-rich repeat receptor kinases mediate signaling, linking cell wall biosynthesis and ACC synthase in Arabidopsis. Plant Cell 20: 3065–3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Y., et al. (2017). Rice sucrose partitioning mediated by a putative pectin methyltransferase and homogalacturonan methylesterification. Plant Physiol. 174: 1595–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A., Ito T., Higaki T., Kutsuna N., Saito T., Ishimizu T., Osada H., Hasezawa S., Matsui M., Demura T.(2010). Cobtorin target analysis reveals that pectin functions in the deposition of cellulose microfibrils in parallel with cortical microtubules. Plant J. 64: 657–667. [DOI] [PubMed] [Google Scholar]
- Zhang T., Tang H., Vavylonis D., Cosgrove D.J.(2019). Disentangling loosening from softening: Insights into primary cell wall structure. Plant J. 100: 1101–1117. [DOI] [PubMed] [Google Scholar]
- Zhang T., Zheng Y., Cosgrove D.J.(2016). Spatial organization of cellulose microfibrils and matrix polysaccharides in primary plant cell walls as imaged by multichannel atomic force microscopy. Plant J. 85: 179–192. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Liao C., Zhao S., Wang C., Guo Y.(2017). The glycosyltransferase QUA1 regulates chloroplast-associated calcium signaling during salt and drought stress in Arabidopsis. Plant Cell Physiol. 58: 329–341. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Wang X., Chen Y., Wagner E., Cosgrove D.J.(2018). Xyloglucan in the primary cell wall: Assessment by FESEM, selective enzyme digestions and nanogold affinity tags. Plant J. 93: 211–226. [DOI] [PubMed] [Google Scholar]
- Zykwinska A., Thibault J.F., Ralet M.C.(2008). Competitive binding of pectin and xyloglucan with primary cell wall cellulose. Carbohydr. Polym. 74: 957–961. [Google Scholar]
- Zykwinska A.W., Ralet M.C., Garnier C.D., Thibault J.F.(2005). Evidence for in vitro binding of pectin side chains to cellulose. Plant Physiol. 139: 397–407. [DOI] [PMC free article] [PubMed] [Google Scholar]