Tagging ROP in the middle of the protein produces a functional protein that provides all ROP function in juvenile moss tissues and marks sites with weakened cell walls long before expansion occurs.
Abstract
Rho of Plants (ROPs) are GTPases that regulate polarity and patterned wall deposition in plants. As these small, globular proteins have many interactors, it has been difficult to ensure that methods to visualize ROP in live cells do not affect ROP function. Here, motivated by work in fission yeast (Schizosaccharomyces pombe), we generated a fluorescent moss (Physcomitrium [Physcomitrella] patens) ROP4 fusion protein by inserting mNeonGreen after Gly-134. Plants harboring tagged ROP4 and no other ROP genes were phenotypically normal. Plants lacking all four ROP genes comprised an unpatterned clump of spherical cells that were unable to form gametophores, demonstrating that ROP is essentially for spatial patterning at the cellular and tissue levels. The functional ROP fusion protein formed a steep gradient at the apical plasma membranes of growing tip cells. ROP also predicted the site of branch formation in the apical cell at the onset of mitosis, which occurs one to two cell cycles before a branch cell emerges. While fluorescence recovery after photobleaching studies demonstrated that ROP dynamics do not depend on the cytoskeleton, acute depolymerization of the cytoskeleton removed ROP from the membrane only in recently divided cells, pointing to a feedback mechanism between the cell cycle, cytoskeleton, and ROP.
INTRODUCTION
Cell polarity is an important process in eukaryotic development. In plants, development occurs in the absence of cell migration and thus requires exquisite control of cell polarity to properly pattern tissues throughout the organism. For example, the establishment of polarized membrane domains with the appropriate auxin efflux carriers sets up the organization of the root (Blilou et al., 2005; Kania et al., 2014; van Dop et al., 2020), while the proper positioning of membrane markers in developing leaves leads to normal stomatal patterning (Zhang et al., 2016; Houbaert et al., 2018; Mansfield et al., 2018). In addition to the complex polarity establishment found in tissues (Zhang and Dong, 2018), seed plants have several cell types, such as root hairs and pollen tubes that undergo highly polarized cell expansion, and this expansion underlies their function. Root hairs are important for nutrient uptake (Gilroy and Jones, 2000), while pollen tubes are critical for sexual reproduction (Chen et al., 2018). In nonflowering plants, polarized cell expansion, also known as tip growth, generates protonemata and rhizoids. Protonemata in mosses establish the plant, as this is the tissue that emerges from the spore, while rhizoids help to anchor the tissue to the soil in both mosses and liverworts (Rounds and Bezanilla, 2013; Shimamura, 2016).

In plants, Rho of Plants (ROP) proteins are small GTPases that share sequence similarity with the Rho/RAC/CDC42 family of G-proteins present in all other eukaryotes (Hall, 2012). Rho family proteins have been extensively studied in mammalian and yeast systems and are known to be critical regulators of cell polarity (Etienne-Manneville and Hall, 2002). For at least the past 20 years, numerous studies have established that ROPs are master regulators of cell polarity in plants (Bloch and Yalovsky, 2013). In flowering plants, ROP is important for patterned cell wall deposition, including the development of tip-growing pollen tubes and root hairs (Lin et al., 1996; Lin and Yang, 1997; Hwang et al., 2005; Gu et al., 2006; Craddock et al., 2012). Besides tip-growing cells, ROP is also important for plant development at the tissue level (Fu et al., 2002; Foucart et al., 2009; Lin et al., 2012; Zhang et al., 2019). The appropriate transcriptional regulation of ROP is essential for patterning the puzzle-shaped pavement cells in Arabidopsis (Arabidopsis thaliana; Fu et al., 2005; Xu et al., 2010; Lin et al., 2015). During secondary cell wall formation, Arabidopsis ROP11 induces the formation of xylem pits via the local disassembly of microtubules (Oda and Fukuda, 2012, 2013; Nagashima et al., 2018; Sugiyama et al., 2019). Various studies have also discovered that ROP genes influence signaling processes involved in pathogen defense, stress responses, and nodule symbiosis in various species (Ke et al., 2012; Poraty-Gavra et al., 2013; Venus and Oelmüller, 2013; Huang et al., 2014; Lei et al., 2015; Miao et al., 2018; Wang et al., 2018). In contrast to flowering plants, which generally have multiple ROP genes grouped into three subfamilies (Bloch and Yalovsky, 2013), the model moss Physcomitrium (Physcomitrella) patens has four highly similar ROP genes within a single subfamily (Eklund et al., 2010; Ito et al., 2014). In fact, the translation of the four P. patens ROP genes results in the production of three distinct proteins that differ by at most two amino acids (ROP1 and ROP4 are identical; ROP1/ROP4 differ from ROP2/ROP3 by one amino acid; ROP2 differs from ROP3 by two amino acids). Loss-of-function studies demonstrated that these ROP genes are functionally redundant in controlling polarized growth (Burkart et al., 2015).
Like the majority of small G-proteins, many ROPs (including all four in P. patens) possess a C-terminal CAAX motif that is important for membrane targeting (Lavy et al., 2002; Yang, 2002; Yalovsky, 2015). The Cys in the CAAX motif serves as a site for prenylation, creating a lipid anchor that can insert into the plasma membrane (Geyer and Wittinghofer, 1997). ROPs also bind to numerous effectors and are regulated by many interacting proteins. For example, guanine exchange factors trigger ROP to release GDP and bind to GTP. GTPase-activating proteins bind to GTP-ROP and activate its intrinsic GTPase activity. Guanine dissociation inhibitors bind to GDP-ROP, interacting with the covalent lipid modification, thereby removing GDP-ROP from the plasma membrane. These proteins interact with ROP directly to precisely regulate its activation both spatially and temporally (Dovas and Couchman, 2005; Feiguelman et al., 2018; Bascom et al., 2019). As a result, the binding sites for these interacting proteins on ROP need to remain accessible for normal ROP function. ROP is a small protein of only 196 amino acids, which is smaller than commonly used fluorescent proteins. Thus, it is challenging to create a fluorescent ROP fusion protein in which all aspects of ROP function remain intact.
Because the covalent lipid modification occurs on the C terminus of ROP, the vast majority of studies in plants have utilized N-terminally tagged ROP proteins. Inspection of ROP’s sequence and comparison to related G-proteins whose structures have been determined revealed that the ROP N terminus is near the nucleotide binding pocket, the switch regions, and the interface known to interact with regulators such as guanine dissociation inhibitor (Hoffman et al., 2000). The switch regions comprise the structural features that define the distinct conformations when ROP is bound to either GTP or GDP (Vetter and Wittinghofer, 2001). Given that several critical ROP features are near the N terminus, tagging at the N terminus could possibly influence normal ROP function. N-Terminally tagged Rho family GTPases have been commonly used for fluorescent live-cell imaging (Wedlich-Soldner et al., 2003). However, in both budding and fission yeasts, gene replacement studies demonstrated that the function of N-terminally tagged CDC42 was affected, introducing temperature-sensitive growth defects (Howell et al., 2012; Bendezú et al., 2015) and negatively influencing CDC42’s association with exocytic vesicles (Watson et al., 2014). As an alternative to N-terminal tagging, Burkart (2014) investigated the possibility of tagging ROP before the CAAX motif in P. patens. The author introduced sequences encoding GFP immediately upstream of the CAAX motif in the endogenous ROP4 genomic locus. However, tagging ROP in this manner was unable to rescue ROP function and could not drive polarized growth (Burkart, 2014).
Besides direct N-terminal tagging, reporter systems for labeling active ROP domains have utilized ROP binding proteins or their specific binding domain (Cdc42- and Rac-interactive binding [CRIB] motif) fused to a fluorescent protein in both animal and plant systems (Nalbant et al., 2004; Zhu and Fu, 2012; Li et al., 2018). Similarly, Förster resonance energy transfer (FRET) sensors that visualize ROP activation dynamically have also been developed (Itoh et al., 2002; Liu et al., 2011; Wong et al., 2018). These reporters are nonetheless based on the direct binding of the conserved CRIB motif (also GTPase binding domain or p21 binding domain) to the Rho family GTPase of interest. The CRIB motif is present in Rho GTPase–activating protein (Wu et al., 2000; Schaefer et al., 2011) and GTPase effectors. The most well-studied effectors include p21-activated kinase (PAK), activated Cdc42-associated kinase (ACK), and Wiscott-Aldrich syndrome protein (WASP) for CDC42 in animal and yeast systems (Hoffman and Cerione, 2000) and RIC for ROP in plant systems (Wu et al., 2001). While these approaches are powerful because they focus on identifying subcellular domains containing the active GTPase, it can be difficult to ensure that all aspects of GTPase function remain unimpaired, since the unregulated expression of reporters could compete with regulators and effectors for the CRIB domain binding sites on the GTPases.
Expressing N-terminally tagged ROP has been the most common approach to study ROP localization in plants (Li et al., 1999, 2018; Yalovsky et al., 2008; Sun et al., 2015; Yi and Goshima, 2020a). Even with efforts to tune down the expression level with weaker or inducible promoters (Le Bail et al., 2019), the expression levels need to be carefully monitored to avoid introducing new phenotypes. Notably, N-terminally tagged ROP fusion proteins exhibit activity when expressed. For example, ROP with an N-terminal GFP tag induces the depolarization of pollen tube growth (Kost et al., 1999), which also occurs when untagged ROP is overexpressed (Fu et al., 2001). Overexpressing constitutively active ROP causes root hair swelling (Molendijk et al., 2001) and narrower leaf shape (Bloch et al., 2005). However, while these activities implicate function, they are not equivalent to demonstrating the replacement of the normal function of untagged ROP. Complementation studies similar to what has been performed in budding and fission yeasts (Howell et al., 2012; Watson et al., 2014; Bendezú et al., 2015) have been challenging in plants. Arabidopsis has 11 ROP genes, many with overlapping functions (Craddock et al., 2012), making it difficult to perform clean genetic complementation in a flowering plant model system.
The moss P. patens can be easily propagated vegetatively, and mutants can be recovered after targeted genome editing. Recent advances in clustered regularly interspaced short palindromic repeats (CRISPR) technology have provided excellent tools to simultaneously edit several genomic loci (Lopez-Obando et al., 2016; Collonnier et al., 2017; Mallett et al., 2019; Yi and Goshima, 2020b), making testing fluorescently fused ROP in a complete knockout background of the whole gene family feasible. In the current study, we inserted the coding sequence (CDS) of mNeonGreen (mNG) fluorescent protein into the native genomic locus of P. patens ROP4 without introducing any other alterations to the genome. We tested fusion proteins with the commonly used N-terminal tag and with an internal “sandwich” tag inspired by previous work in fission yeast with CDC42 (Bendezú et al., 2015). We determined that only the sandwich-tagged ROP4 fusion protein maintained full functionality and was sufficient to drive polarized growth in the absence of all other ROP proteins. Using this functionally tagged ROP fluorescent marker, we investigated ROP localization during growth and its dependence on the cytoskeleton. We discovered that ROP polarization in growing cells is affected by changes in the cytoskeleton in a cell cycle–dependent manner, revealing a potential feedback mechanism between growth, polarity, and the cell cycle.
RESULTS
N-Terminally Tagged ROP4 Is Not Functional
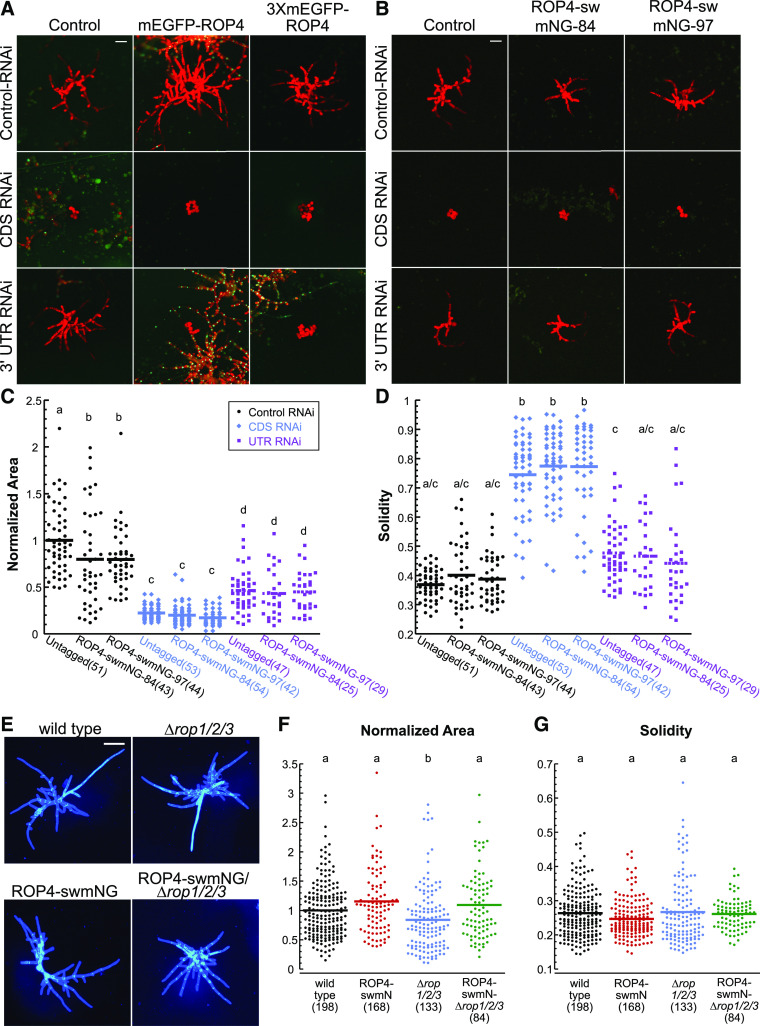
Previously, homologous recombination was used to replace the genomic region containing the ROP4 open reading frame with either a 5′ GFP-tagged cDNA or an untagged cDNA (Burkart et al., 2015). ROP4 was chosen because it encodes the same protein as ROP1. Furthermore, ROP4 and ROP3 exhibit similar expression levels and are the two most highly expressed ROP genes in protonemata. Even though the GFP-tagged protein was expressed, neither the GFP-tagged nor the untagged cDNA was able to replace the function of the wild-type gene, suggesting that the genomic context of the CDS is critical for ROP4 function. To minimally alter gene structure, we used CRISPR-Cas9 mediated homology-directed repair (HDR; Mallett et al., 2019) to insert sequences encoding monomeric enhanced GFP (mEGFP; Vidali et al., 2009) or three tandem mEGFPs (3XmEGFP) immediately downstream of the ROP4 start codon (Supplemental Figure 1A). To determine whether the ROP4 fusion protein was functional, we introduced the mEGFP sequences into a line where the ROP4 3′ untranslated region (UTR) was deleted, rendering the ROP4 locus insensitive to an RNA interference (RNAi) construct targeting the 3′ UTR (Burkart et al., 2015). Thus, using RNAi, we could specifically silence the other three ROP genes that are functionally redundant with ROP4 in the tagged line (Burkart et al., 2015).
Transforming the stably tagged lines with an RNAi construct targeting the CDS regions of all ROP genes (CDS RNAi) and the nuclear GFP:β-glucuronidase (GUS) reporter resulted in plants lacking nuclear GFP that were composed of small spherical cells (Figure 1A). Transformation with a construct that targets the 3′ UTRs of all ROP genes (UTR RNAi) and the nuclear GFP:GUS reporter should not affect ROP4 expression, since the targeting sequence has been deleted from the ROP4 gene. Thus, we would predict that if the tagged ROP4 is functional, plants transformed with the UTR-RNAi construct should exhibit polarized growth. In contrast to this prediction, plants transformed with the UTR-RNAi construct were unpolarized and indistinguishable from the CDS RNAi–transformed plants (Figure 1A). These data demonstrate that the N-terminally tagged ROP proteins are not functional, as neither can maintain polarized growth in moss protonemata in the absence of all other ROP proteins.
Figure 1.
ROP4 Is Functional When the Fluorescent Protein Is Inserted in a Loop within the Protein.
(A) and (B) Transient RNAi assay performed in a P. patens line containing a nuclear GFP:GUS reporter and a deletion of the ROP4 3′ UTR, which renders the ROP4 gene insensitive to the UTR-RNAi construct as described by Burkart et al. (2015). All RNAi constructs simultaneously silence the nuclear GFP:GUS reporter and the target genes, and the control construct only silences GFP:GUS. (A) Representative chlorophyll autofluorescence (red) images of 7-d-old plants regenerated from protoplasts. Control and N-terminally tagged ROP4 lines were transformed with the indicated RNAi constructs. Chlorophyll autofluorescence labels the plant, while loss of nuclear GFP signal indicates successfully silenced plants. Bar for all images = 100 µm. (B) Representative chlorophyll autofluorescence images of 7-d-old plants regenerated from protoplasts from control and sandwich-tagged ROP4 (ROP4-swmNG) transformed with the indicated constructs. Bar for all images = 100 µm.
(C) For the RNAi experiments represented in (B), the area of the chlorophyll autofluorescence was normalized to the chlorophyll autofluorescence area of plants transformed with the control plasmid.
(D) Solidity (convex hull area/area) for RNAi experiments represented in (B). Numbers in parentheses indicate the number of plants for each group. Legend of the graph in (C) also applies to (D) and indicates that black, blue, and purple represent data from protoplasts transformed with the control, CDS, or UTR RNAi constructs, respectively.
(E) Representative images of 7-d-old plants stained with calcofluor and regenerated from protoplasts from the wild type, ∆rop1/2/3, and ROP4-swmNG in the wild-type and ∆rop1/2/3 backgrounds. Bar = 200 µm.
(F) and (G) Normalized area (F) and solidity (G) for the sandwich-tagged ROP4 in the wild type (black) and ∆rop1/2/3 (magenta). Based on the Kolmogorov–Smirnov test, the data in (C), (D), (F), and (G) are normally distributed. Different letters in (C), (D), and (F) indicate groups with significantly different means, as determined by ANOVA with a Tukey’s HSD all pair comparison post test (α = 0.05). Also see Supplemental Tables 2 to 5.
Internally Tagged ROP4 Fully Rescues Polarized Growth
Inspired by previous work in fission yeast (Bendezú et al., 2015) where the ROP homolog CDC42 was functionally tagged in the middle of the protein (sandwich tag), we set out to test whether a similar approach might work for ROP. The fission yeast CDC42 sandwich tag was inserted in a small stretch of amino acids after the α3′ helix. As a result, the fluorescent protein looped out of the protein away from interfaces known to mediate interactions with CDC42. We identified a small stretch of nonconserved amino acids in ROP4 at the analogous CDC42 insertion site and speculated that this region may not be essential for ROP protein function. To test this notion, we generated a sandwich tag for P. patens ROP4 by inserting the CDS of mNG after Gly-134 (Supplemental Figure 1B).
Using CRISPR-mediated HDR, we introduced sequences encoding mNG into the ROP4-∆3′UTR reporter line, rendering the sandwich-tagged ROP4 locus insensitive to the UTR-RNAi construct. Remarkably, in two independently generated sandwich-tagged lines, silencing with the UTR-RNAi construct resulted in polarized plants (Figure 1B) indistinguishable from the parental untagged line. Notably, plant size was reduced to the same extent in sandwich-tagged and untagged control lines (Figure 1C), and solidity, a measure of plant polarity, was similar in all lines (Figure 1D). These data demonstrate that the untagged and sandwich-tagged ROP4 (hereafter referred to as ROP4-swmNG) contribute equally to ROP function. As expected, silencing with the CDS-RNAi construct in all lines resulted in unpolarized plants composed of circular cells with no developmental organization (Figure 1B). The finding that the sandwich-tagged lines transformed with the control RNAi constructs were smaller than the untagged control parental line might reflect differences in expression and/or stability of the sandwich-tagged fusion protein due to the deletion of the 3′ UTR.
To control for possible expression effects resulting from deletion of the 3′ UTR and to ensure that incomplete silencing of ROP did not account for the observed rescue of polarized growth, we isolated several lines where the ROP4 locus with an intact 3′ UTR was tagged appropriately with swmNG and carried null mutations in ROP1, ROP2, and ROP3, indicating that ROP4-swmNG does not affect plant viability. While these lines carried different mutations (Supplemental Figure 2), they all grew normally. Some of the mutants were isolated after cotransforming moss protoplasts with the CRISPR-Cas9 construct and double-stranded DNA oligos containing an in-frame stop codon. These oligos were designed to integrate at the Cas9 cut site via HDR, ensuring that a stop codon would be present soon after the double-strand break (Yi and Goshima, 2020b). One of the lines that incorporated the designed pre-mature stop codons at all three targeted ROP loci was chosen for subsequent experiments (ROP4-swmNG/∆rop1/2/3-196 [Supplemental Figure 2], hereafter referred to as ROP4-swmNG/∆rop1/2/3). As a control, we also generated the same null mutations in the wild-type background (∆rop1/2/3-198 [Supplemental Figure 2], hereafter referred to as ∆rop1/2/3). As expected based on previous RNAi data (Figures 1B to 1D; Burkart et al., 2015), ∆rop1/2/3 plants were smaller than the wild type (Figures 1E and 1F) but exhibited no defects in plant morphology, as measured by solidity (Figures 1E and 1G). Remarkably, the sandwich-tagged ROP4 line grew similarly in the presence or absence of the remaining ROP genes (Figures 1E and 1G), demonstrating that ROP4-swmNG is sufficient to drive polarized growth in P. patens protonemata.
Compared to the N-Terminal Fusion Protein, ROP4-swmNG Localizes to a Smaller Region of the Growing Tip
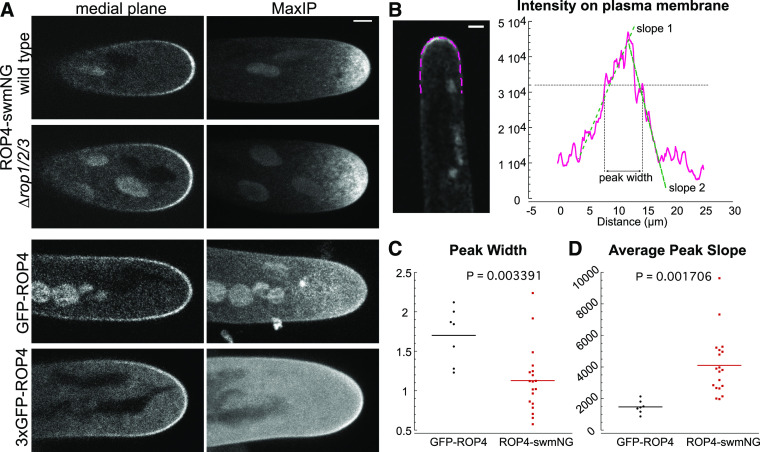
N-Terminal ROP fusion proteins localize to the apical plasma membranes of pollen tubes (Li et al., 1999, 2018; Sun et al., 2015), root hairs (Yalovsky et al., 2008), and moss protonemata (Burkart et al., 2015; Yi and Goshima, 2020a). Since the N-terminal fusion protein is not functional, we wondered whether its localization might be affected. Thus, we used laser-scanning confocal microscopy to compare the localization of the N-terminal or sandwich-tagged ROP4 in growing moss protonemata. Similar to previous studies (Burkart et al., 2015; Yi and Goshima, 2020a), regardless of whether ROP4 was tagged with mEGFP or 3XmEGFP, the N-terminal fusion protein localized to the apical plasma membrane (Figure 2A). Interestingly, the 3XmEGFP tag was not necessarily brighter on the plasma membrane. Instead, cells with this tag had a higher cytosolic background, which might result from cleavage of the N-terminal GFP tag, as mentioned in Yi and Goshima (2020a). Medial plane images of mEGFP-ROP4 revealed a strong plasma membrane signal that covered the entire apical dome of the cell. Maximum projections of confocal Z-stacks demonstrated that this signal often extended 15 to 20 µm back from the cell tip (Figure 2A). While ROP4-swmNG was also enriched at the apical plasma membrane, regardless of the presence or absence of the other three ROP genes, ROP4-swmNG covered a much more tightly focused area of the cell tip. Maximum projections of Z-stacks demonstrated that the ROP4-swmNG signal only reached 5 to 10 µm back from the tip (Figure 2A).
Figure 2.
Comparison of the Localization of the N-Terminally and Sandwich-Tagged ROP4 Proteins.
(A) Representative confocal images of growing moss protonemata of the indicated genotypes. Medial plane shows a single focal plane at the middle of the cell; MaxIP shows the maximum projection of a Z-stack taken through the entire cell volume. Bar for all images = 3 µm.
(B) Example image and measurement of ROP4-swmNG fluorescent signal along the plasma membrane. A 3-pixel-wide trace (magenta) of the plasma membrane was manually drawn and the normalized signal intensity along the line was plotted. Bar = 3 µm.
(C) Measurement of the peak width (indicated by the horizontal gray line) in (B) normalized to the cell width measured 10 µm from the cell tip. n = 7, GFP-ROP4; n = 19, ROP4-swmNG.
(D) Average of the slopes (slopes 1 and 2 in [B], green dashed lines). Slopes were determined by fitting a line to the points in the increasing and decreasing segment of the curve. Statistical analyses were performed using Student’s t test for unpaired data with equal variance with t probability indicated in the graph. Also see Supplemental Tables 6 and 7.
To quantify the differences between the plasma membrane localizations of the N-terminal and sandwich fusion proteins, we drew a line on the plasma membrane of medial sections of apical cells with normalized fluorescence. We obtained fluorescence intensity profiles of the plasma membrane and measured the width of the peak at the same intensity value for all cells (Figure 2B). We discovered that GFP-ROP was spread out over a larger area of the cell apex compared to ROP4-swmNG (Figure 2C), which is consistent with the maximum projections of confocal Z-stacks (Figure 1A). We also fit a line to both the increase and decrease in fluorescence intensity and calculated the absolute value of the slope (Figure 2B). ROP4-swmNG had significantly larger slopes than GFP-ROP (Figure 2D), demonstrating that the sandwich-tagged ROP also formed a steeper gradient on the plasma membrane. Together, these data demonstrate that the functional ROP4-swmNG localizes to a more restricted region of the cell apex than the N-terminal fusion protein.
During Changes in Growth Direction ROP4-swmNG Accumulates at Sites of Cell Expansion
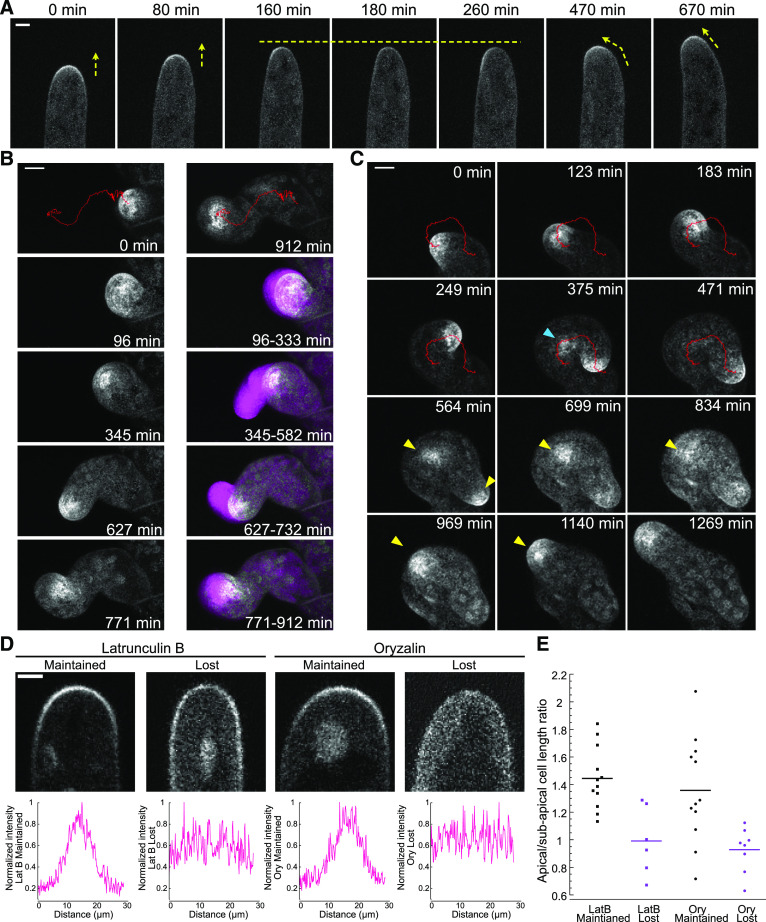
With a functional fluorescent fusion protein in hand, we investigated ROP dynamics during polarized growth. At a growing tip, the apical ROP4-swmNG gradient tracked with the growing tip as it elongated, and no obvious periodic fluctuations were observed in growing cells (Figure 3A; Supplemental Movie 1). In a growing moss plant, not all protonemal filaments are actively growing. During long-term imaging, it is possible to occasionally observe cells that cease to grow and after some time resume growing. In one of these cells, ROP disappeared from the apical plasma membrane when the cell stopped growing (Figure 3A; Supplemental Movie 1). After 100 min, ROP repopulated the left side of the membrane at the cell tip, and when the cell resumed growing, it turned toward the left, indicating that areas of ROP accumulation predict the site of cell expansion (Figure 3A; Supplemental Movie 1). Unfortunately, predicting when a growth site will cease is challenging. Therefore, to examine the behavior of ROP during changes in the direction of growth and the establishment of new growth sites, we used an assay that produces directional changes as well as frequent pauses. When microtubules are disrupted with oryzalin, the growing tip randomly changes direction, and new tips occasionally emerge from the apical cell (Wu and Bezanilla, 2018). Using long-term oryzalin treatment as a tool, we were able to image moss protonemal filaments growing in microfluidic devices for up to 5 d with 12.5 µM oryzalin present in the growth medium. Even with multiple changes in growth direction, ROP consistently associated with the growing tip (Figures 3B and 3C; Supplemental Movies 2 and 3).
Figure 3.
ROP4-swmng Enrichment Anticipates Cell Expansion, And Depends on the Cytoskeleton in Short Apical Cells.
(A) Time-lapse imaging of a ROP4-swmNG/wild-type apical protonemal cell growing and pausing. Yellow arrows indicate growth direction; yellow dashed line indicates pause in growth. Bar for all images = 5 µm. Also see Supplemental Movie 1.
(B) and (C) Time-lapse acquisitions of ROP4-swmNG/wild-type protonemal cells treated with 12.5 µM oryzalin. Images were acquired in cells that grew from 1 to 120 h after drug addition. To accurately track the ROP4-swmNG signal, chloroplast autofluorescence was removed by subtracting the value of a fraction of chlorophyll autofluorescence intensity (*0.4 for [B] and *0.2 for [A] and [C]) from the mNG intensity in the maximum projections of confocal Z-stacks. Red traces were generated by TrackMate using the processed images to track the ROP4-swmNG signal at the growing tip, which mirrored the growth pattern of the cell. (B) Example of one cell switching between isotropic and polarized growth. The images at the top are the first and last time points, with the trace in red of the mNG signal for the entire time course. Four distinct events are depicted in this time course: isotropic swelling (96 to 333 min), polarized growth with small changes in direction (345 to 582 min), large change in direction (627 to 732 min), and pause in growth (771 to 912 min). The first time points for each of these events are shown on the left and are maximum projections of Z-stacks. The right panel shows the overlay of the ROP4-swmNG enrichment in the first time point (gray), with a projection of the subsequent time points (every 3 min) for the subsequent period of time indicated in the bottom right corner of each image (magenta). Bar for all images = 10 µm. Also see Supplemental Movie 2. (C) Example of one cell continuously turning (0 to 471 min) and later developing a new polarized growth site (yellow arrowheads). Blue arrowhead indicates ectopic accumulation of the ROP4-swmNG signal that later diminished and did not lead to subsequent growth. Bar for all images = 10 µm. Also see Supplemental Movie 3.
(D) Example confocal images of ROP4-swmNG/wild type, and signal intensity quantification of protonemal apical cell tips after drug treatment. Graphs were generated by measuring the signal intensity along a manually drawn line tracing the plasma membrane of the cell. Bar for all images = 3 µm. LatB, latrunculin B.
(E) Quantification of the ratio between apical-to-subapical cell length for cells that lost or maintained the apical ROP gradient after drug treatment. Protonemal cells from ROP4-swmNG/wild type were exposed to 25 µM latrunculin B (LatB) or 12.5 µM oryzalin for 5 to 60 min and were imaged using confocal microscopy. Cell lengths were measured during image acquisition. n = 12, 6, 12, and 8 for each category, respectively.
We tracked the focused ROP spot with the TrackMate plugin in Fiji (Tinevez et al., 2017) to generate a trace for ROP movement, which aligned closely with the path of cell growth (Figures 3B and 3C; Supplemental Movie 2). Interestingly, when the cell experienced a period of unfocused growth, we also observed diffuse ROP signal overlapping with the site of cell swelling (Figure 3B, 96 min, 771 min; Supplemental Movie 2). Occasionally, we observed ROP accumulation in an area of the cell that was not expanding (Figure 3C, blue arrowhead). However, in this example (Figure 3C, 375 min), the signal soon decreased, and no growth occurred at that site (Supplemental Movie 3). New polarized growth sites also often formed independently from the original growth site. In this case, as the accumulation of ROP at the old growth site diminished, the cell stopped growing at that site. Concurrently, ROP accumulated near the new growth site where cell expansion took off and a new tip emerged (Figure 3C, yellow arrowheads; Supplemental Movie 3). These data demonstrate that ROP accumulates at actively growing sites. ROP accumulated most strongly during focused tip growth, while unfocused expansion was characterized by diffuse ROP signal along the membrane. Importantly, new sites of polarized expansion always exhibited a strong ROP signal, suggesting that ROP marks the site of maximal expansion.
The Cytoskeleton Affects ROP Polarity in a Cell Cycle–Dependent Manner
As master regulators of polarity, ROP and related GTPases affect both actin and microtubule behavior in plants and other eukaryotes (Gu et al., 2003; Iden and Collard, 2008; Burkart et al., 2015; Sugiyama et al., 2019). To investigate whether ROP localization depends on actin, which is essential for polarized growth (Vidali et al., 2009), we tested whether acute treatment with latrunculin B, which disrupts the actin cytoskeleton, would affect ROP localization. We identified actively growing apical cells and infused them with 0.5% (v/v) DMSO. All actively growing cells retained ROP localization at the cell tip (n = 34 cells). By contrast, after infusion with latrunculin B, roughly 30 to 40% of the cells lost the apical ROP gradient (Figure 3D). Infusion with oryzalin, which disrupts microtubules, also resulted in a similar fraction of cells that lost the apical gradient (Figure 3D). To investigate why only a fraction of cells exhibited this behavior, we quantified the length of the apical cell and normalized it to the length of the subapical cell. Interestingly, short apical cells lost the apical ROP gradient in response to either latrunculin B or oryzalin, while long apical cells maintained this gradient (Figure 3E). Since the apical cell is the dividing cell of the protonemal filament, the length of the cell is indicative of the stage of the cell cycle. The shortest cells are newly generated cells, whereas the longest cells are soon to enter cell division. Thus, these data suggest that actin and microtubules play a role in maintaining ROP localization early in the cell cycle.
To investigate whether ROP dynamics at the cell cortex depend on the cytoskeleton, we used fluorescence recovery after photobleaching (FRAP) to examine ROP turnover in control cells and in cells that maintained the apical ROP gradient in the absence of the cytoskeleton. In the same 5- to 60-min time window after drug addition, cells that maintained normal ROP localization were photobleached at the cell tip (Supplemental Movie 4). In both the ∆rop1/2/3 (Supplemental Figure 3A) and the wild-type (Supplemental Figure 3B) backgrounds, the ROP signal readily recovered within 40 s (Supplemental Figures 3A and 3B, black lines). Treatment with oryzalin (green lines) or latrunculin B (magenta lines) did not cause substantial changes in the rate or level of fluorescence recovery. Only latrunculin B treatment in ∆rop1/2/3, but not in the wild type, slightly decreased the rate of ROP recovery. As a whole, the FRAP data indicate that ROP4-swmNG mobility is not dependent on microtubules or actin in the wild type (Supplemental Figure 3; Supplemental Movie 4).
Long-Term Growth Inhibition Mediated by Actin Depolymerization Leads to Changes in ROP4-swmNG Localization
For a population of cells, ROP accumulation at the tip was not influenced by the acute depolymerization of actin (Figures 3D and 3E). To evaluate whether ROP remained associated with a former growth site in the absence of growth over long time periods, we treated protonemal tissue with 25 µM latrunculin B in a microfluidic imaging device. After identifying apical cells that retained tip-localized ROP accumulation within the first hour of treatment, we continued to image these cells for the next 16 h. Two hours after the addition of latrunculin B, most of the cells that had retained the ROP gradient within the first hour stopped growing completely. In addition, the apical ROP enrichment dissipated in many cells after 2 h, eventually leading to a complete loss of the ROP signal. Interestingly, in a small population of cells, we observed intracellular membranous structures enriched with ROP (Supplemental Figure 4, yellow arrowheads). These membranous structures were morphologically diverse and dynamic, and their appearance seemed to correlate with the reduction of tip-localized ROP signal (Supplemental Figure 4; Supplemental Movie 5). Occasionally, we also observed smaller aggregates with ROP4-swmNG signal (Supplemental Figure 4, blue arrowheads). Similar to cells that naturally pause (Figure 3A), these data demonstrate that in cells that stop growing due to the disruption of actin, the ROP signal disappears from the tip, suggesting that there is a feedback mechanism between growth and ROP localization to help maintain ROP at the site of polarized growth.
ROP Is Essential for Developmental Patterning, Predicting the Future Branching Site during the Apical Cell Division
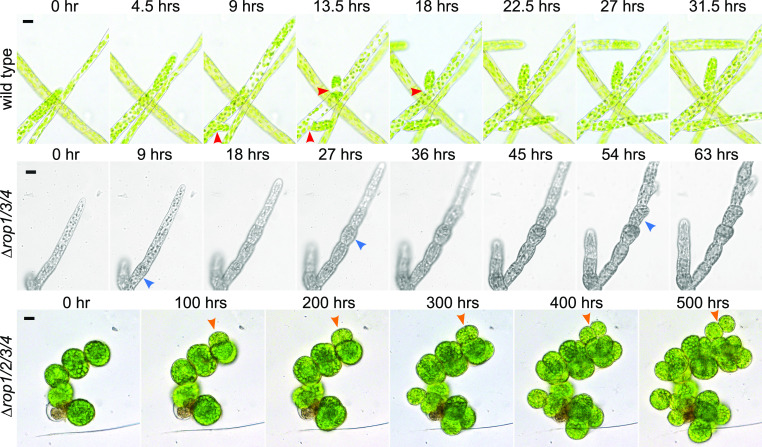
While generating ROP4-swmNG lines in the ∆rop1/2/3 background, we isolated a number of transformants containing lesions in all four ROP genes (Supplemental Figure 5). Despite their extremely slow growth rates, all ∆rop1/2/3/4 mutants could be maintained vegetatively indefinitely. Imaging growing ∆rop1/2/3/4 plants revealed that their cells grew isotropically with no obvious branching pattern (Figure 4; Supplemental Movies 6 and 7). While cell division still generated a flat cell plate between daughter cells, the combination of subsequent isotropic expansion coupled with weak cell adhesion resulted in daughter cells that eventually became loosely associated spheres (Figure 4, orange arrowheads). Furthermore, the ∆rop1/2/3/4 mutants did not form gametophores. We also isolated a triple mutant, ∆rop1/3/4 (Supplemental Figure 2), with an intact ROP2 gene that exhibited an intriguing intermediate phenotype. Apical cells grew slowly but were polarized. However, after cell division, subapical cells swelled and often divided aberrantly, resulting in protonemal filaments composed of rounded cells with few branches and elongated apical cells (Figure 4, blue arrowheads; Supplemental Movie 8), which differed substantially from the other triple mutants (Figure 1E; Yi and Goshima, 2020a) and the wild-type protonemata (Figure 4, red arrowheads; Supplemental Movie 6). In contrast to the complete null mutant, ∆rop1/3/4 formed normal gametophores (Supplemental Figure 5C; Supplemental Movie 9). Taken together, these data suggest that proper ROP levels are essential for ensuring patterning at the subcellular level and at the tissue level.
Figure 4.
ROP Levels Influence Polarized Growth and Developmental Patterning.
Bright-field images of the wild type, ∆rop1/3/4, and ∆rop1/2/3/4 from time-lapse acquisitions of growing protonemata. Red arrowheads in the wild type indicate normal branch divisions, blue arrowheads in ∆rop1/3/4 indicate abnormal cell divisions, and orange arrowheads in ∆rop1/2/3/4 indicate events of diffuse growth forming spherical cells after previous cell division. Bars for all images = 20 µm. Also see Supplemental Movies 6 to 8.
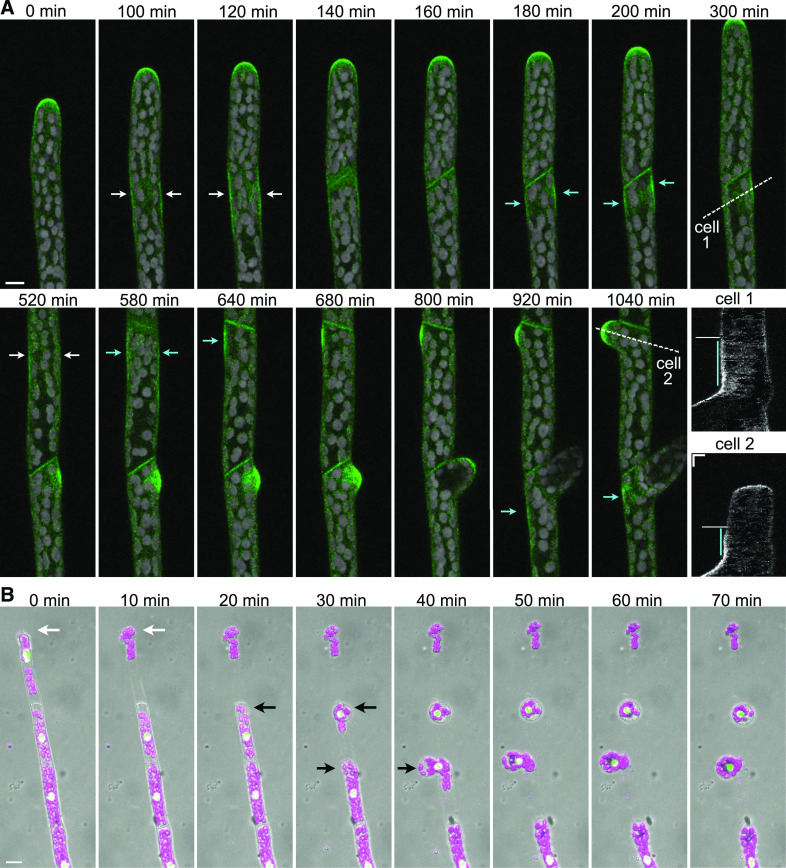
During branch formation, protonemal filaments initiate a new site of polarized cell expansion. Given that ∆rop1/2/3/4 plants did not appear to have clear branching patterns (Figure 4; Supplemental Movie 7) and that ∆rop1/3/4 plants exhibited improperly positioned cell plates in swollen subapical cells (Figure 4; Supplemental Movie 8), resulting in aberrant branch patterning, we investigated the timing of ROP localization during branch formation. ROP4-swmNG localized to the newly formed cell plate in apical cells (Figure 5A), similar to observations made with the nonfunctional N-terminal ROP4 fusion protein (Burkart et al., 2015; Yi and Goshima, 2020a). However, in addition, we discovered that ROP actually accumulates at the cell cortex in the middle of the apical cell 34 ± 10 min (mean ± sd, n = 18 cells) before it is observed in the developing cell plate (Figure 5A, white arrows; Supplemental Movie 10). Interestingly, this timing is consistent with the onset of mitosis. ROP remained at the cell cortex for 307 ± 77 min (mean ± sd, n = 9 cells), with one side accumulating more ROP signal than the other. The cortex with the strongest ROP signal initiated tip growth and a new branch emerged (Figure 5A, cyan arrows, cyan lines in kymograph; Supplemental Movie 10). ROP was also found at the cell cortex across from the branch site marking the site of emergence of a future second branch (Figure 5A, cyan arrows). These data demonstrate that ROP predicts the site of branch formation long before cell expansion occurs at the site.
Figure 5.
ROP4-swmNG Predicts the Site of Branch Formation 5 h before Branches Emerge.
(A) Maximum intensity projections of confocal Z-stacks of a growing protonemal filament in ROP4-swmNG. Bar for all images = 10 µm. Also see Supplemental Movie 10. Green, ROP4-swmNG signal; gray, chlorophyll autofluorescence signal. To reduce the interference from chlorophyll autofluorescence in the green channel, we subtracted autofluorescence collected in the 680 nm channel from the green channel. The merged image was created using the green channel after subtraction and the original 680 nm channel. White arrows indicate ROP4-swmNG cortical enrichment at the onset of mitosis near the middle of a dividing cell in both cell 1 and cell 2. Cyan arrows indicate persistent ROP4-swmNG cortical enrichment at the first branch site and the future second branch site. The dotted white lines in cell 1 and cell 2 were used to generate the kymographs labeled cell 1 and cell 2. The cyan line in the kymographs indicates the time period that ROP localized to the cell cortex before branch initiation; the white horizontal line indicates the time point of the apical cell division. Bars for kymographs = 5 µm (x) and 2 h (y).
(B) The wild-type protonemal filaments expressing a nucleus-localized GFP:GUS fusion protein 5 min after infusion of Driselase into the PDMS imaging device. Images are maximum projections of confocal fluorescence and differential interference contrast (DIC) Z-stacks. Green, GFP signal; magenta, chlorophyll autofluorescence. DIC is shown in gray to indicate the cell outlines. White arrow indicates the apical cell. Black arrows indicate the position where the protoplasts emerge in subapical cells. Bar for all images = 20 µm. Also see Supplemental Movie 11.
Finally, we wondered whether ROP localization is associated with cell wall patterning prior to expansion. We reasoned that areas with the weakest cell wall would be the most sensitive to enzymatic digestion. Thus, we treated growing protonemal filaments in a microfluidic imaging device with Driselase, a cell wall–digesting enzyme mix used to generate moss protoplasts, allowing us to image where protoplasts emerge from the protonemal filament. As expected, protoplasts extruded from the tip of the apical cell where the cell wall is the weakest (Figure 5B, white arrows; Supplemental Movie 11). Surprisingly, 47% of the time (n = 15 cells), protoplasts were expelled from subapical cells with no obvious branch protrusions (Figure 5B, black arrows; Supplemental Movie 11) at the site where we had observed ROP accumulation during branching. By contrast, 53% of the time, the protoplast emerged from the apical cell plate or a position on the side of the filament more basal to the expected future branching site or did not emerge at all. Various factors could contribute to the inability of every protoplast to emerge from a future branch site. The weak cell wall might be positioned facing the cover slip or the top of the imaging chamber, preventing the protoplast from escaping. Alternatively, during normal protonemal development, perhaps not all subapical cells are programmed to branch, and thus weakening of the wall may not have occurred. Nevertheless, the finding that the site of protoplast emergence occurred where cortical ROP accumulated during branching suggests that ROP activity in these cortical domains might modify the cell wall long before cell expansion initiates. These data, together with the observation of ROP at normal and ectopic sites of expansion (Figures 3A and 3C), demonstrate that ROP is recruited to growth sites early on and likely participates in activating cell wall remodeling, ultimately promoting polarized cell expansion.
DISCUSSION
GTPases from the Rho/RAC/CDC42 family are relatively small and have many effectors and regulators that bind to different surfaces of these protein (Berken and Wittinghofer, 2008; Hodge and Ridley, 2016; Bascom et al., 2019). As such, it has been challenging to develop live-cell imaging tools that do not interfere with GTPase function. In seed plants, functional studies have further been complicated due to the presence of multiple redundant small GTPase family members. In contrast to seed plants, moss only requires a single ROP protein to drive growth during the juvenile stages of development (Burkart et al., 2015), providing a system to test the functions of ROP fusion proteins. Here, we showed that the most commonly used fusion protein, an N-terminally tagged ROP, is not functional, as it does not support growth when it is the only ROP present. However, inserting mNG into a loop downstream of Gly-134 in ROP4 resulted in a fully functional protein, as plants containing only the tagged ROP4 grew normally and developed juvenile tissues indistinguishable from those of the wild type.
With this powerful tool in hand, it was possible to probe functional ROP dynamics and localization during growth and development. Here, we found that ROP localizes to the apical plasma membrane of tip-growing protonemata. The highest signal was detected at the tip, and it sharply declined moving away from the tip. During growth, the ROP signal was persistent, with no obvious fluctuations at the cell tip. While the N-terminal ROP fusion protein exhibited a similar localization pattern (Figure 2; Burkart et al., 2015; Yi and Goshima, 2020a), it was quantitatively different, with a shallower tip gradient and populating a larger area of the cell apex. These results suggest that the functional sandwich-tagged fusion protein preferentially associates in a more restricted manner with the plasma membrane of actively growing cells.
Here, we found that ROP localization to the cell apex depends on actin and microtubules in cells that have just completed cell division. However, in cells that are closer to cell division, the acute loss of either microtubules or actin (5 to 60 min) does not affect ROP localization at the plasma membrane (Figure 3). Additionally, our FRAP studies showed that, similar to findings in root hairs (Molendijk et al., 2001), which are fully differentiated, the turnover of ROP at the plasma membrane is largely independent of the cytoskeleton. These results support the finding that in yeast, rather than the cytoskeleton, differences in the diffusion rates of the active and inactive forms of G-protein are critical for concentrating active G-proteins (Bendezú et al., 2015; Woods et al., 2016). If actin depolymerization induces a stress response in moss, similar to what occurs in yeasts (Harrison et al., 2001), perhaps the stress response is cell cycle dependent. Early on in the cell cycle, the stress response is robust, ensuring that ROP is removed. However, in a cell that has passed a threshold size or a particular phase in the cell cycle and is committed to divide, ROP polarity is maintained so that the cell continues to grow until it divides. With sustained actin depolymerization, stress responses may result in changes in gene expression that alter potential cell fates, leading to the removal of ROP from the apical plasma membrane (Supplemental Figure 4).
In contrast to long-term actin depolymerization, long-term treatment with microtubule-depolymerizing drugs does not completely inhibit growth. In cells with reduced levels of cytoplasmic microtubules, the persistent apical actin focus that predicts the site of cell expansion behaves erratically, disassembling and assembling in random spots throughout the cell. Wherever this actin spot is, cell expansion occurs (Wu and Bezanilla, 2018). This assay provides a mechanism to observe ectopic sites of cell expansion. Similar to cells that naturally pause and resume growth, we found that ROP localizes to actively expanding regions where maximal cell expansion occurs. This behavior likely reflects a conserved function of ROP, since it occurs in diverse tip-growing cells, from the protonemal stem cell in mosses to fully differentiated pollen tubes in angiosperms (Luo et al., 2017). When an ectopic site emerges, ROP is found at that site. Often when ROP localization is diffuse, the area of expansion is larger, but as the expansion focuses and forms a tip, ROP localization also focuses, demonstrating that membrane domains containing ROP strongly correlate with active growth sites (Figure 3). A reduction in cytoplasmic microtubule levels disrupts growth directionality, but the actual growth machinery, which depends on actin, is not inhibited.
During branch formation, a physiologically relevant growth initiation process, we found that ROP marks the future expansion site several hours before tip growth occurs (Figure 5A; Supplemental Movie 10). In fact, based on this timing, ROP is recruited to the cell cortex in the apical cell just prior to mitosis, implying that branch patterning is established one to two cell cycles before branch emergence. During division of the apical cell, ROP moves from the cell cortex to the phragmoplast. After division is complete, ROP again is enriched at the cortex adjacent to the new cell plate, but only on the subapical cell side. Cell wall remodeling likely occurs here, as we discovered that the cell wall is frequently weakest at these sites (Figure 5B; Supplemental Movie 11). These findings suggest that ROP is likely required to activate effectors that remodel the wall before tip growth can occur. This mechanism is similar to what occurs at the tips of growing pollen tubes (Luo et al., 2017). However, in contrast to pollen tubes, which are terminally differentiated cells that predominately respond to external signals from the ovule, the recruitment of ROP in protonemata during apical cell division provides a temporal and spatial mechanism linking branch patterning to the cell cycle and ensuring that branches emerge proximal to a cell plate. Correct branch patterning guarantees the appropriate developmental organization and is thus important for the spreading out of moss protonemata in the environment. Future work will focus on identifying how ROP is recruited before mitosis in the apical cell and what effectors ROP activates to remodel the cell wall.
The functional ROP fluorescent fusion protein developed in this study could be used to analyze ROP localization and dynamics throughout development in a variety of plant species. Unlike CRIB4-GFP (Li et al., 2018), this functional fluorescent fusion protein does not necessarily identify active ROP populations. However, by using the ROP fluorescent fusion protein coupled with functional fluorescent guanine exchange factor fusion proteins, it may be possible to image ROP activation domains on the plasma membrane. By analyzing ROP dynamics with the functional sandwich fusion protein in a variety of plant systems and comparing this to studies in animals and yeast, it may be possible to identify evolutionarily conserved mechanisms that regulate small GTPase localization and function.
METHODS
Plasmid Construction and Genotyping
To insert sequences into the genome using CRISPR-Cas9–mediated HDR, two plasmids were generally used: one plasmid contained the Cas9 and the protospacer expression cassettes for introducing double-stranded breaks at the desired genomic locations, and the other plasmid contained the sequence to be inserted and homology regions on either side of that sequence. For N-terminal tagging and for sandwich tagging, protospacers targeting the ROP4 genomic region near the desired insertion site (ATG for the N-terminal tag, and the codon for Gly-134 for the sandwich tag) were incorporated into pMH-ROP4-N and pMH-ROP4-sw as described by Mallett et al. (2019). To generate the homology plasmid for the N-terminal tag of ROP4, primers DC334-337 (Supplemental Table 1) were used to amplify 800 to 1000 bp of genomic sequence up- and downstream of the protospacer target site (5′ and 3′ homology arms). The two homology arms were cloned into pDONR vectors and recombined with mEGFP or 3XmEGFP (Vidali et al., 2009) into a plasmid backbone derived from pGEM-T Easy (Promega), using a three-way recombination reaction (Invitrogen) as described by Mallett et al. (2019). To generate the homology plasmid for the sandwich tag, primers DC773-778 were designed to amplify 5′ and 3′ homology arms. A fragment of the mNG CDS was amplified by DC763,764. For the sandwich tag, we incorporated a linker for CDC42 as described previously by Bendezú et al. (2015). The homology plasmid was assembled by fusing the 5′ homology arm, mNG, and the 3′ homology arm into a plasmid backbone derived from pGEM-T Easy), using NEBuilder HiFi DNA Assembly kit (New England Biolabs). Because the protospacer-targeted sequence was present in the homology plasmid, we introduced a point mutation at the third nucleotide of the protospacer adjacent motif sequence in the homology arm using primers DC771,772.
To generate null mutations in ROP1, ROP2, and ROP3, we constructed a pMH-Cas9-gate plasmid with three protospacers targeting the three genes specifically, according to Mallett et al. (2019). The three protospacers were synthesized as complimentary oligos and annealed to each other. They were ligated into three different entry clones and recombined into pMH-Cas9 using a three-way recombination reaction (Invitrogen) to create pMH-∆ROP123. A plasmid with two protospacers targeting a single ROP1 at two different positions (pMH-ROP1-ps23) was constructed in a similar manner.
Primers DC421-486 were used to genotype N-terminally tagged ROP4. Genotyping of ROP4-swmNG was done using primers DC634,635,767,768. For ROP knockout plants, genotyping was done using competition PCR (Harayama and Riezman, 2017) with primers DC1195-1434 and DC1185,1186. We were able to isolate transformed plants in which all three loci had incorporated the oligo sequences by HDR at very low frequency. However, at high frequency, we isolated mutants with one or two loci edited. To generate more mutants, we either transformed pMH-ROP123 again into single or double mutants or pMH-ROP1-ps23 into lines with the ROP1 and ROP2 loci mutated and selected plants with a deletion between the two protospacers.
Moss Culture, Transformation, Protoplast Regeneration, and RNAi Assays
Physcomitrium (Physcomitrella) patens lines were propagated by homogenization with a tissue homogenizer (Omni International) and cultured on PpNH4 medium (1.03 mM MgSO4, 1.86 mM KH2PO4, 3.3 mM Ca(NO3)2, 2.72 mM (NH4)2-tartrate, 45 μM FeSO4, 9.93 μM H3BO3, 220 nM CuSO4, 1.966 μM MnCl2, 231 nM CoCl2, 191 nM ZnSO4, 169 nM KI, and 103 nM Na2MoO4) supplied with 0.7% (w/v) agar. After homogenization, 5 to 7-d-old tissue was used for protoplast transformation.
For CRISPR-Cas9–mediated HDR, 7.5 µg of the homology plasmid and 7.5 µg of the Cas9/protospacer plasmid were cotransformed into moss protoplasts as previously described (Liu and Vidali, 2011). To generate mutations in the ROP genes, we transformed 10 to 15 µg of pMH-∆ROP123 plasmid and also incorporated 5 μL of each of the three 50- to 60-bp double-stranded homology oligos (Supplemental Table 1) at 50 mM concentration. These oligos were homologous to the region adjacent to the protospacer, with a few mismatches deleting the protospacer adjacent motif sequence and inserting an in-frame stop codon, adapting the methods described in Yi and Goshima (2020b). For transformation with pMH-ROP1-ps23, 15 µg of plasmid was used. To generate plants expressing Ubi:NLS-GFP-GUS, 60 µg of linearized pTK-Ubi:NLS-GFP-GUS (generated according to Wu and Bezanilla, 2014) plasmid was transformed in the wild type and selected with 20 µg/mL G418. Plants with stable integration of the G418 antibiotic resistance and exhibiting clear nuclear-localized GFP were selected.
Transformed protoplasts were plated with liquid plating medium made of PpNH4 supplemented with 8.5% (w/v) mannitol and 10 mM CaCl2 onto cellophane-covered PRM-B plates (PpNH4 medium plus 6% [w/v] mannitol and 10 mM CaCl2). After a 4-d regeneration period, the plants were transferred to PpNH4 plates by moving the entire piece of cellophane. For growth assays, normal PpNH4 was used, and for transient RNAi silencing and stable genomic editing, PpNH4 plates supplemented with 15 µg/mL hygromycin were used. For transient RNAi silencing transformations, plants were imaged 7 d after transformation and discarded. For genomic editing stable transformations, plants were kept on PpNH4 with 15 µg/mL hygromycin selection plates for a week and transferred to normal PpNH4 plates to allow more tissue to be grown for genotyping.
RNA silencing experiments using the ROP4-∆3′UTR reporter line with a nuclear localized GFP:GUS (NLS-GFP:GUS) in its genome were performed according to Bezanilla et al. (2003) and Burkart et al. (2015). Silencing constructs were the same as described by Burkart et al. (2015). N-Terminal and sandwich-tagged ROP4 were stably transformed and incorporated at the ROP4 locus into the ROP4-∆3′UTR/NLS-GFP reporter background to create the tagged plants. Together with the untagged line as a control, 5- to 7-d-old ground tissue was used to generate protoplasts, which were transformed with RNA silencing constructs. The control construct targets the nuclear localization signal (NLS)-GFP transcript, the CDS-RNAi construct targets all four ROP transcripts within their CDSs, and the UTR-RNAi construct targets all 4 ROP transcripts at their 3′ UTRs, but ROP4 is insensitive to this construct, as its 3′ UTR sequences were removed using homologous recombination. Seven days after transformation, plants regenerated from single protoplasts were imaged. Plants lacking NLS-GFP:GUS signal were successfully silenced and were imaged by capturing the chloroplast autofluorescence. A Nikon SMZ25 stereomicroscope with a filter cube (excitation 480/40, dichroic 510, emission 510 long pass) and a color camera (Nikon digital sight DS-Fi2) was used.
To quantify the size and polarity of ROP null mutant plants, 5- to 7-d-old ground tissue was used to generated protoplasts. Plants were regenerated from protoplasts on PRM-B plates for 4 d, followed by PPNH4 plates for 3 d. Seven days after generating protoplasts, individual plants were stained with 0.1 mg/mL calcofluor staining solution on a microscope slide covered with a cover slip and imaged under a Nikon SMZ25 stereomicroscope with a violet filter cube (excitation 420/25, dichroic 455, emission 460 long pass). Images of either chloroplast autofluorescence (red channel) or calcofluor fluorescence (where the combined signal from the red/green/blue channels in the RGB image was converted to a single red channel with Fiji software) were used to analyze plant size and polarity according to Vidali et al. (2007). In brief, images of single plants were manually cropped and highlighted by thresholding the fluorescent signal with Fiji software (Schindelin et al., 2012). Total plant area and circumference were determined from the thresholded images to calculate area and solidity.
Bright-Field Microscopy
To observe plant growth, we loaded plant tissue into a poly(dimethylsiloxane) (PDMS) microfluidic imaging device attached to a glass-bottomed dish (Bascom et al., 2016) filled with Hoagland’s liquid medium. Wild-type and ∆rop1/3/4 plants were homogenized before loading as described by Bascom et al. (2016). However, because ∆rop1/2/3/4 cells were not tightly adhered to one another, no homogenization was required before loading. Time-lapse images were acquired every 10, 20, or 30 min. At each time point, multiple positions in the imaging field were imaged. For ∆rop1/2/3/4, a five-image Z-stack of slices 4 µm apart was captured at each position. For ∆rop1/3/4 gametophore, a three-image Z-stack of slices 2 µm apart was captured. A single image for each position was captured for all other tissue. Extended depth of focus images were created for each Z-stack using Nikon NIS elements. Bright-field illumination light was kept on during the entire acquisition process to provide light for plant growth. Mono-color images were acquired with a DS-Qi2 camera (Nikon). Colored images were acquired with a DS-Vi1 or digital sight DS-Fi2 camera (Nikon).
Confocal Microscopy
For confocal imaging, moss tissue was homogenized and loaded into PDMS devices. After 3 to 5 d of recovery in constant light, actively growing tip cells on newly formed filaments were identified for imaging. The PDMS devices were mounted on a Nikon A1R laser-scanning confocal microscope with a 1.3 numerical aperture (NA) 40× or 1.49 NA 60× oil immersion objective (Nikon). Laser illumination at 488 nm was used for mNG/GFP and chlorophyll autofluorescence excitation. Emission filters were 525/50 nm for mNG/GFP. Chlorophyll autofluorescence was collected at 680 nm after passing through a long-pass dichroic mirror, allowing wavelengths larger than 640 nm to pass through. Time-lapse, Z-stack, and multipoint image acquisition was controlled by NIS-Elements software (Nikon). For long-term imaging of branch initiation events, PDMS devices containing tissue with established actively growing apical cells were placed in far red light for 3 to 4 d prior to imaging. Far red light inhibits branching, but upon exposure to white light, newly growing apical cells branch very regularly, greatly increasing the success rate of capturing branch formation events. Image acquisition occurred at room temperature. White light from the light-emitting diode bright-field light source was kept on for plant growth between image acquisitions.
Cell Wall Digestion and Drug Treatments
Tissue was cultured in PDMS imaging devices for at least 4 d to allow actively growing protonemal tissue to become established. For cell wall digestion, immediately before imaging on a laser-scanning confocal microscope, 0.5% (w/v) Driselase (Sigma-Aldrich) in 8.5% (w/v) mannitol solution was injected into the PDMS device. For pharmacological treatments, Hoagland’s medium was supplemented with 12.5 µM oryzalin or 25 µM latrunculin B and injected into the PDMS device prior to imaging. Within 5 to 10 min after drug injection, apical cells and their corresponding subapical cell lengths were measured using the segmented line measurement tool in NIS-Elements. Meanwhile, Z-stacks of each apical cell were acquired to determine changes in ROP localization.
To measure ROP signal intensity, the single slice image that best represented the medial focal plane was selected for each cell with Fiji software (Schindelin et al., 2012). A region of interest (ROI) was cropped for each image to isolate the same area at the tip of each cell. A segmented line was then manually drawn tracing the plasma membrane at the tip. Intensity values for each pixel along the segmented line were measured. To measure the peak width and slope in Figure 2, individual images were normalized with the enhance contrast function in Fiji with 0.1% saturated pixels before measuring the value along the line. To determine whether the ROP signal was maintained after drug treatment in Figure 3, we measured the image intensity along the manually drawn trace (without normalization). All values along this line were normalized to the largest intensity value along the segmented line and plotted against segmented line length to generate the graph shown in Figures 2 and 3. “Loss” or “maintain” ROP tip gradient was determined manually based the shape of the curve in the graph of each cell. The long-term time-lapse videos were acquired beginning at 1 h after drug injection. Tracking of ROP tip accumulation in the oryzalin-induced curvy cells was done using the TrackMate plugin in Fiji (Tinevez et al., 2017), with a spot diameter of 15 µm.
FRAP Assay
Actively growing apical cells were identified in a PDMS device using a Nikon A1R laser-scanning confocal microscope with a 1.49 NA 60× oil immersion objective. A circular stimulation ROI with a diameter of 2.4 µm was placed at the apical plasma membrane where the ROP signal was the strongest. The 408 nm laser at 50% power was used for stimulation. Before stimulation, images were taken every 1 s for 5 s, followed by stimulation for 1 s, and the imaging continued after stimulation every 1 s for a total of 1.5 min. The mean fluorescence intensity of the ROP-swmNG signal was measured within the stimulation ROI. For each cell, the mean intensity measurements for the first five time points before stimulation were averaged to generate the reference intensity, and the mean intensity value of every time point in the whole movie was divided by the reference intensity value to create a normalized mean intensity. Graphs were generated by averaging the normalized mean intensity of each cells and plotted versus time. For photobleaching during drug treatments, cells that maintained normal ROP tip localization within 1 h of drug injection were identified and photobleached.
Statistical Analysis
All statistical analyses were performed using KaleidaGraph software (Synergy Software). To compare plant sizes and solidity of 7-d-old plants regenerated from protoplasts (Figure 1), all analyses were done using one-way ANOVA with post hoc Tukey honestly significant difference (HSD) test. P values smaller than 0.05 in the Tukey HSD test were reported as significantly different groups (Supplemental Tables 2 to 5). To compare the quantification of the tip gradient signal peak width and slope (Figure 2), Student’s t test for unpaired data with equal variance was used, and P-values smaller than 0.05 were determined as significant (Supplemental Tables 6 and 7).
Accession Numbers
Sequence data from this article can be found in Phytozome under the following accession numbers: ROP1 (Pp3c14_4310), ROP2 (Pp3c2_20700), ROP3 (Pp3c1_21550), and ROP4 (Pp3c10_4950).
Supplemental Data
Supplemental Figure 1. N-terminal and sandwich-tagging strategies.
Supplemental Figure 2. Genomic edits in ROP mutants.
Supplemental Figure 3. FRAP analysis of ROP4-swmNG.
Supplemental Figure 4. Long-term latrunculin treatment reveals aberrant ROP localization.
Supplemental Figure 5. Genomic edits in the ROP4 locus in ∆rop1/2/3/4 mutants.
Supplemental Table 1. Primers used in this study.
Supplemental Table 2. One-way ANOVA for Figure 1C.
Supplemental Table 3. One-way ANOVA for Figure 1D.
Supplemental Table 4. One-way ANOVA for Figure 1F.
Supplemental Table 5. One-way ANOVA for Figure 1G.
Supplemental Table 6. Student t Test for unpaired data with equal variance for Figure 2C.
Supplemental Table 7. Student t Test for unpaired data with equal variance for Figure 2D.
Supplemental Movie 1. ROP localization in a growing apical cell.
Supplemental Movie 2. ROP localization is focused during tip growth but diffuse during cell swelling in oryzalin-treated protonemata.
Supplemental Movie 3. ROP localization correlates with the direction of growth and newly formed growth sites in oryzalin-treated protonemata.
Supplemental Movie 4. Fluorescence recovery after photobleaching of ROP4-swmNG at the cell apex.
Supplemental Movie 5. Formation of ROP-enriched membrane structures in growth-inhibited cells exposed to long-term latrunculin B treatment.
Supplemental Movie 6. Tip growth and branching cell division in wild-type protonemata.
Supplemental Movie 7. Abnormal growth and development in ∆rop1/2/3/4.
Supplemental Movie 8. Abnormal branching and cell division pattern in ∆rop1/3/4.
Supplemental Movie 9. Normal gametophore development in wild type and ∆rop1/3/4.
Supplemental Movie 10. ROP4-swmNG localizes to the future branch site hours before a branch emerges.
Supplemental Movie 11. ROP4-swmNG localization associates with areas of weakened cell walls.
DIVE Curated Terms
The following phenotypic, genotypic, and functional terms are of significance to the work described in this paper:
Acknowledgments
We thank the Barlowe lab at Dartmouth College for providing the DNA template for mNG and Sarah Mischel for helping to genotype ∆rop/1/2/3/4. We thank members of the Bezanilla lab for careful reading of the article. This work was supported the National Science Foundation (grant 1715785 to M.B.), Dartmouth College, and the Dartmouth College Women in Science Program (support to S.R.C.S.).
AUTHOR CONTRIBUTIONS
X.C. and M.B. designed the research. X.C., B.W.M., and S.R.C.S. performed the research. X.C. and M.B. wrote the article.
Footnotes
Articles can be viewed without a subscription.
References
- Bascom C. Jr., Burkart G.M., Mallett D.R., O’Sullivan J.E., Tomaszewski A.J., Walsh K., Bezanilla M.(2019). Systematic survey of the function of ROP regulators and effectors during tip growth in the moss Physcomitrella patens. J. Exp. Bot. 70: 447–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bascom C.S. Jr., Wu S.-Z., Nelson K., Oakey J., Bezanilla M.(2016). Long-term growth of moss in microfluidic devices enables subcellular studies in development. Plant Physiol. 172: 28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezú F.O., Vincenzetti V., Vavylonis D., Wyss R., Vogel H., Martin S.G.(2015). Spontaneous Cdc42 polarization independent of GDI-mediated extraction and actin-based trafficking. PLoS Biol. 13: e1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken A., Wittinghofer A.(2008). Structure and function of Rho-type molecular switches in plants. Plant Physiol. Biochem. 46: 380–393. [DOI] [PubMed] [Google Scholar]
- Bezanilla M., Pan A., Quatrano R.S.(2003). RNA interference in the moss Physcomitrella patens. Plant Physiol. 133: 470–474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blilou I., Xu J., Wildwater M., Willemsen V., Paponov I., Friml J., Heidstra R., Aida M., Palme K., Scheres B.(2005). The PIN auxin efflux facilitator network controls growth and patterning in Arabidopsis roots. Nature 433: 39–44. [DOI] [PubMed] [Google Scholar]
- Bloch D., Lavy M., Efrat Y., Efroni I., Bracha-Drori K., Abu-Abied M., Sadot E., Yalovsky S.(2005). Ectopic expression of an activated RAC in Arabidopsis disrupts membrane cycling. Mol. Biol. Cell 16: 1913–1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloch D., Yalovsky S.(2013). Cell polarity signaling. Curr. Opin. Plant Biol. 16: 734–742. [DOI] [PubMed] [Google Scholar]
- Burkart G.M. (2014). The roles of myosin XI and ROP in moss tip growth. PhD dissertation (Amherst, MA: University of Massachusetts).
- Burkart G.M., Baskin T.I., Bezanilla M.(2015). A family of ROP proteins that suppresses actin dynamics, and is essential for polarized growth and cell adhesion. J. Cell Sci. 128: 2553–2564. [DOI] [PubMed] [Google Scholar]
- Chen W., Gong P., Guo J., Li H., Li R., Xing W., Yang Z., Guan Y.(2018). Glycolysis regulates pollen tube polarity via Rho GTPase signaling. PLoS Genet. 14: e1007373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collonnier C., Epert A., Mara K., Maclot F., Guyon-Debast A., Charlot F., White C., Schaefer D.G., Nogué F.(2017). CRISPR-Cas9-mediated efficient directed mutagenesis and RAD51-dependent and RAD51-independent gene targeting in the moss Physcomitrella patens. Plant Biotechnol. J. 15: 122–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craddock C., Lavagi I., Yang Z.(2012). New insights into Rho signaling from plant ROP/Rac GTPases. Trends Cell Biol. 22: 492–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dovas A., Couchman J.R.(2005). RhoGDI: Multiple functions in the regulation of Rho family GTPase activities. Biochem. J. 390: 1–9.16083425 [Google Scholar]
- Eklund D.M., Svensson E.M., Kost B.(2010). Physcomitrella patens: A model to investigate the role of RAC/ROP GTPase signalling in tip growth. J. Exp. Bot. 61: 1917–1937. [DOI] [PubMed] [Google Scholar]
- Etienne-Manneville S., Hall A.(2002). Rho GTPases in cell biology. Nature 420: 629–635. [DOI] [PubMed] [Google Scholar]
- Feiguelman G., Fu Y., Yalovsky S.(2018). ROP GTPases structure-function and signaling pathways. Plant Physiol. 176: 57–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foucart C., Jauneau A., Gion J.-M., Amelot N., Martinez Y., Panegos P., Grima-Pettenati J., Sivadon P.(2009). Overexpression of EgROP1, a Eucalyptus vascular-expressed Rac-like small GTPase, affects secondary xylem formation in Arabidopsis thaliana. New Phytol. 183: 1014–1029. [DOI] [PubMed] [Google Scholar]
- Fu Y., Gu Y., Zheng Z., Wasteneys G., Yang Z.(2005). Arabidopsis interdigitating cell growth requires two antagonistic pathways with opposing action on cell morphogenesis. Cell 120: 687–700. [DOI] [PubMed] [Google Scholar]
- Fu Y., Li H., Yang Z.(2002). The ROP2 GTPase controls the formation of cortical fine F-actin and the early phase of directional cell expansion during Arabidopsis organogenesis. Plant Cell 14: 777–794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y., Wu G., Yang Z.(2001). Rop GTPase-dependent dynamics of tip-localized F-actin controls tip growth in pollen tubes. J. Cell Biol. 152: 1019–1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geyer M., Wittinghofer A.(1997). GEFs, GAPs, GDIs and effectors: Taking a closer (3D) look at the regulation of Ras-related GTP-binding proteins. Curr. Opin. Struct. Biol. 7: 786–792. [DOI] [PubMed] [Google Scholar]
- Gilroy S., Jones D.L.(2000). Through form to function: Root hair development and nutrient uptake. Trends Plant Sci. 5: 56–60. [DOI] [PubMed] [Google Scholar]
- Gu Y., Li S., Lord E.M., Yang Z.(2006). Members of a novel class of Arabidopsis Rho guanine nucleotide exchange factors control Rho GTPase-dependent polar growth. Plant Cell 18: 366–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Y., Vernoud V., Fu Y., Yang Z.(2003). ROP GTPase regulation of pollen tube growth through the dynamics of tip-localized F-actin. J. Exp. Bot. 54: 93–101. [DOI] [PubMed] [Google Scholar]
- Hall A.(2012). Rho family GTPases. Biochem. Soc. Trans. 40: 1378–1382. [DOI] [PubMed] [Google Scholar]
- Harayama T., Riezman H.(2017). Detection of genome-edited mutant clones by a simple competition-based PCR method. PLoS One 12: e0179165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison J.C., Bardes E.S.G., Ohya Y., Lew D.J.(2001). A role for the Pkc1p/Mpk1p kinase cascade in the morphogenesis checkpoint. Nat. Cell Biol. 3: 417–420. [DOI] [PubMed] [Google Scholar]
- Hodge R.G., Ridley A.J.(2016). Regulating Rho GTPases and their regulators. Nat. Rev. Mol. Cell Biol. 17: 496–510. [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Cerione R.A.(2000). Flipping the switch: The structural basis for signaling through the CRIB motif. Cell 102: 403–406. [DOI] [PubMed] [Google Scholar]
- Hoffman G.R., Nassar N., Cerione R.A.(2000). Structure of the Rho family GTP-binding protein Cdc42 in complex with the multifunctional regulator RhoGDI. Cell 100: 345–356. [DOI] [PubMed] [Google Scholar]
- Houbaert A., et al. (2018). POLAR-guided signalling complex assembly and localization drive asymmetric cell division. Nature 563: 574–578. [DOI] [PubMed] [Google Scholar]
- Howell A.S., Jin M., Wu C.-F., Zyla T.R., Elston T.C., Lew D.J.(2012). Negative feedback enhances robustness in the yeast polarity establishment circuit. Cell 149: 322–333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J.B., et al. (2014). ROP3 GTPase contributes to polar auxin transport and auxin responses and is important for embryogenesis and seedling growth in Arabidopsis. Plant Cell 26: 3501–3518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hwang J.-U., Gu Y., Lee Y.-J., Yang Z.(2005). Oscillatory ROP GTPase activation leads the oscillatory polarized growth of pollen tubes. Mol. Biol. Cell 16: 5385–5399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iden S., Collard J.G.(2008). Crosstalk between small GTPases and polarity proteins in cell polarization. Nat. Rev. Mol. Cell Biol. 9: 846–859. [DOI] [PubMed] [Google Scholar]
- Ito K., Ren J., Fujita T.(2014). Conserved function of Rho-related Rop/RAC GTPase signaling in regulation of cell polarity in Physcomitrella patens. Gene 544: 241–247. [DOI] [PubMed] [Google Scholar]
- Itoh R.E., Kurokawa K., Ohba Y., Yoshizaki H., Mochizuki N., Matsuda M.(2002). Activation of rac and cdc42 video imaged by fluorescent resonance energy transfer-based single-molecule probes in the membrane of living cells. Mol. Cell. Biol. 22: 6582–6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania U., Fendrych M., Friml J.(2014). Polar delivery in plants; commonalities and differences to animal epithelial cells. Open Biol. 4: 140017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ke D., Fang Q., Chen C., Zhu H., Chen T., Chang X., Yuan S., Kang H., Ma L., Hong Z., Zhang Z.(2012). The small GTPase ROP6 interacts with NFR5 and is involved in nodule formation in Lotus japonicus. Plant Physiol. 159: 131–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kost B., Lemichez E., Spielhofer P., Hong Y., Tolias K., Carpenter C., Chua N.-H.(1999). Rac homologues and compartmentalized phosphatidylinositol 4, 5-bisphosphate act in a common pathway to regulate polar pollen tube growth. J. Cell Biol. 145: 317–330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lavy M., Bracha-Drori K., Sternberg H., Yalovsky S.(2002). A cell-specific, prenylation-independent mechanism regulates targeting of type II RACs. Plant Cell 14: 2431–2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Bail A., Schulmeister S., Perroud P.-F., Ntefidou M., Rensing S.A., Kost B.(2019). Analysis of the localization of fluorescent PpROP1 and PpROP-GEF4 fusion proteins in moss protonemata based on genomic “knock-in” and estradiol-titratable expression. Front. Plant Sci. 10: 456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lei M.-J., et al. (2015). The small GTPase ROP10 of Medicago truncatula is required for both tip growth of root hairs and nod factor-induced root hair deformation. Plant Cell 27: 806–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Lin Y., Heath R.M., Zhu M.X., Yang Z.(1999). Control of pollen tube tip growth by a Rop GTPase-dependent pathway that leads to tip-localized calcium influx. Plant Cell 11: 1731–1742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Luo N., Wang W., Liu Z., Chen J., Zhao L., Tan L., Wang C., Qin Y., Li C., Xu T., Yang Z.(2018). The REN4 rheostat dynamically coordinates the apical and lateral domains of Arabidopsis pollen tubes. Nat. Commun. 9: 2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., et al. (2012). A ROP GTPase-dependent auxin signaling pathway regulates the subcellular distribution of PIN2 in Arabidopsis roots. Curr. Biol. 22: 1319–1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D., Ren H., Fu Y.(2015). ROP GTPase-mediated auxin signaling regulates pavement cell interdigitation in Arabidopsis thaliana. J. Integr. Plant Biol. 57: 31–39. [DOI] [PubMed] [Google Scholar]
- Lin Y., Wang Y., Zhu J.K., Yang Z.(1996). Localization of a Rho GTPase implies a role in tip growth and movement of the generative cell in pollen tubes. Plant Cell 8: 293–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y., Yang Z.(1997). Inhibition of pollen tube elongation by microinjected anti-Rop1Ps antibodies suggests a crucial role for Rho-type GTPases in the control of tip growth. Plant Cell 9: 1647–1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu R., Ren D., Liu Y., Deng Y., Sun B., Zhang Q., Guo X.(2011). Biosensors of DsRed as FRET partner with CFP or GFP for quantitatively imaging induced activation of Rac, Cdc42 in living cells. Mol. Imaging Biol. 13: 424–431. [DOI] [PubMed] [Google Scholar]
- Liu Y.-C., Vidali L.(2011). Efficient polyethylene glycol (PEG) mediated transformation of the moss Physcomitrella patens. J. Vis. Exp. 50: e2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Obando M., Hoffmann B., Géry C., Guyon-Debast A., Téoulé E., Rameau C., Bonhomme S., Nogué F.(2016). Simple and efficient targeting of multiple genes through CRISPR-Cas9 in Physcomitrella patens. G3 (Bethesda) 6: 3647–3653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N., Yan A., Liu G., Guo J., Rong D., Kanaoka M.M., Xiao Z., Xu G., Higashiyama T., Cui X., Yang Z.(2017). Exocytosis-coordinated mechanisms for tip growth underlie pollen tube growth guidance. Nat. Commun. 8: 1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mallett D.R., Chang M., Cheng X., Bezanilla M.(2019). Efficient and modular CRISPR-Cas9 vector system for Physcomitrella patens. Plant Direct 3: e00168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mansfield C., Newman J.L., Olsson T.S.G., Hartley M., Chan J., Coen E.(2018). Ectopic BASL reveals tissue cell polarity throughout leaf development in Arabidopsis thaliana. Curr. Biol. 28: 2638–2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miao H., Sun P., Liu J., Wang J., Xu B., Jin Z.(2018). Overexpression of a novel ROP gene from the banana (MaROP5g) confers increased salt stress tolerance. Int. J. Mol. Sci. 19: 3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molendijk A.J., Bischoff F., Rajendrakumar C.S., Friml J., Braun M., Gilroy S., Palme K.(2001). Arabidopsis thaliana Rop GTPases are localized to tips of root hairs and control polar growth. EMBO J. 20: 2779–2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagashima Y., Tsugawa S., Mochizuki A., Sasaki T., Fukuda H., Oda Y.(2018). A Rho-based reaction-diffusion system governs cell wall patterning in metaxylem vessels. Sci. Rep. 8: 11542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalbant P., Hodgson L., Kraynov V., Toutchkine A., Hahn K.M.(2004). Activation of endogenous Cdc42 visualized in living cells. Science 305: 1615–1619. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H.(2012). Initiation of cell wall pattern by a Rho- and microtubule-driven symmetry breaking. Science 337: 1333–1336. [DOI] [PubMed] [Google Scholar]
- Oda Y., Fukuda H.(2013). Spatial organization of xylem cell walls by ROP GTPases and microtubule-associated proteins. Curr. Opin. Plant Biol. 16: 743–748. [DOI] [PubMed] [Google Scholar]
- Poraty-Gavra L., Zimmermann P., Haigis S., Bednarek P., Hazak O., Stelmakh O.R., Sadot E., Schulze-Lefert P., Gruissem W., Yalovsky S.(2013). The Arabidopsis Rho of plants GTPase AtROP6 functions in developmental and pathogen response pathways. Plant Physiol. 161: 1172–1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rounds C.M., Bezanilla M.(2013). Growth mechanisms in tip-growing plant cells. Annu. Rev. Plant Biol. 64: 243–265. [DOI] [PubMed] [Google Scholar]
- Schaefer A., Höhner K., Berken A., Wittinghofer A.(2011). The unique plant RhoGAPs are dimeric and contain a CRIB motif required for affinity and specificity towards cognate small G proteins. Biopolymers 95: 420–433. [DOI] [PubMed] [Google Scholar]
- Schindelin J., et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9: 676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimamura M.(2016). Marchantia polymorpha: Taxonomy, phylogeny and morphology of a model system. Plant Cell Physiol. 57: 230–256. [DOI] [PubMed] [Google Scholar]
- Sugiyama Y., Nagashima Y., Wakazaki M., Sato M., Toyooka K., Fukuda H., Oda Y.(2019). A Rho-actin signaling pathway shapes cell wall boundaries in Arabidopsis xylem vessels. Nat. Commun. 10: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Eklund D.M., Montes-Rodriguez A., Kost B.(2015). In vivo Rac/Rop localization as well as interaction with RhoGAP and RhoGDI in tobacco pollen tubes: Analysis by low-level expression of fluorescent fusion proteins and bimolecular fluorescence complementation. Plant J. 84: 83–98. [DOI] [PubMed] [Google Scholar]
- Tinevez J.-Y., Perry N., Schindelin J., Hoopes G.M., Reynolds G.D., Laplantine E., Bednarek S.Y., Shorte S.L., Eliceiri K.W.(2017). TrackMate: An open and extensible platform for single-particle tracking. Methods 115: 80–90. [DOI] [PubMed] [Google Scholar]
- van Dop M., Fiedler M., Mutte S., de Keijzer J., Olijslager L., Albrecht C., Liao C.-Y., Janson M.E., Bienz M., Weijers D.(2020). DIX domain polymerization drives assembly of plant cell polarity complexes. Cell 180: 427–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venus Y., Oelmüller R.(2013). Arabidopsis ROP1 and ROP6 influence germination time, root morphology, the formation of F-actin bundles, and symbiotic fungal interactions. Mol. Plant 6: 872–886. [DOI] [PubMed] [Google Scholar]
- Vetter I.R., Wittinghofer A.(2001). The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304. [DOI] [PubMed] [Google Scholar]
- Vidali L., Augustine R.C., Kleinman K.P., Bezanilla M.(2007). Profilin is essential for tip growth in the moss Physcomitrella patens. Plant Cell 19: 3705–3722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidali L., van Gisbergen P.A.C., Guérin C., Franco P., Li M., Burkart G.M., Augustine R.C., Blanchoin L., Bezanilla M.(2009). Rapid formin-mediated actin-filament elongation is essential for polarized plant cell growth. Proc. Natl. Acad. Sci. USA 106: 13341–13346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q., Li Y., Ishikawa K., Kosami K.I., Uno K., Nagawa S., Tan L., Du J., Shimamoto K., Kawano Y.(2018). Resistance protein Pit interacts with the GEF OsSPK1 to activate OsRac1 and trigger rice immunity. Proc. Natl. Acad. Sci. USA 115: E11551–E11560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson L.J., Rossi G., Brennwald P.(2014). Quantitative analysis of membrane trafficking in regulation of Cdc42 polarity. Traffic 15: 1330–1343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedlich-Soldner R., Altschuler S., Wu L., Li R.(2003). Spontaneous cell polarization through actomyosin-based delivery of the Cdc42 GTPase. Science 299: 1231–1235. [DOI] [PubMed] [Google Scholar]
- Wong H.L., et al. (2018). In vivo monitoring of plant small GTPase activation using a Förster resonance energy transfer biosensor. Plant Methods 14: 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods B., Lai H., Wu C.-F., Zyla T.R., Savage N.S., Lew D.J.(2016). Parallel actin-independent recycling pathways polarize Cdc42 in budding yeast. Curr. Biol. 26: 2114–2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Gu Y., Li S., Yang Z.(2001). A genome-wide analysis of Arabidopsis Rop-interactive CRIB motif-containing proteins that act as Rop GTPase targets. Plant Cell 13: 2841–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G., Li H., Yang Z.(2000). Arabidopsis RopGAPs are a novel family of rho GTPase-activating proteins that require the Cdc42/Rac-interactive binding motif for rop-specific GTPase stimulation. Plant Physiol. 124: 1625–1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-Z., Bezanilla M.(2014). Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. eLife 3: e03498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S.-Z., Bezanilla M.(2018). Actin and microtubule cross talk mediates persistent polarized growth. J. Cell Biol. 217: 3531–3544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T., Wen M., Nagawa S., Fu Y., Chen J.-G., Wu M.-J., Perrot-Rechenmann C., Friml J., Jones A.M., Yang Z.(2010). Cell surface- and rho GTPase-based auxin signaling controls cellular interdigitation in Arabidopsis. Cell 143: 99–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yalovsky S.(2015). Protein lipid modifications and the regulation of ROP GTPase function. J. Exp. Bot. 66: 1617–1624. [DOI] [PubMed] [Google Scholar]
- Yalovsky S., Bloch D., Sorek N., Kost B.(2008). Regulation of membrane trafficking, cytoskeleton dynamics, and cell polarity by ROP/RAC GTPases. Plant Physiol. 147: 1527–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.(2002). Small GTPases: Versatile signaling switches in plants. Plant Cell 14 (Suppl): S375–S388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi P., Goshima G.(2020a). Rho of plants GTPases and cytoskeletal elements control nuclear positioning and asymmetric cell division during Physcomitrella patens branching. Curr. Biol. 30: 2860–2868. [DOI] [PubMed] [Google Scholar]
- Yi P., Goshima G.(2020b). Transient cotransformation of CRISPR/Cas9 and oligonucleotide templates enables efficient editing of target loci in Physcomitrella patens. Plant Biotechnol. J. 18: 599–601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Bergmann D.C., Dong J.(2016). Fine-scale dissection of the subdomains of polarity protein BASL in stomatal asymmetric cell division. J. Exp. Bot. 67: 5093–5103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Dong J.(2018). Cell polarity: Compassing cell division and differentiation in plants. Curr. Opin. Plant Biol. 45 (Pt A): 127–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Xiong Y., Liu R., Xue H.-W., Yang Z.(2019). The Rho-family GTPase OsRac1 controls rice grain size and yield by regulating cell division. Proc. Natl. Acad. Sci. USA 116: 16121–16126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu L., Fu Y.(2012). Analysis of in vivo ROP GTPase activity at the subcellular level by fluorescence resonance energy transfer microscopy In Plant Signalling Networks: Methods and Protocols, Wang Z.-Y., and Yang Z., eds (Totowa, NJ: Humana Press; ), pp. 145–152. [DOI] [PubMed] [Google Scholar]