Abstract
Ras p21 protein activator 1 (RASA1) is a regulator of Ras GDP and GTP and is involved in numerous physiological processes such as angiogenesis, cell proliferation, and apoptosis. As a result, RASA1 also contributes to pathological processes in vascular diseases and tumour formation. This review focuses on the role of RASA1 in multiple tumours types in the lung, intestines, liver, and breast. Furthermore, we discuss the potential mechanisms of RASA1 and its downstream effects through Ras/RAF/MEK/ERK or Ras/PI3K/AKT signalling. Moreover, miRNAs are capable of regulating RASA1 and could be a novel targeted treatment strategy for tumours.
Keywords: tumour, Ras p21 protein activator 1, microRNA, Ras, therapy
1. Introduction
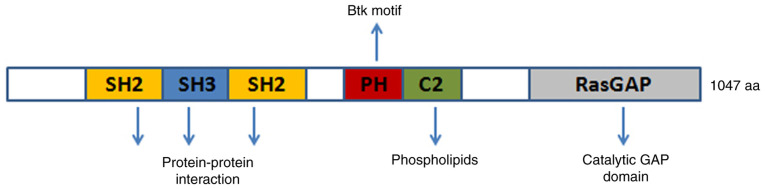
Ras p21 protein activator 1 (RASA1) is located on chromosome 5q14.3 and is a member of the RasGAP family which includes NF1, DAB2IP, and RASAL2 (1). RASA1 contains the following domains: Src homology 2 and 3 (SH2 and SH3), N-terminal C2A and C2B, GTPase-activating protein (GAP), and pleckstrin homology (PH), which is attached to a Bruton's tyrosine kinase (Btk) motif. RASA1 is a GAP with dual-specificity that enhances and accelerates the GTPase activity of Ras and Rap. Notably, intracellular Ca2+ levels regulate the GAP activity of RASA1. When Ca2+ concentrations are high, the C2 domains of Ras and Rap allow for the binding of phospholipids while the PH domain remains inactive and prevents lipid binding. RASA1 is normally located in the cytoplasm as a soluble protein and is recruited to the plasma membrane upon receptor-mediated increases in intracellular Ca2+ concentrations (2). The RasGAP activity of RASA1 is increased when RASA1 associates with the membrane since RasGAP activity is limited in the soluble form of RASA1, although the underlying mechanism is not known (3). SH2-pTyr interaction allows for RASA1 to interact with p190RhoGAP (p190RhoGAP-A, ARHGAP35), which is a GAP for Rho (4). Due to its special structure, RASA1 is involved in physiological functions such as cell growth, proliferation, differentiation, and apoptosis (Fig. 1). RASA1 mutation or epigenetic inactivation has been revealed in numerous human cancers, which has attracted interest for further investigation.
Figure 1.
RASA1 domain and potential function. The GAP domain at RASA1 C-terminus functions mainly as a catalyser for GTP. PH is connected to a Btk motif that facilitates membrane binding upon C2 interaction with the lipid. SH2 and SH3 domains at the N-terminal of RASA1 promote protein-protein interactions. RASA1, Ras p21 protein activator 1; PH, pleckstrin homology domain; Bruton's tyrosine kinase; SH2, Src homology 2; SH3, Src homology 3; GAP, GTPase-activating protein; GTP, guanosine triohosphte.
2. RASA1 involvement in physiological processes
Henkemeyer et al first reported the physiological role of RASA1 is the regulation of embryonic blood vessel development (5). Ephrin type-B receptor 4 (EPHB4), which is a member of the receptor tyrosine kinase family, is a necessary factor for RASA1 during angiogenesis and studies have revealed that EPHB4 recruitment of RASA1 is required to restore blood flow in ischemia-reflow-injury in mice (6). RASA1 has been revealed to suppress RAS phosphorylation, which prevents Ras/MEK1/2/ERK1/2 and PI3K/Akt signalling and leads to a decrease in the growth and migratory capacities of vascular smooth muscle cells (7). The microRNA (miR)-132/212 cluster directly targets RASA1 and Spred1, which modulates Ras-MAPK signalling and promotes arteriogenesis (8,9). The role of RASA1 and CDC42 in the control of endothelial cell (EC) tube network assembly has also been studied (10) and it has been revealed that RASA1 is essential for the survival of EC during developmental angiogenesis (11). Studies in human umbilic vein endothelial cells (HUVECs) have revealed that miR-4530 targeted RASA1 through ERK/MAPK and PI3K/AKT signalling and resulted in inhibition of proliferation and promotion of apoptosis and that miR-132 targeted RASA1 to promote angiogenesis in a myocardial infarction model (12,13).
The inhibition of Ras signal transduction by RASA1 has been revealed to negatively affect the maintenance of lymphatic vasculature by VEGFR-3 in a steady-state situation (14). RASA1 has also been revealed to directly interact with mitogen-activated protein kinase kinase kinase kinase 4 (MAP4K4) for lymphatic vascular development (15–17).
RASA1 is necessary for the normal development and function of T cells and a study has revealed that RASA1-deficiency in mice increased apoptosis of CD4+CD8+ double-positive thymocytes (18). Motility of macrophages and cytoskeletal and adhesion structures have been revealed to be modulated by RASA1-mediated translocation of p190RhoGAP (GRLF1) induced by BCL6 (19). RASA1 has been discovered to play a role in the development of neuron axons (20), dendritic cell differentiation (21), follicular development (22), epididymal development (23), skin wound healing (24,25), and stress (26) (Table I).
Table I.
Role of RASA1 and related factors in various physiological processes.
| Various physiological processes | RASA1 and RASA1-associated factors | (Refs.) |
|---|---|---|
| Angiogenesis | RASA1, EPHB4, miR-132/212, cdc42, miR-4530 | (5–13) |
| Lymphangiogenesis | RASA1,VEGFR-3, MAP4K4 | (14–17) |
| T cell mature | RASA1 | (18) |
| Macrophages motility | RASA1, GRLF1, BCL6 | (19) |
| Development of neuron axon | RASA1, miR-132 | (20) |
| Dendritic cells differentiation | RASA1, mmu-miR-223-3p | (21) |
| Follicular development | RASA1, miR-132 | (22) |
| Epididymal development | RASA1, miR-335 | (23) |
| Skin damage repair | RASA1, miR-132, miR-31 | (24,25) |
| Stress | RASA1, caspase-3, NF-κB | (26) |
RASA1, Ras p21 protein activator 1; EPHB4, ephrin type-B receptor 4; miR, microRNA; MAP4K4, mitogen-activated protein kinase kinase kinase kinase 4.
3. RASA1 involvement in pathological processes
Since RASA1 is a central player of angiogenesis, studies have focused on RASA1 mutation or loss-of-function in vascular diseases such as capillary malformation-arteriovenous malformation (CM-AVM) syndrome, Klippel-Trenaunay-Weber syndrome (KTWS), Sturgeon-Weber syndrome (SWS), vein of Galen aneurysmal malformation (VGAM), MEF2C-related disorders, and Parkes-Weber syndrome (PKWS). Atypical capillary malformation (CM) is a key characteristic of CM-AVM and is frequently paired with multiple arteriovenous malformations (AVMs) and at least one of the following: Rapid vascular malformation, arteriovenous fistulas (AVFs), parks Weber syndrome (PWS), and hereditary haemorrhagic telangiectasia (HHT) (27). CM usually manifests as multifocal wine spots on the skin due to inactivating mutations of RASA1 in 68% of cases (28). p120RASGAP is encoded by RASA1 and directly affects EPHB4, which acts on capillaries, leading to HHT and CM (29,30). KTWS may rarely occur in the affected limb of patients with cutaneous haemangioma, venous varicosity, and hypertrophy of osseous-soft tissue (31). Mutations in RAP1A have been associated with KTWS and interact with RASA1 and Krev interaction trapped protein 1 (KRIT1) (32,33). Patients with SWS harbour vascular malformations in the face, eyes, and brain due to a mutation in RASA1. The underlying mechanism is thought to be Nrf2-mediated oxidative stress, although the causal relationship has not been fully elucidated (34,35). VGAM encompasses a rare vascular malformation in the brain of children caused by RASA1, endoglin, and activin receptor-like kinase 1 (ACVRL1) mutations, which encode for ALK1 and SMAD4 (36,37). Patients with MEF2C-related disorders are severely intellectually impaired and have limited mobility and speech. In addition, they show hypotonia, seizures, and multiple minor anomalies of the brain. Moreover, RASA1 mutation and a decrease in MECP2 and CDKL5 expression have been correlated with this disease (38). PKWS is a rare vascular malformation syndrome with extensive capillary malformations that appear at birth or during early childhood and patients exhibit mutation of RASA1 that leads to a loss of function (39,40). ENG, ACVRL1, and SMAD4 are components of transforming growth factor-beta (TGB-β) signalling and mutations in these genes can cause HHT, which is the most commonly inherited vascular disorder. HHT has also been correlated with RASA1, BMP9, and GDF2 (32,35,41,42).
Cardiovascular and cerebrovascular diseases bring great pressure on countries with an ageing population. A previous study has revealed that RASA1 and its associated factors and vascular smooth muscle cell dysfunction through angiotensin II are central pathways involved in cardiovascular diseases. miR-132 was revealed to prevent CREB activation by angiotensin II via RASA1 (43). Myocardial fibrosis is an important process of cardiac hypertrophy and heart failure, which has been correlated to an upregulation of RASA1 via miR-21 (44,45). Obesity is a risk factor for cardiovascular disease and A study has demonstrated that PPARγ/miR-223 participates in the deposition of adipocytes by targeting RASA1 (46). Another study has linked tricuspid atresia, a congenital heart defect with fatal consequences, with the homozygous RASA1 germline mutation c.1583A>G (p.Tyr528Cys) (47). An animal model of cerebral ischemia has revealed that the GAS5/miR-335/RASA1 axis is regulated to inhibit neuron apoptosis and promote neuroprotection (48).
Chronic kidney disease is related to renal fibrosis and miR-132 has been revealed to inhibit the development of renal fibrosis by targeting RASA1, leading to reduced proliferation of myofibroblasts (49). A leading cause of blindness is age-related macular degeneration (AMD), where studies have revealed abnormal expression of RASA1 in OXYS rats (50). Myelodysplastic syndromes (MDS) encompass a group of haematopoietic stem cell disorders that are characterised by abnormal myeloid cell differentiation and development, ineffective haematopoiesis, refractory hemocytopenia, and haemopoietic failure. Notably, it has been revealed that MDS is related to RASA1 downregulation (51). In a mouse model for CCI-induced neuropathic pain, intrathecal injection of miR-144 mimics reduced the mechanical and thermal pain experienced by mice through the downregulation of RASA1 (52) (Table II).
Table II.
Role of RASA1 and related factors in various pathologic processes.
| Various pathological processes | RASA1 and RASA1-associated factors | (Refs.) |
|---|---|---|
| CM-AVM | RASA1, EPHB4 | (29,30) |
| KTWS | RASA1, KRIT1 | (31,33) |
| SWS | RASA1, Nrf2 | (34,35) |
| VGAM | RASA1, ACVRL1 | (36,37) |
| MEF2C-related disorders | RASA1, MECP2, CDKL5 | (38) |
| PKWS | RASA1 | (39,40) |
| HHT | RASA1, EPHB4, ACVRL1, GDF2 | (32,35,41,42) |
| Cardiovascular and cerebrovascular diseases | RASA1, miR-132, miR-21, miR-223, GAS5/miR-335 | (43–48) |
| Chronic renal disease | RASA1, miR-132 | (49) |
| Age-related macular degeneration | RASA1 | (50) |
| Neuropathic pain | RASA1, miR-144 | (51,52) |
RASA1, Ras p21 protein activator 1; CM-AVM, capillary malformation-arteriovenous malformation; KTWS, Klippel-Trenaunay-Weber syndrome; SWS, Sturgeon-Weber syndrome; VGAM, vein of Galen aneurysmal malformation; MEF2C-related disorders, myocyte enhancer factor 2C-related disorders; PKWS, Parkes-Weber syndrome; HHT, hereditary haemorrhagic telangiectasia; EPHB4, ephrin type-B receptor 4; KRIT, Krev interaction trapped protein 1; ACVRL1, activin receptor-like kinase 1; miR, microRNA.
4. RASA1 and cancers
Investigation into changes in genomic DNA and RNA in HL-60 cells revealed that RASA1 is related to tumorigenesis (53). The Ras-GAP SH3 domain inhibits Rho-GAP activity and inhibits tumour development (54). RASA1 has also been reported as an oncogene through large-scale tumour sequencing studies that are based on 3D mutation clusters in relation to the protein structure (55).
In tumour development, mutations of Ras at residues 12, 13, or 61 affect the activity of intracellular guanosine triohosphte (GTP), which alternates between GDP and GTP forms and activates RasGAP proteins and Ras by Ras GTPase, regulating the guanine nucleotide exchange factors (RasGEFs). The understanding of RASA1 and its relationship with tumour formation is qualitative and is limited to the abnormal expression of RASA1 and several other genes from the sequencing of certain tumours. It has been revealed that abnormal or downregulation of RASA1 expression affects tumorigenesis and the continued technological development in the field of molecular biology allows for more in-depth research, where it has been found that the expression of RASA1 in most tumour cells is associated with intracellular miRNA. Furthermore, a small portion of tumorigenesis may be due to the coupling of RASA1 to its associated protein or gene (56). Nevertheless, the role of RASA1 in tumour formation requires further study.
Lung cancer
Lung cancer incidence and deaths continue to rise and place high pressure on health care with its high incidence (2.09 million new cases in 2018 vs. 1.8 million in 2012) and mortality rate (1.76 million deaths in 2018 vs. 1.6 million in 2012) compared with other tumours (57). The occurrence of lung cancer has been linked to RASA1 mutations by next-generation sequencing (58,59). Zhu et al revealed that hsa-miR-182 downregulated RASA1 to suppress proliferation of lung squamous cell carcinoma through tissue microarray and quantitative PCR (60). Recently, Shi et al demonstrated that miR-30c and miR-21 were significantly upregulated by two KRAS isoforms in small-cell lung cancer and induced drug resistance by inhibiting key tumour suppressor genes such as neurofibromatosis type one (NF1), RASA1, BID, and RASSF8 (61). Mutations of EGFR result in drug resistance and relapse in patients with non-small cell lung cancer (NSCLC) (62). Furthermore, miR-31 directly inhibits the expression of RASA1 and FIH-1, leading to at least partial activation of Raf/MEK/ERK and PI3K/Akt signalling in gefitinib-resistant NSCLC (63). Kitajima and Barbie classified NF1 or RASA1 mutations in small-cell lung carcinoma and proposed the clinical evaluation of MAPK inhibition in an analysis of large genomic datasets of NSCLC [MSK-IMPACT dataset at MSKCC (n=2,004) and TCGA combined lung cancer dataset (n=1,144)] (64). RASA1 and NF1 mutations are strong drivers of NSCLC (65). Fibroblast growth factor receptor-2 (FGFR-2) is a tyrosine kinase receptor, which can selectively bind to fibroblast growth factor (FGF) to promote autophosphorylation and mediate cell response through downstream MAPK and Akt signalling. A previous study has revealed that FGFR-2 mutation is an important driver of lung cancer, which has become a key target of lung cancer drug development. In an FGFR2-mutant resistant model, RASA1 was found to be inactivated (66).
Colorectal cancer
The incidence of colorectal cancer (CRC) is rising and is predicted to increase to 2.5 million new cases by 2035 and is ranked third among malignant tumours across the world and second in some developing areas (67,68). Colorectal specimens including 468 colorectal tumour samples from a large personalised medicine initiative and 17 paired primary-metastatic and 2 metastatic-metastatic specimens from 18 CRC patients were analysed by next-generation sequencing to reveal the presence of RASA1 mutations (69). RASA1 expression has been linked to multiple miRNAs in colorectal cancer. In vivo, miR-31 has been revealed to be negatively correlated with RASA1 protein level and miR-31 was a key player in RAS signalling activation through the inhibition of RASA1 translation, resulting in accelerated growth of colorectal cancer cells and stimulation of tumour formation (70). In rectal cancer cells, expression of miR-21 reduced the expression level of RASA1 resulting in the promotion of cell proliferation, anti-apoptosis, and tumour cell formation (71). RASA1 encoding RAS GTPase is one of the target genes that is continuously downregulated in cells overexpressing miR-21 and upregulated in cells exposed to miR-21 inhibitors (72). These results coincide with a study by Sun et al in which upregulation of miR-223 can be detected in colon cancer cells and promotes tumour growth. Conversely, miR-223 inhibition may reduce or halt the growth of solid tumours (73). miR-335 has been revealed to inhibit the expression of the RASA1 gene by targeting a specific sequence of the 3′UTR of RASA1. Low expression of RASA1 has been revealed to promote tumour cell development and miR-335 expression was increased in patients with CRC compared with normal mucosa, whereas high expression of miR-335 was significantly associated with tumour size and CRC differentiation. miR-335 overexpression in CRC cells was revealed to promote cell proliferation and tumour growth in vivo (74). Antoine-Bertrand et al observed an interaction between p120RasGAP and cancer cells in CRC, which regulates axonal growth mediated by netrin-1 and guides the development of cortical neurons in embryos (75). KRAS mutations are found in 40% of patients with CRC and the effective treatment of CRC with late KRAS mutation is limited at present (76). RASA1 is as an effector of KRAS mutation and may play an important role as a drug treatment target (77). However, studies using CRISPR technology have described that only loss of NF1 promotes resistance to EGFR inhibition (78).
Liver cancer
Primary liver cancer has the seventh-highest incidence (0.84 million new cases in 2018 year) and second highest mortality (0.78 million deaths in 2018) worldwide (79). China has the highest prevalence owing to an increased prevalence of 18.3 per 100,000 and its population of 1.4 billion (80). Mutations of RASA1 have been found by detecting RASA1 and other members of the RasGAP family through sequencing of liver cancer tissue genes (81). The analysis of RASA1 expression and the prognosis of patients revealed that the expression of RASA1 was closely related to tumour size and differentiation and that the prognosis was poor with low RASA1 expression, indicating that RASA1 could be a predictive factor for the prognosis of patients with liver cancer (82). RASA1 is also regulated by miRNA although other potential oncogenes also contribute to the development of liver cancer. Increased miR-31 expression and low RASA1 expression have been detected in cells and tissues from liver cancer patients. Furthermore, it has been confirmed that miR-31 promotes cellular proliferation and inhibits liver cancer cells apoptosis by downregulating RASA1 expression (83). When liver cancer cells are exposed to a hypoxic environment, a significant increase in miR-182 expression occurs, which then promotes angiogenesis via RASA1 and leads to proliferation of cancer cells (84). Thus, RASA1 levels are related to liver cancer cell proliferation. In addition, RASA1 is regulated by pituitary homeobox 1 (PITX1) and protein tyrosine phosphatase 1B (PTP1B). PITX1 has a tumour suppressing function through the regulation of RasGAP expression. PITX1 is downregulated by PTP1B, which drives proliferation and inhibits apoptosis of tumour cells by acting on RASA1 (85). In cells exposed to mild stress, RASA1 is cleaved into an N-terminal fragment (fragment N) by caspase-3, which potently inhibits the activity of NF-κB by augmenting its translocation to the cytoplasm (26). Studies have revealed that liver cancer incidence is reduced by the caspase-3/p120 RasGAP stress-sensing module. However, the overall survival is not affected (26,86).
Breast cancer
Breast cancer is the second leading cause of cancer related-deaths in women (2.08 million new cases and 0.62 million deaths in 2018) and although the genotyping of breast cancers and the subsequent targeted treatment significantly improve the curative rate to more than 65%, it remains necessary to discover more tumour markers for further treatment optimisation (79,87). Some studies have shown that the low expression of RASA1 is related to the occurrence of breast cancer (88,89). This is consistent with a study that demonstrated the presence of 7 genetic mutations in breast cancer, which included RASA1 mutations (90). The expression of RASA1 is prevalent in triple-negative breast cancer (TNBC) tumours and tumour cells with low estrogenic receptor expression (91). Both docking protein 2 (Dok2) and RASA1 function as tumour suppressors and have been detected in several types of solid tumours. Our previous study revealed that a weak Dok2/RASA1 expression was associated with poorly differentiated breast adenocarcinoma. Moreover, a decrease in Dok2 and RASA1 expression was linked to larger tumour size and a heightened chance of metastasis to the axillary lymph node (92). RASA1 is also regulated by miRNA and other potential oncogenes contribute to the development of breast cancer. miR-206/21 has been revealed to promote the growth of hepatoma cells by inhibiting the translation of RASA1 and promoting Ras/Erk signal transduction and cell differentiation (92). miR-421 which targets RASA1, has been revealed to inhibit TNBC tumour growth and metastasis (93). As C2 domains of RASA1 participate in the regulation of calcium signalling, L-type voltage-gated calcium channel gamma subunit 4 (CACNG4) can promote the stability of the internal environment of breast cancer cells and improve the survival and metastasis ability of tumour cells through alternating calcium signalling events and invading key survival and metallic pathway genes including RASA1 (94).
Gynaecologic tumours
RASA1 mutation and abnormal expression is also regarded as a contributing factor to the development of gynaecologic tumours such as cervical and ovarian cancer (95,96). The incidence (98,900 new cases in 2015) of cervical cancer in China has been ranked second in the world after Chile with a clear trend towards younger age of onset (97). However, research examining the relationship between cervical cancer and RASA1 is limited. Zhang et al revealed that miR-21 was highly expressed in the serum of cervical cancer patients compared with healthy control subjects. In addition, the cervical cancer cell lines HeLa and HT-3 were used and it was revealed that miR-21 decreased the expression of RASA1, which led to increased cell proliferation and migration via Ras-induced epithelial-mesenchymal transition (95). In the development of ovarian cancer, the circular RNA circ-ITCH was revealed to extensively inhibit the viability and motility of ovarian cancer cell lines SK-OV-3 and Caov-3 and dampens tumorigenesis in xenografted NOD mice by upregulating RASA1 expression (96).
Leukaemia
Leukaemia (0.43 million new cases and 0.31 million deaths in 2018) is the most common type of cancer in children and young adults, highlighting the importance of discovering novel tumour markers for early diagnosis (79). Scans at DNA and RNA levels using microarray technology in HL-60 cells have demonstrated that RASA1 is a candidate cancer-related gene (53). Lubeck et al confirmed that a lack of RasGAP alone in T cells in RASA1 and NF1 double-deficient mice leads to the development of T cell acute lymphoblastic leukaemia/lymphoma (98). Falconi et al revealed that RASA1 was downregulated and inhibited MSC clone formation in acute myeloid leukaemia patients (51). RASA1 has been revealed to be regulated by miR-223 although the mechanism in leukaemia requires further research (99).
Oral squamous cell carcinomas (OSCCs)
OSCCs are a group of aggressive tumours known for their rapid spread to the lymph nodes and a high treatment failure rate. In addition, the incidence rate of OSCC is increasing (incidence increase ranging from 0.4 to 3.3% per year) among young people below the age of 50 (100). By sequencing the entire genome of 50 paired primary tumours of the tongue and 120 OSCC from male individuals in Taiwan, it has been revealed that RASA1 variants are related to cigarette smoking, betel nut chewing, human papillomavirus infection, and tumour stage (101,102). The development of betel quid chewing-associated tongue carcinomas has been revealed to be related to mutations in RASA1 and CpG islands (103). The expression of miR-182 in OTSCC was revealed to be significantly upregulated, and negatively correlated with RASA1 levels, suggesting that the miR-182/RASA1 axis is a potential target for the treatment of OSCC (104).
Other cancers
While pancreatic cancer is not as common as other tumours, the outcome is markedly poor (105). In pancreatic cancer, several gene mutations including RASA1 are more pronounced (106). Kent et al reported that the expression of RASA1 was decreased in pancreatic cancer cells and further study revealed that RNAi knockdown of RASA1 significantly enhanced the progression of pancreatic cancer for both Capan-1 and MiaPaCa2 cell lines consistent with the miR-31 overexpression phenotype (107).
Prostate cancer incidence is high in Europe and North America with ~0.45 million and 0.23 million new cases in 2018, respectively (108). RASA1 has been discovered to be a potential target gene for advanced drug-resistant recurrent prostate cancer by whole transcriptome sequencing (109). In a genome-wide association study that included 12,518 cases of prostate cancer, RASA1 was revealed to be associated with aggressive prostate cancer while it exhibited no association with nonaggressive prostate cancer (110).
Melanoma samples in patients with metastatic disease often exhibit a loss of RASA1 expression and low RASA1 mRNA has been linked to a decrease in overall survival in patients with BRAF-mutated melanoma (111,112).
In thyroid or gastroenteropancreatic neuroendocrine tumours, the relationship between BRAF and multiple gene mutations including RASA1 has been documented (113,114). Kidney cancer has many subtypes with a clear increasing trend in the incidence in the United States. A similar rise in incidence is also seen in Chinese men. In renal clear cell carcinoma, QKI-5-stable RASA1 mRNA has been found to directly bind to the reactive elemental region of RASA1 QKI and subsequently prevents the activation of the RAS-MAPK signalling, leading to inhibition of cell growth and inducing cell cycle arrest (115). Another study revealed that RASA1 may play a key role in the progression of RCC by decreasing miR-223-3p and subsequently increasing F-box/WD repeat-containing protein 7 (FBXW7) expression (116).
Gastric cancer (GC) is ranked third in mortality among all cancers (79). The incidence of gastric cancer in China is more pronounced in rural areas and is one of the primary malignant tumours which accounts for a marked 42.5% of worldwide GC cases and 45.0% of worldwide deaths that endanger public health (117). Mutations in RASA1 are also present in gastric cancer. Li et al revealed that low miR-335 expression accelerated the invasion and metastasis of cancer cells. Furthermore, miR-335 can be silenced by promoter hypermethylation and is heavily involved in metastatic gastric cancer through its target genes such as RASA1 (118). In oesophageal squamous cell carcinoma cells (ESCC), high expression of miR-21 was revealed to significantly downregulate the expression of RASA1 (119). Mutation or aberrant expression of RASA1 has also been revealed to be related to the development of cutaneous squamous cell carcinoma and sarcoma (120,121). RASA1 and related factors in various cancers are presented in Table III.
Table III.
RASA1 with multiple gene proteins and related cancers.
| Cancers | RASA1 associated gene or protein | (Refs.) |
|---|---|---|
| Lung cancer | RASA1, hsa-miR-182, miR-31, miR-30c, miR-21, NF1 | (58–66) |
| Colorectal cancer | RASA1, miR-31, miR-21, miR-335, miR-223, K-RAS, Netrin-1, NF1 | (69–78) |
| Liver cancer | RASA1, miR-31, miR-182, PITX1, caspase-3 | (26,81–86) |
| Breast cancer | RASA1, miR-206/21, miR-421, CACNG4 | (88–94) |
| Gynecologic tumors | RASA1, miR-21, circ-ITCH, miR-145 | (94,96) |
| Leukemia | RASA1, miR-223 | (51,53,98,99) |
| Oral tongue squamous cell carcinomas | RASA1, miR-182 | (101–104) |
| Pancreatic cancer | RASA1, miR-31 | (106,107) |
| Prostate cancer | RASA1 | (109,110) |
| Melanoma | RASA1, hsa-miR-223, hsa-miR-23b | (111,112) |
| Thyroid cancer | RASA1, BRAF | (113,114) |
| Renal clear cell carcinoma | RASA1, QKI-5, miR-223-3p | (115,116) |
| Gastric cancer | RASA1, miR-335 | (118) |
| Esophageal cancer | RASA1, miR-21 | (119) |
| Cutaneous squamous cell carcinoma | RASA1 | (120) |
| Sarcoma | RASA1, BRAF | (121) |
RASA1, Ras p21 protein activator 1; miR, microRNA; NF1, neurofibromatosis type one; PITX1, pituitary homeobox 1; CACNG4, calcium channel gamma subunit 4.
5. Potential mechanism of RASA1 and tumorigenesis
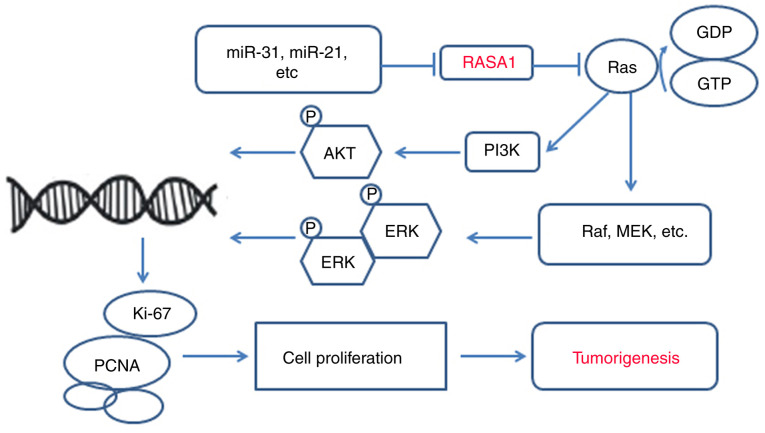
Ras can be activated by inhibiting RasGAPs and increases tumour development risk (122). In this review, we revealed that mutations or aberrant expression of RASA1 are present in almost all cancers and tumour cells and that during the development of tumours, Ras/Raf/MEK/ERK play a central role (59,78,104). This signalling pathway is connected by small GTP protein-membrane binding ligands that initiate a cascade of receptor tyrosine kinases and cytoplasmic proteins. The first protein kinase RAF in the RAS recruitment cascade is activated after GTP binding on the cell membrane through a series of complex processes including changing the phosphorylation state of the protein and binding with the skeleton protein and other kinases. The activated Raf kinase continues to phosphorylate and activate MAPKK protein kinase MEK1/2 and finally phosphorylates and activates ERK1/2, inducing abnormal proliferation, invasion, growth, and distant metastasis of malignant tumours (75,123). In addition, loss of RASA1 function can enhance RAS-ERK signal amplification (66,78). PI3K/AKT have been described as key regulators of cell proliferation and differentiation and are involved in tumour cell proliferation, invasion, and metastasis (124). Some studies have revealed that aberrant RASA1 expression activates PI3K/AKT signalling (51,63,64). Moreover, numerous miRNAs have been revealed to regulate RASA1 expression in several cancers such as lung, colorectal, hepatocellular, and triple-negative breast cancers (60,70,83,91). The miRNA/RASA1 axis is an attractive target for tumour treatment and its effectiveness has been demonstrated in numerous studies (60,70,71,83,92). miRNAs are capable of regulating RASA1 and could be a novel targeted treatment strategy for tumours. Numerous miRNAs have been revealed to be involved in different cancers, such as miR-21 in lung cancer, and thus these miRNAs should be focused on as targeted treatments (125). Various genes or proteins such as RASA1 have fundamental roles at normal levels, however an imbalance of these genes or proteins has been revealed to result in disease or tumors, therefore, targeted therapies such as oncolytic viruses can correct local discordance and treat tumors without affecting overall function (Fig. 2).
Figure 2.
The potential role of RASA1 in tumorigenesis. RASA1 encodes p120RasGAP, which inhibits signal transduction by activating Ras p21, and intrinsic GTPase inhibits Ras-GTP. miRs inhibit RASA1 and it subsequently loses its ability to regulate Ras, leading to activation of the MAPK/MEK/ERK or PI3K/AKT pathways involved in abnormal cell proliferation. RASA1, Ras p21 protein activator 1; miR, microRNA.
6. Conclusion
Although RASA1 primarily participates in angiogenesis and vascular-related diseases, whole-genome sequencing confirmed the importance of RASA1 in tumorigenesis, invasion, metastasis, and drug resistance (120). RASA1 was first reported in blood vessel development or angiogensis and its other functions including antitumor have been gradually revealed, RASA1 play the similar role in physiological process such as angiogenesis, vascular-related diseases or cancers, only the outcome are not same. Normal or balanced expression levels of RASA1 promote normal blood vessel development; aberrant or imbalanced RASA1 expression levels result vascular-related diseases such as CM-AVM or KTWS (40,126). RASA1 has more than one mechanism of action in the development of cancers. In some cancers, the role of RASA1 is more profound and precise (127). From these pathways, we can conduct genetic screening or testing, including RASA1 testing in serum, before the cancer fully develops to achieve early detection and prevention of cancer. Furthermore, these findings may lead to the development of novel drugs that halt cancer progression by blocking the pathways it uses for growth and dissemination. In addition, RASA1 also serves as a good reference for the prognosis of cancer after surgery and treatment. Therefore, for some cancers with a less clear mechanism of action, the aberrant expression of RASA1 may be used to advance cancer treatment strategies.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- RASA1
Ras p21 protein activator 1
- CM-AVM
capillary malformation-arteriovenous malformation
- KTWS
Klippel-Trenaunay-Weber syndrome
- SWS
Sturgeon-Weber syndrome
- VGAM
vein of Galen aneurysmal malformation
- MEF2C-related disorders
myocyte enhancer factor 2C-related disorders
- PKWS
Parkes-Weber syndrome
- HHT
hereditary haemorrhagic telangiectasia
- CM
capillary malformation
- MAPK
mitogen-activated protein kinase
- HUVECs
human umbilic vein endothelial cells
- PI3K
phosphatidylinositol 3-kinase
- PKB
protein kinase B
- EPHB4
ephrin type-B receptor 4
- FGF
fibroblast growth factor
- Dok2
docking protein 2
- MAP4K4
mitogen-activated protein kinase kinase kinase kinase 4
- Ang II
angiotensin II
- Btk
Bruton's tyrosine kinase
- CRC
colorectal cancer
- KRIT1
Krev interaction trapped protein 1
- TGF-β
transforming growth factor-β
- AMD
age-related macular degeneration
- MDS
myelodysplastic syndromes
- NF1
neurofibromatosis type one
- FGFR-2
fibroblast growth factor receptor-2
- PITX1
pituitary homeobox 1
- CACNG4
calcium channel gamma subunit 4
- BQ-TCs
betel quid chewing-associated tongue carcinomas
- GC
gastric cancer
- ESCC
oesophageal squamous cell carcinoma cells
Funding
The present study was supported by the Nature Science Foundation of Hubei Province (grant no. 2017CFB786), the Hubei Province Health and Family Planning Scientific Research Project (grant no. WJ2016Y10), the Jingzhou Science and Technology Bureau Project (grant no. 2017-93), and the National innovation and entrepreneurship training program for College Students (grant no. 202010489017). All funding was provided to XP.
Availability of data and materials
Data sharing is not applicable to this article, as no datasets were generated or analyzed during the present study.
Authors' contributions
XP designed and supervised the study. YZ, YL, BS, HX and YS reviewed the references. YZ, YL and QW performed the research. YZ, XP and JC wrote the manuscript. YZ, PS and RL contributed to tables and figures. XP and JC acquired funding.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing of interests
The authors declare that they have no competing interests.
References
- 1.Harrell Stewart DR, Clark GJ. Pumping the brakes on RAS-negative regulators and death effectors of RAS. J Cell Sci. 2020;133:jcs238865. doi: 10.1242/jcs.238865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dai Y, Walker SA, de Vet E, Cook S, Welch HC, Lockyer PJ. Ca2+-dependent monomer and dimer formation switches CAPRI Protein between Ras GTPase-activating protein (GAP) and RapGAP activities. J Biol Chem. 2011;286:19905–19916. doi: 10.1074/jbc.M110.201301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sot B, Behrmann E, Raunser S, Wittinghofer A. Ras GTPase activating (RasGAP) activity of the dual specificity GAP protein Rasal requires colocalization and C2 domain binding to lipid membranes. Proc Natl Acad Sci USA. 2013;110:111–116. doi: 10.1073/pnas.1201658110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jaber Chehayeb R, Stiegler AL, Boggon TJ. Crystal structures of p120RasGAP N-terminal SH2 domain in its apo form and in complex with a p190RhoGAP phosphotyrosine peptide. PLoS One. 2019;14:e0226113. doi: 10.1371/journal.pone.0226113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Henkemeyer M, Rossi DJ, Holmyard DP, Puri MC, Mbamalu G, Harpal K, Shih TS, Jacks T, Pawson T. Vascular system defects and neuronal apoptosis in mice lacking ras GTPase-activating protein. Nature. 1995;377:695–701. doi: 10.1038/377695a0. [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki J, Aegerter S, Fevurly RD, Mammoto A, Mammoto T, Sahin M, Mably JD, Fishman SJ, Chan J. RASA1 functions in EPHB4 signaling pathway to suppress endothelial mTORC1 activity. J Clin Invest. 2014;124:2774–2784. doi: 10.1172/JCI67084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu X, Wang Z, Wu H, Jiang W, Hu R. Ras ssDNA aptamer inhibits vascular smooth muscle cell proliferation and migration through MAPK and PI3K pathways. Int J Mol Med. 2015;35:1355–1361. doi: 10.3892/ijmm.2015.2139. [DOI] [PubMed] [Google Scholar]
- 8.Lei Z, van Mil A, Brandt MM, Grundmann S, Hoefer I, Smits M, El Azzouzi H, Fukao T, Cheng C, Doevendans PA, et al. MicroRNA-132/212 family enhances arteriogenesis after hindlimb ischaemia through modulation of the Ras-MAPK pathway. J Cell Mol Med. 2015;19:1994–2005. doi: 10.1111/jcmm.12586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Anand S, Majeti BK, Acevedo LM, Murphy EA, Mukthavaram R, Scheppke L, Huang M, Shields DJ, Lindquist JN, Lapinski PE, et al. MicroRNA-132-mediated loss of p120RasGAP activates the endothelium to facilitate pathological angiogenesis. Nat Med. 2010;16:909–914. doi: 10.1038/nm.2186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Norden PR, Kim DJ, Barry DM, Cleaver OB, Davis GE. Cdc42 and k-ras control endothelial tubulogenesis through apical membrane and cytoskeletal polarization: Novel stimulatory roles for GTPase effectors, the small GTPases, Rac2 and Rap1b, and inhibitory influence of Arhgap31 and Rasa1. PLoS One. 2016;11:e0147758. doi: 10.1371/journal.pone.0147758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen D, Teng JM, North PE, Lapinski PE, King PD. RASA1-dependent cellular export of collagen IV controls blood and lymphatic vascular development. J Clin Invest. 2019;130:3545–3561. doi: 10.1172/JCI124917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jing L, Li H, Zhang T, Lu J, Zhong L. MicroRNA-4530 suppresses cell proliferation and induces apoptosis by targeting RASA1 in human umbilical vein endothelial cells. Mol Med Rep. 2019;19:3393–3402. doi: 10.3892/mmr.2019.10000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma T, Chen Y, Chen Y, Meng Q, Sun J, Shao L, Yu Y, Huang H, Hu Y, Yang Z, et al. MicroRNA-132, delivered by mesenchymal stem cell-derived exosomes, promote angiogenesis in myocardial infarction. Stem Cells Int. 2018;2018:3290372. doi: 10.1155/2018/3290372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lapinski PE, Kwon S, Lubeck BA, Wilkinson JE, Srinivasan RS, Sevick-Muraca E, King PD. RASA1 maintains the lymphatic vasculature in a quiescent functional state in mice. J Clin Invest. 2012;122:733–747. doi: 10.1172/JCI46116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roth Flach RJ, Guo CA, Danai LV, Yawe JC, Gujja S, Edwards YJ, Czech MP. Endothelial Mitogen-activated protein kinase kinase kinase kinase 4 is critical for lymphatic vascular development and function. Mol Cell Biol. 2016;36:1740–1749. doi: 10.1128/MCB.01121-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lapinski PE, Lubeck BA, Chen D, Doosti A, Zawieja SD, Davis MJ, King PD. RASA1 regulates the function of lymphatic vessel valves in mice. J Clin Invest. 2017;127:2569–2585. doi: 10.1172/JCI89607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Castorena-Gonzalez JA, Srinivasan RS, King PD, Simon AM, Davis MJ. Simplified method to quantify valve Back-leak uncovers severe mesenteric lymphatic valve dysfunction in mice deficient in connexins 43 and 37. J Physiol. 2020;598:2297–23102. doi: 10.1113/JP279472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lapinski PE, Qiao Y, Chang CH, King PD. A role for p120 RasGAP in thymocyte positive selection and survival of naive T cells. J Immunol. 2011;187:151–163. doi: 10.4049/jimmunol.1100178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pixley FJ, Xiong Y, Yu RY, Sahai EA, Stanley ER, Ye BH. BCL6 suppresses RhoA activity to alter macrophage morphology and motility. J Cell Sci. 2005;118:1873–1883. doi: 10.1242/jcs.02314. [DOI] [PubMed] [Google Scholar]
- 20.Hancock ML, Preitner N, Quan J, Flanagan JG. MicroRNA-132 is enriched in developing axons, locally regulates Rasa1 mRNA, and promotes axon extension. J Neurosci. 2014;34:66–78. doi: 10.1523/JNEUROSCI.3371-13.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bros M, Youns M, Kollek V, Buchmüller D, Bollmann F, Seo EJ, Schupp J, Montermann E, Usanova S, Kleinert H, et al. Differentially tolerized mouse antigen presenting cells share a common miRNA signature including enhanced mmu-miR-223-3p expression which is sufficient to imprint a protolerogenic state. Front Pharmacol. 2018;9:915. doi: 10.3389/fphar.2018.00915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schauer SN, Sontakke SD, Watson ED, Esteves CL, Donadeu FX. Involvement of miRNAs in equine follicle development. Reproduction. 2013;146:273–282. doi: 10.1530/REP-13-0107. [DOI] [PubMed] [Google Scholar]
- 23.Wang J, Ruan K. miR-335 is involved in the rat epididymal development by targeting the mRNA of RASA1. Biochem Biophys Res Commun. 2010;402:222–227. doi: 10.1016/j.bbrc.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 24.Li X, Li D, Wikstrom JD, Pivarcsi A, Sonkoly E, Ståhle M, Landén NX. MicroRNA-132 promotes fibroblast migration via regulating RAS p21 protein activator 1 in skin wound healing. Sci Rep. 2017;7:7797. doi: 10.1038/s41598-017-07513-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shi J, Ma X, Su Y, Song Y, Tian Y, Yuan S, Zhang X, Yang D, Zhang H, Shuai J, et al. MiR-31 mediates inflammatory signaling to promote re-epithelialization during skin wound healing. J Invest Dermatol. 2018;138:2253–2263. doi: 10.1016/j.jid.2018.03.1521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khalil H, Loukili N, Regamey A, Cuesta-Marban A, Santori E, Huber M, Widmann C. The caspase-3-p120-RasGAP module generates a NF-κB repressor in response to cellular stress. J Cell Sci. 2015;128:3502–3513. doi: 10.1242/jcs.174409. [DOI] [PubMed] [Google Scholar]
- 27.Wooderchak-Donahue WL, Johnson P, McDonald J, Blei F, Berenstein A, Sorscher M, Mayer J, Scheuerle AE, Lewis T, Grimmer JF, et al. Expanding the clinical and molecular findings in RASA1 capillary malformation-arteriovenous malformation. Eur J Hum Genet. 2018;26:1521–1536. doi: 10.1038/s41431-018-0196-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Revencu N, Boon LM, Mendola A, Cordisco MR, Dubois J, Clapuyt P, Hammer F, Amor DJ, Irvine AD, Baselga E, et al. RASA1 mutations and associated phenotypes in 68 families with capillary malformation-arteriovenous malformation. Hum Mutat. 2013;34:1632–1641. doi: 10.1002/humu.22431. [DOI] [PubMed] [Google Scholar]
- 29.Yu J, Streicher JL, Medne L, Krantz ID, Yan AC. EPHB4 Mutation implicated in capillary malformation-arteriovenous malformation syndrome: A case report. Pediatr Dermatol. 2017;34:e227–e230. doi: 10.1111/pde.13208. [DOI] [PubMed] [Google Scholar]
- 30.Amyere M, Revencu N, Helaers R, Pairet E, Baselga E, Cordisco M, Chung W, Dubois J, Lacour JP, Martorell L, et al. Germline Loss-of-function mutations in EPHB4 cause a second form of capillary Malformation-arteriovenous malformation (CM-AVM2) Deregulating RAS-MAPK signaling. Circulation. 2017;136:1037–1048. doi: 10.1161/CIRCULATIONAHA.116.026886. [DOI] [PubMed] [Google Scholar]
- 31.Pérez-Alfayate R, Martínez-Moreno N, Rosati SD, Moreu-Gamazo M, Pérez-García C, Martínez-Alvarez R. Klippel-Trenaunay-Weber syndrome associated with multiple cerebral arteriovenous malformations: Usefulness of Gamma Knife stereotactic radiosurgery in this syndrome. World Neurosurg. 2020;141:425–429. doi: 10.1016/j.wneu.2020.06.012. [DOI] [PubMed] [Google Scholar]
- 32.Boutarbouch M, Ben Salem D, Giré L, Giroud M, Béjot Y, Ricolfi F. Multiple cerebral and spinal cord cavernomas in Klippel-Trenaunay-Weber syndrome. J Clin Neurosci. 2010;17:1073–1075. doi: 10.1016/j.jocn.2009.11.013. [DOI] [PubMed] [Google Scholar]
- 33.Karadag A, Senoglu M, Sayhan S, Okromelidze L, Middlebrooks EH. Klippel-Trenaunay-Weber syndrome with atypical presentation of cerebral cavernous angioma: A case report and literature review. World Neurosurg. 2019;126:354–358. doi: 10.1016/j.wneu.2019.03.132. [DOI] [PubMed] [Google Scholar]
- 34.Zhou Q, Zheng JW, Yang XJ, Wang HJ, Ma D, Qin ZP. Detection of RASA1 mutations in patients with sporadic Sturge-Weber syndrome. Childs Nerv Syst. 2011;27:603–607. doi: 10.1007/s00381-010-1258-z. [DOI] [PubMed] [Google Scholar]
- 35.Kadam SD, Gucek M, Cole RN, Watkins PA, Comi AM. Cell proliferation and oxidative stress pathways are modified in fibroblasts from Sturge-Weber syndrome patients. Arch Dermatol Res. 2012;304:229–235. doi: 10.1007/s00403-012-1210-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chida A, Shintani M, Wakamatsu H, Tsutsumi Y, Iizuka Y, Kawaguchi N, Furutani Y, Inai K, Nonoyama S, Nakanishi T. ACVRL1 gene variant in a patient with vein of Galen aneurysmal malformation. J Pediatr Genet. 2013;2:181–189. doi: 10.3233/PGE-13067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Komiyama M, Miyatake S, Terada A, Ishiguro T, Ichiba H, Matsumoto N. Vein of Galen aneurysmal malformation in monozygotic Twin. World Neurosurg. 2016;91:672.e11–e15. doi: 10.1016/j.wneu.2016.04.031. [DOI] [PubMed] [Google Scholar]
- 38.Zweier M, Rauch A. The MEF2C-related and 5q14.3q15 microdeletion syndrome. Mol Syndromol. 2012;2:164–170. doi: 10.1159/000337496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Burrows PE, Gonzalez-Garay ML, Rasmussen JC, Aldrich MB, Guilliod R, Maus EA, Fife CE, Kwon S, Lapinski PE, King PD, Sevick-Muraca EM. Lymphatic abnormalities are associated with RASA1 gene mutations in mouse and man. Proc Natl Acad Sci USA. 2013;110:8621–8626. doi: 10.1073/pnas.1222722110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Revencu N, Boon LM, Dompmartin A, Rieu P, Busch WL, Dubois J, Forzano F, van Hagen JM, Halbach S, Kuechler A, et al. Germline mutations in RASA1 are not found in patients with Klippel-trenaunay syndrome or capillary malformation with limb overgrowth. Mol Syndromol. 2013;4:173–178. doi: 10.1159/000349919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wooderchak-Donahue WL, McDonald J, O'Fallon B, Upton PD, Li W, Roman BL, Young S, Plant P, Fülöp GT, Langa C, et al. BMP9 mutations cause a Vascular-anomaly syndrome with phenotypic overlap with hereditary hemorrhagic telangiectasia. Am J Hum Genet. 2013;93:530–537. doi: 10.1016/j.ajhg.2013.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hernandez F, Huether R, Carter L, Johnston T, Thompson J, Gossage JR, Chao E, Elliott AM. Mutations in RASA1 and GDF2 identified in patients with clinical features of hereditary hemorrhagic telangiectasia. Hum Genome Var. 2015;2:15040. doi: 10.1038/hgv.2015.40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jin W, Reddy MA, Chen Z, Putta S, Lanting L, Kato M, Park JT, Chandra M, Wang C, Tangirala RK, Natarajan R. Small RNA sequencing reveals microRNAs that modulate angiotensin II effects in vascular smooth muscle cells. J Biol Chem. 2012;287:15672–15683. doi: 10.1074/jbc.M111.322669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Diao X, Shen E, Wang X, Hu B. Differentially expressed microRNAs and their target genes in the hearts of streptozotocin-induced diabetic mice. Mol Med Rep. 2011;4:633–640. doi: 10.3892/mmr.2011.489. [DOI] [PubMed] [Google Scholar]
- 45.Queirós AM, Eschen C, Fliegner D, Kararigas G, Dworatzek E, Westphal C, Sanchez Ruderisch H, Regitz-Zagrosek V. Sex- and estrogen-dependent regulation of a miRNA network in the healthy and hypertrophied heart. Int J Cardiol. 2013;169:331–338. doi: 10.1016/j.ijcard.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 46.Ying W, Tseng A, Chang RC, Morin A, Brehm T, Triff K, Nair V, Zhuang G, Song H, Kanameni S, et al. MicroRNA-223 is a crucial mediator of PPARγ-regulated alternative macrophage activation. J Clin Invest. 2015;125:4149–4159. doi: 10.1172/JCI81656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nozari A, Aghaei-Moghadam E, Zeinaloo A, Alavi A, Ghasemi Firouzabdi S, Minaee S, Eskandari Hesari M, Behjati F. A pathogenic homozygous mutation in the pleckstrin homology domain of RASA1 is responsible for familial tricuspid atresia in an Iranian consanguineous family. Cell J. 2019;21:70–77. doi: 10.22074/cellj.2019.5734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dai X, Yi M, Wang D, Chen Y, Xu X. Changqin NO. 1 inhibits neuronal apoptosis via suppressing GAS5 expression in a traumatic brain injury mice model. Biol Chem. 2019;400:753–763. doi: 10.1515/hsz-2018-0340. [DOI] [PubMed] [Google Scholar]
- 49.Bijkerk R, de Bruin RG, van Solingen C, van Gils JM, Duijs JM, van der Veer EP, Rabelink TJ, Humphreys BD, van Zonneveld AJ. Silencing of microRNA-132 reduces renal fibrosis by selectively inhibiting myofibroblast proliferation. Kidney Int. 2016;89:1268–1280. doi: 10.1016/j.kint.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 50.Telegina DV, Korbolina EE, Ershov NI, Kolosova NG, Kozhevnikova OS. Identification of functional networks associated with cell death in the retina of OXYS rats during the development of retinopathy. Cell Cycle. 2015;14:3544–3556. doi: 10.1080/15384101.2015.1080399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Falconi G, Fabiani E, Fianchi L, Criscuolo M, Raffaelli CS, Bellesi S, Hohaus S, Voso MT, D'Alò F, Leone G. Impairment of PI3K/AKT and WNT/β-catenin pathways in bone marrow mesenchymal stem cells isolated from patients with myelodysplastic syndromes. Exp Hematol. 2016;44:75–83.e1-e14. doi: 10.1016/j.exphem.2015.10.005. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X, Guo H, Xie A, Liao O, Ju F, Zhou Y. MicroRNA-144 relieves chronic constriction injury-induced neuropathic pain via targeting RASA1. Biotechnol Appl Biochem. 2020;67:294–302. doi: 10.1002/bab.1854. [DOI] [PubMed] [Google Scholar]
- 53.Ulger C, Toruner GA, Alkan M, Mohammed M, Damani S, Kang J, Galante A, Aviv H, Soteropoulos P, Tolias PP, et al. Comprehensive genome-wide comparison of DNA and RNA level scan using microarray technology for identification of candidate Cancer-related genes in the HL-60 cell line. Cancer Genet Cytogenet. 2003;147:28–35. doi: 10.1016/S0165-4608(03)00155-9. [DOI] [PubMed] [Google Scholar]
- 54.Yang XY, Guan M, Vigil D, Der CJ, Lowy DR, Popescu NC. p120Ras-GAP binds the DLC1 Rho-GAP tumor suppressor protein and inhibits its RhoA GTPase and growth-suppressing activities. Oncogene. 2009;28:1401–1409. doi: 10.1038/onc.2008.498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kamburov A, Lawrence MS, Polak P, Leshchiner I, Lage K, Golub TR, Lander ES, Getz G. Comprehensive assessment of cancer missense mutation clustering in protein structures. Proc Natl Acad Sci USA. 2015;112:E5486–E5495. doi: 10.1073/pnas.1516373112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li L, Fan Y, Huang X, Luo J, Zhong L, Shu XS, Lu L, Xiang T, Chan ATC, Yeo W, et al. Tumor suppression of Ras GTPase-activating protein RASA5 through antagonizing Ras signaling perturbation in carcinomas. Science. 2019;21:1–18. doi: 10.1016/j.isci.2019.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Bade BC, Dela Cruz CS. Lung cancer 2020: Epidemiology, Etiology, and prevention. Clin Chest Med. 2020;41:1–24. doi: 10.1016/j.ccm.2019.10.001. [DOI] [PubMed] [Google Scholar]
- 58.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, Mansukhani M, Koul S, Halmos B, Borczuk AC. Next-generation sequencing of pulmonary sarcomatoid carcinoma reveals high frequency of actionable MET gene mutations. J Clin Oncol. 2016;34:794–802. doi: 10.1200/JCO.2015.62.0674. [DOI] [PubMed] [Google Scholar]
- 59.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, et al. Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat Genet. 2016;48:607–616. doi: 10.1038/ng.3564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu YJ, Xu B, Xia W. Hsa-mir-182 downregulates RASA1 and suppresses lung squamous cell carcinoma cell proliferation. Clin Lab. 2014;60:155–159. doi: 10.7754/clin.lab.2013.121131. [DOI] [PubMed] [Google Scholar]
- 61.Shi L, Middleton J, Jeon YJ, Magee P, Veneziano D, Laganà A, Leong HS, Sahoo S5, Fassan M, Booton R, et al. KRAS induces lung tumorigenesis through microRNAs modulation. Cell Death Dis. 2018;9:219. doi: 10.1038/s41419-017-0243-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sequist LV, Han JY, Ahn MJ, Cho BC, Yu H, Kim SW, Yang JC, Lee JS, Su WC, Kowalski D, et al. Osimertinib plus savolitinib in patients with EGFR mutation-positive, MET-amplified, Non-small-cell lung cancer after progression on EGFR tyrosine kinase inhibitors: Interim results from a multicentre, open-label, phase 1b study. Lancet Oncol. 2020;21:373–386. doi: 10.1016/S1470-2045(19)30785-5. [DOI] [PubMed] [Google Scholar]
- 63.He J, Jin S, Zhang W, Wu D, Li J, Xu J, Gao W. Long Non-coding RNA LOC554202 promotes acquired gefitinib resistance in Non-small cell lung cancer through upregulating miR-31 expression. J Cancer. 2019;10:6003–6013. doi: 10.7150/jca.35097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hayashi T, Desmeules P, Smith RS, Drilon A, Somwar R, Ladanyi M. RASA1 and NF1 are preferentially co-mutated and define a distinct genetic subset of smoking-associated non-small cell lung carcinomas sensitive to MEK inhibition. Clin Cancer Res. 2018;24:1436–1447. doi: 10.1158/1078-0432.CCR-17-2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kitajima S, Barbie DA. RASA1/NF1-Mutant lung cancer: Racing to the Clinic? Clin Cancer Res. 2018;24:1243–1245. doi: 10.1158/1078-0432.CCR-17-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kas SM, de Ruiter JR, Schipper K, Schut E, Bombardelli L, Wientjens E, Drenth AP, de Korte-Grimmerink R, Mahakena S, Phillips C, et al. Transcriptomics and transposon mutagenesis identify multiple mechanisms of resistance to the FGFR Inhibitor AZD4547. Cancer Res. 2018;78:5668–5679. doi: 10.1158/0008-5472.CAN-18-0757. [DOI] [PubMed] [Google Scholar]
- 67.Crockett SD, Nagtegaal I. Terminology, molecular features, epidemiology, and management of serrated colorectal neoplasia. Gastroenterology. 2019;157:949–966. doi: 10.1053/j.gastro.2019.06.041. [DOI] [PubMed] [Google Scholar]
- 68.Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394:1467–1480. doi: 10.1016/S0140-6736(19)32319-0. [DOI] [PubMed] [Google Scholar]
- 69.Kim R, Schell MJ, Teer JK, Greenawalt DM, Yang M, Yeatman TJ. Co-evolution of somatic variation in primary and metastatic colorectal cancer may expand biopsy indications in the molecular era. PLoS One. 2015;10:e0126670. doi: 10.1371/journal.pone.0126670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sun D, Yu F, Ma Y, Zhao R, Chen X, Zhu J, Zhang CY, Chen J, Zhang J. MicroRNA-31 activates the RAS pathway and functions as an oncogenic MicroRNA in human colorectal cancer by repressing RAS p21 GTPase activating protein 1 (RASA1) J Biol Chem. 2013;288:9508–9518. doi: 10.1074/jbc.M112.367763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gong B, Liu WW, Nie WJ, Li DF, Xie ZJ, Liu C, Liu YH, Mei P, Li ZJ. MiR-21 RASA1 axis affects malignancy of colon cancer cells via RAS pathways. World J Gastroenterol. 2015;21:1488–1497. doi: 10.3748/wjg.v21.i5.1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yang Y, Weng W, Peng J, Hong L, Yang L, Toiyama Y, Gao R, Liu M, Yin M, Pan C, et al. Fusobacterium nucleatum increases proliferation of colorectal cancer cells and tumor development in mice by activating Toll-like receptor 4 signaling to nuclear factor-κB, and Up-regulating expression of MicroRNA-21. Gastroenterology. 2017;152:851–866.e24. doi: 10.1053/j.gastro.2016.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sun D, Wang C, Long S, Ma Y, Guo Y, Huang Z, Chen X, Zhang C, Chen J, Zhang J. C/EBP-β-activated microRNA-223 promotes tumour growth through targeting RASA1 in human colorectal cancer. Br J Cancer. 2015;112:1491–1500. doi: 10.1038/bjc.2015.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lu Y, Yang H, Yuan L, Liu G, Zhang C, Hong M, Liu Y, Zhou M, Chen F, Li X. Overexpression of miR-335 confers cell proliferation and tumour growth to colorectal carcinoma cells. Mol Cell Biochem. 2016;412:235–245. doi: 10.1007/s11010-015-2630-9. [DOI] [PubMed] [Google Scholar]
- 75.Antoine-Bertrand J, Duquette PM, Alchini R, Kennedy TE, Fournier AE, Lamarche-Vane N. p120RasGAP protein mediates Netrin-1 protein-induced cortical axon outgrowth and guidance. J Biol Chem. 2016;291:4589–4602. doi: 10.1074/jbc.M115.674846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yarom N, Gresham G, Boame N, Jonker D. KRAS status as a predictor of chemotherapy activity in patients with metastatic colorectal cancer. Clin Colorectal Cancer. 2019;18:e309–e315. doi: 10.1016/j.clcc.2019.05.004. [DOI] [PubMed] [Google Scholar]
- 77.Organ SL, Hai J, Radulovich N, Marshall CB, Leung L, Sasazuki T, Shirasawa S, Zhu CQ, Navab R, Ikura M, et al. p120RasGAP is a mediator of rho pathway activation and tumorigenicity in the DLD1 colorectal cancer cell line. PLoS One. 2014;9:e86103. doi: 10.1371/journal.pone.0086103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Post JB, Hami N, Mertens AEE, Elfrink S, Bos JL, Snippert HJG. CRISPR-induced RASGAP deficiencies in colorectal cancer organoids reveal that only loss of NF1 promotes resistance to EGFR inhibition. Oncotarget. 2019;10:1440–1457. doi: 10.18632/oncotarget.26677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 80.McGlynn KA, Petrick JL, El-Serag HB. Epidemiology of hepatocellular carcinoma. Hepatology. 2020 Apr 22; doi: 10.1002/hep.31288. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Calvisi DF, Ladu S, Conner EA, Seo D, Hsieh JT, Factor VM, Thorgeirsson SS. Inactivation of Ras GTPase-activating proteins promotes unrestrained activity of wild-type Ras in human liver cancer. J Hepatol. 2011;54:311–319. doi: 10.1016/j.jhep.2010.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chen YL, Huang WC, Yao HL, Chen PM, Lin PY, Feng FY, Chu PY. Down-regulation of RASA1 is associated with poor prognosis in human hepatocellular carcinoma. Anticancer Res. 2017;37:781–785. doi: 10.21873/anticanres.11377. [DOI] [PubMed] [Google Scholar]
- 83.Hu C, Huang F, Deng G, Nie W, Huang W, Zeng X. miR-31 promotes oncogenesis in intrahepatic cholangiocarcinoma cells via the direct suppression of RASA1. Exp Ther Med. 2013;6:1265–1270. doi: 10.3892/etm.2013.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Du C, Weng X, Hu W, Lv Z, Xiao H, Ding C, Gyabaah OA, Xie H, Zhou L, Wu J, Zheng S. Hypoxia-inducible MiR-182 promotes angiogenesis by targeting RASA1 in hepatocellular carcinoma. J Exp Clin Cancer Res. 2015;34:67. doi: 10.1186/s13046-015-0182-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tai WT, Chen YL, Chu PY, Chen LJ, Hung MH, Shiau CW, Huang JW, Tsai MH, Chen KF. Protein tyrosine phosphatase 1B dephosphorylates PITX1 and regulates p120RasGAP in hepatocellular carcinoma. Hepatology. 2016;63:1528–1543. doi: 10.1002/hep.28478. [DOI] [PubMed] [Google Scholar]
- 86.Vanli G, Sempoux C, Widmann C. The caspase-3/p120 RasGAP Stress-sensing module reduces liver cancer incidence but does not affect overall survival in gamma-irradiated and carcinogen-treated mice. Mol Carcinog. 2017;56:1680–1684. doi: 10.1002/mc.22624. [DOI] [PubMed] [Google Scholar]
- 87.Sancho-Garnier H, Colonna M. Breast cancer epidemiology. Presse Med. 2019;48:1076–1084. doi: 10.1016/j.lpm.2019.09.022. (In French) [DOI] [PubMed] [Google Scholar]
- 88.Suárez-Cabrera C, Quintana RM, Bravo A, Casanova ML, Page A, Alameda JP, Paramio JM, Maroto A, Salamanca J, Dupuy AJ, et al. A transposon-based analysis reveals RASA1 is involved in triple-negative breast cancer. Cancer Res. 2017;77:1357–1368. doi: 10.1158/0008-5472.CAN-16-1586. [DOI] [PubMed] [Google Scholar]
- 89.Hu X, Stern HM, Ge L, O'Brien C, Haydu L, Honchell CD, Haverty PM, Peters BA, Wu TD, Amler LC, et al. Genetic alterations and oncogenic pathways associated with breast cancer subtypes. Mol Cancer Res. 2009;7:511–522. doi: 10.1158/1541-7786.MCR-08-0107. [DOI] [PubMed] [Google Scholar]
- 90.Liu Y, Liu T, Sun Q, Niu M, Jiang Y, Pang D. Downregulation of Ras GTPase activating protein 1 is associated with poor survival of breast invasive ductal carcinoma patients. Oncol Rep. 2015;33:119–124. doi: 10.3892/or.2014.3604. [DOI] [PubMed] [Google Scholar]
- 91.Huang J, Peng X, Zhang K, Li C, Su B, Zhang Y, Yu W. Co-expression and significance of Dok2 and Ras p21 protein activator 1 in breast cancer. Oncol Lett. 2017;14:5386–5392. doi: 10.3892/ol.2017.6844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Sharma SB, Lin CC, Farrugia MK, McLaughlin SL, Ellis EJ, Brundage KM, Salkeni MA, Ruppert JM. MicroRNAs 206 and 21 cooperate to promote RAS-extracellular signal-regulated kinase signaling by suppressing the translation of RASA1 and SPRED1. Mol Cell Biol. 2014;34:4143–4164. doi: 10.1128/MCB.00480-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Xiao W, Zheng S, Zou Y, Yang A, Xie X, Tang H, Xie X. CircAHNAK1 inhibits proliferation and metastasis of Triple-negative breast cancer by modulating miR-421 and RASA1. Aging (Albany NY) 2019;11:12043–12056. doi: 10.18632/aging.102539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Kanwar N, Carmine-Simmen K, Nair R, Wang C, Moghadas-Jafari S, Blaser H, Tran-Thanh D, Wang D, Wang P, Wang J, et al. Amplification of a calcium channel subunit CACNG4 increases breast cancer metastasis. EBioMedicine. 2020;52:102646. doi: 10.1016/j.ebiom.2020.102646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Zhang L, Zhan X, Yan D, Wang Z. Circulating: MicroRNA-21 is involved in lymph node metastasis in cervical cancer by targeting RASA1. Int J Gynecol Cancer. 2016;26:810–816. doi: 10.1097/IGC.0000000000000694. [DOI] [PubMed] [Google Scholar]
- 96.Hu J, Wang L, Chen J, Gao H, Zhao W, Huang Y, Jiang T, Zhou J, Chen Y. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505:222–228. doi: 10.1016/j.bbrc.2018.09.060. [DOI] [PubMed] [Google Scholar]
- 97.Jiang X, Tang H, Chen T. Epidemiology of gynecologic cancers in China. J Gynecol Oncol. 2018;29:e7. doi: 10.3802/jgo.2018.29.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Lubeck BA, Lapinski PE, Oliver JA, Ksionda O, Parada LF, Zhu Y, Maillard I, Chiang M, Roose J, King PD. GTPase-activating proteins (RasGAPs) Neurofibromin 1 and p120 RasGAP in T cells results in the development of T cell acute lymphoblastic Leukemia. J Immunol. 2015;195:31–35. doi: 10.4049/jimmunol.1402639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jia CY, Li HH, Zhu XC, Dong YW, Fu D, Zhao QL, Wu W, Wu XZ. MiR-223 suppresses cell proliferation by targeting IGF-1R. PLoS One. 2016;6:e27008. doi: 10.1371/journal.pone.0027008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ng JH, Iyer NG, Tan MH, Edgren G. Changing epidemiology of oral squamous cell carcinoma of the tongue: A global study. Head Neck. 2017;39:297–304. doi: 10.1002/hed.24589. [DOI] [PubMed] [Google Scholar]
- 101.Krishnan N, Gupta S, Palve V, Varghese L, Pattnaik S, Jain P, Khyriem C, Hariharan A, Dhas K, Nair J, et al. Integrated analysis of oral tongue squamous cell carcinoma identifies key variants and pathways linked to risk habits, HPV, clinical parameters and tumor recurrence. F1000Res. 2015;4:1215. doi: 10.12688/f1000research.7302.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Su SC, Lin CW, Liu YF, Fan WL, Chen MK, Yu CP, Yang WE, Su CW, Chuang CY, Li WH, et al. Exome sequencing of oral squamous cell carcinoma reveals molecular subgroups and novel therapeutic opportunities. Theranostics. 2017;7:1088–1099. doi: 10.7150/thno.18551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Zhang W, Wang M, Wu Q, Zhu Q, Jiao Y, Zhu Y, Yang B, Ni S, Yu J, Sun H, Zeng YX. Mutational signatures and the genomic landscape of betel quid Chewing-associated tongue carcinoma. Cancer Med. 2019;8:701–711. doi: 10.1002/cam4.1888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Wang J, Wang W, Li J, Wu L, Song M, Meng Q. miR182 activates the Ras-MEK-ERK pathway in human oral cavity squamous cell carcinoma by suppressing RASA1 and SPRED1. Onco Targets Ther. 2017;10:667–679. doi: 10.2147/OTT.S121864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Mizrahi JD, Surana R, Valle JW, Shroff RT. Pancreatic cancer. Lancet. 2020;395:2008–2020. doi: 10.1016/S0140-6736(20)30974-0. [DOI] [PubMed] [Google Scholar]
- 106.Rajamani D, Bhasin MK. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016;8:38. doi: 10.1186/s13073-016-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kent OA, Mendell JT, Rottapel R. Transcriptional regulation of miR-31 by oncogenic KRAS mediates metastatic phenotypes by repressing RASA1. Mol Cancer Res. 2016;14:267–277. doi: 10.1158/1541-7786.MCR-15-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Culp MB, Soerjomataram I, Efstathiou JA, Bray F, Jemal A. Recent global patterns in prostate cancer incidence and mortality rates. Eur Urol. 2020;77:38–52. doi: 10.1016/j.eururo.2019.08.005. [DOI] [PubMed] [Google Scholar]
- 109.Sowalsky AG, Xia Z, Wang L, Zhao H, Chen S, Bubley GJ, Balk SP, Li W. Whole transcriptome sequencing reveals extensive unspliced mRNA in metastatic castration-resistant prostate cancer. Mol Cancer Res. 2015;13:98–106. doi: 10.1158/1541-7786.MCR-14-0273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Berndt SI, Wang Z, Yeager M, Alavanja MC, Albanes D, Amundadottir L, Andriole G, Beane Freeman L, Campa D, Cancel-Tassin G, et al. Two susceptibility loci identified for prostate cancer aggressiveness. Nat Commun. 2015;6:6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Laczny C, Leidinger P, Haas J, Ludwig N, Backes C, Gerasch A, Kaufmann M, Vogel B, Katus HA, Meder B, et al. miRTrail-a comprehensive webserver for analyzing gene and miRNA patterns to enhance the understanding of regulatory mechanisms in diseases. BMC Bioinformatics. 2012;13:36. doi: 10.1186/1471-2105-13-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sung H, Kanchi KL, Wang X, Hill KS, Messina JL, Lee JH, Kim Y, Dees ND, Ding L, Teer JK, et al. Inactivation of RASA1 promotes melanoma tumorigenesis via R-Ras activation. Oncotarget. 2016;7:23885–23996. doi: 10.18632/oncotarget.8127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rusinek D, Swierniak M, Chmielik E, Kowal M, Kowalska M, Cyplinska R, Czarniecka A, Piglowski W, Korfanty J, Chekan M, et al. BRAFV600E-associated gene expression profile: Early changes in the transcriptome, based on a transgenic mouse model of papillary thyroid carcinoma. PLoS One. 2015;10:e0143688. doi: 10.1371/journal.pone.0143688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Park C, Ha SY, Kim ST, Kim HC, Heo JS, Park YS, Lauwers G, Lee J, Kim KM. Identification of the BRAF V600E mutation in gastroenteropancreatic neuroendocrine tumors. Oncotarget. 2016;7:4024–4035. doi: 10.18632/oncotarget.6602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Zhang RL, Yang JP, Peng LX, Zheng LS, Xie P, Wang MY, Cao Y, Zhang ZL, Zhou FJ, Qian CN, Bao YX. RNA-binding protein QKI-5 inhibits the proliferation of clear cell renal cell carcinoma via post-transcriptional stabilization of RASA1 mRNA. Cell Cycle. 2016;15:3094–3104. doi: 10.1080/15384101.2016.1235103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Zhang RL, Aimudula A, Dai JH, Bao YX. RASA1 inhibits the progression of renal cell carcinoma by decreasing the expression of miR-223-3p and promoting the expression of FBXW7. Biosci Rep. 2020;40:BSR20194143. doi: 10.1042/BSR20194143. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 117.Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang Y, Zhou J, Pan K, Sun L, Fang J, et al. A global burden of gastric cancer: The major impact of China. Expert Rev Gastroenterol Hepatol. 2017;11:651–661. doi: 10.1080/17474124.2017.1312342. [DOI] [PubMed] [Google Scholar]
- 118.Li Z, Li D, Zhang G, Xiong J, Jie Z, Cheng H, Cao Y, Jiang M, Lin L, Le Z, et al. Methylation-associated silencing of MicroRNA-335 contributes tumor cell invasion and migration by interacting with RASA1 in gastric cancer. Am J Cancer Res. 2014;4:648–662. [PMC free article] [PubMed] [Google Scholar]
- 119.Chen X, Cai S, Li B, Zhang X, Li W, Liang H, Cao X, Wang L, Wu Z. MicroRNA-21 regulates the biological behavior of esophageal squamous cell carcinoma by targeting RASA1. Oncol Rep. 2019;41:1627–1637. doi: 10.3892/or.2018.6944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Pickering CR, Zhou JH, Lee JJ, Drummond JA, Peng SA, Saade RE, Tsai KY, Curry JL, Tetzlaff MT, Lai SY, et al. Mutational landscape of aggressive cutaneous squamous cell carcinoma. Clin Cancer Res. 2014;20:6582–6592. doi: 10.1158/1078-0432.CCR-14-1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Jouenne F, Reger de Moura C, Lorillon G, Meignin V, Dumaz N, Lebbe C6 Mourah S, Tazi A. RASA1 loss in a BRAF-mutated Langerhans cell sarcoma: A mechanism of resistance to BRAF inhibitor. Ann Oncol. 2019;30:1170–1172. doi: 10.1093/annonc/mdz125. [DOI] [PubMed] [Google Scholar]
- 122.Sears R, Gray JW. Epigenomic inactivation of RasGAPs activates RAS signaling in a subset of luminal b breast cancers. Cancer Discov. 2017;7:131–133. doi: 10.1158/2159-8290.CD-16-1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Degirmenci U, Wang M, Hu J. Targeting Aberrant RAS/RAF/MEK/ERK Signaling for Cancer Therapy. Cells. 2020;9:198. doi: 10.3390/cells9010198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoxhaj G, Manning BD. The PI3K-AKT network at the interface of oncogenic signalling and cancer metabolism. Nat Rev Cancer. 2020;20:74–88. doi: 10.1038/s41568-019-0216-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Markou A, Zavridou M, Lianidou ES. miRNA-21 as a novel therapeutic target in lung cancer. Lung Cancer (Auckl) 2016;7:19–27. doi: 10.2147/LCTT.S60341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Revencu N, Fastre E, Ravoet M, Helaers R, Brouillard P, Bisdorff-Bresson A, Chung CWT, Gerard M, Dvorakova V, Irvine AD, et al. RASA1 mosaic mutations in patients with capillary malformation-arteriovenous malformation. J Med Genet. 2020;57:48–52. doi: 10.1136/jmedgenet-2019-106024. [DOI] [PubMed] [Google Scholar]
- 127.Liu Z, Deng M, Wu L, Zhang S. An integrative investigation on significant mutations and their down-stream pathways in lung squamous cell carcinoma reveals CUL3/KEAP1/NRF2 relevant subtypes. Mol Med. 2020;26:48. doi: 10.1186/s10020-020-00166-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data sharing is not applicable to this article, as no datasets were generated or analyzed during the present study.