Abstract
The mammalian immune system implements a remarkably effective set of mechanisms for fighting pathogens1. Its main components are hematopoietic immune cells, including myeloid cells that control innate immunity and lymphoid cells that constitute adaptive immunity2. However, immune functions are not unique to hematopoietic cells, and many other cell types display basic mechanisms of pathogen defence3–5. To advance our understanding of immunology outside the haematopoietic system, here we systematically investigate the regulation of immune genes in the three major types of structural cells: epithelium, endothelium, and fibroblasts. We characterize these cell types across twelve organs in mice, using cellular phenotyping, transcriptome sequencing, chromatin-accessibility profiling, and epigenome mapping. This comprehensive dataset revealed complex immune gene activity and regulation in structural cells. The observed patterns were highly organ-specific and seem to modulate the extensive interactions between structural cells and haematopoietic immune cells. Moreover, we identified an epigenetically encoded immune potential in structural cells under tissue homeostasis, which was triggered in response to systemic viral infection. This study highlights the prevalence and organ-specific complexity of immune gene activity in non-haematopoietic structural cells, and it provides a high-resolution, multi-omics atlas of the epigenetic and transcriptional networks that regulate structural cells in the mouse.
The structure of most tissues and organs in the mammalian body is shaped by epithelial cells (epithelium), which create internal and external surfaces and barriers; endothelial cells (endothelium), which form the lining of blood vessels; and fibroblasts, which provide essential connective tissue (stroma)6. These three cell types, which we refer to as “structural cells”, have been shown to contribute in important ways to mammalian immunity7–11. However, there has been little systematic investigation across organs, in part because structural cells are difficult to study by genetic ablation due to their essential structural roles in most organs. Multi-omics profiling has emerged as a promising approach to dissecting immune regulation in a systematic, genome-wide manner, as illustrated by recent work on the systems immunology of hematopoietic immune cells12, 13.
In this study, we used multi-omics profiling and integrative bioinformatics to establish a high-resolution atlas of structural cells and of non-hematopoietic immune regulation in the mouse. We observed widespread expression of immune regulators and cytokine signaling molecules in structural cells, organ-specific adaptation to the tissue environment, and unexpectedly diverse capabilities for interacting with hematopoietic cells. These cell-type-specific and organ-specific differences in immune gene activity were reflected by characteristic patterns of chromatin regulation. Most notably, we found evidence of an epigenetically encoded immune potential under homeostatic conditions, and the affected genes were preferentially upregulated in response to an immunological challenge induced by systemic viral infection. We validated and functionally dissected this epigenetic potential of structural cells by further in vivo experiments with recombinant cytokines.
In summary, our study uncovered widespread immune gene regulation in structural cells of the mouse, and it established a multi-organ atlas of the underlying epigenetic and transcription-regulatory programs.
Results
Mapping structural cells across organs
To investigate the regulation of immune genes in structural cells, we performed multi-omics profiling of endothelium, epithelium and fibroblasts from 12 mouse organs (brain, caecum, heart, kidney, large intestine, liver, lung, lymph node, skin, small intestine, spleen and thymus). Single-cell suspensions were analysed by flow cytometry, and sort-purified cell populations were profiled with three genome-wide assays (Fig. 1a): (i) gene expression profiling by low-input RNA sequencing (RNA-seq)14; (ii) chromatin accessibility profiling with the assay for transposase-accessible chromatin using sequencing (ATAC-seq)15; and (iii) epigenome profiling by ChIPmentation16 with an antibody against the promoter and enhancer-linked histone H3K4me2 mark17. All assays produced high-quality data (Supplementary Table 1). This multi-omics dataset is provided as an online resource for interactive browsing and download at http://structural-immunity.computational-epigenetics.org.
Figure 1.
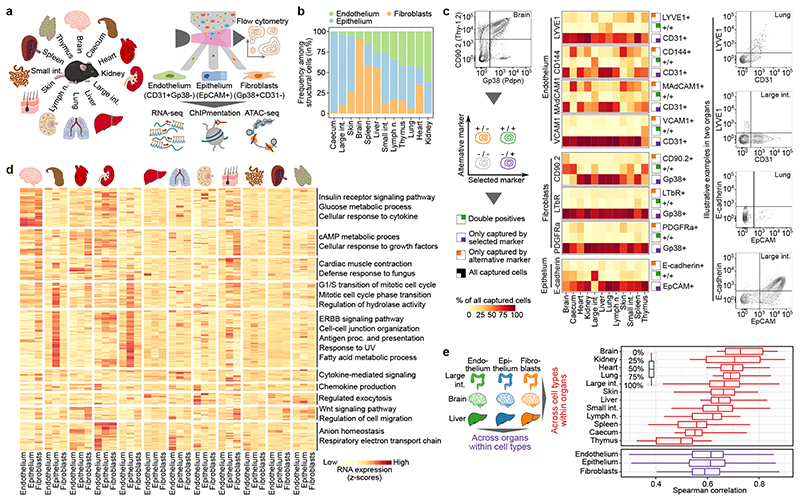
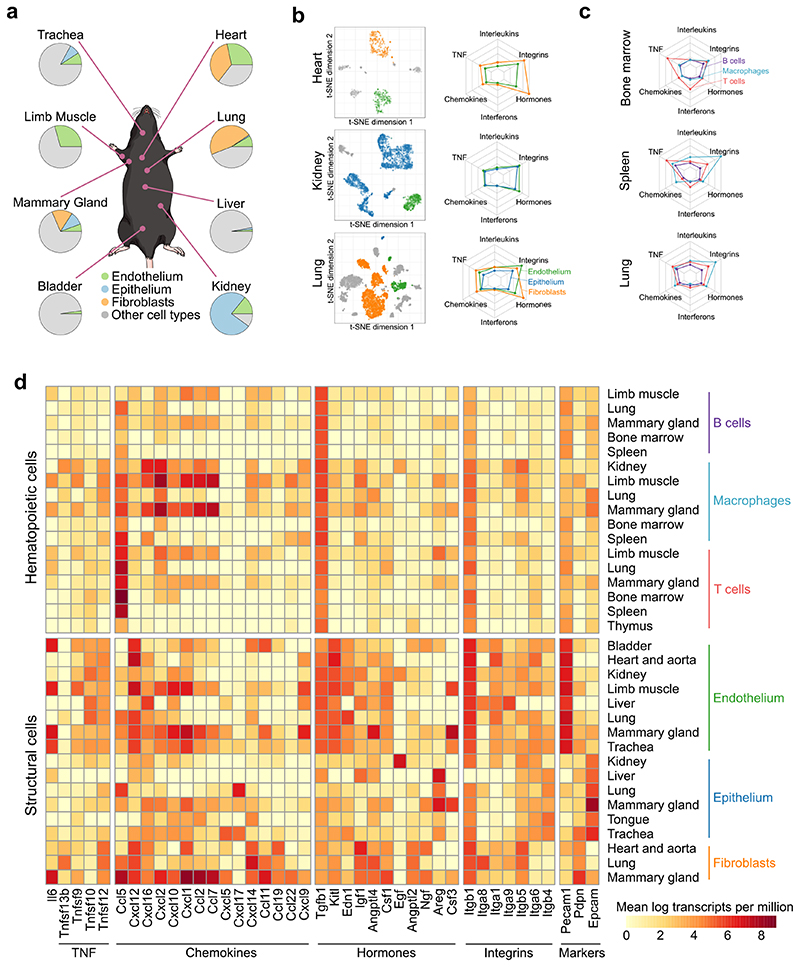
Multi-omics profiling establishes cell-type-specific and organ-specific characteristics of structural cells. a, Schematic outline of the experimental approach. b, Relative frequencies of structural cell types based on flow cytometry. c, Expression of surface markers among structural cells, comparing the standardized sorting of endothelium, epithelium, and fibroblasts to potential alternative markers (left: schematic outline; center: heatmaps showing marker overlap; right: illustrative FACS plots). d, Expression of differentially regulated genes across cell types and organs. Gene clusters are annotated with enriched terms based on gene set analysis. e, Correlation of gene expression across cell types and organs. Sample size: n = 4 (b) and n = 3 (c-e) independent biological replicates.
To maximize comparability across organs, we developed a standardized workflow for tissue dissociation and cell purification (Fig. 1a). Structural cells were purified with an organ-independent sorting scheme that comprised the endothelium marker CD31 (encoded by Pecam1), the epithelium marker EpCAM (Epcam), and the fibroblast marker GP38 (encoded by Pdpn, also known as podoplanin) (Extended Data Fig. 1a, b). All three types of structural cells were detectable in all 12 organs, with strong differences in their relative frequencies (Fig. 1b, Extended Data Fig. 1c). Our standardized tissue dissociation did not cause major technical biases (Extended Data Fig. 1d).
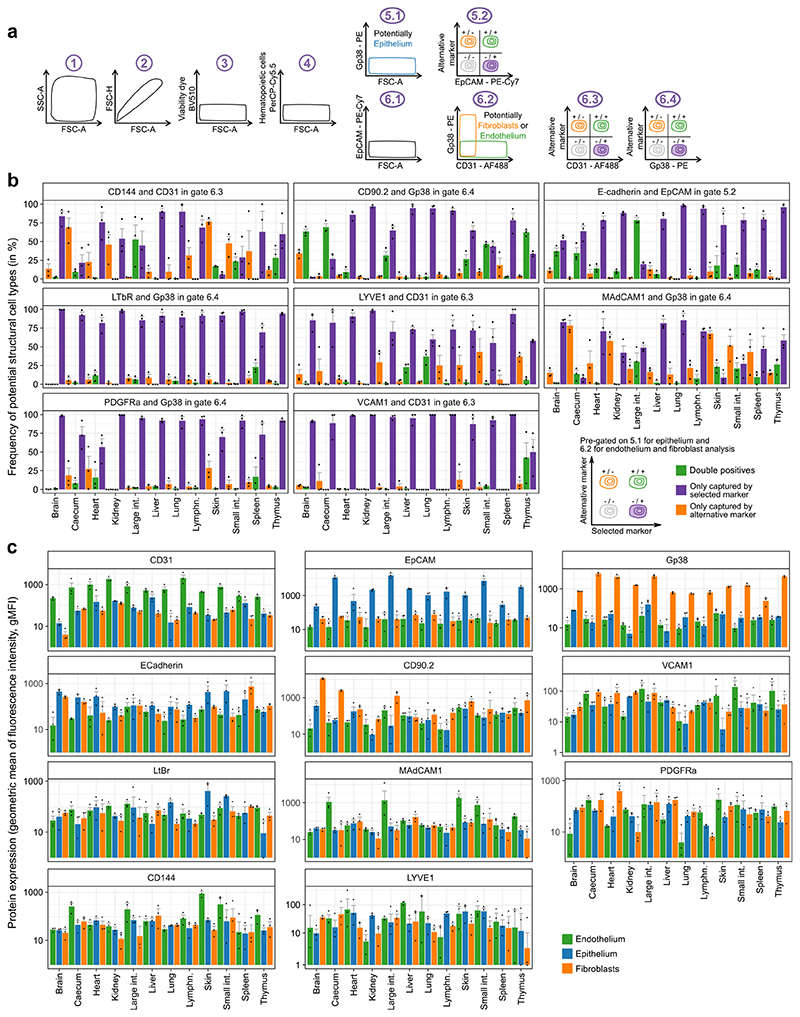
We validated and phenotypically characterized structural cell populations by flow cytometry for additional markers (Fig. 1c, Extended Data Fig. 2). Purification of CD31+GP38− endothelial cells specifically enriched for blood endothelium while excluding the less prevalent CD31+GP38+ cells of lymphatic endothelium, which facilitates the comparison across organs. Alternative endothelial markers such as CD144 (VE-cadherin), MAdCAM1 and VCAM1 (used to assess baseline activation of endothelium under homeostatic conditions) did not improve the identification of blood endothelial cells in our analysis. Similarly, sorting of fibroblasts as GP38+CD31− cells did not miss any major cell populations identified by the alternative markers CD90.2 (Thy-1.2), LTβR or PDGFRα. Finally, E-cadherin could not enhance or replace our sorting of epithelium as EpCAM+ cells.
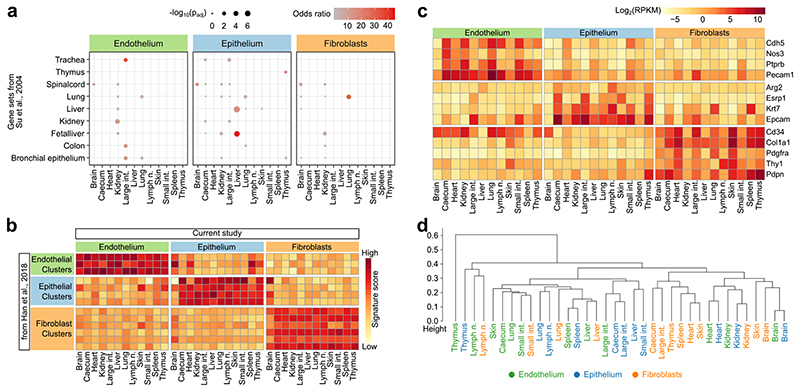
The cellular identity of the sorted endothelium, epithelium and fibroblasts was further confirmed by RNA-seq data analysis, which showed the expected organ-specific (Extended Data Fig. 3a) and cell-type-specific (Extended Data Fig. 3b) patterns of gene expression compared to published multi-tissue expression profiles18, 19. We also observed the expected expression patterns for various cell-type-specific marker genes, but with a high degree of transcriptional heterogeneity across organs (Extended Data Fig. 3c, Supplementary Table 2). Notably, the expression profiles of structural cells within the same organ were globally more similar to each other than structural cells of the same type across organs (Fig. 1d, e, Extended Data Fig. 3d), which suggests that the tissue and organ environment has a major effect on the transcriptomes of structural cells. Our multi-omics profiles for endothelium, epithelium and fibroblasts across 12 mouse organs thus uncover a marked degree of organ-specific differences among structural cells.
Immune gene activity in structural cells
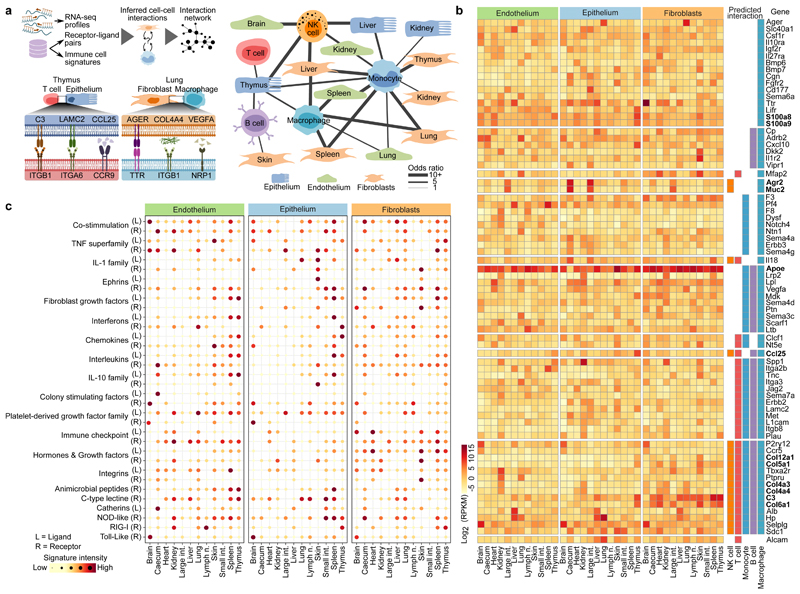
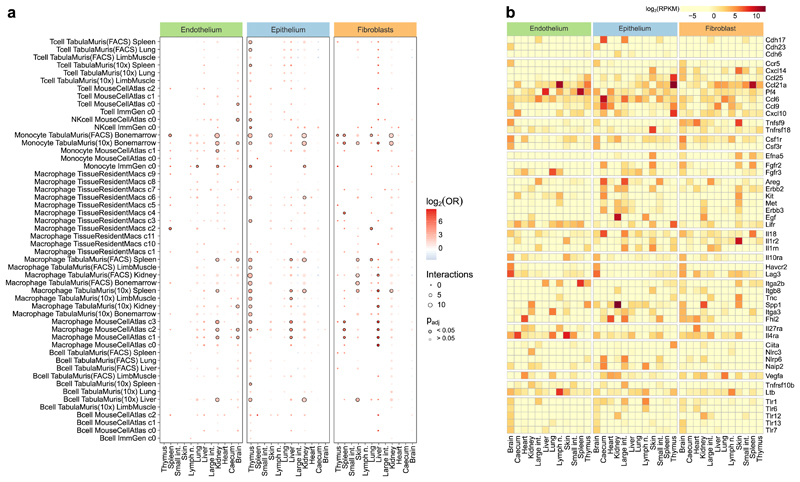
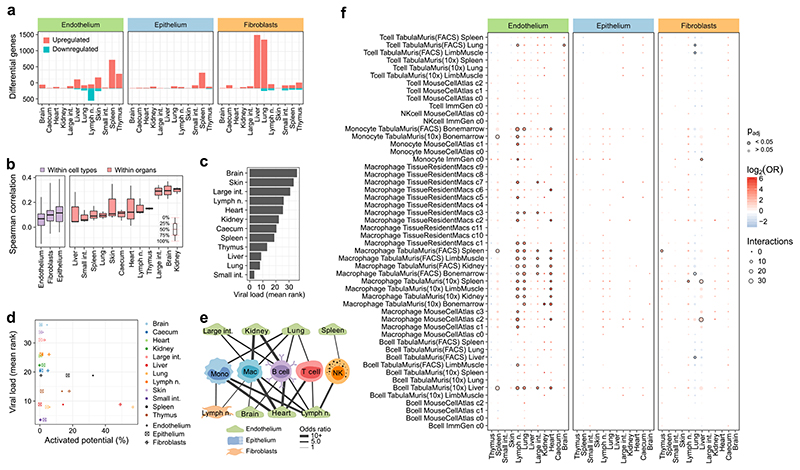
On the basis of our RNA-seq dataset, we investigated the immune gene activity in structural cells. First, we assessed the ability of structural cells to communicate with haematopoietic immune cells, by inferring a network of potential cell–cell interactions based on known receptor–ligand pairs. The resulting cell–cell interaction network (Fig. 2a, Supplementary Table 3) predicted frequent crosstalk between structural cells and haematopoietic cells under homeostasis (Fig. 2a, Extended Data Fig. 4a). Differences across cell types and organs were driven by the characteristic expression patterns of cell-surface proteins and secreted factors in structural cells (Fig. 2b). For example, strong expression of collagens (Col4a3, Col4a4, Col5a1, Col6a1, Col12a1) and of complement component 3 (C3) in fibroblasts is expected to enhance their ability to interact with hematopoietic cells; high levels of Muc2 and Arg2 in the digestive tract fosters interactions with macrophages and NK cells; and Ccl25 expression in thymus epithelium contributes to the maturation of T cells. Shared across all types of structural cells, we observed high expression of the inflammatory mediators Apoe, S100a8, and S100a9.
Figure 2.
Gene expression of structural cells predicts cell-type-specific and organ-specific crosstalk with hematopoietic immune cells. a, Network of potential cell–cell interactions between structural cells and haematopoietic immune cells inferred from gene expression of known receptor–ligand pairs. NK cell, natural killer cell. b, Expression of receptors and ligands in structural cells, annotated with the cell–cell interactions that they may mediate (genes discussed in the text are in bold). RPKM, reads per kilobase of transcript per million mapped reads. c, Gene signatures of receptors (R) and ligands (L) in structural cells. Sample size (all panels): n = 3 independent biological replicates.
Second, we quantified the aggregated activity of various immune gene modules, which were manually curated to capture important components of the immune system (Extended Data Fig. 4b; Supplementary Table 4). We observed widespread activity of these immune gene modules in structural cells, with highly cell-type-specific and organ-specific patterns (Fig. 2c). We sought to validate the unexpectedly strong and diverse immune gene activity of structural cells in a second dataset. To that end, we obtained single-cell transcriptome profiles of mouse tissues from the Tabula Muris resource20, and we identified endothelium, epithelium, and fibroblasts bioinformatically based on marker gene expression. While structural cells were not well covered in this dataset (Extended Data Fig. 5a; Supplementary Table 5), these profiles were sufficient to independently confirm strong and organ-specific activity of immune genes in structural cells (Extended Data Fig. 5b-d). Together, these data reveal widespread activity of immune genes and regulatory modules in structural cells, which are expected to mediate cell-type-specific and organ-specific interactions with haematopoietic immune cells.
Regulatory networks in structural cells
To uncover the regulatory basis of immune gene activity in structural cells, we combined our RNA-seq data with ATAC-seq profiles of chromatin accessibility and ChIPmentation maps for the promoter and enhancer-linked H3K4me2 mark, and we compared gene-regulatory networks across cell types and organs (Fig. 3a).
Figure 3.
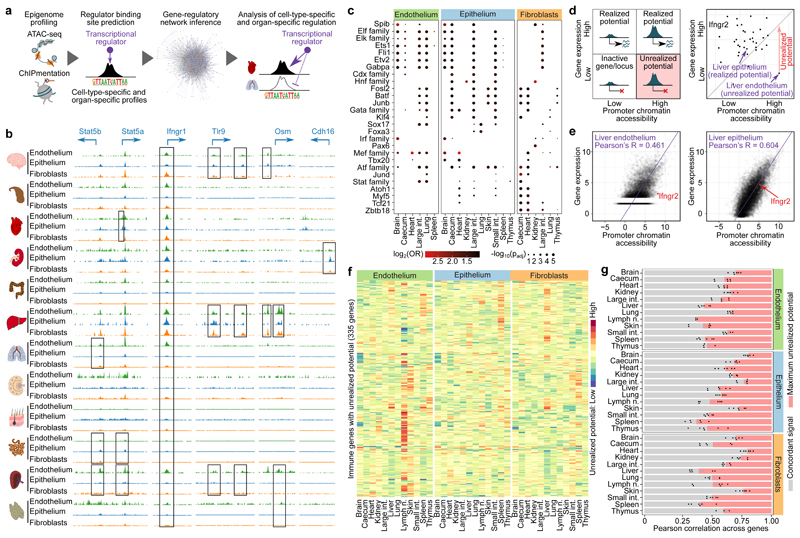
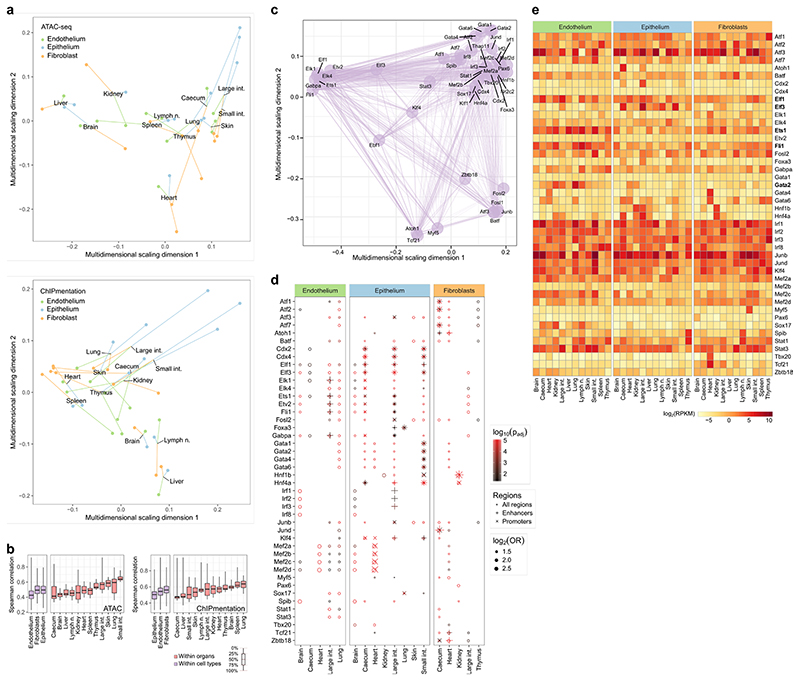
Structural cells implement characteristic gene-regulatory networks and an epigenetic potential for immune gene activation. a, Schematic outline of the gene-regulatory network analysis. b, ATAC-seq signal tracks (average across replicates) for selected genomic regions. Differences between cell types and organs are highlighted by black boxes. c, Motif enrichment for transcriptional regulators in cell-type-specific and organ-specific chromatin marker peaks (one-sided hypergeometric test with multiple-testing correction). d, Schematic outline (left) and a concrete example (right) of the epigenetic potential, based on the comparison of chromatin accessibility (ATAC-seq) in promoter regions with matched gene expression (RNA-seq). e, Scatterplot showing the correlation between promoter chromatin accessibility across all genes in liver endothelium and epithelium with Ifngr2 highlighted by the red dot. f, Immune genes with unrealized epigenetic potential across cell types and organs. g, Pearson correlation between promoter chromatin accessibility and gene expression across cell types and organs (mean and s.e.m across pairwise correlations; red bars indicate the maximum scope for unrealized epigenetic potential). Sample size: ATAC-seq n = 2 (b-g), RNA-seq n = 3 (c-g) independent biological replicates.
We observed extensive cell-type-specific and organ-specific chromatin regulation at immune gene loci (Fig. 3b). For example, the immune-regulatory transcription factors Stat5a and Stat5b showed characteristic patterns of chromatin accessibility; Tlr9 and Osm were characterized by organ-specific heterogeneity in promoter and enhancer regions; and the promoter of the renal cell adhesion molecule Cdh16 was exclusively open in kidney. By contrast, a subset of crucial immune genes (exemplified by Ifngr1) showed high chromatin accessibility in most samples, indicative of a shared core of immune regulation in structural cells. Consistent with our RNA-seq analysis, the chromatin profiles were globally more similar between different types of structural cells in one organ than among the same cell type across organs (Extended Data Fig. 6a, b).
We inferred a gene-regulatory network of structural cells by connecting transcription factors to their target genes, based on predicted binding sites with open chromatin in the respective cells (Extended Data Fig. 6c). Many key regulators of transcription in structural cells showed cell-type-specific and organ-specific activity (Fig. 3c, Extended Data Fig. 6d, Supplementary Table 6). For example, ATF, ELK, ETS and JUND were most active in lung endothelium; KLF and CDX in digestive tract epithelium; and HNF in kidney fibroblasts and in epithelium of caecum, large intestine, and small intestine. We also identified groups of transcription factors that were ubiquitously active in structural cells, such as ELF1, ELF3, ETS1, FLI1 and GATA2 (Extended Data Fig. 6e), which may constitute a shared regulatory basis of immune gene activity in structural cells. Our inferred gene-regulatory networks support a model in which constitutively active regulators establish a shared core of immune functions, while additional factors contribute cell-type-specific and organ-specific adaptions.
Epigenetic potential for gene activation
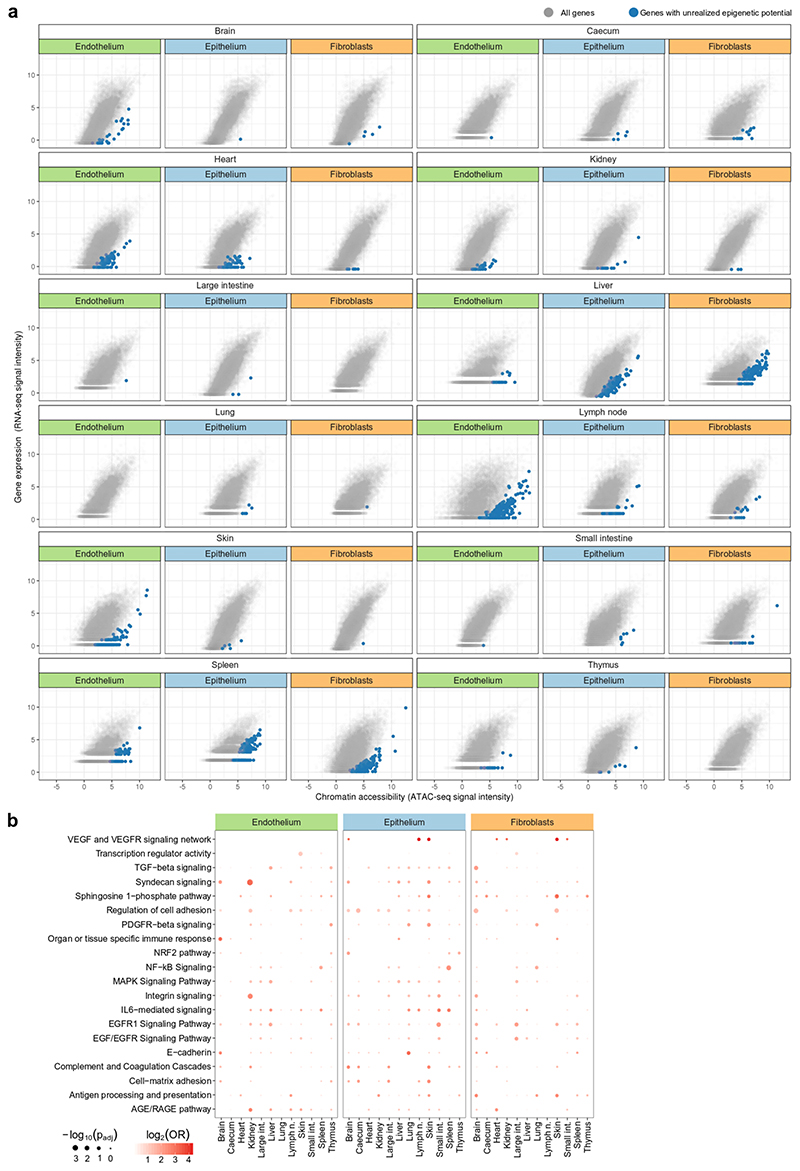
To assess the epigenetically encoded immune potential of structural cells, we quantified the chromatin accessibility of each gene promoter and compared it to the expression level of the corresponding gene. We then scanned for genes with low expression but high promoter accessibility, indicative of an unrealized potential for increased expression (Fig. 3d, e, Extended Data Fig. 7a). For example, chromatin accessibility at the Ifngr2 promoter was high in liver endothelium whereas gene expression was low, which suggests that Ifngr2 has unrealized potential for upregulation without the need to increase promoter accessibility. By contrast, accessibility of the Ifngr2 promoter in liver epithelium was highly consistent with its gene expression, thus constituting a case of realized potential.
The genes with unrealized epigenetic potential in structural cells were enriched for immune functions (Fig. 3f, Extended Data Fig. 7b, Supplementary Tables 7 and 8), consistent with our initial hypothesis. In total, we identified 1,665 genes that fulfilled our definition of unrealized epigenetic potential, of which 335 genes were annotated with at least one immunological term (odds ratio 1.37, P < 10−5, Fisher’s exact test).
To quantify and compare the epigenetic potential across cell types and organs (Fig. 3g), we exploited that genes with strong unrealized potential are outliers when plotting gene expression against promoter accessibility, which results in a reduced correlation between the two data types (Extended Data Fig. 7a). The highest correlation between gene expression and promoter accessibility was observed in brain, caecum, heart, kidney, large intestine and skin, leaving comparatively less room for unrealized epigenetic potential. By contrast, the correlation was notably lower in liver, lymph node, spleen and thymus, which suggests that structural cells in these organs harbour a more pronounced epigenetic potential for gene activation in response to various stimuli.
Our integrative analysis of chromatin accessibility and gene expression thus identified an epigenetically encoded potential for immune gene activation in structural cells. This epigenetic potential is expected to facilitate the rapid response to immunological challenges in a cell-type-specific and organ-specific manner.
Immune genes induced by viral infection
We functionally evaluated the epigenetic potential for immune gene activation by challenging mice with a systemic viral infection model. We infected mice with lymphocytic choriomeningitis virus (LCMV) and collected samples from 12 organs on day 8 after infection (Fig. 4a). We characterized these samples by flow cytometry (Extended Data Fig. 8a, b) and by RNA-seq analysis of sort-purified structural cells (Supplementary Table 1).
Figure 4.
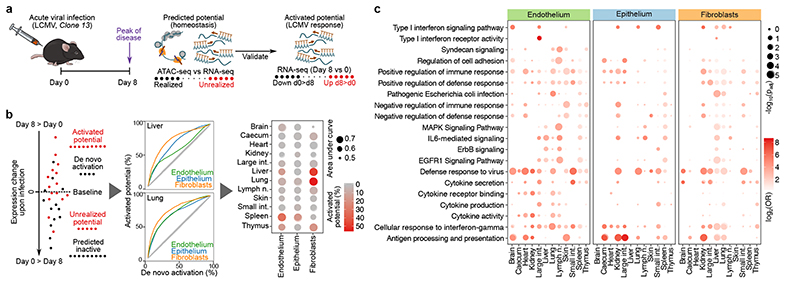
Systemic viral infection activates the immunological potential of structural cells in vivo. a, Schematic outline of the lymphocytic choriomeningitis virus (LCMV) infection analysis. b, Comparison of the changes in gene expression upon LCMV infection (day 8) to the epigenetic potential observed under homeostatic conditions (day 0), using a threshold of zero for differential gene expression (left) or a variable threshold analogous to a ROC curve (center). The area under the curve is interpreted as a measure of the epigenetic potential’s predictiveness for LCMV-induced gene activation, plotted together with the percentage of upregulated genes that carry unrealized epigenetic potential (right). c, Enrichment of immune-related gene sets among the LCMV-induced genes (two-sided Fisher’s exact test with multiple-testing correction). Sample size (all panels): n = 3 independent biological replicates.
LCMV infection resulted in changes of structural cell composition in most organs (Extended Data Fig. 8c), and we observed differential gene expression in a cell-type-specific and organ-specific manner (Extended Data Fig. 9a, Supplementary Table 9). The transcriptional response was globally more similar among structural cells of all three types within the same organ than between structural cells of a given type across organs (Extended Data Fig. 9b), consistent with the patterns of similarity that we observed under homeostatic conditions.
We compared the differential gene expression after LCMV infection with the corresponding epigenetic potential of the genes under homeostatic conditions, and genes with unrealized potential were indeed overrepresented among the LCMV-induced genes (Fig. 4b, left). To quantify how well the epigenetic potential predicts LCMV-induced gene activation, we plotted the percentage of genes with realized potential against the percentage of genes that underwent de novo activation (Fig. 4b, centre), in analogy with receiver operating characteristic (ROC) curves. We detected the strongest association in liver, lung and spleen (Fig. 4b, right). These organ-specific differences showed no clear correlation with differences in viral load (Extended Data Fig. 9c) and appear to constitute intrinsic regulatory differences between cell types and organs (Extended Data Fig. 9d).
The genes that were upregulated in response to systemic LCMV infection showed strong enrichment for immune functions (Fig. 4c), including ‘positive regulation of immune response’, ‘defence response to virus’, ‘cellular response to IFNγ’, and ‘antigen processing and presentation’. We also observed widespread upregulation of interferon-induced as well as interferon-stimulated genes and of key transcriptional regulators in the interferon pathway, which indicates a strong interferon response to LCMV infection in structural cells (Supplementary Table 9).
Finally, we inferred receptor–ligand interactions between structural cells and haematopoietic immune cells, using the transcriptome data of structural cells upon LCMV infection (Extended Data Fig. 9e, f, Supplementary Table 10). We observed an increase in the strength and scope of predicted cell–cell interactions after LCMV infection as compared to homeostatic conditions, largely driven by upregulated gene expression levels for receptors and ligands in structural cells, including B2m, Cd74, Cd47, Cxcl10, Sdc1, Sdc4, Tnfrsf1a and Vcam1.
Systemic LCMV infection thus triggered widespread activation of immune genes that were lowly expressed but epigenetically poised under homeostatic conditions. These results support our model of an epigenetically encoded potential for immune gene activation in structural cells in the context of viral infection.
Cytokine response of structural cells
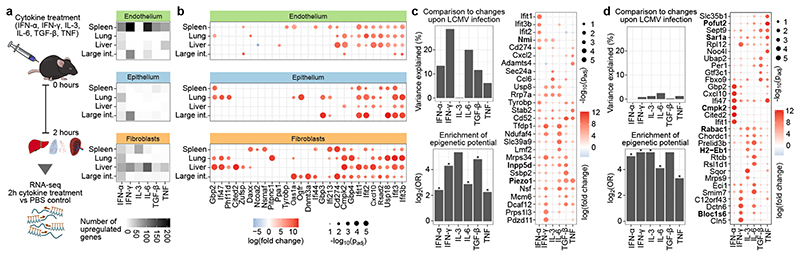
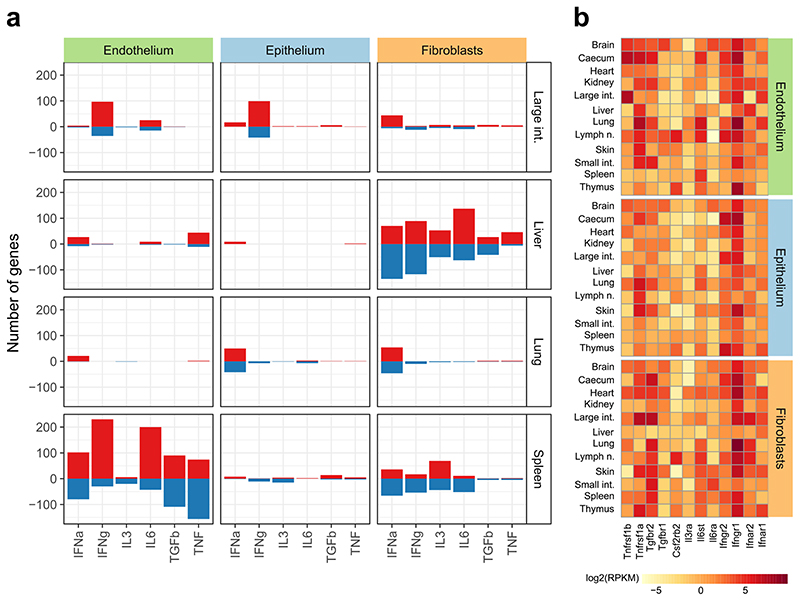
To characterize the effects of individual cytokines that may contribute to the response to LCMV infection, we administered six recombinant cytokines in mice (IFN-α, IFN-γ, IL-3, IL-6, TGF-β, TNF). These cytokines were selected based on our dataset and published results22, while IL-3 was included as a control with no known role in LCMV infection. To focus on the immediate effects of cytokine signaling, structural cells were sort-purified two hours after injection and subjected to RNA-seq profiling (Fig. 5a; Supplementary Table 1). Four organs that responded strongly to LCMV infection were included in the analysis (large intestine, liver, lung, spleen).
Figure 5.
Cytokine treatment induces cell-type-specific and organ-specific changes in structural cells in vivo. a, Schematic outline of the cytokine treatment experiments (left) and number of genes upregulated in each experiment (right). b, Genes upregulated in response to IFN-α treatment. c,d, Cytokine-induced changes in spleen endothelium (c) and liver fibroblasts (d), showing the percentage of LCMV-induced changes that are recapitulated by cytokine treatment (top left), enrichment for genes with unrealized epigenetic potential among the cytokine-induced genes (bottom left), and genes upregulated upon cytokine treatment (genes discussed in the text are in bold). Significant enrichments (two-sided Fisher’s exact test, adjusted p-value < 0.05) are labeled with an asterisk. Differential expression is based on a linear model (two-sided test) with multiple-testing correction (panels b-d). Sample size (all panels): n = 3 independent biological replicates.
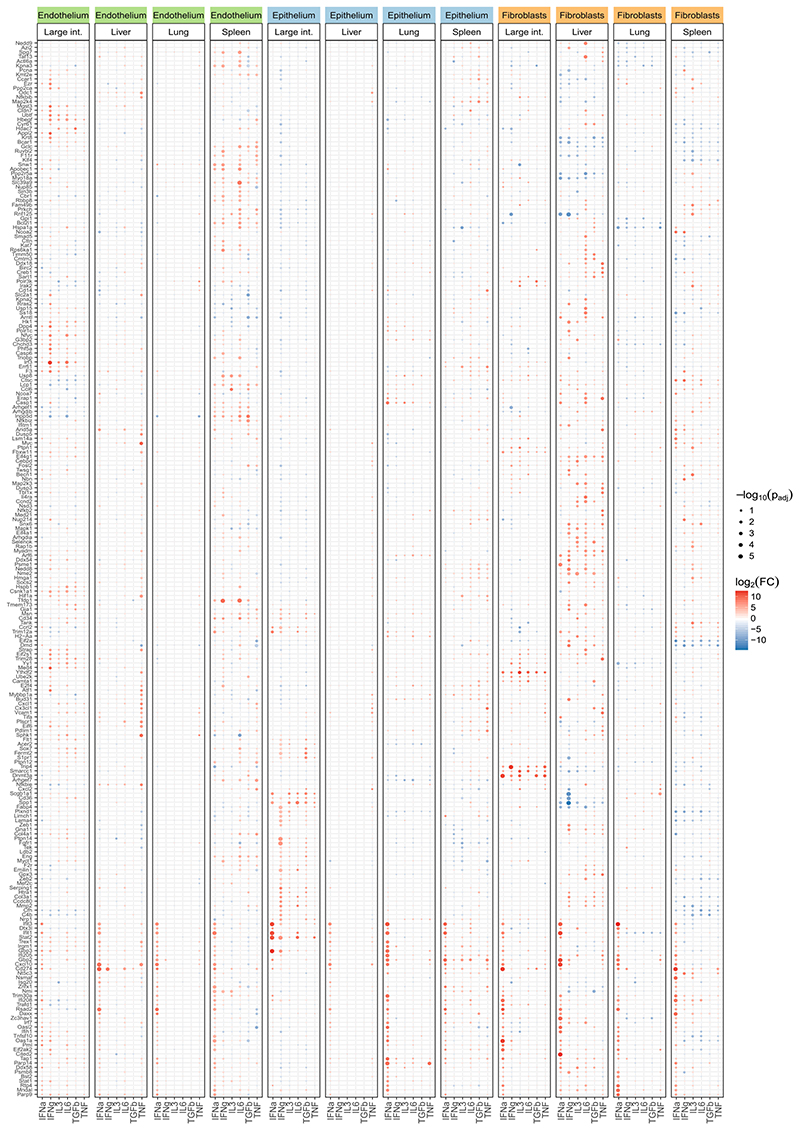
We compared the structural cell transcriptomes between cytokine-treated animals and mock-treated controls, which uncovered various cytokine-induced transcriptional changes. IFN-α treatment had the strongest effect, inducing known interferon target genes and several genes associated with antiviral immunity (Fig. 5b, Extended Data Fig. 10; Supplementary Table 11). We further observed widespread cell-type-specific and organ-specific differences, which were not solely due to differential expression of individual cell surface receptors (Extended Data Fig. 11b), but appear to reflect more general differences in immune gene regulation.
In spleen endothelium (highlighted here because of its strong transcriptional response to LCMV infection), treatment with IFNγ and IL-6 explained a sizable proportion of the LCMV-induced changes (Fig. 5c, top left). The cytokine-induced genes included known interferon target genes, transcriptional regulators such as Nmi and Inpp5d (which encodes SHIP1), and the pro-inflammatory gene Piezo1 (Fig. 5c, right, Supplementary Table 11). For five of the six cytokines (not including IL-3), the upregulated genes showed significant overlap with the genes that carried unrealized epigenetic potential under homeostatic conditions (Fig. 5c, bottom left). These cytokines thus triggered similar aspects of the epigenetic potential in spleen endothelium as observed for systemic LCMV infection.
The results were qualitatively different in liver fibroblasts. We observed little overlap between the transcriptional response to the cytokines and to LCMV infection (Fig. 5d, top left), while there was still a strong association with the epigenetic potential (Fig. 5d, bottom left). Cytokine administration induced genes involved in metabolic processes (Cmpk2, Pofut2) as well as regulators of vesicle or membrane trafficking and antigen presentation (Bloc1s6, H2-Eb1, Rabac1 and Sar1a) (Fig. 5d, right, Supplementary Table 11). Thus, the administration of cytokines triggered different aspects of the epigenetic potential in liver fibroblasts compared with LCMV infection.
In summary, structural cells responded in cell-type-specific and organ-specific ways to in vivo stimulation with individual cytokines, which allowed us to functionally dissect the effects of systemic LCMV infection and provides further validation of the observed epigenetic potential for immune gene activity in structural cells.
Discussion
Structural cells, including endothelium, epithelium and fibroblasts, are important yet underappreciated contributors to mammalian immune responses. Here, we systematically investigated immune gene regulation in these non-haematopoietic cell types, applying multi-omics profiling and integrative bioinformatic analysis to three types of structural cells purified from twelve different organs of the mouse. We observed unexpectedly strong and densely regulated expression of immune genes, both under homeostatic conditions and in response to immunological challenges (systemic viral infection with LCMV, in vivo cytokine treatment).
Immunologists tend to consider structural cells mainly for their barrier function (epithelium, endothelium) and their role as connective tissue (fibroblasts), although important research has identified much more complex roles of structural cells in mammalian immunity23–31. However, comparative multi-organ investigations of structural cells have been lacking. We sought to close this gap in our understanding of mammalian immunity by evaluating the immune regulation of structural cells in a systematic, genome-wide and organism-scale way. We identified three main lines of evidence that highlight the immune-regulatory potential of structural cells.
First, we observed extensive cell-type-specific and organ-specific regulation of genes that influence the capabilities of structural cells to engage in predicted interactions with haematopoietic immune cells. On the basis of our transcriptome data, we inferred an initial network of potential cell–cell interactions between structural cells and haematopoietic cells. It will be an important future goal to dissect these cell–cell interactions and to untangle the precise chain of command in the immunological communication of structural cells and haematopoietic cells.
Second, our genome-wide analysis of chromatin accessibility in structural cells uncovered not only a regulatory basis of their immune functions, but also an initial assessment of the transcriptional regulators that confer cellular identity to endothelial cells, epithelial cells and fibroblasts across 12 organs. Future studies could pursue genetic manipulation of these candidate regulators in a cell-type-specific and organ-specific way, investigating the effect on immune-regulatory functions, epigenetic landscapes and cellular identity.
Third, our integrative analysis of gene expression and chromatin accessibility identified an epigenetically encoded immune potential in structural cells, constituted by genes that were lowly expressed under homeostatic conditions but epigenetically poised for much higher expression. These genes were enriched for immune functions, and they were preferentially upregulated in response to LCMV infection and cytokine administration in vivo. We thus conclude that structural cells are epigenetically pre-programmed for a swift response to a variety of immunological challenges. It will be interesting to explore how the epigenetic potential of structural cells responds to other stimuli and whether it can be modulated for therapeutic purposes, for example in the context of autoimmune diseases or the tumour microenvironment.
In conclusion, our study provides a comprehensive characterization of immune gene regulation in structural cells, and an initial step towards the systematic, organism-scale dissection of immune functions beyond haematopoietic cells. To emphasize the importance of structural cells for mammalian immunity, we tentatively propose the term ‘structural immunity’ for the study of immune functions in the non-haematopoietic, structural cell populations of the body. We see our study and large-scale dataset as a starting point, reference atlas, and a collection of hypotheses for systematic as well as mechanistic explorations in this emerging area of research.
Methods
Mice
C57BL/6J mice were bred and maintained under specific pathogen free conditions at the Institute of Molecular Biotechnology (IMBA) of the Austrian Academy of Sciences in Vienna (Austria). In vivo experiments (systemic viral infection, cytokine treatments) were performed under specific pathogen free conditions at the Anna Spiegel Research Building of the Medical University of Vienna (Austria). Age-matched male mice (8 to 13 weeks old) were used in all experiments. For the characterization of structural cells under homeostatic conditions, mice were sacrificed without any prior treatment. For the systemic viral infection experiments, mice were intravenously infected with 2x106 focus-forming units of lymphocytic choriomeningitis virus (LCMV) strain Clone 13 32, 33 and killed on day 8 after infection. For the cytokine treatment experiments, mice were intravenously injected with 100 μg/kg of the following recombinant cytokines: IFN-α, IFN-γ, IL-3, IL-6, TGF-β or TNF (all from BioLegend) and sacrificed 2 hours after cytokine injection. All mouse experiments were performed in individually ventilated cages according to the respective animal experiment licenses (BMWFW-66.009/0199-WF/V/3v/2015 and BMWFW-66.009/0361-WF/V/3b/2017) approved by the institutional ethical committees and the institutional guidelines at the Department for Biomedical Research of the Medical University of Vienna. Samples numbers are listed in the figure legends. No statistical methods were used to predetermine sample size. The experiments were not randomized, and investigators were not blinded to treatment status during experiments and outcome assessment.
Standardized sample collection and organ dissociation
Different surface markers and sorting schemes were previously used to purify endothelium, epithelium, and fibroblasts in individual organs, whereas our systematic comparison study required standardized cell purification across organs. We therefore tested multiple surface markers across organs and optimized a sorting scheme that produced consistent results for all 12 investigated organs, while excluding cell types that were detectable only in one or a few tissues (most notably lymphatic endothelial cells). The experimental workflow followed the recommendations of the Immunological Genome Project34 regarding sample collection schedule, antibody staining, and sample pooling. At least three same-sex littermate mice were pooled for each biological replicates for homeostatic conditions and for LCMV infection. For the in vivo cytokine treatments, we used individual mice as biological replicates to reduce the total number of mice. Standardized organ harvesting and dissociation protocols were established to obtain single-cell suspension for subsequent cell purification by fluorescence activated cell sorting (FACS). The same digestion solution was used for all organs, to avoid organ-specific technical confounders. The workflow is described in detail below.
Brain
Following decapitation, the skull was cut longitudinally with scissors, and the cranium was opened with tweezers. Both brain hemispheres were carefully collected and placed into cell culture dishes containing cold PBS supplemented with 0.1% BSA (PBS + BSA). White matter was manually removed, the tissue shredded with scissors and added to a 50 ml tube containing 15 ml cold Accumax (Sigma-Aldrich) digestion solution and incubated for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer (Sigma-Aldrich) followed by centrifugation at 300 g for 5 min at 4 °C. Myelin was removed by using density gradient centrifugation. Cells were recovered at the interface between a 80% Percoll layer and a 30% Percoll layer and washed in PBS + BSA to remove excess Percoll.
Caecum, large intestine, small intestine
Luminal contents were removed. The organs were cut longitudinally with scissors, rinsed several times in PBS + BSA to remove mucus, then cut into 0.5 cm pieces and placed in 50 ml tubes containing 20 ml pre-warmed (37 °C) RPMI containing 10% FCS and 5 mM EDTA (RPMI + FCS + EDTA). Samples were incubated for 25 min at 37 °C in a shaking incubator at 200 rpm. Supernatant was collected, samples were resuspended once again in 20 ml pre-warmed RPMI + FCS + EDTA, and incubated for 25 min at 37 °C in a shaking incubator at 200 rpm. These wash steps were performed to dissociate epithelial cells. After the second incubation, supernatants were collected and combined, followed by digestion of the samples in 15 ml cold Accumax for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer. Organ homogenates were combined with the epithelial cell fractions, filtered twice through a 100 μm cell strainer and washed in cold PBS + BSA.
Heart, kidney, lung, spleen, thymus
Organs were rinsed with cold PBS + BSA and shredded with scissors. Tissue fragments were placed into 50 ml tubes containing 20 ml cold Accumax digestion solution and incubated for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer and filtered through a 100 μm cell strainer. After centrifugation, 2 ml cold ACK lysis buffer (Thermo Fisher Scientific) was added for 3 min to lyse red blood cells and the reaction stopped by adding 20 ml of cold PBS + BSA. Supernatants were filtered twice through 100 μm cell strainer and washed once to remove residual ACK lysis buffer.
Liver
Three lobes were removed, rinsed with cold PBS + BSA, and shredded with scissors. Tissue fragments were placed into a 50 ml tube containing 20 ml cold Accumax digestion solution and incubated for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer and filtered through a 100 μm cell strainer. Hepatocytes were removed using density gradient centrifugation. Cells recovered at the interface between an 80% Percoll layer and a 30% Percoll layer were washed in PBS + BSA to remove excess Percoll.
Lymph nodes
Cervical, axillary, and inguinal lymph nodes were combined, carefully pinched with tweezers, and rinsed several times with cold PBS + BSA to release hematopoietic cells. Tissue fragments were placed into a 50 ml tube containing 10 ml cold Accumax digestion solution and incubated for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer and filtered twice through a 100 μm cell strainer.
Skin
Ears were harvested at the base, and the subcutaneous fat layer scrapped off with a scalpel. Tissue fragments were then shredded with scissors, placed into 50 ml tubes containing 15 ml of Accumax digestion solution, and incubated for 45 min at 37 °C while shaking at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer and filtered twice through a 100 μm cell strainer.
Organ-specific sample collection and organ dissociation
Large intestine
The organ was removed and processed as described35. Briefly, luminal contents were removed, the large intestine cut longitudinally with scissors, rinsed several times in PBS + BSA to remove mucus, then cut into 0.5 cm pieces and placed in 50 ml tubes containing 20 ml pre-warmed (37 °C) RPMI containing 10% FCS and 5 mM EDTA (RPMI + FCS + EDTA). Samples were incubated for 40 min at 37 °C in a shaking incubator at 200 rpm after. Supernatant was collected, samples resuspended once again in 20 ml pre-warmed RPMI + FCS + EDTA, and incubated for 20 min at 37 °C in a shaking incubator at 200 rpm. These wash steps were performed to dissociate epithelial cells. After the second incubation, supernatants were collected and combined, followed by incubating samples in RPMI containing 10% FCS and 15 mM HEPES (RPMI + FCS + HEPES) for 10 min at room temperature. Supernatant was discarded and samples digested in RPMI + FCS + HEPES containing 100 U/ml collagenase from Clostridium histolyticum (Sigma) for 1 hour at 37 °C in a shaking incubator at 200 rpm. Remaining tissue fragments were processed with a Dounce homogenizer. Organ homogenates were combined with the epithelial cell fractions, filtered twice through 100 μm cell strainers and washed in cold PBS + BSA.
Lung
The organ was removed and processed as described36. Briefly, the organ was cut into 0.5 cm pieces and placed in gentleMACS C tubes (Miltenyi) containing 160 U/mL collagenase type 1 (Gibco) and 12 U/mL DNAse 1 (Sigma) in RPMI containing 5% FCS, and dissociated using a gentleMACS Dissociator (Miltenyi; program m_lung_01). After incubation at 37°C for 35 min in a shaking incubator at 170 rpm, digested samples were homogenized using the gentleMACS Dissociator (program m_lung_02). Subsequently, cell suspensions were filtered through 70 μM cell strainers and centrifuged for 5 min at 4 °C at 300g. After centrifugation, 1 ml cold ACK lysis buffer (Thermo Fisher) was added for 5 min to lyse red blood cells and the reaction stopped by adding 20 ml of cold PBS + BSA. Supernatants were filtered twice through 100 μm cell strainer and washed once to remove residual ACK lysis buffer.
Liver
Mice were anesthetized (ketamine/xylazine 1:3, 0.1ml/10g mouse, Vetoquinol) before cannulation of the liver and dissociation using a two-step perfusion protocol37. Briefly, the liver was perfused first with 20 mL HBSS (Gibco) containing 0.5 mM EGTA (Sigma) and afterward with 20 mL L15 medium (Gibco) containing 40 mg/L liberase (Roche) at a rate of 5 mL/min. Next, the liver was removed, placed in a Petri dish with 10 ml of the same liberase-containing medium, followed by removal of the liver capsule. Hepatocytes were removed from the resulting cell suspension using density gradient centrifugation. Cells recovered at the interface between an 80% Percoll layer and a 30% Percoll layer were washed in PBS + BSA to remove excess Percoll.
Flow cytometry and FACS
Single-cell suspensions were washed once with PBS containing 0.1% BSA and 5 mM EDTA (PBS + BSA + EDTA). Cells were then incubated with anti-CD16/CD32 (clone 93, BioLegend) to prevent nonspecific binding. Single-cell suspensions were then stained with different combinations of antibodies against CD45 (PerCP-Cy5.5, clone 30-F11), TER-119 (PerCP-Cy5.5, clone TER-119), podoplanin (PE, clone 8.1.1), Ep-CAM (Pe-Cy7, clone 8.8), lymphotoxin beta receptor (APC, clone 5G11), CD31 (FITC, clone MEC13.3), CD90.2 (AF700, clone 30-H12), CD106 (AF647, clone 429), CD144 (BV421, clone BV13), CD324 (APC-Cy7, clone DECMA-1) (all from BioLegend), CD140a (BV605, clone APA5, BD Bioscience), MAdCAM1 (BV421, clone MECA-367, BD Bioscience), and LYVE1 (eFlour 660, clone ALY7, Thermo Fisher Scientific) for 30 min at 4 °C, followed by two washes with PBS + BSA + EDTA. Dead cells were stained either by adding Zombie Red Fixable Viability Dye or Zombie Aqua Fixable Viability Dye (both from BioLegend) immediately prior to flow cytometry characterization or cell sorting. For flow cytometry, cells were acquired with an LSRFortessa (BD Biosciences) cell analyzer using the outlined gating strategies (Extended Data Fig. 1a, 2a, 8a). For FACS, cells were sort-purified with a MoFlo Astrios (Beckman Coulter) or SH800 (Sony) using the outlined gating strategies (Extended Data Fig. 1a, 8a). Data analysis was performed using the FlowJo software (Version 10.5.3, Tree Star).
Transcriptome profiling by Smart-seq2
Smart-seq2 was performed as previously described14, starting from low-input bulk samples. In each experiment, a maximum of 200 cells were sort-purified and deposited in 96-well plates containing 4 μl lysis buffer (1:20 solution of RNase Inhibitor (Clontech) in 0.2% v/v Triton X-100 (Sigma-Aldrich)), spun down, and immediately frozen at -80°C. Reverse transcription was performed using SuperScript II (Invitrogen) followed by PCR amplification with KAPA HiFi HotStart Ready Mix (Kapa Biosystems). cDNA amplification was followed by two rounds of SPRI (Beckman Coulter) purification, and cDNA concentration was measured with a Qubit fluorometer (Life Technologies). Library preparation was conducted on 1 ng of cDNA using the Nextera XT DNA Sample Preparation Kit (Illumina), followed by SPRI (Beckman Coulter) size selection. Libraries were sequenced by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 3000/4000 platform and the 50-bp single-end configuration. Transcriptome profiling by Smart-seq2 was done in three biologically independent experiments. Sequencing statistics are provided in Supplementary Table 1.
Chromatin accessibility mapping by ATAC-seq
ATAC-seq was performed as previously described15, 38, with minor adaptations. In each experiment, a maximum of 50,000 sort-purified cells were collected at 300 g for 5 min at 4 °C. After centrifugation, the pellet was carefully resuspended in the transposase reaction mix (12.5 μl 2xTD buffer, 2 μl TDE1 (Illumina), 10.25 μl nuclease-free water, and 0.25 μl 1% digitonin (Promega)) for 30 min at 37 °C. Following DNA purification with the MinElute kit eluting in 11 μl, 1 μl of eluted DNA was used in a quantitative PCR (qPCR) reaction to estimate the optimum number of amplification cycles. The remaining 10 μl of each library were amplified for the number of cycles corresponding to the Cq value (i.e., the cycle number at which fluorescence has increased above background levels) from the qPCR. Library amplification was followed by SPRI (Beckman Coulter) size selection to exclude fragments larger than 1,200 bp. DNA concentration was measured with a Qubit fluorometer (Life Technologies). Library amplification was performed using custom Nextera primers15. Libraries were sequenced by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 3000/4000 platform and the 50-bp single-end configuration. Chromatin accessibility mapping by ATAC-seq was done in two biologically independent experiments. Sequencing statistics are provided in Supplementary Table 1.
Epigenome mapping by ChIPmentation for H3K4me2
ChIPmentation was performed as previously described16, 39, with minor adaptions. In each experiment, a maximum of 50,000 sort-purified cells were washed once with PBS and fixed with 1% paraformaldehyde for 10 min at room temperature. Glycine (0.125 M final concentration) was added to stop the reaction. Cells were collected at 500 g for 10 min at 4 °C (subsequent work was performed on ice and used cooled buffers and solutions unless otherwise specified) and washed once with ice-cold PBS supplemented with 1 mM phenylmethyl sulfonyl fluoride (PMSF). After centrifugation, cells were lysed in sonication buffer (10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 0.25% SDS, 1x protease inhibitors (Sigma-Aldrich), and 1 mM PMSF) and sonicated (Covaris S220) for 30 min in a microTUBE until the size of most fragments was in the range of 200 to 700 bp. Following sonication, the lysate was adjusted to RIPA buffer conditions (final concentration: 10 mM Tris-HCl pH 8.0, 1 mM EDTA pH 8.0, 140 mM NaCl, 1% Triton X-100, 0.1% SDS, 0.1% sodium deoxycholate, 1x protease inhibitors (Sigma-Aldrich), and 1 mM PMSF).
For each immunoprecipitation, 10 μl magnetic Protein A (Life Technologies) were washed twice and resuspended in PBS supplemented with 0.1% BSA. 1 μg of antibody recognizing H3K4me2 (Aigma-Aldric, clone AW30) was added and bound to the beads by rotating overnight at 4 °C. Beads were added to the sonicated lysate and incubated for 2 h at 4 °C on a rotator followed by washing the beads once with RIPA low-salt buffer (10 mM Tris-HCl, 150 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1 mM EDTA), once with RIPA high-salt buffer (10 mM Tris-HCl, 500 mM NaCl, 0.1% SDS, 0.1% sodium deoxycholate, 1% Triton X-100, and 1 mM EDTA), once with RIPA lithium-chloride buffer (10 mM Tris-HCl, 250 mM LiCl, 0.5% IGEPAL CA-630, 0.5% sodium deoxycholate, and 1 mM EDTA), and once with Tris-Cl pH 8. Bead-bound chromatin was then resuspended in tagmentation mix (5 μl 5xTD buffer, 1 μl TDE1 (Illumina), 19 μl nuclease-free water) and incubated for 10 min at 37 °C.
After tagmentation, the beads were washed once with RIPA and once with cold Tris-Cl pH 8. Bead bound tagmented chromatin was resuspended in 10.5 μl 20 mM EDTA and incubated for 30 min at 50 °C. Then, 10.5 μl 20 mM MgCl2 as well as 25 μl pre-activated 2x KAPA HiFi HotStart Ready Mix (incubation at 98 °C for 45 s, (Kapa Biosystems) were added and incubated for 5 min at 72 °C, followed by incubation for 10 min at 95 °C. Beads are magnetized and 2 μl of each library were amplified in a 10 μl qPCR reaction containing 0.8 mM primers, SYBR Green, and 5 μl Kapa HiFi HotStart ReadyMix to estimate the optimum number of enrichment cycles, using the following program: 72 °C for 5 min, 98 °C for 30 s, 24 cycles of 98 °C for 10 s, 63 °C for 30 s, 72 °C for 30 s, and a final elongation at 72 °C for 1 min. Kapa HiFi HotStart ReadyMix was incubated at 98 °C for 45 s before preparation of all PCR reactions (qPCR and final enrichment PCR), in order to activate the hot-start enzyme for successful nick translation at 72 °C in the first PCR step.
Final enrichment of the libraries was performed in a 50 μl reaction using 0.75 mM primers (custom Nextera primers as described for ATAC-seq) and 25 ml Kapa HiFi HotStart ReadyMix. Libraries were amplified for the for the number of cycles corresponding to the Cq value determined in the qPCR reaction. Enriched libraries were purified using SPRI AMPure XP beads. To prepare input control samples, 3μl of 50mM MgCl2 was added to 15μl sonicated lysate (pool of 5μl of endothelium, epithelium, and fibroblasts lysates from the same organ) to neutralize the EDTA in the SDS lysis buffer; 20μl of tagmentation buffer and 1μl transposase (Illumina) was added, and samples were incubated at 37°C for 10min; chromatin was purified with MinElute PCR purification kit (Qiagen), and 22.5μl of the purified transposition reaction were combined with 25μl of PCR master mix and 0.75 mM primers (custom Nextera). Control libraries were amplified as described above. Libraries were sequenced by the Biomedical Sequencing Facility at CeMM using the Illumina HiSeq 3000/4000 platform and the 50-bp single-end configuration. Epigenome mapping by ChIPmentation was done in two biologically independent experiments, with two exceptions: for endothelium from lymph node and fibroblasts from thymus, only one high-quality profile could be obtained). Sequencing statistics are provided in Supplementary Table 1.
Quantification of LCMV viral RNA
In the LCMV infection experiments, samples were collected at day 8 after infection and snap-frozen in liquid nitrogen. For detection of LCMV viral RNA, pieces of the organs were homogenized with a TissueLyser II (Qiagen), RNA was extracted using QIAzol lysis reagent (Qiagen), and reverse transcription was done using random primers and the First Strand cDNA Synthesis Kit (Thermo Fisher Scientific) according to the manufacturer’s instructions. The expression of the viral gene that encodes the LCMV nucleoprotein (NP) was measured with qPCR using TaqMan Fast Universal PCR Mastermix (Thermo Fisher Scientific) and previously published primers and probes40. The qPCR data were normalized against a reference comprising five established housekeeping genes (TaqMan Gene Expression Assays, Thermo Fisher Scientific): Arf1 (Mm01946109_uH), Rpl37 (Mm00782745_s1), Rab1b (Mm00504046_g1), Ef1a (Mm04259522_g1), and Gapdh (4352339E), in order minimize biases introduced by potential tissue-specific expression of housekeeping genes41, 42. Importantly, this qPCR assay estimates infection levels based on LCMV viral RNA in bulk tissue, and it cannot account for organ-specific differences in the relative frequencies of cells that are susceptible to LCMV infection. The results should therefore be regarded as an indication (rather than as a precise quantitative measure) of the degree to which structural cells in different organs are directly and indirectly affected by the LCMV infections.
RNA-seq data processing and quality control
The RNA-seq data were processed and quality-controlled using established bioinformatics software and custom analysis scripts. Specific emphasis was put on ensuring high purity of the structural cell transcriptomes, while minimizing the risk and impact of potential contaminations (e.g., due to cell-free RNA released by dying cells or as the result of potential impurities during FACS purification of the structural cell populations).
Raw reads were trimmed using trimmomatic (version 0.32) and aligned to the mouse reference genome (mm10) using HISAT2 (version 2.1.0). Gene expression was quantified by counting primary alignments to exons using the function summarizeOverlaps from the GenomicAlignments package (version 1.6.3) in R (version 3.2.3). Gene annotations were based on the Ensembl GENCODE Basic set (genome-build GRCm38.p6).
To detect and remove potential biases arising from contaminating RNA of hematopoietic immune cells among the sorted structural cell populations, we screened all transcriptome profiles for gene expression signatures associated with eight types of hematopoietic immune cells. To that end, we derived gene signatures indicative of B cells, T cells, NK cells, natural killer T cells, macrophages, monocytes, dendritic cells, and neutrophils from a recent mouse single-cell expression atlas19. In addition, a TCR signature was defined by the genes Cd3d, Cd3e, Cd3g, Lat, Lck, Vav1, Tbx21, and Zap70. For each of these gene signatures, aggregated expression values were calculated as follows. First, raw read counts were converted to counts per million (CPM) and log2 transformed using the function voom from the limma package (version 3.26.9) in R. Second, voom-converted values were normalized to z-scores, followed by averaging across all genes in a given gene signature. Samples with detectable contamination by hematopoietic immune cells were automatically discarded, and replacement samples were generated until three uncontaminated samples were available for each cell type in each organ.
As an additional precaution, we computationally corrected for residual contamination by RNA from hematopoietic immune cells, regressing out the gene signatures of hematopoietic immune cells from the matrix of structural cell transcriptomes using the function removeBatchEffect from the limma package in R. With this procedure, we generated corrected log2 counts per million (log2 CPM), which we used in all further analyses unless otherwise stated. Finally, for the comparisons between individual genes, we normalized the corrected expression values by gene length, thereby generating reads per kilobase per million reads (RPKM) values.
Bioinformatic analysis of cell-type-specific and organ-specific gene expression
The processed and quality-controlled RNA-seq profiles of structural cells were analyzed for characteristic differences in gene expression across cell types and organs, and enrichment analysis against public reference data was used to obtain an initial annotation of the identified gene signatures (Fig. 1d, Extended Data Fig 3).
Cell-type-specific and organ-specific marker genes were identified using a two-step procedure. First, we performed pairwise differential expression analysis for each of the three type of structural cells, comparing them across organs. Second, based on the resulting pairwise comparisons, we identified for each organ those genes that were upregulated in a given cell type compared to at least five other organs. Pairwise comparisons between organs were performed using the limma package in R, separately for each of the three structural cell types. Significantly differential genes were selected based on statistical significance (adjusted p-value < 0.05), average expression (log2 CPM > -1), and sequencing coverage (median number of reads greater than 10 in the group with stronger signal). Based on these pairwise comparisons, we counted the total number of times each gene was upregulated in a specific organ compared to all other organs. Genes that were upregulated in comparison to five or more other organs were selected as marker genes of the corresponding organ.
The identified cell-type-specific and organ-specific marker genes were subjected to enrichment analysis with Enrichr (http://amp.pharm.mssm.edu/Enrichr/), using three Enrichr libraries: KEGG_2019_Mouse, GO_Biological_Process_2018, and Human_Gene_Atlas. The enrichment analysis provided an initial biological annotation of the identified marker genes (Fig. 1d) and validated their enrichment for previously reported cell-type-specific and organ-specific marker genes expressed in structural cells (Extended Data Fig 3a). For further confirmation in an independent reference dataset19, we normalized the expression values for each gene within each organ in our dataset, followed by averaging across genes for each set of marker genes from the reference dataset (Extended Data Fig 3b). The resulting structural cell signatures thus reflect gene expression in one cell type compared to the other cell types in the same organ. Finally, we normalized these structural cell gene signatures across organs and cell types to z-scores, resulting in cell-type-specific and organ-specific gene signatures.
Bioinformatic inference of cell-cell interactions
To dissect the cell-type-specific and organ-specific crosstalk of structural cells with hematopoietic immune cells, we quantified the enrichment of known receptor-ligand pairs among the identified marker genes (Fig. 2a). Moreover, we assessed the expression of genes encoding receptors and ligands (Fig. 2b) and aggregated receptor and ligand gene signatures (Fig. 2c). Given the scale of our study, it was not possible to validate the inferred cell-cell interactions experimentally; however, the identification of many well-established interactions and of biologically plausible differences between cell types and organs provides support for these predictions.
Cell-type-specific and organ-specific marker genes of structural cells were obtained from the RNA-seq data as described in the previous section. Marker genes of hematopoietic immune cells were downloaded from Supplementary Table 4 of a recent mouse single-cell expression atlas19; from Supplementary Table 2 of the Tabula Muris paper20, from Dataset_S02 (sheets 8, 12, 14 and 16) of a paper by the ImmGen consortium43, and from Supplementary Table 1 of a large-scale characterization of tissue-resident macrophages44. To aggregate immune cell genes into a consensus set, the union of all identified marker genes was taken.
Receptor-ligand pairs were downloaded from CellPhoneDB45 (as of 21 September 2018) and merged with additional receptor-ligand pairs extracted from Supplementary Data 2 of a recent paper46, retaining only literature-supported pairs. Mouse gene identifiers were mapped to human gene identifiers based on the NCBI HomolGene mapping (build 68) using only the unique mappings. Interactions formed by receptor complexes from CellPhoneDB were transformed to pairwise interactions between individual receptors and ligands, thus including an interaction between each member of a receptor complex and the respective ligand.
Based on these lists of marker genes and receptor-ligand pairs, we inferred potential cell-cell interactions for all pairs of one structural cell type and one hematopoietic immune cell type, quantifying the enrichment for known receptor-ligand pairs among all pairs of marker genes between the structural cell type and the hematopoietic cell type (Fig. 2a). First, we counted all pairs of marker genes for each pair of structural and immune cell types. Second, we calculated the fraction of these gene pairs that were annotated as receptor-ligand pairs. Third, we tested whether this fraction was greater than the fraction of annotated receptor-ligand pairs across all pairs of genes. Fisher’s exact test was used to obtain p-values and odds ratios, as implemented by the function fisher.test in R. P-values were adjusted for multiple testing using the Benjamini-Hochberg method, as implemented the function p.adjust in R. Finally, significantly enriched pairs (adjusted p-value < 0.05) of structural and immune cell types were connected by edges to generate a graph of cell-cell interactions.
In addition to testing for significant enrichment (as described in the previous paragraph), we calculated aggregated receptor and ligand gene signatures (Fig. 2c) based on a manually curated list of immune-related receptors and ligands (Supplementary Table 4). These gene signatures were derived by normalizing expression values for each gene and averaging across all genes in a given set of biologically related receptors and ligands.
Bioinformatic analysis of the Tabula Muris dataset
To assess the expression patterns of immune genes in structural cells in an independent dataset (Extended Data Fig. 5), we obtained single-cell RNA-seq data and t-SNE projections from the Tabula Muris website https://tabula-muris.ds.czbiohub.org/ (21 December 2018) and cell counts from Supplementary Table 1 of the corresponding paper20. Cell types were assigned based on the provided cell ontology class, with stroma cells and basal cells of epidermis both labeled as fibroblasts. Gene expression counts were converted to transcripts per million (TPM) values and log-transformed. Receptor and ligand gene signatures in the Tabula Muris data were based on immune-related receptors and ligands (Supplementary Table 4) and normalized log(TPM) values for each gene, which were averaged across all genes in a given set of biologically related receptors and ligands.
ATAC-seq and ChIPmentation data processing and quality control
The ATAC-seq and H3K4me2 ChIPmentation data were processed using well-established bioinformatics pipelines, followed by quality control following the same approach as for the RNA-seq data (described above). The two data types were processed separately and subsequently integrated as described in the next section.
Raw reads were trimmed with trimmomatic (version 0.32) and aligned to the mouse reference genome (mm10) using bowtie2 (version 2.2.4). Primary alignments with mapping quality greater than 30 were retained. ATAC-seq peaks were called using MACS (version 2.7.6) on each individual sample. H3K4me2 ChIPmentation peaks were called using MACS against the input controls (which were obtained by pooling input control data from the three types of structural cells in equal amounts for each organ). Peaks were aggregated into a list of consensus peaks using the function reduce of the package GenomicRanges (version 1.22.4) in R. Consensus peak that overlapped with blacklisted genomic regions (downloaded from http://mitra.stanford.edu/kundaje/akundaje/release/blacklists/mm10-mouse/) were discarded.
Quantitative measurements were obtained by counting reads within consensus peaks using the function summarizeOverlaps from the GenomicAlignments (version 1.6.3) package in R. Samples with detectable contamination by hematopoietic immune cells were identified in the same way as for the RNA-seq data, using epigenomic signals in promoter peaks to calculate immune cell signatures. Contaminated samples were automatically removed and replaced by new samples. Finally, we computationally corrected for residual contamination in the retained samples by regressed out epigenomic signatures of hematopoietic immune cells from the matrix of signal intensity values across peaks, using the function removeBatchEffect from the limma package in R.
Bioinformatic analysis of cell-type-specific and organ-specific transcription regulation
The processed and quality-controlled RNA-seq, ATAC-seq, and H3K4me2 ChIPmentation data were analyzed for differences in transcription regulation across cell types and organs (Fig. 3a-c). We integrated all three data types to derive sets of genomic regions with characteristic activity patterns (marker peaks), we performed motif enrichment analysis for marker peaks, and we inferred a gene-regulatory network by connecting enriched transcriptional regulators to their target genes. For gene-regulatory network inference, we utilized DNA sequence motifs to connect transcriptional regulators to regulatory regions, and genomic proximity to connect regulatory regions to target genes. To make these coarse approximations more robust and interpretable, we used the ATAC-seq data to exclude motifs that are most likely inactive in the investigated cell types.
First, cell-type-specific and organ-specific marker peaks were identified in analogy to the RNA-seq based identification of marker genes (described above), separately for the ATAC-seq and H3K4me2 ChIPmentation data. For each of the three cell types, we performed pairwise comparisons between organs using the limma package in R. Differential peaks were selected based on statistical significance (adjusted p-value < 0.05), signal intensity (log2 CPM > -1), and sequencing coverage (median number of reads of at least 10 in the group with stronger signal). We then counted – separately for the two data types – the total number of times each peak showed increased signal in a specific organ compared to all other organs. Peaks that were upregulated compared to five or more other organs were selected as marker peaks of the corresponding organ.
Second, we derived a gene-linked list of marker peaks that were marked as promoter-associated peaks, enhancer-associated peaks, and other peaks based on gene annotations as well as the RNA-seq and H3K4me2 ChIPmentation data (for enhancers). To that end, the ATAC-seq peaks were linked to all genes whose transcription start site was located within five kilobases, using the function annotatePeakInBatch of the package ChIPpeakAnno (version 3.4.6) package in R. They were annotated as promoter-associated if they were located within 200 bp from a transcription start site of the respective gene, or as enhancer-associated if they were located within one to five kilobases from a transcription start site, showed a correlation of ATAC-seq and RNA-seq signal greater than 0.3, and overlapped with an H3K4me2 ChIPmentation marker peak.
Third, we performed motif analysis on the gene-linked list of marker peaks to connect regulatory regions to transcriptional regulators. The HOMER software tool (version 4.9.1, http://homer.ucsd.edu/homer/motif/) was used to identify regulator binding motifs in the marker peaks and to determine regulator enrichment. This enrichment analysis was performed separately for all peaks, for promoter-associated peaks, and for enhancer-associated peaks, using the HOMER function findMotifsGenome with a background set consistent of all gene-linked peaks. To avoid biases due to differences in peak size, all regions were standardized to 500 base pairs around the center of the peak. Transcriptional regulators were labeled as significant based on statistical significance (adjusted p-value < 0.001) and the strength of the enrichment (log2 odds ratio > 1.5).
Fourth, we inferred a gene-regulatory network by connecting transcriptional regulators with their target genes. This analysis included only those regulators that were enriched in the motif analysis of gene-linked marker peaks (described in the previous paragraph). No regulators were excluded based on low or undetectable RNA expression levels, given that transcription factors can play a relevant regulatory role despite low expression levels. The gene-regulatory network was constructed as follows: (i) We linked transcriptional regulators to their target peaks using the HOMER function annotatePeaks; (ii) we used the list of gene-linked marker peaks to map these peaks to their target genes; (iii) we constructed the final network based on the links between regulators and target peaks, and between peaks and target genes, using the package igraph (version 1.1.2) in R.
Fifth, we analyzed the inferred gene-regulatory network by determining the network similarity between all pairs of transcriptional regulators using the function similarity from the igraph package in R (with inverse log weighted similarity). Similarities among transcriptional regulators were visualized by multidimensional scaling using the function cmdscale in R, based on similarity measures normalized to a minimum of zero and maximum of one, and converted to distance measures by taking one minus the normalized similarity value.
Epigenetic potential for immune gene activation
To identify genes with unrealized epigenetic potential (i.e., genes whose chromatin state indicates much higher expression than observed under homeostatic conditions), we compared chromatin accessibility (ATAC-seq signal) of promoter regions with the corresponding genes’ expression levels (RNA-seq signal) (Fig. 3d-g).
First, RNA-seq and ATAC-seq read counts were combined into a single table, converted to log2(CPM) values, and quantile normalized using the function voom of the limma package in R. Second, we removed systematic differences between the two assays for each gene using the function removeBatchEffect of the limma package in R. Third, we compared normalized RNA-seq and ATAC-seq signal intensities for each gene, organ, and cell type using differential analysis from the limma package in R. This analysis used biological replicates for RNA-seq and ATAC-seq to statistically assess for a given combination of organ and cell type whether the ATAC-seq signal was disproportionately greater (compared to other organs and cell types) than the RNA-seq signal. Based on this analysis, genes were selected as significant based on the observed difference (log2 fold change > 0.7), statistical significance (adjusted p-value < 0.05), and mean signal intensity (normalized log2 CPM > 0). For ATAC-seq, we further required a minimum sequencing coverage (median number of reads > 5). In addition, we calculated the enrichment of genes with immune functions among genes with unrealized epigenetic potential. To that end, we selected the 200 genes with the greatest difference between ATAC-seq to RNA-seq signal in each organ and cell type, and we calculated their enrichment for our manually curated immune-related gene sets (Supplementary Table 8) using Fisher’s exact test based on the function fisher.test in R.
Bioinformatic analysis of the LCMV infection experiments
To evaluate the epigenetic potential for immune gene activation in structural cells, we compared the RNA-seq profiles of structural cells under homeostatic conditions with those collected on day 8 after infection with LCMV, and we assessed the predictiveness of the unrealized potential for the LCMV-induced changes in gene expression (Fig. 4). This analysis was based on the hypothesis that LCMV infection – which is widely regarded as a systemic infection model – will affect structural cells in most or all investigated organs through direct effects (structural cells may get infected) as well as indirect effects (structural cells respond to aspects of tissue-specific and systemic infection). Our gene expression analysis likely reflects a superposition of several such effects.
First, we identified the genes that were upregulated upon LCMV infection. To that end, we compared samples collected under homeostatic conditions with those collected after LCMV infection, separately for each cell type and organ, using the limma package in R. Differentially expressed genes were selected based on statistical significance (adjusted p-value < 0.05), average expression (log2 CPM > 1), and sequencing coverage (median number of reads > 20 in the upregulated condition). Enrichment analysis was performed by comparing differential genes to our manually curated immune-related gene sets (Supplementary Table 8) using Fisher’s exact test with the function fisher.test in R.
Second, we evaluated whether genes with unrealized epigenetic potential under homeostatic conditions were preferentially upregulated in response to LCMV infection. For each cell type and organ, we split all genes into two groups based on their log2 fold change comparing ATAC-seq signal to RNA-seq signal: Those with unrealized potential (ATAC-seq signal greater than RNA-seq signal) and those without unrealized potential (RNA-seq signal greater than ATAC-seq signal). We then assessed whether the LCMV-induced changes in gene expression (log2 fold change comparing samples collected after LCMV infection to homeostatic conditions) was able to discriminate between the two groups defined by epigenetic potential. For visualization and quantitative comparison across cell types and organs, the data were plotted in diagrams that conceptually resemble receiver operating characteristic (ROC) curves, and the predictiveness of the unrealized potential for gene activation upon LCMV infection was quantified by area-under-the-curve (AUC) values.
Third, the activated potential was calculated for each cell type and organ as the percentage of genes with unrealized potential under homeostatic conditions (ATAC-seq signal greater than RNA-seq signal, p-value < 0.05) that were among the significantly upregulated genes upon LCMV infection (defined as described above).
Bioinformatic analysis of the cytokine treatment experiments
To dissect the in vivo effects of individual cytokines on structural cells (Fig. 5), we analyzed the transcriptional response to the treatment of mice with six cytokines administrated as recombinant proteins, and we obtained RNA-seq profiles (processed and quality-controlled as described above) for gene expression analysis.
First, we compared the gene expression of structural cells between cytokine-treated mice and mock-treated (PBS) controls using the limma package in R, separately for each cell type and organ. Differentially expressed genes were selected based on statistical significance (adjusted p-value < 0.05) and average expression (log2 CPM > 2).
Second, we compared the changes in gene expression upon cytokine treatment to the changes in gene expression upon LCMV infection. To that end, we correlated cytokine-related log fold changes to LCMV-related log fold changes across all genes, separately for each cell type, organ, and cytokine.
Third, we tested whether genes upregulated after cytokine treatment were enriched for genes with unrealized epigenetic potential. To that end, we calculated the enrichment of genes with unrealized epigenetic potential (defined as described above) among genes upregulated by cytokine treatment (adjusted p-value < 0.05, log fold change > 0), separately for each organ, cell type, and cytokine. Fisher’s exact test was used to calculate statistical significance with the function fisher.test in R.
Extended Data
Extended Data Fig. 1.
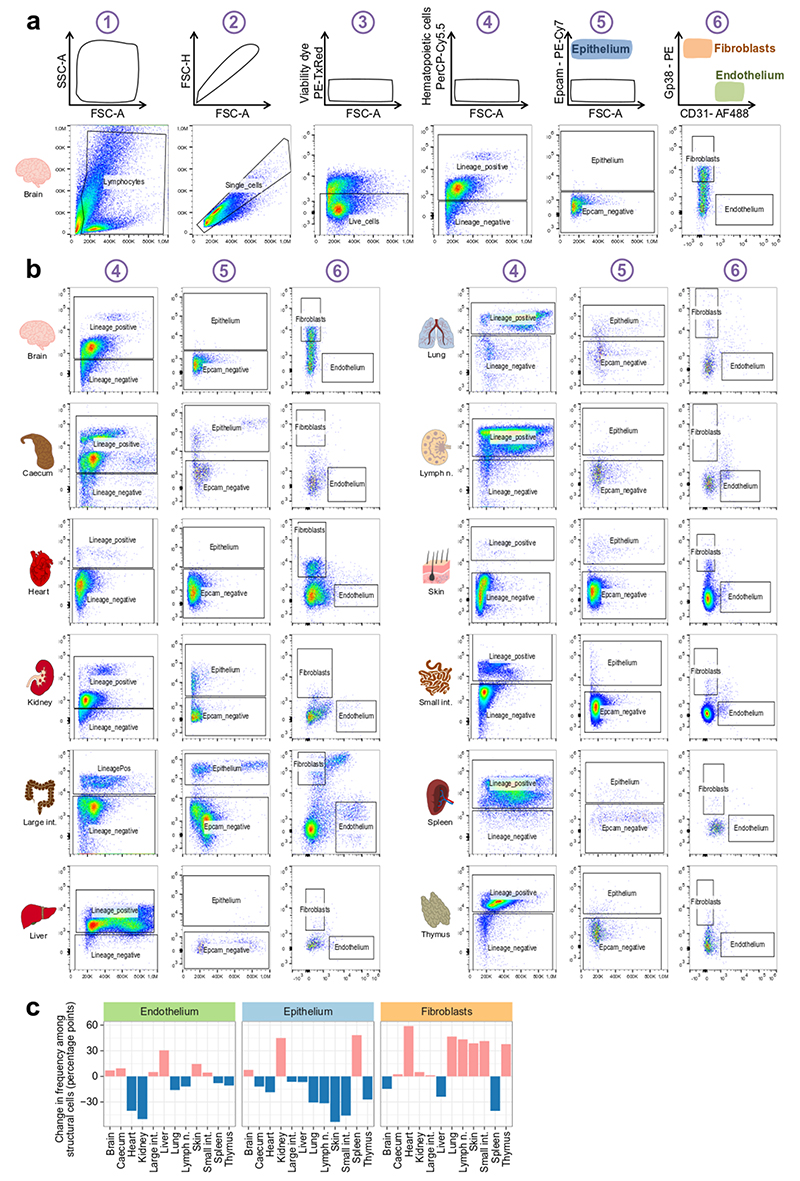
Standardized identification and purification of structural cells across 12 organs. a, Cell-type identification and cell sorting scheme (top row) and representative flow cytometry plots (selected from n=4 independent biologically replicates) in one representative organ (brain) under homeostatic conditions (bottom row). b, Representative plots (selected from n=4 independent biologically replicates) for gating steps 4 to 6 of the standardized cell-type identification and cell sorting scheme (panel a) across the 12 organs under homeostatic conditions. c, Relative frequencies of structural cell types among non-hematopoietic (CD45-) cells across 12 organs, for cell suspensions obtained by standardized organ dissociation. d, Relative frequencies of structural cell types among non-hematopoietic (CD45-) cells across three organs, for cell suspensions obtained by either standardized organ dissociation or organ-specific dissociation protocols. Shown are mean and s.e.m. values. Sample size: n = 4 (c) and n = 3 (d) independent biological replicates.
Extended Data Fig. 2.
Surface marker profiling of structural cells under homeostatic conditions. a, Gating strategy for the flow cytometry-based validation of the structural cell sorting scheme. Identification of structural cells starts with gating for intact cells (1), single cells (2), live cells (3) and non-haematopoietic cells (4). From the resulting non-haematopoietic (CD45-) cell population, potential epithelial cells (5.1) are gated for epithelial cell markers (5.2). Similarly, potential endothelial cells and fibroblasts (6.1, 6.2) are gated for endothelial cell markers (6.3) and fibroblast markers (6.4). b, Relative frequencies of potential structural cell types based on gates 5.2, 6.3 and 6.4 (from a), comparing the selected markers with alternative markers. c, Expression of the selected surface markers of structural cell types (top row) and potential alternative markers for cells gated as in Extended Data Fig. 1a. Shown are mean and s.e.m. values. Sample size (all panels): n = 3 independent biological replicates.
Extended Data Fig. 3.
Comparison of the structural cell transcriptomes to published reference data. a, Overlap of the identified cell-type-specific and organ-specific marker genes (derived from the RNA-seq experiments in the current study) with tissue-specific gene sets from a microarray-based expression atlas (two-sided Fisher’s exact test with multiple-testing correction). b, Gene expression across cell types and organs (from the current study) aggregated across marker genes of structural cell clusters in a single-cell RNA-seq atlas of the mouse19. c, Gene expression across cell types and organs (from the current study) plotted for a manually curated list of commonly used markers of structural cells. d, Hierarchical clustering of structural cells across cell types and organs based on the transcriptome profiles from the current study. Sample size (all panels): n = 3 independent biological replicates.
Extended Data Fig. 4.
Inference of cell-cell interactions across cell types and organs. a, Enrichment analysis for potential cell-cell interactions between structural cells and hematopoietic immune cells, based on gene expression of known receptor-ligand pairs (two-sided Fisher’s exact test with multiple-testing correction). For each combination of one structural cell type and one hematopoietic immune cell type, the analysis assesses whether all pairs of marker genes between the two cell types are enriched for annotated receptor-ligand pairs. b, Differently expressed genes across cell types and organs, based on a manually curated list of receptors and ligands (Supplementary Table 4). Sample size (all panels): n = 3 independent biological replicates.
Extended Data Fig. 5.
Analysis of immune gene expression among structural cells in an independent dataset. a, Relative frequencies of single-cell transcriptomes classified as endothelium, epithelium, and fibroblasts in selected organs according to the Tabula Muris dataset20. b, Expression of immune gene signatures in structural cells according to the Tabula Muris dataset, jointly normalized across all plots (for comparability). c, Expression of immune gene signatures in hematopoietic immune cells according to the Tabula Muris dataset, normalized in the same way as in panel b. d, Expression of selected immune genes in structural cells and in hematopoietic immune cells according to the Tabula Muris dataset. Sample size: n = 7 (all panels) independent biological replicates, comprising 4 male and 3 female mice.
Extended Data Fig. 6.
Analysis of transcription regulation in structural cells. a, Multidimensional scaling analysis of the similarity of chromatin profiles across cell types, organs, and replicates based on ATAC-seq (top) and H3K4me2 ChIPmentation (bottom). b, Correlation of chromatin profiles across cell types and organs for ATAC-seq (left) and H3K4me2 ChIPmentation (right). c, Transcriptional regulators of the inferred gene-regulatory network for structural cells, arranged by similarity using multidimensional scaling. d, Motif enrichment for transcriptional regulators among differential chromatin peaks, shown separately for each regulator (one-sided hypergeometric test with multiple-testing correction). e, Gene expression of the transcriptional regulators across cell types and organs (genes discussed in the text are in bold). Sample size (all panels): n = 2 independent biological replicates.
Extended Data Fig. 7.
Detection and analysis of genes with unrealized epigenetic potential. a, Scatterplot showing the correlation between chromatin accessibility in promoter regions and the corresponding gene expression levels in structural cells across cell types and organs. Genes with significant unrealized epigenetic potential (calculated as the difference between normalized ATAC-seq and RNA-seq signals) are highlighted in blue. b, Enrichment of immune-related gene sets among the genes with unrealized epigenetic potential (two-sided Fisher’s exact test with multiple-testing correction). Sample size (all panels): n = 2 independent biological replicates.
Extended Data Fig. 8.
Standardized identification and purification of structural cells after LCMV infection. a, Cell-type identification and cell sorting scheme (top row) and representative flow cytometry plots (selected from n=3 independent biologically replicates) in one representative organ (brain) after LCMV infection (bottom row). b, Representative plots (selected from n=3 independent biologically replicates) for gating steps 4 to 6 of the standardized cell-type identification and cell sorting scheme (a) across the 12 organs upon LCMV infection. c, Change in the relative frequenct of structural cells upon LCMV infection. size (all panels): n = 3 independent biological replicates.
Extended Data Fig. 9.
Analysis of differential gene expression in response to LCMV infection. a, Number of differentially expressed genes in structural cells upon LCMV infection (this includes not only immune genes but for example also genes associated with the substantial organ-specific tissue damage induced by LCMV infection). b, Correlation of the observed changes in gene expression upon LCMV infection across cell types and organs. c, Organ-specific viral load at day 8 of LCMV infection, measured by qPCR in whole-tissue samples collected from each organ (without FACS purification of individual cell types). Five reference genes were used for normalization and results were ranked across organs, in order to make the analysis robust toward tissue-specific differences in the expression of these housekeeping genes. However, the experimental results do not support an absolute quantification of viral load in each organ nor do they account for differences in the relative frequencies of cells that are susceptible to LCMV infection in each organ. d, Scatterplot illustrating the low correlation between the activated epigenetic potential and the measured viral load across cell types and organs. e, Network analysis (e) and enrichment analysis (f) of potential cell-cell interactions between structural cells and hematopoietic immune cells, inferred from gene expression of known receptor-ligand pairs upon LCMV infection (two-sided Fisher’s exact test with multiple-testing correction). For each combination of one structural cell type and one hematopoietic immune cell type, the analysis assesses whether all pairs of marker genes between the two cell types are enriched for annotated receptor-ligand pairs. Sample size: n=3 (all panels).
Extended Data Fig. 10.
Visualization of differential gene expression in response to in vivo cytokine treatments. The heatmap visualizes changes in the expression of genes associated with immune functions, plotted across cell types, organs, and cytokines (two-sided linear model with multiple-testing correction). Sample size: n = 3 independent biological replicates.
Extended Data Fig. 11.
Analysis of differential gene expression in response to in vivo cytokine treatments. a, Number of differentially expressed genes in response to the individual cytokine treatments. b, Gene expression for the known receptors involved in the response to the individual cytokine treatments, plotted across cell types and organs under homeostatic conditions. Sample size (all panels): n = 3 independent biological replicates.
Supplementary Material
Acknowledgments
We thank the Core Facility Flow Cytometry of the Medical University of Vienna for cell sorting service; the Biomedical Sequencing Facility at CeMM for assistance with next-generation sequencing; Sophie Zahalka and Sylvia Knapp for help and advice with the preparation of lung samples; Sarah Niggemeyer, Jenny Riede, Sabine Jungwirth, and Nicole Fleischmann for animal caretaking; and all members of the Bock lab for their help and advice. This work was conducted in the context of two Austrian Science Fund (FWF) Special Research Programme grants (FWF SFB F6102; FWF SFB F7001). T.K is supported by a Lise Meitner fellowship from the Austrian Science Fund (FWF M2403). N.F. is supported by a fellowship from the European Molecular Biology Organization (EMBO ALTF 241-2017). A.L. is supported by a DOC fellowship of the Austrian Academy of Sciences. A.B. is supported by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement n° 677006). C.B. is supported by a New Frontiers Group award of the Austrian Academy of Sciences and by an ERC Starting Grant (European Union’s Horizon 2020 research and innovation programme, grant agreement n° 679146).
Footnotes
Author contributions:
T.K. designed the project, performed experiments, analyzed data, and cowrote the manuscript. N.F designed and performed the bioinformatic analysis and cowrote the manuscript. V.F.-G., M.S., L.C.S., and A.N. contributed to sample collection and sequencing library preparation. A.L. contributed to the experimental design and performed in vivo experiments (LCMV infections; cytokine treatments). A.F.R. contributed bioinformatic software. A.B. contributed to the experimental design and supervised the in vivo experiments. C.B. supervised the project and cowrote the manuscript. All authors read, contributed to, and approved the final manuscript.
Additional information:
Competing interests: The authors declare no competing financial interests
Supplementary Information is available for this paper.
Data and code availability
Data availability: Raw and processed sequencing data (RNA-seq, ATAC-seq and H3K4me2 ChIPmentation) are available from the NCBI Gene Expression Omnibus (GEO) repository (accession number: GSE134663). In addition, the dataset is provided as an online resource on a supplementary website (http://structural-immunity.computational-epigenetics.org), which includes links to raw and processed sequencing data, further analysis results, and genome browser tracks for interactive visualization of the RNA-seq, ATAC-seq, and ChIPmentation profiles.
Code availability: The analysis source code underlying the final version of the paper is available from the Supplementary Website (http://structural-immunity.computational-epigenetics.org).
References
- 1.Kotas ME, Medzhitov R. Homeostasis, inflammation, and disease susceptibility. Cell. 2015;160:816–827. doi: 10.1016/j.cell.2015.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abbas AK, Lichtman AH, Pillai S. Cellular and Molecular Immunology. 2019 [Google Scholar]
- 3.Buckley CD, Barone F, Nayar S, Benezech C, Caamano J. Stromal cells in chronic inflammation and tertiary lymphoid organ formation. Annu Rev Immunol. 2015;33:715–745. doi: 10.1146/annurev-immunol-032713-120252. [DOI] [PubMed] [Google Scholar]
- 4.Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. 2007;7:803–815. doi: 10.1038/nri2171. [DOI] [PubMed] [Google Scholar]
- 5.Schleimer RP, Kato A, Kern R, Kuperman D, Avila PC. Epithelium: At the interface of innate and adaptive immune responses. J Allergy Clin Immunol. 2007;120:1279–1284. doi: 10.1016/j.jaci.2007.08.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pawlina W, Ross MH. Histology: A Text and Atlas, International Edition: With Correlated Cell and Molecular Biology. Wolters Kluwer Law & Business; 2019. [Google Scholar]
- 7.Humbert M, Hugues S, Dubrot J. Shaping of peripheral T cell responses by lymphatic endothelial cells. Front Immunol. 2016;7:684. doi: 10.3389/fimmu.2016.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Malhotra D, Fletcher AL, Turley SJ. Stromal and hematopoietic cells in secondary lymphoid organs: partners in immunity. Immunol Rev. 2013;251:160–176. doi: 10.1111/imr.12023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nowarski R, Jackson R, Flavell RA. The stromal intervention: Regulation of immunity and inflammation at the epithelial-mesenchymal barrier. Cell. 2017;168:362–375. doi: 10.1016/j.cell.2016.11.040. [DOI] [PubMed] [Google Scholar]
- 10.Perez-Shibayama C, Gil-Cruz C, Ludewig B. Fibroblastic reticular cells at the nexus of innate and adaptive immune responses. Immunol Rev. 2019;289:31–41. doi: 10.1111/imr.12748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Turley SJ, Cremasco V, Astarita JL. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat Rev Immunol. 2015;15:669–682. doi: 10.1038/nri3902. [DOI] [PubMed] [Google Scholar]
- 12.Davis MM, Tato CM, Furman D. Systems immunology: just getting started. Nat Immunol. 2017;18:725–732. doi: 10.1038/ni.3768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Villani AC, Sarkizova S, Hacohen N. Systems immunology: Learning the rules of the immune system. Annu Rev Immunol. 2018;36:813–842. doi: 10.1146/annurev-immunol-042617-053035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Picelli S, et al. Full-length RNA-seq from single cells using Smart-seq2. Nat Protoc. 2014;9:171–181. doi: 10.1038/nprot.2014.006. [DOI] [PubMed] [Google Scholar]
- 15.Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–1218. doi: 10.1038/nmeth.2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmidl C, Rendeiro AF, Sheffield NC, Bock C. ChIPmentation: fast, robust, low-input ChIP-seq for histones and transcription factors. Nat Methods. 2015;12:963–965. doi: 10.1038/nmeth.3542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Y, Li X, Hu H. H3K4me2 reliably defines transcription factor binding regions in different cells. Genomics. 2014;103:222–228. doi: 10.1016/j.ygeno.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 18.Su AI, et al. A gene atlas of the mouse and human protein-encoding transcriptomes. Proc Natl Acad Sci U S A. 2004;101:6062–6067. doi: 10.1073/pnas.0400782101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Han X, et al. Mapping the mouse cell atlas by microwell-seq. Cell. 2018;172:e1017. doi: 10.1016/j.cell.2018.02.001. [DOI] [PubMed] [Google Scholar]
- 20.Schaum N, et al. Single-cell transcriptomics of 20 mouse organs creates a Tabula Muris. Nature. 2018;562:367–372. doi: 10.1038/s41586-018-0590-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bock C, et al. Reference Maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Baazim H, et al. CD8+ T cells induce cachexia during chronic viral infection. Nat Immunol. 2019;20:701–710. doi: 10.1038/s41590-019-0397-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guerrero-Juarez CF, et al. Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat Commun. 2019;10:650. doi: 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kinchen J, et al. Structural remodeling of the human colonic mesenchyme in inflammatory bowel disease. Cell. 2018;175:372–386 e317. doi: 10.1016/j.cell.2018.08.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mizoguchi F, et al. Functionally distinct disease-associated fibroblast subsets in rheumatoid arthritis. Nat Commun. 2018;9:789. doi: 10.1038/s41467-018-02892-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Montoro DT, et al. A revised airway epithelial hierarchy includes CFTR-expressing ionocytes. Nature. 2018;560:319–324. doi: 10.1038/s41586-018-0393-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parikh K, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 28.Plasschaert LW, et al. A single-cell atlas of the airway epithelium reveals the CFTR-rich pulmonary ionocyte. Nature. 2018;560:377–381. doi: 10.1038/s41586-018-0394-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Haber AL, et al. A single-cell survey of the small intestinal epithelium. Nature. 2017;551:333–339. doi: 10.1038/nature24489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rodda LB, et al. Single-cell RNA sequencing of lymph node stromal cells reveals niche-associated heterogeneity. Immunity. 2018;48:1014–1028 e1016. doi: 10.1016/j.immuni.2018.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yan KS, et al. Non-equivalence of Wnt and R-spondin ligands during Lgr5+ intestinal stem-cell self-renewal. Nature. 2017;545:238–242. doi: 10.1038/nature22313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ahmed R, Salmi A, Butler LD, Chiller JM, Oldstone MB. Selection of genetic variants of lymphocytic choriomeningitis virus in spleens of persistently infected mice. Role in suppression of cytotoxic T lymphocyte response and viral persistence. J Exp Med. 1984;160:521–540. doi: 10.1084/jem.160.2.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bergthaler A, et al. Viral replicative capacity is the primary determinant of lymphocytic choriomeningitis virus persistence and immunosuppression. Proc Natl Acad Sci U S A. 2010;107:21641–21646. doi: 10.1073/pnas.1011998107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Immgen Consortium. Final immgen sorting SOP. [Accessed 24 March 2020];2019 http://www.immgen.org/Protocols/ImmGen%20Cell%20prep%20and%20sorting%20SOP.pdf.
- 35.Krausgruber T, et al. T-bet is a key modulator of IL-23-driven pathogenic CD4(+) T cell responses in the intestine. Nat Commun. 2016;7:11627. doi: 10.1038/ncomms11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saluzzo S, et al. First-breath-induced type 2 pathways shape the lung immune environment. Cell Rep. 2017;18:1893–1905. doi: 10.1016/j.celrep.2017.01.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lercher A, et al. Type I interferon signaling disrupts the hepatic urea cycle and alters systemic metabolism to suppress T cell function. Immunity. 2019;51:1074–1087 e1079. doi: 10.1016/j.immuni.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gustafsson C, De Paepe A, Schmidl C, Mansson R. High-throughput ChIPmentation: freely scalable, single day ChIPseq data generation from very low cell-numbers. BMC Genomics. 2019;20:59. doi: 10.1186/s12864-018-5299-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinschewer DD, et al. Innate and adaptive immune control of genetically engineered live-attenuated arenavirus vaccine prototypes. Int Immunol. 2010;22:749–756. doi: 10.1093/intimm/dxq061. [DOI] [PubMed] [Google Scholar]
- 41.Kouadjo KE, Nishida Y, Cadrin-Girard JF, Yoshioka M, St-Amand J. Housekeeping and tissue-specific genes in mouse tissues. BMC Genomics. 2007;8:127. doi: 10.1186/1471-2164-8-127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li B, et al. A Comprehensive Mouse Transcriptomic BodyMap across 17 Tissues by RNA-seq. Scientific Reports. 2017;7:4200. doi: 10.1038/s41598-017-04520-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shay T, et al. Conservation and divergence in the transcriptional programs of the human and mouse immune systems. Proc Natl Acad Sci U S A. 2013;110:2946–2951. doi: 10.1073/pnas.1222738110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lavin Y, et al. Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment. Cell. 2014;159:1312–1326. doi: 10.1016/j.cell.2014.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Vento-Tormo R, et al. Single-cell reconstruction of the early maternal-fetal interface in humans. Nature. 2018;563:347–353. doi: 10.1038/s41586-018-0698-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ramilowski JA, et al. A draft network of ligand-receptor-mediated multicellular signalling in human. Nat Commun. 2015;6:7866. doi: 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data availability: Raw and processed sequencing data (RNA-seq, ATAC-seq and H3K4me2 ChIPmentation) are available from the NCBI Gene Expression Omnibus (GEO) repository (accession number: GSE134663). In addition, the dataset is provided as an online resource on a supplementary website (http://structural-immunity.computational-epigenetics.org), which includes links to raw and processed sequencing data, further analysis results, and genome browser tracks for interactive visualization of the RNA-seq, ATAC-seq, and ChIPmentation profiles.
Code availability: The analysis source code underlying the final version of the paper is available from the Supplementary Website (http://structural-immunity.computational-epigenetics.org).