Abstract
Mg-protoporphyrin IX monomethyl ester (MgPME) cyclase catalyses the formation of the isocyclic ring, the hallmark of chlorins and bacteriochlorins, producing protochlorophyllide a and contributing significantly to the absorption properties of chlorophylls and bacteriochlorophylls. The O2-dependent cyclase is found in oxygenic phototrophs and in some purple bacteria. We overproduced the simplest form of the cyclase, AcsF from Rubrivivax gelatinosus, in Escherichia coli. In biochemical assays the diiron cluster within AcsF is reduced by ferredoxin furnished by NADPH and ferredoxin:NADP+ reductase or by direct coupling to Photosystem I photochemistry, linking cyclase to the photosynthetic electron transport chain. Kinetic analyses yield a k cat of 0.9 min-1, a K M of 7.0 µM for MgPME, and a K d for MgPME of 0.16 µM. Mass spectrometry identified 131-hydroxy-MgPME and 131-keto-MgPME as intermediates in the formation of the isocyclic ring, revealing the reaction chemistry that converts porphyrins to chlorins, and completing the work originated by Sam Granick in 1950.
Chlorophylls, a class of cyclic tetrapyrroles, are among the most abundant natural pigments on Earth. They are the major absorbers of the solar energy that drives photosynthesis, and billions of tonnes of chlorophyll are synthesised annually on land and in the oceans. The decisive biosynthetic step that determines the absorption properties of chlorophyll, and more visually its green color, is the formation of the unique isocyclic fifth ring. This process involves the conversion of Mg-protoporphyrin IX monomethyl ester (MgPME) to 3,8-divinyl protochlorophyllide a (DV PChlide a), and it requires incorporation of an oxygen atom, sourced from either water or O2 (refs. 1,2), indicating the existence of two mechanistically different MgPME cyclases. Most anoxygenic phototrophic bacteria utilise an O2-sensitive radical SAM enzyme containing [4Fe-4S] and cobalamin cofactors to catalyse the reaction3, while oxygenic phototrophs including cyanobacteria, algae and plants, as well as some purple bacteria, adopt an O2-dependent cyclase for the reaction. Three classes of O2-dependent cyclase have been identified4, all with a catalytic subunit AcsF, a putative diiron protein5, but they differ in the requirement for an auxiliary subunit, either Ycf54 for the enzyme found in oxygenic phototrophs6,7, or BciE for the alphaproteobacterial enzyme4. However, there have been no mechanistic studies of the O2-dependent cyclase using purified components, so the details of this important reaction have remained unknown since Granick8 proposed a sequence of reactions that form the isocyclic fifth or E ring.
Early biochemical characterisation of cyclase activity using either intact or fractionated plant developing chloroplasts demonstrated the necessity for O2 (refs. 9–12), and inhibitor studies with specific chelators showed that iron is also required11. Both NADPH and NADH were found to stimulate cyclase activity, with NADPH more effective than NADH (refs. 11,13). These properties are shared by the enzymes from the green alga Chlamydomonas (C.) reinhardtii and the cyanobacterium Synechocystis sp. PCC 6803 (hereafter Synechocystis)14,15 and are characteristic of iron-dependent oxygenases. In addition, the cucumber, Synechocystis and barley enzymes were resolved into soluble and membrane-bound components13,15,16, but it has not been possible to obtain an active, pure cyclase from a native source.
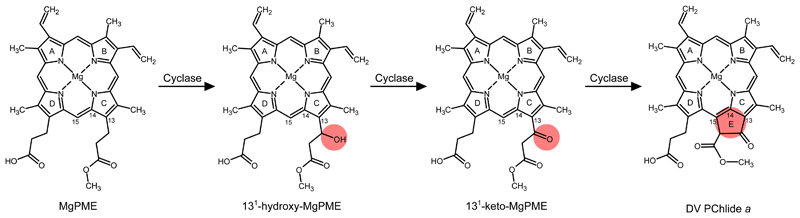
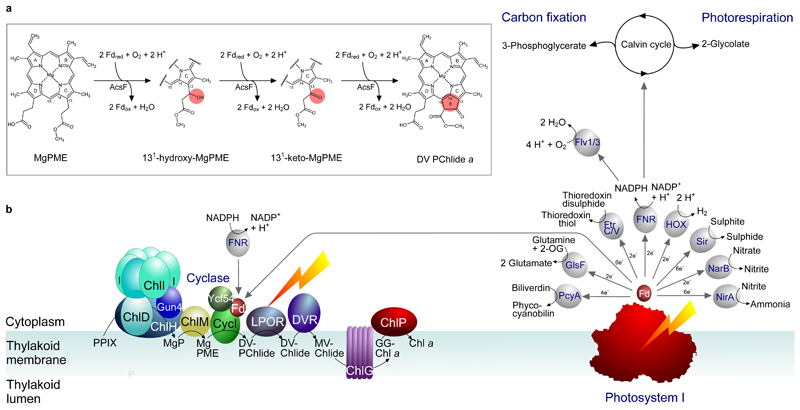
The complexity of E ring formation implies the involvement of multiple sequential reactions. Based on the mechanism of β-oxidation of fatty acids, Granick8 proposed the reaction could proceed through β-oxidation of the C13 methylpropionyl group of MgPME, via 131-132 acrylate, 131-hydroxy and 131-keto intermediates. These proposed reaction intermediates were subsequently detected in some Chlorella mutants17,18, and Castelfranco and co-workers confirmed the 131-hydroxy and 131-keto intermediates using the reconstituted cucumber cyclase system19. The 131-hydroxy derivative of MgPME was detected in these assays and shown to be an active cyclase substrate, and a similar compound was also identified during measurements of cyclase activity in isolated chloroplasts from C. reinhardtii (ref. 14). A chemically synthesised 131-keto analog of MgPME is readily converted to the final cyclase product, thereby validating this intermediate in the reaction sequence19. However, a synthesised acrylate derivative of MgPME was found to be inactive as a cyclase substrate in the reconstituted cyclase system20. It is possible that the acrylate derivative detected in the Chlorella mutants resulted from the 131-hydroxy intermediate undergoing a reverse hydratase reaction1. Based on these findings, the original reaction scheme of Granick and others has been modified to omit the acrylate intermediate, as shown in Fig. 1.
Fig. 1. Proposed reaction intermediates of MgPME cyclase.
Formation of the E ring of chlorophyll is proposed to proceed via hydroxylation, oxidation and cyclisation of the C13 methylpropionyl side chain of MgPME. The chemical change at each step is highlighted. International Union of Pure and Applied Chemistry numbering of the relevant macrocycle carbons is indicated.
Elucidation of the cyclase reaction sequence requires quantities of pure, active enzyme sufficient for biochemical and kinetic analyses. Although there have been no reports of the purification and in vitro reconstitution of the O2-dependent cyclase, an active recombinant cyclase was used successfully as part of a complete ensemble of biosynthetic enzymes that collectively enabled E. coli to synthesise chlorophyll21. Here we report the purification of the single-subunit O2-dependent cyclase AcsF from Rubrivivax (Rvi.) gelatinosus (ref. 5). Cyclase activity was reconstituted using a tricomponent electron transfer system consisting of NADPH, ferredoxin (Fd) and Fd:NADP+ reductase (FNR), and the steady-state kinetic behaviour of the enzyme was characterised. Furthermore, we detected the two previously proposed reaction intermediates in the progress of the reaction and subsequently determined their chemical identity by mass spectrometry. Our work therefore lays the groundwork for future mechanistic and structural study of the O2-dependent cyclase involved in chlorophyll biosynthesis.
Purification of AcsF from Rvi. gelatinosus
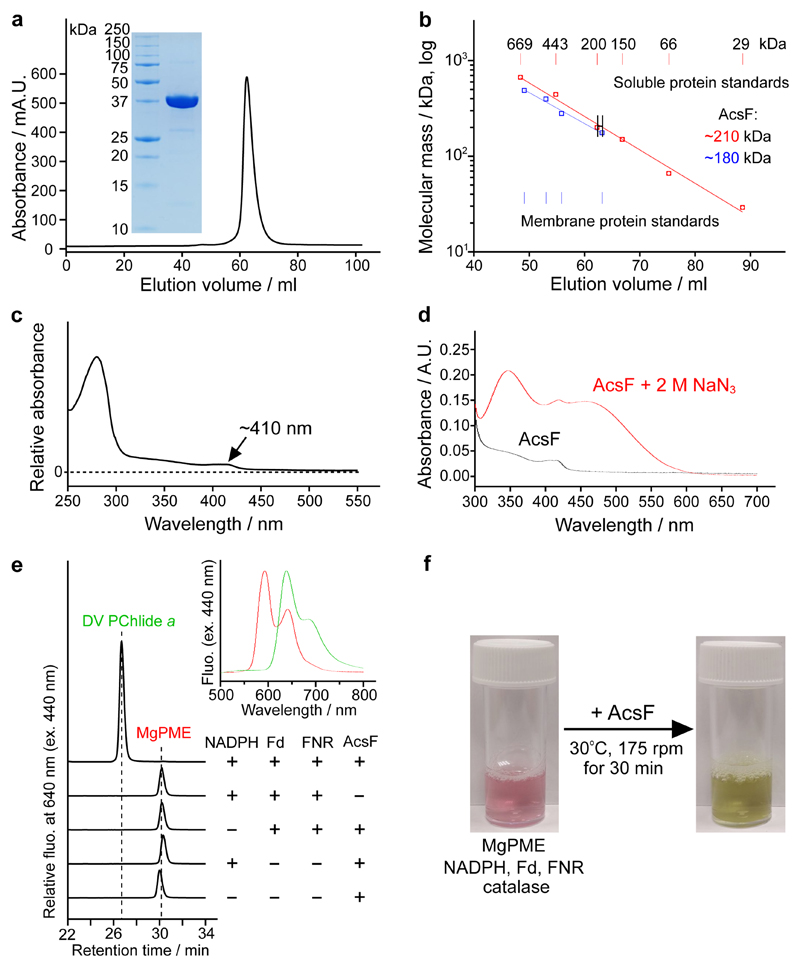
Rvi. gelatinosus AcsF with an N-terminal His6 tag was produced in the E. coli C43(DE3) strain22, supplemented with ferric ammonium citrate (see Materials and Methods). His-tagged AcsF was purified by Ni-IDA affinity chromatography from membrane fractions solubilised with the non-ionic detergent n-dodecyl-β-D-maltoside (β-DDM), followed by an iron reconstitution step with ferrous ammonium sulphate to increase the occupancy of the iron-binding sites. Subsequent gel filtration chromatography, in which a single symmetrical elution peak was observed (Fig. 2a), purified AcsF further. SDS-polyacrylamide gel electrophoresis (PAGE) analysis shows a single polypeptide with an apparent molecular mass of ~37 kDa (Fig. 2a; inset), and migrating further through the gel than expected for a predicted mass of 44 kDa, a common feature of membrane-associated proteins. The protein was over 95% pure, with an overall yield of ~3 mg per litre of culture.
Fig. 2. Purification, spectral characterisation and reconstitution of cyclase activity of AcsF.
a, Gel filtration profile of AcsF on a HiLoad 16/600 Superdex 200 prep grade column monitored by absorbance at 280 nm and SDS-PAGE analysis of 10 µg purified AcsF (inset). Shown are representative of three independently repeated experiments. b, Estimate of the molecular mass of native AcsF from triplicate gel filtration runs (range of elution volume indicated) using calibration curves (logarithm of molecular mass vs elution volume) generated from the data points of soluble (red circles) and membrane (blue squares) protein standards using nonlinear regression analysis. Membrane protein standards of 104, 208, 325 and 416 kDa were used and a value of 72 kDa (size of β-DDM micelles) was added to each molecular mass when generating the calibration curve. The calculated molecular mass values (inset) include the contribution of bound β-DDM molecules. c, Absorbance spectrum of AcsF as isolated. d, Absorbance spectra of 8 µM AcsF in the absence and presence of 2 M sodium azide. e, Cofactor requirements for in vitro cyclase activity of AcsF revealed by end-point HPLC-based assays. A complete assay contained 3.7 µM AcsF, 10 µM MgPME, 2 mM NADPH, 0.2 mg mL-1 spinach Fd, 0.4 units mL-1 spinach FNR and 0.29 mg mL-1 catalase. Retention times and fluorescence spectra (inset) were used to identify pigment species. See Materials and Methods for experimental details. f, Photographs showing the marked colour change showing the activity of AcsF in an assay containing 27 µM MgPME and other assay components at the same concentration as in a.
To investigate the oligomerisation state of the purified AcsF protein, gel filtration calibration curves were produced by analysing commercially available soluble protein standards, and four membrane protein standards available in our laboratory (Fig. 2b). We assumed that the collective mass of β-DDM molecules in complex with membrane proteins is the same as a β-DDM micelle, which is 72 ± 1.4 kDa (ref. 23). The purified AcsF with the associated β-DDM molecules was estimated to have a molecular mass of ~210 kDa with soluble standards, and ~180 kDa using membrane protein standards (Fig. 2b). By subtracting the contribution from detergent molecules, this indicates that AcsF is dimeric or trimeric given the predicted molecular mass of 44 kDa from the primary sequence.
Spectroscopic and biophysical characterisation of AcsF
Purified AcsF has a pale brown colour and exhibits a broad and weak band centred at ~340 nm in the absorbance spectrum (Fig. 2c), which is from an oxo-to-Fe(III) charge transfer transition24. An additional band at ~410 nm was also observed (Fig. 2c). Upon addition of 2 M sodium azide, formation of a chromophore with broad absorbance bands at 345 and 450 nm was observed and the position of the ~410 nm band was unaffected (Fig. 2d). The optical features, including the ~340 nm charge transfer band and the 345 and 450 nm bands when in complex with azide, are characteristic of µ-oxo-bridged diiron clusters, which have been reported with other diiron proteins such as stearoyl-acyl carrier protein Δ9 desaturase24, CmlA in chloramphenicol biosynthesis25, and CLK-1 in ubiquinone biosynthesis26. Assays determined that AcsF contained 2.35 ± 0.04 iron per monomer (Supplementary Fig. 1) providing further evidence for the presence of a diiron cluster in AcsF. We used differential scanning calorimetry to analyse the thermostability of AcsF and the melting point was determined to be 57.2 ºC (see Supplementary Fig. 2 for the melting curve), indicating the protein is stable for activity tests at 30 ºC.
In vitro reconstitution of cyclase activity with AcsF
To test whether the purified AcsF is active, we conducted end-point in vitro assays, followed by pigment analysis using high performance liquid chromatography (HPLC). Apart from the porphyrin substrate, MgPME, and molecular oxygen, an electron donor is also required to reduce the diiron center of AcsF from +3 to +2 during the catalytic cycle as required by most other diiron enzymes. An NADPH electron donor was suggested by early cyclase assays using biochemical fractions from Synechocystis, C. reinhardtii and plants13,15 but no activity was detected with just AcsF and NADPH (Fig. 2e; 4th trace from top), suggesting that it is not directly involved in cyclase activity. Guided by the findings that a few diiron enzymes accept electrons from reduced Fd for activity27,28, we combined spinach Fd and FNR with NADPH to form a tricomponent Fd reduction system, which did support the cyclase activity of AcsF (Fig. 2e; top trace). The activity depends on the presence of NADPH, Fd, FNR and AcsF, as indicated by the control assays (Fig. 2e; 2nd, 3rd, 4th, 5th traces from top). Next, we scaled up the assay and increased the MgPME concentration from 10 to 27 µM. The catalytic activity of AcsF is clearly demonstrated by the dramatic color change of the assay mixture indicating a likely complete conversion with only 30 min incubation (Fig. 2f).
Steady-state kinetic behavior and porphyrin binding of AcsF
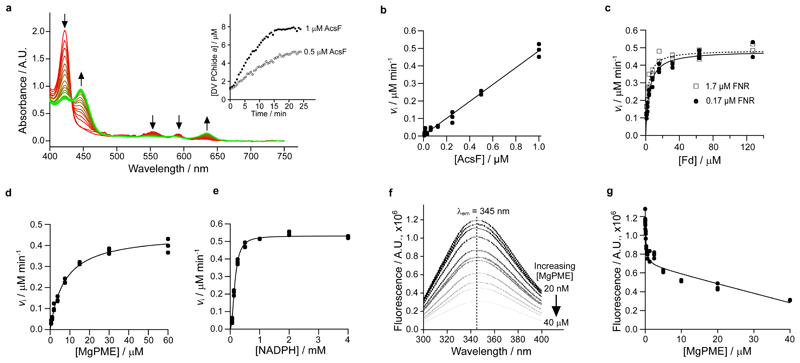
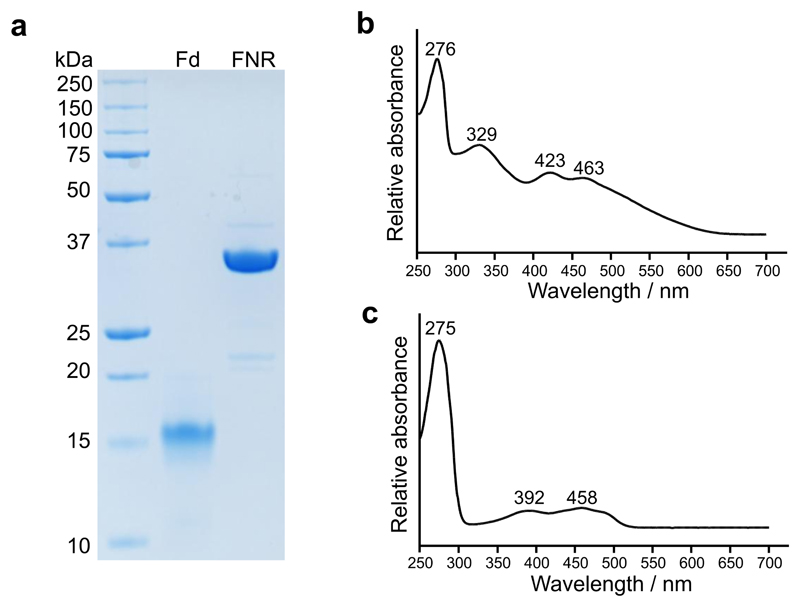
The dramatic color change from red to green upon product formation allowed us to develop an absorption-based continuous assay to investigate the steady-state kinetic behavior of AcsF. We switched to a cyanobacterial source of Fd and FNR, by overexpressing the genes from Anabaena sp. PCC 7119 (hereafter Anabaena) in E. coli (see Extended Data Fig. 1 for electrophoretic analysis and absorbance spectra of purified Anabaena Fd and FNR). Test assays monitored the spectral changes and found a clear trend of decreased absorbance maxima of the substrate at 422, 552 and 592 nm, accompanied by increasing absorbance maxima for the product at 446, 586 and 634 nm (Fig. 3a). The initial rate of product formation was calculated by quantifying DV PChlide a absorbance at 634 nm using a reported extinction coefficient29,30. We observed clear linear dependency of the initial rate with respect to the AcsF concentration (Fig. 3b). As the electron mediator between NADPH and AcsF, Fd was found to be rate-limiting. The initial rate displayed a hyperbolic response to Fd with an apparent K M determined to be 4.05 ± 0.39 µM in the presence of 0.17 µM FNR or 2.41 ± 0.26 µM with 1.7 µM FNR (Fig. 3c). In subsequent assays, Fd was used at a saturating concentration of 31 µM along with 1.7 µM FNR. The dependence of the initial rate on MgPME followed Michaelis-Menten kinetics with the characterising parameters k cat = 0.91 ± 0.02 min-1, K M = 7.03 ± 0.51 µM, k cat/K M = 0.13 ± 0.01 µM-1 min-1 (Fig. 3d). A sigmoidal relationship was found between the initial rate and the NADPH concentration so the Hill equation was used to fit the kinetic data with k cat = 1.06 ± 0.01 min-1, K 0.5 = 0.16 ± 0.01 mM, k cat/K 0.5 = 6.64 ± 0.21 mM-1 min-1 and the Hill coefficient n = 2.1 ± 0.1 (Fig. 3e).
Fig. 3. Steady-state kinetics of AcsF, and binding of MgPME to AcsF analysed by tryptophan fluorescence quenching.
a, The progressive spectral change during a continuous absorbance-based cyclase assay, which contained 1 µM AcsF, 10 µM MgPME, 7.6 µM Anabaena Fd, 0.17 µM Anabaena FNR, 2.5 mM NADPH and 0.29 mg mL-1 catalase. Arrows indicate the direction of change. Inset shows the product (DV PChlide a) evolution with 0.5 and 1 µM AcsF, monitored by absorbance at 634 nm. b–e, The dependence of the initial rate of product formation on AcsF (b), Anabaena Fd (c), MgPME (d) and NADPH (e). Assay conditions were as in a except the following stated differences: b, 7.81 nM to 1 µM AcsF; c, 0.5 µM AcsF, 0.17 or 1.7 µM Anabaena FNR, 0.99 to 127 µM Anabaena Fd; d, 0.5 µM AcsF, 1.7 µM Anabaena FNR, 31 µM Anabaena Fd; e, 0.5 µM AcsF, 30 µM MgPME, 1.7 µM Anabaena FNR, 31 µM Anabaena Fd, 62.5 µM to 4 mM NADPH. Each data point is an independent experiment. The Michaelis-Menten equation (equation 1, see Materials and Methods) was fitted to the kinetic data in c and d, with the characterising parameters K M (apparent) = 4.05 ± 0.39 µM (0.17 µM FNR) or 2.41 ± 0.26 µM (1.7 µM FNR) (c); = 0.91 ± 0.02 min-1, = 7.03 ± 0.51 µM (d). The Hill equation (equation 2, see Materials and Methods) was fitted to the NADPH titration data with = 1.06 ± 0.01 min-1, = 0.16 ± 0.01 mM, n = 2.1 ± 0.1 (e). f, A series of spectra showing quenching of AcsF fluorescence by MgPME. Excitation was set at 280 nm, producing an emission maximum at 345 nm. The average fluorescence spectra of triplicate experiments are shown. g, Plot of AcsF fluorescence against MgPME concentration. Each data point is an independent experiment. The curve fit is described by a modified single-site binding model (equation 3, see Materials and Methods) with K d for MgPME binding of 0.16 ± 0.05 µM.
AcsF is the only required subunit of the cyclase, so we quantified the binding affinity for the porphyrin substrate, MgPME, by measuring the tryptophan fluorescence quenching of AcsF upon binding of MgPME. With increasing concentrations of MgPME, the degree of quenching gradually intensified (Fig. 3f). The titration data fit a single-site binding model with modifications to include the inner filter effect of MgPME (refs. 31,32), giving a dissociation constant K d of 0.16 ± 0.05 µM for MgPME binding (Fig. 3g).
Identification of the reaction intermediates of AcsF
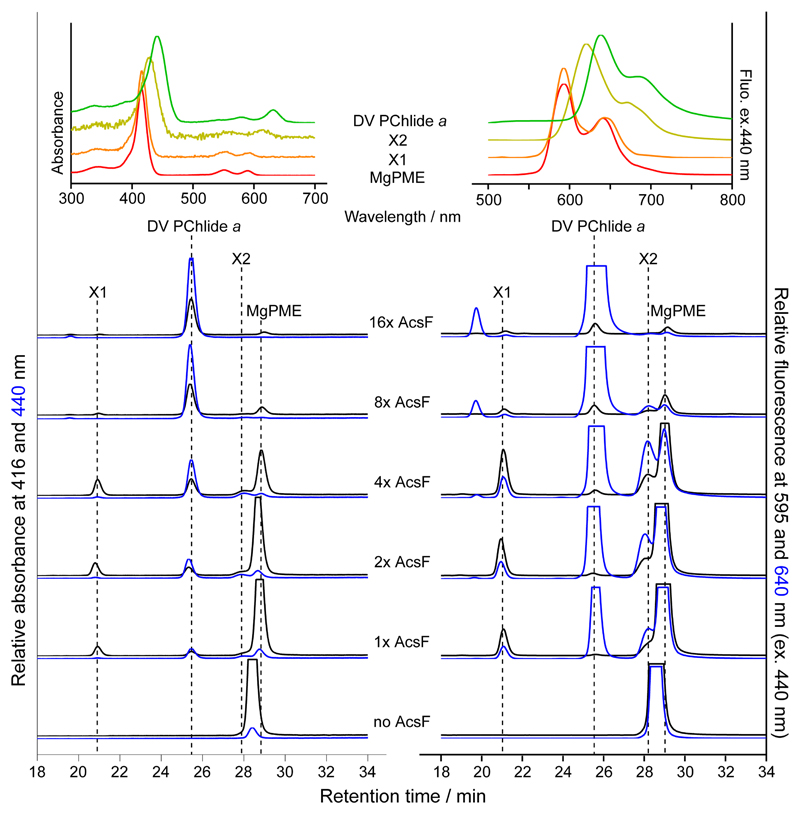
Apart from MgPME substrate and DV PChlide a product, some end-point cyclase assays contained two pigment species provisionally named X1 and X2 for the ~21 and ~28 min HPLC peaks, respectively. Intriguingly, X1 had the same absorbance and fluorescence spectra as MgPME whereas the spectral characteristics of X2 were between those of MgPME and DV PChlide a (Fig. 4; insets). The MgPME we used was highly pure and the other assay components were defined, which was supported by the absence of X1 and X2 in the control assay without AcsF (Fig. 4). Assays with two-fold escalating concentrations of AcsF were terminated after 30 min incubation. As shown in Fig. 4, peaks X1 and X2 became apparent with increasing concentrations of AcsF, reached a maximum level, then gradually disappeared, while the DV PChlide a product continuously accumulated. The dynamics of X1 and X2 during the progress of the reaction are consistent with possible roles as reaction intermediates. Based on their spectral features, we suspected X1 to be the hydroxy and X2 the keto reaction intermediates as previously proposed8,20.
Fig. 4. HPLC elution profiles of pigment extracts from end-point cyclase assays at various AcsF concentrations.
Assay conditions were 20 µM MgPME, 0.2 mg mL-1 spinach Fd, 0.4 units mL-1 spinach FNR, 2 mM NADPH, 0.29 mg mL-1 catalase and AcsF at various concentrations from 0.23 (1x AcsF) to 3.68 (16x AcsF) µM. Assays were initiated by adding AcsF and terminated after 30 min incubation. Pigment extracts from the assays were analysed by HPLC with elution of pigment species monitored by absorbance at 416 and 440 nm and by fluorescence at 595 and 640 nm with excitation at 440 nm. Insets show the acquired absorbance and fluorescence spectra of MgPME, DV PChlide a, and the potential reaction intermediates, X1 and X2.
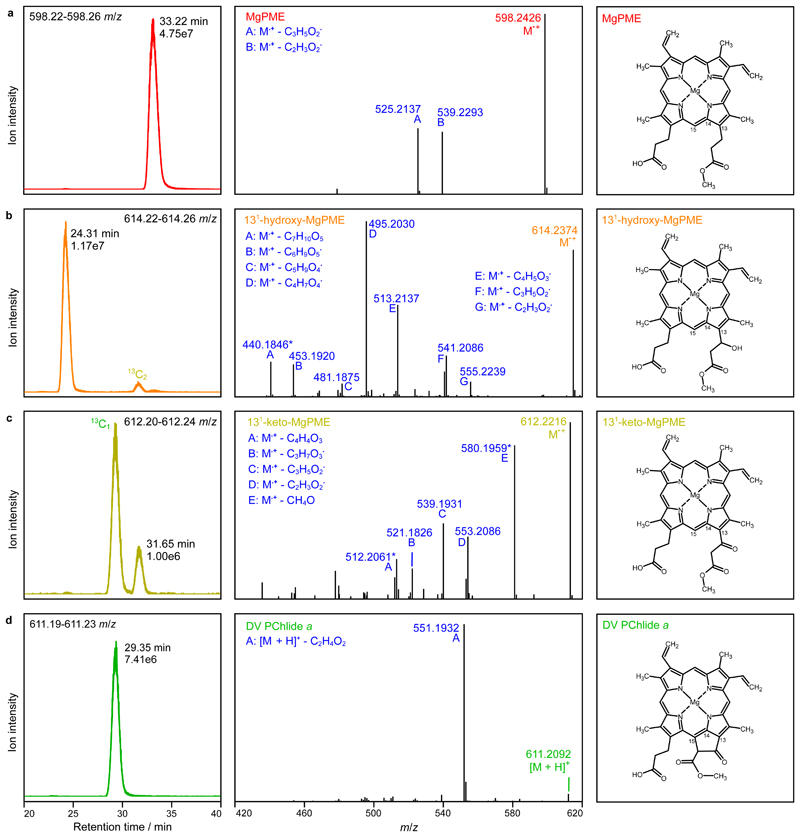
To confirm the identity of X1 and X2, the assay with 0.92 µM AcsF (4x AcsF), which accumulated the two pigment species the most (Fig. 4), was scaled up and the resulting pigment extract analysed by liquid chromatography-electrospray ionisation-tandem mass spectrometry (LC-ESI-MS/MS). Extracted ion chromatograms show that the putative MgPME, 131-hydroxy-MgPME and 131-keto-MgPME intermediates, and DV PChlide a were baseline resolved by reverse phase LC (Fig. 5a–d; left). Consistent with the identification of the extracted ion peak at 614.2 m/z as 131-hydroxy-MgPME (Fig. 5b; left), its retention time at 24 min was significantly earlier than the other three tetrapyrroles, as expected by the greater hydrophilicity conferred by the presence of a hydroxyl group. The non-hydroxylated tetrapyrroles were all clustered in the later 29–33 min eluting region. Mass spectral criteria for validating our identification of MgPME, 131-hydroxy-MgPME, 131-keto-MgPME and DV PChlide a are by comparison of the experimental and theoretical isotopomer m/z values and relative intensities in the full-MS spectra (Supplementary Fig. 3a–d; top left). The monoisotopic ions showed accuracies of 0.49–0.84 ppm, and in three cases there was close agreement in isotopomer patterns. In the case of 131-keto-MgPME (Supplementary Fig. 3c; top left) isotopomer pattern fidelity could not be assessed because the 13C1 and 13C2 ions were merged with coincident unidentified ions. MgPME, 131-hydroxy-MgPME and 131-keto-MgPME (Supplementary Fig. 3a–c; top left) all ionised in the electrospray source to form radical cations. In the case of DV PChlide a, the number of neutral molecules available for radical cation formation was lowered by a dominant population of protonated cations (Supplementary Fig. 3d; top left), presumably as a result of a change in electrochemical properties with cyclisation33. Product ion spectra generated by the higher-energy C-trap dissociation (HCD) of the precursor ions indicate relatively simple neutral loss pathways for MgPME and DV PChlide a (Fig. 5a,d; centre and right) forming the predicted carbocation products in which the positive charge resides on the larger fragment. No side-chain signature product ions at 59 and 73 m/z were detected. Similarly, the putative 131-hydroxy-MgPME and 131-keto-MgPME molecular ions dissociated without generating low m/z signature ions, however they did form a greater number (6–7) of high m/z cation products (Fig. 5b,c; centre). The increased number of potential neutral loss pathways for 131-hydroxy-MgPME and 131-keto-MgPME is an expected consequence of the additional functional group on the C13 methylpropionyl side chain. We further validated these two intermediates (Fig. 5b,c; right) by mapping their product ion spectra to structures generated by these neutral loss pathways (Supplementary Fig. 3b,c; bottom).
Fig. 5. Analysis of extracted pigments by LC-ESI-MS/MS.
The pigment extract from scaled-up in vitro cyclase assays corresponding to 4x AcsF (0.92 µM) in Fig. 4 was analysed. Extracted ion chromatograms (EICs) and product ion spectra derived from HCD of selected monoisotopic molecular ions are shown in the left and centre panels, respectively: a, MgPME, b, 131-hydroxy-MgPME, c, 131-keto-MgPME, d, DV PChlide a. The molecular structures that align with the mass spectral evidence presented here are shown in the corresponding right panels. EICs were generated for the indicated m/z ranges covering the target monoisotopic ions with peaks labelled with their retention times and ion intensities. Peaks mapping to 13C-containing isotopomers that fall within the EIC range are also labelled. Cations generated by gas phase neutral loss reactions are indicated by upper case letters with the eliminated molecular formulas also listed. The majority of product ions are carbocations formed after radical neutral loss; those labelled with an asterisk are radical cations formed after even electron neutral loss. Details of the structures which validate the identifications of the cyclase substrate, intermediates and product are shown in Supplementary Fig. 3.
Coupling the cyclase to reduced Fd produced by Photosystem I
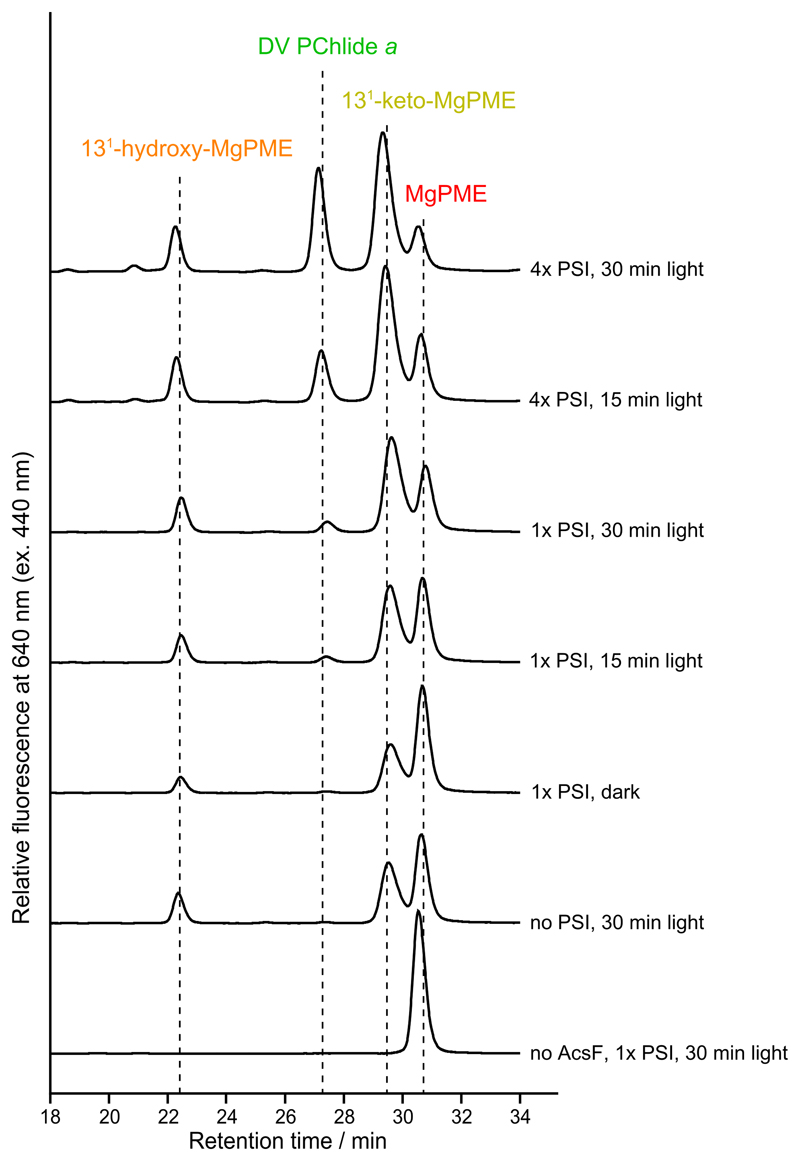
We have shown the cyclase activity requires electrons supplied by reduced Fd (Fig. 2e), which in oxygenic phototrophs is generated by Photosystem I (PSI) using light energy and electrons from plastocyanin (Pc). To test a direct link between the activities of cyclase and PSI, we conducted in vitro assays that coupled the cyclase assay with a reconstituted PSI electron transport system34, containing sodium L-ascorbate (Asc) as the reductant, 2,6-dichlorophenolindophenol (DCPIP) as the electron mediator, spinach Fd, Pc and PSI. It is intriguing that even in the absence of PSI the two cyclase reaction intermediates, 131-hydroxy-MgPME and 131-keto-MgPME, were detected (Extended Data Fig. 2 and Supplementary Fig. 4a). Systematic control assays showed that Asc alone was able to support a low level of cyclase activity, with conversion of 16% of the MgPME substrate to 131-hydroxy-MgPME and 131-keto-MgPME intermediates although with no formation of the final product (Supplementary Fig. 4b). However, the cyclase catalytic cycle was only completed, generating the final product DV PChlide a, in the presence of PSI and with light exposure (Extended Data Fig. 2). The production level of DV PChlide a clearly depended on the PSI level (Extended Data Fig. 2; 1x PSI versus 4x PSI) and was approximately proportional to the duration of light exposure (Extended Data Fig. 2; 15 min light versus 30 min light). These results clearly show the potential for the cyclase step in chlorophyll biosynthesis to be coupled to production of reduced Fd and turnover of PSI, a point that that will be covered in the Discussion.
Discussion
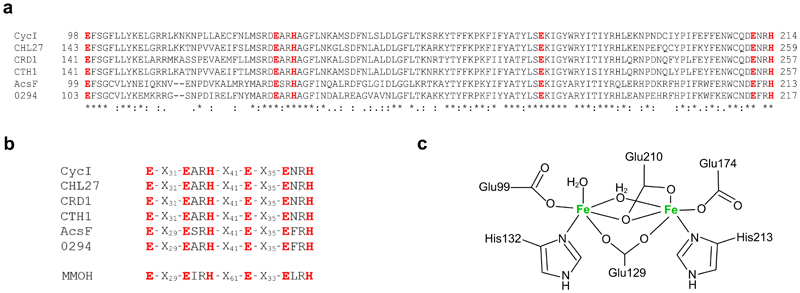
Over 70 years have elapsed since Granick’s proposed reaction sequence for the formation of the isocyclic ring of chlorophyll8. The inability to dissect this sequence represented a large gap in our knowledge of chlorophyll biosynthesis, given that formation of ring E starts to establish the eventual absorption lineshape of chlorophyll. The presence of ring E, a prerequisite for all chlorins and bacteriochlorins, extends the π system along the Qy axis, red-shifting and enhancing the Qy absorption band. The availability of a biochemically pure O2-dependent cyclase represented the only prospect resolving the reaction intermediates, and recent genetic approaches that resolved the enigmatic subunit composition of the O2-dependent cyclase4,21 paved the way for recombinantly producing the enzyme for in vitro analysis. In the present study, we overexpressed a single-subunit cyclase, the Rvi. gelatinosus AcsF, in E. coli and purified the enzyme to homogeneity. The pale brown colour and iron content of the purified AcsF protein, combined with the ~340 nm charge transfer band as well as the distinctive absorbance bands when in complex with azide, provide experimental evidence that AcsF is indeed a diiron protein. Sequence alignments of some AcsF proteins and their putative diiron ligands are shown in Extended Data Fig. 3a, and Extended Data Fig. 3b,c show how the diiron ligands of AcsF might be coordinated, based on motifs shared between AcsF proteins and the soluble methane monooxygenase hydroxylase subunit from Methylococcus capsulatus.
AcsF is a monooxygenase so O2 activation is part of its catalytic cycle, which requires a source of electrons. Our reconstitution tests showed that NADPH can be the ultimate electron source but not the direct electron donor as an oxidoreductase, FNR, and an electron mediator, Fd, were also required to form the electron transfer chain that supports cyclase activity (Fig. 2e). We were able to reconstitute the cyclase activity using either the spinach FNR and Fd (Fig. 2) or recombinantly produced Anabaena counterparts (Fig. 3) which, taken together with the in vivo heterologous activity of AcsF shown in E. coli (ref. 21), indicate that the AcsF reaction likely shares a generic electron transfer chain with other metabolic processes. In its native host, Rvi. gelatinosus, AcsF relies on the reduced Fd generated by the Rnf system35, which utilises the protonmotive force to derive electrons from NADH to Fd, and/or a flavin-based electron bifurcation system36, which oxidises NADH to produce reduced Fd and ubiquinol simultaneously. The cyclase from oxygenic phototrophs is expected to also use reduced Fd as the electron source, given that cyanobacterial, green algal and plant cyclases are active in E. coli (refs. 21,37), and cyanobacterial and plant cyclases are functional in Rvi. gelatinosus (refs. 4,37). Intriguingly, a recent report has already connected the cyclase activity with the plastidal FNR in plants38. Furthermore, while our paper was being reviewed, a report emerged showing that the in vitro activity of barley cyclase is Fd-dependent39. It is conceivable that the requirement for additional electron transfer partners eluded early cyclase characterisations that used complex cellular fractions. The finding of the Fd involvement is significant as it points to a possible connection between the cyclase and the photosynthetic electron transport chain, linking chlorophyll biosynthesis to photosynthetic activity and/or the redox state of thylakoid membranes40. Despite this possible direct link to PSI, the NADPH/FNR/Fd was the most effective source of electrons for assaying the cyclase in vitro (Fig. 4) which, if replicated in vivo, would allow the ‘dark’ synthesis of DV PChlide a. This route for Fd reduction is shown in Fig. 6, in addition to a more direct, PSI-coupled source of electrons. The ultimate source of reduced FNR and Fd is PSI, and the O2-dependent cyclase adds a candidate to the list of metabolic functions supported by PSI (ref. 41). As proof of principle, we devised an assay that couples the catalytic cycle of the cyclase with provision of reduced Fd by PSI (Extended Data Fig. 2). Fig. 6 depicts this dependence of chlorophyll biosynthesis on PSI turnover, and it shows that the light-dependent stage in chlorophyll biosynthesis, catalysed by light-dependent PChlide oxidoreductase (LPOR), is preceded by a cyclase step that, albeit indirectly, is also light-dependent.
Fig. 6. Diagram depicting the Fd-dependent cyclase reaction catalysed by AcsF and the supply of reduced Fd, directly or indirectly, by PSI.
a, The updated sequence of cyclase reactions catalysed by AcsF, with the chemical change of the porphyrin substrate at each step highlighted in the pink circle. Fdred and Fdox represent reduced and oxidised Fd, respectively. b, The chlorophyll biosynthesis pathway is shown on the left, progressing from protoporphyrin IX (PPIX) to chlorophyll a (Chl a), via magnesium protoporphyrin IX (MgP), Mg-protoporphyrin IX monomethyl ester (MgPME), 3,8-divinyl protochlorophyllide a (DV-PChlide), divinyl chlorophyllide a (DV-Chlide), monovinyl chlorophyllide a (MV-Chlide), and geranylgeranyl-chlorophyll a (GG-Chl a). ChlH, D, I are subunits, and Gun4 is an accessory protein, of the magnesium chelatase complex; ChlM is the MgP methyltransferase; CycI is the counterpart of the AcsF cyclase in cyanobacteria and plants, shown here with the accessory protein Ycf54 and Fd; LPOR is the light-dependent PChlide oxidoreductase; DVR is the divinyl reductase, the cyanobacterial version (BciB) of which requires Fd, whereas the plant DVR does not. ChlG is the chlorophyll synthase and ChlP is the geranylgeranyl reductase. The diagram (right) depicts a possible direct link between PSI and chlorophyll biosynthesis, showing that PSI could provide reduced Fd for the cyclase reaction; FNR-based reduction of Fd is also depicted, corresponding to the in vitro assays in Fig. 3 and Extended Data Fig. 2 respectively. Reduced Fd also provides electrons for a variety of cellular functions, shown here for cyanobacterial metabolism and adapted from the diagram in ref. 41. PcyA, phycocyanobilin:Fd oxidoreductase; GlsF, Fd-dependent glutamate synthase; FtrC/V, Fd:thioredoxin reductase; FNR, Fd:NADP+ reductase; Sir, Fd:sulfite reductase; NarB, nitrate reductase; NirA, nitrite reductase; Flv1/3, Flavodiiron 1/3; HOX, bi-directional hydrogenase.
The AcsF cyclase displays classic Michaelis-Menten kinetic behaviour regarding the porphyrin substrate MgPME. The Michaelis constant, K M, was determined to be 7.0 µM (Fig. 3d), typical of chlorophyll biosynthetic enzymes, such as LPOR and magnesium chelatase from Synechocystis with K M values of 8.6 µM for PChlide (ref. 42) and 3.2 µM for deuteroporphyrin IX (ref. 43), respectively. The K d for MgPME binding to AcsF is 0.16 µM (Fig. 3f,g), somewhat lower than for magnesium chelatase (K d of 1.2 µM for deuteroporphyrin IX)31, and the methyltransferase (K d of 2.4 µM for Mg-deuteroporphyrin IX)44. The K M for MgPME greatly exceeds K d, due to the complexity of the reaction undertaken by AcsF, and its reliance on both FNR and Fd during the reaction cycle. This K M value is also likely influenced by β-DDM detergent molecules associated with AcsF and with the hydrophobic MgPME substrate, with consequent effects on product release. Thus, the low turnover number, k cat, of 0.9 min-1 with respect to MgPME (Fig. 3d), reflects the complexity of the catalytic sequence that consists of multiple sequential reactions, each of which requires two electrons supplied by a coupled redox reaction. The value of k cat is comparable with the 0.8 min-1 measured for magnesium chelatase43, both of which are much slower than the 57 s-1 obtained for the methyltransferase44,45, which precedes the cyclase in the chlorophyll biosynthesis pathway. As for NADPH, the kinetic data were best described by the Hill equation and showed positive cooperativity (n = 2.1) (Fig. 3e), which arises from the multiple sequential NADPH-dependent reactions required to form the final product.
The availability of an in vitro cyclase assay enabled us to isolate and identify the progression of chemical species en route to the DV PChlide a product. Amounts of purified enzyme and porphyrin substrate could be varied to adjust the levels of cyclase reaction intermediates and final product, and the MS identification method minimised the possible interference from undefined protein components, pigment impurities and artefacts. We conclusively identified 131-hydroxy-MgPME and 131-keto-MgPME as reaction intermediates on the basis of the m/z of their radical cations and the specific transition of the precursor ion to product ions (Fig. 5 and Supplementary Fig. 3). As the reaction proceeded these two pigment species showed the transition from initial accumulation to subsequent dissipation (Fig. 4), characteristic of reaction intermediates. The hydrophobicity and spectral features of 131-hydroxy-MgPME are consistent with previous studies involving the cucumber and C. reinhardtii cyclase systems (Fig. 4; peak X1)14,46. 131-keto-MgPME was demonstrated to be a cyclase substrate with chemically synthesised pigments14 but was not directly detected in any cyclase system until now. The spectroscopic properties of 131-keto-MgPME (Fig. 4; peak X2) match those of the synthetic version14, and position the pigment between MgPME and DV PChlide a, reminiscent of the ‘longer wavelength metalloporphyrins’ detected in cucumber cotyledons47. We did not detect any pigment species suggestive of a 131-132 acrylate derivative of MgPME in the assay by HPLC or MS analysis, consistent with the report that the synthetic acrylate derivative is not a cyclase substrate20. The reaction sequence, signposted by the reaction intermediates identified by MS, is depicted in Fig. 6a. We note that although there is no final product, DV PChlide a, Asc can support a limited (16%) conversion of MgPME substrate, forming low levels of the 131-hydroxy and 131-keto intermediates (Supplementary Fig. 4b), possibly indicating different redox requirement for the three steps of the catalytic cycle. Although the in vitro assay with FNR/Fd can convert all of the MgPME to DV PChlide a, further work is required to investigate the electron donors in vivo, and the mechanism by which reduced Fd serves as the direct electron donor to the diiron centre of AcsF. Mechanistic details of the activation of molecular oxygen within the AcsF cyclase, a diiron enzyme, require the application of spectroscopic and structural techniques.
In summary, our work removes the last remaining hurdle in the study of the O2-dependent cyclase by showing reduced Fd serves as the direct electron donor to the diiron centre of AcsF. Our approaches for expression, purification and reconstitution of cyclase are transferrable to the study of the other two classes of O2-dependent cyclase, which require auxiliary Ycf54 or BciE subunits4. Our unambiguous identification of the two reaction intermediates provides an insight into the catalytic mechanism, which should be further investigated by mutagenesis and structural studies in order to pinpoint the key residues involved in formation of the isocyclic ring. In addition, a detailed analysis of the interaction between Fd and AcsF should be performed in the future to establish the Fd binding site that promotes electron transfer during the catalytic cycle.
Materials and Methods
Production and purification of AcsF, Fd and FNR
The acsF gene (RGE_33550) was amplified from the genomic DNA of Rvi. gelatinosus using the forward primer 5’-GAGTCTCATATGCTCGCGACCCCGACGATCG-3’ and the reverse primer 5’-GAGTCTGGATCCTCACCATGCCGGGGCCATG-3’, and cloned into the NdeI/BamHI sites of the pET14b vector, resulting in the pET14b-AcsF plasmid. Gene fragments encoding the Anabaena Fd (P0A3C8) and FNR (P21890; lacking the N-terminal 136 aa) were synthesised (Integrated DNA Technologies) with codon optimisation for expression in E. coli, and cloned into the NcoI/HindIII sites of the pET28a vector to get the pET28a-Fd and pET28a-FNR plasmids, respectively. The nucleotide sequences of synthesised genes are listed in Supplementary Table 1. The E. coli C43(DE3), BL21(DE3) and BL21(DE3) ΔiscR (ref. 48) strains were used for overexpression of AcsF, FNR and Fd, respectively. E. coli strains were grown in TB medium (12 g L-1 tryptone, 24 g L-1 yeast extract, 10 g L-1 glycerol, 2.31 g L-1 KH2PO4, 12.54 g L-1 K2HPO4) for AcsF and Fd, and in LB medium for FNR, with 100 µg mL-1 ampicillin or 30 µg mL-1 kanamycin where required. Cultures were grown at 37 ºC with shaking at 220 rpm then shifted to either 30 ºC for AcsF and FNR, or 28 ºC for Fd, with shaking at 175 rpm for induction. For C43(DE3)/pET14b-AcsF, 1/50 of starter culture was inoculated and the culture grown for 5 h before 0.4 mM isopropyl-β-D-thiogalactopyranoside (IPTG) and 0.1 g L-1 ferric ammonium citrate were added. After 24 h induction, cells were harvested and resuspended in buffer A [25 mM MOPS-NaOH, pH 7.5, 10 mM MgCl2, 500 mM NaCl, 10% (vol/vol) glycerol, 20 mM imidazole]. For BL21(DE3)/pET28a-FNR, a 1/100 inoculum was used and the culture grown to an OD600 of 0.6–0.8 before induction with 0.25 mM IPTG. Cells were harvested after 20 h induction and resuspended in 20 mM Tris-HCl pH 9.0. A 1/100 inoculum was used for BL21(DE3) ΔiscR/pET28a-Fd, and the culture grown to an OD600 of 0.6–0.8 before supplementation with 0.25 mM IPTG and 0.1 g L-1 ferric ammonium citrate. After 24 h induction, cells were harvested and resuspended in 20 mM Tris-HCl pH 7.4. All cell suspensions were flash-frozen in liquid N2 and stored at -20 ºC.
For purification of AcsF, cells were defrosted, and supplemented with DNase I, lysozyme and proteinase inhibitor cocktail (Sigma-Aldrich) before incubation at room temperature with shaking for 20 min. Cells were disrupted by one passage through a French pressure cell at 18,000 psi and 8 cycles of 30 s sonication, followed by centrifugation at 43,399 × g at 10 °C for 30 min. The resulting pellet was resuspended in buffer A. Then β-DDM was added at a concentration of 1% (wt/vol) and solubilisation performed at 4 °C on a tube roller for 1 h. Insoluble material was removed by centrifugation at 43,399 × g at 10 °C for 30 min. The soluble fraction was diluted 2× with buffer A and applied to Ni2+-loaded Chelating Sepharose Fast Flow resin (GE Healthcare), washed with buffer B (25 mM MOPS-NaOH, pH 7.5, 10 mM MgCl2, 500 mM NaCl, 10% glycerol, 100 mM imidazole, 0.04% β-DDM) and protein eluted with buffer C (25 mM MOPS-NaOH, pH 7.5, 10 mM MgCl2, 100 mM NaCl, 10% glycerol, 400 mM imidazole, 0.04% β-DDM). Protein-containing fractions were pooled and exchanged into buffer D (50 mM MES-NaOH, pH 6.0, 10 mM MgCl2, 500 mM NaCl, 10% glycerol, 0.04% β-DDM) using a PD-10 column (GE Healthcare). After adding 2 mM ferrous ammonium sulphate, the protein solution was incubated at 4 °C on a tube roller for 1 h, clarified by centrifugation at 21,380 × g at 4 °C for 10 min, and loaded onto a HiLoad 16/600 Superdex 200 prep grade gel filtration column (GE Healthcare) equilibrated with buffer D and eluted at 0.4 mL min-1 using an ÄKTAprime plus instrument monitored by PrimeView 5.0 (hereafter for gel filtration and ion exchange chromatography). Soluble protein standards from the Gel Filtration Markers Kit for 29–700 kDa (Sigma-Aldrich) and membrane protein standards kindly provided by Dr David Swainsbury, University of Sheffield, were analysed using the same gel filtration method. Calibration curves were generated using SigmaPlot 14.0 and used to estimate the molecular mass of AcsF whose concentration was determined by absorbance at 280 nm using an extinction coefficient of 59,820 M-1 cm-1, calculated by the ProtParam tool in the ExPASy portal49.
For purification of FNR, cell breakage was conducted as per the purification of AcsF but the passage through a French pressure cell was omitted. The cell lysate was clarified by centrifugation at 43,399 × g at 10 °C for 30 min, passed through a 0.45 µm filter, and applied to a HiTrap Q HP anion exchange column (GE Healthcare). The column was washed with 20 mM Tris-HCl pH 9.0 and proteins were eluted with a linear gradient of 0–0.25 M NaCl in 20 mM Tris-HCl pH 9.0 over 200 mL. Fractions containing FNR were pooled and concentrated before loading onto a HiLoad 16/600 Superdex 200 prep grade gel filtration column (GE Healthcare) equilibrated with buffer E (20 mM HEPES-NaOH, pH 7.5, 100 mM NaCl) and eluted at 0.5 mL min-1. For purification of Fd, cell breakage was conducted as for the purification of AcsF. The cell lysate was clarified and subjected to anion exchange chromatography as for the purification of FNR except that a linear gradient of 0–0.5 M NaCl in 20 mM Tris-HCl pH 7.4 over 200 mL was used for elution. Fractions containing Fd were pooled and subjected to ammonium sulphate precipitation three times at 0 °C to remove contaminants. Solid ammonium sulphate was directly added to the protein solution until cloudy and protein contaminants were removed by centrifugation. Fd was recovered from the clarified solution by performing ammonium sulphate precipitation twice with 100% saturated solution at 0 °C, reconstituted with ~5 ml 20 mM Tris-HCl pH 7.4 and purified by gel filtration as for FNR. Fd-containing fractions were pooled and subjected to ammonium sulphate precipitation twice with 100% saturated solution at 0 °C to remove potential nucleic acid contamination. The recovered Fd was finally dissolved in buffer E. Concentrations of cofactor-containing FNR and Fd were determined by absorbance using reported extinction coefficients of 9,400 M-1 cm-1 at 458 nm for FNR, and 7,200 M-1 cm-1 at 423 nm for Fd (ref. 50).
Iron quantification
The iron content of the purified AcsF protein was determined using an iron assay kit (Sigma-Aldrich) according to the manufacturer’s instructions. Assays were conducted in triplicate and data was analysed using SigmaPlot 14.0.
Absorbance spectroscopy
Spectra were recorded on either a Cary 60 UV-vis spectrophotometer (Cary WinUV Scan Application 5.1.0.1016) or an Omega FluoStar microplate reader (BMG LABTECH, Reader Control software 5.50 R4) equipped with an LVis plate. To record the AcsF-sodium azide spectrum, 16 µM AcsF was mixed with an equal volume of 4 M sodium azide stock solution prepared in buffer D and the mixture was incubated at room temperature for 30 min. The absorbance spectrum was recorded using 2 M sodium azide in buffer D as blank.
Purification of MgPME
MgPME was extracted from a Rvi. gelatinosus ΔbchEΔacsF mutant as described previously21. The resulting MgPME solution was vacuum dried, reconstituted in a minimal volume of 0.2% (wt/vol) ammonia in methanol and purified on an Agilent 1200 HPLC system (ChemStation for LC 3D systems B.04.02) using a Fortis C18 reverse-phase column (particle size 5 µm; 150 mm × 10 mm). Pigments were eluted at 40 ºC at 2.5 mL min−1 with a linear gradient from 35% (vol/vol) solvent A [350 mM ammonium acetate, 30% (vol/vol) methanol] to 75% (vol/vol) solvent B (methanol) over 35 min and monitored by absorbance at 416 nm. Fractions containing MgPME were collected, mixed with 0.5 vol QH2O, and loaded onto a Discovery DSC-18 SPE tube column (Sigma-Aldrich) for solid phase extraction. The column was washed with QH2O to remove ammonium acetate. MgPME was eluted with methanol, vacuum dried and stored at -20 ºC. MgPME concentration was estimated by absorbance at 589 nm using an extinction coefficient of 18,000 M-1 cm-1 in methanol11.
Differential scanning calorimetry
The purified AcsF was centrifuged at 16,000 × g at 4 °C for 10 min to remove any possible aggregates and diluted to 1 mg mL-1 in buffer D. 300 µL of the diluted AcsF solution was loaded into a NanoDSC (TA Instruments) and subjected to a heat ramp of 1 °C min-1. After removing the start-up hook, the data was converted to molar heat capacity using NanoAnalyze 3.11.0 and exported to Igor Pro 8.04 for processing by a cubic spline baseline fit followed by a Gaussian fit, whose modal value was reported as the melting point.
End-point HPLC-based cyclase assays
Spinach Fd, FNR, bovine catalase and NADPH were from Sigma-Aldrich. Assays were conducted in buffer F (100 mM TES-NaOH, 50 mM HEPES-NaOH, pH 7.7, 10 mM MgCl2, 1 M glycerol, 0.04% β-DDM) with 3.7 µM AcsF, 0.2 mg mL-1 spinach Fd, 0.4 units mL-1 spinach FNR, 2 mM NADPH, 10 µM MgPME (added from 200 µM MgPME stock solution prepared in 0.2% ammonia in methanol) and 0.29 mg mL-1 catalase. To test the cofactor requirement for cyclase activity, AcsF, Fd, FNR and NADPH were omitted where indicated. For reaction intermediate determination, 20 µM MgPME was used and AcsF was added at incrementally doubled concentrations from 0.23 to 3.68 µM. Assays were performed in 50 µL volume in 1.5 mL Eppendorf tubes and initiated with the addition of AcsF, followed by incubation at 30 ºC in the dark with shaking at 175 rpm for 30 min. Then 200 µL of 0.2% ammonia in methanol was added to stop the assay and 35 µL of the clarified pigment extract was analysed by HPLC as described previously37. The elution of pigment species was monitored by absorbance at 416 and 440 nm and fluorescence emission at 595 and 640 nm (excitation at 440 nm).
Coupled PSI-cyclase assays
Asc and DCPIP were from Sigma-Aldrich. Purified spinach PSI and Pc were kindly provided by Dr Guy Mayneord, University of Sheffield. Assays were conducted in buffer F with 2 µM AcsF, 0.04 mg mL-1 spinach Fd, 14 µM MgPME, spinach PSI containing 6 or 22.4 µM Chl a, 20 µM spinach Pc, 2 mM Asc, 60 µM DCPIP and 0.29 mg mL-1 catalase. AcsF, Fd, PSI, Pc, Asc and DCPIP were omitted where indicated to test the cofactor dependency of cyclase activity. Assays were performed in 50 µL volume in 1.5 mL Eppendorf tubes and incubated at 30 ºC either in the dark for 30 min, or under illumination from two red LED bicycle taillights (50 lumens, WQJifv) for 15 or 30 min. Assays were stopped by adding 4 vol 0.2% ammonia in methanol and 20 µL of the clarified pigment extract was analysed by HPLC as described previously21. Pigment elution was monitored by fluorescence emission at 640 nm (excitation at 440 nm).
Continuous absorbance-based cyclase assays
Assays were performed in buffer F with AcsF, Anabaena Fd, Anabaena FNR, NADPH, and MgPME at concentrations specified in the figure legends, and with catalase at 0.29 mg mL-1. Assays were conducted at 30 ºC in 100 µL volume in Greiner µclear F-bottom medium binding 96-well black microplates. Assays were initiated by adding AcsF and the reaction progress was monitored using an Omega microplate reader (BMG LABTECH, Reader Control software 5.50 R4) in absorbance mode for 30 min. Spectra from 400 to 750 nm were recorded for each well every 30 to 60 s (depending on the number of assays). Initial rates (v i) were calculated using the software supplied by the manufacturer (MARS 3.32 R5). Kinetic parameters were determined by fitting equation 1 to the data with nonlinear regression using Igor Pro 8.04. Errors were determined from least squares analysis of the fits. DV PChlide a concentration was estimated by absorbance at 634 nm using an extinction coefficient of 19,796 M-1 cm-1 (refs. 29,30).
| (equation 1) |
| (equation 2) |
Where v i is the initial reaction rate, k cat is the turnover number, K M is the Michaelis constant, n is the Hill coefficient, and K 0.5 is the substrate concentration that gives half-maximal reaction rate in the Hill equation.
Tryptophan fluorescence quenching binding assays
Assays were conducted by mixing 0.2 µM AcsF in buffer D with an equal volume of MgPME solution prepared in buffer G [0.02% (wt/vol) ammonia, 10% (vol/vol) methanol, 90% (vol/vol) buffer D] at concentrations incrementally doubled from 40 nM to 80 µM. The mixture was incubated at 30 °C for 2 min and then fluorescence spectra between 300 and 400 nm (10 nm bandpass) were recorded on a FluroMax 3 fluorimeter (HORIBA Jobin Yvon) (FluorEssence Package 3.9) at 30 °C with excitation at 280 nm (5 nm bandpass). A modified single-site binding equation (equation 3), which takes into account the inner filter effect of light absorbance by MgPME (refs. 31,32), was fitted to the obtained titration data with nonlinear regression using Igor Pro 8.04.
| (equation 3) |
Where F obs is the observed fluorescence, F 0 is the initial fluorescence, F max is the maximum amplitude of fluorescence quenching, [L]T is the total ligand concentration, [E]T is the total enzyme concentration (fixed at 0.1 µM during the fitting procedure), K d is the apparent dissociation constant, and M is the inner filter contribution of ligand.
Pigment analysis by LC-ESI-MS/MS
Scaled-up in vitro assays were conducted as per the end-point HPLC-based cyclase assay with 0.92 µM AcsF. The resulting pigment extract was mixed with 2 vol QH2O and subjected to solid phase extraction as per MgPME purification. Pigments were eluted with methanol, followed by vacuum drying and reconstitution in 50 µL 70% (vol/vol) methanol (LC grade), of which 5 µL was analysed by capillary-flow liquid chromatography (Dionex RSLCnano system, Thermo Scientific) coupled on-line to a Q Exactive HF quadrupole-Orbitrap mass spectrometer (Thermo Scientific). Analytes were separated on a Luna C18 reverse-phase column (particle size 5 µm; pore size 100 Å; 250 mm × 1 mm; Phenomenex) operating at 50 µL min-1 and 40 °C with a linear gradient from 35% (vol/vol) solvent A to 75% (vol/vol) solvent B over 35 min. The mass spectrometer was controlled by Thermo Xcalibur 4.0.27.42 and fitted with a HESI source operating with the following parameters: spray voltage 3,500 V positive, capillary temperature 320 °C, sheath gas 35 units. For full-scan profile MS acquisition, the following parameters were used: range 500–700 m/z, resolution 120,000, automatic gain control target 1e6 and maximum fill time 200 ms. Product ion scans were by centroid parallel reaction monitoring with selection of ions at 598.24, 614.24, 612.22 and 611.21 m/z for MgPME, 131-hydroxy-MgPME, 131-keto-MgPME and DV PChlide a, respectively, and at an isolation width of 1.2 m/z. Other parameters were set to: resolution 30,000, automatic gain control target 2e5, maximum fill time 100 ms and stepped collision energy 30/35/40 eV. Mass spectra were extracted from the output data files and compared with theoretical relative isotopomer ion intensity values using Xcalibur 4.0.27.42. Mapping precursor and product ion masses to their structures was carried out with the aid of ACD/ChemSketch 2019.1.3. These structures were exported to Xara Xtreme 5.1.1.9166 to produce figures.
Extended Data
Extended Data Fig. 1. Analysis of purified Anabaena Fd and FNR.
a, SDS-PAGE analysis of purified Anabaena Fd and FNR. b, Absorbance spectrum of purified Anabaena Fd. c, Absorbance spectrum of purified Anabaena FNR.
Extended Data Fig. 2. HPLC elution profiles of pigment extracts from coupled PSI-cyclase assays.
A complete assay contained 2 µM AcsF, 0.04 mg mL-1 spinach Fd, 14 µM MgPME, spinach PSI containing 6 (1x PSI) or 22.4 µM (~4x PSI) Chl a, 20 µM spinach Pc, 2 mM Asc, 60 µM DCPIP and 0.29 mg mL-1 catalase. Assays were incubated either in the dark for 30 min, or under red light illumination for 15 or 30 min. Pigment extracts from the assays were analysed by HPLC and pigment elution was monitored by fluorescence at 640 nm excited at 440 nm. Pigment species were identified by retention times and fluorescence spectra (as in Fig. 4). See Supplementary Fig. 4a for HPLC analysis of pigment extracts from additional control assays.
Extended Data Fig. 3. The diiron binding motif and proposed diiron ligation of AcsF.
a, Sequence alignments showing the conserved diiron binding motif of AcsF proteins. Sequences are from Synechocystis sp. PCC 6803 (CycI, BAA16583), Arabidopsis thaliana (CHL27, NP_191253), Chlamydomonas reinhardtii (CRD1, XP_001692557; CTH1, XP_001691047), Rubrivivax gelatinosus IL144 (AcsF, BAL96694) and Rhodobacter sphaeroides 2.4.1 (0294, abbreviated for RSP_0294, YP_353369). Conserved, highly similar and similar residues are marked with asterisks, colons and full stops, respectively. The putative diiron ligands are in red and bold. Full-length protein sequences were used for alignments but for clarity, only the putative diiron binding motifs with the residue range indicated, are shown. b, Sequence homologies between the diiron binding motifs of AcsF proteins and the soluble methane monooxygenase hydroxylase subunit from Methylococcus capsulatus Bath (MMOH, P22869). c, Proposed coordination of the diiron ligands of AcsF at the diferrous state based on the crystal structure of MMOH (PDB, 1FYZ)51.
Supplementary Material
Acknowledgements
We thank Dr David Swainsbury, University of Sheffield, for providing membrane protein standards for the gel filtration calibration and Dr Nathan Soulier, The Pennsylvania State University, for supplying the BL21(DE3) ΔiscR strain. G.E.C., N.B.P.A. P.J.J. and C.N.H. gratefully acknowledge financial support from the Biotechnology and Biological Sciences Research Council (BBSRC U.K.), award number BB/M000265/1. C.N.H. is also supported by European Research Council Synergy Award 854126. M.J.D. was supported by BBSRC U.K., award number BB/M012166/1.
Footnotes
Author contributions
G.E.C., N.B.P.A., P.J.J., M.J.D. and C.N.H. designed the research. G.E.C., N.B.P.A. and P.J.J. performed research and analysed data. G.E.C., N.B.P.A., P.J.J., M.J.D. and C.N.H. wrote the manuscript.
Competing interests
The authors declare no competing interests.
Data availability
All supporting data are included in the Supplementary Information. Annotated source data are provided with this paper for Figs. 2b, 2c, 2e, 4, Extended Data Figs. 1, 2, and Supplementary Figs. 1, 4a, 4b. The mass spectrometry raw data-files used for Fig. 5 and Supplementary Fig. 3 have been deposited to https://figshare.shef.ac.uk/articles/dataset/13655774.
Code availability
No custom code was used for the study.
References
- 1.Porra RJ, Schäfer W, Gad’on N, Katheder I, Drews G, Scheer H. Origin of the two carbonyl oxygens of bacteriochlorophyll a Demonstration of two different pathways for the formation of ring E in Rhodobacter sphaeroides and Roseobacter denitrificans and a common hydratase mechanism for 3-acetyl group formation. Eur J Biochem. 1996;239:85–92. doi: 10.1111/j.1432-1033.1996.0085u.x. [DOI] [PubMed] [Google Scholar]
- 2.Porra RJ, Urzinger M, Winkler J, Bubenzer C, Scheer H. Biosynthesis of the 3-acetyl and 131-oxo groups of bacteriochlorophyll a in the facultative aerobic bacterium, Rhodovulum sulfidophilum: The presence of both oxygenase and hydratase pathways for isocyclic ring formation. Eur J Biochem. 1998;257:185–191. doi: 10.1046/j.1432-1327.1998.2570185.x. [DOI] [PubMed] [Google Scholar]
- 3.Wiesselmann M, Hebecker S, Borrero-de Acuña JM, Nimtz M, Bollivar D, Jänsch L, Moser J, Jahn D. Mg-protoporphyrin IX monomethyl ester cyclase from Rhodobacter capsulatus: Radical SAM-dependent synthesis of the isocyclic ring of bacteriochlorophylls. Biochem J. 2020 doi: 10.1042/BCJ20200761. BCJ20200761. [DOI] [PubMed] [Google Scholar]
- 4.Chen GE, Canniffe DP, Hunter CN. Three classes of oxygen-dependent cyclase involved in chlorophyll and bacteriochlorophyll biosynthesis. Proc Natl Acad Sci USA. 2017;114:6280–6285. doi: 10.1073/pnas.1701687114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pinta V, Picaud M, Reiss-Husson F, Astier C. Rubrivivax gelatinosus acsF (previously orf358) codes for a conserved, putative binuclear-iron-cluster-containing protein involved in aerobic oxidative cyclization of Mg-protoporphyrin IX monomethylester. J Bacteriol. 2002;184:746–753. doi: 10.1128/JB.184.3.746-753.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hollingshead S, Kopečá J, Jackson PJ, Canniffe DP, Davison PA, Dickman MJ, Sobotka R, Hunter CN. Conserved chloroplast open-reading frame ycf54 is required for activity of the magnesium protoporphyrin monomethylester oxidative cyclase in Synechocystis PCC 6803. J Biol Chem. 2012;287:27823–27833. doi: 10.1074/jbc.M112.352526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Albus CA, Salinas A, Czarnecki O, Kahlau S, Rothbart M, Thiele W, Lein W, Bock R, Grimm B, Schottler MA. LCAA a novel factor required for magnesium protoporphyrin monomethylester cyclase accumulation and feedback control of aminolevulinic acid biosynthesis in tobacco. Plant Physiol. 2012;160:1923–1939. doi: 10.1104/pp.112.206045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Granick S. The structural and functional relationships between heme and chlorophyll. Harvey Lect Series. 1950;44:220–245. [PubMed] [Google Scholar]
- 9.Chereskin BM, Castelfranco PA. Effects of iron and oxygen on chlorophyll biosynthesis : II. Observations on the biosynthetic pathway in isolated etiochloroplasts. Plant Physiol. 1982;69:112–116. doi: 10.1104/pp.69.1.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chereskin BM, Wong YS, Castelfranco PA. In vitro synthesis of the chlorophyll isocyclic ring: Transformation of magnesium-protoporphyrin IX and magnesium-protoporphyrin IX monomethyl ester into magnesium-2,4-divinyl pheoporphyrin a5 . Plant Physiol. 1982;70:987–993. doi: 10.1104/pp.70.4.987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasrulhaq-Boyce A, Griffiths WT, Jones OT. The use of continuous assays to characterize the oxidative cyclase that synthesizes the chlorophyll isocyclic ring. Biochem J. 1987;243:23–29. doi: 10.1042/bj2430023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walker CJ, Mansfield KE, Smith KM, Castelfranco PA. Incorporation of atmospheric oxygen into the carbonyl functionality of the protochlorophyllide isocyclic ring. Biochem J. 1989;257:599–602. doi: 10.1042/bj2570599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong YS, Castelfranco PA. Resolution and reconstitution of Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase, the enzyme system responsible for the formation of the chlorophyll isocyclic ring. Plant Physiol. 1984;75:658–661. doi: 10.1104/pp.75.3.658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bollivar DW, Beale SI. Formation of the isocyclic ring of chlorophyll by isolated Chlamydomonas reinhardtii chloroplasts. Photosynth Res. 1995;43:113–124. doi: 10.1007/BF00042968. [DOI] [PubMed] [Google Scholar]
- 15.Bollivar DW, Beale SI. The chlorophyll biosynthetic enzyme Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase: Characterization and partial purification from Chlamydomonas reinhardtii and Synechocystis sp. PCC 6803. Plant Physiol. 1996;112:105–114. doi: 10.1104/pp.112.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rzeznicka K, Walker CJ, Westergren T, Kannangara CG, von Wettstein D, Merchant S, Gough SP, Hansson M. Xantha-l encodes a membrane subunit of the aerobic Mg-protoporphyrin IX monomethyl ester cyclase involved in chlorophyll biosynthesis. Proc Natl Acad Sci USA. 2005;102:5886–5891. doi: 10.1073/pnas.0501784102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ellsworth RK, Aronoff S. Investigations on the biogenesis of chlorophyll a: III. Biosynthesis of Mg-vinylpheoporphine a 5 methylester from Mg-protoporphine IX monomethylester as observed in Chlorella mutants. Arch Biochem Biophys. 1968;125:269–277. doi: 10.1016/0003-9861(68)90661-9. [DOI] [PubMed] [Google Scholar]
- 18.Ellsworth RK, Aronoff S. Investigations of the biogenesis of chlorophyll a: IV. Isolation and partial characterization of some biosynthetic intermediates between Mg-protoporphine IX monomethyl ester and Mg-vinylpheoporphine a 5, obtained from Chlorella mutants. Arch Biochem Biophys. 1969;130:374–383. doi: 10.1016/0003-9861(69)90047-2. [DOI] [PubMed] [Google Scholar]
- 19.Wong YS, Castelfranco PA. Properties of the Mg-protoporphyrin IX monomethyl ester (oxidative) cyclase system. Plant Physiol. 1985;79:730–733. doi: 10.1104/pp.79.3.730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walker CJ, Mansfield KE, Rezzano IN, Hanamoto CM, Smith KM, Castelfranco PA. The magnesium-protoporphyrin IX (oxidative) cyclase system: Studies on the mechanism and specificity of the reaction sequence. Biochem J. 1988;255:685–692. [PMC free article] [PubMed] [Google Scholar]
- 21.Chen GE, Canniffe DP, Barnett SFH, Hollingshead S, Brindley AA, Vasilev C, Bryant DA, Hunter CN. Complete enzyme set for chlorophyll biosynthesis in Escherichia coli . Sci Adv. 2018;4:eaaq1407. doi: 10.1126/sciadv.aaq1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miroux B, Walker JE. Over-production of proteins in Escherichia coli: Mutant hosts that allow synthesis of some membrane proteins and globular proteins at high levels. J Mol Biol. 1996;260:289–298. doi: 10.1006/jmbi.1996.0399. [DOI] [PubMed] [Google Scholar]
- 23.Strop P, Brunger AT. Refractive index-based determination of detergent concentration and its application to the study of membrane proteins. Protein Sci. 2005;14:2207–2211. doi: 10.1110/ps.051543805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fox BG, Shanklin J, Somerville C, Münck E. Stearoyl-acyl carrier protein Δ9 desaturase from Ricinus communis is a diiron-oxo protein. Proc Natl Acad Sci USA. 1993;90:2486–2490. doi: 10.1073/pnas.90.6.2486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makris TM, Chakrabarti M, Münck E, Lipscomb JD. A family of diiron monooxygenases catalyzing amino acid beta-hydroxylation in antibiotic biosynthesis. Proc Natl Acad Sci USA. 2010;107:15391–15396. doi: 10.1073/pnas.1007953107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Behan RK, Lippard SJ. The aging-associated enzyme CLK-1 is a member of the carboxylate-bridged diiron family of proteins. Biochemistry. 2010;49:9679–9681. doi: 10.1021/bi101475z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shanklin J, Somerville C. Stearoyl-acyl-carrier-protein desaturase from higher plants is structurally unrelated to the animal and fungal homologs. Proc Natl Acad Sci USA. 1991;88:2510–2514. doi: 10.1073/pnas.88.6.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Choi YS, Zhang H, Brunzelle JS, Nair SK, Zhao H. In vitro reconstitution and crystal structure of p-aminobenzoate N-oxygenase (AurF) involved in aureothin biosynthesis. Proc Natl Acad Sci USA. 2008;105:6858–6863. doi: 10.1073/pnas.0712073105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Helfrich M, Ross A, King GC, Turner AG, Larkum AWD. Identification of [8-vinyl]-protochlorophyllide a in phototrophic prokaryotes and algae: chemical and spectroscopic properties. Biochim Biophys Acta. 1999;1410:262–272. doi: 10.1016/s0005-2728(99)00004-3. [DOI] [PubMed] [Google Scholar]
- 30.Klement H, Helfrich M, Oster U, Schoch S, Rüdiger W. Pigment-free NADPH:protochlorophyllide oxidoreductase from Avena sativa L: Purificaton and substrate specificity. Eur J Biochem. 1999;265:862–874. doi: 10.1046/j.1432-1327.1999.00627.x. [DOI] [PubMed] [Google Scholar]
- 31.Karger GA, Reid JD, Hunter CN. Characterization of the binding of deuteroporphyrin IX to the magnesium chelatase H subunit and spectroscopic properties of the complex. Biochemistry. 2001;40:9291–9299. doi: 10.1021/bi010562a. [DOI] [PubMed] [Google Scholar]
- 32.Meneely KM, Sundlov JA, Gulick AM, Moran GR, Lamb AL. An open and shut case: the interaction of magnesium with MST enzymes. J Am Chem Soc. 2016;138:9277–9293. doi: 10.1021/jacs.6b05134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schäfer M, Drayß M, Springer A, Zacharias P, Meerholz K. Radical cations in electrospray mass spectrometry: formation of open-shell species, examination of the fragmentation behaviour in ESI-MSn and reaction mechanism studies by detection of transient radical cations. Eur J Org Chem. 2007;2007:5162–5174. [Google Scholar]
- 34.Jensen K, Johnston JB, de Montellano PRO, Møller BL. Photosystem I from plants as a bacterial cytochrome P450 surrogate electron donor: terminal hydroxylation of branched hydrocarbon chains. Biotechnol Lett. 2012;34:239–245. doi: 10.1007/s10529-011-0768-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Biegel E, Schmidt S, González JM, Müller V. Biochemistry, evolution and physiological function of the Rnf complex, a novel ion-motive electron transport complex in prokaryotes. Cell Mol Life Sci. 2011;68:613–634. doi: 10.1007/s00018-010-0555-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na+ translocating ferredoxin oxidation. Biochim Biophys Acta. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 37.Chen GE, Hunter CN. Protochlorophyllide synthesis by recombinant cyclases from eukaryotic oxygenic phototrophs and the dependence on Ycf54. Biochem J. 2020;477:2313–2325. doi: 10.1042/BCJ20200221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herbst J, Girke A, Hajirezaei MR, Hanke G, Grimm B. Potential roles of YCF54 and ferredoxin-NADPH reductase for magnesium protoporphyrin monomethylester cyclase. Plant J. 2018;94:485–496. doi: 10.1111/tpj.13869. [DOI] [PubMed] [Google Scholar]
- 39.Stuart D, Sandström M, Youssef HM, Zakhrabekova S, Jensen PE, Bollivar DW, Hansson M. Aerobic barley Mg-protoporphyrin IX monomethyl ester cyclase is powered by electrons from ferredoxin. Plants. 2020;9:1157. doi: 10.3390/plants9091157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steccanella V, Hansson M, Jensen PE. Linking chlorophyll biosynthesis to a dynamic plastoquinone pool. Plant Physiol Biochem. 2015;97:207–216. doi: 10.1016/j.plaphy.2015.10.009. [DOI] [PubMed] [Google Scholar]
- 41.Lea-Smith DJ, Bombelli P, Vasudevan R, Howe CJ. Photosynthetic, respiratory and extracellular electron transport pathways in cyanobacteria. Biochim Biophys Acta. 2016;1857:247–255. doi: 10.1016/j.bbabio.2015.10.007. [DOI] [PubMed] [Google Scholar]
- 42.Heyes DJ, Martin GE, Reid RJ, Hunter CN, Wilks HM. NADPH:protochlorophyllide oxidoreductase from Synechocystis: overexpression, purification and preliminary characterisation. FEBS Lett. 2000;483:47–51. doi: 10.1016/s0014-5793(00)02081-0. [DOI] [PubMed] [Google Scholar]
- 43.Reid JD, Hunter CN. Magnesium-dependent ATPase activity and cooperativity of magnesium chelatase from Synechocystis sp. PCC6803. J Biol Chem. 2004;279:26893–26899. doi: 10.1074/jbc.M400958200. [DOI] [PubMed] [Google Scholar]
- 44.Shepherd M, Hunter CN. Transient kinetics of the reaction catalysed by magnesium protoporphyrin IX methyltransferase. Biochem J. 2004;382:1009–1013. doi: 10.1042/BJ20040661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shepherd M, Reid JD, Hunter CN. Purification and kinetic characterization of the magnesium protoporphyrin IX methyltransferase from Synechocystis PCC6803. Biochem J. 2003;371:351–360. doi: 10.1042/BJ20021394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wong YS, Castelfranco PA, Goff DA, Smith KM. Intermediates in the formation of the chlorophyll isocyclic ring. Plant Physiol. 1985;79:725–729. doi: 10.1104/pp.79.3.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rebeiz CA, Mattheis JR, Smith BB, Rebeiz CC, Dayton DF. Chloroplast biogenesis: Biosynthesis and accumulation of Mg-protoporphyrin IX monoester and longer wavelength metalloporphyrins by greening cotyledons. Arch Biochem Biophys. 1975;166:446–465. doi: 10.1016/0003-9861(75)90408-7. [DOI] [PubMed] [Google Scholar]
- 48.Walters KA, Golbeck JH. Designing a modified clostridial 2[4Fe-4S] ferredoxin as a redox coupler to directly link photosystem I with a Pt nanoparticle. Photosynth Res. 2020;143:165–181. doi: 10.1007/s11120-019-00679-w. [DOI] [PubMed] [Google Scholar]
- 49.Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, et al. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–W603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pueyo JJ, Gómez-Moreno C. Purification of ferredoxin-NADP+ reductase, flavodoxin and ferredoxin from a single batch of the cyanobacterium Anabaena PCC 7119. Prep Biochem. 1991;21:191–204. doi: 10.1080/10826069108018571. [DOI] [PubMed] [Google Scholar]
- 51.Whittington DA, Lippard SJ. Crystal structures of the soluble methane monooxygenase hydroxylase from Methylococcus capsulatus (Bath) demonstrating geometrical variability at the dinuclear iron active site. J Am Chem Soc. 2001;123:827–838. doi: 10.1021/ja003240n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All supporting data are included in the Supplementary Information. Annotated source data are provided with this paper for Figs. 2b, 2c, 2e, 4, Extended Data Figs. 1, 2, and Supplementary Figs. 1, 4a, 4b. The mass spectrometry raw data-files used for Fig. 5 and Supplementary Fig. 3 have been deposited to https://figshare.shef.ac.uk/articles/dataset/13655774.
No custom code was used for the study.