Abstract
STING is essential for control of infections and for tumor immunosurveillance, but can also drive pathological inflammation. STING resides on the endoplasmic reticulum (ER), and traffics following stimulation to ERGIC/Golgi where signaling occurs. Although STING ER exit is the rate-limiting step in STING signaling, the mechanism that drives this process is not understood. Here we identify STEEP as a positive regulator of STING signaling. STEEP was associated with STING and promoted trafficking from the ER. This was mediated through stimulation of phosphatidylinositol-3-phosphate (PI3P) production and ER membrane curvature formation, thus inducing COPII-mediated ER-to-Golgi trafficking of STING. Depletion of STEEP impaired STING-driven gene expression in response to virus infection in brain tissue and in cells from patients with STING-associated diseases. Interestingly, STING gain-of-function mutants from patients interacted strongly with STEEP leading to increased ER PI3P levels and membrane curvature. Thus, STEEP enables STING signaling by promoting ER exit.
Keywords: Innate immunology, DNA sensing, cGAS-STING pathway, intracellular trafficking, SAVI
Introduction
Cytosolic DNA is a potent danger signal, leading to activation of innate immune responses, most notably expression of the cytokines type I interferon (IFN) and interleukin (IL) 1β1. Induction of type I IFN by cytosolic DNA is mediated by cyclic GMP-AMP (cGAMP) synthase (cGAS), which produces 2’3’cGAMP upon DNA binding2. 2’3’cGAMP in turn binds the endoplasmic reticulum (ER)-resident adaptor protein stimulator of IFN genes (STING), which then undergoes conformational changes in a dimeric form and recruits the kinase TBK13, 4, 5. Upon activation, TBK1 phosphorylates STING at serine 366, thus allowing recruitment of the latent transcription factor IFN regulatory factor 3 (IRF3), and positioning of IRF3 for TBK1-mediated phosphorylation6, 7. Upon phosphorylation, IRF3 dissociates from STING to form dimers, and translocate to the nucleus to activate transcription. In addition to type I IFN, the cGAS-STING pathway also induces autophagy and apoptosis, and activates the transcription factor NF-κB, which drives expression of inflammatory genes8, 9.
The cGAS-STING pathway is essential for control of a wide panel of human pathogens10, 11 and for tumor immunosurveillance12. Moreover, several inflammatory diseases are driven by excessive activation of STING signaling13, 14. Therefore, pharmacological activators of the STING pathway are currently being tested for their potential as vaccine adjuvants and as cancer immunotherapy15, 16. Likewise, cGAS-STING antagonism holds promise for treatment of inflammatory diseases17. Given the clinical importance of the cGAS-STING pathway in human diseases, there is an urgent need to obtain in-depth understanding of the mechanisms governing its activity during defense and disease.
Although STING is an ER-resident protein in the resting state, active STING signaling occurs in the Golgi/ERGIC4, 18, 19. Gain-of-function STING mutants from patients with the autoinflammatory disease STING-associated vasculopathy with onset in infancy (SAVI) translocate to the Golgi and signal independent of cGAMP binding14, 19. In fact, it was shown that STING translocation is a rate-limiting event in STING signaling, and proposed that STING is actively retained in the ER, and released through a specific mechanism including the Ca2+ sensor STIM119, 20. Despite this, there is limited knowledge on the mechanisms that govern STING ER exit. Components described to be involved in STING ER exit include translocon proteins4, the autophagy-associated phosphatidylinositol 3-kinase VPS3421, inactive rhomboid protein 2 (iRhom2)22, Sorting nexin 8 (SNX8)23, the ubiquitin regulatory X domain-containing proteins 3B (UBXN3B)24, transmembrane emp24 protein transport domain containing 2 (TMED2)25. However, the actual mechanism of STING ER exit is not known.
Here we identify CxORF56 as a protein that interacts with STING, and demonstrate that this protein is essential for STING ER exit, and for initiation of signaling leading to autophagy and expression of IFN and inflammatory cytokines. We propose the name STING ER exit protein (STEEP) for CxORF56. STEEP mediates DNA-induced STING-dependent increase in ER phosphatidylinositol-3-phosphate (PI3P) production, ER membrane curvature, and COPII-dependent STING ER-to-Golgi trafficking. STEEP deficiency inhibits STING-dependent signaling by dsDNA and human pathogenic microorganisms, and also prevents STING signaling in cells from patients with SAVI and systemic lupus erythematosus (SLE). Thus, STEEP is essential for the critical ER exit step in STING signaling.
Results
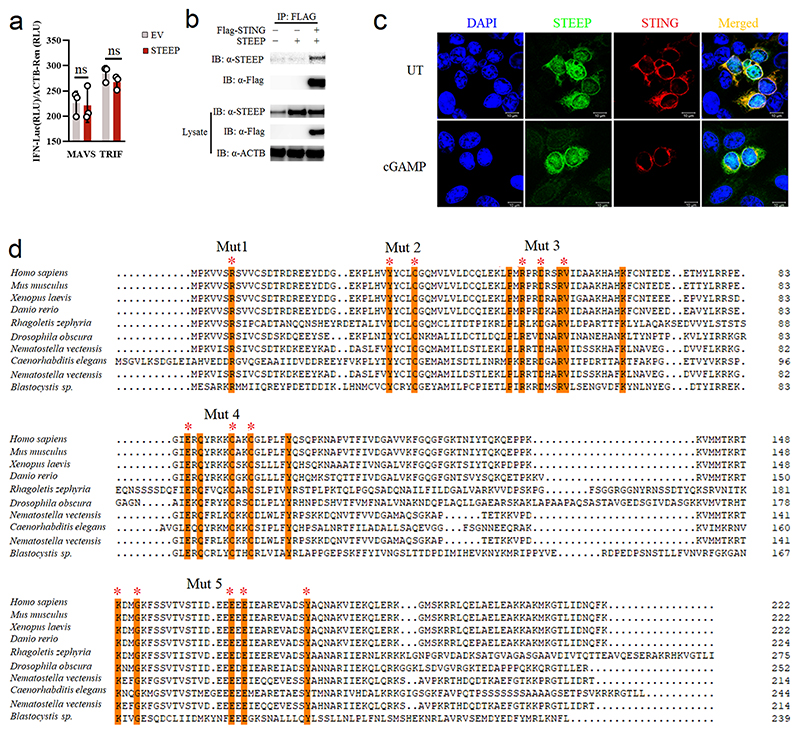
Identification of STEEP as a STING-interacting protein promoting IFNβ induction
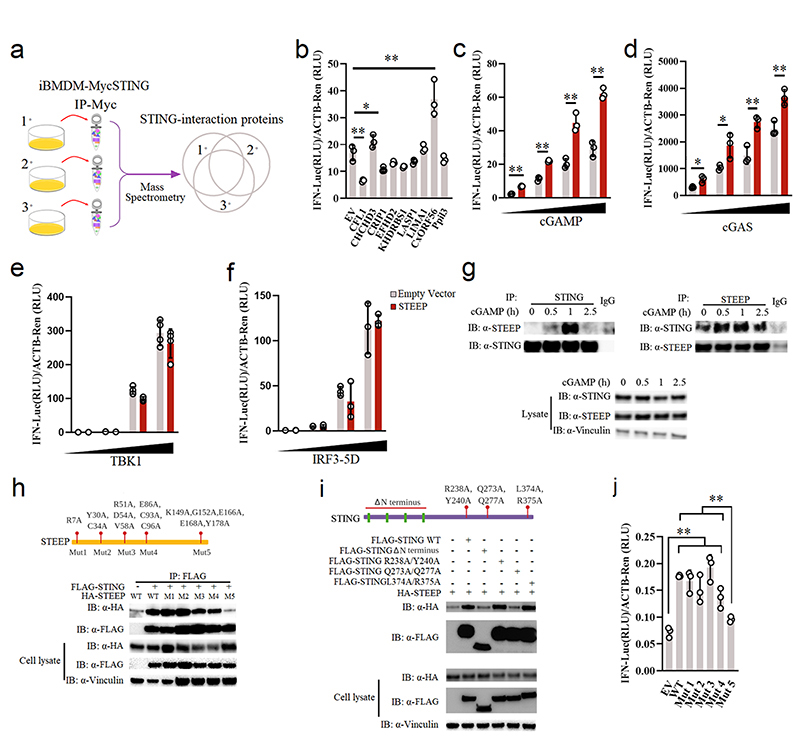
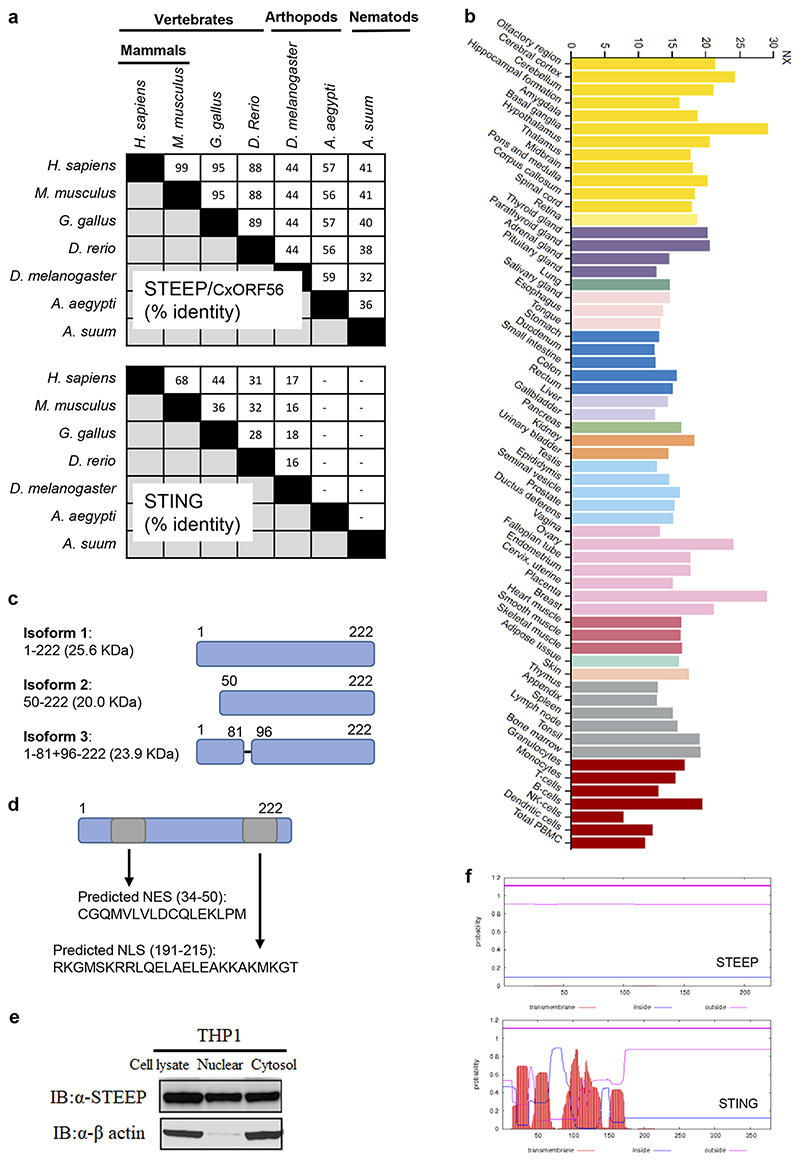
To identify factors that govern early STING signaling, STING was immunoprecipitated from lysates of resting Myc-STING iBMDCs, and interacting proteins were identified by Mass Spectrometry (Fig. 1a). This led to identification of a set of STING-interacting proteins (Supplementary Table 1), many of which were also identified in a previous screen for STING-interacting proteins26. This includes DDX3X and Sarcoplasmic/endoplasmic reticulum calcium ATPase 2; both reported to be involved in STING-dependent responses27, 28, as well as proteins not previously characterized in relation to STING. From this list, nine proteins were selected for further analysis, not using a rigid set of criteria but with focus on potential novelty. The candidates were analyzed by overexpression in HEK-293T-STING cells and evaluated for IFNB promoter activation. Only one of the tested candidate proteins, CxORF56, significantly elevated cGAMP-mediated IFNB promoter activation, while several of the candidates inhibited activation (Fig. 1b). Onwards in this article, we will use the name STEEP for CxORF56. STEEP is highly conserved through evolution, and is more conserved than STING (Extended Data Fig. 1a). The protein belongs to the uncharacterized UPF0428 family, and is highly expressed in most tissues (Extended Data Fig. 1b). Three isoforms of STEEP exist, and the amino acid sequence contains both nuclear entry and nuclear exit signals (Extended Data Fig. 1c-d). Consistently, STEEP was found in both the nuclear and cytoplasmic fraction of THP-1 cell lysates (Extended Data Fig. 1e). Finally, STEEP is predicted not to contain transmembrane regions, unlike STING, which contains four membrane-spanning regions (Extended Data Fig. 1f).
Figure 1. Identification of STEEP as a positive regulator of STING signaling.
(a) Set-up for identification of STING-interacting proteins. (b) Reporter gene assays for IFNB1 promoter activity in STING-expressing HEK-293T cells co-transfected with the indicated expression plasmids for 24 h, and stimulated by 100 nM cGAMP for 6 h (n = 3) RLU, relative light units. (**P = 0.00051, *P = 0.047, **P = 0.0087, left to right). (c) IFNB1 reporter gene assays in STING-expressing HEK-293T cells transfected with STEEP for 24 h, and stimulated with increasing concentrations of cGAMP (n = 3). (**P = 2.37E-05, **P = 0.00014, **P = 0.0024, **P = 0.00049, left to right). (d-f) IFNB1 reporter gene assays in (d) HEK293T-STING or (e-f) HEK-293T cells transfected with STEEP, and increased concentration of cGAS (n = 3) (**P = 0.016, **P = 0.023, **P = 0.0057, **P = 0.0075, left to right), TBK1 (n = 4) or IRF3-5D (n =3) as indicated for 24 h. (g) Immunoprecipitates and lysates from THP1 cells stimulated with cGAMP (100 nM) were immunoblotted with antibodies against STEEP, STING, and vinculin (n = 3). (h) FLAG was immuno-precipitated from HEK-293T lysates transfected with FLAG-STING and HA-tagged STEEP (WT and indicated mutants). Precipitates were immunoblotted with anti-HA (n = 3). (i) FLAG in lysates from HEK-293T cells expressing FLAG-STING (WT and indicated mutants) together with HA-STEEP was immunoprecipitated, and immuno-blotted with anti-HA (n = 3). (j) IFNB1 reporter gene assays for STING-expressing HEK-293T cells transfected with 50 ng of STEEP (WT or mutants), and IFNB1 promoter luciferase reporter, β-actin Renilla reporter for 24 h (n = 3) (**P < 0.01). Data in panel b-f and j are shown as means of cell culture triplicates +/- st.dev. Statistical analysis of data in panel b and j was performed using two-tailed one-way ANOVA test, and panel c and d was performed using two-tailed Student’s t-test.
Further analyses revealed that STEEP overexpression elevated cGAS- and cGAMP-driven IFNB promoter activity in HEK-293T-STING cells in a dose-dependent manner (Fig. 1c-d), whereas STEEP overexpression did not affect TBK1-, IRF3-5D-, MAVS- or TRIF-mediated IFNB promoter activity (Fig. 1e-f, Extended Data Fig. 2a). Co-immunoprecipitation of endogenous STING and STEEP in THP-1 cells confirmed that STEEP and STING interact constitutively, and this was further elevated early after stimulation (Fig. 1g). FLAG-tagged STING also co-precipitated with STEEP (Extended Data Fig. 2b). Consistently, we observed significant colocalization between STEEP and STING in Hela cells before and after cGAMP stimulation (Extended Data Fig. 2c). To map the interaction between STING and STEEP we made a series of STEEP mutants (Fig. 1h, Extended Data Fig. 2d). Five mutants were generated where residues fully conserved in STEEP from all species examined were mutated to alanine. In co-immunoprecipitation experiments with WT STING, we observed that five residues in the C-terminal region of STEEP were essential for interaction with STING (Fig. 1h). Likewise, co-immunoprecipitation of WT STEEP with various STING mutants revealed that the N-terminal transmembrane region, and the STING dimerization interphase were essential for the STING-STEEP interaction (Fig. 1i). By contrast, the capacity to bind cGAMP or to interact with TBK1 was not required. Finally, the inability of the STEEP Mut 5 mutant to interact correlated with its inability to augment STING-driven activation of the IFNB promoter (Fig. 1j). Taken together, these data suggest that the uncharacterized protein STEEP associates with STING at the early stages of signaling to promote type I IFN induction.
STEEP deficiency impairs STING signaling
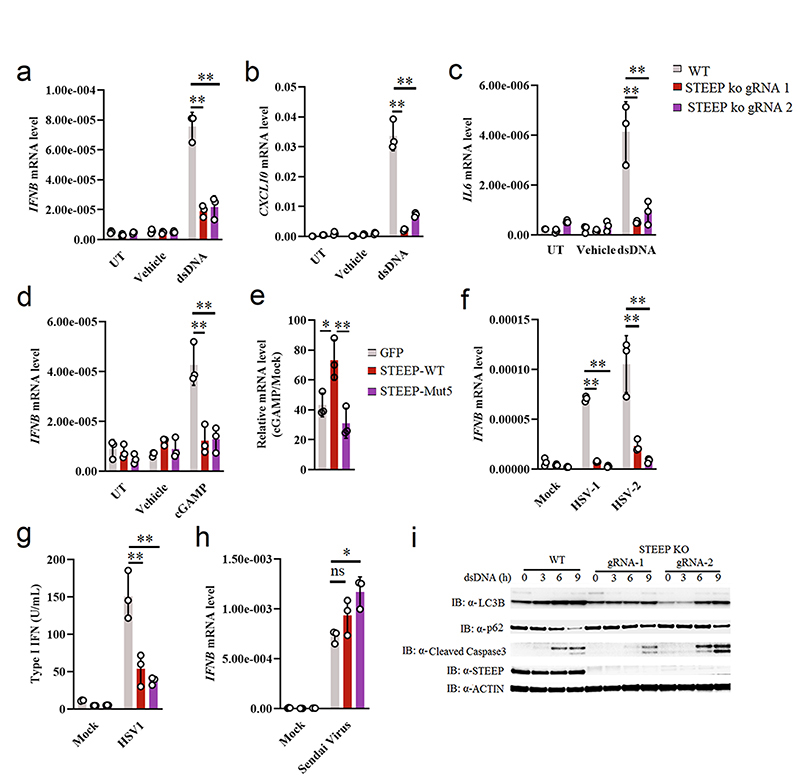
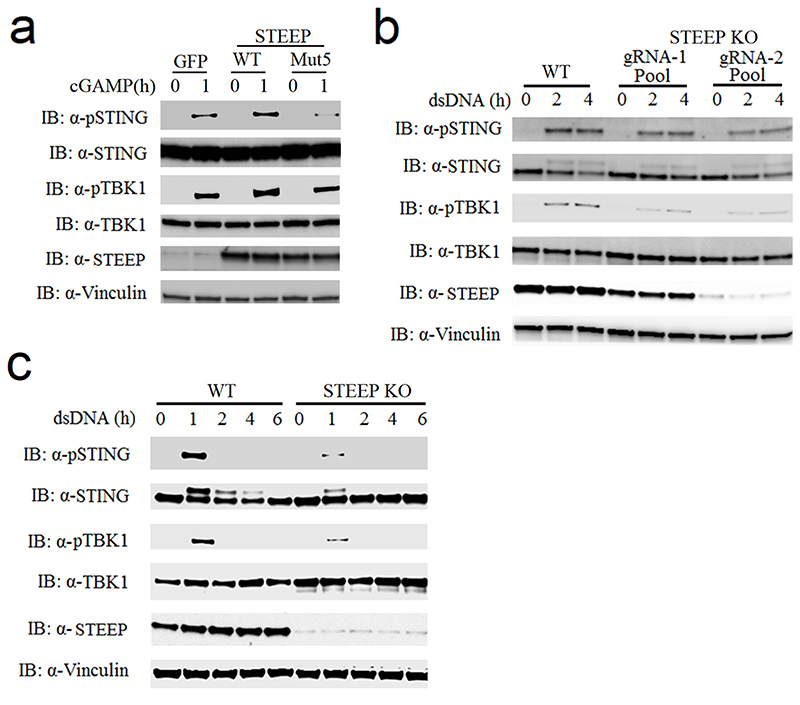
To further characterize the importance of STEEP in the cGAS-STING pathway, two different guide RNAs (gRNAs) were designed targeting different STEEP exons, and clonal THP-1 cell lines were generated using CRISPR-Cas9 technology (Extended Data Fig. 3a-b). Interestingly STEEP-deficient cells exhibited significantly attenuated expression of IFNB, CXCL10, and IL6 after stimulation with dsDNA or cGAMP (Fig. 2a-d). Introduction of mRNA encoding WT STEEP, led to partial rescue of STEEP expression (Extended Data Fig. 3c), and elevated response to cGAMP (Fig. 2e). This was not seen upon expression of STEEP mut5. In addition, STEEP deficiency significantly affected expression of IFNB mRNA induced by the human pathogenic DNA viruses HSV1 and HSV2 (Fig. 2f), as well as induction of type I bioactivity by HSV1 infection (Fig. 2g). By contrast, IFNB expression in response to the RNA virus Sendai virus was not affected in STEEP-deficient cells (Fig. 2h). In addition to IFN responses, STING signaling can also induce NF-κB activation, autophagy, and apoptosis29. As shown in Fig. 2i, STEEP-deficient cells have reduced LC3 conversion and p62 degradation after dsDNA-stimulation, indicating defective activation of autophagy. The effect of STEEP-deficiency on activation of apoptotic responses was modest. This suggests that STEEP is essential for STING-mediated induction of IFN, inflammatory cytokines, and autophagy.
Figure 2. STEEP is essential for IFN induction through the cGAS-STING pathway.
(a-d) WT and STEEP-deficient THP-1 cells were treated with (a-c) dsDNA (500 ng/ml, 6h) (**P < 0.01), (d) 2’3’ cGAMP (100 nM, 2h) (**P = 0.0022, **P = 0.0025, left to right). Total RNA was isolated, and IFNB, CXCL10, and IL6 mRNA were measured as indicated by RT-qPCR. (e) STEEP-deficient THP-1 cells transfected with mRNA encoding GFP, STEEP-WT or STEEP-Mut5, and subsequently stimulated with cGAMP (100 nM, 2h) (**P = 0.032, **P = 0.0068, left to right). IFNB was measured by RT-qPCR. (f-h) WT and STEEP-deficient THP-1 cells were infected with (f-g) HSV1 or HSV2 (both MOI 1, 12 h) (**P < 0.01), or (h) Sendai virus (MOI 1, 12 h) (ns P = 0.19, *P = 0.014, left to right). . Total RNA or culture supernatants were isolated, and IFNB mRNA and type I IFN bioactivity were measured as indicated by RT-qPCR and biological assay, respectively. (n = 3, biologically independent samples) (i) WT and STEEP-deficient THP-1 cells were treated with dsDNA (2 ug/ml) for the indicated time intervals, and levels of LC3, p62, cleaved caspase 3 (CC3), STEEP and actin were determined by immunoblotting. Representative blot from three independent experiments with similar results are shown. Data in panel a-h are shown as means of biological sample triplicates +/- st.dev. (n = 3 biologically independent samples for all data sets in the figure). Statistical analysis of data presented in panel a-h was performed using two-tailed one-way ANOVA test.
STEEP is essential for signaling events between STING dimerization and TBK1 recruitment
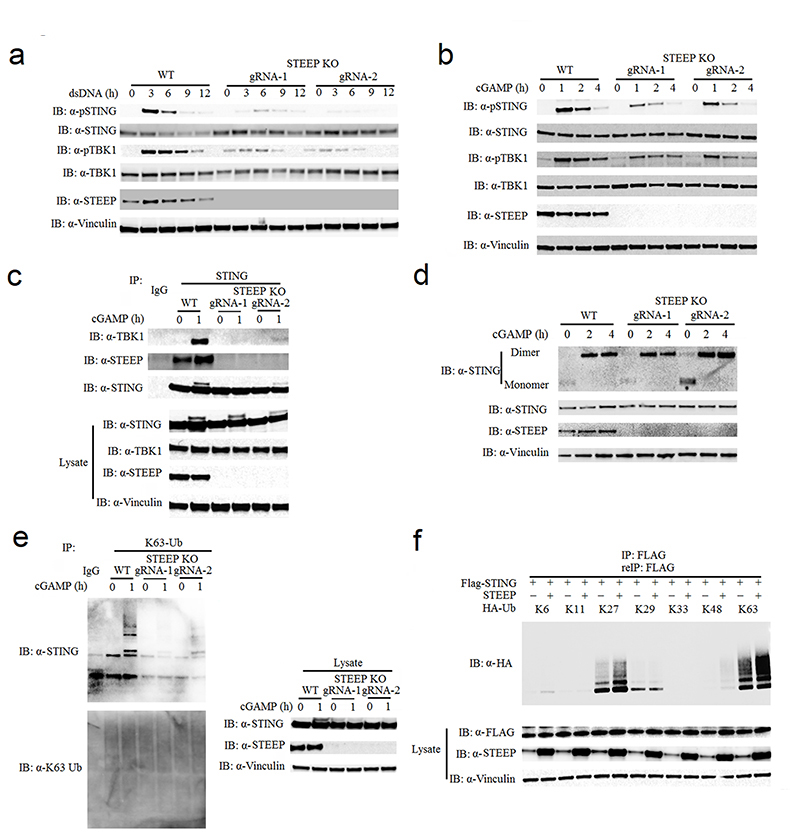
To identify how STEEP regulates cGAS-STING signaling, we compared the activation of different steps in the pathway following stimulation in WT cells and STEEP-deficient cells. Consistent with the reduced IFN induction in STEEP-deficient cells, the phosphorylations of STING and TBK1 were clearly reduced in STEEP-deficient THP-1 cells after stimulation with dsDNA or cGAMP (Fig. 3a-b). Reconstitution of STEEP expression elevated cGAMP-mediated phosphorylation of STING at S366 (Extended Data Fig. 4a). This was paralleled by reduced recruitment of TBK1 to STING in STEEP deficient cells after cGAMP stimulation (Fig. 3c). The observed impairment of STING and TBK1 phosphorylation was also observed in populations of Hela cells and human foreskin fibroblasts transduced with Cas9-STEEP gRNA expressing lentivirus (Extended Data Fig. 4b-c). However, STEEP-deficiency did not affect STING dimerization after cGAMP stimulation (Fig. 3d).
Figure 3. STEEP is essential for STING ubiquitination and recruitment of TBK1.
(a, b) Immunoblot analysis for the indicated proteins from whole cell lysates of WT or STEEP-deficient THP-1 cells after stimulation with (a) dsDNA or (b) cGAMP for the indicated time intervals. (n = 3 biologically independent experiments). (c) Immunoblot analysis of proteins co-immunoprecipitated with endogenous STING probed with the indicated antibodies. The lysates used for the immunoprecipitation were from WT or STEEP-deficient THP-1 cells stimulated with cGAMP for 1 h. (n = 3 biologically independent experiments). (d) STING dimerization assay. Immunoblot analysis with anti-STING probing of lysates from WT or STEEP-deficient THP-1 cells after stimulation with cGAMP for the indicated time intervals. The lysates were run on non-reducing SDS-PAGE prior to blotting. For controls reduced samples were run in parallel and blotted for STING, STEEP, and vinculin. (n = 3 biologically independent experiments). (e) K63-linkage ubiquitin TUBE assay and immunoblotting using anti K63-linkage ubiquitin and anti-STING antibodies. The material used was whole cell lysates from WT or STEEP-deficient THP-1 cells stimulated with cGAMP for 1 h. (n = 3 biologically independent experiments). (f) HEK-293T cells were transfected with FLAG-tagged STING, STEEP, HA-tagged Ub-K11, Ub-K27, Ub-K29, Ub-K33, Ub-K48 or Ub-K63 before co-immunoprecipitation and immunoblot analysis were performed with the indicated antibodies. (n = 3 biologically independent experiments).
Given the essential role of ubiquitination in the STING pathway1, we explored whether STEEP could influence STING ubiquitin levels. Using Tandem Ubiquitin Binding Entity (TUBE) assay, we found that K63-linked ubiquitination of STING was markedly inhibited in STEEP-deficient cells following cGAMP stimulation (Fig. 3e). In HEK293T cells, we co-transfected FLAG-tagged-STING, untagged STEEP, and HA-tagged ubiquitin mutants with all lysines mutated into arginines except for the one indicated. After immunoprecipitation and re-immunoprecipitation with anti-FLAG, we observed that STEEP co-expression significantly enhanced K27- and K63-linked poly-ubiquitination chains on STING, and slightly promoted K48-linked poly-ubiquitination (Fig. 3f). Collectively, these data suggest that STEEP acts in the STING activation pathway downstream of STING dimerization and upstream of STING ubiquitination, phosphorylation and recruitment of TBK1, which all occur after ER exit of STING18, 30.
STEEP promotes STING trafficking from ER to Golgi
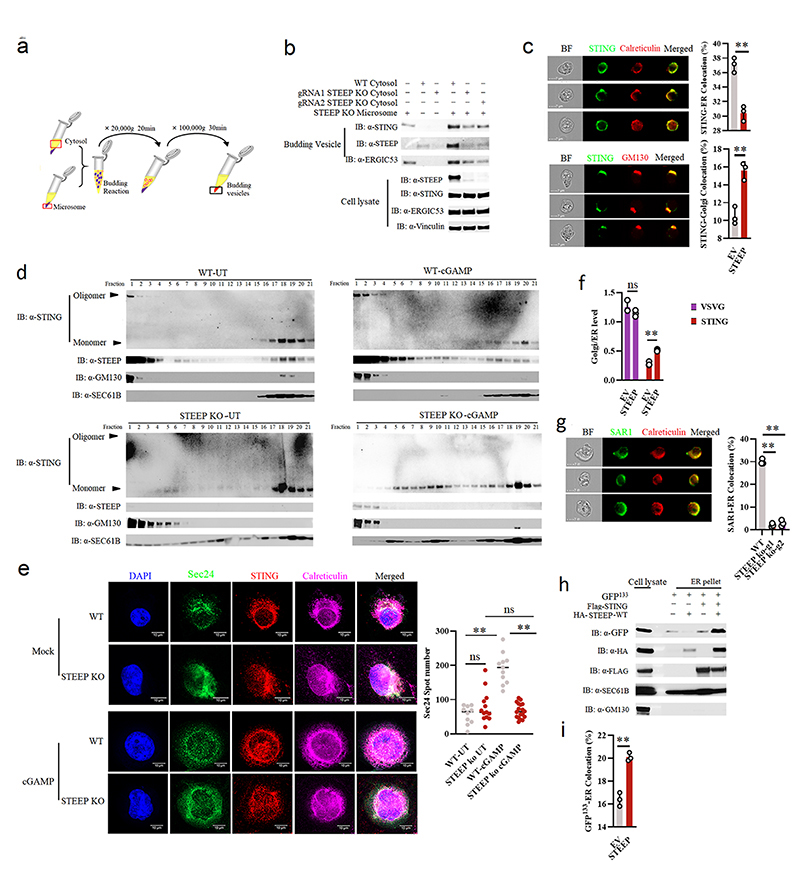
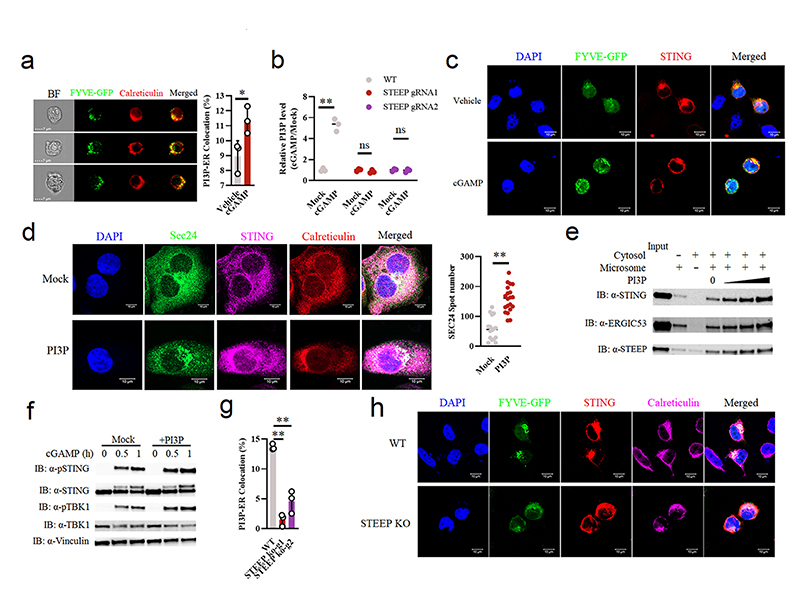
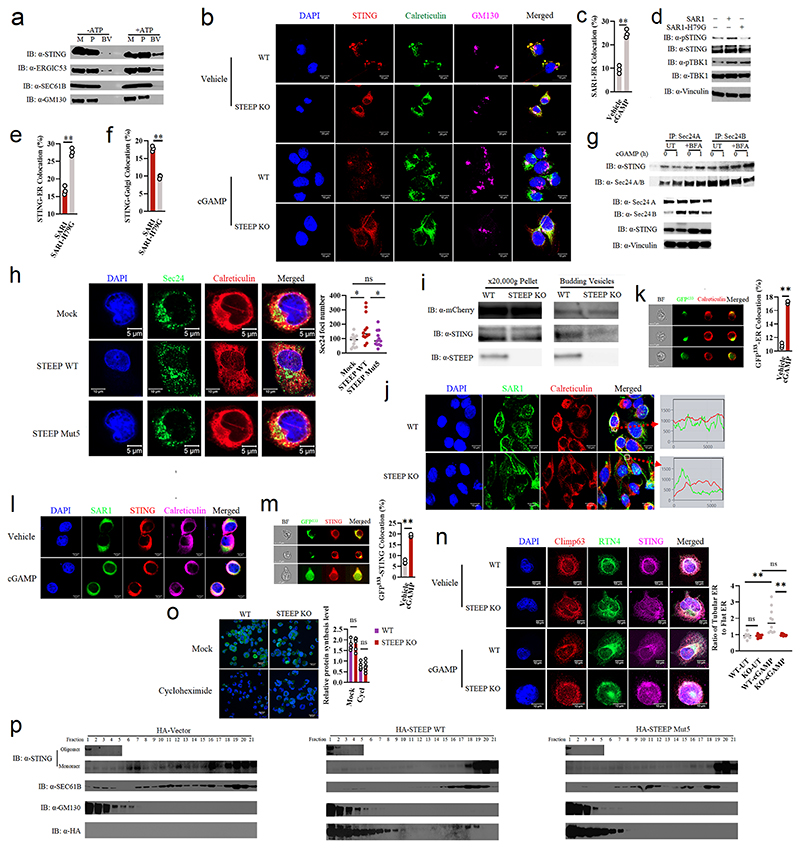
To evaluate whether STEEP promotes STING trafficking, we set up an assay where microsomes from one cell lysate were mixed with cytoplasm from another cell lysate, and co-precipitation of STING with small vesicles after centrifugation was evaluated31, 32 (Fig. 4a). Upon mixture of microsomes from STEEP-deficient THP-1 cells with cytoplasm from WT or STEEP-deficient cells, we observed that more STING was found in the budding vesicle fraction when STEEP was present in the cytoplasmic fraction (Fig. 4b). STING detected in the budding vesicle fraction was not due to contamination from the microsomes, since ATP was required for this to occur (Extended Data Fig. 5a). When STING localization was examined using ImageStream, we observed that expression of STEEP in HEK-293T-STING cells led to reduced amount of STING in the ER while increasing the localization of STING to the Golgi (Fig. 4c). Similarly, cell fractionation experiments and confocal microscopy revealed that STING trafficking from ER to Golgi after cGAMP stimulation was impaired in cells lacking STEEP (Fig. 4d, Extended Data Fig. 5b).
Figure 4. STEEP is essential for STING trafficking from ER to Golgi.
(a) Illustration of the in vitro membrane budding reaction. (b) Immunoblot analyses of budded material from the in vitro membrane budding reactions using cytosolic fractions from WT or STEEP-deficient THP-1 cells and microsomes from STEEP-deficient THP-1 cells. (n = 3 biologically independent experiments). (c) HEK-293T cells were transfected as indicated and probed with mouse anti-FLAG and either rabbit anti-calreticulin (ER marker) or anti-GM130 (Golgi marker). Cells were analysed by ImageStream. (n = 3) (**P = 0.0013, **P = 0.0030, up to down). (d) ER- and Golgi-enriched pellets from mock- or cGAMP-treated WT and STEEP-deficient HeLa cells were fractionated, and immunoblotted for STEEP, STING, GM130 (Golgi), and Sec61B (ER) (n = 2 biologically independent experiments). (e) WT and STEEP-deficient THP-1 cells stimulated with cGAMP for 10min were stained with anti-Sec24, anti-STING, and anti-calreticulin. For quantification of Sec24 foci, 10, 13, 11, and 18 cells from the WT-Mock, STEEP KO-Mock, WT-cGAMP, and STEEP KO-cGAMP, respectively were counted in a blinded fashion. Representative data from one experiment are shown (n = 3 biologically independent experiments). Statistical analysis used: two-tailed unpaired t test with welch’s correction. Each data point represents one cell and are shown as means +/- st.dev. (ns P = 0.15, **P < 0.000001, nsP = 0.41, **P < 0.000001, left to right). (f) HEK-293T cells transfected with STING, VSVG, and STEEP as indicated for 24 h were subjected to ImageStream analysis for colocalization of STING and VSVG with ER and Golgi. The data are shown as Golgi/ER (n = 3) (ns P = 0.18, **P = 0.00050, left to right). (g) FLAG-tagged Sar1 was transfected into WT or STEEP-deficient Hela cells for 24 h, and stimulated with cGAMP (100 nM, 30 min) Cells were probed with rabbit anti-calreticulin (ER marker) and mouse anti-FLAG. (n = 3) (**P = 0.0000040, **P = 0.000012, left to right). (h) HEK-293T cells were transfected as indicated. ER-enriched pellets precipitated with calcium chloride were immunoblotted for GFP, HA, GM130 (Golgi), and Sec61B (ER). (n = 3 biologically independent experiments). (i) GFP-tagged ALPS (GFP133) was co-transfected with the indicated vectors into HEK-293T cells for 24 h. Fixed cells were probed with rabbit anti-calveticulin (ER marker). (n = 3), and analyzed by ImageStream (**P = 0.00080). For data from ImageStream analysis (panels c, f, g, and i), each data point represents the percent of the positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data in panels c, f, g, and i was performed using two-tailed Student’s t-test.
Recent work suggests that STING exits the ER through COPII-mediated export25, 33, 34. Sar1 is a protein involved in COPII vesicle formation on the ER25, 35. cGAMP stimulation promoted recruitment of Sar1 to the ER (Extended Data Fig. 5c), and overexpression of WT, but not an inactive form of Sar1 (Sar1-H79G), led to STING activation and promoted STING trafficking in HEK-293T-STING cells (Extended Data Fig. 5d-f). Additionally, we found that the COPII complex protein Sec24 interacted with STING and the interaction was not inhibited by treatment with brefeldin A (BFA), which blocks ER-to-Golgi trafficking of COPII vesicles (Extended Data Fig. 5g). Moreover, cGAMP stimulation induced elevation in the amounts of Sec24 foci, indicative of COPII vesicle formation (Fig. 4e). The cGAMP-induced Sec24 foci formation was abolished in STEEP-deficient cells (Fig. 4e), and restored upon reintroduction of STEEP expression (Extended Data Fig. 5h). Of note, STEEP-deficiency did not affect the basal amounts of sec24 foci (Fig. 4e). In line with this, the observed redistribution of STING from ER to Golgi upon STEEP expression, was not seen for VSVG, another protein known to traffic via COPII vesicles (Fig. 4f)36. Likewise, STEEP deficiency did not impair VSVG redistribution from microsomes to budding vesicles (Extended Data Fig. 5i). These data suggest that STEEP promotes COPII-mediated trafficking by STING but not VSVG.
When evaluating for Sar1 recruitment to the ER, we found that this cytoplasmic protein was indeed abundantly localized to the ER following STING activation in a STEEP-dependent manner (Fig. 4g and Extended Data Fig. 5j). Based on the knowledge that Sar1 senses and promotes membrane curvature structures through the amphipathic N-terminal helix37, we wanted to examine ER membrane curvature in STING-activated cells, and the dependence on STEEP. Using the ALPS (Amphipathic Lipid Packing Sensors) motif GFP133 membrane curvature probe together with ER-specific staining, we found that ER membrane curvature was indeed induced by STEEP overexpression and cGAMP stimulation (Fig. 4h-i, Extended Data Fig. 5k-l). In addition, STING was observed to cluster at ER curvature-rich regions after cGAMP stimulation (Extended Data Fig. 5m). In line with this, we observed that cGAMP stimulation increased the proportion of the ER being in tubular areas, as measured by the RTN4:Climp63 ratio, and this was dependent on STEEP (Extended Data Fig. 5n). The tubular areas of the ER are assumed to be more curved38.
We noted that lysates from STEEP-deficient cells exhibited a broader distribution of the ER marker Sec61b in the fractionation assay (Fig. 4d), and therefore could have altered ER morphology and function. On the other hand, the basal levels of Sec24 foci and RTN4:Climp63 ratio were not altered in STEEP-deficient cells (Fig. 4e, Extended Data Fig. 5n). In addition, the levels of protein synthesis, which occurs on the ER was not affected by STEEP-deficiency (Extended Data Fig. 5o). Reintroduction of WT STEEP, but not the STEEP mutant 5 with impaired ability to interact with STING, restored the distribution of Sec61b in the fractionation assay (Extended Data Fig. 5p). Importantly, in cells expressing WT STEEP, but not in cells expressing mutant 5, a fraction of the protein localized to the ER. These data suggest that although STEEP deficiency does affect ER morphology, the organelle remains functionally unaffected, based on the parameters measured here. Collectively, STEEP promotes STING trafficking from ER to Golgi by inducing ER membrane curvature, which enables loading into COPII complexes.
STEEP augments ER PI3P production and membrane curvature to promote STING ER exit
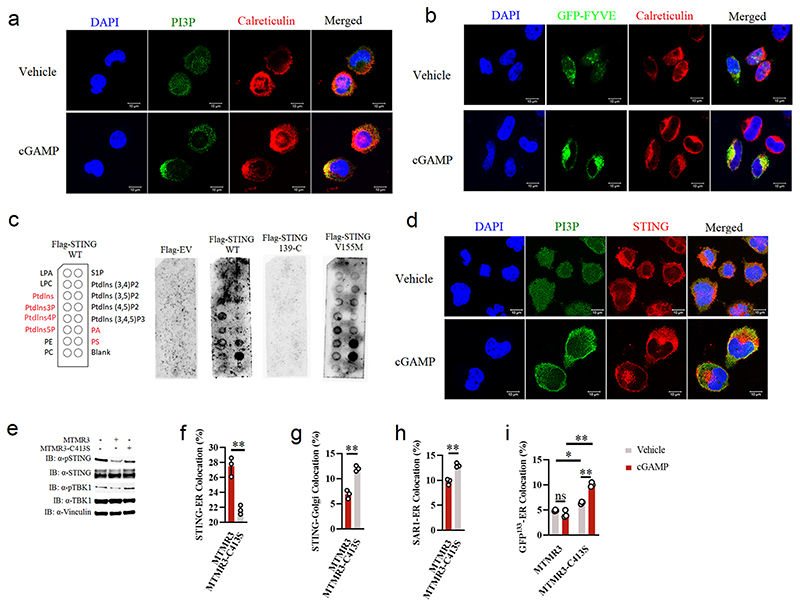
Based on the observation that STEEP promotes ER membrane curvature and STING ER exit, we wanted to explore how this could be mediated. It was previously reported that PI3P but not PI4P is enriched at the ER, and suppression of the PI3P kinase VPS34 has been shown to reduce DNA-induced type I IFN expression21, 39. Accumulation of negatively charged PI3P on one side of a lipid bilayer increases repulsive forces between polar heads, thus promoting membrane curvature40. First, we observed that STING agonist treatment elevated PI3P levels on the ER, as measured by the GFP-FYVE domain reporter, anti-PI3P antibody, and ELISA (Fig. 5a-b, Extended Data Fig. 6a-b), and also that full-length; but not the cytoplasmic region of STING; can interact with PI3P (Extended Data Fig. 6c). In agreement with this, both an anti-PI3P antibody and GFP-FYVE probe exhibited significant co-localization with an anti-STING antibody in activated cells (Fig. 5c, Extended Data Fig. 6d). Addition of exogenous PI3P promoted elevation of the levels of Sec24 foci (Fig. 5d), budding of STING into vesicles (Fig. 5e), and phosphorylation of STING and TBK1 (Fig. 5f). Consistently, overexpression of the PI3P hydrolase MTMR3, but not an inactive MTMR3 mutant, inhibited activation of the STING pathway (Extended Data Fig. 6e). This correlated with impaired STING trafficking from the ER to the Golgi (Extended Data Fig. 6f-g), reduced Sar1 recruitment to the ER (Extended Data Fig. 6h), and significantly decreased the level of ER membrane curvature induced by cGAMP stimulation (Extended Data Fig. 6i). Importantly, the accumulation of PI3P on the ER following STING activation was strongly reduced in STEEP-deficient cells (Fig. 5g-h). Altogether, these data demonstrate that STEEP promotes ER PI3P accumulation and ER membrane curvature to reinforce COPII-mediated STING ER exit.
Figure 5. STEEP governs PI3P accumulation on ER to promote STING ER exit.
(a) HEK-293T cells were co-transfected with GFP-tagged FYVE domain and FLAG-tagged STING for 24 h and stimulated cGAMP (150 nM, 30 min). Cells were analysed by ImageStream for PI3P and ER colocalization. ER, calreticulin. (n = 3) (*P = 0.038). (b) WT and STEEP-deficient HeLa cells were treated with cGAMP (100 nM, 1 h). Acidic lipids were isolated from ER fractions, and PI3P was measured by ELISA. Data are shown as means of biological sample triplicates +/- st.dev. (n = 3 biologically independent samples) (**P = 0.00028, ns P = 0.24, ns P = 0.66, left to right). (c) Confocal microscopy of Hela cells transfected with GFP-FYVE and FLAG-tagged STING, treated with cGAMP (100 nM, 20 min), and immunostained with and anti-FLAG (red). Representative data from one experiment are shown (n = 3 biologically independent experiments). (d) Confocal microscopy of THP-1 cells treated with vehicle or PI3P (100 uM, 2 h), and stained with anti-calreticulin, anti-Sec24, and anti-STING. For quantification of Sec24 foci, 16 and 20 cells from the Mock and PI3P-treated groups, respectively, were counted in a blinded fashion. Representative data from one experiment are shown (n = 3 biologically independent experiments). Statistical analysis was performed using two-tailed unpaired t-test with welch’s correction. Each data point represents one cell and are shown as means +/- st.dev (**P < 0.000001). (e) Immunoblotting of budded material from the in vitro membrane budding reactions incubated with increasing concentrations of PI3P (1 uM, 10 uM and 100 uM). (n = 3 biologically independent experiments). (f) Immunoblotting for the indicated proteins in whole cell lysates from THP-1 cells treated with PI3P (50 uM, 30 min) prior to stimulation with cGAMP, and incubation for the indicated time intervals. (n = 3 biologically independent experiments). (g, h) WT and STEEP-deficient Hela cells were transfected with GFP-tagged FYVE and FLAG-tagged STING for 24 h and stimulated with cGAMP for 20 min. The fixed cells were stained with anti-FLAG, anti-calreticulin and analyzed by ImageStream and Confocal microscopy. (n = 3) (**P = 0.000047, **P = 0.0013, left to right). h, a representative image from 2 biologically independent experiments with similar results is shown. For ImageStream data analysis (panels a, g), each data point represents the percent of the positive cells from one representative sample and shown as means +/- st.dev. Statistical analysis of data presented in panels a, b, and g was performed using two-tailed Student’s t-test.
STEEP recruits the VPS34 complex I to ER to produce PI3P
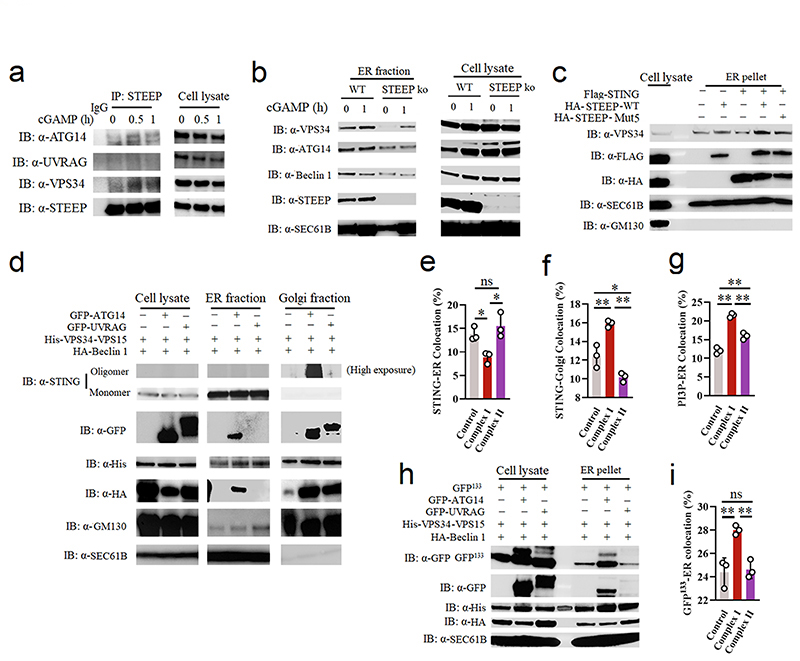
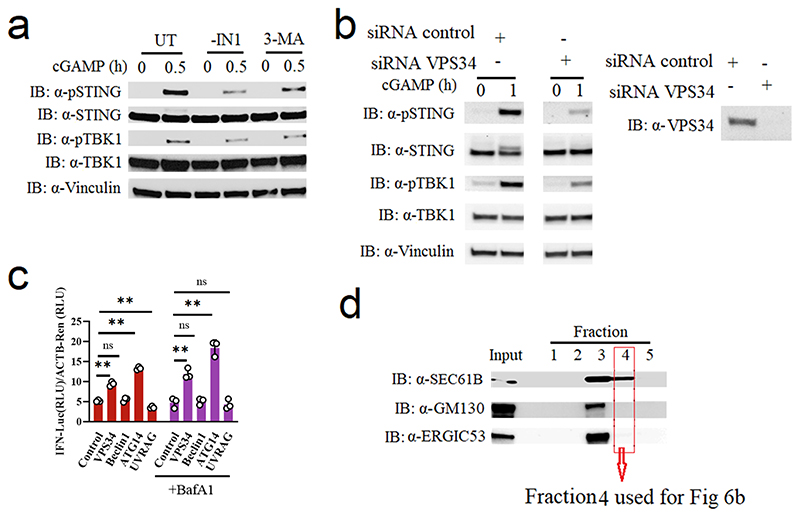
We confirmed previous findings that inhibition of the PI3P kinase VPS34 can suppress STING activation (Extended Data Fig 7a-b). VPS34 can engage in two different complexes to produce PI3P, namely VPS34 complex I (VPS34, Beclin1, VPS15, and ATG14) and Complex II (VPS34, Beclin1, VPS15, and UVRAG)41. Through overexpression in HEK-293T cells with STING stable expression, we found that enforced expression of VPS34 and ATG14 alone promotes IFNB promoter activity (Extended Data Fig. 7c). In addition, endogenous STEEP associated with ATG14 and VPS34, but not UVRAG, and cGAMP stimulation enhanced these interactions (Fig. 6a). Notably, recruitment of VPS34 complex I to the ER after STING activation was reduced in STEEP-deficient cells compared to controls (Fig. 6b, Extended Data Fig. 7d). The STEEP-dependent recruitment of VPS34 to the ER was dependent on expression of STING (Fig. 6c). Finally, we found that overexpression of VPS34 complex I proteins promoted STING trafficking from ER to Golgi (Fig. 6d-f), increased PI3P levels at the ER (Fig. 6g), and induced ER membrane curvature (Fig. 6h-i). These observations demonstrate that the VPS34 complex I is recruited to the ER in a STEEP-dependent manner to produce PI3P, which is essential for STING trafficking.
Figure 6. STEEP recruits the VPS34 complex I to ER to produce PI3P.
(a) Immunoblot analysis for the indicated proteins in anti-STEEP immunoprecipitates from THP1 cell lysates. Cells were stimulated with cGAMP (100 nM, for 0, 0.5, and 1 h). (n = 2 biologically independent experiments). (b) Immunoblot analysis of the indicated proteins from the ER fraction of THP-1 cells treated with cGAMP for 1 h. (n = 3 biologically independent experiments). (c) HEK-293T cells were transfected with Flag-STING and HA-STEEP (WT/mut5). ER-enriched pellets were immunoblotted for VPS34, FLAG, HA, GM130 (Golgi), and Sec61B (ER). (n = 3 biologically independent experiments). (d) ER- and Golgi-enriched pellets from STING-expressing HEK-293T cells transfected with control (His-VPS34, HA-Beclin1, V5-VPS15 and empty vector), Complex I (His-VPS34, HA-Beclin1, V5-VPS15 and GFP-ATG14) or Complex II (His-VPS34, HA-Beclin1, V5-VPS15 and GFP-UVRAG) were fractionated, and immunoblotted for the indicated proteins (n = 3 biologically independent experiments) (e, f) HEK-293T cells were transfected by control, Complex I, or Complex II (as specified under d above) and FLAG-tagged STING for 24 h. Fixed cells were probed with the indicated antibodies, and analyzed by ImageStream. (n = 3) (ns P = 0.54, *P < 0.05, **P = 0.01). (g) ImageStream analysis of PI3P-ER colocalization. HEK-293T cells were transfected with control, Complex I or Complex II (as specified for panel d above), GFP-FYVE and FLAG-tagged STING for 24 h. Fixed cells were probed with the indicted antibodies. (n= 3) (**P < 0.0001, **P = 0.0022, **P = 0.00030, left to right). (h) STING-expressing HEK-293T cells were transfected with GFP-tagged ALPS (GFP133), control, Complex I, or Complex II (as specified in panel d). ER-enriched pellets were immunoblotted for GFP, HA, His, GM130 (Golgi), and Sec61B (ER). (n = 3 biologically independent experiments). (i) HEK-293T cells were transfected with control, Complex I or Complex II, GFP133 and FLAG-tagged STING for 24h. Fixed cells were probed with the indicted antibodies and analyzed by ImageStream. (n = 3) (**P = 0.0053, ns P = 0.94, **P = 0.0074, left to right). For data from ImageStream analysis (panels e, f, g, and i), each data point represents the percent of the positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data in panels e, f, g, and i was performed by two-tailed one-way ANOVA test.
STEEP is important for STING activation in primary human cells and mice
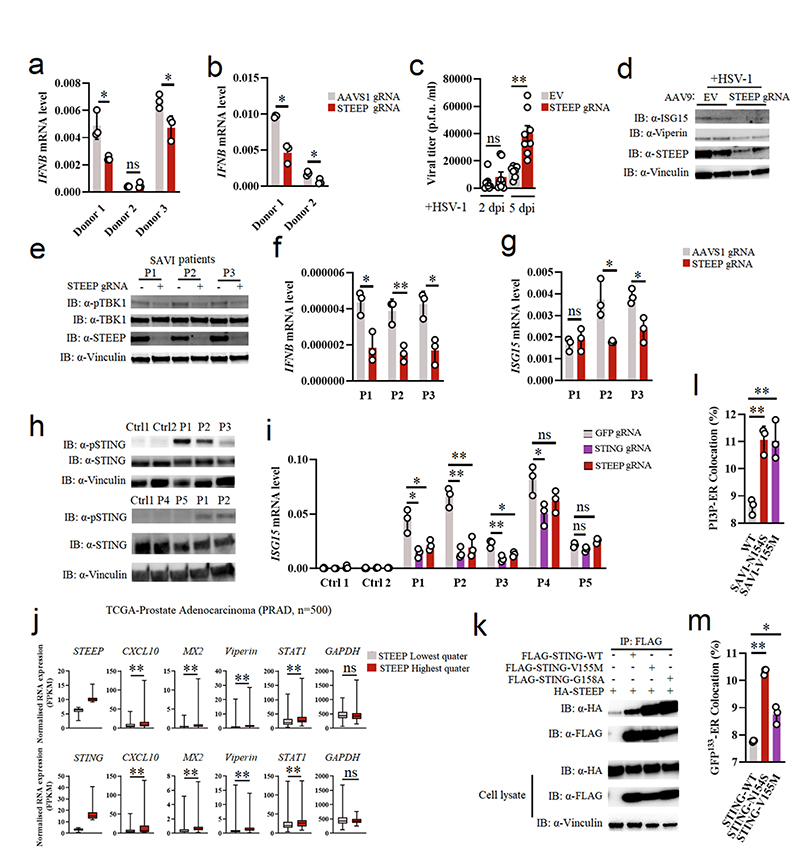
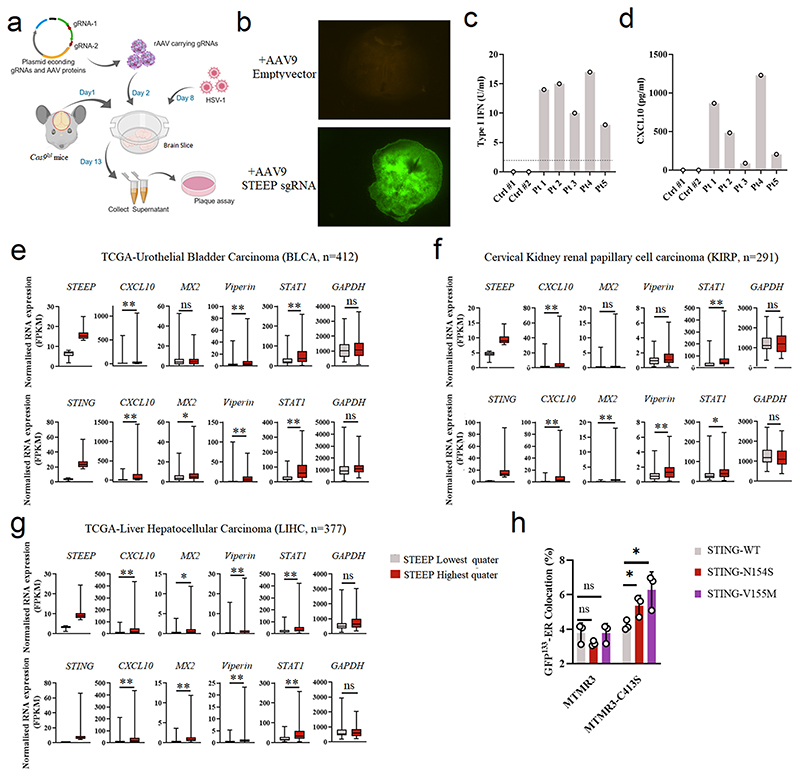
To explore whether STEEP contributes to the activation of STING signaling in primary human cells, we disrupted the STEEP gene using CRISPR/Cas9 in primary macrophages and fibroblasts (Extended Data Fig. 3a, d-f). Under these conditions, we found that suppression of STEEP expression impaired induction of IFNB mRNA expression (Fig. 7a-b). To test whether STEEP was important for immune responses at the tissue level, we targeted the STEEP gene in brain slices from Cas9 Lox-Stop-Lox mice (Cas9lsl) using adeno-associated virus (AAV)-mediated delivery of gRNA (Extended Data Figs. 3g-i, and 8a-8b). We have previously reported that cGAS and STING are essential for control of HSV1 in the mouse brain10. Interestingly, reduced STEEP expression in the brain led to elevated HSV replication in the tissue and impaired ISG responses (Fig. 7c, d). Thus, STEEP promotes STING-dependent antiviral immune responses.
Figure 7. STEEP facilitates STING-dependent antiviral defense and pathological signaling.
(a, b) Primary human fibroblasts and monocyte-derived macrophages treated with Cas9 and gRNAs as indicated were stimulated with vehicle or cGAMP (100 nM, 2h). Total RNA was harvested and IFNB levels measured. (n = 3) (a, *P = 0.049, ns P = 0.59, *P = 0.039, left to right; b, *P = 0.021, *P = 0.014, left to right). (c, d) Brain slices from Cas9+ mice treated with Steep-specific sgRNAs were infected with 5 x 103 PFU HSV1 (c) Viral load in the culture supernatants were determined by plaque assay. Data represent mean +/- st.dev, from n = 8 mice. Statistical analysis: two-tailed Mann-Whitney test (ns P = 0.33, *P = 0.00016, left to right). (d) Immunoblot analysis of ISGs response by indicative proteins in HSV1-infected WT or STEEP KO mouse brain splice (n = 2 mice). Dpi, days post infection. (e-g) Fibroblasts from SAVI patients were treated with Cas9 and the indicated sgRNAs. Whole-cell lysates and total RNA were isolated, and analyzed by immunoblot or RT-qPCR, respectively. e, data are from 3 SAVI patients, and panels f and g data are shown as means +/- st.dev, for n = 3 independent replicates. (ns P = 0.56, *P < 0.05, **P < 0.01). (h) PBMC lysates from healthy controls and five SLE patients were subjected to immunoblotting with antibodies against pSTING (S366), total STING, Vinculin. Data are from n = 2 unrelated healthy donors (Ctrl1, and 2), and n = 5 unrelated SLE patients. (i) Monocytes from two healthy donors and five IFN-high SLE patients were treated with gRNA/Cas9 complexes targeting STING and STEEP. Total RNA was isolated 48 h later and analyzed for ISG15 mRNA levels (n = 3) (ns P > 0.05, *P < 0.05, **P < 0.01). (j) Box plots of TCGA RNA expression profiles in Prostate adenocarcinoma (PRAD). The highest and lowest 25% of expression of STEEP (upper panels) and STING (lower panels) were analyzed by comparing STEEP/STING-high and STEEP/STING-low groups. Statistical analysis: two-tailed Mann-Whitney test. The upper and lower ends of the boxes represent the upper and lower quartiles, and the horizontal line inside the box is median of the dataset. The bar extending parallel from the boxes is the “whiskers”, indicating upper and lower extreme of the dataset (ns P > 0.01, **P < 0.001). (k) Lysates from HEK-293T cells transfected with the indicated STING expression constructs were immunoprecipitated with anti-Flag, and immunoblotted with antibodies directed against HA and FLAG. (n = 3 biologically independent experiments). (l, m) ImageStream analysis of PI3P-ER (l) and ER-GFP133 colocalization (m). HEK-293T cells were transfected with GFP-FYVE/GFP133 and STING/STING-SAVI mutants for 24 h. Fixed cells were incubated with rabbit anti-calreticulin (n = 3 biologically independent experiments) (l, **P = 0.0017, ** P = 0.0053, left to right; m, **P = 3.20E-06, *P = 0.035, left to right).. Data in panels a, b, f, g and i are shown as means of biological technical triplicates +/- st.dev from one representative experiment. For data from ImageStream analysis (panels l and m), each data point represents the percent of the positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data presented in panels a, b, f, and g was used through two-tailed unpaired t-test with welch’s correction, and in panels i, l, and m was performed using two-tailed Student’s t-test.
STEEP is essential for ISG expression in cells from SAVI and SLE patients
SAVI-associated STING gain-of-function mutations promote constitutive type I IFN production without requirement for STING agonists14. Likewise, a subpopulation of SLE patients have elevated cGAMP and ISG levels, which correlates with disease severity42. Given the positive role for STEEP in STING activation, we wondered whether STEEP was important for the activity of SAVI-associated STING mutants and the ISG expression by immune cells from IFN-high SLE patient cells. When the STEEP gene was targeted in cells from SAVI patients (Extended Data Fig 3e), we observed that STING signaling and expression of IFNB and ISG15 was reduced (Fig. 7e-g). In five IFN/ISG-high SLE patients (Extended Data Fig 8c-d), we observed that three had elevated levels of pSTING (Fig. 7h). Targeting of STING or STEEP led to reduced expression of ISG15 selectively in cells from the pSTING-positive patients (Fig. 7i). Finally, through data mining of publicly available gene expression data, we observed a correlation between expression levels of either STING or STEEP and ISG transcripts (Figure 7j, Extended Data Fig 8e-g). These data together support an important role for STEEP in STING activity. At the mechanistic level, SAVI STING variants tended to co-precipitate more abundantly with STEEP than did WT STING (Fig. 7k). Moreover, cells expressing SAVI STING variants had higher levels of PI3P at the ER, and increased ER membrane curvature (Fig. 7l-m). Removal of PI3P with MTMR3 reversed this difference between the variants (Extended Data Fig 8h). These data demonstrate that the STEEP-STING interaction facilitates ligand-independent signaling by SAVI-associated STING mutants by promoting PI3P production, ER membrane curvature, and STING trafficking (Extended Data Fig. 9).
Discussion
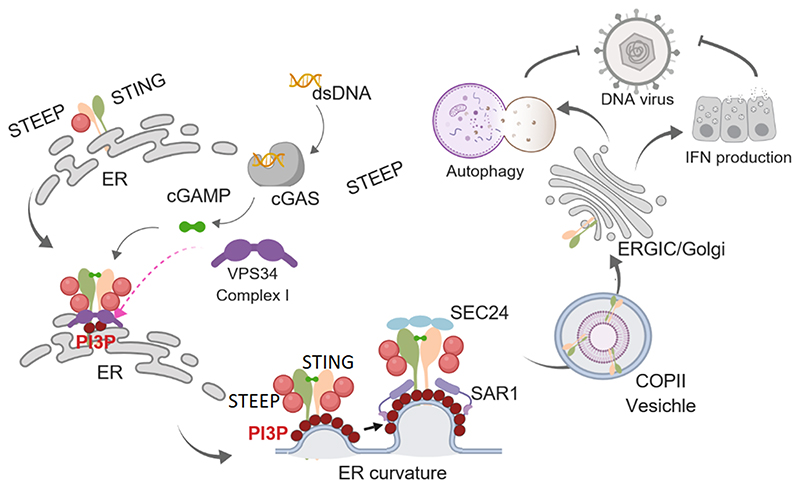
The cGAS-STING pathway is critical for defense against a large panel of infections, and for efficient anti-cancer immune responses1, 10, 11, 12. On the other hand, this pathway is also responsible for the excessive immune response that causes development of some autoinflammatory diseases13, 14. Thus, tight control of STING signaling is essential for ensuring stimulus-specific signaling and avoidance of undesired stimulus-independent activity. STING ER-to-Golgi trafficking is an essential early step in STING signaling18, and has been suggested to be a rate-limiting step in activation of the pathway19. Recently, it was reported that STIM1 plays an essential role in retaining STING in the ER20. Despite this, it is not known what mechanisms govern STING exit from the ER to reach the ERGIC/Golgi where TBK1 is recruited and signaling is activated. We report here that STING interacts with STEEP, a previously uncharacterized protein, and this promotes VPS34-mediated production of PI3P at the ER and membrane curvature. This leads to recruitment of the curvature binding proteins Sar1, association of STING with the COPII vesicle machinery, and traffic out of the ER. STEEP-dependent STING ER exit is essential for STING signaling in antiviral defense and in inflammatory diseases.
Several proteins have been reported to interact constitutively with STING and to be essential for STING trafficking including iRhom222, SNX823, UBXN3B24, and TMED225. Despite this, in-depth understanding of the key question as to how STING leaves the ER to start assembly of the signaling complex at the ERCIG/Golgi has remained unresolved. We found that STEEP interacted constitutively with STING, this interaction was augmented following DNA stimulation, and was also seen for SAVI-associated STING mutants. The STING-STEEP interaction in turn promoted STING trafficking. The cryo-EM structure of cGAMP-bound STING was recently reported and revealed a 180° rotation of the ligand-binding domain relative to the transmembrane domain upon ligand binding5. This rotation involves the region where SAVI-associated mutations have been identified, and it was speculated that these STING mutants have lower threshold for undergoing the domain rotation spontaneously. We found that the STING-STEEP interaction was dependent on the N-terminal transmembrane domains and the dimer interaction surface. It is therefore tempting to speculate that the altered surface of the STING dimer following rotation of the ligand-binding domain relative to the transmembrane domain increases the affinity of STEEP for STING. It still remains to be fully elucidated why STING-mediated TBK1 activation requires trafficking from ER to Golgi. It has been suggested that STING palmitoylation, which is essential for activation, occurs on the Golgi43. However, other mechanisms may also be involved. For instance, some of the Ubiquitin E3 ligases modulating STING may be more abundant on the Golgi, and the composition of bioactive lipids on the Golgi may be more favorable for TBK1 recruitment44.
The trafficking of proteins from the ER can occur through ER-associated protein degradation (ERAD) or COPII. While the former targets misfolded proteins for proteasomal degradation, the latter mediates trafficking of proteins from the ER to the Golgi through coat protein vesicles45. COPII vesicular budding is dependent on the GTPase Sar1 and the coat protein complex consisting of Sec23/24, and Sec13/31. In addition, an ER-associated pathway for control of vesicles in the perinuclear area has been described46. Recent publications reported that STING exits the ER through COPII-mediated export25, 33, 34, and accordingly we found that STING activation lead to recruitment of Sar1 to the ER in a STEEP-dependent process upstream of STING ER exit. Sar1 is recruited from the cytoplasm to the ER in a manner dependent on ER membrane curvature35. High local concentration of PI3P in membranes is well-described to promote membrane curvature, via repulsive forces between the negatively charged polar heads. We observed that the PI3K VPS34 complex I was recruited to STING through a process dependent on STEEP, which also promoted PI3P accumulation on the ER. PI3P was detected by a PI3P binding antibody and a GFP-FYVE domain reporter, while membrane curvature was detected by a curvature-binding probe. Since PI3P and membrane curvature were both measured indirectly, it is in principle possible that unspecific signals were also detected in these measurements. However, our functional data support the conclusions that STEEP promotes ER PI3P production and membrane curvature, although the sequence of these two events is not yet conclusively established. Particularly, Sar1 was recruited to the ER following STING activation in a STEEP-dependent manner and STING ER-exit was prevented by inhibition of PI3K activity. Since ER membranes are often surrounded by endosomes, and sometimes even form contact47, we cannot formally exclude that the PI3P we detect by ImageStream localizes to endosomes. The association of STING with the VPS34 complex I, and the requirement of PI3P for STING ER exit would, however, suggest that a biologically significant proportion of the detected PI3P does localize to the ER. Collectively, our data demonstrate that STEEP promotes PI3P accumulation in the ER and formation of ER membrane curvature to enable COPII-mediated STING trafficking. This fills a significant knowledge gap in the understanding of the rate-limiting step in STING signaling.
STEEP is evolutionarily highly conserved, with strong homology between all vertebrates, and with clear orthologues to human STEEP in arthropods and nematodes. By contrast, nematodes do not have STING and only few arthropods do. This suggests that STEEP is involved in an ancient cellular process, which STING took advantage of as the cGAS-STING pathway developed. Based on the data presented in this work, we suggest that STEEP is involved in ER membrane modulation in relation to the COPII trafficking machinery. However, the observation that the distribution of VSVG between ER and Golgi is not affected by STEEP expression suggests that STEEP is not a general factor in COPII-mediated ER-to-Golgi transport. It should be mentioned that STEEP has a predicted nuclear localization signal, and a pool of STEEP localizes to the nucleus. Thus, it is likely that STEEP also has other functions, which have yet to be discovered. The tight connection between the STING pathway and membrane modulatory systems is further highlighted by the finding that STING induces autophagy34, 48, and that several autophagy-related proteins are involved in both activation and regulation of STING signaling8, 18, 21. This includes VPS34, the activity of which we show was augmented by STEEP upon DNA stimulation. We observed that STING-induced expression of IFN and IL-6, as well as activation of autophagy, were all dependent on STEEP, suggesting a requirement for STING to exit from the ER for these processes. However, STING-dependent caspase-3 cleavage exhibited only modest dependence on STEEP. Unraveling the determinants, which govern whether STING relays signaling to IFN expression or cell death pathways in different immunological contexts deserves urgent attention.
A key observation of the present study is that depletion of STEEP in mouse brain tissue reduces activation of STING signaling and impairs antiviral defense. In addition, we observed that ISG15 expression by cells from SAVI patients and pSTING positive SLE patients was dependent on STEEP, and that levels of several ISG transcripts in cancer patient transcriptome databases correlates with expression of both STING and STEEP. In parallel, STEEP interacted more strongly with SAVI-associated STING than with WT STING, and the patient STING variants also stimulated ER accumulation of PI3P, as well as ER membrane curvature more potently than WT STING. SAVI-associated STING variants can signal independent of cGAMP binding14. Our data suggest that the elevated STEEP interaction of SAVI-associated STING variants, which leads to elevated PI3P production and ER membrane curvature, could allow STING ER exit, and hence signaling independent of cGAMP binding. Since subcellular trafficking is of central importance for activation of signaling by innate immune receptors, including TLRs, RIG-I, and cGAS49, 50, this work provides mechanistic insight into an important principle in immunology. Moreover, our data suggest that pharmacological targeting of the STING-STEEP interaction holds potential for treatment of SAVI, SLE and other STING-dependent inflammatory diseases.
Methods
Cells, Viruses, and Reagents
The cell lines used for the study were iBMDC-MycSTING, THP-1, Vero cells, HEK-293T cells, HaCaT, HeLa cells, and Human foreskin fibroblasts (HFF). The choice of cell models depended on the experiments; e.g. THP-1 cells express high levels of cytokines and were used to evaluate gene expression, while HeLa cells have a large cytoplasm and were often the cell line of choice for immunofluorescence. SAVI patient-derived fibroblasts cells were obtained as described previously14. Human blood-derived monocytes were isolated from normal healthy blood donor buffy coats obtained from Aarhus University Hospital Blood Bank. Human primary fibroblasts were obtained from healthy donors from Department of Dermatology and Venereology, Aarhus University Hospital. The viruses used were HSV-1 (Strain F+, and McKrae), HSV-2 (strain 333), Sendai virus (strain Cantrell). The reagents used were 2’3’-cGAMP (BIOLOG), 60mer dsDNA (DNA technology), PI(3)P diC8 (Echelon), phorbol 12-myristate 13-acetate (PMA, Sigma), Brefeldin A (BFA, Sigma), Bafilomycin A1(BafA1, InvivoGen), 3-Methyladenine (3-MA, InvivoGen), VPS34 Inhibitor 1 (VPS34 IN1, Cayman).
Expression plasmids
For a complete list of plasmids used in this study, please see Supplementary Table 2.
Isolation of monocytes from IFN and CXCL10-high SLE patients
Blood was drawn from SLE patients and healthy donors and used for isolation of serum and peripheral blood mononuclear cell (PBMC)s using Sepmate tubes (Stemcell Technologies) with Ficoll-plaque (GE Healthcare Lifesciences). PBMCs were stored at -180 °C before use in experiments. Serum levels of type I IFN bioactivity and CXCL10 protein, were determined by the HEK-blue IFNα/β assay (InvivoGen), and ELISA (R&D Systems), respectively. For isolation of monocytes, thawed PBMCs were stained with mouse anti–human CD14 (clone M5E) and sorted into cell culture plates. The purity of the cell populations were (Ctrl 1, Ctrl 2, Pt 1, Pt 2, Pt 3, Pt 4, Pt 5): 96.8%, 96.4%, 98.3%, 95.4%, 94.6%, 95,3%, 97.3%. The cells were subjected to genome-editing as described below.
Cell culture and transfection
HEK-293T, Hela and HFFs were cultured in DMEM with 10% fetal bovine serum (FBS) and 100 units/ml penicillin, 100 μg/ml streptomycin. THP-1 cells were cultured in RPMI 1640 media with L-glutamine supplemented with 10% fetal bovine serum, 100 units/ml penicillin, 100 μg/ml streptomycin. THP-1 cells were differentiated into macrophage cells using PMA (150 nM) overnight. The PMA was removed and cells were used for experiments 24 h later. The 60 mer double-stranded DNA (dsDNA) was transfected into cells by Lipofectamine 2000 according to manufacturer’s instructions. The 2’,3’-cGAMP was transfected into cells by digitonin.
Mass spectrometry
Myc was immunoprecipitated from whole cell lysates from iBMDC-MycSTING cells using the protocol described under “Co-Immunoprecipitation” below. The immunoprecipitate was subjected to mass spectrometry analysis.
The samples were denatured, reduced and alkylated in 20 mM Tris-HCl, 8 M Urea, pH 8 containing 5 mM DTT followed by the addition of iodoacetamide to a final concentration of 15 mM. The reduced and alkylated proteins were collected by ultrafiltration (10 kDa cut-off) and washed with 100 mM ammonium bicarbonate on the filter. The proteins were digested with trypsin at 37°C for 16 h (on filter). The tryptic peptides were collected by centrifugation and micro-purified using Empore™ SPE Disks of C18 octadecyl packed in 10 μl pipette tips. NanoLC-MS/MS analyses were performed on an EASY-nLC II system (ThermoScientific) connected to a TripleTOF 5600+ mass spectrometer (AB Sciex). The LC system was setup with a 2 cm x 100 μm trap column and a 15 cm x 75 μm analytical column (pulled emitter). Both columns were packed in-house with RP ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were separated using a 50 min gradient from 5% to 35% phase B (0.1% formic acid and 90% acetonitrile) and a flow rate of 250 nl/min. The collected MS files were converted to Mascot generic format (MGF) using the AB SCIEX MS Data Converter beta 1.1 (AB SCIEX). The generated peak lists were searched against the Uniprot mouse proteome using the Mascot search engine (matrix science). Search parameters were allowing one missed trypsin cleavage site and carbamidomethyl as a fixed modification and oxidation of Met were selected as variable modifications. Peptide tolerance and MS/MS tolerance were set to 10 ppm and 0.1 Da respectively.
IFNB Luciferase reporter gene assay
HEK-293T cells with or without stable STING expression were plated on 96 well plates and transfected with 30 ng constructs harboring IFNB-promoter firefly luciferase reporter genes, and 10 ng β actin-promoter-driven renilla luciferase together with 50 ng constructs encoding either STEEP (WT or mutants), cGAS, TBK1, IRF3-5D, TRIF, MAVS, ATG14, UVRAG, Beclin1, or VPS34. Eighteen hours after transfection, the cells were stimulated with cGAMP or left untreated. Six hours later, the cells were lysed, and the firefly- and renilla luciferase signals were developed with Dual Glo® luciferase assay (Promega) and read on Luminoscan Ascent (Labsystems) according to manufacture’s instructions.
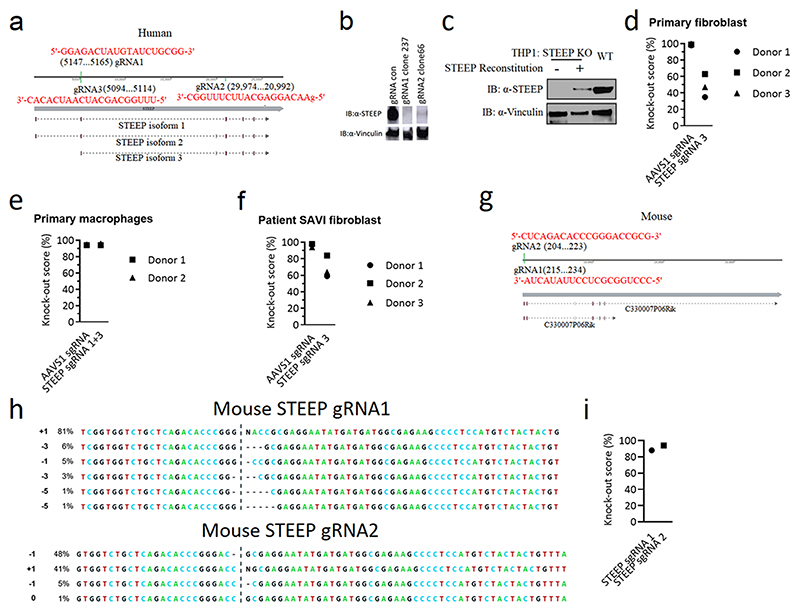
Generation of genome-edited cell lines
The STEEP knockout in THP-1, Hela, and HFF cells was generated by CRISPR/Cas9 technology. The gRNAs were used in human cell lines STEEP sgRNA#1, 5′-GGAGACTATGTATCTGCGG-3′; STEEP sgRNA#2, 5′-gAACAGGAGCATTCTTTGGC-3′ (Extended Data Fig. 3a). The cells were transduced by lentivirus packaged with the targeted sgRNA. After selection by 1ug/ml Puromycin, the knockout efficiency was analyzed by Immuno blotting.
Reconstitution of STEEP expression in cell lines
For the reconstitution of STEEP expression in STEEP-deficient THP-1 cells, STEEP (WT or mut5) mRNA was delivered by Nucleofection. STEEP-deficient THP-1 cells (0.5x106) were electroporated with 2.5 ug GFP, STEEP WT, or STEEP Mut5 mRNA. After electroporation, the cells were differentiated into macrophage cells using PMA (150 nM) overnight. The PMA was removed and cells were used for experiments 24 h later. For the reconstitution of STEEP expression in STEEP-deficient Hela cells, HA-Vector, HA-STEEP WT, or HA-STEEP Mut5 plasmids were transfected into cells through Lipofectamine 2000. After 24 h transfection, the cells were used for the isolation of ER and Golgi fractions.
Genome-editing in human primary cells
CRISPR sgRNAs directed against the negative control locus AAVS1 (5’-GGGGCCACUAGGGACAGGAU-3’) and CXorf56 (sgRNA 1: 5’-GGAGACUAUGUAUCUGCGG-3’, sgRNA 3: 5’-UUUGGCAGCAUCAAUCACAC-3’) were purchased from Synthego with three terminal nucleotides in both ends chemically modified with 2’-O-methyl-3’-phosphorothioate51. Cas9 protein (Alt-R S.p. Cas9 Nuclease 3NLS) was purchased from Integrated DNA Technologies (IDT). Ribonucleoprotein (RNP) complexes were made by complexing Cas9 and sgRNA at a molar ratio of 1:2.5 (Cas9:sgRNA) at 25 °C for 15 min prior to nucleoporation. Cells were resuspended in OptiMEM (Gibco, Thermo Fisher Scientific) and electroporated with a concentration of 300 μg/mL Cas9 protein using the Lonza 4D-Nucleofector System (program CM-138). For primary fibroblasts, 1x105 cells were electroporated for each condition, using the 20 μL Nucleocuvette Strip format (Lonza), and were subsequently seeded into a 24-well plate. For the generation of genetically modified primary macrophages, CD14+ monocytes were initially purified from PBMCs by negative immunomagnetic selection, according to the manufacturer’s instructions (EasySep Human Monocyte Enrichment kit, STEMCELL Technologies). 4x106 isolated monocytes were electroporated for each condition, using the 100 μL Nucleocuvette Vessel format (Lonza), and were subsequently distributed into 8 wells of a 24-well plate (5x105 cells/well). To induce differentiation of monocytes into macrophages, cells were cultured in DMEM supplemented with 10% heat inactivated human AB-positive serum for seven days in the presence of 15 ng/mL M-CSF (R&D Systems). Medium was replenished every third day. Seven days after transfection, frequences of Indel and missense mutations were quantified using the ICE software (http://ice.synthego.com). For this, genomic DNA was extracted from the cells and PCR amplicons spanning the sgRNA target sites were generated using Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific). The following primers were used: AAVS1 forward: 5’-ATTCGGGTCACCTCTCACTCC-3’; AAVS1 reverse: 5’-CAGGGTGGCCACTGAGAACC-3’; CXorf56 forward: 5’-ACCAGGGTAGTAAGAGAGCTGG-3’; CXorf56 reverse: 5’-GGCCGACATGGTGAAACTGC-3’. Purified PCR products were then Sanger-sequenced and Indel frequencies quantified with a reference WT sequence (mock-transfected sample) used as a control.
RNA extraction and quantitative RT–PCR
The RNA isolation and qPCR was performed as described by the manufacturers Roche and Thermo, respectively with the following primers from Applied Biosystems: IFNB1 (Hs01077958_s1), ISG15 (Hs01921425_s1), IL6 (Hs00174131_m1), and CXCL10 (Hs01124251_g1). The mRNA level of the genes were normalized by β-actin using the formula 2Ct(βactin) − Ct(mRNA X). The resulting normalized ratio (NR) was presented directly in the figures.
Immunoblotting
Whole-cell extracts or immunoprecipitation samples were diluted in XT sample buffer and XT reducing agent, and loaded into 4-20% SDS–PAGE gel (Bio-Rad). The proteins were transferred from gel to PVDF membranes through Trans-Blot Turbo™ Transfer System® (Bio-Rad). The membrane was blocked in 5% nonfat skim milk (Sigma) for 1 hour at room temperature. For the STING dimerization assay, cell lysates were prepared by IP lysis buffer (Thermo Fisher Scientific) mixed with XT sample buffer but without XT reducing agent and complete mini inhibitor (Sigma).
The blots were developed by ImageQuant™ LAS 4000 biomolecular imager (GE Healthcare Life Sciences) or ChemiDoc™ Imaging System (Bio-Rad). The information of antibodies used for immunoblotting is listed as follows: rabbit anti-STING (Cell Signaling, D2P2F/#13647, 1:1,000), sheep anti-STING (R&D Systems, AF6516, 1:500), rabbit anti-pSTING (S366) (Cell Signaling Technology, #19781, 1:1,000), rabbit anti-TBK1 (Cell Signaling, 3504, 1:1,000), rabbit anti-pTBK1 (Ser172) (Cell Signaling, D52C2/#5483, 1:1,000), mouse anti-p62 (Cell Signaling, # 88588, 1:1,000), rabbit anti-LC3 (Cell Signaling, #3868, 1:1,000), mouse anti-ERGIC53/LMAN1 (Santa Cruz Biotechnology, sc398893, 1:1000), rabbit anti-calreticulin (Abcam, #ab2907,1:1000), rabbit anti-GM130 (Cell Signaling, #12480, 1:1000), rabbit anti-cleaved caspase3 (Cell Signaling, #9664,1:1000), mouse anti-FLAG M2 (Sigma-Aldrich, F3165, 1:1,000), rabbit anti-ACTB (Cell Signaling, #4970,1:1000), rabbit anti-HA (Cell Signaling, #3274, 1:1,000), rabbit anti-GFP (Cell Signaling, #2956, 1:1,000), mouse anti-His Tag-HRP Conjugate (Cell Signaling, #9991, 1:1000), rabbit anti-Flag Tag-HRP Conjugate (Cell Signaling, #2044, 1:1000), mouse anti-HA Tag-HRP Conjugate (Cell Signaling, #2999, 1:1000), rabbit anti-VPS34 (Cell Signaling, #4263,1:1000), mouse anti-ATG14 (MBL, #M184-3, 1:1000), rabbit anti-UVRAG (MBL, #M160-3, 1:1000), mouse anti-Viperin Antibody (Millipore, #MABF106, 1:1000), rabbit anti-ISG15 (Cell Signaling, #2743, 1:1,000), and mouse anti-vinculin (Sigma, #V9131, 1:10,000). Original blots are shown in Source data numbered according to the figures they refer to.
Co-Immunoprecipitation
For the Co-Immunoprecipitation of endogenous protein, the cells were lysed in IP lysis buffer with 1xProtease Inhibitor Cocktail (Sigma) and 10 mM NaF, and then the cell lysates were centrifuged for 10 min, 2,500 g at 4 °C. The cleared cell lysates were incubated with primary antibody against sheep anti-STING or rabbit anti-STEEP overnight at 4°C. Dynabeads™ Protein G (Invitrogen) was added into the mixtures for extra 2 h. After 4-6 times wash with IP lysis buffer, the immunoprecipitated complexes were eluted by glycine buffer (200 mM glycine, pH 2.5) with Protease Inhibitor Cocktail (Sigma) and 10 mM NaF, and the elutes were neutralized with 1M Tris-Hcl pH 10.4 and then evaluated by Immunoblotting.
For the Co-Immunoprecipitation of overexpressed proteins, the HEK-293T cells were transfected for 24 h by lipofectamine 2000, and then were lysed by IP lysis buffer with 1xProtease Inhibitor Cocktail and 10 mM NaF. After centrifugation at 14000 g for 10 min at 4 °C, the cleared cell lysates were incubated with FLAG® M2 Magnetic Beads (Sigma) for 2 h at RT. After washing 4 times with 1xTBS buffer with 0.05% tween 20 (TBST), the immunoprecipitated complexes were eluted by 3X FLAG® tag peptide (Sigma-Aldrich) with Protease Inhibitor Cocktail (Sigma) and 10mM NaF, and the elutes were evaluated by Immunoblotting.
Purification of ubiquitin conjugates
The endogenous total- or K63 ubiquitylated proteins were isolated through Tandem Ubiquitin Binding Entities (TUBEs, LifeSensors) according to the manufacture’s protocol. The whole cell lysates were centrifuged for 10 min, 14,000 g at 4 °C. The cleared cell lysates were incubated with TUBE magnetic beads for 2 h or overnight followed 4 times wash by TBST. For immunoblot analysis, ~25 μl of 2X SDS reducing sample prep buffer was added to the resin, and heated at ~95°C for 5 min. After 3-5 min on the magnetic rack, the supernatant was carefully harvested without disturbing the beads.
For purification of ectopic ubiquitin, two-step immunoprecipitation was performed as described previously52. Briefly, clear cell lysates after 14000 g centrifugation were incubated with FLAG® M2 Magnetic Beads (Sigma) for 2 h at RT. After washing 4 times with 1xTBST, the immunoprecipitated complexes were eluted by 3X FLAG® tag peptide (Sigma) with Protease Inhibitor Cocktail and 10 mM NaF and the eluates were evaluated by immunoblotting. Then, the eluates in 1% SDS were boiled for 5 min and diluted 1:10 in Lysis buffer. The diluted eluates were re-immunoprecipiated with the FLAG® M2 Magnetic Beads. After washing for 4 times, the immunoprecipitates were eluted by heating at 95 °C for 5 min in SDS reducing sample buffer and then subjected to immunoblot analysis.
Isolation of ER and Golgi fractions
Two 15-cm dishes of confluent Hela cells were collected and spun down at 600 g (5 min). The cell pellet was washed 3 times in 1xPBS and then re-suspended in hypotonic buffer (20 mM HEPES-KOH, pH 7.2, 10 mM KCl, 3 mM MgCl2) plus cocktail protease inhibitors and homogenized by passing through a 25 G needle 25 times. Homogenates were subjected to differential centrifugation at 1000 g (10 min), and 12,000 g (10 min) to remove intact cells, debris, nuclear, mitochondria etc. After centrifugation, the supernatant was made into 30% Opti-Prep and sample mixture through mixing with 50% Opti-Prep (60% Opti-Prep diluted with 20 mM Tricine-KOH, pH 7.4, 42 mM sucrose and 1 mM EDTA). The Opti-Prep step gradient was loaded from bottom to top: 0.5 ml 30% (sample), 0.5 ml 27.5%, 0.5 ml 25%, 1 ml 22%, 1 ml 19%, 0.5 ml 16% and 0.3 ml 12%. Each density of Opti-Prep was prepared by diluting 50% Opti-Prep with a buffer of 20 mM Tricine-KOH, pH 7.4, 250 mM sucrose, and 1 mM EDTA. The Opti-Prep gradient was subjected to centrifugation at 150,000 g for 3 h, using a SW60 Ti Rotor. Each fraction (150 ul) was collected and precipitated by methanol-chloroform before analysis by Immunoblotting.
ER isolation
The ER fractions were purified using either Endoplasmic Reticulum Isolation Kit (Sigma) or Calcium Chloride Solution (CaCl2). THP-1 cells from ten 15 cm dishes were harvested and homogenized by passing through a 22 G needle until ∼85% lysis. The homogenates were centrifuged at 1,000 g for 10 min to remove cell debris and nuclei and then centrifuged at 12,000 g for 15 min to get a post mitochondrial fraction (PMF). For ER isolation with the Endoplasmic Reticulum Isolation Kit, the PMFs were centrifuged at 100,000 g for 1 hour to remove cytosol proteins and the pellets (crude microsome) were re-suspended in 1× Isotonic Extraction Buffer (10 mM HEPES, pH 7.8, 250 mM sucrose, 25 mM potassium chloride, and 1 mM EGTA). The crude microsomes were loaded onto 15-23% Opti-Prep gradient made by Gradient Master and ultracentrifuged for 18 h at 80,000 g. At the end of the run, fractions were collected from the top of the gradient downwards using a 4-inch blunt ended needle. Immunoblotting was applied to identify the fraction(s) most enriched for ER. For ER precipitation with CaCl2, the PMFs were mixed with 8 mM CaCl2 with constant stirring. The stirring was continued for an additional 15 min at 4 °C. The mixture was centrifuged at 8,000 g for 10 min at 4 °C. The pellet contained enriched ER microsomes.
Protein synthesis assay
Newly synthesized proteins were detected with the Click-iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit (Thermo). PMA-differentiated WT and STEEP-deficient THP-1 cells were seeded on coverslips and treated with or without Cycloheximide. The cells were incubated with Click-iT® HPG (L-homopropargylglycine) and fixed with 3.7% formaldehyde in PBS followed by a permeabilization step using 0.5% Triton X-100. The incorporated amino acid were detected based on click-chemistry, with the alkyne-modified proteins being detected with Alexa Fluor® 488 azide. Nuclei were stained by NuclearMask™ Blue Stain working solution to act as internal control. Images were acquired on Zeiss LSM 780 confocal microscope and processed with the Zen Blue software (Zeiss).
Budding assay
The cytosol preparation was carried out as previously described31. The WT or STEEP-deficient THP-1 cells were collected and re-suspended in B88 buffer (20 mM HEPES-KOH, pH 7.2, 250 mM sorbitol, 150 mM potassium acetate and 5 mM magnesium acetate) with protease inhibitors cocktail, phosphatase inhibitors (Roche) and 0.3 mM DTT. The cells were homogenized by passing through a 22 G needle 60 times and the cell homogenates were centrifuged at 160,000 g for 30 min for 4 times. Clarified supernatant fractions were collected.
For the membrane fraction preparation, STEEP-deficient THP-1 cells were lysed in buffer containing 20 mM HEPES-KOH, pH 7.2, 400 mM sucrose and 1 mM EDTA. Homogenates were centrifuged at 1,000 g for 10 min and then the supernatant was further centrifuged at 100,000 g for 1 h. After centrifugation, the pellets were dissolved in B88 buffer which was adjusted to OD600 = 10 for total membrane.
The budding reaction was performed at 30 °C for 1 h with membrane (OD600 = 10), WT/STEEP-deficient THP-1 cytosol (2 mg/ml final concentration), GTP (0.15 mM), with or without a functional ATP regeneration system (40 mM creatine phosphate, 0.2 mg/ml creatine phosphokinase, and 1 mM ATP). After reaction, the solution was centrifuged at 20,000 g for 20 min and the collected supernatant was centrifuged at 100,000 g for 30 min. The pellets were dissolved in the 2X SDS reducing sample prep buffer for immunoblot. All of the above centrifugations was carried out at 4°C.
Extraction of acidic lipids and PI3P ELISA
Acidic lipids from ER pellet made through CaCl2 were extracted using the modified Folch method53. To minimize potential hydrolysis, extraction of acidic lipids was performed on ice. To wash the ER pellet, 3 mL 5% (w/v) trichloroacetic acid (TCA) with 1 mM EDTA was added to the polypropylene tubes containing the precipitated pellet. After being vortexed for 30 sec, the pellet-containing tube was centrifuged at 2,000 g for 5 min and then the supernatant was discarded. To extract the acidic lipids, 3 ml ice-cold chloroform/methanol (1:2) was added. After vigorous vortexing, the sample was incubated on ice for 15 min. To induce a better phase-separation, 1 ml ice-cold chloroform was added, followed by 1ml citric acid/sodium phosphate buffer, consisting of 1.76% (w/v) KCl, 100 mM citric acid, 100 mM Na2HPO4, 5 mM EDTA, 5 mM tetrabutyl ammonium hydrogen sulphate, pH 3.6. After mixing, the sample was incubated on ice for 5 min before centrifugation (2,000 g, 5min) to complete the phase-separation. The lower organic phase was transferred into a clean polypropylene tube using a salinized pipette. The upper aqueous phase was re-extracted with 2 ml synthetic lower phase (1 ml ice-cold chloroform, 1ml citric acid/sodium phosphate buffer) and the resultant lower phase was combined with the previous lower phase extract. Then, 2 ml 0.88% (w/v) KCl was added to the remaining aqueous phase prior to extraction with 2 ml water-saturated butanol. After incubation on ice for 30 min, the sample was centrifuged at 2,000 g for 5 min. The resultant butanol-rich upper phase was combined with the previous organic phase extracts. The combined extracts were dried in a rotary evaporator at 30 °C under 80 mbar vacuum to remove the chloroform and then decreased to 20 mbar to remove the butanol. Subsequently, the solvent was completely removed by a gentle nitrogen purge; and the resulting acidic lipids were stored at -20°C for the later PI3P lipid ELISA. PI3P was detected with the Mass ELISA Kit from Echelon Biosciences Inc. Briefly, the above lipid extraction samples were rehydrated in PBS-T with 3% Protein Stabilizer and then sonicated for 10 min at room temperature. The ELISA was performed according to the instructions of the manufacturer.
Confocal microscopy
Hela or PMA-differentiated THP-1 cells were seeded on coverslips and stimulated as indicated. The cells were fixed with pre-cold methanol for 5 min at -20°C. Cells were blocked in 1xPBS with 1% BSA for 1h. Cells were stained with primary antibodies for 1 h overnight and then stained using secondary antibody (all 1:300, Alexa Fluor; Invitrogen) for 1 h. For the experiments related with PI3P, the cells were fixed with 4% paraformaldehyde (Sigma), permeabilized with Saponin (Sigma), and blocked with 10% Goat serum (Sigma). 1xTBS was used instead of 1xPBS. Images were acquired on Zeiss LSM 780 confocal microscope and processed with the Zen Blue software (Zeiss). The data analysis was performed with Image J. The antibodies used were sheep anti-STING (1:50), rabbit anti-calreticulin (1:100), mouse anti-GM130 (1:3000), mouse anti-FLAG (1:300), rabbit anti-HA (1:50), rabbit anti-Sec24 (1:100), mouse anti-Clim63 (Enzo, #ALX-804-604, 1:100), rabbit anti-RTN4 (Abcam, #ab47085, 1:150), and mouse anti-PtdIns(3)P (5ug/ml, echelon #Z-P003).
ImageStream
The co-localization of STING-ER, STING-Golgi, SAR1-ER, and STING-PI3P was determined by the ImageStream MK II Imaging Flow Cytometer (Amnis, Co., Seattle, WA, USA). The cells transfected with the indicated plasmids for 24 h, were fixed using 4% formalin for 20 min at RT, and then pre-permeabilized with 0.2% Triton X 100 for 6 min. The cells were incubated with primary antibodies for 1 h on ice, and then incubated with the Alexa-Fluor-labeled secondary antibodies for 1 h. After every step, the cells were washed with 1 x PBS 3 times. Finally, the cells were resuspended in 1 x PBS with 2 mM EDTA and 3% BSA. A 60X magnification was used for all samples. The images of 7,000-10,000 single cells with different channels were acquired in the ImageStream and the data was analyzed through IDEAS software v6.2 (Amnis Corporation). The antibodies used were mouse anti-FLAG (1:300), sheep anti-STING (1:50), rabbit anti-Calreticulin (1:50), rabbit anti-GM130 (1:3000), and rabbit anti-HA (1:50). After gating of focused, single, and positive cells with the defined fluorescent markers, the co-localization of STING-Calreticulin (ER marker), STING-GM130 (Golgi marker), STING-GFP133 (ER membrane curvature), PI3P-Calreticulin, or GFP133-Calreticulin were analyzed through the Bright Detail Similarity feature for the corresponding channels. The accuracy of the cell populations gated as representing co-localization was controlled by visual inspection of individual pictures in the gated cell populations.
Preparation and genome-editing of mice brain slices
Brain slices were isolated from Cas9 Lox-Stop-Lox mice (Cas9lsl) and cultured as described previously11. The next day, the brain slices were infected for 6 days with 1 x 1011 vector genomes of Adeno-Associated Virus (AAV) serotype 9 encoding mouse STEEP-targeting sgRNAs and Cre. GFP expression in the brain slices, a measure of expression from the Cas9-containing locus, was evaluated prior to further use (Extended Data Fig 8b). The culture medium with AAV was removed and changed to contain HSV-1 (Mckrae strain; 5 x 103 PFU in 3.5 ml per brain slice). Two and five days post infection culture medium were collected and viral loads were determined by plaque assay on Vero cells following the steps from previous study10. At day five post infection, brain slices were homogenized and the tissue proteins were extracted by RIPA buffer (Thermo) with Protease inhibitor for the Immunoblot as described above.
Analysis of Cancer Genome Atlas expression data
Analysis of correlation between expression of STEEP/STING and the ISGs CXCL10, Mx2, Viperin/RSAD2, and STAT1, was performed based upon data available through the TCGA the Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov). For a given cancer type, tumor patient samples were ranked on the basis of STEEP/STING FPKM (fragments per kilobase of exon model per million reads mapped) value. The ranked samples were evenly divided into four groups, and the statistical comparisons for the genes of CXCL10, Mx2, Viperin/RSAD2, STAT1, and GAPDH (internal control) were performed between the STEEP/STING lowest 25% expression group and the STEEP/STING highest 25% expression group accordingly. Statistical significance was judged by DESeq2 package in R language through two-tailed Mann-Whitney test according to a previous study54. Differentially expressed genes were defined as genes with fold change ≥ 1.5 and adjusted P-value ≤ 0.01.
Statistics and Reproducibility
The data are shown as means of biological replicates ± st.dev. Statistically significant differences between groups were determined using two-tailed Student’s t-test when the data exhibited normal distribution and F Test to compare two variances data p value > 0.1, two-tailed unpaired t-test with welch’s correction when the data exhibited normal distribution but F Test p value < 0.1, two-tailed one-way ANOVA test for multiple comparisons, and two-tailed Mann-Whitney test when the data set did not pass the normal distribution test. The data shown are from single experiments. The majority of experiments were performed at least 3 times with similar results. A few experiments were performed 2 times.
Ethics
Permission to perform the experiment with cells from SLE patients was acquired from the Regional Ethics Committee (1-10-72-288-18). All patients and healthy controls provided informed written consent.
Extended Data
Extended Data Fig. 1. Basic characteristics and properties of CxORF56/STEEP.
(a) Comparison of protein sequence identify between STING and STEEP in different species. (b) mRNA Expression of STEEP in human tissues. The data are from proteinatlas.org. (c) Illustration of the three known isoforms of STEEP. Source, uniprot.org. (d) Predicted nuclear localization and nuclear exit signals in STEEP, based on sequence analyses by cNLS mapper and LocNES, respectively. (e) Whole cell lysates, cytoplasmic fractions, and nuclear fractions from PMA-differnetiated THP-1 cells were immunoplotted for STEEP and β-actin. A representative blot is shown from n = 3 biologically independent experiments with similar results. (f) Predicted transmembrane regions in STEEP and STING. Based on sequence analysis by TMHMM.
Extended Data Fig. 2. STEEP is a positive regulator in STING pathway.
(a) Reporter gene assay for HEK-293T cells transfected with 50 ng STEEP or empty vector, IFNB1 promoter luciferase reporter, β-actin Renilla reporter, and MAVS or TRIF as indicated for 24 h (n = 3) (ns P = 0.86, ns P = 0.23, lift to right). (b) Immunoprecipitation of Flag-tagged STING and STEEP (n = 3 biologically independent experiments). (c) Confocal microscopy of HeLa cells transfected with HA-STEEP and FLAG-STING for 24 h followed by mock treatment or cGAMP stimulation. Representative data from one experiment are shown (n = 3 biologically independent experiments). (d) Alignment of STEEP/CxORF56 from the indicated species, and highlight of residues mutated to alanine in Mut 1-5. Data in panel a are shown as means of biological triplicates +/- st.dev. Statistical analysis of data shown in panel a was performed using two-tailed Student’s t-test.
Extended Data Fig. 3. Targeting of STEEP by CRISPR.
(a) Illustration of gRNAs sequence and location used to target human STEEP. (b) Immunoblot for STEEP in THP-1-derived clones transduced with STING-targeting gRNAs. (c) STEEP KO THP-1 cells were transfected with mRNA encoding WT STEEP. 24 h later, the cells were lysed, and immunoblotted as shown. For comparison, lysates from WT THP-1 cells were included. (d-f) Efficiency of STEEP gene targeting in primary human fibroblasts (d), primary human macrophage (e) and SAVI patient fibroblasts (f). (g) gRNAs used to target STEEP in mice. (h-i) Efficiency of STEEP targeting in MEF cells. The KO scores were calculated based on the ICE Analysis tool from Synthego. In panels b and c, data shown are representative blots from three biologically independent experiments with similar results.
Extended Data Fig. 4. Effect of STEEP KO on STING signaling.
(a) STEEP KO THP-1 cells electroporated with GFP mRNA, WT STEEP mRNA or mut5 STEEP mRNA were stimulated with cGAMP (150 nM, 1 h). Levels of pSTING and pTBK1 were monitored by Immunoblotting. (n = 3 biologically independent experiments). (b-c) Immunoblot analysis of the indicated proteins from whole cell lysates of WT or STEEP KO Hela cells (b) and human foreskin fibroblasts (c) after stimulation with dsDNA for the indicated time intervals. (n = 3 biologically independent experiments). All data shown in this figure are representative blots from three biologically independent experiments.
Extended Data Fig. 5. Impact of STEEP on STING trafficking.
(a) The in vitro membrane budding reaction illustrated in Fig. 4a was performed with material from wild-type THP-1 cells in the presence or absence of ATP. M, membranes; P, post 20.000 g pellet; BV, budding vesicles. Representative blots from three biologically independent experiments with similar results are shown. (b) Confocal microscopy of WT Hela and STEEP KO Hela cells stimulated with/without cGAMP for 0.5 h. The cells were immunostained with anti-STING (red), anti-calreticulin (green), anti-GM130 (purple). Sections were counterstained with DAPI to visualize nuclei. (n = 3 biologically independent experiments). (c) ImageStream analysis of Sar1-ER colocation. FLAG-tagged Sar1 was transfected into HEK-293T with STING expression cells for 24 h, and stimulated with/without cGAMP (100 nM) for 30 min. After fixation and permeabilization, the cells were incubated with rabbit anti-calreticulin (ER marker) and mouse anti-FLAG. (n = 3) (**P = 0.00042). (d) Immunoblot analysis of the indicated proteins from the whole Hela cell lysates transfected with SAR1 or SAR1-H79G. (n = 3 biologically independent experiments). (e, f) Imagestream analysis of STING trafficking from ER (e) to Golgi (f) after transfection of SAR1 or SAR1-H79G for 24 h in HEK-293T with STING expression cells (e, **P = 0.00035; f, **P = 0.00010). (g) Endogenous Sec24 was immunoprecipitated from THP-1 cell lysates isolated after +/-BFA treatment and cGAMP (100 nM) stimulation for the indicated time. Precipitates and lysates were immunoblotted with antibodies against Sec24A/B, STING, and vinculin (n = 3 biologically independent experiments). (h) Confocal microscopy analysis of Sec24 foci in STEEP KO THP-1 cells rescued with WT or mut5 STEEP mRNA and stimulated with cGAMP for 10 min. Nuclei were stained with DAPI. For quantification of Sec24 foci, 12 cells per group were counted in a blinded fashion. Representative data from one experiment are shown (n = 3 biologically independent experiments). Each data point represents one cell and data are shown as means +/- st.dev. Statistical analysis was performed by a two-tailed unpaired t test with welch’s correction (*P = 0.016, ns P = 0.51, *P = 0.045, left to right). (i) Budding vesicle analysis (as illustrated in Fig 4a) for mCherry-VSVG and STING on WT and STEEP KO Hela cells transfected with mCherry-VSVG. The data shown is representative from two biologically independent experiments with similar results. (j) Confocal microscopy imaging of SAR1-Flag (green) and calreticulin (red, ER marker) in WT and STEEP KO Hela cells stimulated with cGAMP (100 nM) for 20 min. Sections were counterstained with DAPI to visualize nuclei. (n = 3 biologically independent experiments). (k) Imagestream analysis of ER membrane curvature in Flag-STING-transfected HEK-293T cells probed with GFP133 with/without 100 nM cGAMP stimulation for 20 min. (n = 3) (**P = 0.000011). (l) Confocal microscopy analysis Hela cells transfected with Flag-tagged STING and GFP133 and treated with 100 nM cGAMP stimulation for 20 min. Cells were probed with antibodies against of STING (Red) and Calreticulin (ER marker, Purple). GFP133 (green) is an ER membrane curvature probe. (n = 3 biologically independent experiments). (m) Imagestream analysis of ER membrane curvature and STING colocation in HEK-293T cells with/without 100 nM cGAMP stimulation for 20 min. (n = 3) (**P = 0.000044). (n) WT and STEEP KO THP-1 cells were stimulated with cGAMP for 10 min, fixed and stained with anti-Clim63, anti-RTN4, and anti-STING. Nuclei were stained with DAPI. For quantification of RTN4:Climp63 ratio, 10 cells per group were counted in a blinded fashion. Representative data from one experiment are shown (n = 3 biologically independent experiments). Each data point represents one cell and data are shown as means +/- st.dev. Statistical analysis was performed by a two-tailed unpaired t test with welch’s correction (ns P = 0.64, **P = 0.0046, **P = 0.0060, ns P = 0.071). (o) Analysis of protein synthesis in WT and STEEP KO THP-1 cells using the Click-iT™ HPG Alexa Fluor™ 488 Protein Synthesis Assay Kit (n = 6) (ns P = 0.62, ns P = 0.10, left to right). (p) ER- and Golgi-enriched pellets from lysates of STEEP-deficient Hela cells transfected with empty vector, HA-STEEP WT, or HA-STEEP mut5 were fractionated by gradient centrifugation. The collected fractions were immunoblotted with anti-STING, anti-HA, anti-GM130 (Golgi), and anti-Sec61B (ER). Representative blots from one experiment are shown (n = 3). For data from ImageStream analysis (panel c, e, f, k, and m), each data point represents the percent of positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data in panels c, e, f, k, m, and o was performed using two-tailed Student’s t-test.
Extended Data Fig. 6. STEEP promotes PI3P production and ER membrane curvature.
(a, b) Confocal microscopy analysis of PI3P level on ER. PI3P was probed with anti-PI3P antibody in THP-1 cells (a), or FYVE-GFP in Hela cells (b) or with/without 100 nM cGAMP stimulation for 20min. ER was stained and visualized using rabbit anti-Calreticulin (ER marker, Red) and mouse anti-flag, and relevant secondary antibodies. (c) PIP Strip membranes blotted with various lipids were incubated sequentially with Flag-tagged WT, 139-C, or V155M STING or with empty vector (EV). An anti-Flag antibody conjugated with HRP was used to visualize the binding. Phospholipids with clear positive binding to WT STING are highlighted in red on the left PIP Strip illustration. (d) Confocal analysis for the colocation of anti-STING and anti-PI3P antibodies in THP-1 cells with/without 100 nM cGAMP stimulation for 15 min. (e) Immunoblot analysis of the indicated proteins from the whole cell lysates of Hela cells transfected MTMR3/MTMR3-C143S, and stimulated for 1h with cGAMP. (n = 3 biologically independent experiments). (f, g) Imagestream analysis of STING trafficking from ER (f) to Golgi (g). HEK-293T cells were transfected by MTMR3/MTMR3-C143S and Flag-tagged STING for 24 h and stimulated cGAMP (150 nM) for 30 min. After fixation and permeabilization, flag and ER were labelled using rabbit anti-calreticulin (ER marker) and mouse anti-flag, and relevant secondary antibodies. (n = 3) (f, **P = 0.0020; g, **P = 0.00088). (h) Imagestream analysis of SAR1-ER colocation. Hela cells were transfected by MTMR3/MTMR3-C143S and Flag-tagged SAR1 for 24 h and stimulated cGAMP (100 nM) for 30 min. After fixation and pre-permeabilization, ER and Flag were labelled using rabbit anti-calreticulin (ER marker) and mouse anti-flag, and relevant secondary antibodies. (n = 3) (**P = 0.0014). (i) Imagestream analysis of ER membrane curvature. HEK cells were co-transfected by MTMR3/MTMR3-C143S, Flag-tagged STING and GFP133 for 24 h and stimulated with/without cGAMP for 30 min. After fixation and pre-permeabilization, the cells were stained with anti-calreticulin and relevant secondary antibodies. (n = 3) (ns P = 0.26, *P = 0.012, **P = 4.3E-05, **P = 1.0E-06, left to right). Panels a-e, representative images from n = 3 biologically independent experiments with at least 2 times similar results are shown. For data from ImageStream analysis (panel f, g, h, i), each data point represents the percent of positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data in panels f-h was performed using two-tailed Student’s t-test, and in panel i was performed using two-tailed one-way ANOVA test.
Extended Data Fig. 7. Vps34 Complex I promotes STING activation.
(a) Immunoblot analysis of the indicated proteins from the cell lysates of THP-1 cells pre-treated with DMSO, 10 uM VPS34 inhibitor (VPS34-IN1) or 5 mM 3-MA and then stimulated with 150 nM cGAMP for the indicated time intervals. (n = 3 biologically independent experiments). (b) Immunoblot analysis of the indicated proteins from the cell lysates of HaCaT cells transfected with siRNA-control or siRNA-vps34 for 36 h and then stimulated with cGAMP for the indicated times. (n = 3 biologically independent experiments). (c) Reporter gene assay. HEK-293T with STING stable expression cells were transfected with 50 ng VPS34, Beclin1, ATG14, UVRAG or empty vector, IFNB1 promoter luciferase reporter, and β-actin Renilla reporter. After transfection for 24 h, the cells were treated with/without BafA1 and then stimulated with 200 nM cGAMP for 6 h (n = 3). Statistical analysis of data in panel c was performed using two-tailed one-way ANOVA test (**P = 1.87E-06, ns P = 0.88, **P = 2.46E-09, **P = 0.0063, **P = 0.00053, ns P = 0.10, **P = 1.74E-06, ns P = 0.99, left to right). (d) The isolation of THP-1 cell fractions were obtained by Opti-prep gradient. After collecting the different layers, immunoblot analysis of the indicated proteins was carried out to probe for organelle enrichment of each fraction. The input was from whole cell lysate. Representative blots from two biologically independent experiments with similar results are shown. Data in panel c are shown as means of biological sample triplicates +/- st.dev.
Extended Data Fig. 8. Role for STEEP in host defense and inflammation.
(a) The workflow for examination of HSV1 replication in genome-edited brain slices from Cas9lsl mice. (b) To evaluate expression from the Cas9 and GFP-containing locus, emission of fluorescence from AAV9-treated Cas9lsl brain slices was monitored. The images represent brain slices 6 days after treatment. Representative images from two biologically independent experiments with similar results are shown. (c-d) Levels of type I IFN bioactivity and CXCL10 protein in serum from five SLE patients and two healthy donors. (e-g) Box plots of TCGA RNA expression profiles in Bladder Urothelial Carcinoma (BLCA, e), Cervical Kidney renal papillary cell carcinoma (KIRP, f), and Liver Hepatocellular Carcinoma (LIHC, g). Samples with the highest 25% and the lowest 25% of STEEP (upper panels) and STING (lower panels) expression were selected and grouped. CXCL10, MX2, Viperin (RSAD2), STAT1 and GAPDH gene expression levels were compared between STEEP/STING-high and STEEP/STING-low groups. The upper and lower ends of the boxes represent the upper and lower quartiles, and the horizontal line inside the box is median of the dataset. The bar extending parallel from the boxes is the “whiskers”, indicating upper and lower extreme of the dataset. Statistical significance was evaluated using two-tailed Mann-Whitney test (ns P > 0.01, *P < 0.01, **P < 0.001). (h) Imagestream analysis of ER membrane curvature. HEK-293T cells were co-transfected by MTMR3/MTMR3-C143S, STING/ STING SAVI mutant and GFP133 for 24 h. After fixation and pre-permeabilization, the cells were stained with anti-calreticulin and relevant secondary antibodies. (n = 3) (ns P = 0.15, ns P = 0.10, *P = 0.038, *P = 0.029, left to right). For data from ImageStream analysis (panel h), each data point represents the percent of positive cells from one representative sample and are shown as means +/- st.dev. Statistical analysis of data panel h was performed using two-tailed Student’s t-test.
Extended Data Fig. 9. Proposed model for action of STEEP in STING signaling.
Interaction between STEEP and STING is observed in the resting state, but this is augmented following STING activation. This may involve the conformational change imposed on STING following cGAMP binding or mediated with low threshold in SAVI gain-of-function STING mutants. The STEEP-STING association increases recruitment of the VPS34 complex I to produce PI3P on the ER. The PI3P accumulation in turn induces ER membrane curvature in the STING containing areas, thus recruiting the COPII complex factors SAR1 and SEC24 to ER to form the ER exit site. This finally sorts STING into COPII vesicles for delivery to ERGIC/Golgi to enable antiviral and inflammatory responses.
Supplementary Material
Acknowledgements
The technical assistance of Kirsten Stadel Petersen, the AU FACS Core facility, and AU Health bioimaging core facility is greatly appreciated. The authors would like to acknowledge professor Thomas Melia, Yale University, for critical reading of the manuscript. This work was funded by the European Research Council (ERC-AdG ENVISION; 786602); The Novo Nordisk Foundation (NNF18OC0030274), the Lundbeck Foundation (R198-2015-171; R268-2016-3927); B.C.Z. is funded by a Postdoc grant from the Danish Council for Independent Research | Medical Sciences (5053-00083B); The postdoc salary to S.A. was funded by the European Union under the Horizon 2020 Research and Innovation Program and Marie Skłodowska-Curie Actions (MSCA) – international fellowship (PathAutoBio 796840); The PhD scholarship to S.J.W. was funded by the European Union under the Horizon 2020 Research and Innovation Program and MSCA-Innovative Training Networks Programme MSCA-ITN (EDGE, 675278); The postdoc grant for A.T. was funded by the Lundbeck Foundation (R264-2017-3344).
Footnotes
Author Contributions
B.C.Z. and S.R.P. conceived the idea and designed the experiments. B.C.Z. unraveled the mechanism. S.B.J. identified STEEP in the mass spectrometry screen. B.C.Z., R.Nandakumar, L.S.R., J.H., A.L., Z.L.G., C.L.S., S.B.J., S.A., M.F.B., C.S., Y.Z., S.J.W., D.O., T.P., C.B., R.Narita, Y.C., and C.G.Z. performed the experiments. A.T., H.S., C.M.D., T.N., and R.G-M provided materials, Z.G., R.H., Z.J.C., J.J.E., R.O.B., M.K.T., and S.R.P. supervised experiments. B.C.Z. and S.R.P. wrote the manuscript.
Competing Interests Statement
The authors declare no competing interests.
Data Availability
The data that support the findings of this study are available from the corresponding author upon request.
References
- 1.Paludan SR. Activation and Regulation of DNA-Driven Immune Responses. Microbiol Mol Biol Rev. 2015;79:225–241. doi: 10.1128/MMBR.00061-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun L, Wu J, Du F, Chen X, Chen ZJ. Cyclic GMP-AMP Synthase Is a Cytosolic DNA Sensor That Activates the Type I Interferon Pathway. Science. 2013;339:786–791. doi: 10.1126/science.1232458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu J, et al. Cyclic GMP-AMP Is an Endogenous Second Messenger in Innate Immune Signaling by Cytosolic DNA. Science. 2013;339:826–830. doi: 10.1126/science.1229963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ishikawa H, Barber GN. STING is an endoplasmic reticulum adaptor that facilitates innate immune signalling. Nature. 2008;455:674–678. doi: 10.1038/nature07317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shang G, Zhang C, Chen ZJ, Bai XC, Zhang X. Cryo-EM structures of STING reveal its mechanism of activation by cyclic GMP-AMP. Nature. 2019;567:389–393. doi: 10.1038/s41586-019-0998-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tanaka Y, Chen ZJ. STING specifies IRF3 phosphorylation by TBK1 in the cytosolic DNA signaling pathway. Sci Signal. 2012;5:ra20. doi: 10.1126/scisignal.2002521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu S, et al. Phosphorylation of innate immune adaptor proteins MAVS, STING, and TRIF induces IRF3 activation. Science. 2015 doi: 10.1126/science.aaa2630. [DOI] [PubMed] [Google Scholar]
- 8.Prabakaran T, et al. Attenuation of cGAS-STING signaling is mediated by a p62/SQSTM1-dependent autophagy pathway activated by TBK1. EMBO J. 2018;37:e97858. doi: 10.15252/embj.201797858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gulen MF, et al. Signalling strength determines proapoptotic functions of STING. Nat Commun. 2017;8 doi: 10.1038/s41467-017-00573-w. 427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Reinert LS, et al. Sensing of HSV-1 by the cGAS-STING pathway in microglia orchestrates antiviral defense in the CNS. Nat Commun. 2016;7 doi: 10.1038/ncomms13348. 13348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li XD, et al. Pivotal roles of cGAS-cGAMP signaling in antiviral defense and immune adjuvant effects. Science. 2013;341:1390–1394. doi: 10.1126/science.1244040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Woo SR, et al. STING-dependent cytosolic DNA sensing mediates innate immune recognition of immunogenic tumors. Immunity. 2014;41:830–842. doi: 10.1016/j.immuni.2014.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Crow YJ, et al. Mutations in the gene encoding the 3 ‘-5 ‘ DNA exonuclease TREX1 cause Aicardi-Goutieres syndrome at the AGS1 locus. Nature Genetics. 2006;38:917–920. doi: 10.1038/ng1845. [DOI] [PubMed] [Google Scholar]
- 14.Liu Y, et al. Activated STING in a vascular and pulmonary syndrome. N Engl J Med. 2014;371:507–518. doi: 10.1056/NEJMoa1312625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fu J, et al. STING agonist formulated cancer vaccines can cure established tumors resistant to PD-1 blockade. Sci Transl Med. 2015;7 doi: 10.1126/scitranslmed.aaa4306. 283ra252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanson MC, et al. Nanoparticulate STING agonists are potent lymph node-targeted vaccine adjuvants. J Clin Invest. 2015;125:2532–2546. doi: 10.1172/JCI79915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haag SM, et al. Targeting STING with covalent small-molecule inhibitors. Nature. 2018;559:269–273. doi: 10.1038/s41586-018-0287-8. [DOI] [PubMed] [Google Scholar]
- 18.Saitoh T, et al. Atg9a controls dsDNA-driven dynamic translocation of STING and the innate immune response. Proc Natl Acad Sci U S A. 2009;106:20842–20846. doi: 10.1073/pnas.0911267106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dobbs N, et al. STING Activation by Translocation from the ER Is Associated with Infection and Autoinflammatory Disease. Cell Host Microbe. 2015;18:157–168. doi: 10.1016/j.chom.2015.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Srikanth S, et al. The Ca(2+) sensor STIM1 regulates the type I interferon response by retaining the signaling adaptor STING at the endoplasmic reticulum. Nat Immunol. 2019;20:152–162. doi: 10.1038/s41590-018-0287-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Konno H, Konno K, Barber GN. Cyclic dinucleotides trigger ULK1 (ATG1) phosphorylation of STING to prevent sustained innate immune signaling. Cell. 2013;155:688–698. doi: 10.1016/j.cell.2013.09.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Luo WW, et al. iRhom2 is essential for innate immunity to DNA viruses by mediating trafficking and stability of the adaptor STING. Nat Immunol. 2016;17:1057–1066. doi: 10.1038/ni.3510. [DOI] [PubMed] [Google Scholar]
- 23.Wei J, et al. SNX8 modulates innate immune response to DNA virus by mediating trafficking and activation of MITA. PLoS Pathog. 2018;14:e1007336. doi: 10.1371/journal.ppat.1007336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L, et al. UBXN3B positively regulates STING-mediated antiviral immune responses. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04759-8. 2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sun MS, et al. TMED2 Potentiates Cellular IFN Responses to DNA Viruses by Reinforcing MITA Dimerization and Facilitating Its Trafficking. Cell Rep. 2018;25:3086–3098. doi: 10.1016/j.celrep.2018.11.048. e3083. [DOI] [PubMed] [Google Scholar]
- 26.Lee MN, et al. Identification of regulators of the innate immune response to cytosolic DNA and retroviral infection by an integrative approach. Nat Immunol. 2013;14:179–185. doi: 10.1038/ni.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szappanos D, et al. The RNA helicase DDX3X is an essential mediator of innate antimicrobial immunity. PLoS Pathog. 2018;14:e1007397. doi: 10.1371/journal.ppat.1007397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu YP, et al. Endoplasmic reticulum stress regulates the innate immunity critical transcription factor IRF3. J Immunol. 2012;189:4630–4639. doi: 10.4049/jimmunol.1102737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paludan SR, Reinert LS, Hornung V. DNA-stimulated cell death: implications for host-defense, inflammatory diseases, and cancer. Nature Reviews Immunology. 2019;19:151–153. doi: 10.1038/s41577-018-0117-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ni G, Konno H, Barber GN. Ubiquitination of STING at lysine 224 controls IRF3 activation. Sci Immunol. 2017;2 doi: 10.1126/sciimmunol.aah7119. eaah7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ge L, Melville D, Zhang M, Schekman R. The ER-Golgi intermediate compartment is a key membrane source for the LC3 lipidation step of autophagosome biogenesis. Elife. 2013;2:e00947. doi: 10.7554/eLife.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ge L, Zhang M, Schekman R. Phosphatidylinositol 3-kinase and COPII generate LC3 lipidation vesicles from the ER-Golgi intermediate compartment. Elife. 2014;3:e04135. doi: 10.7554/eLife.04135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogawa E, Mukai K, Saito K, Arai H, Taguchi T. The binding of TBK1 to STING requires exocytic membrane traffic from the ER. Biochem Biophys Res Commun. 2018;503:138–145. doi: 10.1016/j.bbrc.2018.05.199. [DOI] [PubMed] [Google Scholar]
- 34.Gui X, et al. Autophagy induction via STING trafficking is a primordial function of the cGAS pathway. Nature. 2019;567:262–266. doi: 10.1038/s41586-019-1006-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hanna MGt, et al. Sar1 GTPase Activity Is Regulated by Membrane Curvature. J Biol Chem. 2016;291:1014–1027. doi: 10.1074/jbc.M115.672287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balch WE, McCaffery JM, Plutner H, Farquhar MG. Vesicular stomatitis virus glycoprotein is sorted and concentrated during export from the endoplasmic reticulum. Cell. 1994;76:841–852. doi: 10.1016/0092-8674(94)90359-x. [DOI] [PubMed] [Google Scholar]
- 37.Long KR, et al. Sar1 assembly regulates membrane constriction and ER export. J Cell Biol. 2010;190:115–128. doi: 10.1083/jcb.201004132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibata Y, Voeltz GK, Rapoport TA. Rough sheets and smooth tubules. Cell. 2006;126:435–439. doi: 10.1016/j.cell.2006.07.019. [DOI] [PubMed] [Google Scholar]
- 39.He S, et al. PtdIns(3)P-bound UVRAG coordinates Golgi-ER retrograde and Atg9 transport by differential interactions with the ER tether and the beclin 1 complex. Nat Cell Biol. 2013;15:1206–1219. doi: 10.1038/ncb2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hirama T, et al. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat Commun. 2017;8 doi: 10.1038/s41467-017-01554-9. 1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kihara A, Noda T, Ishihara N, Ohsumi Y. Two distinct Vps34 phosphatidylinositol 3-kinase complexes function in autophagy and carboxypeptidase Y sorting in Saccharomyces cerevisiae. J Cell Biol. 2001;152:519–530. doi: 10.1083/jcb.152.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.An J, et al. Expression of Cyclic GMP-AMP Synthase in Patients With Systemic Lupus Erythematosus. Arthritis Rheumatol. 2017;69:800–807. doi: 10.1002/art.40002. [DOI] [PubMed] [Google Scholar]
- 43.Mukai K, et al. Activation of STING requires palmitoylation at the Golgi. Nat Commun. 2016;7 doi: 10.1038/ncomms11932. 11932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kawasaki T, Takemura N, Standley DM, Akira S, Kawai T. The second messenger phosphatidylinositol-5-phosphate facilitates antiviral innate immune signaling. Cell Host Microbe. 2013;14:148–158. doi: 10.1016/j.chom.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 45.Brandizzi F, Barlowe C. Organization of the ER-Golgi interface for membrane traffic control. Nat Rev Mol Cell Biol. 2013;14:382–392. doi: 10.1038/nrm3588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jongsma ML, et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell. 2016;166:152–166. doi: 10.1016/j.cell.2016.05.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Raiborg C, et al. Repeated ER-endosome contacts promote endosome translocation and neurite outgrowth. Nature. 2015;520:234–238. doi: 10.1038/nature14359. [DOI] [PubMed] [Google Scholar]
- 48.Rasmussen SB, et al. Activation of Autophagy by alpha-Herpesviruses in Myeloid Cells Is Mediated by Cytoplasmic Viral DNA through a Mechanism Dependent on Stimulator of IFN Genes. Journal of Immunology. 2011;187:5268–5276. doi: 10.4049/jimmunol.1100949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lee CC, Avalos AM, Ploegh HL. Accessory molecules for Toll-like receptors and their function. Nat Rev Immunol. 2012;12:168–179. doi: 10.1038/nri3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chan YK, Gack MU. Viral evasion of intracellular DNA and RNA sensing. Nat Rev Microbiol. 2016;14:360–373. doi: 10.1038/nrmicro.2016.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hendel A, et al. Chemically modified guide RNAs enhance CRISPR-Cas genome editing in human primary cells. Nat Biotechnol. 2015;33:985–989. doi: 10.1038/nbt.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang Q, et al. The E3 Ubiquitin Ligase AMFR and INSIG1 Bridge the Activation of TBK1 Kinase by Modifying the Adaptor STING. Immunity. 2014;41:919–933. doi: 10.1016/j.immuni.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 53.Pettitt TR, Dove SK, Lubben A, Calaminus SD, Wakelam MJ. Analysis of intact phosphoinositides in biological samples. J Lipid Res. 2006;47:1588–1596. doi: 10.1194/jlr.D600004-JLR200. [DOI] [PubMed] [Google Scholar]
- 54.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data that support the findings of this study are available from the corresponding author upon request.