Abstract
Angiotensin-converting enzyme 2 (ACE2) is an entry receptor for Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) and a regulator of several physiological processes. ACE2 has recently been proposed to be interferon-inducible, suggesting that SARS-CoV-2 may exploit this phenomenon to enhance viral spread and questioning the efficacy of interferon treatment in Coronavirus disease 2019 (COVID-19). Using a recent de novo transcript assembly that captured previously unannotated transcripts, we describe a novel isoform of ACE2, generated by co-option of intronic retroelements as promoter and alternative exon. The novel transcript, termed MIRb-ACE2, exhibits specific expression patterns across the aerodigestive and gastrointestinal tracts and is highly responsive to interferon stimulation. In stark contrast, canonical ACE2 expression is unresponsive to interferon stimulation. Moreover, the MIRb-ACE2 translation product is a truncated, unstable ACE2 form, lacking domains required for SARS-CoV-2 binding and is therefore unlikely to contribute to or enhance viral infection.
Introduction
Interferons represent the first line of defence against viruses in humans and other jawed vertebrates1. Recognition of viral products in an infected cell results in autocrine and paracrine signalling to induce an antiviral state characterized by expression of a module of interferon-stimulated genes (ISGs) that restrict viral replication and spread1,2. Indeed, recombinant interferon is often given as first-line therapy in viral infection3 and preliminary results suggest that interferon treatment may be effective against Coronavirus disease 2019 (COVID-19)4,5.
Interferon signalling results in rapid upregulation of several hundred ISGs, including genes that inhibit various stages of viral entry and replication as well as transcription factors that further potentiate the interferon response1,2. Given that unchecked interferon signalling and inflammation can result in immunopathology, ISGs are subject to complex regulatory mechanisms6.
At the transcriptional level, long terminal repeats (LTRs), derived from endogenous retroviruses and other LTR retroelements, as well as regulatory sequences in non-LTR retroelements serve as cis-regulatory enhancers for a number of ISGs and are required for their induction7. Adding to this regulatory complexity, many retroelements are themselves interferon-responsive promoters and are upregulated following viral infection or in interferon-driven autoimmunity8–11.
The co-evolution of viruses and hosts has resulted in a number of strategies by which viruses evade or subvert interferon responses12. Compared with other respiratory viruses, Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) elicits a weak interferon response despite strong induction of other chemokines13. Though the mechanism by which SARS-CoV-2 dampens interferon responses remains unclear, the ORF3b, ORF6, and nucleoprotein of the closely-related SARS-CoV function as interferon antagonists14. SARS-CoV-2 uses angiotensin-converting enzyme 2 (ACE2) as its primary receptor15,16 and recent work suggested that SARS-CoV-2 may hijack the interferon response by inducing ACE2 expression17. By integrating multiple human, macaque, and mouse single-cell RNA-sequencing (RNA-seq) datasets, Ziegler et al. identified ACE2 as a primate-specific ISG upregulated following viral infection or interferon treatment17. Use of an ISG as a viral receptor would result in a self-amplifying loop to increase local viral spread, and calls into question the efficacy and safety of recombinant interferon treatment in COVID-19 patients.
Using our recent de novo transcriptome assembly18, we identify a novel, truncated ACE2 transcript, termed MIRb-ACE2, initiated at intronic MIRb and LTR16A1 retroelements that serve as a cryptic promoter, alternative exon and splice site. Notably, we find that the truncated MIRb-ACE2 and not full-length ACE2 is the interferon-inducible isoform, and is strongly upregulated in viral infection and following interferon treatment. Importantly, the protein product of the MIRb-ACE2 transcript does not contain the amino acid residues required for SARS-CoV-2 attachment and entry and is additionally post-translationally unstable. These findings have important implications for the understanding of ACE2 expression and regulation, and thus for SARS-CoV-2 tropism and treatment.
Results
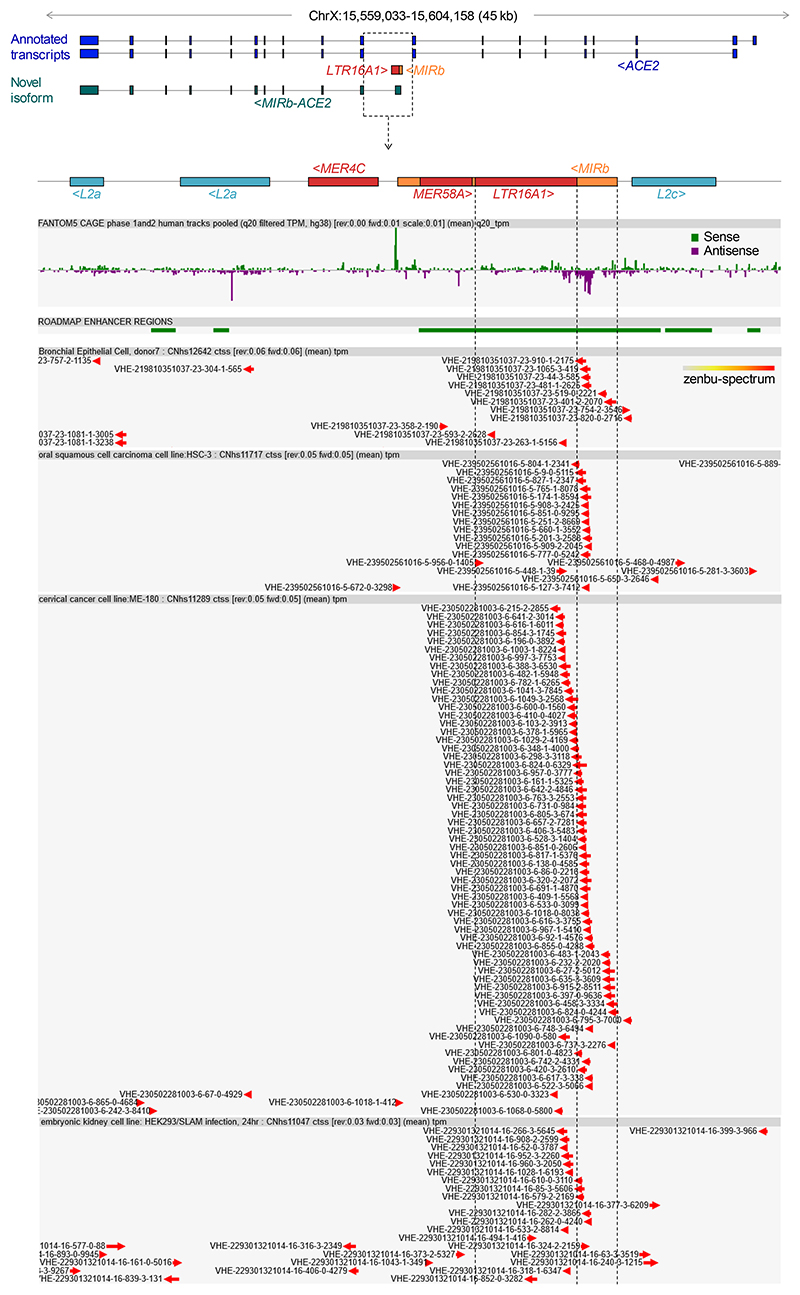
MIRb-ACE2 is a tissue-specific novel isoform of ACE2
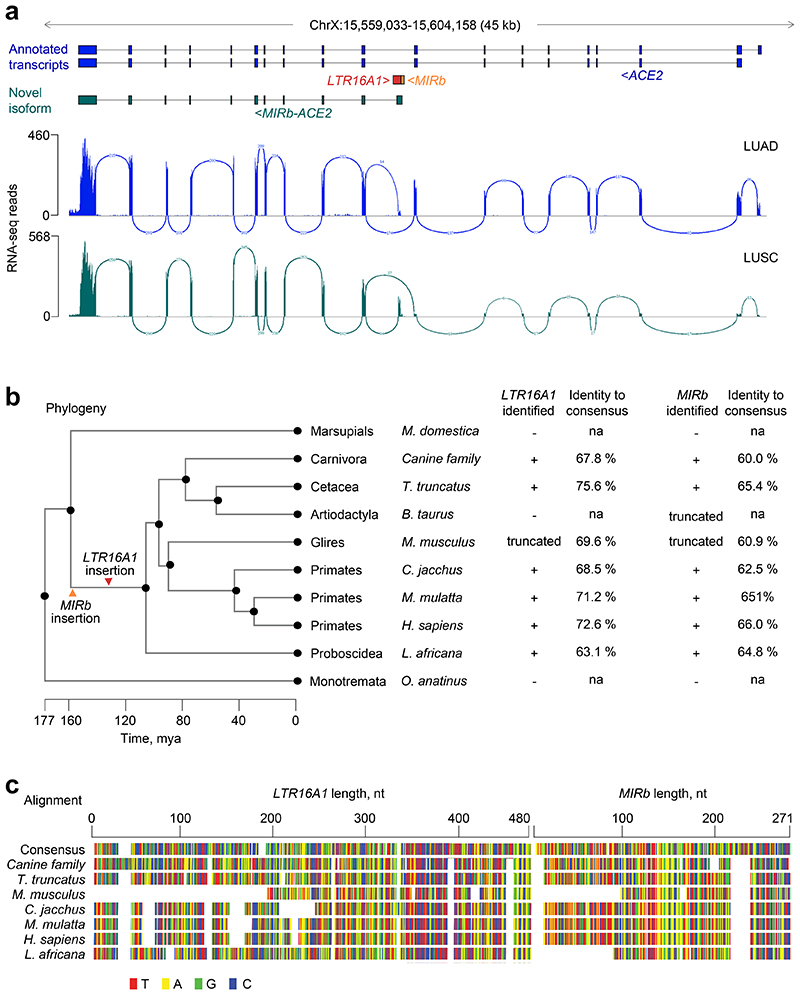
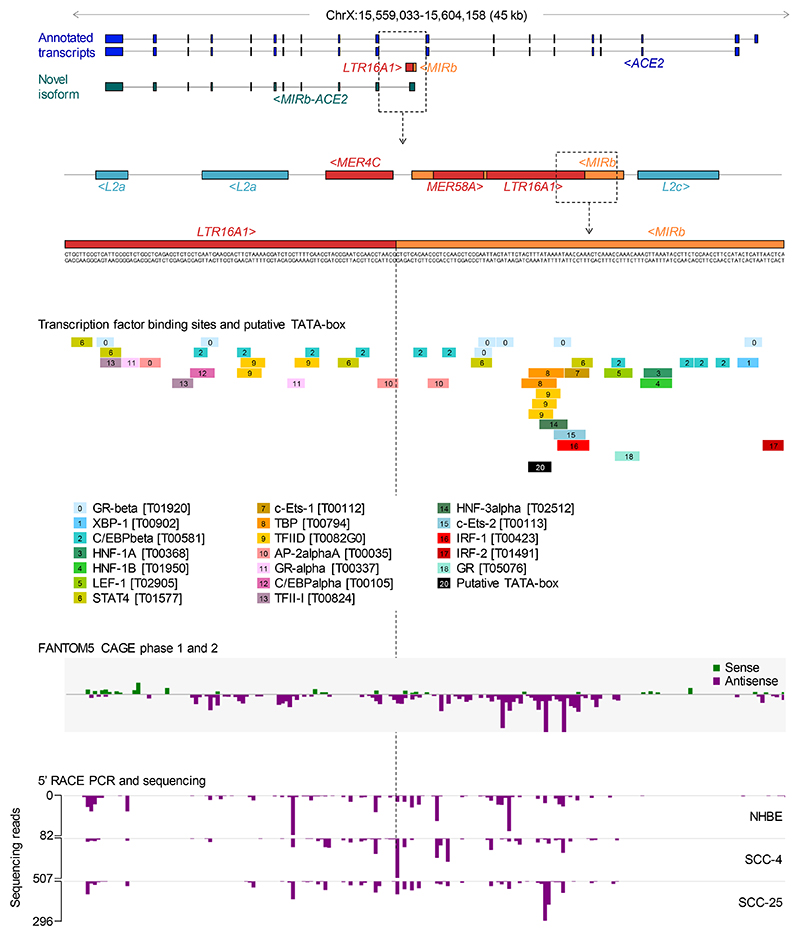
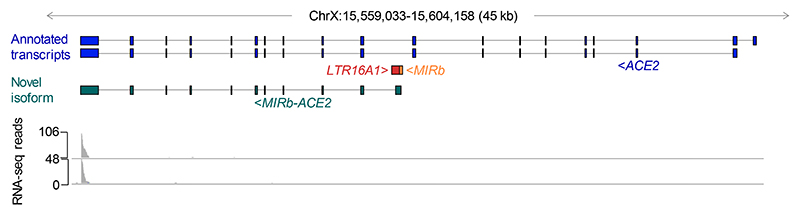
Our recent de novo cancer transcriptome assembly18 identified a chimeric transcript formed by splicing between annotated exons of ACE2 and an LTR16A1 retroelement, integrated in intron 9 of the ACE2 locus. This transcript, which we refer to here as MIRb-ACE2, includes exons 10-19 of ACE2 (Fig. 1a). Splicing between the LTR16A1 retroelement and exon 10 of ACE2 was highly supported by splice junction analysis of RNA-seq data from The Cancer Genome Atlas (TCGA) lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) (Fig. 1a). To identify potential transcription start site(s) of the MIRb-ACE2 transcript, we inspected promoter-based expression analyses of the FANTOM5 data set, which indicated peaks in the LTR16A1 retroelement and the immediately upstream MIRb retroelement in the same intronic region (Extended Data Fig. 1). FANTOM5 CAGE peak distribution over the LTR16A1 and MIRb retroelements exhibited cell-type specificity to a certain degree, with peaks residing almost exclusively in MIRb in bronchial epithelial cells, but extending to LTR16A1 in HEK293 cells (Extended Data Fig. 1). Both LTR16A1 and MIRb retroelements contained multiple transcription factor binding sites, with IRF-1 and IRF-2 binding sites and TATA-box residing in MIRb (Extended Data Fig. 2). To further define the transcription start site(s), we performed 5’ RACE (rapid amplification of cDNA ends) PCR, followed by deep sequencing of the PCR products, amplified from normal human bronchial epithelial (NHBE) cells or human squamous cell carcinoma cell lines SCC-4 and SCC-25, treated with recombinant IFNα (Extended Data Fig. 2). Consistent with FANTOM5 CAGE data, 5’ RACE analysis revealed showed multiple peaks in both LTR16A1 and MIRb, again with evidence of cell-type specificity in their relative utilization (Extended Data Fig. 2). These results suggested that the MIRb and LTR16A1 retroelements acted as a cryptic promoter for the MIRb-ACE2 transcript, with transcription start sites distributed across these two retroelements.
Fig. 1. Identification of the novel MIRb-ACE2 isoform.
a, GENCODE annotated transcripts at the ACE2 locus, intronic position of the MIRb and LTR16A1 elements, structure of the novel MIRb-ACE2 isoform and RNA-seq traces of composite LUAD and LUSC samples. Also shown is splice junction analysis of the same RNA-seq samples. b, Phylogenetic analysis of the MIRb and LTR16A1 sequences in the indicated representative mammalian species and percent sequence identity to the consensus MIRb and LTR16A1 sequences. The arrows indicate the estimated timing of ancestral integrations of the MIRb and LTR16A1 elements, respectively. mya, million years ago. c, Alignment of the MIRb and LTR16A1 sequences in the indicated representative mammalian species and of the consensus MIRb and LTR16A1 sequences.
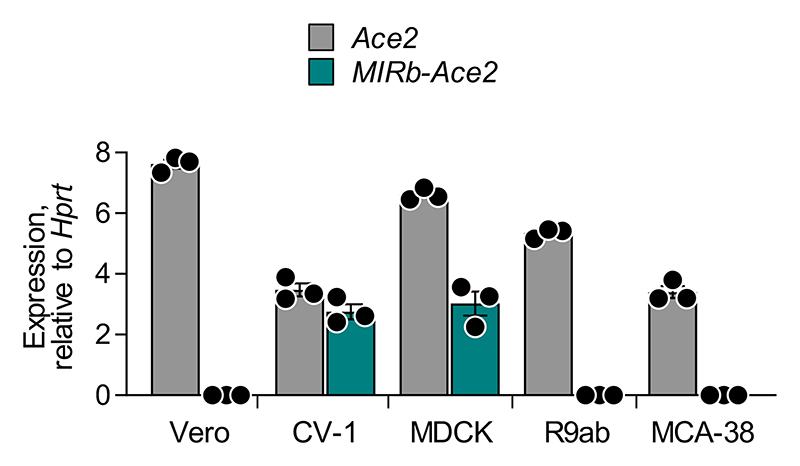
Phylogenetic analysis of the respective LTR16A1 and MIRb elements in the Ace2 loci of representative mammalian species indicated that the ancestral integrations predated estimated dates of mammalian radial divergence (Fig. 1b). Indeed, comparative genomic analysis produced good alignment of the LTR16A1 and MIRb integrations across a variety of species, with humans, dogs, and dolphins showing above 60% sequence identity to the mammalian consensus sequences of LTR16A1 and MIRb (Fig. 1b, c). Of note, the LTR16A1 and MIRb integrations were also present, but truncated in the murine Ace2 locus (Fig. 1b, c), further supported by LASTz Hs-Mm whole genome alignment (Ensembl Compara databases). To further support the intronic initiation of the chimeric MIRb-Ace2 transcript in species other than humans, we performed RT-qPCR analyses using primers complementary to the respective LTR16A1 and Ace2 exon 10 sequences in the African green monkey, dog, rabbit or mouse genome (Extended Data Fig. 3). Whereas all cell lines from these representative species expressed the canonical Ace2 transcript, the MIRb-Ace2 transcript was detected in CV-1, but not Vero cells (both from African green monkey) (Extended Data Fig. 3). The MIRb-Ace2 transcript was undetectable in leporine R9ab and murine MCA-38 cells, but was present in canine MDCK cells (Extended Data Fig. 3), consistent with the high degree of LTR16A1 and MIRb element conservation in the latter species family. Therefore, MIRb-ACE2 expression in humans likely represents a common mammalian feature that has been lost in some, but not all other mammalian species.
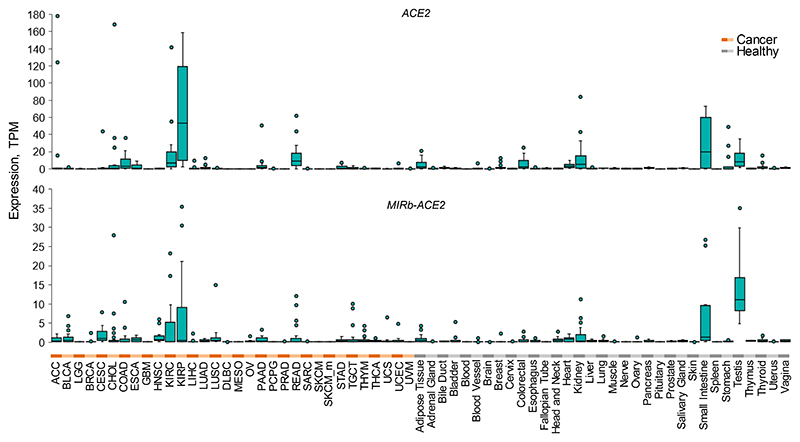
To assess the relative expression of ACE2 and MIRb-ACE2 isoforms, we quantified expression of both transcripts across tissue types in the TCGA and Genotype-Tissue Expression (GTEx) cohorts. Consistent with recent reports17,19, full-length ACE2 was expressed predominantly in the healthy intestine and kidney and tumors of the same histotypes (Extended Data Fig. 4). Expression of MIRb-ACE2 followed a similar overall pattern, but with notable expression also in healthy testis, likely owing to retroelement activation as part of epigenetic reprogramming during spermatogenesis.
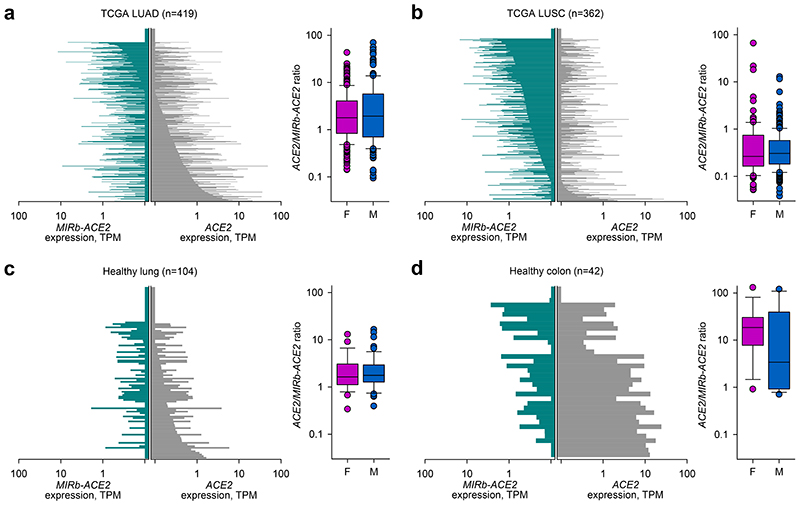
However, despite similar histotype distribution of ACE2 and MIRb-ACE2 expression, the ratio of the two isoforms was characteristically different between distinct histotypes and tumor types. For example, in larger TCGA patient cohorts, LUAD samples expressed higher levels of ACE2 than of MIRb-ACE2 (mean ACE2/MIRb-ACE2 ratio = 5.63), whereas LUSC samples showed the opposite phenotype with higher expression of MIRb-ACE2 (mean ACE2/MIRb-ACE2 ratio = 0.87) (Fig. 2a, b). ACE2 and MIRb-ACE2 expression and their ratios were not affected by patient sex, arguing against a strong effect of the X chromosomal location of ACE2 on either isoform expression (Fig. 2a, b). ACE2 and MIRb-ACE2 exhibited characteristic expression also within tumor types with only weak correlation between the two in the same tumor type (R2=0.252 for LUAD; R2=0.337 for LUSC), suggesting partly independent regulation.
Fig. 2. ACE2 and MIRb-ACE2 isoform expression in cancer and healthy tissues.
a, ACE2 and MIRb-ACE2 isoform expression in LUAD samples (left) and ratio of the two isoforms in female (F, n=225) and male (M, n=161) samples (right). b, ACE2 and MIRb-ACE2 isoform expression in LUSC samples (left) and ratio of the two isoforms in female (F, n=100) and male (M, n=241) samples (right). c, ACE2 and MIRb-ACE2 isoform expression in healthy lung samples (left) and ratio of the two isoforms in female (F, n=29) and male (M, n=54) samples (right). d, ACE2 and MIRb-ACE2 isoform expression in healthy colon samples (left) and ratio of the two isoforms in female (F, n=18) and male (M, n=16) samples (right). In a to d, each bar represents an individual sample. Box plots show the upper and lower quartiles, center lines show the median, whiskers represent the 1.5x interquartile range and individual points represent outliers. TPM, transcripts per million
In healthy lung, expression of ACE2 and MIRb-ACE2 was similar to that in LUAD, with the balance slightly in favor of the full-length form (mean ACE2/MIRb-ACE2 ratio = 2.73) (Fig. 2c). By In contrast, healthy colon expressed considerably higher levels specifically of the full-length isoform (mean ACE2/MIRb-ACE2 ratio = 26.37) (Fig. 2d). These differences in ACE2 and MIRb-ACE2 expression between healthy lung and colon were again independent of gender sex (Fig. 2c, d).
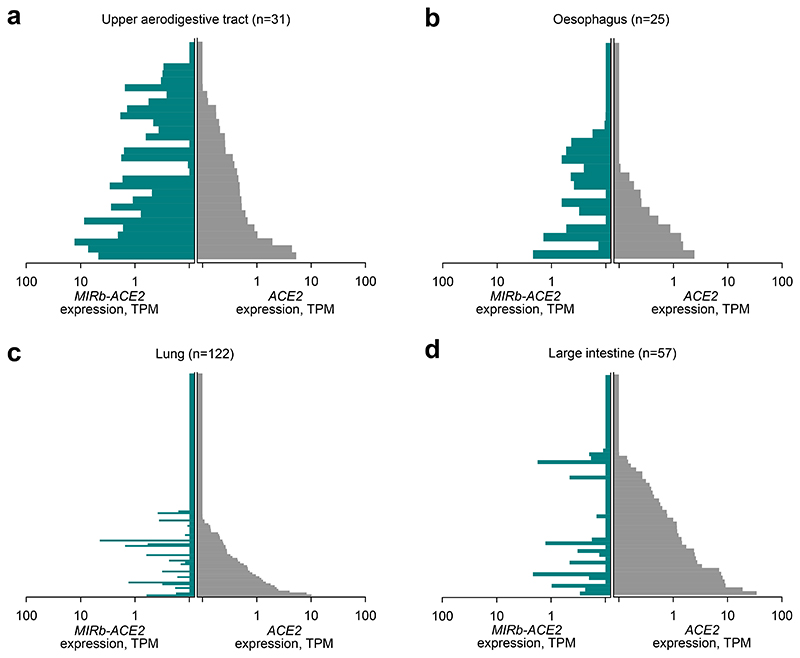
Tissue-specific patterns of ACE2 and MIRb-ACE2 expression suggested dependency on cell lineage or identity. Alternatively, they could reflect transient adaptations to the local microenvironment, such as oxygen or microbiota composition differences between lung and intestine, or even differences in cellular composition between the different compartments. To examine whether patterns of ACE2 and MIRb-ACE2 expression are linked to cell identity, we examined RNA-seq data from 933 cancer cell lines from The Cancer Cell Line Encyclopedia (CCLE). These represent homogenous cell populations, grown under standardized conditions, independently of environmental influences. Again, expression of ACE2 and MIRb-ACE2 was characteristically different between different cell lines and correlated with their anatomical origin (Fig. 3a-d). Cell lines with the highest expression of MIRb-ACE2 were derived from the upper aerodigestive tract, including the mouth and nose (mean ACE2/MIRb-ACE2 ratio = 0.72), followed by esophageal cells lines (mean ACE2/MIRb-ACE2 ratio = 1.66) and lung cell lines (mean ACE2/MIRb-ACE2 ratio = 6.27). Consistent with data from primary biopsies, cells lines from the large intestine exhibited the highest expression of ACE2, with minimal expression of MIRb-ACE2 (mean ACE2/MIRb-ACE2 ratio = 16.97). The low ACE2/MIRb-ACE2 ratio in the upper aerodigestive tract was highly significant when compared with other locations (p=0.0035, when compared with the lung; p=0.0023 when compared with the large intestine, Student’s t-test).
Fig. 3. ACE2 and MIRb-ACE2 isoform expression in cell lines.
a, ACE2 and MIRb-ACE2 isoform expression in cell lines from the upper aerodigestive tract. b, ACE2 and MIRb-ACE2 isoform expression in cell lines from the esophagus. c, ACE2 and MIRb-ACE2 isoform expression in cell lines from the lung. d, ACE2 and MIRb-ACE2 isoform expression in cell lines from the large intestine tract. In a to d, each bar represents an individual sample. TPM, transcripts per million.
Together, these results uncover the transcription of a novel ACE2 isoform, initiated at the intronic MIRb-LTR16A1 retroelements, in a characteristic pattern of expression, forming a gradient from the upper aerodigestive tract (highest MIRb-ACE2 expression) to the large intestine (highest ACE2 expression).
MIRb-ACE2 and not ACE2 transcription is IFN-responsive
ACE2 has recently been described as a human interferon-stimulated gene (ISG), upregulated at the mRNA level following viral infection or interferon treatment17,20. However, this conclusion was based mostly on analysis of single-cell RNA-seq data that might not have sufficient resolution to distinguish the two isoforms. Indeed, inspection of public single-cell RNA-seq data (GSE134355)21, demonstrated the limitation of such technologies, with RNA-seq reads mapping exclusively to the shared 3’ terminal exon of the ACE2 transcripts, and therefore unable to discriminate between the isoforms (Extended Data Fig. 5).
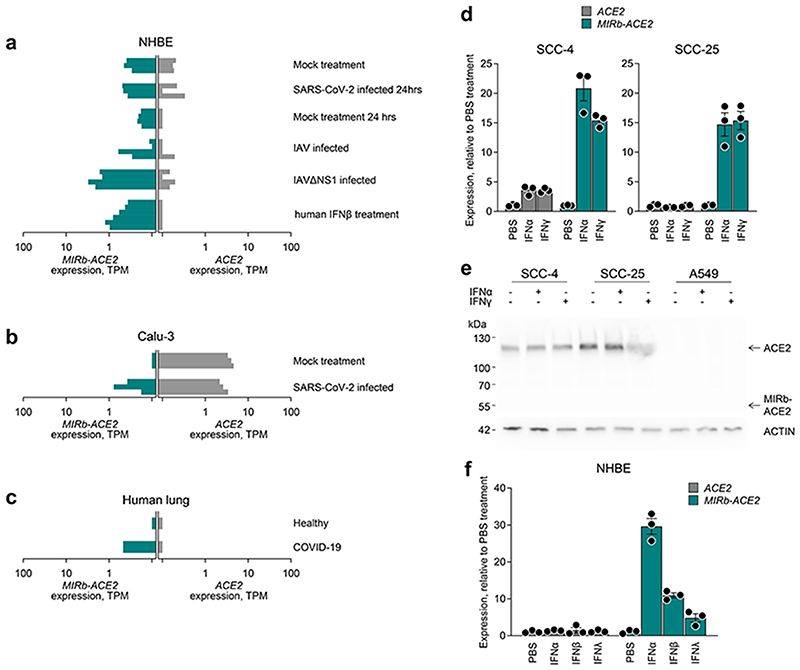
To investigate the inducibility of the two isoforms by IFN or viral infection, we re-analyzsed public RNA-seq data (GSE147507) from NHBE cells, treated with recombinant IFNβ or infected with SARS-CoV-2, Influenza A virus (IAV) or IAV lacking the viral NS1 protein (IAVΔNS1)13. None of the treatments increased expression of full-length ACE2 (Fig. 4a). In stark contrast, MIRb-ACE2 expression was strongly elevated by both IAVΔNS1 infection and recombinant IFNβ treatment, compared with mock treatment (p=0.0005 and p=0.0054, respectively, Student’s t-test). Similar results were also obtained with analysis of lung cancer Calu-3 cells. In the absence of stimulation, Calu-3 cells express exclusively the full-length ACE2 isoform (Fig. 4b). SARS-CoV-2 infection did not affect levels of ACE2 expression, but noticeably induced MIRb-ACE2 expression (Fig. 4b). Lastly, analysis of RNA-seq data from explanted lung tissue from a single COVID-19 patient demonstrated elevated expression of MIRb-ACE2, but not of ACE2, compared with healthy lung tissue (Fig. 4c), albeit statistical comparisons were not possible in this case.
Fig. 4. IFN inducibility of ACE2 and MIRb-ACE2 isoform expression.
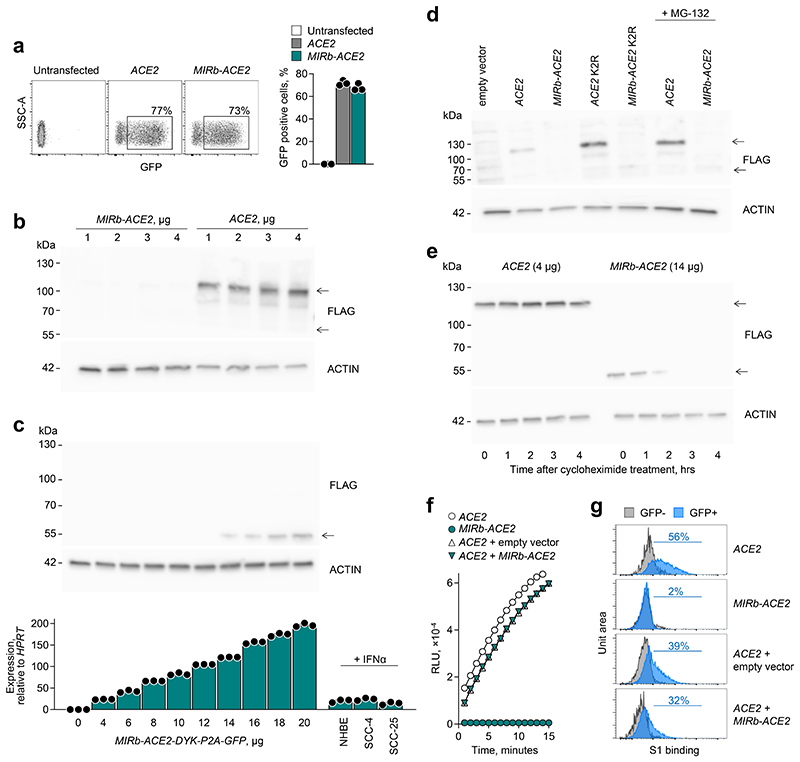
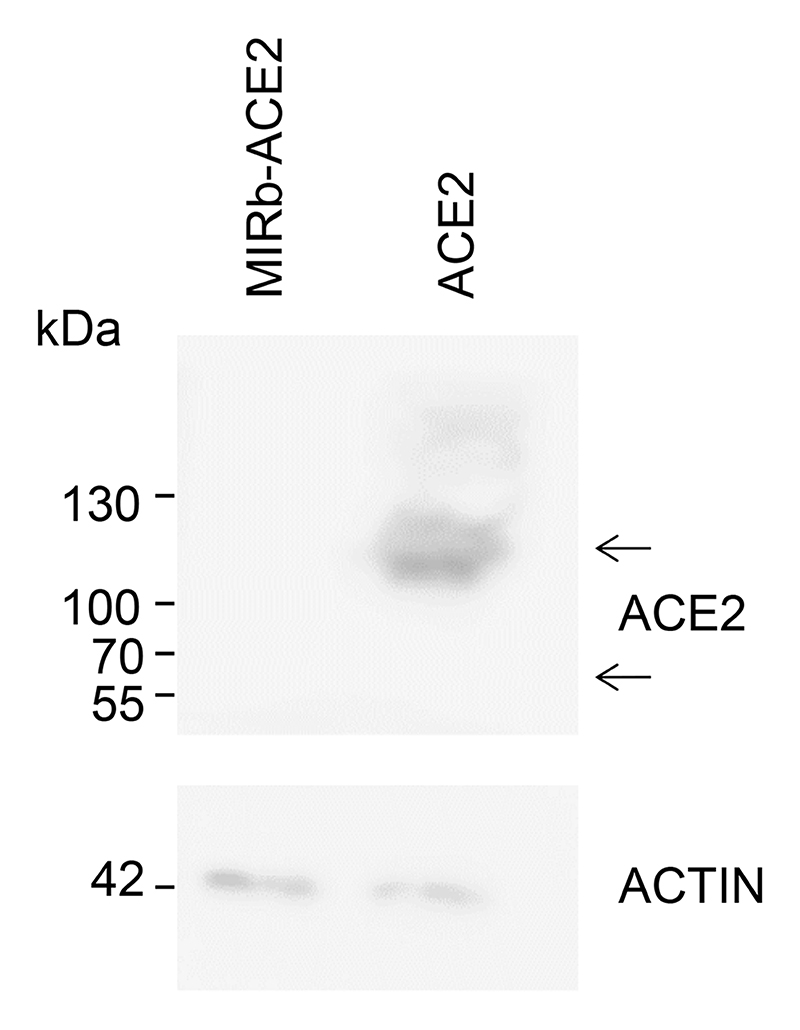
a, ACE2 and MIRb-ACE2 isoform expression NHBE cells following the indicated treatment. b, ACE2 and MIRb-ACE2 isoform expression Calu-3 cells with or without infection with SARS-CoV-2. c, ACE2 and MIRb-ACE2 isoform expression in the lung of a COVID-19 patient and in a healthy lung. In a to c, raw data were obtained form study GSE147507 and each bar represents an individual sample. d, Mean (±SE) ACE2 and MIRb-ACE2 isoform expression, determined by RT-qPCR in SCC-4 and SCC-25 cells with or without IFN stimulation. e, Detection of ACE2 and putative MIRb-ACE2 protein product by Western blotting in cell lysates from the same cells as in d. One representative of 2 experiments is shown. f, Mean (±SE) ACE2 and MIRb-ACE2 isoform expression, determined by RT-qPCR in NHBE cells with or without IFN stimulation. In d and f, each symbol represents the mean value of two technical RT-qPCR replicates of a single culture, and the bars and error bars represent the mean and SE of the three independently-treated cultures in the same experiment. TPM, transcripts per million
To further confirm the IFN-responsiveness exclusively of MIRb-ACE2 expression, we used SCC-4 and SCC-25 cells, which express both isoforms. Compared with mock treatment, addition of recombinant IFNα or IFNγ had a minimal effect on ACE2 expression in SCC-4 cells and no effect in SCC-25 cells (Fig. 4d). This contrasted with very strong induction (~15-fold) of MIRb-ACE2 expression by either type of IFN in both cell lines (Fig. 4d). Lack of ACE responsiveness to IFN stimulation was additionally confirmed at the protein level, where neither IFNα nor IFNγ affected levels of full-length ACE2, detected by Western blotting in SCC-4 and SCC-25 cells or in A549 cells, which express neither isoform and were used as a negative control (Fig. 4e). Splicing from the LTR16A1 retroelement to exon 10 of ACE2 is in-frame and therefore the last 449 amino acids of ACE2 are also present in the putative MIRb-ACE2 protein. Of note, despite strong upregulation at the mRNA level and despite using polyclonal antibodies (ab15348) targeting the C-terminus of ACE2 present in both protein products, we were unable to detect a truncated form that would correspond to the MIRb-ACE2 translation product in SCC-4 or SCC-25 cells (Fig. 4e).
To confirm the differential IFN inducibility of ACE2 and MIRb-ACE2 expression, we stimulated NHBE cells with IFNα, IFNβ or IFNλ, as previously described22. Again, treatment with none of the IFNs had any measurable effect on ACE2 expression in these primary cells (Fig. 4f). This contrasted with robust induction of MIRb-ACE2 expression, particularly by IFNα (Fig. 4f).
Collectively, these data demonstrate that type I, II and III IFNs stimulate transcription of the ACE2 isoform driven by the alternative MIRb-LTR16A1, but not the canonical ACE2 promoter.
The MIRb-ACE2 protein product is not stable
The MIRb-ACE2 isoform is predicted to encode a truncated ACE2 product (amino acids 357-805) and exonization exonization of the LTR16A1 element creates a novel 10 amino acid N-terminal sequence (MREAGWDKGG) in the putative translation product (Extended Data Fig. 6). Importantly, this predicted protein lacks the first 356 amino acids, including the signal peptide, substrate-binding site and domains that interact with SARS-CoV and SARS-CoV-2 spike glycoproteins (Extended Data Fig. 6). Despite sharing the C-terminal half of full-length ACE2, which was readily detectable, endogenously produced MIRb-ACE2 protein was not detectable in SCC-4 and SCC-25 cells naturally expressing the MIRb-ACE2 transcript, by Western blotting using polyclonal anti-ACE2 antibodies (ab15348) (Fig. 4e). To explore the protein-coding potential of the MIRb-ACE2 transcript, we cloned the coding sequences of both isoforms into the pcDNA3.1 mammalian expression vector and transfected HEK293T cells, which do not endogenously express ACE2 that would confound detection of ACE2 produced following transfection16,23. While ACE2-transfected HEK293T cells produced detectable full-length ACE2, no protein of the predicted size was detectable in MIRb-ACE2-transfected cells (Extended Data Fig. 7), in agreement with results using SCC-4 and SCC-25 cells (Fig. 4e). In independently reported findings24, endogenously produced MIRb-ACE2 protein could not be detected by Western blotting using the same polyclonal anti-ACE2 serum (ab15348). However, a Myc-DDK-tagged or GFP-tagged MIRb-ACE2 protein product was detected following overexpression in T24 cells in the same study24. Moreover, a separate study25 reported detection of the putative MIRb-ACE2 protein in primary nasal epithelial cells by Western blotting using the same polyclonal anti-ACE2 serum (ab15348), raising the possibility that the protein can indeed be translated.
To explain the apparent inefficiency of protein production from MIRb-ACE2 transcripts, we cloned the coding sequences of both isoforms into the pcDNA3.1-DYK-P2A-GFP expression vector, which adds both a FLAG tag and P2A peptide-linked GFP as part of the protein product. Expression of GFP was comparable in ACE2-transfected and MIRb-ACE2-transfected cells, suggesting that the single RNA molecule that encodes for both the FLAG-tagged MIRb-ACE2 product and GFP is stable and translated (Fig. 5a). Despite that, following transfection with plasmid concentrations producing readily detectable full-length ACE2 and resulting in MIRb-ACE2 RNA levels comparable with those endogenously produced in IFNα-stimulated cells, we could not detect the predicted MIRb-ACE2 protein with antibodies to the FLAG tag (Fig. 5b). However, the FLAG tagged MIRb-ACE2 protein could be detected in HEK293T cells transfected with much higher plasmid concentrations, resulting in RNA expression levels which were one order of magnitude higher than those observed in IFNα-stimulated NHBE, SCC-4 or SCC2-25 cells (Fig. 5c). Therefore, although the MIRb-ACE2 transcript is efficiently translated (supported by the levels of P2A-linked GFP), the MIRb-ACE2 protein product is much less abundant than the full-length ACE2 at a given RNA transcription level, suggesting post-translational protein instability of the former.
Fig. 5. Stability of the ACE2 and MIRb-ACE2 translation products.
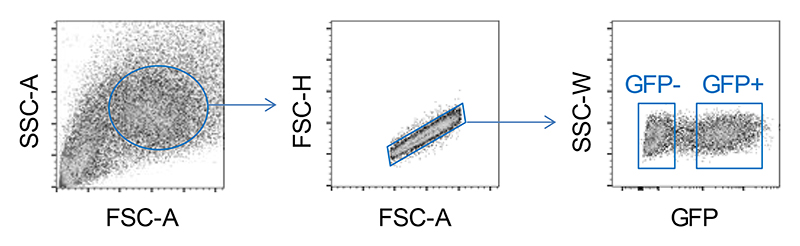
a, Flow cytometric detection of GFP expression (left) and quantitation of mean frequency (±SE) of GFP-expressing cells (right) in HEK293T cells transfected to express either ACE2 or MIRb-ACE2 in conjunction with a FLAG tag and GFP, linked by a P2A peptide. Symbols represent three independently-transfected cultures in the same experiment. One representative of 3 experiments is shown. b, Detection of ACE2 and putative MIRb-ACE2 protein by Western blotting for the FLAG tag in cell lysates from the same cells as in a. Titration the transfection plasmids used is also indicated. One representative of 2 experiments is shown. c, Detection of MIRb-ACE2 protein by Western blotting for the FLAG tag in HEK293T cells transfected with increasing amounts of the expression plasmid (top), and mean (±SE) MIRb-ACE2 expression, determined by RT-qPCR in the same cells, in comparison with MIRb-ACE2 expression in IFNα-stimulated NHBE, SCC-4 and SCC-25 cells (bottom). Each symbol represents the mean value of two technical RT-qPCR replicates of a single culture, and the bars and error bars represent the mean and SE of the three independently-treated cultures in the same experiment. d, Detection of ACE2 and MIRb-ACE2 protein by Western blotting for the FLAG tag in cell lysates from HEK293T cells transfected (with 4 μg of expression plasmids) to express either wild-type isoform or either isoform with the two lysine residues mutated (K2R) (all in conjunction with a FLAG tag and GFP, linked by a P2A peptide). HEK293T cells transfected to express the wild-type isoforms were treated with the MG-132 inhibitor. One representative of 2 experiments is shown. e, Stability of ACE2 and MIRb-ACE2 protein, determined by Western blotting in HEK293T cells transfected to express either isoform, after the indicated times following treatment with cycloheximide. Data from a single experiment are shown. f, Kinetics of mean (±SD) ACE2 enzymatic activity in the supernatant of HEK293T cells transfected to express either ACE2 or MIRb-ACE2 or both (ACE2 + MIRb-ACE2). Expression plasmids were used at 4 μg and 2 μg each for individual transfections and co-transfections, respectively. Symbols represent the mean value of two technical replicates in the same experiment. One representative of 2 experiments is shown. g, Flow cytometric detection of SARS-CoV-2 S1 bindings to HEK293T cells transfected to express either ACE2 or MIRb-ACE2 or both (ACE2 + MIRb-ACE2). ACE2 and MIRb-ACE2 expression plasmids were used at 4 μg and 14 μg for individual transfections, respectively, and at 2 μg and 14 μg and co-transfections, respectively.
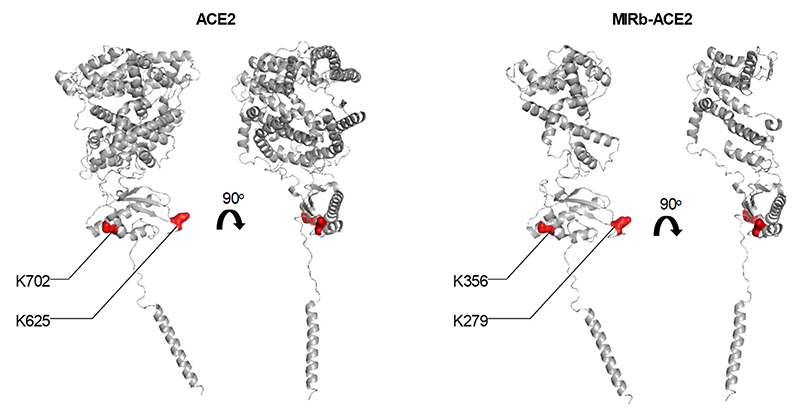
Lysine residues 625 and 702 in the full-length ACE2 protein have been described to be ubiquitinated and may contribute to its proteosomal degradation26. We generated a K625R K702R (K2R) mutant of full-length ACE2, which increased protein levels, compared to the wild-type ACE2 (Fig. 5d). We have introduced the same mutations in the corresponding residues of the predicted MIRb-ACE2 protein product, K279R K356R, which were similarly accessible for ubiquitination (Extended Data Fig. 8). However, we were unable to detect stable protein following transfection with the MIRb-ACE2 K2R-encoding mutant (Fig. 5d). Consistent with this, the addition of the proteasome inhibitor MG-132 was sufficient to increase protein levels of ACE2, but did not rescue the MIRb-ACE2 protein product (Fig. 5d). Moreover, cycloheximide treatment of HEK293T cells transfected with FLAG tagged ACE2 or MIRb-ACE2 constructs led to the rapid loss of MIRb-ACE2 protein, but did not affect levels of full-length ACE2 in the same time frame (Fig. 5e), further supporting reduced stability of the former.
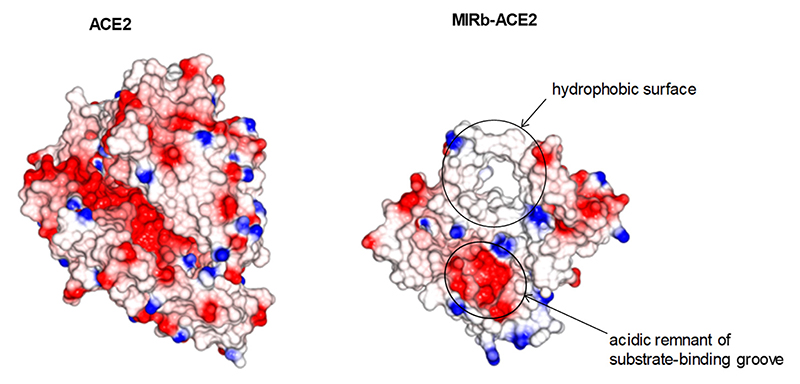
Structural considerations suggested that the MIRb-ACE2 protein product would unlikely retain the partial structure of the canonical ACE2 peptidase fold, as removing most of the this subdomain would expose the remaining component of the highly charged substrate-binding groove as well as large parts of the hydrophobic protein core (Extended Data Fig. 9). Hence, it seems unlikely that a protein coded by the MIRb-ACE2 transcript would form a structure similar to that of the canonical ACE2. Moreover, the MIRb-ACE2 protein product lacks a canonical signal peptide and when an IgG kappa chain-derived signal peptide, which has been successfully used to express the canonical ACE2 ectodomain (residues 15-615)27, was fused to the corresponding domain of the predicted MIRb-ACE2 protein (residues 1-269), there was no detectable secreted protein. These data suggest that the latter protein is subject to post-translational degradation through a proteasome-independent mechanism and therefore unlikely to exert significant biological activity.
Nevertheless, as the MIRb-ACE2 protein was indeed made under certain conditions, it remained possible that it retained some biological function or that it affected the function of canonical ACE2 through heterodimer formation. To examine this possibility, we quantified levels of enzymatically active ACE2, an assay that is considerably more sensitive than Western blotting, and found, as expected, strong enzymatic activity in lysates from ACE2-transfected, but not MIRb-ACE2-transfected cells (Fig. 5f). Furthermore, co-transfection with MIRb-ACE2 did not affect the enzymatic activity conferred by ACE2 transfection (Fig. 5f). To determine any involvement of the predicted MIRb-ACE2 protein in SARS-CoV-2 entry, we measured binding of the S1 subunit of SARS-CoV-2 spike glycoprotein, the first step of viral entry, to cells expressing either or both ACE2 isoforms. HEK293T cells were transfected with the P2A-GFP reporter constructs for ACE2 and MIRb-ACE2 and transfected and untransfected cells were distinguished based on GFP expression (Extended Data Fig. 10). Whereas SARS-CoV-2 S1 efficiently bound HEK293T cells expressing ACE2, it did not bind those expressing MIRb-ACE2 (Fig. 5g). Moreover, co-expression of the two isoforms in the same cells did not alter binding of SARS-CoV-2 S1, beyond the effect of plasmid dilution (Fig. 5g). Collectively, these results argue against significant effect of MIRb-ACE2 expression on ACE2 function or SARS-CoV-2 entry.
Discussion
Regulation of ACE2 expression and function is critical both in physiology and pathology28. The use of ACE2 as a primary receptor for entry by the pandemic coronaviruses, SARS-CoV and SARS-CoV-2 highlighted the potential effect of changes in ACE2 expression, particularly in response to IFN, on the course or severity of COVID-1917. Here, we show that ACE2 transcription and protein production is not responsive to IFN. Instead, we describe a novel RNA isoform, MIRb-ACE2, that is highly responsive to IFN stimulation, but encodes a truncated and unstable protein product. In support of these findings, the new isoform is independently described in two other recent pre-print reports24,25 and matches the sequence recently deposited under GenBank accession number MT505392. We find that the MIRb-ACE2 isoform exhibits distinct patterns of expression along the aerodigestive and gastrointestinal tracts and was likely responsible for the apparent IFN inducibility of ACE2 expression reported by analysis of single-cell RNA-seq data17 and other similar studies20. We further show that transcription of this novel isoform is initiated by intronic retroelements, which function as a cryptic, IFN-responsive promoter, adding further evidence for the widespread involvement of such retroelements in gene regulatory networks.
Indeed, endogenous retroelements comprise nearly half the human genome and can affect many host processes29–31. LTR and non-LTR retroelements represent an abundant source of promoters, enhancers, and polyadenylation sequences that can modulate the expression and structure of neighboring genes32, as with ACE2. For instance, retroelements serve as promoters or enhancers for a number of ISGs, conferring IFN inducibility, exemplified in the case of AIM2 7. Retroelements may further modify the function of ISGs and we have recently described a novel isoform of the ISG CD274 (encoding PD-L1) that produces a truncated form through retroelement exonization33.
The use of the intronic MIRb and LTR16A1 elements as the promoter and alternative exon for the MIRb-ACE2 isoform explains its independent regulation from that of the full-length ACE2 isoform. In addition to IFN inducibility, the cryptic MIRb-LTR16A1 promoter also confers tissue-specific expression, with the highest levels seen in the upper aerodigestive tract, where it can be the predominant isoform. In contrast, the canonical ACE2 isoform far exceeds expression of the MIRb-ACE2 isoform in the lower gastrointestinal tract. It is theoretically possible that the balance of MIRb-ACE2 and full-length ACE2 isoforms plays a role in the spread of SARS-CoV-2, particularly in the upper aerodigestive tract, or that RNA or protein products of MIRb-ACE2 are involved in other pathological or physiological processes. However, the low stability of the MIRb-ACE2 protein product argues that this is unlikely.
Independently of any functional significance, expression of the MIRb-ACE2 isoform needs to be carefully considered in studies examining ACE2 regulation at the transcriptional level17,19,20. The description of this novel isoform highlights the need to validate single-cell RNA-seq data with orthogonal approaches. While single-cell RNA-seq initiatives are an invaluable resource and allow for rapid identification of cell types that express a gene of interest, coverage and read depth are largely insufficient to distinguish between isoforms. Technological advances to improve sequencing depth and bioinformatic tools to impute missing values are rapidly progressing; in the meantime, long-read sequencing techniques to quantify transcript isoforms and confirmation of protein expression levels can be incorporated into existing workflows.
This work established MIRb-ACE2 as the predominantly induced form of ACE2 following viral infection or recombinant interferon treatment, including in the SARS-CoV-2-infected lung. The suggestion that ACE2 is an ISG raised fears that therapeutic interferon could be detrimental17; however, we find that full-length ACE2 is not increased at the mRNA or protein level. The predicted MIRb-ACE2 protein product could be detected in vitro, albeit under high levels of MIRb-ACE2 RNA expression, and it remains possible that the MIRb-ACE2 protein, or fragments thereof, are produced under certain conditions in vivo. Indeed, despite its reduced stability when compared to full-length ACE2, evidence for production of the MIRb-ACE2 protein has also been independently reported24,25. Nevertheless, it is worth noting that the predicted MIRb-ACE2 protein does not contain the residues required for SARS-CoV-2 spike glycoprotein binding15, does not bind recombinant SARS-CoV-2 S1 experimentally and is thus unlikely to contribute to viral spread. These results reconcile the apparent discrepancy between the interferon inducibility of ACE2 with promising data showing improved outcomes in COVID-19 following interferon treatment4,5.
Methods
Cell lines
HEK293T, A549, SCC-4, SCC-25, Vero, CV-1, MDCK, R9ab and MCA-38 cells were obtained from and verified as mycoplasma free by the Cell Services facility at the Francis Crick Institute. Human cell lines were additionally validated by DNA fingerprinting. HEK293T and A549 cells were grown in Iscove’s Modified Dulbecco’s Medium (Sigma-Aldrich) supplemented with 5% fetal bovine serum (Thermo Fisher Scientific), L-glutamine (2 mmol/L, Thermo Fisher Scientific), penicillin (100 U/mL, Thermo Fisher Scientific), and streptomycin (0.1 mg/mL, Thermo Fisher Scientific). SCC-4 and SCC-25 cells were grown in Dulbecco’s Modified Eagle Medium/Nutrient Mixture F-12 (Gibco) supplemented with 10% fetal bovine serum (Thermo Fisher Scientific), L-glutamine (2 mmol/L, Thermo Fisher Scientific), penicillin (100 U/mL, Thermo Fisher Scientific), and streptomycin (0.1 mg/mL, Thermo Fisher Scientific). NHBE cells were cultured as previously described22
Transcript identification, read mapping and quantitation
Transcripts were previously assembled on a subset of the RNA-seq data from The Cancer Genome Atlas (TCGA)18. The alternative promoter within ACE2 was more highly expressed in lung squamous carcinomas than the canonical isoform, prompting us to investigate its biology. RNA-seq data from TCGA, GTEx, CCLE, and other studies were mapped to the cancer-tissue transcriptome assembly and counted as previously described18. Briefly, transcripts per million (TPM) values were calculated for all transcripts in the transcript assembly18 with a custom Bash pipeline (Supplementary Code 1) using GNU parallel34 v3 and Salmon35 v0.12.0, which uses a probabilistic model for assigning reads aligning to multiple transcript isoforms, based on the abundance of reads unique to each isoform35. Splice junctions were visualized using the Integrative Genome Viewer (IGV)36 v2.4.19.
Additional bulk RNA-seq and single-cell RNA-seq data analysis
Bulk RNA-seq data were downloaded from study GSE14750713. Reads were adapter trimmed and filtered for minimal 35nt sequences using Trimmomatic v0.39. Since some samples were infected with SARS-CoV-2 in vitro, we identified and removed viral reads using BowTie2 (seedlength 30nt) to align reads to the Wuhan region reference genome (MN908947). Subsequently, reads were mapped with HISAT2 (optional parameters -p 8 -q -k 5) against GRCh38 reference chromosome assembly and transcripts were quantified against our custom transcriptome assembly using Salmon35 v0.12.0, as described previously18 and above in “Transcript identification, read mapping and quantitation”.
For single-cell RNA-seq data analysis, we downloaded the raw paired end sequencing reads as unmapped bam files from study GSE13435521, which were already demultiplexed, with one individual per tissue per sample. We then used the DropSeq:picard toolbox (v2.3.0) to recapitulate processing of HCL samples as documented on ‘https://github.com/ggjlab/HCL’. In summary, this includes trimming polyA ends from each primary RNA sequencing read and tagging it with the cellular and molecular adapter sequence contained in the secondary read (BASE_RANGE=1-6:22-27:43-48 and BASE_RANGE=49-54, respectively). All reads were then mapped with HISAT2 (optional parameters -p 8 -q -k 5) against GRCh38 reference chromosome assembly. The HISAT2 index here was built with the --exon / --ss option to cover all known splice sites annotated in the GENCODE v34 basic annotation. The cellular and molecular barcode sequences were recovered using the MergeBamAlignment utility in picard.
5’ RACE PCR and sequencing
Total RNA from NHBE, SCC-4 and SCC-25 cells was isolated using the QIAcube (Qiagen) and cDNA synthesis was carried out with the High Capacity Reverse Transcription Kit (Applied Biosystems), with an added RNase inhibitor (Promega). Amplicons were generated using the 5’ RACE System for Rapid Amplification of cDNA Ends (Invitrogen), according to manufacturer’s instructions using primers listed in Supplementary Table 1. Libraries were prepared from amplicons using the NEB Ultra II DNA Library Prep Kit for Illumina (New England Biolabs), according to manufacturer’s instructions and sequenced on a MiSeq system (Illumina). Reads were quality and adapter trimmed in pairs using cutadapt37 v1.18 and aligned with STAR38 v2.7.1a (setting outFilterScoreMinOverLread = 0.1 and outFilterMatchNminOverLread = 0.1) to the a GRCh38 reference with known slice sites from Ensembl release 100. The most 5’ base of reads mapping to the MIRb-ACE2 transcript was taken as the TSS and were obtained from the properly-paired, uniquely-mapping reads using bedtools for visualisation within IGV v2.4.19.
MIRb and LTR16A1 sequence alignments
To identify the integration time of LTR16A1 into the ACE2 locus, we first compared the Homo sapiens LTR16A1 and MIRb to the respective consensus sequences in Dfam 39. Based on sequence identity and the human neutral substitution rate estimated at 2.2 × 10-9 substitutions per site per year, the LTR16A1 insertion is expected to be ~131 million years (with 284 nt matches across 399 nt) and the MIRb insertion ~155 million years (with 159 nt matches across 241 nt). To find evidence for insertion of the LTR16A1 and MIRb elements before the split of the major mammalian lineages, we used the UCSC liftover utility to find the ACE2 gene locus in Rhesus macaque (rheMac10 assembly), marmoset (caljac3 assembly), mouse (mm10 assembly), dog (canFam3 assembly), african elephant (loxAfr3 assembly), bottle-nose dolphin (Turtru2 assembly), cow (bosTau9 assembly), opossum (monDom5 assembly) and platypus (ornAna2). We used the MUSCLE aligner on default settings to build a global alignment of human to rhesus macaque and marmoset, and then aligned all other species to the profile, reverting the strand of the whole sequence for mouse, elephant, cow and opossum due to whole gene inversions. We then used muscle -refine on overlapping 30,000 column blocks to refine the alignment locally. Then we identified the longest potential sequences matching the LTR16A1 and MIRb elements in all species based on the sequences aligning with the repeat sequence in the human genome as annotated by RepeatMasker. These were aligned to LTR16A1 and MIRb consensus sequences from Dfam 3.2 with mafft (--ep 0 --genafpair --maxiterate 1000 options) and intronic sequence clearly distinct from the repeats were trimmed. The two elements are absent from the considerably shorter platypus ACE2 intron. In opossum, the respective intronic sequence is extended but no clear matches with either LTR16A1 or MIRb were found, prompting us to place both insertions ahead of the mammalian radial divergence. The illustration of the lineage tree including node times are taken from timetree.org.
Expression vectors
Open reading frames encoding ACE2, MIRb-ACE2, and respective lysine mutants were synthesized and cloned into the pcDNA3.1-DYK-P2A-GFP mammalian expression vector. Gene synthesis, cloning, and mutagenesis were performed by GenScript and verified by sequencing. Cells were transfected using GeneJuice (EMD Millipore) and harvested 48 hrs post-transfection for downstream assays.
Cell stimulation
For interferon stimulation experiments, 2 × 105 SCC-4 and SCC-25 cells were stimulated with 100 ng/mL IFN-α or IFN-γ (Abcam) or PBS for 48 hrs. For proteasome inhibition experiments, cells were cultured in 20 μM MG-132 (EMD Millipore) 24 hrs after transfection and harvested 48 hrs after transfection. For cycloheximide experiments, cells were treated with 250 μg/mL cycloheximide (Sigma Aldrich) and harvested at denoted time-points. NHBE cells were stimulated for 4 hrs with 1000 ng/ml IFNα, 100ng/ml IFNβ or 100ng/ml IFNλ were used in a previous study22, and stored cDNA was analyzed by RT-qPCR in this study.
Western blot
Cell lysates in RIPA buffer were resuspended in SDS buffer, heat denatured at 95°C for 10 min, run on a 4-20% gel (Biorad), transferred to a PVDF membrane (Biorad), and blocked in 5% (w/v) bovine serum albumin fraction V (Sigma-Aldrich) in TBS-T. Membranes were incubated with primary antibodies to ACE2 (1:1000; ab15348, Abcam), FLAG (1:1000; F1804-50UG clone M2, Sigma-Aldrich), HRP-conjugated secondary antibodies to rabbit IgG or mouse IgG (1:1000; #7074 and #7076, respectively, Cell Signaling Technology), and HRP-conjugated actin (1:25000; ab49900, Abcam). Blots were visualized by chemiluminescence on an Amersham Imager 600 (GE Healthcare).
Reverse transcriptase-based quantitative PCR (RT-qPCR)
Total RNA from cell lines was isolated using the QIAcube (Qiagen), and cDNA synthesis was carried out with the High Capacity Reverse Transcription Kit (Applied Biosystems) with an added RNase inhibitor (Promega). Purified cDNA was used to quantify human ACE2 and MIRb-ACE2, or Ace2 and MIRb-Ace2 in other mammalian species, using variant-specific and species-specific primers (Supplementary Table 1). The IFN-inducible human genes CXCL10 and CD274 were also amplified as controls for the effect of IFN treatment, using transcript-specific primers (Supplementary Table 1). For amplification of a conserved house-keeping gene, primers complementary to HPRT sequences conserved in all species were used (Supplementary Table 1). Values were normalized to HPRT expression using the ΔCT method.
Enzyme assays
ACE2 activity in cell lysates was measured using the SensoLyte 390 ACE2 Activity Assay (AnaSpec) according to manufacturer’s instructions. Recombinant human ACE2 (Sigma-Aldrich) was used as a positive control.
Flow Cytometry
For SARS-CoV-2 S1 binding assays, cells were stained with biotinylated S1 (1:200; Acro Biosystems) for 30 minutes followed by APC-Streptavidin (1:200; Biolegend). For S1 binding assays and for GFP detection, single-cell suspensions were run on a LSR Fortessa (BD Biosciences) running BD FACSDiva v8.0 and analysed with FlowJo v10 (Tree Star Inc.) analysis software.
Statistical analysis
Statistical comparisons were made using GraphPad Prism 7 (GraphPad Software) or SigmaPlot 14.0. Parametric comparisons of normally distributed values that satisfied the variance criteria were made by unpaired Student’s t-tests or One Way Analysis of variance (ANOVA) tests. Data that did not pass the variance test were compared with non-parametric two-tailed Mann–Whitney Rank Sum tests or ANOVA on Ranks tests.
Extended Data
Extended Data Fig. 1. CAGE support for transcriptional initiation of the MIRb-ACE2 transcript.
Normalized data from the FANTOM Consortium and the RIKEN PMI and CLST (DGT) for transcription start sites in the proximity of the intronic MIRb and LTR16A1 elements in the ACE2 locus. Both the sense and antisense orientations are depicted. Data were visualized with the zenbu online viewer (https://fantom.gsc.riken.jp/zenbu) for FANTOM5 Human hg38 promoterome.
Extended Data Fig. 2. 5’ RACE PCR support for transcriptional initiation of the MIRb-ACE2 transcript.
Middle panel: Putative transcription factor binding sites, predicted by PROMO (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3), in the intronic MIRb and LTR16A1 elements in the ACE2 locus A putative TATA-box is also depicted. Lower panel: Mapping of sequencing reads of 5’ RACE PCR products from IFNα-stimulated primary NHBE cells or SCC-4 and SCC-25 cell lines in the intronic MIRb and LTR16A1 elements in the ACE2 locus. A detailed view of CAGE data (from Extended Data Fig. 1) spanning the same region is also included for comparison.
Extended Data Fig. 3. Ace2 and MIRb-Ace2 expression in cells from representative mammalian species.
Mean (±SE) Ace2 and MIRb-Ace2 isoform expression, determined by RT-qPCR, in Vero and CV-1 cells (both from African green monkey), and in MDCK (dog), R9ab (rabbit) and MCA-38 cells (mouse). Expression is normalized to expression of Hprt in each sample. Each symbol represents the mean value of two technical RT-qPCR replicates of a single culture, and the bars and error bars represent the mean and SE of the three individually-treated cultures in the same experiment.
Extended Data Fig. 4. Expression of ACE2 and MIRb-ACE2 isoforms in cancer and healthy tissues.
Box plots of ACE2 and MIRb-ACE2 isoforms expression in cancer patient and healthy control samples from TCGA and GTEx. For each cancer type, 24 samples were included (a total 768 samples), whereas for respective healthy tissues a total of 813 samples were included, varying between 2 and 156 per tissue type. Box plots show the upper and lower quartiles, center lines show the median, whiskers represent the 1.5x interquartile range and individual points represent outliers.
Extended Data Fig. 5. Single-cell RNA-seq coverage of the ACE2 locus.
RNA-seq trace of two multiplexed samples from adult lung, obtained from study GSE134355. Note the lack of coverage across the entire locus with the exception of only the 3’ end of the last exon, shared between the isoforms.
Extended Data Fig. 6. Protein sequence alignment of ACE2 and MIRb-ACE2.
The predicted MIRb-ACE2 translation product is a 459-amino acid protein lacking the indicted single peptide, domains interacting with SARS-CoV spike glycoprotein, but retaining the transmembrane domain. The novel 10-amino acid sequence created by LTR16A1 exonisation is also shown.
Extended Data Fig. 7. Protein production by the ACE2 and MIRb-ACE2 transcript isoforms.
Detection of ACE2 and putative MIRb-ACE2 protein product by Western blotting in cell lysates from HEK293T cells transfected to express either isoform. Proteins were detected using anti-ACE2 rabbit polyclonal antibodies (ab15348) as the primary reagent. One representative of 4 experiments is shown.
Extended Data Fig. 8. Position of the ubiquitin targets in ACE2 and MIRb-ACE2 proteins.
Structure of ACE2 (left) and predicted structure of the MIRb-ACE2 protein product (right) depicting the position of the two mutated K residues, targeted for ubiquitination.
Extended Data Fig. 9. Electrostatic-surface views of ACE2 and MIRb-ACE2 proteins.
Matching projections of the canonical ACE2 (PDB ID: 6M0J) and of the predicted MIRb-ACE2 protein product structures, if the latter were to exist in the same structural arrangement, but truncated. Basic, positively-charged residues shown in blue and acidic, negatively-charged residues in red. Arrows indicate an exposed hydrophobic surface and the open, acidic remnant of the substrate-binding groove.
Extended Data Fig. 10. Identification of cells expressing ACE2 or MIRb-ACE2 based on GFP reporter expression.
Gating strategy for the identification of HEK293T cells transfected with plasmids encoding ACE2 or MIRb-ACE2 in conjunction with a P2A-GFP reporter. Expressing and non-expressing single cells were gated according to GFP expression.
Supplementary Material
Acknowledgments
We are grateful for assistance from the Advanced Sequencing, Scientific Computing, Flow Cytometry and Cell Services facilities at the Francis Crick Institute. The results shown here are in whole or part based upon data generated by The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov). The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. This work benefited from data assembled by the CCLE consortium. This work was supported by the Francis Crick Institute (FC001099, FC001206, FC001078), which receives its core funding from Cancer Research UK, the UK Medical Research Council, and the Wellcome Trust; and by the Wellcome Trust (102898/B/13/Z).
Footnotes
Contributions
K.W.N., J.A., G.R.Y. and G.K. conceived and designed the study. K.W.N., J.A., W.B., G.R.Y., J.M. and A.G.W. performed the experiments. K.W.N., J.A., W.B., G.R.Y., J.M., A.G.W. and G.K. analyzed the data. K.W.N and G.K. wrote the manuscript with contributions from J.A., W.B., G.R.Y., J.M., A.G.W., S.G., and A.W. All authors approved the final manuscript.
Competing interests
The authors declare no competing interests.
Data availability
Data supporting the findings of this study are available within the article and its supplementary information files. All data, plasmids and cell lines are available and from the corresponding author upon reasonable request. Publicly available data were downloaded from the following databases: The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov), The Genotype-Tissue Expression (GTEx) Project (https://gtexportal.org/home); and the Broad Institute Cancer Cell Line Encyclopedia (CCLE) consortium (https://portals.broadinstitute.org/ccle). Additionally, RNA-seq data from individual studies (GSE147507 and GSE134355) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). Source data are provided with this paper.
Code availability
Custom code used in this study is available in the supplementary information.
References
- 1.Sadler AJ, Williams BR. Interferon-inducible antiviral effectors. Nature reviews Immunology. 2008;8:559–568. doi: 10.1038/nri2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Stetson DB, Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- 3.Gibbert K, Schlaak JF, Yang D, Dittmer U. IFN-α subtypes: distinct biological activities in anti-viral therapy. British journal of pharmacology. 2013;168:1048–1058. doi: 10.1111/bph.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hung IF, et al. Triple combination of interferon beta-1b, lopinavir-ritonavir, and ribavirin in the treatment of patients admitted to hospital with COVID-19: an open-label, randomised, phase 2 trial. Lancet (London, England) 2020;395:1695–1704. doi: 10.1016/s0140-6736(20)31042-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang N, et al. Retrospective Multicenter Cohort Study Shows Early Interferon Therapy Is Associated with Favorable Clinical Responses in COVID-19 Patients. Cell host& microbe. 2020 doi: 10.1016/j.chom.2020.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ivashkiv LB, Donlin LT. Regulation of type I interferon responses. Nature reviews Immunology. 2014;14:36–49. doi: 10.1038/nri3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chuong EB, Elde NC, Feschotte C. Regulatory evolution of innate immunity through co-option of endogenous retroviruses. Science (New York, N.Y.) 2016;351:1083–1087. doi: 10.1126/science.aad5497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Young GR, et al. Resurrection of endogenous retroviruses in antibody-deficient mice. Nature. 2012;491:774–778. doi: 10.1038/nature11599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Young GR, Mavrommatis B, Kassiotis G. Microarray analysis reveals global modulation of endogenous retroelement transcription by microbes. Retrovirology. 2014;11:59. doi: 10.1186/1742-4690-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Attig J, Young GR, Stoye JP, Kassiotis G. Physiological and Pathological Transcriptional Activation of Endogenous Retroelements Assessed by RNA-Sequencing of B Lymphocytes. Frontiers in microbiology. 2017;8:2489. doi: 10.3389/fmicb.2017.02489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tokuyama M, et al. ERVmap analysis reveals genome-wide transcription of human endogenous retroviruses. Proceedings of the National Academy of Sciences of the United States of America. 2018;115:12565–12572. doi: 10.1073/pnas.1814589115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.García-Sastre A. Ten Strategies of Interferon Evasion by Viruses. Cell host& microbe. 2017;22:176–184. doi: 10.1016/j.chom.2017.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Blanco-Melo D, et al. Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19. Cell. 2020;181:1036–1045.e1039. doi: 10.1016/j.cell.2020.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kopecky-Bromberg SA, Martínez-Sobrido L, Frieman M, Baric RA, Palese P. Severe acute respiratory syndrome coronavirus open reading frame (ORF) 3b, ORF 6, and nucleocapsid proteins function as interferon antagonists. Journal of virology. 2007;81:548–557. doi: 10.1128/jvi.01782-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shang J, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020;581:221–224. doi: 10.1038/s41586-020-2179-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffmann M, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280.e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ziegler CGK, et al. SARS-CoV-2 Receptor ACE2 Is an Interferon-Stimulated Gene in Human Airway Epithelial Cells and Is Detected in Specific Cell Subsets across Tissues. Cell. 2020;181:1016–1035.e1019. doi: 10.1016/j.cell.2020.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Attig J, et al. LTR retroelement expansion of the human cancer transcriptome and immunopeptidome revealed by de novo transcript assembly. Genome research. 2019;29:1578–1590. doi: 10.1101/gr.248922.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singh M, Bansal V, Feschotte C. A single-cell RNA expression map of human coronavirus entry factors. bioRxiv. 2020 doi: 10.1101/2020.05.08.084806. 2020.2005.2008.084806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith JC, et al. Cigarette Smoke Exposure and Inflammatory Signaling Increase the Expression of the SARS-CoV-2 Receptor ACE2 in the Respiratory Tract. Developmental cell. 2020;53:514–529.e513. doi: 10.1016/j.devcel.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X, et al. Construction of a human cell landscape at single-cell level. Nature. 2020;581:303–309. doi: 10.1038/s41586-020-2157-4. [DOI] [PubMed] [Google Scholar]
- 22.Major J, et al. Type I and III interferons disrupt lung epithelial repair during recovery from viral infection. Science (New York, N.Y.) 2020 doi: 10.1126/science.abc2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng KW, et al. Pre-existing and de novo humoral immunity to SARS-CoV-2 in humans. bioRxiv. 2020 doi: 10.1101/2020.05.14.095414. 2020.2005.2014.095414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Onabajo OO, et al. Interferons and viruses induce a novel primate-specific isoform dACE2 and not the SARS-CoV-2 receptor ACE2. bioRxiv. 2020 doi: 10.1101/2020.07.19.210955. 2020.2007.2019.210955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blume C, et al. A novel isoform of ACE2 is expressed in human nasal and bronchial respiratory epithelia and is upregulated in response to RNA respiratory virus infection. bioRxiv. 2020 doi: 10.1101/2020.07.31.230870. 2020.2007.2031.230870. [DOI] [Google Scholar]
- 26.Stukalov A, et al. Multi-level proteomics reveals host-perturbation strategies of SARS-CoV-2 and SARS-CoV. bioRxiv. 2020 doi: 10.1101/2020.06.17.156455. 2020.2006.2017.156455. [DOI] [Google Scholar]
- 27.Wrobel AG, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nature structural& molecular biology. 2020 doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamming I, et al. The emerging role of ACE2 in physiology and disease. The Journal of pathology. 2007;212:1–11. doi: 10.1002/path.2162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Burns KH, Boeke JD. Human transposon tectonics. Cell. 2012;149:740–752. doi: 10.1016/j.cell.2012.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Feschotte C, Gilbert C. Endogenous viruses: insights into viral evolution and impact on host biology. Nat Rev Genet. 2012;13:283–296. doi: 10.1038/nrg3199. [DOI] [PubMed] [Google Scholar]
- 31.Kassiotis G, Stoye JP. Immune responses to endogenous retroelements: taking the bad with the good. Nat Rev Immunol. 2016;16:207–219. doi: 10.1038/nri.2016.27. [DOI] [PubMed] [Google Scholar]
- 32.Thompson PJ, Macfarlan TS, Lorincz MC. Long Terminal Repeats: From Parasitic Elements to Building Blocks of the Transcriptional Regulatory Repertoire. Molecular cell. 2016;62:766–776. doi: 10.1016/j.molcel.2016.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng KW, et al. Soluble PD-L1 generated by endogenous retroelement exaptation is a receptor antagonist. eLife. 2019;8 doi: 10.7554/eLife.50256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tange O. GNU Parallel: The Command-Line Power Tool. The USENIX Magazine. 2011;36:42–47. [Google Scholar]
- 35.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nature methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Briefings in bioinformatics. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marcel M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:3. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 38.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics (Oxford, England) 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hubley R, et al. The Dfam database of repetitive DNA families. Nucleic acids research. 2016;44:D81–89. doi: 10.1093/nar/gkv1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting the findings of this study are available within the article and its supplementary information files. All data, plasmids and cell lines are available and from the corresponding author upon reasonable request. Publicly available data were downloaded from the following databases: The Cancer Genome Atlas (TCGA) Research Network (http://cancergenome.nih.gov), The Genotype-Tissue Expression (GTEx) Project (https://gtexportal.org/home); and the Broad Institute Cancer Cell Line Encyclopedia (CCLE) consortium (https://portals.broadinstitute.org/ccle). Additionally, RNA-seq data from individual studies (GSE147507 and GSE134355) were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). Source data are provided with this paper.
Custom code used in this study is available in the supplementary information.