Abstract
Mitoribosomes are tethered to the mitochondrial inner membrane to facilitate the co-translational membrane insertion of the synthesized proteins. Here, we report cryo-electron microscopy structures of human mitoribosomes with nascent polypeptide, bound to the insertase OXA1L through three distinct contact sites. OXA1L binding is correlated with a series of conformational changes in the mitoribosomal large subunit that catalyze the delivery of newly synthesized polypeptides. The mechanism relies on the folding of mL45 inside the exit tunnel, forming two specific constriction sites that would limit helix formation of the nascent chain. A gap is formed between the exit and the membrane, making the newly synthesized proteins accessible. Our data elucidate the basis by which mitoribosomes interact with the OXA1L insertase to couple protein synthesis and membrane delivery.
Proteins encoded in the human mitochondrial genome (mtDNA) are essential for life, and aberrations in their synthesis are associated with clinical pathologies (1, 2). Synthesis of these transmembrane proteins of the oxidative phosphorylation complexes occurs on dedicated mitochondrial ribosomes (mitoribosomes) that possess a specialized exit tunnel and regulatory features (3). These mitoribosomes are tightly associated with the inner mitochondrial membrane for efficient protein delivery (4). Co-translational membrane insertion is catalyzed by a putative mitochondrial insertase, the Oxal-Like (OXA1L) protein, a homolog of yeast Oxidase Assembly 1 (Oxal) and bacterial YidC (5–7). In yeast, Oxal is required for the assembly of the oxidative phosphorylation complexes (5), and its C-terminus is associated with the mitoribosome (9, 10). In human cells, depletion of OXA1L impairs the assembly of the complexes I, IV, V (11, 12), and biallelic OXA1L pathogenic mutations present with fatal encephalitis, hypotonia, and developmental delay (12). Mechanistically, the association of the mitoribosome with an insertase would likely protect the hydrophobic membrane proteins during their egression into the lipid bilayer. Based on studies of co-translational folding of membrane proteins into an α-helical conformation in the ribosomal tunnel in bacteria (13), it has been postulated that such secondary structure may also start forming within the mitoribosome tunnel. However, no structure of mitoribosome with a membrane insertase has been reported, thus a mechanism of nascent chain emergence remained unknown.
To capture interactions of a mitoribosomal exit tunnel with its substrate, we used the peptidomimetic actinonin, which structurally resembles formylmethionine, to induce a co-translational accumulation of stalled mitoribosomes during nascent chain synthesis (fig. S1) (14). To sort the particles corresponding to translating mitoribosomes, we collected six cryo-EM data sets and classified them based on the presence of the small subunit (mtSSU) after signal subtraction (15). Then performed a second step of signal subtraction and 3D classification on the A-site tRNA, and a third step on the insertase region (fig. S2A). Subsequent 3D refinement produced a reconstruction that showed fragmented density extending from the tunnel exit. To reduce the flexibility of a putative insertase on the mitoribosome, we subjected the complex to amine-to-amine cross-linking, and repeated the cryo-EM analysis with two additional data sets (fig. S2B-F, see Methods). In silico sorting yielded a subset of 30,744 particles that, upon contrast transfer function refinement and Bayesian polishing (15), resulted in the 2.89 Å reconstruction, revealing densities for tRNAs in the A- and P-sites, continuous density from the peptidyl transferase center (PTC) extending to the tunnel exit, and additional mass outside the exit correlated with the position of the inner mitochondrial membrane (4) (Fig. 1A, fig. S3). The position of the extra density and its overall dimensions were in agreement with the bacterial ribosome:YidC reconstructions (16, 17) that accommodate a single copy of the insertase (fig. S4).
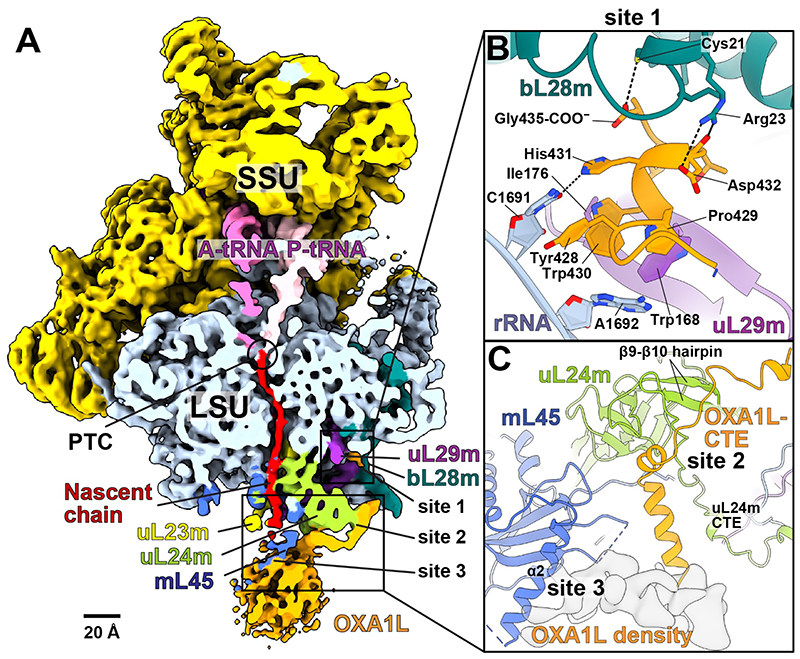
Fig. 1. Human mitoribosome:OXAlL complex.
(A) Density of mitoribosome:OXA1L complex lowpass-filtered to 6 Å resolution for clarity and cut through the tunnel. Contact sites 1, 2, 3 are indicated. (B) Contact site 1 is formed by OXA1L C-terminal helix bound to bL28m, uL29m, and rRNA. Tyr428 is stacked on rRNA C1691 and His431 hydrogen-bonds with C1691. The backbone carbonyl and sidechain of Asp432 form polar interactions with bL28m (Arg23, Pro429, and Trp430) and hydrophobic interactions with uL29m (Trp168, Ile176), and rRNA A1692. (C) The OXA1L-CTE interacts with uL24m (site 2), and the OXA1L core (shown with density) is associated with mL45-α2 (site 3).
Although the local resolution for the additional density is 5-8 Å, reflecting intrinsic flexibility, by combining biochemical stabilization with targeted classification and polishing, we could improve the signal-to-noise ratio and trace ~50-amino acid residues, docked to the surface of the mitoribosomal large subunit (mtLSU) (fig. S5). The quality of the sidechain densities was sufficient for modeling the C-terminal residues of OXA1L (fig. S5D), and mass-spectrometry analysis confirmed the presence of OXA1L (fig. S6). OXA1L binds to the mitoribosome through three distinct contact sites spanning the solvent-exposed side of the mtLSU (Fig. 1A, fig S7). Together, the interface anchoring OXA1L involves three mitoribosomal proteins and rRNA, comprising a total buried surface area of ~1500 Å2.
Site 1 is found in the mitoribosomal cleft towards the L1 stalk, ~70 Å away from the membrane surface (Fig. 1A). It comprises mitochondrial-specific protein extensions of bL28m, uL29m, and the rRNA bypass element H4-H8 (residues 1689-1695) that together form the binding pocket for the OXA1L C-terminal helix (YPWHDTLG-COO–) (fig. S8A,B). A series of interactions involve residues that are generally conserved in vertebrates and underlie a tight association that does not occur in bacteria (fig. S9).
Site 2 is separated from site 1 by 27 Å and consists solely of the mitoribosomal protein uL24m (Fig. 1C). Although the part of OXA1L linking the two contact sites is not well resolved, we could model 40 residues composed of an α-helix, two short 310-helices, and a 10-residue loop. Together it binds to the matrix exposed surface, thus encompassing uL24m, and the loop is stabilized by the β9-β10 hairpin (residues 133-147) at the C-terminal domain of uL24m (Fig. 1C, fig. S9D). Consistent with the shape of uL24m, the OXA1L structure is bent in this region by ~80° (Fig. 1A,C).
Site 3 is ~15 Å away from site 2 and formed by the core domain of mL45, where the globular density of the OXA1L membrane part is attached (Fig. 1C). Although no OXA1L model could be placed here due to the flexibility, continuous density can be traced from the α-helix of OXA1L C-terminal extension (CTE) at site 2 (Fig. 1A). The contact is mostly through the α-helix mL45-α2 (residues 94-128) on top of the core β-sheet of the mL45 Tim44-like domain. Overall, the observed interactions of OXA1L with the mitoribosome are consistent with super-resolution microscopy showing that the CTE truncation (Δ332-433) of OXA1L reduces but not completely abolishes the co-localization with the translating mitoribosome (18).
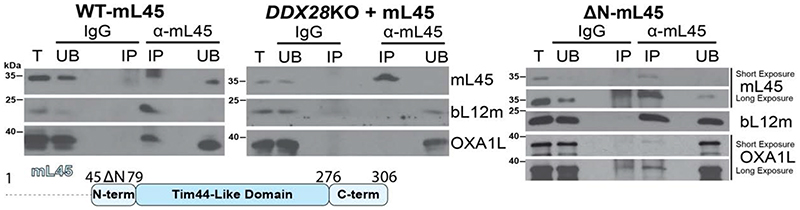
Consistent with the structure, we find that OXA1L forms a stable interaction with mL45 in vitro only when it is assembled into the mitoribosome (Fig 2, fig. S10). To demonstrate this, we used a cell line in which the mtLSU assembly helicase DDX28 has been deleted (19). We ectopically expressed mL45, which associates with the mitochondrial membranes despite the mtLSU assembly defect, but does not interact with OXA1L (Fig. 2, fig. S10). Thus, the interaction between OXA1L and mitoribosomes is mediated by mitoribosome-assembled mL45.
Fig. 2. Biochemical characterization of mL45 and OXA1L interaction.
Immunoprecipitation of mL45 from mitochondrial extracts of T-Rex-293 cells expressing wild-type (WT) or ΔN-mL45, and the DDX28-KO cells overexpressing WT mL45. The cells were probed for mL45, OXA1L, or bL12m as an mtLSU marker, IgG was used as a control. (IP) immunoprecipitated, (UB) unbound, (T) total input.
Next, we examined the cryo-EM data with respect to the exit tunnel interior and identified two distinct states, Class 1a* and Class 1b* (fig. S2B, fig. S11). The particles in Class 1b* represent the OXA1L-unbound (inactive) state. The density in the upper tunnel is separated from the tRNA by 12 Å (fig. S11A). This density extends from mL45 and corresponds to its N-terminal sequence, consistent with a previous study (20). The previous study also proposed that the mL45 N-terminal tail acts to recruit the OXA1L insertase, implying a targeting mechanism in mammalian mitochondria analogous to the signal recognition particle (20). In fungi, mL45 (Mba1) is not a structural component of the mitoribosome (21, 22); whereas, in ciliates, a homolog of the bacterial signal recognition particle binding protein Ffh was identified in the mitoribosomal tunnel (23). To investigate if mL45 N-terminus recruits OXA1L in human mitochondria, we expressed a variant with shortened N-terminus 45-74 (ΔN) and showed that although the relative amount of co-immunoprecipitated OXA1L is lower than for the WT, the truncation of mL45 does not entirely prevent interaction with OXA1L (Fig. 2).
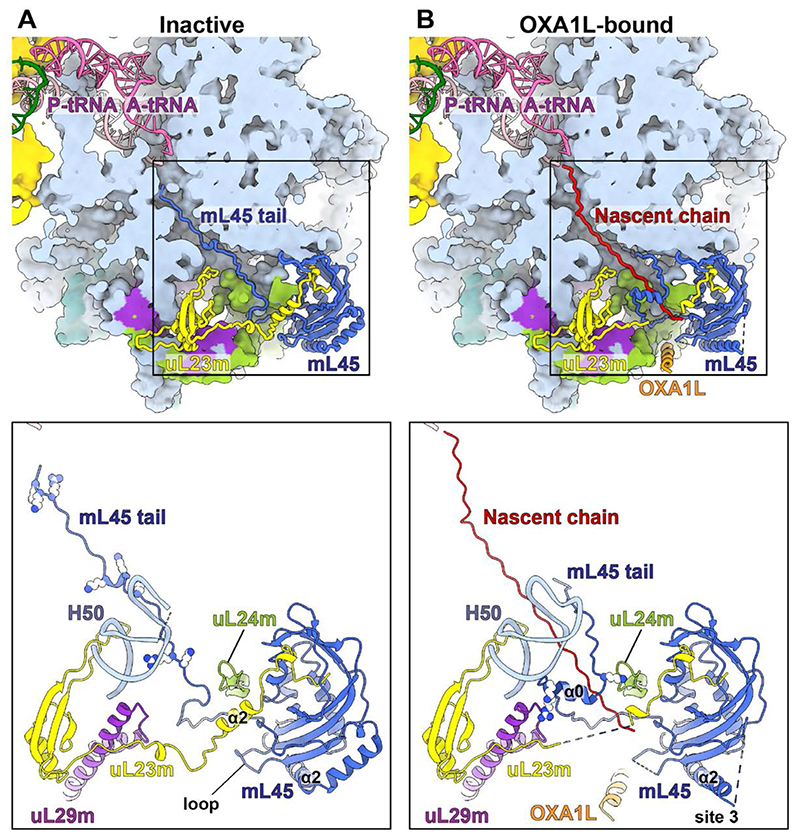
In the inactive state, basic residues in the mL45 N-terminus form electrostatic interactions with the rRNA core that anchor the protein moiety inside the tunnel (Fig. 3A, fig. S11A). In the lower tunnel, mL45 Pro68 is stabilized by the mitochondria-specific rRNA A1804, mL45 Ile76, and uL23m Leu126 (fig. S12A). We found that A1804 is a mtDNA insertion (fig. S8C), which is an unusual feature, as only deletions in the mammalian mitochondrial rRNA have been reported up to date (24–28). This stabilization allows mL45 Arg61 and Lys62 to point towards the rRNA interior (fig. S12A), so that the mL45 N-tail is positioned directly on the nascent polypeptide path (Fig. 3A). This is further correlated with two hydrophobic residues, Val66 and Ile67, forming inter-protein hydrophobic pairs with uL23m-α2 (fig. S12A). These interactions stabilize uL23m-α2 perpendicularly to the tunnel so that it fills the spacings at the exit site, blocking the path.
Fig. 3. Comparison between inactive and OXA1L-bound structures.
(A) In the inactive state, the mL45 N-terminal tail occupies the tunnel. Basic residues in the tunnel are shown as spheres in the bottom panel. uL23m-α2 is stabilized by the mL45 tail and a loop (residues 200-208) of mL45. The tip of H50 is disordered due to flexibility. (B) In the OXA1L-bound state, a nascent polypeptide occupies the tunnel. The mL45 tail is folded and stabilized by uL24m, uL29m, and rRNA H50. The basic residues participating in the stabilization are shown in the bottom panel. The tip of H50 is ordered, uL23m-α2, and the mL45 loop are disordered, and the path through the tunnel is open. The contact site 3 with OXA1L is formed via mL45-α2. The bottom panels show partial structures of uL23m (residues 37-153), uL24m (93-112), and uL29m (94-157).
Class 1a* represent the OXA1L-bound state. A continuous density connected to the P-tRNA is observed in the tunnel, corresponding to a nascent polypeptide of a mitoribosome engaged in protein synthesis and associated with OXA1L (fig. S11B). Glu70-Lys71 of mL45 adopt a conformation related by ~100°, which results in the 12 Å- and 17 Å-displacement of the upstream Val66 and Ile67, respectively. Therefore, the uL23m-α2 is released and destabilized (fig. S12). The loop from the mL45 Tim44-like domain also becomes disordered by losing its interaction with uL23m-α2. The lack of an internal protein scaffold allows the lower tunnel to accommodate a nascent chain. In the upper tunnel, the mL45 membrane-distal tail adopts a folded α-helix (α0), stabilized by the mitochondria-specific insertion of uL29m, β4-β5 hairpin of uL24m, and H50 of rRNA from the opposite sides (Fig. 4A, fig. S13). Thus, a continuous pathway is formed from the tunnel entrance to the mitoribosomal surface at the membrane-facing region that is available for nascent chain conduction (Fig. 4B, movie S1).
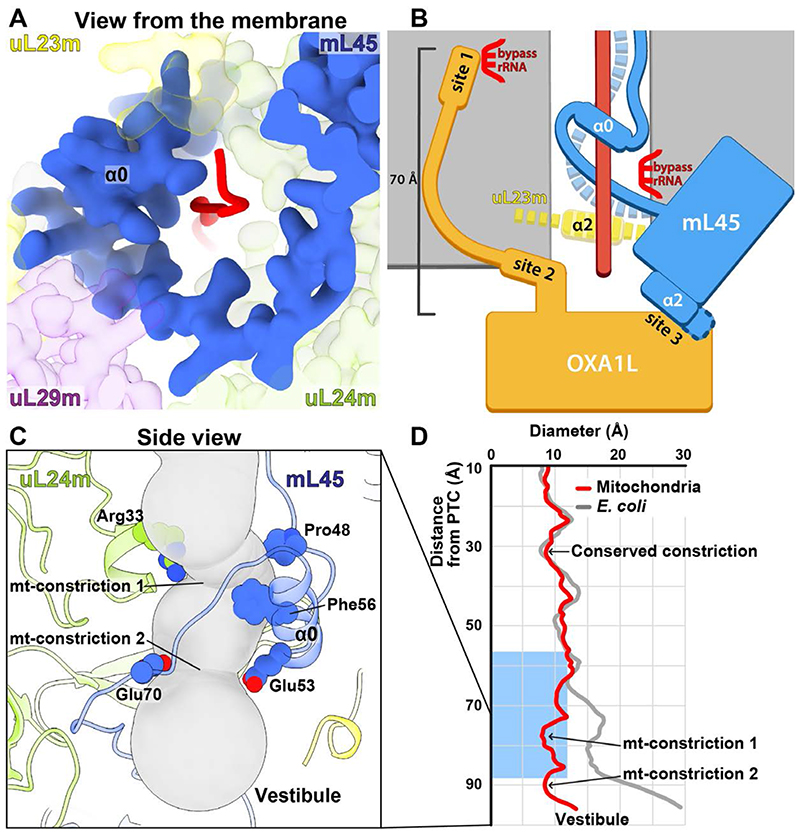
Fig. 4. Mechanism of membrane-tethered nascent polypeptide emergence.
(A) Folding of the mL45 tail stabilized by uL29m and uL24m, resulting in a continuous protein arch coating the interior with the nascent chain. (B) Sideview schematic of the gating mechanism. The folding of mL45-α0 (blue tube) leads to the detachment of its N-terminus from rRNA (dashed blue) and destabilization of uL23m-α2 (dashed yellow) that together open the way for a polypeptide chain (red). OXA1L is docked to the extended mL45-α2 (site 3) for co-translational insertion of the emerging polypeptide. Contact sites 1 and 2 are formed between the OXA1L-CTE and the mitoribosomal surface up to 70 Å away from the membrane surface. (C) Sideview of the lower tunnel, shown as a gray tube. Mitochondria-specific constriction sites are formed by mL45-α0 and uL24m, and the involved residues are shown. (D) The tunnel diameter was calculated for the mitoribosome and E. coli ribosome and plotted along the path. The blue rectangle indicates the position of an α-helical formation within the tunnel in bacteria.
Once folded within the lower tunnel, mL45 forms two mitochondria-specific constriction sites with uL24m (Fig. 4C). The constrictions keep the tunnel width less than 8 Å (Fig. 4D). In contrast, in bacteria, the tunnel increases in diameter to greater than 15 Å, allowing nascent chains to fold in the exit tunnel (29). To assess whether mL45-induced geometry of the exit tunnel determines co-translational protein folding, we employed coarse-grained molecular dynamics simulations with WT and ΔN-mL45 mitoribosomes. Our results show that the observed enclosure of the nascent chain by mL45 decreases helix formation at the constriction sites within the tunnel (fig. S14).
Our data further assigns two additional functions to mL45: 1) directing the nascent polypeptide towards the tunnel exit by compensating for rRNA deletion; 2) positioning the exit at a distance from the membrane surface. The former is achieved by the folded N-terminus of mL45 that supports a specific extended-away conformation of the uL24m β4-β5 hairpin, rather than extruding into the vestibule, as in bacteria (fig. S8D). As a result, the residues at the hairpin tip that were shown to be implicated in bacterial protein folding (30) are 17 Å away, outlining the tunnel walls. This architecture shields an unfolded nascent chain and prevents it from escaping the tunnel prematurely due to rRNA H24 deletion (fig. S8B,D). Then, the positioning of the exit relative to the membrane is mediated by the mL45 Tim44-like domain that allows a gap between the mitoribosomal surface and the insertase, opposed to a closer association as seen in bacteria (fig. S15). This structural arrangement makes the newly synthesized mitochondrial proteins more accessible for peptidyl deformylase, methionine amino peptidase and chaperones prior to the membrane insertion. In yeast, deletion of mL45 (Mba1) is associated with membrane insertion defects (31).
The sequencing of the human mitochondrial genome 40 years ago (32) was a turning point in mitochondrial research, postulating a putative specialized mechanism for the synthesis of these 13 transmembrane proteins. Our discovery of mL45-induced conformational changes represents a gating mechanism of the mitoribosome without similarity in bacterial and cytosolic systems. It forms a cavity secluding unfolded nascent chains and securing their delivery to the membrane. The gating is correlated with the association of the OXA1L insertase, however secondary structure formation is not favored while a nascent chain is at the constriction sites. Together, the data offer a molecular insight into how proteins are synthesized in human mitochondria.
Supplementary Material
One Sentence Summary.
Structural analysis of membrane associated mitoribosome illustrates the mechanism of protein synthesis in human mitochondria.
Acknowledgments
We thank SciLifeLab for access to EM facility (funded by the KAW, EPS, and Kempe foundations), and mass spectrometry facility; Diamond Light Source for access to eBIC (funded by the Wellcome Trust, MRC, and BBSRC) under proposal EM19823-1 with assistance from Y. Chaban; R. Kock Flygaard and N. Fluman for advice.
Funding
Swedish Foundation for Strategic Research (FFL15:0325), Ragnar Söderberg Foundation (M44/16), Cancerfonden (2017/1041), European Research Council (ERC-2018-StG-805230), Knut and Alice Wallenberg Foundation (2018.0080), The Academy of Finland (307431 and 314706 to B.J.B.), NIH-R35 grant (GM118141 to A.B.), MDA Research Grant (MDA-381828 to A.B.). R.B. was supported by the Intramural Research Program of the National Institute of Diabetes and Digestive and Kidney Diseases of the NIH. This work utilized the computational resources of the NIH HPC Biowulf cluster. (http://hpc.nih.gov). Y.I. was supported by H2020-MSCA-IF-2017 (799399-Itohribo). P.M. is the recipient of a postdoctoral fellowship from the American Heart Association (19POST34450174).
Footnotes
Author contributions: B.B., U.R. and A.A. designed the project. J.A., U.R. and A.A. prepared the sample for cryo-EM. J.A. and Y.I. collected and processed the cryo-EM data. Y.I. built the model. A.C., P.M. and A.B. performed biochemical analysis. R.B. performed molecular simulation. A.A. Y.I. and J.A. wrote the manuscript with contributions from all co-authors.
Competing interests: Authors declare no competing interests.
Data and materials availability
The atomic coordinates were deposited in the RCSB Protein Data Bank (PDB) under accession numbers 6ZM5 and 6ZM6. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-11278, EMD-11279, EMD-11280, EMD-11281, EMD-11282, EMD-11283, EMD-11284, EMD-11285, EMD-11286 and EMD-11287.
References and Notes
- 1.De Silva D, Tu Y-T, Amunts A, Fontanesi F, Barrientos A. Mitochondrial ribosome assembly in health and disease. Cell Cycle. 2015;14:2226–2250. doi: 10.1080/15384101.2015.1053672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Suomalainen A, Battersby BJ. Mitochondrial diseases: the contribution of organelle stress responses to pathology. Nat Rev Mol Cell Biol. 2018;19:77–92. doi: 10.1038/nrm.2017.66. [DOI] [PubMed] [Google Scholar]
- 3.Aibara S, Singh V, Modelska A, Amunts A. Structural basis of mitochondrial translation. Elife. 2020;9:1–17. doi: 10.7554/eLife.58362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Englmeier R, Pfeffer S, Förster F. Structure of the Human Mitochondrial Ribosome Studied In Situ by Cryoelectron Tomography. Structure. 2017;25:1574–1581. doi: 10.1016/j.str.2017.07.011. e2. [DOI] [PubMed] [Google Scholar]
- 5.Chen Y, Capponi S, Zhu L, Gellenbeck P, Freites JA, White SH, Dalbey RE. YidC Insertase of Escherichia coli: Water Accessibility and Membrane Shaping. Structure. 2017;25:1403–1414. doi: 10.1016/j.str.2017.07.008. e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.van Bloois E, Jan Haan G, de Gier J-W, Oudega B, Luirink J. F 1 F 0 ATP synthase subunit c is targeted by the SRP to YidC in the E. coli inner membrane. FEBS Lett. 2004;576:97–100. doi: 10.1016/j.febslet.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 7.van der Laan M, Bechtluft P, Kol S, Nouwen N, Driessen AJM. F1F0 ATP synthase subunit c is a substrate of the novel YidC pathway for membrane protein biogenesis. J Cell Biol. 2004;165:213–222. doi: 10.1083/jcb.200402100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildenbeutel M, Theis M, Geier M, Haferkamp I, Neuhaus HE, Herrmann JM, Ott M. The Membrane Insertase Oxa1 Is Required for Efficient Import of Carrier Proteins into Mitochondria. J Mol Biol. 2012;423:590–599. doi: 10.1016/j.jmb.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 9.Jia L. Yeast Oxa1 interacts with mitochondrial ribosomes: the importance of the C-terminal region of Oxa1. EMBO J. 2003;22:6438–6447. doi: 10.1093/emboj/cdg624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szyrach G. Ribosome binding to the Oxa1 complex facilitates co-translational protein insertion in mitochondria. EMBO J. 2003;22:6448–6457. doi: 10.1093/emboj/cdg623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stiburek L, Fornuskova D, Wenchich L, Pejznochova M, Hansikova H, Zeman J. Knockdown of Human Oxa1l Impairs the Biogenesis of F1Fo-ATP Synthase and NADH:Ubiquinone Oxidoreductase. J Mol Biol. 2007;374:506–516. doi: 10.1016/j.jmb.2007.09.044. [DOI] [PubMed] [Google Scholar]
- 12.Thompson K, Mai N, Oláhová M, Scialó F, Formosa LE, Stroud DA, Garrett M, Lax NZ, Robertson FM, Jou C, Nascimento A, et al. OXA 1L mutations cause mitochondrial encephalopathy and a combined oxidative phosphorylation defect. EMBO Mol Med. 2018;10 doi: 10.15252/emmm.201809060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bañó-Polo M, Baeza-Delgado C, Tamborero S, Hazel A, Grau B, Nilsson I, Whitley P, Gumbart JC, von Heijne G, Mingarro I. Transmembrane but not soluble helices fold inside the ribosome tunnel. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07554-7. 5246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter U, Lahtinen T, Marttinen P, Myöhänen M, Greco D, Cannino G, Jacobs HT, Lietzén N, Nyman TA, Battersby BJ. A Mitochondrial Ribosomal and RNA Decay Pathway Blocks Cell Proliferation. Curr Biol. 2013;23:535–541. doi: 10.1016/j.cub.2013.02.019. [DOI] [PubMed] [Google Scholar]
- 15.Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJH, Lindahl E, Scheres SHW. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wickles S, Singharoy A, Andreani J, Seemayer S, Bischoff L, Berninghausen O, Soeding J, Schulten K, van der Sluis EO, Beckmann R. A structural model of the active ribosome-bound membrane protein insertase YidC. Elife. 2014;3 doi: 10.7554/eLife.03035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kedrov A, Wickles S, Crevenna AH, van der Sluis EO, Buschauer R, Berninghausen O, Lamb DC, Beckmann R. Structural Dynamics of the YidC:Ribosome Complex during Membrane Protein Biogenesis. Cell Rep. 2016;17:2943–2954. doi: 10.1016/j.celrep.2016.11.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee RG, Gao J, Siira SJ, Shearwood A-M, Ermer JA, Hofferek V, Mathews JC, Zheng M, Reid GE, Rackham O, Filipovska A. Cardiolipin is required for membrane docking of mitochondrial ribosomes and protein synthesis. J Cell Sci. 2020;133 doi: 10.1242/jcs.240374. jcs240374. [DOI] [PubMed] [Google Scholar]
- 19.Tu Y-T, Barrientos A. The Human Mitochondrial DEAD-Box Protein DDX28 Resides in RNA Granules and Functions in Mitoribosome Assembly. Cell Rep. 2015;10:854–864. doi: 10.1016/j.celrep.2015.01.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kummer E, Leibundgut M, Rackham O, Lee RG, Boehringer D, Filipovska A, Ban N. Unique features of mammalian mitochondrial translation initiation revealed by cryo-EM. Nature. 2018;560:263–267. doi: 10.1038/s41586-018-0373-y. [DOI] [PubMed] [Google Scholar]
- 21.Amunts A, Brown A, Bai X-c, Llacer JL, Hussain T, Emsley P, Long F, Murshudov G, Scheres SHW, Ramakrishnan V. Structure of the Yeast Mitochondrial Large Ribosomal Subunit. Science. 2014;343:1485–1489. doi: 10.1126/science.1249410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Itoh Y, Naschberger A, Mortezaei N, Herrmann JM, Amunts A. Analysis of translating mitoribosome reveals functional characteristics of translation in mitochondria of fungi. Nat Commun. 2020;11 doi: 10.1038/s41467-020-18830-w. 5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tobiasson V, Amunts A. Ciliate mitoribosome illuminates evolutionary steps of mitochondrial translation. Elife. 2020;9 doi: 10.7554/eLife.59264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Greber BJ, Boehringer D, Leibundgut M, Bieri P, Leitner A, Schmitz N, Aebersold R, Ban N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature. 2014;515:283–286. doi: 10.1038/nature13895. [DOI] [PubMed] [Google Scholar]
- 25.Amunts A, Brown A, Toots J, Scheres SHW, Ramakrishnan V. The structure of the human mitochondrial ribosome. Science. 2015;348:95–98. doi: 10.1126/science.aaa1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Greber BJ, Bieri P, Leibundgut M, Leitner A, Aebersold R, Boehringer D, Ban N. The complete structure of the 55S mammalian mitochondrial ribosome. Science. 2015;348:303–308. doi: 10.1126/science.aaa3872. [DOI] [PubMed] [Google Scholar]
- 27.Brown A, Rathore S, Kimanius D, Aibara S, Bai X, Rorbach J, Amunts A, Ramakrishnan V. Structures of the human mitochondrial ribosome in native states of assembly. Nat Struct Mol Biol. 2017;24:866–869. doi: 10.1038/nsmb.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Petrov AS, Wood EC, Bernier CR, Norris AM, Brown A, Amunts A. Structural Patching Fosters Divergence of Mitochondrial Ribosomes. Mol Biol Evol. 2019;36:207–219. doi: 10.1093/molbev/msy221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liutkute M, Samatova E, Rodnina MV. Cotranslational Folding of Proteins on the Ribosome. Biomolecules. 2020;10:97. doi: 10.3390/biom10010097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kudva R, Tian P, Pardo-Avila F, Carroni M, Best RB, Bernstein HD, von Heijne G. The shape of the bacterial ribosome exit tunnel affects cotranslational protein folding. Elife. 2018;7 doi: 10.7554/eLife.36326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bauerschmitt H, Mick DU, Deckers M, Vollmer C, Funes S, Kehrein K, Ott M, Rehling P, Herrmann JM. Ribosome-binding Proteins Mdm38 and Mba1 Display Overlapping Functions for Regulation of Mitochondrial Translation. Mol Biol Cell. 2010;21:1937–1944. doi: 10.1091/mbc.E10-02-0101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, et al. Sequence and organization of the human mitochondrial genome. Nature. 1981;290:457–465. doi: 10.1038/290457a0. [DOI] [PubMed] [Google Scholar]
- 33.Aibara S, Andréll J, Singh V, Amunts A. Rapid Isolation of the Mitoribosome from HEK Cells. J Vis Exp. 2018 doi: 10.3791/57877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75. [PubMed] [Google Scholar]
- 35.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 36.Kim H-J, Barrientos A. MTG1 couples mitoribosome large subunit assembly with intersubunit bridge formation. Nucleic Acids Res. 2018;46:8435–8453. doi: 10.1093/nar/gky672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moggridge S, Sorensen PH, Morin GB, Hughes CS. Extending the Compatibility of the SP3 Paramagnetic Bead Processing Approach for Proteomics. J Proteome Res. 2018;17:1730–1740. doi: 10.1021/acs.jproteome.7b00913. [DOI] [PubMed] [Google Scholar]
- 38.Zheng SQ, Palovcak E, Armache J-P, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tegunov D, Cramer P. Real-time cryo-electron microscopy data preprocessing with Warp. Nat Methods. 2019;16:1146–1152. doi: 10.1038/s41592-019-0580-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Khawaja A, Itoh Y, Remes C, Spåhr H, Yukhnovets O, Höfig H, Amunts A, Rorbach J. Distinct pre-initiation steps in human mitochondrial translation. Nat Commun. 2020;11 doi: 10.1038/s41467-020-16503-2. 2932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Buchan DWA, Jones DT. The PSIPRED Protein Analysis Workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Afonine PV, Poon BK, Read RJ, Sobolev OV, Terwilliger TC, Urzhumtsev A, Adams PD. Real-space refinement in PHENIX for cryo-EM and crystallography. Acta Crystallogr Sect D Struct Biol. 2018;74:531–544. doi: 10.1107/S2059798318006551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity : all-atom structure validation for macromolecular crystallography. Acta Crystallogr Sect D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P, Biedermannova L, et al. CAVER 3.0: A Tool for the Analysis of Transport Pathways in Dynamic Protein Structures. PLoS Comput Biol. 2012;8:e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DeLano WL. The PyMOL Molecular Graphics System, Version 2.3. Hypertens Res. 2014;37:362–387. [Google Scholar]
- 48.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera - A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 49.Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. UCSF ChimeraX: Meeting modern challenges in visualization and analysis. Protein Sci. 2018;27:14–25. doi: 10.1002/pro.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ban N, Beckmann R, Cate JHD, Dinman JD, Dragon F, Ellis SR, Lafontaine DLJ, Lindahl L, Liljas A, Lipton JM, McAlear MA, et al. A new system for naming ribosomal proteins. Curr Opin Struct Biol. 2014;24:165–169. doi: 10.1016/j.sbi.2014.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Troshin PV, Procter JB, Sherstnev A, Barton DL, Madeira F, Barton GJ. JABAWS 2.2 distributed web services for Bioinformatics: protein disorder, conservation and RNA secondary structure. Bioinformatics. 2018;34:1939–1940. doi: 10.1093/bioinformatics/bty045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- 53.Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol. 2011;7:539. doi: 10.1038/msb.2011.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Holmstrom ED, Liu Z, Nettels D, Best RB, Schuler B. Disordered RNA chaperones can enhance nucleic acid folding via local charge screening. Nat Commun. 2019;10 doi: 10.1038/s41467-019-10356-0. 2453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kutzner C, Páll S, Fechner M, Esztermann A, Groot BL, Grubmüller H. More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J Comput Chem. 2019;40:2418–2431. doi: 10.1002/jcc.26011. [DOI] [PubMed] [Google Scholar]
- 56.Flyvbjerg H, Petersen HG. Error estimates on averages of correlated data. J Chem Phys. 1989;91:461–466. [Google Scholar]
- 57.Kumazaki K, Chiba S, Takemoto M, Furukawa A, Nishiyama K, Sugano Y, Mori T, Dohmae N, Hirata K, Nakada-Nakura Y, Maturana AD, et al. Structural basis of Secindependent membrane protein insertion by YidC. Nature. 2014;509:516–520. doi: 10.1038/nature13167. [DOI] [PubMed] [Google Scholar]
- 58.Heller H, Schaefer M, Schulten K. Molecular dynamics simulation of a bilayer of 200 lipids in the gel and in the liquid crystal phase. J Phys Chem. 1993;97:8343–8360. [Google Scholar]
- 59.Zhang J, Pan X, Yan K, Sun S, Gao N, Sui S-F. Mechanisms of ribosome stalling by SecM at multiple elongation steps. Elife. 2015;4 doi: 10.7554/eLife.09684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Noeske J, Wasserman MR, Terry DS, Altman RB, Blanchard SC, Cate JHD. High-resolution structure of the Escherichia coli ribosome. Nat Struct Mol Biol. 2015;22:336–341. doi: 10.1038/nsmb.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mercier E, Rodnina MV. Co-Translational Folding Trajectory of the HemK Helical Domain. Biochemistry. 2018;57:3460–3464. doi: 10.1021/acs.biochem.8b00293. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The atomic coordinates were deposited in the RCSB Protein Data Bank (PDB) under accession numbers 6ZM5 and 6ZM6. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession numbers EMD-11278, EMD-11279, EMD-11280, EMD-11281, EMD-11282, EMD-11283, EMD-11284, EMD-11285, EMD-11286 and EMD-11287.