Abstract
The current paradigm holds that the inhibition of Rho guanosine nucleotide exchange factors (GEFs), the enzymes that stimulate Rho GTPases, can be a valuable therapeutic strategy to treat Rho-dependent tumors. However, formal validation of this idea using in vivo models is still missing. In this context, it is worth remembering that many Rho GEFs can mediate both catalysis-dependent and independent responses, thus raising the possibility that the inhibition of their catalytic activities might not be sufficient per se to block tumorigenic processes. On the other hand, the inhibition of these enzymes can trigger collateral side effects that could preclude the practical implementation of anti-GEF therapies. To address those issues, we have generated mouse models to mimic the effect of the systemic application of an inhibitor for the catalytic activity of the Rho GEF Vav2 at the organismal level. Our results indicate that lowering the catalytic activity of Vav2 below specific thresholds is sufficient to block skin tumor initiation, promotion, and progression. They also reveal that the negative side effects typically induced by the loss of Vav2 can be bypassed depending on the overall level of Vav2 inhibition achieved in vivo. These data underscore the pros and cons of anti-Rho GEF therapies for cancer treatment. They also support the idea that Vav2 could represent a viable drug target.
Introduction
Rho GEFs catalyze the activation step of Rho GTPases, a group of GTP-binding proteins involved in key cancer processes such as proliferation, apoptosis, metastasis, and chemoresistance [1]. Since Rho proteins are seldom mutated in tumors [1], it is widely assumed that the inactivation of the catalytic activity of Rho GEFs could represent a therapeutic avenue to hamper the fitness of Rho GTPase-dependent tumors [1, 2]. This idea is supported by data using loss-of-function approaches on immortalized cancer cell lines and, in much fewer cases, mouse knockout models [1]. However, there are regulatory and functional issues that complicate the full understanding of the therapeutic value of these enzymes. Firstly, most Rho GEFs are multidomain proteins that, in addition to the catalytic Dbl homology (DH) region, can harbor a large variety of catalytic and adaptor domains with additional effector functions [3]. In line with this, it has been shown that a number of Rho GEFs can regulate protumorigenic pathways in cancer cells using GTPase-independent mechanisms [1, 4–8]. To make things even more complex, some of the noncatalytic domains present in these proteins have been associated with tumor suppressor functions [1, 9, 10]. Secondly, the recent discovery of loss-of-function mutations in key GTPases such as RhoA in human tumors indicates that, in some cases, Rho GEFs can play catalysis-dependent tumor suppressor rather than protumorigenic roles [1, 11, 12]. Finally, an issue specially disregarded in the field is the type of negative effects caused by the systemic inactivation of Rho GEFs in healthy tissues that, if dire, might preclude the implementation of Rho GEF-based targeted therapies. Given these lingering questions, it is still unknown whether the inactivation of the catalytic activity of Rho GEFs is sufficient to achieve anti-tumoral effects in vivo and, if that is the case, the level of inhibition of the catalytic activity that has to be achieved to trigger effective therapeutic responses. Likewise, we do not have a clear idea of the side effects that are elicited by the inhibition of the catalytic activity of Rho GEFs in healthy tissues and whether there are therapeutic windows in which such collateral effects can be avoided. All those questions could not be addressed up to now given that the conventional loss-of-function mouse models used in the field have been based on the total depletion of the protein rather than on the catalytic inhibition of the targeted Rho GEFs [1].
Vav2, a member of the Vav subfamily of tyrosine phosphorylation-regulated Rho GEFs [13, 14], represents a good example of the functional complexity of the Rho GEF family. Thus, Vav2 is composed of multiple domains that are involved in regulatory (calponin homology, acidic, SH2, SH3), catalytic (the DH-pleckstrin homology-C1 zinc finger cassette), and adaptor-like (calponin homology, SH3) functions during cell signaling (Fig. 1A) [13, 14]. Recent data using both mouse and 3D organotypic models have revealed that the function of this GEF is essential for in vivo tumorigenic processes such as skin and head and neck cancer [15] (L.F.L.-M. and X.R.B., manuscript submitted). Additional studies performed in cell lines suggest that this GEF is also important for breast cancer and adrenocortical carcinoma [16–18]. However, studies using standard Vav2 knockout mice have revealed that the elimination of Vav2 might trigger a number of physiological dysfunctions. Those include high blood pressure, hypertension-associated comorbidities such as cardiovascular remodeling and renal fibrosis, vascular permeability problems, and glaucoma [19–21]. This information indicates that Vav2 represents a good case scenario to address the pros and cons associated with the systemic inhibition of the catalytic activity of a Rho GEF at the organismal level. To tackle this issue, we have generated cohorts of genetically engineered mice exhibiting varying amounts of Vav2 catalytic activity. With this strategy, we aimed at mimicking the usual conditions found with targeted therapies in which the 100% inactivation of the protein target is hardly achieved. In addition, this strategy enabled us to investigate the effects caused by the systemic inhibition of the catalytic activity of Vav2 in the well-being of the animals. Using this “pharmaco-mimetic” approach, we have found that effective therapeutic impacts can be obtained even under conditions in which some remaining catalytic activity of Vav2 is preserved in cells. Interestingly, we have also unveiled the existence of therapeutic windows in which the fitness of cancer cells, but not of healthy tissues, can be specifically impaired depending on the level of Vav2 catalytic inhibition achieved in vivo.
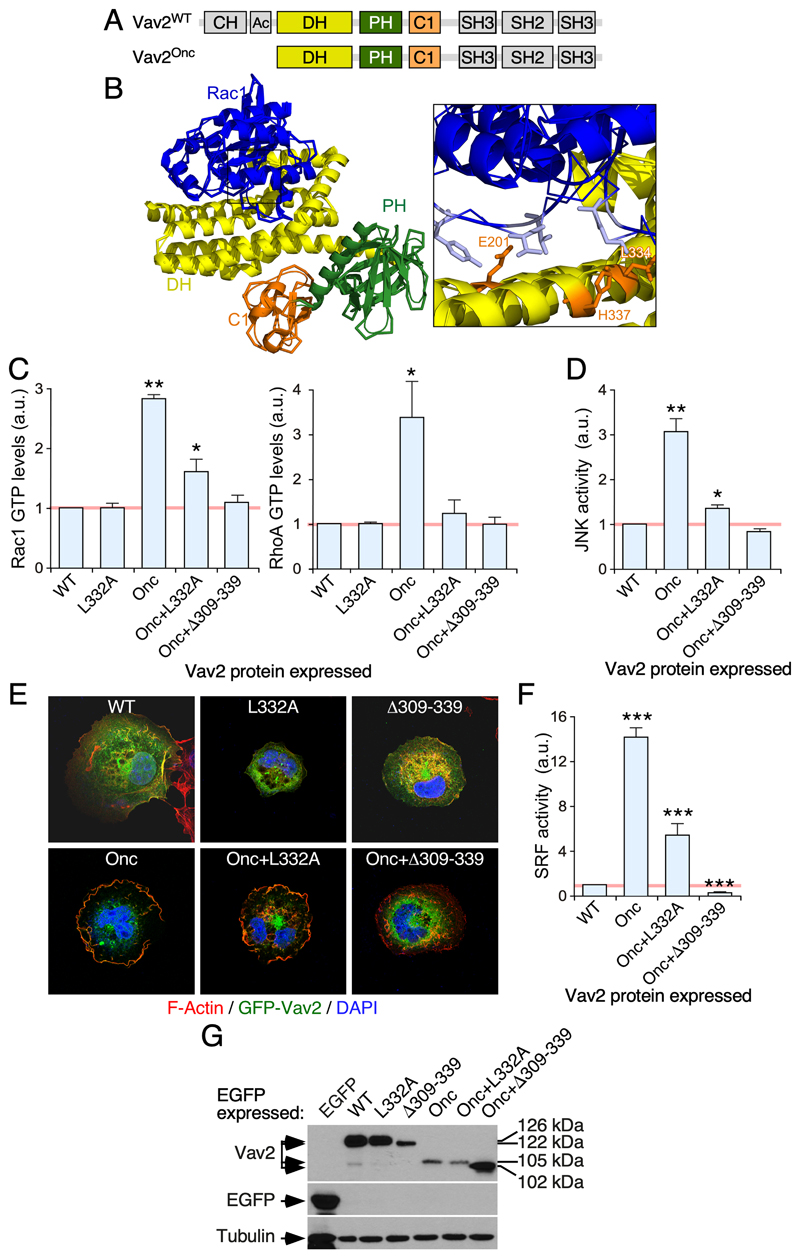
Figure 1. Characterization of the L332A mutation of the Vav2 DH domain.
(A) Schematic representation of the structure of Vav2. Domains forming part of the catalytic cassette are shown in color. The rest of domains are shown in gray. CH, calponin-homology; Ac, acidic; PH, pleckstrin-homology; C1, C1-like zinc finger region; SH, Src-homology.
(B) LEFT, crystal structure of the complex formed by the Vav1 DH-PH-C1 zinc finger catalytic cassette and Rac1 based on previous reports [22, 23]. The color code used of the domains are as in panel A. Rac1 is shown in blue. RIGHT, close up image of the Vav1 DH region containing the Leu334 (orange color) which is analogous to the Leu332 residue that was chosen to impair the catalytic activity of Vav2. Other sites involved in the Vav1-Rac1 interaction are also highlighted in orange color.
(C) Levels of GTP-bound Rac1 (left panel) and RhoA (right panel) in COS1 cells transiently transfected with either EGFP or the indicated EGFP-tagged Vav2 proteins (bottom). Data are represented as fold-change values relative to the level of activity found in EGFP-transfected cells (which was given an arbitrary number of 1, represented as a gray line). The same representation has been used in the panels D and F of this figure. a.u., arbitrary units. *, P < 0.05; **, P < 0.01 (ANOVA and Dunnett’s multiple comparison tests; n = 3, each experiment performed in triplicate).
(D) Levels of JNK activity triggered by the indicated ectopically expressed EGFP-Vav2 versions in COS1 cells (ANOVA and Dunnett’s multiple comparison tests; n = 3, each experiment performed in triplicate).
(E) Example of the membrane ruffling triggered by the indicated ectopic EGFP-Vav2 proteins in COS1 cells (n = 3).
(F) Levels of SRF activity triggered by the indicated ectopic Vav2 proteins in COS1 cells (ANOVA and Dunnett’s multiple comparison tests; n = 3, each experiment performed in triplicate).
(G) Representative immunoblot analyses showing the expression of indicated EGFP-tagged Vav2 versions (top panel) and EGFP (second panel from top) in COS1 cells. The expression of endogenous tubulin α was used as loading control (bottom panel). On the right, we show the molecular weight of the indicated EGFP-tagged proteins. The expected molecular weight of each Vav2 version (without considering the EGFP tag) is: Vav2WT (100 kDa), Vav2Δ309-339 (96 kDa), Vav2Onc (78 kDa), Vav2Onc+Δ309-339 (75 kDa). Similar results were obtained in two other independent experiments.
Results
Identification of a catalytic hypomorphic Vav2 mutant
In order to generate a “pharmaco-mimetic” knock-in Vav2 mouse, we first tried to identify a single point mutation in the catalytic Vav2 DH domain that reduced, but did not abolish, the GEF activity of the protein. On the basis of the known structures of the Tiam1 DH domain and the Vav1 DH-PH-ZF cassette in complex with Rac1 [22–24], we chose the Leu332 residue as a potential site to achieve this effect. This residue is located at a position analogous to the residues of Tiam1 (Leu1194) and Vav1 (Leu334, Fig. 1B) that establish key contacts with the switch II region of Rac1 (residues Gly60, Gln61, Tyr64 and Leu67) [22–24]. In the case of Tiam1, mutation of the Leu1194 residue into alanine severely affects the catalytic activity (≥ 80% reduction) of the protein when tested in biochemical assays. A similar effect is found when the same residue is mutated in the GEF Trio [24, 25]. Using Rho G-LISA assays to investigate the levels of active (GTP-bound) Rho proteins in COS1 cells, we observed that the L332A mutation causes a ≃70% and 100% reduction of the catalytic activity of the oncogenic version of Vav2 (Δ1-186 mutation, referred to hereafter as Vav2Onc; Fig. 1A) towards endogenous Rac1 (Fig. 1C, left panel) and RhoA (Fig. 1C, right panel), respectively. Unlike the case of enhanced green fluorescent protein (EGFP)-tagged wild-type (WT) Vav2, EGFP-Vav2Onc exhibits constitutive and tyrosine phosphorylation-independent exchange activity due to the absence of the autoinhibitory N-terminal calponin-homology and acidic domains. Due to this, the activity of this oncogenic version must reflect the output of Vav2WT under optimal tyrosine phosphorylation conditions according to the current regulatory model for Vav family proteins [13, 14]. The L332A mutation also reduces the ability of Vav2Onc to stimulate the c-Jun N terminal kinase (JNK) (Fig. 1D), a well-known downstream element of the GTPase Rac1 [26–28]. However, the residual catalytic activity of the EGFP-Vav2Onc+L332A protein is sufficient to promote membrane ruffling in COS1 cells (Fig. 1E). As a result, this mutant protein can elicit significant stimulation of the serum responsive factor (Fig. 1F), a transcriptional factor whose activity is regulated by both Rho GTPases and F-actin [29]. As expected, the stimulation of all the foregoing biological readouts is totally abolished when using a version of EGFP-Vav2Onc in which a deleterious internal deletion was created within the catalytic DH domain (Δ309-339 mutation) (Fig. 1C-F). The effect of the L332A and Δ309-339 mutations in the activity of full-length protein could not be tested given that EGFP-Vav2WT has no detectable activity under these conditions (Fig. 1C-F). This is consistent with the lack of activity of the WT versions of Vav family proteins in nonstimulated cells [13, 14, 30]. Immunoblot analyses confirmed that the low activity of EGFP-Vav2Onc+L332A is not due to reduced levels of expression when compared to the EGFP-Vav2Onc protein used as control (Fig. 1G). Taken together, these results indicate that the L332A mutation significantly impairs, but does not abolish, the GEF activity of Vav2 towards Rac1. It does severely impair, however, the ability of Vav2 to stimulate the downstream RhoA GTPase.
Generation of Vav2 L332A knock-in animals and other mouse strain derivatives
We next used standard homologous recombination techniques to replace the exon 11 of the Vav2 locus (which contains the codon for the Leu332 residue) by a mutant one encoding the Ala residue at the same position (Figs. 2A,B and S1). With this approach, we ensured that the mutant Vav2 L332A allele was expressed under the same regulatory sequences and in the very same tissues as the WT counterpart. Consistent with this, we found that the levels of expression of the Vav2L332A protein (Fig. 2C) and Vav2 L332A mRNA (Fig. 2D) in tissues from homozygous Vav2 L332A/L332A mice are similar to those found for their respective WT counterparts in control mice. Further quantitative RT-PCR analyses indicated that the mRNAs encoding the Rac1 GEFs Vav3, Tiam1, P-Rex1, and P-Rex2 show similar expression levels in tissues from Vav2 L332A/L332A and WT mice (Fig. 2E), thus indicating that the missing activity of Vav2 does to lead to the upregulation of other Rac1 GEFs. In line with the COS1 cell-based experiments (Fig. 1), we found using pull-down experiments that the swapping of Vav2WT by the mutant Vav2L332A version leads to a 70-80% reduction in the levels of GTP-bound Rac1 that are typically induced in skeletal muscle upon the in vivo administration of insulin (Fig. 2F). It is important to note that this might not be the case in other signaling contexts due to either lack of functional involvement of Vav2 or to the parallel action of other Rho GEFs with similar enzymatic specificities.
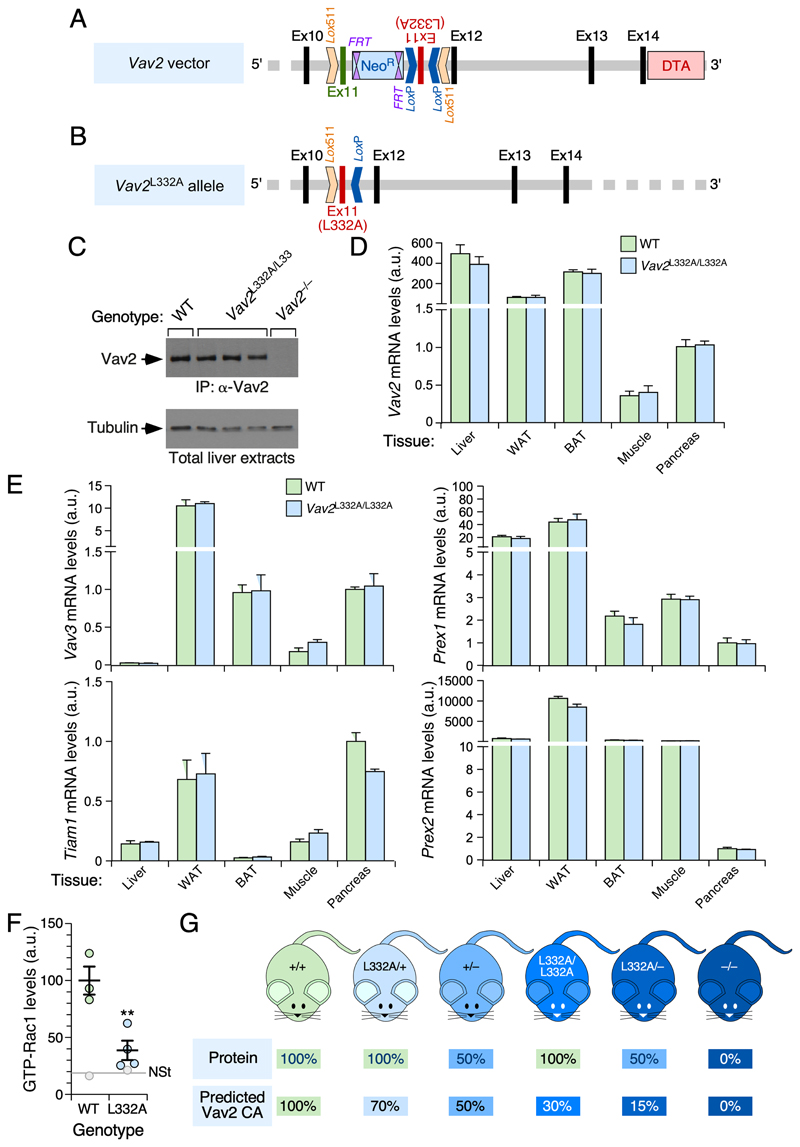
Figure 2. Description and basic characterization of the Vav2 L332A knock-in strain and other mouse models used in this study.
(A) Structure of the targeting vector used in the homologous recombination step in embryonic stem cells. The inverted exon 11 with the mutated codon encoding the L332A residue is shown as a red box. The wild-type exon 11 and other Vav2 gene exons are indicated as green and black boxes, respectively. Intronic sequences are shown as thick gray lanes. Sequences included in the vector to favor positive (Neo R) and negative (Diphtheria toxin A, DTA) selection of embryonic stem cell clones with correctly integrated sequences are indicated as blue and light red boxes, respectively. Lox511 and LoxP recombination sites are also depicted. Ex, exon; FRT, Flippase site for Neo R gene deletion.
(B) Structure of the Vav2L332A allele generated after the Flippase- and Cre-mediated recombination steps described in Figure S1.
(C) Immunoblot analysis showing the expression of Vav2WT and Vav2L332A proteins in liver extracts from the indicated mouse strains (top). The abundance of tubulin α was used as loading control (bottom panel).
(D, E) Relative abundance of the mRNAs for Vav2 (D) and other Rac1 GEFs (E) in the indicated tissues (bottom) from WT and Vav2 L332A/L332A mice. Values were calculated using the ΔΔCt method and normalized to levels found in pancreas (which were given an arbitrary value of 1 in each case). n= 3.
(F) Levels of GTP-bound Rac1 triggered by the in vivo infusion of insulin in skeletal muscle from mice of the indicated genotypes (colored circles). As reference, we show the basal levels of GTP-bound Rac1 found in nonstimulated conditions in WT and Vav2 L332A/L332A mice (gray lane and circles). Data are shown as mean ± SEM. **, P < 0.05 relative to insulin-stimulated WT mice using two-tailed Student’s t-test. n = 3 (WT) and 4 (Vav2L332A/L332A) insulin-infused animals. NSt, nonstimulated.
(G) Predicted levels of Vav2 protein and Vav2 catalytic activity towards Rac1 (based on results from Figure 1C) in mice of the indicated genotypic combinations. CA, catalytic activity.
We next crossed the Vav2 L332A/L332A mice with Vav2–/– and control mice to obtain animals (Vav2 L332A/–, Vav2 L332A/+) with the predicted levels of Vav2 catalytic activity depicted in Figure 2G. It is worth mentioning that this predicted activity only refers to the pool of Rac1 that is directly targeted by Vav2, because the total levels of GTP-bound Rac1 could be much higher in most tissues due to the action of Rho GEFs with Vav2-like patterns of expression (Fig. 2E). Finally, we also generated a compound Vav2 L332A/L332A;Vav3 –/– mouse strain to be used in some of the experiments presented in this work.
Partial impairment of Vav2 GEF activity blocks skin tumorigenesis
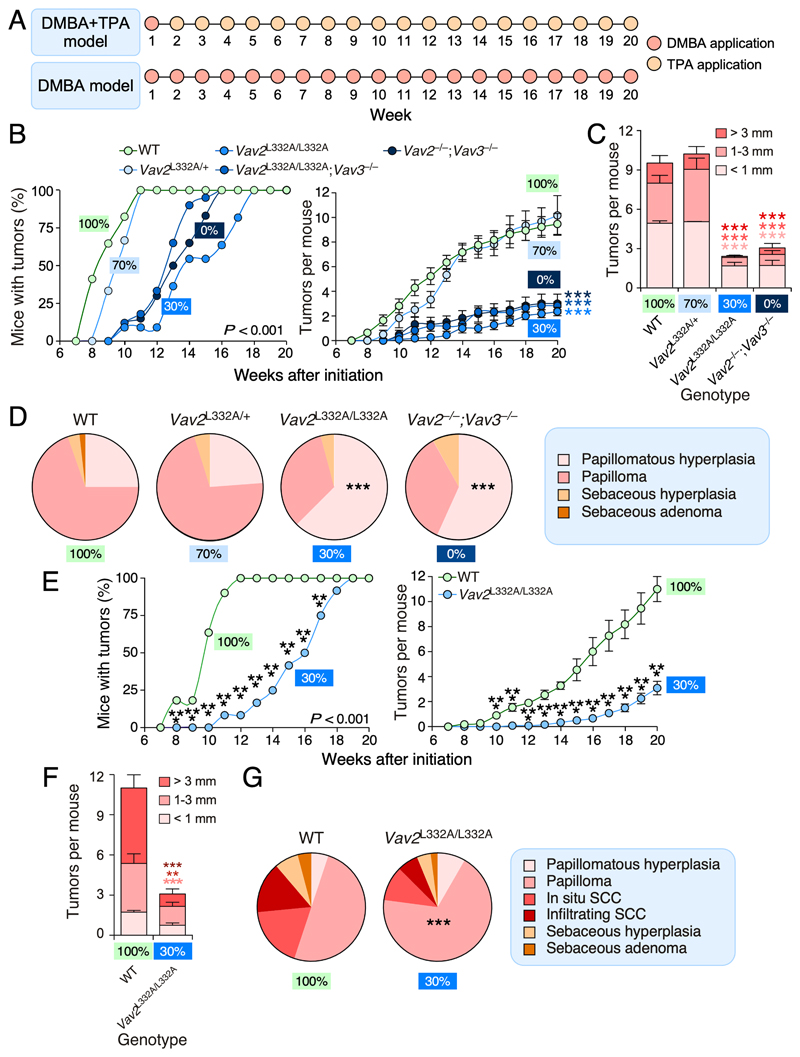
Cohorts of some of the foregoing mouse strains (Fig. 2G) were subjected to standard DMBA+TPA and complete DMBA skin tumorigenesis experiments (Fig. 3A). Previous studies have shown that these two tumorigenic processes are both Vav family- and Rac1-dependent [15, 31]. By contrast, RhoA plays tumor suppressor roles in them [32]. Those studies also demonstrated that such effects are due to intrinsic signaling functions of these proteins in keratinocytes [15, 31] (L.F.L.-M. and X.R.B., manuscript submitted). When tested using the DMBA+TPA skin tumorigenesis protocol (Fig. 3A) [33], we observed that the Vav2L332A/+ mice develop skin tumors with kinetics (Fig. 3B, left panel), multiplicity (Fig. 3B, right panel), size (Fig. 3C), and histopathological features (Fig. 3D) quite similar to those found in WT controls. By contrast, Vav2L332A/L332A and Vav2L332A/L332A;Vav3–/– mice display reduced levels of tumorigenesis that were similar to those found in the Vav2 –/–;Vav3 –/– animals (Fig. 3B-D). We also observed that the Vav2L332A/L332A mice show reduced tumorigenesis when tested using the complete DMBA skin carcinogenesis (Fig. 3A and E), a protocol that allows to monitor cancer progression from the papilloma to the squamous cell carcinoma stage [34]. The few tumors that develop under these experimental conditions in the Vav2 L332A/L332A animals are also smaller (Fig. 3F) and more benign (Fig. 3G) than those found in WT controls. Collectively, these results indicate that the reduced catalytic activity of Vav2L332A affects the initiation, promotion and progression phases of skin tumors as previously seen in the case of Vav2 –/–;Vav3 –/– mice [15].
Figure 3. Partial impairment of Vav2 GEF activity blocks skin tumorigenesis.
(A) Schematic representation of the administration protocol of DBMA and TPA in the case of the DMBA+TPA (top) and DMBA (bottom) carcinogenesis models used in this study.
(B) Penetrance (left graph) and multiplicity (right graph) of tumors in DMBA+TPA-treated mice of the indicated genotypes. On the left panel, statistically significant points include weeks 8 to 14 (in the case of Vav2 L332A/L332A;Vav3 –/– mice), weeks 8 to 15 (in the case of Vav2 –/–;Vav3 –/– mice), and weeks 8 to 17 (in the case of Vav2 L332A/L332A mice) when compared to both WT and Vav2 L332A/+ animals. On the right panel, the differences in tumorigenesis of Vav2L332A/L332A Vav2L332A/L332A;Vav3–/– and Vav2–/–;Vav3–/– mice are statistically significant from weeks 12 to 20 when compared to WT and Vav2L332A/+ animals. ***, P < 0.001 according to ANOVA and Tukey’s HSD tests. n = 17 (WT mice), 6 (Vav2L332A/+ mice), 11 (Vav2L332A/L332A mice), 16 (Vav2L332A/L332A;Vav3–/– mice), 15 (Vav2–/–;Vav3–/– mice).
(C) Size of the tumors generated in WT, Vav2 L332A/+, Vav2 L332A/L332A and Vav2 –/–;Vav3 –/– mice at the endpoint of the experiment shown in panel B. ***, P < 0.001 (Student’s t-test, n size as in panel B). The color of each asterisk has been matched with the corresponding tumor size group.
(D) Type of tumors found in WT, Vav2 L332A/+, Vav2 L332A/L332A, and Vav2 –/–;Vav3 –/– mice at the endpoint of the experiments shown in panel B (left circular plots). The color code for each histological tumor type is explained in the blue box on the right. ***, P < 0.001 (Chi-squared test). n size as in panel B.
(E) Penetrance (left graph) and multiplicity (right graph) of tumors in DMBA-treated adult mice of the indicated genotypes. ***, P < 0.001 (Student’s t-test, n = 11 and 12 in the case of WT and Vav2 L332A/L332A mice, respectively).
(F) Size of the tumors generated in indicated mice at the endpoint of the experiment shown in panel E. **, P < 0.005; ***, P < 0.001 (Student’s t-test, n size as in panel E). The color of each asterisk has been matched with the corresponding tumor size group.
(G) Type of tumors found in indicated mice at the endpoint of the experiments shown in panels E and F (pie chart plots on the left). The color code for each histological tumor type is indicated in the blue box on the right. ***, P < 0.001 (Chi-squared test, n size as in panel E).
In B, C, E and F, values represent the mean ± SEM. In D and G, values represent the relative percentage of samples that belong to the indicated histological subtype. In B to G, the predicted percentage of Vav2 catalytic activity present in each genotype is indicated with colored boxes.
The level of Vav2 inhibition determines the generation of side effects in mice
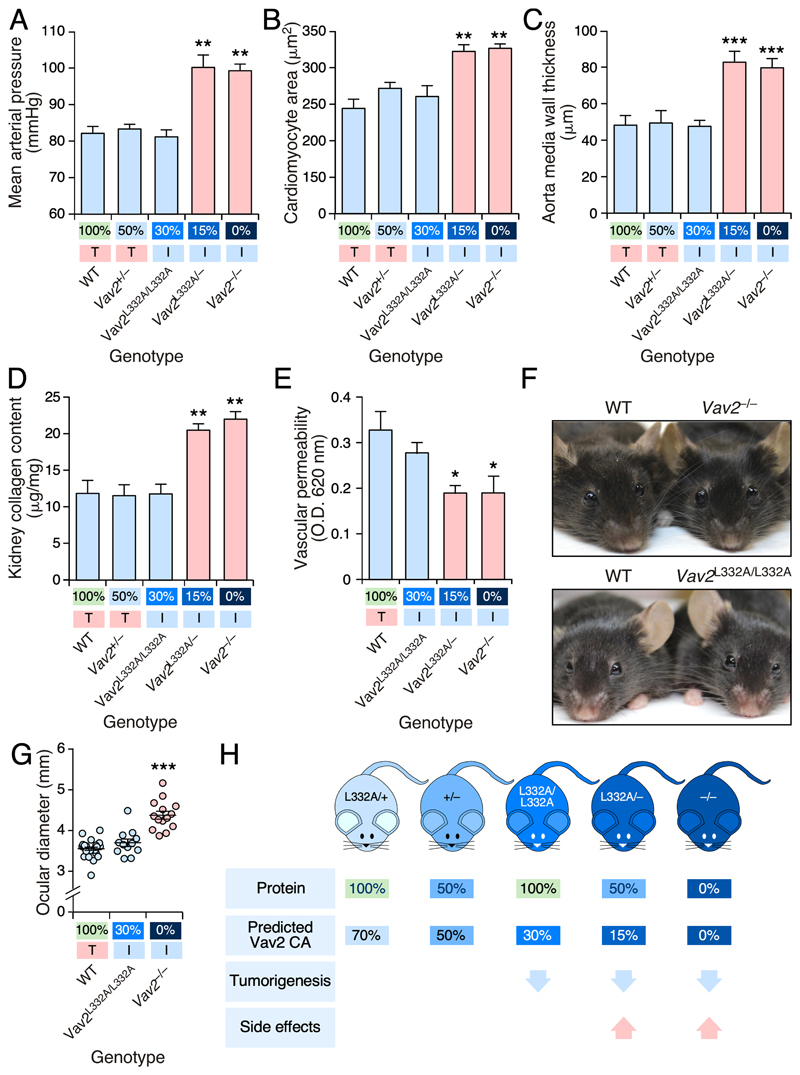
We next analyzed the alterations caused by the inhibition of the catalytic activity of Vav2 in normal tissues. Consistent with previous results [19, 20], we found that Vav2–/– mice develop high blood pressure (Fig. 4A) and several hypertension-associated side effects such as the hypertrophy of the cardiomyocytes located in the left heart ventricle (Fig. 4B), the thickening of the aorta media wall (Fig. 4C), and kidney fibrosis (Fig. 4D). We have previously demonstrated that these defects, which are associated with alterations in the nitric oxide-mediated vasorelaxation responses [20], are caused by defective Rac1 signaling in vascular smooth muscle cells [35]. Unlike the case of knock-out mice, we found that Vav2L332A/L332A animals maintain full cardiovascular and renal homeostasis (Fig. 4A-D). By contrast, the analysis of Vav2L332A/– mice revealed the presence of a hypertensive state and hypertension-associated comorbidities similar to those found in Vav2 –/– mice (Fig. 4A-D). Vav2 +/– mice exhibit normal cardiovascular and renal parameters (Figs. 4A-D), indicating that the defects found in Vav2 L332A/– mice are not due to the reduction in Vav2 protein levels caused by the presence of one copy of the null Vav2 allele. During the characterization of Vav2 –/– mice, we also observed defects in vascular permeability in vivo (S.F. and X.R.B., unpublished data). Although the mechanistic basis of this defect is unknown, evidence from previous publications suggests that it must be Rac subfamily-dependent [36–39]. As in the case of cardiovascular homeostasis, we found that Vav2 L332A/L332A and Vav2 L332A/– animals exhibit vascular permeability responses similar to WT and Vav2 –/– mice, respectively (Fig. 4E).
Figure 4. The level of Vav2 catalytic activity inhibition determines the generation of negative side effects in mice.
(A-D) Mean arterial pressure (A), cardiomyocyte area (B), aorta media wall thickness (C) and kidney collagen content (D) present in mice of the indicated genotypes (A-D, bottom). Normal and pathophysiological values are shown using blue and red bars, respectively. The amount of predicted in vivo Vav2 exchange activity for each genotype is shown at the bottom (the same representation was used in the panels E and G below). In addition, we indicate whether these animals develop normal (T, shaded in red) or impaired (I, shaded in green) skin tumorigenesis according to the results found in Figure 3. **, P < 0.01; ***, P < 0.001 (Student’s t-test, n = 6 per genotype).
(E) Vascular permeability exhibited by mice of the indicated genotypes. Color code of bars and information included in X axis is as in panel A. O.D., optical density. *, P < 0.05 relative to value found in WT mice (Student’s t-test, n = 6 per genotype).
(F and G) Representative example (F) and quantitation (G) of the ocular diameters of mice of the indicated genotypes. ***, P < 0.001 (Student’s t-test, n = 28, 12, and 14 in the case of WT, Vav2 L332A/L332A, and Vav2 –/– mice, respectively).
(H) Summary of the results obtained in this work.
In A-E and G, data represent the mean ± SEM.
To address whether the foregoing observations could be extrapolated outside the vascular system, we investigated the long-term development of glaucoma in mice with impaired Vav2 function [21]. Again, we observed that Vav2 –/– mice, but not the Vav2 L332A/L332A animals, develop buphthalmia (Fig. 4F,G). Collectively, these data indicate that most Vav2-dependent physiological functions can be preserved in mice even when the catalytic activity of the endogenous protein has been significantly reduced (Fig. 4H).
Discussion
A long-standing paradigm in the Rho GTPase field is that the inhibition of the catalytic activity of Rho GEFs must negatively affect both the fitness and malignant properties of Rho-dependent tumors. Up to now, this idea has been demonstrated using standard protein depletion approaches in both cancer cell lines and knockout mice that do not adequately mimic the effects induced by the direct inhibition of the catalytic activity of the interrogated GEF. Another approach has been the administration of chemical inhibitors targeting either the Rho GEF or the GTPase binding interface [1, 40]. However, this experimental avenue has been limited to a reduced number of exchange factors. Furthermore, the results obtained using this approach cannot exclude the possibility that the antitumorigenic effects observed could be caused by the known off-target effects of the inhibitors used [1, 41]. The use of the “pharmaco-mimetic” Vav2 mice has allowed us to demonstrate, for the first time to our knowledge, that targeting the catalytic activity of a Rho GEF is a potential avenue to impair tumorigenic processes in vivo. Perhaps more interestingly, our study has also revealed that effective antitumoral effects can be obtained even under conditions in which significant levels of catalytic activity (30%) of the targeted Rho GEF are maintained (Fig. 4H). We have also found that, depending on the level of catalytic inhibition, these antitumorigenic effects do not have to go along with the generation of negative side effects in healthy tissues (Fig. 4H). In addition to the academic interest, these data offer a proof-of-concept for the potential consideration of Vav2 as a pharmacological target for specific tumor types in the future.
An important issue that remains to be solved in the Rho GTPase field is the lack of functional redundancy of Rho GEFs that are expressed in the same tissues. For example, both the initiation and promotion phases of skin tumors can be indistinctly blocked by inhibiting the catalytic activity of Vav2 (this work) or by depleting the Rac1 GEF Tiam1 [42]. Similar observations have been found with other combinations of Rho GEFs in other tumor types [1, 43, 44]. This suggests that each Rho GEF might target nonoverlapping GTPase pools in an oncogenic insult-, subcellular localization- or cancer stage-specific manner. It is also plausible that Rho GEFs could associate with specific signalosomes that could lead to the engagement of different spectra of signaling pathways and biological responses. Clearly, more work is needed to fully understand the specific role of Rho GEFs that share overlapping expression patterns in tumors. Regardless of the mechanisms involved, these data suggest that the combinatorial administration of Rho GEF-specific drugs could favor a more effective inhibition of specific cancer cell subtypes.
The development of high affinity Rho GEF inhibitors is rather challenging given the shallow nature of the GTPase-binding site of the catalytic DH domain of Rho GEFs. However, one of the most idiosyncratic structural features of the Vav GEF subfamily is the requirement of a DH-PH-C1 zinc finger cassette for full enzymatic activity (Fig. 1A) [13, 14, 22, 23, 45, 46]. This structural feature opens up the possibility of using pockets outside the GTPase binding site for drug development. Other less conventional strategies can be also explored, as exemplified by the development of peptide and RNA aptamers directed against the Rho GEFs Trio and Tiam1, respectively [47, 48]. PROTAC (proteolysis targeting chimera)-mediated degradation avenues represent other possible options to inhibit Vav2 in the near future [49]. According to current evidence, Vav2 inhibitors could be applicable to a variety of tumors such as breast cancer and adrenocortical carcinoma [16–18]. Ongoing data from our lab also indicate that Vav2 plays critical roles in K-Ras-driven non-small cell lung cancer (M.C. and X.R.B, unpublished) and head and neck squamous cell carcinomas (L.F.L.-M. and X.R.B, manuscript submitted), two poor prognosis tumor types that are in acute need of new therapeutics [50]. Although they can be easily eradicated by surgical procedures, cutaneous squamous cell carcinomas also represent a medical challenge when they recur or metastasize [51]. Vav2 becomes deregulated by changes in expression rather than mutations in most of those tumors [16–18] (L.F.L.-M. and X.R.B, manuscript submitted). However, frequent VAV2 mutations have been recently found in hereditary cases of oral squamous cell carcinoma [52].
We have focused on this work on the analysis of known Vav2-dependent tumorigenic and physiological processes. Due to this, we cannot exclude at this moment that other Vav2-dependent physiological responses could exhibit different requirements for the catalytic activity of this GEF. It is also possible that other Vav2-mediated pathobiological processes could utilize catalysis-independent pathways as recently seen in the case of Vav1 and other Rho GEFs [1, 4–8]. The hypomorphic Vav2L332A mouse strain described here will constitute a useful tool to address these potential scenarios in future studies. It will be also worth investigating whether the present results can be extrapolated to other cancer-associated Rho GEFs using analogous mouse genetic models.
Methods
Ethics statement
Animal work was done according to protocols approved by the Bioethics committee of Salamanca University.
Mouse strains
Vav2 –/–, Vav3 –/–, and Vav2 –/–;Vav3 –/– mice have been described elsewhere [19, 53, 54]. The Vav2L332A knock-in mouse strain was generated by GenOway (https://www.genoway.com) using the vector indicated in Figure 2A following standard homologous recombination techniques in embryonic stem cells (Fig. S1). Upon generation of mice from embryonic stem cells, the subsequent steps involved further crosses with transgenic mice to induce the removal of the NeoR cassette (using a Flippase-mediated recombination step) and the swapping of the WT and mutant Vav2 exon 11 (using a Cre-mediated recombination step) (Fig. S1). Vav2 L332A/– and Vav2 L332A/L332A;Vav3 –/– mice were generated using appropriate crosses of Vav2 L332A/L332A animals with the Vav2 –/– and Vav2 –/–;Vav3 –/– strains, respectively. We used male mice in all the experiments.
Cell lines
COS1 cells were obtained from the American Type Culture Collection and grown in DMEM supplemented with 10% fetal calf serum, 1% L-glutamine, penicillin (10 μg/mL) and streptomycin (100 μg/mL). Unless otherwise indicated, all tissue culture reagents used in this work were obtained from Gibco-Thermo Fisher Scientific. Cells were Mycoplasma-free based on periodic determinations using the MycoSEQ™ Mycoplasma detection kit (Thermo Fisher Scientific, Catalog No. 4460626).
Plasmids
The expression plasmid encoding EGFP-Vav2WT (pAA7) has been reported before [55]. The plasmid encoding EGFP-Vav2L332A (pSAF3) was obtained by site-directed mutagenesis of the pAA7 plasmid template using the primers 5’-GGT GCC CAT GCA ACG GGT GGC GAA GTA CCA CCT GCT GCT C-3’ and 5’-GAG CAG CAG GTG GTA CTT CGC CAC CCG TTG CAT GGG CAC C-3’ (the altered nucleotides used to create the L332A mutation are underlined). The plasmid encoding EGFP-Vav2Δ309-339 (pSAF2) was generated by SacI-flanked PCR amplification of the first 1285 bp (amino acids 1-308) of the Vav2 cDNA using the plasmid pAA7 as template and the primers 5’-CCG GGC GAG CTC AAT GGA GCA GTG GCG GCA ATG CGG C-3’ and 5’-CCG GGC GAG CTC TGT GCA CTC CTC CAC TTT CTG-3’ (restriction sites underlined). Upon SacI digestion, the cDNA fragment was cloned into the linearized pNM115 vector containing the Vav2 cDNA sequence from nucleotide 1378 to end (amino acids 340-868) (Fig. S2A). To generate the expression vector encoding EGFP-Vav2Onc (pNM115) (Fig. S2B), the plasmid pKES19 [30] was digested with BstXI, filled-in, and cloned into the SmaI-linearized pEGFP-C2 vector (Clontech-Takara Bio, Catalog No. 632481). The plasmid encoding EGFP-Vav2Onc+L332A (pSAF1) was obtained by site-directed mutagenesis of pNM115 using the primers indicated for pSAF3. The plasmid encoding EGFP-Vav2Onc+Δ309-339 (pSAF4) was generated as indicated for pSAF2 using the plasmid pNM115 as template and the primers 5’-CCG GGC GAG CTC AAT GGG CAT GAC TGA GGA CGA CAA G-3’ and 5’-CCG GGC GAG CTC TGT GCA CTC CTC CAC TTT CTG-3’ (restriction sites underlined) (Fig. S2C). All plasmids used were verified by standard DNA sequencing in our Genomics Facility.
The luciferase plasmids used in promoter activation assays included pFA2-cJun (reporter for JNK activity, Stratagene-Agilent; Catalog No. 219053), pSRE-Luc (reporter for SRF activity; Agilent, Catalog No. 219081), and pRL-SV40 (encoding the Renilla luciferase; Promega, Catalog No. E2231).
Rho GTPase activation assays in COS1 cells
Exponentially growing COS1 cells were transfected using liposomes (Lipofectamine 2000, Thermo Fisher Scientific, Catalog No. 11668019) with 1 μg of the appropriate experimental expression vector. 36 hours post-transfection, the cells were washed with chilled phosphate buffered saline solution and lysed in RIPA buffer [10 mM Tris-HCl (pH 8.0]), 150 mM NaCl, 1% Triton X-100 (Sigma-Aldrich, Catalog No. X100), 1 mM Na3VO4 (Sigma-Aldrich, Catalog No. S6508), 1 mM NaF (Sigma-Aldrich, Catalog No. S7920)], and a mixture of protease inhibitors (CØmplete; Sigma-Aldrich, Catalog No. 11836145001) at 4 °C. Extracts were precleared by centrifugation at 13 200 rpm for 10 min at 4 °C and snap frozen. Upon thawing and determination of total protein concentration (Bradford reactive, Bio-Rad, Catalog No. 5000006), extracts were analyzed using G-LISA assay kits according to the manufacturer’s instructions (Cytoskeleton, Catalog No. BK135). Values are represented as the n-fold change of the experimental sample relative to the activity shown by EGFP-expressing cells (which was given an arbitrary value of 1 in each case).
JNK and SRF activity assays in COS1 cells
For JNK activation assays, exponentially growing COS1 cells were transfected with liposomes (Lipofectamine 2000, Thermo Fisher Scientific, Catalog No. 11668019) with 1, 0.5, and 2 μg of the pFR-Luc, pFA2-cJun, and the appropriate experimental expression vector, respectively. For SRF activation assays, COS1 cells were transfected as above with 1 μg of pSRE-luc, 1 ng of pRL-SV40, and 1 μg of the appropriate experimental vectors. After 24 hours, cells were lysed with Passive Lysis Buffer (Promega, Cat No. E1960) and luciferase activities determined using the Dual Luciferase Assay System (Promega, Cat No. E1960). In all cases, the values of firefly luciferase activity obtained in each experimental point were normalized taking into account the activity of the Renilla luciferase obtained in the same samples. In addition, we analyzed aliquots of the same lysates by Western blot to assess the appropriate expression of the ectopically expressed proteins in each case. Values are represented as the n-fold change of the experimental sample relative to the activity shown by EGFP-expressing cells (which was given an arbitrary value of 1 in each case).
Cytoskeletal change assays in COS1 cells
COS1 cells exponentially growing in 6-well culture plates were transfected with 1 μg of the plasmid of interest with Lipofectamine 2000. After 24 hours, cells were trypsinized and plated onto 12-mm diameter coverslips (Menzel-Glaser, Catalog No. 11778691) pre-treated for 10 min with poly-L-Lysine (1 μg/mL, Sigma-Aldrich, Catalog No. P4707). After 24 hours, the cells were fixed in 4% formaldehyde (Sigma-Aldrich, Catalog No. F8775) in phosphate buffered saline solution for 15 min, washed twice with phosphate buffered saline solution, permeabilized in 25 mM Tris-HCl (pH 8.0) containing 0.5% Triton X-100 for 10 min, washed three times with TBS-T [25 mM Tris-HCl (pH 8.0), 150 mM NaCl, 0.1% Tween-20 (Sigma-Aldrich, Catalog No. P7949)], and blocked for 15 min with 2% bovine serum albumin in TBS-T. After a 20-min long incubation with rhodamine-labeled phalloidin (Molecular Probes-Thermo Fisher Scientific, Catalog No. R415; diluted 1:200 in TBS-T and 2% bovine serum albumin) to visualize the F-actin cytoskeleton, cell preparations were washed thrice with TBS-T, stained with 2-(4-amidinophenyl)-1H -indole-6-carboxamidine (DAPI; Invitrogen-Thermo Fisher Scientific, Catalog No D1306) for 5 min to visualize the nuclei, and mounted onto microscope slides with Mowiol (Calbiochem-Merck, Catalog No 9002-89-5). Samples were analyzed with a Leica TCS SP5 confocal microscope. Images were captured with LAS AF software (version 2.6.0.72266, Leica).
Western blots of total cellular lysates
Transfected COS1 cells were washed with chilled phosphate buffered saline solution and lysed in RIPA buffer at 4 °C. Extracts were precleared by centrifugation at 13 200 rpm for 10 min at 4 °C, denatured by boiling in SDS-PAGE sample buffer, separated electrophoretically, and transferred onto nitrocellulose filters using the iBlot Dry Blotting System (Thermo Fisher Scientific, Catalog No. IB21001). Membranes were blocked in 5% bovine serum albumin (Sigma-Aldrich, Catalog No. A7906) in TBS-T for at least 1 hour and then incubated overnight at 4 °C with the appropriate antibodies to Vav2 (home-made; Lab catalog No. 580-2, 1:1000 dilution), EGFP (1:5000 dilution, Covance, Catalog No. MMS-118P), and tubulin α (1:2000 dilution, Calbiochem-Merck, Catalog No. CP06). After three washes with TBS-T to eliminate the primary antibody, the membranes were incubated with the appropriate secondary antibody (1:5000 dilution, GE Healthcare) for 30 min at room temperature. Immunoreacting bands were developed using a standard chemiluminescent method (Thermo Fisher Scientific, Catalog No. 32106).
Immunoprecipitation of endogenous Vav2 from mouse tissues
Livers from mice of the indicated genotypes were mechanically homogenized in RIPA buffer and CØmplete using a gentleMACS dissociator (Miltenyi Biotec, Catalog No. 130-093-235) and gentleMACS M tubes (Myltenyi Biotec, Catalog No. 130-096-335). Extracts were precleared by centrifugation at 13 200 rpm for 30 min at 4 °C and supernatants quantified using the Bradford reactive (see above). 400 μg of protein extracts in a final volume of 500 μL of RIPA buffer were incubated with 1.5 μL of a homemade antibody to Vav2 for 2 hours at 4 °C in a rotating wheel. Immunocomplexes were collected with 35 μL of Gammabind G-Sepharose beads (GE Healthcare, Catalog No. GE17-0885-01) for 2 hours at 4 °C in a rotating wheel. After three washes with RIPA buffer at 4 °C, the beads were resuspended in SDS-PAGE buffer, boiled for 5 min, and subjected to Western blot analyses as indicated above.
Quantitation of total mRNA levels in tissues
Total RNA was extracted using NZYol (NZYtech, Catalog No. MB18501) and quantitative RT-PCR performed using the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems, Catalog No. 4389986) and the StepOnePlus Real-Time PCR System (Applied BioSystems, Catalog No. 4376600). Raw data were analyzed using the StepOne software (Applied Biosystems). Values were calculated using the ΔΔCt method using the abundance of the endogenous Gapdh mRNA as internal normalization control. In all cases, the data presented in the figures were normalized to the levels found in pancreas, which were given an arbitrary value of 1 in each case. Primers used for transcript quantitation included 5’-AAG CCT GTG TTG ACC TTC CAG-3’ (forward for mouse Vav2), 5’-GTG TAA TCG ATC TCC CGG GAT-3’ (reverse for mouse Vav2), 5’-ATG GAG CCG TGG AAG CAG TG-3’ (forward for mouse Vav3), 5’ -TCC GCC TTC ATC AAG TCT TC-3’ (reverse for mouse Vav3), 5’-AAA GCT CAG GGC GTT CTA CC-3’ (forward for mouse Prex1), 5’ -TAG TAA GGG GCA GGA GGC AT-3’ (reverse for mouse Prex1), 5’-ACA CTG GTT GCC CTG TTT GA-3’ (forward for mouse Prex2), 5’ -CAG CGA TGC GTT TGG ATC TG-3’ (reverse for mouse Prex2), 5’-TGG TTA CAG GAG AGA CTT GGG -3’ (forward for mouse Tiam1), 5’ -GTC CTC CGG GTC TTG TGT G-3’ (reverse for mouse Tiam1), 5’-TGC ACC ACC AAC TGC TTA GC-3’ (forward for mouse Gapdh), and 5’-TCT TCT GGG TGG CAG TGA TG-3’ (reverse for mouse Gapdh).
Infusion of mice with insulin
Mice were fasted for 7 hours, deeply anesthetized with a combination of ketamine (CN #571267.3; Merial) plus xylazine (Bayer, Catalog No. 572126.2), and injected via the inferior vena cava with insulin (0.5 units/kg; Actrapid, Novo Nordisk, Catalog No. 775502). After 5 min, the gastrocnemius muscle was collected and snap frozen until further use.
Determination of GTP-Rac1 levels in skeletal muscle
The frozen gastrocnemius muscle samples were homogenized using a mortar and a pestle and lysed in 1 mL of pulldown buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 5 mM MgCl2, 0.5% Triton X-100, 10 mM β-glycerophosphate, 1 mM DTT and CØmplete). After an incubation of 10 min on ice and a short centrifugation at 4 °C to eliminate cell debris, the supernatants were collected, and protein concentrations quantitated as above. The lysates with equal amount of total protein were incubated with 10 μg of a Glutathione S-transferase (GST) protein fused to the Rac1 binding domain of Pak1 (GST-Pak1 RBD) that was pre-bound to glutathione-Sepharose beads (GE Healthcare, Catalog No. GE17-0756-01) for 2 hours at 4 °C under permanent rotation. The purification of the GST-Pak1 RBD and GST control proteins from E. coli was done as indicated before [55]. After the incubation, the beads were washed three times in pulldown buffer and boiled in SDS-PAGE loading buffer. Released proteins were separated electrophoretically, transferred to nitrocellulose filters, and subjected to immunoblot analyses using antibodies to Rac1 (Cytoskeleton, Catalog No. ARC03). Aliquots of the total cellular lysates used in the above pulldowns were processed in parallel to determine the total amount of Rac1 present in each experimental sample. For quantification, the intensity of the bands was measured with the ImageJ software. Values were normalized taken into consideration the amount of total protein present in each sample.
In vivo carcinogenesis experiments
For the two-step 7,12-dimethylbenz[a]anthracene (DMBA) plus 12-O-tetradecanoylphorbol-13-acetate (TPA) protocol (Fig. 3A, top), the back of 6- to 8-week-old animals of the indicated genotype was shaved and, two days later, carcinogenesis was initiated using a single topic application of DMBA (25 μg diluted in 200 μL of acetone; Sigma-Aldrich, Catalog No. D3254). The promotion phase consisted of biweekly applications of TPA (200 μL of a 1 x 10–4 M solution in acetone; Sigma-Aldrich, Catalog No. P8139) during a 20 week-long period. For complete carcinogenesis (Fig. 3A, bottom), mice were treated biweekly with 5 μg of DMBA alone in 200 μL of acetone for 20 weeks. The number, size (measured with a digital caliper) and incidence of the generated tumors in each protocol was determined weekly.
Pathological analyses of tumors
Tumors from mice of indicated genotypes and treatments were extracted, fixed in 4% paraformaldehyde (Sigma-Aldrich, Catalog No. P6148) and paraffin embedded. Subsequent tissue sectioning (2-3 μm thickness) and staining with hematoxylin-eosin was carried out at the Pathological Unit of our Center. Tumor sections were analyzed blindly by a pathologist (M.C.G.-C.) to classify them according to malignancy grade and level of differentiation.
Blood pressure determination
Blood pressure was recorded in conscious mice using a non-invasive tail cuff method (CODA, Kent Scientific) [54]. To avoid stress-induced changes in those physiological parameters, we subjected mice to similar manipulations during the week previous to the study in order to familiarize them with the experimental procedure.
Histological analyses of heart and aorta
Tissues from animals of indicated genotypes were fixed, cut, and stained as above. Aorta media walls and cardiomyocyte areas were subsequently quantified using the MetaMorph software (Universal Imaging).
Tissue fibrosis determinations
The content of hydroxyproline in kidney was determined with a spectrophotometric method as described [54]. We calculated total collagen assuming that collagen contains a 12.7% of hydroxyproline.
Vascular permeability determinations
Mice of indicated genotypes received topically 32 μL of either TPA (10–4 M in acetone) or vehicle solution alone (acetone) on each ear. A solution of Evans Blue dye (Sigma-Aldrich, Catalog No. E2129) (100 μL of a 1% [weight/volume] solution in phosphate buffered saline) was injected intravenously 30 min later into the lateral tail veins of mice. Upon euthanasia of animals 30 min later, the two ears of each mice were collected and digested for 16 hours in formamide (5 mL/g tissue; Sigma-Aldrich, Catalog No. F9037) with vigorous shaking. After removal of tissue debris by centrifugation, the absorbance of each formamide extract was measured at 620 nm. Acetone-treated ears were used as controls.
Determination of ocular parameters
Buphthalmia was quantified in 6-month-old mice by measuring the diameter of both eyeballs, as indicated before [21].
Statistics
The number of biological replicates (n), the type of statistical tests performed, and the statistical significance are indicated for each experiment in the appropriate figures. Parametric distributions were analyzed using Student’s t-test (when comparing two experimental groups) or ANOVA followed by either Dunnett’s (when comparing more than two experimental groups with a single control group) or Tukey’s HSD test (when comparing more than two experimental groups with every other group). The Chi-squared test was used to determine the significance of the differences between expected and observed frequencies. In all cases, values were considered significant when P ≤ 0.05. Data obtained are given as the mean ± SEM.
Supplementary Material
Acknowledgements
We thank M. Blázquez for lab work and CIC facilities’ personnel for technical assistance. X.R.B. is supported by grants from Worldwide Cancer Research (14-1248), the Castilla-León Government (CSI252P18, CLC-2017-01), the Spanish Ministry of Science and Innovation (MSI) (RTI2018-096481-B-I00), and the Spanish Association against Cancer (GC16173472GARC). X.R.B.’s institution is supported by the Programa de Apoyo a Planes Estratégicos de Investigación de Estructuras de Investigación de Excelencia of the Castilla-León autonomous government (CLC-2017-01). S.F., S.R.-F. and L.F.L.-M. contracts have been supported by funding from the MSI (S.F., BES-2010-031386; S.R.-F., BES-2013-063573), the Spanish Ministry of Universities (L.F.L.-M., FPU13/02923), and the CLC-2017-01 grant (S.R.-F. and L.F.L.-M.). J.R.-V. has been supported by the CIBERONC and, currently, by the Spanish Association against Cancer. Both Spanish and Castilla-León government-associated funding is partially supported by the European Regional Development Fund.
Footnotes
AUTHOR CONTRIBUTIONS
L.F.L.-M. participated in all the experiments shown in Figures 3 and 4B-E, in data analyses, and in artwork design. S.F. carried out the experiments shown in Figures 1 and 4A-E, analyzed the data, and contributed to artwork design. S.R.-F. carried out the experiments shown in Figs. 2C-F and 4F,G, analyzed the data, and contributed to artwork design. A.A. helped in mouse work (crosses, genotyping, carcinogenesis experiments). M.C.G.-M. carried out the histopathological analyses of tumors. M.D. contributed to data analysis and manuscript writing. M.C. and J.R.-V. contributed to the characterization of different phenotypic parameters of Vav2L332A mice. X.R.B. conceived the work, analyzed data, wrote the manuscript, and performed the final editing of figures.
CONFLICTS OF INTEREST
The authors report no conflict of interest associated with this work.
References
- 1.Bustelo XR. RHO GTPases in cancer: known facts, open questions, and therapeutic challenges. Biochem Soc Trans. 2018;46:741–760. doi: 10.1042/BST20170531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–180. doi: 10.1038/nrm1587. [DOI] [PubMed] [Google Scholar]
- 4.Cullis J, Meiri D, Sandi MJ, Radulovich N, Kent OA, Medrano M, et al. The RhoGEF GEF-H1 is required for oncogenic RAS signaling via KSR-1. Cancer Cell. 2014;25:181–195. doi: 10.1016/j.ccr.2014.01.025. [DOI] [PubMed] [Google Scholar]
- 5.Fine B, Hodakoski C, Koujak S, Su T, Saal LH, Maurer M, et al. Activation of the PI3K pathway in cancer through inhibition of PTEN by exchange factor P-REX2a. Science. 2009;325:1261–1265. doi: 10.1126/science.1173569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heidary Arash E, Song KM, Song S, Shiban A, Attisano L. Arhgef7 promotes activation of the Hippo pathway core kinase Lats. EMBO J. 2014;33:2997–3011. doi: 10.15252/embj.201490230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Robles-Valero J, Lorenzo-Martin LF, Fernandez-Pisonero I, Bustelo XR. Rho guanosine nucleotide exchange factors are not such bad guys after all in cancer. Small GTPases. 2018 Jan 24;:1–7. doi: 10.1080/21541248.2018.1423851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lyons LS, Burnstein KL. Vav3, a Rho GTPase guanine nucleotide exchange factor, increases during progression to androgen independence in prostate cancer cells and potentiates androgen receptor transcriptional activity. Mol Endocrinol. 2006;20:1061–1072. doi: 10.1210/me.2005-0346. [DOI] [PubMed] [Google Scholar]
- 9.Khanna N, Fang Y, Yoon MS, Chen J. XPLN is an endogenous inhibitor of mTORC2. Proc Natl Acad Sci USA. 2013;110:15979–15984. doi: 10.1073/pnas.1310434110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Robles-Valero J, Lorenzo-Martin LF, Menacho-Marquez M, Fernandez-Pisonero I, Abad A, Camos M, et al. A paradoxical tumor-suppressor role for the Rac1 exchange factor Vav1 in T cell acute lymphoblastic leukemia. Cancer Cell. 2017;32:608–623 e609. doi: 10.1016/j.ccell.2017.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zandvakili I, Lin Y, Morris JC, Zheng Y. Rho GTPases: Anti- or pro-neoplastic targets? Oncogene. 2017;36:3213–3222. doi: 10.1038/onc.2016.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Svensmark JH, Brakebusch C. Rho GTPases in cancer: friend or foe? Oncogene. 2019;38:7447–7456. doi: 10.1038/s41388-019-0963-7. [DOI] [PubMed] [Google Scholar]
- 13.Rodriguez-Fdez S, Bustelo XR. The Vav GEF family: An evolutionary and functional perspective. Cells. 2019;8 doi: 10.3390/cells8050465. pi: E465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bustelo XR. Vav family exchange factors: an integrated regulatory and functional view. Small GTPases. 2014;5:9. doi: 10.4161/21541248.2014.973757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Menacho-Marquez M, Garcia-Escudero R, Ojeda V, Abad A, Delgado P, Costa C, et al. The Rho exchange factors Vav2 and Vav3 favor skin tumor initiation and promotion by engaging extracellular signaling loops. PLoS Biol. 2013;11 doi: 10.1371/journal.pbio.1001615. e1001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Citterio C, Menacho-Marquez M, Garcia-Escudero R, Larive RM, Barreiro O, Sanchez-Madrid F, et al. The Rho exchange factors Vav2 and Vav3 control a lung metastasis-specific transcriptional program in breast cancer cells. Sci Signal. 2012;5:ra71. doi: 10.1126/scisignal.2002962. [DOI] [PubMed] [Google Scholar]
- 17.Lorenzo-Martin LF, Citterio C, Menacho-Marquez M, Conde J, Larive RM, Rodriguez-Fdez S, et al. Vav proteins maintain epithelial traits in breast cancer cells using miR-200c-dependent and independent mechanisms. Oncogene. 2019;38:209–227. doi: 10.1038/s41388-018-0433-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruggiero C, Doghman-Bouguerra M, Sbiera S, Sbiera I, Parsons M, Ragazzon B, et al. Dosage-dependent regulation of VAV2 expression by steroidogenic factor-1 drives adrenocortical carcinoma cell invasion. Sci Signal. 2017;10 doi: 10.1126/scisignal.aal2464. [DOI] [PubMed] [Google Scholar]
- 19.Sauzeau V, Jerkic M, Lopez-Novoa JM, Bustelo XR. Loss of Vav2 proto-oncogene causes tachycardia and cardiovascular disease in mice. Mol Biol Cell. 2007;18:943–952. doi: 10.1091/mbc.E06-09-0877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sauzeau V, Sevilla MA, Montero MJ, Bustelo XR. The Rho/Rac exchange factor Vav2 controls nitric oxide-dependent responses in mouse vascular smooth muscle cells. J Clin Inv. 2010;120:315–330. doi: 10.1172/JCI38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fujikawa K, Iwata T, Inoue K, Akahori M, Kadotani H, Fukaya M, et al. VAV2 and VAV3 as candidate disease genes for spontaneous glaucoma in mice and humans. PLoS One. 2010;5 doi: 10.1371/journal.pone.0009050. e9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rapley J, Tybulewicz VL, Rittinger K. Crucial structural role for the PH and C1 domains of the Vav1 exchange factor. EMBO Rep. 2008;9:655–661. doi: 10.1038/embor.2008.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chrencik JE, Brooun A, Zhang H, Mathews II, Hura GL, Foster SA, et al. Structural basis of guanine nucleotide exchange mediated by the T-cell essential Vav1. J Mol Biol. 2008;380:828–843. doi: 10.1016/j.jmb.2008.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Worthylake DK, Rossman KL, Sondek J. Crystal structure of Rac1 in complex with the guanine nucleotide exchange region of Tiam1. Nature. 2000;408:682–688. doi: 10.1038/35047014. [DOI] [PubMed] [Google Scholar]
- 25.Liu X, Wang H, Eberstadt M, Schnuchel A, Olejniczak ET, Meadows RP, et al. NMR structure and mutagenesis of the N-terminal Dbl homology domain of the nucleotide exchange factor Trio. Cell. 1998;95:269–277. doi: 10.1016/s0092-8674(00)81757-2. [DOI] [PubMed] [Google Scholar]
- 26.Bustelo XR, Sauzeau V, Berenjeno IM. GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. Bioessays. 2007;29:356–370. doi: 10.1002/bies.20558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Coso OA, Chiariello M, Yu JC, Teramoto H, Crespo P, Xu N, et al. The small GTP-binding proteins Rac1 and Cdc42 regulate the activity of the JNK/SAPK signaling pathway. Cell. 1995;81:1137–1146. doi: 10.1016/s0092-8674(05)80018-2. [DOI] [PubMed] [Google Scholar]
- 28.Crespo P, Bustelo XR, Aaronson DS, Coso OA, Lopez-Barahona M, Barbacid M, et al. Rac-1 dependent stimulation of the JNK/SAPK signaling pathway by Vav. Oncogene. 1996;13:455–460. [PubMed] [Google Scholar]
- 29.Hill CS, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 30.Schuebel KE, Movilla N, Rosa JL, Bustelo XR. Phosphorylation-dependent and constitutive activation of Rho proteins by wild-type and oncogenic Vav-2. EMBO J. 1998;17:6608–6621. doi: 10.1093/emboj/17.22.6608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang Z, Pedersen E, Basse A, Lefever T, Peyrollier K, Kapoor S, et al. Rac1 is crucial for Ras-dependent skin tumor formation by controlling Pak1-Mek-Erk hyperactivation and hyperproliferation in vivo. Oncogene. 2010;29:3362–3373. doi: 10.1038/onc.2010.95. [DOI] [PubMed] [Google Scholar]
- 32.Garcia-Mariscal A, Li H, Pedersen E, Peyrollier K, Ryan KM, Stanley A, et al. Loss of RhoA promotes skin tumor formation and invasion by upregulation of RhoB. Oncogene. 2018;37:847–860. doi: 10.1038/onc.2017.333. [DOI] [PubMed] [Google Scholar]
- 33.Abel EL, Angel JM, Kiguchi K, DiGiovanni J. Multi-stage chemical carcinogenesis in mouse skin: fundamentals and applications. Nat Protoc. 2009;4:1350–1362. doi: 10.1038/nprot.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Luch A. Nature and nurture - lessons from chemical carcinogenesis. Nat Rev Cancer. 2005;5:113–125. doi: 10.1038/nrc1546. [DOI] [PubMed] [Google Scholar]
- 35.Fabbiano S, Menacho-Marquez M, Sevilla MA, Albarran-Juarez J, Zheng Y, Offermanns S, et al. Genetic dissection of the Vav2-Rac1 signaling axis in vascular smooth muscle cells. Mol Cell Biol. 2014;34:4404–4419. doi: 10.1128/MCB.01066-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Amado-Azevedo J, Valent ET, Van Nieuw Amerongen GP. Regulation of the endothelial barrier function: a filum granum of cellular forces, Rho-GTPase signaling and microenvironment. Cell Tissue Res. 2014;355:557–576. doi: 10.1007/s00441-014-1828-6. [DOI] [PubMed] [Google Scholar]
- 37.Garrett TA, Van Buul JD, Burridge K. VEGF-induced Rac1 activation in endothelial cells is regulated by the guanine nucleotide exchange factor Vav2. Exp Cell Res. 2007;313:3285–3297. doi: 10.1016/j.yexcr.2007.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gavard J, Gutkind JS. VEGF controls endothelial-cell permeability by promoting the beta-arrestin-dependent endocytosis of VE-cadherin. Nat cell Biol. 2006;8:1223–1234. doi: 10.1038/ncb1486. [DOI] [PubMed] [Google Scholar]
- 39.Hunter SG, Zhuang G, Brantley-Sieders D, Swat W, Cowan CW, Chen J. Essential role of Vav family guanine nucleotide exchange factors in EphA receptor-mediated angiogenesis. Mol Cell Biol. 2006;26:4830–4842. doi: 10.1128/MCB.02215-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thomas EK, Cancelas JA, Chae HD, Cox AD, Keller PJ, Perrotti D, et al. Rac guanosine triphosphatases represent integrating molecular therapeutic targets for BCR-ABL-induced myeloproliferative disease. Cancer Cell. 2007;12:467–478. doi: 10.1016/j.ccr.2007.10.015. [DOI] [PubMed] [Google Scholar]
- 41.Dutting S, Heidenreich J, Cherpokova D, Amin E, Zhang SC, Ahmadian MR, et al. Critical off-target effects of the widely used Rac1 inhibitors NSC23766 and EHT1864 in mouse platelets. J Thromb Haemost. 2015;13:827–838. doi: 10.1111/jth.12861. [DOI] [PubMed] [Google Scholar]
- 42.Malliri A, van der Kammen RA, Clark K, van der Valk M, Michiels F, Collard JG. Mice deficient in the Rac activator Tiam1 are resistant to Ras-induced skin tumours. Nature. 2002;417:867–871. doi: 10.1038/nature00848. [DOI] [PubMed] [Google Scholar]
- 43.Malliri A, Rygiel TP, van der Kammen RA, Song JY, Engers R, Hurlstone AF, et al. The rac activator Tiam1 is a Wnt-responsive gene that modifies intestinal tumor development. J Biol Chem. 2006;281:543–548. doi: 10.1074/jbc.M507582200. [DOI] [PubMed] [Google Scholar]
- 44.Kawasaki Y, Tsuji S, Muroya K, Furukawa S, Shibata Y, Okuno M, et al. The adenomatous polyposis coli-associated exchange factors Asef and Asef2 are required for adenoma formation in Apc(Min/+)mice. EMBO Rep. 2009;10:1355–1362. doi: 10.1038/embor.2009.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zugaza JL, Lopez-Lago MA, Caloca MJ, Dosil M, Movilla N, Bustelo XR. Structural determinants for the biological activity of Vav proteins. J Biol Chem. 2002;277:45377–45392. doi: 10.1074/jbc.M208039200. [DOI] [PubMed] [Google Scholar]
- 46.Movilla N, Bustelo XR. Biological and regulatory properties of Vav-3, a new member of the Vav family of oncoproteins. Mol Cell Biol. 1999;19:7870–7885. doi: 10.1128/mcb.19.11.7870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bouquier N, Fromont S, Zeeh JC, Auziol C, Larrousse P, Robert B, et al. Aptamer-derived peptides as potent inhibitors of the oncogenic RhoGEF Tgat. Chem Biol. 2009;16:391–400. doi: 10.1016/j.chembiol.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 48.Niebel B, Wosnitza CI, Famulok M. RNA-aptamers that modulate the RhoGEF activity of Tiam1. Bioorg Med Chem. 2013;21:6239–6246. doi: 10.1016/j.bmc.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 49.Verma R, Mohl D, Deshaies RJ. Harnessing the Power of Proteolysis for Targeted Protein Inactivation. Mol Cell. 2020;77:446–460. doi: 10.1016/j.molcel.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 50.Dotto GP, Rustgi AK. Squamous cell cancers: A unified perspective on biology and genetics. Cancer Cell. 2016;29:622–637. doi: 10.1016/j.ccell.2016.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Que SKT, Zwald FO, Schmults CD. Cutaneous squamous cell carcinoma: Incidence, risk factors, diagnosis, and staging. J Am Acad Dermatol. 2018;78:237–247. doi: 10.1016/j.jaad.2017.08.059. [DOI] [PubMed] [Google Scholar]
- 52.Huang Y, Zhao J, Mao G, Lee GS, Zhang J, Bi L, et al. Identification of novel genetic variants predisposing to familial oral squamous cell carcinomas. Cell Discov. 2019;5:57. doi: 10.1038/s41421-019-0126-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Doody GM, Bell SE, Vigorito E, Clayton E, McAdam S, Tooze R, et al. Signal transduction through Vav-2 participates in humoral immune responses and B cell maturation. Nat Immunol. 2001;2:542–547. doi: 10.1038/88748. [DOI] [PubMed] [Google Scholar]
- 54.Sauzeau V, Sevilla MA, Rivas-Elena JV, de Alava E, Montero MJ, Lopez-Novoa JM, et al. Vav3 proto-oncogene deficiency leads to sympathetic hyperactivity and cardiovascular dysfunction. Nat Med. 2006;12:841–845. doi: 10.1038/nm1426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Barreira M, Fabbiano S, Couceiro JR, Torreira E, Martinez-Torrecuadrada JL, Montoya G, et al. The C-terminal SH3 domain contributes to the intramolecular inhibition of Vav family proteins. Sci Signal. 2014;7:ra35. doi: 10.1126/scisignal.2004993. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.