Abstract
The β-barrel assembly machinery (BAM) inserts outer membrane β-barrel proteins (OMPs) in the outer membrane of Gram-negative bacteria. In Enterobacteriacea, BAM also mediates export of the stress sensor lipoprotein RcsF to the cell surface by assembling RcsF-OMP complexes. Here, we report the crystal structure of the key BAM component BamA in complex with RcsF. BamA adopts an inward-open conformation, with the lateral gate to the membrane closed. RcsF is lodged deep inside the lumen of the BamA barrel, binding regions proposed to undergo an outward and lateral opening during OMP insertion. On the basis of our structural and biochemical data, we propose a push-and-pull model for RcsF export upon conformational cycling of BamA and provide a mechanistic explanation for how RcsF uses its interaction with BamA to detect envelope stress. Our data also suggest that the flux of incoming OMP substrates is involved in the control of BAM activity.
Introduction
The vast majority of proteins inserted in the outer membrane of Gram-negative bacteria adopt a β-barrel conformation. Their assembly depends on the activity of the conserved β-barrel assembly machinery (BAM), whose core component is the OMP85-family protein BamA1,2. BamA is an outer membrane 16-stranded β-barrel with a large periplasmic extension consisting of five POlypeptide TRansport-Associated (POTRA) domains at its N-terminus1. Structures of BAM have shown that BamA can adopt two conformations: an outward-open conformation3,4, in which the β-barrel domain opens between strands β1 and β16 to form a lateral gate to the membrane, and an inward-open conformation5,6, in which the lateral gate is sealed while a periplasmic entry pore to the barrel lumen is open. In the bacterium Escherichia coli, four accessory lipoproteins (BamB, BamC, BamD, and BamE) complete BAM, forming a pentameric holocomplex7,8. BamBCDE are anchored to the outer membrane by a lipid moiety but reside in the periplasm. BamB and BamD directly bind the POTRA domains of BamA, while BamC and BamE bind BamD1,2. Although all components are required for efficient assembly of E. coli’s diverse set of OMPs, only BamA and BamD are essential and conserved throughout Gram-negative bacteria1,2. Despite important structural and functional insights during 15 years of intense scrutiny due to the essential activity of BAM in generating and maintaining the outer membrane, crucial questions remain unsolved regarding the mechanism of this molecular machine. In particular, the functional importance of BamA cycling between the outward-open and inward-open conformations remains unclear, as are the respective contributions of the various BAM components to OMP assembly9.
The primary function of BAM is the assembly of OMPs and, when necessary, the translocation of their associated extracellular domains across the outer membrane. More recently, BAM has also been implicated in export of the outer membrane lipoprotein RcsF to the cell surface10,11 via the assembly of complexes between this lipoprotein and three abundant OMPs (OmpA, OmpC, and OmpF)10,11. Support for the involvement of BAM in RcsF export comes from in vivo crosslinking experiments in which a complex between RcsF and BamA, considered to be an intermediate in the formation of RcsF-OmpA/C/F complexes, was trapped10,11. Further, in cells lacking BamB and BamE, RcsF accumulates on BamA and causes a lethal block to BAM-mediated OMP assembly, suggesting that OMPs and surface-exposed RcsF exploit at least partially overlapping assembly routes12,13.
RcsF functions as an envelope stress sensor capable of mounting a protective response when damage occurs in the peptidoglycan or in the outer membrane14,15. Interestingly, we previously determined that sending RcsF to the surface is part of a cellular strategy that enables RcsF to detect damage in the cell envelope. Under stress conditions, newly synthesized RcsF molecules fail to interact with BamA10: they are not exported to the surface and remain exposed to the periplasm, which allows them to trigger the Rcs signaling cascade by reaching the downstream Rcs partner in the inner membrane16. Thus, surface exposure is intimately linked to the function of RcsF. However, the molecular details of the BamA-RcsF interaction, how BAM orchestrates the export of RcsF with OMP assembly, and what prevents RcsF from interacting with BamA under stress conditions remain unknown. Here we sought to address these questions by obtaining structural information about the interaction between BamA and RcsF.
Results
RcsF can be purified with the BAM complex
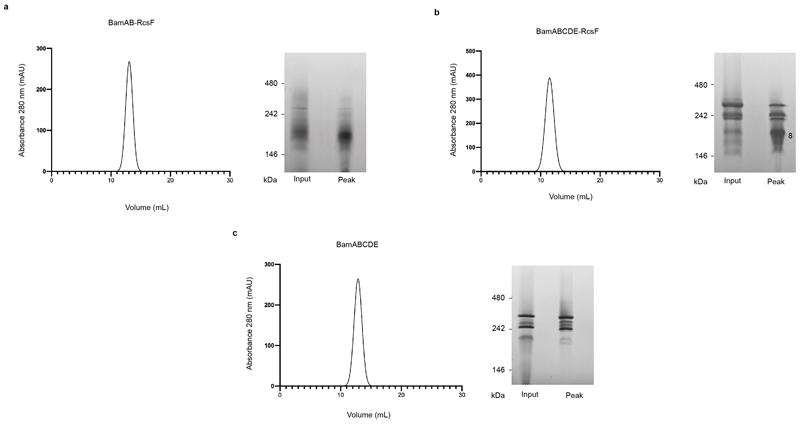
In a series of exploratory experiments, we co-overexpressed RcsF with the BamAB sub-complex, or with the BamABCDE holocomplex; both BamAB-RcsF and BamABCDE-RcsF could be detergent-extracted from the membrane and purified via affinity chromatography using a His-tag on the N-terminus of BamA (Fig. 1a). Using native gel electrophoresis, we confirmed that RcsF binds BamABCDE, and not only BamAB (Fig. 1a, b, c). However, whereas BamAB-RcsF was stable and could be purified to homogeneity by size-exclusion chromatography, BamABCDE-RcsF was unstable (Extended Data Fig. 1a, b). Interesting to note, destabilization of BamABCDE was only observed when RcsF was present (Extended Data Fig. 1c).
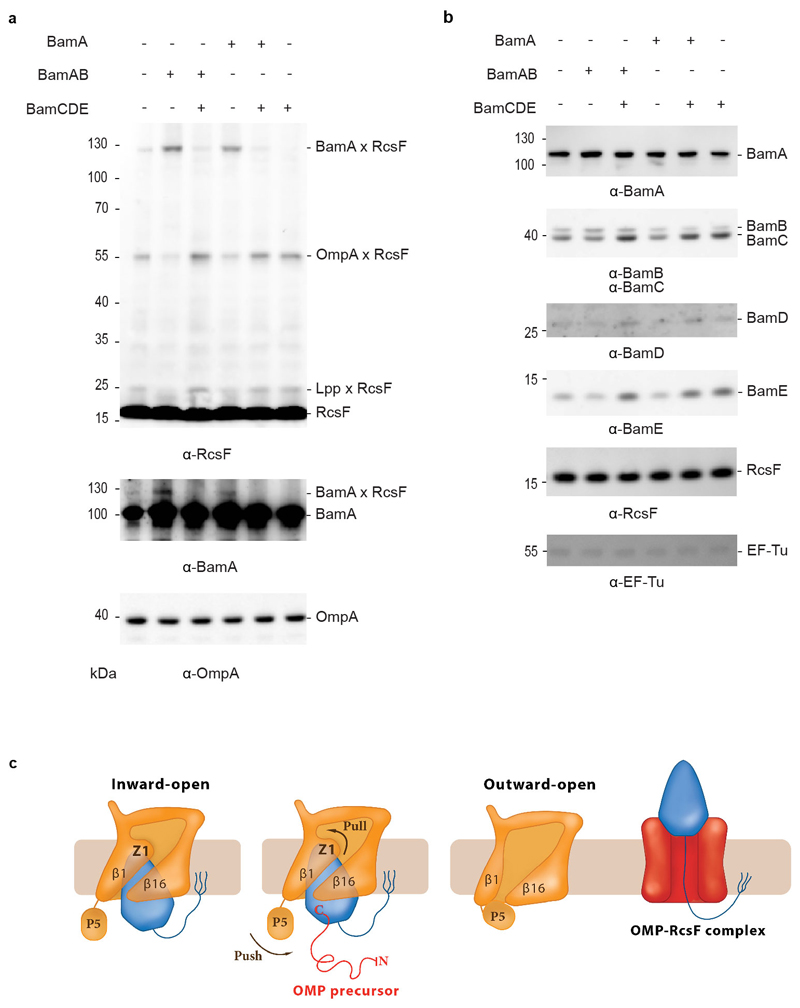
Figure 1. RcsF forms a complex with BamAB and BamABCDE.
(a, b) SDS-PAGE (a) and blue native (b) analysis of purified BAM, BAM-RcsF and BamAB-RcsF complexes obtained via BamA-affinity chromatography. The bands analyzed in (c) are labelled 1 to 8. (c) SDS-PAGE analysis of the complexes shown in panel b (bands 1 to 8). The BAM complex expressed from pRRA1 is a mixture of BamABCDE and BamABDE. n= 4 biologically independent experiments.
BamA is in the inward-open conformation in the structure
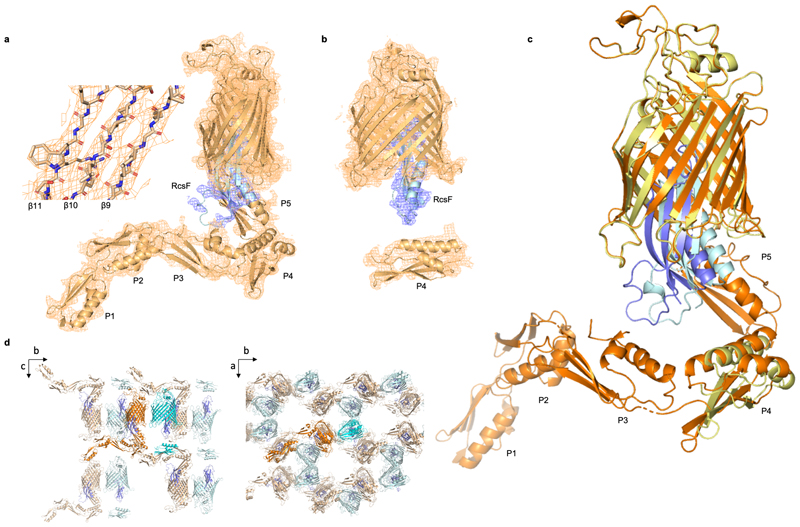
The BamAB-RcsF complex was crystallized and its structure solved to 3.8 Å resolution by molecular replacement using the structures of BamA and RcsF (PDB: 5D0O and 2Y1B, respectively; Supplementary Table 1). While this structure contained BamA and RcsF (Fig. 2), BamB dissociated from the BamA-RcsF complex during crystallization and was absent. The asymmetric unit contained two BamA-RcsF conformers, although for one of them, no unambiguous electron density was observed for POTRA domains 1, 2, 3, and 5 (Extended Data Fig. 2a, b). The β-barrel of BamA was found in an inward-open conformation closely matching that reported in structures of E. coli BamABCDE (6, with a root mean square deviation of 0.9 Å for 383 equivalent Cαatoms in the BamA β-barrel of PDB: 5D0O) or BamA truncates lacking POTRA domains 1-4 or 1-517–20.
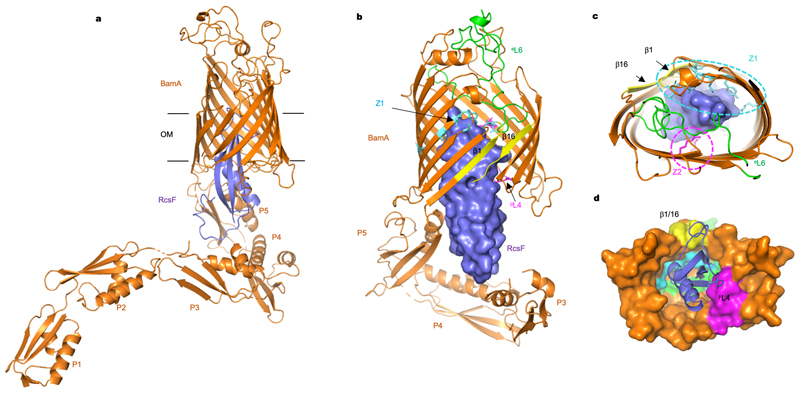
Figure 2. Structure of the BamA-RcsF complex.
(a) Ribbon diagram of the BamA-RcsF complex in side view. BamA, gold; RcsF, blue. (b, c) Front (b) and extracellular (c) views of BamA-RcsF, with RcsF shown as a solvent-accessible surface. POTRA domains 1 and 2 have been omitted for clarity. BamA eL6, green; pL4, magenta. Putative RcsF-interacting residues in contact zones Z1 and Z2 of the BamA β-barrel are colored cyan and magenta, respectively, and shown as sticks. Strands β1 and β16, which form the proposed “lateral gate” of the BamA β-barrel1, are yellow. (d) Periplasmic view of the BamA-RcsF complex, with the BamA β-barrel shown as a solvent-accessible surface and RcsF as a ribbon. Colors are as in panels b and c. POTRA domains were omitted for clarity.
RcsF is located inside the lumen of the BamA β-barrel
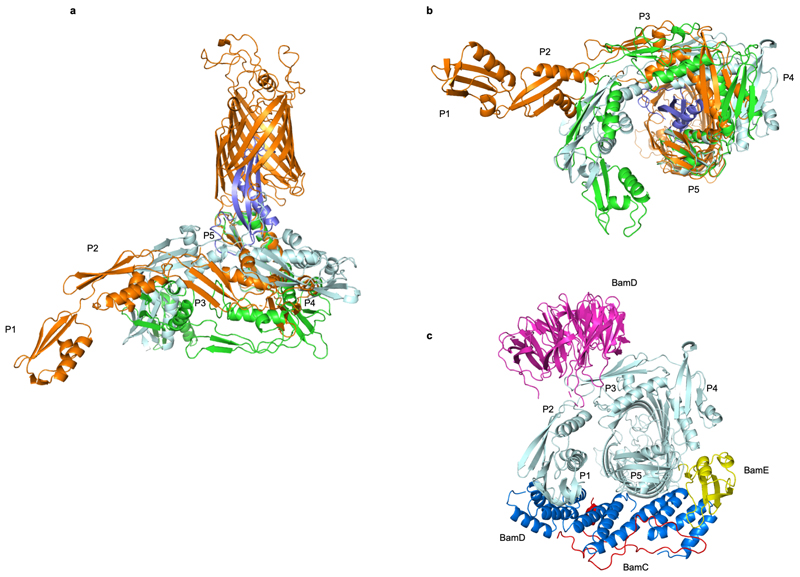
In both BamA copies, RcsF was lodged deep inside the lumen of the BamA β-barrel (Fig. 2a; Extended Data Fig. 2c). RcsF contacts two BamA loops protruding into the β-barrel: (1) extracellular loop L6 (eL6; ~77 Å2 buried surface area, one putative H bond; note that at 3.8 Å resolution, amino-acid sidechain positions cannot be unambiguously determined), and (2) the periplasmic loop connecting strands 7 and 8 (pL4; ~140 Å2 buried surface area, one putative H bond) (Fig. 2, 3a). Although contacting RcsF, these loops retain a conformation closely matching that seen in inward-open BamA structures (Fig. 3b). However, the main BamA-RcsF contact occurs through the luminal wall of the BamA β-barrel, encompassing ~1100 Å2 of buried surface area and comprising up to 15 putative H-bonds (Fig. 2). This RcsF-BamA β-barrel interaction can be divided into three zones. Zone 1 (Z1) consists of perhaps nine H bonds formed by BamA residue 488 and residues 463, 465, and 466 in the loop connecting β3 and β4 and contacting the RcsF loop connecting β1 and β1 (Lβ1-α1) (Fig. 2b, c, Fig. 3a, c). Zone 2 (Z2) is made of perhaps four H bonds formed by BamA residues 592 and 634, located above pL4 (Fig. 2c, 3a). β16, one of the components of the proposed lateral gate of the BamA β-barrel, constitutes the third zone (Fig. 2b, c, 3a). The bottom of RcsF protrudes out of the BamA β-barrel into the periplasm, residing in close proximity to POTRA domains 3-5 (Fig. 2a, b). As a result, RcsF sterically pushes POTRA5 outward, causing a 26° rotation compared to the inward-open conformation found in BamA structures5,6 (Fig. 3b). Although the lipid anchor of RcsF and the N-terminal disordered linker (residues 16-50)21,22 are not apparent in this structure, the position of RcsF is compatible with the lipid anchor residing in the inner leaflet of the outer membrane. Of note, the binding interface between RcsF and BamA does not overlap with the binding sites of BamA for its accessory lipoproteins (Extended Data Fig. 3). Consistent with this observation, the RcsF-BamA interaction is compatible with the binding of BamBCDE, as determined experimentally (Fig. 1; Extended Data Fig. 1).
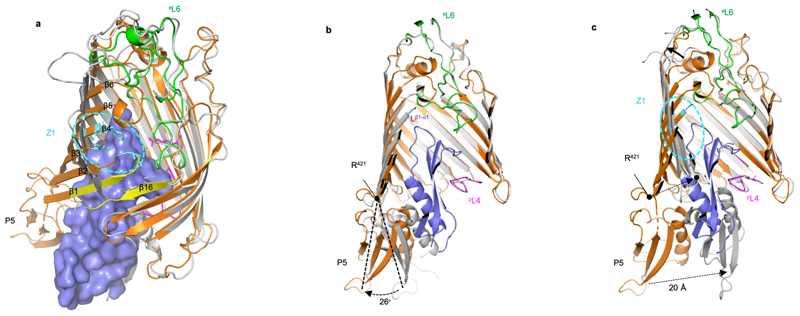
Figure 3. Conformational characteristics of the BamA-RcsF complex.
(a, c) Tilted top view and slabbed side view of the overlay of the BamA-RcsF complex and BamA in the outward-open conformation (grey, taken from BamACDE complex PDB:5EKQ4). The BamA β-barrel undergoes a ~45 outward rotation at strands β1-β6, and a 20 Å inward displacement of POTRA5 compared to the structure of BamA-RcsF presented here. (b) Slabbed side view of the overlay of BamA-RcsF and BamA in the inward-open conformation (grey, taken from BamABCDE complex PDB:5D0O 5). In the structure of BamA-RcsF presented here, POTRA5 makes a 26 outward rotation relative to R421, where it connects to the BamA β-barrel. (a-c) Color scheme for BamA-RcsF is as in Fig. 2b. RcsF is shown as a solvent-accessible surface (a) or a ribbon (b, c). Panels (b, c) show side views, slabbed down to view the interior of the complex. For 5EKQ and 5D0O, the BAM accessory proteins BamB, C, D, and E were omitted for clarity, as were POTRA domains 1-4 in all shown BamA structures.
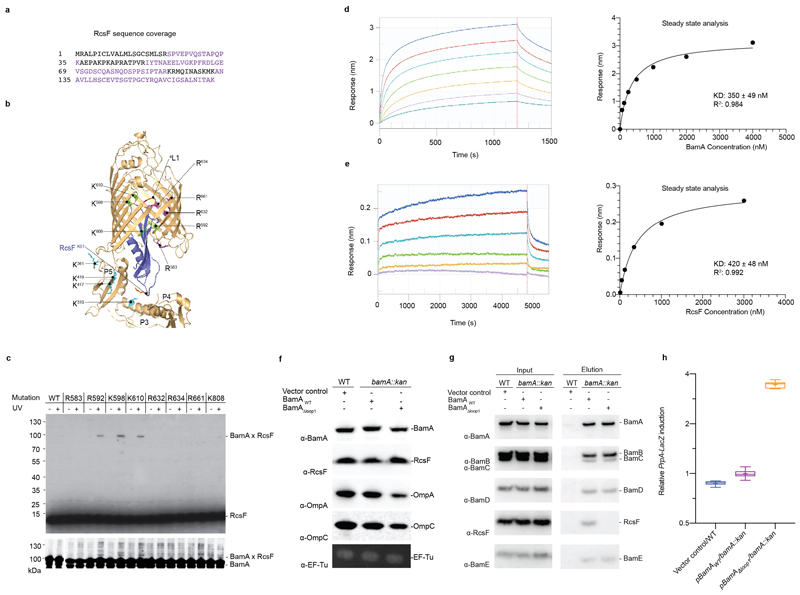
To validate the BamA-RcsF conformation revealed by the X-ray structure, we subjected the complex to crosslinking and analysis via mass spectrometry using the homobifunctional NHS-ester crosslinker disuccinimidyl dibutyric urea23. The sequence coverage of RcsF was about 60% (Extended Data Fig. 4a). Note that one peptide from the N-terminal linker was detected, indicating that the N-terminal disordered region was not degraded during purification. Crosslinks were identified between lysine residues in RcsF (two lysines from the globular domain and one located at the C-terminus of the linker) and those in POTRA4 and POTRA5 (Extended Data Fig. 4b; Supplementary Table 2), providing further support for the architecture of BamA-RcsF determined by crystallography. To confirm that RcsF binds inside the barrel of BamA, we incorporated the photoreactive lysine analog N6-((3-(3-methyl-3H-diazirin-3-yl)propyl)carbamoyl)-L-lysine (DiZPK)24 at multiple positions in the BamA β-barrel domain, selecting residues (R583, R592, K598, K610, R632, R634, R661, K808) whose sidechains face the lumen of the barrel (Extended Data Fig. 4b). After exposure to ultraviolet light, RcsF efficiently crosslinked to BamA when DiZPK was incorporated at three of the selected residues (R592, R598, K610) and to a lower extent at residue K808 (Extended Data Fig. 4b, c), confirming that RcsF binds deep inside the barrel. We measured an equilibrium dissociation constant of 350±49 or 420±48 nM, respectively, depending on whether BamA or RcsF was immobilized (Extended Data Fig. 4d, e). Finally, we deleted loop 1, a short, non-essential25 segment located between the first and second β-strands of the barrel (residues 434 to 437; BamAΔloop1) (Fig. 3a; Extended Data Fig. 4b). BamAΔloop1 is functional 5 and able to rescue the lethality of a ΔbamA deletion mutant, despite the fact that the levels of major OMPs are slightly decreased in cells expressing BamAΔloop1 (Extended Data Fig. 4f). We hypothesized that deleting this loop would destabilize the BamA-RcsF interaction because of the close proximity of loop 1 to the lateral gate area and to loop 6, two regions of interaction between BamA and RcsF. As predicted, RcsF could be pulled down with BamA but not with BamAΔloop1 (Extended Data Fig. 4g). Further, the Rcs signaling cascade, which is turned on when RcsF fails to interact with BamA10, was constitutively induced in ΔbamA cells complemented with BamAΔloop1 (Extended Data Fig. 4h). In sum, these results provide functional evidence for our structure of BamA-RcsF and confirm the presence of RcsF inside the barrel of BamA.
RcsF does not bind BamA when the lateral gate is open
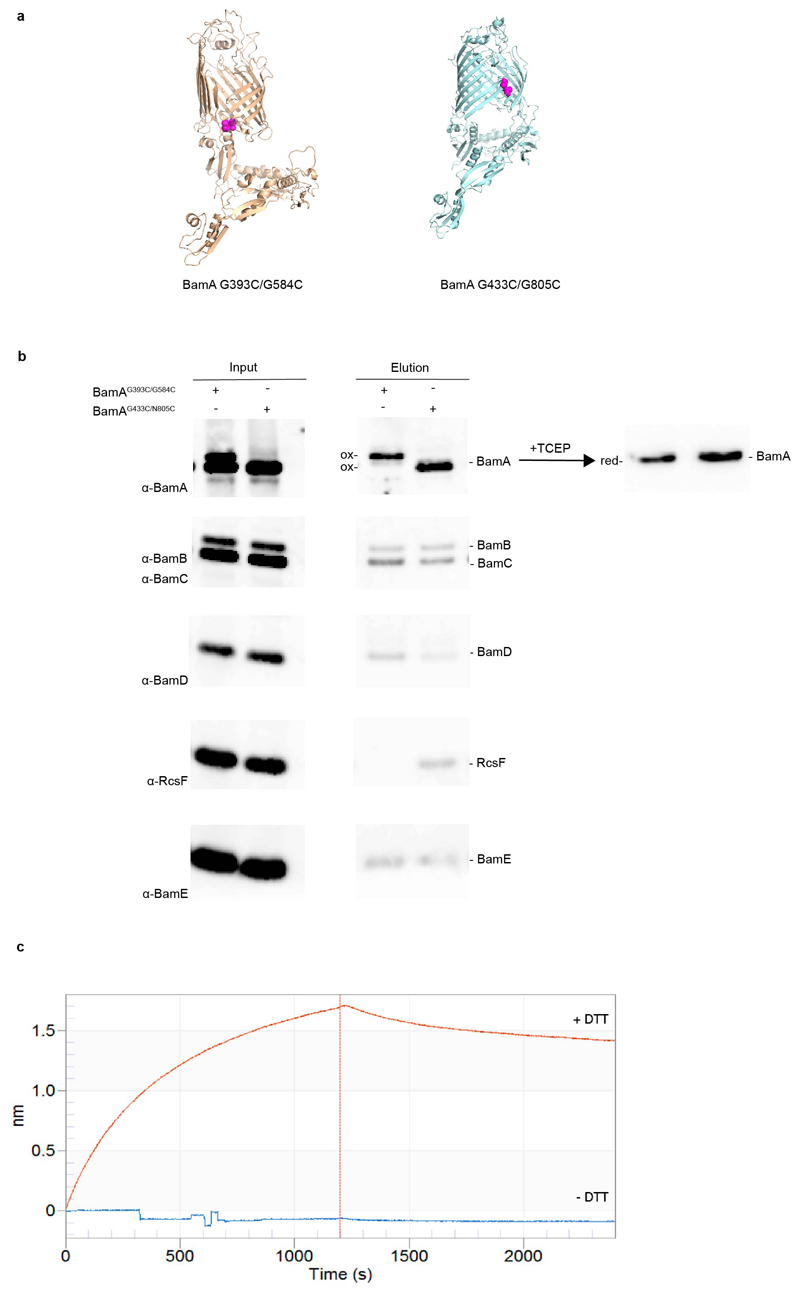
Strikingly, our structure suggests that RcsF binding is incompatible with the BamA β-barrel residing in the outward-open conformation (Fig. 3a). Confirming this hypothesis, RcsF was found to bind BamAG433C/N805C, a mutant in which opening of the lateral gate is prevented by a disulfide bond between β1 and β1626, but not to BamAG393C/G584C, which is locked in the outward-open conformation 5 (Extended Data Fig. 5a, b). However, when reduced, BamAG393C/G584C returned to the inward-open conformation and regained the ability to bind RcsF (Extended Data Fig. 5c). Importantly, given its ability to only bind the inward-open conformation of BamA, the BamA-RcsF complex serves as a proxy for this state. Interestingly, RcsF was recently reported to accumulate on BamA and to jam OMP assembly in the absence of BamB and BamE12,13. Thus, in light of our structural findings, BamA conformational cycling is likely impaired when BamB and BamE are absent. However, binding of these accessory lipoproteins cannot be sufficient to trigger conformational changes in BamA. The structure of BamABCDE has been solved not only in the outward-open conformation but also in the inward-open conformation3–6, despite the presence of all accessory lipoproteins. In addition, we have shown here that RcsF could be co-purified with BamABCDE (Fig. 1; Extended Data Fig. 1), implying that in this purified complex, BamA was in the inward-open conformation. Thus, BamA can remain in the inward-open conformation even when BamBCDE are present, strongly supporting the notion that BAM conformational cycling is triggered by an external signal.
OMP substrates trigger conformational cycling in BamA
What is this trigger? Insights came from in vivo crosslinking experiments carried out in cells in which the expression levels of the BAM components were only moderately increased (~2-fold) compared with wild-type levels. Whereas RcsF can be crosslinked to BamA when the BamA and BamB subunits are slightly over-expressed, the BamA-RcsF complex becomes barely detectable when the moderate over-expression of all BAM components is induced (Fig. 4 a, b). We explain this as follows: the BamAB subcomplex is not functional and does not funnel RcsF to its OMP partners. As a result, BamA-RcsF accumulates and OmpA-RcsF does not form (Fig. 4a). However, if all BAM components (BamABCDE) are moderately overexpressed, BAM activity is restored, RcsF only transiently interacts with BamA, and formation of OmpA-RcsF resumes (Fig. 4a; note that in these experiments, BAM is still expressed at physiological levels from the chromosome—by using plasmids, we manipulate the stoichiometry of the BAM components (Fig. 4b)). Therefore, whether a stable RcsF complex forms with BAM depends critically on the rates of OMP synthesis and delivery to BAM, as well as the ratio of active BAM complexes to the concentrations of OMP and RcsF substrates. When we purified the BamABCDE-RcsF complex (Fig. 1; Extended Data Fig. 1), all BAM components were highly over-expressed, which increased the ratio of active complexes to incoming OMP substrates and allowed accumulation of RcsF on BAM. Thus, our data support a model in which it is the flux of incoming OMP substrates that triggers conformational changes in the BamA barrel and release of RcsF to its OMP partners (see below; Fig. 4c). Although complexes have to date only been observed between RcsF and OmpA/C/F10,11, complexes may form between RcsF and other, less abundant OMPs, depending on the unfolded OMP that is delivered to the BamABCDE-RcsF complex.
Figure 4. BamA-RcsF is a proxy for the inward-open conformation of BamA.
(a) In vivo chemical crosslinking of RcsF with BamA and OmpA. The BamA-RcsF complex accumulates when either BamA alone or BamAB together are moderately over-expressed from a plasmid in cells also expressing BAM at physiological levels from the chromosome. The copies of BamA and BamAB in excess are not functional (BamCDE is required for BAM activity) and do not funnel RcsF to its OMP partners. As a result, RcsF accumulates on BamA and OmpA-RcsF does not form. Over-expression of BamCDE (also from a plasmid) in these cells restores the stoichiometry between the BAM components: BamA-RcsF does not accumulate and the formation of OmpA-RcsF is restored. As shown previously10, levels of OmpA-RcsF are inversely correlated with BamA-RcsF. Overexpression of the BamCDE sub-complex alone does not impact the activity of the BAM complex expressed from the chromosome: wild-type BamA-RcsF and OmpA-RcsF levels are observed. Wild-type BamA-RcsF and OmpA-RcsF levels are also detected when BamA and BamCDE are overexpressed together, as expected given that BamB is not essential. RcsF also forms a complex with the abundant lipoprotein Lpp (Lpp-RcsF), as in10. Protein expression levels of OmpA were analyzed by immunoblot in the non-crosslinked samples, showing no differences. The additional bands that are detected in the lanes where BamA-RcsF is not observed likely correspond to poorly abundant complexes between RcsF and unknown proteins. n= 3 biologically independent experiments. (b) Protein expression levels of BamB, BamC, BamD, BamE and RcsF from no-crosslinked samples overexpressing BamA (pBamA), BamAB (pBamA-B) and BamCDE (pSC263) were analyzed by western blot. EF-Tu expression levels were analyzed as loading control. n= 3 biologically independent experiments. (c) Model proposing that BamA conformational cycling is triggered by incoming OMP substrates on the BAM holocomplex. A BamA inward-to-outward open transition could result in an upward displacement of RcsF via a push-and-pull mechanism, resulting in an OMP-RcsF complex. The push-and-pull mechanism involves BamA POTRA5 (P5) and Z1. The topology of the OMP-RcsF complex remains to be established. For clarity, POTRA1-4 and the BAM lipoproteins have been ommitted.
RcsF is transferred to its partners during BamA cycling
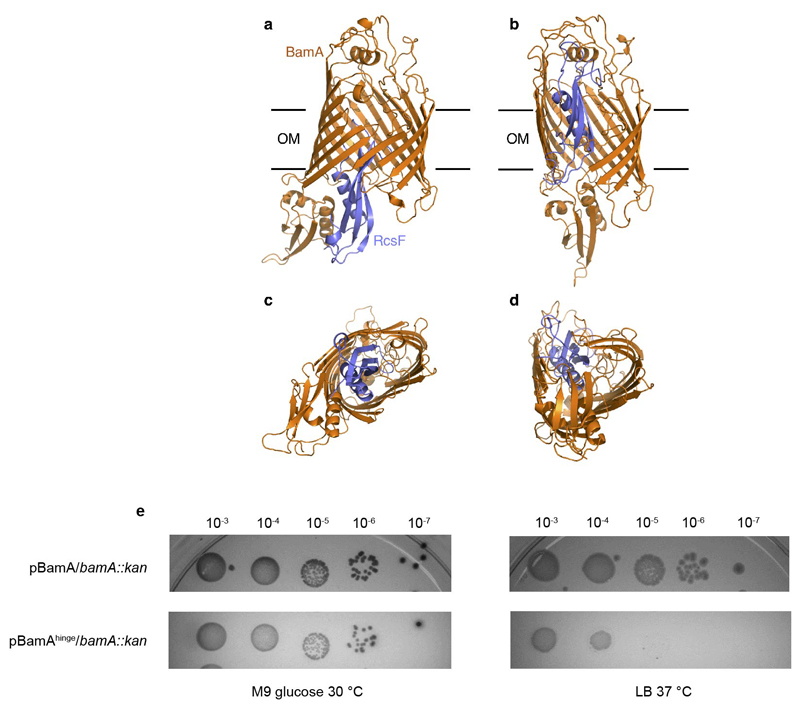
RcsF is not an integral component of BAM; it can bind BamA with high affinity, but it is eventually funneled to OMPs and displayed on the cell surface10,11. It has been proposed that RcsF crosses the outer membrane by being threaded through the lumens of OMPs11. In one possible scenario, RcsF could be transferred from BamA to its OMP partner following opening of the lateral gate and formation of a hybrid barrel (or another non-covalent complex9 between BamA and the nascent OMP), which then buds away from BamA, taking RcsF with it. Available structures show that the transition from the inward-open to the outward-open conformation of BamA encompasses a large outward rotation of strands 1-6 of the BamA β-barrel, as well as a 20 Å inward displacement of POTRA 5 (Fig. 3a, b, c)5,6. Strikingly, BamA strands 1-6 coincide with the main RcsF-BamA interaction zone (Z1) seen in our structure, such that outward rotation of Z1 may exert a pulling force on the tip of RcsF (Fig. 3a, c). Concomitantly, the inward movement of POTRA5 would exert a pushing force on the bottom of RcsF (Fig. 3c). We therefore hypothesize that during the inward-to-outward transition of BamA, this push-and-pull action on RcsF could play a role in the transfer of RcsF to its OMP partners and its translocation to the cell surface (Fig. 4c). Supporting this, dynamic importance sampling simulations in which BamA transitions from the inward-open conformation (as in our structure) to the outward-open conformation (PDB code 5D0Q) show POTRA5 moving towards the periplasmic exit of the lumen, pushing RcsF upwards. The movement of POTRA5 is accompanied later on by the movement of Z1, opening the outward-facing extremity (Extended Data Fig. 6a, b, c, d and Supplementary Video). Furthermore, by introducing a short peptide in the hinge between POTRA5 and the β-barrel domain of BamA (BamAhinge), we could confirm the functional importance of this region for BamA activity. We found indeed that expression of BamAhinge from a plasmid rescues the growth of ΔbamA cells in minimal media at 30°C but not in rich media at 37°C, indicating that BamAhinge is unable to cope with the rate of OMPs folding in these latter conditions (Extended Data Fig. 6e).
Discussion
Our work reveals how BamA interacts with RcsF, providing insights into the mechanism used by BAM to assemble RcsF-OMP complexes, a novel activity by which BAM exports this lipoprotein to the cell surface. It would be surprising if an essential machinery such as BAM—with a global role in formation of the cell envelope—was only dedicated to export RcsF to the surface. Hence, it is tempting to speculate that other lipoproteins may follow the same route and decorate the surface of E. coli, in contrast to the general view that outer membrane lipoproteins face the periplasm27.
By showing that the globular domain of RcsF is lodged deep inside the barrel of BamA, our structure also reveals the remarkable—and unanticipated —finding that the BamA β-barrel can accommodate a lipoprotein “substrate” with a globular domain 12 kDa in size. This finding further establishes BAM as an essential hub that contributes to outer membrane biogenesis by interacting both with nascent OMPs for assembly and lipoproteins for export. Future work will reveal whether other lipoproteins bind BamA in a way similar to RcsF and also clarify the topology of the RcsF-OMP complexes. It has indeed been proposed that the lipid moiety of RcsF is anchored in the outer leaflet of the membrane and that the N-terminal disordered linker is exposed on the cell surface before being threaded through the lumens of the OMPs11. In this model, the globular domain of RscF resides inside the periplasm. Although we cannot rule out that RcsF flips during release from BamA and transfer to its OMP partners, our structure is more consistent with the hypothesis that BamA releases the globular domain of RcsF on the cell surface. Further investigation will therefore be needed to answer this question, and whether the location of RcsF depends on the identity of its OMP partner.
It is also remarkable that RcsF binds the lateral gate area and the outward rotating region of the BamA barrel, sites that sense BAM conformational cycling triggered by incoming OMP substrates. We previously reported that RcsF uses its interaction with BamA to detect stress in the cell envelope: when damage occurs in the peptidoglycan or the outer membrane, newly synthesized RcsF molecules fail to interact with BamA, activating the Rcs stress response10. Our structure provides a possible explanation for this scenario by suggesting that BamA preferentially adopts the outward-open conformation when envelope integrity is impaired, which would de facto prevent RcsF binding and promote Rcs activation. Thus, we propose that cells could monitor envelope integrity via the conformational cycling of BamA.
How the outer membrane of Gram-negative bacteria is assembled remains a long-standing mystery and a crucial question in biology. Here, we focused on BamA, the core component of BAM whose activity is essential to constructing and maintaining the outer membrane. By solving the structure of BamA in complex with its lipoprotein substrate RcsF, our work sheds new light on BAM. It not only provides crucial molecular insights into how BAM exports lipoproteins to the surface, but also uncovers important new features of this essential machinery and its mechanism. Because BAM activity is required for bacterial survival, the complex is an attractive target for new antibiotics28–31. Our work also paves the way to the design of new antibacterials that interfere with BAM conformational cycling, because blocking BAM in the inward-open conformation lethally jams BAM with RcsF.
Methods
Bacterial strains, plasmids, and primers
Bacterial strains and plasmids used in this study are listed in Supplementary Tables 3 and 4, respectively. The parental E. coli strain DH300 is a MG1655 derivative deleted for the lac region and carrying a chromosomal rprA P::lacZ fusion at the λ phage attachment site to monitor Rcs activation32. To delete bamA on the chromosome, a kanamycin resistance (kan) cassette33 with the flanking regions of bamA was PCR amplified from the genomic DNA of a ΔrcsF::kan strain (PL339) using primers “bamA Km del F” and “bamA Km del R”. Then we performed λ-Red recombineering 34 with plasmid pSIM5-tet 35 on the strain containing pSC270 as a bamA-complementing plasmid in DH300. Deletion of bamA was verified by PCR. After preparing P1 lysate from this strain, bamA deletion (by transferring the kan cassette) was performed via P1 phage transduction of the appropriate strains.
We performed site-directed mutagenesis to generate bamA variants on plasmids. For single-codon changes, primer sequences are available upon request; otherwise see Supplementary Table 5. By using pJH114 as a template and performing site-directed mutagenesis (SDM), we introduced a six-histidine tag at the N-terminus of BamA. The C-terminal His-tag of BamE was also removed via SDM, generating pRRA1. The primer pairs were “SDM-HisBamA F” with “SDM-HisBamA R” and “bamE delHis F” with “bamE delHis R”. To add bamB next to bamA in pBamA, both bamA and bamB were PCR amplified as a single DNA fragment from pJH11436 using primers “pTrc-for” and “bamB (NotI) R”. The PCR product and pBamA were digested with NcoI and NotI and then ligated, yielding pBamA-B. pBamAhinge plasmid was generated by SDM using primers “BamA hinge F” and “BamA hinge R” and pBamA as template. To clone bamC, bamD, and bamE as an operon into a low-copy plasmid (pAM238), PCR was performed on pRRA1 as a template using primers “bamC kpnI F” and “pTrc-rev2”. The PCR product and pAM238 were digested with KpnI and XbaI and ligated, generating pSC263. We cloned bamA without the six-histidine tag into the low-copy plasmid pSC2310, yielding pSC270. bamA was PCR amplified from E. coli genomic DNA using primers “BamA (PciI)F” and “BamA (XbaI)R” and ligated with pSC231 predigested with NcoI and XbaI. To generate the bamA variants locked in the closed and open conformations, SDM was performed on pBamA-B. First, the two cysteines in the eL6 loop of BamA were mutated to serines, generating pBamAL6-B. This plasmid was used as template for SDM to generate pBamAG393C/G584C-B and pBamAG433C/N805C-B. To generate bamA variants with amber codons (TAG) to insert 3-(3-methyl-3H-diazirine-3-yl)-propaminocarbonyl-Nε-L-lysine (DiZPK; Artis Chemistry, Shanghai) at selected positions, we performed SDM on pBamA-B and pSC270; primer sequences are available upon request.
Expression and purification of the BamAB-RcsF complex
E. coli PL358 cells harboring pJH118 expressing N-terminal six-histidine-tagged BamA and BamB36 and pSC216 expressing RcsF10 were cultured to overexpress BamA, BamB, and RcsF. Cells were grown in Terrific Broth Auto Inducing Medium (Formedium) supplemented with 0.2% (w/v) L-arabinose at 37 °C (to induce RcsF), ampicillin (200 μg/mL), and chloramphenicol (25 μg/mL). Cells (1 L) were pelleted when they reached OD600 ~ 4, re-suspended in cold phosphate-buffered saline (25 mL) containing a protease-inhibitor cocktail (Complete, Roche), and lysed by two passages through a French pressure cell at 1,500 psi. The cell lysate was centrifuged for 40 min at 40,000 × g and 4 °C. After centrifugation, inner-membrane proteins were solubilized using 0.5% (w/v) N-lauryl sarcosine (Sigma) in a buffer containing 20 mM Tris-HCl [pH 7.5] and 150 mM NaCl for 1.5 h at 4 °C on a roller. The suspension was centrifuged for 40 min at 40,000 x g and 4 °C, after which the inner membrane fraction was in the supernatant while the outer membrane fraction remained in the pellet. Outer-membrane proteins were solubilized using 1% (w/v) n-dodecyl-β-d-maltopyranoside (DDM; Anatrace) in a buffer containing 20 mM Tris-HCl [pH 7.5], 300 mM NaCl, and 20 mM imidazole overnight at 4 °C on a roller. After centrifugation (40,000 x g, 4 °C, 40 min), the supernatant was mixed with Ni-NTA agarose beads (2 mL; IBA Lifescience) equilibrated with 20 mM Tris-HCl [pH 7.5], 300 mM NaCl, 20 mM imidazole, and 1% (w/v) DDM. After washing the resin with 10 column volumes of buffer (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 20 mM imidazole, 0.6% (w/v) tetraethylene glycol monooctyl ether (C8E4; Anatrace), and 0.01% (w/v) DDM), proteins were eluted with 5 column volumes of the same buffer supplemented with 200 mM imidazole. The eluted complex was then concentrated to 1 mL using a Vivaspin 4 Turbo concentrator (Cut-off 5 kDa; Sartorius). A final purification step was performed using size-exclusion chromatography by loading the proteins on a HiLoad 10/300 Superdex 200 column (GE Healthcare) using 20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.6% (w/v) C8E4, and 0.01% (w/v) DDM. Peak fractions were pooled and concentrated to ~30 mg/mL for crystallization.
For co-crystallization with NaI, NaI replaced NaCl in the gel-filtration buffer. Peak fractions were pooled and concentrated to ~30 mg/mL using a Vivaspin 4 Turbo concentrator (Sartorius).
The Blue native electrophoresis analysis of the concentrated complex was carried out on a 3-12% Bis-Tris gel (Life Technologies) following the manufacturer’s instructions. The protein complex bands separated in the native electrophoresis were identified via SDS-PAGE. Briefly, bands of interest were excised, boiled in SDS-PAGE sample buffer, and applied to the top of a polyacrylamide gel.
Expression and purification of BAM (BamABCDE) in complex with RcsF
E. coli BL21 (DE3) was transformed with pRRA1 expressing all five BAM proteins (N-terminal six-histidine-tagged BamA, BamB, BamC, BamD, and BamE) and pSC216 expressing RcsF for BAM and RcsF overexpression. In control cells, only pRRA1 was transformed. Protein expression and purification were performed as described above except that the detergent was exchanged to 0.1% (w/v) DDM during Ni-NTA affinity chromatography and size-exclusion chromatography. Eluted complexes were identified via SDS-PAGE and concentrated to 4 mg/mL. Blue native electrophoresis of the concentrated complexes was carried out as described above.
Crystallization, data collection and structure determination
Crystallization assays were carried out using the hanging drop vapor diffusion method in 48-well plates (Molecular Dimensions) at 20°C. The protein solution was mixed in a 2:1 ratio with the crystallization solution from the reservoir. The best native crystals were grown after 4 to 5 days in C10 and G10 conditions from Morpheus crystallization screen (Molecular dimensions; C10: 0.03 M sodium nitrate, 0.03 M sodium phosphate dibasic, 0.03 M ammonium sulfate, 0.10 M Tris-base [pH 8.5]; BICINE, 20 % (v/v) ethylene glycol; 10 % w/v PEG 8000; G10: 0.02 M sodium formate; 0.02 M ammonium acetate; 0.02 M sodium citrate tribasic dihydrate; 0.02M potassium sodium tartrate tetrahydrate; 0.02 M sodium oxamate; 0.10 M Tris-base [pH 8.5]; BICINE; 20% (v/v) ethylene glycol; 10 % (w/v) PEG 8000).
The crystals were harvested in a nylon loop, flash-cooled and stored in liquid nitrogen for data collection. Crystals were screened on beamlines Proxima-1 and Proxima-2 at Synchrotron Soleil (Gif-sur-Yvettes, France) as well as beamlines I03 and I04-1 at Diamond Light Source (Didcot, UK). Data for structure determination were collected on the Proxima-2 beam-line at Synchrotron Soleil at a wavelength of 1.77 Å. Data were indexed and integrated using XDS37, scaled using XSCALE37 and anisotropically corrected using STARANISO, applying a high resolution cutoff of I/σI = 1.238. The crystals belong to space group C2, with the unit cell dimensions a=158.84, b=142.5300, c=116.4200 Å3 and μ=102.61°. The structure was determined by molecular replacement using Phaser39, with the globular domain of RcsF (residues 51-130, PDB:2Y1B) and the inward open BamA μ-barrel (residues 422-809, PDB: 5D0O) and BamA POTRA domain 4 (residues 265-344; PDB: 5D0O) as search models. Molecular replacement searches identified two copies each of the BamA μ-barrel, RcsF and POTRA domain 4. Following 10 cycles of rigid body refinement POTRA domains 1, 2, 3 and 5 of the first BamA-RcsF copy in the asymmetric unit (Extended Data Figure 2) could be manually placed in the 2FoFc and FoFc difference density and were subjected to an additional 10 rounds of rigid body refinement. The model was refined to 3.8 Å resolution using BUSTER40 and intermittent manual inspection and correction of the model in Coot41. BUSTER was run using Local Structure Similarity Restraints (LSSR) over the two copies in the asymmetric unit, as well as target-based similarity restraints using the inward open BamA structure as reported in PDB:5D0O. The final model shows R and freeR factors of 28.3% and 31.4%, respectively, containing 1388 amino acids, of which 19 are indicated as Ramachandran outliers (1.4%). We note that side chain positioning is frequently ambiguous at 3.8 Å resolution and should therefore not be over-interpreted by users of the deposited model. Side chains for which no unambiguous electron density was observed were not pruned for ease of model interpretation. Such side chains were included in refinement and manually modelled in there most likely rotamer using Coot. Data collection and refinement statistics are found in Supplementary Table 1.
Site-specific photo-crosslinking
We used a site-specific photo-crosslinking method described previously10 with some modifications. To incorporate DiZPK into BamA, we used the pSup-Mb-DIZPK-RS plasmid encoding an evolved Methanosarcina barkeri pyrrolysyl-tRNA synthetase and an optimized suppressor24. DH300 cells were co-transformed with pSup-Mb-DIZPK-RS and one of the plasmids containing an amber codon in BamA in pSC270. Cells were grown in 3-(N-morpholino) propanesulfonic acid (MOPS) minimal medium supplemented with 0.2% glucose, 0.2% (w/v) arabinose, 200 μM IPTG, 0.001% (w/v) casamino acids, and 0.8 mM DiZPK. When cells reached OD600 = 1, 500-μL samples were irradiated with ultraviolet light at 365 nm or left unirradiated for 10 min. Cells were precipitated with trichloroacetic acid, washed with ethanol, and proteins were solubilized in 100 μL SDS-PAGE sample buffer (50 mM Tris-HCl [pH 7.5], 1% (w/v) SDS, 10% (v/v) glycerol, 0.002% (w/v) bromophenol blue) before SDS-PAGE and immunoblotting using anti-RcsF and anti-BamA antibodies.
In vivo BS3 crosslinking
Cells were harvested around mid-log phase (OD600 = ~ 0.5). In vivo crosslinking was performed as described previously10, except that bis(sulfosuccinimidyl)suberate (CovaChem) was used instead of 3,3’-dithiobis(sulfosuccinimidyl propionate).
Chemical crosslinking-mass spectrometry
We first performed buffer exchange of the purified BamAB-RcsF complex using a PD-10 desalting column (GE Healthcare Life Sciences). The complex was eluted with 20 mM HEPES [pH 7.5], 150 mM NaCl, and 0.1% (w/v) DDM. A 30-fold molar excess of the crosslinker disuccinimidyl dibutyric urea (50 mM stock solution in dimethyl sulfoxide, Thermo Scientific) was added to the protein solution and incubated at 37 °C for 1 h. The reaction was quenched by adding Tris-HCl to a final concentration of 20 mM. Crosslinked proteins were precipitated with ethanol and trypsinized, and crosslinked peptides were enriched through cation exchange as described previously42. Briefly, crosslinked proteins (50 μL) were precipitated by adding ice-cold ethanol (150 μL) and 3 M sodium acetate [pH 5.3] (5 μL) prior to incubation at −20 °C for 16 h. The sample was centrifuged (16,200 x g, 4 °C, 30 min), the supernatant was removed, and the pellet was washed by adding 80% (v/v) ice-cold ethanol (200 μL) and vortexing for 30 s. The sample was centrifuged again, the supernatant was removed, and the pellet was dried in a vacuum centrifuge. The pellet was dissolved in 1% (w/v) RapiGest (Waters) (10 μL) and trypsin (Sequencing grade, Promega) solution was added (90 μL, 1:50 trypsin:protein mass ratio) before incubating overnight at 37 °C. Trifluoroacetic acid was added (final concentration 0.5% (v/v)) and the sample was incubated at 37 °C for 1 h to precipitate the Rapigest. The mixture was centrifuged (16,200 x g, 4 °C, 30 min), the supernatant was concentrated using a vacuum centrifuge, and the pellet was dissolved in 20% (v/v) acetonitrile/0.4% (v/v) formic acid (20 μL). Strong cation exchange enrichment was carried out using OMIX 10 μL strong cation exchange pipette tips (Agilent) as previously described42.
Fractionated peptides (5 μL) were injected onto a reverse-phase Acquity M-Class C18, 75 μm x 150 mm column (Waters) and separated via gradient elution of 1-50% (v/v) solvent B (0.1 % (v/v) formic acid in acetonitrile) in solvent A (0.1 % (v/v) formic acid in water) over 60 min at 300 nL/min. The eluate was infused into a Xevo G2-XS (Waters) mass spectrometer operating in positive ion mode. Mass calibration was performed by infusion of aqueous NaI (2 μg/μL). [Glu1]-Fibrinopeptide B was used for the lock mass spray, with a 0.5 s lock spray scan taken every 30 s. The lock mass correction factor was determined by averaging 10 scans. Data acquisition was performed in DDA mode with a 1 s mass-spectrometry scan over m/z 350-2000. Instrument parameters were optimized for the detection of crosslinked peptides, as described previously43. Data processing and crosslink identification were performed using MeroX44.
Expression of BamA mutants and co-purification with RcsF
pBamA and pBamA-B each provide chromosomal-level expression of BamA10. Therefore, we introduced mutations of bamA in these plasmids to test the physiological effects of BamA mutants; plasmids were expressed in the presence or absence of bamA on the chromosome. Cells (2 mL) were harvested at OD600 ~ 0.5 to purify BamA, except during the following experiment. The cysteine mutants of BamA, when oxidized to form a disulfide bond, allow BamA to form an “open” or “closed” lateral gate. Therefore, the efficiency of disulfide-bond formation in these mutants is very important. To enhance the oxidation of cysteines to form disulfide bonds, we added 3 mM tetrathionate as an oxidant45 at OD600 ~ 0.5 and harvested cells (1 mL) at OD600 ~ 1.0.
Since there was a six-histidine tag at the N-terminus of BamA, we used Dynabeads™ His-Tag (Invitrogen) for Ni-affinity purification. After resuspending cells in 350 μL of 25 mM Tris-HCl [pH 7.4], 290 mM NaCl, 1 mM imidazole, and 0.05% (w/v) DDM (buffer A), cells were lysed via mild sonication on ice. Membrane vesicles were further solubilized by increasing the DDM concentration to 1% (w/v). After removing debris via centrifugation at 9,300 × g for 10 min, 5 μL of Dynabeads™ His-Tag (pre-washed with buffer A and resuspended in the same volume) were added to 250 μL of the supernatant, which was incubated for 20 min at 4 °C. The rest of the supernatant was used as the input fraction. The magnetic beads were pulled by a magnet and the supernatant was taken for the flow-through fraction. After washing the beads three times with 750 μL buffer E using the magnet, bound proteins were eluted with 83 μL (three times enrichment compared to the other fractions) of buffer A with 300 mM imidazole. Forty microliters of the input, flow-through, and elution fractions were mixed with SDS-PAGE sample buffer. After denaturation of the three fractions, SDS-PAGE was performed, followed by immunoblotting using rabbit-raised anti-BamA, anti-RcsF10, anti-BamB, anti-BamC, anti-BamD, and anti-BamE.
To determine the redox states of the cysteine-introduced gate mutants of BamA, we added 3 mM N-ethylmaleimide in SDS-PAGE sample buffer to alkylate cysteines to prevent thiol-disulfide exchange. The sample was divided into two aliquots and 10 mM of tris(2-carboxyethyl) phosphine was added to one of them to obtain the reduced state of BamA as a control. Nu-PAGE (4-12% gradient; Novex) was used to separate the oxidized and reduced bands of BamA.
Spotting assay for growth
Cells were grown in M9 minimal glucose medium at 30°C until they reached OD600 =1. Tenfold serial dilutions were made in M9 minimal glucose, plated onto M9 minimal glucose or LB agar, and incubated at 30°C or 37°C. Plates were supplemented with ampicillin (200?μg/ml).
Biolayer interferometry
Untagged BamA was first biotinylated using the EZ-Link NHS-PEG4-biotin kit (Perbio Science). The reaction was stopped by adding Tris [pH 8] to the final concentration of 20 mM. Excess NHS-PEG4-biotin was removed by passing the sample through a Zeba Spin Desalting column (Perbio Science). Biolayer interferometry was performed in black 96-well plates (Greiner) at 25 °C using OctetRed96 (ForteBio). Streptavidin and Ni-NTA biosensor tips (ForteBio) were hydrated with 0.2 mL working buffer (20 mM Tris [pH 8], 150 mM NaCl, 0.03 % (w/v) DDM) and then loaded with biotinylated BamA or 6xHis-tagged RcsF, respectively.
In the forward experiment, purified 6xHis-tagged RcsF (5 μg/mL) was immobilized on Ni-NTA sensors until the signal reached 0.5-0.6 nm. Association and dissociation of BamA to RcsF-coated tips were monitored for 1200 s and 300 s, respectively, by dipping tips into BamA-containing buffer (serial two-fold dilution from 4000 nM to 62.5 nM), and subsequently in buffer only. In the reverse experiment, biotinylated BamA was immobilized on streptavidin sensor tips to a signal of 2.0 nm. The association and dissociation of RcsF (serial 3-fold dilution from 3000 nM to 12.34 nM) to BamA-coated tips were monitored for 4800 s and 700 s, respectively. Dissociation constants were determined using Graphpad Prism by linear regression of the steady-state binding responses in the saturation binding experiment (Extended Data Fig. 4c, d).
For binding of the BamAG393C/G584C mutant (Extended Data Fig. 5c), 6xHis-tagged RcsF was immobilized on Ni-NTA sensors. To follow BamA association and dissociation, RcsF-coated tips were dipped into 0.2 mL of 200 nM BamA solution, with or without 2 mM diothiothreitol, for 1200 s, followed by 1200 s in buffer only.
Antibodies and immunoblotting
Rabbit anti-RcsF antibody was previously generated and used by us10. We newly raised the antibodies against BamA, BamB, BamC, BamD, and BamE in rabbits as follows. Except BamA, the DNA sequences encoding the proteins without the signal sequence were cloned into pET28a (Novagen) using the NcoI and XhoI restriction sites, which allows the expressed proteins to be his-tagged at the C-terminus. For BamA, DNA encoding the POTRA domains (1-4) of BamA with a C-terminal strep-tag (but without the signal sequence) was cloned in pET21a (Novagen). All the proteins above were overexpressed in BL21(DE3) and purified using standard methods for Ni-NTA affinity purification or streptavidin purification (POTRA 1-4). Small aliquots of the purified proteins were sent to the CER group (Marloie, Belgium) to raise antibodies in rabbits. Goat Anti-Rabbit IgG alkaline phosphatase conjugated (Sigma) was used as a secondary antibody at a 1:20,000 dilution.
Antibody specificity was confirmed by comparing the immunoblot of the wild-type strain with that of a mutant using each corresponding antibody. The dilutions of the antibodies for immunoblotting were 1:10,000 (BamA), 1:20,000 (BamB), 1:40.000 (BamC), 1:10,000 (BamD), and 1:20,000 (BamE). The specificity of the antibodies was verified; data are available upon request.
To simplify the detection of Bam components and RcsF after purification of BamA, we used two mixtures of antibodies (anti-BamA plus anti-RcsF; anti-BamB, anti-BamC, anti-BamD, plus anti-BamE). Detection specificity was verified using similar mutants as above but harboring pBamA. Data are available upon request.
The antibody recognizing the transmembrane domain of OmpA is a gift from the Bernstein laboratory46. The rabbit polyclonal OmpC antibody was purchased from EPIGENTEK. The anti-E. coli EF-Tu antibody (mAb 900) was purchased from HycultBiotech. The Goat anti-mouse IgG conjugated with the Cy3 fluorescent dye was used as a secondary antibody for EF-Tu (Amersham).
β-galactosidase assay
Rcs induction was monitored by measuring β-galactosidase activity as described47. Briefly, cells harboring PrprA-lacZ at the attB phage lambda site on the chromosome were diluted 1:100 from overnight cultures in Luria broth (LB), then incubated at 37°C. Cells were harvested at OD600 = 0.6-1. Twenty microliters of cells were harvested and incubated with 80 μl of permeabilization solution (60 mM Na2HPO4·2H2O, 40 mM NaH2PO4·H2O, 10 mM KCl, 1 mM MgSO4·7H2O, 50 mM β-mercaptoethanol) for 30–45 min at room temperature. Then, 600 μl of substrate (1 mg/ml O-nitrophenyl-β-d-galactoside, 50 mM β-mercaptoethanol) were added. The mixture was further incubated at 30°C for 20-90 min. Seven hundred microliters of 1 M Na2CO3 were added to stop the reaction, and the optical density was measured at 420 nm. The standardized amount of β-galactosidase activity was reported in Miller units. The ratio of PrprA-lacZ induction was calculated relative to the basal level in a WT strain. Bar graphs with corresponding statistical analysis were prepared using Prism 7 (GraphPad Software, Inc.).
Statistical methods
The significance of differences among bacterial strains was assessed using GraphPad Prism 8 according to analysis of variance (two ways ANOVA), followed by the application of Tukey’s multiple-comparison test. Normality was assessed using the Shapiro-Wilk test.
Molecular modeling: protein insertion in the lipid bilayer
The initial simulation system was prepared starting from the BamA-RcsF complex determined in this work (PDB code 6T1W) with the POTRA1-4 domains removed. Missing residues and C-terminal residues of BamA and RcsF were completed with MODELLER v9.2248 to generate a protein complex containing residues 347-810 for BamA and 51-134 for RcsF. This complex was preorientated with respect to the membrane normal (z axis) using the structure 5AYW 6 from the OPM database (https://opm.phar.umich.edu)49 as template, then embedded in an asymmetric bilayer to mimic the E. coli outer membrane using CHARMM-GUI (http://www.charmm-gui.org) 50 and following the protocols described for OmpLA51 and BamA52). The inner leaflet was a mixture of 100 lipids: 75 1-palmitoyl(16:0)-2-palmitoleoyl(16:1 cis-9)-phosphatidylethanolamine (PPPE), 20 1-palmitoyl(16:0)-2-vacenoyl(18:1 cis-11)-phosphatidylglycerol (PVPG), and 5 1,10-palmitoyl-2,20-vacenoyl cardiolipin with a net charge of –2e (PVCL2), corresponding to a ratio of 15:4:1. The outer leaflet was composed of 36 LPS (including the lipid A, R1 core and O-antigen polysaccharide fragments). The equilibration was performed according to the standard protocol from CHARMM-GUI Membrane Builder, with restraints that were gradually reduced in 6 steps (2.125 ns overall), using CHARMM version 44b153. A similar protocol was used to build a second system, containing the outward-open conformation of BamA (PDB code 5D0Q)5 with the POTRA1-4 domains removed and the RcsF from 6T1W shifted upwards on the z axis by 30 Å.
Dynamic importance sampling simulations
Dynamic importance sampling (DIMS) uses a biasing with correction approach to improve the sampling efficiency of rare events. The soft ratcheting algorithm generates transitions between states using a stochastic approach54. It should be noted that at any point of the DIMS simulation the intermediate system is realistic (no force is applied and the potential function is not modified) and the simulated pathway is always possible, although it may not be in all cases the lowest energy one. In this work, we carried out DIMS Langevin dynamics (LD) simulations using CHARMM version 44b1 (collision frequency of 25.0) starting from the first system and with the second system as target. The “soft-ratcheting” implemented in DIMS accepts all steps proceeding towards the desired final state and a fraction of steps away from the target. This fraction is defined by DIMS-Cartesian for which we used the recommended value of 10−6 55. Two replicas were simulated, providing similar results, and only the results from the first replica are presented here.
Image and video rendering
The pictures representing molecular data were generated using PyMOL version 2.2.0 (Schrödinger LLC, https://pymol.org). The video was generated from the DIMS simulation (replica 1) using VMD56.
Extended Data
Extended Data Fig. 1. RcsF can be co-purified with the BAM complex.
(a,b,c) Gel filtration profiles of the affinity-purified BamAB-RcsF, BAM-RcsF and BAM complexes. The size exclusion chromatography was performed using a HiLoad 10/300 Superdex 200pg. The input and peak fractions were collected and the samples were analyzed by blue native electrophoresis with Coomassie staining. The migration pattern of BamABCDE-RcsF (b) was modified compared to BamABCDE (c) upon size exclusion chromatography (band 8 increases), reflecting the higher instability of the BamABCDE-RcsF complex. n= 4 biologically independent experiments.
Extended Data Fig. 2. Crystal structure of the BamA-RcsF complex.
(a, b) Final 2Fo-Fc electron map of the BamA-RcsF complex, shown with a map contour level of 0.08 e-/Å3 (root mean square deviation 1.02 Å). The asymmetric unit of the crystals holds two BamA-RcsF copies, one revealing interpretable electron density for the full BamA sequence (a), and a second revealing unambiguous density for POTRA domain 4 only (b). In the second copy (b), the electron density corresponding to POTRA domains 1, 2, 3, and 5 is too weak to allow unambiguous rigid body placement of the domains. All descriptions and images in the main text are based on the first copy (a). (c) Overlay of two BamA-RcsF complexes in the asymmetric unit. The first complex depicts BamA in gold and RcsF in blue, while these molecules are yellow and light blue, respectively, in the second complex. In both copies, RcsF makes an average displacement of 4 Å relative to the BamA β-barrel. (d) Crystal packing of the BamA-RcsF complex viewed along the a- (left) and c-axis (right). For the first copy of the BamA-RcsF complex in the asymmetric unit (orange-slate) the conformation of the POTRA domains is stabilized by the packing along the b-axis, whilst for the second copy (cyan-slate) only POTRA domain 4 is involved in crystal contacts. In the latter, POTRA 5, 3, 2 and 1 are not in contact with neighboring molecules and show weak electron density only due to the lack of conformational stabilization.
Extended Data Fig. 3. Structural dynamics of the BamA POTRA domains.
(a, b) Superimposition of BamA-RcsF (gold and blue, respectively) with the POTRA domains in the inward-open BamABCDE complex (PDB: 5D0O; light blue) or the outward-open BamACDE complex (PDB: 5EKQ; green). Complexes are superimposed based on 400 equivalent Cα atoms in the BamA β-barrel, and shown in side (a) or periplasmic (b) view. For 5d0o and 5ekq, the accessory Bam subunits and the BamA β-barrel are omitted for clarity. (c) Periplasmic view of the inward-open BamABCDE complex, showing binding of the BAM accessory proteins BamB (magenta), BamC (red), BamD (blue), and BamE (yellow). Pulldown experiments showed that RcsF binds the BamABCDE complex (Fig. 1). In agreement with this observation, structural comparisons reveal that RcsF binding would not result in direct steric clashes with any BAM accessory protein. However, the positions of the POTRA domains in the BamA-RcsF and BamABCDE complexes are markedly different. In the BamA-RcsF complex, POTRA5 makes a 26 outward rotation to accommodate RcsF (see also Fig. 3), and a reorganization in the joint between POTRA domains 3 and 2 results in a more extended conformation of the POTRA “arm” and the projection of POTRA domains 2 and 1 further from the BamA β-barrel, a conformation not previously reported in available BamA structures. In the BamABCDE complex, BamD contacts both POTRA5 and the joint of POTRA domains 1 and 2. In the BamA-RcsF complex, POTRA5 and POTRA domains 2 and 1 are too distant to be bridged by BamD; binding of BamD to BamA-RcsF therefore requires a conformational change in the POTRA arm or the dissociation of BamD at either of these two contact points.
Extended Data Fig. 4. Validation of the BamA-RcsF structure.
(a) RcsF aminoacid sequence. The sequence coverage of the XL-MS experiment was about 60% as highlighted in violet (b) Ribbon diagram of the BamA-RcsF structure. Highlighted residues show sites mutated to amber for incorporation of the photoreactive lysine analog DiZPK. Sites that crosslink to RcsF are green, sites that show no crosslinking are magenta. Mutation of extracellular loop 1 (eL1; red) leads to loss of RcsF binding (see panel g). BamA sidechains found to crosslink with RcsF by means of the homobifunctional amine-reactive crosslinker disuccinimidyl dibutyric urea (DSBU) are shown as sticks and colored cyan. Residue K61 from RcsF, which was found to crosslink to BamA using DSBU, is shown as a stick and colored orange. The other two RcsF residues (K42 and K134) that could be crosslinked to BamA are not visible in this structural model. (c) In vivo photocrosslinking experiment in which cells expressing the BamA mutants containing DiZPK at the indicated positions were treated (+) or not (-) with ultraviolet light. Proteins samples were analyzed via SDS-PAGE and immunoblotted with anti-RcsF or anti-BamA antibodies, showing that the photo-crosslinked complexes contain BamA and RcsF. WT, wild type. (d,e) Sensorgrams from biolayer interferometry (left) and corresponding equilibrium binding plots (right) of immobilized RcsF titrated with BamA (d) or immobilized BamA titrated with RcsF (e), n=1 biologically independent experiment. (f) The levels of major OMPs are slightly decreased in cells expressing BamAΔloop1. WT cells harboring the empty plasmid (pAM238) were used as control and EF-Tu expression levels were analyzed as loading control. n= 3 biologically independent experiments. (g) Deletion of loop 1 in BamA prevents RcsF from being pulled down with BamA. WT cells harboring the empty plasmid (pAM238) were used as control. n= 3 biologically independent experiments. (h) Overexpression of pBamAΔLoop1 in a bamA deletion strain activates the Rcs system compared to WT. A chromosomal rprA::lacZ fusion was used to monitor Rcs activity, and specific β-galactosidase activity was measured from cells at mid-log phase (OD600=0.5). Boxplot with whiskers from minimum to maximum. All values were normalized to the average activity obtained for WT cells harbouring the empty plasmid (pET3a) obtained from N=8 biologically independent experiments. Mean is showed as +. WT, wild-type; Kan, kanamycin.
Extended Data Fig. 5. RcsF binds the inward-open conformation of BamA.
Models for the BamAG393C/G584C 5 and BamAG433C/N805C 26 double cysteine mutants, which are locked in the outward-open or inward-open conformation, respectively, when oxidized. Mutated cysteines are shown as atom spheres. (b) BamA barrel locking and RcsF binding. Overexpression of double cysteine mutants pBamAG393C/G584C-B and BamAG433C/N805C-B in a wild-type strain. RcsF can be co-purified with the BamA β-barrel locked in the inward-open conformation (BamAG433C/N805C) by a disulfide bond (ox) but not in the outward-open conformation (BamAG393C/G584C). BamA mutants become reduced (red) following treatment with tris(2-carboxyethyl) phosphine (TCEP) and migrate similarly. The oxidized form of BamAG393C/G584C migrates more slowly than wild-type BamA. As a result, two bands are visible for BamA in the input of BamAG393C/G584C, the lower migrating band corresponding to wild-type BamA expressed from the chromosome. n= 3 biologically independent experiments. (c) Sensorgram from biolayer interferometry of immobilized RcsF titrated with BamAG393C/G584C, without (oxidized; - DTT) or with dithiothreitol (reduced; + DTT). When the β-barrel is locked in the outward-open conformation (- DTT), RcsF is unable to bind BamA. When reduced, BamAG393C/G584C regains binding, demonstrating that BamA reverts to the inward-open conformation in which it can bind RcsF.
Extended Data Fig. 6. The movement of POTRA5 towards the periplasmic exit of the lumen of the BamA barrel could push RcsF upwards.
(a,b) Lateral view of the initial and final conformations, respectively, of the BamA-RcsF complex during the dynamic importance sampling simulation (DIMS) of the BamA-RcsF complex. (c,d) Bottom view (from the periplasm) of the above conformations. BamA and RcsF are colored in orange and blue, respectively. The initial conformation of the system (BamA and RcsF) corresponds to the structure determined in this work (PDB code 6T1W)5 with the POTRA1-4 domains removed. The final conformation of BamA is similar to the outward-open structure (PDB code 5D0Q). The explicit outer membrane and solvent are not shown for clarity. (e) Expression from BamAhinge from a plasmid in ΔbamA cells leads to a severe growth defect when cells are grown at 37°C in rich media, but not when they are grown in minimal media at 30°C. Cells were grown in M9 minimal glucose medium at 30°C until they reached OD600 =1. Tenfold serial dilutions were made in M9 minimal glucose, plated onto M9 minimal glucose or LB agar, and incubated at 30°C or 37°C. Plates were supplemented with ampicillin (200μg/ml). n=3 biologically independent experiments.
Supplementary Material
Acknowledgments
We thank Asma Boujtat for technical help. We are indebted to Dr. Peng R. Chen (Peking University) for sharing DiZPK, to Dr. Harris Bernstein (NIH, Bethesda, USA) for providing strains and plasmids, and to Dr. Michael Deghelt, Dr. Géraldine Laloux, Dr. Camille Goemans (EMBL, Heidelberg, Germany), and Dr. Pauline Leverrier for helpful suggestions and discussions and for providing comments on the manuscript. We thank Pierre Legrand and staff at Soleil Synchrotron France and at Diamond Light Source UK for beamtime and their assistance during data collection. This work was supported, in part, by grants from the Fonds de la Recherche Scientifique – FNRS, from the FRFS-WELBIO grants n° WELBIO-CR-2015A-03 and WELBIO-CR-2019C-03, from the EOS Excellence in Research Program of the FWO and FRS-FNRS (G0G0818N), from the Fédération Wallonie-Bruxelles (ARC 17/22-087), from the European Commission via the International Training Network Train2Target (721484), from the French region Ile-de-France (DIM Malinf) and from the BBSRC (BB/P000037/1, BB/M012573/1).
Footnotes
Author Contributions
J.-F.C., R.R.A., S.E.R., H.R., and S.H.C. wrote the manuscript. J.L., RRA, S.N., G.L., S.H.C., H.R., and J.-F.C. designed the experiments. J.L., R.R.A., S.N., G.L., and S.H.C. performed the experiments, constructed the strains, and cloned the constructs. B.I. performed the dynamic importance sampling simulations. J.L., R.R.A., S.N., S.H.C., H.R., and J.-F.C. analyzed and interpreted the data. A.N.C. and S.E.R. performed and analyzed the crosslinking mass spectrometry experiments. All authors discussed the results and commented on the manuscript.
Author Information
The authors declare no competing financial interests.
Data Availability
Coordinates and structure factors have been deposited in the Protein Data Bank under accession number 6T1W. All other data generated or analysed during this study are included in this published article and its supplementary information file.
References
- 1.Noinaj N, Gumbart JC, Buchanan SK. The beta-barrel assembly machinery in motion. Nat Rev Microbiol. 2017;15:197–204. doi: 10.1038/nrmicro.2016.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hagan CL, Silhavy TJ, Kahne D. beta-Barrel membrane protein assembly by the Bam complex. Annu Rev Biochem. 2011;80:189–210. doi: 10.1146/annurev-biochem-061408-144611. [DOI] [PubMed] [Google Scholar]
- 3.Iadanza MG, et al. Lateral opening in the intact beta-barrel assembly machinery captured by cryo-EM. Nat Commun. 2016;7 doi: 10.1038/ncomms12865. 12865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bakelar J, Buchanan SK, Noinaj N. The structure of the beta-barrel assembly machinery complex. Science. 2016;351:180–186. doi: 10.1126/science.aad3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gu Y, et al. Structural basis of outer membrane protein insertion by the BAM complex. Nature. 2016;531:64–69. doi: 10.1038/nature17199. [DOI] [PubMed] [Google Scholar]
- 6.Han L, et al. Structure of the BAM complex and its implications for biogenesis of outer-membrane proteins. Nat Struct Mol Biol. 2016;23:192–196. doi: 10.1038/nsmb.3181. [DOI] [PubMed] [Google Scholar]
- 7.Wu T, et al. Identification of a multicomponent complex required for outer membrane biogenesis in Escherichia coli. Cell. 2005;121:235–245. doi: 10.1016/j.cell.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Sklar JG, et al. Lipoprotein SmpA is a component of the YaeT complex that assembles outer membrane proteins in Escherichia coli. Proc Natl Acad Sci U S A. 2007;104:6400–6405. doi: 10.1073/pnas.0701579104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schiffrin B, Brockwell DJ, Radford SE. Outer membrane protein folding from an energy landscape perspective. BMC Biol. 2017;15:123. doi: 10.1186/s12915-017-0464-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cho SH, et al. Detecting Envelope Stress by Monitoring beta-Barrel Assembly. Cell. 2014;159:1652–1664. doi: 10.1016/j.cell.2014.11.045. [DOI] [PubMed] [Google Scholar]
- 11.Konovalova A, Perlman DH, Cowles CE, Silhavy TJ. Transmembrane domain of surface-exposed outer membrane lipoprotein RcsF is threaded through the lumen of beta-barrel proteins. Proc Natl Acad Sci U S A. 2014;111:E4350–4358. doi: 10.1073/pnas.1417138111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tata M, Konovalova A. Improper Coordination of BamA and BamD Results in Bam Complex Jamming by a Lipoprotein Substrate. MBio. 2019;10 doi: 10.1128/mBio.00660-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hart EM, Gupta M, Wuhr M, Silhavy TJ. The Synthetic Phenotype of DeltabamB DeltabamE Double Mutants Results from a Lethal Jamming of the Bam Complex by the Lipoprotein RcsF. MBio. 2019;10 doi: 10.1128/mBio.00662-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wall E, Majdalani N, Gottesman S. The Complex Rcs Regulatory Cascade. Annu Rev Microbiol. 2018;72:111–139. doi: 10.1146/annurev-micro-090817-062640. [DOI] [PubMed] [Google Scholar]
- 15.Laloux G, Collet JF. “Major Tom to ground control: how lipoproteins communicate extra-cytoplasmic stress to the decision center of the cell”. J Bacteriol. 2017 doi: 10.1128/JB.00216-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hussein NA, Cho SH, Laloux G, Siam R, Collet JF. Distinct domains of Escherichia coli IgaA connect envelope stress sensing and down-regulation of the Rcs phosphorelay across subcellular compartments. PLoS Genet. 2018;14:e1007398. doi: 10.1371/journal.pgen.1007398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaur H, et al. Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach. J Biomol NMR. 2019;73:375–384. doi: 10.1007/s10858-019-00250-8. [DOI] [PubMed] [Google Scholar]
- 18.Albrecht R, et al. Structure of BamA, an essential factor in outer membrane protein biogenesis. Acta Crystallogr D Biol Crystallogr. 2014;70:1779–1789. doi: 10.1107/S1399004714007482. [DOI] [PubMed] [Google Scholar]
- 19.Ni D, et al. Structural and functional analysis of the beta-barrel domain of BamA from Escherichia coli. FASEB J. 2014;28:2677–2685. doi: 10.1096/fj.13-248450. [DOI] [PubMed] [Google Scholar]
- 20.Hartmann JB, Zahn M, Burmann IM, Bibow S, Hiller S. Sequence-Specific Solution NMR Assignments of the beta-Barrel Insertase BamA to Monitor Its Conformational Ensemble at the Atomic Level. J Am Chem Soc. 2018;140:11252–11260. doi: 10.1021/jacs.8b03220. [DOI] [PubMed] [Google Scholar]
- 21.Leverrier P, et al. Crystal structure of the outer membrane protein RcsF, a new substrate for the periplasmic protein-disulfide isomerase DsbC. J Biol Chem. 2011;286:16734–16742. doi: 10.1074/jbc.M111.224865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rogov VV, Rogova NY, Bernhard F, Lohr F, Dotsch V. A disulfide bridge network within the soluble periplasmic domain determines structure and function of the outer membrane protein RCSF. J Biol Chem. 2011;286:18775–18783. doi: 10.1074/jbc.M111.230185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calabrese AN, Radford SE. Mass spectrometry-enabled structural biology of membrane proteins. Methods. 2018;147:187–205. doi: 10.1016/j.ymeth.2018.02.020. [DOI] [PubMed] [Google Scholar]
- 24.Zhang M, et al. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
- 25.Gu Y, Zeng Y, Wang Z, Dong C. BamA beta16C strand and periplasmic turns are critical for outer membrane protein insertion and assembly. Biochem J. 2017;474:3951–3961. doi: 10.1042/BCJ20170636. [DOI] [PubMed] [Google Scholar]
- 26.Noinaj N, Kuszak AJ, Balusek C, Gumbart JC, Buchanan SK. Lateral opening and exit pore formation are required for BamA function. Structure. 2014;22:1055–1062. doi: 10.1016/j.str.2014.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okuda S, Tokuda H. Lipoprotein sorting in bacteria. Annu Rev Microbiol. 2011;65:239–259. doi: 10.1146/annurev-micro-090110-102859. [DOI] [PubMed] [Google Scholar]
- 28.Storek KM, et al. Monoclonal antibody targeting the beta-barrel assembly machine of Escherichia coli is bactericidal. Proc Natl Acad Sci U S A. 2018;115:3692–3697. doi: 10.1073/pnas.1800043115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Imai Y, et al. A new antibiotic selectively kills Gram-negative pathogens. Nature. 2019;576:459–464. doi: 10.1038/s41586-019-1791-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luther A, et al. Chimeric peptidomimetic antibiotics against Gram-negative bacteria. Nature. 2019;576:452–458. doi: 10.1038/s41586-019-1665-6. [DOI] [PubMed] [Google Scholar]
- 31.Hart EM, et al. A small-molecule inhibitor of BamA impervious to efflux and the outer membrane permeability barrier. Proc Natl Acad Sci U S A. 2019;116:21748–21757. doi: 10.1073/pnas.1912345116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 33.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006 0008 msb4100050 [pii] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yu D, et al. An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A. 2000;97:5978–5983. doi: 10.1073/pnas.100127597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koskiniemi S, Pranting M, Gullberg E, Nasvall J, Andersson DI. Activation of cryptic aminoglycoside resistance in Salmonella enterica. Mol Microbiol. 2011;80:1464–1478. doi: 10.1111/j.1365-2958.2011.07657.x. [DOI] [PubMed] [Google Scholar]
- 36.Roman-Hernandez G, Peterson JH, Bernstein HD. Reconstitution of bacterial autotransporter assembly using purified components. Elife. 2014;3:e04234. doi: 10.7554/eLife.04234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kabsch W. Xds. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tickle IJ, Flensburg C, Keller P, Paciorek W, Sharff A, Vonrhein C, Bricogne G. STARANISO. 2018 [Google Scholar]
- 39.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bricogne G, B E, Brandl M, Flensburg C, Keller P, Paciorek W, Roversi P, S A, Smart OS, Vonrhein C, Womack TO. BUSTER version 2.10.3. 2017 [Google Scholar]
- 41.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. S0907444904019158 [pii] [DOI] [PubMed] [Google Scholar]
- 42.Schmidt C, Robinson CV. A comparative cross-linking strategy to probe conformational changes in protein complexes. Nat Protoc. 2014;9:2224–2236. doi: 10.1038/nprot.2014.144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.James JMB, Cryar A, Thalassinos K. Optimization Workflow for the Analysis of Cross-Linked Peptides Using a Quadrupole Time-of-Flight Mass Spectrometer. Anal Chem. 2019;91:1808–1814. doi: 10.1021/acs.analchem.8b02319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iacobucci C, et al. A cross-linking/mass spectrometry workflow based on MS-cleavable cross-linkers and the MeroX software for studying protein structures and protein-protein interactions. Nat Protoc. 2018;13:2864–2889. doi: 10.1038/s41596-018-0068-8. [DOI] [PubMed] [Google Scholar]
- 45.Osborne AR, Rapoport TA. Protein translocation is mediated by oligomers of the SecY complex with one SecY copy forming the channel. Cell. 2007;129:97–110. doi: 10.1016/j.cell.2007.02.036. [DOI] [PubMed] [Google Scholar]
- 46.Hussain S, Bernstein HD. The Bam complex catalyzes efficient insertion of bacterial outer membrane proteins into membrane vesicles of variable lipid composition. J Biol Chem. 2018;293:2959–2973. doi: 10.1074/jbc.RA117.000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JC. Experiments in Molecular Genetics. Cold Spring Harbor Laboratory Press; 1972. [Google Scholar]
- 48.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 49.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–376. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, et al. CHARMM-GUI Membrane Builder for Complex Biological Membrane Simulations with Glycolipids and Lipoglycans. J Chem Theory Comput. 2019;15:775–786. doi: 10.1021/acs.jctc.8b01066. [DOI] [PubMed] [Google Scholar]
- 51.Wu EL, et al. E. coli outer membrane and interactions with OmpLA. Biophys J. 2014;106:2493–2502. doi: 10.1016/j.bpj.2014.04.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fleming PJ, et al. BamA POTRA Domain Interacts with a Native Lipid Membrane Surface. Biophys J. 2016;110:2698–2709. doi: 10.1016/j.bpj.2016.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brooks BR, et al. CHARMM: the biomolecular simulation program. J Comput Chem. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Perilla JR, Beckstein O, Denning EJ, Woolf TB. Computing ensembles of transitions from stable states: Dynamic importance sampling. J Comput Chem. 2011;32:196–209. doi: 10.1002/jcc.21564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Denning EJ, Woolf TB. Cooperative nature of gating transitions in K(+) channels as seen from dynamic importance sampling calculations. Proteins. 2010;78:1105–1119. doi: 10.1002/prot.22632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. 27-38. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Coordinates and structure factors have been deposited in the Protein Data Bank under accession number 6T1W. All other data generated or analysed during this study are included in this published article and its supplementary information file.