Abstract
Plant roots acquire nutrients and water, while managing interactions with the soil microbiota. Their endodermis provides an extracellular diffusion barrier via a network of lignified cell walls, called Casparian strips, supported by subsequent formation of suberin lamellae. Whereas lignification is thought to be irreversible, suberin lamellae display plasticity, which is crucial for root adaptative responses in the plant. Despite suberin being a major plant polymer, fundamental aspects of its biosynthesis and turnover have remained obscure. Plants shape their root system via lateral root formation, an auxin-induced process requiring local breaking and re-sealing of endodermal lignin and suberin barriers. Here, we show that differentiated endodermal cells have a specific, auxin-mediated transcriptional response, dominated by cell wall remodelling genes. We identified two sets of auxin-regulated GDSL-lipases. One is required for suberin synthesis, while the other can drive suberin degradation. These enzymes constitute novel core players of suberisation, driving root suberin plasticity.
Introduction
Plants require a dynamic and adaptive root system, allowing optimal anchorage and foraging of the soil environment for water and nutrients, while managing interactions with the soil microbiome1–3. Lateral root formation is a key factor modulating root system architecture. In most angiosperms, including Arabidopsis thaliana (Arabidopsis), these organs initiate in xylem-pole associated pericycle cells (XPPs). Auxin is required both for initiation and development of lateral roots4,5. Lateral roots need to traverse the overlying endodermis in order to develop and emerge and this cell layer therefore plays an essential role during lateral root formation, as it has to actively accommodate the expansion growth of the XPP through remodeling of cell shape and volume.
Moreover, in order to minimize both the leakage of nutrients from the stele into the rhizosphere and the entry of soil-borne pathogens, opening and sealing of the lignified and suberized endodermal barriers needs to be tightly-controlled. Therefore, a dynamic de-suberization and re-suberization is bound to play an important role in this process. However, we still lack an understanding of the basic molecular machineries that regulate the dynamics of suberin deposition and degradation during root development6–9. The lignified Casparian Strip (CS) appears to get locally modified in order to allow the growth of the lateral root through this cell layer10 and it was also shown that suberin is deposited in the cell walls of endodermal cells in contact with the later lateral root primordium after emergence8. However, endodermal cells often are already suberized when lateral roots form and it is unknown how suberin is first degraded and later re-synthesized.
All these responses are regulated via auxin-mediated signaling in the endodermis, and expression of a dominant repressor of auxin signaling, short hypocotyl 2-2 (shy2-2) in this cell layer blocks lateral root formation demonstrating the crucial role of SHY2-mediated endodermal auxin signaling in this process10.
Here, we show that differentiated endodermal cells have a distinct auxin-mediated transcriptome. Mining this dataset, we identified a set of 10 GDSL-motif containing enzymes that are differentially regulated after auxin treatment. We confirmed that all of these ten GDSL-motif containing enzymes were indeed expressed in the endodermis, being either repressed or induced during auxin treatment or lateral root formation. We then show that five of the auxin-repressed GDSL-motif containing enzymes are redundantly required for suberin biosynthesis, a full knock-out essentially abrogating suberin accumulation in the endodermis. Among the five auxin-induced GDSL-motif containing genes, we find enzymes that we demonstrate to be sufficient for suberin degradation and required for correct lateral root emergence. The quintuple mutants of the suberin-biosynthetic, GDSL-motif containing enzymes were highly-sensitive to mild salt stress. Single knock-out mutants of members of the suberin degrading class displayed delays in lateral root emergence. The enzymes identified in this work are strong candidates for the currently unidentified suberin polymerases and degradases in plants. This greatly advances our understanding of in vivo suberin formation, as well as the mechanisms underlying its striking developmental plasticity.
Results
A genotype for obtaining specific endodermal auxin responses
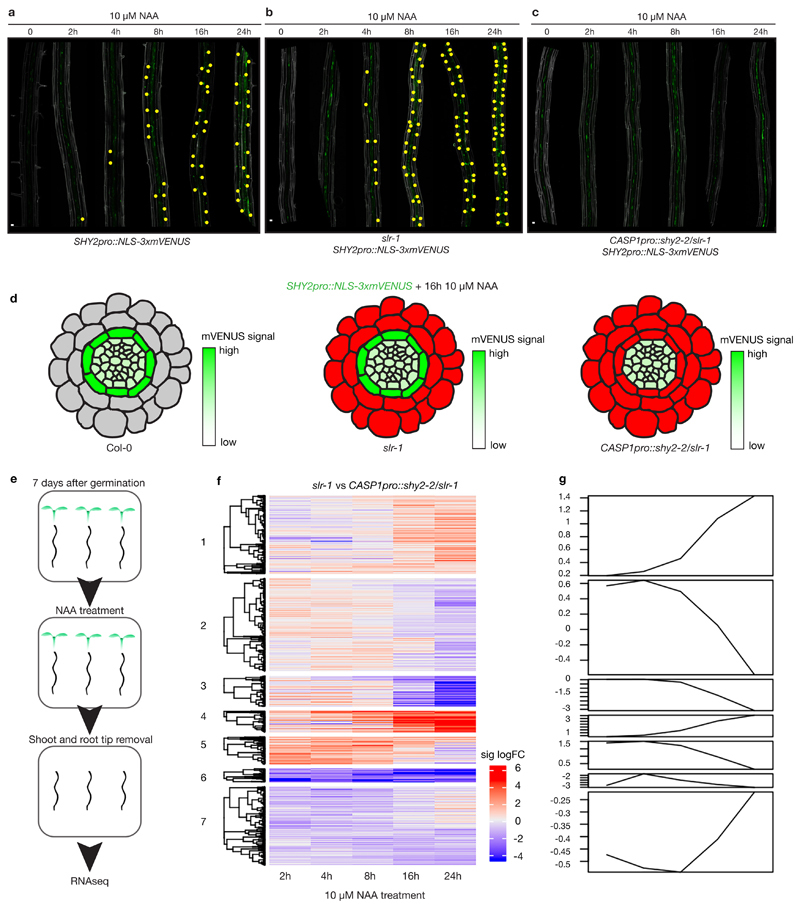
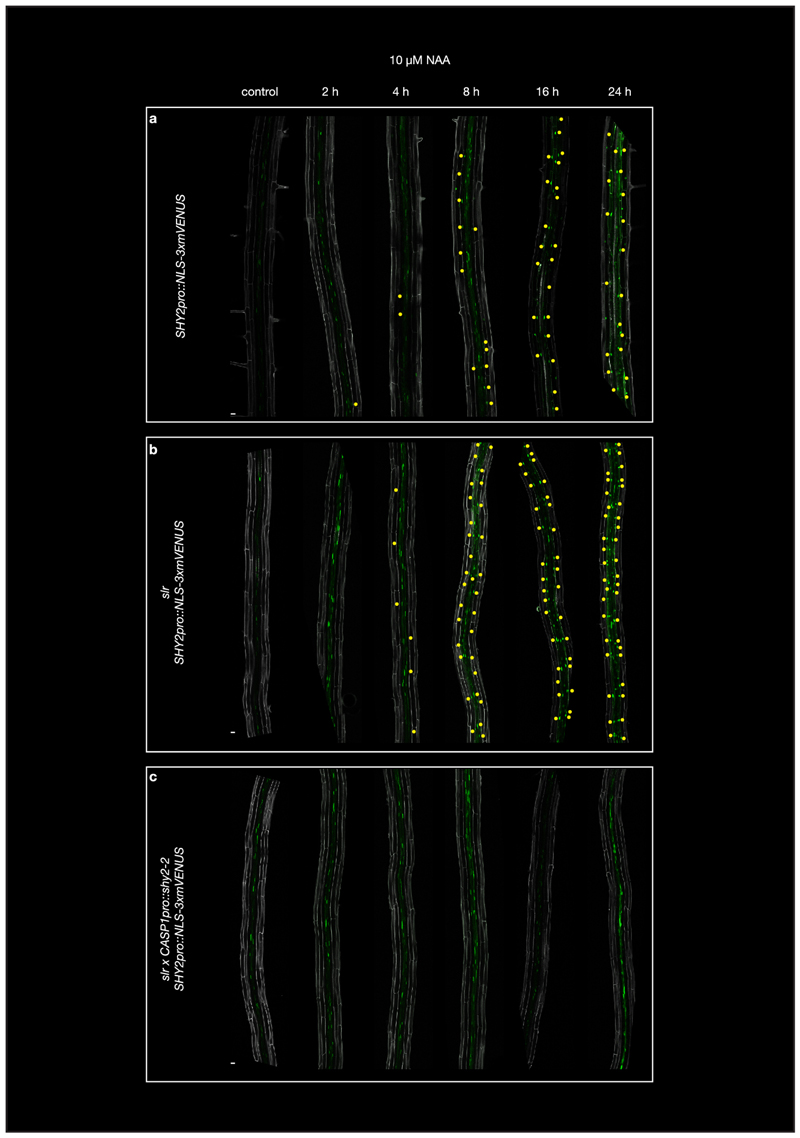
We have previously shown that the drastic changes in endodermal cell volume, as well as CS modification during lateral root emergence10 is mediated by SHORT HYPOCOTYL 2 (SHY2/IAA3)-dependent auxin signaling. Moreover, SHY2 represses its own transcription in a typical, auxin-induced negative feedback loop and is thus also a great, early transcriptional auxin-response marker in the endodermis10,11. However, we ignore the SHY2 targets (direct or indirect) that drive the complex accommodating responses in the endodermis. Therefore, we set out to obtain a SHY2-mediated transcriptional response profile in the endodermis. Generating such a data set comes with particular challenges: First, most endodermal cells at the moment of lateral root emergence are lignified and suberized, making it impossible to employ protoplast isolation used for single cell or cell-type specific sequencing. Secondly, only a subset of endodermal cells, those overlying an auxin-emitting lateral root primordium from stage I and onwards will be stimulated in a SHY2-dependent fashion10. We therefore first thought to simply compare wild-type and CASP1pro::shy2-2 seedlings, that suppresses auxin signaling specifically in the differentiated endodermis, after auxin treatment, in order to obtain an endodermis-specific set of auxin responsive genes. In wild-type, the auxin reporter SHY2pro::NLS-3xmVENUS fluorescence peaks in the endodermis at ~16 hours after NAA treatment and is blocked in the CASP1pro::shy2-2 line (Fig. 1a, d and Extended Data Fig.1). However, since the CASP1pro::shy2-2 transgene also indirectly impairs the auxin-mediated induction of lateral roots10, a simple comparison of the auxin-induced transcriptomes of CASP1pro::shy2-2 roots and wild-type roots after auxin treatment would be dominated by pericycle and cell cycle-related responses, preventing identification of endodermal auxin responses. Therefore, we added a genetic manipulation that would strongly enrich for auxin induced transcriptional changes in the endodermis. We combined the dominant, solitary root 1 (slr-1)/iaa14 mutant with CASP1pro::shy2-2. Lateral root formation is impaired in the Arabidopsis slr-1 mutant12. Importantly, SLR is expressed in the pericycle, cortex and epidermis, but not in the endodermis and the slr-1 mutant should thus specifically block auxin response in the endodermis-surrounding cell layers12,13(Extended Data Fig. 2a). As predicted, we found that auxin-mediated induction of SHY2 in the endodermis was still occurring in slr-1 roots (Fig. 1b, d). We further predicted that, in the combined CASP1pro::shy2-2/slr-1 background, auxin signaling should be largely blocked in all differentiated root cell layers. Indeed, we could not detect induction of SHY2pro::NLS-3xmVENUS in the endodermis in this background. Based on these results, we predicted that a comparison (subtraction) of the NAA-induced transcriptomes of roots from the slr-1 single mutant with the CASP1pro::shy2-2/slr-1 double mutant would allow us to extract a specific endodermal auxin signaling transcriptomic profile, otherwise obscured by the strong, proliferation-inducing auxin responses of the xylem-pole pericycle cells (Fig. 1a-e).
Figure 1. A genetic trick for mapping auxin responses in the differentiated endodermis.
a-c, Maximum images projections of roots expressing SHY2pro::NLS-3xmVENUS treated with 10 μM NAA for 0, 2, 4, 8, 12 and 24hrs. a, Col-0. b, slr-1. c, CASP1pro::shy2-2/slr-1. Yellow dots indicate SHY2pro::NLS-3xmVENUS signal in the endodermis. For more details see Extended Data Figure 1. Images are representative of experiments repeated 3 times. d, Schematic representation of SHY2pro::NLS-3xmVENUS responses in the different genetic backgrounds after 10 μM NAA for 16hrs. SHY2pro::NLS3xmVENUS signal is indicated in green and inhibition of auxin signaling by a slr-1 or CASP1pro::shy2-2 is indicated in red. e, Experimental setup of the RNAseq experiment. f, Heatmap showing the seven clusters containing the significant differentially expressed genes (cut-off fold change > 2, false discovery rate < 0.05) between slr-1 and CASP1pro::shy2-2 / slr-1 during the NAA time course. g, Graphical presentation of the behavior of the seven cluster during the NAA time course depicted in (e). Scale bar in (a), (b) and (c) = 50 μm.
Differentiated endodermal cells have a distinctive transcriptional auxin response
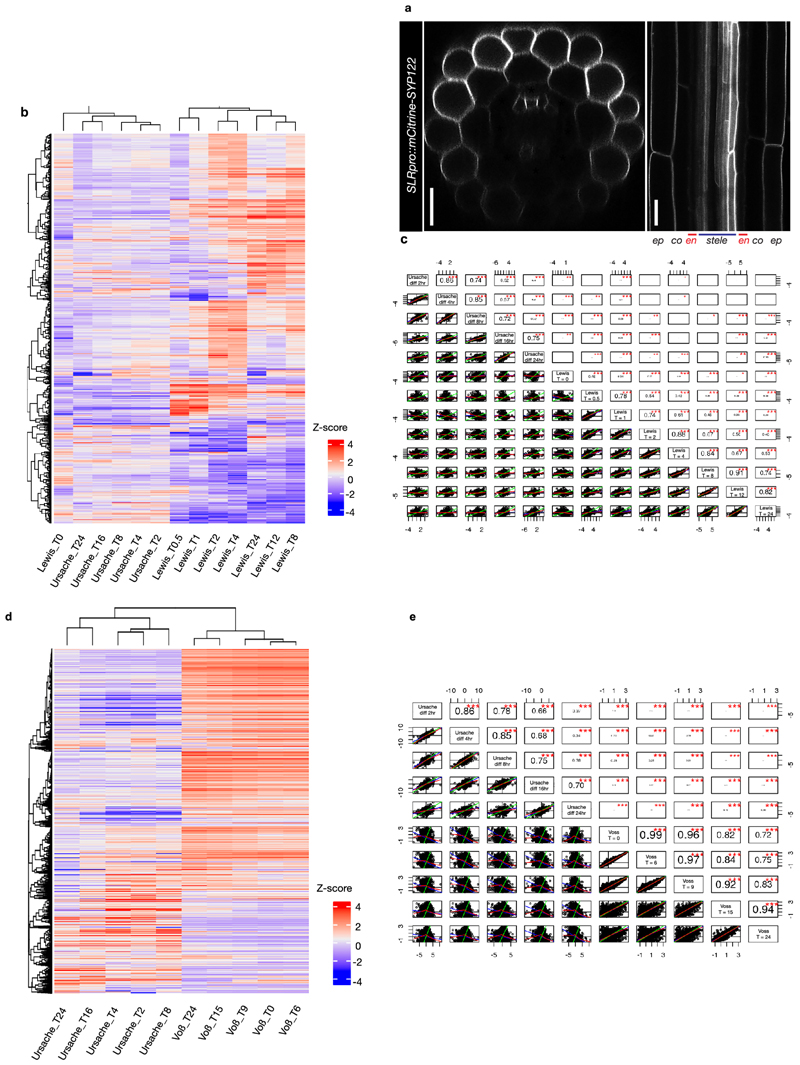
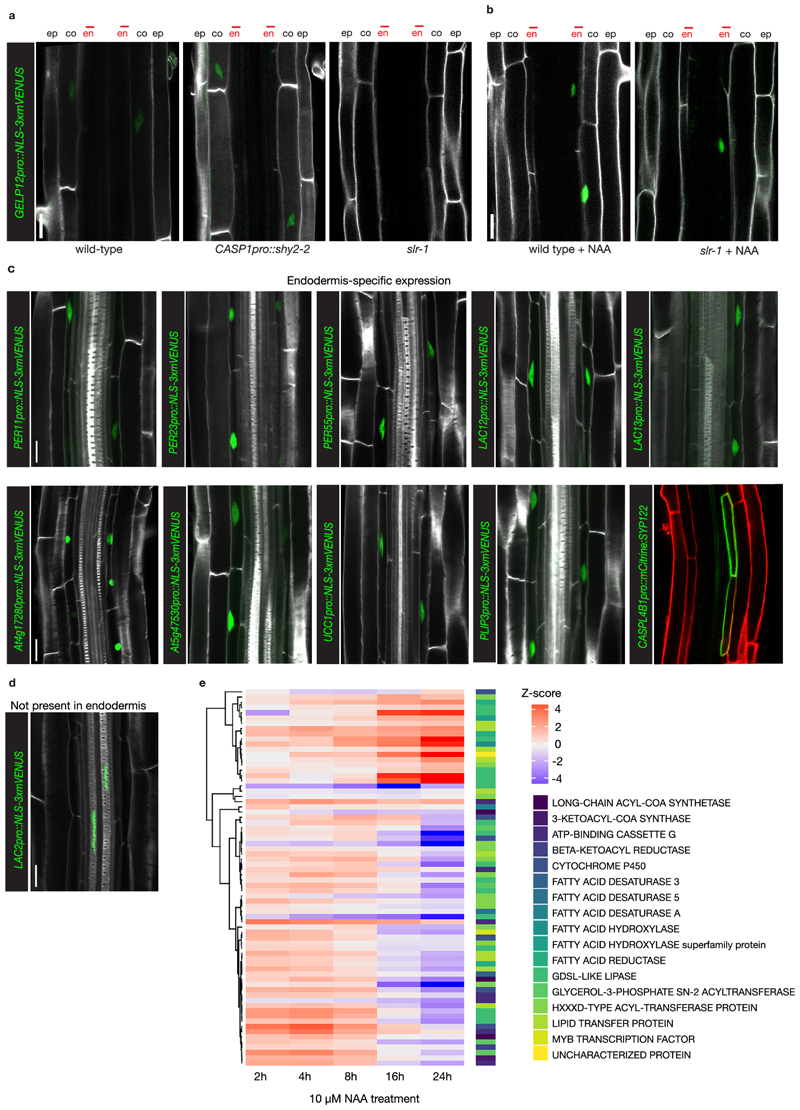
We interrogated the genome-wide transcriptional responses in slr-1 and CASP1pro::shy2-2/slr-1 after NAA treatment at multiple time points. We established that ~800-900 genes are differentially expressed at 2, 4, 8, 16hr after treatment and ~1000 are significantly changed after 24hr treatment compared to the zero timepoint (Supplementary Table 1). Using non-supervised methods and manual tests we settled on 7 clusters to describe the data (Fig. 1f, g). As expected, the data set contained a large number of cell wall-related genes and hardly any cell cycle-related genes. When looking at the gene ontology (GO) annotations, we observed terms linked to auxin signaling and lateral root development (cluster 2 and 5), whereas terms related to lipid transport and fatty acid metabolism were enriched in clusters 3 and 5. The fact that we observed in general little GO terms related solely to auxin signaling and lateral root development is most likely due to our unique experimental design, providing a previously undescribed auxin-response profile focused on a specific, differentiated cell type. To substantiate this impression, we compared the slr-1 versus CASP1pro::shy2-2/slr-1 data with the two other published data sets from transcriptome analyses dealing either with roots treated with auxin or microdissection of root sections after gravistimulation-mediated lateral root induction 14,15. Interestingly, there appeared to be little correlation between the differentially expressed genes in our data set and those in the data sets of Lewis et al., (2013) or Voß et al., (2015) (Extended Data Fig. 2b-c), confirming the unique and specific nature of our transcriptional profile. In order to identify novel genes involved in cell wall modification, as well as to confirm the validity of our transcriptional profile, we selected a wide range of genes possibly related to the observed endodermal responses, including genes linked to lignification, lipid transport and as well as several unknown genes showing particularly strong and high-confidence differential responses. We generated promoter-reporter lines to characterize their expression pattern during root development and lateral root formation. In a strong validation of our approach, 24 out of 27 of the selected genes were found to display auxin-regulated expression in the endodermis (Extended Data Fig. 3 and Supplementary Table 2). A selection of these candidates (with constitutive or induced expression during lateral root formation) is shown in Extended Data Figure 3c.
Many suberization-associated genes are responding to auxin in the endodermis
Since we were interested in possible cell wall modifying enzymes, we searched the list of differentially expressed genes for cell wall-associated functions. We found that many genes with functions attributed to cutin/suberin homeostasis showed highly dynamic, differential expression in our dataset (Extended Data Fig. 3e). Suberin deposition has been shown to be highly plastic and might be continuously turned over, both for adaptation to the soil environment and during lateral root development6–8. Because we still lack an understanding of suberin deposition and turnover in the apoplast, we decided to investigate whether some of the cell wall-related differentially expressed genes could be involved in this process. In particular, we were intrigued by the high number of differentially expressed GELPs in our dataset (Extended Data Fig. 3e), since members of this large family have been shown to be involved in cutin polymerization, but also to be able to degrade both cutin and suberin16–20. Thus, we decided to focus on the differentially regulated set of GELPs.
Concomitant suberin degradation and lateral root cap cuticle formation during lateral root formation
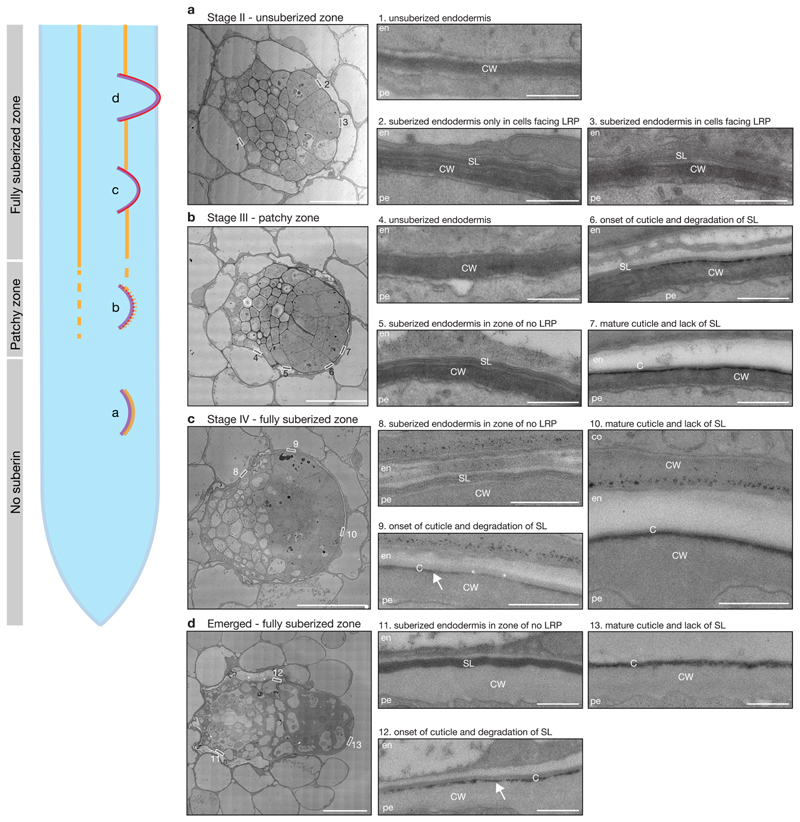
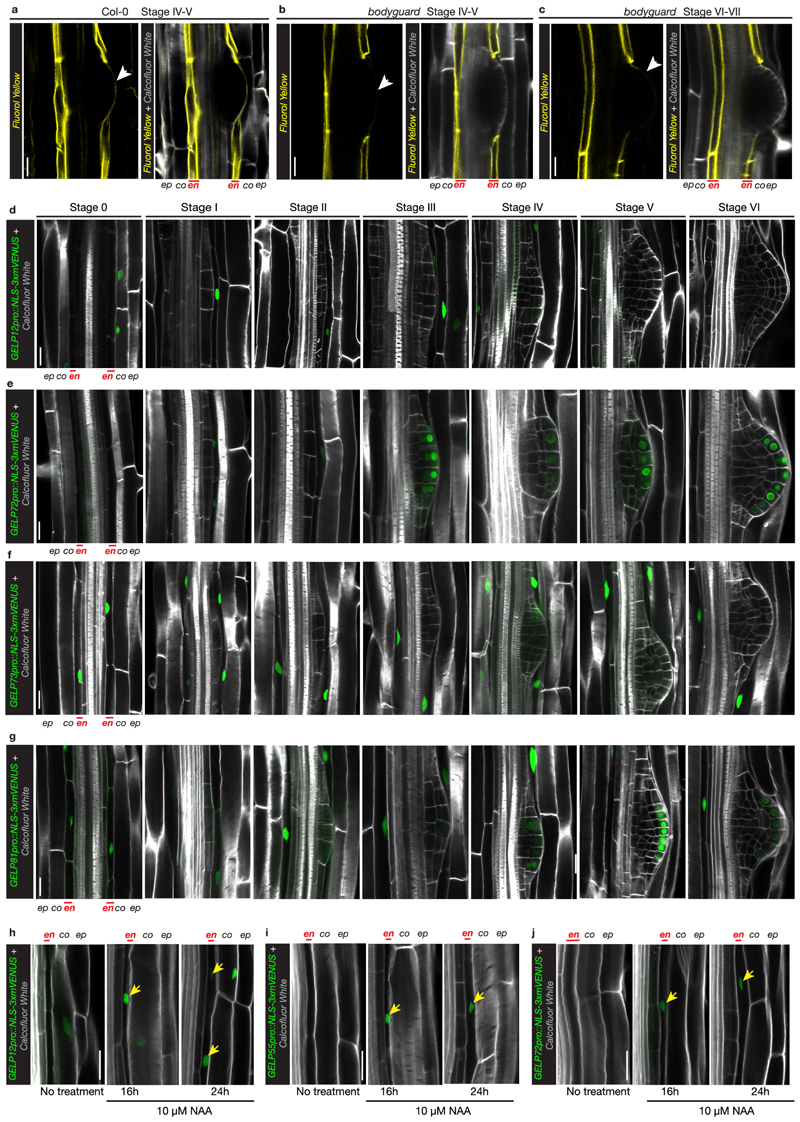
Fluorol Yellow (FY) staining reveals dynamic changes during lateral root formation. However, FY stains both suberin and cutin21 (Extended Data Fig. 6a), and it remains unclear if, how and at which stage endodermal suberin is degraded and a cutin-like structure is formed at the surface of the primordium. Therefore, to get a deeper insight into this process, we analyzed the dynamics of suberin and cutin during lateral root formation using transmission electron microscopy (TEM) (Fig. 2). Analyzing stage II lateral root primordia, which usually form in the unsuberized zone, we could detect suberin lamellae only in the endodermal cell walls facing the lateral root primordia, but not in those on the opposite side of the root (Fig. 2a). Stage III primordia are found in the patchy suberized zone of the root and we expectedly detected both suberized and non-suberized endodermal cells. At this stage, we started to distinguish the onset of the lateral root cap cuticle formation, accompanied by the disappearance of suberin in the endodermal cell walls overlying the primordium (Fig. 2b). In stage IV primordia, which are usually found in the fully suberized zone, we detected suberin deposition in all endodermal cells in zones without a primordium. At the same time, it was difficult to observe any suberin in endodermal cells facing the primordia. Indeed, it appeared as if suberin was degraded in coordination with the formation of the root cap cuticle (Fig. 2c). In fully emerged lateral roots, we could only detect the lateral root cap cuticle, whereas endodermal cells not in contact with the primordium still maintained their suberin lamellae (Fig. 2d). Thus, our analysis reveals that suberin is gradually degraded in cell walls of endodermal cells overlying the lateral root primordium, concomitant with the synthesis of a lateral root cap cuticle in the primordium as a protective coating21.
Figure 2. Suberin is degraded while the lateral root cap cuticle is established during lateral root formation.
a-d, TEM micrographs of root sections containing stage II (a), stage III (b), stage IV (c) lateral root primordia and emerged lateral root (d). The numbered boxed regions are shown as magnifications marked by the same number. Each experiment was repeated 3 times. The root schematic indicates the different stages of lateral root development shown in a-d. Purple = outline of lateral root primordium, yellow = suberin and red = cutin. En = endodermis, pe = pericycle, co = cortex, SL = suberin lamellae, C = lateral root cap cuticle, CW = cell wall. Arrows in panel 9 and 12 indicate the forming lateral root cap cuticle. Scale bars in (a-d) = 10 μm and scale bars in (1-12) panels = 1 μm.
Expression of auxin-repressed GELPs strongly correlates with endodermal suberization
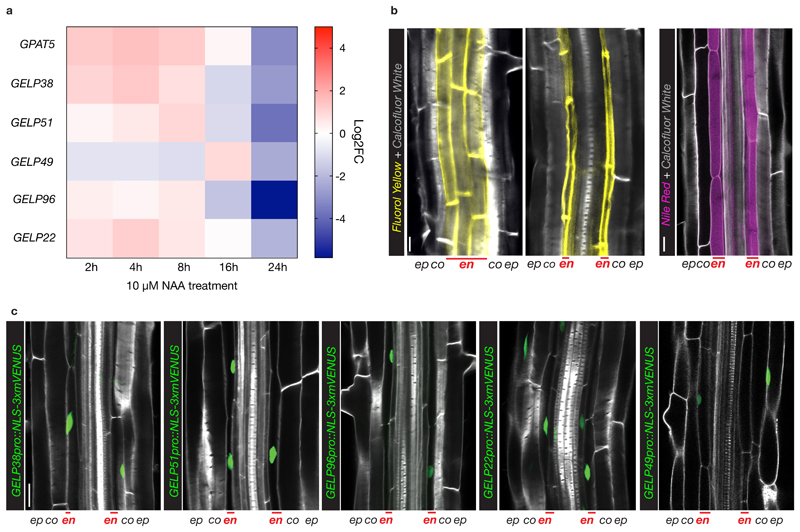
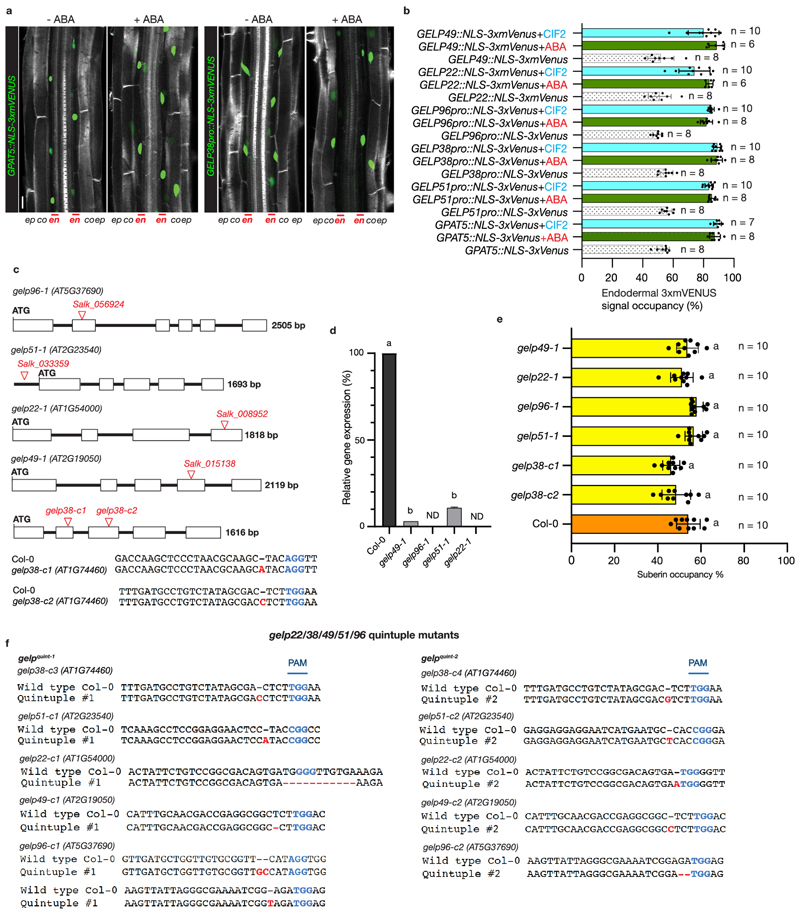
There are still no factors known to mediate suberin polymerization22. Currently, the strongest available interference with suberin biosynthesis in roots relies on either endodermis-specific interference with ABA or cytokinin signaling, artificial overexpression of a cutin-degrading enzyme or tissue-specific manipulation of phenylpropanoid production6,7,18,23. It was previously demonstrated that a member of the large family of GELP proteins20, CUTIN DEFICIENT 1 (CD1), has cutin in vitro synthase activity and CD1 loss-of-function mutants in tomato show partial defects in cuticle formation, but no equivalent evidence exists for suberin synthases. We observed a group of five GELPs (GELP22, 38, 49, 51 and 96) to be downregulated after prolonged auxin treatment (Fig. 3a). Since auxin treatment should down-regulate suberin biosynthetic enzymes during lateral root formation8, we speculated that the five downregulated GELPs might have a role in suberin biosynthesis. This idea was corroborated by the expression of transcriptional reporters for GELP22, 38, 49, 51 and 96. Clearly, GELPXpro::NLS-3xmVENUS reporter lines revealed endodermis-specific expression for GELP38, GELP51 and GELP96, and expression in endodermis and epidermis for GELP22 and GELP49 (Fig. 3c). Since treatment of Arabidopsis seedlings with ABA and CIF2 peptide results in a significant increase in suberin deposition and GPAT5 marker expression7,24,25, we further checked whether GELP22, 38, 49, 51 and 96 would be induced by ABA and CIF2 treatment. All GELP reporter lines were induced in response to these treatments and also expanded their expression domain into the cortex, similar to what has been reported for GPAT5 7 and (Extended Data Fig. 4a,b). Together, our data establishes a strong correlation between suberin biosynthesis and the expression pattern of these five GELPs.
Figure 3. A cluster of five, auxin repressed, GELPs is expressed in the differentiated endodermis.
a, Heatmap showing the differential expression of GPAT5 and GELP38, GELP51, GELP49, GELP96 and GELP22 during the time course of NAA treatment (10 μM). b, Representative image of staining of suberin lamellae in the endodermis using Fluorol Yellow (FY) (yellow) or Nile Red (magenta). c, Confocal images of root sections expressing transcriptional reporters for each of the GELPs mentioned in (a). NLS-3xmVENUS is shown in green, Calcofluor White (CW) staining of cell walls in gray. Each experiment was repeated 3 times. Scale bars in (b) and (c) = 25 μm.
Suberin deposition strongly requires auxin-repressed GELPs
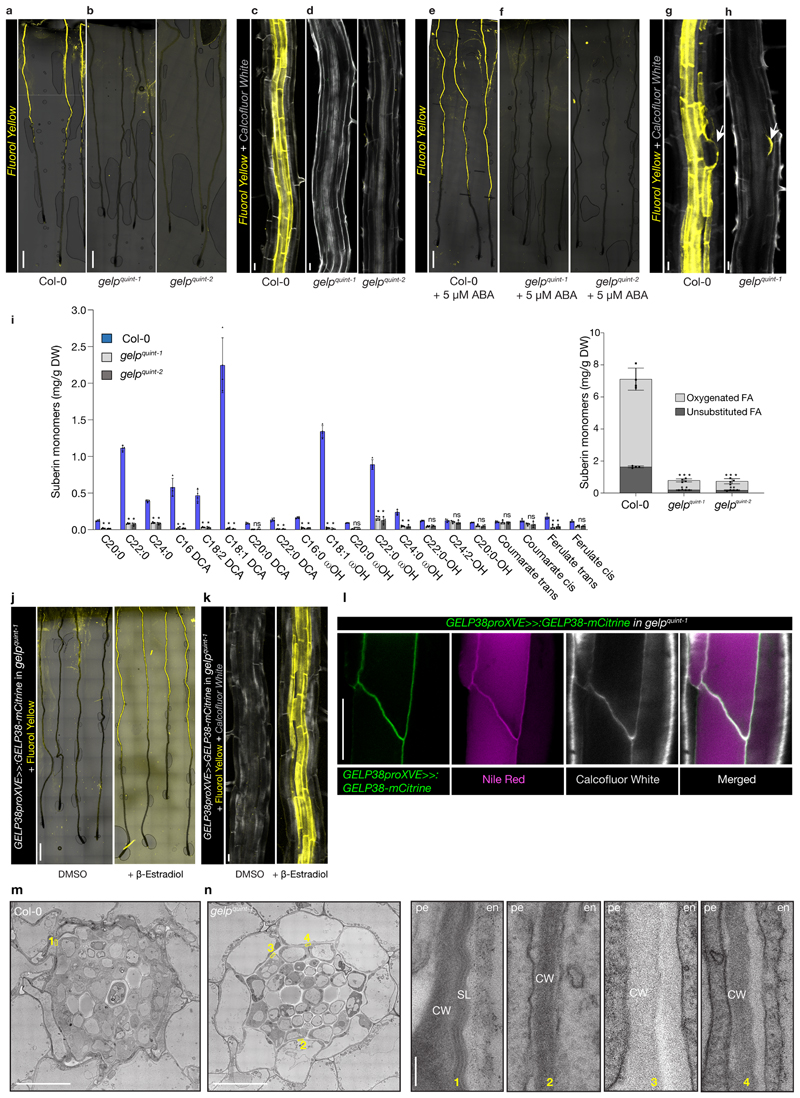
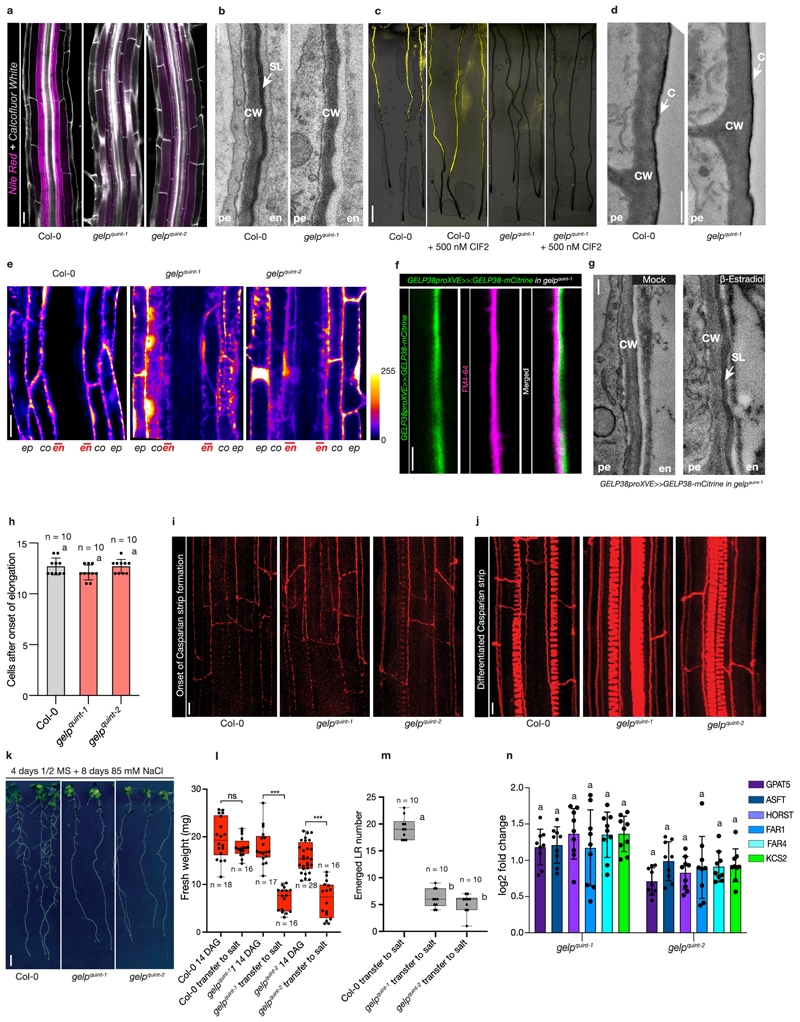
In order to establish a function for these GELPs in suberin biosynthesis, we collected available T-DNA insertion mutants and characterized these for differences in suberin deposition using FY or Nile Red staining, two fluorescent dyes that both stain suberin (Fig. 3b). In the absence of T-DNA insertion lines for GELP38, we generated two loss-of-function mutants using CRISPR/Cas9. None of the single mutants showed any significant difference in suberin occupancy in Arabidopsis roots compared to wild-type (Extended Data Fig. 4c-e). Using CRISPR/Cas9, we then generated two different allelic combinations of the five putative suberin biosynthesis-related GELPs: gelp22-c1/gelp38-c3/gelp49-c1/gelp51-c1/gelp96-c1 and gelp22-c2/gelp38-c4/gelp49-c2/gelp51-c2/gelp96-c2 (hereafter called gelp quint-1 and gelp quint-2) (Extended Data Fig. 4f). To test whether suberin levels in roots of the gelp quint-1 and gelp quint-2 mutants were affected, we stained roots of 5-day old plants with FY and Nile Red. Whereas suberin staining in wild-type resulted in the described patterns18, both quintuple mutants showed a complete absence of suberin staining (Fig. 4a-d and Extended Data Fig. 5a). ABA treatment is known to strongly enhance suberization both in endodermis and cortex7. Yet, even after ABA-induction no suberin deposition could be detected using FY staining in the roots of gelp quint-1 and gelp quint-2 (Fig. 4e, f). CIF2 peptide did also not enhance suberization in gelp quint-1 (Extended Data Fig. 5c). Since cutin and suberin can both be stained by FY, we investigated whether the lateral root cap cuticle was affected in roots of the gelp quint-1 mutant. We found that FY still stained the dome of the emerging lateral root, suggesting that the gelp quint mutants are specifically affected in suberization, leaving the lateral root cap cuticle intact (Fig. 4g,h). This was confirmed by TEM analysis (Extended Data Fig. 5d). Since endodermal suberin was shown to interfere with uptake from the apoplast, we used the fluorescein diacetate (FDA) penetration assay7 to test for the presence of functional suberin lamellae in the endodermis of gelp quint roots. Whereas FDA could only enter the epidermis and cortex in the suberized zone of wild-type roots, it was entering the endodermis of gelp quint roots, demonstrating absence, or a strong deficiency, of the endodermal suberin barrier (Extended Data Fig. 5e). In order to directly measure suberin levels, we performed a chemical analysis of the suberin content in roots of wild-type and gelp quint mutants. This revealed strong reductions in the amount of aliphatic suberin monomers in both gelp quint-1 and gelp quint-2, with nearly identical patterns in both allelic combinations. Dicarboxylic acids were nearly absent (98% reduction), while ω-hydroxy acids and fatty alcohols were reduced by ~ 90% and 50%, respectively (Fig. 4i). Ferulates were reduced by 70%, while coumarates showed only minor reductions. This resulted overall in a ~85% reduction in the amount of suberin monomers compared to wild-type roots (Fig. 4i), correlating well with the FY staining. Next, we complemented the suberin phenotype of the gelp quint-1 mutant by introducing an inducible GELP38proXVE>>GELP38-mCITRINE fusion into the gelp quint-1 mutant. This restored the stereotypical FY staining in roots, demonstrating functionality of the GELP38-mCITRINE protein (Fig. 4j,k). The complementation was further confirmed using TEM analysis (Extended Data Fig. 5g). Moreover, we could show that GELP38-mCITRINE is localized in the apoplast of the endodermis where suberin polymerization takes place (Fig. 4l and Extended Data Fig. 5f). We also tested whether the Casparian strip was unaffected in the gelp quint mutants and found both lignin staining with Basic Fuchsin and uptake of Propidium Iodide (PI) to be unaffected (Extended Data Fig. 5h-j). Thus, a cluster of five auxin-repressed GELPs is essential for normal suberin deposition in the endodermis, but does not affect lateral root cap cuticle formation. Finally, using TEM, we could readily detect suberin lamellae in endodermal cell walls of wild-type, but did not see any indication of lamellae formation in roots of gelp quint-1 (Fig. 4m,n and Extended Data Fig. 5b). Instead, we observed a layer of low electron density in the cell wall of the gelp quint-1 mutant (Fig. 4m,n and Extended Data Fig. 5b). This layer appears amorphous with no resemblance to the lamellar structures in wild-type. We speculate that this layer might be formed due to the accumulation of unpolymerized suberin monomers in the mutant cell wall. Together, our results strongly support an absence of suberin in the endodermis of the gelp quin1 mutant.
Figure 4. Suberin deposition strongly requires a cluster of auxin-repressed GELPs.
a, Confocal images of Col-0 seedling roots stained with FY. b, Confocal images of gelp quint-1 and gelp quint-2 seedling roots stained with FY. Note absence of FY staining. c and d, Close up image of root sections of Col-0 (c) or gelp quint-1 and gelp quint-2 (d) stained by FY for suberin and CW for cell walls. e and f, ABA-induced increased suberin deposition in Col-0 roots (e) whereas (f) shows ABA cannot induce suberin deposition in roots of gelp quint-1 and gelp quint-2 mutants. g and h, FY staining of Col-0 (g) and gelp quint-1 root (h) showing that the cuticle layer protecting emerging lateral roots appears not to be affected in the gelp quint-1 mutant. Each experiment in (a-h) was repeated at least 3 times. i, Chemical analysis of the suberin content in roots of wild-type and gelp quint-1 or gelp quint-2 reveals a ~85% decrease in total suberin monomers. Quantification of aliphatic and aromatic ester-bond suberin monomers isolated from 6-day-old roots of wild-type (Col-0) and gelp quint-1 and gelp quint-2 mutants. The graph shows the analysis of the principal suberin monomers and the inset shows the total monomers per genotype. Values represent the means ± SE, n = 4. Asterisks denote statistically significant differences to wild-type as determined by using ANOVA and Tukey test: *** p < 0.001; ** p < 0.01; * p < 0.05, ns = not significant. j-k, Induction of GELP38-XVEpro::GELP38-mCITRINE restores suberin deposition in the roots of gelp quint-1. l, Confocal image of a root expressing GELP38-XVEpro::GELP38:mCITRINE (green) after β-Estradiol treatment (5 μM) stained with Nile Red (magenta) for suberin and Calcofluor White (gray) for cell walls. m and n, TEM micrographs of root cross sections showing presence of suberin lamellae in wild-type, and absence of suberin lamellae in cross sections of gelp quint-1 roots. Note that the structure of the endodermis of the gelp quint-1 mutant is much better preserved compared to wild-type. The images in (j-n) are representatives of each experiment repeated 3 times. Scale bars in (a, b, e, f, j) = 500 μm, (c, d, g, h, k, l) = 25 μm. Scale bars in (m) and (n) for the whole root sections = 10 μm and for zoomed-in regions = 20 nm.
The gelp quint mutants show expected defects of a suberin-deficient mutant
Since it has been repeatedly demonstrated that roots with non-functional suberin barriers are more susceptible to elevated concentrations of salt, we also subjected 4 DAG seedlings to a mild salt stress (85 mM NaCl) for 8 days. Both gelp quint mutants were more affected by the salt stress compared to wild-type (Extended Data Fig. 5k-m). We observed less emerged lateral roots and the fresh weight of the shoot was also significantly reduced (Extended Data Fig. 5k-m). These observations again strongly support the absence of a functional suberin barrier in the gelp quint mutants. We finally checked whether the expression of known suberin biosynthesis-related genes was altered in the gelp quint mutants. None of them was differentially expressed in the gelp quint mutants (Extended Data Fig. 5n). This excludes the possibility that the observed absence of suberin would be due to an indirect feedback regulation of suberin biosynthesis and supports a direct role for the five GELPs in suberin polymerization in the apoplast.
Some members of auxin-induced GELPs have the capacity to degrade suberin
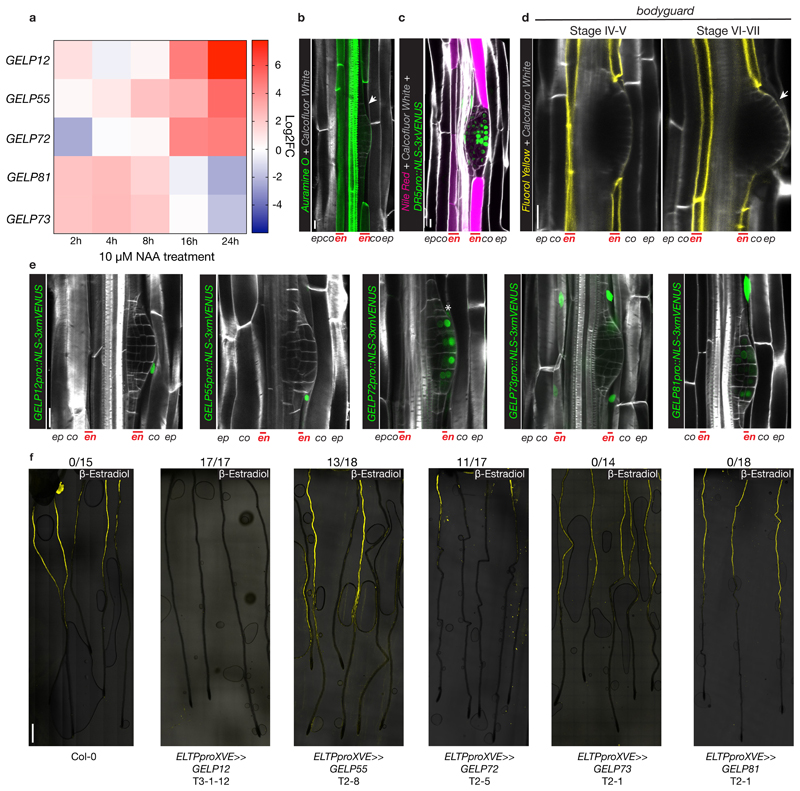
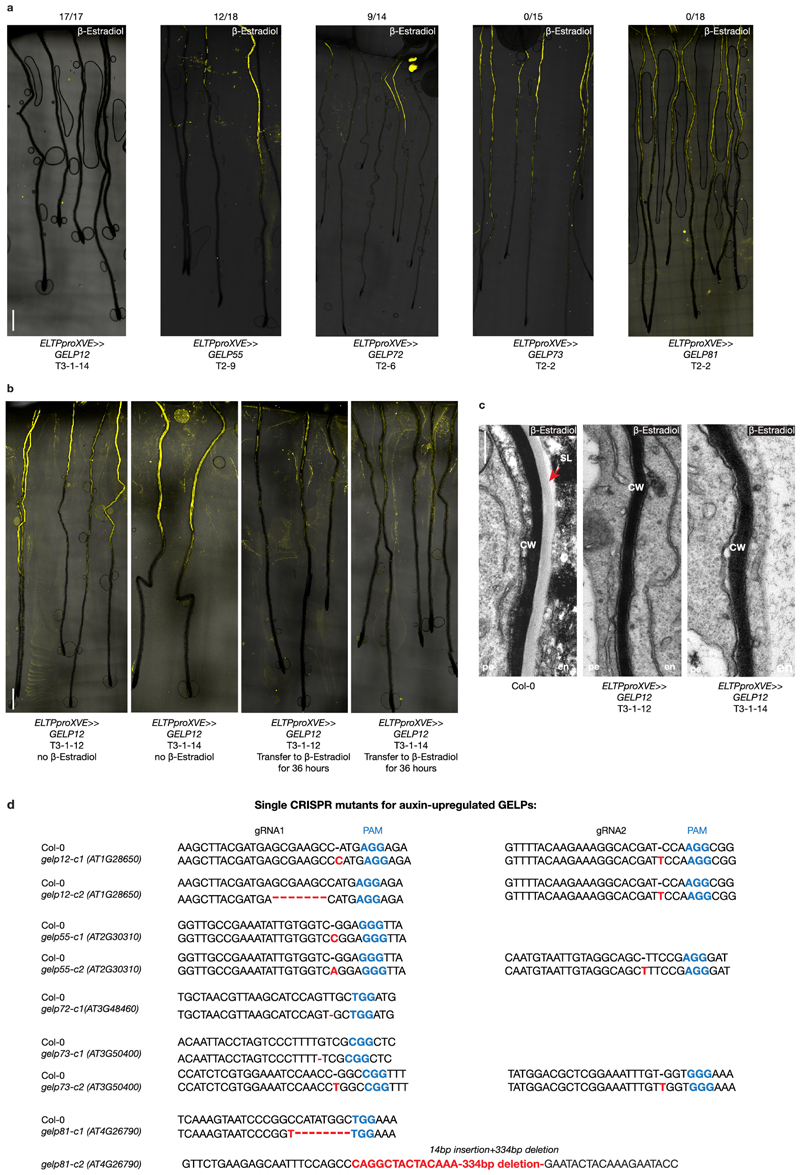
We also identified a group of five GELPs (GELP12, GELP55, GELP72, GELP73 and GELP81) that were induced by auxin (Fig. 5a). Our RNAseq data revealed that GELP12, GELP55 and GELP72 expression peak after 24 hours of NAA treatment, whereas GELP73 and GELP81 peak in expression after ~4 hours (Fig. 5a). Expression of GELP12, GELP55 and GELP72 reporters show specific induction in endodermis during lateral root formation, whereas GELP73 and GELP81reporters display endodermal expression already prior to lateral root formation (Fig. 5e and Extended Data Fig. 6f-g). Since we and others have demonstrated that the endodermal suberin gradually disappears, while a layer of cutin is formed at the primordium (Fig. 2 and Extended Data Fig. 6a)16,21. we hypothesized that, in contrast to the auxin-repressed suberin biosynthetic GELPs, the five auxin-induced GELPs could regulate removal of suberin. First, we confirmed removal of suberin by other fluorescent staining methods besides FY and found that both Auramine O and Nile Red can be used to visualize suberin in differentiated roots and both confirmed local degradation of suberin in cells overlying the lateral root (Fig. 5b, c). We also assessed suberin staining in the bodyguard mutant. This α/β-hydrolase was implicated in the establishment of the root cap cuticle21, but it was not clear whether it affects endodermal suberin accumulation. FY staining in bodyguard roots confirmed the effect on lateral root cap cuticle, but suberin degradation appears to be normal and even more easily observable in this mutant due to the lack of cuticle staining (Fig. 5d and Extended Data Fig. 6b, c). When characterizing the expression patterns of the five putatively suberin degrading GELPs, we observed three different expression patterns. GELP12 and GELP55 marker lines showed expression in the cortex or no signal in absence of lateral root formation, but were induced in endodermal cells overlying the lateral root from stage I to IV (Fig. 5e and Extended Data Fig. 6d). GELP72 was induced in endodermal cells overlying the lateral root primordium and from stage III onwards and was also expressed in the outer layer of the growing primordium (Fig. 5e and Extended Data Fig. 6e). Finally, GELP73 and GELP81 were already expressed in the endodermis prior to lateral root initiation and were induced in the overlying endodermal cells from stage I to IV (Fig. 5e and Extended Data Fig. 6f,g). In addition, from stage IV and onwards GELP81 was induced in the outer layer of the primordium similarly to GELP72 (Extended Data Fig. 6g). Confirming the RNAseq data, GELP12, GELP55 and GELP72 marker lines were induced by auxin in an endodermis-specific fashion (Extended Data Fig. 6h-j). In order to characterize a possible role in suberin degradation, we first undertook a gain-of-function approach by inducibly overexpressing each GELP in the endodermis and assess whether this causes suberin removal using FY staining. Indeed, inducible expression of GELP12, GELP55 and GELP72 in the endodermis caused a disappearance of FY staining, whereas no clear effect on FY staining was observed when inducing GELP81 and GELP73 (Fig. 5f and Extended Data Fig. 7a,c). We also performed a transfer experiment where we germinated the inducible lines with endodermis-specific expression of GELP12 on normal estradiol-free medium for four days to allow suberin formation and then transferred the seedlings to the plates containing estradiol for 36 hours. Also here, we observed a clear reduction in root suberin (Extended Data Fig. 7b). We then tested whether single loss-of-function mutants of each individual GELP affects lateral root formation (Extended data Fig. 7d and 8a-g). We performed root bending assays, in order to synchronously induce lateral roots and to quantify the progression of lateral root development at 18hr and 42hr after gravistimulation26,27. Among the three GELPs that can degrade suberin, only one (gelp72) showed a delayed lateral root development in both alleles and time points (Extended Data Fig. 8d-g). However, the single mutants of the GELP73 and GELP81 genes -that did not affect FY staining when overexpressed in the endodermis -also displayed a delayed lateral root emergence (Extended Data Fig. 8h-k). Thus, some auxin inducible GELPs are able to degrade suberin when overexpressed and some also display single mutant phenotypes during lateral root emergence. Thus, while our data makes auxin-induced GELPs strong candidates for suberin degradases during lateral root emergence and suggests a need for suberin degradation for proper lateral root emergence, further efforts in generating and analyzing multiple mutant combination of these GELPs will be needed to clearly establish their role and the requirement for suberin degradation during this process.
Figure 5. A cluster of auxin-induced GELPs is required for suberin degradation.
a, Heatmap showing the differential expression of GELP12, GELP55, GELP72, GELP81 and GELP73 during the NAA (10 μM) time course. b, Use of Auramine O (green) to visualize degradation of suberin in the cell wall of endodermal cells overlying a lateral root primordium. c, Use of Nile Red staining (magenta) to visualize suberin degradation in the cell wall of endodermal cells overlying a lateral root primordium. Auxin signaling is visualized by DR5pro::NLS-3xVENUS (green). d, FY staining in roots of bodyguard, demonstrating normal presence of suberin lamellae, whereas the cuticle layer surrounding the lateral root primordium is discontinuous (arrow). e, Confocal images showing the expression patterns of the isolated auxin-upregulated GELPs during lateral root formation. NLS-3xmVENUS signal in green and CFW staining of cell walls is in gray. Asterisk indicates endodermal signal. f, FY staining on roots of Col-0 treated with Estradiol results in normal suberin pattern, whereas inducible endodermis-specific overexpression of GELP12, GELP55 or GELP72 results in degradation of suberin highlighted by absence of FY signal. The overexpression of GELP73 and GELP81 results in a normal suberin pattern similar to wild-type. The numbers above the images indicate the ratios between the roots with absence of FY staining and total number of imaged roots. The images in (b-f) are representatives of each experiment repeated at least 3 times. Scale bars in (b, c, d) and (e) = 25 μm. Scale bar in (f) = 500 μm.
Discussion
Our data reveal that differentiated endodermal cells have a distinct auxin-mediated transcriptome. Mining this unique auxin-response dataset allowed us to identify the first suberin synthase candidates (Fig. 1 and Fig. 3) and a set of candidate suberin degradases (Fig. 5). Together these enzymes appear to form a module for regulating suberin plasticity in roots. Knocking out all five suberin synthases resulted in roots without detectable suberin that are hypersensitive to mild stress conditions. The gelp quint mutants now provide the plant community with a powerful tool to test the role of suberin and its plasticity during diverse environmental conditions. It will be exciting to test how the loss of suberin affects interactions with microbiome and susceptibility to pathogen attack1. The strong quintuple mutant and the identification of suberin degradase candidates will also greatly assist to further understand the mechanisms and role of suberin plasticity in plant elemental homeostasis7. The large GELP family in Arabidopsis and other plants certainly provides a rich, but still untapped genetic resource, to better understand how different cell wall modifications affect biological processes and interactions.
Methods
Plant Material
For all experiments, Arabidopsis thaliana ecotype Columbia (Col-0; wild-type) was used. Gene numbers, mutants, and transgenic lines used and generated in this study are described in Supplemental Experimental Procedures. The primers used for genotyping and qPCR-based verification of T-DNA lines are indicated in Supplementary Table 5.
Plant growth conditions
For all experiments, plants were germinated on solid half-strength Murashige and Skoog (MS) medium without addition of sucrose. Seeds were surface sterilized, sown on plates, incubated for 2 days at 4°C for stratification, and grown vertically in growth chambers at 22°C, under continuous light (100 μE). The microscopic analyses (FDA uptake, FY staining, PI uptake, confocal microscopy) were performed on 5 or 7-day-old seedlings.
Bending experiments
Seeds of wild-type Col-0 and gelp mutants were plated on half strength MS containing 120 × 120 x 17 mm square Petri dishes, stratified in the dark at 4°C for 2 days, and grown at 22 °C under constant light (100 μE). Lateral root stages were determined after plates with 4-day-old seedlings were rotated 90° degrees and grown for 18 h and 42 h for synchronized lateral root induction. After bending for 18hr and 42hr, the roots were cleared as described by15 and mounted in 50% glycerol. Determination of lateral root stage in the bent region was done using an upright microscope with differential interference contrast optics. Experiments were repeated three times and each replicate had at least 15 seedlings.
Generation of transgenic lines
CASP1pro::shy2-2 was crossed into slr-1 mutant to produce CASP1pro::shy2-2/slr-1 line. For generating marker lines and overexpression constructs, the In-Fusion Advantage PCR Cloning Kit (Clontech) and Gateway Cloning Technology (Invitrogen) were used. All constructs were transformed by heat shock into Agrobacterium tumefaciens GV3101 strain and then transformed into plants by floral dipping29. At least 10 independent transgenic lines were analyzed for expression patterns, and 1 line showing a representative signal and normal segregation was selected for further studies. For transcriptional reporters, the promoter regions were PCR-amplified from Col-0 genomic DNA and cloned into pDONRP4-P1R (www.thermofisher.com). The resulting plasmids were recombined together with pDONRL1-NLS-3xmVENUS-L230 and the destination vector pFR7m24GW or pFG7m24GW, containing the FastRed or FastGreen cassettes for transgenic seed selection respectively, to create the final PROMOTER::NLS-3xmVENUS expression clones. To be able to induce expression of individual GELPs in the endodermis, the corresponding pDONR221_L1-GELP-L2 clones were created. The resulting clones were recombined with the estradiol-inducible pDONR_P4-ELTPproXVE-P1R23 and destination vector pB7m24GW, to produce ELTPproXVE::GELP overexpression lines. To generate GELP38proXVE>>GELP38:mCitrine, the promoter region of GELP38pro was PCR-amplified from Col-0 genomic DNA and cloned into linearized p1R4-ML:XVE31 with KpnI enzyme by Infusion (Takara) cloning to produce the inducible GELP38proXVE promoter clone. The resulting clone was recombined together with pDONR221_L1-GELP38-L2, pDONR_R2-mCitrine-L3 and pFG7m34GW to produce GELP38proXVE>>GELP38:mCitrine construct. See Supplementary Tables 3 and 4 for details about the primers used for cloning. The spacers to customize sgRNA for SpCas9 were cloned using oligo annealing technique, then ligated into BbsI linearized, Gateway-entry plasmids32. For multiplex targeting of GELP genes, the six sgRNAs utilized were cloned into two vectors, each carrying 3 sgRNAs and co-transformed together into Col-0 ecotype. The primers used for generating single and multiple CRISPR/Cas9 mutants are indicated in the Supplementary Table 7.
Hormonal treatments
Abscisic acid (ABA) was stored as a 50 mM stock solution in methanol. When seedlings were subjected to short-term 10 μM ABA treatment, the transfer was done when the seedlings were 4-days-old. β-Estradiol was prepared as 100 mM stock in DMSO. In case of β-Estradiol treatment, the seedlings were directly germinated on the media containing 5 μM Estradiol. For salt experiments, the seedlings were grown on half-strength MS medium and transferred to 85mM NaCl for 10 days. In case of auxin (NAA) treatment for RNA-seq experiments, the seedlings were first grown on half-strength MS medium and then transferred to 10 μM NAA for 4, 8, 16 and 24 hours. At each time point the, the shoots and the root tips were removed.
Chemical analysis of suberin
Chemical analysis of suberin was performed on six-day old seedlings. Prior to analysis we confirmed that the used growth conditions did not affect the phenotype of gelp quint-1 and gelp quint-2 via FY staining. We used the protocol for the determination of ester-bond lipids as described by Berhin et al. (2019). In brief, 200 mg of seeds were grown on nylon mesh (200 mm pore size). After six days, the roots were shaved off after flash freezing and extracted in isopropanol/0.01% butylated hydroxytoluene (BHT). They were then delipidized three times (1 h, 16 h, 8 h) in each of the following solvents, i.e., chloroform-methanol (2:1), chloroform-methanol (1:1), methanol with 0.01% BHT, under agitation before being dried for 3 days under vacuum. Depolymerization was performed by base catalysis 33. Briefly, dried plant samples were transesterified in 2 mL of reaction medium. 20 mL reaction medium was composed of 3 mL methyl acetate, 5 mL of 25% sodium methoxide in dry methanol and 12 mL dry methanol. The equivalents of 5 mg of methyl heptadecanoate and 10 mg of ω-pentadeca-lactone/sample were added as internal standards. After incubation of the samples at 60 °C for 2 h 3.5 mL dichloromethane, 0.7 mL glacial acetic acid and 1 mL 0.9% NaCl (w/v) Tris 100 mM pH 8.0 were added to each sample and subsequently vortexed for 20 s. After centrifugation (1500 g for 2 min), the organic phase was collected, washed with 2 mL of 0.9% NaCl, and dried over sodium sulfate. The organic phase was then recovered and concentrated under a stream of nitrogen. The resulting cutin monomer fraction was derivatized with BFTSA/pyridine (1:1) at 70 °C for 1 h and injected out of hexane on a HP-5MS column (J&W Scientific) in a gas chromatograph coupled to a mass spectrometer and a flame ionization detector (Agilent 6890N GC Network systems). The temperature cycle of the oven was the following: 2 min at 50 °C, increment of 20 °C/min to 160 °C, of 2 °C/min to 250 °C and 10 °C/min to 310 °C, held for 15 min. 3 independent experiments were performed with 4 replicates for each genotype, respectively, and a representative dataset is presented. The amounts of unsubstituted C16 and C18 fatty acids were not evaluated because of their omnipresence in the plant and in the environment.
Fluorescence Microscopy
Confocal laser-scanning microscopy images were obtained using either a Zeiss LSM 880 (with Zen 2.1 SP3 Black edition), Leica SP8 (with LasX 3.1.5.16308) or Leica SP8-MP (with LasX 3.5.6.21594) microscopes. For green and red fluorophores, the following excitation and detection windows were used: mVENUS/GFP/FY/FDA 488 nm, 500-530 nm; mCITRINE 496 nm, 505-530 nm; PI 520 nm, 590-650 nm; Calcofluor White 405nm, 430-485 nm; Basic Fuchsin/Nile Red 561nm, 600-630 nm. For multiphoton microscopy the following excitation and detection settings were used: mVENUS/GFP/FY/Calcofluor White 960 nm, 435-485 nm (Calcofluor White) and 500-550 nm (mVENUS/GFP/FY). Methods for imaging the CS lignin and PI penetration were previously described25,28. For visualization of FDA transport, chambered cover glasses (Thermo Scientific), were used where the roots were covered with a slice of agar and time lapses were made right after the application of FDA.
Methanol-based Fluorol Yellow staining of suberin in combination with Calcofluor White
For most experiments suberin lamellae were observed in 5 or 7-day-old roots using Fluorol Yellow (FY 088, SANTA CRUZ BIOTECHNOLOGY) staining. Seedlings were incubated in methanol at room temperature for at least three days, stained with FY 088 (0.01%, methanol) for 1 hour at room temperature, rinsed in methanol and counterstained with aniline blue (0.5%, methanol) at room temperature for 1 hour in darkness, washed, and visualized using 1-well chambered cover glass (ThermoFisher Scientific, Catalog Nr. 155361). In order to combine with Calcofluor White for cell wall staining, the seedlings were incubated first in Calcofluor White solution (0.1%, in methanol), for three days and stained with FY as described above.
Electron Microscopy
For chemical fixation, plants were fixed in glutaraldehyde solution (EMS, Hatfield, PA) 2.5% in phosphate buffer (PB 0.1 M [pH 7.4]) for 1h at RT and post fixed in a fresh mixture of osmium tetroxide 1% (EMS) with 1.5% of potassium ferrocyanide (Sigma, St. Louis, MO) in PB buffer for 1h at RT. The samples were then washed twice in distilled water and dehydrated in ethanol solution (Sigma, St Louis, MO, US) at graded concentrations (30% - 40 min; 50% - 40 min; 70% - 40 min; 100% - 2x1h). This was followed by infiltration in Spurr resin (EMS, Hatfield, PA, US) at graded concentrations (Spurr 33% in ethanol - 4h; Spurr 66% in ethanol - 4h; Spurr 100% - 2x8h) and finally polymerized for 48h at 60°C in an oven. For the multiple mutant, ultrathin sections of 50 nm thick were cut transversally at 2 mm below the hypocotyl-root junction, using a Leica Ultracut (Leica Mikrosysteme GmbH, Vienna, Austria), picked up on a copper slot grid 2x1mm (EMS, Hatfield, PA, US) coated with a polystyrene film (Sigma, St Louis, MO, US). For lateral roots, ultrathin sections of 50 nm thick were cut longitudinally (transversally from main root). For High-Pressure freezing, plants were fixed in glutaraldehyde solution (EMS, Hatfield, PA) 2.5% in phosphate buffer (PB 0.1 M [pH 7.4]) for 1h at RT and post-fixed in a fresh mixture of osmium tetroxide 1% (EMS) with 1.5% of potassium ferrocyanide (Sigma, St. Louis, MO) in PB buffer for 1h at RT. The samples were then washed twice in distilled water before a High-Pressure Freezing step (HPF). For the High Pressure Freezing, 2mm long root pieces were cut below the hypocotyl junction region, and then placed in an aluminum planchet of 3mm in diameter with a cavity of 0.2 mm (Art.241, Wohlwend GmbH, Sennwald, Switzerland) filled with Hexadecene (Merck KGaA, Darmstadt, Germany) covered with a tap planchet (Art.353, Wohlwend GmbH, Sennwald, Switzerland) and directly high-pressure frozen using a High-Pressure Freezing Machine HPF Compact 02 (Wohlwend GmbH, Sennwald, Switzerland). The samples were then dehydrated and infiltrated with resin at cold temperature using the Leica AFS2 freeze substitution machine (Leica Mikrosysteme GmbH, Vienna, Austria) with the following protocol: Dehydration in 100% Acetone (Sigma, St Louis, MO, US) at graded temperature (-90°C -10h; from -90°C to -60°C in 2h; -60°C for 8h; from -60°C to -30°C in 2h; -30°C -3h.) This was followed by infiltration in Spurr resin (EMS, Hatfield, PA, US) at graded concentration and temperature (30% -10h from -30°C to 0°C; 66% -10h from 0°C to 20°C; 100% -2X 10h at 20°C) and finally polymerized for 48h at 60°C in an oven. Ultrathin sections of 50 nm thick were cut transversally to the root, using a Leica Ultracut (Leica Mikrosysteme GmbH, Vienna, Austria), picked up on a copper slot grid 2x1mm (EMS, Hatfield, PA, US) coated with a polystyrene film (Sigma, St Louis, MO, US). Micrographs and panoramic were taken with a transmission electron microscope FEI CM100 (FEI, Eindhoven, The Netherlands) at an acceleration voltage of 80kV with a TVIPS TemCamF416 digital camera (TVIPS GmbH, Gauting, Germany) using the software EM-MENU 4.0 (TVIPS GmbH, Gauting, Germany). Panoramic were aligned with the software IMOD34.
RNA-seq experiments
Seeds were surface sterilized, sown on plates, incubated 2 days at 4°C for stratification, and grown vertically in growth chambers at 22°C, under continuous light (100 μE) for 6 days. For each biological replicate (3 in total) 60 seedlings from each genotype were transferred to plates (20 seedlings per plate) containing ½ MS medium supplemented with 10 μM NAA and transferred back into the growth chamber. After the desired incubation period (2, 4, 8, 16 and 24hrs) seedlings were harvested after removal of the root apical meristem (~3mm) and the shoot including hypocotyl and snap frozen in liquid nitrogen. RNA was extracted using a Trizol-based method. After RNase-free DNase (www.qiagen.com) treatment, RNA was cleaned-up using a RNeasy mini-elute kit (www.qiagen.com). RNA-seq libraries were prepared as described 35. In brief, RNA quality was assessed on a Fragment Analyzer (Advanced Analytical Technologies, Inc., Ankeny, IA, USA). RNA-seq libraries were prepared using 1000 ng of total RNA and the Illumina TruSeq Stranded mRNA reagents (Illumina; San Diego, California, USA) on a Sciclone liquid handling robot (PerkinElmer; Waltham, Massachusetts, USA) using a PerkinElmer-developed automated script. Cluster generation was performed with the resulting libraries using the Illumina TruSeq SR Cluster Kit v4 reagents and sequenced on the Illumina HiSeq 2500 using TruSeq SBS Kit v4 reagents. Sequencing data were processed using the Illumina Pipeline Software version 2.2.
RNA-seq data processing and analysis
Data processing was performed by the Lausanne Genomic Technologies Facility using their in-house RNA-seq pipeline. Data analysis was done using an in-house RNA-seq pipeline that performed the following steps. Quality controls were applied for cleaning data for adapters and trimming of low-quality sequence ends. Cleaned data was aligned and read counts computed using two methods: STAR36 + HTSeq37 and STAR + RSEM38. First method generates gene counts and the second method generates isoform counts. TAIR10 genome and Ensembl 21 annotation were used. Additional quality controls were performed using R for inspecting the sample counts summary, pairwise sample correlations, clustering and sample PCA. Statistical analysis was performed for genes and isoforms with the Bioconductor package EdgeR (R version 3.4.0) for normalization and limma (R version 3.18.2) for differential expression. Two types of statistical tests were applied depending on the contrast model tested. A moderated t-test was used for each pairwise comparison in group t0 and group slr-1 vs CASP1pro::shy2-2/slr-1. A moderated F-test was used for each time course model and their interaction. The result files contain one row per gene or transcript. Adjusted p-values have been computed for each comparison by the Benjamini-Hochberg method, controlling for false discovery rate (FDR). Genes were considered significant in further analysis if the adjusted P-value was equal or below 0.05 and the log2 fold change was ≥ 1. Further analysis has been conducted using R (v. 3.4.1). Heatmaps were generated using ComplexHeatmap39 (v1.14.0) using pearson distance, and “average” for clustering. Non-supervised clustering of genes using kmeans (factoextra v1.0.4, https://github.com/kassambara/factoextra) suggested 3 clusters as optimal together. This represented a low resolution and we looked into clusters of size 4-8 which contained slightly lower silhouette values. After testing manually multiple cluster suggestions we settled onto 7 inferred clusters based on the biologically most sensible separation. GO analysis was conducted using the package topGO [v. 2.28.0, weight01 algorithm;40]. GO annotations were obtained through org.At.tairGO (version 3.4.1). For the comparison with Lewis et al. (2013), the published series matrix file was obtained from the GEO archive and the differential gene-expression analysis repeated in order to obtain the expression of all genes. Results were compared to their published table of differentially expressed genes and found to be highly similar. A z-score based on the logFC value was calculated for both, our data-sets and the re-analysed Lewis et al. (2013) data to make the different sets more comparable. For the comparison with Voß et al. (2015) we used directly the published table which included all expressed genes and calculated z-scores for the different time-points. We kept T0, T6, T9, T15 and T24 which resulted in 7145 differentially regulated genes with similar cut-off of p<0.05 and a fold-change of 2.
qPCR analysis
For qPCR quantifications, the plants were grown on plates with half-strength MS medium covered with mesh. In case of quintuple gelp mutants, only root parts (around 100 mg) were collected and total RNA was extracted using a Trizol-adapted ReliaPrep RNA Tissue Miniprep Kit (Promega). For verifying the transcript level in single T-DNA lines, RNA extraction from whole seedlings was performed. Reverse transcription was carried out with PrimeScript RT Master Mix (Takara). All steps were done as indicated in manufacturer’s manual. The qPCR reaction was performed on an Applied Biosystems QuantStudio3 thermocycler using a MESA BLUE SYBR Green kit (Eurogentech). All transcripts were normalized to ADAPTOR PROTEIN-4 MU-ADAPTIN, AP4M (AT4G24550) expression. All primers used for qPCR are indicated in Supplementary Table 6.
HRM analysis of CRISPR mutants
HRM method was employed to screen for the mutants generated using CRISPR/Cas9-based method. Genomic DNA of selected Cas9-free, T2 generation plants, was extracted using CTAB DNA extraction method. The qPCR reaction was performed on Applied Biosystems QuantStudio3 thermocycler using a MeltDoctor HRM Master Mix, according to manufacturer’s indications (Applied Biosystems). HPLC-purified primers were used to generate an amplicon of around 200 base pairs. The results were analyzed using High Resolution Melt Software v3.1 (Thermo Fisher Scientific). The selected candidates were verified by sequencing. Primers used for amplification and sequencing the potential mutation sites are indicated in Supplementary Tables 8 and 9.
Quantification and Statistical Analysis
For quantifying the FY occupancy, confocal images were analyzed with the Fiji package (Version 2.0.0-rc-69/1.52p (build: 269a0ad53f) http://fiji.sc/Fiji)41. Contrast and brightness were adjusted in the same manner for all images. The suberized regions of the roots were measured together with total root lengths to determine the percentage of suberin occupancy. All statistical analyses were done with the GraphPad Prism software version 9.0.0 (86) (https://www.graphpad.com/) or using the R package [version 3.5.1] (http://www.r-project.org). One-way ANOVA was performed, and two-sided t-test was subsequently used as a multiple comparison procedure. For the analysis of lateral root development using the bending assay, we used a Pearson’s χ 2 test. Details about the statistical approaches used can be found in the figure legends. The data are presented as mean ± SD or ±SE where indicated, and “n” represents number of plant roots. Each experiment was repeated at least 3 times.
Extended Data
Extended Data Fig. 1. SHY2pro::NLS-3xmVENUS dynamics in Col-0 (a), slr-1 (b) and CASP1pro::shy2-2/slr-1 treated with NAA.
a. Maximum image projections of roots expressing SHY2pro::NLS-3xmVENUS in Col-0 (a), slr-1 (b) and CASP1pro::shy2-2/slr-1 (c) after 2, 4, 8, 16 and 24 hours of NAA treatment. Yellow dots indicate SHY2pro::NLS-3xmVENUS signal in the endodermis. The images in are representatives of each experiment repeated 3 times. Scale bar in (a) = 50 µm.
Extended Data Fig. 2. Differentiated endodermal cells have a distinct transcriptional response to auxin.
a, Confocal images of a root expressing SLRpro::CITRINE:SYP122 confirming that SLR is expressed in the epidermis, cortex, pericycle and weakly in the stele, but not in the endodermis (indicated by asterisks). The images are representatives of the experiment repeated 3 times. b and c, Comparison of the current dataset (Ursache) with the data set of Lewis et al., (2013). b, Heatmap showing that both datasets cluster separately and do not have significant overlap, which is confirmed by the analysis of correlation between the two data sets (c). d and e, Comparison of the current dataset (Ursache) with the data set of Voß et al., (2015). d, Heatmap showing that both datasets cluster separately and do not have significant overlap, which is confirmed by the analysis of correlation between the two data sets (e). p values in c and e are derived from a Pearson correlation test, are two-sided t-test values and not corrected for multiple testing. The corresponding cut-points (*/**/***) represent here p < 0.05, p < 0.01 and p < 0.001. The r value measures the correlation between 2 sets of data. The p-value states whether there are enough observations to believe that an observed correlation is not appearing by chance. Scale bar = 25 µm.
Extended Data Fig. 3. A high number of differentially regulated genes are expressed in the endodermis.
a, Confocal images of GELP12pro::NLS-3xmVENUS expression in different genetic backgrounds, showing repression in slr-1 under control conditions. b, Auxin treatment (10 µM NAA, 16hrs) results in induction of GELP12pro::NLS-3xmVENUS expression in the endodermis of slr-1 roots. NLS-3xmVENUS signal is shown in green and CFW staining of cell walls is shown in grey. c, Confocal images of roots expressing transcriptional markers of candidate genes differentially expressed between slr-1 and CASP1pro::shy2-2/slr-1 roots and showing specific expression in the endodermis. d, Confocal images showing xylem-specific expression of LAC2pro::NLS-3xmVENUS. e, Heatmap showing the expression dynamics of suberin-related genes significant differentially expressed. NLS-3xmVENUS signal is shown in green, CFW staining of cell walls in gray and cell wall staining by PI in red. The images in (a-d) are representatives of each experiment repeated at least 3 times. Scale bars = 20 µm.
Extended Data Fig. 4. Expression of suberin biosynthesis-related GELPs is induced by ABA and CIF2 treatment.
a, Confocal images showing effect of ABA treatment (1 µM, 24hr) on the expression domain of GPAT5pro::NLS-3xmVENUS and GELP38pro::NLS-3xmVENUS in Arabidopsis roots. The images are representatives of the experiment repeated 3 times. b, Quantification of the effect of ABA (1 µM, 24hr) and CIF2 peptide (500 nM, 24h) treatment on the expression of suberin biosynthesis-related GELPs identified as being repressed by auxin treatment. c, Schematic representation of the different single mutants of the suberin biosynthesis-related GELPs used in this study. d, qPCR results showing the relative gene expression in Col-0 control (100%) and T-DNA insertion lines of the suberin synthesis-related GELPs used in this study. The results are based on three biological replicates. The p value versus the Col-0 control for gelp49-1 is <0.000001 and for gelp51-1 is < 0.000001. ND, not detected. e, Quantification of suberin occupancy in the endodermis of the single mutants of the suberin biosynthesis-related GELPs using FY staining (n = 10 biologically independent samples). The p value versus the Col-0 control for gelp38-c1 is 0.1269, for gelp38-c2 is 0.2616, for gelp51-1 is > 0.9999, for gelp96-1 is 0.9385, for gelp22-1 is > 0.9999 and for gelp49-1 is > 0.9999. Different letters in (d) and (e) (p < 0.05) indicate statistically significant differences between means by ANOVA and two-sided t-test analysis. f, Schematic representation of the mutations in the gelp quint-1 and gelp quint-2 mutants. The mutations are indicated in red and the PAM sites in blue. Error bars in (b), (d) and (e) are SD. Scale bar in (a) = 25 µm.
Extended Data Fig. 5. Extended characterization of the phenotype of gelp quint mutants.
a, Nile Red staining of wild-type and gelp quint-1 and gelp quint-2 roots confirms the absence of suberin in the mutants. b, TEM micrographs of high-pressure frozen roots of wild-type and gelp quint-1 showing the absence of suberin lamella in the mutant. A region without lateral root primordia has been chosen to highlight the absence of suberin lamella. c, FY staining of CIF2 peptide treated Col-0 and gelp quint-1 seedlings showing absence of CIF2-mediated suberin deposition in gelp quint-1. d, TEM micrograph of high-pressure frozen roots of wild-type and gelp quint-1 showing that the lateral root cap cuticle is not affected in the gelp quint-1 mutant. e, Fluorescein di-acetate (FDA) uptake assay in wild-type roots showing a suberin mediated block of uptake at the level of the endodermis. f, Confocal image of a root expressing GELP38-XVEpro::GELP38-mCITRINE (green) in gelp quint-1 after b-Estradiol treatment (5 µM) stained with FM4-64 dye. g, TEM micrographs of roots of gelp quint-1 and the complementation by GELP38-XVEpro::GELP38-mCITRINE showing the complete recovery of endodermis suberin lamella. The images in (a-g) are representatives of each experiment repeated at least 3 times. h, Counting of PI-stained cells as a proxy for Casparian strip barrier in the roots of wild-type and gelp quint-1 and gelp quint-2 seedlings. All individual data points are plotted. No statistically significant difference was detected in using ANOVA and Bonferroni-adjusted paired two-sided t-test. The p value versus the Col-0 control for gelp quint-1 is 0.1680 and for gelp quint-2 is > 0.9999. i-j, Basic Fuchsin staining of the Casparian strip in early and differentiated endodermal cells of wild-type and gelp quint-1 and gelp quint-2 roots. k, Salt stress assay showing that gelp quint-1 and gelp quint-2 mutant seedlings are more sensitive to mild salt stress (85 mM NaCl) compared to wild-type. The images in (i-k) are representatives of each experiment repeated at least 3 times. l, Quantification of the effect of prolonged salt stress on the fresh weight of wild-type and gelp quint-1 and gelp quint-2 seedlings. All individual data points are plotted. The p value versus the Col-0 control for Col-0 transferred to salt is 0.7857, for gelp quint1 versus gelp quint1 transferred to salt is < 0.000001 and for gelp quint-2 versus gelp quint-2 transferred to salt is < 0.000001. m, Quantification of emerged lateral roots in wild-type and gelp quint-1 and gelp quint-2 mutants after 8 days of exposure to salt. All individual data points are plotted (n = 10). n, Quantification of the expression of known suberin biosynthesis-related genes in gelp quint-1 and gelp quint-2 mutants. Results are presented as fold-change compared to their expression levels in Col-0 (n = 9). No statistically significant difference was detected in using ANOVA and Bonferroni-adjusted paired two-sided t-test. For GPAT5, the p value versus Col-0 for gelp quint-1 is > 0.9999 and for gelp quint-2 is 0.4546, for ASFT the p values are > 0.9999 and > 0.9999; for HORST the p values are 0.1122 and > 0.9999; for FAR1 the p values are > 0.9999 and > 0.9999; for FAR4 the p values are 0.1467 and > 0.9999; for KSC2 the p values are 0.1136 and > 0.9999 correspondingly. Different letters in (m) (p < 0.001) and asterisks in (l) (p < 0.001) indicate statistically significant differences between means by ANOVA and two-sided t-test analysis. ns, not significant. For the boxplots in (l) and (m) the center depicts the median while the lower and upper box limits depict the 25th and 75th percentile respectively. Whiskers represent minima and maxima. Closed dots depict individual samples. Data in (h) and (n) are presented as mean +/-SD. Scale bars for (a), (d), (e), (f), (g), (h-j) = 25 µm. Scale bars for (b) and (c) = 1 µm, for (k) = 5 mm.
Extended Data Fig. 6. Auxin-upregulated GELPs show three distinct expression patterns.
a, FY staining of a Col-0 root at the site of lateral root emergence highlighting the presence of cuticle (indicated by arrow). b-c, FY staining of bodyguard mutant root at the site lateral root emergence. The absence of a proper cuticle is highlighted by the gap in FY staining. d-g, Confocal images showing the expression patterns of GELP12 (d), GELP72 (e), GELP73 (f) and GELP81 (g) during lateral root emergence. h-j, Confocal images showing the expression of GELP12 (h), GELP55 (i) and GELP72 (j) after 10 µM NAA treatment. NLS-3xmVENUS signal is in green and Calcofluor White staining of cell walls is in gray. The images are representatives of each experiment repeated at least 3 times. Scale bars = 25 µm.
Extended Data Fig. 7. Overexpression of three auxin-induced GELPs leads to suberin degradation.
a, FY staining on roots of Col-0 treated with b-Estradiol results in normal suberin pattern, whereas inducible endodermis-specific overexpression of GELP12, GELP55 or GELP72 results in degradation of suberin highlighted by absence of FY signal. The overexpression of GELP73 and GELP81 results in a normal suberin pattern similar to wild-type. b, FY staining of ELTPproXVE>>GELP12 lines germinated on normal 1/2MS medium for 4 days and transferred to the plates supplemented with 5 µM β-estradiol for 36 hours to observe the suberin degradation. c, TEM micrographs of ELTPproXVE>>GELP12 grown on plates with and without b-estradiol to highlight the absence of suberin lamella upon induction of GELP12 expression in endodermis. d, Schematic representation of the mutations in the auxin-upregulated single GELP mutants. The mutations are indicated in red and the PAM sites in blue. The images in (a-c) are representatives of each experiment repeated at least 3 times. Scale bars in (a) and (b) = 500 µm. Scale bars in (c) = 1 µm.
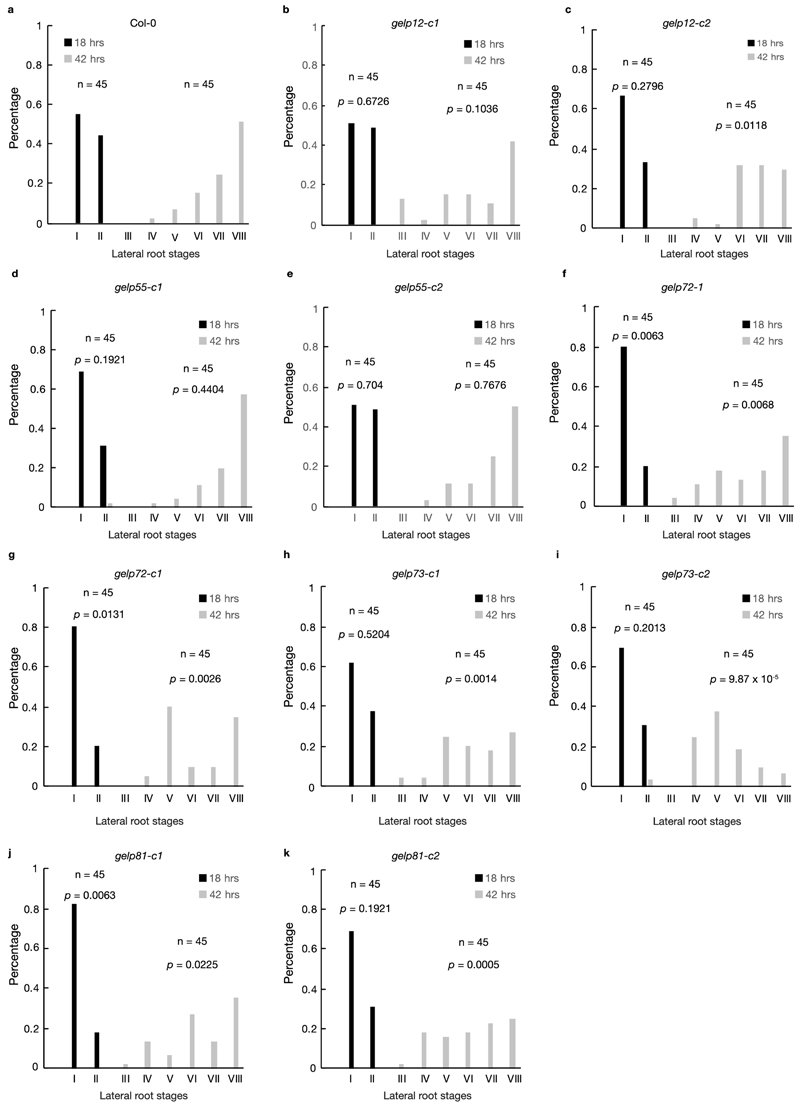
Extended Data Fig. 8. Some auxin-inducible GELPs facilitate lateral root emergence.
a-k, Gravistimulation-mediated induction of lateral root formation to functionally characterize the role of auxin-induced GELPs during lateral root formation. Staging of lateral root development was performed at 18hr and 42hr after gravistimulation. a, Col-0. b and c, gelp12-c1 and gelp12-c2. d and e, gelp55-c1 and gelp55-c2. f and g, gelp72-1 and gelp72-c1. h and i, gelp73-c1 and gelp73-c2. j and k, p values are indicated. A p value below 0.05 indicates a statistically significant difference as determined using a Pearson’s χ2 test. Experiments were repeated three times with a minimal of 15 seedlings per genotype and time point.
Supplementary Material
Acknowledgements
We would to thank Christopher Grefen and Marie Barberon for insightful discussions about GDSL-domain containing proteins, experimental approaches and stimulating discussions. We thank the Electron Microscopy Facility and Imaging Facility of the University of Lausanne and the Center of Microcopy and Image Analysis of the University of Zurich for excellent service and support. Work in the Geldner lab was supported by an ERC Consolidator Grant (GA-N: 616228-ENDOFUN), two consecutive SNSF grants (CRSII3_136278 and 31003A-156261). Robertas Ursache was supported by an EMBO Long-Term Fellowship (EMBO ALTF 1046-2015). Work in the Nawrath lab was supported by SNSF grant (310030_188672/1). Work in the Vermeer lab was supported by grants from the Swiss National Science Foundation (Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung; PP00P3_157524 and 316030_164086), the Netherlands Organisation for Scientific Research (Nederlandse Organ-isatie voor Wetenschappelijk Onderzoek; NWO 864.13.008) and support from the University of Neuchâtel.
Footnotes
Author contributions
R.U., N.G. and J.E.M.V. conceived, designed and coordinated the project. R.U., C.D.J.T.V, V.D.T., D.d.B., K.G., V.S., T.G.A. and J.E.M.V. performed all experimental work. E.S.S., S.C. and S.P. analysed NGS data. J.E.M.V. wrote the first draft of the manuscript. R.U. T.G.A. C.N. N.G. and J.E.M.V. revised the manuscript and all authors were involved in the discussion of the work.
Competing interests
The authors declare no competing interests.
Reporting Summary
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Data availability
All data to support the conclusions of this manuscript are included in the main text and supplementary materials. The full RNA-seq dataset was deposited in GEO (accession GSE153478, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153478).
The following T-DNA tagged and transgenic lines were used in this study: gelp49 (SALK_015138C); gelp51 (SALK_033359C); gelp96 (SALK056924C); gelp96 (SALK056924C) were requested from NASC centre; SHY2pro::NLS-3xmVENUS, CASP1pro::shy2-2, DR5::NLS-3xVENUS10; GPAT5pro::NLS-3xmVENUS28, slr-112 were described previously. The corresponding gene numbers are as follow: SLR, AT4G14550; SHY2, AT1G04240; ELTP, At2g48140; GPAT5, At3g11430; SYP122, At3g52400; BDG, AT1G64670; HORST, At5g58860; ASFT, At5g41040; FAR1, At5g22500; FAR4, At3g44540; KCS2, At1g04220; GELP12, AT1G28650; GELP55, AT2G30310; GELP72, AT3G48460; GELP81, AT4G26790; GELP73, AT3G50400; GELP49, AT2G19050; GELP51, AT2G23540; GELP96, AT5G37690; GELP38, At1G74460; GELP22, AT1G54000; GELP103, At5g45960; PER10, At1g49570; PER11, AT1G68850; PER23, AT2G38390; PER28, At3g03670; PER55, AT5G14130, PER59, At5g19890; LAC2, AT2G29130; LAC12, AT5G05390; LAC13, AT5G07130; LAC16, AT5G58910; UCC1, AT2G32300; PLIP3, AT3G62590; CASPL4B1, AT2G38480; CASPL1A1, AT1G14160; Cytochrome b561 and DOMON domain-containing protein At4g17280; Cytochrome b561 and DOMON domain-containing protein At5g47530.
References
- 1.Castrillo G, et al. Root microbiota drive direct integration of phosphate stress and immunity. Nature. 2017;543:513–518. doi: 10.1038/nature21417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Duran P, et al. Microbial Interkingdom Interactions in Roots Promote Arabidopsis Survival. Cell. 2018;175:973–983 e914. doi: 10.1016/j.cell.2018.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hassani MA, Duran P, Hacquard S. Microbial interactions within the plant holobiont. Microbiome. 2018;6:58. doi: 10.1186/s40168-018-0445-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Banda J, et al. Lateral Root Formation in Arabidopsis: A Well-Ordered LRexit. Trends Plant Sci. 2019;24:826–839. doi: 10.1016/j.tplants.2019.06.015. [DOI] [PubMed] [Google Scholar]
- 5.Stoeckle D, Thellmann M, Vermeer JE. Breakout-lateral root emergence in Arabidopsis thaliana. Curr Opin Plant Biol. 2018;41:67–72. doi: 10.1016/j.pbi.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 6.Andersen TG, et al. Tissue-autonomous phenylpropanoid production is essential for establishment of root barriers. bioRxiv. 2020 doi: 10.1101/2020.06.18.159475. [DOI] [PubMed] [Google Scholar]
- 7.Barberon M, et al. Adaptation of Root Function by Nutrient-Induced Plasticity of Endodermal Differentiation. Cell. 2016;164:447–459. doi: 10.1016/j.cell.2015.12.021. [DOI] [PubMed] [Google Scholar]
- 8.Li B, et al. Role of LOTR1 in Nutrient Transport through Organization of Spatial Distribution of Root Endodermal Barriers. Curr Biol. 2017;27:758–765. doi: 10.1016/j.cub.2017.01.030. [DOI] [PubMed] [Google Scholar]
- 9.Yadav V, et al. ABCG transporters are required for suberin and pollen wall extracellular barriers in Arabidopsis. Plant Cell. 2014;26:3569–3588. doi: 10.1105/tpc.114.129049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vermeer JE, et al. A spatial accommodation by neighboring cells is required for organ initiation in Arabidopsis. Science. 2014;343:178–183. doi: 10.1126/science.1245871. [DOI] [PubMed] [Google Scholar]
- 11.Tian Q, Uhlir NJ, Reed JW. Arabidopsis SHY2/IAA3 inhibits auxin-regulated gene expression. The Plant cell. 2002;14:301–319. doi: 10.1105/tpc.010283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukaki H, Tameda S, Masuda H, Tasaka M. Lateral root formation is blocked by a gain-of-function mutation in the SOLITARY-ROOT/IAA14 gene of Arabidopsis. The Plant journal : for cell and molecular biology. 2002;29:153–168. doi: 10.1046/j.0960-7412.2001.01201.x. [DOI] [PubMed] [Google Scholar]
- 13.Swarup K, et al. The auxin influx carrier LAX3 promotes lateral root emergence. Nat Cell Biol. 2008;10:946–954. doi: 10.1038/ncb1754. [DOI] [PubMed] [Google Scholar]
- 14.Lewis DR, et al. A Kinetic Analysis of the Auxin Transcriptome Reveals Cell Wall Remodeling Proteins That Modulate Lateral Root Development in Arabidopsis. The Plant cell. 2013 doi: 10.1105/tpc.113.114868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Voß U, et al. The circadian clock rephases during lateral root organ initiation in Arabidopsis thaliana. Nature communications. 2015;6:7641. doi: 10.1038/ncomms8641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bakan B, Marion D. Assembly of the Cutin Polyester: From Cells to Extracellular Cell Walls. Plants (Basel) 2017;6 doi: 10.3390/plants6040057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Girard AL, et al. Tomato GDSL1 is required for cutin deposition in the fruit cuticle. Plant Cell. 2012;24:3119–3134. doi: 10.1105/tpc.112.101055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Naseer S, et al. Casparian strip diffusion barrier in Arabidopsis is made of a lignin polymer without suberin. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:10101–10106. doi: 10.1073/pnas.1205726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Philippe G, et al. Ester Cross-Link Profiling of the Cutin Polymer of Wild-Type and Cutin Synthase Tomato Mutants Highlights Different Mechanisms of Polymerization. Plant Physiol. 2016;170:807–820. doi: 10.1104/pp.15.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeats TH, et al. The identification of cutin synthase: formation of the plant polyester cutin. Nat Chem Biol. 2012;8:609–611. doi: 10.1038/nchembio.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berhin A, et al. The Root Cap Cuticle: A Cell Wall Structure for Seedling Establishment and Lateral Root Formation. Cell. 2019;176:1367–1378 e1368. doi: 10.1016/j.cell.2019.01.005. [DOI] [PubMed] [Google Scholar]
- 22.Philippe G, et al. Cutin and suberin: assembly and origins of specialized lipidic cell wall scaffolds. Curr Opin Plant Biol. 2020;55:11–20. doi: 10.1016/j.pbi.2020.01.008. [DOI] [PubMed] [Google Scholar]
- 23.Andersen TG, et al. Diffusible repression of cytokinin signalling produces endodermal symmetry and passage cells. Nature. 2018;555:529–533. doi: 10.1038/nature25976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Doblas VG, et al. Root diffusion barrier control by a vasculature-derived peptide binding to the SGN3 receptor. Science. 2017;355:280–284. doi: 10.1126/science.aaj1562. [DOI] [PubMed] [Google Scholar]
- 25.Fujita S, et al. SCHENGEN receptor module drives localized ROS production and lignification in plant roots. EMBO J. 2020 doi: 10.15252/embj.2019103894. e103894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lucas M, Godin C, Jay-Allemand C, Laplaze L. Auxin fluxes in the root apex co-regulate gravitropism and lateral root initiation. J Exp Bot. 2008;59:55–66. doi: 10.1093/jxb/erm171. [DOI] [PubMed] [Google Scholar]
- 27.Péret B, et al. Auxin regulates aquaporin function to facilitate lateral root emergence. Nat Cell Biol. 2012;14:991–998. doi: 10.1038/ncb2573. [DOI] [PubMed] [Google Scholar]
- 28.Ursache R, Andersen TG, Marhavy P, Geldner N. A protocol for combining fluorescent proteins with histological stains for diverse cell wall components. Plant J. 2018;93:399–412. doi: 10.1111/tpj.13784. [DOI] [PubMed] [Google Scholar]
- 29.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 30.Gasperini D, et al. Multilayered Organization of Jasmonate Signalling in the Regulation of Root Growth. PLoS genetics. 2015;11 doi: 10.1371/journal.pgen.1005300. e1005300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Siligato R, et al. MultiSite Gateway-Compatible Cell Type-Specific Gene-Inducible System for Plants. Plant Physiol. 2016;170:627–641. doi: 10.1104/pp.15.01246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fauser F, Schiml S, Puchta H. Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 2014;79:348–359. doi: 10.1111/tpj.12554. [DOI] [PubMed] [Google Scholar]
- 33.Li-Beisson Y, et al. Acyl-lipid metabolism. The Arabidopsis book / American Society of Plant Biologists. 2013;11 doi: 10.1199/tab.0161. e0161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kremer JR, Mastronarde DN, McIntosh JR. Computer visualization of three-dimensional image data using IMOD. J Struct Biol. 1996;116:71–76. doi: 10.1006/jsbi.1996.0013. [DOI] [PubMed] [Google Scholar]
- 35.Jan M, Gobet N, Diessler S, Franken P, Xenarios I. A multi-omics digital research object for the genetics of sleep regulation. Sci Data. 2019;6:258. doi: 10.1038/s41597-019-0171-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–2849. doi: 10.1093/bioinformatics/btw313. [DOI] [PubMed] [Google Scholar]
- 40.Alexa A, Rahnenfuhrer J. topGO: Enrichment Analysis for Gene Ontology. R package version 2.38.1. 2019 [Google Scholar]
- 41.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data to support the conclusions of this manuscript are included in the main text and supplementary materials. The full RNA-seq dataset was deposited in GEO (accession GSE153478, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE153478).
The following T-DNA tagged and transgenic lines were used in this study: gelp49 (SALK_015138C); gelp51 (SALK_033359C); gelp96 (SALK056924C); gelp96 (SALK056924C) were requested from NASC centre; SHY2pro::NLS-3xmVENUS, CASP1pro::shy2-2, DR5::NLS-3xVENUS10; GPAT5pro::NLS-3xmVENUS28, slr-112 were described previously. The corresponding gene numbers are as follow: SLR, AT4G14550; SHY2, AT1G04240; ELTP, At2g48140; GPAT5, At3g11430; SYP122, At3g52400; BDG, AT1G64670; HORST, At5g58860; ASFT, At5g41040; FAR1, At5g22500; FAR4, At3g44540; KCS2, At1g04220; GELP12, AT1G28650; GELP55, AT2G30310; GELP72, AT3G48460; GELP81, AT4G26790; GELP73, AT3G50400; GELP49, AT2G19050; GELP51, AT2G23540; GELP96, AT5G37690; GELP38, At1G74460; GELP22, AT1G54000; GELP103, At5g45960; PER10, At1g49570; PER11, AT1G68850; PER23, AT2G38390; PER28, At3g03670; PER55, AT5G14130, PER59, At5g19890; LAC2, AT2G29130; LAC12, AT5G05390; LAC13, AT5G07130; LAC16, AT5G58910; UCC1, AT2G32300; PLIP3, AT3G62590; CASPL4B1, AT2G38480; CASPL1A1, AT1G14160; Cytochrome b561 and DOMON domain-containing protein At4g17280; Cytochrome b561 and DOMON domain-containing protein At5g47530.