Abstract
Crop breeding for resistance to pathogens largely relies on genes encoding receptors that confer race-specific immunity. Here we report the identification of the wheat Pm4 race-specific resistance gene to powdery mildew. Pm4 encodes a putative chimeric protein of a serine-threonine kinase and multiple C2-domains and transmembrane regions, a unique domain architecture among known resistance proteins. Pm4 undergoes constitutive alternative splicing generating two isoforms with different protein domain topologies that are both essential for resistance function. Both isoforms interact and localize to the endoplasmatic reticulum (ER) when co-expressed. Pm4 reveals additional diversity of immune receptor architecture to be explored for breeding and suggests an ER-based molecular mechanism of Pm4-mediated race-specific resistance.
Bread wheat (Triticum aestivum) sustains more than one third of humankind1. Around 5% of the total yield losses caused by wheat pathogens and pests is attributable to Blumeria graminis f. sp. tritici (Bgt), the causal agent of wheat powdery mildew2. Host resistance is crucial for controlling the disease and reducing pesticide dependency3. Race-specific resistance is the basis of host resistance in many wheat genotypes, where resistance (R) genes confer strong and mostly complete immunity to some but not all races of a pathogen species. The molecular identification of genetic components of R-mediated resistance contributes to improve disease resistance by tracking R genes with markers and by stacking them4. Moreover, resistance durability benefits from broader R gene pools, allowing more effective gene combination schemes5, by, for instance, combining different molecular modes of resistance6.
Many of the molecularly identified R genes in crops encode nucleotide-binding domain and leucine-rich repeat-containing (NLR) proteins that are intracellular immune receptors that recognize cytoplasmic pathogen-derived effectors7,8. Some wheat immune receptors active against rust pathogens have non-canonical architectures resulting from the fusion of additional domains to the NLR protein (NLR-ID): the wheat stripe rust genes Yr5, Yr7 and YrSP 9 encode proteins with an N-terminal zinc-finger BED domain and the YrU1 10 gene encodes a protein with N-terminal ankyrin-repeat and C-terminal WRKY domains. Although functionally not well characterized11, these integrated domains are believed to act as decoys of virulence effector targets to detect the pathogen, and ultimately, activate immune signalling12,13.
In addition to NLR or NLR-ID receptors, proteins localizing in the plasma membrane such as the Cf receptor-like proteins in tomato against the Cladosporium fulvum pathogen have also been shown to be products of race-specific R genes14. Furthermore, the wheat Stb6 gene encodes a wall-associated receptor kinase (WAK)-like protein15 conferring race-specific resistance against the fungus Zymoseptoria tritici by detecting the presence of a matching apoplastic effector16,17. Finally, tandem kinase-pseudokinases (TKP) have emerged as a new protein family involved in plant immunity18 and include barley and wheat rust resistance genes Rpg1 19, Yr15 18 and Sr60 20 as well as the wheat powdery mildew resistance gene Pm24 21. The diversity of molecular mechanisms resulting in gene-for-gene specificity observed in wheat-pathogen interactions makes the diverse wheat germplasm a promising genetic resource for the identification of novel molecular mechanisms resulting in plant immunity.
We report on cloning the wheat Pm4 race-specific resistance gene to powdery mildew, originally introgressed from tetraploid T. carthlicum 22. Constitutive alternative splicing of Pm4 generates two isoforms, both required for resistance, with different domain architectures forming an ER-associated complex revealing an additional and unique molecular basis for race-specific resistance mechanism in a major crop.
Results
The Pm4 gene provides race-specific resistance to a wide range of Bgt isolates
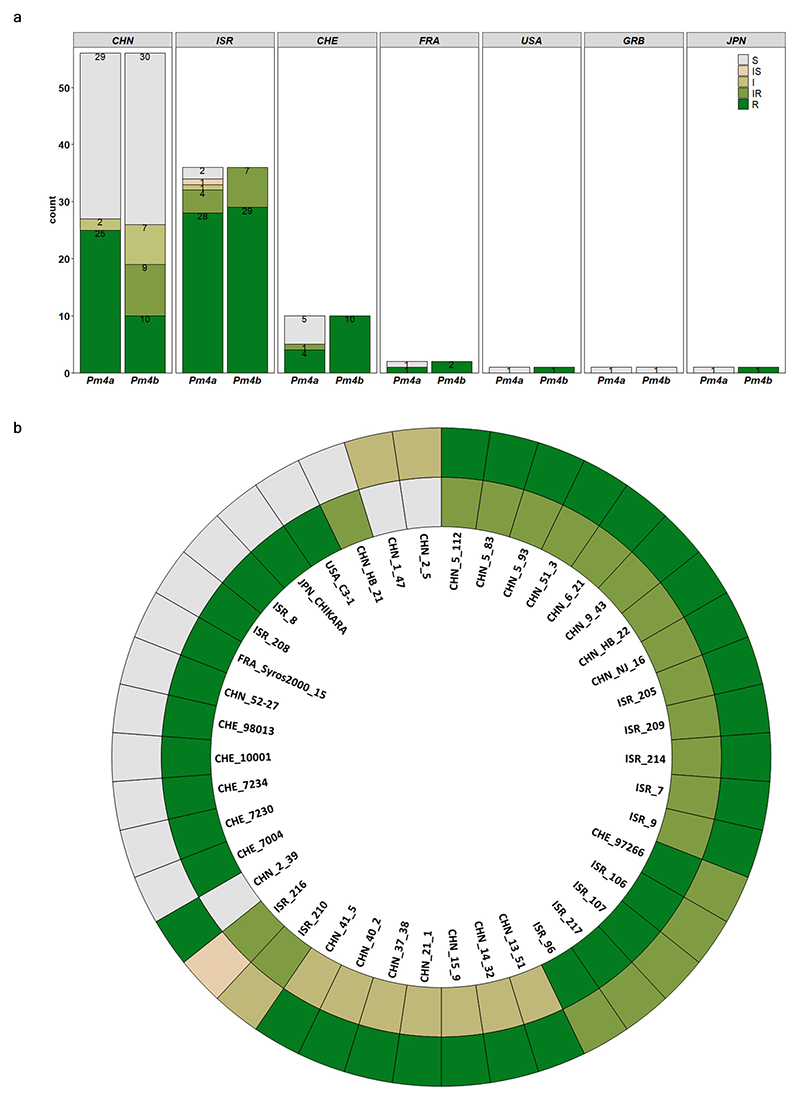
The near-isogenic genetic background of Fed-Pm4a 23 and Fed-Pm4b 22 wheat lines allowed the assessment of the resistance spectra of these two Pm4 alleles. Mildew resistance testing revealed a largely overlapping, yet distinct resistance spectrum (Supplementary Table 1). Both alleles conferred complete resistance to 37 (34.6%) Bgt isolates, mostly from China, Israel and Switzerland, whereas 28 (26.1%) of the Bgt isolates were virulent on both alleles (Extended Data Fig. 1a and Supplementary Table 1). The remaining 42 (39.3%) Bgt isolates showed different reactions on Pm4a and Pm4b, confirming the race-specific nature of the two resistance alleles (Extended Data Fig. 1b and Supplementary Table 1). We evaluated by microscopy the resistance reaction of Fed-Pm4a and Fed-Pm4b lines challenged with a Pm4a/b-avirulent isolate (Bgt96224) and compared it with Fed-Pm2 near-isogenic line (NIL) with the Pm2 gene24. Pm2 encodes a canonical NLR receptor that also confers resistance to Bgt96224. All three genotypes share cv. Federation as recurrent parent, which has no known Pm genes and is susceptible to Bgt96224. At 2 dpi, hypersensitive cell death (HR) was visible in Pm4a/b NILs at significantly lower levels than in the Pm2-containing line (HR 15% Fed-Pm4a and 14% Fed-Pm4b compared to 28% Fed-Pm2). At 6 dpi, almost no fungal microcolonies were observed in both the Pm4a (1%), nor the Pm4b (0%) genotype compared to the Pm2-containing line (26%). Interestingly, Pm4-containing lines showed significantly higher levels of pre-penetration resistance compared to the Pm2 line at 2 and 6 dpi (87% Fed-Pm4a and 88% Fed-Pm4b compared to 49% Fed-Pm2) (Fig. 1a). We conclude that both Pm4 alleles confer rapidly acting resistance mostly at the pre-penetration level but also resulting in some cell death.
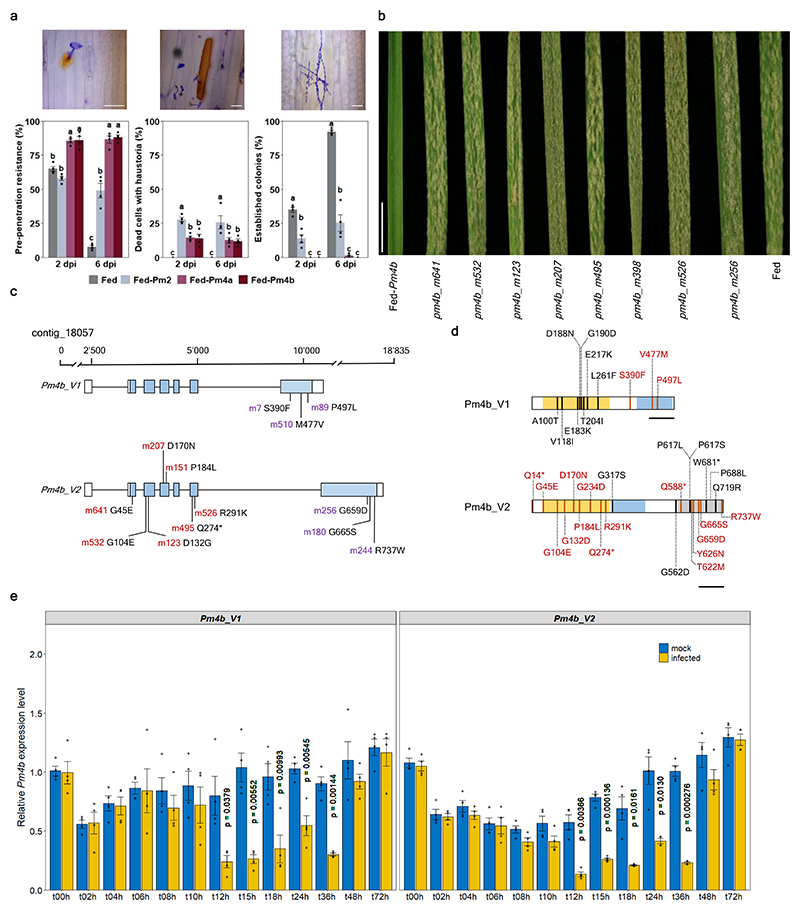
Fig. 1. Molecular identification and characterization of a Pm4b candidate gene.
a, host reactions of Fed-Pm4a, Fed-Pm4b, Fed-Pm2 and Fed challenged with Bgt96224 isolate at 2 and 6 dpi. Left, percentage of pre-penetration resistance arresting conidia growth without hypersensitive cell-death (HR). Middle, percentage of epidermal cells with haustorium associated with HR. Right, percentage of established colonies. Different letters indicate significant differences using ANOVA followed by Tukey honest significant difference (HSD) test (P<0.05). Scale bar, 50μm. b, Powdery mildew infection of seedlings from resistant Pm4b wheat cv. Fed-Pm4b, eight EMS-derived susceptible mutants and the susceptible control Federation. Scale bar, 1 cm c, Gene structure and alternative splicing of the Pm4b gene. Exons are indicated as blue boxes. Mutations identified by MutChromSeq are shown in red. In purple, mutants affected on exons six and seven subjected to expression analysis. Please note that m256 was subjected to flow-sorting and gene expression analysis. d, Pm4b_V1 and Pm4b_V2 protein isoforms with domains indicated by colours: yellow, serine-threonine kinase; light-blue, C2; gray, phosphoribosyltransferase C-terminal. Black and orange vertical lines indicate pm4a and pm4b EMS-derived mutants, respectively. Each mutation, letter after amino acid and its position in the wild-type, is only indicated in one of the two Pm4 isoforms. Asterisks denote early stop codons. Complete information can be found in Supplementary Table 2. Scale bars: 100 aa. e, Transcripts levels of the Pm4_V1 and Pm4_V2 splice variants in mock-inoculated or Bgt-inoculated Fed-Pm4b plants. Error bars denoting s.e.m. are based on four biological replicates. Statistical analysis was done using a two-tailed t-test at p < .05 (mock vs infected) based on n = 4 biological replicates. Exact p values are shown above bars.
Molecular identification and characterization of a Pm4b candidate gene
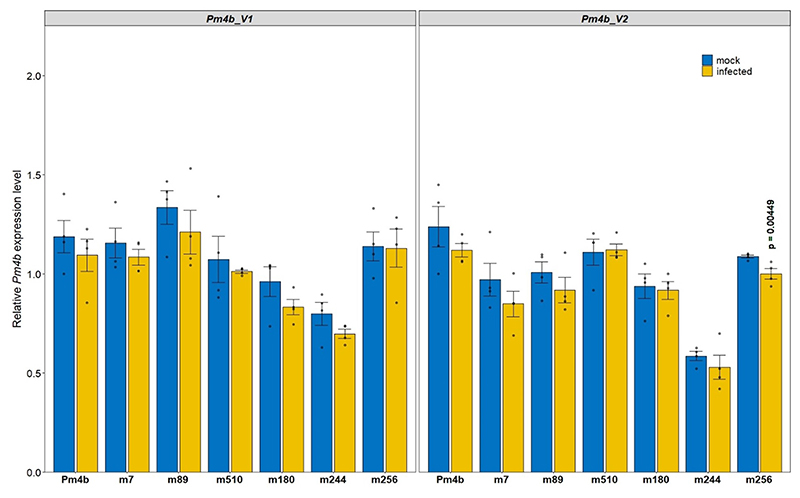
We identified and confirmed 18 EMS-derived pm4b mutants of the Pm4b-containing wheat genotype Fed-Pm4b 22. All these mutants were susceptible to the Pm4a/b-avirulent Bgt96224 isolate (Supplementary Table 2). Chromosome 2A carrying Pm4b was flow-sorted from eight mutants and from the parental genotype (Fig. 1b) and sequenced for gene identification using the MutChromSeq24 approach. After identification of variations in the mutant chromosomes using a Fed-Pm4b de novo assembly, contig_18057 was the only candidate contig for Pm4b. In addition, all of the independent mutations falling within a predicted ORF based on the annotation of the Ae. tauschii Pm4b homologue AET2Gv21296200. Given the multiple splicing variants predicted in AET2Gv21296200, we first clarified the genomic structure and splicing pattern of the Pm4b gene by aligning cDNA products derived from RT-PCR reactions primed with gene-specific primers located on predicted exons 1, 6 and 7, as well as 5′ and 3′ RACE products to the contig_18057 genomic sequence (Fig. 1c). Sequence analysis confirmed that the Pm4b gene consists of seven exons, of which the six and seven exons are alternatively spliced in a mutually exclusive way giving rise to two alternative transcripts, denoted Pm4b_V1 and Pm4b_V2 (Fig. 1c). The two transcripts were also detected in the Pm4a-containing line Fed-Pm4a. Importantly, Pm4-like alternative gene splicing was observed in RNA-seq expression data for the barley Pm4 orthologue HORVU.MOREX.r2.2HG0181350, hereinafter referred as to Hv2HG0181350, where two Pm4_V1- and Pm4_V2-like transcripts translated into two intact ORFs (GenBank: GFJN01021221.1, GFJN01021222.1). Based on the splicing variant Pm4b_V2, seven of the flow-sorted pm4b mutants contained non-synonymous amino acid exchanges, whereas a premature termination codon was introduced in the eight mutant pm4b_m495, possibly resulting in a non-functional protein (Fig. 1c,d and Supplemental Table 2). We confirmed by PCR amplification and Sanger sequencing the mutations identified by MutChromSeq. Further pivotal confirmation of the gene identity was obtained by Sanger sequencing of ten additional pm4b mutants as well as 14 pm4a mutants, which all revealed mutations in the candidate gene. Most mutations were G/C-to-A/T transitions as expected after EMS mutagenesis and caused nonsense (n=4) or missense (n=23) mutations (Fig. 1d and Supplemental Table 2; note that pm4b_m244 has two point mutations). All these mutants were susceptible to the Pm4a/b-avirulent Bgt96224 and Bgt94202 isolates. Motivated by the alternative splicing (AS) exhibited by the Pm4b gene, we focused on mutants affected in exon six (pm4b_m7, pm4b_m89, pm4b_m510) and seven (pm4b_m180, pm4b_m244, pm4b_m256). All these critical mutants did not exhibit significantly different expression levels for splicing variants Pm4b_V1 nor Pm4b_V2 compared to the Pm4b wild type genotype after mock- and Bgt96224-infection at 48 hai (Fig. 1c and Extended Data Fig. 2). Therefore, the loss of resistance was not due to downregulation of Pm4 transcripts. The Pm4_V1 ORF encodes a protein of 560 amino acids, while the Pm4_V2 ORF encodes a predicted protein of 747 amino acids. As mutations in the mutually exclusive exons 6 and 7 both abolished Pm4b-based mildew resistance, we conclude from genetic analysis that both alternatively spliced transcripts and their encoded protein isoforms are needed for Pm4-mediated resistance.
We examined the expression of Pm4_V1 and Pm4_V2 on the wild-type Pm4b wheat genotype Fed-Pm4b after infection with powdery mildew, and the expression of the two transcripts did not significantly differ from each other after mock- and Bgt96224-infection. However, the expression of both transcripts was reduced significantly at early infection stages between 12 and 36 hai, suggesting that mildew infection downregulates Pm4 expression transiently (Fig. 1e). Nearly identical levels of both transcripts suggest that Pm4b_V1 and Pm4b_V2 have a similar contribution to resistance.
Pm4b confers resistance when stably transformed into a susceptible wheat background
To test if the cloned Pm4b candidate gene was sufficient to confer resistance to wheat powdery mildew, we stably co-transformed the Bgt96224-susceptible wheat variety Bobwhite S26 with the two full-length cDNAs of Pm4b_V1 and Pm4b_V2 (Fig. 2a). All tested transgenic T0 plants contained both the Pm4b_V1CDS- and Pm4b_V2CDS transgenes indicating complete co-transformation. The T0 plants were self-fertilized, and four events were chosen at random for T1 family infection with Bgt96224. The three transgenic events T1Pm4b_V1V2CDS-3, T1Pm4b_V1V2CDS-25 and T1Pm4b_V1V2CDS-52.1 showed a 3:1 transgene segregation ratio, suggesting the presence of a single insertion site of Pm4b_V1V2_CDS. In contrast, we detected the presence of both transgenes, Pm4b_V1CDS- and Pm4b_V2CDS, in all T1 plants from family T1Pm4bV1V2CDS-52.2, indicating the presence of the transgene at least at two insertion sites. Importantly, presence of the two transgenes segregated with resistance to Bgt96224 in T1 families (Fig. 2b). We advanced selected T1 plants to the T2 generation for further analysis. T2 plants expressing Pm4b_V1 and Pm4b_V2 also showed resistance to Bgt isolates Bgt96224 and Bgt94202, (Fig. 2c and Supplementary Table 3). The analyzed T2 plants showed higher Pm4 expression levels (Pm4b_V1 between 1.65- and 44.05-fold; Pm4b_V2 between 0.67- and 62.71-fold) compared to the endogenous Pm4b gene in line Fed-Pm4b. However, they were all susceptible to the Pm4a/b-virulent BgtJIW2 and Bgt97251 isolates (Fig. 2c and Supplementary Table 3). These data confirm the race-specific resistance activity provided by the Pm4 gene, which is unaffected by overexpression in the transgenic lines. Transgenic plants overexpressing both Pm4b_V1CDS- and Pm4b_V2CDS transgenes did not significantly differ from Bobwhite S26 with respect to measured agronomic traits (Extended Data Fig. 3), which indicates that ectopic defense activation by the Pm4b_V1CDS- and Pm4b_V2CDS transgenes did not affect plant growth. To further test if both transcript variants are equally needed for Pm4b-mediated resistance as indicated by the mutant analyses, we individually transformed Bobwhite S26 with full-length cDNA of Pm4b_V1 or Pm4b_V2. Transgenic events T1Pm4b_V1CDS-9, T1Pm4b_V1CDS-12 and T1Pm4b_V1CDS-19 were fully susceptible to the Pm4b-avirulent isolates Bgt96224 and Bgt96202 (Extended Data Fig. 4a and Supplementary Table 4). The analyzed T1 plants overexpressing Pm4b_V1 showed higher Pm4b_V1 expression levels (between 1.4- and 3.9-fold) compared to the endogenous Pm4b_V2 transcript in line Fed-Pm4b. Similarly, we selected three transgenic events overexpressing Pm4b_V2: T2Pm4b_V2CDS-6, T1Pm4b_V2CDS-24 and T1Pm4b_V2CDS-29, all of which were fully susceptible to Bgt96224 and Bgt94202. The analyzed T1 plants overexpressing Pm4b_V2 transcript showed higher Pm4b_V2 expression levels (between 1.1- and 20.2-fold) compared to the endogenous Pm4b_V2 transcript in line Fed-Pm4b (Extended Data Fig. 4b and Supplementary Table 4). These data from individual transformation of the two alternative transcripts confirm that both variants must be present to confer resistance, a finding that is in agreement with the mutant analysis.
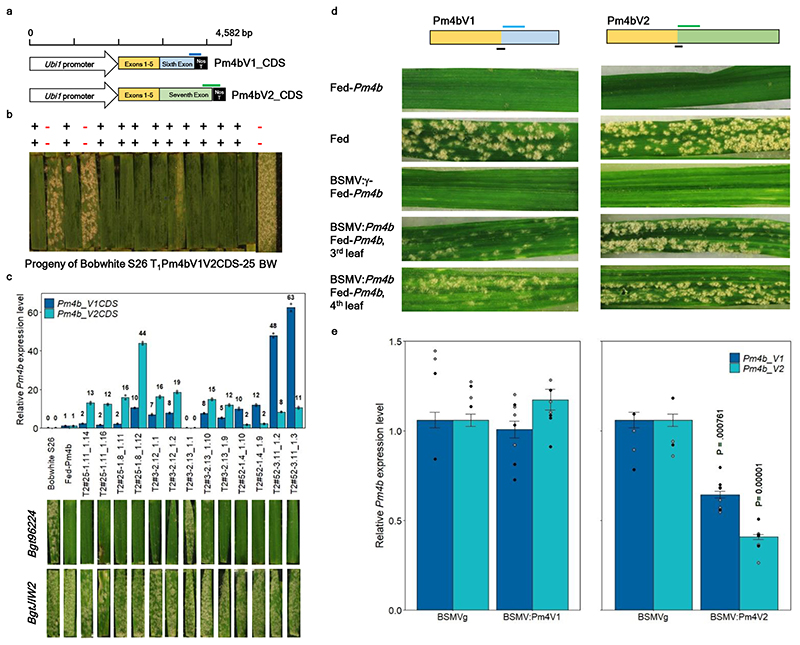
Fig. 2. Confirmation of the functional identity of the Pm4b gene by transgenic complementation and VIGS.
a, Schematic diagram of the two constructs with the coding sequences (CDS) Pm4b_V1CDS and Pm4b_V2CDS, used for transformation of susceptible Bobwhite S26 (BW). Blue and green bars above the schematic diagrams of constructs indicate regions targeted for construct-specific PCR amplification using transgene specific primers displayed in Supplementary Table 7 b, Screening of T1 progeny from T1 family Pm4bV1V2CDS-25. The presence (+) or absence (-) of the Pm4bV1_CDS (top row) and Pm4bV2_CDS (lower row) transgenes corresponded to the resistance/susceptibility phenotype for the individual tested T1 plants. c, Expression levels of Pm4bV1_CDS (blue) and Pm4bV2_CDS (turquoise) transgenes in selected T2 progenies compared to the endogenous Pm4b_V1 and Pm4b_V2 transcripts in the wild-type Fed-Pm4b (second bar). The data points are technical replicates (double quantifications) on single T2 progenies. On top of each bar, number corresponds to the x-fold expression compared to Pm4b_V1 or Pm4b_V2 in the wild-type Fed-Pm4b genotype. Below each T2 progeny, representative images of disease reactions after infection with the Pm4a/b-avirulent Bgt96224 isolate and with the Pm4a/b-virulent BgtJIW2 isolate are shown. d, Schematic diagram of Pm4b_V1 and Pm4b_V2 splicing variants, where blue and green bars indicate regions selected as VIGS targets. Black bars below the diagrams indicate regions targeted for qRT-PCR amplification using transcript-specific primers displayed in Supplementary Table 7. Symptoms of the third and fourth leaves of representative plants subjected to VIGS and after infection with the Pm4b-avirulent Bgt96224 isolate. e. Expression levels of the Pm4bV1 (light green bars) and Pm4bV2 splicing variants (turquoise bars) of BSMV:γ-, BSMV:Pm4b_V1- and BSMV:Pm4b_V2-infected Fed-Pm4b plants assessed by quantitative reverse-transcription PCR (qRT-PCR). Statistical analysis was done using a two-tailed t-test at p < .05 (BSMVg vs BSMV:Pm4V1 or BSMV:Pm4V2) based on n = 4-8 biological replicates, where black and grey dots represent the 3rd and 4th leaves, respectively. Error bars, mean ± s.e.m. Exact P values are shown above bars.
Silencing of Pm4b_V1 or Pm4b_V2 splicing variants compromises powdery mildew resistance in Fed-Pm4b
To further test Pm4b-mediated resistance to powdery mildew through VIGS, we designed silencing constructs for either of the two Fed-Pm4b splicing variants (Fig. 2d). Both constructs targeting Pm4b_V1 or Pm4b_V2 resulted in susceptibility of the Pm4b-containing Fed-Pm4b wheat genotype, visible as large leaf areas covered by sporulating mildew colonies (Fig. 2d). A comparison of mRNA expression by qRT-PCR in Fed-Pm4b leaves infected with BSMV:Pm4b_V2 with Fed-Pm4b plants infected with wild type virus BSMV:γ showed a significant decrease of expression levels of Pm4b_V2 transcripts. Interestingly, the expression of Pm4b_V1 decreased also after silencing of Pm4b_V2, likely because of the formation of secondary siRNA targeting the mRNA sequence shared by both splicing variants26. However, no decrease of Pm4b_V1 or Pmb_V2 expression was observed in BSMV:Pm4b_V1-infected Fed-Pm4b plants, suggesting that this construct was less efficient in directing silencing27 (Fig. 2e). We conclude that the specific targeting of either Pm4b_V1 or Pm4b_V2 expression through VIGS compromised Pm4b-mediated resistance.
The Pm4 gene encodes a putative chimeric kinase-MCTP protein
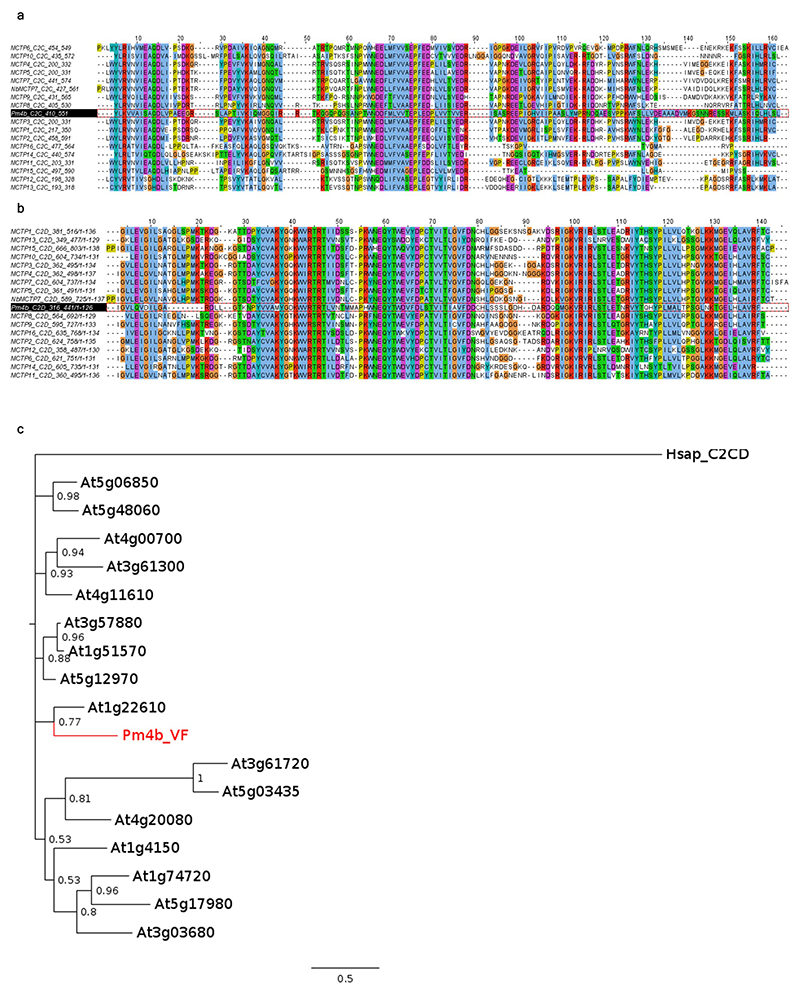
Pm4b_V1 and Pm4b_V2 proteins share the first five exons, predicted to encode a kinase domain with serine/threonine specificity (S_TKc, Fig. 3a,d and Extended Data Fig. 5), but they differ in their C-terminus. Pm4b_V1 isoform has a single C2C domain, while Pm4b_V2 contains a C2D domain coupled to a phosphoribosyl transferase C-terminal domain (PRT_C) with two transmembrane domains (Fig. 3a,c). Pm4b_VF, a hypothetical protein with a combination of all domains of the two isoforms with protein topology S_TKc-C2C-C2D-PRT_C is similar to proteins containing multiple C2-domain and transmembrane region(s) (MCTPs)28,29. However, the S_TKc domain is absent in MCTPs and Pm4b_VF only has the C2C and C2D-PRT_C terminal domains, contrary to the highly conserved domain topology observed in MCTP proteins with three or four C2 domains and a PRT_C domain. Domain Pm4b_C2D is more conserved than Pm4b_C2C compared to Arabidopsis MCTPs C2 domains (Extended Data Fig. 6a,b). The closest Arabidopsis MCTP homologue of Pm4b_VF is MCTP6 (Extended Data Fig. 6c) that contributes to flowering time control cooperatively with MCTP130.
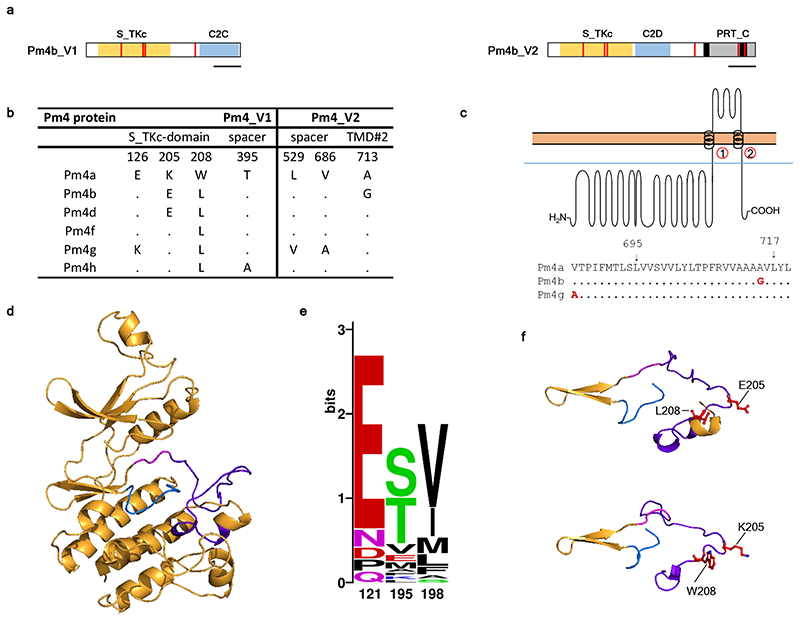
Fig. 3. The Pm4 protein variants differ in the S_TKc and transmembrane domains.
a, Pm4 protein isoforms, Pm4_V1 (left) and Pm4_V2 (right), differ in few amino acid changes (red bars) among the six Pm4 alleles described. Protein domains are indicated by colours corresponding to the ones displayed in Fig. 1d. Scale bar, 100 amino acids. b, Protein sequence comparison of the Pm4 variants, where dots represent identical amino acids to Pm4a. c, Topological model of Pm4b_V2 modified from Protter119 displaying the two transmembrane domains. Below, sequence alignment of the second transmembrane domain of the Pm4a, b and g protein variants, indicating their start and the endpoints at protein level. Dots represent identical amino acids compared to Pm4a. d, Cartoon model of the core domain of the Pm4b S_TKc done using the Phyre2120 server based on the crystal structure of human IKK1 (PDB: 5EBZ, Fold library id: c5ebzF) with 25% of identity and 100.0 % of confidence. In purple, the activation loop, in blue, the catalytic loop and in pink, the DFG motif. e, WebLogo graphical representation of sequence alignment for positions 126, 205 and 208 in Pm4 protein variants compared the kinase-containing resistance proteins described in Extended Data Fig. 5. Note that x-axis numbers correspond to numbers in the alignment of Extended Data Fig. 5. In position 121 (126 in Pm4), kinase-containing resistance proteins mostly have negatively charged amino acids while Pm4g has a Lysine, positively charged. In position 195 (205 in Pm4), Pm4a is the only one, together with BSK1, having a positively charged amino acid. Finally, in position 198 (208 in Pm4) mostly occupied by aliphatic amino acids, Pm4a shows a Tryptophan, which is unique among all the kinases. These amino acid changes might play a fundamental role in differentiating race-specificity among Pm4 protein variants. d, close-up of the catalytic and activation loops of Pm4b (top) and Pm4a (bottom) highlighting the occurring amino acid changes.
The presence of all key conserved residues18,31 in Pm4b-S_TKc (Extended Data Fig. 5) suggests that it is a functional kinase. Besides, four EMS-derived susceptible mutants (pm4b_m207, pm4b_m293, pm4a_m398.1 and pm4b_m291) had missense mutations of key conserved residues, implying that Pm4b-S_TKc is critical for Pm4b-mediated powdery mildew resistance (Extended Data Fig. 5). The closest Arabidopsis homologue to the core kinase domain of Pm4b is CRK6 (AT4G23140), a cysteine-rich receptor-like kinase that confers resistance to Pseudomonas syringae when overexpressed32,33. Interestingly, the barley orthologue of CRK6, HvCRK1, is involved in ROS-mediated basal resistance against powdery mildew34. Furthermore, some of the phylogenetically closest kinase-containing resistance proteins to Pm4b (Supplementary Fig. 1) confer resistance to biotrophic pathogens in wheat and barley18,20,21,35.
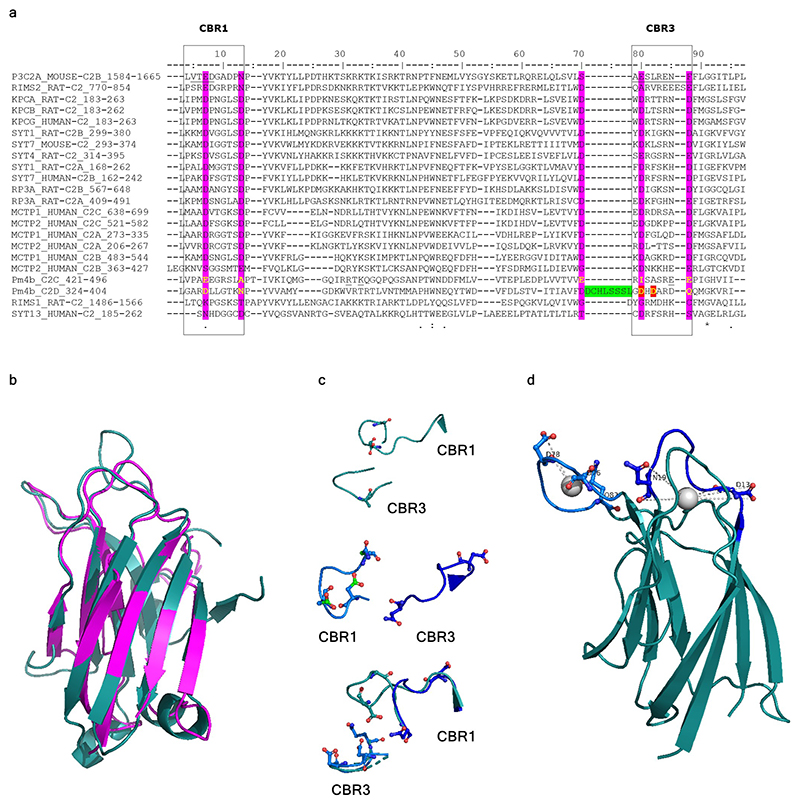
C2 domains are protein signaling motifs with a Ca2+-binding region and a polybasic cluster involved in membrane docking36,37. Only Pm4b_C2D might potentially bind Ca2+ based on the presence of three conserved aspartate residues and two conserved substitutions (glutamine and asparagine) (Extended Data Fig. 7). The C2C domain might be involved in interaction with phosphoinositides, although it does not contain the characteristic positively charged and aromatic residues in the polybasic cluster but conservative substitutions by amino acids with similar physicochemical properties (Supplementary Fig. 2a). Finally, Pm4b_V2 is predicted to have two transmembrane domains highly conserved with Arabidopsis MCTPs-TM domains (Supplementary Fig. 2b). Notably, Pm4b_V2 has a tandem duplication between the transmembrane domains absent in Arabidopsis MCTPs (Supplementary Fig. 2b).
Allelic variations of the Pm4 locus
To facilitate the use of Pm4 in breeding, we designed a diagnostic marker based on Pm4b sequences, and verified the presence of the Pm4 locus and its allelic forms in FedPm4a, Fed-Pm4b and Tm27d2 (Pm4d) after full-length amplification and Sanger sequencing (Fig 3b). We tested the Pm4 haplotype-specific marker in a global wheat collection of 512 accessions, among which the Pm4a allele was absent, whereas Pm4b and Pm4d were detected in 19 and 9 genotypes, respectively. Besides, three new Pm4 alleles, tentatively denoted as Pm4f, Pm4g and Pm4h, were discovered (Fig. 3b). Heterogenic genetic backgrounds with presence of other resistance genes possibly mask the effect of these Pm4 alleles. Nevertheless, we observed that Pm4b- and Pm4d-containing lines are resistant to Bgt94202, Bgt96224, Bgt97223 and Bgt97266 but susceptible to BgtJIW2, the same resistance pattern observed in the Fed-Pm4a and Fed-Pm4b NILs. These phenotyping data suggest the functionality of Pm4b and Pm4d. However, Pm4f- and Pm4g-containing lines were mostly susceptible to the tested Bgt isolates, implying that those are susceptible alleles of Pm4. Finally, the Pm4h allele had a very similar resistance spectrum compared to Pm4b- and Pm4d-containing genotypes and seems to be active (Supplementary Table 5). Pm4 alleles contain single SNPs and/or combinations of shared SNPs affecting mainly the kinase domain (Fig. 3b). Intriguingly, most of the SNP lead to amino acid changes in the S_TKc and transmembrane domains (Fig. 3b,e,f).
Pm4b_V1 and Pm4b_V2 form an ER-associated complex
We examined the subcellular localization of eGFP- and TagRFP-tagged Pm4 individual isoforms co-expressed with characterized markers38–40. eGFP-Pm4b_V2 colocalized with the mCherry-tagged endoplasmic reticulum (ER) marker (Pearson correlation coefficient 0.768 ± 0.02, n = 12) (Fig. 4b and Supplementary Fig. 3). Notably, MCTPs proteins also contain C2C/C2D and PRT-C domains and localize to the ER as well29. This ER-localization has been proposed to be mediated by the presence of transmembrane domains embedded in the PRT_C domain29, which both Pm4V2 and MCTPs share. In contrast, Pm4b_V1 lacks the PRT_C domain and colocalized with the mCherry-tagged cytosol marker (Pearson correlation coefficient 0.765 ± 0.023, n = 12) (Fig. 4a and Supplementary Fig. 3). These results are in line with localization experiments done with truncated MCTPs proteins, where it was demonstrated that the PRT_C domain is essential for the association with the ER network29. Co-infiltration experiments of eGFP- and TagRFP-Pm4b_V1 and Pm4b_V2 revealed a colocalization pattern in the ER (Pearson correlation coefficient 0.765 ± 0.028, n = 12 and 0.782 ± 0.030, n = 10) (Fig. 4c and Supplementary Fig. 3). This suggests that Pm4b_V2 recruits Pm4b_V1 from the cytosol to the ER, possibly by forming an ER-associated complex.
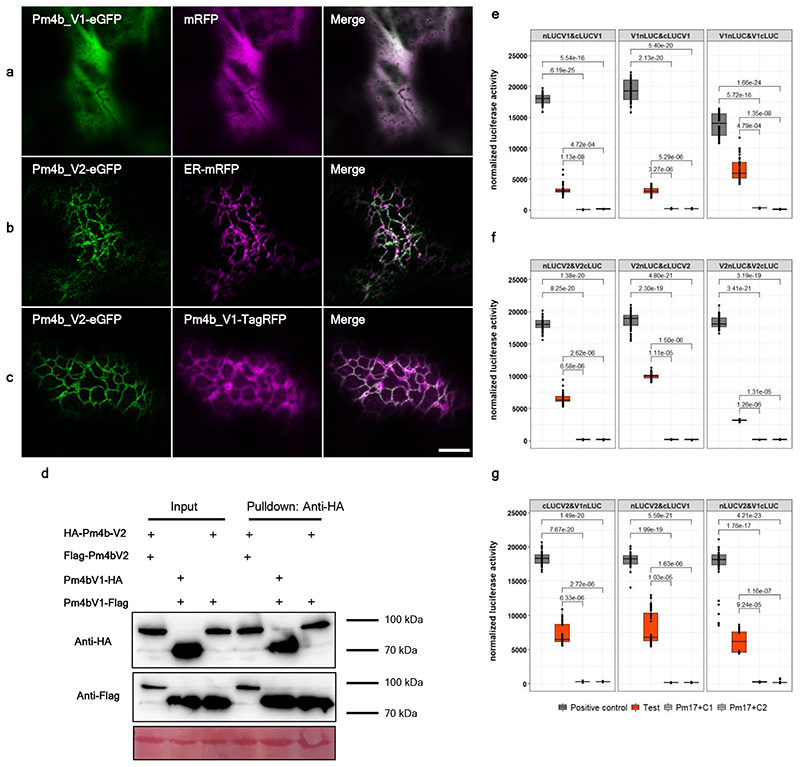
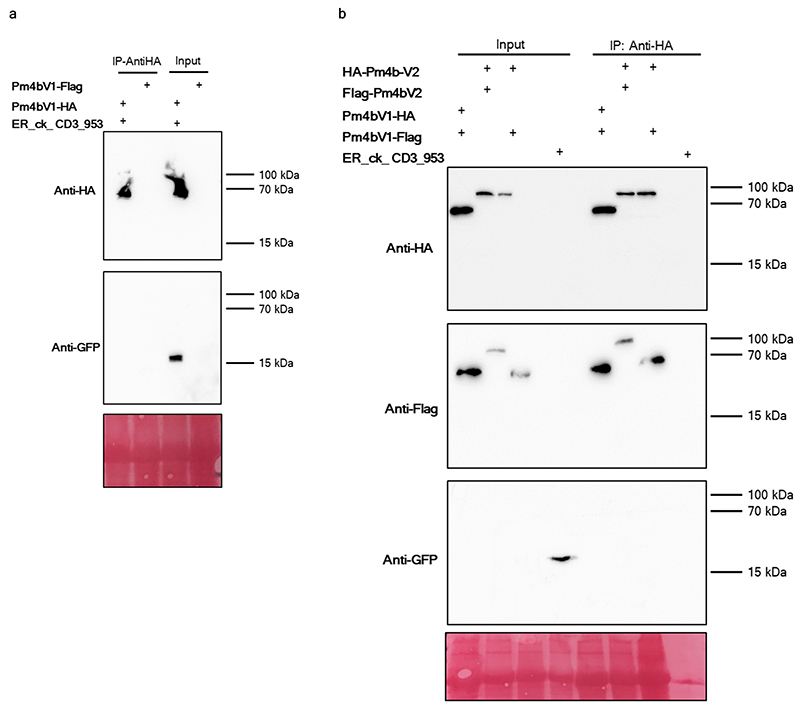
Fig. 4. Pm4_V1 and Pm4_V2 form an ER-associated complex.
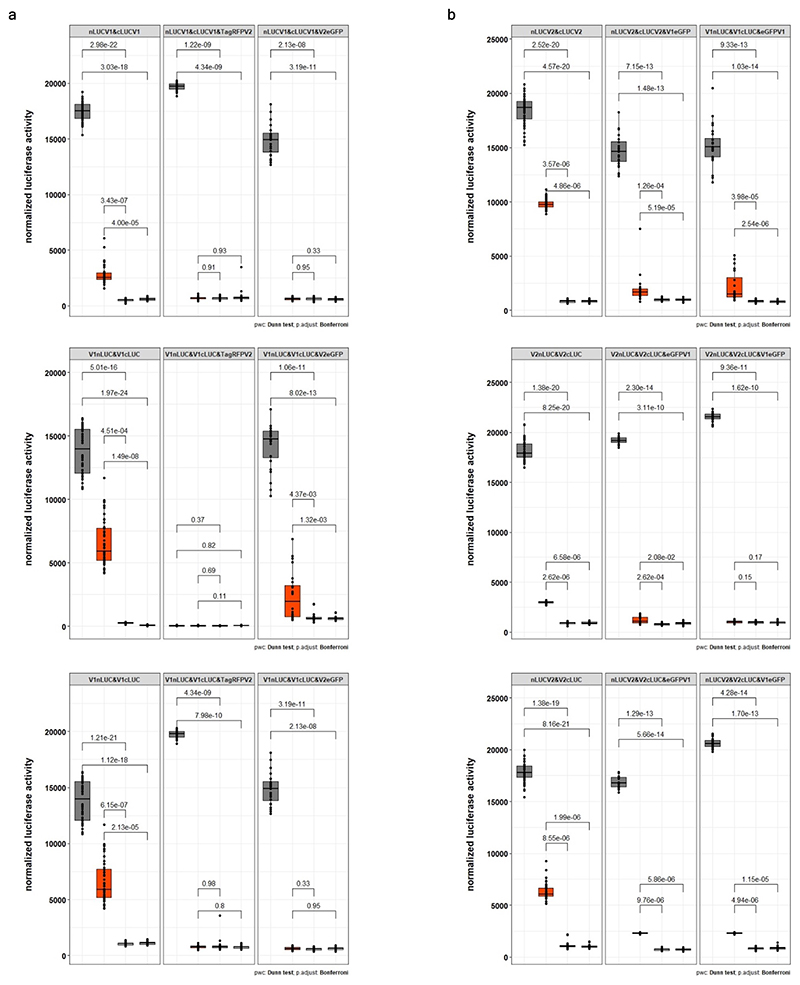
a, Confocal micrographs depicting surface views of N. benthamiana epidermal cells co-expressing Pm4b_V1-eGFP with a marker of the cytosol, b, Pm4b_V2-eGFP with the marker of the endoplasmic reticulum and c, Pm4b_V2-eGFP with Pm4b_V1-TagRFP. Scale bar of 10 μm applies to all images. Localization experiments were repeated five times independently with similar results. d, Identification of potential Pm4b_V1 and Pm4b_V2 homo- and heterodimeric protein interactions via Co-IP. Pm4b_V2 was tagged N-terminally HA- and Flag-tagged. Pm4b_V1 was C-terminally with HA- and Flag-tagged. Representative results of HA pulldown experiments, top panel, where + sign states the presence of the protein. Proteins were detected using anti-HA and anti-Flag antibodies following SDS-PAGE and membrane transfer (bottom panel). First and second columns show homomer formations of Pm4b_V2 and Pm4b_V1, respectively and the third column heteromer formation between Pm4b_V2 and Pm4b_V1. Ponceau staining of the Western blot membrane is depicted at the bottom. Co-immunoprecipitation experiments were repeated three times with similar results. e, Split-luciferase complementation assays showing dimerization of Pm4b_V1 isoform, f, Pm4b_V2 isoform and g, interaction between Pm4b_V1 and Pm4b_V2 isoforms. At the top of each panel the tested combination is displayed, specifying if the N- or C-terminal part of LUC was cloned at the beginning or the end of the protein. For simplicity, V1 and V2 refer to Pm4b_V1 and Pm4b_V2, respectively. The first boxplot corresponds to the positive control, AvrPm3b-AvrPm3b. Second boxplot corresponds to the combination tested, specified at the top in each panel, and the last two to the negative controls used: each component of the test combination with the complementary N-LUC or C-LUC Pm17 tagged. In the boxplots, center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by the geom_boxplot function of the ggplot2 R package; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, individual data points are represented by dots. Significant differences were determined by Krustal-Wallis test followed by Dunn’s multiple comparisons test with two-sided 95.0% confidence interval with Bonferroni correction based on n = 24 (8 technical and 3 biological replicates). Exact P values are shown above bars.
To test for potential Pm4b_V1 and Pm4b_V2 homo and heteromeric protein interactions we first performed co-immunoprecipitation assays. HA-Pm4b_V2 co-immunoprecipitated with the Flag-Pm4b_V2 protein and Pm4b_V1-HA was pulled-down with the Pm4b_V1-Flag tagged protein, suggesting the existence of a multimeric complex. Importantly, the Pm4b_V1 and Pm4b_V2 proteins associated with each other in a specific manner, as HA-Pm4b_V2 and Pm4b_V1-Flag were co-immunoprecipitated (Fig. 4d and Extended Data Fig. 8). These data indicate that Pm4b_V2 and Pm4b_V1 form part of the same complex in vivo. To further test if the two isoforms interact with themselves and each other, we performed luciferase complementation imaging (FLuCI) assays41. We found significantly higher luciferase signals in the Pm4b_V1/Pm4b_V1 and Pm4b_V2/Pm4b_V2 samples compared to the negative controls (Fig. 4e,f). Compared with controls lacking either partner, samples including both Pm4_V1 and Pm4_V2 displayed a significant increase in luciferase signal (Fig. 4g). Interestingly, only N-terminally-tagged N-LUC or C-LUC Pm4b_V2 showed significantly higher luciferase signals, suggesting that domain topology of the C-terminal part of the Pm4b_V2 protein play a critical role in the heteromerisation with Pm4b_V1. To further test whether the two Pm4b variants preferentially establish homo or heteromeric protein interactions, we co-expressed in equal amount the fluorescence tagged Pm4b_V2 protein variant together with Pm4b_V1 / Pm4b_V1 showing high luciferase signal. Similarly, Pm4b_V1 was co-expressed with Pm4b_V2 / Pm4b_V2. In both cases there was a strong reduction of the luciferase signal. This indicates that Pm4b_V1 and Pm4b_V2 protein variants preferentially establish heteromeric rather than homomeric interactions (Extended Data Fig. 9).
Evolutionary origin of the Triticeae-specific Pm4-like gene family
We found 18 Pm4 homologues encoding intact full-length Pm4_V1- and Pm4_V2-like proteins exclusively in various Triticeae species (Supplementary Table 6). Pm4 homologues are present on homeologous group 2 chromosomes of wheat relatives’ rye and barley as well as on A, B and D genomes of diploid, tetraploid and hexaploid wheats (Supplementary Fig. 4a,b and Supplementary Table 6). Pm4 homologues underwent complex evolutionary changes as their clustering did not correspond to 1A, 1B and 1D homologues (Supplementary Fig. 4a,b). Besides, Pm4 is absent in the wheat reference genome sequence of cv. Chinese Spring (CS)1, which also lacks a susceptible Pm4 allele or a homologue, given the low similarity (< 70%) of the CS homologue to Pm4. Finally, among the accessions sequenced in the 10+ Wheat Genomes Project genomes (http://www.10wheatgenomes.com, https://wheat.ipk-gatersleben.de//), cv. SYMattis contained the Pm4d allele at the distal region of 2AL chromosome arm (Supplementary Fig. 5).
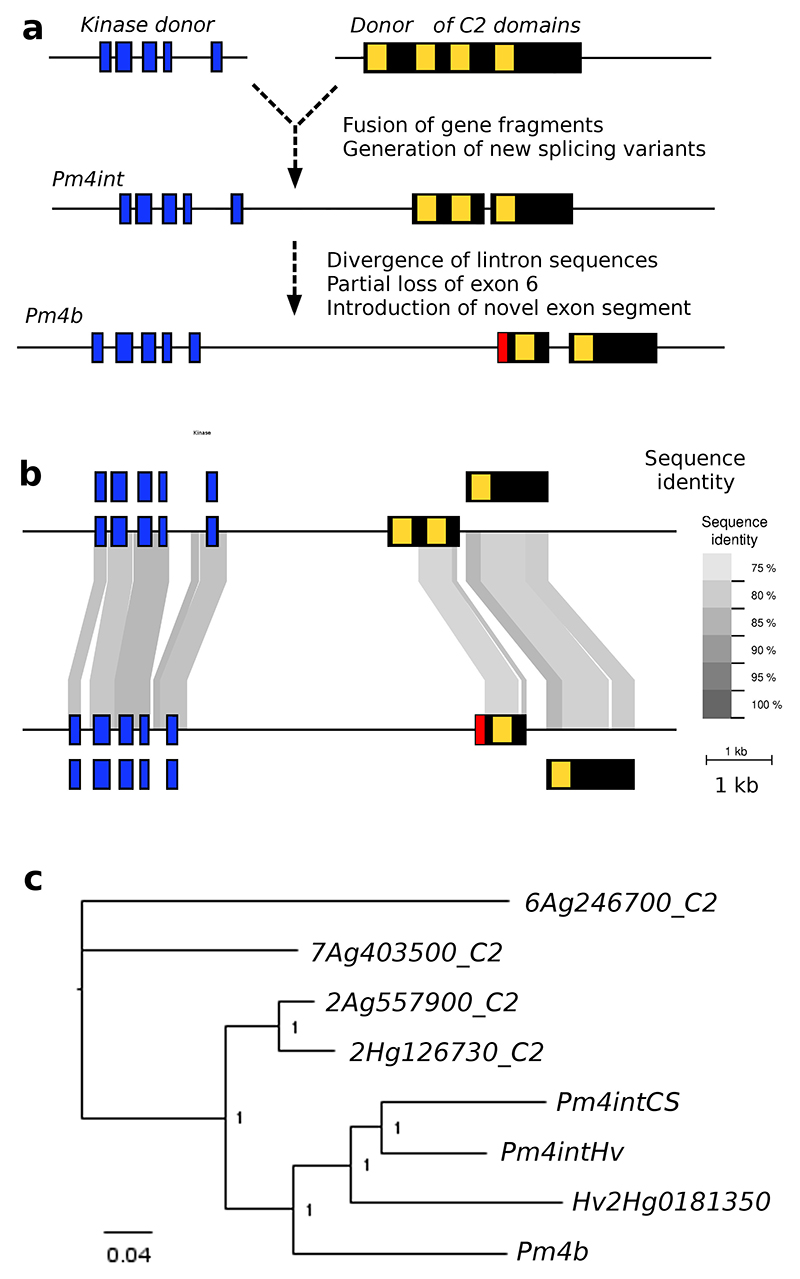
Pm4b apparently evolved in multiple steps, involving a fusion of gene fragments, duplications and subsequent losses and gains of specific sequences. The gene encoding the closest homolog of the C2 domain of Pm4b in Chinese Spring is TraesCS2A01G557900, which is located approximately at position 761 Mb on chromosome 2A, near the position where Pm4b maps in SYMattis, and encodes a canonical MCTP protein. The identification of a Pm4b homolog in barley indicates that the fusion event occurred already in the Triticeae ancestor.
We propose that a 3’ segment of the ancestor of TraesCS2A01G557900 was duplicated and fused to a gene fragment encoding a kinase domain. Such partial gene duplications to nearby loci can be the result of double-strand break repair42. This led to an intermediate form (Pm4int) that encodes a kinase in its 5’ kinase and three C2 domains in its 3’ (Figure 5a). Interestingly, we found this intermediate form on chromosome 2 in both reference genomes for barley43 (cv. Morex) and wheat1 (cv. Chinese Spring). Our data indicate that Pm4int already encodes two different transcripts analogous to those of Pm4b. This is in contrast to the donor C2 TraesCS2A01G557900 which is a single long exon. Pm4int was then duplicated, giving rise to the Pm4b ancestor gene. This gene subsequently lost a segment of exon 6 encoding the first C2 domain and instead acquired a sequence that is unique to Pm4b (Figure 5a,b). Interestingly, all three genes (the donor of the C2 domains, Pm4int and Pm4b) are still all present in a ~1.2 Mb region on barley chromosome 2.
Fig. 5. Evolutionary origin of Pm4b.
a, Model for the evolution of Pm4b. A Kinase domain (blue) was fused to a fragment of a gene encoding a protein with four C2 domains (yellow). The product (Pm4int) encodes two alternative transcripts and comprises 7 exons. Subsequent duplication of Pm4int led to the rise of Pm4b which undergoes re-shuffling of intron 5, leading to loss of the CDS of one C2 domain and to the introduction of a unique sequence in exon 6 (red). b, Comparison of genomic regions of Pm4int (top) and Pm4b (bottom). The two alternative transcripts are depicted on different levels. Sequences that can be aligned at the DNA level are indicated with shaded areas, with sequence identify shown in different shades of grey. c, Phylogenetic tree of the CDS for the C2 domains. Distant homologs 7Ag403500 and 6Ag246700 were used to root the tree. Pm4int and Pm4b from wheat and barley cluster with the descendants of the proposed donor of the C2 domains.
Phylogenetic analysis of the C2 domains shows that Pm4b and Pm4int evolved from the ancestor of TraesCS2A01G557900 (and its barley homolog HORVU2Hr1G126730, Fig. 5c). The emergence of Pm4b from Pm4int apparently occurred soon after, and the phylogenetic tree suggests that there may have been some subsequent gene conversion(s) as the Pm4b and Hv2HG0181350 do not cluster together (Fig. 5c). Molecular dating using fourfold degenerate sites suggest that Pm4int and Pm4b emerged about 20 million years ago. Consequently, sequence conservation between Pm4int and Pm4b is limited to CDS while introns are strongly reshuffled (Fig. 5b). Furthermore, branch lengths in the phylogenetic tree indicate that Pm4b and Pm4int evolved more rapidly than the donor of the C2 domain (Fig. 5c).
Discussion
We cloned through MutChromSeq24 the wheat powdery mildew resistance gene Pm4b, whose functional identity was confirmed by mutagenesis, VIGS and transgenic complementation. While Pm4b is relatively widespread in the hexaploid wheat gene pool, the reference genome of wheat genotype Chinese Spring shows a haplotype with complete absence of a Pm4 allele or homolog.
Pm4 is a valuable gene for use in disease resistance breeding as Pm4 alleles convey resistance to Bgt isolates in economically relevant wheat-growing areas, such as China and USA. The Pm4 haplotype diagnostic marker developed here will facilitate gene deployment in breeding programs aiming at achieving its long-term effectiveness, for instance, by targeted stacking of Pm4 alleles matching the corresponding virulence profile of Bgt isolates44.
Pm4b race-specific action was conserved in transgenic lines, confirming that overexpressing both Pm4b_V1 and Pm4b_V2 did not result in unspecific auto-activity. The molecular basis of race-specificity is well understood in direct or indirect recognition in NLR-based resistance14,45. However, given the novel domain architecture of Pm4, the information on NLR-based specificity cannot be easily applied. However, natural diversity of the alleles at the Pm4 locus reveals some molecular determinants contributing to race-specificity. Possibly, the two amino acid polymorphisms within the activation loop of the S_TKc domain are key determinants of specificity.
Microscopic observations revealed that Pm4-mediated resistance is phenotypically similar to the canonical NLR-based resistance and is associated with epidermal cell death, although at significantly lower levels. HR can be activated via different cellular pathways46, and identification of Pm4 interacting partners and downstream signaling components will support the characterization of Pm4-mediated resistance at the mechanistic level. Pm4 resistance is based to a large extent on pre-penetration resistance suggesting a rapid and efficient host response upon recognition of the mildew pathogen.
Pm4 undergoes constitutive alternative splicing (AS) generating Pm4_V1 and Pm4_V2 splicing variants. While several NLR genes were found to undergo AS under pathogen attack via intron retention or in untranslated regions47,48, in Pm4 we found splicing of mutually exclusive exons. Canonical NLR genes undergoing AS usually generate truncated proteins without a clear biological function. In many of those cases it has been shown that alternative variants are not required for resistance, as in the case of the flax L6 49, tomato BS4 50, rice RGA5 51 or the wheat resistance genes WKS1 52 and Lr10 53. On the other hand, resistance provided by the tobacco N 54, the Arabidopsis RPS4 55 and the Medicago truncatula RCT1 56 resistance genes depends on AS. In these cases, full immunity only occurs when both regular and alternative transcripts are present, which are subjected to a dynamic abundance ratio under pathogen attack (the case of the N 54 or RPS4 55 genes). In contrast, Pm4b_V1 and Pm4b_V2 show identical expression levels, suggesting an equal contribution to resistance. Importantly, based on the mutant analysis, both transcripts and their encoded protein isoforms are needed for resistance. Indeed, the mutations in either Pm4b_V1 or Pm4b_V2 led to full susceptibility whereas in the case of N 54, RPS4 55 or RCT1 56 genes, the absence of alternative splicing variants did not result in susceptibility but in incomplete resistance, or the overexpression of one transcript variant led to full resistance, like the RCT1 case56.
Pm4 encodes a putative kinase-MCTP protein likely resulting from a gene fusion event between a serine/threonine kinase and the C-terminal part of a member of the MCTPs family. Pm4 homologs are found in different Triticeae species but are absent in other grasses within the subfamily Pooideae such as rice and Brachypodium, suggesting a gene fusion event in the ancestor of the Triticeae. Homology-based comparison of the Pm4 core kinase domain with kinase-containing proteins known to be involved in plant immunity points to the functionality of the Pm4 kinase domain. The Pm4 kinase belongs to the RCLK family, many of whose members have been described to be involved in disease resistance57.
RCLK family members such PBS1 and PBS1-like (PBL) proteins transduce immune signals from the plasma membrane58,59 and are also targets of bacterial effectors59–61. Similarly, the kinase domain of Pm4 could be targeted by the specific AvrPm4 effector, inducing a defense reaction. Alternatively, the MCTP domain might be the specific sensor detecting effector manipulation at the ER. In this model, Pm4b_V2 would be the sensor and Pm4b_V1 would be a helper protein, similar to NLR-based interactions with sensor and helper proteins62. Finally, at this stage we cannot exclude the involvement of an NLR, similar to the Prf/Pto system in tomato and the above-mentioned PBS1 guarded by the NLR RPS561,63,64. This NLR might be genetically redundant and functionally non-polymorphic in wheat as it was neither identified by genetic mapping nor by mutagenesis.
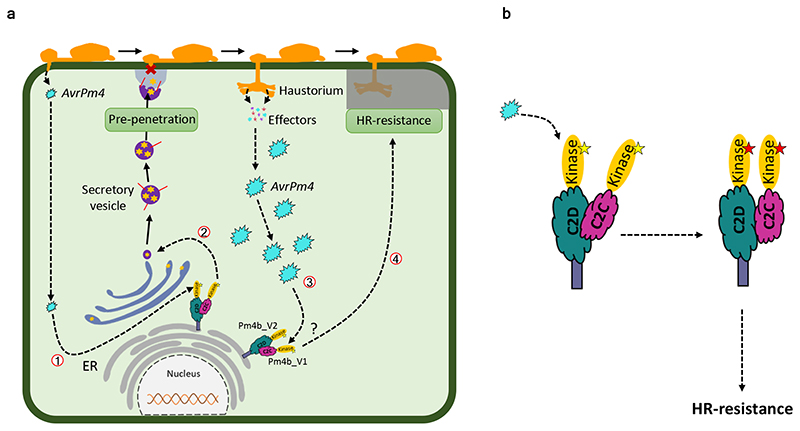
The Arabidopsis protein MCTP1/FTIP interacts via C2 domains with FT, a 175-amino acid length protein part of the mobile flower-promoting signal that promotes the transition from vegetative growth to flowering65. It is known that after a fusion event, the resulting gene may acquire a new function through neofunctionalization66. It is thus tempting to propose that one of the C2 domains present in Pm4 binds the powdery mildew effector to further trigger disease resistance. Indeed, there are experimental data that might support this hypothesis. For instance, the pepper (Capsicum) C2 domain-containing protein SRC2-1 interacts with the Phytophthora capsici INF1 elicitin (PcINF-1) leading to PcINF-1-induced immunity67. Based on the available information along with the work reported here, we present a working model of how Pm4 operates. In this model, Pm4_V1 and Pm4_V2 are in a resting state in the absence of the pathogen forming an ER-associated heterocomplex. After infection by the powdery mildew pathogen (Fig. 6a), there is a rapid, race-specific induction of pre-haustorial resistance in presence of the Pm4b gene. We propose that low levels of the yet unknown AvrPm4 effector released at the early stage of haustorium formation (12-24 hai) results in Pm4b-mediated, papillae-based pre-haustorial resistance (Fig 6a). At the haustorial stage (48 hours), there is a massive release of the AvrPm4 effector inducing a stronger Pm4-mediated defense reaction resulting in HR. In both the early and weak, as well as the later and strong reaction we assume a direct interaction of Pm4 and AvrPm4. However, the signaling output would be different due to different amounts of AvrPm4 which might bind to one of the C2 or S_TKc domains of either Pm4 variant, resulting in conformational changes of the heteromeric complex, leading to activation of the kinase and disease resistance (Fig. 6b). The identification of corresponding effector(s) recognized by Pm4 will be another key element to understand the biological and molecular function of the S_TKc_MCTP based mechanism conferring race-specific resistance to wheat pathogens.
Fig. 6. A possible working model of Pm4-mediated resistance.
a, A schematically drawn wheat epidermal cell attacked by a mature powdery mildew germling. An early release of small amounts of effectors at around 12 hours translates ① into induction of Pm4b-dependent pre-haustorial resistance ②. Later, when large amounts of effectors are present ③, the recognition of AvrPm4 (light blue) by Pm4b protein complex will lead to Pm4b-mediated hypersensitive response (HR) ④. ER, endoplasmic reticulum. b, Schematic model of a possible activation mechanism of Pm4 upon a hypothetical AvrPm4 recognition. In the absence of the AvrPm4, Pm4_V1 and Pm4_V2 are in a resting state, forming a heterocomplex interacting via C2 domains. This heterocomplex is anchored into the membrane of the ER and it is inactive (yellow star in the S_TKc domains). Upon AvrPm4 recognition by the C2C/D or the kinase domains the heterocomplex undergoes conformational changes, leading to activation of the kinase activity (red star in the S_TKc domains) and disease resistance. Numbers indicate the sequence of steps of the proposed model.
ER localization of Pm4b is likely due to the presence of the C-terminal part of a MCTP protein. Extensive work done on Arabidopsis has shown that MCTPs are inserted into the ER via their transmembrane region (TMR)29 as we assume for Pm4b_V2 as well. Likewise, the cytosolic localization of Pm4_V1 (lacking TMR) is in line with the localization observed in MCTPs devoid of TMR29. Finally, we have shown that Pm4b_V1 and Pm4_V2 interact with themselves and each other. We hypothesize that C2 domains play an important role in these interactions. Work done in Arabidopsis has shown that C2 domains are responsible for MCTP physical interaction with other proteins, such as MCTP15/QKY with the receptor-like kinase STRUBBELIG69 and binding to lipids and membrane contact sites29.
The cloning of the Pm4 gene broadens our understanding of both immune receptor architecture and the mechanisms of race-specific activation of the plant immune system. Pyramiding resistance genes that operate by different mechanisms possibly increases the durability of resistance gene combinations70. The chimeric nature of Pm4 with a MCTP domain reveals a potentially novel biochemical context of resistance activation and expands the toolkit available to breeders for the design of resistance breeding strategies.
Online methods
Wheat germplasm, wheat powdery mildew and infection experiments
The susceptible wheat cultivar Federation (GRIN accession number CItr47341; with pedigree Purplestraw 14A/Yandilla), its near-isogenic lines (NILs), Khapli/8*Chancellor//8*Federation (derived from Federation BC8 to Khapli/8*Chancellor) and Federation/W804 (derived from Federation BC7 to W804) were used in the present study to molecularly identify Pm4a and Pm4b. Khapli/8*Chancellor//8*Federation, here denoted as Fed-Pm4a, harbors the Pm4a allele, whose original donor line is Khapli, a tetraploid Triticum turgidum wheat emmer from which the Pm4a gene was transferred to the hexaploid wheat cultivar Chancellor23. Federation/W804, denoted here as Fed-Pm4b, harbors the Pm4b allele introgressed from the original donor line W804, to where the Pm4b allele was transferred from a tetraploid T. carthlicum genotype22. Finally, the wheat genotype Tm27d2, a Triticum monococcum-derived resistant hexaploid line reported to have the Pm4d allele71 was used to study allelic diversity of the Pm4 gene. Federation*4/Ulka (derived from Ulka BC3 to Federation), here denoted as Fed-Pm2, carries the Pm2 resistance gene and was used to compare the resistance reaction at the microscopic level with Fed-Pm4a and Fed-Pm4b. Finally, a global wheat collection of 512 genotypes, the Whealbi collection, representing a wide spectrum of wheat genetic diversity72 was used to study the presence of the Pm4 locus. Detailed passport information is available at https://urgi.versailles.inra.fr/download/iwgsc/IWGSC_RefSeq_Annotations/v1.0/iwgsc_refseqv1.0_Whealbi_GWAS.zip
Blumeria graminis f. sp. graminis (Bgt) isolates Bgt96224, Bgt94202, BgtJIW2 and Bgt97251 were used for infection tests aimed at the molecular identification and further characterization of the Pm4 gene because of their avirulence/virulence pattern on Pm4a and Pm4b. Bgt96224 and Bgt94202 are avirulent (no visible symptoms observed) on the Pm4a/b lines while BgtJIW2 and Bgt97251 are both virulent (leaves fully covered by mycelia). To investigate and compare resistance spectra of Pm4a and Pm4b against a broad variety of globally collected wheat powdery mildew isolates, infection tests were performed on Fed-Pm4a NIL and Pm4b NIL Fed-Pm4b with 108 genetically diverse contemporary Bgt isolates73,74,75 (Supplementary Table 1).
Plants were grown and challenged with appropriated Bgt isolates depending on the experiment as previously described24. Disease levels were assessed 7-9 d after inoculation as one of five classes of host reactions: R = resistance (0-10% of leaf area covered), IR (10-25% of leaf area covered), I (25-50% of leaf area covered), IS (50-75 % of leaf area covered) and S (>75% of leaf area covered).
Microscopic analysis of powdery mildew infection
Infected leaf segments were collected two and six days post infection (dpi) and stained for reactive oxygen species using the 3,3′-diaminobenzidine (DAB)-method76. Leaf segments were then fixed77 and aerial fungal structures were stained for 45 s using 0.25% Coomassie Brilliant Blue (0.15% in EtOH absolute) followed by three washing steps with H2O. Microscopic observations were based on five biological replicates, for each of which 100 A- and B-type epidermal cells78 with only one attempted penetration were used for the evaluation. Using a conventional bright-field microscope (Leica DM LS phase), powdery mildew-wheat interactions were scored based on three categories: (i) early arrest of conidial growth in the absence of hypersensitive cell-death (HR) at the pre-penetration stage without haustorium formation, (ii) epidermal cells penetrated with a visible haustorium and clear signs of HR (iii) established colonies, with haustorium and production of secondary hyphae but not signs of HR.
Generation and screening of EMS-induced Pm4a and Pm4b mutants
Mutants were generated treating Fed-Pm4a and Fed-Pm4b seeds as previously described24. An infection test with the Pm4a/b-avirulent isolate Bgt96224 was done to select potential pm4a,b EMS-induced mutants. From a screen of approximately 6,000 M2 seedlings, we isolated eighteen and twenty-eight putative pm4a and pm4b mutants, respectively. Progeny test to confirm susceptibility to Bgt96224 and genotyping with the previously reported Pm4a co-segregating marker STS-BCD123179 discarded some of mutants as either they turned out to be resistant or they did not amplify for the STS-BCD1231 marker, a sign that a big chromosomal fragment could have been lost after the EMS treatment. At the end, a total of 14 and 18 pm4a and pm4b mutants, respectively, whose susceptibility to the Pm4a/b-avirulent Bgt96224 isolate was confirmed in the M3 generation based on ten different M3 plants from each M2 family.
Primer design and in-house sequencing
All primers used on this study were designed using the Primer blast tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/) and can be found in Supplementary Table 7. In-house Sanger sequencing to check integrity of sequences and constructs was performed on an ABI 3730 (Thermo Fischer Scientific, Waltham, Massachusetts, USA).
Pm4 allele mining
The Whealbi collection was screened for the presence of the Pm4 locus using the Pm4 haplotype-specific marker JS717×JS718. Given the difficulty of amplifying the full-length genomic fragment of Pm4 due to the presence of a 4.5 kb intron between exons 5 and 6 that greatly reduced PCR efficiency, we decided to amplify the gene in two parts. The first part corresponds to the genomic region spanning exons 1 to 5 and the second part to exons 6 to 7. To amplify exons 1 to 5, a long range PCR was performed using the primers JS256×JS257 followed by a nested PCR with JS251xJS257. PCR amplification was done using KAPA Hifi HotStart Polymerase (KK2502, Kapa Biosystems) following manufacturer’s recommendations and with an annealing temperature of 60°C and extension time of 2:00 min. The PCR products were sequenced with the internal primers GH382, GH384, GH385 and JS255. For the amplification of the second part of the gene, a long range PCR using the primers JS278xJS261 followed by a nested PCR with JS278xGH407 was done similarly to the PCR dedicated to amplify the first part of the gene but with an annealing temperature of 63°C and an extension time of 3:00. The PCR products were sequenced with the internal primers JS280, JS292, GH387, GH397 and GH402.
Assessment of alternative splicing of Pm4b mRNA
A first in silico annotation of the Pm4 gene was done based on transcript information from the Ae. tauschii gene AET2Gv21296200, given the lack of RNA-seq data from a Pm4b-containing genotype and the absence of the gene in the Chinese Spring bread wheat reference genome. We elucidated the genomic structure and splicing pattern of the Pm4b gene following a two-steps approach.
First, we perform a rapid amplification of cDNA ends (RACE) to determine the transcriptional start (5’ RACE) and end (3’ RACE) of the Pm4b gene. 3‘- and 5‘-UTR sequences of Pm4b were identified by using the SMARTer™ RACE cDNA Amplification Kit (634923; Clontech) according to the protocol using 40 ng of magnetic bead purified and eluted wheat mRNA as described for RT-qPCR. For reverse transcription of cDNA, the 3’ SMART CDS Primer II A was replaced by primer GH438 in the 5’ RT reaction. Subsequently the same reaction containing the tailed first strand cDNA could be used for both, 3’ and 5’ race PCR. 5' RACE PCR reaction was made with 2 μl of 1:5 diluted cDNA in a 20μl reaction with KAPA2G Robust PCR Kit (KK5501, Sigma-Aldrich, St. Louis, Missouri, USA) and buffer B, gene specific reverse primer GH432 and the provided UPM primer in the Kit. 30 cycles where run according to the touchdown PCR program 1 described in the SMARTer™ RACE Kit manual. On the other hand, 3' race PCR reaction was made with 4 μl of 1:5 diluted cDNA in a 20μl reaction with KAPA2G Robust PCR Kit and buffer B, gene specific forward primer GH377 and a universal reverse primer GH439. After initial denaturation at 95°C for 3 min, a touchdown PCR protocol with 10 cycles of 95°C for 15 secs, 68°C (-0.8°C/cycle) for 30 secs, 72°C for 30 secs, then 25 cycles at 95°C for 15 secs, 61°C for 15 secs, 72° for 30 secs was performed with a final extension at 72° C for 5 min. The obtained 3’ and 5’ race PCR fragments where gel excised, cloned and the sequenced by Sanger sequencing to detect the UTR’s. Based on 5’RACE reactions, we could confirm the presence of at least 182-bp 5’UTR consisting split in two exons. The first one starts spans positions 1’028 to 862 bp before start codon. The second one is a small 16-bp string before start codon. Within this 5’ UTR, no alternative start codons were found. The 3’UTR of Pm4b_V1 is at least 270 bp in length while the one of Pm4b_V2 is 154 bp in length.
Second, guided by the 5’ and 3’ UTRs, we designed primers sitting on both UTRs to study gene structure and splicing. We only found Pm4b_V1 and Pm4b_V2 transcripts variants. The amplification of Pm4b_V1 was achieved using the primers GH398 × GH399 followed by a nested PCR with GH400 × GH401. PCR products were sequenced using primers GH382, GH385, GH387, GH397, JS233 and JS293. For the case of Pm4b_V2, transcript accumulation was confirmed by PCR amplification using the primers GH398 x GH407 followed by a nested reaction with primers GH400 x GH407. PCR product was sequenced with the internal primers GH382, GH385, GH387, JS233, JS280, JS292, JS298 and JS540. PCR amplifications were done using KAPA Hifi HotStart Polymerase (KK2502, Kapa Biosystems) with an annealing temperature of 60°C and extension time of 2:30 min and 3:00 min for amplification of Pm4b_V1 and Pm4b_V2, respectively.
Quantitative Real-Time PCR analysis for detection of Pm4 expression
Expression of Pm4a/b_V1 and Pm4a/b_V2 was quantified in a reverse transcription, quantitative real-time PCR (RT-qPCR) assay, using a CFX96 Real-Time System C1000TM Thermal cycler (Bio-Rad, Hercules, California, USA) and according to MIQE guidelines80. The reference genes ADP and ZFL were selected based on a geNorm study made on eight genes as previously described81. Specificities of amplicons, RT-minus control check, melt curve assessment and efficiency calculation were performed as previously described82. Target-specific amplification efficiencies are given in Supplementary Table 8.
30 mg leaf material was harvested at the specified time points, shock frozen in liquid nitrogen and stored at -80°. RNA extraction was made with the Dynabeads™ mRNA DIRECT™ Purification Kit (61012, Invitrogen) according to the manufacturer’s protocol, with 25 μL of Oligo (dT) 25 per extraction.
First-strand cDNA was synthesized from 40 ng mRNA, using 1/2 reaction of the iScript Advanced cDNA Kit (172-5038, Bio-Rad, Hercules, California, USA). RT-qPCR primers used for the targets Pm4a/b_V1 and Pm4a/b_V2 and the reference genes ZFL and ADP are shown in Supplementary Table 8. RT-qPCR was performed with 4 μL of 20-fold-diluted cDNA in a total reaction volume of 10 μL in technical duplicates using KAPA SYBR® FAST qPCR Master Mix (KK4601, Sigma-Aldrich, St. Louis, Missouri, USA) and 250 μM of each primer. Thermocycling conditions were 95 °C for 20 s, followed by 40 cycles of 95 °C for 3 s, then 63 °C for 20 s for targets Pm4a/b_V1 and ZFL or 60 °C for 20 s for targets Pm4a/b_V2 and ADP. Subsequently a melt curve assessment was performed to exclude detection of potential primer dimers. Relative quantities were calculated and normalized to the reference genes ZFL and ADP revealing the calibrated normalized relative quantities (CNRQ) values, using the program CFX Maestro (Bio-Rad, Hercules, California, USA). To allow comparison of the expression levels between the two splice variants Pm4a/b_V1 and Pm4a/b_V2, the RT-qPCR data were calibrated on the basis of plasmid DNA containing the Pm4_V1 and Pm4_V2 construct, respectively. qPCR on equal plasmid concentration showed equal Cq values for both targets in the range observed usually for technical replicates (< 0.5 Cq).
Wheat transformation
The full-length CDS of both splice variants (Pm4b_V1CDS: 1.6kb and Pm4b_V2CDS: 2.2 kb) were amplified from cDNA with Kapa polymerase (Kapa Biosystems Taq DNA Polymerase (Sigma-Aldrich, St. Louis, Missouri, USA) using the JS274, JS276 (Pm4b_V1CDS) and JS274, JS275 (Pm4b_V2CDS) primers and introducing Asc I and Pac I restriction sites, to be cloned into the pGY1 vector. Pm4b_V1CDS and Pm4b_V2CDS were released from the vector pGY1-Pm4b_V1/V2 by enzymatic digestion using Asc I and Pac I (New England Biolabs, Ipswich, MA), to be subsequently cloned into the Asc I and Pac I sites of the pAHC17 vector under the control of the maize ubiquitin promoter (ubi) with the nopaline synthase terminator (nos)83. Furthermore, Not I restriction sites were introduced into pAHC17 5’ in front of the ubi Promoter and after the nos terminator. The gene cassette ubi:PMI was enzymatically released from the pAHC17 vector backbone using Hind III and Not I, while the gene cassettes ubi:Pm4b_V1CDS and ubi:Pm4b_V2CDS only with Not I. Equimolar amounts of each gene cassette was mixed prior to coating with gold particles. As a selectable marker, the phosphomannose isomerase gene was used84.
The hexaploid spring wheat cultivar Bobwhite S26 was transformed through particle bombardment as previously described81, Briefly, 1617 immature embryos were isolated from freshly harvested wheat seeds (around 0.5mm, and milkish color), and were co-transformed with ubi:Pm4b_V1CDS, ubi:Pm4b_V2CDS and ubi:Pmi gene cassettes by particle bombardment85. Primary T0 transformants were regenerated in tissue culture and selected on mannose-containing media86. We obtained 95 putative transgenic plants, among which, Pm4b_V1CDS and Pm4bV2_CDS were detected in 20 T0 plants using specific primers for the two cDNAs forward primers located in the sixth (JS295) and the seventh exon (JS297), respectively. For both cases, primer HZ010 located in the nos terminator was used as reverse primer. Both PCRs were performed with the following parameters: 30 cycles of 30s at 35°C 95°C, 15s at 61°C, and 40s at 72°C. Transgenic plants with both the Pm4b_V1CDS and Pm4b_V2CDS transgenes were self-fertilized, and four events were chosen at random for T1 family characterization.
Virus Induced Gene Silencing (VIGS)
To specifically silence each splicing variant individually, we focused on exons 6 and 7 of Pm4b to define the VIGS targets. To minimize the possibility of off-target silencing, we blasted the coding sequences of exons six and seven against our own sequencing data obtained from flow-sorted chromosome 2A of Fed-Pm4b as well as against the reference genome assembly of wheat (Chinese Spring1) choosing fragments of 150-250 bp with no homology to other genes. For amplifying Pm4b_V1_target_1 and Pm4b_V2_target_2, primers JS189×JS190 and JS498×499 were used, respectively. Note that primers were designed with Not I and Pac I restriction sites in antisense direction to lead to an antisense insertion in the pBS-BSMV-γ vector. Equimolar amount of pBS-BSMV-α, pBS-BSMV-β and pBS-BSMV-γ transcripts carrying Pm4b_V1_target_1 or Pm4b_V2_target_2 were used to inoculate full-expanded first leaves of Fed-Pm4b seedlings, using the wild type (γ) viral genome as control as previously described87–89. For in vitro synthesis of viral RNA, the Invitrogen™ mMESSAGE mMACHINE™ T7 Transcription Kit (Thermo Fischer Scientific, Waltham, Massachusetts, USA) was used according to the manufacturer’s recommendations. Seeds from Fed-Pm4b cultivar were stratified at 4°C during five days. Seedlings were then placed in a growth chamber (Conviron, Winnipeg, Canada) cycled at 23°C/16°C, 16/8h photoperiod with 60% humidity and a light intensity regime of 350 μmol/(s·m2). Fed-Pm4b plants were inoculated when the first leaf was fully developed as previously described90,91. 14 days after virus infection the 3rd and 4th leaves were detached and infected with the Pm4a/b avirulent isolate Bgt96224, adding 10g/L Benzylaminopurine (BAP)92 to 0.5% agar plates. 7 days later, powdery mildew phenotypes were documented and around 1 cm2 highly mildew infected leaf pieces were sampled for further gene silencing expression analyses as explained before in the section Quantitative Real-Time PCR analysis for detection of Pm4 expression.
Plasmids constructs for protein interaction and localization studies
To generate constructs for the Split-Luciferase complementation assay, cDNA from Fed-Pm4b was used to amplify the full-length Pm4b_V1 CDS with primers JS483 (common forward) and JS486 (stop codon) or JS487 (without stop codon). Likewise, the full-length Pm4b_V2 CDS was amplified using primers JS483 and JS484 (stop codon) or JS485 (without stop codon). All the fragments were cloned into pENTR/D-TOPO vector (Invitrogen) following manufacturer’s recommendations. For the expression clones, the pENTR subclones were recombined into the destination vectors 35S: gwnLUC, 35S: nLUCgw, 35S: gwcLUC, 35S: cLUCgw41, using LR Clonase II (ThermoFisher Scientific) following the manufacturer’s recommendations.
To generate constructs for the co-immunoprecipitation assay, similarly to before, entry clones were generated for full-length Pm4b_V1 CDS using JS483 and JS486 (stop codon) or JS487 (without stop codon) primers. For full-length amplification of Pm4b_V2 CDS primers JS483 and JS484 (stop codon) or JS485 (without stop codon) were used. The subclones were then cloned into expression vector pIPKb00493, using LR Clonase II (ThermoFisher Scientific) and following manufacturer’s recommendations. Introduction of genes encoding fusion proteins into the destination vectors was made by site-directed mutagenesis, amplifying the CDS in the expression clones adding HA/Flag tags by PCR with the Primers, JS589&JS590 (N-terminal Flag), JS593&594 (C-terminal Flag), JS601&JS602 (N-terminal HA), JS488&JS489 (C-terminal HA).
To generate the constructs for fluorescence localization, the pENTR subclones generated for the Split-luciferase complementation assay were recombined into the expression vectors 35S:pGWB50538 and 35S: pMpGWB22894, by LR Clonase II (ThermoFisher Scientific) according to manufacturer’s recommendations. Likewise, the mRFP-fused cytosolic localization sequence (pGWB45538), ER-marker (ER-ck, CD3-95939) and plasma membrane-marker (35S:REM 1.2 m_RFP40) were cloned into A. tumefaciens GV3101.
Agroinfiltrations
Binary plasmids were transformed via freeze-thaw approach95 into Agrobacterium tumefaciens GV3101, which were grown overnight with vigorous shaking (200 rpm) at 28°C in Luria-Bertani (LB) medium supplemented with appropriate selective medium depending on constructs carried. 200μl of this culture was used to inoculate 15 ml LB medium and grown overnight under the same conditions. Bacteria were harvested by centrifugation at 2’500 x g for 15min and then resuspended and diluted in infiltration medium (10 mM MgCl2, 0.1M acetosyringone) to an optimal density at 600 nm = 0.8-1.0. After 2 to 4h of incubation at room temperature, one or more cultures were mixed in a 1:1 ratio with an equally treated Agrobacterium p19-silencing-suppressor strain96 and were infiltrated with a needleless syringe into the abaxial side of leaves from 2- to 4-week-old Nicotiana benthamiana plantlets.
Split – luciferase complementation assay
For the in vivo split-luciferase assay in N. benthamiana, the CDS of Pm4b_V1 and Pm4b_V2 were fused in frame with nLUCgw/gwnLUC and cLUCgw/gwcLUC. As negative controls N- and C-terminal fusions of the Pm17 resistance protein97 to nLUC or cLUC were used. As positive controls, we used the AvrPm3b C-terminally fused to nLUC and cLUC. All the fusion constructs were transformed into A. tumefaciens GV3101 strain. Equal amounts of bacteria producing the nLUC or cLUC, N- or C-terminally-fused proteins were infiltrated in 2-4 weeks old N. benthamiana leaves. The luciferase luminescence signals were imaged 4 days after infiltration using an in vivo plant imaging system (Spark, multimode microplate reader, TECAN, Switzerland).
Plant protein extraction and co-immunoprecipitation
Tissue for co-immunoprecipitation was harvested three days post infiltration and immediately flash frozen in liquid nitrogen. Leaf material (50 mg) was ground to a fine powder and proteins were extracted with Triton-X100 (100mM Tris-HCL pH7.4, 50mM NaCl, 5mM NaF, 5mM NaVo4, 0.5% Triton X-100, PMSF) or Brij-58 (100mM Tris-HCL pH7.4, 50mM NaCl, 5mM NaF, 5mM NaVo4, 0.5% Brij-58, PMSF) lysis buffers (1 mL), and subsequently precipitated by anti HA magnetic beads (10 μl) (mouse, monoclonal, 88837, Thermo Scientific). Precipitates were washed five times with Triton X-100 or Birj-58. Proteins from crude extracts (input) and precipitated proteins were detected by immunoblotting with protein–specific antibodies. The elution, IP, washing and detection were performed at 4°C.
Proteins were separated by SDS-PAGE and transferred to a nitrocellulose membrane (GE Healthcare, Chicago, Illinois, USA). The membrane was then blocked in TBST buffer containing 5% non-fat dry milk under gentle shaking. The blocked membrane was incubated with specific antibodies dissolved in TBST 5% non-fat dry milk powder at a ratio of 1:10’000 (Anti-Flag) or 1:3’000 (Anti-HA-HRP) and incubated at 25°C by shaking at 100rpm for 2 hours, followed by three washes (10 min each) with TBST. The detection of the antibodies was performed with WesternBright ECL HRP substrate (Advansta, San Jose, California, USA), before photographing using the Fusion FX system (Vilber Lourmat, Eberhardzell, Germany). Blotted proteins were stained with Ponceau S. The primary antibodies used in this study were anti-Flag (mouse monoclonal, clone M2, F3165, Sigma-Aldrich, St. Louis, Missouri, USA), anti-HA-HRP (rat monoclonal, clone 3F10, 12013819001, Roche, Basel, Switzerland), and Anti-GFP (mouse monoclonal, clone B34, 902601, BioLegend, San Diego, USA). Anti-mouse immunoglobulin G(IgG) (LabForce, sc2357) was used as a secondary antibody for Flag-tag and GFP detection at a working dilutions of 1:10’000 and 1:5’000, respectively.
Confocal Laser Scanning Microscopy
Confocal images of infiltrated N. benthamiana leaves were taken as previously described98. Briefly, a Leica SP5 confocal laser scanning microscopy system (Leica, Wetzlar, Germany) equipped with Argon and DPSS lasers and hybrid detectors was used. eGFP fluorescence was observed using excitation wavelengths of 488nm and its fluorescence emission was collected at 495 to 550 nm. Tag- and m-RFP fluorescence was observed using excitation wavelengths of 561nm and its fluorescence emission was collected at 575 to 650nm. Leaf samples of 5×5 mm were transferred between a glass slide and a cover slip in a drop of water. Experiments were performed using identical confocal acquisition parameters (e.g. laser power, gain, zoom factor, resolution, and emission wavelengths reception), with detector settings optimized for low background and no pixel saturation.
Pseudo-colored images were obtained using “Green” and “Magenta” look-up-table (LUT) of Fiji software99 (http://rsb.info.nih.gov/ij/). To calculate the most quantitative estimate of co-localization, known as the Pearson correlation coefficient that depends on the amount of colocalized signals in both channels (magenta and green) in a nonlinear manner, we performed the analysis as previously described100 in Image J (http://rsb.info.nih.gov/ij/). In brief, it was made sure that the images acquired have low noise levels and no bleed trough, and that the optical setup used for each color lead to the same point of spread function (PSF). In addition, after splitting the images and removing the blue channel, the background was subtracted and then the Coloc 2 Image J plug in was run.
Chromosome flow sorting, sequencing and MutChromSeq-based identification of a Pm4b candidate gene
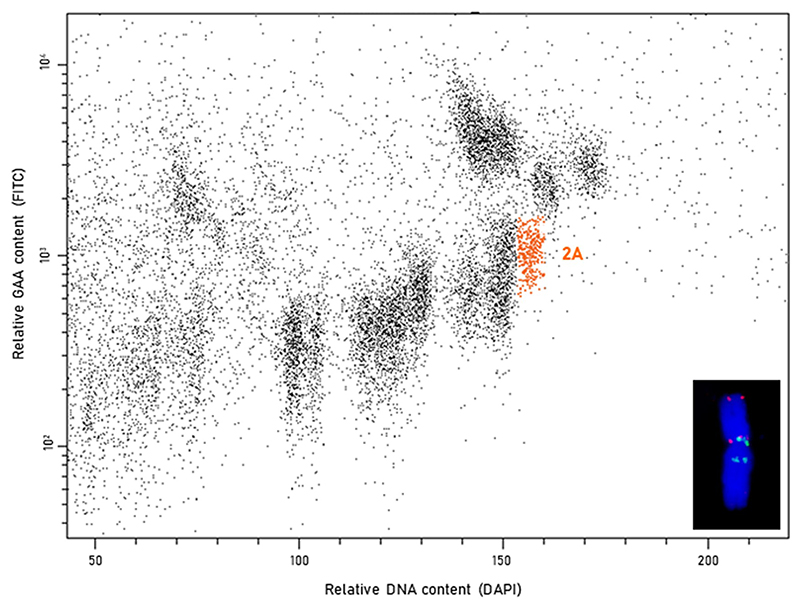
Chromosome flow sorting and sequencing was performed in WT and eight mutants (Supplementary Table 2). Briefly, cycling cells in root tips of young seedlings were accumulated at mitotic metaphase and chromosomes were isolated by mechanical homogenization of formaldehyde-fixed meristem tips as previously described101. Chromosomes in suspension were fluorescently labelled using (GAA)7-FITC as previously described102, chromosomal DNA was stained by DAPI (2 μg/ml) and the suspension was analyzed by FACSAria SORP II flow sorter (BD Biosciences, San Jose, USA). 30,000 copies of chromosome 2A corresponding to 50 ng of DNA were flow-sorted from each line into PCR tube containing 40 μl deionized water using the sort window shown in Extended Data Fig. 10. To estimate the extent of contamination by other chromosomes, 2,000 chromosomes 2A were flow-sorted onto a microscopic slide, labelled by FISH with GAA microsatellite and Afa-family probes (inset of Extended Data Fig. 10) and evaluated microscopically103. The purities in the sorted fractions ranged from 90 to 99% Chromosomal DNA was purified and amplified by Illustra GenomiPhi V2 DNA amplification Kit (GE Healthcare, Piscataway, USA) as previously described104.
MutChromSeq-based identification of a Pm4b candidate gene
Illumina raw reads of flow-sorted chromosomes of EMS-derived mutants were analyzed for their quality using FastQC (http://www.bioinformatics.bbsrc.ac.uk/projects/fastqc). For sequencing adapter removal and quality trimming, cutadapt105 and sickle (https://github.com/najoshi/sickle), with the sickle parameter -q = 25 and -l = 20, were used. MutChromSeq was performed as described previously (https://github.com/steuernb/MutChromSeq)24 with minimum adjustments in the Pileup2XML command (-a 0.1 -c 8) and MutChromSeq command (-a 0 -c 8 -n 3 -z 1). It is important to note, that manual inspection of the MutChromSeq pipeline is advisable. For example, mutations of pm4b_m207 and pm4b_m256 contig_18057 were not identified as such because neither of the two did meet the stringency criteria of the pipeline. pm4b_m207 had a G->A SNP at contig_18057 position 3723, but was only covered by 4 reads. The pm4b_mut256 showed a G ->A SNP at contig_18057 position 11,157 but was only supported by eight out of nine reads, and therefore, not meeting the allele frequency demands of the pipeline.
Protein sequence and domain analysis
Prediction of core domain kinase of Pm4b and resistance proteins displayed in Extended Data Fig. 5 and Supplementary Fig. 1 was done based on Conserved Domain Database (CDD) from NCBI106 (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). Prediction and delimitation of Pm4b C2 domains was done as previously described29. Prediction of transmembrane helices was performed with TMHMM server v.2.0107 (http://www.cbs.dtu.dk/services/TMHMM/) and Phobious108 (http://www.phobius.sbc.su.se). Only transmembrane domains predicted for both applications were considered. 3D structure modelling was done using Phyre2 using intensive modelling mode. Crystal structures served as best templates, % of confidentiality and p-values for each 3D structure modelling are indicated in the legends of the corresponding figures. The structural graphics were generated using PyMOL (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC).
Phylogenetic analysis of Pm4 homologues
To reduce complexity and shorten computation time in the search of Pm4 homologues, we created in silico a hypothetical protein called Pm4_VF, without alternative splicing and with exons 6 and 7 both included in the coding gene (STKc-C2C-C2D-PRT_C). The Pm4b_VF amino acid sequence was used as a query to identify Pm4 homologues via BlastP on genome assemblies of barley H. vulgare 109 (Genome assembly: Barley Pseudomolecules Morex v2.0 2019, https://webblast.ipk-gatersleben.de/barley_ibsc/), goatgrass Ae. tauschii 110 (Genome assembly, Aet_v4.0 https://plants.ensembl.org/Aegilops_tauschii/Info/Index), rye S. cereale (https://webblast.ipk-gatersleben.de/ryeselect/), T. urartu 111 (accession G1812) wild emmer wheat T. turgidum dicoccoides 112 (Genome assembly, Zavitan pseudomolecules), durum wheat T. turgidum durum 113 (Genome assembly, Svevo pseudomolecules) and common wheat1 (Genome assembly, Chinese Spring pseudomolecules, IWGSC RefSeq v1.0). We retrieve a total of 18 Pm4 homologues encoding intact full-length Pm4_V1- and Pm4_V2-like proteins, whose predicted sequences were aligned with Clustalw at default parameters. Phylogenetic trees for Pm4_V1 and Pm4b_V2 homologs were done with MrBayes114, summarized using a burn-in of 25% and visualized with FigTree (http://tree.bio.ed.ac.uk/software/figtree/). All software was obtained from ubuntu repositories (ubuntu.com)
Phylogenetic analysis of kinase domain-containing proteins
A BlastP search of the NCBI non-redundant protein database was used to find proteins described in disease resistance with a kinase domain similar to one present in Pm4b. Considering the increasing evidence of a blurred PTI-ETI dichotomy115, we did not differentiate between PTI- or ETI-related resistance proteins but instead focus on homology. Alignment and phylogenetic tree was conducted in the same way as for the Pm4 homologues described above.
Divergence estimates
Predicted protein sequences were aligned with the program Water. From this alignment, a codon-by-codon DNA alignment was deduced. All protein alignments were inspected by eye and poor alignments were removed. For divergence time estimates, only fourfold degenerate sites were used (i.e. third codon bases for Ala, Gly, Leu, Pro, Arg, Ser, Thr and Val. For Leu, Arg and Ser (which have six possible codons), we used only those codons starting with CT, TC and CG, respectively (where the third base can be exchanged without amino acid change). Divergence time estimates for gene pairs were calculated as previously described116 using a substitution rate of 1.3E-9 substitutions per site per year117.
Statistical analysis
Detailed statistical description is provided in the figure legends, including the type of statistical tests used and the sample size. All analyses were performed using R Statistical Software (R version 3.6.2)118.
Extended Data
Extended Data Fig. 1. Pm4a and Pm4b convey resistance to a wide range of Bgt isolates.
a, Disease reactions of Fed-Pm4a and Fed-Pm4b NILs to 108 genetically diverse contemporary Bgt isolates73,74,121. b, Selection of Bgt isolates for which Fed-Pm4a and Fed-Pm4b NILs showed a differential resistance/susceptibility pattern. The outer and inner circle represent the reaction pattern of Fed-Pm4a and Fed-Pm4b, respectively. Disease reaction was evaluated seven days post-inoculation. Five classes of host reactions were distinguished: R = resistance (0-10% of leaf area covered), IR (10-25% of leaf area covered), I (25-50% of leaf area covered), IS (50-75 % of leaf area covered) and S (>75% of leaf area covered). CHN: China, ISR: Israel; CHE; Switzerland; FRA: France; USA: United States; GRB: Great Britain; JPN; Japan.
Extended Data Fig. 2. Expression profiling of Pm4b mutants following infection with Bgt96224 .
Transcripts levels of the Pm4_V1 and Pm4_V2 splice variants in mock-inoculated or Bgt-inoculated Fed-Pm4b plants. Statistical analysis was done using a two-tailed t-test at p < .05 (mock vs infected) based on n = 4 biological replicates. Error bars, mean ± s.e.m. Exact P values are shown above bars
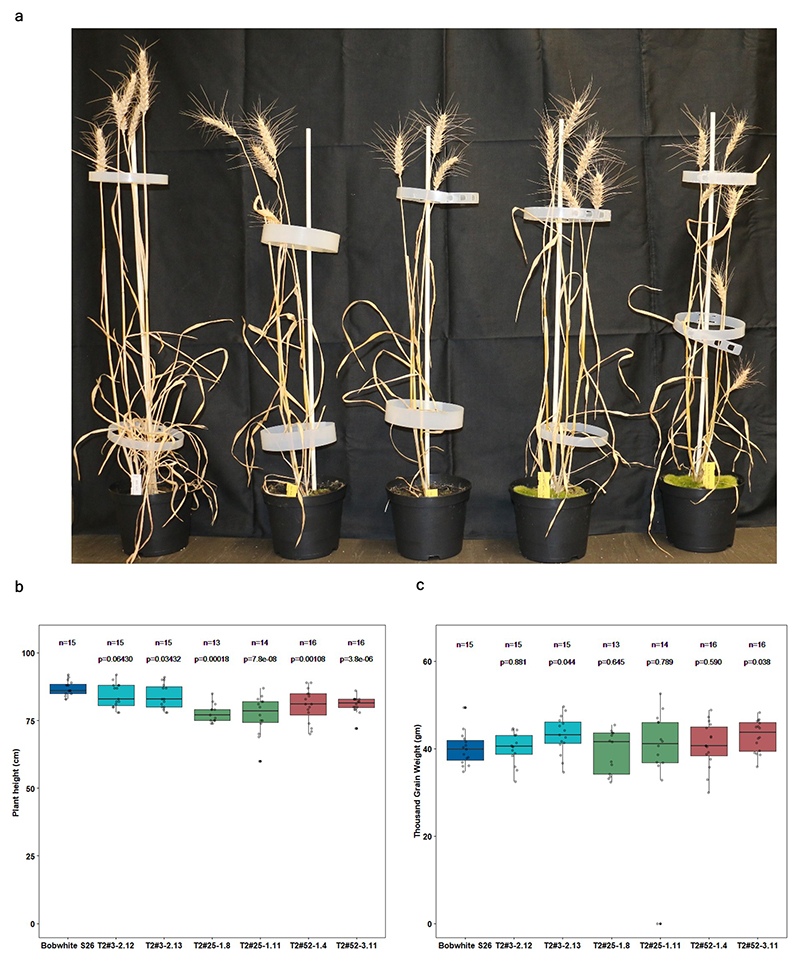
Extended Data Fig. 3. Agronomically-related traits of selected T2 transgenic families overexpressing Pm4b_V1CDS and Pm4b_V2CDS transgenes.
a, Plant growth of representative T2 transgenics from families T2#52-1.4 and T2#52-3.11 compared to Bobwhite S26 in the following order: Bobwhite S26, T2#52-1.4_1.10, T2#52-1.4_1.9, T2#52-3.11_1.2 and T2#52-3.11_1.3 b, Plant height of the T2 families overexpressing Pm4b_V1CDS and Pm4b_V2CDS transgenes presented in Fig 3c and Supplementary Table 3. Names are indicated in the x-axis. c, Thousand Grain Weight for the same T2 families. Selected representative of the same T2 family are displayed with the same color: T2#3 in cyan, T2#25 lime green and T2#52 in magenta. In the boxplots, center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by the geom_boxplot function of the ggplot2 R package; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, individual data points are represented by dots. On top of each boxplot, p values based on two-tailed t-test at p < .05 (transformants versus Bobwhite S26). Above p values, n = the number of T2 progeny.
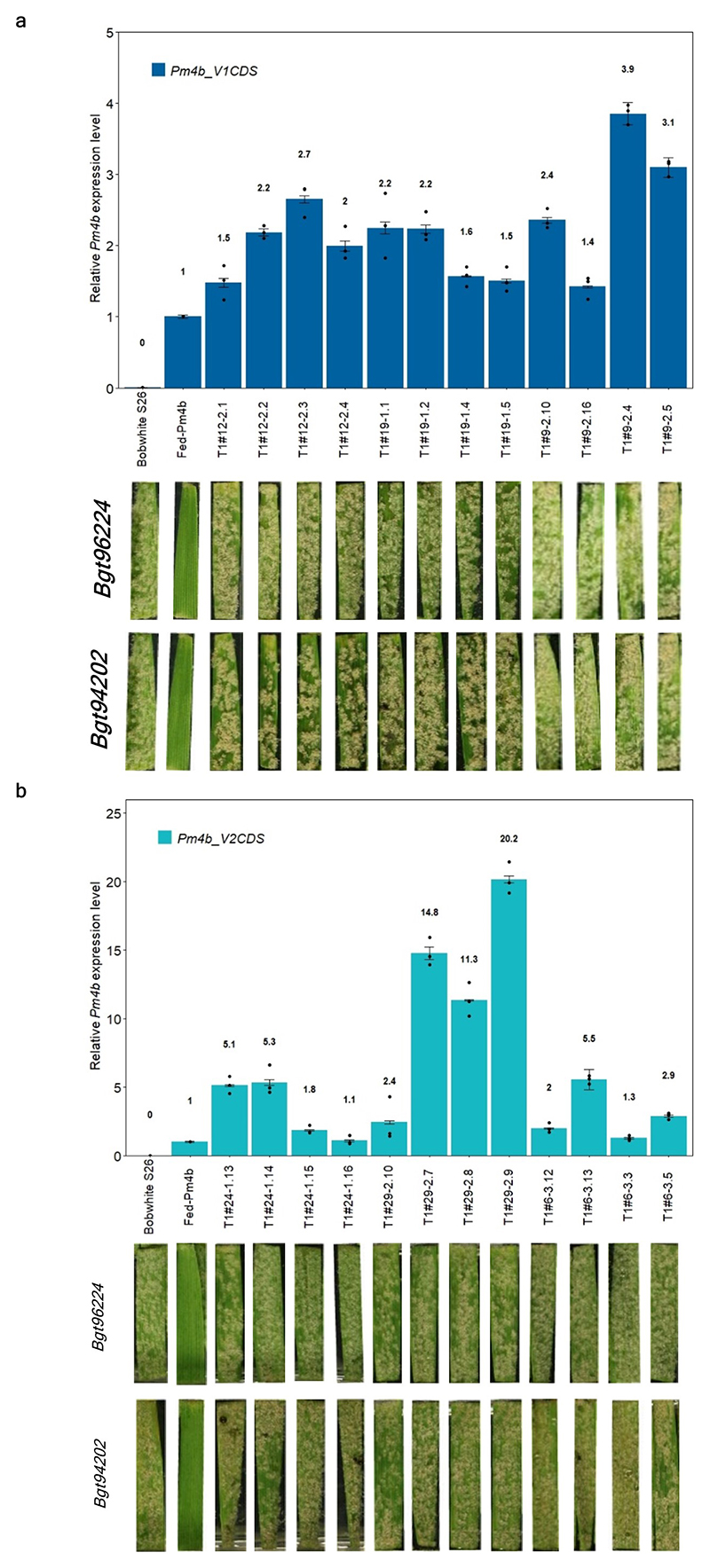
Extended Data Fig. 4. Gene expression in transgenic wheat plants overexpressing single splice variants of the Pm4b gene.
a, Expression levels of Pm4bV1_CDS transgenes in selected T1 progeny for three independent transgenic events (T1#9, T1#12, T1#12) overexpressing full-length cDNA of Pm4b_V1 compared to the endogenous Pm4b_V1 transcripts in the wild-type Fed-Pm4b (second bar). b, Expression levels of Pm4bV2_CDS transgenes in selected T1 progeny for three independent transgenic events (T1#6, T1#24, T1#29) overexpressing full-length cDNA of Pm4b_V2 compared to the endogenous Pm4b_V2 transcripts in the wild-type Fed-Pm4b (second bar). For a and b, data points are technical replicates (triple quantifications) on single T1 progenies. Error bars, mean ± s.e.m. of three technical replicates. On top of each bar, the number corresponds to the x-fold expression compared to Pm4b_V1 or Pm4b_V2 in the wild-type Fed-Pm4 genotype. Below each T1 progeny, representative images of disease reactions after infection with the Pm4a/b-avirulent Bgt96224 and Bgt94202 isolates are shown.
Extended Data Fig. 5. Predicted Pm4 kinase catalytic domain.
A multiple amino acid sequence alignment of 38 protein kinase catalytic domains involved in disease resistance was used to infer the Pm4b kinase domain architecture. In Pm4b (indicated with a red rectangle) all the 14 key conserved residues of protein kinases are present. In the alignment, red arrowheads mark invariant residues (G52, K72, E91, D166, N171, D184, G186, E208, R280), which are numbered with upper case numbers corresponding to their position in the α form of the cAMP-dependent protein kinase catalytic unit (cAPK). Likewise, black arrowheads indicate the mostly invariant residues (G50, V57, F185, D220, G225). Based on the presence of a L residue at position R165 of cAPK in subdomain VI, Pm4 Kinase was classified as a non-RD kinase. Moreover, conserved residues in subdomain VI (D166 -> N171, DLKPAN in Pm4b vs. DLPKPEN in cAPK) and VIII (GTMGYLAPE in Pm4b vs. GT/SXXY/FXAPE in cAPK) indicate that the Pm4 kinase domain is a serine/threonine protein kinase. Labels: red and black arrowheads, key invariant and nearly invariant residues in the protein kinase catalytic domains, respectively. Light blue diamond points to the RD or non-RD kinase determination site. Black asterisks, substrate binding site. Green arrowheads, ATP binding site. Core conserved, diagnostic regions of subdomains I, II, VI, and VIII are highlighted by grey bars labelled with Roman numerals. On top of the wrapped alignment, EMS mutagenized line designations affecting the Pm4 kin domain in Pm4a or Pm4b genes and corresponding amino acid changes are indicated. Violet squares indicate polymorphic amino acids within the kinase domain among the Pm4 allelic variants described in this study. Numbers above violet squares indicate the position on the alignment based on the cAPK sequence.
Extended Data Fig. 6. Sequence alignment of Pm4 C2 domains with homologous C2 domains of Arabidopsis MCTPs.
a, Sequence alignment of Pm4b-C2C with C2C domains from Arabidopsis MCTPs. b, likewise alignment of C2D domains. C2 domains were delimited based on Conserved Domain Database (CDD) from NCBI106. The location of the domain is indicated by the sequence range numbers. C2 domains in Pm4 (black background) are indicated with a red rectangle. c, Phylogenic tree of C2C and C2D domains of Arabidopsis MCTPs and Pm4b-C2C/C2D domains. The human DySF dysferlin C2C/D domains was used as outgroup.
Extended Data Fig. 7. Determination of aspartate residues predicted to be involved in Ca2+-binding in Pm4b C2 domains.
a, Sequence alignment of Pm4b-C2C and Pm4b-C2D domains with C2 domains previously described to bind Ca2+. UniProt entry names followed by the specific C2 domain displayed are located on the left. The region of the C2 domain displayed is indicated by the sequence range numbers. Conserved aspartate residues involved in Ca2+-binding are highlighted in pink. Pm4b_C2C (fourth row from the bottom) does not have conserved aspartate residues and exhibits diverse amino acid substitutions, including D -> E, A or I. However, Pm4b_CD2 (third row from the bottom) has three conserved aspartate residues (positions I, III and IV) and two conservative substitutions, asparagine (position II) and glutamine (position V), both polar and relatively small amino acids. Interestingly, Pm4_C2D contains an insertion of eight amino acids (green) just before the predicted Ca2+ binding region 3 that shifts the position of the conserved aspartate residues at position III and IV (highlighted in red) (see Extended Data Fig. 6). Rectangles denote calcium-binding regions (CBR) 1 and 3, respectively. b, Structured-based alignment of C2D Pm4b_V2 (turquoise) and the C2 domain from PKCα (pink) (Protein kinase C alpha type, PDB: 1DSY). The predicted structural model of the Pm4bC2 domain was done using the Phyre2 server on the basis of the crystal structure of rat otoferlin c2a (PDB: 3L9B, Fold library id: c3l9bA) with 14% of identity and 99.9% of confidence. c, On top, calcium binding regions (CBR) CBR1 and 3 of PKCα. In the middle, CBR1 and 3 of Pm4b_C2D domain. On the bottom part, overall alignment of CBRs 1 and 3 of Pm4b_C2D domain (turquoise) and PKCα (dark blue). d, Three-dimensional structure of C2D domain of Pm4b using the Phyre2120 server based on the crystal structure of rat otoferlin c2a (PDB: 3L9B, Fold library id: c3I9bA) with 14% of identity and 99.9 % of confidence highlighting in blue CBR 1 and 3, with predicted residues involved in Ca2+-binding labelled. Calcium ions are shown as grey balls.
Extended Data Fig. 8. Negative controls for the Pm4b interaction.
a, Pm4b_V1 does not interact with the ER-marker ER_ck_CD3_95339. b, Pull-down with anti-HA beads is specific for the presence of HA-tagged Pm4b variants. Co-immunoprecipitation experiments were repeated two times with similar results.
Extended Data Fig. 9. Binding ability of Pm4b variants for homo- and heteromeric interactions.
a, Split-LUC combinations showing luciferase signal for Pm4b_V1 homomeric interaction in Fig. 4e were co-infiltrated with fluorescence-tagged Pm4b_V2 protein variants. b, Split-LUC combinations showing luciferase signal for Pm4b_V2 homomeric interaction in Fig. 4f were co-infiltrated with fluorescence-tagged Pm4b_V1 protein variants. The data are displayed following the same logic as presented in Figure 4: in each of the 18 panels, the first boxplot corresponds to the positive control, AvrPm3b_N-LUC & AvrPm3b_C_LUC. The second boxplot (orange color) corresponds to the tested combination, displayed at the top of each panel. For simplicity, V1 and V2 refer to Pm4b_V1 and Pm4b_V2, respectively. Finally, the last two boxplots in each panel correspond to the negative controls co-infiltrated. Significant differences were determined by Krustal-Wallis test followed by Dunn’s multiple comparisons test with two-sided 95.0% confidence interval with Bonferroni correction based on n = 24 (8 technical and 3 biological replicates). Exact P values are shown above bars. In the boxplots, center lines show the medians; box limits indicate the 25th and 75th percentiles as determined by the geom_boxplot function of the ggplot2 R package; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles, individual data points are represented by dots.
Extended Data Fig. 10. Bivariate flow karyotype GAA-FITC vs. DAPI obtained after the analysis of chromosomes isolated from mutant pm4b_m256 .
The population representing chromosome 2A, which was flow-sorted, is highlighted in orange. Inset: Flow-sorted chromosomes were identified microscopically after FISH with probes for GAA microsatellites (green) and Afa repeat (red). The fluorescent labeling pattern allowed chromosome identification and estimation of the contamination of sorted fractions by other chromosomes. Chromosomes were counterstained by DAPI (blue).
Supplementary Material
Acknowledgements
We thank Jan Vrána, Zdeňka Dubská, Romana Šperková and Jitka Weiserová for the assistance with chromosome sorting and preparation of chromosomal DNA. This project was financially supported by the University of Zurich, Swiss National Science Foundation grant 310030B_182833 to B.K., the European Research Council under the Grant Agreement 773153 (grant IMMUNO-PEPTALK) to C.Z., and the European Molecular Biology Organization (EMBO Long-Term Fellowships 438-2018) to J.G. M.C.K has received funding from the European Union’s Horizon 2020 research and innovation program under the Marie Skłodowska-Curie grant agreement No 674964. M.K. and J.D. were supported from ERDF project "Plants as a tool for sustainable global development" (No. CZ.02.1.01/0.0/0.0/16_019/0000827). B.K. and J.S.M sincerely thank Dr. Volker Mohler from the Bavarian State Research Center for Agriculture (LfL) for providing seeds from the hexaploid line Tm27d2. J.S.M. sincerely thanks Dr. Nina Chumak from the Department of Plant and Microbial Biology (UZH) for providing the ER-marker (ER-ck, CD3-959).
Footnotes
Author Contributions
J.S.M. and B.K. conceived the project. M.K. and J.D. performed chromosome flow sorting and preparation of chromosomal DNA. T.W., J.S.M., M.H., C.R.P., B.S., and M.C.K. performed bioinformatics analysis. H.Z. performed VIGS. G.H. carried out gene expression studies. J.G. and V.W. performed confocal microscopy. V.W. did validation by transgenic complementation. V.W., J.S.M., L.S., and J.I. performed biochemistry experiments. J.S.M. and L.S. carried out allele mining. C.Z. provided theoretical contributions to the project. J.S.M. and B.K. analyzed the data. J.S.M. and B.K. wrote the manuscript, and all authors revised the manuscript.
Competing Interests statement
The authors declare no competing interests.
Data availability statement
All data is available in the main text or the supplementary materials. Sequence data were deposited at the NCBI GenBank under the accession numbers MT783929 (Pm4b_V1 CDS) and MT783930 (Pm4b_V2 CDS), and at the NCBI short read archive (SRA) database under the accession number PRJNA646941 (flow-sorted chromosome 2A of eight Fed-Pm4b mutants and the wild-type Fed-Pm4b). All Blumeria graminis f. sp. tritici (Bgt) isolates listed in Supplementary Table 1 are kept alive in the Department of Plant and Microbial Biology of the University of Zurich and are available upon request. Any additional data that support the findings of this study are available from the corresponding author upon reasonable request. The databases that we used are all publicly available, please see Methods and the Nature Research Reporting Summary linked to this article.
References
- 1.Appels R, et al. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science (80- ) 2018 doi: 10.1126/science.aar7191. [DOI] [PubMed] [Google Scholar]
- 2.Savary S, et al. The global burden of pathogens and pests on major food crops. Nat Ecol Evol. 2019;3:430–439. doi: 10.1038/s41559-018-0793-y. [DOI] [PubMed] [Google Scholar]
- 3.Singh RP, Rajaram S. Breeding for disease resistance in wheat in Bread wheat: improvement and production. Food and Agriculture Organization of the United Nations. 2002 [Google Scholar]
- 4.Pink DAC. Strategies using genes for non-durable disease resistance. Euphytica. 2002;124:227–236. [Google Scholar]
- 5.Koller T, Brunner S, Herren G, Hurni S, Keller B. Pyramiding of transgenic Pm3 alleles in wheat results in improved powdery mildew resistance in the field. Theor Appl Genet. 2018;131:861–871. doi: 10.1007/s00122-017-3043-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mundt CC. Use of multiline cultivars and cultivar mixtures for disease management. Annu Rev Phytopathol. 2002;40:381–410. doi: 10.1146/annurev.phyto.40.011402.113723. [DOI] [PubMed] [Google Scholar]
- 7.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 8.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 9.Marchal C, et al. BED-domain-containing immune receptors confer diverse resistance spectra to yellow rust. Nat Plants. 2018;4:662–668. doi: 10.1038/s41477-018-0236-4. [DOI] [PubMed] [Google Scholar]
- 10.Wang H, Zou S, Li Y, Lin F, Tang D. An ankyrin-repeat and WRKY-domain-containing immune receptor confers stripe rust resistance in wheat. Nat Commun. 2020 doi: 10.1038/s41467-020-15139-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sarris PF, Cevik V, Dagdas G, Jones JDG, Krasileva KV. Comparative analysis of plant immune receptor architectures uncovers host proteins likely targeted by pathogens. BMC Biol. 2016 doi: 10.1186/s12915-016-0228-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Le Roux C, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015 doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 13.Sarris PF, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015 doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 14.Kourelis J, Van Der Hoorn RAL. Defended to the nines: 25 years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell. 2018 doi: 10.1105/tpc.17.00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saintenac C, et al. Wheat receptor-kinase-like protein Stb6 controls gene-for-gene resistance to fungal pathogen Zymoseptoria tritici. Nat Genet. 2018 doi: 10.1038/s41588-018-0051-x. [DOI] [PubMed] [Google Scholar]
- 16.Kema GHJ, et al. Stress and sexual reproduction affect the dynamics of the wheat pathogen effector AvrStb6 and strobilurin resistance. Nat Genet. 2018;50:375–380. doi: 10.1038/s41588-018-0052-9. [DOI] [PubMed] [Google Scholar]
- 17.Zhong Z, et al. A small secreted protein in Zymoseptoria tritici is responsible for avirulence on wheat cultivars carrying the Stb6 resistance gene. New Phytol. 2017;214:619–631. doi: 10.1111/nph.14434. [DOI] [PubMed] [Google Scholar]
- 18.Klymiuk V, et al. Cloning of the wheat Yr15 resistance gene sheds light on the plant tandem kinase-pseudokinase family. Nat Commun. 2018 doi: 10.1038/s41467-018-06138-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Brueggeman R, et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci U S A. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen S, et al. Wheat gene Sr60 encodes a protein with two putative kinase domains that confers resistance to stem rust. New Phytol. 2020 doi: 10.1111/nph.16169. [DOI] [PubMed] [Google Scholar]
- 21.Lu P, et al. A rare gain of function mutation in a wheat tandem kinase confers resistance to powdery mildew. Nat Commun. 2020 doi: 10.1038/s41467-020-14294-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The TT, McIntosh RA, Bennett FGA. Cytogenetical Studies in Wheat. IX * Monosomic Analyses , Telocentric Mapping and Linkage Relationships of Genes Sr21, Pm4 and Mle Aus. 1979;32:115–126. [Google Scholar]
- 23.Briggle LW. Transfer of Resistance to Erysiphe graminis f. sp. tritici from Khapli Emmer and Yuma Durum to Hexaploid Wheat 1. Crop Sci. 1966 doi: 10.2135/cropsci1966.0011183x000600050020x. [DOI] [Google Scholar]
- 24.Sánchez-Martín J, et al. Rapid gene isolation in barley and wheat by mutant chromosome sequencing. Genome Biol. 2016;17 doi: 10.1186/s13059-016-1082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stam M, Mol JNM, Kooter JM. The silence of genes in transgenic plants. Annals of Botany. 1997 doi: 10.1006/anbo.1996.0295. [DOI] [Google Scholar]
- 26.Carbonell A. Secondary small interfering RNA-based silencing tools in plants: An update. Frontiers in Plant Science. 2019 doi: 10.3389/fpls.2019.00687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang PH, et al. RNase I f -treated quantitative PCR for dsRNA quantitation of RNAi trait in genetically modified crops. BMC Biotechnol. 2018 doi: 10.1186/s12896-018-0413-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Li C, Liang Z, Yu H. Characterization of multiple C2 domain and transmembrane region proteins in arabidopsis. Plant Physiol. 2018 doi: 10.1104/pp.17.01144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Brault ML, et al. Multiple C2 domains and transmembrane region proteins ( MCTP s) tether membranes at plasmodesmata. EMBO Rep. 2019 doi: 10.15252/embr.201847182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu L, et al. FTIP1 is an essential regulator required for florigen transport. PLoS Biol. 2012 doi: 10.1371/journal.pbio.1001313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hanks SK, Quinn AM, Hunter T. The protein kinase family: Conserved features and deduced phylogeny of the catalytic domains. Science (80- ) 1988 doi: 10.1126/science.3291115. [DOI] [PubMed] [Google Scholar]
- 32.Yeh YH, Chang YH, Huang PY, Huang JB, Zimmerli L. Enhanced Arabidopsis pattern-triggered immunity by overexpression of cysteine-rich receptor-like kinases. Front Plant Sci. 2015 doi: 10.3389/fpls.2015.00322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen K, Du L, Chen Z. Sensitization of defense responses and activation of programmed cell death by a pathogen-induced receptor-like protein kinase in Arabidopsis. Plant Mol Biol. 2003 doi: 10.1023/B:PLAN.0000009265.72567.58. [DOI] [PubMed] [Google Scholar]
- 34.Rayapuram C, et al. Regulation of basal resistance by a powdery mildew-induced cysteine-rich receptor-like protein kinase in barley. Mol Plant Pathol. 2012 doi: 10.1111/j.1364-3703.2011.00736.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brueggeman R, et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci U S A. 2002 doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Corbalan-Garcia S, Gómez-Fernández JC. Signaling through C2 domains: More than one lipid target. Biochimica et Biophysica Acta - Biomembranes. 2014 doi: 10.1016/j.bbamem.2014.01.008. [DOI] [PubMed] [Google Scholar]
- 37.Shin OH, Hau W, Wang Y, Südhof TC. Evolutionarily conserved multiple C2 domain proteins with two transmembrane regions (MCTPs) and unusual Ca2+ binding properties. J Biol Chem. 2005 doi: 10.1074/jbc.M407305200. [DOI] [PubMed] [Google Scholar]
- 38.Nakagawa T, et al. Improved gateway binary vectors: High-performance vectors for creation of fusion constructs in transgenic analysis of plants. Biosci Biotechnol Biochem. 2007 doi: 10.1271/bbb.70216. [DOI] [PubMed] [Google Scholar]
- 39.Nelson BK, Cai X, Nebenführ A. A multicolored set of in vivo organelle markers for co-localization studies in Arabidopsis and other plants. Plant J. 2007 doi: 10.1111/j.1365-313X.2007.03212.x. [DOI] [PubMed] [Google Scholar]
- 40.Bücherl CA, et al. Plant immune and growth receptors share common signalling components but localise to distinct plasma membrane nanodomains. Elife. 2017 doi: 10.7554/eLife.25114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gehl C, et al. Quantitative analysis of dynamic protein-protein interactions in planta by a floated-leaf luciferase complementation imaging (FLuCI) assay using binary Gateway vectors. Plant J. 2011 doi: 10.1111/j.1365-313X.2011.04607.x. [DOI] [PubMed] [Google Scholar]
- 42.Wicker T, Buchmann JP, Keller B. Patching gaps in plant genomes results in gene movement and erosion of colinearity. Genome Res. 2010 doi: 10.1101/gr.107284.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mascher M, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544:427–433. doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez J, Beat M. Contribution of recent technological advances to future resistance breeding. Theor Appl Genet. 2019 doi: 10.1007/s00122-019-03297-1. [DOI] [PubMed] [Google Scholar]
- 45.Barsoum M, Sabelleck B, DSpanu P, Panstruga R. Rumble in the Effector Jungle: Candidate Effector Proteins in Interactions of Plants with Powdery Mildew and Rust Fungi. CRC Crit Rev Plant Sci. 2019 doi: 10.1080/07352689.2019.1653514. [DOI] [Google Scholar]
- 46.Greenberg JT, Yao N. The role of regulation of programmed cell death in plant-pathogen interactions. Cellular Microbiology. 2004 doi: 10.1111/j.1462-5822.2004.00361.x. [DOI] [PubMed] [Google Scholar]
- 47.Yang S, Tang F, Zhu H. Alternative splicing in plant immunity. International Journal of Molecular Sciences. 2014 doi: 10.3390/ijms150610424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lai Y, Eulgem T. Transcript-level expression control of plant NLR genes. Molecular Plant Pathology. 2018 doi: 10.1111/mpp.12607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ayliffe MA, et al. Analysis of alternative transcripts of the flax L6 rust resistance gene. Plant J. 1999 doi: 10.1046/j.1365-313X.1999.00377.x. [DOI] [PubMed] [Google Scholar]
- 50.Schornack S, et al. The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 2004 doi: 10.1046/j.1365-313X.2003.01937.x. [DOI] [PubMed] [Google Scholar]
- 51.Cesari S, et al. The Rice Resistance Protein Pair RGA4/RGA5 Recognizes the Magnaporthe oryzae Effectors AVR-Pia and AVR1-CO39 by Direct Binding. Plant Cell. 2013;25:1463–1481. doi: 10.1105/tpc.112.107201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gou JY, et al. Wheat stripe rust resistance protein WKS1 reduces the ability of the thylakoid-associated ascorbate peroxidase to detoxify reactive oxygen species. Plant Cell. 2015 doi: 10.1105/tpc.114.134296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sela H, et al. Ancient diversity of splicing motifs and protein surfaces in the wild emmer wheat (Triticum dicoccoides) LR10 coiled coil (CC) and leucine-rich repeat (LRR) domains. Mol Plant Pathol. 2012 doi: 10.1111/j.1364-3703.2011.00744.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dinesh-Kumar SP, Baker BJ. Alternatively spliced N resistance gene transcripts: Their possible role in tobacco mosaic virus resistance. Proc Natl Acad Sci U S A. 2000 doi: 10.1073/pnas.020367497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang XC, Gassmann W. RPS4-Mediated Disease Resistance Requires the Combined Presence of RPS4 Transcripts with Full-Length and Truncated Open Reading Frames. Plant Cell. 2003 doi: 10.1105/tpc.013474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tang F, Yang S, Gao M, Zhu H. Alternative splicing is required for RCT1-mediated disease resistance in Medicago truncatula. Plant Mol Biol. 2013 doi: 10.1007/s11103-013-0068-6. [DOI] [PubMed] [Google Scholar]
- 57.Liang X, Zhou J-M. Receptor-Like Cytoplasmic Kinases: Central Players in Plant Receptor Kinase-Mediated Signaling. Annu Rev Plant Biol. 2018 doi: 10.1146/annurev-arplant-042817-040540. [DOI] [PubMed] [Google Scholar]
- 58.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci U S A. 2010 doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010 doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 60.Feng F, et al. A Xanthomonas uridine 5’-monophosphate transferase inhibits plant immune kinases. Nature. 2012 doi: 10.1038/nature10962. [DOI] [PubMed] [Google Scholar]
- 61.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science (80- ) 2003 doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 62.Adachi H, Derevnina L, Kamoun S. NLR singletons, pairs, and networks: evolution, assembly, and regulation of the intracellular immunoreceptor circuitry of plants. Current Opinion in Plant Biology. 2019 doi: 10.1016/j.pbi.2019.04.007. [DOI] [PubMed] [Google Scholar]
- 63.Ade J, DeYoung BJ, Golstein C, Innes RW. Indirect activation of a plant nucleotide binding site-leucine-rich repeat protein by a bacterial protease. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0608779104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Qi D, et al. Recognition of the protein kinase Avrpphb Susceptible1 by the disease resistance protein Resistance To Pseudomonas Syringae5 Is dependent on S-acylation and an exposed loop in Avrpphb Susceptible. Plant Physiol. 2014 doi: 10.1104/pp.113.227686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Corbesier L, et al. FT protein movement contributes to long-distance signaling in floral induction of Arabidopsis. Science (80- ) 2007 doi: 10.1126/science.1141752. [DOI] [PubMed] [Google Scholar]
- 66.Freeling M. Bias in Plant Gene Content Following Different Sorts of Duplication: Tandem, Whole-Genome, Segmental, or by Transposition. Annu Rev Plant Biol. 2009 doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 67.Liu ZQ, et al. SRC2-1 is required in PcINF1-induced pepper immunity by acting as an interacting partner of PcINF1. J Exp Bot. 2015 doi: 10.1093/jxb/erv161. [DOI] [PubMed] [Google Scholar]
- 68.Bushnell WR. Aggregation of Host Cytoplasm and the Formation of Papillae and Haustoria in Powdery Mildew of Barley. Phytopathology. 1975 doi: 10.1094/phyto-65-310. [DOI] [Google Scholar]
- 69.Vaddepalli P, et al. The C2-domain protein QUIRKY and the receptor-like kinase STRUBBELIG localize to plasmodesmata and mediate tissue morphogenesis in Arabidopsis thaliana. Dev. 2014 doi: 10.1242/dev.113878. [DOI] [PubMed] [Google Scholar]
- 70.Lowe I, Cantu D, Dubcovsky J. Durable resistance to the wheat rusts: Integrating systems biology and traditional phenotype-based research methods to guide the deployment of resistance genes. Euphytica. 2011 doi: 10.1007/s10681-010-0311-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schmolke M, Mohler V, Hartl L, Zeller FJ, Hsam SLK. A new powdery mildew resistance allele at the Pm4 wheat locus transferred from einkorn (Triticum monococcum) Mol Breed. 2012 doi: 10.1007/s11032-011-9561-2. [DOI] [Google Scholar]
- 72.Pont C, et al. Tracing the ancestry of modern bread wheats. Nat Genet. 2019 doi: 10.1038/s41588-019-0393-z. [DOI] [PubMed] [Google Scholar]
- 73.McNally KE, et al. Distinct domains of the AVRPM3A2/F2 avirulence protein from wheat powdery mildew are involved in immune receptor recognition and putative effector function. New Phytol. 2018 doi: 10.1111/nph.15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Menardo F, et al. Hybridization of powdery mildew strains gives rise to pathogens on novel agricultural crop species. Nat Genet. 2016;48:201–205. doi: 10.1038/ng.3485. [DOI] [PubMed] [Google Scholar]
- 75.Zeng FS, et al. Virulence and diversity of Blumeria graminis f. sp. tritici populations in China. J Integr Agric. 2014;13:2424–2437. [Google Scholar]
- 76.Thordal-Christensen H, Zhang Z, Wei Y, Collinge DB. Subcellular localization of H2O2 in plants. H2O2 accumulation in papillae and hypersensitive response during the barley-powdery mildew interaction. Plant J. 1997 doi: 10.1046/j.1365-313X.1997.11061187.x. [DOI] [Google Scholar]
- 77.Sánchez-Martín J, Rubiales D, Prats E. Resistance to powdery mildew (Blumeria graminis f.sp. avenae) in oat seedlings and adult plants. Plant Pathol. 2011;60:846–856. [Google Scholar]
- 78.Rubiales D, Carver TLW. Defence reactions of Hordeum chilense accessions to three formae speciales of cereal powdery mildew fungi. Can J Bot. 2000 doi: 10.1139/cjb-78-12-1561. [DOI] [Google Scholar]
- 79.Ma ZQ, Wei JB, Cheng SH. PCR-based markers for the powdery mildew resistance gene Pm4a in wheat. Theor Appl Genet. 2004 doi: 10.1007/s00122-004-1605-0. [DOI] [PubMed] [Google Scholar]
- 80.Bustin SA, et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009 doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 81.Brunner S, et al. Transgenic Pm3 multilines of wheat show increased powdery mildew resistance in the field. Plant Biotechnol J. 2012;10:398–409. doi: 10.1111/j.1467-7652.2011.00670.x. [DOI] [PubMed] [Google Scholar]
- 82.Hurni S, et al. The maize disease resistance gene Htn1 against northern corn leaf blight encodes a wall-associated receptor-like kinase. Proc Natl Acad Sci U S A. 2015;112:8780–5. doi: 10.1073/pnas.1502522112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Christensen AH, Quail PH. Ubiquitin promoter-based vectors for high-level expression of selectable and/or screenable marker genes in monocotyledonous plants. Transgenic Res. 1996 doi: 10.1007/BF01969712. [DOI] [PubMed] [Google Scholar]
- 84.Reed J, et al. Phosphomannose isomerase: An efficient selectable marker for plant transformation. Vitr Cell Dev Biol - Plant. 2001 doi: 10.1007/s11627-001-0024-z. [DOI] [Google Scholar]
- 85.Brunner S, et al. Transgenic Pm3b wheat lines show resistance to powdery mildew in the field. Plant Biotechnol J. 2011;9:897–910. doi: 10.1111/j.1467-7652.2011.00603.x. [DOI] [PubMed] [Google Scholar]
- 86.Wright M, et al. Efficient biolistic transformation of maize (Zea mays L.) and wheat (Triticum aestivum L.) using the phosphomannose isomerase gene, pmi, as the selectable marker. Plant Cell Rep. 2001 doi: 10.1007/s002990100318. [DOI] [PubMed] [Google Scholar]
- 87.Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci U S A. 2009;106:9519–9524. doi: 10.1073/pnas.0904152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Holzberg S, Brosio P, Gross C, Pogue GP. Barley stripe mosaic virus-induced gene silencing in a monocot plant. Plant J. 2002 doi: 10.1046/j.1365-313X.2002.01291.x. [DOI] [PubMed] [Google Scholar]
- 89.Loutre C, et al. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 2009;60:1043–1054. doi: 10.1111/j.1365-313X.2009.04024.x. [DOI] [PubMed] [Google Scholar]
- 90.Scofield SR, Huang L, Brandt AS, Gill BS. Development of a virus-induced gene-silencing system for hexaploid wheat and its use in functional analysis of the Lr21-mediated leaf rust resistance pathway. Plant Physiol. 2005 doi: 10.1104/pp.105.061861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Bhullar NK, Street K, Mackay M, Yahiaoui N, Keller B. Unlocking wheat genetic resources for the molecular identification of previously undescribed functional alleles at the Pm3 resistance locus. Proc Natl Acad Sci U S A. 2009 doi: 10.1073/pnas.0904152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Xing L, et al. Pm21 from Haynaldia villosa Encodes a CC-NBS-LRR Protein Conferring Powdery Mildew Resistance in Wheat. Molecular Plant. 2018 doi: 10.1016/j.molp.2018.02.013. [DOI] [PubMed] [Google Scholar]
- 93.Himmelbach A, et al. A set of modular binary vectors for transformation of cereals. Plant Physiol. 2007;145:1192–200. doi: 10.1104/pp.107.111575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ishizaki K, et al. Development of gateway binary vector series with four different selection markers for the liverwort marchantia polymorpha. PLoS One. 2015 doi: 10.1371/journal.pone.0138876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hofgen R, Willmitzer L. Storage of competent cells for Agrobacterium transfonnation. Nucleic Acids Res. 1988;16:9877. doi: 10.1093/nar/16.20.9877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Voinnet O, Rivas S, Mestre P, Baulcombe D. An enhanced transient expression system in plants based on suppression of gene silencing by the p19 protein of tomato bushy stunt virus. Plant J. 2003 doi: 10.1046/j.1365-313X.2003.01676.x. [DOI] [PubMed] [Google Scholar]
- 97.Singh SP, et al. Evolutionary divergence of the rye Pm17 and Pm8 resistance genes reveals ancient diversity. Plant Mol Biol. 2018;98:249–260. doi: 10.1007/s11103-018-0780-3. [DOI] [PubMed] [Google Scholar]
- 98.Gronnier J, et al. Structural basis for plant plasma membrane protein dynamics and organization into functional nanodomains. Elife. 2017 doi: 10.7554/eLife.26404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu J, Abràmoff MD, Magalhães PJ, Ram SJ, et al. Image processing with Image. J Biophotonics international. 2004;11(7):36–42. doi: 10.1201/9781420005615.ax4. Opt. Express 2011. [DOI] [Google Scholar]
- 100.French AP, Mills S, Swarup R, Bennett MJ, Pridmore TP. Colocalization of fluorescent markers in confocal microscope images of plant cells. Nat Protoc. 2008 doi: 10.1038/nprot.2008.31. [DOI] [PubMed] [Google Scholar]
- 101.Vrana J, et al. Flow sorting of mitotic chromosomes in common wheat (Triticum aestivum L.) Genetics. 2000;156:2033–2041. doi: 10.1093/genetics/156.4.2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Giorgi D, et al. FISHIS: Fluorescence In Situ Hybridization in Suspension and Chromosome Flow Sorting Made Easy. PLoS One. 2013;8 doi: 10.1371/journal.pone.0057994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kubaláková M, et al. Analysis and sorting of rye (Secale cereale L.) chromosomes using flow cytometry. Genome. 2003;46:893–905. doi: 10.1139/g03-054. [DOI] [PubMed] [Google Scholar]
- 104.Šimková H, Číhalíková J, Vrána J, Lysák MA, Doležel J. Preparation of HMW DNA from plant nuclei and chromosomes isolated from root tips. Biol Plant. 2003 doi: 10.1023/A:1024322001786. [DOI] [Google Scholar]
- 105.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011 doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 106.Marchler-Bauer A, et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Krogh A, Larsson B, Von Heijne G, Sonnhammer ELL. Predicting transmembrane protein topology with a hidden Markov model: Application to complete genomes. J Mol Biol. 2001 doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 108.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction-the Phobius web server. Nucleic Acids Res. 2007 doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mascher M, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017 doi: 10.1038/nature22043. [DOI] [PubMed] [Google Scholar]
- 110.Luo MC, et al. Genome sequence of the progenitor of the wheat D genome Aegilops tauschii. Nature. 2017 doi: 10.1038/nature24486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ling HQ, et al. Genome sequence of the progenitor of wheat A subgenome Triticum urartu. Nature. 2018 doi: 10.1038/s41586-018-0108-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Avni R, et al. Wild emmer genome architecture and diversity elucidate wheat evolution and domestication. Science (80- ) 2017 doi: 10.1126/science.aan0032. [DOI] [PubMed] [Google Scholar]
- 113.Maccaferri M, et al. Durum wheat genome highlights past domestication signatures and future improvement targets. Nat Genet. 2019 doi: 10.1038/s41588-019-0381-3. [DOI] [PubMed] [Google Scholar]
- 114.Ronquist F, et al. Mrbayes 3.2: Efficient bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012 doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cook DE, Mesarich CH, Thomma BPHJ. Understanding Plant Immunity as a Surveillance System to Detect Invasion. Annu Rev Phytopathol. 2015 doi: 10.1146/annurev-phyto-080614-120114. [DOI] [PubMed] [Google Scholar]
- 116.Buchmann JP, Matsumoto T, Stein N, Keller B, Wicker T. Inter-species sequence comparison of Brachypodium reveals how transposon activity corrodes genome colinearity. Plant J. 2012 doi: 10.1111/j.1365-313X.2012.05007.x. [DOI] [PubMed] [Google Scholar]
- 117.Ma J, Bennetzen JL. Rapid recent growth and divergence of rice nuclear genomes. Proc Natl Acad Sci U S A. 2004 doi: 10.1073/pnas.0403715101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.R Development Core Team R Development Core Team. R: a language and environment for statistical computing. R: A Language and Environmental for Estatistical Computing. 2020 [Google Scholar]
- 119.Omasits U, Ahrens CH, Müller S, Wollscheid B. Protter: Interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014 doi: 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- 120.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015 doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Zeng FS, et al. Virulence and diversity of Blumeria graminis f. sp. tritici populations in China. J Integr Agric. 2014 doi: 10.1016/S2095-3119(13)60669-3. [DOI] [Google Scholar]
- 122.Pont C, et al. Tracing the ancestry of modern bread wheats. Nat Genet. 2019;51:905–911. doi: 10.1038/s41588-019-0393-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. Sequence data were deposited at the NCBI GenBank under the accession numbers MT783929 (Pm4b_V1 CDS) and MT783930 (Pm4b_V2 CDS), and at the NCBI short read archive (SRA) database under the accession number PRJNA646941 (flow-sorted chromosome 2A of eight Fed-Pm4b mutants and the wild-type Fed-Pm4b). All Blumeria graminis f. sp. tritici (Bgt) isolates listed in Supplementary Table 1 are kept alive in the Department of Plant and Microbial Biology of the University of Zurich and are available upon request. Any additional data that support the findings of this study are available from the corresponding author upon reasonable request. The databases that we used are all publicly available, please see Methods and the Nature Research Reporting Summary linked to this article.