Abstract
Recent studies show that molecular convergence plays an unexpectedly common role in the evolution of convergent phenotypes. We exploited this phenomenon to find candidate loci underlying resistance to the emerald ash borer (EAB; Agrilus planipennis), the USA’s most costly invasive forest insect to date, within the pan-genome of ash trees (the genus Fraxinus). We show that EAB-resistant taxa occur within three independent phylogenetic lineages. In genomes from these resistant lineages, we detect 53 genes with evidence of convergent amino acid evolution. Gene tree reconstruction indicates that for 48 of these candidates, the convergent amino acids are more likely to have arisen via independent evolution than by another process, such as hybridisation or incomplete lineage sorting. Seven of the candidate genes have putative roles connected to the phenylpropanoid biosynthesis pathway and 17 relate to herbivore recognition, defence signalling or programmed cell death. Evidence for loss-of-function mutations among these candidates is more frequent in susceptible species, than in resistant ones. Our results on evolutionary relationships, variability in resistance, and candidate genes for defence response within the ash genus could inform breeding for EAB resistance, facilitating ecological restoration in areas this beetle has invaded.
Ash trees (Fraxinus) are key components of temperate forest ecosystems1,2, the health of which affects the provision of ecosystem services including climate change mitigation3. The continued survival of ash in North America and Europe is threatened by a highly destructive invasive insect4,5, the emerald ash borer (EAB; Agrilus planipennis). This wood boring beetle has thus far proved to be highly destructive to the majority of Fraxinus species it has encountered outside of its native range in East Asia4,5. In North America EAB has already killed hundreds of millions of ash trees and billions more are at risk6. In Europe, the beetle has established an invasive range in Moscow from which it is spreading5,7, and there is increasing concern about the threat posed to native F. excelsior populations that have already been severely damaged by ash dieback disease8,9. EAB is a minor pest species within its native range in East Asia10–12 with outbreaks generally associated with the planting of exotic Fraxinus species, such as F. americana and F. velutina in China11–13. Commonly reported natural hosts of EAB (such as F. chinensis and F. mandshurica 13,14) are largely resistant unless otherwise stressed, such as when they are grown along roadsides or in plantations12,13 or under drought conditions15. Fraxinus species from within the native range of EAB may therefore provide a source of genes for resistance breeding4.
Although some genes and compounds that may contribute to resistance in certain Fraxinus taxa have been identified via transcriptomics, proteomics, and the analysis of metabolites16–18, mechanisms of defence response in known EAB resistant species are still not well understood, and some Fraxinus species have not been tested for resistance. We took a genus-wide approach to detect genes related to EAB resistance, inspired by a growing number of studies finding evidence that convergent molecular mutations can provide the genetic basis for independent evolution of phenotypic traits19–22, including cases involving recurrent change at identical amino acid sites23,24. We tested for both phenotypic and molecular convergence relating to EAB-resistance in Fraxinus by assessing 26 taxa with EAB egg bioassays and by assembling de novo and analysing whole genome sequences for 24 diploid species and subspecies, with the aim of identifying candidate genes for resistance.
Results
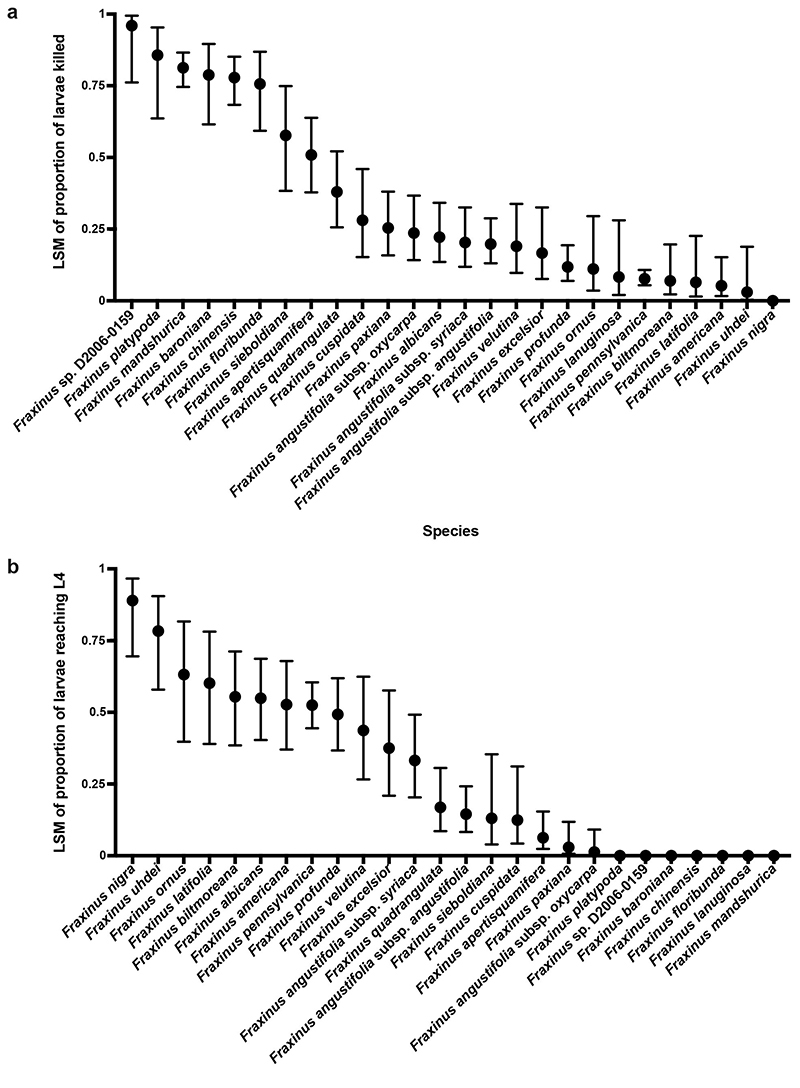
To better understand how resistance to EAB varies across the genus, and to examine evidence for convergent evolution of this trait, we assessed resistance to EAB for 26 Fraxinus taxa (Supplementary Table 1) representing four of the six taxonomic sections and 48% of species25. Tree resistance was scored according to the instar, health and weight of EAB larvae in the stems of inoculated trees eight weeks after infestation26 (Methods; Supplementary Table 1). In F. baroniana, F. chinensis, F. floribunda, F. mandshurica, F. platypoda and Fraxinus sp. D2006-0159, least squares means (LSM) of the proportion of host killed larvae (number of larvae killed by tree defence response divided by total larvae entering the tree) were >0.75 (Fig. 1a, Supplementary Table 2) and LSM estimate of the proportion of larvae successfully entering the tree that reached the L4 instar was zero (Fig. 1b, Supplementary Table 2), indicating that these species are resistant to EAB. In contrast, all other taxa tested had a LSM proportion of larvae killed of 0.58 or less (Fig. 1a) and had LSM for L4 larvae proportion between 0 and 0.89 (Fig. 1b).
Figure 1. Fraxinus species’ resistance to EAB in bioassays.
a, b, Measures of resistance of different Fraxinus taxa to EAB larvae. The x axis shows taxa tested; the y axis shows least squares means (LSM) estimate of the proportion of larvae successfully entering the tree that were killed by a host defence response (a) or LSM estimate of the proportion of larvae successfully entering the tree that reached the L4 instar (b). The error bars represent 95% confidence intervals. Fraxinus sp. D2006-0159 is a genotype from China for which we could not determine a recognised species name. Fraxinus biltmoreana, F. chinensis, F. lanuginosa, F. profunda and F. uhdei are polyploids and were not included in the genomic analyses; F. apertisquamifera was also not included.
To infer a robust phylogenetic framework for Fraxinus, within which to understand the evolution of EAB resistance, and to allow analysis of evidence for molecular convergence, we sequenced and assembled the genomes of 28 individuals from 26 diploid taxa representing all six sections within the genus25, including a common EAB-susceptible accession and a rare putatively EAB-resistant accession26 for F. pennsylvanica (Supplementary Table 3). Estimated genome sizes (1C-values) of the individuals selected for sequencing range between c. 700Mb and 1100Mb (Supplementary Table 4); for all individuals we generated c. 35 to 85X whole genome shotgun coverage with Illumina sequencing platforms (Methods; Supplementary Table 4). On assembly (Methods) these data generated 133,719 to 715,871 scaffolds for each individual, with N50s ranging from 1,987 to 50,545bp (Supplementary Table 4); BUSCO analysis of the genome assemblies (Methods) found between 78.4% and 94.7% of genes were present (either complete or fragmented; Supplementary Table 4). Therefore, despite some of the assemblies being highly fragmented, a sufficient proportion of the gene space had been assembled to allow tests for amino acid convergence to be carried out (see below). We annotated genes in these assemblies via a reference based approach (Methods) using the published genome annotation of F. excelsior 27. We clustered the protein sequences of these genes into putative orthologue groups (OGs; Methods), also including protein sequences from the F. excelsior reference genome and the published genome annotations of Olea europaea 28, Erythranthe guttata 29 and Solanum lycopersicum 30. We found a total of 87,194 OGs, each containing sequences from between two and 32 taxa; 1,403 OGs included a sequence from all 32 taxa.
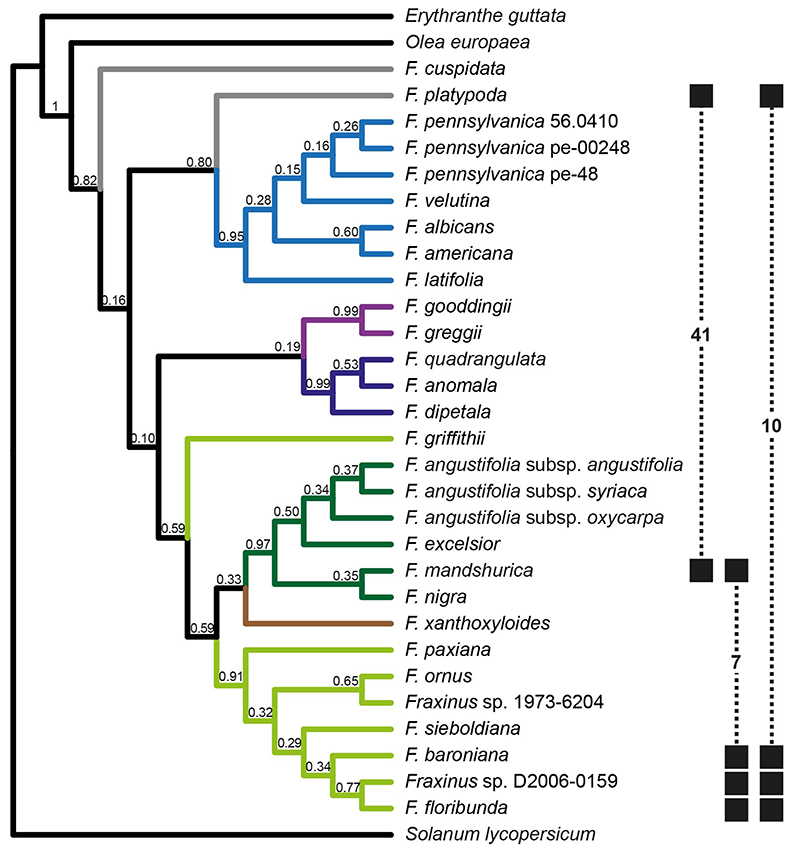
We generated multiple sequence alignments for the 1,403 OGs including all taxa and inferred gene-trees for each (Methods). In order to generate a species-tree estimate for Fraxinus, we conducted Bayesian concordance analysis (Methods). This resulted in a tree based on 272 phylogenetically informative low copy genes (Fig. 2 and Supplementary Note 1). Within this tree, the EAB-resistant taxa identified from our bioassays occurred in three independent lineages: (1) F. mandshurica occurred within a clade corresponding to section Fraxinus that also included susceptible taxa; (2) F. platypoda was sister to a clade corresponding to section Melioides, which includes most of the susceptible American species; (3) F. baroniana, F. floribunda and Fraxinus sp. D2006-0159 clustered together, within a larger clade that included most species in section Ornus, including susceptible F. ornus. Thus, by combining phenotypic data with the most highly-evidenced phylogenetic hypothesis for Fraxinus to date, we show that EAB-resistance has evolved convergently within the genus. A further resistant taxon identified from our bioassay, F. chinensis, was not included in the species-tree analysis because it is a polyploid31.
Figure 2. Species-tree for the genus Fraxinus.
Primary concordance tree from 272 phylogenetically informative loci found in all taxa, inferred via Bayesian concordance analysis with BUCKy. Taxonomic sections within Fraxinus, according to Wallander25, are shown in different colours: dark blue - section Dipetalae; dark green - section Fraxinus; light blue - section Melioides; light green - section Ornus; purple - section Pauciflorae; brown - section Sciadanthus. Fraxinus species not placed into a specific section (incertae sedis) are coloured grey and outgroups black. Numbers above the branches are sample-wide concordance factors. Filled squares (linked by dashed lines) indicate the resistant taxa included in the three pairwise convergence analyses, with the number of candidate genes found from that comparison shown; numbers do not sum to 53 (i.e. the total number of candidate genes) because some genes were identified by more than one pairwise comparison.
We searched for amino acid variants putatively convergent between the resistant lineages using an approach that identifies loci with a level of convergence in excess of that likely to be due to chance alone (grand-conv; Methods). We conducted three pairwise analyses of lineages, with each pair representing two of the three independent lineages of diploid EAB-resistant taxa identified from our egg bioassays and species-tree analysis: (1) F. mandshurica versus F. platypoda, (2) F. mandshurica versus F. baroniana, F. floribunda and Fraxinus sp. D2006-0159, (3) F. platypoda versus F. baroniana, F. floribunda and Fraxinus sp. D2006-0159. In all these analyses we included three outgroups and five Fraxinus species with high susceptibility (Methods). Each analysis was based on alignments of OGs found in all of the included taxa: 3,454 OGs in analysis 1, 3,097 OGs in analysis 2 and 3,026 OGs in analysis 3. Our candidate amino acid variants were those identified by grand-conv as convergent (minimum posterior probability of 0.90) within loci predicted to have the highest excess of convergent over divergent substitutions in the resistant lineages (Methods).
Amino acid states that appear convergent between lineages in a genus could alternatively be due to the unintentional comparison of different gene duplicates (paralogues), or may have arisen from introgressive hybridisation or incomplete lineage sorting (ILS). We checked our candidate loci for the possible confounding effect of paralogy, as well as gene model and alignment errors, leaving a total of 67 amino acid sites in 53 genes (Supplementary Note 2 and Supplementary Table 5). We inferred gene trees for these 53 remaining genes from their CDS alignments. If introgression or ILS were the cause of the shared amino acid states (Supplementary Table 5), we would expect sequences containing apparently convergent residues to group together within their gene-tree, even when nucleotides encoding those residues are removed. In all but one case (OG20252; Supplementary Fig. 1), the pattern of amino acid variation at candidate sites is better explained by a hypothesis of convergent point mutations, rather than introgressive hybridisation or ILS (Supplementary Fig. 1). For four loci (OG11013, OG20859, OG37870 and OG41448) the gene-tree analysis suggests that the state identified as convergent by grand-conv is ancestral within Fraxinus, with change occurring in the other direction (i.e. from the “convergent” state identified by grand-conv to the “non-convergent” state; Supplementary Table 5).
We looked for evidence of loss-of-function of the 53 candidate genes, based on the presence of frameshifts, stop codon gains and start codon losses, in any of the Fraxinus individuals included in our convergence analyses. Six of our 53 candidate genes show evidence of lacking a fully functional allele in a susceptible taxon, compared with one for resistant taxa (Supplementary Note 3 and Supplementary Table 6), suggesting these susceptible taxa may have impaired function of some genes related to defence against EAB.
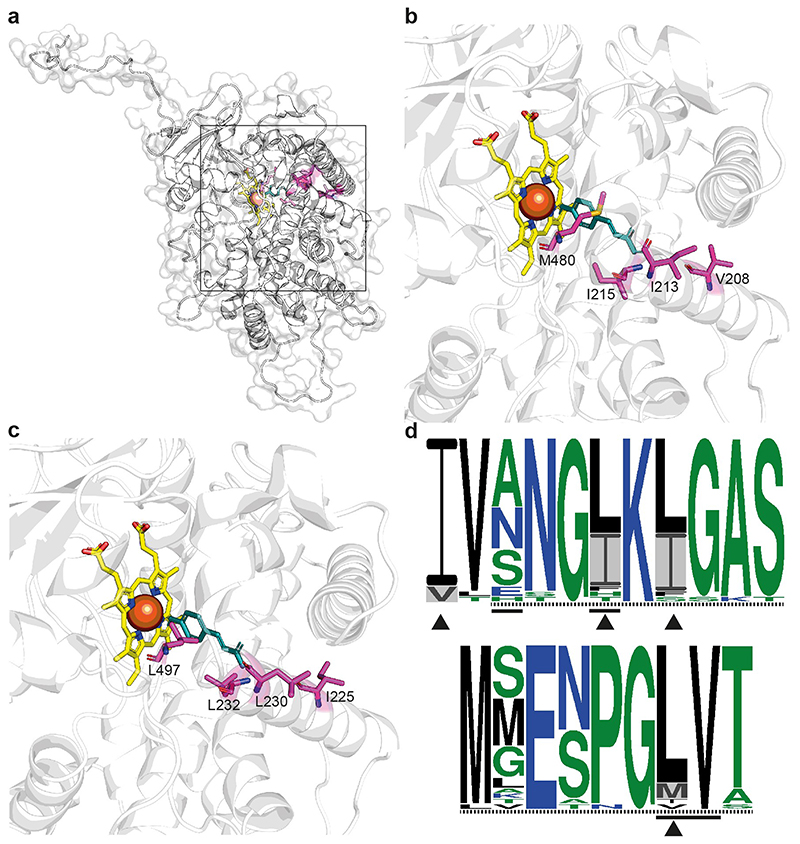
Among our 53 candidate genes, seven have putative roles relating to the phenylpropanoid biosynthesis pathway (Supplementary Note 4). This pathway generates antifeedant and cytotoxic compounds, as well as products involved in structural defence, such as lignin32; it can contribute to indirect defence by producing volatiles which attract parasitoids or predators33. Loci OG15551, OG853 and OG16673 are of particular interest. Four convergent amino acids were identified in OG15551 (Fig. 3), a paralogue of CYP98A3 (Supplementary Fig. 2), which encodes a critical phenylpropanoid pathway enzyme34. Three of the four residues fall within CYP98A3 putative substrate recognition sites, with two at positions predicted to contact the substrate35 including a leucine (sulphur containing)/methionine (non-sulphur containing) variant (Fig. 3), suggesting these variants may affect the protein’s function. OG853 is apparently orthologous to MED5a (also called RFR1; Supplementary Fig. 2), a known regulator of the phenylpropanoid pathway36,37 that seems to be involved in regulation of defence response genes38. OG16673 is a likely glycoside hydrolase; putative Arabidopsis thaliana homologues of OG16673 belong to glycoside hydrolase family 1 and have beta-glucosidase activity, with functions such as chemical defence against herbivory, lignification and control of phytohormone levels39. A role for beta-glucosidases in defence against EAB in individual Fraxinus species has been previously suggested on the basis of chemical40 and transcriptomic18 data, and several metabolomic studies have indicated that products of the phenylpropanoid pathway could be involved41.
Figure 3. Predicted protein structure for OG15551.
a, Predicted structure for OG15551, modelled using the protein sequence for the EAB-resistant species Fraxinus mandshurica. The black box indicates the region containing the active site, which is enlarged in b and c. b, Region containing the predicted active site in F. mandshurica, showing the four amino acid sites at which evidence for convergence between EAB-resistant species was detected. The putative substrate, p-Coumarate, is shown in blue and the heme cofactor in yellow. c, Region containing the predicted active site in the EAB-susceptible F. pennsylvanica pe-48, showing the amino acid states found at the four sites at which evidence for convergence between EAB-resistant species was detected; the putative substrate and cofactor are shown as in b. d, Sequence logos for OG15551 and putatively homologous sequences from other angiosperms for regions containing sites at which evidence of convergence was detected (positions 208-218 (top) and 474-482 (bottom) in the F. excelsior reference protein), showing the degree of sequence conservation across 30 genera. The height of each residue indicates its relative frequency at that site; amino acids are coloured according to their hydrophobicity (blue = hydrophilic; black = hydrophobic; green = neutral). Dashed lines indicate substrate recognition sites and solid lines residues that are predicted to contact the substrate in the A. thaliana CYP98A3 protein35; arrowheads indicate sites at which evidence of convergence between EAB-resistant taxa was detected and grey shading shows the amino acid states associated with resistance.
We found 15 candidate genes (Supplementary Note 4) with possible roles in perception and signalling relevant to defence response against herbivorous insects33. OG4469 is a probable orthologue of AtG-LecRK-1.6, a G-type lectin receptor kinase (LecRK) with ATP binding activity (Supplementary Note 4.3). G-type LecRKs can act as pattern recognition receptors (PRRs) in the perception of feeding insects42; extracellular ATP is a damage-associated molecular pattern (DAMP) whose perception can trigger defence response related genes42. OG38407 appears orthologous to SNIPER4 (Supplementary Fig. 1), a F-box protein encoding gene involved in regulating turnover of defence-response related proteins, for optimal defence activation43; the convergent site is in a leucine rich repeat (LRR) region (Extended Data Fig. 1), which is involved in recognition of substrate proteins for ubiquitination43,44.
Several genes appear to relate to phytohormone biosynthesis and signalling, including those with putative functions in jasmonate (JA; OG41448), brassinosteroid (OG43828), cytokinin (OG39275) and abscisic acid (ABA; OG47560) biosynthesis, and GO terms associated with hormone metabolism and biosynthesis are significantly enriched among our set of candidate genes (Supplementary Note 5; Supplementary Table 7). JA signalling is the central regulatory pathway for defence response against insect herbivores42,45 whereas brassinosteroids and cytokinins can play important roles in insect resistance, via modulation of the JA pathway45,46. ABA is induced by herbivory and is a known modulator of resistance to insect herbivores42,45. OG11720 is putatively orthologous to NRT1.5 (also known as NPF7.3), a member of the NRT1/PTR family47 which is involved in transport of multiple phytohormones (Supplementary Note 4.3); a transcript matching this gene family had decreased expression in response to both mechanical wounding and EAB feeding in F. pennsylvanica 18. Putative functions of further candidates relate to other signalling molecules involved in triggering defence response (Supplementary Note 4.5), including calcium (OG50989)33,48, nitric oxide (NO; OG21033)49,50 and spermine (OG33348)51. Increased resistance to EAB can be artificially induced in Fraxinus species with otherwise high susceptibility52, leading to the suggestion that susceptible species may fail to recognise, or respond quickly enough to, early signs of EAB attack41. Our identification of candidate genes putatively involved in perception and signalling underlines the possibility of differences between EAB-resistant and susceptible Fraxinus species in both their ability to sense and react to attacking insects.
Hypersensitive response (HR), involving programmed cell death (PCD), is associated with effector-triggered immunity in response to microbial pathogens53 but can also be induced by insect herbivory54 and oviposition55. OG16739 and OG37870 are candidates with putative roles related to HR-like effects and PCD. OG16739 has homologues that control cell death in response to wounding, via the induction of ethylene and the expression of defence and senescence related genes56. OG37870 may be orthologous to genes that seem to play a role in controlling PCD of xylem elements57. Candidate loci whose putative functions lack an obvious link to plant defence response (Supplementary Note 4), could be involved in other phenotypic traits shared between EAB resistant species or may play a role in defence response that is not yet understood. We found that 19 of our 53 candidates match the same A. thaliana genes as transcripts that are differentially expressed in response to elm leaf beetle (either in response to simulated egg deposition, or larval feeding)58, including genes, such as OG24969, whose putative A. thaliana homologues lack a clear defence-related function.
We analysed allelic variation at the 67 amino acid sites within the 53 candidate genes for all sequenced taxa assessed for resistance to EAB. Of the 67 sites, seven only have the EAB-resistance associated state in resistant taxa, and another is only homozygous for the EAB-resistance associated state in resistant taxa (Supplementary Table 8). Of the 53 candidate genes, four are only homozygous in resistant taxa for the EAB-resistance associated state at the candidate amino acid site(s) detected within them (OG853, OG21449, OG36502 and OG37560; Supplementary Table 8). If we omit the genomes of F. nigra, F. excelsior and the three F. angustifolia subspecies (sect. Fraxinus), for 24 of the 53 candidate genes we only find the EAB-resistance associated states in resistant taxa, for 48 genes they are only found in taxa with a LS mean proportion of larvae killed of ≥0.25 and the remaining five genes are only homozygous for the EAB-resistance associated states in taxa with a LS mean proportion of larvae killed of ≥0.25 (Supplementary Table 8).
Analysis of previously generated whole genome sequence data for 37 F. excelsior individuals from different European provenances27 revealed that for 50 of the 67 candidate amino acid sites (occurring in 41 of the 53 genes) the EAB-resistance associated state was present, with evidence for polymorphism at seven of these sites (Supplementary Table 9). None of the EAB-resistance associated amino acid states were found in the putatively resistant F. pennsylvanica genotype (Supplementary Table 8), suggesting that different genes, or different variants within these genes, are involved in the intraspecific variation in susceptibility of this species. Despite this, transcripts inferred to be from 11 of our candidate genes showed evidence for differential expression subsequent to EAB-feeding in F. pennsylvanica (Supplementary Note 6 and Supplementary Table 10) and two gene families that were highlighted as potentially important for response to tissue damage in F. pennsylvanica 18 are also represented among our candidates (see above).
Discussion
It has frequently been suggested that EAB has a coevolutionary history with its native Fraxinus hosts within their shared geographic ranges in East Asia, during which defence mechanisms against EAB may have been selected for12,41,59. All six taxa identified as resistant to EAB on the basis of our egg bioassays are native to Asia25 (Fraxinus sp. D2006-0159 originates from material collected in northern China), including known natural EAB host species F. chinensis and F. mandshurica. In addition to F. chinensis and F. mandshurica, the native range of F. platypoda overlaps with that of EAB14,25. The current native ranges of F. baroniana and F. floribunda 25,60 apparently do not overlap with the native range of EAB14, but we cannot discount the possibility that they did in the past and thus that these species also share a coevolutionary history with EAB. Alternatively, it may be the case that the most recent common ancestor of F. baroniana, F. chinensis, F. floribunda and Fraxinus sp. D2006-0159 (all of which belong to sect. Ornus) was an EAB host species and that resistance has been retained in these extant descendants. Among the resistant Asian species are comparatively close relatives of all three major susceptible North American EAB hosts: F. pennsylvanica, F. americana, and F. nigra. It is known that the closely related F. mandshurica and F. nigra can produce hybrids61,62; the phylogenetic proximity of F. platypoda to F. pennsylvanica and F. americana suggests that it may also be possible to increase resistance in these species via hybrid breeding.
By assessing resistance across the genus and testing for molecular convergence we have provided evidence for candidate genes involved in EAB resistance in Fraxinus. Multiple loci, contributing to different defence responses, appear to underlie this trait. In less than 20 years since it was first detected in North America, EAB has caused devastating damage to native ash populations, to the point where Fraxinus risks being lost entirely as a functional component of forest ecosystems4. Our data may help to target future efforts to increase the resistance of American and European ash species to EAB via breeding or gene editing, an intervention that could be required if these species are to persist in the face of the ongoing threat from this invasive beetle. Moreover, these results highlight the potential to use convergence analyses as an approach for identifying candidate genes for traits of interest in organisms where alternative strategies, such as genome-wide association studies or mapping of quantitative trait loci, may be less feasible.
Methods
Data reporting
For the emerald ash borer resistance assays, experiments were conducted using a randomised block design. No statistical methods were used to predetermine sample size. The investigators were not blinded to allocation during experiments and outcome assessment.
Plant material
All plant materials used in this study were sourced from living or seed collections in the UK or USA. Due to biosecurity measures, we were not able to move living materials between the two countries. In our initial selection of material we relied upon species identifications that had already been made in the arboreta or seed banks within which the materials were held. For each of the accessions included in this study, we PCR amplified and Sanger sequenced the nuclear ribosomal internal transcribed spacer (ITS) region, following standard methods; forward and reverse sequences were assembled into contigs using CLC Genomics Workbench v.8.5.1 (QIAGEN Aarhus, Denmark). As far as possible the identity of all materials was verified by Eva Wallander using morphology and ITS sequences, according to her classification of the genus25. This led to some re-designation of samples: of particular note, one of the three accessions of F. pennsylvanica that we genome sequenced was originally sampled as F. caroliniana, and the accession that we originally sampled and genome sequenced as F. bungeana was determined to be F. ornus (Supplementary Table 3). However, subsequent phylogenetic analysis including allele sequences for this latter individual (see below - Distinguishing between different underlying causes of convergent patterns) indicated it to likely be a hybrid between the F. ornus lineage and another lineage within sect. Ornus and therefore we designate it as Fraxinus sp. 1973-6204. We also designated an accession that was originally sampled for genome sequencing as F. chinensis as Fraxinus sp. D2006-0159 due to uncertainty regarding species delimitation. Furthermore, for genotype vel-4, that was redetermined as F. pennsylvanica, we have maintained its original species name (F. velutina). A list of all materials used in the study is shown in Supplementary Table 3, including initial identifications and subsequent identifications by Eva Wallander, as well as details of voucher specimens.
Emerald ash borer resistance assays
Twenty six Fraxinus taxa (species, subspecies and one taxon of uncertain status) were collected for egg bioassay experiments (Supplementary Table 3). We aimed to test three clonal replicates (grafts or cuttings) of at least two genotypes of each species. For some taxa less than two genotypes were available in the US, and occasionally genotypes did not propagate well by graft or cutting so seedlings from the same seedlot were used instead (details for each taxon are included in Supplementary Table 1). The majority of egg bioassays were conducted in 2015 and 2016 and groups of approximately 20 genotypes were conducted in each set (week within year; Supplementary Table 1) with one grafted ramet, cutting or seedling in each block and all ramets, cuttings and seedlings within the block randomised to location/order of assay. To facilitate comprehensive analysis, the same controls were repeated in each week (susceptible F. pennsylvanica genotype pe-37 and/or pe-39 and resistant F. mandshurica genotype ‘mancana’).
Trees were treated as uniformly as possible prior to inoculation. Adult beetles were reared and used for egg production as previously described26. Inoculations were performed in a greenhouse to keep conditions uniform for the duration of the assay, and to minimise predation of the eggs. We followed the EAB egg transfer bioassay method reported by Koch et al.,26 that had previously been used on genotypes of F. pennsylvanica and F. mandshurica with the changes noted below. The egg dose for each tree was determined according to the method of Duan et al.,63, which takes into account the bark surface area. A target density of 400 eggs per m2 was used; this density is above that reported to allow host defences to kill larvae in green ash, but is in the range where competition and cannibalism are minimised63. Twelve individual eggs, on a small strip cut from the coffee filter paper on which they were laid, were taped to each tree. The spacing was varied between eggs to maintain a consistent target dose (e.g. eggs placed 7.5 cm apart on stem 1.0-1.1cm diameter to eggs placed 3 cm apart on stem 2.5-2.6cm diameter). The portion of the tree where eggs were placed was wrapped in medical gauze to protect from jostling and egg predation. Past experiments have shown that egg assay results are not consistent on stems less than 1 cm in diameter. Due to size differences between some species, to achieve the target dose and avoid placing eggs where the stem diameter was <1 cm, occasionally less than 12 eggs were placed. A total of 2199 egg bioassays (each egg represents a bioassay) were conducted on 61 different genotypes and a total of 206 ramets, cuttings or seedlings.
Occasional ramets, cuttings and/or seedlings were considered as assay failures if less than three larvae successfully entered the tree (i.e. the effective egg dose was too low), or if there were other problems with the tree (too small diameter overall, cultivation issues, etc.), and that replicate was excluded from analysis. The final number of ramets/cuttings/seedlings included for each taxon was as follows: F. albicans n = 9, F. americana n = 6, Fraxinus angustifolia subsp. angustifolia n = 10, F. angustifolia subsp. oxycarpa n = 6, F. angustifolia subsp. syriaca n = 8, F. apertisquamifera n = 8, F. baroniana n = 5, F. biltmoreana n = 6, F. chinensis n = 11, F. cuspidata n = 4, F. excelsior n = 5, F. floribunda n = 7, F. lanuginosa n = 3, F. latifolia n = 3, F. mandshurica n = 24, F. nigra n = 4, F. ornus n = 4, F. paxiana n = 7, F. pennsylvanica n = 39, F. platypoda n = 2, F. profunda n = 12, F. quadrangulata n = 6, F. sieboldiana n = 5, Fraxinus sp. Acc #D2006-0159 n = 2, F. udhei n = 3, F. velutina n = 6. Four weeks after egg attachment each egg was inspected to determine if it had successfully hatched, and if there were signs of the larva entering the tree. Larval entry holes, when detected, were marked to assist with future dissection. At eight weeks, dissection of the entry site was performed and galleries made by larval feeding were carefully traced using a grafting knife to determine the outcome of each hatched egg. Health (dead or alive) and weight (in cases when larvae could be recovered intact) was recorded for each larva, and developmental instar was determined using measurements of head capsule and length64,65. Larvae which had been killed by host defence response were distinguished from those that had died from other causes by examining the tissue immediately surrounding a larva for evidence of browning and/or callous formation (indicating a defence response) and by checking for the absence of evidence of any other causes of death, including cannibalism, parasitism and fungal infection. The total number of eggs for which the larvae successfully entered the tree and the outcome recorded was as follows: F. albicans n = 63, F. americana n = 57, Fraxinus angustifolia subsp. angustifolia n = 101, F. angustifolia subsp. oxycarpa n = 55, F. angustifolia subsp. syriaca n = 59, F. apertisquamifera n = 55, F. baroniana n = 33, F. biltmoreana n = 43, F. chinensis n = 95, F. cuspidata n = 32, F. excelsior n = 36, F. floribunda n = 37, F. lanuginosa n = 24, F. latifolia n = 31, F. mandshurica n = 166, F. nigra n = 30, F. ornus n = 27, F. paxiana n = 59, F. pennsylvanica n = 403, F. platypoda n = 21, F. profunda n = 110, F. quadrangulata n = 50, F. sieboldiana n = 26, Fraxinus sp. Acc #D2006-0159 n = 25, F. udhei n = 33, F. velutina n = 42.
Preliminary exploratory data analysis indicated that the proportion of “tree killed” (i.e. larvae killed by tree defence response) and the proportion of live L4 larvae (number divided by the number of larvae that entered the tree) were the best variables to distinguish resistance versus susceptibility at the species level. We fitted a generalised linear mixed model to the proportion tree killed and proportion L4 using the GLIMMIX procedure in SAS v.9.4. The final model specification is proportion as a binomial distribution with a logit link function, species as a fixed effect, and block/replicate nested within sequential week (week within year) as a random effect (this allowed for comprehensive analysis over years and weeks with correct variance/covariance restrictions to account for dependence of eggs within tree and independence of trees in different blocks). Non-significant predictors for propagule type and egg density were eliminated from the final model. Least squares means of tree killed or L4 proportion were calculated with confidence intervals on the data scale (proportion).
Genome size estimation by flow cytometry (FC)
We used FC to estimate the genome size of individuals used for whole genome sequencing (Supplementary Table 4). Fraxinus samples from UK collections were prepared and analysed as described in Pellicer et al.66, with the exception that ‘general purpose isolation buffer’ (GPB67) without the addition of 3% polyvinylpyrrolidone (PVP-40) and LB01 buffer68 were used for some samples. Oryza sativa (‘IR-36’; 1C = 0.50 pg69) was used as an internal standard. For each individual analysed, two samples were prepared (from separate leaves or different parts of the same leaf) and two replicates of each sample run. Fraxinus samples from US collections were analysed using a Sysmex CyFlow Space flow cytometer, as described in Whittemore and Xia70; Pisum sativum (‘Ctirad’; 1C = 4.54 pg71) and Glycine max (‘Williams 82’; 1C = 1.13 pg72) were used as internal standards. For each individual analysed, six samples were prepared (from separate leaves or different parts of the same leaf) and three samples run with each size standard.
DNA extraction
Total genomic DNA was extracted from fresh, frozen or silica-dried leaf or cambial material, using either a CTAB extraction protocol modified from Doyle and Doyle73 or using a Qiagen DNeasy® Plant Mini or Maxi kit.
Genome sequencing and assembly
For each of the 28 diploid individuals selected for whole genome sequencing (Supplementary Table 3), sufficient Illumina sequence data were generated to provide a minimum of c. 30x coverage of the 1C genome size, based on the C-value estimates obtained for the same individuals (see above - Genome size estimation by flow cytometry (FC)), or those of closely related taxa. Libraries with average insert sizes of 300 or 350bp, 500 or 550bp and 800bp or 850bp were prepared from total genomic DNA by the Genome Centre, at Queen Mary University of London, and the Centre for Genomic Research, at the University of Liverpool. Paired-end reads of 125, 150 or 151 nucleotides were generated using the Illumina NextSeq 500, HiSeq 2500 and HiSeq 4000 platforms (Illumina, San Diego, California, USA); see Supplementary Table 4 for the exact combination of libraries, read lengths and sequencing platforms used for each individual. For selected taxa, chosen to represent different sections within the genus, we also generated data from long mate-pair (LMP) libraries (Supplementary Table 4). LMP libraries with average insert sizes of 3kb and 10kb were prepared from total genomic DNA by the Centre for Genomic Research, at the University of Liverpool, and sequenced on an Illumina HiSeq 2500 to generate reads of 125 nucleotides to a depth of c. 10x coverage of the 1C genome size.
Initial assessment of sequence quality was performed for all read pairs from the short-insert libraries (300-850bp inserts) using FastQC v.0.11.3 or v.0.11.5 (www.bioinformatics.babraham.ac.uk/projects/fastqc/). Reads were clipped using the fastx_trimmer tool in the FASTX-Toolkit v.0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/index.html) to remove the first 5-10 nucleotides of each read; for the NextSeq reads, the last 5 nucleotides were also clipped. Adapter trimming was performed using cutadapt v.1.8.174 with the “O” parameter set to 5 and using option “b”; default settings were used for all other parameters. Quality trimming and length filtering was performed using Sickle v.1.3375 with the “pe” option and the following parameter settings: -t sanger -q 20 -l 50 and default settings for other parameters. This yielded quality-trimmed paired and singleton reads with a minimum length of 50 nucleotides; only intact read pairs were used for downstream analyses.
For the LMP libraries, duplicate reads were removed using NextClip v.1.3.1 (https://github.com/richardmleggett/nextclip) with the --remove_duplicates parameter specified and default settings for all other parameters. Adapter trimming was performed using cutadapt v.1.1074; junction adapters were removed from the start of reads by running option “g”, with the adapter sequence anchored to the beginning of reads with the “^” character, and the following settings for other parameters: -O 10 -n 2 -m 25. Other adapter trimming was performed using option “a”, with the further parameters set to the same values as specified above. Quality trimming was performed with PRINSEQ-lite v.0.20.476, with the following parameter settings: -trim_qual_left 20 -trim_qual_right 20 -trim_qual_window 20 -trim_tail_left 101 -trim_tail_right 101 -trim_ns_left 1 -trim_ns_right 1 -min_len 25 -min_qual_mean 20 -out_format 3.
De novo genome assembly was performed for each individual using CLC Genomics Workbench v.8.5.1 (QIAGEN Aarhus, Denmark). All trimmed read pairs from the short-insert libraries were used for assembly under the following parameter settings: automatic optimization of word (k-mer) size; maximum size of bubble to try to resolve=5000; minimum contig length=200bp. Assembled contigs were joined to form scaffolds using SSPACE v.3.077 with default parameters, incorporating data from mate-pair libraries with 3kb and 10kb insert sizes where available. Library insert lengths were specified with a broad error range (i.e. ±40%). Gaps in the SSPACE scaffolds were filled using GapCloser v.1.1278 with default parameters. The average library insert lengths were specified using the estimates produced by SSPACE during scaffolding. Scaffolding and gap filling was not performed for individuals that lacked data from libraries with insert size of ≥800bp (only a single insert size library was available for these taxa; Supplementary Table 4). We did not attempt to extract sequences of organellar origin from the assemblies, or to separately assemble the plastid and mitochondrial genomes.
Sequences within the assemblies that correspond to the Illumina PhiX control library were identified via BLAST. A PhiX bacteriophage reference sequence (GenBank accession number CP004084) was used as a query for BLASTN searches, implemented with the BLAST+ package v.2.5.0+79, against the genome assembly for each taxon with an E value cut-off of 1×10−10. Sequences that matched the PhiX reference sequence at this threshold were removed from the assemblies. We used the assemblathon_stats.pl script (https://github.com/ucdavis-bioinformatics/assemblathon2-analysis/blob/master/assemblathon_stats.pl) with default settings to obtain standard genome assembly metrics, such as N50. BUSCO v.2.080 was used to assess the content of the genome assemblies. The “embryophyta_odb9” lineage was used and analyses run with the following parameter settings: --mode genome -c 8 -e 1e-05 -sp tomato.
Gene annotation and orthologue inference
To annotate genes in the newly assembled Fraxinus genomes, we used a similarity based approach implemented in GeMoMa81, with genes predicted in the F. excelsior BATG0.5 assembly as a reference set. We used the “Full Annotation” gff file for BATG0.5 (Fraxinus_excelsior_38873_TGAC_v2.longestCDStranscript.gff3; available from http://www.ashgenome.org/transcriptomes), which contains the annotation for the single longest splice variant for each gene model. This annotation file also includes preliminary annotations for genes within the organellar sequences (gene models FRAEX38873_v2_000400370-FRAEX38873_v2_000401330) which were not reported in the publication of the reference genome27; none of the sets of putative orthologues used for the species-tree inference or molecular convergence analysis (see below) include these preliminary organellar models from the BATG0.5 reference assembly. The Extractor tool from GeMoMa v.1.3.2 was used to format the data from the reference genome (gff and assembly files), with the following parameter settings: v=true f=false r=true Ambiguity=AMBIGUOUS. To obtain information on similarity between the reference gene models and sequences in the newly assembled Fraxinus genomes, we performed TBLASTN searches of individual exons (i.e. the “cds-parts” file generated by Extractor) against the assembly file for each individual with BLAST+ v.2.2.29+79. makeblastdb was used to format each assembly file into a BLAST database with the following parameter settings: -out ./blastdb -hash_index -dbtype nucl. tblastn was then run with the “cds-parts.fasta” file as the query, with the following parameter settings: -num_threads 24 -db ./blastdb -evalue 1e-5 -outfmt “6 std sallseqid score nident positive gaps ppos qframe sframe qseq sseq qlen slen salltitles” -db_gencode 1 -matrix BLOSUM62 -seg no -word_size 3 -comp_based_stats F -gapopen 11 -gapextend 1 -max_hsps 0. Finally, the GeMoMa tool itself was run for each individual, with the TBLASTN output, cds-parts file and de novo assembly file as input, with the “e” parameter set to “1e-5” and default settings for all other parameters. Because GeMoMa generates predictions for each reference gene model separately, the output may contain gene models that are at identical, or overlapping, positions, especially for genes that belong to multi-gene families (http://www.jstacs.de/index.php/GeMoMa#FAQs). As the presence of these redundant gene models does not prevent the correct inference of sets of orthologues with OMA (see below), we opted to retain the predicted proteins from all gene model predictions generated by GeMoMa for input into OMA. The gffread utility from cufflinks v.2.2.182 was used to generate the CDS for each gene model; getfasta from bedtools v.2.26.083 used to generate full-length gene sequences (i.e. including introns, where present), with the “-name” and “-s” options invoked.
To identify sets of putatively orthologous sequences, we used OMA standalone v.2.0.084,85 to infer OMA groups (OGs) and hierarchical orthologous groups (HOGs). To the protein sets from the 29 diploid Fraxinus genome assemblies (the 28 newly generated assemblies, plus the existing reference assembly for F. excelsior), we added proteomes from three outgroup species: Olea europaea (olive), which belongs to the same family as Fraxinus (Oleaceae); Erythranthe guttata (monkey flower; formerly known as Mimulus guttatus), which belongs to the same order as Fraxinus (Lamiales); Solanum lycopersicum (tomato), which belongs to the same major eudicot clade as Fraxinus (lamiids). For O. europaea, we used the annotation for v6 of the genome assembly28; the file containing proteins for the single longest transcript per gene (OE6A.longestpeptide.fa) was downloaded from: http://denovo.cnag.cat/genomes/olive/download/?directory=.%2FOe6%2F. For E. guttata we used the annotation for v2.0 of the genome assembly29; the file containing proteins for the primary transcript per gene (Mguttatus_256_v2.0.protein_primaryTranscriptOnly.fa) was downloaded from Phytozome 1286. For S. lycopersicum we used the annotation for the vITAG2.4 genome assembly; the file containing proteins for the primary transcript per gene (Slycopersicum_390_ITAG2.4.protein_primaryTranscriptOnly.fa) was downloaded from Phytozome 12.
Fasta formatted files containing the protein sequences from all 32 taxa were used to generate an OMA formatted database. An initial run of OMA was performed using the option to estimate the species-tree from the OGs (option ‘estimate’ for the SpeciesTree parameter); we set the InputDataType parameter to ‘AA’ and left all other parameters with the default settings. The species-tree topology from the initial run was then modified in FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/) to reroot it on S. lycopersicum; nodes within the main clades of Fraxinus species (which corresponded to sections recognised in the taxonomic classification25) were also collapsed and relationships between these major clades and any individual Fraxinus not placed into a clade, were collapsed. OMA was then rerun with the modified species-tree topology specified in Newick format through the SpeciesTree parameter; the species-tree topology is used during the inference of HOGs (https://omabrowser.org/standalone/), and does not influence the OGs obtained.
Species-tree inference
To obtain a more robust estimate of the species-tree for Fraxinus (compared with that estimated by OMA, see above, or existing species-tree estimates based on very few independent loci87,88), we selected clusters of putatively orthologous sequences from the results of the OMA analysis. OGs containing protein sequences from all 32 taxa (29 diploid Fraxinus and three outgroups) were identified and corresponding CDSs aligned with MUSCLE89 via GUIDANCE290 with the following parameter settings: --program GUIDANCE --msaProgram MUSCLE --seqType codon --bootstraps 100, and default settings for other parameters. Datasets where sequences were removed during the alignment process (identified by GUIDANCE2 as being unreliably aligned) or which failed to align due to the presence of incomplete codons (i.e. the sequence length was not divisible by three) were discarded. Alignment files with unreliably aligned codons removed (i.e. including only codons with GUIDANCE scores above the 0.93 threshold for the “colCutoff” parameter) were used for downstream analyses. A custom Perl script was used to identify alignments shorter than 300 characters in length or which included sequences with <10% non-gap characters; these datasets were excluded from further analysis.
The remaining alignment files were converted from fasta to nexus format with the seqret tool from EMBOSS v.6.6.091. MrBayes v. 3.2.692 was used to estimate gene-trees with the following parameter settings: lset nst=mixed rates=gamma; prset statefreqpr=dirichlet(1,1,1,1); mcmc nruns = 2 nchains = 4 ngen = 5000000 samplefreq = 1000; Sumt Burninfrac = 0.10 Contype = Allcompat; Sump Burninfrac = 0.10. Diagnostics (average standard deviation of split frequencies (ASDSF) and for post burnin samples, potential scale reduction factor (PSRF) for branch and node parameters and effective sample size (ESS) for tree length) were examined to ensure that runs for a given locus had reached convergence and that a sufficient number of independent samples had been taken. We discarded datasets where the ASDSF was ≥0.010; all remaining datasets had an ESS for tree length in excess of 500.
We used BUCKy v.1.4.493,94 to infer a species-tree for Fraxinus via Bayesian concordance analysis, which allows for the possibility of gene-tree heterogeneity (arising from biological processes such as hybridisation, that is reported to occur within Fraxinus 87) but which makes no assumptions regarding the reason for discordance between different genes94. First, mbsum v.1.4.4 (distributed with BUCKy) was run on the MrBayes tree files for each locus, removing the trees sampled during the first 500,000 generations of each run as a burn-in. We then used the output from mbsum to select the most informative loci on the basis of the number of distinct tree topologies represented in the sample of trees from MrBayes; loci with a maximum of 2000 distinct topologies were retained. BUCKy was then run on the combined set of mbsum output files for all retained loci under ten different values for the α parameter (0.1, 1, 2, 5, 10, 20, 100, 500, 1000, ∞), which specifies the a priori level of discordance expected between loci. Each analysis with a different α setting was performed with different random seed numbers (parameters -s1 and -s2) and the following other parameter settings: -k 4 -m 10 -n 1000000 -c 2. Run outputs were checked to ensure that the average standard deviation of the mean sample-wide concordance factors (CF; the sample-wide CF is the proportion of loci in the sample with that clade93) was <0.01. The same primary concordance tree (PCT; the PCT comprises compatible clades found in the highest proportion of loci and represents the main vertical phylogenetic signal94) and CFs for each node were obtained for all settings of α.
We also repeated the species-tree inference using full-length sequences (i.e. including introns, where present); alignment and gene-tree inference were carried out as described above, with the exception that the --seqType parameter in GUIDANCE2 was set to “nuc”. BUCKy analyses were performed as described above. The same PCT was obtained for all settings of α, with only minor differences in the mean sample-wide concordance factors. The PCT inferred from the full-length datasets was also identical to that obtained from the CDS analyses; we based our final species-tree estimate on the output of the full-length analyses due to the presence of a larger number of informative loci within these datasets.
One of the informative loci from the CDS analyses (i.e. those with ≤2000 distinct topologies within their gene-tree sample), and three of those from the full-length analyses, were subsequently found to be among our filtered set of candidate loci with evidence of convergence between EAB-resistant taxa (see below). To test whether the signal from these loci had an undue influence on the species-tree estimation, we excluded them and repeated the BUCKy analyses for the CDS and full-length datasets as described above, with the exception that only an α parameter setting of 1 was used. The PCTs obtained from these analyses were identical to those inferred when including all datasets, with minor (i.e. 0.01) differences in CFs.
In addition to the analysis including all taxa, we also performed BUCKy analyses for 13 taxa selected for inclusion in the grand-conv analyses and for the subsets of 10-12 taxa for each of the three grand-conv pairwise comparisons (see below - Analysis of patterns of sequence variation consistent with molecular convergence). OGs containing protein sequences from all 13 taxa (ten Fraxinus and three outgroups) were identified and corresponding CDSs for these 13 taxa aligned with MUSCLE via GUIDANCE2, as described above. We also identified and aligned CDSs for all additional OGs which did not include all 13 taxa, but which contained proteins for all taxa in one of the subsets used for the pairwise comparisons. Filtering of alignments and gene-tree inference were carried out as described above for the full set of 32 taxa. BUCKy was run separately with the MrBayes tree samples for loci including all 13 taxa and for additional loci including taxa for each of the three smaller subsets for the grand-conv pairwise comparisons. BUCKy analyses were performed as described above, with the exception that no filtering of loci on the basis of number of distinct gene-tree topologies was performed, a value of between 2 and 35 was used for the -m parameter, and only a single α parameter setting, of 0.1, was used. Topologies for the PCTs for the set of 13 taxa and subsets of 10-12 taxa were both congruent with each other and with the PCTs inferred from analyses including all taxa, for all nodes with a CF of ≥0.38.
Analysis of patterns of sequence variation consistent with molecular convergence
To test for signatures of putative molecular convergence in protein sequences we used a set of diploid taxa representing the extremes of variation in susceptibility to EAB, as assessed by our egg bioassays. By limiting our analysis at this stage to this subset of taxa we could also maximise the number of genes analysed, as with increasing taxon sampling the number of OGs for which all taxa are represented decreases. This set comprised: five highly susceptible taxa (F. americana, F. latifolia, F. ornus, F. pennsylvanica [susceptible genotype], and F. velutina), five resistant taxa (F. baroniana, F. floribunda, F. mandshurica, F. platypoda and Fraxinus sp. D2006-0159) and three outgroups (O. europaea, E. guttata and S. lycopersicum).
Of the ten Fraxinus genome assemblies included in the convergence analysis, six were from genotypes that were also included in the EAB resistance assays outlined above (F. americana am-6, F. baroniana bar-2, F. floribunda flor-ins-12, F. pennsylvanica pe-48 and F. platypoda spa-1 and Fraxinus sp. D2006-0159 F-unk-1). For the other four, we could not test the exact individual that we sequenced the genome of, but relied upon results of bioassays of other individuals in the same species.
We used the following three pairwise comparisons to screen for loci showing amino acid convergence between resistant taxa:
F. mandshurica (sect. Fraxinus) versus F. platypoda (incertae sedis)
F. mandshurica (sect. Fraxinus) versus F. baroniana, F. floribunda and Fraxinus sp. D2006-0159 (sect. Ornus)
F. platypoda (incertae sedis) versus F. baroniana, F. floribunda and Fraxinus sp. D2006-0159 (sect. Ornus)
The more divergent homologous amino acid sequences are between species, the more likely it is that a convergent amino acid state will occur by chance95. In order to account for this, we compared the posterior expected numbers of convergent versus divergent substitutions across all pairs of independent branches of the Fraxinus species-tree for the selected taxa using a beta-release of the software Grand-Convergence v.0.8.0 (hereafter referred to as grand-conv; https://github.com/dekoning-lab/grand-conv). This software is based on PAML 4.896 and is a development of a method used by Castoe et al95. It has also been recently used, for example, to detect convergence among flowering plant lineages with crassulacean acid metabolism22.
For input into grand-conv, we used the same OGs that were the basis of the BUCKy analyses of the 13 taxa selected for the grand-conv analyses, and analyses of the subsets of taxa for the pairwise comparisons (see above - Species-tree inference). Therefore, as well as meeting the criterion of including all relevant taxa, the OGs analysed with grand-conv had also successfully passed the alignment, filtering and gene-tree inference steps. A set of input files was created for each of the three pairwise comparisons that, where present, removed any taxa from the other resistant lineage from the alignments generated by GUIDANCE2. Alignment files were then converted from fasta to phylip format using the Fasta2Phylip.pl script (https://github.com/josephhughes/Sequence-manipulation). To ensure that sequences from each taxon appeared in a consistent order across all datasets (which is necessary for automating the generation of site-specific posterior probabilities for selected branch pairs with grand-conv), the phylip formatted files were sorted using the unix command “sort” prior to input into grand-conv. Species-tree files for each of the pairwise comparisons were created from the Newick formatted PCTs for the relevant taxon sets generated with BUCKy, with the trees edited to root them on S. lycopersicum.
For the grand-conv analysis, “gc-estimate” was first run on the full set of input alignment files for each pairwise comparison, with the following parameters settings: --gencode=0 --aa-model=lg --free-bl=1, specifying the appropriate species-tree file for each of the pairwise comparisons. Next, “gc-discover” was run to generate site-specific values for the posterior probability (PP) of divergence or convergence for the branch pairs of interest; the numbers for the branch pairs relating to the resistant taxa were established from an initial run of “gc-estimate” and “gc-discover” on a single input file, and then specified when running “gc-discover” on all input files using the --branch-pairs parameter. A custom Perl script was then used to filter the output files containing the site-specific posterior probabilities to identify loci with at least one amino acid site where the PP of convergence was higher than divergence and passed a ≥0.9000 threshold. For this filtered set of datasets with significant evidence of convergence at at least one site, we checked if the “excess” convergence, as measured from the residual values from the non-parametric errors-in-variables regression calculated by grand-conv, was higher for the branch pair of interest than for any other independent pair of branches within the species-tree (i.e. the highest residual was found for the resistant branch pair). Only loci where the highest excess convergence was found in the resistant branch-pair were retained for further analysis.
Refining the initial list of candidate loci identified with grand-conv
For the set of loci with evidence of convergence between at least one pair of resistant lineages from the grand-conv analyses, we applied additional tests to assess the robustness of the pattern of shared amino acid states. Specifically, we checked for the potential impact of alignment uncertainty and orthology/paralogy conflation. For each candidate locus, we identified the Hierarchical Orthologous Group (HOG) from the OMA analysis to which the sequences for the candidate locus belong. These HOGs include sequences for an expanded set of taxa (see above - Species-tree inference) and may represent a single gene for all taxa (i.e. a set of orthologous sequences) or several closely related paralogues84. Protein sequences for HOGs were aligned with GUIDANCE and gene-tree inference conducted with MrBayes, as described above for the OMA putative orthologous groups with the exception that the --seqType parameter in GUIDANCE was set to “aa” and in MrBayes the prset parameter was set to “prset aamodelpr = mixed”. Any MrBayes analyses that had not converged after 5M generations (average standard deviation of split frequencies >0.01) were run for an additional 5M generations. The multiple sequence alignment and gene-tree estimates were then used to refine the initial list of candidate loci. Loci were dropped from initial list of candidates if either of the following applied:
If in the filtered MSA alignment generated by GUIDANCE, the site/sites where convergence was detected were not present, indicating they were in a part of the protein sequences that can not be aligned reliably.
If in the consensus gene-tree estimated by MrBayes, there was evidence that the sequences within which convergence was initially detected (i.e. those belonging to the ten Fraxinus species included in the grand-conv analysis) belong to different paralogues and that the pattern of convergence could be explained by sequences with the “convergent” state belonging to one paralogue and the “non-convergent” state belonging to another paralogue. We also excluded two loci that belong to large gene families (>10 copies) for which the MrBayes analyses failed to reach convergence within a reasonable time (≤10M generations) and for which orthology/paralogy conflation could therefore not be excluded.
Additionally, for the set of loci remaining, we checked for errors in the estimation of gene models (including in the reference models from F. excelsior) that might impact the results of the grand-conv analyses. Specifically, we dropped from our list any loci where the amino acid sites with evidence of convergence were found to be outside of an exon, or outside of the gene itself, following manual correction of the gene model prediction.
Analysis of variants within candidate loci
To assess the possible impact of allelic variants (i.e. those not represented in the genome assemblies) on patterns of amino acid variation associated with the level of EAB susceptibility in Fraxinus, we called variants (SNPs and indels) and predicted their functional effects. For each sequenced Fraxinus individual, trimmed read pairs from the short insert Illumina libraries were mapped to the de novo genome assembly for the same individual using Bowtie 2 v.2.3.097 with the “very-sensitive” preset and setting “maxins” to between 1000 and 1400, depending on the libraries available for that individual. Read mappings were converted to BAM format and sorted using the “view” and “sort” functions in samtools v.1.4.198. Prior to variant calling, duplicate reads were marked and read group information added to the BAM files using the “MarkDuplicates” and “AddOrReplaceReadGroups” functions in picard tools v.1.139 (http://broadinstitute.github.io/picard).
Variant discovery was performed with gatk v.3.899. BAM files were first processed to realign INDELs using the “RealignerTargetCreator” and “IndelRealigner” tools. An initial set of variants was called for each individual using the “HaplotypeCaller” tool, setting the -stand_call_conf parameter to 30. VCF files from the initial variant calling were then hard filtered to identify low confidence calls by running the “VariantFiltration” tool with the -filterExpression parameter set as follows: “QD < 5.0 || FS > 20.0 || MQ < 30.0 || MQRankSum < -8.0 || MQRankSum > 8.0 || ReadPosRankSum < -2.0 || ReadPosRankSum > 2.0” (hard filtering thresholds were selected by first plotting the values for FS, MQ, MQRankSum, QD and ReadPosRankSum from the initial set of variant calls for selected individuals, representing the range of different sequence and library types used, to visualise their distribution and then modifying the default hard filtering thresholds in line with the guidance provided in the gatk document “Understanding and adapting the generic hard-filtering recommendations” (https://software.broadinstitute.org/gatk/documentation/article.php?id=6925)).
Variants passing the gatk hard filtering step (excluding those where alleles had not been called; GT field = ./.) were further analysed using SnpEff v.4.3u100, in order to predict the impact of any variants within genes identified from the convergence analyses. Custom genome databases were built for each individual using the SnpEff command “build” with option “-gtf22”; a gtf file containing the annotation for all genes, as well as fasta files containing the genome assembly, CDS and protein sequences, were used as input. Annotation of the impact of variants was performed by running SnpEff with genes of interest specified using the -onlyTr parameter and the -ud parameter set to “0” to deactivate annotation of up or downstream variants. For each variant predicted to alter the protein sequence, the position of the change was checked to see whether it occurred at a site at which evidence for convergence had also been detected and, if so, whether it involved a change to or from the state identified as being convergent between resistant taxa.
We also used the SnpEff results to check for evidence of mutations that could indicate the presence of non-functional gene copies in certain taxa. Variants annotated as “stop gained”, “start lost” or “frameshift” in the ten ingroup taxa included in the convergence analysis were manually examined to confirm that they would result in a disruption to the expected protein product, and that they were not false positives caused by errors in gene model estimation (e.g. misspecification of intron/exon boundaries). We checked further for evidence of truncation of sequences or errors in the GeMoMa gene model estimation that might be caused by loss-of-function mutations outside of the predicted exon boundaries (e.g. such as the loss of a start codon, which could cause GeMoMa to predict an incomplete gene model if an alternative possible start codon was present downstream). Such putative loss-of-function mutations would not be detected as such by SnpEff because they would be interpreted as low impact intergenic or intron variants.
We used WhatsHap v.0.15101 to perform read-based phasing of alleles for loci with evidence of multiple variants within them. Input files for phasing in each taxon consisted of the fasta formatted genome assembly, VCF file containing variants passing the gatk hard-filtering step and BAM file from Bowtie 2 with duplicates marked and indels realigned (i.e. as input into variant calling with gatk - see above). The WhatsHap “phase” tool was run with the following parameter settings: --max-coverage 20 --indels; only contigs/scaffolds containing the genes of interest were phased (specified using the “--chromosome” option). For loci with evidence of variants within the CDS, we used the output of WhatsHap to generate fully or partially phased allele sequences. The SnpSift tool from SnpEff v.4.3u100 was used to select variants that alter the CDS from the annotated VCF file generated by SnpEff and the positions of these variants checked against the WhatsHap output to see if they fell within phased blocks. For each gene, the number and size (i.e. number of phased variants encompassed) of each phased block was found and a custom Perl script used to select the largest (or joint largest) block for genes with at least one block spanning multiple phased variants within the CDS. Details of phased variants impacting the CDS within the selected blocks, and of variants for genes with only a single variant within the CDS (which were not considered for phasing with WhatsHap, but for which the CDS for separate alleles can be generated), were extracted from the WhatsHap output VCF file; these selected variants were then applied to the gene sequences from each genome assembly to generate individual alleles which are fully or partially phased within the CDS. The “faidx” function in samtools v.1.698 was used to extract the relevant subsequences from the genome assembly files, and the “consensus” command in bcftools v.1.4 (http://www.htslib.org/doc/bcftools-1.4.html) used to obtain the sequence for each allele with the “-H 1” and “-H 2” options. In cases where the selected phased block also spans unphased variants, both sequences output by bcftools will have the state found in the original genome assembly at these sites, as they will for any variants outside of the selected phased block. The revseq and descseq tools from EMBOSS v.6.6.091 were used to reverse complement the sequences for any genes annotated on the minus strand and to rename the output sequences. Phased alleles were used for further phylogenetic analysis of candidate loci (see below); phasing results were also used to check loci with multiple potential loss-of-function mutations within a single individual, to establish whether the mutations are on the same or different alleles. We discounted any cases of potential loss-of-function mutations where multiple frameshifts occurring in close proximity on the same allele resulted in the correct reading frame being maintained.
To check for polymorphism within F. excelsior at sites with evidence of convergence, we examined the combined BAM file generated from mapping Illumina HiSeq reads from 37 individuals from different European provenances (the European Diversity Panel) to the F. excelsior reference genome (BATG0.5) by Sollars et al27. Duplicate reads were removed from the BAM file using the “MarkDuplicates” function in picard tools v.1.139 (http://broadinstitute.github.io/picard), with the REMOVE_DUPLICATES option set to “true”. Selected contigs (containing the genes of interest) were extracted from the BAM file using the “view” function in samtools v.1.698 and visualised with Tablet v.1.17.08.17102; evidence for polymorphism was observed directly from the reads and variants only recorded if supported by at least 10% of reads at that site. Loci OG39275 and OG46977 were excluded from this analysis due to errors in the reference gene models, possibly arising from misassembly, which meant the sites homologous to those with evidence of convergence between EAB-resistant taxa could not be identified (see Supplementary Table 5 for more details).
Distinguishing between different underlying causes of convergent patterns
To test whether evidence of convergence found by grand-conv might actually be due to taxa sharing the same amino acid variant as a result of introgression or incomplete lineage sorting (ILS), we conducted phylogenetic analyses of coding DNA sequences for the candidate loci to infer their gene-trees. We checked to see if sequences with apparently convergent residues group together, even when nucleotides encoding those residues are removed. The CDSs for the refined set of candidate loci were aligned with MUSCLE via GUIDANCE2 and alignment files with unreliably aligned codons removed were used for downstream analyses, as described above (see - Species-tree inference); none of datasets had sequences that were identified by GUIDANCE2 as being unreliably aligned. OG40061 failed to align due to the presence of an incomplete codon at the end of the reference gene model from F. excelsior; we trimmed the final 2bp from the F. excelsior sequence and reran GUIDANCE2 using this modified file.
Phased allele sequences generated using the WhatsHap results (see above - Analysis of variants within candidate loci) were added to the CDS alignments using MAFFT v.7.310103 with the options “--add” and “--keeplength”, in order to splice out any introns present in the phased sequences and maintain the original length of the CDS alignments. For any taxa for which phased sequences had been added, the original unphased sequence was removed from the alignment.
If intragenic recombination has taken place, gene-trees inferred from the CDS alignments may fail to group together the sequences with evidence of convergence even in cases where the convergent pattern is due to ILS or introgressive hybridisation. This is because the phylogenetic signal from any non-recombinant fragments of alleles derived from ILS or introgressive hybridisation may not be sufficiently strong to override that from fragments of alleles that have not been subject to these processes. To account for this possibility, we used hyphy v.2.3.14.20181030beta(MPI)104 to conduct recombination tests with GARD105 with the following parameter settings: 012345 “General Discrete” 3. Where GARD found significant evidence for a recombination breakpoint (p-value < 0.05), we partitioned the alignment into non-recombinant fragments for phylogenetic analysis.
Alignment files were converted to nexus format and gene-trees estimated with MrBayes, as described above (see - Species-tree inference). We checked the ASDSF and used Tracer v.1.6.0 (http://beast.bio.ed.ac.uk/Tracer) to inspect the ESS values for each parameter from the post burnin samples and to confirm that the burnin setting (i.e. discarding the first 10% of samples) was sufficient; in cases where runs had not converged after 5M generations (ASDSF ≥0.010), additional generations were run until an ASDSF of <0.010 was reached. We examined the consensus trees generated by MrBayes to look for evidence that sequences sharing the amino acid states inferred as convergent by grand-conv cluster together in the gene-tree, in conflict with relationships inferred in the species-tree for Fraxinus. In cases where evidence of such clustering was found, the codon(s) corresponding to the amino acid site(s) at which evidence of convergence was detected were excluded and the MrBayes analysis repeated. In cases where sequences that have the “convergent” amino acid group together in the gene-tree even after the codon(s) for the relevant site(s) have been excluded, we concluded that the evidence of convergence detected by grand-conv is more likely due to introgressive hybridisation or ILS. We also examined the gene-tree topologies to assess whether any of the amino acid states identified as convergent by grand-conv is more likely to be the ancestral state for Fraxinus.
Further characterisation of candidate loci
To identify the gene from Arabidopsis thaliana that best matches each of the candidate loci in our refined set, we conducted a TBLASTN search of the F. excelsior protein sequence belonging to the relevant OGs against the A. thaliana sequences in the nr/nt database in GenBank106 and selected the hit with the lowest E value. In cases where the OG lacked a sequence from F. excelsior, we used the protein sequence from F. mandshurica as the query for the TBLASTN search instead.
We also checked for the presence of the F. excelsior sequences within the OrthoMCL clusters generated by Sollars et al.27 to see if they were associated with the same A. thaliana genes as identified by BLAST. We obtained information on the function of the best matching A. thaliana genes from The Arabidopsis Information Resource (TAIR; https://www.arabidopsis.org) and the literature. The OrthoMCL analysis conducted by Sollars et al.27 also included a range of other plant species, including S. lycopersicum (tomato) and the tree species Populus trichocarpa (poplar). As tomato is much more closely related to F. excelsior than is A. thaliana, and poplar is also a tree species, the function of the genes in these taxa may provide a better guide to the function of the F. excelsior genes. We therefore also checked the OrthoMCL clusters containing our candidate F. excelsior genes to identify putative orthologues, or close paralogues, from tomato and poplar. In cases where the OrthoMCL cluster included multiple tomato or poplar genes, we focused attention on the tomato sequence that also belonged to the OMA group, as the putative orthologue of our F. excelsior gene in that species. For poplar, we looked for information on all sequences, unless there were a large number in the cluster (>4). We searched for literature on the function of the tomato and poplar genes, using the gene identifiers from the versions of the genome annotations used for the OrthoMCL analysis27 and also looked for information on PhytoMine, in Phytozome 1286.
To further clarify the orthology/paralogy relationships between our candidates and genes from other species, we conducted phylogenetic analysis of the relevant OrthoMCL clusters from Sollars et al27 for selected loci. Protein sequences belonging to each OrthoMCL cluster were aligned and gene-trees inferred using GUIDANCE2 and MrBayes respectively, as described above for the OGs and HOGs. For the OrthoMCL cluster relating to OG15551, following an initial MrBayes analysis, we removed two incomplete sequences (Migut.O00792.1.p and GSVIVT01025800001, which were missing >25% of characters in the alignment) and two divergent sequences from A. thaliana (AT1G74540 and AT1G74550) which are known to derive from a Brassicales-specific retroposition event and subsequent Brassicaceae-specific tandem duplication107; the alignment and phylogenetic analysis was then repeated for the reduced dataset.
For OG15551, we generated a sequence logo for regions of the protein containing sites at which evidence of convergence was detected. We obtained putatively homologous sequences by downloading the fasta file for the OMA group (OMA Browser fingerprint YGPIYSF108) containing the A. thaliana CYP98A3 gene (AT2G40890); the sequences were filtered to include only those from angiosperms, with a maximum of one sequence per genus retained (29 genera in total). To this dataset, we added the OG15551 protein sequences for F. mandshurica and F. pennsylvanica pe-48 and manually aligned the regions containing the relevant sites (positions 208-218 and 474-482 in the F. excelsior FRAEX38873_v2_000261700 reference protein). We used WebLogo v.3.7.3109 without compositional adjustment to generate logos for each of these regions.
GO term enrichment analysis
To test for the possibility of overrepresentation of particular functional categories among the candidate loci in our refined set, compared with the complete set of genes used as input for the convergence analyses, we conducted gene ontology (GO) enrichment tests. Fisher’s exact tests with the “weight” and “elim” algorithms, which take into account the GO graph topology110, were run using the topGO package111 (v.2.32.0) in R v.3.5.1112. We created a “genes-to-GOs” file for the complete set of F. excelsior gene models included in the grand-conv analyses (n = 3658), using GO terms from the existing functional annotation for the reference genome (Sollars et al.27); only the single longest transcript per gene (see http://www.ashgenome.org/transcriptomes) was included and for any OMA groups that lacked an F. excelsior sequence we used the reference model referred to by the majority of other Fraxinus sequences in the group (i.e. as indicated in the GeMoMa gene model names). We also created a list of F. excelsior reference model genes belonging to our refined set of candidate loci (n = 53); again, for any OMA groups that lacked an F. excelsior sequence, we used the reference model referred to by the majority of other Fraxinus sequences in the group. The complete list of F. excelsior reference genes included in the grand-conv analyses, and their associated GO terms, was used as the background against which the list of gene models from the refined set of candidate loci was tested. Fisher’s exact test was run separately, with each of the algorithms, to check for enrichment of terms within the biological process (BP), molecular function (MF) and cellular component (CC) domains.
Protein modelling
The SignalP 5.0 server113 and Phobius server114 (http://phobius.sbc.su.se/index.html) were used to detect the presence of signal peptides; for SignalP the organism group was set to “Eukarya” and for Phobius the “normal prediction” method was used. All Fraxinus sequences belonging to the OMA groups were used as input for the signal peptide analyses; we only concluded that a signal peptide was present if it was predicted by both methods. The NetPhos 3.1 Server (http://www.cbs.dtu.dk/services/NetPhos/) was used with default settings to identify candidate phosphorylation sites for loci where the amino acid variant observed at a site with evidence of convergence included a serine, threonine or tyrosine. The same protein sequences for low and high susceptibility taxa as used for protein modelling (see below) were input to an initial run of NetPhos 3.1; where evidence for phosphorylation site presence/absence was detected with this initial sequence pair (i.e. present in the sequence with the convergent state and absent from that with the non-convergent state, or vice versa) we reran NetPhos 3.1 on all Fraxinus sequences from the relevant OMA groups to test if this difference was consistently associated with the convergent/non-convergent state. We only counted as potential phosphorylation sites those for which the NetPhos score for phosphorylation potential was ≥0.900 for all sequences with the putative site.
RaptorX-Binding115 (http://raptorx.uchicago.edu/BindingSite/) was used to generate predicted protein models for each of the candidate genes in our refined set, as well as to outline possible binding sites and candidate ligands. Protein sequences for gene models from the F. excelsior reference genome were used for initial protein model and binding site prediction, except in cases where F. excelsior was not present in the OMA group or where comparison with the other ingroup and outgroup taxa indicated the F. excelsior gene model may be incorrect/incomplete; for these loci, the F. mandshurica sequences were used instead as, after the reference, the genome assembly for this taxon is one of the highest quality available. For loci for which a binding site could be successfully predicted (i.e. with at least one potential binding site with a pocket multiplicity value of ≥40), additional models were generated for representative resistant (F. mandshurica or F. platypoda) and susceptible (F. ornus or susceptible F. pennsylvanica) taxa using Swiss-model116 and Phyre2117 (intensive mode), with the exact taxon selection depending on which grand-conv pairwise comparison the locus was detected in (see above - Analysis of patterns of sequence variation consistent with molecular convergence) and which taxa had complete gene models. Where errors were detected in the predicted protein sequences for resistant or susceptible taxa (i.e. due to errors in the predicted gene model, which were detected through comparison with sequences from other species, including those from outgroups) these were corrected prior to modelling (e.g. by trimming extra sequence resulting from incorrect prediction of the start codon). Models predicted by the three independent methods (RaptorX-Binding, Swiss-modeller and Phyre2) were compared by aligning them using PyMOL v.2.0 with the align function to check for congruence; only those loci whose models displayed congruence and where the convergent site was located within/close to the putative active site were taken forward for predictive ligand docking analysis (using the Phyre2 and RaptorX-Binding models for the docking itself). In addition, any loci with congruent models where the site with evidence of convergence is also a putative phosphorylation site presence/absence variant, or which are within a putative functional domain, were analysed further. Ligand candidates were selected based on relevant literature and/or the RaptorX-Binding output, with SDF files for each of the molecules being obtained from PubChem (https://pubchem.ncbi.nlm.nih.gov). SDF files were converted to 3d pdb files using Online SMILES Translator and Structure File Generator (https://cactus.nci.nih.gov/translate/), so they could be used with Autodock. Docking analysis was carried out using Autodock Vina v.1.1.2118 with the GUI PyRx v.0.8119. Following docking, ligand binding site coordinates were exported as SDF files from Pyrex and loaded into PyMOL with the corresponding protein model file for the resistant and susceptible taxa. Binding sites were then annotated and the residues at which evidence for convergence had been detected with grand-conv were labelled.
Evidence for differential expression of candidate loci in F. pennsylvanica
We used published transcriptome assembly and expression data from F. pennsylvanica 18 to look for evidence of differential expression of our candidate loci in response to EAB larval feeding. This dataset comprised six genotypes of F. pennsylvanica, four putatively resistant to EAB and two susceptible to EAB. To identify the orthogues of our genes in the protein sequences of this independently assembled transcriptome18, we repeated the OMA clustering analysis (see above - Gene annotation and orthologue inference) with the addition of these data, available as “Fraxinus_pennsylvanica_120313_peptides” at the Harwood Genomics Project website (https://hardwoodgenomics.org). OMA was run as described above, with the SpeciesTree parameter set to ‘estimate’; because we only intended to use the results for the OGs, and not the HOGs, from this analysis, we did not repeat the clustering with a modified species-tree as was done for our main OMA analysis. Having identified the likely orthologous loci from the F. pennsylvanica transcriptome18, we used the results of the differential expression analysis18 to check whether our candidate loci had significantly increased or decreased expression post-EAB feeding in this dataset.
Extended Data
Extended Data Fig. 1. Predicted protein structures for selected candidate loci.
a, Predicted protein structure for OG36502, modelled using the protein sequence for Fraxinus platypoda. The serine/asparagine variant at the site where convergence was detected is highlighted; the serine is a putative phosphorylation site. b, Predicted protein structure for OG40061, modelled using the protein sequence for F. mandshurica. The asparagine/serine variant at the site where convergence was detected is highlighted; the serine is a putative phosphorylation site. The putative substrate, NADP, is shown docked within the predicted active site. c, Predicted protein structure for OG38407, modelled using the protein sequence for F. mandshurica. The aspartic acid/asparagine variant at the site where convergence was detected is highlighted; the site falls within a leucine rich repeat region (LRR; shaded blue) which is predicted to span from position 111–237 within the protein sequence (detected using the GenomeNet MOTIF tool (www.genome.jp/tools/motif/), searching against the NCBI-CDD and Pfam databases with default parameters; the LRR region was identified as positions 111–237 with an e-value of 1e-05). d, Predicted protein structure for OG21033, modelled using the protein sequence for F. platypoda. The lysine/glutamine at the site where convergence was detected is highlighted. The putative substrate, β-D-Glcp-(1 → 3)-β-D-GlcpA-(1 → 4)-β-D-Glcp, is shown docked within the predicted active site.
Supplementary Material
Acknowledgements
This research utilised Queen Mary’s Apocrita HPC facility, supported by QMUL Research-IT (http://doi.org/10.5281/zenodo.438045). We thank J. Carlson for providing F. pennsylvanica DNA; T. Baxter, S. Brockington, P. Brownless, D. Crowley, S. Honey, R. Irvine, R. Jinks, P. Jones, T. Kirkham, H. McAllister, I. Parkinson and S. Redstone for help with obtaining Fraxinus materials from UK collections; T. Poland for providing EAB eggs; M. Miller for propagating trees for the bioassays; R. Matko for preparation of voucher specimens; J. Pellicer for advice on flow cytometry; P. Howard and M. Struebig for advice on DNA extractions; J. Keilwagen for help with GeMoMa; K. Davies and J. Parker for help with convergence analysis software; the Evolution Labchat group and Rossiter Lab at QMUL for discussions; R. Rose and J. Sayers for advice on protein modelling analyses. This project was funded by the Living with Environmental Change (LWEC) Tree Health and Plant Biosecurity Initiative - Phase 2 (grant number BB/L012162/1) funded jointly by the BBSRC, Defra, ESRC, Forestry Commission, NERC and the Scottish Government. R.J.A.B. acknowledges additional support from the DEFRA Future Proofing Plant Health scheme. R.J.A.B. and L.J.K. acknowledge additional support from the Erica Waltraud Albrecht Endowment Fund. W.J.P. was funded by the Walsh Scholarship Programme of the Department of Agriculture, Food and the Marine, Ireland. E.D.C. was supported by the Marie Skłodowska-Curie Individual Fellowship ‘FraxiFam’ (grant agreement 660003).
Footnotes
Author contributions R.J.A.B. conceived and oversaw the project. L.J.K. and R.J.A.B. wrote the manuscript, with input from J.L.K., W.J.P and S.J.R. L.J.K. conducted gene annotation, orthologue inference, convergence analyses, calling and analysis of variants, GO enrichment analysis and phylogenetic analyses. L.J.K., W.C. and A.T.W. performed genome size estimation by flow cytometry. L.J.K., W.C., E.D.C. and D.W.C. extracted DNA. L.J.K. and E.D.C. assembled the genomes. J.L.K. conceived and oversaw the emerald ash borer (EAB) bioassays. D.W.C. conducted the EAB bioassays. J.L.K. and M.E.M. analysed EAB bioassay data. W.J.P. conducted protein modelling analyses. S.J.R. advised on convergence analyses.
Competing interests The authors declare no competing interests.
Data availability
Underlying data for Figure 1 are available in Supplementary Tables 1 and 2. All trimmed read data and genome assemblies have been deposited in the European Nucleotide Archive under accession number PRJEB20151 (https://www.ebi.ac.uk/ena/browser/view/PRJEB20151). The genome assemblies are also available to download at: http://www.ashgenome.org.
Code availability
The custom scripts used is this study have been deposited in GitHub: https://github.com/lkelly3/eab-ms-scripts.
References
- 1.Pautasso M, Aas G, Queloz V, Holdenrieder O. European ash (Fraxinus excelsior) dieback – A conservation biology challenge. Biological Conservation. 2013;158:37–49. [Google Scholar]
- 2.MacFarlane DW, Meyer SP. Characteristics and distribution of potential ash tree hosts for emerald ash borer. For Ecol Manage. 2005;213:15–24. [Google Scholar]
- 3.Boyd IL, Freer-Smith PH, Gilligan CA, Godfray HCJ. The consequence of tree pests and diseases for ecosystem services. Science. 2013;342 doi: 10.1126/science.1235773. 1235773. [DOI] [PubMed] [Google Scholar]
- 4.Herms DA, McCullough DG. Emerald ash borer invasion of North America: history, biology, ecology, impacts, and management. Annu Rev Entomol. 2014;59:13–30. doi: 10.1146/annurev-ento-011613-162051. [DOI] [PubMed] [Google Scholar]
- 5.Orlova-Bienkowskaja MJ. Ashes in Europe are in danger: the invasive range of Agrilus planipennis in European Russia is expanding. Biol Invasions. 2014;16:1345–1349. [Google Scholar]
- 6.McCullough DG. Challenges, tactics and integrated management of emerald ash borer in North America. Forestry. 2019 doi: 10.1093/forestry/cpz049. [DOI] [Google Scholar]
- 7.Drogvalenko AN, Orlova-Bienkowskaja MJ, Bieńkowski AO. Record of the Emerald Ash Borer (Agrilus planipennis) in Ukraine is Confirmed. Insects. 2019;10 doi: 10.3390/insects10100338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Semizer-Cuming D, Krutovsky KV, Baranchikov YN, Kjær ED, Williams CG. Saving the world’s ash forests calls for international cooperation now. Nature Ecology & Evolution. 2019;3:141–144. doi: 10.1038/s41559-018-0761-6. [DOI] [PubMed] [Google Scholar]
- 9.Evans HF, Williams D, Hoch G, Loomans A, Marzano M. Developing a European Toolbox to manage potential invasion by emerald ash borer (Agrilus planipennis) and bronze birch borer (Agrilus anxius), important pests of ash and birch. Forestry: An International Journal of Forest Research. 2020 doi: 10.1093/forestry/cpz074. [DOI] [Google Scholar]
- 10.Baranchikov Y, Mozolevskaya E, Yurchenko G, Kenis M. Occurrence of the emerald ash borer, Agrilus planipennis in Russia and its potential impact on European forestry. EPPO Bulletin. 2008;38:233–238. [Google Scholar]
- 11.Zhao T, et al. Induced outbreaks of indigenous insect species by exotic tree species. Acta Entomologica Sinica. 2007;50:826–833. [Google Scholar]
- 12.Liu H, et al. Exploratory survey for the emerald ash borer, Agrilus planipennis (Coleoptera: Buprestidae), and its natural enemies in China. Great Lakes Entomol. 2003;36:191–204. [Google Scholar]
- 13.Wei X, Reardon D, Wu Y, Sun J-H. Emerald ash borer, Agrilus planipennis Fairmaire (Coleoptera: Buprestidae), in China: a review and distribution survey. Acta Entomologica Sinica. 2004;47:679–685. Others. [Google Scholar]
- 14.Orlova-Bienkowskaja MJ, Volkovitsh MG. Are native ranges of the most destructive invasive pests well known? A case study of the native range of the emerald ash borer, Agrilus planipennis (Coleoptera: Buprestidae) Biological Invasions. 2018;20:1275–1286. [Google Scholar]
- 15.Showalter DN, Villari C, Herms DA, Bonello P. Drought stress increased survival and development of emerald ash borer larvae on coevolved Manchurian ash and implicates phloem-based traits in resistance. Agricultural and Forest Entomology. 2018;20:170–179. [Google Scholar]
- 16.Whitehill JGA, et al. Interspecific proteomic comparisons reveal ash phloem genes potentially involved in constitutive resistance to the emerald ash borer. PLoS One. 2011;6:e24863. doi: 10.1371/journal.pone.0024863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Whitehill JGA, et al. Interspecific comparison of constitutive ash phloem phenolic chemistry reveals compounds unique to manchurian ash, a species resistant to emerald ash borer. J Chem Ecol. 2012;38:499–511. doi: 10.1007/s10886-012-0125-7. [DOI] [PubMed] [Google Scholar]
- 18.Lane T, et al. The green ash transcriptome and identification of genes responding to abiotic and biotic stresses. BMC Genomics. 2016;17:702. doi: 10.1186/s12864-016-3052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sackton TB, et al. Convergent regulatory evolution and loss of flight in paleognathous birds. Science. 2019;364:74–78. doi: 10.1126/science.aat7244. [DOI] [PubMed] [Google Scholar]
- 20.Arnold BJ, et al. Borrowed alleles and convergence in serpentine adaptation. Proc Natl Acad Sci U S A. 2016;113:8320–8325. doi: 10.1073/pnas.1600405113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hu Y, et al. Comparative genomics reveals convergent evolution between the bamboo-eating giant and red pandas. Proc Natl Acad Sci U S A. 2017;114:1081–1086. doi: 10.1073/pnas.1613870114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X, et al. The Kalanchoë genome provides insights into convergent evolution and building blocks of crassulacean acid metabolism. Nat Commun. 2017;8:1899. doi: 10.1038/s41467-017-01491-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill J, et al. Recurrent convergent evolution at amino acid residue 261 in fish rhodopsin. Proc Natl Acad Sci U S A. 2019;116:18473–18478. doi: 10.1073/pnas.1908332116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhen Y, Aardema ML, Medina EM, Schumer M, Andolfatto P. Parallel molecular evolution in an herbivore community. Science. 2012;337:1634–1637. doi: 10.1126/science.1226630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wallander E. Systematics and floral evolution in Fraxinus (Oleaceae) Belgische Dendrologie Belge. 2012;2012:39–58. [Google Scholar]
- 26.Koch JL, Carey DW, Mason ME, Poland TM, Knight KS. Intraspecific variation in Fraxinus pennsylvanica responses to emerald ash borer (Agrilus planipennis) New Forests. 2015;46:995–1011. [Google Scholar]
- 27.Sollars ESA, et al. Genome sequence and genetic diversity of European ash trees. Nature. 2017;541:212–216. doi: 10.1038/nature20786. [DOI] [PubMed] [Google Scholar]
- 28.Cruz F, et al. Genome sequence of the olive tree, Olea europaea. Gigascience. 2016;5:29. doi: 10.1186/s13742-016-0134-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hellsten U, et al. Fine-scale variation in meiotic recombination in Mimulus inferred from population shotgun sequencing. Proc Natl Acad Sci U S A. 2013;110:19478–19482. doi: 10.1073/pnas.1319032110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tomato Genome Consortium. The tomato genome sequence provides insights into fleshy fruit evolution. Nature. 2012;485:635–641. doi: 10.1038/nature11119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright JW. New chromosome counts in Acer and Fraxinus. Morris Arb Bull. 1957;8:33–34. [Google Scholar]
- 32.Bernards MA, Båstrup-Spohr L. Phenylpropanoid Metabolism Induced by Wounding and Insect Herbivory. In: Schaller A, editor. Induced Plant Resistance to Herbivory. Springer; Netherlands: 2008. pp. 189–211. [Google Scholar]
- 33.Stahl E, Hilfiker O, Reymond P. Plant-arthropod interactions: who is the winner? Plant J. 2018;93:703–728. doi: 10.1111/tpj.13773. [DOI] [PubMed] [Google Scholar]
- 34.Abdulrazzak N, et al. A coumaroyl-ester-3-hydroxylase insertion mutant reveals the existence of nonredundant meta-hydroxylation pathways and essential roles for phenolic precursors in cell expansion and plant growth. Plant Physiol. 2006;140:30–48. doi: 10.1104/pp.105.069690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rupasinghe S, Baudry J, Schuler MA. Common active site architecture and binding strategy of four phenylpropanoid P450s from Arabidopsis thaliana as revealed by molecular modeling. Protein Eng. 2003;16:721–731. doi: 10.1093/protein/gzg094. [DOI] [PubMed] [Google Scholar]
- 36.Dolan WL, Chapple C. Conservation and Divergence of Mediator Structure and Function: Insights from Plants. Plant Cell Physiol. 2017;58:4–21. doi: 10.1093/pcp/pcw176. [DOI] [PubMed] [Google Scholar]
- 37.Bonawitz ND, et al. Disruption of Mediator rescues the stunted growth of a lignin-deficient Arabidopsis mutant. Nature. 2014;509:376–380. doi: 10.1038/nature13084. [DOI] [PubMed] [Google Scholar]
- 38.Dolan WL, Chapple C. Transcriptome Analysis of Four Arabidopsis thaliana Mediator Tail Mutants Reveals Overlapping and Unique Functions in Gene Regulation. G3. 2018;8:3093–3108. doi: 10.1534/g3.118.200573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Xu Z, et al. Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol Biol. 2004;55:343–367. doi: 10.1007/s11103-004-0790-1. [DOI] [PubMed] [Google Scholar]
- 40.Rigsby CM, Herms DA, Bonello P, Cipollini D. Higher Activities of Defense-Associated Enzymes may Contribute to Greater Resistance of Manchurian Ash to Emerald Ash Borer Than A closely Related and Susceptible Congener. J Chem Ecol. 2016;42:782–792. doi: 10.1007/s10886-016-0736-5. [DOI] [PubMed] [Google Scholar]
- 41.Villari C, Herms DA, Whitehill JGA, Cipollini D, Bonello P. Progress and gaps in understanding mechanisms of ash tree resistance to emerald ash borer, a model for wood-boring insects that kill angiosperms. New Phytol. 2016;209:63–79. doi: 10.1111/nph.13604. [DOI] [PubMed] [Google Scholar]
- 42.Erb M, Reymond P. Molecular Interactions Between Plants and Insect Herbivores. Annu Rev Plant Biol. 2019;70:527–557. doi: 10.1146/annurev-arplant-050718-095910. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, Zhu C, Li X. SCFSNIPER4 controls the turnover of two redundant TRAF proteins in plant immunity. Plant J. 2018;95:504–515. doi: 10.1111/tpj.13965. [DOI] [PubMed] [Google Scholar]
- 44.Hua Z, Vierstra RD. The cullin-RING ubiquitin-protein ligases. Annu Rev Plant Biol. 2011;62:299–334. doi: 10.1146/annurev-arplant-042809-112256. [DOI] [PubMed] [Google Scholar]
- 45.Erb M, Meldau S, Howe GA. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012;17:250–259. doi: 10.1016/j.tplants.2012.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Berens ML, Berry HM, Mine A, Argueso CT, Tsuda K. Evolution of Hormone Signaling Networks in Plant Defense. Annu Rev Phytopathol. 2017;55:401–425. doi: 10.1146/annurev-phyto-080516-035544. [DOI] [PubMed] [Google Scholar]
- 47.Lin S-H, et al. Mutation of the Arabidopsis NRT1.5 nitrate transporter causes defective root-to-shoot nitrate transport. Plant Cell. 2008;20:2514–2528. doi: 10.1105/tpc.108.060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huysmans M, Lema AS, Coll NS, Nowack MK. Dying two deaths - programmed cell death regulation in development and disease. Curr Opin Plant Biol. 2017;35:37–44. doi: 10.1016/j.pbi.2016.11.005. [DOI] [PubMed] [Google Scholar]
- 49.Bellin D, Asai S, Delledonne M, Yoshioka H. Nitric oxide as a mediator for defense responses. Mol Plant Microbe Interact. 2013;26:271–277. doi: 10.1094/MPMI-09-12-0214-CR. [DOI] [PubMed] [Google Scholar]
- 50.Zebelo SA, Maffei ME. Role of early signalling events in plant-insect interactions. J Exp Bot. 2015;66:435–448. doi: 10.1093/jxb/eru480. [DOI] [PubMed] [Google Scholar]
- 51.Seifi HS, Shelp BJ. Spermine Differentially Refines Plant Defense Responses Against Biotic and Abiotic Stresses. Front Plant Sci. 2019;10:117. doi: 10.3389/fpls.2019.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Whitehill JGA, Rigsby C, Cipollini D, Herms DA, Bonello P. Decreased emergence of emerald ash borer from ash treated with methyl jasmonate is associated with induction of general defense traits and the toxic phenolic compound verbascoside. Oecologia. 2014;176:1047–1059. doi: 10.1007/s00442-014-3082-8. [DOI] [PubMed] [Google Scholar]
- 53.Nelson R, Wiesner-Hanks T, Wisser R, Balint-Kurti P. Navigating complexity to breed disease-resistant crops. Nat Rev Genet. 2018;19:21–33. doi: 10.1038/nrg.2017.82. [DOI] [PubMed] [Google Scholar]
- 54.Radville L, Chaves A, Preisser EL. Variation in plant defense against invasive herbivores: evidence for a hypersensitive response in eastern hemlocks (Tsuga canadensis) J Chem Ecol. 2011;37:592–597. doi: 10.1007/s10886-011-9962-z. [DOI] [PubMed] [Google Scholar]
- 55.Hilker M, Fatouros NE. Resisting the onset of herbivore attack: plants perceive and respond to insect eggs. Curr Opin Plant Biol. 2016;32:9–16. doi: 10.1016/j.pbi.2016.05.003. [DOI] [PubMed] [Google Scholar]
- 56.Kim CY, Bove J, Assmann SM. Overexpression of wound-responsive RNA-binding proteins induces leaf senescence and hypersensitive-like cell death. New Phytol. 2008;180:57–70. doi: 10.1111/j.1469-8137.2008.02557.x. [DOI] [PubMed] [Google Scholar]
- 57.Bollhöner B, et al. The function of two type II metacaspases in woody tissues of Populus trees. New Phytol. 2018;217:1551–1565. doi: 10.1111/nph.14945. [DOI] [PubMed] [Google Scholar]
- 58.Altmann S, et al. Transcriptomic basis for reinforcement of elm antiherbivore defence mediated by insect egg deposition. Mol Ecol. 2018;27:4901–4915. doi: 10.1111/mec.14900. [DOI] [PubMed] [Google Scholar]
- 59.Rebek EJ, Herms DA, Smitley DR. Interspecific variation in resistance to emerald ash borer (Coleoptera: Buprestidae) among North American and Asian ash (Fraxinus spp.) Environ Entomol. 2008;37:242–246. doi: 10.1603/0046-225X(2008)37[242:IVIRTE]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 60.Wei Z, Green PS. Fraxinus. Flora of China. 1996;15:273–279. [Google Scholar]
- 61.Davidson CG. ‘Northern Treasure’ and ‘Northern Gem’ Hybrid Ash. HortScience. 1999;34:151–152. [Google Scholar]
- 62.Koch JL, et al. Strategies for selecting and breeding EAB-resistant ash. In: McManus Katherine A, Gottschalk Kurt W., editors. Proceedings, 22nd US Department of Agriculture Interagency Research Symposium on Invasive Species; 2011 Jan. 11-14; Annapolis, MD Gen Tech Rep NRS-P-92. US Department of Agriculture, Forest Service, Northern Research Station; Newtown Square, PA: 2011. pp. 33–35. 33-35. [Google Scholar]
- 63.Duan JJ, Larson K, Watt T, Gould J, Lelito JP. Effects of host plant and larval density on intraspecific competition in larvae of the emerald ash borer (Coleoptera: Buprestidae) Environ Entomol. 2013;42:1193–1200. doi: 10.1603/EN13209. [DOI] [PubMed] [Google Scholar]
- 64.Cappaert D, McCullough DG, Poland TM, Siegert NW. Emerald ash borer in North America: a research and regulatory challenge. American Entomologist. 2005;51(3):152–165. 51. [Google Scholar]
- 65.Chamorro ML, Volkovitsh MG, Poland TM, Haack RA, Lingafelter SW. Preimaginal stages of the emerald ash borer, Agrilus planipennis Fairmaire (Coleoptera: Buprestidae): an invasive pest on ash trees (Fraxinus) PLoS One. 2012;7:e33185. doi: 10.1371/journal.pone.0033185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pellicer J, Kelly LJ, Leitch IJ, Zomlefer WB, Fay MF. A universe of dwarfs and giants: genome size and chromosome evolution in the monocot family Melanthiaceae. New Phytol. 2014;201:1484–1497. doi: 10.1111/nph.12617. [DOI] [PubMed] [Google Scholar]
- 67.Loureiro J, Rodriguez E, Dolezel J, Santos C. Two new nuclear isolation buffers for plant DNA flow cytometry: a test with 37 species. Ann Bot. 2007;100:875–888. doi: 10.1093/aob/mcm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Doležel J, Binarová P, Lucretti S. Analysis of Nuclear DNA content in plant cells by flow cytometry. Biol Plant. 1989;31:113–120. [Google Scholar]
- 69.Bennett Michael David, Smith JB. Nuclear DNA amounts in angiosperms. Philos Trans R Soc Lond B Biol Sci. 1991;334:309–345. doi: 10.1098/rstb.1976.0044. [DOI] [PubMed] [Google Scholar]
- 70.Whittemore AT, Xia Z-L. Genome Size Variation in Elms (Ulmus spp.) and Related Genera. HortScience. 2017;52:547–553. [Google Scholar]
- 71.Doležel J, et al. Plant Genome Size Estimation by Flow Cytometry: Inter-laboratory Comparison. Ann Bot. 1998;82:17–26. [Google Scholar]
- 72.Greilhuber J, Obermayer R. Genome size and maturity group in Glycine max (soybean) Heredity. 1997;78:547–551. [Google Scholar]
- 73.Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochemical Bulletin. 1987;19:11–15. [Google Scholar]
- 74.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17:10–12. [Google Scholar]
- 75.Joshi NA, Fass JN. Sickle: A sliding-window, adaptive, quality-based trimming tool for FastQ files. 2011 [Google Scholar]
- 76.Schmieder R, Edwards R. Quality control and preprocessing of metagenomic datasets. Bioinformatics. 2011;27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27:578–579. doi: 10.1093/bioinformatics/btq683. [DOI] [PubMed] [Google Scholar]
- 78.Luo R, et al. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. Gigascience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 81.Keilwagen J, et al. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44:e89. doi: 10.1093/nar/gkw092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Altenhoff AM, Gil M, Gonnet GH, Dessimoz C. Inferring hierarchical orthologous groups from orthologous gene pairs. PLoS One. 2013;8:e53786. doi: 10.1371/journal.pone.0053786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altenhoff AM, et al. The OMA orthology database in 2015: function predictions, better plant support, synteny view and other improvements. Nucleic Acids Res. 2015;43:D240–9. doi: 10.1093/nar/gku1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Goodstein DM, et al. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 2012;40:D1178–86. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wallander E. Systematics of Fraxinus (Oleaceae) and evolution of dioecy. Plant Syst Evol. 2008;273:25–49. [Google Scholar]
- 88.Hinsinger DD, et al. The phylogeny and biogeographic history of ashes (Fraxinus, Oleaceae) highlight the roles of migration and vicariance in the diversification of temperate trees. PLoS One. 2013;8:e80431. doi: 10.1371/journal.pone.0080431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Sela I, Ashkenazy H, Katoh K, Pupko T. GUIDANCE2: accurate detection of unreliable alignment regions accounting for the uncertainty of multiple parameters. Nucleic Acids Res. 2015;43:W7–14. doi: 10.1093/nar/gkv318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Rice P, Longden I, Bleasby A. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet. 2000;16:276–277. doi: 10.1016/s0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 92.Ronquist F, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012;61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ané C, Larget B, Baum DA, Smith SD, Rokas A. Bayesian estimation of concordance among gene trees. Mol Biol Evol. 2007;24:412–426. doi: 10.1093/molbev/msl170. [DOI] [PubMed] [Google Scholar]
- 94.Larget BR, Kotha SK, Dewey CN, Ané C. BUCKy: gene tree/species tree reconciliation with Bayesian concordance analysis. Bioinformatics. 2010;26:2910–2911. doi: 10.1093/bioinformatics/btq539. [DOI] [PubMed] [Google Scholar]
- 95.Castoe TA, et al. Evidence for an ancient adaptive episode of convergent molecular evolution. Proc Natl Acad Sci U S A. 2009;106:8986–8991. doi: 10.1073/pnas.0900233106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- 97.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McKenna A, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly. 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Martin M, et al. WhatsHap: fast and accurate read-based phasing. bioRxiv. 2016 doi: 10.1101/085050. 085050. [DOI] [Google Scholar]
- 102.Milne I, et al. Using Tablet for visual exploration of second-generation sequencing data. Brief Bioinform. 2013;14:193–202. doi: 10.1093/bib/bbs012. [DOI] [PubMed] [Google Scholar]
- 103.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Pond SLK, Frost SDW, Muse SV. HyPhy: hypothesis testing using phylogenies. Bioinformatics. 2005;21:676–679. doi: 10.1093/bioinformatics/bti079. [DOI] [PubMed] [Google Scholar]
- 105.Kosakovsky Pond SL, Posada D, Gravenor MB, Woelk CH, Frost SDW. Automated phylogenetic detection of recombination using a genetic algorithm. Mol Biol Evol. 2006;23:1891–1901. doi: 10.1093/molbev/msl051. [DOI] [PubMed] [Google Scholar]
- 106.Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41:D36–42. doi: 10.1093/nar/gks1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu Z, et al. Evolutionary interplay between sister cytochrome P450 genes shapes plasticity in plant metabolism. Nat Commun. 2016;7:13026. doi: 10.1038/ncomms13026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Altenhoff AM, et al. The OMA orthology database in 2018: retrieving evolutionary relationships among all domains of life through richer web and programmatic interfaces. Nucleic Acids Res. 2018;46:D477–D485. doi: 10.1093/nar/gkx1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 111.Alexa A, Rahnenfuhrer J. topGO: enrichment analysis for gene ontology. 2016 [Google Scholar]
- 112.Team, R. C. R: A language and environment for statistical computing. 2013 Others. [Google Scholar]
- 113.Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 114.Käll L, Krogh A, Sonnhammer ELL. Advantages of combined transmembrane topology and signal peptide prediction--the Phobius web server. Nucleic Acids Res. 2007;35:W429–32. doi: 10.1093/nar/gkm256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Källberg M, et al. Template-based protein structure modeling using the RaptorX web server. Nat Protoc. 2012;7:1511–1522. doi: 10.1038/nprot.2012.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31:455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol. 2015;1263:243–250. doi: 10.1007/978-1-4939-2269-7_19. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Underlying data for Figure 1 are available in Supplementary Tables 1 and 2. All trimmed read data and genome assemblies have been deposited in the European Nucleotide Archive under accession number PRJEB20151 (https://www.ebi.ac.uk/ena/browser/view/PRJEB20151). The genome assemblies are also available to download at: http://www.ashgenome.org.
The custom scripts used is this study have been deposited in GitHub: https://github.com/lkelly3/eab-ms-scripts.