Abstract
Genome-wide association studies (GWAS) have discovered numerous genomic loci associated with Alzheimer’s disease (AD), yet the causal genes and variants remain incompletely identified. We performed an updated genome-wide AD meta-analysis, which identified 37 risk loci, including novel associations near CCDC6, TSPAN14, NCK2, and SPRED2. Using three SNP-level fine-mapping methods, we identified 21 SNPs with greater than 50% probability each of being causally involved in AD risk, and others strongly suggested by functional annotation. We followed this with colocalization analyses across 109 gene expression quantitative trait loci (eQTL) datasets, and prioritization of genes using protein interaction networks and tissue-specific expression. Combining this information into a quantitative score, we find that evidence converges on likely causal genes, including the above four genes, and those at previously discovered AD loci, including BIN1, APH1B, PTK2B, PILRA, and CASS4.
Genome-wide association studies (GWAS) for family history of disease, known as GWAS-by-proxy (GWAX), are a powerful method for performing genetic discovery in large, unselected cohort biobanks, particularly for age-related diseases1. Recent meta-analyses have combined GWAS of diagnosed late-onset Alzheimer’s disease (AD) with GWAX for family history of AD in the UK Biobank2,3, and reported 12 novel disease-associated genomic loci. However, the causal genetic variants and genes which influence AD risk at these and previously discovered loci have only been clearly identified in a few cases. Discovering causal variants has led to deeper insight into molecular mechanisms of multiple diseases, including obesity4, schizophrenia5, and inflammatory bowel disease6. For AD, known causal variants include the 𝜀4 haplotype in APOE, the strongest genetic risk factor for late-onset AD, and a common nonsynonymous variant that strongly alters splicing of CD33 exon 27. Likely causal rare nonsynonymous variants have also been discovered in TREM2 8, PLCG2 and ABI3 9. These findings have strengthened support for a causal role of microglial activation in AD.
Although non-synonymous variants are highly enriched in trait associations, most human trait-associated variants do not alter protein-coding sequence and are thought to mediate their effects via altered gene expression, which is likely to occur in a cell type-dependent manner. A growing number of studies have mapped genetic variants affecting gene expression, known as expression quantitative trait loci (eQTLs), in diverse tissues or sorted cell types10,11. While it has become common to integrate GWAS results with eQTLs, this is often limited to a small number of datasets thought to be relevant.
To identify putative causal genetic variants for AD, we performed a meta-analysis of GWAX in the UK Biobank with the latest GWAS for diagnosed AD12, followed by fine-mapping using three alternative methods. Notably, this updated GWAS tested more genetic variants than the Lambert et al. study13 used in meta-analyses by Jansen et al.3 and Marioni et al.2 (11.5 vs. 7.1 million). The increased power from our meta-analysis revealed four additional AD risk loci, and the higher density genotype imputation identified new candidate causal variants at both novel and established loci. We also performed statistical colocalization analyses with a broad collection of eQTL datasets, including a recent study on primary microglia14, to identify candidate genes mediating risk at AD loci. We find that multiple lines of evidence, including colocalization, tissue- or cell type-specific expression, and information propagation in gene networks, converge on a set of likely causal AD genes.
Results
Meta-analysis discovers 37 loci associated with Alzheimer’s disease risk
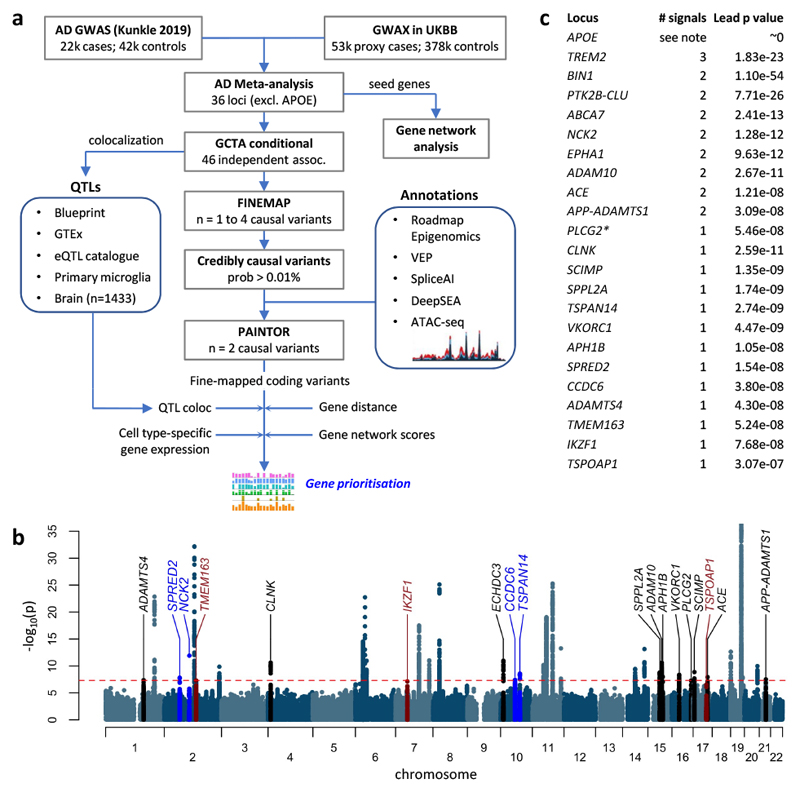
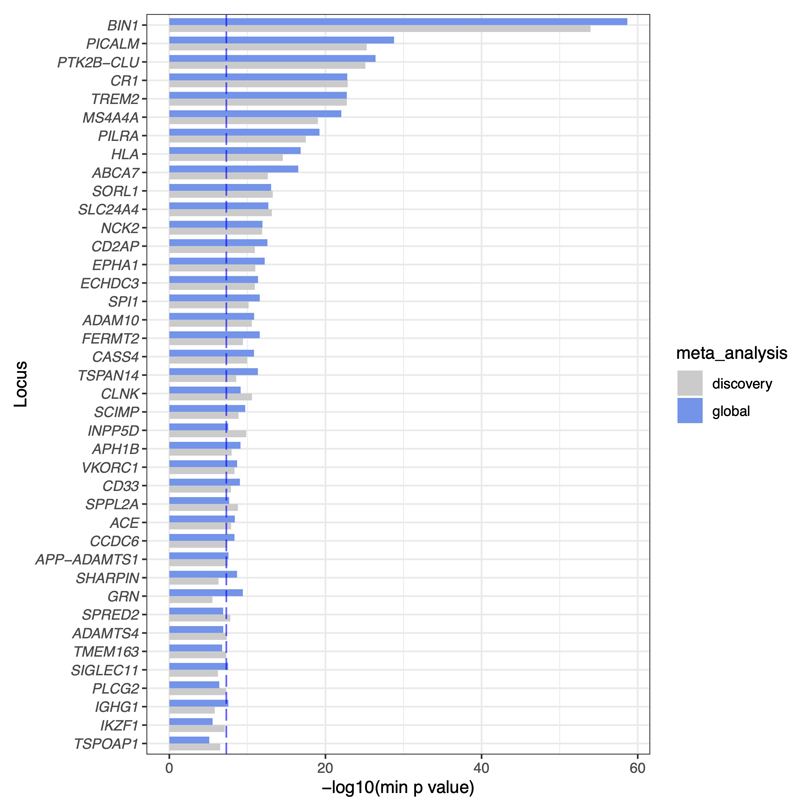
We performed a GWAX in the UK Biobank for family history of AD, based on 53,042 unique individuals who were either diagnosed with AD or who reported a parent or sibling having dementia, and 355,900 controls. This identified 13 risk loci (P < 5 x 10-8), 10 of which have been reported previously. Three novel loci were located near NCK2, PRL, and FAM135B. Notably, PRL has been reported as a CSF biomarker of AD15. We next did a fixed-effects meta-analysis of these GWAX results with the Kunkle et al. stage 1 GWAS meta-analysis of 21,982 cases with diagnosed AD and 41,944 controls12, across 10,687,126 overlapping variants (Fig. 1). This revealed 34 AD risk loci (P < 5 x 10-8), 22 of which were reported in Kunkle et al., while 8 others were reported in either Jansen et al.3 or Marioni et al.2. Four loci were novel, located near NCK2, TSPAN14, SPRED2, and CCDC6. Notably, the PRL and FAM135B regions showed no evidence of association in Kunkle et al. (P > 0.1), and hence were not significant in meta-analysis. We included 37 loci in our follow-up analyses, which included three loci found at suggestive significance (P < 5 x 10-7) near IKZF1, TSPOAP1, and TMEM163 (Fig. 1 and Supplementary Table 1). LD score regression16 showed that most of the inflation in summary statistics was due to the polygenicity of AD rather than confounding by population structure (lambda_GC = 1.140, intercept = 1.0285 with SE = 0.0069; Supplementary Table 2). Of our 37 loci, 16 were nominally replicated (P < 0.05) in either the Gr@ace study17 (4,120 probable AD cases and 3,289 controls) or the FinnGen biobank (v3, 3,697 cases and 131,941 controls) (Supplementary Table 3). Among our four novel loci, only TSPAN14 replicated with P < 0.05 (in FinnGen), although power was limited in these smaller datasets (estimated at 28-76%), and most of the alleles had concordant directions of effect. In a meta-analysis with all four datasets, support for most loci was strengthened (Extended Data Fig. 1), including novel loci TSPAN14, CCDC6 and NCK2, but was weakened for SPRED2 (meta-analysis P = 1.3 x 10-7). Although not included in downstream analyses, four new loci became genome-wide significant, near GRN, IGHG1, SHARPIN, and SIGLEC11 (Supplementary Table 3).
Figure 1. Analysis overview.
a, Summary of AD meta-analysis and data processing steps. b, Manhattan plot of the meta-analysis of GWAS for diagnosed AD and our GWAX in UK Biobank. Novel genome-wide significant loci are labelled in blue, sub-threshold loci in red, and recently discovered loci2,3,12 replicated in our analysis in black. c, The number of independent signals at each locus which is either recently discovered or which has more than one signal, as well as the meta-analysis P value the lead SNP at the locus. *The PLCG2 locus was significant (P < 5 x 10-8) when including Kunkle stage 3 SNPs. Conditional analyses were not done at APOE due to the strength of the signal (see Methods).
Next, we applied stepwise conditioning using GCTA18, with linkage disequilibrium (LD) determined from UK Biobank samples, to identify independent signals at the discovered loci. Apart from APOE, 9 loci had two independent signals, while the TREM2 locus had three signals (Fig. 1c). Interestingly, a number of the loci discovered recently2,3,12 had multiple signals: NCK2, EPHA1, ADAM10, ACE, and APP-ADAMTS1. To extract insight from both new and established AD GWAS discoveries, we performed comprehensive colocalization, annotation, fine-mapping and network analyses to identify causal genes and variants (Fig. 1a).
Colocalization between AD risk loci and gene expression traits
To identify genes whose expression may be altered by risk variants, we performed statistical colocalization19 between each of 36 risk loci (excluding APOE) and a set of 109 eQTL datasets representing a wide variety of tissues, cell types and conditions (Fig. 2 and Supplementary Table 4). The eQTL datasets include a study of primary microglia from 93 brain surgery donors14, a meta-analysis of 1,433 brain cortex samples20, 49 tissues from GTEx11, and 57 eQTL datasets uniformly reprocessed as part of the eQTL catalogue10. The latter include multiple studies in tissues of potential relevance to AD, such as brain, as well as sorted blood immune cell types under different stimulation conditions21–37. For each gene, the colocalization analysis reports the probability that the GWAS and eQTL share a causal variant, referred to as hypothesis 4 (H4).
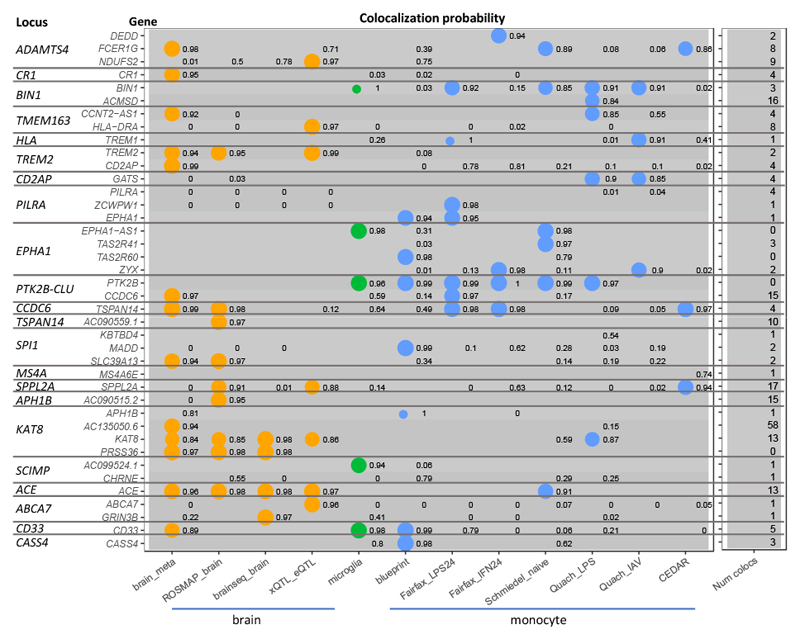
Figure 2. Colocalization with eQTLs.
For genes with the top overall colocalization scores across AD risk loci, the colocalization probability (H4) is shown for selected brain, microglia, and monocyte eQTL datasets. For three loci with multiple signals (BIN1, EPHA1, PTK2B-CLU), scores are shown separately for the conditionally independent signals. The last column shows, for each gene, the number of eQTL datasets with a colocalization probability above 0.8 (Supplementary Tables 5 and 6).
Some studies using colocalization have suggested that there is relatively limited overlap between GWAS associations and eQTLs above that expected by chance6,38. A possible reason is that colocalization analyses can have low sensitivity to detect shared causal variants between traits, which could occur for a number of reasons. First, when a locus has multiple causal variants, and not all causal effects are shared between the two studies, colocalization may not be detected19. Second, if the relevant tissue, cell type, or cellular context has not been assayed, then a colocalization may not be found. Third, differences in LD patterns between studies can reduce the likelihood of a positive colocalization. Lastly, low power in either study can further reduce the colocalization probability. To mitigate the first effect, we performed colocalizations separately for each conditionally independent AD signal, to model the case where not all causal variants are shared, as well as for the combined AD signal at each locus. Problems relating to power, LD mismatch, or missing the relevant cell type or context are partially mitigated by our use of a large number of highly-powered eQTL datasets, which include those with stimulated conditions.
Across the 36 loci, we found 391 colocalizations with at least 80% probability of a shared causal variant between AD and eQTL, representing 80 distinct genes at 27 loci (Supplementary Tables 5 and 6). The genes implicated by colocalization include many that have previously been investigated for roles in AD, such as PTK2B 39,40, BIN1 41,42, PILRA 43, CD33 44,45, and TREM2 46,47, as well as novel candidates including FCER1G, TSPAN14, APH1B, and ACE. However, the presence of multiple genes with colocalization evidence within individual loci suggests that additional lines of evidence are important for prioritizing relevant genes.
Fine-mapping identifies credibly causal variants
Confirming the causal genes underlying AD risk will ultimately require experiments to identify the molecular mechanisms by which gene function is altered. Such experiments must be motivated by strong hypotheses regarding potentially causal variants and their possible effects. To identify candidate causal variants, we used three distinct fine-mapping methods: single causal variant fine-mapping48 on each conditionally independent signal; FINEMAP49, limiting the number of causal variants at each locus to the number of signals determined by GCTA; and PAINTOR50, a method that leverages enrichments in functional genomic annotations to improve causal variant identification (see Methods).
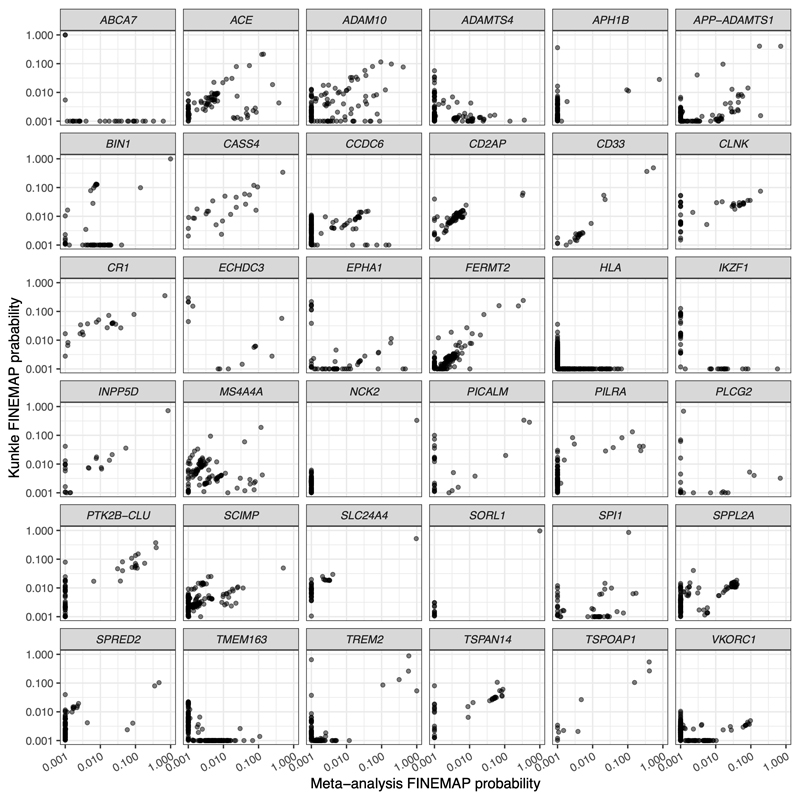
As a reference panel for our analyses, we used LD computed from UK Biobank participants. Previous work has shown that using reference panels that are either too small or poorly matched can result in spurious fine-mapping signals51. For this reason, we conducted a sensitivity analysis (described in the Supplementary Note) by using the same reference panel for conditional analysis and fine-mapping on the non-UK Biobank portion of our meta-analysis (Kunkle et al.). This gave comparable independent signals and SNP probabilities to the full meta-analysis, with the exception of a few loci, namely ABCA7, HLA, EPHA1, and ECHDC3 (Extended Data Fig. 2).
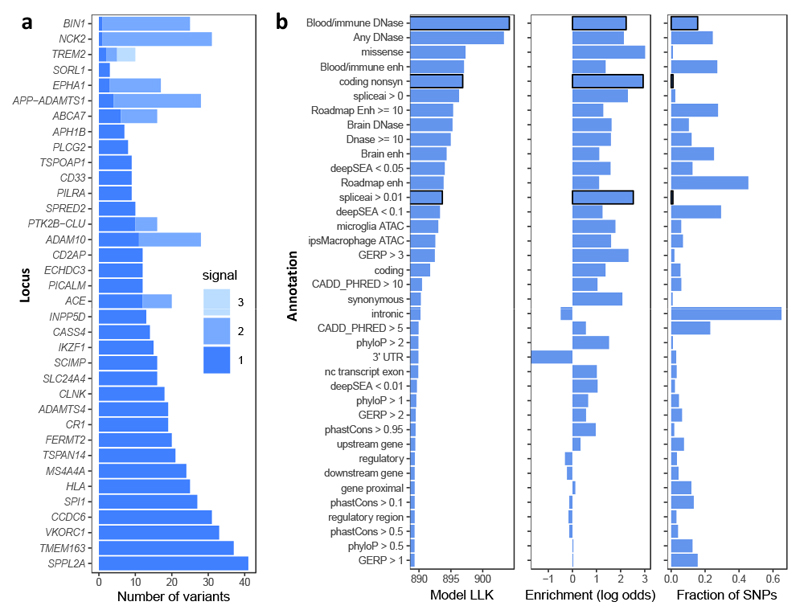
We used 44 annotations individually as input to PAINTOR (Supplementary Table 7); these included ATAC-seq peaks from primary microglia52 or iPSC-derived macrophages53, DNase peaks from the Roadmap Epigenomics project54, variant consequence annotations55, and evolutionary conservation56 (Fig. 3). We also used scores from DeepSEA57 and SpliceAI58, deep-learning methods that predict the effects of variants on transcription factor binding or splicing. Missense mutations were the most enriched annotation, with 19.2-fold increased odds of being causal SNPs, but they comprised only 1% of input SNPs. Blood or immune DNase hypersensitivity peaks merged from 24 Roadmap Epigenomics tissues provided the highest model likelihood, as these peaks covered 16% of SNPs, despite a lower 6.4-fold enrichment. Variants with a non-zero score from SpliceAI, which predicts changes to gene splicing, were also highly enriched (9.3-fold).
Figure 3. Fine-mapping summary.
a, Number of variants with mean causal probability > 1% for each independent signal. Variant counts for independent signals are shown in different shades. b, PAINTOR outputs, showing (left) log-likelihood (LLK) of model for each individual annotation; (middle) log-odds enrichments for individual genomic annotations determined by PAINTOR; (right) fraction of SNPs which are in each annotation (among those selected by FINEMAP probability > 0.01%). Annotations selected for the final model are shown with a black border.
We next built a multi-annotation model in PAINTOR following a stepwise selection procedure, which identified a minimal but informative set of three annotations: blood and immune DNase, nonsynonymous coding variants, and variants with SpliceAI score greater than 0.01. We used probabilities from this PAINTOR model, and computed the mean causal probability per variant across the three fine-mapping methods.
There were 21 variants with mean causal probability above 50% across the fine-mapping methods, and 79 further variants with probabilities from 10-50% (Table 1 and Supplementary Table 8). These include SNPs near established AD risk genes, such as rs6733839 ~20 kb upstream of BIN1, which has recently been shown to alter a microglial MEF2C binding site14 and to regulate BIN1 expression specifically in microglia42. High-confidence variants also include a well-known missense SNP in PILRA 43, and a splice-altering missense SNP in CD33 7. Missense SNP rs4147918 in ABCA7 had 55% causal probability, and ABCA7 harbored 5 further missense SNPs with probabilities greater than 0.01%, at varying allele frequencies. Notably, rs4147918 and 6 other variants within ABCA7, including the lead SNP rs12151021, had positive SpliceAI scores. This is consistent with reports of a burden of deleterious variants at ABCA7 associated with AD59, as well as potential changes to splicing caused by intronic variable tandem repeats60.
Table 1. Top candidate variants.
| ADAMTS4 | rs2070902 | 1.64E-06 | 0.949 | T | 0.2580 | 0.384 | 0.107 | 0.140 | Intronic in candidate gene FCER1G, with predicted splicing change | |
| ADAMTS4 | rs4575098 | 4.30E-08 | 1.063 | A | 0.2350 | 0.339 | 0.033 | 3’ UTR of ADAMTS4, open chromatin | 2 | |
| SPRED2 | rs268120 | 2.08E-08 | 1.063 | A | 0.2502 | 0.556 | 0.033 | Strong DNase peak, predicted by DeepSEA to decrease | ||
| NCK2 | rs143080277 | 1.28E-12 | 0.594 | T | 0.9957 | 1.000 | 0.086 | Enhancer (Roadmap) | ||
| BIN1 | rs6733839 | 1.10E-54 | 1.168 | T | 0.3915 | 0.998 | 0.027 | Microglia ATAC peak. DeepSEA predicts decreased DNaseHS | 14,40 | |
| INPP5D | rs10933431 | 1.41E-10 | 1.080 | C | 0.7817 | 0.833 | 0.022 | 60 | ||
| PILRA | rs1859788 | 3.28E-18 | 0.914 | A | 0.3206 | 0.601 | 0.008 | 0.041 | Known PILRA missense G78R | 41 |
| ECHDC3 | rs7920721 | 1.08E-11 | 0.935 | A | 0.6195 | 0.641 | 0.026 | DNase peak. DeepSEA predicts changed binding of USF, Max, Myc | 61,62 | |
| TSPAN14 | rs1870137 | 2.93E-09 | 0.932 | C | 0.2056 | 0.097 | 0.007 | Top DeepSEA variant, predicting decreased binding of HNF4, FOXA1, SP1 | ||
| TSPAN14 | rs1870138 | 4.51E-09 | 0.933 | A | 0.2057 | 0.068 | 0.004 | Highlighted in text; predicted loss of TAL1 binding | ||
| SORL1 | rs11218343 | 5.59E-14 | 1.205 | T | 0.9630 | 1.000 | 0.209 | 63 | ||
| SORL1 | rs2298813 | 1.52E-04 | 1.089 | A | 0.0470 | 0.451 | 0.054 | 0.003 | Secondary association. Missense; also top DeepSEA variant | |
| APH1B | rs117618017 | 1.05E-08 | 1.089 | T | 0.1395 | 0.895 | 0.007 | 0.019 | Highlighted in text; missense Thr27Ile | 64 |
| PLCG2 | rs12444183 | 5.46E-08 | 0.948 | A | 0.3830 | 0.686 | 0.220 | Near promoter of ncRNA AC099524.1, with strong microglia colocalization | 2 | |
| PLCG2 | rs72824905 | 6.35E-06 | 1.310 | C | 0.9924 | 0.492 | 0.018 | 0.006 | Secondary association; known missense Pro522Arg. Top DeepSEA score | 9 |
| TSPOAP1 | rs2632516 | 3.12E-07 | 0.952 | C | 0.4426 | 0.412 | 0.126 | Overlaps ncRNA containing mir-142, important for hematopoietic development | 62,65 | |
| TSPOAP1 | rs2526377 | 8.45E-07 | 1.049 | A | 0.5579 | 0.169 | 0.006 | Top DeepSEA variant (decreased DNaseHS) in microglial ATAC peak | 66 | |
| ACE | rs4311 | 1.21E-08 | 0.947 | T | 0.4704 | 0.490 | 0.126 | 0.053 | Strong predicted splicing change | 67,68 |
| ACE | rs3730025 | 2.58E-07 | 0.819 | A | 0.9828 | 0.416 | 0.002 | 0.021 | Secondary association; low-frequency missense Tyr244Cys | |
| ABCA7 | rs12151021 | 2.41E-13 | 1.080 | A | 0.3258 | 0.713 | 0.013 | 0.312 | Lead ABCA7 variant | |
| ABCA7 | rs4147918 | 7.63E-07 | 1.128 | A | 0.9587 | 0.552 | 0.071 | 0.045 | Seondary association; missense Gln905Arg; predicted splicing change | 69 |
| CD33 | rs12459419 | 2.02E-08 | 0.944 | T | 0.3256 | 0.662 | 0.001 | 0.070 | Known missense Ala14Val; strong splicing QTL | 7 |
| CASS4 | rs6014724 | 1.07E-10 | 1.116 | A | 0.9122 | 0.548 | 0.083 | Lead CASS4 variant | ||
| CASS4 | rs17462136 | 1.01E-09 | 0.901 | C | 0.0872 | 0.067 | 0.001 | 5’ UTR of CASS4; global top DeepSEA variant predicting decreased TF binding |
A selected list of the most likely causal variants across loci, based on a combination SNP fine-mapping probabilities and annotations. Column ‘SNP prob’ indicates the mean fine-mapping probability for the SNP; the SpliceAI score is the maximum splicing probability for donor gain/loss or acceptor gain/loss, with nonzero values highly enriched for splicing effects; the DeepSEA functional significance score represents the significance above expectation for chromatin feature changes, as well as evolutionary conservation, with lower values more significant. References for specific SNPs are shown2,7,9,14,42,43,61–70.
A number of newly identified AD risk genes had high-confidence fine-mapped variants. These include the NCK2 rare intronic SNP rs143080277 (>99% probability, MAF 0.4%), APH1B missense SNP rs117618017 (90% probability), rs2830489 near ADAMTS1 (72% probability), and rs268120 intronic in SPRED2 (56% probability).
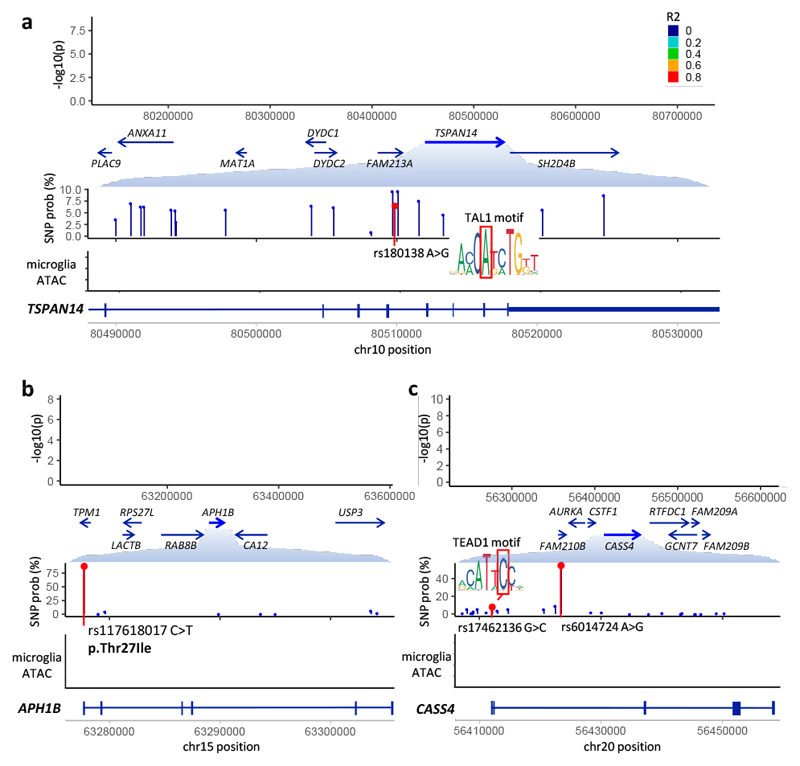
Manual review highlighted a number of candidate causal variants, where the annotation-based SNP probability was higher than that of the other two methods (Fig. 4). Within TSPAN14, rs1870137 and rs1870138 reside within a DNase hypersensitivity peak found broadly across tissues, which is also an ATAC peak in microglia. Of these, rs1870138 lies at the centre of a ChIP-seq peak for binding of multiple transcription factors, including FOS/JUN and GATA1. The AD risk allele rs1870138-G alters an invariant position of a binding motif for TAL1, a gene highly expressed in microglia, and which is a binding partner for GATA1. This allele is also associated with increased monocyte count71 and increased risk for inflammatory bowel disease72. Notably, the AD signal in the region colocalizes with both an eQTL and a splicing QTL for TSPAN14 in multiple datasets, and rs1870138-G associates with higher TSPAN14 expression in brain and in microglia, but with lower expression in some GTEx tissues.
Figure 4. Fine-mapped variants.
a, SNP rs1870138 in an intron of TSPAN14 disrupts an invariant position of a TAL1 motif. b, Missense SNP rs117618017 in exon 1 of APH1B. c, SNP rs17462136 in the 5’ UTR of CASS4 introduces a TEAD1 motif. Each panel shows (top) locus plot with GWAS P-values, SNP color representing LD to the lead SNP; (middle) expanded view of a subregion showing the mean SNP probabilities from fine-mapping; (bottom) read density of ATAC-sequencing assay from primary microglia52.
Missense SNP rs117618017 in exon 1 of APH1B (Thr27Ile) is the likely single causal variant at its locus, with fine-mapping probability of 90% (Fig. 4b). APH1B is a component of the gamma-secretase complex, other members of which (PSEN1, PSEN2) have rare variants associated with early-onset AD73. Interestingly, the AD signal colocalizes with an APH1B eQTL in monocytes, neutrophils and T-cells, and rs117618017-T associates with higher AD risk and higher APH1B expression across datasets. This allele introduces a motif for transcriptional regulator YY1, and is predicted by DeepSEA to increase YY1 binding in multiple ENCODE cell lines. Therefore, it is an open question whether AD risk is mediated by altered APH1B protein structure or altered gene expression.
Finally, the AD association on chromosome 20 colocalizes with an eQTL for CASS4 in Blueprint monocytes and in GTEx whole blood. While intronic lead SNP rs6014724 (55% probability) shows no evidence of transcription factor (TF) binding in ENCODE data, rs17462136 (7% probability) lies in a region of dense TF binding in the 5’ UTR of CASS4 (Fig. 4c). The nucleotide position is highly conserved (GERP score 3.46) and overlaps an ATAC peak in microglia, and the rs17462136-C allele introduces a TEAD1 binding motif. In addition, rs17462136 is more strongly associated with CASS4 expression in multiple eQTL datasets than is rs6014724.
Network evidence prioritizes genes within and beyond GWAS loci
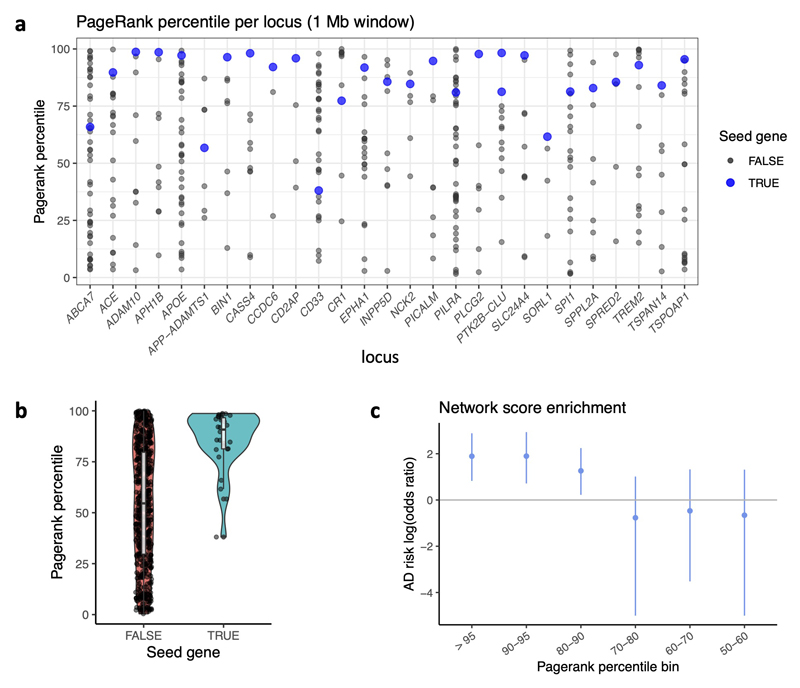
As a further line of evidence, we developed a method that leverages gene network connectivity to prioritize genes at individual loci. We first constructed a gene interaction network combining information from the STRING, IntAct and BioGRID databases. Next, we nominated 32 candidate AD genes (Supplementary Table 9), based on our other evidence sources as well as literature reports, and used these as seed genes similar to the approach used in the priority index for drug discovery74. For each locus in turn, we used as input all seed genes except those at the locus, and propagated information through the network with the page rank algorithm. The “networkScore” for a gene thus represents the degree to which the gene is supported by its interaction with top AD candidate genes at other loci, unbiased by any locus-specific features.
Across AD loci, our selected seed genes were highly enriched for having high network-based gene scores (one-tailed Wilcoxon rank sum test, P = 5 x 10-9; Extended Data Fig. 3). At our four novel AD loci, the nearest gene (NCK2, TSPAN14, SPRED2, CCDC6) in each case was one of the top two highest-scoring genes within 500 kb. Many established or recently discovered AD genes were also the top gene within 500 kb by network score, including ACE, BIN1, CASS4, CD2AP, PICALM, PLCG2, and PTK2B. At the SLC24A4 locus, RIN3 was strongly supported, whereas SLC24A4 was not, in line with evidence from deleterious rare variants that RIN3 may be causal12.
Genes highly ranked by network propagation also include many outside of genome-wide significant AD loci (Supplementary Table 10). Consistent with their involvement in AD, such genes tended to have SNPs with lower P values nearby than did remaining genes (Fig. 5a and Extended Data Fig. 3c), suggesting that numerous AD loci remain to be discovered with larger GWAS sample sizes. Top network-ranked genes include LILRB2 (nearby rs3855678 P = 9.8 x 10-6), which encodes a leukocyte immunoglobulin-like receptor that recognizes multiple HLA alleles, and which may also be involved in amyloid-beta fibril growth75; ABCA1 (rs59237458 P = 4 x 10-6), involved in phospholipid transfer to apolipoproteins and previously associated with AD76; SREBF1 (rs35763683 P = 2 x 10-6), required for lipid homeostasis; and AGRN (rs2710871 P = 4 x 10-6), involved in synapse formation in mature hippocampal neurons. Overall, genes with high network ranks were strongly enriched in biological processes and pathways that have previously been associated with AD, including clathrin-mediated endocytosis, activation of immune response, phagocytosis, Ephrin signaling, and complement activation (Supplementary Table 11).
Figure 5. Genome-wide network and gene expression enrichments.
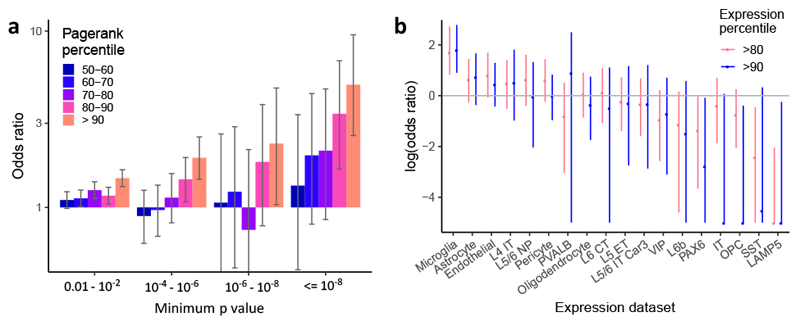
a, Enrichment of low GWAS P values within 10 kb of genes having high vs. low network pagerank percentile (low defined as below 50th percentile). Whiskers represent 95% confidence intervals based on Fisher’s exact test for n = 18,055 genes. b, Enrichment of AD risk near genes with high expression in each brain cell type (above 80th or 90th percentile) relative to the other cell types. Cell types are defined based on single-cell clusters defined in Hodge et al.77. Neuronal cells are defined either by cortical layer (L4, L5, L6), and/or by projection target (IT, intratelencephalic; CT, corticothalamic; ET, extratelencephalic-pyramidal tract; NP, near-projecting), or by binary marker genes (LAMP5, PAX6, PVALB, VIP, SST). OPC, oligodendrocyte precursor cells. Whiskers represent 95% confidence intervals as determined by fgwas.
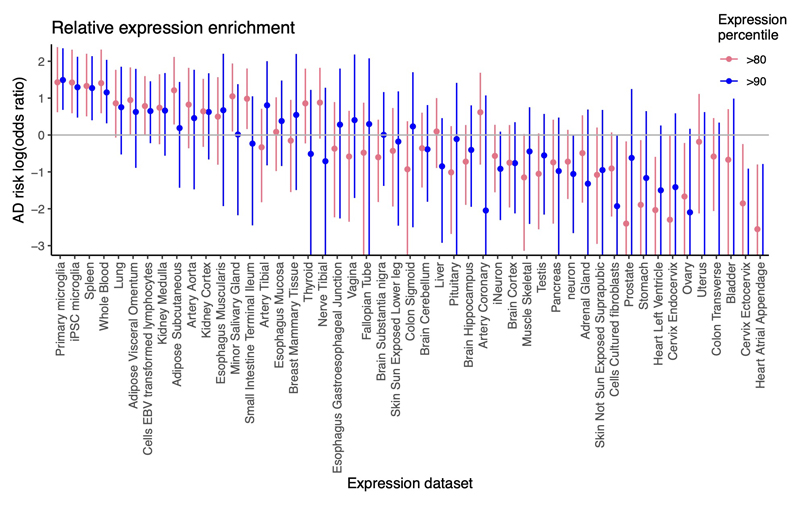
AD risk is enriched near genes with high microglial gene expression
To understand the contribution of cell-type specific gene expression to AD risk, we used fgwas78 to assess the genome-wide enrichment of SNPs near genes highly expressed in specific cell types, based on a single-nucleus sequencing dataset of 49,495 nuclei from six human brain cortical areas77,79. Out of 18 broad cell type clusters, only microglia showed clear enrichment of AD risk (odds ratio (OR) 6.0) near genes with expression above the 90th percentile across cell types (Fig. 5b). We performed a similar analysis looking at bulk gene expression across human tissues from GTEx, along with a small number of additional RNA-seq datasets, including sorted primary microglia from brain surgeries14 (Extended Data Fig. 4 and Supplementary Table 12). This gave consistent results, with microglia showing strong enrichment (OR 4.4), followed by tissues rich in immune cells, including spleen (OR 3.6) and whole blood (OR 3.2). Notably, iPSC-derived microglia showed similar enrichment to primary microglia, while bulk brain tissues (including hippocampus) showed no enrichment.
Integrative gene prioritization from five lines of evidence
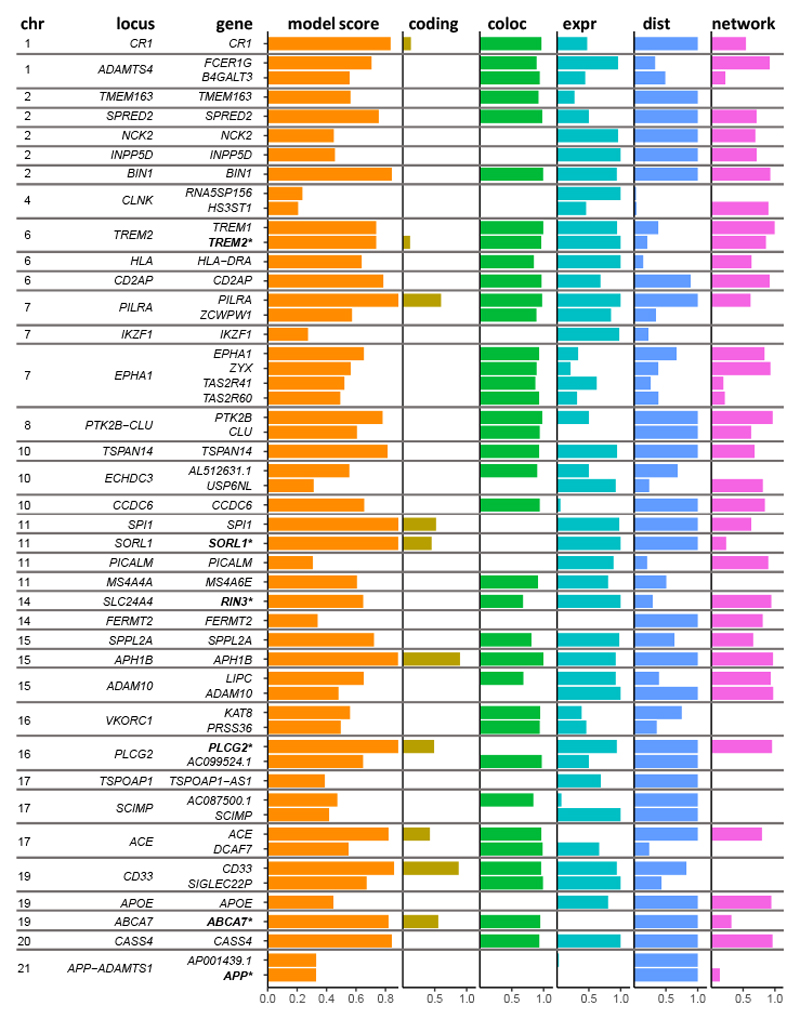
Determining the genes responsible for AD risk across GWAS loci is challenging, in part because few genes have been definitively confirmed as having a causal role. We therefore developed a comprehensive gene prioritization score, which incorporates quantitative information based on five lines of evidence: gene distance to lead SNPs, colocalization, network score, bulk and single-cell gene expression, and the sum of fine-mapped probability for any coding SNPs within a gene (Fig. 6, Extended Data Figs. 5 and 6, and Supplementary Table 13).
Figure 6. Gene evidence summary.
The top gene at each locus is shown, as well as the next 13 top genes by model score; for three loci where a non-coding gene was the top scoring, we also show the top scoring protein-coding gene. Score components for each gene are indicated by colored bars, and points show the distribution of scores for all genes within 500 kb at the locus. Bold gene names are those with evidence of causality based on rare variants from other studies. Scores for all genes are listed in Supplementary Table 13.
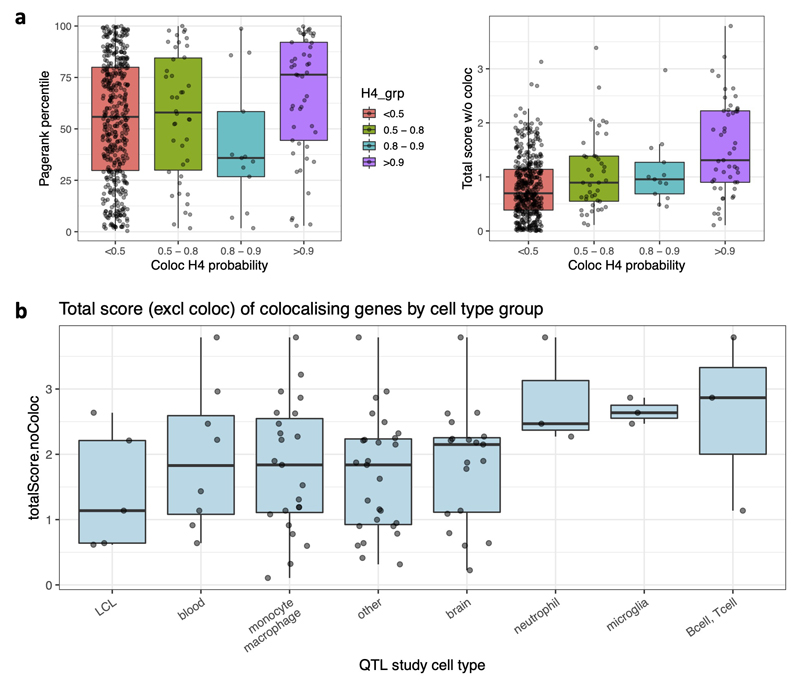
We first explored how best to use colocalization information. We found that genes with maximum colocalization probability (maxH4) above 0.9 had higher prioritization scores based on the other four predictors, but this was not the case for genes with weaker colocalization evidence (Extended Data Fig. 5a). We also examined colocalizations in different cell type or tissue groups, such as brain, microglia, and other GTEx tissues. There was little evidence that colocalizing genes within any specific groups had higher total scores than other groups (Extended Data Fig. 5b), although this conclusion was limited by the low number of studies in some cell types, such as microglia. We therefore based our colocalization score on the maximum colocalization probability across tissues (> 0.9) and normalized this to the range 0-1.
A priori, we do not know which lines of evidence are most important for prioritizing genes. We therefore sought a systematic way to identify appropriate weights for the predictors. Although we do not know the causal AD genes, we selected two independent, unbiased sets of candidate genes for use in supervised learning: genes nearest to the GWAS peaks, and genes with high network scores (>80th percentile). In order to identify weights for our predictive features, we defined two models to discriminate these two gene sets from others within 500 kb, in each case using cross-validated lasso-regularized logistic regression with the remaining variables as predictors. As expected, when predicting genes nearest GWAS peaks, the highest-weight predictor was fine-mapped coding variants; however, only a few loci have such variants. The most informative predictor, determined based on change in mean-squared error when the predictor is left out, was colocalization, followed by coding variants and then network score (Supplementary Table 14). When predicting high network score genes, the most informative predictor was distance to GWAS peak, followed by microglial gene expression, and neither colocalization nor coding variant predictors improved the model. For both models, including hippocampus expression (GTEx) or single-cell astrocyte expression resulted in worse models (increased mean squared error).
We defined our gene prioritization “model score” as the average of the predictions from our two models. The model score identified as top-ranked many AD candidate genes previously suggested as causal (Fig. 6). Exemplifying the importance of integrating genetic evidence sources, ABCA7, SORL1, and CR1 were top-ranked by overall score at their respective loci, despite having only moderate network-based scores, while SORL1, PICALM, and SPI1 were top-ranked despite having limited eQTL colocalization evidence.
While our prioritization further supports many established AD candidate genes, it also implicates novel genes. Among these are FCER1G, which has been reported as a hub gene in microglial gene modules associated with neurodegeneration81,82, and has been experimentally shown to influence microglial phagocytosis83. Another candidate is ZYX, which receives a top network score, is highly expressed in microglia, and which was recently nominated as an AD risk gene based on chromatin interactions between the ZYX promoter and AD risk variants in a ZYX enhancer84.
Discussion
Identifying therapeutic targets for human diseases is a key goal of human genetics research, and is particularly important for neurodegenerative diseases such as AD, for which no disease-modifying therapies yet exist. However, identifying the causal genes and genetic variants from GWAS is challenging, since non-coding associations can act via regulation of distal genes. We approached this challenge for AD by performing comprehensive fine-mapping, eQTL colocalization, network analysis, and quantitative gene prioritization.
Our meta-analysis identified four novel associations near NCK2, SPRED2, TSPAN14, and CCDC6. Each of these was the nearest gene to the association peak and was supported by both eQTL colocalization and network ranking. Yet, despite the large number of eQTL datasets that we used, colocalization of likely AD risk genes was sometimes found in only one or a few datasets; this was the case for SPRED2 (TwinsUK LCL coloc probability 0.99), RIN3 (GTEx frontal cortex probability 0.94), and PILRA (Fairfax LPS-2hr monocyte coloc probability 0.99). Many factors could account for dataset-specific colocalizations, such as biological differences in sample state, differences in LD match between the GWAS and eQTL datasets, and technical differences in the transcriptome annotations used for eQTL discovery. As a result, absence of colocalization provides only weak evidence for lack of an effect in a given tissue type, whereas positive colocalization provides strong support for a shared genetic effect. It is therefore useful to look broadly across eQTL studies for colocalization, which will be facilitated by resources that simplify access to these datasets, such as the eQTL catalogue11.
One of our most confidently prioritized genes was APH1B, encoding a gamma-secretase complex component involved in APP processing. APH1B harbors the likely causal missense variant T27I, yet also has strong colocalization evidence that higher expression correlates with higher AD risk. One possibility is that impaired function of APH1B due to the missense variant leads to upregulation of APH1B transcription. This interpretation would be consistent with evidence from both mice86 and humans87 that loss of APH1B and gamma-secretase function leads to AD. It is noteworthy, however, that recent experiments failed to find an effect of the T27I variant on gamma-secretase activity in HEK cells88.
Among our novel associations, TSPAN14 has a role in defining the localization of ADAM1090, another recently discovered AD gene that is a key component of the alpha-secretase complex and that could thus mediate AD risk via processing of amyloid precursor protein. However, ADAM10 also cleaves the microglia-associated protein TREM2 to generate its soluble ligand-binding domain91. Our fine-mapping showed that the risk SNP rs1870138 is also associated with higher risk for inflammatory bowel disease (IBD), an immune-mediated disease, and with higher monocyte count in UK Biobank participants. Since TSPAN14 is expressed more highly in immune cell types, including microglia, than in brain tissue, it is also plausible that AD risk is mediated by its effect on either immune cell count or activation. Recently proposed AD candidate genes supported by our analyses include RIN3, HS3ST1, and FCER1G. As noted above, FCER1G is a microglial master regulator81–83; RIN3 interacts with both BIN1 and CD2AP in the early endocytic pathway93; HS3ST1 is involved in cellular uptake of tau94 and was recently been associated with AD in an independent Norwegian sample62.
In summary, our study reports quantitative gene prioritization for 36 AD-associated regions, as well as AD-specific gene network scores beyond these loci. Our genetic findings highlight the presence of diverse mechanisms in AD pathogenesis and suggest candidate targets for therapeutic development.
Online Methods
GWAS on family history of AD
Sample QC, variant QC and imputation was performed on all UK Biobank (UKB) participants as described in Bycroft et al.95. After genotype imputation, 93,095,623 variants across 487,409 individuals were available for analysis. To exclude individuals of non-European ancestry, we extracted “White British” ancestry participants as described in Bycroft et al.95. These individuals self-reported their ethnic background as “British” and have similar genetic ancestry based on principal components (PC) analysis. To extract additional individuals of European ancestry, we followed a similar approach to Bycroft et al. and applied Aberrant96 on PCs 1v2, 3v4 and 5v6 across the individuals who self-reported as “Irish” or “Any other white background”. We identified first-degree relatives by applying KING97 v2.0 to 147,522 UKB participants who had at least one relative identified in Bycroft et al. (UKB Field 22021). For each first-degree relative pair, we prioritized AD cases and proxy-cases (see below) for inclusion, and otherwise excluded one of the pair at random. We also excluded variants with low imputation quality (INFO < 0.3) and/or those with minor allele frequencies below 0.0005, resulting in 25,647,815 variants available for analysis.
AD cases were extracted from UKB self-report (field 20002), ICD10 diagnoses (fields 41202 and 41204) and ICD10 cause of death (fields 40001 and 40002) data. UKB participants were asked whether they have a biological father, mother or sibling who suffered from Alzheimer’s disease/dementia (UKB fields 20107, 20110, and 20111, respectively). We extracted all participants with at least one affected relative as proxy-cases. Participants who answered “Do not know” or “Prefer not to answer” were excluded from analyses. All remaining individuals were denoted as controls.
There were 898 AD cases, 52,791 AD proxy cases and 355,900 controls in the combined white British and white non-British cohorts. For association analyses, we lumped the true and proxy-cases together (53,042 unique affected individuals) and used the linear-mixed model implemented in BOLT-LMM98.
AD meta-analysis
To enable meta-analysis combining the UKB cohorts with external case-control studies, we first transformed the AD proxy BOLT-LMM summary statistics from the linear scale to a 1/0 log odds ratio:
with standard error:
where βLMM and seLMM are the SNP effect sizes and standard errors respectively from BOLT-LMM, and f is the fraction of cases in the sample99. Since the affected individuals in our analysis include both true and proxy-cases, we then multiplied the transformed logORs and standard errors by 1.897 to approximate the logORs obtained from a true case/control study1.
We combined the transformed UKB white British cohort, UKB white non-British cohort and the Stage 1 summary statistics from Kunkle et al. using a fixed-effects (inverse variance weighted) meta-analysis across 10,687,126 overlapping variants. For display purposes (Supplementary Table 8), we used CrossMap100 to convert variant positions from GRCh37 to GRCh38.
Replication
To assess replication of our discovered signals, we downloaded the publicly available summary statistics for the Gr@ace study of AD17 from the GWAS catalog, and for the FinnGen GWAS of phenotypes “Alzheimer’s disease, wide definition” and “Alzheimer’s disease (Late onset)” from FinnGen release 3. We extracted summary results for our lead SNPs, or a partner in strong LD when the lead SNP was not found, and present these in Supplementary Table 3. We estimated power to detect our four novel loci at nominal significance (P < 0.05) using the genetic power calculator (zzz.bwh.harvard.edu/gpc/cc2.html) with the genotype relative risks estimated from our meta-analysis, and the allele frequency and case/control count from the GWAS study of interest (Gr@ace or FinnGen), and assuming a disease prevalence of 5%. We performed an inverse variance-weighted meta-analysis of all four studies (Kunkle et al., UKB, Gr@ace, and FinnGen “AD wide”), similar to our discovery meta-analysis.
Conditional analysis and statistical fine-mapping
To run GCTA, we prepared plink input files with genotypes from 10,000 randomly sampled UKB individuals at variants within +/- 5 Mb from each lead SNP. We excluded variants with INFO < 0.85, or which had a P-value from Cochran’s Q test for study heterogeneity < 0.001. We also excluded variants with minor allele frequency (MAF) in UKB below 0.1%, as LD estimates are unreliable at low allele counts. We selected these thresholds after manual examination of fine-mapping results, where we found that more lenient cutoffs led either FINEMAP or PAINTOR to select implausible causal variants at a few loci, such as pairs of very weakly associated rare variants to explain a common variant signal. We ran GCTA (v1.92.1) --cojo-slct with a threshold of P < 10-5 to identify secondary signals at each locus, and then retained only loci with a lead P-value below 5 x 10-8. For the HLA locus, we used a GCTA P-value threshold of 5 x 10-8. We also retained the loci TSPOAP1, IKZF1, and TMEM163 since they had P < 5 x 10-8 in an earlier version of our analysis. We excluded the APOE locus from conditional analysis and fine-mapping because the strength of association in the region would require a more perfect LD panel match to avoid spurious signals.
We then ran FINEMAP (v1.3) at each locus, with --n-causal-snps given as the number of independent SNPs determined by GCTA. For FINEMAP, we excluded variants with MAF < 0.2%. For loci with multiple signals, we also used GCTA --cojo-cond to condition on each independent SNP identified in the previous analysis, and retained SNPs within 500 kb of any conditionally independent SNP at the locus. To fine-map based on GCTA conditional signals, we converted beta and standard error values to approximate Bayes Factors (BF)101 using a prior of W = 0.1 (in Wakefield notation), and used the WTCCC single-causal variant method48, probability = SNP BF / sum(all SNP BFs).
To assess sensitivity of the results to our choice of reference panel, we applied the same steps (GCTA + FINEMAP) to summary statistics from the Kunkle et al. sub-study, which are described further in the Supplementary Note.
Colocalization with eQTLs
For eQTL colocalization, we downloaded summary statistics (see URLs) and determined eQTL genes at FDR 5% for each dataset in a uniform manner, first using Bonferroni correction of lead SNP nominal P values based on the number of variants tested for the gene, and using the Benjamini-Hochberg method to compute FDR. QTL calling for primary microglia was performed with RASQUAL102 with the --no-posterior-update option. For datasets in GRCh38 coordinates, we first used CrossMap100 to convert back to GRCh37 coordinates to match variants between eQTL and GWAS. We used the coloc package19 with default priors to perform colocalization tests between GWAS and eQTLs having lead variants within 500 kb of each other, and passed to coloc all variants within 200 kb of each lead variant. We also ran coloc using P-values for each conditionally independent GWAS signal, obtained with GCTA as described above.
Functional annotations
We used the Ensembl VEP online Web tool (www.ensembl.org/vep)55 to predict variant consequences, and to add selected annotations (Supplementary Table 7). We downloaded bed files based on imputed data for Roadmap Epigenomics DNase and 25-state genome segmentations for 127 epigenomes54. We grouped these into groups “all”, “brain” (epigenomes 7, 9, 10, 53, 54, 67, 68, 69, 70, 71, 72, 73, 74, 81, 82, 125), and “blood & immune” (epigenomes 33, 34, 37, 38, 39, 40, 41, 42, 43, 44, 45, 47, 48, 62, 29, 30, 31, 32, 35, 36, 46, 50, 51, 116). We considered 9 genome segmentation states to represent enhancers: TxReg, TxEnh5, TxEnh3, TxEnhW, EnhA1, EnhA2, EnhAF, EnhW1, EnhW2. We used bedtools103 to determine overlaps, and counted the number of overlaps for each variant with peaks in the above groups. We downloaded FANTOM5104 permissive enhancer annotations from fantom.gsc.riken.jp/5/data/. We downloaded pre-computed SpliceAI scores58 for variants within genes from github.com/Illumina/SpliceAI. We merged filtered whole-genome and exome scores together, and for each AD variant annotated the maximum score across splice donor gain, donor loss, acceptor gain, acceptor loss. We used DeepSEA57 (deepsea.princeton.edu) to annotate variants selected for functional fine-mapping with DeepSEA’s “functional significance” score. BigWig files with PhastCons, PhyloP and GERP RS scores were downloaded from UCSC. We downloaded microglial ATAC-seq based on the study by Gosselin et al.52, aligned reads to GRCh37 with bwa 0.7.15105, and called multisample peaks across all 15 datasets using MACS2106. We prepared bigWig files from alignments by using bedtools genomecov, followed by bedGraphToBigWig. To visualise microglia ATAC-seq tracks we adapted code from wiggleplotr107.
Annotation-based fine-mapping
For fine-mapping with PAINTOR, we first restricted the number of considered variants for computational feasibility, by selecting 3,207 variants which had (i) FINEMAP probability ≥ 0.01% based on the GCTA-identified number of causal variants at the locus, or (ii) had FINEMAP probability ≥ 1% when run with either 1 or 2 causal variants, or (iii) were among the top 20 variants at the locus by FINEMAP probability. We defined binary annotations for input to PAINTOR based on the features described above, thresholding certain scores at multiple levels (e.g. CADD ≥ 5, 10, 20). For Roadmap annotations, we included a category based on whether a variant was in a peak or enhancer in ≥ 10 epigenomes. We ran PAINTOR v3.1 once for each of the 43 annotations (Fig. 2 and Supplementary Table 7), allowing two causal variants per locus.
We built a multi-annotation model using forward stepwise selection. We selected the best annotation by log-likelihood (LLK), Blood & immune DNase, and then ran PAINTOR again for each combination of this annotation and the 42 remaining annotations. We added a top-ranking annotation at each iteration until the model LLK improvement was less than 1. This occurred at iteration 4, and so we kept the first three annotations in the combined model. We computed the mean causal probability for each SNP as the mean of the three fine-mapping methods at loci with two or more signals, or as the mean of the FINEMAP and PAINTOR probabilities for loci with one signal, since FINEMAP gives approximately the same results as WTCCC fine-mapping for a single causal variant.
Network analysis
For network analysis, we created a gene interaction network based on selecting all edges between protein-coding genes from systematic studies (>1,000 interactions) in the IntAct108 and BioGRID databases109, and edges from STRING v10.5110 with edge score > 0.75. This combined network included 18,055 genes and 540,421 edges. We identified 28 top candidate genes across AD loci (Supplementary Table 9) to use as seed genes, and assigned weight to these as the -log10(P value) of the locus lead SNP. We added four genes from the literature (MAPT, PSEN1, PSEN2, ABI3), with a weight (equivalent -log10(P)) of 15. For three loci, the nearest gene was not present in the network (ECHDC3, TMEM163, SCIMP). For each locus, we used all seed genes as input except those at the same locus, and propagated information through the network with the personalized PageRank algorithm111, included in the igraph R package112. Since a gene’s resulting PageRank was highly correlated with its node degree, we compared the PageRank of each gene to the distribution of PageRanks obtained for the same gene in 1,000 iterations of network propagation, where the same number of seed genes were randomly selected. We computed the percentile of a gene’s true PageRank relative to the 1,000 network propagations with randomized inputs. Although the distribution of PageRank percentile was fairly uniform, we further normalised this to a uniform distribution across genes, so that a Pagerank percentile of 90% indicates that a gene’s PageRank relative to permutations is above that of 90% of genes. To determine gene set enrichment, we used the top 1,000 genes by network rank as input to gProfiler113 with default settings, with the set of all genes ranked by the network as a background set. To determine enrichment of low P-value AD SNPs near genes in specific bins of PageRank percentile (Fig. 5a), we first determined for each gene the minimum SNP P value within 10 kb of the gene’s footprint. We excluded genes within 1 Mb of APOE. Then, for genes in each PageRank percentile bin, we used Fisher’s exact test to determine the odds ratio for a gene in that bin (relative to genes with PageRank percentile <50%) to have a minimum SNP P value in the given bin (relative to genes with minimum SNP P > 0.01).
Gene expression
Gene expression values for all tissues were determined in units of transcripts per million (TPM). Both GTEx v8 and the eQTL catalogue provide tables of the median TPM expression across samples for each tissue and gene. For primary microglia, we obtained a table of read counts per gene, computed using FeatureCounts 1.5.3 as described14, from which we computed median TPM. For use in gene prioritization and enrichment analyses, we first selected four GTEx brain tissues (cortex, hippocampus, substantia nigra, cerebellum) to avoid over-representing brain, and then the remaining 41 GTEx tissues, as well primary microglia and in-house expression data from iPSC-derived microglia, iPSC-derived NGN2 cortical neurons, and iPSC-derived neurons from growth factor differentiation. For each gene, we determined the TPM expression relative to all tissues/cell types.
Single-cell gene expression data were obtained from the Allen Brain Institute as a gene-by-cell counts table, based on Smart-seq profiling of six human brain cortical areas77. For each cell type “subclass” as defined in the metadata (but excluding VLMC for having too few cells, and the outlier subclass labelled “exclude”), counts were summed across cells and then normalised to TPM within each subclass. We determined each gene’s TPM expression in each subclass relative to all 18 subclasses.
Genome-wide enrichment
We determined the GRCh37 coordinates of 18,055 genes present in the gene network using the R package annotables 0.1.91. For each AD GWAS SNP, excluding the APOE region (chr19:44-47 Mb), we determined the nearest gene. We defined annotation inputs for fgwas labelling a SNP 1 if it was nearest to a gene with network score in a given percentile bin (50-60, 60-70, 70-80, 80-90, 90-95, >95) and 0 otherwise. We ran fgwas78 (-cc) with all network annotations as input, so that enrichments are with respect to SNPs nearest to genes with network score < 50th percentile. For every bulk gene expression dataset selected above, we defined an annotation for SNPs nearest genes with relative expression above the 80th (or 90th) percentile, and similarly for cell types from single-cell gene expression. We ran fgwas once for each expression annotation to determine enrichment of SNPs near high-expression genes relative to remaining genes (Supplementary Table 12).
Gene prioritization
Five predictors were used for gene prioritization.
The coding score is the sum of the mean fine-mapping probability for missense or LoF variants in a gene.
The expression score is the sum of component scores for bulk and single-cell microglial expression, and rewards genes with expression percentile above the 50th:
Genes without measured expression in a given dataset (bulk, single-cell) are assigned an exprScore of zero for that dataset.
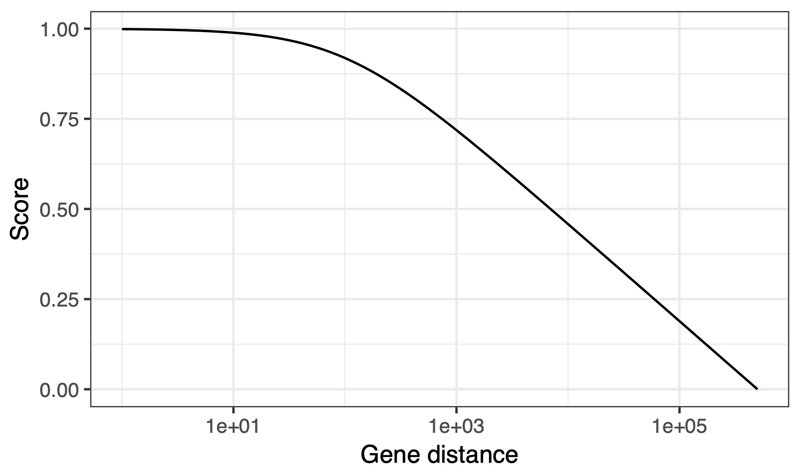
Recent evidence from both eQTLs114 and metabolite GWAS115 suggests that genomic distance from the association peak is a strong predictor of causal target genes. The distance score is defined to give reasonable scores over the main range of interest of 0 - 200 kb:
where x is the minimum distance from the gene’s footprint to the region defined by independent lead SNPs at a GWAS locus, maxDist is 500,000 and distBias is 100 (Extended Data Fig. 6).
The coloc score is defined based on the maximum value across QTL datasets of the “H4” hypothesis probability, and rewards colocalisation probabilities above 0.9:
The network score is determined based on the pagerank percentile for a gene relative to permutations:
Genes not present in the network are assigned a networkScore of zero.
The total score for a gene is the sum of the above five scores.
To give appropriate weight to each component, we trained lasso-regularized logistic regression models with cross-validation using glmnet116. As input we used all protein-coding genes within 500 kb of our AD GWAS peaks, excluding the APOE region due to lack of colocalisation information, and excluding genes not present in the network. For the distance model, genes within 10 kb of each GWAS peak (40 genes) were set as positives, genes 10-100 kb were excluded, and genes >100 kb (394 genes) were set as negatives. These were predicted using the four non-distance predictors. For the network model, genes with pagerank percentile >80% (143 genes) were set as positives, those with pagerank percentile 50-80% were excluded, the 230 other genes were set as negatives, and these were predicted using the four non-network predictors. In each case, we selected the model that minimized mean squared error (MSE), shown in Supplementary Table 14, and used those parameters to generate predictions (in the range 0-1) for all genes at the AD loci. We defined the model score for a gene as the average prediction from the two models. To determine the importance of the predictors to each model (apart from looking at regression coefficients) we ran glmnet models excluding each predictor in turn. If the MSE was lower with a predictor excluded then we removed it from the final model. For each model, we compared the MSE when using our quantitative predictors as defined above, or using categorical predictors by thresholding the predictors into 2-4 bins. For both models, the quantitative predictors gave improved MSE. We also examined models that included as predictors expression scores from astrocytes (based on the single-cell data) and from brain hippocampus (based on the GTEx data), but for both models this resulted in higher MSE and the regularization set the coefficients to zero.
Extended Data
Extended Data Fig. 1. Association of AD loci in discovery + replication (“global”) meta-analysis.
Association of AD loci in discovery + replication dataset (“global”) meta-analysis. For most loci, association significance is increased in the global meta-analysis (blue bars) relative to the discovery analysis (grey bars). The dashed vertical line shows P = 5 x 10-8. P-values were computed by inverse variance weighted meta-analysis, and bars show the -log10(P) for the SNP with minimum P value at the locus in either the discovery or global meta-analysis.
Extended Data Fig. 2. Comparison of fine-mapping in the meta-analysis vs. Kunkle et al.
Comparison of fine-mapping in the meta-analysis vs. Kunkle et al. Scatterplots showing, for each locus, SNP probabilities from FINEMAP applied to either the Kunkle et al. + UK Biobank meta-analysis (x-axis), or to only Kunkle et al. The number of causal variants at each locus was set to the number detected by GCTA in the meta-analysis. For most of the 36 loci, SNP probabilities are well correlated. For a few loci that are well powered in Kunkle et al., this is not the case, namely ABCA7, EPHA1, ECHDC3, and HLA. For these loci, fine-mapping results should be interpreted with caution. Six other loci are not well correlated (ADAMTS4, APH1B, IKZF1, PLCG2, TMEM163, and VKORC1), but these loci are poorly powered in Kunkle et al. (lead P values 2.1 x 10-6 to 2.1 x 10-3).
Extended Data Fig. 3. Network enrichment.
a, The Pagerank percentile of all genes (within 500 kb) at each AD GWAS locus containing a seed gene is shown, with seed genes highlighted in blue. b, A violin/boxplot shows that seed genes have a markedly higher network Pagerank percentile than remaining genes (P = 2.4 x 10-9, one-tailed Wilcoxon rank sum test). c, Log odds ratio enrichment of AD risk among SNPs nearest to genes with network Pagerank percentile in different bins, determined using fgwas (whiskers represent 95% confidence intervals).
Extended Data Fig. 4. Gene expression enrichments.
Expression enrichments for GTEx + microglia. Shown are the log odds ratio enrichments of AD risk among SNPs with relative gene expression in each tissue above the 80th (or 90th) percentile across tissues. Whiskers represent 95% confidence intervals determined by fgwas.
Extended Data Fig. 5. Colocalization scores.
a, Genes with maximum colocalization H4 probability >0.9 have higher Pagerank percentile (left boxplot) and higher total score (sum of the four non-coloc predictors, right boxplot) than do genes without colocalisation (<0.5). Genes with intermediate colocalisation evidence (bins 0.5 - 0.8 and 0.8 - 0.9) show little evidence of having higher scores by the other metrics. Based on this, we chose a maxColoc probability of 0.9 as the lower bound for our colocalization score. b, Boxplot of the total score (excluding coloc) for genes that have a colocalisation probability > 0.9 in at least one QTL dataset within each tissue group. The most significant difference is between totalScore for genes with microglial colocalizations vs. the genes with colocalization in “other” tissues (non-immune GTEx tissues), but the for a difference is weak (P = 0.041, Wilcoxon rank sum test). In all cases, boxplots show the 25th, median, and 75th percentile of the distribution, with whiskers extending to the largest (and smallest) value no further than 1.5 times the interquartile range from the boxplot hinge.
Extended Data Fig. 6. Gene distance score.
The distance score assigned to genes near an AD GWAS peak, which decreases approximately linearly (past a distance of 1 kb) with increasing log-scaled distance up to 500 kb.
Supplementary Material
Acknowledgements
This work was funded by Open Targets (OTAR037). We thank Jeff Barrett for guidance during initiation of the project, and Kaur Alasoo for early access to the eQTL catalogue. We thank Agustín Ruiz for support in using summary results from the Gr@ace study. We acknowledge the participants and investigators of FinnGen study, and of the UK Biobank. R.J.M.F. is supported by grants from the UK Multiple Sclerosis Society (MS 50), the Adelson Medical Research Foundation, and a core support grant from Wellcome and MRC to the Wellcome-MRC Cambridge Stem Cell Institute (203151/Z/16/Z). A.M.H.Y. is supported by a Wellcome Trust PhD for Clinicians fellowship.
Footnotes
Author Contributions
J.S. planned and conducted the analyses, and wrote the paper. J.Z.L. performed the GWAX and meta-analysis. S.C. and E.B. assisted with fine-mapping, variant and gene prioritization. I.B.-H. and P.B. performed and supervised gene network analysis. R.J.M.F. designed and A.M.H.Y. performed the isolation of human microglia from brain biopsies. N.K. performed microglia eQTL mapping. A.B., T.J., D.J.G., and K.E. conceived and supervised the study.
Competing interests
J.Z.L. was an employee of Biogen at the time of the study, and is now an employee of GSK. D.J.G. is an employee of Genomics Plc. T.J. is an employee of GSK. K.E. is an employee of BioMarin Pharmaceutical.
Data availability
Summary statistics from the meta-analysis are available through the NHGRI-EBI GWAS Catalog under accessions GCST90012877 and GCST90012878:
www.ebi.ac.uk/gwas/downloads/summary-statistics
eQTL Catalogue: www.ebi.ac.uk/eqtl
GTEx: www.gtexportal.org
Roadmap Epigenomics: www.roadmapepigenomics.org
DeepSEA: deepsea.princeton.edu
SpliceAI: github.com/Illumina/SpliceAI
FANTOM enhancers: fantom.gsc.riken.jp/5/data/
GERP: hgdownload.cse.ucsc.edu/gbdb/hg19/bbi/All_hg19_RS.bw
PhyloP: hgdownload.cse.ucsc.edu/goldenpath/hg19/phyloP100way
PhastCons: hgdownload.cse.ucsc.edu/goldenpath/hg19/phastCons100way
Brain eQTL meta-analysis summary statistics: www.synapse.org/#!Synapse:syn16984815
Primary microglia eQTL summary statistics, EGA Accession ID: EGAD00001005736
Primary microglia ATAC-seq, dbGaP Study Accession: phs001373.v1.p1
Allen Brain Institute: portal.brain-map.org/atlases-and-data/rnaseq
IntAct database: www.ebi.ac.uk/intact
BioGRID database: thebiogrid.org
STRING database: string-db.org
Code availability
Code for analyses described here can be found at github.com/jeremy37/AD_finemap.
References
- 1.Liu JZ, Erlich Y, Pickrell JK. Case-control association mapping by proxy using family history of disease. Nat Genet. 2017;49:325–331. doi: 10.1038/ng.3766. [DOI] [PubMed] [Google Scholar]
- 2.Marioni RE, et al. GWAS on family history of Alzheimer’s disease. Transl Psychiatry. 2018;8:99. doi: 10.1038/s41398-018-0150-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jansen IE, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer’s disease risk. Nat Genet. 2019;51:404–413. doi: 10.1038/s41588-018-0311-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Claussnitzer M, et al. FTO obesity variant circuitry and adipocyte browning in humans. N Engl J Med. 2015;373:895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sekar A, et al. Schizophrenia risk from complex variation of complement component 4. Nature. 2016;530:177–183. doi: 10.1038/nature16549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang H, et al. Fine-mapping inflammatory bowel disease loci to single-variant resolution. Nature. 2017;547:173–178. doi: 10.1038/nature22969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Malik M, et al. CD33 Alzheimer’s risk-altering polymorphism, CD33 expression, and exon 2 splicing. J Neurosci. 2013;33:13320–13325. doi: 10.1523/JNEUROSCI.1224-13.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guerreiro R, et al. TREM2 variants in Alzheimer’s disease. N Engl J Med. 2013;368:117–127. doi: 10.1056/NEJMoa1211851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sims R, et al. Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat Genet. 2017;49:1373–1384. doi: 10.1038/ng.3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kerimov N, et al. eQTL Catalogue: a compendium of uniformly processed human gene expression and splicing QTLs. bioRxiv. 2020 doi: 10.1101/2020.01.29.924266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.GTEx Consortium. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science. 2020;369:1318–1330. doi: 10.1126/science.aaz1776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kunkle BW, et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat Genet. 2019;51:414–430. doi: 10.1038/s41588-019-0358-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lambert JC, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat Genet. 2013;45:1452–1458. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Young AMH, et al. A map of transcriptional heterogeneity and regulatory variation in human microglia. bioRxiv. 2019 doi: 10.1101/2019.12.20.874099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung YY, et al. Identifying amyloid pathology–related cerebrospinal fluid biomarkers for Alzheimer’s disease in a multicohort study. Alzheimers Dement. 2015;1:339–348. doi: 10.1016/j.dadm.2015.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bulik-Sullivan BK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–295. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moreno-Grau S, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer’s disease and three causality networks: The GR@ACE project. Alzheimers Dement. 2019;15:1333–1347. doi: 10.1016/j.jalz.2019.06.4950. [DOI] [PubMed] [Google Scholar]
- 18.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–75. doi: 10.1038/ng.2213. S1-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sieberts SK, et al. Large eQTL meta-analysis reveals differing patterns between cerebral cortical and cerebellar brain regions. Sci Data. 2020;7:340. doi: 10.1038/s41597-020-00642-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ng B, et al. An xQTL map integrates the genetic architecture of the human brain’s transcriptome and epigenome. Nat Neurosci. 2017;20:1418–1426. doi: 10.1038/nn.4632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmiedel BJ, et al. Impact of genetic polymorphisms on human immune cell gene expression. Cell. 2018;175:1701–1715.e16. doi: 10.1016/j.cell.2018.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jaffe AE, et al. Developmental and genetic regulation of the human cortex transcriptome illuminate schizophrenia pathogenesis. Nat Neurosci. 2018;21:1117–1125. doi: 10.1038/s41593-018-0197-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buil A, et al. Gene-gene and gene-environment interactions detected by transcriptome sequence analysis in twins. Nat Genet. 2015;47:88–91. doi: 10.1038/ng.3162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fairfax BP, et al. Genetics of gene expression in primary immune cells identifies cell type–specific master regulators and roles of HLA alleles. Nat Genet. 2012;44:502–510. doi: 10.1038/ng.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fairfax BP, et al. Innate immune activity conditions the effect of regulatory variants upon monocyte gene expression. Science. 2014;343 doi: 10.1126/science.1246949. 1246949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naranbhai V, et al. Genomic modulators of gene expression in human neutrophils. Nat Commun. 2015;6:7545. doi: 10.1038/ncomms8545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen L, et al. Genetic drivers of epigenetic and transcriptional variation in human immune cells. Cell. 2016;167:1398–141.e24. doi: 10.1016/j.cell.2016.10.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alasoo K, et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gutierrez-Arcelus M, et al. Passive and active DNA methylation and the interplay with genetic variation in gene regulation. Elife. 2013;2:e00523. doi: 10.7554/eLife.00523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lappalainen T, et al. Transcriptome and genome sequencing uncovers functional variation in humans. Nature. 2013;501:506–511. doi: 10.1038/nature12531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kilpinen H, et al. Common genetic variation drives molecular heterogeneity in human iPSCs. Nature. 2017;546:370–375. doi: 10.1038/nature22403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nédélec Y, et al. Genetic ancestry and natural selection drive population differences in immune responses to pathogens. Cell. 2016;167:657–669.e21. doi: 10.1016/j.cell.2016.09.025. [DOI] [PubMed] [Google Scholar]
- 34.Quach H, et al. Genetic adaptation and Neandertal admixture shaped the immune system of human populations. Cell. 2016;167:643–656.e17. doi: 10.1016/j.cell.2016.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schwartzentruber J, et al. Molecular and functional variation in iPSC-derived sensory neurons. Nat Genet. 2018;50:54–61. doi: 10.1038/s41588-017-0005-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.van de Bunt M, et al. Transcript expression data from human islets links regulatory signals from genome-wide association studies for type 2 diabetes and glycemic traits to their downstream effectors. PLoS Genet. 2015;11:e1005694. doi: 10.1371/journal.pgen.1005694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Momozawa Y, et al. IBD risk loci are enriched in multigenic regulatory modules encompassing putative causative genes. Nat Commun. 2018;9:2427. doi: 10.1038/s41467-018-04365-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chun S, et al. Limited statistical evidence for shared genetic effects of eQTLs and autoimmune-disease-associated loci in three major immune-cell types. Nat Genet. 2017;49:600–605. doi: 10.1038/ng.3795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salazar SV, et al. Alzheimer’s disease risk factor Pyk2 mediates amyloid-β-induced synaptic dysfunction and loss. J Neurosci. 2019;39:758–772. doi: 10.1523/JNEUROSCI.1873-18.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raj T, et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat Genet. 2018;50:1584–1592. doi: 10.1038/s41588-018-0238-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Calafate S, Flavin W, Verstreken P, Moechars D. Loss of Bin1 promotes the propagation of Tau pathology. Cell Rep. 2016;17:931–940. doi: 10.1016/j.celrep.2016.09.063. [DOI] [PubMed] [Google Scholar]
- 42.Nott A, et al. Brain cell type–specific enhancer–promoter interactome maps and disease-risk association. Science. 2019;366:1134–1139. doi: 10.1126/science.aay0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rathore N, et al. Paired Immunoglobulin-like Type 2 Receptor Alpha G78R variant alters ligand binding and confers protection to Alzheimer’s disease. PLoS Genet. 2018;14:e1007427. doi: 10.1371/journal.pgen.1007427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chan G, et al. CD33 modulates TREM2: convergence of Alzheimer loci. Nat Neurosci. 2015;18:1556–1558. doi: 10.1038/nn.4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Raj T, et al. CD33: increased inclusion of exon 2 implicates the Ig V-set domain in Alzheimer’s disease susceptibility. Hum Mol Genet. 2014;23:2729–2736. doi: 10.1093/hmg/ddt666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jonsson T, et al. Variant of TREM2 associated with the risk of Alzheimer’s disease. N Engl J Med. 2013;368:107–116. doi: 10.1056/NEJMoa1211103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Claes C, et al. Human stem cell-derived monocytes and microglia-like cells reveal impaired amyloid plaque clearance upon heterozygous or homozygous loss of TREM2. Alzheimers Dement. 2019;15:453–464. doi: 10.1016/j.jalz.2018.09.006. [DOI] [PubMed] [Google Scholar]
- 48.Wellcome Trust Case Control Consortium et al. Bayesian refinement of association signals for 14 loci in 3 common diseases. Nat Genet. 2012;44:1294–1301. doi: 10.1038/ng.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benner C, et al. FINEMAP: efficient variable selection using summary data from genome-wide association studies. Bioinformatics. 2016;32:1493–1501. doi: 10.1093/bioinformatics/btw018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kichaev G, et al. Integrating functional data to prioritize causal variants in statistical fine-mapping studies. PLoS Genet. 2014;10:e1004722. doi: 10.1371/journal.pgen.1004722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Benner C, et al. Prospects of fine-mapping trait-associated genomic regions by using summary statistics from genome-wide association studies. Am J Hum Genet. 2017;101:539–551. doi: 10.1016/j.ajhg.2017.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gosselin D, et al. An environment-dependent transcriptional network specifies human microglia identity. Science. 2017;356 doi: 10.1126/science.aal3222. eaal3222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Alasoo K, et al. Shared genetic effects on chromatin and gene expression indicate a role for enhancer priming in immune response. Nat Genet. 2018;50:424–431. doi: 10.1038/s41588-018-0046-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Roadmap Epigenomics Consortium et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning-based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jaganathan K, et al. Predicting splicing from primary sequence with deep learning. Cell. 2019;176:535–548.e24. doi: 10.1016/j.cell.2018.12.015. [DOI] [PubMed] [Google Scholar]
- 59.Steinberg S, et al. Loss-of-function variants in ABCA7 confer risk of Alzheimer’s disease. Nat Genet. 2015;47:445–447. doi: 10.1038/ng.3246. [DOI] [PubMed] [Google Scholar]
- 60.De Roeck A, et al. An intronic VNTR affects splicing of ABCA7 and increases risk of Alzheimer’s disease. Acta Neuropathol. 2018;135:827–837. doi: 10.1007/s00401-018-1841-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernstein AI, et al. 5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer’s disease modulate Tau-induced neurotoxicity. Hum Mol Genet. 2016;25:2437–2450. doi: 10.1093/hmg/ddw109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Witoelar A, et al. Meta-analysis of Alzheimer’s disease on 9,751 samples from Norway and IGAP study identifies four risk loci. Sci Rep. 2018;8:18088. doi: 10.1038/s41598-018-36429-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jun GR, et al. Transethnic genome-wide scan identifies novel Alzheimer’s disease loci. Alzheimers Dement. 2017;13:727–738. doi: 10.1016/j.jalz.2016.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Andersen OM, Rudolph I-M, Willnow TE. Risk factor SORL1: from genetic association to functional validation in Alzheimer’s disease. Acta Neuropathol. 2016;132:653–665. doi: 10.1007/s00401-016-1615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sassi C, et al. Influence of coding variability in APP-Aβ metabolism genes in sporadic Alzheimer’s disease. PLoS One. 2016;11:e0150079. doi: 10.1371/journal.pone.0150079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lu Q, et al. Systematic tissue-specific functional annotation of the human genome highlights immune-related DNA elements for late-onset Alzheimer’s disease. PLoS Genet. 2017;13:e1006933. doi: 10.1371/journal.pgen.1006933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ghanbari M, et al. A functional variant in the miR‐142 promoter modulating its expression and conferring risk of Alzheimer disease. Hum Mutat. 2019;40:2131–2145. doi: 10.1002/humu.23872. [DOI] [PubMed] [Google Scholar]
- 68.Chung C-M, et al. Fine-mapping angiotensin-converting enzyme gene: separate QTLs identified for hypertension and for ACE activity. PLoS One. 2013;8:e56119. doi: 10.1371/journal.pone.0056119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Nylocks KM, et al. An angiotensin-converting enzyme (ACE) polymorphism may mitigate the effects of angiotensin-pathway medications on posttraumatic stress symptoms. Am J Med Genet B Neuropsychiatr Genet. 2015;168B:307–315. doi: 10.1002/ajmg.b.32313. [DOI] [PubMed] [Google Scholar]
- 70.Kamboh MI, et al. Genome-wide association study of Alzheimer’s disease. Transl Psychiatry. 2012;2:e117. doi: 10.1038/tp.2012.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Canela-Xandri O, Rawlik K, Tenesa A. An atlas of genetic associations in UK Biobank. Nat Genet. 2018;50:1593–1599. doi: 10.1038/s41588-018-0248-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Liu JZ, et al. Association analyses identify 38 susceptibility loci for inflammatory bowel disease and highlight shared genetic risk across populations. Nat Genet. 2015;47:979–986. doi: 10.1038/ng.3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lanoiselée H-M, et al. APP, PSEN1, and PSEN2 mutations in early-onset Alzheimer disease: A genetic screening study of familial and sporadic cases. PLoS Med. 2017;14:e1002270. doi: 10.1371/journal.pmed.1002270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fang H, et al. A genetics-led approach defines the drug target landscape of 30 immune-related traits. Nat Genet. 2019;51:1082–1091. doi: 10.1038/s41588-019-0456-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Amin L, Harris DA. Aβ receptors specifically recognize molecular features displayed by fibril ends and neurotoxic oligomers. bioRxiv. 2019 doi: 10.1101/822361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Nordestgaard LT, Tybjærg-Hansen A, Nordestgaard BG, Frikke-Schmidt R. Loss-of-function mutation in ABCA1 and risk of Alzheimer’s disease and cerebrovascular disease. Alzheimers Dement. 2015;11:1430–1438. doi: 10.1016/j.jalz.2015.04.006. [DOI] [PubMed] [Google Scholar]
- 77.Hodge RD, et al. Conserved cell types with divergent features in human versus mouse cortex. Nature. 2019;573:61–68. doi: 10.1038/s41586-019-1506-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pickrell JK. Joint analysis of functional genomic data and genome-wide association studies of 18 human traits. Am JHum Genet. 2014;94:559–573. doi: 10.1016/j.ajhg.2014.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Bakken TE, et al. Evolution of cellular diversity in primary motor cortex of human, marmoset monkey, and mouse. bioRxiv. 2020 doi: 10.1101/2020.03.31.016972. [DOI] [Google Scholar]
- 80.Chang D, et al. A meta-analysis of genome-wide association studies identifies 17 new Parkinson’s disease risk loci. Nat Genet. 2017;49:1511–1516. doi: 10.1038/ng.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mukherjee S, Klaus C, Pricop-Jeckstadt M, Miller JA, Struebing FL. A microglial signature directing human aging and neurodegeneration-related gene networks. Front Neurosci. 2019;13:2. doi: 10.3389/fnins.2019.00002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhang B, et al. Integrated systems approach identifies genetic nodes and networks in late-onset Alzheimer’s disease. Cell. 2013;153:707–720. doi: 10.1016/j.cell.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Patel KR, et al. Single cell-type integrative network modeling identified novel microglial-specific targets for the phagosome in Alzheimer’s disease. bioRxiv. 2020 doi: 10.1101/2020.06.09.143529. [DOI] [Google Scholar]
- 84.Novikova G, et al. Integration of Alzheimer’s disease genetics and myeloid genomics reveals novel disease risk mechanisms. bioRxiv. 2019 doi: 10.1101/694281. [DOI] [Google Scholar]
- 85.Battle A, et al. Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals. Genome Res. 2014;24:14–24. doi: 10.1101/gr.155192.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Biundo F, Ishiwari K, Del Prete D, D’Adamio L. Deletion of the γ-secretase subunits Aph1B/C impairs memory and worsens the deficits of knock-in mice modeling the Alzheimer-like familial Danish dementia. Oncotarget. 2016;7:11923–11944. doi: 10.18632/oncotarget.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nicolas G, et al. Somatic variants in autosomal dominant genes are a rare cause of sporadic Alzheimer’s disease. Alzheimers Dement. 2018;14:1632–1639. doi: 10.1016/j.jalz.2018.06.3056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zhang X, et al. Negative evidence for a role of APH1B T27I variant in Alzheimer’s disease. Hum Mol Genet. 2020;29:955–966. doi: 10.1093/hmg/ddaa017. [DOI] [PubMed] [Google Scholar]
- 89.Acx H, et al. Inactivation of γ-secretases leads to accumulation of substrates and non-Alzheimer neurodegeneration. EMBO Mol Med. 2017;9:1088–1099. doi: 10.15252/emmm.201707561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Matthews AL, et al. Regulation of leukocytes by TspanC8 tetraspanins and the ‘molecular scissor’ ADAM10. Front Immunol. 2018;9:1451. doi: 10.3389/fimmu.2018.01451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schlepckow K, et al. An Alzheimer‐associated TREM2 variant occurs at the ADAM cleavage site and affects shedding and phagocytic function. EMBO Mol Med. 2017;9:1356–1365. doi: 10.15252/emmm.201707672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Ohkura T, et al. Spred2 regulates high fat diet-induced adipose tissue inflammation, and metabolic abnormalities in mice. Front Immunol. 2019;10:17. doi: 10.3389/fimmu.2019.00017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Juul Rasmussen I, Tybjærg-Hansen A, Rasmussen KL, Nordestgaard BG, Frikke-Schmidt R. Blood-brain barrier transcytosis genes, risk of dementia and stroke: a prospective cohort study of 74,754 individuals. Eur J Epidemiol. 2019;34:579–590. doi: 10.1007/s10654-019-00498-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Zhao J, et al. Rare 3-O-sulfation of heparan sulfate enhances Tau interaction and cellular uptake. Angew Chem Int Ed Engl. 2020;59:1818–1827. doi: 10.1002/anie.201913029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bycroft C, et al. The UK Biobank resource with deep phenotyping and genomic data. Nature. 2018;562:203–209. doi: 10.1038/s41586-018-0579-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Bellenguez C, et al. A robust clustering algorithm for identifying problematic samples in genome-wide association studies. Bioinformatics. 2012;28:134–135. doi: 10.1093/bioinformatics/btr599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–2873. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Loh P-R, et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat Genet. 2015;47:284–290. doi: 10.1038/ng.3190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Pirinen M, Donnelly P, Spencer CCA. Efficient computation with a linear mixed model on large-scale data sets with applications to genetic studies. Ann Applied Stat. 2013;7:369–390. [Google Scholar]
- 100.Zhao H, et al. CrossMap: a versatile tool for coordinate conversion between genome assemblies. Bioinformatics. 2014;30:1006–1007. doi: 10.1093/bioinformatics/btt730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wakefield J. Bayes factors for genome-wide association studies: comparison with P-values. Genet Epidemiol. 2009;33:79–86. doi: 10.1002/gepi.20359. [DOI] [PubMed] [Google Scholar]
- 102.Kumasaka N, Knights AJ, Gaffney DJ. Fine-mapping cellular QTLs with RASQUAL and ATAC-seq. Nat Genet. 2016;48:206–213. doi: 10.1038/ng.3467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Andersson R, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Alasoo K. wiggleplotr: Make read coverage plots from BigWig files. R package version 1101. 2019 [Google Scholar]
- 108.Orchard S, et al. The MIntAct project—IntAct as a common curation platform for 11 molecular interaction databases. Nucleic Acids Res. 2014;42:D358–D363. doi: 10.1093/nar/gkt1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Chatr-Aryamontri A, et al. The BioGRID interaction database: 2017 update. Nucleic Acids Res. 2017;45:D369–D379. doi: 10.1093/nar/gkw1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Szklarczyk D, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fogaras D, Rácz B, Csalogány K, Sarlós T. Towards scaling fully personalized PageRank: algorithms, lower bounds, and experiments. Internet Mathematics. 2005;2:333–358. [Google Scholar]
- 112.Csardi G, Nepusz T. The igraph software package for complex network research. Inter Journal, Complex Systems. 2006;1695:1–9. Others. [Google Scholar]
- 113.Raudvere U, et al. g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update) Nucleic Acids Res. 2019;47:W191–W198. doi: 10.1093/nar/gkz369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Barbeira AN, et al. Widespread dose-dependent effects of RNA expression and splicing on complex diseases and traits. bioRxiv. 2019 doi: 10.1101/814350. [DOI] [Google Scholar]
- 115.Stacey D, et al. ProGeM: a framework for the prioritization of candidate causal genes at molecular quantitative trait loci. Nucleic Acids Res. 2019;47:e3. doi: 10.1093/nar/gky837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Summary statistics from the meta-analysis are available through the NHGRI-EBI GWAS Catalog under accessions GCST90012877 and GCST90012878:
www.ebi.ac.uk/gwas/downloads/summary-statistics
eQTL Catalogue: www.ebi.ac.uk/eqtl
GTEx: www.gtexportal.org
Roadmap Epigenomics: www.roadmapepigenomics.org
DeepSEA: deepsea.princeton.edu
SpliceAI: github.com/Illumina/SpliceAI
FANTOM enhancers: fantom.gsc.riken.jp/5/data/
GERP: hgdownload.cse.ucsc.edu/gbdb/hg19/bbi/All_hg19_RS.bw
PhyloP: hgdownload.cse.ucsc.edu/goldenpath/hg19/phyloP100way
PhastCons: hgdownload.cse.ucsc.edu/goldenpath/hg19/phastCons100way
Brain eQTL meta-analysis summary statistics: www.synapse.org/#!Synapse:syn16984815
Primary microglia eQTL summary statistics, EGA Accession ID: EGAD00001005736
Primary microglia ATAC-seq, dbGaP Study Accession: phs001373.v1.p1
Allen Brain Institute: portal.brain-map.org/atlases-and-data/rnaseq
IntAct database: www.ebi.ac.uk/intact
BioGRID database: thebiogrid.org
STRING database: string-db.org
Code for analyses described here can be found at github.com/jeremy37/AD_finemap.