Abstract
Amplification of MYCN is the driving oncogene in a subset of high-risk neuroblastoma. The MYCN protein and the Aurora-A kinase form a complex during S phase that stabilizes MYCN. Here we show that MYCN activates Aurora-A on chromatin, which phosphorylates histone H3 at serine 10 in S phase, promotes the deposition of histone H3.3 and suppresses R-loop formation. Inhibition of Aurora-A induces transcription-replication conflicts and activates the Ataxia telangiectasia and Rad3 related (ATR) kinase, which limits double-strand break accumulation upon Aurora-A inhibition. Combined inhibition of Aurora-A and ATR induces rampant tumor-specific apoptosis and tumor regression in mouse models of neuroblastoma, leading to permanent eradication in a subset of mice. The therapeutic efficacy is due to both tumor cell-intrinsic and immune cell-mediated mechanisms. We propose that targeting the ability of Aurora-A to resolve transcription-replication conflicts is an effective therapy for MYCN-driven neuroblastoma (141 words).
Introduction
Deregulated expression of one of the three members of the MYC family of oncogenes is observed in the majority of all human tumors and can drive tumorigenesis in many entities1. Tumors driven by multiple oncogenes depend on elevated MYC expression, arguing that strategies that target MYC function may have a large therapeutic impact2. Of the three family members, the neuronal paralogue, MYCN, has been implicated in the genesis of a broad range of predominantly neuroendocrine and pediatric tumors3. A paradigm example is neuroblastoma, in which amplification of MYCN drives the development of a subset of high-risk tumors with poor prognosis, establishing an urgent need for therapeutic strategies that target MYCN function.
MYC proteins are transcription factors that bind to the vast majority of all active promoters and many enhancers, with promoter occupancy paralleling that by RNA polymerase II (RNAPII). Consistent with these observations, they exert widespread effects on both the function and the composition of the basal transcription machinery and can globally release RNAPII from core promoters4–9. In marked contrast, the effects of MYC proteins on gene expression are typically weak and the expression of most MYC-bound genes is not detectably altered. These observations raise the possibility that MYC proteins have critical oncogenic functions that are unrelated to their effects on gene expression10.
We have previously identified a complex of MYCN with the Aurora-A kinase and shown that complex formation stabilizes MYCN in neuroblastoma, since Aurora-A binds immediately adjacent to a major phosphodegron of MYCN and antagonizes recognition of the degron by the FBXW7 ubiquitin ligase11. As consequence, a class of Aurora-A ligands that distort the kinase domain and disrupt the Aurora-A/MYCN complex destabilize MYCN12–14. Conversely, MYCN stabilizes an active conformation of Aurora-A that is capable of substrate recognition, hence binding of MYCN, like that of TPX2, induces Aurora-A kinase activity14. During the cell cycle, MYCN predominantly forms a complex with Aurora-A during S phase and antagonizes the binding of MYCN to co-factors that are required for MYCN-dependent transcriptional activation15. Furthermore, stabilization of MYCN favors promoter-proximal transcription termination via the recruitment of mRNA de-capping factors8. Collectively, the data suggest that complex formation with Aurora-A has a critical role during S phase to limit MYCN-dependent transcription elongation and co-ordinate it with DNA replication15. Here we have clarified the mechanisms underlying these observations and show that they can be exploited for an effective therapy of MYCN-driven neuroblastoma.
Results
Aurora-A binds together with MYCN to chromatin in S phase
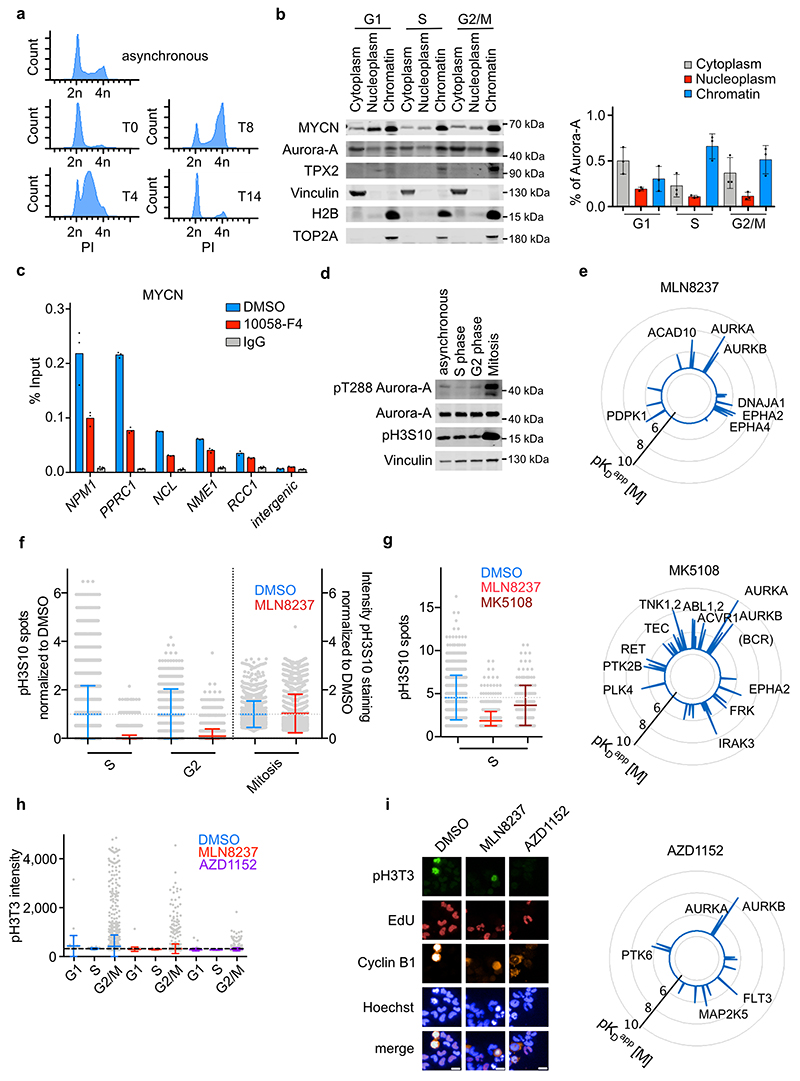
In neuroblastoma cells, MYCN complexes with Aurora-A during S phase15. To determine the subcellular localization of MYCN/Aurora-A complexes, we fractionated extracts of MYCN- amplified IMR-5 neuroblastoma cells that had been synchronized by double-thymidine block and released for 4 h into S, 8 h into G2/M or 14 h into G1 phase (Extended Data Figure 1a). These experiments showed that approximately 70% of the cellular pool of Aurora-A was associated with chromatin during S phase, although expression of the known mitotic chromatin anchor of Aurora-A, TPX2, was low in S compared to G2/M phase (Extended Data Figure 1b)16. Incubation of S phase-synchronized cells for 4 h with 10058-F4, which disrupts heterodimers of MYC or MYCN with MAX6,17,18, reduced chromatin association of both MYCN and Aurora-A by 50%, arguing that MYCN stabilizes a significant fraction of the cellular pool of Aurora-A on chromatin (Figure 1a, Extended Data Figure 1c). Notably, MYCN that was released from chromatin by 10058-F4 did not accumulate in the nucleoplasm, since association with MAX protects MYC from ubiquitin-mediated degradation19.
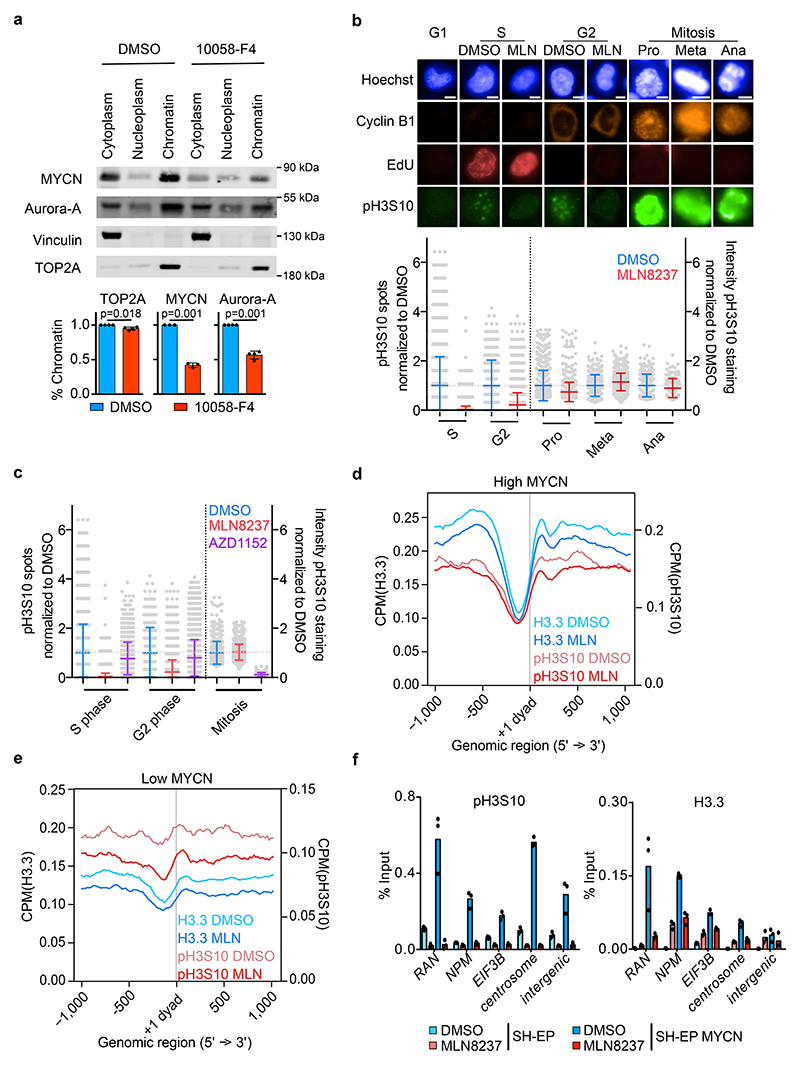
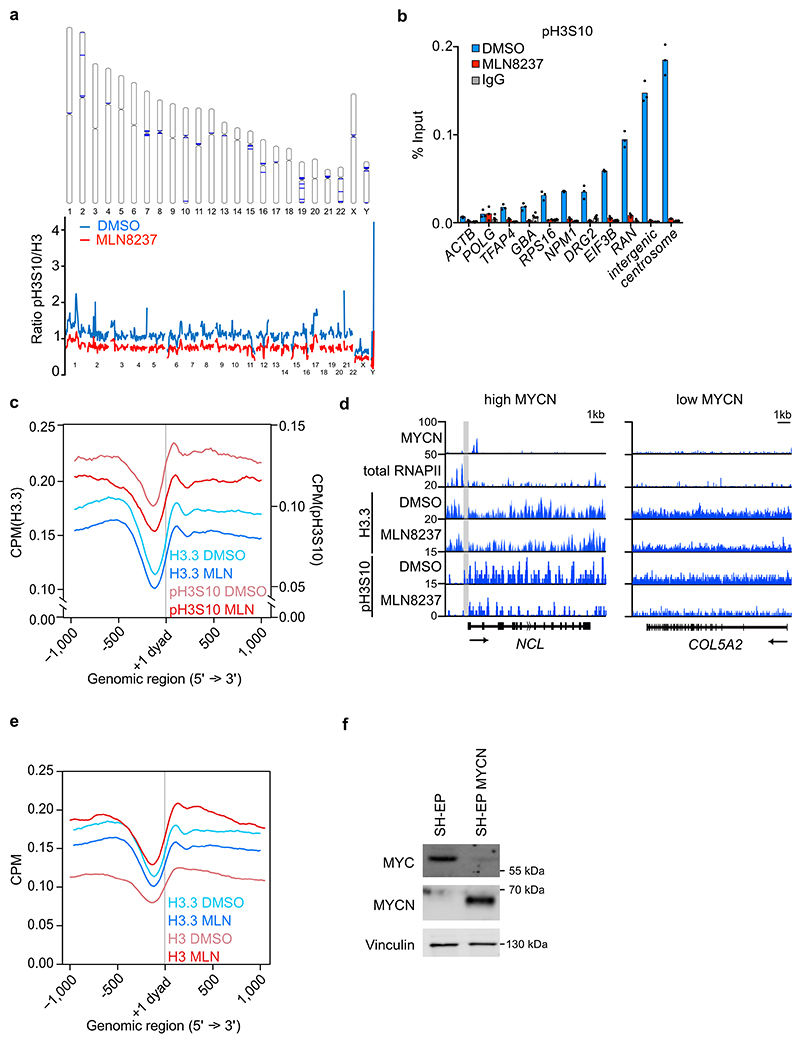
Figure 1. Aurora-A controls histone H3 phosphorylation in S phase.
a. (Top): Immunoblots of indicated proteins from S phase-synchronized IMR-5 cells that were treated for 4 h with 100 μM 10058-F4 or DMSO. (Bottom): Quantitation of relative levels of chromatin-bound proteins. Shown is the mean ± standard deviation (S.D.). P-values were calculated using paired two-tailed t-test relative to DMSO (n=3; unless specified otherwise, n indicates the number of independent biological replicates).
b. Immunofluorescence staining of pH3S10, EdU, Cyclin B1 and Hoechst staining (Top): Pictures illustrating pH3S10 staining in each cell cycle phase. Scale bar is 5 μm. (Bottom): Quantification of pH3S10 staining in IMR-5 cells treated for 8 h with MLN8237 (100 nM) relative to control (DMSO) cells; each grey dot represents a cell. In S and G2 phase number of spots and in mitotic cells intensity of pH3S10 signal compared to DMSO is shown. Shown is the mean ± S.D. (n≥137 cells examined over 3 independent experiments).
c. Quantification of pH3S10 staining in IMR-5 cells treated for 8 h with 100 nM MLN8237 (data are the same as in Figure 1b) or 100 nM AZD1152 relative to control (DMSO) cells; each grey dot represents a cell. In S and G2 phase number of spots and in mitotic cells intensity of pH3S10 signal compared to DMSO is shown. Shown is the mean ± S.D. (n≥390 cells were examined over 3 independent experiments).
d. Metagene plot of ChIP-Rx signal for histone H3.3 and pH3S10 in S phase-synchronized IMR-5 cells treated for 4 h with 1 μM MLN8237 or DMSO. The signal is centered on the first nucleosome (“+1 dyad”) located downstream of the TSS. Shading is ± S.E.M for 3,000 expressed genes with highest MYCN promoter occupancy. (n=3).
e. Metagene plot as in (d) for 3,000 expressed genes with lowest MYCN promoter occupancy.
f. pH3S10 (left) and histone H3.3 (right) ChIP at indicated loci after 4 h incubation with MLN8237 (1 μM) in SH-EP and in SH-EP MYCN cells synchronized in S phase. Shown is the mean ± S.D. of technical triplicates from a representative experiment (n=2).
Aurora-A phosphorylates H3S10 in S phase
Complex formation of Aurora-A with MYCN stabilizes MYCN11 and activates Aurora-A kinase14. Aurora-A localized at spindle poles autophosphorylates at T288 during mitosis 20, but we did not detect T288 phosphorylation during S phase (Extended Data Figure 1d). Aurora-A can phosphorylate Ser10 of histone H321. Indeed, immunofluorescence experiments using a specific Aurora-A inhibitor, MLN8237 (Alisertib; Extended Data Figure 1e)22,23, showed that Aurora-A was required for H3S10 phosphorylation (pH3S10) in S and, to a lesser degree, in G2 phase (Figure 1b and Extended Data Figure 1f). A second Aurora-A inhibitor, MK5108, caused a similar, albeit weaker reduction in pH3S10 in S phase (Extended Data Figure 1g), consistent with its weaker reduction in Aurora-A autophosphorylation12,24. Phosphorylation of H3S10 did not require Aurora-A in mitotic cells (Figure 1b)25. Conversely, inhibition of Aurora-B using AZD1152 (Barasertib)26 blocked pH3S10 in mitosis but had little effect in S and G2 phase (Figure 1c). Furthermore, phosphorylation of histone H3 at threonine 3, which occurs in mitosis and depends on Aurora-B, was blocked by AZD1152, but only weakly affected by MLN8237 (Extended Data Figure 1h,i)27.
Aurora-A controls histone H3.3 incorporation
To determine how Aurora-A-dependent phosphorylation of histone H3S10 impacts on chromatin structure at promoters, we performed spike-in chromatin-immunoprecipitation coupled with sequencing (ChIP-Rx sequencing) experiments from cells that were released from a double-thymidine block for 4 h in the presence of MLN8237. Notably, inhibition of kinase activity and disruption of MYCN/Aurora-A complexes by MLN8237 occur at different IC50 values12. We therefore performed all ChIP-sequencing effects after exposure to 1 μM MLN8237, which is sufficient to induce both effects. This showed that inhibition of Aurora-A broadly suppressed pH3S10 throughout the genome, including notable decreases in pH3S10 levels at peri-centromeric regions (Extended Data Figure 2a). Metagene analysis and ChIP experiments at individual MYCN-bound promoters showed that Aurora-A inhibition also decrease pH3S10 levels immediately adjacent to MYCN binding sites in active promoters (Extended Data Figure 2b,c,d). In neuroblastoma, MYCN, but not MYC, enhances the incorporation of a histone 3 variant, histone H3.3, into promoters, which can also be phosphorylated at S1028. In contrast to bulk histone H3.1, H3.3 is deposited in a replication-independent manner and marks, among others, particularly active genes and their promoters29. Inhibition of Aurora-A led to a decrease in the deposition of histone H3.3, while deposition of bulk histone, which occurs after passage of the replication fork, increased in response to MLN8237 inhibition (Extended Data Figure 2e). Stratification of promoters by MYCN occupancy showed that MYCN-bound promoters were characterized by a nucleosome-depleted zone around the transcription start site and that MLN8237 decreased histone H3.3 occupancy and H3S10 phosphorylation predominantly at the position of the +1 nucleosome (Figure 1d). In contrast, promoters with low MYCN binding showed only small MLN8237-dependent changes in H3.3 and H3S10 occupancy at specific positions around the transcription start site (Figure 1e). To demonstrate directly that these effects were specific for MYCN, we constitutively expressed MYCN in SH-EP cells, which express endogenous MYC; since MYCN suppresses endogenous MYC, this leads to a stable switch from MYC to MYCN expression (Extended Data Figure 2f)8. ChIP experiments showed that MYCN enhanced histone H3S10 phosphorylation and promoted histone H3.3 incorporation and inhibition of Aurora-A blocked both processes at multiple MYCN-bound promoters (Figure 1f).
To determine how these alterations in chromatin structure impact on RNAPII in S phase-synchronized cells, we performed ChIP-Rx sequencing using antibodies directed against total RNAPII. Metagene analyses showed that inhibition of Aurora-A caused a global decrease in the amount of elongating RNAPII (p<0.0001), but not of RNAPII localized proximal to the promoter (Extended Data Figure 3a) and a corresponding increase in the ratio of promoter-bound to elongating RNAPII (“traveling ratio”) (Extended Data Figure 3b). ChIP-Rx using antibodies that specifically recognize elongating RNAPII (RNAPII-pSer2) showed that RNAPII-pSer2 accumulated in a broad zone downstream of the TSS upon Aurora-A inhibition (Extended Data Figure 3c,d). Stratification of promoters by MYCN occupancy showed that RNAPII accumulation in response to MLN8237 was specific for MYCN-bound promoters (Figure 2a,b) and that the degree of accumulation correlated with MYCN occupancy (Figure 2c). The finding agrees with a previous demonstration that H3S10 phosphorylation promotes pause-release of RNAPII30.
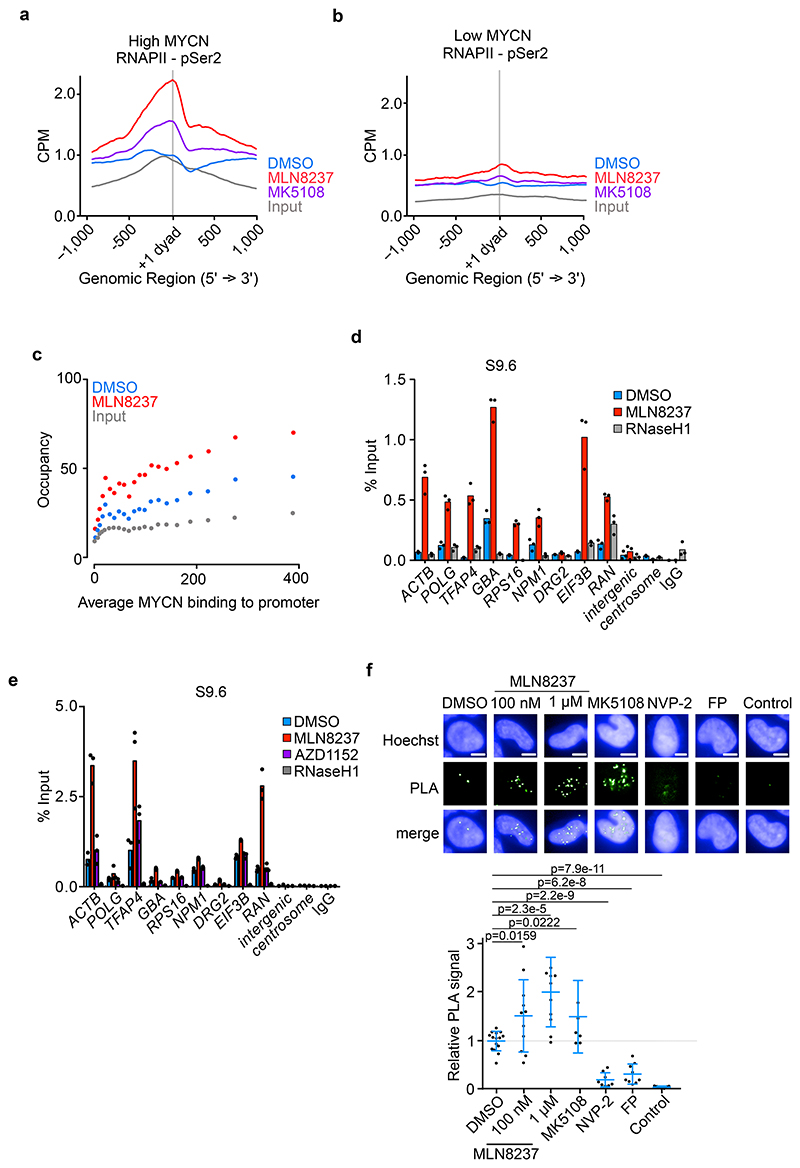
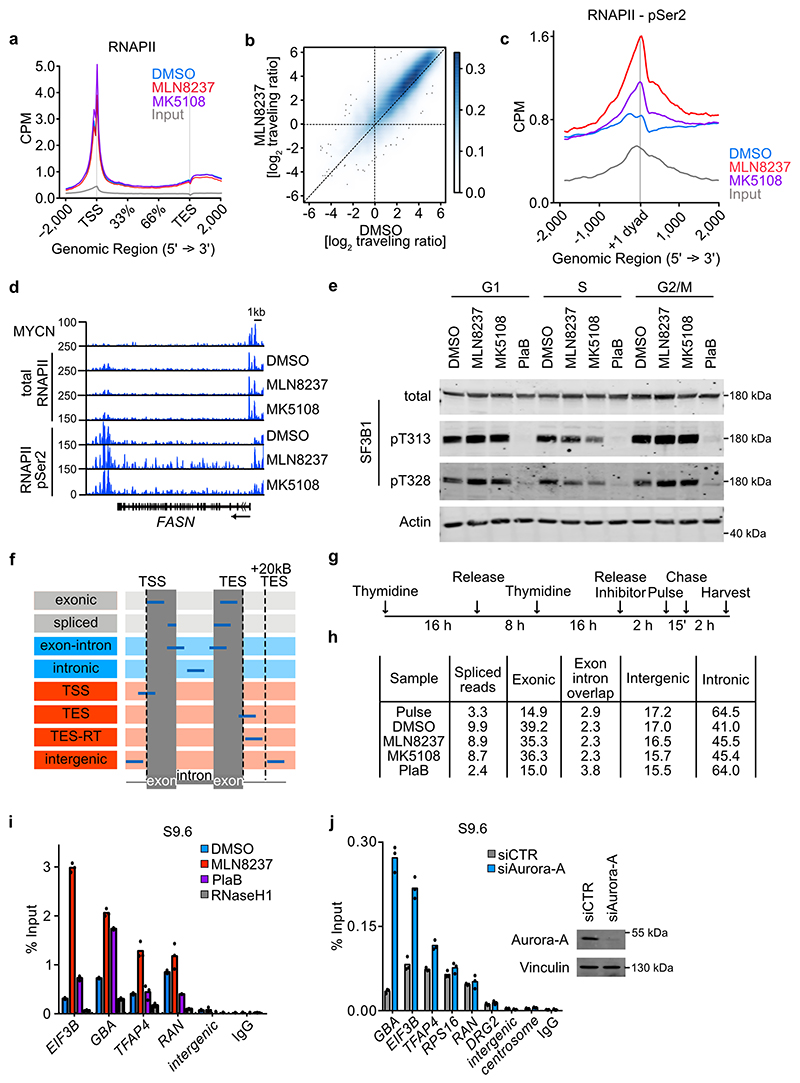
Figure 2. Aurora-A inhibition induces transcription-replication conflicts.
a. Metagene plot of ChIP-Rx signal for RNAPII pSer2 in S phase-synchronized IMR-5 cells treated for 4 h with MLN8237 (1 μM), MK5108 (1 μM) or DMSO. The signal is centered on the first nucleosome (“+1 dyad”) located within 300 nt downstream of the TSS. Data show mean +/- S.E.M. as a shade for the 3,000 expressed genes with highest MYCN promoter occupancy (n=2).
b. Metagene plot as in (a) filtered for the 3,000 expressed genes with lowest MYCN promoter occupancy.
c. Bin plots of average RNAPII pSer2 ChIP-Rx occupancy around the first exon-intron boundary (+/- 200 nt) from S phase-synchronized IMR-5 cells treated for 2 h with MLN8237 (1 μM) or DMSO. Shown is the mean ± S.E.M for the 3,000 genes in each bin. Genes were ordered by MYCN occupancy (n=2).
d. DNA-RNA-Immunoprecipitation (DRIP) using S9.6 antibody, detecting R-loops at the indicated loci after MLN8237 (1 μM, 8 h) treatment. Incubation with RNaseH1 and IgG were used as controls for non-specific chromatin binding. Shown is the mean ± S.D. of technical triplicates from a representative experiment (n=3).
e. DNA-RNA-Immunoprecipitation (DRIP) using S9.6 antibody, indicating R-loops at the indicated loci after 8 h of 1 μM MLN8237 or 1 μM AZD1152 treatment. Incubation with RNaseH1 and IgG were used as controls for non-specific chromatin binding. Shown is the mean ± S.D. of technical triplicates from a representative experiment (n=2).
f. Proximity Ligation Assay between RNAPII and PCNA in asynchronous IMR-5 cells treated for 8 h with the indicated inhibitors (MLN8237, MK5108 (1 μM), NVP-2 (200 nM) or Flavopiridol (FP; 200 nM)). Control shows primary antibodies only in cells treated with 1 μM MLN8237. (Top): Example pictures of PLA in different conditions. (Bottom): Quantification of PLA signals. Each dot represents mean PLA signal of all cells in one well compared to solvent control. Shown is the mean ± S.D.. P-values were calculated comparing treatment to DMSO using unpaired two-tailed t-test. White line indicates 5 μm (n≥6).
Aurora-A inhibition induces transcription-replication conflicts
To understand the mechanisms underlying this perturbance in RNAPII function, we initially tested the impact of Aurora-A inhibition on RNA splicing, since Aurora-A has been implicated in cell cycle-dependent control of mRNA splicing31 and RNAPII accumulates at the first exon/intron boundary upon an impairment of spliceosome function32. Consistently, inhibition of Aurora-A in S phase, but not in other phases of the cell cycle, led to a decrease in SF3B1 phosphorylation at residues pT313 and pT328, which are phosphorylated in active spliceosomes (Extended Data Figure 3e)33. However, compared to an inhibitor of spliceosome, Pladienolide B (PlaB), inhibition of Aurora-A had only a small effect on overall splicing efficiency in S phase, as demonstrated by sequencing nascent RNA immediately after a 15 min pulse of 4-thiouridine (4sU) or after a chase of 2 h (Extended Data Figure 3f,g,h). Since chromatin compaction marked by pH3S10 antagonizes R-loop formation34, we next performed DNA-RNA immunoprecipitation (DRIP)-assays using an antibody that recognizes RNA/DNA-hybrids (S9.6). These assays showed that inhibition of Aurora-A led to large increases of R-loops at all active and MYCN-bound promoters that we tested (Figure 2d). No R-loop accumulation was observed at transcriptionally silent intergenic and centromeric sequences, despite large decreases in pH3S10 levels, consistent with the fact that R-loop formation requires active transcription (Figure 2d). R-loops did not accumulate in response to the Aurora-B inhibitor AZD1152 (Figure 2e) and only weakly in response to spliceosome inhibition by PlaB (Extended Data Figure 3i). siRNA-mediated depletion of Aurora-A also induced R-loop accumulation (Extended Data Figure 3j). To test whether Aurora-A inhibition impedes replication fork progression, we performed proximity-ligation assays (PLAs) using antibodies directed against RNAPII and proliferating cell nuclear antigen (PCNA), which forms the sliding clamp of DNA polymerase35. These assays showed a significant increase in the number of foci upon inhibition of Aurora-A, which was sensitive to inhibition of CDK9, demonstrating that inhibition of Aurora-A causes transcription-replication conflicts (Figure 2f)12.
Aurora-A inhibition activates ATR
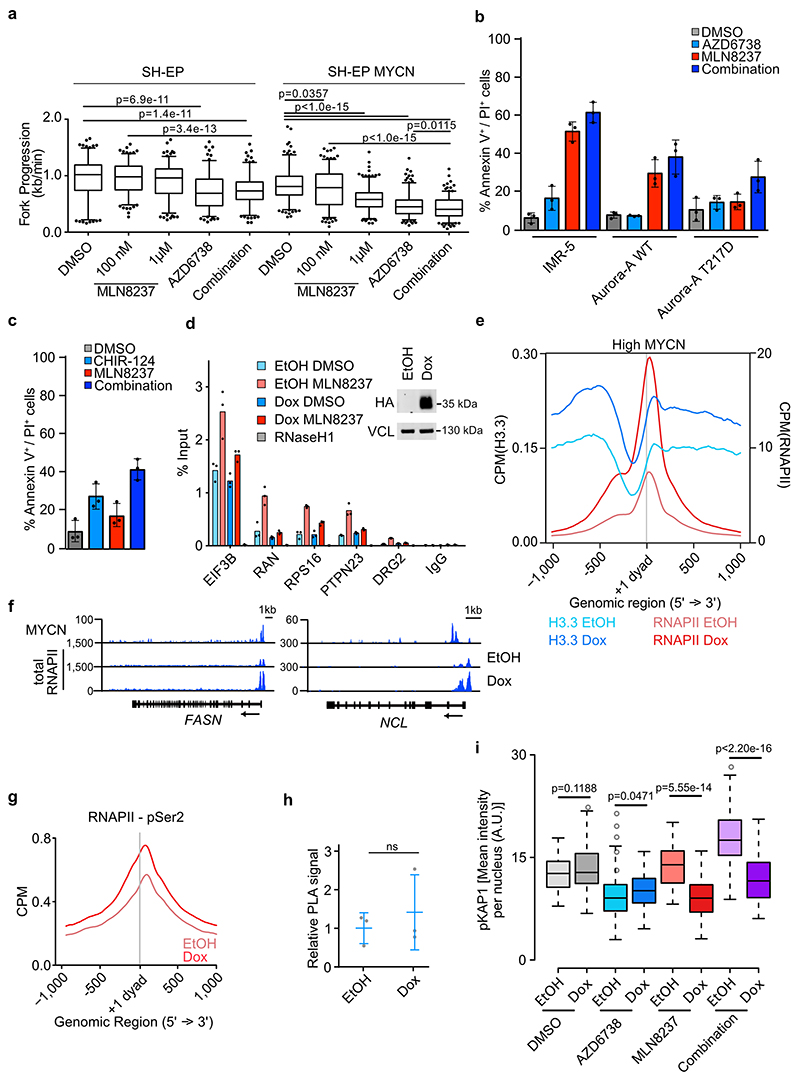
Consistent with these observations, inhibition of Aurora-A in S phase, but much less in G2 phase, induced ATR-dependent phosphorylation of serine 33 of replication protein A (Figure 3a) and of serine 345 of the CHK1 kinase (Figure 3b)36. Notably, these data and our own previous observations15 show that increases in CHK1 phosphorylation required exposure of cells to elevated concentrations of MLN8237. Similar differences between ATR activity towards RPA and CHK1 have been noted before and indicate that MLN8237 at low doses affects the stability of replication forks, but only at elevated concentrations causes CHK1-mediated cell cycle arrest and DNA repair37. We therefore directly assessed the effect of Aurora-A and ATR on replication fork progression and we performed fiber assays, which showed that inhibition of Aurora-A significantly reduced replication fork speed (Figure 3c,d). Concomitant inhibition of Aurora-A and ATR using an inhibitor that is currently being explored in multiple clinical trials, AZD673838,39, led to further slow-down in fork progression, which exceeded that induced by gemcitabine, a known inhibitor of fork progression (Figure 3c,d). Comparison of SH-EP cells with SH-EP-MYCN cells showed that the decrease was much more pronounced in cells expressing MYCN (Extended Data Figure 4a). To support the argument that the induction of transcription-replication conflicts underlies the therapeutic effects in vivo, we used a combination of Aurora-A and PARP1 inhibition as a second strategy to induce transcription-replication conflicts. Inhibition of PARP1, which modulates fork progression and senses replication stress40, caused transcription-replication conflicts (Figure 3e) and decreased fork speed when combined with MLN8237 (Figure 3f,g).
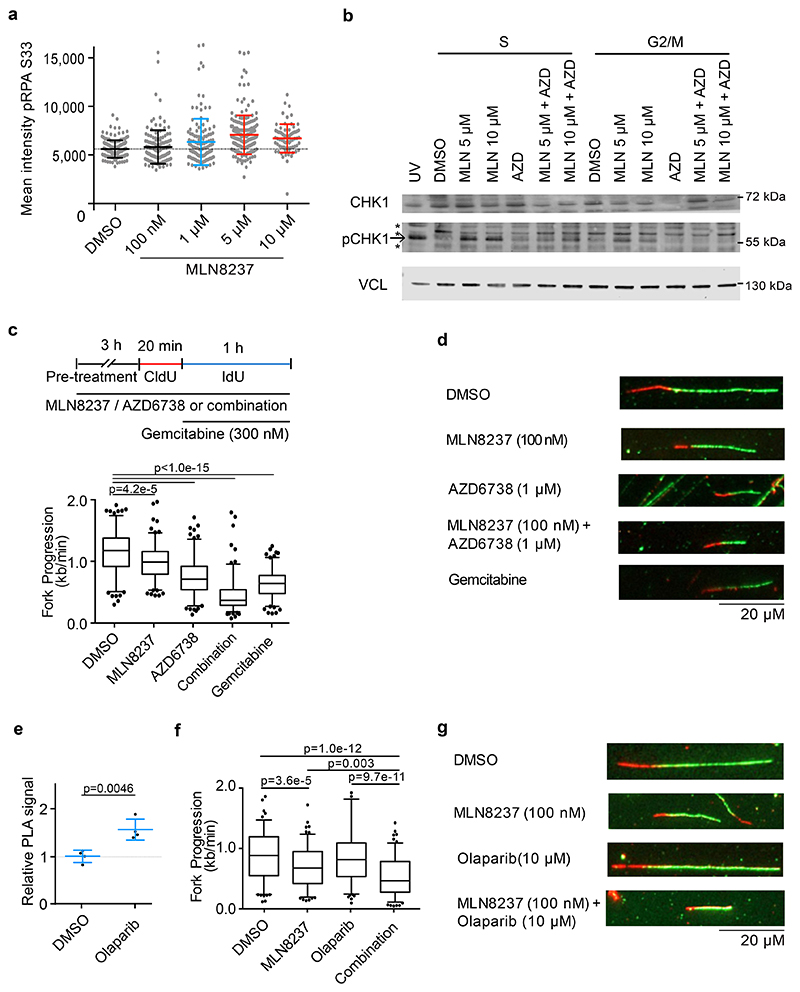
Figure 3. Aurora-A/ATR inhibition reduces replication fork progression.
a. pRPA S33 staining in IMR-5 cells treated for 8 h with indicated concentration of Aurora-A inhibitor. Shown is the mean intensity in each condition ± S.D.; each dot represents one cell (n≥73 cells examined over 3 independent experiments). Color indicates significant difference of mean from control (blue: p=0.0004; red: p<0.0001, DMSO vs. MLN 5 μM p<1.0e-15, DMSO vs. MLN 10 μM p=1.9e-13).
b. Immunoblots of cell cycle synchronized IMR-5 cells treated for 4 h (S phase) or 8 h (G2/M phase) with MLN8237 (MLN) and AZD6738 (AZD, 1μM) or DMSO as control. UV light was used as positive control. Vinculin was used as loading control. Arrow marks specific band while (*) indicate non-specific bands (n=3).
c. Fork progression speed in IMR-5 cells treated for 3 h with 100 nM MLN8237, 1 μM of AZD6738 or a combination of both. Experimental setup is shown on top. Cells were incubated 20 min with CldU, followed by incubation for 1 h with IdU. Gemcitabine (300 nM) was used as positive control. P-value was calculated using unpaired two-tailed t-test comparing two conditions (n≥142 fibers were examined over 2 independent experiments).
d. Representative example pictures of fibers treated as and quantified in (c).
e. Quantification of PLA signal between RNAPII and PCNA in asynchronous IMR-5 cells treated for 8 h with 10 μM Olaparib. Each dot represents mean PLA signal of all cells in one well compared to solvent control. Shown is the mean ± S.D.. P-value was calculated compared to DMSO using unpaired two-tailed t-test (n=4).
f. Quantification of fork progression measured by fiber assays of IMR-5 cells pretreated for 3 h with 100 nM MLN8237, 10 μM of Olaparib or the combination. P-values were calculated using unpaired two-tailed t-test (n≥128 fibers were examined over 2 independent experiments).
g. Representative example pictures of fibers treated as and quantified in (c).
Upon replication stress, ATR inhibits origin firing and stabilizes stalled replication forks; as consequence, inhibition of ATR conflicts in stressed cells leads to formation of double-strand breaks41,42. Microscopy-based cytometry confirmed that inhibition of ATR using AZD6738 strongly enhanced DNA synthesis and accelerated the passage through S phase, irrespective of the presence of MLN8237 (Figure 4a). Co-incubation of cells with MLN8237 and AZD6738 strongly enhanced the number of cells showing elevated phosphorylation of γH2AX, which is a target of several DNA-damage-induced kinases, and of KAP1, a specific substrate of the ATM kinase that is activated at double-strand breaks, in the G2 phase of the cell cycle (Figure 4a,b). Inhibition of ATR enhanced the pro-apoptotic activity of MLN8237 in three MYCN-amplified neuroblastoma cell lines, but had little effect in three MYCN non-amplified lines (Figure 4c). The effect of MLN8237 was on-target, since expression of Aurora-A T217D, a point mutant allele that has reduced affinity to MLN823743,44, suppressed apoptosis in IMR-5 cells (Extended Data Figure 4b). Inhibition of Aurora-A also enhanced the pro-apoptotic effect of a CHK1 inhibitor, CHIR-12445 (Extended Data Figure 4c). We concluded that Aurora-A is required to prevent transcription-replication conflicts in MYCN-amplified neuroblastoma cells and that inhibition of Aurora-A enhances the dependence of MYCN-amplified cells on the ATR kinase for maintaining genomic stability and survival.
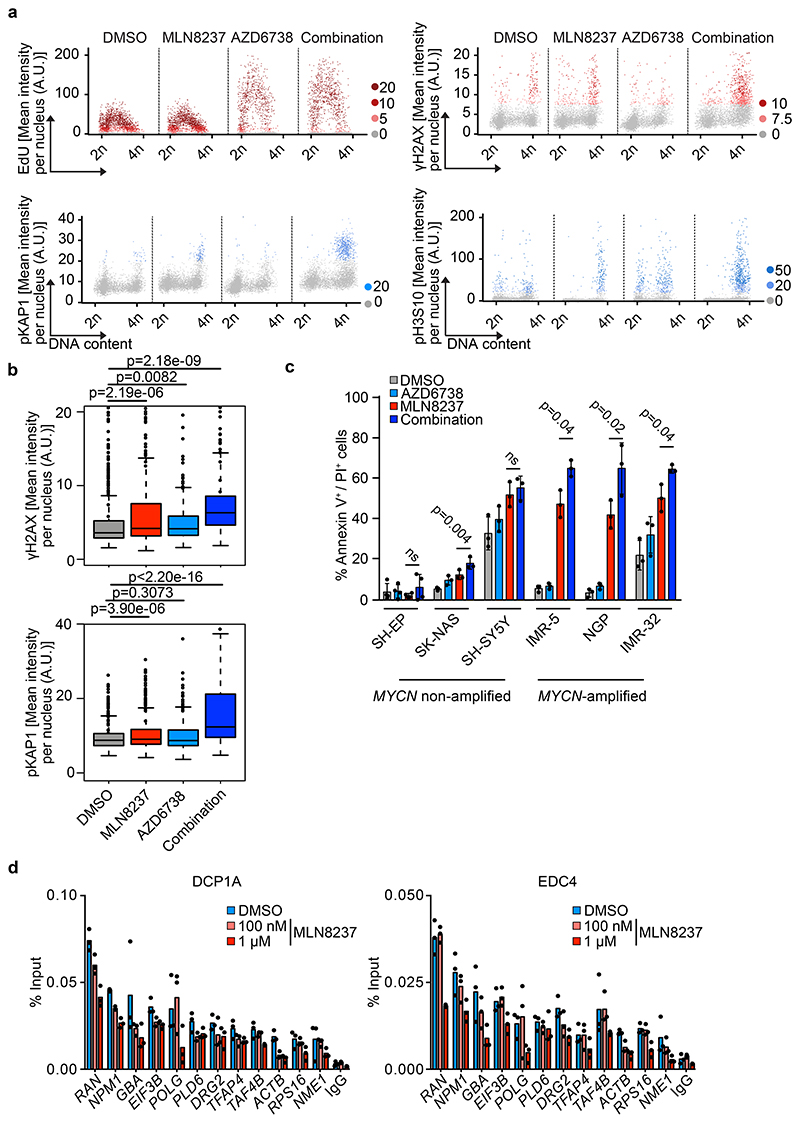
Figure 4. Aurora-A/ATR inhibition induces DNA damage.
a. High-content microscopy-based analysis of MYCN-amplified NGP cells treated for 8 h with 100 nM MLN8237, 1 μM AZD6738 or a combination and assessed for EdU incorporation, γH2AX, pKAP1 and pH3S10. Each dot represents a single cell and is color-coded according to mean fluorescent intensity (n≥3,837 cells were examined over 3 independent experiments). A.U. arbitrary units. Quantification is shown in (b).
b. Quantification of γH2AX (upper panel) and pKAP1 (lower panel) signals shown in (a) (n≥3,837 cells were examined over 3 independent experiments).
c. Annexin-V/PI FACS of the indicated cell lines (MYCN non-amplified: SH-EP, SK-NAS, SH-SY5Y; MYCN-amplified: IMR-5, NGP, IMR-32) treated for 48 h with DMSO, MLN8237 (100 nM), AZD6738 (1 μM) or both. Shown is the mean ± S.D., p-values were calculated using paired two-tailed t-test (n=3).
d. DCP1A (left) and EDC4 (right) ChIP at indicated loci after 4 h of 100 nM or 1 μM MLN8237 treatment in S phase synchronized IMR-5 cells. IgG control was used as control for antibody specificity. Data are presented as mean ± S.D. of technical triplicates from a representative experiment (n=3).
Aurora-A enables transcription termination in S phase
To clarify the role of R-loops in coordinating transcription elongation with DNA replication, we expressed doxycycline-inducible RNAseH1 in MYCN-amplified cells and established that this reduced the MLN8237-induced accumulation of R-loops at active promoters (Extended Data Figure 4d). Removal of R-loops strongly enhanced histone H3.3 incorporation downstream of MYCN-binding sites arguing that the +1 nucleosome is stabilized. This correlated with stalling of RNAPII downstream of the transcription start sites of MYCN-bound promoters (Extended Data Figure 4e,f,g), but had no significant effects on the occurrence of transcription-replication conflicts (Extended Data Figure 4h). However, removal of R-loops by RNAseH1 strongly suppressed the accumulation of cells with high levels of DNA damage in mitosis after Aurora-A or Aurora-A/ATR inhibition, arguing that removal of R-loops facilitates the resolution of these conflicts (Extended Data Figure 4i). To limit R-loop accumulation, MYCN recruits mRNA decapping factors, which terminate transcription close to promoters8,46. Consistently, inhibition of Aurora-A antagonized the association of two de-capping factors, DCP1 and EDC4, with MYCN-bound promoters in S phase (Figure 4d). Collectively, the data argue that MLN8237-induced R-loop formation restricts the accessibility of nascent mRNA for transcription termination factors and thereby undermines the ability of MYCN to terminate transcription and prevent transcription-replication conflicts (see Figure 7e for a model).
Aurora-A/ATR treatment causes tumor-specific DNA damage
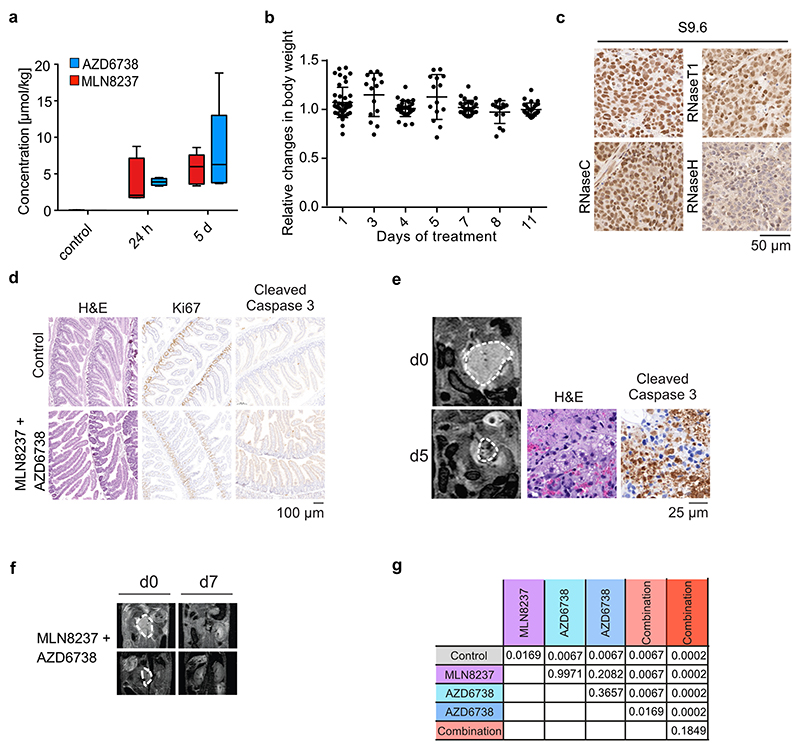
As a single agent, MLN8237 has therapeutic efficacy in the TH-MYCN transgenic model of neuroblastoma47 at a dose of 30 mg/kg, which is the maximum tolerated dose12. Similarly, clinical trials of neuroblastoma and MYCN-driven neuroendocrine prostate carcinoma patients show limited activity of MLN8237 which is in part due to dose-limiting toxicity48–50. To explore whether the MLN8237-induced dependence on ATR can be exploited to improve the efficacy of MLN8237 in MYCN-amplified neuroblastoma, we evaluated the efficacy of 15 mg/kg MLN8237 given in combination with 25-30 mg/kg of AZD6738. Mass spectrometry of tissue samples showed that these dose lead to levels between 4 and 8 μmoles/kg body weight for each drug (Extended Data Figure 5a). At these doses, no toxicity was detectable using either MLN8237 or AZD6738 alone or with these drugs used in combination, in animals with palpable (approximately 1 cm diameter) tumors (Extended Data Figure 5b). We administered drugs for 32 days and assessed responses to treatment at day 1 and 5 on study.
Control tumors in the TH-MYCN model feature the typical architecture of human neuroblastoma with tightly packed cells arranged in a vaguely lobular or nesting pattern, thin fibrovascular septa and occasional areas of hemorrhage (Figure 5a)47. Nuclear chromatin was finely dispersed with few and inconspicuous nuclei and fragmented nuclei were frequently observed. Numerous mitoses and a high Ki67-labelling index illustrate the strong proliferative nature of these neoplasms (Figure 5a). Tumors treated with a combination of MLN8237 and AZD6738 underwent a rapid and profound regressive change with architectural disintegration, expansion of hemorrhage and increased cell death (Figure 5a,b). The latter is evidenced by necrotic cell debris and an increase of tumor cells with fragmented nuclei (Figure 5a).
Figure 5. Treatment responses to combined Aurora-A/ATR inhibition.
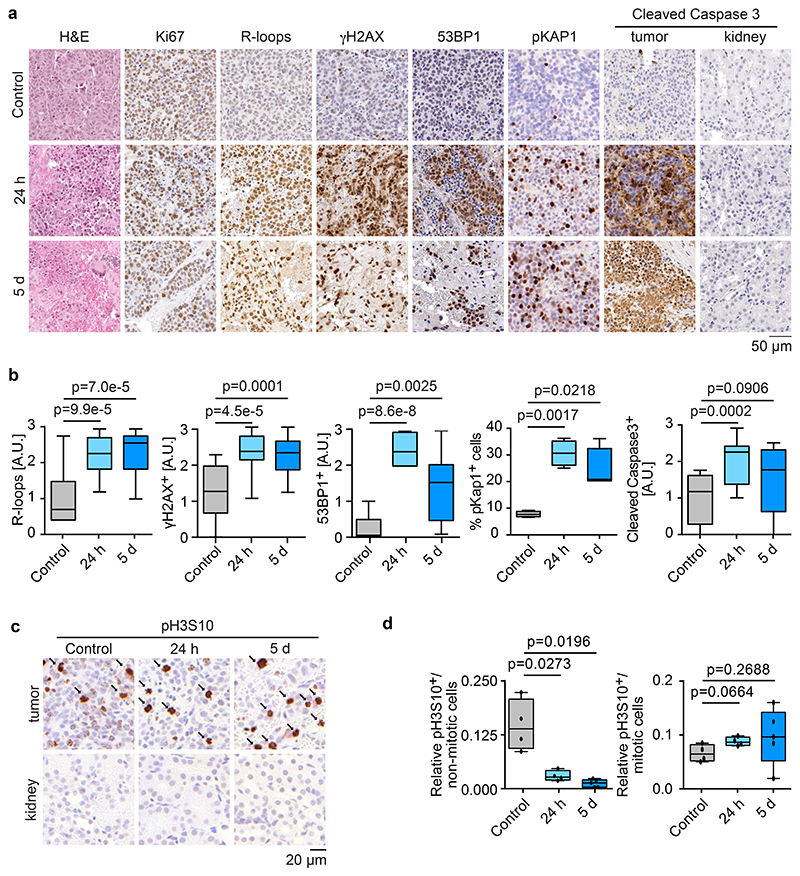
a. Representative sections of tumors of untreated, 24 h treated or 5 days treated TH-MYCN mice showing haemotoxylin and eosin (H&E), Ki67, R-loops, γH2AX, 53BP1, pKAP1 and cleaved caspase 3. The second column on the cleaved caspase staining shows kidney tissue of tumor-bearing mice treated for the same time. Each group has a minimum of four animals.
b. Box plots show quantification of R-loop-, γH2AX-, 53BP1-, pKAP1- and cleaved caspase 3-positive cells in tumor sections. P-values were calculated using unpaired two-tailed t-test using Welch’s correction (n≥12 sections from at least 4 animals from each condition were evaluated).
c. Representative sections of tumors of untreated, 24 h or 5 days treated TH-MYCN mice showing H3S10 phosphorylation. Arrows mark mitotic cells. The second row shows staining of adjacent kidney tissue. Each group has a minimum of four animals.
d. Relative number of pH3S10-positive non-mitotic (left) and mitotic cells (right) shown in panel b. P-value was calculated using an unpaired two-tailed t-test using Welch’s correction (n≥12 sections from at least 4 animals were evaluated for each condition).
Immunohistochemistry confirmed that combined Aurora-A and ATR inhibition strongly decreased pH3S10 levels in non-mitotic cells (Figure 5c,d) and led to a significant increase in R-loop formation (Figure 5a,b and Extended Data Figure 5c). Consistent with the responses observed in tissue culture, Ki67-positivity remained unchanged upon treatment, indicating an absence of cell cycle arrest in response to Aurora-A/ATR inhibition (Figure 5a). In contrast, the number of cells staining positive for phosphorylated γH2AX, of 53BP1 and of phosphorylated KAP1 strongly increased in treated animals (Figure 5a,b). In addition, we noted a large increase in the number of cells that stained positive for cleaved caspase 3, indicating that tumor cells underwent rampant apoptosis in response to Aurora-A/ATR inhibition (Figure 5a,b). Notably, apoptosis induced by Aurora-A/ATR inhibition was tumor cell-specific, since no apoptosis was induced in the adjacent kidney tissue as well as in proliferative gut tissue (Figure 5a and Extended Data Figure 5d).
Aurora-A/ATR treatment causes tumors regression and greatly extends lifespan
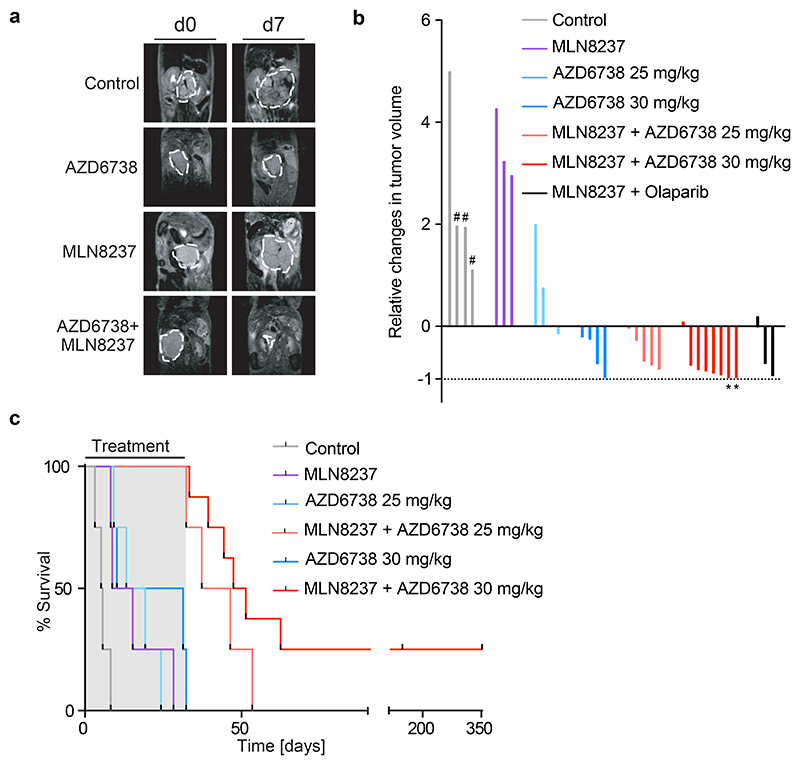
As single agents, both MLN8237 and AZD6738 had variable effects on tumor progression; in contrast, the combination of both drugs led to robust tumor regression in 7/8 treated tumors (Figure 6a,b). Combining MLN8237 with the PARP1 inhibitor, olaparib, also led to widespread apoptosis in tumors and induced tumor regression, supporting the view that replication-transcription conflicts are causative for these effects (Figure 6b and Extended Data Figure 5e). To assess survival benefit, we treated mice for a maximum of 32 days before withdrawal of the drugs. Whereas all mice treated with either MLN8237 or AZD6738 as single agents died while still on treatment, all mice treated with the combination of MLN8237 and AZD6738 survived until the end of treatment. Mice treated with MLN8237 and 25 mg/kg AZD6738 survived for up to 21 days after cessation of therapy. Increasing the dose of AZD6738 to 30 mg/kg as a single agent provided no additional treatment benefit but further prolonged survival in combination with MLN8237, with two out of eight mice remaining tumor-free for up to 150 days after cessation of therapy, indicating a dose-related impact on long-term survival and cure in a subset of treated mice (Figure 6c and Extended Data Figure 5f,g).
Figure 6. Therapeutic efficacy of combined Aurora-A/ATR inhibitor treatment.
a. Representative MRI sections of mice at day 0 and day 7 of treatment with vehicle, MLN8237 (15 mg/kg), AZD6738 (25 mg/kg) or both. Dashed white lines indicate tumor circumference.
b. Waterfall plot showing relative changes in tumor volume during the first seven days of the beginning of treatment with indicated drugs. Control animals marked with hash (#) show measurement at day 4. Mice which survived the treatment for up to 150 days are marked with an asterisk (*).
c. Kaplan Meier survival curve of TH-MYCN mice treated as indicated. AZD6738 was administered at 25-30 mg/kg every day and MLN8237 at 15 mg/kg on a 5 days on, 2 days off schedule. Shaded area indicates duration of the combination treatment (ended at 32 days, 56 doses) (n=8 animals for MLN8237 + AZD6738 (30 mg/kg), n=4 for all other experimental cohorts). P-values are shown in Extended Data Figure 5g.
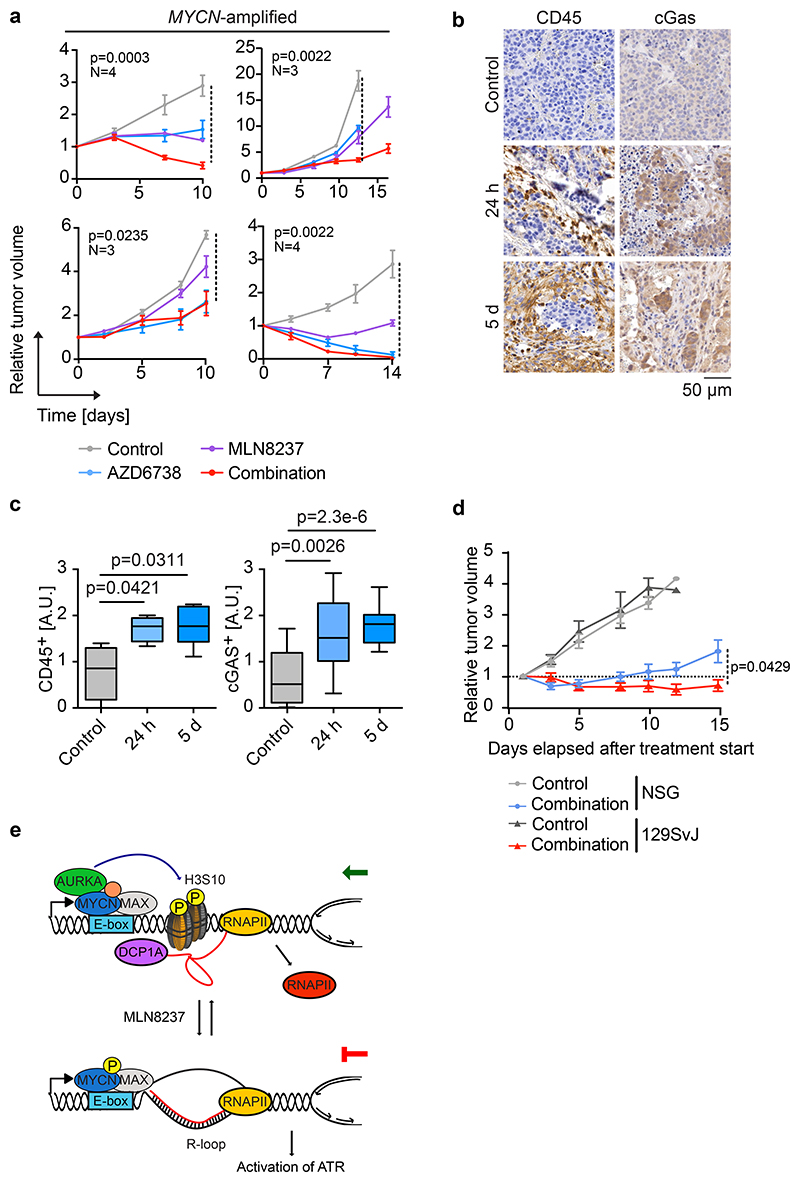
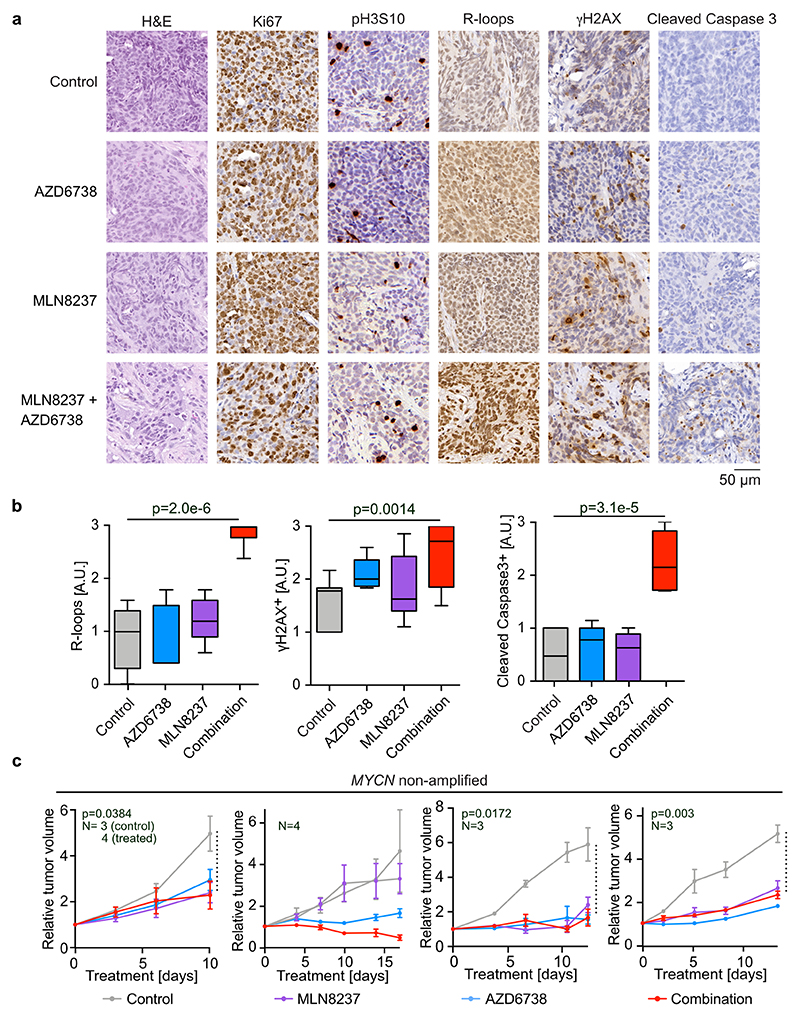
To test this regimen in human tumor samples, we treated four MYCN-amplified and four MYCN non-amplified patient-derived xenografts (PDX) of neuroblastoma with MLN8237, AZD6738 or the combination of both drugs. Tumors recovered at the end of the treatment period showed similar morphological alterations as described for the TH-MYCN mouse model (Extended Data Figure 6a). As in the TH-MYCN model, combining Aurora-A and ATR inhibition led to an accumulation of R-loops (Extended Data Figure 6a,b). Neither single nor combined Aurora-A or ATR inhibition altered the fraction of Ki67-positive cells, while they caused additive or synergistic increases in the number of tumor cells staining positive for phosphorylated γH2AX or undergoing apoptosis (Extended Data Figure 6a,b). In mice bearing MYCN-amplified tumors, the combined inhibition of Aurora-A and ATR suppressed the growth of three out of four tumors better than the best single treatment and caused regression in two tumors (Figure 7a). Combining both inhibitors was more effective than single treatment and cause regression in one of four MYCN non-amplified tumors (Extended Data Figure 6c).
Aurora-A/ATR treatment engages the immune system
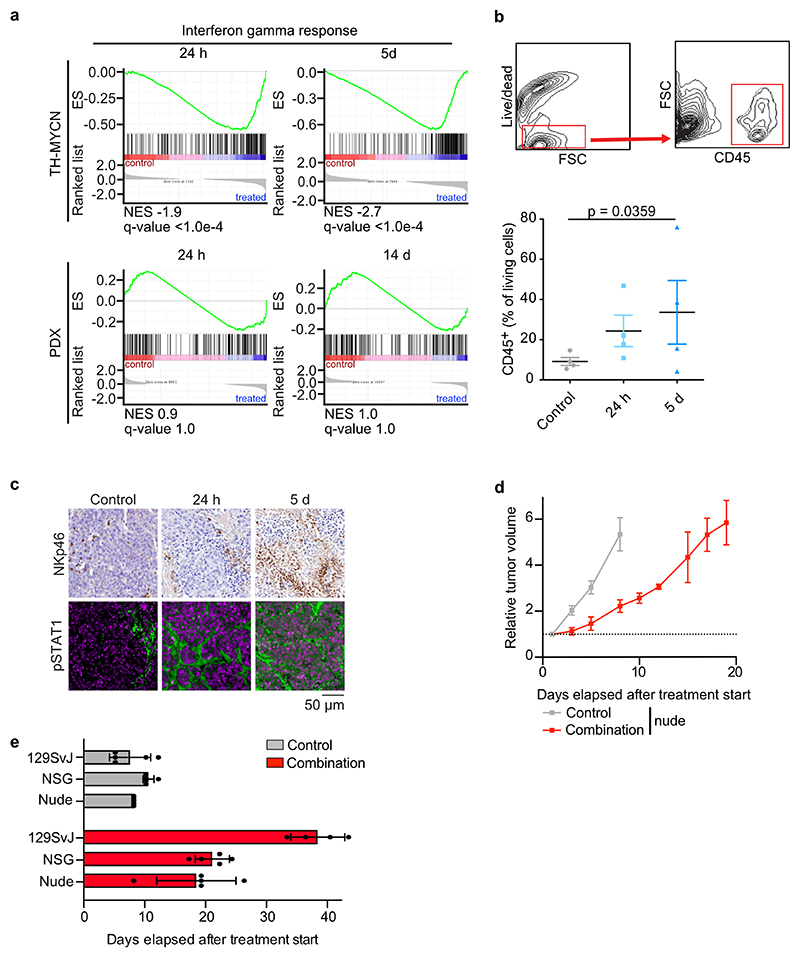
While treatment responses of PDX models reflected the responses in the TH-MYCN models, they were generally weaker in the PDX models. To identify possible causes of this difference, we performed RNA sequencing of control and treated tumors in the transgenic model as well as in the PDX model. Gene set enrichment analysis (GSEA)51 showed a rapid induction of multiple genes involved in cytokine signaling, in particular via interferon-mediated pathways (Extended Data Figure 7a). This transcriptional response was not observed in the PDX model, indicating that it depends on an interaction of tumor cells with cells of the immune system (Extended Data Figure 7a). These observations are consistent with previous demonstrations that MYC proteins exert an immune-suppressive effect via the suppression of cytokine-dependent signaling pathways52. DNA damage activates the interferon and NF-kB pathways via cGAS/STING53 and immunohistochemistry of tumors from the TH-MYCN model documented activation of cGAS/STING in parallel with infiltration of immune cells (Figure 7b,c). FACS analyses and histology confirmed an increased number of immune cells expressing the lymphocyte common antigen (CD45), which included an increase in the number of natural killer cells, in the tumor environment (Extended Data Figure 7b,c). In addition, histology showed activation of STAT1 in response to treatment (Extended Data Figure 7c). To test whether engagement of the host immune system is relevant for the therapeutic response to Aurora-A/ATR inhibition, we injected tumor cells derived from the TH-MYCN mice into either immunocompetent syngeneic 129SvJ mice, immune suppressed nude mice lacking T cells or strongly immunodeficient NSG mice (lacking B-, T and NK cells) and monitored their growth. Tumors grew rapidly upon transplantation in either NSG, nude or 129SvJ mice (Figure 7d and Extended Data Figure 7d). However, whilst treatment with MLN8237/AZD6738 dramatically slowed tumor growth in transplanted syngeneic mice, it had much weaker effects upon transplantation into immune-deficient (NSG) (Figure 7d). Treatment response was also weaker in nude mice, arguing that T-lymphocytes contribute to its efficacy (Extended Data Figure 7d). The differences in tumor growth were reflected in overall survival of treated mice (Extended Data Figure 7e). We concluded that combined Aurora-A/ATR inhibition engages the host immune system for tumor eradication.
Figure 7. Combination therapy engages the immune system.
a. Relative changes in tumor volume of four MYCN-amplified PDX models during treatment with indicated inhibitors. Shown is the mean ± S.E.M.. Each experimental cohort had at least three animals. P-values comparing the control to the combination were calculated using unpaired two-sided t-test.
b. Histology of representative tumor sections showing CD45- and cGAS-positive cells in tumors of TH-MYCN mice treated with combined Aurora-A/ATR inhibition. Each group has a minimum of four animals.
c. Box plots of CD45-(left) and cGAS-positive (right) staining in tumor sections in (d). P-values were calculated using unpaired two-sided t-test using Welch’s correction (n≥12 sections from at least 4 animals from each condition were evaluated).
d. Relative changes in tumor volume of subcutaneous allografts in NSG or 129SvJ mice after treatment with vehicle or a combination of AZD6738 (25 mg/kg every day) and MLN8237 (15mg/kg, 5 days on, 2 days off). Shown is the mean ± S.E.M.. Each group consist of five animals. P-value comparing the relative tumor volume of NSG and 129SvJ treated with the combination at day 15 was calculated using unpaired two-tailed t-test.
e. Model summarizing our findings. For clarity of presentation only one nucleosome is shown.
Discussion
We had previously shown that the MYCN protein forms a complex with the Aurora-A kinase during the S phase in neuroblastoma cells, but its molecular function and how it can be exploited for therapy had remained unclear. Here we have shown that MYCN recruits Aurora-A to chromatin in S phase, where it phosphorylates H3S10, and promotes the incorporation of histone H3.3 into promoters, thereby antagonizing R-loop accumulation (Figure 7e). R-loop formation limits the accessibility of nascent mRNA for splicing and for mRNA decapping factors, which enable promoter-proximal termination46. In addition, stabilization of the first nucleosome may promote premature polyadenylation and transcription termination in the first intron54. In neuroblastoma, Aurora-A has catalytic functions, but also binds to and stabilizes MYCN complexes by antagonizing degradation by FBXW711,13,14. Inhibition of Aurora-A by MLN8237 abrogates histone H3S10 phosphorylation at low drug concentrations (100 nM), which parallel inhibition of Aurora-A catalytic activity12. In contrast, the MLN8237-dependent decrease in association of mRNA decapping factors with promoters, the increase in transcription-replication conflicts and activation of ATR required elevated concentrations of MLN8237, which correspond to levels that destabilize MYCN12. Since stabilization of MYCN enables it to recruit BRCA1 and transcription termination factors8, we suggest that both phosphorylation of H3S10 and the ability to stabilize MYCN enable Aurora-A to prevent transcription-replication conflicts. The findings are in line with recent observations that Aurora-A has kinase-independent functions during DNA replication55. We note that transcription-replication conflicts are stochastic and rare events, hence the expected MYCN- and Aurora-A-dependent effects on histone occupancy and gene expression averaged over a large cell population are expected to be small, consistent with both our global and gene-specific analyses10.
While Aurora-A monotherapy is effective at high doses in models of neuroblastoma and other MYCN-driven tumors, its efficacy in human patients is limited, which is in part due to a dose-limiting toxicity that is also see in mice48. We found that inhibition of ATR strongly increased DNA replication in MYCN-amplified neuroblastoma cells and thus aggravated double-strand break formation and apoptosis caused by Aurora-A inhibition42,56. In vivo, the combined treatment with low and non-toxic doses of Aurora-A and ATR inhibitors rapidly induced tumor-specific DNA damage and greatly extended survival, with treatment responses differing between immuno-competent and deficient models. One reason for this difference in response is that the responses are both due to cell-intrinsic DNA damage and due to recognition of tumor cells by the host immune system, hence an intact immune response is critical for an efficient therapeutic response. Additional factors, such as the different vascularization of orthotopically growing tumors relative to transplanted tumors or mechanisms that promote tumor cell plasticity57 may affect the therapeutic outcome. Multiple strategies are currently being explored that target the transcription machinery of pediatric tumors for therapy. Our finding that a subset of treated mice remains tumor-free for long periods of time and appears to be cured of neuroblastoma establishes that targeting transcription-replication conflicts is an effective strategy for the treatment of MYCN-driven neuroblastomas and possibly other MYCN-driven tumors that can be realized with currently available inhibitors. Finally, we note that the Aurora-A interaction domain of MYCN is not conserved in MYC, and the Aurora-A dependent histone H3.3-deposition is specifically catalyzed by MYCN, not MYC; since individual tumors almost always express and depend on a single MYC family member, expression of MYCN may provide a straightforward strategy for identifying patients that are susceptible to this treatment.
Methods
Further information on research design is available in the Nature Research Reporting Summary linked to this article.
Cell culture
Cell lines derived from human neuroblastoma (IMR-5, SH-EP, NGP, SH-EP, SH-SY5Y, SK-NAS) were verified by STR profiling and grown in RPMI-1640 (Thermo Fisher Scientific). Murine NIH-3T3 cells were grown in DMEM (Thermo Fisher Scientific). Media were supplemented with 10% fetal calf serum (Sigma-Aldrich and Capricorn Scientific GmbH) and penicillin/streptomycin (Sigma-Aldrich). All cells were routinely tested for mycoplasma contamination. For double-thymidine block, cells were treated for 16 h with 2 mM thymidine (Sigma-Aldrich), released for 8 h into normal medium and then blocked again (2 mM, 16 h). For release, cells were washed with PBS before medium was added. For nocodazole block in mitosis, cells were treated for 16 h with 0.6 μg/ml nocodazole (Sigma-Aldrich).32For pulsed 5-ethynyl-2’-deoxyuridine (EdU, Thermo Fisher Scientific) incorporation, cells were incubated for 30 min in medium containing 10 μM EdU. The Click-iT EdU Alexa Fluor Imaging Kit (Thermo Fisher Scientific) was used for EdU detection. Inhibitors were used in the following concentrations according to previous publications as well as length of treatment in our study. 10058-F4: 100 μM; 18, MLN8237: 100 nM or 1 μM12; MK5108: 1 μM12; AZD6738: 1μM38; AZD1152: 100 nM or 1 μM26; PlaB: 1 μM32,58; NVP-2: 200 nM59; Flavopiridol: 200 nM8,60.
Immunofluorescence staining
IMR-5 cells were plated in 96-well plates (Greiner). Inhibitors were added for 8 h depending on the experiment. Cells were fixed and permeabilized with methanol for 20 min. After removing methanol, cells were blocked with 5% BSA in PBS. Samples were stained with primary antibody against pRPA S33 (rabbit, A300-246A, Bethyl, 1:400), pH3S10 (rabbit, 06-570, Sigma, 1:500), pH3T3 (rabbit, Cell Signaling, 1:100) or Cyclin B1 (mouse, sc-245, Santa Cruz, 1:500) in 5% BSA in PBS overnight at 4 °C and after washing incubated with secondary antibody (Alexa Fluor 488 and Alexa Fluor 568 from Invitrogen, 1:400) for 1 h at room temperature (RT). Nuclei were counterstained using Hoechst 33342 (Sigma-Aldrich). High-throughput pictures were taken with an Operetta® High-Content Imaging System with 20-fold or 40-fold magnification. Images were analyzed using Harmony® High Content Imaging and Analysis Software.
Proximity Ligation Assay (PLA)
IMR-5 cells were plated in 384-well plates (Greiner), treated for 8 h with the indicated inhibitors and fixed with methanol for 20 min. After blocking for 30 min with 5% BSA in PBS, cells were incubated over night at 4 °C with primary antibody against PCNA (rabbit, ab92552, Abcam, 1:1,000) and total RNA Polymerase II (mouse, F12, Santa Cruz, 1:1,000). PLA was performed using Duolink® In Situ Kit (Sigma-Aldrich) according to the manufacturer’s protocol. Nuclei were counterstained using Hoechst 33342 (Sigma-Aldrich). Pictures were taken with an Operetta® High-Content Imaging System with 40-fold magnification and analyzed using Harmony® High Content Imaging and Analysis Software. Sixteen image fields per well were acquired with a total of at least 1,000 cells per sample.
Quantitative image-based cytometry (QIBC)
Asynchronously growing MYCN-amplified NGP neuroblastoma cells were grown on sterile 12 mm glass coverslips until they reached a cell density of 70 to 90%. Following an EdU pulse of 20 min, cells were fixed in 4% formaldehyde for 15 min, washed once in PBS, permeabilized for 5 min in 0.2% Triton X-100 (Sigma-Aldrich) in PBS, washed twice with PBS and incubated in blocking solution (filtered DMEM containing 10% FBS and 0.02% sodium azide) for 15 min. EdU Click-iT reactions were performed prior to primary antibody incubations according to manufacturer’s recommendations (Thermo Fisher Scientific). Primary antibodies (γH2AX (mouse, 613402, BioLegend, 1:1,000); pKAP1 S824 (rabbit, ab70369, Abcam, 1:500); pH3S10 (rabbit, ab5176, Abcam, 1:2,000)) were diluted in blocking solution and incubated at RT for 2 h. Secondary antibodies (Alexa Fluor 488, 568, and 647 anti-mouse and anti-rabbit IgG from Thermo Fisher Scientific) were diluted 1:500 in 5% BSA in PBS and incubated at RT for 1 h. Cells were washed once with PBS and incubated for 10 min with 4’,6-Diamidino-2-Phenylindole Dihydrochloride (DAPI, 0.5 μg/ml) in PBS at RT. Following three washing steps in PBS, coverslips were briefly washed with distilled water and mounted on 5 μl Mowiol-based mounting media (Mowiol 4.88 (Calbiochem) in glycerol/TRIS).
Automated multichannel wide-field microscopy for high-content imaging and quantitative image-based cytometry (QIBC) was performed using the Olympus ScanR System as described previously61. Images were analyzed with the inbuilt Olympus ScanR Image Analysis Software Version 3.0.1, a dynamic background correction was applied, and nuclear segmentation was performed using an integrated intensity-based object detection module based on the DAPI signal. All downstream analyses were focused on properly detected cell nuclei containing a 2C-4C DNA content as measured by total and mean DAPI intensities. Fluorescence intensities were quantified and were depicted as arbitrary units. Within one experiment, similar cell numbers were compared for the different conditions and representative single cell scatter plots are shown. Fluorescence intensities were quantified and are depicted as arbitrary units. Color-coded scatter plots of asynchronous cell populations as well as statistical analysis were generated using R.
Immunoblots and immunoprecipitations
Whole cell extracts were prepared using RIPA buffer (50 mM HEPES, 140 mM NaCl, 1 mM EDTA; 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich). Lysates were cleared by centrifugation and protein concentrations were determined by Bradford or BCA-Assay.
For fractionation, cells were treated as indicated, washed with TBS containing protease and phosphatase inhibitor cocktails (Sigma-Aldrich) and harvested by centrifugation (300 g, 20 min, 4 °C). Lysis was carried out in sucrose buffer I (10 mM HEPES pH 7.9, 0.34 M sucrose, 3 mM CaCl2, 2 mM magnesium acetate, 0.1 mM EDTA, 0.5% NP-40) for 20 min at 4 °C with rotation. Nuclei were pelleted (3,900 g, 20 min, 4 °C) and washed once with sucrose buffer I without NP-40. Lysis of nuclei was carried out in nucleoplasmic lysis buffer (20 mM HEPES pH 7.9, 3 mM EDTA, 10% glycerol, 150 mM potassium acetate, 1.5 mM MgCl2) by performing 20 strokes with a dounce-homogenizer on ice. Lysed nuclei were incubated 30 min on ice and homogenized with 30 strokes. Benzonase (25 units, Merck) was added and incubated for 1 h at 16 °C. Unsolubilized chromatin was pelleted by centrifugation (18,000 g, 30 min 4 °C), resuspended in Lämmli buffer containing Benzonase (5 units) to release chromatin bound proteins, incubated at RT for 1 h and heated for 15 min at 95 °C.
Protein samples were separated on Bis-Tris gels and transferred to a PVDF membrane (Millipore). Protein expression was analyzed by immunoblotting with the indicated primary antibodies listed in the Reporting Summary. Membranes were scanned and analyzed using a Licor Odyssey scanner and Image Studio (LI-COR Biosciences) or LAS4000 Mini Imaging system (Fuji).
For immunoblots showing multiple proteins with similar molecular weight, one representative loading control is shown. Vinculin or actin were used as loading control.
Flow cytometry analysis (FACS)
For PI-FACS cells were harvested by trypsinization, washed with cold PBS and fixed in 80% ethanol overnight at -20 °C. After washing with PBS, the cells were resuspended in PBS with RNase A (24 μg/ml) and propidium iodide (PI, 54 μM) and incubated for 30 min at 37 °C. For AnnexinV/PI-FACS, the supernatant of the respective culture was combined with cells harvested by trypsinization and washed with cold PBS. The cell pellet was re-suspended in 100 μl 1x AnnexinV-binding buffer (10 mM HEPES pH 7.4, 140 mM NaCl, 2.5 mM CaCl2) with AnnexinV/Pacific Blue dye and incubated for 15 min at RT in the dark. Afterwards 400 μl 1x binding buffer containing PI (54 μM) was added and the samples were stored cold and dark until analysis. Subsequent analysis of all cell cycle related FACS experiments was performed on a BD FACSCanto II flow cytometer using BD FACSDIVA™ Software.
Tumors were dissociated using Collagenase IV (3.2 mg/ml, Worthington # LS004209), Deoxyribonuclease I (1 mg/ml, Worthington # LS002007), Soybean trypsin inhibitor (2 mg/ml, Worthington #LS003587) in Ca/Mg-free PBS, and were incubated with CD45 PE/Cy7 (109830, Biolegend), Live-dead marker (L34975, Invitrogen) and F4/80 block (743282, BDBioscience). FACS experiments of tumor samples were performed on a BDFACSAria ll cytometer.
DNA-RNA-Immunoprecipitation (DRIP)
DRIP was performed as described62. Briefly, cells were digested with 0.5% SDS and proteinase K overnight. DNA was extracted with phenol/chloroform and precipitated with ethanol. DNA was digested using a cocktail of restriction enzymes (Bsrg1, EcoR1, HindIII, SspI, XbaI (NEB)) overnight at 37 °C. For RNaseH-treated sample DNA was additionally incubated with RNaseH1 (NEB) overnight. DNA was purified as described above. S9.6 antibody (Merck, MABE1095 or Absolute, Ab01137-2.0), which detects RNA/DNA hybrids63, was coupled to A/G-Dynabeads® (Invitrogen). DNA in 1 x binding buffer (10 mM NaPO4 pH 7.0, 140 mM NaCl, 0.05% Triton X-100) was added to the antibody-coupled beads overnight. After extensive washing, DNA was eluted with elution buffer (50 mM Tris-HCl pH 8.0, 10 mM EDTA, 0.5% SDS) and treated for 2 h at 45 °C with proteinase K. After DNA extraction, locus-specific DRIP signals were assessed by RT-PCR (for primers see section below).
High-throughput sequencing
ChIP and ChIP-sequencing was performed as described previously4. For spike-in experiments (ChIP-Rx) 10% of fixed NIH-3T3 mouse cell lines were added before lysis. Cells were treated with 1% formaldehyde for 5 min at RT following 5 min of incubation with glycine. After cell lysis (5 mM PIPES pH 8.8, 5 mM KCl, 0.5% NP40), nuclei were resuspended in RIPA buffer (50 mM HEPES pH 7.9, 140 mM NaCl, 1% Triton-X-100, 0.1% deoxycholic acid (DOC), 0.1% SDS, 1 mM EDTA containing protease and phosphatase inhibitor cocktail) and DNA was fragmented to a size <500 bp using a Covaris M220 sonifier. Antibodies (total RNA-Polymerase II (mouse, Santa Cruz, A10), RNA-Polymerase pSer2 (rabbit, Abcam, ab5095), H3 (rabbit, Abcam, ab1791), pH3S10 (rabbit, Abcam, ab177218), H3.3 (rabbit, Abcam, ab176840)) were bound to Protein A/G-Dynabeads® (Invitrogen) and immunoprecipitated. After extensive washing, chromatin was eluted with 1% SDS and crosslinking was reverted overnight. Chloroform/phenol extraction was used for DNA purification. After DNA extraction occupancy of different proteins were assessed by RT-PCR. Primers were used for GBA (forward: AGCCCTTCCTCAAGTCTCAT; reverse: ACTGTGGGAATTCAATCGCC), EIF3B (forward: TGGGTGTGCTGTGAGTGTAG; reverse: ATGGACAATTCTGAGGGGCA), ACTB (forward: GAGGGGAGAGGGGGTAAA; reverse: AGCCATAAAAGGCAACTTTCG), TFAP4 (forward: CCGGGCGCTGTTTACTA; reverse: CAGGACACGGAGAACTACAG), RAN (forward: CCGTGACTCTGGGATCTTGA; reverse: CAAGGTGGCTGAAACGGAAA), POLG (forward: CTTCTCAAGGAGCAGGTGGA; reverse: TCATAACCTCCCTTCGACCG), NPM1 (forward: TTCACCGGGAAGCATGG; reverse: CACGCGAGGTAAGTCTACG), RPS16 (forward: CCGAGCGTGGACTAGACAA; reverse: GTTAGCCGCAACAGAAGCC), DRG2 (forward: CGTGGGCCAGTACAGCAT; reverse: CCGGAAGCCAAAGAGAACAG), Centrosome (forward: TCATTCCCACAAACTGCGTTG; reverse: TCCAACGAAGGCCACAAGA), Intergenic region (forward: TTTTCTCACATTGCCCCTGT; reverse: TCAATGCTGTACCAGGCAAA), NCL (forward: CTACCACCCTCATCTGAATCC; reverse: TTGTCTCGCTGGGAAAGG), RCC1 (forward: AGTGGTCGCTTCTTCTCCTT; reverse: GCATTAGACCCACAACTCCG), NME1 (forward: GGGGTGGAGAGAAGAAAGCA; reverse: TGGGAGTAGGCAGTCATTCT), PPRC1 (forward: GTGAGGATTAGCGCTTGGAG; reverse: TGCTGTACGTTCCTTTCACC), PTPN23 (forward: CCAGTCTCCGGTCAGTGATT; reverse: CGTATTGTCAAGAGCCGTGG), and PLD6 (forward: GCTGTGGGTCCCGGATTA; reverse: CCTCCAGAGTCAGAGCCA).
Shown analysis of RT-PCR show mean and standard deviation of technical triplicates as well as an overlay of each data point to indicate the distribution of the data.
ChIP sequencing was performed as described in64. Using the NEBNext® ChIP-Seq Library Prep Master Kit or the NEBNext® UltraII DNA Library Prep Kit for Illumina, purified DNA was end-repaired, A-tailed, ligated to Illumina adaptors, size-selected (200 bp) and purified with a gel extraction kit. DNA fragments were amplified by 15 to 18 cycles of PCR and library size and amount of library was specified with the Fragment Analyzer (Agilent).
RNA sequencing was performed as described previously65 using an Illumina NextSeq 500. RNA was extracted using RNeasy mini columns (Qiagen) including on-column DNase I digestion. mRNA was isolated using the NEBNext® Poly(A) mRNA Magnetic Isolation Module (NEB) and library preparation was performed with the NEBNext® Ultra™ RNA Library Prep Kit for Illumina following the instruction manual. Libraries were amplified with 12 PCR cycles and purified using Agencourt AMPure XP Beads (Beckman Coulter). Library quantification and size determination was performed with the Fragment Analyzer (Agilent).
For 4sU labelled nascent RNA sequencing, IMR-5 cells were cell cycle synchronized using double thymidine block. At timepoint of release indicated inhibitors were added. 2 h before harvest cells were incubated with 500 μM of 4sU (Sigma-Aldrich) in RPMI to label nascent RNA. After 15 min medium was changed. RNA was harvested using Qiagen miRNeasy kit. After extraction and quantification of total RNA by Nanodrop, an equal amount was labelled with biotin (Pierce) in presence of DMF-HPDP buffer. Free biotin removal was carried out by chloroform-isoamyl alcohol extraction after which RNA was resuspended into nuclease free water. Dynabeads™ MyOne™ Streptavidin C1 beads (Life Technologies) were used for enrichment of biotinylated RNA, which was then eluted by 100 mM DTT and cleaned by RNeasy MinElute cleanup kit. Nascent RNA concentration was then measured using RiboGreen RNA assay kit and equal amount was used for library preparation. Before library preparation, rRNA was depleted using NEBNext® rRNA Depletion Kit and then all eluted material was used for NEBNext® Ultra Directional kit with 14 PCR cycles.
All libraries were sequenced for 75 cycles using Illumina NextSeq 500 system. Following base calling with Illumina’s FASTQ Generation Software v1.0.0 high quality PF clusters were selected for further analysis.
DNA fiber assay
DNA fiber assays were carried out as previously described66. Newly synthesized DNA was labeled via treatment with 5-chloro-2-deoxyuridine (CldU, 25 μM, Sigma-Aldrich) for 20 min, followed by 5-iodo-2-deoxyuridine (IdU, 50 μM, Sigma-Aldrich) for 1 h, in the presence of inhibitors as indicated. Cells were lysed by spreading buffer (200 mM Tris pH 7.4, 50 mM EDTA, 0.5% SDS) and DNA fibers spread on glass slides prior to fixation in a methanol:acetic acid solution. After DNA denaturation by 2.5 M HCl, CldU- and IdU-labelled tracts were detected by immunostaining using mouse anti-BrdU (B44, BD), rat anti-BrdU (BU1/75, ICR1, Abcam) antibodies and Alexa Fluor 488-conjugated goat anti-mouse IgG, Alexa Fluor 555-conjugated goat anti-rat IgG (Thermofisher) as secondary antibodies. DNA fibers were visualized with fluorescence microscopy (Axio Scope A1 microscope, Zeiss) and analyzed with ImageJ. Statistical testing was performed using Graph Pad Prism v.6. Unpaired Student’s t-test was calculated with an assumed significance for p-values ≤ 0.05.
Animal experiments
All animal experiments with transgene TH-MYCN mice or tissue were approved by The Institute of Cancer Research Animal Welfare and Ethical Review Body and performed in accordance with the UK Home Office Animals (Scientific Procedures) Act 1986, the United Kingdom National Cancer Research Institute guidelines for the welfare of animals in cancer research and the ARRIVE (animal research: reporting in vivo experiments) guidelines.
Transgenic TH-MYCN mice were genotyped to detect the presence of human MYCN transgene47. The study was performed using both male and female homozygous mice, which developed palpable tumors at 35 to 45 days with a 100% penetrance. Tumor development was monitored weekly by palpation by an experienced animal technician. Mice with palpable tumors were then enrolled (day 0) in the study. The tumor volume was subsequently monitored by MRI at day 0 and 7. Mice were treated with 25 mg/kg or 30 mg/kg of AZD6738 (p.o., daily), 15 mg/kg MLN8237 (p.o., in a ‘five days on, two days off’ schedule) or vehicle control, to a maximum of 56 doses. MLN8237 was dissolved in 10% (2-Hydroxypropyl)-β-cyclodextrin,1% Sodium hydrogen carbonate and AZD6738 was dissolved in 10% DMSO, 40% propylene glycol, 50% water. Mice were allowed access to sterile food and water ad libitum.
For combination studies of MLN8237 with PARP inhibitor, Olaparib was dosed at 45 mg/kg (p.o., daily for 5 days). Prior to dosing, Olaparib was made fresh from stock solution (50 mg/ml in DMSO) and diluted with 10% 2-hydroxy-propyl-beta-cyclodextrin [HPCD]. Tumors were collected 2 hours after last dose on Day 5.
MR images were acquired on a 1 Tesla M3 small animal MRI scanner (Aspect Imaging, Shoham, Israel) or a 7 Tesla Bruker MRI. Mice were anaesthetized using isoflurane delivered via oxygen gas and their core temperature was maintained at 37 °C. Anatomical fat-suppressed T2-weighted coronal images (TE=9 ms (1T) or 36 ms (7T), TR=4500 ms) were acquired from 20 contiguous 1-mm-thick slices through the mouse abdomen. Tumor volumes were determined using segmentation from regions of interest drawn on each tumor-containing slice using Horos medical image viewer.
One million tumor cells from a TH-MYCN tumor were injected subcutaneously into the right flank of immunocompetent 129SvJ mice, immunodeficient NSG or nude mice (female, 6 weeks old) to established murine allograft model. Mice bearing allografts with a mean diameter of 4-5 mm were treated as in the TH-MYCN model. Studies were terminated when the mean diameter of the tumor reached 15 mm. Tumor volumes were measured by Vernier caliper across two perpendicular diameters, and volumes were calculated according to the formula: V = 4/3π [(d1 + d2) / 4]3.
All experiments with patient derived xenografts were conducted according to the institutional animal protocols and the national laws and regulations. The experiments were conducted as previously described in four replicates67. In short, NOD.Cg-Prkdcscd Il2rgtm1Sug/JicTac mice (Taconic) or Rj:NMRI-Foxn1 nu/nu (Janvier labs) mice (female, 6-10 weeks old) were used to perform all patient-derived xenograft experiments. Prior to the experiments, patient tumors were serially transplanted in mice at least three times. Caliper measurement was used to monitor tumor growth. Tumor volume was calculated with the formula length x width2 / 2. Mice were sacrificed with cervical dislocation when tumor size exceeded 2,000 mm3. Drugs were dissolved in DMSO/Tween/0.9%NaCl. Mice were treated with 50 mg/kg of AZD6738 (p.o., daily), 7.5 mg/kg MLN8237 (p.o., in a ‘five days on, two days off’ schedule), the combination or vehicle control for two weeks.
All mice were housed in pathogen-free barrier conditions under 12 h light-dark cycles and with temperature and humidity set points at 20–25 °C and 30–70%, respectively.
Immunohistochemistry
For immunohistochemical analysis, samples were embedded in paraffin and sectioned at 6 μm using a microtome (Leica). Sections were deparaffinized, re-hydrated, and subjected to high-temperature antigen retrieval at pH 9.0 for pKAP1 or pH 6.0 for all other stainings. Sections were washed, and species-appropriate secondary antibody reagents were applied (rabbit: Super Boost goat anti-rabbit poly HRP, mouse: MOM Kit, rat: Vectastatin Elite ABC Peroxidase Kit). HRP-conjugated secondary antibodies were visualized by DAB staining (Vector Laboratories). Stainings were recorded using Panoramic Desk scanner and analyzed using Case Viewer software (3D HISTECH). For scoring of tumor sections at least five pictures per tumor section were extracted and blindly scored according to a scale from 0-3.
Bioinformatical analysis and statistics
All bioinformatical analysis were done with commercial tools. Base calling was performed using Illumina’s FASTQ Generation software v1.0.0 and sequencing quality was tested using the FastQC script. For ChIP-sequencing, reads were mapped independently to the human hg19 and murine mm10 (spike-in) genome using Bowtie168 with default parameters. A spike-in normalization factor was calculated by dividing the number of mapped reads of the spike-in of the smallest sample by the number of mapped reads of the spike-in for each sample. For each sample, this factor was multiplied by the number of reads that map to the human genome and all bam files for subsequent analysis were adjusted to this read count. SAMtools69 were used for manipulating bam-file (indexing, subsampling and generation of bedgraph-files). Peak calling was performed using MACS1470 and bedgraph files were generated using the genomecov function from BEDtools.
Traveling ratios for RNAPII ChIP-seq were calculated by counting reads with BEDtools “intersectBed”71 around the TSS (-30 to +300 bp) and within gene bodies (+300 bp to TES) of Ensembl genes. Read density graphs were obtained using the computeMatrix function from DeepTools72. Gene body counts were normalized to the length of the gene and TSS counts were divided by gene body counts. Metagene window plots were generated with ngs.plot.r73. The shaded area corresponds to standard errors. First exon/intron boundaries were extracted from the GRCh37 annotation and nucleosome coordinates from the published data sets GSM1838910 and GSM183891174. Pause sites and MYCN-activated and repressed genes were as defined in Herold et al. (2019). Ideogram showing distribution of ChIP-seq data on chromosomes were visualized using RIdeogram75.
For mRNA-sequencing, reads were mapped to hg19 using Tophat2 76 and Bowtie2 68 and samples were normalized to the number of mapped reads in the smallest sample. Reads per gene were counted using the “summarizeOverlaps” function from the R package “GenomicAlignments” using the “union”-mode and Ensembl genes. Non- and weakly expressed genes were removed (mean count over all samples <1). Differentially expressed genes were called with edgeR and p-values were adjusted for multiple-testing using the Benjamini-Höchberg procedure. Gene set enrichment analysis (GSEA)51 were done with the “Hallmark” databases from MSigDB77, 1,000 permutations and default settings. Browser tracks were created using Integrated Genome Browser.
4sU-sequencing analysis was performed as previously described32. To determine the effect of drug treatment on splicing efficiency, all experimental repeats for each condition were combined, and for each gene the fraction of spliced reads relative to total reads was calculated. Each experimental condition (drug treatment) was then compared to the DMSO control using both t test and Wilcoxon matched-pairs signed rank test in GraphPad Prism.
To determine the effect of MLN8237 treatment on total and pSer2 RNA Pol2 distribution over the downstream pause site (n=7,760) or the first exon-intron boundary of expressed genes (n=64,764), spike-normalized samples were processed with bedtools intersect to count the number of reads over a 400-nt window centered on the downstream pause site or the exon-intron boundary, respectively. The corresponding MLN8237- and DMSO-samples were then compared to each other using the Mann-Whitney test in GraphPad Prism. To stratify genes for affinity to MYCN, MYCN ChIPseq reads from S phase synchronized and DMSO treated IMR-5 cells were counted in a 600 bp window centered at the TSS using BEDtools intersect. Lists of downstream pause sites belonging to the top 3,000 or bottom 3,000 MYCN bound genes were obtained by intersecting the corresponding lists, restricted to genes with minimal expression in IMR-5 cells.
In box plots central line shows median and the borders of the boxes show the interquartile range of the plotted data. The whiskers extend to 1.5 x the interquartile range and outliers are shown as dots.
Bioinformatical analysis and statistics
Information on statistical tests used, numbers of samples, definitions of error bars and statistical measures displayed in the graphs are provided in the caption of the figure.
Sample size depended on experiments. Microscopy experiment aimed to acquire at least 45 cells per conditions. Fiber assay aimed to record at least 100 fibers per condition. Analysis of this number of cells or fibers was sufficient to obtain normal distribution of the data and reliable mean.
All animal experiments were done with at least 3 animals per condition. Animals were randomized for treatment to ensure each group has the same starting point. For evaluation of immunohistochemistry pictures were blindly scored by several independent people.
No data were excluded from the analyses. Statistical tests were performed using Prism (GraphPad) or R.
Extended Data
Extended Data Figure 1. Aurora-A phosphorylates H3S10 in S phase.
a. Propidium-Iodide stained FACS profiles of IMR-5 cells synchronized at the G1/S boundary by a double-thymidine block (T0). Asynchronous cells are shown as controls. Cells were released from the block for 4 h into S phase, for 8 h into G2/M phase and for 14 h into G1 phase Data representative of 3 independent experiments with similar results.
b. Fractionation of synchronized MYCN-amplified IMR-5 neuroblastoma cells. (Left): Immunoblots of equal aliquots of each fraction were probed for the indicated proteins. Data representative of 3 independent experiments with similar results. (Right): Quantitation of Aurora-A levels in each cell cycle phase. Shown is mean ± S.D. (n=3 independent experiments).
c. MYCN ChIP at indicated loci in asynchronous IMR-5 cells treated for 4 h with 10058-F4 (100 μM). IgG control was used as control for antibody specificity. Data are presented as mean of technical triplicates. Data representative of 3 independent experiments with similar results.
d. Immunoblots of indicated proteins of IMR-5 cells either asynchronous, synchronized by double thymidine block into S or G2 phase or synchronized into mitosis by incubation with nocodazole for 16 h. Data representative of 2 independent experiments with similar results.
e. Radar blots documenting specificity of Aurora kinase inhibitors: MLN8237, MK5108, AZD1152; data are from22.
f. Quantification of pH3S10 staining in IMR-5 cells treated for 8 h with MLN8237 (1 μM) relative to control (DMSO) cells; each grey dot represents a cell. In S and G2 phase number of spots and in mitotic cells intensity of pH3S10 signal relative to DMSO is shown. Data are presented as mean ± S.D. (n≥614 cells per condition examined over 3 independent experiments).
g. Quantification of pH3S10 staining of IMR-5 cells in S phase treated for 8 h with 100 nM MLN8237, 1 μM MK5108 or DMSO as control. Each dot represents one cell. Data are presented as mean ± S.D. (n≥147 cells per condition examined over 3 independent experiments).
h. Quantification of pH3T3 intensity in IMR-5 cells treated for 8 h with 100 nM MLN8237, 100 nM AZD1152 or DMSO as control; each grey dot represents a cell. Shown is the mean ± S.D. (n≥47 cells per condition examined over 3 independent experiments).
i. Representative example pictures of 3 independent experiments with similar results. White line indicates 15 μm.
Extended Data Figure 2. Effects of Aurora-A inhibition on H3.3 and pH3S10.
a. (Top): Ideogram illustrating the 50 bins with the strongest reduction in pH3S10 MLN8237 as compared to pH3S10 DMSO (blue rectangles). (Bottom): Levels of pH3S10 relative to total H3 along all chromosomes of S phase-synchronized IMR-5 cells treated for 4 h with 1 μM MLN8237 (red line) and DMSO (blue line) (n=2 independent experiments).
b. pH3S10 ChIP at indicated loci in S phase synchronized IMR-5 cells treated for 4 h with 1 μM MLN8237. IgG control was used as control for antibody specificity. Data are presented as mean of technical triplicates Data representative of 3 independent experiments with similar results
c. Metagene plot of ChIP-Rx signal for histone H3.3 and pH3S10 in S phase-synchronized IMR-5 cells treated for 4 h with MLN8237 (1 μM) or DMSO. The graph is centered on the first nucleosome (“+1 dyad”) downstream of the TSS for N=14,340 genes. (n=2 independent replicates).
d. Browser tracks of MYCN, total RNAPII, H3.3 and pH3S10 of ChIP-Rx at a MYCN high (left) and low (right) bound locus. Nucleosome-free zone is indicated in grey.
e. Metagene plot of ChIP-Rx signal for histone H3 and histone H3.3 in S phase-synchronized IMR-5 cells treated for 4 h with 1 μM MLN8237 or DMSO. The signal is centered on the first nucleosome (“+1 dyad”) located downstream of the TSS for N=14,340 genes (n=3 independent experiments).
f. Immunoblot of asynchronous SH-EP and SH-EP MYCN cells. Vinculin was used as loading control. Data representative of 3 independent experiments with similar results.
Extended Data Figure 3. Aurora-A impacts RNAPII, splicing and R loop formation in S phase.
a. Metagene plot of all expressed genes (N=17,533) illustrating distribution of ChIP-Rx signal of total RNAPII within transcribed regions in S phase-synchronized IMR-5 cells treated for 4 h with 1 μM MLN8237, 1 μM MK5108 or solvent control (DMSO) (n=2 independent experiments).
b. 2D Kernel density plot showing RNAPII traveling ratio in S phase-synchronized cells treated for 4 h with MLN8237 (1 μM) and in control (DMSO) cells (n=2 independent experiments).
c. Metagene plot of ChIP-Rx signal for RNAPII pSer2 in S phase-synchronized IMR-5 cells treated for 2 h with MLN8237 (1 μM), MK5108 (1 μM) or DMSO. The signal is centered on the first nucleosome (“+1 dyad”) located within 300 nt downstream of the TSS for n=14,340 genes (n=2 independent replicates).
d. Browser tracks of MYCN, total RNAPII and RNAPII pSer2 of ChIP-Rx at FASN gene.
e. Immunoblots of IMR-5 cells that were synchronized for the indicated cell cycle phase and treated with Pladienolide B (PlaB;1 μM), MLN8237 (1 μM), MK5108 (1 μM) or DMSO (4 h). Actin was used as loading control. Data representative of 3 independent experiments with similar results.
f. Definition of read categories; blue lines show where reads were mapped to; grey categories (exonic, spliced) reads represent mature mRNA, blue reads categories (exon-intron, intronic) represents non-spliced pre-mRNA, red categories (TSS, TES, TES-RT, intergenic) represents RNA without coding sequence. TSS: Transcription start site, TES: transcription end site.
g. Diagram showing the setup of the 4sU-sequencing experiments in S phase-synchronized IMR-5 cells.
h. Mean percentage of reads recovered in each category described in (f) (n=3 independent experiments). All treatments (PlaB, MK, MLN) significantly reduce the percentage of spliced reads relative to the “DMSO” control (p<1.0e-15 using paired two-tailed t test and Wilcoxon matched-pairs signed rank test).
i. DRIP using S9.6 antibody after 8 h of 1 μM MLN8237 or 1 μM PlaB treatment. Incubation with RNaseH1 and IgG were used as controls. Shown is the mean of technical triplicates. Data representative of 2 independent experiments with similar results.
j. (Left) DRIP using S9.6 antibody after 48 h of siAurora-A treatment. Shown is the mean of technical triplicates. (Right) Immunoblot of siRNA-transfected IMR-5 cells. Vinculin was used as loading control. Data representative of 2 independent experiments with similar results.
Extended Data Figure 4. Characterization of DNA replication and R-loop formation.
a. Quantification of fork progression of SH-EP and SH-EP MYCN cells pretreated for 3 h with 100 nM or 1 μM MLN8237, 1 μM of AZD6738 or a combination (100 nM MLN8237 and 1 μM AZD6738). P-values were calculated using unpaired two-tailed t-test comparing two conditions (n≥168 fibers per condition were examined over 2 independent experiments).
b. Annexin-V/PI FACS of IMR-5 or IMR-5 cells with inducible Aurora-A WT and T217D. After pre-treatment for 24 h with doxycycline to induce Aurora-A WT and T217D, cells were treated for 48 h with MLN8237 (100 nM), AZD6738 (1 μM) or both. Shown is the mean ± S.D. (n=3 independent experiments).
c. Annexin-V/PI FACS of IMR-5 treated for 48 h with MLN8237 (100 nM), CHIR-124 (1 μM) or both. Shown is the mean ± S.D. (n=3 independent experiments).
d. DNA-RNA-Immunoprecipitation (DRIP) using S9.6 antibody of IMR-5 cells expressing a doxycycline inducible HA-RNaseH1. After 24 h doxycycline treatment, cells were treated for 8 h with 1 μM MLN8237. Incubation with RNaseH1 and IgG were used as controls for non-specific chromatin binding. Shown is the mean of technical triplicates. Data representative of 2 independent experiments with similar results. Insert shows expression of HA-RNaseH1 in asynchronous IMR-5 cells upon doxycycline treatment for 24 h. Vinculin (VCL) was used as loading control. Data representative of 3 independent experiments with similar results.
e. Metagene plot of ChIP-Rx signal for total RNAPII and H3.3 in S phase-synchronized RNaseH1-IMR-5 cells treated for 24 h with doxycycline to induce RNaseH1 expression or EtOH (as control). Data show mean for N=3,731 genes. The signal is centered on the first nucleosome (“+1 dyad”) located within 300 nt downstream of the TSS (n=2 independent experiments).
f. Browser tracks of MYCN and total RNAPII of ChIP-Rx in (e) at FASN and NCL gene.
g. Metagene plot of ChIP-Rx signal for RNAPII pSer2 in S phase-synchronized RNaseH1-IMR-5 cells treated for 24 h with doxycycline (to induce RNaseH1 expression) or EtOH (as control). Data shown for n=14,340 genes. The signal is centered on the first nucleosome ("+1 dyad") located within 300 nt downstream of the TSS for 14,340 genes (n=2 independent experiments).
h. Quantification of PLA signals between RNAPII and PCNA in asynchronous RNaseH1-IMR-5 cells treated for 24 h with doxycycline to induce RNaseH1 expression. Each dot represents mean PLA signal of all cells in one well compared to solvent control. Shown is the mean ± S.D.. P-value was calculated using unpaired two-tailed t-test relative to DMSO control (n=3 independent experiments).
i. Boxplot showing intensity of pKAP1 staining in mitotic cells upon the treatment with the indicated drugs (8 h) and induction of RNaseH1 expression for 24 h. P-value was calculated using two-tailed t-test between EtOH and Dox of each condition (n≥50 cells per condition were examined over 2 independent experiment).
Extended Data Figure 5. Treatment is not toxic and tumor specific.
a. Boxplot showing the concentration of indicated inhibitors in the tumor tissue of mice treated for 24 h or 5 days. N=4 mice (control, 24 h treatment), 5 mice (5 days treatment).
b. Relative changes in body weight of mice treated with MLN8237 and AZD6738 compared to mice treated with vehicle control. Shown is the mean ± S.D. N=42 mice (control), 43 mice (treated).
c. Staining of tumor tissue with S9.6 incubated with different RNases to document specificity of S9.6. staining for R-loops (n=1 section for each experimental condition).
d. Histology of proliferative gut tissue in untreated (top) and treated (bottom) mice showing H&E, Ki-67 and cleaved caspase 3 staining. N=2 mice (control), 3 mice treated).
e. Representative MRI as well as sections of a tumor treated with a combination of MLN8237 and olaparib (N=3 animals were evaluated).
f. MRI sections of the two long term-surviving mice at day 0 and after 7 days of treatment with the combination of AZD6738 (30 mg/kg) and MLN8237 (15 mg/kg). Dashed white lines indicate tumor circumference.
g. P-values calculated using Mantel-Cox log-rank test comparing the survival of different groups shown in Figure 6c.
Extended Data Figure 6. Therapeutic efficacy in Patient-derived xenograft models.
a. Histology of representative tumor sections of MYCN-amplified PDX models treated as indicated. AZD6738 was administered at 50 mg/kg every day and MLN8237 at 7.5 mg/kg on a 5 days on, 2 days off schedule. Shown are stainings from tumors recovered after treatment for 14 days. Each group comprises three animals.
b. Box plot showing quantification of R-loop-, γH2AX- and cleaved caspase 3-positive cells in tumor sections. P-values were calculated using unpaired two-tailed t-test using Welch’s correction (n≥9 sections from animals described in panel (a) were evaluated).
c. Relative changes in tumor volume of four MYCN non-amplified PDX models upon treatment with the indicated inhibitors. Shown is the mean ± S.E.M. N indicates the animal number for each experimental cohort. P-values comparing control and combination (indicated with a black dashed line) were calculated using unpaired two-sided t-test.
Extended Data Figure 7. Aurora-A/ATR inhibition engages the immune system.
a. GSEA signatures showing response of a hallmark gene set indicating Interferon gamma response in the TH-MYCN (top) and a PDX (bottom) model.
b. (Top): Representative pictures by flow cytometry showing cell gating. (Bottom): Populations of CD45+ immune cells in the tumor microenvironment after treatment of TH-MYCN mice with the combination of AZD6738 (25 mg/kg) and MLN8237. Shown is the mean ± S.E.M.. Significance was calculated using unpaired two-tailed t-test (N=4 animals from each condition were evaluated).
c. Histology of representative tumor sections showing NKp46-positive cells and pSTAT1 in tumors of TH-MYCN mice treated with combined Aurora-A/ATR inhibition. N=4 mice (control, 24 h treatment), 5 mice (5 days treatment).
d. Relative changes in tumor volume of subcutaneous xenografts in nude mice after treatment with vehicle or the combination of MLN8237 and AZD6738 (25 mg/kg). Shown is the mean ± S.E.M. (N=5 animals per group).
e. Bar graph showing the survival of allograft mice treated with MLN8237 and AZD6738. Shown is the mean ± S.E.M. (N=4 animals per group).
Acknowledgements
This work was supported by grants from the European Research council (AuroMYC), the German Cancer Aid (Enable), the Federal Ministry of Education and Research (DKTK), and the German Research Foundation via the DFG Research Group 2314 to M.E. and the German Cancer Aid via the Mildred Scheel Early Career Center to G.B.. Cancer Research UK support the Cancer Imaging Centre at ICR, in association with the MRC and Department of Health (England) (C1060/A16464) and a Children with Cancer UK Research Fellowship (Y.J.). A.G.H. is supported by the German Research Foundation and the Wilhelm Sander Stiftung and participates in the BIH-Charité Clinical Scientist Program. E. P. and L. C. received Children with Cancer UK Project Grant (2014/174). L. C. received Cancer Research UK Program Grants (C34648/A18339 and C34648/A14610). R.B. received Cancer Research UK Programme Award (C24461/A23302). We thank Barbara Bauer for technical help with immunohistochemistry, Andreas Schlosser for proteomic analyses, Werner Schmitz for mass spectrometry, Barbara Martins da Costa and Jana Rolff for help with animal experiments, Stefanie Heinzlmayr and Bernhard Küster for providing the graphics on Aurora-A inhibitors, and members of the Eilers laboratory for evaluating immunohistochemical images.
Footnotes
Author Contributions
I.R. and G.B. performed most experiments, E.P. and Y.J. performed all in vivo experiments and MRI measurements in TH-MYCN mice, H.D.G. analyzed in vivo experiments in PDX models, M.G. performed high content microscopy experiments, C.G. performed fiber assays, C.S.-V. analyzed immunofluorescence experiments, M.R. performed in vitro kinase assays. P.G. analyzed ChIP and RNA sequencing data, M.R. evaluated the pathology, C.P.A. measured high-throughput data, P.B. analyzed proteomic data, G.B., E. P., A.G.H., M.A., M.D., R.B., A.E., J.A, L.C and M.E. devised and supervised experiments and G.B., L.C. and M.E. wrote the paper.
Conflict of Interest
The authors declare no competing interests.
Data Availability
NGS data is available at the Gene Expression Omnibus under the accession number GSE144288. Source data for all Figures and Extended Data Figures have been provided as Source Data files. All other data supporting the findings of the study are available from the corresponding authors on request.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149:22–35. doi: 10.1016/j.cell.2012.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Soucek L, et al. Modelling Myc inhibition as a cancer therapy. Nature. 2008;455:679–683. doi: 10.1038/nature07260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rickman DS, Schulte JH, Eilers M. The Expanding World of N-MYC-Driven Tumors. Cancer Discov. 2018;8:150–163. doi: 10.1158/2159-8290.CD-17-0273. [DOI] [PubMed] [Google Scholar]
- 4.Walz S, et al. Activation and repression by oncogenic MYC shape tumour-specific gene expression profiles. Nature. 2014;511:483–487. doi: 10.1038/nature13473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sabo A, et al. Selective transcriptional regulation by Myc in cellular growth control and lymphomagenesis. Nature. 2014;511:488–492. doi: 10.1038/nature13537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nie Z, et al. c-Myc Is a Universal Amplifier of Expressed Genes in Lymphocytes and Embryonic Stem Cells. Cell. 2012;151:68–79. doi: 10.1016/j.cell.2012.08.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin CY, et al. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Herold S, et al. Recruitment of BRCA1 limits MYCN-driven accumulation of stalled RNA polymerase. Nature. 2019;567:545–549. doi: 10.1038/s41586-019-1030-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zeid R, et al. Enhancer invasion shapes MYCN-dependent transcriptional amplification in neuroblastoma. Nature genetics. 2018;50:515–523. doi: 10.1038/s41588-018-0044-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baluapuri A, Wolf E, Eilers M. Target gene-independent functions of MYC oncoproteins. Nature reviews. 2020;21:255–267. doi: 10.1038/s41580-020-0215-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto T, et al. Stabilization of N-Myc is a critical function of Aurora A in human neuroblastoma. Cancer cell. 2009;15:67–78. doi: 10.1016/j.ccr.2008.12.005. [DOI] [PubMed] [Google Scholar]
- 12.Brockmann M, et al. Small molecule inhibitors of aurora-a induce proteasomal degradation of N-myc in childhood neuroblastoma. Cancer cell. 2013;24:75–89. doi: 10.1016/j.ccr.2013.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gustafson WC, et al. Drugging MYCN through an allosteric transition in Aurora kinase A. Cancer cell. 2014;26:414–427. doi: 10.1016/j.ccr.2014.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richards MW, et al. Structural basis of N-Myc binding by Aurora-A and its destabilization by kinase inhibitors. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:13726–13731. doi: 10.1073/pnas.1610626113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buchel G, et al. Association with Aurora-A Controls N-MYC-Dependent Promoter Escape and Pause Release of RNA Polymerase II during the Cell Cycle. Cell reports. 2017;21:3483–3497. doi: 10.1016/j.celrep.2017.11.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kufer TA, et al. Human TPX2 is required for targeting Aurora-A kinase to the spindle. J Cell Biol. 2002;158:617–623. doi: 10.1083/jcb.200204155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, et al. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- 18.Zirath H, et al. MYC inhibition induces metabolic changes leading to accumulation of lipid droplets in tumor cells. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:10258–10263. doi: 10.1073/pnas.1222404110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mathsyaraja H, et al. Max deletion destabilizes MYC protein and abrogates Emicro-Myc lymphomagenesis. Genes & development. 2019 doi: 10.1101/gad.325878.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng K, Bastos RN, Barr FA, Gruneberg U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol. 2010;191:1315–1332. doi: 10.1083/jcb.201008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim SR, Kim KB, Chae YC, Park JW, Seo SB. H3S10 phosphorylation-mediated transcriptional regulation by Aurora kinase A. Biochem Biophys Res Commun. 2016;469:22–28. doi: 10.1016/j.bbrc.2015.11.063. [DOI] [PubMed] [Google Scholar]
- 22.Klaeger S, et al. The target landscape of clinical kinase drugs. Science. 2017;358 doi: 10.1126/science.aan4368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Manfredi MG, et al. Characterization of Alisertib (MLN8237), an Investigational Small-Molecule Inhibitor of Aurora A Kinase Using Novel In Vivo Pharmacodynamic Assays. Clin Cancer Res. 2011;17:7614–7624. doi: 10.1158/1078-0432.CCR-11-1536. [DOI] [PubMed] [Google Scholar]
- 24.Shimomura T, et al. MK-5108, a highly selective Aurora-A kinase inhibitor, shows antitumor activity alone and in combination with docetaxel. Mol Cancer Ther. 2010;9:157–166. doi: 10.1158/1535-7163.MCT-09-0609. [DOI] [PubMed] [Google Scholar]
- 25.Shannon KB, Salmon ED. Chromosome dynamics: new light on Aurora B kinase function. Curr Biol. 2002;12:R458–460. doi: 10.1016/s0960-9822(02)00945-4. [DOI] [PubMed] [Google Scholar]
- 26.Wilkinson RW, et al. AZD1152, a selective inhibitor of Aurora B kinase, inhibits human tumor xenograft growth by inducing apoptosis. Clin Cancer Res. 2007;13:3682–3688. doi: 10.1158/1078-0432.CCR-06-2979. [DOI] [PubMed] [Google Scholar]
- 27.Wang F, et al. Histone H3 Thr-3 phosphorylation by Haspin positions Aurora B at centromeres in mitosis. Science. 2010;330:231–235. doi: 10.1126/science.1189435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zeineldin M, et al. MYCN amplification and ATRX mutations are incompatible in neuroblastoma. Nat Commun. 2020;11:913. doi: 10.1038/s41467-020-14682-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nakatani Y, Ray-Gallet D, Quivy JP, Tagami H, Almouzni G. Two distinct nucleosome assembly pathways: dependent or independent of DNA synthesis promoted by histone H3.1 and h3.3 complexes. Cold Spring Harb Symp Quant Biol. 2004;69:273–280. doi: 10.1101/sqb.2004.69.273. [DOI] [PubMed] [Google Scholar]
- 30.Ivaldi MS, Karam CS, Corces VG. Phosphorylation of histone H3 at Ser10 facilitates RNA polymerase II release from promoter-proximal pausing in Drosophila. Genes & development. 2007;21:2818–2831. doi: 10.1101/gad.1604007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore MJ, Wang Q, Kennedy CJ, Silver PA. An alternative splicing network links cell-cycle control to apoptosis. Cell. 2010;142:625–636. doi: 10.1016/j.cell.2010.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cossa G, et al. Localized Inhibition of Protein Phosphatase 1 by NUAK1 Promotes Spliceosome Activity and Reveals a MYC-Sensitive Feedback Control of Transcription. Mol Cell. 2020;77:1322–1339 e1311. doi: 10.1016/j.molcel.2020.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Girard C, et al. Post-transcriptional spliceosomes are retained in nuclear speckles until splicing completion. Nat Commun. 2012;3:994. doi: 10.1038/ncomms1998. [DOI] [PubMed] [Google Scholar]
- 34.Castellano-Pozo M, et al. R loops are linked to histone H3 S10 phosphorylation and chromatin condensation. Mol Cell. 2013;52:583–590. doi: 10.1016/j.molcel.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 35.Hamperl S, Bocek MJ, Saldivar JC, Swigut T, Cimprich KA. Transcription-Replication Conflict Orientation Modulates R-Loop Levels and Activates Distinct DNA Damage Responses. Cell. 2017;170:e719. doi: 10.1016/j.cell.2017.07.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nature reviews. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shiotani B, et al. Two distinct modes of ATR activation orchestrated by Rad17 and Nbs1. Cell reports. 2013;3:1651–1662. doi: 10.1016/j.celrep.2013.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Foote KM, et al. Discovery and Characterization of AZD6738, a Potent Inhibitor of Ataxia Telangiectasia Mutated and Rad3 Related (ATR) Kinase with Application as an Anticancer Agent. Journal of Medicinal Chemistry. 2018;61:9889–9907. doi: 10.1021/acs.jmedchem.8b01187. [DOI] [PubMed] [Google Scholar]
- 39.Lecona E, Fernandez-Capetillo O. Targeting ATR in cancer. Nat Rev Cancer. 2018;18:586–595. doi: 10.1038/s41568-018-0034-3. [DOI] [PubMed] [Google Scholar]
- 40.Maya-Mendoza A, et al. High speed of fork progression induces DNA replication stress and genomic instability. Nature. 2018;559:279–284. doi: 10.1038/s41586-018-0261-5. [DOI] [PubMed] [Google Scholar]
- 41.Hamperl S, Cimprich KA. Conflict Resolution in the Genome: How Transcription and Replication Make It Work. Cell. 2016;167:1455–1467. doi: 10.1016/j.cell.2016.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sollier J, et al. Transcription-coupled nucleotide excision repair factors promote R-loop-induced genome instability. Mol Cell. 2014;56:777–785. doi: 10.1016/j.molcel.2014.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sloane DA, et al. Drug-Resistant Aurora A Mutants for Cellular Target Validation of the Small Molecule Kinase Inhibitors MLN8054 and MLN8237. ACS Chem Biol. 2010 doi: 10.1021/cb100053q. [DOI] [PubMed] [Google Scholar]
- 44.Dodson CA, et al. Crystal structure of an Aurora-A mutant that mimics Aurora-B bound to MLN8054: insights into selectivity and drug design. Biochem J. 2010;427:19–28. doi: 10.1042/BJ20091530. [DOI] [PubMed] [Google Scholar]
- 45.Tse AN, et al. CHIR-124, a novel potent inhibitor of Chk1, potentiates the cytotoxicity of topoisomerase I poisons in vitro and in vivo. Clin Cancer Res. 2007;13:591–602. doi: 10.1158/1078-0432.CCR-06-1424. [DOI] [PubMed] [Google Scholar]
- 46.Brannan K, et al. mRNA decapping factors and the exonuclease Xrn2 function in widespread premature termination of RNA polymerase II transcription. Mol Cell. 2012;46:311–324. doi: 10.1016/j.molcel.2012.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Weiss WA, Aldape K, Mohapatra G, Feuerstein BG, Bishop JM. Targeted expression of MYCN causes neuroblastoma in transgenic mice. The EMBO journal. 1997;16:2985–2995. doi: 10.1093/emboj/16.11.2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.DuBois SG, et al. Phase II Trial of Alisertib in Combination with Irinotecan and Temozolomide for Patients with Relapsed or Refractory Neuroblastoma. Clin Cancer Res. 2018;24:6142–6149. doi: 10.1158/1078-0432.CCR-18-1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beltran H, et al. A Phase II Trial of the Aurora Kinase A Inhibitor Alisertib for Patients with Castration-resistant and Neuroendocrine Prostate Cancer: Efficacy and Biomarkers. Clin Cancer Res. 2019;25:43–51. doi: 10.1158/1078-0432.CCR-18-1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mosse YP, et al. A Phase II Study of Alisertib in Children with Recurrent/Refractory Solid Tumors or Leukemia: Children’s Oncology Group Phase I and Pilot Consortium (ADVL0921) Clin Cancer Res. 2019;25:3229–3238. doi: 10.1158/1078-0432.CCR-18-2675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Subramanian A, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kortlever RM, et al. Myc Cooperates with Ras by Programming Inflammation and Immune Suppression. Cell. 2017;171:1301–1315 e1314. doi: 10.1016/j.cell.2017.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Barber GN. STING: infection, inflammation and cancer. Nature reviews Immunology. 2015;15:760–770. doi: 10.1038/nri3921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chiu AC, et al. Transcriptional Pause Sites Delineate Stable Nucleosome-Associated Premature Polyadenylation Suppressed by U1 snRNP. Mol Cell. 2018;69:648–663 e647. doi: 10.1016/j.molcel.2018.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Guarino Almeida E, Renaudin X, Venkitaraman AR. A kinase-independent function for AURORA-A in replisome assembly during DNA replication initiation. Nucleic Acids Res. 2020;48:7844–7855. doi: 10.1093/nar/gkaa570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Merigliano C, Mascolo E, Cesta A, Saggio I, Verni F. A new role for Drosophila Aurora-A in maintaining chromosome integrity. Chromosoma. 2019;128:41–52. doi: 10.1007/s00412-018-00687-0. [DOI] [PubMed] [Google Scholar]
- 57.Debruyne DN, et al. BORIS promotes chromatin regulatory interactions in treatment-resistant cancer cells. Nature. 2019;572:676–680. doi: 10.1038/s41586-019-1472-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kotake Y, et al. Splicing factor SF3b as a target of the antitumor natural product pladienolide. Nat Chem Biol. 2007;3:570–575. doi: 10.1038/nchembio.2007.16. [DOI] [PubMed] [Google Scholar]
Additional References to Methods
- 59.Olson CM, et al. Pharmacological perturbation of CDK9 using selective CDK9 inhibition or degradation. Nat Chem Biol. 2018;14:163–170. doi: 10.1038/nchembio.2538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chao SH, et al. Flavopiridol inhibits P-TEFb and blocks HIV-1 replication. The Journal of biological chemistry. 2000;275:28345–28348. doi: 10.1074/jbc.C000446200. [DOI] [PubMed] [Google Scholar]
- 61.Michelena J, et al. Analysis of PARP inhibitor toxicity by multidimensional fluorescence microscopy reveals mechanisms of sensitivity and resistance. Nat Commun. 2018;9:2678. doi: 10.1038/s41467-018-05031-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ginno PA, Lott PL, Christensen HC, Korf I, Chedin F. R-loop formation is a distinctive characteristic of unmethylated human CpG island promoters. Mol Cell. 2012;45:814–825. doi: 10.1016/j.molcel.2012.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boguslawski SJ, et al. Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. Journal of immunological methods. 1986;89:123–130. doi: 10.1016/0022-1759(86)90040-2. [DOI] [PubMed] [Google Scholar]
- 64.Chen X, et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell. 2008;133:1106–1117. doi: 10.1016/j.cell.2008.04.043. [DOI] [PubMed] [Google Scholar]
- 65.Jaenicke LA, et al. Ubiquitin-Dependent Turnover of MYC Antagonizes MYC/PAF1C Complex Accumulation to Drive Transcriptional Elongation. Mol Cell. 2016;61:54–67. doi: 10.1016/j.molcel.2015.11.007. [DOI] [PubMed] [Google Scholar]
- 66.Kopper F, et al. Damage-induced DNA replication stalling relies on MAPK-activated protein kinase 2 activity. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:16856–16861. doi: 10.1073/pnas.1304355110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Henssen AG, et al. Therapeutic targeting of PGBD5-induced DNA repair dependency in pediatric solid tumors. Science translational medicine. 2017;9 doi: 10.1126/scitranslmed.aam9078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Quinlan AR. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. In: Baxevanis Andreas D, et al., editors. Current protocols in bioinformatics. Vol. 47. 2014. pp. 11 12 11–11 12 34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Ramirez F, Dundar F, Diehl S, Gruning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Shen L, Shao N, Liu X, Nestler E. ngs.plot: Quick mining and visualization of next-generation sequencing data by integrating genomic databases. BMC Genomics. 2014;15:284. doi: 10.1186/1471-2164-15-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Devaiah BN, et al. BRD4 is a histone acetyltransferase that evicts nucleosomes from chromatin. Nat Struct Mol Biol. 2016;23:540–548. doi: 10.1038/nsmb.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hao Z, et al. RIdeogram: drawing SVG graphics to visualize and map genome-wide data on the idiograms. PeerJ Preprints. 2019;7 doi: 10.7717/peerj-cs.251. e27928v27919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liberzon A, et al. Molecular signatures database (MSigDB) 3.0. Bioinformatics. 2011;27:1739–1740. doi: 10.1093/bioinformatics/btr260. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
NGS data is available at the Gene Expression Omnibus under the accession number GSE144288. Source data for all Figures and Extended Data Figures have been provided as Source Data files. All other data supporting the findings of the study are available from the corresponding authors on request.