Abstract
For life to emerge, confinement of catalytic reactions within protocellular environments has been proposed as a decisive aspect to regulate chemical activity in space.1 Today, cells and organisms adapt to signals2–6 by processing them through reaction networks that ultimately provide downstream functional responses and structural morphogenesis.7,8 Re-enacting such signal processing in de-novo designed protocells is a profound challenge, but of high importance for understanding the design of adaptive systems with life-like traits. We report on engineered all-DNA protocells9 harbouring an artificial metalloenzyme10 whose olefin metathesis activity leads to downstream morphogenetic protocellular responses with varying levels of complexity. The artificial metalloenzyme catalyses the uncaging of a pro-fluorescent signal molecule, that generates a self-reporting fluorescent metabolite designed to weaken DNA duplex interactions. This leads to pronounced growth, intra-particular functional adaptation in the presence of a fluorescent DNA mechanosensor,11 or inter-particle protocell fusion. Such processes mimic chemically transduced processes found in cell adaptation and cell-to-cell adhesion. Our concept showcases new opportunities to study life-like behaviour via abiotic bioorthogonal chemical and mechanical transformations in synthetic protocells. Furthermore, it reveals a strategy for inducing complex behaviour in adaptive- and communicating soft-matter microsystems, and illustrates how dynamic properties can be upregulated and sustained in micro-compartmentalized media.
In living cells, a diversity of different signals are processed in a crowded environment using reaction networks, that may even be organized spatially in liquid/liquid phase-segregated membraneless organelles.12–15 In recent years, (bio)polymer coacervates have emerged as a preferable protocell model system16,17 over liposomes18–21 for emulating some of the critical features that are essential to cellular functioning: macromolecular crowding22 and phase-segregation.2 Some advanced mimics include DNA-containing protocells that can exchange DNA as information and may allow for simple communication.23,24 However, it is of critical importance to develop strategies able to convert signals from diverse origin to allow for intra- or inter-protocell downstream processes such as functional adaptation, communication, and simple morphogenesis.
Artificial metalloenzymes (ArMs) are emerging biohybrid catalysts containing an organometallic moiety as synthetic co-factor embedded in a protein scaffold. ArMs can be evolved by genetic means, and hold great promise to impart new types of abiotic activities in biological systems.25,26 ArMs based on the biotin-streptavidin (Sav) technology have been shown to operate in cells and allow for spatially-controlled bioorthogonal activation of functional molecules such as prodrugs10,27,28,29
Inspired by such conversion strategies foreign to the host system, we set out to compartmentalize an artificial metathase based on the biotin-Sav technology inside crowded all-DNA protocells. This introduces new-to-nature catalytic activities, beyond the commonly used DNA-tethered enzymes30 or DNAzymes, allowing processing of non-DNA signals within DNA nano-environments, and enabling access for downstream reactions using non-DNA, but DNA-interacting metabolites. For this purpose, we rely on a “close-to-release” reaction cascade that leads to a “gain-of-function” via ring-closing metathesis (RCM)-triggered uncaging of a pro-fluorescent signal. The uncaged product is reactive as a downstream metabolite by interacting with duplex DNA. The strategy allows to both (i) monitor the compartmentalized abiotic reaction and the effect of crowding and (ii) modulate signal transduction leading to downstream morphological responses (Fig. 1). We show that the accumulated product can dynamize DNA duplexes, ultimately resulting in protocellular growth, mechanical activation, adhesion and fusion, thus mimicking rudimentary, functional and morphological adaptation of (proto)cells. We prepared all-DNA based protocells (PCs) with liquid ssDNA interior and DNA hydrogel shells to compartmentalize the ArMs. The PCs are formed by a heat-induced self-compartmentalization of two ssDNA multiblock copolymers, p(A20-m) and p(T20-n) (Supplementary Fig. 1)9. A20 and T20 represent homo-repeats of 20 adenine and 20 thymine nucleobases, while m and n are defined barcode sequences for functionalization (Fig. 1). Immobilization of the ArM is achieved by functionalizing the m core-barcodes with a complementary biotinylated Biot-m* ssDNA, that binds to Sav. Thanks to its homotetrameric nature, immobilized Sav can additionally accommodate a biotinylated Hoveyda-Grubbs catalyst (Biot-Ru; Supplementary Fig. 2). For efficient immobilization, we used a molar ratio of Sav/Biot-m* = 1:1, providing, on average, 75% free binding sites for reaction with Biot-Ru. An appealing feature of such ArMs is the possibility to improve their catalytic performance via site-directed mutagenesis (Supplementary Note 1).10 We surmised that this strategy might provide a versatile tool for the optimization of catalysts within the crowded environment provided by the PCs.
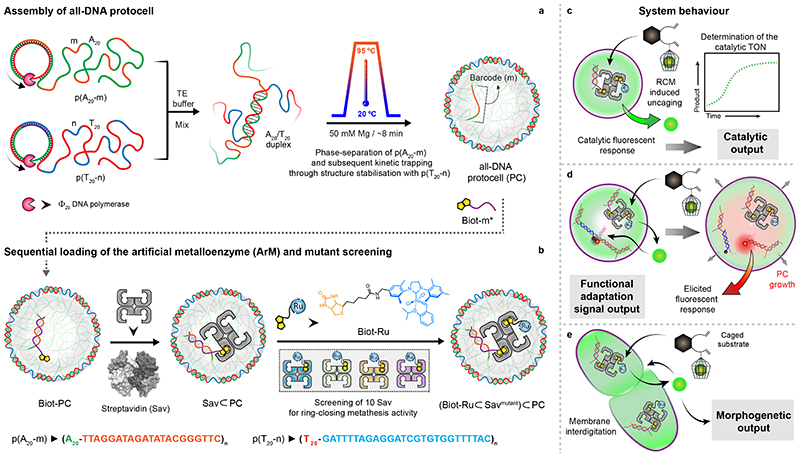
Fig. 1. Design concept, strategy, and system behaviour of artificial metalloenzyme (ArM)-catalysed signal conversion and downstream adaptation inside all-DNA protocells (PCs).
(a) Synthesis of sequence-controlled multiblock ssDNA polymers via rolling circle amplification (RCA). The mixture of the ssDNA polymers is subjected to a heating ramp (3 °C/min) in the presence of Mg2+. Thermally-induced phase-separation of p(A20-m) during heating and duplex (A20/T20) formation (at the coacervate surface) during cooling, leads to the self-assembly of kinetically-trapped all-DNA PCs. The core of the PCs is composed of liquid p(A20-m) while the shell is constituted with p(T20-n) crosslinked via A20/T20 duplexes. The p(A20-m) dissolves below its cloud point temperature, but remains entrapped in a liquid state under high osmotic pressure and macromolecular crowding in the tight hydrogel-like shell. In addition to their straightforward assembly, such PCs allow selective functionalization of the core and the shell using the barcodes (m,n).(b) Sequential loading and assembly of the artificial metalloenzyme (ArM) is achieved by attaching Biot-m* to the core barcode m, followed by addition of streptavidin (Sav) and a biotinylated olefin metathesis catalyst (Biot-Ru), to afford a PC-loaded ArM ((Biot-Ru⊂Savmutant)⊂PC, or ArM⊂PC hereafter). The catalytic activity of the ArM⊂PC was optimized by screening 10 Sav mutants. (c-e) Schematic representation of three adaptive responses of the RCM inside the PCs. (c) The immobilized ArM catalyses a DNA-orthogonal uncaging reaction giving rise to a primary fluorescent signal. (d) The uncaged product instigates swelling of the PCs and destabilizes a mechanofluorescent force module (installed in PCs) which, triggers a secondary fluorescence output. (e) The accumulated uncaged product also induces membrane dynamization of the PCs, resulting in pronounced morphological transformations, ultimately leading to PC fusion.
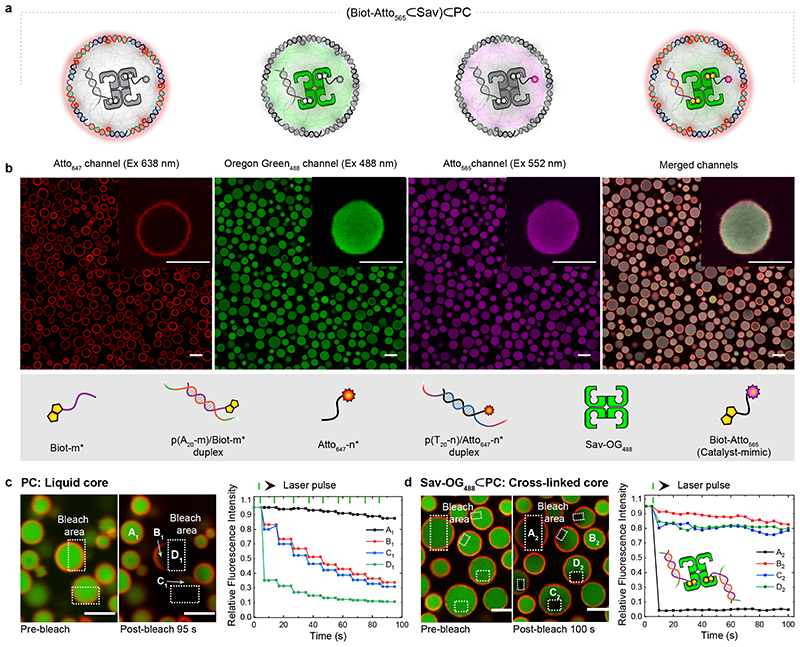
First, we demonstrate the uptake and selective functionalization of the individual compartments using fluorescent model compounds: (i) Atto647-n* (red channel) for the PC shell, (ii) Oregon-green488 labelled Sav-OG488 (green channel) and (iii) biotinylated-Atto565 (magenta channel) as a catalyst mimic (Fig. 2a,b). After functionalization of the PC core-barcodes (m) with Biot-m*, Sav-OG488 – that binds to Biot-m* – and the catalyst-mimic (Biot-Atto565) – that fills the remaining biotin binding sites of Sav-OG488 – were added sequentially. The loading protocol, adjuvants and stoichiometry were fine-tuned to ensure a homogeneous loading of the PCs. Confocal laser scanning microscopy (CLSM) of the loaded PCs reveals their spatially programmed assembly via the respective fluorescence. This confirms that the PC shells are sufficiently porous for Sav, the largest entity in our functionalization scheme (ca. 5·5·5 nm3).
Fig. 2. Compartment-selective functionalization of PCs.
(a) Schematic representation of (Biot-Atto565⊂Sav)⊂PC depicting all of its components. (b) CLSM images of (Biot-Atto565⊂Sav)⊂PC: The shell of the PCs is functionalized with Atto647-n* (red channel). The Sav-OG488 (green channel) is immobilized in the PC core via Biot-m* (hybridized with barcodes m), followed by biotin-Atto565 (magenta channel) binding to the Sav-OG488. (c) CLSM images of pristine PCs before and after photo-bleaching within the dotted rectangles. The corresponding fluorescence intensities at positions A1, B1, C1, and D1 (during nine sequential photo-bleaching events) reveal that the core is liquid (B1, C1 partially bleached) and that the shells are cross-linked (i.e., only bleached in the irradiated areas). (d) CLSM images of Sav-loaded PCs before and after photo-bleaching in the dotted rectangles. The corresponding fluorescence during photobleaching at position A2, B2, C2, and D2 highlight the gelled core, due to the multivalent cross-linking of the Sav-OG488 with Biot-m* (hybridized to the p(A20-m) polymer). The green ticks on the top indicate the photo-bleaching events. Scale bars: 3 μm.
The physical state of the pristine and Sav-loaded (Sav⊂PCs) PCs was elucidated by fluorescence recovery after photobleaching (FRAP; Fig. 2c,d, Supplementary movies 1 and 2). The pristine PCs exhibit near-simultaneous bleaching of both the irradiated and non-irradiated core areas within single PCs (Fig. 2c). Repeated photobleaching leads to stepwise bleaching of the full PC core, confirming a liquid-like interior of the PCs surrounded by a hydrogel-like shell.9 In contrast, the Sav-OG488⊂PC cores and shells show almost no fluorescence recovery within the exposed area after a single photo-bleaching pulse (Fig. 2d). Although the non-irradiated areas lose 20% of their initial intensity after bleaching, a complete reorganization and homogenization of the interior is absent. This suggests that the Sav is a multivalent crosslinker of the p(A20-m)/Biot-m* interior. The interior is in a gel state.
To monitor the catalytic RCM reaction prompted by the metathase (Biot-Ru⊂Sav) anchored in the PC cores and the signal translation resulting from the uncaging of the pro-fluorescent cargos, we evaluated two diene-containing substrates. Both display self-reporting functions and a downstream metabolite interaction with DNA was expected: (i) a naphthalene precursor which releases umbelliferone (Subs-I), and (ii) a dimethoxynaphthalene precursor (Subs-II) which eliminates water upon RCM (Fig. 3a,f). Uncaged umbelliferone and dimethoxynaphthalene allow monitoring the RCM activity by fluorescence spectroscopy and GC-MS, respectively. Importantly, with respect to downstream response, coumarin-derivatives, such as umbelliferone, are well-known dsDNA intercalators (in particular A/T).31 We further hypothesized that dimethoxynaphthalene might also interact with DNA due to its hydrophobicity and π-stacking ability.32
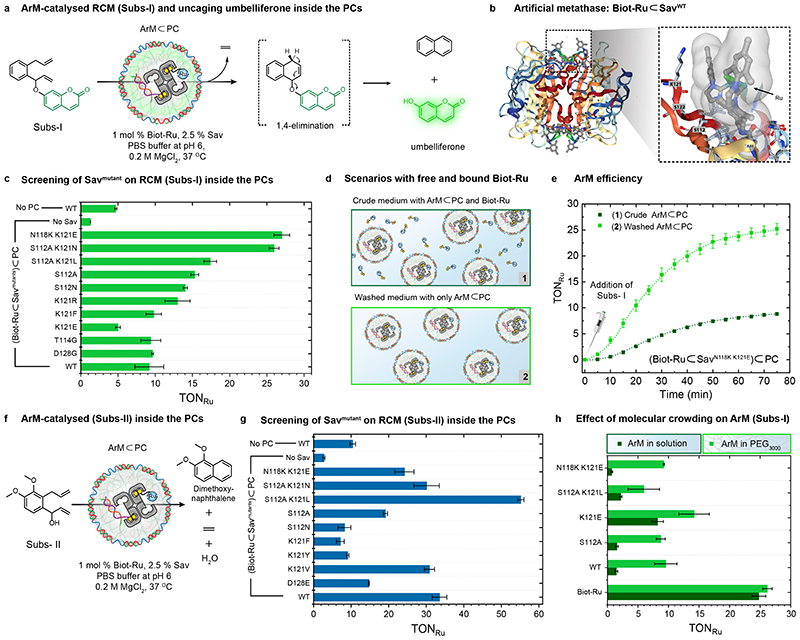
Fig. 3. Intraprotocellular ring-closing metathesis (RCM) of Subs-I and Subs-II catalysed by genetically-engineered ArMs.
(a) Schematic representation of the RCM-triggered uncaging of umbelliferone inside the PCs from a caged precursor (Subs-I) (b) X-ray crystal structure (PDB: 5IRA) of the artificial metathase Biot-Ru⊂SavWT. Biot-Ru and proximal protein residues are depicted as ball-and-stick and stick, respectively. (c) Genetic optimization of ArM for the uncaging of umbelliferone from Subs-I. The TONRu are based on yields of umbelliferone determined by fluorescence spectroscopy. Data are the means and standard deviation of duplicate reactions. (d) Schematic representation of two experiments: (1) RCM is performed using a crude mixture of free Biot-Ru (1 mol% catalyst loading with respect to the [Subs-I]) and ArM⊂PC, (2) RCM is performed using ArM⊂PC after a washing step to remove unbound Biot-Ru (i.e., centrifugation followed by re-dispersion). The Ru content of these samples was determined by ICP-MS (Supplementary Table 2). (e) Comparison of the turnover number (TONRu) of ArM⊂PC vs. free Biot-Ru, revealing the effect of the environment resulting from compartmentalization within Sav⊂PCs. (f) Schematic representation of the ArM-catalysed RCM of a dimethoxynaphthalene precursor (Subs-II) inside PCs. (g) Genetic optimization of the ArM for the RCM of Subs-II inside PCs. The TONRu are based on yields of dimethoxynaphthalene determined by GC-MS using an internal standard. Data are the means and standard deviation of duplicate reactions. (h) Crowding effect on the TONRu resulting from addition of 50 mg/mL PEG (3000 Da) for different mutants, the WT and the free co-factor Biot-Ru.
After assembling ArM⊂PCs ((Biot-Ru⊂Sav)⊂PCs), we compared the turnover numbers of the ArM (TONRu) in solution and encapsulated state for both Subs-I and Subs-II (Fig. 3c,g). The TONRus in PCs are normalized to the Biot-Ru content determined by inductively coupled plasma mass spectrometry (ICP-MS) and to purified PCs. For Subs-I and Subs-II, the TONRus of ArM⊂PC are 2-fold and 3-fold higher respectively (in case of wild type SavWT) than the free ArM in solution. To highlight the effect of the PC environment on metathase kinetics, we compared the catalytic activity for the most active mutant SavN118K K121E with Subs-I for (i) unwashed ArM⊂PCs containing excess Biot-Ru in solution, and (ii) washed ArM⊂PCs, thus removing excess Biot-Ru (Fig. 3d,e). According to ICP-MS, ca. 24% of the added Biot-Ru catalyst is immobilized in the PC core (Supplementary Note 2; Table S2). Interestingly, Subs-I affords only 8.8 TONRu in (i), while 25 TONRu are detected after purification in (ii). Hence, the genetically-optimized ArMs are at least threefold more active within the PC interior (Fig. 3c,e). Consequently, compartmentalization of the ArM in a crowded PC provides a propitious environment for catalysis.
Next, we screened a focused library of 10 Sav mutants, identified in previous campaigns,26 for RCM activity inside and outside the PCs (Fig. 3c,g; Supplementary Fig. 3b,d). For Subs-I, with the notable exception of (Biot-Ru⊂SavK121E)⊂PC, most compartmentalized ArMs lead to higher TONs compared to free Biot-Ru⊂Sav (Supplementary Fig. 3b). Strikingly, while Biot-Ru⊂SavK121E is the most active mutant screened in solution, (Biot-Ru⊂SavK121E)⊂PC is the least active in the PC. Introduction of an additional mutation, thus relocating a cationic lysine from position 121 to 118, afforded the most active compartmentalized ArM (Biot-Ru⊂SavN118K K121E)⊂PC and the least active ArM in solution. For Subs-II, a different mutant exhibits the highest TONRu (Biot-Ru⊂SavS112A K121L, Fig. 3g and Supplementary Fig. 3d). While challenging to rationalize, these results highlight how subtle changes in the second coordination sphere around the cofactor (i.e. mutation and/or presence of ssDNA) dramatically affect the RCM activity.
We also investigated the effect of MgCl2 in RCM with Subs-II. For (Biot-Ru⊂SavWT)⊂PC, the TONRu is twofold higher in the presence of 200 mM MgCl2 compared to 50 mM MgCl2, but shows no further increase above 200 mM (Supplementary Fig. 3c,e). Higher [Cl−] have been shown to stabilize the Ru catalyst under physiological conditions,29 which is also confirmed in this protocellular environment.
We surmised that the marked increase in TON observed for the compartmentalized ArM may be caused by the following effects: i) presence of p(A20-m), ii) high [Mg2+], iii) accumulation of Subs-I within the PCs or iv) crowding. HPLC analysis revealed a modest accumulation of Subs-I in empty PCs and in Sav-loaded PCs, Sav⊂PCs (<20 %, compared to the surroundings). Gratifyingly, addition of PEG3000 leads to a fivefold increase in TON vs. free Biot-Ru⊂Sav N118K K121E for Subs-I, as well as for all other Sav isoforms. In stark contrast, the free co-factor Biot-Ru does not experience a change in the crowded environment (Fig. 3h). Accordingly, the increased TON for compartmentalized ArMs is primarily caused by crowding.33 Differences in the relative effect of crowding between PEG and the ArM inside the PCs likely relate to the chemical nature of p(A20-m) ssDNA inside the PCs.
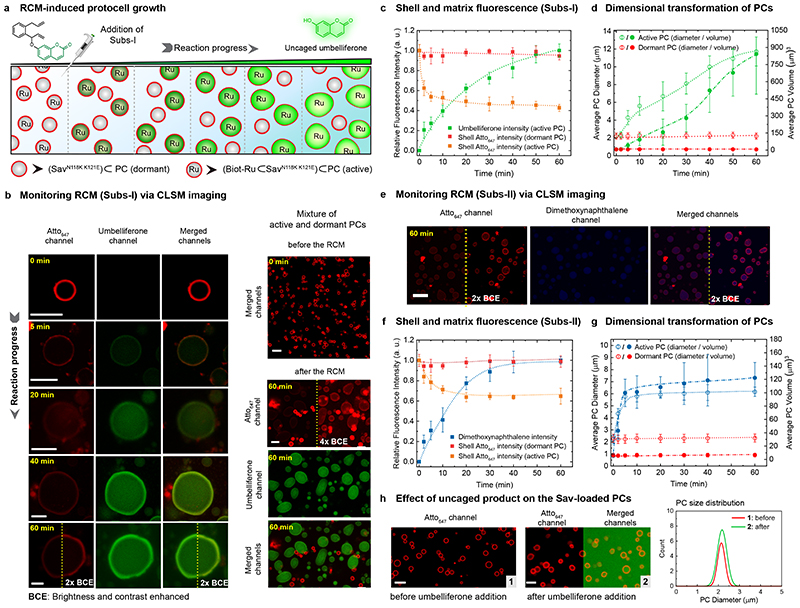
We selected the most active ArM PC/substrate systems ─ (Biot-Ru⊂SavN118K K121E)⊂PC/Subs-I and (Biot-Ru⊂SavS112A K121L)⊂PC/Subs-II ─ for in situ microscopy studies (Figure 4). To monitor downstream responses resulting from the catalytic activity, we prepared a mixture of active ((Biot-Ru⊂SavN118K K121E)⊂PC) and dormant PCs (SavN118K K121E⊂PC), which are initially indistinguishable with red shells (Atto647-n* labelling of shell-barcode n) and colourless cores (t=0 min, Fig. 4 a,b; Supplementary Fig. 5). Once Subs-I is injected, green fluorescence appears in the core of the active PCs, confirming that the compartmentalized ArM catalyses the RCM and uncages the umbelliferone inside the PCs (Fig. 4b, Supplementary Fig. 4). The rate of umbelliferone release inside the active PCs (Fig. 4c; calculated from CLSM) correlates with the macroscopic fluorescence spectroscopy (Fig. 3e). Strikingly, a substantial expansion of the active PCs occurs (up to 5-fold in diameter and 125-fold in volume). The green fluorescence persists in the core and accumulates in the shell. It does not dilute visibly into the surrounding indicating tight binding to dsDNA-domains, which are located in the shell and at the core/shell interface (A20/T20 and Atto647-n*/n). Concomitantly, the red shell fluorescence decreases by about 50% after an hour of RCM activity. This is due to a thickness increase of the shell by swelling and potentially due to a loss of some Atto647-n* by duplex breakage (see below). Since the PCs are under high osmotic pressure due to the entrapped p(A20-m) with all its counterions9 and only stabilized by the A20/T20 duplexes at the core/shell interface, the swelling strongly suggests that the RCM products (i) intercalate into the dsDNA31, thus (ii) weaken the dsDNA interactions substantially, and (iii) lead to the dynamization of the membrane layer. The PC swelling caused by the release of dimethoxynaphthalene from Subs-II is less pronounced (3-fold in diameter, Fig. 4e-g; Supplementary Fig. 6). This relates to the poorer intercalating properties of dimethoxynaphthalene vs. umbelliferone.32 To further underscore the intercalation, Supplementary Fig. 7 displays melting curves of A20-m/T20-m* dsDNA in the presence of the intercalators. The melting curve flattens out in presence of umbelliferone, and the duplexes do not re-hybridize upon cooling. The perturbation of the duplex hysteresis is less pronounced for dimethoxynaphthalene.
Fig. 4. CLSM monitoring of ArM-catalysed RCM inside PCs and ensuing morphological transformations.
(a) RCM-induced umbelliferone uncaging in a mixture of active and dormant PCs (i.e., (Biot-Ru⊂SavN118K K121E)⊂PC and, SavN118K K121E⊂PC). Addition of Subs-I leads to umbelliferone accumulation within the active PCs, accompanied by a gradual increase in their volume. The dormant PCs remain unaltered, both in fluorescence and size. (b) Time-dependent CLSM images for RCM-induced umbelliferone uncaging in a single PC (left array) and a mixed PC system (right array). The shells of all PCs are labelled with Atto647-n* (red channel). At t=0 (before the addition of Subs-I), all PCs are visible as red circles and indistinguishable. The RCM-induced uncaging of umbelliferone (green channel; excitation at 405 nm) is recorded over a 60 min period. The gradual increase in fluorescence intensity (green channel) in the core of the active PCs results from the RCM-induced uncaging of the umbelliferone. Brightness and contrast of some of the images (red and merged channels) are increased to visualize the shell of the active PCs. (c) Normalized time-dependent fluorescence intensities of umbelliferone (green trace), the shell of the active PCs (orange trace), and the shell of the dormant PCs (red trace). The fluorescence intensities are averaged from the CLSM images using ~25 PCs. (d) Time-dependent increase in dimension for active and dormant PCs (averaged over 25 PCs). (e) CLSM images of the RCM-induced dimethoxynaphthalene formation inside the PCs. (f) Normalized time-dependent fluorescence intensities of the RCM product (blue trace), the shell of the active PCs (orange trace), and the shell of the dormant PCs (red trace). (g) Time-dependent RCM-induced change in the dimensions for active and dormant PCs (averaged over 25 PCs). The dimethoxynaphthalene product has less influence on the morphological transformation of PCs, due to weak interaction with the duplexes. (h) CLSM images of SavN118K K121E⊂PCs (dormant-PC) before and after the addition of umbelliferone (500 μM). The size distribution (shown on the right) indicates that the morphology of the PCs remains unaltered when the intercalator was added in solution even though the diffusion of umbelliferone is observed into the PC core. Scale bar: 5 μm. Conditions: 1% catalyst loading, 0.2 − 0.3 mM of substrates, 10 mM phosphate buffer, pH 6, 0.1 M MgCl2.
Control experiments underscore the importance of compartmentalizing the catalytic event to induce PC growth. The expansion is absent in dormant PCs after the addition of Subs-I (Fig. 4d). This confirms that Subs-I is not a morphogen, and that the metabolite (umbelliferone) produced in an active PC does not induce changes in a dormant PC via diffusion. We also added a high concentration of umbelliferone to dormant PCs and observed no morphological transformation, although umbelliferone (green fluorescence) accumulates in the core and shell (Fig. 4h). Moreover, the morphology of dormant PCs remains unaltered upon addition of a mixture of both uncaged products, umbelliferone and naphthalene (Supplementary Fig. 8). These observations clearly demonstrate that the conversion inside the PCs is the decisive aspect to trigger downstream changes. Although unexpected, these observations highlight the possibility of emergent, self-inflicted behaviour of active PCs.
Next, we showcase signal-transduction cascades – a distinctive feature of living systems – that result from the compartmentalized catalytic event using Subs-I. Therein, the release of the abiotic metabolite produced from Subs-I is exploited to drive the metamorphosis of the PCs, leading to distinct functional intraparticle adaptation, and ultimately, interparticle interaction.
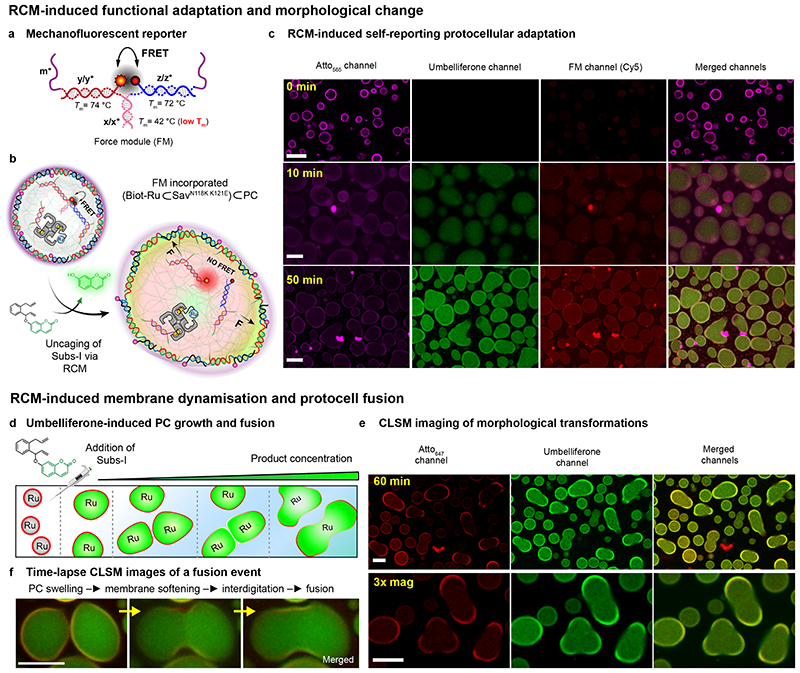
First, we supposed that the swelling and internal stress may be harnessed to break specifically engineered mechano-fluorescent DNA force-sensing modules (FM), hybridized as crosslinkers to the m-barcodes in the PC interior (Fig. 5a, Supplementary Fig. 9a,b).11 The FM consists of a short dsDNA (x/x*) equipped with a fluorophore (Cy5, yellow-red dot) and a quencher (red-black dot, Fig. 5a), and is designed as weakest network link. Its zipper geometry facilitates mechanical opening. Upon addition of Subs-I, the catalytic production of umbelliferone leads to swelling and the mechanical stress induces indeed the rupture of the FM, as revealed by a new red Cy5 fluorescence (Fig. 5c, Supplementary Fig. 9c). In line with the signal-transduction cascade mechanism, a delayed appearance (ca. 20 min) of the Cy5 fluorescence compared to the umbelliferone fluorescence is found (Supplementary Fig. 9e). This reflects the critical stretching threshold that the FM requires for unzipping. Correspondingly, the signal transduction cascade allows for functional adaptation revealed by the appearance of a new colour (a simple PC phenotype) and liberates two previously masked ssDNA sequences for future DNA-based reaction schemes.
Fig. 5. RCM-induced, self-reporting, downstream functional adaptation and morphological output.
(a) Structure of the mechanofluorescent force-sensing module (FM). The red-emitting fluorophore (Cy5) and the quencher (Iowa RQ) are in close proximity, thus quenching the red fluorescence by FRET. The FM has two m* domains, which are used to cross-link the PC core at core barcodes m. The double-stranded x/x* is the weakest link (−ΔGy/y* = 25.8 kcal mol−1 > −ΔGz/z* = 25.2 kcal mol−1 > −ΔGm/m* = 24 kcal mol−1 > −ΔGx/x* = 9 kcal mol−1) in the FM. Homogeneous loading of such FMs was confirmed by functionalization with a FM without the quencher (Supplementary Fig. 10). (b) Schematic representation of the FM-loaded active PCs ((Biot-Ru⊂SavN118K K121E)⊂PC), in which 70% of the barcodes (m) inside the PCs are used for catalyst loading and 30% are used to attach the FM. RCM-induced uncaging of umbelliferone triggers PC swelling and FM activation as revealed by red fluorescence. (c) Time-dependent CLSM. The magenta channel represents the Atto565-n* conjugated PC shells; the green channel reports on the uncaging of umbelliferone inside the PC core, and the red channel reveals the second fluorescent output from the dissociated FM. The isolated fluorescent “hotspots” are contamination, while the PC interior turns homogeneously red. (d) Schematic representation of RCM-induced morphological transformation in PCs. The PCs undergo swelling and membrane rupture and PC fusion upon increasing the concentration of the Subs-I. (e) CLSM images of swollen and fused PCs after 60 min following Sub-I addition. (f) Time-lapse fusion event of protocells. Larger overview in Supplementary Fig. 11. Scale bars: 5 μm. 3x mag: three times magnified. For the fusion experiments, [Subs-I] = 0.6 − 0.8 mM, 10 mM phosphate buffer, pH 6, 0.1 M MgCl2.
We further hypothesized that higher substrate concentrations and extended reaction times may lead to shell rupture and promote PC interactions. Indeed, when subjecting active PCs to 3-fold Subs-I concentration, fusion of PCs occurs. Fusion occurs by growth-induced symmetry-breaking of the original spherical PCs (most likely due to defects in the shell) and ultimately shell rupture, whereupon interaction between the cores follows (Fig. 5d-f; Supplementary Fig. 11). Since the interior is gelled due to tetravalent Sav, a secretion of the interior as liquid phase cannot occur. We suggest that the fusion into rather homogeneous assemblies is assisted by (i) gaining additional inter-protocellular biotin-Sav interactions and (ii) minimizing the interface with the water.
In summary, we introduced a generic strategy to encapsulate ArMs inside the macromolecularly-crowded core of all-DNA PCs. We then highlighted how ‘abiotic’ catalytic signal conversion strategies can be used for downstream morphological adaptation on different levels. The key step relies on selecting a synthetic, non-DNA substrate that can be turned into a DNA duplex-weakening metabolite, which triggers the swelling of the PCs trapped in a metastable state (being under high osmotic pressure), resulting in (i) PC growth, (ii) activation of mechanophores for functional adaptation (e.g. colour), and (iii) PC fusion. Such features are reminiscent of the behaviour of living cells – albeit on simplistic levels. We unravelled that the ArMs are more active inside the crowded environment and that site-directed mutagenesis of the ArMs improves the catalytic performance. Remarkably, all morphological changes in the PCs are observed only when performing catalysis inside the PCs: The external addition of substrates or products to inactive PCs does not induce any morphological change.
These findings reveal the opportunities of endowing PCs with abiotic catalytic activity, here ‘abiotic’ in the sense of being foreign to typical DNA-systems, to generate chemical signals compartmentalized within hybrid protocellular environments. These PCs display advanced adaptive and emergent behaviour. While we highlighted pathways towards downstream functions on a morphological level, we believe that the merger of artificial metalloenzymes and colloidal coacervates can provide avenues for adaptive functional systems in sensor applications or as colloidal factories for functional compounds. Although cross-disciplinary approaches to explore the design, structuration, function and evolutionary potential of metabolic PCs with genetically evolved proteinaceous catalysts are in their infancy, our approach provides valuable insights (i) into the acquisition of chemically-triggered adaptive behaviour of prebiotic coacervates and (ii) towards a minimalistic design of life-like abiotic systems. Capitalizing on the interaction between dsDNA and abiotic metabolites presented herein, we envision fascinating opportunities to further engineer interactive PC systems, that can communicate, translate and process various types of signals in more complex sensory environments.
Methods
Synthesis of circular ssDNA template and multiblock DNA polymer
The 5’-phosphorylated template and its corresponding ligation strand (see Supplementary Table 1 and Supplementary Fig. 1) were mixed (in equimolar amounts) to a final concentration of 1 μM in TE buffer (Invitrogen; 10 mM Tris(hydroxymethyl)aminomethane pH=8 and 1 mM EDTA) containing additionally 100 mM NaCl (total 100 μL). The low concentration of the strands is required to prevent inter-strand hybridization. The buffered solution was heated to 85°C (for 5 min) at a rate of 3°C s═1 and cooled to 20°C at 0.01°C s═1. After annealing the strands, 20 μL of 10 X commercial ligase buffer (Lucigen; 500 mM TRIS-HCI, 100 mM MgCl2, 50 mM dithiothreitol, and 10 mM ATP), 70 μL of water and 10 μL of T4 Ligase (2 U μL═1) were added to the tube containing 100 μL of the template strand, stirred (10 min, 400 rpm) and left to react for 4 hours at room temperature. The T4 Ligase was then denatured by heating the reaction mixture for 20 min at 70°C. Then, 10 μL of Exonuclease I (Lucigen; 40 U μL═1) and 10 μL Exonuclease III (Lucigen; 200 U μL═1) were added, and the mixture was left overnight at 37°C on a thermo-shaker with gentle stirring (300 rpm) to remove unreacted ligation strands and non-circularized templates in solution. The exonucleases were subsequently deactivated by heating the reaction mixture at 80°C for 40 min. The circular templates were purified by filtration through the Amicon Ultracentrifugal filters with a 10 kDa cut-off (Merck Millipore) and washed 3 times using TE buffer over the same filter. The ssDNA concentrations were measured using a ScanDrop (Jena Analytic) spectrophotometer, and the solutions were diluted to 1 μM using TE buffer. The template synthesis was repeated multiple times, and a stock solution of the circular template was prepared to avoid batch-to-batch discrepancy.
To synthesize multiblock ssDNA polymers via rolling circle amplification (RCA), 10 μL circular template (1 μM) were mixed with 134 μL of ultrapure nuclease-free water, 20 μL of commercial 10 X polymerase buffer (Lucigen; 500 mM TRIS-HCl, 100 mM (NH4)2SO4, 40 mM Dithiothreitol, 100 mM MgCl2), 2 μL of exonuclease resistant primer (10 μM in TE buffer), 4 μL of Φ29 Polymerase (Lucigen; 10 U μL−1), 20 μL of pyrophosphatase (New England Biolabs; 0.1 U μL−1) and 10 μL of an adjusted dNTP mixture (total dNTP concentration of 100 mM, the percentage of each base corresponds to the expected sequence composition in the ssDNA polymer). The reaction mixtures were kept for 60 hours at 30 °C on a thermo-shaker with gentle stirring (300 rpm). The resulting polymer was then purified by filtration through Amicon Ultra-centrifugal filters with a 30 kDa cut-off (Merck Millipore) and washed thoroughly (3 times) using 400 μL of TE buffer. The concentration of the ssDNAs were determined using a ScanDrop (Jena Analytic) spectrophotometer. It is highly recommended to use the pyrophosphatase enzyme to prevent the formation of insoluble magnesium pyrophosphate as a side product that interferes with the polymerization process and is also responsible for the appearance of so-called nanoflowers.
Standard protocol for protocells (PCs) synthesis
The PCs were prepared by mixing the starting ssDNA multiblock polymers at the target concentration (for p(A20-m), typically, around 0.15 g L−1, which corresponds to the repeating unit (r.u.) concentration of 10 μM and for p(T20-n), around 0.04 g L−1, which corresponds to [r.u.] = 2.5 μM) in TE buffer (pH 8). The ratio of p(A20-m) : p(T20-n) = 1 : 4 (or 1 : 3.5) represents an optimized empirical condition, in order to obtain well-defined PCs with minimized free p(T20-n) in solution. Generally, PCs were synthesized in 200 μL PCR tubes, and the volume of the PC stock solution varied from 50 to 100 μL. Freshly prepared PCs were used for all the measurements. The mixture was heated to 95 °C for 15 min for homogenization and thermal cleavage of long ssDNA polymer chains (no phase-separation). A solution of MgAc2 (1 M) was added to the mixture to attain a final concentration of 50 mM (for phase-separation of p(A20-m)) and subsequently heated to 95 °C for 5 min with heating and cooling ramps of 3 °C min−1. After the formation of the PCs, a stoichiometric amount of corresponding fluorescently-labelled barcode* oligomeric sequences (Atto488-m*, Atto565-n* or Atto647-n*) was added to the solution. The resulting colloidal solution was left for 1 h at room temperature with constant shaking (450 − 500 rpm) temperature to hybridize with the PC-barcodes before monitoring them via CLSM imaging. The thermal reduction of the molecular weight of the RCA polymers is critical to attaining spherical PCs instead of unwanted agglomerates. The PCs are stable for a week, even though they slowly sediment to the bottom of the tube. The PCs can be sedimented and redispersed numerous times after the sequential functionalization of the core and the shell.
Encapsulation of the catalyst in the PC interior
A stock solution of Biot-m* (0.5 mM) was added to freshly prepared PC medium (50 μL, [r.u.] or [barcode-m] = 10 μM) to entirely functionalize the m-barcodes inside the PCs with biotin. The Biot-PCs were left 1 h at room temperature with gentle shaking (300 rpm). An aqueous solution of Sav (1.3 mM in Milli-q water) was added to the Biot-PC medium to a final concentration of 40 μM. The mixture was vortexed and left 2 h to incubate at room temperature. The Sav-loaded protocells (Sav⊂PC) were centrifuged (12000 rpm, ~8 min) and redispersed with 10 mM phosphate buffer containing 200 mM MgCl2. A stock solution of Biot-Ru (5 mM in DMSO) was added to the Sav PCs to a final concentration of catalyst 25-30 μM (depending on the experiments). The mixture was left 1 h to incubate at room temperature with gentle stirring (400 rpm) to prevent sedimentation of the protocells. Finally, the (Biot-Ru⊂Sav)⊂PCs were washed (2 times) and redispersed to remove unbound Biot-Ru from the reaction medium (see Supplementary Fig. 2).
Fluorescence recovery after photobleaching (FRAP) experiments
The PCs were first imaged using a low intensity of the corresponding laser. Photobleaching was attained using 100% intensity on both the 488 nm and 638 lines (≤10 mW in the focal plane). The liquid/solid core-shell PCs were photobleached with repetitive single pulses (1 s at each step), and the images were recorded. On the other hand, single-step photobleaching (6 s) was used for the protein-loaded PCs (Sav⊂PCs) to deplete the fluorescence fully at the region of interest, and the images were compared before and after the photobleaching steps. The data are presented in Fig. 2c,d. In both cases, Atto647-n* was used to visualize the shell of the PCs. The relative fluorescence intensities of the region of interests (obtained from the image stacks) are plotted against the timeframe of the experiments.
Quantification of RCM-induced uncaging and catalytic assay in the PC interior
The stock solution containing catalyst-loaded protocells ((Biot-Ru⊂Sav)⊂PCs) was aliquoted into two batches (20 μL each). The reaction medium was diluted to 200 μL containing a buffered solution of either Subs-I or Subs-II in order to reach 1% - 2% catalyst loading (depending on the specific experiment) and the substrate concentration was maintained 100 μM. In the case of Subs-I, the reaction mixture was loaded (in duplicates or triplicates) on a 96-well plate and the fluorescence intensities were recorded in a plate reader over 1 h. The plate was analysed in a Tecan reader (Infinite M1000Pro) by fluorescence of the uncaged product (temperature 37 °C, kinetic interval: 5 min, excitation 325nm, emission 450 nm, bandwidth interval 5.0 nm, settle time 0.1 s). The well-plate was shaken for 10 seconds before every cycle of measurements. The concentration of the uncaged product throughout the reaction was extrapolated from a calibration curve using umbelliferone at various concentrations. For Subs-II, the aliquots from the reaction mixture were collected at multiple time-points, and the aliquots were extracted with ethyl acetate (3x). The ethyl acetate extract was then subjected to GC-MS to quantify the product concentration and TON.
ICP-MS analysis and activity assay for the RCM with protocells
The biotin-loaded PCs (700 μL) were loaded with Sav (40 μM, 21 μL of a 1.33 mM stock solution), and incubated for 1 h at room temperature. After incubation, the solution was centrifuged and washed with PBS buffer. The catalyst Biot-Ru was then added to the Sav⊂PCs (25 μM, 3.5 μL of a 5 mM stock solution in DMSO), and the resulting mixture was incubated for 1 h at room temperature. After incubation, the solution was divided into two; one aliquot was subjected to centrifugation followed by washing with PBS buffer, while the other aliquot was not washed. The washed and unwashed PCs (20 μL) were then placed into a microplate reader, followed by the addition of a solution of Subs-I (180 μL of a 111.11 μM solution in PBS buffer containing 200 mM MgCl2, pH 6). The plate was inserted in a Tecan reader (Infinite M1000Pro) and analysed by fluorescence (temperature 37 °C, kinetic cycle: 15, kinetic interval: 5.00 min, excitation 325nm, emission 450 nm, bandwidth interval 5.0 nm, settle time 0.1 s). The remaining volume of the PCs were split into 3x 100 μL aliquots, diluted to 250 μL with milli-q water, and subjected to ICP-MS analysis.
CLSM monitoring of RCM-induced uncaging inside the PC interior
The shells of the freshly prepared, washed (Biot-Ru⊂Sav)⊂PCs were hybridized with Atto647-n* in order the visualize (63x objective, excitation: 638 line, 364 well-plate) the protocells before the addition of the substrates (Fig. 4b,e). A stock solution of Subs-I or Subs-II (20 mM) was added to the reaction medium (40 μL) to a final concentration of 0.2 to 0.5 mM (a higher concentration was used for the fusion experiments). The (Biot-Ru⊂Sav)⊂PCs were then visualized by excitation with two lasers: 405 nm (for the RCM-product) and 638 nm (for the shell). The area of interest for the CLSM measuring was changed after capturing one image to prevent the photobleaching of umbelliferone or dimethoxynaphthalene (except the single PC measurements). The stock solutions of the substrates were centrifuged before the addition of the clear supernatant solution to the reaction medium. During the reaction, the well-plate was kept at 25 °C using a temperature-controlled microscopic stage. All the raw images are processed using ImageJ (Fiji) software. The brightness and contrast of a few images are enhanced (mentioned in the corresponding figure) to visualize the different parts of the protocells.
Assembly of mechanofluorescent sensor modules (FM)
The different HPLC-purified oligomeric sequences forming the mechanosensing modules (e.g., for module m*-y*-x, x*-z*-m*, Cy5-y, and z-Q (or z) dissolved in TE buffer at 100 μM) were mixed with 10 × folding buffer (containing 120 mM MgAc2 and 1 M NaCl) and diluted with TE buffer to a final concentration of 25 μM of the module (i.e., 25 μM in TE buffer containing 100 mM of NaCl and 12 mM of MgAc2) (Supplementary Note S4). With the aim of minimizing the fluorescence at rest, the quencher strand was introduced in slight excess (1.1 eq.) and the fluorophore strand in a slight lack (0.9 eq.) with respect to the sacrificial duplex strand. This ensures that every fluorophore couples in a module containing a quencher. The mixture was heated to 85 °C and cooled to 25 °C at 0.01 °C s–1 to assemble the module to anneal all the strands. We ensured the correct assembly of the mechanosensing force modules (FM) by 2% agarose gel electrophoresis (Supplementary Fig. 7b). In Supplementary Fig. 7b, the modules are folded using the quencher (Iowa Black® RQ)-functionalized strand (z-Q), which hinders the observation of a fully folded module due to the quenching of DNA stains (lanes 5 and 6). The fully folded FM displays a faint fluorescent band (lane 6), which also confirms the self-assembled duplexes with Cy5 and the quencher positioning themselves in close proximity. There was no self-dimerization observed in the assembly process.
Loading of FM in the core of the PCs before the catalysis
A fresh stock solution of PCs (50 μL, [barcode-m] = 13-15 μM) was prepared and were kept at room temperature on a thermoshaker with constant shaking (400 rpm). To obtain efficient and uniform loading of the mechanofluorescent module inside the PCs, the solution of FM must be added before the addition of Sav. 60-65 % of the barcodes-m were functionalized with Biot-m*, while 30-35% of the barcodes were cross-linked with FM. A stock solution of Biot-m* (2.5 μL; 200 μM) and freshly prepared stock solution of FM (5.25 μL; 25 μM) were mixed in a PCR tube and added to the PC solution. The PCs were vortexed after the addition and left on a thermoshaker at room temperature for 60 min with constant shaking (450-500 rpm). The FM-loaded PCs were washed before the subsequent addition of Sav and Biot-Ru.
Determination of melting temperatures (Tm)
Temperature-dependent UV–vis spectra were recorded on a ScanDrop (Jena Analytics) with Peltier heating setup. The ssDNA sequences were mixed and heated to 85 °C and cooled down to 25 °C at 0.01 °C s−1. The final concentration of the duplexes was set to 2 μM in the reaction buffer (10 mM phosphate buffer containing 200 mM MgAc2). Heating ramps were performed on 50 μl of a solution in a 45 μl Quartz cuvette (Hellma Analytics) and heated at 1 °C min–1, and the extinction at 260 nm was recorded every 60s.
Supplementary Material
Acknowledgments
We acknowledge support by the European Research Council Starting grant to AW (TimeProSAMat, (agreement 677960) and Advanced Grant to TRW (DrEAM, agreement 694424), the DFG Cluster of Excellence livMatS “Living, Adaptive and Energy-Autonomous Materials Systems” and the NCCR Molecular Systems Engineering. A.S. acknowledges the support by the Alexander von Humboldt Foundation.
Footnotes
Author contribution statement
A.S. and A.W. conceived the project. A.S., V.S., A. W. and T. R. W. designed and performed all the experiments. A.S. analysed the data and prepared the preliminary draft. A.W., T.R.W supervised the project. All authors contributed to writing the manuscript.
Financial and Non-Financial Competing Interests
The authors declare no competing interests.
Additional Information
Supplementary information is available in the online version of the paper. Reprints and permission information is available online at www.nature.com/reprints. Correspondence and requests for materials should be addressed to A. Walther or T. R. Ward.
Data availability statement
The data of the catalysis is experiments is available online. Any other data can be made available upon reasonable request to the corresponding authors.
References
- 1.Oparin AI. Origin of Life. Dover; 1952. [Google Scholar]
- 2.Brangwynne CP, et al. Germline P granules are liquid droplets that localize by controlled dissolution/condensation. Science. 2009;324:1729–1732. doi: 10.1126/science.1172046. [DOI] [PubMed] [Google Scholar]
- 3.Pawson T. Specificity in signal transduction: from phosphotyrosine-SH2 domain interactions to complex cellular systems. Cell. 2004;116:191–203. doi: 10.1016/s0092-8674(03)01077-8. [DOI] [PubMed] [Google Scholar]
- 4.Pawson T, Nash P. Assembly of cell regulatory systems through protein interaction domains. Science. 2003;300:445–452. doi: 10.1126/science.1083653. [DOI] [PubMed] [Google Scholar]
- 5.You L, Cox RS, Weiss R, Arnold FH. Programmed population control by cell–cell communication and regulated killing. Nature. 2004;428:868–871. doi: 10.1038/nature02491. [DOI] [PubMed] [Google Scholar]
- 6.Bacchus W, et al. Synthetic two-way communication between mammalian cells. Nat Biotechnol. 2012;30:991–996. doi: 10.1038/nbt.2351. [DOI] [PubMed] [Google Scholar]
- 7.Na S, et al. Rapid signal transduction in living cells is a unique feature of mechanotransduction. Proc Natl Acad Sci USA. 2008;105:6626–6631. doi: 10.1073/pnas.0711704105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trepat X, et al. Universal physical responses to stretch in the living cell. Nature. 2007;447:592–595. doi: 10.1038/nature05824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Merindol R, Loescher S, Samanta A, Walther A. Pathway-controlled formation of mesostructured all-DNA colloids and superstructures. Nat Nanotechnol. 2018;13:730–738. doi: 10.1038/s41565-018-0168-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jeschek M, et al. Directed evolution of artificial metalloenzymes for in vivo metathesis. Nature. 2016;537:661–665. doi: 10.1038/nature19114. [DOI] [PubMed] [Google Scholar]
- 11.Merindol R, Delechiave G, Heinen L, Catalani LH, Walther A. Modular design of programmable mechanofluorescent DNA hydrogels. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08428-2. 528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin N. Dynamic synthetic cells based on liquid–liquid phase separation. ChemBioChem. 2019;20:2553–2568. doi: 10.1002/cbic.201900183. [DOI] [PubMed] [Google Scholar]
- 13.Sokolova E, et al. Enhanced transcription rates in membrane-free protocells formed by coacervation of cell lysate. Proc Natl Acad Sci USA. 2013;110:11692–11697. doi: 10.1073/pnas.1222321110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zwicker D, Seyboldt R, Weber CA, Hyman AA, Jülicher F. Growth and division of active droplets provides a model for protocells. Nat Phys. 2017;13:408–413. [Google Scholar]
- 15.Changeux J-P, Christopoulos A. Allosteric Modulation as a Unifying Mechanism for Receptor Function and Regulation. Cell. 2016;166:1084–1102. doi: 10.1016/j.cell.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 16.Strulson CA, Molden RC, Keating CD, Bevilacqua PC. RNA catalysis through compartmentalization. Nat Chem. 2012;4:941–946. doi: 10.1038/nchem.1466. [DOI] [PubMed] [Google Scholar]
- 17.Booth R, Qiao Y, Li M, Mann S. Spatial positioning and chemical coupling in coacervate-in-proteinosome protocells. Angew Chem Int Ed. 2019;58:9120–9124. doi: 10.1002/anie.201903756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mansy SS, et al. Template-directed synthesis of a genetic polymer in a model protocell. Nature. 2008;454:122–125. doi: 10.1038/nature07018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Adamala K, Szostak JW. Nonenzymatic template-directed RNA synthesis inside model protocells. Science. 2013;342:1098–1100. doi: 10.1126/science.1241888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Adamala K, Szostak JW. Competition between model protocells driven by an encapsulated catalyst. Nat Chem. 2013;5:495–501. doi: 10.1038/nchem.1650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langton MJ, Scriven LM, Williams NH, Hunter CA. Triggered release from lipid bilayer vesicles by an artificial transmembrane signal transduction system. J Am Chem Soc. 2017;139:15768–15773. doi: 10.1021/jacs.7b07747. [DOI] [PubMed] [Google Scholar]
- 22.Fulton AB. How crowded is the cytoplasm? Cell. 1982;30:345–347. doi: 10.1016/0092-8674(82)90231-8. [DOI] [PubMed] [Google Scholar]
- 23.Gobbo P, et al. Programmed assembly of synthetic protocells into thermoresponsive prototissues. Nat Mater. 2018;17:1145–1153. doi: 10.1038/s41563-018-0183-5. [DOI] [PubMed] [Google Scholar]
- 24.Joesaar A, et al. DNA-based communication in populations of synthetic protocells. Nat Nanotechnol. 2019;14:369–378. doi: 10.1038/s41565-019-0399-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wilson ME, Whitesides GM. Conversion of a protein to a homogeneous asymmetric hydrogenation catalyst by site-specific modification with a diphosphinerhodium(I) moiety. J Am Chem Soc. 1978;100:306–307. [Google Scholar]
- 26.Schwizer F, et al. Artificial metalloenzymes: reaction scope and optimization strategies. Chem Rev. 2018;118:142–231. doi: 10.1021/acs.chemrev.7b00014. [DOI] [PubMed] [Google Scholar]
- 27.Okamoto Y, et al. A cell-penetrating artificial metalloenzyme regulates a gene switch in a designer mammalian cell. Nat Commun. 2018;9 doi: 10.1038/s41467-018-04440-0. 1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan X, et al. Optimized tetrazine derivatives for rapid bioorthogonal decaging in living cells. Angew Chem Int Ed. 2016;55:14046–14050. doi: 10.1002/anie.201608009. [DOI] [PubMed] [Google Scholar]
- 29.Sabatino V, Rebelein JG, Ward TR. “Close-to-release”: spontaneous bioorthogonal uncaging resulting from ring-closing metathesis. J Am Chem Soc. 2019;141:17048–17052. doi: 10.1021/jacs.9b07193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilner OI, et al. Enzyme cascades activated on topologically programmed DNA scaffolds. Nat Nanotechnol. 2009;4:249–254. doi: 10.1038/nnano.2009.50. [DOI] [PubMed] [Google Scholar]
- 31.Ou C-N, Tsai C-H, Tapley KJ, Song P-S. Photobinding of 8-methoxypsoralen and 5,7-dimethoxycoumarin to DNA and its effect on template activity. Biochemistry. 1978;17:1047–1053. doi: 10.1021/bi00599a017. [DOI] [PubMed] [Google Scholar]
- 32.Banks TM, Clay SF, Glover SA, Schumacher RR. Mutagenicity of N-acyloxy-N-alkoxyamides as an indicator of DNA intercalation part 1: evidence for naphthalene as a DNA intercalator. Org Biomol Chem. 2016;14:3699–3714. doi: 10.1039/c6ob00162a. [DOI] [PubMed] [Google Scholar]
- 33.Ellis RJ. Macromolecular crowding: obvious but underappreciated. Trends Biochem Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data of the catalysis is experiments is available online. Any other data can be made available upon reasonable request to the corresponding authors.