Abstract
Eukaryogenesis is one of the most enigmatic evolutionary transitions, during which simple prokaryotic cells gave rise to complex eukaryotic cells. While evolutionary intermediates are lacking, gene duplications provide information on the order of events by which eukaryotes originated. Here we use a phylogenomics approach to reconstruct successive steps during eukaryogenesis. We found that gene duplications roughly doubled the proto-eukaryotic gene repertoire, with families inherited from the Asgard archaea-related host being duplicated most. By relatively timing events using phylogenetic distances we inferred that duplications in cytoskeletal and membrane trafficking families were among the earliest events, whereas most other families expanded predominantly after mitochondrial endosymbiosis. Altogether, we infer that the host that engulfed the proto-mitochondrion had some eukaryote-like complexity, which drastically increased upon mitochondrial acquisition. This scenario bridges the signs of complexity observed in Asgard archaeal genomes to the proposed role of mitochondria in triggering eukaryogenesis.
Compared to prokaryotes, eukaryotic cells are tremendously complex. Eukaryotic cells are larger, contain more genetic material, have multiple membrane-bound compartments and operate a dynamic cytoskeleton. Although certain prokaryotes have some eukaryote-like complexity, such as a large size, internal membranes and even phagocytosis-like cell engulfment1,2, a fundamental gap remains. The last eukaryotic common ancestor (LECA) already had the intracellular organisation and gene repertoire characteristic of present-day eukaryotes3, making the transition from prokaryotes to eukaryotes – eukaryogenesis – one of the main unresolved puzzles in evolutionary biology1,4.
Most eukaryogenesis scenarios posit that a host, related to the recently discovered Asgard archaea5,6, took up an Alphaproteobacteria-related endosymbiont7,8 that gave rise to the mitochondrion. However, the timing and impact of this endosymbiosis event in the evolution of eukaryotic complexity are hotly debated and at the heart of different scenarios on eukaryogenesis9.
Besides the acquisition of genes via the endosymbiont, the proto-eukaryotic genome expanded through gene inventions, duplications and horizontal gene transfers during eukaryogenesis10,11. Previous work suggested that gene duplications nearly doubled the ancestral proto-eukaryotic genome11. Gene families such as small GTPases, kinesins and vesicle coat proteins greatly expanded, which enabled proto-eukaryotes to employ an elaborate intracellular signalling network, a vesicular trafficking system and a dynamic cytoskeleton12–15.
Uncovering the order in which these and other eukaryotic features emerged is complicated due to the absence of intermediate life forms. However, duplications occurred during the transition and are likely to yield valuable insights into the intermediate steps of eukaryogenesis. In this study we attempt to reconstruct the successive stages of eukaryogenesis by systematically analysing large sets of phylogenetic trees inferred from prokaryotic and eukaryotic sequences. We determined the scale of gene inventions and duplications during eukaryogenesis and how different functions and phylogenetic origins had contributed to these eukaryotic innovations. Furthermore, we timed the prokaryotic donations and duplications relative to each other using information from phylogenetic branch lengths.
Results
Unprecedented resolution of duplications during eukaryogenesis
To obtain a comprehensive picture of duplications during eukaryogenesis we made use of the Pfam database16 (see Methods). We took a phylogenomics approach inspired by the ‘ScrollSaw’ method14, which limits phylogenetic analyses to slowly evolving sequences and collapses duplications after LECA and thereby increases the resolution of deep tree nodes. We constructed phylogenetic trees and detected 10,233 nodes in these trees that represent a single Pfam domain in LECA (‘LECA families’) (Fig. 1a). These 10,233 LECA families do not include genes having only small Pfam domains, which we excluded for computational reasons, or genes without any domains. Therefore, we used a linear regression analysis to obtain an estimated LECA genome containing 12,753 genes (95% prediction interval: 7,447 – 21,840) (Extended Data Fig. 1).
Fig. 1. Characterisation of duplications during eukaryogenesis.
a, Density plot showing the distribution of the number of Pfam domains in present-day prokaryotes (green) and eukaryotes (purple) in comparison with the acquisition, invention and LECA estimates obtained from phylogenetic trees (see inset). b, Number of acquisitions or inventions that gave rise to a particular number of LECA families, demonstrating the skewedness of duplications across protein families. c, Odds of duplication for LECA families according to KOG functional categories. 81% of pairwise comparisons were significantly different (Supplementary Fig. 1). The poorly characterised categories and functions of very few families (cell motility, extracellular structures and nuclear structure) are not depicted. d, Odds of duplication for LECA families according to cellular localisation. 54% of pairwise comparisons were significantly different (Supplementary Fig. 2). c-d, Numbers on the right side indicate the number of LECA families and dashed lines indicate the odds of all LECA families in total.
Comparing the number of inferred LECA families to extant eukaryotes showed that the genome size of LECA reflected that of a typical present-day eukaryote (Fig. 1a), which is in line with the inferred complexity of LECA, but in contrast with lower estimates obtained previously11,17. We used the split between Opimoda and Diphoda as root position of the eukaryotic tree of life18. As the exact position of the eukaryotic root is under debate19, we tested alternative root positions and obtained very similar numbers of LECA families, except for the root positions at the base of and within the excavates (15 – 46% fewer families compared with an Opimoda-Diphoda root; Extended Data Fig. 2a). In case of a true excavate root, this could reflect fewer genes in LECA. However, given the sampling imbalance between both sides of an excavate root and the reduced nature of sampled excavate genomes, we consider a gene-rich LECA and subsequent gene losses a more likely scenario.
The multiplication factor – the number of LECA families divided by the number of acquired and invented genes or domains – was 1.8, approximating the near doubling reported before11. The observed doubling was validated in an additional data set (Supplementary Table 1), despite a recent study that inferred very few duplications during eukaryogenesis (see Supplementary Information)20. Although on average genes duplicated once, the distribution of duplications is heavily skewed with many acquisitions from prokaryotes or eukaryotic inventions not having undergone any duplication (Fig. 1b). The enormous expansion of the proto-eukaryotic genome was dominated by massive duplications in a small set of families (Supplementary Table 2).
Duplicated and non-duplicated LECA families differed considerably in their functions and cellular localisations. Metabolic LECA families rarely had a duplication history, whereas LECA families involved in information storage and processing, and cellular processes and signalling were more likely to descend from a duplication (χ2 = 572, df = 2, P = 7.7 × 10−125; Fig. 1c, Supplementary Fig. 1). Notable exceptions to this pattern were families involved in cell wall or membrane biogenesis and translation, which were rarely duplicated. The observed differences in functions were reflected by differences between cellular localisations, with proteins in the endomembrane system and cytoskeleton mostly resulting from a duplication (χ2 = 262, df = 4, P = 1.6 × 10−55; Fig. 1d, Supplementary Fig. 2). Like duplications, inventions primarily occurred to families involved in informational and cellular processes (χ2 = 226, df = 2, P = 8.8 × 10−50 (function); χ2 = 186, df = 4, P = 4.9 × 10−39 (localisation); Extended Data Fig. 3, Supplementary Fig. 3–6). For complex eukaryotes to emerge, most innovations occurred in nuclear processes, the endomembrane system, intracellular transport and signal transduction, especially due to gene duplications.
Relatively large contribution of the host to duplicated LECA families
For the Pfams that were donated to the eukaryotic stem lineage we identified the prokaryotic sister group, which represents the best candidate for the Pfam’s phylogenetic origin (Extended Data Fig. 4a). Most acquisitions had a bacterial sister group (77%), of which only a small proportion was alphaproteobacterial (7% of all acquisitions), in agreement with previous analyses10,21,22. The acquisitions from archaea (16%) predominantly had an Asgard archaeal sister (7% of all acquisitions). Moreover, the most common Asgard archaeal sister group was solely comprised of Heimdallarchaeota (Extended Data Fig. 4b); especially Heimdallarchaeote LC3 was frequently the sister group. This is in line with previous analyses providing support for either all Heimdallarchaeota or LC3 being the currently known archaeal lineage most closely related to eukaryotes23,24. The species in alphaproteobacterial sister groups, on the other hand, came from different orders (Extended Data Fig. 4c), consistent with the recently proposed deep phylogenetic position of mitochondria8. The remaining acquisitions (7%) had an unclear prokaryotic ancestry (see Supplementary Discussion).
Families with different sister clades varied substantially in the number of gene duplications they experienced during eukaryogenesis (χ2 = 50, df = 5, P = 1.2 × 10-9 (duplication tendency); χ2 = 190, df = 5, P = 4.3 × 10-39 (LECA families from duplication); Fig. 2). The multiplication factor of 2.2 for families likely inherited from the Asgard archaea-related host was strikingly high compared with the invented families and families acquired from bacteria (between 1.3 and 1.8). Especially duplications related to the ubiquitin system and trafficking machinery contributed to the relatively large number of host-related paralogues (Supplementary Table 2). In contrast, there was a clear deficit of duplications in families with an alphaproteobacterial sister group (multiplication factor of 1.3). Hence, the endosymbiont marginally contributed to the near doubling of the genetic material via duplications during eukaryogenesis, whereas the host contributed relatively the most.
Fig. 2. Contribution of different phylogenetic origins to duplications during eukaryogenesis.
a, Duplication tendency as fraction of clades having undergone at least one duplication. b, Multiplication factors, defined as the number of LECA families divided by the number of acquisitions or inventions. These numbers are shown beside the corresponding bar. a, b, Dashed lines indicate the duplication tendency and multiplication factor for all acquisitions and LECA families. The four (a) and three (b) pairwise comparisons that did not give a significant P value (χ2 contingency table test) are shown. Prokaryotic: unclear prokaryotic ancestry (could not be assigned to a domain or lower taxonomic level).
Using branch lengths to time acquisitions and duplications
The remarkable differences in duplication dynamics between families with different affiliations could tentatively stem from differences in timing of these acquisitions and subsequent duplications. For example, the low number of alphaproteobacterial-associated duplications could be the result of a late mitochondrial acquisition. To research this, branch lengths in phylogenetic trees can be used. They serve as a good proxy for relative time and have previously been used to time the acquisition of genes from the different prokaryotic donors10. Shorter branch lengths, corrected for differences in evolutionary rates across families, reflect more recent acquisitions. Duplications were not included in the previous analysis, but they can be timed in a similar way with the length of the branch connecting the duplication and LECA nodes (Fig. 3). Although the measure has been criticised for its assumption that evolutionary rates pre- and post-LECA are correlated25,26, it yielded correct timings for specific post-LECA events10,27. The observed trends can either be created by a common rate change in proteins of the same phylogenetic origin or can be due to different time points of acquisitions. Previous studies10,27 showed that the latter explanation is most plausible.
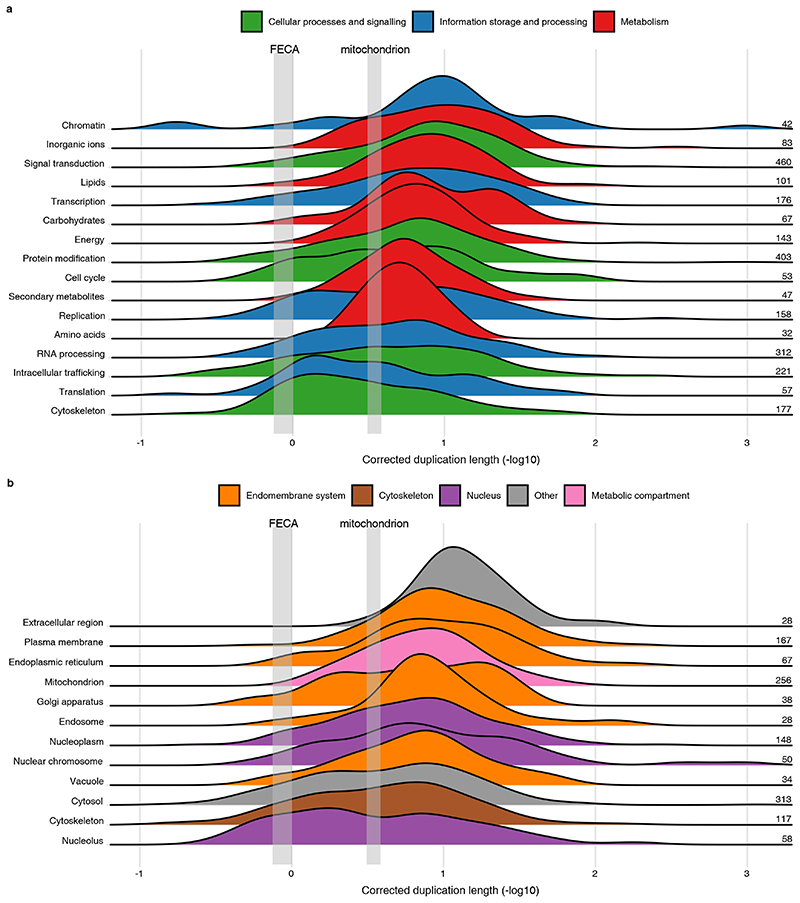
Fig. 3. Timing of acquisitions and duplications from different phylogenetic origins during eukaryogenesis.
Ridgeline plot showing the distribution of corrected stem or duplication lengths, depicted as the additive inverse of the log-transformed values. Consequently, longer branches have a smaller value and vice versa. For clarity, a peak of near-zero branch lengths is not shown (see Extended Data Fig. 6). Numbers indicate the number of acquisitions or duplications for which the branch lengths were included. Groups of stem and duplication lengths are ordered based on the median value. The tree illustrates how the stem and duplication lengths were calculated; the symbols and colour schemes are identical to Fig. 1a. The phylogenetic distances between the acquisition or duplication and LECA were normalised by dividing it by the median branch length between LECA and the eukaryotic terminal nodes. In case of duplications the shortest of the possible normalised paths was used. Pairwise comparisons that did not give a significant P value (Mann-Whitney U test) are shown.
Although the inclusion of duplications in branch length analyses provides potentially valuable information, duplications could have affected the branch lengths by causing a shift in evolutionary rate. The stem lengths of acquisitions that happened simultaneously should approximate the same value, enabling us to assess the effect of duplication on branch lengths. We observed slight but notable increases in stem lengths for duplicated families from alphaproteobacterial origin (Extended Data Fig. 5a) and for more recent duplications in vertebrates (Extended Data Fig. 5f), but not for duplicated families from Asgard archaeal origin (Extended Data Fig. 5b). It is therefore possible that in some families an accelerated rate could result in a slightly too early inferred duplication event according to our branch length analysis. We further checked if there was a rate change after duplication in different functional groups of proteins and looked for an effect of homomer-to-heteromer transitions but we could not detect a clear pattern of rate shifts for different groups of proteins (Extended Data Fig. 5c-e). We validated the use of duplication lengths by examining phylogenetic trees containing more recent duplications in the primate lineage, for which we have multiple intermediate speciation events. The distributions of duplication lengths followed the speciation events (Extended Data Fig. 5g), demonstrating the validity of using duplication lengths to obtain an order of events. We also observed a small effect of function but the effect of time was much larger (Extended Data Fig. 5h). Although duplications themselves and function can have an influence, time is the predominant factor explaining the differences in branch lengths. Thus, analysing branch lengths, also in duplicated families, is a valid and effective approach to infer an order of events.
Branch lengths point to a mitochondria-intermediate scenario
For the timing of acquisitions we obtained similar results as before10, with archaeal stems being longer than bacterial stems (P = 4.5 × 10−98, two-sided Mann-Whitney U-test; Fig. 3). Among the archaeal stem lengths the Asgard archaeal stems were shortest, as were the alphaproteobacterial stems among the bacterial stems, although for the first the difference failed to reach statistical significance (P = 0.88 and P = 4.0 × 10−4, respectively). This pattern is independent of the normalisation by post-LECA branches, the presence of duplications and functional divergence between the acquisition and LECA (Extended Data Fig. 6). Figure 3 shows that there is a wide distribution of host-related duplication lengths, with a substantial number of duplication lengths both longer and shorter than (alphaproteo)bacterial stem lengths. Bacteria-affiliated, endosymbiont-related and invented families showed the shortest duplication lengths. These duplication lengths were not affected by the position of the eukaryotic root (Extended Data Fig. 2b). The differences in branch lengths indicate that an increase in genomic complexity via duplications likely had already occurred prior to the mitochondrial acquisition.
To shed light on the evolution of cellular complexity we categorised the duplications according to their functional annotations and cellular localisations. A marked distinction in duplication lengths between different functions can be observed, with duplications in metabolic functions corresponding to shorter branches (P = 8.0 × 10−5, Kruskal-Wallis test; Fig. 4a, Supplementary Fig. 7). Moreover, a substantial number of duplication lengths in information storage and processes, and cellular processes and signalling functions were longer than the alphaproteobacterial stem length and duplications related to energy production, which mainly involve the mitochondria. These long duplication lengths include multiple duplications assigned to the cytoskeleton and intracellular trafficking. Duplications in signal transduction and transcription families mainly had shorter branch lengths, indicating that these regulatory functions evolved and diversified relatively late. With respect to cellular localisation, nucleolar and cytoskeletal duplication lengths were longest. Most duplications related to the endomembrane system had duplication lengths similar to those of mitochondrial duplications (Fig. 4b, Supplementary Fig. 8). These findings indicate that the increase in cellular complexity before the mitochondrial acquisition mainly comprised the evolution of cytoskeletal, intracellular trafficking and nucleolar components.
Fig. 4. Timing of duplications during eukaryogenesis according to function and localisation.
a, b, Ridgeline plots showing the distribution of duplication lengths for different functional categories (a) and cellular localisations (b). To enable a comparison with the timing of acquisitions, the binomial-based 95% confidence interval of the median of the Asgard archaeal (FECA) and alphaproteobacterial stem lengths (mitochondrion) are depicted in grey, indicating the divergence of eukaryotes from their Asgard archaea-related and Alphaproteobacteria-related ancestors, respectively. Groups are ordered based on the median value. For significant differences between groups, see Supplementary Fig. 7–8.
Discussion
This large-scale analysis of duplications during eukaryogenesis provides compelling evidence for a mitochondria-intermediate eukaryogenesis scenario. The results suggest that the Asgard archaea-related host already had some eukaryote-like cellular complexity, such as a dynamic cytoskeleton and membrane trafficking. Upon mitochondrial acquisition there was an even further increase in complexity with the establishment of a complex signalling and transcription regulation network and by shaping the endomembrane system. These post-endosymbiosis innovations could have been facilitated by the excess of energy allegedly provided by the mitochondrion28,29.
A relatively complex host is in line with the presence of homologues of eukaryotic cytoskeletal and membrane trafficking genes in Asgard archaeal genomes5,6,30. Moreover, some of them, including ESCRT-III homologues, small GTPases and (loki)actins, have duplicated in these archaea as well, either before eukaryogenesis or more recently5,6,30. This indicates that there has already been a tendency for at least the cytoskeleton and membrane remodelling to become more complex in Asgard archaeal lineages. A dynamic cytoskeleton and trafficking system, perhaps enabling primitive phagocytosis31, might have been essential for the host to take up the bacterial symbiont. Molecular and cell biology research in these archaea, from which the first results have recently become public32,33, is highly promising to yield more insight into the nature of the host lineage. In addition to a reconstruction of the host, further exploration of the numerous acquisitions, inventions and duplications during eukaryogenesis is key to fully unravelling the origin of eukaryotes.
Methods
In this study we inferred and analysed two different sets of phylogenetic trees. The first set (‘Pfam-ScrollSaw’) was used for the main analysis, whereas the second set (‘KOG-to-COG clusters’) was used to verify our method to infer duplications during eukaryogenesis. We also used a third, already existing set of gene trees (‘human phylome’) to validate the use of branch lengths in case of duplications. Below we describe how we created and analysed the main set of phylogenetic trees. The second and third sets of gene trees are described in the Supplementary Methods.
Data
We used 209 eukaryotic (predicted) proteomes from an in-house dataset that has been used and described before34. Prokaryotic proteomes (3,457 in total) were extracted from eggNOG 4.535. The prokaryotic dataset was supplemented with nine predicted proteomes from the recently described Asgard superphylum6.
Pfam assignment
We used hmmsearch (HMMER v3.1b236) with the Pfam 31.0 profile hidden Markov models (HMMs)16 and the corresponding gathering thresholds to assess to which Pfam what part of each prokaryotic and eukaryotic sequence should be assigned. We opted for Pfam profile HMMs to collect homologous sequences because of their sensitivity to detect homology. The domains that were hit were extracted from the sequences based on the envelope coordinates. If a sequence had hits to multiple Pfams and these hits were overlapping for at least 15 amino acids only the best hit was used. If the same Pfam had multiple hits in the sequence due to an insertion relative to the model the different hits were artificially merged. Since the latter is more prone to errors for short models and short sequences contain less phylogenetic signal, profile HMMs shorter than fifty amino acids were not considered for further analysis.
Reduction of sequences
For each Pfam, the number of prokaryotic sequences was reduced with kClust v1.037 using a clustering threshold of 2.93, which corresponds to a sequence identity of 60%. We chose this threshold because we expect it to retain sufficient prokaryotic diversity while removing sequences from related species to keep the analysis computationally feasible. However, because of horizontal gene transfer (HGT), it will also remove sequences from more distantly related species in some cases.
The number of eukaryotic sequences was reduced with a novel method38 based on the ScrollSaw approach14. The idea behind ScrollSaw is that instead of selecting a species subset a priori, the slowest evolving sequences are selected. In that way the resolution of deep nodes in trees from expanded families is drastically improved. Although in the original paper14 the distances between sequences were calculated with a maximum likelihood method, we used the bit score in BLAST39 as a proxy to obtain genetic distances. For each Pfam an all species versus all species BLAST was performed. Because we were only interested in the best hit the max_target_seqs option was set to 1. Although this option has raised some attention recently40,41, we only used it as a proxy for evolutionary distance and our analysis would not be seriously impacted by this option given the overall small sizes of our databases. Subsequently, bidirectional best hits (BBHs) between sequences from different eukaryotic groups were identified. Eukaryotic species can be grouped into different ‘supergroups’, whose names and definitions have changed following new findings19,42. The species in our dataset are from the following six groups: Archaeplastida + Cryptista, SAR + Haptista, Discoba, Metamonada, Obazoa and Amoebozoa. For our main analysis we used BBHs between sequences from two groups, because that provided the best resolution38. Although the exact position of the root of the eukaryotic tree of life is uncertain19, a likely position is between Opimoda (Obazoa and Amoebozoa in our set) and Diphoda (other supergroups)18. Therefore, BBHs between Opimoda and Diphoda sequences were identified and the corresponding sequences were used for phylogenetic analysis.
To assess the impact of a different position of the eukaryotic root, we also identified BBHs between five groups, merging Metamonada and Discoba into Excavata, and four groups, in which Archaeplastida + Cryptista and SAR + Haptista were combined as Diaphoretickes and Obazoa and Amoebozoa were together as Amorphea (see ‘Effect of the position of the eukaryotic root’).
Phylogenetic analysis
Multiple sequence alignments were made with MAFFT v7.31043 (auto option) and trimmed with trimAl v1.4.rev1544 (gap threshold 10%). Phylogenetic trees were inferred with IQ-TREE v1.6.445 (LG4X model46, 1000 ultrafast bootstraps47). If the consensus tree had a higher likelihood than the best tree from the search, the first was used for further analysis. Because inferring trees for PF00005 (ABC transporter), PF00072 (response regulator receiver domain), PF00528 (binding-protein-dependent transport system inner membrane component), PF02518 (histidine kinase-, DNA gyrase B-, and HSP90-like ATPase) and PF07690 (major facilitator superfamily) in this way was too computationally demanding, we used FastTree v2.1.1048 with the LG model to construct trees for these Pfams. These Pfams were not considered for branch length analysis.
Tree analyses
Removal of interspersing prokaryotes
Trees were analysed with an in-house ETE349 script. We examined whether the tree contained prokaryotic sequences that probably reflect recent HGT events and that might interfere with our analysis. Prokaryotic sequences from a single genus that were in between eukaryotic sequences were pruned from the tree. If there was only one prokaryotic sequence in the tree it was kept only if it was an Asgard archaeal sequence, because it has been reported that sometimes only a single sequenced Asgard archaeon contains a homologue to sequences otherwise only present in eukaryotes6. This was the case for 16 trees containing LECA families (see below), including RPL28/MAK16, Sec23/24, UFM1 and the C-terminal domain of tubulins, for which the Asgard archaeal origin has been shown before. Because another prokaryotic outgroup to root these trees was lacking, they were not used to calculate stem lengths (see ‘Branch length analysis’).
Annotation of eukaryotic nodes
For each eukaryotic clade the nodes were annotated as duplications prior to LECA, LECA nodes, post-LECA nodes or unclassified. Only clades that contained at least one LECA node were of interest. The node combining the eukaryotic clade with the rest of the tree (if present) was annotated as acquisition node.
For the annotation of nodes in trees the information from the eukaryotic sequences that were not in the BBHs were included, since the number of eukaryotic sequences in the trees had been reduced. To correctly assign in-paralogues we additionally performed an own species versus own species BLAST for each Pfam (max_target_seqs 2). The sequences belonging to a Pfam that were not in the tree were mapped onto their best hits in the tree according to the BLAST score.
In order to infer reliable duplication nodes in the tree, duplication consistency scores were calculated for all internal nodes starting from the root of a eukaryotic clade. This score is the overlap of species at both sides of a node divided by the total number of species at both sides, taking both sequences in the tree and assigned sequences (as described above) into account. If the duplication consistency score was at least 0.2 and both daughter nodes fulfilled the LECA criteria, this node was annotated as a duplication node. The first LECA criterion was that a node had to have both Opimoda and Diphoda tree sequences in the clade. Secondly, to take care of post-LECA HGT events among eukaryotes and of tree uncertainties, the mean presence of a potential LECA family in the five different supergroups (Obazoa, Amoebozoa, SAR+Haptista, Archaeplastida+Cryptista, Excavata) had to be at least 15%. If a node did not fulfil the LECA criteria it was annotated as a post-LECA node.
The abovementioned thresholds were chosen based on manual inspection of a selection of trees. Using different thresholds for duplication consistency (0, 0.1, 0.2, 0.3) and LECA coverage scores (0, 5, 10, 15, 20 and 25%) had a gradual impact on the absolute numbers and quality measures, such as the fraction of well-supported LECA and duplication nodes (Supplementary Table 3). This underlines that the reported results were not contingent on the specific set of thresholds chosen and that for most nodes the duplication consistency and LECA coverage was high.
After this first annotation round all LECA nodes in the trees were re-evaluated. If there were duplication nodes in both daughters, the node connecting these duplications had to be a duplication node as well even though its duplication consistency score was below the threshold. This was only the case for two nodes in total. If there were duplication nodes in only one daughter lineage, the LECA node was annotated as unclassified. It could reflect a duplication event or a tree artefact due to rogue taxa. If there were no duplication nodes in either daughter lineages, all LECA nodes in the daughter lineages of this LECA node were reannotated as post-LECA nodes.
Rooting eukaryote-only trees
For trees with only eukaryotic sequences and trees for which all prokaryotic sequences had been removed, inferring the root poses a challenge. For these trees duplication and LECA nodes were called in unrooted mode. The distances between the LECA nodes were calculated and the tree was rooted in the middle of the LECA nodes that were furthest apart, resulting in an additional duplication node at this root. If there were no duplications found in this way, because there were less than two duplications in the tree, rooting was tried on each internal node. The node that fulfilled the duplication criteria and that maximised the species overlap was chosen. If none fulfilled the criteria, it was checked if the entire tree fulfilled the LECA criteria. For Pfams for which we could not infer a tree because there were only two or three sequences selected, we also checked if this Pfam in itself fulfilled the LECA criteria. These Pfams correspond to eukaryote-specific families that did not duplicate.
Sister group identification
For each eukaryotic clade in trees also containing prokaryotic sequences the sister group was identified in an unrooted mode. By doing so, the eukaryotic clade initially had two candidate sister groups. Eukaryotic sequences in a sister group, if present, were ignored, as they could reflect HGT events, contaminations, tree artefacts or true additional acquisitions. To infer the actual sister group it was first checked if one of the two candidate sister groups was more likely by checking if one of them consisted only of Asgard archaea, TACK archaea, Asgard plus TACK archaea, alphaproteobacteria, beta/gammaproteobacteria, or alpha/beta/gammaproteobacteria. If so, that clade was chosen as the actual sister group. If both sister groups had the same identity or if both groups had another identity than the ones described above, the tree was rooted on the farthest leaf from the eukaryotic clade. In many cases the last common ancestor of the taxa in the sister group was Bacteria, Archaea or cellular organisms (“LUCA”) according to the NCBI taxonomy. Such wide taxonomic assignments likely reflect extensive HGT among distantly related prokaryotes. In these cases it was checked if one of the previously mentioned groups or otherwise a particular phylum or proteobacterial class comprised a majority of the prokaryotic taxa to get a more precise sister group classification.
We observed that in a substantial number of cases there was another eukaryotic clade with LECA nodes in the sister group of a eukaryotic clade. These cases could reflect a duplication and subsequent loss in prokaryotes but probably reflect tree artefacts. Therefore these clades were ignored for the branch length analysis. Acquisitions that were nested, i.e. they shared exactly the same prokaryotic sister group because one acquisition had in its sister clade only one prokaryotic clade and one or multiple other acquisitions, were merged for further analysis.
Branch length analysis
Multiple branch lengths were calculated in clades containing LECA nodes. For the stem length (sl) the distance to the acquisition node – the node uniting the eukaryotic clade and its prokaryotic sister – was calculated for each LECA node. This distance was divided by the median of the distances from the LECA node to the eukaryotic leaves (eukaryotic branch lengths (ebl)) to correct for rate differences between orthologous groups as done before10. In case of multiple possible paths due to duplications, the minimum of these distances was used as the sl, since it was closest to sl values from zero-duplication clades. To calculate the duplication length (dl) a similar approach was followed, using the duplication node instead of the acquisition node.
To investigate the impact of rates after duplication in both paralogue lineages within a family, we also calculated for all duplication nodes in Asgard archaea-derived families the minimal sl going through these duplications (Extended Data Fig. 5c, d). In this way, we obtained an sl value for each duplication, in addition to the aforementioned sl value for each acquisition. These values were also divided into duplications in families that had undergone a transition from homomers to heteromers (proteasome, Snf7, TRAPP, Vps36 and OST3/OST6) and the rest (Extended Data Fig. 5e).
Combining eukaryote-only Pfam families with prokaryotic donations in their clan
The classification of protein families into Pfams is not based on taxonomic levels. A Pfam present only in eukaryotes can therefore be the result of a duplication event instead of a bona fide invention. To distinguish these possible scenarios we used the Pfam clans, in which related Pfam families are combined. If there were only eukaryote-only Pfams in a clan based on our analysis, these Pfams were merged into one invention event. If there was only one Pfam with an acquisition from prokaryotes and for this Pfam there was only one acquisition, the eukaryote-only Pfams were combined with this acquisition. If there were multiple acquisitions in a clan, a profile-profile search with HH-suite3 v3.0.350 was performed to assign eukaryote-only Pfams to an acquisition. Per acquisition in a clan an alignment was made from the tree sequences in the corresponding eukaryotic clade with MAFFT L-INS-i v7.31043. Profile HMMs were made of these alignments (hhmake –M 50) and they were combined in a database (ffindex_build). The eukaryote-only Pfam HHMs were searched against the acquisition HHM database per clan with hhsearch. Each Pfam was assigned to the acquisition that had the best score.
Functional annotation
Functional annotation of sequences was performed using emapper-1.0.351 based on eggNOG orthology data35. Sequence searches were performed using DIAMOND v0.8.22.8452.
The most common KOG functional category among the tree sequences of a LECA node was chosen as the function of the LECA node. If there was not one function most common, the node was annotated as S (function unknown). For the functional annotation of duplication nodes a Dollo parsimony approach was used. For this we checked if there was one single annotation shared between LECA nodes at both sides, ignoring unknown functions. If this was not the case but the parent duplication node (if present) had a function, this function was also used for the focal duplication node. The functional annotation of the prokaryotic sister group was performed the same way as for a LECA node. In the figures the names of most categories were shortened for increased readability: Translation (Translation, ribosomal structure and biogenesis), RNA processing (RNA processing and modification), Replication (Replication, recombination and repair), Chromatin (Chromatin structure and dynamics), Cell cycle (Cell cycle control, cell division, chromosome partitioning), Signal transduction (Signal transduction mechanisms), Cell wall/membrane (Cell wall/membrane/envelope biogenesis), Intracellular trafficking (Intracellular trafficking, secretion, and vesicular transport), Protein modification (Posttranslational modification, protein turnover, chaperones), Energy (Energy production and conversion), Carbohydrates (Carbohydrate transport and metabolism), Amino acids (Amino acid transport and metabolism), Nucleotides (Nucleotide transport and metabolism), Coenzymes (Coenzyme transport and metabolism), Lipids (Lipid transport and metabolism), Inorganic ions (Inorganic ion transport and metabolism), Secondary metabolites (Secondary metabolites biosynthesis, transport and catabolism).
The same approach was used to assign cellular components to LECA and duplication nodes, using a custom set of gene ontology terms: extracellular region (GO:0005576), cell wall (GO:0005618), cytosol (GO:0005829), cytoskeleton (GO:0005856), mitochondrion (GO:0005739), cilium (GO:0005929), plasma membrane (GO:0005886), endosome (GO:0005768), vacuole (GO:0005773), peroxisome (GO:0005777), cytoplasmic vesicle (GO:0031410), Golgi apparatus (GO:0005794), endoplasmic reticulum (GO:0005783), nuclear envelope (GO:0005635), nucleoplasm (GO:0005654), nuclear chromosome (GO:0000228) and nucleolus (GO:0005730).
Predicting the number of genes in LECA
We used a linear regression model to predict the number of genes in LECA based on the inferred number of Pfam domains in LECA. For this we used the number of sufficiently long Pfam domains (see ‘Pfam assignment’ above) and the number of protein-coding genes in the eukaryotes in our dataset. The assumptions of a normal distribution of gene values and equal variance at each Pfam domain value were reasonably met after log transformation. Based on the relationship between the number of Pfam domains and genes in present-day eukaryotes, the number of protein-coding genes in LECA was estimated.
Effect of the position of the eukaryotic root
The eukaryotic phylogeny and the position of its root are incorporated in our analysis at two points: in the ScrollSaw step during the identification of BBHs between eukaryotic taxa and in the LECA criteria in the tree analyses. For computational reasons we limited the analysis on the impact of the eukaryotic phylogeny on our results to the Pfams that were only present in eukaryotes. In addition to the Opimoda-Diphoda BBHs, we selected the sequences from BBHs between either five or four supergroups, as described before, and inferred phylogenetic trees. The three different sets of trees were analysed using all seven root possibilities, given the monophyly of Amorphea, Diaphoretickes, Discoba and Metamonada. To fulfil the LECA criteria a node had to contain tree sequences from both sides of the root and the mean presence of a potential LECA family in the four different groups had to be at least 15%.
Statistical analysis
Overrepresentations of functions and localisations in duplications, inventions and innovations, and overrepresentations of sister groups in duplications and duplication tendencies were tested by comparing odds ratios with Fisher’s exact tests (only pairwise comparisons of functions for inventions and localisations for innovations due to small sample sizes) or χ2 contingency table tests (rest). Differences in branch lengths were assessed with a Kruskal-Wallis test, followed by Mann-Whitney U tests upon a significant outcome of the Kruskal-Wallis test. Only one Kruskal-Wallis test did not give a significant result (Extended Data Fig. 2b). Differences between two groups were assessed with Mann-Whitney U tests. All performed tests were two-sided. In all cases of multiple comparisons, the P values were adjusted to control the false discovery rate.
The ridgeline plots were drawn with the ggridges v0.5.1 R package (https://github.com/wilkelab/ggridges).
Extended Data
Extended Data Fig. 1. Estimating the number of LECA genes from the number of Pfam domains with linear regression.
Scatter plot showing the number of Pfam domains and protein-coding genes in present-day eukaryotes, with each dot representing one genome. The regression line (black) and its 95% confidence (filled grey) and prediction intervals (dashed grey) are depicted. The vertical line corresponds to the obtained number of LECA Pfam domains.
Extended Data Fig. 2. Effect of a different phylogenetic position of the eukaryotic root.
a, Number of inferred LECA families considering different root positions. These numbers are based on phylogenetic trees from Pfams that are only present in eukaryotes. Besides the Opimoda and Diphoda groups, two other group definitions were used to identify bidirectional best hits (BBHs) and select sequences for tree inference. Names of root positions indicate either the lineage at one side of the root or the position of the split (ADis-DiaM: Amorphea+Discoba – Diaphoretickes+Metamonada; AM-DiaDis: Amorphea+Metamonada – Diaphoretickes+Discoba). Excavate sequences, especially from Metamonada species, are rarely involved in BBHs, unless specifically searched for (Excavata in BBHs 5 groups; Discoba and Metamonada in BBHs 4 groups). b, Distribution of duplication lengths obtained using different root positions for eukaryote-only trees based on the four group BBHs. The difference between distributions is not statistically significant according to the Kruskal-Wallis test.
Extended Data Fig. 3. Fraction of LECA families resulting from inventions.
a, Contribution of inventions to LECA families performing different functions. 82% of pairwise comparisons were significantly different (Supplementary Fig. 3). b, Fraction of LECA families resulting from either an invention or duplication – a eukaryotic innovation – according to functional category. 84% of pairwise comparisons were significantly different (Supplementary Fig. 5). c, Contribution of inventions to LECA families performing their function in different cellular components. 51% of pairwise comparisons were significantly different (Supplementary Fig. 4). d, Fraction of LECA families resulting from an innovation according to cellular localisation. 74% of pairwise comparisons were significantly different (Supplementary Fig. 6). a-d, Dashed lines indicate the overall invented or innovated fraction.
Extended Data Fig. 4. Phylogenetic origin of acquired Pfams.
a, b, Phylogeny of the prokaryotes (a) and Asgard archaea (b) present in our dataset based on the NCBI taxonomy. The branch widths and numbers indicate the number of acquisitions from a group. c, Number of acquisitions from different alphaproteobacterial orders or a combination of multiple orders (‘Alphaproteobacteria’).
Extended Data Fig. 5. Effect of duplications on branch lengths.
a, b, Distribution of alphaproteobacterial (a) and Asgard archaeal (b) stem lengths (sl’s) for acquisitions without and with duplications. Two alphaproteobacterial sl’s from acquisitions with Magnetococcales as sister group were removed based on the previously inferred phylogenetic position of mitochondria8. c, d, Distribution of Asgard archaeal sl’s for information storage and processing (c) and cellular processes and signalling families (d), comparing those without and with duplications. Upon removal of the outliers, the difference in cellular processes and signalling families no longer reached statistical significance. e, Distribution of Asgard archaeal sl’s for duplicated acquisitions, in which homomer-to-heteromer transitions had occurred compared to the other duplicated acquisitions. f, Distribution of vertebrate sl’s for families without and with duplications. g, Distribution of duplication lengths (dl’s) grouped according to the lineage in which the duplication occurred. All pairwise comparisons were significantly different (Mann-Whitney U tests). h, Distribution of differences in log-transformed dl values for all pairwise comparisons between chordate duplications according to age and functional annotation. All groups are significantly different (Mann-Whitney U tests). a-f, P values of Mann-Whitney U tests are shown. c-e, The minimal sl via each duplication node is plotted.
Extended Data Fig. 6. Effect of branch length normalisation and functional divergence.
a, Ridgeline plot showing the distribution of uncorrected stem (rsl) or duplication lengths (rdl). Numbers indicate the number of acquisitions or duplications for which the branch lengths were included. The low peaks at very short branch lengths are an artefact from near-zero branch lengths. Groups are ordered based on the median value of rsl’s and rdl’s. b, Ridgeline plot showing the distribution of sls for non-duplicated acquisitions that share the same functional annotation of the prokaryotic sister group and are therefore expected to have undergone little functional divergence during eukaryogenesis. a, b, Branch lengths are depicted as the additive inverse of the log-transformed values. Pairwise comparisons that did not give a significant P value (Mann-Whitney U tests) are shown.
Supplementary Material
Acknowledgements
We thank K. S. Marakova and E. V. Koonin for sharing their KOG-to-COG protein clusters with us. We are grateful to T. J. P. van Dam, E. S. Deutekom and G. J. P. L. Kops for useful advice and discussions. This work is part of the research programme VICI with project number 016.160.638, which is (partly) financed by the Netherlands Organisation for Scientific Research (NWO). T.G. acknowledges support from the Spanish Ministry of Science and Innovation for grant PGC2018-099921-B-I00 and from the European Union’s Horizon 2020 research and innovation programme under grant agreement ERC-2016-724173.
Footnotes
Author contributions
J.J.E.v.H., T.G. and B.S. conceived the study. J.V. and J.J.E.v.H. performed the research. J.V., J.J.E.v.H., T.G. and B.S. analysed and interpreted the results. M.M.H. performed the analysis on the human phylome. M.M.H. and A.v.V. aided in the development of the tree analysis pipeline. L.M.v.W. implemented the ScrollSaw-based method. J.V., J.J.E.v.H. and B.S. wrote the manuscript, which was edited and approved by all authors.
Competing interests
The authors declare no competing interests.
Data availability
Fasta files, phylogenetic trees and their annotations are available in figshare with the identifier53 doi:10.6084/m9.figshare.10069985.
Code availability
The code used to annotate the phylogenetic trees can be accessed in Github (https://github.com/JulianVosseberg/feca2leca).
References
- 1.Dacks JB, et al. The changing view of eukaryogenesis – fossils, cells, lineages and how they all come together. J Cell Sci. 2016;129:3695–3703. doi: 10.1242/jcs.178566. [DOI] [PubMed] [Google Scholar]
- 2.Shiratori T, Suzuki S, Kakizawa Y, Ishida K. Phagocytosis-like cell engulfment by a planctomycete bacterium. Nat Commun. 2019;10:1–11. doi: 10.1038/s41467-019-13499-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koumandou VL, et al. Molecular paleontology and complexity in the last eukaryotic common ancestor. Crit Rev Biochem Mol Biol. 2013;48:373–396. doi: 10.3109/10409238.2013.821444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Szathmáry E. Toward major evolutionary transitions theory 2.0. Proc Natl Acad Sci U S A. 2015;112:10104–10111. doi: 10.1073/pnas.1421398112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Spang A, et al. Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. 2015;521:173–179. doi: 10.1038/nature14447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zaremba-Niedzwiedzka K, et al. Asgard archaea illuminate the origin of eukaryotic cellular complexity. Nature. 2017;541:353–358. doi: 10.1038/nature21031. [DOI] [PubMed] [Google Scholar]
- 7.Roger AJ, Muñoz-Gómez SA, Kamikawa R. The origin and diversification of mitochondria. Curr Biol. 2017;27:R1177–R1192. doi: 10.1016/j.cub.2017.09.015. [DOI] [PubMed] [Google Scholar]
- 8.Martijn J, Vosseberg J, Guy L, Offre P, Ettema TJG. Deep mitochondrial origin outside the sampled alphaproteobacteria. Nature. 2018;557:101–105. doi: 10.1038/s41586-018-0059-5. [DOI] [PubMed] [Google Scholar]
- 9.Poole AM, Gribaldo S. Eukaryotic origins: how and when was the mitochondrion acquired? Cold Spring Harb Perspect Biol. 2014;6:a015990. doi: 10.1101/cshperspect.a015990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pittis AA, Gabaldón T. Late acquisition of mitochondria by a host with chimaeric prokaryotic ancestry. Nature. 2016;531:101–104. doi: 10.1038/nature16941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Makarova KS, Wolf YI, Mekhedov SL, Mirkin BG, Koonin EV. Ancestral paralogs and pseudoparalogs and their role in the emergence of the eukaryotic cell. Nucleic Acids Res. 2005;33:4626–4638. doi: 10.1093/nar/gki775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jékely G. Small GTPases and the evolution of the eukaryotic cell. BioEssays. 2003;25:1129–1138. doi: 10.1002/bies.10353. [DOI] [PubMed] [Google Scholar]
- 13.Wickstead B, Gull K, Richards TA. Patterns of kinesin evolution reveal a complex ancestral eukaryote with a multifunctional cytoskeleton. BMC Evol Biol. 2010;10:110. doi: 10.1186/1471-2148-10-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elias M, Brighouse A, Gabernet-Castello C, Field MC, Dacks JB. Sculpting the endomembrane system in deep time: high resolution phylogenetics of Rab GTPases. J Cell Sci. 2012;125:2500–2508. doi: 10.1242/jcs.101378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dacks JB, Field MC. Evolutionary origins and specialisation of membrane transport. Curr Opin Cell Biol. 2018;53:70–76. doi: 10.1016/j.ceb.2018.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fritz-Laylin LK, et al. The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- 18.Derelle R, et al. Bacterial proteins pinpoint a single eukaryotic root. Proc Natl Acad Sci U S A. 2015;112:E693–E699. doi: 10.1073/pnas.1420657112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Burki F, Roger AJ, Brown MW, Simpson AGB. The new tree of eukaryotes. Trends Ecol Evol. 2020;35:43–55. doi: 10.1016/j.tree.2019.08.008. [DOI] [PubMed] [Google Scholar]
- 20.Tria FDK, et al. Gene duplications trace mitochondria to the onset of eukaryote complexity. 2019 doi: 10.1101/781211v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esser C, et al. A genome phylogeny for mitochondria among α-proteobacteria and a predominantly eubacterial ancestry of yeast nuclear genes. Mol Biol Evol. 2004;21:1643–1660. doi: 10.1093/molbev/msh160. [DOI] [PubMed] [Google Scholar]
- 22.Pisani D, Cotton JA, McInerney JO. Supertrees disentangle the chimerical origin of eukaryotic genomes. Mol Biol Evol. 2007;24:1752–1760. doi: 10.1093/molbev/msm095. [DOI] [PubMed] [Google Scholar]
- 23.Narrowe AB, et al. Complex evolutionary history of translation elongation factor 2 and diphthamide biosynthesis in archaea and parabasalids. Genome Biol Evol. 2018;10:2380–2393. doi: 10.1093/gbe/evy154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Williams TA, Cox CJ, Foster PG, Szöllősi GJ, Embley TM. Phylogenomics provides robust support for a two-domains tree of life. Nat Ecol Evol. 2020;4:138–147. doi: 10.1038/s41559-019-1040-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin WF, et al. Late mitochondrial origin is an artifact. Genome Biol Evol. 2017;9:373–379. doi: 10.1093/gbe/evx027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lane N. Serial endosymbiosis or singular event at the origin of eukaryotes? J Theor Biol. 2017;434434:58–67. doi: 10.1016/j.jtbi.2017.04.031. [DOI] [PubMed] [Google Scholar]
- 27.Pittis AA, Gabaldón T. On phylogenetic branch lengths distribution and the late acquistion of mitochondria. 2016 doi: 10.1101/064873v1. [DOI] [Google Scholar]
- 28.Lane N, Martin W. The energetics of genome complexity. Nature. 2010;467:929–934. doi: 10.1038/nature09486. [DOI] [PubMed] [Google Scholar]
- 29.Lane N. Bioenergetic constraints on the evolution of complex life. Cold Spring Harb Perspect Biol. 2014;6:a015982. doi: 10.1101/cshperspect.a015982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Klinger CM, Spang A, Dacks JB, Ettema TJG. Tracing the archaeal origins of eukaryotic membrane-trafficking system building blocks. Mol Biol Evol. 2016;33:1528–1541. doi: 10.1093/molbev/msw034. [DOI] [PubMed] [Google Scholar]
- 31.Martijn J, Ettema TJG. From archaeon to eukaryote: the evolutionary dark ages of the eukaryotic cell. Biochem Soc Trans. 2013;41:451–457. doi: 10.1042/BST20120292. [DOI] [PubMed] [Google Scholar]
- 32.Akıl C, Robinson RC. Genomes of Asgard archaea encode profilins that regulate actin. Nature. 2018;562:439–443. doi: 10.1038/s41586-018-0548-6. [DOI] [PubMed] [Google Scholar]
- 33.Imachi H, et al. Isolation of an archaeon at the prokaryote–eukaryote interface. Nature. 2020;577:519–525. doi: 10.1038/s41586-019-1916-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deutekom ES, Vosseberg J, van Dam TJP, Snel B. Measuring the impact of gene prediction on gene loss estimates in Eukaryotes by quantifying falsely inferred absences. PLOS Comput Biol. 2019;15:e1007301. doi: 10.1371/journal.pcbi.1007301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huerta-Cepas J, et al. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eddy SR. Accelerated Profile HMM Searches. PLOS Comput Biol. 2011;7:e1002195. doi: 10.1371/journal.pcbi.1002195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hauser M, Mayer CE, Söding J. kClust: fast and sensitive clustering of large protein sequence databases. BMC Bioinformatics. 2013;14:248. doi: 10.1186/1471-2105-14-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van Wijk LM, Snel B. The first eukaryotic kinome tree illuminates the dynamic history of present-day kinases. 2020 doi: 10.1101/2020.01.27.920793v2. [DOI] [Google Scholar]
- 39.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 40.Shah N, Nute MG, Warnow T, Pop M. Misunderstood parameter of NCBI BLAST impacts the correctness of bioinformatics workflows. Bioinformatics. 2019;35:1613–1614. doi: 10.1093/bioinformatics/bty833. [DOI] [PubMed] [Google Scholar]
- 41.González-Pech RA, Stephens TG, Chan CX. Commonly misunderstood parameters of NCBI BLAST and important considerations for users. Bioinformatics. 2019;35:2697–2698. doi: 10.1093/bioinformatics/bty1018. [DOI] [PubMed] [Google Scholar]
- 42.Adl SM, et al. Revisions to the classification, nomenclature, and diversity of eukaryotes. J Eukaryot Microbiol. 2019;66:4–119. doi: 10.1111/jeu.12691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics. 2009;25:1972–1973. doi: 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32:268–274. doi: 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Le SQ, Dang CC, Gascuel O. Modeling protein evolution with several amino acid replacement matrices depending on site rates. Mol Biol Evol. 2012;29:2921–2936. doi: 10.1093/molbev/mss112. [DOI] [PubMed] [Google Scholar]
- 47.Minh BQ, Nguyen MAT, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price MN, Dehal PS, Arkin AP. FastTree 2 – approximately maximum-likelihood trees for large alignments. PLOS ONE. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huerta-Cepas J, Serra F, Bork P. ETE 3: reconstruction, analysis, and visualization of phylogenomic data. Mol Biol Evol. 2016;33:1635–1638. doi: 10.1093/molbev/msw046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Steinegger M, et al. HH-suite3 for fast remote homology detection and deep protein annotation. BMC Bioinform. 2019;20:473. doi: 10.1186/s12859-019-3019-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huerta-Cepas J, et al. Fast genome-wide functional annotation through orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34:2115–2122. doi: 10.1093/molbev/msx148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 53.Vosseberg J, et al. Data for: Timing the origin of eukaryotic cellular complexity with ancient duplications. figfigshare. 2020 doi: 10.6084/m9.figshare.10069985.v3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Fasta files, phylogenetic trees and their annotations are available in figshare with the identifier53 doi:10.6084/m9.figshare.10069985.
The code used to annotate the phylogenetic trees can be accessed in Github (https://github.com/JulianVosseberg/feca2leca).