Abstract
Nosocomial acquisition and transmission of vancomycin-resistant Enterococcus faecium (VREfm) is the driver for E. faecium carriage in hospitalized patients which, in turn, is a risk factor for invasive infection in immune-compromised patients. Here, we provide a comprehensive picture of E. faecium transmission in an entire sampled patient population using a sequence-driven approach. We prospectively identified and followed 149 hematology patients admitted to a hospital in England for 6 months. Patient stools (n=376) and environmental swabs (n=922) were taken at intervals and cultured for E. faecium. We sequenced 1,560 isolates (1,001 stool, 559 environment) and focused our genomic analyses on 1,477 isolates (95%) in the hospital-adapted clade A1. Out of 101 patients who provided ≥2 stool samples, forty (40%) developed E. faecium carriage after admission based on culture, compared with 64 patients (63%) based on genomic analysis (73% VREfm). Half of 922 environmental swabs (447, 48%) were positive for VREfm. Network analysis showed that, out of 111 patients positive for the A1 clade, 67 had strong epidemiological and genomic links with at least 1 other patient and/or their direct environment, supporting nosocomial transmission. Six patients (3.4%) developed an invasive E. faecium infection from their own gut-colonizing strain, which was preceded by nosocomial acquisition of the infecting isolate in half of these. Two informatics approaches (subtype categorization to define phylogenetic clusters and the development of a SNP cut-off for transmission) were central to our analyses, both of which will inform the future translation of E. faecium sequencing into routine outbreak detection and investigation. In conclusion, we showed that carriage and environmental contamination by the hospital-adapted E. faecium lineage was hyperendemic in our study population and that improved infection control measures will be needed to reduce hospital acquisition rates.
Introduction
Enterococcus faecium is a gut commensal and frequent cause of nosocomial infection in immunocompromised and critically ill patients.1 Healthcare-associated E. faecium is commonly resistant to numerous antibiotics including ampicillin and vancomycin, limiting treatment options.2 The 30-day crude mortality rate for bacteremia with vancomycin-resistant E. faecium (VREfm) is 35%, with higher mortality and hospital stay for bacteremia caused by VREfm versus vancomycin-susceptible E. faecium. 3 The World Health Organization has classified VREfm as high priority in their list of pathogens for research on new antibiotics.4 The development of VREfm carriage in the gut is the single most important risk factor for VREfm bacteremia.5,6 Preventing E. faecium acquisition requires an understanding of reservoirs and transmission routes. It has been known since the 1990’s that VREfm is shed into the hospital environment where it may persist despite standard cleaning,7,8 and that being admitted to a room previously occupied by a VRE-positive patient is a risk factor for acquiring VRE.9 This evidence is based on culture and bacterial typing techniques with low discrimination,10 which has limited ability to establish relatedness between isolates from patients and their environment, the frequency with which patients carry more than one strain, and patterns and frequency of transmission. Whole genome sequencing has been successfully used to study transmission of E. faecium between livestock and humans11,12, in national surveillance programs13,14, across health-care networks15 and within hospitals.16–18 While previous hospital studies have largely focused on isolates associated with bacteremia, a recent study19 sequenced both carriage and clinical isolates prospectively. However, it did not measure acquisition rates, ascertain within patient diversity, as a single isolate was sequenced per patient, or define the role of the hospital environmental as a reservoir, as it lacked environmental sampling.
Here, we address existing knowledge gaps by undertaking longitudinal genomic surveillance of E. faecium carriage, environmental contamination and transmission in a defined patient cohort.
Results
Study patients commonly carried vancomycin-resistant E. faecium
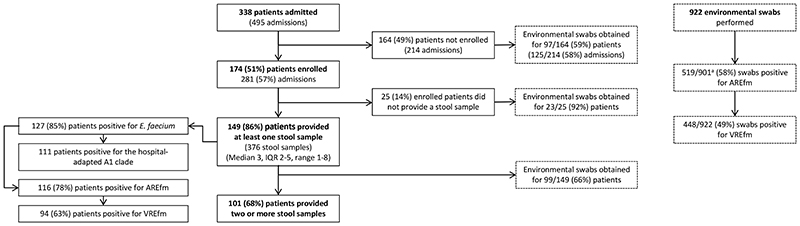
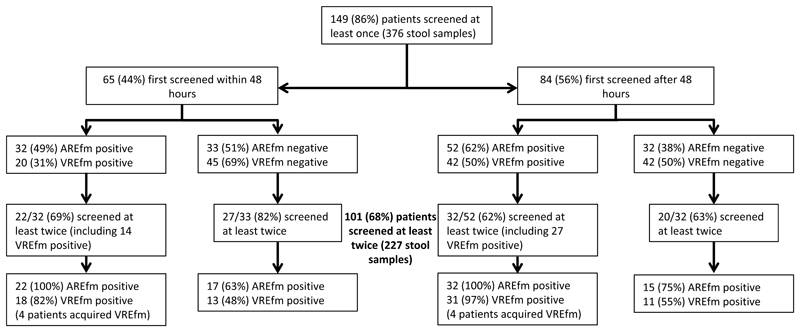
First, we determined the extent to which a putatively high-risk population of patients on two hematology wards carried and acquired E. faecium based on culture methods. The study ward characteristics, antimicrobial stewardship, infection control and cleaning policies are described in Supplementary Methods. We recruited 174 of 338 patients (51%) admitted to the two wards over 6 months (Figure 1). Study participants were a median age of 61 years (IQR 49 to 69, range 19-94), were admitted a median of once (IQR 1 to 2, total 281 admissions), and stayed a median of 16 days (IQR 7 to 27 days) (Supplementary Table 1). 149/174 participants provided at least one stool for culture, which resulted in the isolation of any E. faecium, ampicillin-resistant E. faecium (AREfm) and VREfm from 85%, 78% and 63% of the 149 cases, respectively. Based on 101 patients who provided ≥2 stools, 40/101 (40%) cases acquired E. faecium (changed from culture-negative to culture-positive) following admission (Extended Data Figure 1).
Figure 1. Study participants and E. faecium culture positivity.
The number of patients sampled is shown as a subset of those enrolled, which in turn are a subset of those admitted to the two hematology wards. The overall number of environmental swabs taken from the hospital environment and for non-sampled patients is given. Culture positivity values for E. faecium, ampicillin-resistant E. faecium and vancomycin-resistant E. faecium are shown for both sampled participants and environmental swabs at the left and right-hand side of the figure, respectively. Supplementary Table 2 provides a breakdown of positivity rates for different environmental sources. a 21 swabs were negative for VREfm and were not cultured for AREfm.
Vancomycin-resistant E. faecium was ubiquitous in the ward environment
In parallel, we evaluated the extent to which the patient environment contained E. faecium. Nearly half of 922 environmental swabs (447, 48%) taken over 6 months were culture-positive for VREfm, the positive proportion ranging from 36% for medical devices to 76% for non-touch areas (see Supplementary Table 2 for full details). No E. faecium was isolated by air testing. Environmental swab VREfm positivity was similar between the two wards (237/457 (52%) vs. 166/327 (51%), for ward A and B respectively; chi2 p=0.76), and between individual rooms versus multiple-bed bays (119/255 (47%) vs. 196/374 (52%), respectively; chi2 p=0.16). Of 41 swabs taken from bedroom/bathroom areas after patient discharge and routine cleaning, 13 (32%) and 8 (20%) were positive for AREfm and VREfm, respectively. Deep cleaning was undertaken on ward B over a 3-day period during the study, when patients were moved elsewhere. This failed to eradicate E. faecium in 4/43 (9%) sampled locations prior to patients returning to the ward. These four environmental isolates collected immediately after deep cleaning were all genetically related (0 to 1 SNP) to isolates collected just before cleaning, demonstrating that bacteria in these four sites persisted through decontamination. Any benefit of deep cleaning was short-lived, as around half of sampled sites were positive within 3 days of patient return (Supplementary Table 2). Isolates from positive sites within 3 days of patient return were mostly related (14/18, 0 to 4 SNPs) to isolates collected immediately before cleaning, demonstrating that reestablishment of environmental contamination was mostly caused by bacteria that were likely reintroduced by colonized patients.
Delineation of E. faecium subtypes
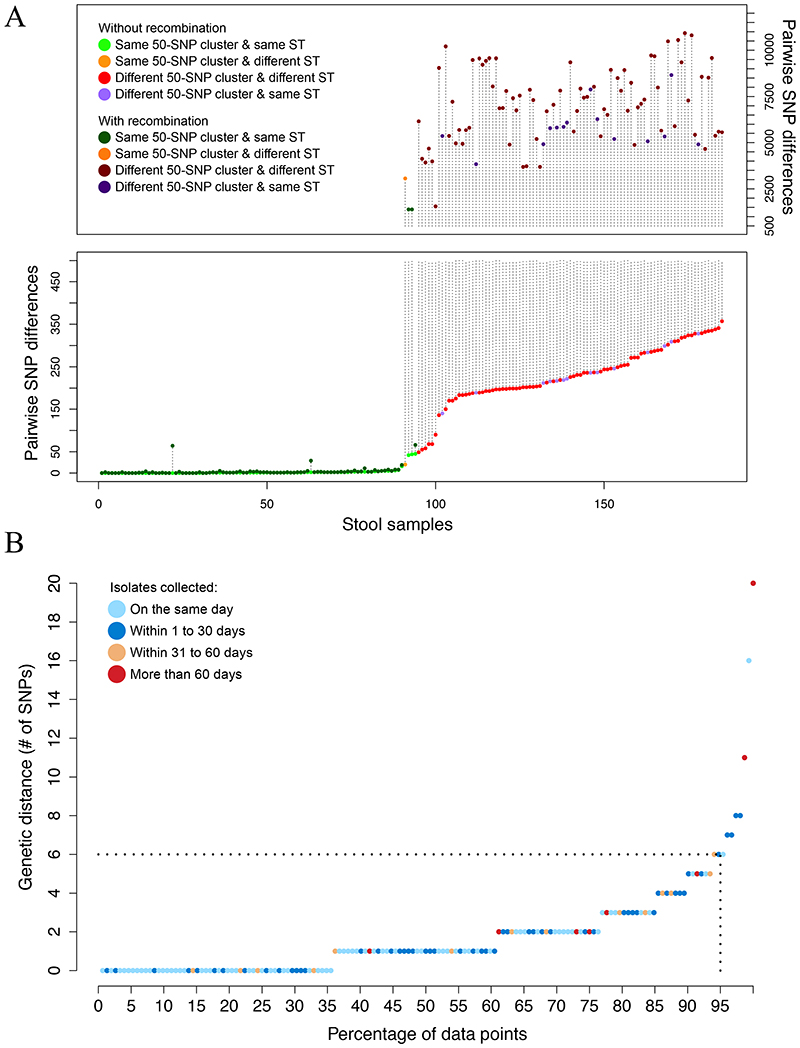
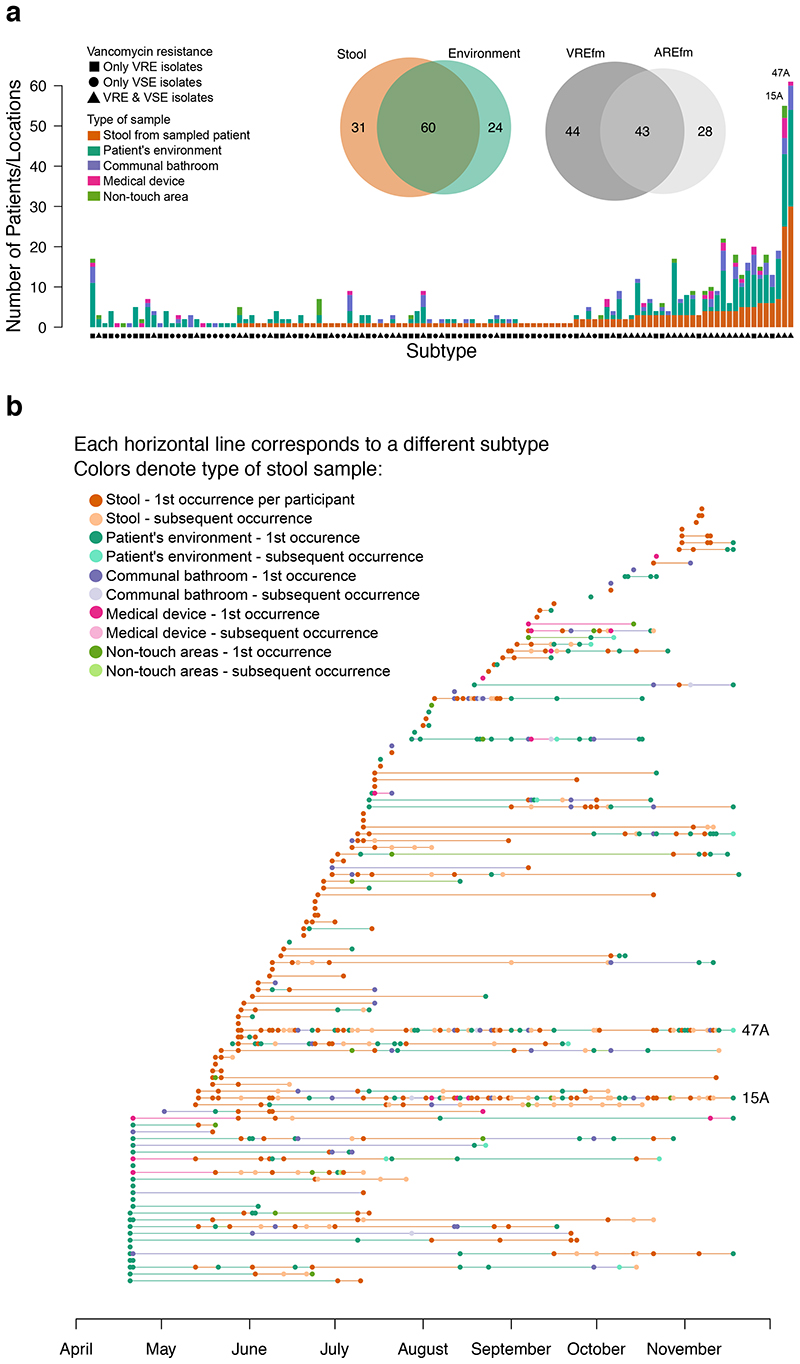
Having established that carriage of drug-resistant E. faecium was highly endemic and that environmental contamination was ubiquitous, we sought to update evidence for E. faecium relatedness in the two reservoirs using a contemporary, genome-based approach. We sequenced 1,560 isolates (1,001 from stool, 559 from the environment) and confined further analyses to 1,477 isolates (95%) assigned to the hospital-adapted clade A1 (see Supplementary Methods for details of how clade A1 isolates were defined). Of these, 943 were from 263 stools/111 patients (Figure 1), and 534 were from the environment. Clade A1 is an ampicillin-resistant E. faecium lineage which frequently acquires vancomycin resistance.20 We restricted our genomic analysis to this clade because it accounts for the vast majority of invasive infections.14 We further divided the clade A1 population into discrete and non-overlapping genetic clusters based on monophyletic groups in the whole-genome phylogeny, such that isolates within each group were no more than 20 SNPs different from each other, which were referred to as ‘subtypes’. We first chose an arbitrary cut-off of 50 SNPs to define monophyletic groups in the phylogeny. We counted pairwise SNP differences between isolates of the same and different 50-SNP clusters within each stool sample (Figure 2A), before and after removing recombination. With one exception, all isolates differing by less than 20 SNPs had limited recombination and always belonged to the same sequence type (ST). Based on these results, we selected 20 SNPs as the threshold to define E. faecium subtypes (see Supplementary Methods and Extended Data Figure 2 for details). We identified 115 genetically distinct E. faecium subtypes distributed across patient and environmental sources (31 from stool alone, 24 from the environment alone, and 60 from both) (Figure 3A & B). We found a very good correlation between the clustering provided by subtypes (n=115), STs (n=55) and BAPS21 clusters (n=25), where subtypes provided the highest discrimination and BAPS clusters the lowest (Supplementary Data 1).
Figure 2. E. faecium within host diversity.
(A) Pairwise SNP differences between isolates of the same stool before and after removing recombination for a total of 185 stool samples. In samples with all isolates belonging to the same 50-SNP cluster, the maximum distance is plotted. In samples with multiple 50-SNP clusters, the minimum SNP distance between isolates from different clusters is plotted. Colors are used to indicate whether isolates belong to the same cluster and ST. The length of the vertical dotted lines denote the number of SNPs attributable to recombination. (B) Maximum pairwise SNP distance among isolates of the same subtype and in the same patient (n=152). The color of the dot denotes the time span between isolates.
Figure 3. Frequency and time span of E. faecium subtypes.
(A) Frequency of E. faecium subtypes in stool and the environment. Each of 115 bars represent a different subtype ordered by increasing frequency in patient stool. Venn diagrams show distribution of vancomycin resistance (upper) and place of isolation (lower). (B) Isolation of 115 E. faecium subtypes over time. Each horizontal line represents a distinct subtype. Circles represent instances when subtypes were isolated, and the color of circles the source of isolation (stool or environmental). The number of dark brown circles for each subtype denotes the number of subjects colonised with it. Horizontal lines joining circles of the same subtype are colored based on the source of the preceding sample.
Carriage of multiple E. faecium subtypes was common
Carriage of multiple E. faecium strains is an important confounder for outbreak investigations that include an evaluation of stool carriage isolates, since a non-outbreak strain could be erroneously selected. Furthermore, standard typing methods may not distinguish between even distantly related strains carried by the same individual. We used genomic data to re-evaluate the question of mixed-strain carriage, sequencing numerous isolates cultured from 185 stools (109 patients) that had two or more primary plate colonies sequenced (median 5, IQR 3 to 5, total 865 colonies) (see Supplementary Methods for further details). Within the limits of detection of our methods, we found that just over half of all stools (94 stools from 63 patients) contained at least two (two (n=83), three (n=10) or four subtypes (n=1)), providing clear evidence that mixed strain E. faecium carriage is common. When patients were colonized with multiple subtypes, isolate pairs from the same stool belonging to different subtypes had a median of 235 SNPs (IQR 198 to 289), whereas isolate pairs of the same subtype had a median of 0 SNPs (IQR 0 to 1).
E. faecium carried by patients were commonly shared with their environment
Having defined subtypes in the stool of individual patients, we then defined the frequency with which subtypes were shared between patients. 36/91 subtypes (40%) identified in stools were isolated from two or more patients. This included two highly dominant subtypes (denoted here as 47A (sequence type (ST)78) and 15A (ST80)), which were isolated from 25 and 30 participants, respectively, spanned the entire study, and accounted for 243/943 (26%) of all stool isolates (Figure 3A). E. faecium subtypes in stool were often present in the ward environment (60/91, 66%). This was particularly the case for subtypes isolated from multiple patients, which were over-represented in the environment compared to subtypes isolated from single patients (32/36, 89%, versus 28/55, 51%, Fisher’s exact test, p<0.001). Subtypes carried by multiple patients were particularly associated with contamination of communal bathrooms and medical devices (23/36 vs. 9/55, and 11/36 vs. 2/55, respectively; p<0.001 for both).
Nosocomial acquisition of E. faecium and vancomycin-resistant E. faecium was common
Almost two thirds of patients (64/101) acquired one or more E. faecium subtype through a total of 111 acquisition events (changed from subtype negative to subtype positive), which is nearly three times the number of acquisitions detected by culture alone (n=40). Culture underestimated E. faecium acquisition because it could not detect the acquisition of new subtypes by individuals already colonised with E. faecium. This equates to an acquisition rate of 59.4/100 admissions compared to 21.4/100 admissions based on the number of admissions/readmissions by study patients over 6 months (n=187). The two most common subtypes (47A (ST78) and 15A (ST80)) accounted for 28% of acquisition events. Eighty-one acquisitions (in 52 patients) were VREfm and 30 acquisitions (in 26 patients) were AREfm. In addition to the 81 VREfm subtype acquisitions, four patients had a vancomycin-susceptible subtype detected in stool that switched to vancomycin-resistant due to vanA, which can be explained by gene acquisition or presence of a mixed population of susceptible/resistant isolates. This indicates that the dominant mode of VREfm acquisition within this study was through transfer of already vancomycin-resistant subtypes rather than horizontal gain of vancomycin resistance genes.
Common E. faecium subtypes were not more tolerant to hospital disinfectants
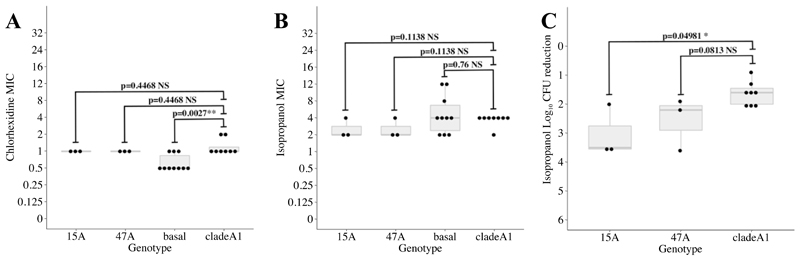
Because E. faecium may persist in the hospital environment through increased tolerance to hospital disinfectants such as chlorhexidine or alcohol,22,23 we investigated whether the two most common subtypes had higher minimum inhibitory concentrations (MICs) to these two agents, or higher tolerance to isopropanol as previously described.23 MICs to chlorhexidine and alcohol were not higher on average (Extended Data Figure 3A and B) than that of other clade A1 subtypes, and neither were more tolerant to isopropanol (Extended Data Figure 3C). These results suggest that factors other than increased tolerance to disinfectants may be responsible for the higher frequency of these subtypes.
SNP cut-off to detect recent E. faecium transmission
Although acquisition implies transmission, we applied a more stringent cut-off to each subtype to quantify recent E. faecium transmission between patients and/or their environment. This was based on the rationale that the maximum genetic diversity found in the same patient defines the amount of diversity that could potentially be transferred from one person to another. We thus quantified the amount of diversity in subtypes with at least two available isolates from the same patient (total of 152 patient-subtype combinations from 104 patients). Within-host subtype diversity in our study population was ≤6 SNPs in 95% of comparisons (Figure 2B). Applying this cut-off to the acquisition events showed that 78 (70%) of the acquired subtypes were highly related (median 0, IQR 0 to 2 SNPs, maximum 6 SNPs) to an isolate from a previously sampled patient, supporting recent transmission. Epidemiological analysis of these 78 putative donor-recipient pairs demonstrated that 61 (78%) pairs had resided in the same location (bay, room or ward) at the same time or within 7 days, providing strong epidemiological evidence for transmission (Table 1, Supplementary Table 3, Supplementary Data 2).
Table 1. Genomic and epidemiological evidence of nosocomial E. faecium transmission.
| Number of subtypes (patients) |
Median SNP distance (IQR) |
Strong environme links3 |
|
|---|---|---|---|
| Acquired subtypes | 111 (64)1 | - | 41/111 |
| Genetically unlinked | 22 (19) | - | 1/22 |
| Genetically linked to | 89 (56) | - | 40/89 |
| Future sampled patients | 11 (9) | - | 1/11 |
| Previously sampled patients | 78 (52) | 0 (0-2) | 39/78 |
| With weak epidemiological links | 17 (15) | 2 (0-2) | 7/17 |
| With strong epidemiological links | 61(43) | 0 (0-1) | 32/61 |
| Same bay or room, same time | 16 (11) | 0 (0-0) | 13/16 |
| Same bay within 7 days | 5 (5) | 0 (0-1) | 4/5 |
| Same ward, same time | 32 (24) | 0 (0-1) | 11/32 |
| Same ward within 7 days | 8 (8) | 2.5 (0-3) | 4/8 |
| Index subtypes | 116 (80)4 | - | 24/116 |
| Genetically unlinked2 | 37 (32) | - | 4/37 |
| Genetically linked to | 79 (50) | - | 20/79 |
| Future sampled patients | 28 (24) | - | 5/28 |
| Previously sampled patients | 51 (43) | 1 (0-3) | 15/51 |
| With weak epidemiological links | 20 (20) | 2 (0-3) | 5/20 |
| With strong epidemiological links | 31 (25) | 1 (0-2) | 10/31 |
| Same bay or room, same time | 13 (11) | 0 (0-2) | 4/13 |
| Same bay within 7 days | 2 (2) | 1 (0-2) | 1/2 |
| Same ward, same time | 16 (14) | 1 (0-2.5) | 5/16 |
| Same ward within 7 days | - | - | - |
A total of 227 unique subtype-patient combinations were identified in 111 patients positive for clade A1, 38 of whom carried a single subtype and 73 carried multiple subtypes. Of the 227 subtype-patient combinations, 111 were acquired based on consecutive sampling and 116 were detected in the first available stool sample. For each subtype in each patient, evidence of nosocomial transmission was supported by genetic links to E. faecium isolates sampled in previous patients or environmental locations. Epidemiological data provided a second level of evidence of hospital transmission.
Among patients with at least two available stool samples (n=101).
Using a cut-off of 6 SNPs to detect recent transmission of E. faecium subtypes during the study period.
Genetically linked and with strong epidemiological links to previously sampled environmental sites.
Among all patients positive for the hospital- adapted clade (n=111).
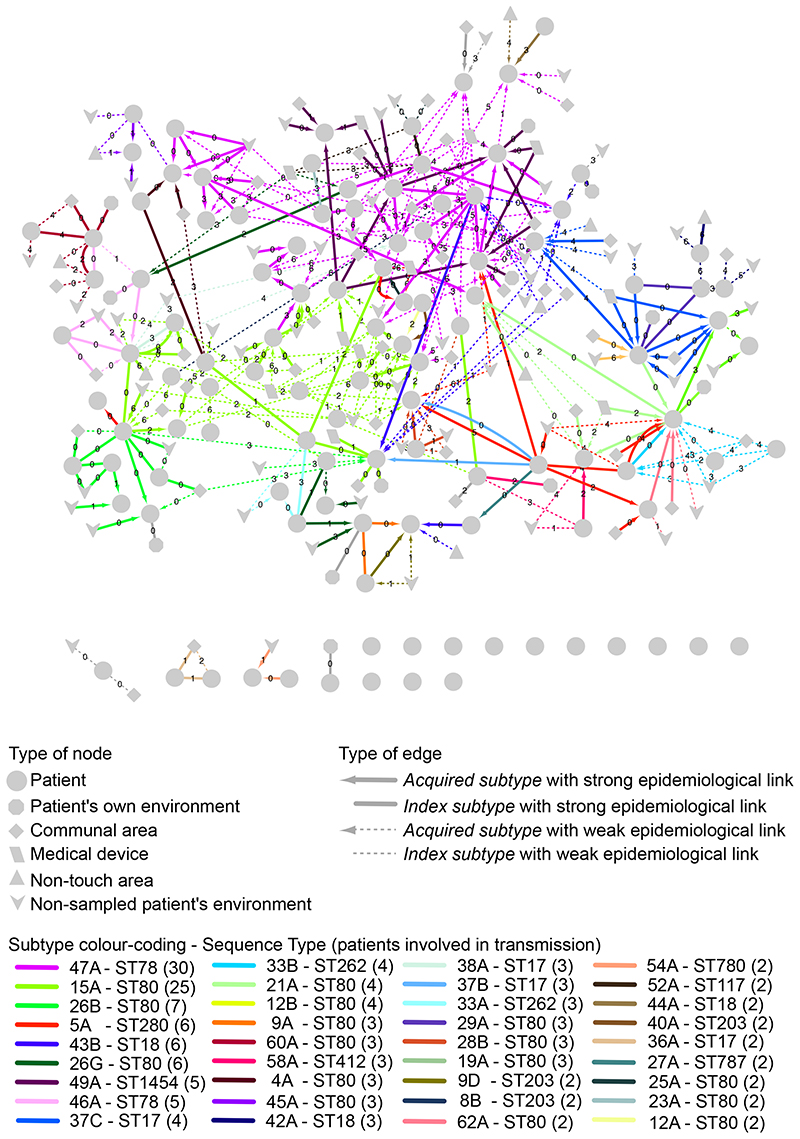
We then applied the 6 SNP cut-off to all patients positive for the hospital-adapted clade A1 (n=111). Figure 4 shows a visual representation of transmission between patients and their environment using a network that combined bacterial relatedness and strength of epidemiological links. A high proportion of patients (67/111) had strong genetic (≤6 SNPs) and epidemiological links to one or more patients and/or their direct environment, supporting nosocomial transmission. There were four cases where two subtypes transmitted between the same donor-recipient pair, which could arise from a single event involving more than one strain or repeated transmission events. We also found evidence of multiple variants of the same subtype being transmitted in the same transmission event, as revealed by the range of SNP distances between isolates of the same transmitted subtype in the donor and recipient patients (Supplementary Data 2).
Figure 4. E. faecium transmission network.
Transmission of clade A1 E. faecium subtypes between patients (n=111) and their environment represented as a network. Only those environmental locations from which E. faecium was cultured that was genetically related (isolate within 0 to 6 SNPs) to at least one patient isolate are shown. For each patient node, lines show the shortest genetic link (within 0 to 6 SNPs) to a previously sampled patient or environmental location (putative transmission). Numbers on network edges show SNP distances. Edge colors show the subtype being transmitted. Acquired subtype refers to subtypes not present in previously collected stool samples, whereas subtypes present in the first available stool are termed index subtypes.
Genetic and epidemiological links were used to reconstruct the temporal and spatial spread of each E. faecium transmission cluster (Supplementary Table 4). The size of transmissions clusters ranged from 2 to 8 patients. Eleven clusters consisted of a single transmission event, i.e. involving only two patients, and, with one exception, did not involve the hospital environment. The remaining 15 transmission clusters involved 3 to 8 patients and in most cases (13/15), the hospital environment was found to be a plausible source of transmission (see Extended Data Figure 4 for two examples).
Invasive E. faecium infections were associated with new VREfm acquisition
A serious consequence of E. faecium carriage is the development of invasive infection. This occurred in 6 study patients (3.4%), equating to 21 invasive infections per 1,000 admissions (see Table 2 and Supplementary Table 5 for details). Five of these patients had at least one stool cultured, all 5 of whom were positive for E. faecium. Comparison of stool and disease-associated E. faecium genomes in each patient showed that the invasive and stool subtype was highly related in all 5 cases (0 to 5 SNPs). The invasive subtype was acquired after admission in three cases based on an earlier negative stool for the invasive subtype. Two patients had strong epidemiological links with another study case with whom they shared an identical isolate based on a core genome comparison.
Table 2. Relatedness of E. faecium associated with carriage and invasive disease.
| Patient ID |
Clinical samples | Stool samples | Relatedness analysis | |||||
|---|---|---|---|---|---|---|---|---|
| Sample type |
No. of samples1 (colonies sequenced) |
Isolate subtype (Sequence type) (SNP range) |
No. of stool samples1 (colonies sequenced) |
Stool subtype(s) (Sequence type)2 |
SNP distance: matching infection/stool subtype |
Classification of stool subtype |
Epidemiological link3 (SNP distance with isolate from linked case) |
|
| C016 | Blood | 8 (56) | 12B (80)(0-3) | 6 (38) | 12B (80), 32A (262), 15A (80), 25A (80), 47A (78) | 0 to 5 | Index | None |
| C027 | Blood | 1 (1) | - | 0 | - | - | - | - |
| D041 | Blood | 1 (10) | 47A (78) (0-0) | 3 (15) | 47A (78), 49A (1454), 5A (280) | 0 to 4 | Acquired | Strong link with patient D034 (0) |
| C076 | Surgical Biopsy |
1 (1) | 15A (80) | 2 (6) | 15A (80), 37B (17) | 0 to 2 | Index | Weak link with D012 (6) |
| D049 | Blood | 1 (10) | 15A (80) (0-1) | 2 (12) | 15A (80), 28B (80), 5A (280), 37B (17) | 1 to 2 | Acquired | Weak link with C084 (1) |
| C095 | Blood | 2 (20) | 45A (80) (0-3) | 1 (5) | 45A (80) | 1 to 4 | Acquired | Strong link with C092 (0) |
Samples positive for E. faecium.
Subtypes identified across all stool samples from the same patient.
Strong link, defined as admission to the same bay, room or ward at the same time or within 7 days following discharge of the previous occupant; admission to the same ward separate by more than 7 days or admission to the study hospital but to different wards were considered weak epidemiological link.
Discussion
Our study is the first prospective observational study to quantify hospital acquisition rates for E. faecium, by combining in depth longitudinal sampling and use of whole-genome sequencing. This approach allowed us to demonstrate that E. faecium acquisition rates were significantly higher than indicated by culture alone. We were able to establish the hospital location of such acquisitions, thanks to the density of clinical and environmental sampling and integration of genomic and epidemiological data. Underpinning the above was the finding that mixed strain carriage was common and the description of within-patient strain diversity at the core genome. We also provided clear evidence of invasive E. faecium infections originating from patients’ own gut-colonizing strains, and demonstrated that nosocomial transmission is a key risk factor for subsequent colonization and infection. Our study confirms previous observations that VREfm can persist in the hospital environment despite standard cleaning7,8, and that sharing hospital wards previously occupied by a VREfm-positive patient is a risk factor for acquiring VREfm, although we did not demonstrate direct acquisition from a previously contaminated room.
Carriage and environmental contamination by the hospital-adapted E. faecium lineage was hyperendemic in our study population, rates of which exceeded those reported previously from CUH20 and elsewhere.22,24–27 This occurred despite the use of cleaning products and procedures with proven efficacy against VREfm based on effectiveness in reducing rates of infection.26,27 These high transmission rates were in sharp contrast to those detected for Klebsiella pneumoniae (n=0 patients)25 and Escherichia coli (n=20 patients, data not published) in the same setting and patient population. Our study design supported the development of two informatics approaches (subtype categorization to define phylogenetic clusters and the development of a SNP cut-off for transmission) that will inform future genomic epidemiology studies of E. faecium as well as translation of bacterial sequencing into routine outbreak detection and investigation. The use of subtype provided important new insights into the rate at which patients carried more than one strain. Carriage of multiple E. faecium subtypes was common, a finding that can be explained by repeated acquisition events and/or a single transmission of a mixed population. Mixed strain carriage indicates that the sensitivity of transmission detection based on stool testing will depend on the number of primary plate colonies tested from each sample. Subtype data also provided evidence for extensive overlap in the E. faecium populations residing in the patient gut and in their environment, which is consistent with a highly dynamic pattern of two-way spread between these reservoirs. The most prevalent E. faecium subtypes in patients were the ones most often detected in the hospital environment and, related to this, subtypes associated with large transmission clusters were over-represented in the environment, suggesting environmental surveillance could be an alternative to patient screening in this setting.
Our study found that the two most common subtypes accounted for 28% of acquisition events. In vitro susceptibility testing of representative isolates ruled out increased tolerance to disinfectants as one the factors responsible for the higher frequency of these subtypes. A recent genomic epidemiological study conducted in a German hospital19 similarly found that the increase observed in VREfm was mostly attributable to the expansion of two major clones that were present throughout the hospital, where both intrahospital patient-to-patient transmission and reintroduction from local hospitals had occurred. Future work is needed to elucidate the bacterial and epidemiological factors driving the expansion of particular E. faecium clones.
Our study has several limitations. First, we sampled less than 50% of patients admitted to the two hematology wards, and we did not sample healthcare workers. Unsampled carriers can explain, in part, why some acquired subtypes were not detected in any other patient (genetically unlinked), or why some patient pairs lacked strong epidemiological links despite carrying highly related isolates. Altogether, unsampled carriers would result in an under-estimation of transmission reported here. Another limitation is that we did not sequence the full diversity of E. faecium in stool samples but a maximum of five colonies. This can lead to some subtypes being wrongly classified as acquired but instead being present at low abundance in previous samples. Future studies will need to sequence directly from plate sweeps to capture the full heterogeneity within individuals. On the other hand, subtypes classified as index could have been potentially acquired between admission and the time of sampling.
In conclusion, we presented the most detailed genomic study of E. faecium hospital transmission. Whereas rates of acquisition and degree of endemicity of the global clade A1 E. faecium may vary between hospitals, and regions, the mechanisms of transmission, infection sources and methodological developments presented here are likely to be generalizable to other settings. This is particularly true for hemato-oncology units, which are often associated with high rates of VREfm infection and colonization in the USA, Europe and Australia. The high endemicity and acquisition rates of drug-resistant E. faecium in hematology wards poses an important challenge to infection control. Patient screening, adequate provision of isolation and en-suite toilet facilities, improved and more frequent cleaning procedures, and stricter health-care worker hygiene practices will all be needed, in addition to antimicrobial stewardship interventions to curtail this global epidemic.
Methods
Setting and study design
The study protocol was approved by the National Research Ethics Service (ref: 14/EE/1123), and the Cambridge University Hospitals NHS Foundation Trust Research and Development Department (ref: A093285). We conducted a prospective observational study of consecutive patients admitted to two hematology wards at the Cambridge University Hospitals NHS Foundation Trust (CUH) in the UK between 13 May and 13 Nov 2015. Patients were enrolled following informed written consent, after which stool samples were requested on admission, every week and at discharge, and cultured for E. faecium. Dates of hospital admission, ward transfers and bed positions were extracted electronically using the hospital bed tracking system. Blood cultures taken for clinical reasons were recorded and cultures positive for E. faecium retrieved from the routine laboratory. Three weeks before the study started, environmental sampling for E. faecium was performed on both wards to establish baseline levels of contamination. Environmental sampling was also conducted throughout the study, in which communal bathrooms, toilets, non-touch surfaces (air vents and HEPA filters) and a range of medical devices were swabbed every fortnight. The day ward (used for outpatient chemotherapy administration) was swabbed at the start and midpoint of the study. In addition, two pooled swabs were taken in each of the patient bedside and bathroom areas on the day of discharge for participants if no discharge stool sample was provided, and for non-participants. Air sampling was performed on three occasions on both wards. Supplementary Methods provide details of bed layout, swabbing and air sampling methodology and locations, and infection control, cleaning and antibiotic prescribing policies.
Microbiology, DNA sequencing and phylogenetic analyses
Isolation, identification and susceptibility testing of E. faecium from stool, environmental samples and blood cultures are described in Supplementary Methods. Antibiotic selective media were used to isolate ampicillin-resistant E. faecium (AREfm) and VREfm from stool and the environment, with additional non-antibiotic media used in first stool samples to isolate any E. faecium. Multiple E. faecium colonies were picked from primary cultures of positive stool samples to detect genetic diversity. Supplementary Methods describe the rationale for selecting isolates for sequencing. DNA was extracted, libraries prepared and 125-bp paired end sequences determined on an Illumina HiSeq2000. Isolate genomes belonging to the hospital-adapted clade A1 (previously known as clonal complex (CC) 17)28 were mapped to E. faecium Aus0004 strain (GenBank accession number CP003351) using SMALT v0.7.4 (http://www.sanger.ac.uk/science/tools/smalt-0). Single nucleotide polymorphisms (SNPs) were identified from BAM files using SAMtools v0.1.1929 to create a whole-genome alignment. Mobile genetic elements and recombination events detected by Gubbins v1.4.1030 were removed to define the core genome. RAxML v8.2.831 with 100 bootstraps was used to create a maximum likelihood tree from the core genome alignment. Pairwise genetic distances between isolates were calculated based on core genome SNPs. Isolates sequenced from blood cultures and stool from the same patient were compared to determine genetic relatedness and origin of the invasive isolate.
Genomic and epidemiological analyses to quantify nosocomial transmission
Genomic and epidemiological analysis was limited to the hospital-adapted A1 clade (see Extended Data Figure 5 and Supplementary Methods for how these were identified). E. faecium acquisition was defined based on culture (transition from culture-negative to culture-positive stool), and genetic criteria (acquisition of a new subtype) for patients with at least two available stool samples (n=101). Acquisition rates were calculated as the number of E. faecium acquisitions divided by the number of admissions to hematology wards by these 101 patients (n=187). Subtypes present in the first available stool based on sequencing of a median of 5 independent colonies (IQR 5-6) were termed index subtypes as opposed to acquired subtypes. For each subtype in every patient, we considered previously sampled patients and environmental locations as possible sources and identified the putative donor as the one with the genetically closest E. faecium isolate (within the 6 SNP cut-off). Admission to the same bay, room or ward at the same time or within 7 days were classified as strong epidemiological links, while admissions in the same ward separated by more than 7 days or to the study hospital but to different wards were classified as weak epidemiological links (see Supplementary Methods for a further explanation of this classification). A transmission network was constructed using R v3.4.132 and visualized in Cytoscape v3.2.0.33 Transmission plots were drawn using R to visualize the spatial and temporal spread of E. faecium subtypes.
Isopropanol and chlorhexidine MIC testing
Isolates were grown from -70°C storage in glycerol onto Columbia blood agar (CBA) plates overnight in 37°C air. MIC testing was done using the broth microdilution method.34 Isolates had a final dilution of 5 x 105 CFU/ml in iso-sensitest broth in a 96-well flat-bottomed microtiter plate. Isopropanol concentrations were tested at 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 12, 16, 24 and 32% (v/v). Chlorhexidine concentrations were tested at 0, 0.125, 0.25, 0.5, 1, 2, 4, 8, 12, 16, 24 and 32 mg/L. Isolates were tested in triplicate, and the MIC was found by visually inspecting the microtiter plate for the well containing the lowest concentration of biocide where no visible growth had occurred. The resulting MIC values are presented in Supplementary Table 6.
Isopropanol tolerance assay
Isolates were grown from -70°C storage in glycerol onto CBA plates for overnight incubation in 37°C air. A colony was then incubated overnight in 10 mL brain heart infusion (BHI) broth at 37°C in air. The overnight cultures were diluted to 0.5 at optical density 600 nm (OD600nm) in PBS. 23% (v/v) isopropanol was added to 1 mL of the dilution, and 23% PBS was added to another 1 mL dilution. Both were vortexed thoroughly then incubated at room temperature for 5 minutes. The samples were then serially diluted between 10 and 1000000-fold in 7.5% Tween 80 in PBS, to inactivate the isopropanol. 50 μL of each dilution was evenly spread onto Muller Hinton agar plates using an L-shaped spreader. Once dried, plates were incubated overnight at 37°C in air. Each isolate was tested in triplicate (biological replicate) and each dilution was plated in triplicate (technical replicate). Colonies were counted and averaged across technical triplicates, and the log10 CFU reduction calculated for each isolate between exposure to PBS to exposure to 23% isopropanol.23 The log10 CFU reduction values are presented in Supplementary Table 7.
Statistical methods
The null hypothesis (no difference between means) of median (across replicates) MICs and log10 CFU reduction values between lineage and subtype groups (that is, clade A1 vs. basal, clade A1 vs. 15A (ST80) and clade A1 vs. 47A (ST78)) in Extended Data Figure 2 was rejected for P < 0.05 and was assessed using an unpaired Mann-Whitney test with a two-tailed P value. This test was performed using the wilcox.test function from R package stats (v3.6.3).
Extended Data
Extended Data Fig. 1. E. faecium stool culture positivity during study.
Diagram showing E. faecium positivity in patients who provided stool samples within (left branch) or after (right branch) 48 hours from index admission. Subsequent boxes show numbers of patients positive or negative for AREfm and VREfm, and, for patients screened at least twice, whether their positivity status changed, suggestive of E. faecium acquisition. A total of 40 cases acquired E. faecium based on culture, either by acquiring any type of E. faecium after being negative for it (17 and 15 patients in the left and right arms, respectively) or VREfm after being already positive for AREfm (4 and 4 patients in the left and right arms, respectively). Abbreviations: AREfm, ampicillin-resistant E. faecium (which may be vancomycin susceptible or resistant); VREfm, vancomycin-resistant E. faecium.
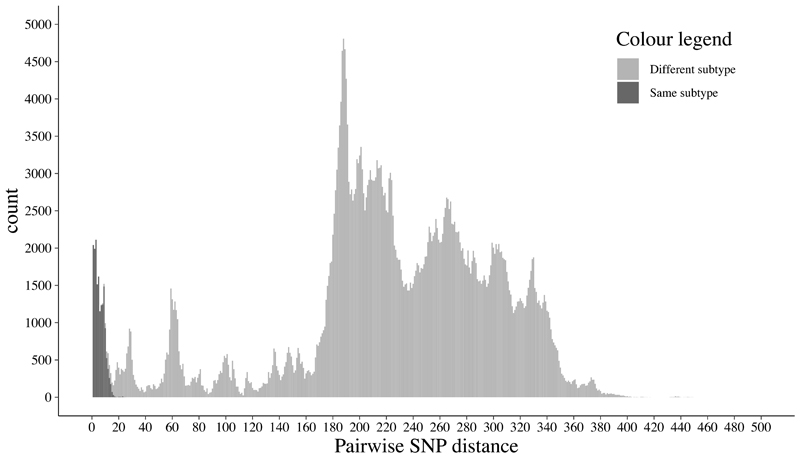
Extended Data Fig. 2. Histogram of pairwise SNP differences between isolates of the same and different subtypes.
Histogram of pairwise SNP differences between 943 clade A1 isolates from stool samples. SNP differences between isolates from the same subtype are shown in dark grey, and between isolates in different subtypes in light grey.
Extended Data Fig. 3. Chlorhexidine and isopropanol susceptibility among selected E. faecium isolates.
Chlorhexidine and isopropanol susceptibility testing results for a subset of phylogenetically representative E. faecium isolates (n=24 biologically independent samples) from the two major subtypes (15A/ST80 (n=3) and 47A/ST78 (n=3)), rest of subtypes in ‘clade A1’ (n=8) and ‘basal’ isolates (n=10) to clade A1. Each dot denotes the median MIC value (panels A and B) or median reduction in colony forming units (CFU) (panel C) across three independent replicates for each isolate tested. In the boxplots, the lower and upper hinges correspond to the first and third quartiles (the 25th and 75th percentiles), and the middle horizontal line to the median. P-values for two-tailed, unpaired Mann-Whitney are showed as NS (non-significant, P > 0.05), * (P < 0.05) or ** (P < 0.01).
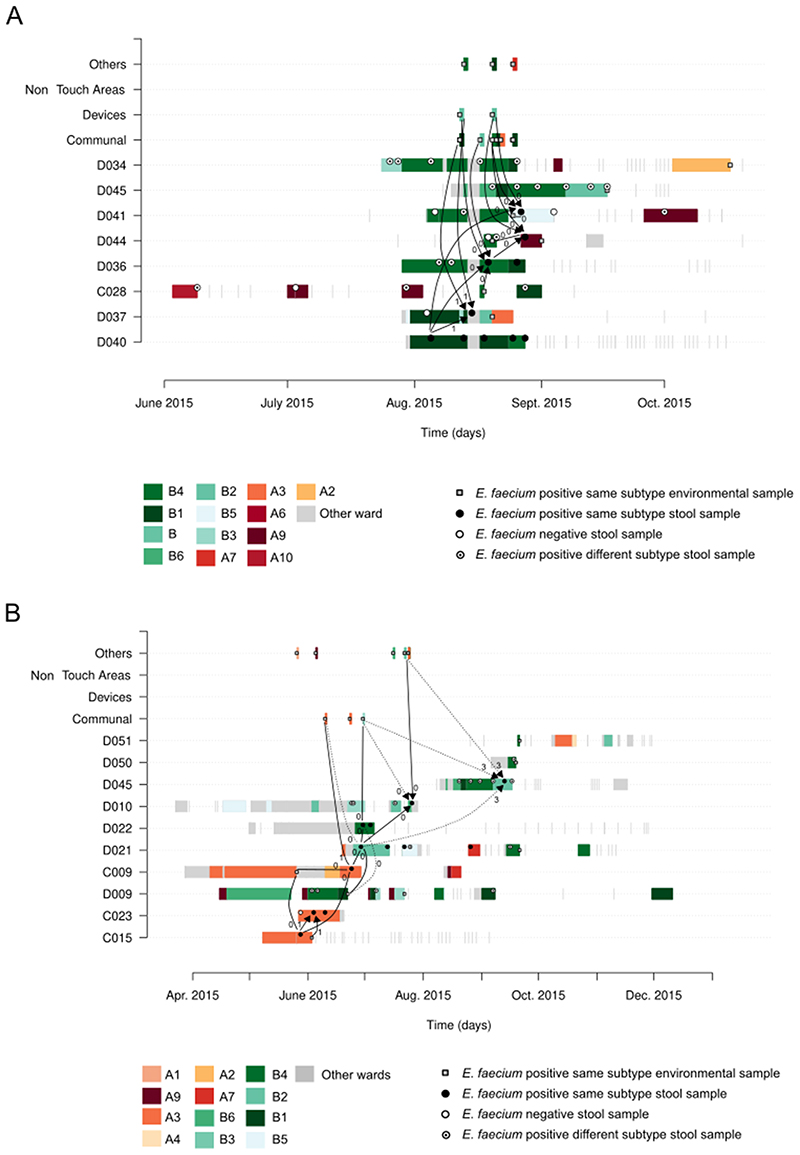
Extended Data Fig. 4. Exemplars of E. faecium transmission clusters.
Each row represents the hospital admission period(s) of patients with the exception of the top four rows, which show different environmental sources. Ward of admission is denoted as A or B, and the room numbered and color-coded. Visits to other hospital wards or areas are colored in grey. Positivity results for stool and environmental samples are shown as circles and squares, respectively. Blunt lines and arrowed lines are drawn to point to the putative sources of index and acquired subtypes respectively, the numbers adjacent to these lines indicating the minimum genetic distance observed between connected samples, which ranged from 0 to 6 SNPs. Solid and dotted lines denote strong and weak epidemiological links, respectively. (A) Exemplar of transmission cluster in the same ward (subtype 49A – ST1454). Strong genetic and epidemiological links point to transmission of this subtype in different rooms of ward B among patients D040, D037, D036, D044 and D041. Strong links to the hospital environment, including communal bathrooms and medical devices, suggest their involvement as reservoirs for onward transmission to patients. (B) Exemplar of transmission cluster spanning both hematology wards and involving 7 patients (subtype 26B – ST80). Strong genetic and epidemiological links point to transmission of this subtype in room A3 among patients C015, C023, C009 and D021, followed by spread in different rooms of ward B among patients D021, D022, D010 and D045.
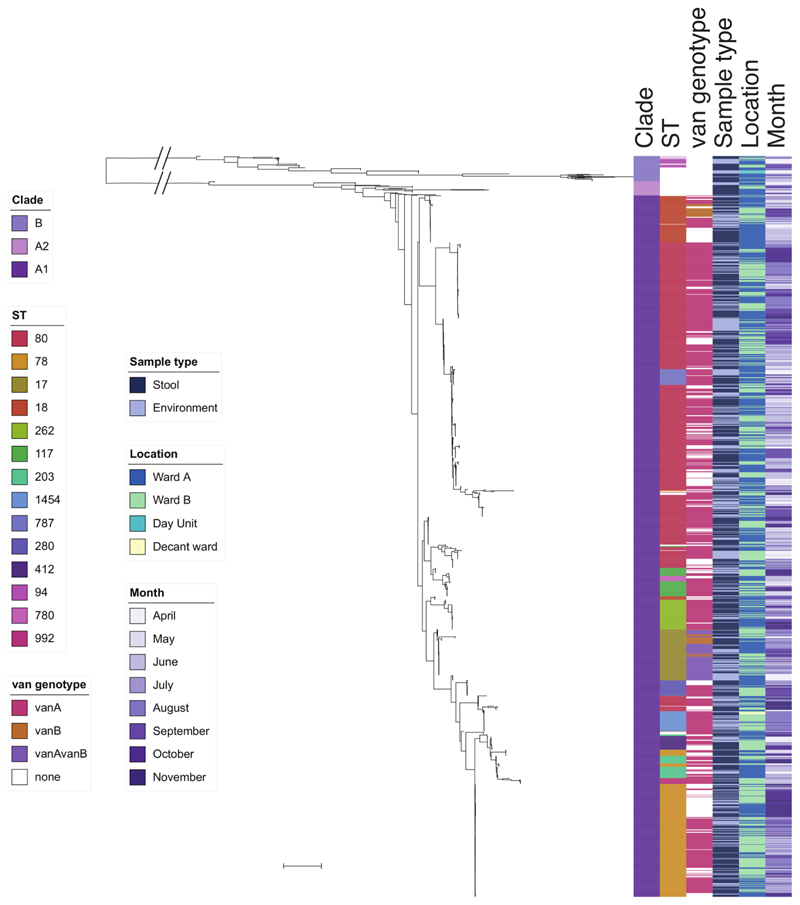
Extended Data Fig. 5. Midpoint rooted maximum likelihood tree based on SNPs in the core genes of 1,560 E. faecium isolates.
E. faecium genomes (1,001 stool, 559 environmental) labeled by clade (B, A2, and A1), commonest sequence types (STs) (only those with more than 10 isolates shown), van genotype, source, ward of origin and month of isolation. Scale bar, ~10,000 SNPs.
Supplementary Material
Acknowledgments
We gratefully acknowledge the contribution of the nurses and healthcare workers on both wards at CUH for assistance with sample collection and the support of the ward matrons, Anne Green and Claire Cowling. We thank Lois Chaparadza and Dr Rosie Swayne for assistance with sample and clinical data collection. We thank Dr Lydia Drumright, Dr Afzal Chaudhry and the EPIC and Clinical Informatics teams for providing patient movement data. We are grateful to the library construction, sequencing and Pathogen Informatics teams at the Wellcome Trust Sanger Institute for assistance with Illumina sequencing. The flocked swabs were donated by Copan Italia spa. This publication presents independent research supported by the Health Innovation Challenge Fund (WT098600, HICF-T5-342), a parallel funding partnership between the Department of Health and Wellcome Trust. The views expressed in this publication are those of the author(s) and not necessarily those of the Department of Health or Wellcome Trust. T.G. is a Wellcome Trust Research Training Fellow (103387/Z/13/Z). F.C. is a Wellcome Trust Sir Henry Postdoctoral Fellow (201344/Z/16/Z). C.L. is a Wellcome Trust Sir Henry Postdoctoral Fellow (110243/Z/15/Z). MET is a Clinician Scientist Fellow supported by the Academy of Medical Sciences, the Health Foundation and the NIHR Cambridge Biomedical Research Centre. S.J.P. is a National Institute for Health Research (NIHR) Senior Investigator.
Footnotes
Competing interests:
N.M.B. is on the advisory board for Discuva Ltd. S.J.P. is a consultant to Specific Technologies. S.J.P., J.P. and F.C. are consultants for Next Gen Diagnostics. All other authors declare that they have no conflicts of interest.
Author contributions
T.G. and S. J. P. designed the study, wrote the study protocol and case record forms, obtained ethical and research and development approvals for the study, and supervised the data collection. M.E.T. supported ethics approvals. T.G., C.L. and C.C. were responsible for collecting samples, clinical and epidemiological data. T.G., C.L., B.B. and P.N. isolated and identified E. faecium, and B.B. and P.N. undertook susceptibility testing and extracted genomic DNA. N.M.B. and D.A.E. provided access to E. faecium cultures in the routine diagnostic microbiology laboratory, and provided expert opinion relating to infection control. T.G. and F.C. undertook the epidemiological and bioinformatic analyses with contributions from J.P. and K.R. B.B. undertook susceptibility testing to disinfectants with contributions from E.M.H. J.P. supervised the genomic sequencing. F.C. and S.J.P. wrote the first draft of the manuscript. S.J.P. supervised and managed the study. All authors had access to the data and read, contributed and approved the final manuscript.
Data availability
The whole genome sequences from this study have been deposited at the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) under the study numbers PRJEB12937, PRJEB13191 and PRJEB13192. Individual accession numbers and isolate metadata are listed in Supplementary Data 1. Supplementary Data 2 includes the genetic and epidemiological links characterized in this study. Source data for Figures and Extended Data Figures are provided with this manuscript.
References
- 1.Arias CA, Murray BE. The rise of the Enterococcus: beyond vancomycin resistance. Nature Reviews Microbiology. 2012;10:266–78. doi: 10.1038/nrmicro2761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Werner G, et al. Emergence and spread of vancomycin resistance among Enterococci in Europe. Eurosurveillance. 2008;13:1–11. [PubMed] [Google Scholar]
- 3.Prematunge C, et al. VRE and VSE Bacteremia Outcomes in the Era of Effective VRE Therapy: A Systematic Review and Meta-analysis. Infection Control & Hospital Epidemiology. 2015:1–10. doi: 10.1017/ice.2015.228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tacconelli E, et al. Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. The Lancet Infectious Diseases. 2017;3099:1–10. doi: 10.1016/S1473-3099(17)30753-3. [DOI] [PubMed] [Google Scholar]
- 5.Taur Y, et al. Intestinal domination and the risk of bacteremia in patients undergoing allogeneic hematopoietic stem cell transplantation. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2012;55:905–14. doi: 10.1093/cid/cis580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moradigaravand D, et al. Within-host evolution of Enterococcus faecium during longitudinal carriage and transition to bloodstream infection in immunocompromised patients. Genome medicine. 2017;9:119. doi: 10.1186/s13073-017-0507-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Falk PS, Winnike J, Woodmansee C, Desai M, Mayhall CG. Outbreak of Vancomycin-Resistant Enterococci in a Burn Unit. Infection Control & Hospital Epidemiology. 2000;21:575–582. doi: 10.1086/501806. [DOI] [PubMed] [Google Scholar]
- 8.Neely AN, Maley MP. Survival of enterococci and staphylococci on hospital fabrics and plastic. Journal of clinical microbiology. 2000;38:724–6. doi: 10.1128/jcm.38.2.724-726.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mitchell BG, Dancer SJ, Anderson M, Dehn E. Risk of organism acquisition from prior room occupants: a systematic review and meta-analysis. Journal of Hospital Infection. 2015;91:211–217. doi: 10.1016/j.jhin.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Turabelidze D, Kotetishvili M, Kreger A, Morris JG, Sulakvelidze A. Improved pulsed-field gel electrophoresis for typing vancomycin-resistant enterococci. Journal of Clinical Microbiology. 2000;38:4242–4245. doi: 10.1128/jcm.38.11.4242-4245.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gouliouris T, et al. Genomic surveillance of enterococcus faecium reveals limited sharing of strains and resistance genes between livestock and humans in the United Kingdom. mBio. 2018;9:1–15. doi: 10.1128/mBio.01780-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Arredondo-Alonso S, et al. Plasmids shaped the recent emergence of the major nosocomial pathogen Enterococcus faecium. mBio. 2020;11:1–17. doi: 10.1128/mBio.03284-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T, et al. A three-year whole genome sequencing perspective of Enterococcus faecium sepsis in Australia. PLoS ONE. 2020;15:1–15. doi: 10.1371/journal.pone.0228781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Raven KE, et al. A decade of genomic history for healthcare-associated Enterococcus faecium in the United Kingdom and Ireland. Genome Research. 2016;26:1388–1396. doi: 10.1101/gr.204024.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pinholt M, et al. WGS of 1058 Enterococcus faecium from Copenhagen, Denmark, reveals rapid clonal expansion of vancomycin-resistant clone ST80 combined with widespread dissemination of a vanA-containing plasmid and acquisition of a heterogeneous accessory genome. Journal of Antimicrobial Chemotherapy. 2019;74:1776–1785. doi: 10.1093/jac/dkz118. [DOI] [PubMed] [Google Scholar]
- 16.van Hal SJ, et al. Evolutionary dynamics of Enterococcus faecium reveals complex genomic relationships between isolates with independent emergence of vancomycin resistance. Microbial Genomics. 2016;2:1–11. doi: 10.1099/mgen.0.000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang G, et al. Evolution and mutations predisposing to daptomycin resistance in vancomycin-resistant Enterococcus faecium ST736 strains. PLoS ONE. 2018;13:1–17. doi: 10.1371/journal.pone.0209785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raven KE, et al. Complex Routes of Nosocomial Vancomycin-Resistant Enterococcus faecium Transmission Revealed by Genome Sequencing. Clinical Infectious Diseases. 2017;64:886–893. doi: 10.1093/cid/ciw872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liese J, et al. Expansion of Vancomycin-Resistant Enterococcus faecium in an Academic Tertiary Hospital in Southwest Germany: A large-scale whole-genome-based outbreak investigation. Antimicrobial Agents and Chemotherapy. 2019;63:1–13. doi: 10.1128/AAC.01978-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brown DFJ, Walpole E. Evaluation of selective and enrichment media for isolation of glycopeptide-resistant enterococci from faecal specimens. Journal of Antimicrobial Chemotherapy. 2003;51:289–296. doi: 10.1093/jac/dkg076. [DOI] [PubMed] [Google Scholar]
- 21.Tonkin-Hill G, Lees JA, Bentley SD, Frost SDW, Corander J. Fast hierarchical Bayesian analysis of population structure. Nucleic Acids Research. 2019;47:5539–5549. doi: 10.1093/nar/gkz361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Prieto AMG, et al. The two-component system ChtRS contributes to chlorhexidine tolerance in Enterococcus faecium. Antimicrobial Agents and Chemotherapy. 2017;61:1–9. doi: 10.1128/AAC.02122-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pidot SJ, et al. Increasing tolerance of hospital Enterococcus faecium to handwash alcohols. Science Translational Medicine. 2018;10:eaar6115. doi: 10.1126/scitranslmed.aar6115. [DOI] [PubMed] [Google Scholar]
- 24.de Regt MJa, et al. High acquisition and environmental contamination rates of CC17 ampicillin-resistant Enterococcus faecium in a Dutch hospital. The Journal of antimicrobial chemotherapy. 2008;62:1401–6. doi: 10.1093/jac/dkn390. [DOI] [PubMed] [Google Scholar]
- 25.Ludden C, et al. A One Health Study of the Genetic Relatedness of Klebsiella pneumoniae and Their Mobile Elements in the East of England. Clinical Infectious Diseases. 2020;70:219–226. doi: 10.1093/cid/ciz174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Passaretti CL, et al. An Evaluation of Environmental Decontamination With Hydrogen Peroxide Vapor for Reducing the Risk of Patient Acquisition of Multidrug-Resistant Organisms. Clinical infectious diseases: an official publication of the Infectious Diseases Society of America. 2013;56:27–35. doi: 10.1093/cid/cis839. [DOI] [PubMed] [Google Scholar]
- 27.Grabsch EA, et al. Significant reduction in vancomycin-resistant enterococcus colonization and bacteraemia after introduction of a bleach-based cleaning-disinfection programme. The Journal of hospital infection. 2012;82:234–42. doi: 10.1016/j.jhin.2012.08.010. [DOI] [PubMed] [Google Scholar]
- 28.Lebreton F, et al. Emergence of Epidemic Multidrug-Resistant Enterococcus faecium from Animal and Commensal Strains. mBio. 2013;4:1–10. doi: 10.1128/mBio.00534-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croucher NJ, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Research. 2015;43:e15–e15. doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–1313. doi: 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing; 2017. [Google Scholar]
- 33.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27:431–432. doi: 10.1093/bioinformatics/btq675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiegand I, Hilpert K, Hancock REW. Agar and broth dilution methods to determine the minimal inhibitory concentration (MIC) of antimicrobial substances. Nature protocols. 2008;3:163–75. doi: 10.1038/nprot.2007.521. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The whole genome sequences from this study have been deposited at the European Nucleotide Archive (ENA) (www.ebi.ac.uk/ena) under the study numbers PRJEB12937, PRJEB13191 and PRJEB13192. Individual accession numbers and isolate metadata are listed in Supplementary Data 1. Supplementary Data 2 includes the genetic and epidemiological links characterized in this study. Source data for Figures and Extended Data Figures are provided with this manuscript.