Abstract
Animal behavior is encoded in neuronal circuits in the brain. To elucidate the function of these circuits, it is necessary to identify, record from and manipulate networks of connected neurons. Here we present BAcTrace (Botulinum Activated Tracer), a genetically encoded, retro-grade, transsynaptic labelling system. BAcTrace is based on C. botulinum neurotoxin A, Botox, which we have engineered to travel retrogradely between neurons to activate an otherwise silent transcription factor. We validate BAcTrace at three neuronal connections in the Drosophila olfactory system. We show that BAcTrace-mediated labeling allows electrophysiological recordings of connected neurons. Finally, in a challenging circuit with highly divergent connections, BAcTrace correctly identifies 12 out of 16 connections, which were previously observed by electron microscopy.
Introduction
The development of genetic tools to elucidate connectivity and manipulate neurons and circuits has advanced our understanding of how the brain works. Increasingly, these tools are being used to study diseases of the nervous system and develop effective treatments [1, 2].
In the context of circuit research, the ability to identify and manipulate pre- or post-synaptic cells to neurons of interest is of crucial importance. Tools to label downstream neurons are called anterograde, while retrograde tools reveal the input neurons to a given population.
Drosophila melanogaster is a key model organism to study the genetic and circuit basis of animal behavior. The fly has a rich behavioral repertoire encoded in a relatively small nervous system. This simplicity is paired with extensive collections of genetic reagents, both to investigate gene function and to label and manipulate most neuronal classes [3]. While these reagents offer genetic access to neurons, until recently tools to map synaptic connections in Drosophila have been lacking. This has recently changed with the development of electron microscopy methods to map connections in larval and adult brains [4, 5, 6]. Furthermore, contact-based, genetically encoded systems for anterograde tracing such as trans-Tango and TRACT have been developed [7, 8]. Despite these additions to the experimental toolbox, a retrograde labelling system for Drosophila is still missing. While rabies virus and its modifications constitute examples of retrograde transsynaptic tools for mice [1], their applicability to flies is far from simple, both because of the experimental difficulties of delivering viruses via brain injections and because the virus neurotropism may not extend to flies.
Here, we introduce BAcTrace (Botulinum Activated Tracer), a genetically encoded tracing tool, designed for retrograde tracing in Drosophila. We first established the system in tissue culture and then implemented and refined BAcTrace in flies, showing that it can reveal the connectivity between olfactory Projection Neurons (PNs) and 3 different classes of presynaptic neurons: Olfactory Receptor Neurons, Kenyon Cells of the Mushroom Bodies and Lateral Horn Neurons. BAcTrace provides a way to test connectivity and manipulate components in circuits with high sensitivity and specificity.
Results
System design
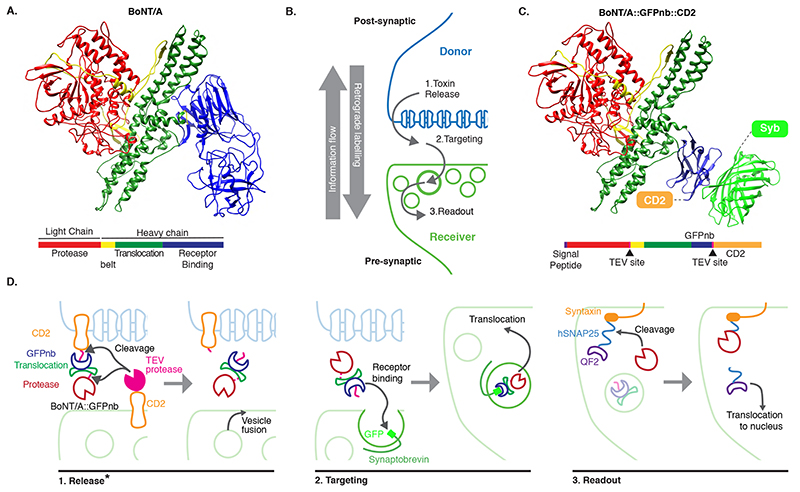
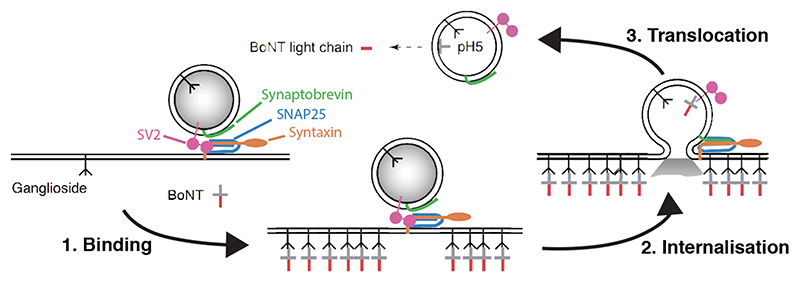
In contrast to contact-based systems, BAcTrace shares two key feature with the rabies virus-based methods. First, labelling is triggered by protein transfer between connected neurons and second, this transfer is followed by a signal amplification step. Clostridium botulinum neurotoxin A1 (BoNT/A) is at the core of our system. BoNT/A is a modular protein (Figure 1A and Supplementary Figure 1) with a well-studied mechanism of action (Extended Data Figure 1). BoNT/A is made as a single polypeptide which gets cleaved by proteases generating a Light Chain (LC) and a Heavy Chain (HC). In vertebrates, the receptor binding domain (RBD), located in the C-terminal half of the HC, enables enrichment on neuronal membranes by interacting with a neuron-specific lipid (polysialoganglioside GT1b). Upon neurotransmitter vesicle fusion in the axon, the RBD gains access to the vesicle lumen and binds with high affinity to a second partner, synaptic protein SV2 [9]. When the vesicle is recycled and acidified, the translocation domain (TD) undergoes a conformational change injecting the LC across the vesicle membrane. The LC is a protease highly specific for the human protein SNAP25 (hSNAP25) and once in the cytosol it cleaves its target, preventing further neurotransmitter release [10, 11].
Figure 1. System design.
(A) Molecular structure of BoNT/A (PDB id 3BTA, [40]). Toxin domains are colored red (protease), yellow (belt), green (Translocation, TD) and blue (Receptor Binding, RBD). (B) Schematic showing the steps leading to labelling. (C) Retargeted BoNT/A: the RBD from (A) is replaced by an anti-GFP nanobody (blue). Also shown in light green the bound GFP (PDB id 3OGO, [13]). (D) Proteins and molecular mechanisms mediating each step from (B). Colors as in (C) except pink sections in BoNT/A::GFPnb::CD2 which indicate TEV cleavage sites. * The release step was shown to be TEV independent later in the project.
In BAcTrace, BoNT/A is expressed in post-synaptic “Donor” neurons and transferred, similarly to rabies virus, to connected pre-synaptic “Receiver” cells where it triggers expression of an effector gene. BAcTrace can be broken down into three steps (Figure 1B). In the first step, BoNT/A is made in Donor neurons attached to the extracellular portion of a transmembrane protein (CD2). The Tobacco Etch Protease (TEV) [12] is made in Receiver neurons attached to the extracellular portion of a second transmembrane protein. At synapses both proteins interact and TEV cleaves two recognition sites, releasing the toxin from the membrane, and allowing separation of the LC and HC after translocation (Figure 1C,D). While we initially considered TEV cleavage essential, we later found TEV dispensable for toxin transfer (see below). Because flies lack the vertebrate BoNT/A receptor SV2, we switched the RBD for a single-chain anti-GFP nanobody (GFPnb) [13] to create BoNT/A::GFPnb::CD2 (Figure 1C and Supplementary Figure 2). In the second step, modified BoNT/A is targeted to neurotransmitter vesicles of Receiver neurons that express a Synaptobrevin::GFP fusion, oriented with GFP inside the vesicle. Re-targeting prevents binding of the toxin to vertebrate neurons and renders it safe for researchers handling the flies. In the third step, we made a toxin sensor by linking the QF2 transcription factor [14] to Drosophila Syntaxin via amino acids 141-206 of hSNAP25 [15] (QF2::hSNAP25::Syx). Syntaxin targets the transcriptionally inactive sensor to the synaptic membrane. However, after translocation cytosolic LC releases QF2, triggering QUAS-reporter expression. BoNT/A LC is highly specific and it does not cleave Drosophila SNAP25 [15], therefore toxin expression should not be harmful to flies.
BAcTrace is active in Drosophila cells
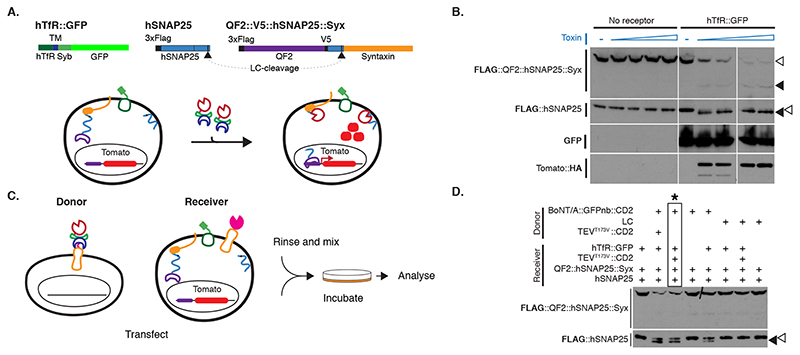
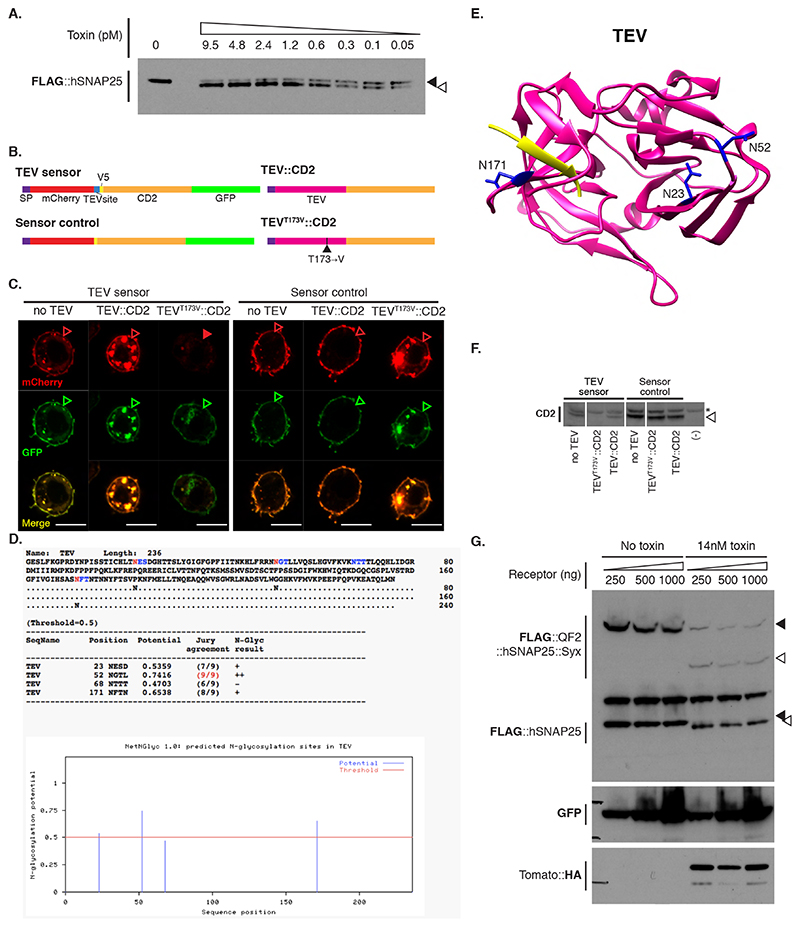
We tested BAcTrace feasibility in Drosophila S2 cells. We made BoNT/A::GFPnb in bacteria and added the purified toxin to S2 cells rendered sensitive by expression of a hTfR::GFP receptor; this chimeric hTfR constitutively cycles between the plasma membrane and a low pH cellular compartment ([16] and Supplementary Note 1). We also transfected the cells with a FLAG-tagged hSNAP25 (Figure 2A). We determined cleavage of FLAG::hSNAP25 by Western blot as a small shift of 0.9kDa (9 amino acids). All tested toxin concentrations induced efficient cleavage (Figure 2B, Extended Data Figure 2A and Supplementary Note 2). Furthermore, hTfR::GFP was strictly required for cleavage (Figure 2B).
Figure 2. BAcTrace in tissue culture.
(A) Experiment for testing the activity of E. coli purified toxin. (B) Western blot analysis of S2 cell extracts. Empty arrowheads indicate un-cleaved and solid arrowheads cleaved toxin sensors. Toxin concentrations (nM): 0, 1.7, 3.4, 6.9, 14, 0, 1.7, 3.4, 6.9 and 14. Smaller, faint band in TdTomato::HA panels is a C-terminal degradation fragment. (C) Cell mixing experiments to test the activity of toxin produced in insect cells. (D) Western blot analysis of S2 cell extracts from cell-mixing experiments. The lane marked with * contains the components shown in (C). Epitopes detected by antibodies are indicated in bold in (B) and (D). Experiments (B) and (D) were done once as shown in the figure. No-receptor controls were included more than three times in similar experiments with the same results shown in B.
We also tested and confirmed that the sensor used in flies, QF2::hSNAP25::Syx, is cleaved by BoNT/A::GFPnb (Figure 2B). Furthermore, the released QF2 induces TdTomato expression, confirming that the transcription factor activity of QF2 is not inhibited by the 62 amino acids of hSNAP25 left on its C-terminus following cleavage.
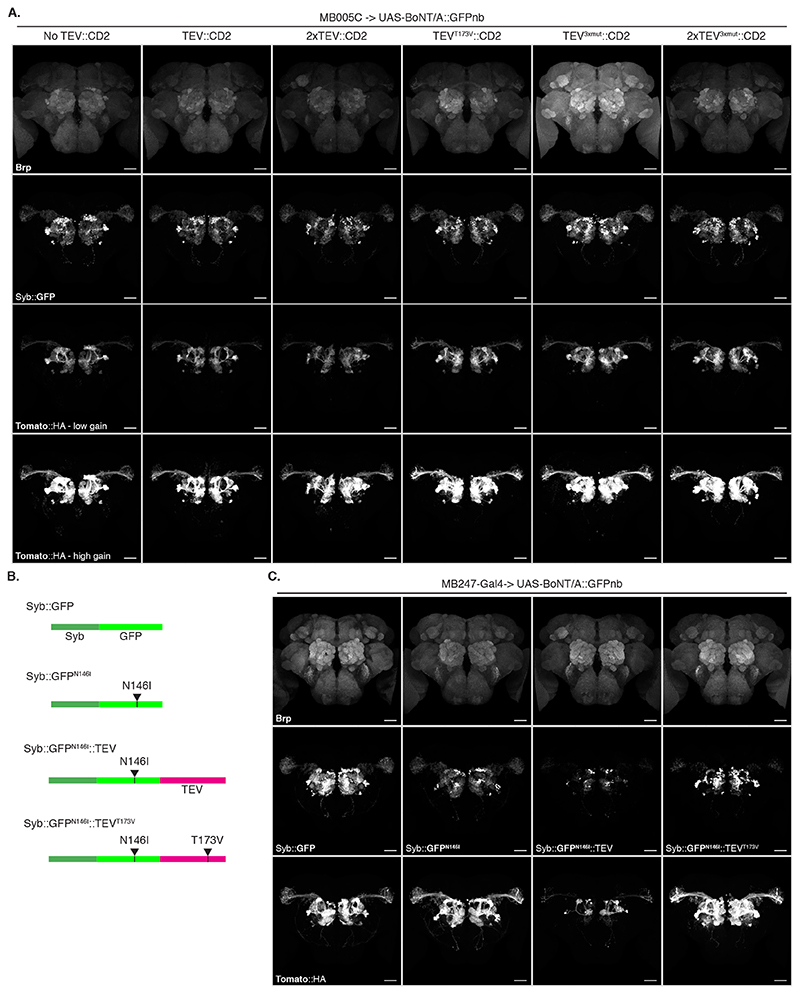
Both TEV and BoNT/A are originally cytosolic proteins. We therefore verified that they remained active when produced as extracellular membrane fusions in Drosophila cells. We found TEV was inactive unless we removed a predicted glycosylation site (TEVT173V) (Extended Data Figure 2B-F and Supplementary Note 3). Next, we tested the function of plasma membrane-targeted BoNT/A using a cell-mixing experiment. We transfected two cell populations: Donor cells with either membrane-targeted BoNT/A::GFPnb::CD2 or cytosolic LC protease (negative control) and Receiver cells with receptor and sensors (Figure 2C). One day later we mixed the cell populations and two days later, we could detect hSNAP25 cleavage, confirming that the BoNT/A::GFPnb::CD2 fusion protein was able to pass from the Donor cell to the cytoplasmic compartment of the Receiver cell and cleave the hSNAP25-based sensor proteins (Figure 2D). Curiously, we found that TEVT173V::CD2 was not essential for this transfer, perhaps due to TEV-independent cleavage and release of BoNT/A::GFPnb from the Donor cell membrane. Critically, just as for the experiments with bacterial BoNT/A::GFPnb (Figure 2B), we observed a strict requirement for TfR::GFP in Receiver cells. Increasing the amount of receptor passed a certain threshold did not increase its efficiency (Extended Data Figure 2G).
BAcTrace works as a transsynaptic system in flies
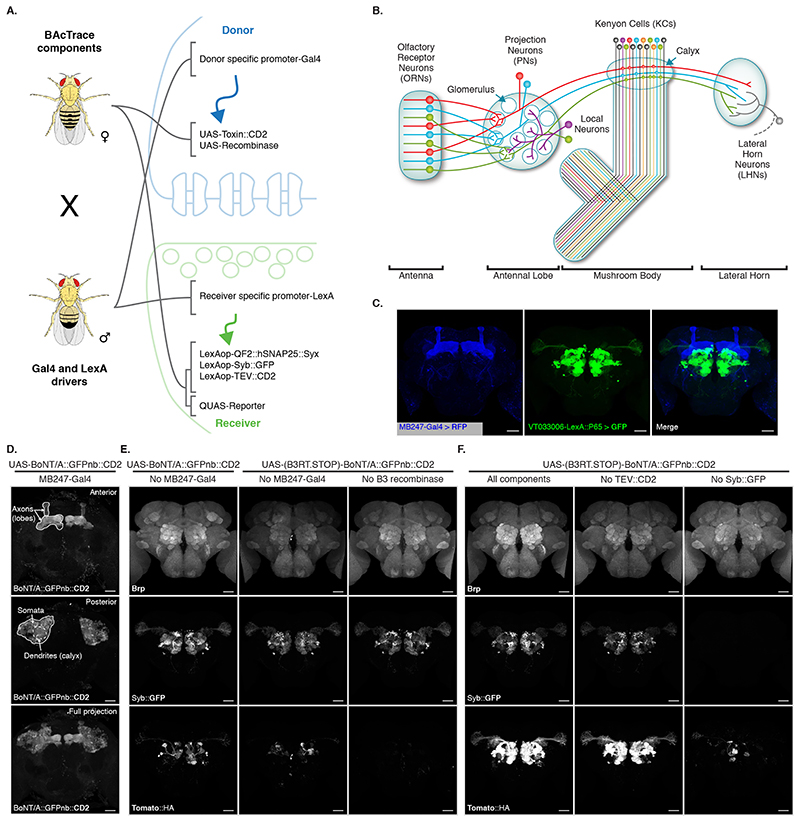
Having established BAcTrace in cell culture, we implemented the system in flies. We encoded Donor components in UAS vectors, which were driven by Gal4, with Receiver components driven by LexA. For most experiments, we crossed flies containing all BAcTrace components to flies with Gal4 and LexA drivers (Figure 3A). One important limitation of BAcTrace is that LexA and Gal4 expression cannot overlap. If they do, activation will take place in the cells with overlapping expression, producing a false positive result. Furthermore, the toxin is likely to get internalized and become unavailable for transsynaptic labelling.
Figure 3. BAcTrace in vivo.
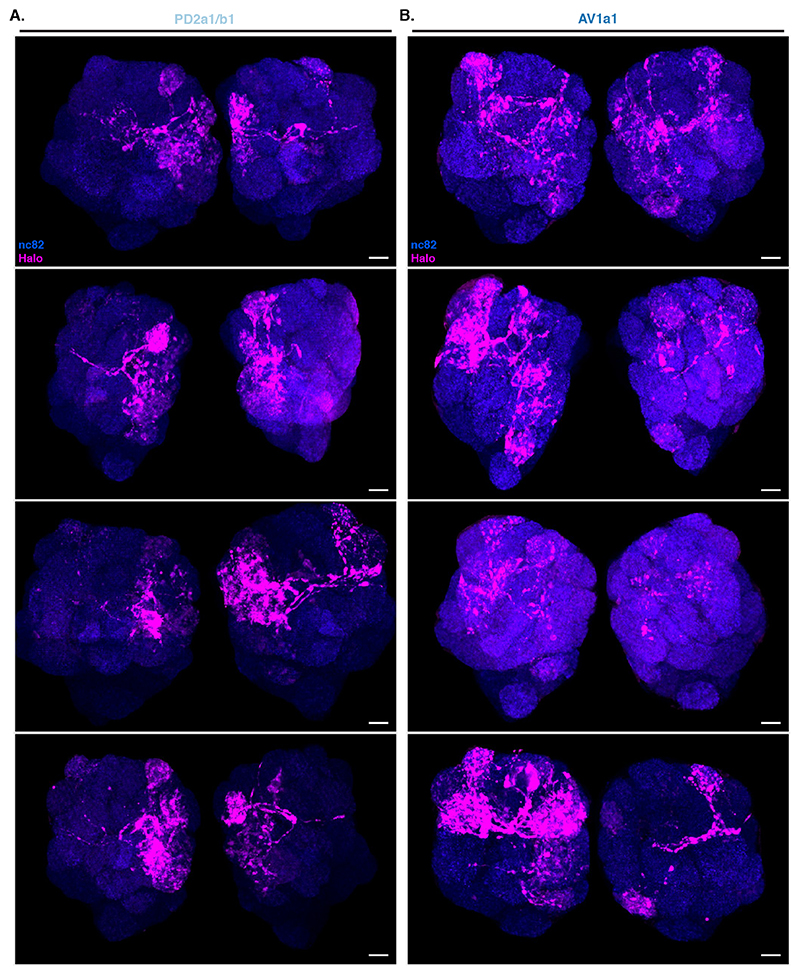
(A) Genetic strategy for expressing BAcTrace components. The female fly contributes all UAS, LexAop and QUAS components while the male fly contributes a Gal4 driver defining the Donor cells and a LexA driver defining the Receiver cells. (B) First three layers of the fly olfactory system; adapted from [41]. (C) Expression patterns of MB247-Gal4 (blue) and VT033006-LexA::P65 (green). (D) BoNT/A::GFPnb::CD2 expression in the mushroom bodies. (E) BAcTrace negative controls. Left panels, No Gal4 driver. Middle panels, added a B3 DNA recombinase dependent transcriptional stop cassette in front of the toxin transgene; UAS-B3 present. Right panels, stop cassette and MB247-Gal4 present but UAS-B3 absent. (F) BAcTrace experiments. Left panels, all components present as described in Figure 1D. Middle panels, TEV transgene absent. Right panels, Syb::GFP receptor transgene absent. (E) and (F) are full and (C) and (D) include partial maximum intensity projections of confocal stacks. Epitopes detected by antibodies are indicated in bold in (C-F). All animals are 3-4 days old. Genotypes for each panel are described in Supplementary Table 16. Results shown in C-F are representative of at least 4 brains. Scale bars 30μm.
We chose the well characterized fly olfactory system for initial testing of BAcTrace in vivo (Figure 3B). Briefly, Drosophila has 50 types of peripheral Olfactory Receptor Neurons (ORNs). ORNs of each type express a general co-receptor, Odorant receptor Co-Receptor (Orco) and one of 50 different olfactory receptor genes, conferring responses to a specific set of odors. ORNs expressing the same receptor relay information to one of 50 glomeruli in the brain’s Antennal Lobe (AL). In each glomerulus, the axons of 20-100 ORNs make strong connections (e.g. ~1215 synapses per Projection Neurons (PN) in glomerulus DM6 [17]) onto the dendrites of 1-8 PNs. These in turn make a modest number of reciprocal synapses (e.g.~40 synapses per PN in glomerulus DM6 [17]) onto ORNs. PN axons project onto Kenyon Cells (KCs) in the calyx of the Mushroom Bodies (MB) and to the Lateral Horn (LH) [18]. It is important to note that PNs connect to KCs almost randomly [19, 20, 21, 22]; therefore, expressing toxin even in a small number of KCs should label most or all PNs.
We expressed Donor components in KCs using the MB247-Gal4 driver and Receiver components in PNs using the broad LexA driver VT033006-LexA::P65 (Figure 3C, Extended Data Figure 3). We confirmed that BoNT/A::GFPnb::CD2 can be expressed and trafficked throughout KCs, detecting expression in soma, dendrites and axons (Figure 3D).
We first established the background signal of the system in vivo by bringing together all components except the Gal4 driver. We found low-frequency, stochastic labelling of PNs (Left panels in Figure 3E) likely due to weak, Gal4-independent toxin expression in PNs (BAcTrace working in cis instead of trans). To reduce this background, we added a transcriptional stop cassette to BoNT/A::GFPnb::CD2 which can be removed by the B3 DNA recombinase [23]. This stop cassette reduces background in the absence of Gal4 (Middle panels in Figure 3E). Furthermore, in the absence of the B3 recombinase transgene (Right panels in Figure 3E) there is no stochastic PN labelling, even in the presence of MB247-Gal4, indicating that the remaining stochastic background in the middle panels of Figure 3E is due to Gal4-independent expression of B3, possibly during development. A much lower, non-stochastic background was not ameliorated by the stop cassette (Right panels in Figure 3E). We mapped the source of this signal to the V5 tag present in the sensor (Supplementary Figure 3 and Supplementary Note 4). Except when indicated, in further experiments we used a sensor without V5.
Next, we repeated the experiment including Gal4 and found that toxin expression in the MB induced strong labelling of most PNs in the VT033006 line even in the absence of wild type TEV (Figure 3F). Although further experiments hinted at a small improvement in efficiency with mutant TEVT173V (Extended Data Figure 4 and Supplementary Note 5), for simplicity we decided to omit it from the remaining experiments. Critically, the Syb::GFP receptor was required for in vivo transfer of toxin (Right panels in Figure 3F), indicating that the system should select for synaptic rather than non-synaptic cell contacts.
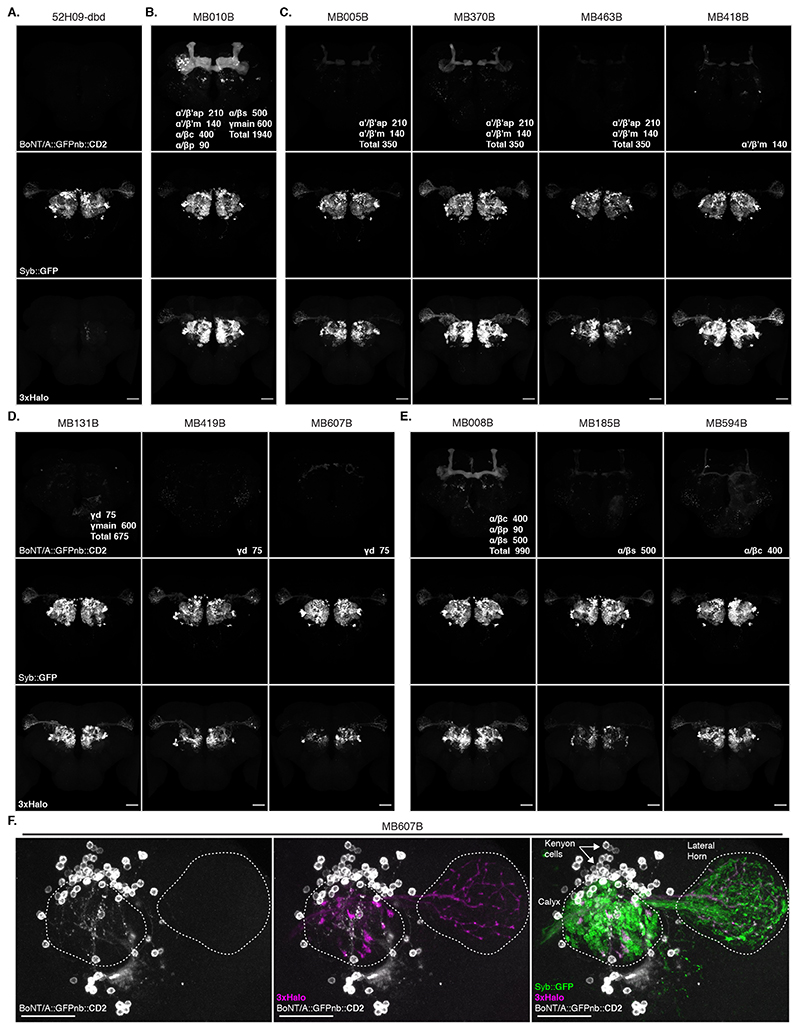
We confirmed our MB247-Gal4 results using a panel of split Gal4 lines [24] and found that toxin expression in KC subtypes also labelled presynaptic PNs (Extended Data Figure 5 and Supplementary Note 6).
BAcTrace expression in Olfactory Receptor Neurons labels connected PNs
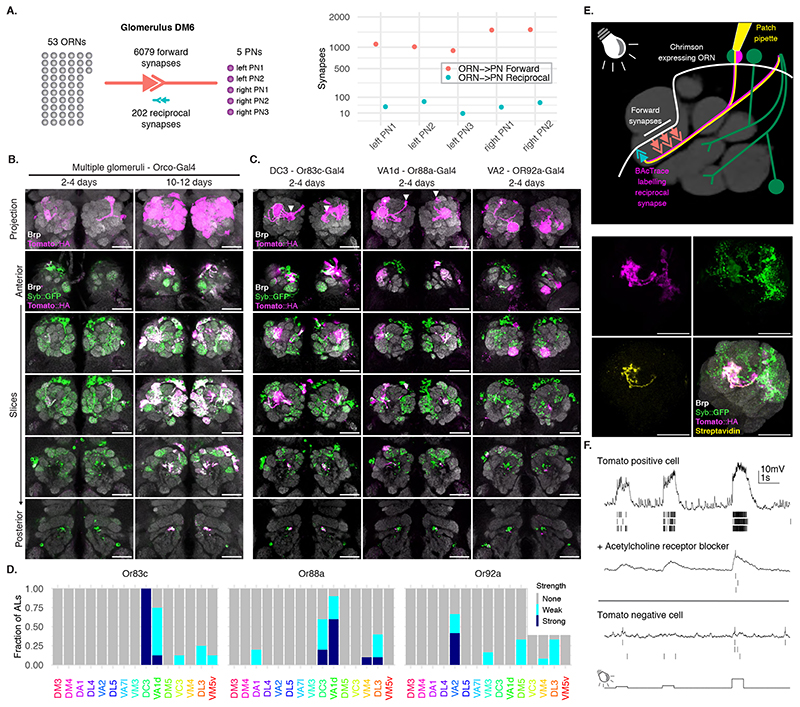
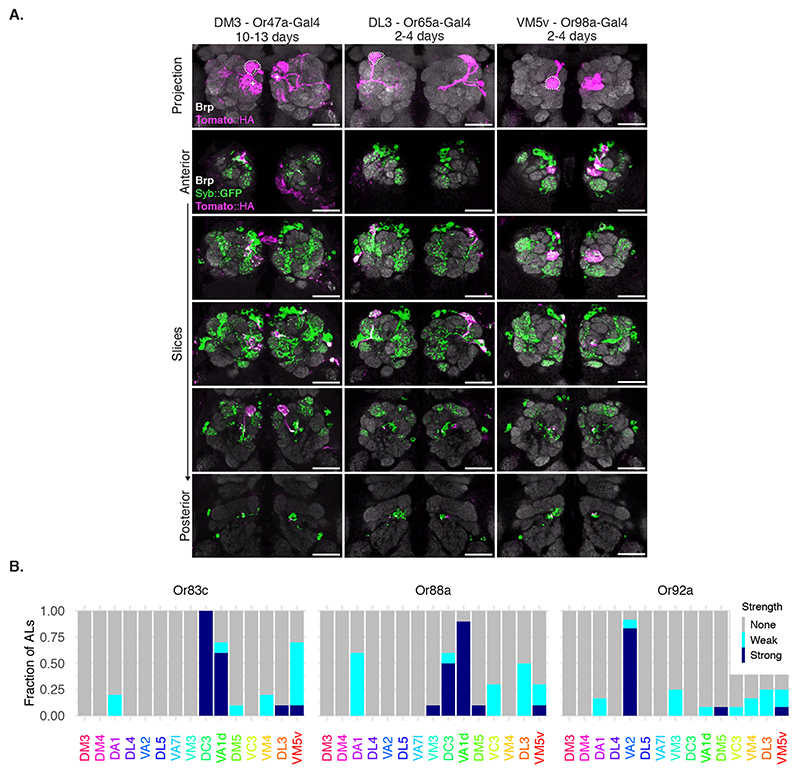
Next, we examined the labelling specificity of BAcTrace using an experimental configuration where only a subset of Receiver cells are connected to toxin-expressing Donor cells. We selected the reciprocal synapses from PNs to ORNs (Figure 3B) for two reasons. First, highly specific Gal4 driver lines are available for ORN subtypes [25, 26] and second, while ORN axons make strong connections to PN dendrites, EM connectomics identified a moderate-strength reciprocal connection between these two cell types [27], e.g. DM6 glomerulus PNs make ~40 reciprocal synapses onto ORNs (Figure 4A)[17].
Figure 4. BAcTrace expression in ORNs labels connected PNs.
(A) ORN->PN forward and reciprocal connectivity [17]. Left: aggregated numbers of forward and reciprocal synapses between PNs and ORNs in the DM6 glomerulus. Right: Forward and reciprocal synapses per PN. (B) Time-dependent labelling in PNs triggered by BAcTrace expression in most ORNs. Images shown are representative of 5 animals per time point. (C) BAcTrace expression in ORN types induces labelling in connected PNs (dotted circles). Solid white arrowheads indicate neuronal soma and + indicates glomerulus not innervated by the ORNs. (D) Quantification of the labeling shown in (C) for the 15 glomeruli with highest expression level in VT033006-LexA::P65 (Extended Data Figure 3). Sample size: Or83c n=8ALs, Or88a n=10ALs and Or92a n=11ALs. (E) Whole-cell patch clamp recording of a BAcTrace-labelled PN. Top: Experimental set up: BAcTrace labelled DC3 PNs (magenta) and Chrimson expressing Donor ORNs (white). Recorded cell is filled and labelled with biocytin-streptavidin (yellow). Bottom: Maximum intensity projection of a confocal stack showing the recorded and filled DC3 PN. (F) Representative voltage traces and spikes extracted from three light presentations. Top: Traces from the cell shown in (C). Middle: traces after the addition of the nicotinic acetylcholine receptor blocker mecamylamine (200μM). Bottom: Traces from a GFP+ and TdTomato-neuron. The animal in (E) was 14 days old. The same result was observed in 3 animals. Epitopes detected by antibodies are indicated in bold in (B), (C) and (E). Genotypes for each panel are described in Supplementary Table 16. Scale bars 30μm.
We expressed toxin in the majority of ORNs using Orco-Gal4 (Figure 4B, for the reverse experiment see Supplementary Note 7). In 2-4 days old animals we detected little labelling above background. However, at nine to ten days after eclosion we found consistent labelling in most glomeruli covered by the VT033006-LexA::P65 line (Figure 4B). Given this time dependence, BAcTrace labelling could be used to characterize the strength of a connection under different conditions (e.g. mutant backgrounds, level of stimulation, etc).
Toxin expression in single ORN types (Figure 4C,D and Extended Data Figure 6) resulted in strong labelling in connected PNs in animals as young as 2 days old, e.g. Or83c, Or88a, Or92a, Or65a and Or98a, while in other cases labelling took longer, e.g. 10-13 days for Or47a. While these differences may be due to cell type-specific differences in the number of ORN->PN reciprocal synapses, they could also be the result of expression level variability of BAcTrace components in the different cell types (i.e. detection system components in PNs or toxin in ORNs).
In some cases BAcTrace labelled PNs targeting glomeruli known not to be innervated by Donor cells (Figure 4C,D and Extended Data Figure 6). This could be artifactual background labelling, as characterized before (Figure 3E). It might be triggered by non-synaptic contacts from the ORN axons as they traverse the AL or it could be due to unexpected, synaptic contacts. For instance, some ORNs and PNs have small processes in neighboring glomeruli.
For an initial assessment of neuronal health, we used BAcTrace to label DC3 PNs while expressing the light-gated cation channel CsChrimson in connected Donor Or83c ORNs [28](Figure 4E). We stimulated the ORNs using light while performing electrophysiological recordings from the TdTomato labelled PNs in 14 days old flies (we obtained similar results on 5 days old animals, not shown).
Graded light stimulation induced increasing spiking responses in PNs indicating that ORNs are able to release neurotransmitter and stimulate connected PNs, and that BoNT/A::GFPnb::CD2 was not overtly toxic (Figure 4F). These responses could be partially blocked by the nicotinic acetylcholine receptor blocker mecamylamine, indicating the responses are due to synaptic transmission from ORNs. Furthermore, light responses were absent from TdTomato negative control neurons.
Mapping connections in the Lateral Horn
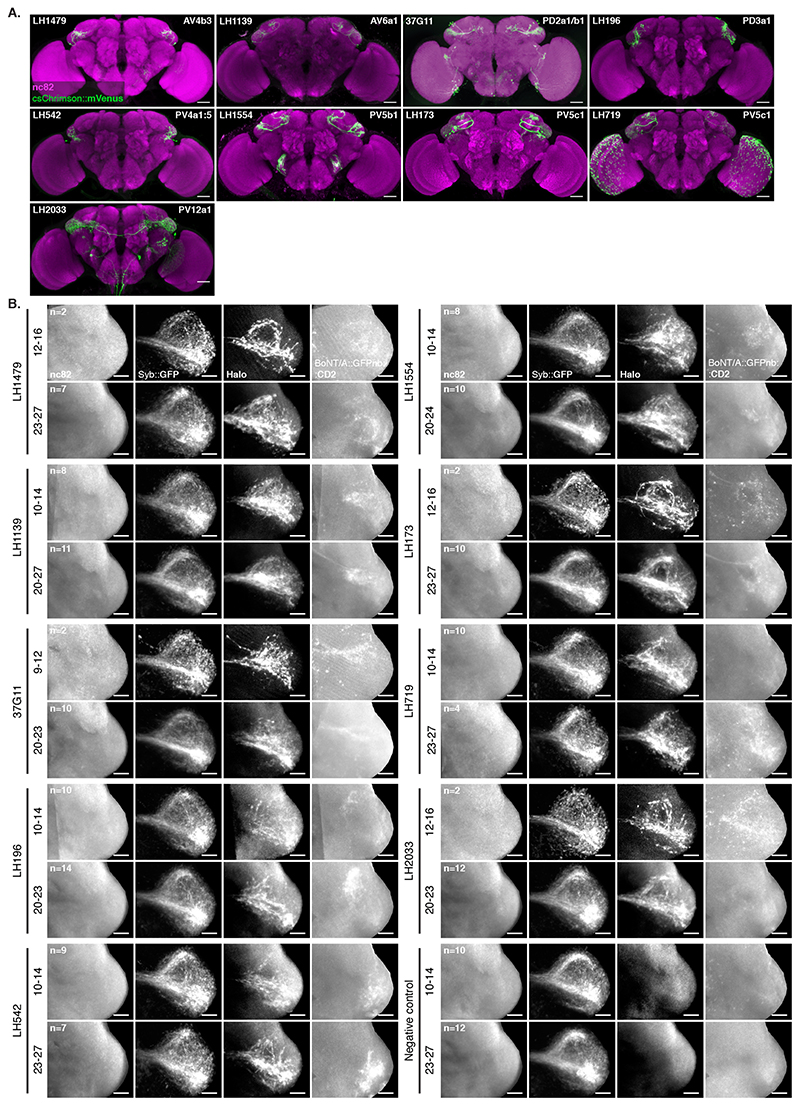
In the Lateral Horn (LH), connectivity is less well understood and projections are more divergent; each olfactory PN type having many post-synaptic Lateral Horn Neuron (LHN) partners. These partners share fewer synapses than those we assessed in our previous experiments (Figure 3B)[29, 30]. We expressed Receiver components in PNs using the broad VT033006-LexA::P65 and toxin using a panel of 9 Gal4 and SplitGal4 lines that express in 8 Donor LHN cell types (Figure 5A,B and Extended Data Figure 7A) [31]. All 8 Donors induced labelling of Receiver PNs (Extended Data Figure 7B). Consistent with our expectations of weaker connectivity from individual Receiver cells, labelling was only observed in older animals (16-18d but not 2-3d, not shown).
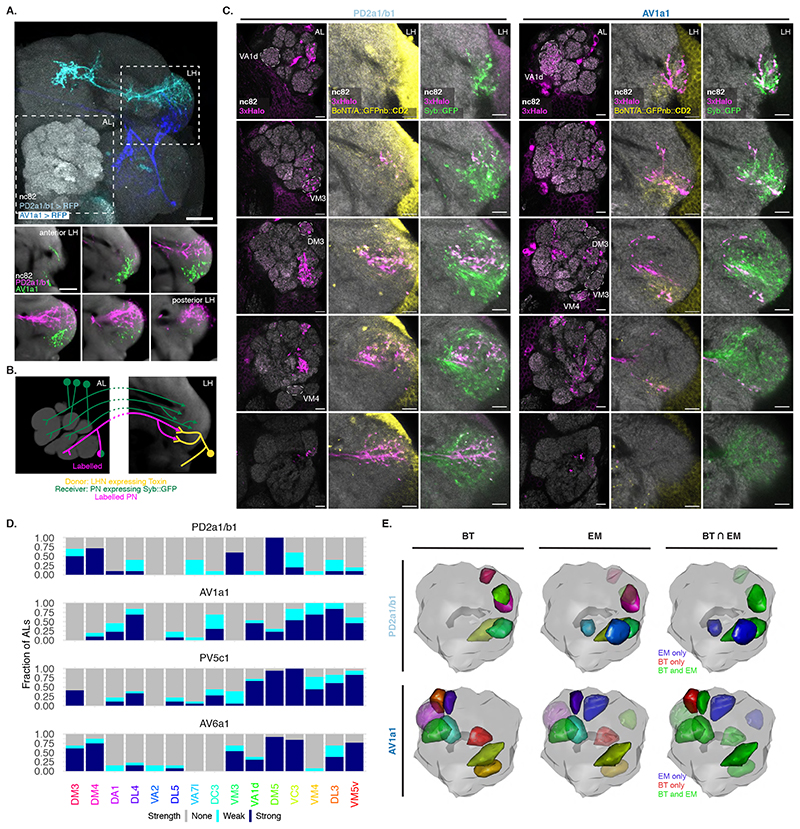
Figure 5. BAcTrace reveals connections between PNs and LHNs.
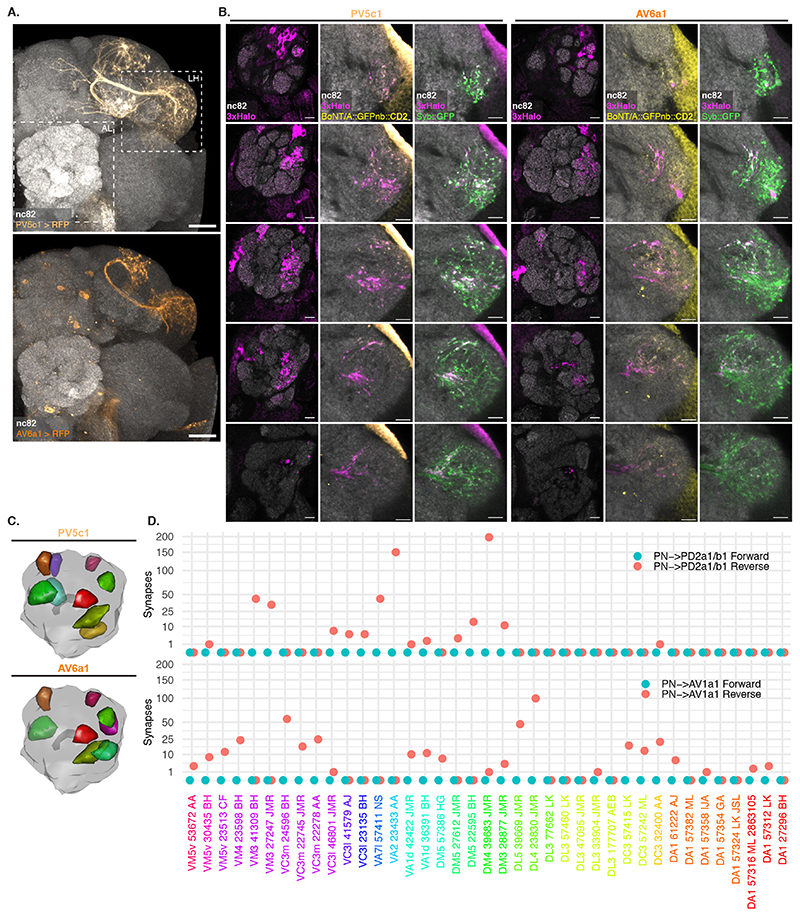
(A) Top: Split Gal4 lines LH989 and LH1983 drive GFP expression in LHN cell types PD2a1/b1 and AV1a1, respectively. Bottom: single slices of the LH shown in the top panel. nc82 staining from an average brain. AL: Antennal Lobe, LH: Lateral Horn. Images for a representative brain of 5 animals for each line. (B) Schematic of the experiment shown in (C). (C) Single slices from a representative AL and corresponding LH showing PNs labelled by expression of toxin in PD2a1/b1 and AV1a1 (see D for sample sizes). Examples of glomeruli differentially labelled between the 2 lines (VA1d, VM3, DM3 and VM4) are indicated with dotted lines. (D) Stacked bar plot showing labelling frequency per glomerulus for PD2a1/b1 (n=10 ALs), AV1a1 (n=13 ALs), PV5c1 (n=18 ALs) and AV6a1 (n=13 ALs). Glomeruli coloring same as in Figure 4D. (E) 3D renderings of ALs. BT: glomeruli with BAcTrace signal >3, EM: glomeruli with more than 10 synapses and BT⋂EM: meeting both conditions. Glomeruli coloring in BT and EM same as in (D); in BT⋂ EM green satisfies BT and EM criteria, blue EM only and red BT only. Epitopes detected by antibodies are indicated in bold in A and C. Genotypes for each panel are described in Supplementary Table 16. Scale bars: (A) 50μm and (C) 10μm.
Unlike in our previous experiments, there were noticeable differences in Receiver PN labelling across animals having the same Donor LHNs and even among left and right sides of the same brain (Extended Data Figure 8). We focused on 2 LHN types for which EM connectivity data shows lack of reciprocal synapses: PD2a1/b1 [29] and AV1a1 [30] as well as two types for which more limited information is available: PV5c1 and AV6a1 (Figure 5, Extended Data Figure 9 and Supplementary Note 8 and 9). PD2a1/b1 and AV1a1 target the dorsal and ventral LH respectively, with little spatial overlap (Figure 5A). Each Donor LHN cell type labelled Receiver PNs targeting different glomeruli. For example, VM3 and DM3 are labelled only in PD2a1/b1 while VM4 and VA1d are only labelled in AV1a1. Labelling differences can also be seen in the LH neuropil where the Receiver signal (Halo) has minimal overlap between the lines (Figure 5B,C). When comparing PD2a1/b1 and AV1a1, 12 out of the 15 glomeruli scored were labelled by at least one of the two Donor cell types; 11 of those 12 were labelled with different strengths, suggesting differences in connectivity (Figure 5D).
Having established that BAcTrace shows consistent differences in PN labelling when triggered by different Donor LHNs, we compared these results with previously published data (Extended Data Figure 10). When comparing pairs of methods, none were linearly correlated for the two LHN cell types studied (Pearson coefficient probability p<0.05, Supplementary Figure 4A). Nevertheless, when we binarized the presence or absence of connections by using a threshold of 10 synapses for the EM dataset and 3 arbitrary units for BAcTrace we found that for PD2a1/b1 neurons, 5 out of the 7 PNs connected in the EM dataset are also identified as connected by BAcTrace (Figure 5E). In the case of AV1a1, 7 out of 9 connected PN types in the EM dataset are also identified as connected by BAcTrace.
Discussion
In this study we present BAcTrace, a genetically encoded retrograde labelling system as well as the application of C. botulinum neurotoxin as a circuit tracer.
We used the fly’s olfactory system [17, 32, 18, 30, 33, 29, 5] to explore three important and related questions about BAcTrace. First, does labelling occur in the retrograde direction? Second, is labelling specific? Third, how many synapses are required? Support for the retrograde direction of labelling comes from the PN->LHN experiments; PD2a1/b1 and AV1a1 have no reciprocal synapse in the PN->LHN connection that could mediate anterograde transfer from LHNs to PNs. While BAcTrace labelling observed at these particular connections must be retrograde, and by design BAcTrace should have a strong retrograde bias, our results do not conclusively prove BAcTrace works exclusively in the retrograde direction. BAcTrace is specific since it most frequently labelled the correct PNs when toxin was expressed in ORNs. This was confirmed by the good match between our PN->LHN results and EM connectivity data. Regarding BAcTrace’s sensitivity, in our experiments we could detect labelling in connections ranging from 10 synapses (PN->LHN) to >200 synapses (PN->KC). Does this range encompass functional connections? Recent work has found that seemingly low connection strengths, constituting only a handful of synaptic contacts, can generate measurable effects on postsynaptic activity [29, 34, 4, 35, 17, 36]. While these numbers are tentative and could be different for different neuronal classes, they indicate that BAcTrace sensitivity falls within the range of known functional connections.
Current BAcTrace limitations include the lack of panneuronal coverage related to the impossibility of Gal4 and LexA drivers to overlap and the toxicity of the Syntaxin sensor when expressed widely. Furthermore, the presence of false positives in some of our experiments suggests that when used as a discovery tool other methods should be considered to confirm the identified connections; we expect this to become less of a requirement as community usage extends our understanding of the system’s performance.
Given rapid advances in the field of EM connectomics [37], which can reveal dense connectivity for all the neurons within a brain, one might wonder if it will supplant transsynaptic tracing going forwards. However, transsynaptic labelling has numerous advantages, suggesting that these approaches should remain complementary rather than competitive for the foreseeable future. First, EM connectomics is limited to post-mortem specimens, while transsynaptic labelling methods can rapidly reveal connectivity in living animals, enabling neurons to be targeted for recording or manipulation or be followed over time; furthermore connection-based labelling can enable more precise labelling of neurons than can be achieved with genetic drivers alone, for instance in cases where neurons that are genetically similar, such as olfactory PNs, but make connections with different partners, such as specific ORNs, are to be functionally manipulated. Second, EM connectomics will continue to remain prohibitively expensive and resource-intensive for many laboratories and for studies requiring the comparison of multiple specimens. Third, densely reconstructed connectomics datasets are still missing important information e.g. identification of excitatory vs inhibitory synapses, electrical synapses and extra-synaptic communication. In contrast, genetic tools can provide a readout for these properties, for example by using BAcTrace with neurotransmitter or peptidergic specific LexA lines or in combination with methods such as electrophysiology or effectors such as Crimson and/or GCaMP. There are also cases in which the two approaches are synergistic. Transsynaptic tools are particularly well suited to unequivocally link neurons identified in EM volumes to those labelled by genetic drivers since they simultaneously reveal neuronal morphology and connectivity. This process will be essential to the functional exploitation of the small number of reference connectomes that will become available over the next few years [38, 39].
Online Methods
Molecular cloning and transgenic flies
Backbones of plasmids used in S2 cell experiments were based on the Drosophila Gateway vector collection (Drosophila Genomics Resource Center, cat. 1071-1138). Backbones of plasmids used for making transgenic flies were derived from pJFRC19 [42], pJFRC81 [43] and pJFRC161 [23]. Synthesized DNA sequences were codon-optimized for Drosophila expression and made by GeneArt (Thermo Fisher Scientific, Inc) or IDT (Integrated DNA Technologies, Inc).
Plasmids were made using Gibson assembly [44]. All fragments were PCR amplified with overlapping primers yielding scarless products. All fusions were sequenced to control for mutations introduced during the cloning. GenBank accession numbers for all constructs can be found in Supplementary Table 9, Supplementary Table 12, Supplementary Table 13 and Supplementary Table 14. Transgenic flies were made by BestGene Inc.
S2 cell transfections
S2 cells were acquired from ThermoFisher Scientific (cat no R69007) and cultured according to the manufacturer recommendations. Once the culture was established the cells were transferred from Serum containing Schneider’s medium into medium containing increasing proportions of serum free Express 5 medium (ThermoFisher Scientific, cat no 10486025).
Plasmid DNA for S2 cell transfections was prepared using a MidiPrep DNA purification kit according to the manufacturer instructions (QIAGEN, cat no 12143).
S2 cell transfections were done in 6-well plates, each well containing 2ml of 1x106cells/ml. A total of 2μg of plasmid DNA was diluted with culture medium to a volume of 100μl and the mixture was vortexed. 3μl of FuGENE-HD transfection reagent (Promega, cat no E2311) were added to the diluted DNA, mixed gently and incubated for 10 minutes at room temperature. This mixture was then added drop-wise to the well with cells and the plate was put back into the incubator for 24h. Cells were then rinsed to remove transfection mix. Depending on the experiment either toxin was added as required (as shown in Figure 2A) or cells were mixed 1:1 (as shown in Figure 2B) followed by further 48h incubation before staining or western blot analysis. Plasmids for experiments shown in Figure 2B are listed in Supplementary Table 3, Figure 2D in Supplementary Table 4, Extended Data Figure 2A in Supplementary Table 5, Extended Data Figure 2G in Supplementary Table 6 and those from Extended Data Figure 2F in Supplementary Table 7.
Western blot analysis
Cells for Western blot analysis were resuspended in culture medium, pelleted, rinsed in PBS and pelleted again. Pellets were resuspended in 1x sample buffer (ThermoFisher Scientific, cat no NP0007) with a reducing agent (ThermoFisher Scientific, cat no NP0004) and heat denatured.
Samples were then loaded in 12% bis-tris gels (ThermoFisher Scientific, cat no NP0342BOX) and run using MOPS buffer (ThermoFisher Scientific, cat no NP0001). Gels were transferred to PVDF membranes (Millipore Inc, cat no IPVH00010) and developed using the antibodies shown in Supplementary Table 8 and ECL reagents (GE Healthcare Ltd, cat no RPN2232) following the manufacturer recommendations. HRP-conjugated secondary antibodies were purchased from Cell Signaling Technology, Inc.
Image acquisition and processing
Confocal stacks of fly brains were imaged at 768 × 768 pixels every 1 μm (voxel size of 0.46 × 0.46 × 1 μm; 0.6 zoom factor) using an EC Plan-Neofluar 40×/1.30 Oil DIC M27 objective and 16-bit color depth. For LHN glomeruli scoring higher magnification images were taken at 1024 x 1024 pixels every 0.5 μm (voxel size of 0.19 x 0.19 x 0.5 μm; 1-1.1 zoom factor). All images were acquired using Zen software on Zeiss LSM710 and Zeiss LSM880 confocal microscopes. For batch processing of images (e.g. brain registration) we used a combination of R [45], Fiji [46] and CMTK (www.nitrc.org).
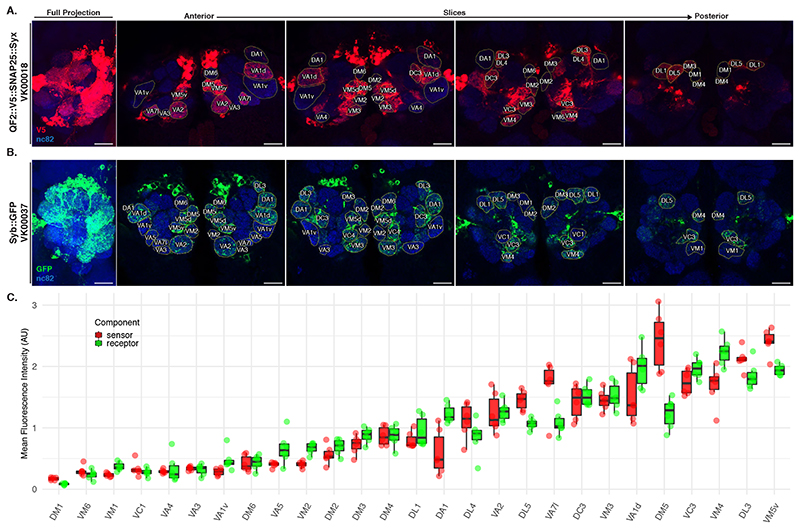
Fluorescence quantification
To quantify the expression levels of Syb::GFP and QF2::V5::hSNAP25::Syx driven by the VT033006-LexA::P65 driver (Extended Data Figure 3) we co-stained brains for the neuropil marker Bruchpilot and GFP or Bruchpilot and V5, respectively. We then acquired high-magnification confocal stacks of the stained ALs. Next we split the Zeiss LSM files into NRRD files using FIJI [46] followed by segmentation of each glomerulus by drawing a region of interest in the centre of the glomerulus (the plane which captured most of its area). Both glomerulus identification and segmentation were done using the nc82 channel, blind in relation to the GFP and V5 fluorescence intensities. Segmentations were saved as ROI files. GFP and V5 mean intensities were obtained using FIJI and were normalised to the mean intensity of the nc82 channel for each glomerulus. This normalisation is intended to counter the drop in intensity due to light scatter when imaging deeper into the tissue.
Drosophila stocks
Fly stocks were maintained at 25°C on Iberian food. The driver lines used in this study are summarized in Supplementary Table 15, LexA responsive transgenes in Supplementary Table 14, Gal4 ones in Supplementary Table 13 and QUAS ones in Table Supplementary Table 12. All brain images are from female flies.
LHN BAcTrace labelling quantification
We took high-magnification confocal stacks of antennal lobes and used the nc82 channel to identify and annotate the 15 glomeruli with the highest expression level of Receiver components (Extended Data Figure 3C) using regions of interest in FIJI. We assigned a value of 0 (absent), 3 (present but weak) and 10 (strong) to annotated glomeruli by examining the intensity of the QUAS-Halo reporter channel. Once all antennal lobes were blindly annotated in relation to LHN type, we used the toxin labelling (CD2) channel to assign each specimen to the correct LHN cell type. Stacks are available online [47] for readers to examine.
Light microscopy PN-LHN overlap score
In order to quantify the overlap between neuronal skeletons for PNs and LHNs, derived from both light-level and EM data, we employed the ‘overlap score’ from [33]:
Skeletons were resampled so that we considered ’points’ in the neuron at 1 μm intervals and an ’overlap score’ calculated as the sum of f(is, jk) over all points s of i. Here, i is the axonal portion of a neuron, j is the dendritic portion of a putative target, δ is the distance between two points at which a synapse might occur (e.g. 1 μm), and d is the Euclidean distance between points s and k. The sum was taken of the scores between each point in i and each point in j.
Overlap scores were calculated between light-level reconstructions from stochastic labelling experiments [31, 48], that have been previously been registered from hundreds of brains to a common template, categorized and identified [49, 33]. We also made use of a complete set of uniglomerular PNs, reconstructed from a single EM dataset comprising a whole fly brain [50].
Electrophysiology
Electrophysiological recordings were carried out as described in [33] with minor modifications. Briefly, one day after eclosion flies were anesthetized with CO2, females of the correct genotype were selected, transferred to all trans-retinal fly food and kept in the dark. Five days later, flies were cold-anesthetized, placed in the recording chamber and dissected under dim light for recording as described in [51]. Data acquisition was performed as previously described with the only difference that a pco.edge 4.2 CMOS camera was used. For CsChrimson excitation of ORNs, a short (0.5 sec) pulse of light (550nm) was applied via a Cairn OptoLED controller.
Extended Data
Extended Data Fig. 1.
Extended Data Fig. 2.
Extended Data Fig. 3.
Extended Data Fig. 4.
Extended Data Fig. 5.
Extended Data Fig. 6.
Extended Data Fig. 7.
Extended Data Fig. 8.
Extended Data Fig. 9.
Extended Data Fig. 10.
Supplementary Material
Acknowledgments
This work was supported by a Dorothy Hodgkin Fellowship from the Royal Society to S.C. (DH120072), ERC Starting (211089) and Consolidator (649111) grants and core support from the MRC (MC-U105188491) to G.S.X.E.J. We are grateful to Bazbek Davletov (University of Sheffield), Enrico Ferrari (University of Lincoln) and Jason Arsenault (University of Toronto) for sharing reagents and advice at early stages of the project. We acknowledge the Bloomington Stock Center for fly stocks, the Developmental Studies Hybridoma Bank for antibodies and the Drosophila Genomics Resource Center (NIH grant 2P40OD010949) for plasmids. Finally, we would like to thank Richard Benton and all members of the Jefferis group for many insightful comments on the manuscript.
Footnotes
Authors Contributions
S.C. and G.S.X.E.J. conceived the project, obtained funding and supervised the research. S.C., L.B., A.M., and A.S. designed, cloned, purified proteins, conducted tissue culture experiments and analyzed the results. S.C., M.G. and B.S. designed and conducted BAcTrace experiments in flies, stained, imaged and analyzed the results. S.F. designed, conducted and analyzed the results of electrophysiology experiments. Y.A. made VT033006-LexA::P65 and obtained MFCO data. A.S.B. summarized EM connectivity data for LHNs. S.C. and G.S.X.E.J wrote the manuscript with input from all authors.
Competing Interests
The authors declare no competing interests.
Data availability
All data necessary for confirming the conclusions presented in the article are represented fully within the article. In addition all raw and processed brain images and blots are freely available from the authors. All fly strains used are listed in Tables Supplementary Table 12, Supplementary Table 13, Supplementary Table 14 and Supplementary Table 15 and are available either from Bloomington stock center (Supplementary Table 17) or from the authors. All plasmids are available upon request from the authors and their sequences deposited in GenBank as described in Tables Supplementary Table 9, Supplementary Table 12, Supplementary Table 13 and Supplementary Table 14. Brain confocal stacks used for making ORN and LHN plots are available from the Zenodo repository [47].
References
- [1].Luo Liqun, Callaway Edward M, Svoboda Karel. Genetic dissection of neural circuits: A decade of progress. Neuron. 2018 Apr;98(2):256–281. doi: 10.1016/j.neuron.2018.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Ugur Berrak, Chen Kuchuan, Bellen Hugo J. Drosophila tools and assays for the study of human diseases. Dis Model Mech. 2016 Mar;9(3):235–44. doi: 10.1242/dmm.023762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Venken Koen JT, Simpson Julie H, Bellen Hugo J. Genetic manipulation of genes and cells in the nervous system of the fruit fly. Neuron. 2011 Oct;72(2):202–30. doi: 10.1016/j.neuron.2011.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Ohyama Tomoko, Schneider-Mizell Casey M, Fetter Richard D, Aleman Javier Valdes, Franconville Romain, Rivera-Alba Marta, Mensh Brett D, Branson Kristin M, Simpson Julie H, Truman James W, Cardona Albert, et al. A multilevel multimodal circuit enhances action selection in Drosophila. Nature. 2015 Apr;520(7549):633–9. doi: 10.1038/nature14297. [DOI] [PubMed] [Google Scholar]
- [5].Zheng Zhihao, Lauritzen J Scott, Perlman Eric, Robinson Camenzind G, Nichols Matthew, Milkie Daniel, Torrens Omar, Price John, Fisher Corey B, Sharifi Nadiya, Calle-Schuler Steven A, et al. A complete electron microscopy volume of the brain of adult Drosophila melanogaster. Cell. 2018 Jul; doi: 10.1016/j.cell.2018.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Takemura Shin-Ya, Lu Zhiyuan, Meinertzhagen Ian A. Synaptic circuits of the Drosophila optic lobe: the input terminals to the medulla. J Comp Neurol. 2008 Aug;509(5):493–513. doi: 10.1002/cne.21757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Talay Mustafa, Richman Ethan B, Snell Nathaniel J, Hartmann Griffin G, Fisher John D, Sorkaç Altar, Santoyo Juan F, Chou-Freed Cambria, Nair Nived, Johnson Mark, Szymanski John R, et al. Transsynaptic mapping of second-order taste neurons in flies by trans-tango. Neuron. 2017 Nov;96(4):783–795.e4. doi: 10.1016/j.neuron.2017.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang Ting-Hao, Niesman Peter, Arasu Deepshika, Lee Donghyung, De La Cruz Aubrie L, Callejas Antuca, Hong Elizabeth J, Lois Carlos. Tracing neuronal circuits in transgenic animals by transneuronal control of transcription (tract) Elife. 2017;6 doi: 10.7554/eLife.32027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dong Min, Yeh Felix, Tepp William H, Dean Camin, Johnson Eric A, Janz Roger, Chapman Edwin R. Sv2 is the protein receptor for botulinum neurotoxin a. Science. 2006 Apr;312(5773):592–6. doi: 10.1126/science.1123654. [DOI] [PubMed] [Google Scholar]
- [10].Montal Mauricio. Botulinum neurotoxin: a marvel of protein design. Annu Rev Biochem. 2010;79:591–617. doi: 10.1146/annurev.biochem.051908.125345. [DOI] [PubMed] [Google Scholar]
- [11].Pirazzini Marco, Rossetto Ornella, Eleopra Roberto, Montecucco Cesare. Botulinum neurotoxins: Biology, pharmacology, and toxicology. Pharmacol Rev. 2017;69(2):200–235. doi: 10.1124/pr.116.012658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Pauli Andrea, Althoff Friederike, Oliveira Raquel A, Heidmann Stefan, Schuldiner Oren, Lehner Christian F, Dickson Barry J, Nasmyth Kim. Cell-type-specific tev protease cleavage reveals cohesin functions in Drosophila neurons. Dev Cell. 2008 Feb;14(2):239–51. doi: 10.1016/j.devcel.2007.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Kubala Marta H, Kovtun Oleksiy, Alexandrov Kirill, Collins Brett M. Structural and thermodynamic analysis of the gfp:gfp-nanobody complex. Protein Sci. 2010 Dec;19(12):2389–401. doi: 10.1002/pro.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Riabinina Olena, Luginbuhl David, Marr Elizabeth, Liu Sha, Wu Mark N, Luo Liqun, Potter Christopher J. Improved and expanded q-system reagents for genetic manipulations. Nat Methods. 2015 Mar;12(3):219–22. doi: 10.1038/nmeth.3250. 5 p following 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Washbourne P, Pellizzari R, Baldini G, Wilson MC, Montecucco C. Botulinum neurotoxin types a and e require the snare motif in snap-25 for proteolysis. FEBS Lett. 1997 Nov;418(1-2):1–5. doi: 10.1016/s0014-5793(97)01328-8. [DOI] [PubMed] [Google Scholar]
- [16].Gupta Gagan D, Swetha MG, Kumari Sudha, Lakshminarayan Ramya, Dey Gautam, Mayor Satyajit. Analysis of endocytic pathways in Drosophila cells reveals a conserved role for gbf1 in internalization via geecs. PLoS One. 2009 Aug;4(8):e6768. doi: 10.1371/journal.pone.0006768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Tobin William F, Wilson Rachel I, Lee Wei-Chung Allen. Wiring variations that enable and constrain neural computation in a sensory microcircuit. Elife. 2017 May;6 doi: 10.7554/eLife.24838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Masse Nicolas Y, Turner Glenn C, Jefferis Gregory SXE. Olfactory information processing in Drosophila. Curr Biol. 2009 Aug;19(16):R700–13. doi: 10.1016/j.cub.2009.06.026. [DOI] [PubMed] [Google Scholar]
- [19].Turner Glenn C, Bazhenov Maxim, Laurent Gilles. Olfactory representations by Drosophila mushroom body neurons. J Neurophysiol. 2008 Feb;99(2):734–46. doi: 10.1152/jn.01283.2007. [DOI] [PubMed] [Google Scholar]
- [20].Murthy Mala, Fiete Ila, Laurent Gilles. Testing odor response stereotypy in the Drosophila mushroom body. Neuron. 2008 Sep;59(6):1009–23. doi: 10.1016/j.neuron.2008.07.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Caron Sophie JC, Ruta Vanessa, Abbott LF, Axel Richard. Random convergence of olfactory inputs in the Drosophila mushroom body. Nature. 2013 May;497(7447):113–7. doi: 10.1038/nature12063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Gruntman Eyal, Turner Glenn C. Integration of the olfactory code across dendritic claws of single mushroom body neurons. Nat Neurosci. 2013 Dec;16(12):1821–9. doi: 10.1038/nn.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Nern Aljoscha, Pfeiffer Barret D, Svoboda Karel, Rubin Gerald M. Multiple new site-specific recombinases for use in manipulating animal genomes. Proc Natl Acad Sci U S A. 2011 Aug;108(34):14198–203. doi: 10.1073/pnas.1111704108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Aso Yoshinori, Hattori Daisuke, Yu Yang, Johnston Rebecca M, Iyer Nirmala A, Ngo Teri-TB, Dionne Heather, Abbott LF, Axel Richard, Tanimoto Hiromu, Rubin Gerald M. The neuronal architecture of the mushroom body provides a logic for associative learning. Elife. 2014 Dec;3:e04577. doi: 10.7554/eLife.04577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Couto Africa, Alenius Mattias, Dickson Barry J. Molecular, anatomical, and functional organization of the Drosophila olfactory system. Curr Biol. 2005 Sep;15(17):1535–47. doi: 10.1016/j.cub.2005.07.034. [DOI] [PubMed] [Google Scholar]
- [26].Fishilevich Elane, Vosshall Leslie B. Genetic and functional subdivision of the Drosophila antennal lobe. Curr Biol. 2005 Sep;15(17):1548–53. doi: 10.1016/j.cub.2005.07.066. [DOI] [PubMed] [Google Scholar]
- [27].Rybak Jürgen, Talarico Giovanni, Ruiz Santiago, Arnold Christopher, Cantera Rafael, Hansson Bill S. Synaptic circuitry of identified neurons in the antennal lobe of Drosophila melanogaster. J Comp Neurol. 2016 Jun;524(9):1920–56. doi: 10.1002/cne.23966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Klapoetke Nathan C, Murata Yasunobu, Kim Sung Soo, Pulver Stefan R, Birdsey-Benson Amanda, Cho Yong Ku, Morimoto Tania K, Chuong Amy S, Carpenter Eric J, Tian Zhijian, Wang Jun, et al. Independent optical excitation of distinct neural populations. Nat Methods. 2014 Mar;11(3):338–46. doi: 10.1038/nmeth.2836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Dolan Michael-John, Belliart-Guérin Ghislain, Bates Alexander Shakeel, Frechter Shahar, Lampin-Saint-Amaux Aurélie, Aso Yoshinori, Roberts Ruairí JV, Schlegel Philipp, Wong Allan, Hammad Adnan, Bock Davi, et al. Communication from learned to innate olfactory processing centers is required for memory retrieval in Drosophila. Neuron. 2018 Nov;100(3):651–668.e8. doi: 10.1016/j.neuron.2018.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Huoviala Paavo, Dolan Michael-John, Love Fiona M, Frechter Shahar, Roberts Ruairí JV, Mitrevica Zane, Schlegel Philipp, Bates Alexander Shakeel, Aso Yoshinori, Rodrigues Tiago, Cornwall Hannah, et al. Neural circuit basis of aversive odour processing in Drosophila from sensory input to descending output. bioRxiv. 2018 [Google Scholar]
- [31].Dolan Michael-John, Frechter Shahar, Bates Alexander Shakeel, Dan Chuntao, Huoviala Paavo, Roberts Ruairí Jv, Schlegel Philipp, Dhawan Serene, Tabano Remy, Dionne Heather, Christoforou Christina, et al. Neurogenetic dissection of the Drosophila lateral horn reveals major outputs, diverse behavioural functions, and interactions with the mushroom body. Elife. 2019 May;8 doi: 10.7554/eLife.43079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Butcher Nancy J, Friedrich Anja B, Lu Zhiyuan, Tanimoto Hiromu, Meinertzhagen Ian A. Different classes of input and output neurons reveal new features in microglomeruli of the adult Drosophila mushroom body calyx. J Comp Neurol. 2012 Jul;520(10):2185–201. doi: 10.1002/cne.23037. [DOI] [PubMed] [Google Scholar]
- [33].Frechter Shahar, Bates Alexander Shakeel, Tootoonian Sina, Dolan Michael-John, Manton James, Jamasb Arian Rokkum, Kohl Johannes, Bock Davi, Jefferis Gregory. Functional and anatomical specificity in a higher olfactory centre. Elife. 2019 May;8 doi: 10.7554/eLife.44590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Felsenberg Johannes, Jacob Pedro F, Walker Thomas, Barnstedt Oliver, Edmondson-Stait Amelia J, Pleijzier Markus W, Otto Nils, Schlegel Philipp, Sharifi Nadiya, Perisse Emmanuel, Smith Carlas, et al. Integration of parallel opposing memories underlies memory extinction. Cell. 2018;175(3):709–722.e15. doi: 10.1016/j.cell.2018.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sayin Sercan, De Backer Jean-Francois, Siju KP, Wosniack Marina E, Lewis Laurence P, Frisch Lisa-Marie, Gansen Benedikt, Schlegel Philipp, Edmondson-Stait Amelia, Sharifi Nadiya, Fisher Corey B, et al. A neural circuit arbitrates between persistence and withdrawal in hungry Drosophila. Neuron. 2019 Nov;104(3):544–558.e6. doi: 10.1016/j.neuron.2019.07.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Kazama Hokto, Wilson Rachel I. Origins of correlated activity in an olfactory circuit. Nat Neurosci. 2009 Sep;12(9):1136–44. doi: 10.1038/nn.2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kornfeld Jörgen, Denk Winfried. Progress and remaining challenges in high-throughput volume electron microscopy. Curr Opin Neurobiol. 2018;50:261–267. doi: 10.1016/j.conb.2018.04.030. [DOI] [PubMed] [Google Scholar]
- [38].Schlegel Philipp, Costa Marta, Jefferis Gregory Sxe. Learning from connectomics on the fly. Curr Opin Insect Sci. 2017;24:96–105. doi: 10.1016/j.cois.2017.09.011. [DOI] [PubMed] [Google Scholar]
- [39].Bates Alexander Shakeel, Janssens Jasper, Jefferis Gregory Sxe, Aerts Stein. Neuronal cell types in the fly: single-cell anatomy meets single-cell genomics. Curr Opin Neurobiol. 2019;56:125–134. doi: 10.1016/j.conb.2018.12.012. [DOI] [PubMed] [Google Scholar]
- [40].Lacy DB, Tepp W, Cohen AC, DasGupta BR, Stevens RC. Crystal structure of botulinum neurotoxin type a and implications for toxicity. Nat Struct Biol. 1998 Oct;5(10):898–902. doi: 10.1038/2338. [DOI] [PubMed] [Google Scholar]
- [41].Cachero Sebastian, Jefferis Gregory SXE. Drosophila olfaction: the end of stereotypy? Neuron. 2008 Sep;59(6):843–5. doi: 10.1016/j.neuron.2008.09.017. [DOI] [PubMed] [Google Scholar]
- [42].Pfeiffer Barret D, Ngo Teri-TB, Hibbard Karen L, Murphy Christine, Jenett Arnim, Truman James W, Rubin Gerald M. Refinement of tools for targeted gene expression in Drosophila. Genetics. 2010 Oct;186(2):735–55. doi: 10.1534/genetics.110.119917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Pfeiffer Barret D, Truman James W, Rubin Gerald M. Using translational enhancers to increase transgene expression in Drosophila. Proc Natl Acad Sci U S A. 2012 Apr;109(17):6626–31. doi: 10.1073/pnas.1204520109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Gibson Daniel G, Young Lei, Chuang Ray-Yuan, Venter J Craig, Hutchison Clyde A, 3rd, Smith Hamilton O. Enzymatic assembly of dna molecules up to several hundred kilobases. Nat Methods. 2009 May;6(5):343–5. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- [45].R Core Team. R: A language and environment for statistical computing. 2016 [Google Scholar]
- [46].Schindelin Johannes, Arganda-Carreras Ignacio, Frise Erwin, Kaynig Verena, Longair Mark, Pietzsch Tobias, Preibisch Stephan, Rueden Curtis, Saalfeld Stephan, Schmid Benjamin, Tinevez Jean-Yves, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012 Jun;9(7):676–82. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Cachero Sebastian, Gkantia Marina, Bates Alexander S, Frechter Shahar, Blackie Laura, McCarthy Amy, Sutcliffe Ben, Strano Alessio, Aso Yoshinori, Jefferis Gregory SXE. BAcTrace a new tool for retrograde tracing of neuronal circuits. Zenodo; 2020. Jan 25, [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Chiang Ann-Shyn, Lin Chih-Yung, Chuang Chao-Chun, Chang Hsiu-Ming, Hsieh Chang-Huain, Yeh Chang-Wei, Shih Chi-Tin, Wu Jian-Jheng, Wang Guo-Tzau, Chen Yung-Chang, Wu Cheng-Chi, et al. Three-dimensional reconstruction of brain-wide wiring networks in Drosophila at single-cell resolution. Curr Biol. 2011 Jan;21(1):1–11. doi: 10.1016/j.cub.2010.11.056. [DOI] [PubMed] [Google Scholar]
- [49].Costa Marta, Manton James D, Ostrovsky Aaron D, Prohaska Steffen, Jefferis Gregory SXE. Nblast: Rapid, sensitive comparison of neuronal structure and construction of neuron family databases. Neuron. 2016;91(2):293–311. doi: 10.1016/j.neuron.2016.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Bates AS, Schlegel P, Roberts RJV, Drummond N, Tamimi IFM, Turnbull R, Zhao X, Marin EC, Popovici PD, Dhawan S, Jamasb A, et al. Complete connectomic reconstruction of olfactory projection neurons in the fly brain. bioRxiv. 2020 doi: 10.1016/j.cub.2020.06.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Kohl Johannes, Ostrovsky Aaron D, Frechter Shahar, Jefferis Gregory SXE. A bidirectional circuit switch reroutes pheromone signals in male and female brains. Cell. 2013 Dec;155(7):1610–23. doi: 10.1016/j.cell.2013.11.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Davletov Bazbek, Bajohrs Mark, Binz Thomas. Beyond botox: advantages and limitations of individual botulinum neurotoxins. Trends Neurosci. 2005 Aug;28(8):446–52. doi: 10.1016/j.tins.2005.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data necessary for confirming the conclusions presented in the article are represented fully within the article. In addition all raw and processed brain images and blots are freely available from the authors. All fly strains used are listed in Tables Supplementary Table 12, Supplementary Table 13, Supplementary Table 14 and Supplementary Table 15 and are available either from Bloomington stock center (Supplementary Table 17) or from the authors. All plasmids are available upon request from the authors and their sequences deposited in GenBank as described in Tables Supplementary Table 9, Supplementary Table 12, Supplementary Table 13 and Supplementary Table 14. Brain confocal stacks used for making ORN and LHN plots are available from the Zenodo repository [47].