Abstract
Changes in gene expression play a fundamental role in phenotypic evolution. Transcriptome evolutionary dynamics have so far mainly been compared among distantly related species and remain largely unexplored during rapid organismal diversification, in which gene regulatory changes have been suggested as particularly effective drivers of phenotypic divergence. Here, we studied gene expression evolution in a model system of adaptive radiation, the cichlid fishes of African Lake Tanganyika. By comparing gene expression profiles of six different organs in 74 cichlid species representing all subclades of this radiation, we demonstrate that the rate of gene expression evolution varies among organs, transcriptome parts, and the subclades of the radiation, indicating different strengths of selection. We found that the non-coding part of the transcriptome evolved more rapidly than the coding part, and that the gonadal transcriptomes evolved more rapidly than the somatic ones, with the exception of liver. We further show that the rate of gene-expression change was not constant over the course of the radiation but accelerated at its later phase. Finally, we show that – at the per-gene level – the evolution of expression patterns is dominated by stabilizing selection.
During the second half of the last century, it has become increasingly clear that changes in gene expression play a fundamental role in phenotypic evolution1–8. Previous large-scale studies of gene expression evolution comparing distantly related vertebrate species revealed substantial variation in the rate of transcriptome evolution among organs and evolutionary subclades4,9, between the coding and non-coding part of the transcriptome10, as well as across developmental timepoints11. However, little is known about the dynamics of gene expression evolution during adaptive radiations, which are characterised by the unusually rapid ecological and morphological diversification of an organismal lineage into distinct ecological niches12,13. Yet, precisely for such outbursts of organismal diversity, gene regulatory changes have been proposed as a key mechanism promoting rapid phenotypic divergence13–15, making the study of transcriptome evolution of particular interest in the context of adaptive radiations16.
Here, we examined the dynamics of gene expression evolution in one of the most striking examples of adaptive radiation, the cichlid fishes of African Lake Tanganyika16–18. This species flock comprises about 240 endemic cichlids that evolved in less than 10 Myr and show an extraordinary degree of eco-morphological divergence17–19. We sequenced the transcriptomes of five organs (brain, gill, liver, ovary, and testis) in three males and three females of 74 cichlid species, representing all phylogenetic subclades – so-called ‘tribes’ – and all major ecological guilds of the cichlid fauna of Lake Tanganyika17 (Fig. 1a, Supplementary Tables 1 and 2). In addition, we sequenced the transcriptomes of the lower pharyngeal jaw bone (LPJ) in the same set of 445 specimens. The LPJ is the central component of the cichlids’ pharyngeal jaw apparatus, that is, a second set of functional jaws in the pharynx used to masticate food20, and hypothesized to be a key-innovation triggering cichlid adaptive radiations18,21–24. These target organs were selected because of their involvement in ecological, physiological, and behavioural adaptations during cichlid adaptive radiations16,18,20,25,26 and to enable comparisons to previous studies3,11,27.
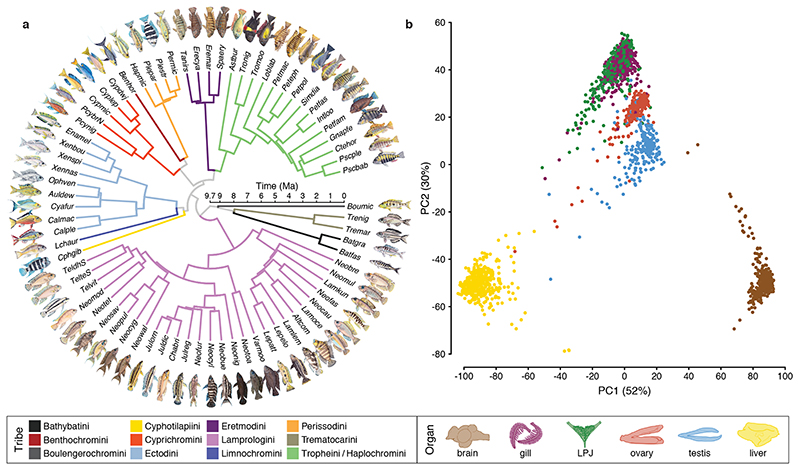
Fig. 1. Gene expression patterns across the adaptive radiation of cichlid fishes in African Lake Tanganyika.
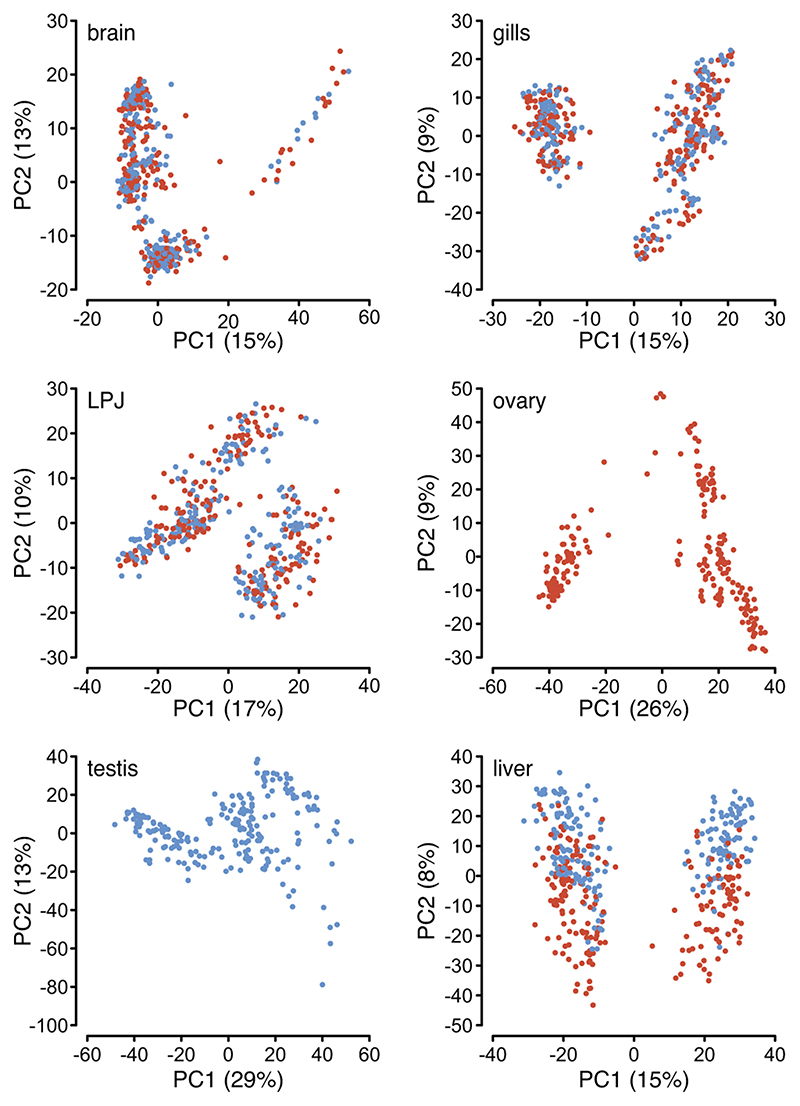
a, Time-calibrated species tree of the adaptive radiation of cichlid fishes in African Lake Tanganyika based on genome-wide data18, pruned to the 74 taxa used in this study. Species names are abbreviated using a six-letter code, whereby the first three letters represent the genus and the last three letters the species name (see Extended Data Fig. 9 and Supplementary Table 2 for full species names). Branches are colour-coded according to phylogenetic subclades, i.e., tribes, as indicated in the lower panel. b, Principal component analyses of overall gene expression levels. Samples (n = 2,131) are coloured according to organ type as indicated in the lower panel. The proportions of variance explained by the first two principal components (PC1 and PC2) are indicated in parenthesis at the X- and Y-axes, respectively.
Results
Patterns of gene expression
To study gene expression evolution during rapid organismal diversification, we generated a total of 2,131 transcriptome profiles (equivalent to individual RNAseq libraries) from typically five organs in six adult specimens of 74 species of cichlid fishes from African Lake Tanganyika (median sequencing depth per tissue: 9.6-9.9 million reads per library; 125 bp strand-specific single-end reads; mapped against the phylogenetically equidistant Oreochromis niloticus reference genome; median of read mapping ca. 76%). A time-calibrated species tree based on genome-wide data taken from18 and pruned to the taxon set of this study is shown in Fig. 1a; details on individual samples including sampling dates and locations are available in Supplementary Table 1; information on sequencing and mapping coverage is provided in Supplementary Figure 1; and the variance within and between species is shown in Supplementary Figures 2–4.
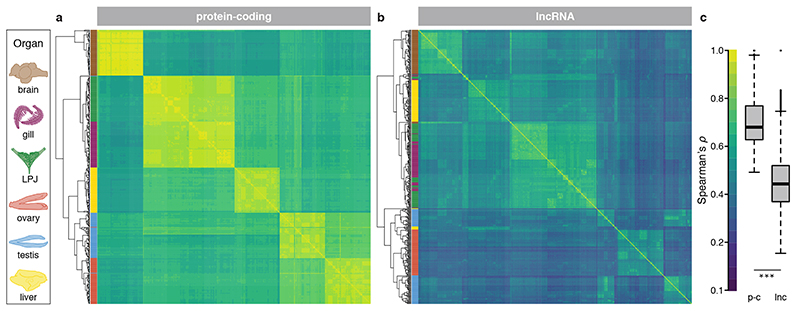
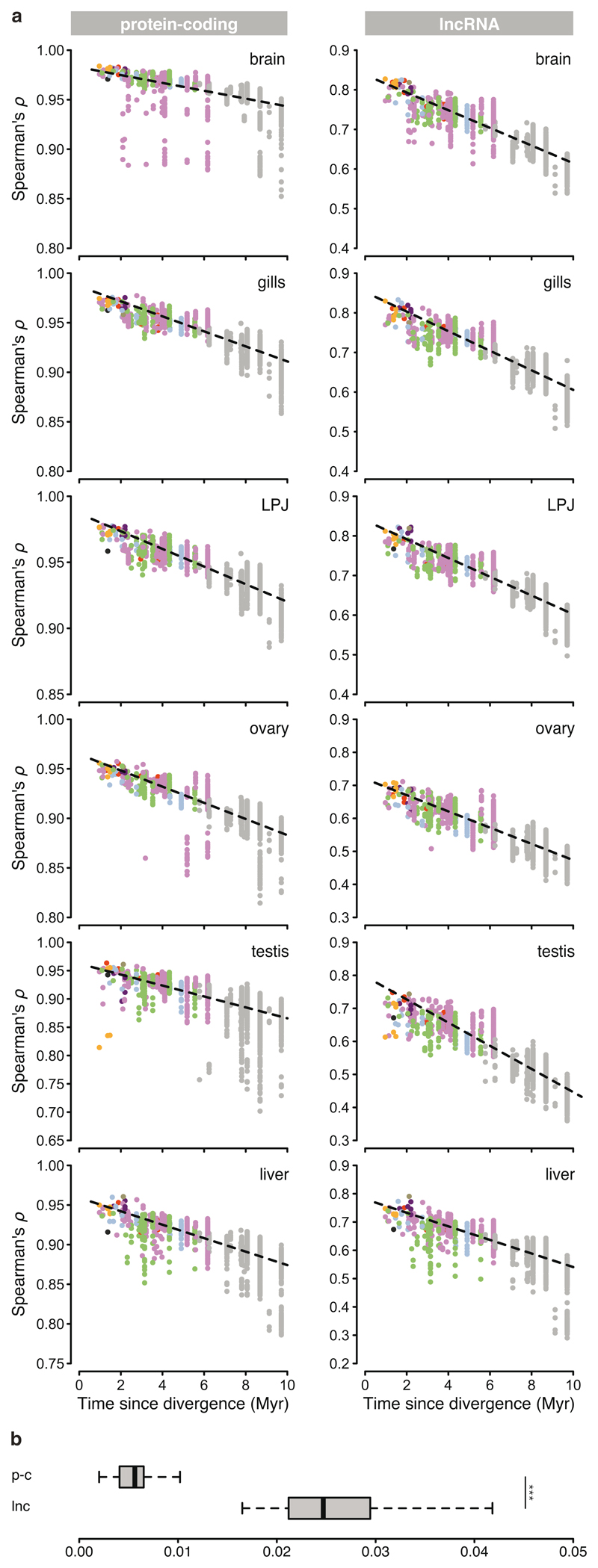
As a first step, to explore the global patterns of gene expression differentiation among species and across organs, we performed a principal component analysis (PCA) on the entire data set. The PCA clearly separated the expression profiles according to organ type (Fig. 1b) – with the exception of gill and LPJ transcriptomes, which showed largely overlapping gene expression profiles. This similarity in gene expression profiles is not surprising, given their common developmental origin (the LPJ is derived from the fusion of the left and right fifth ceratobranchials20). Within organs, on the other hand, the species-specific transcriptome profiles clustered by tribe (Fig. 2), indicating a strong phylogenetic signal in the data. This in turn suggests that, similar to mammals4, gene expression changes have accumulated over the course of the adaptive radiation of cichlid fishes in Lake Tanganyika, resulting in – overall – more similar gene expression profiles between more closely related species, irrespective of their adaptations to particular ecological niches. When comparing between the two largest classes of RNAs in the cichlids’ transcriptomes – that is, between protein-coding and long non-coding (lnc) RNAs – we found that the correlations in gene expression levels among the 74 cichlid species were significantly higher for protein-coding genes (Fig. 3a) than for lncRNAs (Fig. 3b) (pairwise Spearman’s rank correlation coefficients [Spearman’s ρ] between gene expression levels measured as transcripts per million [TPM]; two-sided t-test: P < 10-8; Fig. 3c). Also, the hierarchical clustering of gene expression levels revealed a much more pronounced grouping according to organ type in protein-coding genes compared to lncRNAs (Fig. 3a, b). This suggests that, also during the short evolutionary timescale spanned by the cichlid adaptive radiation in Lake Tanganyika (< 10 Myr18), the non-coding part of the transcriptome has undergone more rapid turnovers in expression trajectories than the coding part. The observed accelerated evolution of lncRNAs can be interpreted as a sign of more relaxed selection regimes in lncRNAs compared to protein-coding genes3.
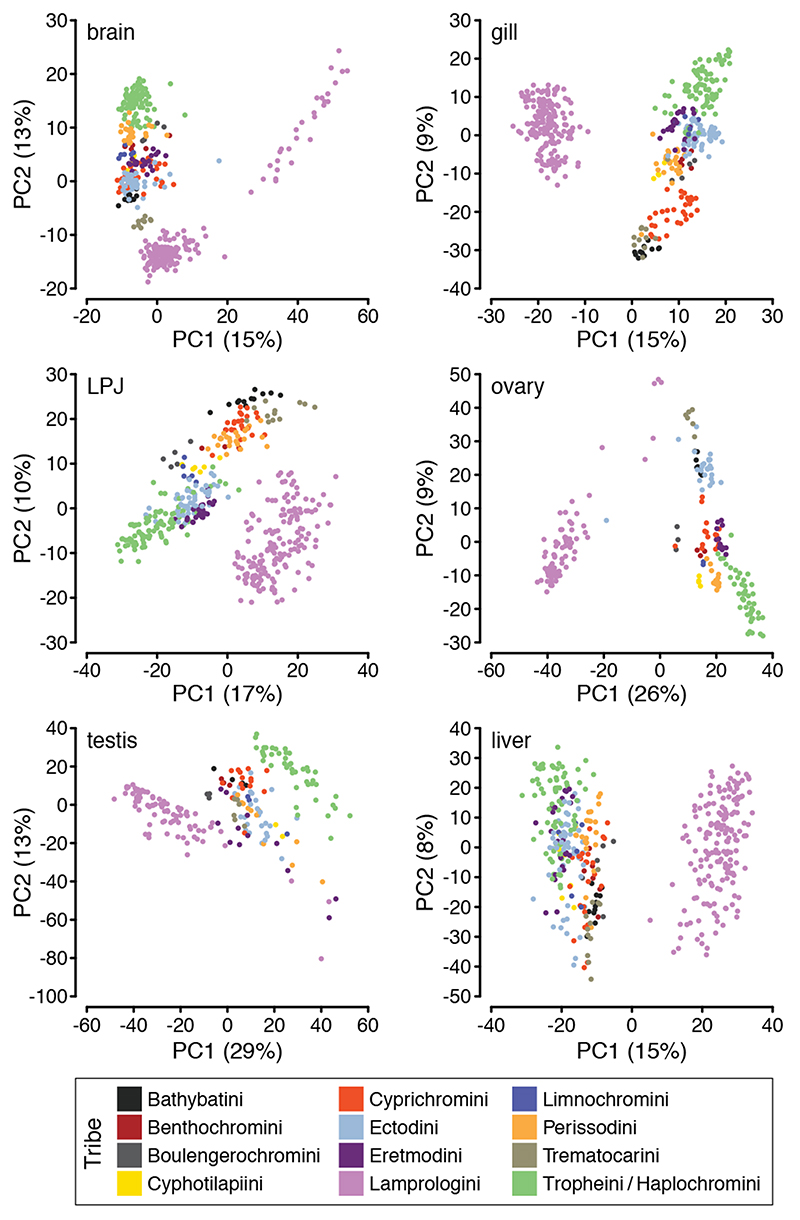
Fig. 2. Principal component analyses of overall gene expression levels per organ.
The proportion of variance explained by the first two principal components (PC1 and PC2) is shown for brain, gill, lower pharyngeal jaw bone (LPJ), ovary, testis, and liver and indicated in parenthesis at the X- and Y-axes, respectively. Samples (brain: n = 428; gills: n = 434; LPJ: n = 425; ovary: n = 219; testis: n = 213; liver: n = 412) are coloured according to tribe as shown in the lower panel.
Fig. 3. Gene expression similarities among species and transcriptome parts.
Hierarchical clustering of expression levels estimated as transcripts per million (TPM) for protein-coding genes (n = 27,105) (a) and lncRNAs (n = 4,719) (b). The heatmaps represent Spearman’s rank correlation coefficients between pairs of species (n = 442 samples). Sample clustering is represented as a tree on the left side of each heatmap, with organs colour-coded as indicated in the organ illustrations on the left side. c, Spearman’s rank correlation coefficients between pairs of species for protein-coding genes (p-c) and lncRNAs (lnc). Boxplot centre lines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5x interquartile range; two-sided t-test: P < 10-8.
When examining the transcriptome profiles of the somatic organs for sex-specific differences, we found a pronounced difference between females and males in liver but not in the other organs (Extended Data Fig. 1). Sex-biased gene expression in liver has been reported before4, albeit with a smaller magnitude than observed here. These sex differences in liver gene expression can mainly be attributed to its function as the main metabolic organ secreting hormones and maintaining homeostasis, and being responsive to sex steroids28–33 (for an in-depth analysis of sex-specific differences in Tanganyikan cichlid fishes as well as of sex chromosome evolution see ref.34).
Transcriptome evolution
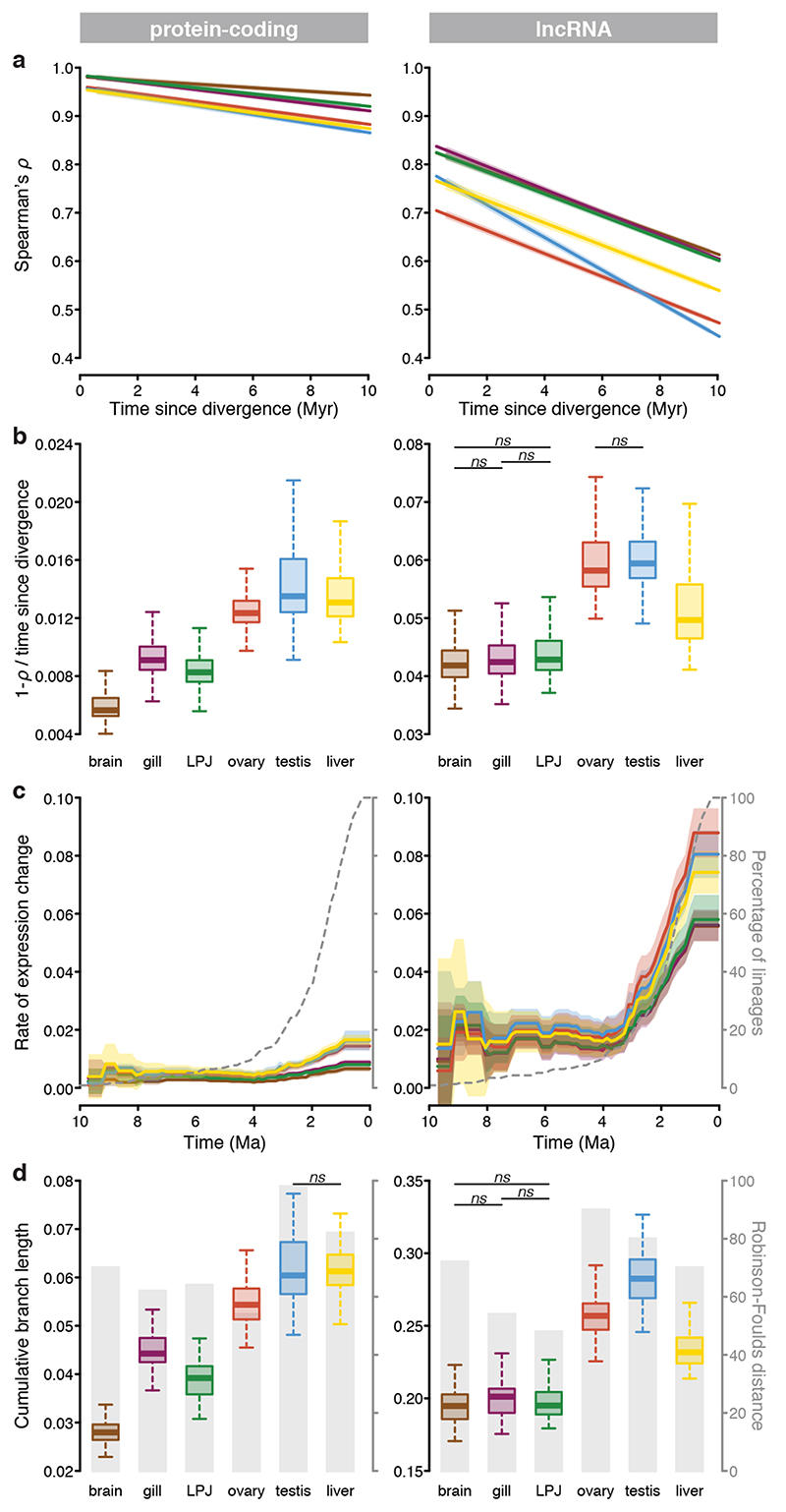
To compare the rate of evolution of the coding and non-coding part of the transcriptome in the different organs, we correlated Spearman’s ρ of gene expression levels with divergence times (taken from ref.18) for all pairs of cichlid species examined (Fig. 4a; Extended Data Fig. 2a). We found that, similar to the pattern observed in tetrapods featuring much deeper phylogenetic splits4, the rate of transcriptome evolution (measured as [1 – ρ] / divergence time) differed significantly among organs (Fig. 4b; ANOVA: P < 10-8 for protein-coding genes and lncRNAs), as well as between the transcriptome parts in cichlids (Fig. 4b; Extended Data Fig. 2; two-sided t-test: P < 10-8). The expression levels of protein-coding genes evolved significantly slower in brain, LPJ, and gill, than in testis, liver, and ovary (left panel in Fig. 4b; ANOVA: P < 10-8; Supplementary Table 3a), indicating that – taken as a whole – the organ-specific transcriptomes evolved under different selection regimes during the course of the Tanganyikan cichlid radiation.
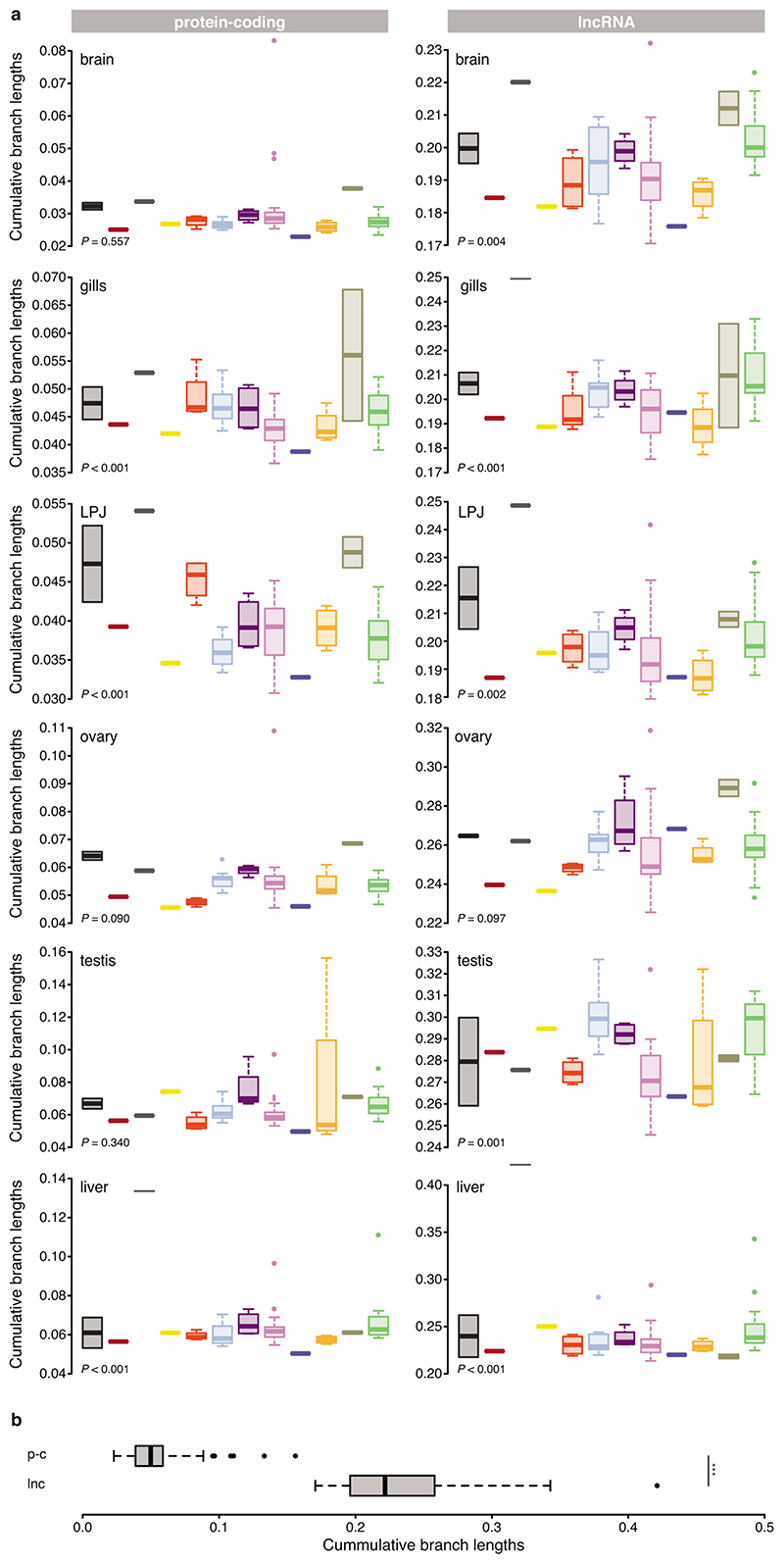
Fig. 4. Rate of gene expression evolution across organs for protein-coding genes (left panel) and lncRNAs (right panel).
a, Regression lines of gene expression similarities (Spearman’s ρ) of species pairs (brain, ovary, gills and testis: n = 74 taxa; LPJ and liver: n = 73 taxa) as a function of their divergence time18 for each organ (colour-coded as labelled in Fig. 1b). 95% confidence intervals are represented in shadow lines. b, Rate of expression changes (measured as [1 – ρ] / divergence time) within each organ (n = 73 taxa). c, Mean rate of expression change (measured as expression branch length calculated along the fixed species tree topology (n = 73 taxa) in relation to its branch length (Extended Data Figs. 5–7) over time, sampled in steps of 0.15 Myr along the phylogeny (as in ref.18) for each organ (colour-coded as labelled in Fig. 1b). The dotted line represents the accumulation of species over time (taken from Ronco et al. 18). 95% confidence intervals are represented in shadow lines. d, Cumulative branch length (calculated from root to tips of expression trees with a fixed species tree topology (n = 73 taxa, Extended Data Figs. 5, 6) for each organ. Grey barplots represent the Robinson-Foulds distances58 between expression trees with fixed topologies (as in18) and expression trees with no topological constraints (topology only depending on the expression data, see method section for more details). In all boxplots (b, d) centre lines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5x interquartile range. Outliers are not shown. All pairwise comparisons among organs were significant (one-sided Tukey HSD test, adjusted for multiple testing: P < 0.05) unless indicated in the plots (mentioned as n.s.). LPJ = lower pharyngeal jaw bone.
The correlations between Spearman’s ρ and divergence times were weaker in lncRNAs than in protein-coding genes (Extended Data Fig. 2b; two-sided t-test: P < 10-8), suggesting more relaxed selective constraints in lncRNAs. The lncRNA expression levels of the gonads evolved fastest compared to the other organs, followed by liver and then the remaining three organs (brain, gill, and LPJ), which showed similar patterns of expression evolution (right panel in Fig. 4b; ANOVA: P < 10-8, Supplementary Table 3b). Organ-specific evolutionary rates thus emerge as a transcriptome-wide trend in cichlids, with protein-coding genes being subject to stronger conservation – indicating stronger purifying selection and/or weaker positive selection – in gene expression levels compared to lncRNAs3.
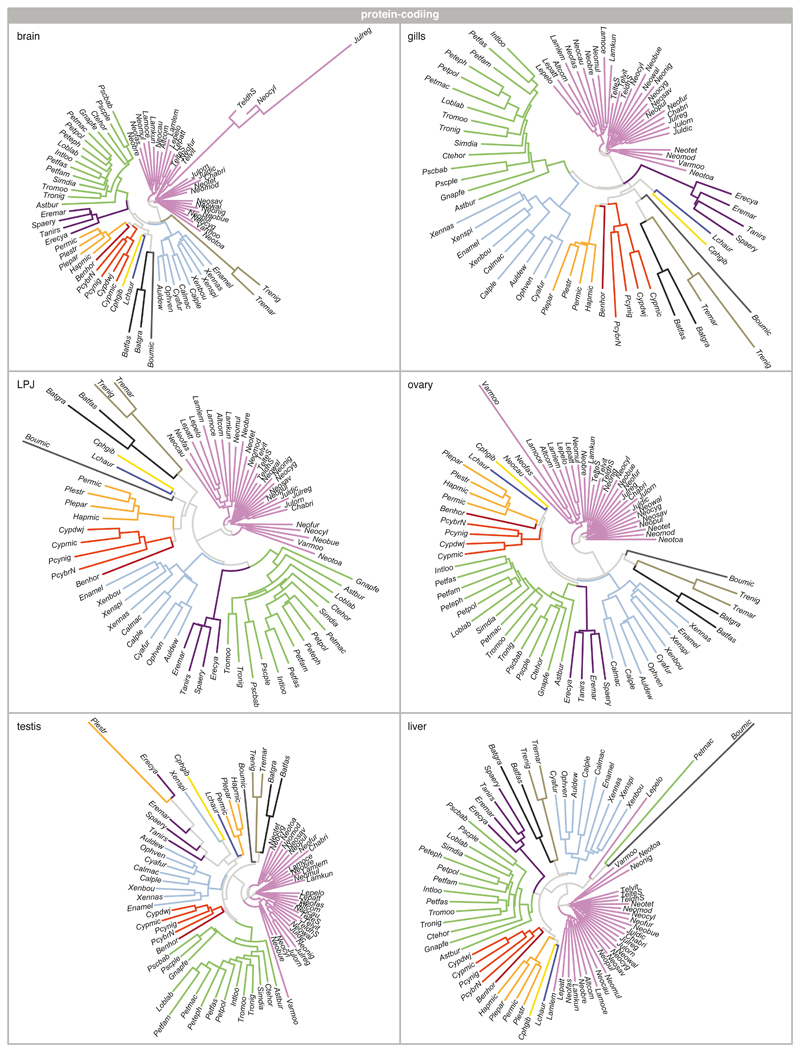
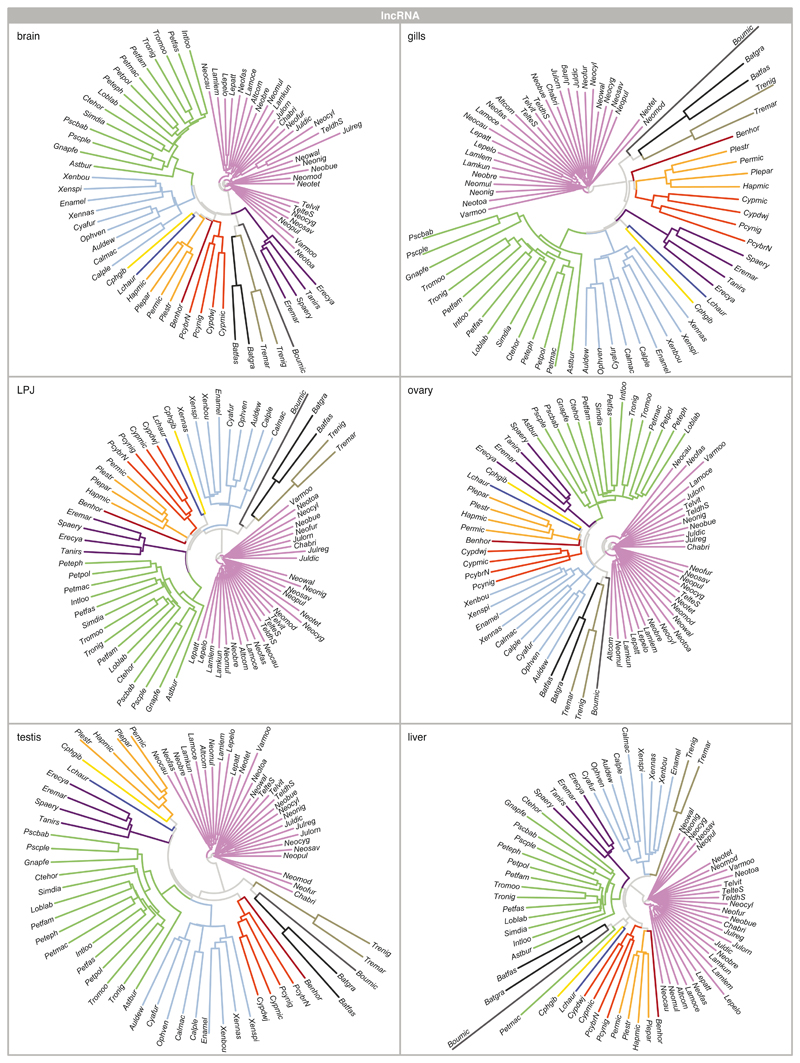
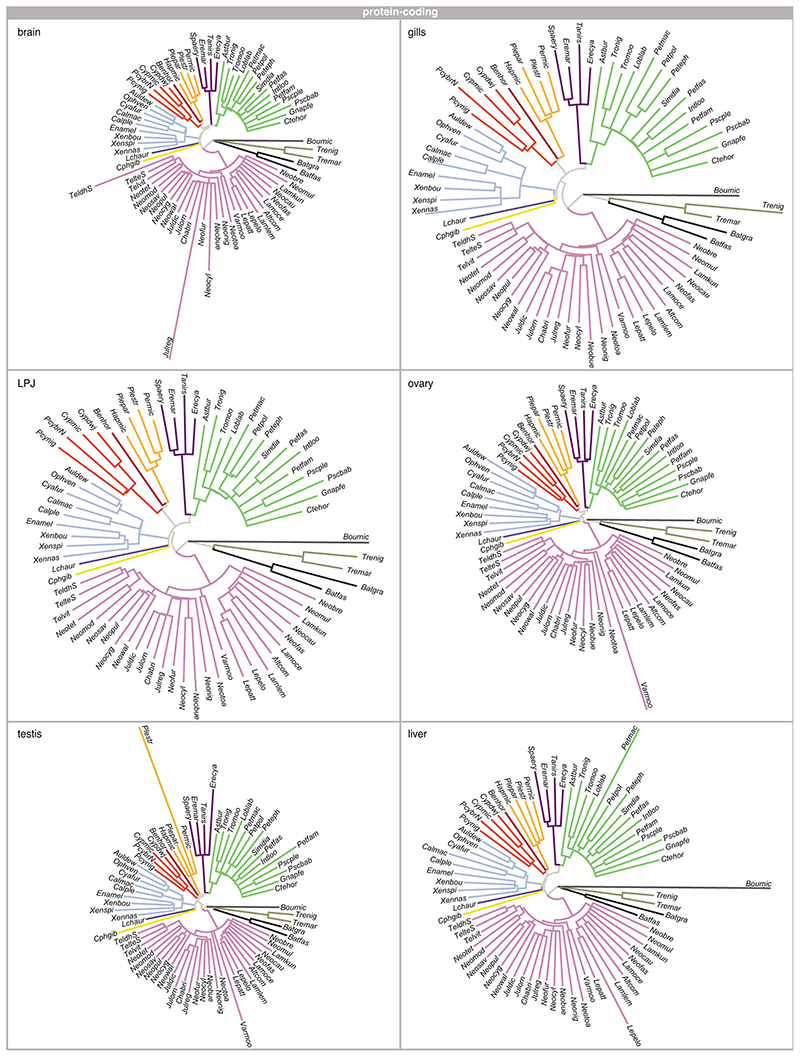
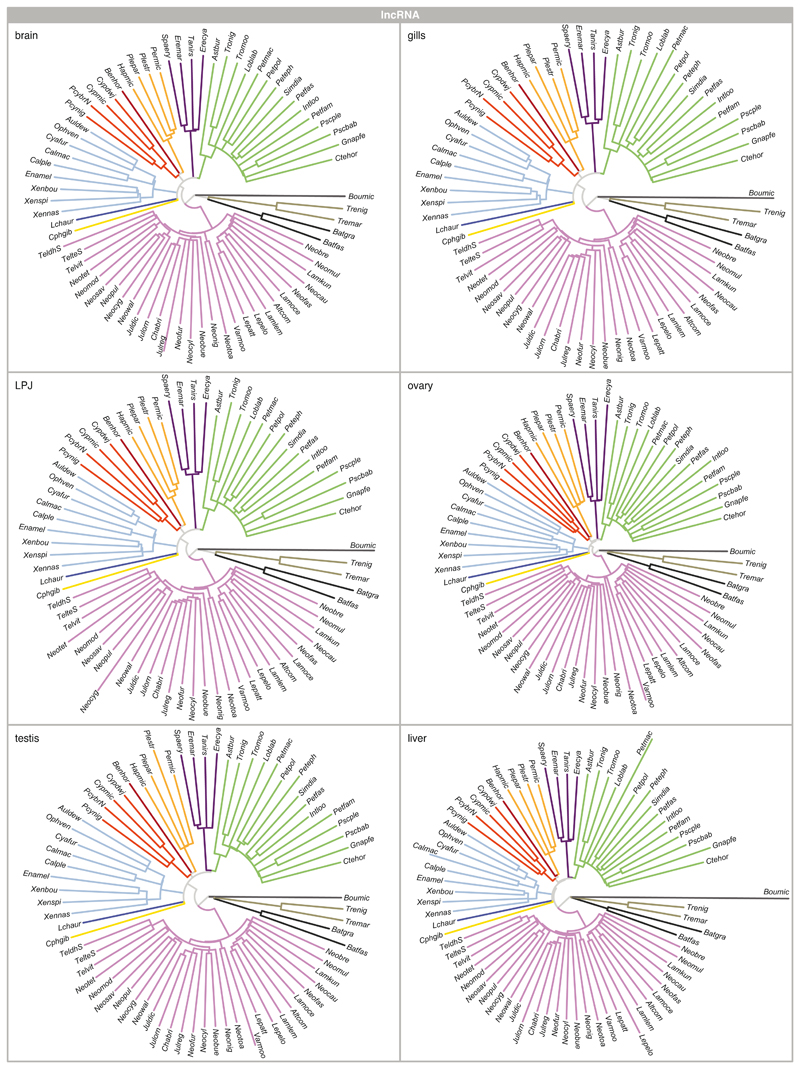
As a complementary approach to the comparison of Spearman’s ρ, whole-transcriptome evolutionary trajectories can also be explored with gene expression trees based on pairwise gene expression distances – a strategy less sensitive to outliers and potential inaccuracies in the normalization process4,35. The expression trees based on protein-coding and lncRNA transcriptome profiles of all six organ samples recovered, in most cases, the major phylogenetic clustering of the species into tribes (protein-coding: Extended Data Fig. 3; lncRNA; and Extended Data Fig. 4). This once more illustrates that most changes at the level of whole transcriptomes have accumulated over evolutionary time, such that closely related species show more similar gene expression profiles. However, the relative positions of the tribes to one another were not congruent among the expression trees and differed in all cases from the phylogenetic relationships derived from genome-wide data18 (Fig. 1a).
The most obvious and consistent difference between the expression trees and the species tree concerns the position of the most species-rich tribe of cichlid fishes in Lake Tanganyika, the Lamprologini, which were placed as sister clade to all remaining tribes in the expression trees (Extended Data Figs. 3, 4) but are clearly nested within the cichlid radiation according to the species tree18 (Fig. 1a). That the Lamprologini show characteristic global transcriptome profiles in all organs that are different from those of all other tribes already became apparent in the organ-specific PCAs (Fig. 2). This pattern is not an artefact of the use of a greater number of Lamprologini species in our analyses (reflecting the de facto greater number of Lamprologini species17), since repeated re-analyses of the data with a balanced number of representatives per tribe yielded similar results (Supplementary Figure 5).
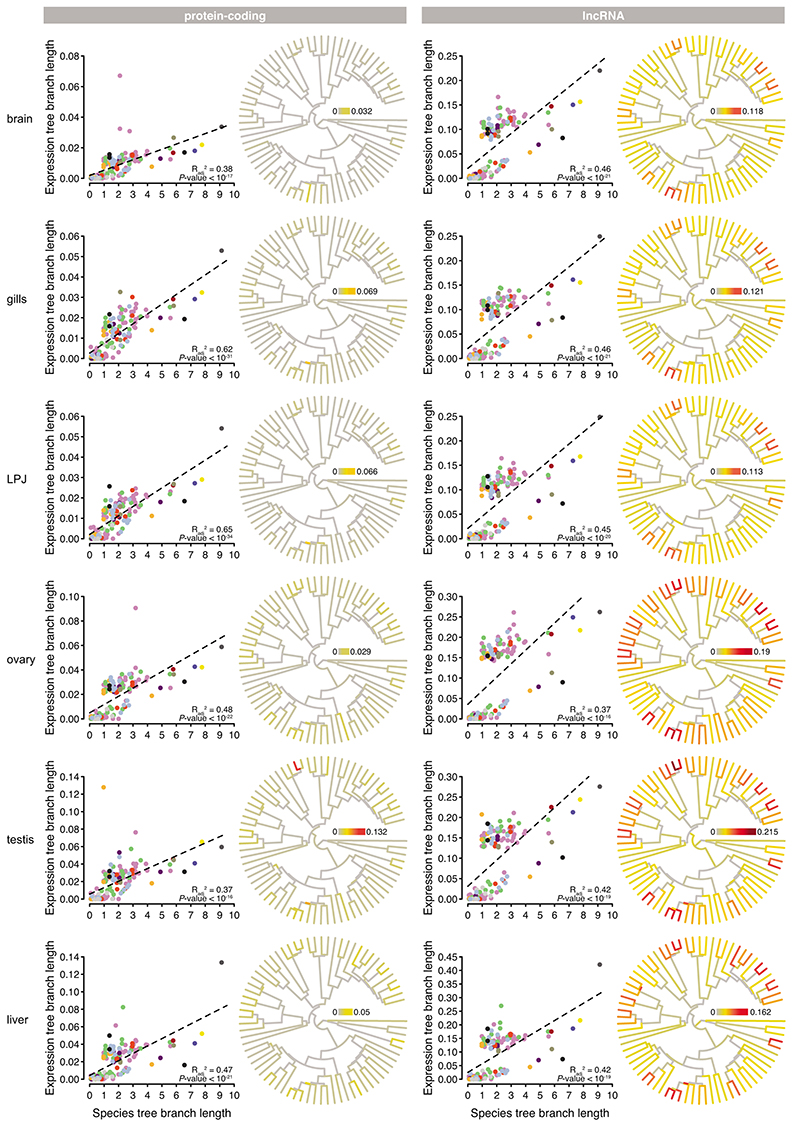
Next, to reconstruct gene expression changes along the phylogeny, we projected expression branch lengths on a time-calibrated species tree18 using the Fitch and Margoliash method36 (protein-coding: Extended Data Fig. 5; lncRNA: Extended Data Fig. 6). The branch lengths of these expression trees correlated positively with the branch lengths of the time-calibrated species tree in all organs and in both transcriptome parts (linear model: P < 10-8; Extended Data Fig. 7). We then estimated the rate of expression change for each branch in the species tree (measured as the expression trees’ branch lengths divided by the species tree’s branch lengths; Extended Data Fig. 7) and quantified rate changes through time (mean rates sampled in steps of 0.15 Myr along the phylogeny as in Ronco et al. 18). This revealed that transcriptome evolution was not constant over the course of the cichlid adaptive radiation in Lake Tanganyika, but became accelerated in the late phase of the radiation, coinciding with a high number of speciation events18 (Fig. 4c, Extended Data Fig. 7). This effect was more pronounced in lncRNAs compared to protein coding genes (Fig. 4c).
Using the cumulative branch lengths (from root to tip) of the expression trees as a proxy for the rate of transcriptome evolution, irrespective of the temporal signals reported above, we corroborated that gene expression levels evolved differently among organs and between the transcriptome parts (Fig. 4d; Supplementary Table 7). Similar to the results based on Spearman’s ρ (Fig. 4a, b), the organ-specific topology-fixed expression trees of protein-coding genes (Extended Data Fig. 5) showed differences in branch lengths, with liver and testis evolving fastest (Fig. 4d; ANOVA: P < 10-8; Supplementary Table 4a). In the lncRNA expression trees (Extended Data Fig. 6), the cumulative branch lengths were similar for brain, gill, and LPJ, but significantly longer for ovary, testis, and liver (Fig. 4d; ANOVA: P < 0. 10-8, Supplementary Table 4b). By calculating Robinson-Foulds distances between the trees obtained from the expression data and the time-calibrated species tree18 (Fig. 4d), we show that the majority of changes in gene expression follow the species tree. However, there is substantial variation among the organs. For example, in agreement with Warnefors and Kaessmann37, brain experienced fewer changes in the overall levels of gene expression than testis.
Finally, by comparing the rates of expression change (represented by the cumulative branch length) among the tribes, we found substantial differences between the radiation’s subclades in all organs (ANOVA: P < 10-8, Extended Data Fig. 8; Supplementary Table 5). For example, Trematocarini showed a high rate of expression changes compared to the other tribes in brain, gill, LPJ, and ovary, Cyprichromini had a high rate of expression changes in gill and LPJ, while Eretmodini featured a high rate of expression changes in testis. The most specie-rich tribe of cichlids in Lake Tanganyika, the Lamprologini, showed intermediate rates of expression change in most organs. Overall, the observed differences in rate of expression changes among the subclades for the different organs might reflect the organ’s lineage-specific involvement in ecological, physiological, and behavioural adaptations.
Organ-specific expression patterns
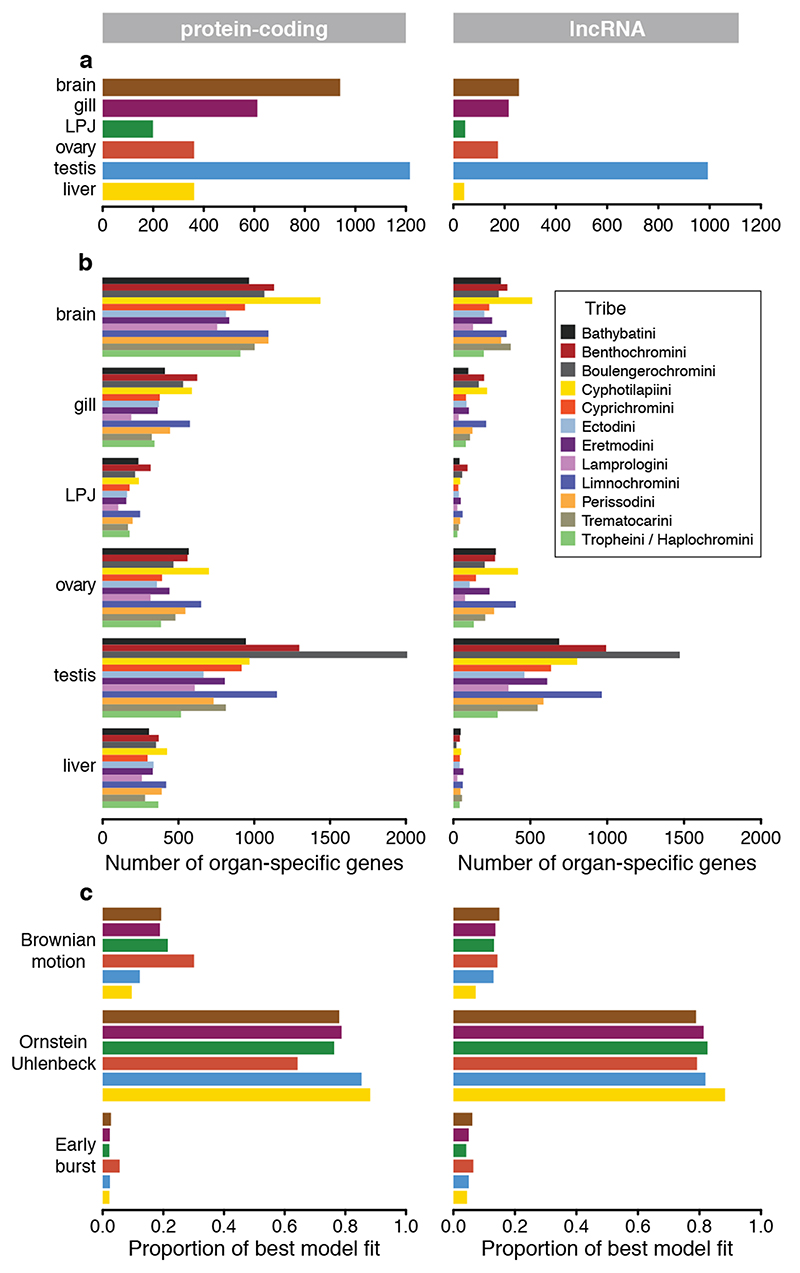
As gene expression evolution might be constrained by core organ functions2–4,38, we determined the degree of organ specificity in gene expression across the different organs using the organ specificity index τ39. The number of organ-specific genes varied substantially, also with respect to the two investigated transcriptome parts (Fig 5a). In testis, which exhibited the fastest evolving transcriptome at the coding level (Fig. 4), we also found most organ-specific genes (Fig. 5a, b), closely followed by brain (for protein-coding genes only, Fig. 5a), which showed the slowest evolving transcriptome (Fig. 4). We also found substantial differences in the number of organ-specific genes among the different tribes (for protein-coding genes and lncRNAs; Fig. 5b).
Fig. 5. Organ-specific expression and expression dynamics of protein-coding genes (left panel) and lncRNAs (right panel).
a, Number of organ-specific genes in each organ. b, Number of organ-specific genes in each organ that are shared across species of the same tribe. Bars are colour-coded according to tribes as defined in Fig. 1a. c, Proportion of per-gene expression patterns that have evolved under a Brownian motion, the Ornstein-Uhlenbeck or the Early burst model of trait evolution. Proportions are shown per organ and colour-coded according to organ type as labelled in Fig 1a.
Gene expression dynamics
To examine, in more detail, the dynamics of gene expression evolution on the per-transcript level, we tested, for each organ and for each expressed gene separately, the model fit to three common models of trait evolution along the time-calibrated species tree. More specifically, we asked whether the gene expression levels (TPM) of a particular protein-coding gene or lncRNA are more likely to have evolved under a Brownian Motion (BM), a single-optimum Ornstein-Uhlenbeck (OU), or an Early Burst (EB) model of trait evolution. We found that, for the majority of protein-coding genes (64%-88%, depending on the organ; Fig. 5c and Supplementary Table 6a) and lncRNAs (79%-88%, depending on the organ; Fig. 5c and Supplementary Table 6b), the gene expression levels evolved according to the OU model of trait evolution, suggesting that (stabilizing) selection has shaped the expression patterns of these genes (this is similar to mammals, see ref.40). The expression levels of 9-30% of the protein-coding genes and 7-15% of the lncRNAs (depending on the organ) are most compatible with a BM model of trait evolution, suggesting that these transcripts have evolved more or less neutrally. The smallest fraction of transcripts (2-5% for protein-coding genes, and 4-6% for lncRNAs) showed expression patterns that fit the EB model of trait evolution, suggesting rapid divergence in gene expression near the onset of the radiation. When comparing between organs and transcriptome parts, we found that the ovary transcriptomes contained a comparatively larger fraction of protein-coding genes with BM-like expression dynamics than other organs along the phylogeny, whereas the transcriptomes of testis and liver featured the largest fractions of protein-coding genes compatible with the OU model. For lncRNA, the liver transcriptomes contained the largest proportion of genes with OU-like expression dynamics (Fig. 5c and Supplementary Table 6).
Discussion
Through the inspection of 2,131 transcriptome profiles from a set of 74 closely related species representative of the adaptive radiation of cichlid fishes in African Lake Tanganyika, we show that the rate of gene expression evolution varies among organs and among the subclades of the radiation, and also between protein-coding genes and lncRNAs. Using several different approaches, we demonstrate that the transcriptomes of brain, gill, and LPJ evolve significantly slower than gonadal and liver transcriptomes. This holds true for protein-coding genes as well as for lncRNAs, suggesting that this pattern represents a transcriptome-wide trend in Tanganyikan cichlids.
Our results on gene expression dynamics over the course of the cichlid adaptive radiation in Lake Tanganyika are only partially consistent with previous work on transcriptome evolution at much deeper phylogenetic levels. Similar to earlier studies3,4,27,41,42, we found that the rate of gene expression evolution (for protein-coding genes and for lncRNAs) was slowest in the brain. The comparatively slow levels of transcriptome evolution in the brain have previously been attributed to the greater degree of specialisation in neuronal organs3,4,43. It thus appears plausible that also in cichlids, organ complexity may explain the differences in transcriptome evolution among brain, gills, and LPJ on one side, and the gonadal organs and liver on the other side.
The consistently fastest rates of gene expression evolution as well as the largest number of organ-specific transcripts have so far been reported for testis (in protein-coding genes and lncRNAs)3,4,27,42, and it has been suggested that this is due to sex-related selective forces3 including sperm competition44, as well as to the particular permissive chromatin conformation during spermatogenesis43, leading to greater transcriptional activity and reduced transcriptional constraints, potentially facilitating transcriptional noise3,43,45. We corroborate here that also during rapid adaptive radiation of cichlid fishes in Lake Tanganyika, testis features the single most rapidly evolving transcriptome at the level of protein-coding genes (Fig. 4) and contains the largest number of organ-specific genes (Fig. 5). At the level of lncRNAs, however, the transcriptomes of both gonadal organs, testis, and ovary, appear to have evolved equally rapid (Fig. 4). This argues against transcriptional noise as explanation for the high rates of gene expression evolution in testis, but rather attests overall high rates of gene expression evolution in gonads in cichlids.
The reconstruction of the mean rate of gene-expression change along the time-calibrated species tree (Fig. 4c) revealed that transcriptome evolution was not constant over the course of the radiation, but was accelerated – in all organs and transcriptome parts – in the radiation’s late phase. This pattern was particularly evident in lncRNAs (Fig. 4c). We have recently shown that the later phase of the cichlid adaptive radiation in Lake Tanganyika is characterized by an increase in the number of speciation events as well as accelerated phenotypic evolution in the ecologically relevant LPJ and in a signalling trait (body pigmentation)18. The increase in the rate of gene-expression change in this later phase of the radiation is in line with the putative role of gene-expression evolution during taxonomic and phenotypic diversification.
A main difference in terms of the rate of transcriptome evolution between our study and previous work on mammals (or tetrapods) concerns the liver. While previous studies reported moderate levels of gene expression evolution in liver4,27,42, we found that the rate of transcriptome evolution in this organ (at the level of protein-coding genes) is nearly as fast as the one observed in testis (Fig. 4). Since some of the most important functions of the liver are connected to the digestive system, it is possible that the accelerated rate of transcriptome evolution in this organ in Tanganyikan cichlids reflects rapid dietary adaptations characteristic for this adaptive radiation18,46,47. On the other hand, the transcriptome of the other feeding-related trait in our study, the LPJ, evolved comparably slowly, despite being similarly transcriptionally active as other organs. That gonads and liver, which show relatively little morphological variation among species (within each organ type), contain the most rapidly evolving transcriptomes in the cichlid adaptive radiation in Lake Tanganyika, whereas the morphologically highly diverse LPJ18 features a slowly evolving transcriptome, indicates that the overall rate of gene expression evolution in an adult organ is not related to its rate of morphological evolution. We note, however, that in order to understand the relationship between transcriptome evolution and varying morphological evolutionary rates, comparative gene-expression analyses across different ontogenetic stages are necessary3,11. This developmental perspective is not covered in our study targeting adult transcriptomes and should be in the focus of future investigations.
The patterns of gene-expression evolution also differed among the sub-clades of the cichlid adaptive radiation in Lake Tanganyika, for example in the number of organ-specific genes (Fig. 5b). The most consistent difference in gene expression patterns occurred between the most species-rich tribe of cichlids in Lake Tanganyika, the Lamprologini, and the remaining tribes (Fig. 2, Extended Data Figs. 3–6). We can only speculate that the deviating overall gene expression trajectories of the Lamprologini are somehow connected to their unique lifestyle compared to other cichlid tribes in Lake Tanganyika. For example, all Lamprologini are substrate spawners, whereas all but one (Boulengerochromis microlepis) of the non-Lamprologini species in Lake Tanganyika are mouth brooders48. Alternatively, the distinct gene expression profiles of the Lamprologini could be due to particular features of genome evolution. For example, we have recently shown that the Lamprologini are characterized by the highest levels of per-genome heterozygosity of all Tanganyikan cichlid tribes18.
Overall, the observed differences in the rate of gene expression evolution between organs (Fig. 4 and 5), transcriptome parts (Fig. 3 and 5), and the subclades of the radiation (Fig. 5, Extended Data Fig. 8) suggest that differing strengths of selection have shaped transcriptome evolution in the course of the cichlid adaptive radiation in Lake Tanganyika. This is further supported by the observation that the expression levels of the majority of protein-coding genes and lncRNAs are in line with an OU model of trait evolution with varying strengths of selection (Fig. 5c).
Methods
Sampling
Sampling was performed between 2014 and 2017 at 31 locations (see Supplementary Table 1 for sampling locations and GPS coordinates) around Lake Tanganyika, under research permits issued by the University of Burundi and the Ministère de l’Eau, de l’Environnement, de l’Aménagement du Territoire et de l’Urbanisme, Republic of Burundi; the Tanzania Commission for Science and Technology (COSTECH), the Tanzania National Parks Authority (TANAPA), and the Tanzania Wildlife Research Institute (TAWIRI), United Republic of Tanzania; the Lake Tanganyika Research Unit, Department of Fisheries, Mpulungu, and the Department of Immigration, Kasama Regional Office, Republic of Zambia. This study included RNA samples from six different sources (brain, gills, liver, ovary, testis and LPJ) of six adult specimens (three males and three females, except for one species for which we had three male and four females, Supplementary Table 2) each of 76 cichlid species. In most cases, the six specimens per species were collected from the same location (Supplementary Table 1). Two species (Tylochromis polylepis and Oreochromis tanganicae) were excluded for all downstream analyses, because these are not part of the endemic adaptive radiation of cichlid fishes in Lake Tanganyika but belong to more ancestral lineages that have colonized the lake secondarily18. One species (Cyprichromis leptosoma) was only used for parts of the analyses, because we lacked brain and LPJ samples for this species (see Extended Data Fig. 9). Thus, for the comparative analyses, we only used species that are part of the radiation18 and for which all organs were collected. Our sampling covers the entire phylogenetic spectrum of the adaptive radiation of cichlids in Lake Tanganyika (tribes Bathybatini, Benthochromini, Boulengerochromini, Cyphotilapiini, Cyprichromini, Ectodini, Eretmodini, Lamprologini, Limnochromini, Perissodini, Trematocarini, and Tropheini). The number of species sampled per tribe scales with the tribe’s total number of species17,18 (Supplementary Table 1 and 2). Organs were derived from adult wild-caught specimens dissected in the field immediately upon capture. Entire organs (with the exception of liver from very large specimens, which were only partially sampled) and the entire LPJ were stored individually in RNAlater.
Extraction, library preparation, and Illumina sequencing
Organs and LPJ were homogenized (FastPrep-24; MP Biomedicals) and total RNA was extracted using the Direct-zol RNA kit (Zymo) according to the manufacturer’s protocol. Individual libraries were constructed using the Illumina TruSeq stranded-protocol including RiboZero Gold rRNA depletion (Illumina, San Diego, CA) and sequenced on an Illumina HiSeq 2500 in SE 125 bp mode at approximately 10 million reads per library. Library construction and sequencing was conducted at the Genomics Facility Basel (GFB), University of Basel and ETH Zurich Department of Biosystems Science and Engineering (D-BSSE). RNA extraction, library preparation and sequencing were randomized with respect to species, organ and sex to avoid batch effects. Library preparation failed for 43 samples (Supplementary Table 7 and 8 for more information on the samples).
Quality filtering, mapping and read counting
Illumina strand-specific single-end sequences were quality-filtered using Trimmomatic49 (v.0.33) with a four bp window size, a required window quality of 15, and a minimum read length of 80 bp (2/3 of the initial read length), followed by adapter removal in the same software. In the absence of well-assembled and annotated reference genomes for the vast majority of the cichlid species under investigation, we opted for a read mapping strategy against the Nile tilapia (Oreochromis niloticus), a closely related and well-annotated cichlid genome. This is also the only cichlid reference genome that has been assembled to chromosomal level. Mapping of all transcriptomes against a common, phylogenetically equidistant, and closely related (see Matschiner et al. 50) reference genome has the additional advantage of facilitating ortholog assignments.
Cleaned reads were mapped against the Nile tilapia genome assembly (RefSeq assembly version GCF_001858045.1_ASM185804v24) with STAR51 (v.2.5.2a), applying the following settings: --outFilterMultimapNmax 1 --outFilterMatchNminOverLread 0.4 --outFilterScoreMinOverLread 0.4. Unique alignments were reported in sorted BAM format and assigned to genes using the HTSeq-count script from the HTSeq52 framework (v.0.6.1p1) (Supplementary Table 7a). Prior to further analyses and following current recommendations (Deseq253 v.1.24.0.), we excluded 5,829 reference genes from the total of 38,425 annotated Nile tilapia genes (Supplementary Figure 7b) on the basis of very low expression levels in our data (five or less counts in less than three samples). The amount of reads per species and library kept at each step of the pipeline is reported in Supplementary Tables 9 and 10. Mapping statistics are reported in Supplementary Figure 6.
All subsequent analyses were performed on two classes of RNA, protein-coding RNAs and long non-coding (lnc) RNAs. The latter are the best-represented class of non-protein coding RNAs annotated in the Nile tilapia genome and have been studied in detail in other organisms42,54,55. Our final gene dataset contained 27,105 protein-coding genes and 4,719 lncRNAs (Supplementary Table 7b), across all organs and species. Outlier samples for each organ were identified via a k-mean clustering approach using the function fviz_cluster from the R package factoextra (v1.0.6) (https://www.rdocumentation.org/collaborators/name/Alboukadel%20Kassambara). Samples that did not cluster with any other sample were removed (n = 52 samples; Supplementary Table 8). The sample exclusion did not change the number of species included in the analysis (Supplementary Table 7).
Global expression patterns and normalisation
Expression counts were normalised with the R (v.3.5.0) package DESeq253 (v.1.24.0.). We used variance stabilizing transformations (VST) to transform the data, resulting in a matrix with values having constant variances along the range of mean values. Multivariate between-group principal components analysis (PCA) was then used to illustrate global patterns of gene expression differences among samples and across organs with the DESeq2 plotPCA function. The expression values were transformed into transcripts per million (TPM) values56 and the biological replicates of each species and each organ were grouped by calculating the median of TPM values. Variances within species and within sexes are represented in Supplementary Figure 1. TPM values were then split into two categories (protein-coding genes, n = 27,105; and lncRNA, n = 4,719), representing two different parts of the transcriptome (coding versus non-coding). Genes were placed in each group based on the Nile tilapia NCBI annotation file (GCF_001858045.1_ASM185804v24). The resulting TPM (summarised and split) values were used for all downstream analyses.
Pairwise expression similarities
Similarity of gene expression between pairs of species was estimated (separately for protein-coding genes and lncRNAs) using Spearman’s ρ, and the pairwise distances between all pairs of species was computed as 1-ρ. Heatmaps of expression similarities among samples were produced using the pheatmap function in the R package pheatmap (v.1.0.12, https://cran.r-project.org/web/packages/pheatmap/index.html). To investigate how well the expression patterns reflect the phylogenetic relatedness of the samples, we used a two-sided Student’s t-test. In order to test for a possible sample size effect when comparing protein-coding genes and lncRNAs, we performed permutation tests (n = 10,000) in which we randomly sampled the same number of protein-coding genes out of the total set of protein coding genes (n = 27,105) as there are annotated lncRNAs (n = 4,719), and calculated correlation coefficients on these random subsets.
Expression divergence through time within organs
As in Brawand et al. 4 and in Necsulea & Kaessmann10, we measured the relationship between gene expression and divergence time (separately for protein-coding genes and lncRNA) with a linear regression between Spearman’s ρ (as x variable) and divergence times18 (as y variable) for all pairs of species. The rate of evolution within each organ was then measured as [(1- ρ) / divergence time] for all pairs of species. A one-way ANOVA was used to determine if there were any differences between the organs. Whenever significant effects were detected, post-hoc evaluations were performed using Tukey’s Honestly Significant Difference (HSD) test.
Gene expression trees
Following Brawand et al. 4, gene expression phylogenies were constructed using the neighbour-joining approach on the pairwise distance between species (computed as 1 - ρ, separately for both transcriptome parts and for each organ) with the NJ function in the R package ape57 (v.5.3). Topological dissimilarities (measured as Robinson-Foulds distance58) between the expression trees and the time-calibrated species tree based on genome-wide data (taken from Ronco et al. 18) were calculated using the treedist function in the R package phangorn59 (v. 2.5.3). To test whether the rate of gene expression change along the species tree was similar among organs, we estimated, for every branch in the species tree, expression distances (computed as 1 - ρ, separately for both transcriptome parts and each organ), using the Fitch and Margoliash36 method as implemented in PHYLIP (v.3.697, http://evolution.genetics.washington.edu/phylip.html). Cumulative branch lengths from root to tips were calculated per species within each organ and then compared across organs. The rate of transcriptome change was then reported as the branch length estimated with expression data divided by the corresponding branch length of the species tree. To illustrate the temporal dynamics of transcriptome evolution, we plotted – per organ – the mean rate of expression change sampled in 0.15 million year steps (as in ref.18) along the time-calibrated species tree. Cumulative branch lengths from root to tips (based on a fixed topology) were calculated per species and organ and then compared across organs and tribes using an ANOVA.
Organ-specific expression
Organ-specificity indexes (τ) were calculated for the two parts of the transcriptome following a modified version60 of the initial Tau formula39. The Tau of a gene is defined as following:
where n H is the number of organs examined (in our case n H = 6) and SH(i, max) is the highest expression signal of gene i across the nH organs. As proposed in Guschanski et al. 38, organ-specific indexes were calculated using the normalized but not the transformed gene expression matrix. Tau values were then calculated using the median gene expression values per organ and tribe. Tau indices over 0.8 were considered as indicative of organ-specific expression38. The number of organ-specific genes was reported per organ and tribe.
Gene expression dynamics
To examine the dynamics of gene expression changes along the phylogeny for each gene individually, we fitted models of trait evolution to the TPM gene expression values (summarised as median per species). To do so, we used the fitContinuous function within the R (v.3.5.0) package Geiger61 (v.2.0.6.1). Specifically, we fitted a Brownian Motion (BM), an Ornstein-Uhlenbeck (OU), and an Early Burst (EB) model of trait evolution along the time-calibrated species tree (see above). Note that the EB model was tested because of the prediction that trait evolution should be rapid early in an adaptive radiation and to slow down through time as the available niche space becomes filled. We applied 10,000 iterations and default parameter bounds except for the alpha parameter in the OU model (attraction strength to central value), which was set to a lower limit of exp(-500) and an upper limit of 20. The approach was applied for each organ and for each transcriptome part separately (protein-coding genes, lncRNA). Genes that were not expressed (TPM = 0) within an organ were removed prior to the analyses. The fraction of expressed genes per organ used for this analysis was between 96-99% of all protein-coding genes (n = 27,105) and between 81-99% of all lncRNA (n = 4,719). We then compared the models by calculating the difference in Akaike information criterion (AIC) and reported for each gene the best model fit (the model with the lowest AIC). The number of genes per model is reported in Supplementary Table 6.
Extended Data
Extended Data Fig. 1. Gene expression patterns per organ and sex.
Principal component analyses of overall gene expression levels in brain, gill, lower pharyngeal jaw bone (LPJ), ovary, testis, and liver. Samples (brain: n = 428; gills: n = 434; LPJ: n = 425; ovary: n = 219; testis: n = 213; liver: n = 412) are coloured according to sex (red: female, blue: male). The proportion of variance explained by the first two principal components (PC1 and PC2) for each organ are indicated in parenthesis at X- and Y-axes, respectively.
Extended Data Fig. 2. Expression variation through time within organs and transcriptome parts.
a, Pairwise Spearman’s rank correlation coefficient (ρ) of per species (brain, ovary, gills and testis: n = 74 taxa; LPJ and liver: n = 73 taxa) as a function of divergence time18 for protein-coding genes (left panel) and lncRNAs (right panel) in brain, gill, LPJ, ovary, testis, and liver. Samples are colour-coded according to tribe as defined in Fig. 1a; pairs of species belonging to two different tribes are indicated in grey. The regression line is represented with a dashed black line. b, Comparison of rate of expression change (measured as [1 – ρ] / divergence time18) between protein-coding genes and lncRNAs (two-sided t-test: P < 10-16). Boxplot centre lines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5x interquartile range. Outliers are not shown.
Extended Data Fig. 3. Protein-coding expression trajectories.
Neighbour-joining trees based on pairwise distance matrices of gene expression between pairs of species (n = 73 taxa) for protein-coding genes for brain, gill, LPJ, ovary, testis, and liver. All trees are coloured according to tribe as defined in Fig. 1a (see Extended Data Fig. 9 and Supplementary Table 2 for full species names).
Extended Data Fig. 4. lncRNAs expression trajectories.
Neighbour-joining trees based on pairwise distance matrices of gene expression between pairs of species (n = 73 taxa) for lncRNAs for brain, gill, LPJ, ovary, testis, and liver. All trees are colour-coded according to tribe as defined in Fig. 1a (see Extended Data Fig. 9 and Supplementary Table 2 for full species names).
Extended Data Fig. 5. Rate of protein-coding gene expression evolution along the phylogeny.
Species tree with branch lengths estimated along the fixed species tree topology36 (n = 73 taxa) based on pairwise correlations of gene expression of protein-coding genes in brain, gill, LPJ, ovary, testis, and liver. All trees are colour-coded according to tribe as defined in Fig. 1a (see Extended Data Fig. 9 and Supplementary Table 2 for full species names).
Extended Data Fig. 6. Rate of lncRNA gene expression evolution along the phylogeny.
Species tree with branch lengths estimated along the fixed species tree topology36 (n = 73 taxa) based on pairwise correlations of gene expression of lncRNAs in brain, gill, LPJ, ovary, testis, and liver. All trees are colour-coded according to tribe as defined in Fig. 1a (see Extended Data 9 and/or Supplementary Table 2 for full species names).
Extended Data Fig. 7. Rate of transcriptome evolution within organs for protein-coding genes (left panel) and lncRNAs (right panel).
Linear regression of the expression tree branch length (calculated along the fixed species tree (n = 73 taxa) topology, Extended Data Fig. 3c, d) as a function of species tree branch lengths (Fig. 1a) for brain, gill, LPJ, ovary, testis, and liver. Data points representing branches within tribes are colour-coded according to the tribe as defined in Fig. 1a, and data points representing branches that link species from different tribes are coloured in grey. Dashed lines represent linear model fits. Next to each plot, a time-calibrated species tree is shown, with branches coloured according to the rate of transcriptome evolution (measured as expression tree branch length divided by species tree branch length).
Extended Data Fig. 8. Level of expression variation within organs.
a, Cumulative branch lengths (from root to tip of expression tree branch length calculated along the fixed species tree (n = 73 taxa) topology; Extended Data Fig. 3c, d) for protein-coding genes (left panel) and lncRNAs (right panel) in brain, gill, LPJ, ovary, testis, and liver calculated per species and summarised per tribe (n = 12 tribes). Boxplots are colour-coded according to tribe as defined in Fig. 1a; boxplot centre lines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5x interquartile range. Differences among the tribes were assessed using an ANOVA (see Supplementary Table 5 for the P-values for all pairwise comparisons). b, Comparison of cumulative branch lengths between protein-coding genes and lncRNAs (two-sided t-test: P < 10-8). Boxplot centre lines represent the median, box limits the upper and lower quartiles, and whiskers the 1.5x interquartile range.
Extended Data Fig. 9. Species information.
List of species used in this experiment with abbreviation code, full species name and tribe information.
Supplementary Material
Acknowledgements
We would like to thank the University of Burundi, the Ministère de l’Eau, de l’Environnement, de l’Aménagement du Territoire et de l’Urbanisme, Republic of Burundi, the Tanzania Commission for Science and Technology (COSTECH), the Tanzania Fisheries Research Institute (TAFIRI), the Tanzania National Parks Authority (TANAPA), the Tanzania Wildlife Research Institute (TAWIRI), and the Lake Tanganyika Research Unit, Department of Fisheries, Republic of Zambia, for research permits; G. Banyankimbona (University of Burundi), I. Kimirei (TAFIRI, Kigoma, Tanzania), and T. Banda and L. Makasa (Department of Fisheries, Mpulungu, Zambia) for assistance with research permits; the boat crews of the ‘Chomba’ (D. Mwanakulya, J. Sichilima, and H. D. Sichilima Jr.) and the ‘Maji Makubwa II’ (G. Kazumbe and family) for help during field work; V. Huwiler (Kalambo Lodge, Zambia), ‘Charity’ (Nkupi Lodge, Zambia), and the Zytkow family (Ndole Bay Lodge, Zambia) for lodging; C. Zytkow (Conservation Lake Tanganyika, Zambia), and P. Lassen and V. Huwiler (Kalambo Lodge, Zambia) for logistic support; C. Beisel and E. Burcklen at the Genomics Facility Basel for library preparation and sequencing; and J. Himes for fish illustrations. We also would like to thank A. Necsulea for discussions and advice on the project. Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing centre at University of Basel (with support by the SIB/Swiss Institute of Bioinformatics). This work was funded by the European Research Council (ERC, Consolidator Grant Nr. 617585 ‘CICHLID~X’) and the Swiss National Science Foundation (SNF) to W.S.
Footnotes
Author Contributions
A.E., F.R., A.I., N.B., L.W., and W.S. collected and/or dissected the specimens in the field. A.E. and N.B. organised the RNA-seq data production. A.E. processed and mapped the reads. A.E. performed all data analyses except for the temporal dynamics of transcriptome evolution that F.R. performed. A.B. contributed ideas and supervised data analyses. F.R. formatted the final figures. A.E. and W.S. wrote the manuscript with inputs from all authors. The project was originally designed by W.S., with inputs from A.E., F.R., A.B., and A.I.
Competing interest declaration
The authors declare no competing interests.
Additional Information
Reporting summary
Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability
The datasets generated during and/or analysed during the current study are available in the NCBI repository under the BioProject accession number PRJNA550295. All related metadata are available on Dryad under the project accession number https://doi.org/10.5061/dryad.fj6q573sj.
Code availability
All custom codes generated during and/or analysed in the current study are available on Dryad under project accession number https://doi.org/10.5061/dryad.fj6q573sj.
References
- 1.King M, Wilson CA. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 2.Romero IG, Ruvinsky I, Gilad Y. Comparative studies of gene expression and the evolution of gene regulation. Nat Rev Genet. 2012;13:505–516. doi: 10.1038/nrg3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet. 2014;15:734–748. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 4.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 5.Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- 6.Barrier M, Robichaux RH, Purugganan MD. Accelerated regulatory gene evolution in an adaptive radiation. Proc Natl Acad Sci. 2001;98:10208–10213. doi: 10.1073/pnas.181257698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Whittall JB, Voelckel C, Kliebenstein DJ, Hodges SA. Convergence, constraint and the role of gene expression during adaptive radiation: Floral anthocyanins in Aquilegia. Mol Ecol. 2006;15:4645–4657. doi: 10.1111/j.1365-294X.2006.03114.x. [DOI] [PubMed] [Google Scholar]
- 8.Stern DL, Orgogozo V. The loci of evolution: How predictable is genetic evolution? Evolution. 2008;62:2155–2177. doi: 10.1111/j.1558-5646.2008.00450.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Naqvi S, et al. Conservation, acquisition, and functional impact of sex-biased gene expression in mammals. Science. 2019;365:55–61. doi: 10.1126/science.aaw7317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Necsulea A, Kaessmann H. Evolutionary dynamics of coding and non-coding transcriptomes. Nat Rev Genet. 2014;15:734–748. doi: 10.1038/nrg3802. [DOI] [PubMed] [Google Scholar]
- 11.Cardoso-Moreira M, et al. Gene expression across mammalian organ development. Nature. 2019;571:505–509. doi: 10.1038/s41586-019-1338-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schluter D. The ecology of adaptive radiation. Oxford University Press; 2000. [Google Scholar]
- 13.Berner D, Salzburger W. The genomics of organismal diversification illuminated by adaptive radiations. Trends Genet. 2015;31:491–499. doi: 10.1016/j.tig.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 14.Brawand D, et al. The genomic substrate for adaptive radiation in African cichlid fish. Nature. 2014;513:375–381. doi: 10.1038/nature13726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jones FC, et al. The genomic basis of adaptive evolution in threespine sticklebacks. Nature. 2012;484:55–61. doi: 10.1038/nature10944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salzburger W. Understanding explosive diversification through cichlid fish genomics. Nat Rev Genet. 2018;19:705–717. doi: 10.1038/s41576-018-0043-9. [DOI] [PubMed] [Google Scholar]
- 17.Ronco F, Büscher HH, Indermaur A, Salzburger W. The taxonomic diversity of the cichlid fish fauna of ancient Lake Tanganyika, East Africa. J Gt Lakes Res. 2020;46:1067–1078. doi: 10.1016/j.jglr.2019.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ronco F, et al. Drivers and dynamics of a massive adaptive radiation in African cichlid fishes. Nature. doi: 10.1038/s41586-020-2930-4. in press. [DOI] [PubMed] [Google Scholar]
- 19.Fryer G, Iles TD. The cichlid fishes of the great lakes of Africa: their biology and evolution. Oliver and Boyd; 1972. [Google Scholar]
- 20.Liem KF. Evolutionary strategies and morphological innovations: Cichlid pharyngeal jaws. Syst Zool. 1973;22:425–441. [Google Scholar]
- 21.Hulsey CD. Function of a key morphological innovation: fusion of the cichlid pharyngeal jaw. Proc Biol Sci. 2006;273:669–675. doi: 10.1098/rspb.2005.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meyer A. Ecological and evolutionary consequences of the trophic polymorphism in Cichlasoma citrinellum (Pisces: Cichlidae) Biol J Linn Soc. 1990;39:279–299. [Google Scholar]
- 23.Salzburger W, Mack T, Verheyen E, Meyer A. Out of Tanganyika: Genesis, explosive speciation, key-innovations and phylogeography of the haplochromine cichlid fishes. BMC Evol Biol. 2005;5:1–15. doi: 10.1186/1471-2148-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Muschick M, Barluenga M, Salzburger W, Meyer A. Adaptive phenotypic plasticity in the Midas cichlid fish pharyngeal jaw and its relevance in adaptive radiation. BMC Evol Biol. 2011;11:116. doi: 10.1186/1471-2148-11-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salzburger W. The interaction of sexually and naturally selected traits in the adaptive radiations of cichlid fishes. Mol Ecol. 2009;18:169–185. doi: 10.1111/j.1365-294X.2008.03981.x. [DOI] [PubMed] [Google Scholar]
- 26.Ronco F, Roesti M, Salzburger W. A functional trade-off between trophic adaptation and parental care predicts sexual dimorphism in cichlid fish. Proc R Soc B Biol Sci. 2019;286:20191050. doi: 10.1098/rspb.2019.1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Khaitovich P, Enard W, Lachmann M, Pääbo S. Evolution of primate gene expression. Nat Rev Genet. 2006;7:693–702. doi: 10.1038/nrg1940. [DOI] [PubMed] [Google Scholar]
- 28.Fernández-Pérez L, Mirecki-Garrido M, Recio C, Guerra B. In: Chemistry and biological activity of steroids. Ribeiro Salvador JA, Cruz Silva MM, editors. IntechOpen; 2019. pp. 1–25. [DOI] [Google Scholar]
- 29.Van Thiel DH, Gavaler JS. In: Thomson ABR, Shaffer E, editors. Springer; US: 1992. pp. 337–353. [DOI] [Google Scholar]
- 30.Shen M, Shi H. Sex hormones and their receptors regulate liver energy homeostasis. Int J Endocrinol. 2015;2015:1–12. doi: 10.1155/2015/294278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qiao Q, et al. Deep sexual dimorphism in adult medaka fish liver highlighted by multi-omic approach. Sci Rep. 2016;6:1–12. doi: 10.1038/srep32459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rose E, Flanagan SP, Jones AD. The effects of synthetic estrogen exposure on the sexually dimorphic liver transcriptome of the sex-role-reversed gulf pipefish. PLoS One. 2015;10:1–20. doi: 10.1371/journal.pone.0139401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zheng W, et al. Transcriptomic analyses of sexual dimorphism of the zebrafish liver and the effect of sex hormones. PLoS One. 2013;8:1–12. doi: 10.1371/journal.pone.0053562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.El Taher A, Ronco F, Matschiner M, Salzburger W. Dynamics of sex chromosome evolution in a rapid radiation of cichlid fishes. Co-submitted. doi: 10.1126/sciadv.abe8215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meisel RP, Malone JH, Clark AG. Faster-X evolution of gene expression in Drosophila. PLoS Genet. 2012;8:1–19. doi: 10.1371/journal.pgen.1003013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fitch W, Margoliash E. Construction of phylogenetic trees. Science. 1967;155:279–284. doi: 10.1126/science.155.3760.279. [DOI] [PubMed] [Google Scholar]
- 37.Warnefors M, Kaessmann H. Evolution of the correlation between expression divergence and protein divergence in mammals. Genome Biol Evol. 2013;5:1324–1335. doi: 10.1093/gbe/evt093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Guschanski K, Warnefors M, Kaessmann H. The evolution of duplicate gene expression in mammalian organs. Genome Res. 2017;27:1461–1474. doi: 10.1101/gr.215566.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yanai I, et al. Genome-wide midrange transcription profiles reveal expression level relationships in human tissue specification. Bioinformatics. 2005;21:650–659. doi: 10.1093/bioinformatics/bti042. [DOI] [PubMed] [Google Scholar]
- 40.Chen J, et al. A quantitative framework for characterizing the evolutionary history of mammalian gene expression. Genome Res. 2019;29:53–63. doi: 10.1101/gr.237636.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chan ET, et al. Conservation of core gene expression in vertebrate tissues. J Biol. 2009;8:33. doi: 10.1186/jbiol130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Necsulea A, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505:635–640. doi: 10.1038/nature12943. [DOI] [PubMed] [Google Scholar]
- 43.Soumillon M, et al. Cellular source and mechanisms of high transcriptome complexity in the mammalian testis. Cell Rep. 2013;3:2179–2190. doi: 10.1016/j.celrep.2013.05.031. [DOI] [PubMed] [Google Scholar]
- 44.Nielsen R, et al. A scan for positively selected genes in the genomes of humans and chimpanzees. PLoS Biol. 2005;3:0976–0985. doi: 10.1371/journal.pbio.0030170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eling N, Morgan MD, Marioni JC. Challenges in measuring and understanding biological noise. Nat Rev Genet. 2019;20:536–548. doi: 10.1038/s41576-019-0130-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muschick M, Indermaur A, Salzburger W. Convergent evolution within an adaptive radiation of cichlid fishes. Curr Biol. 2012;22:2362–2368. doi: 10.1016/j.cub.2012.10.048. [DOI] [PubMed] [Google Scholar]
- 47.Wagner CE, McIntyre PB, Buels KS, Gilbert DM, Michel E. Diet predicts intestine length in Lake Tanganyika’s cichlid fishes. Funct Ecol. 2009;23:1122–1131. [Google Scholar]
- 48.Konings A. Tanganyika Cichlids in their natural habitat. Cichlid Press; 2015. [Google Scholar]
- 49.Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matschiner M, Böhne A, Ronco F, Salzburger W. The genomic timeline of cichlid fish diversification across continents. Nat Commun. 2020 doi: 10.1038/s41467-020-17827-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dobin A, et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sarropoulos I, Marin R, Cardoso-moreira M, Kaessmann H. Developmental dynamics of lncRNAs across mammalian organs and species. Nature. 2019;571:510–514. doi: 10.1038/s41586-019-1341-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kutter C, et al. Rapid turnover of long noncoding RNAs and the evolution of gene expression. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li B, Ruotti V, Stewart RM, Thomson JA, Dewey CN. RNA-Seq gene expression estimation with read mapping uncertainty. Bioinformatics. 2009;26:493–500. doi: 10.1093/bioinformatics/btp692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paradis E, Claude J, Strimmer K. APE: Analyses of phylogenetics and evolution in R language. Bioinformatics. 2004;20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 58.Robinson DF, Foulds LR. Comparison of phylogenetic trees. Math Biosci. 1981;53:131–147. [Google Scholar]
- 59.Schliep KP. phangorn: Phylogenetic analysis in R. Bioinformatics. 2011;27:592–593. doi: 10.1093/bioinformatics/btq706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Liao B, Zhang J. Low rates of expression profile divergence in highly expressed genes and tissue-specific genes during mammalian evolution. Mol Biol Evol. 2006;23:1119–1128. doi: 10.1093/molbev/msj119. [DOI] [PubMed] [Google Scholar]
- 61.Pennell MW, et al. Geiger v2.0: An expanded suite of methods for fitting macroevolutionary models to phylogenetic trees. Bioinformatics. 2014;30:2216–2218. doi: 10.1093/bioinformatics/btu181. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during and/or analysed during the current study are available in the NCBI repository under the BioProject accession number PRJNA550295. All related metadata are available on Dryad under the project accession number https://doi.org/10.5061/dryad.fj6q573sj.
All custom codes generated during and/or analysed in the current study are available on Dryad under project accession number https://doi.org/10.5061/dryad.fj6q573sj.