Abstract
The calcium release activated calcium (CRAC) channel is activated by the endoplasmic reticulum-resident calcium sensor protein STIM1. Upon activation, STIM1 C-terminus changes from an inactive, tight to an active, extended conformation. A coiled-coil (CC) clamp involving the CC1 and CC3 domains is essential in controlling STIM1 activation, with CC1 as key entity. The NMR-derived solution structure of the CC1 domain represents a three-helix bundle stabilized by interhelical contacts, which are absent in the Stormorken disease-related STIM1 R304W mutant. Two interhelical sites between CC1α1 and CC1α2 helices are key in controlling STIM1 activation, affecting the balance between tight and extended conformations. NMR-directed mutations within these interhelical interactions restore the physiological, store-dependent activation behavior of the gain-of-function STIM1 R304W mutant. This study reveals the functional impact of interhelical interactions within the CC1 domain for modifying the CC1-CC3 clamp strength to control the activation of STIM1.
1. Introduction
The calcium ion is one of the most important second messengers within a eukaryotic cell. Ca2+ controls a broad number of processes such as gene expression, cell division, apoptosis, muscle contraction and secretion. Incorrect regulation of cytosolic Ca2+ concentrations can result in the development of diseases.1, 2, 3, 4
Store Operated Calcium Entry (SOCE) is a prominent way to allow Ca2+ influx into a cell. The two molecular key proteins mediating SOCE are (i) the Orai protein, which represents the Ca2+ selective channel in the plasma membrane (PM), and (ii) the endoplasmic reticulum (ER) resident STIM protein, which senses the ER luminal Ca2+ concentration. Lower ER luminal Ca2+ concentrations, i.e. store depletion, lead to STIM1 activation. Activated STIM1 clusters at ER-PM junctions for coupling to Orai culminating in Ca2+ influx.5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15 In recent years, a sophisticated model of this store-depletion triggered STIM1 activation has been developed, emphasizing the role of both the N-terminal ER luminal as well as the C-terminal cytosolic part of STIM1. The C-terminal segment of STIM1 contains three coiled-coil (CC) domains (CC1, CC2 and CC3), with CC1 playing a central regulatory role in controlling both the inactive, resting state and the activated state of the protein.14, 15 The CC2 and CC3 domains represent the STIM Orai activating region (SOAR) /CRAC activating domain (CAD) which is the minimal STIM1 fragment to activate Orai.16, 17 Further studies identified an Orai activating small fragment (OASF) including CC1, CC2 and CC3 crucial for STIM1 homomerization and concurrent elongation. These conformational events are decisive steps for Orai activation.18 OASF can adopt different structures, in particular a tight form in the resting state and an extended form in the activated state. The tight state involves an intramolecular CC1-CC3 interaction (“clamp”), which is released in the extended state resulting in exposure of the SOAR / CAD domain for Orai activation.14, 15, 16, 17 The recently discovered gain of function (GOF) STIM1 R304W mutant causes the Stormorken disease. The R304W mutation located within the CC1 domain of STIM1, leads to store-independent extension of the STIM1 cytosolic portion resulting in constitutive STIM1 R304W puncta formation and Orai channel activation.19, 20, 21, 22
In this study we report the first NMR solution structure of the CC1 domain of STIM1, a three-helix bundle with the α1, α2 and α3 helices connected by flexible loops. We identified intramolecular, interhelical contacts between CC1 α1 and α2 that are absent in the Stormorken related CC1 R304W domain. The functional relevance of these interhelical contact sites within CC1 is assessed by cell electrophysiology and FRET analyses of NMR guided specific mutations of full-length STIM1. By mutation of these interhelical sites in the Stormorken STIM1 R304W, the pathophysiological constitutive, store-independent activation of CRAC channels is reversed into store-dependent activation behaviour, revealing the essential regulatory impact of the CC1 α1 – α2 interaction sites on the STIM1 activation state.
2. Results
2.1. CC contacts stabilize STIM1 CC1 three-helix bundle
CC1 fragments were over-expressed in both 15N singly and 15N,13C doubly isotopically labeled minimal media and purified for NMR-investigations according to a recently developed protocol.23 More than 95% of the primary sequence of STIM1 CC1 is predicted to assume a coiled-coil fold.24 By their nature coiled-coil motifs consist of repeats of only a few amino acid types in similar environments.25, 26, 27 Therefore, high resolution NMR structure analyses of coiled-coil proteins is very challenging. We overcame the difficulties imposed by extensively overlapping signals by application of carbon detected 3D NMR (see Methods) achieving a sequence specific assignment of more than 83% of the backbone amide and 44% of the side chain resonances. The chemical shifts assignments of the STIM1 CC1 domain were deposited in the BMRB28 under ID 50114.
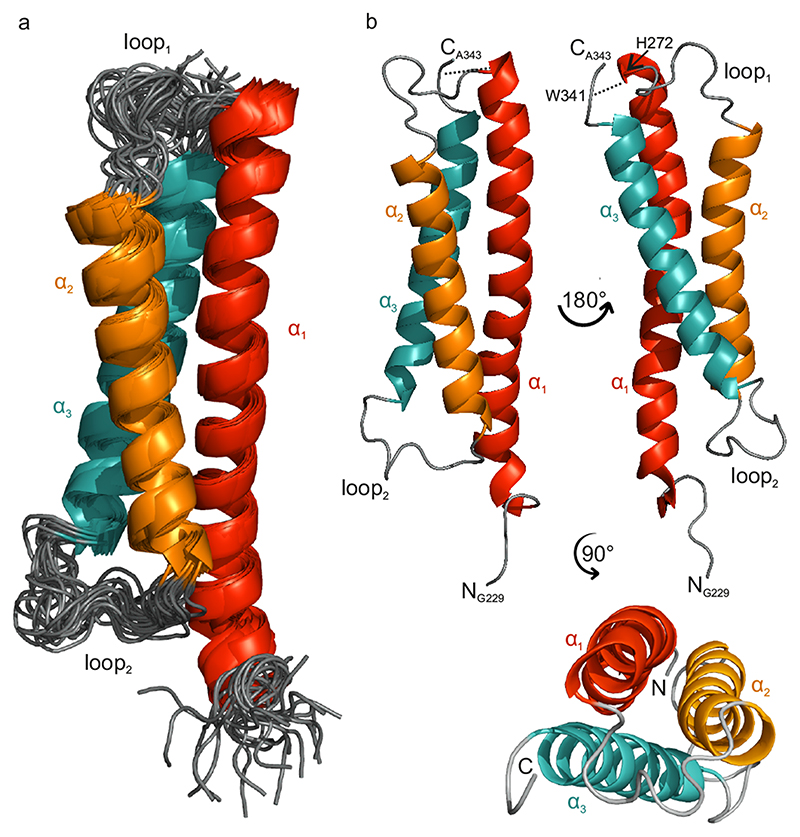
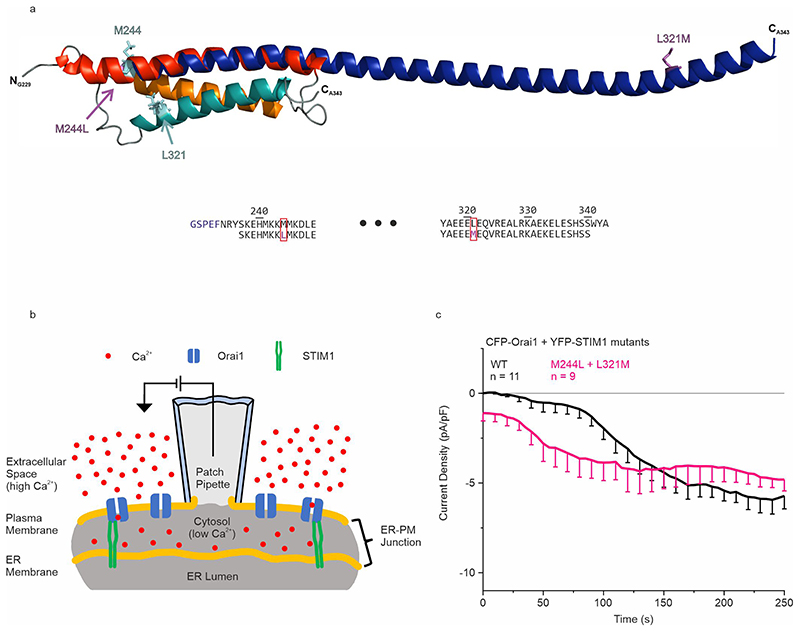
Based on these assignments, high resolution monomeric structures of STIM1 CC1 in solution were computed as a bundle of the 20 lowest energy conformations by CYANA29, 30 from NOE restraints and hydrogen bond restraints from CS-Rosetta31 as shown in Fig. 1a, with three views of the lowest restraint violation structure from this bundle in Fig. 1b. These structures were water-refined and deposited in the protein database (PDB ID: 6YEL). They are in striking contrast to the only available crystallographic structure of this domain32, which consists of a single extended helix shown in Extended Data Fig. 1a with the superimposed NMR result. This difference may be related to two point mutations (M244L and L321M) in the crystallographic structure located in positions, where interhelical interactions seen in the NMR structure would be affected. Addressing the functional behavior of the STIM1 M244L + L321M double mutant when co-expressed with Orai1 in life cell patch-clamp experiments (detailed in Extended Data Fig. 1b), revealed indeed partially pre-activated Orai1 currents (Extended Data Fig. 1c), in line with the stretched CC1 conformation observed in the crystallographic study.14, 15, 32
Fig. 1. STIM1 CC1 forms a compact three-helix bundle.
a Ribbon representation of 20 spatially aligned conformers of STIM1 CC1 fragment (backbone RMSD 1.5 Å for all residues, 0.51 Å for the α-helices, see Methods). b The lowest restraint violation energy structure (α1: H240-E270, α2: L282-L303 and α3: A317-S340). The dashed line indicated the NOE contact between H272 and W341. The molecular graphics were created by PyMOL (v. 2.3, Schrödinger, LLC).
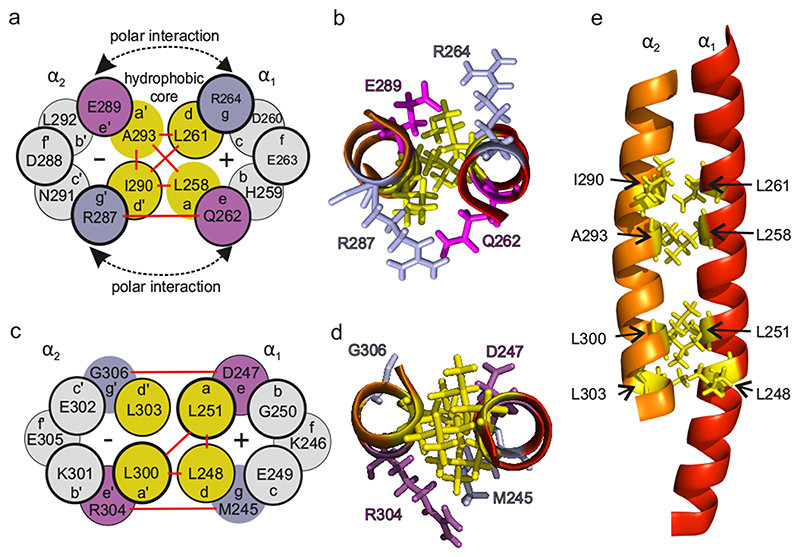
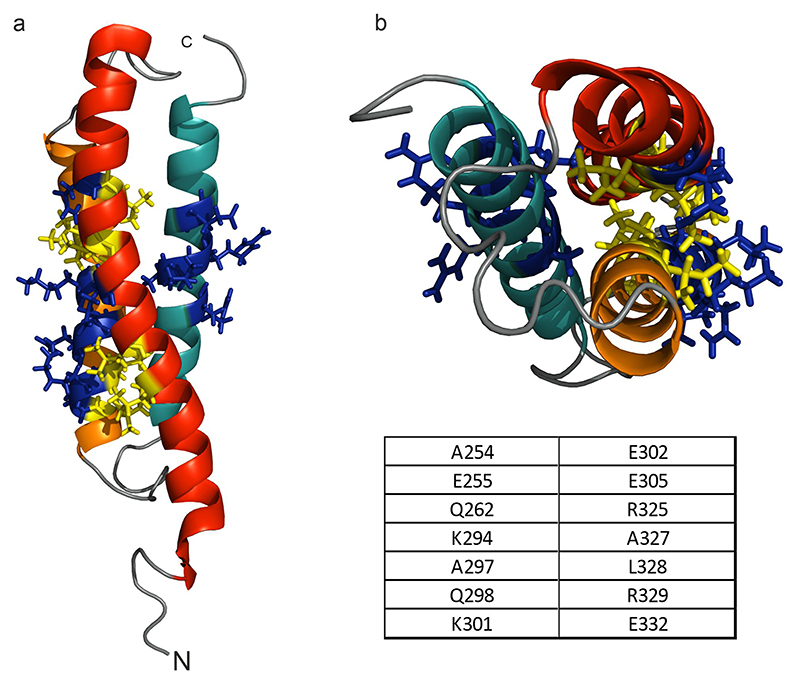
NOE contacts between hydrophobic residues in CC1 α1 and α2, notably interconnecting L261 (α1) to I290 and A293 (α2) as well as L258 (α1) to the same residues in (α2), all within typical coiled-coil moieties, determine the antiparallel alignment of the α1 and α2 helices forming a characteristic coiled-coil interaction pattern (Fig. 2a,b,e). This is also corroborated by a NOE correlation connecting HN-G306 (loop2) and Hα-D247 (α1) located near the C- and N-terminal parts of α2 and α1 (see Fig. 2), respectively. The inter-helix contact is stabilized by another coiled-coil interaction motif connecting the hydrophobic residue pairs L248 and L251 of α1 to L300 and L303 of α2, respectively (Fig. 2c, d, e). The position of CC1 α3 helix is less well defined, but a distinct NOE contact of W341 NE1/HE1 to H272 HA corroborates a 3-helix bundle (see Fig. 1b).
Fig. 2. Coiled-coil epitopes between α1 and α2 helices of CC1.
a, c Scheme of both coiled-coil regions identified in NMR; red lines denote the observed NOE contacts. b, d Top-views of these two coiled-coil motifs in the NMR solution model. e Side-view of the entire α1-α2 segment highlighting the coiled-coil interaction sites.
To avoid non-specific CC1 homo-oligomerization23 NMR experiments were recorded in presence of 7.0 mM SDS (sodium dodecyl sulfate), just below the CMC (critical micelle concentration) of 8.2 mM (25 °C),33 where SDS does not cause secondary structure changes, as proven by CD spectra (Supplementary Fig. 1). Intermolecular NOEs between surface residues of STIM1 CC1 and SDS indicate protection of specific regions (Extended Data Fig. 2). NMR relaxation experiments revealed an overall rotational correlation time of 6.11±0.67 ns, in accordance with a near spherical ~100 aa protein.34 Together with previous light scattering studies23, this proves that the dominant CC1 species under these conditions is monomeric. Therefore, one can rule out that the observed NOE contacts originate from a CC1 dimer or oligomer.
In summary, experimental NMR data indicate a tight anti-parallel alignment of the α1 and α2 helices of STIM1 CC1 connected by two pairs of coiled-coil interaction sites. Strikingly, both of them involve residues from the CC1 α1 helix (L248, L251, L258, L261), the mutation of which is well known to switch STIM1 into an activated state.14, 15, 35 The potential impact of these coiled-coil contacts within CC1 on STIM1 function will be later on probed by NMR structure-directed mutations in the CC1 α2 helix as described in 2.4.
2.2. Stormorken mutation alters CC contacts of STIM1 CC1
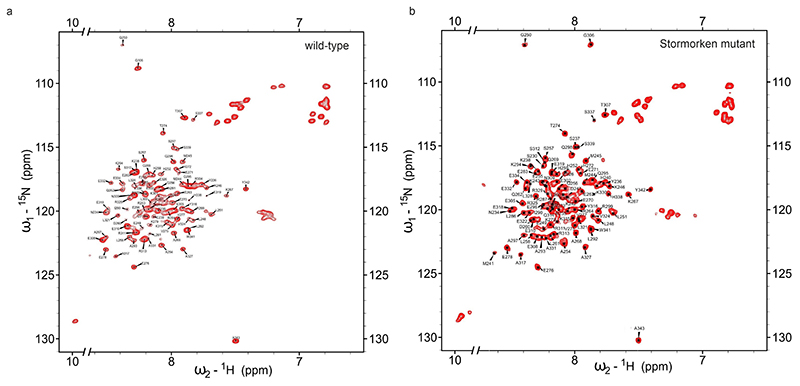
NMR spectra of the Stormorken (R304W) mutant of the STIM1 CC1 fragment revealed major differences compared to the corresponding wild-type fragment. The NMR assignment of the Stormorken mutant CC1 domain (BMRB28 ID 50118) is not as complete as of the wild-type due to more severe overlap and fewer unambiguous NOE cross-peaks. Non-sequential NOE signals of L258, I290, A293 and G306, which are clearly observable for the wild-type CC1 but not in its R304W mutant indicate a less compact structure with fewer interhelical contacts. Consistently, the R304W mutation is known to induce an extended conformation and homomerization of STIM1 that results in constitutive binding to and activation of Orai1 channels in the cell.20, 21 Homo-oligomerization of CC1 R304W was again prevented by SDS detergent to allow the NMR investigation of monomeric species with reduced signal overlap and manageable spectral resolution (Extended Data Fig. 3).
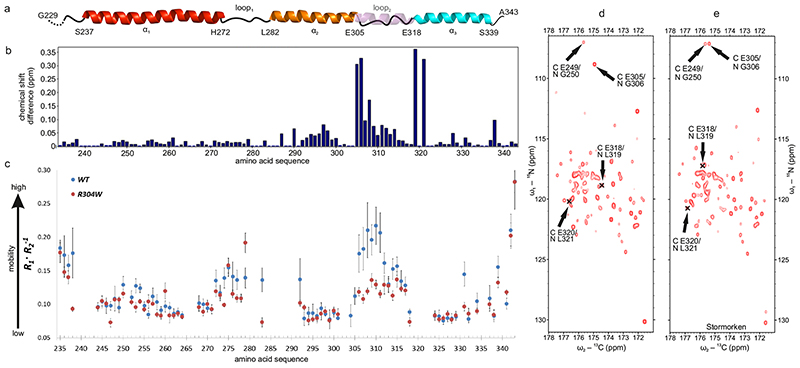
Large chemical shift differences (CSDs) between monomeric wild-type and mutant R304W CC1 domains (Fig. 3a, b) indicate that the mutation disturbs the close-by interhelical interactions occurring in the wild-type (see Fig. 2), likely due to absent charge, increased bulkiness, and rigidity of Trp 304. As the backbone chemical shifts of the N-terminal residues (N234-L286) of the Stormorken CC1 domain are nearly identical to the wild-type in all 2D and 3D spectra, their structure is hardly affected. At residue I290, the peptide bond NH and HN chemical shifts start to deviate (Fig. 3b). The largest CSDs, also evident from carbon detected CON correlation experiments shown in Fig. 3d, e, can be observed directly downstream from the mutated position (R304W) at E305 and G306. Further, the chemical shifts of E319 and L321, which are located remote in sequence from residue 304, are heavily affected by the mutation indicating a change in their positions relative to the α1 interhelical interaction sites. Notably, no sequence remote NOEs are observed for the W341 side chain, in contrast to wild-type CC1 (see above), which is a further manifestation of a less compact helix arrangement in the mutant.
Fig. 3. Stormorken mutation alters CC contacts and increases loop2 rigidity of CC1.
a CC1 secondary structure predicted from backbone chemical shifts The transparent helix (in violet) represents the predicted α-helical propensity in the Stormorken mutant. b Chemical shift differences (1H, 15N) between STIM1 CC1 wild-type and Stormorken mutant. c Backbone relaxation dynamic profiles for wild-type (blue dots) and Stormorken mutant (red dots) of CC1. R1/R2 values were computed from 5 independent measurements each with error propagation (mean ± S.D.). d and e show carbon detected 2D CON 700 MHz NMR spectra of wild-type and Stormorken STIM1 CC1, respectively.
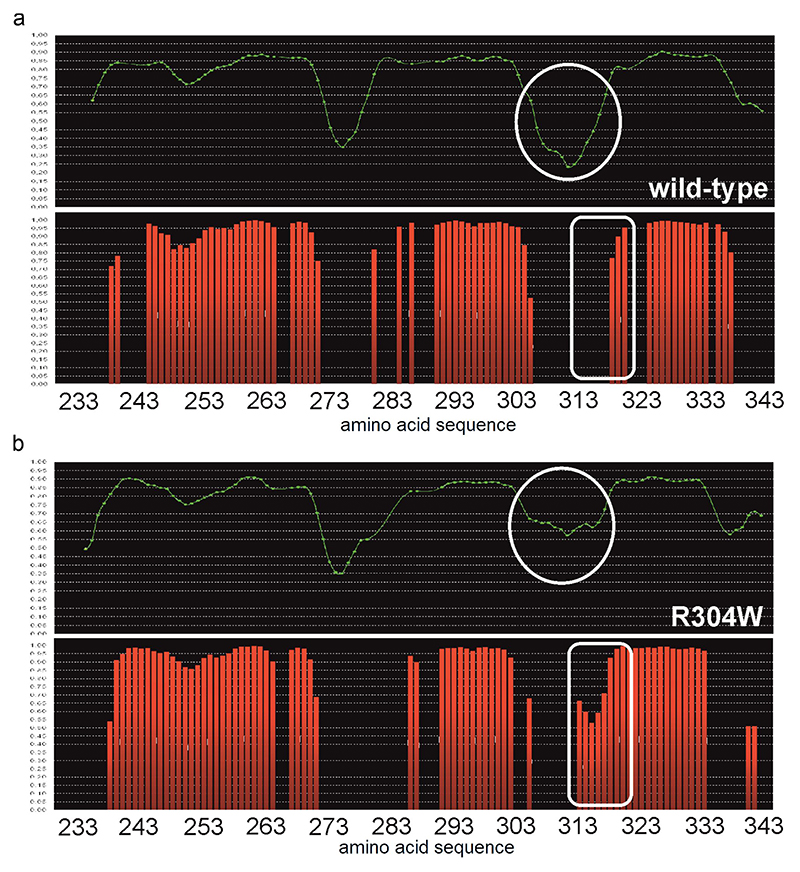
According to chemical shift analysis by TalosN36 (Fig. 3a and Extended Data Fig. 4), the propensities of the 52 N-terminal amino acids to form an α-helical structure (α1) are nearly identical for the wild type and the Stormorken CC1 mutant, with residues H272-L282 being dynamic (loop1). In the Stormorken fragment, the R304W mutation increases the α-helical propensity for residues R313-E318, a region following α2, which is dynamic (loop2) in the wild-type (Extended Data Fig. 4). Further confirmation of these experimental results comes from spin relaxation data, as outlined in the next section.
2.3. Stormorken mutation increases loop2 rigidity of CC1
Sequence specific protein backbone mobility can be assessed via the ratio of longitudinal (R1) and transverse (R2) NMR relaxation rates of peptide bond 15N atoms.34 In general, higher R1/R2 ratios indicate greater mobility, which enables us to discriminate flexible regions from more rigid secondary structure elements. Relaxation analysis revealed that the wild-type STIM1 CC1 consists of three rigid segments connected by two mobile regions (Fig. 3c). This result, which is independent from the chemical shift analysis, is consistent with the solution structure (Fig. 1) featuring three distinct α-helices connected by two flexible loops. Relaxation analysis assigns the three rigid secondary structural segments of CC1 to H240-E270, L282-L303 and A317-S340, corresponding to the α1, α2 and α3 helices, respectively.
Comparing the R1·R2 -1 graphs34 in Fig. 3c one can clearly recognize high mobility in the STIM1 CC1 wild-type within residues E271-H281 (i.e. loop1) and even higher mobility for R304-Y316 (i.e. loop2). The observed local dynamic behavior is consistent with the chemical shift derived data (Extended Data Fig. 4), but contradicts the crystallographic structure of CC1 (Extended Data Fig. 1a) with a single α-helix (S237-W340) and no disordered regions32. The previously published NMR solution structure of the CC1[TM-distal]-CC2 STIM1 fragment (S312-R387)37 matches 30 C-terminal residues of the STIM1 CC1 fragment. For this overlapping region, our structure data corroborate the earlier results (Supplementary Fig. 2).
Comparison of the R1·R2 -1 relaxation ratios of the α-helical regions of the STIM1 CC1 Stormorken mutant to the wild-type reveals similar overall rotational correlation times (τc)38 of 6.27±0.62 ns and 6.11±0.67 ns, respectively, consistent with prevailing monomeric species.34 In detail, the R1·R2 -1 ratios for the first ca. 70 and the last ca. 25 residues were very similar between wild-type and Stormorken mutant. However, downstream from the Stormorken mutation site (R304W), i.e. residues R305-Y316, we found a significant increase in R1·R2 -1 compared to the wild-type. Apparently, the introduction of the bulky, rigid and hydrophobic residue Trp reduces the backbone mobility considerably. This corroborates the CSDs observed for these 11 residues (Fig. 3b). The CSDs and chemical shift derived order parameters (Extended Data Fig. 4) as well as the backbone NHHN relaxation analysis (Fig. 3c) indicate local structural and dynamics changes in loop2 of CC1 upon R304W mutation. Summarizing, the NMR data for the Stormorken CC1 mutant prove that the R304W mutation causes increased rigidity and promotes elongation of the α2 helix towards the C-terminus by formation of ~3.5 semi-stable additional helix turns. Owed to the more complete NMR assignments and relaxation data of both the wild-type CC1 and its R304W mutant (Fig. 3 and Extended Data Fig. 4), the experimentally confirmed helix extension exceeds the previously reported 1.5 turns.21
Based on these NMR data of the CC1 wild-type as well as R304W Stormorken fragment and the known constitutive activity of full-length STIM1 R304W, we focused next on the functional impact of interhelical contact sites of the wild-type CC1 and particularly the role of CC1 α2 in affecting the activation status of STIM1.
2.4. CC contact mutations of CC1 reduce Orai1 activation
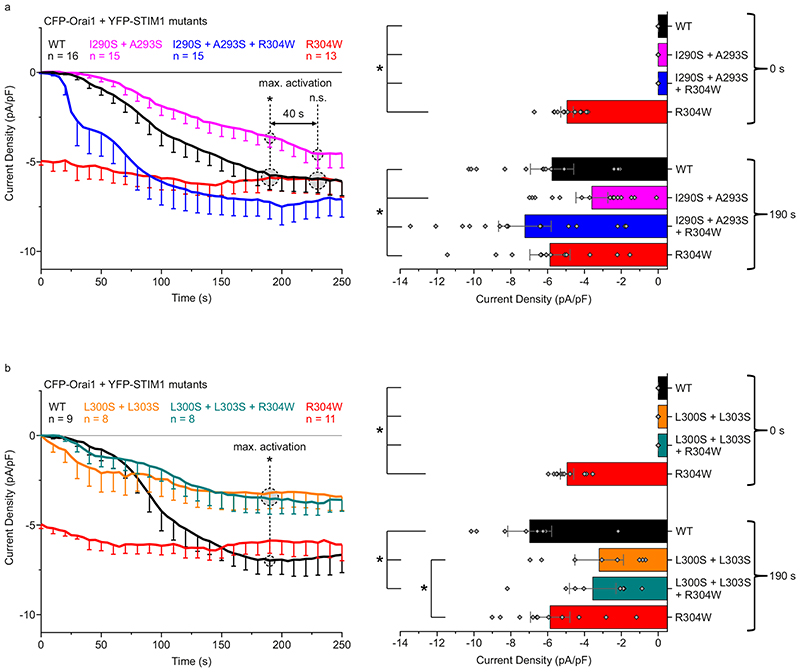
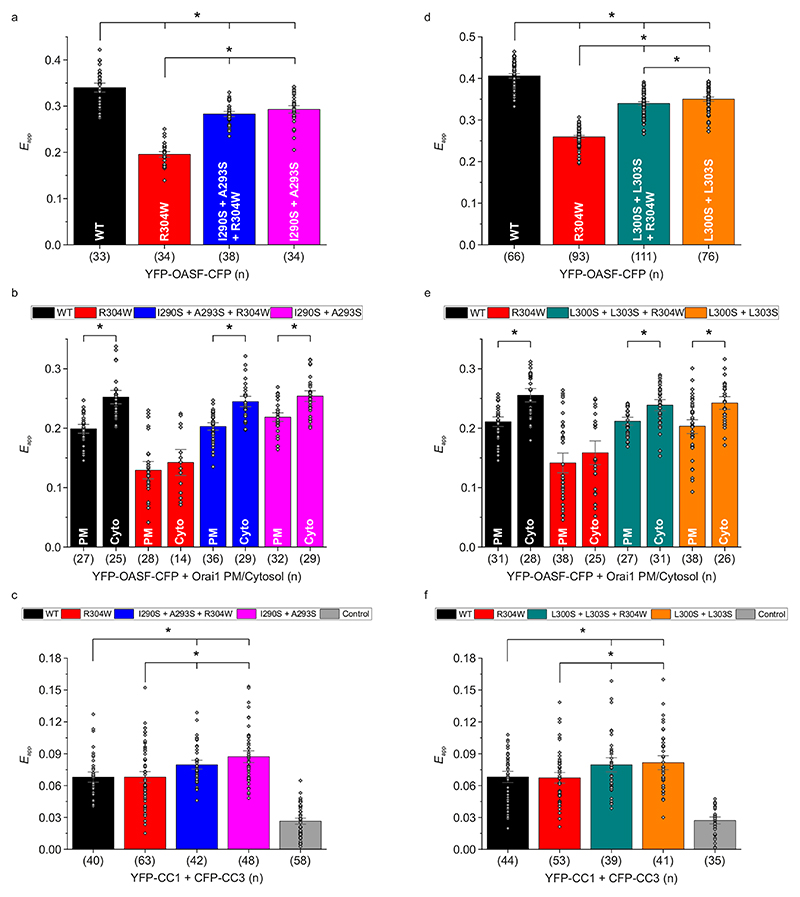
In order to assess the relevance of the CC1 NMR solution structure and especially the impact of the two observed coiled-coil interaction sites between the CC1 α1 and α2 helices on STIM1 activation, we employed live-cell patch-clamp electrophysiology (see Extended Fig. 1b) to probe Orai1 current activation in the context of multiple STIM1 point mutations within the CC1 α2 helix. The rationale for preferential mutation in the α2 rather than the α1 helix of the CC1 domain is based on the fact that several CC1 α1 helix residues are involved in controlling the resting, tight state of STIM114, 35. The hydrophobic residues I290 and A293, which showed distinct NOE close contacts with L258 and L261, were both mutated to more hydrophilic serines in order to weaken15 this coiled-coil interaction site in the full-length STIM1 protein. Another serine mutation was applied to interfere with the hydrophobic contacts of residues L300 and L303 of the second coiled-coil interaction site, where L300 was found to exhibit NOE contacts to both L248 and L251. Additionally, we studied the effect of two double mutations, I290S + A293S and L300S + L303S, on the constitutively active Stormorken STIM1 R304W mutant.18
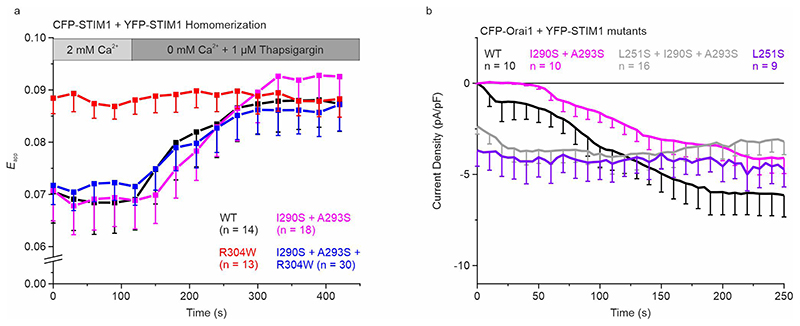
To assess the impact of I290S + A293S on the early STIM1 activation step leading to puncta formation, we carried out FRET-detected STIM1 homomerization experiments.39 We observed a slightly delayed FRET increase for STIM1 I290S + A293S after Ca2+ store depletion with 1 μM thapsigargin that reached a similar maximum homomerization FRET as with STIM1 wild-type (WT) (Extended Data Fig. 5a). In patch-clamp recordings using the whole cell configuration (Fig. 4a), HEK293 cells co-expressing CFP-Orai1 and YFP-STIM1 I290S + A293S similarly showed slower activation kinetics of store-dependent Orai1 currents in comparison to those co-expressing WT STIM1. Maximum current activation was delayed by approximately 40 s (Fig. 4a left). For quantitative analysis, we compared the current densities at time-points 0 s and 190 s to evaluate potentially different behaviors of the distinct STIM1 mutated forms. At 190 s (close to the current maximum reached with WT STIM1), STIM1 I290S + A293S yielded significantly smaller currents than the other WT and mutant STIM1 forms studied (Fig. 4a right). When introducing the I290S + A293S mutations into the Stormorken STIM1 R304W mutant, a striking change in behavior was observed with current levels at the start of the experiment. At 0 s, the STIM1 I290S + A293S + R304W triple mutant exhibited no initial current activation in stark contrast to the constitutive currents of the Stormorken STIM1 R304W mutant. Thus, store-dependent activation behavior has been re-stored with the STIM1 I290S + A293S + R304W triple mutant, albeit activation kinetics occurred faster compared to WT STIM1 (Fig. 4a left and right). The store-dependent activation behavior of the triple mutant was further supported by STIM1 FRET homomerization experiments. Contrary to the initial high homomerization FRET of STIM1 R304W, the STIM1 I290S + A293S + R304W triple mutant showed a store-dependent homomerization FRET increase similar to STIM1 WT Extended Data Fig. 5). Summarizing, the weakening of one interhelical interaction site between CC1 α1 and α2 obviously favors a STIM1 state of lower activatability and even successfully antagonizes the R304W induced activated state of STIM1 linked to Stormorken disease.
Fig. 4. CC contact mutations of CC1 reduce Orai1 activation.
a Mutation of the first (I290S + A293S) interhelical site. Left: Orai1 current activation shown by patch clamp recordings of N-terminally tagged CFP-Orai1 co-expressed with YFP-STIM1 I290S + A293S ± R304W. Right: Comparison of current activation levels at Time 0 vs. 190 seconds. b Mutation of the second (L300S + L303S) interhelical site. Left and Right: Same as in a but for CFP-Orai1 co-expressed with YFP-STIM1 L300S + L303S ± R304W. Color code: WT (black), R304W (red), I290S + A293S + R304W / L300S + L303S + R304W (blue / cyan), and I290S + A293S / L300S + L303S (magenta / orange). HEK293 cells were exclusively used for all experiments. Student’s two-tailed t-test was employed for statistical analyses with differences considered statistically significant at p<0.05. Asterisks (*) indicate significant difference. Exact p values are listed in Supplementary Table 4. Experiments were replicated on at least two different days using independent transfections with the indicated number of cells (n). Data represent mean values ± SEM.
In an analogous approach interfering with the second interhelical coiled-coil interaction, HEK293 cells co-expressing CFP-Orai1 and YFP-STIM1 L300S + L303S led us to observe a significantly reduced overall Orai1 current activation plateau when compared to STIM1 WT (Fig. 4b left). Impressively, the constitutive activity obtained with STIM1 R304W was fully abolished with the triple mutant STIM1 L300S + L303S + R304W exhibiting store-dependent activation behavior. The maximum of the Orai1 current levels reached (at 190 s) was reduced compared to that obtained with STIM1 WT, and Orai1 currents densities were almost indistinguishable between the STIM1 triple and double mutant which lacked R304W (Fig. 4b left and right). Hence, interference with the second α1-α2 coiled-coil interaction site of the CC1 domain completely abrogates the effect of the Stormorken mutation by restoring store-dependent activation behavior of Orai1 currents.
2.5. CC contact mutations restore OASF R304W tight state
In an attempt to mechanistically understand the reversal of the pathophysiological behavior of the GOF STIM1 Stormorken mutant into an almost physiological store-dependent activation, we utilized the doubly fluorescent-labeled soluble C-terminal OASF portion (aa 233-474 including CC1, CC2 and CC3 domains of STIM1) as STIM1 conformational sensor14 for detecting changes of intramolecular FRET (Supplementary Fig. 3a). High FRET of YFP-OASF-CFP conformational sensor represents the inactive, tight state of STIM114, 15, while introducing the Stormorken R304W mutation switches YFP-OASF-R304W-CFP into an activated, extended state exhibiting a reduced FRET21. Additional introduction of the respective double mutations I290S + A293S or L300S + L303S led to a significant increase in FRET when compared to that of the Stormorken OASF R304W construct (Fig. 5a,d). The higher FRET of OASF I290S + A293S + R304W or OASF L300S + L303S + R304W suggests a switch from an extended state to a more tight, inactive state characteristic for the WT OASF fragment. Furthermore, both I290S + A293S and L300S + L303S induced conformational changes on their own that were not significantly altered by the presence or absence of R304W (Fig. 5a, d). Likely, mutation of either interhelical coiled-coil interaction site provokes conformational rearrangements that result in a slightly altered phenotype. Overall, the OASF FRET measurements reflect the impact of the CC1α2 mutations on the full-length STIM1 activation state. The electrophysiological measurements show store-dependent Orai1 activation with both STIM1 triple mutants (R304W + I290S + A293S and R304W + L300S + L303S, respectively). Correspondingly, the OASF triple mutants reveal the expected high FRET close to WT OASF, in line with a STIM1 tight state.
Fig. 5. CC contact mutations restore OASF R304W tight state by reinforcing CC1-CC3 clamp.
a-c Mutation of the first (I290S + A293S) interhelical site. a and b Intramolecular FRET showing conformational changes of YFP-OASF-CFP I290S + A293S ± R304W in the absence (a) and presence (b) of unlabeled Orai1. c Intermolecular FIRE interaction indicating heteromerization of CFP-CC3 with YFP-CC1 I290S + A293S ± R304W. d-f Mutation of the second (L300S + L303S) interhelical site. Same as in a-c but for YFP-OASF-CFP L300S + L303S ± R304W (d) and (e) as well as CFP-CC3 with YFP-CC1 L300S + L303S ± R304W (f). For FIRE control experiments, a CFP construct lacking the CC3 domain was co-expressed with YFP-CC1. Color code: WT (black), R304W (red), I290S + A293S + R304W / L300S + L303S + R304W (blue / cyan), I290S + A293S / L300S + L303S (magenta / orange), and control (gray). HEK293 cells were exclusively used for all experiments. Student’s two-tailed t-test was employed for statistical analyses with differences considered statistically significant at p<0.05. Asterisks (*) indicate significant difference. Exact p values are listed in Supplementary Table 5. Experiments were replicated on at least two different days using independent transfections with the indicated number of cells (n). Data represent mean values ± SEM.
In a further attempt to observe the respective conformational changes upon interaction with Orai1, we co-expressed the double-labeled OASF constructs with unlabeled Orai1 and probed for differences of FRET between the plasma membrane associated fraction (where interaction with Orai1 occurs) and that located in the cytosol. Binding to Orai1 causes an extension of the conformational sensor, which translates into a decrease of FRET as reported previously14. Both OASF I290S + A293S + R304W and OASF L300S + L303S + R304W exhibited a significant conformational switch into an extended state upon interaction with Orai1 (Fig. 5b, e), similar to WT OASF. In contrast, OASF R304W did not undergo such a switch and generally exhibited a much lower FRET, as it assumed already and maintained an extended state.
Taken together, the OASF triple and double mutants behave similar to the WT OASF upon steady-state interaction with Orai1, while the Stormorken mutant OASF 304W lacks a change of its already extended conformation. Furthermore, the Stormorken mutant forms of both full-length STIM1 and OASF are markedly affected by mutations in the CC1 α2 helix in a way that the constitutive activity is reversed into store-operated behavior and the extended conformation is markedly shifted towards the tight one, respectively.
2.6. CC contact mutations reinforce STIM1 CC1-CC3 clamp
In order to further investigate the mechanisms of action of these CC1 α2 double mutations I290S + A293S and L300S + L303S as well as the significance of residues I290, A293, L300, and L303 for the structure and activation of STIM1, we employed our FIRE (FRET-derived interactions in a restricted environment) system15 (Supplementary Fig. 3b). The robust interaction between CC1 and CC3 fragments reflects formation of the CC1-CC3 clamp controlling the tight, resting state of STIM115 via an interaction of CC1 with CAD/SOAR35. As published previously, the Stormorken mutant R304W acts differently at fragment level than in the context of a larger molecule, such as OASF or full length STIM121. The reason for this is that per se the affinity between CC1 and CC3 fragments is not reduced by the R304W mutation, but CC1-CC3 clamp formation in OASF or full-length R304W STIM1 is sterically hindered both by increased CC1 homomerization and the increased rigidity of the loop2 between CC1α2 and α3 21. It is therefore not surprising that in FIRE experiments CC1 R304W + CC3 generated an equally strong FRET signal as with the wild type CC1 (Fig. 5c, f). Strikingly, co-expression of CC1 I290S + A293S or CC1 L300S + L303S with CC3 revealed a significantly increased FRET, which indicates that the tight state of STIM1 is favored by these substitutions. Even more impressive is the interaction found between the triple mutants CC1 I290S + A293S + R304W or CC1 L300S + L303S + R304W and CC3 (Fig. 5c, f), suggesting a reinforced CC1-CC3 clamp that stabilizes the resting state of STIM1. It appears that these hydrophilic mutations within the CC1 α2 helix increase the availability of the hydrophobic residues in the CC1 α1 helix (L248, L251, L258 and L261, see Fig. 2) that are involved in the formation of the CC1-CC3 clamp15 or CC1-SOAR/CAD interface40 to control the STIM1 tight, resting state14, 35. Overall, the FIRE results suggest that weakening of either interhelical CC1 α1-α2 interactions strengthens the CC1-CC3 clamp, favoring the resting state of STIM114 and functionally slowing activation kinetics of or reducing CRAC channel activation. The stronger interaction of the CC3 fragment becomes particularly evident with the investigated CC1 triple mutants, which provides a mechanistic understanding of the reversal of constitutive activity of the Stormorken STIM1 mutant into a store-operated activation observed in patch clamp experiments (Fig. 6).
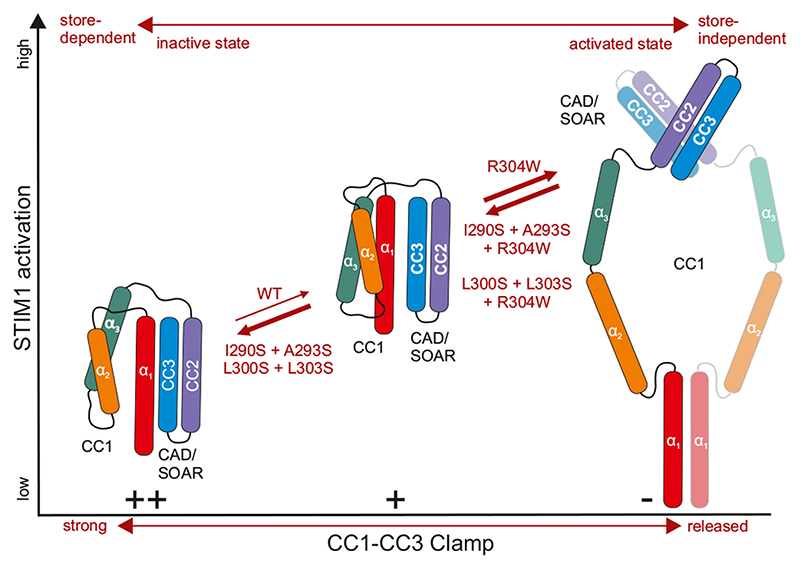
Fig. 6. Simplified model of STIM1 activation.
The CC1-CC3 clamp provides control over the release of CAD/SOAR from the CC1 domain. Interhelical interactions within the CC1 domain affect the strength of the CC1-CC3 clamp formation, the release of which leads to the exposure of the CAD/SOAR domain. The extended, activated state (right) can be switched to the tight, inactive states (left, middle) by weakening of CC1 interhelical interactions concomitantly strengthening CC1-CC3 interactions. Mutations stated at the bold arrows lead to the respective STIM1 activation states. For simplicity, the dimeric state of the CC1-CAD/SOAR fragments in the tight, store-dependent states is omitted, and the activated state might as well exhibit a crossing of the CC1 α3.
Discussion
The solution NMR structure of the monomeric STIM1 CC1 domain represents a tight three-helix bundle stabilized by two prominent interhelical coiled-coil interactions between the helices CC1α1 and CC1α2. The divergence from the X-ray CC1 structure32, which is essentially composed of dimers of long straight α-helical protein molecules, from our solution structure may be induced by crystal packing forces41 and/or by the two mutations (M244L, L321M) that have been introduced to achieve crystallization. Indeed, the STIM1 M244L + L321M double mutant constitutively induced Orai1 currents – in line with the stretched conformation observed in the crystallographic study32. The constitutive activation was not as strong as with the Stormorken mutant STIM1 R304W as the double mutant still partly responded to store-depletion (Extended Data Fig. 1c).
In the case of the Stormorken disease-causing STIM1 CC1 R304W mutant, NMR data did not provide evidence for CC1α1 and CC1α2 interhelical interactions. This finding corroborates the suggested conformational extension of the Stormorken CC140 as well as of the respective full-length STIM1 molecule21 in the activated state. One key reason for this conformational extension is increased rigidity of CC1 loop2, which has been mimicked by multiple alanine (305-311)A mutations in full-length STIM1 that lead to constitutive activation of STIM1/Orai currents similar to STIM1 R304W21. Mutations (I290S A293S or L300S L303S) within the CC1α2 interhelical interaction sites derived from the NMR structure interfere with hydrophobic interactions between CC1α1 and CC1α2. Impressively, they reverse the effect of the Stormorken mutation on the STIM1 activation state by stabilizing the resting, store-dependent behavior of STIM1. Functionally, live cell electrophysiological experiments revealed restoration of the store-dependent activation behavior of the usually constitutively active Stormorken STIM1 mutant, with either double mutations introduced in each of these interhelical interaction sites. On a mechanistic basis (Fig. 6), these mutations within the CC1α2 helix significantly restored the tight conformational state of STIM1 R304W, as derived from the OASF conformational sensor. In molecular terms, the CC1-CC3 clamp that provides a powerful control of the STIM1 activation state15 is markedly stronger. Interference with the CC1α1-CC1α2 interhelical contact sites has a stabilizing effect on the resting, inactive state of STIM1 in both fragment and full-length STIM1 proteins. The weakening of the coiled-coil contacts between L258-L261 and I290-A293 as well as L248-L251 and L300 seems to favor a CC1 state with enhanced freedom for establishing the binding interface between CC1 and CAD/SOAR (Fig. 6). The binding interface at the CC1 domain particularly involves L248, L251, L258 and L261 located in the CC1α1 domain35, 40, 42, all of which contribute also to the two interhelical interaction sites identified in the CC1 NMR structure. To probe for a potential antagonistic effect of the I290S + A293S mutations on another STIM1 GOF mutation, i.e. L251S14, located in the CC1 α1 dimerization domain40, we performed patch-clamp experiments using the corresponding STIM1 triple mutant. When combined with L251S, the double mutation I290S + A293S was not able to reverse the constitutive activation behavior of STIM1 L251S. In contrast to R304W, this distinct behavior might occur from the additional disturbance of the binding interface between CC1α1-CC3 by L251S (Extended Data Fig. 5b).
Shifting the balance from interhelical interactions between CC1α1 -CC1α2 to CC1α1 – CC3 apparently reinforces the CC1-CAD/SOAR interaction thereby stabilizing the STIM1 resting state (Fig. 6).15 A limitation of our study might be seen in the fact that the regulatory interhelical interactions were identified in the monomeric CC1 NMR structure. However, the CC1 domain as an autonomous entity replicates molecular characteristics in its interaction with the CAD/SOAR domain also when not covalently linked to each other as shown in several reports.15, 35 This is in line with our data obtained with fragments in FIRE experiments. Moreover, our results further validate the regulatory effect of the interhelical interactions of the CC1 domain in the larger OASF STIM1 fragment and in the full-length STIM1 form.
In summary, we established the three dimensional structure of the monomeric wild-type STIM1 CC1 protein domain in solution and further characterized the conformational and dynamic changes caused by the Stormorken disease causing R304W mutant based on extended NMR assignments and relaxation analysis. Analysis of this STIM1 CC1 solution structure led to the identification of essential intramolecular interhelical interaction sites as NMR-directed mutation targets. Their manipulation enables control over the Stormorken disease–induced STIM1 activation state and reveals a fine-tuned balance between intramolecular, interhelical CC1 interactions and contacts to the CC3 domain of CAD/SOAR in the control of CRAC channel activation.
Online Methods
Protein expression for NMR analysis
The pGEX 4T-1 GST-tagged expression vector including a thrombin cleavage site was kindly provided by Mitsuhiko Ikura’s laboratory (University of Toronto, CAN). The 15N and 15N-13C isotopically labeled STIM1 CC1 protein fragments (amino acids 234-343) were expressed and purified following our published protocol (see Supplementary Fig. 4. In addition, “ILKV” selectively unlabeled43 and perdeuterated (~70%2H) samples of STIM1 CC1 were prepared to resolve assignment ambiguities. The point-mutated R304W fragment was prepared using the QuikChange XL site-directed mutagenesis kit. The correctness of all described plasmids was confirmed by sequence analysis (Eurofins Genomics GmbH). Unless otherwise stated, STIM1 CC1 wild-type as well as the Stormorken mutant (R304W) protein fragments were swapped to Tris-HCl buffer (20 mM Tris-HCl, 0.2% SDS, pH 7.25) and concentrated to 0.3 mM for further NMR analysis. The sub-CMC concentration of SDS protects hydrophobic sites involved in oligomerization while leaving intramolecular contacts intact.
NMR spectroscopy
NMR experiments were recorded using the 700 MHz Bruker Avance III spectrometer (equipped with a cryogenically cooled TCI probe) at the Austro-Czech RERI-uasb NMR center in Linz. Further proton and carbon detected NMR experiments were obtained using Bruker spectrometers at 950 MHz and 700 MHz proton frequency at CERM in Sesto Fiorentino. Spectrometers were running Topspin software (versions 3.5 and 3.6). The protein samples were measured at 310 K in 5 mm NMR tubes (Wilmad), 5 mm shaped tubes (Bruker) and 5 mm water susceptibility matched Shigemi tubes (Shigemi).
Backbone correlation data were acquired by standard1H-15N-HSQC, HNCO, HNCO, HNCA, HN(CA)CO, HN(CO)CA, HNCACB and HN(CO)CACB experiments.44 To confirm correctness and resolve ambiguities of the resonance assignment, carbon detected CON, CBCACON, CANCO, CACO and CBCACO spectra were acquired.45 The side-chain resonances were assigned using two double resonance experiments [(H)CCH-TOCSYali and HC(C)H-COSYali] and their cross-peaks were mapped into the 1H-13C HSQCali spectrum.44
In 950 MHz 15N-edited NOESY-HSQC spectra (mixing time 80 ms) a relatively small number of NOE cross peaks could be resolved and assigned unequivocally within the coiled-coil contact regions (Extended Data Fig. 6). The apparent scarcity of NOE restraints is owed to missing or ambiguous side-chain assignments caused by the severe peak overlap of the hydrophobic side chain resonances which, as mentioned above, is typical of NMR spectra of coiled-coil proteins. 15N-T1 and 15N-T2 relaxation times were determined by exponential fitting of peak heights in sets of different 15N-HSQC phase sensitive experiments with different relaxation delays as described in the Supplementary Table 1. Chemical shifts of the STIM1 CC1 domain and the corresponding Stormorken mutant were assigned with the help of CARA software (DISS. ETH No. 15947, version 1.9.1) and deposited under BMRB IDs 50114 and 50118, respectively.
Structure calculation and refinement
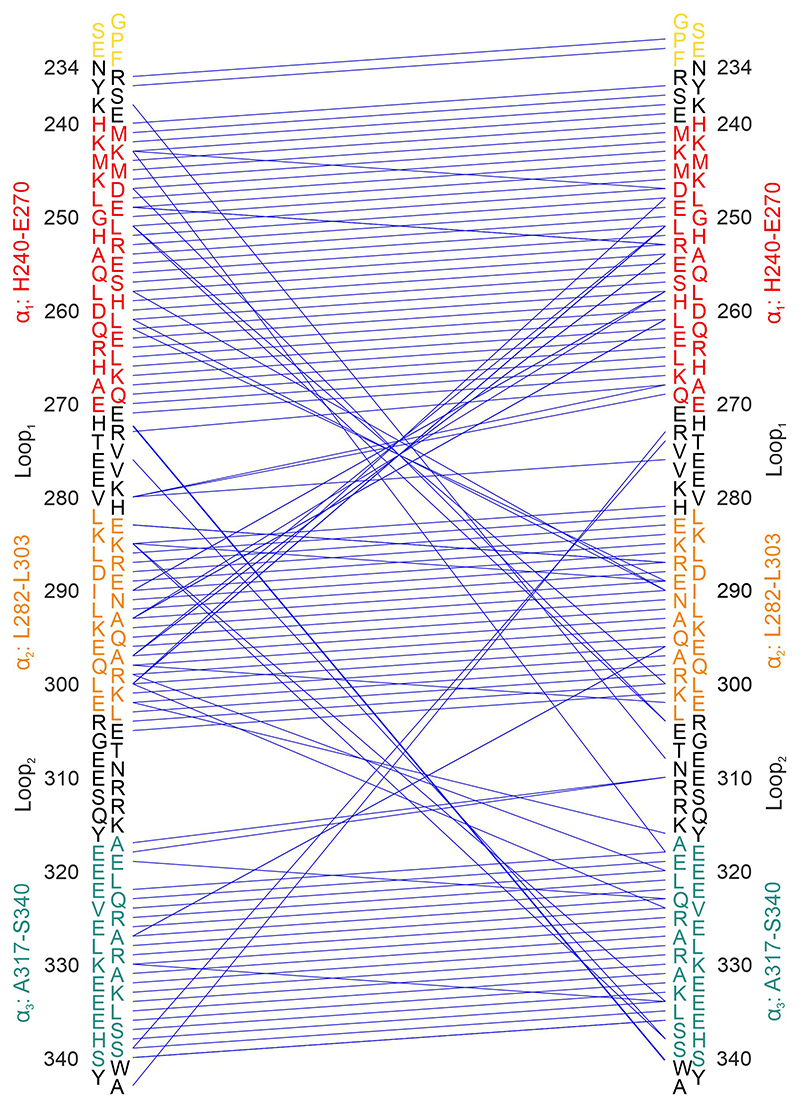
Initial structural model for STIM1 CC1 wild-type fragment was assembled by CYANA v3.0 calculation.30 For this calculation 138 dihedral angle restraints were extracted from TalosN estimation36 and merged with 118 hydrogen bond upper limit constraints obtained from CS-Rosetta web server (csrosetta.bmrb.wisc.edu) based on assigned chemical shifts. 36 distinct long-range constraints (summarized in Supplementary Table 2) were derived from 15N-edited NOESY-HSQC spectra (mixing time 80 ms) assuming an upper limit of 6 Å. The NOE contact map is summarized in Extended Data Fig. 6. In total, 400 structures were calculated by CYANA in 50,000 annealing steps. The 20 conformers with lowest CYANA target function values were selected, merged into a structure bundle and water-refined by YASARA (Yet Another Scientific Artificial Reality Application) version 18.12.1646 together with 107 distance and 138 dihedral angle restraints. A two-step probabilistic refinement protocol was applied using YASARA macros (nmr_refinevacuo.mcr and nmr_refinesolv.mcr).
The first step, in vacuo, uses the Nova47 force field, and the second step uses the YASARA force field46 for final refinement embedded in explicit water molecules. Ten cycles of this protocol were used to generate a total of 200 refined structures and from these the 20 structures with lowest total energy shown in Fig. 1. were deposited at the PDB (6YEL). The PROCHECK39 analysis of ordered residues ((S(ϕ)+S(ψ) ≥1.8) reveals 98.6% residue in the most favored region, 1.2% in additional allowed region, 0.1% in the generously allowed region and 0.1% residue in the disallowed region in the Ramachandran plot. Additional structure computation statistics are reported in Supplementary Table 3.
Molecular cloning and mutagenesis
Human Orai1 (hOrai1, accession number NM_032790) was kindly provided by A. Rao (Harvard Medical School). N-terminally tagged Orai1 constructs were engineered by using SalI and SmaI restriction sites of the vectors pECFP-C1 and pEYFP-C1 (Clontech laboratories, CA, USA). N-terminally labeled human STIM1 (hSTIM1, accession number NM_003156) were made available by T. Meyer (Stanford University). pECFP-STIM1 mutants (I290 + A293S, R304W, I290 + A293S + R304W) and pEYFP-STIM1 mutants (M244L + L321M, L251S, L251S + I290S + A293S, I290 + A293S, L300S + L303S, R304W, I290 + A293S + R304W, L300S + L303S + R304W) were built with the aid of the QuikChange XL site-directed mutagenesis kit (Stratagene, CA, USA). For the generation of double-tagged STIM1-OASF constructs, CFP was introduced into pEYFP-C2 via SacII and Xba1 and the OASF fragment (aa233–474) of STIM1 was inserted via restriction sites EcoRI and SacII. Double-tagged YFP-OASF-CFP mutants (I290 + A293S, L300S + L303S, R304W, I290 + A293S + R304W, L300S + L303S + R304W) were built with the aid of the QuikChange XL site-directed mutagenesis kit (Stratagene). FIRE system constructs (Y-TMG as well as C-TMG) contained the STIM1 signal peptide followed by EYFP (Y) or ECFP (C), a 29 aa linker, the STIM1 TM domain, and a 32 glycine linker followed by the protein fragment of interest (CC1 233–343, CC3 388–430). The QuikChange XL site-directed mutagenesis kit (Stratagene) was employed to generate Y-TMG point mutants (CC1 I290 + A293S, CC1 L300S + L303S, CC1 R304W, CC1 I290 + A293S + R304W, CC1 L300S + L303S + R304W). The correctness of all constructs was verified by sequence analysis (Eurofins Genomics GmbH).
Confocal FRET microscopy
Confocal FRET microscopy of HEK293 cells was carried out at room temperature 24–36 hours after transfection. The experimental setup consisted of a CSU-X1 Real-Time Confocal System (Yokogawa Electric Corporation, Japan) combined with two CoolSNAP HQ2 CCD cameras (Photometrics, AZ, USA). The installation was also fitted with a dual port adapter (dichroic: 505lp, cyan emission filter: 470/24, yellow emission filter: 535/30, Chroma Technology Corporation, VT, USA). An Axio Observer.Z1 inverted microscope (Carl Zeiss, Oberkochen, Germany) and two diode lasers (445 and 515 nm, Visitron Systems, Puchheim, Germany) were connected to the described configuration. All described components were positioned on a Vision IsoStation anti-vibration table (Newport Corporation, CA, USA). Image recording and control of the confocal system were carried out with the VisiView software package (v2.1.4, Visitron Systems). Cross-excitation and spectral bleed-through necessitate image correction prior to any FRET calculation. Cross-excitation calibration factors were therefore determined for all expressed DNA constructs on each measurement day. After threshold determination as well as background signal subtraction, the apparent FRET efficiency E app was calculated on a pixel-to-pixel basis. This was performed with a custom program48 integrated into MATLAB (v7.11.0, The MathWorks, Inc., MA, USA) that implements a microscope-specific constant G parameter of 2.75.49
Patch Clamp Whole Cell Experiments
Patch clamp electrophysiology experiments of HEK293 (DSMZ) cells were carried out at room temperature 24–36 hours after transfection with two Ag/AgCl electrodes serving as both recording as well as reference electrodes. Every five seconds, a voltage ramp in the range of -90 to +90 mV was applied over one second; the holding potential was adjusted to 0 mV. The current amplitudes observed at -74 mV were used for data evaluation. Passive store depletion was achieved with the intracellular pipette solution, which consisted of (in mM): 145 Cs methane sulfonate, 20 EGTA, 10 HEPES, 8 NaCl, 5 MgCl2 pH 7.2. The standard extracellular solution was comprised of (in mM) 145 NaCl, 10 HEPES, 10 CaCl2, 10 glucose, 5 CsCl, 1 MgCl2 pH 7.4. The junction potential (-12 mV) caused by combination of a sulfonate-based pipette solution with a Cl--based extracellular solution was not considered. Leak correction was applied to all recordings: the current amplitude recorded during the initial voltage ramp applied immediately after whole-cell break-in was subtracted from all subsequent current amplitudes for store-dependent experiments. Constitutively active currents were corrected with a current amplitude obtained during a voltage ramp applied after perfusion with 10 μM La3+ at the end of the experiment. Individual recordings were normalized by dividing the current amplitudes by the whole-cell capacitance. The WinWCP software package (v4.2.2, University of Strathclyde, UK) was used for data recording and processing.
Statistics
OriginPro 2019 (OriginLab Corporation, MA, USA) was used for statistical analyses and graph plots. Results are depicted as mean values ± SEM determined from the indicated number n of repeats. Student’s two-tailed t-test together with the F-test of equality of variances was employed for statistical analyses with differences considered statistically significant at p<0.05. The one-sample Kolmogorov-Smirnov test was used to verify the presence of a normal distribution for the analyzed datasets. The Grubbs test was applied to eliminate outliers.
Extended Data
Extended Data Fig. 1. CC1 crystallographic structure and corresponding patch-clamp-data.
a Top: Comparison of the published crystallographic structure fragment (PDB: 4O9B, blue) to the STIM1 CC1 NMR model.
The positions of the residues mutated in the X-ray structure are labelled. The first five N-terminal residues (G229-F233) of the recombinant fragment, the remainder of the thrombin cleavage sequence, are not part of the native sequence.
Bottom: Partial sequence alignment of the fragments used for NMR (top) and crystallography (bottom) with mutations indicated by red boxes. The molecular graphics were created by PyMOL (v. 2.3, Schrödinger, LLC). b Depiction of a patch clamp experiment using the whole cell configuration. The entry of Ca2+ ions through Orai1 channels generates an electrical current that is registered by an Ag/AgCl electrode. This electrode is inserted into a glass pipette that is sealed to the plasma membrane of a target cell. c Orai1 current activation shown by patch clamp recordings of N-terminally tagged CFP-Orai1 co-expressed with YFP-STIM1 M244L + L321M (pink). HEK293 cells were exclusively used for all recordings. The patch clamp experiment was replicated on two different days using independent transfections with the indicated number of cells (n). Data represent mean values ±SEM.
Extended Data Fig. 2. The solution NMR structure of STIM1 CC1.
Extended Data Fig. 2 Side a and top b views of the solution structure of STIM1 CC1. Blue color represents residues that are in close NOE contact with the SDS detergent (the residues are listed in the table insert). The NMR experiments were recorded in presence of 7.0 mM SDS to avoid non-specific CC1 homo-oligomerization occurring in absence of the detergent. We note that at this concentration below the CMC (critical micelle concentration) of 8.2 mM (25 °C), SDS does not cause any secondary structure changes, as proven by CD spectra (Supplementary Fig. 1). The intermolecular NOEs observed between the surface-exposed residues of STIM1 CC1 and SDS molecules are consistent with protection of the highlighted (blue) residues listed in the table insert by the detergent, thus preventing CC1 homo-oligomerization while leaving intramolecular coiled-coil contacts intact.
Extended Data Fig. 3. Spectra of STIM1 CC1 wild-type and Stormorken mutant.
Assigned 700 MHz1H-15N HSQC spectra of 0.3 mM 15N-STIM1 CC1 wild-type a and Stormorken mutant b.
Extended Data Fig. 4. Secondary structure prediction of STIM1 CC1 wild-type and Stormorken mutant.
Secondary structure prediction for STIM1 CC1 wild-type a and Stormorken mutant b from Talos-N36. Green lines represent the order parameter S2 predicted from the chemical shifts. Red bars indicate the probability (in %*100) for residue to adapt a helical secondary structure.
Extended Data Fig. 5. STIM1 homomerization and Orai1 activation by STIM1 mutants.
a STIM1 homomerization experiments of N-terminally tagged CFP- and YFP-STIM1 I290S + A293S ± R304W. Ca2+ store depletion was induced by perfusion with 1 μM thapsigargin in Ca2+ free solution. b Orai1 current activation shown by patch clamp recordings of N-terminally tagged CFP-Orai1 co-expressed with YFP-STIM1 L251S ± I290S + A293S. Color code: WT (black), L251S (purple), R304W (red), L251S + I290S + A293S (gray), I290S + A293S + R304W (blue), and I290S + A293S (magenta). HEK293 cells were exclusively used for all experiments. Experiments were replicated on at least two different days using independent transfections with the indicated number of cells (n). Data represent mean values ± SEM.
Extended Data Fig. 6. Graphical representation of NOE distance and hydrogen bond restraints.
Graphical representation of NOE distance and hydrogen bond restraints used for the structure calculation. The parallel closely spaced lines indicate intra-helical (i to i+4) restraints from CS-Rosetta31. The lines crossing each other near the center are characteristic of anti-parallel alignment of helices. The secondary structure ranges are indicated, as well as the non-native residues (G1-F5), by color coding corresponding to the Figures in the main text.
Supplementary Material
Acknowledgement
This work has been supported by iNEXT, grant number 653706, funded by the Horizon 2020 programme of the European Commission and by the Austrian Science Fund (FWF) PhD program W1250 “NanoCell”, P32947 to M.Fa. as well as P27263 to C.R., by the Fondazione Cassa di Risparmio di Firenze and the Italian Ministero dell’Istruzione, dell’Università e della Ricerca through the “Progetto Dipartimenti di Eccellenza 2018-2022” to the Department of Chemistry “Ugo Schiff” of the University of Florence. L.C., M.Fr., C. L. and E.R. also acknowledge the support of the University of Florence and the Recombinant Proteins JOYNLAB. Instruct-ERIC, a Landmark ESFRI project, and specifically the CERM/CIRMMP Italy Centre is also acknowledged. The NMR experiments were performed at the NMR laboratory of the Upper Austrian - South Bohemian Research Infrastructure Center in Linz, “RERI-uasb”, supported by the European Union through the ERDF INTERREG IV (RU2-EU-124/100--2010) program (ETC Austria-Czech Republic 2007-2013, project M00146, “RERI-uasb” for N.M.) and at the CERM institute in Sesto Fiorentino (iNEXT NMR HEDC infrastructure, PID: 6162 and 3428). Inspiring discussions with members of the COST (European Cooperation in Science and Technology) Action CA15209 EURELAX are acknowledged.
Footnotes
Author contributions
C.L., C.R., and N.M. conceived and oversaw the study. P.R., M.Fa., H.G., C.R., and N.M. wrote the manuscript. M.Fa. carried out molecular biology experiments. P.R., L.C., A.G., E.R., M.Fr., and M.B. conducted and analyzed NMR experiments. H.G. performed and analyzed electrophysiological as well as fluorescence microscopy experiments. F.H., H.K., and T.R. contributed to writing and mechanistic concepts of protein function. All authors reviewed and approved the final version of the manuscript.
Competing interests
The authors declare no competing interests.
Data availability statement
The 20 lowest total energy structures and the NOE constraints of wild type CC1 were deposited at the PDB database under accession code 6YEL. The chemical shifts assignments of the wild type STIM1 CC1 and its Stormorken mutant were deposited in the BMRB database under ID 50114 and ID 50118, respectively. All other relevant data are available in this article and its supplementary information files, or from the corresponding authors upon reasonable request.
References
- 1.Bootman MD, Lipp P, Berridge MJ. The organisation and functions of local Ca(2+) signals. Journal of cell science. 2001;114:2213–2222. doi: 10.1242/jcs.114.12.2213. [DOI] [PubMed] [Google Scholar]
- 2.Berridge MJ, Bootman MD, Roderick HL. Calcium signalling: dynamics, homeostasis and remodelling. Nat Rev Mol Cell Biol. 2003;4:517–529. doi: 10.1038/nrm1155. [DOI] [PubMed] [Google Scholar]
- 3.Berridge MJ. Inositol trisphosphate and calcium signalling mechanisms. Biochimica et biophysica acta. 2009;1793:933–940. doi: 10.1016/j.bbamcr.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 4.Lacruz RS, Feske S. Diseases caused by mutations in ORAI1 and STIM1. Annals of the New York Academy of Sciences. 2015;1356:45–79. doi: 10.1111/nyas.12938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Putney JW., Jr A model for receptor-regulated calcium entry. Cell Calcium. 1986;7:1–12. doi: 10.1016/0143-4160(86)90026-6. [DOI] [PubMed] [Google Scholar]
- 6.Zhang SL, et al. STIM1 is a Ca2+ sensor that activates CRAC channels and migrates from the Ca2+ store to the plasma membrane. Nature. 2005;437:902–905. doi: 10.1038/nature04147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 8.Yeromin AV, Zhang SL, Jiang W, Yu Y, Safrina O, Cahalan MD. Molecular identification of the CRAC channel by altered ion selectivity in a mutant of Orai. Nature. 2006;443:226–229. doi: 10.1038/nature05108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Baba Y, et al. Coupling of STIM1 to store-operated Ca2+ entry through its constitutive and inducible movement in the endoplasmic reticulum. Proc Natl Acad Sci U S A. 2006;103:16704–16709. doi: 10.1073/pnas.0608358103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luik RM, Wang B, Prakriya M, Wu MM, Lewis RS. Oligomerization of STIM1 couples ER calcium depletion to CRAC channel activation. Nature. 2008;454:538. doi: 10.1038/nature07065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Navarro-Borelly L, Somasundaram A, Yamashita M, Ren D, Miller RJ, Prakriya M. STIM1-Orai1 interactions and Orai1 conformational changes revealed by live-cell FRET microscopy. J Physiol. 2008;586:5383–5401. doi: 10.1113/jphysiol.2008.162503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stathopulos PB, Ikura M. Partial unfolding and oligomerization of stromal interaction molecules as an initiation mechanism of store operated calcium entry. Biochem Cell Biol. 2010;88:175–183. doi: 10.1139/o09-125. [DOI] [PubMed] [Google Scholar]
- 14.Muik M, et al. STIM1 couples to ORAI1 via an intramolecular transition into an extended conformation. The EMBO journal. 2011;30:1678–1689. doi: 10.1038/emboj.2011.79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fahrner M, et al. A coiled-coil clamp controls both conformation and clustering of stromal interaction molecule 1 (STIM1) J Biol Chem. 2014;289:33231–33244. doi: 10.1074/jbc.M114.610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yuan JP, Zeng W, Dorwart MR, Choi YJ, Worley PF, Muallem S. SOAR and the polybasic STIM1 domains gate and regulate Orai channels. Nat Cell Biol. 2009;11:337–343. doi: 10.1038/ncb1842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Park CY, et al. STIM1 clusters and activates CRAC channels via direct binding of a cytosolic domain to Orai1. Cell. 2009;136:876–890. doi: 10.1016/j.cell.2009.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Muik M, et al. A Cytosolic Homomerization and a Modulatory Domain within STIM1 C Terminus Determine Coupling to ORAI1 Channels. J Biol Chem. 2009;284:8421–8426. doi: 10.1074/jbc.C800229200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Morin G, et al. Gain-of-Function Mutation in STIM1 (P.R304W) Is Associated with Stormorken Syndrome. Human mutation. 2014;35:1221–1232. doi: 10.1002/humu.22621. [DOI] [PubMed] [Google Scholar]
- 20.Nesin V, et al. Activating mutations in STIM1 and ORAI1 cause overlapping syndromes of tubular myopathy and congenital miosis. Proc Natl Acad Sci U S A. 2014;111:4197–4202. doi: 10.1073/pnas.1312520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fahrner M, et al. A dual mechanism promotes switching of the Stormorken STIM1 R304W mutant into the activated state. Nat Commun. 2018;9:825–825. doi: 10.1038/s41467-018-03062-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Misceo D, et al. A dominant STIM1 mutation causes Stormorken syndrome. Human mutation. 2014;35:556–564. doi: 10.1002/humu.22544. [DOI] [PubMed] [Google Scholar]
- 23.Rathner P, Stadlbauer M, Romanin C, Fahrner M, Derler I, Müller N. Rapid NMR-scale purification of (15)N,(13)C isotope-labeled recombinant human STIM1 coiled coil fragments. Protein Expr Purif. 2018;146:45–50. doi: 10.1016/j.pep.2018.01.013. [DOI] [PubMed] [Google Scholar]
- 24.Lupas A, Van Dyke M, Stock J. Predicting coiled coils from protein sequences. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 25.Crick FHC. Is α-Keratin a Coiled Coil? Nature. 1952;170:882–883. doi: 10.1038/170882b0. [DOI] [PubMed] [Google Scholar]
- 26.Crick FHC. The packing of α-helices: simple coiled-coils. Acta Crystallographica. 1953;6:689–697. [Google Scholar]
- 27.Lupas AN, Gruber M. The structure of alpha-helical coiled coils. Adv Protein Chem. 2005;70:37–78. doi: 10.1016/S0065-3233(05)70003-6. [DOI] [PubMed] [Google Scholar]
- 28.Ulrich EL, et al. BioMagResBank. Nucleic Acids Research. 2007;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Güntert P, Mumenthaler C, Wüthrich K. Torsion angle dynamics for NMR structure calculation with the new program DYANA. Journal of molecular biology. 1997;273:283–298. doi: 10.1006/jmbi.1997.1284. [DOI] [PubMed] [Google Scholar]
- 30.Güntert P. Automated NMR structure calculation with CYANA. Methods in molecular biology (Clifton, NJ) 2004;278:353–378. doi: 10.1385/1-59259-809-9:353. [DOI] [PubMed] [Google Scholar]
- 31.Lange OF, et al. Determination of solution structures of proteins up to 40 kDa using CS-Rosetta with sparse NMR data from deuterated samples. Proc Natl Acad Sci U S A. 2012;109:10873–10878. doi: 10.1073/pnas.1203013109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui B, et al. The inhibitory helix controls the intramolecular conformational switching of the C-terminus of STIM1. PloS one. 2013;8 doi: 10.1371/journal.pone.0074735. e74735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Limbach H-H, Ratajczak H, Orville-Thomas WJ. Molecular interactions. Vol. 1 John Wiley & Sons Ltd; Chichester, New York, Brisbane, Toronto: 1980. [Google Scholar]
- 34.Palmer AG., 3rd Nmr probes of molecular dynamics: overview and comparison with other techniques. Annual review of biophysics and biomolecular structure. 2001;30:129–155. doi: 10.1146/annurev.biophys.30.1.129. [DOI] [PubMed] [Google Scholar]
- 35.Ma G, et al. Inside-out Ca signalling prompted by STIM1 conformational switch. Nat Commun. 2015;6 doi: 10.1038/ncomms8826. 7826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. Journal of Biomolecular NMR. 2013;56:227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stathopulos PB, et al. STIM1/Orai1 coiled-coil interplay in the regulation of store-operated calcium entry. Nat Commun. 2013;4 doi: 10.1038/ncomms3963. 2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kay LE, Torchia DA, Bax A. Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 39.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 40.Hirve N, Rajanikanth V, Hogan PG, Gudlur A. Coiled-Coil Formation Conveys a STIM1 Signal from ER Lumen to Cytoplasm. Cell Rep. 2018;22:72–83. doi: 10.1016/j.celrep.2017.12.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Luo J, Liu Z, Guo Y, Li M. A structural dissection of large protein-protein crystal packing contacts. Scientific Reports. 2015;5 doi: 10.1038/srep14214. 14214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma G, et al. Optogenetic engineering to probe the molecular choreography of STIM1-mediated cell signaling. Nat Commun. 2020;11:1039–1039. doi: 10.1038/s41467-020-14841-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnarjuna B, Jaipuria G, Thakur A, D’Silva P, Atreya HS. Amino acid selective unlabeling for sequence specific resonance assignments in proteins. J Biomol NMR. 2011;49:39–51. doi: 10.1007/s10858-010-9459-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kanelis V, Forman-Kay JD, Kay LE. Multidimensional NMR methods for protein structure determination. IUBMB life. 2001;52:291–302. doi: 10.1080/152165401317291147. [DOI] [PubMed] [Google Scholar]
- 45.Bermel W, Felli I, Kümmerle R, Pierattelli R. 13C Direct-detection biomolecular NMR. Concepts in Magnetic Resonance Part A. 2008;32A:183–200. [Google Scholar]
- 46.Krieger E, Vriend G. YASARA View - molecular graphics for all devices - from smartphones to workstations. Bioinformatics (Oxford, England) 2014;30:2981–2982. doi: 10.1093/bioinformatics/btu426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA--a self-parameterizing force field. Proteins. 2002;47:393–402. doi: 10.1002/prot.10104. [DOI] [PubMed] [Google Scholar]
- 48.Derler I, et al. Dynamic but not constitutive association of calmodulin with rat TRPV6 channels enables fine tuning of Ca2+-dependent inactivation. J Physiol. 2006;577:31–44. doi: 10.1113/jphysiol.2006.118661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zal T, Gascoigne NR. Photobleaching-corrected FRET efficiency imaging of live cells. Biophys J. 2004;86:3923–3939. doi: 10.1529/biophysj.103.022087. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The 20 lowest total energy structures and the NOE constraints of wild type CC1 were deposited at the PDB database under accession code 6YEL. The chemical shifts assignments of the wild type STIM1 CC1 and its Stormorken mutant were deposited in the BMRB database under ID 50114 and ID 50118, respectively. All other relevant data are available in this article and its supplementary information files, or from the corresponding authors upon reasonable request.