Abstract
The human proteome is a major source of therapeutic targets. Recent genetic association analyses of the plasma proteome enable systematic evaluation of the causal consequences of variation in plasma protein levels. Here we estimated the effects of 1,002 proteins on 225 phenotypes using two-sample Mendelian randomization (MR) and colocalization. Of 413 associations supported by evidence from MR, 130 (31.5%) were not supported by results of colocalization analyses, suggesting that genetic confounding due to linkage disequilibrium (LD) is widespread in naïve phenome-wide association studies of proteins. Combining MR and colocalization evidence in cis-only analyses, we identified 111 putatively causal effects between 65 proteins and 52 disease-related phenotypes (www.epigraphdb.org/pqtl/). Evaluation of data from historic drug development programs showed that target-indication pairs with MR and colocalization support were more likely to be approved, evidencing the value of this approach in identifying and prioritizing potential therapeutic targets.
Despite increasing investment in research and development (R&D) in the pharmaceutical industry1, the rate of success for novel drugs continues to fall2. Lower success rates make new therapeutics more expensive, reducing availability of effective medicines and increasing healthcare costs. Indeed, only one in ten targets taken into clinical trials reaches approval2, with many showing lack of efficacy (˜50%) or adverse safety profiles (˜25%) in late stage clinical trials after many years of development3,4. For some diseases, such as Alzheimer’s disease, the failure rates are even higher5.
Thus, early approaches to prioritize target-indication pairs that are more likely to be successful are much needed. It has previously been shown that target-indication pairs for which genetic associations link the target gene to related phenotypes are more likely to reach approval6. Consequently, systematically evaluating the genetic evidence in support of potential target-indication pairs is a potential strategy to prioritize development programs. While systematic genetic studies have evaluated the putative causal role of both methylome and transcriptome on diseases7,8, studies of the direct relevance of the proteome are in their infancy9,10.
Plasma proteins play key roles in a range of biological processes and represent a major source of druggable targets11,12. Recently published genome-wide association studies (GWAS) of plasma proteins have identified 3,606 conditionally independent single nucleotide polymorphisms (SNPs) associated with 2,656 proteins (‘protein quantitative trait loci’, pQTL)9,13,14,15,16. These genetic associations offer the opportunity to systematically test the causal effects of a large number of potential drug targets on the human disease phenome through Mendelian randomization (MR)17. In essence, MR exploits the random allocation of genetic variants at conception and their associations with disease risk factors to uncover causal relationships between human phenotypes, and has been described in detail previously18,19.
For MR analyses of proteome, unlike more complex exposures, anintuitive way to categorize protein-associated variants is into cis-acting pQTLs located in the vicinity of the encoding gene (defined as ≤500kb from the leading pQTL of the test protein in this study) and trans-acting pQTLs located outside this window. The cis-acting pQTLs are considered to have a higher biological prior and have been widely employed in relation to some phenome-wide scans of drug targets such as CETP 20 and IL6R 21. Trans-acting pQTLs may operate via indirect mechanisms and are therefore more likely to be pleiotropic22, although they may support causal inference where they are likely to be non-pleiotropic.
Here we pool and cross-validate pQTLs from five recently published GWAS and use them as instruments to systematically evaluate the causal role of 968 plasma proteins onthe human phenome, including 153 diseases and 72 risk factors available in the MR-Base database23. Results of all analyses are available in an open online database (www.epigraphdb.org/pqtl/), with a graphical interface to enable rapid and systematic queries.
Results
Characterizing genetic instruments for proteins
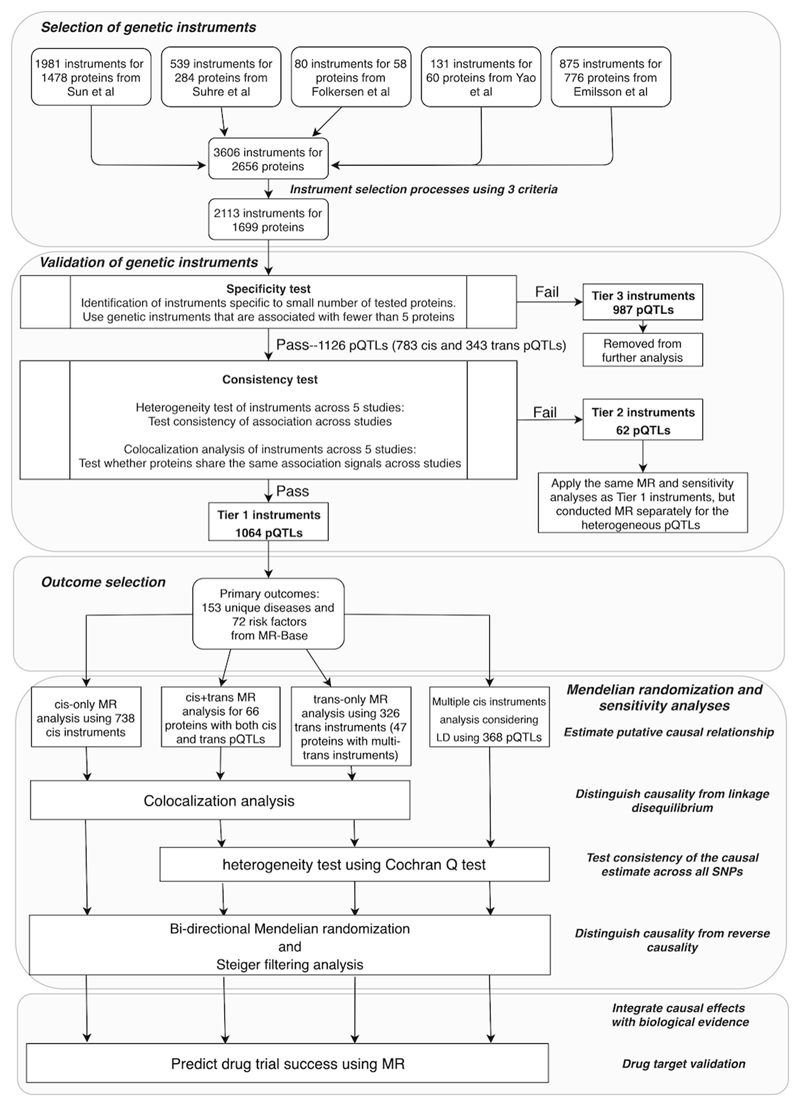
Figure 1 summarizes the genetic instrument selection and validation process. Briefly, we curated 3,606 pQTLs associated with 2,656 proteins from five GWAS9,13,14,15,16. After removing proteins and SNPs using criteria such as LD-pruning listed in Online Methods (Instrument selection), we retained 2,113 pQTLs for 1,699 proteins as instruments for the MR analysis (Supplementary Table 1). Among these instruments, we conducted further validation by categorizing them into three tiers based on their likely utility for MR analysis (Online Methods, Instrument validation): 1,064 instruments of 955 proteins with the highest relative level of reliability (tier 1); 62 instruments that exhibited SNP effect heterogeneity across studies (Supplementary Figs. 1 and 2), indicating uncertainty in the reliability of one or all instruments for a given protein (tier 2; Supplementary Tables 2 and 3); and 987 non-specific instruments that were associated with more than five proteins (tier 3). For the 263 tier 1 instruments associated with between two and five proteins, 68 of them influenced multiple proteins in the sample biological pathway and thus are likely to reflect vertical pleiotropy and remain valid instruments (Supplementary Note, Distinguishing vertical and horizontal pleiotropic instruments using biological pathway data)22.
Figure 1. Study design of this phenome-wide MR study of the plasma proteome.
The study included instrument selection and validation, outcome selection, four types of MR analyses, colocalization, sensitivity analyses, and drug target validation.
Among the 1,126 tier 1 and 2 instruments, 783 (69.5%) were cis-acting (within 500kb of the leading pQTL) and 343 were trans-acting. Of 1,002 proteins with a valid instrument, 765 had only a single cis or trans instrument, 66 were influenced by both cis and trans SNPs (Supplementary Table 4), and 153 had multiple conditionally distinct cis instruments (381 cis instruments shown in Supplementary Table 5).
Estimated effects of plasma proteins on human phenotypes
We undertook two-sample MR to systematically evaluate evidence for the causal effectsof 1,002 plasma proteins (with tier 1 and tier 2 instruments) on 153 diseases and 72 disease-related risk factors (Supplementary Table 6 and Online Methods, Phenotype selection). Overall, we observed 413 protein-trait associations with MR evidence (P < 3.5 x 10-7at aBonferroni-corrected threshold) using either cis or trans instruments (or both for proteins with multiple instruments).
Genetically filtering out predicted associations between proteins and phenotypes may indicate four explanations: causality, reverse causality, confounding by LD between the leading SNPs for proteins and phenotypes, or horizontal pleiotropy (Supplementary Fig.3). Given these alternative explanations, we conducted a set of sensitivity analyses to establish whether the MR association reflects a causal effect of protein on phenotype: tests of reverse causality using bi-directional MR24and MR Steiger filtering25,26; heterogeneity analyses for proteins with multiple instruments27, and colocalization analyses28to investigate whether the genetic associations with both protein and phenotype shared the same causal variant(Fig.1). To avoid unreliable inference from colocalization analysis due to the potential presence of multiple neighboring association signals, we also developed and performed pairwise conditional and colocalization analysis (PWCoCo) of all conditionally independent instruments against all conditionally independent association signals for the outcome phenotypes (Online Methods, Pairwise conditional and colocalization analysis; Fig. 2). For this study, MR and colocalization were the two methods filtering reliable associations. After the colocalization analysis, 283 of the 413 protein-phenotype associations had profiles supportive of causality.
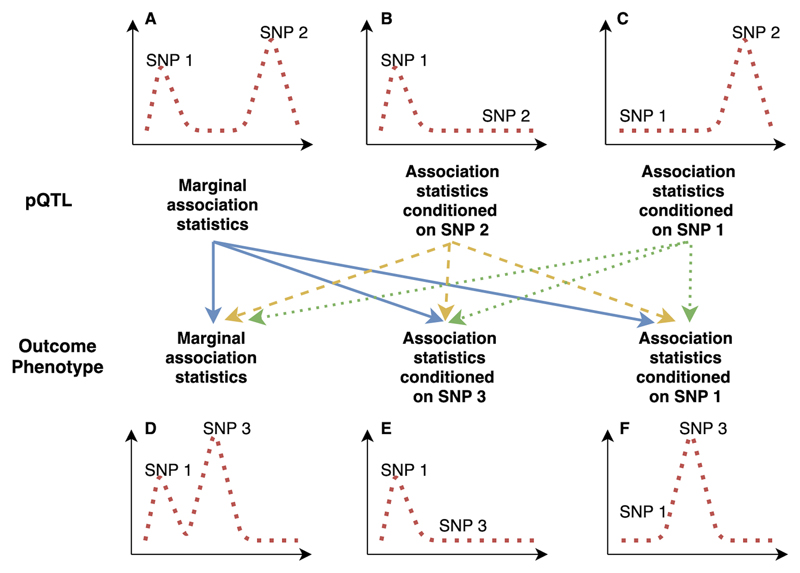
Figure 2. A demonstration of pairwise conditional and colocalization (PWCoCo) analysis.
Assume there are two conditional independent association pQTL signals (SNP 1 and SNP 2) and two conditional independent outcome signals (SNP 1 and SNP3) in the tested region. A naïve colocalization analysis using marginal association statistics will return weak evidence of colocalization (showed in regional plots A and D). By conducting the analyses conditioning on SNP 2 (plot B) and 1 (plot C) for the pQTLs and conditioning on SNP 1 (plot E) and 3 (plot F) for the outcome phenotype, each of the ninepairwise combinations of pQTL and outcome association statistics (represented as lines with different colors in the middle of this figure) will be tested using colocalization. In this case, the combination of plot B and plot E shows evidence of colocalization but the remaining eightdo not.
Estimating protein effects on human phenotypes using cis pQTLs
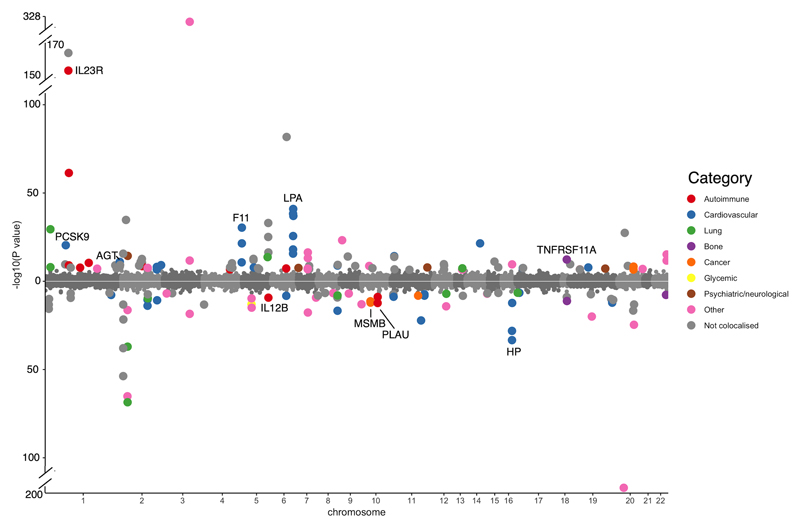
In the MR analyses using cis-pQTLs, we identified 111 putatively causal effects of 65 proteins on 52 phenotypes, with strong evidence of MR (P < 3.5 x 10-7) and colocalization (posterior probability> 80%; after applying PWCoCo) between the protein- and phenotype-associated signals (Fig.3 and Supplementary Table 7). A further 69 potential associations had evidence from MR but did not have strong evidence of colocalization (posterior probability < 80%; Supplementary Table 8), highlighting the potential for confounding by LD and the importance of colocalization analyses in MR of proteins. Evidence of potentially causal effects supported by colocalization was identified across a range of disease categories, including anthropometric phenotypes and cardiovascular and autoimmune diseases (Supplementary Note, Disease areas ofprotein-trait associations), and our findings replicated some previous reported associations (Supplementary Note, MR results replicated previous findings).
Figure 3. Miami plot for the cis-only analysis, with circles representing the MR results for proteins on human phenotypes.
The labels refer to top MR findings with colocalization evidence, with each protein represented by one label. The color refers to top MR findings with P < 3.09 x 10-7, where red refers to immune-mediated phenotypes, blue refers to cardiovascularphenotypes, green refers to lung-related phenotypes, purple refers to bone phenotypes, orange refers to cancers, yellow refers to glycemic phenotypes, brown refers to psychiatric phenotypes, pink refers to other phenotypesand grey refers to phenotypes that showed less evidence of colocalization. The x-axis is the chromosome and position of each MR finding in the cis region. The y-axis is the -log10 P value of the MR findings, MR findings with positive effects (increased level of proteins associated with increasing the phenotype level) are represented by filled circles on the top of the Miami plot, while MR findings with negative effects (decreased level of proteins associated with increasing the phenotype level) are on the bottom of the Miami plot.
Of 437 proteins with tier 1 or tier 2 cis instruments from Sunet al. 9and Folkersen et al. 14, 153 (35%) had multiple conditionally independent SNPs in the cis region identified by GCTA-COJO29(Supplementary Table 5). We applied an MR model that takes into account the LD structure between conditionally independent SNPs in these cis regions30. In this analysis, we identified 10 additional associations thathad not reached our Bonferroni corrected P-value threshold in the single-variant cis analysis. Generally, the MR estimates from the multi-cis MR analyses were consistent with the single-cis instrumented analyses (Supplementary Table 9).
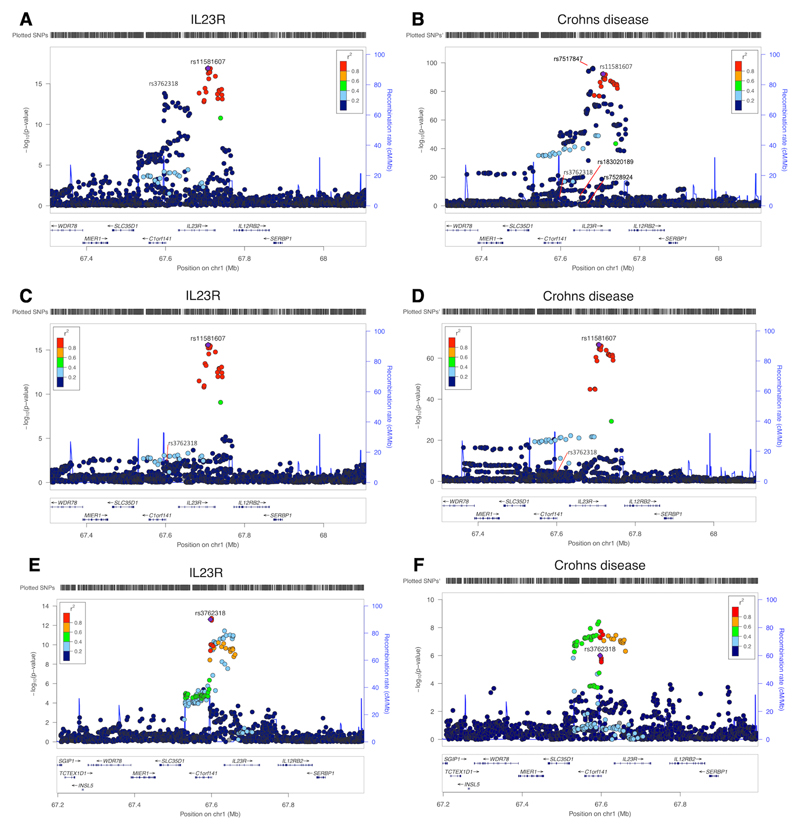
In regions with multiple cis instruments, 16 of the 111 top cis MR associations only showed evidence of colocalization after conducting PWCoCo analysis for both the proteins and the human phenotypes, where none was observed between marginal results (Supplementary Table 7). For example, interleukin 23 receptor (IL23R) had two conditionally independent cis instruments: rs11581607 and rs37623189. ConventionalMR analysis combining both instruments showed a strong association of IL23R with Crohn’s disease (OR = 3.22, 95% CI = 2.93 to 3.53, P = 6.93 x 10-131; Supplementary Table 9b). There were four conditionally independent signals (conditional P < 1 x 10-7) predicted for Crohn’s disease in the same region (data from de Lange et al. 31). In the marginal colocalization analyses, we observed no evidence of colocalization (Fig.4 and Supplementary Fig. 4, colocalization probability = 0). After performing PWCoCo with each distinct signal in an iterative fashion, we observed compelling evidence of colocalization between IL23R and one of the Crohn’s disease signals for the top IL23R signal (rs11581607) (colocalization probability = 99.3%), but limited evidence for the second conditionally independent IL23R hit (rs7528804) (colocalization probability = 62.9%). Additionally, for haptoglobin, which showed MR evidence for LDL-cholesterol (LDL-C), there were two independentcis instruments. There was little evidence of colocalization between the two using marginal associations (colocalization probability=0.0%). However, upon performing PWCoCo, we observed strong evidence of colocalization for both instruments (colocalization probabilities = 99%; Supplementary Table 10 and Supplementary Fig. 5). Both examples demonstrate the complexity of the associations in regions with multiple independent signals and the importance of applying appropriate colocalization methods in these regions. Of the 413 associations with MR evidence (using cis and trans instruments), 283 (68.5%) also showed strong evidence of colocalization using either a traditional colocalization approach (260 associations) or after applying PWCoCo (23 associations), suggesting that one third of the MR findings could be driven by genetic confounding by LD between pQTLs and other causal SNPs.
Figure 4. Regional association plots of IL23R plasma protein level and Crohn’s disease in theIL23R region.
a, b, Regional plots of IL23R protein level and Crohn’s disease without conditional analysis. Plot in b lists the sets of conditionally independent signals for Crohn’s disease in this region: rs7517847, rs7528924, rs183020189, rs7528804 (a proxy for the second IL23R hit rs3762318, r 2=0.42 in the 1000 Genome Europeans) and rs11209026 (a proxy for the top IL23R hit rs11581607, r 2=1 in the 1000 Genome Europeans), conditional P value < 1x10-7. c, Regional plot of IL23R with the joint SNP effects conditioned on the second hit (rs3762318) for IL23R. d, Regional plot of Crohn’s disease with the joint SNP effects adjusted for other independent signals except the top IL23R signal rs11581607. e, Regional plot of IL23R with the joint SNP effects conditioned on the top hit (rs11581607) for IL23R. f, Regional plot of Crohn’s disease with the joint SNP effects adjusted for other independent signals except the second IL23R signal rs3762318. The heatmap ofthe colocalization evidence for IL23R association on Crohn’s disease (CD) in the IL23R region is presented in Supplementary Figure 4.
Due to potential epitope-binding artefacts driven by protein-altering variants32, we also flag putatively causal links where the lead instrument is a protein-altering variant or is in high LD (r 2>0.8) with one (Supplementary Tables 7 and 8 filtered by column “VEP_pQTL_Ldproxy” including missense, stop-lost/gained, start-lost/gained and splice-altering variants).
Using trans-pQTLs as additional instrument sources
Trans pQTLs are more likely to influence targets though pleiotropic pathways. Among the 1,316 trans instruments we identified from five studies, 73.5% were associated with more than five proteins, compared with1.8 % of cis instruments(Supplementary Table 1). However, in the context of MR, includingnon-pleiotropic trans-pQTLs may increase the reliability of the protein-phenotype associations since (i) they will increase variance explained of the tested protein and increase power of the MR analysis; (ii) the causal estimate will not be reliant on a single locus, where multiple instruments exist; and (iii) further sensitivity analyses, such as heterogeneity test of MR estimatesacross multiple instruments, can be conducted. Therefore, we extendedour MR analyses to include 343 non-pleiotropic trans instruments (Supplementary Fig.6).
To utilize trans instruments, we first combined cis and trans instruments for 66 proteins that had both cis and trans instruments (noted as cis + trans analysis). However, none reached our pre-defined Bonferroni-corrected threshold, and only two protein-phenotype associations showed even suggestive evidence (P < 1 x 10-5) (Supplementary Table 11). Further, after including trans instruments, 17 of the cis-only signals were attenuated. Secondly, we performed trans-only MR analyses of 293 proteins and identified 158 associations with 44 phenotypes that also had strong evidence (posterior probability > 0.8) of colocalization (Supplementary Table 12). A further 54 trans-only MR associations did not have strong evidence of colocalization (Supplementary Table 13).
Some of the trans analyses with MR and colocalization evidence suggest causal pathways that are confirmed by evidence from rare pathogenic variants or existing therapies. For example, although we had no cis instrument for Protein C (Inactivator Of Coagulation Factors Va And VIIIa) (PROC) (Supplementary Fig.7a), we found evidence for a causal association between PROC levels and deep venous thrombosis (P = 1.27 x 10-10; colocalization probability > 0.9) using a trans pQTL, rs867186(Supplementary Fig. 7b), which is a missense variant in PROCR 33, the gene encoding the endothelial protein C receptor (EPCR). Individuals with mutations in PROC have protein C deficiency, a condition characterized by recurrent venous thrombosis for which replacement protein C is an effective therapy.
From 47 proteins with multiple trans instruments, we identified four additional MR associations, but none showed strong evidence of colocalization (Supplementary Table 13) and little evidence of heterogeneity (Supplementary Table 14).
Estimating protein effects on human phenotypes using pQTLs with heterogeneous effects across studies
Among the 2,113 selected instruments, we checked whether the 1,062 instruments with association information in at least two studies showed consistent effect size across studies (Supplementary Table 15). For these SNPs, we found that 62 showed evidence of difference in effect size across studies (tier 2 instruments), for which we performed MR analyses using the most significant SNP across studies and report the findings with caution. Some proteins that are targets of approved drugs were found to have potential causal effects in this analysis, such as interleukin-6 receptor (IL6R) on rheumatoid arthritis (RA)34, and coronary heart disease(CHD)21(Supplementary Table 16). Tocilizumab, a monoclonal antibody against IL6R, is used to treat RA, while canakinumab, a monoclonal antibody against interleukin-1 beta (an upstream inducer of interleukin-6), has been shown to reduce cardiovascular events specifically among patients who showed reductions in interleukin-635.
As another test of heterogeneity across studies, where the same protein was measured in two or more studies, we performed colocalization analysis of each pQTL (in one study) against the same pQTL (in another study) for the two studies in which we had access to full summary results (Sun et al. 9 and Folkersen et al. 14). Of the 41 proteins measured in both studies, 76pQTLs could be tested using conventional colocalization and PWCoCo (Supplementary Table 15). We found weak evidence of colocalization for 51 pQTLs (posterior probability < 0.8), which suggested either two different signals were present within the test region or the protein has a pQTL in one study but not in the other. In either case, as one of the two distinct signals may be genuine, we performed MR analysis of these 25 pQTLs using instruments from each study separately. Eight associations had MR evidence, but only one showed colocalization evidence (IL27 levels on human height; Supplementary Table 17).
Sensitivity analyses to evaluate reverse causality
For potential associations between proteins and phenotypes identified in the previous analyses, we undertook two sensitivity analyses to highlight results due to reverse causation: bi-directional MR24and Steiger filtering25(Online Methods, Distinguishing causal effects from reverse causality). In general, we found little evidence of reverse causality for genetic predisposition to diseases on protein level changes (more details in Supplementary Note, Bi-directional MR and Steiger filtering results; Supplementary Data 1).
Drug target prioritization and repositioning using phenome-wide MR
Given that human proteins represent the major source of therapeutic targets, we sought to mine our results for targets of molecules already approved as treatments or in ongoing clinical development. We first compared MR findings for 1,002 proteins against 225 phenotypes with historic data on progression of target-indication pairs in Citeline’s PharmaProjects (downloaded on 9th May 2018). Of 783 target-indication pairs with an instrument for the protein and association results for a phenotype similar to the indication for which the drug had been trialled, 9.2% (73 pairs) had successful (approved) drugs, 69.1% had failed drugs (including 195 failed drugs in the clinical stage and 354 drugs that failed in the preclinical stage) and 20.3% were for drugs still in development (161 pairs). The 268 pairs for successful (73) or failed (195) drugs were included in further analyses (Supplementary Table 18). We observed eight target-indication pairs of successful drugs with MR and colocalization evidence of a potentially causal relationship between protein and disease (Supplementary Table 19). After removing duplicate genetic evidence for related indications for the same therapy (Online Methods, Drug target validation and repositioning), six successful drugs remained from 214 pairs (Supplementary Table 20). In addition to the PROC and IL6R examples discussed earlier, we found Proprotein convertase subtilisin/kexin type 9 (PCSK9) (target for evolocumab) for hypercholesterolemia and hyperlipidaemia, Angiotensinogen (AGT) for hypertension, IL12B for psoriatic arthritis and psoriasis, and TNF Receptor Superfamily Member 11a (TNFRSF11A) for osteoporosis. For each of these examples, the direction of effect between circulating protein and disease risk was consistent with the therapeutic mechanism, except IL6R and PROC at first sight. However, for IL6R and PROC, the alleles associated with higher soluble protein levels have been shown to also lead to lower intracellular pathway activation36,37, indicating consistency of direction with the therapeutic approach. These examples highlight the importance of careful examination of the biological mechanisms underlying plasma pQTLs to enable translation. Further removing associations potentially driven by protein-altering variants, as well as drugs that were in large part motivated by genetic evidence (e.g. PCSK9 fits both exclusion criteria), comparisons of the remaining 191 pairs indicated that protein-phenotype associations with MR and colocalization evidence remained more likely tobecome successful target-indication pairs (Table 1). Although we acknowledge the limited sample size of the test set, this raises enthusiasm for the utility of pQTL MR analyses with colocalization as a method for target prioritization.
Table 1. Enrichment analysis comparing target-indication pairs with or without MR and colocalization evidence.
| YES | NO | |
|---|---|---|
| YES | 4 | 40 |
| NO | 0 | 147 |
The protein-phenotype association pairs were grouped into four categories: (i) pairs with both MR/colocalization indications of causality and drug trial success; (ii) pairs with MR and colocalization evidence but no drug trial evidence; (iii) pairs with no strong MR or colocalization evidence but with drug trial evidence; and (iv) pairs with no strong MR, colocalization or drug trial evidence. The cut-off for MR evidence was P < 3.5 x 10-7; the cut off for colocalization evidence was posterior probability > 80%. The drug trial evidence was obtained from PharmaProjects database. The MR and colocalization analysis results involved in this analysis including both tier 1 and tier 2 instruments in both cis and trans region. More results comparing MR and trial evidence for cis-only and tier 1 instruments can be found in Supplementary Table 20.
Previous efforts have highlighted the opportunities and challenges of using genetics for drug repositioning38. Weidentified three approved drugsfor which we found pQTL MR and colocalization evidence for five phenotypes other than the primary indication and 23 drug targets under development for 33 alternative phenotypes (Supplementary Table 21). An example of urokinase-type plasminogen activator (PLAU) levels associated with lower inflammatory bowel disease (IBD) risk is presented in the Supplementary Note (Case study for drug repurposing) and Supplementary Figure 8.
We also evaluated drugs in current clinical trials and identified eight additional protein-phenotype associations with MR and colocalization evidence (Supplementary Table 22), for which we observe MR evidence implicating an increased likelihood of success.
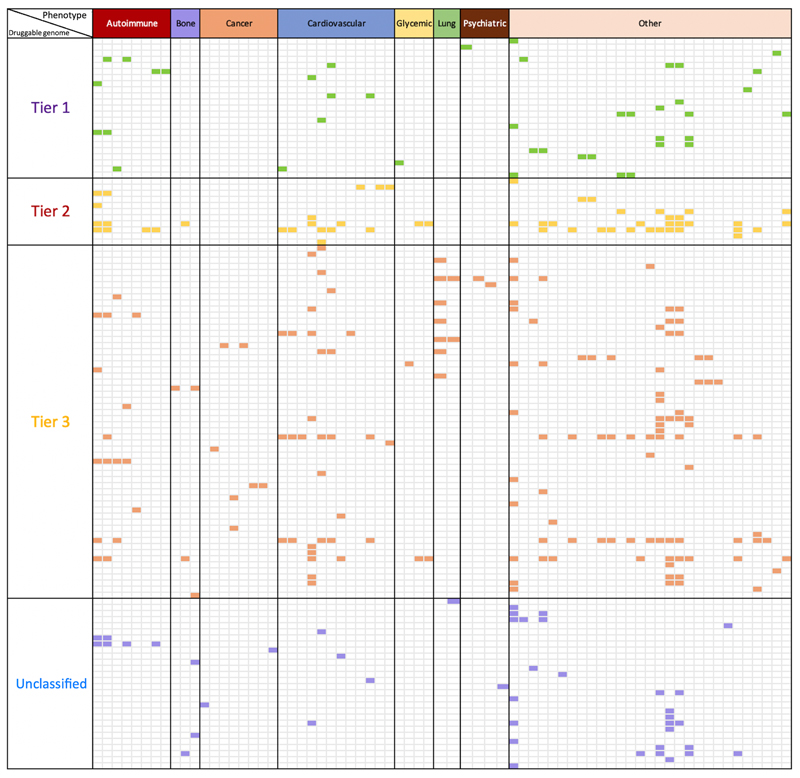
Finally, we compared the 1,002 instrumentable proteins (i.e. those that passed our instrument selection procedure) against the druggable genome39, and found that 682 of the 1,002 (68.1%) instrumentable proteins overlapped with the druggable genome (Supplementary Table 23 and Online Methods, Enrichment of proteome-wide MR with the druggable genome). We conducted a further enrichment analysis to assess the overlap between putative causal protein-phenotype associations and the druggable genome (Supplementary Table 24). Of the 295 top findings (120 proteins on 70 phenotypes) with both MR and colocalization evidence, 250 of them (87.7%) overlapped with the druggable genome (Fig.5). This enrichment analysis will become more valuable with the continuous evolution of the druggable genome38.
Figure 5. Enrichment of phenome-wide MR of the plasma proteome with the druggable genome.
In this figure, we only show proteins with convincing MR and colocalization evidence with at least one of the 70 phenotypes. The x-axis shows the categories of 70 human phenotypes, where the phenotypes have been grouped into 8 categories: 8 autoimmune diseases (red), 3 bone phenotypes (purple), 8 cancers (orange), 12 cardiovascular phenotypes (blue), 4 glycemic phenotypes (yellow), 2 lung phenotypes (green), 4 psychiatric phenotypes (brown), and 29 other phenotypes (pink). The y-axis presents the tiers of the druggable genome (as defined by Finan et al.39) of 120 proteins under analysis, where the proteins have been classified into 4 groups based on their druggability: tier 1 contains 23 proteins that are efficacy targets of approved small molecules and biotherapeutic drugs, tier 2 contains 11 proteins closely related to approved drug targets or with associated drug-like compounds, tier 3 contains58 secreted or extracellular proteins or proteins distantly related to approved drug targets, and 28 proteins have unknown druggable status (Unclassified). The cells with colors are protein-phenotype associations with strong MR and colocalization evidence. Cells in green are associations overlapping with the tier 1 druggable genome, while cells in yellow, red or purple were associations with tier 2, tier 3 or unclassified. More detailed information is shown in Supplementary Table 24.
Discussion
MR analysis of molecular phenotypes against disease phenotypes provides a promising opportunity to validate and prioritizenovel or existing drug targets through prediction of efficacy and potential on-target beneficial or adverse effects40. Our phenome-wide MR study of the plasma proteome employed fivepQTL studies to robustly identify and validate genetic instruments for thousands of proteins. We used these instruments to evaluate the potential effects of modifying protein levels on hundreds of complex phenotypes available in MR-Base23in a hypothesis-free approach17. We confirmed that protein-phenotype associations with both MR and colocalization evidence predicted a higher likelihood of a particular target-indication pair being successful and highlight 283 potentially causal associations. Collectively, we underline the important role of pQTL MR analyses as an evidence source to support drug discovery and development and highlight a number of key analytical approaches to support such inference.
In particular, we note the distinct opportunities and methodological requirements for MR of molecular phenotypes, such as transcriptomics and proteomics, compared to other complex exposures. For example, the number of instruments is often limited for proteins, restricting the opportunity to apply recently developed pleiotropy robust approaches27,41. New methods such as MR-robust adjusted profile scoring (MR-RAPS)42 allow inclusion of many weak instruments in the MR analysis and have been applied to a recent proteome-wide MR study10. However, we note some examples where inclusion of multiple weaker instruments can reduce power and yield different results to those based on cis instruments alone40,43, and we note very limited additional gain from inclusion of trans instruments. A major advantage of proximal molecular exposures is the ability to include cis instruments (or interpretable trans instruments) with high biological plausibility, limiting the likelihood of horizontal pleiotropy22,44. Further, we note the limited gain from inclusion of trans instruments in our analysis. However, undue focus on single SNP MR approaches brings susceptibility to other pitfalls, such as the inability to examine heterogeneity of effect and to evaluate and remove potential epitope artefacts.
To provide robust MR estimates for proteins, we note the important role of a number of sensitivity analyses following the initial MR in order to distinguish causal effects of proteins from those driven by horizontal pleiotropy, genetic confounding through LD45and/or reverse causation25. Of note, only two-thirds of our putative causal associations had strong evidence of colocalization, suggesting that a substantial proportion of the initial findings were likely to be driven by genetic confounding through LD between pQTLs and other disease-causal SNPs. To avoid misleading results, we suggest that for regions with multiple molecular trait QTLs, it is important to consider methods such as PWCoCo, which can avoid the assumptions of traditional colocalization approaches of just a single association signal per region46. In the current study, application of PWCoCo identified evidence of colocalization for 23 additional protein-phenotype associations hidden to marginal colocalization46. We note that recent recommendations support the use of colocalization as a follow up analysis to reduce false positives47.
An important limitation of this work is that protein levels are known to differ between cell types48. In this study, we have estimated the role of protein measured in plasma on a range of complex human phenotypes but are unable to assess the relevance of protein levels in other tissues. WhileeQTL studies highlight a large proportion of eQTLs being shared across tissues37, there are many which show cell type and state specificity49, highlighting the potential value of applying the current approach to data from proteomics analyses in other cell types and tissues. We also hypothesize that, in instances with multiple conditionally distinct pQTLs but where we observe colocalization of only certain conditionally distinct pQTL-phenotype pairs, this may reflect underlying cell- and state-specific heterogeneity in bulk plasma pQTLs, among which only certain cell-types or states are causal50. Although pQTL studies have not yet been performed as systematically across tissues or states as eQTL studies, it remains encouraging that our analyses using plasma proteins identify associations across a range of disease categories, including for psychiatric diseases for which we may expect key proteins to function primarily in the brain.
Evaluating the potential of MR to inform drug target prioritization, we demonstrated that the presence of pQTL MR and colocalization evidence for a target-indication pair predicts a higher likelihood of approval. One of the limitations of our approach is the lack of comprehensive coverage of genetic data for all phenotypes for which drugs are in development, as well as our inability to instrument the entire proteome through pQTLs. As such, ongoing expansions in the scale, diversity and availability of GWAS will be important in providing more precise estimates of the value of MR and colocalization in drug target prioritization and in enabling its broader application.
Another potential limitation of our work is the presence of epitope-binding artefacts driven by coding variants that may yield artefactual cis-pQTLs32. In particular, such instances may lead to false negative conclusions where, in the presence of a silent missense variant causing an artefactual pQTL but with no actual effect on protein function or levels, we do not correctly instrument the target protein. In instances where the missense variant appears to be driving the association with the phenotype, we suggest that causal inference may remain valid but inference on direction of association is challenged. Finally, the limited coverage of the proteome afforded by current technologies leavesthe possibility of undetected pleiotropy of instruments. While cis-pQTLs are less likely to be prone to horizontal pleiotropy than trans-pQTLs, it is well known from studies of gene expression that cis variants can influence levels of multiple neighboring genesand hence the same is likely to be true for proteins. Future larger GWAS of the plasma proteome are likely to uncover many more variant-protein associations, increasing the apparent pleiotropy of many pQTLs.
In conclusion, this study identified 283 putatively causal effects between the plasma proteome and the human phenome using the principles of MR and colocalization. These observations support, but do not prove, causality, as potential horizontal pleiotropy remains an alternative explanation. Our study provides both an analytical framework and an open resource to prioritize potential new targetsand a valuable resource for evaluation of both efficacy and repurposing opportunities by phenome-wide evaluation of on-target associations.
Methods
Instrument selection
pQTLs from five GWAS9,13-16 were used for the instrument selection (Fig. 1). We first mapped SNPs to genome build GRCh37.p13 coordinates and then used the following criteria to select instruments:
We selected SNPs that were associated with any protein (using a P-value threshold ≤ 5 x 10-8) in at least one of the five studies, including both cis and trans pQTLs.
Due to the complex LD structure of SNPs within the human Major Histocompatibility Complex (MHC) region, we removed SNPs and proteins coded for by genes within the MHC region (chr6: from 26Mb to 34Mb).
We then conducted linkage disequilibrium (LD) clumping for the instruments with the TwoSampleMR R package23 to identify independent pQTLs for each protein. We used r 2< 0.001 as the threshold to exclude dependent pQTLs in the cis (or trans) gene region.
After instrument selection, 2,113 instruments were kept for further instrument validation (Supplementary Table 1). The instrument selection process, and the number of instruments for proteins at each step in the process, is illustrated in Figure 1.
We incorporated conditionally distinct signals from protein association data through systematic conditional analysis. Of the fivestudies, Sun et al. 9 reported conditionally distinct results for both cis and trans pQTLs, which have been used in our study. Folkersen et al.14 have shared summary statistics, with which we performed approximate conditional analyses ourselves using GCTA-COJO29, with genotype data from mothers in the Avon Longitudinal Study of Parents and Children (ALSPAC) as the LD reference panel51,52(a description of the ALSPAC cohort can be found in Supplementary Note, Description of ALSPAC study). Conditionally independent signals in the cis region for Sun et al. and Folkersen et al. are reported inSupplementary Table 5.
Instrument validation
For the 2,113 instruments, we further classified them into three groups (noted as tier 1, tier 2 and tier 3 instruments) using two major instrument-filtering steps: a specificity test and a consistency test. More details of instrument validation, including harmonization of proteins and instruments and statistical tests for consistency can be found in the Supplementary Note (The protocol of the instrument validation).
Test estimating instrument specificity
Absence of horizontal pleiotropy is one of the core assumptions for MR. This assumes that the genetic variant should only be related to the outcome of interest through the instrumented exposure. We noted that some SNPs were associated with more than one protein. For example, APOE SNP rs7412 is associated with a set of proteins such as ADAM11, APBB2, and APOB. We plotted a histogram of the number of proteins each instrument was associated with (Supplementary Fig.6) and considered instruments associated with more than 5 proteins as highly pleiotropic and assigned them as tier 3 instruments (which were excluded from all analyses). For instruments associated with fewer than (or equal to) five proteins, we reported the number of proteins each of them (and their proxies with LD r 2>0.5) was associated with to indicate the level of potential pleiotropy.
To further distinguish vertical and horizontal pleiotropy for these instruments, we used biological pathway information from Reactome (https://reactome.org/) and protein-protein interaction information from STRING DB (https://string-db.org/) implemented in EpiGraphDB (www.epigraphdb.org; Supplementary Note, Distinguishing vertical and horizontal pleiotropic instruments using biological pathway data). After this analysis, 68 instruments associated with multiple proteins were mapped to the same pathway (or same PPI) and were considered as valid instruments. Given there are other pathways and PPIs that may be not included in Reactome and STRING, we kept tier 1 and 2 instruments associated with 1 to 5 proteins for the main MR analysis, but we recorded the number of proteins and number of pathways these instruments are associated with as an indication of potential pleiotropy.
Consistency test estimating instrument heterogeneity across studies
Among the 2,113 pQTLs selected as instruments, we looked up available protein GWAS results (Sun et al. 9, Suhre et al. 13 and Folkersen et al. 14 with full GWAS summary statistics; Yao et al. 15 and Emilsson et al. 16 with pQTLs only) and found 1,062 pQTLs (or proxies with r2>0.8) with association information in at least two studies (Supplementary Table 15). We then tested the beta-beta correlation using the Pearson correlation function in R. The results of the beta-beta correlations of SNP effects for each pair of studies and the number of SNPs included in each correlation analysis can be found in Supplementary Table 2.
We further performed two consistency tests on the instruments thatwere present across studies: (i) pairwise Z test; (ii) colocalization analysis of proteins across studies (details of the analyses in Supplementary Note, The protocol of the instrument validation). Instruments showing evidence of high heterogeneity across studies using either the pair-wise Z test (pairwise Z > 5) or colocalization analysis (PP < 80%), were flagged as tier 2 instruments. Recognizing that lack of replication and effect heterogeneity does not preclude at least one of these effects being genuine, we used these instruments separately for the follow-up genetic analyses (Supplementary Table 3) and reported the findings with caution.
We designated instruments passing both pleiotropy and consistency tests as tier 1instruments and used them as primary instruments for the MR analysis.
Identifying cis and trans instruments
We further split tier 1 instruments into two groups: (i) cis-acting pQTLs within a 500-kb window from each side of the leading pQTL of the protein were used for the initial MR analysis (defined as the cis-only analysis)45; (ii) trans-acting pQTLs outside the 500-kb window of the leading pQTL were designated as trans instruments. While trans instruments may be more prone to pleiotropy, their inclusion could increase statistical power as well as the scope of downstream sensitivity analyses (e.g. tests for heterogeneity between instruments). Therefore, for the proteins with cis instruments, we also looked for additional trans instruments, and if these were available, we conducted further MR analyses using both sets of instruments (defined as the "cis + trans" analysis).
Forcis instruments, we looked up their predicted consequence via Variant Effect Predictor53hosted by Ensembl. We identified coding variants (including missense, stop-lost/gained, start-lost/gained and splice-altering variants) sinceepitope-binding artefacts driven by coding variants may yield artefactual cis pQTLs32. We then conducted a sensitivity MR analysis that excluded cis instruments thatare in the coding region to further avoid the potential issue of epitope-binding artefacts driven by coding variants.
Phenotype selection
We obtained effect estimates for the association of the pQTLs with complex human phenotypes using GWAS summary statistics that were included in the MR-Base database (http://www.mrbase.org). We selected GWAS with the greatest excepted statistical power when multiple GWAS records for the same phenotype were available in MR-Base. Diseases were defined as primary outcomes. Risk factors were defined as secondary outcomes. After selection, 153 diseases and 72 risk factors (such as lipids and glucose phenotypes) were included as outcomes for the MR analyses (Supplementary Table 6).
Causal inference and sensitivity analyses
The following sections describe the two-sample MR analyses using single or small numbers of instruments on 153 diseases and 72 risk factors. To identify possible violations of assumptions of MR and to distinguish between the aforementioned scenarios in Supplementary Figure 3, we therefore conducted the following sensitivity analyses: colocalization analysis28, tests for heterogeneity between instrumental SNPs27, bi-directional MR24, and Steiger filtering25,26(Fig.1).
Estimating the causal effects of proteins on human phenotypes using MR
In the initial MR analysis, proteins were treated as the exposures and 225 complex human phenotypes as the outcomes (Fig. 1, Estimate putative causal relationship). Due to high correlation among some of the tested phenotypes (e.g. coronary heart disease (CHD) and myocardial infarction), we used the PhenoSpD method54,55to provide a more appropriate estimate of the number of independent tests. We selected a P-value threshold of 0.05, corrected for the number of independent tests, as our threshold for prioritizing MR results for follow up analyses (number of tests= 142,857; P < 3.5 x 10-7).
MR analysis using single locus instruments
First, the strongest cis pQTL variants for each protein were used as the instrumental variable (described as ‘single cis’ analysis). The Wald ratio56method was used to obtain MR effect estimates. In this analysis, the MR effect estimates were sensitive to the particular choice of pQTLs, since only the most strongly associated SNPs within each genomic region were used as instruments. Burgess et al. recently suggested that more precise causal estimates can be obtained using multiple genetic variants from a single gene region, even if the variants are correlated30,57. We used multiple conditional independent cis SNPs (Supplementary Table 5) against all 225 phenotypes to further evaluate the MR findings from our initial MR analysis (described as ‘multiple cis’ analysis). A generalized inverse variance weighted (IVW) model considering the LD pattern between the multiple cis SNPs was used to estimate the MR effects, where the pairwise LD (r 2) were obtained from the 1000 Genomes European ancestry reference samples.
MR analysis using multi-locus instruments
Among the measured proteins reported in Sun et al. 9, 34% had both cis and trans pQTLs and 30% had only trans pQTLs. We also conducted MR on proteins with both cis and trans pQTLs (noted as the cis + trans MR analysis) and proteins with only trans pQTLs (noted as trans-only analysis). In the cis + trans MR analysis, we tested the protein-phenotype associations of 66 proteins with both cis and trans instruments. The IVW method was used to obtain MR effect estimates. In the trans-only MR analysis, we used 351 trans instruments for 298 proteins. The IVW method was used when two or more trans instruments were included in the analysis, whereas the Wald ratio method was used when only one trans instrument was included in the analysis.
MR analysis software
The majority of MR analyses (including Wald ratio, IVW, bi-directional MR, MR Steiger filtering and heterogeneity test across multiple instruments) were conducted using the MR-Base Two Sample MR R package (github.com/MRCIEU/TwoSampleMR)23. The IVW analysis considering LD pattern was conducted using the MendelianRandomization R package58. The MR results were plotted as forest plots and Miami plots using code derived from the ggplot2 package in R.
Distinguishing causal effects from genomic confounding due to linkage disequilibrium
Results that survived the multiple testing threshold in the MR analysis were evaluated using a stringent Bayesian model (colocalization analysis) to estimate the posterior probability (PP) of each genomic locus containing a single variant affecting both the protein and the phenotype28. For protein and phenotype GWAS lacking sufficient SNP coverage or missing key information (e.g. allele frequency or effect size), we conducted the “LD check” analysis(more details of the two methods in Supplementary Note, Linkage disequilibrium check).
Pairwise conditional and colocalization analysis
The presence of multiple conditionally distinct association signals within the same genomic region will influence the performance of colocalization analysis. We therefore developed an analysis pipeline to integrate conditional and colocalization approaches for regions with multiple conditionally independent pQTLs. Where there was convincing MR evidence below the P-value threshold of 3.5 x 10-7, but no good evidence of colocalization using the marginal SNP effects of the exposures and outcomes (in total 148 MR associations in both cis and trans regions), we performed pairwise colocalization analyses of all conditionally distinct pQTLs against all identified conditionally distinct association signals in the outcome data (noted as pair-wise conditional and colocalization analysis: PWCoCo). The conditional analysis for proteins and human phenotypes was conducted using the GCTA-COJO package29, with genotype data from mothers in the Avon Longitudinal Study of Parents and Children (ALSPAC) as the LD reference panel51,52 (a description of the ALSPAC cohort can be found in Supplementary Note, Description of ALSPAC study). Figure 2 demonstrates the ninepossible pair-wise combinations of various conditional signals for proteins and phenotypes at which there are two independent signals in the region (Supplementary Table 27).
For protein-phenotype associations that only showed colocalization evidence after we applied PWCoCo, we recorded the PWCoCo model that showed colocalization evidence in a new column “PWCoCo_model”, in Supplementary Tables 7, 8, 11, 12, 13, 16 and 17.
Heterogeneity test and directionality test of MR findings
For MR analyses using two or more instruments, we conducted heterogeneity tests to estimate the variability in the causal estimates obtained for each SNP (i.e. how consistent is the causal estimate across all SNPs used as separate instruments) (Fig. 1, Consistency of the causal estimate across all SNPs). Cochran’s Q test statistic was calculated for the IVW analyses, which is expected to be chi-squared distributed with number of SNPs minus one degrees of freedom27. Lower heterogeneity suggests a lower chance of violations of assumptions in MR estimates, such as the presence of confounding through horizontal pleiotropy59.
In order to mitigate the potential impact of reverse causality (i.e. the hypothesised outcome actually has a causal effect on the hypothesised exposure and not vice versa), we used two approaches to identify directions of causality: bi-directional MR and Steiger filtering (more details in Supplementary Note, Directionality test).
Drug target validation and repositioning
Approved drug targets have previously been shown to be enriched for gene-phenotype associations6. We therefore wished to assess whether approved drug targets were enriched for protein-phenotype associations, as obtained in the present study using MR. We assessed the support for approved drug targets among our MR findings using Fisher’s exact test. Target-indication pairs for successful and failed drugs were identified using a manually annotated version of PharmaProjects database from Citeline (https://pharmaintelligence.informa.com/). The phenotypes used in the MR analyses and the indications listed in Citeline’s PharmaProjects (downloaded on 9th May 2018) were then manually mapped to MeSH headings as a common ontology. This allowed us to match the protein-phenotype associations with corresponding target-indication pairs. To improve this matching, we implemented a similarity matrix, derived from all MeSH headings in the manual mapping, and retained matches with a relative similarity greater than 0.7 for our analyses (the similarity matrix has been previously described in Nelson et al. 6). We then compared whether the target-indication pair represented a successful or failed drug against whether there was a signal or not for the corresponding protein-phenotype pair among our MR findings. For the purposes of this test, a signal was defined as an MR result with P < 3.5 x 10-7 (which is the Bonferroni P-value threshold of the MR analysis) with supporting evidence from colocalization analysis. We further conducted a set of sensitivity analyses based on the following criteria to increase the reliability of the enrichment analysis:
We checked the direction of effect of MR findings and drug trial results for the eightapproved drugs using therapeutic direction information from PharmaProjects.
For target-indication pairs linked to similar phenotypes (for example, the same target associated with angina and myocardial infarction), we removed one of them to avoid double counting the same association.
To avoid the influence of epitope-binding artefacts, we removed MR results estimated using missense variants as an instrument.
We checked whether approved drugshad been motivated by genetics from Drug Bank (https://www.drugbank.ca/), which may have inflated the OR estimate.
In total, we removed 75 target-indication pairs based on criteria2 (45 pairs), 3 (23 pairs) and 4 (2 pairs; some pairs appeared in multiple situations) and conducted the comparison between protein-phenotype associations using MR and target-indication pairs from PharmaProjects, both on each criterion separately and on all criteria together (Supplementary Table 20).
Phenome-wide MR has demonstrated the potential to validate, repurpose and predict on-target side effects of drug targets. Of the protein-phenotype associations that showed evidence of colocalization identified in the cis-only, cis+trans, trans-only or MR analyses using pQTLs with heterogeneous effects across studies (noted as tier 2 instruments), we first looked up how many proteins with MR evidence were established drug targets in the Informa PharmaProjects database. We then looked up how many of the associations were established target-indication pairs in the PharmaProjects database. More importantly, we predicted the potential adverse effects and repositioning opportunities of all marketed drugs and drugs under development using phenome-wide MR.
Enrichment of proteome-wide MR with the druggable genome
Previously, Finan et al. 39systematically identified 4479 genes as the newest druggable genome compendium. This study stratified the druggable genome set into three tiers. Tier 1 (1,427 genes) included efficacy targets of approved small molecules and biotherapeutic drugs, as well as targets modulated by clinical-phase drug candidates;tier 2 was composed of 682 genes encoding proteinsclosely related to drug targets, or with associated drug-like compounds;and tier 3 contained 2,370 genes encoding secreted or extracellular proteins, distantly related proteins to approved drug targets, and members of key druggable gene families not already included in tier 1 or tier 2. We assessed whether the 1,002 proteins we selected for the MR analyses overlapped with the 4,479 genes from the druggable genome (Supplementary Table 23). The proteins were mapped based on the HGNC name of the encoding genes. We further assessed the overlap based on whether the protein had cis or trans instruments and based on the druggable genome tiers.
In addition to the above comparison between instrumentable and druggable genome, we also assessed the enrichment of top pQTL MR findings with the druggable genome. 295 protein-phenotype associations (120 proteins on 70 phenotypes) with both MR and colocalization evidence were selected for this analysis. We stratified the 120 proteins into 4 groups based on their druggability: tier 1 contained 23 proteins, tier 2 contained 11 proteins, tier 3 contained 58 proteins, and 28 proteins remained unclassified. The 70 phenotypes were stratified into 8 groups: 8 autoimmune diseases, 3 bone phenotypes, 8 cancer phenotypes, 12 cardiovascular phenotypes, 4 glycemic phenotypes, 2 lung phenotypes, 4 psychiatric phenotypes and 29 other phenotypes. The protein-phenotype associations with MR and colocalization evidence were colored separately based on their druggability tiers. More details of this enrichment analysis are shown in Figure 5 and Supplementary Table 24.
Supplementary Material
Acknowledgements
We are extremely grateful to all the families who took part in the ALSPAC study, the midwives for their help in recruiting them, and the whole ALSPAC team, which includes interviewers, computer and laboratory technicians, clerical workers, research scientists, volunteers, managers, receptionists and nurses. We acknowledge Jack Bowden for statistical support and advice relating to MR-Egger regression.
This publication is the work of the authors, and Jie Zheng will serve as guarantor for the contents of this paper. J. Z. is funded by a Vice-Chancellor Fellowship from the University of Bristol. This research was also funded by the UK Medical Research Council Integrative Epidemiology Unit(MC_UU_00011/1 and MC_UU_00011/4), GlaxoSmithKline, Biogen and the Cancer Research Integrative Cancer Epidemiology Programme (C18281/A19169). The UK Medical Research Council and Wellcome (Grant ref: 102215/2/13/2) and the University of Bristol provide core support for ALSPAC. T. R. G. holds a Turing Fellowship with the Alan Turing Institute. A comprehensive list of grants funding is available on the ALSPAC website (http://www.bristol.ac.uk/alspac/external/documents/grant-acknowledgements.pdf). G. H. is funded by the Wellcome Trust and the Royal Society [208806/Z/17/Z]. M. V. H. is supported by a British Heart Foundation Intermediate Clinical Research Fellowship (FS/18/23/33512) and the National Institute for Health Research Oxford Biomedical Research Centre. This study was funded/supported by the NIHR Biomedical Research Centre at University Hospitals Bristol NHS Foundation Trust and the University of Bristol (GDS and TRG)[*]. This work was supported by the Elizabeth Blackwell Institute for Health Research, University of Bristol and the Medical Research Council Proximity to Discovery Award. P. E. is supported by CRUK [C18281/A19169]. S. L. is funded by the Bau Tsu Zung Bau Kwan Yeun Hing Research and Clinical Fellowship (200008682.920006.20006.400.01) from the University of Hong Kong. J. D. is funded by the National Institute for Health Research [Senior Investigator Award]. J. D.sits on the International Cardiovascular and Metabolic Advisory Board for Novartis (since 2010), the Steering Committee of UK Biobank (since 2011), the MRC International Advisory Group (ING) member, London (since 2013), the MRC High Throughput Science ‘Omics Panel Member, London (since 2013), the Scientific Advisory Committee for Sanofi (since 2013), the International Cardiovascular and Metabolism Research and Development Portfolio Committee for Novartis, and the Astra Zeneca Genomics Advisory Board (2018). P. C. H. is supported by CRUK Population Research Postdoctoral Fellowship C52724/A20138.
Participants in the INTERVAL randomized controlled trial were recruited with the active collaboration of NHS Blood and Transplant England (www.nhsbt.nhs.uk), which has supported field work and other elements of the trial. DNA extraction and genotyping was co-funded by the National Institute for Health Research (NIHR), the NIHR BioResource (http://bioresource.nihr.ac.uk) and the NIHR [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] [*]. The academic coordinating centre for INTERVAL was supported by core funding from: NIHR Blood and Transplant Research Unit in Donor Health and Genomics (NIHR BTRU-2014-10024), UK Medical Research Council (MR/L003120/1), British Heart Foundation (SP/09/002; RG/13/13/30194; RG/18/13/33946) and the NIHR [Cambridge Biomedical Research Centre at the Cambridge University Hospitals NHS Foundation Trust] [*]. A complete list of the investigators and contributors to the INTERVAL trial is provided in Di Angelantonioet al. (Lancet 390, 2360-2371, 2017). The academic coordinating centre would like to thank blood donor centre staff and blood donors for participating in the INTERVAL trial.
We gratefully acknowledge all studies and databases that have made their GWAS summary data available for this study: arcOGEN (Arthritis Research UK Osteoarthritis Genetics), BCAC (the Breast Cancer Association Consortium), C4D (Coronary Artery Disease Genetics Consortium), CARDIoGRAM (Coronary ARtery DIsease Genome wide Replication and Meta-analysis), CKDGen (Chronic Kidney Disease Genetics consortium), DIAGRAM (DIAbetes Genetics Replication And Meta-analysis), EAGLE (EArly Genetics and Lifecourse Epidemiology Consortium), EAGLE Eczema (EArly Genetics and Lifecourse Epidemiology Eczema Consortium), EGG (Early Growth Genetics Consortium), ENIGMA (Enhancing Neuro Imaging Genetics through Meta Analysis), GCAN (Genetic Consortium for Anorexia Nervosa), GEFOS (GEnetic Factors for OSteoporosis Consortium), GIANT (Genetic Investigation of ANthropometric Traits), GIS (Genetics of Iron Status consortium), GLGC (Global Lipids Genetics Consortium), GliomaScan (cohort-based genome-wide association study of glioma), GPC (Genetics of Personality Consortium), GUGC (Global Urate and Gout consortium), HaemGen (haemotological and platelet traits genetics consortium), IGAP (International Genomics of Alzheimer's Project), IIBDGC (International Inflammatory Bowel Disease Genetics Consortium), ILCCO (International Lung Cancer Consortium), IMSGC (International Multiple Sclerosis Genetic Consortium), ISGC (International Stroke Genetics Consortium), MAGIC (Meta-Analyses of Glucose and Insulin-related traits Consortium), MDACC (MD Anderson Cancer Center), MESA (Multi-Ethnic Study of Atherosclerosis), Neale’s lab (a team of researchers from Benjamin Neale’s group, who made the UK Biobank GWAS summary statistics publically available),OCAC (Ovarian Cancer Association Consortium), IPSCSG (the International PSC study group), NHGRI-EBI GWAS catalog (National Human Genome Research Institute and European Bioinformatics Institute Catalog of published genome-wide association studies), PanScan (Pancreatic Cancer Cohort Consortium), PGC (Psychiatric Genomics Consortium), Project MinE consortium, ReproGen (Reproductive ageing Genetics consortium), SSGAC (Social Science Genetics Association Consortium), TAG (Tobacco and Genetics Consortium), and UK Biobank.
J. Z. acknowledges his grandmother ChenZhu for all her support, may she rest in peace.
*The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health and Social Care.
Footnotes
Author contributions
J. Z., V. H. andD. B. performed the Mendelian randomization analysis. J. Z. and D. B. performed the colocalization analysis. J. Z. performed the conditional analysis. V. H., Y. L., B. E., and T. R. G. developed the database and web browser. J. Z., V. W., and M. R. H. performed the drug target prioritization and enrichment analysis. J. Z. and R. S. conducted the druggable genome analysis. J. Z. and P. E. conducted the pathway and protein-protein interaction analysis. M. R. H., A. G., T. G. R., B. E., H. M. M., J. Y., C. L., S. L., and J. R. conducted supporting analyses. J. R. S., B. B. S., J. D., H. R., and J. C. M. provided key data and supported the MR analysis. M. R. H., S. B., J. Z. L., K. E., L. M., M. V. H., D. W., and M. R. N. reviewed the paper and provided key comments. J. Z., V. H., D. B., V. W., P. C. H., A. S. B., G. D. S., G. H., R. A. S., and T. R. G. wrote the manuscript. J. Z., T. R. G., and R. A. S. conceived and designed the study and oversaw all analyses.
Competing Interests Statement
A. G., L. M., M. R. H., D. W., M. R. N., R. S., and R. A. S.are employees and shareholders in GlaxoSmithKline. H. R., J. Z. L., and K. E. are employees and shareholders in Biogen. J. Z and V. H. is employed on a grant funded by GlaxoSmithKline. D. B. is employed on a grant funded by Biogen. T. R. G., G. H., and G. D. S. receive funding from GlaxoSmithKline and Biogen for the work described here. A. S. B. has received grants from Merck, Novartis, Biogen, Pfizer and AstraZeneca. M. V. H. has collaborated with Boehringer Ingelheim in research, and in accordance with the policy of the Clinical Trial Service Unit and Epidemiological Studies Unit (University of Oxford), did not accept any personal payment.
This work was supported by Health Data Research UK, which is funded by the UK Medical Research Council, Engineering and Physical Sciences Research Council, Economic and Social Research Council, Department of Health and Social Care (England), Chief Scientist Office of the Scottish Government Health and Social Care Directorates, Health and Social Care Research and Development Division (Welsh Government), Public Health Agency (Northern Ireland), British Heart Foundation and Wellcome.
Data availability
The data (GWAS summary statistics) used in the analyses described here are freely accessible in the MR-Base platform (www.mrbase.org). All our analysis results for 989 proteins against 225 human phenotypes are freely available to browse, query and download in EpiGraphDB (http://www.epigraphdb.org/pqtl/). An application programming interface (API) and R package documented on the website enable users to programmatically access data from the database.
Code availability
The code used in the Mendelian randomization and colocalization analyses described here are freely accessiblevia our GitHub repo (https://github.com/MRCIEU/epigraphdb-pqtl). The MR analysis was conducted using TwoSampleMR R package (https://github.com/MRCIEU/TwoSampleMR). We implemented the colocalization analysis using the coloc R package (created by Chris Wallaceet al.), which can be downloaded here (https://cran.r-project.org/web/packages/coloc/index.html).
References
- 1.Plenge RM, Scolnick EM, Altshuler D. Validating therapeutic targets through human genetics. Nat Rev Drug Discov. 2013;12:581–594. doi: 10.1038/nrd4051. [DOI] [PubMed] [Google Scholar]
- 2.Hay M, Thomas DW, Craighead JL, Economides C, Rosenthal J. Clinical development success rates for investigational drugs. Nat Biotechnol. 2014;32:40–51. doi: 10.1038/nbt.2786. [DOI] [PubMed] [Google Scholar]
- 3.Arrowsmith J, Miller P. Phase II and Phase III attrition rates 2011-2012. Nat Rev Drug Discov. 2013;12:569. doi: 10.1038/nrd4090. [DOI] [PubMed] [Google Scholar]
- 4.Harrison RK. Phase II and phase III failures: 2013-2015. Nat Rev Drug Discov. 2016;15:817. doi: 10.1038/nrd.2016.184. [DOI] [PubMed] [Google Scholar]
- 5.Cummings JL, Morstorf T, Zhong K. Alzheimer’s disease drug-development pipeline: few candidates, frequent failures. Alzheimers Res Ther. 2014;6:37. doi: 10.1186/alzrt269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 7.Zhu Z, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48:481–487. doi: 10.1038/ng.3538. [DOI] [PubMed] [Google Scholar]
- 8.Richardson TG, et al. Systematic Mendelian randomization framework elucidates hundreds of CpG sites which may mediate the influence of genetic variants on disease. Hum Mol Genet. 2018;27:3293–3304. doi: 10.1093/hmg/ddy210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun BB, et al. Genomic atlas of the human plasma proteome. Nature. 2018;558:73–79. doi: 10.1038/s41586-018-0175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chong M, et al. Novel drug targets for ischemic stroke identified through Mendelian randomization analysis of the blood proteome. Circulation. 2019;140:819–830. doi: 10.1161/CIRCULATIONAHA.119.040180. [DOI] [PubMed] [Google Scholar]
- 11.Santos R, et al. A comprehensive map of molecular drug targets. Nat Rev Drug Discov. 2017;16:19–34. doi: 10.1038/nrd.2016.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Imming P, Sinning C, Meyer A. Drugs, their targets and the nature and number of drug targets. Nat Rev Drug Discov. 2006;5:821–834. doi: 10.1038/nrd2132. [DOI] [PubMed] [Google Scholar]
- 13.Suhre K, et al. Connecting genetic risk to disease end points through the human blood plasma proteome. Nat Commun. 2017;8:14357. doi: 10.1038/ncomms14357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Folkersen L, et al. Mapping of 79 loci for 83 plasma protein biomarkers in cardiovascular disease. PLoS Genet. 2017;13:e1006706. doi: 10.1371/journal.pgen.1006706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yao C, et al. Genome-wide association study of plasma proteins identifies putatively causal genes, proteins, and pathways for cardiovascular disease. Nat Commun. 2018;9:3268. doi: 10.1038/s41467-018-05512-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Emilsson V, et al. Co-regulatory networks of human serum proteins link genetics to disease. Science. 2018;361:769–773. doi: 10.1126/science.aaq1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evans DM, Davey Smith G. Mendelian randomization: new applications in the coming age of hypothesis-free causality. Annu Rev Genomics Hum Genet. 2015;16:327–350. doi: 10.1146/annurev-genom-090314-050016. [DOI] [PubMed] [Google Scholar]
- 18.Davey Smith G, Ebrahim S. ‘Mendelian randomization’: can genetic epidemiology contribute to understanding environmental determinants of disease? Int J Epidemiol. 2003;32:1–22. doi: 10.1093/ije/dyg070. [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, et al. Recent developments in Mendelian randomization studies. Curr Epidemiol Rep. 2017;4:330–345. doi: 10.1007/s40471-017-0128-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Millwood IY, et al. Association of CETP gene variants with risk for vascular and nonvascular diseases among Chinese adults. JAMA Cardiol. 2018;3:34–43. doi: 10.1001/jamacardio.2017.4177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Interleukin-6 Receptor Mendelian Randomisation Analysis (IL6R MR) Consortium et al. The interleukin-6 receptor as a target for prevention of coronary heart disease: a mendelian randomisation analysis. Lancet. 2012;379:1214–1224. doi: 10.1016/S0140-6736(12)60110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swerdlow DI, et al. Selecting instruments for Mendelian randomization in the wake of genome-wide association studies. Int J Epidemiol. 2016;45:1600–1616. doi: 10.1093/ije/dyw088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hemani G, et al. The MR-Base platform supports systematic causal inference across the human phenome. Elife. 2018;7:e34408. doi: 10.7554/eLife.34408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Timpson NJ, et al. C-reactive protein levels and body mass index: elucidating direction of causation through reciprocal Mendelian randomization. Int J Obes. 2011;35:300–308. doi: 10.1038/ijo.2010.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hemani G, Tilling K, Davey Smith G. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. PLoS Genet. 2017;13:e1007081. doi: 10.1371/journal.pgen.1007081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hemani G, et al. Automating Mendelian randomization through machine learning to construct a putative causal map of the human phenome. bioRxiv. 2017 doi: 10.1101/173682. [DOI] [Google Scholar]
- 27.Bowden J, et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med. 2017;36:1783–1802. doi: 10.1002/sim.7221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Giambartolomei C, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014;10:e1004383. doi: 10.1371/journal.pgen.1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang J, et al. Conditional and joint multiple-SNP analysis of GWAS summary statistics identifies additional variants influencing complex traits. Nat Genet. 2012;44:369–375. doi: 10.1038/ng.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Burgess S, Dudbridge F, Thompson SG. Combining information on multiple instrumental variables in Mendelian randomization: comparison of allele score and summarized data methods. Stat Med. 2016;35:1880–1906. doi: 10.1002/sim.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Lange KM, et al. Genome-wide association study implicates immune activation of multiple integrin genes in inflammatory bowel disease. Nat Genet. 2017;49:256–261. doi: 10.1038/ng.3760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Solomon T, et al. Identification of common and rare genetic variation associated with plasma protein levels using whole-exome sequencing and mass spectrometry. Circ Genom Precis Med. 2018;11:e002170. doi: 10.1161/CIRCGEN.118.002170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Taylor FB, Jr, Peer GT, Lockhart MS, Ferrell G, Esmon CT. Endothelial cell protein C receptor plays an important role in protein C activation in vivo. Blood. 2001;97:1685–1688. doi: 10.1182/blood.v97.6.1685. [DOI] [PubMed] [Google Scholar]
- 34.Hashizume M, et al. Tocilizumab, a humanized anti-IL-6R antibody, as an emerging therapeutic option for rheumatoid arthritis: molecular and cellular mechanistic insights. Int Rev Immunol. 2015;34:265–279. doi: 10.3109/08830185.2014.938325. [DOI] [PubMed] [Google Scholar]
- 35.Ridker PM, et al. Modulation of the interleukin-6 signalling pathway and incidence rates of atherosclerotic events and all-cause mortality: analyses from the Canakinumab Anti-Inflammatory Thrombosis Outcomes Study (CANTOS) Eur Heart J. 2018;39:3499–3507. doi: 10.1093/eurheartj/ehy310. [DOI] [PubMed] [Google Scholar]
- 36.Ferreira RC, et al. Functional IL6R 358Ala allele impairs classical IL-6 receptor signaling and influences risk of diverse inflammatory diseases. PLoS Genet. 2013;9:e1003444. doi: 10.1371/journal.pgen.1003444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stacey D, et al. Elucidating mechanisms of genetic cross-disease associations: an integrative approach implicates protein C as a causal pathway in arterial and venous diseases. medRxiv. 2020 doi: 10.1101/2020.03.16.20036822. [DOI] [Google Scholar]
- 38.Sanseau P, et al. Use of genome-wide association studies for drug repositioning. Nat Biotechnol. 2012;30:317–320. doi: 10.1038/nbt.2151. [DOI] [PubMed] [Google Scholar]
- 39.Finan C, et al. The druggable genome and support for target identification and validation in drug development. Sci Transl Med. 2017;9:eaag1166. doi: 10.1126/scitranslmed.aag1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Holmes MV, Ala-Korpela M, Smith GD. Mendelian randomization in cardiometabolic disease: challenges in evaluating causality. Nat Rev Cardiol. 2017;14:577–590. doi: 10.1038/nrcardio.2017.78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44:512–525. doi: 10.1093/ije/dyv080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao Q, Wang J, Hemani G, Bowden J, Small DS. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. aRxiv. 2018 [Google Scholar]
- 43.Evans DM, et al. Mining the human phenome using allelic scores that index biological intermediates. PLoS Genet. 2013;9:e1003919. doi: 10.1371/journal.pgen.1003919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Timpson NJ. One size fits all, are there standard rules for the use of genetic instruments in Mendelian randomization? Int J Epidemiol. 2016;45:1617–1618. doi: 10.1093/ije/dyw197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hemani G, Bowden J, Davey Smith G. Evaluating the potential role of pleiotropy in Mendelian randomization studies. Hum Mol Genet. 2018;27:R195–R208. doi: 10.1093/hmg/ddy163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wu Y, et al. Colocalization of GWAS and eQTL signals at loci with multiple signals identifies additional candidate genes for body fat distribution. Hum Mol Genet. 2019;28:4161–4172. doi: 10.1093/hmg/ddz263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wainberg M, et al. Opportunities and challenges for transcriptome-wide association studies. Nat Genet. 2019;51:592–599. doi: 10.1038/s41588-019-0385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uhlén M, et al. Tissue-based map of the human proteome. Science. 2015;347:1260419. doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 49.GTEx Consortium et al. Genetic effects on gene expression across human tissues. Nature. 2017;550:204–213. doi: 10.1038/nature24277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim-Hellmuth S, et al. Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations. Nat Commun. 2017;8:266. doi: 10.1038/s41467-017-00366-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Boyd A, et al. Cohort Profile: the 'children of the 90s'—the index offspring of the Avon Longitudinal Study of Parents and Children. Int J Epidemiol. 2013;42:111–127. doi: 10.1093/ije/dys064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fraser A, et al. Cohort Profile: the Avon Longitudinal Study of Parents and Children: ALSPAC mothers cohort. Int J Epidemiol. 2013;42:97–110. doi: 10.1093/ije/dys066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McLaren W, et al. The Ensembl Variant Effect Predictor. Genome Biol. 2016;17:122. doi: 10.1186/s13059-016-0974-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nyholt DR. A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet. 2004;74:765–769. doi: 10.1086/383251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cichonska A, et al. metaCCA: summary statistics-based multivariate meta-analysis of genome-wide association studies using canonical correlation analysis. Bioinformatics. 2016;32:1981–1989. doi: 10.1093/bioinformatics/btw052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lawlor DA, Harbord RM, Sterne JA C, Timpson N, Davey Smith G. Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med. 2008;27:1133–1163. doi: 10.1002/sim.3034. [DOI] [PubMed] [Google Scholar]
- 57.Burgess S, Zuber V, Valdes-Marquez E, Sun BB, Hopewell JC. Mendelian randomization with fine-mapped genetic data: Choosing from large numbers of correlated instrumental variables. Genet Epidemiol. 2017;41:714–725. doi: 10.1002/gepi.22077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yavorska OO, Burgess S. MendelianRandomization: an R package for performing Mendelian randomization analyses using summarized data. Int J Epidemiol. 2017;46:1734–1739. doi: 10.1093/ije/dyx034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Haycock PC, et al. Best (but oft-forgotten) practices: the design, analysis, and interpretation of Mendelian randomization studies. Am J Clin Nutr. 2016;103:965–978. doi: 10.3945/ajcn.115.118216. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data (GWAS summary statistics) used in the analyses described here are freely accessible in the MR-Base platform (www.mrbase.org). All our analysis results for 989 proteins against 225 human phenotypes are freely available to browse, query and download in EpiGraphDB (http://www.epigraphdb.org/pqtl/). An application programming interface (API) and R package documented on the website enable users to programmatically access data from the database.
The code used in the Mendelian randomization and colocalization analyses described here are freely accessiblevia our GitHub repo (https://github.com/MRCIEU/epigraphdb-pqtl). The MR analysis was conducted using TwoSampleMR R package (https://github.com/MRCIEU/TwoSampleMR). We implemented the colocalization analysis using the coloc R package (created by Chris Wallaceet al.), which can be downloaded here (https://cran.r-project.org/web/packages/coloc/index.html).