Abstract
Follicular helper T cells (Tfh) are implicated in type 1 diabetes (T1D) and their development has been linked to CD28 costimulation. We tested whether Tfh were decreased by costimulation blockade using the CTLA-4-Ig fusion protein (Abatacept) in a mouse model of diabetes and in individuals with new onset T1D. Unbiased bioinformatic analysis identified that ICOS+ Tfh, and other ICOS+ populations including T-peripheral helper cells, were highly sensitive to costimulation blockade. We were able to use pre-treatment Tfh profiles to derive a model that could predict clinical response to Abatacept in individuals with T1D. Using two independent approaches we demonstrated that higher frequencies of ICOS+ Tfh at baseline were associated with a poor clinical response following Abatacept administration. Tfh analysis may therefore represent a new stratification tool, permitting the identification of individuals most likely to benefit from costimulation blockade.
CD28 costimulation licenses T cells for effective activation and is a key therapeutic target in autoimmunity. The natural regulator of CD28 is the inhibitory receptor CTLA-4, and a soluble version of this molecule has been developed for therapeutic use. Soluble CTLA-4 (a fusion protein with human immunoglobulin; CTLA-4-Ig) is widely used in autoimmune conditions including rheumatoid arthritis (RA), psoriatic arthritis and juvenile idiopathic arthritis1, 2, 3. Studies in the NOD mouse model of type 1 diabetes (T1D) suggested a protective effect of CTLA-4-Ig in this disease setting4 leading to a trial of the clinically licensed CTLA-4-Ig molecule Abatacept (Orencia®; Bristol-Myers Squibb) in individuals with new onset T1D. A randomised double-blind placebo-controlled trial showed that adjusted C-peptide levels were 59% higher in recipients of Abatacept at 2 years compared with placebo5, and the beneficial effects were largely maintained a year following therapy cessation6, although it was clear that some individuals benefited more than others. Thus, CTLA-4-Ig-based costimulation blockade in both mice and humans implicates the CD28 pathway in T1D pathogenesis, however the precise CD28-dependent processes involved remain ill-defined. Identifying and monitoring these could potentially help explain, and even predict, why certain individuals make a better response to costimulation blockade than others.
Although T1D has classically been considered to be a TH1-mediated pathology, a signature of follicular helper T cell (Tfh) differentiation was identified in this disease setting7. Tfh support B cell responses within the germinal centers (GC) of secondary lymphoid tissues and are characterised by markers such as CXCR5, ICOS and PD-1 as well as the transcription factor Bcl68, 9, 10. Memory Tfh in the blood share T cell receptor (TCR) clonotypes with their lymphoid tissue counterparts11, 12 and can home to GC in response to secondary immunisation13, 14. Murine T cells responding to a pancreatic self-antigen adopted a Tfh phenotype and GC were formed in the pancreatic lymph nodes (PanLN) of mice developing diabetes7. Likewise, in humans with T1D a higher proportion of blood-borne Tfh were observed within the memory compartment than in matched non-diabetic individuals7, and similar data were obtained in two independent T1D patient cohorts15, 16. Subsequent studies showed that circulating cells with a Tfh phenotype were increased in children with multiple islet autoantibodies at risk of developing T1D17, 18. Thus, circulating Tfh-like cells have been associated with T1D in multiple patient cohorts, and increases in these cells may precede the development of overt disease.
CD28 has long been implicated in the development of Tfh and the provision of T cell help for antibody responses. Mice deficient in the CD28 ligands, CD80 and CD86, fail to form GC19, associated with an inability of their T cells to upregulate the chemokine receptor CXCR5 that guides T cells towards B cell follicles20. More recently, it was reported that Tfh differentiation was sensitive to the strength of CD28 engagement, and that this could be modulated by CTLA-421. A clear prediction of these studies is that use of CTLA4-Ig (Abatacept) to inhibit CD28 costimulation would be expected to decrease Tfh differentiation, and there are already suggestions that this may be the case in primary Sjogren’s syndrome22, rheumatoid arthritis23 and multiple sclerosis24. We were therefore interested to assess the impact of Abatacept on Tfh populations in T1D.
Here we show that Tfh are a sensitive biomarker of costimulation blockade in both mice and humans with T1D and reveal that pre-treatment Tfh profiles can be used to predict response to Abatacept. These data ascribe new value to Tfh analysis and suggest its potential as a stratification tool prior to immunotherapy.
Results
Abatacept decreases immunisation-induced Tfh in mice
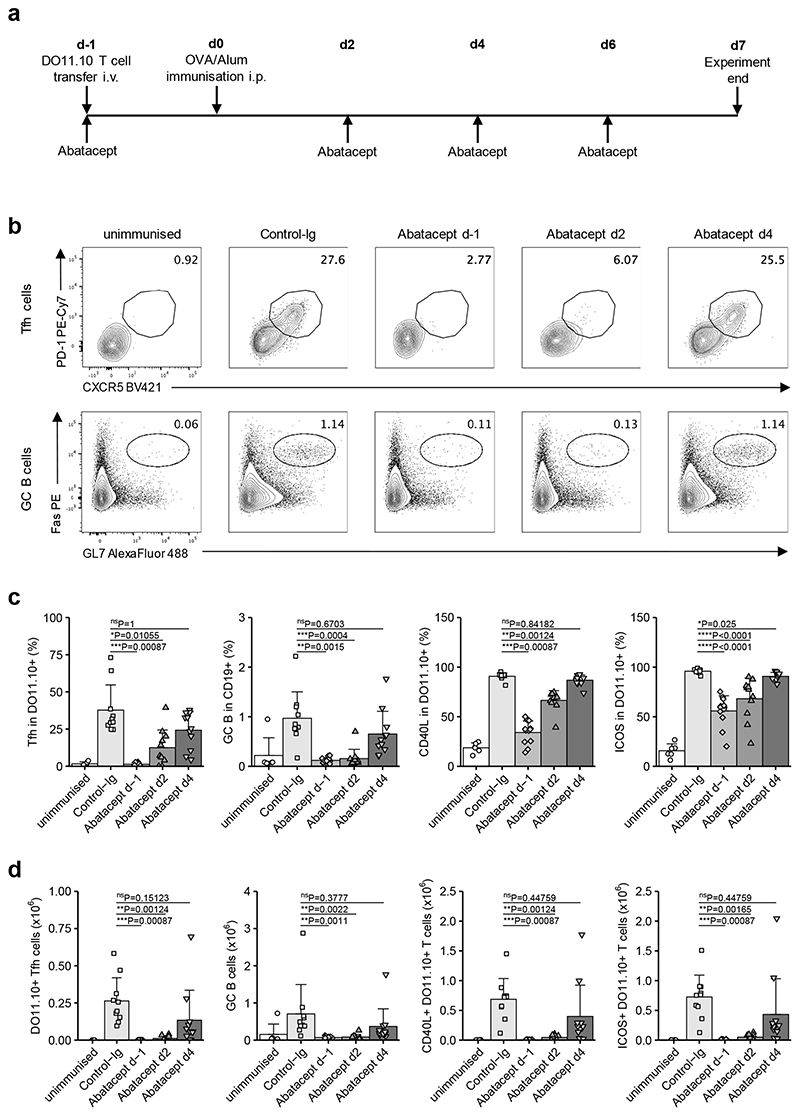
Costimulation blockade using antibodies against CD80 and CD86 decreased the capacity of adoptively transferred TCR transgenic T cells to differentiate into Tfh following immunisation21. We predicted that costimulation blockade with Abatacept would yield similar results, since this reagent binds to CD80 and CD86 in a manner that inhibits their engagement of CD28. We sought to confirm this, and concurrently to probe the kinetic requirements for effective Tfh inhibition, by providing Abatacept at different timepoints (Fig. 1a). We reasoned that the timing of costimulation blockade was an important consideration given that in autoimmune settings, treatment is likely to be delivered when the T cell response is already underway. Initiation of Abatacept treatment one day prior to immunisation (d-1) with ovalbumin (OVA) resulted in DO11.10 T cells exhibiting a significantly lower frequency (Fig. 1b,c) and absolute number (Fig. 1d) of Tfh than in Control-Ig treated animals. This was associated with inhibition of GC B cell formation as well as a decrease in expression of molecules involved in T cell / B cell collaboration such as CD40L and ICOS (Fig. 1b-d). Delaying costimulation blockade until 2 days after immunisation partially reduced its capacity to inhibit Tfh, and delaying it until day 4 abrogated the effects on Tfh and GC B cells (Fig. 1). Thus, costimulation blockade with Abatacept reduced Tfh, but delaying its administration rendered it less effective.
Figure 1. Time-sensitive inhibition of Tfh by Abatacept in immunised mice.
DO11.10 T cells (2 x 105) were injected i.v. into Cd28−/− mice that were immunised i.p. with 200 μg of OVA/alum 24 h later. Abatacept or Control-Ig were administered i.p. every two to three days starting at the indicated time points. Control-Ig treatment was initiated at d-1. At day 7 after immunisation, spleens were harvested for analysis. (a) Representation of treatment scheme. (b) Representative flow cytometry plots for CXCR5+PD-1+ Tfh cells in gated CD4+DO11.10+ cells (top) and Fas+GL7+ GC B cells in gated CD19+ cells (bottom). (c) Collated data for Tfh cells, GC B cells, and CD40L and ICOS frequencies in gated CD4+DO11.10+ cells. (d) Collated data for absolute numbers of DO11.10+ Tfh, GC B cells, CD40L+CD4+DO11.10+ cells and ICOS+CD4+DO11.10+ cells. Data are compiled from five independent experiments; n=6 mice unimmunised, 10 mice Control-Ig, 12 mice Abatacept d-1, 11 mice each Abatacept d2 and Abatacept d4. Mean + SD are shown. (c) and (d) Kruskal-Wallis test for multiple comparisons followed by pairwise two-tailed Mann-Whitney U test with Bonferroni correction; ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant.
Abatacept decreases Tfh in a mouse model of diabetes
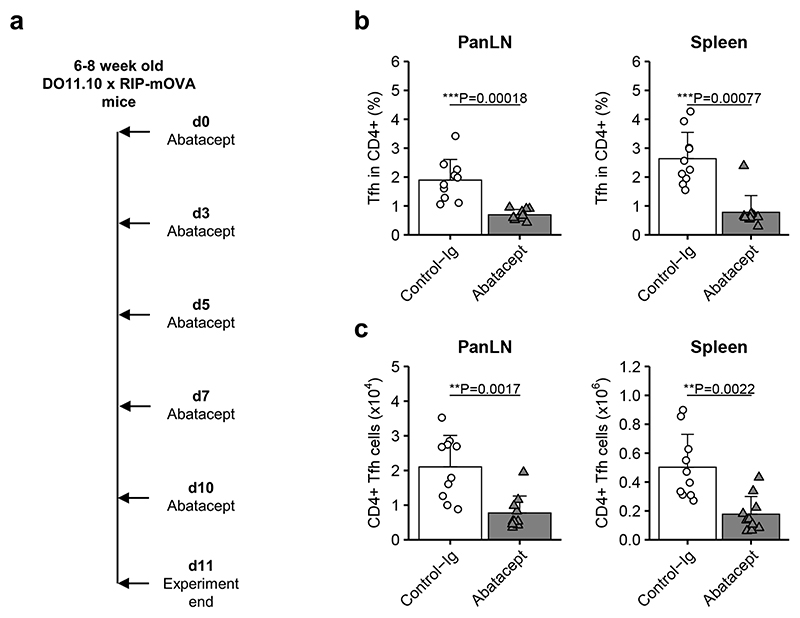
The inability of Abatacept to inhibit Tfh when delivered 4 days after immunisation raised the possibility that T cells already engaged in a response to autoantigen may be resistant to the Tfh modulating effects of this drug. To explore this idea, we examined the same TCR transgenic T cells responding to a pancreas-expressed, rather than immunised, protein. Mice that express the DO11.10 TCR transgene in conjunction with its cognate antigen in pancreatic beta cells (DO11.10 x RIP-mOVA mice) develop spontaneous islet autoimmunity and diabetes with 100% penetrance25. In these mice, islet-expressed OVA is presented to T cells in the PanLN26, and this is associated with T cell differentiation to a Tfh phenotype7. All mice manifest autoimmune islet infiltration by 5 weeks of age and we have established that CD28 costimulation is required for diabetes development (data not shown). To assess the impact of costimulation blockade on Tfh cells in the setting of an ongoing immune response to pancreatic autoantigen, we administered a short course of Abatacept to DO11.10 x RIP-mOVA mice (Fig. 2a). The results of this experiment revealed a decrease in Tfh at the site of antigen presentation (PanLN) as well as the spleen (Fig. 2b,c). Thus, even though T cell priming and Tfh differentiation were already underway prior to treatment, Abatacept was able to suppress the Tfh response.
Figure 2. Abatacept decreases Tfh during an ongoing autoimmune response in mice.
Abatacept or Control-Ig were injected every two to three days i.p. into 6-8 week old DO11.10 x RIP-mOVA mice. At day 11, pancreas-draining lymph nodes (panLN) and spleens were harvested for analysis. (a) Representation of treatment scheme. (b,c) Collated data for frequencies (b) and absolute numbers (c) of Tfh cells in gated CD4+ cells. Data are compiled from two independent experiments; n=10 mice in each treatment group. Mean + SD are shown. Two-tailed Mann-Whitney U test; ***, p < 0.001; **, p < 0.01.
Abatacept decreases circulating Tfh in type 1 diabetes patients
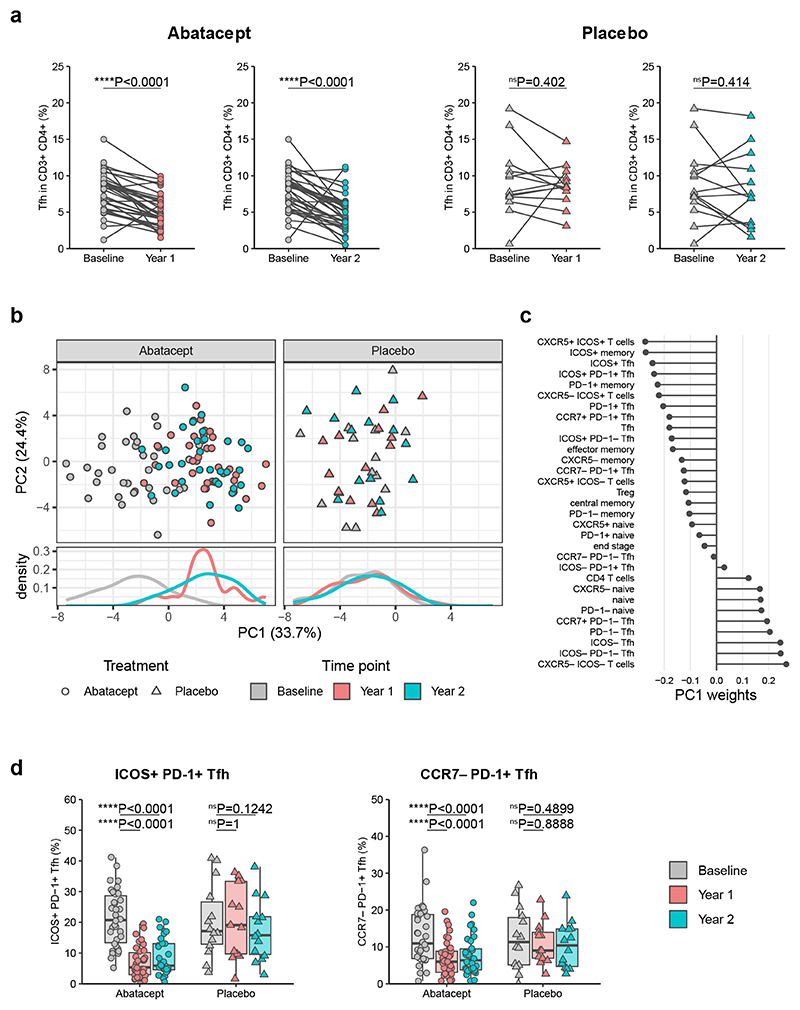
To assess whether Abatacept decreased circulating Tfh in humans with T1D, we obtained access to frozen samples from individuals with new onset T1D treated with Abatacept or placebo via Trialnet Study TN09 (NCT00505375). We were provided with samples from 36 Abatacept-treated individuals and 14 placebo-treated individuals, with 3 samples typically being available for each individual: baseline, and 1 and 2 years post treatment. Associated clinical data revealed a relative preservation of C-peptide in Abatacept-treated individuals compared with placebo-treated individuals (Supplementary Fig. 1), in line with the original trial results from the entire cohort5. Samples were stained with a panel of T cell markers including ones associated with a Tfh phenotype (for gating strategy see Supplementary Fig. 2). Since we previously showed that circulating CD4+CD45RA−CXCR5+ cells (Tfh) were overrepresented in humans with T1D7, we first examined whether this population was Abatacept-sensitive. Our analysis revealed that Tfh were significantly decreased after Abatacept treatment at both 1 and 2 year timepoints, whereas this was not the case in the placebo-treated control group (Fig. 3a). Principal component analysis of gated flow cytometry data revealed that the highest proportion of variance in this dataset is explained by Abatacept-induced changes, since treated samples are separated from untreated samples along PC1 for Abatacept treatment but not placebo treatment (Fig. 3b). The major cell population contributing to this separation was T cells expressing CXCR5 and ICOS (Fig. 3c). CCR7loPD-1+CXCR5+ cells, previously identified as circulating Tfh precursors that correlate with disease activity in autoimmunity27, also contributed to PC1 and were decreased by Abatacept treatment (here called CCR7−PD-1+ Tfh) (Fig. 3c). Graphed datapoints for the ICOS+PD-1+ Tfh and CCR7−PD-1+ Tfh populations are provided for illustrative purposes, and depict the Abatacept-induced change in cell frequency (Fig. 3d). To study the impact on Tfh subsets, additional trial samples were analysed with a panel incorporating the chemokine receptors CXCR3 and CCR628. The Abatacept-induced reduction of Tfh, and particularly ICOS+PD-1+Tfh, was corroborated, however there was no obvious skewing of the Tfh subpopulations defined by CXCR3 and/or CCR6 expression (Supplementary Fig. 3). Overall, these findings demonstrated that cells expressing Tfh markers were amongst the populations most affected by costimulation blockade.
Figure 3. Abatacept decreases Tfh in patients with new onset type 1 diabetes.
Frozen PBMC samples from patients with recent onset T1D that received Abatacept or placebo were thawed and stained for flow cytometry analysis. Samples were taken at baseline, one year and two years after treatment initiation. (a) Collated data for Tfh (CD45RA−CXCR5+) frequencies in CD3+CD4+ cells from recipients of Abatacept (left) or placebo (right). (b) Principal component analysis on population frequencies obtained from flow cytometry analysis. Analysis was performed on all samples simultaneously and split into treatment groups for visualisation purposes. (c) Contributions of individual populations to PC1. (d) Collated data for ICOS+PD-1+ and CCR7−PD-1+ frequencies in CD4+CD45RA−CXCR5+ cells. Shown are boxplots with black horizontal line denoting median value, while box represents interquartile ranges (IQR, Q1-Q3 percentile) and whiskers show minimum (Q1– 1.5 * IQR) and maximum (Q3 + 1.5 * IQR) values. Abatacept, n = 34 patients; Placebo, n = 13 patients (Year 1) or 14 patients (Baseline and Year 2). Two-tailed Wilcoxon signed-rank test; ****, p < 0.0001; ns, not significant.
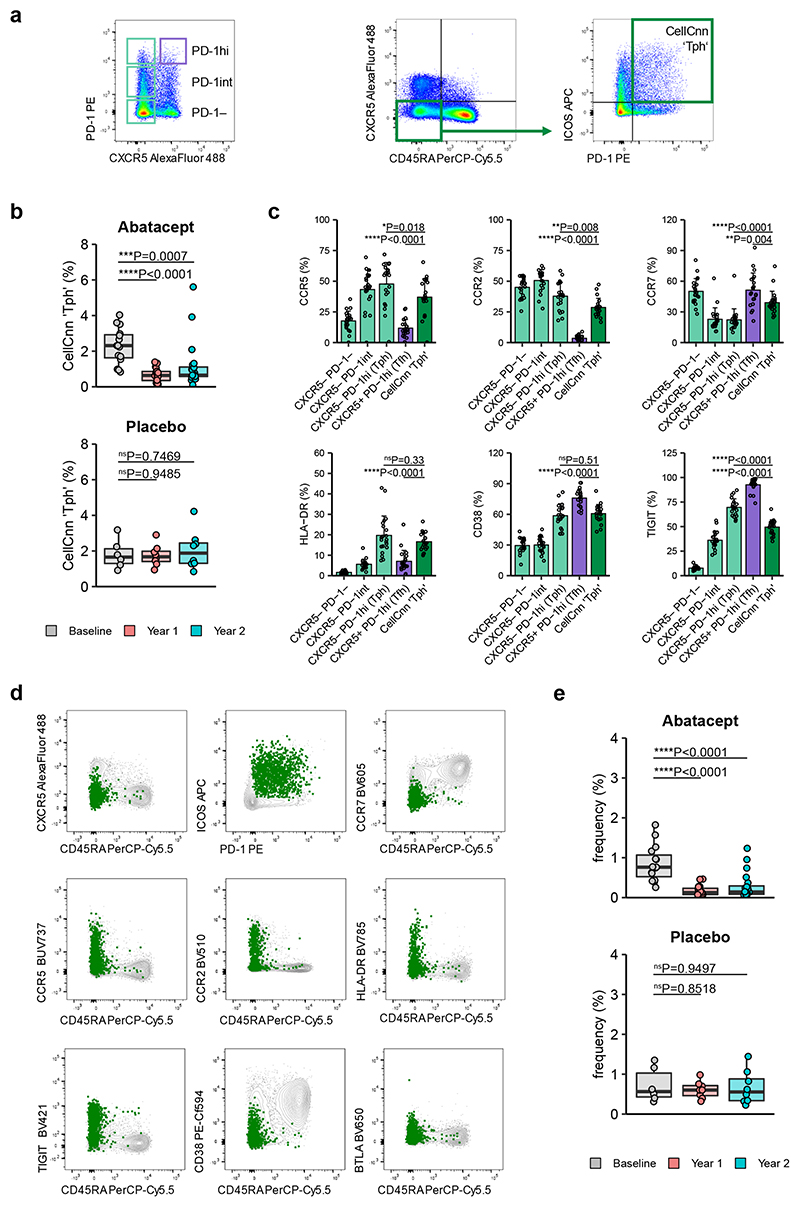
Additional Abatacept-sensitive populations in type 1 diabetes revealed by CellCnn
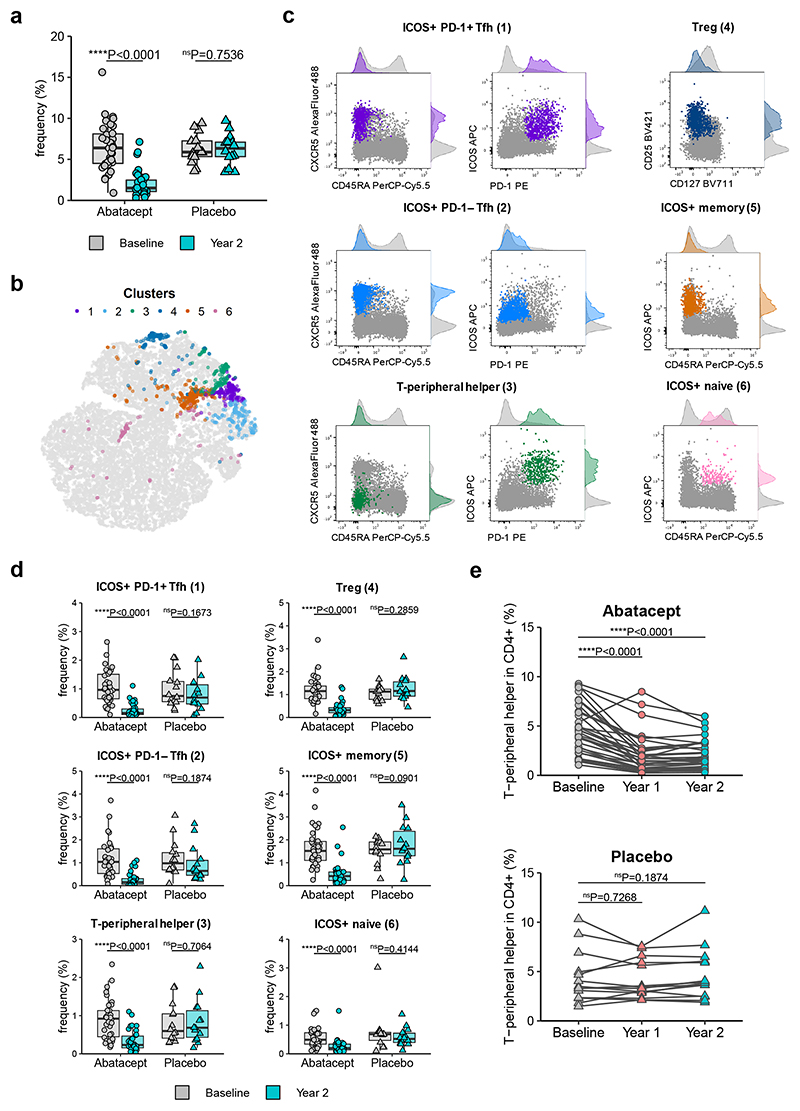
Given the bias associated with manual gating, we tested whether unbiased analysis would also identify a change in Tfh-like cells following Abatacept treatment. We used the machine-learning algorithm CellCnn29, a representation learning approach using convolutional neural networks designed to identify rare cell subsets associated with disease status in a data-driven way. When samples are split into 2 groups (e.g. Abatacept versus placebo), this approach is able to establish marker expression profiles (termed filters) of individual cells whose frequency is associated with the assigned group. In our analysis, CellCnn identified a filter whose corresponding cells were present at high frequencies in all samples at baseline and in placebo treated samples, but were significantly reduced in Abatacept-treated samples after two years of treatment (Fig. 4a), indicating that this particular filter was associated with Abatacept-induced changes. Since filters detected by CellCnn do not necessarily represent a homogenous cell population, k-means clustering was applied to identify individual cell types affected by Abatacept treatment. In total, 6 clusters were found (Fig. 4b) that showed distinct expression profiles of the selected markers. By overlaying these cell clusters on flow cytometry data (Fig. 4c, Supplementary Fig. 4) we ascribed names to them that we believe reflect their identity, and assessed the change in the frequency of these populations in Abatacept or placebo treated individuals (Fig. 4d). Consistent with our original manual gating approach, CellCnn identified both ICOS+PD-1+ Tfh (cluster 1) and ICOS+PD-1− Tfh (cluster 2) to be decreased by Abatacept. A third cluster, comprising memory cells that lack CXCR5 but co-express ICOS and PD-1 (cluster 3), was also identified as Abatacept responsive (Fig. 4d). This phenotype is reminiscent of T-peripheral helper cells (Tph) that were identified in the rheumatoid joint and are increased in individuals with higher disease activity30. Manual gating of Tph confirmed a significant reduction in this population in people receiving Abatacept but not placebo at both year 1 and year 2 (Fig. 4e). CellCnn also identified Treg (cluster 4) to be Abatacept-sensitive, consistent with published literature31, 32, in addition to two other clusters characterised by ICOS expression (ICOS+ memory; cluster 5, ICOS+ naive; cluster 6). Note that the term “naive” is used as shorthand to reflect the fact that the cells in cluster 6 are CD45RA+, however their CD45RA expression level is slightly lower than bona fide naive T cells (Fig. 4c, cluster 6), suggesting they are antigen experienced. Thus machine-learning identified 2 Tfh populations and 4 additional populations to be Abatacept-sensitive, all of which expressed ICOS.
Figure 4. Data-driven analysis reveals additional Abatacept-sensitive populations in type 1 diabetes patients.
CellCnn analysis followed by k-means clustering of filter-specific cells was applied to flow cytometry data of samples taken at baseline and two years after Abatacept or placebo treatment initiation. (a) Frequency of filter specific cells in each analysed sample. (b) t-SNE projection of down-sampled, pooled flow cytometry data of all samples used for CellCnn analysis. K-means clusters of filter-specific cells are highlighted. (c) Representative flow cytometry overlays of cluster-specific cells (colour) on original flow cytometry data (grey). Examples shown are from a baseline sample. (d) Frequency of cluster-specific cells in each analysed sample. (e) Collated data for frequency of manually gated T-peripheral helper cells (ICOS+PD-1+CXCR5−CD45RA− in CD4+CD3+). Abatacept, n = 34 patients; Placebo, n = 13 patients (Year 1) or 14 patients (Baseline and Year 2). In (a) and (d) boxplots are shown with black horizontal line denoting median value, while box represents interquartile ranges (IQR, Q1-Q3 percentile) and whiskers show minimum (Q1– 1.5 * IQR) and maximum (Q3 + 1.5 * IQR) values. Two-tailed Wilcoxon signed-rank test; ****, p < 0.0001; ns, not significant.
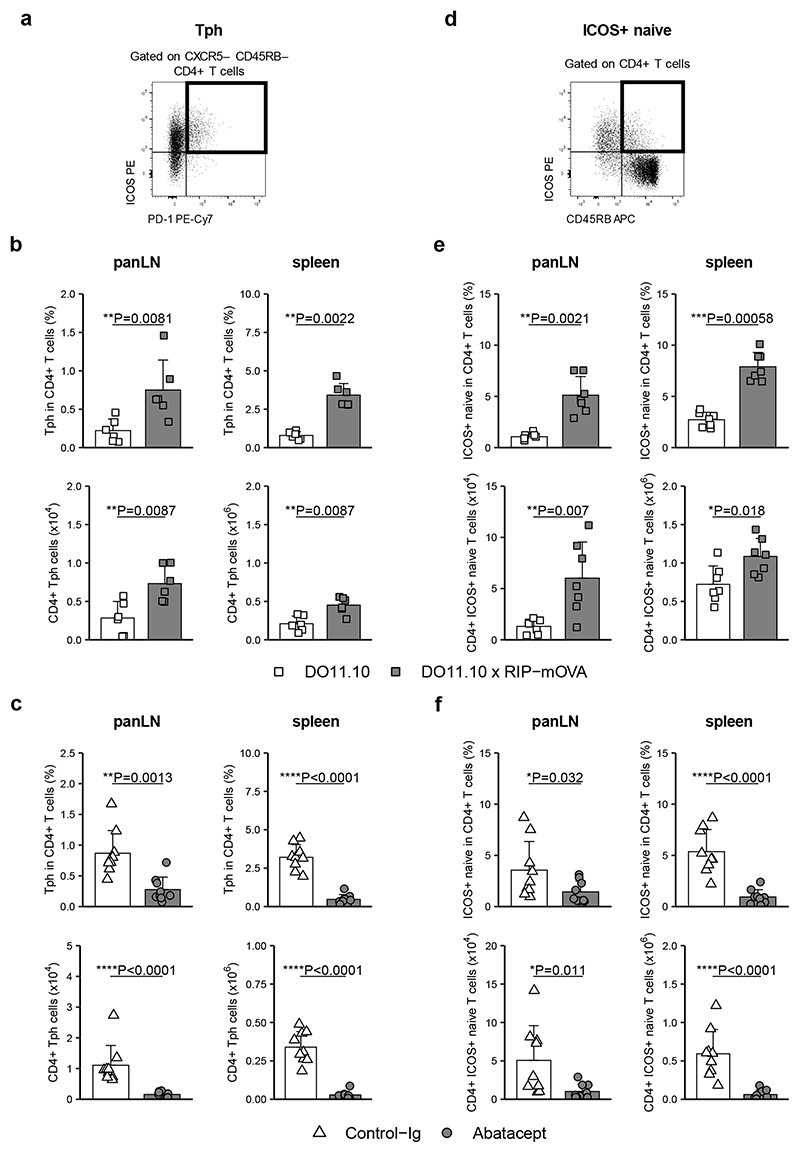
Since Tph have not previously been reported to be costimulation dependent, and ICOS+ naive cells have not previously been described, we explored these populations further in our mouse model of diabetes. Cells with a “Tph” or “ICOS+ naive” phenotype could be identified in mice (Fig. 5a,d), were enriched in autoimmune animals (Fig. 5b,e), and were reduced following Abatacept treatment (Fig. 5c,f). These murine data provide additional support for the costimulation sensitivity of these 2 populations.
Figure 5. “Tph” and “ICOS+ naive” cells are elevated in a mouse model of diabetes and sensitive to costimulation blockade.
Cells isolated from panLN and spleens were stained with a panel of markers to identify Tph (CD4+CD45RB−CXCR5−ICOS+PD-1+) and ICOS+ naive T cells (CD4+CD45RB+ICOS+). Representative flow cytometry plots for gating strategy of Tph (a) and ICOS+ naive T cells (d) in spleen. Collated data for frequencies (top) and absolute numbers (bottom) of Tph (b) and ICOS+ naive T cells (e) in panLN (left) and spleen (right) of DO11.10 and DO11.10 x RIP-mOVA mice. (c,f) DO11.10 x RIP-mOVA mice were treated with Abatacept and Control-Ig according to treatment scheme depicted in Fig. 2a. Shown are collated data for frequencies (top) and absolute numbers (bottom) of Tph (c) and ICOS+ naive T cells (f) in panLN (left) and spleen (right). Data are compiled from 2 (c, f), 3 (b) or 4 (e) independent experiments; n=6 (b), 7 (e) or 9 (c, f) mice. Mean + SD are shown. Two-tailed Mann-Whitney U test; ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05.
To further explore the identity of the “Tph” population identified by CellCnn, additional trial samples were analysed. “Tph” cells were also decreased by Abatacept in this set of samples, and their expression of markers such as CCR5, CCR2, HLA-DR and CD38 was similar to that of Tph identified by standard gating (CXCR5−PD-1hi)30 (Fig. 6a-c). Applying CellCnn to these data identified a cluster of cells expressing Tph markers to be costimulation-sensitive (Fig. 6d,e). Overall, machine learning approaches indicated that populations with the characteristics of Tfh and Tph, as well as additional ICOS+ populations, were strongly reduced after Abatacept treatment.
Figure 6. Tph cells identified through CellCnn display marker expression consistent with a Tph profile.
Frozen PBMC samples from recent onset T1D patients that received Abatacept or placebo were thawed and analysed by flow cytometry for Tph and Tfh markers. (a) Representative gating strategy for CXCR5 vs PD-1 populations (left) and Tph previously identified through CellCnn analysis (right). (b) Collated data for frequency of cells in the CellCnn ‘Tph’ gate. (c) Expression of Tph markers on “Tph” identified by CellCnn compared with classically identified CXCR5−PD1hi Tph gated as shown in (a). Data was obtained from baseline samples and shown are mean + SD. (d, e) CellCnn analysis of samples identifies a cluster of Tph-phenotype cells. Shown is expression of indicated markers within cluster (green) and all cells (grey) of representative sample (d) and frequency of cluster-specific cells in Abatacept- or Placebo-treated T1D patients (e). (b, e) Abatacept, n=15 (Baseline) or 20 (Year 1 and Year 2) patients; Placebo, n=6 (Baseline) or 8 (Year 1 and Year 2) patients; (c) n=21 patients. In (b) and (e) boxplots are shown with black horizontal line denoting median value, while box represents interquartile ranges (IQR, Q1-Q3 percentile) and whiskers show minimum (Q1– 1.5 * IQR) and maximum (Q3 + 1.5 * IQR) values. Two-tailed Mann-Whitney U test; ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant.
Baseline Tfh phenotype is associated with clinical response to Abatacept
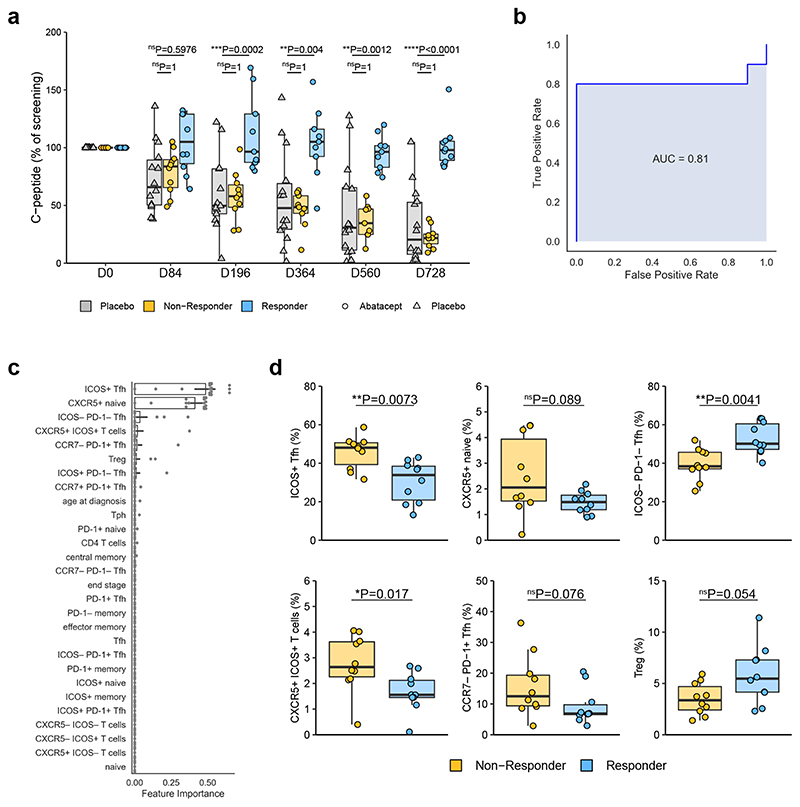
We next explored whether an individual’s clinical response following Abatacept treatment could be predicted from their T cell phenotype at baseline. Clinical response was assessed by relative C-peptide retention at the 2-year timepoint. Gated flow cytometry data were used, with a Tph gate and an ICOS+ naive gate being added on the basis of their identification in the above analysis (Supplementary Fig. 5a). Age at diagnosis was also included since there is evidence that diagnosis at a young age is associated with a more rapid loss of beta cells33. Within the Abatacept-treated subjects, the 10 with the best clinical response (responders) and the 10 with the poorest response (non-responders) (Fig. 7a) were used to build a predictive model using gradient boosting34, 35. Pairwise correlation comparisons were conducted between features to identify and remove features that were highly correlated (Pearson correlation coefficient greater than 0.95), ensuring feature importance could be legitimately interpreted from our gradient boosting model (Supplementary Fig. 5b): where two features were shown to be highly correlated, the one least correlated with outcome was removed from the set of features used to build the predictive model. The gradient boosting model was constructed using nested leave-one-out cross validation. Each of the n patients was iteratively removed from the dataset and kept aside for testing purposes. The remaining n-1 baseline samples were used for model training and hyperparameter (learning rate, maximum depth and number of estimators) tuning using 3-fold cross validation. The optimal model from this training process was then used to make a prediction on the “left-out” sample, and feature weights were recorded.
Figure 7. Baseline Tfh phenotype is associated with clinical response to Abatacept.
(a) C-peptide AUC (as % of screening C-peptide AUC) of placebo treated and top 10 (at day 728) responder and non-responder Abatacept-treated patients. Responder, n=9 (D196, D364, D560) or 10 (all other time points) patients; Non-Responder, n=9 (D364, D560) or 10 (all other time points) patients; Placebo, n=13 (D196) or 14 (all other time points) patients. (b) A gradient boosting model was constructed using nested leave-one-out cross validation to predict clinical response following Abatacept treatment. ROC curve of the predictive model is shown. (c) Features ranked by importance for predicting clinical response following Abatacept treatment. Bar shows mean and black lines represent 95% confidence intervals, n=20 patients. (d) Frequencies of indicated flow cytometry gated populations at baseline (n=10 patients in each group). In (a) and (d) boxplots are shown with black horizontal line denoting median value, while box represents interquartile ranges (IQR, Q1-Q3 percentile) and whiskers show minimum (Q1– 1.5 * IQR) and maximum (Q3 + 1.5 * IQR) values. (a) Two-way ANOVA with Bonferroni correction; (d) Two-tailed Mann-Whitney U test; ****, p < 0.0001; ***, p < 0.001; **, p < 0.01; *, p < 0.05; ns, not significant.
We were able to predict response to Abatacept with 85% accuracy and an area under curve (AUC) of 0.81 (Fig. 7b). The two features that emerged as being most important in predicting C-peptide retention following Abatacept treatment were ICOS+ Tfh (CD3+CD4+CD45RA−CXCR5+ICOS+) and CXCR5+ naive cells (CD3+CD4+CXCR5+CD45RA+) (Fig. 7c,d). Again, the term “naive” is used as shorthand for CD45RA+, however cells in this gate have lower expression of CD45RA than naive T cells (see “CXCR5+ naive” quadrant in Supplementary Fig. 2). ICOS−PD-1− Tfh also contribute to predictive power in this model, with opposing directionality to ICOS+ Tfh as expected (Fig. 7d). The CCR7loPD-1+CXCR5+ cells shown previously27 to correlate with an active Tfh program were also identified in the model (CCR7−PD-1+ Tfh) (Fig. 7c,d). Grouped time-series plots illustrate the dynamic change in the frequencies of these cell populations over time (Supplementary Fig. 5c) illustrating that responder and non-responder populations are broadly non-overlapping both before and during Abatacept treatment. Note that only baseline data were used to generate the model, avoiding the caveat that Abatacept treatment directly alters the frequencies of some of these populations.
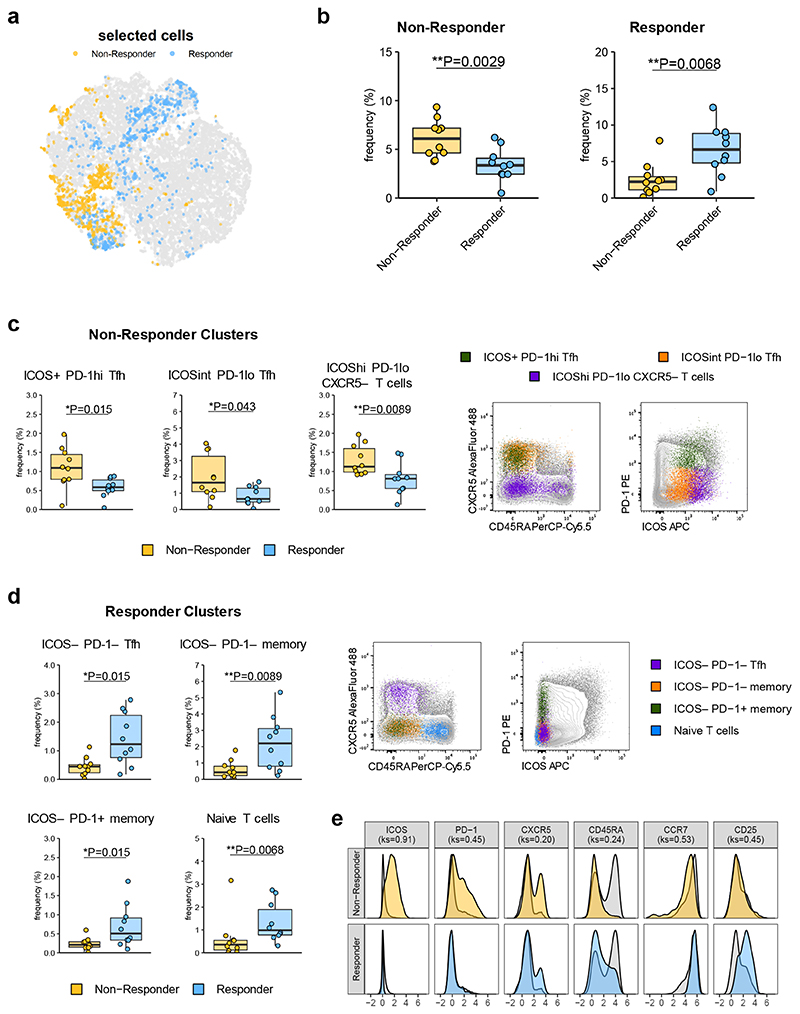
As an independent approach, we were interested in whether data-driven analysis would detect similar cell subsets at baseline that differed between individuals who went on to make good or poor clinical responses following Abatacept therapy. Using CellCnn we were able to identify two filters, one of which shows higher frequencies of corresponding cells in samples from the 10 individuals exhibiting the poorest clinical response, while the other exhibits an inverse relationship, leading us to label these filters as “Non-Responder” and “Responder”, respectively (Fig. 8a,b). In the non-responder filter, k-means clustering revealed 3 statistically significant T cell clusters; ICOS+PD-1hiTfh, ICOSintPD-1loTfh and ICOShiPD-1loCXCR5− T cells (Fig. 8c, Supplementary Fig. 6a,b). The first 2 of these provide independent support for the predictive power of the ICOS+ Tfh population identified in our gradient boosting model. Indeed, cells identified by CellCnn in those clusters overlaid the manual gates used for the predictive model (Supplementary Fig. 7a). ICOS+PD-1hi Tfh partially encompasses the CCR7−PD-1+ Tfh population also identified by the model (Supplementary Fig. 7b). Conversely, the clusters identified in the filter found for responder patients were dominated by ICOS− cell populations, including ICOS−PD-1− Tfh, ICOS−PD-1− memory cells, ICOS−PD-1+ memory cells and naive T cells (Fig. 8d, Supplementary Fig. 6c,d, Supplementary Fig. 7c).
Figure 8. Data-driven analysis identifies cell signatures linked to clinical response to Abatacept.
CellCnn analysis followed by k-means clustering of filter-specific cells was applied to flow cytometry data of samples taken at baseline from top 10 responder and non-responder Abatacept treated patients. (a) t-SNE projection of down-sampled, pooled flow cytometry data of all samples used for CellCnn analysis. Filter-specific cells for responder and non-responder filter are highlighted. (b) Frequencies of filter-specific cells in each sample for responder and non-responder filter. (c) Frequencies and representative flow cytometry overlays for clusters found in non-responder filter-specific cells. (d) Frequencies and representative flow cytometry overlays for clusters found in responder filter-specific cells. (e) Histograms of marker expression of filter-specific cells (yellow; non-responder, blue; responder) or all cells (grey). n=10 patients in each group. In (b), (c) and (d) boxplots are shown with black horizontal line denoting median value, while box represents interquartile ranges (IQR, Q1-Q3 percentile) and whiskers show minimum (Q1– 1.5 * IQR) and maximum (Q3 + 1.5 * IQR) values. (b), (c) and (d) two-tailed Mann–Whitney U test; (e) two-tailed Kolmogorov-Smirnov (ks) test; **, p < 0.01; *, p < 0.05. All representative overlay plots are from the same baseline sample.
The difference in ICOS expression between patients that go on to be Abatacept responders versus non-responders is clear from a combined analysis of all cells contributing to clusters identified in both filters (Fig. 8e). Although overall PD-1 expression also differed between the two groups (Fig. 8e), its relationship with clinical response is more complex since certain PD-1+ populations are associated with a good response (e.g. ICOS−PD-1+ memory) and others with a poor response (e.g. ICOS+PD-1+ Tfh). Notably, both ICOS and PD-1 can influence Tfh migration and function36, 37. Analysis of Abatacept-treated mice showed that an analogous staining panel could be used to build a predictive model of clinical response with 84% accuracy and an AUC of 0.83 (Supplementary Fig. 8a,b). CellCnn identified filters that were enriched in mice that went on to be responders or non-responders, with ICOS being expressed at higher levels in the cells within the non-responder clusters (Supplementary Fig. 8c,d,e). Overall, both the predictive model and the CellCnn algorithm suggested that analysis of Tfh markers in baseline blood samples could predict clinical response following Abatacept immunotherapy.
Discussion
Heterogeneity in the response to costimulation inhibitors like Abatacept limits their utility as first line therapies, and therefore the ability to predict response to these reagents would have significant impact on how they are deployed. However, a fine-grained understanding of CD28-sensitive immune subsets, and their link to pathogenicity, has been lacking.
We show here that in both mice and humans experiencing ongoing autoimmune responses, costimulation blockade reduced Tfh frequencies, with principal component analysis identifying the loss of CXCR5+ICOS+ T cells as the biggest contributor to Abatacept-induced change. Since the majority of CXCR5+ T cells are central memory cells7, 28, it is plausible that the Abatacept-induced decrease in central memory T cells previously reported32 reflects the loss of Tfh.
Importantly, we identified several new Abatacept-sensitive populations, including a population resembling Tph cells, thought to provide T cell help to B cells in the rheumatoid synovium30. Emerging data suggest these cells are expanded in children with islet autoantibodies who go on to develop diabetes38, and are associated with disease activity in SLE39 and RA40, 41, suggesting insights into their drug sensitivity could have broad applicability. Furthermore, we found that cells resembling circulating Tfh precursors27 also exhibit Abatacept sensitivity, as do a population of T cells expressing CD45RA and intermediate levels of ICOS. These could comprise recently activated T cells that have not yet lost CD45RA, or alternatively revertants that have lost, then re-expressed, this marker. Such revertants were first described in rodent models42,43, where it was shown that they retained the capacity to provide B cell help42. ICOS+CD45RA+ T cells may therefore warrant further investigation in T-cell dependent autoimmune diseases featuring autoantibody production.
Tfh that remain post-Abatacept treatment exhibit an altered phenotype with a decrease in the frequency of Tfh expressing ICOS, PD-1 or both, and a reciprocal increase in Tfh expressing neither marker. The impact on ICOS expression appears dominant; accordingly, changes in ICOS+PD-1− Tfh were a clear contributor to the Abatacept-induced variation in principal component analysis, while changes in ICOS−PD-1+ Tfh were not. The ability of CD28 signaling to promote ICOS expression is consistent with the original identification of ICOS as an “Inducible Costimulator”, responsive to CD28 engagement44, 45. Our new data suggest a continuous requirement for CD28 signalling to sustain ICOS expression, implying that this hierarchy is perpetuated even after T cell activation. Consistent with this, RNAseq analysis identified Icos to be highly CD28-sensitive in human memory T cells46.
ICOS signalling is critical for maintaining Tfh characteristics, with loss of ICOS permitting upregulation of Klf2 and a reversion of Tfh phenotype47. Our data suggest that while CD28 blockade primarily inhibits early Tfh differentiation, prolonged CD28 blockade may effectively inhibit ICOS signalling, by ensuring that remaining Tfh are ICOS-negative. Since ICOS is required for Tfh maintenance, this may explain why CD28 blockade remains capable of decreasing Tfh during an ongoing autoimmune response. Alternatively, the ability of CD28 blockade to inhibit differentiation of new Tfh would lead to a decrease over time if turnover of Tfh was high.
Using gradient boosting, an ensemble machine learning method, on gated flow cytometry outputs from pre-treatment samples, we were able to build a predictive model of Abatacept sensitivity that could assign the clinical response at year 2 with 85% accuracy. There are two caveats to this model; first, it is built on data from a relatively small number of patients and second, it intentionally focuses on the best and worst responders. Thus, while it may work with high accuracy in these patient groups, it may be less effective in individuals showing a borderline response. Notwithstanding these caveats, the model highlights several T cell populations whose collective frequencies appear to inform the clinical response to Abatacept. Chief among these is the ICOS+Tfh population, for which higher frequencies are associated with a poor clinical response. Reciprocally, ICOS−PD-1− Tfh contribute to the model, with a higher frequency being associated with a better clinical response following Abatacept treatment.
Since Abatacept has only been trialled once in new onset T1D, we were unable to apply our predictive model to an independent dataset. We therefore sought an alternative means of validation. Using CellCnn we obtained independent corroboration for key aspects of our model. Notably this approach confirmed that a poor clinical response was associated with higher frequencies of ICOS+ Tfh at baseline, while a good response was associated with higher frequencies of ICOS−PD-1− Tfh. In addition, this analysis also revealed an effect of ICOS expression on CXCR5-negative cells. Thus, ICOS appears to be the most discerning cellular marker associated with preservation of beta-cell function following Abatacept treatment as assessed by two independent approaches, with data from a mouse model providing additional support.
Robust predictive markers of responsiveness to Abatacept are currently lacking, although there are suggestions that individuals with greater inflammatory activity exhibit a better clinical response48. A recent study using whole blood RNASeq detected changes in expression of B cell genes that were associated with clinical response in subjects with T1D treated with Abatacept49, however these were not apparent until 84 days post treatment initiation. Since interventions that target Tfh inevitably alter B cell phenotype, it is tempting to speculate that such changes could be secondary to the altered T cell phenotypes observed here.
Our report is the first to suggest that baseline Tfh phenotypes have the potential to predict clinical response to an immunotherapy. It will be important to confirm these findings in a separate cohort of patients and to explore their wider applicability. For example, it remains to be established whether the T cell phenotypes we have identified can predict the response to Abatacept in other clinical settings, such as rheumatoid arthritis, or whether they are specific to T1D. Similarly, it will be important to ascertain whether these populations predict clinical response to other immunotherapies targeting costimulatory pathways or T cell / B cell collaboration. Broader implications aside, the incorporation of Tfh analysis could alter the landscape for the rational use of Abatacept and novel versions of this molecule with improved affinity, stability and pharmacokinetics that are emerging.
Methods
Patients
Cryopreserved PBMC samples from a clinical trial (NCT00505375) that has previously been published5 were provided by Type 1 Diabetes TrialNet as part of the Effects of CTLA-4 IG (Abatacept) on the Progression of Type 1 Diabetes in New Onset Subjects (TN-09) study. Briefly, in this study individuals with recent onset T1D (diagnosed within the past 100 days) were randomised to receive CTLA4-Ig (Abatacept) (10mg/kg) or placebo (saline) intravenously on days 1, 14, 28 and subsequently once monthly for 2 years. The protocol and consent document of this trial were approved by appropriate independent ethics committees or institutional review boards. All participants (or parents) provided written, informed consent; in addition to their parents providing consent, participants younger than 18 years of age signed a study assent form Samples were provided from study participants at the time of screening and 12 and 24 months following treatment initiation. Data from 36 Abatacept-treated and 14 placebo-treated patients were acquired. Samples from two Abatacept-treated individuals were excluded from the analysis due to low data quality. For one placebo-treated patient, no 12-month sample was acquired. Samples were supplied in a blinded and randomised way in two batches separated by a break of 9 months. A further set of samples from 20 Abatacept-treated and 8 placebo-treated patients were obtained and analysed during revision of the manuscript (Fig. 6, Fig. S3). Demographic and clinical data were only provided following submission of raw data files to TrialNet. To assess stimulated C-peptide secretion, four-hour mixed meal tolerance tests (MMTTs) were performed at screening and at 24 months. Additional two-hour MMTTs were conducted at 3, 6, 12 and 18 months, although for some patients C-peptide data was not available for all timepoints. For comparison across all timepoints only the first 2 hours of the 4-hour MMTTs were used.
Mice
BALB/c DO11.10 TCR transgenic mice were obtained from The Jackson Laboratory and BALB/c Cd28 −/− mice from Taconic Laboratories. BALB/c RIP-mOVA mice (expressing the ovalbumin transgene under control of the rat insulin promoter, from line 296-1B) were a gift from W. Heath (Doherty Institute, Melbourne, Australia). DO11.10 mice were crossed with RIP-mOVA mice to generate DO11.10 x RIP-mOVA mice. Mice were housed according to Home Office guidelines in individually vented cages with environmental enrichment (e.g. cardboard tunnels, paper houses, chewing blocks) in a temperature and humidity-controlled facility with a 14 h light–10h dark cycle and ad libitum feeding at University College London Biological Services Unit. Experiments were performed in accordance with the relevant Home Office project and personal licenses following approval from the University College London Animal Welfare Ethical Review Body.
In Vivo Experiments
For adoptive transfer experiments, 2 x105 T cells from DO11.10 mice were injected i.v. into Cd28 −/− recipients. 24h later, recipients were immunised i.p. with 200 μg of OVA/alum (Sigma). Where indicated mice were injected i.p. with 500 μg Abatacept (Royal Free Hospital Pharmacy) or control antibody (human IgG1, BioXCell) at the same time as adoptive transfer (d-1). Subsequently, mice received 250 μg Abatacept or control antibody every 2-3 days over the course of the experiment (see Fig. 1a). For experiments using DO11.10 x RIP-mOVA mice, 6-13 week old animals were injected i.p. with 500 μg Abatacept or control antibody. Mice were subsequently treated with 250 μg Abatacept or control antibody every 2-3 days over a period of 11 days. For experiments in Fig. S8, DO11.10 x RIP-mOVA mice with a blood glucose reading between 180 and 290 mg/dL were injected i.p. with Abatacept, 500 μg for the initial dose then 250 μg three times weekly, for four weeks and blood glucose was monitored. Mouse spleen and lymph nodes were mashed to create single cell suspensions and 2-10 x 106 cells were used for flow cytometry staining. All injections were carried out in the absence of anesthesia and analgesia, and mice were returned immediately to home cages following the procedure. The welfare of experimental animals was monitored regularly (typically immediately post procedure, then at least every 2–3 days). No unexpected adverse events were noted during the course of these experiments.
Human sample preparation
Cryopreserved samples were thawed in a 37°C water bath and vial contents transferred to a 15 mL Falcon tube. Pre-warmed defrost media (RPMI (Glutamax with HEPES) (Life Technologies (Thermo Fisher)), 5% human AB serum (Sigma), 20 nM TAPI-2 (Sigma), 50 U/mL Benzonase (Sigma) was added dropwise to 10 mL. Cells were rested in 4 mL resting media (RPMI with 10% human AB serum, 20 nM TAPI-2) for 1 hour at 37°C. 2 x 106 cells were used for subsequent flow cytometry staining.
Flow Cytometry
Mouse cells were surface stained with Fas PE (BD Biosciences, clone: Jo2, 1/50), CD19 BUV395 (BD Biosciences, clone: 1D3, 1/50), CD4 BUV395 (BD Biosciences, clone: GK1.5, 1/100), CD4 PerCP-Cy5.5 (BD Biosciences, clone: RM4-5, 1/100), GL7 AlexaFluor 488 (Biolegend, clone: GL7, 1/200), CXCR5 BV421 (Biolegend, clone: L138D7, 1/20), PD-1 PE-Cy7 (Biolegend, clone: RMP1-30, 1/50), ICOS PE (eBioscience (Thermo Fisher), clone: 7E.17G9, 1/100), CD45 BUV395 (BD Biosciences, clone: 30-F11, 1/100), CD45RB APC (used in mouse panels in place of CD45RA, eBioscience (Thermo Fisher), clone: C363.16A, 1/100) and DO11.10 TCR APC (eBioscience (Thermo Fisher), clone: KJ126, 1/100) for 30 minutes at 4°C. Cells were fixed and permeabilised using the Foxp3/Transcription Factor Staining Buffer Set (eBioscience (Thermo Fisher)) and stained intracellularly with CD40L PE (BD Biosciences, clone: MR1, 1/100) for 30 minutes at 4°C. For experiments involving Abatacept blockade in DO11.10 x RIP-mOVA mice, cells were stained with fixable viability dye eFluor 780 (eBioscience (Thermo Fisher)) in PBS for 10 minutes at 4°C. After washing once with PBS containing 2% fetal calf serum, samples were preincubated with purified anti-CD16/32 (BD Biosciences, clone: 2.4G2, 1 μL) for 5 minutes at 37°C. In Fig. S8, mouse cells were preincubated with purified anti-CD16/32 for 5 minutes at 37°C and stained with CXCR5 BV421 for 30 minutes at 37°C. Subsequently, an antibody cocktail containing CD3 BUV395 (BD Biosciences, clone: 145-2C11, 1/50), CD4 PerCP-Cy5.5, CD45RB APC, CCR7 AlexaFluor 488 (Biolegend, clone: 4B12, 2.5 μL), PD-1 PE-Cy7, ICOS PE, CD25 PE-Cf594 (BD Biosciences, clone: PC61, 1/200) and fixable viability dye eFluor 780 was added and cells were incubated for 30 minutes at 37°C.
Human cells were washed once in PBS and stained for 15 minutes at 37°C with CCR7 BV605 (Biolegend, clone: G043H7, 3 μL) in Brilliant Stain Buffer (BD Biosciences). An antibody cocktail containing CD3 BUV395 (BD Biosciences, clone: SK7, 3 μL), CD4 PE-Cy7 (BD Biosciences, clone: SK3, 3 μL), CD25 BV421 (BD Biosciences, clone: M-A251, 3 μL), CD45RA PerCP-Cy5.5 (eBioscience (Thermo Fisher), clone: HI100, 1 μL), CD62L AlexaFluor 700 (Biolegend, clone: DREG-56, 3 μL), CD127 BV711 (BD Biosciences, clone: HIL-7R-M21, 3 μL), CXCR5 AlexaFluor 488 (BD Biosciences, clone: RF8B2, 5 μL), PD-1 PE (eBioscience (Thermo Fisher), clone: ebioJ105, 3 μL) and ICOS biotin (eBioscience (Thermo Fisher), clone: ISA-3, 3 μL) was subsequently added and cells were incubated for another 15 minutes at 4°C. Cells were then washed in PBS, streptavidin APC (BD Biosciences, 2 μL) was added to the residual volume and cells were incubated for 10 minutes at 4°C. Cells were resuspended in fixable viability dye eFluor 780 in PBS and incubated for 10 minutes at 4°C before being washed in PBS twice. Due to technical issues with CD62L staining this marker was not considered in any of the downstream analysis. In Fig. 6, human cells were sequentially stained with CCR2 BV510 (Biolegend, clone: K036C2, 3 μL), CCR5 BUV737 (BD Biosciences, clone: 2D7, 1 μL) and CCR7 BV605 at 37°C for 30, 20 and 15 minutes, respectively. Subsequently, an antibody cocktail containing CD3 BUV395, CD4 PE-Cy7, CXCR5 AlexaFluor 488, CD45RA PerCP-Cy5.5, HLA-DR BV785 (Biolegend, clone: L243, 3 μL), CD38 PE-Cf594 (BD Biosciences, HIT2, 1 μL), TIGIT BV421 (Biolegend, clone: A15153G, 3 μL) and BTLA BV650 (BD Biosciences, clone: J168-540, 2 μL) was added and cells were incubated for 15 minutes at 4°C. In Fig. S3, human cells were stained with CD3 BUV395, CD4 PE-Cy7, CXCR5 AlexaFluor 488, CD45RA PerCP-Cy5.5, CXCR3 BV785 (Biolegend, clone: G025H7, 3 μL) and CCR6 APC-R700 (BD Biosciences, clone: 11A9, 3 μL) for 15 minutes at 4°C.
All data was acquired on a BD LSRFortessa (BD Biosciences) using BD FACSDiva (BD Biosciences). For manual analysis, data was analysed using FlowJo software version 10. For automated analysis, data was pre-gated on live CD3+CD4+ cells in FlowJo, loaded into R using the Bioconductor package flowCore and underwent quality control using Bioconductor package FlowAI with standard configurations50. Low-quality events were removed and marker expression was transformed using arcsinh transformation using the Bioconductor package flowVS. CellCnn was run using a filter difference threshold of 0.5, maximum epochs of 100 and otherwise standard configurations. Filter specific cells were identified as cells having a filter response value in the upper 5% of the overall filter response. K-means clustering was performed using the CRAN package Stats, and optimal number of clusters were chosen using the Elbow method. Cluster information was added to fcs files using Bioconductor packages CytoML and flowWorkspace. The CRAN package Rtsne was used to compute t-SNE.
Statistics and Predictive Modelling
Statistical analysis was performed using R v3.5.1 and Python v3.7. Two-sided Mann–Whitney U was used for comparison of two unpaired means. For comparison of paired means two-sided Wilcoxon signed-rank test was used. Comparison of more than two means was performed using two-sided ANOVA or Kruskal-Wallis test with Bonferroni correction. Equality of histograms in Fig. 6e and Fig. S8e was assessed using the Kolmogorov-Smirnov test. Normality was tested using Shapiro-Wilk test and homogeneity of variance was tested using Levene’s test. All measurements were taken from distinct samples. For boxplots, the black horizontal line indicates the median, the boxes represent interquartile range (IQR, Q1-Q3 percentile) and whiskers show minimum (first quartile - 1.5 * IQR) and maximum (third quartile + 1.5 * IQR). Principal component analysis was performed on scaled and centered data. Plots were produced using either CRAN packages ggplot2, ggpubr, ggsignif, RColourBrewer and scales in R or matplotlib and seaborn in Python. All predictive modelling was conducted using Python v3.7. Data cleaning and formatting was carried out using either CRAN packages plyr, stringr and tidyr in R or pandas and numpy in Python. The gradient boosting algorithm was implemented using sklearn. Feature correlation was calculated using Pearson’s r to ensure that the model did not contain sets of highly correlated variables which can make predictions unstable and dilute feature importance effects. Feature pairs which were correlated with r>=0.95 were determined, and only the most predictive feature of each pair was included in the gradient boosting model.
Supplementary Material
Acknowledgements
This work was funded by Diabetes UK, MedImmune Ltd (now AstraZeneca plc), the Medical Research Council, and the Rosetrees Trust. The authors received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement No 675395. Diabetes research in the Walker lab is supported by Type One Mission. We acknowledge the support of the Type 1 Diabetes TrialNet Study Group, which identified study participants and provided samples and follow-up data for this study. The Type 1 Diabetes TrialNet Study Group is a clinical trials network funded by the National Institutes of Health (NIH) through the National Institute of Diabetes and Digestive and Kidney Diseases, the National Institute of Allergy and Infectious Diseases, and The Eunice Kennedy Shriver National Institute of Child Health and Human Development, through the cooperative agreements U01 DK061010, U01 DK061016, U01 DK061034, U01 DK061036, U01 DK061040, U01 DK061041, U01 DK061042, U01 DK061055, U01 DK061058, U01 DK084565, U01 DK085453, U01 DK085461, U01 DK085463, U01 DK085466, U01 DK085499, U01 DK085505, U01 DK085509, and JDRF. The contents of this article are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or JDRF. We thank A. Pesenacker for helpful comments on the manuscript.
Footnotes
Author contributions
N.M.E. performed experiments, analysed data, prepared figures and co-wrote the manuscript. F.H. performed experiments, analysed data and edited the manuscript. N.T. performed predictive modelling, prepared figures and co-wrote the manuscript. C.J.W., L.P., R.K., A.K., V.O., E.M.R., E.N., Y.E., M.E., and R.B., assisted with experiments and edited the manuscript. P.A., and L.J., provided expertise and funding. M.P. provided expertise and facilitated sample sharing. M.R. provided expertise and was co-applicant for funding. L.S.K.W. conceptualised and supervised the study, applied for funding and wrote the manuscript.
Competing interests
AstraZeneca plc contributed to the funding of the study. P.A. and L.J. declare an interest in developing costimulation blockade reagents at AstraZeneca plc. L.S.K.W. and N.T. are inventors on a patent application related to these findings.
Code Availability
Computer code is available from the corresponding author on request.
Data Availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.
References
- 1.Blair HA, Deeks ED. Abatacept: A Review in Rheumatoid Arthritis. Drugs. 2017;77:1221–1233. doi: 10.1007/s40265-017-0775-4. [DOI] [PubMed] [Google Scholar]
- 2.Mease PJ, et al. Efficacy and safety of abatacept, a T-cell modulator, in a randomised, double-blind, placebo-controlled, phase III study in psoriatic arthritis. Ann Rheum Dis. 2017;76:1550–1558. doi: 10.1136/annrheumdis-2016-210724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brunner HI, et al. Subcutaneous Abatacept in Patients With Polyarticular-Course Juvenile Idiopathic Arthritis: Results From a Phase III Open-Label Study. Arthritis & rheumatology. 2018;70:1144–1154. doi: 10.1002/art.40466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lenschow DJ, et al. Differential effects of anti-B7-1 and anti-B7-2 monoclonal antibody treatment on the development of diabetes in the nonobese diabetic mouse. The Journal of experimental medicine. 1995;181:1145–1155. doi: 10.1084/jem.181.3.1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Orban T, et al. Co-stimulation modulation with abatacept in patients with recent-onset type 1 diabetes: a randomised, double-blind, placebo-controlled trial. Lancet. 2011;378:412–419. doi: 10.1016/S0140-6736(11)60886-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Orban T, et al. Costimulation modulation with abatacept in patients with recent-onset type 1 diabetes: follow-up 1 year after cessation of treatment. Diabetes Care. 2014;37:1069–1075. doi: 10.2337/dc13-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kenefeck R, et al. Follicular helper T cell signature in type 1 diabetes. Journal of Clinical Investigation. 2015;125:292–303. doi: 10.1172/JCI76238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu D, et al. The transcriptional repressor Bcl-6 directs T follicular helper cell lineage commitment. Immunity. 2009;31:457–468. doi: 10.1016/j.immuni.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 9.Johnston RJ, et al. Bcl6 and Blimp-1 are reciprocal and antagonistic regulators of T follicular helper cell differentiation. Science. 2009;325:1006–1010. doi: 10.1126/science.1175870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nurieva RI, et al. Bcl6 mediates the development of T follicular helper cells. Science. 2009;325:1001–1005. doi: 10.1126/science.1176676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Heit A, et al. Vaccination establishes clonal relatives of germinal center T cells in the blood of humans. J Exp Med. 2017;214:2139–2152. doi: 10.1084/jem.20161794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hill DL, et al. The adjuvant GLA-SE promotes human Tfh cell expansion and emergence of public TCRbeta clonotypes. J Exp Med. 2019;8:1857–1873. doi: 10.1084/jem.20190301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schmitt N, Bentebibel SE, Ueno H. Phenotype and functions of memory Tfh cells in human blood. Trends in Immunology. 2014;35:436–442. doi: 10.1016/j.it.2014.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sage PT, Alvarez D, Godec J, von Andrian UH, Sharpe AH. Circulating T follicular regulatory and helper cells have memory-like properties. Journal of Clinical Investigation. 2014;12:5191–204. doi: 10.1172/JCI76861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu X, et al. Inhibition of increased circulating Tfh cell by anti-CD20 monoclonal antibody in patients with type 1 diabetes. PLoS One. 2013;8:e79858. doi: 10.1371/journal.pone.0079858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferreira RC, et al. IL-21 production by CD4 effector T cells and frequency of circulating follicular helper T cells are increased in type 1 diabetes patients. Diabetologia. 2015;58:781–790. doi: 10.1007/s00125-015-3509-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Serr I, et al. miRNA92a targets KLF2 and the phosphatase PTEN signaling to promote human T follicular helper precursors in T1D islet autoimmunity. Proc Natl Acad Sci U S A. 2016;113:E6659–E6668. doi: 10.1073/pnas.1606646113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viisanen T, et al. Circulating CXCR5+PD-1+ICOS+ Follicular T Helper Cells Are Increased Close to the Diagnosis of Type 1 Diabetes in Children With Multiple Autoantibodies. Diabetes. 2017;66:437–447. doi: 10.2337/db16-0714. [DOI] [PubMed] [Google Scholar]
- 19.Borriello F, et al. B7-1 and B7-2 have overlapping, critical roles in immunoglobulin class switching and germinal center formation. Immunity. 1997;6:303–313. doi: 10.1016/s1074-7613(00)80333-7. [DOI] [PubMed] [Google Scholar]
- 20.Walker LS, et al. Compromised OX40 function in CD28-deficient mice is linked with failure to develop CXCR5-positive CD4 cells and germinal centers. J Exp Med. 1999;190:1115–1122. doi: 10.1084/jem.190.8.1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang CJ, et al. CTLA-4 controls follicular helper T-cell differentiation by regulating the strength of CD28 engagement. Proc Natl Acad Sci U S A. 2015;112:524–529. doi: 10.1073/pnas.1414576112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Verstappen GM, et al. Attenuation of Follicular Helper T Cell-Dependent B Cell Hyperactivity by Abatacept Treatment in Primary Sjogren’s Syndrome. Arthritis & rheumatology. 2017;69:1850–1861. doi: 10.1002/art.40165. [DOI] [PubMed] [Google Scholar]
- 23.Piantoni S, Regola F, Scarsi M, Tincani A, Airo P. Circulating follicular helper T cells (CD4+CXCR5+ICOS+) decrease in patients with rheumatoid arthritis treated with abatacept. Clin Exp Rheumatol. 2018;36:685. [PubMed] [Google Scholar]
- 24.Glatigny S, et al. Abatacept Targets T Follicular Helper and Regulatory T Cells, Disrupting Molecular Pathways That Regulate Their Proliferation and Maintenance. J Immunol. 2019 doi: 10.4049/jimmunol.1801425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clough LE, et al. Release from Regulatory T Cell-Mediated Suppression during the Onset of Tissue-Specific Autoimmunity Is Associated with Elevated IL-21. J Immunol. 2008;180:5393–5401. doi: 10.4049/jimmunol.180.8.5393. [DOI] [PubMed] [Google Scholar]
- 26.Walker LS, Chodos A, Eggena M, Dooms H, Abbas AK. Antigen-dependent proliferation of CD4+ CD25+ regulatory T cells in vivo. J Exp Med. 2003;198:249–258. doi: 10.1084/jem.20030315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.He J, et al. Circulating precursor CCR7(lo)PD-1(hi) CXCR5(+) CD4(+) T cells indicate Tfh cell activity and promote antibody responses upon antigen reexposure. Immunity. 2013;39:770–781. doi: 10.1016/j.immuni.2013.09.007. [DOI] [PubMed] [Google Scholar]
- 28.Morita R, et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity. 2011;34:108–121. doi: 10.1016/j.immuni.2010.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arvaniti E, Claassen M. Sensitive detection of rare disease-associated cell subsets via representation learning. Nature communications. 2017;8 doi: 10.1038/ncomms14825. 14825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rao DA, et al. Pathologically expanded peripheral T helper cell subset drives B cells in rheumatoid arthritis. Nature. 2017;542:110–114. doi: 10.1038/nature20810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pieper J, et al. CTLA4-Ig (abatacept) therapy modulates T cell effector functions in autoantibody-positive rheumatoid arthritis patients. BMC Immunol. 2013;14:34. doi: 10.1186/1471-2172-14-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Orban T, et al. Reduction in CD4 central memory T-cell subset in costimulation modulator abatacept-treated patients with recent-onset type 1 diabetes is associated with slower C-peptide decline. Diabetes. 2014;63:3449–3457. doi: 10.2337/db14-0047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leete P, et al. Differential Insulitic Profiles Determine the Extent of beta-Cell Destruction and the Age at Onset of Type 1 Diabetes. Diabetes. 2016;65:1362–1369. doi: 10.2337/db15-1615. [DOI] [PubMed] [Google Scholar]
- 34.Breiman L. Technical Report 486. Dept. Statistics; Univ. California, Berkeley: 1997. Arcing the edge. Available at www.stat.berkeley.edu. [Google Scholar]
- 35.Friedman JH. Technical Report. Dept. Statistics; Stanford University: 1999. Greedy Function Approximation: A Gradient Boosting Machine. [Google Scholar]
- 36.Xu H, et al. Follicular T-helper cell recruitment governed by bystander B cells and ICOS-driven motility. Nature. 2013;496:523–527. doi: 10.1038/nature12058. [DOI] [PubMed] [Google Scholar]
- 37.Shi J, et al. PD-1 Controls Follicular T Helper Cell Positioning and Function. Immunity. 2018;49:264–274 e264. doi: 10.1016/j.immuni.2018.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ekman I, et al. Circulating CXCR5(-)PD-1(hi) peripheral T helper cells are associated with progression to type 1 diabetes. Diabetologia. 2019;62:1681–1688. doi: 10.1007/s00125-019-4936-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bocharnikov AV, et al. PD-1hiCXCR5-T peripheral helper cells promote B cell responses in lupus via MAF and IL-21. JCI insight. 2019;4 doi: 10.1172/jci.insight.130062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang F, et al. Defining inflammatory cell states in rheumatoid arthritis joint synovial tissues by integrating single-cell transcriptomics and mass cytometry. Nat Immunol. 2019 doi: 10.1038/s41590-019-0378-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fortea-Gordo P, et al. Two populations of circulating PD-1hiCD4 T cells with distinct B cell helping capacity are elevated in early rheumatoid arthritis. Rheumatology (Oxford) 2019;58:1662–1673. doi: 10.1093/rheumatology/kez169. [DOI] [PubMed] [Google Scholar]
- 42.Bell EB, et al. Both CD45R(low) and CD45R(high) “revertant” CD4 memory T cells provide help for memory B cells. Eur J Immunol. 2001;31:1685–1695. doi: 10.1002/1521-4141(200106)31:6<1685::aid-immu1685>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 43.Merica R, Khoruts A, Pape KA, Reinhardt RL, Jenkins MK. Antigen-experienced CD4 T cells display a reduced capacity for clonal expansion in vivo that is imposed by factors present in the immune host. J Immunol. 2000;164:4551–4557. doi: 10.4049/jimmunol.164.9.4551. [DOI] [PubMed] [Google Scholar]
- 44.Hutloff A, et al. ICOS is an inducible T-cell co-stimulator structurally and functionally related to CD28. Nature. 1999;397:263–266. doi: 10.1038/16717. [DOI] [PubMed] [Google Scholar]
- 45.McAdam AJ, et al. Mouse inducible costimulatory molecule (ICOS) expression is enhanced by CD28 costimulation and regulates differentiation of CD4+ T cells. J Immunol. 2000;165:5035–5040. doi: 10.4049/jimmunol.165.9.5035. [DOI] [PubMed] [Google Scholar]
- 46.Glinos DA, Soskic B, Jostins L, Sansom DM, Trynka G. Genomic profiling of T cell activation reveals dependency of memory T cells on CD28 costimulation. 2019 doi: 10.1038/s41435-020-00118-0. Preprint at https://www.biorxiv.org/content/biorxiv/early/2018/09/18/421099.full.pdf. [DOI] [PMC free article] [PubMed]
- 47.Weber JP, et al. ICOS maintains the T follicular helper cell phenotype by down-regulating Kruppel-like factor 2. J Exp Med. 2015;212:217–233. doi: 10.1084/jem.20141432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cabrera SM, et al. Innate immune activity as a predictor of persistent insulin secretion and association with responsiveness to CTLA4-Ig treatment in recent-onset type 1 diabetes. Diabetologia. 2018;61:2356–2370. doi: 10.1007/s00125-018-4708-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Linsley PS, Greenbaum CJ, Speake C, Long SA, Dufort MJ. B lymphocyte alterations accompany abatacept resistance in new-onset type 1 diabetes. JCI insight. 2019;4 doi: 10.1172/jci.insight.126136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Monaco G, et al. flowAI: automatic and interactive anomaly discerning tools for flow cytometry data. Bioinformatics. 2016;32:2473–2480. doi: 10.1093/bioinformatics/btw191. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Computer code is available from the corresponding author on request.
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files.