Abstract
Organoid technology holds great promise for regenerative medicine but has not yet been applied to humans. Here, we address this challenge in the context of cholangiocyte organoids and cholangiopathies, which represent a leading indication for liver transplantation. Using single-cell RNA sequencing we show that primary human cholangiocytes display transcriptional diversity which is lost in organoid culture. However, cholangiocyte organoids remain plastic and resume their in vivo signatures when transplanted back in the biliary tree. We then utilize a new model of cell engraftment in human livers undergoing ex vivo normothermic perfusion to demonstrate that this property allows extrahepatic organoids to repair human intrahepatic ducts after transplantation. Our results provide proof-of-principle that cholangiocyte organoids can be used to repair human biliary epithelium.
Organoids have a unique potential for tissue repair as they retain key functions and characteristics of their tissue of origin. Nevertheless, their ability to repair native epithelia and restore their complexity has not been established in humans, while organoid engraftment and survival in vivo has only been demonstrated in a limited number of animal studies (1). The bile duct epithelium presents an archetypal and clinically important system for addressing this challenge and for developing proof-of-concept studies in human. Indeed, disorders of the biliary system, which transfers bile from the liver to the duodenum, account for 70% of paediatric and up to a third of adult liver transplantation (2). This results in a pressing need for therapeutic alternatives, such as cell-based therapy. Furthermore, organoids suitable for regenerative medicine applications can be easily derived from biliary epithelial cells, known as cholangiocytes (3). Finally, the bile ducts also recapitulate the epithelial diversity found in other hollow-lumen organs (4). Indeed, different regions along the biliary tree display distinct transcriptional profiles and functional properties, such as the chemical modification of bile (5, 6), as well as variation in disease susceptibility between the intrahepatic ducts, extrahepatic ducts and the gallbladder. Nevertheless, the impact of this regional variation on the characteristics and regenerative potential of the organoids derived from different regions of the biliary tree remains to be characterized. To address these questions and demonstrate the value of organoids for regenerative medicine in humans, we first characterize cholangiocyte diversity in vivo using single-cell transcriptomics and confirm that different regions of the human biliary tree contain cells with distinct transcriptional profiles. We then show that cholangiocytes lose these differences in organoid culture and become interchangeable, but their regional identity can be restored in vitro by environmental stimuli. We subsequently use a biliary injury mouse model and a novel model for cell transplantation in human organs undergoing ex vivo normothermic perfusion to prove that this plasticity allows cholangiocytes from one region to repair a different region of the biliary tree paving the way for cell-based therapy using organoids.
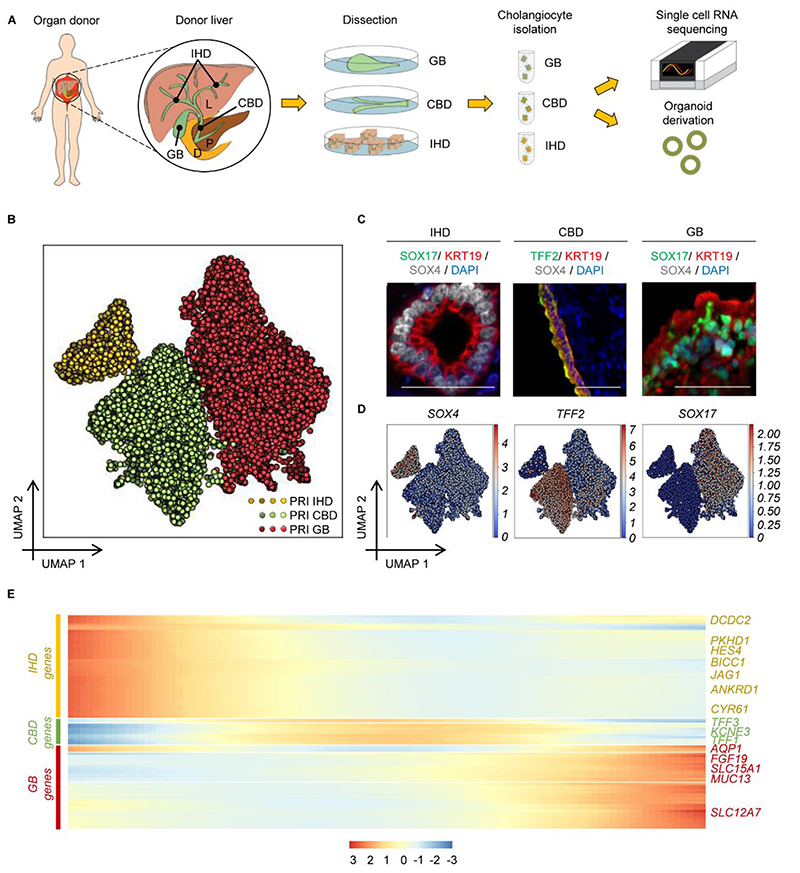
To characterize the cellular composition of the human biliary epithelium, cholangiocytes from different regions (Intrahepatic Bile Ducts (IHD): 5 patients, 7295 cells; Common Bile Duct (CBD): 3 patients, 3006 cells; Gallbladder (GB): 3 patients, 3702 cells) were isolated using magnetic bead sorting and their transcriptome was determined using droplet encapsulation single-cell RNA sequencing (scRNAseq) (Fig. 1A-B, Fig. S1A-C). The isolated cells expressed key cholangiocyte markers, including KRT7, KRT19, SOX9, and GGT (Fig. S2A). The transcriptomes of all three biliary cell populations shared a core transcriptional profile, illustrated by their proximity in UMAP space and high connectivity in Partition-based Graph Abstraction (PAGA) analysis when compared to different liver cell types, such as stellate cells and liver sinusoidal endothelial cells (LSECs, Fig. S2B-S2E). However, more detailed analysis after sub-clustering of cholangiocytes revealed non-overlapping expression modules of the three populations (Fig. 1B). This suggests that, despite their similarities, cholangiocytes from different regions exhibit unique gene expression signatures (6). Accordingly, Differentially Expressed Genes (DEG) analysis (Data S1) identified known region-specific markers, including aquaporins (7), mucins (8), FGF19 (9), SOX17 (10) in the extrahepatic biliary tree, JAG1 (11), TACSTD2 (12) and YAP target genes in intrahepatic cholangiocytes (18, 14), as well as novel markers including DCDC2, TFF1-3, SLC15A1 (Fig. 1C-1D, Fig. S3A-S3D). Most of these genes correspond to functional markers such as bile acid receptors or channels modifying bile composition (Fig. S3C). Thus, the transcriptional divergence among cholangiocytes from different regions could reflect adaptation to their microenvironment, such as variation in bile properties along the biliary tree (15). Accordingly, cholangiocytes from anatomically adjacent and hence environmentally similar regions (e.g. intrahepatic and common bile duct vs. gallbladder) displayed higher transcriptional similarity. This was illustrated by PAGA analysis (Fig. S3E-S3F), in agreement with results from diffusion pseudotime (DPT) and single-cell consensus clustering (SC3) analyses (Fig. S4-S5). These results point towards a progressive change in the expression of region-specific markers (Fig. 1E, Data S2), and a gradual transition in the transcriptional signature of cholangiocytes from adjacent regions (Fig. S4A-S4C) rather than distinct subpopulations (Fig. 1E, S4-S5). This gradient in gene expression is likely to support adjustment of the cells to environmental conditions, such as the gradual change in bile composition from the intrahepatic ducts to the gallbladder. In conclusion, our results show that the human biliary epithelium is comprised of cholangiocytes displaying a gradual shift in their transcriptional profile along the biliary tree, which is likely imposed by region-specific microenvironments.
Fig. 1. Transcriptional profiling of primary cholangiocytes.
(A) Schematic representation of the methodology used for single cell RNA sequencing (scRNAseq). (B) UMAP plot (7295 primary cells, n=10 individuals) illustrating distinct primary cholangiocyte populations in different regions of the biliary tree. (C-D) Immunofluorescence images (C) and UMAP representation of normalized gene expression (D) of primary cholangiocytes illustrating differential expression of representative region markers. Scale bars: 50μm. (E) Heatmap of top 100 differentially expressed genes (DEGs) in pseudotime (Data S2) demonstrating a gradual transition in the transcriptional profile of cholangiocytes between different regions of the biliary tree. PRI, Primary; IHD, IntraHepatic Ducts; CBD, Common Bile Duct; GB, Gallbladder; P, Pangreas; D, Duodenum.
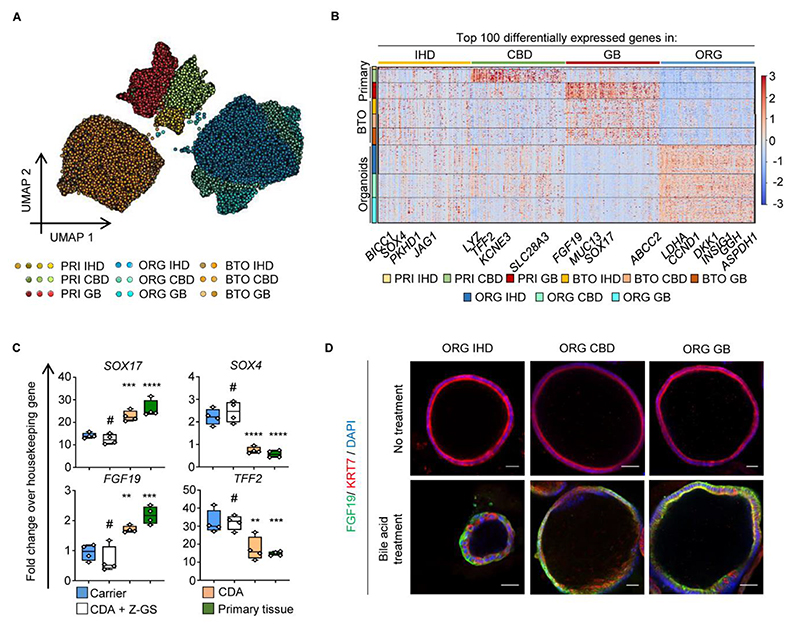
We subsequently used this single-cell map of the human biliary tree as a framework to characterise cholangiocyte organoids. To this end, a fraction of the primary cholangiocytes isolated for scRNAseq from each region (IHD, CDB, GB) were propagated as organoids using our established conditions (3, 16). The resulting organoids expressed cholangiocyte markers (KRT7, KRT19, SOX9, HNF1B, CFTR; Fig. S6A-S6B); displayed comparable functionality (ALP, GGT activity; Fig. S6C-S6D) and similar expansion potential regardless of their region of origin (Fig. S6E). To further explore these similarities, we performed scRNAseq on these organoids (2 lines per region; GB: 5859 cells; CBD 5321 cells; IHD 6641 cells; Fig. S1A-C). UMAP and PCA analyses demonstrated that organoids exhibited overlapping transcriptomic profiles (Fig. 2A, Fig. S7A-S7D) indicating that cholangiocytes grown in vitro assume a similar transcriptional signature independent of their region of origin. Of note, regressing cell cycle related genes did not change these observations excluding that a common “proliferation” signature could mask differences between organoids of different spatial origins (Fig. S7A-S7C, S7E). Furthermore, we did not detect any cells co-expressing known somatic stem cell markers (LGR5, PROM1, TACSTD2, NCAM), excluding the possibility that organoid similarities reflect a common progenitor/stem cell identity (Fig. S7F).
Fig. 2. Cholangiocyte Organoid (CO) identity is controlled by niche stimuli.
(A) UMAP (35,603 cells) of primary cholangiocytes and their corresponding organoids before and after gallbladder bile treatment, illustrating similarities between different region organoids and changes in their signature in response to bile. PRI, Primary; IHD, IntraHepatic Ducts; CBD, Common Bile Duct; GB, Gallbladder; ORG, Organoids; BTO, Bile-Treated Organoids. (B) Heatmap of top 100 Differentially Expressed Genes (DEGs) between primary regions, organoids and BTOs (Data S1–S2), illustrating that organoids lose regional differences and upregulate culture-related genes, but re-acquire gallbladder markers following bile treatment. (C-D) QPCR (C) (n=4 samples per group; center line, median; box, interquartile range (IQR); whiskers, range; housekeeping gene, HMBS; #P>0.05, **P<0.01, ***P<0.001, ****P<0.0001); and immunofluorescence (D) demonstrating upregulation of gallbladder markers and bile acid target genes following treatment with chenodeoxycholic acid (CDA), in the absence of the FXR inhibitor Z-GS. Z-GS, Z-guggulsterone. Scale bars, 50μm.
We then compared organoids from different regions with primary cholangiocytes to explore if these similarities corresponded to loss of their original regional identity in vitro (Fig. 2A). Organoids and primary cells following cell cycle regression shared a core transcriptional profile reflecting their common cholangiocyte nature, which was illustrated by their proximity in UMAP space and high PAGA connectivity when compared to different liver cell types, such as stellate cells and LSECs (Fig. S7C-D). However, DEG analyses highlighted downregulation of region specific markers, such as SLC13A1 and SLC26A3 (Fig. 2B, Fig. S7G); while Gene Ontology (GO) and Gene Set Enrichment Analyses (GSEA) identified these DEGs as factors facilitating the adaptation of cholangiocytes to their respective microenvironments, e.g. bile acid vs. culture medium processing genes (Fig. S8A-S8C). Furthermore, we confirmed upregulation of YAP target genes (Data S3) in organoids, in accordance with previous reports (14). Consequently, primary cholangiocytes propagated as organoids adapt to their new microenvironment by maintaining their core transcriptional signature, while losing the expression of markers specific to their region of origin.
To explore the mechanisms controlling cholangiocyte identity, we decided to add bile in our culture conditions as the principal determinant of the cholangiocyte microenvironment. Different organoids (IHD, CBD, GB) were treated with human gallbladder bile for 72 hours and then characterized using scRNAseq (Fig. 2A, S1A-S1C) (GB: 3815 cells; CBD 3224 cells; IHD 3653 cells). UMAP and PCA revealed that treated organoids assumed a new overlapping gene expression profile (Fig. 2A, S9A) confirming a shared capacity to adapt to exposure to bile. Importantly, PAGA and DEG analyses showed that this transcriptional profile was shifted towards a gallbladder identity (Fig. 2B, Fig. S9B-S9C). To characterise the factors controlling this transition, we interrogated differentially expressed genes in bile-treated organoids. GO, GSEA and UMAP analyses (Fig. S9D-S9F) confirmed the induction of region-specific markers (SOX17, MUC13, FGF19; Fig. 2B, S9F) and revealed upregulation of bile acid receptor pathways and downstream targets (NR1H4/FXR, NR1I2, NR0B2, SLC51A, FGF19, ABCA1, PPARG; Fig. 2B, S9D-S9F). Of note, these results were validated through activation and inhibition of the Farnesoid X receptor (FXR), using chenodeoxycholic acid and z-guggulsterone respectively (Fig. 2C-2D), thereby confirming that regardless of their origin, cholangiocytes grown in vitro can respond and adapt to environmental stimuli. Together, these results suggest that cholangiocyte organoids could assume different regional identities when instructed by the appropriate niche factors.
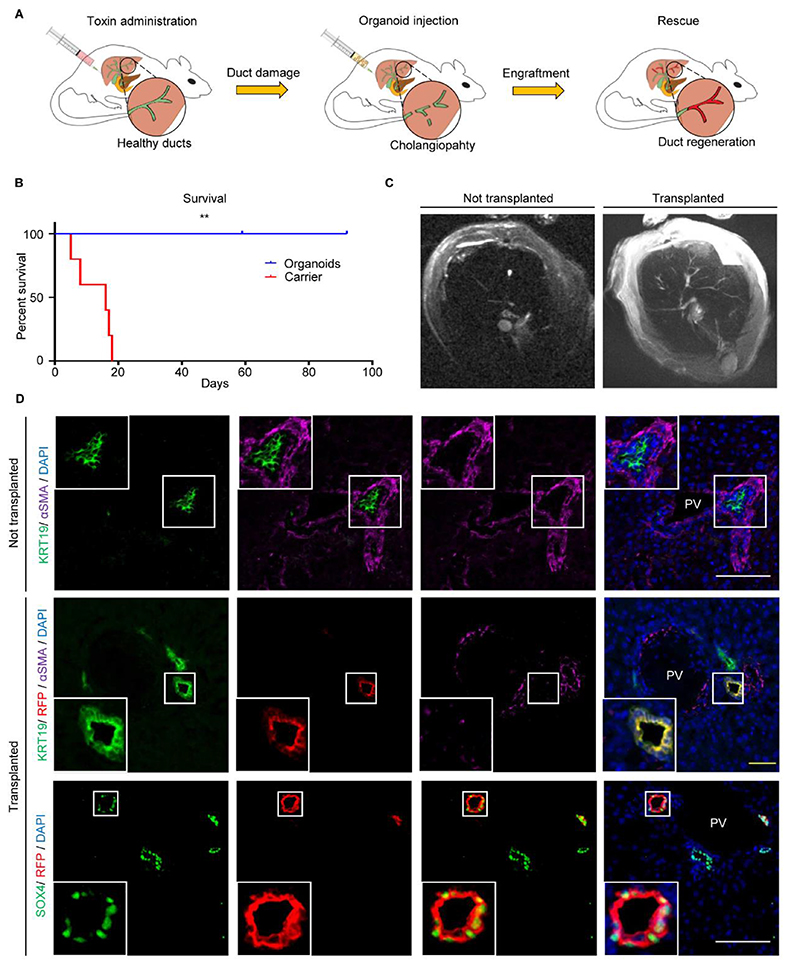
To validate cholangiocyte plasticity and explore its functional implications, we decided to assess if organoids from one region of the biliary tree could repair a different region following transplantation. For this, we induced cholangiopathy in immunodeficient mice using 4,4’ methylenedianiline (MDA) (17) (Fig. 3A-3B, S10, S11) and attempted to rescue the phenotype with intraductal delivery (18) of human gallbladder organoids expressing Red Fluorescent Protein-expressing (RFP). Control animals receiving carrier medium without cells lost weight (Fig. S10A) and died within 3 weeks (Fig. 3B, Table S1), developing cholestasis (Fig. S10B) and cholangiopathy demonstrated by IF (Fig. S10C), histology (Fig. S10D) and Magnetic Resonance Cholangio-Pancreatography (MRCP) (Fig. 3C, S11A-S11C, Movie S1–S6). On the contrary, animals receiving organoids were electively culled at the end of the experiment and survived for up to 3 months with resolution of cholangiopathy and normal serum biochemistry (Fig. 3B-3C, S10A-S10B, S11B-S11C, Movie S3–S4, S6). The transplanted gallbladder cholangiocytes engrafted in various size intrahepatic ducts (Fig. 3D, S12A-C, Movie S7–S9) corresponding to ~25-55% of the regenerated biliary epithelium (Fig. S12C), and assumed an intrahepatic identity by losing gallbladder (SOX17) and expressing intrahepatic markers (SOX4, DCDC2, BICC1) (Fig. 3D, Fig. S12A-S12B). Core biliary markers (KRT7, KRT19, CFTR) were also expressed (Fig. S12A), while we observed YAP activation both in engrafted and native cells (Fig. S12B, S12E) in accordance with previous reports (13). Of note, we never observed expression of other hepatic lineage markers such as albumin indicating that cholangiocyte organoid plasticity is likely to be limited to their biliary lineage (Fig. S12A). Furthermore, the engrafted cells expressed proliferation markers (Fig. S12B, S12D) at similar levels to native mouse cholangiocytes; while abnormal growth or tumour formation was never noticed in all the analyses performed (Fig. 3C, 3D, S10D, S12A-S12B), including T1 weighed body MR imaging at the end of the experiment (Movie S1, S3). Thus, organoid transplantation provides the healthy cells required to repair the damaged epithelium and rescue acute injury.
Fig. 3. Cholangiocyte organoids (COs) rescue cholangiopathy following transplantation and assume an identity corresponding to the site of engraftment.
(A) Experimental outline schematic. (B) Kaplan-Meier curve (Table S1: number of animals at risk) demonstrating animal rescue following gallbladder organoids injection; P=0.0018(**), log-rank test. (C) Magnetic Resonance Cholangiopancreatography (MRCP) demonstrating rescue of cholangiopathy following organoid injection. (D) Immunofluorescence demonstrating engraftment of Red Fluorescent Protein (RFP)-expressing gallbladder organoids in portal triads, with upregulation of intrahepatic (SOX4) markers. Scale bars; yellow, 50μm; white, 100μm. PV, portal vein.
To ensure that animal rescue and resolution of cholangiopathy was specific to cholangiocyte organoids, we repeated the experiment using Mesenchymal Stem Cells (MSCs), as a different cell type known to provide anti-inflammatory effects following transplantation through paracrine signalling (Fig. S13A-S13C). This experiment allowed us to explore if organoids are essential for duct regeneration and animal rescue; and if some of the observed effects could be attributed to paracrine signals which are not unique to our cells. In sum, MSCs failed to engraft (Fig. S13C) and rescue the transplanted animals, which exhibited no difference in survival compared to controls (P>0.05; Fig. S13A) and no resolution of cholestasis on serum biochemistry (Fig. S13B). Consequently, cholangiopathy resolution is specific to the engraftment of cholangiocyte organoids; and although additional therapeutic effects of our cells through growth factor and cytokine secretion cannot be completely excluded, these effects are unique to cholangiocyte organoids.
We then explored if organoid culture is required to ‘unlock’ the cells’ plasticity or if this reflects an inherent property of primary cholangiocytes. To achieve this, we transplanted primary gallbladder cholangiocytes (Fig. S13A-S13C) directly post isolation without in vitro culture. Under these conditions very few primary cholangiocytes engrafted in the mouse bile ducts (Fig. S13C) most likely due to the cumulative stress of isolation and transplantation; and failed to rescue the animals or resolve cholestasis (Fig. S13A, S13B). Nonetheless, the engrafted cells expressed intrahepatic markers and lost expression of gallbladder markers (Fig. S13C). In conclusion, cholangiocyte plasticity is not limited to organoids grown in vitro; however, organoid culture is necessary for the cholangiocytes to recover from the stress of isolation and for large scale expansion providing the cell numbers required for engraftment and biliary repair.
Finally, to ensure that our results are not specific to the intrahepatic compartment or gallbladder organoids, we used our established methodology (3) to transplant common bile duct-derived cholangiocyte organoids in the gallbladder of immunocompromised mice. The engrafted cells exhibited loss of common bile duct makers and upregulation of gallbladder markers (Fig. S14), confirming that our previous findings apply to different compartments of the biliary tree and to organoids of different origin. Taken together, these results establish that cholangiocytes from different regions of the biliary tree are interchangeable and suggest that extrahepatic cells can be used to repair acute intrahepatic duct injury.
Cell transplantation experiments in mouse models are extremely useful but are not always predictive of therapeutic outcome (19). Furthermore, the mouse liver microenvironment is different to human, raising the possibility that our results may not translate between species. To address these challenges, we developed a new model for cell-based therapy in human utilizing ex vivo organ perfusion (20). Ex-vivo Normothermic Perfusion (NMP) was developed to improve organ preservation and reduce ischaemia-reperfusion injury by circulating warm oxygenated blood through liver grafts prior to transplantation. Importantly, the biliary tree is particularly susceptible to ischaemia which results in duct damage (21, 22). Low bile pH (< 7.5) during NMP is used as a predictor of this type of cholangiopathy (23).
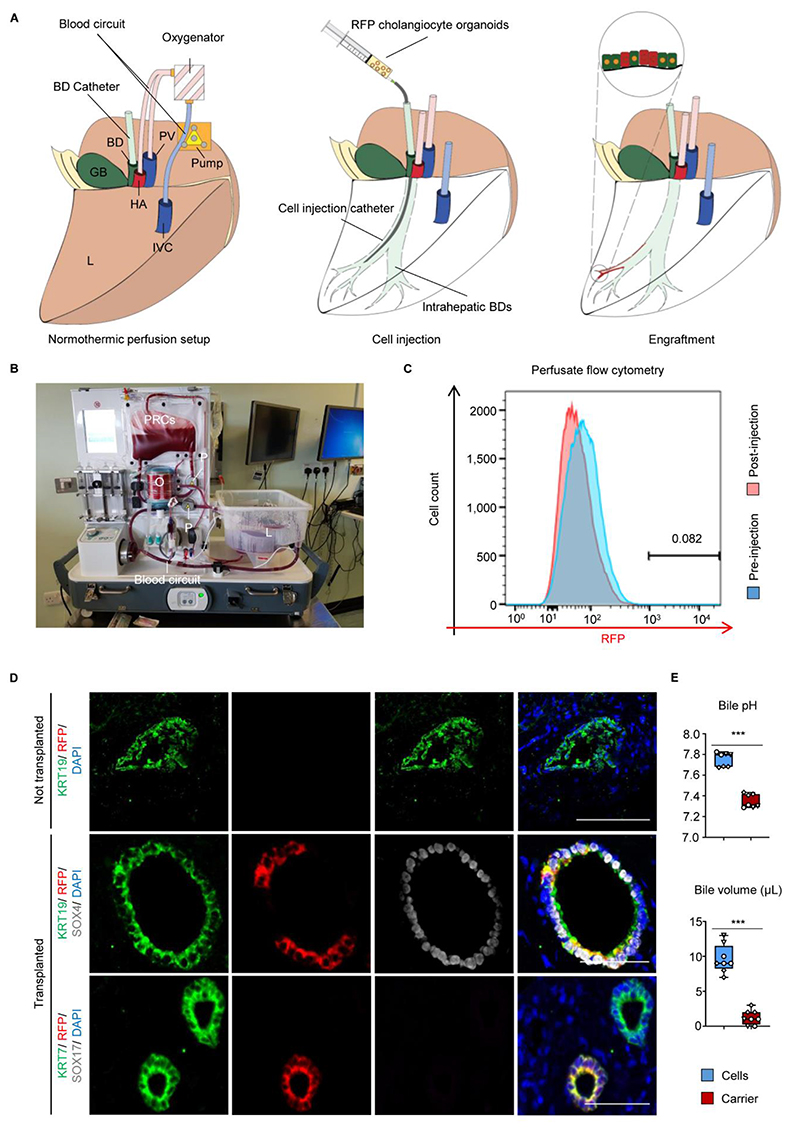
To assess the therapeutic potential of our cells for repairing human bile ducts, RFP gallbladder organoids were injected in the intrahepatic ducts of deceased transplant donor livers (n=3) with a bile pH<7.5 at the start of the experiment, signifying ischaemic duct injury. The organs were perfused with oxygenated blood and nutrients at normal body temperature (20); Fig. 4A-4B, S15A) for up to 100 hours in order to maintain a near-physiological microenvironment. Importantly, the organoids were delivered in a terminal branch of the intrahepatic ducts under fluoroscopic guidance to minimize the area of distribution of the cells and maximize cell density (Fig. S15B). At the end of the experiment, ultrasound imaging revealed no evidence of duct dilatation or obstruction (Fig. S15C), while RFP-expressing cells were not detected in the perfusate by flow cytometry, confirming that the injected cells remained in the biliary compartment (Fig. 4C). More importantly, the transplanted organoids engrafted in the intrahepatic biliary tree (Fig. 4D, S16A), with RFP cells regenerating ~40-85% of the injected ducts (Fig. 16B); and expressing key biliary markers (KRT7, KRT19, CFTR, GGT). Furthermore, engrafted gallbladder organoids exhibited loss of gallbladder (SOX17) and upregulation of intrahepatic (SOX4, BICC1, DCDC2) markers without differentiation to other hepatic lineages (Fig. 4D, S15D, S16A-S16B). Thus, at the end of the experiment, the injected ducts consisted of a mixture of native and transplanted cholangiocytes (Fig. S16A-S16B), with multiple transition points between donor and recipient cells and no evidence of cholangiopathy (Fig 4D, S15D, S16A).
Fig. 4. Cholangiocyte organoids (COs) engraft in a human liver receiving Normothermic Perfusion (NMP) and improve bile properties.
(A) Schematic representation of the technique for organoid injection and (B) photograph of the NMP circuit used. BD, Bile Duct; GB, Gallbladder; HA, Hepatic Artery; PV, Portal Vein; IVC, Inferior Vena Cava; L, Liver RFP, Red Fluorescent Protein; P, pump; O, oxygenator; PRC, Packed Red Cells. (C) Flow cytometry revealing absence of RFP cells in the perfusate. (D) Immunofluorescence revealing engraftment of RFP gallbladder organoids with upregulation of intrahepatic (SOX4) and loss of gallbladder (SOX17) markers. Scale bars, 50μm. (E) Organoid injection improves bile pH and choleresis. ***P<0.001. N=3 NMP livers. Each measurement is represented by a different data point, each organ is represented by a different symbol.
Conversely, control ducts not receiving cells demonstrated evidence of ischaemic injury with loss of epithelial continuity and sloughing of cells in the duct lumen (Fig. 4D). We subsequently characterised the impact of engraftment on organ function. Physiologically, cholangiocytes modify the composition and pH of bile through water transfer and bicarbonate secretion (6). Therefore, we compared the bile from organoid-injected vs. carrier-injected ducts. Accordingly, bile aspirated from ducts injected with cells exhibited higher pH and volume (Fig. 4E) confirming that transplanted cholangiocytes retain their function to modify bile composition. Together, these results provide the first proof-of-principle that perfused organs can be used to ascertain functional engraftment of human cells and validate our mouse data by showing that cholangiocytes are interchangeable for transplantation in human organs.
Our results show that the biliary epithelium is composed of cholangiocytes with diverse transcriptional profiles which are determined by their local environment. This diversity is lost in organoid culture due to the lack of niche stimuli. However, organoids can adapt appropriately to local environmental cues both in vitro and following transplantation, restore the expression of region-specific markers and assume different regional identities. Thus, organoids from a single region could potentially repair the entirety of the biliary tree. This plasticity could have significant implications for regenerative medicine. Indeed, although autologous cell-based therapy potentially avoids the need for immunosuppression its application for primary organoids is limited by the impact of disease on the epithelium. However, cholangiopathies belong to a family of localising diseases, affecting predominantly specific regions of an organ (24). Consequently, our results provide proof-of-concept that cholangiocytes from spared regions, such as the gallbladder, could be used for autologous cell-based therapy to repair human intrahepatic bile ducts, which constitute the most common site of injury in cholangiopathies. Moreover, our novel model for cell engraftment in human perfused organs paves the road for the use of ex vivo cell-based therapy to improve graft function prior to transplantation, which could ultimately increase the number of useable organs and reduce pressure on the transplant waiting list. In this context, quality controlled and readily available allogeneic cholangiocyte organoids from a cell bank could be used routinely in the future to prevent ischaemic cholangiopathy in organs at risk of biliary injury (e.g. low bile pH), since the organ recipients will receive immunosuppression as part of their standard care. Importantly, our results provide proof-of-principle for the transplantation of organoids in human organs which could expedite regulatory approval and fast-tack first-in-man trials. Ultimately, the same approach could also be applied to a variety of ex vivo perfused organs and cell types to validate functional cell engraftment, demonstrate safety, improve cell transplantation technique and efficacy and accelerate clinical translation of new cell-based therapies.
Supplementary Material
One sentence summary.
Single-cell RNA sequencing analyses combined with a novel model for cell transplantation in human livers reveal that intra- and extra-hepatic cholangiocytes are interchangeable for regenerative medicine applications.
Acknowledgements
The authors would like to thank A. Petrunkina and the NIHR Cambridge BRC Cell Phenotyping Hub for their help with flow cytometry and cell sorting; K Burling and the MRC MDU Mouse Biochemistry Laboratory [MRC_MC_UU_12012/5] for processing mouse serum samples; the Cambridge Biorepository for Translational Medicine for the provision of human tissue used in the study, Pedro Madrigal and Chichau Miau for bioinformatic support; Carlos Costa for his help with the NMP experiments; Kate Reid and Rachel Clarke for their help with radiological imaging during NMP experiments; Peter Humphreys, Darran Clements and Simon McCallum for their help with confocal imaging. The monoclonal antibody TROMA-III developed by R Kemler was obtained from the Developmental Studies Hybridoma Bank, created by the NICHD of the NIH and maintained at The University of Iowa, Department of Biology, Iowa City, IA 52242.
Funding
F.S. was supported by an NIHR Clinical Lectureship, the Academy of Medical Sciences Starter Grant for Clinical Lecturers, the Addenbrooke’s Charitable Trust and the Rosetrees Trust. T.B. was supported an EASL Juan Rodes fellowship. The L.V. lab is funded by the ERC advanced grant New-Chol, the Cambridge University Hospitals National Institute for Health Research Biomedical Research Centre and the core support grant from the Wellcome Trust and Medical Research Council of the Wellcome-Medical Research Council Cambridge Stem Cell Institute. L.G. and Sa.S were supported by a BHF Senior Research Fellowship (FS/18/46/33663).
Footnotes
Author contributions F.S. conceived and designed the study, performed experiments, acquired, interpreted and analysed the data, developed and validated the protocols described, performed bioinformatic analysis, generated the figures, wrote and edited the manuscript. D.M. Performed bioinformatics analysis. O.C.T. contributed to cell culture, harvesting and processing of tissue, single-cell RNA sequencing (scRNAseq) experiments and animal experiments. T.E.B. contributed to animal experiments. St.S. performed the Magnetic Resonance Imaging (MRI) experiments. E.M.G. performed the 3D reconstruction of the mouse biliary tree from MRI images. E.M.G. and S.S.U. reviewed and reported the MRI images. T.B. contributed to tissue culture, QPCR, immunofluorescence, tissue clarification and wholemount staining experiments. B.T.W. contributing to tissue dissociation and scRNAseq experiments. J.G.B. contributed to scRNAseq experiments. K.M. contributed to primary tissue harvesting tissue. Gi.C. contributed to scRNAseq experiments, flow cytometry and magnetic associated cell sorting. R.L.G. contributed to animal experiments, IF, and tissue histology. N.L.B. contributed to animal experiments. V.L.M. contributed to harvesting primary tissue. K.C., C.F., S.R, L.S. contributed to ex-vivo normothermic perfusion (NMP) experiments. J.B. contributed to imaging. L.G. contributed to tissue clarification and wholemount staining experiments as well as critical review of this data. D.O. and A.O. contributed to flow cytometry analyses and contributed through critical revision of the manuscript for important intellectual content. P. H. contributed to 3D reconstruction and rendering of immunofluorescence images. S.E.B. contributed to experimental design and critical revision of the manuscript for important intellectual content. Ga.C provided primary human samples. S. E. D. reviewed and reported the histology images and contributed through critical revision of the manuscript for important intellectual content. I.A. contributed to bile aspiration using microdialysis catheters. A.J.B., C.J.E.W. contributed to NMP experiments and critical revision of the manuscript for important intellectual content. T.C.S performed fluoroscopic cannulation of peripheral ducts in NMP experiments and contributed through critical revision of the manuscript for important intellectual content. M.P.M., W.T.H.G, G.F.M, Sa.S, S.T., P.G., E.M. contributed through critical revision of the manuscript for important intellectual content. K.S.-P. provided primary tissue, performed animal and ex-vivo normothermic perfusion experiments, contributed to the design and concept of study, interpreted the data and edited the manuscript. L.V. Designed and conceived the study, interpreted the data, wrote and edited the manuscript. All the authors approved the manuscript.
Competing interests FS, KSP and LV are founders and shareholders of BILITECH. LV is a founder and shareholder of DEFINIGEN. The remaining authors have no competing interests to disclose.
Data and materials availability
All data is available in the main text or the supplementary materials. Single-cell RNA sequencing data are available on ArrayExpress. Accession number: E-MTAB-8495
References
- 1.Drost J, Clevers H. Translational applications of adult stem cell-derived organoids. Dev. 2017;144:968–975. doi: 10.1242/dev.140566. [DOI] [PubMed] [Google Scholar]
- 2.Squires RH, Ng V, Romero R, Ekong U, Hardikar W, Emre S, Mazariegos GV. Evaluation of the pediatric patient for liver transplantation: 2014 practice guideline by the American Association for the Study of Liver Diseases, American Society of Transplantation and the North American Society for Pediatric Gastroenterology, Hepatology and Nutrition. Hepatology. 2014;60:362–98. doi: 10.1002/hep.27191. [DOI] [PubMed] [Google Scholar]
- 3.Sampaziotis F, Justin AW, Tysoe OC, Sawiak S, Godfrey EM, Upponi SS, Gieseck RL, De Brito MC, Berntsen NL, Gómez-Vázquez MJ, Ortmann D, et al. Reconstruction of the mouse extrahepatic biliary tree using primary human extrahepatic cholangiocyte organoids. Nat Med. 2017;23:954–963. doi: 10.1038/nm.4360. [DOI] [PubMed] [Google Scholar]
- 4.Parikh K, Antanaviciute A, Fawkner-Corbett D, Jagielowicz M, Aulicino A, Lagerholm C, Davis S, Kinchen J, Chen HH, Alham NK, Ashley N, et al. Colonic epithelial cell diversity in health and inflammatory bowel disease. Nature. 2019;567:49–55. doi: 10.1038/s41586-019-0992-y. [DOI] [PubMed] [Google Scholar]
- 5.Rimland CA, Tilson SG, Morell CM, Tomaz RA, Lu W-Y, Adams SE, Georgakopoulos N, Otaizo-Carrasquero F, Myers TG, Ferdinand JR, Gieseck RL, et al. Regional differences in human biliary tissues and corresponding in vitro derived organoids. Hepatology. 2020 doi: 10.1002/hep.31252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tabibian JH, Masyuk AI, Masyuk TV, O’Hara SP, LaRusso NF. Physiology of cholangiocytes. Compr Physiol. 2013;3:541–565. doi: 10.1002/cphy.c120019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Masyuk AI, LaRusso NF. Aquaporins in the hepatobiliary system. Hepatology. 2006;43:S75–S81. doi: 10.1002/hep.20996. [DOI] [PubMed] [Google Scholar]
- 8.Yoo K-S, Choi HS, Jun DW, Lee HL, Lee OY, Yoon BC, Lee KG, Paik SS, Kim YS, Lee J. MUC Expression in Gallbladder Epithelial Tissues in Cholesterol-Associated Gallbladder Disease. Gut Liver. 2016;10:851–8. doi: 10.5009/gnl15600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zweers SJLB, Booij KAC, Komuta M, Roskams T, Gouma DJ, Jansen PLM, Schaap FG. The human gallbladder secretes fibroblast growth factor 19 into bile: Towards defining the role of fibroblast growth factor 19 in the enterobiliary tract. Hepatology. 2012;55:575–583. doi: 10.1002/hep.24702. [DOI] [PubMed] [Google Scholar]
- 10.Zong Y, Stanger BZ. Molecular mechanisms of bile duct development. Int J Biochem Cell Biol. 2011;43:257–64. doi: 10.1016/j.biocel.2010.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Si-Tayeb K, Lemaigre FP, Duncan SA. Organogenesis and Development of the Liver. Dev Cell. 2010;18:175–189. doi: 10.1016/j.devcel.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 12.Aizarani N, Saviano A, Sagar, Mailly L, Durand S, Herman JS, Pessaux P, Baumert TF, Grün D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pepe-Mooney BJ, Dill MT, Alemany A, Ordovas-Montanes J, Matsushita Y, Rao A, Sen A, Miyazaki M, Anakk S, Dawson PA, Ono N, et al. Single-Cell Analysis of the Liver Epithelium Reveals Dynamic Heterogeneity and an Essential Role for YAP in Homeostasis and Regeneration. Cell Stem Cell. 2019;25:23–38.e8. doi: 10.1016/j.stem.2019.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Planas-Paz L, Sun T, Pikiolek M, Cochran NR, Bergling S, Orsini V, Yang Z, Sigoillot F, Jetzer J, Syed M, Neri M, et al. YAP, but Not RSPO-LGR4/5, Signaling in Biliary Epithelial Cells Promotes a Ductular Reaction in Response to Liver Injury. Cell Stem Cell. 2019;25:39–53.e10. doi: 10.1016/j.stem.2019.04.005. [DOI] [PubMed] [Google Scholar]
- 15.Lanzini A. BILE. Encycl Food Sci Nutr. 2003:471–478. [Google Scholar]
- 16.Tysoe OC, Justin AW, Brevini T, Chen SE, Mahbubani KT, Frank AK, Zedira H, Melum E, Saeb-Parsy K, Markaki AE, Vallier L, et al. Isolation and propagation of primary human cholangiocyte organoids for the generation of bioengineered biliary tissue. Nat Protoc. 2019;14:1884–1925. doi: 10.1038/s41596-019-0168-0. [DOI] [PubMed] [Google Scholar]
- 17.Lee M-O, Kim JH, Yoon B-I, Kwon S-B, Kang K-S, Kim M, Kong G, Hwang J-W, Lee B-H, Kim H-L, Yi J-Y, et al. Time- and Dose-based Gene Expression Profiles Produced by a Bile-duct-damaging Chemical, 4,4’- methylene Dianiline, in Mouse Liver in an Acute Phase. Toxicol Pathol. 2008;36:660–673. doi: 10.1177/0192623308320272. [DOI] [PubMed] [Google Scholar]
- 18.Berntsen NL, Fosby B, Valestrand L, Tan C, Reims HM, Schrumpf E, Karlsen TH, Line PD, Melum E. Establishment of a surgical bile duct injection technique giving direct access to the bile ducts for studies of the murine biliary tree. Am J Physiol - Gastrointest Liver Physiol. 2018;314:G349–G359. doi: 10.1152/ajpgi.00124.2017. [DOI] [PubMed] [Google Scholar]
- 19.Arber C, Brenner MK, Reddy P. Mouse models in bone marrow transplantation and adoptive cellular therapy. Semin Hematol. 2013;50:131–144. doi: 10.1053/j.seminhematol.2013.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nasralla D, Coussios CC, Mergental H, Akhtar MZ, Butler AJ, Ceresa CDL, Chiocchia V, Dutton SJ, García-Valdecasas JC, Heaton N, Imber C, et al. A randomized trial of normothermic preservation in liver transplantation. Nature. 2018;557:50–56. doi: 10.1038/s41586-018-0047-9. [DOI] [PubMed] [Google Scholar]
- 21.Skaro AI, Jay CL, Baker TB, Wang E, Pasricha S, Lyuksemburg V, Martin JA, Feinglass JM, Preczewski LB, Abecassis MM. The impact of ischemic cholangiopathy in liver transplantation using donors after cardiac death: The untold story. Surgery. 2009;146:543–553. doi: 10.1016/j.surg.2009.06.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Enestvedt CK, Malik S, Reese PP, Maskin A, Yoo PS, Fayek SA, Abt P, Olthoff KM, Shaked A. Biliary complications adversely affect patient and graft survival after liver retransplantation. Liver Transpl. 2013;19:965–72. doi: 10.1002/lt.23696. [DOI] [PubMed] [Google Scholar]
- 23.Watson CJE, Kosmoliaptsis V, Pley C, Randle L, Fear C, Crick K, Gimson AE, Allison M, Upponi S, Brais R, Jochmans I, et al. Observations on the ex situ perfusion of livers for transplantation. Am J Transplant. 2018;18:2005–2020. doi: 10.1111/ajt.14687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Farmer RG, Hawk WA, Turnbull RB, Farmer RG, Hawk WA, Turnbull RB. ‘Clinical patterns in Crohn’s disease: a statistical study of 615 cases.’. [Accessed: 11 September 201];Gastroenterology. 1975 68(4 Pt 1):627–35. Gastroenterology. 68, 627-35(1975) [PubMed] [Google Scholar]
- 25.MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, et al. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383. doi: 10.1038/s41467-018-06318-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McCarthy DJ, Campbell KR, Lun ATL, Wills QF. Scater: pre-processing, quality control, normalization and visualization of single-cell RNA-seq data in R. Bioinformatics. 2017;33:1179–1186. doi: 10.1093/bioinformatics/btw777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–420. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Haghverdi L, Lun ATL, Morgan MD, Marioni JC. Batch effects in single-cell RNA-sequencing data are corrected by matching mutual nearest neighbors. Nat Biotechnol. 2018;36:421–427. doi: 10.1038/nbt.4091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wolf FA, Angerer P, Theis FJ. SCANPY: Large-scale single-cell gene expression data analysis. Genome Biol. 2018;19 doi: 10.1186/s13059-017-1382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Haghverdi L, Büttner M, Wolf FA, Buettner F, Theis FJ. Diffusion pseudotime robustly reconstructs lineage branching. Nat Methods. 2016;13:845–848. doi: 10.1038/nmeth.3971. [DOI] [PubMed] [Google Scholar]
- 31.Qiu X, Mao Q, Tang Y, Wang L, Chawla R, Pliner HA, Trapnell C. Reversed graph embedding resolves complex single-cell trajectories. Nat Methods. 2017;14:979–982. doi: 10.1038/nmeth.4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kiselev VY, Kirschner K, Schaub MT, Andrews T, Yiu A, Chandra T, Natarajan KN, Reik W, Barahona M, Green AR, Hemberg M. SC3: Consensus clustering of single-cell RNA-seq data. Nat Methods. 2017;14:483–486. doi: 10.1038/nmeth.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. 2018;7 doi: 10.1093/gigascience/giy083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, et al. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90–W97. doi: 10.1093/nar/gkw377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sampaziotis F, De Brito MC, Madrigal P, Bertero A, Saeb-Parsy K, Soares FAC, Schrumpf E, Melum E, Karlsen THTH, Bradley JAA, Gelson WTH, et al. Cholangiocytes derived from human induced pluripotent stem cells for disease modeling and drug validation. Nat Biotechnol. 2015;33:845–852. doi: 10.1038/nbt.3275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sampaziotis F, de Brito MC, Geti I, Bertero A, Hannan NR, Vallier L. Directed differentiation of human induced pluripotent stem cells into functional cholangiocyte-like cells. Nat Protoc. 2017;12:814–827. doi: 10.1038/nprot.2017.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravikumar R, Jassem W, Mergental H, Heaton N, Mirza D, Perera MTPR, Quaglia A, Holroyd D, Vogel T, Coussios CC, Friend PJ. Liver Transplantation After Ex Vivo Normothermic Machine Preservation: A Phase 1 (First-in-Man) Clinical Trial. Am J Transplant. 2016;16:1779–1787. doi: 10.1111/ajt.13708. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data is available in the main text or the supplementary materials. Single-cell RNA sequencing data are available on ArrayExpress. Accession number: E-MTAB-8495