Abstract
The contribution of ribosome heterogeneity and ribosome-associated proteins to the molecular control of proteomes in health and disease remains enigmatic. We demonstrate that Survival Motor Neuron (SMN) protein, loss of which causes the neuromuscular disease spinal muscular atrophy (SMA), binds to ribosomes and that this interaction is tissue-dependent. SMN-primed ribosomes are preferentially positioned within the first five codons of a set of mRNAs which are enriched for translational enhancer sequences in the 5’UTR and rare codons at the beginning of their coding sequence. These SMN-specific mRNAs are associated with neurogenesis, lipid metabolism, ubiquitination, chromatin regulation and translation. Loss of SMN induces ribosome depletion, especially at the beginning of the coding sequence of SMN-specific mRNAs, leading to impairment of proteins involved in motor neuron function and stability, including acetylcholinesterase. Thus, SMN plays a crucial role in the regulation of ribosome fluxes along mRNAs which encode proteins relevant to SMA pathogenesis.
Introduction
Translation is the most energy consuming process in cells (1, 2) and represents a core mechanism coordinating multiple post-transcriptional processes. Hence, it is not surprising that several mRNAs are largely controlled at the translational, rather than transcriptional, level (3–5). Indeed, loss of post-transcriptional and translational control has been linked to cancer (6, 7), autism (8) and neurodegenerative disease (9–11), highlighting the critical contribution of translation to a broad spectrum of disease pathogenesis.
Ribosomes have been placed in the spotlight as putative direct influencers of translation by acting as mRNA regulatory elements, or “filters” (12, 13). Recent findings also suggest that ribosome composition is not fixed and uniform, but rather is heterogeneous and can be modulated at the level of ribosomal protein composition (14–16), rRNA variants (17, 18), and/or by ribosome-associated proteins (RAPs) (12, 19), which exert a direct role on mRNA selection (15) and function (19, 20). Although this represents an exciting potential mechanism for ribosome-based control of gene expression, at present it remains unclear whether direct or indirect defects in ribosome heterogeneity can contribute to disease pathogenesis.
Depletion of Survival Motor Neuron (SMN) protein, following homozygous deletion or mutations in SMN1, causes spinal muscular atrophy (SMA) (21, 22). The human genome contains a second SMN gene (SMN2), almost identical to SMN1. Aberrant splicing of SMN2 transcripts mostly results in a truncated and unstable protein. The remaining 10-20% of SMN2-derived mRNAs are translated into a full-length, stable SMN protein, rescuing the lethality of SMN1 loss in humans. SMA is primarily characterized by loss of lower motor neurons, leading to muscle atrophy and wasting. However, the molecular mechanisms leading to motor neuron death in SMA remain complex and unresolved (22–26). Although classically known to play a role in the biogenesis of ribonucleoparticles (RNPs) (27), SMN is also a strong candidate to be directly implicated in the control of translation: it is thought to associate with polysomes in cell cultures (11, 28), as well as rat and mouse spinal cords (11, 29) and mouse brain (11). Moreover, SMN influences translation in vitro (11, 28, 30) and in vivo (11). Hence, it is possible that, in addition to its canonical roles (27), SMN protein functions as a ribosome modulator leading to early and local dysfunction of translation when levels of SMN are decreased. In line with this hypothesis, genome-wide defects occurring in mRNA recruitment onto polysomes have previously been observed in SMA (11), but the mechanism(s) linking SMN to these defects have yet to be elucidated.
Here, we present evidence suggesting that SMN is a ribosome-associated protein acting as a master regulator of translation on a specific subset of mRNAs relevant to SMA pathogenesis.
Results
SMN binds ribosomes in vitro and in vivo, with tissue specificity
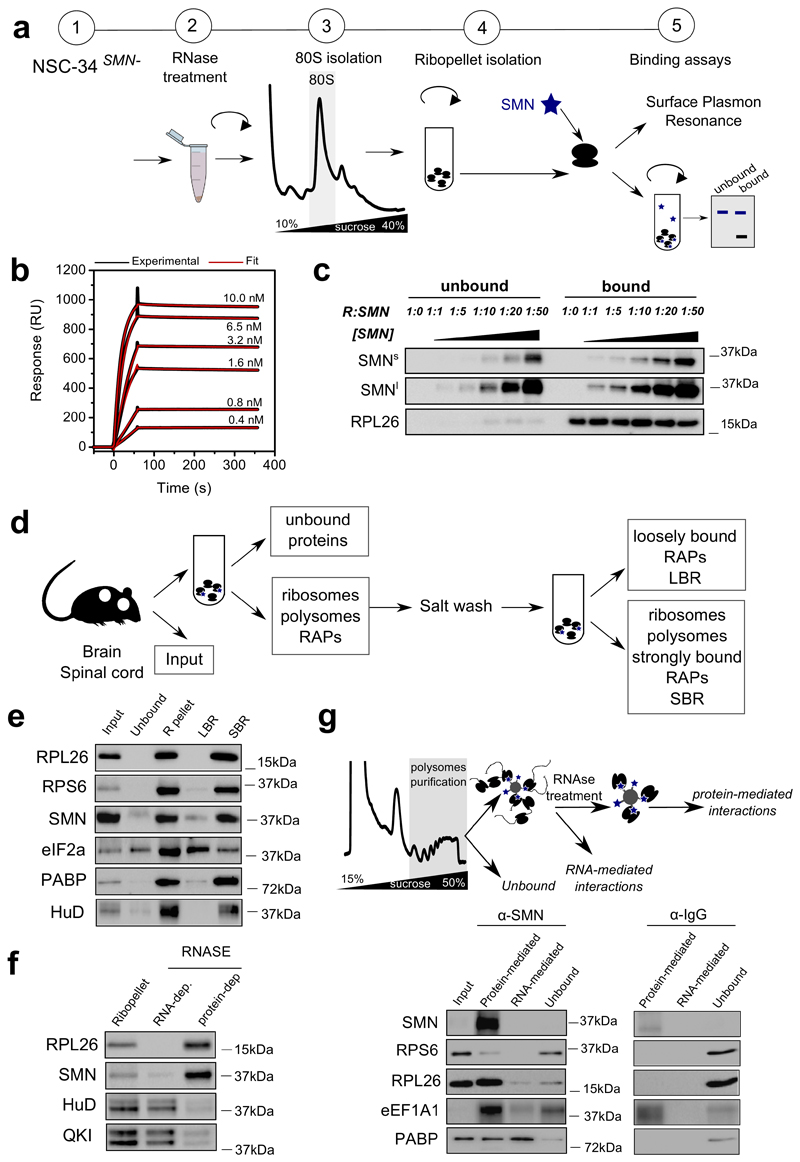
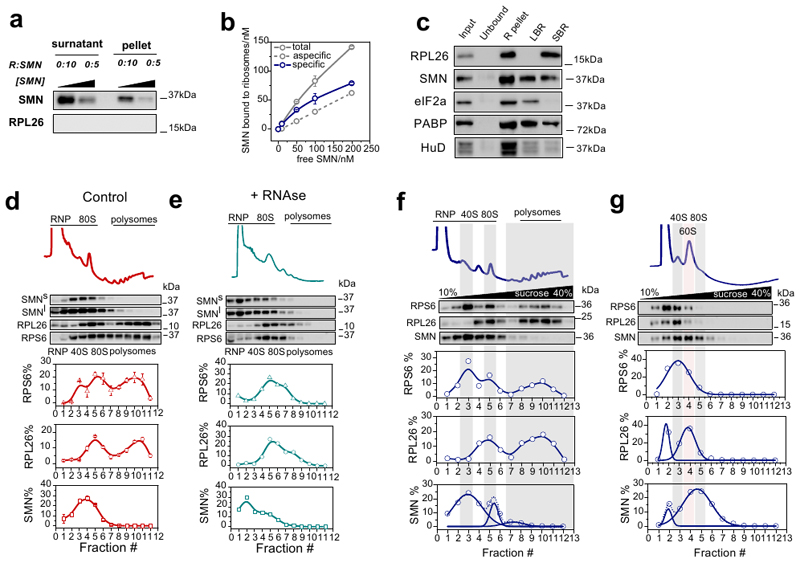
Guided by previous evidence suggesting an association of SMN with polysomes (11, 28–30), we hypothesized that SMN may play an as yet uncharacterized role in regulating translation by acting as a ribosome-associated protein. To detail the interaction between SMN and ribosomes, we characterized the binding of recombinant SMN to purified SMN-free ribosomes obtained from cells which do not express the full length SMN (11). Using two complementary approaches (Fig. 1a) we found that SMN strongly binds ribosomes in vitro (Fig. 1b and c, Supplementary Table 1, Extended data Fig. 1a and b). Next, we performed subcellular fractionation coupled to high salt wash (31) (Fig. 1d) in mouse tissues. Before and after salt washes, SMN remained tightly associated with ribosomes in brain (Fig. 1e) and spinal cord (Extended data Fig. 1c). To monitor if the interaction of SMN with ribosomes/polysomes is mRNA dependent, we treated the ribosome/polysome pellet with RNase I and observed that SMN still sedimented with ribosomes/polysomes, suggesting that this association is mRNA-independent (Fig. 1f, Extended data Fig. 1d and e).
Fig. 1. SMN interacts with the translation machinery in vitro and in vivo in an RNA-independent manner.
a, Schematic overview of experimental design for studying the binding of recombinant SMN to purified SMN-free ribosomes, used for downstream binding analysis: i) Surface Plasmon Resonance (SPR) and ii) ultracentrifugation analysis to separate bound and unbound SMN which were analyzed by western blotting. b, Typical SPR titration curves were used to estimate the KD (ka=9.7±4.8 × 106 M-1s-1, kd=9.4±5 × 10-5 s-1, KD=9.8±2.8 × 10-12 M, values are the average ± SD, n=6 independent experiments) Typical SPR curve: black line indicates the titration, the red line indicates fitting for 1:1 binding model. c, Western blotting of unbound and ribosome-associated SMN at different SMN concentrations. The concentration of ribosomes was constant in all experiments. Representative example of 3 independent experiments. Immunoblots were acquired at short (SMNs) and long (SMNl) exposure times. d, Schematic overview of the subcellular fractionation protocol used to study the association of SMN to ribosomes in brain and spinal cord. e, Western blot analysis on cytoplasmic lysates from brain; input; ribosome-free cytoplasmic components (unbound); ribosomal subunits, ribosomes and polysomes (R-pellet); loosely ribosome-bound proteins (LBR); strongly ribosome-bound proteins (SBR). PABP and eIF2a are proteins associated to polysomes and HuD is an RNA binding protein associated with polysomes through RNA interactions. The ribosomal proteins L26 and S6 were used as control of ribosome sedimentation. The results are representative of 3 independent experiments. f, The ribo-pellet was treated with RNase I and ultracentrifuged to separate proteins interacting with ribosomes through RNA-dependent or independent interactions. The RNA binding proteins HuD and QKI were used as controls for RNA-dependent interactions. g, Schematic of immunoprecipitation of SMN from purified polysomal fractions (upper panel). The first wash corresponds to the “unbound” lane. After on beads RNase treatment, proteins were extracted from beads (Protein-mediated) or from washes (RNA-mediated). The IP was performed on sucrose fractions corresponding to polysomes in brain P5 with anti-SMN or mouse IgG as control (lower panel). The results are representative of 2 independent experiments. Statistical source data and unprocessed blots are provided in Source data Fig. 1.
Secondly, we co-immunoprecipitated SMN with ribosomal proteins and translation factors from purified polysomes, and found that SMN is associated with ribosomal proteins through protein-protein interactions and with the Poly(A) binding protein mainly via mRNA-dependent interactions (Fig. 1g). Next, to understand whether SMN is preferentially associated to the large or small subunit of the ribosome, we induced the dissociation of the ribosomal subunits and found that SMN primarily co-sediments with the 60S subunit (Extended data Fig. 1f and g).
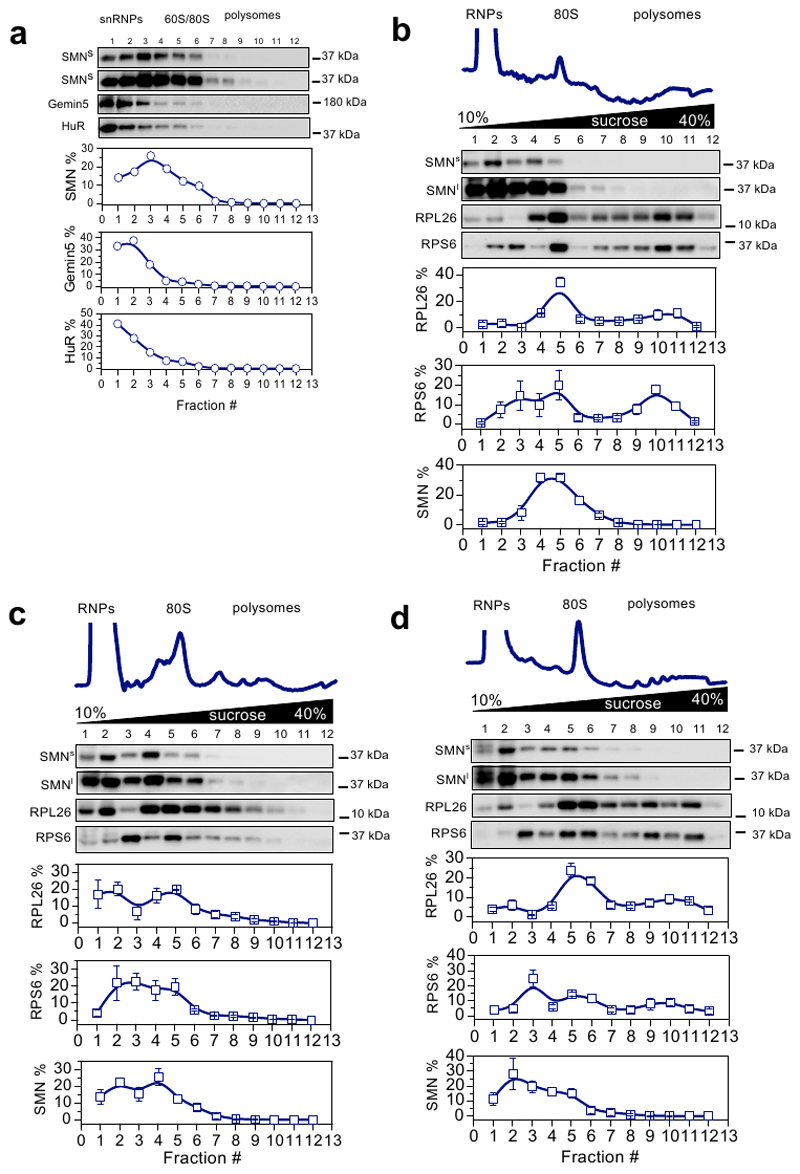
Thirdly, to rule out the possibility that the observed interaction of SMN with ribosomes and polysomes derived from Gemin-granules (32, 33), we compared the co-sedimentation profile of SMN, Gemin- and RNA-granules after sucrose gradient fractionation of cell lysates. We found that SMN co-sediments independently from HuR, a marker of mRNA-granules, and from Gemin5, a marker of Gemin-granules, which has also been proposed to bind ribosomes (31) (Extended data Fig. 2a). These findings suggest that, in addition to being part of Gemin-granules, SMN is a bona fide ribosome-associated protein.
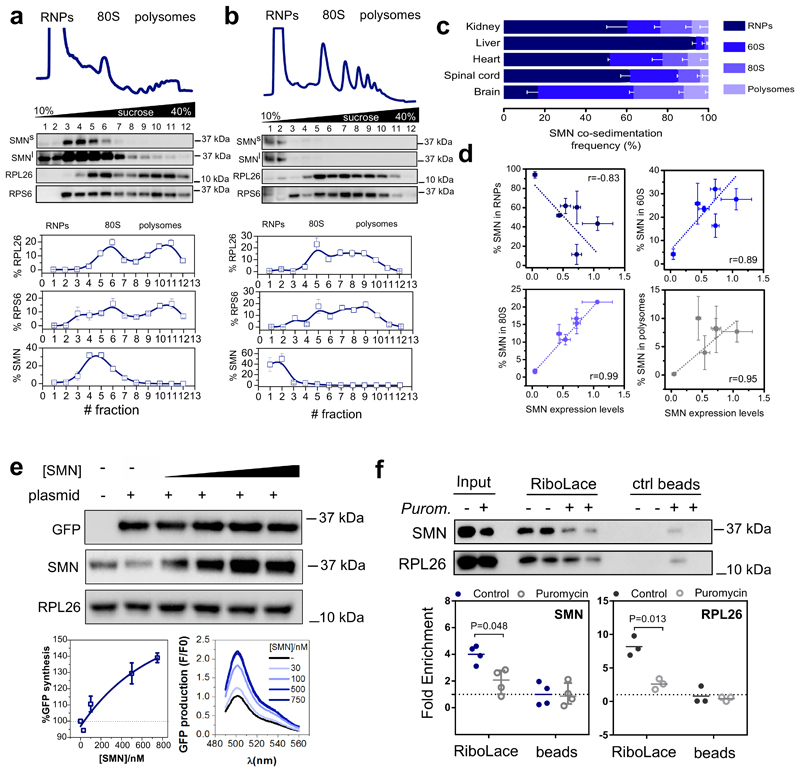
Since SMN expression levels are known to be tissue-dependent (34), we next wanted to establish whether SMN displays distinctive ribosome binding ability in a tissue-specific manner. To test this hypothesis, we established the relative co-sedimentation of SMN with ribonucleoparticles (RNPs), ribosomal subunits, ribosomes and polysomes in spinal cord, brain, kidney, liver and heart from wild-type mice (Fig. 2a and b and Extended data Fig. 2b-d). We observed a tissue-dependent association of SMN with the translation machinery (Fig. 2c). Interestingly, this variability was proportional with the overall abundance of SMN, as SMN levels negatively correlated with RNP association (r=-0.83 and positively correlated with 60S, 80S and polysome association (r = 0.89, 0.99 and r=0.95 respectively) (Fig. 2d). This confirmed that the association of SMN with ribosomes and polysomes is tissue-specific and dependent on SMN concentration in vivo, similar to what we observed in vitro (Fig. 1b and c). This finding suggests that a subset of ribosomes are associated with SMN in a concentration and tissue-dependent manner in vivo. We termed these SMN-associated ribosomes “SMN-primed ribosomes”.
Fig. 2. SMN interacts with the translation machinery in a concentration dependent manner across different tissues and is associated to actively translating ribosomes positively regulating translation.
Co-sedimentation profiles of SMN in a, brain and b, liver. The relative distributions of SMN, and markers of the small (RPS6) and large (RPL26) subunits of the ribosome were used as controls for sedimentation (upper panels). The relative distribution of each protein along the profile is shown as the average ± SEM of n=3 biologically independent experiments. Immunoblots were acquired at short (SMNs) and long (SMNl) exposure time. c, Summary of SMN co-sedimentation with RNPs, 60S, 80S and polysomes in different tissues. The percentages are shown as the average ± SEM of n=3 biologically independent experiments and were obtained using co-sedimentation profiles shown in panels (a, b and Extended data Fig.2b-d). d, Relationship between the relative expression level of SMN in different tissues from (34) and the relative distribution of SMN in RNPs, 60S, 80S and polysomes obtained from (c). Data are presented as the average ± SEM of n=3 biologically independent experiments. e, In vitro translation of reporter GFP in the presence of different concentrations of recombinant SMN. As a negative control a reaction in the absence of the GFP reporter was run in parallel. RPL26 was used as a loading control. Left lower panel, semi-quantitative analysis of GFP level in the presence of different concentrations of recombinant SMN. Plotted are the averages ± SEM from n=3 independent experiments. Right lower panel, the production of GFP was monitored by measuring the appearance of fluorescence in independent assays. f, Western blot analysis of SMN association to active ribosomes using RiboLace (36) in human cells (upper panels) before and after treatment with the translation inhibitor puromycin (100 μM, 1h). RPL26 is used as a marker of ribosomes. The enrichment of SMN and RPL26 with respect to the not-functionalized beads is shown. Plotted are the average ± SEM for n=4 (SMN) and n=3 (RPL26) biologically independent experiments. Significant changes were assessed using a two-sided t-test. Statistical source data and unprocessed blots are provided in Source data Fig. 2.
SMN positively regulates translation and is associated with actively translating ribosomes
It has previously been proposed that SMN may act as a repressor of cap-dependent translation in vitro (28), but this result is not in complete agreement with recent findings, including in vivo studies where SMN loss leads to translation defects (11, 30, 35). Therefore, we wanted to establish whether SMN might positively regulate cap-independent translation and be associated with stalled or actively translating ribosomes. To test this, we took advantage of an in vitro transcription translation assay using different concentrations of recombinant SMN and a reporter gene whose translation is controlled by a viral translational enhancer element, in particular an IRES. We observed that higher concentrations of SMN led to higher production of reporter protein (Fig. 2e), suggesting that SMN is a positive regulator of translation in vitro.
Next, we investigated the association of SMN to active ribosomes in cellulo using the RiboLace method (36), which facilitates the specific isolation of actively translating ribosomes. We confirmed that SMN is associated with active ribosomes and that this association is lost when translation is inhibited by puromycin, a translation inhibitor that releases active ribosomes from mRNAs (Fig. 2f). These findings show that SMN positively regulates translation by binding to actively translating ribosomes.
SMN-primed ribosomes are positioned within the first five codons of a specific subset of mRNAs
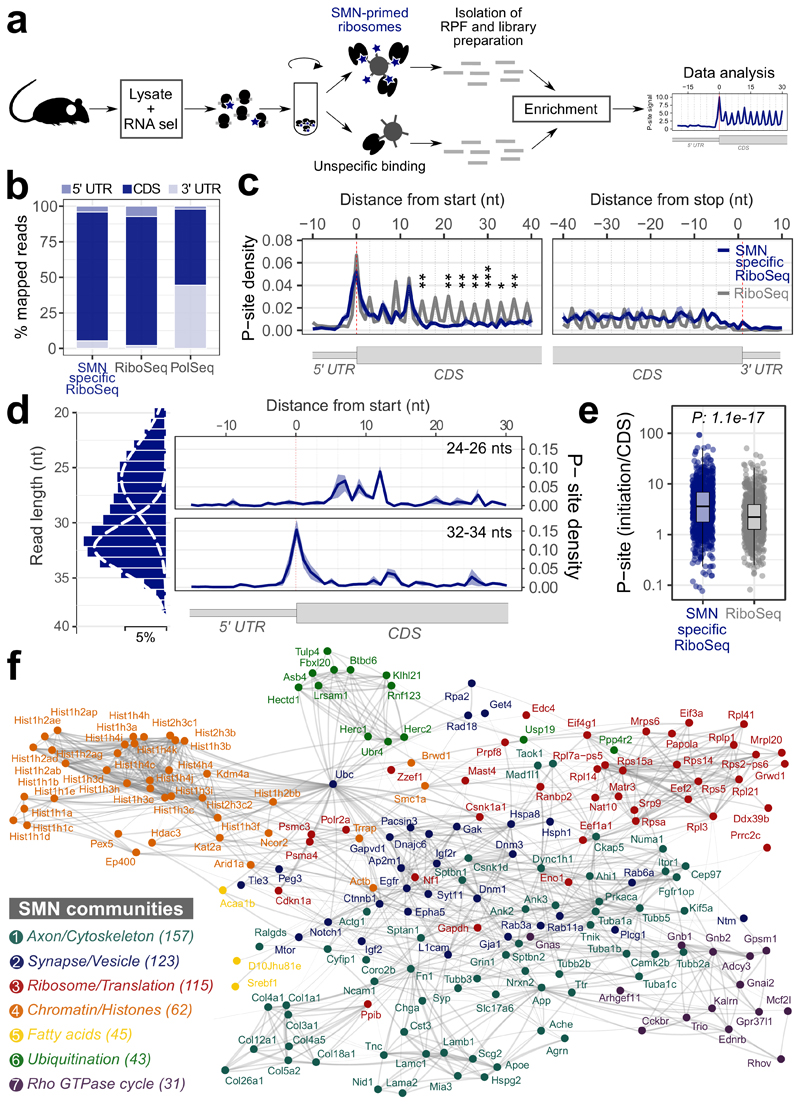
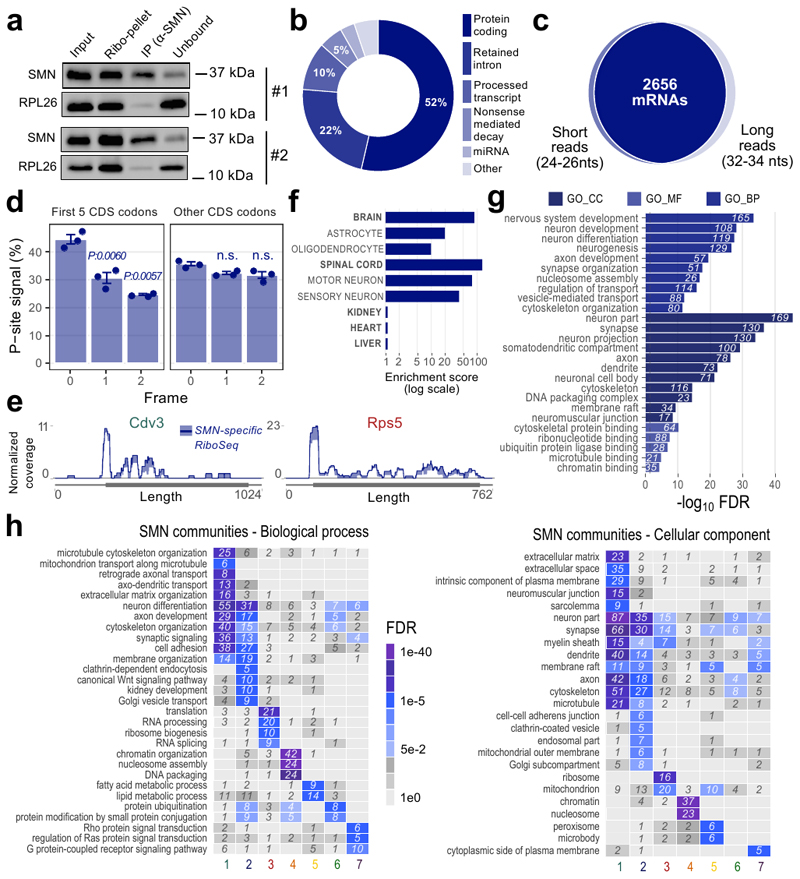
To establish whether SMN-primed ribosomes control a specific subset of mRNAs and where SMN-primed ribosomes are preferentially positioned along transcripts, we isolated SMN-primed ribosomes by immunoprecipitation and performed ribosome profiling in wild-type mouse brains (Fig. 3a). SMN co-immunoprecipitated with RPL26 (Extended data Fig. 3a), further demonstrating that SMN binds to ribosomes by RNA-independent interactions. By comparing RNA fragments protected by SMN-primed ribosomes to both control IgG and classical ribosome profiling performed on the same tissue (see Materials and Methods), we identified a set of 5587 transcripts, corresponding to 2842 genes (Supplementary Table 2). The vast majority of these transcripts (52%) were protein-coding genes (Extended data Fig. 3b). SMN-primed ribosome protected fragments (RPFs) map prevalently to the coding sequence (CDS), similar to control ribosome profiling (RiboSeq) data and distinct from RNA-Seq of polysomal RNA (Fig. 3b).
Fig. 3. Ribosome profiling of SMN-primed ribosomes reveals enriched mRNAs organized in functionally well-defined communities.
a, Schematic representation of the protocol used to isolate RNA fragments protected by ribosomes associated with SMN. b, Positional enrichment along the three mRNA regions of SMN-specific RiboSeq reads. The bar plots display the percentage of reads aligning on 5’ UTR, CDS and 3’ UTR for SMN-specific RiboSeq, RiboSeq and classical sequencing of polysomal transcripts (PolSeq from (11)) as control. c, Overlay meta-profiles for mRNAs enriched in SMN-bound RPFs based on the P-site position of SMN-primed ribosomes (blue) and total ribosomes (gray) for n=3 and n=2 biologically independent samples, respectively. The line represents the average ± SEM. Differences were tested using the two-sided t-test (p-values corresponding to codons 6 and 8-13 are: 0.0021, 0.0051, 0.0018, 0.0006, 0.0480, 0.0017, 0.0280). d, Left panel: distribution of read lengths, fitted with two Gaussian curves. Right panels: meta-profiles based on the P-sites of short reads (24-26 nucleotides, upper panel) and long reads (32-34 nucleotides, lower panel). Data are presented as the average ± SEM of n=3 biologically independent samples. e, Dot plots showing the distributions of the ratios between the average number of P-sites on the first five codons (initiation) and the average number of P-sites on the whole coding sequence (CDS) for SMN-specific RiboSeq (n=874 transcripts) and classical RiboSeq (n=704 transcripts) in control mouse brain. Statistical significance was determined using the two-sided Wilcoxon-Mann-Whitney test. f, Representative protein interaction network of SMN-specific transcripts. Connections are based on STRING annotation and weighted by interaction score. Connectivity analysis of the full network identified seven communities characterized by high intra connectivity and color-labelled. The insert reports the name and the number of SMN-specific genes in each community. Statistical source data are provided in Source data Fig. 3.
Next, we analyzed in detail the P-site position of SMN-primed RPFs near the start and stop codon of transcripts associated with SMN-primed ribosomes. Compared to classical ribosome profiling from control mouse brain, we observed a significant accumulation of signal within the first five codons of the CDS (Fig. 3c). Two distinct populations of RPFs with different lengths contributed to this effect: shorter fragments (24-26 nucleotides) peaking on the fifth codon (Fig. 3d, upper panel), and longer fragments (32-34 nucleotides) peaking on the first codon (Fig. 3d, lower panel). These two populations are shared among the selected transcripts (Extended data Fig. 3c) and may be associated with different ribosome conformations (37). We further confirmed that SMN-primed ribosomes preferentially occupy the beginning of the CDS by determining for each transcript the ratio between P-sites on the first 5 codons (initiation region) and the whole CDS (Fig. 3e). Since the P-site signal was lower and increasingly out of frame after the 5th codon (Extended data Fig. 3d), we considered the set of 874 protein coding transcripts (corresponding to 619 genes) displaying a signal within the first codons as bona fide SMN-specific transcripts for further analysis. Representative coverage profiles of SMN-specific transcripts are shown in Extended data Fig. 3e.
Expression analysis of cell types within compartments of the nervous system revealed the highest enrichment for motor neurons, followed by sensory neurons, astrocytes and oligodendrocytes (Extended data Fig. 3f). Gene Ontology analysis further highlighted the significant association of neuron-specific functions with mRNAs enriched in SMN-primed ribosomes (Extended data Fig. 3g).
Finally, we charted known interactions between proteins coded by SMN-specific transcripts. Structural analysis of the network revealed the presence of seven communities, (Fig. 3f). Each community is characterized by a distinct functional identity (Extended data Fig. 3h). Ordered by decreasing gene number, the communities are related to: Axon/Cytoskeleton, Synapse/Vesicle, Ribosome/Translation, Chromatin/Histones, Fatty Acids, Ubiquitination, Rho GTPase cycle (Fig. 3f and Extended data Fig. 3h). These communities resemble several roles linked to SMN and themes that are known to be defective in SMA (11, 38, 47, 39–46).
Transcripts bound by SMN-primed ribosomes display defects in ribosome recruitment at the beginning of the CDS during early stages of SMA
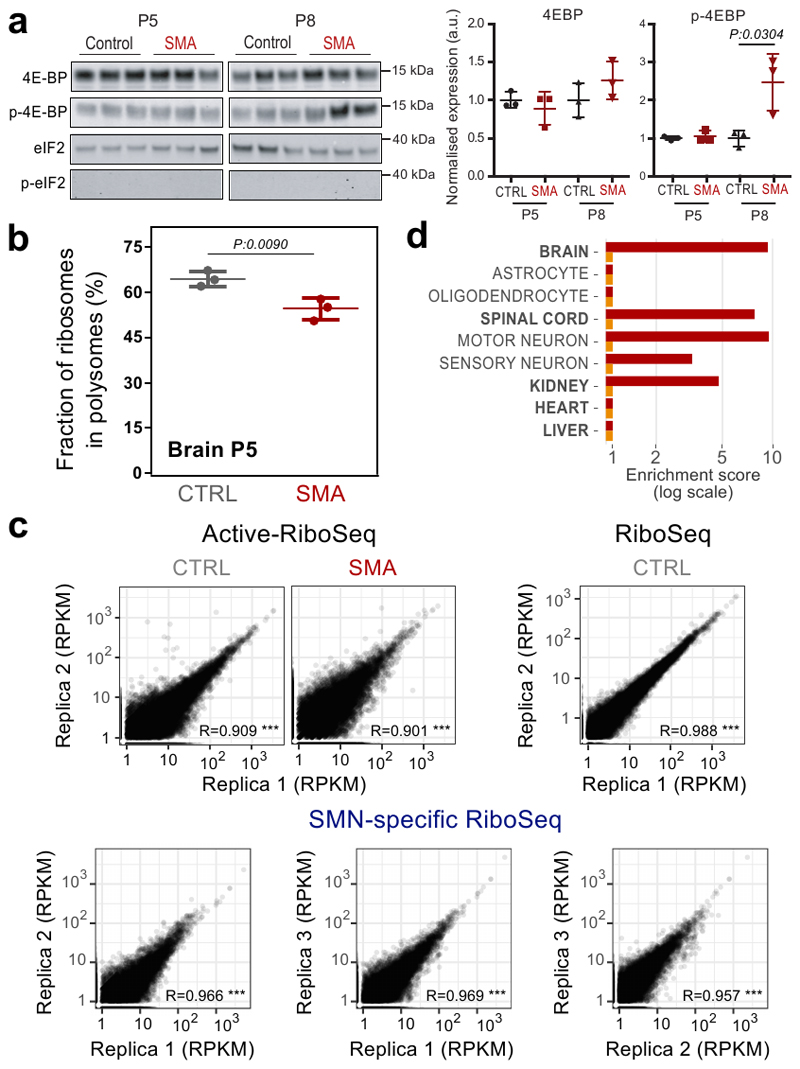
To establish whether loss of SMN interaction with ribosomes gives rise to translational defects in SMA at early stages of disease (11), we first ruled out the possibility that translational changes are caused by pathways controlling translation, in particular the PERK (Unfolded Protein Response) and mTORC1 pathways. (Extended data Fig. 4a and 4B).
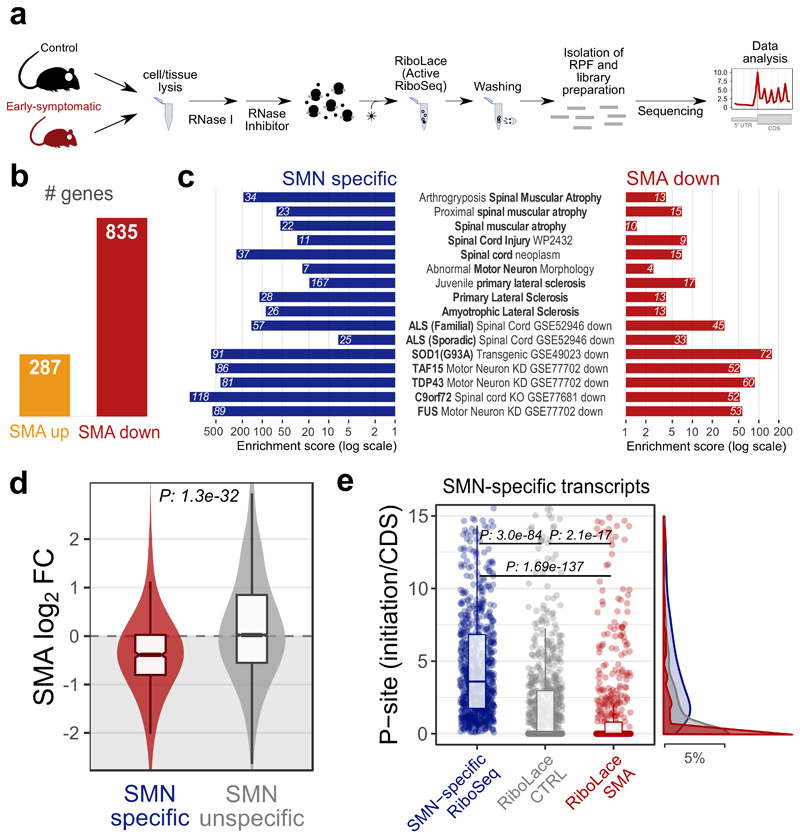
Next, we analyzed the positioning of active ribosomes in SMA mouse brains and age-matched controls using the Active-RiboSeq method based on RiboLace (36) (Fig. 4a and Extended data Fig. 4c). Notably, the majority of genes with significantly altered translation (76%) were characterized by a decreased ribosome occupancy in SMA (Fig. 4b), in agreement with previously reported translation defects (11). As with SMN-specific transcripts, the 835 genes with defects in active translation were strongly enriched for both brain and spinal cord compartments, particularly motor neurons (Extended data Fig. 4d). Importantly, transcripts bound by SMN-primed ribosomes and with defects in active translation in SMA match transcripts previously associated with SMA and other motorneuron diseases such as ALS (Fig. 4c).
Fig. 4. Transcripts bound by SMN-primed ribosomes display defects in positioning of active ribosomes at early stages of SMA.
a, Experimental design for active ribosome profiling (RiboLace) of control and early symptomatic SMA brains. b, Number of genes with significantly increased (up) or decreased (down) active ribosome occupancies in early symptomatic SMA mouse brain. c, Over-representation analysis of terms associated with motor neuron diseases among genes enriched in SMN-primed ribosomes (SMN specific, blue) and genes with decreased translation occupancy in SMA (SMA down, red). Genes with increased translation occupancy in SMA did not show any significant enrichment for terms associated with motor neuron diseases. d, Comparison between SMA active ribosome occupancy changes in SMN-specific genes (red, n=554 genes) and SMN unspecific genes (grey, n=13178 genes). SMN specific genes show a significant shift towards a reduction in SMA active ribosome occupancy (two-sided Wilcoxon rank-sum test). e, Dot plots showing the ratio distribution between the average number of P-sites on the first five codons (initiation) and the average number of P-sites on the whole coding sequence (CDS) for SMN-specific transcripts based on signal from SMN-specific RiboSeq (blue, n=874 transcripts), control and SMA Active-RiboSeq (gray and red, n=859 and n=774 transcripts respectively). Significant differences were determined using the two-sided Wilcoxon-Mann-Whitney test. Corresponding distributions are displayed on the right of the plot. Statistical source data are provided in Source data Fig. 4.
Prompted by this finding, we further verified that SMN-specific transcripts display significantly decreased signal from Active-RiboSeq in SMA, with respect to SMN-unspecific transcripts (P: 1.3e-32, Fig. 4d), and that the loss of signal is located preferentially at the beginning of their CDS, (Fig. 4e, P: 2.1e-17).
These results suggest that loss of SMN induces defects in ribosome occupancy within the first 5 codons of mRNAs bound by SMN-primed ribosomes, and that SMN is required during early stages of translation, when the nascent chain is short and not yet deep inside the exit tunnel of the ribosome.
Translationally defective transcripts in SMA display deficient use of rare codons in the CDS and enrichment in translational enhancer sequences in the 5’UTR
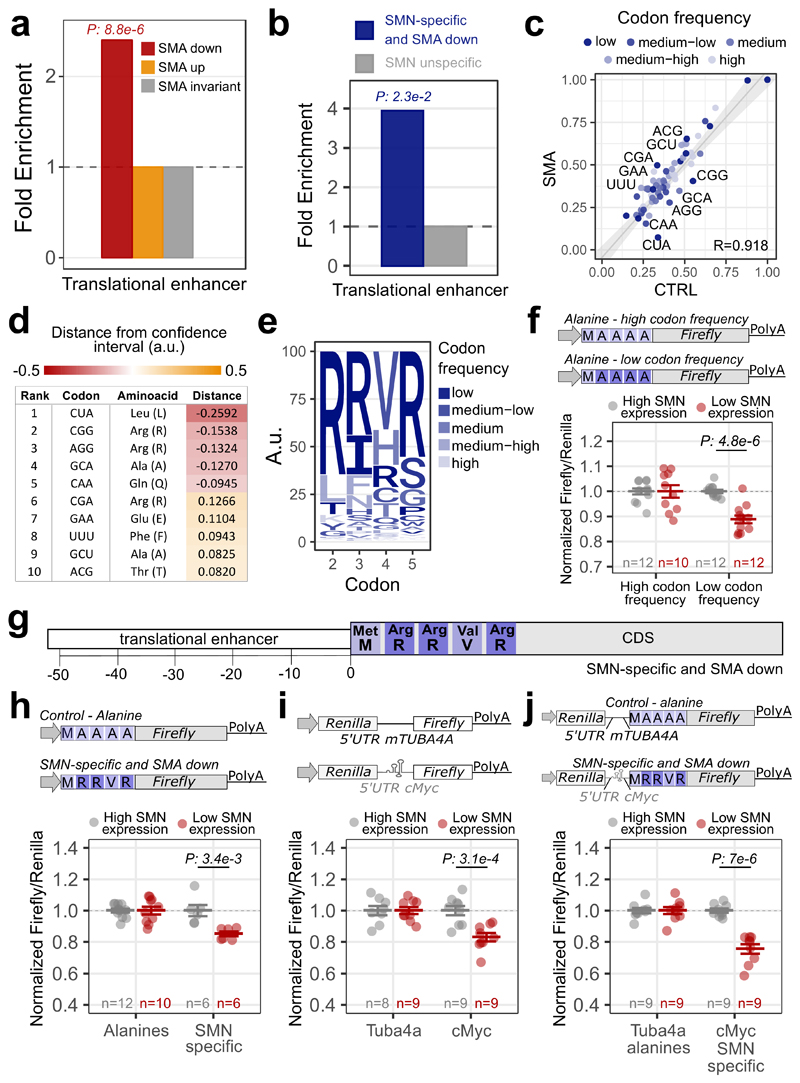
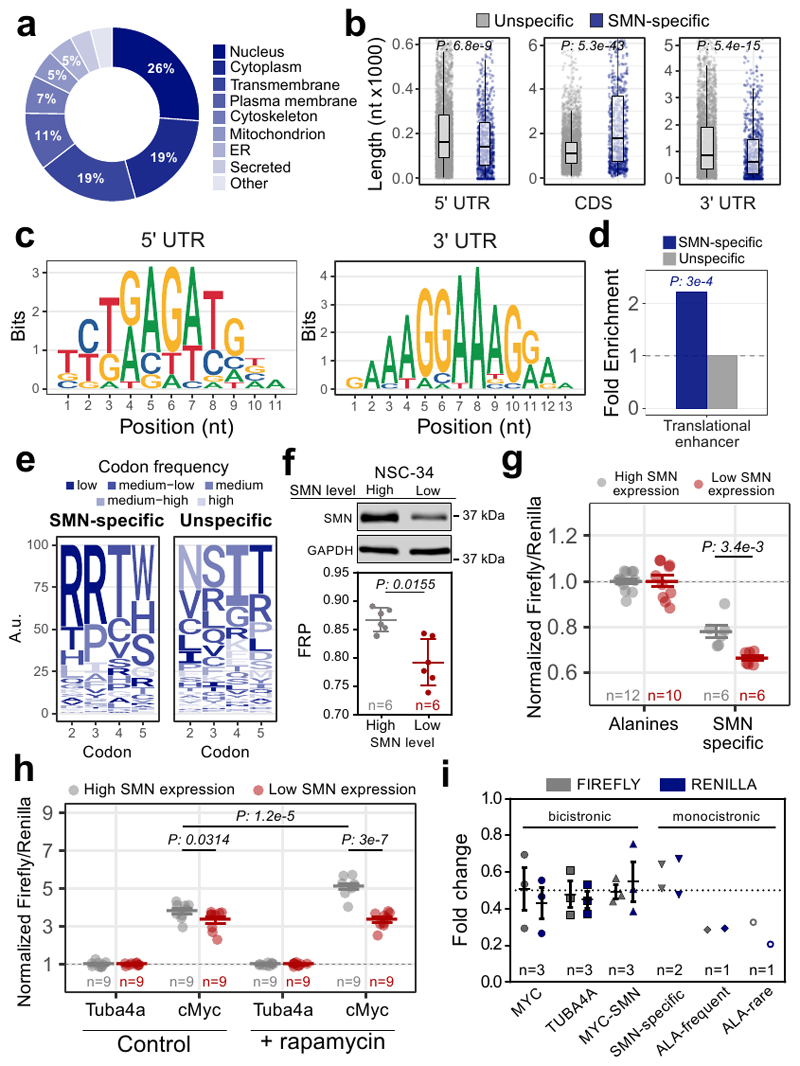
The association of SMN-primed ribosomes with the first five codons of a subset of mRNAs (Fig. 3c–f) involved in SMA and ALS (Fig. 4c) and defectively translated in SMA (Fig. 4d and e), suggests a ribosome-based mechanism underlying the pathophysiology and cell-type specificity of these diseases. Exploring in more detail the molecular and functional features of these mRNAs, we found that they largely encode proteins either localized to the nucleus or mitochondria, including both transmembrane and secreted proteins (Extended data Fig. 5a). These transcripts also display longer CDSs and shorter UTRs (Extended data Fig. 5b). Consistently with the reported propensity of SMN to bind RNA sequences rich in G and A (48, 49), GA-rich consensus motifs were found as enriched in both the 5’ and 3’UTRs (Extended data Fig. 5c). The 5’ UTRs of SMN-specific transcripts are also enriched for sequences previously associated (50) with a translation regulatory function, that we termed translational enhancer sequences (Extended data Fig. 5d), in line with our in vitro results (Fig. 2e). Consistently, transcripts with translational enhancer sequences in their 5’ UTRs are more apt to show translational defects in SMA (Fig. 5a), especially SMN-specific mRNAs (Fig. 5b). These results suggest that mRNAs bearing translational enhancer sequences in their 5’UTR might be more susceptible to translational defects upon SMN loss.
Fig. 5. Translationally defective transcripts in SMA display specific features.
a, Over-representation of translational enhancers in genes with significantly increased (yellow, 287) or decreased (red, 835) ribosome occupancies in SMA. Two-sided Fisher’s test p-value is shown. b, Over-representation of translational enhancers in SMN-specific genes with defects in ribosome occupancy in SMA (blue, 52). The two-sided Fisher’s test p-value is shown. c, Comparison between the codon usage index, based on the P-sites signal in control and SMA Active-RiboSeq. Each dot represents a codon and is colored according to the amino-acid frequency in the transcriptome, divided in 5 classes (low: rare codons; high: frequent codons). Regression line, its 99% confidence level interval and Pearson correlation coefficient are displayed. The 10 furthest points from the confidence level are labelled. d, List of triplet, corresponding amino-acid and distance from the regression line of codons. Negative values correspond to codons whose P-site coverage is lower in SMA than in CTRL, positive values correspond to opposite coverage. e, Logo-like representation of the most frequent amino-acids in SMN-specific mRNAs with significant alterations in ribosome occupancy in SMA at the beginning of the CDS. Letters are colored as in b. f, Luciferase assays are shown as the ratio between Fluc and Rluc normalized to the results obtained with the frequent alanine repeats vector. g, Diagram summarizing the feature combinations in the 5’UTR and CDS of SMN-specific transcripts with translational defects in SMA. h, Luciferase assays are shown as the ratio between Fluc and Rluc normalized to the alanine vector. i, Luciferase assays for the translational enhancer sequence within the 5’UTR of cMyc. j, Luciferase assays for testing the synergic contribution of the features in h and i. For all the luciferase assays the results are normalized for the values in the SMN high expression cells, that were set to 1. The number of biologically independent experiments is reported in the graph. Results are shown as the average ± SEM. Significant changes were assessed using one-sided t-test. Statistical source data are provided in Source data Fig. 5.
By analyzing the composition of the CDS of SMN-specific mRNAs, we found that the first five codons are enriched for rare arginine codons (Extended data Fig. 5e). Furthermore, mRNAs with translational defects at the early stages of SMA are also enriched in rare codons (Fig. 5c), primarily encoding leucine and arginine amino-acids (Fig. 5d). Strikingly, arginine was the most frequent amino-acid within the first 5 codons in SMN-specific transcripts with significant translational defects in SMA (Fig. 5e).
These results suggest that mRNAs with rare codons at the beginning of the CDS are more prone to translational defects upon SMN loss than those with frequent codons. To confirm this, we used a luciferase assay and compared two reporters with either frequent or rare codons encoding for alanines at the 5’ of the luciferase CDS. We tested the translation efficiency of these reporters in motor neuron-like cells with low SMN expression levels (Extended data Fig. 5f), that recapitulate an in cellulo model of SMA. Using this assay, we confirmed that rare codons are more translationally sensitive to SMN depletion than frequent codons (Fig. 5f).
Taken together these analyses suggest that mRNAs with decreased ribosome occupancy in SMA and enriched in SMN-primed ribosomes are characterized by the combination of translational enhancer sequences in the 5’UTR and by rare codons, especially arginines, at the beginning of the CDS (Fig. 5g).
A luciferase reporter assay confirmed that the presence of the “arginine-rich” motif associated with SMN-specific transcripts induces a translational repression in cells expressing low levels of SMN (Fig. 5h and Extended data Fig. 5g). Similarly, a reporter gene under the control of the c-Myc translational enhancer sequence (Extended data Fig. 5h) was less efficiently translated when cells expressed low levels of SMN (Fig. 5i). This decrease was still present by treating cells with rapamycin (an inhibitor of cap-dependent translation), confirming a positive role of SMN in cap-independent translation (Extended data Fig. 5h). A reporter containing both the “arginine-rich” motif and translational enhancer sequences revealed an additive effect of the motifs, resulting in an increased translational defect in cells expressing low levels on SMN (Fig. 5j and Extended data 5i). In summary, these findings demonstrate that a combination of two features is required for SMN-specific mRNAs to be controlled translationally: i) translational enhancer sequences in the 5’UTR, and ii) rare codons in the first five codons of the CDS.
SMN-specific transcripts belonging to all SMN-communities show alterations in ribosome recruitment
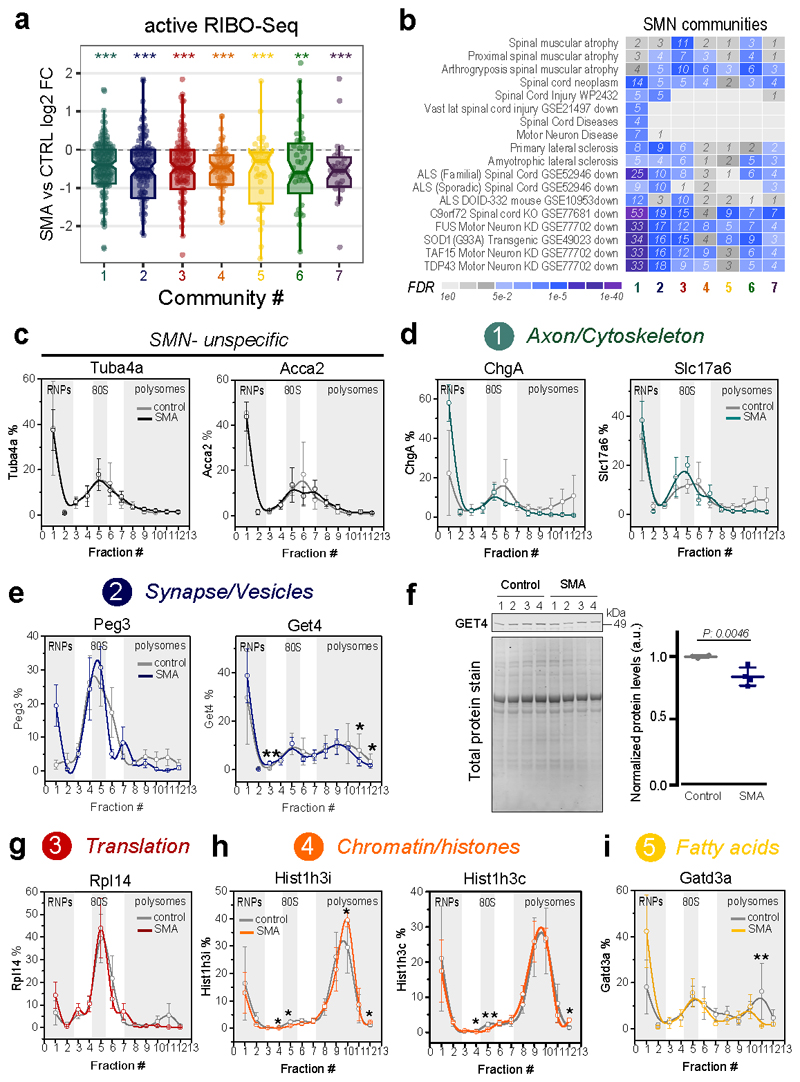
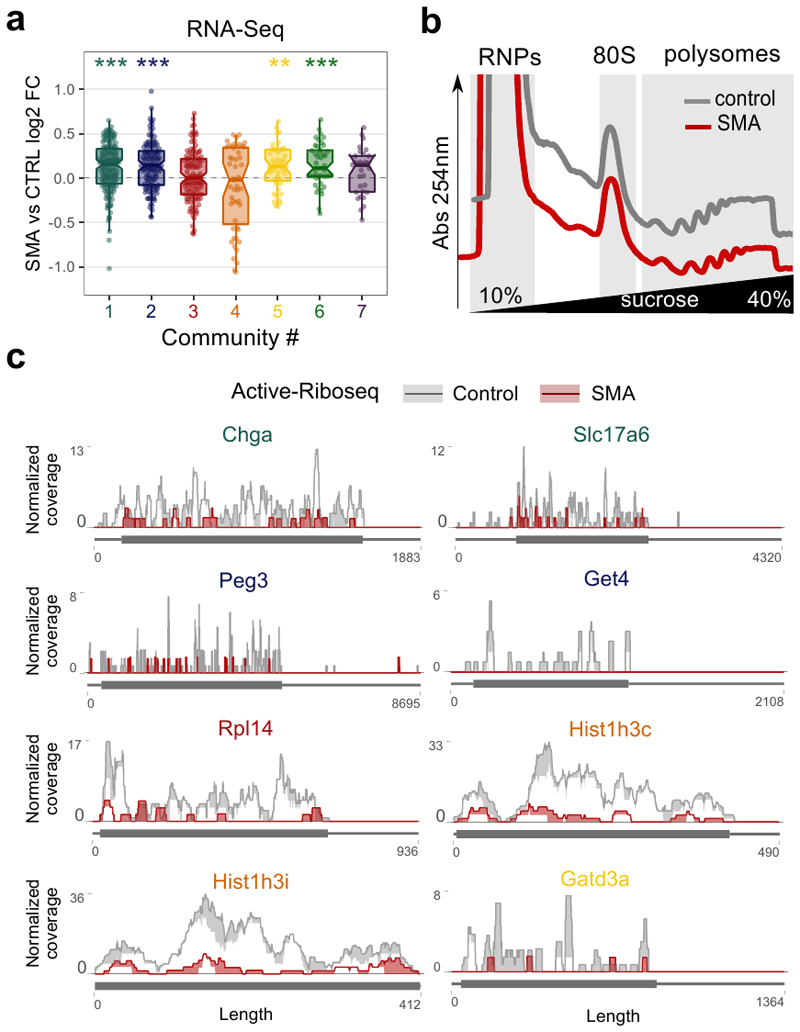
To further validate the role of ribosome-associated defects in SMA pathogenesis, we explored the effect of SMN loss on transcripts belonging to the seven SMN-specific communities previously identified (Fig. 3f). Transcripts belonging to all the seven communities have a significantly decreased ribosome occupancy at early stage of the disease (Fig. 6a). This effect is not caused by transcriptional down-regulation (Extended data Fig. 6a). The communities are also enriched in transcripts reported to be down-regulated in SMA (community 3, Translation/Ribosome) and ALS (community 1, Axon/Cytoskeleton and 2 Synapse/Vesicle) (Fig. 6b).
Fig. 6. Communities of mRNAs bound by SMN-primed ribosomes show reduced ribosome occupancy.
a, Comparison between SMA active ribosome occupancy changes in SMN-specific genes, binned in the 7 SMN-communities. All the communities show a significant shift towards a reduction in SMA active ribosome occupancy (Two-sided one-sample Wilcoxon rank-sum test, ** p-value <0.01; *** p-value <0.001). The number of genes for each community is reported in Fig. 3f. b, Over-representation analysis of terms associated with motor neuron diseases among SMN-specific genes belonging to the 7 SMN-communities. The heatmap is colored according to the significance of the enrichments. c, Relative co-sedimentation profile of SMN unspecific transcripts, Tuba4a and Acca2 along the sucrose gradient fractions of control (gray lines) and early symptomatic (red lines) brains. Data are represented as average ± SEM among n=3 biologically independent samples. d-e, Relative co-sedimentation profile of mRNAs bound by SMN-primed ribosomes and belonging to the community 1 (ChgA and Slc17a6) and 2 (Peg3 and Get4). Data are represented as average ± SEM among n=3 biologically independent samples. One-sided t-test, * p <0.05; ** p <0.01. f, Lysates of brain of late-symptomatic SMA mice were compared to age-matched littermate controls using western blot. Quantification of immunoblot for GET4 was normalized to total protein stain. SMA expression values are normalized and compared to control values for each of the tissues. Data are represented as average ± SEM among n=4 biologically independent samples. Significant changes were assessed using a two-sided t-test. g-i, Relative co-sedimentation profile of mRNAs bound by SMN-primed ribosomes and belonging to the community 3 (Rpl14), 4 (Hist1h3i and Hist1h3c) and 5 (Gatd3a). Data are represented as average ± SEM among n=3 biologically independent samples. One-sided t-test, * p-value <0.05; ** p-value <0.01. Statistical source data and unprocessed blots are provided in Source data Fig. 6.
According to these and previous results (Fig. 4 and 5), SMN-dependent translational defects are compatible with a loss of SMN-primed ribosomes along the CDS of SMN-specific transcripts. To test this further, we performed a co-sedimentation analysis of SMN-specific mRNAs along sucrose gradients of control and early symptomatic mouse brain (Extended data Fig. 6b), selected from the five most abundant communities (Supplementary Table 3 and Extended data Fig. 6c). Tuba4a and Acca2, chosen as controls, did not show any change (Fig. 6c). Indeed, we confirmed that at early stage of disease transcripts belonging to communities 1, 2, 3 and 5 are depleted from polysomal fractions, shifting towards ribosome-free fractions (Fig. 6d–g and i) and leading to possible changes at the protein level (Fig. 6f). Two histone transcripts, representatives of community 4, showed an accumulation on polysomes in parallel to a decrease in the occupancy in active ribosomes by using RiboLace (Fig. 6a). A plausible explanation is that inactive ribosomes might be stalled along the histone transcripts, leading to a decrease in protein production in SMA, as previously shown for several histones (39).
Overall, these results support our genome-wide findings and suggest that SMN-specific transcripts are depleted of ribosomes in SMA.
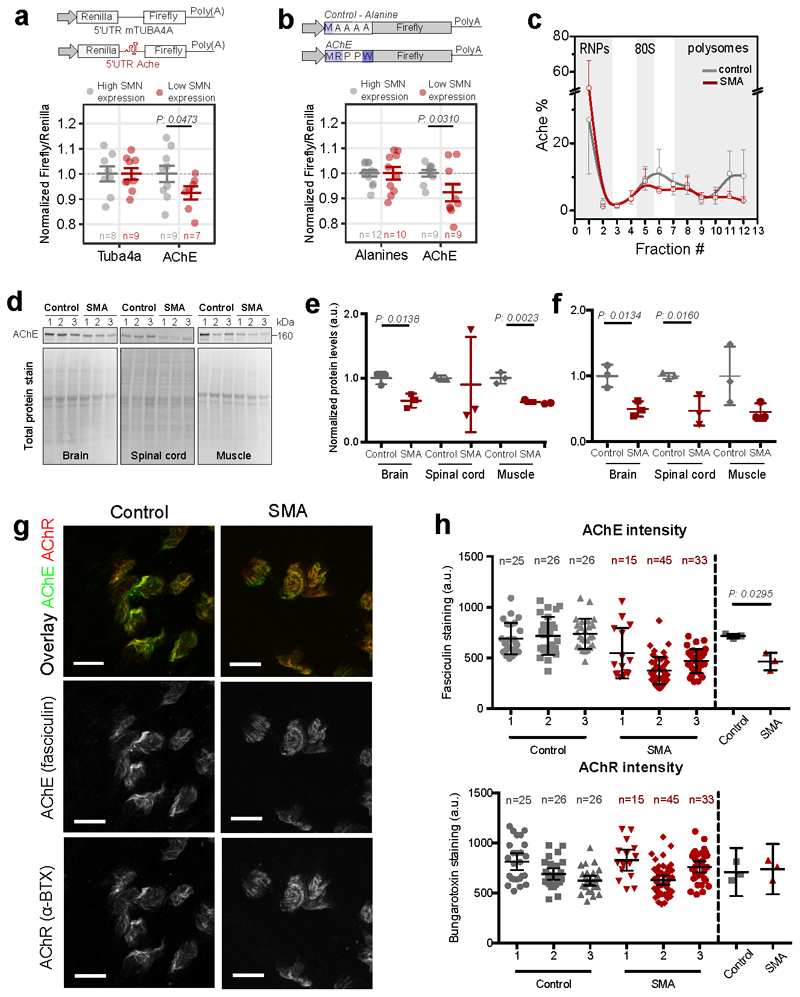
Acetylcholinesterase mRNA is an SMN-specific transcript and an early marker of local defects at the NMJ in SMA
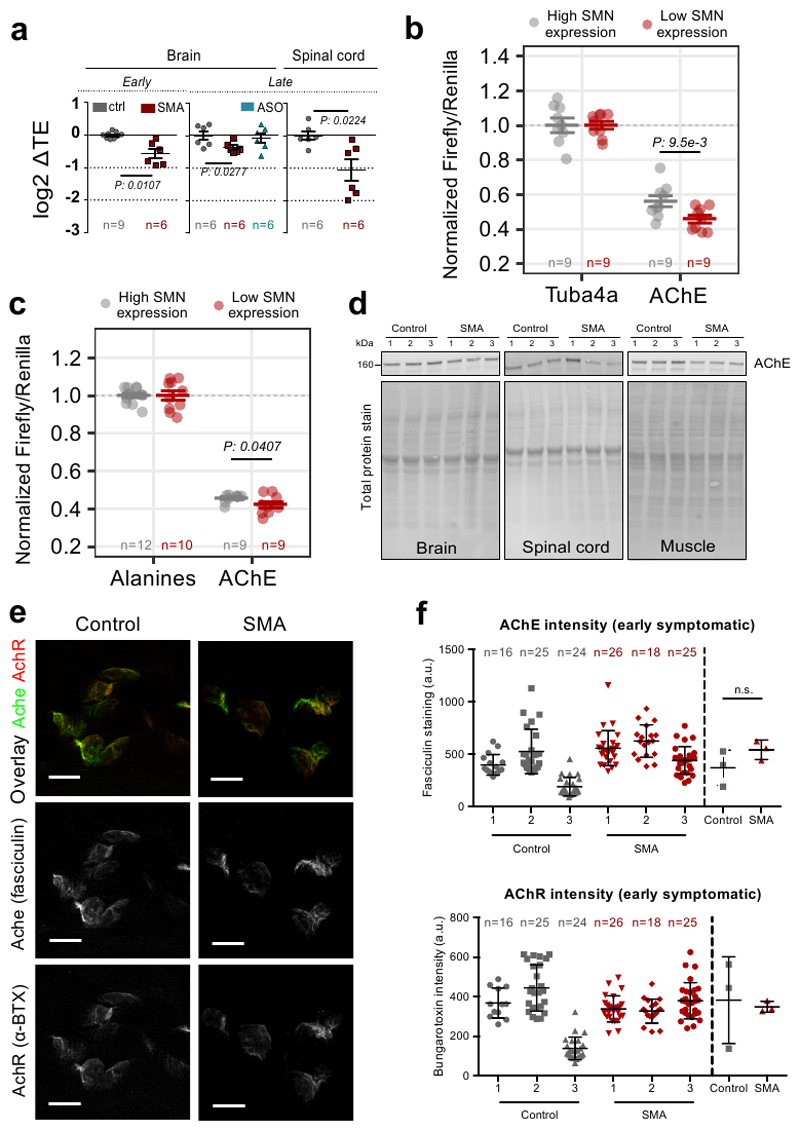
Next, we further characterized a neuron-specific gene, belonging to the larger SMN community (Axon/Cytoskeleton): acetylcholinesterase (AChE). This transcript displays a significant decrease in translational efficiency in both brain and spinal cord (Extended data Fig. 7a). Importantly, in SMA mice treated with a single injection of ASO which restores SMN levels (11), AChE expression was restored to control levels, supporting a direct relationship between AChE and SMN levels (Extended data Fig. 7a). To confirm that the 5’ UTR and the sequence of the first five amino-acids of the AChE transcript are associated with translational defects under SMA-like conditions in cellulo, we performed two luciferase assays (Fig. 7a-b and Extended data Fig. 7b-c). For both these functional motifs, we confirmed that loss of SMN expression causes translational defects in protein production of the reporter genes (Fig. 7a–b). In addition, we observed that at an early stage of disease the AChE transcript is depleted from polysomal fractions, shifting towards the ribosome-free fractions (Fig. 7c). In agreement with these results, we observed that AchE protein expression was decreased at both early and late stages of SMA in several tissues (Fig. 7d–f and Extended data Fig. 7d). Moreover, local expression of AChE protein was significantly impaired at the neuromuscular junction (NMJ) in symptomatic SMA mice (Fig. 7g and h), whilst acetylcholine receptor (AChR) expression remained unchanged (Fig. 7g and h). Downregulation of AChE protein at the NMJ temporally follows the translational defect in ribosome occupancy at earlier stages (Extended data Fig. 7e-f), serving as a molecular marker of impairment at the NMJ in SMA.
Fig. 7. The acetylcholinesterase transcript shows ribosome drop-off and defective production of protein at the NMJ in SMA.
a, Luciferase assays with a reporter bearing the 5’UTR of Ache. The 5’UTR of Tuba4a was used as control. Results are shown as the average ± SEM, n is shown in the graph Significant changes between cell lines were assessed using the one-sided t-test. b, Luciferase assays for testing the contribution of the first five codons of AChE. Upper panel, representation of the reporter. The experiment was performed as in Fig. 5g and 5h. Results are shown as the average ± SEM, n is shown in the graph. Significant changes were assessed using a one-sided t-test. c, Relative co-sedimentation profile of AChE mRNA in control (gray) and early symptomatic (red) brains. Data are represented as average ± SEM among n=3 biologically independent samples. d, Protein levels in brain, spinal cord and muscle of late-symptomatic SMA mice were compared to controls using western blot. e, Quantification of immunoblots for AChE normalized to total protein stain at early-symptomatic stage. Data are represented as average ± SEM among n=3 biologically independent samples. Significant changes were assessed using a two-sided t-test. f, Quantification of immunoblots for AChE normalized to total protein stain at late-symptomatic stage SMA Data are represented as average ± SEM among n=3 biologically independent samples. Significant changes were assessed using the two-sided t-test. g, g, Representative images for control and late-symptomatic SMA mouse neuromuscular endplates. Acetylcholine receptors were labelled using alpha-bungarotoxin (BTX) and AChE was labelled using fasciculin-2 (FCC). A total of 77 endplates from control and 93 endplates from SMA mice taken from 6 muscles and 3 mice per genotype were imaged and analysed. Scale bar: 10 μm. h, FCC and BTX average intensity were determined for 2 FDB muscles in 3 control and 3 SMA biologically independent mice. N in graph indicates the number of analysed endplates per mouse. Data are represented as average ± SEM. Significant changes were assessed using a two-sided t-test. Statistical source data and unprocessed blots are provided in Source data Fig. 7.
Discussion
This study provides experimental evidence supporting the hypothesis that SMN is a ribosome-associated protein, demonstrating that SMN can be found in two distinct functional ‘populations’ within the cytoplasm. One population is associated with the well-known, canonical role of SMN in RNP biogenesis via its association with Gemin granules (40, 51). In line with Jablonka et al. (52) we found a second population that was not associated with Gemin granules, but rather with ribosomes. We show that SMN as a ribosome-associated protein in vitro and in vivo, in agreement with the known ability of SMN to co-sediment with polysomes in vivo, in cellulo and in vitro (11, 28–30), as well as with general factors of translation such as eEF1A (53). Strikingly, the population of ribosomes associated with SMN (SMN-primed ribosomes), only made up a small fraction of the total ribosome pool. SMN-primed ribosomes display two unique characteristics: i) they are associated with a specific subset of mRNAs which form functionally related communities, and; ii) they are preferentially placed within the first five codons of the CDS.
SMN-primed ribosomes bind mRNAs characterized by strong enrichment in rare codons at the beginning of the CDS, particularly arginine-codons, and in translational enhancer sequences in the 5’UTR. The ability to associate with a subset of mRNAs has previously been observed for ribosomes containing particular ribosomal proteins (15, 20). Notably, we found that a significant number of mRNAs bound by SMN-primed ribosomes have previously been linked to the pathogenesis of SMA, as well as to related neuromuscular conditions such as ALS. This provides a possible molecular explanation as to why defects in ubiquitously-expressed proteins, such as SMN, can lead to the specific sensitivity to degeneration of motor neurons in conditions such as SMA and ALS.
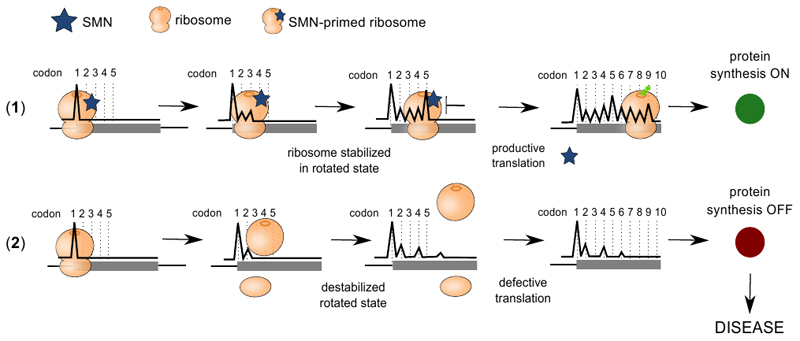
The fact that SMN-primed ribosomes are located within the first codons of SMN-specific transcripts suggests a highly specific, local function for this defined subpopulation of ribosomes. It has been observed that a ribosome-pause at these first codons acts as a translational checkpoint to ensure productive ribosome elongation and protein synthesis (54). Accordingly, SMN binds to the elongation factor eEF1A (53). We found that SMN is required for productive translation and that the positive regulation of translation is lost when SMN expression is reduced (11, 28, 30). In addition, we found two distinct populations of mRNA fragments protected by SMN-primed ribosomes characterized by different lengths. Similar bimodal distributions in read lengths have been observed in yeast, and have been associated with diverse structural conformations of the ribosome during translation elongation (37, 55). Further investigation of these ribosome conformations may require the use of diverse translation inhibitors, similar to Wu et al. (56). By profiling actively translating ribosomes (36) in control and early symptomatic SMA tissues we found that the vast majority of genes associated with a significant variation in ribosome occupancy displayed a strong decrease in active ribosomes numbers upon SMN depletion. The mRNAs associated with SMN-primed ribosomes showed profound positional defects at beginning of the CDS, suggesting a possible loss of specific ribosome recruitment or pausing mediated by SMN-primed ribosomes at a critical initial step during translation, leading to ribosome drop off (54). Thus, we propose a model whereby SMN regulates the translation of rare codons by acting as a stabilizer of specific ribosome conformations at the beginning of the CDS, where it can induce a functional ribosome slowdown that ensures productive translation (Extended data Fig. 8).
Among the mRNAs bound by SMN-primed ribosomes and characterized by translational defects in SMA, we identified acetylcholinesterase (AChE) whose 5’UTR and first 5 codons are translationally sensitive to low levels of SMN expression. AChE performs a central role in NMJ function by turning over acetylcholine after it has been used for signal transduction by motor neurons. Dysfunction and denervation of the NMJ is one of the earliest pathological features of SMA (57–59). In agreement with our proposed model, we found that, in early symptomatic SMA, the recruitment of AChE mRNA on polysomes is reduced, resulting in defects at the protein level in SMA-related tissues and at the NMJ at later stages of disease. Previously, an absence of the asymmetric A12 form of AChE was observed in the serum of SMA Type I patients (60). Moreover, mutations affecting AChE in humans cause congenital endplate acetylcholinesterase deficiency, a disease displaying a number of clinical features overlapping with those observed in SMA (61). Thus, SMN-primed ribosomes play a crucial role in regulating AChE levels that are likely to contribute to NMJ defects at the core of SMA pathogenesis.
The robust influence of SMN levels on ribosome binding, alongside the higher relative concentration of SMN protein found in the nervous system (34), supports a model whereby a stronger effect on transcripts bound by SMN-primed ribosomes should be observed in these tissues. Thus, a different scenario for better understanding the molecular pathogenesis of SMA can be generated, in which tissue- and concentration-specific regulation of SMN concertedly tune the correct translation of mRNAs bound by SMN-primed ribosomes, as illustrated here by the effect observed on AChE.
Taken together, our findings demonstrate a central role for SMN in the regulation of ribosome heterogeneity, acting as a master modulator of ribosome fluxes on a disease-specific subset of disease-relevant mRNAs characterized by specific sequence features. This reveals an important role for ribosome-associated proteins in the regulation of tissue-specific disease pathogenesis in SMA and related conditions.
Materials and Methods
Animal models
Animal procedures and breeding were performed in accordance with University of Edinburgh institutional guidelines and under appropriate project and personal licenses granted by the UK Home Office (PPL: P92BB9F93). The ‘Taiwanese’ mouse model of severe SMA (62), on a congenic FVB background, was established from breeding pairs originally purchased from Jackson Labs and maintained following an established breeding strategy (63). Phenotypically normal littermates (Smn+/-;SMN2tg/0) were used as controls. Litters were genotyped using standard protocols. All mice were housed within the animal care facilities in Edinburgh under standard SPF conditions. For clarity, throughout the paper we refer to the time points at which tissue was collected as early and late symptomatic. Early symptomatic was postnatal day 5 (P5) and late symptomatic was P7. All tissues were quickly dissected, snap-frozen and stored at -80°C until use.
Subcellular fractionation from tissues
The method was adapted from Francisco-Velilla et al., 2016 (31). Lysates from brain or spinal cord were obtained as in (11). A few μL of the lysate was kept for protein extraction (input). The sample was centrifuged for 67 min at 100,000 rpm using a TLA100.2 rotor (Beckman). Supernatant corresponds to proteins not associated to ribosomes or polysomes (unbound). The pellet containing ribosomes, (R pellet) was solubilized in 15 mM Tris-HCl pH 7.4, 100 mM KCl, 5 mM MgCl2, 2 mM DTT, 290 mM sucrose and few μL were kept for protein extraction. The remaining volume was adjusted to 500 mM KCl, to detach mildly associated proteins. The sample was loaded on a discontinuous sucrose gradient (720 μL buffer 40% (w/v) sucrose, 15 mM Tris-HCl pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT (bottom layer) and 480 μl buffer 20% (w/v) sucrose, 15 mM Tris-HCl pH 7.4, 500 mM KCl, 5 mM MgCl2, 2 mM DTT (top layer) and ultra-centrifuged at 100,000 rpm or 1.5 h using a TLA100.2 rotor (Beckman). Supernatants contain proteins loosely associated to ribosomes and polysomes (LBR), the pellet contains washed ribosomes (RBR). The pellet was dissolved in sample buffer, proteins in the other fractions were extracted using a Methanol/Chloroform protocol. RNAse treatment: the R-pellet was prepared as described above was treated with RNAse I (1.5U/1Abs260) for 45 min at rt. The lysate was ultracentrifuged as above to isolate the proteins associated to ribosomes/polysomes via RNA-dependent interactions and proteins associated via protein-dependent interactions. Protein extraction was performed as above before analysis by western blotting. The following primary antibodies were used: RPS6 (Cell Signaling; AB_331355 (1:1000)); RPL26 (Abcam; AB_945306 (1:1500)); SMN (BD Transduction Laboratories; 610646 (1:1000)); HuD (Santa Cruz Biotechnology (1:1000)); QKI (Abcam Ab-126742 (1:1000)). The following secondary antibodies were used: anti-Mouse HRP (Santa Cruz Biotechnology; Sc-2357 (1:3000)); anti-Rabbit HRP (Santa Cruz Biotechnology; Sc-2004 (1:3000)).
Western blotting from tissues
Western blot on tissues from SMA and control mice was performed exactly as described before (64). The following primary antibodies were used 4E-BP (Abcam; Ab-2606 (1:1000)); P-4E-BP (Abcam; Ab-27792 (1:1000)); eIF2α (Santa Cruz Biotechnology; Sc-11386 (1:1000)); p-eIF2α (Abcam; Ab32157 (1:1000)); Get4 (Proteintech 27768-1-AP (1:1000)); AE2 (acetylcholinesterase) (DSHB AE2DSHB (1:1000)). Antibody detection was performed using fluorescent secondary antibodies (LI-COR) and protein loading was normalized to a fluorescent total protein stain (LI-COR). Quantification was performed as described previously (34).
Ribosome purification and ribosome-binding assay
Purification of 80S ribosomes was performed from NSC-34 depleted from SMN using CRISPR-Cas9 technology (11). The lysate was treated with RNase I (7.5 Units /1 Abs260 lysate) at RT for 45 min and analyzed by sucrose gradient (10-40%) (65). The fraction corresponding to the 80S was collected and 2mM DTT was added. After centrifugation at 90,000 rpm for 4 h using a TLA100.2 rotor (Beckmann) the pellet was resuspended in 10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 2 mM DTT, 100 μg/mL cycloheximide and stored at −80°C. Ribosome concentration was calculated as in (66). Recombinant SMN was purchased (ENZO) and incubated with ribosomes for 2 h at 4°C. SMN bound ribosomes were purified from unbound SMN by ultracentrifugation at 90,000 rpm for 4 h using the TLA100.2 rotor on 30% sucrose cushion. The supernatant was kept for protein purification by using the chloroform/methanol protocol and the pellet was directly dissolved in sample buffer (Santa Cruz), heated at 99°C for 10 min and resolved by SDS-PAGE. SMN and RPL26, were detected using primary and secondary antibodies described above.
Surface Plasmon Resonance
Surface plasmon resonance (SPR) experiments were carried out using a Biacore X100 (GE Healthcare, US). Binding experiments were done at 22 °C in 20 mM Hepes-NaOH, pH 7.4, 150 mM NaCl, 5mM MgCl2, and 0.005 % surfactant P20 as a running buffer. SMN protein was immobilized on C1 sensor chip (GE Healthcare, US) via amine coupling following manufacturer’s recommended procedure. The carboxymethyl dextran surface was activated with a 7-min injection at 5 μl/min of a 1:1 ratio of 0.4 M 1-ethy-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and 0.1 M N-hydroxy succinimide (NHS). Buffer without the protein was injected over the flow cell 1, as a reference for subtraction from the binding signal obtained in experiments on flow cell 2, where SMN was immobilized. Residual reactive sites on the surface of the sensor chip were deactivated with a 7-min injection of 1 M ethanolamine, pH 8.5. A series of ribosome concentrations, typically ranging from 0.4-10 nM were injected over immobilized SMN protein at a constant flow rate 80 μl/min for 1 min. Ribosome dissociation was monitored for 300 s, followed by the injection of 0.1 M glycine-NaOH, pH 12, and 0.3 % Triton X100 for 11 s at 80 μl/min for the regeneration of the surface. Sensorgrams were processed with BiaEvaluation software (GE Healthcare, US). The average response of blank injections was subtracted from all analyte injections. Kinetic parameters were determined from the processed data by globally fitting ka and kd to a 1:1 binding model. Six independent estimations of KD were done and the results shown are the average KD ± SD, n = 6.
In vitro transcription-translation assays
In vitro transcription-translation was performed using the 1-Step Human Coupled IVT Kit HeLa lysates and pCFE-GFP as reporter (Thermo Scientific). GFP protein quantifications were performed by western blotting and fluorescence spectroscopy. For western blotting, after incubation of each reaction at 30°C for 1.5 h, proteins were extracted, solubilized in sample buffer and analyzed by SDS-PAGE. The production of EGFP was monitored using an TurboGFP Thermo Scientific (1:1000). SMN was monitored using SMN BD Transduction Laboratories 6 (1:1000). RPL26 was used as control. Protein production was quantified by densitometric analysis using ImageJ. The EGFP signal was normalized to the RPL26 signal. The GFP production by fluorescence spectroscopy was monitored following the height of the emission spectra maximum at 502 nm. 10 μL sample after 1.5 h incubation at 30°C in the presence of different SMN concentrations was added to a 1-cm quartz cuvette filled with 990 μL of buffer. Spectra were acquired on a Fluoromax-4 (Horiba Jobin-Yvon) with lex=482 nm.
Immunoprecipitation of SMN-associated proteins from polysomal fractions and ribosomal pellets
IP from polysomal fractions was performed on pooled polysomal fractions from control brains (11). Samples were diluted 3x using 30 mM Tris-HCl (pH 7.5), 100 mM NaCl, 10 mM MgCl2, 20 μL/mL cycloheximide. One mL was kept as input, and the remaining sample was divided into two parts and incubated for 2 h at 4 °C with either 2 μg of SMN antibody (BD Biosciences) or, as negative control, 2 μg of anti-Mouse IgG (Abcam) while rotating. Dynabeads Protein G and Dynabeads Protein A (Life Technologies) were added and kept rotating for 2h at 4 °C. The supernatant was stored as “Unbound”. Beads were washed 3x with 500 μL Washing Buffer (10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 10 mM NaCl, 1% Triton X-100, 1 mM DTT, 0.2 mg/mL cycloheximide, 1% Na-deoxycholate, 2.5 μL/mL Protease Inhibitor Cocktail). RNase A/T1 was added (200 μg/mL final concentration) and the samples kept under rotation for 1.5 h at 4 °C. Samples were placed on the magnetic stand and the supernatant stored. Next proteins (“RNA-mediated”) were extracted. After extensive washing, proteins on the beads (“Protein-mediated”) were dissolved in sample buffer for western blotting. All other samples (“Input”, “RNA mediated” and “Unbound”) were purified by methanol/chloroform extraction and analyzed by western blotting. The primary antibodies described against the following proteins were used: SMN, RPS6; RPL26; eIF4A1 (Abcam; AB_732122 (1:1000)); PABP (Abcam; AB_777008 1:1000).
Polysomal profiling and co-sedimentation analysis of proteins
Cytoplasmic lysates from cell lines (NSC-34 and Hek-293) or frozen mouse tissues or cells were prepared as described previously (11). Lysates were loaded on a sucrose gradient (10-40% sucrose [m/v], in 100 mM NaCl, 10 mM MgCl2, 10 mM Tris/HCl pH 7.5) and ultracentrifuged for 1.5 h at 198,000 g at 4°C in a Beckman Optima™ LE-80K Ultracentrifuge. One ml fractions were collected monitoring the absorbance at 254 nm with an ISCO UA-6 UV detector. For Mg2+ depletion, cells were lysed in 10 mM NaCl, 10 mM Tris/HCl pH 7.5, 1% Triton-X, 1% Na-deoxycholate and loaded on sucrose gradients prepared in 100 mM NaCl, 10 mM Tris/HCl pH 7.5.
Proteins were extracted using the methanol/chloroform protocol (67) and solubilized in Sample Buffer (Santa Cruz Biotechnology) for the SDS-PAGE and western blotting. The following primary antibodies were used anti RPS6; RPL26, SMN and Gemin5 (Novusbio; NB100-61049 (1:1000)); HuR (Santa Cruz Biotechnology Sc-71290 (1:1000)). The secondary antibodies were used as above.
The co-sedimentation profiles of proteins along the sucrose gradient, was obtained as described in (11). The relative percentage of the protein intensity of each fraction along the sucrose gradient was calculated as follows:
where % Proteinn is the percentage of the protein in the fraction n; [Protein]n is the density of the protein in the fraction n and 12 is the total number of fractions.
Cloning of 5’UTRs and SMN-specific motifs and Luciferase assay
pGL3 plasmid was used to generate monocistronic reporters. Sequences for the first five amino-acids of the Firefly luciferase CDS were introduced by complementary oligonucleotides designed to form overhangs corresponding to HindIII and NcoI restriction sites. pRuF plasmid was used to create bicistronic luciferase reporters to study AchE, Tuba4a, or cMYC 5’-UTRs, that were PCR-amplified from cDNAs from mouse brain (P5), or from an available plasmid, using suitable cloning primers (Supplementary Table 4). In the pRuF vector, Renilla and Firefly cDNAs are transcribed as a single transcript separated by the cloned 5′-UTR sequences. pRuF plasmids were further modified to study the combined effect of 5’UTR and first five amino-acids, inserted by exploiting the restriction sites PmlI and NarI. All plasmid clones were checked by DNA sequencing.
NSC-34 expressing high or low levels of SMN (11) were seeded in 24-well plates and co-transfected with 3:1 ratio (300ng + 100ng) of pGL3 and pRLSV40 plasmid -to normalize for transfection efficiency- or with 400ng pRuF plasmids using TurboFect Transfection Reagent (Thermo Scientific). Luciferase assays were run 48h after transfection, using the dual-luciferase reagent (Promega) as in (65). Total RNA was extracted by TRIZOL from lysates from the lysates used for dual luciferase assays to determine the expression level of luciferase reporters. cDNA was synthesized from 100 ng RNA using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific). qPCR was carried out using Kapa Syber Fast qPCR Mastermix (Kapa Biosystems) and specific primers (Supplementary Table 5). 18S was used as reference gene.
Immunoprecipitation of SMN with active ribosomes
Control MCF7 cells and MCF7 cells treated with puromycin 100 μM for 1h were lysed according to (36). Active ribosomes were isolated using RiboLace kit (IMMAGINA Biotechnology). Proteins were extracted from beads using sample buffer (Santa Cruz). Five μL of lysate were kept as input. Proteins were resolved using SDS-PAGE and western blotting as in (36).
Active and classical Ribosome profiling
Cytoplasmic lysates from P5 control and early symptomatic SMA mouse brains were prepared as in (11). For Active-ribosome profiling, RiboLace kit (IMMAGINA Biotechnology) was used following manufacturer’s instructions. The libraries quality and quantity were assessed by using the high-sensitivity DNA chip on the BioAnalyzer (Agilent) according to the manufacturer’s protocol and Qubit® 2.0 (ThermoFisher Scientific). The libraries were sequenced on an Illumina HiSeq2500. Ribosome profiling from control P5 brains was performed after polysomal purification. Polysomes were purified as described above. Polysomal fractions were pooled and digested with Rnase I (150U/unit of area of polysomes, calculated from the polysomes profile) for 2h at 4°C. The digestion was stopped by 400 U SUPERase-In RNase inhibitor (Thermo Fisher Scientific). RNA was extracted as in Bernabò et al., 2017(11). Ribosome Protected Fragments (RPF) were isolated and the libraries were prepared following the Ingolia protocol (68).
The nuclease digestion was performed on the same brain lysates used for Ribo-Seq. Two biological replicates were performed. Indexes for library preparation are listed in Supplementary Table 6. Active ribosome profiling was performed in parallel to ribosome profiling, starting from 25 μL of tissue lysates treated with RNAseI (5U/ absorbance at 260 nm).
Ribosome profiling of SMN-primed ribosomes
Cytoplasmic lysates from P5 brains were obtained as before (11). Endonuclease digestion was performed with RNase I (5U/Unit of absorbance at 260nm in the lysate) at rt for 45 min. The reaction was stopped with SUPERase-In RNase inhibitor (Thermo Fisher Scientific). The digested lysates were centrifuged for 70 min at 100,000 rpm (TLA100.2 rotor, 4°C) and the ribosome pellet solubilized in 10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 1% Triton X-100, 600 U/mL RiboLock RNase Inhibitor (Thermo Scientific), 0.2 mg/mL cycloheximide, Protease Inhibitor Cocktail. Ribosomes associated to SMN were purified by immunoprecipitation using mouse anti-SMN antibody or anti-IgG (Abcam; Ab-18443) as a control for unspecific binding. Briefly, the ribosome suspension was incubated with 2 μL of antibody for 1 h and 40 min in orbital rotation at 4°C. Dynabeads Protein G (Life technologies) were added and incubated for 1 hr at 4°C in orbital rotation. The supernatant was removed using a magnetic stand and the beads were washed 2 times for 5 min with washing buffer (10 mM Tris-HCl pH 7.5, 10 mM MgCl2, 150 mM NaCl, 1% Triton X-100, 0.2 mg/mL cycloheximide, Protease Inhibitor Cocktail) before extraction of RNA with Trizol. The ribosome protected fragments from both SMN and IgG immunoprecipitation were isolated and used for library preparation as described above. Experiments were performed in triplicate.
Co-sedimentation profiles of proteins and of mRNA
Polysomal profiling from cell lines or tissues as in (11). Proteins from each fraction were extracted and analysed by SDS-PAGE and western blotting as above.
RNA from each sucrose fraction was extracted using TRIZOL or Phenol/chloroform as in (5). Equal volumes of RNA were used for cDNA synthesis using iScript cDNA synthesis kit. EvaGreen-based ddPCR reaction mixtures were composed of 1x QX200 EvaGreen ddPCR Supermix, 150 nM forward and reverse primers (Supplementary Table 5) and 1:10 diluted cDNA. For droplet-PCR, a 20 μL aliquot from each of the assembled ddPCR mixtures and 70 μL Droplet Generation Oil for EvaGreen were loaded into droplet generation cartridge (Bio-Rad). After sample partitioning with QX200 Droplet Generator (Bio-Rad), the entire droplet emulsion volume was transferred in a T100 thermal cycler (Bio-Rad). After PCR, droplets were read individually by QX200 Droplet Reader (Bio-Rad) and the data analysed by QuantaSoft (Bio-Rad). The % of each transcript along the profile was as follows:
where n is the number of the fraction.
For qRT-PCR, the retrotranscription reaction was performed starting from the same volume of RNA obtained from each fraction (1-3 μL/polysomal profiling experiment) using the RevertAid First Strand cDNA synthesis kit (Thermo Scientific). qPCR was carried out using the CFX Connect Real-Time PCR Detection System (BioRad) using Kapa Syber Fast qPCR Mastermix (Kapa Biosystems) or qPCRBIO SyGreen Mix Separate-ROX (PCRBiosystem). Primer sequences are provided in Supplementary Table 5. The percentage of each transcript distribution along the profile was obtained using the following formula in the case qPCR:
where n is the number of the fraction, %[mRNA]n is the percentage of mRNA of choice in each fraction.
NMJ fluorescence microscopy
For NMJ analysis, flexor digitorum brevis (FDB) muscle was dissected from early- and late-symptomatic mice using procedures similar to those described previously (69) and fixed in 4% PFA for 20 min at RT. Muscles were stained for 30 minutes at RT on a rotating platform using alpha-bungarotoxin (BTX) conjugated to Alexa Fluor 594 (Invitrogen) and fasciculin-2 (FCC) conjugated to Alexa Fluor 488 (Invitrogen custom production, kind gift from Prof David Beeson, University of Oxford), both at 1:1,000. FDB muscles were mounted in mowiol on microscope slides and imaged using a Nikon A1R confocal system at the IMPACT Facility, University of Edinburgh. Complete, en face neuromuscular endplates were identified based on BTX labelling (70), and BTX and FCC intensity were determined using FIJI.
Data Analysis
Preprocessing of ribosome profiling data
Reads were processed by removing 5’ adapters, discarding reads shorter than 20 nucleotides and trimming the first nucleotide of the remaining ones (Trimmomatic v0.36). Reads mapping on M. musculus rRNAs (SILVA rRNA database, release 119) and tRNAs (Genomic tRNA database: gtrnadb.ucsc.edu/) were removed. Remaining reads were mapped on the mouse transcriptome (Gencode M6 transcript annotations). Antisense and duplicate reads were removed. All alignments were performed with Bowtie2 (v2.2.6) employing default settings.
Identification of SMN-specific transcripts and differential analyses
Transcript counts were normalized using the trimmed mean of M-values normalization method (TMM) implemented in the edgeR Bioconductor package. Transcripts with FPKM > 1 in all the replicates of at least 1 condition were kept. Differential analyses were performed with edgeR (glmQLFTest function).
SMN-specific transcripts in healthy mouse brains were selected based on 2 comparisons:
SMN-specific RiboSeq vs IgG control, to remove any possible non-specific signal.
SMN-specific RiboSeq vs total RiboSeq to select transcripts specifically enriched in SMN-primed ribosomes over total ribosomes.
Enrichment p-values for each comparison were combined with the Fisher method and SMN-specific transcripts were selected based on the following thresholds: combined p-value < 0.05, average log2 SMN-specific fold enrichment > 0.25, SMN-specific RiboSeq FPKM > 1.
Genes with significant alterations in translation in SMA were selected comparing CTRL vs SMA Active-RiboSeq, with the following thresholds: absolute log2 fold change > 0.50, p-value < 0.05, Active-RiboSeq FPKM > 1.
The cellular localization of the protein codified by SMN-specific transcript was downloaded from UniProt.
Identification of SMN-specific communities
Protein-protein interactions between SMN-specific genes were downloaded from the STRING database (v 11.0, Mus Musculus dataset, interaction score >= 0.3). Network analysis was performed with the igraph R package (https://igraph.org/r/). SMN-specific communities within the network were identified with the cluster_fast_greedy function and ranked by gene size. The representative network shown in figure 3f was generated with the geomnet R package using the Fruchterman-Reingold Algorithm force-directed layout.
Identification of brain-specific transcripts
Brain-specific transcripts were identified by merging the mRNAs selected for Active-RiboSeq and RiboSeq, using a double threshold on their signal: average FPKM and CPM values > 80th percentile. This step ensures to work with transcripts with sufficient coverage for further analysis.
Positional analysis of ribosome profiling data
The identification of the P-site position within the reads was performed using riboWaltz (v1.1.0) (71) with the automatic detection of the optimal extremity and P-site offsets. For all samples of Active-RiboSeq and classical RiboSeq the optimal offset was set to 16 nucleotides from the 3’ end of the reads. For SMN-specific RiboSeq the optimal offset was set to 12 nucleotides from the 5’ end of the reads. Most of the downstream analyses were performed using the following functions included in riboWaltz (71):
region_psite for the percentage of P-sites falling in the three annotated transcript regions (5’ UTR, CDS and 3’ UTR);
rlength_distr for the distribution of read lengths. The distribution was fitted with two Gaussian with the MASS R package;
metaprofile_psite for the metaprofiles based on the P-site position. To overlay and compare metaprofiles from different sample, the area under the curve was set to 1;
frame_psite for the percentage of P-sites falling in the three possible translation reading frames;
codon_usage_psite for the comparison between the codon usage index, based on the sum of in-frame P-sites from control and SMA. Stop codons were removed and the dots were colored according to the amino-acid frequency in the mouse transcriptome (downloaded from https://www.kazusa.or.jp/codon/).
The ratios between the average number of P-sites on the first five codons (initiation) and the average number of P-sites on the whole coding sequence (CDS) was computed as follows. First, each transcript was split in two regions, including respectively the nucleotides from 0 to 14 and from 15 to the end of the CDS. Second, the average number of P-site falling in the two regions and the ratio between the first and the second region was computed for each transcript. Logo-like representations of the most frequent amino-acids codified at the beginning of the CDS are based on the number of occurrences of each codon, using brain-specific transcripts previously identified as background. Triplets with fold enrichments > 1 were selected and the weighted sum among synonymous codons was computed. The resulting values are displayed as percentages.
Functional enrichment analyses
Functional annotation enrichment analyses of SMN-specific transcripts, SMN-communities and genes with alterations in active translation in SMA were performed with the Enrichr resource (http://amp.pharm.mssm.edu/Enrichr/) and the enrichR R package.
Enrichment analysis of translational enhancer sequences was performed based on annotation retrieved from Weingarten-Gabbay et al., 2016 (50).
Statistics and Reproducibility
All box plots show the first quartile, median and third quartile and the whisker extends from the smallest value to the larger value at most 1.5 * IQR from the hinge. If present, notches display a confidence interval corresponding to the median +/- 1.58 * IQR/sqrt(n) where n is the number of data.
Results from luciferase assays are normalized for the values in the SMN high expression cells, that were set to 1. Results are shown as the average ± SEM. Significant changes were assessed using one-sided t-test.
All the other assays have been performed 2 times unless specified in the legends. For all assays, quantification and statistics were derived from n = 3 independent experiments unless specified in the legends.
Extended Data
Extended Data Fig. 1. SMN interacts with the translation machinery in vitro and in vivo in an RNA-independent manner.
a, Samples were ultracentrifuged to evaluate the aspecific sedimentation of SMN. The experiment was performed as in Fig.1c, in the absence of ribosomes. The presence of SMN in the pellet was determined by western blot and densitometric analysis. b, Saturation curve obtained using experiments as in Fig.1. The unspecific binding was obtained as in panel (a). The data represent the average ± SEM among n=3 independent experiments. c, Western blot analysis on P7 cytoplasmic lysates from control P7 spinal cord. The experiment was performed as in Fig.1e and was repeated independently 2 times with similar results. d-e, Polysomal profiling and co-sedimentation profiles of lysates obtained from control brain before (d) and after (e) RNAse I treatment. RPL26 and RPS6 were used as sedimentation controls for the large and small ribosomal subunits, respectively. The sedimentation distribution of proteins along the sucrose gradient is expressed as percentage in each fraction considering as 100% the sum of all fractions. Data in (d) report the average ± SEM among n=3 biologically independent samples. f, Polysomal profile and co-sedimentation analysis of SMN and ribosome markers RPL26 and RPS6 in Hek-293 using a lysis buffer with high 10 mM Mg2+. The distribution of SMN along the profile was fitted with three Gaussian curves to identify different populations. g, Polysomal profile and co-sedimentation analysis of SMN and ribosome markers in Hek-293 lysed in the absence of Mg2+ to release the large and small subunits. RPL26 and RPS6 were used as sedimentation controls for the large and small ribosomal subunits, respectively. The sedimentation distribution of proteins along the sucrose gradient is expressed as in (d). The distributions of RPS6, RPL26 and SMN along the profile were fitted with one or two Gaussian curves. Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 1.
Extended Data Fig. 2. SMN interacts with the translation machinery in a concentration dependent manner across different tissues and is associated to actively translating ribosomes positively regulating translation.
a, Co-sedimentation of SMN proteins with RiboNucleoParticles (snRNP/RNPs) and ribosomal subunits in Hek-293 lysates. Each lane of the western blot corresponds to a fraction along the profile. Gemin 5 was used as marker of the Gemin granules and HuR of RNA-granules. b-d, Polysomal profiling and co-sedimentation profiles of lysates obtained from P3 control mouse spinal cord (b), heart (c) and kidney (d). The ribosomal proteins RPL26 and RPS6 were used as sedimentation controls for the large and small ribosomal subunits, respectively. The sedimentation distribution of proteins along the sucrose gradient is expressed as percentage in each fraction considering as 100% the sum of all fractions. The percentage shown are average ± SEM from n=3 biologically independent experiments. Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 2.
Extended Data Fig. 3. Ribosome profiling of SMN-primed ribosomes reveals enriched mRNAs organized in functionally well-defined communities.
a, Examples of immunoprecipitation of SMN-ribosomes from ribo-pellet in control mouse brain lysates after RNaseI treatment. RPL26 was used as control. The experiment was repeated independently three times. b, Transcript types enriched in fragments protected by SMN-primed ribosomes that protect predominantly RNAs associated with protein coding genes. c, Venn diagram showing the intersection of SMN-specific protein coding transcripts covered by short (24-26 nucleotides) and long (32-34 nucleotides) reads. d, Percentage of P-sites according to the three reading frames for the first five codons of the CDSs and for the remaining region of the sequences. A clear trinucleotide periodicity in the correct frame is detectable only at the beginning of the coding sequence. Results are shown as the average ± SEM among n=3 biologically independent samples. The statistical significance from two-sided t-test comparing frames 1 and 2 with respect to frame 0 are reported. e, Representative RPF coverage tracks for SMN-specific transcripts in SMN-specific RiboSeq. The average normalized coverage and the standard error among replicates are represented in each profile. The structure of the transcript, showing the boundaries of CDS and UTR regions, is outlined below each profile. f, Over-representation of tissue-specific markers among genes enriched in SMN-primed ribosomes. Tissues where SMN levels were previously measured are displayed. The enrichment score was calculated with enrichR. g, Gene ontology and pathway enrichment analysis of genes enriched in SMN-primed ribosomes. The number of genes is displayed on the right of the bars. (GO_BP:biological process, GO_CC:cellular component, GO_MF: molecular function). h, Comparative annotation enrichment analysis of transcripts belonging to the seven SMN-specific communities. The heatmaps are colored according to the significance of the enrichments. The analysis was performed on Gene Ontology terms: Biological Process (left) and cellular component (right). Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 3.
Extended Data Fig. 4. Transcripts bound by SMN-primed ribosomes display defects in positioning of active ribosomes at early stages of SMA.
a, mTORC1 is activated only at late stages of SMA. Upper panel, total proteins from brains at early and late stages of disease (SMA) and from littermate controls (Ctrl) were probed for phosphorylation responses by immunoblotting of known mTORC1 downstream targets (4E-BP) and PERK (eIF2a). In the right panels the quantification for 4E-BP is shown as the average signal ± SEM among n=3 biologically independent samples (normalized to total protein stain, not shown). The statistical significance from one-way ANOVA with Tukey’s multiple comparisons test is reported. b, Fraction of ribosomes in polysomes obtained from polysomal profiling of control and SMA mouse brain at early stage of disease (P5). The data represent the average ± SEM (n = 3 mice). Significant decreases were tested by two-sided t-test. c, Pairwise correlations of RPKM per protein coding transcripts (n=49825 mRNAs) between the two replicas of control and SMA Active-RiboSeq and control RiboSeq (upper panels) and between the three replicas of control SMN-specific RiboSeq (lower panels). Pearson’s correlation coefficients and statistical significance from two-sided Williams’ test are shown (*** pvalue < 1e-16). d, Over-representation of tissue-specific markers among genes with significantly increased (yellow) or decreased (red) active ribosome occupancies in SMA. Tissues where SMN levels were previously measured are displayed in bold. The enrichment score was calculated with enrichR. Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 4.
Extended Data Fig. 5. Translationally defective transcripts in SMA display specific features.
a, Localization of proteins synthesized by mRNAs enriched in SMN-primed ribosomes. Manually annotated and reviewed proteins from the UniProt database were considered. b, Lengths comparison of regions of SMN-specific transcripts (n=874) compared with unspecific transcripts (n=4010). Statistical significance was determined with one-sided Wilcoxon-Mann-Whitney tests. c, Logo representation of top enriched motifs in the 5`UTR (left panel) and 3’ UTR (right panel) of SMN-specific transcripts, by discriminative analysis of k-mer composition. d, Over-representation of translational enhancer sequences among SMN-specific transcripts (619, in blue). Annotation of translational enhancer was retrieved from (50). two-sided Wilcoxon rank-sum test to calculate the p-value shown. e, Logo-like representation of the most frequent amino-acids in SMN-specific (left) and unspecific RNAs (right) at the beginning of the CDS. f, SMN levels in NSC-34 expressing high and low SMN levels. Comparison between the fraction of ribosomes in polysomes (FRP) in NSC-34 expressing high and low levels of SMN. The results are the average ± SEM. Significant differences were determined using a two-sided t-test. g, Luciferase assays were performed in NSC-34 expressing high or low levels of SMN. Alanine repeats values were set to 1. h, The 5’UTR of Tuba4a was used as control. Results for luciferase assays with or without treatment with rapamycin are reported. In panels (g) and (h) the number of biologically independent experiments is reported. Results are shown as the average ± SEM. Significant changes were assessed using one-sided t-test. i, Fold change of Renilla and Firefly luciferase in cells expressing high vs low levels of SMN. The level of the two transcripts in cells expressing high levels of SMN was set to 1. The number of biologically independent samples is reported and the data represent the average ± SEM. Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 5.
Extended Data Fig. 6. Communities of mRNAs bound by SMN-primed ribosomes show reduced ribosome occupancy.
a, Comparison between total RNA fold changes (SMA vs CTRL) in SMN-specific genes, binned in the 7 SMN-communities. Data were retrieved from Bernabò et al., 2017 (11). Significant shifts in each community were identified with the two-sided one-sample Wilcoxon rank-sum test. The number of genes for each community is reported in Fig. 3f. b, Representative polysomal profiles obtained from CTRL and SMA mouse brains (early-symptomatic). c, Representative active RiboSeq coverage tracks for SMN-specific transcripts selected for validation by polysomal profiling. The average normalized coverage and the standard error among replicates are represented in each profile. The structure of the transcript, showing the boundaries of CDS and UTR regions, is outlined below each profile. Statistical source data are provided in Source data Extended data Fig. 6.
Extended Data Fig. 7. The acetylcholinesterase transcript shows ribosome drop-off and defective production of protein at the NMJ in SMA.
a, Translation efficiency of Acetylcholinesterase from brain at early and late stage and in spinal cord at late stage, and after SMN-restoring Antisense Oligonucleotides treatment (11). The average ± SEM from n=3 biologically replicates in n=2 technical replicates is shown. For control brain early stage the average ± SEM from n=3 biologically replicates in n=3 technical replicate is shown. Translation efficiencies were obtained as the ratio of changes in polysomal and total RNA normalized to Tuba4a. Significant differences were determined using a two-sided t- test. b, Luciferase assays were performed in NSC34 expressing high or low levels of SMN in n=9 biologically independent replicates. c, Luciferase assays for testing the contribution of the first five codons of AChE with respect to four alanines. In (b) and (c) the number of biologically independent experiments is reported. Results are the average ± SEM. Significant changes were assessed using one-sided t-test. d, Brain, spinal cord and muscle lysates of early-symptomatic SMA mice were compared to controls using western blot. Experiments were repeated independently 3 times with similar results. e, Representative images for control (left) and early-symptomatic SMA mouse (right) neuromuscular endplates. Acetylcholine receptors (AChR) were labelled using alpha-bungarotoxin (BTX) and acetylcholinesterase (AChE) using fasciculin-2 (FCC). Top panels show FCC / BTX overlap, middle and bottom panels individual channels in greyscale. Endplates n=65 from control and n=73 endplates from SMA mice from 6 muscles and 3 mice/genotype were imaged and analysed. Scale bar: 10 μm. f, FCC and BTX average intensity were determined for 2 FDB muscles in 3 control and 3 SMA mice. The number of endplates per mouse is reported. Results are the average fluorescence intensity ± SEM. Significant differences were determined using a two-sided t-test. Statistical source data and unprocessed blots are provided in Source data Extended data Fig. 7.
Extended Data Fig. 8. Schematic representation of the role of SMN-primed ribosomes in translation based on SMN-specific ribosome profiling.
(1) After recruitment of ribosomes at the AUG, SMN-primed ribosomes are translating the first codons and stop at the fifth codon in a rotated state, according to the results shown in Figure 3 and to Laureau et al., 2014 (37). The ribosome pause at the fifth codon ensures productive translation as proposed by Han et al., 2014 (54) and in accordance with our results (Fig. 3 and 4). The blue star represents SMN. (2) When SMN expression is lost in SMA, SMN-specific transcripts are bound by SMN-free ribosomes. These ribosomes cannot properly pause within the first 5 codons, causing ribosomes to drop-off (as observed by Han et al., 2014 (54), leading to disruption of normal protein synthesis.
Supplementary Material
Acknowledgements
We thank Prof Norbert Polacek, University of Bern, for the careful reading of the manuscript and useful suggestions. We thank Prof David Beeson, University of Oxford, for kindly providing us with the fasciculin-II (FCC)-Alexa Fluor 488 conjugate; the IMPACT imaging facility at the University of Edinburgh for assistance with imaging and Haiyan Zhou and Prof. Francesco Muntoni, University College London, for kindly providing us with the ASO tissues. We thank the Core Facilities Next Generation Sequencing Facility (NGS) and High Throughput Screening (HTS) at Department CIBIO University of Trento for technical support.
Funding
This work was supported by Provincia Autonoma di Trento, Italy (AxonomiX research project), the UK SMA Research Consortium, SMA Europe, the Wellcome Trust (106098/Z/14/Z), Stichting Spieren voor Spieren (the Netherlands), Slovenian Research Agency program grant P1-0391, COST action CA15126, AFM-Telethon (reference number 22129), Caritro Foundation (young Post-doc funding grant) and by Telethon (reference number GGP19115). In addition, we acknowledge financial support from IMMAGINA Biotechnology (Italy).
Footnotes
Author contributions
F.L. and T.T. performed all the high-throughput computational analyses. P.B. performed the SMN-specific Ribo-Seq and sub-cellular fractionation experiments. P.B., E.P. and M.C. performed all other Ribo-Seq library preparation. E.G. and T.G. generated and maintained all experimental animals, performed mouse tissue collection and related western blotting and fluorescence microscopy of NMJs. F.M., A.I., J.O. and A.R. performed the cloning for dual luciferase experiments and the dual luciferase analysis. F.M. and A.R. performed all qRT-PCR analysis. D.D., N.O. and G.A. performed the SPR analysis. M.M, M.D.S. and G.V. performed the IVTT experiments and data analysis. G.V. performed all polysomal purifications, RNA and protein extractions, western blotting and data analysis; F.L., T.T., E.G. and G.V. prepared the figures; G.V. conceived experiments and directed the research; A.Q., T.G. and G.V. obtained the funding. F.L., T.T., E.G., T.G and G.V wrote the manuscript. All authors contributed during preparation, revision and writing of the manuscript.
Conflict of Interest
M.C. is CEO of IMMAGINA Biotechnology; G.V. is scientific advisor to IMMAGINA Biotechnology; T.H.G. has served on SMA advisory boards for Roche. The remaining authors declare no competing financial interests.
Data availability
Ribosome profiling data generated by the current study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE154106. Classical and active ribosome profiling data of healthy mouse brains that were reanalyzed in the current study were retrieved from GEO: GSE102318 (RiboSeq), GSE102354 (Active-RiboSeq). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.
References
- 1.Buttgereit F, Brand MD, Buttgereit MD, Brand F, Buttgereit F, Brand MD. A hierarchy of ATP-consuming processes in mammalian cells. Biochem J. 1995;312(Pt1):163–167. doi: 10.1042/bj3120163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Russell JB, Cook GM. Energetics of bacterial growth:balance of anabolic and catabolic reactions. Microbiol. 1995;59:48–62. doi: 10.1128/mr.59.1.48-62.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vogel C, de Abreu RS, Ko D, Le SYY, Shapiro BA, Burns SC, Sandhu D, Boutz DR, Marcotte EM, Penalva LO, De Sousa Abreu R, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. TL - 6. Mol Syst Biol. 2010;6VN-re:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schwanhüusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M, Schwanhäusser B, Busse D, Li N, et al. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 5.Tebaldi T, Re A, Viero G, Pegoretti I, Passerini A, Blanzieri E, Quattrone A. Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics. 2012;13:220. doi: 10.1186/1471-2164-13-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhat M, Robichaud N, Hulea L, Sonenberg N, Pelletier J, Topisirovic I. Targeting the translation machinery in cancer. Nat Rev Drug Discov. 2015 doi: 10.1038/nrd4505. [DOI] [PubMed] [Google Scholar]
- 7.Topisirovic I, Sonenberg N. Translation and cancer. Biochim Biophys Acta-GeneRegul Mech. 2015 doi: 10.1016/j.bbagrm.2015.05.004. [DOI] [PubMed] [Google Scholar]
- 8.Darnell JC, VanDriesche SJ, Zhang C, Hung KYS, Mele A, Fraser CE, Stone EF, Chen C, Fak JJ, Chi SW, Licatalosi DD, Richter JD, Darnell RB. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khalil B, Morderer D, Price PL, Liu F, Rossoll W. mRNP assembly,axonal transport,and local translation in neurodegenerative diseases. Brain Res. 2018 doi: 10.1016/j.brainres.2018.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Costa CJ, Willis DE. To the end of the line:Axonal mRNA transport and local translationin health and neurodegenerative disease. Dev. 2018 doi: 10.1002/dneu.22555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bernabò P, Tebaldi T, Groen EJNN, Lane FM, Perenthaler E, Mattedi F, Newbery HJ, Zhou H, Zuccotti P, Potrich V, Shorrock HK, Muntoni F, et al. In Vivo Translatome Profiling in Spinal Muscular Atrophy Reveals a Role for SMN Proteinin Ribosome Biology. Cell Rep. 2017;21:953. doi: 10.1016/j.celrep.2017.10.010. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Simsek D, Barna M. An emergingrole for theribosome as a nexus for post-translational modifications. Curr Opin Cell Biol. 2017 doi: 10.1016/j.ceb.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mauro VP, Edelman GM. The ribosome filter hypothesis. Proc Natl Acad Sci. 2002 doi: 10.1073/pnas.192442499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xue S, Barna M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat Rev Mol Cell Biol. 2012;13:355–369. doi: 10.1038/nrm3359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi Z, Fujii K, Kovary KM, Genuth NR, Röst HL, Teruel MN, Barna M. Heterogeneous Ribosomes Preferentially Translate Distinct Subpools of mRNAs Genome-wide. Mol Cell. 2017 doi: 10.1016/j.molcel.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Slavov N, Semrau S, Airoldi E, Budnik B, van Oudenaarden A. Differential Stoichiometry among Core Ribosomal Proteins. Cell Rep. 2015 doi: 10.1016/j.celrep.2015.09.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kurylo CM, Parks MM, Juette MF, Zinshteyn B, Altman RB, Thibado JK, Vincent CT, Blanchard SC. Endogenous rRNA Sequence Variation Can Regulate Stress Response Gene Expression and Phenotype. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.08.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Parks MM, Kurylo CM, Dass RA, Bojmar L, Lyden D, Vincent CT, Blanchard SC. Variant ribosomal RNA alleles are conserved and exhibit tissue-specific expression. Sci Adv. 2018 doi: 10.1126/sciadv.aao0665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fujii K, Susanto TT, Saurabh S, Barna M. Decoding the Function of Expansion Segments in Ribosomes. Mol Cell. 2018 doi: 10.1016/j.molcel.2018.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kondrashov N, Pusic A, Stumpf CR, Shimizu K, Hsieh AC, Xue S, Ishijima J, Shiroishi T, Barna M. Ribosome-mediated specificity in Hox mRNA translation and vertebrate tissue patterning. Cell. 2011;145:383–397. doi: 10.1016/j.cell.2011.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lefebvre S, Burglen L, Reboullet S, Clermont O, Burlet P, Viollet L, Benichou B, Cruaud C, Millasseau P, Zeviani M et al. Identification and characterization of a spinal muscular atrophy-determining gene[seecomments] Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 22.Groen EJN, Talbot K, Gillingwater TH. Advances in therapy for spinal muscularatrophy:Promises and challenges. Nat Rev Neurol. 2018 doi: 10.1038/nrneurol.2018.4. [DOI] [PubMed] [Google Scholar]
- 23.Zhang Z, Lotti F, Dittmar K, Younis I, Wan L, Kasim M, Dreyfuss G. SMN Deficiency Causes Tissue-Specific Perturbations in the Repertoire of sn RNAs and Widesprea Defects in Splicing. Cell. 2008 doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lotti F, Imlach WL, Saieva L, Beck ES, Hao LT, Li DK, Jiao W, Mentis GZ, Beattie CE, McCabe BD, Pellizzoni L. An SMN-dependent U12 splicing event essential for motor circuit function. Cell. 2012 doi: 10.1016/j.cell.2012.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bäumer D, Lee S, Nicholson G, Davies JL, Parkinson NJ, Murray LM, Gillingwater TH, Ansorge O, Davies KE, Talbot K. Alternative splicing events are a late feature of pathology in a mouse model of spinal muscula atrophy. PLoS Genet. 2009 doi: 10.1371/journal.pgen.1000773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Simon CM, Dai Y, VanAlstyne M, Koutsioumpa C, Pagiazitis JG, Chalif JI, Wang X, Rabinowitz JE, Henderson CE, Pellizzoni L, Mentis GZ. Converging Mechanisms of p53 Activation Drive MotorNeuron Degeneration in Spinal Muscular Atrophy. Cell Rep. 2017 doi: 10.1016/j.celrep.2017.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Price PL, Morderer D, Rossoll W. Advances in Neuro biology. 2018 doi: 10.1007/978-3-319-89689-2_6. [DOI] [PubMed] [Google Scholar]
- 28.Sanchez G, Dury AY, Murray LM, Biondi O, Tadesse H, El fatimy R, Kothary R, Charbonnier F, Khandjian EW, Côté J. A novel function for the survival motoneuron proteinas a translational regulator. Hum Mol Genet. 2013;22:668–684. doi: 10.1093/hmg/dds474. [DOI] [PubMed] [Google Scholar]
- 29.Béchade C. Sub cellular distribution of survival motorneuron(SMN)protein:Possible involvement in nucleocytoplasmic and dendritic transport. Eur J Neurosci. 1999;11:293–304. doi: 10.1046/j.1460-9568.1999.00428.x. [DOI] [PubMed] [Google Scholar]
- 30.Fallini C, Donlin-Asp PG, Rouanet JP, Bassell GJ, Rossoll W. Deficiency of the Survival of MotorNeuron Protein Impairs mRNA Localization and Local Translation in the Growth Cone of Motor Neurons. J Neurosci. 2016;36:3811–20. doi: 10.1523/JNEUROSCI.2396-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Francisco-Velilla R, Fernandez-Chamorro J, Ramajo J, Martinez-Salas E. The RNA-binding protein Gemin5 binds directly to the ribosome and regulates global translation. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Burghes AHM, Beattie CE. Spinal muscular atrophy:Why do low levels of survival motorneuron protein make motorneurons sick? Nat Rev Neurosci. 2009 doi: 10.1038/nrn2670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007 doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Groen EJN, Perenthaler E, Courtney NL, Jordan CY, Shorrock HK, Van Der Hoorn D, Huang YT, Murray LM, Viero G, Gillingwater TH. Temporal and tissue-specific variability of SMN protein levels in mouse models of spinalmuscular atrophy. Hum Mol Genet. 2018 doi: 10.1093/hmg/ddy195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tapia O, Narcís JO, Riancho J, Tarabal O, Piedrafita L, Calderó J, Berciano MT, Lafarga M. Cellular bases of the RNA metabolism dysfunction in motorneurons of a murine model of spinal muscular atrophy:Role of Cajal bodies and the nucleolus. Neurobiol. 2017 doi: 10.1016/j.nbd.2017.08.004. [DOI] [PubMed] [Google Scholar]
- 36.Clamer M, Tebaldi T, Lauria F, Bernabò P, Gómez-Biagi RF, Marchioretto M, Kandala DT, Minati L, Perenthaler E, Gubert D, Pasquardini L, et al. Active ribosome profiling with RiboLace. Cell Rep. 2018 doi: 10.1016/j.celrep.2018.09.084. [DOI] [PubMed] [Google Scholar]
- 37.Lareau LF, Hite DH, Hogan GJ, Brown PO. Distinct stages of the translation elongationcycle revealed by sequencing ribosome-protected mRNA fragments. Elife. 2014;2014 doi: 10.7554/eLife.01257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hensel N, Claus P. The Actin Cytoskeleton in SMA and ALS:How Does It Contribute to Motoneuron Degeneration? Neuroscientist. 2018 doi: 10.1177/1073858417705059. [DOI] [PubMed] [Google Scholar]
- 39.Tisdale S, Lotti F, Saieva L, VanMeerbeke JP, Crawford TO, Sumner CJ, Mentis GZ, Pellizzoni L. SMN is essential for the biogenesis of U7 Small nuclear ribonucleoprotein and 3’-end formation of Histone mRNAs. Cell Rep. 2013 doi: 10.1016/j.celrep.2013.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Singh RN, Howell MD, Ottesen EW, Singh NN. Diverse role of survival motorneuron protein. Biochim Biophys Acta-Gene Regul Mech. 2017 doi: 10.1016/j.bbagrm.2016.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nölle A, Zeug A, Vanbergeijk J, Tönges L, Gerhard R, Brinkmann H, Alrayes S, Hensel N, Schill Y, Apkhazava D, Jablonka S, et al. The spinal muscular atrophy disease protein SMN is linked to the Rho-kinase pathway via profilin. Hum Mol Genet. 2011 doi: 10.1093/hmg/ddr425. [DOI] [PubMed] [Google Scholar]
- 42.Ramser J, Ahearn ME, Lenski C, Yariz KO, Hellebrand H, vonRhein M, Clark RD, Schmutzler RK, Lichtner P, Hoffman EP, Meindl A, Baumbach-Reardon L. Rare Missense and Synonymous Variants in UBE1 Are Associated with X-Linked Infantile Spinal Muscular Atrophy. Am J Hum Genet. 2008 doi: 10.1016/j.ajhg.2007.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aghamaleky Sarvestany A, Hunter G, Tavendale A, Lamont DJ, Llavero Hurtado M, Graham LC, Wishart TM, Gillingwater TH. Label-free quantitative proteomic profiling identifies disruption of ubiquitin homeostasis as a key driver of Schwann Cell defects in spinalmuscular atrophy. J Proteome Res. 2014 doi: 10.1021/pr500492j. [DOI] [PubMed] [Google Scholar]
- 44.Fuller HR, Gillingwater TH, Wishart TM. Commonality amid diversity:Multi-study proteomic identification of conserved disease mechanismsin spinal muscular atrophy. Neuromuscul disord. 2016 doi: 10.1016/j.nmd.2016.06.004. [DOI] [PubMed] [Google Scholar]
- 45.Murray LM, Beauvais A, Gibeault S, Courtney NL, Kothary R. Transcriptional profiling of differentially vulnerable motorneurons at pre-symptomatic stage inthe Smn(2b/-) mousemodel of spinal muscular atrophy. Acta Neuropathol Commun. 2015 doi: 10.1186/s40478-015-0231-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saal L, Briese M, Kneitz S, Glinka M, Sendtner M. Subcellular transcriptome alterations in a Cellculture model of spinal muscular atrophy point to widespread defects in axonal growth and presynaptic differentiation. RNA. 2014 doi: 10.1261/rna.047373.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossoll W, Jablonka S, Andreassi C, Kröning AK, Karle K, Monani UR, Sendtner Smn M. the spinal muscular atrophy-determining gene product,modulates axon growth and localization ofβ-actin mRNA in growth cones of motoneurons. J Cell Biol. 2003 doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bertrandy S, Burlet P, Clermont O, Huber C, Fondrat C, Thierry-Mieg D, Munnich A, Lefebvre S. The RNA-binding properties of SMN:Deletion analysis of the zebrafish orthologue defines domainsconserved in evolution. Hum Mol Genet. 1999 doi: 10.1093/hmg/8.5.775. [DOI] [PubMed] [Google Scholar]
- 49.Ottesen EW, Singh NN, Luo D, Singh RN. High-affinity RNA targets of the Survival MotorNeuron protein reveal diverse preferences for sequence and structural motifs. Nucleic Acids Res. 2018 doi: 10.1093/nar/gky770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Weingarten-Gabbay S, Elias-Kirma S, Nir R, Gritsenko AA, Stern-Ginossar N, Yakhini Z, Weinberger A, Segal E. Comparative genetics: Systematic discovery of cap-independent translation sequences in human andviral genomes. Science(80-. ) 2016 doi: 10.1126/science.aad4939. [DOI] [PubMed] [Google Scholar]
- 51.Zhang HL, Pan F, Hong D, Shenoy SM, Singer RH, Bassell GJ. Active transport of the survival motorneuron protein and the roleof exon-7 in cytoplasmic localization. J Neurosci. 2003 doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jablonka S. Co-regulation of survival of motorneuron(SMN)protein and its interactor SIP1 during development and in spinal muscular atrophy. Hum Mol Genet. 2001 doi: 10.1093/hmg/10.5.497. [DOI] [PubMed] [Google Scholar]
- 53.Fuller HR, Morris GE. SMN complexes of nucleus and cytoplasm:A proteomic study for SMA therapy. Transl. 2010;1:261–267. [Google Scholar]
- 54.Han Y, Gao X, Liu B, Wan J, Zhang X, Qian SB. Ribosome profiling reveals sequence-independent post-initiation pausing as a signature of translation. Cell Res. 2014;24:842–851. doi: 10.1038/cr.2014.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Matsuo Y, Ikeuchi K, Saeki Y, Iwasaki S, Schmidt C, Udagawa T, Sato F, Tsuchiya H, Becker T, Tanaka K. Ubiquitination of stalled ribosome triggers ribosome-associated quality control. Nat. 2017;8:159. doi: 10.1038/s41467-017-00188-1. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wu CCC, Zinshteyn B, Wehner K, Green R. High-Resolution Ribosome Profiling Defines Discrete Ribosome Elongation States and Translational Regulation during Cellular Stress. Mol Cell. 2019 doi: 10.1016/j.molcel.2018.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Murray LM, Comley LH, Thomson D, Parkinson N, Talbot K, Gillingwater TH. Selective vulnerability of motorneurons and dissociation of pre-and post-synaptic pathology at the neuromuscular junction in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddm367. [DOI] [PubMed] [Google Scholar]
- 58.Kariya S, Park GH, Maeno-Hikichi Y, Leykekhman O, Lutz C, Arkovitz MS, Landmesser LT, Monani UR. Reduced SMN protein impairs maturation of the neuromuscular junctions in mouse models of spinal muscular atrophy. Hum Mol Genet. 2008 doi: 10.1093/hmg/ddn156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Martínez-Hernández R, Bernal S, Also-Rallo E, Alías L, Barcelõ M, Hereu M, Esquerda JE, Tizzano EF. Synaptic defects in type i spinal muscular atrophy in human development. J Pathol. 2013 doi: 10.1002/path.4080. [DOI] [PubMed] [Google Scholar]
- 60.Niebroj‐Dobosz I, Hausmanowa‐Petrusewicz I. Serum cholinesterase activity in infantile and juvenile spinal muscular atrophy. Acta Neurol Scand. 1989 doi: 10.1111/j.1600-0404.1989.tb03864.x. [DOI] [PubMed] [Google Scholar]
- 61.Hutchinson DO, Walls TJ, Nakano S, Camp S, Taylor P, Harper CM, Groover RV, Peterson HA, Jamieson DG, Engel AG. Congential endplate acetylocholinesterase deficiency. Brain. 1993 doi: 10.1093/brain/116.3.633. [DOI] [PubMed] [Google Scholar]
- 62.Hsieh-Li HM, Chang JG, Jong YJ, Wu MH, Wang NM, Tsai CH, Li H. A mouse model for spinal muscular atrophy. Nat. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 63.Riessland M, Ackermann B, Förster AF, Jakubik M, Hauke J, Garbes L, Fritzsche I, Mende Y, Blumcke I, Hahnen E, Wirth B. SAHA ameliorates the SMA phenotype in two mousemodels for spinal muscular atrophy. Hum Mol Genet. 2010 doi: 10.1093/hmg/ddq023. [DOI] [PubMed] [Google Scholar]
- 64.Huang YT, vanderHoorn D, Ledahawsky LM, Motyl AAL, Jordan CY, Gillingwater TH, Groen EJN. Robust comparison of prote in levels acrosst issues and throughout development using standardized quantitative western blotting. J Vis Exp. 2019 doi: 10.3791/59438. [DOI] [PubMed] [Google Scholar]
- 65.Viero G, Lunelli L, Passerini A, Bianchini P, Gilbert RJ, Bernabò P, Tebaldi T, Diaspro A, Pederzolli C, Quattrone A, Bernab?? P, et al. Three distinc tribosome assemblies modulated by translation are the building blocks of polysomes. J Cell Biol. 2015;208:581–596. doi: 10.1083/jcb.201406040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chen E, Sharma MR, Shi X, Agrawal RK, Joseph S. Fragile X mental retardation protein regulates translation by binding directly to the ribosome. Mol Cell. 2014 doi: 10.1016/j.molcel.2014.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wessel D, Flügge UI. A method for the quantitative recovery of protein indilute solution in the presence of detergents and lipids. Anal biochem. 1984 doi: 10.1016/0003-2697(84)90782-6. [DOI] [PubMed] [Google Scholar]
- 68.Ingolia NT, Brar GA, Rouskin S, McGeachy AM, Weissman JS. The ribosome profiling strategy for monitoring translation in vivo by deep sequencing of ribosome-protected mRNA fragments. Nat. 2012;7:1534–1550. doi: 10.1038/nprot.2012.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sleigh JN, Gillingwater TH, Talbot K. The contribution of mousemodels to understanding the pathogenesis of spinal muscular atrophy. Dis Model Mech. 2011 doi: 10.1242/dmm.007245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jones RA, Harrison C, Eaton SL, LlaveroHurtado M, Graham LC, Alkhammash L, Oladiran OA, Gale A, Lamont DJ, Simpson H, Simmen MW, Soeller C, Wishart TM, Gillingwater TH. Cellular and Molecular Anatomy of the Human Neuromuscular Junction. Cell Rep. 2017 doi: 10.1016/j.celrep.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lauria F, Tebaldi T, Bernabò P, Groen EJN, Gillingwater TH, Viero G. riboWaltz:Optimization of ribosome P-site positioning in ribosome profiling data. PLoS Comput Biol. 2018 doi: 10.1371/journal.pcbi.1006169. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Ribosome profiling data generated by the current study have been deposited in the Gene Expression Omnibus (GEO) under the accession code GSE154106. Classical and active ribosome profiling data of healthy mouse brains that were reanalyzed in the current study were retrieved from GEO: GSE102318 (RiboSeq), GSE102354 (Active-RiboSeq). Source data are provided with this paper. All other data supporting the findings of this study are available from the corresponding author on reasonable request.