Abstract
Porphyromonas gingivalis, an asaccharolytic member of the Bacteroidetes, is a keystone pathogen in human periodontitis that may also contribute to the development of other chronic inflammatory diseases. P. gingivalis utilizes protease-generated peptides derived from extracellular proteins for growth, but how those peptides enter the cell is not clear. Here we identify RagAB as the outer membrane importer for peptides. X-ray crystal structures show that the transporter forms a dimeric RagA2B2complex, with the RagB substrate-binding surface-anchored lipoprotein forming a closed lid on the RagA TonB-dependent transporter. Cryo-electron microscopy structures reveal the opening of the RagB lid and thus provide direct evidence for a "pedal bin" nutrient uptake mechanism. Together with mutagenesis, peptide bindingstudies and RagAB peptidomics, our work identifies RagAB as a dynamic,selective OM oligopeptide acquisition machine that is essential for the efficient utilisation of proteinaceous nutrients by P. gingivalis.
Introduction
The Gram-negative Bacteroidetes are abundant members of thehuman microbiota, especially in the gut. Outside the gut, Bacteroidetes often cause disease, with the best-known examples being the oral Bacteroidetes Porphyromonas gingivalis and Tannerella forsythia that are part of the "red complex" involved in periodontitis1, the most prevalent infection-driven chronic inflammation in the Western world2. Accumulating evidence suggests a link between periodontitis and other chronic inflammatory diseases, including rheumatoid arthritis, Alzheimer's disease, chronic obstructive pulmonary disease and cardiovascular disease3–7.Given this link, and its fastidious growth requirements, it is important to understand how P. gingivalis, thrives and causes dysbiosis of the oral microbiota, leading to inflammation and periodontal tissue destruction.
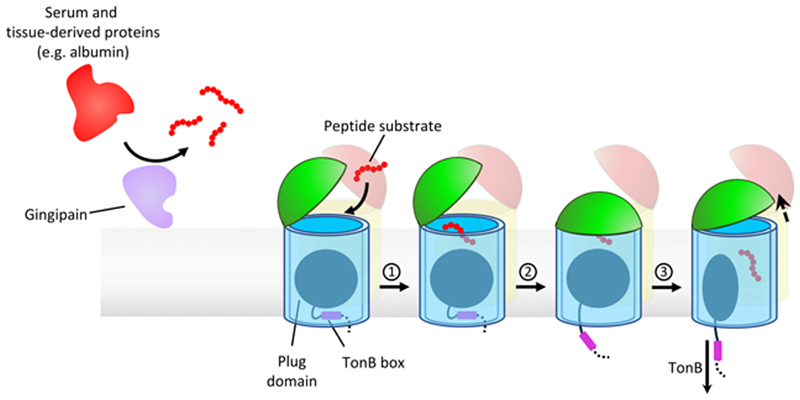
Unlike many human gut Bacteroidetes that specialise in degrading glycans, P. gingivalis is asaccharolytic and exclusively utilises peptides for growth8. Those peptides are generated by multiple proteases9,10, the best-known of which are the gingipains. These large and abundant surface-anchored cysteine endoproteases possess cumulative trypsin-like activity, and are essential for P. gingivalis virulence and growth on proteins as the sole source of carbon11. Crucially, it is not clear how gingipain-generated peptides are taken up by P. gingivalis, although a role for the RagAB outer membrane (OM) protein complex has been proposed12. RagAB consists of a TonB dependent transporter (TBDT) RagA (PG_0185) and a substrate-binding surface lipoprotein RagB (PG_0186). However, a recent crystal structure of P. gingivalis RagB was claimed to contain bound monosaccharides13, and a role in polysaccharide utilisation was proposed based on sequence similarity with SusCD systems from gut Bacteroidetes13–15. Recent structures suggest that SusC and SusD proteins from Bacteroides thetaiotaomicron form stable dimeric complexes (SusC2D2), with SusD capping the extracellular face of the SusC transporter. Molecular dynamics (MD) simulations and electrophysiology studies led to the proposal that nutrient uptake likely occurs via a hinge-like opening of the SusD lid, a so-called pedal bin mechanism16. However, the molecular details of nutrient acquisition by SusCD-like complexes remain unclear, as do any differences between putative OM peptide and glycan transporters.
Here we report integrated structural and functional studies of RagAB purified from P. gingivalis W83 via X-ray crystallography and cryo-electron microscopy (cryo-EM). The crystal structure shows a dimeric RagA2B 2complex in which RagB caps the RagA transporter, forming a large internal chamber that is occupied by co-purified bound peptides. Remarkably, and in sharp contrast to the crystal structure of RagAB (and indeed of SusCDs), cryo-EM reveals large conformational changes, with three distinct states found in a single dataset of the detergent-solubilized RagA2B2 transporter. Together with RagAB peptidomics, structure-based site-directed mutagenesis, and peptide binding studies, we show that RagAB is a dynamic OM oligopeptide acquisition machine, with considerable substrate selectivity that is essential for the efficient utilisation of protein substrates by P. gingivalis.
Results
Purification and X-ray crystal structure determination of RagAB from P. gingivalis
Since RagA does not express in the E. coli OM, we purified RagAB directly from P. gingivalis W83 KRAB (Δkgp/ΔrgpA/ΔrgpB). This strain lacks gingipains, reducing proteolysis of many OM proteins. RagAB is one of the most abundant OM proteins inP. gingivalis, and with the exception of the 230 kDa Hemagglutinin A (HagA), no co-purifying proteins are present (Fig. 1a). This indicates that the RagAB complex represents the complete transporter. Diffracting crystals were obtained by vapour diffusion, and the structure was solved by molecularreplacement using data to 3.4 Å resolution (Methods and Supplementary Table 1).
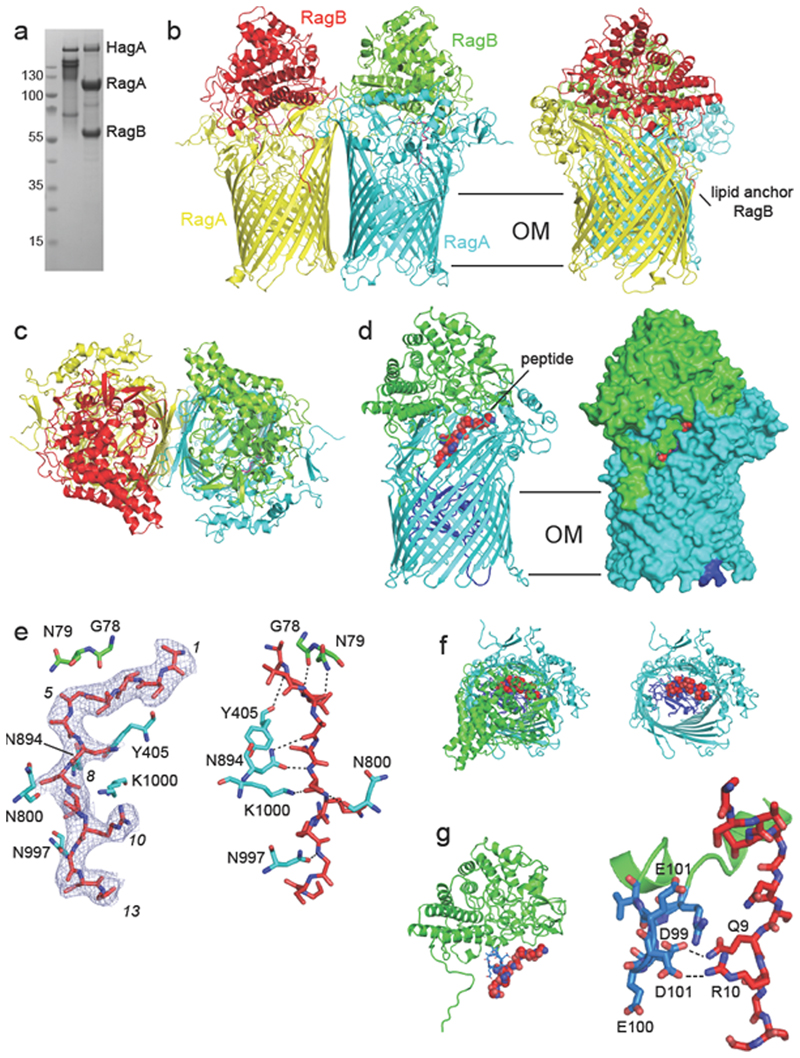
Figure 1. Crystal structure of the RagA2 B 2 transporter suggests bound peptides.
a, SDS-PAGE gel showing purified RagAB from W83 KRAB before (left lane) and after boiling in SDS-PAGE sample buffer. The gel is representative of three independent purifications. b,c, Views of RagA2B 2from the plane of the OM (b) and from the extracellular space (c). d, Side views of RagAB (right panel; surface representation) showing the bound peptide as a red space filling model. The plug domain of the RagA TBDT (cyan) is dark blue. e, 2Fo-Fc density (blue mesh; 1.0 σ, carve = 1.8) of the peptide after final refinement (left panel). The right panel shows potential hydrogen bonds (distance < 3.6 Å) between RagAB and the peptide backbone. Arbitrary peptide residue numbering is indicated in italics. f, Extracellular views of RagAB with (left panel) and without the RagB lid (green). g, Side view (top panel) and close-up of the RagB acidic loop (blue) and the bound peptide. Potential hydrogen bonds are indicated by dashed lines. Structural figures were made with Pymol69.
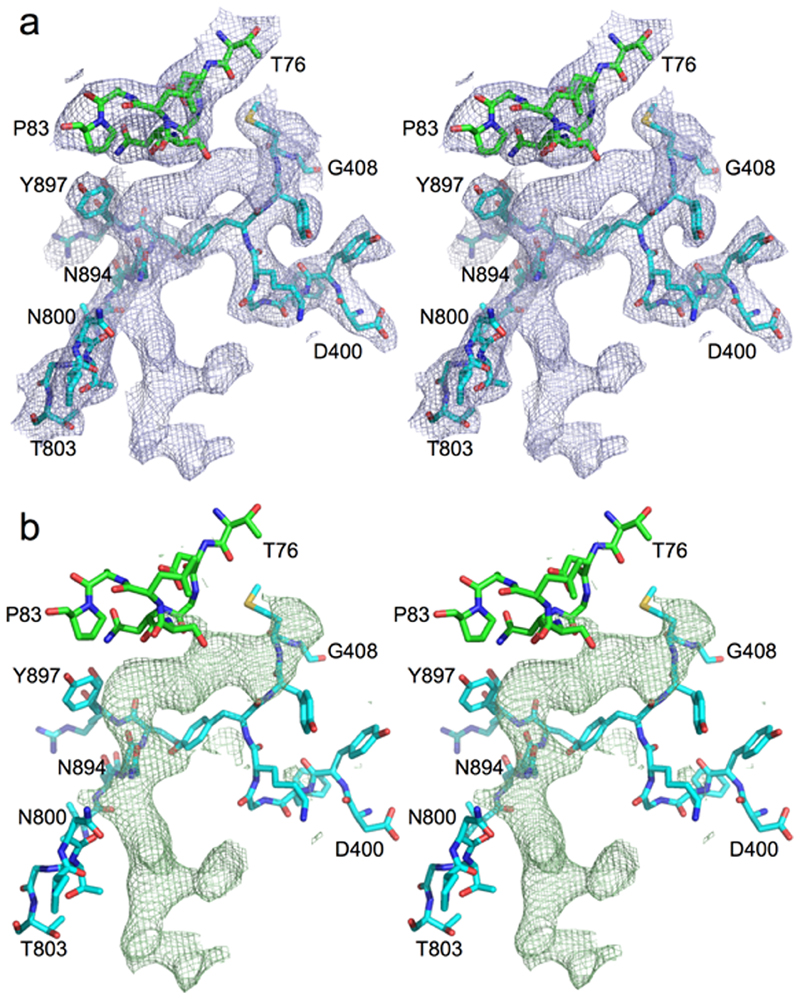
RagAB is dimeric (RagA2B2), with the same subunit arrangement and architecture (Fig. 1) observed in two SusCD complexes from Bacteroides thetaiotaomicron 16. RagB caps RagA, burying an extensive surface area (~3800 Å2) and forming a large, closed internal cavity which contains clearly resolved density for an elongated, ~40 Å long molecule that is bound at the RagAB interface (Extended Data Fig. 1). The electron density strongly suggests that the bound molecule is a peptide of ~13 residues in length (Figs. 1d-g), although most side chains are truncated after the Cβ or Cγ positions. Exceptions are residues 9-10 of the peptide, where well-defined side chain density comes close to residues D99/D101 in RagB.The main chain of the peptide has somewhat higher average B-factors than the neighbouring RagA (~95 A2 and 73 A2 respectively), suggesting that it corresponds to an ensemble of different peptides with similar backbone conformations, hereafter referred to as "the peptide". We modelled the peptide arbitrarily as A1STTG5ANSQR10GSG13 (Fig. 1e), which decreased the Rfree of the model by about 0.25%. D99 and D101 of RagB are part of an acidic loop insertion (Asp99-Glu-Asp-Glu102) into an α-helix in RagB’s ligand binding site that protrudes into the RagAB binding cavity (Fig. 1g). The acidic nature of this loop suggests that P. gingivalis W83 RagAB may prefer to take up basic peptides. Interestingly, this acidic loop is absent in several RagB orthologs, including that from strain ATCC 33277 (Supplementary Figs. 1 and 2). Several RagAB residues likely form hydrogen bonds with the backbone of the peptide (Fig. 1e), e.g. the side chain of N894 in RagA forms hydrogen bonds with residues 6 and 8 of the peptide, and may therefore be important for substrate binding. Collectively, RagAB residues interact with the backbone of 8 out of the 13 visible peptide residues, and most of those interactions are provided by RagA.
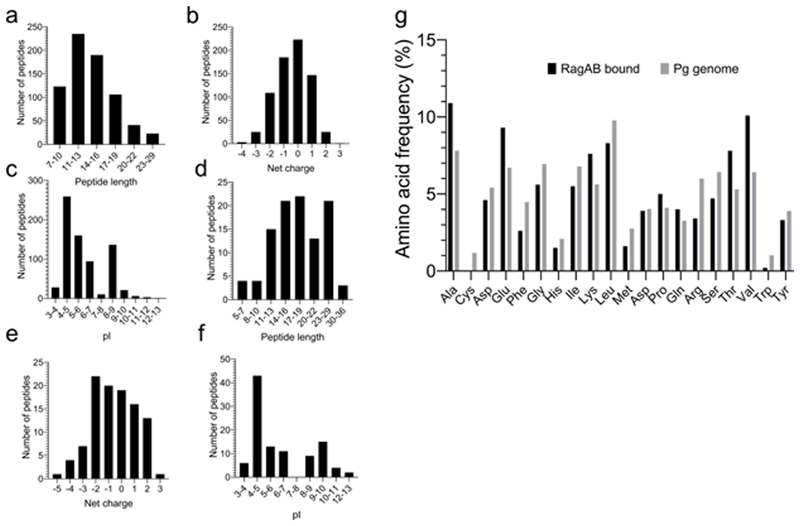
To confirm the presence of co-purified peptides in RagAB we employed a LC-MS/MS peptidomics approach. RagAB from W83 KRAB was associated with several hundred unique peptides (Supplementary Table 2) with none present in the negative control Omp40-41 sample (Methods), confirming our structure-based hypothesis that RagAB contains an ensemble of peptides. The peptides originate mostly from abundant P. gingivalis proteins representing all cellular compartments. As a comparison, we also analysed peptides bound to RagAB purified from the gingipain-expressing W83 wild-type strain. This complex is identical to that of the KRAB strain but, due to gingipain activity, the RagAB-associated peptides in wild-type W83 are different from those in the gingipain-deficient KRAB strain (Supplementary Table 2). Any in-depth analysis of RagAB-bound peptides is difficult because of the non-quantitative nature of MS. Assuming equal abundance of each detected peptide, there are no dramatic differences in length distribution, pI profiles and amino acid frequencies within the RagAB-bound peptides from W83 KRAB and wild-type strains (Extended Data Fig. 2).
Conformational changes in RagAB
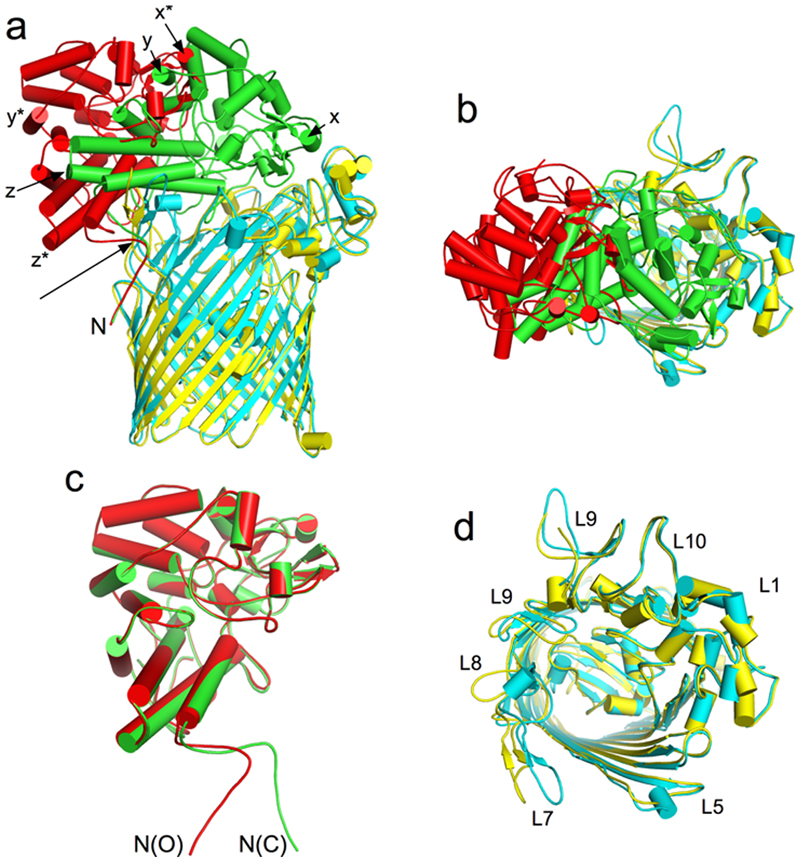
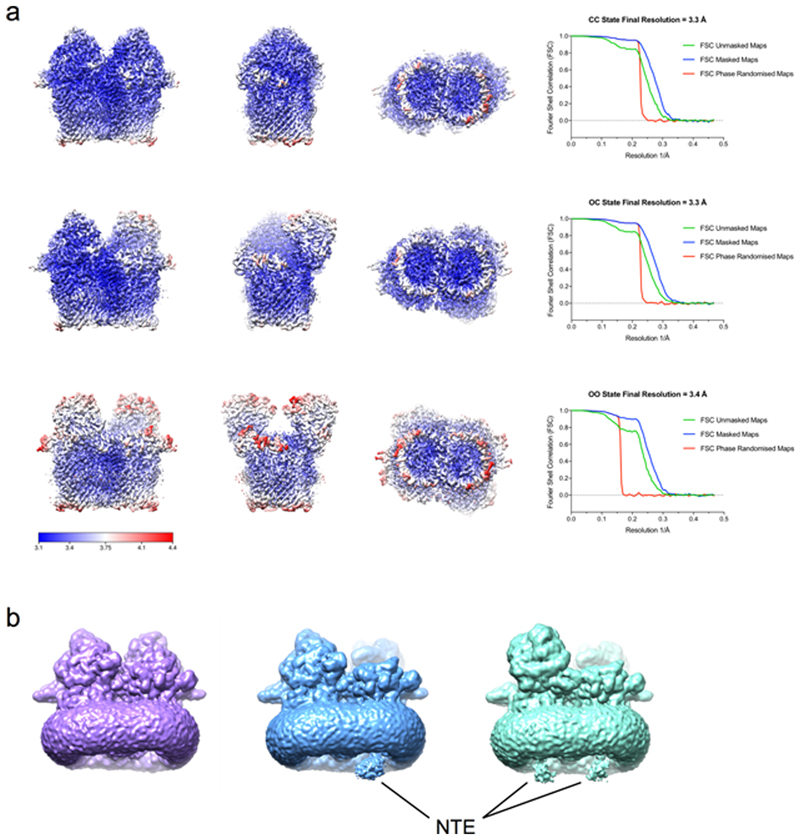
The crystal structure of RagAB is very similar to previously determined SusCD structures. Both TBDT barrels are closed on the extracellular side by their RagB (or SusD) caps, even when no substrate is bound16. This suggests either that the closed state is energetically favourable, or that crystallisation selects for closed states from a wider conformational ensemble. We therefore investigated the structure of detergent-solubilized RagAB in solution using single particle cryo-EM. RagAB is unambiguously dimeric in solution. Strikingly, following initial 2D classification it was immediately apparent that multiple conformations of the RagA2B2 dimer were present, and after further classification steps, three distinct conformations were identified. Following 3D reconstruction and refinement, three structures were obtained at near-atomic resolution, corresponding to the three possible combinations of open and closed dimeric transporters: closed-closed (CC; 3.3 Å), open-closed (OC; 3.3 Å) and open-open (OO; 3.4 Å) (Figs. 2 and 3; Supplementary Table 3).
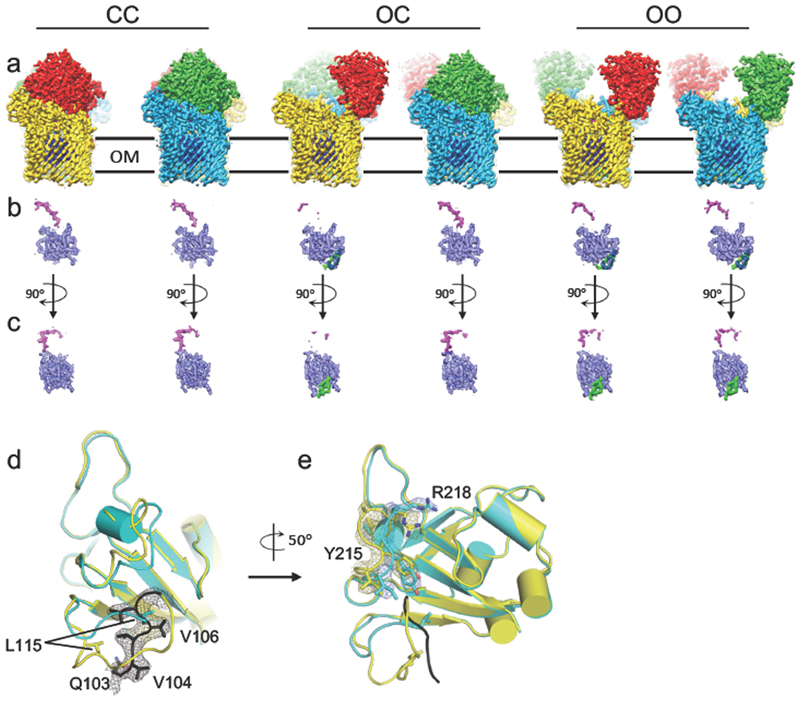
Figure 2. Different conformations of the RagA2B2 transporter revealed by cryo-EM.
a-c, Cartoon views for closed-closed RagA2B2 (CC; a), open-closed (OC; b) and open-open (OO; c). Views are from the plane of the OM (left and middle panels) and from the extracellular space (right panel). Bound peptide is shown as space filling models in magenta. The plug domains o RagA are coloured dark blue.
Figure 3. DPeptide and plug dynamics in RagA2 B 2 revealed via cryo-EM.
a, Density maps viewed from the membrane plane of the closed-closed, open-closed and open-open RagA2B2 complexes coloured as in Figure 2. b, c. Isolated density for the plug domain and the bound peptide ensemble viewed as in (a) and after a 90 degree clockwise rotation (c). Additional density for the plug domains of open RagAB complexes is shown in green. d,e, Superposition of RagA plug domains in the open (yellow) and closed (cyan) RagAB complexes of the OC state, showing different dynamics (d;view approximately as inc) and conformational changes (e) in going from the open to the closed state. The Ton box region Q103-G108, visible only in the open state, is coloured black with electron density shown in (d). In e, density for open and closed complexes is shown for the region I214-R218.
The CC state is essentially identical to the X-ray crystal structure (backbone r.m.s.d. values of ~0.6 Å), and the bound peptide occupies the same position. In the OO state, each RagB lid has undergone a substantial conformational rearrangement that swings RagB upward, exposing the peptide binding site and plug domain within the barrel interior. The bound peptide is present in both barrels of the OO state, at the same position as in the CC state, but the density is weaker (Fig. 3), suggesting lower occupancy or greater mobility for the bound peptide in the open state. The presence of peptide in open complexes is consistent with a PISA interface analysis17 that shows the observed peptide binding site is mainly formed by RagA.
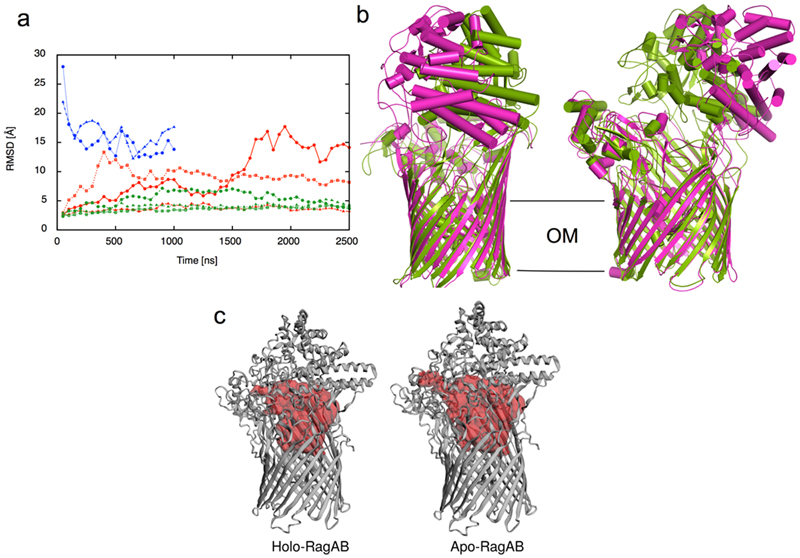
The most interesting state is the open-closed transporter (OC), in which the internal symmetry of the RagA2B2 complex is broken. The density for bound peptide in the open barrel of the OC RagAB is much weaker than in either barrel of the OO state (Fig. 3) even when both maps are generated without symmetry, suggesting cross-talk between RagAB units. The OC state unambiguously shows that the RagB lids in the dimeric complex can open and close separately. Fascinatingly, no "intermediate" states in the opening of the transporter are observed: essentially all of the individual RagAB pairs are either open or closed, and these are dimerised to give each of the three possible states observed (CC, OC and OO). This suggests that either the dynamicsof lid opening and closing are very rapid, and/or that the energy landscape for opening and closing is very rugged, with distinct stable minima only existing for the three discrete states observed here. MD simulations performed priorto obtaining the EM structures show a similar, albeit less wide opening of the RagB lid upon removal of the peptide (Extended Data Fig. 3). Superposition of the RagA subunits of the open and closed states reveals that the N-terminal ~10 residues of the RagB lid and the lipid anchor at the back of the complex remain stationary during lid opening (Extended Data Fig. 4), acting as a hinge about which the rest of the protein moves as a rigid body (Supplementary Movie).
The open-closed complex reveals changes in the RagA plug domain
In the consensus model of TonB-dependent transport14, extracellular substrate binding induces a conformational change that is transmitted through the plug domain of the TBDT and results in disordering of its N-terminal Ton box. This increases accessibility of the Ton box from the periplasm, permitting interaction with TonB in the inner membrane. The disordered state of the Ton box can therefore be considered a signal, ensuring that only substrate-loaded transporters form productive complexes with TonB, avoiding futile transport cycles. In crystal structures of TBDTs, Ton box conformation varies. Where visible, this region interacts with other parts of the plug domain and is assumed to be inaccessible to TonB. In other structures, electron density for the Ton box is missing, indicating increased mobility. Importantly, the correlation between ligand binding and visibility of the Ton box is poor18, likely owing to crystallisation19. In contrast, the OC state observed by cryo-EM provides an unbiased structure that relates ligand binding site occupancy to changes in the plug domain (Fig. 3). In the open side, the first visible residue is Q103 at the start of the TonB box, whereas in the closed side (and in the crystal and CC EM structures), the density starts at L115. The conformation of L115-S119 is also different from that in the open complex, with shifts for L115 as large as 10 Å (Fig. 3d). Interestingly, the plug region A211-A219 is also different in the two states. Notably, the pronounced shift for R218 (~9 Å for the head group; Fig. 3e) may be important given its location at the bottom of the binding cavity. A211-A219 contacts the region following the Ton box, highlighting a potential allosteric route to communicate binding site occupancy to the Ton box. This is a mechanism that would be consistent with previous AFM data suggesting that the plugs of TBDTs consist of two domains20: an N-terminal, force-labile domain that is removed by TonB to form a channel and a more stable C-terminal domain that would include R218 in RagA. The difference in dynamics of the N-terminus of RagA is most evident in unsharpened maps of the OC state contoured at low levels, which clearly show globular density connected to the plug domain only in the open state (Extended Data Fig. 5). This density corresponds to an ~80-residue N-terminal extension (NTE; Pfam 13715, Carboxypeptidase D regulatory-like domain) that is present in a relatively small subset of TBDTs21 and which precedes the Ton box. In the closed state, the NTE and Ton box are not resolved in the cryo-EM maps, presumably because of increased mobility.
RagAB is important for growth ofP. gingivalis onproteins as carbon source
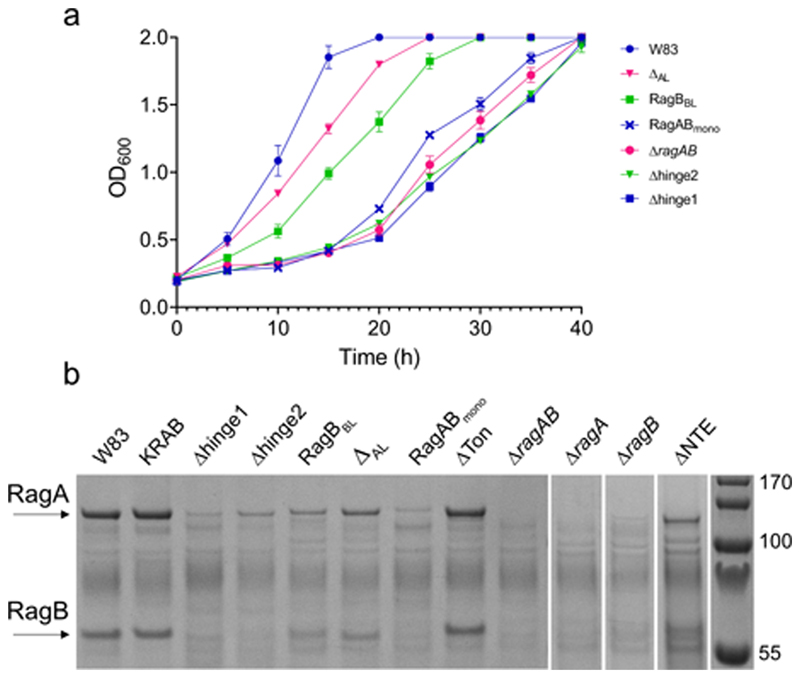
We initially assessed whether the P. gingivalis strains W83 and ATCC 33277 (hereafter named ATCC) could grow on minimal medium supplemented with BSA as a sole carbon source (BSA-MM; Methods). Interestingly, while growth on rich medium is identical, robust growth on BSA-MM is observed only for the rag-1 locus W83 strain22. By contrast, the rag-4 strain ATCC grows slowly, and after a substantial lag phase (Figs. 4a,c). Given the high sequence similarity of the RagA and RagB proteins (Supplementary Figs. 1 and 2), the distinct growth profiles of W83 and ATCC suggest that relatively small differences in RagAB could alter substrate uptake sufficiently to dramatically affect growth on the relatively small set of peptides that can be generated from BSA.
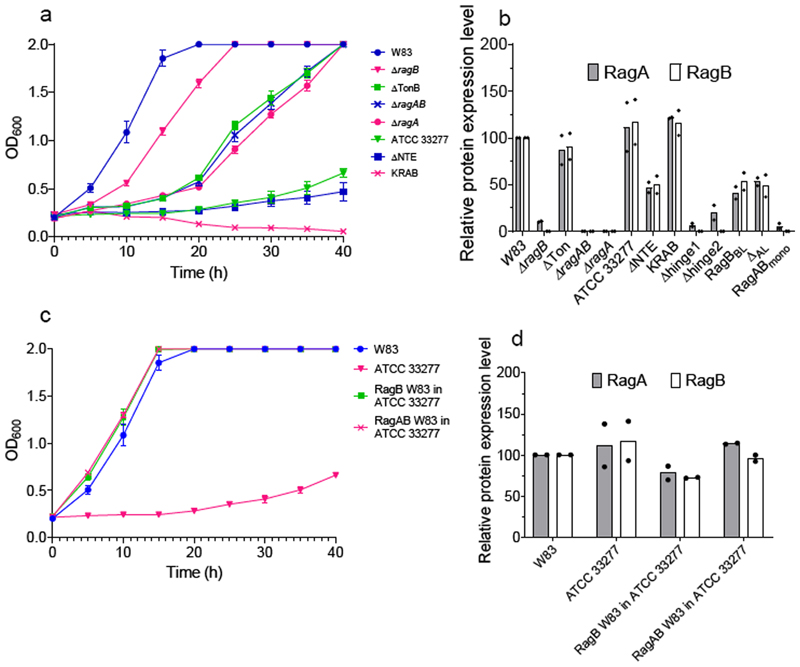
Figure 4. RagAB is important for growth on BSA.
a, Representative growth curves (n = 3, mean ± standard error of the mean) for different P. gingivalis and ATCC strains and mutant W83 ragAB variants on BSA-MM. b, Outer membrane RagAB protein expression levels for strains and mutants determined by SDS-PAGE densitometry. c, Representative growth curves (n = 3, mean ± standard error of the mean) for growth on BSA-MM for wild type P. gingivalis W83 and ATCC and strains in which RagAB or RagB from ATCC was replaced with the corresponding ortholog from W83. d, Corresponding OM protein expression levels of RagAB. Graphs in b and d show the mean of two independent replicates. The dots show the individual replicates.
To further investigate potential functional differences between the RagAB complexes from W83 and ATCC we constructed an ATCC strain in which W83 ragABwas expressed from a single-copy plasmid in the ATCC ΔragAB background. We also made a strain in which the genomic copy of ATCCragB was replaced by W83 ragB. Remarkably, replacement of either RagB or RagAB from ATCC with the corresponding orthologs from W83 results in robust growth of ATCC on BSA-MM (Fig. 4c). Since the OM levels of RagAB for all strains are similar (Fig. 4d), these results have several important implications. Firstly, they confirm that RagAB is required for growth on extracellular protein-derived oligopeptides. Secondly, they suggest that different RagB lipoproteins can form functional complexes with the same RagA transporter. Finally, and perhaps most surprisingly, RagB appears to determine the substrate specificity of the complex. We speculate that TonB-dependent transporters such as SusC and RagA may have acquired SusD and RagB substrate binding lipoproteins to confer substrate specificity.
The long lag phase during growth of ATCC on 1% BSA-MM is consistent with previous data showing that this strain requires either high substrate concentrations (3% BSA) or supplementation with rich medium for growth on BSA or human serum albumin (HSA) 23–25. This may seem surprising, given that HSA is an abundant component of gingival crevicular fluid (GCF), an inflammatory exudate which is dominated by abundant plasma proteins such as albumin26. However, GCF does also contain many other proteins that could be substrates for ATCC, and many other bacteria are present and could generate different substrates from those proteins. Thus, slow growth on a single substrate in vitro does not translate to poor fitness in vivo. Poor growth on BSA also does not imply that ATCC RagAB does not take up any BSA peptides, as the phenotype could be caused by inefficient uptake of a particular BSA peptide that provides one or more essential amino acids. Our Supplementary Table 2 identifies only 63 BSA-derived peptides after trypsin digest, so the substrate pool is relatively small. This notion is supported by previous data showing that while ATCC efficiently cleaves human albumin, transferrin, immunoglobulin G and collagen, the two latter substrates don’t support growth24.
To better define the role of RagAB in peptide utilisation, we constructed clean deletions in P. gingivalis W83 for ragA, ragB, and ragAB and grew the resulting strains on BSA-MM. Compared to wild-type W83 RagAB, growth of the ragA and ragAB deletion strains exhibits long (~15-20 hrs) lag periods (Fig. 4a). Unlike the gingipain-null KRAB strain11, both mutant strains grow eventually, most likely due to passive, RagAB-independent uptake of small peptides produced after prolonged BSA digestion. Interestingly, the ΔragB strain grows better on BSA than the ΔragAB strain. Since the strain still produces RagA (albeit at very low levels; Fig. 4b and Extended Data Fig. 6) this demonstrates that RagB, in contrast to RagA, is not required for growth on BSA-derived oligopeptides in vitro, in accordance with previous work12. Collectively, the growth data suggest that RagAB mediates uptake of extracellular oligopeptides produced by gingipains, and help explain data showing that the transporter is important for in vivo fitness and virulence of P. gingivalis 22,27.
We next constructed mutant strains for structure-function studies (see Methods), and tested growth on BSA-MM (Figs. 4a and Extended Data Fig. 6). Most mutant strains show very low OM levels of RagAB, complicating conclusions about functionality (Extended Data Fig. 6). Exceptions are the Ton box deletion (ΔTon) and the NTE deletion (ΔNTE), which are expressed at reasonable levels (Fig. 4b). The ΔTon strain has a similar phenotype as ΔRagAB, demonstrating that this variant is inactive and therefore that RagAB is a bona-fide TBDT. Strikingly, the ΔNTE variant resembles the KRAB strain and does not grow even after a prolonged period (Fig. 4a). Since the ΔRagAB strain (which also lacks the NTE) does grow after a lag phase, the results suggest that the NTE is important. In Proteobacteria, structures of TBDTs with a different N-terminal domain (Pfam 07660; STN) have been reported28. This domain interacts with an anti-sigma factor in the inner membrane to stimulate TBDT expression in response to the presence of their cognate substrates, exemplified by the E. coli FecA-FecIR system29–31 in which the anti-sigma factor fecR and the sigma factor fecI are located in the same operon as fecA. By contrast, while P. gingivalis has five anti-sigma factors, none of these are located near the ragAB locus. Moreover, deletion of the N-terminal domain in FecA only affects FecA upregulation and has no effect on transport activity29, contrasting with the dramatic phenotype for the RagAΔNTE mutant. Thus, while an involvement of the RagA NTE in signalling seems likely, establishing the signalling partner(s) and mechanism remains the subject of further work.
Peptide binding by RagAB is selective
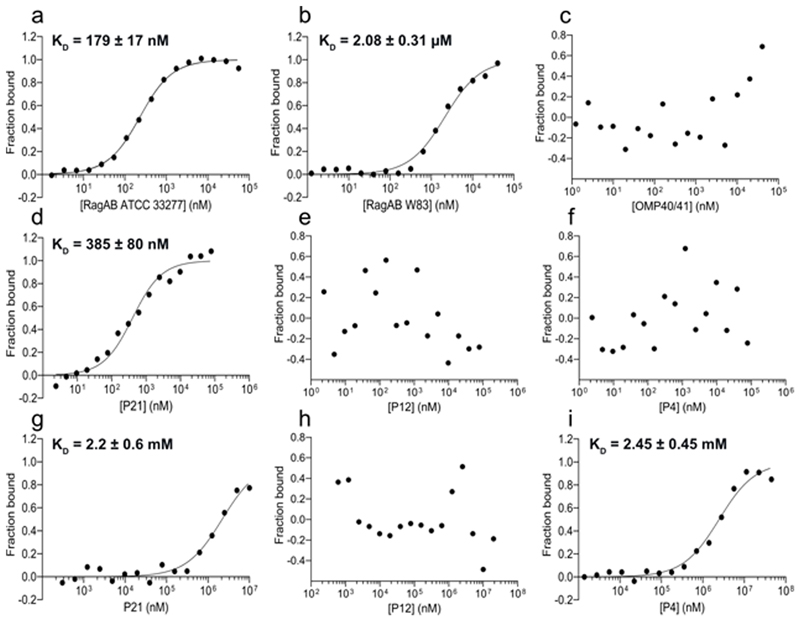
We next characterised peptide binding to RagAB in more detail. Since RagAB is purified with an ensemble of peptides that cannot be removed without denaturing the complex, added peptides compete with bound peptides, resulting in the measurement of apparent rather than true dissociation constants. The 21-residue peptide KATAEALKKALEEAGAEVELK (henceforth named P21; charge -1) from the C-terminus of ribosomal protein L7 was abundant in W83 KRAB RagAB (Methods), and was synthesised in addition to its 12-residue core sequence (DKATAEALKKAL, denoted P12; charge +1) that is also present in a number of similar peptides but, crucially, is not identified as a RagAB-bound peptide. We also tested an 11-residue peptide (named P4; sequence NIFKKNVGFKK) of the arginine deiminase ArcA from Streptococcus cristatus that was proposed to bind to RagB from ATCC32.
We used microscale thermophoresis (MST) to asses peptide binding to RagAB. Initially, we used N-terminally fluorescein-labelled P4 peptide (P4-FAM) and unlabelled RagAB to maximise the signal-to-noise, and obtained apparent dissociation constants of ~2 μM for W83 RagAB and ~0.2 μM for ATCC RagAB (Extended Data Fig. 7). The negative control Omp40-41 did not show binding, demonstrating that these results are not due to non-specific partitioning of fluorescently-labelled peptide into detergent micelles. However, the fluorophore may affect peptide binding in an unpredictable way, and therefore we next performed MST experiments for unlabelled P21, P12 and P4 peptides and His-tag labelled RagAB from wild-type W83. Remarkably, we observed robust binding only for P21, with an apparent Kd (~0.4 μM; Extended Data Fig. 7) similar to that observed for substrate binding to classical TBDTs33–34. While the fit to a single binding site model is reasonable, models that assume two binding sites yield significantly better fits (Supplementary Fig. 3). This provides an indication that the two RagA2B2 binding sites may not be equivalent, perhaps due to cooperativity. The data for P12 and P4 suggest that these peptides cannot displace the bound endogenous peptides. Notably, the result for unlabelled P4 contrasts with that for P4-FAM (Discussion). We next asked whether the three peptides bind to RagB, purified from E. coli. W83 RagB produced good-quality binding curves with P21 and P4, with similar apparent dissociation constants of ~2 mM, but no binding was observed for P12 (Extended Data Fig. 7). The relatively low apparent affinities of RagB for oligopeptides are in good agreement with literature values for oligosaccharide binding to SusD proteins determined via ITC16,35,36. Collectively, the P21 data suggest that peptides bind with lower affinities to RagB (P21 Kd app. ~2 mM) than to RagAB (P21 Kd app. ~0.4 μM), which makes sense assuming that after initial capture by RagB, the peptide needs to be transferred to RagA. The data also show that the P4 and P12 peptides are not good substrates for W83 RagAB, confirming that the transporter has considerable substrate selectivity.
Since we can measure peptide binding to purified RagAB in vitro by MST, added peptides compete successfully with co-purified endogenous peptides. This, together with the robust growth observed in BSA-MM, led us to ask whether we could detect acquisition of BSA tryptic peptides by W83 RagAB in vitro (Methods). Indeed, the sample incubated with the BSA digest revealed six bound BSA peptides in addition to endogenous peptides, suggesting that the BSA peptides only partly replace the co-purified ensemble (Supplementary Table 2). This is due to the fact that the BSA digest contains 63 different peptides, such that the concentration of BSA "binder" peptides is not high enough to replace all endogenous peptides. By contrast, a 100-fold excess of P21 completely displaced the co-purified peptides (Supplementary Table 2). The fact that only a subset of BSA peptides binds to RagAB demonstrates that the transporter is selective. We also incubated the BSA tryptic digest experiment with W83 RagB. Prior to incubation, RagB contains only one bound co-purified peptide. After incubation and post-SEC, two BSA peptides are detected, demonstrating that at least some peptides bind to RagB with sufficient affinity to survive SEC (Supplementary Table 2).
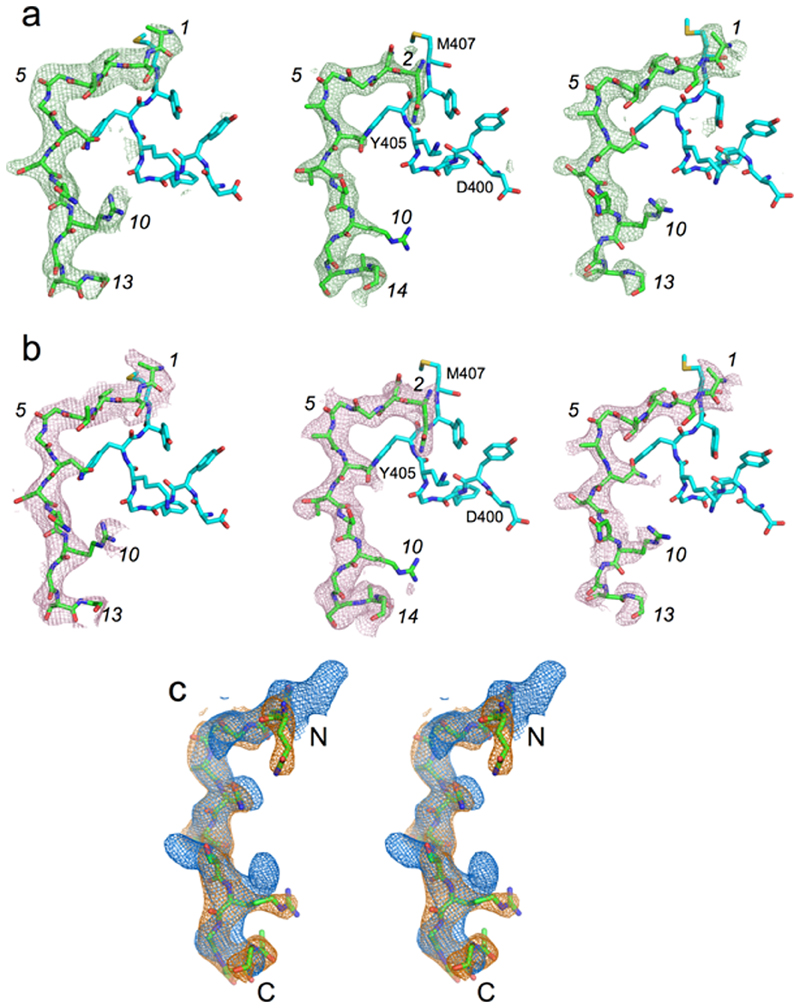
In an attempt to build a unique peptide sequence into the electron density maps, we crystallised RagAB purified from wild-type W83 in the absence and presence of a 50-fold molar excess of P21. Comparison of the maps reveals peptide density at the same site, but with some differences particularly at the N-terminus (Extended Data Fig. 8). However, the P21 sequence does not fit the density unambiguously, and we therefore speculate that P21 and other peptides are bound with register shifts and perhaps different chain directions. In all three structures the same RagAB residues hydrogen bond with the backbones of the modelled substrates, providing a clear rationale how the transporter can bind many oligopeptides. Future studies will be required to define precisely how peptides bind to RagAB, perhaps by collecting anomalous X-ray data on P21-like peptides labelled at unique positions with heavy atoms.
Discussion
Our combined data show that RagAB is a dynamic OM oligopeptide transporter that is important for growth of P. gingivalis on extracellular protein substrates and possibly for peptide-mediated signalling32. We propose a transport model in which the open RagB lid binds substrates before delivering them to RagA via lid closure (Extended Data Fig. 9). This would make the RagA Ton box accessible for interaction with TonB, permitting formation of a transport channel into the periplasmic space. To test the premise that substrate binding induces lid closure we collected cryo-EM data on RagAB in the absence and presence of excess P21. Particle classification indeed shows a decrease in OO states and a clear increase in CC states in the P21 sample, in accordance with our model (Extended Data Fig. 10).
How does signalling to TonB occur, and how are unproductive interactions of TonB with "empty" transporters avoided? In the classical, smaller TBDTs such as FecA and BtuB, the ligand binding sites involve residues of the plug domain, suggesting that ligand binding is allosterically communicated to the periplasmic face of the plug. However, there are no direct interactions between the visible, well-defined part of the peptide substrates and the RagA plug. What, then, causes the observed conformational changes in the plug? One possibility is that parts of peptide substrates that are invisible (e.g. due to mobility) contact the plug. Given the very large solvent-excluded RagAB cavity even with the modelled 13-residue peptide (~ 9800 Å3; ~11500 Å3 without peptide; Extended Data Fig. 3), there is enough space to accommodate the long substrates identified by the peptidomics, and these could contact the plug directly. However, the presence of substrate density in the open states of the cryo-EM structures suggests that it may not be the occupation of the binding site per se that is important for TonB interaction, but closure of the RagB lid (Extended Data Fig. 9). Thus, we hypothesise that certain peptides may bind to RagAB, but do not generate the closed state of the complex and signal occupancy of the binding site to TonB. This may provide an alternative explanation for the different MST results for the P4 and P4-FAM peptide titrations to RagAB (Extended Data Fig. 7). Given the nature of the MST signal, titrating unlabelled P4 to labelled RagAB is likely to give a signal only if the binding causes a conformational change (e.g. lid closure in the case of RagAB). In the "reverse" experiment, the readout is on the labelled peptide (P4-FAM), and the large change in mass upon binding could generate a thermophoretic signal in the absence of any conformational change. Thus, P4 may be an example of a substrate that binds non-productively to RagAB.
A fascinating question is why RagAB and related transporters are dimeric. To our knowledge, there are no other TBDTs that function as oligomers, and there is no obvious reason why dimerisation would be beneficial. There are few clues in the structures, but when RagA in, e.g., the closed complexes of the CC and OC states is superposed, RagB shows a rigid-body shift of ~3-5 Å, and vice versa. A similar trend is observed when the open complexes of the OC and OO states are considered. Moreover, the peptide densities in the OO state are stronger than that of the open complex in the OC state (Fig. 3). Together, this suggests that the individual RagAB complexes could exhibit some kind of cross-talk such as cooperative substrate binding. This notion is supported by the MST data for P21 binding to RagAB, suggesting the presence of two, non-equivalent ligand binding sites.
Methods
Bacterial strains and general growth conditions
Porphyromonas gingivalis strains (listed in Table S4) were grown in enriched tryptic soy broth (eTSB per liter: 30 g tryptic soy broth, 5 g yeast extract; further supplemented with 5 mg hemin; 0.25 g L-cysteine and 0.5 mg menadione) or on eTSB blood agar (eTSB medium containing 1.5% [w:v] agar, further supplemented with 5% defibrinated sheep blood) at 37 °C in an anaerobic chamber (Don Whitley Scientific, UK) with an atmosphere of 90% nitrogen, 5% carbon dioxide and 5% hydrogen. Escherichia coli strains (listed in Supplementary Table 4), used for all plasmid manipulations, were grown in Luria–Bertani (LB) medium and on 1.5% agar LB plates. For antibiotic selection in E. coli, ampicillin was used at 100 μg/ml. P. gingivalis mutants weregrown in the presence of erythromycin at 5 μg/ml and/or tetracycline at 1 μg/ml.
Growth of P. gingivalis in minimal medium supplemented with BSA
An important general limitation of studies on TonB-dependent transport is that it has not yet been reconstituted in vitro from purified components, due to the involvement of two membranes, one of which has to be energised and contain the ExbBD-TonB complex. This limits the ability to directly link any TBDT to substrate import. Therefore, to determine the role of RagAB in the transport of peptides in vivo, a growth assay with minimal medium containing BSA as the sole carbon source (BSA-MM) was used. Notably, the peptides generated by extracellular P. gingivalis proteases such as gingipains must be intracellular to allow growth, giving a qualitative measure of peptide import. P. gingivalis strains were grown overnight in eTSB. The cultures were washed twice with enriched Dulbecco's Modified Eagle's Medium (eDMEM per liter: 10 g BSA; further supplemented with 5 mg hemin; 0.25 g L-cysteine and 0.5 mg menadione) and finally resuspended in eDMEM. The OD600 in each case was adjusted to 0.2 and bacteria were grown for 40 hours. OD600 was measured at 5 hourintervals.
Mutant construction
For RagA, the following mutants were made: a Ton box deletion (ΔTon; residues V100-Y109 deleted), a NTE domain deletion (ΔNTE; residues V25-K99 deleted) a putatively monomeric RagAB version made via introduction of a His6-tag between residues Q570-G571; RagABmono), and deletions of the two loops (L7 and L8) that remain associated with RagB during lid opening as based on the cryo-EM structures (Δhinge1, residues Q670-G691 deleted and replaced by one Gly; Δhinge2, residues L731-N748 deleted and replaced by one Gly). For RagB, two variants were made, both involving the acidic loop: a deletion of this loop (ΔAL; residues R97-S104) and conversion of the acidic loop into a basic loop (RagBBL; D99/E100/D101/E102 replaced by R99/K100/R101/K102). All P. gingivalis mutants were prepared using wild-type W83 strain or its derivatives (unless stated otherwise) and constructed by homologous recombination37. Mutants were generated in wild type W83 rather than KRAB W83 due to the presence of multiple antibiotic cassettes in the latter strain, resulting from removal of the gingipain genes. The “swap” mutants were prepared using the ATCC33277 strain as background. For deletion strains, 1 kb regions upstream (5’) and downstream (3’) from the ragA, ragBand both ragAB genes, as well as chosen antibiotic resistance cassettes, were amplified by PCR. Obtained DNA fragments were cloned into pUC19 vector using restriction digestion method and/or Gibson method38. For the construction of master plasmids for RagA and RagB mutants the whole gene sequences were amplified with addition of antibiotic cassettes, 1kb downstream fragments and cloned into pUC19 vector. Desired mutations were introduced into master plasmids by SLIM PCR39. A similar method was used for the “swap” RagB-W83inATCC and RagB-ATCCinW83 strains. In both deletion RagB plasmids we inserted the gene of the other strain (i.e. ragB from W83 into ΔRagB-ATCC). We also obtained a RagAB “swap” strains using the pTIO-1 plasmid40. The DNA sequences of ragAB from both strains were cloned with the addition of their promoters into pTIO plasmids and further conjugated with the opposite deletion strains using E. coli S-17 λpir (i.e. RagAB-W83-pTIO into delRagAB-ATCC)41. Primers used for plasmid construction and mutagenesis are listed in Supplementary Table 4. All plasmids were analyzed by PCR and DNA sequencing. P. gingivalis competent cells 42 were electroporated with chosen plasmids and plated on TSBY with appropriate antibiotics – erythromycin (5 μg/ml) or tetracycline (1 μg/ml) and grown anaerobically for approximately 10 days. Clones were selected and checked for correct mutations by PCR and DNA sequencing. Bacterial strains generated and used in this study are listed in Supplementary Table 4.
W83 KRAB RagAB production and purification
The non-His tagged RagAB complex from P. gingivalis W83 KRAB was isolated from cells grown in rich media. In brief, cells from 6 l of culture were lysed by 1 pass through a cell disrupter (0.75 kW; Constant Biosystems) at 23,000 psi, followed by ultracentrifugation at 200,00 x g for 45 minutes to sediment the total membrane fraction. The membranes were homogenised and pre-extracted with 100 ml 0.5% sarkosyl in 20 mM Hepes pH 7.5 (20 min gentle stirring at room temperature) followed by ultracentrifugation (200,000 x g; 30 min) to remove inner membrane proteins43. The sarkosyl wash step was repeated once, after which the pellet (enriched in OM proteins) was extracted with 100 ml 1% LDAO (in 10 mM Hepes/50 mM NaCl pH 7.5) for 1 hour by stirring at 4 °C. The extract was centrifuged for 30 min at 200,000 x g to remove insoluble debris. The solubilised OM was loaded on a 6 ml Resource-Q column and eluted with a linear NaCl gradient to 0.5 M over 20 column volumes. Fractions containing RagAB were ran on analytical SEC (Superdex 200 Increase GL 10/300) in 10 mM Hepes/100 mM NaCl/0.05% LDAO pH 7.5 in order to obtain RagAB of sufficient purity. Finally, the protein was detergent-exchanged to C8E4 using two rounds of ultrafiltration (100 kDa MWCO), concentrated to 15-20 mg/ml and flash-frozen in liquid nitrogen.
Wild type W83 RagAB production and purification
Homologous recombination was used to add a 8×His-tag to the C terminus of genomic ragB in the P. gingivalis W83 strain. The mutant was grown about 20 h in rich medium under anaerobic conditions. The cells from 6 l of culture were collected and processed as outlined above. The insoluble material was homogenised with 1.5% LDAO (in 20 mM Tris-HCl/300 mM NaCl pH 8.0) and the complex was purified by nickel-affinity chromatography (Chelating Sepharose; GE Healthcare) followed by gel filtration using a HiLoad 16/60 Superdex 200 column in 10 mM Hepes/100 mM NaCl, 0.05% DDM pH 8.0.
Purification of RagB from W83 and ATCC 33277 expressed in E. coli
Genes encoding for the mature parts of RagB from W83 and ATCC 33277 (with His6-tags at the C-terminus) were amplified by PCR from genomic DNA extracted from P. gingivalis. The DNA fragments were purified and cloned into the arabinose-inducible pB22 expression vector44 using NcoI/XbaI restriction enzymes. The obtained expression plasmid was transformed into E. coli strain BL21 (DE3). Transformed E. coli cells were grown in LB media containing ampicillin (100 μg/ml) at 37 °C to an OD 600 ~ 0.6 and expression of the recombinant protein was induced with 0.1% arabinose. After ~2.5 h at 37 °C, cells were collected by centrifugation (5,000 × g; 15 min), resuspended in 20 mM Tris-HCl/300 mM NaCl pH 8.0 and lysed by 1 pass through a cell disrupter (0.75 kW; Constant Biosystems) at 23,000 psi, followed by ultracentrifugation at 200,000 × g for 45 minutes. The supernatant was loaded on nickel-affinity resin (Chelating Sepharose; GE Healthcare) and after washing with 30 mM imidazole, protein was eluted with buffer containing 250 mM imidazole. Protein was further purified by gel filtration in 10 mM Hepes/100 mM NaCl pH 7.5 using a HiLoad 16/60 Superdex 200 column.
Crystallisation and structure determination of RagAB from W83 KRAB
Sitting drop vapour diffusion crystallisation trials were set up using a Mosquito crystallisation robot (TTP Labtech) using commercially available crystallisation screens (MemGold 1 and 2; Molecular Dimensions). Initial hits were optimised manually by hanging drop vapour diffusion. Crystals were cryoprotected by transferring them for 5-10 s in mother liquor containing an additional 10% PEG400. A few crystals optimised from MemGold 2 condition C8 (18% PEG200, 0.1 M KCl, 0.1 M K-phosphate pH 7.5) diffracted anisotropically to below 4 Å at the Diamond Light Source (DLS) synchrotron at Didcot, UK (space group C2221; cell dimensions ~190 x 377 x 369 Å, with four RagAB complexes in the asymmetric unit). Data were processed via Xia245 or Dials46. The structure was solved by molecular replacement with Phaser47, using data to 3.4 Å resolution. The structures of RagB (PDB 5CX8) and a Sculptor-modified model of BT2264 SusC (PDB 5FQ8) were used as search models. The RagAB model was built iteratively by a combination of manually building in COOT48 and the AUTOBUILD routine within Phenix49, and was refined with Phenix50 using TLS refinement with 1 group per chain. Given that the R values are reasonable and the R-Rfree gaps are not excessively large we did not use NCS in the final rounds of refinement. Using NCS also did not improve the peptide density. The final R and Rfree factors of the RagAB structure are 20.5 and 25.5%, respectively (Supplementary Table 1). Structures of RagAB purified from wild type W83 (+/- P21) were solved via molecular replacement using Phaser, using the best-defined RagAB complex as search model. Structures were refined within Phenix as above (Supplementary Table 1) and structure validation was carried out with MolProbity51.
Crystallisation and structure determination of wild type RagAB W83 in the absence and presence of P21
Crystallisation trials were performed as outlined above with the following modifications: the protein was not detergent-exchanged after gel filtration in DDM; two other commercially available crystallisation screens were used (MemChannel and MemTrans; Molecular Dimensions); For co-crystallisation of RagAB W83 with P21 peptide, RagAB W83 at a concentration of 17 mg/ml (~0.1 mM) was incubated overnight at 4 °C with 5 mM P21 peptide. For RagAB W83 the crystals were optimised from MemTrans condition F6 (22% PEG400, 0.07 M NaCl, 0.05 M Na-citrate pH 4.5), for RagAB W83 + P21 the crystals were optimised from MemChannel condition D3 (15% PEG1000, 0.05 M Li-sulfate, 0.05 M Na-phosphate monobasic, 0.08 M citrate pH 4.5). Crystals were cryoprotected by transferring them for 5-10 s in mother liquor containing additional PEG400 to generate a final concentration of ~25%.
Microscale thermophoresis
Initial isothermal titration calorimetry (ITC) experiments showed that P4 addition to buffer without protein generated very large heats, precluding ITC as a method to assess peptide binding. For MST, the Monolith NT.115 instrument (NanoTemper Technologies GmbH, Munich, Germany) was used to analyse the binding interactions between P4, P12 and P21 peptides and RagAB from W83 as well as RagB from W83 and ATCC 33277. Proteins were labelled with Monolith His-tag Labeling Kit RED-tris-NTA 2nd Generation (NanoTemper Technologies). For all interactions the concentrations of both fluorescently labelled molecule and ligand were empirically adjusted using Binding Check mode (MO.Control software, NanoTemper Technologies). Experiments were performed in assay buffer (10 mM Hepes/100 mM NaCl pH 7.5) with addition of 0.03% DDM in the case of RagABs. For the measurements, sixteen 1:1 ligand dilutions were prepared and then mixed with one volume of labelled protein followed by loading into Monolith NT.115 Capillaries. Initial fluorescence measurements followed by thermophoresis measurement were carried out using 100% LED power and medium MST power, respectively. Data for three independently pipetted measurements were analysed (MO.Affinity Analysis software, NanoTemper Technologies), allowing for determination of apparent dissociation constants (KD app.). The data was presented using GraphPad Prism 8. The interactions between unlabelled W83 and ATCC 33277 RagABs and FAM-labelled P4 peptide were determined in the same way.
Isolation of the outer membrane fractions for quantitation of RagAB
Outer membrane fractions from 1 l of culture were isolated using sarkosyl extraction method (see RagAB W83 KRAB production and purification). After extraction in LDAO, the samples in 10 mM Hepes/50 mM NaCl pH 7.5 containing 1% LDAO were diluted 3 times and loaded on SDS-PAGE. The bands were analysed quantitatively using Image Lab 6.0.1 software (BIO-RAD). The band at ~70 kDa was used as a reference sample (loading control). This analysis allows comparison of the levels of RagAB integrated into the OM and, in combination with the growth assays, a qualitative assessment of function.
Peptide identification by LC-MS
Bound peptides were isolated by precipitation via addition of trichloroacetic acid to a final concentration of 30% and incubation at 4 ºC for 2 hrs. Subsequently the peptide-containing supernatants were collected by centrifugation at 17,000xg. The isolated peptides were micropurified using Empore™ SPE Disks of C18 octadecyl packed in 10 μl pipette tips.
LC-MS/MS was performed using an EASY-nLC 1000 system (Thermo Scientific) connected to a QExactive+ Hybrid Quadrupole-Orbitrap Mass Spectrometer (Thermo Scientific). Peptides were dissolved in 0.1 % formic acid and trapped on a 2 cm ReproSil-Pur C18-AQ column (100 μm inner diameter, 3 μm resin; Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). The peptides were separated on a 15-cm analytical column (75 μm inner diameter) packed in-house in a pulled emitter with ReproSil-Pur C18-AQ 3 μm resin (Dr. Maisch GmbH, Ammerbuch-Entringen, Germany). Peptides were eluted using a flow rate of 250 nl/min and a 20-minute gradient from 5% to 35% phase B (0.1% formic acid and 90% acetonitrile or 0.1 % formic acid, 90 % acetonitrile and 5% DMSO). The collected MS files were converted to Mascot generic format (MGF) using Proteome Discoverer (Thermo Scientific).
The data were searched against the P. gingivalis proteome (UniRef at uniprot.org) or the Swiss-prot database using a bovine taxonomy. Database searches were conducted on a local mascot search engine. The following settings were used: MS error tolerance of 10 ppm, MS/MS error tolerance of 0.1 Da, and either non-specific enzyme or trypsin. Based on chromatogram peak heights and the number of MS/MS spectra observed for a particular peptide in the mass spectrometry data, the 21-residue peptide KATAEALKKALEEAGAEVELK (henceforth named P21; charge -1) from ribosomal protein L7 is very abundant in W83 KRAB RagAB.
Generation of BSA tryptic mixture
BSA was solubilized in 100 mM ammonium bicarbonate /8M urea pH 8.0. DTT was added to a final concentration of 10 mM and the mixture was incubated for 60 min at RT. Protein was alkylated by addition of iodoacetamide to a final concentration of 30 mM and incubation for 60 min at RT in the dark. Next, the concentration of DTT was adjusted to 35 mM followed by dilution of the sample to 1 M urea. For cleavage, trypsin from bovine pancreas (Sigma) was added at 1:25 (trypsin : BSA) mass ratio and the sample was incubated overnight at 37 ºC. Digestion was stopped by acidifying the sample to pH < 2.5 with formic acid. Peptides were purified using Peptide Desalting Spin Columns (Thermo Scientific) and dried using Speed Vacuum Concentrator Savant SC210A (Thermo Scientific).
Acquisition of BSA-derived peptides and P21 by RagAB and RagB in vitro
W83 RagAB and W83 RagB were incubated overnight at 4 °C with an ~100-fold molar excess of tryptic BSA digest (see Generation of BSA tryptic mixture) or P21 peptide. The samples were then ran on Superdex 200 Increase GL 10/300 in 10 mM Hepes/100 mM NaCl pH 7.5 with addition of 0.03% DDM for RagAB W83. Fractions containing protein were pooled, acidified by addition of formic acid to a final concentration 0.1% and analysed by MS.
qPCR
Samples of 1 ml of bacterial cultures (OD600 = 1.0) were centrifuged (5,000 x g; 5 min) at 4 °C, pellets were resuspended in 1 ml Tri Reagent (Ambion), incubated at 60 °C for 20 minutes, cooled to room temperature and total RNA was isolated according to manufacturer instructions. Genomic DNA was removed from samples by digestion with DNAse I (Ambion); 2 μg of RNA was incubated with 2 U of DNAse for 60 minutes at 37 °C. Following digestion, RNA was purified using Tri Reagent. Reverse transcription of 50 ng of RNA was performed with High-Capacity cDNA Reverse Transcription Kit (Life Technologies), and reaction mixture was then diluted 20 times. Real-time PCR was done in 10 μl reaction volume, using KAPA SYBR FAST qPCR Master Mix (Kapa Biosystems) with 2 μl of diluted reverse transcription mixture as template. Primers used are listed in Table S4. Reaction conditions were 3 minutes at 95 °C, followed by 40 cycles of denaturation for 3 seconds at 95 °C and annealing/extension for 20 seconds at 60 °C. The reaction was carried out with a CFX96 thermal cycler (Bio-Rad), and data was analysed in Bio-Rad CFX Manager software.
CryoEM sample preparation and data collection
A sample of purified RagAB solubilised in a DDM-containing buffer (10 mM HEPES pH 7.5, 100 mM NaCl, 0.03 % DDM) was prepared at 1.75 mg/ml (principle dataset) or 3 mg/ml (P21 addition experiment). For the P21 addition experiment, control and P21-doped grids (with 50-fold molar excess of P21 peptide) were prepared at the same time from the same purified stock of RagAB for consistency. In all cases, a 3.5 μL aliquot was applied to holey carbon grids (Quantifoil 300 mesh, R1.2/1.3), which had been glow discharged at 10 mA for 30 s before sample application. Blotting and plunge freezing were carried out using a Vitrobot Mark IV (FEI) with chamber temperature set to 6 °C and 100 % relative humidity. A blot force of 6 and a blot time of 6 s were used prior to vitrification in liquid nitrogen-cooled liquid ethane.
Micrograph movies were collected on a Titan Krios microscope (Thermo Fisher) operating at 300 kV with a GIF energy filter (Gatan) and K2 summit direct electron detector (Gatan) operating in counting mode. Data acquisition parameters for each data set can be found in Supplementary Table 3.
Image processing
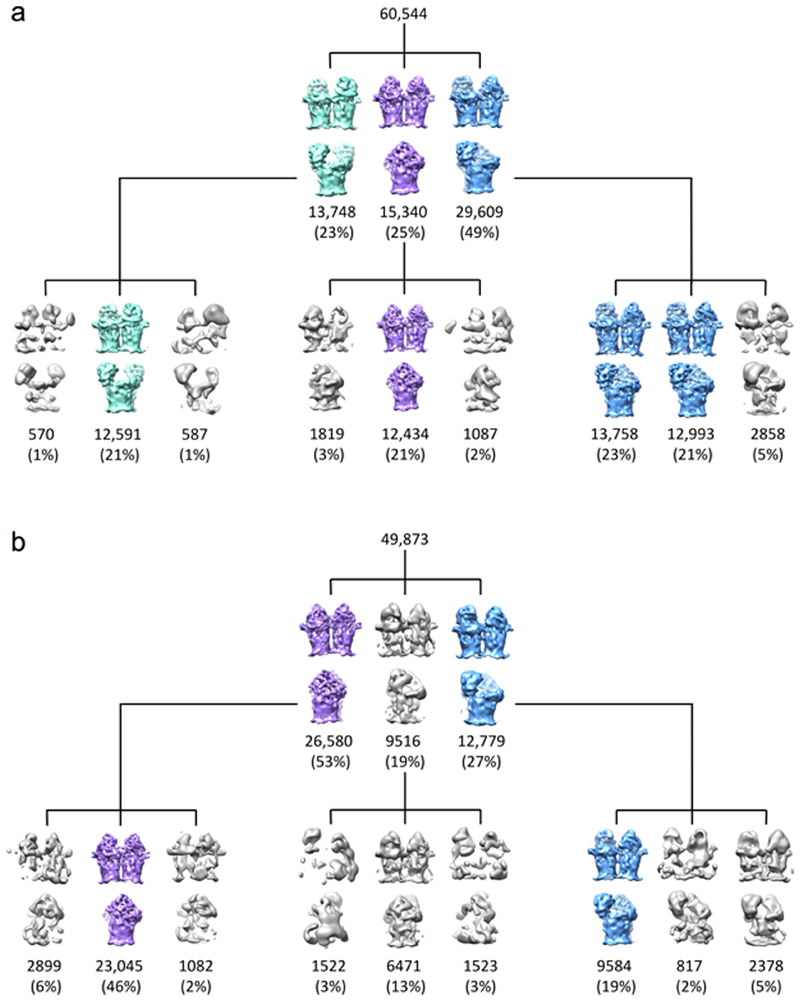
Image processing was carried out using RELION (v2.1 and v3.0)52,53. Drift correction was performed using MotionCor254 and contrast transfer functions were estimated using gCTF55. Micrographs with estimated resolutions poorer than 5 Å and defocus values >4 μm were discarded using a python script56. For the principle RagAB dataset, particles were autopicked in RELION using a gaussian blob with a peak value of 0.3. Control and experimental datasets for the P21 addition experiment were autopicked using the ‘general model’ in crYOLO57. In both cases, particles were extracted in 216 x 216 pixel boxes and subjected to several rounds of 2D classification in RELION52. 3D starting models were generated de novofrom the EM data by stochastic gradient descent in RELION. Processing of RagAB control and P21-doped datasets was only taken as far as 3D classification.
For the principle RagAB dataset, three conformational states representing the CC, OC and OO states were apparent in the first round of 3D classification and the corresponding particle stacks were treated independently in further processing. C2 symmetry was applied to both the CC and OO reconstructions. Post-processing was performed using soft masks and yielded reconstructions for the CC, OC and OO states with resolutions of 3.7 Å, 3.7 Å and 3.9 Å respectively, as estimated by gold standard Fourier Shell Correlations using the 0.143 criterion. The original micrograph movies were later motion corrected in RELION 3.053. Particles contributing to the final reconstructions were re-extracted from the resulting micrographs. Following reconstruction, iterative rounds of per-particle CTF refinement, with beam tilt estimation, and Bayesian particle polishing were employed which improved the resolution of post-processed CC, OC and OO maps to 3.3 Å, 3.3 Å and 3.4 Å respectively.
Model building into cryoEM maps
Examination of the maps revealed that their handedness was incorrect. Maps were therefore Z-flipped in UCSF Chimera58. The RagAB W83 KRAB crystal structure was rigid-body fit to the CC density map and subjected to several iterations of manual refinement in COOT and ‘real space refinement’ in Phenix50. The asymmetric unit was symmetrised in Chimera after each iteration. Starting models for the OC and OO states of the complex were obtained from the CC structure by rigid-body fitting of one or both RagB subunits to their cognate open density in the OC and OO maps respectively. These too were subjected to several iterations of manual refinement in COOT and ‘real space refinement’ in Phenix. Molprobity was used for model validation51.
Molecular Dynamics simulations
The bound peptide in the refined crystal structure of a RagAB "monomer" was removed in silico and the system was inserted into a palmitoyloleoyl-phosphatidylethanolamine (POPE) bilayer using the CHARMM-GUI Membrane Builder59. The systems were solvated using a TIP3P water box and neutralized by adding the required counter-ions. Simulations were performed using GROMACS 5.1.260 and the all-atom CHARMM36 force fields61,62. For the long-range Coulomb interactions, the partice-mesh Ewald (PME) summation method63 has been employed with a short-range cutoff of 12 Å and a Fourier grid spacing of 0.12 nm. In addition, the Lennard-Jones interactions were considered up to a distance of 10 Å and a switch function was used to turn off interactions smoothly at 12 Å. Achieved by semi-isotropic coupling to a Parrinello-Rahman barostat64 at 1 bar with a coupling constant of 5 ps, the final unbiased simulations were performed in the isothermal-isobaric (NPT) ensemble. A Nosé-Hoover thermostat65,66was used to keep the temperature at 300 K with a coupling constant of 1 ps. A total of three simulations of 2500 ns were carried out for the apo complex with a time step of 2 fs by applying constraints on hydrogen atom bonds using the LINCS algorithm67. Similarly, the dimeric complex of RagA2B2 in the OO state was simulated for 500 ns. Cavity calculations were performed using CASTp68.
Extended Data
Extended Data Fig. 1. Unbiased peptide density in RagAB.
Stereo diagrams showing 2Fo-Fc (a) and Fo-Fc density (b) in RagAB KRAB before any modelling and refinement of the peptide. Selected segments of RagA (cyan) and RagB (green) neighbouring the peptide are shown. Map contouring parameters are 1.0σ, carve = 2 for the 2Fo-Fc and 3.0σ, carve = 2 for the Fo-Fc map. The extensive contacts of RagA with the peptide are confirmed by a PISA interface analysis17 which shows that 26 RagA residues form an interface with the peptide compared to only 8 for RagB, generating interface areas of 620 and 240 Å2with RagA and RagB respectively. The PISA CSS (complexation significance score) is maximal (1.0) for peptide-RagA while it is only 0.014 for peptide-RagB. This suggests that the observed co-crystal structure represents a state where the ligand has been partially transferred from an initial, presumably low(er)-affinity binding site on RagB to a high(er)-affinity binding site in the RagAB complex, allowing co-purification.
Extended Data Fig. 2. RagAB binds a wide range of oligopeptides.
a-c, LC-MS/MS analysis of peptides bound to RagAB W83 KRAB, showing length distribution (a), total charge (b) and pI (c). d-f, Analysis of peptides bound to RagAB W83 wild-type, showing length distribution (d), total charge (e) and pI (f). For charge calculations, the pH was assumed to be 7.0 and contributions of any His residues were ignored. g, Amino acid frequency of RagAB-bound peptides (KRAB and wild-type combined; black) vs. the amino acid composition in the P. gingivalis proteome (gray), showing a substantial enrichment of Ala, Glu, Lys, Thr and Val. By contrast, aromatics (Phe, Trp) and bulky hydrophobics (Leu) appear to be under-represented. The peptides bound to RagAB from W83 KRAB vary in length from 7 to 29 residues, with a broad maximum of around 13 residues that fits well with the peptide density observed in the structures. Assuming equal abundance of each detected peptide, there is a slight preference for neutral to slightly acidic peptides, and the pI distribution has a bimodal shape, with maxima for acidic and slightly basic peptides. Analysis of the smaller RagAB-bound peptide set from wild-type W83 (d-f) yields a slightly wider size range from 5-36 residues, but overall there are no dramatic differences in the collective length, net charge and pI of the RagAB-bound peptide populations from W83 KRAB and wild-type strains.
Extended Data Fig. 3. Molecular dynamics simulations of RagAB show lid opening.
a, Cα-rmsd values of RagB lids in apo-RagAB (red) and peptide-bound RagAB (green) with reference to the starting crystal structure in the closed conformation. The Cα-rmsd values of the RagB lids in RagA2B2 are shown in blue with reference to the OO EM state. Each point indicates an average of 50 ns simulation trajectory. b, Comparison of the RagA2B2 open conformation from EM (magenta) with the snapshot of the most open simulation at 2500 ns (green). c, Internal surface of peptide binding cavities in closed holo-RagAB and apo-RagAB, generated with CASTp68. The bound peptide from a RagAB subunit in the crystal structure was removed in silico to generate a closed apo-complex, and three independent MD simulations were performed. For one of the simulations, a clear opening of the RagB lid was observed, reminiscent of recent results for a SusCD transporter and supporting the notion that ligand removal resets the transporter to favour the open state16. None of the peptide-bound complexes shows lid opening on the timescale of the simulations. While this suggests that lid opening is less favourable in the ligand-bound state, it does not contradict our observation of open, ligand-bound complexes via EM. The EM structures allowed us to compare both open states, which showed that the RagB lid in the simulation opens less wide than that in the EM structure, at least during the timescale of the simulation. We also observed a partial closing of both RagB lids during a 1000 ns simulation starting from the OO EM state (a, blue curves). The r.m.s.d. values of both RagB subunits decrease from ~30 Å in the EM structure (t = 0 ns) to ~15 Å, which is similar to the opening observed in one of the apo-RagAB simulations starting from the closed structure. Thus, it appears that the energy minimum for the open state in the simulations is different from that in solution, for reasons that are not clear.
Extended Data Fig. 4. RagB moves as a rigid body during lid opening.
a, Superposition of RagA subunits in the open (yellow) and closed (cyan) RagAB complexes in the OC state, showing the rigid-body movement of RagB. Equivalent points are indicated by x,y,z (closed RagB; green) and by x*, y*, z* (open RagB; red). The arrow indicates the approximate pivot point in the N-terminus of RagB at the back of the complex. b, Superposition as in (a), viewed from the extracellular side. Lid opening results in displacements of up to 45 Å for main chain atoms at the front of the complex, furthest away from the RagB N-terminus. c, Superposition of the open and closed states of RagB, with the N-termini indicated. d, Extracellular view of superposed RagA, with selected loops labelled. The conformational changes upon lid movement are mostly confined to those parts of the protein that continue to interact with RagB at the back of the complex (L7-L9).
Extended Data Fig. 5. Local resolution-filtered cryoEM maps and evidence for NTE density.
a, CC, OC and OO states of RagAB filtered and coloured by local resolution. Corresponding Fourier Shell Correlation (FSC) curves are shown (right). b, Unsharpened maps of RagAB displayed at low contour levels to reveal diffuse density attributed to the NTE. CC, OC and OO states are coloured purple, blue and green respectively.
Extended Data Fig. 6. Analysis of RagAB mutants.
a, Representative growth curves (n = 3, mean ± standard error of the mean) for mutant W83 ragAB variants on BSA-MM. For comparison, W83 WT and ΔragAB strains are shown as well. b, Representative SDS-PAGE gel (n = 2) showing OM protein expression levels following removal of inner membrane proteins by sarkosyl treatment. The RagABmono and both RagA hinge loop mutant strains have a similar phenotype as ΔRagAB, suggesting they cannot take up oligopeptides produced by gingipains. However, the OM protein levels show that very little RagAB is present, so that no conclusions about functionality can be drawn. The RagB acidic loop mutants show intermediate growth on BSA-MM, suggesting that oligopeptide uptake is somewhat impaired. However, the OMP levels of the acidic loop mutants are substantially lower than wild type, suggesting that both acidic loop mutants are likely functional and arguing that the slow growth of rag-4 P. gingivalis strain ATCC 33277 on BSA-MM is not due to the absence of the acidic loop in RagB.
Extended Data Fig. 7. RagAB and RagB bind peptides selectively.
MST titration curves with the P4-FAM peptide for (a) RagAB from ATCC 33277, (b) RagAB from W83 and (c) Omp40-41 from W83 (negative control). The other panels show MST profiles for unlabelled P21, P12 or P4 binding to His-tag labelled W83 RagAB (d-f) and His-tag labelled W83 RagB (g-i). Experiments and listed Kd values represent the mean of three independent experiments ± SD.
Extended Data Fig. 8. Electron density comparison for different peptide ensembles.
a, Fo-Fc electron density maps (contoured at 3.0 σ) for the modelled peptide following final refinement for RagAB purified from W83 KRAB (left panel; 3.4 Å resolution), RagAB from W83 wild type co-crystallised with excess P21 peptide (middle panel; 2.6 Å), and RagAB from W83 wild type (right panel; 3.0 Å). Peptide sequence of the P21 co-crystal structure is arbitrarily modelled as QNGGANTSRGSAG, with numbering in italics according to the W83 KRAB peptide. Neighbouring residues D400-M407 of RagA are shown as cyan stick models for orientation purposes. b, Simulated annealing composite 2Fo-Fc omit maps for peptides bound to RagAB complexes as in (a). An annealing temperature of 500 K was used, with 5% of the models omitted. The fact that two different peptide ensembles produce similar maps as the P21 peptide, together with the inability to model the P21 sequence, suggests that the substrates are bound with register shifts and perhaps different chain directions. c, Stereo view of superposed 2Fo-Fc maps (made within Phenix; 1.0 σ, carve = 2.0) for WT RagAB in the absence (blue) and presence (orange) of P21 peptide, generated with the same high resolution cutoff (3.0 Å). For orientation purposes, the P21 peptide model is shown as sticks.
Extended Data Fig. 9. Schematic demonstrating the proposed mechanism of substrate capture and translocation by RagAB.
Peptide ligands to be imported by the RagAB system are predominantly generated by the action of gingipains on serum and tissue-derived proteins. 1. A lid-open state of RagAB permits peptide binding. 2. Contributions from both RagA and RagB to peptide binding elicits closure of the lid, forming the transport-competent state of the complex. This is signalled across the OM by perturbation of the TonB box region on the periplasmic side of the plug domain, making it accessible to TonB. 3. According to the literature consensus, TonB-mediated disruption of the plug permits substrate translocation and a return to the open state of RagAB.
Extended Data Fig. 10. Cryo-electron microscopy data support substrate-induced lid closure in RagAB.
a,b, 3D classes for RagAB ‘as purified’ (a) and in the presence of 50-fold excess P21 peptide (b). Classes corresponding to the CC, OC and OO states are coloured purple, blue and green respectively. Junk or ambiguous classes e.g. where RagA barrels are incomplete are coloured grey. In the presence of P21 there was no clear OO state whilst the the proportion of the CC state increased, supporting the proposed mechanism of substrate capture.
Supplementary Material
Acknowledgements
This work was supported by a Welcome Trust Investigator award (214222/Z/18/Z to BvdB). We would further like to thank personnel of the Diamond Light Source for beam time (Block Allocation Group numbers mx-13587 and mx-18598) and assistance with data collection. All EM was performed at the Astbury Biostructure Laboratory which was funded by the University of Leeds and the Wellcome Trust (108466/Z/15/Z). We thank Drs. Rebecca Thompson, Emma Hesketh and Dan Maskell for EM support. We would also like to thank T. Kantyka for help in designing MS experiments. This study was supported in part by National Science Centre, Poland grants UMO-2015/19/N/NZ1/00322 and UMO-2018/28/T/NZ1/00348 to MM, UMO-2016/23/N/NZ1/01513 to ZN, UMO-2018/29/N/NZ1/00992 to GPB, and UMO-2018/31/B/NZ1/03968 and NIDCR/DE 022597 (NIH) to JP. LC-MS/MS analysis was supported by the Novo Nordisk Foundation (BioMS). JBRW was supported by a Wellcome Trust 4 year PhD studentship (215064/Z/18/Z).
Footnotes
Author contributions
MM, JP and BvdB initiated the project. MM cultured cells, purified and crystallised proteins and performed MST binding experiments, with guidance from JP and BvdB. JBRW and SR determined cryo-electron microscopy structures, supervised by NR. ZN performed cloning and strain construction. GB carried out qPCR experiments, and CS and JJE performed the peptidomics analysis. KP performed the MD simulations, supervised by UK. BvdB purified and crystallised proteins and determined the RagAB crystal structures. AB collected crystallography data. The manuscript was written by BvdB with input from MM, JBRW, NR and JP.
Ethics declarations
Competing interests
The authors declare no competing interests.
Data availability
The data supporting the findings of this study are available from the corresponding authors upon reasonable request. Coordinates and structure factors that support the findings of this study have been deposited in the Protein Data Bank with accession codes 6SLI (KRAB RagAB), 6SLJ (WT RagAB) and 6SLN (WT RagAB + P21). EM structure coordinates have been deposited in the Electron Microscopy Data Bank with accession codes 6SM3 (CC), 6SMQ (OC) and 6SML (OO). The raw cryoEM movie mode micrographs for the primary dataset containing the CC, OC and OO structures, will be deposited in the EMPIAR database. Source data for Figs. 1a, 4a-d and Extended Data Figs. 2, 6a,b and 7 are included in this article and its Supplementary Information files.
References
- 1.Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr Microbial complexes in subgingival plaque. J Clin Periodontol. 1998;25:134–44. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- 2.Eke PI, et al. Update on prevalence of periodontitis in adults in the United States: NHANES 2009 to 2012. J Periodontol. 2015;86:611–22. doi: 10.1902/jop.2015.140520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominy SS, et al. Porphyromonas gingivalis in Alzheimer's disease brains: evidence for disease causation and treatment with small-molecule inhibitors. Sci Adv. 2019;5:eaau3333. doi: 10.1126/sciadv.aau3333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Potempa J, Mydel P, Koziel J. The case for periodontitis in the pathogenesis of rheumatoid arthritis. Nat Rev Rheumatol. 2017;13:606–620. doi: 10.1038/nrrheum.2017.132. [DOI] [PubMed] [Google Scholar]
- 5.Tonetti MS, et al. Treatment of periodontitis and endothelial function. N Engl J Med. 2007;356:911–920. doi: 10.1056/NEJMoa063186. [DOI] [PubMed] [Google Scholar]
- 6.Usher AK, Stockley RA. The link between chronic periodontitis and COPD: a common role for the neutrophil? BMC Med. 2013;11:241. doi: 10.1186/1741-7015-11-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bui FQ, et al. Association between periodontal pathogens and systemic disease. Biomed J. 2019;42:27–35. doi: 10.1016/j.bj.2018.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mayrand D, Holt SC. Biology of asaccharolytic black-pigmented Bacteroides species. Microbiol Rev. 1988;52:134–152. doi: 10.1128/mr.52.1.134-152.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nemoto TK, Ohara-Nemoto Y, Bezerra GA, Shimoyama Y, Kimura SA. Porphyromonas gingivalis periplasmic novel exopeptidase, acylpeptidyl oligopeptidase, releases N-acylated di- and tripeptides from oligopeptides. J Biol Chem. 2016;291:5913–5925. doi: 10.1074/jbc.M115.687566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potempa J, Banbula A, Travis J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontol. 2000;24:153–192. doi: 10.1034/j.1600-0757.2000.2240108.x. 2000. [DOI] [PubMed] [Google Scholar]
- 11.Grenier D, et al. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect Immun. 2001;69:5166–5172. doi: 10.1128/IAI.69.8.5166-5172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagano K, et al. Characterization of RagA and RagB in Porphyromonas gingivalis study using gene-deletion mutants. J Med Microbiol. 2007;56:1536–1548. doi: 10.1099/jmm.0.47289-0. [DOI] [PubMed] [Google Scholar]
- 13.Goulas T, et al. Structure of RagB, a major immunodominant outer-membrane surface receptor antigen of Porphyromonas gingivalis . Mol Oral Microbiol. 2016;31:472–485. doi: 10.1111/omi.12140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bolam DN, van den Berg B. TonB-dependent transport by the gut microbiota: novel aspects of an old problem. Curr Opin Struct Biol. 2018;51:35–43. doi: 10.1016/j.sbi.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 15.Bolam DN, Koropatkin NM. Glycan recognition by the Bacteroidetes Sus-like systems. Curr Opin Struct Biol. 2012;22:563–569. doi: 10.1016/j.sbi.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 16.Glenwright AJ, et al. Structural basis for nutrient acquisition by dominant members of the human gut microbiota. Nature. 2017;541:407–411. doi: 10.1038/nature20828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 18.Noinaj N, Guillier M, Barnard TJ, Buchanan SK. TonB-dependent transporters: regulation, structure, and function. Annu Rev Microbiol. 2010;64:43–60. doi: 10.1146/annurev.micro.112408.134247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Freed DM, Horanyi PS, Wiener MC, Cafiso DS. Conformational exchange in a membrane transport protein is altered in protein crystals. Biophys J. 2010;99:604–10. doi: 10.1016/j.bpj.2010.06.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hickman SJ, Cooper REM, Bellucci L, Paci E, Brockwell DJ. Gating of TonB-dependent transporters by substrate-specific forced remodelling. Nat Commun. 2017;8 doi: 10.1038/ncomms14804. 14804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gómez-Santos N, Glatter T, Koebnik R, Świątek-Połatyńska MA, Søgaard-Andersen LA. TonB-dependent transporter is required for secretion of protease PopC across the bacterial outer membrane. Nat Commun. 2019;10 doi: 10.1038/s41467-019-09366-9. 1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hall LM, et al. Sequence diversity and antigenic variation at the rag locus of Porphyromonas gingivalis . Infect Immun. 2005;73:4253–4262. doi: 10.1128/IAI.73.7.4253-4262.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Milner P, Batten JE, Curtis MA. Development of a simple chemically defined medium for Porphyromonas gingivalis: requirement for alpha-ketoglutarate. FEMS Microbiol Lett. 1996;140:125–30. doi: 10.1016/0378-1097(96)00159-0. [DOI] [PubMed] [Google Scholar]
- 24.Grenier D, et al. Role of gingipains in growth of Porphyromonas gingivalis in the presence of human serum albumin. Infect Immun. 2001;69:5166–72. doi: 10.1128/IAI.69.8.5166-5172.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagano K, et al. Trimeric structure of major outer membrane proteins homologous to OmpA in Porphyromonas gingivalis. J Bacteriol. 2005;187:902–11. doi: 10.1128/JB.187.3.902-911.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lamster IB, Ahlo JK. Analysis of gingival crevicular fluid as applied to the diagnosis of oral and systemic diseases. Ann N Y Acad Sci. 2007;1098:216–29. doi: 10.1196/annals.1384.027. [DOI] [PubMed] [Google Scholar]
- 27.Curtis MA, Hanley SA, Aduse-Opoku J. The rag locus of Porphyromonas gingivalis: a novel pathogenicity island. J Periodontal Res. 1999;34:400–405. doi: 10.1111/j.1600-0765.1999.tb02273.x. [DOI] [PubMed] [Google Scholar]
- 28.Josts I, Veith K, Tidow H. Ternary structure of the outer membrane transporter FoxA with resolved signalling domain provides insights into TonB-mediated siderophore uptake. Elife. 2019;8:e48528. doi: 10.7554/eLife.48528. pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim I, Stiefel A, Plantör S, Angerer A, Braun V. Transcription induction of the ferric citrate transport genes via the N-terminus of the FecA outer membrane protein, the Ton system and the electrochemical potential of the cytoplasmic membrane. Mol Microbiol. 1997;23:333–44. doi: 10.1046/j.1365-2958.1997.2401593.x. [DOI] [PubMed] [Google Scholar]
- 30.Braun V, Mahren S, Ogierman M. Regulation of the FecI-type ECF sigma factor by transmembrane signalling. Curr Opin Microbiol. 2003;6:173–80. doi: 10.1016/s1369-5274(03)00022-5. [DOI] [PubMed] [Google Scholar]
- 31.Koebnik R. TonB-dependent trans-envelope signalling: the exception or the rule? Trends Microbiol. 2005;13:343–7. doi: 10.1016/j.tim.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 32.Ho MH, Lamont RJ, Xie H. Identification of Streptococcus cristatus peptides that repress expression of virulence genes in Porphyromonas gingivalis . Sci Rep. 2017;7 doi: 10.1038/s41598-017-01551-4. 1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moynié L, et al. The complex of ferric-enterobactin with its transporter from Pseudomonas aeruginosa suggests a two-site model. Nat Commun. 2019;10 doi: 10.1038/s41467-019-11508-y. 3673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Balhesteros H, et al. TonB-Dependent Heme/Hemoglobin Utilization by Caulobacter crescentus HutA. J Bacteriol. 2017;199:e00723–16. doi: 10.1128/JB.00723-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Koropatkin NM, Martens EC, Gordon JI, Smith TJ. Starch catabolism by a prominent human gut symbiont is directed by the recognition of amylose helices. Structure. 2008;16:1105–15. doi: 10.1016/j.str.2008.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rogowski A, et al. Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun. 2015;6 doi: 10.1038/ncomms8481. 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nguyen KA, Travis J, Potempa J. Does the importance of the C-terminal residues in the maturation of RgpB from Porphyromonas gingivalis reveal a novel mechanism for protein export in a subgroup of Gram-Negative bacteria? J Bacteriol. 2007;189:833–843. doi: 10.1128/JB.01530-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gibson DG, et al. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 2009;6:343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- 39.Chiu J, March PE, Lee R, Tillett D. Site-directed, Ligase-Independent Mutagenesis (SLIM): a single-tube methodology approaching 100% efficiency in 4 h. Nucleic Acids Res. 2004;32:e174. doi: 10.1093/nar/gnh172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tagawa J, et al. Development of a novel plasmid vector pTIO-1 adapted for electrotransformation of Porphyromonas gingivalis . J Microbiol Methods. 2014;105:174–179. doi: 10.1016/j.mimet.2014.07.032. [DOI] [PubMed] [Google Scholar]
- 41.Belanger M, Rodrigues P, Progulske-Fox A. Genetic manipulation of Porphyromonas gingivalis . Curr Protoc Microbiol. 2007;5 doi: 10.1002/9780471729259.mc13c02s05. 13C.2.1–13C.2.24. [DOI] [PubMed] [Google Scholar]
- 42.Smith CJ. Genetic transformation of Bacteroides spp. using electroporation. Methods Mol Biol. 1995;47:161–169. doi: 10.1385/0-89603-310-4:161. [DOI] [PubMed] [Google Scholar]
- 43.Filip C, Fletcher G, Wulff JL, Earhart CF. Solubilization of the cytoplasmic membrane of Escherichia coli by the ionic detergent sodium-lauryl sarcosinate. J Bacteriol. 1973;115:717–722. doi: 10.1128/jb.115.3.717-722.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winter G, Lobley CM, Prince SM. Decision making in xia2. Acta Crystallogr D Biol Crystallogr. 2013;69:1260–73. doi: 10.1107/S0907444913015308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Winter G, et al. DIALS: implementation and evaluation of a new integration package. Acta Crystallogr D Struct Biol. 2018;74:85–97. doi: 10.1107/S2059798317017235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 49.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen VB, et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Scheres SHW. RELION: implementation of a Bayesian approach to cryo-EM structure determination. J Struct Biol. 2012;180:519–530. doi: 10.1016/j.jsb.2012.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7:e42166. doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Li X, Zheng SQ, Egami K, Agard DA, Cheng Y. Influence of electron dose rate on electron counting images recorded with the K2 camera. J Struct Biol. 2013;184:251–260. doi: 10.1016/j.jsb.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Thompson RF, Iadanza MG, Hesketh EL, Rawson S, Ranson NA. Collection, pre-processing and on-the-fly analysis of data for high-resolution, single-particle cryo-electron microscopy. Nat Protoc. 2019;14:100–118. doi: 10.1038/s41596-018-0084-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Wagner T, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:218. doi: 10.1038/s42003-019-0437-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petterson EF, et al. USCF Chimera – A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 59.Jo S, Kim T, Iyer VG, Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J Comput Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 60.Abraham MJ, et al. GROMACS: high performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1:19–25. [Google Scholar]
- 61.Best RB, et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi1 and chi2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Darden T, York D, Pedersen L. Particle mesh ewald: an N log (N) method for ewald sums in large systems. J Chem Phys. 1993;98:10089–10092. [Google Scholar]
- 64.Parrinello M, Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J Appl Phys. 1981;52:7182–7190. [Google Scholar]
- 65.Nosé SA. Molecular Dynamics Method for Simulations in the Canonical Ensemble. Mol Phys. 1984;52:255–268. [Google Scholar]
- 66.Hoover WG. Canonical dynamics: equilibrium phase-space distributions. Phys Rev A. 1985;31:1695. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 67.Hess B, Bekker H, Berendsen HJC, Fraaije JGEM. LINCS: A linear constraint solver for molecular simulations. J Comput Chem. 1997;18:1463–1472. [Google Scholar]
- 68.Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: computed atlas of surface topography of proteins. Nucleic Acids Res. 2018;46:W363–W367. doi: 10.1093/nar/gky473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schrödinger LLC. The PyMOL Molecular Graphics System, Version 1.8 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data supporting the findings of this study are available from the corresponding authors upon reasonable request. Coordinates and structure factors that support the findings of this study have been deposited in the Protein Data Bank with accession codes 6SLI (KRAB RagAB), 6SLJ (WT RagAB) and 6SLN (WT RagAB + P21). EM structure coordinates have been deposited in the Electron Microscopy Data Bank with accession codes 6SM3 (CC), 6SMQ (OC) and 6SML (OO). The raw cryoEM movie mode micrographs for the primary dataset containing the CC, OC and OO structures, will be deposited in the EMPIAR database. Source data for Figs. 1a, 4a-d and Extended Data Figs. 2, 6a,b and 7 are included in this article and its Supplementary Information files.