Abstract
Studying within-species variation has traditionally been limited to culturable bacterial isolates and low-resolution microbial community fingerprinting. Metagenomic sequencing and technical advances have enabled culture-free, high-resolution strain and subspecies analyses at high throughput and in complex environments. This holds great scientific promise but has also led to an overwhelming number of methods and terms to describe infraspecific variation. This Review aims to clarify these advances by focusing on the diversity within bacterial and archaeal species in the context of microbiomics. We cover foundational microevolutionary concepts relevant to population genetics and summarise how within-species variation can be studied and stratified directly within microbial communities with a focus on metagenomics. Finally, we describe how common applications of within-species variation can be achieved using metagenomic data. We aim to guide the selection of appropriate terms and analytical approaches to facilitate researchers in benefiting from the increasing availability of large, high-resolution microbiome genetic sequencing data.
Introduction
For over a century, bacterial cultivation has enabled the isolation and classification of thousands of bacterial strains. Through these efforts, a species concept was translated in the bacterial context as a group of individuals that form a coherent genomic cluster1 (see below for details and disagreements). Despite this genetic similarity, it was also established that a large magnitude of phenotypic variance is possible among strains from the same species (’conspecific’ strains). The importance of variability within species has been particularly well studied in the context of pathogenicity, and many species have been found to have both pathogenic and commensal strains (for example, Escherichia coli 2 and Bacteroides fragilis 3). Indeed, a classic example are E. coli strains, which can be pathogenic, commensal, host- associated or environmental2. The relationship between strain identity and host health demonstrates how it can be insufficient to study microbial communities at species level resolution, and the same applies in many other areas, such as drug response4, nutrient cycling5, nitrogen fixation6 and host association7.
Cultivation-based approaches have a fundamental8 and continued9 role in studying within- species variation but, despite their recent methodological progress10, they have important limitations. Few microorganisms can be easily cultivated under isolated, laboratory conditions, and cultivation is typically low-throughput. Even when culturing is possible, organisms are then studied in isolation and not their natural community setting. Culture-free, strain-level analysis of entire microbiomes has been possible for over 15 years11–16, but it has been limited due to shallow read depths and small sample sizes. With recent technological and algorithmic innovations in metagenomics (Box 1) and decreasing sequencing costs, large-scale metagenomic analysis of variation within species has become feasible. There is great promise in these approaches17–20 and they have vastly increased the rate of discovery, but they are also leading to scientific and semantic challenges.
Box 1. Molecular approaches to characterize variation within species.
A wide range of methods are available for studying within-species variation, either based on cultured isolates or directly in microbial communities.
Microbiome-based methods are less established but are not limited to culturable microbiota. Foundational community-fingerprinting methods like DGGE, TRFLP and ARISA181,182 enabled some species to be studied at high resolution without culturing. Due to their low- throughput and limited resolution, these methods have largely been superseded by genetic sequencing approaches. Despite its origin as a low-resolution method, 16S rRNA gene amplicon analysis methods can sometimes now differentiate within some species using Oligotyping183,184, amplicon sequence variants (ASV)185–187 and SNVs in full gene sequences188. However, 16S rRNA approaches remain extremely limited in resolution for within-species analysis and can be confounded by multiple, non-identical copies of the 16S rRNA gene per genome188.
Shotgun metagenomic sequencing provides more information by considering more marker genes or whole genomes. Many tools have been developed to analyse metagenomic data to describe variation within species19,20. The major approaches include: SNV-based profiling, either within predefined marker genes56,90,98,105 or across whole species-reference genomes103,104,119, overall similarity to strain-reference genomes92,93, sequence typing116 and gene content-based profiling95. Metagenomic assembled genomes (MAGs) can be recovered by binning and assembling of co-abundant genes189; however, these come with important limitations (Box 3).
Non-microbiomic but culture-free methods include microfluidics-based techniques that enable organism-specific enrichment prior to sequencing190,191; and single-cell sequencing, which produces single amplified genomes (SAGs)192. Culturing is becoming possible for a growing number of bacteria due to methodological advances, such as culturomics, which combines the use of multiple culture conditions with rapid bacterial identification10.
Non-genomic approaches, such as cryo-electron microscopy-based imaging and transcriptome, proteome- and metabolome-based profiling methods can capture phenotypic differences within species and can be used both separately and in conjunction with genomic approaches. These methods range from well-established, such as serotyping and functional profiling, to more recent and high-throughput, such as thermal proteome profiling140.
In the traditional cultivation approach, ’strain’ refers to a pure culture or isolate, denoting a taxonomic entity rather than a natural concept21. This operational definition cannot be transferred directly to the modern culture-free approaches and a widely accepted, biologically meaningful definition of strain remains elusive. Exacerbating this situation, and perhaps in response to the lack of generally accepted terminology, a plethora of overlapping terms have been coined in high-resolution microbiome studies and are often poorly defined. The resulting confusion impedes communication and synergy among researchers both in microbiome fields and beyond. To place new operational definitions in the correct context of existing conceptual definitions of within-species variation, it is essential to understand the microevolutionary processes that create and constrain variation within species.
In this Review, we summarise the processes that produce and constrain variation within species, and describe how the balance of these forces shapes the magnitude and structure of the variation. We then provide an overview of the major ways in which this variation can be studied and stratified into categories using metagenomic data and define commonly used terminology, which we then put into the context of applications. We use ’within-species variant’ to refer to any grouping below the species level. Throughout this Review, we highlight the advances and challenges that are resulting from the use of metagenomic data to study within-species diversity.
Variation and cohesion within species
Processes leading to within-species variation.
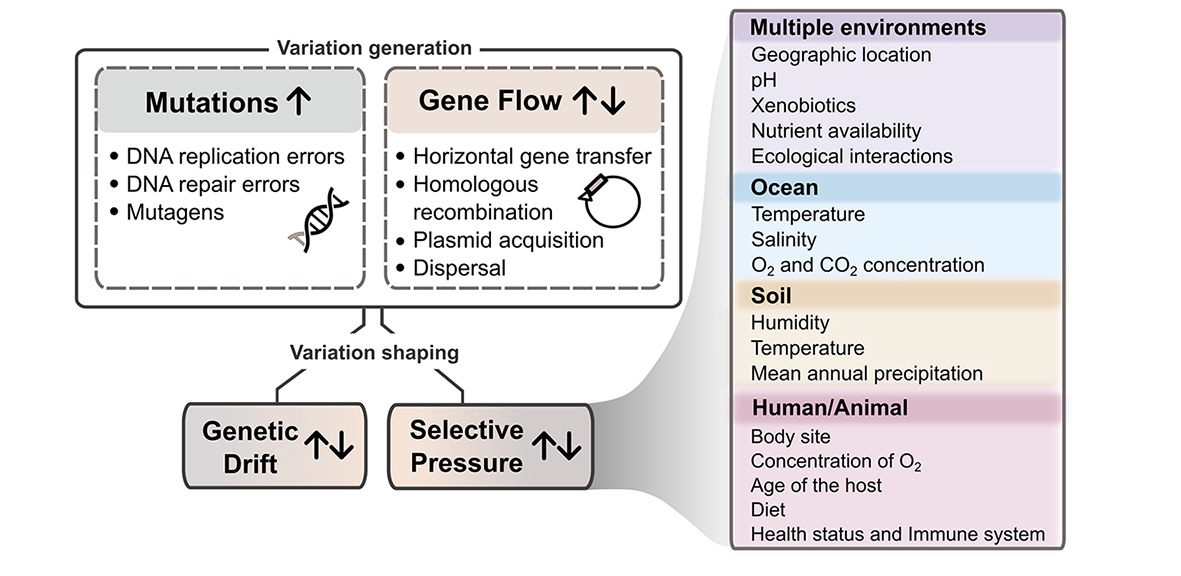
Diversity within species is the result of continuous processes of variation generation and subsequent selection and drift (Figure 1). Mutations and gene flow introduce genetic variability into otherwise identical lineages of clonal daughter cells.
Figure 1.
Drivers of variability within bacterial species. Within-species variability is introduced by mutations, which usually increase the amount of variations within a species (up arrow), and gene flow mechanisms, which can increase or decrease the amount of variation within a species. This variability is shaped by genetic drift and selective pressure, which can also increase or decrease the amount of variation. Selective pressures are shaped by many biotic and abiotic factors, some of which are known to drive adaptation in particular habitats more than others.
Mutations (that is, substitutions, insertions, deletions and inversions) arise continuously in the genome due to errors in the DNA replication process, damages caused by mutagens or errors in the DNA repair and recombination mechanisms22. Although the typical mutation rate for double-helix DNA-based organisms is approximately 1 nucleotide change per 109 nucleotides per replication23, mutation rates can vary across and within species by orders of magnitude24. Selection for lower or higher rates balances the metabolic cost of reducing mutation frequency versus the impact of deleterious mutations25. The direction of this balancing depends on habitat conditions, population size and mutator allele strength25. The rate of accumulation of mutations within a lineage of bacteria depends on the mutation rate, as well as natural selection and genetic drift that act upon the mutations. This further diversifies the observed rates of mutation. For example, non-lethal rates of mutation have been observed from 10-9 to 10-3 mutations per genome per generation in Vibrio species26,27. Further, not all portions of the bacterial genome are equally subject to mutations. Mutation accumulation rates are higher in accessory genes than in core genes, unless a core gene is located near accessory genes or mobile genetic elements, and higher in secondary chromosomes than in primary chromosomes28,29. In general, deletions are more frequent than insertions and non-functional sequences are readily lost from bacterial genomes30,31. Mutations that arise in one genome can be passed vertically to descendants or horizontally to neighbouring cells.
The transfer of genetic variation from one population to another (gene flow) can cause rapid and large-scale additions and rearrangements of genomic regions32. DNA can be transferred between cells by horizontal gene transfer (HGT) via transformation, transduction, conjugation, gene transfer agents and membrane vesicles33,34. Newly acquired donor DNA can stay separate within the acceptor cell (for example, as a plasmid or lytic phage) or can be incorporated into the genome of the acceptor through a number of mechanisms34, including homologous recombination 35. HGT is more frequent within species, but it can also occur between species36. It can result in replacement of genetic segments with donor homologs, often within species via homologous recombination, or in acquisition of new genetic material. In terms of impact on within-species variation, the most important factor of HGT is not the mechanism (for example, homologous recombination) but rather whether or not the genetic material being transferred is novel to the recipient population or species (discussed below). The main processes limiting HGT include lack of surface compatibility for the conjugative process, CRISPR-mediated microbial immunity37 and restricted host specificity of bacteriophages34. Notorious examples of HGT between conspecific [G ] variants include two cases where toxin genes were transferred from toxigenic to non-toxigenic strains in Clostridioides difficile 38 and in E. coli39, with the latter causing 54 deaths in 2011 in Germany.
Natural selection and genetic drift determine the fate of within-species variation introduced through mutation and gene flow. Genetic drift randomly eliminates genetic variations within a population, whereas natural selection maintains or eliminates variations that confer a fitness advantage or disadvantage, respectively. In this context, the effect of natural selection is limited by the background noise of genetic drift40. Natural selection is driven by a multitude of biotic and abiotic factors that differentially influence the survival and replicative capability of species subpopulations (Figure 1). These factors can shape the composition of microbial communities at the species and within-species levels through community assembly41 and classic evolutionary forces. Selective pressure factors vary from habitat to habitat and can include pH, temperature, oxygen and other gas concentration, nutrient availability, direct competition or commensalism with other bacteria, predation by phages and eukaryotes, and presence of stress-inducing xenobiotics such as drugs, anti-microbial compounds and heavy metals.
Species definitions and mechanisms of species cohesion.
With the vertical accumulation of mutations and the horizontal acquisition of genes, variation among the descendants of one cell could constantly increase, creating a continuous landscape of genetic variation across bacterial genomes. However, when genomic similarities are compared across bacteria, distinct clusters are observed. These clusters are thought of as species in bacteria42, though the applicability of a ’species’ concept is contested43. In this Review, we use the word ’species’ to reflect these clusters of genetic similarity.
For many decades, bacterial species delineation based on genome similarity has been measured using DNAߝDNA hybridization (DDH). According to the bacterial nomenclature code, conspecific genomes have >70% similarity by DDH. Increasingly, DDH is complemented or replaced by DNA sequencing of isolates and average nucleotide identity (ANI) comparisons8,44, with approximately >70% similarity in DDH corresponding to >94% of ANI in the core genome and >96% in universal marker genes 7,45–49 The approximation in these correspondences can affect classification, as in the case of Fusobacterium nucleatum, for which subspecies were defined based on DNA–DNA hybridization50 but then suggested to be reclassified as separate species after reassessment with in silico measurements of ANI51. As suggested by early studies13,52,53, the presence of a distinctive bacterial species boundary is identifiable using metagenomic data and was recently confirmed by large-scale studies, which identified this boundary at ANI thresholds based on whole genomes (~95%)54,55 and on markers genes (96.5%)48,56 and also described a drastic drop in gene flow in core genomes.
Despite the overall consistency of genomic ANI data, defining bacterial and archaeal species remains controversial, with over twenty conceptual definitions of ’species’57–60 and some researchers questioning the concept altogether43. The biological and the phylogenetic concepts of species are the most applicable for bacteria and archaea61. The former defines species as a group of individuals that can interbreed resulting in viable offspring, which translates to the possibility for homologous recombination in Prokaryotes, whereas the latter defines species as clades that are characterized by distinctive phenotypic properties. Both concepts predict a decline in the homologous recombination36,62 and HGT63 rates between different species. The multitude of potential species definitions are not necessarily well served by ANI-based genome comparisons alone. Instead, other methods can be used to operationally define species, in addition to, or in place of ANI, such as by phenotype, similarity in universal-single copy marker genes (for example, 16S rRNA), and gene content46,64.
The genomic similarity within species is called ’cohesion’. This is maintained predominantly through within-species recombination and selection against lower-fitness alleles55,65. If an allele is more beneficial than all others in a population, it can spread completely through that population, resulting in a ’hard selective sweep’ 66,67. When recombination rates are low, it is likely that the whole genome will hitchhike to prevalence along with this adaptive allele, resulting in a ’genome-wide selective sweep’68. When hard, whole-genome selective sweeps occur, they can reduce diversity within a species and maintain dissimilarity between species65,69,70.
Determinants of magnitude and structure of variation within a species.
Diversity within species is generated, maintained and purged to different extents, such that some species are highly heterogenous whereas others are tightly cohesive. These features of within-species variation depend on the populations observed (Box 3) and can be described globally or locally. The balance between the forces that increase diversity and those that maintain cohesion shapes both the magnitude and the structure of variation within a species.
Box 3. Challenges in studying variation within species in metagenomics.
Investigations of variation within species in microbial communities are faced with study- design, technical and methodological challenges. A main study-design challenge is the ’unobserved variation’ paradigm: you do not see what you do not sample. If low variability is seen within a species, it is difficult to prove that it is not due to under-sampling or sampling bias. This bias can be temporal (for example, due to strain turnover or extinctions) or spatial (for example, due to proximate sampling areas, such as soil or skin, harbouring substantially different infraspecific profiles). Shallow sequencing depth also biases against observing low abundance within-species variants. These biases are mitigated by the increasing number of deeply sequenced metagenomic samples. However, integration of these samples across studies is still faced with technical challenges well-known in metagenomics195,197–199.
Although undoubtedly useful for investigating unknown and under-represented species, metagenomic assembled genomes (MAGs) have important limitations. MAGs are population consensus genomes, thus, loci may be polyallelic and unlinked164,196. When compared to isolate genomes, MAGs often have low assembly quality, are less complete and are more likely chimeric103,164,192,196,200. For these reasons, and due to the difficulty in detecting chimeras below the species level, MAGs should not be considered equivalent to genomes sequenced from isolates196. The use of the term ’complete MAG’ (CMAG) should be adopted only for MAGs that are analogous to complete isolate genomes, which are usually a single circular contig with no gaps.
To avoid confusing isolate genomes and MAGs, the growing practice of uploading MAGs to public genome databases196 should be discouraged and the phrase ’genome-resolved metagenomics’ should not be used for MAG studies that do not directly assess heterogeneity within MAGs. Single-cell sequencing approaches provide a promising alternative to MAGs for recovering genomes from metagenomes, but are limited by high cost, low throughput, potential contamination and quality issues due to using a single molecule of DNA201.
Continued technical advances, decreasing sequencing costs, and increasing integration of complementary methodologies will be necessary to counteract these challenges in data generation and integration.
The amount of variation generated within a species depends on the mutation rate, generation time, tendency for inter-species HGT, and population size, whereas the amount of variation that persists depends on the stringency of selective pressures in its habitats, the population size71, and the frequency and severity of selective sweeps. The balance between divergence and cohesion is modulated by selection and drift, which are shaped by biotic and abiotic factors of the ecological niche (Figure 1). HGT can increase the genetic variation within a population if the material being transferred is novel to the receiving population, for example if the donor cell was dispersed from a foreign population or is distantly related. Conversely, HGT can homogenise a population in terms of specific gene content or single nucleotide variant (SNV) presence if it spreads this genetic material throughout the population, resulting in a gene-specific hard selective sweep 72.
Within a species, a structured population can arise due to a combination of soft selective sweeps — when multiple alternative adaptive alleles spread and coexist in a population73— along with drift and dispersal into new locations with similar or new ecological niches. For instance, when the rate of mutation generation is high and the rate of within-species recombination is low, strains may diverge into subgroups that are more internally cohesive relative to one another. Specifically, reduction of the ratio between recombination to mutation (r/m) events below 0.25 seems to enable subpopulations to diverge freely36. This may result in the establishment of subspecies74,75, which are groups of strains with partially disrupted gene flow that might be in the process speciation.
Sub-speciation can be caused or accelerated by physical or geographic barriers that block gene flow between sub-speciating groups (’allopatric’), which leads to divergence of subspecies either due to natural selection or drift76. However, sub-speciation can also happen without spatial separation (’sympatric’). In this case, it is likely that there is a selective advantage to specialization, for example, to diminish competition for resources. Due to the extreme dispersibility of bacteria and archaea, complete physical blocks to gene flow may be rare and there might be in-between scenarios. When occasional gene flow occurs and niches overlap, purifying selection can maintain partial cohesion between subspecies, which can prevent divergence from establishing stable subspecies77.
At one extreme, species can be monotypic; that is they have a uniform or ’smeared’ distribution of genetic similarities across their entire population. Monotypic species with low diversity are more likely to be specialists, with narrow geographic distributions or host ranges, or are the product of recent speciation78,79. Chlamydia trachomatis is an example of a monotypic low diversity intracellular pathogenic species80. At the other extreme, species with subspecies (’polytypic’) and high diversity are more likely to be free-living generalists with multiple adaptations to distinct and fluctuating environments, with broad geographic ranges or many partially overlapping niches77,79. For example, E. coli has at least six phylogroups that tend to be more prevalent in different habitats81.
Much of the fundamental knowledge described above was obtained on a species-by-species basis through culture- and isolation-based experiments. The rise of microbiomic approaches enables the characterisation of variation across many species at a large scale and offers promising new research avenues (Box 2). To meaningfully place these new findings into context it is important to adapt concepts and terminology from this body of knowledge appropriately for use in metagenomic studies.
Box 2. Culturing isolates versus metagenomics for analysis of variation within species.
Traditionally, investigations below the species level have relied on studying cultured isolates. With the rise of metagenomics, the amount of high-resolution genetic data has increased. Generally, this data is analysed based on variation within specific genetic segments (for example, marker genes) or within genomes recovered through assembly (MAGs) (Box 1). Although this enables unprecedented discoveries due to the large scale of data produced, these new methods also have important limitations and introduce new complexity (see the table). Although metagenomics provides important new benefits over studying isolates, the two methods remain complementary9,193. To ensure future synergy between the two approaches, isolate genome and metagenomic assembled genome (MAG) data quality must be readily available and comparable, and a common vocabulary should be maintained.
| Criteria | Culturing isolates | Metagenomic sequencing |
|---|---|---|
| Scope of microorganisms that can be studied below the species level | Must be culturable in isolation but can be low abundance in original sample | Must be abundant or deeply sequenced |
| Ability to describe multiple species variants within one sample (of the same or of different species) | Requires multiple rounds of isolation Intractable for low abundance variants | Can be determined from sequencing data from one sample, but sufficient sequencing depth required to distinguish from sequencing error |
| Ability to determine whether genetic variants originate from same organism (genetic linkage) | Possible (as long as variation within isolate colony is low, which is normally the case) | Very difficult or impossible in current typical approach but improvements are possible; for example, long reads, time- series data and Hi-C sequencing194 |
| Ability to put species variant in context of community | Limited and work intensive | Implicitly supported, though biases exist147,161,195 |
| Ability to describe phenotypic differences between within- species variants | Heterogeneity can be assessed133 with clinical, environmental and industrial relevance | Limited to description of potential phenotypes |
| Support for follow-up study | Isolates can be directly experimented on (for example, response to drug exposures) | Extracted DNA can be further tested molecularly (for example, PCR) |
| Main method for genome recovery | Isolate shotgun genomic sequencing and assembly | Shotgun metagenomic DNA sequencing followed by assembly (MAG; Box 1) |
| Quality of the recovered genomes | Often remain at draft level but usually are high quality with little contamination | May have higher error rates and be chimeric, contaminated and incomplete192,196 |
| Quality assessment of the recovered genomes | Provided by central repositories, with various guidelines developed (see, for example, Ref.42) | Routinely assessed but ad hoc by authors Recommendations are emerging192 |
| Determining presence or absence of gene in the recovered genome | Usually simple and correct | Difficult to be certain |
| Expected impact of long read sequencing | Longer contigs, less challenged by repetitive regions | Better genomes for the most abundant organisms, low abundance fraction still hard to access |
Stratification of within-species variation
Within-species variation often needs to be stratified into meaningful groups to be studied and associated with categorical variables, such as health status, geographic location or metabolic capability. The theory described above can support conceptual definitions of such groups, but these generally cannot be used directly in microbiological studies. Instead, operational definitions of variant groups must be devised based on criteria that can be measured. Typically, this is done on genetic or phenotypic scales. The appropriate metrics to use to operationally define variant groups, such as ’strains’, depends on the biological questions being asked and the methodology being used (Figure 2A).
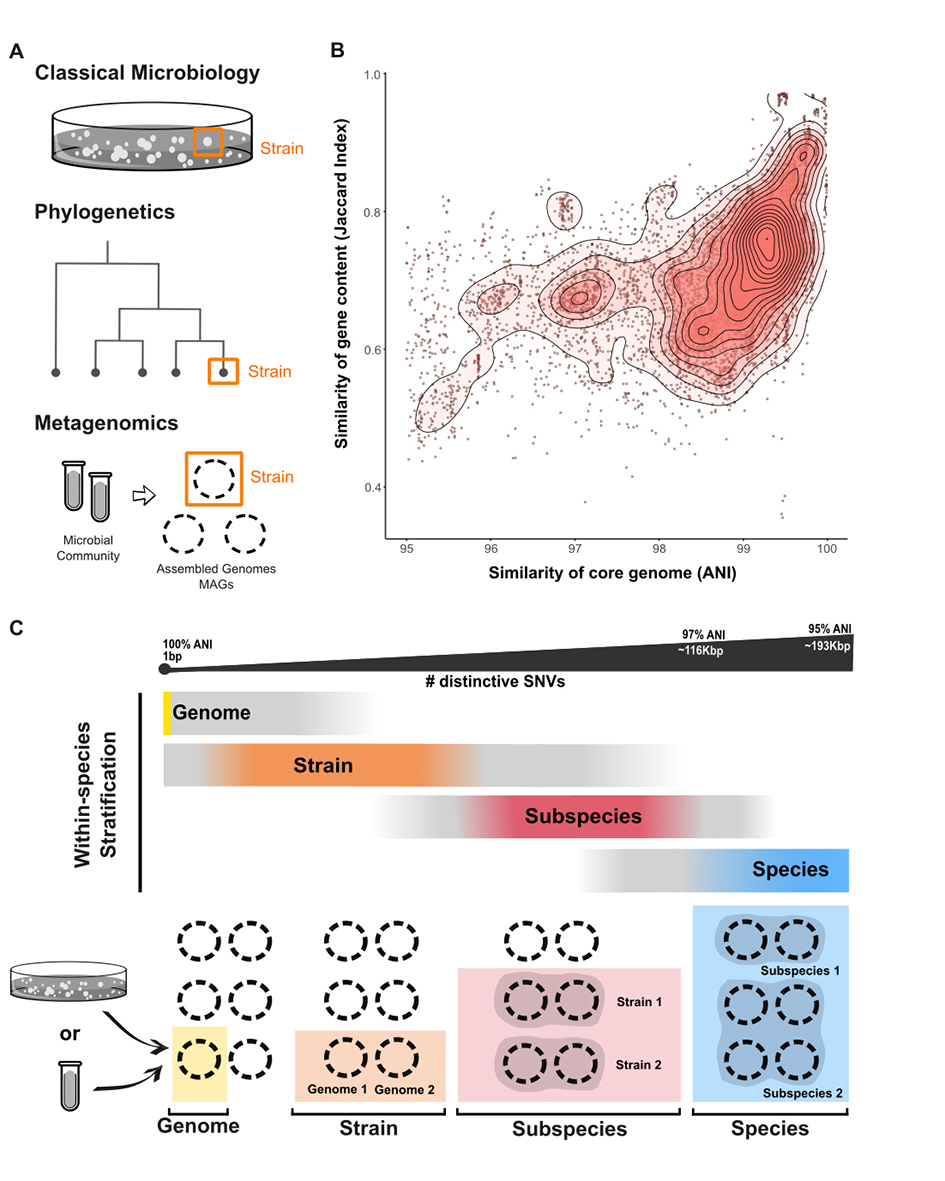
Figure 2.
Within-species stratifications (a) Different operational definitions of ’strain’, based on the field of investigation: a cultured isolate in classic microbiology, a leaf node in a phylogenetic tree, and a metagenomic assembled genome (MAG) in metagenomics(b) Each point is a pairwise-comparison of one isolate genome versus all other conspecific isolate genomes. The data99 is from 155 bacterial species, each with at least 10 sequenced isolate genomes. Opacity of red-coloured topographical overlay indicates density of points. The plot shows the relationship between the similarity of the core genome, measured by average nucleotide identity (ANI), versus the similarity of gene content, measured by Jaccard Index. Genomes with higher similarity between their core gene sequences tend to have more genes in common (Spearman correlation R=0.57, p < 2.2e-16). However, high ANI does not necessarily imply highly similar gene content, with many genomes with over 99% core genome ANI having less than 70% of genes in common. Most within-species ANI values are greater than 97%, the few data points below 95% ANI are not shown (83% and 4% of data points, respectively). The data are adapted from Ref".(c) Spatial distribution of key terminology used to stratify variation within bacterial species, ranging from a single nucleotide variation in the whole genome to the species-level threshold (97% ANI). The coloured portions of the bars reflect the recommended scope of use for each term, and the grey portions indicate the common, often unspecific, scope of use. Broadly speaking, conspecific genomes have identical nucleotides at homologous positions across 97% of their genome (97% ANI), which corresponds to differing on the order of 116,000 SNVs based on an average bacterial genome (3.87Mb180). The bottom panel illustrates the hierarchy of these terms, with a species potentially containing multiple subspecies, a subspecies containing multiple strains, and a strain containing multiple (non-identical) genomes. These genomes can be sequenced from cultured isolates or through assembly of a metagenomic sample, creating a MAG which represents the consensus genome of a population of cells.
Genetic stratification using metagenomic data.
Within-species genetic variation can be measured in many ways, some common metrics being overall genome similarity, the number of shared and unique genes, and/or the number and nature of SNVs. In this section, we discuss how these measures are taken, and explore their strengths and limitations. When these analytical approaches are applied to the large amount of data produced by metagenomic sequencing, within-species profiling can be performed in a high-throughput manner simultaneously for many species (see, for example, Refs 75,82–90 and examples below). However, this also raises various data quality issues, such as incomplete and partially erroneous data, as well as technical challenges, such as large computational and storage requirements.
Overall genome similarity at infra-specific levels can be assessed from metagenomic data either directly from reads and reference genomes91–93 or through comparisons of metagenomic assembled genomes (MAGs)54. Reference-genome based approaches can be limited by low availability of appropriate reference genomes, especially in non-human microbiomes. Large sets of MAGs are now available and methods to calculate ANI have improved in efficiency94. However, calculating ANI for large genomic cohorts remains computationally challenging54. Further, using MAGs in ANI comparisons can introduce inaccuracies due to data quality limitations and incompleteness (Box 3).
Decline of ANI and recombination rate can be indicators of ongoing subdivision of a species57. However, in contrast to species boundaries, within-species variants do not seem to display a universal threshold based on genome or marker genes that would categorise them into groups. Instead, the range and distribution of ANI values within species varies by taxon and population54, which limits its utility for broad stratification. Further, genetic differences that are coded by a small number of nucleotides relative to the size of the genome, and thus have a small impact on ANI, can have a very large impact on phenotype. Therefore, at the small scale of ANI differences that occur within species, measures of gene content, SNVs and indels are more informative than ANI for defining biologically relevant within-species variants.
Gene content is the sum of all genes in a genome, including core genes (which are present in almost all conspecific variants) and accessory genes (which are only present in a subset). Differences in accessory gene content between variants can arise at the single-gene level95 or at the genetic-segment level82, which can include multiple genes (’structural variation’). Gene content differences can be calculated based either on the presence or absence of a gene96, or additionally on the number of copies of that gene97. Gene order (’synteny’) is considered within structural variation, but has not yet been addressed directly by metagenomic methods. Metagenomic data can be used to study within-species gene content variation by looking for gene clusters95 or by associating gene content with variants defined by SNV profiles75,98. The relationship between gene content similarity and phylogeny is complicated by HGT. However, comparative studies of conspecific genomes have shown that pairwise similarity based on gene content is correlated with pairwise similarities based on core genome ANI99,100 (Figure 2B), and that distinct SNV profiles can correspond to distinct gene profiles75.
SNV differences can be used to compare conspecific variants at high resolution. These comparisons can consider the number of variant positions, their locations (for example, in core genes, accessory genes or intergenic regions), their spread across the genome (clustered or disperse) and their potential phenotypic impact (for example, synonymous or nonsynonymous mutations). In metagenomes, the identification of SNVs can be de novo98,101, based on MAGs102, or based on pre-existing reference genes or genomes103–105. The degree of similarity between the references and the actual community members can have a big impact on the accuracy of the results106. Identifying SNVs based on MAGs can reveal population dynamics, such as hard and soft selective sweeps in populations of lake bacteria102, but can also introduce errors due to the potential low quality of MAG references (Box 3). Groups of conspecific genomes can be defined from metagenomic data based on the distinctive presence of SNVs (for example, ’SNP-types’ (Ref. 107)); from thousands of SNVs indicating population structure by defining subspecies75 and subpopulations108, to tens of SNVs delimiting strains107. Isolate data has been used to show that single SNV differences can determine phenotype, such as pathogenicity109,110 or antimicrobial drug resistance111,112. The ability to detect low abundance SNVs in microbiomic data is limited when sequencing depths are shallow and population sizes are large. When SNVs are likely to have been vertically transferred, then they can be used to define haplotypes and lineages. Extending this approach, SNVs can be used to reconstruct phylogeny within a species113; however, care must be taken to use loci that are unlikely to have been in an HGT region, such as housekeeping genes114.
When multiple genetic variants are in one chromosome they are ’linked’. Linked variants are inherited together, but this linkage can be disrupted by recombination or mutation. Determining the linkage between alleles can be used to track lineages, reconstruct haplotypes (’phasing variants’) and detect HGT. However, metagenomic data is inherently limited in providing linkage data when the typical short-read, shotgun sequencing approach is used because this method breaks up DNA. Assembling short reads may be able to recover linkage information; however chimerism is common when there are multiple highly similar genomes within one sample, such as multiple conspecific strains (’strain heterogeneity’). Instead of exact profiles of linked alleles, shotgun metagenomics is usually limited to providing sets of multiallelic loci with allele frequency information. These can still be useful for many applications, as described in the final section of this Review. They can also be used to perform population genetic analyses for a species, such as to calculate estimates of population diversity (for example, n (Pi) - diversity or average pairwise genetic difference (between individuals)), population structure (for example, fixation index (Fst) or allele similarity between populations) and selection pressure (dN/dS, pN/pS, Tadjima’s D, or Fay and Wu's H)115.
Many software tools have been developed to measure and categorise diversity within species using metagenomic data. Generally, these have two broad aims: classification and discovery. Classification-oriented tools (for example, metaMLST)116, PathoScope93, MetaPhlAn2117, StrainSifter118, Sigma92, SPARSE91 StrainEst119) aim to detect if a known, characterised, within-species group (for example, a target genome, named strain, classic typed subspecies or MLST type) is present in a sample. Discovery-oriented tools typically group within-species variation into clusters of similarity using one of three measures: gene content (for example, PanPhlAn95), SNVs in whole or core genomes (for example, metaSNV104) or SNVs in marker genes (for example, Lineages algorithm120, ConStrains107, StrainPhlAn105, DESMAN98, StrainFinder121, mOTUs256), which might be followed up with detection of distinctive gene content (for example, DESMAN98). Although many tools claim to provide ’strain level’ resolution, the term ’strain’ is defined differently across software (see next section for discussion of definitions). The tools that can recover SNV linkage information de novo from SNV abundances across samples include ConStrains107, DESMAN98, StrainFinder121, and the Lineages algorithm120. When the assumption can be made that samples contain a single dominant within-species group, tools like StrainPhlAn105 and metaSNV104 can also be used to cluster SNVs into within-species groups (’strains’ and ’subspecies’, respectively).
Although these tools enable many applications of metagenomic data to study within-species variation (see below) they have some important limitations. For example, tools that rely on mapping reads to reference genomes or marker genes are inherently limited by the availability of appropriate reference genomes, which in some environments is very low (for example, freshwater and soil). This limitation can be circumvented by building and using MAGs (for example, as in DESMAN), but MAG quality concerns must be considered, especially if time-series data is not available (Box 3). Other logistical limitations include requiring an extremely high depth of coverage (for example, reported87,98 limitation for ConStrains) and not being able to handle large magnitudes of data (for example, reported98,104 limitation for Lineages algorithm). These selected examples demonstrate how limitations of foundational software can arise as the metagenomic field progresses towards larger and more complex datasets. These and other limitations result in tools being difficult or impossible to run, or not feasible to use with current reasonably sized datasets, preventing results from being reproducible or extendible.
The software referenced in this Review are examples of tools that reportedly perform the methodological approaches described. These references are not endorsements or reports of accuracy or usability. The reported features of many tools have been compared in recent reviews19,20, but a thorough comparison of accuracies has not yet been completed (although are expected to be addressed in the Critical Assessment of Metagenome Interpretation (CAMI)122 framework). Future work is expected to make comparisons for within-species analysis software; however, what exactly is meant by the specific terminology of each tool (for example, ’SNV-type’, ’strain populations’ etc.) and their mapping to common terms (for example, strain, subspecies) will have to be carefully handled.
Terms for genetic stratification.
There are many terms that stratify variation within species (Table 1). Out of the terms that are both most commonly used and recognised by the International Code of Nomenclature of Prokaryotes44, we highlight three terms to cover the range of genetic variation within species: genome, strain and subspecies (Figure 2C). In this section, we discuss conflicts in the usage of these terms in culture-based microbiology and metagenomics and suggest solutions.
Table 1. Definitions of terms used to stratify or describe variation within species.
| Term | Definition | Notes |
|---|---|---|
| Genotype | The set of alleles of an organism | Variable throughout time due to mutation and recombination |
| Haplotype | Set of alleles or single nucleotide variants (SNVs) that are inherited together from a single parent165 | Genetic signature of a lineage or clonal line, which can be disrupted through recombination |
| Haplogroup | Group of similar haplotypes with a common ancestor that has a clade-specific SNV or SNVs166 | In human context, used to describe a group of people that share a common ancestor |
| Lineage and sublineage | Unbranched sequence of ancestral and descendant entities. Each ancestor may have multiple descendants, but only one is included in the lineage. Each entity could be an organism, clade, population or subspecies, among others. 167 A sublineage is a subsection of a lineage | Can be visualised as an unbranched path through an evolutionary tree |
| Clone | Population of bacterial cells derived from a single parent cell129 | In evolutionary terms, it is assumed to include all the descendants of the parent cell (monophyletic)21. Cultured isolates are samples of clones |
| Isolate | A pure culture obtained from a single colony separated from others in vitro168 | Presumed to be and usually is derived from a single organism |
| Clade | Group of taxonomic entities composed of one ancestor and all of its evolutionary descendants167 | Synonym: monophyletic group |
| Strain | Set of genetically similar descendants of a single colony or cell44. Depending on the field, it can be genetic- or phenotypic- based | Descriptive subdivision of a species. Used widely but often with loose and/or inconsistent definitions. Can be described as ’taxonomic’ or ’natural’21 |
| Within-species variant | Any sub-classification of a species | General term that does not imply a level of resolution or phylogeny |
| Classic or typed subspecies | Set of strains that are genetically or phenotypically distinct and have a type strain available in a culture collection169; for example, Lactococcus lactis subspecies lactis and L. lactis ssp. cremoris | The name of a classic subspecies cannot be validly published if the description is based on studies of a mixed culture44. Variety was used as synonym of subspecies (now deprecated)44 |
| Population subspecies | Set of local populations of strains that live Species with subspecies are in a subdivision of a species’ spatial range and differs from other populations of the same species by phenotypic or genotypic characteristics74,128 | ‘polytypic’, without are ’monotypic’ |
| Population | Group of organisms, which live in a particular location or ecological niche at a given time | Can be used to refer to all members of a species or to a subset of the entire population |
| Subpopulation | Portion of a population that is partially isolated from others and in which allele frequencies evolve independently170 | A ‘metapopulation’ is a group of subpopulations |
| Strain population | A set of strains living simultaneously in the same spatial location or niche | Distinct from population subspecies, which can include multiple populations or ecotypes |
| Ecotype | An ecologically homogeneous population72. A clade within a species that has adapted to a particular environment. The scale of genetic dissimilarity between ecotypes can vary greatly | Ecotypes must be ecologically distinct enough that they can coexist indefinitely171. A mutant within an ecotype can outcompete the other strains in its own ecotype, but not those from a different ecotype69 |
| Phylotype | Clade in which all members contain a homologous sequence (marker gene or marker genes, genetic or inter-genic regions) that are distinctively similar | The threshold level of similarity may be arbitrarily chosen. Not limited to within species |
| SNV-type or SNP type | Set of genomes that share a distinctive set of SNVs.107 | Also used to describe the type of a SNV (for example, the exact switch in nucleotides) |
| Structural variant | Set of genomes that share distinctive structural variations172 | Structural variations can be defined as insertions, deletions and inversions greater than 50bp in size172 |
| Pathotype | Set of genomes that cause the same disease using the same set of virulence factors173 | Based on observational data; phenotypic and genotypic. It is not necessarily a clade |
| Serotype and serovar | Cells or viruses classified together based on their cell surface antigens, allowing the epidemiologic classification of organisms to the subspecies level174–176 | Different strains can belong to the same serotype177. Certain serotypes are often associated with specific pathotypes178 |
| Phagotype (or phage type) | Set of genomes susceptible to a particular bacteriophage and demonstrated by phage typing179 | Also called ‘lysotype’179 |
For decades, the most common source of microbial genomes was sequencing of isolates. Recently, this rate of production has been overtaken by rapid production of MAGs. A barrier to synergy between isolate-based and metagenomic research stems from the misinterpretation of MAGs as equivalent to isolate genomes (Box 3). The former might represent a population containing considerable diversity, whereas the latter usually represents a cultured isolate with little diversity. Considering also the rise in single cell sequencing, it will be useful to increasingly qualify the term genome’ as: cellular, isolate, or metagenomic.
The term ’strain’ is widely used across fields in microbiology and has many contrasting definitions (Figure 2A). In bacteriology, a strain is the ’’descendants of a single isolation in pure culture, and usually is made up of a succession of cultures ultimately derived from an initial single colony”8 founded by one or more cells 44 This is a strain in the taxonomic sense21 (’taxonomic strain’), used for type strains and culture collections. In this case, the origin of a strain is at isolation. An alternative definition, used for example in epidemiology, recognises a strain as an entity existing in nature21. This ’natural strain’ is defined as a set of conspecific isolates with distinctive genotypic and/or phenotypic characteristics123. A ’taxonomic strain’ can be thought of as an isolated, cultured sample of a ’natural strain’21. Operationally, the boundaries of natural and taxonomic strains vary. For example, taxonomic strains can become phenotypically heterogeneous with as few as three mutations124, but would still be called the same strain. By contrast, in some cases, isolates need to have less than three SNV differences125 to be considered to come from the same natural strain. This demonstrates that the genetic thresholds for strain delineation have not been universally set in culture-centric microbiology.
These two definitions of ’strain’, among others126, continue to coexist in culture-centric microbiology, and adoption of the term in microbiomics has extended this complexity. The disambiguating prefixes ’taxonomic’ versus ’natural’ are rarely used; however, this duality can clarify the mixed usage of the term ’strain’ in metagenomics. Strain-level metagenomics often poses two types of questions: classification and discovery. Classification questions ask if genetic segments (sequencing reads) belong to a particular ’taxonomic strain’, such as detecting if the probiotic strain Bifidobacterium bifidium BB12 is present in a stool sample. Discovery questions ask if there are subgroups within a species that form ’natural strains’, for example by clustering the genetic variation of genomes or of genetic segments. Conflict can arise among metagenomic tools for strain discovery that use different definitions of a natural strain - and will implicitly therefore give different results.— for example, defining natural strains based on differential gene content95 versus based on SNVs in shared genes105.
A universally applicable, operational definition of strain with strong biological basis has not been established and may not exist. In theory, genomes with as few as one SNV difference could be referred to as different strains. However, this practice is not recommended due to the unmanageable number of strains it would produce from metagenomic data. There are no rules on how many SNVs define a separate strain and whether such SNVs need to be fixed in the population or need to effect phenotype. In practice, the choice of how to set this cut-off is implicit in the choice of the strain-level profiling tool (for example, more than 0.1% of the nucleotides on species-specific marker genes, as set in StrainPhlAn) or is set by the analysis authors (for example, greater than 98% ANI127). Given such variability in the operational definition of strain, it becomes particularly valuable to use more specific terminology, instead of the generic term ’strain’ (see Table 1 and the section entitled ‘Microbiome applications of within-species variation’ for guidelines).
Subspecies group conspecific strains and many definitions of the term exist128. In classic microbiology, subspecies are clusters of strains that are genetically or phenotypically distinct, have a type strain available129, and are named (for example, Bacillus subtilis subsp. subtilis). Over time, the basis for classification of subspecies has shifted from qualitative phenotypic measures to genomic similarities between isolates130. This change has resulted in classification switches, such as the demotion of species to subspecies (for example, in Bifidobacterium longum 131) and vice versa for example, in Polynucleobacter necessarius 132). Thus, classic named subspecies do not (yet) necessarily align with distinct genomic clusters. By contrast, in a population biology context, a subspecies is a set of local populations that live in a subdivision of a species’ spatial range and that differ from other populations of the same species74; for example by genotype or phenotype128. Adapting the term subspecies for microbiomics implies the same usage dichotomy as described for strains: classification of reads to an existing ’classic subspecies’ and discovery of ’population subspecies’ by clustering within-species genetic variation observed across spatial scales.
Although the strict definitions of these terms do not limit the relative amounts of variation they can each contain, in practice, it is useful to put them in context of each other and use them in the suggested ranges (Figure 2C). As these ranges are guidelines, actual thresholds for group delineations should be included in reports when each term is used. Importantly, ’ strain’ is subordinate to ’ subspecies’ and thus should not be used to refer generally to any grouping subordinate to species (as it sometimes is). We also discourage using the term ’subspecies’ due to its different definition but visual similarity to ’subspecies’. Instead, we recommend using the terms ’infraspecific’ or simply ’within-species’. For example, inappropriate usage of ’strain-level analysis’ or ’sub-species analysis’ would be replaced with ’infraspecific analysis’ or ’within-species analysis’. Additionally, non-specific groupings within species can be referred to as ’within-species variants.
Phenotypic stratification in microbial communities.
Genetic variation within a species can manifest as phenotypic differences in complex ways. Different genetic variants can manifest as the same phenotype, whereas the same genetic variant can manifest as different phenotypes under different conditions133. The scale of genetic differences and their phenotypic impact are also not necessarily correlated, such as dramatically increased antibiotic resistance being conferred with as little as one SNV111,112. Further, different phenotypes can be observed when bacteria are cultured in isolation or in coculture or are within their natural community. For example, Pseudomonas aeruginosa has distinct gene expression profiles in vitro versus during human infection, including genes involved in antibiotic resistance, cell-cell communication and metabolism, which have implications for therapy development134. Differences in phenotype can also be seen within species — for example, two strains of the halophilic bacterium Salinibacter ruber had similar expression patterns when cultured in isolation but had distinct patterns when grown in co- culture135. These examples highlight the importance of studying phenotypic variation within species directly in microbiomes, and several methods exist (Box 1). For example, metatranscriptomics has been used to reveal functional diversity between conspecific symbionts in mussels136 and metagenomically inferred replication rates have distinguished between infraspecific subpopulations of Citrobacter koseri in infants137.
The complicated relationship between genotype and phenotype implies that phenotypic classification schemes can be at odds with genetic stratifications, and specialised vocabulary exists (Table 1). In medicine and epidemiology, it has been useful to categorise bacteria into (possibly polyphyletic) groups based on differential pathogenicity (pathotypes) or cell- surface antigens (serotypes). For example, the enteric E. coli group includes both commensal and pathogenic strains, which are divided into at least seven pathotypes45. In ecology, groups can also be defined based on behaviour and their functional role in a community, for example, based on the type of resources exploited and the way in which they are exploited138,139. Species grouped this way are called ’guild s’, a concept and term which could similarly be used to describe groups of strains. This kind of grouping was designed to give an appropriate resolution for the analysis of competition within ecosystems and generalisation of findings across communities. Although phenotype is the most relevant to many biological questions, it is hard to measure at large scale (though methods are progressing4,140). With microbiome genetic sequencing, genotypes are much easier to measure in high throughput but linking them to phenotype is challenging as phenotype can change drastically with habitat and small genotypic differences.
Microbiome applications of within-species variation
The many scales and dimensions of variation within species reflect the wide range of biological questions that a ‘within-species investigation’ can address. Isolate based approaches have been used to investigate many biological questions that involve within- species variation141,142. With the rise of metagenomic approaches, some of the same questions can now be investigated in high throughput and for many species in the community simultaneously (with important limitations; Box 2; Box 3). Below we describe how many of the important biological applications that were pioneered using isolate-based methods can now be investigated using a metagenomics approach. We summarise common examples of such investigations into five major themes, built around key biological questions (Figure 3). For each theme, we summarise methodological approaches and appropriate terminology and provide examples of relevant studies or software.
Figure 3.
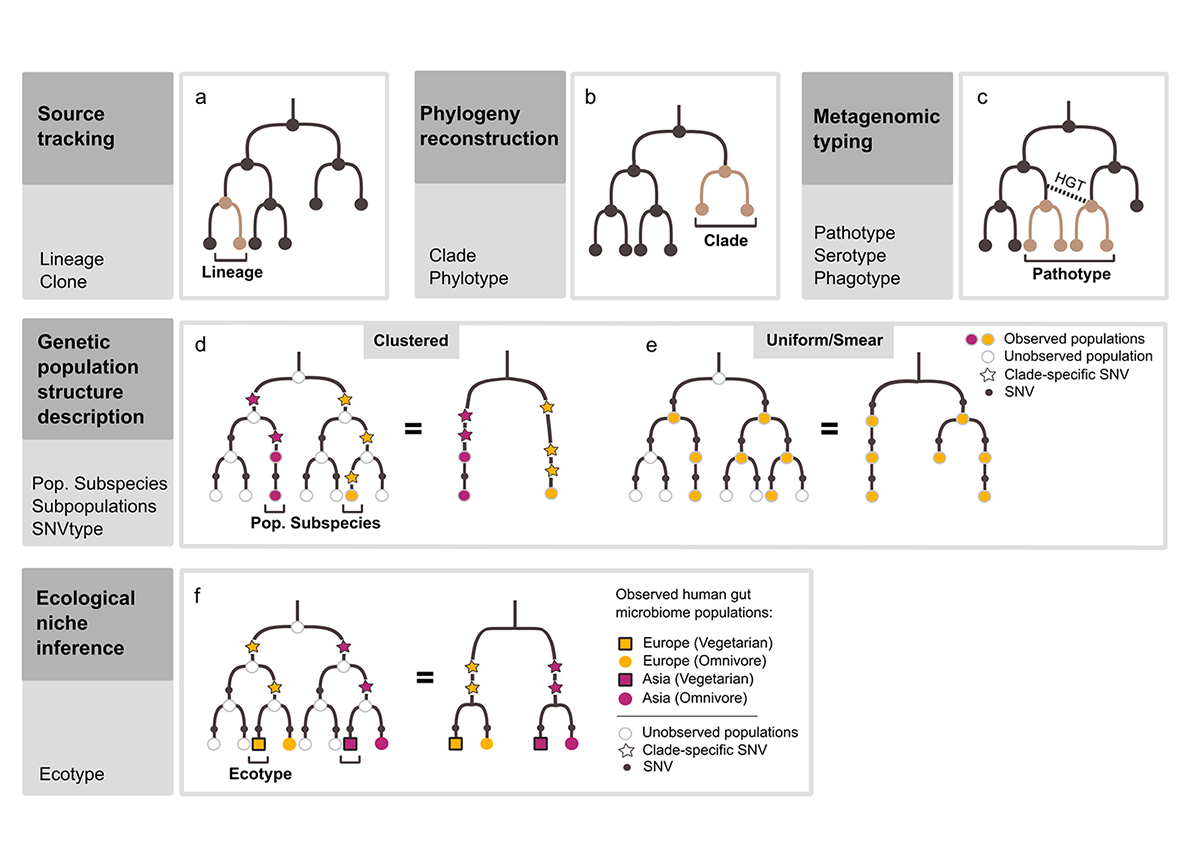
Applications of within-species variation. Five major areas of investigation for within-species-oriented metagenomic data analysis are illustrated, paired with corresponding appropriate terminology. Trees depict the genetic similarity and ancestry of potentially coexisting populations, with nodes representing populations and edges representing genetic differences accumulating from top to bottom. (a) Source tracking is concerned with identifying an unbranched path through a tree of ancestors and descendants (a ‘lineage’, pink edges and nodes). (b) Phylogeny reconstruction aims to build a tree which reflects the history of within-species variants based on their genetic similarity. A phylogeny might be cut into complete sub-trees (‘clades’) which may be called ‘phylotypes’. (c) Metagenomic typing detects the presence of a previously identified signature of interest within a species. For example, the presence of a gene associated with pathogenicity could be the criteria for detecting a ‘pathotype’. This gene may have been transferred between clades via horizontal gene transfer (HGT), so may be at odds with the within-species phylogeny. (d, e) The genetic population structure of a species can be described from the distribution of the genetic similarities across observed variants. (d) A ‘clustered’ structure occurs when there is a discontinuity across genetic similarities, enabling clades to be grouped into distinct clusters. Such a non-uniform structure is created by unobserved (extinct or unsampled) intermediate populations. A hypothetical within-species history with unobserved populations (white nodes) can be simplified (=), showing how unobserved populations can lead to a clustered genetic distribution, which may include distinct population subspecies. As SNVs (black dots) accumulate through this history, some might be specific to a particular set of populations (coloured dots). (e) When unobserved intermediate populations are rare or when they are spread widely through a species, the genetic distribution appears uniform or smeared and distinct groups of populations are not seen. (f) Ecological niche inference combines population observational data with phenotypic and/or habitat data to identify populations that have adapted to particular niches (‘ecotypes’). Adaptive traits might be identified by comparing populations but potential geographic confounds must also be considered.
Source tracking.
Where did the cells in this sample originally come from? To determine patterns of transmission or dispersal of microbial cells, their exact source population must be identified. The probability that a cell was dispersed from or is a direct descendant of a particular source population can be calculated by comparing genetic material from the target cell or population against genetic material from its potential source population or ancestors (‘source tracking’, ‘transmission tracking’ or ‘lineage tracking’) (Figure 3a). Strategies to determine source populations from metagenomic data include detecting the presence of shared SNVs87,88,90,143, CRISPR signals144, or strain-specific genes89 and genome reconstruction145. These approaches have been used to assess, for example, whether there is transmission of bacterial cells from the human oral cavity to the gut87, from mother to infant88,143, from probiotic treatment to the consumer89, or from faecal microbiome transplant (FMT) donor to recipient90. These strategies can be complicated by metagenomic disruption of allele linkage, multiple source populations, and evolution of the target population since dispersal from its source. Thus, although lineage tracking approaches can be useful for pathogen source detection145, they can also be insufficient for epidemiological outbreak analysis146. In the context of source tracking, the general term ‘strain’ could be replaced by the more specific term ‘lineage’, which can be characterised by a haplotype. Determining genomic haplotypes from metagenomic data remains a challenge147; however long-read sequencing of single DNA molecules provides promise as error rates decline148,149.
Phylogeny reconstruction.
What is the evolutionary history of variants within this species? In phylogeny reconstruction (Figure 3b), the relative ancestry of multiple lineages within a species is inferred from genetic similarity. This similarity can be based on full genomes or genetic segments (for example, marker genes). Due to HGT and homologous recombination, the phylogeny that would be reconstructed can vary based on the loci chosen and the phylogeny of genetic segments may not reflect overall genomic phylogeny150. Alternatively, within-species phylogenetic studies might focus on reconstruction of the history of a particular gene or plasmid within a species. Phylogeography puts these histories in the context of observed geographic distributions151,152. Phylogenetic analysis using isolate genomes is well established153 and these methods can be applied to microbial communities if high quality genomes are recovered, for example using MAGs or single amplified genomes (SAGs)86. However, data quality issues must be considered before this application (Box 3). Alternatively, a typical approach is to identify conspecific, homologous genetic segments in metagenomes (for example, through alignment to reference sequences), detect SNVs in them56,103–105 and then infer their most probable history105. Groups within species can be defined based on phylogeny by cutting the resultant tree at an arbitrary level of similarity, creating ‘phylotypes’. In this context, the general term ‘strain’ could be replaced by the more specific terms clade or phylotype.
Genetic population structure description.
Does this species have distinct subpopulations and/or subspecies? Describing a species’ genetic population structure can, for example, suggest its geographic history or explain heterogeneous associations with host disease states75. A species’ population structure can be determined by overlaying genetic data with observational data to describe the distribution of genetic similarities between variants within and across populations154. A uniform structure (‘smear’) occurs when there is a smooth distribution in genetic similarity across the observed species variants. This occurs when populations of ancestral and sister clades exist; that is, there are few unobservable (extinct or undetectable) branches within a tree (Figure 3e). By contrast, a ’clustered’ structure occurs when there is a discontinuity across genetic similarities, enabling clades to be grouped into distinct clusters. Such a non-uniform structure is created by extinct branches within a tree (Figure 3d). This manifests as subpopulations, which are subsets of a whole population that have distinct frequencies of genetic variations (for example, alleles or SNVs).
Metagenomics can be used to study population genetics of species within microbiomes19 by looking for clustering of genetic similarities across potential subpopulations. Detecting subpopulations is sensitive to sampling effort, as discontinuities in genetic similarity can be due to failure to observe intermediates (Box 3). Assessing such genetic similarities can be based on SNV allele frequencies in whole genomes75,104,108, SNVs in marker genes56,105 or gene content differences155. When MAGs or SAGs are produced, genome-based ANI clustering can also be used156. MAGs can also be used to track SNV and gene content differences, such as changes in populations of lake bacteria over time102. In this context, ‘strain’ is sometimes inappropriately used to refer to a subpopulation or subspecies. Subpopulations might be ecotypes if they have adapted to different niches, for example, through genome-wide sweep s instead of gene-specific sweep s72,157.
Ecological niche inference.
Have the variants within this species adapted to different conditions? Looking at within- species variants in conjunction with their habitats can provide information about their niche specificity (Figure 3f). When genetic data is used to make inferences about uncharacterised habitats, this is sometimes referred to as ‘reverse ecology’158. These inquiries often aim to identify the genetic segments (for example, genes, operons, plasmids) that are key to adapting to particular environments. Acquisition of these segments might be from vertical or horizontal transmission and thus can be in contrast with the phylogenetic history of the species. For example, a gene can rapidly become ubiquitous across populations due to frequent HGT under selective conditions (gene-specific sweep) for example, in the presence of antibiotics)72. A common approach to investigate these questions using metagenomic data is to look at conspecific subpopulations of cells that are known to have adapted to different conditions, for example, different human host diets84, soil versus plant-host associated159, or shifts in lake water habitats102, and identify distinctive genes82,95,98’104. Methods used in metagenome-wide association studies (MWAS) can also be applied here, though these are not often focused on adaptive evolution of populations160. In this context, the general term ‘strain’ could be replaced by the more specific term ecotypes12.
In the example shown in Figure 3, ‘genetic population structure’ investigations would focus on the allele frequency differences between European and Asian populations to decide whether these are distinct subpopulations or belong to one continuous population. Investigations on ‘ecological niche inference’ would focus on the gene differences in the gut- associated microbiome species associated with different diets, regardless of whether the European and Asian populations are distinct subpopulations.
Typing.
Does this species variant belong to a previously described sub-group of the species? Typing analyses assess the presence of genetic features (for example, SNVs, genes, operons or plasmids.) of specific interest in conspecific species variants (Figure 3c). In this context, within-species groups are not defined based on evolutionary history or habitat ranges, but simply on the presence or absence of specific genetic features. Such features may confer habitat fitness, may be transient and may only be expressed under rare or artificial conditions, such as antimicrobial resistance genes, pathogenicity genes (for example, enteropathogenic E. coli (EHEC)), or flagella. In this case, HGT is a major consideration; presence of a genetic feature does not necessarily reflect phylogeny. For example, serogroups are potentially polyphyletic groups within a species that are defined based on the presence of cell surface antigens, which allows their epidemiological classification.
Metagenomic approaches can be used to detect the genetic features that defined a type. SNVs of known116 or novel104 importance can be detected based on reference sequences. Detecting the presence of type-defining genes based on homology to reference sequences is well established in metagenomics141,161, but determining with certainty that these detected genes are present in a specific strain is more difficult due to the possibility of HGT within the community. In metagenomic data, HGT can be studied directly, with162 or without163 assembling genomes (reviewed in Ref 164).
Comparative analyses of within-species variants with the same phenotype can be used to discover the specific genetic features that are associated (and may be causing) the phenotype (such as in MWAS160). For example, conspecific cells could be grouped into a pathogenic ‘variant’ based only on their presence within hosts that are displaying similar symptoms, without knowing the evolutionary relationship of the cells or their typical habitats. In this context, the general term ‘strain’ could be replaced by the more specific term pathotype.
The themes described above have traditionally been investigated using isolate genomic approaches or low-resolution molecular methods (Box 1). As metagenomic studies increasingly create large amounts of data, dozens of new methods have been established to investigate the same questions, often with their own novel vocabulary. Considering how these new methods map back to the fundamental biological questions they are addressing and the history of research in the area will help to control the explosion of terminology. Many studies will include a combination of these themes, but considering the fundamental units separately facilitates breaking down complex questions and selecting the most appropriate methodology and terminology.
Conclusions
Despite often being the highest resolution taxonomic category considered in microbiome surveys, species can contain extreme phenotypic variability. Studying such variability used to be relatively limited in scope, with a few key isolate-based methods and a limited pool of culturable bacteria. With the development of metagenomic sequencing, the number of species that can be studied and the number of methods that can be used have increased substantially. The possibility to stratify variation within species according to many criteria, and at many scales, has also led to a growing and frequently imprecise terminology. Understanding how the variability within a species arose and identifying the central biological question being asked can help to determine the correct terminology and methodology to use. In some cases, the most appropriate term may have an operational definition, and its details and cut-off thresholds might vary across studies. To facilitate communication and collaboration, and enable future comparative meta-studies, vocabulary that does not have strict and widely- known definitions should be avoided when possible or explicitly described both in terms of the criteria and the thresholds being used. This Review aims to guide such descriptions and support a more informed development and application of within-species investigation techniques to metagenomic data.
Acknowledgements
Funding for research in the authors’ laboratories was provided by the European Research Council (ERC) (grant ERC-AdG-669830 MicrobioS), the European Union’s Horizon 2020 Research and Innovation Programme (grant 825694 MICROB-PREDICT), the German Federal Ministry of Education and Research (Bundesministerium fur Bildung und Forschung, BMBF) (grant 01GL1746B PRIMAL), and the European Molecular Biology Laboratory (EMBL).
Glossary
- Metagenomics
The study of all genomes present in a sample from a microbial community. Often performed as shotgun metagenomics, in which extracted DNA is fragmented before sequencing.
- Population
A set of individuals that occupy a particular spatial area
- Mutator allele
Genetic variation (allele) that results in an increased mutation rate.
- Horizontal gene transfer (HGT)
The movement of genetic information between organisms. This is in contrast to vertical gene transfer from parent to offspring.
- Homologous recombination
Type of genetic recombination in which genetic material is exchanged between two similar or identical regions of DNA.
- Conspecific
Belonging to the same species. For example, conspecific strains are strains that belong to the same species.
- Genetic drift
Change of allele frequencies in a population caused by stochastic factors
- Marker genes
In microbiome context: genes or genetic segments whose presence or specific DNA sequence is distinctive of a category of interest, such as a species or clade.
- Selective sweep
Reduction of the genetic variation in a population due to selection acting on novel mutations or existing alleles.
- Hard selective sweep
One beneficial allele at a locus replaces most other alleles in the population.
- Soft selective sweep
Multiple beneficial alleles at a locus gain prevalence, replacing standing genetic variation in the population.
- Metagenomic assembled genome (MAG)
A genome sequence recoveredfrom metagenomic data, usually fragmented, and potentially incomplete or contaminated. Typically, shotgun metagenomic sequencing produces short DNA sequences that are then assembled and binned into ’genomes ’ using k-mer frequencies and abundance information.
- Type strain
A living culture that serves as a fixed reference point for the assignment of bacterial and archaeal names. It is descended from the original isolate used in a species ’ description and shares all of its relevant phenotypic and genotypic properties.
- Microbiomics
The study of microbial communities (microbiomes) using one or more ’-omic ’ approaches (e.g. genomics, transcriptomics, proteomics, etc.)
- Infraspecific
Below species level, that is, at a higher resolution than ’species ’.
- Polyphyletic
Describes a group of organisms that do not share an immediate common ancestor. Not a clade.
- Guilds
A guild is a group of species that use the same type of resources in a similar way. Originally defined as a group of species (Root, 1967) but concept could be applied to strains or subspecies.
- Genome-wide sweep
Alleles at the locus under selection cause other linked loci (for example, genome, plasmid) to gain or lose abundance across the population. Also known as a ’broad’ sweep.
- Gene-specific sweep
Only alleles at the locus under selection gain or lose abundance across the population. Also known as a ’narrow’ or ’locus-specific ’ sweep.
Footnotes
Competing interests
The authors declare no competing interests.
Author contributions
The authors contributed equally to all aspects of the article.
Bibliography
- 1.Moore WEC, et al. Report of the Ad Hoc Committee on Reconciliation of Approaches to Bacterial Systematics. Int J Syst Evol Microbiol. 1987;37:463–464. [Google Scholar]
- 2.Leimbach A, Hacker J, Dobrindt U. E. coli as an all-rounder: The thin line between commensalism and pathogenicity. Curr Top Microbiol Immunol. 2013;358:3–32. doi: 10.1007/82_2012_303. [DOI] [PubMed] [Google Scholar]
- 3.Pierce JV, Bernstein HD. Genomic Diversity of Enterotoxigenic Strains of Bacteroides fragilis. PLoS One. 2016;11:e0158171. doi: 10.1371/journal.pone.0158171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maier L, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–628. doi: 10.1038/nature25979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Neuenschwander SM, Ghai R, Pernthaler J, Salcher MM. Microdiversification in genome-streamlined ubiquitous freshwater Actinobacteria. ISME J. 2018;12:185–198. doi: 10.1038/ismej.2017.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Triplett E. Genetics of Competition for Nodulation of Legumes. Annu Rev Microbiol. 1992;46:399–428. doi: 10.1146/annurev.mi.46.100192.002151. [DOI] [PubMed] [Google Scholar]
- 7.Nowrouzian FL, Adlerberth I, Wold AE. Enhanced persistence in the colonic microbiota of Escherichia coli strains belonging to phylogenetic group B2: role of virulence factors and adherence to colonic cells. Microbes Infect. 2006;8:834–840. doi: 10.1016/j.micinf.2005.10.011. [DOI] [PubMed] [Google Scholar]
- 8.Whitman WB, Bergey’s Manual Trust . Bergey’s Manual of Systematics of Archaea and Bacteria. Bergey’s Manual of Systematics of Archaea and Bacteria. Wiley; 2015. [Google Scholar]
- 9.Zhao S, et al. Adaptive Evolution within Gut Microbiomes of Healthy People. Cell Host Microbe. 2019;25:656–667.e8. doi: 10.1016/j.chom.2019.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lagier J-C, et al. Culturing the human microbiota and culturomics. Nat Rev Microbiol. 2018;16:540–550. doi: 10.1038/s41579-018-0041-0. [DOI] [PubMed] [Google Scholar]
- 11.Tyson GW, et al. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature. 2004;428:37–43. doi: 10.1038/nature02340. [DOI] [PubMed] [Google Scholar]
- 12.Allen EE, et al. Genome dynamics in a natural archaeal population. Proc Natl Acad Sci U S A. 2007;104:1883–1888. doi: 10.1073/pnas.0604851104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eppley JM, Tyson GW, Getz WM, Banfield JF. Genetic exchange across a species boundary in the archaeal genus ferroplasma. Genetics. 2007;177:407–16. doi: 10.1534/genetics.107.072892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppley JM, Tyson GW, Getz WM, Banfield JF. Strainer: Software for analysis of population variation in community genomic datasets. BMC Bioinformatics. 2007;8:398. doi: 10.1186/1471-2105-8-398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lo I, et al. Strain-resolved community proteomics reveals recombining genomes of acidophilic bacteria. Nature. 2007;446:537–541. doi: 10.1038/nature05624. [DOI] [PubMed] [Google Scholar]
- 16.Denef VJ, et al. Proteogenomic basis for ecological divergence of closely related bacteria in natural acidophilic microbial communities. Proc Natl Acad Sci U S A. 2010;107:2383–2390. doi: 10.1073/pnas.0907041107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Segata N. On the Road to Strain-Resolved Comparative Metagenomics. mSystems. 2018;3:e00190–17. doi: 10.1128/mSystems.00190-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Suez J, Zmora N, Segal E, Elinav E. The pros, cons, and many unknowns of probiotics. Nat Med. 2019;25:716–729. doi: 10.1038/s41591-019-0439-x. [DOI] [PubMed] [Google Scholar]
- 19.Denef VJ. Peering into the Genetic Makeup of Natural Microbial Populations Using Metagenomics. Population Genomics: Microorganisms. 2018:49–75. [Comprehensive review on application of metagenomic approaches for microbial population genomics.] [Google Scholar]
- 20.Bobay L-M, Raymann K. Population Genetics of Host-Associated Microbiomes. Curr Mol Biol Reports. 2019;5:128–139. [Google Scholar]
- 21.Dijkshoorn L, Ursing BM, Ursing JB. Strain, clone and species: Comments on three basic concepts of bacteriology. J Med Microbiol. 2000;49:397–401. doi: 10.1099/0022-1317-49-5-397. [Compares and summarises definitions of key terminology in bacteriological (isolate-based) context.] [DOI] [PubMed] [Google Scholar]
- 22.Brown T. Genomes 2nd edition. Oxford: Wiley-Liss; 2002. Genomes. [Google Scholar]
- 23.Alberts B, Johnson A, Lewis J, et al. Molecular Biology of the Cell Garland Science. Garland Science; 2002. [Google Scholar]
- 24.Fijalkowska IJ, Schaaper RM, Jonczyk P. DNA replication fidelity in Escherichia coli: a multi-DNA polymerase affair. FEMSMicrobiol Rev. 2012;36:1105–21. doi: 10.1111/j.1574-6976.2012.00338.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- 26.Dillon MM, Sung W, Sebra R, Lynch M, Cooper VS. Genome-Wide Biases in the Rate and Molecular Spectrum of Spontaneous Mutations in Vibrio cholerae and Vibrio fischeri. Mol Biol Evol. 2017;34:93–109. doi: 10.1093/molbev/msw224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Strauss C, Long H, Patterson CE, Te R, Lynch M. Genome-Wide Mutation Rate Response to pH Change in the Coral Reef Pathogen Vibrio shilonii AK1. MBio. 2017;8:e01021–17. doi: 10.1128/mBio.01021-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cooper VS, Vohr SH, Wrocklage SC, Hatcher PJ. Why Genes Evolve Faster on Secondary Chromosomes in Bacteria. PLoS Comput Biol. 2010;6:e1000732. doi: 10.1371/journal.pcbi.1000732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bobay L-M, Traverse CC, Ochman H. Impermanence of bacterial clones. Proc Natl Acad Sci. 2015;112:8893–8900. doi: 10.1073/pnas.1501724112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Andersson JO, Andersson SGE. Pseudogenes, Junk DNA, and the Dynamics o Rickettsia Genomes. Mol Biol Evol. 2001;18:829–839. doi: 10.1093/oxfordjournals.molbev.a003864. [DOI] [PubMed] [Google Scholar]
- 31.Mira A, Ochman H, Moran NA. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–96. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- 32.Lawrence JG, Retchless AC. Horizontal Gene Transfer Methods in molecular biology. Vol. 532. Humana Press; 2009. The interplay of homologous recombination and horizontal gene transfer in bacterial speciation; pp. 29–53. [DOI] [PubMed] [Google Scholar]
- 33.Lerner A, Matthias T, Aminov R. Potential Effects of Horizontal Gene Exchange in the Human Gut. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.01630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas CM, Nielsen KM. Mechanisms of, and Barriers to, Horizontal Gene Transfer between Bacteria. Nat Rev Microbiol. 2005;3:711–721. doi: 10.1038/nrmicro1234. [Reviews the major concepts and mechanisms of HGT and their implications for genome flux across populations.] [DOI] [PubMed] [Google Scholar]
- 35.Rocha EPC, Cornet E, Michel B. Comparative and evolutionary analysis of the bacterial homologous recombination systems. PLoS Genet. 2005;1:0247–0259. doi: 10.1371/journal.pgen.0010015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fraser C, Hanage WP, Spratt BG. Recombination and the Nature of Bacterial Speciation. Science. 2007;315:476–480. doi: 10.1126/science.1127573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gasiunas G, Sinkunas T, Siksnys V. Molecular mechanisms of CRISPR-mediated microbial immunity. Cell Mol Life Sci. 2014;71:449–465. doi: 10.1007/s00018-013-1438-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brouwer MSM, et al. Horizontal gene transfer converts non-toxigenic Clostridium difficile strains into toxin producers. Nat Commun. 2013;4 doi: 10.1038/ncomms3601. 2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kaper JB, O’Brien AD. Enterohemorrhagic Escherichia coli and Other Shiga Toxin-Producing E. coli. American Society of Microbiology; Overview and Historical Perspectives; pp. 3–13. [Google Scholar]
- 40.Hallatschek O, Hersen P, Ramanathan S, Nelson DR. Genetic drift at expanding frontiers promotes gene segregation. Proc Natl Acad Sci. 2007;104:19926–19930. doi: 10.1073/pnas.0710150104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nemergut DR, et al. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev. 2013;77:342–56. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chun J, et al. Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes. Int J Syst Evol Microbiol. 2018;68:461–466. doi: 10.1099/ijsem.0.002516. [DOI] [PubMed] [Google Scholar]
- 43.Doolittle WF. Population Genomics: How Bacterial Species Form and Why They Don’t Exist. Curr Biol. 2012;22:R451–R453. doi: 10.1016/j.cub.2012.04.034. [DOI] [PubMed] [Google Scholar]
- 44.International Code of Nomenclature of Prokaryotes. Int J Syst Evol Microbiol. 2019;69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 45.Croxen MA, et al. Recent Advances in Understanding Enteric Pathogenic Escherichia coli. Clin Microbiol Rev. 2013;26:822–880. doi: 10.1128/CMR.00022-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Konstantinidis KT, Tiedje JM. Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sc U S A. 2005;102:2567–72. doi: 10.1073/pnas.0409727102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. Proc Natl Acad Sci. 2009;106:19126–19131. doi: 10.1073/pnas.0906412106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mende DR, Sunagawa S, Zeller G, Bork P. Accurate and universal delineation of prokaryotic species. Nat Methods. 2013;10:881–884. doi: 10.1038/nmeth.2575. [DOI] [PubMed] [Google Scholar]
- 49.Goris J, et al. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int J Syst Evol Microbiol. 2007;57:81–91. doi: 10.1099/ijs.0.64483-0. [DOI] [PubMed] [Google Scholar]
- 50.Dzink JL, Sheenan MT, Socransky SS. Proposal of Three Subspecies of Fusobacterium nucleaturn Knorr 1922: Fusobacterium nucleatum subsp. nucleatum subsp. nov. , comb. nov.; Fusobacterium nucleatum subsp. polymorphum subsp. nov. , norn. rev. , comb. nov.; and Fusobacterium nucleatum subsp. vincentii subsp. nov., norn. rev., comb. nov. Int J Syst Bacteriol. 1990;40:74–78. doi: 10.1099/00207713-40-1-74. [DOI] [PubMed] [Google Scholar]
- 51.Kook JK, et al. Genome-Based Reclassification of Fusobacterium nucleatum Subspecies at the Species Level. Curr Microbiol. 2017;74:1137–1147. doi: 10.1007/s00284-017-1296-9. [DOI] [PubMed] [Google Scholar]
- 52.Konstantinidis KT, Delong EF. Genomic patterns of recombination clonal divergence and environment in marine microbial populations. ISME J. 2008;2:1052–1065. doi: 10.1038/ismej.2008.62. [DOI] [PubMed] [Google Scholar]
- 53.Caro-Quintero A, Konstantinidis KT. Bacterial species may exist, metagenomics reveal. Environmental Microbiology. 2012;14:347–355. doi: 10.1111/j.1462-2920.2011.02668.x. [DOI] [PubMed] [Google Scholar]
- 54.Jain C, Rodriguez-R LM, Phillippy AM, Konstantinidis KT, Aluru S. High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat Commun. 2018;9 doi: 10.1038/s41467-018-07641-9. 5114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Olm MR, et al. Consistent Metagenome-Derived Metrics Verify and Delineate Bacterial Species Boundaries. mSystems. 2020;5 doi: 10.1128/mSystems.00731-19. 647511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Milanese A, et al. Microbial abundance, activity and population genomic profiling with mOTUs2. Nat Commun. 2019;10 doi: 10.1038/s41467-019-08844-4. 1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mayden RL. A hierarchy of species concepts: the denouement in the saga of the species problem. Species The units of biodiversity. 1997:381–423. [Google Scholar]
- 58.Wilkins JS. How to be a chaste species pluralist-realist: the origins of species moc and the synapomorphic species concept. Biol Philos. 2003;18:621–638. [Google Scholar]
- 59.Hey J. The mind of the species problem. Trends Ecol Evol. 2001;16:326–329. doi: 10.1016/s0169-5347(01)02145-0. [DOI] [PubMed] [Google Scholar]
- 60.Bapteste E, et al. Prokaryotic evolution and the tree of life are two different things. Biol Direct. 2009;4:34. doi: 10.1186/1745-6150-4-34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Konstantinidis KT, Ramette A, Tiedje JM. The bacterial species definition ii the genomic era. Philos Trans R Soc B Biol Sci. 2006;361:1929–1940. doi: 10.1098/rstb.2006.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bobay L-M, Ochman H. Biological Species Are Universal across Life’s Domai. Genome Biol Evol. 2017;9:491–501. doi: 10.1093/gbe/evx026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Moldovan MA, Gelfand MS. Pangenomic Definition of Prokaryotic Species a the Phylogenetic Structure of Prochlorococcus spp. Front Microbiol. 2018;9:428. doi: 10.3389/fmicb.2018.00428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Snel B, Bork P, Huynen MA. Genome phylogeny based on gene content. Nat Genet. 1999;21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 65.Achtman M, Wagner M. Microbial diversity and the genetic nature of microbial species. Nat Rev Microbiol. 2008;6:431–440. doi: 10.1038/nrmicro1872. [DOI] [PubMed] [Google Scholar]
- 66.Barton NH. The effect of hitch-hiking on neutral genealogies. Genet Res. 1998;72:123–133. [Google Scholar]
- 67.Hermisson J, Pennings PS. Soft sweeps and beyond: understanding the pattern and probabilities of selection footprints under rapid adaptation. Methods Ecol Evol. 2017:700–716. [Google Scholar]
- 68.Shapiro BJ, et al. Population genomics of early events in the ecological differentiation of bacteria. Science. 2012;336:48–51. doi: 10.1126/science.1218198. [Demonstrates that gene-specific selective sweeps followed by gradually decreasing gene flow can lead to ecologically differentiated conspecific subpopulations.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Cohan FM. Bacterial Species and Speciation. Syst Biol. 2001;50:513–524. doi: 10.1080/10635150118398. [DOI] [PubMed] [Google Scholar]
- 70.Cohan FM. Selective Sweep. Springer US; 2007. Periodic Selection and Ecological Diversity in Bacteria; pp. 78–93. [Google Scholar]
- 71.Charlesworth B. Effective population size and patterns of molecular evolution and variation. Nat Rev Genet. 2009;10:195–205. doi: 10.1038/nrg2526. [DOI] [PubMed] [Google Scholar]
- 72.Cohan FMM. Bacterial Speciation: Genetic Sweeps in Bacterial Species. Curr Biol. 2016;26:R112–R115. doi: 10.1016/j.cub.2015.10.022. [DOI] [PubMed] [Google Scholar]
- 73.Hermisson J, Pennings PS. Soft sweeps: molecular population genetics of adaptation from standing genetic variation. Genetics. 2005;169:2335–52. doi: 10.1534/genetics.104.036947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Monroe B. A Modern Concept of the Subspecies. Auk. 1982;99:608–609. [Google Scholar]
- 75.Costea P, et al. Subspecies in the global human gut microbiome. MolSystBiol. 2017;13:960–960. doi: 10.15252/msb.20177589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Retchless AC, Lawrence JG. Temporal Fragmentation of Speciation in Bacteria. Science. 2007;317:1093–1096. doi: 10.1126/science.1144876. [DOI] [PubMed] [Google Scholar]
- 77.Shapiro BJ. What Microbial Population Genomics Has Taught Us About Speciation. In: Polz MF, Rajora Editors OP, editors. Population Genomics: Microorganisms. Springer Nature; 2018. pp. 31–47. [Google Scholar]
- 78.Sheppard SK, Guttman DS, Fitzgerald JR. Population genomics of bacterial host adaptation. Nat Rev Genet. 2018;19:549–565. doi: 10.1038/s41576-018-0032-z. [An extensive review about origins of genetic population structure in Prokaryotes and how to study it in context of host-microbiome interactions and adaptations.] [DOI] [PubMed] [Google Scholar]
- 79.Bobay L-M, Ochman H. Factors driving effective population size and pan-genome evolution in bacteria. BMC Evol Biol. 2018;18:153. doi: 10.1186/s12862-018-1272-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Smelov V, et al. Chlamydia trachomatis Strain Types Have Diversified Regionally and Globally with Evidence for Recombination across Geographic Divides. Front Microbiol. 2017;8:2195. doi: 10.3389/fmicb.2017.02195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tenaillon O, Skurnik D, Picard B, Denamur E. The population genetics of commensal Escherichia coli. Nat Rev Microbiol. 2010;8:207–217. doi: 10.1038/nrmicro2298. [DOI] [PubMed] [Google Scholar]
- 82.Zeevi D, et al. Structural variation in the gut microbiome associates with host health. Nature. 2019;568:43–48. doi: 10.1038/s41586-019-1065-y. [DOI] [PubMed] [Google Scholar]
- 83.Lloyd-Price J, et al. Strains, functions and dynamics in the expanded Human Microbiome Project. Nature. 2017;550:61–66. doi: 10.1038/nature23889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.De Filippis F, et al. Distinct Genetic and Functional Traits of Human Intestinal Prevotella copri Strains Are Associated with Different Habitual Diets. Cell Host Microbe. 2019;25:444–453.e3. doi: 10.1016/j.chom.2019.01.004. [DOI] [PubMed] [Google Scholar]
- 85.Ferretti P, et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Stewart RD, et al. Compendium of 4,941 rumen metagenome-assembled genomes for rumen microbiome biology and enzyme discovery. Nat Biotechnol. 2019;37:953–961. doi: 10.1038/s41587-019-0202-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Schmidt TS, et al. Extensive transmission of microbes along the gastrointestinal tract. Elife. 2019;8 doi: 10.7554/eLife.42693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Asnicar F, et al. Studying Vertical Microbiome Transmission from Mothers to Infants by Strain-Level Metagenomic Profiling. mSystems. 2017;2 doi: 10.1128/mSystems.00164-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zmora N, et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell. 2018;174:1388–1405.e21. doi: 10.1016/j.cell.2018.08.041. [DOI] [PubMed] [Google Scholar]
- 90.Smillie CS, et al. Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplantation Article Strain Tracking Reveals the Determinants of Bacterial Engraftment in the Human Gut Following Fecal Microbiota Transplanta. Cell Host Microbe. 2018;23:229–240.e5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zhou Z, Luhmann N, Alikhan NF, Quince C, Achtman M. Accurate Reconstruction of Microbial Strains from Metagenomic Sequencing Using Representative Reference Genomes. Research in Computational Molecular Biology RECOMB 2018 Lecture Notes in Computer Science. 2018:225–240. 10812 LNBI. [Google Scholar]
- 92.Ahn T-H, Chai J, Pan C. Sigma: Strain-level inference of genomes from metagenomic analysis for biosurveillance. Bioinformatics. 2015;31:170–177. doi: 10.1093/bioinformatics/btu641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hong C, et al. PathoScope 2.0: a complete computational framework for strain identification in environmental or clinical sequencing samples. Microbiome. 2014;2:33. doi: 10.1186/2049-2618-2-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ondov BD, et al. Mash: Fast genome and metagenome distance estimation using MinHash. Genome Biol. 2016;17 doi: 10.1186/s13059-016-0997-x. 029827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Scholz M, et al. Strain-level microbial epidemiology and population genomics from shotgun metagenomics. Nat Methods. 2016;13:435–438. doi: 10.1038/nmeth.3802. [DOI] [PubMed] [Google Scholar]
- 96.Zhu A, Sunagawa S, Mende DR, Bork P. Inter-individual differences in the gene content of human gut bacterial species. Genome Biol. 2015;16:82. doi: 10.1186/s13059-015-0646-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Greenblum S, Carr R, Borenstein E. Extensive Strain-Level Copy-Number Variation across Human Gut Microbiome Species. Cell. 2015;160:583–594. doi: 10.1016/j.cell.2014.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Quince C, et al. DESMAN: a new tool for de novo extraction of strains from metagenomes. Genome Biol. 2017;18:181. doi: 10.1186/s13059-017-1309-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Maistrenko OM, et al. Disentangling the impact of environmental and phylogenetic constraints on prokaryotic within-species diversity. ISME J. 2020;1 doi: 10.1038/s41396-020-0600-z. 735696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Andreani NA, Hesse E, Vos M. Prokaryote genome fluidity is dependent on effective population size. ISME J. 2017;11:1719–1721. doi: 10.1038/ismej.2017.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nayfach S, Shi ZJ, Seshadri R, Pollard KS, Kyrpides NC. New insights from uncultivated genomes of the global human gut microbiome. Nature. 2019;568:505–510. doi: 10.1038/s41586-019-1058-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bendall ML, et al. Genome-wide selective sweeps and gene-specific sweeps in natural bacterial populations. ISME J. 2016;10:1589–1601. doi: 10.1038/ismej.2015.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Nayfach S, Rodriguez-Mueller B, Garud N, Pollard KS. An integrated metagenomics pipeline for strain profiling reveals novel patterns of bacterial transmission and biogeography. Genome Res. 2016;26:1612–1625. doi: 10.1101/gr.201863.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Costea PI, et al. metaSNV: A tool for metagenomic strain level analysis. PLoS One. 2017;12:e0182392. doi: 10.1371/journal.pone.0182392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Truong DT, Tett A, Pasolli E, Huttenhower C, Segata N. Microbial strain- level population structure and genetic diversity from metagenomes. Genome Res. 2017;27:626–638. doi: 10.1101/gr.216242.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bush SJ, et al. Genomic diversity affects the accuracy of bacterial single-nucleotide polymorphism-calling pipelines. Gigascience. 2020;9 doi: 10.1093/gigascience/giaa007. giaa007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Luo C, et al. ConStrains identifies microbial strains in metagenomic datasets. Nat Biotechnol. 2015;33:1045–1052. doi: 10.1038/nbt.3319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Delmont TO, et al. Single-amino acid variants reveal evolutionary processes that shape the biogeography of a global SAR11 subclade. Elife. 2019;8 doi: 10.7554/eLife.46497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Jackson RW, et al. Identification of a pathogenicity island, which contains genes for virulence and avirulence, on a large native plasmid in the bean pathogen Pseudomonas syringae pathovar phaseolicola. Proc Natl Acad Sci. 1999;96:10875–10880. doi: 10.1073/pnas.96.19.10875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Scholz BK, Jakobek JL, Lindgren PB. Restriction fragment length polymorphism evidence for genetic homology within a pathovar of Pseudomonas syringae. Appl Environ Microbiol. 1994;60:1093–1100. doi: 10.1128/aem.60.4.1093-1100.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Pan XS, Yague G, Fisher LM. Quinolone resistance mutations in Streptococcus pneumoniae gyrA and parC proteins: Mechanistic insights into quinolone action from enzymatic analysis, intracellular levels, and phenotypes of wild-type and mutant proteins. Antimicrob Agents Chemother. 2001;45:3140–3147. doi: 10.1128/AAC.45.11.3140-3147.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Forslund K, Sunagawa S, Coelho LP, Bork P. Metagenomic insights into the human gut resistome and the forces that shape it. BioEssays. 2014;36:316–329. doi: 10.1002/bies.201300143. [DOI] [PubMed] [Google Scholar]
- 113.Petkau A, et al. SNVPhyl: a single nucleotide variant phylogenomics pipeline for microbial genomic epidemiology. Microb Genomics. 2017;3:e000116. doi: 10.1099/mgen.0.000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Jain R, Rivera MC, Lake JA, Lake J. Horizontal gene transfer among genomes: The complexity hypothesis. Proc Natl Acad Sci. 1999;96:3801–3806. doi: 10.1073/pnas.96.7.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Polz MF, Rajora OP. Population genomics : microorganisms. 2019 [Google Scholar]
- 116.Zolfo M, Tett A, Jousson O, Donati C, Segata N. MetaMLST: Multi-locus strain-level bacterial typing from metagenomic samples. Nucleic Acids Res. 2017;45:e7. doi: 10.1093/nar/gkw837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Truong DT, et al. MetaPhlAn2 for enhanced metagenomic taxonomic profiling. Nature Methods. 2015;12:902–903. doi: 10.1038/nmeth.3589. [DOI] [PubMed] [Google Scholar]
- 118.Tamburini FB, et al. Precision identification of diverse bloodstream pathogens in the gut microbiome. Nat Med. 2018;24:1809–1814. doi: 10.1038/s41591-018-0202-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Albanese D, Donati C. Strain profiling and epidemiology of bacterial species from metagenomic sequencing. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02209-5. 2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.O’Brien JD, et al. A Bayesian approach to inferring the phylogenetic structure of communities from metagenomic data. Genetics. 2014;197:925–37. doi: 10.1534/genetics.114.161299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Smillie CS, et al. Strain Tracking Reveals the Determinants of Bacterial Engraftmen in the Human Gut Following Fecal Microbiota Transplantation. Cell Host Microbe. 2018;23:229–240.e5. doi: 10.1016/j.chom.2018.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Sczyrba A, et al. Critical Assessment of Metagenome Interpretation - A benchmark o: metagenomics software. Nat Methods. 2017;14:1063–1071. doi: 10.1038/nmeth.4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Struelens MJ, et al. Consensus guidelines for appropriate use and evaluation of microbial epidemiologic typing systems. Clin Microbiol Infect. 1996;2:2–11. doi: 10.1111/j.1469-0691.1996.tb00193.x. [DOI] [PubMed] [Google Scholar]
- 124.Spira B, De Almeida Toledo R, Maharjan RP, Ferenci T. The uncertain consequences of transferring bacterial strains between laboratories - RpoS instability as an example. BMC Microbiol. 2011;11 doi: 10.1186/1471-2180-11-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Kong LY, et al. Clostridium difficile: Investigating Transmission Patterns between Infected and Colonized Patients Using Whole Genome Sequencing. Clin Infect Dis. 2019;68:204–209. doi: 10.1093/cid/ciy457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Saak CC, Gibbs KA. The Self-Identity Protein IdsD Is Communicated between Cells in Swarming Proteus mirabilis Colonies. J Bacteriol. 2016;198:3278–3286. doi: 10.1128/JB.00402-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Brooks B, et al. Strain-resolved analysis of hospital rooms and infants reveals overlap between the human and room microbiome. Nat Commun. 2017;8 doi: 10.1038/s41467-017-02018-w. 1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Patten MA. Subspecies and the philosophy of science. Auk. 2015;132:481–485. [Google Scholar]
- 129.International Committee on Systematics of Prokaryotes. International Code of Nomenclature of Prokaryotes: Prokaryotic Code (2008 Revision) Int J Syst Evol Microbiol. 2019;69:S1–S111. doi: 10.1099/ijsem.0.000778. [DOI] [PubMed] [Google Scholar]
- 130.Meier-Kolthoff JP, et al. Complete genome sequence of DSM 30083(T), the type strain (U5/41(T)) of Escherichia coli, and a proposal for delineating subspecies in microbial taxonomy. Stand Genomic Sci. 2014;9:2. doi: 10.1186/1944-3277-9-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fukuyama M, et al. Unification of Bifidobacterium infantis and Bifidobacterium suis as Bifidobacterium longum. Int J Syst Evol Microbiol. 2002;52:1945–1951. doi: 10.1099/00207713-52-6-1945. [DOI] [PubMed] [Google Scholar]
- 132.Hahn MW, Schmidt J, Pitt A, Taipale SJ, Lang E. Reclassification of four Polynucleobacter necessarius strains as representatives of Polynucleobacter asymbioticus comb. nov., Polynucleobacter duraquae sp. nov., Polynucleobacter yangtzensis sp. nov. and Polynucleobacter sinensis sp. nov., and emended description of Polynucleobacter necessarius. Int J Syst Evol Microbiol. 2016;66:2883–2892. doi: 10.1099/ijsem.0.001073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ackermann M. A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol. 2015;13:497–508. doi: 10.1038/nrmicro3491. [DOI] [PubMed] [Google Scholar]
- 134.Cornforth DM, et al. Pseudomonas aeruginosa transcriptome during human infection. Proc Natl Acad Sci U S A. 2018;115:E5125–E5134. doi: 10.1073/pnas.1717525115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Gonzalez-Torres P, et al. Interactions between closely related bacterial strains are revealed by deep transcriptome sequencing. Appl Environ Microbiol. 2015;81:8445–56. doi: 10.1128/AEM.02690-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Ansorge R, et al. Functional diversity enables multiple symbiont strains to coexist in deep-sea mussels. Nat Microbiol. 2019;4:2487–2497. doi: 10.1038/s41564-019-0572-9. [DOI] [PubMed] [Google Scholar]
- 137.Olm MR, et al. Identical bacterial populations colonize premature infant gut, skin, and oral microbiomes and exhibit different in situ growth rates. Genome Res. 2017;27:601–612. doi: 10.1101/gr.213256.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Pedros-Alio C. Plankton Ecology. Springer; Berlin, Heidelberg: 1989. Toward an Autecology of Bacterioplankton; pp. 297–336. [Google Scholar]
- 139.Root RB. The Niche Exploitation Pattern of the Blue-Gray Gnatcatcher. Ecol Monogr. 1967;37:317–350. [Google Scholar]
- 140.Mateus A, et al. Thermal proteome profiling in bacteria: probing protein state in vivo. Mol Syst Biol. 2018;14:e8242. doi: 10.15252/msb.20188242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Land M, et al. Insights from 20 years of bacterial genome sequencing. Funct Integr Genomics. 2015;15:141–161. doi: 10.1007/s10142-015-0433-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Gutleben J, et al. The multi-omics promise in context: from sequence to microbial isolate. Critical Reviews in Microbiology. 2018;44:212–229. doi: 10.1080/1040841X.2017.1332003. [DOI] [PubMed] [Google Scholar]
- 143.Ferretti P, et al. Mother-to-Infant Microbial Transmission from Different Body Sites Shapes the Developing Infant Gut Microbiome. Cell Host Microbe. 2018;24:133–145.e5. doi: 10.1016/j.chom.2018.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Lam TJ, Ye Y. CRISPRs for Strain Tracking and Their Application to Microbiota Transplantation Data Analysis. Cris J. 2019;2:41–50. doi: 10.1089/crispr.2018.0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Mu A, et al. Reconstruction of the Genomes of Drug-Resistant Pathogens for Outbreak Investigation through Metagenomic Sequencing. mSphere. 2019;4 doi: 10.1128/mSphere.00529-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Didelot X, Walker AS, Peto TE, Crook DW, Wilson DJ. Within-host evolution of bacterial pathogens. Nat Rev Microbiol. 2016;14:150–162. doi: 10.1038/nrmicro.2015.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Quince C, Walker AW, Simpson JT, Loman NJ, Segata N. Shotgun metagenomics, from sampling to analysis. Nat Biotechnol. 2017;35:833–844. doi: 10.1038/nbt.3935. [Reviews how microbial communities can be studied using metagenomic sequencing, with comments on sources of bias and comparisons of analytical methods.] [DOI] [PubMed] [Google Scholar]
- 148.Koren S, Phillippy AM. One chromosome, one contig: complete microbial genomes from long-read sequencing and assembly. Curr Opin Microbiol. 2015;23:110–120. doi: 10.1016/j.mib.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 149.Somerville V, et al. Long-read based de novo assembly of low-complexity metagenome samples results in finished genomes and reveals insights into strain diversity and an active phage system. BMC Microbiol. 2019;19:143. doi: 10.1186/s12866-019-1500-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Jiang X, et al. Dissemination of antibiotic resistance genes from antibiotic producers to pathogens. Nat Commun. 2017;8 doi: 10.1038/ncomms15784. 15784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Linz B, et al. An African origin for the intimate association between humans and Helicobacter pylori. Nature. 2007;445:915–918. doi: 10.1038/nature05562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Thorell K, et al. Rapid evolution of distinct Helicobacter pylori subpopulations in the Americas. PLoS Genet. 2017;13 doi: 10.1371/journal.pgen.1006546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Gardy JL, et al. Whole-Genome Sequencing and Social-Network Analysis of a Tuberculosis Outbreak. N Engl J Med. 2011;364:730–739. doi: 10.1056/NEJMoa1003176. [DOI] [PubMed] [Google Scholar]
- 154.Gregory AC, et al. Marine DNA Viral Macro- and Microdiversity from Pole to Pole. Cell. 2019;177:1109–1123.e14. doi: 10.1016/j.cell.2019.03.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Arevalo P, VanInsberghe D, Elsherbini J, Gore J, Polz MF. A Reverse Ecology Approach Based on a Biological Definition of Microbial Populations. Cell. 2019;178:820–834.e14. doi: 10.1016/j.cell.2019.06.033. [DOI] [PubMed] [Google Scholar]
- 156.Garcia SL, et al. Contrasting patterns of genome-level diversity across distinct cooccurring bacterial populations. ISME J. 2018;12:742–755. doi: 10.1038/s41396-017-0001-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kopac S, et al. Genomic Heterogeneity and Ecological Speciation within One Subspecies of Bacillus subtilis. Appl Environ Microbiol. 2014;80:4842–4853. doi: 10.1128/AEM.00576-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Levy R, Borenstein E. Evolutionary Systems Biology. Vol. 751. Springer; New York, NY: 2012. Reverse Ecology: From Systems to Environments and Back; pp. 329–345. [DOI] [PubMed] [Google Scholar]
- 159.Burghardt LT, et al. Select and resequence reveals relative fitness of bacteria in symbiotic and free-living environments. Proc Natl Acad Sci. 2018;115:2425–2430. doi: 10.1073/pnas.1714246115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Wang J, Jia H. Metagenome-wide association studies: fine-mining the microbiome. Nat Rev Microbiol. 2016;14:508–522. doi: 10.1038/nrmicro.2016.83. [DOI] [PubMed] [Google Scholar]
- 161.Knight R, et al. Best practices for analysing microbiomes. Nat Rev Microbiol. 2018;16:410–422. doi: 10.1038/s41579-018-0029-9. [DOI] [PubMed] [Google Scholar]
- 162.Song W, Wemheuer B, Zhang S, Steensen K, Thomas T. MetaCHIP: community-level horizontal gene transfer identification through the combination of best-match and phylogenetic approaches. Microbiome. 2019;7:36. doi: 10.1186/s40168-019-0649-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Seiler E, Trappe K, Renard BY. Where did you come from, where did you go: Refining metagenomic analysis tools for horizontal gene transfer characterisation. PLOS Comput Biol. 2019;15:e1007208. doi: 10.1371/journal.pcbi.1007208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Douglas GM, Langille MGI. Current and promising approaches to identify horizontal gene transfer events in metagenomes. Genome Biol Evol. 2019;11:2750–2766. doi: 10.1093/gbe/evz184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Cox C Barry, Moore Peter D., L R. Biogeography: An Ecological and Evolutionary Approach. 2016 [Google Scholar]
- 166.Arora D, Singh A, Sharma V, Bhaduria HS, Patel RB. HgsDb: Haplogroups Database to understand migration and molecular risk assessment. Bioinformation. 2015;11:272–275. doi: 10.6026/97320630011272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Cantino P, de Queiroz K. PhyloCode: A Phylogenetic Code of Biological Nomenclature. PhyloCode. 2010 www.ohiou.edu/phylocode. [Google Scholar]
- 168.Tenover FC, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed- field gel electrophoresis: Criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Schloter M, Lebuhn M, Heulin T, Hartmann A. Ecology and evolution of bacterial microdiversity. FEMS Microbiology Reviews. 2000;24:647–660. doi: 10.1111/j.1574-6976.2000.tb00564.x. [DOI] [PubMed] [Google Scholar]
- 170.Hamilton M. Population Genetics. Wiley-Blackwell; 2009. [Google Scholar]
- 171.Cohan FM. Transmission in the Origins of Bacterial Diversity, From Ecotypes to Phyla. Microbiol Spectr. 2017;5 doi: 10.1128/microbiolspec.mtbp-0014-2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Alkan C, Coe BP, Eichler EE. Genome structural variation discovery and genotyping. Nat Rev Genet. 2011;12:363–376. doi: 10.1038/nrg2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Kaper JB, Nataro JP, Mobley HLT. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 174.Samuel B. Medical Microbiology. Univ of Texas Medical Branch; 1996. [Google Scholar]
- 175.Kenneth R, George R, Sherris JC. Medical microbiology: an introduction to infectious diseases. McGraw-Hill Medical; 2004. [Google Scholar]
- 176.The American Heritage Medical Dictionary - Serovar. Houghton Mifflin; 2007. [Google Scholar]
- 177.Silva NA, et al. Genomic Diversity between Strains of the Same Serotype and Multilocus Sequence Type among Pneumococcal Clinical Isolates. Infect Immun. 2006;74:3513–3518. doi: 10.1128/IAI.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Fratamico PM, et al. Advances in Molecular Serotyping and Subtyping of Escherichia coli|. Front Microbiol. 2016;7 doi: 10.3389/fmicb.2016.00644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Miller-Keane, Marie O. Miller-Keane Encyclopedia and Dictionary of Medicine, Nursing, and Allied Health, Seventh Edition. Saunders, an imprint of Elsevier, Inc. 2003 [Google Scholar]
- 180.diCenzo GC, Finan TM. The Divided Bacterial Genome: Structure, Function, and Evolution. Microbiol Mol Biol Rev. 2017;81 doi: 10.1128/MMBR.00019-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Hamady M, Knight R. Microbial community profiling for human microbiome projects: Tools, techniques, and challenges. Genome Res. 2009;19:1141–1152. doi: 10.1101/gr.085464.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Nocker A, Burr M, Camper AK. Genotypic Microbial Community Profiling: A Critical Technical Review. Microb Ecol. 2007;54:276–289. doi: 10.1007/s00248-006-9199-5. [Reviews foundational methods that enabled microbial diversity to be assessed directly within a microbial community, sometimes at within-species resolution.] [DOI] [PubMed] [Google Scholar]
- 183.Eren AM, Borisy GG, Huse SM, Mark Welch JL. Oligotyping analysis of the human oral microbiome. Proc Natl Acad Sci. 2014;111:E2875–E2884. doi: 10.1073/pnas.1409644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Eren AM, et al. Minimum entropy decomposition: Unsupervised oligotyping for sensitive partitioning of high-throughput marker gene sequences. ISME J. 2015;9:968–979. doi: 10.1038/ismej.2014.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Amir A, et al. Deblur Rapidly Resolves Single-Nucleotide Community Sequence Patterns. mSystems. 2017;2 doi: 10.1128/mSystems.00191-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Tikhonov M, Leach RW, Wingreen NS. Interpreting 16S metagenomic data without clustering to achieve sub-OTU resolution. ISME J. 2015;9:68–80. doi: 10.1038/ismej.2014.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Johnson JS, et al. Evaluation of 16S rRNA gene sequencing for species and strain- level microbiome analysis. Nat Commun. 2019;10 doi: 10.1038/s41467-019-13036-1. 5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Nielsen HB, et al. Identification and assembly of genomes and genetic elements in complex metagenomic samples without using reference genomes. Nat Biotechnol. 2014;32:822–828. doi: 10.1038/nbt.2939. [DOI] [PubMed] [Google Scholar]
- 190.Yu FB, et al. Microfluidic-based mini-metagenomics enables discovery of novel microbial lineages from complex environmental samples. Elife. 2017;6 doi: 10.7554/eLife.26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Shi X, et al. Microfluidics-Based Enrichment and Whole-Genome Amplification Enable Strain-Level Resolution for Airway Metagenomics. mSystems. 2019;4 doi: 10.1128/mSystems.00198-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.Bowers RM, et al. Minimum information about a single amplified genome (MISAG) and a metagenome-assembled genome (MIMAG) of bacteria and archaea. Nat Biotechnol. 2017;35:725–731. doi: 10.1038/nbt.3893. [Establishes minimal quality reporting requirements for metagenome-assembled genomes (MAGs).] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Almeida A, et al. A new genomic blueprint of the human gut microbiota. Nature. 2019;568:499–504. doi: 10.1038/s41586-019-0965-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Beitel CW, et al. Strain- and plasmid-level deconvolution of a synthetic metagenome by sequencing proximity ligation products. Peer J. 2014;2:e415. doi: 10.7717/peerj.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Costea PI, et al. Towards standards for human fecal sample processing in metagenomic studies. Nat Biotechnol. 2017;35:1069–1076. doi: 10.1038/nbt.3960. [DOI] [PubMed] [Google Scholar]
- 196.Shaiber A, Eren AM. Composite Metagenome-Assembled Genomes Reduce the Quality of Public Genome Repositories. MBio. 2019;10 doi: 10.1128/mBio.00725-19. [Provides an example of how assembling genomes from metagenomes (creating MAGs) can lead to poor quality genomic data and why these genomes should not be considered the same as genomes from isolates.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Schmidt TSB, Raes J, Bork P. The Human Gut Microbiome: From Association to Modulation. Cell. 2018;172:1198–1215. doi: 10.1016/j.cell.2018.02.044. [Reviews the known connections between human gut microbiome and health, including discussion of strain-level variation.] [DOI] [PubMed] [Google Scholar]
- 198.Salter SJ, et al. Reagent and laboratory contamination can critically impact sequence-based microbiome analyses. BMC Biol. 2014;12:87. doi: 10.1186/s12915-014-0087-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199.Goldstein S, Beka L, Graf J, Klassen JL. Evaluation of strategies for the assembly of diverse bacterial genomes using MinION long-read sequencing. BMC Genomics. 2019;20:23. doi: 10.1186/s12864-018-5381-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 200.Alneberg J, et al. Genomes from uncultivated prokaryotes: a comparison of metagenome-assembled and single-amplified genomes. Microbiome. 2018;6:173. doi: 10.1186/s40168-018-0550-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Gawad C, Koh W, Quake SR. Single-cell genome sequencing: current state of the science. Nat Rev Genet. 2016;17:175–88. doi: 10.1038/nrg.2015.16. [DOI] [PubMed] [Google Scholar]