Abstract
Open microfluidic cell culturing devices offer new possibilities to simplify loading, culturing, and harvesting of individual cells or microtissues due to the fact that liquids and cells/microtissues are directly accessible. We present a complete workflow for microfluidic handling and culturing of individual cells and microtissue spheroids, which is based on the hanging-drop network concept: The open microfluidic devices are seamlessly combined with fluorescence-activated cell sorting (FACS), so that individual cells, including stem cells, can be directly sorted into specified culturing compartments in a fully automated way and at high accuracy. Moreover, already assembled microtissue spheroids can be loaded into the microfluidic structures by using a conventional pipet. Cell and microtissue culturing is then performed in hanging drops under controlled perfusion. On-chip drop size control measures were applied to stabilize the system. Cells and microtissue spheroids can be retrieved from the chip by using a parallelized transfer method. The presented methodology holds great promise for combinatorial screening of stem-cell and multicellular-spheroid cultures.
Microfluidics technology has become an important tool in cell biology research, as cells and tissue structures, liquid volumes, as well as the culturing environments can be precisely controlled.1–3 The large majority of microfluidic devices feature a “closed” format, meaning that cells and liquids are kept and transferred in micropatterned channel networks and specific microcompartments, all of which feature ceilings. Loading of liquids and cells into closed compartments, as well as their removal after a successfully conducted experiment, e.g., in order to perform downstream analysis, often includes several tedious handling steps, which limit the reproducibility of the experiments and robustness of the results. A plethora of microfluidic cell preparation, loading, sorting, and trapping concepts has been described in literature.1–5 Another challenge arises from the need for continuous long-term operation (>3 days) of microfluidic setups, as flow control needs to be robust, tube connections need to be reliable over time, and the occurrence of bubbles that perturb the flow needs to be prevented at any time.
Open microfluidic systems potentially allow to overcome many of these issues.6 As the liquid network and compartments are open and, therefore, much better accessible, liquid introduction, liquid sampling, as well as cell loading and removal can be largely simplified. Examples include micro-devices, in which the culturing compartments are open to air, so that cells and medium can be easily loaded and manipulated with a micropipet. People have used open compartments to expose cells to controlled flows,7 concentration gradients of compounds,8,9 or to patch cells at the wall of wells.10 Moreover, open microwells, in which single or multiple cells have been cultured, were turned upside down, so that the cells were relocated to the liquid–air interface at the bottom of the liquid volume in order to conduct dynamic optical imaging from the bottom.11 Open microfluidic conduits feature physical conditions different from those of closed ones,12 so that new concepts of pumping by, e.g., using evaporation, capillary pumps,13 or surface-force-driven methods14 become possible. In more sophisticated devices, the fluid was “suspended” between two or more air phases or between air and a second immiscible liquid.15 Additionally, channels were directly imprinted on superhydrophobic substrates.16 In summary, open microfluidic systems offer several advantages, however, for gaining more acceptance of such systems in the field of cell and microtissue culturing, simple and robust culturing concepts are required.
Here, we present a workflow for microfluidic handling and culturing of cells and microtissue spheroids in suspension. We employed the previously presented technology of hanging-drop networks,17 a fully open microfluidic concept, and engineered new functions, which, in our opinion, considerably simplify key steps of microfluidic cell culturing. The first and most important step includes (i) precise loading of single cells by using a fluorescence-activated cell sorting (FACS) unit and loading of microtissue structures by using a pipet into the culturing compartments of a prefilled microfluidic network. Subsequent steps include (ii) robust long-term perfusion in a controlled cell-culturing environment, and (iii) parallel harvesting of cells and microtissue structures from the microfluidic device compartments for subsequent analysis. Operation protocols and device technology render the system attractive for a broad community and useable for a wide range of applications, including combinatorial studies of stem-cell and multicellular-spheroid cultures under constant perfusion.
Experimental Section
Microfluidic Platform
The microfluidic platform is based on a recently presented hanging-drop network technology.17 In the present study, we used a 4 × 6-drop array with a single inlet and single outlet (Figure 1A). Rim-like micropatterns define wetted and nonwetted areas and were fabricated by using standard soft lithography methods.17 In short, the casting mold was produced from a 4-in. silicon wafer with two patterned SU-8 layers (both 250 μm thick) on top. The mold was used to produce polydimethylsiloxane (PDMS) replica structures. To improve handling of the flexible PDMS devices, they were bonded to a standard microscopy slide containing through-holes to connect the tubing from the top. The substrate was operated in an upside-down configuration so that hanging drops were formed below the circular areas. The hanging drops were interconnected through channels for liquid circulation, to form a bottom-open microfluidic network.
Figure 1. Microfluidic cell culturing workflow.
(A) Layout of the microfluidic device, consisting of a 4-by-6 array of interconnected drops with a single inlet and single outlet. Liquid is loaded from the top and automatically distributed all over the network due to capillary forces. (B) Single-cell loading into standing drops by using FACS and spheroid loading by using pipets. (C) Culturing under controlled perfusion and a close-up of the outlet: state 1, larger drop = liquid removal; state 2, smaller drop = no liquid outflow. (D) Parallel cell and spheroid transfer from the hanging-drop network to a receiver plate.
Preparation and Liquid Loading
A first critical step consists of loading liquid into the empty microfluidic system. Prior to loading, the drop structures of the fabricated chip were rendered hydrophilic, while the rim structures remained hydrophobic. This was achieved by a selective oxygen plasma treatment of the surface with the help of a PDMS mask. A thin PDMS layer with holes at the drop locations was placed onto the PDMS patterns, so that the plasma did only activate the area in between the rims.
Liquid loading was then carried out by simply pipetting the required medium volume from the top into the network not later than 30 min after plasma activation (Figure 1A). This step was performed by placing the system downside-up on a support so that the micropatterns face upward. The liquid is then automatically drawn through the whole network due to capillary forces.
Cell Loading
For loading single cells, we developed a method to directly sort the respective cells into the fluidic system by using FACS. The microfluidic system was placed in a BD FACSAria III flow cytometer (BD Biosciences), configured with a 100 μm nozzle, a sheath pressure of 20 psi, and 30-kHz droplet generation. All sorts were carried using the single-cell sort precision mode. A regular holder for microscopy slides was used, and the chip filled with medium was placed inside the FACS device with the drops facing upward (Figure 1B, left). The sorting chamber was then closed, and a HEPA filter was used to prevent contamination during the sorting procedure. Evaporation during the sorting process was reduced by cooling the stage to 4 °C using the internal cooling system. The preprogrammed positions of each drop were aligned with the chip features. Once set up, FACS-assisted cell loading was executed fully automatically by using the motorized xy-stage.
Fluorescence-labeled (GFP+) HCT-116 cells were loaded as a suspension in PBS with a concentration of 106 cells/mL. The following sorting criteria and parameters were chosen: 1% of GFP+ cell population (maximum fluorescence) at 600 events/s. In average, 6 cells per second were sorted, resulting in a sorting time of 16.6 s for 100 cells.
Granulocyte/macrophage progenitor (GMP) cells were sorted from Pu.1eYFP reporter mice,21 defined as lin−/Sca1−/c-Kit+/CD34+/CD16/32+. Before antibody staining, the cells were filtered through a 40-μm nylon mesh (Becton Dickinson AG, Switzerland) and incubated with anti-CD16/32 antibody (BD2.4G2; BD Pharmingen, Switzerland) or CD16/32 antibody conjugated to AlexaFluor 700. For the lineage (lin) depletion step, biotinylated antibodies for CD3e (145-2C11), CD11b (M1/70), CD19 (1D3), CD41 (MWReg30), B220 (RA3-6B2), Gr-1 (TER-119), TER-119 (RA3-6B2) and streptavidin-conjugated beads Roti-MagBeads (Carl Roth AG, Switzerland) were used on a EasySep magnet (Stem Cell Technologies).
Spheroid Loading
Preformed microtissue spheroids were transferred through simple pipetting steps (Figure 1B, right). Spheroids were taken up into a pipet tip with 20 μL of medium and were allowed to settle at the bottom of the tip. As all liquid compartments are directly accessible, the spheroids were loaded into the microfluidic system by simply bringing the tip into contact with the specific target drop. The two liquid–air interfaces, the one of the tip and the other one of the standing drop, merged, and the spheroid was transferred to the drop by gravitational force. No pipet action was required, and no additional liquid was added to the standing drop during this procedure. Multiple spheroids could be simultaneously loaded by using a multichannel pipet.
Culturing
After cell loading, the microfluidic device had to be flipped upside down and connected to tubing (Figure 1C). The chip flipping could be easily done by hand in approximately 1 s. No additional cell transfer, closure of the microfluidic system, or any other preparation step were required. Important characteristics of hanging-drop networks include that liquid inflow and liquid outflow have to be carefully balanced, so that the total liquid volume in the system remains constant, and the drops do not drip off or dry out. Further, liquid evaporation from the open network has to be limited through a humid environment (incubator) and compensated for via the applied flow. Both issues could be addressed by using a simple but highly effective procedure (details in Figure 1C): The network outlet is also realized as a hanging drop, and the tube inlet position is adjusted to be precisely at the position of the liquid–air interface, a measure through which the drop size is inherently kept constant and stable over time. If the drop volume increases, the drop height increases as well and the liquid–air interface moves below the tube inlet (state 1). Liquid is then withdrawn from the drop until the liquid–air interface and tube inlet are at the same height or until the tube inlet is lower (state 2). Thereby, a segmented flow is generated at the outlet. This flow behavior results in an inherent self-regulation of the size of the outlet drop. Because of Young–Laplace’s law, the size and radius of the outlet drop is defining the radii of all drops in the network, so that any dripping off or dry-out can be prevented during perfusion culturing. The concept allows for operating the system fully independently of any active control, so that it can be placed in a conventional incubator or on a fully automated time-lapse microscope stage. For the inflow side we used a syringe pump and for the outflow side a peristaltic pump, which was operated at a higher flow-rate than the inflow pump.
The cultivation was done on a Leica DMI6000B microscope (Leica AG, Germany) or a Nikon Ti Eclipse microscope (Nikon Instruments Europe B.V., Switzerland), both with stage-top incubators for continuous time-lapse studies, or inside a Teco-20 incubator (Selutec GmBH, Germany). A software-based autofocusing was executed before every image acquisition. Fluorescence micrographs were acquired using a SPECTRA X Light Engine (Lumencor Inc.) and the respective filters for GFP and YFP.
The drop size was regulated as previously explained. After loading, the device was placed onto a custom holder, and the in- and outlets were connected to the pumps via Teflon tubes of different size. The inlet tube had an internal diameter of 0.5 mm and was connected to the device by a 21-gauge 90-degree-bent needle (Techcon Systems). The outlet tube had an internal diameter of 0.3 mm and was connected to the chip by a 32-gauge 90-degree-bent needle, which was used for the drop height regulation. The vertical distance between needle tip and hydrophobic rim was set to ∼600 μm by using a small adjustment tool. The inlet was fed by a neMESYS syringe pump system (cetoni GmbH, Germany), while the outflow was controlled by an ISMATEC IPC peristaltic pump (Idex Health and Science). The use of a peristaltic pump for the outflow was important, as a large volume of air is drawn out, when the drop is smaller than the outlet tubing. The chosen flow-rates for the inlet pump included 2 μL/min for spheroid cultivations and 0.5 μL/min for single cells. The exact value of the flow-rate at the outlet is less critical but has to be higher than that at the inlet. A flow-rate of twice the value at the inlet was usually chosen.
Image Analysis
Image analysis was done by using the Nikon NIS Elements Advanced Research software. Fluorescence-signal quantification was performed after background subtraction.
Biochemical Assays
The ATP content of the microtissue spheroids was determined using the CellTiter-Glo 3D Cell Viability Assay (Promega, Switzerland) according to the manufacturer’s instructions.
Harvesting
All cells and spheroids were harvested in parallel onto a specially designed receiver plate, which comprised of an array of hydrophilic spots arranged at the same pitch as the hanging drops (Figure 1D). Super-hydrophobic areas surrounded the hydrophilic spots. The hanging-drop chip and the receiver plate were both placed into a custom-assembled harvesting platform. The platform ensured a precise alignment and parallel movement of the chips. The hanging drop network was approached to the receiver plate, so that all drops got into short contact (∼1 s) with the hydrophilic areas, and so that small volumes of liquid including all cells and spheroids were transferred in parallel. Cells and spheroids remained isolated from each other and were accessible for further analysis. The receiver plate was fabricated by photo-lithographic patterning of a hydrophobic silazane layer on a 4-in. glass substrate.
Results and Discussion
Liquid Loading
Initial filling of the microfluidic device was straightforward and was carried out within less than a minute by using a conventional pipet. The presented design holds a total liquid volume of 200 μL. The open nature of the network prevented any entrapment of bubbles, a problem that exists in many closed microfluidic systems and led to the development of several “antibubble” strategies.18,19 All drops have the same size due to pressure equilibration, even though liquid filling was applied to only a few of them.
Spheroid Loading
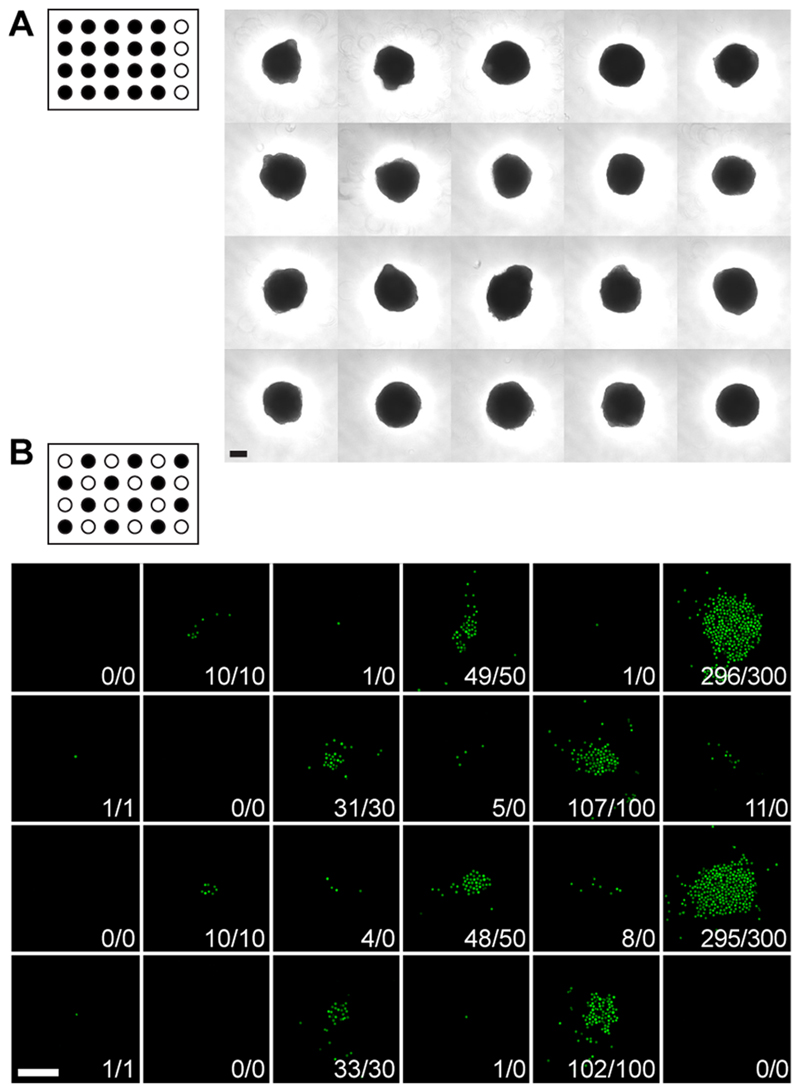
Spheroids were loaded directly from the production well plate into the chip with a multichannel pipet (always 2 or 3 at a time). A total of 20 spheroids could easily be loaded within less than 10 min. The flipping of the chip was surprisingly simple and straightforward to perform manually within a second, while the drops remained stable. No cross-talk was observed. After the flipping of the chip, the spheroids did sediment to the bottom center of the hanging drop (Figure 2A). The simplicity of the procedure allows for a very flexible arrangement of spheroids; drops can either be left without or filled with different spheroid types according to experimental requirements.
Figure 2.
(A) Bright-field picture of the hanging-drop chip, loaded with cellular spheroids, after turning it upside down and cell sedimentation. (B) Green-fluorescence micrographs of cells, sorted into the hanging drop chip, after turning it upside down and sedimentation. The resulting and intended cell numbers per drop are indicated. (The target drops are marked in black at the top of each image. Both scale bars are 200 μm).
FACS-Assisted Cell Loading
Cells were repeatedly sorted into the standing drops by using FACS. Sorting parameters were optimized with respect to accuracy, potential splashing, and speed. Figure 2B shows a result of loading GFP positive HCT-116 cells that were kept in suspension in PBS. For illustration and accuracy assessment, a chess-like pattern has been chosen with different numbers of cells per drop (1, 10, 30, 50, 100, and 300 cells). Only cells with a high GFP expression level (top 1%) were sorted, cells with lower expression levels were routed to waste. The duration of such a sorting run depends on the number and appearance frequency of available cells in the sorting gate and the desired number of cells per drop and generally included between 10 and 20 min for a 4-by-6 drop array. No splashing or bubbles were observed during the loading procedure, and the drop network remained stable. For low numbers of cells (<10), no cross-contamination between neighboring drops was observed and the obtained cell numbers corresponded to the predefined values. For more than 10 cells sorted per drop, cross-contamination could not be completely eliminated and a fluctuation of ∼5% was observed. Transfer of cells to neighboring drops can have two reasons: (1) We observed that cells are intrinsically spread in an area of ∼2 mm in diameter within the drop after the sort. Cells off center may be dragged more easily to neighboring drops. (2) For larger cell numbers, the additional liquid volume that has been added to the standing drops by the sorting drops (∼2 nL) induces liquid exchange between the neighboring drops, so that cells may be dragged along with the liquid flow. The maximal number of cells per drop was around 2000; for larger numbers, we observed air bubbles remaining in the drop.
Culturing of Spheroids
Figure 3A shows a side view of the chip filled with red-colored liquid. The precise position of the tubing in the outlet drop (close-up view in Figure 3B) determines and levels the drop size throughout the whole array. All drops had the same size and were stable over the whole duration of the experiments (Figure 3C). Evaporation was inherently counteracted through the pumping protocol, so that the drop size did not depend on the humidity level of the culturing environment. For stable operation, it is important to notice that the outflow controlled by the peristaltic pump always has to be larger than the inflow controlled by the syringe pump. Further, the inflow should be larger than the total evaporation rate (typically ∼1% of the total chip volume per hour in a humidified environment). As a result of the continuous alternation between air and liquid withdrawn at the outflow, a segmented flow is generated, which can be handled more robustly by a peristaltic pump than by a syringe pump.
Figure 3.
(A) Picture of the hanging-drop chip filled with red food dye for showing the hanging drop network. (B) Close-up view of the outlet drop with the tubing tip placed at the liquid–air interface defining the drop height. (C) Measured drop height variation over 10 h at a flow-rate 5 μL/min. (D) Bright-field and fluorescence micrographs of eight spheroids at the beginning and after 3 days in culture (loaded drops are marked in black, scale bars are 200 μm). (E) Relative spheroid growth in perfusion mode and (F) ATP content at the start and after 3 days in a static, D3(s), and perfused chip culture, D3(p). (Medians are marked in black).
Continuous-flow cell culturing was demonstrated in two different experimental setups: (i) growth and viability of HCT-116 spheroids over several days in culture and (ii) formation and culturing of spheroids from HCT-116 cells loaded by using FACS.
Eight GFP-positive HCT-116 spheroids were loaded into the chip using a standard pipet. The spheroids have been formed in a GravityPLUS hanging-drop system (InSphero AG, Switzerland) over 3 days. After loading, the spheroids were exposed to constant perfusion at a flow-rate of 2 μL/min. As a control, eight spheroids were cultured in conventional, low-adhesion wells with medium exchange every 2 days (“static” conditions). After 3 days of cultivation, all spheroids were collected and an ATP-viability-assay was performed. Figure 3D shows all spheroids after loading and after 3 days in culture (bright field and fluorescence images). The measured relative growth was consistently 38 ± 6% (n = 7, excluding the outlier, Figure 3E). The measured ATP content increased for both, the control and the perfused cultures as a result of cell proliferation (Figure 3F). Generally, larger values were measured for the perfused cultures, however, also with larger variation, which can be attributed to the variations in the initial size of the spheroids before starting the experiments.
Loading and Culturing of a HCT-116 Cancer Cell Line
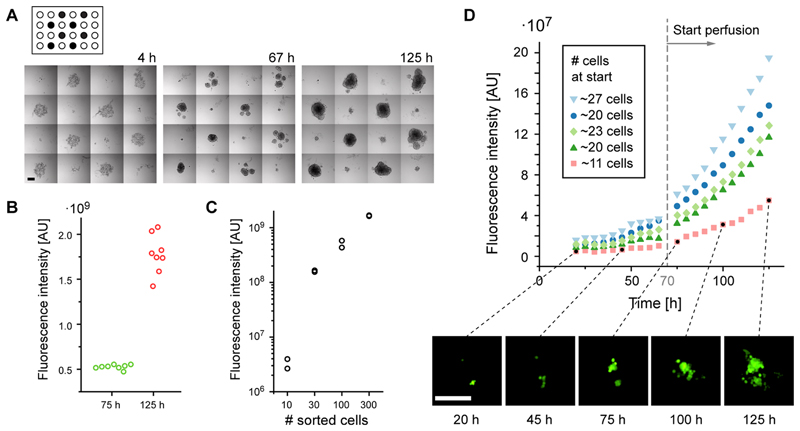
Fluorescent HCT-116 colon cancer cells were sorted into the hanging-drop chip in different cell numbers and configurations. HCT-116 cells are adherent cells and form cell clusters when cultured on nonadherent substrates. Figure 4A shows bright-field images of hanging drops at different time points, where 300 cells have been sorted into 8 of the drops in a chess-board-like pattern. As observed previously, sorting relatively large numbers of cells entailed a fluctuation of ∼5% in neighboring drops. Cells then sedimented to the hanging drop bottom after sorting (4 h), aggregated into cell clusters (67 h) and proliferated in all drops (125 h). Similar total fluorescence intensities in all 8 drops after 75 and 125 h in the continuously perfused culture evidenced the reproducibility of the sorting and culturing method and proved viability of the cells after (Figure 4B). Further, in the case of sorting different cell numbers into droplets, the resulting cell cluster size and total fluorescence intensity nicely correlated with the initially sorted cell numbers, as it is shown in Figure 4C. Intensities were measured 125 h after sorting of 10, 30, 100, and 300 cells (each in duplicates) in a similar configuration as shown in Figure 4A. Finally, cell aggregation and cell culturing has been continuously monitored by using automated fluorescence time-lapse microscopy. Figure 4D shows the fluorescence intensity versus time for five different low cell numbers of less than 30. The initial cell number was determined from bright-field images. In all cases, continuous medium exchange through perfusion has a clear effect on the growth rate of cells cultured in hanging drops. The growth rate increased visibly after 70 h when culturing was switched from static to perfusion mode. Again, the fluorescence intensities correlated with the initial cell number. The variations observed for cell numbers close to ∼20 might be attributed to variations in cell viability, cell state, and initial GFP expression.
Figure 4.
(A) Bright-field images of 300 fluorescent HCT-116 cells, sorted in a chess-board-like pattern into hanging drops, and of subsequent spheroid formation during 125 h (target drops are marked in black, scale bar is 200 μm, see Supplementary Movie 1 for full time-lapse sequence). (B) Fluorescence intensity increase of 8 spheroids derived from 300 sorted, fluorescent HCT-116 cells over time. (C) Fluorescence intensity of cell clusters after 125 h in culture in dependence of the initially sorted cell number (n = 2). (D) Fluorescence intensity development versus time for clusters consisting of low numbers of cells, and fluorescence micrographs at different time points (scale bar is 200 μm, contrast has been increased for better visibility).
Loading and Culturing of Primary Blood Progenitor Cells
Freshly isolated mouse granulocyte and macrophage progenitor (GMP) cells were sorted from bone marrow suspension directly into the hanging-drop chip, containing a permissive culture medium (10% FCS, 100 ng/mL SCF, 100 ng/mL TPO and 20 ng/mL IL3, 10 ng/mL IL6). A target number of 50 GMP cells per drop was sorted, and the cells were cultured in the hanging-drops for 5 days. Medium was exchanged by using flow intervals every 2 days. Figure 5A shows phase-contrast images of all 24 hanging drops following sedimentation of the cells 1 day after sorting (day 1) and at day 5. The number of cells per drop after sorting amounted to 52.3 ± 4.7 (n = 24) cells, once again demonstrating the high accuracy and precision of FACS-based device loading. Cells expanded in all drops during the 5 days in continuous-flow culture. In contrast to adherent cells (cf. Figure 4), the nonadherent blood progenitor cells did not form clusters or aggregates, as expected. Compared to expansion cultures in wells, where the movement of cells across the surface of the cell culture vessel can be frequently observed, all cells stayed in the center of the image at the lowest point of the liquid–air interface and did not move. The GMPs were derived from a knock-in reporter line expressing a fluorescently tagged version of the transcription factor PU.1 (PU.1eYFP). Following culturing for 5 days, some GMP differentiated into macrophages and formed clusters of adherent cells with high levels of PU.1eYFP expression (Figure 5C). This demonstrates that the hanging-drop approach enables one to simultaneously observe transitions in cellular behavior and to assess fluorescent reporter genes using time-lapse imaging. After 5 days in culture, all cells were retrieved from the microfluidic hanging-drop device and further characterized by flow cytometry (Figure 5B). To this end the retrieved cells were stained with antibodies against Ly-6G a marker for granulocytes and F4/80, a marker for macrophages. Flow-cytometric analysis suggests that 15.9% of the cells differentiated into macrophages and 15.6% into granulocytes. This finding demonstrates that the hanging-drop approach can be conveniently combined with standard immunophenotypic analysis procedures that are routinely used in hematology.
Figure 5.
(A) Phase contrast images (10×) of all 24 drops at day 1, after loading 50 granulocyte/macrophage progenitor (GMP) cells from bone marrow suspension into each drop using a FACS unit, and at day 5 after subsequent culturing in the hanging drops (target drops are schematically marked in black, scale bar is 200 μm). (B) Flow-cytometric characterization of retrieved cells after 5 days in culture. (C) Yellow fluorescence micrographs of the hanging-drop culture taken every day (scale bar is 200 μm, see Supplementary Movie 2 for full time-lapse sequence).
Flow Rates
Possible flow rates for spheroid cultivation ranged between 0 and 10 μL/min in order to maintain stable chip operation. Higher flow rates increased the chance of liquid losses, as the drop height control needed to react much faster then. For good cell viability and reasonable medium consumption in long-term experiments, a flow rate of 2 μL/min has been chosen. Dragging and transfer of spheroids into other droplets has never been observed for this flow regime.
For experiments with individual nonadherent or adherent cells before spheroid formation, drag-away is more likely to occur. For those cases, the flow rates were set below 1 μL/min. Finally, pulsed flow, driven by the inlet syringe pump, was tested and proved to be a viable option, as liquid is only drawn away, when the drops increase in size, even when the peristaltic pump was continuously running. Pulsed flow enables the settlement of single cells at the drop bottom during low-flow phases.
Harvesting
Spheroids could be easily transferred in parallel by using the fabricated receiver plate and a custom-made alignment and transfer setup (Figure 6A). The cell transfer as described in the Experimental Section is similar to a previously published concept. Whereas Cavnar et al.20 transferred spheroids from isolated hanging drops into wells, we show here that this concept is also applicable for hanging-drop networks by using a flat surface-patterned receiver plate. Figure 6B shows the results, which include a plate containing eight spheroids in a chess-board like pattern after transfer. We repeatedly harvested 100% of the spheroids in the designated locations. No cross-contamination was observed. In the receiver plate, the spheroids are individually accessible, as liquid connections through channels do no more exist. Therefore, all spheroids can be further assayed without liquid and cellular cross-talk. The pitch of the hanging drops and receiving structures is 4.5 mm, which is compatible with the 384-well format.
Figure 6.
(A) Photograph of the harvesting platform including the hanging drop chip at the top and the receiver plate below. (B) Spheroids imaged after harvesting on the receiver plate. Gray shadows are optical artifacts in transmission-light microscopy mode, which are produced by the curved drop liquid–air interface. Initially loaded drops are marked in black in the top schematic. Scale bar is 200 μm.
Conclusions
The open format of the hanging-drop network technology has been fully exploited to implement new features for loading, culturing, and harvesting of single cells, multiple cells, and microtissue spheroids into, in, and from a microfluidic environment. The most important characteristics of this format include the fully open design and the resulting accessibility of any part of the fluidic network. FACS has been used since decades for sorting cells and has been made compatible for use with microscopy glass slides.22 Here, we developed a method, with which cells can be sorted directly into compartments of a microfluidic perfusion network, so that the accuracy and sorting capability of FACS is directly combined with subsequent microfluidic culturing. Whereas most cell loading protocols follow stochastic mechanisms, FACS-loading allows for positioning the cells of interest at a defined location of the microfluidic chip. Further, handling and manipulation steps of potentially delicate cells are reduced. To the best of our knowledge, it is the first time that cells have been directly sorted into a ready-to-use microfluidic network without the need of any intermediary manipulation. A wide range of cell numbers (here, between 1 and 300 cells) can be sorted into each drop. It is worth mentioning that cells can also be sorted according to their light-scattering characteristics (forward and side scattering), independent of the emission of fluorescence signals. This feature enables to distinguish single cells from cell clusters and to perform qualitative assessments of the morphology and intrinsic structure of the cells during loading. Adherent cells then form aggregates and tissue-like structures and have been demonstrated to remain viable over at least 5 days. The seamless combination of FACS and the microfluidic device opens new perspectives and applications in cell-based research, where only a small and defined selection of cells out of a large population is of interest. Hematopoietic progenitor cells are a prime example. We were able to sort and load a defined population of GMPs sorted from a freshly isolated bone marrow cell suspension directly into the chip and to subsequently culture, expand, and phenotypically characterize them for more than 5 days.
On the other hand, the open nature of the system greatly simplifies the loading of microtissue spheroids, in particular with respect to closed systems, where special traps23 or compartments24 are required. Moreover, the open system offers great flexibility in how different types of spheroids are arranged in a microfluidic network and keeps them in their original environment, in which any cell adhesion is inherently prevented. These advantages hold great potential for a wide range of applications including the use of stem cells,25–28 embryonic bodies,29,30 and the large variety of microtissue spheroids.31–34 The loading procedures described here constitute important alternatives to the previously published method, where defined cell suspensions were supplied during filling the chip with liquid.17
The new drop-size regulation substantially simplifies the setup and its operation and opens the route to highly parallelized experiments. Several chips can be run in parallel without any need for microscope- or computer-based control. The liquid–air interface at the bottom of the culturing compartments provides ideal culturing conditions for suspension cell cultures without adhesive interactions to surfaces that might affect their morphology or cellular behavior. The absence of a solid support surface is even more important for microtissue structures, for which cell adhesion has to be prevented by any means, as it would entail loss of tissue morphology, integrity, and functionality. Closed systems require the application of special antiadhesion coatings. Further, gravity and the drop shape ensure that cells and spheroids are automatically centered at the bottom of the drop and keep their position also under controlled perfusion.
In addition to cell and tissue loading also harvesting of the cellular material from the device is very simple due to the open system format. Postculture analysis and experiments including viability assays, ELISAs, staining, or determination of end point values can be performed with the individual cell aggregates or tissues.
In summary, all the new features described here substantially extend the number of possible applications of the hanging-drop network format. Handling steps are reduced to a minimum without compromising the microfluidic functionality, which is at the basis of robust and efficient operation.
Supplementary Material
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.5b03513.
Time-lapse sequences of Figure 5c (phase contrast and fluorescence, scale bar is 200 μm) (AVI)
Acknowledgments
This work was supported by the European Union FP7 grant under the project “Body on a chip” Grant ICT-FET-296257 and an individual Ambizione Grant 142440 of the Swiss National Science Foundation for Olivier Frey. Martin Etzrodt and Timm Schroeder are supported by an EMBO long-term fellowship (Grant ALTF 825-2012) and a Swiss National Science Foundation grant (Grant 31003A_156431), respectively.
Footnotes
The authors declare no competing financial interest.
References
- (1).Mu X, Zheng W, Sun J, Zhang W, Jiang X. Small. 2013;9:9–21. doi: 10.1002/smll.201200996. [DOI] [PubMed] [Google Scholar]
- (2).Kovarik ML, Gach PC, Ornoff DM, Wang Y, Balowski J, Farrag L, Allbritton NL. Anal Chem. 2012;84:516–540. doi: 10.1021/ac202611x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).El-Ali J, Sorger PK, Jensen KF. Nature. 2006;442:403–411. doi: 10.1038/nature05063. [DOI] [PubMed] [Google Scholar]
- (4).Lindström S, Andersson-Svahn H. Lab Chip. 2010;10:3363–3372. doi: 10.1039/c0lc00150c. [DOI] [PubMed] [Google Scholar]
- (5).Mehling M, Tay S. Curr Opin Biotechnol. 2014;25:95–102. doi: 10.1016/j.copbio.2013.10.005. [DOI] [PubMed] [Google Scholar]
- (6).Kaigala GV, Lovchik RD, Delamarche E. Angew Chem Int Ed. 2012;51:11224–11240. doi: 10.1002/anie.201201798. [DOI] [PubMed] [Google Scholar]
- (7).Hsu C-H, Chen C, Folch A. Lab Chip. 2004;4:420–424. doi: 10.1039/b404956j. [DOI] [PubMed] [Google Scholar]
- (8).Keenan TM, Hsu CH, Folch A. Appl Phys Lett. 2006;89:114103. [Google Scholar]
- (9).Wright Ga, Costa L, Terekhov A, Jowhar D, Hofmeister W, Janetopoulos C. Microsc Microanal. 2012;18:816–828. doi: 10.1017/S1431927612000475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Lau AY, Hung PJ, Wu AR, Lee LP. Lab Chip. 2006;6:1510–1515. doi: 10.1039/b608439g. [DOI] [PubMed] [Google Scholar]
- (11).Bocchi M, Rambelli L, Faenza A, Giulianelli L, Pecorari N, Duqi E, Gallois J-C, Guerrieri R. Lab Chip. 2012;12:3168–3176. doi: 10.1039/c2lc40124j. [DOI] [PubMed] [Google Scholar]
- (12).Melin J, van der Wijngaart W, Stemme G. Lab Chip. 2005;5:682–686. doi: 10.1039/b501781e. [DOI] [PubMed] [Google Scholar]
- (13).Zimmermann M, Bentley S, Schmid H, Hunziker P, Delamarche E. Lab Chip. 2005;5:1355–1359. doi: 10.1039/b510044e. [DOI] [PubMed] [Google Scholar]
- (14).Kim S-J, Paczesny S, Takayama S, Kurabayashi K. Lab Chip. 2013;13:2091–2098. doi: 10.1039/c3lc50187f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Casavant BP, Berthier E, Theberge AB, Berthier J, Montanez-Sauri SI, Bischel LL, Brakke K, Hedman CJ, Bushman W, Keller NP, Beebe DJ. Proc Natl Acad Sci U S A. 2013;110:10111–10116. doi: 10.1073/pnas.1302566110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Oliveira NM, Neto AI, Song W, Mano JF. Appl Phys Express. 2010;3 085205. [Google Scholar]
- (17).Frey O, Misun PM, Fluri DA, Hengstler JG, Hierlemann A. Nat Commun. 2014;5:4250. doi: 10.1038/ncomms5250. [DOI] [PubMed] [Google Scholar]
- (18).Lochovsky C, Yasotharan S, Gunther A. Lab Chip. 2012;12:595–601. doi: 10.1039/c1lc20817a. [DOI] [PubMed] [Google Scholar]
- (19).Sung JH, Shuler ML. Biomed Microdevices. 2009;11:731–738. doi: 10.1007/s10544-009-9286-8. [DOI] [PubMed] [Google Scholar]
- (20).Cavnar SP, Salomonsson E, Luker KE, Luker GD, Takayama S. J Lab Autom. 2014;19:208–214. doi: 10.1177/2211068213504296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Kirstetter P, Anderson K, Porse BT, Jacobsen SEW, Nerlov C. Nat Immunol. 2006;7:1048–1056. doi: 10.1038/ni1381. [DOI] [PubMed] [Google Scholar]
- (22).Stovel RT, Sweet RG. J Histochem Cytochem. 1979;27:284–288. doi: 10.1177/27.1.374588. [DOI] [PubMed] [Google Scholar]
- (23).Ruppen J, Cortes-Dericks L, Marconi E, Karoubi G, Schmid Ra, Peng R, Marti TM, Guenat OT. Lab Chip. 2014;14:1198–1205. doi: 10.1039/c3lc51093j. [DOI] [PubMed] [Google Scholar]
- (24).Kim J-Y, Fluri Da, Marchan R, Boonen K, Mohanty S, Singh P, Hammad S, Landuyt B, Hengstler JG, Kelm JM, Hierlemann A, Frey O. J Biotechnol. 2015;205:24–35. doi: 10.1016/j.jbiotec.2015.01.003. [DOI] [PubMed] [Google Scholar]
- (25).Banerjee M, Bhonde RR. Cytotechnology. 2006;51:1–5. doi: 10.1007/s10616-006-9001-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (26).Seita J, Weissman IL. Wiley Interdiscip Rev Syst Biol Med. 2010;2:640–653. doi: 10.1002/wsbm.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Gutiérrez L, Lindeboom F, Ferreira R, Drissen R, Grosveld F, Whyatt D, Philipsen S. Exp Hematol. 2005;33:1083–1091. doi: 10.1016/j.exphem.2005.06.014. [DOI] [PubMed] [Google Scholar]
- (28).Etzrodt M, Endele M, Schroeder T. Cell Stem Cell. 2014;15:546–558. doi: 10.1016/j.stem.2014.10.015. [DOI] [PubMed] [Google Scholar]
- (29).Medvinskii LA, Kolesnichenko TS. Bull Exp Biol Med. 1981;91:408–410. [Google Scholar]
- (30).Seiler AEM, Spielmann H. Nat Protoc. 2011;6:961–978. doi: 10.1038/nprot.2011.348. [DOI] [PubMed] [Google Scholar]
- (31).Kelm JM, Timmins NE, Brown CJ, Fussenegger M, Nielsen LK. Biotechnol Bioeng. 2003;83:173–180. doi: 10.1002/bit.10655. [DOI] [PubMed] [Google Scholar]
- (32).Kelm JM, Fussenegger M. Trends Biotechnol. 2004;22:195–202. doi: 10.1016/j.tibtech.2004.02.002. [DOI] [PubMed] [Google Scholar]
- (33).Hsiao AY, Tung Y-C, Qu X, Patel LR, Pienta KJ, Takayama S. Biotechnol Bioeng. 2012;109:1293–1304. doi: 10.1002/bit.24399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Bartosh TJ, Ylöstalo JH, Mohammadipoor A, Bazhanov N, Coble K, Claypool K, Lee RH, Choi H, Prockop DJ. Proc Natl Acad Sci U S A. 2010;107:13724–13729. doi: 10.1073/pnas.1008117107. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Time-lapse sequences of Figure 5c (phase contrast and fluorescence, scale bar is 200 μm) (AVI)