Abstract
Ovarian function is central to female fertility, and several genome-wide association studies (GWAS) have been carried out to elucidate the genetic background of traits and disorders that reflect and affect ovarian physiology. While GWAS have been successful in reporting numerous genetic associations and highlighting involved pathways relevant to reproductive aging, for ovarian disorders, such as premature ovarian insufficiency and polycystic ovary syndrome, research has lagged behind due to insufficient study sample size. Novel approaches to study design and analysis methods that help to fit GWAS findings into biological context will improve our knowledge about genetics governing ovarian function in fertility and disease, and provide input for clinical tools and better patient management.
Genetics of Ovarian Biology
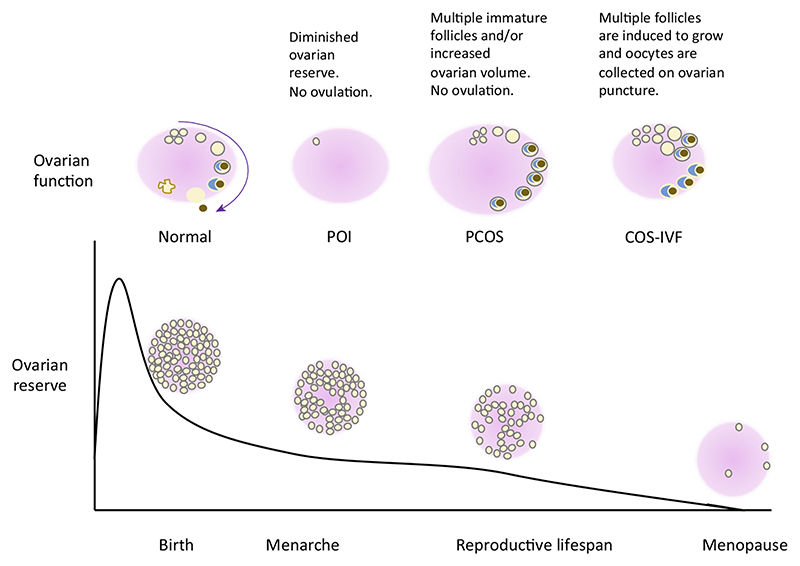
The ovarian reserve (see Glossary), one of the key elements of female fertility, is influenced by many factors, including genetics. As a result, much attention has been focused on elucidating the genetic background of both normal reproduction and various disorders that affect and reflect the ovarian reserve and healthy folliculogenesis. Ovarian function revolves around folliculogenesis, a cyclic process responsible for the maturation and release of oocytes from the ovaries. The follicle reserve and the effectiveness of folliculogenesis affect female reproductive potential, which directly depends on natural reproductive aging, delineated by menarche and menopause-the two time-points in a woman’s life that open and close the reproductive window, respectively. However, in many women the normal course of events is disturbed by pathologies such as premature ovarian insufficiency (POI) and polycystic ovary syndrome (PCOS). Both POI and PCOS have a significant impact on female fertility because compromised reproduction is an inherent feature of POI, while PCOS is the most common cause of anovulatory infertility [1]. In addition to being essential for natural conception, ovarian reserve and response also remain the key limiting steps in assisted conception because one of the prerequisites for successful in vitro fertilization (IVF) treatment is the availability of multiple good-quality oocytes (Figure 1).
Glossary.
- Anti-Müllerian hormone (AMH)
a hormone produced by granulosa cells of small antral follicles. AMH levels reflect the number of small antral follicles (oocytes) and decline with age. Lower AMH levels may indicate a decreased dynamic ovarian reserve.
- Antral follicle count (AFC)
the number of (2–10 mm) antral follicles visible in the ovaries by ultrasonography early in the menstrual cycle.
- Controlled ovarian stimulation (COS)
refers to the growth of multiple ovarian follicles when induced by stimulating the ovaries with exogenous hormones.
- Early menopause (EM)
menopause before the age of 45 years.
- Follicle-stimulating hormone (FSH)
a pituitary-derived hormone that stimulates the growth of ovarian follicles. Elevated FSH levels in early menstrual cycle may indicate a decreased ovarian reserve.
- Genome-wide association study (GWAS)
a study to test the association between millions of genetic markers and a phenotype of interest. Owing to the large number of association tests carried out, a P value threshold of P = <5 × 10−8 is used to avoid false-positive results.
- In vitro fertilization (IVF)
a methodology used to treat infertility. A typical IVF treatment cycle involves COS, in vitro fertilization of the obtained oocytes, and culture of the embryos, which are then transferred into the uterus.
- Luteinizing hormone (LH)
a pituitary-derived hormone that triggers ovulation.
- Ovarian reserve
a term used to describe female reproductive potential as defined by the total number of oocytes in the ovaries.
- Polycystic ovary syndrome (PCOS)
the most common endocrine disorder among reproductive-aged women (prevalence ~10%) that encompasses menstrual cycle disturbances, hyperandrogenism, and in some cases also obesity and insulin resistance.
- Premature ovarian insufficiency (POI)
menopause before the age of 40 years; affects about 1% of reproductive-aged women.
Figure 1. Ovarian Reserve Throughout Life, Normal Ovarian Function, And the Impact of Various Pathologies.
The ovarian reserve is established before birth, and thereafter the number of follicles containing the oocytes decreases through controlled atresia until menopause when the ovarian reserve is virtually exhausted. Normally, during the reproductive lifespan ovarian follicles go through distinct developmental stages and one oocyte is released monthly. However, in women with polycystic ovary syndrome (PCOS), hormonal disturbances result in follicular arrest and anovulation, while in premature ovarian insufficiency (POI) the ovarian reserve is depleted prematurely. During ovarian stimulation (COS) used for in vitro fertilization (IVF), exogenous hormones are used to stimulate the growth and release of several oocytes.
Normal reproductive aging has a strong genetic component, and the heritability of menopausal age can be as high as 90% [2]. Similarly, genetic factors have been implicated both in the pathogenesis of POI [3] and PCOS (heritability up to 70%) [4], justifying the search for genetic determinants. Although epidemiological evidence points to a complex interplay between reproductive aging and various ovary-related traits or conditions [5,6], the extent to which these phenotypes share genetic determinants has only recently begun to be clarified.
Over the past decade, advances in array-based genotyping technologies and the concurrent development of a wide variety of software tools have led to increased interest in genome-wide association studies (GWAS). These studies have broadened our horizon regarding the genetic architecture of complex traits and have resulted in new discoveries concerning the genetics governing female reproduction. This review summarizes current knowledge and addresses the impact that GWAS have on our understanding of ovarian biology as a whole, while concomitantly discussing potential new developments in study group selection, data analysis, and interpretation, and finally the translational potential of the findings.
Designing the Study–Biobanks or Patient-Based Cohorts?
Both hypothesis-driven candidate gene studies and hypothesis-free GWAS begin by defining the phenotype and selecting the appropriate study group, a crucial step that defines the success of a study [7]. Traditionally the research centers conducting the study have recruited very precisely defined participants to ensure homogeneity of the study group. However, the power of a study to detect genetic associations, that each have a small effect on the complex trait or disease of interest, depends directly on the study size; the larger the study the more associations, and smaller effects are robustly identified [8]. The establishment of large populationbased biobanks that collect biological material, phenotype, and lifestyle data has made it possible to dramatically increase the sample size for some traits. For example, recent GWAS meta-analyses for age at menarche/menopause have included tens of thousands of women and revealed dozens of associated loci [9–11]. However, the question remains of whether population-based biobanks with healthcare records and questionnaire-based data can be used for large-scale studies on phenotypes that are usually defined using rigorous clinical criteria. One of the main advantages of recruiting individuals for a specific study is the possibility to collect additional biological material (urine, follicular cells, and other tissues) or information on diseasespecific sub-phenotypes, such as hormonal values or data from ultrasound scans, which are usually not collected from individuals recruited into population-based biobanks. Biobanks have proved to be extremely useful for studying anthropometric traits (such as height) that can be extracted from self-reported data [12]. Several biobanks have established female healthspecific questionnaires, thereby creating valuable datasets for reproductive health-related phenotypes which could also provide insight into ovarian biology. For example, a recent study proposed that menstrual cycle length could be a proxy marker for ovarian reserve and oocyte quality [13], and recently a GWAS for this trait was published, highlighting variants near the FSHB gene that encodes the ß subunit of follicle-stimulating hormone (FSH) [14]. Because menstrual cycle length is among the phenotypes that some biobanks collect data for (relevant questions are included in the UK Biobanki, the Estonian Biobankii, and the LifeLines Biobankiii female health questionnaires), we will probably soon have more information about the genetics of menstrual cycle. Moreover, phenotypes such as parity also reflect ovarian function to some extent, and GWAS conducted thus far suggest that associated genetic variants are there to be found with the help of larger study cohorts [15,16].
In addition, biobanks collect data on medical history and current health status and, depending on regional legislation, also have the possibility to link with national or hospital databases, creating the opportunity to confirm diagnoses or identify cases. An algorithm combining various inclusion and exclusion criteria has successfully been used to extract patients with suspected POI from a population-based biobank, to explore the prevalence and epidemiology of the condition [17], and a similar approach could also be used in studies aimed at finding genetic associations with POI. Furthermore, the usability of text-mining algorithms for the detection of women with PCOS has also been explored [18], providing additional means for increasing sample size of this phenotype.
Successful reproductive aging GWAS meta-analyses have demonstrated the value of population-based biobanks. Whether these resources could also be used to progress genetic studies on traits and disorders associated directly with ovarian biology remains to be established.
Analyzing the Data–Quality or Quantity?
The main concern related to using biobanks for studying various disorders is the question whether increase in quantity comes at the price of quality. As mentioned in the previous section, specifically recruited patients enable seemingly homogenous study groups to be put together. For example, all three criteria used for diagnosing PCOS (Box 1) overlap to some degree, but also encompass some different characteristics, resulting in varying phenotypes under the umbrella of PCOS diagnosis. Until now it has been suggested that all diagnostic criteria should be treated as separate entities in genetic association studies in the interests of clinical precision and study group homogeneity. However, no significant genetic heterogeneity across the National Institutes of Health (NIH) and Rotterdam criteria, and in self-reported disease status, was observed in the latest PCOS GWAS [19]. This suggests that, although important from a clinical perspective, less-stringent clinical criteria, International Classification of Diseases (ICD) codes, or even self-reported status can be used for some conditions to increase the power to detect associations in large-scale studies. However, this approach needs to be validated for each phenotype either by assessing genetic heterogeneity in comparison with clinically confirmed cases, or by replicating established genotype—phenotype associations [20].
Box 1. PCOS Diagnostic Criteria.
Owing to the heterogeneous clinical manifestation of PCOS, which results in nearly 20 different phenotypes [71], the diagnosis cannot be made based on a single characteristic. Over the years many different definitions have been used, but currently three sets of criteria have been proposed for diagnosing PCOS that use in various combinations the following characteristics: hyperandrogenism (presenting as excessive body hair, acne, or baldness), hyperandrogenemia (elevated male sex hormone levels in blood), oligomenorrhea (less than nine menstrual periods a year), amenorrhea (no menstrual periods), and polycystic ovaries (increased ovarian volume or at least 12 follicles measuring 2-9 mm in diameter in at least one ovary). According to the NIH/National Institute of Child Health and Human Development (NICHD) criteria established in 1990, PCOS is defined as the presence of hyperandrogenism and/or hyperandrogenemia and menstrual dysfunction [72]. The so-called Rotterdam criteria proposed in 2003 state that two of the following three characteristics must be present: hyperandrogenism and/or hyperandrogenemia, menstrual dysfunction, and polycystic ovaries visualized on ultrasound [73]. Use of the Rotterdam criteria is also suggested by the Endocrine Society [74]. Finally, the Androgen Excess Society Criteria published in 2009 proposed defining PCOS as the presence of both hyperandrogenism (clinical and/or biochemical) and ovarian dysfunction (oligo- or anovulation and/or polycystic ovaries) [71]. All three criteria also state that other conditions that might mimic these symptoms need to be excluded.
In addition to larger sample size, study power can be increased by other means of expanding the dataset, for example by taking advantage of repeated measurements [21] such as hormone values or other quantitative traits that vary in time and have been measured at different timepoints. Repeated measurements are, for example, generated in IVF treatment cycles, where women often undergo more than one treatment cycle, resulting in multiple data-points for hormonal measurements and controlled ovarian stimulation (COS) outcome (the number of oocytes retrieved in each cycle). In data analysis, usually all but one cycle is discarded; however, more complex statistical models that make full use of the data at hand are necessary here [22].
The novel approaches to study group formation and the multi-layered structure of collected data for certain traits discussed in this section provide a means to increase analysis power. Potentially, this could lead to interesting new discoveries regarding the genetics of PCOS, POI, and ovarian reserve.
What Have We Learned So Far about Ovarian Biology from GWAS?
The list of traits related to the ovarian function and ovarian reserve interrogated by the GWAS approach includes not only those that are markers of ovarian reserve but also the folliculogenesis-related pathologies POI and PCOS as well as reproductive aging parameters, such as age at menarche and menopause, that can provide clues about the processes governing ovarian physiology. The primary outcome of a GWAS analysis—a list of significantly associated variants, is a starting point to unravel the physiological mechanisms underlying a trait. Because GWAS takes advantage of linkage disequilibrium (LD) between variants, and the majority of GWAS hits lie in intergenic (regulatory) regions [23], the use of different approaches is necessary to find the most likely causal genes and associated biological mechanisms. A plethora of analytical tools have been developed to take the list of variants forward and fit them into a larger biological picture by aiding the identification of likely causal genes and pathways, and the dissection of underlying mechanisms and genetic correlations to other comorbidities and traits (Box 2).
Box 2. What Can We Do with GWAS Findings In Silico?
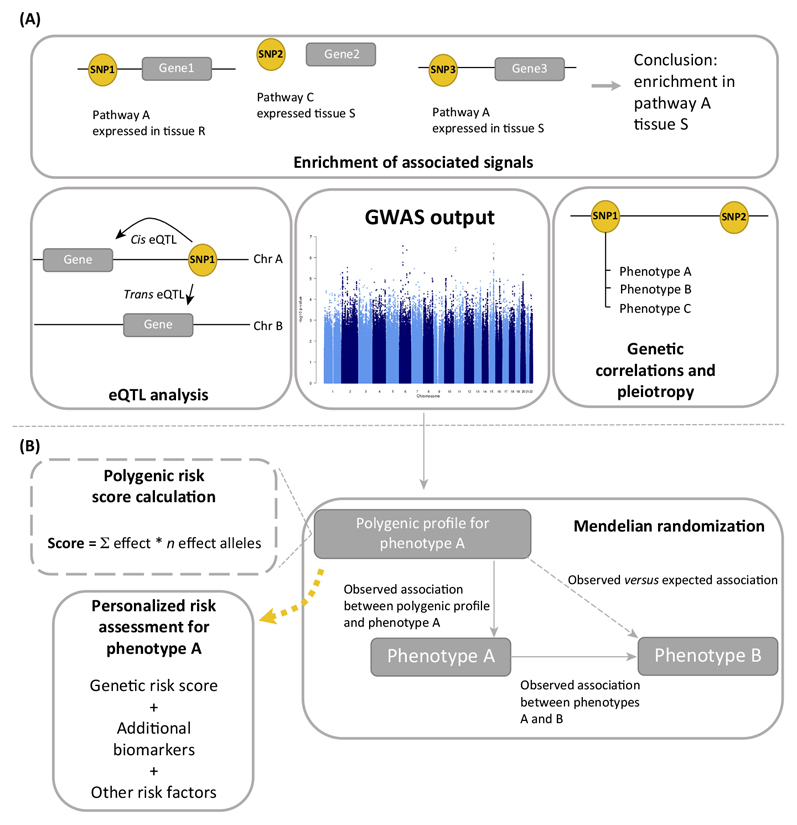
Because the variants identified in GWAS are not always directly causal, but are instead surrogates for true causal variants, different approaches (Figure IA) are used to fit GWAS findings into a larger biological picture and provide possible explanations for how genetic variation could influence the phenotype, for example by modifying gene expression. We list here some of the analytical methods that have been most used in studies related to ovarian function.
Expression quantitative trait locus (eQTL) analysis aims to find genetic variants that influence gene expression. eQTLs can be local, near the SNP of interest (cis-eQTL), or distant. The latter are usually called trans-eQTLs and are spatially separated from the SNP of interest, for example on another chromosome. Several freely accessible databases have been established for various human tissues. The most comprehensive public eQTL databases are currently the whole blood eQTL browseriv [75] and the Genotype-Tissue Expression (GTEx) databasev [76].
’Pathway and enrichment analyses’ fit GWAS hits into a functionally more meaningful context by evaluating the joint effect of many genes, thereby highlighting those biological processes or functional domains most affected by the associated variants. To facilitate such analyses, tools such as Meta-Analysis Gene-set Enrichment of variaNT Associations (MAGENTAvii) [77] and Data-driven Expression-Prioritized Integration for Complex Traits (DEPICTviii) [78] have been developed. These programs use GWAS summary statistics (P value and chromosomal positions) as an input and propose causal genes, find enriched biological processes/pathways, gene sets, and also tissues or cell types where genes from associated loci show high expression.
In addition to looking for enrichment in pre-defined pathways, tissues, or functional elements, text-mining tools such as Gene Relationships Across Implicated Loci (GRAILix) offer the possibility to prioritize genes by scanning published papers for keywords that are similar for associated markers [79]. In addition, GWAS data can be used to explore the shared genetic component between traits by assessing pleiotropy (one locus influences multiple phenotypes) or by using the recently developed LD Score Regression method that takes advantage of the enrichment of heritability in particular genomic regions that is shared across many traits [80].
From a clinical perspective, polygenic profiles generated based on the associations observed in GWAS can be used for assessing the causal relationships between phenotypes using Mendelian randomization [81], similarly to clinical randomization studies, only using genotype data as instruments (Figure IB). In the future, polygenic profiles could also be used for personalized risk assessment and counseling.
Figure I.
How To Fit the GWAS Findings in a Larger Biological Picture? Various approaches (A) contextualizing GWAS findings, and (B) harnessing GWAS findings for translational output.
Ovarian Reserve
The ovarian reserve can be indirectly estimated by hormonal or ultrasound markers [FSH, anti-Müllerian hormone (AMH), antral follicle count (AFC)], and some studies have been conducted to identify genes associated with these markers, resulting in several variants that associated with AMH or early follicular phase FSH values in Caucasian and African American women [24]. However, none of these hits were later confirmed in studies specifically aimed at finding the genetic regulators of sex hormones, including FSH [25] and AMH [26]. Instead a signal near FSHB that influenced FSH as well as luteinizing hormone (LH) levels was reported in a study group consisting mainly of female Europeans [25] (Figure 2A), while variants directly in the AMH locus were found to regulate AMH levels in males but not females [26]. In addition, variants associated with AFC were also reported [27], several of which were also associated with AMH levels [27], indicating an overlap between the genetic determinants for these traits. However, the variants reported in [24] and [27] are near genes (LRRC61, GPR12, KLRAP1, BLK, MACROD2) that have previously not been linked to female reproduction and, considering the limited sample size in the original studies, validation in larger independent cohorts and functional studies will be necessary to confirm their role in ovarian biology.
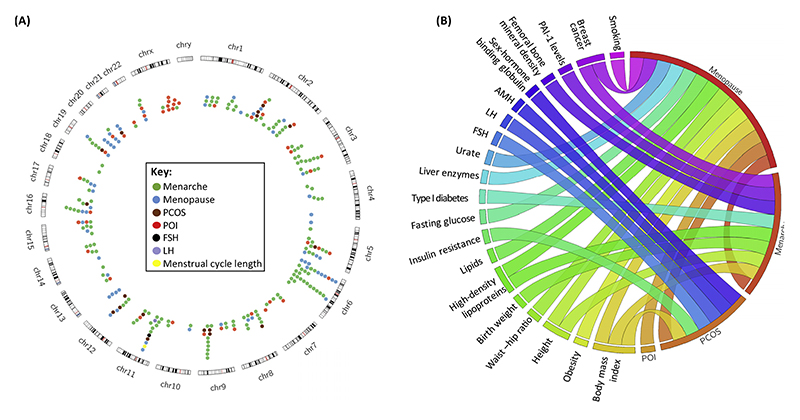
Figure 2. Reported Genetic Associations and Genetic Correlations between Phenotypes.
(A) Genome-wide significant loci associated with menarche, menopause, polycystic ovary syndrome, FSH and LH levels, and menstrual cycle length; and genes associated with monogenic forms of POI. (B) Genetic correlations between phenotypes reflecting ovarian physiology and overall health status. Data are from published studies [9,10,19] that report genetic correlations based on observed pleiotropy between loci or LD score regression analysis. Abbreviations: AMH, anti-Müllerian hormone; FSH, follicle-stimulating hormone; LD, linkage disequilibrium; LH, luteinizing hormone; PAI-1, plasminogen activator inhibitor type 1; PCOS, polycystic ovary syndrome; POI, premature ovarian insufficiency.
Another trait closely related to the ovarian reserve is the response to COS used in IVF. One very small GWAS (n = 92) has been done on IVF-related traits (oocyte yield, the total amount of FSH used during stimulation, the amount of FSH needed for retrieving one oocyte, embryo quality, and likelihood of pregnancy), but likely due to power this study only yielded a few sub-significant associations [28].
Apart from the few studies conducted for hormone levels, GWAS related to ovarian reserve parameters have suffered from a lack of sufficiently sized study groups. Further studies, especially for COS parameters, will be necessary to reveal the genetic determinants of ovarian response.
Reproductive Aging
For natural reproductive aging, several GWAS have been conducted, both for the timing of menarche [9,11,29–31] and menopause [10,32–36]. The largest studies, involving more than 180 000 and nearly 70 000 women, respectively, have identified altogether approximately 150 associated loci in women of European descent [37] (Figure 2A). Collectively, these explain about 3-4% [9,11] and 6% [10], respectively, of the variance in menarche and menopause timing. The reported effect sizes range from 2 weeks up to 1.25 years for menarche (largest effect for a low-frequency variant in TACR3) [9,11], and from ~4 weeks to nearly 1 year for menopause (largest effects for common variants in MCM8 and low-frequency variants in HELB) [10]. Several regions initially identified in one population have also shown consistent associations with menarche/menopause timing in other populations [30,31,38–41], pointing to significant genetic overlap of reproductive timing across various populations and races. In addition, variants associated with menarche and menopause timing are enriched in regions that contain genes for monogenic puberty disorders [9,10]. Furthermore, menopause associations tend to lie near genes responsible for monogenic forms of POI [10]. This again highlights not only the genetic overlap between distinct disorders and normal variation observed in a population but also between the two time-points that define the reproductive lifespan. Additionally, both menarche and menopause show genetic correlations with several other phenotypes (Figure 2B). Tissue enrichment analysis with DEPICT (Box 2) and previously published loci ([9,10] and Tables S1-S4 in the supplemental information online) showed no statistically significant enriched tissues for menarche loci (top-ranking terms included musculoskeletal system, connective tissue cells, and central nervous system), but highlighted the endocrine, blood, and stem cells together with the immune system and urogenital tissues (including ovary) for menopause loci.
Biological pathways highlighted in studies include DNA repair and immune response for menopause [10,34], and energy homeostasis, pituitary function, nuclear hormone receptor signaling, steroidogenesis, and gene silencing for menarche [9]. Interestingly, for menopause signals, no enrichment was seen in genes associated with ovarian function (the tested list included 130 genes associated with POI, folliculogenesis, ovarian dysgenesis, etc.) [10]. Nevertheless, several of the identified candidate genes are directly involved in processes governing ovarian function. For example, variation in FSHB is associated with both age at menarche [9] and menopause [10], and FSH plays a central role in folliculogenesis. MCM8, that participates in DNA replication and is reported to have the biggest effect on menopause timing, was found to be associated with follicle count and is also expressed in human ovarian follicles [27], and furthermore is one of the genes responsible for monogenic forms of POI [42,43]. Several of the other known menopause loci have been associated with traits reflecting the ovarian function [44–46], and a recent study showed that SYCP2L (encoding an oocyte centromere protein), which is associated with menopausal age [10] and with COS and IVF outcome [44], is expressed in oocytes, regulates primordial oocyte survival [47], and is associated with reduced fertility in aged female mice [47]. These results demonstrate that, in addition to identifying the genetic variants associated with reproductive aging in the general population, GWAS of these traits also offer valuable leads for further research into ovarian biology.
Taken together, GWAS for natural reproductive aging are a perfect example of how this approach can improve our understanding of reproductive health-associated phenotypes because these studies have not only highlighted the mechanisms behind ovarian aging but have also demonstrated a complex network of interactions between different (reproductive) phenotypes.
Premature Ovarian Insufficiency
To date, six studies using a GWAS approach have been conducted to find variants associated with POI in various populations ([48–53], reviewed in [3]). Numerous sub-significant associations [50–53] have been reported, and none of these associations were replicated in subsequent studies. Furthermore, no associations were observed in regions previously associated with monogenic forms of POI [3]. This is probably caused by the low prevalence of POI, making it difficult to collect sufficient numbers of cases. However, there is evidence that the same variants associated with age at normal menopause contribute to both early menopause and POI [54–56], indicating a considerable genetic overlap between POI and (early) menopause. This also underlines the possibility of studying the reproductive aging and ovarian insufficiency as a continuum without setting any cut-offs for menopausal age. Nevertheless, to fully understand the POI phenotype and its place in the reproductive aging continuum, a well-powered study for this phenotype is also highly anticipated.
Although not within the scope of this review, in recent years another hypothesis-free approach, namely next-generation sequencing (NGS), has identified several novel genes implicated in idiopathic POI. NGS encompasses high-throughput DNA sequencing technologies including whole-genome sequencing (WGS), whole-exome sequencing (WES), and other targeted NGS. The respective findings have been reviewed in more detail previously [3,57], but of interest is the fact that candidate genes identified by WES (including MCM8 and MCM9) are involved in meiosis, DNA repair, and chromosome stability, mirroring the findings from reproductive aging GWAS. Furthermore, a targeted NGS among potential POI candidate genes showed that mutations in ADAMTS19 are associated with POI [58], confirming a previous sub-significant finding from a POI GWAS [52]. In this respect, GWAS and NGS represent two complementary approaches. On the one hand, NGS can determine the exact sequence of DNA, and therefore will give information about all genetic variants, including rare variants. However, it is still not feasible to sequence the large quantities of samples necessary for a genome-wide analysis, hence NGS is well suited for analyzing familial cases. On the other hand, GWAS samples are genotyped using a genome-wide array, and variants not directly genotyped are inferred or ‘imputed’ based on sequenced reference data and known haplotype structure. This makes GWAS a more cost-effective method most suitable for the analysis of common genetic variants because the quality of imputation is much lower for rare genetic variants (and much larger study groups are needed for rare variant analysis).
PCOS
Recent years have seen a rapid emergence of PCOS GWAS studies and, since 2011, five studies have been published [19,59–62]. Together, these studies have identified 16 loci that are significantly associated with PCOS (Table 1). Moreover, four regions (THADA, C9orf3, FSHB, and YAP1) have been replicated in more than one study, and in women of Caucasian and Chinese Han backgrounds, suggesting common pathogenesis mechanisms across different populations. The first two studies identified 11 loci in Chinese Han PCOS patients [59,60]. The third PCOS GWAS conducted among Koreans did not identify any significant associations, but reported replication of seven of the previously found PCOS associations [61]. The study by Hayes et al. was the first PCOS GWAS among Caucasians and, as a result, two novel signals for PCOS risk were reported [62]. The most recent study included a total of ~7000 PCOS cases (both self-reported and clinically confirmed) and reported three novel hits [19]. Notably, this provides evidence the Erb-B pathway might be implicated in PCOS, and this pathway is involved in primordial germ cell development in mice [63]. In addition, ERBB3 interacts with YAP1 in the Hippo pathway, which regulates organ size by controlling cell proliferation and apoptosis, and has been associated with primordial follicle pool in mice [64]. Taken together, these five studies provide considerable evidence for the role of the Hippo pathway, neuroendocrine changes (mediated by LHCGR, FSHR and FSHB), and EGFRs in PCOS pathophysiology. In addition, the PCOS susceptibility variants found by Day et al. and Hayes et al. were also found to be associated with AMH, FSH, and LH levels [19,62], offering some insight into the genetics of these traits as well. Finally, variants associated with menopause also associate with PCOS risk, suggesting a shared genetic background for ovarian aging and PCOS [19,46], and providing an evolutionary explanation for the high prevalence of PCOS-the primordial follicle pool is larger or more efficiently used in PCOS patients [19].
Table 1. GWAS Loci Associated with Polycystic Ovary Syndrome.
| Marker | Location | Nearest Gene(s) | Comments | Refs |
|---|---|---|---|---|
| rs1351592 | 2q33.3-q34 | ERBB4 (Erb-B2 receptor tyrosine kinase 4) | PCOS susceptibility allele also associates with higher AMH levels in girls. Nominal associations were found for two other members of the EGFR family (ERBB2/HER2 and ERBB3 HER3). | [19] |
| rs7563201 rs13429458 |
2p21 | THADA (thyroid adenoma associated) | PCOS susceptibility allele also associates with higher AMH levels in girls. | [19,59] |
| rs13405728 | 2p21 | LHCGR (luteinizing hormone/choriogonadotropin receptor) | [59] | |
| rs2268361 | 2p21-p16 | FSHR (follicle-stimulating hormone receptor) | [60] | |
| rs13164856 | 5q31 | RAD50 (RAD50 double-strand break repair protein) | PCOS susceptibility allele also associates with higher AMH levels in girls. | [19] |
| rs804279 | 8p23.1-p22 | GATA4 (GATA-binding protein), NEIL2 (nei-like DNA glycosylase 2) | [62] | |
| rs10993397 rs3802457 |
9q22.32 | C9orf3, FANCC (Fanconi anemia complementation group C) | [60,62] | |
| rs2479106 | 9q33.3 | DENND1A (DENN domain-containing 1A) | [59] | |
| rs11031006 | 11p13 | FSHB (follicle-stimulating hormone ß subunit) | PCOS susceptibility allele also associates with higher AMH levels in girls. Marker also associated with FSH and LH levels. | [19,62] |
| rs1894116 rs11225154 |
11q13 | YAP1 (Yes-associated protein 1) | PCOS susceptibility allele also associates with higher AMH levels in girls. | [19,60] |
| rs705702 | 12q13 | RAB5B (RAB5B, member RAS oncogene family), SUOX (sulfite oxidase) | [60] | |
| rs2272046 | 12q15 | HMGA2 (high mobility group AT-hook 2) | [60] | |
| rs1275468 | 12q21.2 | KRR1 [KRR1, small subunit processome component, homolog (yeast)] | PCOS susceptibility allele also associates with higher AMH levels in girls. | [19] |
| rs4784165 | 16q12.1 | TOX3 (TOX high mobility group box family member 3) | [60] | |
| rs2059807 | 19p13.3-p13.2 | INSR (insulin receptor) | [60] | |
| rs6022786 | 20q13.2 | SUMO1P1 (SUMO 1 pseudogene 1) | [60] |
In conclusion, the GWAS approach has proven to be relatively successful for finding genetic susceptibility factors of PCOS. While some of the findings (such as the neuroendocrine component) were to be expected, others are novel and warrant further studies to understand their role in disease development.
What Can GWAS of Traits Related to Ovarian Biology Provide to the Patients?
From a clinical point of view, additional tools facilitating disease prediction, early diagnosis, informed interventions, or personalized treatment would substantially improve patient management, and there is accumulating evidence that GWAS findings can provide the necessary means for this. Addition of genetic markers to known risk factors was shown to improve the performance of clinical breast cancer risk prediction models [65] or help to predict Parkinson’s disease progression [66], while drug mechanisms with genetic support from GWAS have better success rates [67]. Although individual associated genetic markers identified in GWAS confer only a modest disease risk, the combined effect of tens or thousands of markers can be sufficient to be clinically useful. Polygenic risk scores with genome-wide data can be used to summarize the effects of many genetic variants, and proof of concept for their use in disease prediction has been shown in simulations [68]. Ideally, risk models should consider environmental, lifestyle, and genetic risk factors, as well as their interactions.
In the context of ovarian biology, such risk-prediction models would be useful for the timely detection of women at risk of PCOS [69] orearly/premature menopause [54,56]. Because the most significantly associated genetic variants identified in menopausal timing studies show potential for predicting reproductive lifespan [54,56], it could be expected that the use of polygenic risk scores involving more markers would also improve prediction accuracy. Furthermore, genetic variants associated with natural menopausal timing also seem to have an impact on the onset of menopause in women undergoing chemo- or radiotherapy [45]. This means that genetic risk prediction models could provide a basis for more personalized counseling regarding family planning, lifestyle choices, or the use of modern technologies for maintaining fertility, such as oocyte cryopreservation. For PCOS, genetic risk profiles have been proposed for individualizing treatment approaches depending on risk categories [69]; however, currently proposed genetic risk profiles show poor predictive value [69] and cannot be used as stand-alone clinical tools.
Input for individualizing treatment can also come from Mendelian randomization analyses, which use GWAS data to explore the causality between phenotypes. For example, the causal roles of increased body mass index and insulin resistance in the pathophysiology of PCOS were confirmed using this approach [19], highlighting the importance of lowering weight and reducing insulin resistance as a part of PCOS treatment. As the causal relationships between other phenotypes are revealed, this could highlight additional ways for more personalized patient counseling to decrease disease risk.
Finally, a marker profile associated with ovarian stimulation outcome would be a valuable pharmacogenomic tool for personalizing and optimizing treatment protocols because there is evidence that individual genetic variation could be related to COS response in IVF [70]. Considering the genetic overlap between reproductive aging and other ovarian-related phenotypes, genetic variants associated with reproductive aging could also be potential candidates for assessing ovarian function and response in infertility treatment. Personalized COS-IVF protocols could lower the risk of side effects from treatment (such as ovarian hyperstimulation syndrome or poor ovarian response), and could potentially lead to more efficient treatment in terms of oocytes received or even pregnancy rate. However, further studies will be necessary to determine how these genetic variants associate with known markers of ovarian reserve and IVF outcome, and how they perform individually or in combination.
In summary, genetic markers are stable and detectable throughout life, and this supports their use as prognostic biomarkers. The main factors currently hindering their application in the clinical setting include the lack of information on functional significance for the majority of identified variants, and on how genetic variants interact with each other or with environmental factors. Hopefully the coming years will see both novel analytical methods for modeling these interactions and also the application of genetic markers in the clinical setting.
Concluding Remarks and Future Perspectives
It is evident that, although for some phenotypes discussed in this review (such as age at menarche and menopause) GWAS have been true success stories, for others (PCOS, POI, ovarian reserve and response) the best is yet to come. The studies conducted so far have shown that some findings will support what is previously known or suspected (such as the involvement of DNA repair mechanisms in ovarian aging, or the importance of neuroendocrine mechanisms in PCOS susceptibility), while others will prompt new investigations. As analytical methods for finding causative genes or biological context improve, so will our knowledge on the genetics governing ovarian physiology. However, functional studies for validating the GWAS findings and for understanding how associated variants modify biological mechanisms largely remain an untouched territory in the context of ovarian biology.
While many gaps in our knowledge remain (see Outstanding Questions), we hope that new and innovative approaches to study design, valuable lessons from other phenotypes, and close collaboration between clinicians and scientists will pave the way for new discoveries and, more importantly, for novel means to harness GWAS findings on ovarian function for the benefit of the patients.
Outstanding Questions.
How exactly are reproductive aging, ovarian reserve and ovarian disorders related at a genetic level? Can well-designed GWAS in phenotypes where this approach has not been used successfully, together with analytical methods for assessing genetic correlations, provide a sufficient answer?
How do the identified genetic associations exert their biological effects on these traits and conditions? Gene expression, methylation, and protein level datasets for follicular cell subpopulations, ovarian stromal cells, or even oocytes may complement our knowledge on how genetic variation modifies gene expression in ovarian tissue.
How can the findings from GWAS be successfully translated to individualizing healthcare in prevention, early diagnosis, and treatment?
Supplementary Material
Supplemental information associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.tem.2016.04.011.
Trends.
Large population-based biobanks can be harnessed for genetic studies in ovary-related phenotypes to take research efforts to the next level.
New analytical methods that use GWAS summary statistics can be used to identify the most likely causal genes, pathways, underlying mechanisms, genetic correlations, and causal relationships between phenotypes.
There is significant genetic overlap between traits and disorders reflecting ovarian function, such as age at natural menopause, polycystic ovary syndrome, and premature ovarian insufficiency.
Results from large-scale genetic association studies can provide information for more personalized patient management.
Acknowledgments
The work has received funding from the European Commission Horizon 2020 research and innovation programme under grant agreement 692065 (project WIDENLIFE) and EC-FP7 IAPP Project grant SARM EU324509, and has also been supported by grant IUT34-16 from the Estonian Ministry of Education and Research and grant EU48695 from Enterprise Estonia.
Footnotes
References
- 1.Teede H, et al. Polycystic ovary syndrome: a complex condition with psychological, reproductive and metabolic manifestations that impacts on health across the lifespan. BMC Med. 2010;8:41. doi: 10.1186/1741-7015-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.de Bruin JP, et al. The role of genetic factors in age at natural menopause. Hum Reprod. 2001;16:2014–2018. doi: 10.1093/humrep/16.9.2014. [DOI] [PubMed] [Google Scholar]
- 3.Qin Y, et al. Genetics of primary ovarian insufficiency: new developments and opportunities. Hum Reprod Update. 2015;21:787–808. doi: 10.1093/humupd/dmv036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vink JM, et al. Heritability of polycystic ovary syndrome in a Dutch twin-family study. J Clin Endocrinol Metab. 2006;91:2100–2104. doi: 10.1210/jc.2005-1494. [DOI] [PubMed] [Google Scholar]
- 5.Sadrzadeh S, et al. Birth weight and age at menarche in patients with polycystic ovary syndrome or diminished ovarian reserve, in a retrospective cohort. Hum Reprod. 2003;18:2225–2230. doi: 10.1093/humrep/deg409. [DOI] [PubMed] [Google Scholar]
- 6.Welt CK, Carmina E. Clinical review: lifecycle of polycystic ovary syndrome (PCOS): from in utero to menopause. J Clin Endocrinol Metab. 2013;98:4629–4638. doi: 10.1210/jc.2013-2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zondervan KT, Cardon LR. Designing candidate gene and genome-wide case-control association studies. Nat Protoc. 2007;2:2492–2501. doi: 10.1038/nprot.2007.366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Visscher PM, et al. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Perry JRB, et al. Parent-of-origin-specific allelic associations among 106 genomic loci for age at menarche. Nature. 2014;514:92–97. doi: 10.1038/nature13545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Day FR, et al. Large-scale genomic analyses link reproductive aging to hypothalamic signaling, breast cancer susceptibility and BRCA1-mediated DNA repair. Nat Genet. 2015;47:1294–1303. doi: 10.1038/ng.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lunetta KL, et al. Rare coding variants and X-linked loci associated with age at menarche. Nat Commun. 2015;6:7756. doi: 10.1038/ncomms8756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wood AR, et al. Defining the role of common variation in the genomic and biological architecture of adult human height. Nat Genet. 2014;46:1173–1186. doi: 10.1038/ng.3097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vassena R, et al. Menstrual cycle length in reproductive age women is an indicator of oocyte quality and a candidate marker of ovarian reserve. Eur J Obstet Gynecol Reprod Biol. 2014;177:130–134. doi: 10.1016/j.ejogrb.2014.03.027. [DOI] [PubMed] [Google Scholar]
- 14.Ruth KS, et al. Genetic evidence that lower circulating FSH levels lengthen menstrual cycle, increase age at menopause and impact female reproductive health. Hum Reprod. 2016;31:473–481. doi: 10.1093/humrep/dev318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aschebrook-Kilfoy B, et al. Genome-wide association study of parity in Bangladeshi women. PLoS ONE. 2015;10:e0118488. doi: 10.1371/journal.pone.0118488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tropf FC, et al. Human fertility, molecular genetics, and natural selection in modern societies. PLoS ONE. 2015;10:e0126821. doi: 10.1371/journal.pone.0126821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Haller-Kikkatalo K, et al. The prevalence and phenotypic characteristics of spontaneous premature ovarian failure: a general population registry-based study. Hum Reprod. 2015;30:1229–1238. doi: 10.1093/humrep/dev021. [DOI] [PubMed] [Google Scholar]
- 18.Castro V, et al. Identification of subjects with polycystic ovary syndrome using electronic health records. Reprod Biol Endocrinol. 2015;13:116. doi: 10.1186/s12958-015-0115-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Day FR, et al. Causal mechanisms and balancing selection inferred from genetic associations with polycystic ovary syndrome. Nat Commun. 2015;6:8464. doi: 10.1038/ncomms9464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–572. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tin A, et al. Using multiple measures for quantitative trait association analyses: application to estimated glomerular filtration rate. J Hum Genet. 2013;58:461–466. doi: 10.1038/jhg.2013.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Missmer SA, et al. Analysis of multiple-cycle data from couples undergoing in vitro fertilization: methodologic issues and statistical approaches. Epidemiology. 2011;22:497–504. doi: 10.1097/EDE.0b013e31821b5351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schuh-Huerta SM, et al. Genetic variants and environmental factors associated with hormonal markers of ovarian reserve in Caucasian and African American women. Hum Reprod. 2012;27:594–608. doi: 10.1093/humrep/der391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruth KS, et al. Genome-wide association study with 1000 genomes imputation identifies signals for nine sex hormone-related phenotypes. Eur J Hum Genet. 2016;24:284–290. doi: 10.1038/ejhg.2015.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perry JRB, et al. Genome-wide association study identifies common and low-frequency variants at the AMH gene locus that strongly predict serum AMH levels in males. Hum Mol Genet. 2016;25:382–388. doi: 10.1093/hmg/ddv465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schuh-Huerta SM, et al. Genetic markers of ovarian follicle number and menopause in women of multiple ethnicities. Hum Genet. 2012;131:1709–1724. doi: 10.1007/s00439-012-1184-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van Disseldorp J, et al. Genome-wide analysis shows no genomic predictors of ovarian response to stimulation by exogenous FSH for IVF. Reprod Biomed Online. 2011;22:382–388. doi: 10.1016/j.rbmo.2010.12.006. [DOI] [PubMed] [Google Scholar]
- 29.Elks CE, et al. Thirty new loci for age at menarche identified by a meta-analysis of genome-wide association studies. Nat Genet. 2010;42:1077–1085. doi: 10.1038/ng.714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Demerath EW, et al. Genome-wide association study of age at menarche in African-American women. Hum Mol Genet. 2013;22:3329–3346. doi: 10.1093/hmg/ddt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanikawa C, et al. Genome wide association study of age at menarche in the Japanese population. PLoS ONE. 2013;8:e63821. doi: 10.1371/journal.pone.0063821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He C, et al. Genome-wide association studies identify loci associated with age at menarche and age at natural menopause. Nat Genet. 2009;41:724–728. doi: 10.1038/ng.385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stolk L, et al. Loci at chromosomes 13,19 and 20 influence age at natural menopause. Nat Genet. 2009;41:645–647. doi: 10.1038/ng.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stolk L, et al. Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat Genet. 2012;44:260–268. doi: 10.1038/ng.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pyun J-A, et al. Genome-wide association studies and epistasis analyses of candidate genes related to age at menarche and age at natural menopause in a Korean population. Menopause. 2014;21:522–529. doi: 10.1097/GME.0b013e3182a433f7. [DOI] [PubMed] [Google Scholar]
- 36.Perry JRB, et al. DNA mismatch repair gene MSH6 implicated in determining age at natural menopause. Hum Mol Genet. 2014;23:2490–2497. doi: 10.1093/hmg/ddt620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry JRB, et al. Molecular insights into the aetiology of female reproductive ageing. Nat Rev Endocrinol. 2015;11:725–734. doi: 10.1038/nrendo.2015.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen CTL, et al. Meta-analysis of loci associated with age at natural menopause in African-American women. Hum Mol Genet. 2014;23:3327–3342. doi: 10.1093/hmg/ddu041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rahmani M, et al. Shared genetic factors for age at natural menopause in Iranian and European women. Hum Reprod. 2013;28:1987–1994. doi: 10.1093/humrep/det106. [DOI] [PubMed] [Google Scholar]
- 40.Shen C, et al. Evaluating GWAS-identified SNPs for age at natural menopause among Chinese women. PLoS ONE. 2013;8:e58766. doi: 10.1371/journal.pone.0058766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carty CL, et al. Replication of genetic loci for ages at menarche and menopause in the multi-ethnic Population Architecture using Genomics and Epidemiology (PAGE) study. Hum Reprod. 2013;28:1695–1706. doi: 10.1093/humrep/det071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tenenbaum-Rakover Y, et al. Minichromosome maintenance complex component 8 (MCM8) gene mutations result in primary gonadal failure. J Med Genet. 2015;52:391–399. doi: 10.1136/jmedgenet-2014-102921. [DOI] [PubMed] [Google Scholar]
- 43.AlAsiri S, et al. Exome sequencing reveals MCM8 mutation underlies ovarian failure and chromosomal instability. J Clin Invest. 2015;125:258–262. doi: 10.1172/JCI78473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Laisk-Podar T, et al. Genetic variants associated with female reproductive ageing—potential markers for assessing ovarian function and ovarian stimulation outcome. Reprod Biomed Online. 2015;31:199–209. doi: 10.1016/j.rbmo.2015.05.001. [DOI] [PubMed] [Google Scholar]
- 45.van Dorp W, et al. Genetic variation may modify ovarian reserve in female childhood cancer survivors. Hum Reprod. 2013;28:1069–1076. doi: 10.1093/humrep/des472. [DOI] [PubMed] [Google Scholar]
- 46.Saxena R, et al. Gene variants associated with age at menopause are also associated with polycystic ovary syndrome, gonadotrophins and ovarian volume. Hum Reprod. 2015;30:1697–1703. doi: 10.1093/humrep/dev110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou J, et al. Accelerated reproductive aging in females lacking a novel centromere protein SYCP2L. Hum Mol Genet. 2015;24:6505–6514. doi: 10.1093/hmg/ddv359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Caburet S, et al. Genome-wide linkage in a highly consanguineous pedigree reveals two novel loci on chromosome 7 for non-syndromic familial Premature Ovarian Failure. PLoS ONE. 2012;7:e33412. doi: 10.1371/journal.pone.0033412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Oldenburg RA, et al. A genome-wide linkage scan in a Dutch family identifies a premature ovarian failure susceptibility locus. Hum Reprod. 2008;23:2835–2841. doi: 10.1093/humrep/den278. [DOI] [PubMed] [Google Scholar]
- 50.Pyun J-A, et al. LAMC1 gene is associated with premature ovarian failure. Maturitas. 2012;71:402–406. doi: 10.1016/j.maturitas.2012.01.011. [DOI] [PubMed] [Google Scholar]
- 51.Qin Y, et al. Association of 8q22.3 locus in Chinese Han with idiopathic premature ovarian failure (POF) Hum Mol Genet. 2012;21:430–436. doi: 10.1093/hmg/ddr462. [DOI] [PubMed] [Google Scholar]
- 52.Knauff EAH, et al. Genome-wide association study in premature ovarian failure patients suggests ADAMTS19 as a possible candidate gene. Hum Reprod. 2009;24:2372–2378. doi: 10.1093/humrep/dep197. [DOI] [PubMed] [Google Scholar]
- 53.Kang H, et al. Parathyroid hormone-responsive B1 gene is associated with premature ovarian failure. Hum Reprod. 2008;23:1457–1465. doi: 10.1093/humrep/den086. [DOI] [PubMed] [Google Scholar]
- 54.Murray A, et al. Common genetic variants are significant risk factors for early menopause: results from the Breakthrough Generations Study. Hum Mol Genet. 2011;20:186–192. doi: 10.1093/hmg/ddq417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin Y, et al. ESR1, HK3 and BRSK1 gene variants are associated with both age at natural menopause and premature ovarian failure. Orphanet J Rare Dis. 2012;7:5. doi: 10.1186/1750-1172-7-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Perry JRB, et al. A genome-wide association study of early menopause and the combined impact of identified variants. Hum Mol Genet. 2013;22:1465–1472. doi: 10.1093/hmg/dds551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chapman C, et al. The genetics of premature ovarian failure: current perspectives. Int J Womens Health. 2015;7:799–810. doi: 10.2147/IJWH.S64024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fonseca DJ, et al. Next generation sequencing in women affected by nonsyndromic premature ovarian failure displays new potential causative genes and mutations. Fertil Steril. 2015;104:154–162. doi: 10.1016/j.fertnstert.2015.04.016. [DOI] [PubMed] [Google Scholar]
- 59.Chen Z-J, et al. Genome-wide association study identifies susceptibility loci for polycystic ovary syndrome on chromosome 2p16.3, 2p21 and 9q33.3. Nat Genet. 2011;43:55–59. doi: 10.1038/ng.732. [DOI] [PubMed] [Google Scholar]
- 60.Shi Y, et al. Genome-wide association study identifies eight new risk loci for polycystic ovary syndrome. Nat Genet. 2012;44:1020–1025. doi: 10.1038/ng.2384. [DOI] [PubMed] [Google Scholar]
- 61.Lee H, et al. Genome-wide association study identified new susceptibility loci for polycystic ovary syndrome. Hum Reprod. 2015;30:723–731. doi: 10.1093/humrep/deu352. [DOI] [PubMed] [Google Scholar]
- 62.Hayes MG, et al. Genome-wide association of polycystic ovary syndrome implicates alterations in gonadotropin secretion in European ancestry populations. Nat Commun. 2015;6:7502. doi: 10.1038/ncomms8502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Toyoda-Ohno H, et al. Members of the ErbB receptor tyrosine kinases are involved in germ cell development in fetal mouse gonads. Dev Biol. 1999;215:399–406. doi: 10.1006/dbio.1999.9482. [DOI] [PubMed] [Google Scholar]
- 64.Xiang C, et al. Hippo signaling pathway reveals a spatiotemporal correlation with the size of primordial follicle pool in mice. Cell Physiol Biochem. 2015;35:957–968. doi: 10.1159/000369752. [DOI] [PubMed] [Google Scholar]
- 65.Dite GS, et al. Breast cancer risk prediction using clinical models and 77 independent risk-associated SNPs for women aged under 50 years: Australian Breast Cancer Family Registry. Cancer Epidemiol Biomarkers Prev. 2016;25:359–365. doi: 10.1158/1055-9965.EPI-15-0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pihlstrøm L, et al. A cumulative genetic risk score predicts progression in Parkinson’s disease. Mov Disord. 2016;31:487–490. doi: 10.1002/mds.26505. [DOI] [PubMed] [Google Scholar]
- 67.Nelson MR, et al. The support of human genetic evidence for approved drug indications. Nat Genet. 2015;47:856–860. doi: 10.1038/ng.3314. [DOI] [PubMed] [Google Scholar]
- 68.Wray NR, et al. Prediction of individual genetic risk to disease from genome-wide association studies. Genome Res. 2007;17:1520–1528. doi: 10.1101/gr.6665407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee H, et al. A genetic risk score is associated with polycystic ovary syndrome-related traits. Hum Reprod. 2016;31:209–215. doi: 10.1093/humrep/dev282. [DOI] [PubMed] [Google Scholar]
- 70.Altmäe S, et al. Genetic predictors of controlled ovarian hyperstimulation: where do we stand today? Hum Reprod Update. 17:813–828. doi: 10.1093/humupd/dmr034. [DOI] [PubMed] [Google Scholar]
- 71.Azziz R, et al. The Androgen Excess and PCOS Society criteria for the polycystic ovary syndrome: the complete task force report. Fertil Steril. 2009;91:456–488. doi: 10.1016/j.fertnstert.2008.06.035. [DOI] [PubMed] [Google Scholar]
- 72.Zawadzki J, Dunaif A. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. In: Dunaif A, et al., editors. Diagnostic criteria for polycystic ovary syndrome: towards a rational approach. Blackwell Scientific; 1992. pp. 377–384. [Google Scholar]
- 73.Rotterdam ESHRE/ASRM-Sponsored PCOS Consensus Workshop Group. Revised 2003 consensus on diagnostic criteria and long-term health risks related to polycystic ovary syndrome. Fertil Steril. 2004;81:19–25. doi: 10.1016/j.fertnstert.2003.10.004. [DOI] [PubMed] [Google Scholar]
- 74.Legro RS, et al. Diagnosis and treatment of polycystic ovary syndrome: an Endocrine Society clinical practice guideline. J Clin Endocrinol Metab. 2013;98:4565–4592. doi: 10.1210/jc.2013-2350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Westra H-J, et al. Systematic identification of trans eQTLs as putative drivers of known disease associations. Nat Genet. 2013;45:1238–1243. doi: 10.1038/ng.2756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ardlie KG, et al. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Segrè AV, et al. Common inherited variation in mitochondrial genes is not enriched for associations with type 2 diabetes or related glycemic traits. PLoS Genet. 2010;6:e1001058. doi: 10.1371/journal.pgen.1001058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pers TH, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Raychaudhuri S, et al. Identifying relationships among genomic disease regions: predicting genes at pathogenic SNP associations and rare deletions. PLoS Genet. 2009;5:e1000534. doi: 10.1371/journal.pgen.1000534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bulik-Sullivan B, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–1241. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Do R, et al. Common variants associated with plasma triglycerides and risk for coronary artery disease. Nat Genet. 2013;45:1345–1352. doi: 10.1038/ng.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.