Myeloproliferative neoplasm (MPN)-associated myelofibrosis is a clonal disorder characterised by decreased blood cell concentrations, bone marrow fibrosis and a risk of transformation to acute myeloid leukemia (AML). Persons who become RBC-transfusion-dependent have a poor prognosis with limited therapeutic options to reverse their anemia (1). Most persons with MPN-associated myelofibrosis have driver mutations in JAK2, CALR or MPL resulting in aberrant JAK/STAT signaling (2). Non-driver mutations are common in spliceosome components, epigenetic regulators and notably in ASXL1, EZH2, IDH1/2, SRSF2 and U2AF1Q157, which are associated with adverse outcomes and are termed high-risk molecular mutations (3, 4).
RESUME was a randomized, double-blind, parallel group, controlled trial, assessing rates of RBC-transfusion-independence in subjects with MPN-associated myelofibrosis and RBC-transfusion-dependence receiving pomalidomide or placebo (5). Although response rates were similar, the study identified some differences between responders in the pomalidomide and placebo cohorts, including some subjects with a sustained response to pomalidomide not observed in the placebo cohort. We reasoned the mutational landscape of subjects in the RESUME study might correlate with these pomalidomide responses.
Baseline DNA samples with corresponding clinical data were available for 205 of the 252 randomized subjects. Median age was 70 years (range, 44–90 years). Additional subject co-variates are reported and displayed in Table S1. We performed targeted DNA sequencing on samples from the 205 subjects using a Fluidigm Access Array 33 gene panel followed by next generation sequencing. Specific methods and analysis pipelines are described in the Supplement.
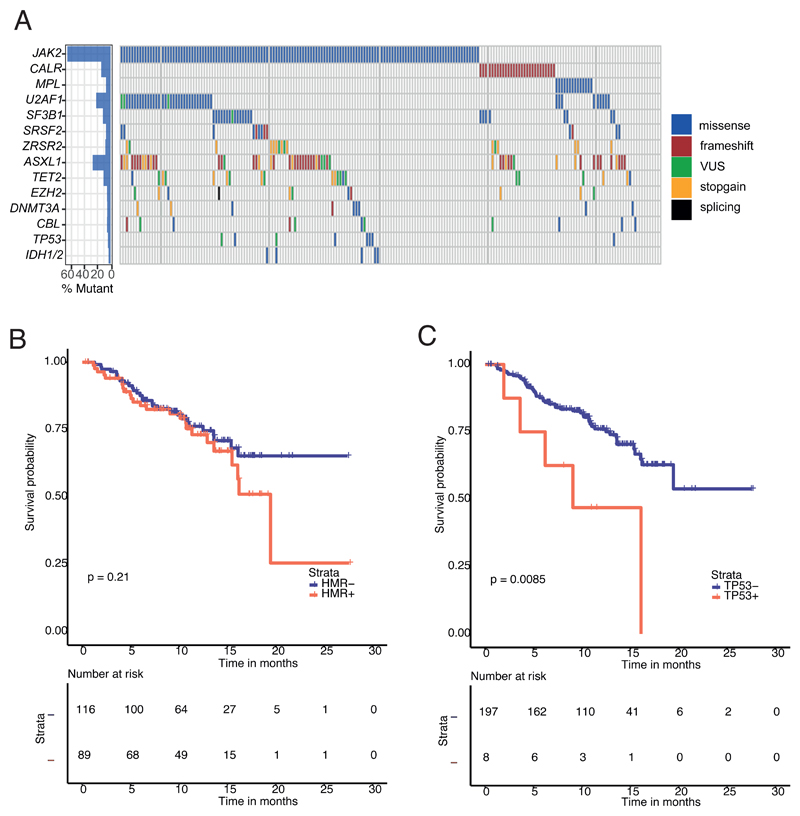
Four hundred and seventy two mutations were detected (Table S2). 198 of the 205 subjects (97%; 95% Confidence Interval [CI], 95, 99%) had a mutation in ≥ 1 targeted gene. 2 mutations were detected in 37% (28, 41%) and ≥ 3 in 40% (33, 46%; Figure 1A). 7 subjects had no detectable mutation. JAK2V617F, CALR and MPL mutations were identified in 136 (66% [59, 72%]), 29 (14% [8, 19%]) and 14 (7% [4, 11%]) subjects (Figure 1A). There was no detectable driver mutation in 26 subjects with primary myelofibrosis (13% [8, 18%]; Table S1). Non-driver mutations were detected in 78% of subjects (70, 82%) with a higher frequency in males (85% [79, 91%] versus 57% [44, 70%] for females; P = 0.0001). 45% of subjects (38, 52%) had a spliceosome mutation including U2AF1 (22% [16, 28%]); SF3B1 (12% [8, 16%]) SRSF2 (7% [3, 14%]) and ZRSR2 (8% [4, 12%]). More spliceosome mutations were detected in men compared with women (52% [44, 60%] versus 27% [15, 38%]; P = 0.002; Table S1). Spliceosome mutations were mutually exclusive in 85%. 7 subjects had > 1 spliceosome mutation (Figure 1). Spliceosome mutations were also less common in subjects with prior polycythemia vera (14% [2, 32%]) compared with subjects with prior essential thrombocythemia (38% [22, 55%]) and those with primary myelofibrosis (51% [43, 59%]; P 0.003; Table S1). Mutations in ASXL1 (27% [21, 33%]); TET2 (11% [7, 15%]), DNMT3A (5% [2, 8%]) and EZH2 (5% [2, 8%]) were detected at rates similar to those reported (3, 6). 43% (36, 50%) of subjects had a high molecular risk mutation. Subjects with JAK2 V617F were significantly more likely than subjects with a CALR mutation to have a non-driver mutation (80% [73, 87%] versus 62% [(44, 80%] P = 0.036), a spliceosome mutation (47% [36, 53%] versus 28% [6, 36%]; P = 0.055), a U2AF1 mutation (26% [17, 32%] versus none; P = 0.002) and a high-risk molecular mutation (46% [38, 54%] versus 24% [8, 40%]; P = 0.028). Some mutations were positively correlated including JAK2 V617F and U2AF1 (Odds Ratio [OR], 2.1 [0.9, 4.4]; P = 0.07), ASXL1 and EZH2 (OR = 5.2 [1.4, 18.4]; P = 0.011) and SF3B1 and TP53 (OR = 5 [1.1, 22.2]; P = 0.035; Figure S1).
Figure 1. The mutational landscape of transfusion dependent myelofibrosis.
(A) Frequency of mutations detected at baseline by targeted gene sequencing (n= 205 patients). Each column represents an individual patient. Only genes with pathogenic mutations detected in at least 5 patients are shown. Each colour corresponds to mutation type. VUS=variant of uncertain significance (see methods). (B) Kaplan-Meier curve of leukemia-free survival (LFS) stratified by HMR at 1-year; HMR did not influence 12-month LFS; HMR-mutated (73.1% [95% CI 64–80%]) versus HMR-wild type (WT) patients (76.2% [95% CI 69–82%]), P=0.21. (C) Kaplan-Meier curve of LFS stratified by TP53 at 1-year showing a poorer LFS in TP53-mutated patients (47% [95% CI 29–63%]) versus TP53-WT (76% [95% CI 70–81%]), p=0.0085. HMR (high molecular risk - ASXL1, EZH2, SRSF2, IDH1/2, U2AF1 Q157)
High variant allele frequency (VAF, >l/=50%) JAK2 mutations were common in this cohort, occurring in 76 of 136 (56%) JAK2 mutated patients (Table S2). High VAF JAK2 mutation was more often detected in post-PV MF (83%, 95% CI 67-98%) than PMF (47%, 95% CI 37-57%), P = 0.006, which has been widely reported previously(7). There was no association between presence of high VAF JAK2 mutation and frequency of NDM, specific mutation subgroups or treatment response in this study. The median VAF for spliceosome point mutations was 44% (interquartile range, IQR 38-52%), in keeping with these mutations occurring as an early event in disease pathogenesis, consistent with the literature (8, 9). TP53 mutations were frequently more likely to be subclonal with a median VAF of 29% (IQR 18-41%), in line with clonal evolution in these patients who were at increased risk of leukemic transformation. Non driver mutations in other signalling genes (CBL, HRAS, KRAS, NRAS, SH2B3) were frequently subclonal with a median VAF of 20% (IQR 16-32%), which is suggestive that such mutations typically occur as secondary events in patients undergoing clonal evolution (10).
Median follow-up was 12 months (range, 11-14 months). 52 subjects died (25%) including 8 transforming to AML. 1-year survival was 76% (70, 80%). There were no significant associations between mutation topography and survival including high-risk mutations (Figure 1B). An exception was TP53 mutation which was associated with a 1-year leukemia-free survival (LFS) of 47% (29, 63%) versus 76% (70, 81%) in those without a TP53 mutation (P = 0.0085; Figure 1C).
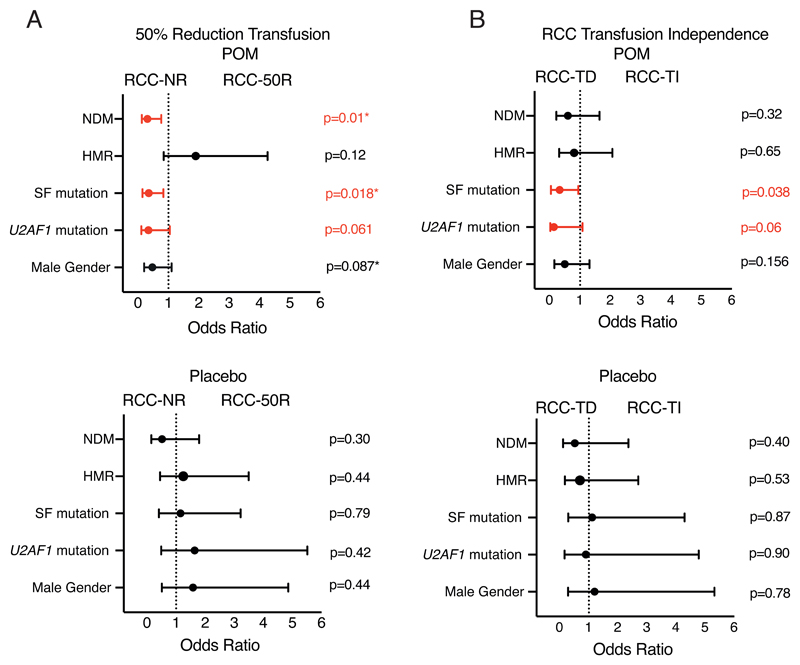
There was no significant correlation between the probability of becoming RBC-transfusion-independent and driver mutation state in either cohort. However, subjects with a non-driver mutation were less likely to achieve a 50 percent decrease in RBC-transfusions (OR=0.33 [0.17, 0.7]; P = 0.002). When analysed by treatment arm, this reduction was only seen in subjects receiving pomalidomide (OR = 0.3 [0.1, 0.8]; P = 0.01; Figure 2A) but not in those receiving placebo (OR = 0.5 [0.2, 1.8]; P = 0.3; Figure 2A). In the entire cohort, we detected a correlation between having a spliceosome mutation and a lower probability of becoming RBC-transfusion-independent in all subjects (OR = 0.5 [0.2, 1.1]; P = 0.096). However, this association was significant only in subjects receiving pomalidomide (OR = 0.3 [0.11, 0.94] P = 0.038 versus OR = 1.1 [0.29, 4.3] P = 0.87; Figure 2B). This association was largely attributable to a lower rate of RBC-transfusion-independence in subjects with a U2AF1 mutation receiving pomalidomide (OR = 0.1 [0.02, 1.1]; P = 0.06 versus placebo OR = 0.9 [0.2, 4.8]; P = 0.90; Figure 2B). A similar correlation with receiving pomalidomide was detected when the endpoint was a 50 percent decrease in RBC-transfusions in subjects with a spliceosome mutation (OR = 0.35 [0.15, 0.84]; P = 0.018; versus OR = 1.1 [0.4, 3.2]; P = 0.79; Figure 2A). No other specific mutation or combination significantly correlated with either response category. Re-calculated RBC-transfusion-independence rates in the pomalidomide and placebo cohorts, censoring subjects with splicing factor mutations, showed no treatment effect on RBC-transfusion-independence rates (22% [12, 31%] versus 15% [3, 27%], P = 0.40) or 50 percent reduction in RBC-transfusions (35% [34, 70%] versus 32% [8, 64%], P = 0.75).
Figure 2. Impact of mutational status on transfusion responses.
(A) Forest plot depicting influence of mutational status on achievement of 50% reduction in red blood cell transfusion requirement; patients with SF mutations were less likely to achieve this in the POM arm only (OR=0.35 [0.15, 0.84]; P=0.018). This association with poorer transfusion responses in SF-mutated patients was restricted to POM-treated patients. (B) Forest plot depicting the influence of mutational status on achievement of red blood cell transfusion independence (RBC-TI) by treatment arm; patients with SF mutations were less likely to achieve RBC-TI in the POM arm (OR=0.33 [0.11, 0.94] P=0.038) but not in the placebo arm; (OR=1.1 [0.29, 4.3] P=0.87); predominantly attributable to patients with U2AF1 mutations; (OR=0.1 [0.021, 1.1]; P=0.06). Univariate logistic regression was performed for each group and outcomes with significant (P<0.05) odd ratios (OR) are denoted in red. Figure 2A were adjusted for male gender (denoted by a star) which was independently significant on univariate analysis. RCC-NR = Red Cell Count No Response; RCC 50-R = Red Cell Count 50% Reduction in Transfusion Requirement; RCC-TD = Red Cell Count-Transfusion Dependent; RCC-TI = Red Cell Count-Transfusion Independent; NDM = Non Driver Mutation; HMR = High Molecular Risk Mutation; SF=Splice Factor.
Although considerable data indicate a correlation between high-risk mutations and survival, we found no such correlation (3). There are some likely explanations. 1st, all of our subjects were high-risk with 2 independent adverse risk co-variates, anemia and RBC-transfusion-dependence and 2nd, median follow-up was only 1-year. Of note, we detected a correlation between TP53 mutation and LFS. TP53 mutation is not typically classified as a high-risk molecular mutation in MPN-associated myelofibrosis but this may need reconsideration (11).
The conclusion of the RESUME study was there was no significant difference in response rates to pomalidomide and placebo in achieving RBC-transfusion-independence or achieving a 50 percent reduction in RBC-transfusion frequency. This conclusion was perplexing as there were several subjects in the pomalidomide cohort responding to pomalidomide who lost their response when pomalidomide was stopped and regained their response when pomalidomide was re-started. There were no similar subjects in the placebo cohort. Randomization is intended to balance cohorts for known and unknown (latent) co-variates correlated with outcome(s). When numbers of subjects randomized is small, such as the 252 subjects in the RESUME study, the substantial likelihood of an imbalance for ≥ 1 co-variates is presumed to be encompassed in the P-value. However, this adjustment is imperfect. We found a correlation between likelihood of response to pomalidomide and non-driver mutations including spliceosome mutations, a correlation not found in the placebo cohort. However, after censoring for spliceosome mutations, receiving pomalidomide still had no significant impact on rates of RBC-transfusion-independence or a 50 percent reduction in RBC-transfusions. The study was not powered to detect such difference and these analyses were not pre-specified. Consequently, our conclusions are hypothesis-generating requiring experimental validation.
Although it has been established that U2AF1 mutated patients with myelofibrosis are associated with anemia and inferior survival, an association with resistance to immunomodulators (IMiDs) has not be reported. The target protein for Pomalidamide is Cereblon, a gene with multiple alternatively spliced isoforms. In both myeloma and del (5q) myelodysplastic syndromes, it has been established that high levels of full length cereblon mRNA are required for IMiD activity (12). We speculate that aberrant cereblon splicing in patients with a spliceosome mutations might influence response to Pomalidomide. Our data also suggest a need for mutation topography analyses and stratification pre-randomization in intervention trials in MPN-associated myelofibrosis.
Acknowledgements
Supported in part by a National Institute for Health Research (NIHR) academic clinical lectureship (OC), the John Fell Fund, a Medical Research Council Senior Clinical Fellowship (AJM, MR/l006340/1), a Cancer Research UK (CRUK) Senior Cancer Research Fellowship. RPG acknowledges support from the National Institute of Health Research (NIHR) Biomedical Research Centre funding scheme. Supported in part by an unrestricted educational grant from Celgene Corp.
Footnotes
Conflicts of interest
GS has participated in speakers bureau and received research funding from Celgene.
MFM has participated in advisory boards and speakers bureaus for Celgene and Novartis. AJM has participated in advisory boards and speakers bureaus for Celgene and Novartis.
Author contributions
OC designed and analyzed experiments and prepared the typescript; JO performed the statistical analysis and contributed to preparing the typescript. NB and GW performed bioinformatic analysis; GB designed the sequencing panels, processed samples and performed experiments; AH contributed to the design of the panels and advised on data analyses; AT, HKA, GB, TD, HG, QJ, J-JK, RM, FP, VR, GS, AV, DZ, MFM and JZ contributed samples. RPG supervised the project and helped prepare the typescript. AJM conceived and supervised the project, designed experiments and help prepare the typescript. All authors read and approved the typescript.
References
- 1.Tefferi A. Anemia in myelofibrosis-prevalence, the U2AF1 connection, new treatments. Blood Cancer J. 2017;7(12):648. doi: 10.1038/s41408-017-0032-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rampal R, Al-Shahrour F, Abdel-Wahab O, Patel JP, Brunel JP, Mermel CH, et al. Integrated genomic analysis illustrates the central role of JAK-STAT pathway activation in myeloproliferative neoplasm pathogenesis. Blood. 2014;123(22):e123–33. doi: 10.1182/blood-2014-02-554634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vannucchi AM, Lasho TL, Guglielmelli P, Biamonte F, Pardanani A, Pereira A, et al. Mutations and prognosis in primary myelofibrosis. Leukemia. 2013;27:1861. doi: 10.1038/leu.2013.119. [DOI] [PubMed] [Google Scholar]
- 4.Tefferi A, Finke CM, Lasho T, Hanson CA, Ketterling R, Gangat N, et al. U2AF1 mutation types in primary myelofibrosis: phenotypic and prognostic distinctions. Leukemia. 2018;32 doi: 10.1038/s41375-018-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tefferi A, Al-Ali HK, Barosi G, Devos T, Gisslinger H, Jiang Q, et al. A randomized study of pomalidomide vs placebo in persons with myeloproliferative neoplasm-associated myelofibrosis and RBC-transfusion dependence. Leukemia. 2017;31(5):1252. doi: 10.1038/leu.2017.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tefferi A, Lasho TL, Finke CM, Elala Y, Hanson CA, Ketterling RP, et al. Targeted deep sequencing in primary myelofibrosis. Blood Adv. 2016;1(2):105–11. doi: 10.1182/bloodadvances.2016000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vannucchi AM, Antonioli E, Guglielmelli P, Pardanani A, Tefferi A. Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal. Leukemia. 2008;22(7):1299–307. doi: 10.1038/leu.2008.113. [DOI] [PubMed] [Google Scholar]
- 8.Papaemmanuil E, Cazzola M, Boultwood J, Malcovati L, Vyas P, Bowen D, et al. Somatic SF3B1 Mutation in Myelodysplasia with Ring Sideroblasts. New Engl J Med. 2011;365(15):1384–95. doi: 10.1056/NEJMoa1103283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shirai Cara L, Ley James N, White Brian S, Kim S, Tibbitts J, Shao J, et al. Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo. Cancer Cell. 2015;27(5):631–43. doi: 10.1016/j.ccell.2015.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Santos FPS, Getta B, Masarova L, Famulare C, Schulman J, Datoguia TS, et al. Prognostic impact of RAS-pathway mutations in patients with myelofibrosis. Leukemia. 2020;34(3):799–810. doi: 10.1038/s41375-019-0603-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tefferi A, Guglielmelli P, Nicolosi M, Mannelli F, Mudireddy M, Bartalucci N, et al. GIPSS: genetically inspired prognostic scoring system for primary myelofibrosis. Leukemia. 2018;32(7):1631–42. doi: 10.1038/s41375-018-0107-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonasova A, Bokorova R, Polak J, Vostry M, Kostecka A, Hajkova H, et al. High level of full-length cereblon mRNA in lower risk myelodysplastic syndrome with isolated 5q deletion is implicated in the efficacy of lenalidomide. Eur J Haematol. 2015;95(1):27–34. doi: 10.1111/ejh.12457. [DOI] [PubMed] [Google Scholar]