Summary
SARS-CoV-2 Spike protein is critical for virus infection via engagement of ACE21, and is a major antibody target. Here we report chronic SARS-CoV-2 with reduced sensitivity to neutralising antibodies in an immune suppressed individual treated with convalescent plasma, generating whole genome ultradeep sequences over 23 time points spanning 101 days. Little change was observed in the overall viral population structure following two courses of remdesivir over the first 57 days. However, following convalescent plasma therapy we observed large, dynamic virus population shifts, with the emergence of a dominant viral strain bearing D796H in S2 and ΔH69/ΔV70 in the S1 N-terminal domain NTD of the Spike protein. As passively transferred serum antibodies diminished, viruses with the escape genotype diminished in frequency, before returning during a final, unsuccessful course of convalescent plasma. In vitro, the Spike escape double mutant bearing ΔH69/ΔV70 and D796H conferred modestly decreased sensitivity to convalescent plasma, whilst maintaining infectivity similar to wild type. D796H appeared to be the main contributor to decreased susceptibility but incurred an infectivity defect. The ΔH69/ΔV70 single mutant had two-fold higher infectivity compared to wild type, possibly compensating for the reduced infectivity of D796H. These data reveal strong selection on SARS-CoV-2 during convalescent plasma therapy associated with emergence of viral variants with evidence of reduced susceptibility to neutralising antibodies.
Keywords: SARS-CoV-2; COVID-19; antibody escape, Convalescent plasma; neutralising antibodies; mutation; evasion; resistance; immune suppression
Clinical case history of SARS-CoV-2 infection in setting of immune-compromised host
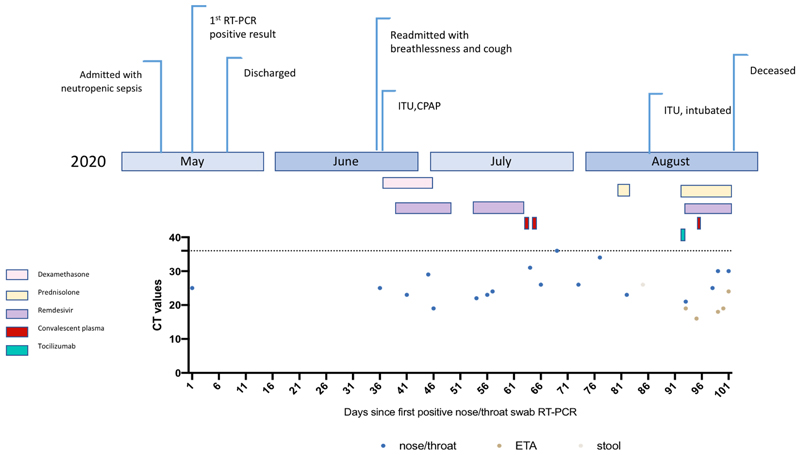
A septuagenarian male was admitted to a tertiary hospital in summer of 2020 and had tested positive for SARS-CoV-2 RT-PCR 35 days previously on a nasopharyngeal swab (Day 1) at a local hospital (Extended data 1 and 2). His past medical history was significant for marginal B cell lymphoma diagnosed in 2012, with previous chemotherapy including vincristine, prednisolone, cyclophosphamide and anti-CD20 B cell depletion with rituximab. It is likely that both chemotherapy and underlying lymphoma contributed to B and T cell combined immunodeficiency (Extended data 2 and 3, Supplementary Table 1). Computed tomography (CT) of the chest showed widespread abnormalities consistent with COVID-19 pneumonia (Supplementary Figure 1). Treatment included two 10-day courses of remdesivir with a five day gap in between (Extended data 1). Two units of convalescent plasma were administered on days 63 and 65 (Extended data 3). Following clinical deterioration, remdesivir and a unit of convalescent plasma were administered on day 95, but the individual unfortunately died on day 102 (Supplementary text).
Virus genomic comparative analysis of 23 sequential respiratory samples over 101 days
The majority of samples were respiratory samples from nose and throat or endotracheal aspirates during the period of intubation (Supplementary Table 3). Ct values ranged from 16-34 and all 23 respiratory samples were successfully sequenced by standard single molecule sequencing approach as per the ARTIC protocol implemented by COG-UK; of these 20 additionally underwent short-read deep sequencing using the Illumina platform (Supplementary table 4). There was general agreement between the two methods (Extended data 4). However due to the higher reliability of Illumina for low frequency variants, this was used for formal analysis2,3. Additionally, single genome amplification and sequencing of Spike using extracted RNA from respiratory samples was used as an independent method to detect mutations observed (Extended data 4). Finally, we detected no evidence of recombination, based on two independent methods.
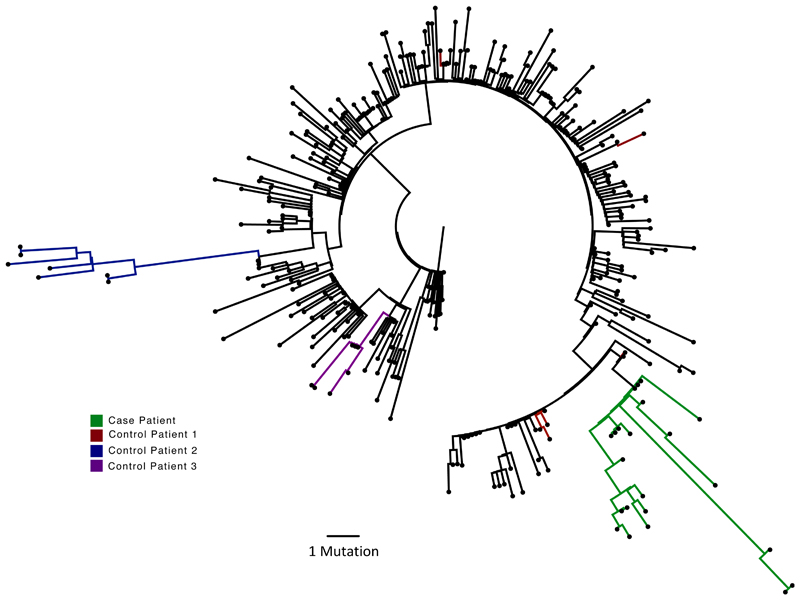
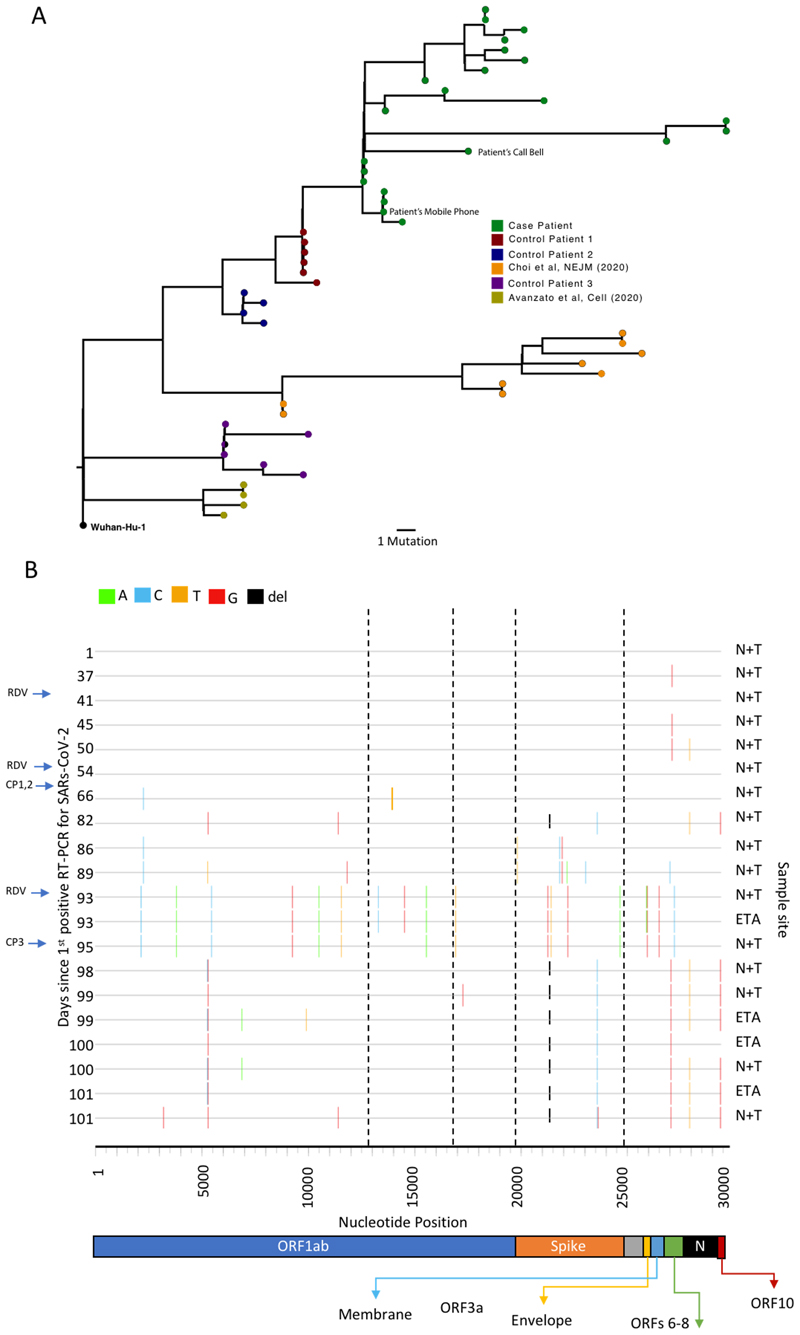
Maximum likelihood analysis of patient-derived whole genome consensus sequences demonstrated clustering with other local sequences from the same region (Figure 1). The infecting strain was assigned to lineage 20B bearing the D614G Spike variant. Environmental sampling showed evidence of virus on surfaces such as telephone and call bell. Sequencing of these surface viruses showed clustering with those derived from the respiratory tract (Extended data 2). All samples were consistent with having arisen from a single underlying viral population. In our phylogenetic analysis, we included sequential sequences from three other local patients identified with persistent viral RNA shedding over a period of 4 weeks or more as well as two long term immunosuppressed SARS-CoV-2 ‘shedders’ recently reported4,5, (Extended data 2, Supplementary Table 2). While the sequences from the three local patients as well as from Avanzato et al5 showed little divergence with no amino acid changes in Spike over time, the case patient showed significant diversification. The Choi et al report4 showed similar degree of diversification as the case patient. Further investigation of the sequence data suggested the existence of an underlying structure to the viral population in our patient, with samples collected at days 93 and 95 being rooted within, but significantly divergent from the original population (Extended data 5 and 6). The relationship of the divergent samples to those at earlier time points argues against superinfection.
Figure 1.
Analysis of 23 Patient derived whole SARS-CoV-2 genome sequences in context of local sequences and other cases of chronic SARS-CoV-2 shedding. Circularised maximum-likelihood phylogenetic tree rooted on the Wuhan-Hu-1 reference sequence, showing a subset of 250 local SARS-CoV-2 genomes from GISAID. This diagram highlights significant diversity of the case patient (green) compared to three other local patients with prolonged shedding (blue, red and purple sequences). All “United Kingdom / English” SARS-CoV-2 genomes were downloaded from the GISAID database and a random subset of 250 selected as background.
SARS-CoV-2 viral diversity
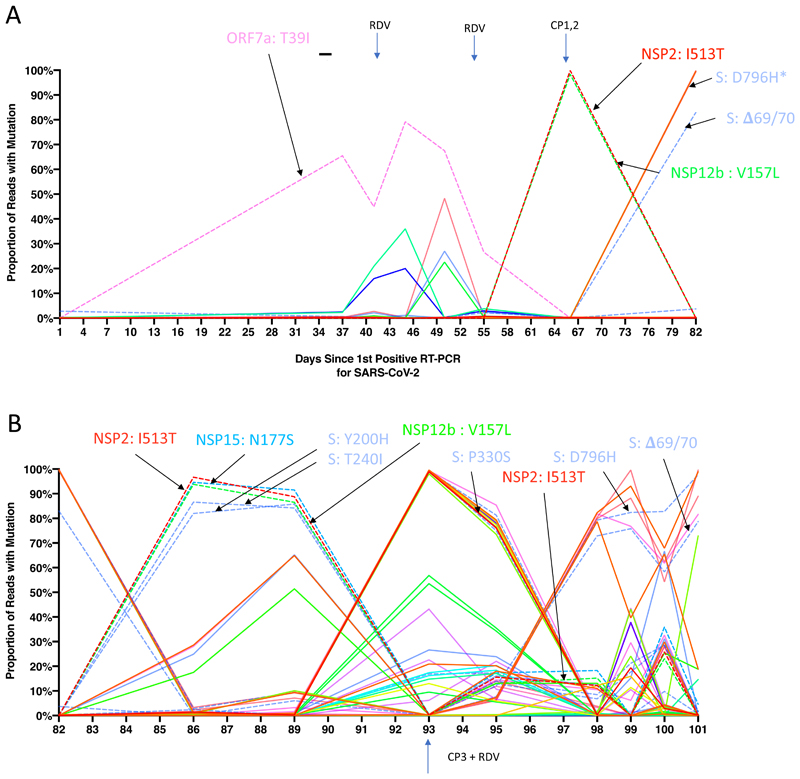
All samples tested positive by RT-PCR and there was no sustained change in Ct values throughout the 101 days following the first two courses of remdesivir (days 41 and 54), or the first two units of convalescent plasma with polyclonal antibodies (days 63 and 65, Extended data 3). Of note we were not able to culture virus from stored swab samples. Consensus sequences from short read deep sequence Illumina data revealed dynamic population changes after day 65, as shown by a highlighter plot (Extended data 6). In addition, we were also able to follow the dynamics of virus populations down to low frequencies during the entire period (Figure 2, Supplementary Table 4). Following remdesivir at day 41 the low frequency variant analysis allowed us to observe transient amino acid changes in populations at below 50% abundance in Orf 1b, 3a and Spike, with a T39I (C27509T) mutation in ORF7a reaching 79% on day 45 (Figure 2, pink, supplementary information). At day 66 we noted I513T in NSP2 (T2343C) and V157L (G13936T) in RdRp had emerged from undetectable at day 54 to almost 100% frequency (Figure 2, red and green dashed lines), with the polymerase being the more plausible candidate for driving this sweep. Notably, spike variant N501Y, which can increase the ACE2 receptor affinity6, and which is present in the new UK B1.1.7 lineage7, was observed on day 55 at 33% frequency, but was eliminated by the sweep of the NSP2/RdRp variant.
Figure 2. Whole genome variant trajectories showing amino acids and relationship to treatments.
Data based on Illumina short read ultra deep sequencing at 1000x coverage. Variants shown reached a frequency of at least 10% in at least 2 samples. Treatments indicated are convalescent plasma (CP) and Remdesivir (RDV). Variants described in the text are designated by labels using the same colouring as the position in the genome. Variants labelled are represented by dashed lines. A. Variants detected in the patient from days 1-82. *D796H (light blue) is at the same frequency as NSP3 K902N (orange) therefore it is hidden beneath B. Variants detected in the patient from days 82-101.
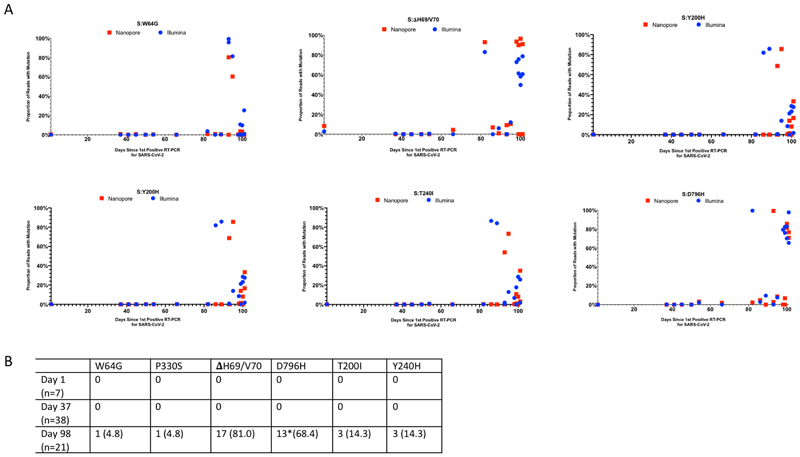
In contrast to the early period of infection, between days 66 and 82, following the first two administrations of convalescent sera, a shift in the virus population was observed, with a variant bearing D796H in S2 and ΔH69/ΔV70 in the S1 N-terminal domain (NTD) becoming the dominant population at day 82. This was identified in a nose and throat swab sample with high viral load as indicated by Ct of 23 (Figure 3A). The deletion was detected transiently at baseline according to short read deep sequencing. ΔH69/ΔV70 was due to an out of frame six nucleotide deletion resulting in the sequence of codon 68 changing from ATA to ATC.
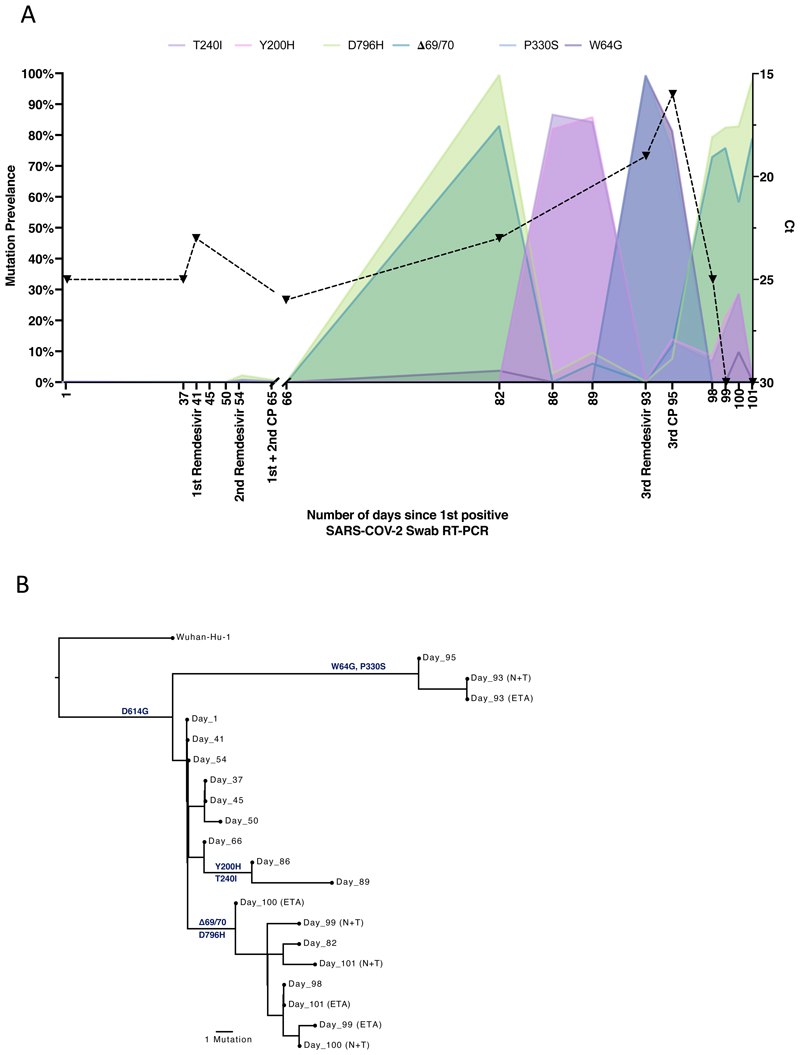
Figure 3. Longitudinal variant frequencies and phylogenetic relationships for virus populations bearing six Spike (S) mutations.
A. At baseline, all six S variants (Illumina sequencing) except for ΔH69/V70 were absent (<1% and <20 reads). Approximately two weeks after receiving two units of convalescent plasma (CP), viral populations carrying ΔH69/V70 and D796H mutants rose to frequencies >80% but decreased significantly four days later. This population was replaced by a population bearing Y200H and T240I, detected in two samples over a period of 6 days. These viral populations were then replaced by virus carrying W64G and P330S mutations in Spike, which both dominated at day 93. Following a 3rd course of remdesivir and an additional unit of convalescent plasma, the ΔH69/V70 and D796H virus population re-emerged to become the dominant viral strain reaching variant frequencies of >75%. Pairs of mutations arose and disappeared simultaneously indicating linkage on the same viral haplotype. CT values from respiratory samples are indicated on the right y-axis (black dashed line and triangles). Where there were duplicate readings on the same day, to remain consistent, N+T samples were plotted B. Maximum likelihood phylogenetic tree of the case patient with day of sampling indicated. Spike mutations defining each of the clades are shown ancestrally on the branches on which they arose. On dates where multiple samples were collect, these are indicated as endotracheal aspirate (ETA) and Nose + throat swabs (N+T).
On Days 86 and 89, viruses obtained from upper respiratory tract samples were characterised by the Spike mutations Y200H and T240I, with the deletion/mutation pair observed on day 82 having fallen to frequencies of 10% or less (Figure 2 and 3). The Spike mutations Y200H and T240I were accompanied at high frequency by two other non-synonymous variants with similar allele frequencies, coding for I513T in NSP2, V157L in RdRp and N177S in NSP15 (Figure 2A). Both of these were also previously observed at >98% frequency in the sample on day 66 (Figure 2A, red and green lines), arguing that this new lineage emerged out of a previously existing population.
Sequencing of a nose and throat swab sample at day 93 identified viruses characterised by Spike mutations P330S at the edge of the RBD and W64G in S1 NTD at close to 100% abundance, with D796H along with ΔH69/ΔV70 at <1% abundance and the variants Y200H and T240I at frequencies of <2%. Viruses with the P330S variant were detected in two independent samples from different sampling sites, arguing against the possibility of contamination. The divergence of these samples from the remainder of the population (Figure 2, 3B and Extended data 5 and 6) suggests the possibility that they represent a compartmentalised subpopulation.
Patterns in the variant frequencies suggest competition between virus populations carrying different mutations, viruses with the D796H/ΔH69/ΔV70 deletion/mutation pair rising to high frequency during CP therapy, then being outcompeted by another population in the absence of therapy. Specifically, these data are consistent with a lineage of viruses with the NSP2 I513T and RdRp V157L variant, dominant on day 66, being outcompeted during therapy by the mutation/deletion variant. With the lapse in therapy, the original strain, having acquired NSP15 N1773S and the Spike mutations Y200H and T240I, regained dominance, followed by the emergence of a separate population with the W64G and P330S mutations.
In a final attempt to reduce the viral load, a third course of remdesivir (day 93) and third dose of CP (day 95) were administered. We observed a re-emergence of the D796H + ΔH69/ΔV70 viral population (Figure 2, 3). The inferred linkage of D796H and ΔH69/ΔV70 was maintained as evidenced by the highly similar frequencies of the two variants, suggesting that the third unit of CP led to the re-emergence of this population under renewed positive selection. In further support of our proposed idea of competition, noted above, frequencies of these two variants appeared to mirror changes in the NSP2 I513T mutation (Figure 2), suggesting these as markers of opposing clades in the viral population. Ct values remained low throughout this period with hyperinflammation, eventually leading to multi-organ failure and death at day 102. The repeated increase in frequency of the viral population with CP therapy strongly supports the hypothesis that the deletion/mutation combination conferred selective advantage.
Spike mutants emerging post convalescent plasma impair neutralising antibody potency
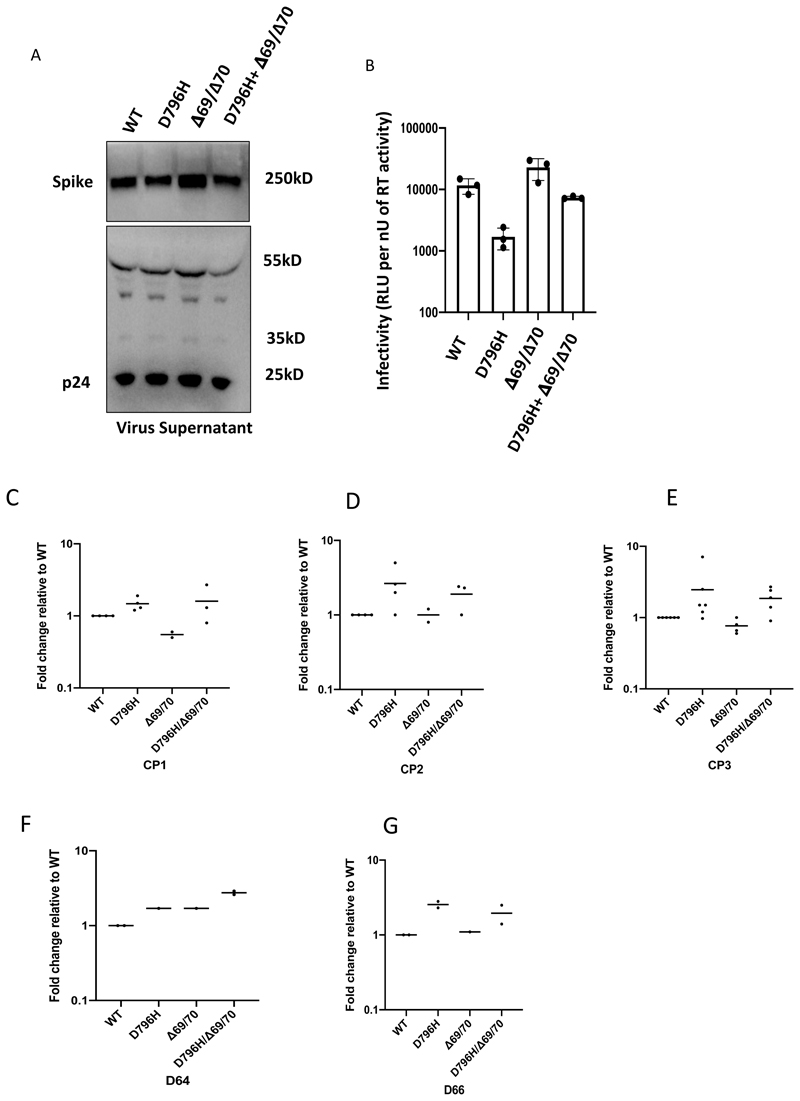
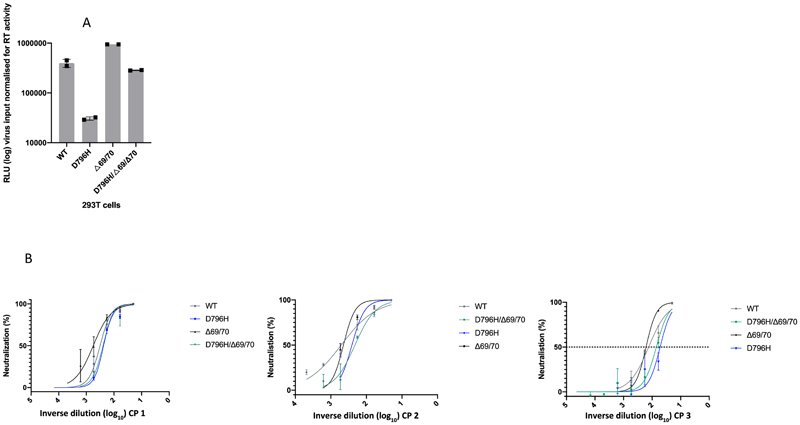
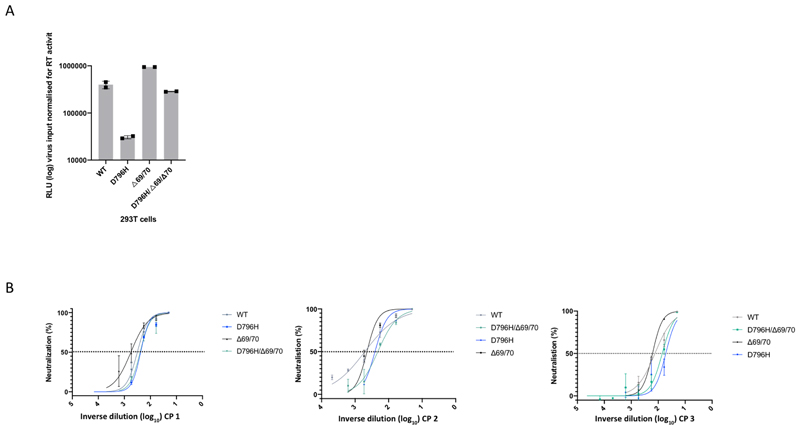
Using lentiviral pseudotyping we generated wild type, ΔH69/ΔV70 + D796H and single mutant Spike proteins in enveloped virions in order to measure neutralisation activity of CP against these viruses (Figure 4). This system has been shown to give generally similar results to replication competent virus8,9. Spike protein from each mutant was detected in pelleted virions (Figure 4A). We also probed with an HIV-1 p24 antibody to monitor levels of lentiviral particle production (Figure 4A, Supplementary Figure 2). We then measured infectivity of the pseudoviruses, correcting for virus input using reverse transcriptase activity measurement, and found that ΔH69/ΔV70 appeared to have two-fold higher infectivity over a single round of infection compared to wild type (Figure 4B, Extended data 7). By contrast, the D796H single mutant had significantly lower infectivity as compared to wild type and double mutant had similar infectivity to wild type (Figure 4B, Extended data 7).
Figure 4. Spike mutant D796H + ΔH69/V70 infectivity and sensitivity convalescent plasma (CP).
A. western blot of virus pellets after centrifugation of supernatants from cells transfected with lentiviral pseudotyping plasmids including Spike protein. Blots are representative of two independent transfections. B. Single round Infectivity of luciferase expressing lentivirus pseudotyped with SARS-CoV-2 Spike protein (WT versus mutant) on 293T cells co-transfected with ACE2 and TMPRSS2 plasmids. Infectivity is corrected for reverse transcriptase activity in virus supernatant as measured by real time PCR. Data points represent technical replicates (n=3) with mean and error bars representing standard error of mean; data are representative of two independent experiments C-E. convalescent plasma (CP units 1-3) neutralization potency against pseudovirus virus bearing Spike mutants D796H, ΔH69/V70 and D796H + ΔH69/V70 F, G patient serum neutralisation potency against pseudovirus virus bearing Spike mutants D796H, ΔH69/V70 and D796H + ΔH69/V70. Patient serum was taken at indicated Day (D). Indicated is serum dilution required to inhibit 50% of virus infection (ID50), expressed as fold change relative to WT. Data points represent means of technical replicates and each data point is an independent experiment (n=2-6). Mean of data points in C-G is shown by horizontal bars.
We found that D796H alone and the D796H + ΔH69/ΔV70 double mutant were less sensitive to neutralisation by convalescent plasma samples (Figure 4C-E, Extended data 7). By contrast the ΔH69/ΔV70 single mutant did not reduce neutralisation sensitivity. In addition, patient derived serum from days 64 and 66 (one day either side of CP2 infusion) similarly showed lower potency against the D796H + ΔH69/ΔV70 mutants (Figure 4F, G).
A panel of nineteen monoclonal antibodies (mAbs) isolated from three donors was previously identified to neutralize SARS-CoV-2. To establish if the mutations incurring in vivo (D796H and ΔH69/ΔV70) resulted in a global change in neutralization sensitivity we tested neutralising mAbs targeting the seven major epitope clusters previously described (excluding non-neutralising clusters II, V and small [n =<2] neutralising clusters IV, X). The eight RBD-specific mAbs (Extended data 8) exhibited no major change in neutralisation potency and non-RBD specific COVA1-21 showing 3-5 fold reduction in potency against ΔH69/ΔV70+D796H and ΔH69/ΔV70, but not D796H alone9 (Extended data 8). We observed no differences in neutralisation between single/double mutants and wild type, suggesting that the mechanism of escape was likely outside these epitopes in the RBD. These data confirm the specificity of the findings from convalescent plasma and suggest that mutations observed are related to antibodies targeting regions outside the RBD. Interestingly, ΔH69/ΔV70 containing viruses showed reduced neutralisation sensitivity to the mAb COVA1-21, targeting an as yet undefined epitope outside the RBD. 10.
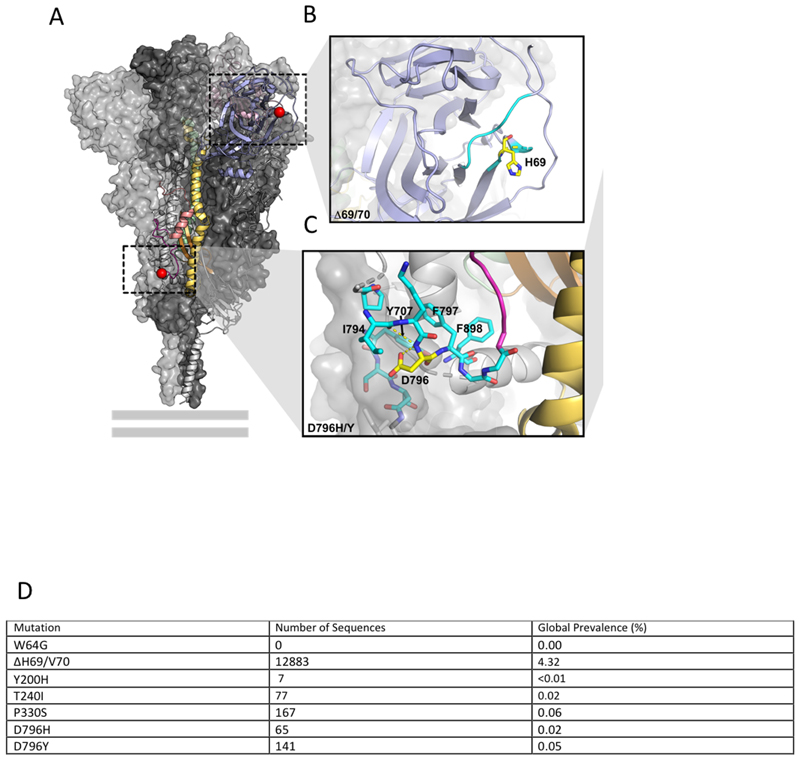
To understand how the ΔH69/ΔV70 and D796H might confer antibody resistance, we assessed how they might affect the Spike structure (Extended data 9). We based this analysis primarily on a structure lacking stabilising modifications (PDB 6xr8)11, but also referred to stabilised structures determined at different pH values12. ΔH69/ΔV70 is located in a disordered, glycosylated loop at the distal surface of the NTD, near the binding site of polyclonal antibodies derived from COV57 plasma13,14 (Extended data 9). As this loop is flexible and highly accessible, ΔH69/V70 could in principle affect antibody binding in this region. D796 is located near the base of Spike, in a surface loop that is structurally somewhat disordered in the prefusion conformation and becomes part of a large disordered region in the post fusion S2 trimer11 (Extended data 9). The loop containing residue 796 is proposed to be targeted by antibodies15, despite mutations at position 796 being relatively uncommon (Extended data 9). In the RBD-down Spike structures11,12, D796 forms contacts with residues in the neighbouring protomer, including the glycosylated residue N709 (Extended data 9).
Discussion
Here we have documented a repeated evolutionary response by SARS-CoV-2 in the presence of antibody therapy during the course of a persistent infection in an immunocompromised host. The observation of potential selection for specific variants coinciding with the presence of antibodies from convalescent plasma is supported by the experimental finding of two-fold reduced susceptibility of these viruses to convalescent plasma containing polyclonal antibodies. In this case the emergence of the variant was not the primary reason for treatment failure. We have noted in our analysis signs of compartmentalised viral replication based on the sequences recovered in upper respiratory tract samples. Both population genetic and small animal studies have shown a lack of reassortment between influenza viruses within a single host during an infection, suggesting that acute respiratory viral infection may be characterised by spatially distinct viral populations16,17. In the analysis of data, it is important to distinguish genetic changes which occur in the primary viral population from apparent changes that arise from the stochastic observation of spatially distinct subpopulations in the host. While the samples we observe on days 93 and 95 of infection are genetically distinct from the others, the remaining samples are consistent with arising from a consistent viral population. We note that Choi et al reported the detection in post-mortem tissue of viral RNA not only in lung tissue, but also in the spleen, liver, and heart4. Mixing of virus from different compartments, for example via blood, or movement of secretions from lower to upper respiratory tract, could lead to fluctuations in viral populations at particular sampling sites.
This is a single case report and therefore limited conclusions can be drawn about generalisability.
An important limitation is that the data were derived from sampling from the upper respiratory tract and not the lower tract, thus limiting the inferences that can be drawn regarding viral populations in this single case.
In addition to documenting the emergence of SARS-CoV-2 Spike ΔH69/ΔV70 in vivo, we show that this mutation modestly increases infectivity of the Spike protein in a pseudotyping assay. The deletion was observed contemporaneously with the rare S2 mutation D796H after two separate courses of CP, with other viral populations emerging. D796H, but not ΔH69/ΔV70, conferred reduction in susceptibility to polyclonal antibodies in the units of CP administered, though we cannot speculate as to their individual impacts on sera from other individuals. It is intriguing that the ΔH69/ΔV70 + D796H double mutant diminished in between CP courses, suggesting that there were other selective forces at play in the intervening period, possibly driven by the inflammation observed in the individual. This includes the possibility that the haplotype with ΔH69/ΔV70 + D796H may have carried mutations in other regions deleterious during that intervening period. Although ΔH69/V70 is expanding at a high rate18, D796 mutations are also increasing. D796H has been documented in 0.02% of global sequences and D796Y appears in 0.05% of global sequences (Extended data 9).
The effects of CP on virus evolution seen here are unlikely to apply in immune competent hosts where viral diversity is likely to be lower due to better immune control. Our data highlight that infection control measures may need to be tailored to the needs of immunocompromised patients and also caution in interpretation of CDC guidelines that recommend 20 days as the upper limit of infection prevention precautions in immune compromised patients who are afebrile19. Due to the difficulty with culturing clinical isolates, use of surrogates are warranted20. However, where detection of ongoing viral evolution is possible, this serves as a clear proxy for the existence of infectious virus. In our case we detected environmental contamination whilst in a single occupancy room and the patient was moved to a negative-pressure high air-change infectious disease isolation room.
Clinical efficacy of convalescent plasma in severe COVID-19 has not been demonstrated21, and its use in different stages of infection and disease remains experimental; as such, we suggest that it should be reserved for use within clinical trials, with rigorous monitoring of clinical and virological parameters. The data from this single case report might warrant caution in use of convalescent plasma in patients with immune suppression of both T cell and B cell arms; in such cases, the antibodies administered have little support from cytotoxic T cells, thereby reducing chances of clearance and theoretically raising the potential for escape mutations. Whilst we await further data, where clinical trial enrolment is not possible, convalescent plasma administered for clinical need in immune suppression should ideally only be considered as part of observational studies, undertaken preferably in single occupancy rooms with enhanced infection control precautions, including SARS-CoV-2 environmental sampling and real-time sequencing. Understanding of viral dynamics and characterisation of viral evolution in response to different selection pressures in the immunocompromised host is necessary not only for improved patient management but also for public health benefit.
Methods
Clinical Sample Collection and Next generation sequencing
Serial samples were collected from the patient periodically from the lower respiratory tract (sputum or endotracheal aspirate), upper respiratory tract (throat and nasal swab), and from stool. Nucleic acid extraction was done from 500μl of sample with a dilution of MS2 bacteriophage to act as an internal control, using the easyMAG platform (Biomerieux, Marcy-l'Étoile) according to the manufacturers’ instructions. All samples were tested for presence of SARS-CoV-2 with a validated one-step RT q-PCR assay developed in conjunction with the Public Health England Clinical Microbiology22. Amplification reaction were all performed on a Rotorgene™ PCR instrument. Samples which generated a CT of ≤36 were considered to be positive.
Sera from recovered patients in the COVIDx study23 were used for testing of neutralisation activity by SARS-CoV-2 mutants.
SARS-CoV-2 serology by multiplex particle-based flow cytometry (Luminex)
Recombinant SARS-CoV-2 N, S and RBD were covalently coupled to distinct carboxylated bead sets (Luminex; Netherlands) to form a 3-plex and analyzed as previously described (Xiong et al. 2020). Specific binding was reported as mean fluorescence intensities (MFI).
Whole blood T cell and innate stimulation assay
Whole blood was diluted 1:5 in RPMI into 96-well F plates (Corning) and activated by single stimulation with phytohemagglutinin (PHA; 10 μg/ml; Sigma-Aldrich), or LPS (1 μg/ml, List Biochemicals) or by co-stimulating with anti-CD3 (MEM57, Abcam, 200 ng/ml, 1:1000) and IL-2 (Immunotools, 1430U/ml, 1:1000). Supernatants were taken after 24 hours. Levels (pg/ml) are shown for IFNg, IL17, IL2, TNFa, IL6, IL1b and IL10. Cytokines were measured by multiplexed particle based Flow cytometry on a Luminex analyzer (Bio-Plex, Bio-Rad, UK) using an R&D Systems custom kit (R&D Systems, UK).
For viral genomic sequencing, total RNA was extracted from samples as described. Samples were sequenced using MinION flow cells version 9.4.1 (Oxford Nanopore Technologies) following the ARTICnetwork V3 protocol (https://dx.doi.org/10.17504/protocols.io.bbmuik6w) and BAM files assembled using the ARTICnetwork assembly pipeline (https://artic.network/ncov-2019/ncov2019-bioinformatics-sop.html). A representative set of 10 sequences were selected and also sequenced using the Illumina MiSeq platform. Amplicons were diluted to 2 ng/μl and 25 μl (50 ng) were used as input for each library preparation reaction. The library preparation used KAPA Hyper Prep kit (Roche) according to manufacturer’s instructions. Briefly, amplicons were end-repaired and had A-overhang added; these were then ligated with 15mM of NEXTflex DNA Barcodes (Bio Scientific, Texas, USA). Post-ligation products were cleaned using AMPure beads and eluted in 25 μl. Then, 20 μl were used for library amplification by 5 cycles of PCR. For the negative controls, 1ng was used for ligation-based library preparation. All libraries were assayed using TapeStation (Agilent Technologies, California, USA) to assess fragment size and quantified by QPCR. All libraries were then pooled in equimolar accordingly. Libraries were loaded at 15nM and spiked in 5% PhiX (Illumina, California, USA) and sequenced on one MiSeq 500 cycle using a Miseq Nano v2 with 2x 250 paired-end sequencing. A minimum of ten reads were required for a variant call.
Bioinformatics Processes
For long-read sequencing, genomes were assembled with reference-based assembly and a curated bioinformatics pipeline with 20x minimum coverage across the whole-genome24. For short-read sequencing, FASTQs were downloaded, poor-quality reads were identified and removed, and both Illumina and PHiX adapters were removed using TrimGalore v0.6.625. Trimmed paired-end reads were mapped to the National Center for Biotechnology Information SARS-CoV-2 reference sequence MN908947.3 using MiniMap2-2.17 with arguments -ax and sr26. BAM files were then sorted and indexed with samtools v1.11 and PCR optical duplicates removed using Picard (http://broadinstitute.github.io/picard). A consensus sequences of nucleic acids with a minimum whole-genome coverage of at least 20× were generated with BCFtools using a 0% majority threshold.
Variant calling
Variant frequencies were validated using custom code as part of the AnCovMulti package (github.com/PollockLaboratory/AnCovMulti). The main idea behind this validation was to identify and remove consistent potential amplification errors and mutability near the end of Illumina reads. Furthermore, stringent filtering was applied to remove biased amplification of early laboratory-induced mutations or very low copy variations.
Filtering consisted of requiring exact initiation at a primer within two bp of the start of a read, a minimum of 247 bp length read, fewer than four well-separated sites divergent from the reference sequence, a maximum insertion size of three nucleotides, a maximum deletion size of 11 bp, and resolution of conflicting signal from different primers.
Single Genome Amplification and sequencing
Viral RNA extracts were reverse transcribed from each sample to sufficiently capture the diversity of the viral population without introducing resampling bias. SuperScript IV (Thermofisher Scientific) and the gene specific primers were used for reverse transcription. Template RNA was degraded with RNAse H (Thermofisher Scientific). All primers used were ‘in-house’ primers designed using the multiple sequence alignment of the patient’s consensus NGS sequences. Partial Spike (amino acids 21-800) was amplified as 1 continuous length of DNA (Spike ~ 1.8 kb) by nested PCR. Terminally diluted cDNA was PCR-amplified using Platinum® Taq DNA Polymerase High Fidelity (Invitrogen, Carlsbad, CA) so that 30% of reactions were positive27. By Poisson statistics, sequences were deemed ≥80% likely to be derived from HIV-1 single genomes. We obtained between 20–60 single genomes at each sample time point to achieve 90% confidence of detecting variants present at ≥8% of the viral population in vivo28,29. Partial spike amplicons obtained from terminal dilution PCR amplification were Sanger sequenced to form a contiguous sequence using another set of 8 in-house primers. Sanger sequencing was provided by Genewiz UK and manual sequence editing was performed using DNA Dynamo software (Blue Tractor Software Ltd, UK).
Phylogenetic Analysis
All available full-genome SARS-CoV-2 sequences were downloaded from the GISAID database (http://gisaid.org/)30 on 16th December. Duplicate and low-quality sequences (>5% N regions) were removed, leaving a dataset of 212,297 sequences with a length of >29,000bp. All sequences were sorted by name and only sequences sequenced with United Kingdom / England identifiers were retained. From this dataset, sequences were de-duplicated and where background sequences were required in figures, randomly subsampled using seqtk (https://github.com/lh3/seqtk). All sequences were aligned to the SARS-CoV-2 reference strain MN908947.3, using MAFFT v7.475 with automatic flavour selection31. Major SARS-CoV-2 clade memberships were assigned to all sequences using both the Nextclade server v0.9 (https://clades.nextstrain.org/) and Phylogenetic Assignment Of Named Global Outbreak Lineages (pangolin)32.
Maximum likelihood phylogenetic trees were produced using the above curated dataset using IQ-TREE v2.1.233. Evolutionary model selection for trees were inferred using ModelFinder34 and trees were estimated using the GTR+F+I model with 1000 ultrafast bootstrap replicates35. All trees were visualised with Figtree v.1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/), rooted on the SARS-CoV-2 reference sequence and nodes arranged in descending order. Nodes with bootstraps values of <50 were collapsed using an in-house script.
In-depth allele frequency variant calling
The SAMFIRE package version 1.06 36 was used to call allele frequency trajectories from BAM file data. Reads were included in this analysis if they had a median PHRED score of at least 30, trimming the ends of reads to achieve this if necessary. Nucleotides were then filtered to have a PHRED score of at least 30; reads with fewer than 30 such reads were discarded. Distances between sequences, accounting for low-frequency variant information, was also conducted using SAMFIRE. The sequence distance metric, described in an earlier paper37, combines allele frequencies across the whole genome. Where L is the length of the genome, we define q(t) as a 4 x L element vector describing the frequencies of each of the nucleotides A, C, G, and T at each locus in the viral genome sampled at time t. For any given locus i in the genome we calculate the change in allele frequencies between the times t1 and t2 via a generalisation of the Hamming distance
where the vertical lines indicate the absolute value of the difference. These statistics were then combined across the genome to generate the pairwise sequence distance metric
The Mathematica software package was to conduct a regression analysis of pairwise sequence distances against time, leading to an estimate of a mean rate of within-host sequence evolution. In contrast to the phylogenetic analysis, this approach assumed the samples collected on days 93 and 95 to arise via stochastic emission from a spatially separated subpopulation within the host, leading to a lower inferred rate of viral evolution for the bulk of the viral population.
All variants were indecently validated using custom code as part of the AnCovMulti package, found at https://github.com/PollockLaboratory/AnCovMulti.
Western blot analysis
Forty-eight hours after transfection of cells with plasmid preparations, the culture supernatant was harvested and passed through a 0.45-μm-pore-size filter to remove cellular debris. The filtrate was centrifuged at 15,000 rpm for 120 min to pellet virions. The pelleted virions were lysed in Laemmli reducing buffer (1 M Tris-HCl [pH 6.8], SDS, 100% glycerol, β-mercaptoethanol, and bromophenol blue). Pelleted virions were subjected to electrophoresis on SDS–4 to 12% bis-Tris protein gels (Thermo Fisher Scientific) under reducing conditions. This was followed by electroblotting onto polyvinylidene difluoride (PVDF) membranes. The SARS-CoV-2 Spike proteins were visualized by a ChemiDoc> MP imaging system (Biorad) using anti-Spike S2 (Invitrogen at 1:1000 dilution) and anti-p24 Gag antibodies (NIH AIDS Reagents 1:1000 dilution).
Recombination Detection
All sequences were tested for potential recombination, as this would impact on evolutionary estimates. Potential recombination events were explored with nine algorithms (RDP, MaxChi, SisScan, GeneConv, Bootscan, PhylPro, Chimera, LARD and 3SEQ), implemented in RDP5 with default settings38. To corroborate any findings, ClonalFrameML v1.1239 was also used to infer recombination breakpoints. Neither programs indicated evidence of recombination in our data.
Structural Viewing
The Pymol Molecular Graphics System v2.4.0 (https://github.com/schrodinger/pymol-open-source/releases) was used to map the location of the four spike mutations of interested onto a SARS-CoV-2 spike structure visualised by Wrobel et al (PDB: 6ZGE)40.
Testing of convalescent plasma for antibody titres
The Anti-SARS-CoV-2 ELISA (IgG) assay used to test CP for antibody titres was Euroimmun Medizinische Labordiagnostika AG. This indirect ELISA based assay uses a recombinant structural spike 1 (S1) protein of SARS-CoV-2 expressed in the human cell line HEK 293 for the detection of SARS-CoV2 IgG.
Generation of Spike mutants
Amino acid substitutions were introduced into the D614G pCDNA_SARS-CoV-2_Spike plasmid as previously described41 using the QuikChange Lightening Site-Directed Mutagenesis kit, following the manufacturer’s instructions (Agilent Technologies, Inc., Santa Clara, CA).
Pseudotype virus preparation
Viral vectors were prepared by transfection of 293T cells by using Fugene HD transfection reagent (Promega). 293T cells were transfected with a mixture of 11ul of Fugene HD, 1μg of pCDNAp19Spike-HA, 1ug of p8.91 HIV-1 gag-pol expression vector42,43, and 1.5μg of pCSFLW (expressing the firefly luciferase reporter gene with the HIV-1 packaging signal). Viral supernatant was collected at 48 and 72h after transfection, filtered through 0.45um filter and stored at -8°C. The 50% tissue culture infectious dose (TCID50) of SARS-CoV-2 pseudovirus was determined using Steady-Glo Luciferase assay system (Promega).
Standardisation of virus input by SYBR Green-based product-enhanced PCR assay (SG-PERT)
The reverse transcriptase activity of virus preparations was determined by qPCR using a SYBR Green-based product-enhanced PCR assay (SG-PERT) as previously described44. Briefly, 10-fold dilutions of virus supernatant were lysed in a 1:1 ratio in a 2x lysis solution (made up of 40% glycerol v/v 0.25% Trition X-100 v/v 100mM KCl, RNase inhibitor 0.8 U/ml, TrisHCL 100mM, buffered to pH7.4) for 10 minutes at room temperature.
12μl of each sample lysate was added to thirteen 13μl of a SYBR Green master mix (containing 0.5μM of MS2-RNA Fwd and Rev primers, 3.5pmol/ml of MS2-RNA, and 0.125U/μl of Ribolock RNAse inhibitor and cycled in a QuantStudio. Relative amounts of reverse transcriptase activity were determined as the rate of transcription of bacteriophage MS2 RNA, with absolute RT activity calculated by comparing the relative amounts of RT to an RT standard of known activity.
Serum/plasma pseudotype neutralization assay
Spike pseudotype assays have been shown to have similar characteristics as neutralisation testing using fully infectious wild type SARS-CoV-28.Virus neutralisation assays were performed on 293T cell transiently transfected with ACE2 and TMPRSS2 using SARS-CoV-2 Spike pseudotyped virus expressing luciferase45. Pseudotyped virus was incubated with serial dilution of heat inactivated human serum samples or convalescent plasma in duplicate for 1h at 37°C. Virus and cell only controls were also included. Then, freshly trypsinized 293T ACE2/TMPRSS2 expressing cells were added to each well. Following 48h incubation in a 5% CO2 environment at 37°C, the luminescence was measured using Steady-Glo Luciferase assay system (Promega).
mAb pseudotype neutralisation assay
Virus neutralisation assays were performed on HeLa cells stably expressing ACE2 and using SARS-CoV-2 Spike pseudotyped virus expressing luciferase as previously described46. Pseudotyped virus was incubated with serial dilution of purified mAbs9 in duplicate for 1h at 37°C. Then, freshly trypsinized HeLa ACE2-expressing cells were added to each well. Following 48h incubation in a 5% CO2 environment at 37°C, the luminescence was measured using Bright-Glo Luciferase assay system (Promega) and neutralization calculated relative to virus only controls. IC50 values were calculated in GraphPad Prism.
Extended Data
Extended Data Figure 1.
Clinical time line of events with longitudinal respiratory sample CT values. CT – cycle threshold.
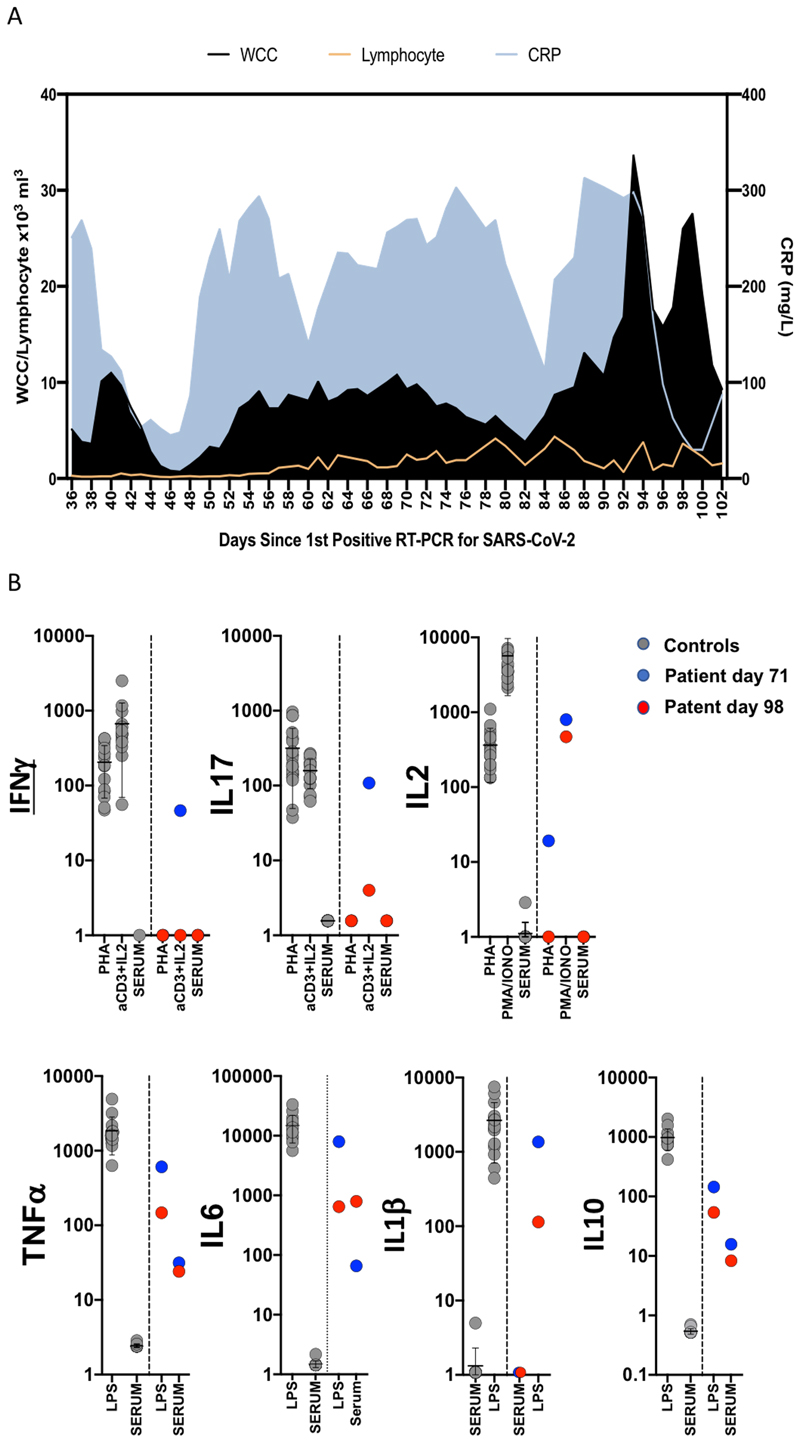
Extended data 2.
A. Blood parameters over time in patient case: White cell count (WCC) and lymphocyte counts are expressed as x103 Cells/mm3. CRP: C reactive protein. B. Assessment of T cell and innate function. Whole blood cytokines were measured in whole blood after 24 hours stimulation either after T-cell stimulation with PHA or anti CD3/IL2 or innate stimulation with LPS. Healthy controls are shown as grey circles (N=15), Patient at d71 and d98 is shown as blue circles or red circles respectively. Cytokine levels are shown as pg/ml stimulation. Mean is shown by line and whiskers representing standard deviation.
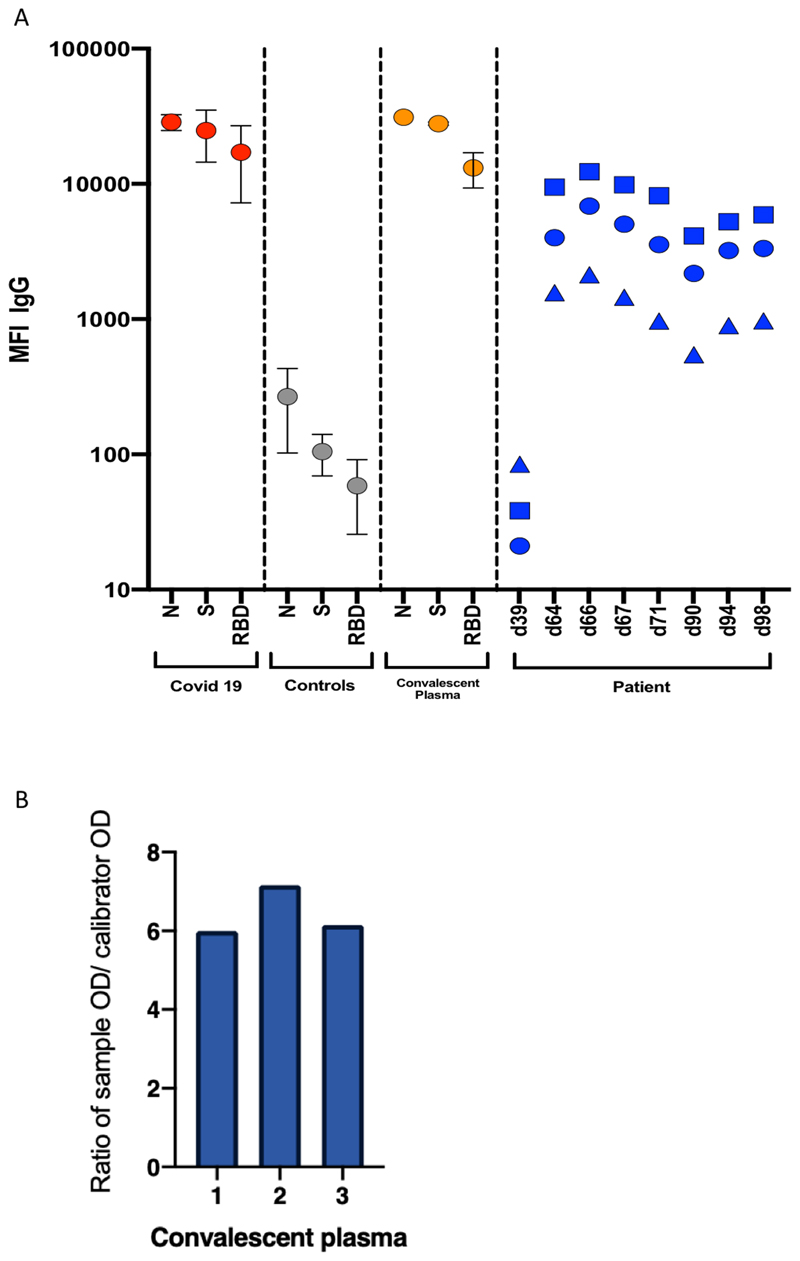
Extended Data Figure 3.
A. Serum SARS-CoV-2 antibody levels and virus population changes in chronic SARS-CoV-2 infection. Anti SARS-CoV2 IgG antibodies in patient and pre/post convalescent plasma compared to RNA+ Covid19 patients and prepandemic healthy controls: Red, grey and gold: IgG antibodies to SARS-CoV2 nucleocapsid protein (N), trimeric S protein (S) and the receptor binding domain (RBD) were measured by multiplexed particle based flow cytometry (Luminex) in RNA+ COVID-19 patients (N=20, red dots), Pre-pandemic healthy controls (N=20, grey dots) and in the convalescent donor plasma (orange dots); Results are shown as mean fluorescent intensity (MFI) +/- SD. Patient sera over time in blue: Anti SARS-CoV2 IgG to N (blue squares), S (blue circles) and RBD (blue triangles). Timing of CP units is also shown. B. SARS-CoV-2 antibody titres in patient and in convalescent plasma. Measurement of SARS-CoV-2 specific IgG antibody titres in three units of convalescent plasma (CP) by Euroimmun assay.
Extended data 4. Comparison between short-read (Illumina) and long-read single molecule (Oxford Nanopore) sequencing methods for the six observed Spike mutations.
Concordance was generally good between the majority of timepoints, however due to large discrepancies in a number of timepoints, we suggest that due to the high base calling error rate, Nanopore is not yet suitable for calling minority variants. As such, all figures in the main paper were produced using Illumina data only. B. Single genome sequencing (SGS) data from respiratory samples at indicated days. Indicated are the number of single genomes obtained at each time point with the mutations of interest (identified by deep sequencing). *denominator is 19 as for 2 samples the primer reads were poor quality at amino acid 796 at day 98. Amino acid variant and corresponding nucleotide position: S:W64G = 21752, S:Δ69 = 21765-21770, S:Y200H = 22160, S:T240I = 22281, S:P330S = 22550, S:D795H = 23948
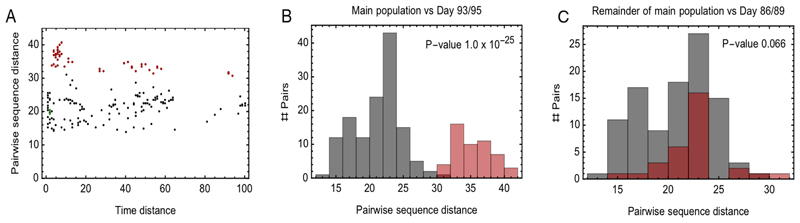
Extended Data Figure 5. Evidence for within-host cladal structure.
A. Pairwise distances between samples measured using the all-locus distance metric plotted against pairwise distances in time (measured in days) between samples being collected. Internal distances between samples in the proposed main clade are shown in black, distances between samples in the main clade and samples collected on days 93 and 95 are shown in red, and internal distances between samples collected on days 93 and 95 are shown in green. B. Pairwise distances between samples in the larger clade (black) and between these samples and those collected on days 93 and 95 (red). The median values of the distributions of these values are significantly different according to a Mann Whitney test. C. Pairwise distances between samples in the main clade, once those collected on days 86, 89, 93, 95 have been removed (black) and between these samples and those collected on days 86 and 89 (red). The median values of the distributions of these values are not significantly different at the 5 level according to a Mann Whitney test.
Extended Data Figure 6.
A. Close-view maximum-likelihood phylogenetic tree indicating the diversity of the case patient and three other long-term shedders from the local area (red, blue and purple), compared to recently published sequences from Choi et al (orange) and Avanzato et al (gold). Control patients generally showed limited diversity temporally, though the Choi et al sequences were highly divergent. Environmental samples (patient’s call bell, and patient’s mobile phone) are indicated. Tree branched have been collapsed where bootstrap support was <60.
B. Highlighter plot indicating nucleotide changes at consensus level in sequential respiratory samples compared to the consensus sequence at first diagnosis of COVID-19. Each row indicates the timepoint the sample was collected (number of days from first positive SARS-CoV-2 RT-PCR). Black dashed lines indicate the RNA-dependent RNA polymerase (RdRp) and Spike regions of the genome. There were few nucleotide substitutions between days 1-54, despite the patient receiving two courses of remdesivir. The first major changes in the spike genome occurred on day 82, following convalescent plasma given on days 63 and 65. The amino acid deletion in S1, ΔH69/V70 is indicated by the black lines. Sites: Endotracheal aspirate (ETA) or Nose/throat swabs (N+T).
Extended Data 7. In vitro infectivity and neutralisation sensitivity of Spike pseudotyped lentiviruses.
A. infection of target 293T cells expressing TMPRSS2 and ACE2 receptors using equal amounts of virus as determined by reverse transcriptase activity. Data points represent technical replicates (n=2), with mean shown with error bars representing standard deviation. Data are representative of n=2 independent experiments (n=2). B. Representative Inverse dilution plots for Spike variants against convalescent plasma units 1-3. Data points represent mean neutralisation of technical replicates and error bars represent standard error of the mean of replicates. Data are representative of two independent experiments (n=2).
Extended Data Figure 8.
A. Neutralization potency of a panel of monoclonal antibodies targeting the RBD is not impacted by Spike mutations D796H or ΔH69/V70. Lentivirus pseudotyped with SARS-CoV-2 Spike protein: WT (D614G background), D796H, ΔH69/V70, D796H+ΔH69/V70 were produced in 293T cells and used to infect target Hela cells stably expressing ACE2 in the presence of serial dilutions of indicated monoclonal antibodies. Data are means of technical replicates with error bars representing SD. Data are representative of at least two independent experiments. RBD: receptor binding domain. B. Classes of RBD binding antibodies and fold changes for Spike mutations D796H or ΔH69/V70 are indicated based Bouwer et al. Clusters II, V contain only non-neutralising mAbs, smaller neutralising mAb clusters IV (n=2) and X (n=1) were not tested. Red indicates significant fold changes.
Extended Data 9. Location of Spike mutations ΔH69/Y70 and D796H.
A. The SARS-CoV-2 spike trimer (PDB ID: 6xr8) with two protomers represented as surfaces and one protomer represented as a ribbon. The NTD is coloured in light blue, the RBD in light pink, the fusion peptide in dark pink, the HR1 domain in yellow, the CH domain in pale green, and the CD domain in brown. The location of D796 and H69 are indicated by red spheres. The loop connecting D796 to the fusion peptide is coloured magenta to improve visibility. The double grey lines provide orientation relative to the membrane. B. A close-up of the region defined by the box around H69 in panel A. H69 is highlighted in yellow. Residues containing atoms that are within 6 Å of H69 are highlighted in cyan. C. A close-up of the region defined by the box around D796 in panel A. D796 is highlighted in yellow. Residues containing atoms that are within 6 Å of D796 are highlighted in cyan. Hydrogen bonds are indicated by dashed yellow lines. Hydrophobic residues in the vicinity of D796 have been labelled. Y707 is from the neighbouring protomer. D. Global prevalence of selected spike mutations detailed in this paper. All high coverage sequences were downloaded from the GISAID database on 6th January and aligned using MAFFT; as of this date there were 298254 sequences available. The global prevalence of each of the six spike mutations W64G, ΔH69/V70, Y200H, T240I, P330S and D796H were assessed by viewing the multiple sequence alignment in AliView, sorting by the column of interest, and counting the number of mutations.
Supplementary Material
Acknowledgements
We are immensely grateful to the patient and his family. We would also like to thank the staff at CUH and the NIHR Cambridge Clinical Research Facility. We would like to thank Dr Ruthiran Kugathasan and Professor Wendy Barclay for helpful discussions and Dr Martin Curran, Dr William Hamilton, and Dr. Dominic Sparkes. We would like to thank Prof Andres Floto and Prof Ferdia Gallagher. We thank Dr James Voss for the kind gift of HeLa cells stably expressing ACE2. We would like to thank James Nathan for RBD protein and Leo James for N protein. COG-UK is supported by funding from the Medical Research Council (MRC) part of UK Research & Innovation (UKRI), the National Institute of Health Research (NIHR) and Genome Research Limited, operating as the Wellcome Sanger Institute. RKG is supported by a Wellcome Trust Senior Fellowship in Clinical Science (WT108082AIA). LEM is supported by a Medical Research Council Career Development Award (MR/R008698/1). SAK is supported by the Bill and Melinda Gates Foundation via PANGEA grant: OPP1175094. DAC is supported by a Wellcome Trust Clinical PhD Research Fellowship. CJRI acknowledges MRC funding (ref: MC_UU_00002/11). This research was supported by the National Institute for Health Research (NIHR) Cambridge Biomedical Research Centre, the Cambridge Clinical Trials Unit (CCTU) and by the UCL Coronavirus Response Fund and made possible through generous donations from UCL’s supporters, alumni, and friends (LEM). JAGB is supported by the Medical Research Council (MC_UP_1201/16). IG is a Wellcome Senior Fellow and supported by the Wellcome Trust (207498/Z/17/Z). DDP is supported by NIH GM083127.
Footnotes
Competing interests: the authors declare no competing interests
Ethics
The study was approved by the East of England – Cambridge Central Research Ethics Committee (17/EE/0025). Written informed consent was obtained from both the patient and family. Additional controls with COVID-19 were enrolled to the NIHR BioResource Centre Cambridge under ethics review board (17/EE/0025).
Author contributions
Conceived study: RKG, SAK, DAC, AS, TG, EGK
Designed experiments: RKG, SAK, DAC, LEM, JAGB, EGK, AC, NT, AC, CS, RD, RG, DDP, YM
Performed experiments: SAK, DAC, LEM, RD, CRS, AJ, IATMF, KS, TG, CJRI, BB, JS, MJvG, LGC, GBM, LK
Interpreted data: RKG, SAK, DAC, PM, LEM, JAGB, PM, SG, KS, TG, JB, KGCS, IG, CJRI, JAGB, IUL, DR, JS, BB, RAG. DDP, RD, LCG, GBM
Contributor Information
The CITIID-NIHR BioResource COVID-19 Collaboration:
Stephen Baker, Gordon Dougan, Christoph Hess, Nathalie Kingston, Paul J. Lehner, Paul A. Lyons, Nicholas J. Matheson, Willem H. Owehand, Caroline Saunders, Charlotte Summers, James E.D. Thaventhiran, Mark Toshner, Michael P. Weekes, Ashlea Bucke, Jo Calder, Laura Canna, Jason Domingo, Anne Elmer, Stewart Fuller, Julie Harris, Sarah Hewitt, Jane Kennet, Sherly Jose, Jenny Kourampa, Anne Meadows, Criona O’Brien, Jane Price, Cherry Publico, Rebecca Rastall, Carla Ribeiro, Jane Rowlands, Valentina Ruffolo, Hugo Tordesillas, Ben Bullman, Benjamin J Dunmore, Stuart Fawke, Stefan Gräf, Josh Hodgson, Christopher Huang, Kelvin Hunter, Emma Jones, Ekaterina Legchenko, Cecilia Matara, Jennifer Martin, Federica Mescia, Ciara O’Donnell, Linda Pointon, Nicole Pond, Joy Shih, Rachel Sutcliffe, Tobias Tilly, Carmen Treacy, Zhen Tong, Jennifer Wood, Marta Wylot, Laura Bergamaschi, Ariana Betancourt, Georgie Bower, Chiara Cossetti, Aloka De Sa, Madeline Epping, Stuart Fawke, Nick Gleadall, Richard Grenfell, Andrew Hinch, Oisin Huhn, Sarah Jackson, Isobel Jarvis, Daniel Lewis, Joe Marsden, Francesca Nice, Georgina Okecha, Ommar Omarjee, Marianne Perera, Nathan Richoz, Veronika Romashova, Natalia Savinykh Yarkoni, Rahul Sharma, Luca Stefanucci, Jonathan Stephens, Mateusz Strezlecki, Lori Turner, Eckart M.D.D. De Bie, Katherine Bunclark, Masa Josipovic, Michael Mackay, Federica Mescia, Alice Michael, Sabrina Rossi, Mayurun Selvan, Sarah Spencer, Cissy Yong, Ali Ansaripour, Alice Michael, Lucy Mwaura, Caroline Patterson, Gary Polwarth, Petra Polgarova, Giovanni di Stefano, Codie Fahey, Rachel Michel, Sze-How Bong, Jerome D. Coudert, Elaine Holmes, John Allison, Helen Butcher, Daniela Caputo, Debbie Clapham-Riley, Eleanor Dewhurst, Anita Furlong, Barbara Graves, Jennifer Gray, Tasmin Ivers, Mary Kasanicki, Emma Le Gresley, Rachel Linger, Sarah Meloy, Francesca Muldoon, Nigel Ovington, Sofia Papadia, Isabel Phelan, Hannah Stark, Kathleen E Stirrups, Paul Townsend, Neil Walker, and Jennifer Webster
The COVID-19 Genomics UK (COG-UK) Consortium:
Samuel C Robson, Nicholas J Loman, Thomas R Connor, Tanya Golubchik, Rocio T Martinez Nunez, Catherine Ludden, Sally Corden, Ian Johnston, David Bonsall, Colin P Smith, Ali R Awan, Giselda Bucca, M. Estee Torok, Kordo Saeed, Jacqui A Prieto, David K Jackson, William L Hamilton, Luke B Snell, Catherine Moore, Ewan M Harrison, Sonia Goncalves, Derek J Fairley, Matthew W Loose, Joanne Watkins, Rich Livett, Samuel Moses, Roberto Amato, Sam Nicholls, Matthew Bull, Darren L Smith, Jeff Barrett, David M Aanensen, Martin D Curran, Surendra Parmar, Dinesh Aggarwal, James G Shepherd, Matthew D Parker, Sharon Glaysher, Matthew Bashton, Anthony P Underwood, Nicole Pacchiarini, Katie F Loveson, Alessandro M Carabelli, Kate E Templeton, Cordelia F Langford, John Sillitoe, Thushan I de Silva, Dennis Wang, Dominic Kwiatkowski, Andrew Rambaut, Justin O’Grady, Simon Cottrell, Matthew T.G. Holden, Emma C Thomson, Husam Osman, Monique Andersson, Anoop J Chauhan, Mohammed O Hassan-Ibrahim, Mara Lawniczak, Alex Alderton, Meera Chand, Chrystala Constantinidou, Meera Unnikrishnan, Alistair C Darby, Julian A Hiscox, Steve Paterson, Inigo Martincorena, David L Robertson, Erik M Volz, Andrew J Page, Oliver G Pybus, Andrew R Bassett, Cristina V Ariani, Michael H Spencer Chapman, Kathy K Li, Rajiv N Shah, Natasha G Jesudason, Yusri Taha, Martin P McHugh, Rebecca Dewar, Aminu S Jahun, Claire McMurray, Sarojini Pandey, James P McKenna, Andrew Nelson, Gregory R Young, Clare M McCann, Scott Elliott, Hannah Lowe, Ben Temperton, Sunando Roy, Anna Price, Sara Rey, Matthew Wyles, Stefan Rooke, Sharif Shaaban, Mariateresa de Cesare, Laura Letchford, Siona Silveira, Emanuela Pelosi, Eleri Wilson-Davies, Myra Hosmillo, Áine O'Toole, Andrew R Hesketh, Richard Stark, Louis du Plessis, Chris Ruis, Helen Adams, Yann Bourgeois, Stephen L Michell, Dimitris Gramatopoulos, Jonathan Edgeworth, Judith Breuer, John A Todd, Christophe Fraser, David Buck, Michaela John, Gemma L Kay, Steve Palmer, Sharon J Peacock, David Heyburn, Danni Weldon, Esther Robinson, Alan McNally, Peter Muir, Ian B Vipond, John BoYes, Venkat Sivaprakasam, Tranprit Salluja, Samir Dervisevic, Emma J Meader, Naomi R Park, Karen Oliver, Aaron R Jeffries, Sascha Ott, Ana da Silva Filipe, David A Simpson, Chris Williams, Jane AH Masoli, Bridget A Knight, Christopher R Jones, Cherian Koshy, Amy Ash, Anna Casey, Andrew Bosworth, Liz Ratcliffe, Li Xu-McCrae, Hannah M Pymont, Stephanie Hutchings, Lisa Berry, Katie Jones, Fenella Halstead, Thomas Davis, Christopher Holmes, Miren Iturriza-Gomara, Anita O Lucaci, Paul Anthony Randell, Alison Cox, Pinglawathee Madona, Kathryn Ann Harris, Julianne Rose Brown, Tabitha W Mahungu, Dianne Irish-Tavares, Tanzina Haque, Jennifer Hart, Eric Witele, Melisa Louise Fenton, Steven Liggett, Clive Graham, Emma Swindells, Jennifer Collins, Gary Eltringham, Sharon Campbell, Patrick C McClure, Gemma Clark, Tim J Sloan, Carl Jones, Jessica Lynch, Ben Warne, Steven Leonard, Jillian Durham, Thomas Williams, Sam T Haldenby, Nathaniel Storey, Nabil-Fareed Alikhan, Nadine Holmes, Christopher Moore, Matthew Carlile, Malorie Perry, Noel Craine, Ronan A Lyons, Angela H Beckett, Salman Goudarzi, Christopher Fearn, Kate Cook, Hannah Dent, Hannah Paul, Robert Davies, Beth Blane, Sophia T Girgis, Mathew A Beale, Katherine L Bellis, Matthew J Dorman, Eleanor Drury, Leanne Kane, Sally Kay, Samantha McGuigan, Rachel Nelson, Liam Prestwood, Shavanthi Rajatileka, Rahul Batra, Rachel J Williams, Mark Kristiansen, Angie Green, Anita Justice, Adhyana I.K Mahanama, Buddhini Samaraweera, Nazreen F Hadjirin, Joshua Quick, Radoslaw Poplawski, Leanne M Kermack, Nicola Reynolds, Grant Hall, Yasmin Chaudhry, Malte L Pinckert, Iliana Georgana, Robin J Moll, Alicia Thornton, Richard Myers, Joanne Stockton, Charlotte A Williams, Wen C Yew, Alexander J Trotter, Amy Trebes, George MacIntyre-Cockett, Alec Birchley, Alexander Adams, Amy Plimmer, Bree Gatica-Wilcox, Caoimhe McKerr, Ember Hilvers, Hannah Jones, Hibo Asad, Jason Coombes, Johnathan M Evans, Laia Fina, Lauren Gilbert, Lee Graham, Michelle Cronin, Sara Kumziene-SummerhaYes, Sarah Taylor, Sophie Jones, Danielle C Groves, Peijun Zhang, Marta Gallis, Stavroula F Louka, Igor Starinskij, Chris Jackson, Marina Gourtovaia, Gerry Tonkin-Hill, Kevin Lewis, Jaime M Tovar-Corona, Keith James, Laura Baxter, Mohammad T Alam, Richard J Orton, Joseph Hughes, Sreenu Vattipally, Manon Ragonnet-Cronin, Fabricia F Nascimento, David Jorgensen, Olivia Boyd, Lily Geidelberg, Alex E Zarebski, Jayna Raghwani, Moritz UG Kraemer, Joel Southgate, Benjamin B Lindsey, Timothy M Freeman, Jon-Paul Keatley, Joshua B Singer, Leonardo de Oliveira Martins, Corin A Yeats, Khalil Abudahab, Ben EW Taylor, Mirko Menegazzo, John Danesh, Wendy Hogsden, Sahar Eldirdiri, Anita Kenyon, Jenifer Mason, Trevor I Robinson, Alison Holmes, James Price, John A Hartley, Tanya Curran, Alison E Mather, Giri Shankar, Rachel Jones, Robin Howe, Sian Morgan, Elizabeth Wastenge, Michael R Chapman, Siddharth Mookerjee, Rachael Stanley, Wendy Smith, Timothy Peto, David Eyre, Derrick Crook, Gabrielle Vernet, Christine Kitchen, Huw Gulliver, Ian Merrick, Martyn Guest, Robert Munn, Declan T Bradley, Tim Wyatt, Charlotte Beaver, Luke Foulser, Sophie Palmer, Carol M Churcher, Ellena Brooks, Kim S Smith, Katerina Galai, Georgina M McManus, Frances Bolt, Francesc Coll, Lizzie Meadows, Stephen W Attwood, Alisha Davies, Elen De Lacy, Fatima Downing, Sue Edwards, Garry P Scarlett, Sarah Jeremiah, Nikki Smith, Danielle Leek, Sushmita Sridhar, Sally Forrest, Claire Cormie, Harmeet K Gill, Joana Dias, Ellen E Higginson, Mailis Maes, Jamie Young, Michelle Wantoch, Sanger Covid Team, Dorota Jamrozy, Stephanie Lo, Minal Patel, Verity Hill, Claire M Bewshea, Sian Ellard, Cressida Auckland, Ian Harrison, Chloe Bishop, Vicki Chalker, Alex Richter, Andrew Beggs, Angus Best, Benita Percival, Jeremy Mirza, Oliver Megram, Megan Mayhew, Liam Crawford, Fiona Ashcroft, Emma Moles-Garcia, Nicola Cumley, Richard Hopes, Patawee Asamaphan, Marc O Niebel, Rory N Gunson, Amanda Bradley, Alasdair Maclean, Guy Mollett, Rachel Blacow, Paul Bird, Thomas Helmer, Karlie Fallon, Julian Tang, Antony D Hale, Louissa R Macfarlane-Smith, Katherine L Harper, Holli Carden, Nicholas W Machin, Kathryn A Jackson, Shazaad SY Ahmad, Ryan P George, Lance Turtle, Elaine O'Toole, Joanne Watts, Cassie Breen, Angela Cowell, Adela Alcolea-Medina, Themoula Charalampous, Amita Patel, Lisa J Levett, Judith Heaney, Aileen Rowan, Graham P Taylor, Divya Shah, Laura Atkinson, Jack CD Lee, Adam P Westhorpe, Riaz Jannoo, Helen L Lowe, Angeliki Karamani, Leah Ensell, Wendy Chatterton, Monika Pusok, Ashok Dadrah, Amanda Symmonds, Graciela Sluga, Zoltan Molnar, Paul Baker, Stephen Bonner, Sarah Essex, Edward Barton, Debra Padgett, Garren Scott, Jane Greenaway, Brendan AI Payne, Shirelle Burton-Fanning, Sheila Waugh, Veena Raviprakash, Nicola Sheriff, Victoria Blakey, Lesley-Anne Williams, Jonathan Moore, Susanne Stonehouse, Louise Smith, Rose K Davidson, Luke Bedford, Lindsay Coupland, Victoria Wright, Joseph G Chappell, Theocharis Tsoleridis, Jonathan Ball, Manjinder Khakh, Vicki M Fleming, Michelle M Lister, Hannah C Howson-Wells, Louise Berry, Tim Boswell, Amelia Joseph, Iona Willingham, Nichola Duckworth, Sarah Walsh, Emma Wise, Nathan Moore, Matilde Mori, Nick Cortes, Stephen Kidd, Rebecca Williams, Laura Gifford, Kelly Bicknell, Sarah Wyllie, Allyson Lloyd, Robert Impey, Cassandra S Malone, Benjamin J Cogger, Nick Levene, Lynn Monaghan, Alexander J Keeley, David G Partridge, Mohammad Raza, Cariad Evans, Kate Johnson, Emma Betteridge, Ben W Farr, Scott Goodwin, Michael A Quail, Carol Scott, Lesley Shirley, Scott AJ Thurston, Diana Rajan, Iraad F Bronner, Louise Aigrain, Nicholas M Redshaw, Stefanie V Lensing, Shane McCarthy, Alex Makunin, Carlos E Balcazar, Michael D Gallagher, Kathleen A Williamson, Thomas D Stanton, Michelle L Michelsen, Joanna Warwick-Dugdale, Robin Manley, Audrey Farbos, James W Harrison, Christine M Sambles, David J Studholme, Angie Lackenby, Tamyo Mbisa, Steven Platt, Shahjahan Miah, David Bibby, Carmen Manso, Jonathan Hubb, Gavin Dabrera, Mary Ramsay, Daniel Bradshaw, Ulf Schaefer, Natalie Groves, Eileen Gallagher, David Lee, David Williams, Nicholas Ellaby, Hassan Hartman, Nikos Manesis, Vineet Patel, Juan Ledesma, Katherine A Twohig, Elias Allara, Clare Pearson, Jeffrey K. J. Cheng, Hannah E Bridgewater, Lucy R Frost, Grace Taylor-Joyce, Paul E Brown, Lily Tong, Alice Broos, Daniel Mair, Jenna Nichols, Stephen N Carmichael, Katherine L Smollett, Kyriaki Nomikou, Elihu Aranday-Cortes, Natasha Johnson, Seema Nickbakhsh, Edith E Vamos, Margaret Hughes, Lucille Rainbow, Richard Eccles, Charlotte Nelson, Mark Whitehead, Richard Gregory, Matthew Gemmell, Claudia Wierzbicki, Hermione J Webster, Chloe L Fisher, Adrian W Signell, Gilberto Betancor, Harry D Wilson, Gaia Nebbia, Flavia Flaviani, Alberto C Cerda, Tammy V Merrill, Rebekah E Wilson, Marius Cotic, Nadua Bayzid, Thomas Thompson, Erwan Acheson, Steven Rushton, Sarah O'Brien, David J Baker, Steven Rudder, Alp Aydin, Fei Sang, Johnny Debebe, Sarah Francois, Tetyana I Vasylyeva, Marina Escalera Zamudio, Bernardo Gutierrez, Angela Marchbank, Joshua Maksimovic, Karla Spellman, Kathryn McCluggage, Mari Morgan, Robert Beer, Safiah Afifi, Trudy Workman, William Fuller, Catherine Bresner, Adrienn Angyal, Luke R Green, Paul J Parsons, Rachel M Tucker, Rebecca Brown, Max Whiteley, James Bonfield, Christoph Puethe, Andrew Whitwham, Jennifier Liddle, Will Rowe, Igor Siveroni, Thanh Le-Viet, Amy Gaskin, and Rob Johnson
Data Availability
Long-read sequencing data that support the findings of this study have been deposited in the NCBI SRA database with the accession codes SAMN16976824 - SAMN16976846 under BioProject PRJNA682013 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA682013). Short reads and data used to construct figures were deposited at https://github.com/Steven-Kemp/sequence_files. All data are also available from the corresponding author.
Code Availability
The SAMFIRE package Version 1.06 was used for filtering and calling variants from the Illumina data. It is available at https://github.com/cjri/samfire/ for review. Additional code was used to validate the variant frequencies and can be found at https://github.com/PollockLaboratory/AnCovMulti.
References
- 1.Hoffmann M, et al. SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor. Cell. 2020;181:271–280 e278. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kim KW, et al. Respiratory viral co-infections among SARS-CoV-2 cases confirmed by virome capture sequencing. 2020 doi: 10.1038/s41598-021-83642-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bull RA, et al. Analytical validity of nanopore sequencing for rapid SARS-CoV-2 genome analysis. Nat Commun. 2020;11:6272. doi: 10.1038/s41467-020-20075-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choi B, et al. Persistence and Evolution of SARS-CoV-2 in an Immunocompromised Host. The New England journal of medicine. 2020;383:2291–2293. doi: 10.1056/NEJMc2031364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Avanzato VA, et al. Case Study: Prolonged infectious SARS-CoV-2 shedding from an asymptomatic immunocompromised cancer patient. Cell. 2020 doi: 10.1016/j.cell.2020.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Starr TN, et al. Deep Mutational Scanning of SARS-CoV-2 Receptor Binding Domain Reveals Constraints on Folding and ACE2 Binding. Cell. 2020;182:1295–1310 e1220. doi: 10.1016/j.cell.2020.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rambaut ALN, Pybus O, Barclay W, Carabelli AC, Connor T, Peacock T, Robertson DL, Volz E, COVID-19 Genomics Consortium UK (CoG-UK) Preliminary genomic characterisation of an emergent SARS-CoV-2 lineage in the UK defined by a novel set of spike mutations. 2020 < https://virological.org/t/preliminary-genomic-characterisation-of-an-emergent-sars-cov-2-lineage-in-the-uk-defined-by-a-novel-set-of-spike-mutations/563>. [Google Scholar]
- 8.Schmidt F, et al. Measuring SARS-CoV-2 neutralizing antibody activity using pseudotyped and chimeric viruses. bioRxiv. 2020 doi: 10.1101/2020.06.08.140871. 2020.2006.2008.140871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brouwer PJM, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. 2020;369:643–650. doi: 10.1126/science.abc5902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zussman ME, Bagby M, Benson DW, Gupta R, Hirsch R. Pulmonary vascular resistance in repaired congenital diaphragmatic hernia vs. age-matched controls. Pediatr Res. 2012;71:697–700. doi: 10.1038/pr.2012.16. [DOI] [PubMed] [Google Scholar]
- 11.Cai Y, et al. Distinct conformational states of SARS-CoV-2 spike protein. Science. 2020 doi: 10.1126/science.abd4251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou T, et al. Cryo-EM Structures of SARS-CoV-2 Spike without and with ACE2 Reveal a pH-Dependent Switch to Mediate Endosomal Positioning of Receptor-Binding Domains. Cell Host & Microbe. 2020;28:867–879.e865. doi: 10.1016/j.chom.2020.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Robbiani DF, et al. Convergent antibody responses to SARS-CoV-2 in convalescent individuals. Nature. 2020;584:437–442. doi: 10.1038/s41586-020-2456-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barnes CO, et al. Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies. Cell. 2020;182:828–842 e816. doi: 10.1016/j.cell.2020.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shrock E, et al. Viral epitope profiling of COVID-19 patients reveals cross-reactivity and correlates of severity. Science. 2020 doi: 10.1126/science.abd4250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sobel Leonard A, et al. The effective rate of influenza reassortment is limited during human infection. PLoS Pathog. 2017;13:e1006203. doi: 10.1371/journal.ppat.1006203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Richard M, Herfst S, Tao H, Jacobs NT, Lowen AC. Influenza A Virus Reassortment Is Limited by Anatomical Compartmentalization following Coinfection via Distinct Routes. J Virol. 2018;92 doi: 10.1128/JVI.02063-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kemp S, et al. Recurrent emergence and transmission of a SARS-CoV-2 Spike deletion H69/V70. bioRxiv. 2021 doi: 10.1101/2020.12.14.422555. 2020.2012.2014.422555. [DOI] [Google Scholar]
- 19.CDC. Discontinuation of Transmission-Based Precautions and Disposition of Patients with COVID-19 in Healthcare Settings (Interim Guidance) 2020 < https://www.cdc.gov/coronavirus/2019-ncov/hcp/disposition-hospitalized-patients.html>.
- 20.Boshier FAT, et al. Remdesivir induced viral RNA and subgenomic RNA suppression, and evolution of viral variants in SARS-CoV-2 infected patients. medRxiv. 2020 doi: 10.1101/2020.11.18.20230599. 2020.2011.2018.20230599. [DOI] [Google Scholar]
- 21.Simonovich VA, et al. A Randomized Trial of Convalescent Plasma in Covid-19 Severe Pneumonia. N Engl J Med. 2020 doi: 10.1056/NEJMoa2031304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meredith LW, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. The Lancet Infectious Diseases. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collier DA, et al. Point of Care Nucleic Acid Testing for SARS-CoV-2 in Hospitalized Patients: A Clinical Validation Trial and Implementation Study. Cell Rep Med. 2020 doi: 10.1016/j.xcrm.2020.100062. 100062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loman N, Rowe W, Rambaut A. 2020 (v1) [Google Scholar]
- 25.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet journal. 2011;17:10–12. [Google Scholar]
- 26.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics (Oxford, England) 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jordan MR, et al. Comparison of standard PCR/cloning to single genome sequencing for analysis of HIV-1 populations. J Virol Methods. 2010;168:114–120. doi: 10.1016/j.jviromet.2010.04.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palmer S, et al. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. Journal of clinical microbiology. 2005;43:406–413. doi: 10.1128/JCM.43.1.406-413.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Keele BF, et al. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:7552–7557. doi: 10.1073/pnas.0802203105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shu Y, McCauley J. GISAID: Global initiative on sharing all influenza data - from vision to reality. Euro surveillance: bulletin Europeen sur les maladies transmissibles = European communicable disease bulletin. 2017;22:30494. doi: 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Katoh K, Standley DM. MAFFT Multiple Sequence Alignment Software Version-7-Improvements in Performance and Usability. Molecular Biology and Evolution. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rambaut A, et al. A dynamic nomenclature proposal for SARS-CoV-2 lineages to assist genomic epidemiology. Nature Microbiology. 2020;5:1403–1407. doi: 10.1038/s41564-020-0770-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Minh BQ, et al. IQ-TREE 2: New Models and Efficient Methods for Phylogenetic Inference in the Genomic Era. Molecular Biology and Evolution. 2020;37:1530–1534. doi: 10.1093/molbev/msaa015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. ModelFinder: fast model selection for accurate phylogenetic estimates. Nature Methods. 2017;14:587–589. doi: 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Minh BQ, Nguyen MA, von Haeseler A. Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol. 2013;30:1188–1195. doi: 10.1093/molbev/mst024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Illingworth CJ. SAMFIRE: multi-locus variant calling for time-resolved sequence data. Bioinformatics. 2016;32:2208–2209. doi: 10.1093/bioinformatics/btw205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lumby CK, Zhao L, Breuer J, Illingworth CJ. A large effective population size for established within-host influenza virus infection. Elife. 2020;9 doi: 10.7554/eLife.56915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Martin DP, Murrell B, Golden M, Khoosal A, Muhire B. RDP4: Detection and analysis of recombination patterns in virus genomes. Virus evolution. 2015;1 doi: 10.1093/ve/vev003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Didelot X, Wilson DJ. ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput Biol. 2015;11:e1004041. doi: 10.1371/journal.pcbi.1004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wrobel AG, et al. SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nature Structural & Molecular Biology. 2020;27:763–767. doi: 10.1038/s41594-020-0468-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gregson J, et al. HIV-1 viral load is elevated in individuals with reverse transcriptase mutation M184V/I during virological failure of first line antiretroviral therapy and is associated with compensatory mutation L74I. Journal of Infectious Diseases. 2019 doi: 10.1093/infdis/jiz631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naldini L, Blomer U, Gage FH, Trono D, Verma IM. Efficient transfer, integration, and sustained long-term expression of the transgene in adult rat brains injected with a lentiviral vector. Proc Natl Acad Sci U S A. 1996;93:11382–11388. doi: 10.1073/pnas.93.21.11382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gupta RK, et al. Full length HIV-1 gag determines protease inhibitor susceptibility within in vitro assays. AIDS. 2010;24:1651. doi: 10.1097/qad.0b013e3283398216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vermeire J, et al. Quantification of reverse transcriptase activity by real-time PCR as a fast and accurate method for titration of HIV, lenti- and retroviral vectors. PloS one. 2012;7:e50859–e50859. doi: 10.1371/journal.pone.0050859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mlcochova P, et al. Combined point of care nucleic acid and antibody testing for SARS-CoV-2 following emergence of D614G Spike Variant. Cell Rep Med. 2020 doi: 10.1016/j.xcrm.2020.100099. 100099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seow J, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020;5:1598–1607. doi: 10.1038/s41564-020-00813-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Long-read sequencing data that support the findings of this study have been deposited in the NCBI SRA database with the accession codes SAMN16976824 - SAMN16976846 under BioProject PRJNA682013 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA682013). Short reads and data used to construct figures were deposited at https://github.com/Steven-Kemp/sequence_files. All data are also available from the corresponding author.
The SAMFIRE package Version 1.06 was used for filtering and calling variants from the Illumina data. It is available at https://github.com/cjri/samfire/ for review. Additional code was used to validate the variant frequencies and can be found at https://github.com/PollockLaboratory/AnCovMulti.