Abstract
Highly selective gene expression is a key requirement for antigenic variation in several pathogens, allowing evasion of host immune responses and maintenance of persistent infections 1. African trypanosomes, parasites that cause lethal diseases in humans and livestock, employ an antigenic variation mechanism that involves monogenic antigen expression from a pool of >2600 antigen-coding genes 2. In other eukaryotes, the expression of individual genes can be enhanced by mechanisms involving the juxtaposition of otherwise distal chromosomal loci in the three-dimensional nuclear space 3–5. However, trypanosomes lack classical enhancer sequences or regulated transcription initiation 6,7. In this context, it has remained unclear how genome architecture contributes to monogenic transcription elongation and transcript processing. Here, we show that the single expressed antigen coding gene displays a specific inter-chromosomal interaction with a major mRNA splicing locus. Chromosome conformation capture (Hi-C) revealed a dynamic reconfiguration of this inter-chromosomal interaction upon activation of another antigen. Super-resolution microscopy showed the interaction to be heritable and splicing dependent. We find a specific association of the two genomic loci with the antigen exclusion complex, whereby VEX1 occupied the splicing locus and VEX2 the antigen coding locus. Following VEX2 depletion, loss of monogenic antigen expression was accompanied by increased interactions between previously silent antigen genes and the splicing locus. Our results reveal a mechanism to ensure monogenic expression, where antigen transcription and mRNA splicing occur in a specific nuclear compartment. These findings suggest a new means of post-transcriptional gene regulation.
Keywords: 3D genome architecture, RNA maturation, antigenic variation, monoallelic, Trypanosoma brucei.
Monogenic expression, the expression of a single gene from a large gene family, is essential for several important biological processes. One of the most striking examples of such regulation is the expression of a single odorant receptor from more than 1400 genes in mammalian olfactory sensory neurons 3. Likewise, monogenic expression is a key feature of antigenic variation, an immune evasion strategy used by pathogens such as Plasmodium falciparum or Trypanosoma brucei. Antigenic variation refers to the capacity of an infecting organism to systematically alter the identity of proteins displayed to the host immune system 1. How pathogens ensure the exclusive expression of only one antigen from a large pool of antigen coding genes remains one of the most intriguing questions in infection biology. Through a combination of chromosome conformation capture and imaging techniques, here we investigate the role of genome architecture and show that the spatial integration of transcription and mRNA splicing in a dedicated sub-nuclear compartment underpins monogenic antigen expression in trypanosomes.
In T. brucei, a unicellular parasite responsible for lethal and debilitating diseases in humans and animals, 10 million copies of a single variant surface glycoprotein (VSG) isoform are exposed on the surface of the parasite. The exclusive expression of only one VSG gene per cell and the periodic switching of the expressed VSG gene allow the parasite to evade the host immune system and to maintain persistent infections 2,8. While the T. brucei genome encodes for >2600 VSG isoforms, in the bloodstream of the mammalian host, a VSG gene can only be transcribed when located in one of ~15 VSG expression sites. Those bloodstream expression sites are polycistronic transcription units located adjacent to telomeres on different chromosomes. Each bloodstream expression site contains an RNA polymerase I (Pol I) promoter, followed by several expression site associated genes and a single VSG gene 7.
Notably, Pol I transcription initiates at all VSG expression site promoters, but transcription elongation and transcript processing are highly selective and limited to just one expression site at a time 9,10. As a result, the single active VSG gene is expressed as the most abundant mRNA and protein in the cell; 5-10% of the total in each case. Why transcription is aborted at all but one expression site is not known. In trypanosomes, mRNA maturation involves trans-splicing, a process that adds a common spliced leader sequence to each pre-mRNA and is coupled to polyadenylation 11. In addition, the proximity of individual genes to nuclear condensates composed of splicing factors has recently been proposed to play a role in gene expression regulation in mammals 12,13. Thus, regulated access to RNA maturation compartments may represent an evolutionary conserved strategy for gene expression control.
One mechanism to ensure monogenic expression is the juxtaposition of otherwise distal chromosomal loci in the three-dimensional nuclear space. In particular, specific interactions between promoter and enhancer sequence elements can ensure the selective regulation of individual genes. Although classic enhancer structures appear to be absent in many unicellular eukaryotes such as trypanosomes, several observations suggest that a specific genome organization is required for monogenic VSG expression. The single active VSG gene is transcribed in an extranucleolar Pol I compartment known as the expression site body 14. In those very rare cases (<10-8) where two VSG genes are simultaneously active, both co-localize at the expression site body 15,16. In addition, the transcribed chromosome core regions and the subtelomeric regions coding for the large reservoir of silent VSG genes, appear to fold into structurally distinct compartments 7, similar to active A and silent B compartments described in mammalian cells 5. While the nature of the expression site body has remained enigmatic, a protein complex specifically associated with the active VSG gene was identified recently. VSG-exclusion 1 (VEX1) emerged from a genetic screen for allelic exclusion regulators 17 while VEX2 was affinity-purified in association with VEX1 18. The bipartite VEX protein complex maintains mutually exclusive VSG expression 18 but it remains unclear how these proteins exert their function. In this study we aimed to identify the mechanism that connects RNA maturation, genome architecture and the VEX complex to ensure monogenic antigen expression.
Given the well-characterized role of promoter-enhancer interactions in the selective regulation of genes, we set out to identify specific DNA-DNA interactions with a regulatory role in monogenic VSG expression. To this end we used a T. brucei culture homogenously expressing a single VSG gene for chromosome conformation capture (Hi-C) analysis. In addition, we employed the mHi-C analysis pipeline, which allowed us to retain many multi-mapping reads and greatly increased the read coverage across repetitive regions of the genome 19.
In order to visualize specific interaction patterns of loci of interest (viewpoints) in the Hi-C dataset, we applied a virtual 4C analysis pipeline to extract genome-wide interaction profiles for chosen viewpoints. To identify VSG gene specific interaction patterns, we chose the active and several inactive VSG genes located in expression sites as viewpoints and plotted the extracted virtual 4C interaction data onto the genome. As expected, we observed a distance-dependent decay of intra-chromosomal interactions between each viewpoint and its upstream and downstream genomic region (Fig. 1a; Extended Data Fig. 1a). For the VSG-2 gene located on chr. 6 in expression site 1 (Fig. 1a, top panel), the distance-dependent decay was not as characteristic as for other viewpoints. As we have published previously 7, this expression site is separated from the core region of chr. 6 by a centromere, which can serve as a boundary element, inhibiting frequent interactions between the two chromosomal arms.
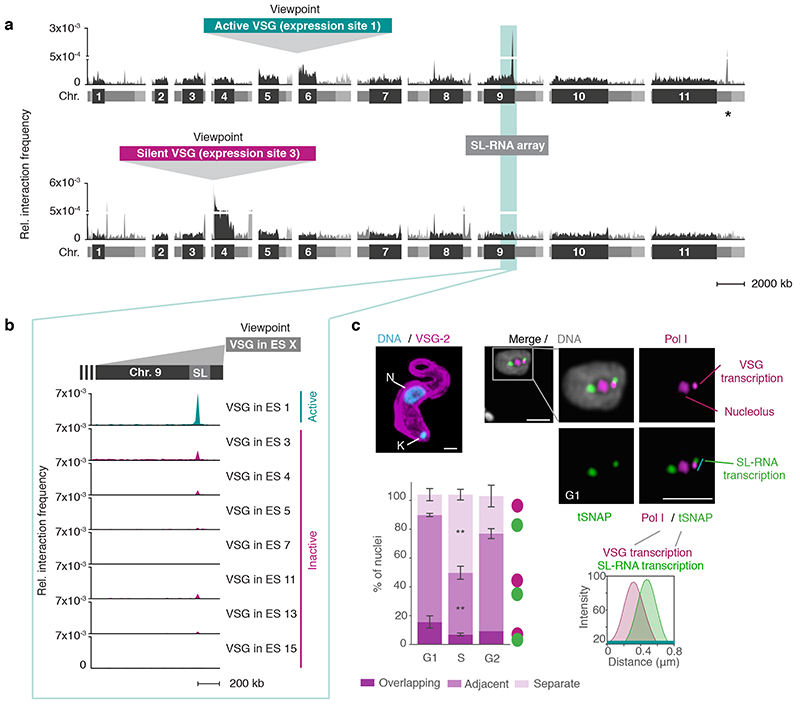
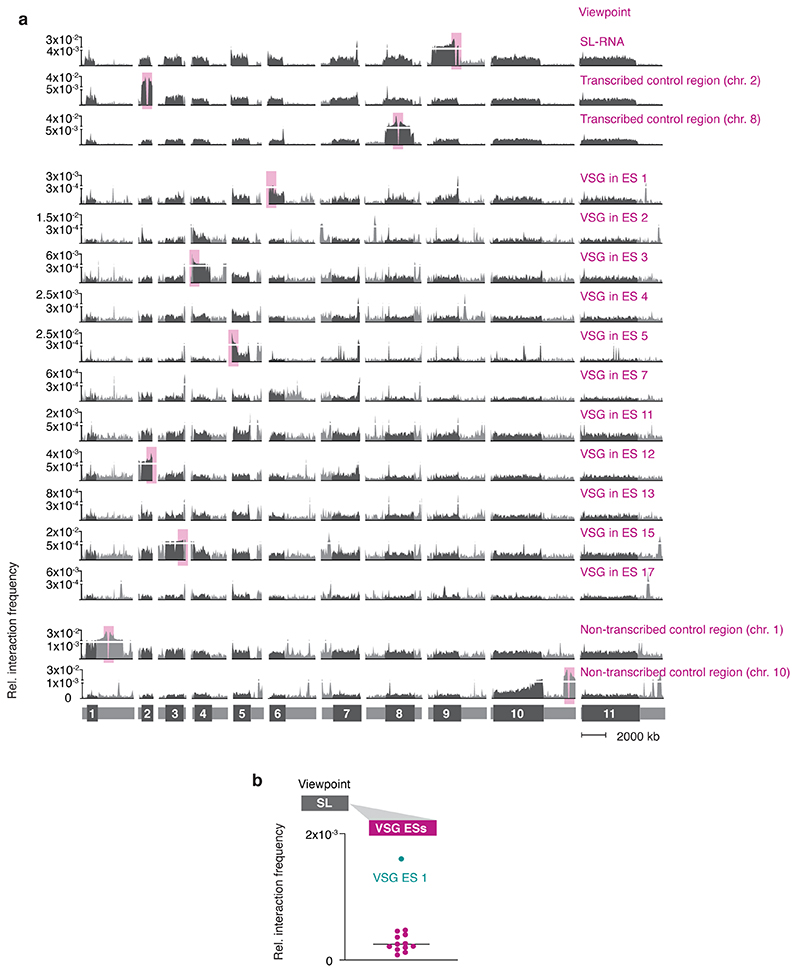
Fig. 1. The active VSG expression site (ES) stably interacts with the spliced leader RNA (SL) array.
a, Hi-C (virtual 4C) analysis, viewpoints: active VSG gene in ES 1 (VSG-2, top panel) and silent VSG gene in ES 3 (VSG-6, bottom panel). Relative interaction frequencies between the viewpoint and the 11 megabase chromosomes are shown. Chromosome cores, dark grey; subtelomeric regions, light grey. The hemizygous subtelomeric regions are displayed in the following order: 5′(haplotype A)–5′(haplotype B)–diploid chromosome core–3′(haplotype A)–3′(haplotype B). Bin size 50 kb. * marks the centromere on chr. 11. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 2. b, Virtual 4C analysis, viewpoints: active VSG gene in ES 1 and inactive VSG genes in ES 3, 4, 5, 7, 11, 13 and 15. Relative interaction frequencies between the viewpoint and the SL-RNA locus on the right arm of chr. 9 is plotted. Bin size 20 kb. The analyses in a-b are based on Hi-C experiments with VSG-2 expressing cells (n=2, average interaction frequencies are shown). c, Immunofluorescence-based colocalization studies of tSNAPmyc (SL-RNA locus marker – SL-RNA transcription compartment) and a nucleolar and active-VSG transcription compartment marker (Pol I, largest subunit) using super resolution microscopy. The stacked bar graph depicts proportions of G1, S phase or G2 nuclei with overlapping, adjacent or separate signals for the SL-RNA and VSG transcription compartments (these categories were defined by thresholded Pearson’s correlation coefficients – see methods). A two-tailed paired Student’s t-test was used to compare S or G2 nuclei versus G1 nuclei; statistical significance is highlighted where applicable: **, p < 0.01. Values are averages of three independent experiments and representative of two independent biological replicates (≥100 nuclei); error bars, SD. Detailed n and p values are provided in Source Data Fig. 1. DNA was counter-stained with DAPI; the images correspond to maximal 3D projections of stacks of 0.1 μm slices; scale bars 2 μm. N, nucleus; K, kinetoplast (mitochondrial genome).
Strikingly, we found the active VSG-2 gene located on chr. 6 in expression site 1 to very frequently interact with a distinct locus on chr. 9 (Fig. 1a-b). Levels of interaction frequency were higher than intra-chromosomal interactions of VSG-2 with its genomic location on chr. 6, pointing to a strong and stable inter-chromosomal interaction. The locus on chr. 9 interacting with the active VSG gene is the SL-RNA array, a genomic locus essential for RNA maturation. This locus contains a cluster of ~150–200 tandemly repeated genes encoding the spliced leader RNA (SL-RNA). SL-RNA is an RNA Pol II-transcribed ncRNA that is trans-spliced to the 5′-end of all trypanosome mRNAs, conferring the 5′-cap structure required for RNA maturation, export and translation 11. Conversely, just like arbitrarily chosen control regions, VSG genes residing in inactive expression sites interacted less frequently or at background levels with the SL-RNA locus (Fig. 1a-b; Extended Data Fig. 1a). In agreement with these observations, when we chose the SL-RNA locus as viewpoint, we found it to interact more frequently with the active VSG expression site than with any inactive VSG expression site (Extended Data Fig. 1b). Looking for further genomic loci that made inter-chromosomal interactions with the VSG-2 gene of at least 20% of the VSG-2 – SL-RNA interaction frequency, we identified a second locus: the centromere of chr. 11 interacted with VSG-2 at 37% of the frequency observed for the VSG-2 – SL-RNA interaction (Fig. 1a, top panel). Since the VSG-2 gene is located next to the centromere of chr. 6, we suspect these interactions to be a consequence of previously observed centromere – centromere interactions, and not to be related to the active expression of VSG-2. Thus, the Hi-C analysis revealed a strong and selective interaction between the Pol I-transcribed active VSG gene and the Pol II-transcribed SL-RNA locus located on a different chromosome.
To visualize the spatial proximity between the active VSG gene and the SL-RNA locus at the level of individual cells and with an independent assay, we performed super-resolution immunofluorescence microscopy assays (IFAs). The site of VSG transcription is characterized by an extra-nucleolar accumulation of RNA Pol I 14. The site of Pol II-transcribed SL-RNA is marked by an accumulation of the small nuclear RNA-activating protein complex (tSNAPc), an RNA Pol II promoter-binding transcription factor 20. Both the VSG transcription and the SL-RNA transcription compartments appear to have diameters of approx. 300 nm, within a T. brucei nucleus with a diameter of approx. 2 μm. Super-resolution microscopy revealed one VSG transcription compartment, reflecting the hemizygous active subtelomeric VSG-2, and two separate SL-RNA transcription compartments, reflecting the diploid SL-RNA arrays located in the core of chr. 9 (Fig. 1c; Extended Data Fig. 2). By scoring nuclei for overlapping, adjacent or separate VSG and SL-RNA transcription compartments, we found that one of the SL-RNA transcription compartments was adjacent to the VSG transcription compartment in the majority of cells (Fig. 1c). The SL-RNA transcription compartments were always extra-nucleolar (Extended Data Fig. 2d), but those adjacent to the VSG transcription compartment were significantly less intense and significantly closer to the nucleolus (Extended Data Fig. 2e-f). Further, during DNA replication in S phase, the VSG and SL-RNA transcription compartments were detected in separate locations in >50% of nuclei. Therefore, throughout this study, IFAs were subsequently performed in G1 cells, unless indicated otherwise. Notably, VSG and SL-RNA transcription compartments were once again adjacent in most G2 nuclei (Fig. 1c; Extended Data Fig. 2b-c), indicating that the interaction is resolved during S phase and successfully re-established after replication. Taken together, IFAs supported the findings made by Hi-C, suggesting that the Pol I VSG transcription compartment interacts with one of the Pol II SL-RNA transcription compartments.
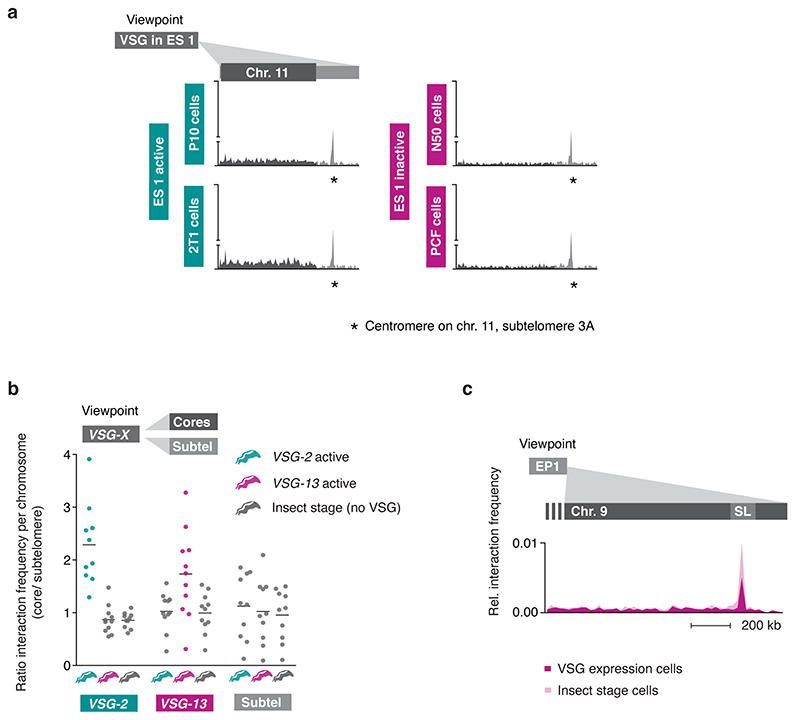
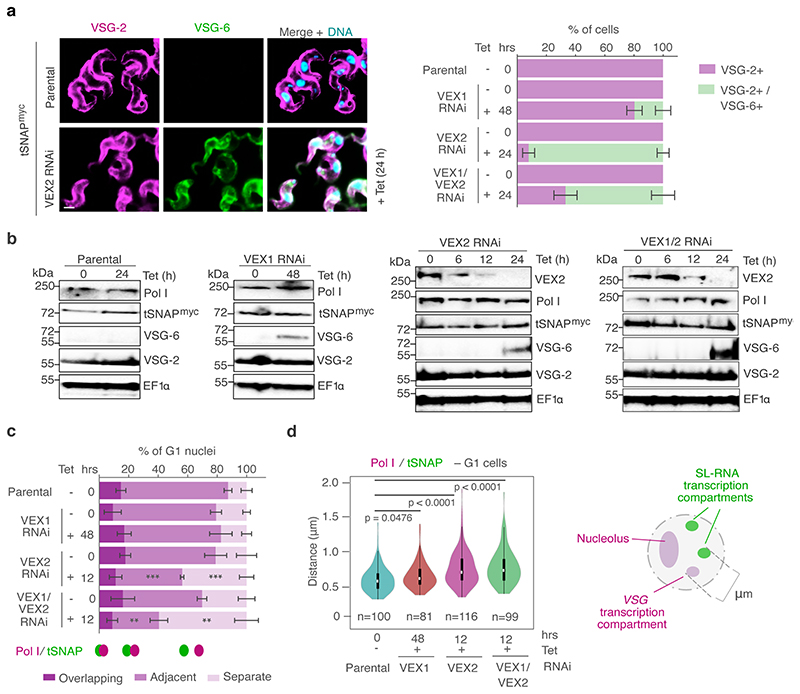
To determine whether the interaction with the SL-RNA transcription compartment is specific for the active VSG gene, and therefore changes following a VSG switching event, we performed Hi-C experiments using an isogenic T. brucei cell line expressing a different VSG isoform, VSG-13 (Fig. 2a) 21. VSG-13 resides within expression site 17, which is located on one of five intermediate-sized chromosomes. The presence of co-transcribed resistance markers upstream of VSG-2 in expression site 1 and VSG-13 in expression site 17 allowed us to specifically select for parasites expressing VSG-13 through drug selection (Fig. 2a). The exclusive activity of expression site 1 or 17 was verified by RNA-seq (Fig. 2a).
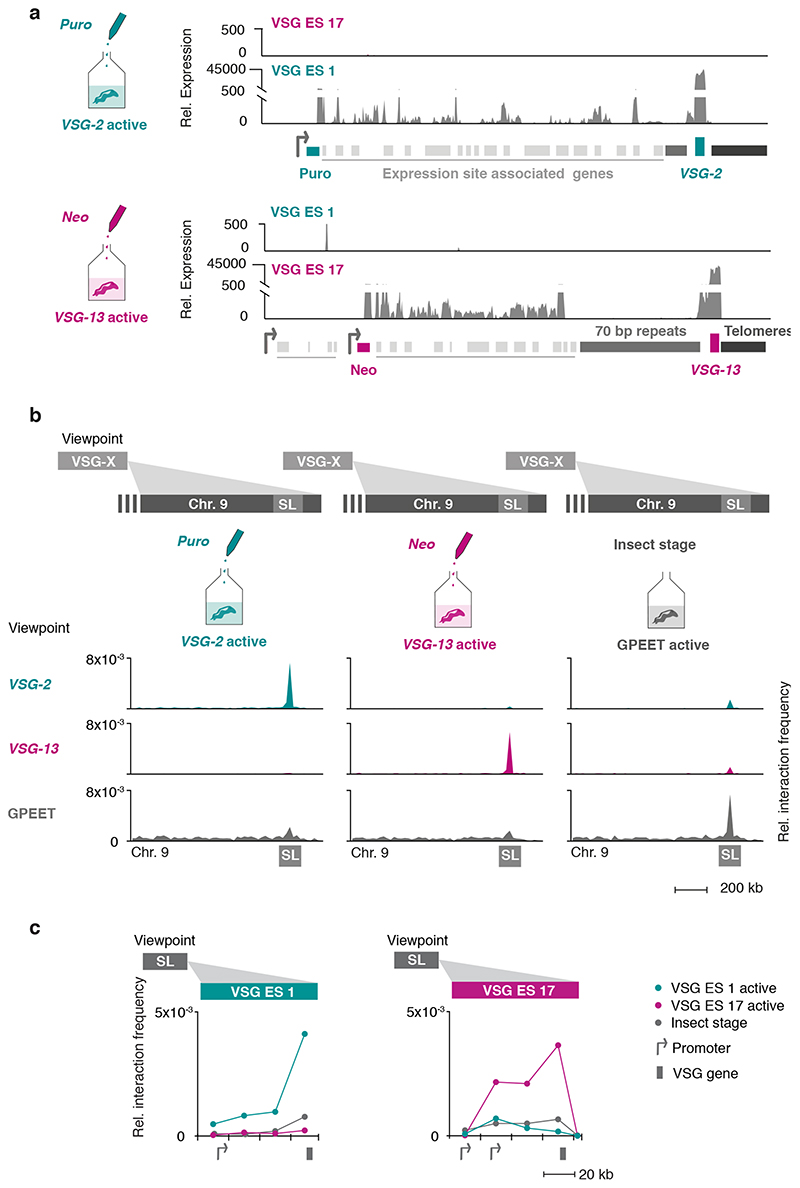
Fig. 2. The interaction between the active VSG gene and the SL-RNA locus is dynamic and changes during a switch in VSG expression.
a, Transcriptome analyses of isogenic cell lines after selection with puromycin (active: VSG-2), top panel, or neomycin (active: VSG-13), bottom panel. For each condition, the average of three biological replicates is plotted. b, Hi-C (virtual 4C) analysis, viewpoints: VSG-2, VSG-13, GPEET gene array (chr. 6). Relative interaction frequencies between the viewpoint and the SL-RNA locus on chr. 9 are plotted. Bin size 20 kb. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 2. c, Hi-C (virtual 4C) analysis, viewpoint: SL-RNA locus (chr. 9). Relative interaction frequencies between the viewpoint and VSG ES 1 or ES 17 are shown. Bin size 20 kb. The analyses in b and c are based on Hi-C experiments with cells expressing VSG-2, VSG-13 or insect stage cells expressing procyclin genes (the average of n=2 biologically independent replicates is shown).
Hi-C analysis revealed that VSG-2 - SL-RNA interactions dropped 20-fold to average inter-chromosomal interaction levels in parasites expressing VSG-13, while interactions between the newly activated VSG-13 and the SL-RNA locus increased 36-fold (Fig. 2b, left and middle panel). As suspected, VSG-2 – centromere (chr. 11) interactions remained unchanged after inactivation of VSG-2 (Extended Data Fig 3a), suggesting that this is indeed a consequence of centromere – centromere interaction. Further, we found that upon activation of each expression site, the bin harboring the respective active VSG gene displayed the strongest interaction with the SL-RNA locus, suggesting that the VSG gene itself, not its promoter, interacts with the splicing locus (Fig. 2c). In addition, we detected decreased interaction of the inactivated VSG-2 gene with the transcribed chromosome cores, while the activated VSG-13 gene displayed increased interaction with chromosome cores (Extended Data Fig. 3b). This observation indicates that activation of a VSG gene is intimately linked to a transition from a silent to an actively transcribed compartment within the nucleus. Conversely, VSG gene inactivation results in a transition from an active to an inactive nuclear compartment.
To further explore the relationship between SL-RNA interaction frequency and gene expression, we performed Hi-C analyses using insect stage parasites that do not express any VSGs, but instead express a different group of surface antigens called procyclin genes. Confirming the importance of the SL-RNA interaction, the GPEET and EP1 procyclin genes displayed an increased interaction frequency with the SL-RNA locus upon activation in insect stage cells (Fig. 2b, right panel; Extended Data Fig. 3c). Like VSG genes, procyclin genes are transcribed by RNA Pol I at high levels and require efficient trans-splicing for mRNA maturation. Thus, Hi-C analyses of T. brucei cell lines expressing different antigens indicate that interactions with the SL-RNA locus are dynamic and specific for actively transcribed antigen-coding genes.
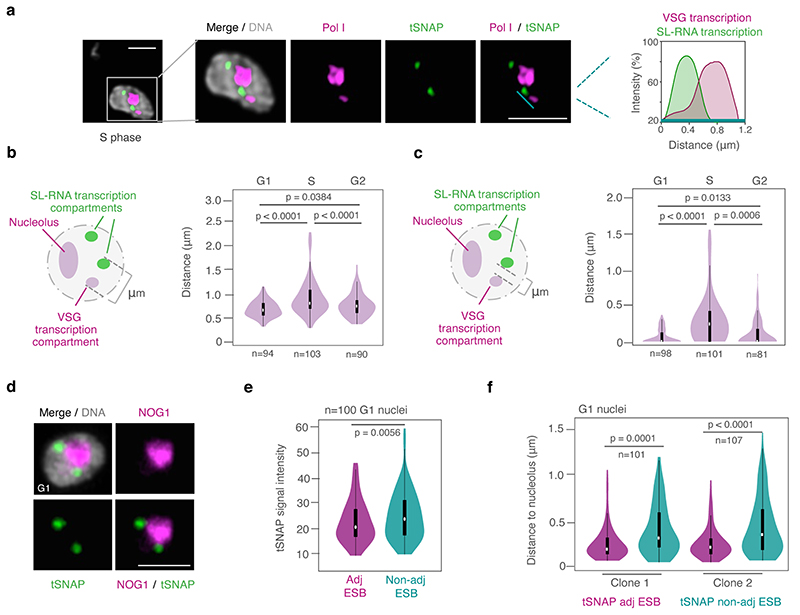
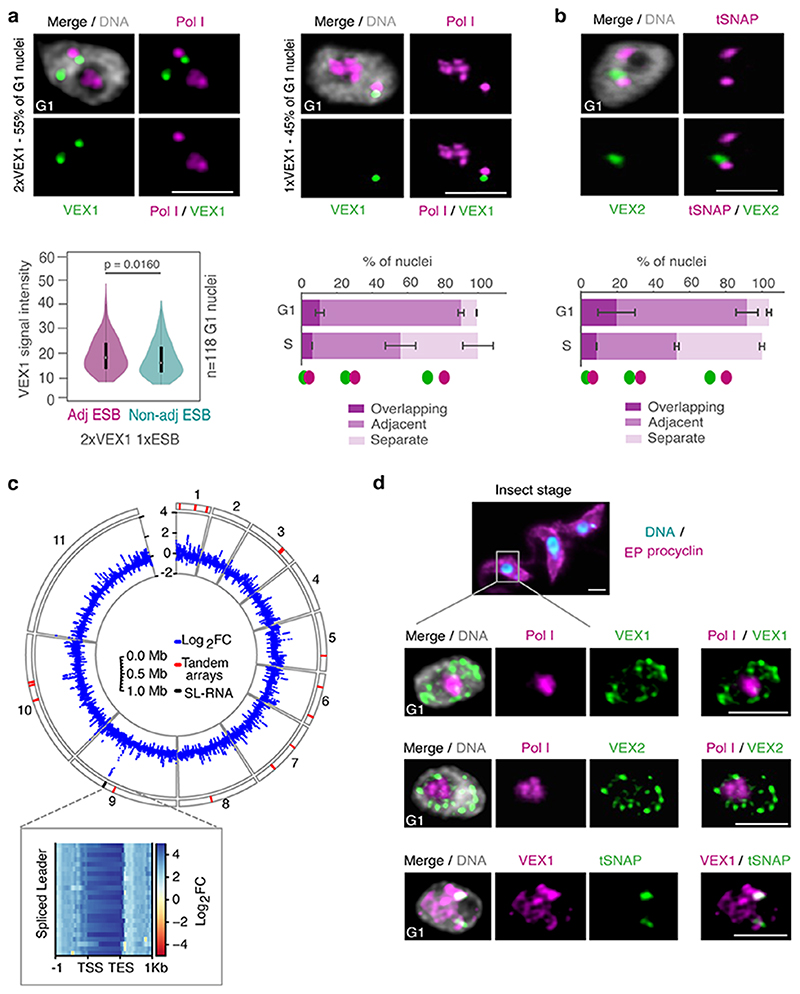
Previously, we had shown that the bipartite VEX-complex is associated with the actively transcribed VSG gene and maintains monogenic VSG expression but that by microscopy VEX1 and VEX2 signals only partially overlap each other 18. Given a similar juxtaposition of the VSG transcription and the SL-RNA transcription compartments, we sought to investigate the relationship between the VEX complex and these transcription compartments in more detail. Using optimized immunofluorescence staining protocols and super-resolution microscopy, we were able to detect two VEX1 foci in the majority of G1 nuclei (55 +/-4 %); one VEX1 focus was detected in the remainder. These VEX1 signals specifically co-localized with the SL-RNA transcription compartments (Fig. 3a). In contrast, the majority of G1 cells (97 +/-1 %) only had one VEX2 focus, which specifically co-localized with the VSG transcription compartment (Fig. 3b). As expected, one VEX1 focus was adjacent to the VSG transcription compartment (Extended Data Fig. 4a) while the VEX2 focus was adjacent to one of the two SL-RNA transcription compartments (Extended Data Fig. 4b). Thus, our IFAs revealed association of VEX1 with the SL-RNA transcription compartments and VEX2 with the VSG transcription compartment. Moreover, VEX1 signal intensity was significantly higher at the focus adjacent to the VSG transcription compartment (Extended Data Fig. 4a), indicating that all nuclei may have two VEX1 foci, but the second focus may be below the detection limit in some cells. Further, to verify a specific interaction between VEX1 and the SL-RNA locus, we reanalyzed published VEX1-ChIP-seq data, previously only mapped to VSG expression sites 18. The ChIP-seq data revealed a striking enrichment of VEX1 at the SL-RNA locus that was greater than at any other gene, including the active VSG gene (Fig. 3c; Extended Data Fig. 4c; Supplementary Information, sheet 1). These data suggest that the VEX-complex is specifically associated with the VSG and SL-RNA transcription compartments.
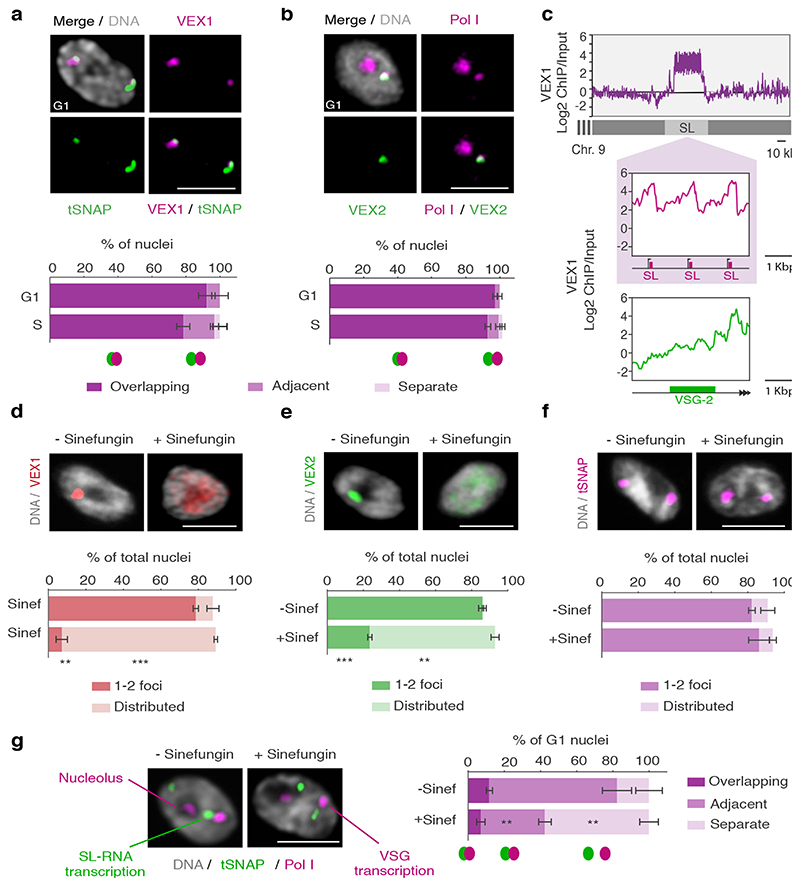
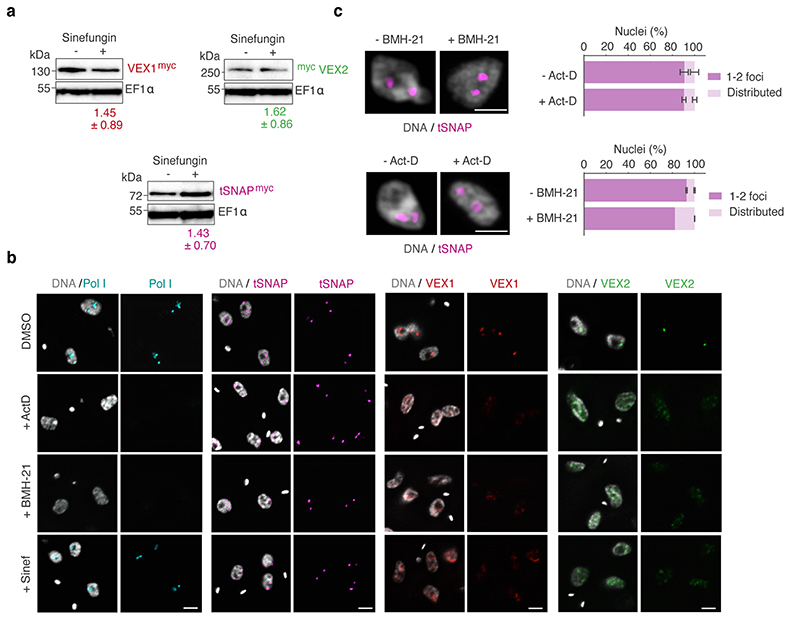
Fig. 3. The VEX complex associates with both the active VSG gene and the Spliced Leader (SL) locus in a splicing-dependent manner.
a-b, Immunofluorescence-based colocalization studies of VEX1myc / tSNAPGFP and GFPVEX2 / Pol I. tSNAP and Pol I are used as markers for the SL-RNA and VSG transcription compartments, respectively. The stacked bar graphs depict proportions of nuclei with overlapping, adjacent or separate signals (these categories were defined by thresholded Pearson’s correlation coefficient – see methods); values are averages of four (a) or two (b) independent experiments (≥100 G1 or S phase nuclei). c, VEX1myc chromatin immunoprecipitation followed by next generation sequencing (ChIP-Seq) analysis. The top graph depicts log2 fold changes of ChIP signal versus input sample across the SL-RNA locus (bin size 300 bp). The inset shows a zoom on three SL repeats (arrows, promoter; magenta boxes, SL-RNA; bin size 10 bp); the bottom plot represents log2 fold enrichment across the active VSG gene (VSG-2, green box, bin size 10 bp). d-f, Immunofluorescence analyses of VEX1myc (d), mycVEX2 (e) and tSNAPmyc (f) before and after sinefungin treatment (5 μg ml-1 for 30 min at 37 °C). Cells displaying no detectable signal (<10%) were excluded. Values are averages of two independent experiments (≥200 nuclei each). g, Immunofluorescence-based colocalization studies of the SL-RNA transcription (tSNAPmyc) and the VSG transcription (Pol I, large subunit) compartments following treatment with sinefungin. The stacked bar graph depicts proportions of G1 nuclei with overlapping, adjacent or separate signals. Values are averages of two independent experiments and two biological replicates (≥100 G1 nuclei). The studies in a-b and d-g were undertaken using super resolution microscopy and the images correspond to maximal 3D projections of stacks of 0.1 μm slices; DNA was counter-stained with DAPI; scale bars 2 μm. In a-b and d-g, a two-tailed paired Student’s t-test was used to compare G1 versus S nuclei and non-treated versus treated nuclei, respectively, for each category; statistical significance is highlighted where applicable: **, p < 0.01; ***, p < 0.001. Experiments in a-b and d-g are representative of at least two independent biological replicates; error bars, SD. Detailed n and p values are provided in Source Data Fig. 3.
While VSG and SL-RNA transcription compartments separate during S phase (Fig. 1c; Extended Data Fig. 2a-c), VEX1 does not separate from the SL-RNA transcription compartment and VEX2 does not separate from the VSG transcription compartment (Fig. 3a and b). Also, consistent with the loss of VSG expression in insect-stage cells, the extranucleolar Pol I focus is lost, while SL-RNA transcription compartments can still be identified through tSNAP localization; the VEX proteins also redistribute but a pool of VEX1 remains detectable at SL-RNA transcription compartments (Extended Data Fig. 4d). These results indicate that VEX2 marks the VSG transcription compartment in a developmental stage specific manner, which in bloodstream stage cells may facilitate the re-establishment of compartment connectivity to propagate expression of a specific antigen. Consistent with this idea, VEX complex reassembly after DNA replication is dependent upon CAF-1 histone chaperone function 18.
Given the close spatial proximity between the site of VSG transcription and the site of SL-RNA transcription, we next questioned whether the splicing process itself impacts the connection between these compartments (Fig. 3d-g; Extended Data Fig. 5). Sinefungin inhibits the cap guanylyltransferase-methyltransferase Cgm1 22 and subsequent methylation of the SL-RNA cap, required for trans-splicing 23,24. We found that inhibition of trans-splicing with sinefungin 25 disrupted both VEX1 (Fig. 3d) and VEX2 (Fig. 3e) localization within 30 min, while the tSNAP transcription factor was not affected under the same conditions (Fig. 3f). Notably, inhibition of splicing by sinefungin also disrupted the connection between the VSG and SL-RNA transcription compartments, revealed by separation of the Pol I and tSNAP signals (Fig. 3g); neither the VEX, nor the tSNAP protein levels were substantially affected by sinefungin treatment (Extended Data Fig. 5a). The delocalization of the VEX proteins following splicing inhibition suggests that the process of RNA maturation or mature RNAs from the active VSG expression site play a role in the assembly and/or maintenance of these protein condensates. Consistent with this idea, inhibition of Pol I or both Pol I and Pol II transcription with BMH-21 or actinomycin D, respectively, also similarly disrupted VEX1 and VEX2 localization without affecting tSNAP localization (Extended Data Fig. 5b-c). Thus, VEX protein localization is dependent on Pol I transcription or VSG expression site RNAs and mRNA splicing activity.
Next, we aimed to investigate the mechanism by which the VEX complex ensures monogenic VSG expression. Previously, we found that VEX2 depletion leads to a strong activation of VSG genes located in previously silent expression sites 18. Thus, following our observation that the VEX-complex spans the VSG transcription and the SL-RNA transcription compartments, we sought to determine whether VEX2 functions as a connector or as an exclusion factor. That is, whether following VEX2 depletion the active VSG gene loses connectivity to the SL-RNA transcription compartment or whether previously inactive expression sites start to interact with the SL-RNA transcription compartment. To test these models, we analyzed the distance between the extranucleolar Pol I focus (expression site body) and the SL-RNA transcription compartment following depletion of VEX-complex components (Extended Data Fig. 6a-b). IFA data revealed a dissociation of Pol I from the SL-RNA transcription compartments in 45% of G1 nuclei following 12 hours of VEX2 knockdown (P <0.001) and 60% following VEX1 - VEX2 double-knockdown (P <0.01) (Fig. 4a-b; Extended Data Fig. 6c-d). Association between the extranuclear Pol I focus and the SL-RNA transcription compartment was not significantly disrupted following VEX1-knockdown. Therefore, in the absence of VEX2, Pol I initially separates from the SL-RNA compartment (12 h) and at later time points it disperses 18, indicating that VEX2 sustains a local reservoir of Pol I at the active VSG gene. Pol I dispersion temporally overlaps with activation of previously ‘silent’ VSG expression sites.
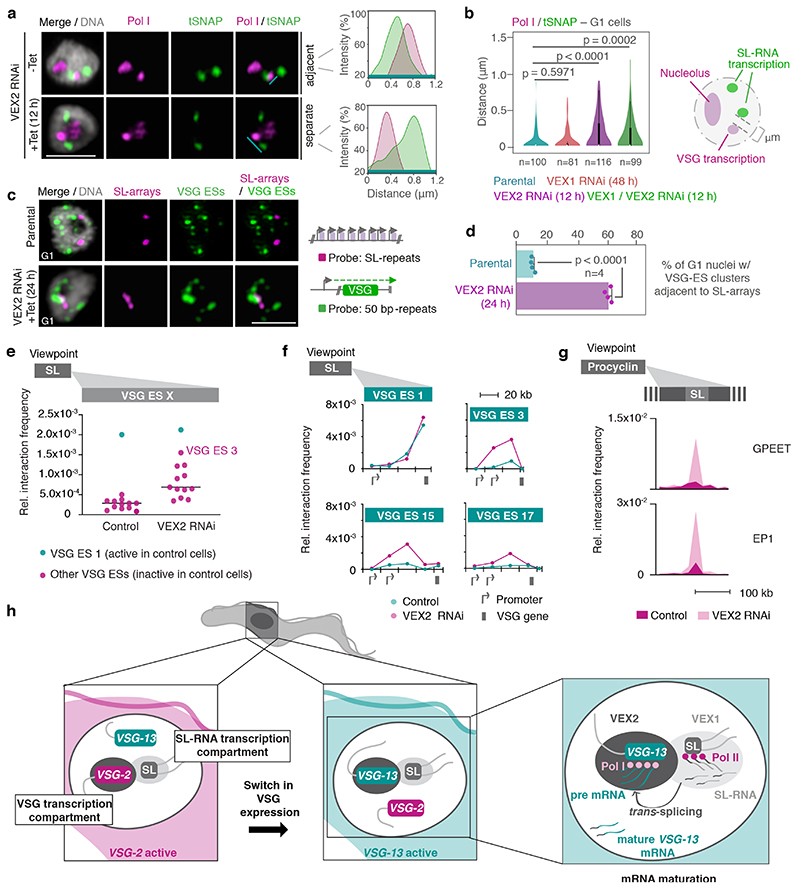
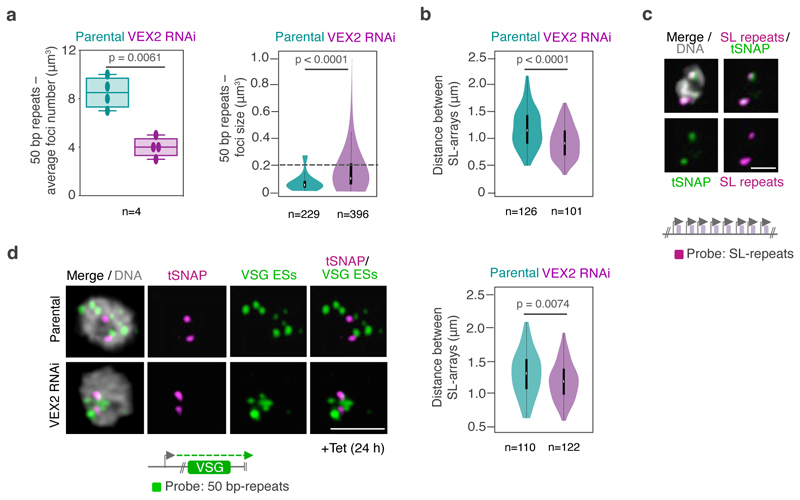
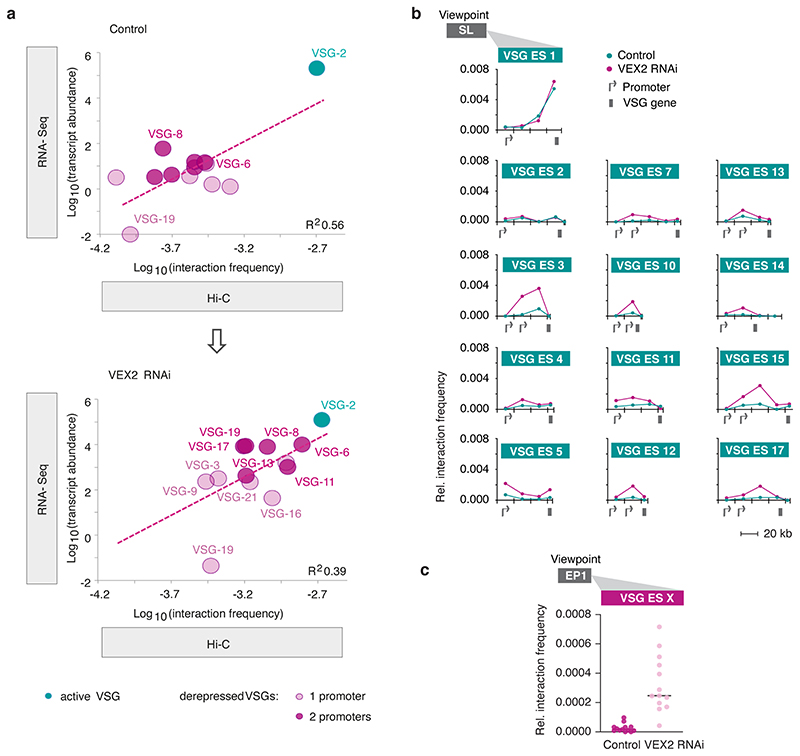
Fig. 4. The exclusive association between the active VSG gene and the SL-locus is VEX2-dependent.
a-b, Immunofluorescence and super resolution microscopy based colocalization studies of tSNAPmyc (SL-RNA transcription compartment) and Pol I (nucleolus and extranucleolar reservoir) following VEX1, VEX2 and VEX1/VEX2 knockdown. On the right-hand side, two representative histograms depict the distribution of signal intensity across the distance indicated by the cyan lines. b, The violin plot depicts the ‘inner’ distance between the Pol I extranucleolar reservoir and the nearest SL-RNA transcription compartment (≥ 81 G1 nuclei) following VEX1, VEX2 and VEX1/VEX2 knockdown. White circles show the medians; box limits indicate the 25th and 75th percentiles as determined by R software; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. c-d, DNA fluorescence in situ hybridization (FISH) and super resolution microscopy based colocalization studies of the SL-RNA transcription compartments (probe: digoxigenin labeled SL repeats) and VSG expression sites (probe: biotin-labeled 50 bp repeats) following VEX2 knockdown. The bar graph (d) depicts the % of G1 nuclei with VSG expression site clusters (size > 0.2 μm3) overlapping or adjacent to the SL-arrays (within 60 nm) before and after VEX2 knockdown; error bars, SD. The data are average of two biological replicates and two independent experiments. Representative images in a and c: all nuclei are G1; images correspond to maximal 3D projections of stacks of 0.1 μm slices; DNA was counter-stained with DAPI; scale bars 2 μm; images are representative of two biological replicates and two independent experiments. Statistical analysis was undertaken using a two-tailed unpaired (b) or paired (d) Student’s t-test. Detailed n and p values are provided in Source Data Fig. 4. e, Hi-C (virtual 4C) analysis, viewpoint: SL-RNA locus (chr. 9). Relative interaction frequencies between the viewpoint and the VSG expression sites are shown before and after VEX2 knockdown. Each dot represents the average value for one expression site. Bin size 20 kb. f, Virtual 4C analysis, viewpoint: SL-RNA locus (chr. 9). Relative interaction frequencies between the viewpoint and VSG expression sites is shown. Bin size 20 kb. g, Virtual 4C analysis, viewpoint: EP1 (chr. 10) or GPEET gene array (chr. 6). Relative interaction frequencies between the viewpoint and the SL-RNA locus are plotted. Bin size 20 kb. The analyses in e-g are based on Hi-C experiments with cells before and 24 h after VEX2 knockdown (the average of three biological replicates is shown). The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 2. h, Schematic model for monogenic VSG expression. A strong inter-chromosomal interaction between the SL-array and the active VSG gene facilitates spatial integration of transcription and mRNA maturation. VEX1 and VEX2 are primarily SL- and active-VSG associated, respectively, and sustain monogenic VSG expression by excluding other VSGs. The VSG-SL organelle is reconfigured upon activation of a different VSG.
Next, we performed DNA FISH to visualize all VSG expression sites in the 3D nuclear space and their position relative to the SL-RNA compartments before and after VEX2 depletion (Fig. 4c-d; Extended Data Fig. 7). We used one probe that recognized the 50 bp repeats, present immediately upstream of all VSG expression sites, and another probe that recognized the SL-RNA repeats. In unperturbed cells, expression sites were distributed throughout the nucleus (Fig. 4c), as described previously 15,16. Following VEX2 depletion (24 h), the number of individual VSG expression site signals decreased while their size increased (Extended Data Fig. 7a), consistent with the formation of VSG expression site clusters. Indeed, the proportion of G1 nuclei displaying VSG expression site clusters overlapping or adjacent to an SL-RNA compartment increased upon VEX2 knockdown (Fig. 4d). We also observed a decrease in the distance between SL-arrays (Extended Fig. 7b-d). Therefore, in the absence of VEX2, previously ‘silent’ VSG expression sites displayed increased spatial proximity to SL-RNA compartments and were derepressed.
To further explore the role of VEX2 in controlling interactions between antigen-coding genes and SL-RNA loci, we performed Hi-C analyses in VEX2-depleted cells. After 24 hours of VEX2 depletion, all previously silent expression sites displayed increased interaction frequencies with the SL-RNA locus (Fig. 4e), consistent with the DNA FISH analysis (Fig. 4c-d). The interaction between VSG expression site 3 and the SL-RNA locus showed the strongest increase. Notably, this is the expression site containing the most de-repressed VSG (VSG-6) following VEX2 depletion 18. Interaction frequencies of the active VSG expression site 1 with the SL-RNA locus remained unchanged, correlating with sustained and dominant VSG-2 expression. Thus, both Hi-C and DNA FISH analyses point to close spatial proximity between the active-VSG locus and one of the SL-RNA loci before and after VEX2 depletion. Indeed, overall SL-RNA interactions correlated with VSG transcript levels before and after VEX2 knockdown (Extended Data Fig. 8a).
Besides the VSG genes located in previously silent expression sites, expression site associated genes were also strongly upregulated following VEX2 knockdown 18. In line with this finding, we observed the largest increase in SL-RNA interactions for the regions upstream of the VSG gene in each de-repressed expression site, where expression site associated genes are located (Fig. 4f, Extended Data Fig. 8b).
As a third group of RNA Pol l transcribed genes, insect stage specific procyclin genes are upregulated upon VEX2 depletion 18. Correlating with these data, following VEX2-knockdown, we found GPEET and EP1 procyclin genes to exhibit strongly increased interaction frequencies with the SL-RNA array and also with VSG expression sites (Fig. 4g, Extended Data Fig. 8c). Thus, our data suggest that VEX2 has a dual function: specifically enhancing mRNA splicing of the VSG gene that is connected to the SL-RNA transcription compartment and, at the same time, excluding all other VSG expression sites and procyclin genes from the SL-RNA compartment to ensure monogenic VSG expression.
By combining proximity ligation and super-resolution microscopy, we were able to demonstrate spatial integration of the active VSG expression site and a genomic locus important for RNA maturation. Our data show that this supramolecular assembly is composed of a VSG transcription compartment with the active VSG gene, RNA Pol l and VEX2 and an SL-RNA transcription compartment with the SL-RNA array, RNA Pol II, VEX1 and the tSNAP complex, presumably together with other factors important for mRNA trans-splicing 20. Based on these findings we propose a model in which VSG choice is intimately associated with an inter-chromosomal interaction, bringing together two nuclear compartments to ensure efficient VSG mRNA processing at only one expression site (Fig. 4h). In the VSG transcription compartment, the VSG gene is transcribed by highly processive RNA Pol I, generating large amounts of VSG pre-mRNA that requires efficient processing to prevent premature degradation. In the SL-RNA transcription compartment, SL-RNA, an essential substrate for maturation of every mRNA, is produced. The close spatial proximity of the two compartments in a single organelle provides a sufficiently high concentration of trans-splicing substrate to ensure the efficient maturation of highly abundant VSG transcripts.
Our data indicate that VEX2 is not simply a tether. Instead, they led us to propose a temporal ‘choose and consolidate’ component to our model: VEX2 is recruited to expression sites in a stochastic and competitive manner by Pol I transcription (‘choose’), as supported by Pol I inhibition experiments that resulted in dispersion of VEX2 ( 18 and Extended Data Fig. 5b). Subsequently, the presence of VEX2 at one expression site ‘consolidates’ expression site transcription, as supported by VEX2 depletion experiments that resulted in delocalization of the Pol I focus (Fig. 4a-b). Consolidation by VEX2 also involves the formation of a particularly prominent interaction between the active VSG and the SL-RNA locus, as supported by interactions primarily with the regions upstream of de-repressed VSGs following VEX2 depletion (Fig. 4f). Increased interaction with the SL-RNA array is accompanied by increased transcription of expression sites, as supported by Hi-C and FISH (Fig. 4c-f), also leading to efficient VSG mRNA processing. We propose that these initial interactions occur in a stochastic manner, providing opportunities to overcome exclusion. The probability of forming and consolidating a new stable interaction with the SL-RNA locus is likely enhanced when the VEX-complex is no longer effectively sequestered; when the active VSG expression site is damaged 26, or when chromatid cohesion 27 or telomeric chromatin 28 are disrupted, for example. According to this model, VEX2 is the key molecule excluding all but one VSG expression site from the SL-RNA transcription compartment, thereby ensuring expression of a single VSG gene per cell.
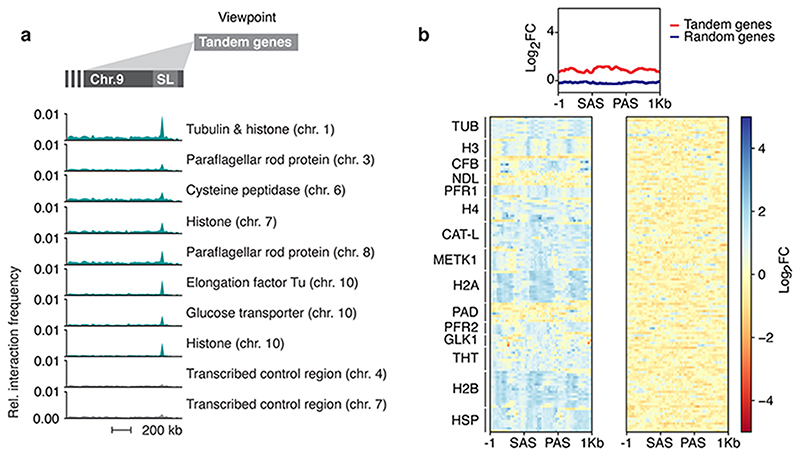
RNA processing as the limiting factor in monogenic VSG expression has been proposed previously, based on the observations that all VSG expression site promoters are active, yet processive polycistronic transcription and mature transcripts are specific to the single active expression site 9,10. Given the observation that co-transcriptional RNA processing can affect elongation rates in other organisms 29, it will be interesting to determine whether efficient VSG mRNA processing will exert a positive feedback on transcription elongation along expression sites. Also, it was shown recently that splicing can activate transcription 30. It remains to be shown if factors located in the SL-RNA transcription compartment recruit the transcription machinery to the interacting VSG expression site and thereby enhance transcription. Notably, we also find other highly expressed housekeeping genes, such as core histones and tubulin, to associate with the SL-RNA locus (Extended Data Fig. 9a) and with VEX1 (Extended Data Fig. 9b). Thus, interactions with the SL-RNA locus, which is thought to be >250 kbp in length 31, may play a broader role for the regulation of gene expression in T. brucei.
While the importance of intra-chromosomal interactions in regulating gene expression has been shown in many complex eukaryotes, the significance of inter-chromosomal interactions has been questioned. Interactions between different chromosomes were thought to require complicated, possibly error-prone mechanisms to be re-established following mitosis. Yet, our data demonstrate a stable interaction between the active VSG gene, located either on chr. 6 or on an intermediate-sized chromosome, and the SL-RNA array locus on chr. 9; an interaction that is also stably propagated during cell-division. To our knowledge, these findings represent the first report of a selective inter-chromosomal interaction that connects transcription and mRNA splicing.
Protein condensates have recently emerged as important features that compartmentalize nuclear functions; transcription control by RNA polymerases, for example 32. Regulated switching between adjacent transcriptional and splicing condensates has been described in mammalian cells 33 and RNAs are major actors in facilitating genomic interactions and phase transitions 34. Given the recent finding that members of a family of helicases are global regulators of RNA-containing, phase-separated organelles 35, it is tempting to speculate that maintenance of VSG transcription-maturation compartment connectivity is similarly regulated by the putative RNA helicase VEX2 18. We show here that in T. brucei, the assembly of two membrane-less nuclear condensates, each with a specific function in VSG gene transcription and RNA maturation, and both containing protein and RNA molecules 15,20, regulates monogenic VSG expression. By shaping a highly selective and specific genome architecture, VEX2 allows only one VSG gene to productively interact with the mRNA splicing compartment.
Methods
No statistical methods were used to predetermine sample size. The experiments were not randomized and investigators were not blinded to allocation during experiments and outcome assessment.
T. brucei growth and manipulation
Bloodstream-form T. brucei, Lister 427 and 2T1 cells 36, both wild-type with respect to VEX1, VEX2 and SNAP42 subunits, were grown in HMI-11 medium and genetically manipulated using electroporation 37; cytomix was used for all transfections. Puromycin, phleomycin, hygromycin and blasticidin were used at 2, 2, 2.5 and 10 μg ml-1 for selection of recombinant clones; and at 1, 1, 1 and 2 μg ml-1 for maintaining those clones, respectively. RNAi experiments were undertaken through tetracycline induction at 1 μg ml-1. A double selection T. brucei cell line was used that derived from the Lister 427 bloodstream-form MITat 1.2 isolate 21. A neomycin resistance gene in VSG expression site 17 and a puromycin resistance gene in VSG expression site 1 allowed the selection for a homogenous cell population that either expressed VSG-2 from expression site 1 or VSG-13 from expression site 17. Cells were cultivated with either 10 μg ml-1 of neomycin (also referred to as N50 cells) or 0.1 μg ml-1 of puromycin (referred to as P10 cells). Established procyclic-form T. brucei, Lister 427 cells were grown in SDM-79 at 27 °C and genetically manipulated using electroporation as above. Blasticidin or hygromycin were used at 10 or 50 and 2 or 1 μg ml-1 for selection and maintenance, respectively.
Plasmids
The VEX1 (Tb927.11.16920, 574 bp) 17, VEX2 (Tb927.11.13380, 471 bp) 18 and VEX1/VEX2 18 RNAi cassettes were excised prior to electroporation by digesting with AscI. The VEX112myc 17 and SNAP4212myc 38 C-terminal tagging vectors were linearised with SphI. The 6mycVEX2 and GFPVEX2 N-terminal tagging vectors were linearised with XhoI. The SNAP42 GFP C-terminal tagging vector was made by replacing the 12 x c-myc tag and as also linearised with SphI, respectively. Linearised RNAi constructs, under the control of tetracycline-inducible promoters, were transfected into 2T1 cells, which allow for targeting to a single genomic locus validated for robust inducible expression 36.
ChIP-seq
ChIP-Seq was carried out as described in 18. Reads were aligned to the 11 curated megabase chromosomes from the TREU927 strain genome sequence 39, and a non-redundant set of BES and mVSG contigs from the Lister 427 strain 40–42 using bowtie2 43 in very-sensitive alignment mode, and alignments were compressed and sorted using samtools 44. Bowtie2 attempted to align 54.0 and 49.9 million read pairs with 70.84 and 82.43 % success rates, respectively, resulting in 38.3 and 41.1 million aligned read pairs. PCR duplicate reads were removed using Picard MarkDuplicates (http://broadinstitute.github.io/picard/) resulting in 26.8 and 41.3 million aligned read pairs for analysis. Alignments were visually inspected with the Artemis genome browser 45. Circular plots (Extended Data Fig. 4c) were generated using the R library circlize 46 and bedgraph files for log2 fold change (Fig. 3c; Extended Data Fig. 9b) were generated using deeptools2 47. Bedgraphs were generated with 1kb, 300 bp or 10 bp bins and the option smoothLength 5000 (see figure legends for details).
Spliced leader RNA sequences were annotated using the sequences: Promoter: CGTTTCTGGCACGACAGTAAAATATGGCAAGTGTCTCAAAACTGCCTGTACAGCTTATTTTTGGGACACACCCATGCTTTC…Transcript…AACTAACGCTATTATTAGAACAGTTTCTGTACTATATTGGTATGAGAAGCTCCCAGTAGCAGCTGGGCCAACACACGCATTTGTGCTGTTGGTTCCCGCCGCATACTGCGGGAATCTGGAAGGTGGGGTCGGATGACCTC and the ‘transcript’ features were plotted with transcription start site (TSS) and transcription end site (TES) denoting the 5′ and 3′ extremities. Fold enrichment traces covering the spliced leader locus were calculated directly using deeptools bamCompare. Heat maps (Extended Data Fig. 4c; Extended Data Fig. 8b) were generated using deeptools2 bamCompare, computeMatrix and plotHeatmap 47 and resulting vector graphics files were then assembled into figures using Adobe Illustrator. Genomic regions for tandem genes and arbitrarily selected genes with a paralog count of 0 were assembled in bed files, using annotated mRNA sequences from TriTrypDB v5.1 of the TREU927 genome sequence. All scripts necessary to reproduce the ChIP-Seq analyses have been deposited together with the results of those analyses under https://doi.org/10.5281/zenodo.3628212.
Protein blotting
Protein samples were run according to standard protein separation procedures, using SDS-PAGE. However, for VEX2 detection, the use of Bis-Tris gels with a neutral pH environment and a Bis-Tris/Bicine based transfer buffer (containing a reducing agent and 10% methanol) were critical for protein separation and transfer, respectively (NuPAGE, Invitrogen). Otherwise, western blotting was carried out according to standard protocols. The following primary antibodies were used: rabbit α-VEX2 (1:1,000), rabbit α-pol-I largest subunit 17 (1:500), rabbit α-VSG-2 (1:20,000), rabbit α-VSG-6 (1:20,000), mouse α-c-myc (Millipore, clone 4A6, 1:7,000), rabbit α-GFP (Abcam Ab290, 1:1,000) and mouse α-EF1α (Millipore, clone CBP-KK1, 1:20,000). We used horseradish peroxidase coupled secondary antibodies (α-mouse and α-rabbit, Biorad, 1:2,000). Blots were developed using an enhanced chemiluminescence kit (Amersham) according to the manufacturer’s instructions. Densitometry was performed using Fiji v. 2.0.0. Uncropped blots are provided as Source Data Extended Data Figs 5 and 6.
Immunofluorescence
Immunofluorescence microscopy was carried out according to standard protocols. For wide field microscopy (Extended Data Fig. 6a), the cells were attached to 12-well 5 mm slides (Thermo Scientific). For super resolution microscopy, the cells were attached to poly-L-lysine treated coverslips (thickness 1.5 mm), stained and only then mounted onto glass slides. For colocalization studies with Pol I we used antigen-retrieval. Prior to permeabilization, fixed cells were rehydrated in PBS for 5 min at RT, held at 95 °C for 60 s in freshly prepared antigen retrieval buffer (100 mM Tris, 5% urea, pH 9.5) and then washed 3 x 5 min in PBS at RT. Cells were mounted in Vectashield with DAPI (wide field) or stained with 1 μg ml-1 DAPI for 10 min and then mounted in Vectashield without DAPI (super resolution). In T. brucei, DAPI-stained nuclear and mitochondrial DNA were used as cytological markers for cell-cycle stage; one nucleus and one kinetoplast (1N:1K) indicates G1, one nucleus and an elongated kinetoplast (1N:eK) indicates S phase, one nucleus and two kinetoplasts (1N:2K) indicates G2/M and two nuclei and two kinetoplasts (2N:2K) indicates post-mitosis 48,49. Primary antisera were rat α-VSG-2 (1:10,000), rabbit α-VSG-6 (1:10,000), rabbit α-GFP (Invitrogen, 1:250; Abcam, 1:500), mouse α-myc (New England Biolabs, clone 9B11, 1:2,000), rabbit α-pol-I largest subunit 17 (1:100), rabbit α-NOG1 50 (1:500) and mouse α-EP procyclin (1:1,000). The secondary antibodies were Alexa Fluor conjugated goat antibodies (Thermo Scientific): α-mouse, α-rat and α-rabbit, AlexaFluor 488 or Alexa Fluor 568 (1:1,000 or 1:2,000, for super resolution or wide field microscopy, respectively). Sinefungin, actinomycin and BMH-21 were applied at 2 μg ml-1, 10 μg ml-1 and 2 μM, respectively, for 30 minutes at 37 °C.
Fluorescence in situ hybridization (FISH)
For DNA FISH experiments, biotin- and digoxigenin-labeled DNA probes were generated by PCR using standard conditions with OneTaq polymerase (NEB), with the exception that a 1:2 ratio of biotin-16-dUTP (Roche) or digoxigenin-11-dUTP (Roche) and dTTP were used in the reaction. 50 bp repeats and SL repeats were amplified from T. brucei L427 genomic DNA (Supplementary Information, sheet 3), a smear of products with various sizes was generated but only fragments of 400 bp or less were gel extracted and purified. DNA probes were co-precipitated with herring sperm DNA (Sigma) at 10 μg/mL and yeast tRNA (Invitrogen) at 10 μg/mL. Probes were then resuspended to a concentration of 1000 ng/mL in hybridization buffer (50% formamide, 10% dextran sulfate, 2×SSC). Before hybridization, cells were prepared similarly as for immunofluorescence: trypanosomes were fixed in 3% PFA for 15 min at 37 °C, washed three times with PBS and finally resuspended in 1% BSA. The cells were attached to poly-L-lysine treated slides, then permeabilized with 0.5% Triton X-100 in PBS for 15 min at RT, washed three times with PBS and then treated with 1 mg/mL of RNAse A (Invitrogen) in PBS for 1 h at RT. This was followed by a blocking step with 10 μg/mL of herring sperm DNA and 10 μg/mL of yeast tRNA in hybridization buffer (50% formamide, 10% dextran sulfate, 2×SSC) for 40 min at RT. After adding the probe mix to slides, the samples were sealed with gene frames and denatured on an inverted heat block at 85 °C for 5 min, followed by overnight incubation at 37 °C. After hybridization, the slides were washed with 50% formamide, 2×SSC for 30 min at 37 °C, followed by three 10-min washes in 1×SSC, 2×SSC, and 4×SSC at 50 °C. Samples were then incubated with an anti-digoxigenin antibody (Abcam, clone 21H8) diluted 1:10,000 in 1% BSA in PBS for 2 h at RT. After washing 3 times for 10 min in TBS with 0.01% Tween, the slides were incubated for 1 h with a streptavidin-Alexa Fluor 488 conjugate (Invitrogen) and a goat anti-mouse Alexa Fluor 568 antibody (Invitrogen), both diluted to 1:500 in 1% BSA. Samples were washed in TBS with 0.01% Tween as before and mounted in Vectashield with DAPI. For all experiments involving RNAi, the knockdown was always verified by western-blot and the VSG derepression phenotype confirmed by IFA prior to FISH analysis.
Imaging and image analysis
For wide field microscopy, cells were analyzed using a Zeiss Axiovert 200M microscope with an AxioCam MRm camera and the ZEN Pro software (Carl Zeiss, Germany). The images were acquired as z-stacks (0.1-0.2 μm) and further deconvolved using the fast iterative algorithm in Zen Pro.
For super resolution microscopy, cells were analyzed using a Leica TCS SP8 confocal laser scanning microscope in Hyvolution Mode and the Leica Application Suite X (LASX) software (Leica, Germany) or a Zeiss 880 Airyscan and the Zeiss ZEN software (Carl Zeiss, Germany). The Hyvolution mode allows super resolution level images, general used settings: highest resolution / lowest speed; pinhole 0.5. All representative images obtained by super resolution microscopy correspond to maximal 3D projections by brightest intensity of stacks of approximately 30 slices of 0.1 μm - Images with DNA in grey.
VEX1, VEX2 and tSNAP foci and Pol I nucleolar and ESB signals could be detected in over 85-90% of nuclei. Counts in total cells or specific cell cycle phases were performed typically in >200 or >100 nuclei, respectively. All quantifications are averages or representative of at least two biological replicates and independent experiments (details for specific experiments can be found in the Source Data file). Pearson’s correlation coefficient (PCC) was applied as a statistical measure of colocalization 51. Overlapping, adjacent and separate foci presented a PCC in the following ranges: ≥ 0.5 / ≤ 1, ≥ -0.5 / < 0.5, ≥ -1 / < -0.5, respectively. Regarding the distance measurements between the ESB and tSNAP compartment (Extended Data Fig. 2c and 4b, ‘inside edge’ distance), a control measurement (Extended Data Fig. 2b; Extended Data Fig. 6d, ‘outside edge’ distance) was performed to ensure that changes in the distance between the two protein condensates following cell cycle progression or VEX RNAi were not a consequence of changes in the diameter of the foci. For the ESB / tSNAP localization following VEX2 or VEX1 / VEX2 RNAi (Fig. 4a-b; Extended Data Fig. 6c-d), all the imaging and analyses were performed at 12 h post-induction, a timepoint where there was sufficient VEX2 knockdown (Extended Data Fig. 6b) but both nucleolar Pol I and the ESB could be detected in > 85% of cells; the ESB is not detectable at later time points 18. Moreover, the ESB / tSNAP localization analyses following VEX RNAi or sinefungin treatment were restricted to G1 cells to exclude any cell cycle bias, as these protein condensates can separate during S phase (Fig. 1c; Extended Data Fig. 2 a-c). In approximately 75% of G2 cells, two ESBs could be detected; the n values provided in the figures (Fig. 1c; Extended Data Fig. 2b-c) correspond to ESB / tSNAP pairs (the number of nuclei is stated in the legend). All signal quantifications were performed as follows: corrected fluorescence = integrated density − (selected area x mean fluorescence of background readings). For all quantifications, images were acquired with the same settings and equally processed. All the images were processed and scored using Fiji v. 2.0.0. 52, using stacks of approximately 30 slices of 0.1 μm; except Fig. 4d and Extended Data Fig. 7b, where the analysis was performed using Imaris 9.5 (Oxford Instruments). Briefly, 3D composites were loaded into Imaris 9.5, the DAPI channel was used to segment the nuclei; VSG expression site foci were segmented in the 50 bp repeats channel; diameter, area, signal intensity and volume were determined for all foci in all nuclei, the foci average number and volume distribution in the parental population versus VEX2 RNAi is depicted in Extended Data Fig. 7b. Foci with a size ≥0.2 μm3 were defined as VSG ES clusters.
For all experiments involving RNAi, the knockdown was always verified by western-blot and the VSG derepression phenotype confirmed by IFA and/or FACS analysis.
RNA-Seq
RNA isolation: The RNA-seq experiment and data analysis was performed as described previously 53, using three replicates each for VSG-2 and VSG-13 expressing cells. 45 million cells were harvested per replicate at 1,500 x g and 4 °C for 10 min. Cells were washed with 1× TDB (5 mM KCl, 80 mM NaCl, 1 mM MgSO4, 20 mM Na2HPO4, 2 mM NaH2PO4, 20 mM glucose pH 7.4). RNA isolation was performed using the NucleoSpin RNA kit (Macherey-Nagel; cat. no. 740955.10) according to the manufacturer’s instructions with minor changes. 3.8 μl of 1 M RNAse-free dithiothreitol (Sigma-Aldrich; cat. no. 10197777001) and 1 μl of 1:10 Ambion ERCC RNA Spike-In Mix (ThermoFisherScientific; cat. no. 4456739) was added to the cell lysis buffer prior to use. Removal of ribosomal RNA: rRNA was removed by hybridization as described previously 53. All solutions were kept free from nucleases. For each hybridization reaction, 2 μg of total RNA was mixed with 10 μl of formamide (SigmaAldrich; cat. no. F9037-100ML), 2.5 μl of 20× SSC (3 M NaCl, 0.3 M sodium citrate, the pH was adjusted to 7.0 with HCl), 5 μl of 0.005 M EDTA pH 8 (stock solution 0.5 M; ThermoFisherScientific; cat. no. AM9260G), 2.48 μl of 100 μM rRNA depletion mix (total 4 μg of oligos) and RNAse-free water (ThermoFisherScientific; cat. no. AM9938) to a total volume of 50 μl. Hybridization was performed for 5 min at 80 °C, ramp down to 25 °C at intervals of 5 °C per minute. Subsequently, 2 μl of RNAse-OUT (ThermoFisherScientific; cat. no. 10777019) and 50 μl of 1x SCC containing 20% formamide were added. Dynabead MyOne Streptavidin C1 beads (ThermoFisherScientific; cat. no. 65001) were prepared as recommended by the manufacturer for RNA applications and immobilization of nucleic acids. Three rounds of oligo capture were performed, using 120 μl (1.2 mg) of magnetics beads per round. The resulting supernatant, containing rRNA-depleted RNA, was purified using RNeasy MinElute CleanUp Kit (QIAGEN; cat. no. 74204). Depletion of rRNAs was evaluated on a 1.2% TBE-agarose gel. cDNA synthesis, library preparation and sequencing: Synthesis of cDNA was performed using NEBNext Ultra Directional RNA Library Prep Kit from Illumina (New England Biolabs; cat. no. E7420) according to the manufacturer’s instruction. The concentration of cDNA was measured using Qubit dsDNA HS Assay Kit (Invitrogen, cat. no. Q32854) and a Qubit 2.0 Fluorometer (Invitrogen; cat. no. Q32866). To generate strand-specific RNA-seq libraries, uracil excision and removing of the second strand was performed prior to conversion of Y-shaped adapters. Therefore, 3 μl of USER enzyme (New England Biolabs; cat. no. M5505) were mixed with 16 μl of adapter-ligated DNA, 1 μl of TruSeq PCR primer cocktail (50 μM) and 20 μl of KAPA HiFi HotStart ReadyMix (KAPA Biosystems, cat. no. KK2601). USER digestion was performed at 37 °C for 15 min, followed by the published amplification protocol. Library concentrations were determined in duplicate using Qubit dsDNA HS Assay Kit (Invitrogen, cat. no. Q32854) and a Qubit 2.0 Fluorometer (Invitrogen, cat. no. Q32866) and quantified using the KAPA Library Quantification Kit (KAPA Biosystems, cat. no. KK4824) according to the manufacturer’s instruction. Strand-specific RNA-sequencing libraries were sequenced in paired-end mode on an Illumina NextSeq 500 sequencer (2 × 75 cycles). Processing of sequencing data: The sequencing datasets were mapped to the TbruceiLister427_2018 genome assembly (release 43, downloaded from TriTrypDB 54) using BWA-mem 55. The alignments were converted from SAM to BAM format, sorted, indexed and filtered by alignment quality (q>0) using SAMtools version 1.9 44. To visualize read coverage, the number of reads was normalized per billion mapped reads and coverage files were generated in the wiggle format using COVERnant version 0.3.1with the subcommand ratio 56.
In situ Hi-C
In situ Hi-C was performed as previously described 7. 2 × 108 cells were collected per replicate and resuspended in 40 ml of 1× trypanosome dilution buffer (1× TDB; 0.005 M KCl, 0.08 M NaCl, 0.001 M MgSO4 × 7H2O, 0.02 M Na2HPO4, 0.002 M NaH2PO4 × 2H2O, 0.02 M glucose) or 1× PBS (insect stage cells). Cells were fixed in the presence of 1% formaldehyde for 20 min at room temperature by addition of 4 ml of 11% formaldehyde solution (50 mM Hepes-KOH pH 7.5, 100 mM NaCl, 1 mM EDTA pH 8.0, 0.5 mM EGTA pH 8.0, 11% formaldehyde). The reaction was stopped by addition of 3 ml of 2 M glycine and incubation for 5 min at room temperature and 15 min on ice. Cells were washed twice in 1× TDB for bloodstream form cells or 1× PBS for insect stage cells, respectively, and the cell pellet was snap-frozen in liquid nitrogen. Cells were resuspended in 1 ml of permeabilization buffer (100 mM KCl, 10 mM Tris pH 8.0, 25 mM EDTA) supplemented with protease inhibitors (1.5 mM pepstatin A, 4.25 mM leupeptin, 1.06 mM PMSF, 1.06 mM TLCK) and digitonin (200 μM final concentration) and incubated for 5 min at room temperature. Cells were washed twice in 1× NEBuffer3.1 (NEB, B7003S) and resuspended in 342 μl of 1× NEBuffer3.1. After addition of 38 μl of 1% SDS, and an incubation at 65 °C for 10 min, SDS was quenched by addition of 43 μl of 10% Triton-X 100 (Sigma). Incubation was continued at room temperature for 15 min. Another 35 μl of water, 13 μl of 10× NEBuffer3.1 and 100 units of MboI (NEB, R0147M) were added and the chromatin was digested at 37 °C overnight while shaking. To inactivate MboI, the sample was incubated at 65 °C for 20 min. Restriction fragments were biotinylated by supplementing the reaction with 60 μl of fill-in mix (0.25 mM biotin-14-dATP (Life Technologies, 19524016), 0.25 mM dCTP, 0.25 mM dGTP, 0.25 mM dTTP (Fermentas), 40 U of DNA polymerase I, large (Klenow) fragment (NEB, M0210)) and incubation at 23 °C for 4 h. The end-repaired chromatin was transferred to 665 μl of ligation mix (1.8% Triton-X 100, 0.18 mg BSA, 1.8× T4 DNA Ligase Buffer (Invitrogen, 46300018) and 5 μl of T4 DNA ligase (invitrogen, 15224025) were added. The ligation was performed for 4 h at 16 °C with interval shake. Crosslinks were reversed by adding 50 μl of 10 mg/ml proteinase K (65 °C for 4 h) following addition of another 50 μl of 10mg/ml proteinase K, 80 μl of 5 M NaCl and 70 μl of 10% SDS (65 °C, overnight). DNA was precipitated with ethanol and resuspended in 257 μl of TLE (10 mM Tris-HCl, 0.1 mM EDTA, pH 8.0). SDS was added to a final concentration of 0.1% and the sample was split among two tubes for sonication (Covaris S220; microtubes, 175 W peak incident power, 10% duty factor, 200 cycles per burst, 240 s treatment). The samples were recombined and the volume was adjusted to 300 μl with TLE. Fragments between 100 and 400 bp in size were selected using Agencourt AMPure XP beads (Beckman Coulter), according to the manufacturer’s instructions. The DNA fragments were eluted off the beads in 55 μl of TLE. For end-repair and biotin removal from un-ligated ends, 70 μl of end-repair mix was added (1× Ligation buffer (NEB), 357 μM dNTPs, 25U T4 PNK (NEB, M0201), 7.5U T4 DNA polymerase I (NEB, M0203), 2.5U DNA polymerase I, large (Klenow) fragment (NEB, M0210)) and incubated for 30 min at 20 °C and 20 min at 75 °C. To inactivate the enzymes, EDTA was added to a final concentration of 10 mM. To isolate biotin-labelled ligation junctions, 50 μl of 10 mg/ml Dynabeads MyOne Streptavidin C1 (Life Technologies, 65001) were washed with 400 μl of 1× Tween washing buffer (TWB; 5 mM Tris-HCl pH 7.5, 0.5 mM EDTA, 1 M NaCl, 0.05% Tween-20), collected with a magnet, resuspended in 400 μl of 2× binding buffer (10 mM Tris-HCl pH 7.5, 1 mM EDTA, 2 M NaCl) and added to the sample suspended in 330 μl TLE. Biotinylated DNA was bound to the beads by incubating the sample for 15 min at room temperature with slow rotation. Subsequently, the DNA-bound beads were captured with a magnet, washed twice with 400 μl of 1× binding buffer, washed once in 100 μl of 1× TLE T4 ligase buffer and resuspeded in 41 μl of TLE. For polyadenylation, 5 μl of 10× NEBuffer2.1, 1 μl of 10 mM dATP and 3 μl of 5 U/μl of Klenow fragment (3′→ 5′ exo (-)) (NEB, M0212) were added and the sample was incubated for 30 min at 37 °C followed by a deactivation step at 65 °C for 20 min. Beads were collected with a magnet, washed once with 400 μl 1× Quick ligation buffer (NEB, M2200) and resuspended in 46.5 μl of 1× Quick ligation buffer (NEB, M2200). 2.5 μl of DNA Quick ligase (NEB, M2200) and 0.5 μl of 50 μM annealed TruSeq adapters were added and incubated for 1 h at room temperature. Beads were separated on a magnet, resuspended in 400 μl of 1× TWB and washed for 5 min at room temperature with rotation. Beads were washed on the magnet with 200 μl 1× binding buffer and 200 μl of 1× NEBuffer2.1 and resuspended in 20 μl of 1× NEBuffer2.1. The library was amplified in eight separate reactions of 50 μl. Per reaction, 1.5 μl of 25 μM TruSeq PCR primer cocktail (TruSeq PCR primer cocktail_F, 5′-AATGATACGGCGACCACCGAG-3′; TruSeq PCR primer cocktail_R; 5′-CAAGCAGAAGACGGCATACGAG-3′), 25 μl of 2× Kapa HiFi HotStart Ready Mix (Kapa Biosystems, KR0370) and 21.5 μl of water were added to 2 μl of library bound to the beads. Amplification was performed as follows: 3 min at 95 °C, 5 cycles of 20 s at 98 °C, 30 s at 63 °C and 30 s at 72 °C, 1 cycle of 1 min at 72 °C, hold at 4 °C. The PCR reactions were pooled and the beads were removed from the supernatant using a magnet. The library was purified by addition of 1.5 volumes of Agencourt AMPure XP beads (Beckman Coulter), according to the manufacturer’s instructions. The sample was eluted off the beads using 25 μl of 1× TLE buffer, transferred to a fresh tube and the concentration was determined using Qubit (Qubit dsDNA HS Assay Kit, Thermo Fisher) and qPCR (KAPA SYBR FAST qPCR Master Mix, Kapa Biosystems), according to the manufacturer’s instructions. Library size distributions were determined on a 5% polyacrylamide gel. Paired-end 75-bp sequencing was carried out using the Illumina NextSeq 500 system with mid or high output NextSeq 500/550 kits v.2.5 according to the manufacturer’s instructions.
Mapping of Hi-C reads and generation of interaction matrices
Reads were mapped to a modified version of the TbruceiLister427_2018 genome assembly (downloaded from TriTrypDB, release 43) containing the following modifications. For all Hi-C experiments, we masked a newly discovered misassembly in bloodstream expression site 2 (BES2) with Ns. For Hi-C experiments in 2T1-control 36 and VEX2 knockdown cells, we added the transfected constructs as separate contigs to the genome. The construct sequences, as well as the modified genome have been deposited together with the results of the analyses under https://doi.org/10.5281/zenodo.3628212. Mapping, filtering, normalization and read counting were performed by the mHi-C pipeline as described in 19. We modified the pipeline to be compatible with the T. brucei genome assembly and also incorporated a merging step for the individual replicates after the removal of duplicate reads, but before data normalization (step 4) in order to avoid the introduction of any bias by the merge. We chose ICEing as the normalization method and finally filtered the mHi-C outcome by the posterior probability of 0.6 (i.e. reads are assigned to a bin with a likelihood of at least 60%). Downstream analyses such as normalizing for the different ploidy within the T. brucei genome assembly, have been implemented with in-house scripts. The digestion of the reference genome with the restriction site has been implemented using HiC-Pro Utilities 57. All scripts necessary to reproduce the Hi-C analyses can be found at: https://github.com/bgbrink/PRJEB35632.
Virtual 4C analysis
To visualize interactions between one genomic region (viewpoint) and all other genomic sites, relevant bins were extracted from a 20-kb or 50-kb Hi-C matrix. An average interaction value for every genomic bin was calculated if the viewpoint regions spanned more than one bin. The coordinates that define the different viewpoints used in this study are shown in Supplementary Information sheet 2. To determine the relative interaction frequency of a viewpoint with chromosome cores and subtelomeres, the average interaction frequency of the viewpoint with each chromosome core and subtelomere was calculated based on the relative interaction frequencies extracted by virtual 4C analysis. The ratio between the average interaction frequency (core) and the average interaction frequency (subtelomeres) was calculated for each chromosome and plotted as one dot. The virtual 4C analysis has been implemented using HiC sunt dracones (https://doi.org/10.5281/zenodo.3570496). All scripts necessary to reproduce the Hi-C analyses can be found at: https://github.com/bgbrink/PRJEB35632.
Statistical analysis
All statistical analysis was performed using GraphPad Prism Software (version 7.0). A detailed summary of n and p values for all the analyses performed in this study is provided as a Source Data file.
Resources and Reagents
All unique materials are available on request.
Extended Data
Extended Data Fig. 1. Genome-wide interaction frequencies of VSG genes in expression sites and the SL-RNA locus.
a, Hi-C (virtual 4C) analysis with locations of viewpoints marked by pink boxes. Viewpoints VSG ES 4, 7, 11 and 17 are located on intermediate chromosomes that are not depicted in this figure. Interaction frequencies between each viewpoint and the 11 mega-base chromosomes are shown. Chromosome cores, dark grey; subtelomeric regions, light grey. The hemizygous subtelomeric regions of each chromosome are displayed in the following order: 5′(haplotype A)–5′(haplotype B)–diploid chromosome core–3′(haplotype A)–3′(haplotype B). Bin size 50 kb. Virtual 4C analyses in a-b are based on Hi-C experiments of VSG-2 expressing cells (n=2, the average is shown). As viewpoint control regions, two actively transcribed regions from the diploid cores of chr. 2 and chr 8 and two non-transcribed regions from the subtelomeres of chr. 1 and chr. 10 were chosen. All four control regions were arbitrarily chosen. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 2. b, Virtual 4C analysis, viewpoint: SL-RNA locus (chr. 9). Relative interaction frequencies between the viewpoint and the active VSG ES 1 (cyan) and inactive VSG ESs (magenta) is shown. Each dot represents the average value for one expression site. Bin size 20 kb.
Extended Data Fig. 2. Dynamic association between the active VSG gene and the spliced leader RNA (SL) array transcription compartments.
a, Immunofluorescence-based colocalization studies of tSNAPmyc (SL-RNA transcription compartment) and a nucleolar and active VSG transcription compartment marker (Pol I, largest subunit) using super resolution microscopy - a plot showing signal intensity across a defined area (cyan line) is presented. b-c, The violin plot shows ‘outside edge’ (b) or ‘inside edge’ distance (c) measurements between the SL-RNA and VSG transcription compartments in G1, S phase or G2 nuclei – n values correspond to number of nuclei except in G2 where the n value corresponds to the number of expression-site body (ESB) / tSNAP pairs detected (in a total of 68 and 61 nuclei). d, Immunofluorescence-based colocalization studies of tSNAPmyc and a nucleolar compartment marker (NOG1) using super resolution microscopy. e, The plot shows signal intensity measurements of the SL-RNA compartments, adjacent or non-adjacent to the expression-site-body (ESB). f, The plot shows ‘inside edge’ distance measurements between the SL-RNA compartments, adjacent or non-adjacent to the ESB and the nucleolus. ESB: Pol I extranucleolar reservoir and VSG transcription compartment. a, d, DNA was counter-stained with DAPI; the images correspond to maximal 3D projections of stacks of 0.1 μm slices and are representative of two biological replicates and three independent experiments; scale bars 2 μm. Violin plots (b, c, e, f): white circles show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. P values were determined using a two-tailed unpaired (b-c) or paired (e-f) Student’s t-test. Detailed n and p values are provided in Source Data Extended Data Fig.2.
Extended Data Fig. 3. Changes in DNA-DNA interactions following a change in VSG isoform expression.
a, Virtual 4C analysis, viewpoint: VSG-2 in expression site 1. Relative interaction frequencies between the viewpoint and chr. 11 are plotted. Bin size 20 kb. * marks the centromere on chr. 11 (located on subtelomere 3A). The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 2. b, Hi-C (virtual 4C) analysis, viewpoints VSG-2, VSG-13 and a subtelomeric control region. Each dot represents the ratio of: the average interaction frequency of the viewpoint with the chromosome core / the average interaction frequency with the subtelomeres. One dot per chromosome is plotted. The black bar marks the median ratio per viewpoint. Bin size 50 kb. c, Virtual 4C analysis, viewpoint: EP1 gene array (chr. 10). Relative interaction frequencies between the EP1 array and the SL-RNA locus are plotted. Bin size 20 kb.
Extended Data Fig. 4. The VEX complex associates with both the active VSG gene and the Spliced Leader (SL) locus in a cell cycle and developmental stage-dependent manner.
a-b, Immunofluorescence-based colocalization studies of VEX1myc / Pol I and GFPVEX2 / tSNAPmyc in bloodstream form cells. tSNAP and Pol I were used as markers for the SL-RNA and VSG transcription compartments, respectively. The stacked bar graphs depict proportions of nuclei with overlapping, adjacent or separate signals (these categories were defined by thresholded Pearson’s correlation coefficient – see methods); values are averages of two independent experiments (≥100 nuclei for G1 and S phase cells); error bars, SD. The violin plot (a) shows signal intensity measurements of the VEX1 foci, adjacent or non-adjacent to the expression-site-body (ESB / VSG transcription compartment / Pol I extranucleolar reservoir) – in cells with 2 VEX1 foci and 1 ESB. White circles show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. P values were determined using a paired Student’s t-test; detailed n and p values are provided in Source Data Extended Data Fig. 4. c, VEX1myc chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) analysis. The circle plot represents log2 fold change of ChIP versus Input of non-overlapping 1 kb bins of the 11 megabase chromosomes; outside track shows tandem arrays (red) and the SL-RNA locus (black). An inset zooming on the SL-RNA locus is depicted: heat-map of SL-gene loci. Bin size 300 bp. d, Localization of tSNAPGFP and colocalization studies of VEX1myc or mycVEX2 and Pol I and VEX1myc and tSNAPGFP in procyclic forms (insect-stage), using immunofluorescence. Procyclic forms do not express VSGs whereas procyclins are the major surface glycoprotein. Images in a-b and d were obtained using super resolution microscopy and correspond to maximal 3D projections of stacks of 0.1 μm slices; DNA was counter-stained with DAPI; scale bars 2 μm; images are representative of independent experiments using two different biological replicates.
Extended Data Fig. 5. VEX1 and VEX2, but not tSNAP, delocalize following transcription or splicing inhibition.
a, Western-blot analysis of VEX1myc, mycVEX2 and tSNAPmyc before and after sinefungin treatment (5 μg ml-1 for 30 min at 37 °C), which blocks trans-splicing in trypanosomes. The values in red, green and magenta correspond to the fold change in VEX1, VEX2 and tSNAP abundance, respectively, between non-treated and treated samples (normalized against EF1α, loading control) in four independent experiments (not-statistically significant). b, Immunofluorescence analysis of Pol I, VEX1myc, mycVEX2 and tSNAPmyc localization following actinomycin D (ActD, Pol I + Pol II inhibitor, 10 μg ml-1 for 30 min at 37°C), BMH-21 (Pol I inhibitor, 1 μM for 30 min at 37°C) or sinefungin treatment. c, tSNAPmyc before and after ActD and BMH-21 treatment. Cells displaying no detectable signal (<10%) were excluded. Values are averages of two independent experiments (≥200 nuclei each); error bars, SD. Images in b-c were obtained using super resolution microscopy, correspond to maximal 3D projections of stacks of 0.1 μm slices and are representative of multiple biological replicates and independent experiments; DNA was counter-stained with DAPI; scale bars 2 μm. Uncropped blots (a) and detailed n and p values (c) are provided as Source Data Extended Data Fig 5.
Extended Data Fig. 6. Pol I and tSNAP expression and localization following knockdown of the VEX complex.
a, Immunofluorescence-based analysis of VSG expression following tetracycline (Tet) inducible VEX1 knockdown, VEX2 knockdown or VEX1/VEX2 knockdown. In unperturbed cells (parental strain), VSG-2 (magenta) is the active VSG and VSG-6 (green) is a silent VSG used to monitor derepression. The stacked bar graph depicts percentages of VSG-2 single positive cells and VSG-2/VSG-6 double positive cells; values are averages of two independent experiments and two biological replicates. DNA was counter-stained with DAPI; scale bar 2 μm. b, Western-blot analysis of VEX2, Pol I, tSNAPmyc, VSG-6 and VSG-2 expression following VEX1, VEX2 or VEX1/VEX2 knockdown. EF1α was used as a loading control. The data is representative of two independent experiments and two biological replicates. c-d, Immunofluorescence-based colocalization studies of tSNAPmyc (SL-RNA transcription compartment) and Pol I (nucleolus and extranucleolar reservoir). The stacked bar graph in c depicts proportions of G1 nuclei with tSNAPmyc / Pol I overlapping, adjacent or separate signals (these categories were defined by thresholded Pearson’s correlation coefficient – see methods) following tetracycline (Tet) inducible VEX1 (48 h), VEX2 (12 h) or VEX1/VEX2 knockdown (12 h). tSNAPmyc / extranucleolar Pol I localization were not monitored beyond 12 h following VEX2 and VEX1/2 knockdown as Pol I signal drops below detection at later time-points. The values are averages of two independent experiments and two biological replicates (≥100 G1 nuclei). In the violin plot in d, the ‘outside edge’ distance between the Pol I extranucleolar focus and tSNAP foci was measured in > 81 G1 nuclei. White circles show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. In a/c, error bars, SD. In c-d, knockdown conditions were compared to parental cells using two-tailed paired (c) or unpaired (d) Student’s t-tests; in c, statistical significance is highlighted when applicable: **, p < 0.01; ***, p < 0.001. Uncropped blots (b) and detailed n and p values (c/d) are provided in Source Data Extended Data Fig. 6.
Extended Data Fig. 7. The exclusive association between the active VSG and the SL-locus is VEX2-dependent.
a-b, DNA fluorescence in situ hybridization (FISH) and super resolution microscopy based colocalization studies of the SL-RNA transcription compartments (probe: digoxigenin labeled SL repeats) and VSG expression sites (probe: biotin-labeled 50 bp repeats) following VEX2 knockdown. a, the box and violin plots depict the average number and the size of the 50 bp repeats foci, respectively, before and after VEX2 knockdown. b, the violin plot represents the distance between both SL-arrays (before and after VEX2 knockdown. c-d, DNA FISH combined with immunofluorescence colocalization studies of the SL-RNA transcription compartments and VSG expression sites before and after VEX2 knockdown using super resolution microscopy. tSNAPmyc (protein marker) and a DNA probe for the SL repeats (biotin-labeled) or DNA probes for the 50 bp repeats (VSG ESs, biotin labeled) were used (c and d, respectively). The violin plot (d) represents the distance between both tSNAP foci before and after VEX2 knockdown. For all violin plots: white circles show the medians; box limits indicate the 25th and 75th percentiles; whiskers extend 1.5 times the interquartile range from the 25th and 75th percentiles; polygons represent density estimates of data and extend to extreme values. For the box plot in a, centerlines show the medians; box limits indicate the 25th and 75th percentiles whiskers extend between the minimum and maximum values. Two-tailed paired (a, left hand side) or unpaired Student’s t-tests (a, right hand side; b,d) were applied for statistical analysis. Detailed n and p values are provided in Source Data Extended Data Fig. 7. Representative images (c-d): all nuclei are G1; images correspond to maximal 3D projections of stacks of 0.1 μm slices; DNA was counter-stained with DAPI; scale bars 2 μm. The data correspond to (a, box plot) or are representative of (a, violin plot & b-d) two biological replicates and two independent experiments.
Extended Data Fig. 8. Genome-wide changes in VEX2 depleted cells.
a, Correlation between the average interaction frequency of VSG expression-sites as viewpoint with the SL-RNA locus and VSG expression in reads per kilobase per million (RNA-seq data from 18) in control and VEX2-depleted cells. b, Hi-C (virtual 4C) analysis, viewpoint: SL-RNA locus (chr. 9). Relative interaction frequencies between the viewpoint and VSG expression sites are shown. Bin size 20 kb. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 3. c, Virtual 4C analysis, viewpoint: EP1 gene array (chr.10). Relative interaction frequencies between the viewpoint and the VSG expression sites are shown. Each dot represents the average value for one expression site. Bin size 20 kb.
Extended Data Fig. 9. Tandem arrays interact with the Spliced Leader (SL) locus.
a, Hi-C (virtual 4C) analysis, viewpoint: different tandem gene arrays and control sites. Relative interaction frequencies between the different viewpoints and the SL-RNA locus are plotted. Bin size 20 kb. As viewpoint control regions, two actively transcribed regions from the diploid cores of chr. 4 and chr 7 were arbitrarily chosen. The coordinates of all viewpoints used for virtual 4C analyses are listed in Supplementary Information sheet 3. b, VEX1myc chromatin immunoprecipitation followed by next generation sequencing (ChIP-seq) analysis. Top panel, metagene plot for tandem genes compared to randomly selected genes with no paralogues. Lower left, heat map of tandem genes. Lower right, randomly selected genes with no paralogues.
Supplementary Material
Acknowledgements
We thank the Dundee Imaging Facility and J. Rouse for access to the Zeiss 880 Airyscan and Leica Confocal SP8 Hyvolution microscope, respectively, and S. Alsford (London School of Hygiene & Tropical Medicine) for the SNAP42 tagging construct. We thank N. Jones (Oxford Instruments) for advice on image analysis using Imaris 9.5. We further thank R. Cosentino and all members of the Siegel, Ladurner, Meissner and Boshart labs for valuable discussion, T. Straub (Core facility Bioinformatics, BMC) for providing server space and help with the data analysis, the Core Unit Systems Medicine, University of Würzburg for NGS. We also thank the reviewers for insightful comments and suggestions, which added value to the work. The work was funded by a Wellcome Trust Investigator Award to D.H. [100320/Z/12/Z], by the German Research Foundation [SI 1610/3-1 and 213249687 – SFB 1064], the Center for Integrative Protein Science (CIPSM) and by an ERC Starting Grant [3D_Tryps 715466] to T.N.S.. The University of Dundee Imaging Facility is supported by the MRC Next Generation Optical Microscopy award [MR/K015869/1]. L.S.M.M. was supported by a grant of the German Excellence Initiative to the Graduate School of Life Science, University of Würzburg.
Footnotes
Author contributions
Experiments were designed by J.F., V.L., L.S.M.M., D.H. and T.N.S. and carried out by J.F., V.L. and L.S.M.M., unless indicated otherwise. The initial observation that the active VSG gene interacts with the SL-RNA locus was made by L.S.M.M. based on Hi-C assays. The initial observation that VEX1 interacts with the SL-RNA locus was made by L.G. based in IFAs and ChIP-seq. RNAi, chemical inhibition, FISH and super resolution and other microscopy experiments were performed and analyzed by J.F. Hi-C experiments were performed by V.L. and L.S.M.M. and computational analysis was carried out by B.G.B., V.L. and L.S.M.M. RNA-seq experiments were performed by V.L.; data analysis was carried out by V.L. and B.G.B. ChIP-seq data analysis was carried out by S.H. Funding was acquired by D.H. and T.N.S. The work was supervised by D.H. and T.N.S. The manuscript was written by J.F., V.L., D.H. and T.N.S. and edited by all other co-authors.
Competing interests
The authors declare no competing interests.
Data and materials availability
High-throughput sequencing data (Hi-C and RNA-Seq) generated for this study have been deposited at GitHub (https://github.com/bgbrink/PRJEB35632) and in the European Nucleotide Archive (ENA) under primary accession number PRJEB35632, respectively. Previously published ChIP-seq and RNA-Seq data that were used for this study are publicly available at the European Nucleotide Archive www.ebi.ac.uk/ena (accession no. PRJEB25352 and PRJEB21615, respectively). Processed data and results are available under https://doi.org/10.5281/zenodo.3628212. All statistical analysis and unprocessed western blots are provided in the Source Data files.
Code availability
Code and processed data generated for this study have been deposited at GitHub (https://github.com/bgbrink/PRJEB35632) and Zenodo https://doi.org/10.5281/zenodo.3628212.
References
- 1.Deitsch KW, Lukehart SA, Stringer JR. Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol. 2009;7:493–503. doi: 10.1038/nrmicro2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horn D. Antigenic variation in African trypanosomes. Mol Biochem Parasitol. 2014;195:123–129. doi: 10.1016/j.molbiopara.2014.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bashkirova E, Lomvardas S. Olfactory receptor genes make the case for inter-chromosomal interactions. Curr Opin Genet Dev. 2019;55:106–113. doi: 10.1016/j.gde.2019.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dekker J, Mirny L. The 3D genome as moderator of chromosomal communication. Cell. 2016;164:1110–1121. doi: 10.1016/j.cell.2016.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schoenfelder S, Fraser P. Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet. 2019;20:437–455. doi: 10.1038/s41576-019-0128-0. [DOI] [PubMed] [Google Scholar]
- 6.Clayton C. Regulation of gene expression in trypanosomatids: living with polycistronic transcription. Open Biol. 2019;9 doi: 10.1098/rsob.190072. 190072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller LSM, et al. Genome organization and DNA accessibility control antigenic variation in trypanosomes. Nature. 2018 doi: 10.1038/s41586-018-0619-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Aresta-Branco F, Erben E, Papavasiliou FN, Stebbins CE. Mechanistic similarities between antigenic variation and antibody diversification during Trypanosoma brucei infection. Trends Parasitol. 2019;35:302–315. doi: 10.1016/j.pt.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Kassem A, Pays E, Vanhamme L. Transcription is initiated on silent variant surface glycoprotein expression sites despite monoallelic expression in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2014;111:8943–8948. doi: 10.1073/pnas.1404873111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vanhamme L. Differential RNA elongation controls the variant surface glycoprotein gene expression sites of Trypanosoma brucei. Mol Microbiol. 2000;36:328–340. doi: 10.1046/j.1365-2958.2000.01844.x. [DOI] [PubMed] [Google Scholar]
- 11.Michaeli S. Trans-splicing in trypanosomes: machinery and its impact on the parasite transcriptome. Future Microbiol. 2011;6:459–474. doi: 10.2217/fmb.11.20. [DOI] [PubMed] [Google Scholar]
- 12.Bertero A, et al. Dynamics of genome reorganization during human cardiogenesis reveal an RBM20-dependent splicing factory. Nat Commun. 2019;10:1538. doi: 10.1038/s41467-019-09483-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khanna N, Hu Y, Belmont AS. HSP70 transgene directed motion to nuclear speckles facilitates heat shock activation. Curr Biol. 2014;24:1138–1144. doi: 10.1016/j.cub.2014.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Navarro M, Gull K. A pol I transcriptional body associated with VSG mono-allelic expression in Trypanosoma brucei. Nature. 2001;414:759–763. doi: 10.1038/414759a. [DOI] [PubMed] [Google Scholar]
- 15.Budzak J, et al. Dynamic colocalization of 2 simultaneously active VSG expression sites within a single expression-site body in Trypanosoma brucei. Proc Natl Acad Sci U S A. 2019;116:16561–16570. doi: 10.1073/pnas.1905552116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chaves I, et al. Subnuclear localization of the active variant surface glycoprotein gene expression site in Trypanosoma brucei. Proc Natl Acad Sci U S A. 1998;95:12328–12333. doi: 10.1073/pnas.95.21.12328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Glover L, Hutchinson S, Alsford S, Horn D. VEX1 controls the allelic exclusion required for antigenic variation in trypanosomes. Proc Natl Acad Sci U S A. 2016;113:7225–7230. doi: 10.1073/pnas.1600344113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faria J, et al. Monoallelic expression and epigenetic inheritance sustained by a Trypanosoma brucei variant surface glycoprotein exclusion complex. Nat Commun. 2019;10:3023. doi: 10.1038/s41467-019-10823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zheng Y, Ay F, Keles S. Generative modeling of multi-mapping reads with mHi-C advances analysis of Hi-C studies. Elife. 2019;8 doi: 10.7554/eLife.38070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hury A, Goldshmidt H, Tkacz ID, Michaeli S. Trypanosome spliced-leader-associated RNA (SLA1) localization and implications for spliced-leader RNA biogenesis. Eukaryot Cell. 2009;8:56–68. doi: 10.1128/EC.00322-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueiredo LM, Janzen CJ, Cross GA. A histone methyltransferase modulates antigenic variation in African trypanosomes. PLoS Biol. 2008;6:e161. doi: 10.1371/journal.pbio.0060161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takagi Y, Sindkar S, Ekonomidis D, Hall MP, Ho CK. Trypanosoma brucei encodes a bifunctional capping enzyme essential for cap 4 formation on the spliced leader RNA. J Biol Chem. 2007;282:15995–16005. doi: 10.1074/jbc.M701569200. [DOI] [PubMed] [Google Scholar]
- 23.McNally KP, Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol Cell Biol. 1992;12:4844–4851. doi: 10.1128/mcb.12.11.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ullu E, Tschudi C. Trans splicing in trypanosomes requires methylation of the 5’ end of the spliced leader RNA. Proc Natl Acad Sci U S A. 1991;88:10074–10078. doi: 10.1073/pnas.88.22.10074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McNally KP, Agabian N. Trypanosoma brucei spliced-leader RNA methylations are required for trans splicing in vivo. Mol Cell Biol. 1992;12:4844–4851. doi: 10.1128/mcb.12.11.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Glover L, Alsford S, Horn D. DNA break site at fragile subtelomeres determines probability and mechanism of antigenic variation in African trypanosomes. PLoS Pathog. 2013;9 doi: 10.1371/journal.ppat.1003260. e1003260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Landeira D, Bart JM, Van Tyne D, Navarro M. Cohesin regulates VSG monoallelic expression in trypanosomes. J Cell Biol. 2009;186:243–254. doi: 10.1083/jcb.200902119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang X, Figueiredo LM, Espinal A, Okubo E, Li B. RAP1 is essential for silencing telomeric variant surface glycoprotein genes in Trypanosoma brucei. Cell. 2009;137:99–109. doi: 10.1016/j.cell.2009.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kornblihtt AR, de la Mata M, Fededa JP, Munoz MJ, Nogues G. Multiple links between transcription and splicing. Rna. 2004;10:1489–1498. doi: 10.1261/rna.7100104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiszbein A, Krick KS, Begg BE, Burge CB. Exon-mediated activation of transcription starts. Cell. 2019;179:1551–1565. doi: 10.1016/j.cell.2019.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.De Lange T, et al. Tandem repetition of the 5’ mini-exon of variant surface glycoprotein genes: a multiple promoter for VSG gene transcription? Cell. 1983;34:891–900. doi: 10.1016/0092-8674(83)90546-9. [DOI] [PubMed] [Google Scholar]
- 32.Hnisz D, Shrinivas K, Young RA, Chakraborty AK, Sharp PA. A phase separation model for transcriptional control. Cell. 2017;169:13–23. doi: 10.1016/j.cell.2017.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo YE, et al. Pol II phosphorylation regulates a switch between transcriptional and splicing condensates. Nature. 2019;572:543–548. doi: 10.1038/s41586-019-1464-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li X, Fu XD. Chromatin-associated RNAs as facilitators of functional genomic interactions. Nat Rev Genet. 2019;20:503–519. doi: 10.1038/s41576-019-0135-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hondele M, et al. DEAD-box ATPases are global regulators of phase-separated organelles. Nature. 2019;573:144–148. doi: 10.1038/s41586-019-1502-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Alsford S, Kawahara T, Glover L, Horn D. Tagging a T. brucei RRNA locus improves stable transfection efficiency and circumvents inducible expression position effects. Mol Biochem Parasitol. 2005;144:142–148. doi: 10.1016/j.molbiopara.2005.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover L, et al. Genome-scale RNAi screens for high-throughput phenotyping in bloodstream-form African trypanosomes. Nat Protoc. 2015;10:106–133. doi: 10.1038/nprot.2015.005. [DOI] [PubMed] [Google Scholar]
- 38.Alsford S, Horn D. Elongator protein 3b negatively regulates ribosomal DNA transcription in african trypanosomes. Mol Cell Biol. 2011;31:1822–1832. doi: 10.1128/MCB.01026-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Berriman M, et al. The genome of the African trypanosome Trypanosoma brucei. Science. 2005;309:416–422. doi: 10.1126/science.1112642. [DOI] [PubMed] [Google Scholar]
- 40.Becker M, et al. Isolation of the repertoire of VSG expression site containing telomeres of Trypanosoma brucei 427 using transformation-associated recombination in yeast. Genome Res. 2004;14:2319–2329. doi: 10.1101/gr.2955304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hertz-Fowler C, et al. Telomeric expression sites are highly conserved in Trypanosoma brucei. PLoS One. 2008;3 doi: 10.1371/journal.pone.0003527. e3527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolev NG, Ramey-Butler K, Cross GA, Ullu E, Tschudi C. Developmental progression to infectivity in Trypanosoma brucei triggered by an RNA-binding protein. Science. 2012;338:1352–1353. doi: 10.1126/science.1229641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rutherford K, et al. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 46.Gu Z, Gu L, Eils R, Schlesner M, Brors B. circlize Implements and enhances circular visualization in R. Bioinformatics. 2014;30:2811–2812. doi: 10.1093/bioinformatics/btu393. [DOI] [PubMed] [Google Scholar]
- 47.Ramirez F, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165. doi: 10.1093/nar/gkw257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siegel TN, Hekstra DR, Cross GA. Analysis of the Trypanosoma brucei cell cycle by quantitative DAPI imaging. Mol Biochem Parasitol. 2008;160:171–174. doi: 10.1016/j.molbiopara.2008.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Woodward R, Gull K. Timing of nuclear and kinetoplast DNA replication and early morphological events in the cell cycle of Trypanosoma brucei. J Cell Sci. 1990;95(Pt 1):49–57. doi: 10.1242/jcs.95.1.49. [DOI] [PubMed] [Google Scholar]
- 50.Park JH, Jensen BC, Kifer CT, Parsons M. A novel nucleolar G-protein conserved in eukaryotes. J Cell Sci. 2001;114:173–185. doi: 10.1242/jcs.114.1.173. [DOI] [PubMed] [Google Scholar]
- 51.Barlow AL, Macleod A, Noppen S, Sanderson J, Guerin CJ. Colocalization analysis in fluorescence micrographs: verification of a more accurate calculation of Pearson’s correlation coefficient. Microsc Microanal. 2010;16:710–724. doi: 10.1017/S143192761009389X. [DOI] [PubMed] [Google Scholar]
- 52.Schindelin J, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kraus AJ, Brink BG, Siegel TN. Efficient and specific oligo-based depletion of rRNA. Sci Rep-Uk. 2019;9:12281. doi: 10.1038/S41598-019-48692-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aslett M, et al. TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res. 2010;38:D457–D462. doi: 10.1093/nar/gkp851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wedel C, Forstner KU, Derr R, Siegel TN. GT-rich promoters can drive RNA pol II transcription and deposition of H2A.Z in African trypanosomes. Embo J. 2017;36:2581–2594. doi: 10.15252/embj.201695323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High-throughput sequencing data (Hi-C and RNA-Seq) generated for this study have been deposited at GitHub (https://github.com/bgbrink/PRJEB35632) and in the European Nucleotide Archive (ENA) under primary accession number PRJEB35632, respectively. Previously published ChIP-seq and RNA-Seq data that were used for this study are publicly available at the European Nucleotide Archive www.ebi.ac.uk/ena (accession no. PRJEB25352 and PRJEB21615, respectively). Processed data and results are available under https://doi.org/10.5281/zenodo.3628212. All statistical analysis and unprocessed western blots are provided in the Source Data files.
Code and processed data generated for this study have been deposited at GitHub (https://github.com/bgbrink/PRJEB35632) and Zenodo https://doi.org/10.5281/zenodo.3628212.