Abstract
Primary cilia are microtubule-based organelles that are important for signaling and sensing in eukaryotic cells. Unlike the thoroughly studied motile cilia, the three-dimensional architecture and molecular composition of primary cilia are largely unexplored. Yet, studying these aspects is necessary to understand how primary cilia function in health and disease. We developed an enabling method for investigating the structure of primary cilia isolated from MDCK-II cells at molecular resolution by cryo-electron tomography. We show that the textbook ‘9 + 0’ arrangement of microtubule doublets is only present at the primary cilium base. A few microns out, the architecture changes into an unstructured bundle of EB1-decorated microtubules and actin filaments, putting an end to a long debate on the presence or absence of actin filaments in primary cilia. Our work provides a plethora of insights into the molecular structure of primary cilia and offers a methodological framework to study these important organelles.
Cilia are important organelles for most eukaryotic cells with critical motile and sensory functions1–3. These microtubulebased organelles are exposed to the extracellular environment and act as signal transducers and/or actuators. Primary cilia have fundamental roles in development and homeostasis; genetic disorders that affect their assembly, structure and function result in a plethora of diseases4,5.
Cilia are traditionally classified by their ability or inability to beat as motile or nonmotile cilia. Motile cilia contain a ninefold symmetric arrangement of microtubule doublets around a central pair of microtubules, giving rise to the so-called ‘9+2’ conformation. In nonmo-tile cilia, the central pair is absent and their axonemal configuration is therefore referred to as ‘9+0’ Motile cilia are mostly known for their beating activity6, which allows cells to swim or move fluids over tissues4,5. Motile cilia have been extensively studied using biochemistry, genetics, light and electron microscopy7. This was facilitated by the ease of use of several model organisms such as Chlamydomonas 8–10, sea urchins8,11 and Tetrahymena 8, which provide large numbers of motile cilia with simple deciliation protocols12. Structural studies of motile cilia have been revolutionized particularly by the advent of cryo-electron microscopy (cryo-EM)13 and subtomogram averaging (StA). Cryo-electron tomography (cryo-ET) has enabled the unambiguous localization of protein complexes within motile cilia and the visualization of their structure and conformational changes required for ciliary activity. This has provided many insights into the mechanisms underlying their function10,14–16.
In stark contrast to these success stories, we have little mechanistic understanding of how nonmotile primary cilia perform their functions. Proteomic data of primary cilia are available17,18 and fluorescence imaging has been used to validate protein localization and dynamics,19. EM images of chemically fixed primary cilia show the lack of distinctive elements, such as dynein arms, radial spokes and a central pair complex, which are typical of motile cilia20,21. Thus, primary cilia have long been falsely regarded as simplified versions of motile cilia. Some cilia, such as the ones of Caenorhabditis elegans and IMCD-3 cells, have been reported to show noncanonical axonemal structures with microtubule doublets in the axoneme core20–22. This suggests that the canonical structure of the cilium might be modified in different cell types according to the specific function performed by the cilium. However, high-resolution information on the molecular structure and composition of primary cilia was thus far unobtainable. This is because sample preparation of primary cilia for cryo-ET was not feasible. Although there are protocols for primary cilia isolation23,24, they are not suitable for cryo-ET because they destroy the structural details. We reasoned that this methodological gap needed to be addressed to uncover the molecular mechanisms that govern the various functions of primary cilia. Primary cilia perform fundamental roles in cells, which are known to be important for human health. They are signaling hubs for photoreception25 and olfaction22 and play key roles in multiple developmental pathways26,27 (for example, Hedgehog or Wnt28,29). They also act as flow and chemo-sensors, for example, in the collecting ducts of renal epithelia, where they project into the lumen of the duct and sense bulk filtrate physicochemical properties and flow30,31. When these primary cilia are defective, cell growth and proliferation become unregulated, resulting in one of the many possible ciliopathies4,5 known as polycystic kidney disease32,33.
To enable cryo-ET studies of primary cilia in all these contexts, we present a method that provides the first molecular resolution cryo-EM reconstructions of primary cilia. Our sample preparation method enables cryo-ET investigations of primary cilia from cultured cell monolayers by mechanical isolation directly onto an EM grid. By StA of axonemal components of primary cilia from tissue culture cells, we revealed remarkable differences between the structures of motile and primary cilia, and the presence of previously undescribed axonemal structures.
Results
The MDCK-II primary cilium axoneme does not conform to the ‘9+0’ microtubule arrangement
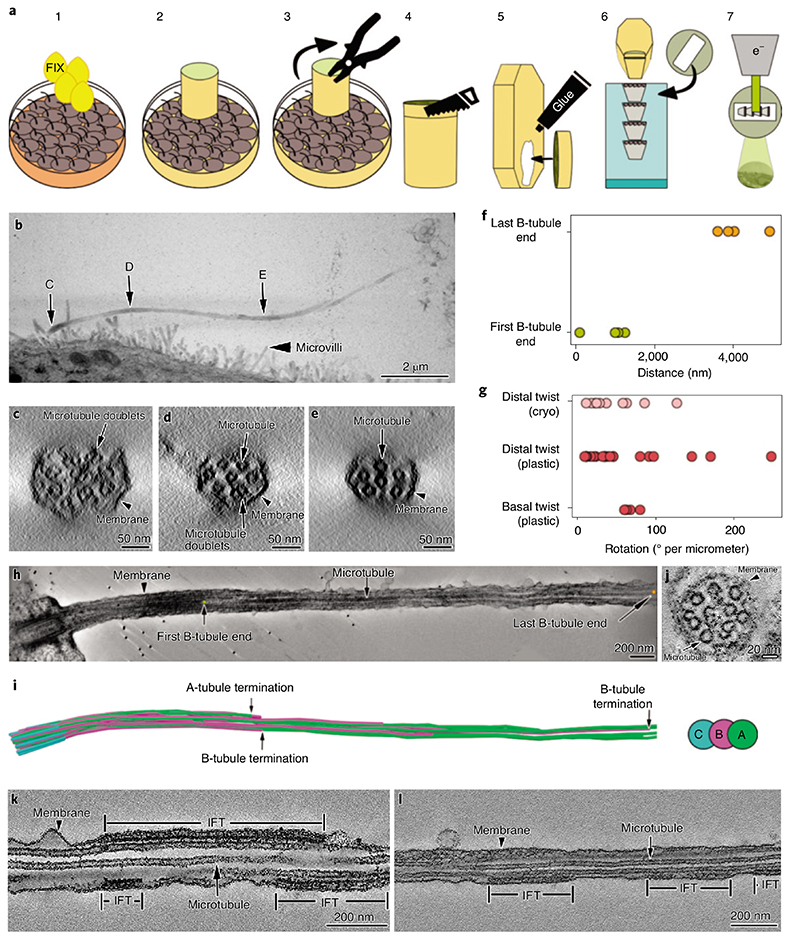
To gain an overview of the axonemal architecture of the primary cilium, we performed transmission ET on serial sections of resin-embedded MDCK-II cells (Fig. 1a, steps 1-7 and Fig. 1b). We found that axonemal microtubules did not arrange in the canonical ‘9 + 0’ configuration. While moving from the basal body toward the tip, the 9 + 0 configuration was initially present but already within the first micrometer of the axoneme and microtubules migrated sequentially toward the center of the axoneme (Fig. 1c–e and Supplementary Video 1). Between 2 and 5 μm distal to the basal body most B-tubules terminated (Fig. 1f, h,i). Therefore, the distal three-quarters of the axoneme were composed solely of singlets (Fig. 1e,f,h) that terminated at different distances from the basal body. This was accompanied by a concomitant reduction in axonemal diameter since the base always showed a consistent diameter of 220 nm whereas the distal regions shrank to about half the basal thickness (Fig. 1b–e). To further characterize the three-dimensional (3D) organization of primary cilia axonemal microtubules, we measured the axonemal twist, defined as the collective rotation of the microtubules around the cilium’s central axis (Fig. 1g). Different from motile cilia, which have a highly consistent organization of their microtubules and show no twist while at rest8, primary cilia axonemes do not have a defined geometry and are twisted. There was a consistent right-handed rotation of the microtubule doublets along the ciliary baso-proximal region of approximately 56° per micrometer (n=4, mean=56.4,s.d.=7.3) (Fig. 1g, Supplementary Table 1 and Supplementary Video 2). However, the axonemal twist varied widely along the distal segments of the cilium where only singlets are present (Fig. 1g and Supplementary Table 1). Moreover, we observed that the microtubule twist was decoupled from the axonemal twist (Supplementary Video 3), suggesting the absence of a strong intermicrotubule cross-linking. Therefore, the main part of the primary cilium axoneme appeared as a loosely organized microtubule bundle rather than a consistently ordered structure as in the case of motile cilia.
Fig. 1. I Room temperature ET of MDCK-II primary cilia.
a, Description of the steps for resin-embedding and serial sectioning of MDCK-II monolayers. b, Montage of projections from five serial sections covering the entire primary cilium. C, D and E are the areas of the cilium that were digitally sectioned to obtain the images shown in panels c,d and e, respectively. c-e, Representative proximodistal tomographic slices from the same cilium shown in b, showing the axonemal organization of microtubule doublets and singlets. c, Cross-section of the cilium proximal segment showing that 9-fold symmetrical arrangement of the microtubule doublets is already lost. d,e, The number of microtubule doublets (d) and microtubules singlets (e) decreases in the distal part of the cilium. f, Distance measured from the distal side of the basal body to the first and last B-tubule terminations. g, Axonemal twist measured at the basal and distal regions of the cilium. H,i, Tomographic section along a cilium (H), and its microtubule segmentation (i), showing representative positions of first and last B-tubule terminations. J, Ciliary cross-section showing the variety of axonemal structures other than microtubules. k,l, Longitudinal sections through cilia containing IFT train-like particles sandwiched between the ciliary membrane and microtubule singlets. Asterisks indicate locations of unidentified densities.
Analysis of our tomograms also revealed the presence of electron-dense material along the ciliary axis, sandwiched between the microtubules and the plasma membrane (Fig. 1k,l), reminiscent of the intraflagellar transport (IFT) trains found in motile cilia34; these are specialized bidirectional transport machines required for ciliary assembly, moving components into and out of the cilium35. Further characterization of IFT-related and other unidentified electron densities (Fig. 1j) could not be performed by room temperature EM due to insufficient resolution and other methodological limitations. This was additional motivation to develop a method to enable cryo-ET studies of primary cilia.
Cryo-peel off method to prepare primary cilia fit for cryo-electron tomography
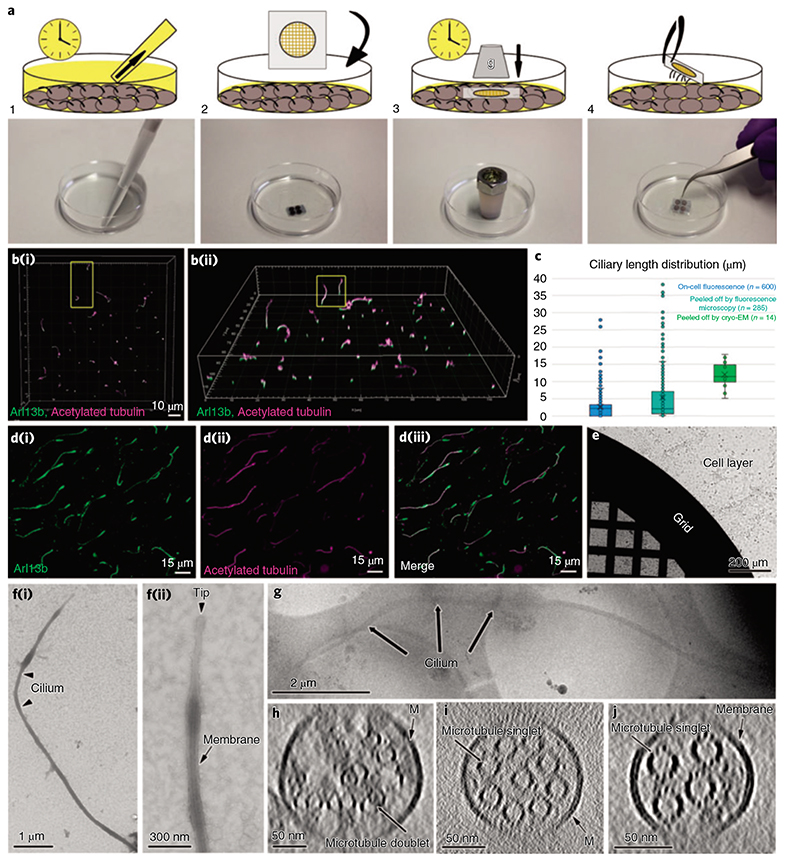
While motile cilia are routinely imaged with cryo-ET, so far this was not possible for primary cilia. Inspired by a previously published method for mechanical isolation of primary cilia24, we found a way that enables sample preparation for cryo-ET (Fig. 2a, steps 1-4). Briefly, cells were grown on Petri dishes to total confluence for about 3 weeks and starved up to 2d to induce cilia elongation (Fig. 2b, steps 1 and 2). Then, a poly-L-lysine-treated surface was pressed against the apical membrane and retrieved together with cilia that adhered to it (see Methods for a detailed description).
Fig. 2. I Cryo-peel off: a method to prepare primary cilia for cryo-ET.
a, Description of the steps followed for the peel off of primary cilia from MDCK-II monolayers. Step 1: removal of the deciliation buffer from the cell culture; step 2: application of poly-L-lysine-coated EM grids, supported by a glass slide, on the apical surface of the cell layer; step 3: pressure was applied over time on the glass slide to favor the adhesion of cilia to the EM grid; step 4: retrieval of the glass slide/EM grid from the cells’ apical surface and consequent primary cilia peel off. b, Laser scanning confocal fluorescence microscopy of the MDCK-II monolayer showing the variability of ciliary lengths after 2d of cell starvation; (b(i)), top view; (b(ii)), tilted view. c, Distribution of lengths of cilia associated with cells (blue), cilia peeled-off by fluorescence microscopy (cyan) and cilia peeled-off by cryo-EM (green). d, Immunofluorescence staining of peeled-off cilia on a glass slide, showing the colocalization of ciliary membrane (d(i)) and microtubules (d(ii) and the combined images (d(iii)).e, A Quantifoil grid on a cell monolayer during the peel off procedure as depicted in a (steps 2 and 3). f(i), Negative staining TEM of a peeled-off primary cilium; f(ii), the zoomed-in view of the ciliary tip from the same cilium shows the preservation of the ciliary membrane. g, Low-magnification cryo-EM image of a peeled-off and plunge-frozen cilium. h-J, Representative proximodistal cryo-tomographic slices of plunge-frozen cilia (h) close to the base central shaft (i) and distal segment (J).
Before the preparation of the cryo-ET samples, we characterized the overall structural integrity of the peeled-off cilia by light microscopy and transmission EM (TEM). For immunofluorescence imaging, glass coverslips were used as the substrate (see Methods for a detailed description) (Fig. 2d(i-iii)). Colocalization of acetylated tubulin and Arl13b, which are markers for axonemal microtubules and ciliary membrane, respectively, showed that peeled-off cilia preserved their membranes (Fig. 2d(i-iii)). The average length of peeled-off cilia was comparable to the one of cilia still connected to cells (Fig. 2c and Supplementary Data 1). For TEM, we used EM grids as the substrate (Fig. 2e) and performed negative staining at room temperature (Fig. 2f(i,ii)). Electron micrographs showed intact membranated cilia without obvious morphological alterations, with consistent lengths and tolerable levels of contamination from cell apical material (Fig. 2f(i,ii)). Having verified that peeled-off primary cilia maintain overall structural integrity, we used cryo-EM grids as the substrate for cryo-peel off. Contact force and time were optimized to reduce cell debris contamination and still allow for the capture of cilia. After peel off, grids were plunge-frozen and about 20 tomography tilt series were obtained from the segments of cilia lying over carbon holes (Fig. 2g). Cilia on cryo-EM grids were on average longer and with a narrower length distribution compared to cilia on cells or after peel off on glass slides (Fig. 2c and Supplementary Data 1). This might indicate that short assembling cilia were not caught by the peel off method on cryo-EM grids. However, visualization of short cilia at low magnification in cryo-EM is challenging; thus, short cilia that in fact were present on the grid might have escaped our search. Regardless, the cryo-peel off method enabled cryo-ET imaging of properly preserved primary cilia.
While the ET reconstruction of resin-embedded samples allowed the visualization of full-length cilia, only selected segments of the structure were imaged by cryo-ET. To understand which areas of the cilium these segments corresponded to, we compared the cryo-tomograms with the room temperature full cilia reconstructions. Only a small subset of cryo-tomograms showed the presence of microtubule doublets (Fig. 2h). This might be caused by the predisposition of MDCK-II cilia to break at a weak spot where the axoneme transitions from microtubule doublets to singlets. Additionally, most axonemes contained fewer than nine microtubule singlets (Fig. 2i,j and Supplementary Video 4), which formed heterogeneous bundles with diameters below 220 nm. These features suggest that most regions imaged by cryo-ET corresponded to centrodistal segments of primary cilia. Our (cryo-)ET investigations confirmed that the unconventional 3D organization of the primary cilium axoneme is, for the most part, formed by a loosely structured bundle of microtubule singlets. This architecture raises several questions regarding ciliary assembly and IFT.
Anterograde IFT trains of primary cilia might move along A-microtubules
In Chlamydomonas, anterograde and retrograde IFT trains are spatially segregated as they move exclusively along the B- and A-tubules of the microtubule doublet, respectively34. This spatial segregation enables efficient transport by avoiding collisions between trains that move in opposite directions. The fact that primary cilia are in large parts ‘singlet only’ raises questions about the way anterograde and retrograde IFT coordinates their collision-free motion. Similar to motile cilia, the IFT of the primary cilium comprises about 23 adapter proteins that organize into 2 large complexes, namely IFT-A and IFT-B. Kinesin and dynein motors are responsible for the anterograde and retrograde directions, respectively. IFT has been visualized in primary cilia using fluorescence microscopy36, but it is unclear how anterograde trains are structured and how they proceed from the end of the B-tubules into the singlet-only area to reach the ciliary tip.
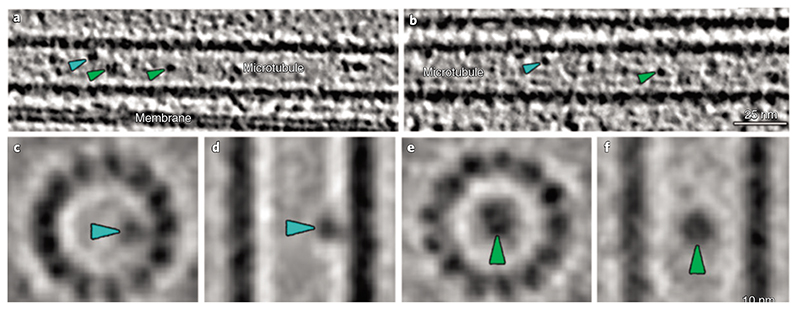
In room temperature and cryo-tomograms, we observed elongated densities in the space between microtubules and the ciliary membrane (Fig. 1k,l, Fig. 3a-c and Supplementary Videos 4 and 5). These elongated structures were as long as 900 nm and showed morphological similarities, with the IFT trains recently resolved by cryo-ET in motile Chlamydomonas cilia10. In cryo-tomograms, the IFT-like structures showed subunit repeat (approximately 6 nm) and morphology similar to those of the IFT-B complexes in the anterograde trains of Chlamydomonas (Fig. 3d,e). Thus, they could represent IFT-B components of anterograde trains in primary cilia. Additionally, particles that resembled IFT-A and IFT-dyneins were visible in the vicinity of these IFT-B-like structures (Fig. 3a,b). Using StA, we generated a 3D model of the IFT-B-like subunit, which we directly compared to the one from the Chlamydomonas anterograde trains10. The remarkable morphological similarity between these two models suggests that MDCK-II IFT-B organizes into polymers in primary cilia as in the Chlamydomonas motile cilia10. Furthermore, we consistently found anterograde-like IFT trains along microtubule singlets (A-tubules) (Fig. 3a-cand Extended Data Videos 4 and 5). All these observations seem to suggest that coordination of anterograde and retrograde trains in motile and primary cilia may be achieved in different ways.
Fig. 3. I IFT-B-like polymers are visible in cryo-ET of primary cilia.
a, Slice through a denoised tomogram of a primary cilium showing an IFT train (green frame). The train subunits show a similar repeat to the one described for the IFT-B complexes in Chlamydomonas anterograde IFT trains (d). The yellow arrowheads in a indicate a second row of particles placed between the IFT-B-like polymer and the membrane. This location is typical of IFT-A complexes in the anterograde trains of Chlamydomonas. Ring-shaped objects (blue arrowheads) were also present in the area expected for dynein motor cargoes. IFT-A-like (yellow arrowheads) and IFT dynein-like particles (blue arrowheads) appeared partially dissociated from the IFT-B-like polymer, indicating that the structure of the polymer might be altered during the peel off procedure. b, Slice through the same IFT train shown in a after rotating the tomogram by 90° around the microtubule long axis. c, 3D visualization of the relative position of microtubule singlets and two anterograde-like IFT trains from a cryo-tomogram of a primary cilium. d, Subtomogram-averaged model showing the repeat of IFT-B, IFT-A and IFT dynein in the anterograde IFT trains in Chlamydomonas (modified from Jordan et al.10). e, StA of IFT-B-like particle repeat from MDCK-II primary cilia. Masked cross-correlation coefficient between the structures shown in e and IFT-B-like in d was approximately 0.59.
An EB1-like protein decorates A-tubules in primary cilia
Axonemal microtubules in motile cilia show prominent periodical decoration of their inner and outer surfaces by several protein complexes other than IFT. Our cryo-ET investigation showed that large, periodic macromolecular complexes are not present on primary cilia microtubules; therefore, confirming that radial spokes, axonemal dyneins and other large microtubule-associated proteins are not components of primary cilia. Instead, we found the sporadic presence of distinct particles within the lumen of primary cilia microtubules (Figs. 4a,b and 5a). Some of these microtubule inner proteins (MIPs) were associated with the internal wall of the microtubules (Fig. 4c, d), while others seemed to be floating in the microtubule lumen (Fig. 4e,f). In both cases, their arrangement differed from the periodic MIP decoration typical of motile cilia8,13,37,38. The lack of an obvious periodical decoration by MIPs and large axonemal complexes raises the question about how the stability of the primary cilium axoneme is maintained.
Fig. 4. Proteins are present in the microtubule lumen in MDCK-II primary cilia (MIPs).
a,b, Cryo-electron tomograms of primary cilia show MIPs associated with the internal walls of ciliary microtubules (blue arrowheads) and ‘floating’ in the lumen of the microtubule (green arrowheads).c,d, Subtomogram-averaged structure of microtubule wall-associated MIPs. c, Digital slice perpendicular to the microtubule axis. d, Digital slice parallel to the microtubule axis. e,f, Subtomogram-averaged structure of MIPs located in the center of the microtubule. e, Digital slice perpendicular to the microtubule axis. f, Digital slice parallel to the microtubule axis.
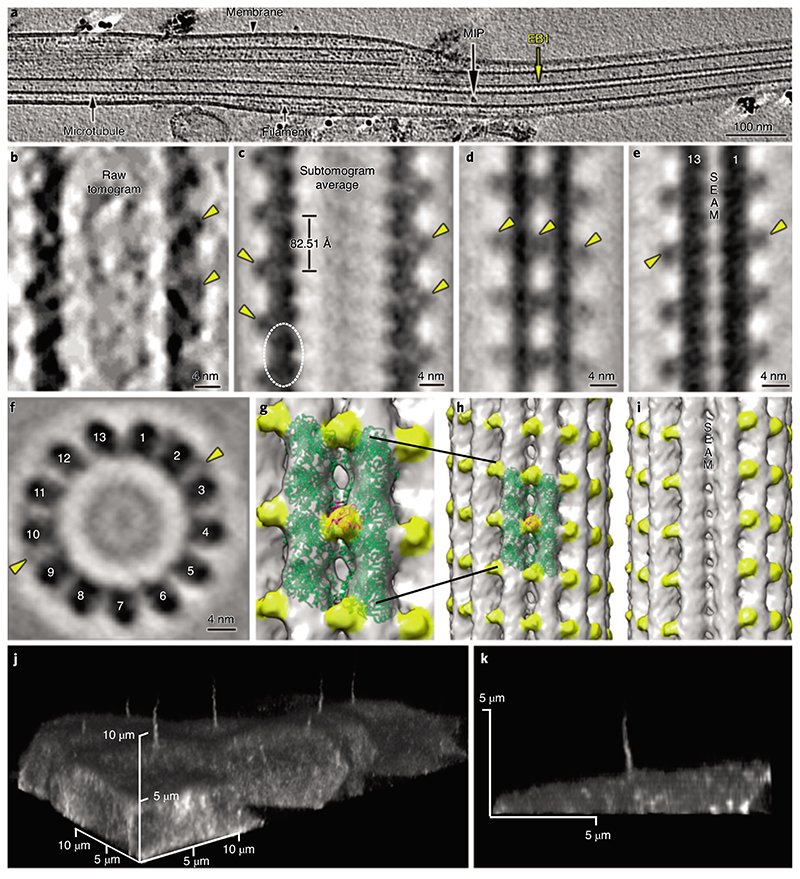
Fig. 5. I Cryo-ET of primary cilia shows decorations of microtubule singlets by EB1.
a, Representative longitudinal tomography section of a plunge-frozen primary cilium showing EB1-decorated microtubule singlets. Also visible are some MIPs and filaments. b, Close-up view of the EB1 microtubule singlet decoration as seen in a raw tomogram. c-i, Longitudinal slice along a subtomogram-averaged model (c) of the electron density map of microtubule singlets from primary cilia showing the localization (dashed line) and repeat of the tubulin dimer and the associated EB1 pattern. The digital slice (d) and isosurface visualization (h) show the localization of EB1 between protofilaments, recapitulating a microtubule B-type lattice. The digital slice (e) and isosurface visualization (i) show the absence of EB1 along the microtubule seam. f, Orthogonal slice showing 13 protofilaments and some EB1 particles. g, Zoom-in on two protofilaments of the microtubule shown in h. J,k, Live confocal microscopy of MDCK-II cells stably expressing mNeonGreen-tagged EB1 (j). The EB1 signal is visible in the cilium and cytoplasm. Lateral view of a single cilium extending from the apical surface of a MDCK-II cell (k). The continuous EB1 signal is stronger at the base and progressively decreases towards the tip, probably because of the reduction in the number of microtubules.
To address this question, we inspected the outer surface of the primary cilium microtubule singlet, searching for any periodic decorations. We found a periodic decoration bysmall globular densities,similarly sized to tubulin monomers (Fig. 5a,b), with a periodicity matching that of tubulin dimers (mean = 82.51 Å, s.d. = 1.5 Å) (Fig. 5a,b, Extended Data Fig. 1b and Supplementary Video 6). Using StA, we resolved a microtubule singlet at about 18.5 Å (Fourier shell correlation (FSC) coefficient = 0.143; Fig. 5c-i and Extended Data Fig. 1a) and observed that the decorating protein is placed every other tubulin tetrameric contact along the microtubule length, that is, binding to 4 tubulin monomers every 82.51 Å. The pattern of the decorating protein recapitulated the handedness of the underlying 13-protofilament B-type lattice (Fig. 5g–i). The protein decoration was not present at the microtubule seam (Fig. 5e,i), suggesting that its docking requires a particular tubulin arrangement with a longitudinal shift of about 9 Å between protofilaments. Our model also suggests a specific interaction between this protein and four distinct tubulin dimers (Fig. 5c). However, this observation is based on minute differences in densities between what appear to be intradimeric and interdimeric spaces and must be validated in the future with higher-resolution models. The diameter of the decorating protein is around 25 Å and its volume could accommodate a polypeptide of approximately 25 kDa. Recent proteomic analysis of primary cilia from IMCD-3 cells using APEX technology for proximity labeling revealed candidate proteins associated with microtubules18. We reasoned that among all of them only EB1 (encoding gene: Maprel; molecular weight = 30 kDa) showed a molecular weight and microtubule binding regime coherent with our model (Fig. 5g,h). In line with this hypothesis, we know that in the in vitro reconstituted microtubule-Mal3 complex (Protein Data Bank no. 5MJS), Mal3 (EB1 homolog in yeast) is absent at the seam, shows a periodicity of 82.09 Å and is located at tetrameric contacts involving 4 tubulin dimers39. Since these features match the ones observed in our model, we fitted a 20-Å resolution map of the tubulin-Mal3 complex to our average (Fig. 5g,h), showing a high cross-correlation coefficient (approximately 0.9). To validate the ciliary localization of EB1, we generated an MDCK-II cell line stably expressing mNeonGreen-tagged EB1 (mNG-EB1). Confocal microscopy confirmed the presence of EB1 along the length of the cilium (Fig. 5j,k and Supplementary Video 7). The mNG-EB1 signal appears more intense at the base and progressively decreases toward the tip, probably because of the reduced number of microtubules toward the ciliary tip. This suggests that EB1 decorates both microtubule doublets and singlets and that the amount of ciliary EB1 is therefore proportional to the number of microtubules present in a given segment of the axoneme. The localization of EB1 in both isolated and in situ primary cilia was additionally confirmed by immunofluorescence microscopy (Extended Data Fig. 2). These results show that, in primary cilia, EB1 decoration is not restricted to the microtubule tips but extends along most of the ciliary shaft, suggesting that EB1 contributes to microtubule stabilization.
Filamentous actin as a new axonemal component of MDCK-II primary cilia
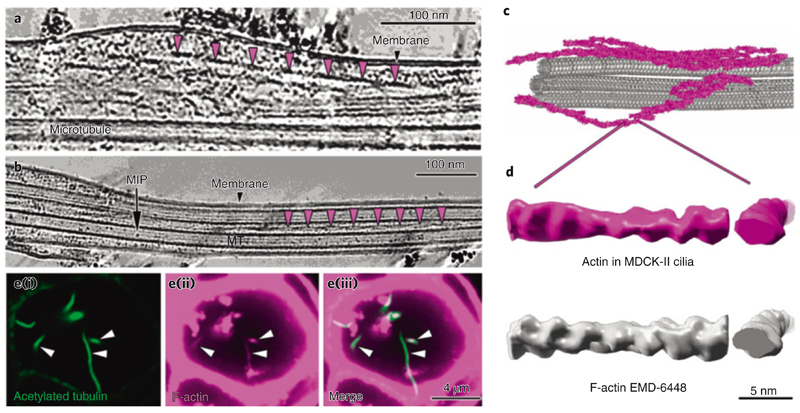
During our search for other structures that might contribute to the stability of the axoneme of primary cilia, we observed the presence of periodical helical and filamentous structures. These structures appeared around and intertwined with EB1-decorated microtubules (Figs. 5a and 6a-c), had lengths ranging between 120 and 375 nm and an approximate pitch of 75 nm (Fig. 6a,b and Supplementary Video 6). The periodic nature of the filaments indicated that they might be a polymer and their shape and repeat suggested that they could be filamentous actin. To address this exciting hypothesis, we used StA and obtained a higher resolved 3D model of this new filamentous structure (Fig. 6d), which we compared by electron density map fitting to the structure of F-actin (Electron Microscopy Data Bank no. EMDB-6448). Their comparison showed a high similarity between the two models (Fig. 6d and Supplementary Video 8), with a cross-correlation coefficient of about 0.89, strongly suggesting that the investigated filaments are indeed F-actin. We additionally confirmed this finding by immunofluorescence confocal microscopy, which revealed colocalization of acetylated tubulin and F-actin (phalloidin) along the primary cilia of MDCK-II cysts (Fig. 6e(i)-(iii)). Together these results show that actin filaments are constitutive components of the axoneme in the primary cilia of MDCK-II cells.
Fig. 6. I Primary cilia contain actin filaments.
a,b, Slice through a denoised tomogram of a primary cilium showing numerous actin filaments in the space between the axoneme and the membrane. The repeats of actin filament half-twists are indicated by the magenta arrowheads in a and b. b, Actin filaments are also found in between microtubule singlets. c, 3D visualization of microtubule singlets and some actin filaments from the tomogram in a. d, Comparison of a subtomogram-averaged model of F-actin from the primary cilium (magenta) with a deposited structure (EMDB-6448) (left, longitudinal view; right, cross view). e, Immunofluorescence microscopy of MDCK-II cysts showing the colocalization of acetylated tubulin (green) (i) and F-actin (magenta) (ii) in primary cilia. The merged images are shown in (iii).
Discussion
In this study, we showed that the application of our new cryo-peel off method provides unprecedented insights into the molecular structure of primary cilia. We discovered unexpected differences in the molecular composition and organization of the axoneme of primary cilia compared to motile cilia. We showed that the primary cilium of MDCK-II cells does not conform to the ‘9+0’ axonemal microtubule arrangement. The peripheral ring of nine microtubule doublets is disrupted within micrometers from the transition zone as some microtubule doublets migrate to the center of the axo-neme. These findings agree with previous ultrastructural studies of IMCD-3 primary cilia from chemically fixed cells20,21. We additionally discovered that the major part of the axoneme is composed of a reduced number of microtubule singlets. According to a number of EM investigations of resin-embedded and sectioned samples, the microtubular organization we described for the MDCK-II cilium is probably relevant for the function of primary cilia in other mammalian tissues also20,21,40,41.
The remarkable difference between the 3D organization of axonemal microtubules in motile and primary cilia poses natural questions about the adaptation of ciliary processes, such as assembly, function and IFT. We have previously shown that in the motile cilia of Chlamydomonas, IFT makes differential use of A-tubules and B-tubules for retrograde and anterograde transport, respectively34. In primary cilia, IFT has been imaged by fluorescence microscopy while traversing the whole length of the cilium36, the cytoskeleton of which, as we showed, mostly contains A-tubule singlets. Our cryo-ET investigations provide the first data showing IFT trains in mammalian primary cilia. Since anterograde train-like structures were observed along A-tubules, we speculate that in primary cilia anterograde IFT uses microtubule singlets to travel toward the tip. Thus, we propose that the choice of microtubules by anterograde and retrograde IFT in primary cilia probably does not conform to the rules followed by motile cilia34. However, we cannot exclude a spatial segregation of anterograde and retrograde IFT in primary cilia perhaps through the establishment of direction-specific sets of microtubule singlets or even protofilaments.
The structural similarity between IFT-B-like polymers in primary cilia and the anterograde IFT-B polymers in motile cilia strongly suggests that train assembly is conserved from algae to mammals. Structural and length similarities also indicate that the gliding activity, which is typical of Chlamydomonas, is not the sole function of the polymeric nature of IFT trains. Since IFT transport also takes place in the unstructured axoneme of primary cilia, the evolution of the ninefold symmetric arrangement of axonemal microtubules does not seem to be related to IFT but might have evolved to enable the beating of motile cilia.
The ‘9 + 2’ structure of motile cilia is very robust, withstanding high mechanical loads during beating. Since primary cilia do not beat, they might not require the same degree of internal regularity and structure. The microtubule arrangement we found in the primary cilia of MDCK-II cells might be optimized for sensing extracellular fluid flows. Primary cilia regulate their bending rigidity and stiffness according to the flow regime they are exposed to42 but how this is achieved is not understood. The most common factors known to affect ciliary stiffness are tubulin modifications, nucleotide binding states, decoration by microtubule-associated proteins, microtubule cross-linkers and active force generation by molecular motors. The periodical arrangement of a large number of MIPs in motile cilia has been shown to substantially contribute to microtubule doublet stabilization43. However, microtubules in motile and primary cilia strongly differ in the number and type of MIPs11’13.
Our data show that the MIPs of the primary cilium are sparse, do not have any apparent periodicity and are more similar to particles previously identified in the microtubule lumen of neurons, astrocytes and stem cells44. Because several modifying enzymes are assumed to interact with tubulin residues lying on the inner surface of the microtubule lattice, some of the luminal proteins visible in primary cilia microtubules could be acetyltransferases and deacetylases migrating along the lumen45. However, it is unlikely that these proteins physically contribute to the mechanical stability of the microtubules. Therefore, the stabilization and tuning of the mechanical properties of long-lived primary cilium microtubules may rely on factors other than periodically arranged MIPs. Possibilities are tubulin posttranslational modifications or the EB1 microtubule decoration that we discovered in this work.
In primary cilia, EB1 has been detected by proteomic analysis18 and immunofluorescence microscopy46. In motile mammalian cilia, EB1 and EB3 were reported to localize to the ciliary tip47, while in Chlamydomonas they localize to the proximal part of the basal bodies48. Thus, EB1 is commonly thought to be associated with the microtubule +end. However, similar to our observation in primary cilia, EB1 is also found along the walls of both in vitro reconsti-tuted49 and cytoplasmic50 microtubules. The presence of an EB1 scaffold (approximately 3,500 copies of EB1 in a 6-μm long cilium) on the microtubules may stabilize specific lattice parameters, directly affecting primary cilia microtubule stiffness and stability. Accordingly, EB1 knockdown in fibroblasts results in the assembly of short ciliary stumps47,51. Despite difficulties in discerning whether this effect arises from the absence of EB1 on cytoplasmic microtubules or from a direct effect on ciliary microtubules, the assembly of stumpy cilia in EB1 mutants seems to indicate a reduction in microtubule stability. This hypothesis is supported by in vitro studies showing that the bending stiffness of the microtubule is modulated by EB1 binding in a concentration-dependent manner52.
The combination of EB1 and different tubulin-nucleotide binding states can also affect microtubule stiffness by changing tubulin lattice parameters, such as dimer repeat distance53. We measured that the tubulin dimer repeat of primary cilia microtubules is about 82.5 Å. Remarkably, this tubulin dimer repeat distance is very close to the one found in motile axonemes extracted from Chlamydomonas (approximately 82.6Å)13. This suggests that this specific lattice configuration is conserved across ciliated cells and has physiological relevance for the function of axonemal microtubules. In this context, EB1 microtubule decoration and the consequent stabilization of a specific lattice configuration is required for proper assembly and function of primary cilia.
Another possible function of the EB1 microtubule decoration in the primary cilium could be the regulation of the binding of other microtubule-associated proteins54, potentially influencing axonemal mechanics and IFT. Additionally, EB1 is known to interact in cells with proteins that link microtubules with actin filaments, such as GAS2 (refs. 54,55), G2L2 (refs. 54,55) and MACF54,56, some of which are found in primary cilia18. Therefore, EB1 may also link axonemal microtubules to the actin filaments we discovered within MDCK-II cilia.
Previous studies provided indirect and contradictory pieces of evidence regarding the presence of F-actin inside primary cilia17,18,57. Traces of actin-related proteins such as Arpc3 (ref. 17), the inverted formin FHDC1 (ref. 58), myosin17, microtubule-actin cross-linking factors18 and other actin-interacting proteins57,59 have been found. Recently, actin has been shown to form puncta at ciliary excision sites, thus being involved in cilia decapitation60. However, a direct visualization of the actin filament network in cilia has been missing. Through fluorescence microscopy and cryo-ET analysis, we clearly showed the presence of bundles of actin filaments underneath the ciliary membrane.
Sporadically, we observed the colocalization of actin filaments and extracellular vesicles that appear to be tethered to the ciliary membrane (Extended Data Fig. 3). These vesicles are reminiscent of urocrine signaling exocyst-containing vesicles that were recently found attached to MDCK primary cilia61. Since actin has been linked to ciliary exocytic vesicle formation62, the filaments we observed close to vesicles could be directly involved in their formation and trafficking.
We also observed actin filaments intertwined with axonemal microtubules and not directly associated with the ciliary membrane. These filaments might also be part of the exocytic vesicle formation machinery, remaining at the center of the axoneme while being actively transported. Accordingly, it has been shown that LifeAct-stained filaments can move along the cilium to reach the tip before ciliary decapitation60. Alternatively, centrally located actin filaments might represent a constitutive component of the primary cilium axoneme, contributing to its mechanical stability and dynamics in the same way F-actin does in the cell body. Actin polymerization and ciliary growth have an antagonistic relation-ship58,63. However, whether the contradictory effects seen on ciliary dynamics upon F-actin perturbations58 result from a specific effect on ciliary actin or from the disruption of the acto-myosin cortex has not been elucidated. Therefore, the potential role of actin filaments as modulators of ciliary stiffness in the cilium require further investigations.
Our results show the suitability of the cryo-peel off method to image primary cilia from cell monolayers by cryo-ET, which allowed the identification and structural analysis of new ciliary components. Our work reveals a plethora of previously unknown facts and surprising differences between well-studied motile cilia and much less understood primary cilia. Our method promises to enable the ultra-structural analysis of genetically engineered ciliary phenotypes for the study of complex processes, such as primary ciliary assembly and function in different cell types and experimental setups. The investigative approach we describe in this study will pave the way for many additional and insightful investigations of primary cilia and in turn allow us to better understand these important organelles in animal models and humans, both in health and disease.
Supplementary Material
© 2021 Springer Nature
To order reprints, please contact:
Tel +1 212 726 9278; reprints@us.nature.com
Printed by The Sheridan Press
Footnotes
Online content
Any methods, additional references, Nature Research reporting summaries, source data, extended data, supplementary information, acknowledgements, peer review information; details of author contributions and competing interests; and statements of data and code availability are available at https://doi.org/10.1038/s41594-020-0507-4.
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Badano JL, Mitsuma N, Beales PL, Katsanis N. The ciliopathies: an emerging class of human genetic disorders. Annu Rev Genomics Hum Genet. 2006;7:125–148. doi: 10.1146/annurev.genom.7.080505.115610. [DOI] [PubMed] [Google Scholar]
- 2.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 3.Waters AM, Beales PL. Ciliopathies: an expanding disease spectrum. Pediatr Nephrol. 2011;26:1039–1056. doi: 10.1007/s00467-010-1731-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Afzelius BA. A human syndrome caused by immotile cilia. Science. 1976;193:317–319. doi: 10.1126/science.1084576. [DOI] [PubMed] [Google Scholar]
- 5.Mitchison HM, Valente EM. Motile and non-motile cilia in human pathology: from function to phenotypes. J Pathol. 2017;241:294–309. doi: 10.1002/path.4843. [DOI] [PubMed] [Google Scholar]
- 6.Satir P, Heuser T, Sale WS. A structural basis for how motile cilia beat. Bioscience. 2014;64:1073–1083. doi: 10.1093/biosci/biu180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Harris EH, Stern DB, Witman GB. The Chlamydomonas Sourcebook. 2nd. Academic Press; 2009. [Google Scholar]
- 8.Pigino G, et al. Comparative structural analysis of eukaryotic flagella and cilia from Chlamydomonas, Tetrahymena, and sea urchins. J Struct Biol. 2012;178:199–206. doi: 10.1016/j.jsb.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 9.Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T. Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol. 2009;186:437–446. doi: 10.1083/jcb.200903082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jordan MA, Diener DR, Stepanek L, Pigino G. The cryo-EM structure of intraflagellar transport trains reveals how dynein is inactivated to ensure unidirectional anterograde movement in cilia. Nat Cell Biol. 2018;20:1250–1255. doi: 10.1038/s41556-018-0213-1. [DOI] [PubMed] [Google Scholar]
- 11.Nicastro D, et al. The molecular architecture of axonemes revealed by cryoelectron tomography. Science. 2006;313:944–948. doi: 10.1126/science.1128618. [DOI] [PubMed] [Google Scholar]
- 12.Witman GB. In: Methods in Enzymology. Vallee RB, editor. Vol. 134. Academic Press; 1986. pp. 280–290. [DOI] [PubMed] [Google Scholar]
- 13.Ma M, et al. Structure of the decorated ciliary doublet microtubule. Cell. 2019;179:909–922.:e12. doi: 10.1016/j.cell.2019.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin J, Nicastro D. Asymmetric distribution and spatial switching of dynein activity generates ciliary motility. Science. 2018;360:eaar1968. doi: 10.1126/science.aar1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pigino G, Ishikawa T. Axonemal radial spokes: 3D structure, function and assembly. Bioarchitecture. 2012;2:50–58. doi: 10.4161/bioa.20394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Oda T, Yanagisawa H, Kamiya R, Kikkawa M. A molecular ruler determines the repeat length in eukaryotic cilia and flagella. Science. 2014;346:857–860. doi: 10.1126/science.1260214. [DOI] [PubMed] [Google Scholar]
- 17.Ishikawa H, Thompson J, Yates JR, III, Marshall WF. Proteomic analysis of mammalian primary cilia. Curr Biol. 2012;22:414–419. doi: 10.1016/j.cub.2012.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mick DU, et al. Proteomics of primary cilia by proximity labeling. Dev Cell. 2015;35:497–512. doi: 10.1016/j.devcel.2015.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ott C, Lippincott-Schwartz J. Visualization of live primary cilia dynamics using fluorescence microscopy. Curr Protoc Cell Biol. 2012;57 doi: 10.1002/0471143030.cb0426s57. 4.26.1-4.26.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun S, Fisher RL, Bowser SS, Pentecost BT, Sui H. Three-dimensional architecture of epithelial primary cilia. Proc Natl Acad Sci USA. 2019;116:9370–9379. doi: 10.1073/pnas.1821064116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gluenz E, et al. Beyond 9+0: noncanonical axoneme structures characterize sensory cilia from protists to humans. FASEB J. 2010;24:3117–3121. doi: 10.1096/fj.09-151381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doroquez DB, Berciu C, Anderson JR, Sengupta P, Nicastro D. A high-resolution morphological and ultrastructural map of anterior sensory cilia and glia in Caenorhabditis elegans. Elife. 2014;3:e01948. doi: 10.7554/eLife.01948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ishikawa H, Marshall WF. In: Methods in Enzymology. Marshall WF, editor. Vol. 525. Academic Press; 2013. pp. 311–325. [DOI] [PubMed] [Google Scholar]
- 24.Huang B, Masyuk T, LaRusso N. In: Methods in Cell Biology. Sloboda RD, editor. Vol. 94. Academic Press; 2009. pp. 103–115. [DOI] [PubMed] [Google Scholar]
- 25.Bujakowska KM, Liu Q, Pierce EA. Photoreceptor cilia and retinal ciliopathies. Cold Spring Harb Perspect Biol. 2017;9:a028274. doi: 10.1101/cshperspect.a028274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yuan S, Zhao L, Brueckner M, Sun Z. Intraciliary calcium oscillations initiate vertebrate left-right asymmetry. Curr Biol. 2015;25:556–567. doi: 10.1016/j.cub.2014.12.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Essner JJ, Amack JD, Nyholm MK, Harris EB, Yost HJ. Kupffer’s vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development. 2005;132:1247–1260. doi: 10.1242/dev.01663. [DOI] [PubMed] [Google Scholar]
- 28.Wheway G, Nazlamova L, Hancock JT. Signaling through the primary cilium. Front Cell Dev Biol. 2018 doi: 10.3389/fcell.2018.00008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anvarian Z, Mykytyn K, Mukhopadhyay S, Pedersen LB, Christensen ST. Cellular signalling by primary cilia in development, organ function and disease. Nat Rev Nephrol. 2019;15:199–219. doi: 10.1038/s41581-019-0116-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goetz JG, et al. Endothelial cilia mediate low flow sensing during zebrafish vascular development. Cell Rep. 2014;6:799–808. doi: 10.1016/j.celrep.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 31.Praetorius HA. The primary cilium as sensor of fluid flow: new building blocks to the model. A review in the theme: cell signaling: proteins, pathways and mechanisms. Am J Physiol Cell Physiol. 2015;308:C198–C208. doi: 10.1152/ajpcell.00336.2014. [DOI] [PubMed] [Google Scholar]
- 32.Yoder BK. Role of primary cilia in the pathogenesis of polycystic kidney disease. J Am Soc Nephrol. 2007;18:1381–1388. doi: 10.1681/ASN.2006111215. [DOI] [PubMed] [Google Scholar]
- 33.Kathem SH, Mohieldin AM, Nauli SM. The roles of primary cilia in polycystic kidney disease. AIMS Mol Sci. 2014;1:27–46. doi: 10.3934/molsci.2013.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stepanek L, Pigino G. Microtubule doublets are double-track railways for intraflagellar transport trains. Science. 2016;352:721–724. doi: 10.1126/science.aaf4594. [DOI] [PubMed] [Google Scholar]
- 35.Kozminsky KG, Johnson KA, Forscher P, Rosenbaum JL. A motility in the eukaryotic flagellum unrelated to flagellar beating. Proc Natl Acad Sci USA. 1993;90:5519–5523. doi: 10.1073/pnas.90.12.5519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ishikawa H, Marshall WF. Efficient live fluorescence imaging of intraflagellar transport in mammalian primary cilia. Methods Cell Biol. 2015;127:189–201. doi: 10.1016/bs.mcb.2015.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sui H, Downing KH. Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature. 2006;442:475–478. doi: 10.1038/nature04816. [DOI] [PubMed] [Google Scholar]
- 38.Song K, et al. In situ structure determination at nanometer resolution using TYGRESS. Nat Methods. 2020;17:201–208. doi: 10.1038/s41592-019-0651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Loeffelholz O, et al. Nucleotide- and Mal3-dependent changes in fission yeast microtubules suggest a structural plasticity view of dynamics. Nat Commun. 2017;8:2110. doi: 10.1038/s41467-017-02241-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fisch C, Dupuis-Williams P. Ultrastructure of cilia and flagella—back to the future! Biol Cell. 2011;103:249–270. doi: 10.1042/BC20100139. [DOI] [PubMed] [Google Scholar]
- 41.Hess RA. Small tubules, surprising discoveries: from efferent ductules in the turkey to the discovery that estrogen receptor alpha is essential for fertility in the male. Anim Reprod. 2015;1:7–23. [PMC free article] [PubMed] [Google Scholar]
- 42.Nguyen AM, Young Y-N, Jacobs CR. The primary cilium is a self-adaptable, integrating nexus for mechanical stimuli and cellular signaling. Biol Open. 2015;4:1733–1738. doi: 10.1242/bio.014787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Owa M, et al. Inner lumen proteins stabilize doublet microtubules in cilia and flagella. Nat Commun. 2019;10:1143. doi: 10.1038/s41467-019-09051-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Garvalov BK, et al. Luminal particles within cellular microtubules. J Cell Biol. 2006;174:759–765. doi: 10.1083/jcb.200606074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Coombes C, et al. Mechanism of microtubule lumen entry for the α-tubulin acetyltransferase enzyme αTAT1. Proc Natl Acad Sci USA. 2016;113:E7176-E7184. doi: 10.1073/pnas.1605397113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Boehlke C, et al. Differential role of Rab proteins in ciliary trafficking: Rab23 regulates Smoothened levels. J Cell Sci. 2010;123:1460–1467. doi: 10.1242/jcs.058883. [DOI] [PubMed] [Google Scholar]
- 47.Schrøder JM, et al. EB1 and EB3 promote cilia biogenesis by several centrosome-related mechanisms. J Cell Sci. 2011;124:2539–2551. doi: 10.1242/jcs.085852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Pedersen LB, Geimer S, Sloboda RD, Rosenbaum JL. The microtubule plus end-tracking protein EB1 is localized to the flagellar tip and basal bodies in Chlamydomonas reinhardtii. Curr Biol. 2003;13:1969–1974. doi: 10.1016/j.cub.2003.10.058. [DOI] [PubMed] [Google Scholar]
- 49.Roth D, Fitton BP, Chmel NP, Wasiluk N, Straube A. Spatial positioning of EB family proteins at microtubule tips involves distinct nucleotide-dependent binding properties. J Cell Sci. 2018;132:jcs219550. doi: 10.1242/jcs.219550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Leterrier C, et al. End-binding proteins EB3 and EB1 link microtubules to ankyrin G in the axon initial segment. Proc Natl Acad Sci USA. 2011;108:8826–8831. [Google Scholar]
- 51.Schrøder JM, Schneider L, Christensen ST, Pedersen LB. EB1 is required for primary cilia assembly in fibroblasts. Curr Biol. 2007;17:1134–1139. doi: 10.1016/j.cub.2007.05.055. [DOI] [PubMed] [Google Scholar]
- 52.Lopez BJ, Valentine MT. Mechanical effects of EB1 on microtubules depend on GTP hydrolysis state and presence of paclitaxel. Cytoskeleton (Hoboken) 2014;71:530–541. doi: 10.1002/cm.21190. [DOI] [PubMed] [Google Scholar]
- 53.Zhang R, LaFrance B, Nogales E. Separating the effects of nucleotide and EB binding on microtubule structure. Proc Natl Acad Sci USA. 2018;115:E6191-E6200. doi: 10.1073/pnas.1802637115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Stroud MJ, et al. GAS2-like proteins mediate communication between microtubules and actin through interactions with end-binding proteins. J Cell Sci. 2014;127:2672–2682. doi: 10.1242/jcs.140558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nazgiewicz A, Atherton P, Ballestrem C. GAS2-like 1 coordinates cell division through its association with end-binding proteins. Sci Rep. 2019;9:5805. doi: 10.1038/s41598-019-42242-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Slep KC, et al. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lee S, Tan HY, Geneva II, Kruglov A, Calvert PD. Actin filaments partition primary cilia membranes into distinct fluid corrals. J Cell Biol. 2018;217:2831–2849. doi: 10.1083/jcb.201711104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Copeland SJ, McRae A, Guarguaglini G, Trinkle-Mulcahy L, Copeland JW. Actin-dependent regulation of cilia length by the inverted formin FHDC1. Mol Biol Cell. 2018;29:1611–1627. doi: 10.1091/mbc.E18-02-0088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kohli P, et al. The ciliary membrane-associated proteome reveals actin-binding proteins as key components of cilia. EMBO Rep. 2017;18:1521–1535. doi: 10.15252/embr.201643846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Phua SC, et al. Dynamic remodeling of membrane composition drives cell cycle through primary cilia excision. Cell. 2017;168:264–279.:e15. doi: 10.1016/j.cell.2016.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zuo X, et al. Primary cilia and the exocyst are linked to urinary extracellular vesicle production and content. J Biol Chem. 2019;294:19099–19110. doi: 10.1074/jbc.RA119.009297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nager AR, et al. An actin network dispatches ciliary GPCRs into extracellular vesicles to modulate signaling. Cell. 2017;168:252–263.:e14. doi: 10.1016/j.cell.2016.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mirvis M, Stearns T, James Nelson W. Cilium structure, assembly, and disassembly regulated by the cytoskeleton. Biochem J. 2018;475:2329–2353. doi: 10.1042/BCJ20170453. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.