Abstract
ABC (ATP binding cassette) transporters, ubiquitous in all kingdoms of life, carry out essential substrate transport reactions across cell membranes. Their transmembrane domains bind and translocate substrates and are connected to a pair of nucleotide binding domains, which bind and hydrolyze ATP to energize import or export of substrates. Over four decades of investigations into ABC transporters have revealed numerous details from atomic-level structural insights to their functional and physiological roles. Despite all these advances, a comprehensive understanding of the mechanistic principles of ABC transporter function remains elusive. The human multidrug resistance transporter ABCB1, also referred to as P-glycoprotein (P-gp), is one of the most intensively studied ABC exporters. Using ABCB1 as the reference point, we aim to compare the dominating mechanistic models of substrate transport and ATP hydrolysis for ABC exporters and to highlight the experimental and computational evidence in their support. In particular, we point out in silico studies that enhance and complement available biochemical data. “This article is part of a Special Issue entitled: Beyond the Structure Function Horizon of Membrane Proteins edited by Ute Hellmich, Rupak Doshi and Benjamin McIlwain.”
Keywords: ABC transporter, ABCB1, P-glycoprotein, Molecular dynamics simulations, Mechanistic models, Transport cycle
1. Introduction to ABC transporter function and architecture
This review discusses the architecture and function of ATP-binding cassette (ABC) exporters with a focus on ABCB1. We will present an overview of available transport cycle and ATP hydrolysis models, review their similarities and differences and place them into context of available data for ABCB1. Despite more than forty years of intensive research on ABCB1 and its relatives, many fundamental questions regarding substrate recognition, translocation and coupling of ATP binding and hydrolysis at the nucleotide binding domains (NBDs) to conformational changes in the transmembrane domains (TMDs) have not yet been satisfyingly resolved. Advancements of in silico approaches have shown great promise to complement experimental approaches and to address these questions. We will highlight available computational approaches, discuss how they were used to study ABCB1 transporter function and to test different aspects of proposed ABC transport cycle models.
ABC proteins are found in all kingdoms of life [1,2] where they mediate translocation of diverse substrates under ATP consumption. The ABC protein superfamily can be divided into four major groups, the first three of which constitute membrane proteins: (i) importers of nutrients (mainly bacterial ABC transporters) (ii) exporters of diverse compounds including peptides, metabolites or xenobiotics (such as ABCB1) (iii) ABC proteins that act as ion channels or channel regulators and (iv) ABC proteins that lack TMDs and are involved in DNA repair and translation. Most of the 48 human ABC proteins are membrane transporters (subfamilies ABCA-D, and ABCG) that facilitate directional transport of small molecules, possibly against a concentration gradient. The ABCC subfamily includes ABCC7 (CFTR: cystic fibrosis conductance regulator), which is an ion channel, as well as ABCC8 and ABCC9 that are regulators of the Kir6.2 potassium ion channel [3]. In contrast, the subfamilies E and F participate in DNA repair and translation and consist only of NBDs [4,5].
The minimal functional unit of all ABC transporters comprises two TMDs and two NBDs. Full transporters have all four domains expressed as a single polypeptide chain, while half transporters have one NBD and one TMD expressed as a single peptide chain and two of these polymers assemble to form homo- or heterodimers. All structural elements are present twice in symmetric homodimers; pseudo-symmetry is maintained (Fig. 1) in heterodimers and full transporters despite sequence divergence [6,7]. The ABCB subfamily is heterogeneous, as it includes both full length (ABCB1, ABCB4, ABCB5 and ABCB11), as well as hetero- and homodimeric half transporters (ABCB2/B3 and ABCB6-10, respectively).
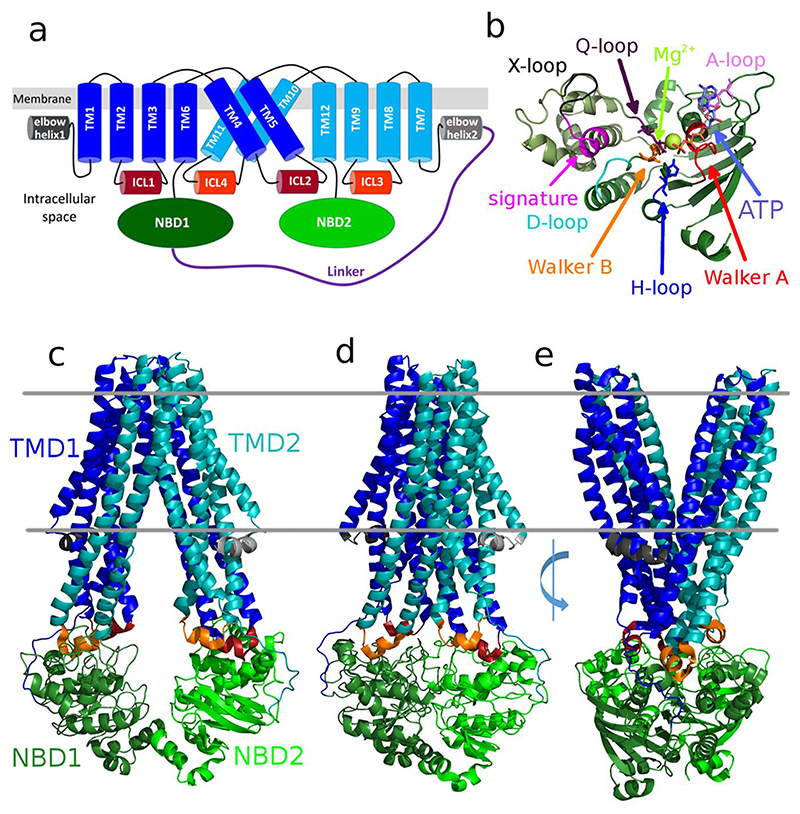
Fig. 1. Structural features of ABCB1.
a) Topological model of the ABCB fold. The elbow helices preceding TMH1/TMH7 anchor the TMDs in the membrane, the long ICL2/4 cross-over and interact with the trans-NBD, while ICL1/3 bind to the cis-NBD. b) Focused view on the nucleotide binding domain looking at the NBD-NBD interface, highlighting the conserved motifs with bound Mg2+ and ATP. Crystal structures of c) inward open, d) occluded and e) outward open conformation of MsbA (PDB ID: 5TV4, 5TTP, 3B60, respectively) colored according to the topological model. The inward open structure shows separated NBDs, the occluded structure is closed on the intracellular and the extracellular side. The outward open structure shows a large separation of the TMDs towards the extracellular space allowing for substrate release.
For all ABC transporters, the TMDs are the site of substrate recognition and translocation; they begin at their N-terminus with the elbow helix, which is a helical structure oriented parallel to and by its amphipathic nature partially inserted into the membrane from the intracellular side (Fig. 1). Beyond their putative role in anchoring the TMDs into the membrane, in MsbA the elbow helix was also shown to interact with the substrates daunorubicin [8], suggesting a role at the beginning of the substrate translocation process.
As the ABC transporter undergoes its substrate translocation/ATP hydrolysis cycle, the TMDs go through conformational changes to bind substrate on one side of the membrane and to release it to the other. TMD conformational changes are connected to changes in the NBDs which energize the transport cycle through ATP binding and hydrolysis and induce conformational changes in the TMDs. The NBD dimer comprises two distinct nucleotide binding sites (NBSs) [9–11], each capable of binding one nucleotide. ATP acts as the “molecular glue” that holds the NBDs together in a head-to-tail conformation. The NBSs are the most conserved parts of the protein and harbor a set of distinct motifs, such as Walker A, Walker B and ABC signature motif (C-loop), which signify ABC transporter family membership at the sequence level (Table 1, Fig. 1c). The conserved motifs are essential for nucleotide binding, hydrolysis and NBD-NBD as well as NBD-TMD communication. The NBDs can be divided into two subdomains, the core or RecA-like subdomain that includes the Walker A motif and the helical subdomain, which contains the signature motif. A plethora of in silico work deals with the motions of NBD dimers (Section 4) and the role of the conserved motifs. Most ABCB transporters have two canonical NBSs that bind ATP, however the heterodimeric ABCB2/B3 peptide transporter and the bile salt transporter ABCB11 contain one degenerate NBS. Non-canonical NBSs hydrolyze ATP only at a very low rate [12], because residues essential for a high hydrolysis rate deviate from the classical (or canonical) sequence (Table 1).
Table 1. Consensus sequence and function of conserved motifs in the NBDs listed according to their occurrence from N- to C-terminus.
| Motif | Consensus sequence |
Function |
|---|---|---|
| A-loop [13] | (F/K)xYa | ATP binding (base stacking) |
| Walker A or P-loop [14] |
GxxGxGK(S/T)a | ATP binding (phosphate binding) |
| Q-loop [15] | hV(S/P)Qb | TMD-NBD communication |
| X-loop [16,17] | TRVGDKGTQ | TMD-NBD communication |
| signature motif or C-loop [18] |
LSGGQ(K/R)Q | Phosphate binding, NBD-NBD communication |
| Walker B [19] | hhhhDEb | ATP hydrolysis |
| D-loop [20,21] | SALD | NBD-NBD communication, unidirectionality of transport |
| H-loop [22] | hAHRLb | ATP hydrolysis |
x = any residue.
h = hydrophobic residue.
1.1. Short introduction to the ABCB family
Members of the human ABCB subfamily exhibit a wide variety of physiological functions. ABCB1 (multidrug resistance protein 1 (MDR1), P-glycoprotein, or P-pg) is a major player in cellular detoxification and multidrug resistance [23–25]. ABCB2 and ABCB3 (Transporter associated with Antigen Processing (TAP1 and TAP2)) form a heterodimer that is essential in adaptive immunity for loading short antigen peptides onto the major histocompatibility complex (MHC class 1) [26–28]. ABCB4 (multidrug resistance protein 2 or MDR2) [29,30] and ABCB11 (bile salt export pump or BSEP) [31,32] transport phosphatidylcholine and bile salts, respectively, into the bile duct. ABCB5 is involved in melanogenesis, the production of the pigment melanin by melanocytes [33–35]. The half transporters ABCB7 (Fe/S cluster transport) [36,37], ABCB8 (intracellular peptide trafficking) [38,39], and ABCB10 (peptide transporter) [39,40] are mitochondrial exporters. The peptide transporter ABCB9 (TAP-like or TAPL) [41–43] shows a lysosomal localization, while the cellular localization (lysosome, endo-lysosome, Golgi and/or mitochondria) and function of ABCB6 (heme and porphyrin transport) remains disputed [44–47].
The main role of ABCB1 seems to be cellular detoxification by expelling a large number of chemically unrelated hydrophobic compounds from the plasma membrane. ABCB1 is expressed at tissue barriers such as skin, intestine [48], blood brain barrier, placental barrier, and blood testis barrier [23] to limit the entry of environmental toxins and keep the level of xenobiotics below a harmful level. While normally showing a protective function, ABCB1 frequently interferes with chemotherapy treatment by actively removing the medication, when cancer cells express the transporter [49,50]. Besides its problematic role in chemotherapy, ABCB1 is of further clinical relevance since it regulates bioavailability and pharmacokinetics of many drugs, e.g. by preventing brain penetration, uptake from the intestine or permeation through the placenta [51]. ABCB1 transporter activity has also been exploited e.g. in second generation antihistamines to prevent adverse sedative effects by limiting penetration through the blood brain barrier [52].
Despite the large empirical knowledge on ABCB1 function, transported compounds and residues involved, several fundamental questions remain poorly understood at the molecular level: e.g. the molecular basis for the promiscuity of substrate recognition, the route substrates take upon extraction from the membrane through the TMDs into the extracellular space, the conformations and sequence of transport cycle states, as well as coupling between ATP binding/hydrolysis and substrate transport. Available experimental and in silico data will be reviewed in the following sections.
2. High-resolution views of the ABCB fold
Knowledge of protein structure is essential for understanding dynamics and function on the atomic level and to link specific interactions, conformations and structural changes to transport. Important breakthroughs in ABC transporter structural biology have been achieved: for instance, there are now structures available for human members of the ABCA [53], ABCB [54], ABCC [55] and ABCG [56,57] subfamilies. Transporters from the ABCB subfamily have been determined in three conformations: (i) most structures showed an inward open (or inward facing) conformation, where the NBDs separated by a variable range from less than a nanometer to up to a few nanometers [7,58]; (ii) The outward open conformation was found for the bacterial Sav1866 [9] and MsbA transporters [59] and showed nucleotide stabilized NBD dimers and extracellularly separated TMDs; (iii) Occluded conformations have been determined for the bacterial transporters McjD [60], PglK [61], and MsbA [62] revealing closely associated NBDs and TMDs with a fully occluded substrate binding side. Distance measurements using electron paramagnetic resonance (EPR) spin probes attached to the NBDs showed a distribution of NBD separation ranging over 2 nm in the absence of nucleotides for MsbA and ABCB1 [63,64]. The distance between Walker A motifs of ABCB1 without bound nucleotide was approximately 1.6 nm in a native lipid membrane, and 2.2 nm in a detergent micelle [65]. The addition of nucleotides induced NBD association [64,66] that resulted in a single distance peak. These data demonstrate the strong dependence of transporter conformation on the presence of nucleotides and the membrane environment and show that a large NBD separation is possible in detergents, but might be suppressed in the membrane environment. The lipid A bound cryo electron microscopy (Cryo-EM) structure of MsbA (PDB ID: 5TTP) [62], determined in lipid nanodiscs, shows an intermediate conformation in the absence of nucleotides, in which the NBSs are separated, while the NBDs are in contact through their respective C-termini. The degree and functional relevance of NBD separation is an ongoing debate in the field and has implications for ATP hydrolysis and transport models (see below).
ABCB transporters contain six core transmembrane helices per TMD that form the substrate translocation path in their center of a functional dimeric assembly. In the ABCB, ABCC and ABCD transporter subfamilies, the intracellular loops (ICLs) assemble into a tetra-helical bundle that stabilizes a long protrusion into the intracellular space (Fig. 1b), thereby positioning the NBDs far from the membrane. The transmembrane helices 4 and 5 reach over to the trans NBD in a domain swap arrangement and interact through their coupling helices with the conserved binding grooves of NBDs using a ball-and-socket type structure [9,67] (Fig. 1). A network of salt-bridges and hydrophobic interactions stabilizes the ICL helix bundle, which has a role in transmitting nucleotide-dependent conformational changes [68], although the mechanical-functional implications of the long protrusions and the domain swapped architecture are not well understood.
Available crystal structures in different conformations show that the TMDs of ABC transporters do not move in a purely rigid body fashion during the transport cycle. Structural flexibility is encoded by helix bending, helical kinking and unwinding caused by helix breaking residues (prolines, glycines) [69]. Their mutation in ABCB1 was shown to affect coupling of the substrate translocation to ATPase activity, e.g. the G346C mutation in TM6 changed communication between the TMDs and NBDs [70], while the G185V mutation stiffened TM3 and enhanced coupling [71]. In both cases, the reduced ATP turnover and basal AT-Pase activity was attributed to increased helicity and an ensuing TMD rigidification.
In addition to inward and outward open conformation, transporter structures in an occluded state are available for the ABCB subfamily. These are believed to represent a transition state in the transport cycle, where the extracellular and the intracellular sides are closed, which seems to be a necessary intermediate conformation to prevent a channel like function during the transition between the outward- and the inward facing state. The known exception is ABCC7, which is an ATP dependent Cl- channel. Structures of the transporters McjD [60] and PglK [61] show an occluded cavity in the center of the membrane, to which a substrate could bind. In contrast, in the cryo-EM structure of MsbA [62] the cavity is collapsed, leaving no space for binding of a lipid A substrate. It thus remains unclear, if the collapsed conformation may be part of the empty return step after substrate release and ATP hydrolysis.
Full transporters of the ABCB subfamily have a linker region of approximately 75 amino acids connecting NBD1 with TMD2 (Fig. 1a). The segment is presumably very flexible as no structural information has been obtained yet and secondary structure predictions suggest complete absence of stable structural elements. Cleavage of the linker slightly increased ATP hydrolysis rates [72], thus suggesting that it has a limited functional role. Dephosphorylation of the typically phosphorylated [73] linker decreased substrate induced ATPase activity, but maximal activity at higher substrate concentrations was maintained, suggesting that phosphorylation allows for regulation of transporter activity. Shortening of the linker by 34 residues resulted in increased basal ATPase activity in mouse ABCB1, abrogation of substrate stimulation of ATPase activity and drastically decreased transport [74]. The same linker deletion in human ABCB1 abolished function, indicating that a minimal linker length of 40–45 residues is required.
ABC transporters couple ATP binding and hydrolysis at the NBDs to conformational changes in the TMDs for substrate translocation across the membrane. Binding of substrates to the TMDs of ABCB1 increases the ATPase activity [75–77], while ATP hydrolysis and conformational changes in the NBDs lead to structural changes in the TMD [78], thus showing that the TMDs and NBDs can communicate bidirectionally. Transporter architecture implies that the ICLs and their four coupling helices are the structural elements connecting the TMDs with the NBDs and must be involved in transmitting conformational changes. Within the NBDs, the conserved motifs of the Q- and X-loops are assumed to be essential for this interdomain communication [20,79,80]. Interestingly, in ABCB1 redundancy was found for the Q-loop, where only one of the conserved glutamines was needed for active transport [80–82], while removing both residues (Q475A/Q1118A) abrogated transport and trapped ABCB1 in the inward facing state. Involvement of these ATP interacting loops indicates that the NBS are essential elements in this process. However, while the phenomenon of inter-domain communication is well established, the precise molecular movements required for and triggered by substrate translocation and ATP hydrolysis remain unresolved. The available structures of ABCB exporters have provided a starting point for modelling and hypothesis generation for biochemical studies. Importantly, they are also essential starting structures for simulations that have been carried out to investigate several aspects of transporter function, as outlined in Section 4.
3. ABC transporter function
3.1. General models for substrate transport
The first and very general model of substrate translocation, the ‘alternating access model’ [83], proposed that a transporter alternates between an inward and an outward facing state with two different substrate affinities. Access to the substrate binding site can be locked by two gates, one of which must always be closed to prevent a channel-like function, implying the existence of an occluded transition state in which both gates are closed. The TMDs of ABCB1 assume an inward open conformation in the absence of nucleotides, with a high affinity substrate binding pocket [84]. After substrate and ATP binding at the TMDs and NBDs, respectively, the TMDs undergo structural rearrangements that allow them to first reach an occluded and then an outward open state with a decreased affinity for substrate [85,86].
Based on experimental results, the alternating access model was further developed into the ‘hydrophobic vacuum cleaner’ model [87]: (i) ABCB1 substrates were found to be structurally diverse but hydrophobic and thus to accumulate in the lipid bilayer, and (ii) spontaneous flipping of drugs between membrane leaflets is relatively slow [88]. The vacuum cleaner model requires ABCB1 to retrieve its substrate from the inner leaflet of the membrane and then to flip them into the outer leaflet or to expel them directly to the extracellular space. This model can also incorporate polyspecificity by the proposed two-step mechanism in which substrates are first enriched (depending on logP, up to several orders of magnitude) in the membrane. The high substrate concentration then allows for low affinity and thus non-selective binding. Substrates stimulate the ABCB1 ATPase activity up to ten fold [75,89–92], thereby showing that substrate binding triggers the transport activity of ABCB1. Removal of substrates enriched in the membrane allows for maintaining a low intracellular concentration in the presence of a high extracellular concentration of substrates. This model implicitly emphasizes the importance of the membrane environment for the transport process. Transport is energized by ATP, which also imposes unidirectionality [20].
3.2. Mechanistic models of the transport cycle
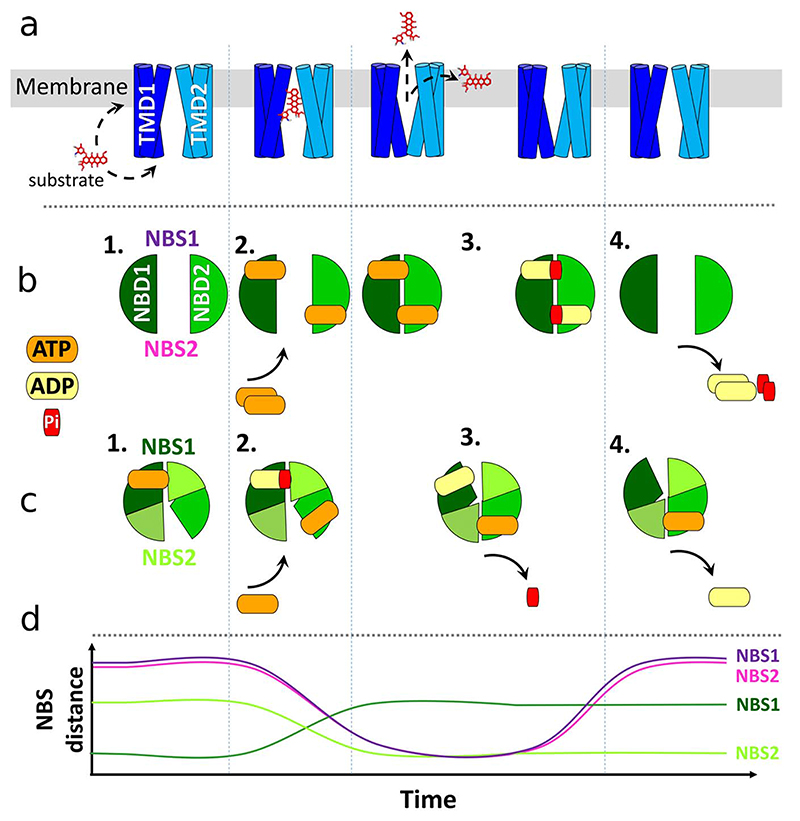
Models describing the steps of the transport cycle minimally include, for the NBDs, ATP binding and hydrolysis, ADP and inorganic phosphate (Pi) release and nucleotide dependent structural changes. These steps are linked to a conformational switch of the TMDs, which interchange between the high-affinity inward-facing state and the low-affinity outward-facing state. The wide opening of the TMDs as observed in the outward facing crystal structures (e.g. Sav1866 [93]) and the large separation of the NBDs seen in the inward facing crystal structures (of e.g. mouse ABCB1 [58]) have shaped the perception of the conformational changes during the transport cycle of ABCB1, though not fully consistent with all biological evidence. Many transport cycle models have been put forward which may only minimally differ in details, or might propose drastically different conformations and steps for the transport. Here, we focus only on the most recognized models. Overall, models of the catalytic cycle can be divided into two main categories; either (i) predicting complete NBD separation or (ii) requiring the NBDs to remain in contact throughout the transport cycle. Asymmetrical transporter conformations are then associated with asymmetric binding states for nucleotide. The best known models that include NBD separation are the Tweezers-like model [94], ATP switch model [95] and processive clamp model [96–98]. In contrast, the alternating sites model [99], constant contact model [100] or nucleotide occlusion model [101–103] propose continuous NBD-NBD contact of at least one NBS throughout the transport cycle. All models implicitly agree on a tight coupling between NBDs and TMDs, with the closed NBD dimer corresponding to the outward open TMD conformation and the inward facing TMD conformation showing looser NBD association. The catalytic cycle models are summarized in Table 2 and Fig. 2. There is experimental support for all these models (see Section 3.3). When considering different models, it should also be kept in mind that ATP concentrations in cells are 10-fold above KD for ABCB1 (0.3–0.5 mM) [104], while the concentration of ADP is similar to its KD [103]. Transporters might therefore almost always be nucleotide bound, though affinity is transporter conformation dependent [103] and still debated, as outlined below.
Table 2. Different catalytic models summarized briefly in four steps. (The Tweezer model that has been proposed based on the maltose importer is included for further reference and comparison).
| Steps | Nucleotide occlusion [101–103] |
ATP switch [95] |
Processive clamp [96] |
Alternating sites [99] |
Constant contact [100] |
Tweezers-like (in MalFGK2)[94] |
|---|---|---|---|---|---|---|
| 1 | NBDs: | NBDs: | NBDs: | NBDs: | NBDs: | NBDs: |
| semi-open dimer | separated, empty | ATP binding, dimer | semi-open dimer | semi-open dimer | semi-open dimer | |
| NBS1: | formation | NBS1: | NBS1: | NBSs: | ||
| occluded ATP | ATP binding | occluded, ATP-bound | empty | |||
| NBS2: | NBS2: | NBS2: | ||||
| loosely ATP binding | empty | occluded, empty | ||||
| TMDs: | ||||||
| inward open | TMDs: | |||||
| inward open, bound substrate | ||||||
| TMDs: | TMDs: | |||||
| inward open, substrate binding | inward open | |||||
| 2 | TMDs: | NBDs: | NBS1a: hydrolysis, maybe | NBS2: | NBS1: | Maltose loaded MBP binds |
| substrate binding | ATP binding, dimer | release of Pi | ATP binding | occluded, hydrolysis | to protein | |
| NBS1: hydrolysis → high energy | NBS2: | NBSs: | ||||
| TMDs: | ADP + Pi state | open | ATP binding | |||
| outward open, | TMDs: inward open | TMDs: | ||||
| substrate release | outward open | |||||
| 3 | NBS1: | NBDs: hydrolysis | NBS2a: | NBS1: | NBS1: | NBDs: |
| hydrolysis | ATP hydrolysis, release of | Pi release | Pi release, open | dimerize | ||
| NBS2: | Pi | NBS2: | MBP: | |||
| occluded TMDs: |
binding ATP | releases maltose | ||||
| outward open → substrate release |
TMD: outward open |
|||||
| 4 | NBS1: | NBD: | NBD: | NBS1: | NBS1: | NBSs: |
| ADP, Pi release | ADP, Pi release, | separated, ADP release | ADP release | ADP release, empty, | ATP hydrolysis | |
| separated | occluded | NBD: | ||||
| NBS2: ATP, occluded |
separated TMDs: |
|||||
| TMDs: | TMDs: | TMDs: | inward open | |||
| inward open | inward open | inward open → substrate release | release of maltose and MBP |
MBP: Maltose-binding protein.
Pi: Inorganic phosphate or HPO4 2−.
Sequential hydrolysis in NBS1 and NBS2 with unknown order.
Fig. 2. Cartoon summarizing the two main types of transport models (with/without constant NBD contact).
a) Motions of the TMDs during the transport cycle, including the two possible paths (between membrane sides or between membrane leaflets) of substrate binding and release. b) NBD and nucleotide motions including full NBD separation (Tweezers-like [94], ATP switch model [95] and processive clamp [96–98]). c) NBD motions predicted for models assuming constant contact (alternating sites [99], constant contact [100] or nucleotide occlusion model [101,102]). d) Schematic illustration of the change in distances across the ATP binding site (from Walker A to ABC signature motif as a function of the transport cycle (purple hues: NBS distances in NBD full-separation models; green hues: NBS distances in NBD constant contact models).
3.3. Experimental support for different catalytic models
The large differences in transporter architecture between type I (e.g. MalEFGK2 [105]) and II importers (e.g. BtuCDF [106]) as well as type I (e.g. ABCB1 [7]) and II exporters (e.g. ABCG5/8 [56]) seem incompatible with a single model for the transport cycle, as deduced from the mechanical implications of these structures. It also remains unresolved whether even all transporters of the ABCB family share a common transport mechanism, as they all translocate different substrates and some transporters for instance have two canonical NBS, while others harbor a degenerate site. It is well possible that degeneration of one ATPase implies a change in the mechanism of harvesting the energy from ATP binding and hydrolysis and thereby leading to an alternative route through the transport cycle. Interestingly, a comparison of supporting experimental evidence reveals that switch and processive clamp models are mainly supported by measurements on bacterial transporters. In contrast, the alternating sites, constant contact and nucleotide occlusion models are mainly supported by data from mammalian transporters. It remains unresolved, if these differences i) represent a common trend with the bacterial and mammalian transporters using different mechanisms, ii) are due to experimental underdetermination, thereby leaving room for interpretation, or iii) can be attributed to the particularities of each transporter.
3.3.1. Asymmetric NBS occupation
Un-synchronized ATP binding or hydrolysis lead to asymmetric nucleotide binding states in the NBSs, which may induce structural and functional asymmetry for the entire transporter. Asymmetric conformations are predicted by the alternating sites, the nucleotide occlusion and the constant contact models. Asymmetric conformations may seem expected for ABC transporters with a non-canonical NBS, but for transporters with two canonical, fully ATPase competent sites, structural symmetry could be preserved. Models supporting an asymmetric mechanism like the constant contact model [107] emphasize relative motions of the helical vs. the core (RecA-like) sub-domains within the NBDs. This extra flexibility would allow nucleotide exchange in the open NBS, while keeping the second NBS closed and ATP bound in an asymmetric conformation [107]. The coupling helices of the TMDs are attached to the NBDs at the boundary between the core and helical domain, suggesting that communication between the sub-domains could allow for cross-talk to the TMDs by allostery. Moreover, intra-NBD interactions between motifs and nucleotides (e.g. the formation of the catalytic dyad in the closed NBS [22]) have been used to explain the cooperative nature of ATP hydrolysis.
Functional asymmetry and differences in affinity for ATPγS (a non- hydrolyzable ATP analogue) to both NBS have been determined for Chinese hamster ABCB1 and revealed two very different KD values of 6±4μ M and 740 ± 420 μ M depending on the occlusion of the NBS [103]. An asymmetric intermediate was captured by trapping Chinese hamster ABCB1 in the post-hydrolytic state by replacing the inorganic phosphate (Pi) with orthovanadate (Vi) leading to a ~ 1:1 stoichiometry, i.e. ATP hydrolysis in one NBS resulting in a very stable transporter conformation [75]. In contrast, in bacterial ABC exporters, the pre-hydrolytic trapped state by BeFx [108], the Vi trapped state with Mn2+ (replacing Mg2+) [109] and the coordination of Mn2+ in the ATP-bound state were observed to be symmetric [110]. Mutation of the Walker B catalytic glutamate also trapped the protein in an asymmetric state with tight binding of a single ATP to one of the NBSs and referred to as nucleotide occlusion, while the second NBS took a more open conformation with a lower ATP affinity [103,111]. From the asymmetric ATP trapping it may be inferred that ATP hydrolysis is most likely not synchronized between the NBSs of ABCB1. However, the functional implications for the number of ATPs hydrolyzed (see below) remain unresolved, because asymmetric nucleotide trapping with Vi is consistent with a single ATP hydrolysis as well as with two sequential ATP hydrolysis events per transport cycle.
3.3.2. Number of ATP hydrolyzed per cycle
Despite numerous studies, it is still debated whether hydrolysis of one or two ATPs is needed for a transport event and different experiments showed different outcomes (see e.g [81,101,112–114]). In the NBD dimer of the yeast intracellular peptide transporter Mdl1p, ATP hydrolysis of one ATP is followed by formation of a stable intermediate with one ATP and one ADP bound, which then allows for hydrolysis of the second ATP. However, under ATP limiting conditions, the dimer dissociates before hydrolysis of the second ATP [96]. Similarly, in the bacterial NBD-only system MJ0796, the NBDs dissociate after hydrolysis of one ATP [115] suggesting that a single ATP hydrolysis event is sufficient for NBD separation. In case of mouse ABCB1, single Walker B catalytic glutamate mutants were found to still allow for conformational changes, while the double catalytic mutant trapped the transporter in the outward facing conformation [66]. Of note, transporters with a degenerate NBS (e.g. bacterial TM287/288, or mammalian ABCB2/B3 and ABCB11 as well as all transporters of the ABCC subfamily) are able to support transport with only one canonical NBS, suggesting that a single ATP hydrolysis event can in principle suffice for sustaining transport.
3.3.3. NBD separation
Experimental results suggest that two ATPs bound to the NBDs are needed for complete NBD dimerization, which agrees with all models [96,114,116]. However, it still remains debated, if the NBDs fully separate during the transport cycle. Widely inward open crystal structures of eukaryotic ABCB1 suggested that this can indeed be the case [7,58,74]. Similarly, EPR, lanthanide-based resonance energy transfer (LRET) and cryo-EM data on bacterial ABC transporters showed that their NBDs can undergo full separation in the absence of nucleotides when purified in detergent micelles [66,116,117]. In spite of the observed separation between the NBDs in ABCB1, cross-linking experiments showed that human ABCB1 cross-linked at its ICLs (L175C/ N820C) remained ATPase active [118] and restored ATPase activity of a Q-loop double mutant Q475A/Q1118A [82], suggesting that ATPase activity does not per se require a wide separation of NBDs.
It is very difficult to quantify the transport cycle with sufficiently high temporal and spatial resolution. Answering the questions discussed above, i.e. NBD separation, number of ATP hydrolyzed per translocation event, symmetry for transporter conformation and nucleotides occupancy should enable clarification of the essential steps of the power stroke of the transport cycle. Simulations can in principle address these questions by enhanced sampling methods, but rely heavily on empirical input from models and experiments (for details, see Section 4). While challenging, a combination of simulations and experiments with repeated hypothesis generation and testing should be able to explore the framework provided by the transport models and further define the transport cycle.
4. In silico assessment of substrate transport and ATP hydrolysis
To fully understand substrate recognition or conversion of chemical energy stored in the ATP phosphate bonds to conformational changes that lead to substrate transport, insights into the structural, mechanical, dynamic and energetic properties are needed throughout the transport cycle at the atomic level. Experimental approaches, despite their impressive advancements, have limitations: EPR and FRET-based approaches for instance yield information about protein structure and dynamics, but rely on probes added to specific positions within the protein, and require protein modification. X-ray crystallography yields high-resolution all-atom snapshots of proteins, but requires crystal formation. Cryo-EM can obtain near atom resolution structures, but requires averaging of many images and sorting of particles. Molecular dynamics (MD) simulations are very useful for structural and dynamic investigations at the atomic level and can in principle provide detailed information on conformations, dynamics, populations, energies, barriers and allostery for the entire transport cycle. They are therefore well placed to complement and enhance wet lab experimental approaches. Classical MD simulations use a force field to describe matter. A force field consists of a collection of functions combined with empirical parameters tailored to optimally approximate the underlying chemical and physical principles without the need of solving computationally extremely expensive quantum mechanical equations. Force fields include bonded terms (bonds, angles and torsions) that describe the interactions through covalently bonded atoms, as well as non-bonded through-space electrostatic (Coulomb) and van der Waals interactions. MD simulations solve Newton's equation of motion. They start with a system consisting of a defined number of atoms and molecules in an initial configuration and explore the dynamics of these atoms and their motions through the underlying energy landscape (or hypersurface). Temperature is represented by atom velocities at the molecular level and thus connected to energy barriers and to the extent of conformational changes that can be achieved. Quantification of free energy profiles of paths connecting different states become accessible when all relevant conformations are visited with a frequency that corresponds to the underlying free energy profile, thereby providing Boltzmann distributions.
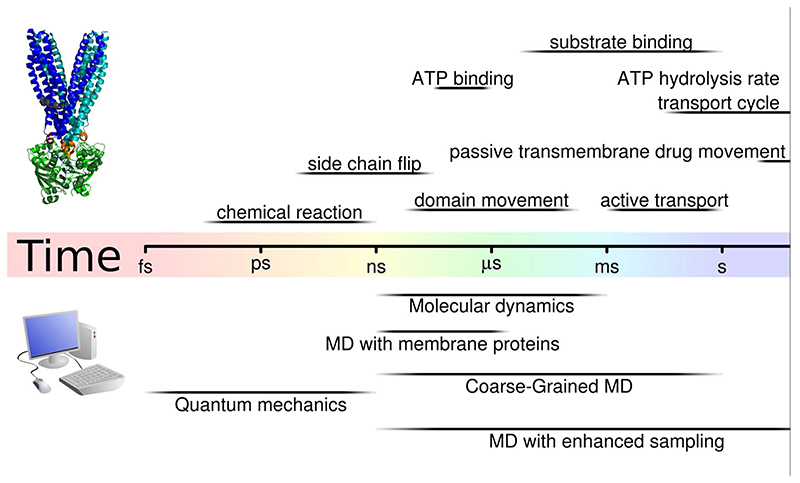
For the study of protein function, high resolution structures with representative conformations are an essential input, because non-re-presentative or high energy conformations can lead to un-physiological conformational changes and structures. Even when a high resolution, membrane embedded protein structure is available, MD simulations are no free lunch. A single time step (the time resolution of the simulation)is limited to 2 fs, thus 2 μs of simulation time require 1 billion repeating steps of solving Newton's equation of motions for all particles. Sampling, which is the exploration of the energy landscape to reach representative conformations and populations, requires massive computer time, which is only available on dedicated hardware or large high-performance computer clusters. The time windows that are now accessible for investigations with atomic details are in the microsecond range, which still falls orders of magnitude short of the time required for completing the transport cycle of ABCB1. While ATP hydrolysis happens in the millisecond timescale within the NBDs [119], the full transport cycle is a fraction of a second – a timeframe that is currently unattainable with unbiased MD simulations [120–122]. Fig. 3 shows time scales of biological processes relevant for transport by ABC proteins and compares these to simulation times accessible with different MD techniques (see below).
Fig. 3.
Comparison of time scales: The top rows indicate the time scales of different biochemical processes, including events that are relevant for the transport cycle of ABC transporters. The bottom rows show times scales that are currently accessible to in silico techniques.
The sampling limitation can be amended by non-equilibrium methods and biased techniques. Bias or external forces can be applied to accelerate the search in conformational space or to limit the exploration to the interesting low energy regions [123–126]. This allows to investigate the low energy conformations (e.g. metadynamics [124]) or to push the system across energy barriers (e.g. by steered MD [126]). These methods can keep track of the bias and use this information to reconstruction of the original underlying free energy landscape by postprocessing analysis or to estimate the work (or activation energy) needed for a conformational transition. Reaction coordinates represent the interesting degrees of freedom and need typically to be defined beforehand. They can be very general, consisting of a simple linear distance between two atoms, or very complex geometric operations describing domain rearrangements and conformational changes. Reaction coordinates can also represent a chemical reaction, e.g. ATP hydrolysis. Definition of reaction coordinates remains a challenge and a deep understanding of the molecular mechanism and the conformational changes are required, which is often not known at the necessary level of detail.
Another way to speed up simulations is to decrease the number of particles used to explicitly describe the system. This can be obtained by merging three or four atoms into a single larger virtual particle using a technique called coarse graining, but requires the time-consuming development of a new tailored force field [127]. A speedup of 10,000–40,000 [128] is achievable and puts the millisecond time window at reach. This performance boost is possible, because the calculation time step is extended from 2 fs to 20–40 fs, the number of calculated interactions is decreased by 10-fold and the energy hypersurface becomes much smoother. Coarse graining is a very successful model for membrane dynamics and recapitulates most membrane properties [129], but its application to proteins [130] is limited, as hydrogen bonds, the peptide bond, protein secondary structure or structural changes cannot be fully described. The coarse grained Martini force field was successfully applied to study protein-lipid interactions, protein diffusion and protein aggregation [131,132]. It can be expected that coarse grained simulations will become a widely used technique for the study of interactions of membrane components such as cholesterol or substrates with membrane proteins including ABC transporters.
Several focused questions regarding ABCB1 (and homologues) function have been addressed with simulations, revealing details of the transport cycle (discussed below). Simulations of the large and slowly moving ABC transporter systems need experimental data to develop reaction coordinates for enhanced sampling approaches, which are needed to ameliorate the sampling challenge (as outlined above). However, incorporation of experimental data can be challenging due to the diversity of models proposed for the ABC transport cycle and hence the requirement to translate these into different reaction coordinates required for enhanced sampling techniques. In addition, the frequently observed inward facing transporter structures have been difficult to use as starting structures for ABC transporter simulations. Strikingly, they appeared to be relatively unstable when inserted into membrane [133], which makes it difficult to discriminate between interesting conformational changes and the initial relaxation of the starting coordinates during the equilibration phase of any simulation. Despite these challenges, a number of interesting studies have been put forward that investigate ABC transporter/ABCB1 function and dynamics with in silico approaches:
4.1. Dynamics of the nucleotide binding domains
A key distinction between the two main types of transport models is the degree of NBD separation during the transport cycle. MD simulations should be able to identify the correct model by exploring conformations, dynamics and energetics of NBD separation. Several MD simulations of full length ABC transporters have been carried out starting from the inward or the outward facing conformations, thus beginning to elucidate structural changes and their dependence on the nucleotide bound state.
The NBD dimer stabilizing effect of ATP binding to the Walker A and ABC signature motifs is well characterized and accepted [66,134–139]. A driving force for NBD association is charge complementation: Walker A and the ABC signature motif are both positively charged on the NBS surface, and structurally in close proximity within each NBS, but sequentially located on opposing NBDs, therefore repelling each other in the absence of nucleotides. Binding of ATP, which carries a charge of - 4, complements their electrostatic fields and exerts an attractive force on both NBDs, thereby promoting NBD dimerization.
Simulation of full length transporters (Sav1866, MsbA, ABCB1) starting from a closed NDB conformation [79] [136] indicate that two ATPs are necessary to maintain the outward open conformation and the closed NBD dimer geometry, which is in agreement with crystal structure showing two bound nucleotides in the Sav1866 and MsbA crystal structure [59,93]. The NBDs of ATP-bound ABCB1 remained associated, but became asymmetric in the presence of two ATP and Mg2+[136]. In simulations of Sav1866, bound ATP stably maintained the symmetric dimer conformation [140]. For Sav1866, during simulations of the posthydrolysis state, neither ADP nor Pi dissociated from the NBDs over the course of sub-microsecond long simulations [140]. Opening of the TMD intracellular gate was reported, while closure of the extracellular side was not contemporaneously observed. However, it seems unlikely that this is a stable conformation, as a structure with both gates in the open conformation would represent a channel. Simulations of asymmetric nucleotide bound states will be discussed in the next section. In contrast, in the absence of nucleotides, the Sav1866 NBD-NBD interface became more mobile and hydrated, but did not dissociate, while closure of the outward open TMDs conformation was seen [141]. These results suggest that NBD separation might not be necessary to reach an occluded conformation. Comparable conformations were also observed in the crystal structures of McjD [60] and PglK [61]. This indicates that an occluded TMD conformation is plausible when the NBD are closely associated.
MD simulations studying nucleotide induced NBD associations were also carried out and started from inward facing conformations. MD simulations of the T. maritima TM287/288 heterodimeric exporter indicate that a single bound ATP is not enough to promote dimerization [134] as two nucleotides were needed to induce dimerization. Consistently, EPR data [137] showed that TM287/288 populates both inward- and outward facing conformations at hydrolysing conditions in the presence of ATP, while when trapped by Vi or by mutation of the catalytic glutamate, an associated NBD dimer geometry was observed, showing that the presence of nucleotide is required. The same conclusion was reached for the bacterial transporter MJ0796 (from M. jan-naschii) by LRET examining the association behavior of isolated NBDs [116].
Simulations starting from full-length ABC transporter structures with larger NBD separation turned out to be more challenging. Starting structures of MsbA [142–144] and ABCB1 [69] resulted unstable and changed conformation towards smaller NBD separation. Thereby, the structures spontaneously moved along a naturally existing energy gradient towards a lower energy state, which is a shallow but wide energy basin leading to very dynamic conformations, in line with EPR data [144]. For simulations starting from the inward open mouse ABCB1 structure [145] inserted in a lipid bilayer membrane, two metastable states could be reached in the presence of ATP: either i) ATP interacted only with the corresponding Walker A motif or ii) it also connected with the ABC signature motif of the opposite NBD, forming a partial dimer [121]. A canonical dimer structure was not observed, possibly due to the absence of substrate [121], or alternatively, sampling might have been insufficient for reaching a canonical dimer geometry in these unbiased simulations.
In summary, simulations suggest that bacterial homodimeric ABC transporters might reach the closed NBD dimer in a symmetric conformation in the presence of two bound ATPs. These results are in line with the ATP switch or the processive clamp model, which include/ imply symmetry in the ATP bound state, but also indicate the presence of fully separated NBDs after ATP hydrolysis. In contrast, the results from simulations of ABCB1 are less well converged, therefore not being able to rule out that the observed asymmetry could also be the result of limited sampling or the consequence of unstable (and therefore nonequilibrium) starting structures (models).
4.2. Asymmetric nucleotide binding and hydrolysis
Structural asymmetry in the NBDs has frequently been observed in simulations using an asymmetric nucleotide binding configuration. Opening of NBSs was seen when devoid of nucleotide, in line with most transport cycle models which predict that a closed, but empty NBS should not exist. A simulation starting from the symmetric Sav1866 transporter structure, while containing only one ATP, showed opening of the apo site and in parallel a further reduction of the Walker A - signature sequence distance in the ATP bound NBS [146], when compared to the crystal structure. Similar observations were made in simulations of Sav1866 with ATP in both NBSs or with ATP in NBS1 and ADP in NBS2. Here, ATP maintained strong interactions between the NBDs, while ADP could not stabilize the NBS bound geometry, leading to asymmetric NBS opening [147]. At the same time, the interactions within the tetra-helix bundle of the ICLs became weaker and increased flexibility was apparent in both NBSs. Closure of the TMD extracellular region was observed in one of the ATP + ADP bound replicas accompanied by twisting motions of the NBDs, suggesting that a NBD dissociation might not be necessary for reaching the closed TMD conformation, which is in line with alternating sites and constant contact models. In contrast, experimental data on bacterial ABC exporters, such as MsbA and LmrA showed that the NBDs are very mobile in the absence of nucleotide, and separate by over 2 nm [63,64,148], in line with full NBD separation during the transport cycle.
Another important distinction between ABC transporter models are the number of ATP molecules hydrolyzed per transport cycle. This has implications for mechanical coupling between ATP hydrolysis and transport. Binding of ATP to an NBS exerts an attractive force on both NBDs, whereby the γ-phosphate (Pγ) predominantly forms hydrogen bonds with the ABC signature sequence, while the Pα and Pβ mainly interact with the Walker A and B motifs. Breaking of the bond between Pβ and Pγ by ATP hydrolysis leads to a dramatic change in the equilibrium of forces. Now, the connection between the NBDs in the form of the ATP phosphate chain no longer exists, because ADP and Pi are two separate molecules. Importantly, within the NBDs, the NBSs are symmetrically arranged, but displaced from the geometric center. Thus, hydrolysis of a single ATP in one NBS will lead to rotational forces promoting asymmetric opening of one NBS, unless ATP hydrolysis is synchronized between the two NBSs. Synchronized hydrolyses of both ATPs would not generate such torque as both NBSs exert equal and therefore symmetrically balanced forces leading to linear NBD separation without rotation. Asymmetric models, which require only one ATP hydrolysis event to proceed through the transport cycle, therefore also need to include NBD rotation. In contrast, the ATP switch and the processive clamp models predict hydrolysis of two ATPs. Hydrolysis might then be synchronized, or proceed in two sequential steps, which leads to a new challenge: in the former situation, a mechanism is required for synchronized hydrolysis. In the latter, the symmetric complex (which is a high energy configuration [66,149]) needs to retain the first ADP + Pi until the second ATP is hydrolyzed or alternatively, energy conversion might proceed through two sequential rotational steps with opposing rotational direction. One of the supporting arguments for a two-step ATP hydrolysis mechanism is that both NBSs of ABCB1 are equally capable for ATP hydrolysis and that ABCB1 activity is fully blocked when one site is degenerated by a mutation [150]. In contrast, for ABCB11 a single ATP hydrolysis event might suffice, because it has only one canonical NBS which can hydrolyze ATP at a sufficient rate to sustain transport.
In summary, simulations of ABCB1 and its homologues have shown that the geometry of the NBDs are sensitive to the state of nucleotide binding, showing that ATP stabilizes the bound state, while ADP, ADP + Pi or the apo NBS induce separation, thereby recapitulating and complementing experimental results. Difficulties arise from limited sampling or unstable starting conformations, which need to be overcome to discriminate between the various transport models. There is accumulating (but not yet fully conclusive) support to indicate that bacterial homodimeric transporters might largely follow geometrically symmetric steps through the transport cycle. In contrast, the asymmetry observed in several ABCB1 simulations indicates that ABCB1 possibly follows a largely asymmetric path though the transport cycle.
4.3. Molecular mechanism of ATP hydrolysis
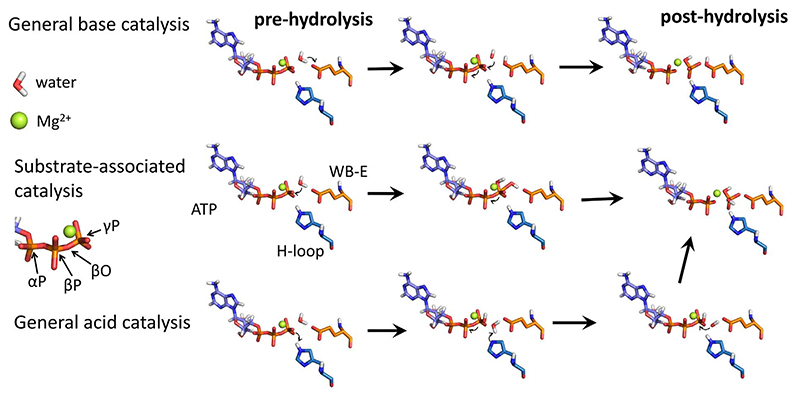
ATP binding and hydrolysis are processes that involve most of the conserved motifs within the NBD (Fig. 1c and Table 1): Conserved motifs such as A-loop, Walker A and ABC signature motif stabilize ATP, while the Mg2+ ion is positioned by ATP supported by the Q-loop and the Walker B glutamate. Both the Walker B glutamate and the H-loop histidine are necessary for effective ATP hydrolysis [22,151]. The first proposed catalytic mechanism was a ‘general base model’: the Walker B glutamate first accepts a proton from the lytic water molecule. The resulting hydroxide ion then attacks the γ-phosphate of ATP, which results in PβO-Pγ bond breakage and subsequent Pi release [152–154] (Fig. 4). The ‘general acid model’ assumes that a proton is abstracted from the H-loop histidine by Pγ of ATP, which in turn removes a hydrogen from the lytic water. The resulting hydroxide ion can then attack the PβO-Pγ bond of ATP. As a third alternative, the ‘substrate-assisted catalysis’ mechanism is built on the role of the H-loop histidine, which was proposed to maintain the conformation of the catalytic dyad, while water spontaneously attacked the Pγ of ATP [22].
Fig. 4.
ATP hydrolysis mechanisms showing the essential steps of the catalysis reaction of: top) the “general base catalysis”, middle) the “Substrate-associated catalysis” and bottom) the “general acid catalysis” reaction.
Recently, ATP hydrolysis in the NBDs (MalK2) of in the Maltose transporter MalEFGK2 was directly investigated using a mixed quantum mechanics/molecular mechanics (QM/MM) simulation approach. The atoms involved in the chemical reaction itself were described by a computationally very expensive quantum mechanical (QM) treatment [155] to describe chemical reactions of bond formation and bond breaking. This inner QM core of the simulations was then surrounded by all other atoms of the system described at the molecular mechanic (MM) classical force field level, which are orders of magnitude less computationally expensive. The three ATP hydrolysis scenarios were tested by combining the QM/MM simulations with a metadynamic biased sampling technique to steer the exploration of the energy hypersurface along the preset reaction coordinates of each hydrolysis model. The calculations showed the lowest activation barrier for the general base catalysis mechanism (43.9 kJ/mol), followed by the general acid catalysis mechanism (92.5 kJ/mol), while the substrate-assisted catalysis mechanism would require 134.3 kJ/mol. Experiments found an activation energy of 72.3 ± 4.2 kJ/mol [156] (Fig. 4). These results show that QM/MM approaches have the potential to reveal individual steps of the ATP hydrolysis event with atomic resolution.
5. Global conformational changes throughout the transport cycle
The ABC transporter transmembrane domains form the substrate translocation path across the membrane. The substrate binding pocket is of higher affinity for compounds, when accessible from the intracellular side and of lower affinity when exposed to the extracellular side [86]. The conformational changes of the transport cycle lead to changes between the substrate translocation path and also the residues constituting the substrate binding site explaining the changing affinity. The conformational transition between outward and inward facing states of MsbA was simulated, where the overall work and the reaction path were optimized to develop a low energy and therefore representative transition path [142,143]. As the protein is pushed from one conformation to the other along reaction coordinates, energy input (i.e. work) is required. Since this work is path dependent, it is presumed that paths requiring the least work are the most representative. The selected reaction coordinates were TMD angles, NBD center of mass distance and angle and were applied symmetrically to both half transporters. The least work was needed when the transporter was transformed from the outward to the inward open conformation in four distinct transition phases: (i) closing the periplasmic gate of the TMDs, and (ii) twisting the NBDs. The NBD rotation was suggested to be a necessary component of the conformational transition that has to twist before the cytoplasmic gate can be opened. The NBD twist was followed by (iii) increasing the NBD-NBD distance and finally (iv) opening the substrate binding pocket towards the intracellular side (see [142,143] for details).
TMD flexibility was found to decrease upon binding of ATP to the NBDs of ABCB1 in MD simulations [136], in agreement with experimental observations [66,137,157]. Similarly, MD simulations showed that the outward closed conformation of the ABCB1 was stabilized by bound ATP [79]. This finding was unexpected, because it had been generally assumed that the TMDs reach the outward open conformation upon ATP binding and NBD dimerization, as observed e.g. in the structure of Sav1866 [9]. However, the crystal structures of McjD [60], MsbA [62] and PglK [61] also showed an occluded TMD conformation in the presence of nucleotides and dimerized NBDs. Based on the MD simulations of ABCB1 [79], it was speculated that the ATP hydrolysis step, rather than ATP binding, might push the TMDs from the outward closed to the outward open conformation. These findings are supported by experimental results from EPR measurements on ABCB1 [66], while EPR data on MsbA [144], LmrA [63], ABCB1 [62] and structures of the bacterial ABC exporters Sav1866 and MsbA [59,93] showed outward open structures in the presence of AMPPNP.
The study of the full transport cycle, which takes approximately 0.3 s for ABCB1, requires enhanced sampling techniques and new methodological developments as applied for the study of MsbA [142,143]. Computational studies revealing the dynamics of the transport process at all atom resolution will remain a challenge, because of (i) the large difference in the time window (simulations now reach low μs, while the transport cycle is in the range of hundreds of milliseconds), (ii) the complexity of the conformational changes requiring extensive exploration of conformational space and (iii) the uncertainty of the sequence of steps implies the comparison of multiple paths. Simulations of the transport cycle will continue to rely on the guidance from experimental data to be able to address these challenges and to contribute a dynamic all atom view.
5.1. Membrane effects
Conformation and function of membrane proteins is strongly dependent on their environment, the lipid bilayer. Since many ABC transporters recruit their cargo from the membrane, with ABCB1 being a prime example, such a dependence on the lipid environment may be exceptionally pronounced. As recognized early on, compounds interacting with ABCB1 are typically lipophilic or amphipathic and accumulate in the membrane [18]. It was observed that, due to this accumulation, the measured affinities for substrate binding from the membrane are in the millimolar range [84,85,158,159]. This may also have facilitated the development of polyspecificity, as observed in ABCB1 (and other multidrug transporters), because the typically high enthalpic component of the binding energy, which also leads to high specificity and selectivity, is not needed and is possibly low. This expected small contribution of enthalpy to substrate binding is a burden for docking studies, because scoring functions used in docking are optimized for high affinity binders and largely rely on the enthalpic contribution of binding.
Beyond accumulating substrates, the membrane environment plays a second important role by exerting mechanical and structural constraints on the transporter. These are caused by lateral membrane pressure [160,161], membrane thickness [162] and fluidity [163], all of which are important for transporter stabilization and functionality. Experiments showed that ABCB1 activity can be modulated by changes in membrane thickness [117,159] and fluidity, membrane melting temperature [164], membrane composition [75,117], while detergents reduced or possibly eliminated ATPase activity [159]. Loo and Clarke [82] observed that ATPase and transport function of ICL cross-linked mutant transporters differed between native membrane and liposomes. We would like to refer our reader to the detailed summaries on the topic of experimental approaches studying lipid/membrane dependency of ABCB1 [165,166]. The effects of the environment on ABCB1 have also been addressed by MD simulations. Here, the conformational behavior of ABCB1 in conditions resembling the ionic detergent sodium cholate conditions used for crystallization were compared with a membrane environment containing cholesterol, POPC (palmitoyloleoyl-phospha-tidylcholine) and 150 mM NaCl [133]. Interestingly, the transporter remained close to the conformation observed in the crystal only if the environment also resembled the conditions used for crystallization. In contrast, ABCB1 showed large conformational changes in the membrane environment leading to a decrease of the NBD-NBD distance. Similarly, LRET studies on MsbA observed differences in NBD separation based on the membrane environment [119]. Such effects can potentially be rationalized by comparing the highly dynamic and adjustable detergent environment of a micelle with the membrane environment that forms a two-dimensional fluid imposing constraint.
5.2. Substrate binding
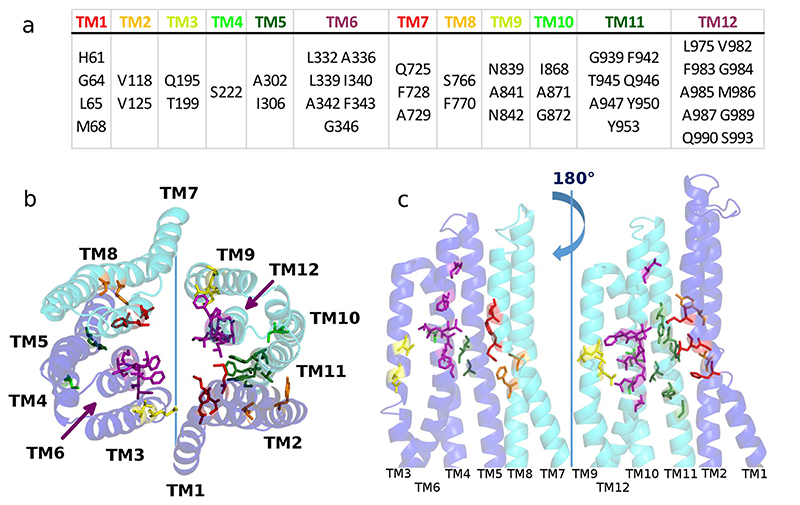
The TMD dimer in ABCB1 contains highly promiscuous binding site (s) for substrate and forms a translocation path in its center (Fig. 5). The interaction of the TMDs with substrates has been thoroughly investigated, and many residues involved in ligand binding were identified [167–169]. Biochemical analysis showed that all transmembrane helices are involved in substrate binding, covering a volume within the TMDs that is larger than any known ABCB1 substrate. ABCB1 is known to bind at least 300 compounds [170] and distinct binding sites for Hoechst 33324, rhodamine 123 [171] and prazosin [76] have been mapped. ABCB1 substrates and inhibitors have apparent affinities ranging from 37 nM to 160 mM [166], with compounds inhibiting ABCB1 showing a slightly higher affinity [135,172]. Binding is dominated by unspecific hydrophobic interactions, but specific hydrogen bonds have also been identified [173]. The residues identified to be involved in substrate interactions are oriented towards the inner cavity of the TMD and cluster halfway through the membrane, close to the restriction area as apparent in the crystal structures of inward facing ABCB1 (Fig. 5) [167,169,174]. Binding of the ligands QZ59-RRR and QZ59-SSS to ABCB1 in crystal structures (PDB ID: 4M2S and 4M2T) [58] and of lipid A to MsbA (PDB ID: 5TV4) [62] in a cryo-EM structure has been observed. The substrate binding sites overlap to a large extend with the biochemically identified ligand binding sites as highlighted in Fig. 5. Polyspecificity is attributed to the abundance of hydrophobic residues in the binding pocket, which provide a large and mainly hydrophobic patch for binding of various compounds. These residues adjust their conformation upon transition to the outward facing conformation, thereby leading to a decrease in affinity [74,168]. Free energy calculations were used to study ligand binding and to identify the binding site of nicardipin and morphine to inward facing ABCB1 [175]. These calculations showed an energy minimum for substrate binding when interacting with residues known to be involved in ligand binding. Enhanced sampling methods were used to study the substrate transport event of ABCB1 by inducing a transition from the inward to the outward facing state. Daunorubicin and verapamil were then found to move over 1 nm towards the extracellular compartment [176]. Substrates did not show specificity for one substrate translocation path, when comparing repeated simulation, suggesting that the process could be stochastic [172]. In contrast, functional data indicate that ABCB1 might contain two at least partially separated translocation paths [173,177], because substrates (rhodamine 123, verapamil, propafenones) associated with the R (for rhodamine binding) and the H (for Hoechst binding) site [171] were differently affected by charged residues positioned within the substrate translocation path.
Fig. 5. Substrate interacting residues in ABCB1 [167,169,174].
a) Table lists interacting residues grouped by transmembrane helices. b) These interacting residues are highlighted on an outward facing model of ABCB1 viewed from the extracellular space. The model was built with modeller [178] using Sav1866 (PDB ID: 2HYD) [9] as template (c) Open book representation of TMD1 and TMD2 viewed from the substrate binding pocked (as indicated by the blue line in panel b).
Promiscuity and weak binding are challenges for experimental as well as computational approaches. For correct in silico treatment, all ligand binding poses and their binding energy must be known. The hydrophobic nature of ABCB1-drug interactions requires treatment of entropy at high precision, which implies extensive sampling to obtain Boltzmann weighted distributions. With the availability of massive computer resources, it should now become possible to identify those properties that characterize e.g. the R and the H site.
6. Conclusions and outlook
Although many details of substrate recognition and translocation, ATP binding and hydrolysis as well as ABC transporter structures and structural changes have been elucidated, the mechanism of the overall transport cycle of ABCB1 remains elusive at the molecular level. TMDs and NBDs are linked by structural cross-talk as substrate binding increases the ATP hydrolysis rate and ATP hydrolysis leads to substrate translocation. The ICLs and some NBD motifs seem to have an essential role in this allosteric coupling. The NBDs form a sandwich dimer upon ATP binding, while the NBSs open to an uncertain extent after hydrolysis. Changes in the NBD-NBD conformation (distance and/or orientation) allow for the conversion of the chemical energy of ATP into mechanical energy by structural dynamics and conformation changes, also ensuring unidirectionality of transport. Environmental factors have an additional impact on substrate transport by ABC transporters. The membrane can increase the local substrate concentrations by several orders of magnitude; hence a high affinity substrate binding site might not be necessary to support the barrier function of ABCB1. The membrane also provides mechanical and structural support by affecting transporter conformation, conformational equilibria, confining motions and consequently impacting on the height of energy barriers encountered throughout the transport cycle. Increasing experimental and computational data suggest that the widely inward open conformations captured by some crystal structures might be too extreme, while some seemingly contradictory observations (e.g. differential effects of nucleotide binding and hydrolysis and NBD closure as observed by spectroscopic techniques) await elucidation.
Accumulating data suggests that the transport cycle of ABCB1 includes asymmetric states and that the NBDs carry an intrinsic tendency to adopt an asymmetrically occluded conformation. A similar asymmetry has until now rarely been observed in bacterial (homodimeric) half transporters. The available information still leaves room for both main types of models of the catalytic cycle (with/without NBD separation). It has also not yet been conclusively answered, whether all ABC transporters from the ABCB, ABCC and ABCD subfamilies, from archaea to humans, follow the same path through the transport cycle, especially because some transporters have two canonical NBS that efficiently hydrolyze ATP, while others carry a degenerate site. Consensus exists that ATP brings the two NBDs together. It remains debated however, whether the concentration of ATP, which in the cytosol is tenfold above KD, suffices to continuously bind and maintain ABC transporters in the ATP-bound conformation; this question is intimately linked to how far the NBDs can separate when the transporter is in its native membrane environment. The importance of the membrane in affecting protein conformations and equilibria is often underestimated, maybe because it is so difficult to quantify. For simulations starting from detergent solubilized transporter crystal structures, these effects are of pivotal importance and need to be correctly addressed to draw conclusions. The use of more native like environments in experiments, such as nanodiscs in cryo-EM or lipid cubic phase in crystallographic studies, is expected to improve the situation in the near future.
Several in silico studies provide important observations of the conformational changes of ABC transporters. Knowledge of forces and energies will be required to fully comprehend ABCB1 function from simulations. Structurally encoded dynamic movements of NBDs and TMDs have been captured. The next important step will be to connect NBDs and TMDs movements to nucleotides and substrate interactions with the aim to study substrate translocation directly as well as to elucidate the cross-talk between these domains. Computational studies have the advantage to contemporaneously provide dynamic information for all atoms, but also carry substantial limitations. In contrast, experimental data are often coarser, frequently exclusively probing e.g. one specific interaction or distance with high precision, but typically do not suffer from sampling problems. Building on the high complementary of obtainable results, it will most likely be a well-placed combination of several methods that can assemble and add the last piece to the tricky puzzle of the transport cycle of ABC transporters. in silico approaches are clearly part of the method portfolio to shed light on the remaining ABC transporter mysteries and to push our understanding of ABC transporter biology beyond the structure-function horizon.
Acknowledgements
Financial support by the Austrian Science Fund (FWF) for the stand alone project P23319 and the SFB subproject F3524 to TS is gratefully acknowledged. UAH acknowledges support by the Carl-Zeiss Foundation, the Center of Biomolecular Magnetic Resonance (BMRZ) funded by the state of Hesse and the 2017 Fulbright-Cottrell Award (funded by the German-American Fulbright commission, the Research Corporation for Science Advancement (RCSA) and the Bundesministerium für Bildung und Forschung (BMBF)). DRS acknowledges support from a Hans Böckler Stiftung PhD fellowship. We thank Franziska von Hammerstein, Erika Diehl and Luca Lauth for critical reading and insightful comments. We would like to apologize to all researchers whose work was not cited due to the sheer mass of publications in this field, but are thankful to everyone who has shaped our understanding of ABC transporters.
Transparency document
The http://dx.doi.org/10.1016/j.bbamem.2017.10.028 associated with this article can be found in online version.
References
- [1].ABC Proteins: From Bacteria to Man. Academic Press; 2015. [Google Scholar]
- [2].ABC Transporters - 40 Years on. Springer International Publishing; 2016. [Google Scholar]
- [3].Bryan J, Munoz A, Zhang X, Dufer M, Drews G, Krippeit-Drews P, Aguilar-Bryan L. ABCC8 and ABCC9: ABC transporters that regulate K + channels. Pflugers Arch. 2007;453:703–718. doi: 10.1007/s00424-006-0116-z. [DOI] [PubMed] [Google Scholar]
- [4].Barthelme D, Dinkelaker S, Albers SV, Londei P, Ermler U, Tampe R. Ribosome recycling depends on a mechanistic link between the FeS cluster domain and a conformational switch of the twin-ATPase ABCE1. Proc Natl Acad Sci U S A. 2011;108:3228–3233. doi: 10.1073/pnas.1015953108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Boel G, Smith PC, Ning W, Englander MT, Chen B, Hashem Y, Testa AJ, Fischer JJ, Wieden HJ, Frank J, Gonzalez RL, Jr, et al. The ABC-F protein EttA gates ribosome entry into the translation elongation cycle. Nat Struct Mol Biol. 2014;21:143–151. doi: 10.1038/nsmb.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Hohl M, Briand C, Grutter MG, Seeger MA. Crystal structure of a heterodimeric ABC transporter in its inward-facing conformation. Nat Struct Mol Biol. 2012;19:395–402. doi: 10.1038/nsmb.2267. [DOI] [PubMed] [Google Scholar]
- [7].Jin MS, Oldham ML, Zhang Q, Chen J. Crystal structure of the multidrug transporter P-glycoprotein from Caenorhabditis elegans. Nature. 2012;490:566–569. doi: 10.1038/nature11448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Smriti P, Zou HS. McHaourab, mapping daunorubicin-binding sites in the ATP-binding cassette transporter MsbA using site-specific quenching by spin labels. J Biol Chem. 2009;284:13904–13913. doi: 10.1074/jbc.M900837200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Dawson RJP, Locher KP. Structure of a bacterial multidrug ABC transporter. Nature. 2006;443:180–185. doi: 10.1038/nature05155. [DOI] [PubMed] [Google Scholar]
- [10].Hobson AC, Weatherwax R, Ames GF. ATP-binding sites in the membrane components of histidine permease, a periplasmic transport system. Proc Natl Acad Sci U S A. 1984;81:7333–7337. doi: 10.1073/pnas.81.23.7333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jones PM, George AM. A new structural model for P-glycoprotein. J Membr Biol. 1998;166:133–147. doi: 10.1007/s002329900455. [DOI] [PubMed] [Google Scholar]
- [12].Procko E, Ferrin-O’Connell I, Ng SL, Gaudet R. Distinct structural and functional properties of the ATPase sites in an asymmetric ABC transporter. Mol Cell. 2006;24:51–62. doi: 10.1016/j.molcel.2006.07.034. [DOI] [PubMed] [Google Scholar]
- [13].Ambudkar SV, Kim IW, Xia D, Sauna ZE. The A-loop, a novel conserved aromatic acid subdomain upstream of the Walker A motif in ABC transporters, is critical for ATP binding. FEBS Lett. 2006;580:1049–1055. doi: 10.1016/j.febslet.2005.12.051. [DOI] [PubMed] [Google Scholar]
- [14].Walker JE, Saraste M, Runswick MJ, Gay NJ. Distantly related sequences in the alpha-subunits and Beta-subunits of Atp synthase, myosin, kinases and other Atp-requiring enzymes and a common nucleotide binding fold. EMBO J. 1982;1:945–951. doi: 10.1002/j.1460-2075.1982.tb01276.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Ambudkar SV, Kim IW, Sauna ZE. The power of the pump: mechanisms of action of P-glycoprotein (ABCB1) Eur J Pharm Sci. 2006;27:392–400. doi: 10.1016/j.ejps.2005.10.010. [DOI] [PubMed] [Google Scholar]
- [16].Davidson AL, Laghaeian SS, Mannering DE. The maltose transport system of Escherichia coli displays positive cooperativity in ATP hydrolysis. J Biol Chem. 1996;271:4858–4863. [PubMed] [Google Scholar]
- [17].Kluth M, Stindt J, Droge C, Linnemann D, Kubitz R, Schmitt L. A mutation within the extended X loop abolished substrate-induced ATPase activity of the human liver ATP-binding cassette (ABC) transporter MDR3. J Biol Chem. 2015;290:4896–4907. doi: 10.1074/jbc.M114.588566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Higgins CF. Abc transporters - from microorganisms to man. Annu Rev Cell Biol. 1992;8:67–113. doi: 10.1146/annurev.cb.08.110192.000435. [DOI] [PubMed] [Google Scholar]
- [19].Geourjon C, Orelle C, Steinfels E, Blanchet C, Deleage G, Di Pietro A, Jault JM. A common mechanism for ATP hydrolysis in ABC transporter and helicase superfamilies. Trends Biochem Sci. 2001;26:539–544. doi: 10.1016/s0968-0004(01)01907-7. [DOI] [PubMed] [Google Scholar]
- [20].Grossmann N, Vakkasoglu AS, Hulpke S, Abele R, Gaudet R, Tampe R. Mechanistic determinants of the directionality and energetics of active export by a heterodimeric ABC transporter. Nat Commun. 2014;5 doi: 10.1038/ncomms6419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- [22].Zaitseva J, Jenewein S, Jumpertz T, Holland IB, Schmitt L. H662 is the linchpin of ATP hydrolysis in the nucleotide-binding domain of the ABC transporter HlyB. EMBO J. 2005;24:1901–1910. doi: 10.1038/sj.emboj.7600657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Cordon-Cardo C, O’Brien JP, Casals D, Rittman-Grauer L, Biedler JL, Melamed MR, Bertino JR. Multidrug-resistance gene (P-glycoprotein) is expressed by endothelial cells at blood-brain barrier sites. Proc Natl Acad Sci U S A. 1989;86:695–698. doi: 10.1073/pnas.86.2.695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hoffmeyer S, Burk O, von Richter O, Arnold HP, Brockmoller J, Johne A, Cascorbi I, Gerloff T, Roots I, Eichelbaum M, Brinkmann U. Functional polymorphisms of the human multidrug-resistance gene: multiple sequence variations and correlation of one allele with P-glycoprotein expression and activity in vivo. Proc Natl Acad Sci U S A. 2000;97:3473–3478. doi: 10.1073/pnas.050585397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Immunohistochemical localization in normal tissues of different epitopes in the multidrug transport protein P170: evidence for localization in brain capillaries and crossreactivity of one antibody with a muscle protein. J Histochem Cytochem. 1989;37:159–164. doi: 10.1177/37.2.2463300. [DOI] [PubMed] [Google Scholar]
- [26].de la Salle H, Zimmer J, Fricker D, Angenieux C, Cazenave JP, Okubo M, Maeda H, Plebani A, Tongio MM, Dormoy A, Hanau D. HLA class I deficiencies due to mutations in subunit 1 of the peptide transporter TAP1. J Clin Invest. 1999;103:R9–R13. doi: 10.1172/JCI5687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Neefjes JJ, Momburg F, Hammerling GJ. Selective and Atp-dependent translocation of peptides by the Mhc-encoded transporter. Science. 1993;261:769–771. doi: 10.1126/science.8342042. [DOI] [PubMed] [Google Scholar]
- [28].Scholz C, Tampe R. The peptide-loading complex—antigen translocation and MHC class I loading. Biol Chem. 2009;390:783–794. doi: 10.1515/BC.2009.069. [DOI] [PubMed] [Google Scholar]
- [29].Deleuze JF, Jacquemin E, Dubuisson C, Cresteil D, Dumont M, Erlinger S, Bernard O, Hadchouel M. Defect of multidrug-resistance 3 gene expression in a subtype of progressive familial intrahepatic cholestasis. Hepatology. 1996;23:904–908. doi: 10.1002/hep.510230435. [DOI] [PubMed] [Google Scholar]
- [30].Smit JJ, Schinkel AH, Mol CA, Majoor D, Mooi WJ, Jongsma AP, Lincke CR, Borst P. Tissue distribution of the human MDR3 P-glycoprotein. Lab Investig. 1994;71:638–649. [PubMed] [Google Scholar]
- [31].Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, Hofmann AF, Meier PJ. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- [32].Stieger B. Role of the bile salt export pump, BSEP, in acquired forms of cholestasis. Drug Metab Rev. 2010;42:437–445. doi: 10.3109/03602530903492004. [DOI] [PubMed] [Google Scholar]
- [33].Chen KG, Szakacs G, Annereau JP, Rouzaud F, Liang XJ, Valencia JC, Nagineni CN, Hooks JJ, Hearing VJ, Gottesman MM. Principal expression of two mRNA isoforms (ABCB5 alpha and ABCB5 beta) of the ATP-binding cassette transporter gene ABCB5 in melanoma cells and melanocytes. Pigment Cell Res. 2005;18:102–112. doi: 10.1111/j.1600-0749.2005.00214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Frank NY, Margaryan A, Huang Y, Schatton T, Waaga-Gasser AM, Gasser M, Sayegh MH, Sadee W, Frank MH. ABCB5-mediated doxorubicin transport and chemoresistance in human malignant melanoma. Cancer Res. 2005;65:4320–4333. doi: 10.1158/0008-5472.CAN-04-3327. [DOI] [PubMed] [Google Scholar]
- [35].Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, Fuhlbrigge RC, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Allikmets R, Raskind WH, Hutchinson A, Schueck ND, Dean M, Koeller DM. Mutation of a putative mitochondrial iron transporter gene (ABC7) in X-linked sideroblastic anemia and ataxia (XLSA/A) Hum Mol Genet. 1999;8:743–749. doi: 10.1093/hmg/8.5.743. [DOI] [PubMed] [Google Scholar]
- [37].Bekri S, Kispal G, Lange H, Fitzsimons E, Tolmie J, Lill R, Bishop DF. Human ABC7 transporter: gene structure and mutation causing X-linked sideroblastic anemia with ataxia with disruption of cytosolic iron-sulfur protein maturation. Blood. 2000;96:3256–3264. [PubMed] [Google Scholar]
- [38].Hogue DL, Liu L, Ling V. Identification and characterization of a mammalian mitochondrial ATP-binding cassette membrane protein. J Mol Biol. 1999;285:379–389. doi: 10.1006/jmbi.1998.2259. [DOI] [PubMed] [Google Scholar]
- [39].Shirihai OS, Gregory T, Yu C, Orkin SH, Weiss MJ. ABC-me: a novel mitochondrial transporter induced by GATA-1 during erythroid differentiation. EMBO J. 2000;19:2492–2502. doi: 10.1093/emboj/19.11.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Young L, Leonhard K, Tatsuta T, Trowsdale J, Langer T. Role of the ABC transporter Mdl1 in peptide export from mitochondria. Science. 2001;291:2135–2138. doi: 10.1126/science.1056957. [DOI] [PubMed] [Google Scholar]
- [41].Demirel O, Waibler Z, Kalinke U, Grunebach F, Appel S, Brossart P, Hasilik A, Tampe R, Abele R. Identification of a lysosomal peptide transport system induced during dendritic cell development. J Biol Chem. 2007;282:37836–37843. doi: 10.1074/jbc.M708139200. [DOI] [PubMed] [Google Scholar]
- [42].Wolters JC, Abele R, Tampe R. Selective and ATP-dependent translocation of peptides by the homodimeric ATP binding cassette transporter TAP-like (ABCB9) J Biol Chem. 2005;280:23631–23636. doi: 10.1074/jbc.M503231200. [DOI] [PubMed] [Google Scholar]
- [43].Yamaguchi Y, Kasano M, Terada T, Sato R, Maeda M. An ABC transporter homologous to TAP proteins. FEBS Lett. 1999;457:231–236. doi: 10.1016/s0014-5793(99)01042-x. [DOI] [PubMed] [Google Scholar]
- [44].Kiss K, Brozik A, Kucsma N, Toth A, Gera M, Berry L, Vallentin A, Vial H, Vidal M, Szakacs G. Shifting the paradigm: the putative mitochondrial protein ABCB6 resides in the lysosomes of cells and in the plasma membrane of erythrocytes. PLoS One. 2012;7 doi: 10.1371/journal.pone.0037378. e37378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Krishnamurthy PC, Du G, Fukuda Y, Sun D, Sampath J, Mercer KE, Wang J, Sosa-Pineda B, Murti KG, Schuetz JD. Identification of a mammalian mitochondrial porphyrin transporter. Nature. 2006;443:586–589. doi: 10.1038/nature05125. [DOI] [PubMed] [Google Scholar]
- [46].Leighton J, Schatz G. An ABC transporter in the mitochondrial inner membrane is required for normal growth of yeast. EMBO J. 1995;14:188–195. doi: 10.1002/j.1460-2075.1995.tb06989.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhang C, Li D, Zhang J, Chen X, Huang M, Archacki S, Tian Y, Ren W, Mei A, Zhang Q, Fang M, et al. Mutations in ABCB6 cause dyschromatosis universalis hereditaria. J Invest Dermatol. 2013;133:2221–2228. doi: 10.1038/jid.2013.145. [DOI] [PubMed] [Google Scholar]
- [48].Cordon-Cardo C, O’Brien JP, Boccia J, Casals D, Bertino JR, Melamed MR. Expression of the multidrug resistance gene product (P-glycoprotein) in human normal and tumor tissues. J Histochem Cytochem. 1990;38:1277–1287. doi: 10.1177/38.9.1974900. [DOI] [PubMed] [Google Scholar]
- [49].Chen CJ, Chin JE, Ueda K, Clark DP, Pastan I, Gottesman MM, Roninson IB. Internal duplication and homology with bacterial transport proteins in the mdr1 (P-glycoprotein) gene from multidrug-resistant human cells. Cell. 1986;47:381–389. doi: 10.1016/0092-8674(86)90595-7. [DOI] [PubMed] [Google Scholar]
- [50].Szakacs G, Paterson JK, Ludwig JA, Booth-Genthe C, Gottesman MM. Targeting multidrug resistance in cancer. Nat Rev Drug Discov. 2006;5:219–234. doi: 10.1038/nrd1984. [DOI] [PubMed] [Google Scholar]
- [51].Leslie EM, Deeley RG, Cole SP. Multidrug resistance proteins: role of P-gly-coprotein, MRP1, MRP2, and BCRP (ABCG2) in tissue defense. Toxicol Appl Pharmacol. 2005;204:216–237. doi: 10.1016/j.taap.2004.10.012. [DOI] [PubMed] [Google Scholar]
- [52].Obradovic T, Dobson GG, Shingaki T, Kungu T, Hidalgo IJ. Assessment of the first and second generation antihistamines brain penetration and role of P-glycoprotein. Pharm Res. 2007;24:318–327. doi: 10.1007/s11095-006-9149-4. [DOI] [PubMed] [Google Scholar]
- [53].Qian H, Zhao X, Cao P, Lei J, Yan N, Gong X. Structure of the human lipid exporter ABCA1. Cell. 2017;169:1228–1239. doi: 10.1016/j.cell.2017.05.020. e1210. [DOI] [PubMed] [Google Scholar]
- [54].Shintre CA, Pike AC, Li Q, Kim JI, Barr AJ, Goubin S, Shrestha L, Yang J, Berridge G, Ross J, Stansfeld PJ, et al. Structures of ABCB10, a human ATP-binding cassette transporter in apo- and nucleotide-bound states. Proc Natl Acad Sci U S A. 2013;110:9710–9715. doi: 10.1073/pnas.1217042110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Liu F, Zhang Z, Csanady L, Gadsby DC, Chen J. Molecular structure of the human CFTR ion channel. Cell. 2017;169:85–95. doi: 10.1016/j.cell.2017.02.024. e88. [DOI] [PubMed] [Google Scholar]
- [56].Lee JY, Kinch LN, Borek DM, Wang J, Urbatsch IL, Xie XS, Grishin NV, Cohen JC, Otwinowski Z, Hobbs HH, Rosenbaum DM. Crystal structure of the human sterol transporter ABCG5/ABCG8. Nature. 2016;533:561–564. doi: 10.1038/nature17666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Taylor NMI, Manolaridis I, Jackson SM, Kowal J, Stahlberg H, Locher KP. Structure of the human multidrug transporter ABCG2. Nature. 2017;546:504–509. doi: 10.1038/nature22345. [DOI] [PubMed] [Google Scholar]
- [58].Li J, Jaimes KF, Aller SG. Refined structures of mouse P-glycoprotein. Protein Sci. 2014;23:34–46. doi: 10.1002/pro.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Choudhury HG, Tong Z, Mathavan I, Li Y, Iwata S, Zirah S, Rebuffat S, van Veen HW, Beis K. Structure of an antibacterial peptide ATP-binding cassette transporter in a novel outward occluded state. Proc Natl Acad Sci U S A. 2014;111:9145–9150. doi: 10.1073/pnas.1320506111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Perez C, Gerber S, Boilevin J, Bucher M, Darbre T, Aebi M, Reymond JL, Locher KP. Structure and mechanism of an active lipid-linked oligosaccharide flippase. Nature. 2015;524:433–438. doi: 10.1038/nature14953. [DOI] [PubMed] [Google Scholar]
- [62].Mi W, Li Y, Yoon SH, Ernst RK, Walz T, Liao M. Structural basis of MsbA-mediated lipopolysaccharide transport. Nature. 2017;549:233–237. doi: 10.1038/nature23649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Hellmich UA, Lyubenova S, Kaltenborn E, Doshi R, van Veen HW, Prisner TF, Glaubitz C. Probing the ATP hydrolysis cycle of the ABC multidrug transporter LmrA by pulsed EPR spectroscopy. J Am Chem Soc. 2012;134:5857–5862. doi: 10.1021/ja211007t. [DOI] [PubMed] [Google Scholar]
- [64].Zou P, Bortolus M, Mchaourab HS. Conformational cycle of the ABC transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. J Mol Biol. 2009;393:586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Qu Q, Sharom FJ. FRET analysis indicates that the two ATPase active sites of the P-glycoprotein multidrug transporter are closely associated. Biochemistry. 2001;40:1413–1422. doi: 10.1021/bi002035h. [DOI] [PubMed] [Google Scholar]
- [66].Verhalen B, Dastvan R, Thangapandian S, Peskova Y, Koteiche HA, Nakamoto RK, Tajkhorshid E, McHaourab HS. Energy transduction and alternating access of the mammalian ABC transporter P-glycoprotein. Nature. 2017;543:738–741. doi: 10.1038/nature21414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Zolnerciks JK, Wooding C, Linton KJ. Evidence for a Sav1866-like architecture for the human multidrug transporter P-glycoprotein. FASEB J. 2007;21:3937–3948. doi: 10.1096/fj.07-8610com. [DOI] [PubMed] [Google Scholar]
- [68].Doshi R, Ali A, Shi W, Freeman EV, Fagg LA, van Veen HW. Molecular disruption of the power stroke in the ATP-binding cassette transport protein MsbA. J Biol Chem. 2013;288:6801–6813. doi: 10.1074/jbc.M112.430074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Wen PC, Verhalen B, Wilkens S, McHaourab HS, Tajkhorshid E. On the origin of large flexibility of P-glycoprotein in the inward-facing state. J Biol Chem. 2013;288:19211–19220. doi: 10.1074/jbc.M113.450114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Storm J, O’Mara ML, Crowley EH, Peall J, Tieleman DP, Kerr ID, Callaghan R. Residue G346 in transmembrane segment six is involved in interdomain communication in P-glycoprotein. Biochemistry. 2007;46:9899–9910. doi: 10.1021/bi700447p. [DOI] [PubMed] [Google Scholar]
- [71].Omote H, Figler RA, Polar MK, Al-Shawi MK. Improved energy coupling of human P-glycoprotein by the glycine 185 to valine mutation. Biochemistry. 2004;43:3917–3928. doi: 10.1021/bi035365l. [DOI] [PubMed] [Google Scholar]
- [72].Sato T, Kodan A, Kimura Y, Ueda K, Nakatsu T, Kato H. Functional role of the linker region in purified human P-glycoprotein. FEBS J. 2009;276:3504–3516. doi: 10.1111/j.1742-4658.2009.07072.x. [DOI] [PubMed] [Google Scholar]
- [73].Szabo K, Bakos E, Welker E, Muller M, Goodfellow HR, Higgins CF, Varadi A, Sarkadi B. Phosphorylation site mutations in the human multidrug transporter modulate its drug-stimulated ATPase activity. J Biol Chem. 1997;272:23165–23171. doi: 10.1074/jbc.272.37.23165. [DOI] [PubMed] [Google Scholar]
- [74].Esser L, Zhou F, Pluchino KM, Shiloach J, Ma J, Tang WK, Gutierrez C, Zhang A, Shukla S, Madigan JP, Zhou T, et al. Structures of the multidrug transporter P-glycoprotein reveal asymmetric ATP binding and the mechanism of polyspecificity. J Biol Chem. 2017;292(2):446–461. doi: 10.1074/jbc.M116.755884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Senior AE, Al-Shawi MK, Urbatsch IL. ATP hydrolysis by multidrug-resistance protein from Chinese hamster ovary cells. J Bioenerg Biomembr. 1995;27:31–36. doi: 10.1007/BF02110328. [DOI] [PubMed] [Google Scholar]
- [76].Shapiro AB, Fox K, Lam P, Ling V. Stimulation of P-glycoprotein-mediated drug transport by prazosin and progesterone. Evidence for a third drug-binding site. Eur J Biochem. 1999;259:841–850. doi: 10.1046/j.1432-1327.1999.00098.x. [DOI] [PubMed] [Google Scholar]
- [77].Sharom FJ, DiDiodato G, Yu X, Ashbourne KJ. Interaction of the P-glycoprotein multidrug transporter with peptides and ionophores. J Biol Chem. 1995;270:10334–10341. doi: 10.1074/jbc.270.17.10334. [DOI] [PubMed] [Google Scholar]
- [78].Watchko JF, Daood MJ, Mahmood B, Vats K, Hart C, Ahdab-Barmada M. P-glycoprotein and bilirubin disposition. J Perinatol. 2001;21(1):S43–47. doi: 10.1038/sj.jp.7210633. discussion S59-62. [DOI] [PubMed] [Google Scholar]
- [79].Xu Y, Seelig A, Berneche S. Unidirectional transport mechanism in an ATP dependent exporter. ACS Cent Sci. 2017;3:250–258. doi: 10.1021/acscentsci.7b00068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Zolnerciks JK, Akkaya BG, Snippe M, Chiba P, Seelig A, Linton KJ. The Q loops of the human multidrug resistance transporter ABCB1 are necessary to couple drug binding to the ATP catalytic cycle. FASEB J. 2014;28:4335–4346. doi: 10.1096/fj.13-245639. [DOI] [PubMed] [Google Scholar]
- [81].Barsony O, Szaloki G, Turk D, Tarapcsak S, Gutay-Toth Z, Bacso Z, Holb IJ, Szekvolgyi L, Szabo G, Csanady L, Szakacs G, et al. A single active catalytic site is sufficient to promote transport in P-glycoprotein. Sci Rep. 2016;6 doi: 10.1038/srep24810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Loo TW, Clarke DM. Attachment of a ‘molecular spring’ restores drug-stimulated ATPase activity to P-glycoprotein lacking both Q loop glutamines. Biochem Biophys Res Commun. 2017;483:366–370. doi: 10.1016/j.bbrc.2016.12.137. [DOI] [PubMed] [Google Scholar]
- [83].Jardetzky O. Simple allosteric model for membrane pumps. Nature. 1966;211:969–970. doi: 10.1038/211969a0. [DOI] [PubMed] [Google Scholar]
- [84].Callaghan R, Ford RC, Kerr ID. The translocation mechanism of P-glycoprotein. FEBS Lett. 2006;580:1056–1063. doi: 10.1016/j.febslet.2005.11.083. [DOI] [PubMed] [Google Scholar]
- [85].Clay AT, Sharom FJ. Lipid bilayer properties control membrane partitioning, binding, and transport of P-glycoprotein substrates. Biochemistry. 2013;52:343–354. doi: 10.1021/bi301532c. [DOI] [PubMed] [Google Scholar]
- [86].Rees DC, Johnson E, Lewinson O. ABC transporters: the power to change. Nat Rev Mol Cell Biol. 2009;10:218–227. doi: 10.1038/nrm2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Higgins CF, Gottesman MM. Is the multidrug transporter a flippase? Trends Biochem Sci. 1992;17:18–21. doi: 10.1016/0968-0004(92)90419-a. [DOI] [PubMed] [Google Scholar]
- [88].Eytan GD, Regev R, Oren G, Assaraf YG. The role of passive transbilayer drug movement in multidrug resistance and its modulation. J Biol Chem. 1996;271:12897–12902. doi: 10.1074/jbc.271.22.12897. [DOI] [PubMed] [Google Scholar]
- [89].Ambudkar SV, Lelong IH, Zhang J, Cardarelli CO, Gottesman MM, Pastan I. Partial purification and reconstitution of the human multidrug-resistance pump: characterization of the drug-stimulatable ATP hydrolysis. Proc Natl Acad Sci U S A. 1992;89:8472–8476. doi: 10.1073/pnas.89.18.8472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Loo TW, Clarke DM. Drug-stimulated ATPase activity of human P-glycoprotein requires movement between transmembrane segments 6 and 12. J Biol Chem. 1997;272:20986–20989. doi: 10.1074/jbc.272.34.20986. [DOI] [PubMed] [Google Scholar]
- [91].Sarkadi B, Muller M, Homolya L, Hollo Z, Seprodi J, Germann UA, Gottesman MM, Price EM, Boucher RC. Interaction of bioactive hydrophobic peptides with the human multidrug transporter. FASEB J. 1994;8:766–770. doi: 10.1096/fasebj.8.10.7914178. [DOI] [PubMed] [Google Scholar]
- [92].Urbatsch IL, Al-Shawi MK, Senior AE. Characterization of the ATPase activity of purified Chinese hamster P-glycoprotein. Biochemistry. 1994;33:7069–7076. doi: 10.1021/bi00189a008. [DOI] [PubMed] [Google Scholar]
- [93].Dawson RJ, Locher KP. Structure of the multidrug ABC transporter Sav1866 from Staphylococcus aureus in complex with AMP-PNP. FEBS Lett. 2007;581:935–938. doi: 10.1016/j.febslet.2007.01.073. [DOI] [PubMed] [Google Scholar]
- [94].Chen J, Lu G, Lin J, Davidson AL, Quiocho FA. A tweezers-like motion of the ATP-binding cassette dimer in an ABC transport cycle. Mol Cell. 2003;12:651–661. doi: 10.1016/j.molcel.2003.08.004. [DOI] [PubMed] [Google Scholar]
- [95].Higgins CF, Linton KJ. The ATP switch model for ABC transporters. Nat Struct Mol Biol. 2004;11:918–926. doi: 10.1038/nsmb836. [DOI] [PubMed] [Google Scholar]
- [96].Janas E, Hofacker M, Chen M, Gompf S, van der Does C, Tampe R. The ATP hydrolysis cycle of the nucleotide-binding domain of the mitochondrial ATP-binding cassette transporter Mdl1p. J Biol Chem. 2003;278:26862–26869. doi: 10.1074/jbc.M301227200. [DOI] [PubMed] [Google Scholar]
- [97].Smith PC, Karpowich N, Millen L, Moody JE, Rosen J, Thomas PJ, Hunt JF. ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer. Mol Cell. 2002;10:139–149. doi: 10.1016/s1097-2765(02)00576-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].van der Does C, Tampe R. How do ABC transporters drive transport? Biol Chem. 2004;385:927–933. doi: 10.1515/BC.2004.121. [DOI] [PubMed] [Google Scholar]
- [99].Senior AE, Al-Shawi MK, Urbatsch IL. The catalytic cycle of P-glycoprotein. FEBS Lett. 1995;377:285–289. doi: 10.1016/0014-5793(95)01345-8. [DOI] [PubMed] [Google Scholar]
- [100].Jones PM, George AM. Opening of the ADP-bound Active Site in the ABC Transporter ATPase Dimer: Evidence for a Constant Contact, Alternating Sites Model for the Catalytic Cycle. Proteins-structure Function and Bioinformatics. 2009;75:387–396. doi: 10.1002/prot.22250. [DOI] [PubMed] [Google Scholar]
- [101].Sauna ZE, Ambudkar SV. About a switch: how P-glycoprotein (ABCB1) harnesses the energy of ATP binding and hydrolysis to do mechanical work. Mol Cancer Ther. 2007;6:13–23. doi: 10.1158/1535-7163.MCT-06-0155. [DOI] [PubMed] [Google Scholar]
- [102].Sauna ZE, Kim IW, Nandigama K, Kopp S, Chiba P, Ambudkar SV. Catalytic cycle of ATP hydrolysis by P-glycoprotein: evidence for formation of the E.S reaction intermediate with ATP-gamma-S, a nonhydrolyzable analogue of ATP. Biochemistry. 2007;46:13787–13799. doi: 10.1021/bi701385t. [DOI] [PubMed] [Google Scholar]
- [103].Siarheyeva A, Liu R, Sharom FJ. Characterization of an asymmetric occluded state of P-glycoprotein with two bound nucleotides: implications for catalysis. J Biol Chem. 2010;285:7575–7586. doi: 10.1074/jbc.M109.047290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Beis I, Newsholme EA. The contents of adenine nucleotides, phosphagens and some glycolytic intermediates in resting muscles from vertebrates and invertebrates. Biochem J. 1975;152:23–32. doi: 10.1042/bj1520023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Oldham ML, Chen J. Crystal structure of the maltose transporter in a pretranslocation intermediate state. Science. 2011;332:1202–1205. doi: 10.1126/science.1200767. [DOI] [PubMed] [Google Scholar]
- [106].Hvorup RN, Goetz BA, Niederer M, Hollenstein K, Perozo E, Locher KP. Asymmetry in the structure of the ABC transporter-binding protein complex BtuCD-BtuF. Science. 2007;317:1387–1390. doi: 10.1126/science.1145950. [DOI] [PubMed] [Google Scholar]
- [107].Jones PM, O’Mara ML, George AM. ABC transporters: a riddle wrapped in a mystery inside an enigma. Trends Biochem Sci. 2009;34:520–531. doi: 10.1016/j.tibs.2009.06.004. [DOI] [PubMed] [Google Scholar]
- [108].Hellmich UA, Monkemeyer L, Velamakanni S, van Veen HW, Glaubitz C. Effects of nucleotide binding to LmrA: a combined MAS-NMR and solution NMR study. Biochim Biophys Acta. 2015;1848:3158–3165. doi: 10.1016/j.bbamem.2015.10.003. [DOI] [PubMed] [Google Scholar]
- [109].Wiegand T, Lacabanne D, Keller K, Cadalbert R, Lecoq L, Yulikov M, Terradot L, Jeschke G, Meier BH, Bockmann A. Solid-state NMR and EPR Spectroscopy of Mn2 + -substituted ATP-fueled protein engines. Angew Chem Int Ed Eng. 2017;56:3369–3373. doi: 10.1002/anie.201610551. [DOI] [PubMed] [Google Scholar]
- [110].Collauto A, Mishra S, Litvinov A, McHaourab HS, Goldfarb D. Direct spectroscopic detection of ATP turnover reveals mechanistic divergence of ABC exporters. Structure. 2017;25:1264–1274. doi: 10.1016/j.str.2017.06.014. e1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Tombline G, Senior AE. The occluded nucleotide conformation of P-glycoprotein. J Bioenerg Biomembr. 2005;37:497–500. doi: 10.1007/s10863-005-9498-4. [DOI] [PubMed] [Google Scholar]
- [112].Martin C, Higgins CF, Callaghan R. The vinblastine binding site adopts high- and low-affinity conformations during a transport cycle of P-glycoprotein. Biochemistry. 2001;40:15733–15742. doi: 10.1021/bi011211z. [DOI] [PubMed] [Google Scholar]
- [113].Tombline G, Bartholomew LA, Urbatsch IL, Senior AE. Combined mutation of catalytic glutamate residues in the two nucleotide binding domains of P-glycoprotein generates a conformation that binds ATP and ADP tightly. J Biol Chem. 2004;279:31212–31220. doi: 10.1074/jbc.M404689200. [DOI] [PubMed] [Google Scholar]
- [114].Urbatsch IL, Tyndall GA, Tombline G, Senior AE. P-glycoprotein catalytic mechanism: studies of the ADP-vanadate inhibited state. J Biol Chem. 2003;278:23171–23179. doi: 10.1074/jbc.M301957200. [DOI] [PubMed] [Google Scholar]
- [115].Zoghbi ME, Altenberg GA. Hydrolysis at one of the two nucleotide-binding sites drives the dissociation of ATP-binding cassette nucleotide-binding domain dimers. J Biol Chem. 2013;288:34259–34265. doi: 10.1074/jbc.M113.500371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Zoghbi ME, Altenberg GA. ATP binding to two sites is necessary for dimerization of nucleotide-binding domains of ABC proteins. Biochem Biophys Res Commun. 2014;443:97–102. doi: 10.1016/j.bbrc.2013.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Moeller A, Lee SC, Tao H, Speir JA, Chang G, Urbatsch IL, Potter CS, Carragher B, Zhang Q. Distinct conformational spectrum of homologous multidrug ABC transporters. Structure. 2015;23:450–460. doi: 10.1016/j.str.2014.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Loo TW, Bartlett MC, Clarke DM. Human P-glycoprotein is active when the two halves are clamped together in the closed conformation. Biochem Biophys Res Commun. 2010;395:436–440. doi: 10.1016/j.bbrc.2010.04.057. [DOI] [PubMed] [Google Scholar]
- [119].Zoghbi ME, Cooper RS, Altenberg GA. The lipid bilayer modulates the structure and function of an ATP-binding cassette exporter. J Biol Chem. 2016;291:4453–4461. doi: 10.1074/jbc.M115.698498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kandt C, Monticelli L. Membrane protein dynamics from femtoseconds to seconds. Methods Mol Biol. 2010;654:423–440. doi: 10.1007/978-1-60761-762-4_22. [DOI] [PubMed] [Google Scholar]
- [121].O’Mara ML, Mark AE. Structural characterization of two metastable ATP-bound states of P-glycoprotein. PLoS One. 2014;9:e91916. doi: 10.1371/journal.pone.0091916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Stockner T, Mullen A, MacMillan F. Investigating the dynamic nature of the ABC transporters: ABCB1 and MsbA as examples for the potential synergies of MD theory and EPR applications. Biochem Soc Trans. 2015;43:1023–1032. doi: 10.1042/BST20150138. [DOI] [PubMed] [Google Scholar]
- [123].Schlitter J, Engels M, Kruger P, Jacoby E, Wollmer A. Targeted molecular-dynamics simulation of conformational change - application to the T–R transition in insulin. Mol Simul. 1993;10:291. [Google Scholar]
- [124].Bussi G, Laio A, Parrinello M. Equilibrium free energies from nonequilibrium metadynamics. Phys Rev Lett. 2006;96 doi: 10.1103/PhysRevLett.96.090601. 090601. [DOI] [PubMed] [Google Scholar]
- [125].Dama JF, Hocky GM, Sun R, Voth GA. Exploring valleys without climbing every peak: more efficient and forgiving metabasin metadynamics via robust on-the-fly bias domain restriction. J Chem Theory Comput. 2015;11:5638–5650. doi: 10.1021/acs.jctc.5b00907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Isralewitz B, Izrailev S, Schulten K. Binding pathway of retinal to bacterio-opsin: a prediction by molecular dynamics simulations. Biophys J. 1997;73:2972–2979. doi: 10.1016/S0006-3495(97)78326-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Marrink SJ, Risselada HJ, Yefimov S, Tieleman DP, de Vries AH. The MARTINI force field: coarse grained model for biomolecular simulations. J Phys Chem B. 2007;111:7812–7824. doi: 10.1021/jp071097f. [DOI] [PubMed] [Google Scholar]
- [128].Arnarez C, Uusitalo JJ, Masman MF, Ingolfsson HI, de Jong DH, Melo MN, Periole X, de Vries AH, Marrink SJ. Dry Martini, a coarse-grained force field for lipid membrane simulations with implicit solvent. J Chem Theory Comput. 2015;11:260–275. doi: 10.1021/ct500477k. [DOI] [PubMed] [Google Scholar]
- [129].Wassenaar TA, Ingolfsson HI, Bockmann RA, Tieleman DP, Marrink SJ. Computational Lipidomics with insane: a versatile tool for generating custom membranes for molecular simulations. J Chem Theory Comput. 2015;11:2144–2155. doi: 10.1021/acs.jctc.5b00209. [DOI] [PubMed] [Google Scholar]
- [130].Monticelli L, Kandasamy SK, Periole X, Larson RG, Tieleman DP, Marrink SJ. The MARTINI coarse-grained force field: extension to proteins. J Chem Theory Comput. 2008;4:819–834. doi: 10.1021/ct700324x. [DOI] [PubMed] [Google Scholar]
- [131].Pluhackova K, Gahbauer S, Kranz F, Wassenaar TA, Bockmann RA. Dynamic cholesterol-conditioned dimerization of the G protein coupled chemokine receptor type 4. PLoS Comput Biol. 2016;12 doi: 10.1371/journal.pcbi.1005169. e1005169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Wassenaar TA, Pluhackova K, Moussatova A, Sengupta D, Marrink SJ, Tieleman DP, Bockmann RA. High-throughput simulations of dimer and trimer assembly of membrane proteins. The DAFT approach. J Chem Theory Comput. 2015;11:2278–2291. doi: 10.1021/ct5010092. [DOI] [PubMed] [Google Scholar]
- [133].O’Mara ML, Mark AE. The effect of environment on the structure of a membrane protein: P-glycoprotein under physiological conditions. J Chem Theory Comput. 2012;8:3964–3976. doi: 10.1021/ct300254y. [DOI] [PubMed] [Google Scholar]
- [134].Furuta T, Sato Y, Sakurai M. Structural dynamics of the Heterodimeric ABC transporter TM287/288 induced by ATP and substrate binding. Biochemistry. 2016;55:6730–6738. doi: 10.1021/acs.biochem.6b00947. [DOI] [PubMed] [Google Scholar]
- [135].Pan C, Weng J, Wang W. Conformational dynamics and protein-substrate interaction of ABC transporter BtuCD at the occluded state revealed by molecular dynamics simulations. Biochemistry. 2016;55:6897–6907. doi: 10.1021/acs.biochem.6b00386. [DOI] [PubMed] [Google Scholar]
- [136].Pan L, Aller SG. Equilibrated atomic models of outward-facing P-glycoprotein and effect of ATP binding on structural dynamics. Sci Rep. 2015;5:7880. doi: 10.1038/srep07880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Timachi MH, Hutter CA, Hohl M, Assafa T, Bohm S, Mittal A, Seeger MA, Bordignon E. Exploring conformational equilibria of a heterodimeric ABC transporter. elife. 2017;6 doi: 10.7554/eLife.20236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Wen PC, Tajkhorshid E. Dimer opening of the nucleotide binding domains of ABC transporters after ATP hydrolysis. Biophys J. 2008;95:5100–5110. doi: 10.1529/biophysj.108.139444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Zaitseva J, Jenewein S, Wiedenmann A, Benabdelhak H, Holland IB, Schmitt L. Functional characterization and ATP-induced dimerization of the isolated ABC-domain of the haemolysin B transporter. Biochemistry. 2005;44:9680–9690. doi: 10.1021/bi0506122. [DOI] [PubMed] [Google Scholar]
- [140].Oliveira AS, Baptista AM, Soares CM. Conformational changes induced by ATP-hydrolysis in an ABC transporter: a molecular dynamics study of the Sav1866 exporter. Proteins: Struct, Funct, Bioinf. 2011;79:1977–1990. doi: 10.1002/prot.23023. [DOI] [PubMed] [Google Scholar]
- [141].Becker JP, Van Bambeke F, Tulkens PM, Prevost M. Dynamics and structural changes induced by ATP binding in SAV1866, a bacterial ABC exporter. J Phys Chem B. 2010;114:15948–15957. doi: 10.1021/jp1038392. [DOI] [PubMed] [Google Scholar]
- [142].Moradi M, Tajkhorshid E. Mechanistic picture for conformational transition of a membrane transporter at atomic resolution. Proc Natl Acad Sci U S A. 2013;110:18916–18921. doi: 10.1073/pnas.1313202110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Moradi M, Tajkhorshid E. Computational recipe for efficient description of large-scale conformational changes in biomolecular systems. J Chem Theory Comput. 2014;10:2866–2880. doi: 10.1021/ct5002285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Dong J, Yang G, McHaourab HS. Structural basis of energy transduction in the transport cycle of MsbA. Science. 2005;308:1023–1028. doi: 10.1126/science.1106592. [DOI] [PubMed] [Google Scholar]
- [145].Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Jones PM, George AM. Molecular-dynamics simulations of the ATP/apo state of a multidrug ATP-binding cassette transporter provide a structural and mechanistic basis for the asymmetric occluded state. Biophys J. 2011;100:3025–3034. doi: 10.1016/j.bpj.2011.05.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Gyimesi G, Ramachandran S, Kota P, Dokholyan NV, Sarkadi B, Hegedus T. ATP hydrolysis at one of the two sites in ABC transporters initiates transport related conformational transitions. Biochim Biophys Acta Biomembr. 2011;1808:2954–2964. doi: 10.1016/j.bbamem.2011.07.038. [DOI] [PubMed] [Google Scholar]
- [148].Siarheyeva A, Lopez JJ, Lehner I, Hellmich UA, van Veen HW, Glaubitz C. Probing the molecular dynamics of the ABC multidrug transporter LmrA by deuterium solid-state nuclear magnetic resonance. Biochemistry. 2007;46:3075–3083. doi: 10.1021/bi062109a. [DOI] [PubMed] [Google Scholar]
- [149].Scian M, Acchione M, Li M, Atkins WM. Reaction dynamics of ATP hydrolysis catalyzed by P-glycoprotein. Biochemistry. 2014;53:991–1000. doi: 10.1021/bi401280v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Urbatsch IL, Sankaran B, Bhagat S, Senior AE. Both P-glycoprotein nucleotide-binding sites are catalytically active. J Biol Chem. 1995;270:26956–26961. doi: 10.1074/jbc.270.45.26956. [DOI] [PubMed] [Google Scholar]
- [151].Ernst R, Koch J, Horn C, Tampe R, Schmitt L. Engineering ATPase activity in the isolated ABC cassette of human TAP1. J Biol Chem. 2006;281:27471–27480. doi: 10.1074/jbc.M601131200. [DOI] [PubMed] [Google Scholar]
- [152].Fersht A. Structure and Mechanism in Protein Science: A Guide to Enzyme Catalysis and Protein Folding. 1999 [Google Scholar]
- [153].Orelle C, Dalmas O, Gros P, Di Pietro A, Jault JM. The conserved glutamate residue adjacent to the Walker-B motif is the catalytic base for ATP hydrolysis in the ATP-binding cassette transporter BmrA. J Biol Chem. 2003;278:47002–47008. doi: 10.1074/jbc.M308268200. [DOI] [PubMed] [Google Scholar]
- [154].Payen LF, Gao M, Westlake CJ, Cole SP, Deeley RG. Role of carboxylate residues adjacent to the conserved core Walker B motifs in the catalytic cycle of multidrug resistance protein 1 (ABCC1) J Biol Chem. 2003;278:38537–38547. doi: 10.1074/jbc.M305786200. [DOI] [PubMed] [Google Scholar]
- [155].Zhou Y, Ojeda-May P, Nagaraju M, Pu J. Toward determining ATPase mechanism in ABC transporters: development of the reaction path-force matching QM/MM method. Methods Enzymol. 2016;577:185–212. doi: 10.1016/bs.mie.2016.05.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Hsu WL, Furuta T, Sakurai M. ATP hydrolysis mechanism in a maltose transporter explored by QM/MM metadynamics simulation. J Phys Chem B. 2016;120:11102–11112. doi: 10.1021/acs.jpcb.6b07332. [DOI] [PubMed] [Google Scholar]
- [157].Mishra S, Verhalen B, Stein RA, Wen PC, Tajkhorshid E, McHaourab HS. Conformational dynamics of the nucleotide binding domains and the power stroke of a heterodimeric ABC transporter. elife. 2014;3 doi: 10.7554/eLife.02740. e02740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [158].Li-Blatter X, Beck A, Seelig A. P-glycoprotein-ATPase modulation: the molecular mechanisms. Biophys J. 2012;102:1383–1393. doi: 10.1016/j.bpj.2012.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Sharom FJ, Yu X, Chu JW, Doige CA. Characterization of the ATPase activity of P-glycoprotein from multidrug-resistant Chinese hamster ovary cells. Biochem J. 1995;308(Pt 2):381–390. doi: 10.1042/bj3080381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Cantor RS. The lateral pressure profile in membranes: a physical mechanism of general anesthesia. Biochemistry. 1997;36:2339–2344. doi: 10.1021/bi9627323. [DOI] [PubMed] [Google Scholar]
- [161].Jerabek H, Pabst G, Rappolt M, Stockner T. Membrane-mediated effect on ion channels induced by the anesthetic drug ketamine. J Am Chem Soc. 2010;132:7990–7997. doi: 10.1021/ja910843d. [DOI] [PubMed] [Google Scholar]
- [162].Johnson SM. The effect of charge and cholesterol on the size and thickness of sonicated phospholipid vesicles. Biochim Biophys Acta. 1973;307:27–41. doi: 10.1016/0005-2736(73)90022-9. [DOI] [PubMed] [Google Scholar]
- [163].Chapman D. Fluidity and phase transitions of cell membranes. Biomembranes. 1975;7:1–9. doi: 10.1007/978-1-4684-7668-2_1. [DOI] [PubMed] [Google Scholar]
- [164].Romsicki Y, Sharom FJ. The ATPase and ATP-binding functions of P-glycoprotein—modulation by interaction with defined phospholipids. Eur J Biochem. 1998;256:170–178. doi: 10.1046/j.1432-1327.1998.2560170.x. [DOI] [PubMed] [Google Scholar]
- [165].Neumann J, Rose-Sperling D, Hellmich UA. Diverse relations between ABC transporters and lipids: an overview. Biochim Biophys Acta. 2017;1859:605–618. doi: 10.1016/j.bbamem.2016.09.023. [DOI] [PubMed] [Google Scholar]
- [166].Sharom FJ. Complex interplay between the P-glycoprotein multidrug efflux pump and the membrane: its role in modulating protein function. Front Oncol. 2014;4:41. doi: 10.3389/fonc.2014.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Chufan EE, Sim HM, Ambudkar SV. Molecular basis of the polyspecificity of P-glycoprotein (ABCB1): recent biochemical and structural studies. Adv Cancer Res. 2015;125:71–96. doi: 10.1016/bs.acr.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Gutmann DA, Ward A, Urbatsch IL, Chang G, van Veen HW. Understanding polyspecificity of multidrug ABC transporters: closing in on the gaps in ABCB1. Trends Biochem Sci. 2010;35:36–42. doi: 10.1016/j.tibs.2009.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Loo TW, Clarke DM. Mutational analysis of ABC proteins. Arch Biochem Biophys. 2008;476:51–64. doi: 10.1016/j.abb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- [170].Callaghan R. Providing a molecular mechanism for P-glycoprotein; why would I bother? Biochem Soc Trans. 2015;43:995–1002. doi: 10.1042/BST20150131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Shapiro AB, Ling V. Positively cooperative sites for drug transport by P-glycoprotein with distinct drug specificities. Eur J Biochem. 1997;250:130–137. doi: 10.1111/j.1432-1033.1997.00130.x. [DOI] [PubMed] [Google Scholar]
- [172].McCormick JW, Vogel PD, Wise JG. Multiple drug transport pathways through human P-glycoprotein. Biochemistry. 2015;54:4374–4390. doi: 10.1021/acs.biochem.5b00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [173].Donmez Cakil Y, Khunweeraphong N, Parveen Z, Schmid D, Artaker M, Ecker GF, Sitte HH, Pusch O, Stockner T, Chiba P. Pore-exposed tyrosine residues of P-glycoprotein are important hydrogen-bonding partners for drugs. Mol Pharmacol. 2014;85:420–428. doi: 10.1124/mol.113.088526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [174].Frank GA, Shukla S, Rao P, Borgnia MJ, Bartesaghi A, Merk A, Mobin A, Esser L, Earl LA, Gottesman MM, Xia D, et al. Cryo-EM analysis of the conformational landscape of human P-glycoprotein (ABCB1) during its catalytic cycle. Mol Pharmacol. 2016;90:35–41. doi: 10.1124/mol.116.104190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [175].Subramanian N, Condic-Jurkic K, Mark AE, O’Mara ML. Identification of possible binding sites for morphine and Nicardipine on the multidrug transporter P-glycoprotein using umbrella sampling techniques. J Chem Inf Model. 2015;55:1202–1217. doi: 10.1021/ci5007382. [DOI] [PubMed] [Google Scholar]
- [176].Wise JG. Catalytic transitions in the human MDR1 P-glycoprotein drug binding sites. Biochemistry. 2012;51:5125–5141. doi: 10.1021/bi300299z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Parveen Z, Stockner T, Bentele C, Pferschy S, Kraupp M, Freissmuth M, Ecker GF, Chiba P. Molecular dissection of dual pseudosymmetric solute translocation pathways in human P-glycoprotein. Mol Pharmacol. 2011;79:443–452. doi: 10.1124/mol.110.067611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]