Abstract
Nanopore sequencers can be used to selectively sequence certain DNA molecules in a pool by reversing the voltage across individual nanopores to reject specific sequences, enabling enrichment and depletion to address biological questions. Previously, we achieved this using dynamic time warping to map the signal to a reference genome, but the method required substantial computational resources and did not scale to gigabase-sized references. Here we overcome this limitation by using GPU base calling. We show enrichment of specific chromosomes from the human genome and of low-abundance organisms in mixed populations without a priori knowledge of sample composition. Finally, we enrich targeted panels comprising 25,600 exons from 10,000 human genes and 717 genes implicated in cancer, identifying PML-RARA fusions in the NB4 cell line in <15 hours sequencing. These methods can be used to efficiently screen any target panel of genes without specialised sample preparation using any computer and suitable GPU. Our toolkit, readfish, is available at https://www.github.com/looselab/readfish.
Introduction
Selective sequencing, or “Read Until”, refers to the ability of a nanopore sequencer to reject individual molecules whilst they are being sequenced. This requires the rapid classification of current signal from the first part of the read to determine whether the molecule should be sequenced or removed and replaced with a new molecule. We first demonstrated this using dynamic time warping (DTW) to compare the signal with a simulated current trace derived from a reference sequence 1. Although DTW enabled a small set of use cases, it required significant computational resources, preventing its generalised use 2. Another recent method using raw signal, UNCALLED 3, has a lighter computational footprint than previous signal-based methods, but is limited in search space and still requires significant computational resources. An alternative approach, which uses direct base calling of signal chunks 4, demonstrated benefit compared with sequencing without Read Until as it filtered out unwanted reads, but did not provide any enrichment and required significant CPU resources.
Our goal was to work with nucleotide sequences rather than raw signals to exploit existing tools, utilise reasonable computational resources and show enrichment of targets. To do this, we used Oxford Nanopore Technologies (ONT) base calling software. ONT have developed a number of base callers for nanopore sequence data, initially utilising Hidden Markov Models and available through the metrichor cloud service 5. They replaced these with neural network models running on CPU and then Graphical Processing Units (GPU). For real time base calling, ONT provide a range of computational platforms with integrated GPUs (minIT, Mk1C, GridION and PromethION). These devices enable real time base calling sufficient to keep pace with flow cells generating data. Most recently, these base callers acquired a server-client configuration, such that raw signal can be passed to the server and a nucleotide sequence returned. Using this, we show that GPU base calling can be used to deliver a real time stream of nucleotide data from flow cells sequencing with up to 512 channels simultaneously. At the same time, the GPU can base call completed reads, and optimised tools such as minimap2 6 can therefore be used to map reads as they are generated, enabling dynamic updating of both the targets and the reference genome as results change.
As our method does not use raw signal comparison, we do not have to convert reference genomes into signal space as in DTW or other signal methods 1,3. We are constrained by access to a sufficiently powerful GPU. The results presented here mainly utilise the ONT GridION MK1, which includes an NVIDIA GV100 GPU, but we also use an NVIDIA 1080, showing that this approach works on any device capable of real time base calling. We apply this approach to a range of model problems. First, we select specific human chromosomes, illustrating gigabase references are not a constraint. Second, we enrich low-abundance genomes from a mixed population and find we reduce the time required to answer a biological question (time-to-answer) and improve the ability to assemble low-copy genomes. Adaptive sampling is the process by which the software changes what is being sequenced in response to what has been seen during an experiment. To illustrate this, we use Centrifuge to identify the most abundant species present in a metagenomic sample, monitor depth of coverage for each in real time and enrich for the least abundant genomes without a priori knowledge of content 7. This method is necessarily limited by the composition of the reference database and also requires network access to retrieve references once identified. Finally, we enrich panels of human genes, including 25,600 target regions corresponding to ~10,000 genes and 717 genes from the COSMIC (Catalogue of Somatic Mutations in Cancer) panel 8. We demonstrate how Read Until can be used to capture information on key targets without the need for custom library preparation and show we can identify a known translocation in the NB4 cell line in <15 hours 9.
We provide a configurable toolkit, readfish, enabling targeted sequencing of gigabase-sized genomes. This includes depletion of host sequences as well as example methods giving minimum coverage depth for specific sequences in a population. Configuration of these tools is relatively straightforward and requires no additional computation as long as a sufficiently powerful GPU capable of base calling multiple flow cells in real time is available.
Results
Methods Overview
Selective sequencing requires bidirectional communication with a nanopore sequencer through the Read Until API 10. The API provides a stream of raw current samples from every sequencing pore on the flow cell and allows the user to respond in real time, either rejecting a read from a specific pore or allowing a read to finish naturally. Previous API implementations served any signal seen as a potential read and so required processing many signals that were not genuine reads, causing analysis challenges 4. The current API discriminates true DNA signal from background more efficiently and is configured to only provide signals identified as DNA reducing the analysis burden. We reasoned that the signal served by the API should be compatible with the Guppy basecaller and so retrieve short sequences that are processed in base space.
Supplementary Figure 1A illustrates the workflow for base calling reads as they are being sequenced. Briefly, data chunks of signal are served from the Read Until API. Chunks default to one second duration but can be configured by the user. We found 0.4s chunk durations (~180 bases, see methods) balanced the need for small chunks with API performance (Supplementary Table 1 and Supplementary Figure 2). The data chunk (up to 512 reads from a MinION flow cell) is converted to a Guppy compatible format and base called using pyguppyclient 11. Base called data are then mapped to a reference with minimap2 6. Reads may uniquely map, map to multiple locations, or may not map at all. In response the user can choose to reject a read (unblock), acquire more data for that read (proceed) or stop receiving data for the remainder of that read (stop receiving).
Read Until Performance
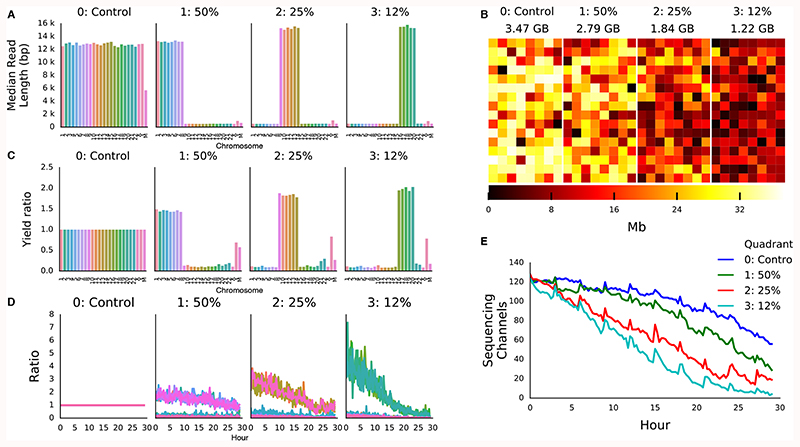
To test performance of our real time base calling approach on enrichment and depletion, we sequenced the well-studied NA12878 reference cell line 12. The flow cell was configured to operate in quadrants each sequencing: a control (all reads accepted), chromosomes 1-8 (50% of reads accepted), chromosomes 9-14 (25% of reads accepted), and finally chromosomes 16-20 (12.5% of reads accepted). Reads are base called and mapped to the reference regardless of quadrant. Median read lengths per chromosome in each quadrant indicate those sequenced or rejected (Figure 1A). Selectively sequenced reads have a median read length of ~15 kb. Rejected reads have a median length of ~500 bases, equating to ~1.1 seconds of sequencing time at 450 bases per second, although median data collected was closer to 1.5 seconds. Reads are base called, mapped and the unblock action sent and actioned within ~1s of the read starting. This run generated 9.5 Gb of sequence data, unevenly distributed across the quadrants; 3.47 Gb in the control, 2.79 Gb at 50% acceptance, 1.84 at 25% acceptance and only 1.22 Gb at 12% (Figure 1B, Supplementary Table 2). For each quadrant the optimal enrichment is 2-fold, 4-fold and 8-fold but we see lower enrichments by the end of the experiment, presumably due to reduced yield (Figure 1C). We observe enrichment of target sequences in all cases compared with control. Relative enrichment is closer to the theoretical maximum at the beginning of the sequencing run (Figure 1D). Analysis of available channels contributing to data generation shows that sequencing capacity is lost faster as more reads are rejected (Figure 1E). For this experiment, we did not nuclease flush the flow cell, but anticipate improvements in both yield and enrichment if we did. We were able to call all batches within our 0.4 second window (Supplementary Figure 3E).
Figure 1. Human Genome Scale Selective Sequencing.
A) Median read lengths for reads sequenced from GM12878 and mapped against HG38 excluding alt chromosomes. The four panels each represent a quadrant of the flow cell. In the control all reads are sequenced, in the second reads mapping to chromosomes 1-8, in the third reads mapping to chromosomes 9-14 and the fourth reads mapping to chromosomes 16-20. The combined length of each of these target sets equates to approximately ½, ¼ and ⅛ of the human genome respectively. B) Heatmap of throughput per channel in each quadrant from the flow cell illustrating reduced yield as the proportion of reads rejected is increased. C) Yield ratio for each chromosome in each condition normalised against yield observed for each chromosome in the control quadrant. D) Yield of on target reads calculated in a rolling window over the course of the sequencing run showing the loss of enrichment potential. E) Plot of the number of channels contributing sequence data over the course of the sequencing run. Channels are lost at a greater rate when more reads are rejected.
A common goal in sequencing library preparation is to remove host DNA to enrich for a metagenomic subpopulation 13,14. Selective sequencing may be beneficial in conjunction with library preparation methods. We considered metagenomics applications as a similar class of problem. Nicholls et. al. generated a reference dataset using the ZymoBIOMICS Microbial Community Standards 15. They were able to generate sufficient data to assemble several of the bacteria into single contigs (without binning). Notably, eukaryotic genomes that were present at lower abundance (2%) did not generate high contiguity assemblies. This is not surprising as the coverage depth for Saccharomyces cerevisiae was 17x and Cryptococcus neoformans 10x when sequencing on a single GridION flow cell 15. Enriching for these low abundance components is conceptually similar to depleting host material from a sample. In our experiments we utilise the ZymoBIOMICS high molecular weight DNA standard (D6322). This sample will a priori improve assemblies due to the longer read lengths and further differs from Nicholls et al. as it excludes C. neoformans.
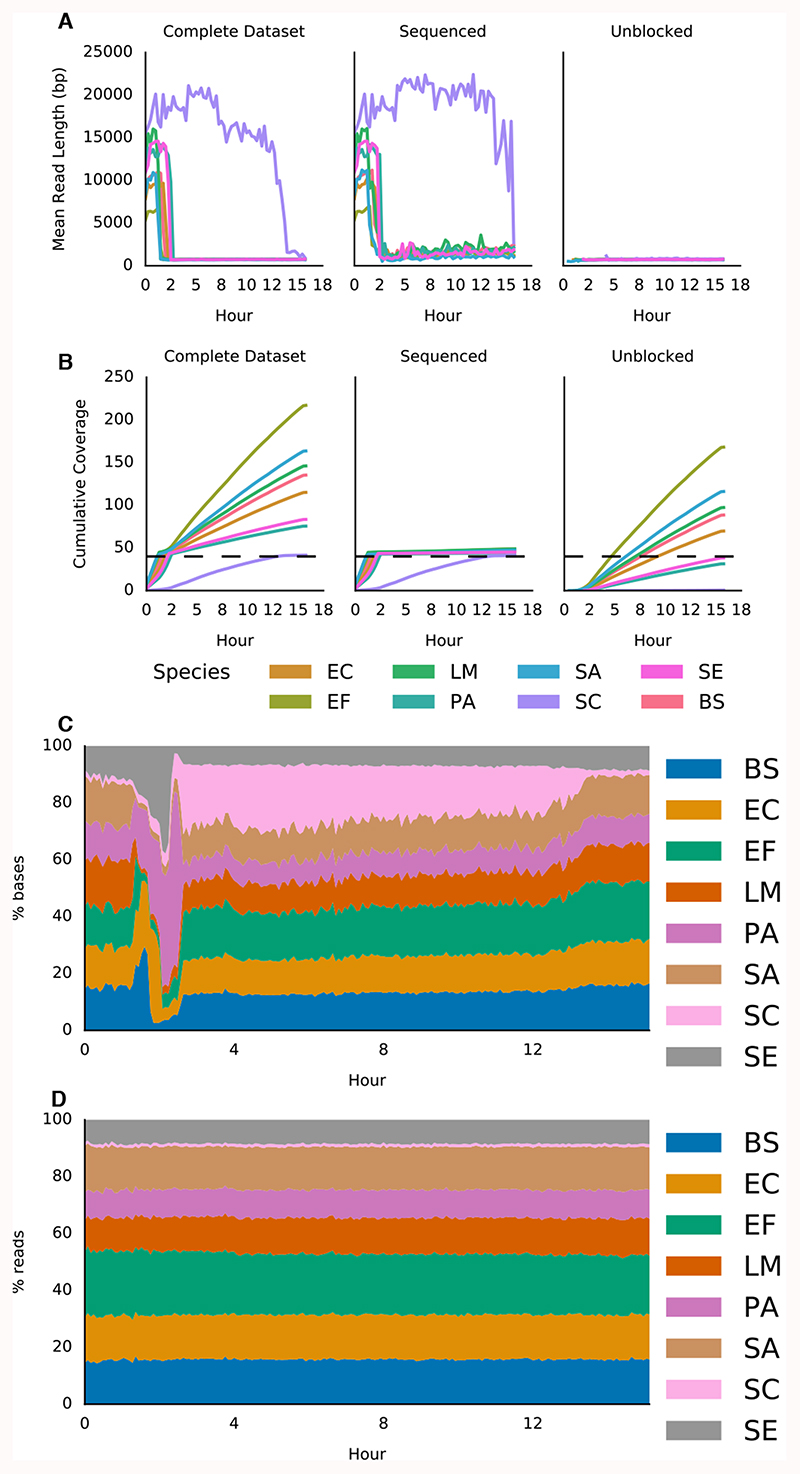
To see if selective sequencing could improve the relative coverage of low abundance material we developed a simple pipeline (readfish align) to drive our selective sequencing decisions (Supplementary Figure 1B). This pipeline aligns completed reads against a reference as they are written to disc, then calculates coverage depth. Once an individual species reaches the desired coverage depth, new reads mapping to that species are rejected. We simultaneously base call both the real time stream from Read Until and completed reads. Finally, we implemented Run Until to stop the run once all targets had reached sufficient coverage. These experiments used a community specific reference file. Mean read lengths for target genomes reduce as they are added to the rejection list and the mean read length becomes dominated by short, rejected reads (Figure 2A). Plotting coverage over time for reads not rejected by Read Until shows a decrease in coverage accumulation for completed genomes (i.e those at the desired coverage level) with an increase in sequencing potential for the least abundant sample, S. cerevisiae (Figure 2B). The proportion of bases mapping to each genome reveals the shift in sequencing capacity to S. cerevisiae (Figure 2C). Relative abundance can still be determined when running Read Until as the proportion of reads mapping to each genome does not change (Figure 2D). The run automatically stops once each genome reaches 40x, taking ~16 hours and 4.4 Gb of sequence data (Supplementary Figure 4).
Figure 2. Adaptive sequencing enriching for the least abundant genome and ensuring uniform 40x coverage.
A) Mean read lengths for reads sequenced from the ZymoBIOMICS mock metagenomic community mapped against the provided references (ZymoBIOMICS, USA). Read lengths are reported for the whole run, the deliberately sequenced reads and those which were actively unblocked. B) Shows cumulative coverage of each ZymoBIOMICS genome during the sequencing run. The total coverage still accumulated as unblocked reads, though short, still map. Sequencing was automatically terminated once each sample reached 40x. C) Stacked area graph illustrating how the proportion of bases mapping to each species changes over time. D) In contrast, the proportion of reads mapping to each species over time doesn’t change significantly. Species and composition are: bs - Bacillus subtilis (14%), ef - Enterococcus faecalis (14%), ec - Escherichia coli (14%), lm - Listeria monocytogenes (14%), pa - Pseudomonas aeruginosa (14%), sc - Saccharomyces cerevisiae (2%), se - Salmonella enterica (14%), sa - Staphylococcus aureus (14%).
This sample should be 2% S. cerevisiae by bases, typically yielding ~88 Mb or 7x of sequence data. Using selective sequencing we see 40x coverage, naively a 5.7 fold increase in on target data. However, a flow cell not implementing selective sequencing would have higher yield, so real world enrichment is lower. Nicholls et al. report 16 Gb on a similar sample generated in 48 hours, which would result in ~25x of S. cerevisiae bringing enrichment closer to 1.6x 15,16. Theoretically enrichment of a 2% subset should be greater, but there is a cost to rejecting an individual read. Even so we could enrich the least abundant element compared with that expected from the sample composition in multiple experiments (n=3). Thus we accelerate time-to-answer for a particular coverage depth (16 hours vs 48 hours). This approach assumes knowledge of the sample a priori and so is of limited practical relevance. By integrating a metagenomics classifier into our pipeline (readfish centrifuge) we avoid this requirement 7. As strains are identified within the sample they can be dynamically tracked and added to a rejection list illustrating the principle of adaptive sequencing.
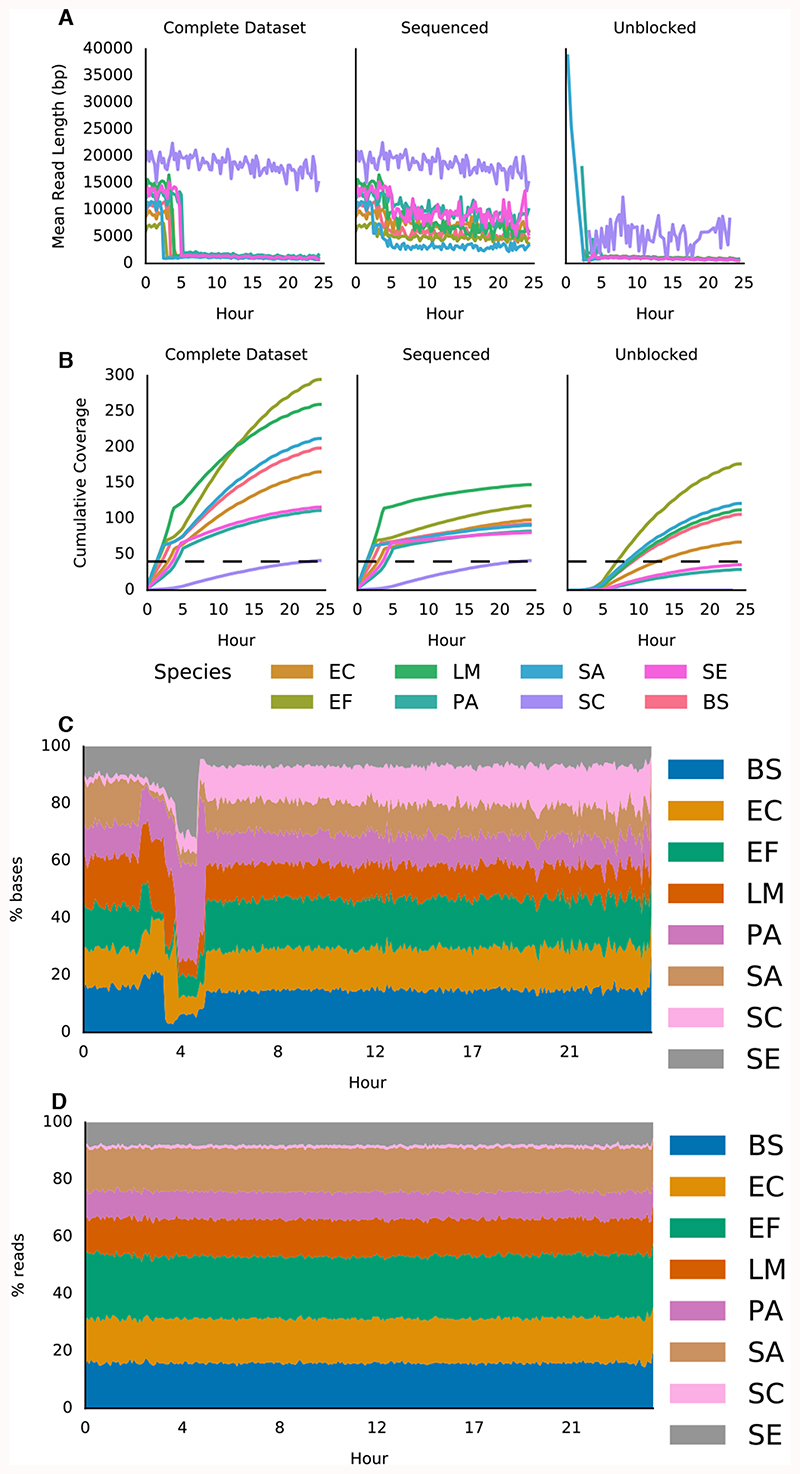
Using this approach we generated 5.995 Gb of sequence data and identified all bacterial genomes in the sample, although we observed enrichment, the flow cell became completely blocked before reaching target coverage (Figure 3, Supplementary Table 2, Supplementary Figures. 5 and 6). 6 Gb of sequence should result in ~10x coverage; here we obtained 41x coverage (Figure 3B). In this case, we considered the entire read as a candidate for read until, consequently some reads are rejected later into the read. This results in a wider range of mean rejected read lengths, particularly for S. cerevisiae (Figure 3A). This experiment was completed within 24 hours, illustrating the benefits in terms of time-to-answer. As expected, improved coverage depth results in almost complete assemblies using MetaFlye compared to that achieved by Nicholls et. al (Supplementary Figure 7); in part a consequence of improved read lengths here 15,17. Subsequent nuclease flushing of the flow cell would increase effective throughput, but this was not our goal.
Figure 3. Adaptive sequencing enriching for the least abundant genome with centrifuge read classification and ensuring uniform 50x coverage.
A) Mean read lengths for reads sequenced from the ZymoBIOMICS mock metagenomic community mapped against the provided references. Read lengths are reported for the whole run, the deliberately sequenced reads and those which were actively unblocked. B) Shows cumulative coverage of each ZymoBIOMICS genome during the sequencing run. The total coverage still accumulated as unblocked reads, though short, still map. Sequencing was automatically terminated once each sample reached 50x. The small overshoot in sequenced reads coverage is likely caused by the centrifuge step lagging as reads are not instantly written to disk. C) Stacked area graph illustrating how the proportion of bases mapping to each species changes over time. D) In contrast, the proportion of reads mapping to each species over time doesn’t change significantly. Species and composition as in Figure 2.
Methods for target panel enrichment include PCR amplification, bait capture methods and CRISPR-Cas9 approaches 18–21. These methods are reliable and cost effective at scale, but have development, instrument and consumable costs. Unlike methods that capture native DNA 20, PCR based methods cannot capture methylation information without additional processing. Such panels cannot be altered easily.
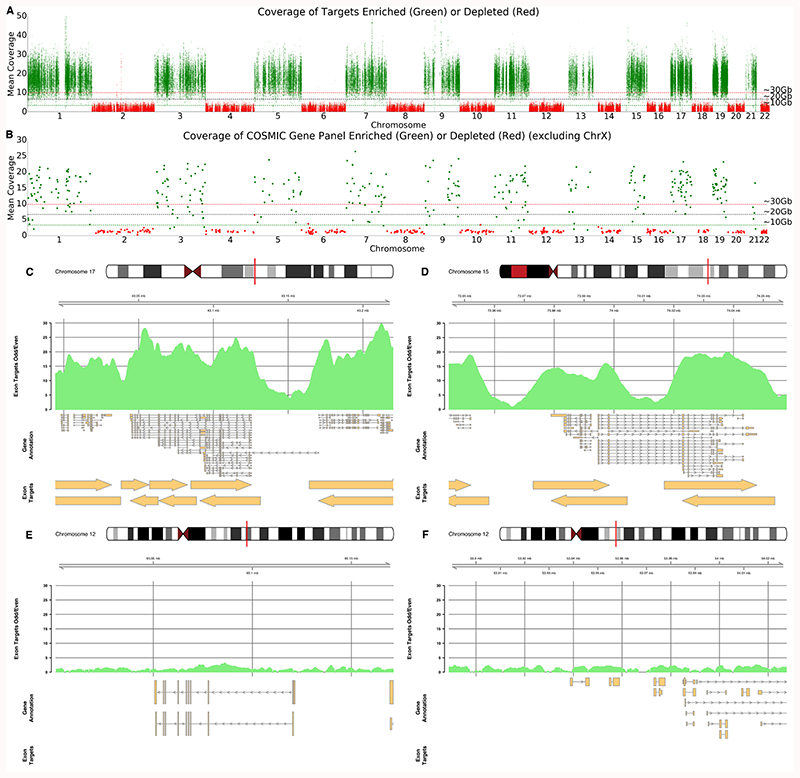
Selective sequencing provides an alternative and so we identified 19,296 target genes annotated as protein coding with Transcript Name IDs (see methods) from the human genome (GRCh38) excluding those on X and Y and ignoring alt chromosomes 22. We extracted exon coordinates, extended 3kb either side and collapsed overlapping targets. We enriched for targets found on odd numbered chromosomes, rejecting all reads from outside these targets. This results in a total search space of 176 Mb (~5%) containing 25,600 targets covering ~10,000 genes (Figure 4A). A single GridION flow cell with 1,660 pores gave 6.1 Gb of sequence data in 24 hours. After nuclease flushing, loading additional library and 24 hours more sequencing gave 5.573 Gb (total yield: 11.675 Gb, N50 9 kb, Supplementary Table 2). Exon targets had median coverage of 17.23x (mean 17.39x) with 75%>14.15x, 25%>20.42x. On “control” even chromosomes, median coverage was 0.98x (mean 1.2x). Detailed coverage plots of targets on ODD (Figures. 4C and D) and EVEN (Figures 4E and F) chromosomes correlate with the target regions. Controlling for these experiments is complicated by flow cell variability. We compare with theoretical yields of 10, 20 and 30 Gb resulting in approximately 3-10x coverage. Our effective enrichment is from 2.7-5.4x consistent with our earlier observations. Nuclease flushing significantly assists enrichment and flow cell efficiency (Supplementary Figure 8).
Figure 4. Half Exome Panel Targeted Sequencing.
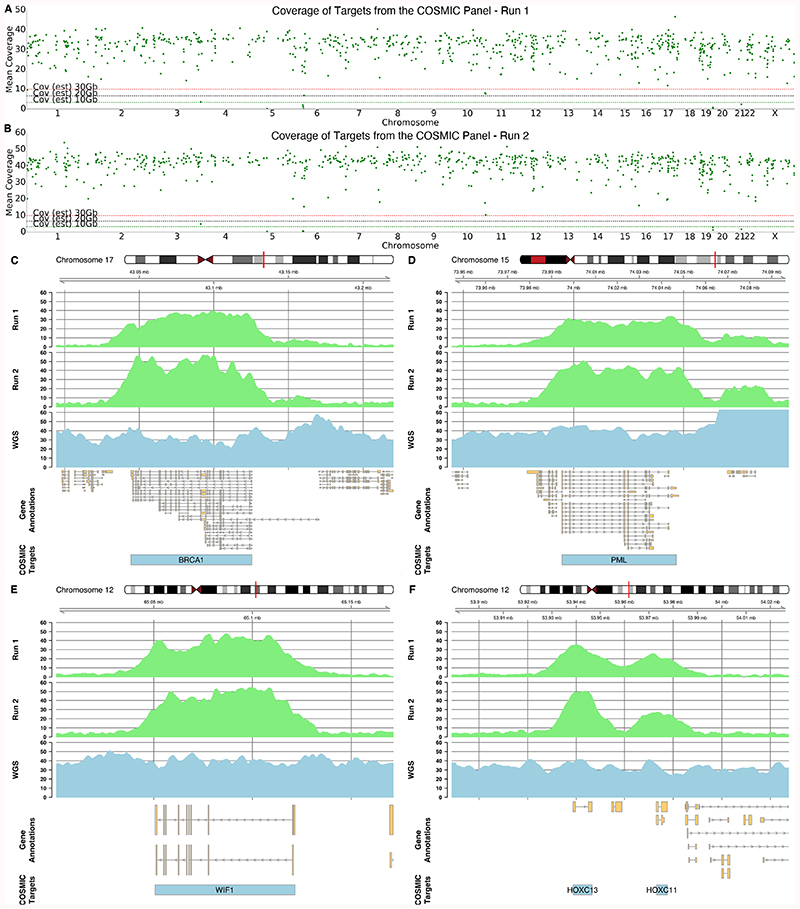
A) Mean coverage across each exon target in the genome ordered by chromosome. Exons on odd numbered chromosomes are enriched (green) and depleted on even numbered chromosomes (red). B) Mean coverage across each exon for genes within the COSMIC panels. For A and B, horizontal lines represent approximate mean expected coverage for flow cells yielding 10, 20 or 30 Gb of data in a single run. Mean coverage calculated by mosdepth 33. C,D,E,F) Coverage plots for highlighted genes including BRCA1 (C), PML (D), WIF1 (E) and HOXC13 and HOXC11 (F). C and D are enriched as they are found on chromosome 17 and 15 whilst E and F are depleted as genes are on chromosome 12. Exon target regions indicated by arrows. In this experiment, different targets were used for the Watson and Crick strands as illustrated by the offsets. Note the absence of target regions for panels E and F.
Our exon panel contains 371 genes from COSMIC with median coverage of 13.7x (Figure 4B) 8. Figures 4C and 4D show coverage for BRCA1, PML and surrounding targets. Although preferable to include introns, here we excluded intronic sequences to reduce the total search space (although not required). To further explore this and illustrate the flexibility of our approach, we targeted the entire COSMIC panel (717 genes) excluding those with no given genomic coordinates (Supplementary File 1). Including flanking 5 kb sequences, our search space was 89.9 Mb (~2.7% of the genome). Using a flow cell with 1,724 pores we generated 3.7 Gb within 24 hours. Nuclease flush and reload generated a further 6.03 Gb giving a total of 9.73 Gb, with a read N50 of 940 bases (Figure 5, Supplementary Figure 9, Supplementary Table 2). Deliberately rejected reads had an N50 of 515 bases; sequenced reads had an N50 of 11,564 bases. Gene targets had median coverage 32.2x (mean 30.7x) (Figure 5A, Supplementary File 1), with 75% of genes >28x, 25% of genes > 35x. Figure 5C-F shows coverage for BRCA1, PML, WIF1, HOXC11/C13. The specificity of selective sequencing is clear, particularly where neighbouring genes in the HOXC cluster are not sequenced. A second run, utilising three flushes, one every 24 hours, generated a total of 17.87 Gb with a read N50 of 793 bases (Supplementary Figure 10, Supplementary Table 2). Gene targets had median coverage 42.3x (mean 40.5x) (Figure 5B), with 75% of genes >44x, 25% of genes >38x. To test performance of readfish on non-ONT hardware, we ran the same experiment using an NVIDIA GeForce GTX 1080 GPU using the fast model of the basecaller. This run generated only 6.7 Gb of data with a read N50 of 799 bases (Supplementary Figure 11, Supplementary Table 2). Median coverage of genes was 19.6x (mean 19.1x), with 75% of genes >20.99x, 25% of genes >17.78x.
Figure 5. COSMIC Panel Targeted Sequencing.
A & B) Mean coverage across the selected COSMIC gene regions ordered by chromosome for two independent sequencing runs of NA12878. Horizontal lines represent approximate mean expected coverage for flow cells yielding 10, 20 or 30 Gb of data in a single run. Mean coverage calculated by mosdepth 33. C,D,E,F) Coverage plots from each run (light green) for highlighted genes including BRCA1 (C), PML (D), WIF1 (E) and HOXC13 and HOXC11 (F). For comparison, coverage in the same regions for a 35X whole genome sequenced nanopore run shown in blue. COSMIC Target regions indicated by blue bars and include intronic sequence.
The difference in yield between these runs is largely due to flow cell variation, particularly the third run which showed unusual flow cell activity (Supplementary Figure 12). However, normalising enrichment to the total yield of each flow cell shows similar performance in each experiment for a selection of target genes including PML, WIF1, HoxC11/C13, RARA and BRCA1 (Supplementary Figures 13-17). This suggests that any steps taken to maximise yield, such as flushing, will result in enhanced enrichment. As with any native nanopore sequence data, these data can be used to assess structural variants and nucleotide variation. As shown in Supplementary Table 3, these data show recall and precision equivalent to, or better than, reference Nanopore whole genome data at similar coverage without targeting 12. Structural variants within the targeted regions can be detected with high recall (Supplementary Table 4). Crucially, between 5-10 typical flow cells would be required to generate equivalent coverage without read until.
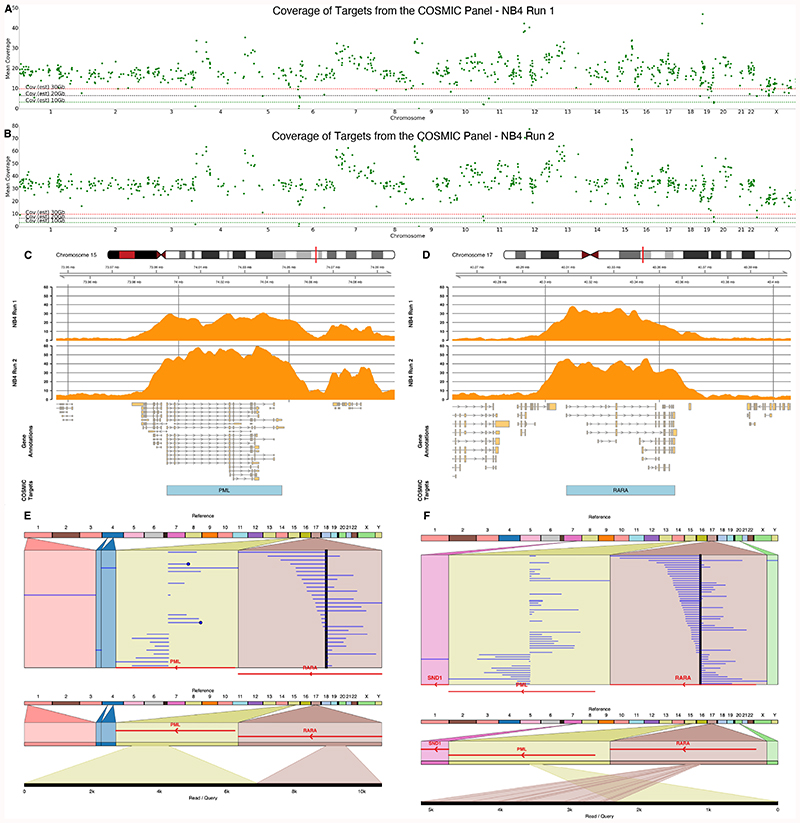
To test screening for structural variants we used the NB4 acute promyelocytic leukemia (APL) cell line 9. Using the same COSMIC panel we identified the translocation using a flow cell with only 1,196 pores, generating 4.5 Gb of sequence data in under 15 hours (Supplementary Figure 18. Median coverage of targets was 11.46x (mean 11.78x) (Figures 6A,C,D), with 75% of genes >9.5x, 25% of genes > 13.4x. Analysis with svim looking for breakpoint ends, ignoring in/dels, identified two candidates passing default filtering (see methods) 23. The breakpoint can also be detected with sniffles (data not shown) 24. Of these candidates, one captured the known breakpoint supported by six reads. A further 24 hours of sequencing (~3Gb) resulted in median coverage of 17.37x (mean 18x) and 9 reads supporting the variant (Figure 6E, Supplementary Table 5). No complex rearrangements were reported in NA12878 using the same COSMIC panel (Supplementary Table 5). A subsequent repeat of this experiment (Supplementary Figure 19), with flushing every 24 hours, generated 15.9 Gb of sequence data. Median coverage of targets was 34x (mean 35.5x) (Figure 6B), with 75% of genes > 38x, 25% of genes > 30x and 23 reads supporting the breakpoint (Figure 6F, Supplementary Table 5).
Figure 6. COSMIC Panel Targeted Sequencing of NB4.
A & B) Mean coverage across each of the COSMIC target regions ordered by chromosome for two independent sequencing runs of the NB4 cell line. Horizontal dashed line indicates expected coverage from a flow cell yielding 10, 20 or 30 Gb of sequence data in a single run. C & D) Coverage plots for each NB4 sequencing run shown in orange for PML (C) and RARA (D). E & F) Reads mapping to chromosomes 15 and 17 derived from the NB4 cell line runs 1 and 2 respectively indicating the fusion between PML and RARA. Mappings of example individual reads are shown. Breakpoints identified using svim, visualisations using Ribbon 23,31.
Discussion
The idea of selectively sequencing (‘Read Until’) individual molecules using only computational methods is a unique capability of nanopore sequencing 1. Here we exploit ONT tools to provide a true real time stream of sequence data as nucleotide bases and provide a toolkit to design and control selective sequencing experiments called readfish. This approach removes the need for complex signal mapping algorithms but does require a sufficiently fast base caller. Prior work illustrated that this method was feasible, but required extensive additional computation and did not show significant enrichment over throughput achieved without running ‘Read Until’ 4. Here we demonstrate real enrichment over that expected from a similar control flow cell. We also show that standard techniques for enhancing flow cell yield such as nuclease flushing and loading additional library are similarly beneficial for read until experiments. Although not extensively exploited here, nuclease flushing and reuse of flow cells does increase yield and enrichment and we have taken to flushing read until experiments every 24 hours.
We find that increased rejection of reads on a flow cell negatively impacts sequencing yield and so observed enrichment. The main benefit of selective sequencing in metagenomics and host depletion is to improve time-to-answer. For samples which sequence well (i.e do not tend to block the flow cell), additional enrichment benefits may be observed. Notably, running selective sequencing does not disrupt the proportion of reads by count that map to a specific reference. Thus, for metagenomics, it is still possible to assess relative abundance whilst focussing sequencing length on specific subsets of reads. Future methods proposed by ONT to address blocking, such as onboard nucleases, might increase throughput in future.
Key benefits of our approach are that we utilise only computational resources available in the GridION Mk1. As we use current commercially provided base callers, we can utilise new algorithms and pores as they are developed. Thus, although not yet tested, we could use this method on RNA if sufficiently long reads require depletion. Similarly. we could use methylation-aware base callers to sequence regions of DNA starting from either high or low methylation regions. As we obtain sequence, rather than signal, we greatly simplify the construction of pipelines for downstream analysis of reads. Although we focus on results for the GridION Mk1 we show this method can be used with any MinION configuration provided sufficient available GPU to base call a sequencing run in real time (Supplementary Note 1). As we show here, it is possible to utilise the fast base calling model and obtain effective enrichment using a single Nvidia GeForceGTX 1080 GPU. Other users have reported success with the high accuracy model on systems configured with NVIDIA 2080 GPUs (J Tyson, Pers. Comm.). In cost terms, any platform capable of real time base calling will be compatible with our approach. In principle this method should scale to the PromethION.
We demonstrate that selective sequencing of arbitrary targeted regions of the human genome results in actionable coverage and can identify SNVs and SVs in the COSMIC panel. For SV analysis, DNA extraction, library preparation, sequencing and analysis could be completed within 24 hours. When sequencing a subset of a large genome, large numbers of off-target reads are sampled whilst detecting those of interest and the precise parameters of optimal target size and coverage have yet to be defined. Consequently, library preparation methods enriching for regions of interest will result in higher coverage than ‘Read Until’. But the design of such panels is relatively costly and inflexible once developed. Methods relying on amplification result in the loss of methylation data, which can be found using the methods presented here.
In readfish selective sequencing, targets can be updated by a single configuration file. Developing a new panel is as straightforward as compiling a list of target regions. Here we also illustrate the concept of adaptive sequencing, as in our metagenomics examples, where targets can be dynamically adjusted during a run. In theory a panel could be updated in response to observations of the data in real time, perhaps adding targets where candidate novel structural variants have been identified or removing targets where sufficient evidence is available to eliminate the possibility of an SV existing.
Of course, throughput achievable on platforms such as the PromethION at scale provides efficient whole genome sequencing 25. Thus, any effective method for enrichment must be as efficient, including the additional computation required. By utilising the available GPU computational capacity during the sequencing run, we address this issue. There is no reason, in theory, why samples could not be multiplexed on a single flow cell as long as sufficient yield can be obtained to address the biological question.
Although we have focussed exclusively on applications for ‘Read Until’, we believe that a real time sequence data stream as bases has significant advantages for future pipelines. If sequence data can be streamed directly into an analysis pipeline and conclusions drawn without the requirements for data storage, then field deployment of sequencing for detection of specific sequences might be accelerated. Ultimately, it may be possible to stream sequence data for calling of structural variants and further analysis in real time.
Online Methods
Library preparation and sequencing
Standard LSK-109 (ONT) sequencing libraries were prepared from either the ZymoBIOMICS HMW DNA Standard (DS6322 ZymoBIOMICS USA) or DNA extracted from GM12878 cells (Coriell), or NB4 cells (gift from M. Hubank) as described in Jain et al 12. Human DNA for exon enrichment or gene targeting was sheared to approximately 12kb using g-TUBE (Covaris). Sequencing runs used either the GridION Mk1 or a MinION with NVIDIA GeForceGTX 1080 GPU (see Supplementary Table S2). Standard scripts for sequencing were used with one modification, namely that the size of data chunk delivered by MinKNOW was reduced from 1 second to 0.4 seconds by changing the value of the break_reads_after_seconds parameter in the relevant TOML file (located in ../minknow/conf/package/sequencing/ for MinKNOW core version 3.6). All sequencing used FLO-MIN106 R9.4.1 flow cells.
When running read until experiments seeking to maximise yield, throughput on the flow cell should be monitored closely. Our practice has been to nuclease flush flow cells every 24 hours to maximise throughput. For maximising occupancy on the flow cell, users should experiment with loading more library than they might otherwise do. For example, where a user might load 400 ng of library with a read length N50 of 10-15 kb, we would recommend loading 600 ng of library. This assumes R9.4 flow cells. This protocol has not yet been tested on R10.
Single Nucleotide Variant Detection
SNPs in NA12878 read data were called using Nanopolish in methylation aware mode 26. Reads were mapped to hg38 removing ALTs with minimap2 using standard settings for ONT reads 6. High confidence gold standard SNPs were identified from the Genome In A Bottle (GIAB) truth set 27. SNPs were compared with a 35x WGS NA12878 reference set recalled using the same guppy basecaller model 12. SNP comparisons were made using HAP.PY using default settings and the same target sites used for selective sequencing 28.
Structural Variant Detection and Concordance
Reads were mapped to the hg38 primary assembly with minimap2 and standard ONT settings. Variants were called using SVIM and Sniffles with default settings and minimum variant length set as 50 23,24. Only SVIM variant calls with QUAL above 10 and longer than 50bp were kept. Variants of the same type present in both SVIM and Sniffles callsets were selected as the final call set using SURVIVOR and a maximal distance between breakpoints was set to 500 29. Only insertions and deletions intersecting the COSMIC Target Panel were considered for concordance calculations in WGS, Run1 and Run2. Concordance calculations were performed with Truvari 30 with reference distance set as 1.5Kb, percent size similarity as 0.3 and only insertions and deletions larger than 50bp within the COSMIC Target Panel were considered. For analysis of the translocation in the NB4 cell lines, variant calls were filtered with quality 10 and non BND (Breakpoint End) structural variant types were ignored. SVs were visualised with Ribbon 31.
Target Lists
The exact target list used to configure exon capture can be obtained at the following link: http://www.ensembl.org/biomart/martview/454f99b3f65c7e62669229fd48de8e47?VIRTUALSCHEMANAME=default&ATTRIBUTES=hsapiens_gene_ensembl.default.structure.ensembl_gene_id|hsapiens_gene_ensembl.default.structure.ensembl_gene_id_version|hsapiens_gene_ensembl.default.structure.ensembl_transcript_id|hsapiens_gene_ensembl.default.structure.ensembl_transcript_id_version|hsapiens_gene_ensembl.default.structure.chromosome_name|hsapiens_gene_ensembl.default.structure.exon_chrom_start|hsapiens_gene_ensembl.default.structure.exon_chrom_end&FILTERS=hsapiens_gene_ensembl.default.filters.with_hgnc_trans_name.only|hsapiens_gene_ensembl.default.filters.chromosome_name.‘1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20,21,22,X,Y’|hsapiens_gene_ensembl.default.filters.biotype.‘protein_coding’&VISIBLEPANEL=attributepanel
Read Until Cache Configuration and Chunk Size
A read begins with adapter sequences as well as optional barcodes. Additionally read starts sometimes stall as DNA engages with the pore before signal containing sequence data are available. The first chunk of data may not provide an optimal base call and additional data may be required. Calling any single fragment of data in isolation is less informative than calling the entire signal and so we implement a read cache concatenating adjacent signal data from the same read. This enables base calling the complete signal for each read since it started. As of MinKNOW version 3.6, the sequencing platform is effectively limited to a lower bound chunk size of 0.4s. As shown in Supplementary Figure. S2 and Supplementary Table S1, more than 80% of human reads can be base called and aligned within 2 chunks or 0.8s worth of data. For bacterial sequences more than 40% of reads can be base called and aligned within a single chunk or 0.4s worth of data. So by observation, the smallest possible chunk size will enable the fastest decision making for any given sequence. In a typical experiment we find that 90% of reads can be processed (called, mapped, and decision made) within three chunks (1.2 s, Supplementary Figure. S2, Supplementary Table S1).
Base caller Configuration
The Guppy basecaller contains several models for base calling that trade speed (fast) for accuracy (high accuracy model, hac) and can optionally call methylation. For selective sequencing, the goal is speed and so we investigated the efficacy of both the fast and hac models finding the GridION Mk1 easily powerful enough to use the hac model. Across all experiments shown here the average batch of reads called in 0.28s and contained 30 reads. At maximum load, individual reads are processed in less than 0.002s. Thus we call at least 100 read fragments per second and even at peak load can typically call all 512 reads (see Supplementary Figures. S3-7, Supplementary Figure S10).
Experiment Configuration
Depending on experiment configuration, the response to read mapping varies (see online methods). If depleting contaminants (host depletion) then reads mapping to that reference should be rejected. For enrichment, reads mapping to a target should be sequenced. The action for non mapping reads will depend on the experiment. If the experimental goal is enriching low abundance or unknown targets, non mapping reads should be sequenced. If enriching for subsets of a known reference, non mapping reads might be rejected in favour of sampling more. Given the variety of options, we provide a configuration file allowing any mapping result to trigger any action. We include the option to dynamically update this file during sequencing enabling target switches whilst sequencing. The configuration also allows different experiments on regions of the same flow cell (see https://github.com/LooseLab/readfish/blob/master/TOML.md).
readfish Code availability
The ONT Read Until API is required for running Read Until 10. The results presented here used an updated version of this API, available from our GitHub (https://github.com/LooseLab/read_until_api_v2; Git commit cff0f52). These changes were required for Python3 compatibility and also change the behaviour of the read cache enabling consecutive chunks of data to be stored for calling. As the ONT tool chain matures to Python3 such changes will no longer be required. pyguppyclient (v0.0.5), a python interface to the Guppy base calling server is currently available on PyPI. Our code is available open source at http://www.github.com/LooseLab/readfish and installable via PyPI.
Read Until Implementation
ReadFish scripts
ReadFish is a set of scripts that control sequencing in real time. Each script is accessed as a sub-command, and a description is given below.
targets
This script runs the core Read Until process as specified in the experiment TOML file. It can select specific regions of a genome, mapping reads in real time using minimap2 and rejecting reads appropriately. This script should be started once the initial mux scan has completed. The experiment TOML file can be updated during a sequencing run to change the configuration of the Read Until process. It is through this mechanism that the align and centrifuge commands can change Read Until behaviour during a run. Configuration parameters are available under the help flag. Tables 1 and 2 describe the mapping parameters and configuration options for various possible experiment types.
Table 1. Description of possible read mapping conditions.
| Mapping Condition | Description |
|---|---|
| multi_on | Read fragment maps multiple locations including region of interest. |
| multi_off | Read fragment maps to multiple locations not including region of interest. |
| single_on | Read fragment only maps to region of interest. |
| single_off | Read fragment maps to one location but it is not a region of interest. |
| no_map | Read fragment does not map to the reference. |
| no_seq | No sequence was obtained for the signal fragment. |
Table 2.
Example configurations for different experiment types. “Unblock” causes a read to be ejected from the pore, “proceed” means that a read continues to sequence and serve data through the API for later decisions, “stop receiving” allows the read to continue sequencing with no further data served through the API.
| Experiment Type | Region of Interest for Alignments | Mapping Condition | |||||
|---|---|---|---|---|---|---|---|
| multi_on | multi_off | single_on | single_off | no_map | no_seq | ||
| Host Depletion | Known Host Genome | unblock | proceed | unblock | proceed | proceed | proceed |
| Targeted Sequencing | Known regions from one or more genomes. | stop receiving | proceed | stop receiving | unblock | proceed | proceed |
| Target Coverage Depth (known sample composition) | All known genomes within the sample, tracked for coverage depth. | stop receiving | proceed | stop receiving | unblock | proceed | proceed |
| Low Abundance Enrichment (unknown sample composition) | All genomes within the sample that can be identified as well as those that cannot. | stop receiving | proceed | stop receiving | unblock | proceed | proceed |
align
This script runs an instance of the “Run Until” monitoring system that watches as completed reads are written to disc. When new data is detected this pipeline will map the data against the target reference genome (specified in the experiment TOML file) and compute the cumulative coverage for the sequencing run. Once a genomic target reaches sufficient coverage, it will be added to the unblock list. Optionally, the user can provide additional targets from the start of the run to implement “host depletion”. Finally, the user can configure align to stop the entire run if all samples have reached the required coverage depth. At present, this coverage depth is uniform for all samples, so it is not possible to have variable coverage over a target set.
centrifuge
This script runs an instance of the “Run Until” monitoring system. As completed reads are written to disc this programme (Supplementary Figure. S1C) will classify the reads using centrifuge and a user defined index. When 2000 reads are uniquely classified the corresponding reference genome is downloaded from RefSeq 32 and incorporated into a minimap2 index. At this point the same process as in align is used to determine coverage depth. The new alignment index is passed to the core Read Until script (targets) by updating the experiment TOML file allowing dynamic updates for both the unblock list and the genomic reference.
unblock-all
This script is provided as a test of the Read Until API where all incoming read fragments are immediately unblocked. It allows a user to quickly determine if their MinKNOW instance is able to provide and process unblock signals at the correct rate. Users should provide a bulk FAST5 file for playback for this testing process.
validate
This script is a standalone tool for validating an experiment TOML file. We provide a ru_schema.json (https://github.com/LooseLab/readfish/blob/master/ru/static/ru_toml.schema.json) that describes the required configuration format.
Reporting Summary
For further details see the Reporting Summary for this manuscript.
Supplementary Material
Acknowledgments
The authors thank J Quick, J Tyson, J Simpson and N Loman for helpful comments and (mainly) criticisms. E Birney, N Goldman and A Senf for helpful insights and discussion on these approaches. We thank M Hubank and L Gallagher for access to materials and reagents as well as general boundless enthusiasm. We thank M Jain for assisting in manipulating data. The authors also thank S Reid, C Wright, C Seymour, J Pugh and G Pimm from Oxford Nanopore Technologies for advice on MinKNOW and Guppy operations as well as extensive troubleshooting!
Funding
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/N017099/1 RM, ML, BB/M020061/1 ML,, BB/M008770/1, 1949454 AP], and the Wellcome Trust [grant number 204843/Z/16/Z, NH, ML], and the Defence Science and Technology Laboratory [grant number DSTLX-1000138444 RM, ML]..
Footnotes
Author contributions
M.L and A.P conceived the study. A.P, N.H and M.L acquired data. T.C and R.M designed and implemented metagenomics applications. A.P, B.J.D and M.L analysed and interpreted data. All authors discussed the results and contributed to the final manuscript.
Competing interests
ML was a member of the MinION access program and has received free flow cells and sequencing reagents in the past. ML has received reimbursement for travel, accommodation and conference fees to speak at events organized by Oxford Nanopore Technologies.
Data Availability
All reads generated in the course of this study are available from the ENA under project id PRJEB36644.
Code availability
Our code is available open source at http://www.github.com/LooseLab/readfish
References
- 1.Loose M, Malla S, Stout M. Real-time selective sequencing using nanopore technology. Nat Methods. 2016;13:751–754. doi: 10.1038/nmeth.3930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Masutani B, Morishita S. A framework and an algorithm to detect low-abundance DNA by a handy sequencer and a palm-sized computer. Bioinformatics. 2019;35:584–592. doi: 10.1093/bioinformatics/bty663. [DOI] [PubMed] [Google Scholar]
- 3.Kovaka S, Fan Y, Ni B, Timp W, Schatz MC. Targeted nanopore sequencing by real-time mapping of raw electrical signal with UNCALLED. doi: 10.1101/2020.02.03.931923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Edwards HS, Krishnakumar R, Sinha A, Bird SW, Patel KD, Bartsch MS. Real-Time Selective Sequencing with RUBRIC: Read Until with Basecall and Reference-Informed Criteria. Sci Rep. 2019;9 doi: 10.1038/s41598-019-47857-3. 11475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rang FJ, Kloosterman WP, de Ridder J. From squiggle to basepair: computational approaches for improving nanopore sequencing read accuracy. Genome Biol. 2018;19:90. doi: 10.1186/s13059-018-1462-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li H. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics. 2018;34:3094–3100. doi: 10.1093/bioinformatics/bty191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D, Song L, Breitwieser FP, Salzberg SL. Centrifuge: rapid and sensitive classification of metagenomic sequences. Genome Res. 2016;26:1721–1729. doi: 10.1101/gr.210641.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tate JG, Bamford S, Jubb HC, Sondka Z, Beare DM, Bindal N, Boutselakis H, Cole CG, Creatore C, Dawson E, Fish P, et al. COSMIC: the Catalogue Of Somatic Mutations In Cancer. Nucleic Acids Res. 2019;47:D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mozziconacci M-J, Rosenauer A, Restouin A, Fanelli M, Shao W, Fernandez F, Toiron Y, Viscardi J, Gambacorti-Passerini C, Miller WH., Jr Molecular cytogenetics of the acute promyelocytic leukemia-derived cell line NB4 and of four all-trans retinoic acid--resistant subclones. Genes Chromosomes Cancer. 2002;35:261–270. doi: 10.1002/gcc.10117. Others. [DOI] [PubMed] [Google Scholar]
- 10.read_until_api. [Accessed: 4th August 2020]; Available at: https://github.com/nanoporetech/read_until_api.
- 11.pyguppyclient. [Accessed: 4th August 2020]; Available at: https://github.com/nanoporetech/pyguppyclient.
- 12.Jain M, Koren S, Miga KH, Quick J, Rand AC, Sasani TA, Tyson JR, Beggs AD, Dilthey AT, Fiddes IT, Malla S, et al. Nanopore sequencing and assembly of a human genome with ultra-long reads. Nat Biotechnol. 2018;36:338–345. doi: 10.1038/nbt.4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wain J, Leggett RM, Livermore DM, O’Grady J. Nanopore metagenomics enables rapid clinical diagnosis of bacterial lower respiratory infection. Biotechnology. 2019 doi: 10.1038/s41587-019-0156-5. [DOI] [PubMed] [Google Scholar]
- 14.Marotz CA, Sanders JG, Zuniga C, Zaramela LS, Knight R, Zengler K. Improving saliva shotgun metagenomics by chemical host DNA depletion. Microbiome. 2018;6 doi: 10.1186/s40168-018-0426-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicholls SM, Quick JC, Tang S, Loman NJ. Ultra-deep, long-read nanopore sequencing of mock microbial community standards. Gigascience. 2019;8 doi: 10.1093/gigascience/giz043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.mockcommunity. [Accessed: 17th January 2020]; Available at: https://github.com/LomanLab/mockcommunity.
- 17.Kolmogorov M, Yuan J, Lin Y, Pevzner PA. Assembly of long, error-prone reads using repeat graphs. Nat Biotechnol. 2019;37:540–546. doi: 10.1038/s41587-019-0072-8. [DOI] [PubMed] [Google Scholar]
- 18.Kozarewa I, Armisen J, Gardner AF, Slatko BE, Hendrickson CL. Overview of Target Enrichment Strategies. Curr Protoc Mol Biol. 2015;112:7.21.1–7.21.23. doi: 10.1002/0471142727.mb0721s112. [DOI] [PubMed] [Google Scholar]
- 19.Gnirke A, Melnikov A, Maguire J, Rogov P, LeProust EM, Brockman W, Fennell T, Giannoukos G, Fisher S, Russ C, Gabriel S, et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat Biotechnol. 2009;27:182–189. doi: 10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowen R, Heron A, Sedlazeck F, Timp W. Targeted Nanopore Sequencing with Cas9 for studies of methylation, structural variants and mutations. BioRxiv. 2019 [Google Scholar]
- 21.Loose M. Finding the Needle: Targeted Nanopore Sequencing and CRISPR-Cas9. The CRISPR Journal. 2018;1:265–267. doi: 10.1089/crispr.2018.29028.mlo. [DOI] [PubMed] [Google Scholar]
- 22.Cunningham F, Achuthan P, Akanni W, Allen J, Amode MR, Armean IM, Bennett R, Bhai J, Billis K, Boddu S, Cummins C, et al. Ensembl 2019. Nucleic Acids Res. 2019;47:D745–D751. doi: 10.1093/nar/gky1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heller D, Vingron M. SVIM: structural variant identification using mapped long reads. Bioinformatics. 2019;35:2907–2915. doi: 10.1093/bioinformatics/btz041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sedlazeck FJ, Rescheneder P, Smolka M, Fang H, Nattestad M, von Haeseler A, Schatz M. Accurate detection of complex structural variations using single molecule sequencing. bioRxiv. 2017 doi: 10.1101/169557. 169557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beyter D, Ingimundardottir H, Eggertsson HP. Long read sequencing of 1,817 Icelanders provides insight into the role of structural variants in human disease. BioRxiv. 2019 doi: 10.1038/s41588-021-00865-4. [DOI] [PubMed] [Google Scholar]
- 26.Simpson JT, Workman RE, Zuzarte PC, David M, Dursi LJ, Timp W. Detecting DNA cytosine methylation using nanopore sequencing. Nat Methods. 2017;14:407–410. doi: 10.1038/nmeth.4184. [DOI] [PubMed] [Google Scholar]
- 27.Zook JM, McDaniel J, Olson ND, Wagner J, Parikh H, Heaton H, Irvine SA, Trigg L, Truty R, McLean CY, De La Vega FM, et al. An open resource for accurately benchmarking small variant and reference calls. Nat Biotechnol. 2019;37:561–566. doi: 10.1038/s41587-019-0074-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.hap.py. [Accessed: 4th August 2020]; Available at: https://github.com/Illumina/hap.py.
- 29.Jeffares DC, Jolly C, Hoti M, Speed D, Shaw L, Rallis C, Balloux F, Dessimoz C, Bähler J, Sedlazeck FJ. Transient structural variations have strong effects on quantitative traits and reproductive isolation in fission yeast. Nat Commun. 2017;8 doi: 10.1038/ncomms14061. 14061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.truvari. [Accessed: 7th August 2020]; Available at: https://github.com/spiralgenetics/truvari.
- 31.Nattestad M, Chin C-S, Schatz MC. Ribbon: Visualizing complex genome alignments and structural variation. bioRxiv. 2016 doi: 10.1101/082123. 082123. [DOI] [Google Scholar]
- 32.Pruitt KD, Maglott DR. RefSeq and LocusLink: NCBI gene-centered resources. Nucleic Acids Res. 2001;29:137–140. doi: 10.1093/nar/29.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pedersen BS, Quinlan AR. Mosdepth: quick coverage calculation for genomes and exomes. Bioinformatics. 2018;34:867–868. doi: 10.1093/bioinformatics/btx699. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All reads generated in the course of this study are available from the ENA under project id PRJEB36644.
Our code is available open source at http://www.github.com/LooseLab/readfish