Abstract
Purpose
There is potential for faecal microbiome profiling to improve CRC screening. This has been demonstrated by research studies, but it has not been quantified at scale using samples collected and processed routinely by a national screening programme.
Experimental Design
Between 2016-2019, the largest of the NHS Bowel Cancer Screening Programme (NHSBCSP) hubs prospectively collected processed gFOBT with subsequent colonoscopy-outcomes: blood-negative (n=491 (22%)); CRC (n=430 (19%)); adenoma (n=665 (30%)); colonoscopy-normal (n=300 (13%)); non-neoplastic (n=366 (16%)). Samples were transported and stored at room temperature. DNA underwent 16S rRNA gene V4 amplicon sequencing. Taxonomic profiling was performed to provide features for classification via random forests (RFs).
Results
Samples provided 16S amplicon-based microbial profiles, which confirmed previously described CRC-microbiome associations. Microbiome-based RF models showed potential as a first-tier screen, distinguishing CRC or neoplasm (CRC or adenoma) from blood-negative with AUC 0.86 (0.82-0.89) and AUC 0.78 (0.74-0.82), respectively. Microbiome-based models also showed potential as a second-tier screen, distinguishing from among gFOBT blood-positive samples, CRC or neoplasm from colonoscopy-normal with AUC 0.79 (0.74-0.83) and AUC 0.73 (0.68-0.77), respectively. Models remained robust when restricted to fifteen taxa, and performed similarly during external validation with metagenomic datasets.
Conclusions
Microbiome features can be assessed using gFOBT samples collected and processed routinely by a national CRC screening programme to improve accuracy as a first or second-tier screen. The models required as few as fifteen taxa, raising the potential of an inexpensive qPCR test. This could reduce the number of colonoscopies in countries that use faecal occult blood test screening.
Keywords: CRC, neoplasm, accuracy, microbiome, gFOBT
Introduction
Globally, CRC is the third most common cause of cancer deaths. 1 Screening reduces mortality by detecting asymptomatic adenomas or early-stage CRC. 2 Countries have adopted different screening approaches. In England, the NHS Bowel Cancer Screening Programme (NHSBCSP) tests for occult faecal blood; if detected, participants are referred for colonoscopy. Until June 2019, the NHSBCSP used the guaiac faecal occult blood test (gFOBT). Specificity is limited, with only 40% of screening colonoscopies detecting adenoma and 10% CRC; 3 4 this represents a significant cost, resource, and patient burden.
Research suggests that faecal microbiome analysis may serve as an improvement or adjunct to current CRC screening. 5 However, previous studies have not yet bridged the gap between pre-clinical, basic scientific discovery and the population-scale necessary for translation to a national screening programme. These limitations were outlined in a systematic review: many had small numbers of participants (the largest had 490, of which 120 were CRC patients); many collected samples in a manner incompatible with national screening (refrigerated/frozen samples); some used post-colonoscopy samples (bowel preparation alters the microbiome); and few had the opportunity to externally validate their models. 5
We aimed to quantify the utility of integrating microbiome analysis into a national CRC screening programme by analysing microbiome features from large numbers of routinely processed NHSBCSP gFOBT samples. Technical studies have shown that it is possible to measure a subset of clinically-relevant microbiome features from gFOBT stored at room temperature. 6–13 Two studies have analysed large numbers of bowel-preparation naïve individuals, but neither performed microbiome analysis directly from screening samples; one study has performed preliminary analysis of screening faecal immunochemical test (FIT) samples, but did not determine diagnostic performance of the microbiome. 14 15 16 To our knowledge, our study is the first to analyse microbiome features from large numbers of routinely processed gFOBT screening samples.
To reflect the aims of the NHS Bowel Cancer Screening Programme, we explored the potential of microbiome-based RF models to detect CRC alone, or to detect CRC and adenoma (a group we term ‘neoplasm’). We investigated the potential to use these microbiome-based RF models as a first-tier screen, equivalent to the use of gFOBT; we used gFOBT blood-negative samples as the control group, as 98% of screening gFOBT yield a blood-negative result. Additionally, we explored the potential to use the microbiome-based RF models as a second-tier screen; a second-tier represents an opportunity to triage those samples with a blood-positive gFOBT result, in order to reduce the number of unnecessary screening colonoscopies. As a second-tier screen, we explored the potential of microbiome-based RF models to distinguish gFOBT blood-positive samples associated with CRC or neoplasm, from gFOBT blood-positive samples associated with a normal colonoscopy result. We used ‘colonoscopy-normal’ samples as the control group, as although a proportion of screening colonoscopies yield a ‘non-neoplastic’ diagnosis (e.g. diverticulosis, non-dysplastic polyp), this is a heterogeneous group. We found that microbiome-based RF models show potential as a first-tier screen for the detection of CRC (AUC 0.86 (0.82-0.89)) or neoplasm (AUC 0.78 (0.74-0.82)), and as a second-tier screen, for the detection of CRC (AUC 0.79 (0.74-0.83)) or neoplasm (AUC 0.73 (0.68-0.77).
Materials and Methods
Study design and participants
The NHSBCSP Southern Hub (Guildford, UK) prospectively collected a convenience series of routinely processed gFOBT October 2016-August 2019: this included all ‘blood-positive’ gFOBT (blue discolouration affecting five or six squares) processed by the Southern Hub (n=3700), and a random sample of ‘blood-negative’ (no blue discolouration) gFOBT (n=530). Of the samples collected, 3601 (85%) had complete basic clinical data recorded on the NHS Bowel Cancer Screening Programme database at the time of the final data extract. From this group, we selected samples to achieve sample sizes that were approximately equal across the different clinical groups (Fig.1, Supplementary_Methods).
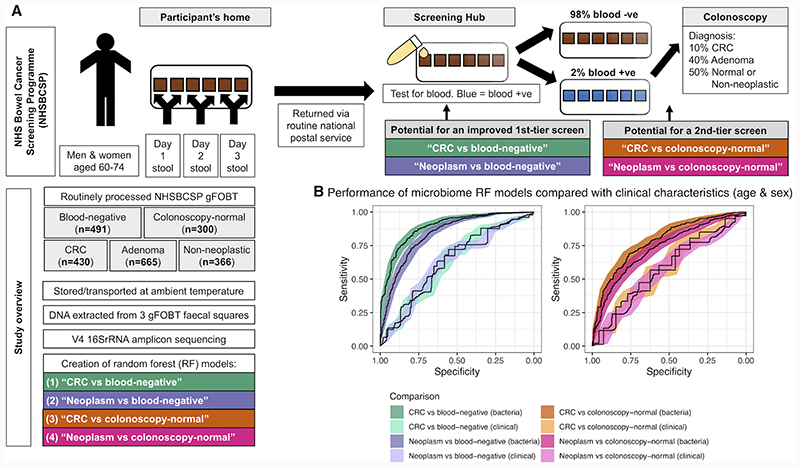
Figure 1. Microbiome taxonomic profiling demonstrates potential to improve CRC screening accuracy.
(A) Overview of the NHS Bowel Cancer Screening Programme (NHSBCSP) and the design of this study. Briefly, we used 16S amplicon-based microbiome profiling from routinely collected gFOBT specimens to supplement first-tier (CRC/neoplasm vs. blood-negative) or second-tier (CRC/neoplasm vs. colonoscopy-normal) opportunities for early cancer screening. (B) Microbiome profiles improve CRC or neoplasm classification versus blood-negative gFOBT samples (first-tier screening application) or blood-positive colonoscopy-normal samples (second-tier screening application) relative to purely clinical characteristics (age and sex). Classification used random forest (RF) models and shows the performance of the ‘total’ RF models bootstrapped from the total datasets. Shading represents the 95% CI. Clinical = RF models based on age & sex. Bacteria = RF models based on relative abundances of genera. Neoplasm = a group comprising an approximately equal ratio of CRC, low-risk adenoma, intermediate-risk adenoma and high-risk adenoma samples.
This enabled profiling of 2,252 samples: samples whereby haemoglobin was not detected i.e. ‘blood-negative’(n=491 (22%)) and ‘blood-positive’(n=1,761 (78%)). Blood-positive samples had the following colonoscopy-diagnoses: CRC (n=430 (19%)), adenoma (n=665 (30%)), colonoscopy-normal (n=300 (13%)), non-neoplastic condition (n=366 (16%)). Whilst the composition of our overall study group does not reflect the composition of the NHS Bowel Cancer Screening Programme population (2% of gFOBT are blood-positive; 10% of screening colonoscopies reveal CRC, 40% adenoma and 50% reveal a normal colon or non-neoplastic condition), we required these respective sample numbers in order to adequately profile the CRC and neoplasm-associated microbiome and to train RF models. 3 4 Test statistics that are affected by disease prevalence would be different in the NHS Bowel Cancer Screening Programme population, for example PPV would be lower.
Samples were transported to the University of Leeds at room temperature, and stored at room temperature prior to DNA extraction. The NHSBCSP asks participants to record the date of faecal collection; this information was available for 2,167 samples. Of these, 1,363 recorded three consecutive days; 95 recorded a single date (implying a single stool), and maximum duration between collections was 16 days. Time between faecal collection and DNA extraction was 46-706 days (median 374 days) (Supplementary_Methods). To determine whether prolonged storage at room temperature prior to DNA extraction altered results, a set of DNA extraction replicates was created. Three squares were dissected and combined to make a sample and, after a period of time (6-23 months), the alternate three squares were dissected and combined to make a replicate (n=26 pairs). For comparison, a set of ‘same-day’ DNA extraction replicates were created, whereby three squares of faecally-loaded card were dissected and combined to make a sample and, at the same time, the alternate three squares were dissected and combined to make a replicate (n=48 pairs).
Data was extracted from the NHSBCSP database: age, sex, screening-round, episode-outcome, and for blood-positive gFOBT: diagnosis (normal, adenoma (low, intermediate or high-risk) 17 , CRC, non-neoplastic condition), and lesion location. In cases of more than one lesion, only the most advanced was recorded. Data is based on information collected and quality assured by Public Health England (PHE) Population Screening Programmes. Access to the data was facilitated by the PHE Office for Data Release.
The screening age is 60-74 inclusive. People aged over 74 can self-refer to the programme. The study cohort contained 35 older participants (ages 75-89) and one younger participant (aged 59, one week before their birthday). A power calculation was performed using the R package pwr (based on a variance-stabilised linear model) using effect sizes from the Human Microbiome Project (RRID:SCR_012956) with Bonferroni correction. 18 Assuming 900 samples with 50 thousand reads/sample, we anticipated power 0.95 to detect a 0.055-unit difference in common taxa (0.003 relative abundance), and a 0.022-unit difference in rare taxa (0.0004 relative abundance).
Ethical approval: Tyne & Wear South REC(IRAS:188007; REC:16/NE/0210), BCSP Research Committee(BCSPID_160), Office for Data Release(ODR1617_126). Patients and the public were not involved in the study design but have since been involved in the study and will be involved in the dissemination of results.
Laboratory methods
From each developed gFOBT (Hema Screen, Immunostics, Inc), three alternate squares of faecally-loaded card were dissected and processed as a combined sample. This approach subsamples a larger volume of stool, ensuring adequate material even from thinly-smeared cards, and leaves three residual squares for alternative analysis or extraction replicates. DNA was extracted using a modified version of the QIAamp DNA Mini Kit protocol (Qiagen, Germany) (detailed in Supplementary_Methods). DNA extraction was performed in batches of up to 24 samples; to limit batch effects, batches were designed to contain samples representing the different clinical groups. Library preparation was according to the Earth Microbiome Project (EMP) 16S Illumina Amplicon methodology with single PCR reactions of 20ng DNA/sample and additional indexes to increase multiplexing capacity. 19 Samples were pooled and sequenced across two runs, each comprising one lane of an Illumina HiSeq3000, for 2x150bp sequencing, with a 10bp single index read.
Bioinformatic and statistical analysis
During quality control, 16 samples had fewer than 10,000 reads and were removed from analysis. With these samples removed, read count/sample was 14,635-555,465 (median 123,265).
Reads were stripped of adaptors using cutadapt and trimmed to maximum 145bp. 20 Pairs were merged, denoised and representative sequences chosen using DADA2. 21 Further processing was conducted in QIIME2 (version 2019.4). 22 Differences of Shannon index were assessed by Kruskal-Wallis test. Taxa were assigned by the QIIME2 feature classifier using the BLAST+ algorithm 23 24 using the SILVA version 132 99% similarity database (RRID:SCR_006423). 25 Principle coordinate analysis (PCoA) of Bray-Curtis distances was performed. Further analysis was performed using R (version 3.5.1). Differences in beta diversity were assessed by PERMANOVA analysis of Bray-Curtis distances using Adonis. 26 Differences in beta diversity between sample groups were further explored by PERMANOVA analysis of Bray-Curtis distances performed using the beta-group-significance function within QIIME2. 27 Taxa differing significantly between groups were obtained using LEfSe (Linear discriminant analysis Effect Size) (RRID:SCR_014609). 28
Random Forest (RF) models and AUC were generated using randomForest (RRID:SCR_015718) and pROC. 29–31 For the neoplasm models, the neoplasm group contained an approximately equal number of randomly selected low, intermediate and high-risk adenomas and CRC. Alternate samples were assigned to test or validation models (Supplementary_Table.3); when used, total sample sets were also bootstrapped by randomForest during training. Each forest was built with 1,000 trees. Mtry was determined based on the lowest out-of-bag error. 95% confidence intervals for the receiver operating characteristic (ROC) curves and AUC were created using 2,000 stratified bootstrap replicates. AUC were compared using roc.test, using the method of DeLong. 32 Confusion matrices were created using the predict function of randomForest using the default vote proportion cutoff of 50%.
Taxa were compared to nine CRC faecal metagenomic datasets 33–40 , processed using MetaPhlAn version 3.0 (RRID:SCR_004915). 41 42 43 The majority of the datasets have been comprehensively profiled in two recent meta-analyses. 33 34 Datasets were collapsed to genus-level for comparison. The Thomas_c 34 and Yachida 35 datasets were merged as they originated from the same cohort. RF models were built as above, using taxa present in all datasets. For within-dataset comparisons, each study was randomly split 20 times into equal sized training and validation sets, and mean AUC recorded. For the leave-one-dataset-out (LODO), models were built using all but one dataset, and validated on the missing dataset. For each test/validation pair of cohorts, confusion matrices were created using the predict function of randomForest using the default vote proportion cutoff of 50%. Sensitivity was calculated as the proportion of CRC samples called as CRC within the validation dataset, based on the test dataset RF model. Specificity was calculated as the proportion of control samples called as control. For the self-validation comparisons, the mean sensitivity and specificity of the 20 repetitions was recorded.
To compare our gFOBT-derived biomarker with microbial taxonomic biomarkers from existing datasets, we used the genus-summarised profiles to calculate a single, meta-analysed biomarker. This used the ‘metafor’ R package with a random effects model incorporating standardised mean differences from these taxonomic profiles and sample sizes from all ten datasets (including either gFOBT CRC vs blood-negative or CRC vs colonoscopy-normal).
Data is available: PRJEB37635 (http://www.ebi.ac.uk/ena/data/view/PRJEB37635).
Role of the funding source
The funders had no role in study design, data collection, analysis, interpretation, or writing. The corresponding author had full access to all the data and final responsibility for the decision to submit for publication.
Results
Summary of population characteristics and microbiome profiling
We profiled the faecal microbiomes of 2,252 NHSBCSP participants using gFOBT samples, confirming that NHSBCSP gFOBT contained adequate material for V4 16S rRNA gene amplicon sequencing. Samples retained after quality control represented phenotypes of blood-negative gFOBT (n=491 (22%)) and blood-positive (n=1761 (78%)). The blood-positive samples were grouped according to subsequent colonoscopy diagnosis: CRC (n=430 (19%)), adenoma (n=665 (30%)), colonoscopy-normal (n=300 (13%)), non-neoplastic diagnosis (n=366 (16%))(Table.1). The male preponderance of CRC and adenoma samples (67% and 65%) likely reflects the male-preponderance of colorectal neoplasia; 44 in later analysis we show that sex has minimal effect on overall microbiome structure.
Table 1. Table of participant characteristics.
| Clinical group | Mean age (SD) | Number of samples | ||
|---|---|---|---|---|
| Total | Male (%) | Female (%) | ||
| gFOBT blood-negative | 67.0 (4.5) | 491 (22%) | 205 (42%) | 286 (58%) |
| gFOBT blood-positive, with the following diagnosis at colonoscopy: | ||||
| CRC | 68.1 (5.0) | 430 (19%) | 289 (67%) | 141 (33%) |
| Adenoma | 66.3 (4.7) | 665 (30%) | 432 (65%) | 233 (35%) |
| Normal colonoscopy | 66.6 (4.3) | 300 (13%) | 155 (52%) | 145 (48%) |
| Non-neoplastic diagnosis | 66.7 (4.7) | 366 (16%) | 188 (51%) | 178 (49%) |
Of the CRC samples, lesion data was available for 359/430 (83%), corresponding to 378 colorectal cancers (342 (95%) samples resulted in a single colorectal cancer being detected at colonoscopy; 17 (5%) samples resulted in more than one synchronous colorectal cancer being detected at colonoscopy). Where type was recorded (n=298 (79%)), the majority were adenocarcinoma (n=297 (99%)); and one rectal tumour was a squamous cell carcinoma (<1%). Where grade was recorded (n=253 (67%)), the majority were well/moderately differentiated (n=224 (89%)); 29 (11%) were poorly differentiated. The commonest tumour location was sigmoid/rectum (Table.2). Unfortunately, tumour stage was not available. Of the non-neoplastic samples, lesion data was available for 333/366 (91%). Many had more than one diagnosis, the commonest being ‘diverticulosis’ (Supplementary_Methods).
Table 2. Table of CRC locations.
| CRC tumour location | Number |
|---|---|
| Ileum | 1 (<1%) |
| Caecum | 43 (11%) |
| Ascending colon | 40 (11%) |
| Hepatic flexure | 21 (6%) |
| Transverse colon | 32 (8%) |
| Splenic flexure | 15 (4%) |
| Descending colon | 12 (3%) |
| Sigmoid | 90 (24%) |
| Recto-sigmoid | 27 (7%) |
| Rectum | 96 (25%) |
| Anus | 1 (<1%) |
Pairs of technical DNA extraction replicates extracted after prolonged storage had similar microbiome structures, equivalent to ‘same-day’ DNA extraction replicates, confirming that time until DNA extraction has minimal effect on results (Supplementary_Fig.1).
Gut microbiome profiles of the NHSBCSP cohort
While the amount of biomass and resolution of amplicon-based taxonomic profiling from these samples was limited, it was more than sufficient to establish overall faecal microbiome structure, as well as to subsequently classify by phenotype. As expected, microbial structure was dominated by a gradient trade-off between Bacteroidetes versus Firmicutes phylum members, with beta diversity minimally influenced by clinical group (~1% variation in microbiome structure, by Bray-Curtis PERMANOVA), and even less by sex and age (Supplementary_Table.1 & Fig.2). Microbiome structure differed significantly between individual clinical groups by Bray-Curtis PERMANOVA (Supplementary_Table.2). Similarly, alpha diversity was significantly higher in blood-negative and CRC samples, although with very small effect size difference between groups (Kruskal-Wallis p = 4.50 x 10-25)(Supplementary_Table.3 & Fig.2). This suggested a combination of both global and taxon-specific differences in the microbiome during CRC, in agreement with previous studies. 45
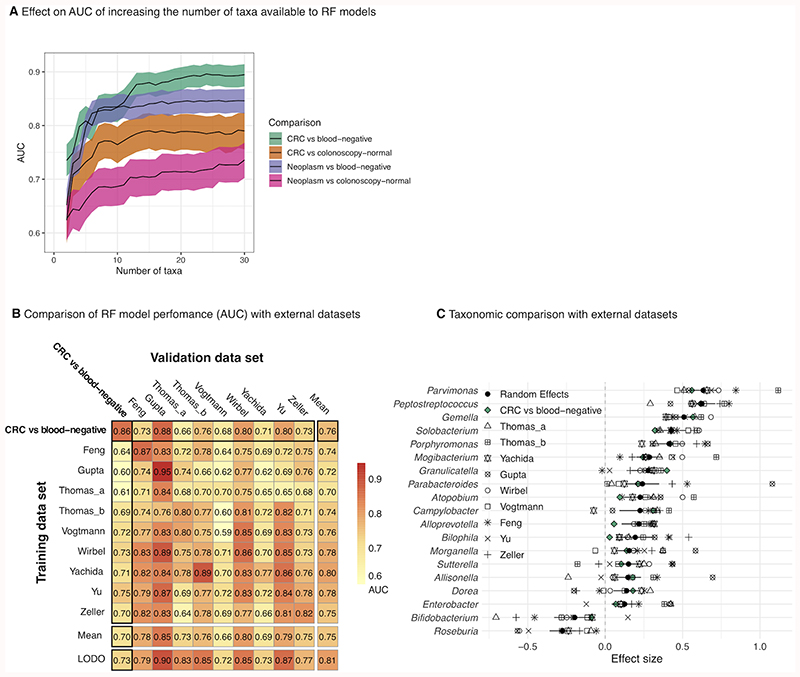
Figure 2. Microbiome-based gFOBT CRC/neoplasm classification requires as few as 15 taxa and compares favourably with models built using external shotgun metagenomic datasets.
(A) Genus-level bacteria only ‘total’ RF classification models were built using an increasing number of taxa of decreasing RF importance score. Shading represents the 95% CI of the AUC. Neoplasm = a group comprising an approximately equal ratio of CRC, low-risk adenoma, intermediate-risk adenoma and high-risk adenoma samples. For each model, the AUC plateaus at approximately 15 taxa. (B) Performance of the amplicon-based “CRC vs blood-negative” total RF model compared to models built using external faecal shotgun metagenomic datasets. The matrix displays cross-prediction AUCs. LODO (leave-one-dataset-out) denotes AUC generated by training a model using all but the dataset of the associated column and testing it using the dataset of that column. Within-study and cross-study performance of the “CRC vs blood-negative” model falls within the range of performances of the external models, indicating a degree of generalisability. (C) Specific taxa prioritised by gFOBT amplicon-based regression models (at the genus level) are strikingly similar to genera prioritised from shotgun metagenomic taxonomic profiles in complementary populations.
We thus went on to identify specific taxa that were significantly enriched/depleted between clinical groups, which proved to include CRC-microbiome associations described in the existing literature. Both inflammation-associated and oral microbes were enriched, such as Escherichia-Shigella, Peptostreptococcus, Porphyromonas, Fusobacterium and Parvimonas (Supplementary_Fig.3). Interestingly, 43 taxa were significantly enriched and 43 depleted in the blood-negative group compared with the blood-positive colonoscopy-normal group. Existing studies usually compare CRC to either healthy volunteers (equivalent to the blood-negative group) or controls with a normal colonoscopy; it is rare for both groups to be available within a study. Thus, notably, choice of control group was shown to affect which taxa were CRC-enriched relative to controls (Supplementary_Fig.3). Of the CRC-enriched taxa, seven featured in both comparisons (including Porphyromonas, Parvimonas and Peptostreptococcus), and of the CRC-depleted taxa, only one featured in both comparisons (Anaerotruncus). An inverse association with CRC was shown for 25 taxa between the two choices of control group (including Fusobacterium and Escherichia-Shigella). These findings indicate that choice of control group can have an important bearing on results, and suggest that certain taxa (especially typically oral taxa e.g. Porphyromonas, Parvimonas and Peptostreptococcus) may have an association with CRC that is independent of the presence of faecal-blood (at least at the level detectable by gFOBT), whereas others (Fusobacterium and Escherichia-Shigella) may not.
Microbiome analysis of NHSBCSP samples has the potential to improve CRC screening
To determine whether microbiome profiles from NHSBCSP gFOBT samples could improve screening accuracy, we created random forest (RF) classifiers using relative abundances of genera (Fig.1). Whilst LEfSe analysis indicates taxa which are significantly enriched or depleted between groups, RF classifiers identify taxa which have predictive associations. 28 29 30 We assessed four models, the first two of which investigated whether microbiome analysis could be used as a first-tier screen - that is, to distinguish CRC or neoplasm from blood-negative gFOBT. Based on a randomly selected 50% training-validation split, CRC outcomes were separated from blood-negative gFOBTs (“CRC vs blood-negative”) with AUC 0.86 (0.82-0.89)(Supplementary_Table.4-6). The second model distinguished neoplasm (a group comprising an approximately equal ratio of CRC, low, intermediate and high-risk adenoma) from blood-negative gFOBTs (“Neoplasm vs blood-negative”) with AUC 0.78 (0.74-0.82)(Supplementary_Table.5&6). Neither model showed a significant difference between AUCs of the test or validation sets (Supplementary_Table.5).
The next two models assessed whether microbiome profiles could distinguish, strictly among the blood-positive samples, CRC or neoplasm from subsequently colonoscopy-normal samples (i.e. a second-tier screen, to identify gFOBT false positives). As expected, these more biologically similar outcomes were more difficult to differentiate, but were still accessible via microbiome measures. The third model distinguished CRC from colonoscopy-normal gFOBT (“CRC vs colonoscopy-normal”) with AUC 0.79 (0.74-0.83)(Supplementary_Table.5 & 6 & Fig.4). The last model differentiated neoplasms from colonoscopy-normal gFOBT (“Neoplasm vs colonoscopy-normal”) with AUC 0.73 (0.68-0.77)(Supplementary_Table.5 & 6 & Fig.4). Again, neither model showed a significant difference between AUCs of the test or validation sets (Supplementary_Table.5).
All of the models performed significantly better than models generated for comparison which used age and sex. Combining age and sex with relative abundances of genera led to a small improvement in AUC for three of the models (Supplementary_Table.5). Model performance remained similar after restricting the models to a small number of taxa, mimicking what might be possible by qPCR; for all four models, AUC increased as the number of taxa increased up to fifteen, after which the AUC approximately stabilised (Fig.2, Supplementary_Table.5 & Fig.4). Interestingly, the fifteen most important taxa for the “CRC vs blood-negative” and “CRC vs colonoscopy-normal” models featured eight of the same taxa, including Fusobacterium, Peptostreptococcus, Parvimonas, Gemella, Odoribacter and Faecalibacterium, and three taxa (Faecalibacterium, Akkermansia and Escherichia-Shigella) were shared between the “Neoplasm vs blood-negative” and “Neoplasm vs colonoscopy-normal” models (Supplementary_Fig.4). Several of the same taxa appeared in the fifteen taxa most important to the “CRC vs blood-negative” and “Neoplasm vs blood-negative”, and “CRC vs colonoscopy-normal” and “Neoplasm vs colonoscopy-normal” models respectively (Supplementary_Fig.4).
Finally, we compared the performance of these 16S-based RF models to similar models using existing faecal shotgun metagenomic datasets (Fig.2, Supplementary_Fig.5). 33–40 As the majority of these existing studies had only profiled CRC, we restricted the comparison to the two CRC RF models. Within-study cross-validation of the “CRC vs blood-negative” model produced an AUC of 0.86, which compared favourably with the AUCs of the external datasets (range 0.59-0.95)(Fig.2, Supplementary_Fig.5). Between-study performance of the model also fell within the range of performances of the models built using the external datasets, and the majority of the most important taxa paralleled those of the external studies, indicating a degree of generalisability. The “CRC vs. colonoscopy-normal” model had a within-study cross-validation AUC that was within the range of the models built using external datasets, but between-study validation performance was lower (Fig.2, Supplementary_Fig.5). Taxa which were of highest importance to the model were shared by many of the models built using external datasets, indicating both their potential underlying biological importance and their ability to be consistently detected by a variety of assays.
For completeness, we also explored the ability of microbial RF models to detect adenoma. Performance was generally comparable; models distinguished CRC from adenoma with AUC 0.71 (0.66-0.76), adenoma from colonoscopy-normal with AUC 0.72 (0.67-0.77) and adenoma from blood-negative with AUC 0.84 (0.80-0.87) (Supplementary_Table.7-10). The taxa of greatest importance to the RF models included several ‘CRC-associated’ taxa. Lastly, we investigated the performance of bacteria RF models using a ‘colonoscopy-control’ group, comprising an approximately equal ratio of non-neoplastic and colonoscopy-normal samples (Supplementary_Table.7-10). CRC was detected with an AUC 0.76 (0.72-0.80), similar to the RF model which used colonoscopy-normal samples alone as the control group. However, the models designed to detect adenoma and neoplasm performed inferiorly compared with RF models built using colonoscopy-normal samples alone. This could reflect the heterogeneous nature of the non-neoplastic group, or greater microbiome similarity between the adenoma and non-neoplastic groups.
Discussion
To our knowledge, this is the first study to profile the microbiome of large numbers of CRC screening samples, collected and processed routinely by a national screening programme, and to demonstrate the potential of microbiome analysis as an accurate adjunct to early screening. We profiled the faecal microbiome of 2,252 processed NHSBCSP gFOBT samples, representing blood-negative results, colonoscopy-normal outcomes, CRC, adenomas and non-neoplastic diagnoses. Using random forest models as a simple classification method, microbiome taxonomic profiles were able to serve as accurate first and second-tier screens, the former separating CRC/neoplasm from blood-negative results, and the latter separating CRC/neoplasm from normal-colonoscopy results. All four microbiome-based models performed significantly better than models built using the only clinical data available - age and sex - and were robust to hold-out validation and in comparison to external data.
As a baseline for translational applications, the first-tier “CRC vs blood-negative” model performed similarly to existing screening methods. This includes those that rely on low-dimensional or high-dimensional biomarkers. For example, a meta-analysis of FIT and a separate study of FIT for CRC screening reported an AUC for the detection of CRC as high as 0.95. 46 47 Separately, a trial of the FDA-approved Cologuard reached an AUC of 0.94 for the discrimination of CRC vs ‘non-advanced neoplasia/lesser findings’, and with FIT an AUC of 0.89. 48 Our microbiome-based “Neoplasm vs blood-negative” model again performed similarly (possibly superiorly) to existing methods (AUCs from the aforementioned studies of 0.72(FIT), 0.67(FIT) and 0.73(Cologuard)), 47 48 although differences in the composition of the case and control groups between the studies should be borne in mind. Importantly, in comparison with Cologuard, which requires whole stool and costs approximately $600/test, amplicon-based microbiome profiling requires very little biomaterial and would be easier to translate to a national screening programme. The fact that model performance required as few as fifteen taxa, in agreement with existing studies, raises the potential of a rapid qPCR-based test which could be integrated into a screening programme at low cost. 34 49–52 Although we were not able to assess it in our study, it has been shown that microbiome-analysis is able to detect lesions missed by FIT, suggesting a potential role as an adjunct to FIT for the detection of non-bleeding CRC. 53
The second-tier models perhaps showed the greatest clinical potential, as they were able to identify CRC and neoplasms from among the blood-positive gFOBT cohort. Currently all NHSBCSP participants with a blood-positive gFOBT are referred for colonoscopy, yet 50% reveal a normal bowel or non-neoplastic condition. The high number of unnecessary colonoscopies carries associated risks and strains endoscopy capacity. There are limited examples of second-tier screens in the existing literature. A study from the NHSBCSP programme demonstrated second-tier performance for the detection of neoplasm by FIT with AUC 0.63, improved to 0.66 by incorporating screening data. 54 A similar study reported an equivalent AUC of 0.69 (FIT), improved to 0.76 by questionnaire-collected data. 55 The advantage of a microbiome-based second-tier screen that could be performed using existing screening samples is that it would not require additional tests, nor would it place extra burden on screening participants, something which can potentially jeopardise screening uptake.
Given that we profiled the microbiome directly from gFOBT screening samples, we were interested to compare the performance of our models with the existing microbiome literature, most of which has used shotgun metagenomics and/or frozen whole stool. Performance compared favourably: meta-analyses and a systematic review reported AUCs of 0.68-0.95 (detection of CRC), and AUCs of 0.59-0.94 (detection of neoplasm - many studies, like ours, report inferior detection of neoplasms compared with CRC, due to the reduced discriminatory power of microbiome-based models to detect adenomas 34 ). 5 33 34 49 50 56–59 It is remarkable that our models performed so well in light of the fact that samples were prepared routinely by screening participants in their own homes (in the majority of instances over three days), transported through the routine post, stored at room temperature (for on average one year prior to DNA extraction), and the following variables, all of which affect the microbiome, were unknown: antibiotic/medication-use, diet, comorbidities, smoking status, and BMI. 60 While this technical variability and missing information will unavoidably affect the precision of microbiome measurements feasible from gFOBT, and their applicability to general microbiome epidemiology, it is noteworthy that they do not impede gFOBT microbiome use for CRC screening. We further confirmed this in a quantitative manner, by comparing the performance of our CRC models with models built using nine external metagenomic datasets. Validation of the gFOBT-based models among studies showed similar performance and, interestingly, identification of many of the same discriminatory taxa.
These taxa included those previously described as CRC-associated, including Fusobacterium, Escherichia-Shigella, Peptostreptococcus, Porphyromonas, Parvimonas, Alistipes, and Gemella, and those that have previously been shown to be inversely associated with CRC, including Faecalibacterium 61 and Lactobacillus. 49 Although we limited ourselves to analysis at the genus level for simplicity, these genera contain species which have been associated with CRC, including inflammation-associated and oral-taxa: Fusobacterium nucleatum, 49 pks+Escherichia coli, 62 Peptostreptococcus stomatis, 36 Peptostreptococcus anaerobius, 35 Porphyromonas asaccharolytica, 49 Porphyromonas somerae, 33 Porphyromonas uenonis, 33 Parvimonas micra, 49 Alistipes finegoldii, 49 and Gemella morbillorum. 33 It is hypothesised that oral taxa may increase colonic mucosal permeability, allowing bacterial invasion, with resulting inflammation, and subsequent epithelial proliferation. 63 64 65 Certain taxa have also been shown to be capable of inducing and/or promoting tumourigenesis: colibactin, produced by pks+Escherichia coli, is able to damage DNA, 62 whilst Fusobacterium nucleatum promotes tumour proliferation and a pro-tumour inflammatory state. 66 It was interesting that some (but not all) of these taxa remained CRC-enriched even in comparisons with the blood-positive colonoscopy-normal group, suggesting that certain CRC-microbiome associations may act independently of the presence of faecal blood.
Among this study’s potential limitations, two stand out. The first is that participants in the blood-negative group did not undergo colonoscopy, as this would disrupt routine screening. As the sensitivity of gFOBT for CRC is estimated to be 50%, the blood-negative group may have included undiagnosed adenomas or CRC. 67–69 However, because the incidence of CRC is low, the absolute number of undiagnosed CRC is predicted to have been small, with little effect on the performance of the RF models, except perhaps to have made the result more conservative. This leads to an arguably minor, but still systematic, difference between these controls and a broader population: the specific models evaluated here will under-predict non-bleeding cancers and should be further generalised prior to application. The second is that the majority of the blood-negative samples were collected within a short time-frame at the beginning of the study. However, any effect due to prolonged storage prior to DNA extraction is likely to have been minimal, as DNA extraction replicates created after 6-23 months storage at room temperature demonstrated similar microbiome structures, equivalent to ‘same-day’ DNA extraction replicates.
In addition to the refinements that would be necessary to translate these results into a screening product, including investigation of sensitivity, consistency and cost-effectiveness analysis, future work aims to replicate the study using NHSBCSP FIT samples. The advantage of having performed the current study is that, should microbiome analysis of FIT (which collects a much smaller volume of faeces) not produce adequate accuracy, a gFOBT-based microbiome screening test could still be used as an adjunct to the NHSBCSP. We also plan to investigate whether screening accuracy could be improved further by the incorporation of additional clinical data, FIT concentration, and faecal mutation, bacterial virulence-factor or toxin testing. 33 34 49 52 70 71 In conclusion, this study has confirmed that microbiome analysis can be performed on samples collected and processed routinely by a national CRC screening programme to improve accuracy. Models required as few as fifteen taxa, making this practical to implement as an inexpensive qPCR-based test. This could reduce the number of unnecessary colonoscopies in countries which use faecal occult blood test screening.
Supplementary Material
Translational Relevance.
To assess the utility of microbiome profiles for national-scale colorectal cancer (CRC) screening, we assessed 2,252 routinely processed NHS Bowel Cancer Screening Programme guaiac faecal occult blood test (gFOBT) samples. We generated four microbiome-based random forest classification models, each showing potential to improve accuracy. Two distinguished either CRC or neoplasm (CRC or adenoma) from gFOBT blood-negative samples (equivalent to first-tier screening). Two distinguished CRC or neoplasm from samples that had tested positive for blood by gFOBT, with participants referred for colonoscopy, but at colonoscopy no-lesion was found (second-tier screening to rule out gFOBT false positives). Each model remained robust to validation and when restricted to fifteen taxa, raising the possibility of an inexpensive qPCR-test. The models performed favourably compared with existing microbiome studies, FIT and Cologuard. These results suggest that microbiome analysis could be integrated into national CRC screening to improve accuracy and reduce the number of unnecessary screening colonoscopies.
Acknowledgements
This work was funded by a Wellcome Trust Clinical Research Training Fellowship (203524/Z/16/Z) to C Young, a Pathological Society of Great Britain & Ireland ‘Visiting Fellowship’ (2234) to C Young and a Cancer Research UK Grand Challenge Initiative (OPTIMISTICC C10674/A27140) to P Quirke and C Huttenhower. P Quirke is a National Institute of Health Research Senior Investigator.
Funding
This work was funded by a Wellcome Trust Clinical Research Training Fellowship (203524/Z/16/Z) to CY, a Pathological Society of Great Britain & Ireland ‘Visiting Fellowship’ (2234) to CY and a Cancer Research UK Grand Challenge Initiative (OPTIMISTICC C10674/A27140) to PQ and CH. PQ is a National Institute of Health Research Senior Investigator.
Footnotes
Contributors
CY, HW, EM, PQ: Study design and supervision. SB, MB, CJ, CB, EM: Acquisition of data and samples. CY, AFB, DB, NG, LW: Sample processing. CY, HW, JB, KT, YY, CH: Data analysis. CY, HW, KT, YY, CH: Drafting of the manuscript. CY, HW, AFB, DB, NG, LW, SB, MB, CJ, CB, KT, YY, EM, CH, JB, PQ: Critical revision of the manuscript. CY, PQ: Fund raising for the study. All authors approved the final version of the manuscript.
Competing interests
None declared.
References
- 1.Ferlay J, E M, Lam F, Colombet M, Mery L, Piñeros M, Znaor A, Soerjomataram I, Bray F. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer. 2018. [accessed 9.8.20 2020]. Available from: https://gco.iarc.fr/today .
- 2.Koo S, Neilson LJ, Von Wagner C, et al. The NHS Bowel Cancer Screening Program: current perspectives on strategies for improvement. Risk Manag Healthc Policy. 2017;10:177–87. doi: 10.2147/rmhp.S109116. [published Online First: 2017/12/23] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bowel cancer screening: the facts (FOB test kit) [accessed 24.9.19]. Available from: https://www.gov.uk/government/publications/bowel-cancer-screening-benefits-and-risks .
- 4.Scottish Bowel Screening Programme Statistics for invitations between 1 May 2016 and 30 April 2018. 2019. [accessed 20.7.19]. https://www.isdscotland.org/Health-Topics/Cancer/Publications/2019-02-05/2019-02-05-Bowel-Screening-Publication-Summary.pdf Available from: https://www.isdscotland.org/Health-Topics/Cancer/Publications/2019-02-05/2019-02-05-Bowel-Screening-Publication-Summary.pdf .
- 5.Amitay EL, Krilaviciute A, Brenner H. Systematic review: Gut microbiota in fecal samples and detection of colorectal neoplasms. Gut Microbes. 2018;9(4):293–307. doi: 10.1080/19490976.2018.1445957. [published Online First: 2018/03/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vogtmann E, Chen J, Amir A, et al. Comparison of Collection Methods for Fecal Samples in Microbiome Studies. Am J Epidemiol. 2017;185(2):115–23. doi: 10.1093/aje/kww177. published Online First: 2016/12/18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha R, Chen J, Amir A, et al. Collecting Fecal Samples for Microbiome Analyses in Epidemiology Studies. Cancer Epidemiol Biomarkers Prev. 2016;25(2):407–16. doi: 10.1158/1055-9965.epi-15-0951. [published Online First: 2015/11/26] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dominianni C, Wu J, Hayes RB, et al. Comparison of methods for fecal microbiome biospecimen collection. BMC Microbiol. 2014;14:103. doi: 10.1186/1471-2180-14-103. [published Online First: 2014/04/25] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wong WSW, Clemency N, Klein E, et al. Collection of non-meconium stool on fecal occult blood cards is an effective method for fecal microbiota studies in infants. Microbiome. 2017;5(1):114. doi: 10.1186/s40168-017-0333-z. [published Online First: 2017/09/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Taylor M, Wood HM, Halloran SP, et al. Examining the potential use and long-term stability of guaiac faecal occult blood test cards for microbial DNA 16S rRNA sequencing. Journal of Clinical Pathology. 2017;70(7):600–06. doi: 10.1136/jclinpath-2016-204165. [DOI] [PubMed] [Google Scholar]
- 11.Vogtmann E, Chen J, Kibriya MG, et al. Comparison of Fecal Collection Methods for Microbiota Studies in Bangladesh. Appl Environ Microbiol. 2017;83(10) doi: 10.1128/aem.00361-17. published Online First: 2017/03/05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.von Huth S, Thingholm LB, Bang C, et al. Minor compositional alterations in faecal microbiota after five weeks and five months storage at room temperature on filter papers. Scientific Reports. 2019;9(1):19008. doi: 10.1038/s41598-019-55469-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Byrd DA, Sinha R, Hoffman KL, et al. Comparison of Methods To Collect Fecal Samples for Microbiome Studies Using Whole-Genome Shotgun Metagenomic Sequencing. mSphere. 2020;5(1):e00827–19. doi: 10.1128/mSphere.00827-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Amitay EL, Werner S, Vital M, et al. Fusobacterium and colorectal cancer: Causal factor or passenger? Results from a large colorectal cancer screening study. Carcinogenesis. 2017;38(8):781–88. doi: 10.1093/carcin/bgx053. [published Online First: 2017/06/06] [DOI] [PubMed] [Google Scholar]
- 15.Eklof V, Lofgren-Burstrom A, Zingmark C, et al. Cancer-associated fecal microbial markers in colorectal cancer detection. International Journal of Cancer. 2017;141(12):2528–36. doi: 10.1002/ijc.31011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grobbee EJ, Lam SY, Fuhler GM, et al. First steps towards combining faecal immunochemical testing with the gut microbiome in colorectal cancer screening. United European gastroenterology journal. 2020;8(3):293–302. doi: 10.1177/2050640619890732. [published Online First: 2020/03/28] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Logan RFA, Patnick J, Nickerson C, et al. Outcomes of the Bowel Cancer Screening Programme (BCSP) in England after the first 1 million tests. Gut. 2011:1439–46. doi: 10.1136/gutjnl-2011-300843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.A framework for human microbiome research. Nature. 2012;486(7402):215–21. doi: 10.1038/nature11209. [published Online First: 2012/06/16] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Earth Microbiome Project. [accessed 11.2.19]. Available from: http://www.earthmicrobiome.org .
- 20.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnetjournal. 2011;17(1):10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 21.Callahan BJ, McMurdie PJ, Rosen MJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature methods. 2016;13:581. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bolyen E, Rideout JR, Dillon MR, et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nature Biotechnology. 2019;37(8):852–57. doi: 10.1038/s41587-019-0209-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bokulich NA, Kaehler BD, Rideout JR, et al. Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome. 2018;6(1):90. doi: 10.1186/s40168-018-0470-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho C, Coulouris G, Avagyan V, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quast C, Pruesse E, Yilmaz P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic acids research. 2012;41(D1):D590–D96. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oksanen J, Blanchet FG, Friendly M, et al. vegan: Community Ecology Package. 2018. [accessed 13.8.19]. R package version 2.5-3. 2018 [Available from: https://CRAN.R-project.org/package=vegan].
- 27.Anderson MJ. A new method for non-parametric multivariate analysis of variance. 2001;26(1):32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x. [DOI] [Google Scholar]
- 28.Segata N, Izard J, Waldron L, et al. Metagenomic biomarker discovery and explanation. Genome biology. 2011;12(6):R60–R60. doi: 10.1186/gb-2011-12-6-r60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Breiman L. Random Forests. Machine Learning. 2001;45(1):5–32. doi: 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 30.Wiener ALaM. Classification and Regression by randomForest. R News. 2002;2(3):18–22. [Google Scholar]
- 31.Robin X, Turck N, Hainard A, et al. pROC: an open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics. 2011;12(1):77. doi: 10.1186/1471-2105-12-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. [published Online First: 1988/09/01];Biometrics. 1988 44(3):837–45. [PubMed] [Google Scholar]
- 33.Wirbel J, Pyl PT, Kartal E, et al. Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. [published Online First: 2019/04/03];Nat Med. 2019 25(4):679–89. doi: 10.1038/s41591-019-0406-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Thomas AM, Manghi P, Asnicar F, et al. Metagenomic analysis of colorectal cancer datasets identifies cross-cohort microbial diagnostic signatures and a link with choline degradation. Nature Medicine. 2019;25(4):667–78. doi: 10.1038/s41591-019-0405-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yachida S, Mizutani S, Shiroma H, et al. Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. [published Online First: 2019/06/07];Nat Med. 2019 25(6):968–76. doi: 10.1038/s41591-019-0458-7. [DOI] [PubMed] [Google Scholar]
- 36.Gupta A, Dhakan DB, Maji A, et al. Association of Flavonifractor plautii, a Flavonoid-Degrading Bacterium, with the Gut Microbiome of Colorectal Cancer Patients in India. mSystems. 2019;4(6):e00438–19. doi: 10.1128/mSystems.00438-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Feng Q, Liang S, Jia H, et al. Gut microbiome development along the colorectal adenoma-carcinoma sequence. [published Online First: 2015/03/12];Nat Commun. 2015 6:6528. doi: 10.1038/ncomms7528. [DOI] [PubMed] [Google Scholar]
- 38.Vogtmann E, Hua X, Zeller G, et al. Colorectal Cancer and the Human Gut Microbiome: Reproducibility with Whole-Genome Shotgun Sequencing. [[published Online First: 2016/05/14]];PLoS One. 2016 11(5):e0155362. doi: 10.1371/journal.pone.0155362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yu J, Feng Q, Wong SH, et al. Metagenomic analysis of faecal microbiome as a tool towards targeted non-invasive biomarkers for colorectal cancer. Gut. 2017;66(1):70–78. doi: 10.1136/gutjnl-2015-309800. [published Online First: 2015/09/27] [DOI] [PubMed] [Google Scholar]
- 40.Zeller G, Tap J, Voigt AY, et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. [[published Online First: 2014/11/30]];Mol Syst Biol. 2014 10(11):766. doi: 10.15252/msb.20145645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Segata N, Waldron L, Ballarini A, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nature methods. 2012;9(8):811–14. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pasolli E, Truong DT, Malik F, et al. Machine Learning Meta-analysis of Large Metagenomic Datasets: Tools and Biological Insights. PLoS computational biology. 2016;12(7):e1004977–e77. doi: 10.1371/journal.pcbi.1004977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bernau C, Riester M, Boulesteix AL, et al. Cross-study validation for the assessment of prediction algorithms. [[published Online First: 2014/06/17]];Bioinformatics. 2014 30(12):i105–12. doi: 10.1093/bioinformatics/btu279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White A, Ironmonger L, Steele RJC, et al. A review of sex-related differences in colorectal cancer incidence, screening uptake, routes to diagnosis, cancer stage and survival in the UK. BMC cancer. 2018;18(1):906–06. doi: 10.1186/s12885-018-4786-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yan Y, Drew DA, Markowitz A, et al. Structure of the Mucosal and Stool Microbiome in Lynch Syndrome. [[published Online First: 2020/04/03]];Cell Host Microbe. 2020 27(4):e4. doi: 10.1016/j.chom.2020.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JK, Liles EG, Bent S, et al. Accuracy of fecal immunochemical tests for colorectal cancer: systematic review and meta-analysis. [[published Online First: 2014/03/25]];Annals of internal medicine. 2014 160(3):171. doi: 10.7326/m13-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Brenner H, Chen H. Fecal occult blood versus DNA testing: indirect comparison in a colorectal cancer screening population. Clin Epidemiol. 2017;9:377–84. doi: 10.2147/CLEP.S136565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Imperiale TF, Ransohoff DF, Itzkowitz SH, et al. Multitarget stool DNA testing for colorectal-cancer screening. [[published Online First: 2014/03/22]];N Engl J Med. 2014 370(14):1287–97. doi: 10.1056/NEJMoa1311194. [DOI] [PubMed] [Google Scholar]
- 49.Dai Z, Coker OO, Nakatsu G, et al. Multi-cohort analysis of colorectal cancer metagenome identified altered bacteria across populations and universal bacterial markers. Microbiome. 2018;6(1):70. doi: 10.1186/s40168-018-0451-2. [published Online First: 2018/04/13] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sze MA, Schloss PD. Leveraging Existing 16S rRNA Gene Surveys To Identify Reproducible Biomarkers in Individuals with Colorectal Tumors. mBio. 2018;9(3):e00630–18. doi: 10.1128/mBio.00630-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ai D, Pan H, Li X, et al. Identifying Gut Microbiota Associated With Colorectal Cancer Using a Zero-Inflated Lognormal Model. [[published Online First: 2019/05/10]];Front Microbiol. 2019 10:826. doi: 10.3389/fmicb.2019.00826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gao R, Wang Z, Li H, et al. Gut microbiota dysbiosis signature is associated with the colorectal carcinogenesis sequence and improves the diagnosis of colorectal lesions. Journal of Gastroenterology and Hepatology. 2020 doi: 10.1111/jgh.15077. n/a(n/a) [DOI] [PubMed] [Google Scholar]
- 53.Baxter NT, Ruffin MT, Rogers MAM, et al. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Medicine. 2016;8(1):37. doi: 10.1186/s13073-016-0290-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cooper JA, Parsons N, Stinton C, et al. Risk-adjusted colorectal cancer screening using the FIT and routine screening data: development of a risk prediction model. British journal of cancer. 2018;118(2):285–93. doi: 10.1038/bjc.2017.375. [published Online First: 2017/11/02] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stegeman I, de Wijkerslooth TR, Stoop EM, et al. Combining risk factors with faecal immunochemical test outcome for selecting CRC screenees for colonoscopy. Gut. 2014;63(3):466–71. doi: 10.1136/gutjnl-2013-305013. published Online First: 2013/08/22. [DOI] [PubMed] [Google Scholar]
- 56.Zhang B, Xu S, Xu W, et al. Leveraging Fecal Bacterial Survey Data to Predict Colorectal Tumors. Frontiers in genetics. 2019;10:447–47. doi: 10.3389/fgene.2019.00447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Shah MS, DeSantis TZ, Weinmaier T, et al. Leveraging sequence-based faecal microbial community survey data to identify a composite biomarker for colorectal cancer. Gut. 2018;67(5):882–91. doi: 10.1136/gutjnl-2016-313189. published Online First: 2017/03/28. [DOI] [PubMed] [Google Scholar]
- 58.Huang Q, Peng Y, Xie F. Fecal fusobacterium nucleatum for detecting colorectal cancer: a systematic review and meta-analysis. The International journal of biological markers. 2018 doi: 10.1177/1724600818781301. 1724600818781301. [published Online First: 2018/07/04] [DOI] [PubMed] [Google Scholar]
- 59.Zhang X, Zhu X, Cao Y, et al. Fecal Fusobacterium nucleatum for the diagnosis of colorectal tumor: A systematic review and meta-analysis. [[published Online First: 2019/01/14]];Cancer Med. 2019 8(2):480–91. doi: 10.1002/cam4.1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhernakova A, Kurilshikov A, Bonder MJ, et al. Population-based metagenomics analysis reveals markers for gut microbiome composition and diversity. Science. 2016;352(6285):565–9. doi: 10.1126/science.aad3369. [published Online First: 2016/04/30] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Guo S, Li L, Xu B, et al. A simple and novel fecal biomarker for colorectal cancer: Ratio of Fusobacterium nucleatum to probiotics populations, based on their antagonistic effect. Clinical Chemistry. 2018;64(9):1327–37. doi: 10.1373/clinchem.2018.289728. [DOI] [PubMed] [Google Scholar]
- 62.Pleguezuelos-Manzano C, Puschhof J, Huber AR, et al. Mutational signature in colorectal cancer caused by genotoxic pks+ E. coli. Nature. 2020;580(7802):269–73. doi: 10.1038/s41586-020-2080-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Dejea CM, Wick EC, Hechenbleikner EM, et al. Microbiota organization is a distinct feature of proximal colorectal cancers. Proceedings of the National Academy of Sciences of the United States of America; 2014. pp. 18321–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Drewes JL, White JR, Dejea CM, et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ biofilms and microbiomes. 2017;3:34. doi: 10.1038/s41522-017-0040-3. [published Online First: 2017/12/08] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomkovich S, Dejea CM, Winglee K, et al. Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. [[published Online First: 2019/03/12]];J Clin Invest. 2019 130:1699–712. doi: 10.1172/jci124196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Brennan CA, Garrett WS. Fusobacterium nucleatum - symbiont, opportunist and oncobacterium. Nature reviews Microbiology. 2019;17(3):156–66. doi: 10.1038/s41579-018-0129-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Moss S, Mathews C, Day TJ, et al. Increased uptake and improved outcomes of bowel cancer screening with a faecal immunochemical test: results from a pilot study within the national screening programme in England. Gut. 2017;66(9):1631–44. doi: 10.1136/gutjnl-2015-310691. [published Online First: 2016/06/09] [DOI] [PubMed] [Google Scholar]
- 68.Blanks R, Burón Pust A, Alison R, et al. Screen-detected and interval colorectal cancers in England: Associations with lifestyle and other factors in women in a large UK prospective cohort. International journal of cancer. 2019;145(3):728–34. doi: 10.1002/ijc.32168. [published Online First: 2019/02/15] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Morris EJA, Whitehouse LE, Farrell T, et al. A retrospective observational study examining the characteristics and outcomes of tumours diagnosed within and without of the English NHS Bowel Cancer Screening Programme. British Journal of Cancer. 2012;107(5):757–64. doi: 10.1038/bjc.2012.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhao D, Liu H, Zheng Y, et al. A reliable method for colorectal cancer prediction based on feature selection and support vector machine. Medical & biological engineering & computing. 2019;57(4):901–12. doi: 10.1007/s11517-018-1930-0. [published Online First: 2018/11/28] [DOI] [PubMed] [Google Scholar]
- 71.Zhai RL, Xu F, Zhang P, et al. The Diagnostic Performance of Stool DNA Testing for Colorectal Cancer: A Systematic Review and Meta-Analysis. Medicine (Baltimore) 2016;95(5):e2129. doi: 10.1097/md.0000000000002129. [published Online First: 2016/02/06] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.