Abstract
Predicting the impact of non-coding genetic variation requires interpreting it in the context of 3D genome architecture. We have developed deepC, a transfer learning based deep neural network that accurately predicts genome folding from megabase-scale DNA sequence. DeepC predicts domain boundaries at high-resolution, learns the sequence determinants of genome folding and predicts the impact of both large-scale structural and single base pair variations.
Introduction
Most genetic variants associated with common diseases affect gene regulatory regions distal to target genes1,2. Genome 3D structure is central to mediating these functional interactions, but its intricately convoluted and large-scale nature renders it challenging to understand and predict. Proposed machine learning and polymer modelling approaches to predict 3D genome structure have produced promising results, but none effectively integrates across resolutions. Methods that use information at the base pair level focus on window-to-window-based predictions3–5, while methods that incorporate a large genomic context do so by coarse segregation into genomic features6–8 or polymer beads9,10, thus compromising their ability to predict the impact of variation at base pair resolution.
We propose that to accurately predict topologically associated domains (TADs) a model needs to capture sequence patterns across large genomic distances. Regulatory elements can interact over megabase distances and boundary elements lying in between may alter chromatin contacts significantly. A chromatin interaction model should thus integrate information at the megabase-scale. However, to predict the impact of genetic variation the model must also learn to interpret DNA sequence at base pair resolution. Since 3D genome interactions are determined by genomic regulatory elements such as CTCF bound domain boundaries11, a model that has learned the grammar of regulatory elements could help guide the prediction of 3D genome structure.
Based on these ideas, we developed deepC, a deep neural network that bridges the gap from base pairs to TADs. DeepC uses a transfer learning approach and tissue-specific Hi-C data to train models that predict genome folding from megabase (Mb) windows of DNA sequence (Fig. 1a). The trained models can then be used to predict chromatin domain boundaries at high resolution and to identify the sequence determinants of genome folding. Importantly, they allow us to predict the impact of genetic variants from large structural variations down to single-nucleotide polymorphisms.
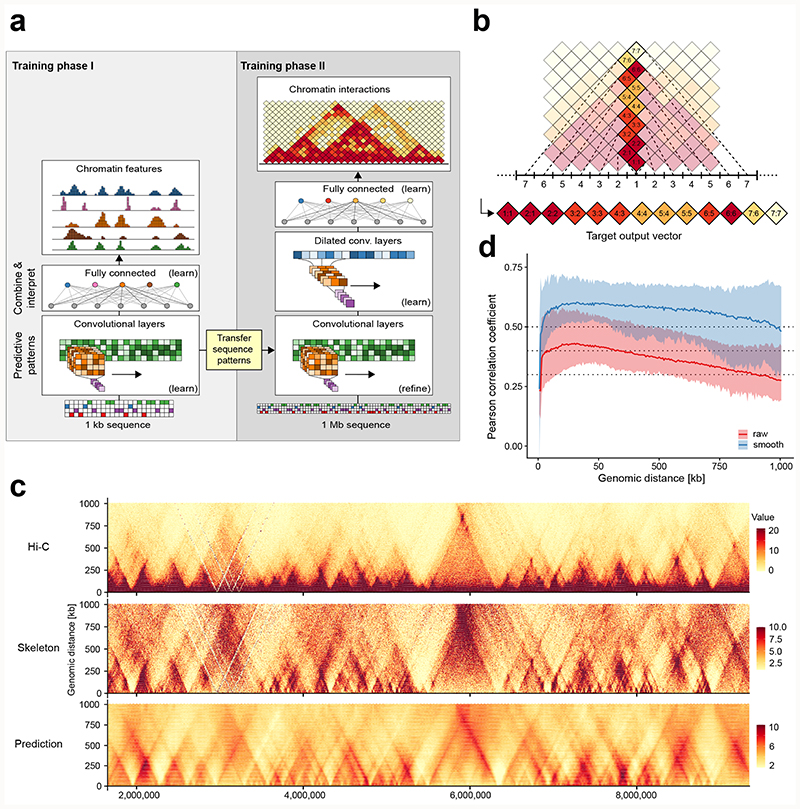
Figure 1. Predicting Hi-C interactions from DNA sequence.
a) Overview of the deepC architecture and training workflow. b) Encoding of Hi-C data as target vector for prediction given a 1 Mb window of DNA sequence. c) Comparison of Hi-C data, the derived Hi-C skeleton and the interactions predicted from DNA sequence using deepC. Shown is a ~ 7 Mb region on hold out chromosome 17. d) Distance-stratified Pearson correlation between the Hi-C skeleton and the deepC predictions in a cross-validation scheme across all chromosomes. Solid lines indicate the mean correlation value and the area indicates the space between the maximum and the minimum values over all chromosomes. Red shows the correlation with the raw and blue with the (5x5) mean filter smoothed skeleton values. Dotted lines at 0.3, 0.4 and 0.5.
Results
A deep learning model for predicting chromatin interactions from megabase-scale DNA
We encode Hi-C data as a vector of pairwise interaction values between 5 kilobase (kb) genomic bins at distances of up to ~1 Mb (Fig. 1b). DeepC learns to predict these contact frequencies taking as input the underlying ~1 Mb window of DNA sequence. The deepC network architecture is constructed from a convolutional module with max-pooling that has proven powerful for predicting chromatin features from DNA sequence12,13. This is followed by a dilated convolutional module that excels at incorporating large-scale context while maintaining resolution14–16. Finally, a fully connected layer integrates the detected patterns over a megabase of DNA sequence to predict chromatin folding.
We found two factors to be crucial for deepC’s effective learning and generalization. First, we percentile-normalize the raw contact frequency signal in Hi-C data by genomic distance (Extended Data Figure 1, Methods), termed the “skeleton”. This normalization reveals informative longer-range interactions and enhances the contrast at domain boundaries. Second, we employ transfer learning17 (Fig. 1a, Extended Data Figure 2), a concept that has proven powerful in deep learning applications for image analysis and natural language processing. In a first phase of training the initial convolutional module learns to predict a compendium of chromatin features such as open chromatin regions and CTCF binding sites across cell types12,18,19. Next, the convolutional module is stripped of the fully connected layer responsible for interpretation. Only the learned sequence patterns are transferred to the second training phase where they are refined, and the dilated module and fully connected layer are trained ab initio to predict chromatin interactions. The same weights pre-trained on a chromatin feature compendium across cell types are used for transfer learning irrespective of the cell type of the Hi-C data source.
We trained deepC models on seven human20 and one mouse21 Hi-C data sets with different sequencing depths and at different resolutions (Supplementary Figure 1 – 4). We focused our analysis on the primary GM12878 (~3.6 B reads) and K562 (~1.3 B reads) data, training models at 5 kb resolution. DeepC yields smooth but detailed predictions that resolve the hierarchical nature of TADs and insulated domains (Fig. 1c, Supplementary Figure 1). In a cross-validation scheme across all chromosomes in GM12878 (Fig. 1d), deepC achieves an average, distance-stratified Pearson correlation between predictions and Hi-C skeleton of ~0.36 on raw skeleton data and ~0.57 when applying a small smoothing filter to the discrete and noisy skeleton (~0.28 and ~0.51 in K562, Supplementary Figure 5). We compared deepC to a recently proposed random-forest-based method HiC-Reg8, that predicts chromatin interactions up to 1 Mb distance using chromatin features of the interacting windows and the window in between rather than using DNA sequence as input. We observed that deepC generalizes better in predicting the domain structure of unseen chromosomes (Supplementary Figure 6 and 7).
Although we focused our main analysis on the deeply sequenced Hi-C data from Rao et al.20 we hypothesized that deepC is able to predict chromatin interactions after training on data with significantly lower sequencing depth. To this end we trained GM12878 models with Hi-C data downsampled from originally ~26 B to 1 B, 100 M and 10 M valid Hi-C contacts respectively (Supplementary Figure 8). The 1 B and 100 M contact models still learned to predict chromatin structure, with a mean Pearson correlation of 0.46 and 0.4 between the 1 B and 100 M model on hold out chromosomes 16 and 17 respectively. In contrast the 10 M model failed to learn chromatin structure. While deepC can predict chromatin interactions from less deeply sequenced samples, we do note that dedicated methods have been proposed for increasing the resolution of Hi-C maps that take as input low resolution maps directly22,23 rather than predicting from DNA sequence.
We train separate models for each Hi-C data set derived from a different cell type. These distinct models learn tissue-specific chromatin interactions (Extended Data Figure 3, Supplementary Figure 9). We also trained a deepC model to predict chromatin interactions in multiple cell types jointly, but we found that the jointly trained network captured the tissue-specific Hi-C patterns less well compared to the individually trained models (Supplementary Figure 10).
Validating deepC with high sensitivity chromosome confirmation capture
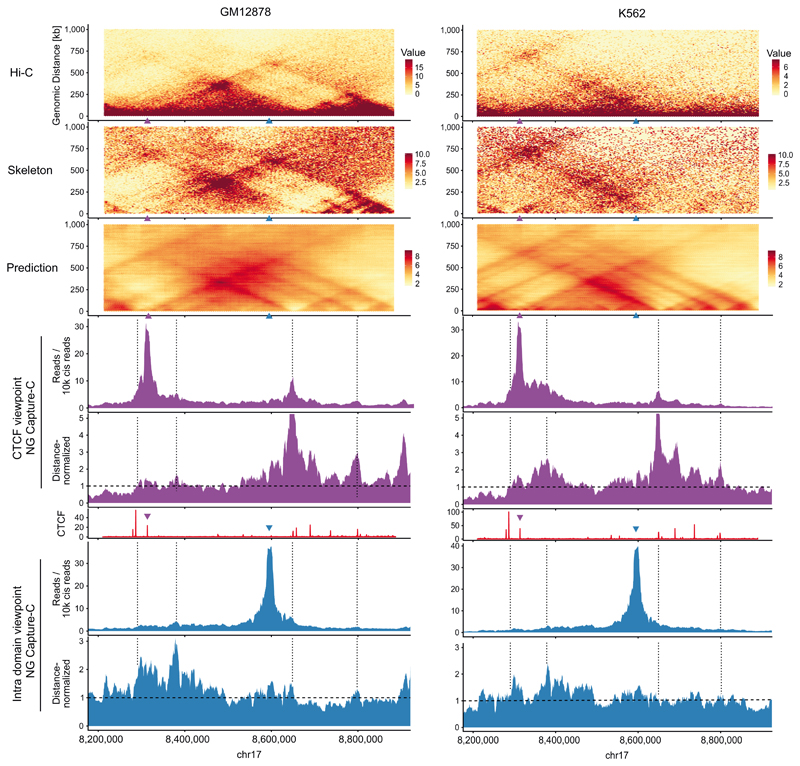
We next sought to validate deepC predictions with an independent set of chromatin interactions. To this end, we utilized NG Capture-C24 (Methods), which generates high-resolution interaction data from targeted viewpoints and identifies chromatin interactions at higher sensitivity than Hi-C. We captured the interactions of 220 viewpoints in two cell types (GM12878 and K562) covering 81 CTCF sites and 139 sites lying within insulated domains, not overlapping with regulatory elements to capture the domain structure. We observed good agreement between the predicted domain structure and interaction peaks in the NG Capture-C tracks (Extended Data Figure 4), showing deepC is capable of predicting true biophysical boundaries that are evident in these sensitive 3C assays but poorly captured by the original Hi-C, especially at lower sequencing depth.
Although deepC effectively captures the positions of boundary elements some aspects of interactions are not captured fully by the model. When comparing the virtual 4C track from the Hi-C skeleton, the predictions and distance-normalized NG Capture-C tracks from CTCF viewpoints (Supplementary Figure 11) we saw that the NG Capture-C correlated better with the Hi-C skeleton than with the predictions (Fig. 2a, Pearson correlation in GM12878: 0.59 vs 0.30; K562: 0.55 vs 0.37). This was due to the tendency of deepC predictions to de-emphasize the characteristic punctate nature of signal at the apex of interacting CTCF elements. This may be explained by an inability of deepC to model detailed characteristics of the loop extrusion mechanism such as cohesin processivity, which may not be encoded in the local DNA sequence and more dependent on factors such as nuclear concentration of extruding factors25. In contrast, for intra domain viewpoints deepC correlates equally well with NG Capture-C data as NG Capture-C correlates with the Hi-C skeleton (Fig. 2a, GM12878: 0.30 vs 0.36; K562: 0.46 vs 0.42). Therefore, even though deepC is predicting interactions from sequence in these instances it performs as well as comparing two different experimental sources of 3C data.
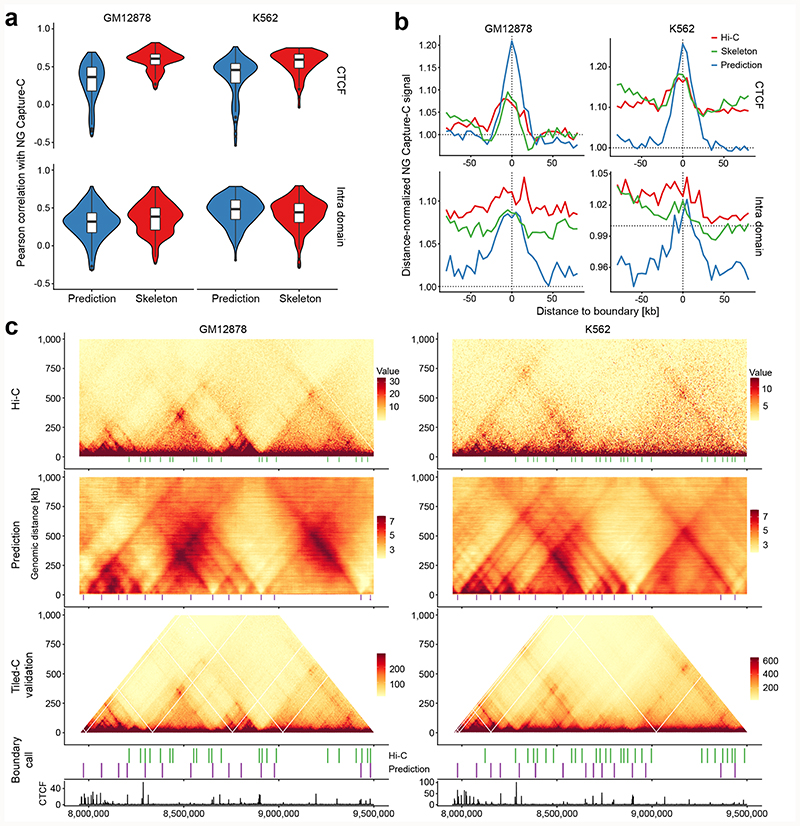
Figure 2. Validation of deepC predictions.
a) Comparing the correlation between the validation NG Capture-C profiles and the virtual 4C profiles derived from the Hi-C skeleton (red); and the deepC prediction map (blue) from all viewpoints in two cell types. Compared are n = 81 CTCF and n = 139 intra domain viewpoints. Boxplots: median middle thick line, 25th and 75th percentile left and right hinge respectively, whiskers stretch up 1.5 times the IQR (inter quartile range). b) Meta-profiles of the average NG Capture-C signal over domain boundaries called at high resolution from the Hi-C data, the skeleton and the deepC predicted interaction map respectively. Shown is the mean distance-normalized NG Capture-C signal relative to the boundary centre. The labels “CTCF” and “Intra domain” refer to the NG Capture-C viewpoint fragments. These were designed to overlap either CTCF sites or to lie within insulated domains but not overlap with regulatory genomic elements, so as to capture the domain structure and not the interactions of specific genomic elements. c) Shown are Hi-C data, the deepC predicted interaction map and the Tiled-C high sensitivity map over a locus on chr17, a hold-out chromosome. Boundaries called at high resolution from Hi-C (green) and deepC predictions (purple) are aligned under the respective map and the Tiled-C map. Cell-type-specific CTCF ChIP-seq tracks are visualized below. For contrast, Hi-C and Tiled-C data were bounded between 5 and 95 % of coverage and deepC predictions were bounded between 2 and 8 predicted regression score.
Taken together, these analyses suggested that deepC is capable of modelling the DNA encoded signals that determine the activity and position of boundary elements. To test this, we called boundaries within 1 Mb from the NG Capture-C viewpoints using the Hi-C data, the skeleton and the deepC predictions respectively (Supplementary Figure 12 and 13) using the established insulation-score-based approach26 with parameters for high-resolution calling (Methods). We then compared the called boundaries from these three sources with the high-sensitivity NG Capture-C 3C data to quantify the enrichment of chromatin interactions (Fig. 2b, Supplementary Figure 14). We observed clear enrichment over the deepC predicted boundaries indicating that they on average represent biophysical barriers to genome interactions. In contrast, the boundaries called directly from the Hi-C data and from the Hi-C skeleton showed less pronounced enrichment suggesting that calling directly from the data, on average, captures boundaries less effectively at the available sequencing depths. We confirmed these results with boundaries called using TopDom27 (Supplementary Figure 15), a TAD caller that showed best overall robustness in a recent bench marking study28.
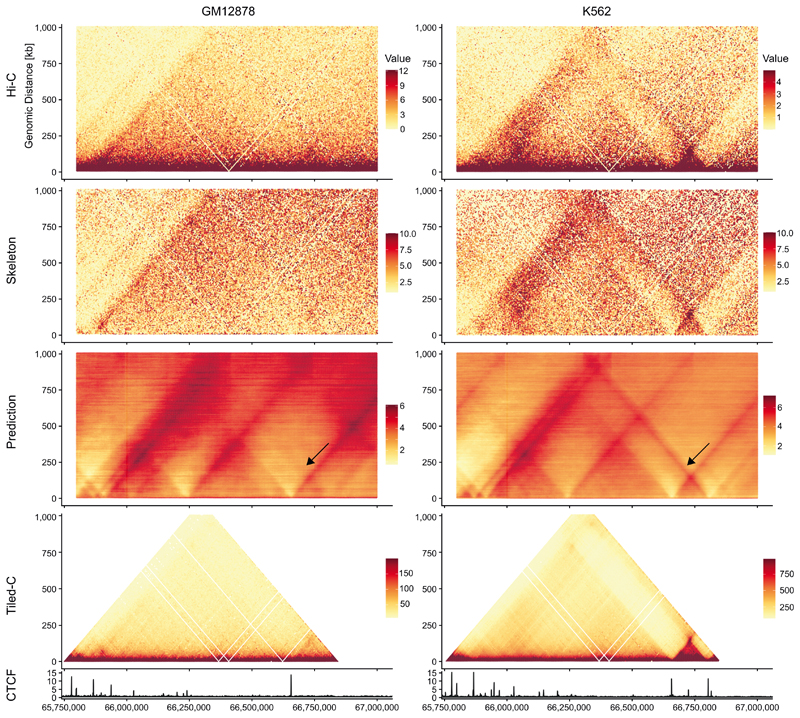
To visualize the coherence of the deepC predictions and called boundaries across specific loci at a sensitivity higher than the available Hi-C data, we utilized Tiled-C29 (Methods), which generates Hi-C like data for specific loci at high sensitivity and resolution. We performed Tiled-C for a selection of loci where deepC predicted fine-grained boundaries (Fig. 2c, Supplementary Figure 16) or cell-type-specific patterns (Extended Data Figure 3, Supplementary Figure 9). In line with the enrichment analysis, we confirmed that when called at high resolution, deepC boundaries are evident and align well with the boundaries in this highly sensitive 3C data. The added benefit for boundary calling is particularly striking in the comparatively lower-coverage K562 Hi-C data (~1.3 B reads) (Fig, 2c, Supplementary Figure 16).
When comparing the overall structure of deepC predictions to the Hi-C and Tiled-C data we observed that deepC tends to predict inter domain interactions in the form of stripes and dots more pronounced, some of which are only faintly detectable in Hi-C and Tiled-C data and some appear novel (Fig. 1c, 2c, Supplementary Figure 9). This suggests that deepC tends to underestimate the insulation between domains. Future refinements to the model architecture might be able to better capture the inter domain insulation. The effect appears amplified when comparing skeleton transformed to raw data as necessary for Tiled-C.
Dissecting the sequence determinants of genome folding
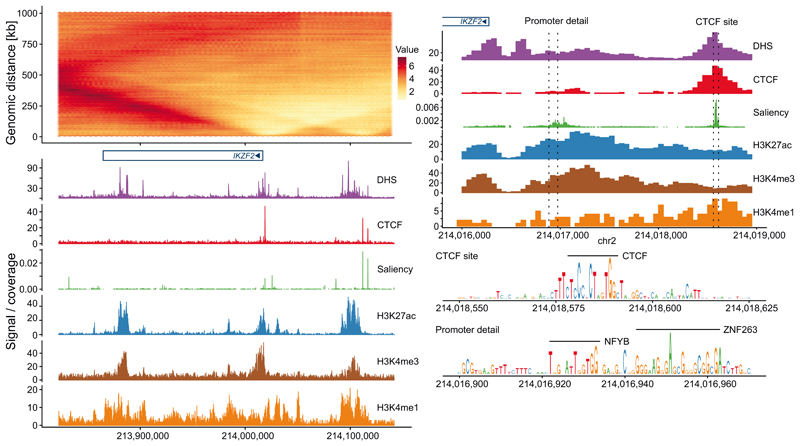
DeepC allows us to dissect the sequence determinants of genome folding at base pair resolution. To estimate the relative importance of every base pair for predicting chromatin interactions we employed the saliency score as a computationally efficient method adapted from image analysis30. The saliency score estimates how much the interaction prediction depends on each single base pair by calculating the gradient of the model output with respect to the sequence input (Methods). The saliency score predicts important regions and highlights transcription factor motifs within them (Extended Data Figure 5).
Genome-wide we identify sharp saliency peaks at CTCF sites and broader saliency peaks at active promoters (Fig. 3a). As bases with high saliency scores mark positions predicted to be important for chromatin architecture, we hypothesized that mutations within these regions would be enriched for those affecting gene expression. To test this, we retrieved 6607 GM12878 cell-type-specific eQTLs (GTEx v7) that are located in open chromatin (DNase-seq) or CTCF sites (CTCF ChIP-seq, ENCODE) and are thus likely to lie in regulatory elements. We found that these eQTLs have significantly higher saliency scores than SNPs randomly re-sampled from the same regions (p-value < 1e-85 using a two-sample Kolmogorov-Smirnow test) (Supplementary Figure 17). This suggests that the deepC saliency score can be used to fine map eQTLs when expression changes are mediated through an impact on chromatin architecture.
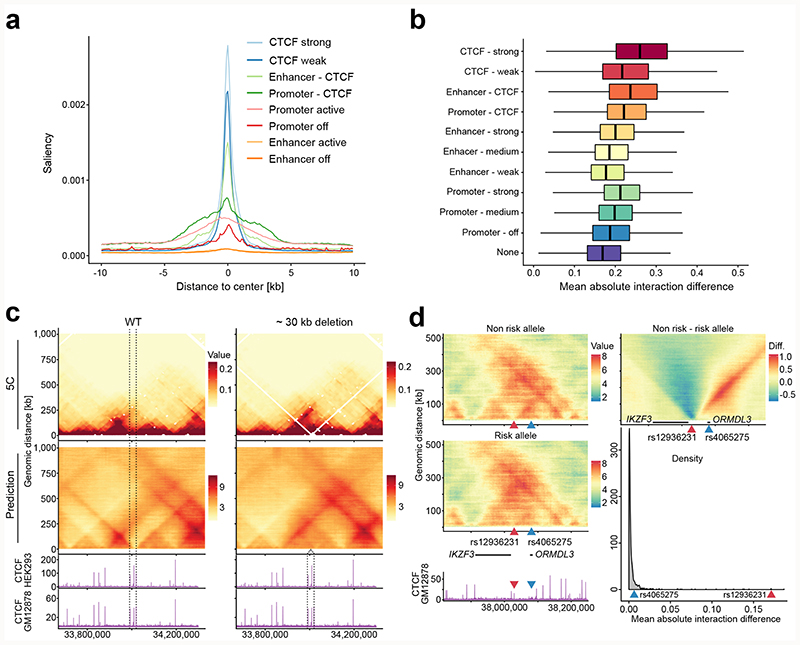
Figure 3. DeepC for dissecting the determinants of genome folding and predicting the impact of variation.
a) Mean saliency enrichment in 10 bp bins over different classes of regulatory elements genome-wide in GM12878 cells. b) Mean predicted chromatin interaction difference between reference and variant when deleting all open chromatin and CTCF sites and shuffled background sequences individually, genome-wide in GM12878 cells. Differences were quantified as the mean absolute interaction difference per bin to bin interaction. Each deletion was classified based on the overlap with genome segmentation classes. A total of n = 96805 was analysed. Boxplots show the median as thick line, the 25th and 75th percentile as left and right hinge respectively. The whiskers stretch up 1.5 times the IQR (inter quartile range) away from the respective whisker. Outliers outside this range were omitted from plotting. c) Effect of deleting a ~ 30 kb fragment containing four CTCF sites. Shown is the effect as validated by 5C data in wild-type (WT) and mutant in HEK293 cells by Hnisz et al. and the predicted effect using the GM12878 model. CTCF ChIP-seq tracks from HEK293 and GM12878 cells are visualized below. Dotted lines mark the deleted fragment. The mutant deepC prediction was shifted to be centred on the deleted site. d) Predicting the effect of two Asthma associated SNPs with the GM12878 model. Shown is the prediction for non risk and the risk allele, the differential map (non risk – risk) and a GM12878 CTCF ChIP-seq track. Location of the SNPs is indicated by triangles (red – rs12936231, blue – rs4065275). Black lines indicate the location of ORMDL3 and IKZF3 and only those two genes are highlighted for clarity. Comparing the predicted SNP effects against 1000 randomly sampled SNPs within CTCF sites places rs4065275 in the top 11 % and rs12936231 in the top 1% of predicted mean absolute interaction difference.
A long-standing question has been which functional elements within the genome underlie the patterns of genome folding. To investigate this, we performed an in silico deletion screen of all active elements genome-wide and used deepC to assess their importance for chromatin interactions (Fig. 3b). As expected, we find that deleting CTCF sites as well as enhancers and promoters with proximal CTCF binding has the strongest average predicted impact. We also find promoter and enhancers without proximal CTCF binding sites to be important, with deletions of promoters on average having a stronger effect. In addition, deletions of promoters and enhancers with strong activity-associated histone marks have a higher predicted impact than those without such marks.
Our analysis suggests that in addition to known factors such as CTCF binding and orientation, active regulatory elements, in particular promoters, are critical elements for effectively predicting genome interactions. Furthermore, deepC predicts boundaries not associated with CTCF sites. Taken together this indicates that deepC has learned aspects of a complex grammar of genome folding beyond CTCF motifs and their relative orientation and suggests a causal role for regulatory elements in defining and stabilizing 3D genome structure.
Predicting the impact of sequence variation on genome folding
To test deepC’s ability to predict the impact of sequence variation on genome folding we utilized two well-characterized examples from the literature. Hnisz et al. showed that a ~30 kb CRISPR-mediated deletion in HEK293T cells, encompassing four CTCF sites at the LMO2 locus, leads to a local rearrangement of the chromatin structure as confirmed by 5C31. Computationally reproducing this deletion in GM12878 (Fig. 3c) and K562 cells (Supplementary Figure 18) recapitulates the domain fusion observed by Hnisz and colleagues. Furthermore, using deepC we computationally deleted the CTCF sites individually, predicting that no single deletion alone is sufficient for causing the rearrangement (Supplementary Figure 19), suggesting multiple redundant boundaries at this region.
Crucially, our sequence-based model can predict the impact of single base pair variants. To demonstrate this, we tested two asthma-risk associated SNPs32 shown to impact ORMDL3 expression in immune cells. Schmiedel et al. demonstrated that the SNPs impact CTCF binding sites, disrupting enhancer-promoter interactions of ORMDL3 in CD4-positive T-cells. DeepC predictions recapitulate the loss of a boundary element, insulating ORMDL3 from downstream interactions (Fig. 3d). When testing the individual SNPs Supplementary Figure 20 a and b), deepC predicts the rs12936231 risk allele to have a strong effect on genome folding (mean absolute interaction difference 0.176). In contrast, although the rs4065275 risk-allele suggests a boundary creating effect, the predicted strength is weak (0.006). To put these predicted SNP effects into context, we compared them to the in silico deletion screen effects (Supplementary Figure 20c). The effect of rs12936231 lies above the 25th percentile of the 500 bp, weak CTCF site deletions. In addition, we sampled 1000 SNPs from CTCF sites (Fig. 3d) as well as from promoters, enhancers and background sequences (Supplementary Figure 20d). In comparison to sampled 1000 CTCF SNPs, rs4065275 lies within the top 11 % and rs12936231 within the top 1 %. Taken together, deepC prioritizes rs12936231 as the likely causal variant. Interestingly, deepC predicts this effect in GM12878 (immortalized B-cells) but not in K562 (myeloid leukemia cell line with erythroid characteristics) or IMR90 (human embryonic lung fibroblasts), pointing to a potential lymphoid specific effect (Supplementary Figure 21).
Discussion
Mammalian chromatin architecture folds at the megabase and sub-megabase scale constraining distal regulatory interactions within TADs and smaller insulated domains11,33,34. Ultimately, chromatin interactions are encoded in the DNA sequence through an intricate interplay of protein binding sites and other sequence determinants. Understanding the link between individual sequences and large-scale chromatin interactions at base pair resolution is a key challenge for understanding chromatin architecture and its role in gene regulation35,36. We developed deepC to traverse the gap between base pair sequences and megabase structures. DeepC is the first sequence-based deep learning model that predicts chromatin interactions from DNA sequence while integrating a context of megabase scale. This scale of analysis is necessary for accurate prediction of chromatin interactions, which in turn allowed for the determination of the elements driving these interactions and assessment of mutations disrupting them.
We found deepC models to yield substantially better predictions when we pre-seeded the model with hidden layers optimized to predict a compendium of chromatin features. This allows deepC to predict intricate chromatin interactions even when trained on low depth, low resolution Hi-C data. By validating the results with NG Capture-C and Tiled-C, each 3C methods capable of extreme depth and sensitivity, we showed that the deepC approach effectively increases the resolution of Hi-C data. We demonstrated that deepC can be used to fine-map the sequence determinants of chromatin architecture at base pair resolution and link these with effects on gene expression. Additionally, our genome wide deletion screen of potential regulatory elements shed light on the mechanics of chromatin interactions. It confirmed that CTCF binding site deletions are most likely to cause strong chromatin interaction changes. Importantly, deepC also indicates that both promoters and enhancers contribute to genome folding, in addition to CTCF. Generally, promoter deletions have a higher predicted effect on genome organization than enhancer deletions, and we find that deletions of enhancer and promoters associated with active chromatin marks have a higher predicted impact than those without such marks. Our observations are in line with findings from orthologous methods, that find CTCF binding, open chromatin, active histone marks and RNA-seq to be most predictive4–8,10. The finding that identifying active promoters and enhancers, in addition to CTCF binding sites, is required to accurately predict 3D genome structures suggests that these elements play an important role in establishing these structures, possibly via recruitment of its components and by actively stabilizing certain loops.
We believe deep learning-based genome folding predictions will facilitate chromatin architecture research. In a parallel study, Fudenberg et al.37 have developed an alternative model (Akita) to accurately predict interaction in megabase scale loci. DeepC and Akita have a similar convolutional module as network base but vary significantly in the remaining network structure as well as the data encoding and training scheme. We believe that future comparative study and consolidation between these advances will bring further insights into genome function.
Here we present deepC as a valuable tool for dissecting the functional elements that shape chromatin architecture and for predicting the impact of sequence changes from single base pair to structural variants. Furthermore, deepC represents a step towards predictive models of gene regulation that integrate the intricate and long-ranged chromatin landscape of mammalian genomes.
Online Methods
Chromatin feature data
As human chromatin feature compendium the ENCODE18 and Roadmap19 chromatin data utilized in DeepSEA12 were used. Narrow peak calls (hg19) for 918 experiments were downloaded. The data was supplemented with additional erythroid linage data. Five sets of ATAC-seq data from Corces et al.38, two DNase-seq experiments39 and ten ATAC-seq and one CTCF ChIP-seq experiment from Downes et al. and in house erythroid differentiation40 were used. All data used are listed in Supplementary Table 1. All additional data was aligned to hg19 using the NGseqBasic pipeline41. Peaks were called with macs242 (default parameters, -q 0.01). The peak signals were aggregated following the procedure described in Zhou et al.12. In brief, the genome was split into 200 bp bins. All peak calls were intersected with these bins. If a bin overlaps a peak call to at least 50 % (100 bp), the bin was labelled as belonging to that dataset class. All genomic bins that do not intersect with at least one peak call were discarded. Only autosomes were used for all analysis.
Mouse chromatin data were retrieved from ENCODE18. Histone modification peak calls were downloaded from the ENCODE data portal. For DNase-seq, ATAC-seq and transcription factor ChIP-seq data the aligned bam files were downloaded and peak called with macs242 as described above. Replicates were collapsed into unions. All mouse data used are listed in Supplementary Table 1.
Hi-C data
Publicly available, deeply sequenced Hi-C data from Rao et al.20 was used. The available 5 and 10 kb resolution intra chromosomal contacts maps of 7 cell lines (and 1kb data from GM12878) were downloaded and normalized using the provided KRnorm factors. Four replicates of mouse ES cell data21 were retrieved as raw fastqs from (GSE96107).
Hi-C encoding for deep learning
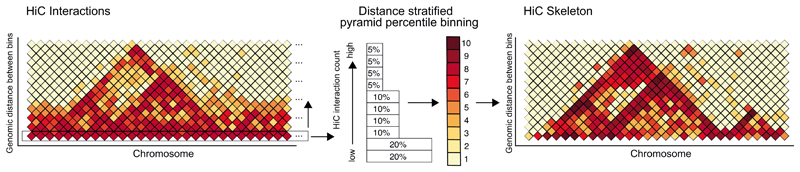
The genome was divided into bins matching the bin size of the respective Hi-C data resolution used for training (1 kb, 5 kb, 10kb). For every stretch of DNA of size 1 Mb + bin size bps (e.g. 1005000 for 5kb bins), the chromatin interactions associated with the window were assigned as squares in a vertical, zig-zag pole over the centre of the sequence window (see Fig. 1b). Every square encodes the Hi-C interactions observed between two bin sized windows of increasing distance (up to 1 Mb away). By sliding the large DNA stretch over a chromosome with a bin sized increment, this encoding recovers the chromosome wide Hi-C map up to an interaction distance of 1 Mb. Regions with a median interaction count along this pole of 0 where excluded from training. The Hi-C data was percentile normalized across individual chromosomes for every interaction distance in bin sizes. It is of particular interest to resolve high levels of Hi-C interactions at high resolution and only a low percentage of chromatin interactions is expected to yield strong interactions at larger distances for example the corners of TAD triangular structures. Thus, the percentile normalization was designed to better resolve these high interaction levels at larger distances by using uneven percentiles in a pyramid like scheme (from low to high: 2 x 20 %, 4 x 10 %, 4 x 5 %, see Extended Data Figure 1). The identifier of the respective pyramid percentile (1 – 10) was stored. The chromatin interaction network was then trained to predict the percentile identifier as a regression problem (see below).
Deep neural network architectures and training
A two-step training process with transfer learning (Extended Data Figure 2) was used. First a convolutional neural network was trained to predict chromatin features from 1 kb of DNA sequence, using the compendium of 936 datasets described above. The principle network architecture was adapted from DeepSEA12. Five convolutional layers (hidden units: 300, 600, 600, 900, 900; filter widths: 8, 8, 8, 4, 4, 4) with ReLU activation, max pooling (widths: 4, 5, 5, 5, 2) and dropout (rate: 0.2) were used followed by a fully connected layer with sigmoid activation to output individual probabilities for each chromatin feature class (multi-label classification). The network parameters were trained by minimizing the sum of the binary cross entropies using the ADAM optimizer (epsilon 0.1) in batches of 100. Batch size, dropout rate, learning rate and filter size were optimized by grid search.
Second, a chromatin interaction network was trained to predict Hi-C interaction from DNA sequence. The chromatin interaction network takes as input 1 Mb + 1x Hi-C bin size [bp], (e.g. 1005000 for a 5 kb bin network). The first module consists of five convolutional layers, with ReLU, max pooling and dropout with the dimensions and hyperparameters matching the chromatin feature network. The hidden weights were initialized by seeding with the weights of the trained chromatin feature network from step one. All chromatin features were used for pre-training and the same weights were used for seeding the chromatin interaction network training independent of the Hi-C data cell type.
The second module is a series of ten dilated, gated 1D convolutional layers with residuals16. Gated convolutional layers require training double the amount of filter parameters but have the potential of modelling more complex functions through their multiplicative units. The residual units allow information to propagate more easily through the network without having to necessarily pass through convolutions43. 100 hidden units were used, and dilation rates were increased exponentially to reach the full sequence context in the last layer (1, 2, 4, 8, 16, 32, 64, 128, 256, 1). The dilated layers were followed by a fully connected layer. Output are the predicted interaction strengths (in units matching the percentile normalization). The model was trained with ADAM (epsilon 0.1) to minimize the mean square error between the outputs and the true percentiles. GPU memory limited us to using a batch size of 1. Hidden units (for dilated layers), dropout rate, learning and ADAM epsilon were optimized using grid search.
Network training, computational resources and limitations
For both training procedures the data were split into training, validation and test set based on chromosomes. For the chromatin feature network chr11 and 12 were used for validation and chr15, 16 and 17 for testing. For the chromatin interaction network, to increase the number of training examples the same validation chromosomes were used but only chr16 and 17 were used as test chromosomes.
All models were trained on NVIDIA Titan V cards with 12 GB of video memory. Training on smaller cards is possible but slower. The final models have ~ 60 M parameters. Scaling the models to larger DNA inputs will likely benefit from network pruning or a refined architecture.
Fully training the chromatin feature network required 14 epochs with about 8 hours per epoch. The training set order was reshuffled after every epoch. To minimize the amount of times large chunks of DNA sequence had to be loaded into memory the network was trained on one chromosome at a time. Within a chromosome, the order in which training batches were drawn was random. Interestingly, we observed that the chromatin interaction network, when seeded with the pre-trained weights in the first convolutional filters, converged quickly, after training on ~ 3 - 6 chromosomes and only marginally improved after training for an entire epoch or longer. Models were trained for one full epoch as we have not observed significant improvement after training for longer and the limited batch size as well as the network complexity make training slow. For cross-validation, we trained multiple iterations holding out different chromosomes from training.
While training networks is only feasible with GPU support, predictions with trained models can be run on CPU only. For example, predicting the impact of a variant requires ~ 5 min with GPU and ~ 2h with only CPU support.
Predicting changes in chromatin interactions
For calculating differences in chromatin interactions, the interactions over the reference sequence were predicted for every position that is within 1 Mb (plus 1x Hi-C bin size) of the sequence variant. This matches the respective models spatial reach. The reference sequence was then modified to match the sequence variant of interest. After predicting the chromatin interactions over the variant sequence, the difference was quantified by calculating the absolute difference between reference and variant prediction at each interaction bin and summarized as the mean absolute difference over all covered interactions.
Distance-stratified correlation
The Pearson correlation coefficient was calculated between the Hi-C skeleton and the deepC predicted regression score. Note that the skeleton percentiles are discrete (percentile tag 1 – 10), while the regression score is continuous. The Hi-C skeleton is noisy even at very deep sequencing depths (e.g. GM12878). Therefore, a small mean filter was employed using a 5x5 window to smooth the skeleton and the distance-stratified correlation was calculated between the prediction and the raw or the smoothed skeleton respectively.
Comparison against HiC-Reg8
The available CrossChrom predictions, trained on chr14 and predicted on chr17, were downloaded from the supplementary material. HiC-Reg predictions were distance-normalized as described above.
Re-aligning and downsampling Hi-C data
The primary GM12878 replicate was realigned from raw fastqs using HiCPro44. The valid Hi-C interactions were downsampled to achieve 1 billion, 100 million and 10 million valid interactions, respectively. Mouse ES cell Hi-C data were aligned and processed from raw fastq data using HiCPro44.
Selection of validation capture probes
A total of 220 viewpoints were selected for validating the deepC predictions, specifically selecting genomic locations where the Hi-C data and deepC predictions differed in detail or where the deepC predicted structures were only very faintly noticeable in the Hi-C data. Two sets were designed, one targeting 81 CTCF sites and one targeting 139 intra domain viewpoints that lie within a distinct Hi-C/deepC domain but are not intersecting with any potential functional elements. Capture probes were designed using CapSequm (http://apps.molbiol.ox.ac.uk/CaptureC/cgi-bin/CapSequm.cgi), filtering out repetitive probe regions as described in the online documentation. For the final probe design see Supplementary Table 2.
Cell culture and fixation
Human GM12878 lymphocyte cell line, were obtained from the NIGMS Human Genetic Cell Repository at the Coriell Institute for Medical Research and cultured in RPMI 1640 supplemented with 15% FBS, 2mM L-Glutamine and 100U/ml Pen-Strep at 37 °C in a 5% CO2 incubator. K562 cells were supplied by the WIMM transgenics facility. Cells were maintained in RPMI 1640 media supplemented with 10 % FCS at 37 °C in a 5% CO2 incubator. Both cell types were fixed and processed using the same protocol. Cells were resuspended at 1x106 cells per ml and fixed at room temperature with 2% v/v formaldehyde for 10 minutes. Fixation was quenched with 120 mM glycine. Cells were washed with ice cold PBS. Cells resuspended in cold lysis buffer (10 mM Tris, 10 mM NaCl, 0.2% Igepal CA-630 and complete proteinase inhibitor (Roche)) and snap frozen to −80 °C. See Life Science Reporting Summary for additional details.
3C library preparation
3C libraries were prepared as described previously24 with the following modifications: Centrifugation’s were performed at 500 rcf, thermomixer incubations were set to 500 rpm, and following ligation chromatin was pelleted by centrifugation (15 min, 4°C, 500 rcf) and the supernatant discarded. To increase sequencing depth and minimize PCR duplicates experiments were performed in technical triplicates and four unique adapters were used per replicate.
NG Capture-C
Double capture was performed as described previously24 with biotinylated oligonucleotides (IDT xGen Lockdown Probes) in two pools (Supplementary Table 2) with 3 pg of each oligonucleotide per 3C library. The generated NG Capture-C libraries were sequenced using Illumina sequencing platforms (V2 chemistry; 150-bp paired-end reads) and data collected using the NextSeq System Suite (v2). To resolve even subtle changes in chromatin interaction domains at high resolution, the libraries were deeply sequenced (GM12878 CTCF - 128 M; GM12878 intra domain -118 M reads; K562 CTCF – 302 M; K562 intra domain – 289 M reads). All technical replicates were merged for the analysis.
Tiled-C
Tiled-C generates Hi-C like data focused on loci of interest at greater depth by using an oligonucleotide capture enriching a 3C library for viewpoints tiled over the regions of interest. Tiled oligonucleotides were designed using the design approach and tool described in Oudelaar and Beagrie et al. 29 (https://oligo.readthedocs.io/en/latest/). A panel of double-stranded capture oligonucleotides from Twist Bioscience (Custom probes for NGS target enrichment) was used. The Tiled-C procedure was performed as described in Oudelaar and Beagrie et al. using the 3C libraries from GM12878 and K562 cells. All biological and technical replicates were merged for the analysis. For a summary of the regions of interest and designed probes see Supplementary Table 3.
NG Capture-C analysis
NG Capture-C data were mapped, quality controlled and visualized using CcseqBasicS45 following the procedure described previously24.
Tiled-C analysis
Tiled-C data were mapped and quality controlled using the tiled mode of CcseqBasicS described above. Each region was ICE normalized using the ICE implementation of HiCPro with default parameters.
Distance-normalize NG Capture-C tracks
To compare them to the Hi-C skeleton, NG Capture-C tracks were normalized for distance dependence per viewpoint. The number of interactions with each restriction enzyme fragment was extracted and the distance to the viewpoint recorded. The interactions were then normalized for the total number of cis interactions for the respective viewpoint. Pooling this information across all viewpoints we observed that the distance decay approximately follows a log – log linear trend when we split the data into three distance bins (close, intermediate and far). The distance thresholds for these bins were empirically optimized for every NG Capture-C set (see Supplementary Table 4), excluding all interactions closer than 2.5 kb to the respective viewpoint. The distance decay is then approximated by three linear regression fits, one for each distance bin. The distance-normalized interactions were calculated per viewpoint by dividing the observed cis normalized interactions with the expected interactions from the linear fit at the respective distance.
Insulation score boundary calling
Interaction domain boundaries were called using the Hi-C data, the Hi-C skeleton and the deepC predicted interactions using an insulation score-based approach that was adapted from Crane et al.26. Using the 5 kb bin sized data, the mean insulation score profile was calculated based on a 25 kb window to allow for a more intricate boundary call. The first derivative, or delta vector, of the insulation score profile was approximated using a 1D Sobel operator. Zero crossings in this delta vector represent local minima and maxima of the insulation score. Maxima were discarded. The remaining boundaries were further filtered by calculating the approximation of the second derivative of the insulation profile using the same procedure described above. The height of this delta2 vector reflects the change in delta, with sharper boundaries having a higher delta2 score. Boundaries with a delta2 score smaller than 0.1 were removed and the remaining boundaries were stratified based on their delta2 score.
TopDom
TopDom was retrieved from the gitHub implementation (https://github.com/HenrikBengtsson/TopDom). Boundaries were called using the window parameters 5, 10 and 20. Boundaries between all types of called domains were used.
Distance-normalized NG Capture-C signal over boundaries
The mean, distance-normalized NG Capture-C signals over boundaries were calculated. In NG Capture-C tracks from single viewpoints, domain boundaries can be subtle and get harder to detect the further away from the viewpoint they are located. Therefore, boundaries further then 1 Mb away from a viewpoint were excluded. The mean normalized Capture-C signal over boundaries relative to their centre was calculated.
Virtual4C from Hi-C skeleton and deepC maps
By extracting all interacting windows with a viewpoint of interest, Hi-C data can be transformed into virtual 4C profiles. For this work, virtual 4C profiles were derived from the Hi-C skeleton and the deepC predictions yielding distance-normalized profiles. Virtual 4C profiles from the Hi-C skeleton and deepC predictions were compared to distance-normalized NG Capture-C tracks by calculating the respective Pearson correlation of all interactions within 1 Mb from a given viewpoint. Because the skeleton percentiles are discrete and punctuate, a running mean smoothing window of 25 kb was applied. In contrast, the deepC predictions are smooth and therefore no additional smoothing was applied.
Chromatin segmentation
GM12878 and K562 chromatin data were downloaded from the ENCODE data portal (see Supplementary Table 5). Filtered alignments to hg19 were downloaded and replicates were merged. Peaks were called using macs242 with default settings and -q 0.01. Deeptools46 was used to create bigwig coverage tracks. DNase-seq and CTCF ChIP-seq peaks were merged to a union set merging peaks within 10 bp of each other using bedtools47 (bedtools merge -d 10). Union peaks were formatted to 600 bp elements centred on the peaks. Deeptools was then used to extract the read coverage for each chromatin dataset over each peak union element. For this, elements were extended to 1000 bps to better capture flanking histone modifications. Using the derived count matrix, chromatin classes were segmented using GenoSTAN48 running on the elements rather than entire chromosome stretches. The HMM model was trained using the Poisson log-normal distributions. Twelve classes were fitted and merged into eleven classes based on similarity of the chromatin signatures. The classes were manually curated and classified into promoter, enhancer and CTCF sites with varying activity levels based on H3K27ac coverage.
Saliency score
Adapted from image analysis30 the saliency score serves as a proxy for the importance of every base pair to the interaction predictions. Explicitly, the saliency score was calculated as the dot product of the gradient of the model output with respect to the sequence input and the one-hot encoded DNA sequence input. This effectively masks the impact of non-present bases. For a given window the saliency score relates to the interaction pole on the center. To visualize saliency tracks, the sequence window was moved in bin sized steps and the saliency per base pair was averaged over all sequence windows (sized 1 Mb + bin size) that include the respective base pair. To simplify visualization and interpretation the absolute value of the saliency score was used. Metaplots were computed with deepTools.
eQTL data analysis
EBV transformed lymphocyte specific eQTLs were retrieved from GTEx (v7 accessed from the GTEx portal 01/03/2019). A union of DNase-seq and CTCF ChIP-seq peaks was created using bedtools merge. The eQTL SNPs were filtered for intersection with the union of GM12878 open chromatin and CTCF peaks. Indels were removed. A background SNP set was constructed by shuffling the eQTL SNPs on the respective same chromosome and forcing them to stem from within the union peaks (bedtools shuffle -chrom -incl). Absolute saliency scores of the SNP bases derived from the 5 kb resolution GM12878 model were extracted. Empirical cumulative distributions were derived and tested for significance using a two sample Kolmogorov-Smirnov test (R, ks.test, reshuffled SNP saliency (n = 6607) vs. eQTL set (n = 6607) saliency, alternative hypothesis: “less”).
Deletion screen
Separately, GM12878 DNase-seq and CTCF ChIP-seq peaks were merged if multiple peaks were found within 1.5 kb of each other (bedtools merge -d 1500). Peaks were extended to at least 300 bp. All DNase peaks that overlapped with CTCF peaks were removed. For every remaining CTCF (n=45635) and DNase (n=47320) site the impact on chromatin interactions upon deleting the respective site was predicted the 5 kb GM12878 model. Chromatin classes were assigned based on overlap with the GenoSTAN chromatin segmentation described above. In addition, n = 3850 background sites were selected by shuffling all CTCF and open chromatin sites on chr16 forcing no overlap (bedtools shuffle -chrom -noOverlapping).
5C data
Processed 5C data from Hnisz et al.31 were downloaded, binned into 5kb bins and visualized.
SNP sampling
1000 random SNPs each were sampled from strong CTCF sites, strong promoters and strong enhancers as classified by the chromatin segmentation procedure described above. For a background set, 1000 SNPs were sampled from the 400 bp regions flanking these regulatory elements while avoiding any overlap with other elements. SNPs positions were sampled using bedtools shuffle and variant bases were randomly selected from the three bases not present in hg19 at the respective position.
Statistics and Replication
Statistical analysis war performed in R. Statistical tests are described in the relevant subsection of the Online Methods. NG Capture-C and Tiled-C experiments were performed once, technical replicates were pooled for maximum read depth (see Life Science Reporting Summary for additional details).
Additional software and packages
All neural networks were implemented in python (v3.5) and tensorflow49 (developed under 1.8.0).
Additional Tools
samtools50 (v1.3)
FastQC (v0.11.4) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/)
Bowtie51 (v1.1.2)
Additional R packages
cowplot (v0.6.2, https://github.com/wilkelab/cowplot)
GenomicRanges52 – (v1.30.3)
ggplot253 (v3.1.0)
RcolorBrewer (v1.1.1-2, https://cran.r-project.org/web/packages/RColorBrewer/index.html)
rtracklayer54 (v1.30.4)
tidyverse (v1.3.0) (https://www.tidyverse.org)
zoo55 (v1.8.1)
Additional Python libraries
numpy56 (1.16.4)
h5py (v2.9.0, http://www.h5py.org)
pysam (0.15.2, https://github.com/pysam-developers/pysam)
Extended Data
Extended Data Fig. 1. Percentile normalizing Hi-C data for deep learning.
The Hi-C interactions are percentile-binned in a distance-stratified manner. For every genomic distance, in steps equal to the bin size, the Hi-C signal is split into unequal percentiles ranging from 20 % bottom to 5 % top. The percentiles are attributed the values 1 to 10 yielding the Hi-C skeleton. The unequal percentile sizes ensure a finer distinction of the differences at the high Hi-C interaction value range, while minor differences in the low interaction value range are squished. Effectively, this procedure reduces the proximity signal and enhances domains and domain boundaries.
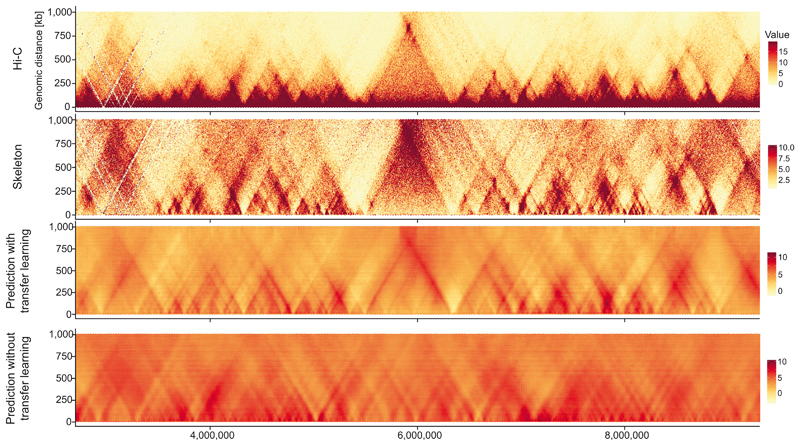
Extended Data Fig. 2. Comparison of deepC training with and without transfer learning.
Training a deepC model with the same architecture but without pre-seeding the lower convolutional layers with the chromatin feature model weights results in the emergence of triangular structures. Their positioning however does not match with the Hi-C structures. In contrast, with pre-seeding the predicted domains overlap well with the Hi-C skeleton.
Extended Data Fig. 3. Tissue-specific deepC predictions.
Shown is a region on chromosome 2 around the MEIS1 locus. DeepC predicts a small domain with insulation to the upstream regions (black arrow) in a tissue specific manner. The domain is only visible in K562 Hi-C data and matches with tissue-specific CTCF binding. Tiled-C confirms the tissue-specific domain. For contrast, Tiled-C data were bounded between the 5 and 95 percentiles.
Extended Data Fig. 4. NG Capture-C validation of deepC predictions.
a) Example region with overlap of GM12878: Hi-C, skeleton and deepC prediction; NG Capture-C tracks, distance-normalized NG Capture-C tracks and CTCF ChIP-seq track (red). Shown is a CTCF viewpoint (purple triangle) and an intra domain viewpoint (blue triangle) not overlapping with any active elements. Dashed lines in the distance-normalized NG Capture-C tracks indicate the expected interaction value. Dotted black lines highlight deepC prediction details that correspond to boundaries in the NG Capture-C tracks. b) K562 data of the same region.
Extended Data Fig. 5. Mapping important features for genome folding.
Shown are GM12878 deepC predictions over the IKZF2 locus (a) on chromosome 2 and focused on the IKZF2 promoter (b). Aligned are DHS as well as ChIP-seq tracks for CTCF and histone modifications. Shown in green is the saliency score which is a proxy for the importance every base has in predicting the chromatin interactions of that region. The saliency score shows sharp peaks overlapping CTCF binding sites and broader peaks overlapping active gene promoters. Resolving the saliency score at base pair resolution (b) highlights CTCF and general transcription factor binding motifs.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Robert Beagrie for help in refining the manuscript. This work was supported by the MRC (MC_UU_12009/14 to J.R.H.) and the Wellcome Trust via Strategic Award (106130/Z/14/Z to J.R.H.) and Institutional Strategic Support Fund (reference 105605/Z/14/Z to J.R.H.). The Wellcome Trust Genomic Medicine and Statistics PhD Programme (203728/Z/16/Z to R.S. & 203141/Z/16/Z to R.B.). The Stevenson Junior Research Fellowship at University College, Oxford (to A.M.O). G.L. is supported by the Wellcome Trust supporting award (090532/Z/09/Z to G.L.). Y.W.T. is supported by the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013 to Y.W.T.) ERC grant agreement no. 617071.
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS.
Footnotes
Author contributions
R.S., G.L. and J.R.H. conceived the project. R.S, R.B., Y.W.T. and G.L designed the neural network architectures. R.S. optimized and trained the neural networks and performed downstream analysis. R.S. M.G. D.D. A.M.O. and J.R.H. designed and evaluated the validation strategy. M.G. performed NG Capture-C experiments. D.D. performed Tiled-C experiments. R.S., A.M.O. and J.T. performed bioinformatic analysis of NG Capture-C and Tiled-C. R.S. performed integrative analysis and prepared the figures. R.S., G.L and J.R.H. wrote the manuscript with inputs from all authors.
Competing financial interests
The authors declare no competing financial interests.
Data availability
Hi-C data from Rao et al. is available under GSE63525. Chromatin feature data from ENCODE, Roadmap and other publicly available data are listed in detail with accession numbers in Supplementary Table 1. Additional ENCODE data used for chromatin segmentation and visualization are listed with accession numbers in Supplementary Table 5. Tiled-C and NG Capture-C validation data are available under the GEO super series GSE137437.
Code availability
All code for training and employing deepC networks as well as trained models are available under: https://github.com/rschwess/deepC; All code for training and employing chromatin feature networks is available under: https://github.com/rschwess/deepHaem
References
- 1.Hindorff LA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc Natl Acad Sci U S A. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Maurano MT, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012;337:1190–1195. doi: 10.1126/science.1222794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schreiber J, Libbrecht M, Bilmes J, Noble WS. Nucleotide sequence and DNaseI sensitivity are predictive of 3D chromatin architecture. 2017 doi: 10.1101/103614v5. [DOI] [Google Scholar]
- 4.Whalen S, Truty RM, Pollard KS. Enhancer–promoter interactions are encoded by complex genomic signatures on looping chromatin. Nat Genet. 2016;48:488–496. doi: 10.1038/ng.3539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li W, Wong WH, Jiang R. DeepTACT: predicting 3D chromatin contacts via bootstrapping deep learning. Nucleic Acids Res. 2019;47:e60. doi: 10.1093/nar/gkz167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Qi Y, Zhang B. Predicting three-dimensional genome organization with chromatin states. PLOS Comput Biol. 2019;15:e1007024. doi: 10.1371/journal.pcbi.1007024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Belokopytova PS, Nuriddinov MA, Mozheiko EA, Fishman D, Fishman V. Quantitative prediction of enhancer–promoter interactions. Genome Res. 2020;30:72–84. doi: 10.1101/gr.249367.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhang S, Chasman D, Knaack S, Roy S. In silico prediction of high-resolution Hi-C interaction matrices. Nat Commun. 2019;10:5449. doi: 10.1038/s41467-019-13423-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buckle A, Brackley CA, Boyle S, Marenduzzo D, Gilbert N. Polymer Simulations of Heteromorphic Chromatin Predict the 3D Folding of Complex Genomic Loci. Mol Cell. 2018;72:786–797.:e11. doi: 10.1016/j.molcel.2018.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bianco S, et al. Polymer physics predicts the effects of structural variants on chromatin architecture. Nat Genet. 2018;50:662–667. doi: 10.1038/s41588-018-0098-8. [DOI] [PubMed] [Google Scholar]
- 11.Hnisz D, Day DS, Young RA. Insulated Neighborhoods: Structural and Functional Units of Mammalian Gene Control. Cell. 2016;167:1188–1200. doi: 10.1016/j.cell.2016.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhou J, Troyanskaya OG. Predicting effects of noncoding variants with deep learning–based sequence model. Nat Methods. 2015;12:931–934. doi: 10.1038/nmeth.3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kelley DR, Snoek J, Rinn JL. Basset: learning the regulatory code of the accessible genome with deep convolutional neural networks. Genome Res. 2016;26:990–999. doi: 10.1101/gr.200535.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelley DR, et al. Sequential regulatory activity prediction across chromosomes with convolutional neural networks. Genome Res. 2018;28:739–750. doi: 10.1101/gr.227819.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu F, Koltun V. Multi-Scale Context Aggregation by Dilated Convolutions. 2015 Preprint at: http://arxiv.org/abs/1511.07122. [Google Scholar]
- 16.Oord, van den A, et al. WaveNet: A Generative Model for Raw Audio. 2009 IEEE Int Conf Acoust Speech Signal Process; 2016. pp. 3437–3440. [Google Scholar]
- 17.Yosinski J, Clune J, Bengio Y, Lipson H. How transferable are features in deep neural networks? Adv Neural Inf Process Syst. 2014;4:3320–3328. [Google Scholar]
- 18.Bernstein BE, et al. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74. doi: 10.1038/nature11247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kundaje A, et al. Integrative analysis of 111 reference human epigenomes. Nature. 2015;518:317–330. doi: 10.1038/nature14248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rao SSP, et al. A 3D Map of the Human Genome at Kilobase Resolution Reveals Principles of Chromatin Looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonev B, et al. Multiscale 3D Genome Rewiring during Mouse Neural Development. Cell. 2017;171:557–572.e24. doi: 10.1016/j.cell.2017.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang Y, et al. Enhancing Hi-C data resolution with deep convolutional neural network HiCPlus. Nat Commun. 2018;9 doi: 10.1038/s41467-018-03113-2. 750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu Q, Lv H, Jiang R. hicGAN infers super resolution Hi-C data with generative adversarial networks. Bioinformatics. 2019;35:i99–i107. doi: 10.1093/bioinformatics/btz317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Davies JOJ, et al. Multiplexed analysis of chromosome conformation at vastly improved sensitivity. Nat Methods. 2016;13:74–80. doi: 10.1038/nmeth.3664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–2049. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Crane E, et al. Condensin-driven remodelling of X chromosome topology during dosage compensation. Nature. 2015;523:240–244. doi: 10.1038/nature14450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shin H, et al. TopDom: An efficient and deterministic method for identifying topological domains in genomes. Nucleic Acids Res. 2015;44:e70. doi: 10.1093/nar/gkv1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zufferey M, Tavernari D, Oricchio E, Ciriello G. Comparison of computational methods for the identification of topologically associating domains. Genome Biol. 2018;19:217. doi: 10.1186/s13059-018-1596-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Oudelaar AM, et al. Dissection of the 4D chromatin structure of the α-globin locus through in vivo erythroid differentiation with extreme spatial and temporal resolution. 2019 doi: 10.1101/763763v2. [DOI] [Google Scholar]
- 30.Simonyan K, Vedaldi A, Zisserman A. Deep Inside Convolutional Networks: Visualising Image Classification Models and Saliency Maps. 2nd Int Conf Learn Represent ICLR 2014 - Work Track Proc; 2013. [Google Scholar]
- 31.Hnisz D, et al. Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science. 2016;351:1454–1458. doi: 10.1126/science.aad9024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmiedel BJ, et al. 17q21 asthma-risk variants switch CTCF binding and regulate IL-2 production by T cells. Nat Commun. 2016;7:13426. doi: 10.1038/ncomms13426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robson MI, Ringel AR, Mundlos S. Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol Cell. 2019;74:1110–1122. doi: 10.1016/j.molcel.2019.05.032. [DOI] [PubMed] [Google Scholar]
- 34.Dixon JR, Gorkin DU, Ren B. Chromatin Domains: The Unit of Chromosome Organization. Mol Cell. 2016;62:668–680. doi: 10.1016/j.molcel.2016.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Marti-Renom MA, et al. Challenges and guidelines toward 4D nucleome data and model standards. Nat Genet. 2018;50:1352–1358. doi: 10.1038/s41588-018-0236-3. [DOI] [PubMed] [Google Scholar]
- 36.Spielmann M, Lupiáñez DG, Mundlos S. Structural variation in the 3D genome. Nat Rev Genet. 2018;19:453–467. doi: 10.1038/s41576-018-0007-0. [DOI] [PubMed] [Google Scholar]
- 37.Fudenberg G, Kelley DR, Pollard KS. Predicting 3D genome folding from DNA sequence. 2019 doi: 10.1101/800060v1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Corces MR, et al. Lineage-specific and single-cell chromatin accessibility charts human hematopoiesis and leukemia evolution. Nat Genet. 2016;48:1193–1203. doi: 10.1038/ng.3646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwessinger R, et al. Sasquatch: predicting the impact of regulatory SNPs on transcription factor binding from cell- and tissue-specific DNase footprints. Genome Res. 2017;27:1730–1742. doi: 10.1101/gr.220202.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Downes DJ, et al. An integrated platform to systematically identify causal variants and genes for polygenic human traits. 2019 doi: 10.1101/813618v1. [DOI] [Google Scholar]
- 41.Telenius J, Consortium TW, Hughes JR. NGseqBasic - a single-command UNIX tool for ATAC-seq, DNaseI-seq, Cut-and-Run, and ChIP-seq data mapping, high-resolution visualisation, and quality control. 2018 doi: 10.1101/393413v1. [DOI] [Google Scholar]
- 42.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He K, Zhang X, Ren S, Sun J. Deep Residual Learning for Image Recognition. Multimed Tools Appl. 2015;77:10437–10453. [Google Scholar]
- 44.Servant N, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16:259. doi: 10.1186/s13059-015-0831-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Telenius JM, et al. CaptureCompendium: a comprehensive toolkit for 3C analysis. 2020 Preprint at: http://biorxiv.org/content/early/2020/02/18/2020.02.17.952572. [Google Scholar]
- 46.Ramírez F, Dündar F, Diehl S, Grüning BA, Manke T. deepTools: a flexible platform for exploring deep-sequencing data. Nucleic Acids Res. 2014;42:W187–W191. doi: 10.1093/nar/gku365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zacher B, et al. Accurate Promoter and Enhancer Identification in 127 ENCODE and Roadmap Epigenomics Cell Types and Tissues by GenoSTAN. PLoS One. 2017;12:e0169249. doi: 10.1371/journal.pone.0169249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Abadi M, et al. TensorFlow: A System for Large-Scale Machine Learning TensorFlow: A system for large-scale machine learning. 12th USENIX Symp Oper Syst Des Implement (OSDI ’16); 2016. pp. 265–284. [Google Scholar]
- 50.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lawrence M, et al. Software for Computing and Annotating Genomic Ranges. PLoS Comput Biol. 2013;9:e1003118. doi: 10.1371/journal.pcbi.1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 54.Lawrence M, Gentleman R, Carey V. rtracklayer: An R package for interfacing with genome browsers. Bioinformatics. 2009;25:1841–1842. doi: 10.1093/bioinformatics/btp328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zeileis A, Grothendieck G. Zoo: S3 infrastructure for regular and irregular time series. J Stat Softw. 2005;14:1–27. [Google Scholar]
- 56.van der Walt S, Colbert SC, Varoquaux G. The NumPy Array: A Structure for Efficient Numerical Computation. Comput Sci Eng. 2011;13:22–30. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Hi-C data from Rao et al. is available under GSE63525. Chromatin feature data from ENCODE, Roadmap and other publicly available data are listed in detail with accession numbers in Supplementary Table 1. Additional ENCODE data used for chromatin segmentation and visualization are listed with accession numbers in Supplementary Table 5. Tiled-C and NG Capture-C validation data are available under the GEO super series GSE137437.
All code for training and employing deepC networks as well as trained models are available under: https://github.com/rschwess/deepC; All code for training and employing chromatin feature networks is available under: https://github.com/rschwess/deepHaem