Abstract
The ability to incorporate a desired functionality into proteins of interest in a site-specific manner can provide powerful tools for investigating biological systems and creating therapeutic conjugates. However, there are not any universal methods that can be applied to all proteins, and it is thus important to explore the chemical strategy for protein modification. In this paper, we developed a new reactive peptide tag/probe pair system for site-specific covalent protein labeling. This method relies on the recognition-driven reaction of a peptide tag and a molecular probe, which comprises the lysine-containing short histidine tag (KH6 or H6K) and a binuclear nickel (II)-nitrilotriacetic acid (Ni2+-NTA) complex probe containing a lysine-reactive N-acyl-N-alkyl sulfonamide (NASA) group. The selective interaction of the His-tag and Ni2+-NTA propeles a rapid nucleophilic reaction between a lysine residue of the tag and the electrophilic NASA group of the probe by the proximity effect, resulting in the tag-site-specific functionalization of proteins. We characterized the reactive profile and site-specificity of this method using model peptides and proteins in vitro, and demonstrated the general utility for production of a nanobody-chemical probe conjugate without compromising its binding ability.
1. Introduction
Covalent modification of proteins with designer chemical probes is a powerful approach to provide many exciting tools for life science research, biotechnology and drug development. This technique allows for visualization of biomolecules, controlling protein functions, and generating antibody-drug conjugates for treatment of various diseases.1–3 For such biological/therapeutic applications, methodologies that enable site-specific modification of proteins are required.4 It is widely recognized that conventional N-hydroxysuccinimide (NHS)-based probes generate heterogenous bioconjugates due to their random modification of all surface-exposed lysine residues in proteins. Cysteine-maleimide conjugation coupled with site-directed mutagenesis is a standard approach for site-specific protein modification. However, this may be ill-suited for proteins having several cysteines essential for their activity. Nowadays, the most commonly used techniques to define the position and number of modifications represent self-labeling protein tags, such as SNAP tag and Halo tag. While these are undoubtedly powerful, the large protein tag domain may perturb the intrinsic behaviors and/or functions of proteins of interest, especially for small or multimeric proteins.4
To circumvent these issues, short peptide tag/probe pair systems offer promising alternatives for site-specific protein modification.5–7 In this approach, a peptide tag-fused protein can be labeled with a designed probe through the specific molecular recognition. Since the sizes of the tag and probe are small, the modification is less likely to impair protein function. The tetra-cysteine tag and FlAsH probe pair is one of the successful examples of the short peptide tag approach, while it has a limitation to use for biomedical applications due to the toxicity of arsenicbased probe.8 Our group has previously developed a reactive tag system that relies on binding-induced nucleophilic reactions between Cys-appended oligo His-tags and nickel (II)-nitrilotriacetic acid (Ni2+-NTA) probes containing α-chloroacetamide, and utilized for site-specific labeling of proteins in vitro and in living cells.6,9 On the other hand, the strategy dependent on the cysteine thiol requires a reduction process prior to the modification to keep it reactive form and is not suitable for disulfide-containing proteins.9–12 Also, Ni2+-NTA moiety remains covalently on the tag, which may have an unfavorable effect on the quenching of the fluorophore, the surface charge of the protein and concern about the intrinsic toxicity of Ni2+. To tackle this challenge, several methods using non-cysteine amino acids as reactive sites of the tag and/or probes containing cleavable linker have recently been developed by us and other groups.13–16 However, all of these suffer from sluggish reaction rates and low modification efficiency.
Here, we report a rapid and site-specific protein labeling with a novel short-peptide tag/probe pair system. This method employs a lysine containing His-tag (that is, KH6 or H6K) with Ni2+-NTA-tethered N-acyl-N-alkyl sulfonamide (NASA) reagent, recently reported by our group as a cleavable electrophile for modifying the amino group of lysine side chain.17,18 Upon complexation of Ni2+-NTA with the H6 sequence, the lysine residue in the tag reacts with the NASA group to introduce a reporter moiety via the proximity effect, and concurrently, the Ni2+-NTA part is cleaved off (Figure 1a). In this work, we characterized reaction properties of the method using model peptides and His-tagged enhanced green fluorescent protein (EGFP) in vitro. Furthermore, with this method, we prepared a chemically functionalized nanobody and applied it for imaging of membrane surface proteins in live cells.
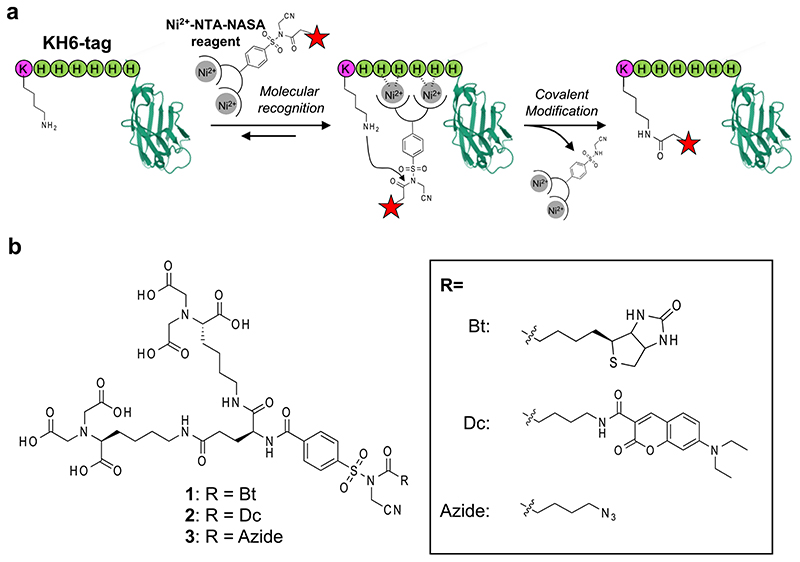
Figure 1. Site-specific covalent modification of KH6-tagged protein with Ni2+-NTA-NASA reagent.
(a) Schematic illustration of the reaction. (b) Molecular structures of NTA-NASA reagents (1–3) used in this study.
2. Results and discussion
2.1. Design of NTA-tethered NASA reagents and optimization of tag sequence
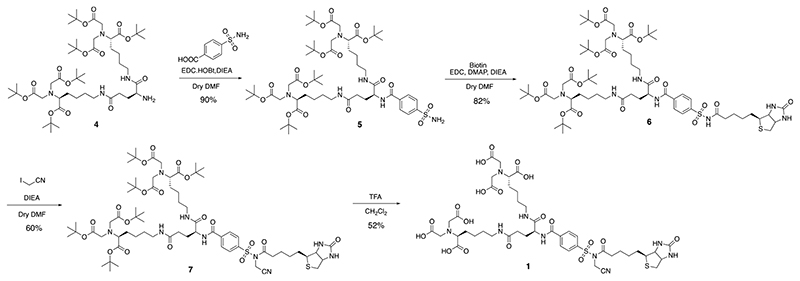
According to a guideline for ligand-directed covalent modification of proteins with NASA chemistry, an affinity ligand with sub μM order K d value is required to achieve rapid and efficient labeling.17 We thus employed bis-NTA (K d of the nickel complex for H6 tag = 270 nM) as an affinity moiety and connected it with a biotin tag through a NASA reactive linker (Figure.1b, compound 1).19 We also prepared several N-terminal acetylated oligo-histidine peptides containing an amino acid having nucleophilic side chain, AcXH6AW (X = K, C, Y, S, T, E), to examine amino-acid selectivity of the present method. Synthetic scheme of 1 is shown in Figure 2. The t-butyl protected bis-NTA part was connected with benzenesulfonamide to yield a core intermediate 5 of the labeling reagent. 5 was then coupled with biotin (or any desired probes) followed by activation via alkylation of the acyl sulfonamide group with iodoacetonitrile. Finally, deprotection of the t-butyl group afforded NTA-NASA reagent 1. Synthesis for other compounds and peptides are shown in the Supplementary Information.
Figure 2. Synthesis scheme of compound 1.
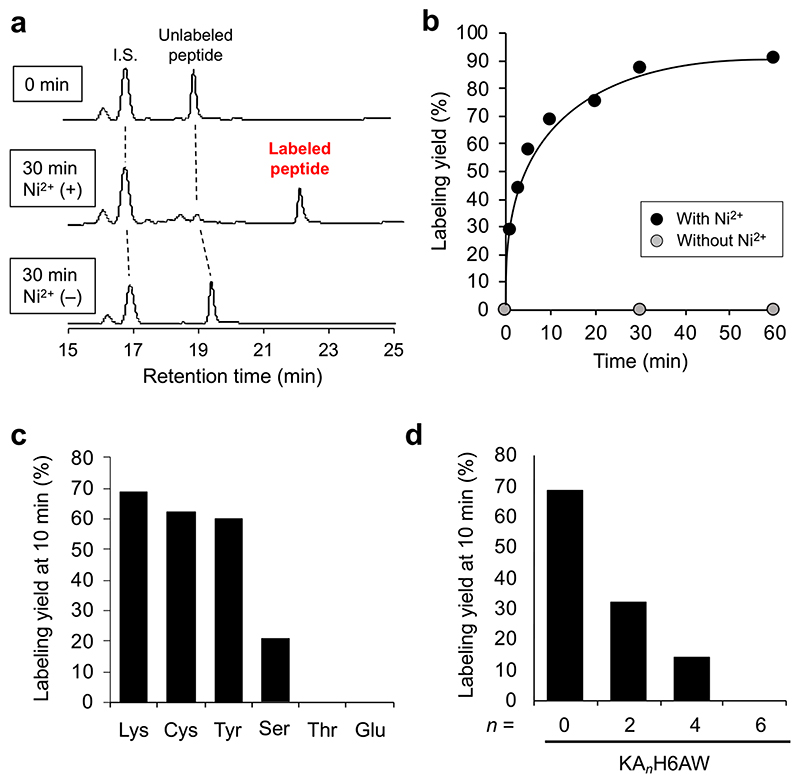
The peptide labeling was initially performed in vitro by incubating 1 (4 μM) in an aqueous solution of Ac-KH6AW peptide (1 μM in PBS, pH 7.4) at 37 °C in the presence and absence of Ni2+. Reverse-phase HPLC analysis coupled with matrix-assisted desorption/ionization time-of-flight mass/mass spectrometry (MALDI-TOF MSMS) showed that, rapid reaction occurred between Ni2+-complexed 1 and the ε-amino group of lysine residue in Ac-KH6AW (~90% yield at 30 min) to give the biotinylated peptide without any covalent adducts of the Ni2+-NTA moiety (Figure 3a, b and Figure S1). The covalent modification by 1 did not proceed in the absence of Ni2+, indicating that the reaction is facilitated by the proximity effect based on the H6-tag/Ni2+-NTA complexation (Figure 3a, b). Regarding the amino acid selectivity, it was found that cysteine, tyrosine and serine also react with the NASA group to form a thioester, phenol ester and an alkyl ester, respectively, while other amino acids, including threonine and glutamic acid, does not give the labeled product (Figure 3c, Figure S2). Although the labeling efficiency of cysteine and tyrosine are comparable to that of lysine (Figure 3c), the stability of the formed covalent bond in aqueous solution is much lower for cysteine (thioester) and tyrosine (phenol ester) than for lysine (amide bond).20–22 We thus used the tags containing lysine in the following experiments.
Figure 3. Covalent labeling of model peptides with 1.
(a) HPLC analysis of the labeling reaction between Ac-KH6AW peptide (1 μM) and 1 (4 μM) in the presence and absence of Ni2+ (8 μM) at 30 min. (b) Time profile of the reaction between AcKH6AW peptide (1 μM) and 1 (4 μM). (c) Amino acid selectivity of this method. Ac-XH6AW peptides (X = K (Lys), C (Cys), Y (Tyr), S (Ser), T (Thr), E (Glu)) were incubated with Ni2+-complexed 1. The yields at 10 min reaction time were plotted. (d) Summary of the labeling yields at 10 min with the Ac-KAnH6AW peptides (n = 0, 2, 4, 6) in the presence of Ni2+.
To evaluate the spacer length dependency of the proximity-driven covalent labeling, we prepared lysine-containing oligo-histidine peptides with several alanine spacers, Ac-Lys-Alan-His6-Ala-Trp (Ac-KAn-H6AW, n = 0, 2, 4, 6). Importantly, the reaction efficiency was significantly dropped in a spacer length dependent manner (Figure 3d, Figure S3), highlighting that the labeling selectively proceeds on a lysine residue spatially adjacent to His-tag, not on distant one. We also confirmed that changing the position of lysine to the C-terminal side of oligo histidine (that is, Ac-H6KAW) does not significantly alter the labeling kinetics (Figure S4). Based on these results, we decided that a H6 sequence flanking a single lysine (without any spacer), that is, KH6 or H6K, is the optimal tag design for site-specific labeling with Ni2+-NTA-NASA probes.
2.2. Site-specific labeling of His-tag fused proteins
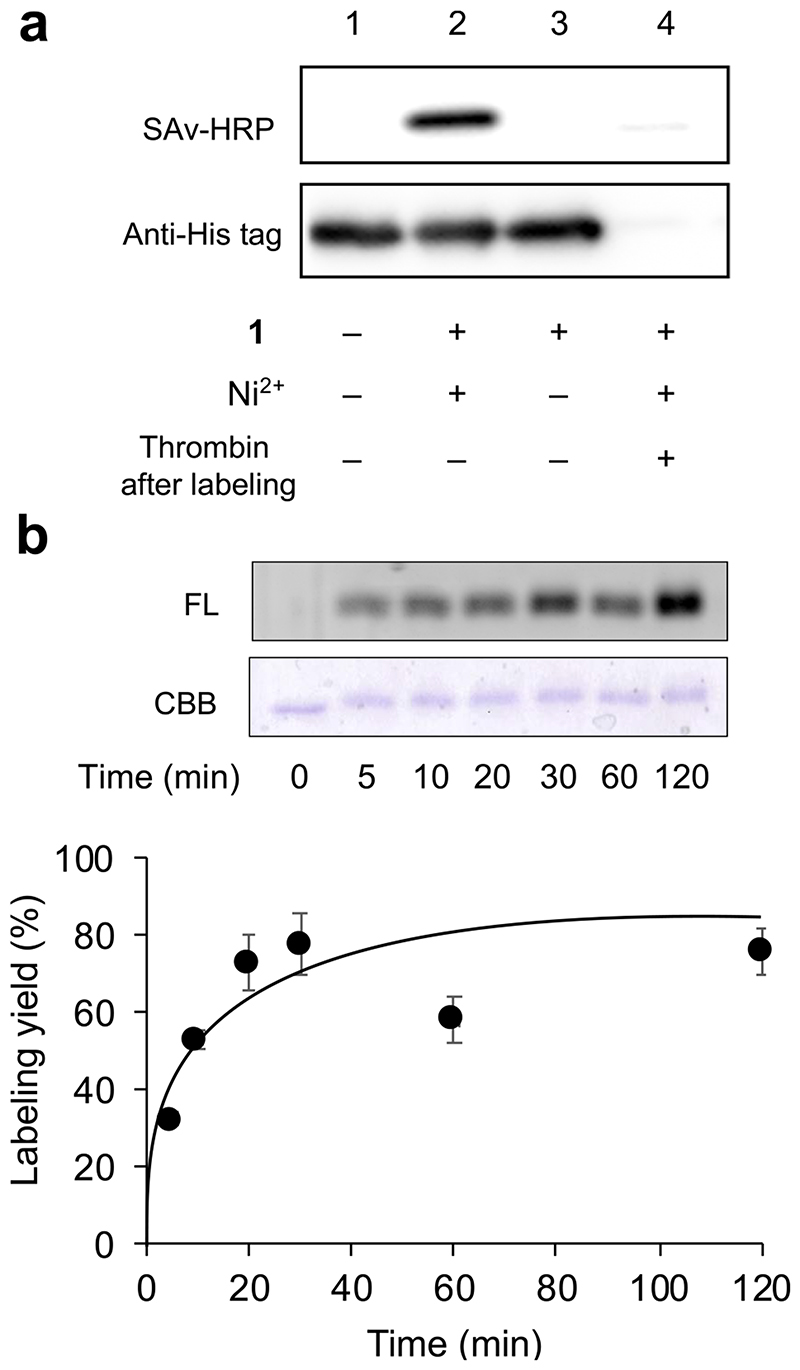
Having established the optimal tag/NTA-NASA reagent pair, we proceeded to investigate the labeling of tag-fused protein. The KH6 tag was fused with EGFP at its N-terminus (KH6-EGFP, the primary sequence is shown in Figure S5). A thrombin cleavage site was inserted between the tag and EGFP to confirm the labeling site specificity. The labeling reaction with 1 was monitored by western blotting using StreptAvidin-HRP. As shown in Figure 4a, Ni2+-dependent biotinylation of EGFP was clearly observed. This labeling signal was substantially abrogated when the biotinylated EGFP was treated with thrombin, indicating that the modification exclusively occurs at N-terminal peptide tag region, not protein scaffold, though the EGFP has 21 Lys residues (Figure S5b).
Figure 4. Labeling of KH6-tagged EGFP.
(a) Western blotting analysis of the tag-site specific labeling with 1. Reaction conditions: The KH6-EGFP (0.5 μM) was incubated in PBS containing 1 (0.5 μM) and NiCl2 (1 μM) at 37 °C for 1 h, followed by treatment with thrombin for 16 h at 22 °C. (b) SDS-PAGE analysis of the time course of the labeling reaction with 2. Reaction conditions: The KH6-EGFP (1 μM) was incubated in PBS containing 2 (3 μM) in the presence of NiCl2 (6 μM) at 37 °C for 5 min-2 h.
The modular design of our probe allows for easy exchange of reporter elements to be introduced into KH6-tagged protein. Indeed, from the core intermediate 5, we can readily synthesize compound 2 and 3 containing a fluorescent coumarin dye and a clickable azide handle, respectively (Figure.1b and SI). Both compounds are capable of labeling of KH6-EGFP (Figure S6), as well as 1, proving the applicability of our strategy to introduce a variety of probe into proteins. Time profiling of KH6-EGFP labeling was evaluated with 2. In gel fluorescence analysis revealed that the reaction reached a plateau at approximately 30 min, and the labeling yield was estimated to be 80% (Figure 4b, Figure S7). This protein labeling kinetics is similar to the case of peptide-based experiment.
Overall, these results clearly demonstrated that KH6-tag/NTA-NASA reagent pair system provides a generic tool for rapid, high-efficient, and site-specific protein modification with desired chemical probes.
2.3. Site-specific modification of nanobody and its usage for live-cell imaging of a membrane protein
Finally, we applied this method to modify a nanobody, single antigen-binding domain of camelid antibody. Chemically functionalized nanobodies are widely applied for their targeting abilities with high specificity, thermal stabilities and small sizes (~14 kDa) in the fields of biotechnology, medicine, and diagnostics.23 However, the production of modified nanobodies with well-controlling position and number of synthetic probes still remains a challenge because there are many of lysines and two preserved cysteins in the nanobody scaffold. Given that most of nanobodies are expressed in E.coli with His-tag for purification, we envisioned that our labeling method should provide a powerful means for site-specific modification of nanobody by the simple introduction of only one lysine into adjacent sites of the tag.
As a proof-of-principle, we used a nanobody against GluD2 (Glutamate receptor ionotropic, delta-2) that plays a key role in synaptogenesis and synaptic plasticity.24 The GluD2 nanobody was expressed in E.coli with a C-terminal H6K tag and purified followed by conjugation with 1. As shown in Figure S8b, a single-labeled product (~40% yield) was observed with 1 equivalent of 1 to protein by MALDI-TOF MS analysis. Under 2 or 3 equivalent of 1, yields were improved (~80%) with the single-labeled nanobody remaining major, though a double-labeled product was slightly observed (Figure S8b). As a negative control, a nanobody tagged with a H6A sequence gave no labeled products (Figure S8c), highlighting that the labeling specifically proceeds on the lysine residue in the H6K tag.
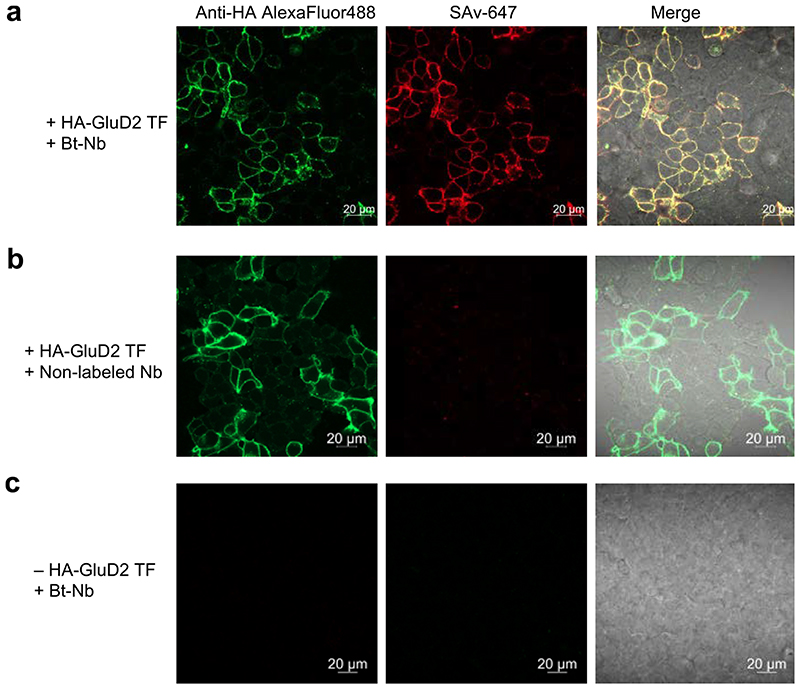
We then investigated the binding ability of the labeled GluD2 nanobody to GluD2 expressing cells. The biotinylated GluD2 nanobody with 1 was incubated with live HEK293T cells transiently transfected with HA-tagged GluD2 (HA-GluD2) followed by treatment with StreptAvidin-HiLyte647 (SAv-647) and analysis with confocal microscopy. As shown in Figure 5a, clear fluorescence of SAv-647 was observed predominantly at plasma membrane in the transfected cells. This signal was well merged with immunofluorescence signal for HA-tagged protein with anti-HA antibody conjugated with AlexaFluor488. No significant SAv-647 signals were detected in cases of the nanobody without modification and cells without expressing GluD2 (Figure 5b, c). These results proved that the nanobody modified by this method retain its binding ability. Furthermore, we also succeeded in direct conjugation of fluorescent dye to the nanobody using azide-type probe 3 and visualization of GluD2 in live cells using it (Figure S9).
Figure 5. Confocal imaging of HEK293T cells transiently expressing with HA-tagged GluD2.
(a) Membrane surface GluD2 was visualized by treatment of an anti-HA antibody conjugated with AlexaFluor488 (green) and the biotinylated nanobody (Bt-Nb) followed by SAv-647 (red). (b) Using non-biotinylated nanobody or (c) non-transfected cells, any SAv-647 signals were not observed. TF, transfection. Conditions: HEK293T cells (2 x 105 cells) expressing HA-GluD2 were incubated in PBS (-) containing Bt-Nb (1 μM) on ice for 3 min. The cells were washed one time, followed by incubation in HEPES-buffered Dulbecco’s Modified Eagle’s Medium (DMEM-HEPES) containing Streptavidin-HiLyte647 (5 μg/mL)/Anti-HA alexa488 (5 μg/mL) on ice for 3 min. After washing twice with DMEM-HEPES, the cells were analyzed with a confocal microscope (Excitation/Detection wavelength: 488 nm/410–617 nm for anti-HA AlexaFlur488, 640 nm/650–700 nm for SAv-647).
Overall, this study demonstrated the utility of the peptide tag-based site-specific modification with NTA-NASA probes for engineering proteins without impairing protein functions.
3. Conclusion
In summary, we demonstrated a versatile labeling method for site-selective covalent modification of His-tagged proteins. The chief advantages of this method include (i) a simple labeling protocol with a short reaction time (~1h); (ii) flexible availability of the functional molecular probes introducing proteins; (iii) small molecular size of the tag and probe; (iv) good compatibility for conventional His-tag based expression/purification system, which allows easy access to nanobody bioconjugation. Importantly, this labeling method has a potential for direct modification of His-tagged proteins expressing in live cell contexts, as our group has reported with other several strategies.9,11,12 Moreover, given the compatibility of short-peptide tags with emerging technologies for genome editing such as CRISPR/Cas9 system, this method would allow in situ labeling of “endogenously expressed” KH6/H6K-tagged proteins.25 Research along these lines is currently underway.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bmc.2020.115947.
Acknowledgment
We thank Dr. Muneo Tsujikawa (Kyoto University) for plasmid construction. This work was funded by MEXT, Japan to V.T., Grant-in-Aid for Young Scientists (18 K14334) and Grant-in-Aid for Scientific Research on Innovative Areas “Integrated Bio-metal Science” (19H05764) to T.T., and Japan Science and Technology Agency (JST) ERATO Grant JPMJER1802 to I.H. This work was also supported by a Grant-in-Aid for Scientific Research on Innovative Areas “Chemistry for Multimolecular Crowding Biosystems” (17H06348).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- 1.Tamura T, Hamachi I. Chemistry for Covalent Modification of Endogenous/Native Proteins: From Test Tubes to Complex Biological Systems. J Am Chem Soc. 2019;141:2782. doi: 10.1021/jacs.8b11747. [DOI] [PubMed] [Google Scholar]
- 2.Spicer CD, Davis BG. Selective Chemical Protein Modification. Nat Commun. 2014;5:4740. doi: 10.1038/ncomms5740. [DOI] [PubMed] [Google Scholar]
- 3.Takaoka Y, Ojida A, Hamachi I. Protein Organic Chemistry and Applications for Labeling and Engineering in Live-Cell Systems. Angew Chem, Int Ed. 2013;52:4088. doi: 10.1002/anie.201207089. [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto S, Hamachi I. Recent Progress in Chemical Modification of Proteins. Anal Sci. 2019;35:5. doi: 10.2116/analsci.18R003. [DOI] [PubMed] [Google Scholar]
- 5.Lotze J, Reinhardt U, Seitz O, Beck-Sickinger AG. Peptide-Tags for Site-Specific Protein Labelling in Vitro and in Vivo. Mol BioSyst. 2016;12:1731. doi: 10.1039/c6mb00023a. [DOI] [PubMed] [Google Scholar]
- 6.Uchinomiya S, Ojida A, Hamachi I. Peptide Tag/Probe Pairs Based on the Coordination Chemistry for Protein Labeling. Inorg Chem. 2014;53(4):1816–1823. doi: 10.1021/ic401612z. [DOI] [PubMed] [Google Scholar]
- 7.Tamura T, Shigemitsu H, Hamachi I. In Situ Protein Labeling in Complex Environments. In: Atwood JL, editor. Comprehensive Supramolecular Chemistry II. Vol. 4. Elsevier; Oxford: 2017. p. 409. [Google Scholar]
- 8.Gaietta G, Deerinck TJ, Adams SR, et al. Multicolor and Electron Microscopic Imaging of Connexin Trafficking. Science. 2002;296:503. doi: 10.1126/science.1068793. [DOI] [PubMed] [Google Scholar]
- 9.Uchinomiya S, Nonaka H, Wakayama S, Ojida A, Hamachi I. In-Cell Covalent Labeling of Reactive His-Tag Fused Proteins. Chem Commun. 2013;49:5022. doi: 10.1039/c3cc41979g. [DOI] [PubMed] [Google Scholar]
- 10.Nonaka H, Tsukiji S, Ojida A, Hamachi I. Non-Enzymatic Covalent Protein Labeling Using a Reactive Tag. J Am Chem Soc. 2007;129:15777. doi: 10.1021/ja074176d. [DOI] [PubMed] [Google Scholar]
- 11.Nonaka H, Fujishima S, Uchinomiya S, Ojida A, Hamachi I. Selective Covalent Labeling of Tag-Fused GPCR Proteins on Live Cell Surface with a Synthetic Probe for Their Functional Analysis. J Am Chem Soc. 2010;132:9301. doi: 10.1021/ja910703v. [DOI] [PubMed] [Google Scholar]
- 12.Zenmyo N, Tokumaru H, Uchinomiya S, et al. Optimized Reaction Pair of the Cyshis Tag and Ni(II)-Nta Probe for Highly Selective Chemical Labeling of Membrane Proteins. Bull Chem Soc Jpn. 2019;92:995. [Google Scholar]
- 13.Uchinomiya S, Nonaka H, Fujishima S, Tsukiji S, Ojida A, Hamachi I. Site-Specific Covalent Labeling of His-Tag Fused Proteins with a Reactive Ni(II)–NTA Probe. Chem Commun. 2009;39:5880. doi: 10.1039/b912025d. [DOI] [PubMed] [Google Scholar]
- 14.Hintersteiner M, Weidemann T, Kimmerlin T, Filiz N, Buehler C, Auer M. Covalent Fluorescence Labeling of His-Tagged Proteins on the Surface of Living Cells. ChemBioChem. 2008;9:1391. doi: 10.1002/cbic.200800089. [DOI] [PubMed] [Google Scholar]
- 15.Mortensen MR, Mortensen MR, Skovsgaard MB, et al. Introduction of an Aldehyde Handle on Nanobodies by Affinity-Guided Labeling. Bioconjug Chem. 2020;31:1295. doi: 10.1021/acs.bioconjchem.0c00151. [DOI] [PubMed] [Google Scholar]
- 16.Mortensen MR, Nielsen NL, Palmfeldt J, Gothelf KV. Imidazole Carbamate Probes for Affinity Guided Azide-Transfer to Metal-Binding Proteins. Org Biomol Chem. 2019;17:1379. doi: 10.1039/c8ob03017k. [DOI] [PubMed] [Google Scholar]
- 17.Tamura T, Ueda T, Goto T, et al. Rapid Labelling and Covalent Inhibition of Intracellular Native Proteins Using Ligand-Directed N-Acyl-N-Alkyl Sulfonamide. Nat Commun. 2018;9:1870. doi: 10.1038/s41467-018-04343-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ueda T, Tamura T, Hamachi I. Development of a Cell-Based Ligand-Screening System for Identifying Hsp90 Inhibitors. Biochemistry. 2020;59(2):179–182. doi: 10.1021/acs.biochem.9b00781. [DOI] [PubMed] [Google Scholar]
- 19.Lata S, Reichel A, Brock R, Tampé R, Piehler J. High-Affinity Adaptors for Switchable Recognition of Histidine-Tagged Proteins. J Am Chem Soc. 2005;127(29):10205. doi: 10.1021/ja050690c. [DOI] [PubMed] [Google Scholar]
- 20.Takaoka Y, Nishikawa Y, Hashimoto Y, Sasaki K, Hamachi I. Ligand-Directed Dibromophenyl Benzoate Chemistry for Rapid and Selective Acylation of Intracellular Natural Proteins. Chem Sci. 2015;6:3217. doi: 10.1039/c5sc00190k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hughes CC, Yang Y-L, Liu W-T, Dorrestein PC, La Clair JJ, Fenical W. Marinopyrrole A Target Elucidation by Acyl Dye Transfer. J Am Chem Soc. 2009;131(34):12094. doi: 10.1021/ja903149u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bracher PJ, Snyder PW, Bohall BR, Whitesides GM. The Relative Rates of Thiol-Thioester Exchange and Hydrolysis for Alkyl and Aryl Thioalkanoates in Water. Orig Life Evol Biosph. 2011;41:399. doi: 10.1007/s11084-011-9243-4. [DOI] [PubMed] [Google Scholar]
- 23.Muyldermans S. Nanobodies: Natural Single-Domain Antibodies. Annu Rev Biochem. 2013;82:775–797. doi: 10.1146/annurev-biochem-063011-092449. [DOI] [PubMed] [Google Scholar]
- 24.Yuzaki M, Aricescu AR. A GluD Coming-Of-Age Story. Trends Neurosci. 2017;40:138. doi: 10.1016/j.tins.2016.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Willems J, de Jong APH, Scheefhals N, et al. Orange: A CRISPR/Cas9-Based Genome Editing Toolbox for Epitope Tagging of Endogenous Proteins in Neurons. PLoS Biol. 2020;18:e3000665. doi: 10.1371/journal.pbio.3000665. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.