Abstract
The cytokine IL-6 controls the survival, proliferation and effector characteristics of lymphocytes through activation of the transcription factors STAT1 and STAT3. While STAT3 activity is an ever-present feature of IL-6 signaling in CD4+ T cells, prior T-cell receptor activation limits the IL-6 control of STAT1 in effector and memory populations. Here we show that STAT1 phosphorylation in response to IL-6 was regulated by protein tyrosine phosphatases (PTPN2, PTPN22) expressed in response to the activation of naïve CD4+ T cells. Transcriptomic and chromatin immunoprecipitation-sequencing of IL-6 responses in naïve and effector memory CD4+ T cells showed how the suppression of STAT1 activation shaped the functional identity and effector characteristics of memory CD4+ T cells. Thus, protein tyrosine phosphatases induced by activation of naïve T cells determined the way activated or memory CD4+ T cells sensed and interpreted cytokine signals.
Naïve, activated and memory T cells display differences in their ability to respond to antigen. These include changes in proliferation, survival, sensitivity to antigen, dependence on co-stimulatory signals and alterations in T cell homing1. Cytokines responsible for the control of these activities often signal through receptor-associated Janus kinases (Jak proteins) that regulate cytoplasmic transcription factors termed signal transducers and activators of transcription (STAT)2. Thus, the Jak-STAT pathway senses and interprets environmental signals essential for proliferation and functional identity2. Here, we examined whether cytokine cues delivered by the Jak-STAT pathway can be adapted to fine-tune the effector properties of individual CD4+ T cell subsets.
Studies of infection, inflammation, autoimmunity and cancer demonstrate that the cytokine IL-6 is essential for the generation of adaptive immunity3. Activities include the maturation and maintenance of antibody secreting B cells, and responses that shape the effector characteristics of CD4+ T helper (TH) cells3. In this regard, mice lacking IL-6 often show deficiencies in T cell effector function and memory recall4, 5, 6, 7, 8, 9. Studies also suggest that CD4+ T cells display differences in IL-6 responsiveness that may reflect the activation status of the T cell7, 10, 11, 12. How these differences arise is currently unclear.
The receptor complex responsible for IL-6 signaling consists of a type-1 cytokine receptor (IL 6R, CD126) and a signal-transducing β-receptor (gp130, CD130) subunit3. IL-6R is shed in response to CD4+ T cell activation, and inflammatory T cells from sites of disease often display low IL 6R expression7, 10, 12, 13, 14, 15, 16, 17. IL 6 activates the latent transcription factors STAT1 and STAT33. IL-6 control of STAT3 is essential for T cell recruitment and survival and the maintenance of activated T cells within inflamed tissues11, 14, 16. These STAT3-driven responses include the transactivation of anti-apoptotic regulators and genes that determine the effector or regulatory characteristic of CD4+ T cells3, 11, 18. In contrast, IL-6 activation of STAT1 plays a more regulatory role and often determines the transcriptional output of STAT318, 19, 20, 21. These studies illustrate a complex interplay between STAT1 and STAT3, and emphasize how STAT1 signaling may shape the biological properties of IL-618, 20, 21, 22, 23, 24. Significantly, CD4+ T cell activation has been shown to alter IL-6 signaling through STAT19, 11. Here we show that STAT1 phosphorylation in response to IL-6 is suppressed in activated and memory CD4+ T cells and identified protein tyrosine phosphatases as regulators of STAT1 activity. Our data further showed how this re-programming mechanism may influence the way effector memory CD4+ T cells sense and interpret IL-6 signals in disease.
Results
Infiltrating synovial CD4+ T cells have altered IL-6-mediated STAT1 activation
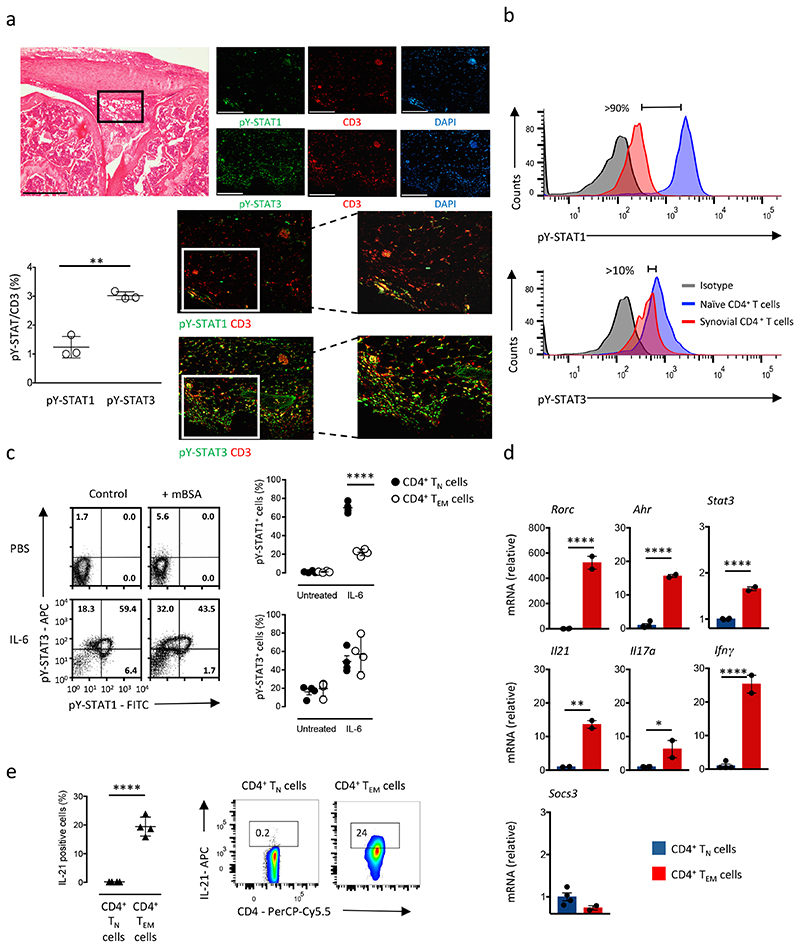
Previous studies suggest that CD4+ T cell activation alters cytokine signaling through the Jak-STAT pathway4, 9, 11. To further these findings we established antigen-induced arthritis (AIA) in C57Bl/6 wild-type mice through immunization with methylated BSA (mBSA). Histological joint sections from day 10 of AIA were evaluated for tyrosine phosphorylated STAT1 and STAT3 (hereafter pY-STAT1 and pY STAT3) using immunofluorescence (Fig. 1a). While pY-STAT3 co-localized with CD3+ T cells, pY STAT1 showed weak co-localization with the CD3 stain (Fig. 1a). To verify these observations, CD4+ T cells were isolated at day 10 of AIA from the inflamed synovium of wild-type mice and stimulated ex vivo with IL-6 (20ng/ml). When compared to IL-6-treated CD4+CD25−CD44loCD62LhiCD127hi naïve T cells (TN cells) from the spleen of wild-type mice, intracellular flow cytometry showed that synovial CD4+ T cells displayed reduced pY STAT1 staining (Fig. 1b). Both CD4+ T cell populations displayed comparable pY STAT3 responses to IL-6 (Fig. 1b). To test whether prior antigenic challenge with mBSA restricted the ability of IL-6 to signal through STAT1, we extracted total CD4+ T cells from the inguinal draining lymph nodes of wild-type mice immunized with mBSA, stimulated them ex vivo for 30 min with 20ng/ml IL-6 and monitored changes in pY-STAT1 and pY-STAT3 by intracellular flow cytometry. When compared to wild-type CD4+ T cells from the inguinal lymph node of non-challenged mice, CD4+ T cells from mBSA-immunized mice showed impaired pY-STAT1 detection in response to IL 6 (Fig. 1c). This reduction in pY STAT1 was particularly evident in activated or memory CD4+ T cells (Fig. 1c).
Figure 1. Infiltrating T-cells showed impaired STAT1 activity in response to arthritis induction.
(a) Representative H&E staining of knee joints at day 10 post disease induction (antigen-induced arthritis, AIA) (bar: 500μm); boxed area shows the location of the immunofluorescence. Representative immunofluorescence with antibodies against CD3 (red), pY-STAT1 or pY-STAT3 (green) is shown together with DAPI counterstaining (blue) (bar: 100μm). Graph shows the proportion of CD3+ T cells displaying either pY STAT1 or pY-STAT3 (n=3). (b) Phosphorylation of STAT1 and STAT3 by flow cytometry of infiltrating synovial CD4+ T cells during AIA after stimulation with 20ng/ml IL-6 compare to CD4+ TN cells. (c) Representative flow cytometry of pY-STAT1 and pY-STAT3 in CD4+ T cells extracted from inguinal lymph nodes of mBSA challenged (n=4) and non-challenged mice (control) (n=3) following stimulation with 20ng/ml IL-6 for 30 min. Graphs show quantification of pY-STAT1 and pY-STAT3 activity in CD4+ TN and CD4+ TEM cells (n=4). (d) Quantitative PCR of Ahr, Ifng, Il17a, Il21, Rorc, Socs3 and Stat3 in CD4+ TN (n=4) and CD4+ TEM cells (n=2) extracted from inguinal lymph nodes of mBSA challenged mice. (e) Intracellular flow cytometry analysis of IL-21 production in CD4+ TN and CD4+ TEM cells extracted from inguinal lymph nodes after 4 hours stimulation with PMA, ionomycin and monensin (n=4). Data are representative of three independent experiments (c,e), two independent experiments (a,b) and one experiment involving biological replicates (d). ****P<0.0001, **P<0.01, *P<0.05 (Two-tailed unpaired Student’s t test (a,b,d,e) and one-way ANOVA test with Tukey’s multiple comparison test (c). Data are shown as mean ± s.d.)
STAT1 is an important determinant of T cell effector function20, 21, 22. We therefore used quantitative PCR to evaluate the effector characteristics of CD4+ T cells from wild-type mice with AIA. Analysis was performed on CD4+ TN cells and CD4+CD25−CD44hiCD62LloCD127int-hi effector memory T cells (TEM cells) from the inguinal lymph nodes of wild-type mice with AIA. Ahr, Il21, Rorgt, Il17a, Ifng and Stat3 were all highly expressed in CD4+ TEM cells compared to CD4+ TN cells (Fig. 1d). In contrast, the expression of Socs3, a negative regulator of Jak-STAT signaling, remained comparable in both CD4+ T cell populations (Fig. 1d). In addition, intracellular cytokine staining of CD4+ TN and CD4+ TEM cells showed that CD4+ TEM cells generated increased amounts of IL-21 (Fig. 1e and Supplementary Fig. 1a). Thus, CD4+ TN and CD4+ TEM cells showed differences in IL-6 responsiveness.
Control of STAT1 activity is not determined by IL-6R signaling in CD4+ T cells
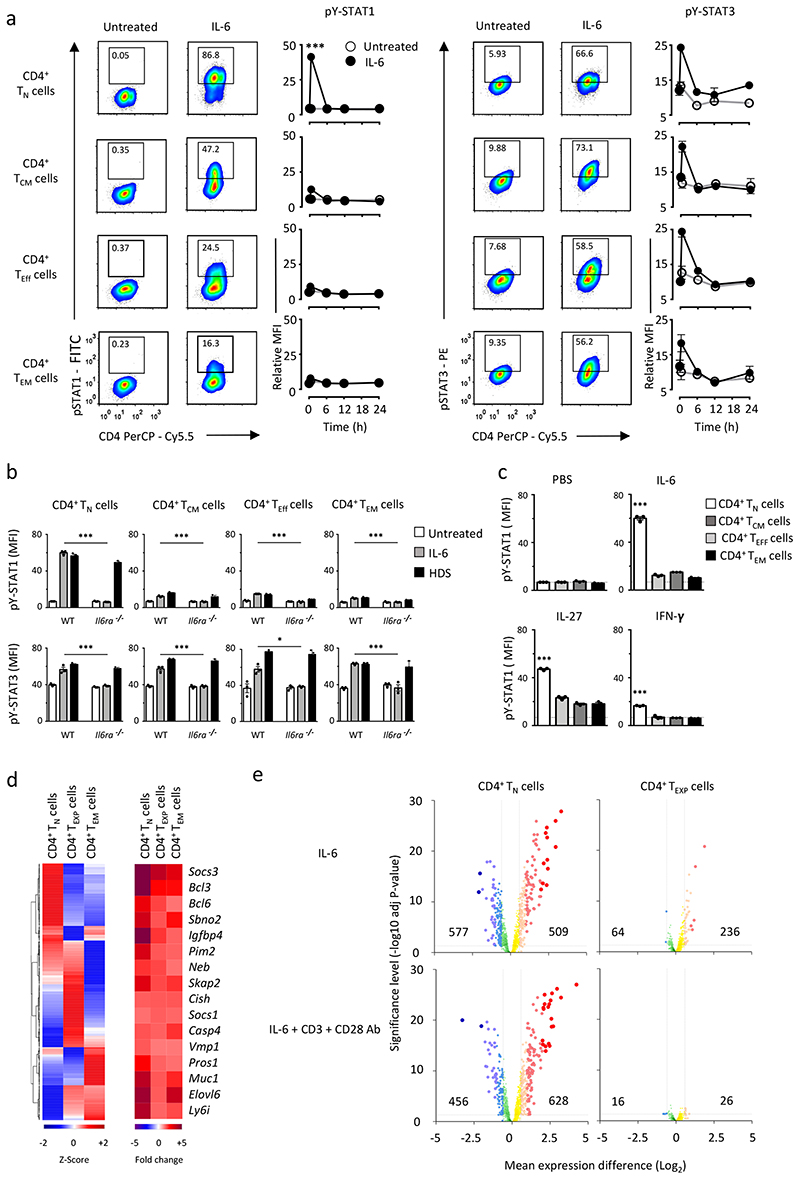
To determine if T cell subsets display different IL-6 signaling properties, splenic CD4+ TN cells, CD4+CD25−CD44hiCD62LhiCD127hi central memory T cells (TCM cells), CD4+CD25−CD44loCD62LloCD127lo-int effector T cells (TEff cells) and CD4+ TEM cells were purified from wild-type mice. These populations showed differences in IL-6R and gp130 expression, but displayed a similar transient activation of pY STAT3 in response to IL-6 (Fig. 2a and Supplementary Fig. 1b). A strong induction of pY-STAT1 was observed in IL-6-treated CD4+ TN cells, while CD4+ TCM, TEff and TEM cells showed impaired pY-STAT1 activation (Fig. 2a). A similar regulation of pY STAT1 was also observed in human CD4+ T cells (Supplementary Fig. 1c).
Figure 2. CD4+ T cell subsets show different response to IL-6.
(a) Representative flow cytometry analysis of STAT1 and STAT3 responses in naïve (TN), central memory (TCM), effector (TEff) and effector memory (TEM) CD4+ T cells after 30 min IL-6 stimulation (20ng/ml). Numbers indicate the percentage of pY-STAT1 or pY STAT3 staining. Temporal changes in pY-STAT1 and pY-STAT3 are shown for each T cell subset following IL 6 stimulation (n=3). (b) Detection of pY-STAT1 and pY-STAT3 in CD4+ TN, CD4+ TCM, CD4+ TEff and CD4+ TEM cells from WT and IL6ra-/- mice. CD4+ T cells were stimulated for 30 min with an equimolar concentration of IL-6 or an IL-6-sIL-6R fusion protein (HDS) (n=3). (c) Intracellular flow cytometry analysis of pY-STAT1 in CD4+ T cells following 30 min stimulation with IL-6, IL-27 or IFNγ (20ng/ml) (n=3). (d) Microarray expression data is presented for CD4+ TN (n=3), CD4+ TEM (n=3), and in vitro expanded CD4+ effector-like T cells (See Supplementary Fig.2a, CD4+ TEXP) (n=4) treated with 20ng/ml IL-6 for 6 hours. Analysis was confined to genes displaying both a relative signal intensity of >150 and >1.5-fold alteration in expression following IL-6 treatment (P<0.05). Heat map is hierarchically clustered based in the relative expression (Z-score) (left panel) or Fold change (right panel). (e) Volcano plots displaying IL-6 regulated gene expression in CD4+ TN and CD4+ TEXP cells stimulated with IL 6 (20ng/ml) or in combination with antibodies against CD3 and CD28. An interactive figure can be found on-line (http://jones-cytokinelab.co.uk/NI2019/figure2d.shtml). Data are representative of two independent experiments (a,c) and one experiment involving biological replicates (b,d,e). ***P<0.001 *P<0.05 (Two-tailed unpaired Student’s t test (a) and one-way ANOVA with Tukey’s multiple comparison test (b,c). Data are shown as mean ± s.e.m).
Given that activation-induced shedding of IL-6R may affect IL-6 receptor signaling, we investigated the IL 6 control of pY-STAT1 and pY-STAT3 in splenic CD4+ TN, TCM, TEff and TEM cells from wild-type and Il6ra-/- mice. IL-6 signaling in Il6ra-/- CD4+ T cells was triggered by IL-6 trans-signaling using an IL 6 sIL 6R chimeric fusion protein (HDS)7, 8. Il6ra-/- CD4+ T cells were not activated by IL-6 alone, but responded to an equimolar concentration of HDS, and all Il6ra-/- CD4+T cells showed increased pY STAT3 following HDS stimulation (Fig. 2b). While HDS induced pY-STAT1 in Il6ra-/- CD4+ TN cells, changes in pY-STAT1 was not seen in HDS-treated CD4+ TCM, TEff and TEM cells from Il6ra-/- mice (Fig. 2b). Thus, the loss of STAT1 signaling in activated CD4+ T cells is independent of changes in IL-6R regulation. A similar pattern of pY STAT1 regulation was noted in wild-type CD4+ T cells stimulated with IL-27 or IFN-γ (Fig. 2c and Supplementary Fig. 2d), suggesting the control of STAT1 phosphorylation was not unique to IL-6.
TCR activation regulates IL-6R signaling in CD4+ T cells
To investigate whether prior TCR engagement contributed to the changes in STAT1 activation in activated or memory CD4+ T cells we compared IL-6 signaling in CD4+ TN cells and CD4+ T cells previously activated with antibodies against CD3 and CD28. Wild-type CD4+ TN cells were cultured for 72h with antibodies against CD3 and CD28 followed by a 48h culture in fresh media, in the absence of exogenous stimulation. This rest period restored the surface expression of IL-6R on these TCR-experienced effector-like CD4+ T cells (TEXP cells; Supplementary Fig. 2a). While IL 6 stimulation of CD4+ TEXP cells induced pY STAT3, the activation of pY STAT1 was minimal when compared to the IL-6-dependent induction of pY-STAT1 in CD4+ TN cells (Supplementary Fig. 2b,c), indicating CD4+ T cell activation altered the subsequent activation of STAT1 by IL-6.
To test if TCR signaling altered the IL-6-induced activation of STAT1, we generated CD4+ TEXP cells from wild-type CD4+ TN cells using varying concentrations of co-stimulatory CD3 (0.1-10 μg/ml) and CD28 (0.5-15 μg/ml) antibodies (Supplementary Fig. 3a) and stimulated the expanded CD4+ TEXP cells with IL-6. The IL-6-dependent induction of pY-STAT3 was not affected by differences in antibody concentration. However the suppression of pY-STAT1 in response to IL-6 was sensitive to anti-CD3 antibody treatment with increasing doses of antibody leading to a repression of pY-STAT1 (Supplementary Fig. 3a). Thus, TCR signaling affected the inhibition of pY-STAT1 by IL-6.
We next determined whether changes in STAT1 activation altered the transcriptional output of IL-6. CD4+ TN, TEXP and TEM cells were stimulated for 6 h with IL-6 followed by transcriptomic analysis. This time point was selected based on the temporal profile of pY-STAT1 and pY-STAT3 detection (Fig. 2a), and the optimal expression of STAT-target genes (Ahr, Bcl3, Bcl6, Kat2b, Il10, Il21, Pim1, Stat3), as determined by Q-PCR (Supplementary Fig. 3b). CD4+ TN, TEXP, and TEM cells expressed a series of genes that were both common (e.g., Socs1, Sbno2, Bcl6) and unique (e.g., CD4+ TN – Cxcr1, Tnfrsf14; CD4+ TEXP – Gzma, Ajuba; CD4+ TEM – Ahr, Il10, Il21) to all three CD4+ T cell populations (Fig. 2d and Supplementary Fig. 3c). The number of transcripts enhanced by IL-6 in CD4+ TEXP cells (236) was markedly reduced when compared to the IL-6-dependent induction of transcripts in CD4+ TN cells (509) (Fig. 2e). The inclusion of anti-CD3+CD28 co-stimulatory antibodies further suppressed the number of genes induced by IL-6 in CD4+ TEXP cells (26), indicating the capacity of TCR activation to replace the signal delivered by IL-6 (Fig. 2e and Supplementary Fig. 3d). Thus, IL-6 controls very distinct patterns of gene regulation in CD4+ TN, TEXP and TEM cells that may shape the functional properties of these CD4+ T cells.
STAT1 phosphorylation is regulated by PTPN2
We next determined whether TCR activation altered the expression of genes linked with IL-6 receptor signaling in CD4+ TN, TEXP and TEM cells. No significant differences in the expression of IL 6 receptor subunits (Il6ra, Il6st), Janus kinases (Jak1, Jak2, Tyk2), STATs (Stat1, Stat3, Stat5a, Stat5b) and regulators of IL-6 or Jak-STAT signaling (Socs1, Socs3, Pias1, Dusp2, Cish, Arid5a, Arid5b) were observed between these CD4+ T cells (Supplementary Fig. 4a). Thus, the immediate regulation of STAT1 activity in CD4+ TEXP cells and CD4+ TEM cells was not attributed to changes in the make-up of the IL-6 signaling cascade.
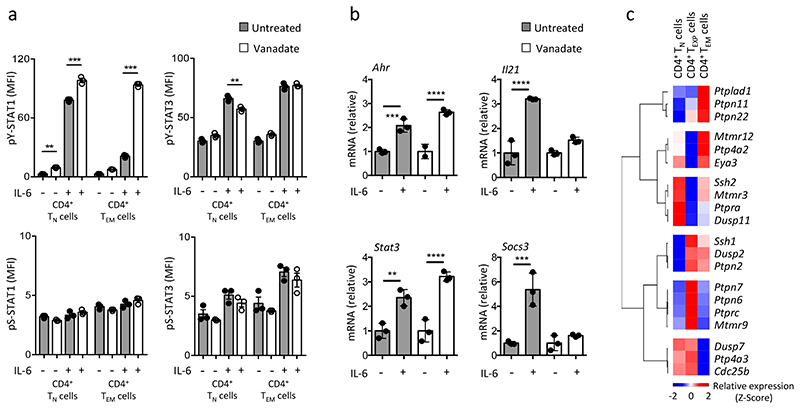
To examine how CD4+ T cell activation affected STAT1 signaling we assessed IL-6 responses in wild-type CD4+ TN and TEM cells using antibodies for serine phosphorylated STAT1 and STAT3 (hereafter pS-STAT1 and pS STAT3). Intracellular flow cytometry showed that the IL-6 activation of pS-STAT1 and pS STAT3 was comparable in both CD4+ T cells (Fig. 3a). However, CD4+ TEM cells showed suppressed pY-STAT1 in response to IL-6 (Fig. 3a and Supplementary Fig. 1c). We therefore addressed whether protein phosphatases controlled the tyrosine phosphorylation of STAT1. Treatment of CD4+ TN and TEM cells with the protein tyrosine phosphatase inhibitor sodium orthovanadate reversed the loss of pY-STAT1 activation in IL-6-stimulated CD4+ TEM cells (Fig. 3a). IL 6 control of pY STAT3 was unaltered by sodium orthovanadate (Fig. 3a). Analysis of Ahr, Il21, Socs3 and Stat3 expression in IL-6-treated CD4+ TEM cells showed that the inclusion of sodium orthovanadate reduced expression of Il21 and Socs3 (Fig. 3b). Thus, protein tyrosine phosphatase activity appears integral to the IL-6 control of STAT1 in CD4+ TEM cells.
Figure 3. Induction of protein tyrosine phosphatases following T-cell activation limits STAT1 signalling.
(a) CD4+ TN and CD4+ TEM cells were pre-treated for 5 min with 5mM sodium orthovanadate (vanadate) prior to IL-6 (20ng/ml) stimulation for 30 min. Changes in pY-STAT1 and pY-STAT3 activity were monitored by intracellular flow cytometry (MFI). A comparable analysis of pS-STAT1 and pS-STAT3 is shown as a control (n=3). (b) Quantitative PCR for Ahr, Il21, Stat3 and Socs3 after vanadate pre-treatment and 20ng/ml IL-6 stimulation in CD4+ TEM cells (n=3). (c) Heatmap analysis of Affymetrix transcriptomic data identifies the top 20 genes (P<0.05; relative signal intensity of >150; 1.5-fold alteration) associated with protein tyrosine phosphatase enzyme family. Data is presented as a hierarchical cluster using the average linkage method (row 1-pearson rank correlation). Data are representative of two independent experiment (a,b) and one experiment involving biological replicates (c). ***P<0.001; ** P<0.01 (one-way ANOVA with Tukey’s multiple comparison test (a) and two-way ANOVA with Sidak multiple comparison test (b). Data are shown as mean ± s.e.m (a) and mean ± s.d (b).
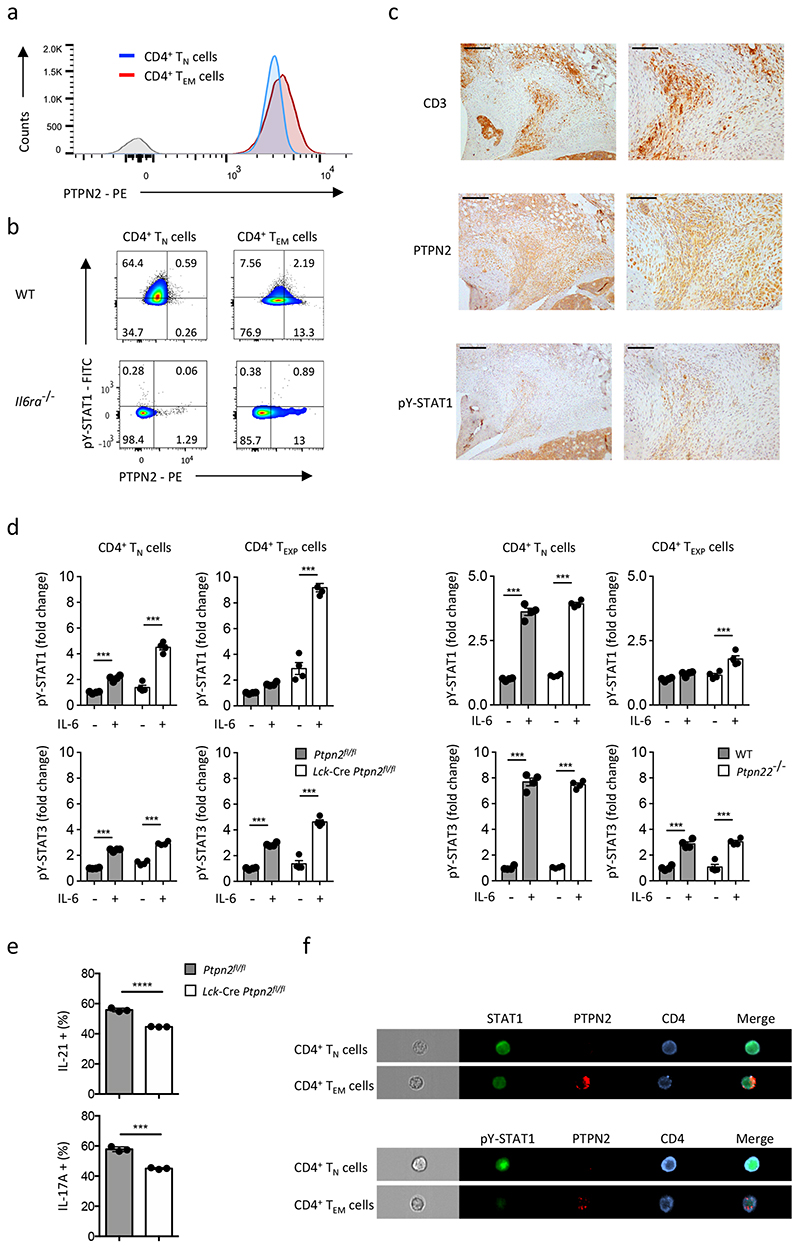
To identify protein tyrosine phosphatases responsible for the regulation of STAT1 in CD4+ TEXP and TEM cells we compared transcriptomic data from CD4+ TN, TEXP and TEM cells activated with anti-CD3+CD28 co-stimulatory antibodies. Several protein phosphatases were more highly expressed in CD4+ TEXP and TEM cells than CD4+ TN cells, and included the protein tyrosine phosphatases Ptpn2 and Ptpn22 (Fig. 3c). Flow cytometry showed that PTPN2 was more highly expressed in CD4+ TEM cells than CD4+ TN cells (Fig. 4a). Il6ra-/- CD4+ TEM cells had comparable expression of PTPN2 to wild-type CD4+ TEM cells (Fig. 4b), indicating that PTPN2 expression was independent of IL-6 signaling. To confirm the relevance of these findings to pathology we conducted immunohistochemistry of joint tissue from wild-type mice with AIA. Synovial CD3+ T cells showed enhanced expression of PTPN2 and low pY-STAT1 staining (Fig. 4c). Similarly, flow cytometry of IL-6-stimulated CD4+ TEM cells revealed that PTPN2 expression correlated with suppression of pY-STAT1 (Fig. 4b, Supplementary Fig. 4b). We next investigated IL 6 signaling in CD4+ TN and TEXP cells from whole genome C57Bl/6 Ptpn22 -/- mice and Lck-Cre Ptpn2 fl/fl mice, which lack PTPN2 in CD4+ T cells. As controls we used IL-6-treated CD4+ TN and TEXP cells isolated from C57Bl/6 wild-type mice (for PTPN22) or C57Bl/6 Ptpn2 fl/fl littermates. IL-6 activated pY-STAT1 in both Lck-Cre Ptpn2 fl/fl and Ptpn22 -/- CD4+ TEXP cells (Fig. 4d). When compared to IL-6 stimulated CD4+ TEXP cells from Lck-Cre Ptpn2 fl/fl mice, the recovery of pY-STAT1 activity in IL-6 treated Ptpn22 -/- CD4+ TEXP cells was less obvious (Fig. 4d). This possibly reflected a lower expression of Ptpn22 in CD4+ TEXP cells (Fig. 3c). Thus, PTPN2 acted as a repressor of IL-6-induced STAT1 tyrosine phosphorylation in activated CD4+ T cells.
Figure 4. STAT1 activity is regulated by PTPN2.
(a) Representative histogram of PTPN2 staining in CD4+ TN and CD4+ TEM cells by flow cytometry. (b) Flow cytometry analysis of STAT1 phosphorylation and PTPN2 expression in CD4+ TN and CD4+ TEM cells analyzed 30 min after stimulation with 20 ng/ml IL-6. (c) Immunohistochemistry of the inflamed synovium from wild-type mice with antigen-induced arthritis (day-10 post disease induction) in tissue sections stained with antibodies against CD3, Ptpn2 and pY STAT1. Scale bar, 100μm (left panel) and 200μm (right panel). (d) Analysis of pY-STAT1 and pY-STAT3 in CD4+ TN and CD4+ TEXP cells derived from Ptpn2 fl/fl, Lck-Cre Ptpn2 fl/fl (left panel) or wild-type and Ptpn22 -/- mice (right panel) (n=4) exposed to IL-6 (20 ng/ml) for 30 min in combination with antibodies against CD3 and CD28. Fold change relative to the untreated controls are compared. (e) IL-21 and IL-17A quantification by flow cytometry in CD4+ TEM cells from Ptpn2 fl/fl and Lck-Cre Ptpn2 fl/fl mice (n=3). (f) ImageStream analysis of STAT1 and PTPN2 localization in CD4+ TN and CD4+ TEM cells stained with antibodies against STAT1, pY-STAT1, PTPN2 and CD4. Data are representative of three independent experiments (a,b), two independent experiments (f) and one experiment involving biological replicates (c,d,e). ****P<0.0001; *** P<0.001 (One-way ANOVA with Tukey’s multiple comparison test (d) and Two-tailed unpaired Student’s test (e). Data are shown as mean ± s.d.).
We next explored whether PTP activity affected the production of IL-17A and IL-21 in CD4+ TEM cells. Intracellular flow cytometry indicated that Lck-Cre Ptpn2 fl/fl and Ptpn22 -/- CD4+ TEM cells mice generated less IL-17A and IL 21 than CD4+ TEM cells from Ptpn2 fl/fl littermates or wild-type mice (Fig. 4e, Supplementary Fig. 4c). Moreover, ImageStream analysis showed the co-localization of PTPN2 with non-phosphorylated STAT1 in wild-type CD4+ TEM cells (Fig. 4f). Thus, PTPN2, and to a lesser extent PTPN22, regulated STAT1 signaling in activated and memory CD4+ T cells.
T cell activation re-tunes the transcriptional output of IL-6 in CD4+ TEM cells
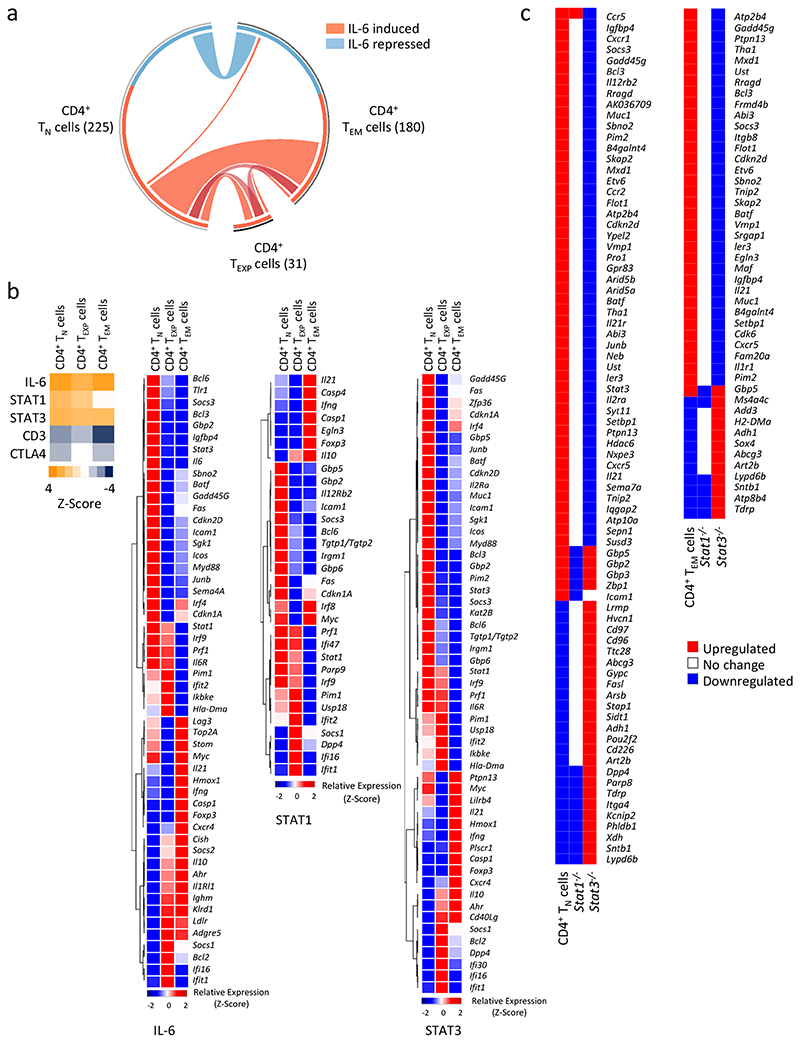
Next, we compared the transcriptional output of IL-6 in CD4+ TN, TEXP and TEM cells. Analysis was confined to significantly regulated genes (p<0.05) that displayed both a relative signal intensity of >150 and >1.5 fold alteration in expression. Circos visualization identified a number of genes that were under IL-6 control in CD4+ TN (225), TEXP (31) and TEM (180) cells (Fig. 5a). Hierarchical clustering and validation of selected gene targets identified IL-6 gene signatures that were either common to all three CD4+ T cells or uniquely expressed by a particular population (Supplementary Fig. 3c). Genes regulated by IL-6 in CD4+ TN, TEXP and TEM cells were mainly STAT3 target genes23, 24 and included genes that encoded transcriptional regulators (e.g., Bcl3, Bcl6, Etv6), co-repressors (e.g., Sbno2, Muc1) and negative regulators (e.g., Socs3, Pim1, Batf) of transcription factors such as STAT3, NF-κB and AP1 (Supplementary Fig. 3c).
Figure 5. Transcriptomic analysis of IL-6 responses in CD4+ T cells.
(a) Circos visualisation details the IL-6 regulated gene changes in CD4+ TN and CD4+ TEXP cells (See Supplemental Figure-2), and ex vivo sorted CD4+ TEM cells. Total number of IL-6 regulated genes is presented in parenthesis for each population (P< 0.05, Chip Intensity 150+, and > 1.5-fold change). Lines coloured in red represent up-regulated genes and all down-regulated gene changes are blue. Connecting lines highlight common genes that are IL-6 regulated in two or more of the populations. (b) IPA analysis of genes associated with IL-6, STAT1 and STAT3 upstream regulators. Top left heat map shows the predicted activated state (orange) and the predicted inhibited state (blue) of transcription regulators. Upstream regulator analysis for CTLA4 and CD3 are presented as controls. Relative expression heat maps are presented as a hierarchical cluster using the average linkage method (row 1-pearson rank correlation). The differential expression of genes being regulated by IL-6, STAT1 or STAT3 is shown for CD4+ TN, CD4+ TEXP and CD4+ TEM cells. (c) IL-6 regulated gene changes derived from transcriptomic analysis were directly compared with datasets derived from IL-6 stimulated Stat1 -/- and Stat3 -/- CD4+ T cells (GSE65621).
Next, we conducted a molecular pathway analysis of the transcriptomic data from IL-6-treated CD4+ TN, TEXP and TEM cells (Fig. 5b). We also mapped these datasets against publicly-available transcriptomic data from IL 6-stimulated Stat1 -/- and Stat3 -/- CD4+ T cells (Fig. 5c)20. This analysis identified a series of STAT1-regulated genes commonly associated with interferon (e.g., Irf8, Gbp2, Gbp5, Gbp6, Stat1, Parp9). In contrast, STAT3-regulated genes displayed greater functional diversity and were implicated in interleukin signaling, immune activation, proliferation, catabolism and metabolism (e.g., Socs3, Bcl3, Il6r, Kat2b) (Fig. 5b,c). This collective approach demonstrated that STAT1 and STAT3 regulated very distinct patterns of gene expression in IL-6 stimulated CD4+ TN, TEXP and TEM cells (Fig. 5b,c). For example, when compared with IL-6 stimulated CD4+ TN cells, activation with IL-6 enhanced the expression of genes associated with prolonged lymphocyte survival and memory (e.g., Hmox1, Myc, Cd83), and regulatory (e.g., Lag3, Il10, Foxp3) or effector (e.g., Ahr, Il21, Ifng) characteristics in CD4+ TEM cells.
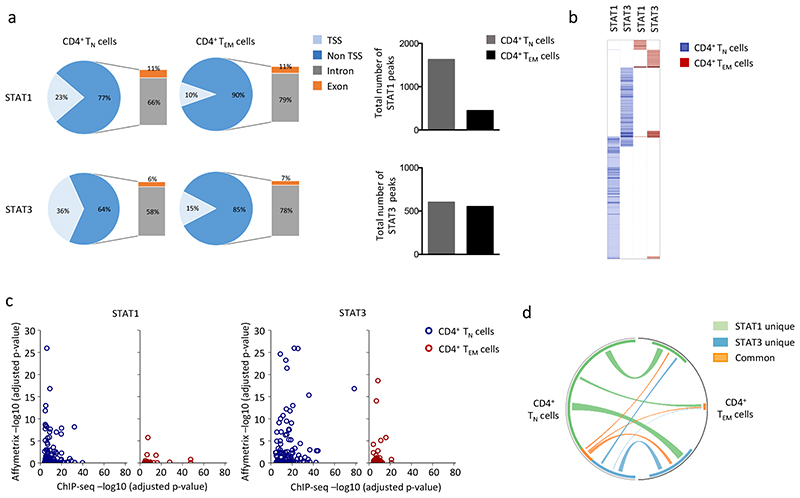
We next performed ChIP-seq in IL-6-stimulated CD4+ TN and TEM cells. Sequencing peaks displaying a 4-fold increase above input (P<0.0001; FDR 0.05) were aligned to the genome and assigned to transcription start sites (TSS), exons and introns (Fig. 6 and Supplementary Fig. 5). Peaks located in undefined intergenic regions were excluded from the analysis. ChIP-seq of IL-6-stimulated CD4+ TN cells identified 1625 peaks associated with STAT1 and 602 peaks for STAT3 (Fig. 6a). The number of STAT1 (446) and STAT3 (552) peaks was reduced in CD4+ TEM cells (Fig. 6a). To understand the regulatory properties of STAT1 and STAT3 we first confined our analysis to peaks residing within TSS regions (Fig. 6b and Supplementary Fig. 5). Sequencing peaks that mapped to defined gene promoters were correlated against corresponding transcriptomic data from IL-6-treated CD4+ TN and TEM cells (Fig. 6c and Supplementary Fig. 5). STAT1 and STAT3 showed substantially reduced binding to TSS regions in CD4+ TEM cells compared to CD4+ TN cells (Fig. 6b,c,d). Very few genes shared STAT1 and STAT3 binding (e.g., Stat3, Stat5b, Icam1, Socs3, Sigirr, and Akt2) (Fig. 6b,d), and these co-regulated genes were largely restricted to CD4+ TN cells (Fig 6b,d). Stat3 was the only gene that was bound by both STAT1 and STAT3 in CD4+ TEM cells (Fig. 6c). To determine the specificity of these DNA-transcription factor interactions we used computational tools to identify consensus DNA motifs for STAT1 and STAT3 binding (Supplementary Fig. 5c). Analysis of ChIP-seq datasets from IL-6-treated CD4+ TN and TEM cells identified sequences resembling an IFN-regulated STAT responsive element (ISRE; E-value 5.9e-040) for STAT1 binding, and sequences homologous to a gamma-activated sequence (GAS; E-value 1.2e-115) for STAT3 (Supplementary Fig. 5c). We also identified consensus motifs for other transcription factors including SP1 and C2H2 Zn-finger transcription factor proteins (Supplementary Fig. 5c) and ChIP-qPCR showed STAT1 and STAT3 bound to SP1 consensus binding sequences (Supplementary Fig. 5d and Supplementary Table 2). Thus, transcriptional differences between IL-6 stimulated CD4+ TN and TEM cells are shaped by STAT1 and STAT3 docking to both classical STAT-responsive elements and DNA motifs that suggested a regulatory interplay with other transcription factors in CD4+ TN and TEM cells.
Figure 6. ChIP-seq analysis of STAT1 and STAT3 binding in IL-6 stimulated CD4+ T cells.
ChIP-seq was performed on genomic DNA extracted from sorted CD4+ TN and CD4+ TEM cells following 1-hour stimulation with IL-6 in presence of antibodies against CD3 and CD28. Peak calling and downstream data processing are described in Materials & Methods. (a) Pie charts show the proportion of peaks associated with STAT1 and STAT3 binding to defined genomic regions. The total number of peaks identified is displayed graphically. All datasets residing outside TSS regions were only included if located to exonic or intronic sites. (b) Analysis of gene clusters regulated by binding STAT1 and STAT3 in TSS promoter regions. The heat map shows the score value for each gene identified with Homer for STAT1 and STAT3 ChIP-seq data in CD4+ TN (blue) and CD4+ TEM (red) cells. (c) Comparison of ChIP-seq datasets against Affymetrix gene expression (relative significance; -(log10 (adjusted P-value)). Analysis of STAT1 and STAT3 datasets is shown for CD4+ TN (blue) and CD4+ TEM (red) subsets. An interactive figure of additional information can be found on-line (http://jones-cytokinelab.co.uk/NI2019/figure6c.shtml) (d) Circos visualization of STAT1 and STAT3 binding to TSS regions of genes under IL-6 regulation in CD4+ TN and CD4+ TEM cells. Connecting lines are color coded to reflect involvement of STAT1 (green), STAT3 (blue) or both STAT1 and STAT3 (orange).
Genes under IL-6 control in CD4+ TEM cells are regulated at distal promoter regions
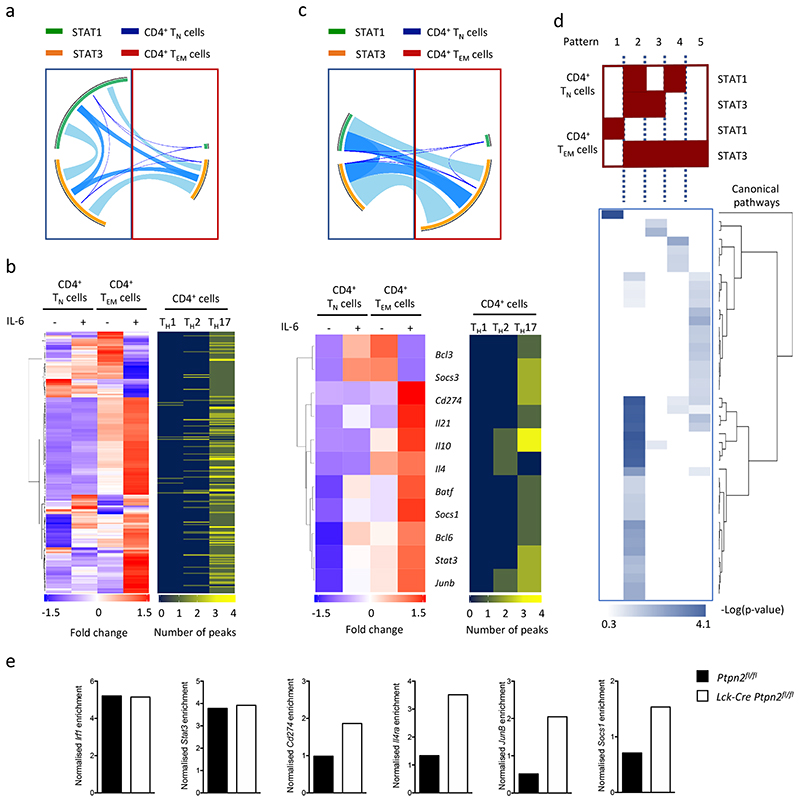
Next we investigated the canonical pathways associated with the genes that were bound with STAT1 or STAT3 in IL-6-treated CD4+ TN and TEM cells. Bioinformatic analysis identified a selective enrichment of genes involved in disease processes, catabolism and cytokine signaling in CD4+ TEM cells as compared to CD4+ TN cells (Supplementary Fig. 6a and Supplementary Table 3). Many of the genes associated with these pathways were distinct from those showing STAT1 or STAT3 binding to TSS (Fig. 6b), suggesting that genes under IL-6 regulation in CD4+ TEM cells may be controlled by STAT1 and STAT3 binding to distal promoter regions. To identify possible mechanisms that may explain this distal regulation we evaluated whether STAT1 and STAT3 peaks aligned with enhancer regions displaying enriched binding of the histone acetyl-transferase P300 (hereafter P300 enhancer elements25, 26, 27, 28). STAT1 and STAT3 ChIP-seq datasets from IL-6-treated CD4+ TN or TEM cells were mapped against publicly-available P300 ChIP-seq data from mouse CD4+ T cells polarized in vitro into TH1, TH2 or TH17 cells28 (Supplementary Fig. 6b and Supplementary Table 4). A combined analysis of IL-6-treated CD4+ TN and TEM cells identified 558 genes that bound P300 in association with either STAT1 or STAT3 (Fig. 7a,b). In IL 6-stimulated CD4+ TN and TEM cells we identified 215 genes that displayed alternate binding of STAT1 or STAT3 in CD4+ TN and TEM cells (Fig. 7b). These genes were selectively induced or repressed by IL-6 in CD4+ TEM cells (Fig. 7b, Supplementary Table 5). Among these, 208 genes aligned with genes bound by P300 in TH17 cells28. These included genes linked with proliferation and survival (e.g., Vmp1, Rbpj, Fasl), immune regulation (e.g., Cd200, Cish, Ctla4, Cd69), alternate lineage fates (e.g., Ahr, Batf, Bcl6, Cxcr5, Etv6, Fosl2, Irf4, Stat3) or differences in T cell effector function (e.g., Il10, Il21, Il4ra, Il17a, Il17ra, Il21r) (Supplementary Table 5). This analysis indicated that genes controlled by IL-6 in CD4+ TN and TEM cells are associated with P300 enhancer elements that are potentially activated or suppressed by changes in the pattern of STAT1 or STAT3 binding.
Figure 7. IL-6 regulates the interaction of STAT1 and STAT3 with P300 enhancer sites.
(a) Circos plot shows the co-localisation of STAT1 (blue) and STAT3 (orange) binding to genomic regions sharing P300 enrichment in CD4+ TN and CD4+ TEM cells. The connecting lines show the relationship of STAT1 and STAT3 binding between CD4+ TN and CD4+ TEM cells. P300 ChIP-seq datasets (Accession number GSE40463, GSE60482) are derived from TH1, TH2 and TH17 cells. (b) Heat map showing the expression of all IL-6 regulated genes linked with P300 binding in CD4+ TN and CD4+ TEM cells (positioned left). The correspondingly aligned heatmap (positioned right) shows the relationship to P300 sites in TH1, TH2 and TH17 cells and shows the number of clustered P300 sites affiliated to an individual gene (blue=0, yellow=4). Specific examples of individual genes are shown. (c) Circos visualisation of 135 genes that display P300 binding in association with either STAT1 or STAT3 in CD4+ TN versus CD4+ TEM cells. (d) IPA predictions of the five distinct patterns of STAT binding identified from panel c. Hierarchical clustering of canonical pathways was performed using -Log (P-value). Supplemental Table 6 lists the canonical pathways represented in the heatmap. (e) STAT1 binding enrichment quantification by ChIP-qPCR in Ptpn2fl/fl and Lck-Cre:Ptpn2fl/fl CD4+ TEM cells (one experiment with pool samples from 12 Ptpn2fl/fl and 8 Lck-Cre Ptpn2fl/fl mice).
Among the 215 genes regulated by IL-6 in CD4+ TEM cells, 80 displayed STAT1 or STAT3 binding in ChIP-seq data from IL-6 activated CD4+ TN cells, but not in IL-6 treated CD4+ TEM cells (Fig. 7c). This suggested that the binding of these transcription factors to these sites acted as repressors of gene activation. ChIP-seq analysis of the other 135 genes showed that some form of STAT1 or STAT3 binding to P300 enhancer elements was retained in both IL-6 stimulated CD4+ TN and TEM cells (Fig. 7c and Supplementary Fig. 6b). These included Il10, Il21, Il21r, Bcl3, Batf, Junb, Socs1 and Cd274 (Fig.7c). Circos visualization illustrated how the binding of STAT1 and STAT3 to these promoters differed between IL-6 stimulated CD4+ TN and TEM cells (Fig. 7c). We identified 5 discrete patterns of STAT1 and STAT3 binding – pattern 1 (CD4+ TN cells: no STAT binding; TEM cells: STAT1), pattern 2 (CD4+ TN cells: STAT1, STAT3; TEM cells: STAT3), pattern 3 (CD4+ TN cells: STAT3; TEM cells: STAT3), pattern 4 (CD4+ TN cells: STAT1; TEM cells: STAT3) and pattern 5 (CD4+ TN cells: no STAT binding; TEM cells: STAT3) (Fig.7d). Computational analysis of the genes affiliated to each pattern revealed links with the cytokine control of T cell proliferation, differentiation and survival (Fig. 7d, Supplementary Fig. 6b and Supplementary Table 6). This was particularly apparent in pattern 2, and to a lesser extent, pattern 3 and 5. Promoter regions assigned to pattern 2 displayed binding of STAT1 and STAT3 in IL-6 stimulated CD4+ TN cells, but showed a loss of STAT1 in IL-6 activated CD4+ TEM cells. Genes identified with this form of STAT regulation included Junb, Il4ra, Cd274 and Socs1 (Fig.7c and Supplementary Fig.6b) and suggested a potential link to the PTPN2-regulated of pY-STAT1. We therefore conducted a ChIP qPCR analysis of STAT1 binding to the promoters of Junb, Il4ra, Cd274 and Socs1 in IL 6-stimulated Lck-Cre Ptpn2fl/fl and Ptpn2fl/fl CD4+ TEM cells. STAT1 binding to these promoters was specifically enriched in DNA samples from Lck-Cre Ptpn2fl/fl CD4+ TEM cells as compared to control Ptpn2fl/fl CD4+ TEM cells (Fig. 7e). In contrast, the binding of STAT1 to the TSS regions of Stat3 and Irf1 remained unaltered, and DNA samples from Lck-Cre Ptpn2fl/fl CD4+ TEM cells and control Ptpn2fl/fl CD4+ TEM cells showed similar enrichment for STAT1 (Fig. 7e). Thus, PTPN2 activity determined STAT1 binding to specific gene promoter regions in IL-6 activated CD4+ TEM cells.
PTPN2 correlates with indices of synovial pathology in rheumatoid arthritis patients
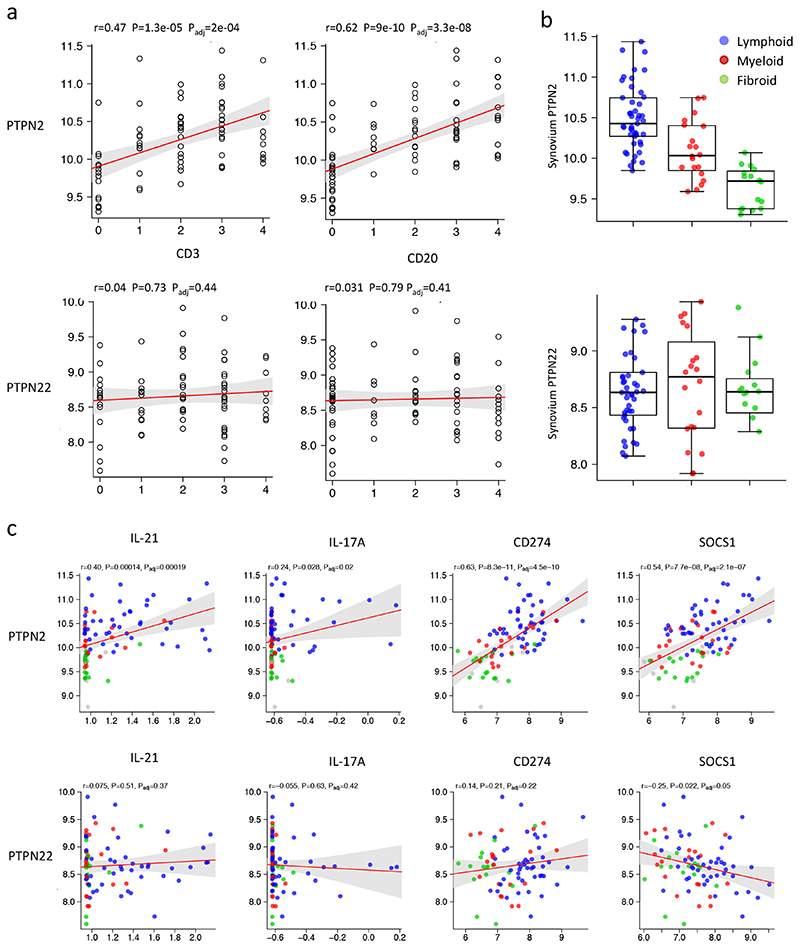
Many of the genes identified in IL-6 stimulated CD4+ TEM cells contribute to the generation and maintenance of effector T cells associated with autoimmunity (Supplementary Table 6). We therefore investigated the relationship between PTPN2 and PTPN22 and these IL-6-regulated genes in RNA-seq datasets from synovial tissues biopsies of 87 patients with rheumatoid arthritis. Expression of PTPN2 and PTPN22 in these biopsy samples was compared against corresponding histological staining of the inflamed synovium for the lymphocyte markers CD3 and CD20 by immunohistochemistry. Analysis revealed a close correlation between PTPN2 and CD3 and CD20 (Fig 8a). This association was particularly evident in synovial biopsies displaying evidence of ectopic lymphoid-like structures (lymphoid-rich) and synovitis with a prominent mononuclear cell infiltrate (myeloid-rich) (Fig. 8b)29. In contrast, PTPN22 displayed a more uniform pattern of expression within the inflamed synovium and showed no correlation with lymphocyte markers or the type of synovial pathology (Fig. 8a,b). To establish a possible link between PTPN2 and the synovial expression of genes controlled by IL-6 in activated or memory CD4+ T cells PTPN2 was compared against the synovial expression of IL21, IL17A, CD274 and SOCS1. Analysis of synovial RNA-seq datasets showed a correlation between PTPN2 and IL21, IL17A, CD274 and SOCS1 (Fig. 8c). This relationship was particularly evident in both lymphoid-rich and myeloid-rich synovitis (Fig. 8c). No clear correlation was observed between PTPN22 and these inflammatory markers (Fig. 8c). Thus, in human synovial pathology, PTPN2 associates with the involvement of lymphocytes in the disease process and corresponds with the expression of several inflammatory mediators linked to the regulation of STAT1 by PTP enzymes.
Figure 8. Association of PTPN2 with rheumatoid arthritis.
(a) Correlations of PTPN2 and PTPN22 with lymphocyte cell markers CD3 and CD20 (left). (b) Distribution of PTPN2 and PTPN22 in patients stratified according synovial pathology (lymphoid – blue, myeloid – red and fibroid – green) (right). (c) Pearson correlations of synovium PTPN2 and PTPN22 with inflammatory markers including IL21, IL17A, CD274 and SOCS1 in lymphoid (blue), myeloid (red) and fibroid (green) phenotypes. P values were adjusted using false discovery rate (FDR) correction (Benjamini-Hochberg).
Discussion
Through analysis of Jak-STAT signalling in CD4+ TN and TEM cells we identified that protein tyrosine phosphatases induced as a response to CD4+ TN cell activation altered the transcriptional output of IL 6 in CD4+ TEM cells. Our investigation showed that inhibition of STAT1 phosphorylation by PTPN2 affected the expression of certain STAT-regulated target genes in activated or memory CD4+ T cells. Thus, protein tyrosine phosphatases have the capacity to modify the way particular T cell subsets sense and interpret common cytokine cues. Whilst the study focussed on the biology of IL 6, this mechanism may also shape the transcriptional output of other lymphokines in CD4+ T cells.
Protein phosphatases including dual specificity protein phosphatases (DUSP) and protein tyrosine phosphatases (PTP) can regulate Jak-STAT signaling30, 31. For example, PTPN2 and PTPN11 control the phosphorylation of STAT1 in fibroblasts32, 33. We showed that protein tyrosine phosphatases restrained IL 6 signaling through STAT1 in activated and memory CD4+ T cells. Transcriptional profiling of PTP expression in CD4+ T cells identified several candidate enzymes that were induced following CD4+ TN activation, including PTPN2, PTPN22 and DUSP2. Because DUSP2 inhibits signaling through STAT3 and restricts TH17 differentiation30, we assessed whether PTPN2 and PTPN22 affected STAT1 phosphorylation. PTPN2 and PTPN22 inhibited STAT1 activity, but had a less obvious impact on STAT3 activity. This observation might indicate a physical interaction between these PTP and STAT1.
PTPN2 and PTPN22 control various lymphocyte responses, and individuals with genetic polymorphisms in PTPN2 or PTPN22 frequently show increased susceptibility to autoimmune disease34, 35. The ability of PTPN2 and PTPN22 to control Jak-STAT signaling may contribute to these outcomes through the control of immune activation, tolerance and autoimmunity. Our data showed that synovial PTPN2 was highly expressed in lymphoid-rich synovitis. This form of joint pathology is defined by the presence of functional ectopic lymphoid aggregates within the inflamed synovium36. Significantly, PTPN2 has been linked with the regulation of follicular TH cells and the activation of T and B cell responses37, 38, 39, 40. Our investigation showed that PTPN2 controlled the expression of genes commonly associated with ectopic lymphoid-like structures. For example, PTPN2-control of STAT1 phosphorylation was shown to affect the transactivation of inflammatory cytokines (e.g., IL-17A, IL 21), transcription factors (e.g., Bcl6), immune checkpoint regulators (e.g., CD274) and homeostatic chemokine receptors (e.g., CXCR4, CXCR5) involved in the activity or spatial organization of lymphoid aggregates36. The gene signature identified through our screen may therefore predict the efficacy of adoptive immunotherapy and vaccination strategies, or response to biological drug therapies.
ChIP-seq of STAT1 and STAT3 in IL-6-treated CD4+ TN and TEM cells showed that both transcription factors bound to consensus motifs for STAT proteins (e.g., ISRE, GAS), and sequences specific for other transcription factors (e.g., SP1-like proteins)41, 42, 43, 44. While the binding of STATs to these genomic sites requires further analysis, our results suggested that the induction of PTPN2 in activated CD4+ T cells affected the expression of STAT-regulated genes controlled by P300 enhancer elements. Many of the genes associated with these enhancers were induced by IL-6 in CD4+ TEM cells and included genes commonly associated with TH1, TH2, TH17 or TFH cells27, 28. These activities fit with the capacity of IL-6 to govern CD4+ T cell memory4, 5, 45, 46. For example, IL-6 renders antigen-specific T cells refractory to suppression by regulatory T cells56, 57. However, IL-6 signaling is not critical for the generation or maintenance of CD4+ memory cells4, 6. Instead, our data revealed that IL-6 promotes the effector or functional characteristics of CD4+ TEM cells. This contrasts with the activities of IL-23, which regulates memory recall through the control of cell-cycle progression and proliferation7, 47, 48. Thus, PTPN2 control of STAT1 may support CD4+ T cell memory responses by shaping effector memory functions or prolonging lymphocyte survival. Such findings may be relevant to our understanding of how T cells become released from anergy and might explain how T cells become directed down a commitment pathway as a response to specific TCR antigens39, 49, 50.
Materials and methods
Recombinant Cytokines
Recombinant mouse IL-6 (IL-6), IL-27, IL-10, IL-7 and IFNγ were purchased from R&D Systems. The IL-6-sIL-6R fusion protein HDS (Mw: 63.5kDa) was expressed in CHO cells and purified through a partnership with the CRO Biovian OY (Turku, Finland). HDS was engineered by coupling the entire coding sequence (amino acid residues 1-364) for the differentially spliced variant of human IL-6R (containing the unique COOH-terminal amino acid sequence: GSRRRGSCGL) to IL-6 (amino acid residues 29-212) via a flexible glycine-serine rich linker sequence (single amino sequence: GGGGSGGGGSLE)8.
Antibodies
Mouse specific antibodies against CD3ε/γ (17A2; Biolegend), CD4 (RM4-5; eBioscience), CD25 (PC61.5; eBioscience), CD44 (IM7; BD Biosciences), CD62L (MEL-14; Life Technologies), CD126 (D7715A7; eBioscience), CD127 (25-1271-82; eBioscience),βTCR (H57-597), gp130 (125623; R&D Systems), IFNγ (XMG1.2; eBioscience), IL-4 (11B11; eBioscience), IL-17A (TC11-18H10.1; Biolegend), IL-21 (Recombinant mouse IL-21R Fc Chimera protein; R&D and IL-21 receptor antibody; Jackson Immuno Research) and PTPN2 (AF1930; R&D) were used. For detection of human antigens, we used antibodies specific to CD3 (UCHT1; BioLegend), CD4 (RPA-T4; eBioscience), CD45RA (HI100; BioLegend), CD45RO (UCHL1; BioLegend), CD62L (DREG-56; BD Biosciences), CD197 (CCR7; G043H7; BioLegend). Human and mouse cross-specific antibodies to pY STAT1 (pY701; 4a), pY-STAT3 (pY705; 4/P-STAT3), pS-STAT1 (pS727; K51-856) and pS STAT3 (pS727, 49/p-Stat3) were from BD Biosciences.
Mice
Inbred wild type C57BL/6 male mice were purchased from Charles River UK. C57BL/6 IL-6 receptor deficient mice (Cd126 -/-) mice have been described previously and were bred under approved UK Home Office guidelines in Cardiff University7. Ptpn2 fl/fl , Lck-Cre:Ptpn2 fl/fl, and Ptpn22 -/- mice were bred and housed at the Peter MacCallum Cancer Centre (Melbourne, Australia). All mice were 8-12 weeks of age. For T cell stimulation experiments eight-week-old male Lck-Cre;Ptpn2fl/fl mice and Ptpn2fl/fl littermate controls were used38. All procedures were performed in accordance with the NHMRC Australian Code of Practice for the Care and Use of Animals, and approved by the Peter MacCallum Animal Ethics and Experimentation Committee (Ethics number: AEEC 570). Antigen-induced arthritis was performed under the UK Home Office-approved project licences PPL 30/2928 and PB3E4EE13 as previously described51. Briefly, mice were immunized (s.c.) with 100 μl mBSA (1 mg/ml emulsified in Complete Freund’s Adjuvant; CFA) and 160 ng Bordetella pertussis toxin (i.p.) (all from Sigma-Aldrich). Mice were administered with mBSA and CFA (s.c.) one week later. Inflammatory arthritis was triggered 21 days following the initial immunization by intra-articular administration of mBSA (10 μl; 10 mg/ml) into the right knee joint. Animals were monitored daily for wellbeing and clinical signs of arthritis, and killed at indicated time points for evaluation of joint-infiltrating T cells by flow cytometry and immunofluorescence. For flow cytometric analysis of synovial CD4+ T cells, inflamed synovium was first dissected and digested in Collagenase type IV (37°C, 1 hour) before passing through a 40μm cell strainer to generate single cell suspensions.
Human synovial samples
Synovial samples were acquired through a minimally invasive ultrasound-guided synovial biopsy (see Reference52) from 87 patients presenting with early rheumatoid arthritis (RA) naïve to therapy from the Pathobiology of Early Arthritis Cohort (PEAC). Ethical approval was granted by the King’s College Hospital Research Ethics Committee (REC 05/Q0703/198). Paraffin embedded sections (3μm) of each biopsy was stained with haematoxylin and eosin. Immune cell infiltration was determined in sequentially cut sections by staining for B-cells (CD20), T cells (CD3), macrophages (CD68) and plasma cells (CD138) as previously reported, categorising samples into Lympho-myeloid, Diffuse-Myeloid and Pauci-immune Fibroid pathotypes53.
CD4+ T cell cultures
Murine CD4+ T cells were enriched by negative magnetic selection (Miltenyi Biotec) before purification of naïve (CD4+CD25−CD44loCD62LhiCD127hi), central memory (CD4+CD25−CD44hiCD62LhiCD127hi), effector (CD4+CD25−CD44loCD62LloCD127lo-int) or effector memory (CD4+CD25−CD44hiCD62LloCD127int-hi) T cells using a BD FACS ARIA II (BD Biosciences). T cell subset purity was >98%. Naïve CD4+ T cells were cultured in RPMI-1640 supplemented with 10% (v/v) FCS, 2mM L-glutamine, 100U/ml penicillin, 100μg/ml streptomycin, 1mM sodium pyruvate and 50μM 2-mercaptoethanol. 1 x 105 CD4 T cells were activated by plate bound anti-CD3 (1μg/ml; 145-2C11, R&D Systems) and soluble anti-CD28 (5μg/ml; 37.51, eBioscience). Where indicated, CD4+ T cells were rested for 48 hours in the absence of stimulatory antibodies or cytokines (see Supplementary Fig. 2a). CD4+ T cells from the inguinal lymph nodes of mBSA-immunized and non-immunized mice were derived using a CD4+ T cell isolation kit (Miltenyi Biotec) and treated with IL-6 for 30 min with anti-CD3/CD28 stimulation. Human peripheral blood mononuclear cells (PBMC) were isolated from fresh whole blood as previously described54. Naïve (CD3+CD4+CD45ROloCD62LhiCCR7hi), central memory (CD3+CD4+CD45ROhiCD62LhiCCR7hi), effector (CD3+CD4+CD45ROloCD62LloCCR7lo) or effector memory (CD3+CD4+CD45ROhiCD62LloCCR7lo) CD4+ T cells were then purified using a BD FACS ARIA II (BD Biosciences). To investigate the involvement of protein tyrosine phosphatases CD4 T cells were pre-treated (5 min) with 5mM sodium orthovanadate (New England BioLabs (UK) Ltd) prior to subsequent stimulation.
Histological analysis
Formalin-fixed paraffin-embedded knee joints from AIA-challenged mice were prepared for immunofluorescent and immunohistochemical detection of antigens as described previously51. For immunofluorescence, sections were rehydrated and antigen retrieval performed in 10mM sodium citrate buffer containing 0.05% (v:v) Tween 20 (95°C, 40 min). Sections were incubated with 10% (v:v) goat or swine serum appropriate to the secondary antibody. Cells positive for CD3 and intracellular phospho-STAT1 and STAT3 were detected using CD3 (A0452, Dako), and pY-STAT1 (Tyr701; 58D6) or pY-STAT3 (Tyr705; D3A7) specific antibodies from Cell Signaling Technologies. For CD3 staining, primary antibody detection was performed using biotinylated swine anti-rabbit IgG (E0431, Dako) with streptavidin-APC (BD Biosciences). For pY-STAT1 and pY-STAT3 detection a secondary rabbit anti-goat IgG Alexa Fluor 488 antibody (Life Technologies) was used. Slides were mounted with Prolong Gold Antifade with DAPI nuclear counterstain (Invitrogen). Images were collected using a Zeiss Apotome microscope and analyzed using ImageJ software. For immunohistochemistry, antigens were detected in paraffin sections using antibodies against CD3 (A0452, Dako), pY-STAT1 (Tyr701; 58D6) and PTPN2 (AF1930, R&D Systems). Antigen retrieval was performed as above, and endogenous peroxidase activity blocked using 3% (v:v) H2O2. Antibody labelling was detected using biotinylated secondary antibodies (Dako, E0431), the Vectastain ABC kit and diaminobenzidine (Vector Laboratories). Sections were counterstained with haematoxylin. Images were collected using Leica DM 2000 Led and quantification of staining performed using the Leica QWin microscope imaging software.
Flow cytometry
Analysis was performed as described previously7, 8, 15. For the intracellular detection of STAT1 and STAT3 phosphorylation, purified CD4+ T cells were fixed in 2% (w:v) paraformaldehyde for 15 min at 37°C, followed by permeabilization in 90% (v:v) methanol at -20°C for 3hrs. Cells were stained for CD4 and phosphorylated STAT1, STAT3 7. To evaluate effector cytokine production, CD4+ T cells were stimulated with 50ng PMA (phorbol 12-Myristate 13-Acetate), 500ng ionomycin and 3μM monesin for 4 hours prior to flow cytometric analysis7, 8, 15. Cells were acquired on a CyAn ADP analyzer (Beckman-Coulter) and analysed using Summit (software v4.3, Beckman-Coulter) or FlowJo 10 (TreeStar). For imaging flow cytometry, cells were resuspended in 100 μl of PBS and acquired using the ImageStream imaging flow cytometer (Amnis). For co-localization analysis ImageStream software IDEAS (Amnis) was used.
RNA purification and Q-PCR
For quantitative real-time PCR (Q-PCR), and Affymetrix gene chip analysis, total RNA was extracted from purified or cultured CD4 T cells using the RNeasy Mini Kit (Qiagen) and QIAshredders (Qiagen). Contaminating genomic DNA was removed by on-column DNase digestion (Qiagen). RNA was converted to cDNA using the High Capacity cDNA Reverse Transcription Kit with RNase Inhibitor kit (Life Technologies). Gene expression was determined by Q-PCR8, 55 using the QuantStudio 12K Flex Real-Time PCR system and the following TaqMan probes from Thermofisher: Ahr (Mm00478932_m1), Bcl3 (Mm00504306_m1), Bcl6 (Mm01342164_m1), Il10 (Mm00439614_m1), Il21 (Mm00517640_m1), Irf1 (Mm01288580_m1), Socs3 (Mm00545913_s1), Stat3 (Mm01219775_m1), Pim1 (Mm00435712_m1) and Actb (Mm01205647_g1) as a housekeeping gene. Relative mRNA expression was determined by the comparative cycle threshold (CT) method and normalised to the gene Actb.
Affymetrix microarray and transcriptomic analysis
Purification of high quality RNA (RNA integrity number >8.5) was confirmed using Agilent RNA Nano microfluidic chips using a 2100 Bioanalyzer Instrument (Agilent Technologies). Expression profiling was performed in triplicate using Affymetrix Mouse GeneChip® 2.0ST microarrays (Affymetrix). Single-stranded cDNA was synthesized using the Ambion® WT (Whole Transcript) Expression Kit with the Affymetrix® Genechip® Poly-A RNA Control Kit and Terminal Labelling kit. Arrays were scanned using the GeneChip Scanner 3000 7G (Affymetrix). Raw Affymetrix data files (CEL files) were imported into an in-house analysis pipeline written in R (version 3.1.1) using Bioconductor packages, namely limma, affy and oligo56, 57, 58, 59. Data were background corrected, log2 transformed and quantile normalized using the oligo package (RMA) “best practice”. Differentially expressed genes and transcripts were identified using the limma package “best practice” workflow and P-values were corrected by multiple testing using Benjamini-Hochberg (false discovery rate)57.
Bespoke coding (Perl) (Code available on request) was used to unite data over all conditions. To identify differentially expressed genes over all experiments, we selected the genes that were classified as having altered expression (either decreasing or decreasing) by a difference of 1.5-fold or greater, with a significant value P≤0.05 and a minimal expression value of 150 relative intensity units. Only transcripts fulfilling all these selection criteria in three independent microarray experiments were included in the analysis. Files were also created in the input format required for molecular and pathway analysis using Metacore integrated software suite (Thomson Reuters)59 and Ingenuity Pathway Analysis (IPA, http://www.ingenuity.com/products/ipa). Transcriptome matrix visualization and hierarchical clustering were performed using Morpheus software (https://software.broadinstitute.org/morpheus/). Circos plot were obtained using the Circos software (http://circos.ca/software/)60. Networks were visualization and analysed using the open sourced program Gephi (0.9.1) (https://gephi.org/). Microarray data have been deposited in ArrayExpress under Accession code E-MTAB-7682.
RNA-sequencing
Open access datasets from IL-6 treated Stat1-/- and Stat3-/- T cells (GSE65621) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) and aligned and processed using an in-house bioinformatic pipeline. Briefly, RNA-seq single-end fastq files were mapped to mouse assembly GRCm38 using STAR61. Transcript counts were produced with FeatureCounts62 and data normalised using the Bioconductor package, DeSeq263 obtaining gene expression values as FPKM (Fragments Per Kilobase Million).
For the human samples, RNA from homogenised synovial tissue was extracted in Trizol. 1μg total RNA was used as input material for library preparation using TruSeq RNA Sample Preparation Kit v2 (Illumina). Generated libraries were amplified with 10 cycles of PCR. Library size was confirmed using 2200 TapeStation and High Sensitivity D1K screen tape (Agilent Technologies) and concentration was determined by Q-PCR based method using Library quantification kit (KAPA). Libraries were multiplexed (five per lane) and sequenced on Illumina HiSeq2500 (Illumina) to generate 50 million paired-end 75 base pair reads. Transcript abundance was derived using Kallisto v0.43.0 with GENCODE v24/GRCh38 as reference64. Transcript abundances were summarised over transcript isoforms using Bioconductor package tximport 1.4.0. Imported abundances were processed using DESeq2 1.14.1 and transformed as regularised log expression (RLE). Statistical analysis of gene-gene correlations was performed using Pearson correlation. P values were adjusted using false discovery rate (FDR) correction (Benjamini-Hochberg). RNA-seq data have been deposited in ArrayExpress under Accession code E-MTAB-6141.
Chromatin Immunoprecipitation (ChIP)-seq
STAT1 and STAT3 chromatin Immunoprecipitation was performed as previously described20. Briefly, 1x107 naïve (TN) and effector memory (TEM) CD4+ T cells were activated for 1h with 20ng/ml IL-6 in the presence of antibodies against CD3 and CD28 (as described earlier). Genomic DNA was extracted, cross-linked and fragmented by sonication prior to overnight incubation with 5ug of anti-STAT1 (sc-592, Santa Cruz Biotechnology) or anti-STAT3 (sc-482, Santa Cruz Biotechnology) antibodies. The quality of the immunoprecipitation was confirmed by ChIP-qPCR for Irf1 and Socs3. To minimize systematic biases in the downstream data, an input reference control sample (chromatin taken before ChIP) was used to correct for genomic copy number variations, sonication-induced fragmentation bias, and chromatin accessibility. ChIP-seq libraries for Ion Torrent sequencing were prepared according to manufacturer’s instructions (# 4473623, Ion ChIP-Seq Library Preparation on the Ion Proton TM System). Briefly, DNA fragments were end-repaired and ligated to ion-compatible adapters. Libraries were amplified and size-selected for insert lengths of approximately 100-250bp. Between 40M to 70M reads were obtained for each sample and mapped to Murine Genome Build GRCm38 (mm10) using the Ion Proton recommended mapper, Bowtie265, 66. Reads were removed where mapping quality was less than q20 (phred score) and peaks called using HOMER (Hypergeometric Optimization of Motif EnRichment). To identify putative peaks in both ChIP and input, we first used HOMER findPeaks with a False Discovery Rate (FDR) value of 0.05. To identify sample peaks in the context of input, we then used HOMER findPeaks with the default parameters (Fold Change > 4-fold, P-value < 0.0001). Peaks were visualised using Integrative Genomics Viewer (IGV 2.3.8867). Available p300 ChIP-seq fastq files from CD4+ T cells, TH1, TH2 (GSE40463) and TH17 (GSE60482) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/) and aligned and processed using an in-house pipeline. Reads were mapped to the same mm10 assembly using BWA. All reads with a mapping quality less than q20 were removed. HOMER isoftware was used to locate P300 enhancer elements (using the Homer option, –style super) and parameters Fold Change > 2-fold, P value < 0.0001. To align STAT1 or STAT3 peaks with SE regions, overlapping loci identified by STAT1 and STAT3 ChIP-seq and p300 ChIP-seq were identified using bedtools (http://bedtools.readthedocs.io/en/latest/)68. ChIP-Seq data have been deposited in ArrayExpress under Accession code E-MTAB-6273.
ChIP-qPCR
To validate STAT-binding to promoter regions, Taqman custom assays were designed (see Supplementary Table 2 for oligonucleotide primer sequences) and qPCR performed using a QuantStudio 12K Flex Real-Time PCR System. For analysis of SP1, chromatin immunoprecipitation was conducted as previously described using 5ug of SP1 antibody (#17-601, Millipore) or isotype specific IgG control. Analysis by qPCR used oligonucleotide primer sequences to the promoter regions of Irf1, Socs3, Stat3, Cd274, Il4ra, Junb and Socs1 (Supplementary Table 2). Specific enrichment was normalised by subtracting the IgG control values from those derived for the input and antibody specific immunoprecipitation samples. Value were expressed as 2^ΔCT.
Motif finding
MEME Suite 4.11.2 software was used to discover de novo enriched DNA consensus sequences present in peaks identified within the STAT1 or STAT3 ChIP-seq datasets. All sequence predictions derived from MEME based on the interaction of known transcription factors with target DNA sequences where substantiated using STAMP69 (http://benoslab.pitt.edu/stamp/) and JASPAR database (http://jaspar.genereg.net/cgi-bin/jaspar_db.pl)70.
Statistics
To determine the statistical significance of differences between data sets, Two-tailed unpaired Student’s t-test were performed when two populations were compared. One-way ANOVA followed Tukey’s comparison test was used for multiple comparisons, unless otherwise specified, conducted using GraphPad Prism 5 (GraphPad Software). Statistical significance is also highlighted with the following notations: *P<0.05; **P<0.01; ***P<0.001, ****P<0.0001. P ≤ 0.05 was considered significantly different.
Reporting Summary
Further information on experimental design is available in the Life Sciences Reporting Summary linked to this article.
Supplementary Material
Acknowledgements
This manuscript is dedicated in fond memory of J. Uceda Fernandez, a dearly loved friend and colleague who was tragically taken from us on the 29th August 2018. Remembered forever.
Versus Arthritis (Reference 20770, 19796 awarded to SAJ, NMW & GWJ), Hoffmann la Roche (to SAJ), Kidney Research UK (to SAJ) and the National Health and Medical Research Council of Australia (to TT) provided grant support for this project. GWJ is recipient of a Versus Arthritis Career Development Fellowship (Reference 20305). JUF held a la Caixa PhD Studentship administered through the British Council, and BCC is supported by a PhD studentship from the Systems Immunity University Research Institute at Cardiff. JL is recipient of a Rutherford Fellowship Grant. Analysis of synovial tissues was conducted with support from the Medical Research Council (to CJP; Reference 36661) and the Versus Arthritis funded Experimental Arthritis Treatment Centre (to CJP; Reference 20022). PRT is recipient of a Wellcome Trust Investigator Award (107964/Z/15/Z) and receives funding through the UK Dementia Research Institute. Bioinformatic analysis was developed with support from the Systems Immunity University Research Institute in Cardiff. The authors would like to thank Yuka Kanno, Chris Hunter, Stephen Turner, Toshi Nakayama, Magdalena Czubala, Rachel Errington, Atsushi Onodera, Kiyoshi Hirahara and John O’Shea for their advice, constructive opinions and kind support.
Footnotes
Author contributions
SAJ, GWJ, ACF and JPT wrote the text and prepared the figures for the paper. All authors reviewed and approved the final manuscript draft. SAJ, GWJ, TT, NMW, CP, PRT, CJP, JPT, ACF designed the study. BCC, RA, ML, MJT, BS, ACF, JPT and NMW conducted bioinformatic, biostatistical, and molecular pathway analyses. Laboratory based studies were performed by JPT, ACF, FW, MJT, XL, JUF, ADS, JL, DH and DM.
Competing interests
SAJ has received funding support from Hoffman-La Roche, GlaxoSmithKline, Ferring Pharmaceuticals and NovImmune SA, and during the last 5 years he has acted as an advisory consultant for Roche, Chugai Pharmaceuticals, NovImmune SA, Genentech, Sanofi Regeneron, Johnson & Johnson, Janssen Pharmaceuticals, Eleven Biotherapeutics.
Data Availability
Microarray, ChiP-seq and RNA-seq data have been deposited in ArrayExpress under Accession code E MTAB-7682, E-MTAB-6273 and E-MTAB-6141, respectively. Available p300 ChIP-seq fastq files from CD4 T cells, Th1, Th2 (GSE40463) and Th17 (GSE60482), and Stat1-/- and Stat3-/- T cells (GSE65621) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/). Access to interactive data sets can be found at www.jones-cytokinelab.co.uk (see relevant Figure Legends for additional information).
References
- 1.MacLeod MK, Kappler JW, Marrack P. Memory CD4 T cells: generation, reactivation and re-assignment. Immunology. 2010;130:10–15. doi: 10.1111/j.1365-2567.2010.03260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Shea JJ, et al. The JAK-STAT pathway: impact on human disease and therapeutic intervention. Annu Rev Med. 2015;66:311–328. doi: 10.1146/annurev-med-051113-024537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hunter CA, Jones SA. IL-6 as a keystone cytokine in health and disease. Nat Immunol. 2015;16:448–457. doi: 10.1038/ni.3153. [DOI] [PubMed] [Google Scholar]
- 4.Nish SA, et al. T cell-intrinsic role of IL-6 signaling in primary and memory responses. Elife. 2014;3:e01949. doi: 10.7554/eLife.01949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Longhi MP, et al. Interleukin-6 is crucial for recall of influenza-specific memory CD4 T cells. PLoS Pathog. 2008;4:e1000006. doi: 10.1371/journal.ppat.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Strutt TM, et al. Direct IL-6 Signals Maximize Protective Secondary CD4 T Cell Responses against Influenza. The Journal of Immunology. 2016 doi: 10.4049/jimmunol.1600033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones GW, et al. Loss of CD4+ T cell IL-6R expression during inflammation underlines a role for IL-6 trans signaling in the local maintenance of Th17 cells. J Immunol. 2010;184:2130–2139. doi: 10.4049/jimmunol.0901528. [DOI] [PubMed] [Google Scholar]
- 8.Fielding CA, et al. Interleukin-6 signaling drives fibrosis in unresolved inflammation. Immunity. 2014;40:40–50. doi: 10.1016/j.immuni.2013.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harker JA, Lewis GM, Mack L, Zuniga EI. Late interleukin-6 escalates T follicular helper cell responses and controls a chronic viral infection. Science. 2011;334:825–829. doi: 10.1126/science.1208421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Briso EM, Dienz O, Rincon M. Cutting edge: soluble IL-6R is produced by IL-6R ectodomain shedding in activated CD4 T cells. J Immunol. 2008;180:7102–7106. doi: 10.4049/jimmunol.180.11.7102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Teague TK, et al. Activation-induced inhibition of interleukin 6-mediated T cell survival and signal transducer and activator of transcription 1 signaling. J Exp Med. 2000;191:915–926. doi: 10.1084/jem.191.6.915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hong C, et al. Interleukin-6 expands homeostatic space for peripheral T cells. Cytokine. 2013;64:532–540. doi: 10.1016/j.cyto.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chtanova T, et al. Identification of T cell-restricted genes, and signatures for different T cell responses, using a comprehensive collection of microarray datasets. J Immunol. 2005;175:7837–7847. doi: 10.4049/jimmunol.175.12.7837. [DOI] [PubMed] [Google Scholar]
- 14.Curnow SJ, et al. Inhibition of T cell apoptosis in the aqueous humor of patients with uveitis by IL-6/soluble IL-6 receptor trans-signaling. J Immunol. 2004;173:5290–5297. doi: 10.4049/jimmunol.173.8.5290. [DOI] [PubMed] [Google Scholar]
- 15.Nowell MA, et al. Therapeutic targeting of IL-6 trans signaling counteracts STAT3 control of experimental inflammatory arthritis. J Immunol. 2009;182:613–622. doi: 10.4049/jimmunol.182.1.613. [DOI] [PubMed] [Google Scholar]
- 16.Atreya R, et al. Blockade of interleukin 6 trans signaling suppresses T-cell resistance against apoptosis in chronic intestinal inflammation: evidence in crohn disease and experimental colitis in vivo. Nat Med. 2000;6:583–588. doi: 10.1038/75068. [DOI] [PubMed] [Google Scholar]
- 17.Liao W, Lin JX, Wang L, Li P, Leonard WJ. Modulation of cytokine receptors by IL-2 broadly regulates differentiation into helper T cell lineages. Nat Immunol. 2011;12:551–559. doi: 10.1038/ni.2030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Villarino AV, Kanno Y, O’Shea JJ. Mechanisms and consequences of Jak-STAT signaling in the immune system. Nat Immunol. 2017;18:374–384. doi: 10.1038/ni.3691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Costa-Pereira AP, et al. Mutational switch of an IL-6 response to an interferon-gamma-like response. Proc Natl Acad Sci U S A. 2002;99:8043–8047. doi: 10.1073/pnas.122236099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hirahara K, et al. Asymmetric Action of STAT Transcription Factors Drives Transcriptional Outputs and Cytokine Specificity. Immunity. 2015;42:877–889. doi: 10.1016/j.immuni.2015.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Peters A, et al. IL-27 Induces Th17 Differentiation in the Absence of STAT1 Signaling. J Immunol. 2015;195:4144–4153. doi: 10.4049/jimmunol.1302246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jones GW, et al. Exacerbated inflammatory arthritis in response to hyperactive gp130 signalling is independent of IL-17A. Ann Rheum Dis. 2013;72:1738–1742. doi: 10.1136/annrheumdis-2013-203771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson AE, et al. IL-6-driven STAT signalling in circulating CD4+ lymphocytes is a marker for early anticitrullinated peptide antibody-negative rheumatoid arthritis. Ann Rheum Dis. 2016;75:466–473. doi: 10.1136/annrheumdis-2014-205850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pratt AG, et al. A CD4 T cell gene signature for early rheumatoid arthritis implicates interleukin 6-mediated STAT3 signalling, particularly in anti-citrullinated peptide antibody-negative disease. Ann Rheum Dis. 2012;71:1374–1381. doi: 10.1136/annrheumdis-2011-200968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hnisz D, et al. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Whyte WA, et al. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vahedi G, et al. Super-enhancers delineate disease-associated regulatory nodes in T cells. Nature. 2015;520:558–562. doi: 10.1038/nature14154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ciofani M, et al. A validated regulatory network for Th17 cell specification. Cell. 2012;151:289–303. doi: 10.1016/j.cell.2012.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Humby F, et al. Evaluation of Minimally Invasive, Ultrasound-guided Synovial Biopsy Techniques by the OMERACT Filter--Determining Validation Requirements. J Rheumatol. 2016;43:208–213. doi: 10.3899/jrheum.141199. [DOI] [PubMed] [Google Scholar]
- 30.Lu D, et al. The phosphatase DUSP2 controls the activity of the transcription activator STAT3 and regulates T17 differentiation. Nat Immunol. 2015 doi: 10.1038/ni.3278. [DOI] [PubMed] [Google Scholar]
- 31.Bohmer FD, Friedrich K. Protein tyrosine phosphatases as wardens of STAT signaling. JAKSTAT. 2014;3:e28087. doi: 10.4161/jkst.28087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.ten Hoeve J, et al. Identification of a nuclear Stat1 protein tyrosine phosphatase. Mol Cell Biol. 2002;22:5662–5668. doi: 10.1128/MCB.22.16.5662-5668.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu TR, et al. SHP-2 is a dual-specificity phosphatase involved in Stat1 dephosphorylation at both tyrosine and serine residues in nuclei. J Biol Chem. 2002;277:47572–47580. doi: 10.1074/jbc.M207536200. [DOI] [PubMed] [Google Scholar]
- 34.Hinks A, et al. Association between the PTPN22 gene and rheumatoid arthritis and juvenile idiopathic arthritis in a UK population: further support that PTPN22 is an autoimmunity gene. Arthritis Rheum. 2005;52:1694–1699. doi: 10.1002/art.21049. [DOI] [PubMed] [Google Scholar]
- 35.Sharp RC, Abdulrahim M, Naser ES, Naser SA. Genetic Variations of PTPN2 and PTPN22: Role in the Pathogenesis of Type 1 Diabetes and Crohn’s Disease. Front Cell Infect Microbiol. 2015;5:95. doi: 10.3389/fcimb.2015.00095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pitzalis C, Jones GW, Bombardieri M, Jones SA. Ectopic lymphoid-like structures in infection, cancer and autoimmunity. Nat Rev Immunol. 2014;14:447–462. doi: 10.1038/nri3700. [DOI] [PubMed] [Google Scholar]
- 37.Wiede F, Sacirbegovic F, Leong YA, Yu D, Tiganis T. PTPN2-deficiency exacerbates T follicular helper cell and B cell responses and promotes the development of autoimmunity. J Autoimmun. 2017;76:85–100. doi: 10.1016/j.jaut.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 38.Wiede F, et al. T cell protein tyrosine phosphatase attenuates T cell signaling to maintain tolerance in mice. J Clin Invest. 2011;121:4758–4774. doi: 10.1172/JCI59492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Salmond RJ, Brownlie RJ, Morrison VL, Zamoyska R. The tyrosine phosphatase PTPN22 discriminates weak self peptides from strong agonist TCR signals. Nat Immunol. 2014;15:875–883. doi: 10.1038/ni.2958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wiede F, La Gruta NL, Tiganis T. PTPN2 attenuates T-cell lymphopenia-induced proliferation. Nat Commun. 2014;5:3073. doi: 10.1038/ncomms4073. [DOI] [PubMed] [Google Scholar]
- 41.Look DC, Pelletier MR, Tidwell RM, Roswit WT, Holtzman MJ. Stat1 depends on transcriptional synergy with Sp1. J Biol Chem. 1995;270:30264–30267. doi: 10.1074/jbc.270.51.30264. [DOI] [PubMed] [Google Scholar]
- 42.Cantwell CA, Sterneck E, Johnson PF. Interleukin-6-specific activation of the C/EBPdelta gene in hepatocytes is mediated by Stat3 and Sp1. Mol Cell Biol. 1998;18:2108–2117. doi: 10.1128/mcb.18.4.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z, Jones S, Hagood JS, Fuentes NL, Fuller GM. STAT3 acts as a co-activator of glucocorticoid receptor signaling. J Biol Chem. 1997;272:30607–30610. doi: 10.1074/jbc.272.49.30607. [DOI] [PubMed] [Google Scholar]
- 44.Zhu M, John S, Berg M, Leonard WJ. Functional association of Nmi with Stat5 and Stat1 in IL-2-and IFNgamma-mediated signaling. Cell. 1999;96:121–130. doi: 10.1016/s0092-8674(00)80965-4. [DOI] [PubMed] [Google Scholar]
- 45.Kasahara Y, et al. Role of interleukin 6 for differential responsiveness of naive and memory CD4+ T cells in CD2-mediated activation. J Exp Med. 1990;172:1419–1424. doi: 10.1084/jem.172.5.1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beagley KW, et al. Peyer’s patch B cells with memory cell characteristics undergo terminal differentiation within 24 hours in response to interleukin-6. Cytokine. 1991;3:107–116. doi: 10.1016/1043-4666(91)90030-h. [DOI] [PubMed] [Google Scholar]
- 47.Lee YK, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haines CJ, et al. Autoimmune memory T helper 17 cell function and expansion are dependent on interleukin-23. Cell Rep. 2013;3:1378–1388. doi: 10.1016/j.celrep.2013.03.035. [DOI] [PubMed] [Google Scholar]
- 49.Mohrs M, Lacy DA, Locksley RM. Stat signals release activated naive Th cells from an anergic checkpoint. J Immunol. 2003;170:1870–1876. doi: 10.4049/jimmunol.170.4.1870. [DOI] [PubMed] [Google Scholar]
- 50.van Panhuys N, Klauschen F, Germain RN. T-cell-receptor-dependent signal intensity dominantly controls CD4(+) T cell polarization In Vivo. Immunity. 2014;41:63–74. doi: 10.1016/j.immuni.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jones GW, et al. Interleukin-27 inhibits ectopic lymphoid-like structure development in early inflammatory arthritis. J Exp Med. 2015;212:1793–1802. doi: 10.1084/jem.20132307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kelly S, et al. Ultrasound-guided synovial biopsy: a safe, well-tolerated and reliable technique for obtaining high-quality synovial tissue from both large and small joints in early arthritis patients. Ann Rheum Dis. 2015;74:611–617. doi: 10.1136/annrheumdis-2013-204603. [DOI] [PubMed] [Google Scholar]
- 53.Humby F, et al. Ectopic lymphoid structures support ongoing production of class-switched autoantibodies in rheumatoid synovium. PLoS medicine. 2009;6:e1. doi: 10.1371/journal.pmed.0060001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jones GW, et al. Naive and activated T cells display differential responsiveness to TL1A that affects Th17 generation, maintenance, and proliferation. FASEB J. 2011;25:409–419. doi: 10.1096/fj.10-166843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Greenhill CJ, et al. Interleukin-10 regulates the inflammasome-driven augmentation of inflammatory arthritis and joint destruction. Arthritis Res Ther. 2014;16:419. doi: 10.1186/s13075-014-0419-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy--analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 59.Carvalho BS, Irizarry RA. A framework for oligonucleotide microarray preprocessing. Bioinformatics. 2010;26:2363–2367. doi: 10.1093/bioinformatics/btq431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobin A, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30:923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 63.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nature biotechnology. 2016;34:525–527. doi: 10.1038/nbt.3519. [DOI] [PubMed] [Google Scholar]
- 65.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Robertson G, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 67.Robinson JT, et al. Integrative genomics viewer. Nature biotechnology. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253-258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mathelier A, et al. JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2016;44:D110-115. doi: 10.1093/nar/gkv1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Microarray, ChiP-seq and RNA-seq data have been deposited in ArrayExpress under Accession code E MTAB-7682, E-MTAB-6273 and E-MTAB-6141, respectively. Available p300 ChIP-seq fastq files from CD4 T cells, Th1, Th2 (GSE40463) and Th17 (GSE60482), and Stat1-/- and Stat3-/- T cells (GSE65621) were obtained from GEO (https://www.ncbi.nlm.nih.gov/geo/). Access to interactive data sets can be found at www.jones-cytokinelab.co.uk (see relevant Figure Legends for additional information).