Abstract
Membrane trafficking is an essential cellular process conserved across all eukaryotes, which regulates the uptake or release of macromolecules from cells, the composition of cellular membranes and organelle biogenesis. It influences numerous aspects of cellular organisation, dynamics and homeostasis, including nutrition, signalling and cell architecture. Not surprisingly, malfunction of membrane trafficking is linked to many serious genetic, metabolic and neurological disorders. It is also often hijacked during viral infection, enabling viruses to accomplish many of the main stages of their replication cycle, including entry into and egress from cells. The appropriation of membrane trafficking by viruses has been studied since the birth of cell biology and has helped elucidate how this integral cellular process functions. In this Review, we discuss some of the different strategies viruses use to manipulate and take over the membrane compartments of their hosts to promote their replication, assembly and egress.
Introduction
Viruses are obligate intracellular parasites that are completely dependent on their hosts for their continued survival, replication and spread. Consequently, they have evolved exquisite strategies to manipulate different cellular systems not only to replicate but also to get in and out of cells, while at the same time avoiding or suppressing the intrinsic defence mechanisms of the host. Every virus has their own unique cellular challenges, which vary depending on if they have an RNA or DNA genome, are enveloped or not and whether they replicate in the nucleus or cytoplasm. Although the precise mechanisms vary between different virus families, there are many common themes, including how viruses hijack and repurpose membrane organisation and trafficking, the topic of this Review.
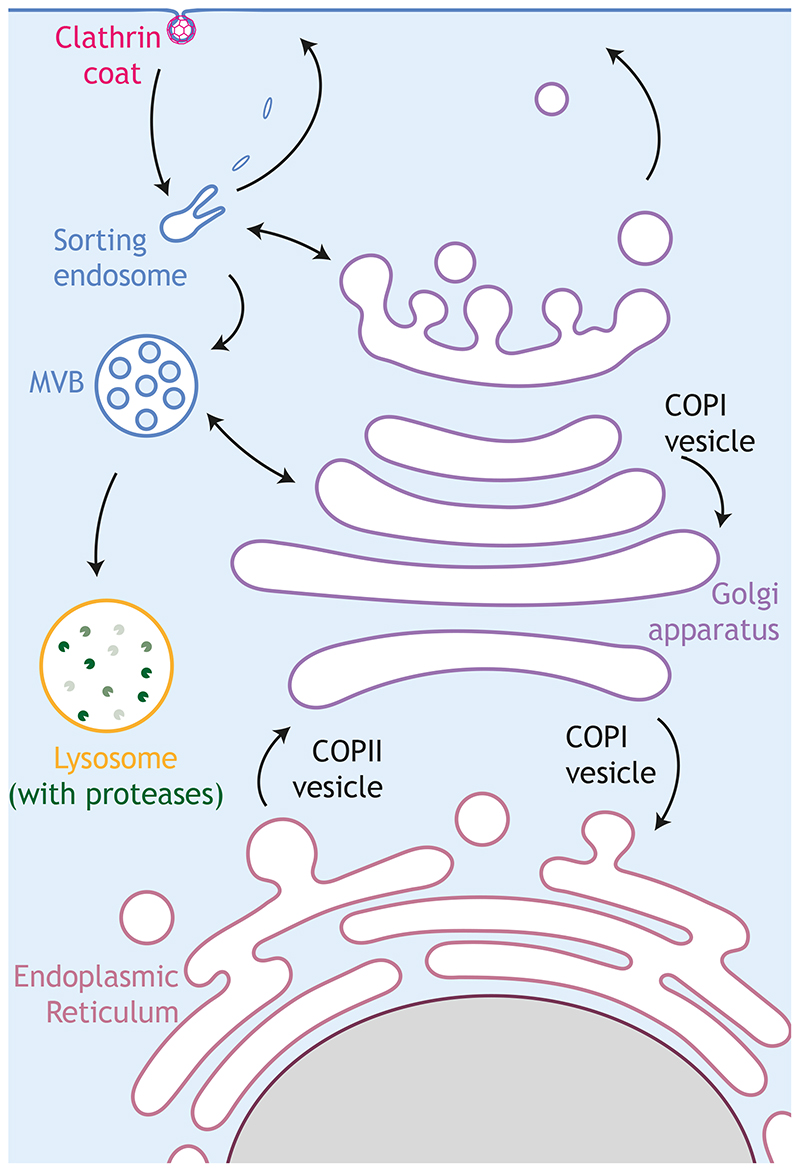
Membrane trafficking is a dynamic and versatile process consisting of the interconnected secretory and endocytic pathways (Fig. 1). In the secretory pathway, proteins destined to be sent out of the cell or delivered to the plasma membrane or intermediate membrane compartments are first translocated into the endoplasmic reticulum (ER) (Gemmer and Forster, 2020), where new lipids are also synthesised (Balla et al., 2020; Yang et al., 2018). ER cargoes are subsequently transported to the Golgi apparatus, which acts as a central transport hub, sorting them into distinct transport carriers in the trans-Golgi network (TGN), to ensure they are correctly delivered to their final destinations (Glick and Luini, 2011; Guo et al., 2014; Miller and Schekman, 2013; Pantazopoulou and Glick, 2019). In the endocytic pathway, extracellular or plasma-membrane bound cargoes are internalised into endosomes and sorted to be recycled back to the plasma membrane, sent to other destinations such as the Golgi or degraded in lysosomes (Grant and Donaldson, 2009; Kirchhausen et al., 2014). The identity of all these different membrane compartments is determined by their lipid composition and by Rab and Arf GTPases, which recruit specific effector proteins (Donaldson et al., 2016; Langemeyer et al., 2018; Mizuno-Yamasaki et al., 2012; Thomas and Fromme, 2020).
Figure 1. Main intracellular trafficking pathways.
Specialised regions of the endoplasmic reticulum (ER) serve as a departure station for newly synthesised cargo that travel to the Golgi apparatus in COPII vesicles. At the Golgi, cargo proteins mature and leave from the trans-Golgi network towards the plasma membrane or endosomes. COPI vesicles help Golgi organisation by trafficking within the Golgi and from the Golgi to the ER. The protein composition at the plasma membrane is also regulated by the endocytic pathway. Clathrin-coated vesicles bring internalised cargoes to sorting endosomes where these cargoes are either sorted in recycling endosomes en route for the plasma membrane, or in late endosomes/multivesicular bodies (MVBs), going towards proteases-containing lysosomes for degradation. Secretory and endocytic pathways are highly interconnected, which is represented by a bidirectional arrow.
Studies on the involvement and impact of viral infection on the regulation, dynamics and function of membrane trafficking exist for almost every viral family, although some of them are more studied than others (Ketter and Randall, 2019; Robinson et al., 2018). Understanding how viruses take advantage of membrane trafficking offers the promise of obtaining fundamental mechanistic insights into the regulation and function of this essential cellular process. It can also provide a critical understanding into the underlying cause of disease and help identify potential targets for therapeutic interventions. In this Review, we discuss examples from different viral families to illustrate how viruses subvert and reorganise membrane trafficking to suit their own needs.
Transport and access to replication sites
Enveloped viruses gain access to the cell by direct fusion of their envelope with the plasma membrane, which gives immediate access to the cytoplasm, or by piggybacking on cellular internalisation pathways (Helenius, 2018; Mercer et al., 2020; Yamauchi and Helenius, 2013). In the second strategy, the viral capsid or genome is subsequently released into the cytosol by fusion of the viral envelope with the limiting endosomal membrane, in a process driven by viral fusion proteins (Kielian, 2014). In the case of non-enveloped viruses, after endocytosis, cytoplasmic access is achieved by the disruption of the endosomal membrane (Daussy and Wodrich, 2020; Moyer and Nemerow, 2011).
Once in the cytoplasm, the capsid or genome often needs to be transported to a particular cellular location to initiate replication (Fig. 2). In general, RNA viruses replicate in the cytoplasm, frequently in specialised structures assembled on the surface of membrane-bound compartments (Miller and Krijnse-Locker, 2008; Neufeldt et al., 2018). In contrast, DNA viruses typically replicate in the nucleus, which provides easy access to the replication and transcription machinery of the host, the possibility of genome integration and some degree of protection against cellular defence mechanisms (El-Jesr et al., 2020; Rathinam and Fitzgerald, 2011). As an exception, giant DNA viruses, including poxviruses, encode their own replication machinery and generally replicate in the cytoplasm (Schramm and Locker, 2005), although there is increasing evidence that they still require host factors from the nucleus (Postigo et al., 2017; Senkevich et al., 2017).
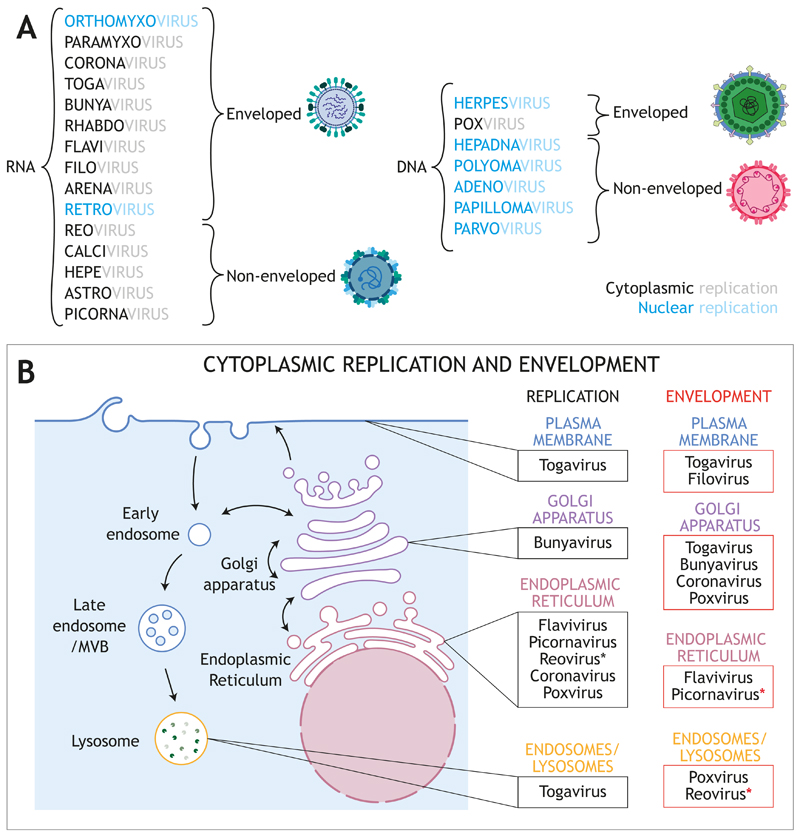
Figure 2. Virus classification.
A Main RNA and DNA viral families causing human diseases. Enveloped and non-enveloped classification is shown. Enveloped viruses are those in which at least one type of infectious form of the virion is enveloped by a lipid membrane, which can be acquired from a variety of cellular origins. Blue and black indicates nuclear and cytoplasmic replication, respectively. B Sites of replication and envelopment / budding for the viral families that replicate in the cytoplasm. In Reoviridae, association with ER membranes of viral factories has been reported. Red asterisks indicate that these families are classified as non-enveloped, although a non-lytic egress has been described (see text for details).
For viruses that undergo nuclear replication, the immediate task after entry is actually getting to the nucleus, which can be a considerable distance away from the site of infection, especially in neurons (Miranda-Saksena et al., 2018). Fortunately, the cell provides a convenient transport system and viruses encode proteins that recruit the minus-end-directed microtubule motor dynein to be actively transported to a perinuclear location (Dodding and Way, 2011; Naghavi and Walsh, 2017; Wang et al., 2018). The next challenge is the physical barrier imposed by the nuclear envelope. Viruses bypass this in different ways (Fay and Pante, 2015). Some retroviruses, such as murine leukemia virus, gain access during mitosis when the nuclear envelope is absent (Goff, 2007). The drawback of this strategy is that during interphase they are unable to integrate into the host’s genome to establish long term viral replication (Goff, 2007; Katz et al., 2005). Penetration through one of the many nuclear pores circumvents this limitation. However, the nuclear pore complex (NPC) has an upper size limit of ~39 nm (Lin and Hoelz, 2019; Pante and Kann, 2002), and only some viruses, such as hepatitis B virus, are small enough to penetrate intact (Jiang and Hildt, 2020). In contrast, larger viruses such as Herpesvirus simplex 1 (HSV1) and adenoviruses disassemble their capsid in the cytoplasm or in association with the NPC, allowing the viral genome to pass through (Fay and Pante, 2015; Li et al., 2019).
Replication and assembly on membrane compartments
Cytoplasmic viral replication often takes place at specialised replication organelles (ROs), which are assembled on the surface of a membrane-bound compartment (Miller and Krijnse-Locker, 2008; Neufeldt et al., 2018). Viral infection alters these membranes to generate invaginations or cavities in which the replication machinery is concentrated and organised to efficiently perform viral replication in a controlled location. They also serve as a framework to coordinate replication and assembly. The cellular source and location of membranes for RO formation are diverse, varying between different virus families (Fig. 2). For example, Togaviridae form ROs at the plasma membrane and endosomes, although endosomal ROs may not be fully functional in the Togavirus genus alphavirus (Frolova et al., 2010). The structure of ROs also varies depending on the viral family. Flaviviridae such as Dengue virus (DENV) and Zika virus (ZKV) form slightly opened invaginations at the ER, with a 11-nm pore connecting the lumen to the cytosol, providing access to cytosolic components which are essential for replication (Miorin et al., 2013; Neufeldt et al., 2018; Welsch et al., 2009). In contrast, hepatitis C virus (HCV) forms cup-shaped ROs called double membrane vesicles (DMVs) that protrude from ER membranes (Cortese et al., 2017; Neufeldt et al., 2018; Romero-Brey et al., 2012). Although the lumina of some of these DMVs are connected to the cytosol, most of them are closed (Romero-Brey et al., 2012). It is possible that DMVs close after replication and therefore represent a later stage, such as assembly or envelopment. Coronaviridae also induce characteristic DMVs that likely contain newly synthesised RNA genomes (Cortese et al., 2020; Gosert et al., 2002; Klein et al., 2020; Snijder et al., 2020). These are also connected to the cytosol through a pore, suggesting the existence of pore-stabilising viral proteins such as the nsp3 protein in mouse hepatitis coronavirus (Wolff et al., 2020). Finally, lipid droplets are hijacked at the ER by flaviviruses and SARS-CoV-2 for efficient replication, suggesting a general role of these lipid storage organelles in early stages of virus replication (Cloherty et al., 2020; Dias et al., 2020; Paul and Bartenschlager, 2015).
ROs assemble with the assistance of host proteins and a limited number of non-structural viral proteins, but how they are initiated and organised remains a mystery. It is also not understood how membrane curvature is induced to form ROs, but the answer may lie in specialised viral proteins. In the case of DENV, the integral viral membrane proteins NS4A and NS4B drive RO formation (Paul and Bartenschlager, 2015). These two proteins contain amphipathic helices which, when inserted in a single leaflet of the ER membrane, would induce membrane curvature to facilitate the formation of ROs. Membrane curvature may also be increased by the oligomerisation of these proteins (Stern et al., 2013; Zou et al., 2014). The ER-tubulation factor reticulon 3.1 A (Voeltz et al., 2006) and atlastins (dynamin-related GTPases) have been linked to membrane remodelling in Flaviviridae (Aktepe et al., 2017; Monel et al., 2019; Neufeldt et al., 2019), highlighting the role of cellular membrane-bending proteins in RO formation.
In many cases, RO formation substantially modifies the organisation and function of cellular membranes, resulting in the establishment of a new status quo that favours virus replication and assembly. A good example of this is seen in infections caused by Picornaviridae members, such as poliovirus and other enteroviruses, that heavily affect Golgi organisation (Belov, 2016; Belov and Sztul, 2014; Jackson, 2014). In normal conditions, Golgi maintenance depends on a continuous supply of membranes from the ER, mainly in the form of COPII vesicles, whose formation is driven by the Sar1 GTPase (Peotter et al., 2019; Ward et al., 2001). In contrast, Golgi-localised Arf GTPases, which are activated by different GTPase exchange factors, including GBF1, recruit the COPI complex to control Golgi-to-ER and intra-Golgi trafficking (Fig. 1), as well as other effectors such as the lipid modifying enzyme phosphatidylinositol-4-kinase IIIβ (PI4KIIIβ) (Godi et al., 1999). Impairing Sar1 or Arf1 functions disrupts the Golgi, which is partly reabsorbed by the ER (Ward et al., 2001). Despite work from many groups, it is still not clear how the Golgi organisation is maintained (Glick and Luini, 2011; Pantazopoulou and Glick, 2019) and understanding how viruses manipulate the Golgi may provide additional molecular insights.
During Picornaviridae infection, ER-to-Golgi trafficking is shut down and the COPII and Golgi-organising machinery, including Arf1, GBF1 and PI4KIIIβ, is recruited to ROs (Doedens et al., 1997; Doedens and Kirkegaard, 1995; Rust et al., 2001). This machinery, together with Sar1, is then hijacked and used during viral replication: Sar1 is required as its loss inhibits foot-and-mouth disease virus infection, while a dominant negative Sar1 impairs poliovirus RNA replication (Hsu et al., 2010; Midgley et al., 2013). However, the precise function of Sar1 in the replication cycle of Picornaviridae and other RNA viruses is still not fully clear. In contrast, we have a much better understanding of the role of Arf1, GBF1 and PI4KIIIβ (Hsu et al., 2010; Lanke et al., 2009). It is proposed that, early in infection, the conserved enteroviral protein 3A localises to the Golgi, where it enhances the recruitment of PI4KIIIβ by Arf1/GBF1 at the expense of COPI. The increase in PI4KIIIβ activity leads to higher PI4P levels in Golgi and ER-Golgi intermediate compartment (ERGIC) membranes and the displacement of COPI disrupts the normal organisation of the Golgi apparatus (Hsu et al., 2010; Melia et al., 2019; Wessels et al., 2006). This rise in PI4P triggers the recruitment of viral RNA polymerase 3Dpol, contributing to the formation of functional ROs, which associate with ER membranes, although they contain Golgi components (Hsu et al., 2010; Moghimi et al., 2020). A high-PI4P microenvironment is also important for efficient replication of the Flaviviridae member HCV (Harak et al., 2014; Hsu et al., 2010; Reiss et al., 2011). In fact, the formation of RO membranes rich in PI4P and cholesterol with mixed ER-Golgi identity seems common in RNA viruses that replicate in association with the ER (Belov, 2016). This PI4P and cholesterol enrichment is typical of Golgi membranes in non-infected cells and may serve for recruitment of viral and cellular proteins required to generate ROs. Finally, hijacking the Golgi machinery for RO formation may be also a common theme for RNA viruses, as GBF1 is also crucial for the replication of HCV, Coronaviridae and Phlebovirus (Goueslain et al., 2010; Lebsir et al., 2019; Martinez and Arias, 2020; Uckeley et al., 2019; Verheije et al., 2008). Although manipulation of the Golgi is common, there are likely degrees of Golgi disruption as enveloped RNA viruses such as HCV or SARS-Cov-2 still need to traverse the Golgi to exit the cell (Coller et al., 2012; Ghosh et al., 2020). In fact, Arf1 is required for replication of mouse hepatitis coronavirus, even though it is not as drastically displaced to ROs as is the case in cells infected with Picornavirus (Belov et al., 2005; Verheije et al., 2008).
The investigation of ROs is an exciting and rapidly growing field, with new membrane-associated ROs continuously being described for a number of viruses, including orthoreoviruses, which were initially thought to replicate in membrane-less viral inclusions (Fernandez de Castro et al., 2014). In all cases, further extensive molecular, morphological and functional characterisation is still required, because we lack a complete understanding of the composition and organisation of these essential replication structures.
Egress - getting the best out of the cell
To spread the infection, new viral progeny need to get out of the cell. Virus release can occur after cell lysis, but this is a dangerous option, since it is likely to trigger an increased immune response. Alternatively, viruses can leave the cell by budding at the plasma membrane or after getting enveloped by a cellular membrane compartment (Fig. 3). Historically, the term budding has been preferred for small viruses, while envelopment or wrapping is more often used for bigger viruses, such as HSV1 or poxviruses. In this Review, we use the term budding when viruses acquire membrane at the plasma membrane during their release. In contrast, we utilise envelopment when the membrane is obtained from an intracellular compartment. During envelopement, which can occur at a number of organelles, including the ER, the Golgi apparatus or even secretory autophagosomes, viruses acquire a number of membranes, the outermost of which will eventually fuse with the plasma membrane, releasing an enveloped virion (Fig. 2). Independently of which membrane is used for envelopment, the process presents similar challenges. First, it frequently requires the presence of viral proteins in the acceptor membrane to mediate the initial interaction with the non-enveloped virion. Second, the membrane must be deformed to accommodate the virus. Finally, the virus has to close the membrane, which may involve viral manipulation of the ESCRT (endosomal sorting complex required for transport) machinery of the host (Box 1) (Barouch-Bentov et al., 2016; Corless et al., 2010; Tabata et al., 2016). Once the enveloped virus is in the lumen of an organelle, it can behave like a cellular cargo that is sorted and secreted by exocytosis either via the canonical pathway (ER-Golgi-exocytic vesicles) or through less coventional pathways, involving secretory lysosomes or even autophagosomes.
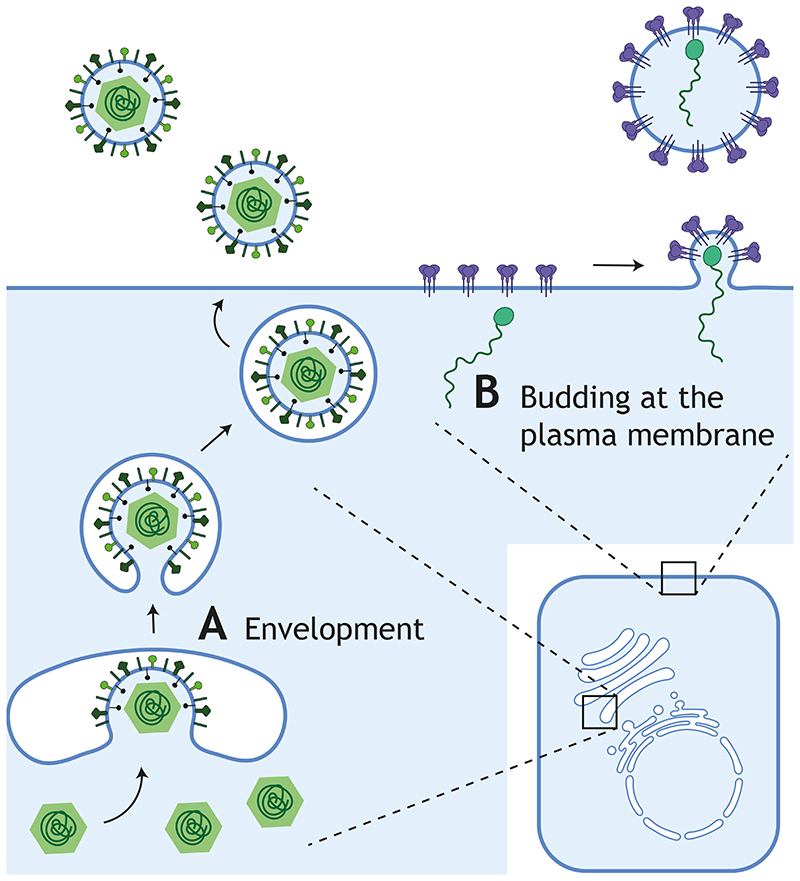
Figure 3. Envelopment and Budding during viral egress.
Viral particles assembled in the nucleus or the cytoplasm are enveloped by a cellular membrane to leave the cell in the absence of cell lysis. There are two different ways to acquire this membrane. A Virions can be enveloped using an intracellular compartment, such as the Golgi apparatus, and travel through the membrane trafficking system to be released by fusion of its outer membrane with the cell surface. B In contrast, budding at the plasma membrane directly releases extracellular viral particles. Both strategies involve similar processes that require membrane bending toward the luminal or extracellular space and scission of the neck to generate membrane-bound virions.
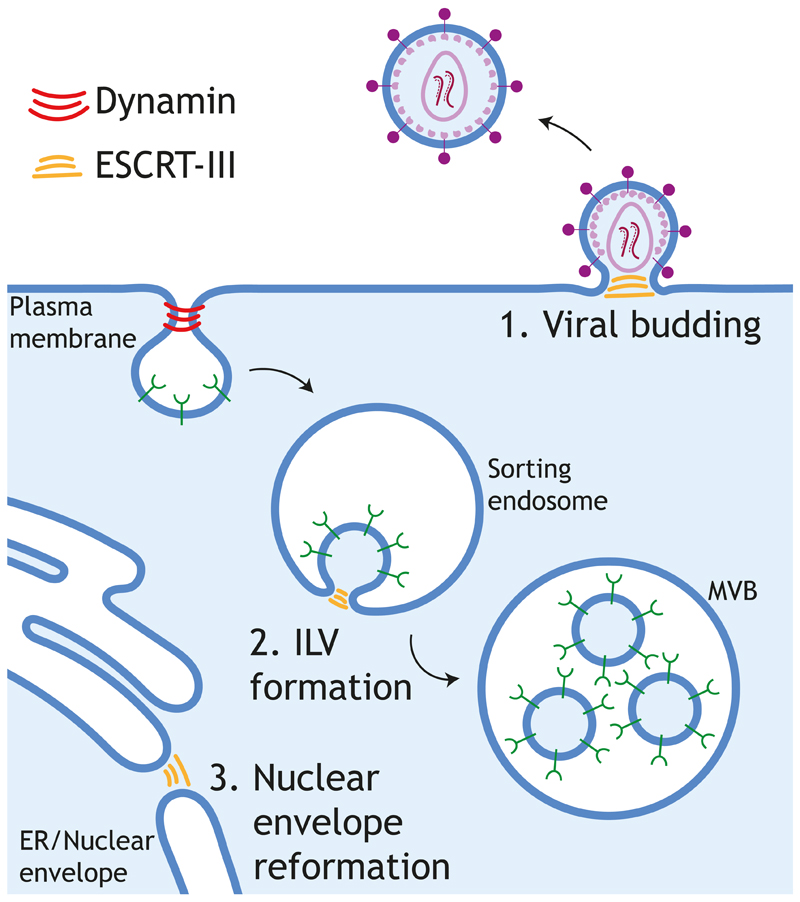
Box 1. The Escrt Pathway.
The Endosomal Sorting Complex Required for Transport (ESCRT) pathway participates in essential processes such as the formation of intraluminal vesicles in multivesicular bodies (MVBs), abscission during cell division and reformation of the nuclear envelope (Carlton and Martin-Serrano, 2009; McCullough et al., 2015; Remec Pavlin and Hurley, 2020). The function of ESCRT is to catalyse the scission of a cytoplasm-filled vesicle or to close a cytoplasm-filled hole at a specific membrane-bound compartment. The scission catalysed by ESCRT is topologically opposed to that of dynamin. Both components act from the cytoplasm; however, dynamin encircles and constricts a membrane neck whereas ESCRT acts from the inside of a topologically opposed and cytoplasm-filled neck or stalk. Mechanistically, the site of action of ESCRT is determined by the recruitment of adaptor proteins to specific membranes, which in turn recruit early-acting ESCRT factors via conserved motifs known as late domains. These early factors include Bro1 domain proteins, such as Alix, and ESCRT-I and -II complexes. Afterwards, the ESCRT-III complex is recruited. ESCRT-III forms filaments that, together with VPS4 ATPases, drive membrane remodelling and scission. ESCRT is involved in the budding of HIV at the plasma membrane and in the envelopment of a number of viral families at different cellular organelles, such as the nuclear envelope, the ER and the Golgi apparatus (Votteler and Sundquist, 2013). In HIV, the viral polyprotein Gag acts as an adaptor at the plasma membrane to activate the ESCRT pathway to facilitate virus budding (McCullough et al., 2018; Votteler and Sundquist, 2013). Gag contains YPXL and P(T/S)AP late domains that bind the early-acting factor Alix and TSG101, respectively. PPXY motif in other retroviral Gag proteins (e.g. Murine Leukemia Virus) also helps initiate the ESCRT pathway by interacting with E3 ubiquitin-protein ligase NEDD4. Ubiquitin could also act as a late domain that recruits early-acting factors that contain ubiquitin-binding domains. At the end of the process, ESCRT-III and VPS4 ATPases are recruited and catalyse the scission of virions budding at the plasma membrane. Proteins containing these late domains have been found in many enveloped viruses and we are just starting to understand the importance of the ESCRT pathway in virus maturation and egress (Chen and Lamb, 2008; Votteler and Sundquist, 2013).
Envelopment at the early secretory pathway
The early secretory pathway comprises the ER, the ERGIC and the cis-Golgi apparatus (Gomez-Navarro and Miller, 2016). RNA viruses, such as Flaviviridae and Coronaviridae, which replicate in ROs associated with the ER or the ER/ERGIC respectively, are enveloped at the early secretory pathway (Klein et al., 2020; Knoops et al., 2008; Krijnse-Locker et al., 1994; Stertz et al., 2007), suggesting there is a strong coupling between the sites of replication and envelopment. In fact, in the Flavivirus genus, the pore of ROs is adjacent to enveloping particles at the ER. In this genus, which includes Dengue and Zika viruses, the nucleocapsids are enveloped with ER membranes decorated with the viral proteins prM and E, forming enveloped particles in the lumen of the ER. Expression of prM and E in non-infected cells leads to formation of subviral particles, suggesting that prM and E drive envelopment at the ER (Ferlenghi et al., 2001). From their site of envelopment, flaviviruses and coronaviruses use the host trafficking machinery to get to the Golgi before leaving the cell. Ultrastructural analysis demonstrated that viral particles from β-coronaviruses (a group that includes SARS-CoV-2) accumulate at the Golgi and in nearby vesicles, leading to the suggestion that they travel through the Golgi and then leave the cell by vesicular transport (Stertz et al., 2007; Ulasli et al., 2010). However, more recent work found that β-coronaviruses travel through the Golgi and the TGN to reach the endosomal system and leave the cell from secretory lysosomes (Ghosh et al., 2020), similar to the non-lytic egress pathway of the non-enveloped genus Orthoreovirus ((Fernandez de Castro et al., 2020); see below). In contrast to β-coronaviruses, HCV, an enveloped flavivirus, does not seem to travel through lysosomes. Instead, after trafficking from the ER to the Golgi in COPII vesicles, HCV uses the membrane trafficking machinery to leave the cell using the secretory pathway, similar to cellular cargoes (Coller et al., 2012; Syed et al., 2017). In fact, egress of HCV is tightly linked to the secretion of very-low-density lipoproteins (VLDL) (Bartenschlager et al., 2011; Gastaminza et al., 2008; Huang et al., 2007). Furthermore, before envelopment at the ER, HCV assembly, which takes place in association with lipid droplets, has parallels with VLDL production as both require triglycerides and cholesterol (Aizaki et al., 2008; Maillard et al., 2011; Shelness and Sellers, 2001). Both are also rich in apolipoproteins, such as apoE, which is essential for HCV assembly (Bartenschlager et al., 2011). Finally, the restricted production of VLDL in the liver could contribute to HCV hepatotropism (Huang et al., 2007), suggesting that membrane trafficking requirements may influence organ tropisms of viral infections.
As with cellular cargoes, once inside the Golgi lumen, viruses frequently undergo additional maturation steps, which can result in morphological changes appreciable by electron microscopy, such as in the case of Coronaviridae, Togaviridae and Bunyaviridae (Risco et al., 2003; Salanueva et al., 1999; Salanueva et al., 2003). This maturation is probably the consequence of physical rearrangements of the viral structure due to posttranslational modifications, including the action of pro-convertases such as furin, an essential step for activating the fusion machinery of many enveloped viruses (Braun and Sauter, 2019; Thomas, 2002).
Envelopment at the late secretory pathway
Envelopment at late compartments takes place in a number of viral families. Here, we focus on Poxviridae and Herpesviridae to illustrate how this process occurs. Vaccinia virus (Poxviridae) undergoes a complex replication and assembly process in cytoplasmic ROs — known as viral factories — located close to the nucleus at the microtubule organising centre of the cell (Leite and Way, 2015; Ploubidou et al., 2000). Replication initially results in the assembly of Intracellular Mature Virions (IMVs), consisting of a core of genomic DNA and viral proteins, surrounded by a single membrane (Chichon et al., 2009; Chlanda et al., 2009). This single membrane bilayer is not acquired by envelopment, but is derived from the ER membrane by a mechanism that is still not fully understood (Chlanda et al., 2009; Krijnse Locker et al., 2013; Moss, 2015; Weisberg et al., 2017). A limited number of studies suggest that IMVs, which are infectious, are capable of leaving infected cells by directly budding at the plasma membrane (Meiser et al., 2003; Tsutsui, 1983). However, it is more generally accepted that the majority of IMVs are released when infected cells lyse. In addition, some IMVs are capable of leaving the cell by an alternative route that involves their envelopment with a membrane cisterna derived from the TGN or an endosomal compartment (Schmelz et al., 1994; Smith et al., 2002; Tooze et al., 1993). The identity of these membranes is modified by the presence of integral and peripheral viral membrane proteins, some of which, such as B5 and the lipid-modifying enzyme F13 are essential for IMV envelopment (Blasco and Moss, 1991; Engelstad and Smith, 1993; Smith et al., 2002; Wolffe et al., 1993). The molecular basis of IMV envelopment is still largely obscure, although recent observations point to the possible involvement of ESCRT-III components and the ATPase vacuolar protein sorting-associated protein 4B (VPS4B) (Huttunen et al., 2020). ESCRT-associated proteins Alix and Tumor susceptibility gene 101 protein (TSG101) may also have a role, as their depletion leads to a reduction in virus release (Honeychurch et al., 2007). The hijacking of the ESCRT machinery by vaccinia is maybe not surprising given it is frequently used by multiple viral families during their envelopment or budding (Box 1). However, some viruses, such as influenza, seem to carry out these processes in an ESCRT-independent manner (Chen et al., 2008; Rossman et al., 2010). Interestingly, F13 contains a conserved YXXL motif that may be recognised by Alix to facilitate the recruitment of ESCRT components (Honeychurch et al., 2007). It is possible that ESCRT mediates the final closing step in IMV envelopment by the TGN. However, it has been proposed that ESCRT brings about envelopment of IMVs by multivesicular bodies (MVBs), which subsequently fuse with the plasma membrane to release viral particles (Huttunen et al., 2020). Interestingly, the use of MVBs for envelopment has also been recently reported for the herpesvirus human cytomegalovirus (HCMV) (Flomm et al., 2021). It is of course possible that ESCRT participates in budding of IMVs at the plasma membrane, as it does for HIV (McCullough et al., 2018; Meiser et al., 2003; Tsutsui, 1983; Votteler and Sundquist, 2013).
Envelopment of IMVs by TGN-derived cisternae results in the addition of two membranes and the formation of Intracellular Enveloped Virions (IEVs) (Smith et al., 2002). Once formed, the integral IEV membrane protein A36 recruits kinesin-1 to drive microtubule-dependent transport of virions to the cell periphery (Dodding et al., 2011; Rietdorf et al., 2001; Ward and Moss, 2001). It is thought that the outermost membrane of the IEV fuses with the plasma membrane, but the molecular basis of this process remains unknown. During this fusion step, A36 relocates to the plasma membrane beneath the cell-associated extracellular virion (CEV) attached to the outside of the cell (van Eijl et al., 2000). These CEV locally activate Src and Abl family kinases, resulting in the phosphorylation of A36 and the induction of a signalling network that activates Arp2/3-driven actin polymerisation (Donnelly et al., 2013; Frischknecht et al., 1999; Moreau et al., 2000; Newsome et al., 2004; Reeves et al., 2005), which enhances the cell-to-cell spread of the virus (Cudmore et al., 1995; Doceul et al., 2010; Horsington et al., 2013; Ward and Moss, 2001). Interestingly, immediately after fusion with the plasma membrane, both septins and clathrin are recruited beneath CEVs prior to induction of actin polymerisation (Pfanzelter et al., 2018; Snetkov et al., 2016). Septins, a family of cytoskeletal proteins, act as a restriction factor to suppress release of CEVs from plasma membrane invaginations (Pfanzelter et al., 2018; Spiliotis and McMurray, 2020). In contrast, clathrin promotes A36 clustering to enhance actin assembly (Humphries et al., 2012; Snetkov et al., 2016). Although beneficial to promoting vaccinia spread, the main function of clathrin is most likely to promote endocytosis of A36 and associated IEV proteins from the plasma membrane back to the TGN to facilitate additional viral assembly once CEV are released from the cell surface (Husain and Moss, 2003).
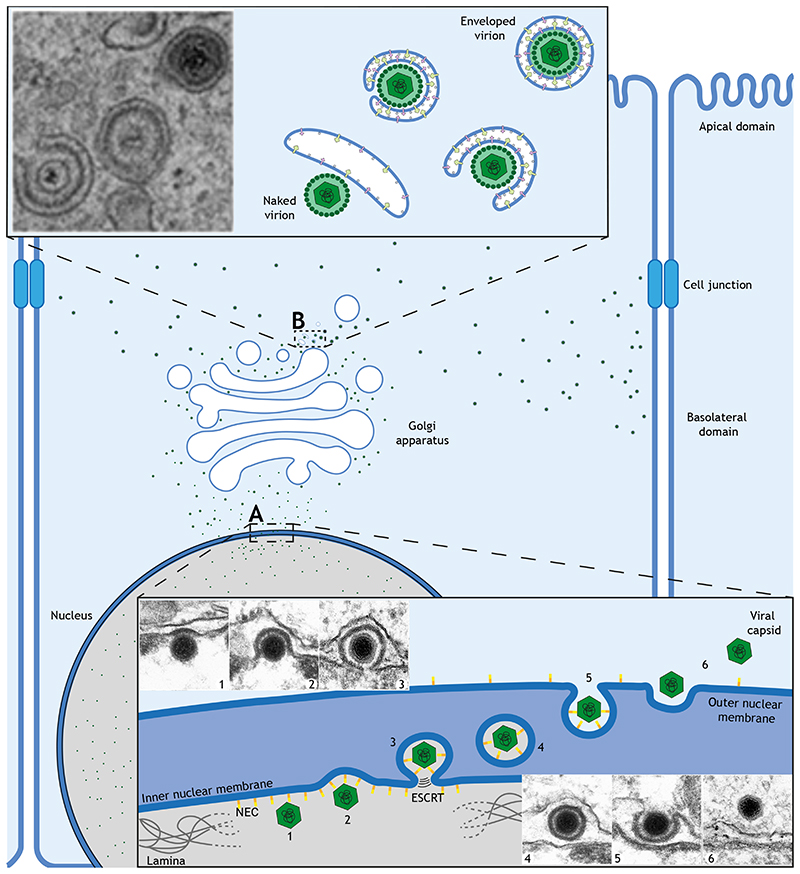
In contrast to vaccinia, HSV1 egress has the additional hurdle of traversing the nuclear envelope, since viral replication and initial assembly occur in the nucleus (Bigalke and Heldwein, 2016; Lv et al., 2019). To achieve this task the virus disassembles the nuclear lamina before enveloping itself in nuclear membrane only to shed this envelope to gain access to the cytoplasm (Fig. 4). Members of the Herpesviridae family accomplish this feat using the Nuclear Egress Complex (NEC), consisting of two viral proteins (pUL31 and pUL34 in HSV1). The NEC, which is anchored to the inner nuclear membrane (INM), promotes both viral envelopment and disruption of the nuclear lamina (Bigalke and Heldwein, 2016). During mitosis, phosphorylation of lamins results in disassembly of the nuclear lamina (Margalit et al., 2005). A similar process occurs during Herpesviridae infection, as the NEC recruits protein kinase C, which is thought to phosphorylate lamin B, while the viral kinase pUS3 phosphorylates lamin A/C (Mou et al., 2007; Park and Baines, 2006). Lamin-associated proteins such as emerin also get phosphorylated, contributing to nuclear lamina disassembly (Leach et al., 2007; Leach and Roller, 2010). After local lamina dissolution, the virus is enveloped by the INM in a process that topologically resembles the formation of intraluminal vesicles during the formation on MVBs (Fig. 4). The NEC controls viral envelopment by the INM, since it is sufficient to induce membrane budding in vitro and forms perinuclear vesicles when expressed in the absence of other viral proteins (Bigalke et al., 2014; Draganova et al., 2020; Klupp et al., 2007; Lorenz et al., 2015). The ESCRT complex, which is also involved in the formation of intraluminal vesicles in MVBs, is recruited by the NEC to facilitate INM envelopment closure, although molecular details are still lacking (Arii et al., 2018; Lee et al., 2012). Finally, to reach the cytosol, enveloped capsids in the nuclear envelope lumen fuse with the outer nuclear membrane in a process reminiscent of cell entry (Fig. 4). During the initial establishment of HSV1 infection, the viral proteins gB, gD and the heterodimer gH-gL are required for membrane fusion during entry (Connolly et al., 2020). These proteins are also present in the nuclear envelope and are incorporated into viral particles when they get enveloped by the INM (Ahmad and Wilson, 2020; Cai et al., 1987; Forrester et al., 1992). Loss of gB and gH results in the accumulation of enveloped viral particles in the perinuclear space, whereas single deletions impair viral entry, but not nuclear egress (Farnsworth et al., 2007b). This suggests that both fusion mechanisms are subtly different even though they both release capsids into the cytoplasm. The main difference is that during the initial establishment of infection incoming non-enveloped capsids have a set of viral proteins between the capsid and the envelope that form the tegument, whereas those undergoing egress only acquire tegument in the cytosol before being enveloped at the TGN. As with vaccinia, the TGN cisternae that envelope egressing HSV-1 capsids require the presence of viral membrane proteins, including gE-gI and gD (Farnsworth et al., 2003; Farnsworth et al., 2007a; McMillan and Johnson, 2001) as well as the ESCRT complex (Barnes and Wilson, 2019). Recent biochemical and structural analysis of the herpesvirus tegument complex pUL7:pUL51, which is involved in envelopment (Albecka et al., 2017; Roller and Fetters, 2015), reveals that the pUL51 structure resembles that of the ESCRT-III component CHMP4B and also forms similar filaments (Butt et al., 2020). This homology suggests that pUL51 may promote membrane scission in a similar way to ESCRT, thus providing a redundant pathway to guarantee envelopment (Butt et al., 2020). Once enveloped, the virus follows the route used by cellular cargoes to leave the cell.
Figure 4. Nuclear and cellular egress of HSV-1.
A Viral capsids assembled in the nucleus interact with the inner nuclear membrane (INM) via the viral nuclear egress complex (NEC). During primary envelopment, the NEC locally disassembles the nuclear lamina (1) and helps the INM envelop the capsid (2), which is sealed with the help of the ESCRT complex (3-4). Next, enveloped capsids fuse with the outer nuclear membrane (ONM) (5) to release the virions into the cytoplasm, in a process called de-envelopment (6). B In the cytoplasm, capsids acquire tegument before getting enveloped by trans-Golgi or endosomal membranes enriched in viral proteins and sealed by ESCRT during this secondary envelopment. Enveloped virions are transported to the basolateral domain of the plasma membrane, including cell junctions, for release. Electron micrographs in A: from (Mettenleiter et al., 2013) and B: from (Maringer et al., 2012).
When exiting polarised cells, it is important to leave at the right plasma membrane domain as this determines the accessibility to surrounding tissues which will influence virus invasiveness and pathogenicity (Cong and Ren, 2014; Tamhankar and Patterson, 2019; Tucker and Compans, 1993). In polarised cells, the cargo sorting machinery ensures proteins destined for export are directed to follow either basolateral or apical routes (Deborde et al., 2008; Weisz and Rodriguez-Boulan, 2009). Herpesviridae and other families have evolved to use the same cargo sorting machinery to ensure they spread from the right plasma membrane domain. In the case of HSV1, exit occurs at cell junctions, a part of the basolateral domain (Johnson et al., 2001; Sugimoto et al., 2008). This exit strategy is controlled by the gE-gI viral complex at the TGN; when gE function is impaired, viral release occurs at the apical plasma membrane and spread is reduced (Johnson et al., 2001). Interestingly, gE contains a YXXФ (where Ф is a bulky hydrophobic amino acid) motif in its cytoplasmic tail that is required for its TGN localisation (Alconada et al., 1999). When recognised by the plasma membrane adaptor AP2, this endocytic motif YXXФ would recycle gE back to the TGN. However, this scenario would not explain how gE directs HSV1 to the basolateral domain. Hypothetically, the YXXФ motif could be recognised at the TGN by other AP complexes and/or accessory factors to direct viral egress to lateral membranes. In fact, basolateral sorting of cargoes involve the recognition of the YXXФ motif by AP complexes (Bonifacino, 2014). It is also noteworthy that, during HSV1 infection, many TGN markers relocate to cell junctions, whereas other trafficking components disperse cytoplasmically (McMillan and Johnson, 2001; Wisner and Johnson, 2004). This redistribution is not a byproduct of envelopment or egress, as it also occurs in the absence of viral envelopment (Wisner and Johnson, 2004), suggesting the TGN function and organisation is altered to favour the virus in infected cells.
HCMV, another member of the Herpesviridae family, also induces extensive remodelling of cellular membranes (Das and Pellett, 2007; Das and Pellett, 2011; Das et al., 2007; Villinger et al., 2015). HCMV envelopment takes place in a complex perinuclear assembly compartment, consisting of a set of cisternae and vesicles that have membrane identities associated with cis-Golgi, TGN, endosomes and ER (Alwine, 2012; Buchkovich et al., 2009; Das and Pellett, 2011; Das et al., 2007). Exactly how HCMV manipulates the cell to form this complex assembly compartment is still a mystery, while membrane organisation is understudied in other Herpesviridae infections. Future studies into the molecular mechanisms that govern these complex membrane reorganisations will uncover if they constitute a general effect of Herpesviridae infection or whether each member has its own unique strategy.
Egress of non-enveloped viruses in the absence of cell lysis
Non-enveloped viruses typically spread after cell lysis; however, there are examples of nonlytic egress. In the case of Picornaviridae, such as poliovirus and Coxsackievirus, this pathway involves manipulation of autophagosomes (Mutsafi and Altan-Bonnet, 2018) (Fig. 5). Autophagosomes normally capture organelles, cellular components and intracellular pathogens to direct their recycling or destruction in lysosomes (Yu et al., 2018). However, poliovirus particles found in autophagosomes labelled with lipidated microtubule-associated protein light chain 3 (LC3-II) do not follow this degradative pathway (Bird et al., 2014; Chen et al., 2015; Robinson et al., 2014). Rather, autophagosomes that surround and capture virions at the ER become secretory, fusing with the plasma membrane to release a vesicle containing viral particles (Chen et al., 2015; Ponpuak et al., 2015; Robinson et al., 2014). This mechanism explains how the bulk of poliovirus release occurs before cell lysis (Jackson et al., 2005). Importantly, hepatitis A virus, a distant member of the Picornaviridae family, has also been shown to leave cells with a membranous envelope (Feng et al., 2013). These data appear to blurr the distinction between enveloped and non-enveloped viruses.
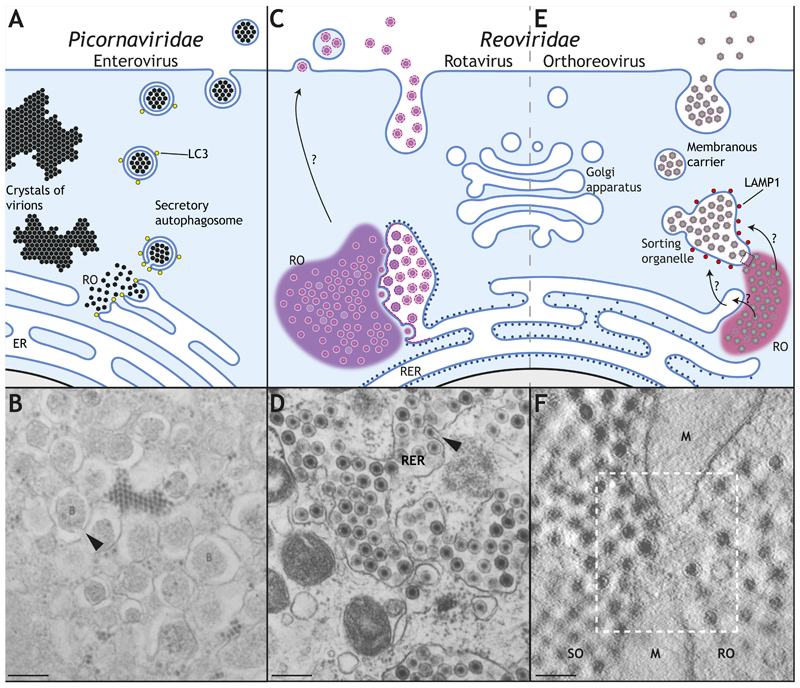
Figure 5. Non-lytic egress of non-enveloped viruses.
A Enteroviruses replicate in cytoplasmic replication organelles (ROs) associated with the endoplasmic reticulum (ER). A proportion of mature virions are packaged into LC3-positive secretory autophagosomes (arrow head in B (Dales et al., 1965)), whereas others accumulate intracellularly. C Rotavirus also replicates and assemble in ROs associated with ER membranes, which are used for envelopment (see arrow head in D (Poruchynsky et al., 1991)). This membrane is subsequently removed in the ER lumen before virions are released, possibly by a non-conventional secretory pathway (Jourdan et al., 1997). E Orthoreovirus replicates in ROs associated with the ER and other membranes before accumulating inside a sorting organelle (SO) with lysosomal identity (LAMP1 positive) by an unknown mechanism. Membrane carriers (MC) then bud from the SO to transport virions to the plasma membrane. F The electron micrograph shows the close proximity between the RO and the SO in orthoreovirus-infected cells, also associated with mitochondrial membranes (M) (Fernandez de Castro et al., 2020). Scale bars, 200 nm.
Other non-enveloped viruses have evolved alternative strategies. These include orthoreoviruses, which replicate and assemble in ROs known as viral inclusions (Fernandez de Castro et al., 2014; Garces Suarez et al., 2019) (Fig. 5). These replication sites are associated with ER and mitochondrial membranes, as well as lipid droplets (Fernandez de Castro et al., 2014; Tenorio et al., 2018). More recently, non-lytic egress of mammalian orthoreovirus has been found to occur via membranous carriers, which fuse with the plasma membrane to release non enveloped viral particles (Fernandez de Castro et al., 2020). These membrane carriers seem to bud from larger membrane-bound organelles with a lysosomal identity (LAMP1 positive) that are associated with viral inclusions (Fernandez de Castro et al., 2020) (Fig. 5). In contrast to Picornaviridae, these structures lack autophagic identity. It is not immediately obvious how the lysosomal identity of this compartment is maintained in the absence of virion degradation. Moreover, exactly how assembled orthoreovirus virions translocate from the cytoplasm into the lumen of these larger sorting organelles without acquiring an envelope remains to be established. The answer may be found in the analysis of the close relative rotavirus, which replicates and assembles in RNA-protein condensates known as cytoplasmic inclusions (Garces Suarez et al., 2019; Geiger et al., 2020). Rotaviruses induce and hijack the early stages of autophagy and the anterograde COPII machinery to transport viral proteins from the ER to the sites of viral assembly (Crawford et al., 2019). This blocks ER-to-Golgi traffic, as seen with many other RNA viruses (Doedens et al., 1997; Xu et al., 2000). Rotavirus virions access the lumen of the ER by envelopment. However, the envelope is thought to be subsequently removed by an unknown mechanism in the lumen of the ER (Tian et al., 1996) (Fig. 5). It is possible that orthoreoviruses enter sorting organelles (or a previous compartment, like the ER) by envelopment and, similarly to rotaviruses, shed this envelope once they are luminal. The situation actually appears more complex as extracellular vesicles containing infectious rotavirus have been recently described (Santiana et al., 2018). This observation suggests that rotavirus release occurs via multiple mechanisms, and may be cell type dependent.
Conclusions and future perspectives
Historically, early insights into the organisation and function of membrane trafficking were linked to analysis of viral infections or the trafficking of viral proteins (Butt et al., 2020; Fries and Rothman, 1980; Helenius, 2020; Rothman and Fine, 1980). Today, we have a far greater molecular understanding of how membrane trafficking regulates the movement of cellular cargoes and lipids and the maintenance of organelles and cellular architecture (Emr et al., 2009; Guo et al., 2014; Pantazopoulou and Glick, 2019). However, it is clear from the few examples we have discussed that, in the majority of cases, we still lack a full molecular understanding of how even relatively simple viruses manipulate cellular membranes for their own ends. It is also evident that viruses can still provide additional molecular insights into the regulation and function of membrane trafficking.
Ideally, we should all be examining viral infections at a sub-cellular level in a living organism. However, this presents many technical problems including the ability to image the right tissue with sufficient resolution and speed. Going forward, the use of organoids offers the possibility of examining the cellular impact of viral infection in a more complex 3D situation that is closer to the real physiological situation than a cell monolayer in a culture dish (Cugola et al., 2016; Ettayebi et al., 2016; Garcez et al., 2016; Ramani et al., 2018). Another frustration in working with viruses is their size, which limits our ability to fully resolve what is happening in the light microscope. Because of this, studies have traditionally relied heavily on morphological analysis using electron microscopy. However, these static snap shots do not provide the full picture as they lack true dynamic information. The use of super-resolution microscopy combined with image analysis is now revolutionising multiple aspects of cellular virology by allowing us to see dynamic events in ever increasing detail (Gray et al., 2019; Scherer et al., 2020; Sekine et al., 2017). In addition to light microscopy, improvements in electron microscopy techniques, such as focused ion beam scanning electron microscopy (FIB-SEM), are allowing us to image whole cells or even tissues, whereas cryo-electron tomography is providing even higher resolution snap shots and unprecedented molecular insights into viral structures and membrane rearrangements in their native state in the cell (Calder and Rosenthal, 2016; Ibiricu et al., 2011; Quemin et al., 2020). These technological advances in combination with functional assays, such as CRISPR screens, are undoubtedly driving a renaissance for membrane trafficking and virus research.
Summary.
This review at the intersection of membrane trafficking and virology discusses how viruses hijack the organisation and function of cellular membranes to accomplish their replication, assembly and egress.
Acknowledgements
We thank Jeremy Carlton for his critical reading of this manuscript. We apologise to all who have helped advance the field of membrane trafficking of viruses and whose work could not be cited due to space limitation. MW is funded by Cancer Research UK [FC001209], the UK Medical Research Council [FC001209] and the Wellcome Trust [FC001209] at the Francis Crick Institute. M.H-G and G.L. are funded by the Postdoctoral Training Programme of the Francis Crick Institute. Figures were made with Biorender and Adobe Illustrator.
References
- Ahmad I, Wilson DW. HSV-1 Cytoplasmic Envelopment and Egress. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21175969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizaki H, Morikawa K, Fukasawa M, Hara H, Inoue Y, Tani H, Saito K, Nishijima M, Hanada K, Matsuura Y, et al. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J Virol. 2008;82:5715–24. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aktepe TE, Liebscher S, Prier JE, Simmons CP, Mackenzie JM. The Host Protein Reticulon 3.1A Is Utilized by Flaviviruses to Facilitate Membrane Remodelling. Cell Rep. 2017;21:1639–1654. doi: 10.1016/j.celrep.2017.10.055. [DOI] [PubMed] [Google Scholar]
- Albecka A, Owen DJ, Ivanova L, Brun J, Liman R, Davies L, Ahmed MF, Colaco S, Hollinshead M, Graham SC, et al. Dual Function of the pUL7-pUL51 Tegument Protein Complex in Herpes Simplex Virus 1 Infection. J Virol. 2017;91 doi: 10.1128/JVI.02196-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alconada A, Bauer U, Sodeik B, Hoflack B. Intracellular traffic of herpes simplex virus glycoprotein gE: characterization of the sorting signals required for its trans-Golgi network localization. J Virol. 1999;73:377–87. doi: 10.1128/jvi.73.1.377-387.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alwine JC. The human cytomegalovirus assembly compartment: a masterpiece of viral manipulation of cellular processes that facilitates assembly and egress. PLoS Pathog. 2012;8:e1002878. doi: 10.1371/journal.ppat.1002878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arii J, Watanabe M, Maeda F, Tokai-Nishizumi N, Chihara T, Miura M, Maruzuru Y, Koyanagi N, Kato A, Kawaguchi Y. ESCRT-111 mediates budding across the inner nuclear membrane and regulates its integrity. Nat Commun. 2018;9:3379. doi: 10.1038/s41467-018-05889-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balla T, Sengupta N, Kim YJ. Lipid synthesis and transport are coupled to regulate membrane lipid dynamics in the endoplasmic reticulum. Biochim Biophys Acta Mol Cell Biol Lipids. 2020;1865 doi: 10.1016/j.bbalip.2019.05.005. 158461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnes J, Wilson DW. Seeking Closure: How Do Herpesviruses Recruit the Cellular ESCRT Apparatus? J Virol. 2019;93 doi: 10.1128/JVI.00392-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barouch-Bentov R, Neveu G, Xiao F, Beer M, Bekerman E, Schor S, Campbell J, Boonyaratanakornkit J, Lindenbach B, Lu A, et al. Hepatitis C Virus Proteins Interact with the Endosomal Sorting Complex Required for Transport (ESCRT) Machinery via Ubiquitination To Facilitate Viral Envelopment. mBio. 2016;7 doi: 10.1128/mBio.01456-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartenschlager R, Penin F, Lohmann V, Andre P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- Belov GA. Dynamic lipid landscape of picornavirus replication organelles. Curr Opin Virol. 2016;19:1–6. doi: 10.1016/j.coviro.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Fogg MH, Ehrenfeld E. Poliovirus proteins induce membrane association of GTPase ADP-ribosylation factor. J Virol. 2005;79:7207–16. doi: 10.1128/JVI.79.11.7207-7216.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belov GA, Sztul E. Rewiring of cellular membrane homeostasis by picornaviruses. J Virol. 2014;88:9478–89. doi: 10.1128/JVI.00922-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigalke JM, Heldwein EE. Nuclear Exodus: Herpesviruses Lead the Way. Annu Rev Virol. 2016;3:387–409. doi: 10.1146/annurev-virology-110615-042215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigalke JM, Heuser T, Nicastro D, Heldwein EE. Membrane deformation and scission by the HSV-1 nuclear egress complex. Nat Commun. 2014;5:4131. doi: 10.1038/ncomms5131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird SW, Maynard ND, Covert MW, Kirkegaard K. Nonlytic viral spread enhanced by autophagy components. Proc Natl Acad Sci U S A. 2014;111:13081–6. doi: 10.1073/pnas.1401437111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R, Moss B. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000-Dalton outer envelope protein. J Virol. 1991;65:5910–20. doi: 10.1128/jvi.65.11.5910-5920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonifacino JS. Adaptor proteins involved in polarized sorting. J Cell Biol. 2014;204:7–17. doi: 10.1083/jcb.201310021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun E, Sauter D. Furin-mediated protein processing in infectious diseases and cancer. Clin Transl Immunology. 2019;8:e1073. doi: 10.1002/cti2.1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchkovich NJ, Maguire TG, Paton AW, Paton JC, Alwine JC. The Endoplasmic Reticulum Chaperone BiP/GRP78 Is Important in the Structure and Function of the Human Cytomegalovirus Assembly Compartment. Journal of Virology. 2009;83:11421–11428. doi: 10.1128/JVI.00762-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butt BG, Owen DJ, Jeffries CM, Ivanova L, Hill CH, Houghton JW, Ahmed MF, Antrobus R, Svergun DI, Welch JJ, et al. Insights into herpesvirus assembly from the structure of the pUL7:pUL51 complex. Elife. 2020;9 doi: 10.7554/eLife.53789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai WZ, Person S, Warner SC, Zhou JH, DeLuca NA. Linkerinsertion nonsense and restriction-site deletion mutations of the gB glycoprotein gene of herpes simplex virus type 1. J Virol. 1987;61:714–21. doi: 10.1128/jvi.61.3.714-721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calder LJ, Rosenthal PB. Cryomicroscopy provides structural snapshots of influenza virus membrane fusion. Nat Struct Mol Biol. 2016;23:853–8. doi: 10.1038/nsmb.3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JG, Martin-Serrano J. The ESCRT machinery: new functions in viral and cellular biology. Biochem Soc Trans. 2009;37:195–9. doi: 10.1042/BST0370195. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372:221–32. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BJ, Leser GP, Jackson D, Lamb RA. The influenza virus M2 protein cytoplasmic tail interacts with the M1 protein and influences virus assembly at the site of virus budding. J Virol. 2008;82:10059–70. doi: 10.1128/JVI.01184-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YH, Du W, Hagemeijer MC, Takvorian PM, Pau C, Cali A, Brantner CA, Stempinski ES, Connelly PS, Ma HC, et al. Phosphatidylserine vesicles enable efficient en bloc transmission of enteroviruses. Cell. 2015;160:619–630. doi: 10.1016/j.cell.2015.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chichon FJ, Rodríguez MJ, Risco C, Fraile-Ramos A, Fernandez JJ, Esteban M, Carrascosa JL. Membrane remodelling during vaccinia virus morphogenesis. Biol Cell. 2009;101:401–14. doi: 10.1042/BC20080176. [DOI] [PubMed] [Google Scholar]
- Chlanda P, Carbajal MA, Cyrklaff M, Griffiths G, Krijnse-Locker J. Membrane rupture generates single open membrane sheets during vaccinia virus assembly. Cell Host Microbe. 2009;6:81–90. doi: 10.1016/j.chom.2009.05.021. [DOI] [PubMed] [Google Scholar]
- Cloherty APM, Olmstead AD, Ribeiro CMS, Jean F. Hijacking of Lipid Droplets by Hepatitis C, Dengue and Zika Viruses-From Viral Protein Moonlighting to Extracellular Release. Int J Mol Sci. 2020;21 doi: 10.3390/ijms21217901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller KE, Heaton NS, Berger KL, Cooper JD, Saunders JL, Randall G. Molecular determinants and dynamics of hepatitis C virus secretion. PLoS Pathog. 2012;8:e1002466. doi: 10.1371/journal.ppat.1002466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong Y, Ren X. Coronavirus entry and release in polarized epithelial cells: a review. Rev Med Virol. 2014;24:308–15. doi: 10.1002/rmv.1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connolly SA, Jardetzky TS, Longnecker R. The structural basis of herpesvirus entry. Nat Rev Microbiol. 2020 doi: 10.1038/s41579-020-00448-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corless L, Crump CM, Griffin SD, Harris M. Vps4 and the ESCRT-III complex are required for the release of infectious hepatitis C virus particles. J Gen Virol. 2010;91:362–72. doi: 10.1099/vir.0.017285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M, Goellner S, Acosta EG, Neufeldt CJ, Oleksiuk O, Lampe M, Haselmann U, Funaya C, Schieber N, Ronchi P, et al. Ultrastructural Characterization of Zika Virus Replication Factories. Cell Rep. 2017;18:2113–2123. doi: 10.1016/j.celrep.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortese M, Lee JY, Cerikan B, Neufeldt CJ, Oorschot VMJ, Kohrer S, Hennies J, Schieber NL, Ronchi P, Mizzon G, et al. Integrative Imaging Reveals SARS-CoV-2-Induced Reshaping of Subcellular Morphologies. Cell Host Microbe. 2020;28:853–866 e5. doi: 10.1016/j.chom.2020.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford SE, Criglar JM, Liu Z, Broughman JR, Estes MK. COPII Vesicle Transport Is Required for Rotavirus NSP4 Interaction with the Autophagy Protein LC3 II and Trafficking to Viroplasms. J Virol. 2019;94 doi: 10.1128/JVI.01341-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cudmore S, Cossart P, Griffiths G, Way M. Actin-based motility of vaccinia virus. Nature. 1995;378:636–8. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- Cugola FR, Fernandes IR, Russo FB, Freitas BC, Dias JL, Guimaraes KP, Benazzato C, Almeida N, Pignatari GC, Romero S, et al. The Brazilian Zika virus strain causes birth defects in experimental models. Nature. 2016;534:267–71. doi: 10.1038/nature18296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dales S, Eggers HJ, Tamm I, Palade GE. Electron Microscopic Study of the Formation of Poliovirus. Virology. 1965;26:379–89. doi: 10.1016/0042-6822(65)90001-2. [DOI] [PubMed] [Google Scholar]
- Das S, Pellett PE. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology. 2007;361:263–73. doi: 10.1016/j.virol.2006.11.019. [DOI] [PubMed] [Google Scholar]
- Das S, Pellett PE. Spatial relationships between markers for secretory and endosomal machinery in human cytomegalovirus-infected cells versus those in uninfected cells. J Virol. 2011;85:5864–79. doi: 10.1128/JVI.00155-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S, Vasanji A, Pellett PE. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J Virol. 2007;81:11861–9. doi: 10.1128/JVI.01077-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daussy CF, Wodrich H. “Repair Me if You Can”: Membrane Damage, Response, and Control from the Viral Perspective. Cells. 2020;9 doi: 10.3390/cells9092042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deborde S, Gravotta D, Lakkaraju A, Rodriguez-Boulan E. Golgi apparatus and epithelial cell polarity. In: Mironov AA, Pavelka M, editors. The Golgi Apparatus: State of the art 110 years after Camillo Golgi’s discovery. Springer Vienna; Vienna: 2008. pp. 563–579. [Google Scholar]
- Dias SSG, Soares VC, Ferreira AC, Sacramento CQ, Fintelman-Rodrigues N, Temerozo JR, Teixeira L, Nunes da Silva MA, Barreto E, Mattos M, et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pat hog. 2020;16:e1009127. doi: 10.1371/journal.ppat.1009127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doceul V, Hollinshead M, van der Linden L, Smith GL. Repulsion of superinfecting virions: a mechanism for rapid virus spread. Science. 2010;327:873–876. doi: 10.1126/science.1183173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP, Mitter R, Humphries AC, Way M. A kinesin-1 binding motif in vaccinia virus that is widespread throughout the human genome. EMBO J. 2011;30:4523–38. doi: 10.1038/emboj.2011.326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodding MP, Way M. Coupling viruses to dynein and kinesin-1. EMBO J. 2011;30:3527–39. doi: 10.1038/emboj.2011.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens JR, Giddings TH, Jr, Kirkegaard K. Inhibition of endoplasmic reticulum-to-Golgi traffic by poliovirus protein 3A: genetic and ultrastructural analysis. J Virol. 1997;71:9054–64. doi: 10.1128/jvi.71.12.9054-9064.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doedens JR, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaldson JG, Johnson DL, Dutta D. Rab and Arf G proteins in endosomal trafficking and cell surface homeostasis. Small GTPases. 2016;7:247–251. doi: 10.1080/21541248.2016.1212687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly SK, Weisswange I, Zettl M, Way M. WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr Biol. 2013;23:999–1006. doi: 10.1016/j.cub.2013.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Draganova EB, Zhang J, Zhou ZH, Heldwein EE. Structural basis for capsid recruitment and coat formation during HSV-1 nuclear egress. Elife. 2020;9 doi: 10.7554/eLife.56627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Jesr M, Teir M, Maluquer de Motes C. Vaccinia Virus Activation and Antagonism of Cytosolic DNA Sensing. Front Immunol. 2020;11 doi: 10.3389/fimmu.2020.568412. 568412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emr S, Glick BS, Linstedt AD, Lippincott-Schwartz J, Luini A, Malhotra V, Marsh BJ, Nakano A, Pfeffer SR, Rabouille C, et al. Journeys through the Golgi--taking stock in a new era. J Cell Biol. 2009;187:449–53. doi: 10.1083/jcb.200909011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstad M, Smith GL. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–37. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- Ettayebi K, Crawford SE, Murakami K, Broughman JR, Karandikar U, Tenge VR, Neill FH, Blutt SE, Zeng XL, Qu L, et al. Replication of human noroviruses in stem cell-derived human enteroids. Science. 2016;353:1387–1393. doi: 10.1126/science.aaf5211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth A, Goldsmith K, Johnson DC. Herpes simplex virus glycoproteins gD and gE/gl serve essential but redundant functions during acquisition of the virion envelope in the cytoplasm. J Virol. 2003;77:8481–94. doi: 10.1128/JVI.77.15.8481-8494.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth A, Wisner TW, Johnson DC. Cytoplasmic residues of herpes simplex virus glycoprotein gE required for secondary envelopment and binding of tegument proteins VP22 and UL11 to gE and gD. J Virol. 2007a;81:319–31. doi: 10.1128/JVI.01842-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnsworth A, Wisner TW, Webb M, Roller R, Cohen G, Eisenberg R, Johnson DC. Herpes simplex virus glycoproteins gB and gH function in fusion between the virion envelope and the outer nuclear membrane. Proc Natl Acad Sci U S A. 2007b;104:10187–92. doi: 10.1073/pnas.0703790104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fay N, Pante N. Nuclear entry of DNA viruses. Front Microbiol. 2015;6:467. doi: 10.3389/fmicb.2015.00467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Z, Hensley L, McKnight KL, Hu F, Madden V, Ping L, Jeong SH, Walker C, Lanford RE, Lemon SM. A pathogenic picornavirus acquires an envelope by hijacking cellular membranes. Nature. 2013;496:367–71. doi: 10.1038/nature12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferlenghi I, Clarke M, Ruttan T, Allison SL, Schalich J, Heinz FX, Harrison SC, Rey FA, Fuller SD. Molecular organization of a recombinant subviral particle from tick-borne encephalitis virus. Mol Cell. 2001;7:593–602. doi: 10.1016/s1097-2765(01)00206-4. [DOI] [PubMed] [Google Scholar]
- Fernandez de Castro I, Tenorio R, Ortega-Gonzalez P, Knowlton JJ, Zamora PF, Lee CH, Fernandez JJ, Dermody TS, Risco C. A modified lysosomal organelle mediates nonlytic egress of reovirus. J Cell Biol. 2020;219 doi: 10.1083/jcb.201910131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez de Castro I, Zamora PF, Ooms L, Fernandez JJ, Lai CM, Mainou BA, Dermody TS, Risco C. Reovirus forms neo-organelles for progeny particle assembly within reorganized cell membranes. mBio. 2014;5 doi: 10.1128/mBio.00931-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flomm FJ, Soh TK, Schneider C, Britt HM, Thalassinos K, Pfitzner S, Reimer R, Grünewald K, Bosse JB. Intermittent bulk release of human cytomegalovirus through multivesicular bodies. bioRxiv. 2021 doi: 10.1371/journal.ppat.1010575. 2020.12.31.424954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrester A, Farrell H, Wilkinson G, Kaye J, Davis-Poynter N, Minson T. Construction and properties of a mutant of herpes simplex virus type 1 with glycoprotein H coding sequences deleted. J Virol. 1992;66:341–8. doi: 10.1128/jvi.66.1.341-348.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries E, Rothman JE. Transport of vesicular stomatitis virus glycoprotein in a cell-free extract. Proc Natl Acad Sci U S A. 1980;77:3870–4. doi: 10.1073/pnas.77.7.3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frischknecht F, Moreau V, Rottger S, Gonfloni S, Reckmann I, Superti-Furga G, Way M. Actin-based motility of vaccinia virus mimics receptor tyrosine kinase signalling. Nature. 1999;401:926–9. doi: 10.1038/44860. [DOI] [PubMed] [Google Scholar]
- Frolova EI, Gorchakov R, Pereboeva L, Atasheva S, Frolov I. Functional Sindbis virus replicative complexes are formed at the plasma membrane. J Virol. 2010;84:11679–95. doi: 10.1128/JVI.01441-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garces Suarez Y, Martinez JL, Torres Hernandez D, Hernandez HO, Perez-Delgado A, Mendez M, Wood CD, Rendon-Mancha JM, Silva-Ayala D, Lopez S, et al. Nanoscale organization of rotavirus replication machineries. Elife. 2019;8 doi: 10.7554/eLife.42906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcez PP, Loiola EC, Madeira da Costa R, Higa LM, Trindade P, Delvecchio R, Nascimento JM, Brindeiro R, Tanuri A, Rehen SK. Zika virus impairs growth in human neurospheres and brain organoids. Science. 2016;352:816–8. doi: 10.1126/science.aaf6116. [DOI] [PubMed] [Google Scholar]
- Gastaminza P, Cheng G, Wieland S, Zhong J, Liao W, Chisari FV. Cellular determinants of hepatitis C virus assembly, maturation, degradation, and secretion. J Virol. 2008;82:2120–9. doi: 10.1128/JVI.02053-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geiger F, Papa G, Arter WE, Acker J, Saar KL, Erkamp N, Qi R, Bravo J, Strauss S, Krainer G, et al. Rotavirus Replication Factories Are Complex Ribonucleoprotein Condensates. bioRxiv. 2020 2020.12.18.423429. [Google Scholar]
- Gemmer M, Forster F. A clearer picture of the ER translocon complex. J Cell Sci. 2020;133 doi: 10.1242/jcs.231340. [DOI] [PubMed] [Google Scholar]
- Ghosh S, Dellibovi-Ragheb TA, Kerviel A, Pak E, Qiu Q, Fisher M, Takvorian PM, Bleck C, Hsu VW, Fehr AR, et al. beta-Coronaviruses Use Lysosomes for Egress Instead of the Biosynthetic Secretory Pathway. Cell. 2020 doi: 10.1016/j.cell.2020.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick BS, Luini A. Models for Golgi traffic: a critical assessment. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a005215. a0052l5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godi A, Pertile P, Meyers R, Marra P, Di Tullio G, lurisci C, Luini A, Corda D, De Matteis MA. ARF mediates recruitment of PtdIns-4-OH kinase-beta and stimulates synthesis of PtdIns(4,5)P2 on the Golgi complex. Nat Cell Biol. 1999;1:280–7. doi: 10.1038/12993. [DOI] [PubMed] [Google Scholar]
- Goff SP. Host factors exploited by retroviruses. Nat Rev Microbiol. 2007;5:253–63. doi: 10.1038/nrmicro1541. [DOI] [PubMed] [Google Scholar]
- Gomez-Navarro N, Miller E. Protein sorting at the ER-Golgi interface. J Cell Biol. 2016;215:769–778. doi: 10.1083/jcb.201610031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gosert R, Kanjanahaluethai A, Egger D, Bienz K, Baker SC. RNA replication of mouse hepatitis virus takes place at double-membrane vesicles. J Virol. 2002;76:3697–708. doi: 10.1128/JVI.76.8.3697-3708.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goueslain L, Alsaleh K, Horellou P, Roingeard P, Descamps V, Duverlie G, Ciczora Y, Wychowski C, Dubuisson J, Rouille Y. Identification of GBF1 as a cellular factor required for hepatitis C virus RNA replication. J Virol. 2010;84:773–87. doi: 10.1128/JVI.01190-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant BD, Donaldson JG. Pathways and mechanisms of endocytic recycling. Nat Rev Mol Cell Biol. 2009;10:597–608. doi: 10.1038/nrm2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RDM, Albrecht D, Beerli C, Huttunen M, Cohen GH, White IJ, Burden JJ, Henriques R, Mercer J. Nanoscale polarization of the entry fusion complex of vaccinia virus drives efficient fusion. Nat Microbiol. 2019;4:1636–1644. doi: 10.1038/s41564-019-0488-4. [DOI] [PubMed] [Google Scholar]
- Guo Y, Sirkis DW, Schekman R. Protein sorting at the trans-Golgi network. Annu Rev Cell Dev Biol. 2014;30:169–206. doi: 10.1146/annurev-cellbio-100913-013012. [DOI] [PubMed] [Google Scholar]
- Harak C, Radujkovic D, Taveneau C, Reiss S, Klein R, Bressanelli S, Lohmann V. Mapping of functional domains of the lipid kinase phosphatidylinositol 4-kinase type III alpha involved in enzymatic activity and hepatitis C virus replication. J Virol. 2014;88:9909–26. doi: 10.1128/JVI.01063-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A. Virus Entry: Looking Back and Moving Forward. J Mol Biol. 2018;430:1853–1862. doi: 10.1016/j.jmb.2018.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helenius A. Standing on the Shoulders of Viruses. Annu Rev Biochem. 2020;89:21–43. doi: 10.1146/annurev-biochem-011320-103928. [DOI] [PubMed] [Google Scholar]
- Honeychurch KM, Yang G, Jordan R, Hruby DE. The vaccinia virus F13L YPPL motif is required for efficient release of extracellular enveloped virus. J Virol. 2007;81:7310–5. doi: 10.1128/JVI.00034-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsington J, Lynn H, Turnbull L, Cheng D, Braet F, Diefenbach RJ, Whitchurch CB, Karupiah G, Newsome TP. A36-dependent actin filament nucleation promotes release of vaccinia virus. PLoS Pathog. 2013;9:e1003239. doi: 10.1371/journal.ppat.1003239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu NY, Ilnytska O, Belov G, Santiana M, Chen YH, Takvorian PM, Pau C, van derSchaar H, Kaushik-Basu N, Balla T, et al. Viral reorganization of the secretory pathway generates distinct organelles for RNA replication. Cell. 2010;141:799–811. doi: 10.1016/j.cell.2010.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Sun F, Owen DM, Li W, Chen Y, Gale M, Jr, Ye J. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc Natl Acad Sci U S A. 2007;104:5848–53. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphries AC, Dodding MP, Barry DJ, Collinson LM, Durkin CH, Way M. Clathrin potentiates vaccinia-induced actin polymerization to facilitate viral spread. Cell Host Microbe. 2012;12:346–59. doi: 10.1016/j.chom.2012.08.002. [DOI] [PubMed] [Google Scholar]
- Husain M, Moss B. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J Virol. 2003;77:11754–66. doi: 10.1128/JVI.77.21.11754-11766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huttunen M, Yakimovich A, White IJ, Kriston-Vizi J, Martin-Serrano J, Sundquist WI, Mercer J. Vaccinia virus hijacks ESCRT-mediated multivesicular body formation for virus egress. bioRxiv. 2020 doi: 10.26508/lsa.202000910. 2020.07.15.203935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibiricu I, Huiskonen JT, Dohner K, Bradke F, Sodeik B, Grunewald K. Cryo electron tomography of herpes simplex virus during axonal transport and secondary envelopment in primary neurons. PLoS Pathog. 2011;7:e1002406. doi: 10.1371/journal.ppat.1002406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT. Poliovirus-induced changes in cellular membranes throughout infection. Curr Opin Virol. 2014;9:67–73. doi: 10.1016/j.coviro.2014.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson WT, Giddings TH, Jr, Taylor MP, Mulinyawe S, Rabinovitch M, Kopito RR, Kirkegaard K. Subversion of cellular autophagosomal machinery by RNA viruses. PLoS Biol. 2005;3:e156. doi: 10.1371/journal.pbio.0030156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang B, Hildt E. Intracellular Trafficking of HBV Particles. Cells. 2020;9 doi: 10.3390/cells9092023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson DC, Webb M, Wisner TW, Brunetti C. Herpes simplex virus gE/gI sorts nascent virions to epithelial cell junctions, promoting virus spread. J Virol. 2001;75:821–33. doi: 10.1128/JVI.75.2.821-833.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jourdan N, Maurice M, Delautier D, Quero AM, Servin AL, Trugnan G. Rotavirus is released from the apical surface of cultured human intestinal cells through nonconventional vesicular transport that bypasses the Golgi apparatus. J Virol. 1997;71:8268–78. doi: 10.1128/jvi.71.11.8268-8278.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz RA, Greger JG, Skalka AM. Effects of cell cycle status on early events in retroviral replication. J Cell Biochem. 2005;94:880–9. doi: 10.1002/jcb.20358. [DOI] [PubMed] [Google Scholar]
- Ketter E, Randall G. Virus Impact on Lipids and Membranes. Annu Rev Virol. 2019;6:319–340. doi: 10.1146/annurev-virology-092818-015748. [DOI] [PubMed] [Google Scholar]
- Kielian M. Mechanisms of Virus Membrane Fusion Proteins. Annu Rev Virol. 2014;1:171–89. doi: 10.1146/annurev-virology-031413-085521. [DOI] [PubMed] [Google Scholar]
- Kirchhausen T, Owen D, Harrison SC. Molecular structure, function, and dynamics of clathrin-mediated membrane traffic. Cold Spring Harb Perspect Biol. 2014;6 doi: 10.1101/cshperspect.a016725. a016725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, Stanifer ML, Boulant S, Bartenschlager R, Chlanda P. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. 2020;11:5885. doi: 10.1038/s41467-020-19619-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klupp BG, Granzow H, Fuchs W, Keil GM, Finke S, Mettenleiter TC. Vesicle formation from the nuclear membrane is induced by coexpression of two conserved herpesvirus proteins. Proc Natl Acad Sci U S A. 2007;104:7241–6. doi: 10.1073/pnas.0701757104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoops K, Kikkert M, Worm SH, Zevenhoven-Dobbe JC, van der Meer Y, Koster AJ, Mommaas AM, Snijder EJ. SARS-coronavirus replication is supported by a reticulovesicular network of modified endoplasmic reticulum. PLoS Biol. 2008;6:e226. doi: 10.1371/journal.pbio.0060226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse Locker J, Chlanda P, Sachsenheimer T, Brugger B. Poxvirus membrane biogenesis: rupture not disruption. Cell Microbiol. 2013;15:190–9. doi: 10.1111/cmi.12072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krijnse-Locker J, Ericsson M, Rottier PJ, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langemeyer L, Frohlich F, Ungermann C. Rab GTPase Function in Endosome and Lysosome Biogenesis. Trends Cell Biol. 2018;28:957–970. doi: 10.1016/j.tcb.2018.06.007. [DOI] [PubMed] [Google Scholar]
- Lanke KH, van der Schaar HM, Belov GA, Feng Q, Duijsings D, Jackson CL, Ehrenfeld E, van Kuppeveld FJ. GBF1, a guanine nucleotide exchange factor for Arf, is crucial for coxsackievirus B3 RNA replication. J Virol. 2009;83:11940–9. doi: 10.1128/JVI.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach N, Bjerke SL, Christensen DK, Bouchard JM, Mou F, Park R, Baines J, Haraguchi T, Roller RJ. Emerin is hyperphosphorylated and redistributed in herpes simplex virus type 1-infected cells in a manner dependent on both UL34 and US3. J Virol. 2007;81:10792–803. doi: 10.1128/JVI.00196-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach NR, Roller RJ. Significance of host cell kinases in herpes simplex virus type 1 egress and lamin-associated protein disassembly from the nuclear lamina. Virology. 2010;406:127–37. doi: 10.1016/j.virol.2010.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebsir N, Goueslain L, Farhat R, Callens N, Dubuisson J, Jackson CL, Rouille Y. Functional and Physical Interaction between the Arf Activator GBF1 and Hepatitis C Virus NS3 Protein. J Virol. 2019;93 doi: 10.1128/JVI.01459-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CP, Liu PT, Kung HN, Su MT, Chua HH, Chang YH, Chang CW, Tsai CH, Liu FT, Chen MR. The ESCRT machinery is recruited by the viral BFRF1 protein to the nucleus-associated membrane for the maturation of Epstein-Barr Virus. PLoS Pathog. 2012;8:e1002904. doi: 10.1371/journal.ppat.1002904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leite F, Way M. The role of signalling and the cytoskeleton during Vaccinia Virus egress. Virus Res. 2015;209:87–99. doi: 10.1016/j.virusres.2015.01.024. [DOI] [PubMed] [Google Scholar]
- Li G, Qi X, Hu Z, Tang Q. Mechanisms Mediating Nuclear Trafficking Involved in Viral Propagation by DNA Viruses. Viruses. 2019;11 doi: 10.3390/v11111035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin DH, Hoelz A. The Structure of the Nuclear Pore Complex (An Update) Annu Rev Biochem. 2019;88:725–783. doi: 10.1146/annurev-biochem-062917-011901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz M, Vollmer B, Unsay JD, Klupp BG, Garcia-Saez AJ, Mettenleiter TC, Antonin W. A single herpesvirus protein can mediate vesicle formation in the nuclear envelope. J Biol Chem. 2015;290:6962–74. doi: 10.1074/jbc.M114.627521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lv Y, Zhou S, Gao S, Deng H. Remodeling of host membranes during herpesvirus assembly and egress. Protein Cell. 2019;10:315–326. doi: 10.1007/s13238-018-0577-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maillard P, Walic M, Meuleman P, Roohvand F, Huby T, Le Goff W, Leroux-Roels G, Pecheur EI, Budkowska A. Lipoprotein lipase inhibits hepatitis C virus (HCV) infection by blocking virus cell entry. PLoS One. 2011;6:e26637. doi: 10.1371/journal.pone.0026637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margalit A, Vlcek S, Gruenbaum Y, Foisner R. Breaking and making of the nuclear envelope. J Cell Biochem. 2005;95:454–65. doi: 10.1002/jcb.20433. [DOI] [PubMed] [Google Scholar]
- Maringer K, Stylianou J, Elliott G. A network of protein interactions around the herpes simplex virus tegument protein VP22. J Virol. 2012;86:12971–82. doi: 10.1128/JVI.01913-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JL, Arias CF. Role of the Guanine Nucleotide Exchange Factor GBF1 in the Replication of RNA Viruses. Viruses. 2020;12 doi: 10.3390/v12060682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Clippinger AK, Talledge N, Skowyra ML, Saunders MG, Naismith TV, Colf LA, Afonine P, Arthur C, Sundquist WI, et al. Structure and membrane remodeling activity of ESCRT-III helical polymers. Science. 2015;350:1548–51. doi: 10.1126/science.aad8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Frost A, Sundquist WI. Structures, Functions, and Dynamics of ESCRT-III/Vps4 Membrane Remodeling and Fission Complexes. Annu Rev Cell Dev Biol. 2018;34:85–109. doi: 10.1146/annurev-cellbio-100616-060600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMillan TN, Johnson DC. Cytoplasmic domain of herpes simplex virus gE causes accumulation in the trans-Golgi network, a site of virus envelopment and sorting of virions to cell junctions. J Virol. 2001;75:1928–40. doi: 10.1128/JVI.75.4.1928-1940.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meiser A, Sancho C, Krijnse Locker J. Plasma membrane budding as an alternative release mechanism of the extracellular enveloped form of vaccinia virus from HeLa cells. J Virol. 2003;77:9931–42. doi: 10.1128/JVI.77.18.9931-9942.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melia CE, Peddie CJ, de Jong AWM, Snijder EJ, Collinson LM, Koster AJ, van der Schaar HM, van Kuppeveld FJM, Barcena M. Origins of Enterovirus Replication Organelles Established by Whole-Cell Electron Microscopy. mBio. 2019;10 doi: 10.1128/mBio.00951-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer J, Lee JE, Saphire EO, Freeman SA. SnapShot: Enveloped Virus Entry. Cell. 2020;182:786–786 e1. doi: 10.1016/j.cell.2020.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mettenleiter TC, Muller F, Granzow H, Klupp BG. The way out: what we know and do not know about herpesvirus nuclear egress. Cell Microbiol. 2013;15:170–8. doi: 10.1111/cmi.12044. [DOI] [PubMed] [Google Scholar]
- Midgley R, Moffat K, Berryman S, Hawes P, Simpson J, Fullen D, Stephens DJ, Burman A, Jackson T. A role for endoplasmic reticulum exit sites in foot- and-mouth disease virus infection. J Gen Virol. 2013;94:2636–2646. doi: 10.1099/vir.0.055442-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller EA, Schekman R. COPII - a flexible vesicle formation system. Curr Opin Cell Biol. 2013;25:420–7. doi: 10.1016/j.ceb.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S, Krijnse-Locker J. Modification of intracellular membrane structures for virus replication. Nat Rev Microbiol. 2008;6:363–74. doi: 10.1038/nrmicro1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miorin L, Romero-Brey I, Maiuri P, Hoppe S, Krijnse-Locker J, Bartenschlager R, Marcello A. Three-dimensional architecture of tick-borne encephalitis virus replication sites and trafficking of the replicated RNA. J Virol. 2013;87:6469–81. doi: 10.1128/JVI.03456-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda-Saksena M, Denes CE, Diefenbach RJ, Cunningham AL. Infection and Transport of Herpes Simplex Virus Type 1 in Neurons: Role of the Cytoskeleton. Viruses. 2018;10 doi: 10.3390/v10020092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno-Yamasaki E, Rivera-Molina F, Novick P. GTPase networks in membrane traffic. Annu Rev Biochem. 2012;81:637–59. doi: 10.1146/annurev-biochem-052810-093700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghimi S, Viktorova E, Zimina A, Szul T, Sztul E, Belov GA. Enterovirus infection induces massive recruitment of all isoforms of small cellular Arf GTPases to the replication organelles. J Virol. 2020 doi: 10.1128/JVI.01629-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monel B, Rajah MM, Hafirassou ML, Sid Ahmed S, Burlaud-Gaillard J, Zhu PP, Nevers Q, Buchrieser J, Porrot F, Meunier C, et al. Atlastin Endoplasmic Reticulum-Shaping Proteins Facilitate Zika Virus Replication. J Virol. 2019;93 doi: 10.1128/JVI.01047-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau V, Frischknecht F, Reckmann I, Vincentelli R, Rabut G, Stewart D, Way M. A complex of N-WASP and WIP integrates signalling cascades that lead to actin polymerization. Nat Cell Biol. 2000;2:441–8. doi: 10.1038/35017080. [DOI] [PubMed] [Google Scholar]
- Moss B. Poxvirus membrane biogenesis. Virology. 2015;479–480:619–26. doi: 10.1016/j.virol.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mou F, Forest T, Baines JD. US3 of herpes simplex virus type 1 encodes a promiscuous protein kinase that phosphorylates and alters localization of lamin A/C in infected cells. J Virol. 2007;81:6459–70. doi: 10.1128/JVI.00380-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moyer CL, Nemerow GR. Viral weapons of membrane destruction: variable modes of membrane penetration by non-enveloped viruses. Curr Opin Virol. 2011;1:44–9. doi: 10.1016/j.coviro.2011.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutsafi Y, Altan-Bonnet N. Enterovirus Transmission by Secretory Autophagy. Viruses. 2018;10 doi: 10.3390/v10030139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naghavi MH, Walsh D. Microtubule Regulation and Function during Virus Infection. J Virol. 2017;91 doi: 10.1128/JVI.00538-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Cortese M, Acosta EG, Bartenschlager R. Rewiring cellular networks by members of the Flaviviridae family. Nat Rev Microbiol. 2018;16:125–142. doi: 10.1038/nrmicro.2017.170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neufeldt CJ, Cortese M, Scaturro P, Cerikan B, Wideman JG, Tabata K, Moraes T, Oleksiuk O, Pichlmair A, Bartenschlager R. ER-shaping atlastin proteins act as central hubs to promote flavivirus replication and virion assembly. Nat Microbiol. 2019;4:2416–2429. doi: 10.1038/s41564-019-0586-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Scaplehorn N, Way M. SRC mediates a switch from microtubule-to actin-based motility of vaccinia virus. Science. 2004;306:124–9. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- Pantazopoulou A, Glick BS. A Kinetic View of Membrane Traffic Pathways Can Transcend the Classical View of Golgi Compartments. Front Cell Dev Biol. 2019;7:153. doi: 10.3389/fcell.2019.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pante N, Kann M. Nuclear pore complex is able to transport macromolecules with diameters of about 39 nm. Mol Biol Cell. 2002;13:425–34. doi: 10.1091/mbc.01-06-0308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park R, Baines JD. Herpes simplex virus type 1 infection induces activation and recruitment of protein kinase C to the nuclear membrane and increased phosphorylation of lamin B. J Virol. 2006;80:494–504. doi: 10.1128/JVI.80.1.494-504.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paul D, Bartenschlager R. Flaviviridae Replication Organelles: Oh, What a Tangled Web We Weave. Annu Rev Virol. 2015;2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- Peotter J, Kasberg W, Pustova I, Audhya A. COPII-mediated trafficking at the ER/ERGIC interface. Traffic. 2019;20:491–503. doi: 10.1111/tra.12654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfanzelter J, Mostowy S, Way M. Septins suppress the release of vaccinia virus from infected cells. J Cell Biol. 2018;217:2911–2929. doi: 10.1083/jcb.201708091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ploubidou A, Moreau V, Ashman K, Reckmann I, Gonzalez C, Way M. Vaccinia virus infection disrupts microtubule organization and centrosome function. EMBO J. 2000;19:3932–44. doi: 10.1093/emboj/19.15.3932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ponpuak M, Mandell MA, Kimura T, Chauhan S, Cleyrat C, Deretic V. Secretory autophagy. Curr Opin Cell Biol. 2015;35:106–16. doi: 10.1016/j.ceb.2015.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poruchynsky MS, Maass DR, Atkinson PH. Calcium depletion blocks the maturation of rotavirus by altering the oligomerization of virus-encoded proteins in the ER. J Cell Biol. 1991;114:651–6. doi: 10.1083/jcb.114.4.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A, Ramsden AE, Howell M, Way M. Cytoplasmic ATR Activation Promotes Vaccinia Virus Genome Replication. Cell Rep. 2017;19:1022–1032. doi: 10.1016/j.celrep.2017.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quemin ERJ, Machala EA, Vollmer B, Prazak V, Vasishtan D, Rosch R, Grange M, Franken LE, Baker LA, Grunewald K. Cellular Electron Cryo-Tomography to Study Virus-Host Interactions. Annu Rev Virol. 2020 doi: 10.1146/annurev-virology-021920-115935. [DOI] [PubMed] [Google Scholar]
- Ramani S, Crawford SE, Blutt SE, Estes MK. Human organoid cultures: transformative new tools for human virus studies. Curr Opin Virol. 2018;29:79–86. doi: 10.1016/j.coviro.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rathinam VA, Fitzgerald KA. Cytosolic surveillance and antiviral immunity. Curr Opin Virol. 2011;1:455–62. doi: 10.1016/j.coviro.2011.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves PM, Bommarius B, Lebeis S, McNulty S, Christensen J, Swimm A, Chahroudi A, Chavan R, Feinberg MB, Veach D, et al. Disabling poxvirus pathogenesis by inhibition of Abl-family tyrosine kinases. Nat Med. 2005;11:731–9. doi: 10.1038/nm1265. [DOI] [PubMed] [Google Scholar]
- Reiss S, Rebhan I, Backes P, Romero-Brey I, Erfle H, Matula P, Kaderali L, Poenisch M, Blankenburg H, Hiet MS, et al. Recruitment and activation of a lipid kinase by hepatitis C virus NS5A is essential for integrity of the membranous replication compartment. Cell Host Microbe. 2011;9:32–45. doi: 10.1016/j.chom.2010.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remec Pavlin M, Hurley JH. The ESCRTs - converging on mechanism. J Cell Sci. 2020;133 doi: 10.1242/jcs.240333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rietdorf J, Ploubidou A, Reckmann I, Holmstrom A, Frischknecht F, Zettl M, Zimmermann T, Way M. Kinesin-dependent movement on microtubules precedes actin-based motility of vaccinia virus. Nat Cell Biol. 2001;3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- Risco C, Carrascosa JL, Frey TK. Structural maturation of rubella virus in the Golgi complex. Virology. 2003;312:261–9. doi: 10.1016/S0042-6822(03)00384-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson M, Schor S, Barouch-Bentov R, Einav S. Viral journeys on the intracellular highways. Cell Mol Life Sci. 2018;75:3693–3714. doi: 10.1007/s00018-018-2882-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson SM, Tsueng G, Sin J, Mangale V, Rahawi S, McIntyre LL, Williams W, Kha N, Cruz C, Hancock BM, et al. Coxsackievirus B exits the host cell in shed microvesicles displaying autophagosomal markers. PLoS Pathog. 2014;10:e1004045. doi: 10.1371/journal.ppat.1004045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roller RJ, Fetters R. The herpes simplex virus 1 UL51 protein interacts with the UL7 protein and plays a role in its recruitment into the virion. J Virol. 2015;89:3112–22. doi: 10.1128/JVI.02799-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romero-Brey I, Merz A, Chiramel A, Lee JY, Chlanda P, Haselman U, Santarella-Mellwig R, Habermann A, Hoppe S, Kallis S, et al. Three-dimensional architecture and biogenesis of membrane structures associated with hepatitis C virus replication. PLoS Pathog. 2012;8:e1003056. doi: 10.1371/journal.ppat.1003056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossman JS, Jing X, Leser GP, Lamb RA. Influenza virus M2 protein mediates ESCRT-independent membrane scission. Cell. 2010;142:902–13. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothman JE, Fine RE. Coated vesicles transport newly synthesized membrane glycoproteins from endoplasmic reticulum to plasma membrane in two successive stages. Proc Natl Acad Sci U S A. 1980;77:780–4. doi: 10.1073/pnas.77.2.780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust RC, Landmann L, Gosert R, Tang BL, Hong W, Hauri HP, Egger D, Bienz K. Cellular COPII proteins are involved in production of the vesicles that form the poliovirus replication complex. J Virol. 2001;75:9808–18. doi: 10.1128/JVI.75.20.9808-9818.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva IJ, Carrascosa JL, Risco C. Structural maturation of the transmissible gastroenteritis coronavirus. J Virol. 1999;73:7952–64. doi: 10.1128/jvi.73.10.7952-7964.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salanueva IJ, Novoa RR, Cabezas P, Lopez-Iglesias C, Carrascosa JL, Elliott RM, Risco C. Polymorphism and structural maturation of bunyamwera virus in Golgi and post-Golgi compartments. J Virol. 2003;77:1368–81. doi: 10.1128/JVI.77.2.1368-1381.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiana M, Ghosh S, Ho BA, Rajasekaran V, Du WL, Mutsafi Y, De Jesus-Diaz DA, Sosnovtsev SV, Levenson EA, Parra GI, et al. Vesicle-Cloaked Virus Clusters Are Optimal Units for Inter-organismal Viral Transmission. Cell Host Microbe. 2018;24:208–220 e8. doi: 10.1016/j.chom.2018.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherer J, Hogue IB, Yaffe ZA, Tanneti NS, Winer BY, Vershinin M, Enquist LW. A kinesin-3 recruitment complex facilitates axonal sorting of enveloped alpha herpesvirus capsids. PLoS Pathog. 2020;16:e1007985. doi: 10.1371/journal.ppat.1007985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol. 1994;68:130–47. doi: 10.1128/jvi.68.1.130-147.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm B, Locker JK. Cytoplasmic organization of POXvirus DNA replication. Traffic. 2005;6:839–46. doi: 10.1111/j.1600-0854.2005.00324.x. [DOI] [PubMed] [Google Scholar]
- Sekine E, Schmidt N, Gaboriau D, O’Hare P. Spatiotemporal dynamics of HSV genome nuclear entry and compaction state transitions using bioorthogonal chemistry and super-resolution microscopy. PLoS Pathog. 2017;13:e1006721. doi: 10.1371/journal.ppat.1006721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Katsafanas GC, Weisberg A, Olano LR, Moss B. Identification of Vaccinia Virus Replisome and Transcriptome Proteins by Isolation of Proteins on Nascent DNA Coupled with Mass Spectrometry. J Virol. 2017;91 doi: 10.1128/JVI.01015-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shelness GS, Sellers JA. Very-low-density lipoprotein assembly and secretion. Curr Opin Lipidol. 2001;12:151–7. doi: 10.1097/00041433-200104000-00008. [DOI] [PubMed] [Google Scholar]
- Smith GL, Vanderplasschen A, Law M. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol. 2002;83:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- Snetkov X, Weisswange I, Pfanzelter J, Humphries AC, Way M. NPF motifs in the vaccinia virus protein A36 recruit intersectin-1 to promote Cdc42:N-WASP-mediated viral release from infected cells. Nat Microbiol. 2016;1 doi: 10.1038/nmicrobiol.2016.141. 16141. [DOI] [PubMed] [Google Scholar]
- Snijder EJ, Limpens R, de Wilde AH, de Jong AWM, Zevenhoven-Dobbe JC, Maier HJ, Faas F, Koster AJ, Barcena M. A unifying structural and functional model of the coronavirus replication organelle: Tracking down RNA synthesis. PLoS Biol. 2020;18:e3000715. doi: 10.1371/journal.pbio.3000715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiliotis ET, McMurray MA. Masters of asymmetry - lessons and perspectives from 50 years of septins. Mol Biol Cell. 2020;31:2289–2297. doi: 10.1091/mbc.E19-11-0648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern O, Hung YF, Valdau O, Yaffe Y, Harris E, Hoffmann S, Willbold D, Sklan EH. An N-terminal amphipathic helix in dengue virus nonstructural protein 4A mediates oligomerization and is essential for replication. J Virol. 2013;87:4080–5. doi: 10.1128/JVI.01900-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stertz S, Reichelt M, Spiegel M, Kuri T, Martinez-Sobrido L, Garcia-Sastre A, Weber F, Kochs G. The intracellular sites of early replication and budding of SARS-coronavirus. Virology. 2007;361:304–15. doi: 10.1016/j.virol.2006.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimoto K, Uema M, Sagara H, Tanaka M, Sata T, Hashimoto Y, Kawaguchi Y. Simultaneous tracking of capsid, tegument, and envelope protein localization in living cells infected with triply fluorescent herpes simplex virus 1. J Virol. 2008;82:5198–211. doi: 10.1128/JVI.02681-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed GH, Khan M, Yang S, Siddiqui A. Hepatitis C Virus Lipoviroparticles Assemble in the Endoplasmic Reticulum (ER) and Bud off from the ER to the Golgi Compartment in COPII Vesicles. J Virol. 2017;91 doi: 10.1128/JVI.00499-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabata K, Arimoto M, Arakawa M, Nara A, Saito K, Omori H, Arai A, Ishikawa T, Konishi E, Suzuki R, et al. Unique Requirement for ESCRT Factors in Flavivirus Particle Formation on the Endoplasmic Reticulum. Cell Rep. 2016;16:2339–47. doi: 10.1016/j.celrep.2016.07.068. [DOI] [PubMed] [Google Scholar]
- Tamhankar M, Patterson JL. Directional entry and release of Zika virus from polarized epithelial cells. Virol J. 2019;16:99. doi: 10.1186/s12985-019-1200-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tenorio R, Fernandez de Castro I, Knowlton JJ, Zamora PF, Lee CH, Mainou BA, Dermody TS, Risco C. Reovirus sigmaNS and muNS Proteins Remodel the Endoplasmic Reticulum to Build Replication Neo-Organelles. mBio. 2018;9 doi: 10.1128/mBio.01253-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G. Furin at the cutting edge: from protein traffic to embryogenesis and disease. Nat Rev Mol Cell Biol. 2002;3:753–66. doi: 10.1038/nrm934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas LL, Fromme JC. Extensive GTPase crosstalk regulates Golgi trafficking and maturation. Curr Opin Cell Biol. 2020;65:1–7. doi: 10.1016/j.ceb.2020.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian P, Ball JM, Zeng CQ, Estes MK. The rotavirus nonstructural glycoprotein NSP4 possesses membrane destabilization activity. J Virol. 1996;70:6973–81. doi: 10.1128/jvi.70.10.6973-6981.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol. 1993;60:163–78. [PubMed] [Google Scholar]
- Tsutsui K. Release of vaccinia virus from FL cells infected with the IHD-W strain. J Electron Microsc (Tokyo) 1983;32:125–40. [PubMed] [Google Scholar]
- Tucker SP, Compans RW. Virus infection of polarized epithelial cells. Adv Virus Res. 1993;42:187–247. doi: 10.1016/S0065-3527(08)60086-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uckeley ZM, Moeller R, Kuhn LI, Nilsson E, Robens C, Lasswitz L, Lindqvist R, Lenman A, Passos V, Voss Y, et al. Quantitative Proteomics of Uukuniemi Virus-host Cell Interactions Reveals GBF1 as Proviral Host Factor for Phleboviruses. Mol Cell Proteomics. 2019;18:2401–2417. doi: 10.1074/mcp.RA119.001631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulasli M, Verheije MH, de Haan CA, Reggiori F. Qualitative and quantitative ultrastructural analysis of the membrane rearrangements induced by coronavirus. Cell Microbiol. 2010;12:844–61. doi: 10.1111/j.1462-5822.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eijl H, Hollinshead M, Smith GL. The vaccinia virus A36R protein is a type Ib membrane protein present on intracellular but not extracellular enveloped virus particles. Virology. 2000;271:26–36. doi: 10.1006/viro.2000.0260. [DOI] [PubMed] [Google Scholar]
- Verheije MH, Raaben M, Mari M, Te Lintelo EG, Reggiori F, van Kuppeveld FJ, Rottier PJ, de Haan CA. Mouse hepatitis coronavirus RNA replication depends on GBF1-mediated ARF1 activation. PLoS Pathog. 2008;4:e1000088. doi: 10.1371/journal.ppat.1000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villinger C, Neusser G, Kranz C, Walther P, Mertens T. 3D Analysis of HCMV Induced-Nuclear Membrane Structures by FIB/SEM Tomography: Insight into an Unprecedented Membrane Morphology. Viruses. 2015;7:5686–704. doi: 10.3390/v7112900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voeltz GK, Prinz WA, Shibata Y, Rist JM, Rapoport TA. A class of membrane proteins shaping the tubular endoplasmic reticulum. Cell. 2006;124:573–86. doi: 10.1016/j.cell.2005.11.047. [DOI] [PubMed] [Google Scholar]
- Votteler J, Sundquist WI. Virus budding and the ESCRT pathway. Cell Host Microbe. 2013;14:232–41. doi: 10.1016/j.chom.2013.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang IH, Burckhardt CJ, Yakimovich A, Greber UF. Imaging, Tracking and Computational Analyses of Virus Entry and Egress with the Cytoskeleton. Viruses. 2018;10 doi: 10.3390/v10040166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward BM, Moss B. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J Virol. 2001;75:11651–63. doi: 10.1128/JVI.75.23.11651-11663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward TH, Polishchuk RS, Caplan S, Hirschberg K, Lippincott-Schwartz J. Maintenance of Golgi structure and function depends on the integrity of ER export. J Cell Biol. 2001;155:557–70. doi: 10.1083/jcb.200107045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisberg AS, Maruri-Avidal L, Bisht H, Hansen BT, Schwartz CL, Fischer ER, Meng X, Xiang Y, Moss B. Enigmatic origin of the poxvirus membrane from the endoplasmic reticulum shown by 3D imaging of vaccinia virus assembly mutants. Proc Natl Acad Sci U S A. 2017;114:E11001–E11009. doi: 10.1073/pnas.1716255114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weisz OA, Rodriguez-Boulan E. Apical trafficking in epithelial cells: signals, clusters and motors. J Cell Sci. 2009;122:4253–66. doi: 10.1242/jcs.032615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welsch S, Miller S, Romero-Brey I, Merz A, Bieck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. Composition and threedimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe. 2009;5:365–75. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessels E, Duijsings D, Niu TK, Neumann S, Oorschot VM, de Lange F, Lanke KH, Klumperman J, Henke A, Jackson CL, et al. A viral protein that blocks Arfl-mediated COP-I assembly by inhibiting the guanine nucleotide exchange factor GBF1. Dev Cell. 2006;11:191–201. doi: 10.1016/j.devcel.2006.06.005. [DOI] [PubMed] [Google Scholar]
- Wisner TW, Johnson DC. Redistribution of cellular and herpes simplex virus proteins from the trans-golgi network to cell junctions without enveloped capsids. J Virol. 2004;78:11519–35. doi: 10.1128/JVI.78.21.11519-11535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolff G, Limpens R, Zevenhoven-Dobbe JC, Laugks U, Zheng S, de Jong AWM, Koning RI, Agard DA, Grunewald K, Koster AJ, et al. A molecular pore spans the double membrane of the coronavirus replication organelle. Science. 2020;369:1395–1398. doi: 10.1126/science.abd3629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolffe EJ, Isaacs SN, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–41. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu A, Bellamy AR, Taylor JA. Immobilization of the early secretory pathway by a virus glycoprotein that binds to microtubules. EMBO J. 2000;19:6465–74. doi: 10.1093/emboj/19.23.6465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi Y, Helenius A. Virus entry at a glance. J Cell Sci. 2013;126:1289–95. doi: 10.1242/jcs.119685. [DOI] [PubMed] [Google Scholar]
- Yang Y, Lee M, Fairn GD. Phospholipid subcellular localization and dynamics. J Biol Chem. 2018;293:6230–6240. doi: 10.1074/jbc.R117.000582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Chen Y, Tooze SA. Autophagy pathway: Cellular and molecular mechanisms. Autophagy. 2018;14:207–215. doi: 10.1080/15548627.2017.1378838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou J, Xie X, Lee le T, Chandrasekaran R, Reynaud A, Yap L, Wang QY, Dong H, Kang C, Yuan Z, et al. Dimerization of flavivirus NS4B protein. J Virol. 2014;88:3379–91. doi: 10.1128/JVI.02782-13. [DOI] [PMC free article] [PubMed] [Google Scholar]