Abstract
Meiotic crossovers are tightly restricted in most eukaryotes, despite an excess of initiating DNA double-strand breaks. The majority of plant crossovers are dependent on Class I interfering repair, with a minority formed via the Class II pathway. Class II repair is limited by anti-recombination pathways, however similar pathways repressing Class I crossovers are unknown. We performed a forward genetic screen in Arabidopsis using fluorescent crossover reporters, to identify mutants with increased or decreased recombination frequency. We identified HIGH CROSSOVER RATE1 (HCR1) as repressing crossovers and encoding PROTEIN PHOSPHATASE X1. Genome-wide analysis showed that hcr1 crossovers are increased in the distal chromosome arms. MLH1 foci significantly increase in hcr1 and crossover interference decreases, demonstrating an effect on Class I repair. Consistently, yeast two-hybrid and in planta assays show interaction between HCR1 and Class I proteins, including HEI10, PTD, MSH5, and MLH1. We propose that HCR1 plays a major role in opposition to pro-recombination kinases to restrict crossovers in Arabidopsis.
Keywords: Meiosis, crossover, interference, phosphatase, PPX1, PP4, Arabidopsis
Meiosis is a specialized cell division occurring in eukaryotes, where a single round of DNA replication is coupled to two rounds of chromosome segregation, to generate haploid cells that can undergo sexual fusion1,2. During meiotic prophase I, homologous chromosomes pair and undergo programmed recombination, which can produce reciprocal crossovers between chromosomes1,2. Meiotic recombination and chromosome segregation cause the haploid gametes to be genetically mosaic1,2. As a consequence, sex has a profound effect on genetic variation and adaptation1,2.
Meiotic recombination initiates with formation of DNA double strand breaks (DSBs), via a conserved topoisomerase-related protein SPO111,2. In plants, SPO11-1 and SPO11-2 form a heterotetramer with MTOPVIB to generate meiotic DSBs3–5. Mutation of spo11-1, spo11-2 or mtopvib prevents homolog pairing, causing univalent segregation at metaphase I and aneuploid gametes in Arabidopsis3–5. Meiotic DSBs are resected to generate single-stranded DNA (ssDNA) that is bound by the RecA-related proteins DMC1 and RAD516. DMC1/RAD51 nucleofilaments mediate interhomolog strand invasion to form displacement loops2,6. In wild type Arabidopsis, ~150-250 DSB-associated foci are evident along the meiotic chromosome axis, when DMC1, RAD51, RPA1a and γH2A.X are immunostained during early meiotic prophase I2,7. In wild type Arabidopsis, only ~10 of these DSBs are ultimately repaired as interhomolog crossovers2,8. The remaining strand invasion events are disassembled by non-crossover pathways, which include FANCM, RECQ4A, RECQ4B and FIGL12,9. Meiotic DSBs may also be repaired using the sister chromatid2,10.
In plants, the major pathway generating crossovers is termed Class I (also known as the ZMM pathway)2. Class I crossovers show interference, meaning they are more widely spaced than expected by chance11. In plants, ~80-85% of crossovers are dependent on the Class I pathway, which includes MSH4, MSH5, ZIP4, SHOC1, PTD, HEI10, HEIP1, MER3, MLH1 and MLH3 2,12. The Class I pathway functions to stabilize interhomolog joint molecules and promotes crossover resolution via double Holliday junctions13. Within this pathway; MSH4 and MSH5 form the MutSy heterodimer that associates with meiotic chromosomes and stabilizes interhomolog joint molecules13, SHOC1 and PTD form a catalytically inactive XPF:ERCC1 endonuclease-related complex that has affinity for joint molecules13, HEI10 belongs to a family of ubiquitin and SUMO E3 ligases13, MER3 is a DNA helicase14, and MLH1 and MLH3 form the MutLy heterodimer, which has endonuclease activity13. A minority (~15-20%) of crossovers in plants are non-interfering and dependent on the Class II pathway2. In anti-recombination pathway mutants, for example recq4a recq4b, large increases in Class II crossovers occur15,16.
Progression of the meiotic cell cycle and recombination are regulated by multiple protein kinases pathways, whose targets include DSB proteins, the Class I pathway and the chromosome axis17. For example, cell division kinase CDK1;A promotes Class I crossovers in Arabidopsis, and directly targets MLH1 in vitro 18. In mammals and budding yeast, the ATM/ATR (Mec1/Tel1) DNA damage kinases are activated by meiotic DSBs, and mediate feedback signalling on recombination in cis and trans 19–21. Zip3, the budding yeast HEI10 ortholog has been shown to be an Mec1/Tel1 target in budding yeast22, which is antagonized by the PPH3 PP4 protein phosphatase complex22. The Dbf4/Drf1-dependent kinase Cdc7 complex (DDK) phosphorylates a MSH4 degron to stabilize its association with recombination sites in budding yeast23. Proteins of the chromosomes axis, including ASY1 and REC8, are also extensively phosphorylated during meiosis24,25. How protein kinases and phosphatases are balanced to control meiotic crossovers in plants remains unknown.
To identify new factors that control meiotic recombination we performed a forward genetic screen using a fluorescent crossover reporter. This screen identified the high crossover rate1 (hcr1) mutant in PROTEIN PHOSPHATASE X1, which functions in the nuclear PP4 protein phosphatase complex26–28. Crossovers increased most strongly in distal euchromatic regions in hcr1 and the strength of interference decreased. As MLH1 foci significantly increase in hcr1, this shows that HCR1 represses the Class I crossover pathway. Consistently, yeast two-hybrid and co-immunoprecipitation assays show that HCR1 interacts with the Class I proteins HEI10, PTD, MSH5 and MLH1. We also observed two-hybrid interactions between HCR1 and chromosome axis proteins, DSB factors and recombinases, indicating a potential broader regulatory role during meiosis. We propose that HCR1/PPX1 PP4 phosphatases act in opposition to pro-recombination kinase pathways, in order to limit crossovers in Arabidopsis.
Results
A forward genetic screen for mutants with altered meiotic crossover frequency
To isolate new factors controlling meiotic crossover frequency we performed a forward genetic screen in Arabidopsis thaliana (Fig. 1a). Fluorescent reporters of crossover frequency are available in Arabidopsis, which consist of linked FTL/CTL T-DNA insertions expressing different colors of fluorescent protein in the seed (NapA promoter) or pollen (LAT52 promoter)29–31 (Fig. 1b). When FTLs are hemizygous, inheritance of fluorescence can be used to score crossover frequency within the interval defined by the T-DNAs29–32 (Fig. 1b). We selected the 420 FTL for mutagenesis, which defines a 5.1 megabase interval located in the left sub-telomeric region of chromosome 330,32 (Fig. 1a,b). 420 was chosen for mutagenesis, as crossover frequency in this region is known to be sensitive to multiple recombination and chromatin pathways32–36.
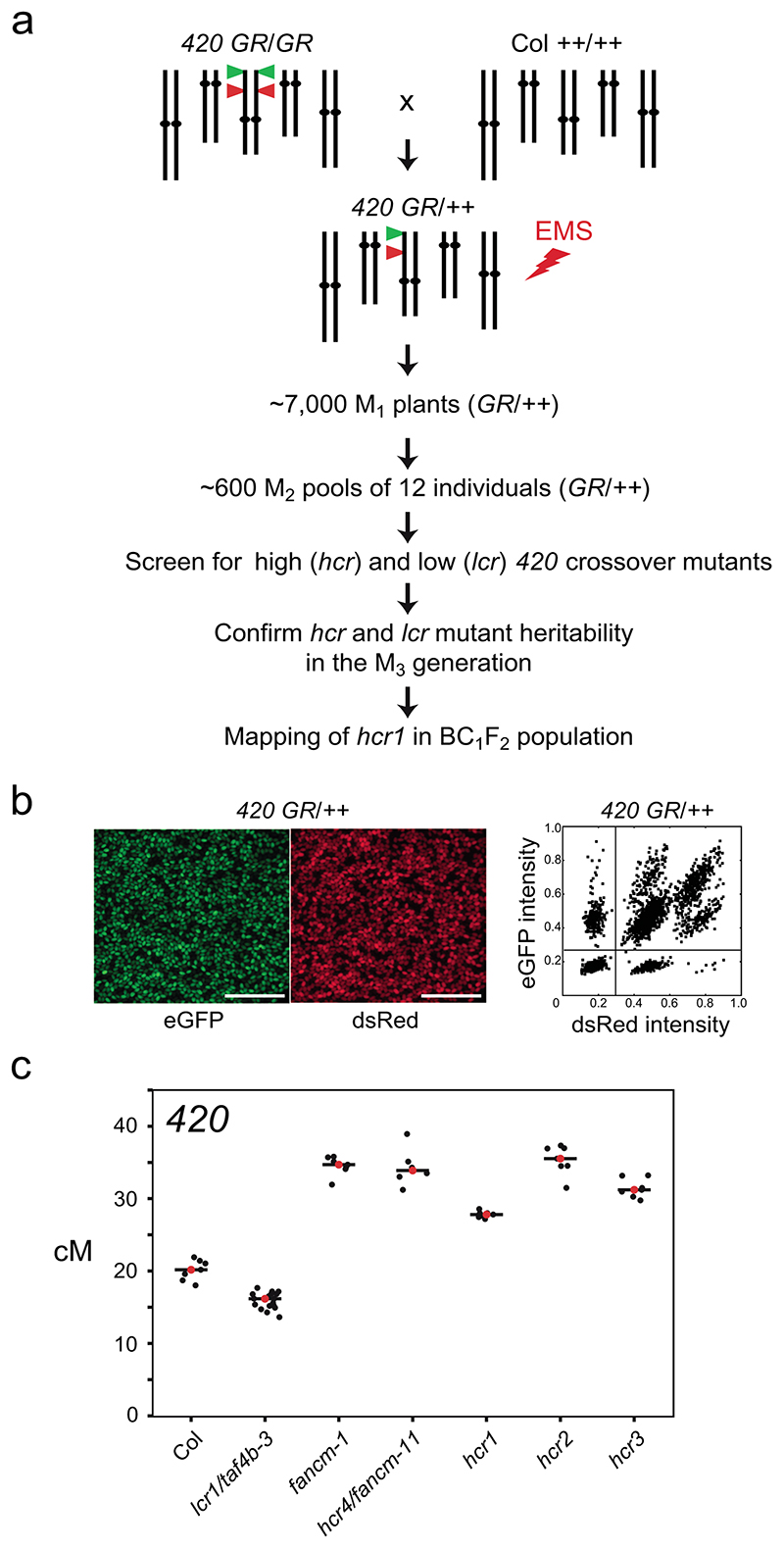
Figure 1. A forward genetic screen for mutants with changed 420 crossover frequency.
a, Schematic diagram of a forward genetic screen for identifying high (hcr) or low (lcr) crossover mutants, using the 420 FTL crossover reporter interval (green and red triangles on chromosome 3). 420/++ seed were EMS treated and the subsequent steps followed to identify the hcr1 mutant. b, Representative fluorescent micrographs of 420/++ seeds. Scale bars=5 mm. A representative plot of red (dsRed) and green (eGFP) fluorescence values from 420/++ seed is shown alongside. Vertical and horizontal lines indicate thresholds for colour:non-colour classifications used for crossover frequency estimation. c, 420 crossover frequency (cM) in wild type, fancm, hcr and lcr mutants. Mean values are indicated by red dots and the black horizontal bar.
We generated ~10,000 420/++ hemizygous seed via crossing and used this for ethyl methanesulfonate (EMS) mutagenesis (Fig. 1a). From these seed, ~7,000 M1 plants were grown and M2 seed was collected (Fig. 1a). The seed from 12 independent M1 plants were combined to generate ~600 M2 pools (Fig. 1a), and seed within these pools were pre-selected to be red-green fluorescent (420/++ hemizygous). Approximately 150 pre-selected seeds were grown from each M2 pool and allowed to self-fertilize (Fig. 1a). Seed from individual M2 plants were used to score crossover frequency within 420 (Fig. 1a,b). In our growth conditions, 420 in self-fertilized wild type Col/Col inbred plants shows a mean crossover frequency of 20.19 cM (standard deviation=1.43) (Fig. 1c and Table S1). In total, 2,883 M2 individuals were screened and the majority (81.4%) showed 420 crossover frequency within the range of 18-22 cM (Extended Data Fig. 1). 19 putative high or low crossover frequency mutants were self-fertilized and M3 progeny tested for 420 crossover frequency, of which 5 were confirmed to show a heritable recombination phenotype in the next generation (Fig. 1a).
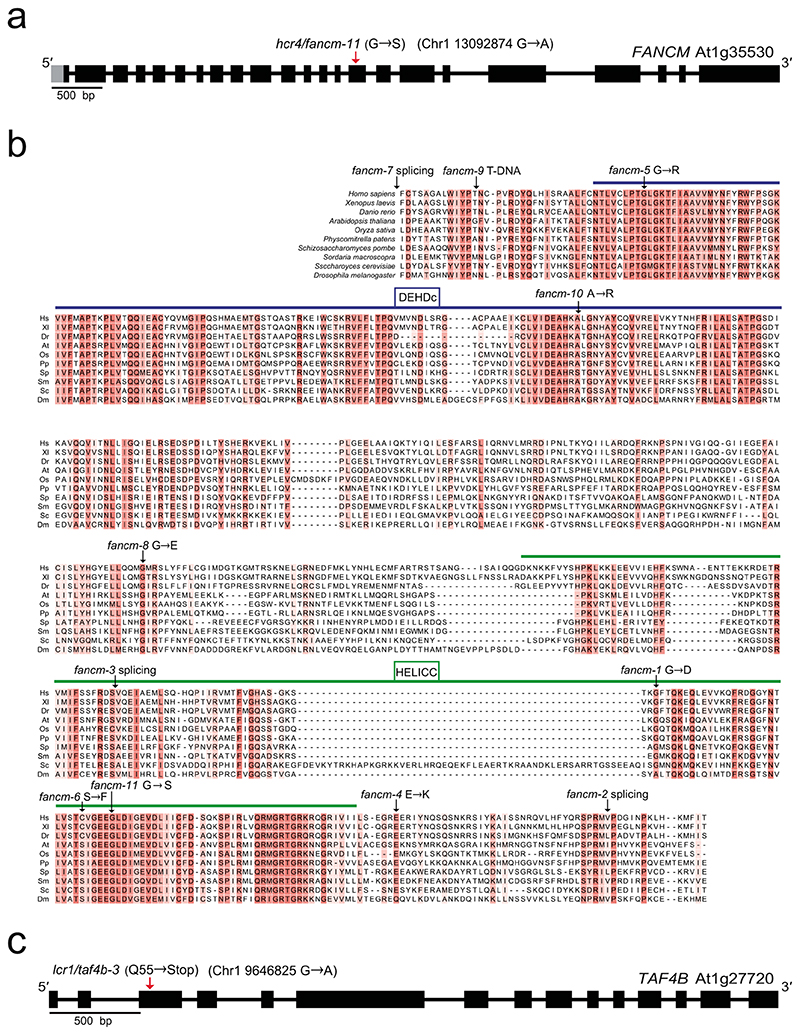
We identified four mutants with high and one with low 420 crossover frequency (Fig. 1c and Supplementary Table 1), which we term high crossover rate1 (hcr1), hcr2, hcr3, hcr4 and low crossover rate1 (lcr1). The hcr4 mutant was shown to be allelic to fancm (Extended Data Fig. 2 and Supplementary Table 1), which is a known repressor of Class II crossovers37. The hcr4 (fancm-11) allele is caused by a non-synonymous amino acid substitution (G540S) in the conserved SF2 helicase domain and shows a comparable effect on 420 crossover frequency to the fancm-1 allele37 (Fig. 1c, Supplementary Table 1 and Extended Data Fig. 2). We identified that lcr1 was allelic with the taf4b mutant (taf4b-3) (Supplementary Table 2 and Extended Data Fig. 2), which was previously shown to promote crossovers in the distal chromosome arms34. In this study we focused on identification and functional characterization of hcr1 (Fig. 1c).
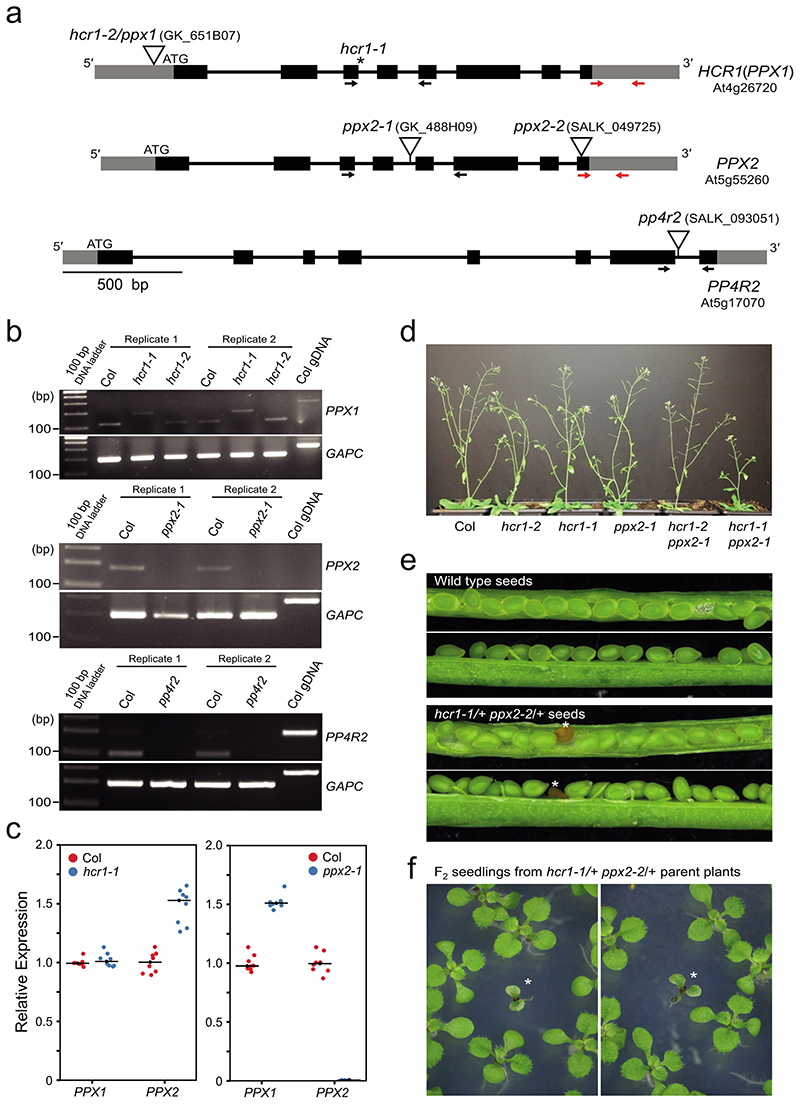
To map the hcr1 mutation we produced a BC1F2 mapping population by backcrossing M3 hcr1 420 (GR/GR) plants to wild type (Col) (Fig. 1a and Fig. 2a). 420 crossover frequency was not significantly different between hcr1/+ BC1F1 and wild type, showing that hcr1 is recessive (Welch’s t-test, P=0.241) (Fig. 2a and Supplementary Table 3). The hcr1/+ 420/++ BC1 hybrid plants were then self-fertilized to generate a 300 individual BC1F2 population, which were scored for 420 crossover frequency (Fig. 1a). Material from the 60 BC1F2 plants with highest 420 crossover frequency was pooled and used for genomic DNA extraction and short-read sequencing (Fig. 2a). We applied the SHORE38 mapping pipeline in order to identify candidate EMS mutations in the high crossover BC1F2 sequencing library (Fig. 2b). The candidate mutation with highest frequency was a G to A substitution in a splice donor site of the 3rd intron of At4g26720, which encodes PROTEIN PHOSPHATASE X1 (PPX1)28 (Fig. 2b,c and Supplementary Table 4).
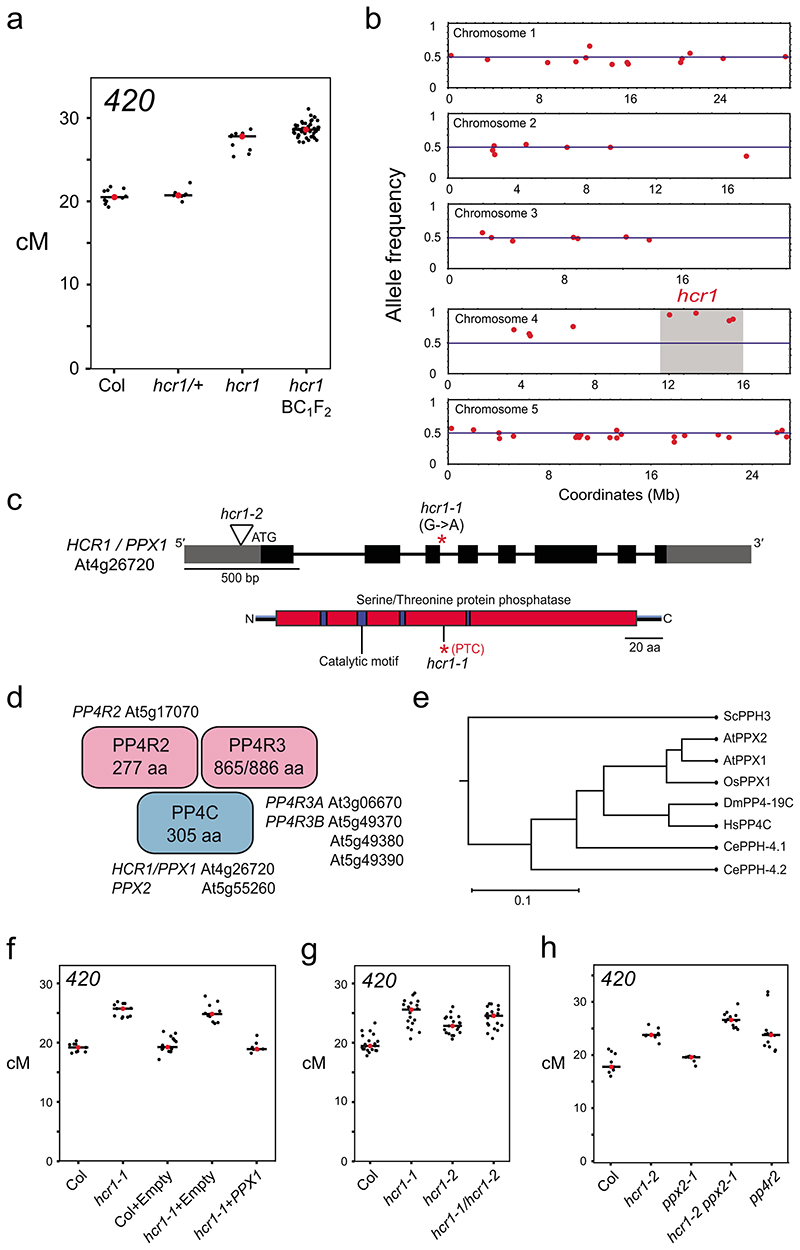
Figure 2. HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1.
a, 420 crossover frequency (cM) in wild type, hcr1-1, hcr1-1/+ and high recombination hcr1-1 BC1F2 individuals used for DNA extraction and mapping-by-sequencing. Mean cM values are indicated by red dots and horizontal lines. b, Allele frequency of EMS mutations (red) identified by SHOREmap in high recombination hcr1-1 BC1F2 individuals. The blue horizontal line indicates 0.5 allele frequency. The hcr1-1 candidate region and mutations are highlighted on chromosome 4 with grey shading. c, HCR1/PPX1 gene with exons shown as boxes (black=CDS, grey=UTR) and the position of the hcr1-1 substitution. The hcr1-2 T-DNA insertion (triangle) is located in the gene 5’-UTR (triangle). A diagram of the HCR1/PPX1 protein is shown indicating the serine/threonine protein phosphatase domain (red), catalytic motifs (blue) and the position of the premature stop codon (*,PTC) caused by hcr1-1. d, A representation of the PP4 phosphatase complex with subunits (PP4C, PP4R2, PP4R3) shown and cognate Arabidopsis homologous genes. e, PPX/PP4C neighbor joining phylogenetic tree based on an alignment of amino acid sequences. The scale bar represents the number of changes per amino acid position. f, As for a, but showing 420 crossover frequency in hcr1-1 after transformation with PPX1 or empty vector constructs. g, As for a, but showing 420 crossover frequency in hcr1-1, hcr1-2 and hcr1-1/hcr1-2 F1 hybrids. h, 420 crossover frequency in the hcr1-2, ppx2 and pp4r2 mutants.
HIGH CROSSOVER RATE1 encodes PROTEIN PHOSPHATASE X1
We used RT-PCR to amplify and sequence PPX1 mRNA from hcr1 plants, which revealed intron 3 retention, causing a premature stop codon (Fig. 2c and Extended Data Fig. 3). The stop codon is predicted to truncate PPX1 (143 of 305 residues) and remove conserved metal-binding histidine residues in the C-terminal region39 (Fig. 2c and Extended Data Fig. 4a). However, the truncated protein has the potential to encode three of four conserved PPX1 catalytic motifs (GDXHG, GDXVDRG and GNHE) in the N-terminal region39 (Fig. 2C and Extended Data Fig. 4a). PPX1 is the catalytic subunit of the hetero-multimeric PP4 serine/threonine protein phosphatase complex, which includes two additional regulatory subunits (PP4R2 and PP4R3)40 (Fig. 2d). PP4 complexes have multiple roles in mitotic and meiotic DNA recombination and repair in diverse eukaryotes26,41–49.
To prove whether the splice acceptor mutation in PPX1 causes the hcr1 420 crossover phenotype, we performed a complementation test (Fig. 2f and Supplementary Table 5). A 4,515 bp genomic fragment containing the PPX1 gene was PCR amplified from wild type (Col) and inserted into an Agrobacterium binary vector and used to transform hcr1 420/++ plants. We observed that the hcr1 plants transformed with PPX1, but not empty vector, showed 420 crossover frequency not significantly different to wild type (Welch’s t-test, P=0.357) (Fig. 2f and Supplementary Table 5). We obtained a second T-DNA insertion (GK_651B07) mutation in PPX1, using a located in the 5′-UTR, which we term hcr1-2, and term the EMS allele hcr1-1 (Fig. 2c,g and Supplementary Table 6). We measured 420 crossover frequency in hcr1-2 homozygotes and observed a significant increase compared to wild type (Welch’s t-test, P=5.43×10-8) (Fig. 2g and Supplementary Table 6), although the phenotype was weaker than hcr1-1. We crossed hcr1-1 with hcr1-2 to generate hcr1-1/hcr1-2 F1 hybrids, which showed significantly higher 420 crossovers compared to wild type, demonstrating allelism (Welch’s t-test, P=4.91×10-10) (Fig. 2g and Supplementary Table 6). Together, these genetic data identify PPX1 as HCR1.
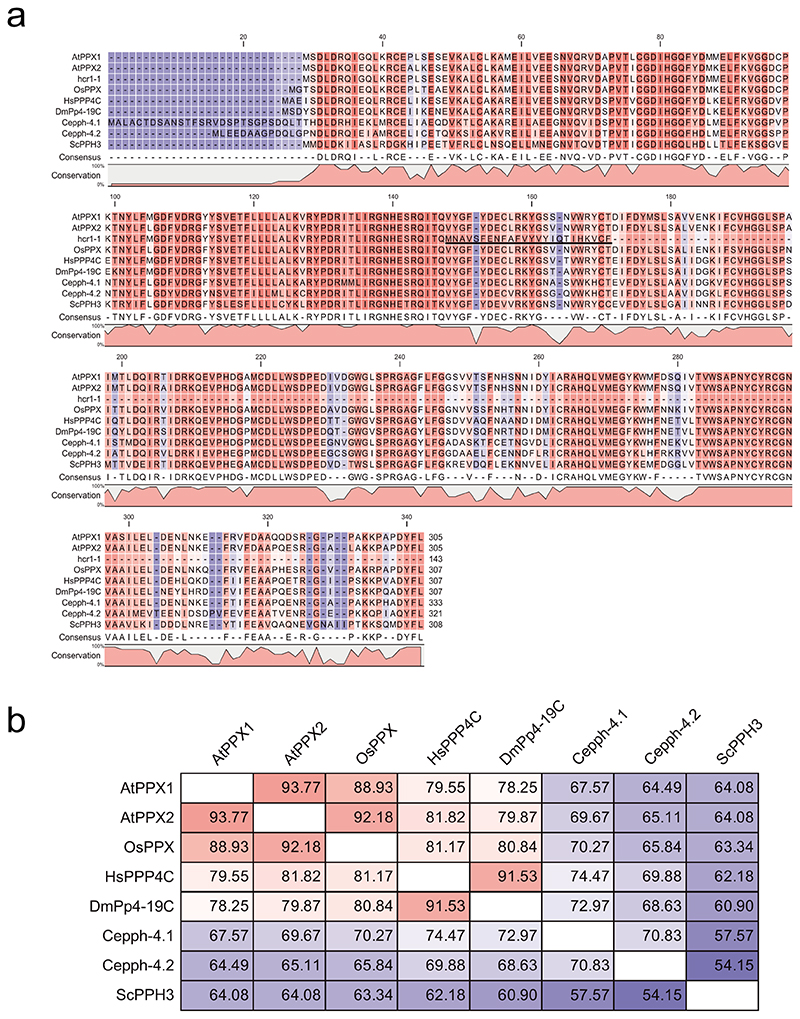
The Arabidopsis genome encodes a second PP4C catalytic subunit gene PPX2 (At5g55260) which shows 93.8% amino acid sequence identity to PPX150–52 (Fig. 2d and Extended Data Fig.4). Functional redundancy between Arabidopsis PPX1 and PPX2 has been observed previously28. We obtained a T-DNA insertion in PPX2 (GK_488H09), which disrupts mRNA expression, but did not observe a significant effect on 420 crossovers, compared to wild type (Welch’s t-test, P=0.119) (Fig. 2h, Extended Data Fig. 3a,b and Supplementary Table 7). However, hcr1-2 ppx2-1 double mutants showed a significant increase in 420 crossovers, compared to hcr1-2 (Welch’s t-test, P=1.42×10-4) (Fig. 2h and Supplementary Table 7). We also crossed hcr1-1 with a second ppx2 T-DNA insertion allele (ppx2-2) and failed to identify hcr1-1 ppx2-2 double mutants in the F2 generation. As the siliques of hcr1-1/+ ppx2-2/+ plants contained aborted seed not seen in wild type controls, this supports that the double mutant is embryo or seedling lethal (Extended Data Fig. 3d-3f). Arabidopsis encodes a single gene for the PP4R2 regulatory subunit (At1g17070), and we obtained a T-DNA insertion that disrupts mRNA expression of this gene (Extended Data Fig. 3a,b). We observed that pp4r2 shows a significant increase in 420 crossover frequency, compared to wild type (Welch’s t-test, P=4.24×10-5), with a similar phenotypic strength to hcr1-2 (Fig. 2h and Supplementary Table 7). As pp4r2 mutants are viable, this indicates that the T-DNA insertion is likely to be hypomorphic. Together, this is consistent with HCR1/PPX1 and PPX2 acting in PP4 complexes with PP4R2 to repress meiotic crossovers in Arabidopsis. We also note that recent mass spectroscopy data from Arabidopsis has confirmed the presence of HCR1/PPX1, PPX2, PP4R2L and PP4R3A complexes in vivo 28.
Meiosis-specific knockdown of HCR1/PPX1 and PPX2 using meiMIGS
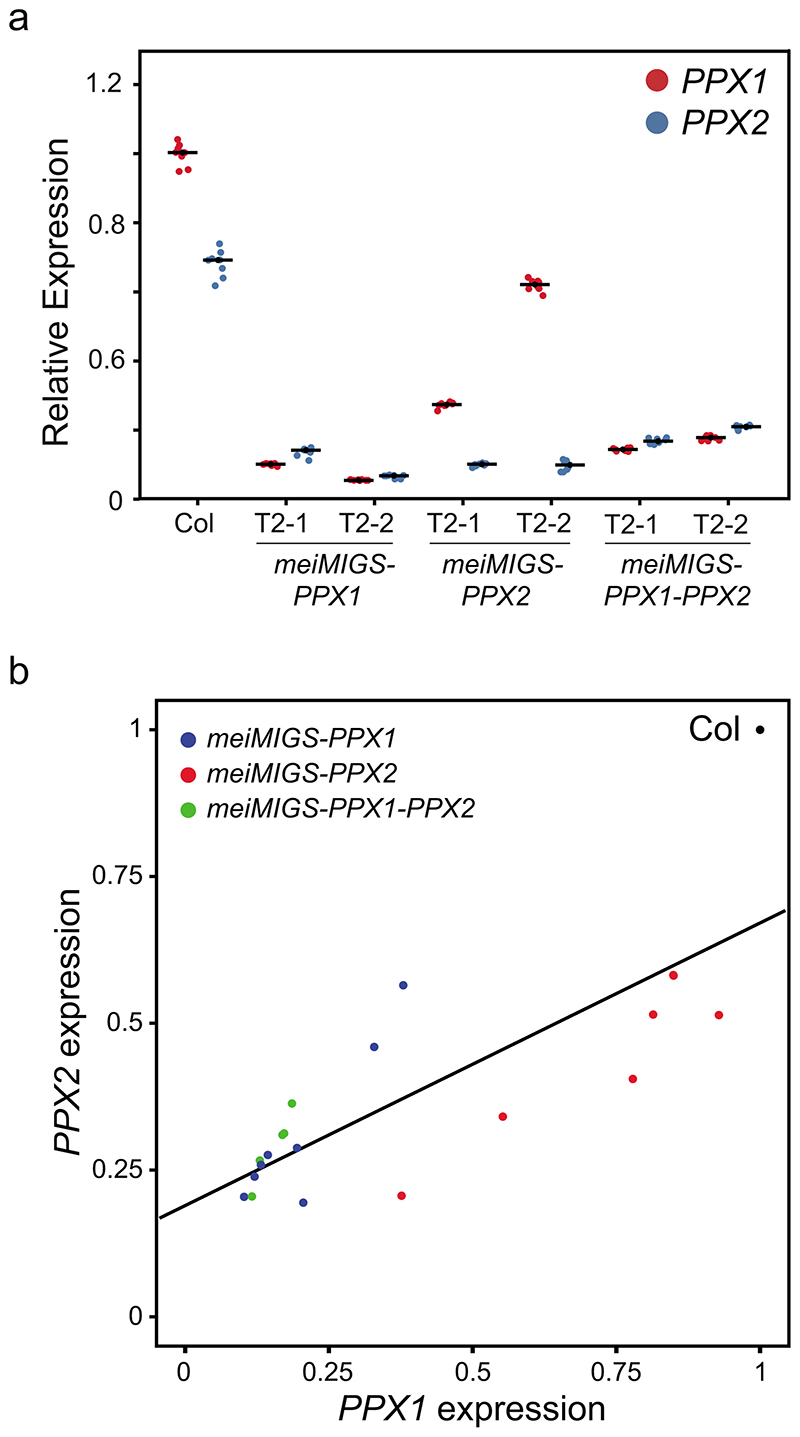
Our genetic analysis indicates functional redundancy between PPX1 and PPX2 (Fig. 2h). This is consistent with null ppx1 ppx2 double mutants causing severe developmental phenotypes, not observed in the single mutants28. Therefore, we sought to silence both PPX1 and PPX2 specifically during meiosis. For this purpose we adapted miRNA-induced gene silencing (MIGS) for use during meiosis53. MIGS constructs fuse a microRNA173 (miR173) target site upstream of target transcript sequences53. Transcript cleavage of the fusion RNA by endogenous miR173 is an efficient trigger of 22 nucleotide trans-acting siRNAs (tasiRNAs), which act to silence endogenous gene transcripts that share sequence homology in trans 53. To drive MIGS specifically during meiosis (meiMIGS), we expressed miRNA173-target PPX1 and PPX2 gene fusions from the DMC1 promoter54 (Fig. 3a). We measured PPX1 and PPX2 transcripts levels from meiotic stage floral buds in meiMIGS transformed plants and observed a significant reduction of both genes in all tested lines, compared to wild type (Welch’s t-test, all P<1.51×10-9) (Extended Data Fig. 5). Cross-silencing of PPX1 and PPX2 by the meiMIGS constructs is expected, as these genes share 86.6% nucleotide identity. The constructs were transformed into 420/++ plants and we observed a significant increase in crossover frequency compared to wild type (Welch’s t-test, all P<1.01×10-4) (Fig. 3b and Supplementary Table 8). We correlated relative expression of PPX1 and PPX2 in these backgrounds with 420 crossover frequency and observed a significant negative correlation in both cases (PPX1 r=-0.76 P=6.73×10-5, PPX2 r=-0.64 P=1.81×10-3) (Fig. 3b-3c and Extended Data Fig. 5). Together, this demonstrates quantitative increases in crossover frequency that correlate with the degree of PPX1 and PPX2 silencing.
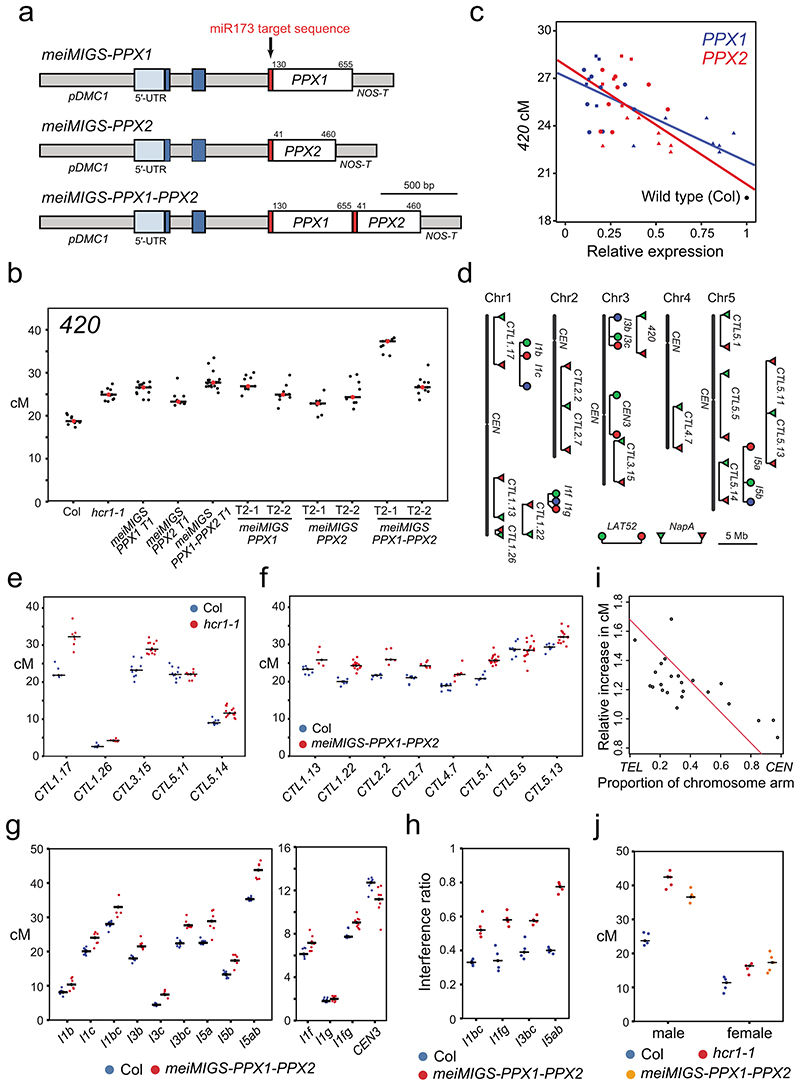
Figure 3. Euchromatic crossover frequency increases and crossover interference decreases in hcr1 and meiMIGS-PPX1-PPX2.
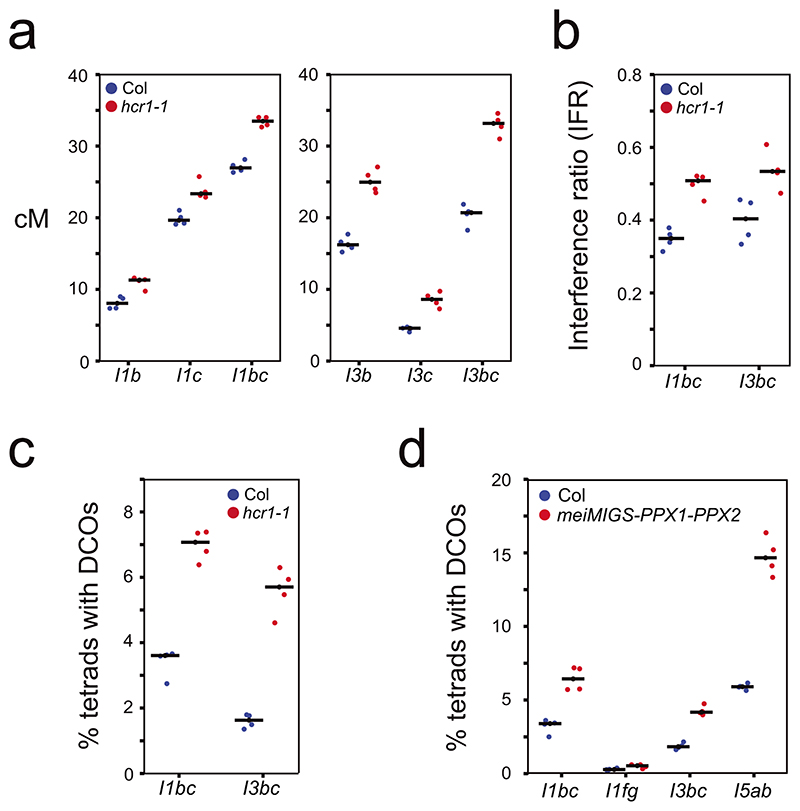
a, Graphical representation of the meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 constructs. b, 420 crossover frequency (cM) in wild type, meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 T1 and T2 transgenic lines. c, Correlation between 420 cM and PPX1/HCR1 and PPX2 transcript levels in floral buds of wild type and meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 T2 transgenic lines. The y axis represents 420 cM and x axis indicates fold-enrichment of PPX1 (blue) and PPX2 (red) transcript levels compared to PPX1 and PPX2 in wild type in RT-qPCR analysis. DMC1 was used as a meiotic gene for normalization. Mean values of triple replicate RT-qPCRs were used. Wild type (Col), meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 plants are shown as a black circle, red or blue-circles, -triangles and -squares, respectively. d, FTL T-DNA intervals throughout the Arabidopsis genome used to measure crossover frequency. Circles indicate LAT52-driven, and triangles indicate NapA-driven FTL transgenes. e, As for c, but showing FTL crossover frequency in wild type (blue) and hcr1-1 (red). Mean values are indicated by horizontal black lines. f, As for c, but showing FTL crossover frequency in wild type (blue) and meiMIGS-PPX1-PPX2 (red). g, As for c, but showing pollen-based FTL crossover frequency in wild type (blue) and meiMIGS-PPX1-PPX2 (red). h, Crossover interference ratio measured using FTL pollen tetrads in wild type (blue) compared with meiMIGS-PPX1-PPX2 (red). i, Correlation between FTL cM change in hcr1-1 or meiMIGS-PPX1-PPX2 and the midpoint of the FTL interval analysed relative to the telomere (TEL) and centromere (CEN). j, 420 crossover frequency (cM) in male and female meiosis of wild type (blue), hcr1-1 (red) and meiMIGS-PPX1-PPX2 (orange).
Euchromatic crossovers increase and the strength of interference decreases in hcr1 and meiMIGS-PPX1-PPX2
To investigate the effect of hcr1 and meiMIGS-PPX1-PPX2 on crossover frequency in other genomic regions, we crossed these lines with additional FTL/CTL recombination reporters29–31 (Fig. 3d), expressing fluorescent proteins using either seed (Fig. 3e,f), or pollen promoters (Fig. 3g). Plants carrying seed-based CTL reporters were self-fertilized and measure both male and female meiosis (Fig. 3e,f). We observed that distal FTL intervals CTL1.17, CTL1.26, CTL3.15 and CTL5.4 showed significantly higher crossover frequency in hcr1-1, compared to wild type (Welch’s t-test, all P<1.08×10-4) (Fig. 3e and Supplementary Table 9). In contrast, the centromere spanning interval CTL5.11 did not significantly change in hcr1-1 (Fig. 3e and Supplementary Table 9). The same patterns were confirmed using meiMIGS-PPX1-PPX2, which showed significant crossover increases in the distal and interstitial FTL intervals CTL1.13, CTL1.22, CTL2.2, CTL2.7, CTL4.7, CTL5.1 and CTL5.13, compared to wild type (Welch’s t-test, all P < 1.71×10-3), whereas the centromeric interval CTL5.5 did not significantly change (Fig. 3f and Supplementary Table 10).
We crossed meiMIGS-PPX1-PPX2 with pollen-based FTL intervals, which are combined with the quartet1 mutation31 (Fig. 3d, 3g,h and Supplementary Table 11-12). This assay measures crossover frequency and interference specifically in male meiosis31. For analysis we used a deep learning pipeline DeepTetrad, which enables high-throughput analysis of fluorescent tetrads55. We tested four three-color FTL intervals located in distal chromosome regions; I1bc, I1fg, I3bc and I5ab. All intervals, except the relatively narrow I1g, showed significant crossover increases in meiMIGS-PPX1-PPX2 compared to wild type (Welch’s t-test, all P<7.28×10-3) (Fig. 3g, Extended Data Fig. 6 and Supplementary Table 11). We also tested the centromere-spanning FTL CEN3, which significantly decreased in meiMIGS-PPX1-PPX2 (Welch’s t-test, P=5.05×10-3) (Fig. 3g and Supplementary Table 12). Across all FTL data, we correlated the proximity of each interval midpoint to the centromere, with the change in crossover frequency that occurred in hcr1-1 or meiMIGS-PPX1-PPX2 relative to wild type (Fig. 3i and Supplementary Tables 9-12). This analysis revealed a significant negative correlation (r=-0.709 P=1.48×10-4) between the crossover increase and proximity to the centromere (Fig. 3i). These results show that the distal chromosome regions significantly increase crossovers in hcr1 and meiMIGS-PPX1-PPX2 when measured in male meiosis alone, or in both male and female meiosis. To specifically compare male and female recombination, we backcrossed wild type, hcr1 and meiMIGS-PPX1-PPX2 plants that were 420/++ hemizygous, as either male or female parents. The 420 interval is heterochiasmic and shows significantly higher crossover frequency in male (24.23 cM), compared with female (10.98 cM) (Welch’s t-test P=2.92x10-6) (Figure 3j and Supplementary Table 13). We observed that both hcr1 and meiMIGS-PPX1-PPX2 showed significant crossover increases in male (Welch’s t-test P=6.25×10-7 and 2.15×10-7) and female (Welch’s t-test P=2.81×10-3 and 1.75×10-3) meiosis, compared to wild type (Figure 3j and Supplementary Table 13).
For three-color, pollen-based FTL intervals we are able to measure crossovers in adjacent regions and thereby measure interference31,55. (Fig. 3h, Extended Data Fig. 6b and Supplementary Table 14). Crossover interference ratios (IFR) are calculated using the genetic map distance in the test interval, with and without a crossover occurring in the adjacent interval. An IFR of 1 indicates an absence of interference31,55. We observed that meiMIGS-PPX1-PPX2 causes an increase in crossover frequency, but a decrease in the strength of interference in FTLs I1bc, I1fg, I3bc and I5ab (Welch’s t-test, all P<3.05×10-3) (Fig. 3g,h and Supplementary Table 14). Therefore, a higher incidence of double crossovers in adjacent intervals occurs in meiMIGS-PPX1-PPX2, compared to wild type (Extended Data Fig. 6d). We repeated three-color analysis using FTL intervals I1bc and I3bc in hcr1-1 and again observed significantly increased crossover frequency and decreased crossover interference (higher IFR) (Welch’s t-tests, P=2.7×10-4 , P=8.1×10-3) (Extended Data Fig. 6a-c and Supplementary Table 14).
Genome-wide mapping of crossovers in meiMIGS-PPX1-PPX2
Our FTL data indicate that the euchromatic chromosome arms undergo an increase in crossover frequency in hcr1 and meiMIGS-PPX1-PPX2. Notably, these FTL experiments were performed in a Col/Col inbred background. Therefore, we sought to test the effect of meiMIGS-PPX1-PPX2 on crossovers in a hybrid background (Fig. 4a). We crossed wild type (Col), or a meiMIGS-PPX1-PPX2 transgenic line in the Col background carrying the 420 FTL, to Ler and generated Col/Ler F1 hybrids (Fig. 4a and Supplementary Table 15). We measured 420 crossover frequency in wild type and meiMIGS-PPX1-PPX2 Col/Ler F1 hybrids and observed a significant increase in meiMIGS-PPX1-PPX2 (Welch’s t-test, P=6.55×10-11) (Fig. 4b and Supplementary Table 15). This demonstrates that PPX1 and PPX2 repress crossovers in both inbred and hybrid backgrounds.
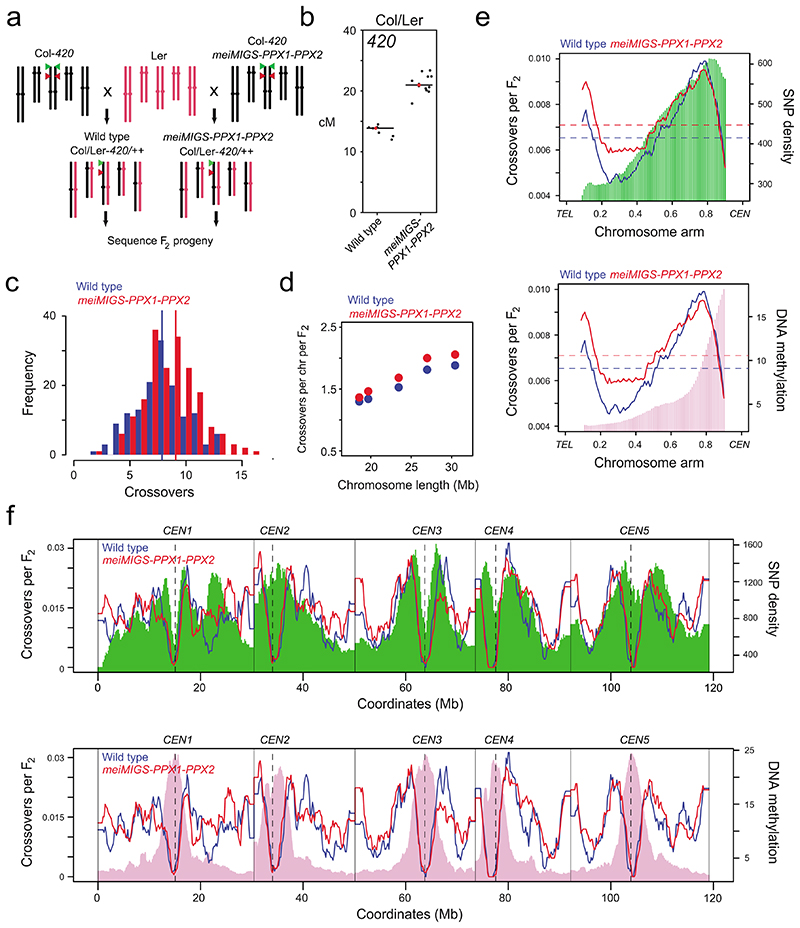
Figure 4. Genome-wide mapping of crossovers in meiMIGS-PPX1-PPX2.
a, Schematic diagram showing crossing of meiMIGS-PPX1-PPX2 Col-420 (black) and wild type Col-420 (black), to Ler (red) to generate F2 populations for genotyping-by-sequencing. Green and red triangles indicate 420 T-DNAs on chromosome 3. b, 420 crossover frequency (cM) in wild type and meiMIGS-PPX1-PPX2 Col/Ler F1 hybrids. c, Histogram of crossover number per F2 individual in wild type (blue) Col/Ler and meiMIGS-PPX1-PPX2 (red) populations. Vertical dashed lines indicate mean values. d, Crossovers per chromosome per F2 compared with chromosome length in wild type (blue) and meiMIGS-PPX1-PPX2 (red). e, Normalized crossover frequency plotted along chromosome arms orientated from telomere (TEL) to centromere (CEN) in wild type (blue) and meiMIGS-PPX1-PPX2 (red) F2 populations. Mean values are indicated by horizontal dashed lines. Also plotted is Col/Ler SNP frequency (green, upper) and DNA methylation (pink, lower). f, As for e, but without telomere-centromere scaling. Vertical solid lines indicate telomeres and vertical dotted lines indicate centromeres.
We self-fertilized wild type and meiMIGS-PPX1-PPX2 Col/Ler F1 plants and generated 144 wild type and 192 meiMIGS-PPX1-PPX2 F2 plants, from which genomic DNA was extracted. This DNA was sequenced and data was analysed using the TIGER pipeline8,56, in order to identify crossover locations in each wild type and meiMIGS-PPX1-PPX2 F2 individual (Fig. 4a, c-f). Crossovers were mapped to an average of 962 bp and 936 bp in wild type and meiMIGS-PPX1-PPX2 F2 populations, respectively (Supplementary Table 15). We observed a significant increase in crossovers per F2 from 7.86 in wild type, to 8.57 in meiMIGS-PPX1-PPX2 (Welch’s t-test, P=7.7×10-3) (Fig. 4c). We observed increased crossover numbers on each chromosome in meiMIGS-PPX1-PPX2 compared to wild type (Fig. 4d), and a positive correlation between crossover number and chromosome length (wild type r=0.986, meiMIGS-PPX1-PPX2 r=0.983) (Fig. 4d and Supplementary Table 16).
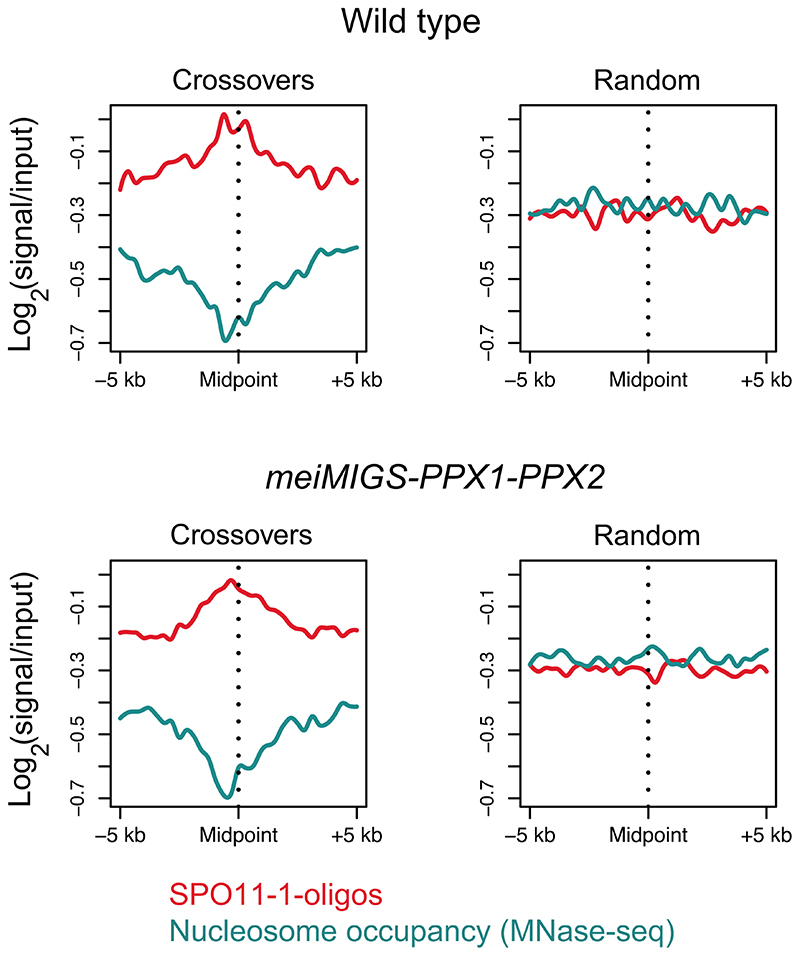
We analysed the crossover landscape in wild type and meiMIGS-PPX1-PPX2 (Fig. 4e,f). We averaged all chromosome arms along their telomere-centromere axes and plotted crossover frequency per F2 in wild type and meiMIGS-PPX1-PPX2 (Fig. 4e,f). Wild type and meiMIGS-PPX1-PPX2 show a U-shaped distribution of crossover frequency along the chromosomes, with high recombination in the distal sub-telomeres and pericentromeres (Fig. 4e,f). We observed that the first 60-70% of the chromosome arms from the telomeres showed elevated crossovers in meiMIGS-PPX1-PPX2 compared to wild type, whereas the pericentromeres and centromeres showed a similar level of recombination (Fig. 4e,f), which is consistent with our previous FTL analysis (Fig. 3d-g and 3i). DNA methylation is highest in the centromeric region33, where recombination is suppressed in both wild type and meiMIGS-PPX1-PPX2 (Fig. 4e,f). We compared crossover frequency to Col/Ler SNP frequency, which follows an ascending gradient from the telomeres to the centromeres (Fig. 4e,f). The distal regions of the chromosomes with lowest SNP density and lowest DNA methylation underwent the greatest crossover increase in meiMIGS-PPX1-PPX2,s compared to wild type (Fig. 4e,f). We analysed nucleosome occupancy (MNase-seq) and SPO11-1-oligos (a marker of meiotic DSBs) around crossover locations in wild type and meiMIGS-PPX1-PPX2, compared to the same number of randomly chosen locations57,58. We observed that crossovers in both genotypes showed a similar depletion of nucleosome occupancy and enrichment of SPO11-1-oligos, compared to random positions (Extended Data Fig. 7). This indicates that while distal regions increase crossovers in meiMIGS-PPX1-PPX2, recombination retains a local bias for accessible DNA that experiences higher DSB levels.
hcr1 and meiMIGS-PPX1-PPX2 show elevated Class I MLH1 foci at diakinesis stage
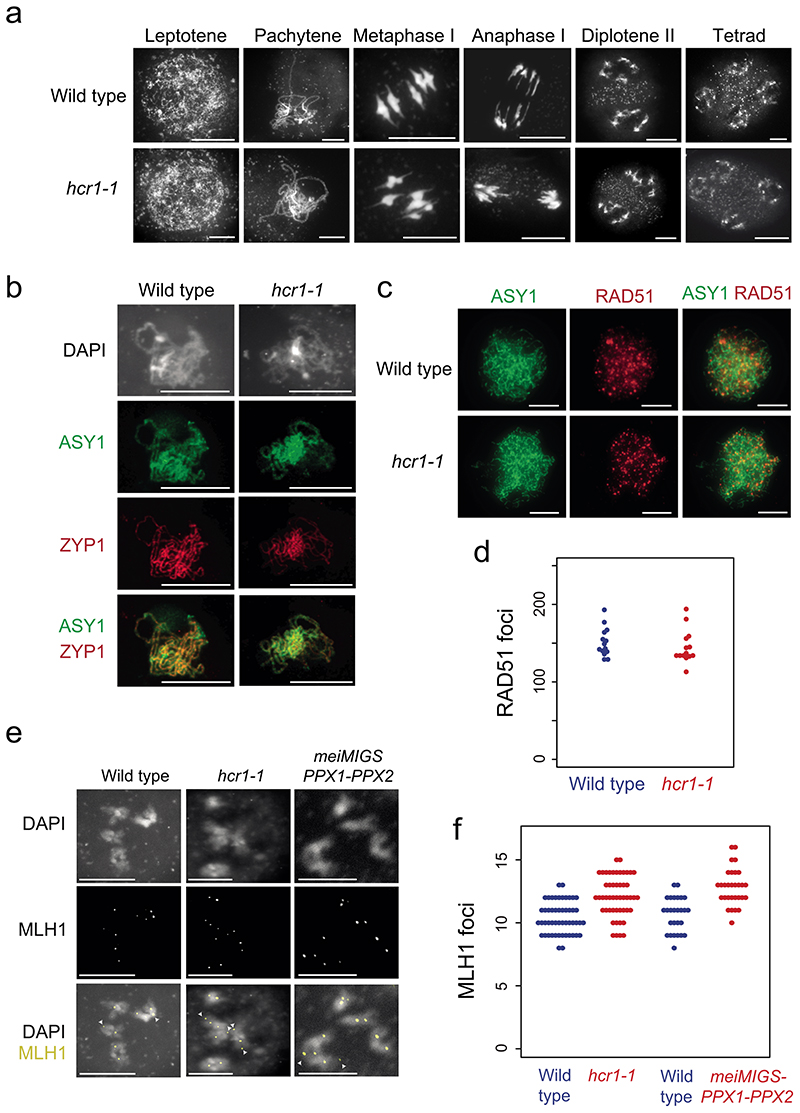
We used cytological analysis to analyze meiosis in hcr1-1 compared to wild type. We spread wild type and hcr1-1 male meiocytes and stained chromosomes using 4’,6-diamidino-2-phenylindole (DAPI) (Fig. 5a). We observed normal chromosome morphology during prophase I (leptotene and pachytene) in hcr1-1, normal bivalent morphology at metaphase I and chromosome segregation during anaphase I and meiosis II (Fig. 5a). This is consistent with hcr1-1 showing no difference in fertility compared to wild type (Supplementary Table 16). To investigate formation of the chromosome axis and homolog synapsis, we immunostained wild type and hcr1-1 meiocytes for the HORMA domain protein ASY1 and the synaptonemal complex protein ZYP1, during prophase I (Fig. 5b). Wild type and hcr1-1 showed normal homolog synapsis and immunostaining of ASY1 and ZYP1 (Fig. 5b).
Figure 5. Meiotic MLH1 foci are elevated in hcr1 whereas RAD51, ASY1 and ZYP1 immunostaining are unchanged.
a, Representative images of male meiocytes spread and stained with DAPI in wild type (Col) and hcr1-1, at the labeled stages of meiosis. Scale bars=10 μm. b, Representative images of ASY1 (green) and ZYP1 (green) immunostaining of wild type (Col-0) and hcr1-1 male meiocytes at pachytene. Nuclei spreads were also stained with DAPI. Scale bars=10 μM. c, Representative images of ASY1 (green) and RAD51 (red) co-immunostaining on wild type (Col-0) and hcr1-1 male meiocytes during early prophase I. Scale bars=10 μM. d, Quantification of RAD51 foci number per cell in wild type and hcr1-1. e, Representative images of MLH1 (red) immunostaining of male meiocytes at diakinesis stage in wild type, hcr1-1 and meiMIGS-PPX1-PPX2. Cells were also DNA stained with DAPI (blue). Arrows represent MLH1 foci at distal locations on the chromosomes. Scale bars=10 μM. f, Quantification of MLH1 foci number per cell scored at diakinesis stage in wild type (blue), hcr1-1 (red) and meiMIGS-PPX1-PPX2 (red). Scale bars=10 μM. All cytological experiments represent data collected from at least two biological replicates.
We immunostained meiocytes in early prophase I for ASY1 and the DSB marker RAD51 and observed no significant difference in RAD51 foci number between wild type and hcr1-1 (Fig. 5c,d and Supplementary Table 18) (Wilcoxon t-test, P=0.32). This is consistent with normal levels of meiotic DSBs forming in hcr1 relative to wild type. Finally, we immunostained for the MLH1 Class I protein at diakinesis stage on DAPI-stained male meiocyte spreads (Fig. 5e,f and Supplementary Table 19). Quantification of MLH1 foci numbers per nucleus showed a significant increase in hcr1-1 (mean=12.1 foci), compared to wild type (mean=10.4 foci) (Wilcoxon test, P=5.3×10-7) (Fig. 5e,f and Supplementary Table 19). We also measured MLH1 foci in wild type (Col) and meiMIGS-PPX1-PPX2, using the same transgenic line as for genotyping-by-sequencing. We observed that meiMIGS-PPX1-PPX2 showed significantly higher MLH1 foci (mean=12.8), compared to wild type (mean=10.7) (Wilcoxon test P=2.5×10-6) (Supplementary Table 20). Together, this is consistent with the crossover increases observed in hcr1 and meiMIGS-PPX1-PPX2 being mediated mainly via the Class I repair pathway.
HCR1 interacts with the Class I crossover pathway proteins HEI10, PTD, MSH5 and MLH1
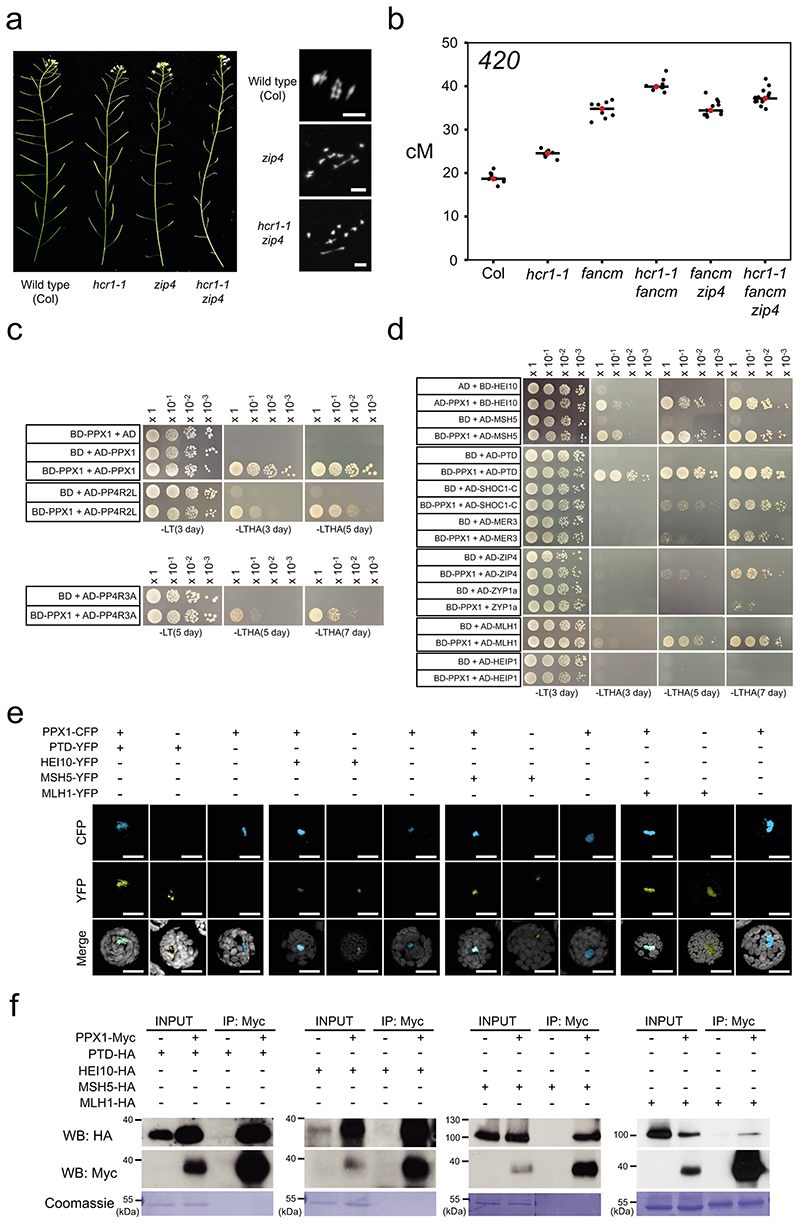
As we observed elevated MLH1 foci in hcr1 and meiMIGS-PPX1-PPX2 (Fig. 5e,f), we sought to investigate genetic interactions with the Class I and Class II repair pathways. Class I pathway mutants, for example zip4, have low fertility due to reduced crossovers, unbalanced chromosome segregation and aneuploid gametes14 (Fig. 5a). Fertility of Class I mutants can be restored by mutations that block non-crossover formation and increase Class II crossovers, for example fancm 37. We generated zip4 hcr1 double mutants and observed that fertility was not restored (Fig. 6a). We performed meiotic chromosome spreads and counted chiasma, bivalents and univalents in wild type (Col), zip4 and zip4 hcr1 (Supplementary Table 21). We observed that zip4 and zip4 hcr1 showed strongly reduced bivalents (zip4 mean=0.8, zip4 hcr1 mean=1.3), compared to wild type (mean=5) (Wilcoxon test, Col vs zip4 P=5.22×10-12, Col vs zip4 hcr1 P=1.43×10-11). The bivalent counts for zip4 and zip4 hcr1 were not significantly different from one another (Wilcoxon test P=0.11). This is further consistent with a major effect for hcr1 on the Class I pathway. We also generated hcr1 fancm double mutants carrying the 420 FTL interval, and observed an additive increase in genetic distance in the double mutant compared to hcr1 and fancm single mutants (Welch’s t-tests, P=2.7×10-11, P=6.77×10-6) (Fig. 6b and Supplementary Table 20). The hcr1 fancm zip4 triple mutant showed lower 420 crossover frequency than hcr1 fancm, but higher than fancm zip4 (Welch’s t-test, P=5.60×10-4, P=9.93×10-4) (Fig. 6b and Supplementary Table 22). This suggests that hcr1 may also increase the number of Class II crossovers, at least in a fancm zip4 mutant background (Fig. 6b and Supplementary Table 22).
Figure 6. HCR1 genetically and physically interacts with the Class I crossover pathway.
a, Representative siliques from wild type, hcr1-1, zip4 and hcr1-1 zip4 plants. Shown alongside are representative metaphase I chromosome spreads stained with DAPI from wild type (Col), zip4 and zip4 hcr1. This was repeated with three biological replicates. Scale bars=10μM. b, 420 crossover frequency (cM) in wild type, hcr1-1, fancm, fancm zip4, hcr1-1 fancm and hcr1-1 fancm zip4. Mean values are indicated by red dots and horizontal lines. c, Yeast two hybrid assays showing interactions of HCR1 with PP4R2 and PP4R3. Yeast co-transformants were grown until OD600=1, diluted 10-, 100- and 1,000-fold, spotted on synthetic dropout media (SD) lacking leucine/tryptophan (-LT) and leucine/trptophan/histidine/adenine (-LTHA) and grown for 3 to 5 days. d, As for c, but showing interactions of HCR1 with HEI10, PTD and MSH5, and weaker interactions with SHOC1, MER3, ZIP4, MLH1 and ZYP1a. Yeast transformants were grown on SD (-LTHA) for 3, 5 or 7 days. e, Co-localization of fluorescent protein fusions with HCR1 and HEI10, PTD, MSH5 and MLH1 in Arabidopsis protoplasts. All scale bars=20 μm. Experiments were repeated at least three times. f, Coimmunoprecipitation analyses of HCR1 and HEI10, PTD, MSH5 and MLH1. IB=immunoblot; IP=immunoprecipitation. Experiments were repeated at least three times.
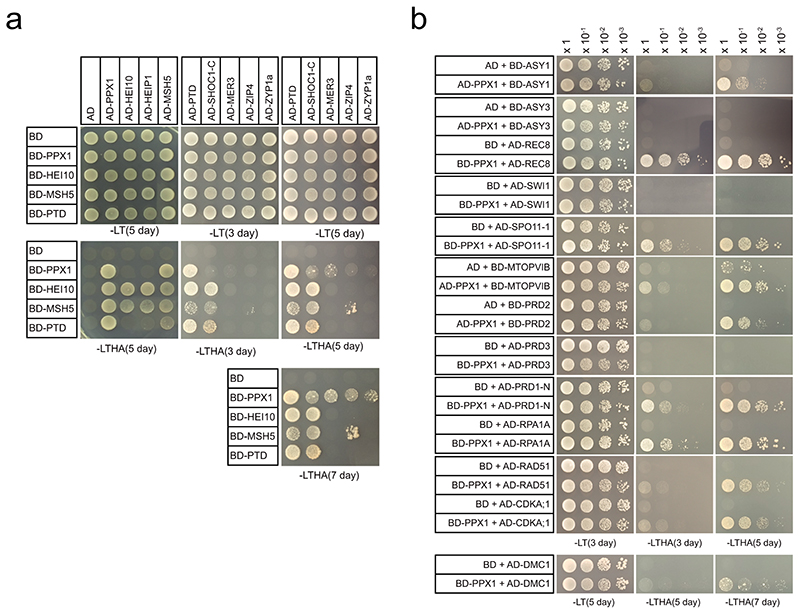
We investigated whether HCR1 physically interacts with known components of the meiotic recombination pathways. We cloned HCR1/PPX1 into yeast 2-hybrid (Y2H) AD and BD vectors and tested interactions with Class I proteins, in addition to the PP4 regulatory subunits PP4R2L and PP4R3A (Fig. 6c,d, Extended Data Fig. 8 and Supplementary Table 23). As expected28, HCR1 interacts strongly with the PP4 regulatory subunits PP4R2L and PP4R3A (Fig. 6c and Supplementary Table 23). Of the tested Class I combinations we observed strong Y2H interactions between HCR1 and HEI10, MSH5 and PTD (Fig. 6c,d). We also detected weaker interactions between HCR1 and the Class I pathway proteins MER3, ZIP4, SHOC1 and MLH1 (Extended Data Fig. 8d and Supplementary Table 22). Within the Class I pathway we observed strong interactions between HEI10, HEIP1 and MSH5, and between SHOC1 and PTD (Extended Data Fig. 8a,b), consistent with data in rice and Arabidopsis12,59,60. We additionally tested a wider set of 13 meiotic proteins that included the synaptonemal complex protein ZYP1a, DNA repair factors (DMC1, RAD51, RPA1A), DSB proteins (PRD1, PRD2, PRD3, SPO11-1, MTOPVIB) and meiotic chromosome axis proteins (ASY1, ASY3, SWI1 and REC8). Using serial dilutions, we observed that HCR1 shows strong interactions with REC8, SPO11-1, PRD1, RPA1A, MTOPVIB and PRD2 and weaker interactions with ASY1, RAD51, DMC1, ZYP1a and CDKA;1 (Fig. 6c,d and Extended Data Fig. 8a,b). Hence, although HCR1 represses the Class I crossover pathway, it may play a more widespread role regulating protein phosphorylation during Arabidopsis meiosis.
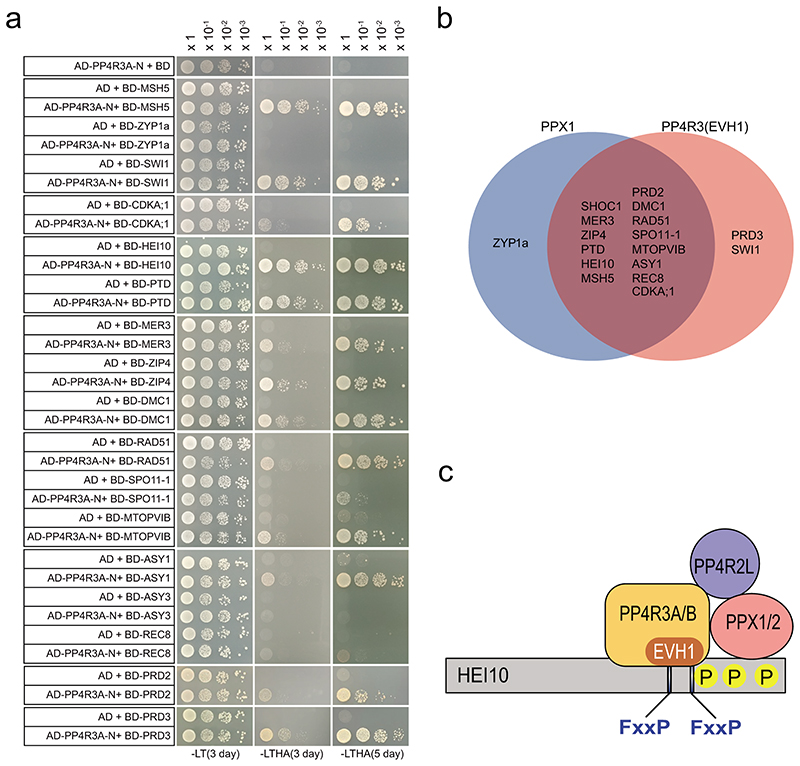
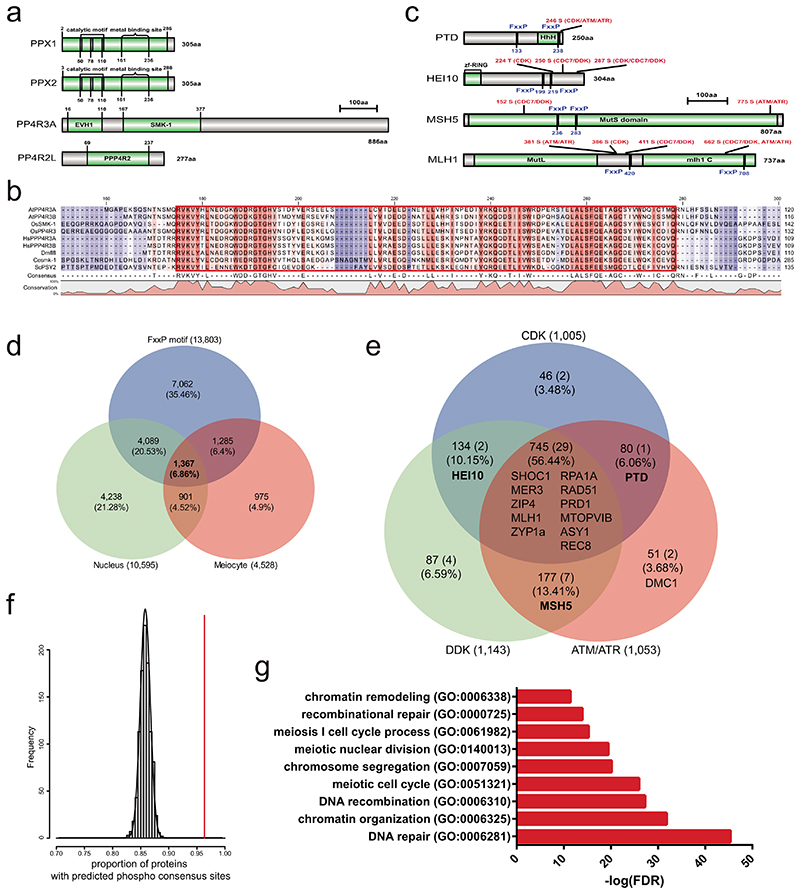
The human PP4 complex targets multiple proteins by recognizing a short motif (FxxP) via the PP4R3 Ena/Vasp Homology1 (EVH1) domain61. To explore whether a similar mechanism is relevant in Arabidopsis we performed yeast two-hybrid experiments using the Arabidopsis PP4R3A (At3g06670) EVH1 domain (residues 1-166) (Extended Data Figure 9). The PP4R3A-EVH1 domain interacts with 14 of 15 proteins observed as HCR1 interactors (Extended Data Figure 9). Additionally, PP4R3A showed two-hybrid interactions with PRD3 and SWI1 (Extended Data Figure 9). These data are consistent with HCR1/PPX1 and PP4R3A PP4 subunits interacting with a diverse set of proteins that regulate meiotic chromosomes and recombination, including Class I factors.
We sought to further test protein-protein interactions between HCR1 and Class I proteins in planta, using transient transfection and co-localization studies in Arabidopsis protoplasts (Fig. 6e). As reported52, expression of a HCR1-CFP fusion protein showed nuclear localization (Fig. 6e). We co-expressed PPX1-CFP with PTD-YFP, HEI10-YFP, MSH5-YFP and MLH1-YFP fusion proteins and observed nuclear co-localization in all cases (Fig. 6e). We confirmed physical association of PPX1 using co-immunoprecipitation following transient expression in Arabidopsis protoplasts of PPX1-Myc, together with PTD-HA, HEI10-HA, MSH5-HA or MLH1-HA (Fig. 6f). In each case, these experiments confirmed that these proteins interact in planta (Fig. 6f).
As discussed, human protein phosphatase 4 (PP4) complexes bind the consensus motif FxxP, via the PP4R3A EVH1 domain61 (Extended Data Fig. 9a,b and 10). Interestingly, 15 of 18 PPX1 interactors, and 12 of 16 PP4R3A-EVH1 interactors, identified using Y2H assays contain at least one FxxP motif (Extended Data Fig. 9, 10 and Supplementary Table 20). The PPX1 and PP4R3A interactors also possess multiple consensus sites used by CDK, DDK and ATM/ATR kinases (Extended Data Fig. 10c and Supplementary Table 20). We searched genome-wide for potential meiotic PP4 substrates according to the criteria of; (i) FxxP motifs (n=13,803), (ii) predicted nuclear location (n=10,595) and (iii) meiocyte-specific expression34,61 (n=4,528). This search identified 1,367 candidate targets for the PP4 complex during meiosis (Extended Data Fig. 10d). 1,315 of these proteins (96.2%) have at least one phosphorylation consensus site (Extended Data Fig. 10e). Furthermore, 15 of 18 PPX1 Y2H interactors, 12 of 16 PP4R3A-EVH1 Y2H interactors and 49 of 84 known meiotic proteins were included in this list of candidate PP4 substrates (Extended Data Fig. 10e and Supplementary Table 23). The proportion of candidate PP4 substrates (1,367) with at least one phosphorylation site is significantly higher than the random expectation (comparing to numbers of phosphorylation sites in 1,000 random sets of 1,367 proteins, Z-test P=7.02×10-31) (Extended Data Fig. 10f). The 1,367 predicted meiotic PP4 substrates are also significantly enriched in GO terms for DNA repair, DNA recombination, chromatin organization and meiosis I cell cycle (Extended Data Fig. 10g). Together this indicates the wide potential for PP4 regulation of meiosis and recombination in Arabidopsis.
Discussion
We identified the HCR1/PPX1 phosphatase as a repressor of crossover frequency in Arabidopsis. We provide genetic, cytological and protein-protein interaction data that a major target of HCR1/PPX1 is the Class I crossover pathway, with a minor role repressing Class II crossovers (Fig. 7). Our protein interaction data indicate that HEI10, PTD, MSH5 and MLH1 are likely direct targets for HCR1/PPX1 PP4 phosphatase activity within the Class I pathway. However, we also observed that HCR1/PPX1 and PP4R3A interact in a two-hybrid assay with components of the chromosome axis (ASY1, ASY3, REC8, SWI1), DSB proteins (SPO11-1, MTOPVIB, PRD1, PRD2) and recombinases (RPA1A, RAD51, DMC1), consistent with a broader regulatory role during meiosis.
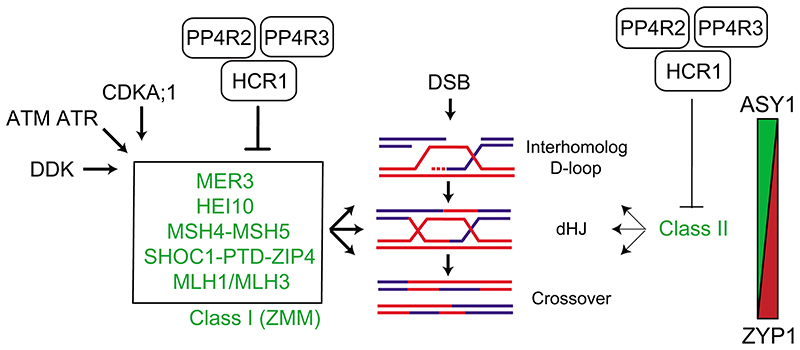
Figure 7. HCR1/PPX1 PP4 control of meiotic crossover recombination in Arabidopsis.
During meiosis, recombination is initiated by DNA double strand breaks (DSB) that can be resected to form single stranded DNA (ssDNA). In the central diagram a resected ssDNA end (blue) from one homolog has invaded the second homolog (red), to form an interhomolog displacement loop (D-loop). A subset of IH D-loops are further processed to form double Holliday junctions (dHJs), which may be resolved into a crossover. The Class I (also known as ZMM) pathway proteins (green) acts at multiple steps within the formation and stabilization of IH D-loops and dHJs and their resolution into interfering crossovers. The activity of the Class I pathway has been shown to be promoted by independent kinase pathways, including CDK, DDK and Mec1/Tel1 (ATM/ATR). We propose that HCR1 acts with PP4R2 and PP4R3 in PP4 phosphatase complexes that antagonize one or more of the pro-recombination kinase pathways on Class I targets and thereby restrict the number of interfering crossovers that form per meiosis. The Class II pathway contributes to ~10% of crossovers in wild type. Our data also indicate a minor role for repression of the Class II pathway by HCR1. On the right is a diagram indicating that during progression of meiotic recombination, the abundance of axis protein ASY1 (green) is depleted, as the synaptonemal complex protein ZYP1 (red) increases.
In the absence of HCR1/PPX1, we propose that the action of pro-recombination kinases on the Class I pathway promotes stabilization of interhomolog strand invasion and crossover formation (Fig. 7). The crossover increases observed in hcr1 and meiMIGS-PPX1-PPX2 were most pronounced in the distal chromosome ends. Notably, distal crossover increases are characteristic of situations with elevated Class I activity in Arabidopsis, including male meiosis, HEI10 and CDKA;1 36,62,63, although distal increases are also observed in mutants that increase Class II crossovers (e.g recq4a recq4b)9,16,62. The causes of distal biases in crossover formation in these backgrounds remain incompletely understood. Chromatin may be an important influence, as meiotic DSBs are elevated in gene-associated nucleosome-free regions, and there are positive associations with euchromatic chromatin marks, including H3K4me3 and H2A.Z35,58,64,65. In contrast, heterochromatic modifications including H3K9me2 and dense DNA methylation are associated with crossover suppression33,66. Additionally, Class I crossovers are subject to interference, which inhibits formation of adjacent crossovers in a distance-dependent manner11. A complete understanding of the crossover landscape in hcr1 will require further investigation of how chromatin, chromosome structure and interference co-operate spatially and temporally during meiosis.
Within the Class I pathway, HEI10 belongs to a family of conserved ubiquitin or SUMO E3 ligases that promote interfering crossover formation in diverse eukaryotes2,13. In Arabidopsis, HEI10 is a dosage-sensitive promoter of Class I crossover repair16,36. HEI10 shows a dynamic localization pattern along plant meiotic chromosomes, initially showing numerous foci along the axis, which become restricted to a small number of foci that overlap MLH1 foci during late prophase I12,67,68. In budding yeast, the HEI10 ortholog Zip3 is phosphorylated in a DSB-dependent manner by Mec1 (ATR), which is antagonized by PPH322. This is of particular interest as PPH3 is a HCR1/PPX1 ortholog, indicating that repression of the Class I pathway by PP4 phosphatases may be conserved between plants and fungi.
In mice, orthologs of HEI10 (e.g. RNF212) act to regulate association of the MutSy Msh4-Msh5 heterodimer with meiotic chromosomes69,70. Msh4-Msh5 heterodimers are capable of forming sliding clamps on DNA in vitro and associate with recombination foci along meiotic chromosomes in vivo 71,72. MutSy is proposed to bind nascent joint molecules and protect them from dissolution by anti-recombinases, including Sgs1-Top3-Rmi1 in budding yeast71,73,74. MutSy can also directly or indirectly recruit the MutLy (Mlh1-Mlh3) endonuclease heterodimer to promote crossover resolution75–77. Budding yeast Msh4 was recently identified as an intrinsically unstable protein that is degraded by the proteasome via an N-terminal degron23. Phosphorylation of the degron by the cell cycle kinase Cdc7-Dbf4 (DDK) inhibits Msh4 degradation and thereby promotes crossover repair23. As Arabidopsis HCR1/PPX1 physically interacts with MSH5 and MLH1 this may promote MutSy and MutLy dephosphorylation and thereby repress Class I crossover repair.
We observed physical interaction between HCR1/PPX1 and PTD, which is the partner protein of SHOC1, which together form a XPF-ERCC1-related complex60,78–80. Orthologs of the SHOC1-PTD complex include budding yeast Zip2-Spo16, which bind branched DNA molecules in vitro, lacks endonucleolytic activity and acts with Zip4 to promote crossover formation80,81. However, phosphorylation of Zip2-Spo16-Zip4 has been not reported in budding yeast or other organisms. Since Arabidopsis PTD interacts with HCR1 and PP4R3A-EVH1 and contains consensus phosphorylation sites, it is possible that plant SHOC1-PTD-ZIP4 complexes may be regulated by phosphorylation.
It is also possible that HCR1/PPX1 may regulate phosphorylation of the DSB machinery, or components of the meiotic chromosome axis, as observed in Caenorhabditis elegans 82. Furthermore, orthologs of ASY1 (Hop1), REC8 (Rec8) and ZYP1 (Zip1) proteins in budding yeast are known to be regulated via phosphorylation24,25. Hence, it is possible that Arabidopsis ASY1, REC8 and ZYP1 may be dephosphorylated by PP4. However, we did not observe significant changes to RAD51 foci or ASY1 and ZYP1 immunostaining during meiosis in hcr1 at the cytological level.
We consider three pro-recombination kinase pathways as candidates for HCR1/PPX1 PP4 antagonism (Fig. 7). First, cell division kinase (Cdk)-cyclin complexes are drivers of cell cycle progression, including during meiosis and are known to regulate recombination18,63,83. Second, Dbf4-dependent kinase (DDK) (Cdc7-Ddf4) plays a prominent role in the initiation of DNA replication, but also in regulation of recombination and kinetochore behaviour during meiosis84–89. Third, the ATM/ATR phosphatidylinositol 3-kinase-related kinases (PIKKs) are activated by DSBs and regulate meiotic DSB number and distribution in yeast and mammals19–21. Together these kinase pathways play complex and interacting roles in the promotion of crossovers during meiosis90.
In Arabidopsis, CDKA;1 (the homolog of human Cdk1 and Cdk2) plays a role in promoting Class I crossovers63,91. Hence, HCR1/PPX1 may remove phosphorylation from CDKA;1 targets within the Class I pathway and thereby limit crossovers (Fig. 7). Interestingly, mutation of CDK consensus motifs (S/T-P) in budding yeast Zip3 had no effect on phosphorylation, whereas mutation of Tel1/Mec1 sites (S/T-Q) did22. As noted earlier, Zip3 phosphorylation has been shown to be regulated by PP422, meaning that HCR1 may regulate HEI10 phosphorylation in an analogous manner in Arabidopsis (Fig. 7). Indeed, it has been shown that many Mec1 phospho-targets, including Zip1, are also PP4 substrates in budding yeast49. In Arabidopsis ATM and ATR are redundantly required for DSB repair92. The atm single mutant is partially sterile with increased meiotic DSBs, chromosomal fragmentation and moderately increased Class I crossovers93,94. In budding yeast, DDK is responsible for Msh4 degron phosphorylation and stabilization23. Hence, it is possible that HCR1 could remove phosphorylation from MutSy and thereby promote its destabilization and repress crossovers (Fig. 7). However, the meiotic function of DDK kinases in plants is currently unknown.
Studies in diverse systems and contexts have identified PP4 phosphatase complexes as key regulators of DNA repair and recombination. For example, the DNA damage response involves kinase regulation, which is balanced with antagonising phosphatases41. Defined roles for PP4 complexes include; (i) dephosphorylation of gamma-H2AX during recovery from DNA damage checkpoints in Drosophila, budding yeast and human42–45, (ii) prevention of Rad53 hyperphosphorylation during DSB repair and promoting DNA end resection in budding yeast95, (iii) dephosphorylating RPA2 to promote DNA repair via homologous recombination27, (iv) promoting NHEJ-mediated DSB repair, which occurs partially via KRAB-associated protein1 (KAP1)48, (v) regulation of Mec1 during DSB repair and at sites of replication fork collapse26, and (vi) regulating Zip1 phosphorylation during meiosis to control homology-independent centromere pairing49. Our work identifies PPX1-PP4 phosphatase complexes as repressing the Class I crossover pathway during Arabidopsis meiosis. We propose that PP4 complexes may generally act in opposition to pro-recombination kinases to regulate meiotic crossovers in eukaryotes.
Methods
Plant materials
Arabidopsis plants were grown under controlled conditions of 22°C, 50-60% humidity and 16/8 hour light-dark cycles. Seeds were incubated at 4°C in the dark for 3-4 days in order to stratify germination. Seed-expressed FTL/CTL and pollen-expressed FTL lines were used29,30. T-DNA insertion lines in ppx1 (GK_651B07), ppx2 (GK_488H09), pp4r2 (SALK_093051), zip4-2 96 (SALK_068052) and the fancm-1 EMS mutant37 were provided by Nottingham Arabidopsis Stock Centre. Genotyping of hcr2-1 was performed by PCR amplification using oligonucleotides ppx1-F and ppx1-R for wild type, and ppx1-F and GABI_LB for the T-DNA allele. Genotyping of ppx2-1 was carried out by PCR amplification using primers ppx2-F and ppx2-R for wild type, and ppx2-R and GABI_LB for the T-DNA allele. Genotyping of pp4r2 was performed by PCR amplification using oligonucleotides pp4r2-F and pp4r2-R for wild type, and pp4r2-R and LBb1.3 for the T-DNA allele. Genotyping of hcr1-1 was performed by PCR amplification using hcr1-F and hcr1-R dCAPs markers, followed by FokI restriction endonuclease digestion. zip4-2 and fancm-1 genotyping was performed as previously described33. Genotyping oligonucleotide sequences can be found in Supplementary Table 24.
Ethyl-methyl sulfonate mutagenesis of Arabidopsis seed
Approximately 10,000 seeds from 420 GR/++ hemizygote plants were obtained by crossing 420 (GR/GR) homozyotes to wild type (Col-0). These seed were soaked in 40 ml of 100 mM phosphate buffer (pH 7.5) in a 50 ml tube for 1 hour. Seeds were washed with fresh 100 mM phosphate buffer and then treated with 0.3% (v/v) ethyl-methyl sulfonate (EMS) and incubated for 12 hours at room temperature. EMS treated seeds were washed 10 times with distilled water and immediately sown on soil. From these seed, ~7,000 M1 plants were germinated and grown. The seeds from 12 independent M1 plants were combined to generate ~600 M2 pools. From each M2 pool, approximately ~150 seeds were pre-selected as 420/++ hemizygotes, based on red and green fluorescence, grown and self-fertilized. The resulting seed were analysed for 420 crossover frequency.
Measurement of crossover frequency and interference using fluorescent seed and pollen
Crossover frequency was measured by analyzing counts of fluorescent and non-fluorescent seeds from FTL/++ hemizygote plants using a CellProfiler image analysis pipeline97,98. CellProfiler enables the quantification of green-alone fluorescent seeds (NGreen), red-alone fluorescent seeds (NRed) and total seeds (NTotal). Crossover frequency (cM) is calculated using the formula: cM = 100×(1-[1-2(NGreen+NRed)/NTotal]1/2)30,32. To test whether crossover frequency was significantly different between genotypes we used Welch’s t-tests.
Pollen FTLs were generated in qrt-1 mutant background, where the four-pollen products of male meiosis are attached to one another31. FTLs express eYFP (Y), dsRed (R) or eCFP (C) fluorescent proteins under the post-meiotic LAT52 promoter. Pollen tetrad FTL-based measurement of crossover frequency and interference were carried out using DeepTetrad, as described31,55. DeepTetrad is a deep learning-based image analysis pipeline that recognizes pollen tetrad classes of two or three‐color FTL intervals. The two color FTL interval CEN3 produces parental ditype (PD), tetra type (T), and non-parental ditype (NPD) tetrads, and crossover frequency was calculated using the Perkin’s equation:
Three‐color FTL intervals (I1bc, I1fg, I2fg, I3bc and I5ab) produce 12-tetrad classes: no recombination (A), single crossover interval 1 (B; SCO-i1), single crossover interval 2 (C; SCO-i2), two-strand double crossover (D; 2stDCO), three-strand double crossover a (E; 3st DCOa), three-strand double crossover b (F; 3st DCOb), four-strand double crossover (G; 4st DCO), non-parental ditype interval 1, non-crossover interval 2 (H; NPD-i1 NCO-i2), non-crossover interval 1, non-parental ditype interval 2 (I; NCO-i1 NPD-i2), non-parental ditype interval 1, single crossover interval 2 (J; NPD-i1 SCO-i2), single crossover interval 1, non-parental ditype interval 2 (K; SCO-i1 NPD-i2) and non-parental ditype interval 1, non-parental ditype interval 2 (L; NPD-i1 NPD-i2)31. Fluorescent tetrad states were identified using DeepTetrad and crossover frequency (cM) was calculated using the Perkin’s equation.
Crossover interference ratio (IFR=σ) in two linked intervals, which is the ratio of the genetic map distance with an adjacent crossover XY to the genetic map distance without an adjacent crossover Xδ, was calculated by DeepTetrad using the formulae:
Identification of candidate hcr1-1 mutations using DNA sequencing and SHOREmap
Sixty hcr1 BC1F2 individuals with high (>27 cM) 420 crossover frequency were identified and 5 mg of seed from each BC1F2 individual were pooled. Sterilized seed were germinated on ½ MS agar plates and bulk 7-day old seedlings collected. ~3 grams of pooled seedlings were ground in liquid N2 using a mortar and pestle. The leaf powder was transferred into a pre-chilled mortar with 40 ml of fresh nuclear isolation buffer (25 mM Tris-HCl, pH 7.5, 0.44 M sucrose, 10 mM MgCl2, 0.5% Triton X-100, 10 mM β-mercaptoethanol, 2 mM spermine, EDTA-free Protease Inhibitor Cocktail) and the contents were homogenized. The tissue lysate was kept on ice and incubated for 30 minutes with rocking. The filtered contents were centrifuged at 4°C at 3,000g for 25 minutes. The supernatant was removed and the pellet was subjected to DNA extraction using CTAB. CTAB-extracted and purified DNA was sheared to a size range 200-500 bp using a Bioruptor sonicator. 1 μg of input DNA was diluted in 150 μl of TE buffer and sonicated for 22 minutes using high voltage with 30 second ON/OFF cycles. The sonicated DNA was concentrated in a 60 μl volume and DNA in the size range ~300-400 bp from a 2% agarose gel stained with 1×SYBR gold using a UV transilluminator. 50 ng of purified DNA in 60 μl volume was used as input for library construction using an Illumina Truseq Nano DNA LT library prep kit. The hcr1-1 BC1F2 library was sequenced using an Illumina Genome Analyser (100 bp paired) Hiseq 2000 instrument.
SHOREmap (v.3.0) was applied to align paired-end reads to the TAIR10 reference genome using the GenomeMapper tool38. Raw reads were trimmed according to quality values with a cut-off Phred score of +33 or +64, using the function SHORE import. SHORE function consensus was used to detect sequence variation between the hcr1 BC1F2 and the TAIR10 reference assembly. Single nucleotide polymorphisms (SNPs) with high quality marker scores (>40), supported by at least 10 unique reads, were applied using SHOREmap backcross for analysis of allele frequency. Using SHOREmap annotate we compared the TAIR10 gene annotation and obtained a list of EMS-derived that included predicted effects on gene expression and function. Mutations were screened for those with (i) greater than 80% allele frequency, and (ii) non-synonymous, splice site or premature stop codon changes in predicted genes. Additionally, candidate mutations were examined based on their location within genes with predicted or known functions relevant to meiosis, protein location in the nucleus, and known molecular functions provided in the TAIR database.
Genetic complementation of hcr1-1 by PPX1
A 4.5 kb genomic DNA fragment containing HCR1/PPX1 was PCR amplified using primers PPX1-F and PPX1-R (Supplementary Table 24). The PCR product was digested by PstI and SmaI restriction enzymes and cloned into the binary vector pGREEN0029. The pGREEN0029-PPX1 and empty vector constructs were electroporated into Agrobacterium strain GV3101-pSOUP and transformed into Arabidopsis plants by floral dipping99. T1 plants were selected for kanamycin resistance and genotyped using primers designed from left and right borders of the HCR1/PPX1 transgene (Supplementary Table 24).
Construction of PPX/PP4 phylogenetic tree
The neighbour-joining method was used to construct a PPX/PP4 phylogenetic tree. Amino acid sequences of AtPPX1 (NP_194402.1), AtPPX2 (NP_200337.1), OsPPX (XP_015612628), DmPp4-19C (NP_001285489), HsPPP4C (NP_001290432), Cepph-4.1 (NP_499603), Cepph-4.2 (NP_001022898), and ScPPH3 (AJV04101) were used for multiple sequence alignments.
Generation of meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 transgenic plants
To generate meiosis-specific microRNA mediated gene silencing (meiMIGS) transgenic plants, 1.5 kb of genomic DNA including the DMC1 promoter, 5’-UTR, two introns and the third exon were PCR amplified from Col genomic DNA using primers DMC1-1p_1.5kb-Lv0-GGAG-F and DMC1-1p_1.5kb-Lv0-CATT-R (Supplementary Table 24). PCR products was cloned into the universal Level 0 (Lv0) vector (pAGM9121) using the Golden Gate cloning system. PPX1 and PPX2 cDNA regions were cloned into the Lv0 vector (pAGM9121) following amplification using forward primers that included the miR173 target sequence and reverse primers (Supplementary Table 21). PPX1-PPX2 fusion cDNA was generated by overlap PCR and cloning into Lv0 vector pAGM9121. The DMC1 promoter and MIGS-PPX1/2/1-2 Lv0 vectors were assembled into Lv1 position 2 vector pICH47742 with the NOPALINE SYNTHASE GENE (NOS) terminator (pICH41421). Each Lv1 vector containing meiMIGS cassettes was assembled into a Level 2 (Lv2) binary vector (pAGM4723) with the antibiotic resistant gene BAR containing Lv vector (pICSL11017) and linker (pICH41744). The Lv2 binary vectors were electroporated into Agrobacterium strain GV3101-pSOUP and transformed into Arabidopsis by floral dipping.
Genotyping-by-sequencing of F2 plants and crossover identification
Genomic DNA from wild type and meiMIGS-PPX1-PPX2 Col/Ler F2 individuals was extracted using CTAB to prepare sequencing libraries, as described56. 150 ng of DNA was fragmented using 0.3 units of dsDNA Shearase (Zymo Research) in a final volume of 15 μl. The digested DNA was end-repaired for 30 minutes at 20°C in a reaction volume of 30 μl (3 units of T4 DNA polymerase (New England Biolabs), 10 units of T4 polynucleotide kinase (Thermo Fisher Scientific), 1.25 units of Klenow fragment (New England Biolabs) and 0.4 mM dNTPs). DNA fragments were cleaned using AMPure XP magnetic SPRI beads (Beckman-Coulter, A63881), as described56. DNA was A-tailed, and then ligated with barcoded Illumina adaptors in a reaction volume of 20 μl, as described56. Eight DNA libraries were pooled, washed and eluted in 30 μl elution buffer (10 mM Tris-HCl, pH 8.0). The 30 μl mixture was combined in a tube containing 16 μl of AMPure XP magnetic SPRI beads (Beckman-Coulter). After 5 minutes of incubation at room temperature, the samples were placed in a magnetic rack for 2 minutes and the supernatant (42 μl) was transferred to a fresh tube and mixed with 0.23 volumes (9.5 μl) of SPRI beads. After 5 minutes of incubation at room temperature, the tubes were placed on a magnetic rack for 2 minutes. The supernatant was discarded, and the beads washed twice with 80% ethanol for 30 seconds. The beads were air-dried for 10 minutes and DNA was eluted in 20 μl of 10 mM Tris (pH 8.0). 12 μl of the eluate was amplified using twelve cycles of PCR in a reaction volume of 50 μl using KAPA HiFi Hot-Start ReadyMix PCR kit (Kapabiosystems) and the reported DNA oligonucleotides56. The PCR products were then purified using SPRI beads and quantified using a Bioanalyzer. The 96 barcoded libraries were subjected to paired-end 150 bp sequencing using an Illumina HiSeqX instrument.
Immunocytological analysis of wild type and hcr1 meiocytes
Chromosome spreads of Arabidopsis pollen mother cells was prepared using fixed buds and DAPI-stained, as described100. Pachytene cells were immunostained for ASY1 and ZYP1, and diakinesis cells were immunostained for MLH1, using fixed buds, as described100,101. Leptotene-stage meiocytes were immunostained for ASY1 and RAD51 using fresh buds, as described102. The following antibodies were used: α-ASY1 (rat, 1:200 or 1:500 dilution), α-ZYP1 (rabbit, 1:200 dilution), α-MLH1 (rabbit, 1:200 dilution) and α-RAD51 (rabbit, 1:300 dilution)100,102,103. Microscopy was performed using a DeltaVision personal DV microscope (Applied precision/GE Healthcare) equipped with a CDD Coolsnap HQ2 camera (Photometrics). Image capture was carried out using SoftWoRx software version 5.5 (Applied precision/GE Healthcare). For ASY1 and RAD51 co-immunostaining of leptotene-stage nuclei, individual cell images were acquired as Z-stacks of 10 optical sections of 0.2 μM each, and the maximum intensity projection for each cell was decided using ImageJ. Number of MLH1 foci per meiotic cell and RAD51 foci per cell associated with the axis protein ASY1 were manually scored. Wilcoxon tests were used to assess significant differences between wild type and hcr1-1 MLH1 and RAD51 foci counts.
Yeast two hybrid assays
For yeast two-hybrid (Y2H) assays the open reading frames of Arabidopsis genes were cloned into pGBKT7 BD and pGADT7 AD vectors (Clontech, 630490) using BamHI and StuI sites, using a Gibson assembly cloning system (NEB #E2621L). Information of all oligonucleotides used for Y2H assays are in Supplementary Table 24. Both BD and AD vectors were co-transformed into S. cerevisiae strain AH109 and selected on synthetic dropout medium lacking leucine (-L) and tryptophan (-T). The colonies of yeast transformant cells were streaked onto both (-LT) and (-LTH (histidine) A (adenine)) synthetic mediums and grown for 3 to 5 days at 30°C. The cells grown in synthetic medium (-LT) were grown until OD600 = 1 and diluted 10-, 100- and 1,000-fold in water and spotted on synthetic medium (-LTHA) for 3 to 7 days.
Transient expression of fusion proteins in Arabidopsis protoplasts for co-localization and co-immunoprecipitation analysis
Transient expression vectors in protoplasts were constructed using Golden Gate cloning. The full-length coding regions of PPX1/HCR1 and meiotic genes were PCR amplified from cDNA and cloned into Lv0 universal vector (pICH41331). For epitope and fluorescent protein tagging, the Lv0 vectors with coding regions lacking stop codon were assembled in the Lv1 transient expression vector (pICH47742), using the 35S promoter vector (pICH51266), C-terminal vectors (YFP, CFP, Myc tag/pICSL50010 and HA tag/pICSL50009) and NOPALINE SYNTHASE GENE (NOS) terminator vector (pICH41421). Information of all oligonucleotides for protoplast transient expression is provided in Supplementary Table 24.
Plasmid DNA and mesophyll protoplasts were prepared, as described104. 20×103 protoplasts were transfected with 20 μg of total plasmid DNA and incubated for 6-12 hours at room temperature. To detect colocalization of PPX1-CFP and meiotic protein-YFP, 20 μg of total plasmid DNA (a mixture of PPX1-CFP with YFP fusion constructs HEI10-YFP, PTD-YFP or MSH5-YFP) were co-transfected into 20×103 protoplasts and incubated at room temperature for 12 hours. As a negative control, PPX1-CFP alone or YFP-fusion plasmid alone were transfected. The fluorescence of transfected mesophyll protoplasts was detected using a confocal microscope (LSM 800, Zeiss).
For co-immunoprecipitation analysis, 40 μg of PPX1-Myc tag and meiotic gene-HA tag DNA plasmids were co-transfected into protoplasts, or individually transfected as a negative control. Total protein was extracted using extraction buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 5 mM EDTA, 1 mM dithiothreitol, protease inhibitor cocktail (Roche) and 1% Triton X-100). The extracted proteins were separated by SDS–PAGE using 8% polyacrylamide gels, transferred to a nitrocellulose membrane and immunodetected with anti-HA (1:2,000 Roche 12013819001) or anti-Myc (1:2,000 Santa Cruz sc-9E10) antibodies. For co-immunoprecipitation (Co-IP) analysis, transfected protoplasts were lysed with IP buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 1 mM EDTA, 0.5% Triton X-100, 10% glycerol and protease inhibitor cocktail). Lysates were incubated with 1 μg anti-myc antibody for 12 hours with rotation at 4 °C. Then, the protoplast lysate and antibody mixture were incubated with 50% protein G-coated agarose beads (Millipore 16-201), pre-cleared with IP buffer, for an additional 2 hours. Protein-coated agarose beads were washed with IP buffer three times. Proteins were extracted using extraction buffer and subjected to western blotting using anti-HA antibodies.
Prediction of PP4 complex target proteins in Arabidopsis
To predict PP4 target proteins during meiosis, FxxP motif containing proteins were identified by searching protein sequences from TAIR. Nuclear proteins were obtained from the TAIR10 GO cellular compartment annotation by selecting terms “nucleus”, “other cellular components: host cell nucleus”, “other cellular components: nucleus-vacuole junction”. We used a previous RNA-seq dataset, which identified genes that showed significantly higher expression in male meiocytes compared to leaf34. The meiotically expressed, nuclear proteins with FxxP motifs were further classified according to the presence of predicted phosphorylation consensus sites of CDK, DDK and ATM/ATR, predicted using GPS 5.0105. To test for significant enrichment of phosphorylation consensus motifs in the predicted PP4 target proteins, we generated random sets of the same number of genes which were analysed for predicted phosphosites. The observed phosphosite overlaps were compared with the random using a Z-test.
Extended Data
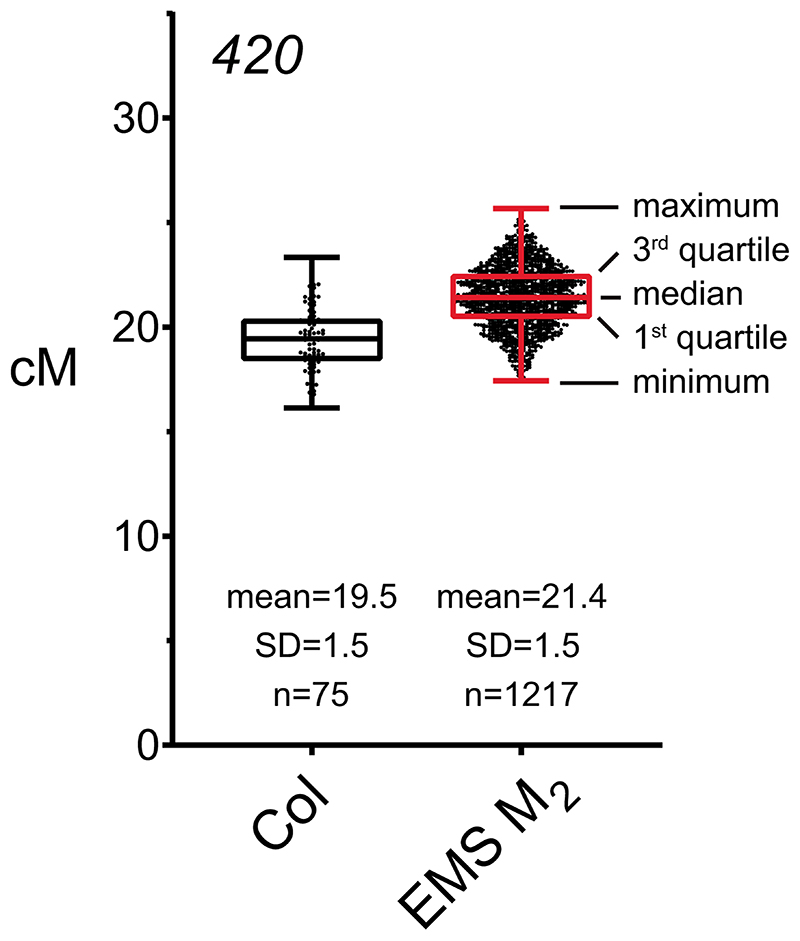
Extended Data Fig. 1. 420 crossover frequency in wild type and M2 plants derived from the EMS population.
Box and whisker plot showing 420 crossover frequency (cM) for wild type (Col/Col) 420/++ plants (n=75) and EMS-treated M2 420/++ plants (n=1,217). Black dots indicate 420 crossover frequency in individual plants. Horizontal lines of black (wild type, Col) and red (EMS M2) box plots represent maximum, 3rd quartile, median, 1st quartile and minimum in 420 cM. In this study, wild type plants show a mean value of 19.5 cM (standard deviation=1.5) within 420, and the majority (81.4%, 991/1,217) of M2 plants display 420 crossover frequency within the range of 18-22 cM (Mean=21.4 cM, SD=1.5). 420 crossover frequency in M2 plants was significantly increased compared to wild type (Welch’s t-test P=2.2×10-16), which may have been caused by heterozygous EMS polymorphisms.
Extended Data Fig. 2. EMS mutations identified in FANCM (hcr4) and TAF4b (lcr1).
a, FANCM gene structure is shown, including the EMS mutation site in hcr4/fancm-11. The red arrow indicates the G to A substitution within exon 15, which causes a G to S amino acid substitution. Exons are shown as boxes (black=CDS, grey=UTR). Scale bar=0.5 kb. b, Multiple sequence alignment of the DEHDc (blue line) and HELICC (green line) domains of FANCM in different species. The mutation positions of the fancm-1 to fancm-10 alleles that were previously identified1,2, and fancm-11 (hcr4), are shown. The fancm-11 mutation is located in a conserved motif within the SF2 helicase domain (bold arrow). c, Gene structure of TAF4b is shown with the location of the lcr1 (taf4b-3) mutation indicated in exon 3 (red arrow), which causes a premature stop codon.
Extended Data Fig. 3. T-DNA insertions in Arabidopsis PP4/PPX complex genes.
a, The gene structures of PPX1 (At4g26720), PPX2 (At5g55260) and PP4R2 (At5g17070) are shown. Exons are shown as boxes (black=CDS, grey=UTR). Scale bar=0.5 kb. The EMS induced hcr1-1 mutation is located at the splice donor site of the 3rd intron, shown by the asterisk. The red arrows indicate the location of primers for RT-qPCR in PPX1 and PPX2. The hcr1-2 T-DNA (GK_651B07) insertion position in the 5’-UTR is indicated. The position of the ppx2-1 (GK_488H09), ppx2-2 (SALK_049725), and pp4r2 (SALK_093051) T-DNA insertions are shown, which are located in the 4th intron, 8th exon and 7th intron, respectively. The arrows spanning the ppx2 and pp4r2 T-DNA insertions indicate primer positions used for RT-PCR. b, RT-PCR amplification and quantification for PPX1, PPX2 and PP4R2 mRNA expression in wild type Col, hcr1-1, ppx1-2, ppx2-1 and pp4r2. Floral cDNA from two biological replicates were evaluated by RT-PCR amplification for PPX1, PPX2, PP4R2 (shown in a) and GAPC expression. RT-PCR amplicon sizes for wild type, hcr1-1, ppx1-2, ppx2-1, pp4r2 cDNAs and wild type genomic DNA (positive/negative control) are shown. c, Plot showing RT-qPCR enrichment of PPX1 and PPX2 in hcr1-1 and ppx2-1. Relative transcript levels of PPX1 and PPX2 were measured in wild type, hcr1-1, and ppx2-1 using qRT-PCR. TUB2 was used for normalization. The y axis indicates fold-enrichment of PPX1 and PPX2 transcript levels, compared to PPX1 and PPX2 in wild type. RT-qPCR reactions of two technical replicates for each of four biological samples were shown as dots. Mean values are indicated by horizontal lines. Significance between wild type and mutants was assessed by Welch’s t-test. Asterisks indicate P<0.001. d, Photograph showing developmental phenotypes of wild type, hcr1-2, hcr1-1, ppx2-1, hcr1-2 ppx2-1 and hcr1-1 ppx2-1 grown alongside one another. e, Photograph showing seeds of wild type and hcr1-1/+ ppx2-2/+ plants. Asterisks indicate defective seeds. f, Photograph showing F2 seedlings grown from self-fertilization of F1 hcr1-1/+ ppx2-2/+ plants, with asterisks indicating developmentally delayed seedlings.
Extended Data Fig. 4. Alignment of PP4 homolog protein sequences from diverse eukaryotes.
a, Amino acid sequence alignment of AtPPX1, the predicted hcr1-1 truncated protein, AtPPX2 and PP4 homologs from different eukaryotic species. The predicted hcr1-1 truncated protein consisting of 143 residues is shown. The underlined region indicates amino acids generated due to the retention of the 3rd intron. Hash symbols indicate the locations of conserved PP4 catalytic motifs (GDXHG, GDXVDRG and GNHE) and the histidine (H) residues required for metal binding in C-terminal region. b, As for a, but showing percent identity of amino acid sequence between PP4 homologs.
Extended Data Fig. 5. Meiosis-specific knockdown of PPX1 and PPX2 in meiMIGS transgenic plants.
a, qRT-PCR analysis of PPX1/HCR1 and PPX2 transcripts in floral buds of wild type and meiMIGS-PPX1, meiMIGS-PPX2 and meiMIGS-PPX1-PPX2 T2 transgenic lines. The y axis indicates fold-enrichment of PPX1 and PPX2 transcripts, compared to PPX1 in wild type. DMC1 was used as a meiotic gene for normalization. Replicate measurements are shown as dots and mean values shown by horizontal lines. b, Correlation between PPX1 and PPX2 transcript levels in wild type, meiMIGS-PPX1, meiMIGS-PPX2, and meiMIGS-PPX1-PPX2 lines. The x and y axis indicate relative PPX1 and PPX2 transcript levels in meiMIGS-PPX1 (blue), meiMIGS-PPX2 (red), and meiMIGS-PPX1-PPX2 (green) lines respectively, compared to PPX1 and PPX2 expressions in wild type Col plant (r=0.80, P value=1.21×10-5).
Extended Data Fig. 6. Crossover frequency and interference measured in wild type and hcr1-1 using fluorescent pollen.
a, Crossover frequency measured using the pollen FTLs I1bc and I3bc from wild type and hcr1-1. Crossover frequency in each interval of the three-color FTLs was measured using the DeepTetrad pipeline3 (Supplementary Table 20). b, Crossover interference ratio measured using FTL pollen tetrads in wild type and hcr1-1. Crossover interference ratio (IFR) were calculated using the DeepTetrad pipeline3,4. c, Plots showing the % of tetrads containing double crossovers, using data from the three-color FTL intervals in wild type and hcr1-1. d, As for c, but showing FTL data from the I1bc, I1fg, I3bc and I5ab intervals in wild type and meiMIGS-PPX1-PPX2. Tetrads were classified into 12 fluorescence classes (A-L) by DeepTetrad, as described3,4. Mean values are indicated by horizontal lines.
Extended Data Fig. 7. SPO11-1-oligonucleotides and nucleosome occupancy around wild type and meiMIGS-PPX1-PPX2 crossovers.
10 kb windows surrounding crossover midpoints identified from wild type or meiMIGS-PPX1-PPX2 plants, or the same number of randomly selected positions, were analysed for SPO11-1-oligos (log2(SPO11-1-oligos/gDNA), red) or nucleosome occupancy (log2(MNase-seq/gDNA), blue)5.
Extended Data Fig. 8. Yeast two hybrid assays showing interactions of HCR1/PPX1 with meiotic proteins.
a, Yeast two hybrid assays testing interaction between HCR1/PPX1 and Class I (ZMM) proteins. The yeast co-transformants were grown until OD600 = 1 and spotted on synthetic dropout media (SD) lacking leucine/tryptophan (-LT) and leucine/trptophan/histidine/adenine (-LTHA) for 3, 5 or 7 days. b, Yeast two hybrid assays of HCR1/PPX1 and meiotic proteins involved in axis formation, DSB formation and DNA repair. The yeast transformants were grown until OD600 = 1, then diluted 10-, 100- and 1,000-fold in water, and spotted on SD (-LT) and SD (-LTHA) plates to examine growth in 3, 5, or 7 days (Supplementary Table 23).
Extended Data Fig. 9. The EVH1 domain of Arabidopsis PP4R3A interacts with meiotic proteins.
a, Yeast two-hybrid assays testing interaction between the PP4R3A EVH1 domain and meiotic proteins. PP4R3A-N indicates the PP4R3A N-terminal region (1-166 aa) containing the EVH1 domain. The yeast co-transformants were grown until OD600 = 1 and spotted on synthetic dropout media (SD) lacking leucine/tryptophan (-LT) and leucine/trptophan/histidine/adenine (-LTHA) for 3 and 5 days. The yeast transformants were grown until OD600 = 1, then diluted 10-, 100- and 1,000-fold in water, and spotted on SD (-LT) and SD (-LTHA) plates to examine growth. b, Venn diagram summarizing yeast two hybrid assays of meiotic proteins that interact with HCR1/PPX1 and the PP4R3A EVH1 domain. c, A schematic model of Arabidopsis PP4 holoenzyme complex that recognizes target protein HEI10 for dephosphorylation via the PP4R3A EVH1 domain and PPX1.
Extended Data Fig. 10. Genome-wide prediction of PP4 complex target proteins during meiosis.
a, Protein domain (green) structure of Arabidopsis PP4 subunits PPX1, PPX2, PP4R2 and PP4R3. b, Amino acid alignment of the PP4R3A homolog EVH1 domain (red box). Hash symbols (#) indicate conserved tyrosine (Y) and tryptophan (W) residues. c, As for a, but showing the positions of FxxP motifs and phosphorylation consensus sites in PTD, HEI10, MSH5 and MLH1. d, Venn diagram showing overlap of meiotically expressed, nuclear proteins with FxxP motifs. e, Venn diagram showing overlap of candidate PP4 target proteins with CDK, DDK or ATM/ATR kinase consensus motifs, predicted using GPS 5.06. The location of HCR1 Y2H interactors are indicated within the Venn diagram. f, Histogram showing a significant enrichment of proteins containing phosphorylation sites in the predicted 1,367 PP4 targets, compared to 1,000 sets of randomly chosen genes (n=1,367). The vertical red line indicates observed predicted PP4 target proteins containing phosphorylation sites, compared to the random sets (black lines). g, Gene ontology (GO) enrichment analysis of the predicted PP4 targets, using PANTHER (http://pantherdb.org/). Benjamini-Hochberg False Discovery Rate (FDR) correction was used for enrichment test.
Supplementary Material
Acknowledgments
We thank Gregory Copenhaver (University of North Carolina), Avraham Levy (The Weizmann Institute), and Scott Poethig (University of Pennsylvania) for FTLs/CTLs, Raphael Mercier (Max Planck Institute, Cologne) for fancm-1, Liliana Ziolkowska and Charles Underwood (Max Planck Institute, Cologne) for helping grow the EMS population, Mathilde Grelon (INRA, Versailles) for MLH1 antibodies, Chris Franklin (University of Birmingham) for ASY1, ZYP1 and RAD51 antibodies and the Gurdon Institute for access to microscopes. This work was funded by the Suh Kyungbae Foundation (JaK, JuK, JP, EK, HK, DB, YMP, KC), Next-Generation BioGreen 21 Program PJ01337001 (JaK, JuK, JP, EK, HK, DB, YMP, KC) and PJ01342301 (HSC, SL, IH), Rural Development Administration, Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education NRF-2020R1A2C2007763 (HK, DB, KC), Marie-Curie International Training Network ‘COMREC’ (DN), BBSRC grant EpiSpiX BB/N007557/1 (XZ, IH), BBSRC ERA-CAPs grant BB/M004937/1 (CL, IH) and ERC Consolidator Award ERC-2015-CoG-681987 ‘SynthHotSpot’ (CL, AT, IH).
Footnotes
Author Contributions Statement
Design and conception of experiments: DN, JaK, CL, JuK, JP, EK, PK, KC, IH.
Acquisition and analysis of data: DN, JaK, CL, JuK, JP, HSC, HK, DB, YMP, PK, SL, AT, XZ, IH, KC.
Wrote the manuscript: DN, JaK, CL, JuK, KC, IH.
Competing Interests Statement
The authors declare no competing financial or non-financial interests in relation to this work.
Data Availability
Genome sequencing data of F2 plants can be found at the ArrayExpress repository hosted by the European Bioinformatics Institute (EBI) (https://www.ebi.ac.uk/arrayexpress/). The data can be found at accessions E-MTAB-9621 and E-MTAB-10168.
References
- 1.Villeneuve AM, Hillers KJ. Whence meiosis? Cell. 2001;106:647–50. doi: 10.1016/s0092-8674(01)00500-1. [DOI] [PubMed] [Google Scholar]
- 2.Mercier R, Mézard C, Jenczewski E, Macaisne N, Grelon M. The molecular biology of meiosis in plants. Annu Rev Plant Biol. 2015;66:297–327. doi: 10.1146/annurev-arplant-050213-035923. [DOI] [PubMed] [Google Scholar]
- 3.Grelon M, Vezon D, Gendrot G, Pelletier G. AtSPO11-1 is necessary for efficient meiotic recombination in plants. EMBO J. 2001;20:589–600. doi: 10.1093/emboj/20.3.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robert T, et al. The TopoVIB-Like protein family is required for meiotic DNA double-strand break formation. Science (80-) 2016;351:943–949. doi: 10.1126/science.aad5309. [DOI] [PubMed] [Google Scholar]
- 5.Hartung F, et al. The catalytically active tyrosine residues of both SPO11-1 and SPO11-2 are required for meiotic double-strand break induction in Arabidopsis. Plant Cell. 2007;19:3090–9. doi: 10.1105/tpc.107.054817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hunter N. Meiotic Recombination: The Essence of Heredity. Cold Spring Harb Perspect Biol. 2015;7 doi: 10.1101/cshperspect.a016618. a016618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferdous M, et al. Inter-homolog crossing-over and synapsis in Arabidopsis meiosis are dependent on the chromosome axis protein AtASY3. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002507. e1002507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rowan BA, et al. An Ultra High-Density Arabidopsis thaliana Crossover Map That Refines the Influences of Structural Variation and Epigenetic Features. Genetics. 2019;213:771–787. doi: 10.1534/genetics.119.302406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Girard C, et al. AAA-ATPase FIDGETIN-LIKE 1 and Helicase FANCM Antagonize Meiotic Crossovers by Distinct Mechanisms. PLoS Genet. 2015;11 doi: 10.1371/journal.pgen.1005369. e1005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cifuentes M, Rivard M, Pereira L, Chelysheva L, Mercier R. Haploid Meiosis in Arabidopsis: Double-Strand Breaks Are Formed and Repaired but Without Synapsis and Crossovers. PLoS One. 2013;8 doi: 10.1371/journal.pone.0072431. e72431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berchowitz LE, Copenhaver GP. Genetic interference: don’t stand so close to me. Curr Genomics. 2010;11:91–102. doi: 10.2174/138920210790886835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Y, et al. HEIP1 regulates crossover formation during meiosis in rice. Proc Natl Acad Sci. 2018;115:10810–10815. doi: 10.1073/pnas.1807871115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pyatnitskaya A, Borde V, De Muyt A. Crossing and zipping: molecular duties of the ZMM proteins in meiosis. Chromosoma. 2019;128:181–198. doi: 10.1007/s00412-019-00714-8. [DOI] [PubMed] [Google Scholar]
- 14.Mercier R, et al. Two meiotic crossover classes cohabit in Arabidopsis: one is dependent on MER3, whereas the other one is not. Curr Biol. 2005;15:692–701. doi: 10.1016/j.cub.2005.02.056. [DOI] [PubMed] [Google Scholar]
- 15.Séguéla-Arnaud M, et al. Multiple mechanisms limit meiotic crossovers: TOP3α and two BLM homologs antagonize crossovers in parallel to FANCM. Proc Natl Acad Sci U S A. 2015;112:4713–8. doi: 10.1073/pnas.1423107112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serra H, et al. Massive crossover elevation via combination of HEI10 and recq4a recq4b during Arabidopsis meiosis. Proc Natl Acad Sci U S A. 2018;115:2437–2442. doi: 10.1073/pnas.1713071115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Marston AL, Amon A. Meiosis: Cell-cycle controls shuffle and deal. Nature Reviews Molecular Cell Biology. 2004;5:983–997. doi: 10.1038/nrm1526. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, et al. The Arabidopsis Cdk1/Cdk2 homolog CDKA ;1 controls chromosome axis assembly during plant meiosis. EMBO J. 2020;39:1–19. doi: 10.15252/embj.2019101625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lange J, et al. The Landscape of Mouse Meiotic Double-Strand Break Formation, Processing, and Repair. Cell. 2016;167:695–708.e16. doi: 10.1016/j.cell.2016.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia V, Gray S, Allison RM, Cooper TJ, Neale MJ. Tel1(ATM)-mediated interference suppresses clustered meiotic double-strand-break formation. Nature. 2015;520:114–118. doi: 10.1038/nature13993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lange J, et al. ATM controls meiotic double-strand-break formation. Nature. 2011;479:237–40. doi: 10.1038/nature10508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Serrentino M-E, Chaplais E, Sommermeyer V, Borde V. Differential association of the conserved SUMO ligase Zip3 with meiotic double-strand break sites reveals regional variations in the outcome of meiotic recombination. PLoS Genet. 2013;9 doi: 10.1371/journal.pgen.1003416. e1003416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.He W, et al. Regulated Proteolysis of MutSγ Controls Meiotic Crossing Over. Mol Cell. 2020;78:168–183.e5. doi: 10.1016/j.molcel.2020.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carballo JA, Johnson AL, Sedgwick SG, Cha RS. Phosphorylation of the axial element protein Hop1 by Mec1/Tel1 ensures meiotic interhomolog recombination. Cell. 2008;132:758–70. doi: 10.1016/j.cell.2008.01.035. [DOI] [PubMed] [Google Scholar]
- 25.Brar GA, et al. Rec8 phosphorylation and recombination promote the step-wise loss of cohesins in meiosis. Nature. 2006;441:532–536. doi: 10.1038/nature04794. [DOI] [PubMed] [Google Scholar]
- 26.Hustedt N, et al. Yeast PP4 interacts with ATR homolog Ddc2-Mec1 and regulates checkpoint signaling. Mol Cell. 2015;57:273–289. doi: 10.1016/j.molcel.2014.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee DH, et al. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010;17:365–372. doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, et al. The PROTEIN PHOSPHATASE4 Complex Promotes Transcription and Processing of Primary microRNAs in Arabidopsis. Plant Cell. 2019;31:486–501. doi: 10.1105/tpc.18.00556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G, Rossidivito G, Hu T, Berlyand Y, Poethig RS. Traffic lines: new tools for genetic analysis in Arabidopsis thaliana. Genetics. 2015;200:35–45. doi: 10.1534/genetics.114.173435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Melamed-Bessudo C, Yehuda E, Stuitje AR, Levy AA. A new seed-based assay for meiotic recombination in Arabidopsis thaliana. Plant J. 2005;43:458–66. doi: 10.1111/j.1365-313X.2005.02466.x. [DOI] [PubMed] [Google Scholar]
- 31.Berchowitz LE, Copenhaver GP. Fluorescent Arabidopsis tetrads: a visual assay for quickly developing large crossover and crossover interference data sets. Nat Protoc. 2008;3:41–50. doi: 10.1038/nprot.2007.491. [DOI] [PubMed] [Google Scholar]
- 32.Ziolkowski PA, et al. Juxtaposition of heterozygous and homozygous regions causes reciprocal crossover remodelling via interference during Arabidopsis meiosis. Elife. 2015;4 doi: 10.7554/eLife.03708. e03708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yelina NE, et al. DNA methylation epigenetically silences crossover hot spots and controls chromosomal domains of meiotic recombination in Arabidopsis. Genes Dev. 2015;29:2183–202. doi: 10.1101/gad.270876.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lawrence EJ, et al. Natural Variation in TBP-ASSOCIATED FACTOR 4b Controls Meiotic Crossover and Germline Transcription in Arabidopsis. Curr Biol. 2019;29:2676–2686. doi: 10.1016/j.cub.2019.06.084. [DOI] [PubMed] [Google Scholar]
- 35.Choi K, et al. Arabidopsis meiotic crossover hot spots overlap with H2A.Z nucleosomes at gene promoters. Nat Genet. 2013;45:1327–36. doi: 10.1038/ng.2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ziolkowski PA, et al. Natural variation and dosage of the HEI10 meiotic E3 ligase control Arabidopsis crossover recombination. Genes Dev. 2017;31:306–317. doi: 10.1101/gad.295501.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crismani W, et al. FANCM limits meiotic crossovers. Science. 2012;336:1588–90. doi: 10.1126/science.1220381. [DOI] [PubMed] [Google Scholar]
- 38.Allen R, Nakasugi K, Doran RL, Millar AA, Waterhouse PM. Facile mutant identification via a single parental backcross method and application of whole genome sequencing based mapping pipelines. Front Plant Sci. 2013;4:1–8. doi: 10.3389/fpls.2013.00362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi Y. Serine/Threonine Phosphatases: Mechanism through Structure. Cell. 2009;139:468–484. doi: 10.1016/j.cell.2009.10.006. [DOI] [PubMed] [Google Scholar]
- 40.Gingras AC, et al. A novel, evolutionarily conserved protein phosphatase complex involved in cisplatin sensitivity. Mol Cell Proteomics. 2005;4:1725–1740. doi: 10.1074/mcp.M500231-MCP200. [DOI] [PubMed] [Google Scholar]
- 41.Ramos F, Villoria MT, Alonso-Rodríguez E, Clemente-Blanco A. Role of protein phosphatases PP1, PP2A PP4 and Cdc14 in the DNA damage response. Cell Stress. 2019 doi: 10.15698/cst2019.03.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakada S, Chen GI, Gingras AC, Durocher D. PP4 is a γH2AX phosphatase required for recovery from the DNA damage checkpoint. EMBO Rep. 2008 doi: 10.1038/embor.2008.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chowdhury D, et al. A PP4-Phosphatase Complex Dephosphorylates γ-H2AX Generated during DNA Replication. Mol Cell. 2008 doi: 10.1016/j.molcel.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Merigliano C, et al. A role for the twins protein phosphatase (PP2A-B55) in the maintenance of Drosophila genome integrity. Genetics. 2017 doi: 10.1534/genetics.116.192781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Keogh MC, et al. A phosphatase complex that dephosphorylates γH2AX regulates DNA damage checkpoint recovery. Nature. 2006 doi: 10.1038/nature04384. [DOI] [PubMed] [Google Scholar]
- 46.O’Neill BM, et al. Pph3-Psy2 is a phosphatase complex required for Rad53 dephosphorylation and replication fork restart during recovery from DNA damage. Proc Natl Acad Sci U S A. 2007 doi: 10.1073/pnas.0703252104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee DH, et al. A PP4 phosphatase complex dephosphorylates RPA2 to facilitate DNA repair via homologous recombination. Nat Struct Mol Biol. 2010 doi: 10.1038/nsmb.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, et al. Protein phosphatase PP4 is involved in NHEJ-mediated repair of DNA double-strand breaks. Cell Cycle. 2012 doi: 10.4161/cc.20957. [DOI] [PubMed] [Google Scholar]
- 49.Falk JEJ, Chan ACA, ho A, Hoffmann E, Hochwagen A. A Mec1- and PP4-Dependent checkpoint couples centromere pairing to meiotic recombination. Dev Cell. 2010;19:599–611. doi: 10.1016/j.devcel.2010.09.006. [DOI] [PubMed] [Google Scholar]
- 50.Pérez-Callejón E, et al. Identification and molecular cloning of two homologues of protein phosphatase X from Arabidopsis thaliana. Plant Mol Biol. 1993;23:1177–1185. doi: 10.1007/BF00042351. [DOI] [PubMed] [Google Scholar]
- 51.Moorhead GBG, De Wever V, Templeton G, Kerk D. Evolution of protein phosphatases in plants and animals. Biochem J. 2009;417:401–409. doi: 10.1042/BJ20081986. [DOI] [PubMed] [Google Scholar]
- 52.Su C, et al. The Protein Phosphatase 4 and SMEK1 Complex Dephosphorylates HYL1 to Promote miRNA Biogenesis by Antagonizing the MAPK Cascade in Arabidopsis. Dev Cell. 2017;41:527–539.e5. doi: 10.1016/j.devcel.2017.05.008. [DOI] [PubMed] [Google Scholar]
- 53.de Felippes FF, Wang J, Weigel D. MIGS: miRNA-induced gene silencing. Plant J. 2012;70:541–547. doi: 10.1111/j.1365-313X.2011.04896.x. [DOI] [PubMed] [Google Scholar]
- 54.Klimyuk VI, Jones JD. AtDMC1, the Arabidopsis homologue of the yeast DMC1 gene: characterization, transposon-induced allelic variation and meiosis-associated expression. Plant J. 1997;11:1–14. doi: 10.1046/j.1365-313x.1997.11010001.x. [DOI] [PubMed] [Google Scholar]
- 55.Lim EC, et al. DeepTetrad: high-throughput image analysis of meiotic tetrads by deep learning in Arabidopsis thaliana. Plant J. 2020;101:473–483. doi: 10.1111/tpj.14543. [DOI] [PubMed] [Google Scholar]
- 56.Rowan BA, Patel V, Weigel D, Schneeberger K. Rapid and Inexpensive Whole-Genome Genotyping-by-Sequencing for Crossover Localization and Fine-Scale Genetic Mapping. G3 (Bethesda) 2015;5:385–98. doi: 10.1534/g3.114.016501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Choi K, et al. Recombination Rate Heterogeneity within Arabidopsis Disease Resistance Genes. PLoS Genet. 2016;12 doi: 10.1371/journal.pgen.1006179. e1006179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Choi K, et al. Nucleosomes and DNA methylation shape meiotic DSB frequency in Arabidopsis thaliana transposons and gene regulatory regions. Genome Res. 2018;28:532–546. doi: 10.1101/gr.225599.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang J, et al. A multiprotein complex regulates interference-sensitive crossover formation in rice. Plant Physiol. 2019;181:221–235. doi: 10.1104/pp.19.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Macaisne N, Vignard J, Mercier R. SHOC1 and PTD form an XPF-ERCC1-like complex that is required for formation of class I crossovers. J Cell Sci. 2011;124:2687–91. doi: 10.1242/jcs.088229. [DOI] [PubMed] [Google Scholar]
- 61.Ueki Y, et al. A Consensus Binding Motif for the PP4 Protein Phosphatase. Mol Cell. 2019 doi: 10.1016/j.molcel.2019.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Fernandes JB, Seguela-Arnaud M, Larcheveque C, Lloyd AH, Mercier R. Unleashing meiotic crossovers in hybrid plants. Proc Natl Acad Sci U S A. 2017;115:2431–2436. doi: 10.1073/pnas.1713078114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wijnker E, et al. The Cdk1/Cdk2 homolog CDKA;1 controls the recombination landscape in Arabidopsis. Proc Natl Acad Sci U S A. 2019 doi: 10.1073/pnas.1820753116. 201820753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.He Y, et al. Genomic features shaping the landscape of meiotic double-strand-break hotspots in maize. Proc Natl Acad Sci U S A. 2017;114:12231–12236. doi: 10.1073/pnas.1713225114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Liu S, et al. Mu Transposon Insertion Sites and Meiotic Recombination Events Co-Localize with Epigenetic Marks for Open Chromatin across the Maize Genome. PLoS Genet. 2009;5 doi: 10.1371/journal.pgen.1000733. e1000733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Underwood CJ, et al. Epigenetic activation of meiotic recombination near Arabidopsis thaliana centromeres via loss of H3K9me2 and non-CG DNA methylation. Genome Res. 2018;28:519–531. doi: 10.1101/gr.227116.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chelysheva L, et al. The Arabidopsis HEI10 is a new ZMM protein related to Zip3. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002799. e1002799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang K, et al. The role of rice HEI10 in the formation of meiotic crossovers. PLoS Genet. 2012;8 doi: 10.1371/journal.pgen.1002809. e1002809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Reynolds A, et al. RNF212 is a dosage-sensitive regulator of crossing-over during mammalian meiosis. Nat Genet. 2013;45:269–78. doi: 10.1038/ng.2541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qiao H, et al. Antagonistic roles of ubiquitin ligase HEI10 and SUMO ligase RNF212 regulate meiotic recombination. Nat Genet. 2014;46:194–9. doi: 10.1038/ng.2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Woglar A, Villeneuve AM. Dynamic Architecture of DNA Repair Complexes and the Synaptonemal Complex at Sites of Meiotic Recombination. Cell. 2018;173:1678–1691.e16. doi: 10.1016/j.cell.2018.03.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Snowden T, Acharya S, Butz C, Berardini M, Fishel R. hMSH4-hMSH5 recognizes Holliday Junctions and forms a meiosis-specific sliding clamp that embraces homologous chromosomes. Mol Cell. 2004;15:437–51. doi: 10.1016/j.molcel.2004.06.040. [DOI] [PubMed] [Google Scholar]
- 73.Jessop L, Rockmill B, Roeder GS, Lichten M. Meiotic chromosome synapsis-promoting proteins antagonize the anti-crossover activity of sgs1. PLoS Genet. 2006;2:e155. doi: 10.1371/journal.pgen.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Oh SD, Lao JP, Taylor AF, Smith GR, Hunter N. RecQ Helicase, Sgs1, and XPF Family Endonuclease, Mus81-Mms4, Resolve Aberrant Joint Molecules during Meiotic Recombination. Mol Cell. 2008;31:324–336. doi: 10.1016/j.molcel.2008.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Manhart CM, et al. The mismatch repair and meiotic recombination endonuclease Mlh1-Mlh3 is activated by polymer formation and can cleave DNA substrates in trans. PLOS Biol. 2017;15 doi: 10.1371/journal.pbio.2001164. e2001164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zakharyevich K, Tang S, Ma Y, Hunter N. Delineation of joint molecule resolution pathways in meiosis identifies a crossover-specific resolvase. Cell. 2012;149:334–47. doi: 10.1016/j.cell.2012.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ranjha L, Anand R, Cejka P. The Saccharomyces cerevisiae Mlh1-Mlh3 heterodimer is an endonuclease that preferentially binds to holliday junctions. J Biol Chem. 2014;289:5674–5686. doi: 10.1074/jbc.M113.533810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wijeratne AJ, Chen C, Zhang W, Timofejeva L, Ma H. The Arabidopsis thaliana PARTING DANCERS gene encoding a novel protein is required for normal meiotic homologous recombination. Mol Biol Cell. 2006;17:1331–43. doi: 10.1091/mbc.E05-09-0902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lu P, Wijeratne AJ, Wang Z, Copenhaver GP, Ma H. Arabidopsis PTD is required for type I crossover formation and affects recombination frequency in two different chromosomal regions. J Genet Genomics. 2014;41:165–75. doi: 10.1016/j.jgg.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 80.De Muyt A, et al. A meiotic XPF–ERCC1-like complex recognizes joint molecule recombination intermediates to promote crossover formation. Genes Dev. 2018;32:283–296. doi: 10.1101/gad.308510.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Arora K, Corbett KD. The conserved XPF:ERCC1-like Zip2:Spo16 complex controls meiotic crossover formation through structure-specific DNA binding. Nucleic Acids Res. 2019;47:2365–2376. doi: 10.1093/nar/gky1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sato-Carlton A, et al. Protein Phosphatase 4 Promotes Chromosome Pairing and Synapsis, and Contributes to Maintaining Crossover Competence with Increasing Age. PLoS Genet. 2014;10 doi: 10.1371/journal.pgen.1004638. e1004638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Henderson KA, Kee K, Maleki S, Santini PA, Keeney S. Cyclin-dependent kinase directly regulates initiation of meiotic recombination. Cell. 2006;125:1321–32. doi: 10.1016/j.cell.2006.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lam I, Keeney S. Mechanism and Regulation of Meiotic Recombination Initiation. Cold Spring Harb Perspect Biol. 2014;7 doi: 10.1101/cshperspect.a016634. a016634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Valentin G, Schwob E, Della Seta F. Dual role of the Cdc7-regulatory protein Dbf4 during yeast meiosis. J Biol Chem. 2006;281:2828–2834. doi: 10.1074/jbc.M510626200. [DOI] [PubMed] [Google Scholar]
- 86.Sasanuma H, et al. Cdc7-dependent phosphorylation of Mer2 facilitates initiation of yeast meiotic recombination. Genes Dev. 2008;22:398–410. doi: 10.1101/gad.1626608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wan L, et al. Cdc28-Clb5 (CDK-S) and Cdc7-Dbf4 (DDK) collaborate to initiate meiotic recombination in yeast. Genes Dev. 2008;22:386–397. doi: 10.1101/gad.1626408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Matos J, et al. Dbf4-Dependent Cdc7 Kinase Links DNA Replication to the Segregation of Homologous Chromosomes in Meiosis I. Cell. 2008;135:662–678. doi: 10.1016/j.cell.2008.10.026. [DOI] [PubMed] [Google Scholar]
- 89.Chen X, et al. Phosphorylation of the Synaptonemal Complex Protein Zip1 Regulates the Crossover/Noncrossover Decision during Yeast Meiosis. PLOS Biol. 2015;13 doi: 10.1371/journal.pbio.1002329. e1002329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Keeney S, Lange J, Mohibullah N. Self-organization of meiotic recombination initiation: general principles and molecular pathways. Annu Rev Genet. 2014;48:187–214. doi: 10.1146/annurev-genet-120213-092304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Nowack MK, et al. Genetic Framework of Cyclin-Dependent Kinase Function in Arabidopsis. Dev Cell. 2012;22:1030–1040. doi: 10.1016/j.devcel.2012.02.015. [DOI] [PubMed] [Google Scholar]
- 92.Culligan KM, Britt AB. Both ATM and ATR promote the efficient and accurate processing of programmed meiotic double-strand breaks. Plant J. 2008;55:629–638. doi: 10.1111/j.1365-313X.2008.03530.x. [DOI] [PubMed] [Google Scholar]
- 93.Garcia V, et al. AtATM is essential for meiosis and the somatic response to DNA damage in plants. Plant Cell. 2003;15:119–32. doi: 10.1105/tpc.006577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Yao Y, et al. ATM Promotes RAD51-Mediated Meiotic DSB Repair by Inter-Sister-Chromatid Recombination in Arabidopsis. Front Plant Sci. 2020;11 doi: 10.3389/fpls.2020.00839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Villoria MT, et al. PP4 phosphatase cooperates in recombinational DNA repair by enhancing double-strand break end resection. Nucleic Acids Res. 2019 doi: 10.1093/nar/gkz794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Chelysheva L, et al. Zip4/Spo22 is required for class I CO formation but not for synapsis completion in Arabidopsis thaliana. PLoS Genet. 2007;3:e83. doi: 10.1371/journal.pgen.0030083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.van Tol N, Rolloos M, van Loon P, van der Zaal BJ. MeioSeed: a CellProfiler-based program to count fluorescent seeds for crossover frequency analysis in Arabidopsis thaliana. Plant Methods. 2018;14:32. doi: 10.1186/s13007-018-0298-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Carpenter AE, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat Protoc. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 100.Chelysheva L, et al. An easy protocol for studying chromatin and recombination protein dynamics during Arabidopsis thaliana meiosis: immunodetection of cohesins, histones and MLH1. Cytogenet Genome Res. 2010;129:143–53. doi: 10.1159/000314096. [DOI] [PubMed] [Google Scholar]
- 101.Lambing C, Kuo PC, Tock AJ, Topp SD, Henderson IR. ASY1 acts as a dosage-dependent antagonist of telomere-led recombination and mediates crossover interference in Arabidopsis. Proc Natl Acad Sci U S A. 2020;24:13647–13658. doi: 10.1073/pnas.1921055117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sanchez-Moran E, Santos J-L, Jones GH, Franklin FCH. ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 2007;21:2220–33. doi: 10.1101/gad.439007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Higgins JD, Sanchez-Moran E, Armstrong SJ, Jones GH, Franklin FCH. The Arabidopsis synaptonemal complex protein ZYP1 is required for chromosome synapsis and normal fidelity of crossing over. Genes Dev. 2005;19:2488–2500. doi: 10.1101/gad.354705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Hwang I, Sheen J. Two-component circuitry in Arabidopsis cytokinin signal transduction. Nature. 2001;413:383–389. doi: 10.1038/35096500. [DOI] [PubMed] [Google Scholar]
- 105.Xue Y, et al. GPS 2.1: Enhanced prediction of kinase-specific phosphorylation sites with an algorithm of motif length selection. Protein Eng Des Sel. 2011;24:255–260. doi: 10.1093/protein/gzq094. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome sequencing data of F2 plants can be found at the ArrayExpress repository hosted by the European Bioinformatics Institute (EBI) (https://www.ebi.ac.uk/arrayexpress/). The data can be found at accessions E-MTAB-9621 and E-MTAB-10168.