Abstract
Based on clinical trials using first generation EGFR tyrosine kinase inhibitors (TKI) it became a doctrine that K-RAS mutations drive resistance to EGFR inhibition in non-small cell lung cancer (NSCLC). Conversely, we provide evidence that EGFR signaling is engaged in K-RAS driven lung tumorigenesis in humans and in mice. Specifically, genetic mouse models revealed that deletion of Egfr quenches mutant K-ras activity and transiently reduces tumor growth. However, Egfr inhibition initiates a rapid resistance mechanism involving non-Egfr ErbB family members. This tumor-escape mechanism clarifies the disappointing outcome of first generation TKI, and suggests high therapeutic potential of pan-ERBB inhibitors. Indeed, based on various experimental models including genetically engineered mouse models, (patient derived-) xenografts and in vitro experiments, we demonstrate that the FDA approved pan-ERBB inhibitor afatinib effectively impairs K-RAS driven lung tumorigenesis. Our data strongly suggests reconsidering the use of pan-ERBB inhibition in clinical trials to treat K-RAS mutated NSCLC.
Introduction
Lung cancer is still the number one cancer related killer in men and women, with less than 20 % of patients surviving more than 5 years. (1) Lung adenocarcinomas (AC), the most common non-small cell lung cancer (NSCLC) subtype, are stratified by different driver mutations, with activating mutations in V-Ki-ras2 Kirsten rat sarcoma viral oncogene homolog (K-RAS) and in epidermal growth factor receptor (EGFR) being the most abundant ones. (2, 3) While treatment with EGFR targeting small molecule tyrosine kinase inhibitors (TKI) such as erlotinib, gefitinib or afatinib is initially effective in lung cancer patients with common activating EGFR mutations (i.e. L858R point mutation and exon 19 deletions), acquisition of resistance almost inevitably occurs. (4–7) In contrast and despite extensive research over the last three decades, there is no effective inhibitor targeting mutated K-RAS protein available in the clinics. (8) Traditionally, oncogenic K-RAS mutations were thought to render the protein constitutively active and independent from its upstream activator EGFR. (9) Therefore, K-RAS mutations have been proposed as a mechanism of primary resistance to EGFR TKI, and many studies demonstrated poor clinical outcomes using erlotinib and gefitinib in patients with NSCLC harboring K-RAS mutations. (10–13) In contrast, more recent work demonstrated that mutated K-RAS is not completely locked in its active form. The combination of an irreversible inhibitor specific to K-RASG12C together with erlotinib or gefitinib in K-RAS G12C-mutated lung cancer cell lines showed synergistic effects, demonstrating that mutated K-RAS is indeed activated by upstream receptor tyrosine kinase EGFR. (14, 15) Our finding that K-RAS driven lung AC display increased expression of EGFR and its ligands as well as downstream targets supports this discovery and prompted us to clarify if EGFR signaling contributes to the development of this detrimental disease.
Results
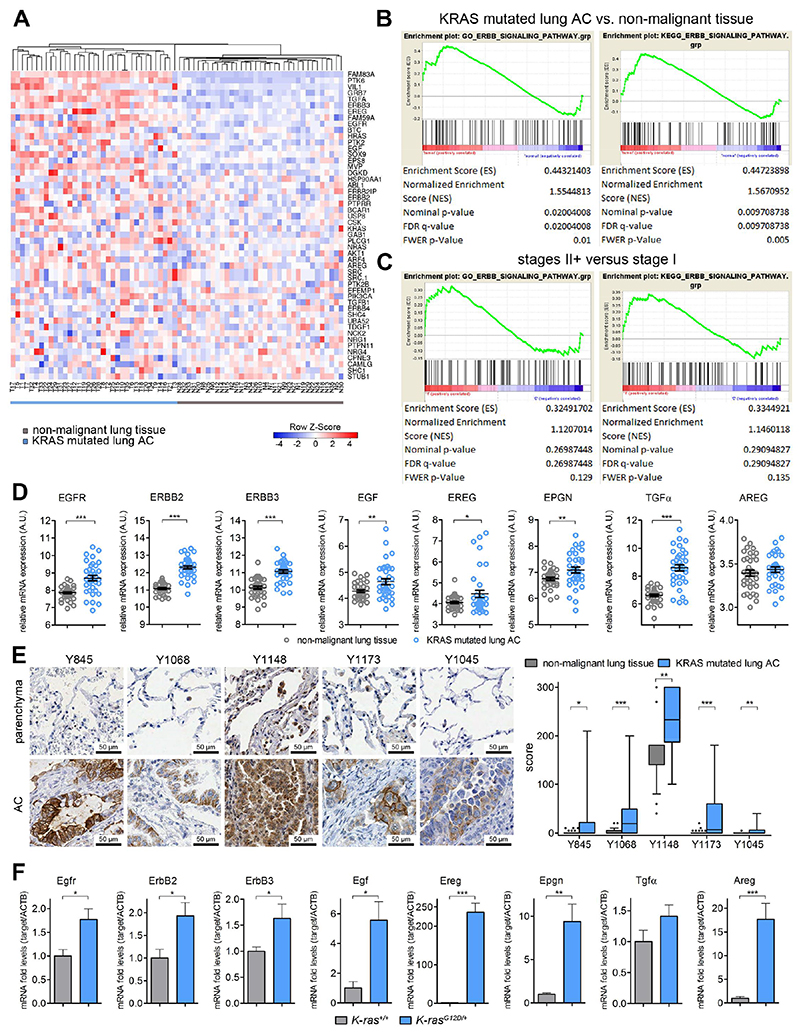
ERBB signaling is activated in human K-RAS mutated lung AC
We analyzed publically available mRNA expression data from the Gene Expression Omnibus (GSE75037) by transcriptional profiling and hierarchical clustering of human K-RAS mutated tumor biopsies versus adjacent non-tumorous lung tissue using gene signatures of ERBB activation. K-RAS driven lung AC tissue showed a uniform expression pattern of genes involved in ERBB signal transduction (Fig. 1A and Fig. S1A) (16), and mRNA expression of these genes in the tumor tissue was significantly enriched as compared to healthy tissue (Fig. 1B) (17). Furthermore, gene ratios of K-RAS mutated tumor versus adjacent lung parenchyma in patients suffering from stage II and higher advanced lung AC exhibited a significantly higher imprint of ERBB signature as compared to patients with stage I tumors, suggesting an impact of ERBB signaling during malignant progression (Fig. 1C). In more detail, we observed mRNA upregulation of the ERBB family members EGFR, ERBB2 and ERBB3 and several of its ligands (EGF, EREG, EPGN, TGFα, AREG) in human K-RAS mutated lung AC tissue compared to adjacent parenchyma (Fig. 1D). This analysis of bulk tumor samples suggest the activation of ERBB signaling in K-RAS driven tumorigenesis, but it does not allow us to discriminate whether this activation stems from the tumor cells or the stroma. Hence, we performed immunohistochemistry of resections of K-RAS mutated mucinous human lung AC and paired non-tumorous lung parenchyma, probing for the activating tyrosine phosphorylation sites Y845, Y1068, Y1148, Y1173 and inactivating site Y1045 (Fig. 1E and Fig. S2A). Analysis of the respective sections by our board certified pathologists (HP, LB, KD and JM) revealed positive tumor cells for all phosphorylation sites, whereas staining for Y1068 and Y1173 was completely absent in the stromal compartment, and staining intensity of Y1148 was much stronger in tumor cells compared to the stroma. Phosphorylation of Y1045, which counteracts EGFR activation by tagging the receptor for ubiquitination and subsequent proteasomal degradation (18), was restricted to tumor cells and endothelial cells, but absent on most tumor samples. Importantly, grading of phospho-EGFR expression demonstrated significantly higher activation of EGFR in tumor cells as compared to healthy lung parenchyma (Fig. 1E). Moreover, when we compared the phosphorylation status of EGFR in K-RAS mutated lung AC to lung AC samples harboring either wildtype (wt) K-RAS or mutated EGFR we did not observe significant differences in the activation of EGFR in tumor cells, indicating that EGFR is activated in lung AC independently of the oncogenic driver. (Fig. S2B). We then took advantage of a second cohort of K-RAS mutated patients harboring lung AC and confirmed expression of EGFR and its activation as marked by an additional activating phosphorylation site at Y1086 in tumor cells. In addition, we found that ERBB2 was expressed and activated in those tumors (Fig. S2C and S2D). Since mucinous lung AC often lack the transcription factor NKX2-1, we checked NKX2-1 expression in the human lung AC samples used in the tissue microarrays. In our cohorts the distribution of NKX2-1 expression was heterogeneous and did not correlate with EGFR activation (Fig. S2E and S2F). Altogether, these data demonstrates that ERBB signaling is activated in human K-RAS driven lung AC in tumor and to some extent in stroma cells.
Fig. 1. K-RAS mutated lung AC display increased EGFR activity.
(A) Heat map for mRNA expression in K-RAS mutated tumor biopsies (T1-T35) and adjacent non-malignant, healthy lung parenchyma (N1-N35) of the same patients. Displayed are the top 50 differentially regulated genes within the GO ERBB signaling pathway (GO: 0038127) and hierarchical clustering was performed using heatmapper.ca online tool (B) GSEA for GO and KEGG ERBB pathway signatures in K-RAS mutant tumors versus healthy “normal” tissue and (C) and in K-RAS mutated tumors of stage II and higher versus stage I. (D) Relative mRNA expression of indicated genes in healthy lung tissue and K-RAS tumors. n=35 per group, error bars, mean ± s.d. Data in (A)-(D) was retrieved from the Gene Expression Omnibus (GSE75037) (E) High resolution pictures of representative immunohistochemical stainings for indicated EGFR phosphorylation sites in human non-malignant lung parenchyma and K-RAS mutated lung AC and boxplot (min to max) of scoring values comparing EGFR phosphorylation specifically in tumor cells versus healthy tissue. n≥30 per group. (F) Relative mRNA expression in wildtype (K-ras+/+, n=5) and tumor bearing mouse lungs (K-rasG12D/+, n=6) at 10 weeks post tumor induction via Ad.Cre treatment. Actb was used for normalization. Data presented as mean ± s.d. (D) – (F) *p<0.05, **p<0.01, ***p<0.001.
Genetic EGFR ablation impairs growth of K-RAS mutated lung AC
Seeking for an adequate animal model to address the role of EGFR in K-RAS driven lung tumorigenesis, we analyzed the mRNA expression profile of tumor bearing lungs derived from a widely used mouse model for autochthonous lung tumors, where oncogenic K-ras is activated via intranasal inhalation with Ad.Cre. (19, 20) In line with the data for human lung AC, we observed increased expression of several members of the Egfr-ErbB signaling pathway upon K-ras G12D driven lung tumorigenesis in this model. Indeed, elevated mRNA levels of Egfr, ErbB2, ErbB3 and their ligands Egf, Ereg, Epgn, Tgfα, Areg in tumor harboring lung lysates versus healthy lung controls 10 weeks after oncogenic onset indicates a key role of Egfr-mediated signaling in this genetically engineered mouse model (GEMM, Fig. 1F).
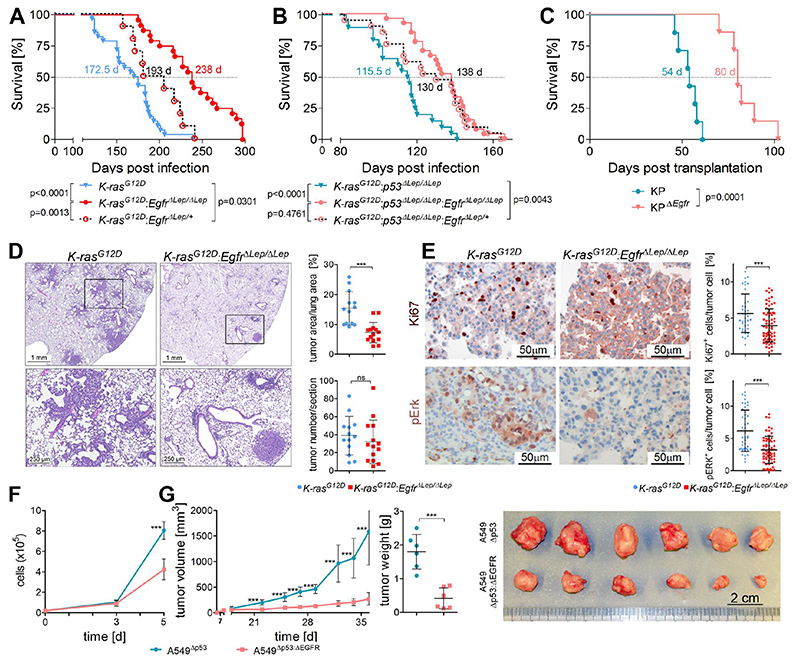
To test the impact of EGFR mediated signal transduction in K-RAS driven lung tumorigenesis, we crossed K-ras LSL-G12D (K) mice (19) with Egfr floxed/floxed (21) mice. Inhalation with Ad.Cre allowed for concomitant K-ras G12D activation and Egfr deletion in rising lung tumors in the K-ras G12D:Egfr ΔLep/ΔLep (KE) mice and resulted in significantly prolonged survival of mice as compared to K mice (Fig. 2A). Similarly, in mice with K-ras G12D activation and simultaneous deletion of p53 (22) in lung epithelial cells (K-ras G12D:p53 ΔLep/ΔLep, hereafter KP), representing a GEMM for advanced lung adenocarcinomas (23), we also observed a significant survival benefit when Egfr was deleted in K-rasG12D:p53ΔLep/ΔLep:EgfrΔLep/ΔLep (KPE) mice (Fig. 2B). Intriguingly, this survival effect was dose dependent, since mice harboring tumors with heterozygote deletion of Egfr also exhibited advanced survival times compared to K and KP mice, but decreased survival compared to KE mice (Fig. 2A and 2B). In order to validate efficient and complete recombination of the floxed Egfr allele in the rising tumor cells, we isolated tumor cells from the lungs of KP and KPE mice used in the survival analysis after death of the animals. Genotyping of these KP and KPE cells after passaging them 5-10 times in vitro ruled out incomplete recombination of Egfr (Fig. S3A).
Fig. 2. Genetic EGFR ablation in K-RAS mutated lung AC reduces tumor growth.
(A) and (B) Kaplan-Meier analysis of K (K-rasG12D, n=24), KE (K-rasG12D:EgfrΔLep/ΔLep, n=28), KP (K-rasG12D:p53ΔLep/ΔLep, n=20) and KPE (K-rasG12D:p53ΔLep/ΔLep:EgfrΔLep/ΔLep, n=27) mice following intranasal infection with Ad.Cre. (C) Survival analysis of immunocompetent recipient mice following orthotopic transplantation of syngeneic K-ras G12D mutated and p53 deficient KP cells, with and without Egfr deletion (n=7 per group). (A) – (C) The median survival times of the respective groups are indicated. Differences in survival of groups were tested using the Log-rank test, and respective p values are shown. (D) Representative pictures of H&E staining including higher magnification of indicated area (bottom) of tumor bearing lungs 10 weeks post Ad.Cre inhalation of mice with specified genotypes. For quantitation, the mean values of two sections per mouse were used. Graphs represent mean of ratios ± s.d. of tumor area versus healthy lung area and mean of tumor numbers ± s.d. per section (n=13 mice for K-rasG12D and n=14 mice for K-rasG12D:EgfrΔLep/ΔLep). (E) Representative pictures of immunohistochemical staining of mouse lungs 10 weeks post tumor induction using antibodies specific for Ki67 and pErk. Tumor cell intrinsic expression of the respective genes in at least 5 individual tumors per mouse was evaluated using TissueGnostics software. Graphs represent mean ± s.d. of Ki67 and pErk positive tumor cells normalized to all tumor cells (n=5-7 mice per group). (F) Cell count of p53 deficient versus p53/EGFR double knockout A549 cells following standard in vitro cultivation. Graph represents mean ± s.d. of three individual clones per group. (G) Mean volumes ± s.d. of xenografted tumors comparing EGFR expressing versus EGFR deficient p53 knockout A549 cells, monitored over the course of the experiment as well as the endpoint tumor weight ± s.d. (n=6 per group). Picture illustrates the tumors after finalization of the experiment. (D) to (G) *p<0.05, ***p<0.001.
In these experimental models, stromal cells in addition to the epithelial cells also have the potential to be transduced by Ad.Cre, resulting in recombination of transgenes within these cells and eventually triggering S100 positive Langerhans cell histiocytosis-like neoplasms in the lungs. (24, 25) Although the majority of tumor cells stained positive for the alveolar type II (AT2) marker Surfactant protein C (SP-C, Fig. S3B), whereas S100 expression was restricted to single stromal cells, we cannot exclude that Egfr deletion in stromal cells might contribute to the observed phenotype. Therefore we performed orthotopic transplantation of KP and KPE tumor cells isolated from lungs of KP and KPE mice at 6 weeks post Ad.Cre inhalation. These cells were delivered to the lungs of syngeneic immunocompetent mice via tail vein injection. As in the Ad.Cre model, mice harboring Egfr deficient K-ras mutated transplanted tumors exhibited a significant survival advantage compared to mice injected with Egfr expressing K-ras mutated lung AC cells, further demonstrating that tumor cell intrinsic deletion of Egfr impairs growth of K-ras mutated lung AC (Fig. 2C).
For histopathologic analysis, we first verified Egfr knockout in lung tumor sections of KE mice 10 weeks post Ad.Cre inhalation (Fig. S3C). Mice harboring Egfr knockout tumors exhibited reduced tumor burden, which was also reflected by reduced lung to body weight ratios (Fig. 2D and Fig. S3D). Furthermore, the increase on SP-C mRNA expression in lungs of K mice compared to healthy lungs reflects higher abundance of AT2 cells, the main cell type of origin of K-ras driven lung AC, and was reduced in KE mice. (Fig. S3E). (26–28) Intriguingly, there was no significant difference of total tumor numbers per area and grade in KE as compared to K mice (Fig. 2D and Fig. S3F). KE mice showed significantly reduced tumor cell proliferation but no changes in apoptosis when compared to K mice (Fig. 2E and Fig. S3G). Downstream of K-ras signaling, we found decreased activation of Extracellular Signal Regulated Kinase 1 and 2 (Erk1/2) in tumors of KE mice (Fig. 2E). On the other hand, activated Serine-Threonine Protein Kinase Akt levels were higher in the KE group, most likely to compensate loss of Egfr (Fig. S3H). On the mRNA level, loss of Egfr also reduced mRNA expression of Egfr ligands Areg, Ereg and Epgn in mouse lungs 10 weeks post tumor induction (Fig. S3I).
Next we used CRISPR-Cas9 technology to generate EGFR-deficient isogeneic clones of the human lung AC cell line A549 (which harbors a homozygous K-RAS G12S mutation), with and without concomitant p53 deletion (Fig S4A). EGFR-deficiency reduced in vitro growth of A549 cells significantly (Fig. 2F and Fig. S4B). Furthermore, EGFR-deficiency interfered with A549 tumor growth after xenotransplantation into immunodeficient NOD SCID gamma (NSG) mice, regardless of the p53 status (Fig. 2G and Fig S4C). Together, these data demonstrates that growth of K-RAS mutated lung AC depends on expression of upstream EGFR, both in vitro and in vivo.
Mutant K-RAS activity in lung AC depends on upstream EGFR activation
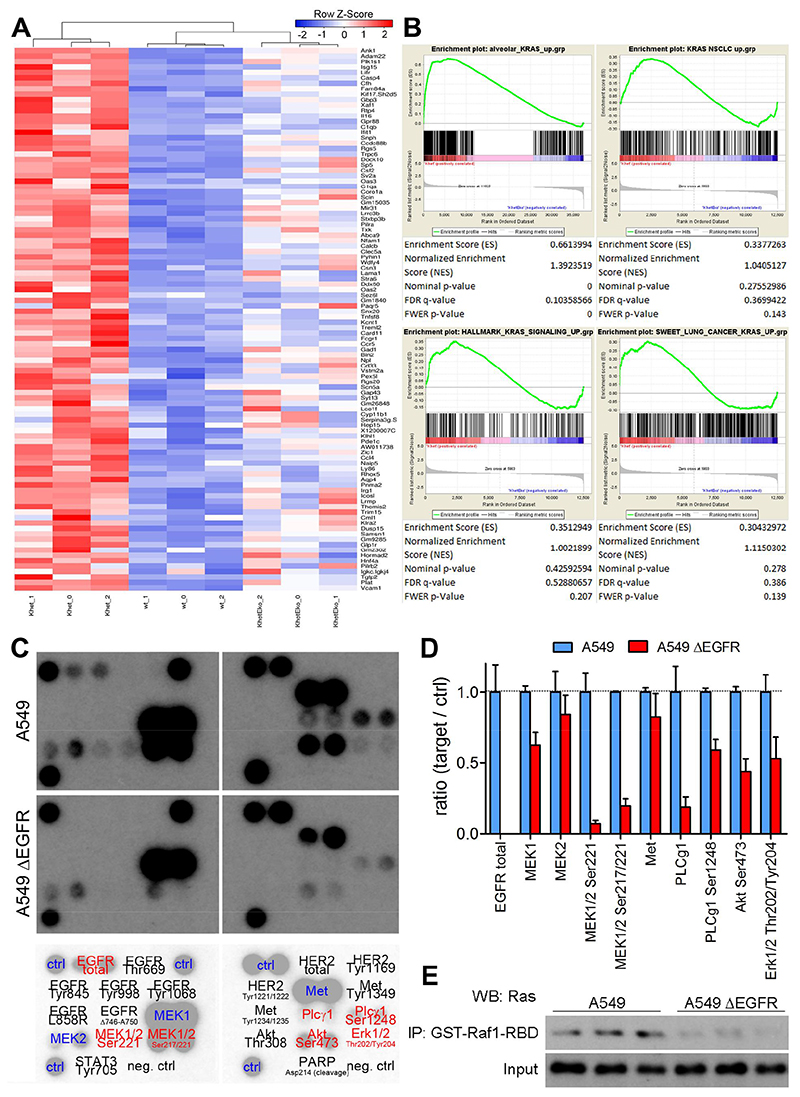
Next we aimed to identify the key signaling nodes affected by loss of EGFR in K-RAS mutated lung AC cells. Therefore, we isolated primary mouse alveolar type 2 pneumocytes from the lungs of wt, K and KE mice and transduced cells in vitro with Ad.Cre. We confirmed purity of cell isolates, activation of mutated K-ras and loss of Egfr in K and KE cells two days following transduction (Fig. S5A and S5B). Five days post Ad.Cre treatment we retrieved RNA of wt, K and KE cells and subjected it to RNAseq analysis. As in bulk tumor tissue of human and mouse origin we detected a significant increase of Egfr expression in primary cell isolates upon activation of the mutant K-ras G12D allele in K cells (Fig S5C), as well as an overall increase in the expression of ErbB signature genes (Fig. S5D). Based on that data we generated a mutant K-ras gene signature dataset which includes the 500 most upregulated genes in K cells versus wt cells and hence depicts the most prominent alterations upon K-ras G12D activation in type II pneumocytes, (alveolar_KRAS_up, Supplementary Table 1). Using the top 100 genes of this signature to perform unsupervised clustering analysis revealed that KE cells grouped closer to wt than to K cells, suggesting that K-ras activity is significantly impaired upon Egfr knockout (Fig. 3A). Indeed, enrichment of alveolar_KRAS_up gene signature in K cells compared to KE cells was highly significant (Fig. 3B). We validated this finding by probing for additional published and unpublished (KRAS NSCLC_up: top 500 genes significantly upregulated in KRAS mutated NSCLC compared to adjacent non-malignant lung tissue) K-RAS signature data sets. Probing for all analyzed datasets showed enrichment in K cells when compared to KE cells (Fig. 3B) indicating that deletion of Egfr abrogates K-RAS activity. Egfr knockout in KE also significantly abrogated ErbB signaling signatures as evidenced by the GO and KEGG ERBB pathway gene sets, as compared to K cells (Fig. S5E). Further, at this early time point after K-rasG12D activation and Egfr knockout the PI3K-Akt-mTOR signaling signature was decreased in KE cells and Egfr deficiency resulted in a better prognostic index (Fig. S5E). In agreement with other studies (14, 15), our gene expression analysis points out that mutant K-ras activity depends on upstream EGFR expression. However, the primary cells used for RNAseq analysis were heterozygous for the K-ras G12D transgene, and we could not rule out an impact of the wt K-ras allele. Hence we analyzed A549 cell lysates using an antibody array and found decreased phosphorylation of virtually all downstream mediators of EGFR in A549ΔEGFR cells when compared to EGFR expressing cells (Fig. 3C and 3D). Downregulation of Mitogen-activated protein kinase kinases MEK1/2 and ERK1/2 further indicates a decrease in K-RAS activity in EGFR-deficient A549 cells despite the homozygous G12S mutation. Indeed, using the RAS binding domain of the RAF proto-oncogene serine/threonine-protein kinase (RAF1-RBD) for pulldown of active, GTP bound RAS, we perceived reduced levels of activated RAS in EGFR-deficient A549 cells (Fig. 3E). To further validate inhibition of mutant K-RAS activity upon EGFR knockout we took advantage of mouse KP cells and an antibody specifically recognizing mutant K-rasG12D RAF1-RBD pulldown revealed reduced active K-rasG12D in KPE cells compared to KP cells, whereas both cell lines lacked active forms of H-ras and N-ras (Fig. S5F). Taken together, these data indicates that activity of mutated K-RAS relies on activation of upstream mediators including EGFR and may represent therapeutic intervention opportunities in K-RAS driven tumors.
Fig. 3. Inhibition of EGFR signaling downregulates K-RAS mutated activity.
(A) Heat map of top 100 upregulated genes in K-rasG12D versus wildtype type II alveolar cells and hierarchical clustering of wildtype (wt_0-2), K-rasG12D (Khet_0 – 2) and K-rasG12D:EgfrΔ/Δ (KhetEko_0-2) mouse pneumocytes. (B) GSEA of K-rasG12D versus K-rasG12D:EgfrΔ/Δ mouse pneumocytes for indicated gene sets. (C), Representative picture of antibody array of cell lysates of A549 and A549ΔEGFR cells (n=2 clones per group with 2 spots each). Antibody probes are decoded in lower panel, black color indicates that proteins were not detected in this assay and red color specifies proteins downregulated in the A549ΔEGFR group. (D) Densitometric quantitation of microarray films (n=2 clones per groups). (E) Western blot probing for RAS following GST-RAF1-RBD mediated pulldown in A549 and A549ΔEGFR cell lysates and respective input samples (n=3 per group).
Afatinib, but not erlotinib or gefitinib, reduces growth of K-RAS mutated lung AC
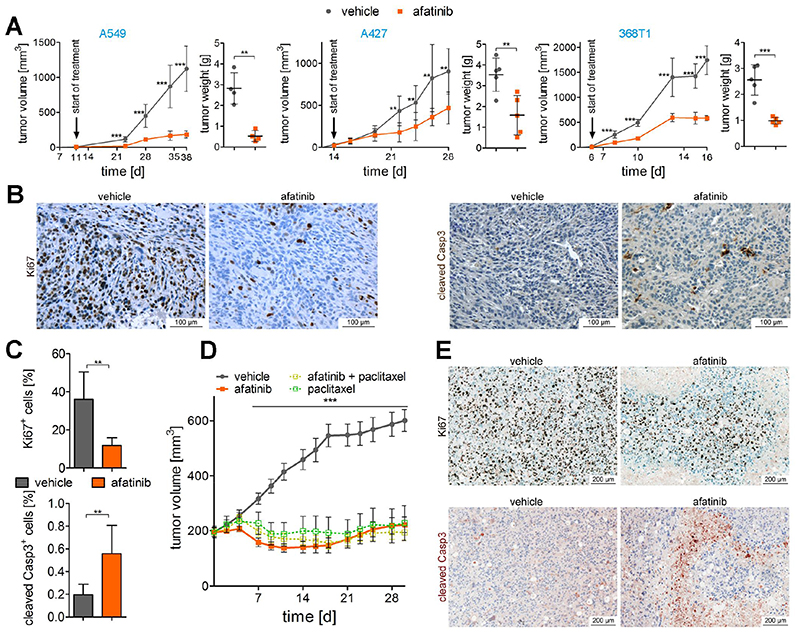
Accordingly, we tested in vitro efficacy of the EGFR TKI inhibitors afatinib, erlotinib and gefitinib in several K-RAS mutated human and mouse lung AC cell lines and included two EGFR mutated cell lines as controls (Fig. S6A to S6C). As expected, HCC827 cells were particularly sensitive to EGFR TKI treatment, since these cells harbor the EGFR ΔE746-A750 mutation. In H1975 cells, the T790M gatekeeper mutation on top of the EGFR L858R prevented cytotoxic effects of erlotinib and gefitinib (29), whereas afatinib inhibited cell growth in the nano-molar range. In K-RAS mutated cell lines, afatinib exhibited an IC50 in the low micro-molar range, whereas the IC50 values of erlotinib and gefitinib were about 10 – 20 fold higher (Fig. S6A to S6C). When checking downstream mediators of K-RAS, we found decreased activation of ERK and MEK as well as AKT following short term afatinib treatment of A549 cells in vitro, but there was no effect when treated with erlotinib (Fig S6D). In vivo, we found that afatinib treatment of mice significantly reduced growth of xenografted human cell lines A549 and A427 and of transplanted p53 deficient mouse 368T1 cells, which all exhibit K-RAS mutations (Fig. 4A). As evidenced by IHC, afatinib treatment significantly reduced proliferation rate and induced apoptosis of grafted cells (Fig. 4B and 4C). Further, we detected significantly decreased phospho-Erk levels but no difference in phospho-Akt levels in engrafted 368T1 cells when harvested at the end of the experiment (Fig. S6E &F).
Fig. 4. Afatinib reduces growth of K-RAS mutant lung AC in vivo.
(A) Graphs display tumor volumes of (xeno-)grafts using indicated cell lines monitored over the experimental period and tumor volumes at the end of experiment. Mice were treated with vehicle alone or afatinib at 5 mg/kg body weight via oral gavage, 5 times per week, and the start of treatment is indicated. Means ± s.d. are shown. n=4 per group in the A549 experiment and n=5 per group in A427 and 368T1 experiment. Unpaired two-tailed t-test for individual time points and tumor weight. (B) Representative pictures of Ki67 and cleaved Caspase 3 staining of 368T1 cell line derived grafts upon vehicle and afatinib treatment and (C) quantitation of positive cells for respective staining (n=5). Positive tumor cells where determined using TissueGnostics software. (D) Mean tumor volumes ± s.d. of patient derived xenografts of lung AC tissue with KRASG12C mutation. Mice were treated with afatinib (15 mg/kg body weight, daily), paclitaxel (15 mg/kg body weight, once per week) or a combination of both treatments (n=8 per group). Unpaired two-tailed t-test for individual time points. (E) Representative Ki67 and cleaved caspase 3 stainings of sections of vehicle treated versus afatinib treated patient derived xenografts. (A), (C), (D), **p<0.01, ***p<0.001.
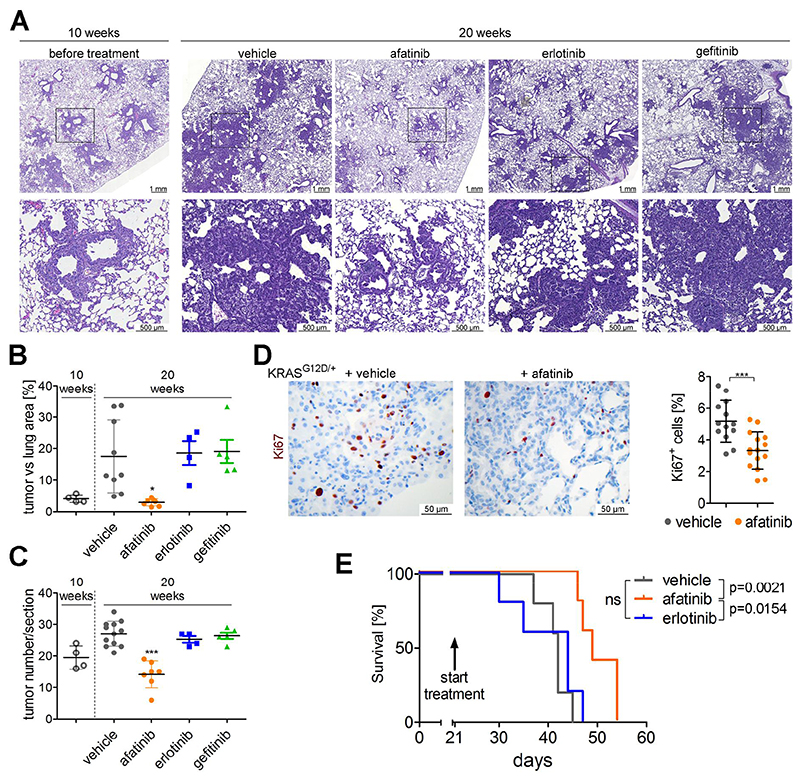
Encouraged by these results that afatinib inhibits K-RAS driven growth of lung AC cell lines in vitro and in vivo we then performed patient derived xenografts (PDX) using tumor tissue of a K-RAS G12C mutated patient. Strikingly, the engrafted tumors were highly sensitive to afatinib treatment, with an efficacy that was similar or even higher than that of the chemotherapeutic agent paclitaxel (Fig. 4D). However, combination therapy with afatinib and paclitaxel could not further diminish tumor volume. As in the cell line derived xenografts we observed reduced cell proliferation and increase in apoptosis in the afatinib treated group (Fig. 4E). We complemented these studies by using the K-ras G12D based GEMMs harboring autochthonous tumors. First, we treated K mice with afatinib (and vehicle controls) for a period of 9 weeks, starting treatment one week post-tumor induction. When we analyzed the lungs of these mice, we noticed significantly reduced tumor burden and number in lungs of afatinib treated mice as compared to vehicle treated mice (Fig. S7A and S7B). In accordance to reduced tumor area we also detected reduced oncogenic K-ras G12D levels in total lung lysates of the afatinib treated group, which reflects the decreased amount of tumor cells in the lungs (Fig. S7C). Having demonstrated that afatinib reduced tumorigenesis at early stages, we investigated whether EGFR targeting TKI may retain growth of more advanced tumors. Therefore we treated mice with already established K-ras G12D lung AC with afatinib, erlotinib, gefitinib or vehicle control, starting 10 weeks post-tumor initiation. Following another 10 weeks of treatment, mice were sacrificed and lungs subjected to analysis. Intriguingly, afatinib treatment, but not erlotinib or gefitinib impaired growth of K-ras G12D driven lung tumors. Indeed, tumor areas and numbers in lungs of afatinib treated mice (20 weeks post tumor onset) were slightly reduced compared to lungs 10 weeks after K-ras G12D induction (before treatment), whereas tumor areas and numbers in control and erlotinib/gefitinib treated groups were significantly higher (Fig. 5A to 5C). These data was paralleled by a significantly reduced lung to body weight ratio in the afatinib treated group compared to the other groups, as well as by reduced SP-C mRNA expression in the afatinib treated group compared to the other groups (Fig S7D and S7E). Furthermore, all afatinib treated tumors were classified as grade I tumors, whereas the majority of tumors in the control group were of grade II or III as graded by board certified pathologists (HP & KD; Fig 5A). (30) As in xenograft experiments, long term afatinib treatment significantly reduced tumor cell proliferation and Erk activation, but not activation of Akt (Fig 5D and S7F). Ultimately, the beneficial effects of afatinib were also highlighted in a survival analysis using the syngeneic transplant model, where 368T1 lung cancer cells (K-ras G12D:p53 Δ/Δ) were orthotopically transplanted into immunocompetent mice. Indeed, afatinib administration to mice starting 3 weeks post transplantation significantly prolonged survival of these mice in comparison to vehicle treated mice, whereas erlotinib treatment did not exhibit any beneficial effects (Fig. 5E).
Fig. 5. Afatinib, but not first generation EGFR TKI, inhibits growth of autochthonous K-ras tumors.
(A) Representative pictures of H&E stained lung sections of KrasG12D/+ mice 10 weeks post Ad.Cre inhalation (left panel) or 20 weeks post Ad.Cre inhalation and treatment over the last 10 weeks with vehicle, afatinib, erlotinib or gefitinib (5 mg/kg body weight, 5 times per week via oral gavage). Lower panels represent magnifications of indicated sections in top panel. n≥4 per group. (B), (C) Graphs represent mean ± s.d. of tumor area versus total lung area ratios and mean of tumor numbers ± s.d. per section of lung of mice. Each data point represents the mean value of two sections derived from one mouse. One way ANOVA and Tukey’s Multiple Comparison Test. (D) Representative pictures of Ki67 staining of lung tumors 20 weeks post Ad.Cre induction and 10 weeks of vehicle versus afatinib treatment. Ki67 positive tumor cells in at least 3 tumors per mouse were quantitated using TissueGnostic software, and blot indicates mean ± s.d. of Ki67 positive tumor cells. Student t-test, n= 4 mice per group. (E) Survival analysis of immunocompetent mice following orthotopic transplantation of syngeneic 368T1 lung AC cells. 3 weeks post injection, treatment with vehicle, afatinib or erlotinib (5 mg/kg body weight, 5 times per week via oral gavage) was started. Median survival times were 42 days for vehicle group, 49 days for afatinib group and 44 days for erlotinib group. Log-rank test, n=5. (B) to (E) *p<0.05, **p<0.01, ***p<0.001.
Afatinib blocks a tumor-escape mechanism mediated by non-EGFR ERBB family members
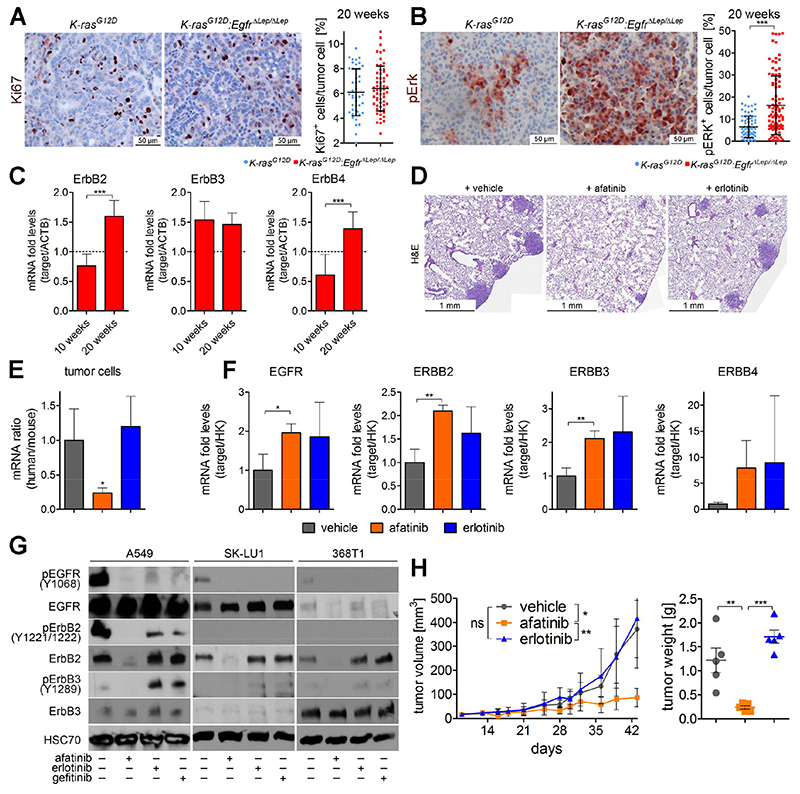
Puzzled by the fact that K-RAS driven lung AC cells and tumors were sensitive to genetic EGFR knockout and to treatment with EGFR TKI afatinib, but not to erlotinib or gefitinib, we reanalyzed the EGFR-deficient A549 cell line. As stated above, we noticed that A549ΔEGFR cells in early passages after gene knockout and monoclonal expansion exhibited severely reduced proliferation rates in vitro (Fig. S4B). However, over time A549ΔEGFR regained their proliferative capacity and in higher passages there was no noticeable difference in cell growth between wt A549 and A549ΔEGFR cells (Fig. S8A). We performed gene expression analysis of these cells and found that in high passage A549ΔEGFR cells the ERBB family members ERBB2, -3 and -4 were significantly upregulated (Fig. S8B), suggesting that these ERBB family members compensate for the loss of EGFR. Next, we checked mouse tumors at later time points in K and KE mice 20 weeks post tumor-induction. Intriguingly, proliferation rate of KE tumors was similar to K tumors (Fig. 6A), indicating the activation of a compensatory program in Egfr deficient tumor cells at later time points. This was also illustrated by elevated Erk phosphorylation in KE versus K lungs at 20 weeks (Fig. 6B). Notably, proliferation and Erk activation were downregulated at 10 weeks (Fig. 2E). On mRNA level, expression analysis revealed significantly higher ErbB2 and ErbB4 expression in KE versus K lungs compared to the ten week time point (Fig. 6C). ErbB3 expression was already increased at 10 weeks in KE lungs, maybe as a vanguard of the compensation machinery (Fig. 6C). These data was of particular interest, since ERBB2 and ERBB3 were also significantly upregulated in human K-RAS lung AC tissue confirming the implication of other non-EGFR ErbB family members in K-RAS mutated lung tumorigenesis (Fig. 1D).
Fig. 6. ERBB family members mediate resistance to EGFR inhibition, which can be blocked by afatinib.
(A) Representative picture of Ki67 and (B) of pErk in lung tumors of indicated mice 20 weeks post Ad.Cre administration. The percentage of tumor cells expressing the respective proteins were quantitated in at least 8 individual tumors per mouse using TissueGnostics software. Graphs represent mean ± s.d. of percentage of Ki67 and pErk positive tumor cells (n=6 mice per group). (C) mRNA expression of indicated genes in lungs of K-rasG12D:EgfrΔLep/ΔLep mice 10 weeks and 20 weeks post Ad.Cre inhalation. Actb was used as a housekeeper gene and relative expression is normalized to expression levels of K-rasG12D mice at respective time points (dotted line). n≥6 per group. (D) Representative photographs of H&E stained mouse lung sections, 5 weeks post orthotopic transplantation of A549Δp53 cells by tail vein injection and 3 weeks of treatment with vehicle, afatinib or erlotinib (5 mg/kg body weight, 5 times per week via oral gavage). (E) Relative mRNA expression ratios of human versus mouse housekeeping genes (ACTB and 28S) and in mouse lungs 5 weeks following orthotopic transplantation of A549Δp53 cells and 3 weeks of indicated treatment, and (F) relative mRNA expression levels of human variants of indicated genes normalized to human housekeeping genes (ACTB and 28S). (G) Western blot probing for indicated proteins in A549, SK-LU1 and 368T1 cell lysates following treatment with 1 μM of afatinib, erlotinib or gefitinib for 48 h. (H) Tumor volumes of A549ΔEGFR xenografts in mice receiving vehicle, afatinib or erlotinib treatment (5 mg/kg body weight, 5 times per week via oral gavage), starting 14 days post transplantation, monitored over the experimental period and tumor weights at the end of experiment. n≥5. (A) - (H) *p<0.05, **p<0.01, ***p<0.001.
Similar to genetic knockout of Egfr, afatinib treatment induced significant upregulation of the ErbB receptors in tumors of K mice (Fig. S8C). To test whether this compensatory upregulation of ERBB family members stems from the tumor cells or the stroma, we orthotopically transplanted human A549Δp53 cells into NSG mice and treated the experimental mice with afatinib or erlotinib. Complimentary to the other models used, we noticed significantly decreased tumor burden in lungs of afatinib treated mice compared to vehicle and erlotinib treated mice, which was quantitated by RT-PCR using primers specific for human housekeeping genes and thus specific for xenografted A549 cells (Fig. 6D and 6E). Further gene expression analysis revealed enhanced expression of human (i.e. tumor cell derived) EGFR, ERBB2 and ERBB3 in the tumors of afatinib treated mice, which was comparable to upregulation in the erlotinib treated group (Fig. 6F). However, on protein level, afatinib could utterly abrogate the ERBB2 and ERBB3 mediated compensatory mechanism. In fact, afatinib triggered a dramatic reduction in ERBB2 protein expression and completely blocked ERBB3 activation by phosphorylation, as demonstrated in three different K-RAS mutated cell lines (Fig 6G). Importantly, neither erlotinib nor gefitinib had any effect on ERBB2 protein expression, and both TKI exacerbated compensatory mechanisms via ERBB3 activation (ERBB4 protein expression levels were not detectable). Altogether, these data suggested that both the genetic knockout of EGFR, as well as the treatment with EGFR TKI engages a tumor-compensatory mechanism via other non-EGFR ERBB family members, which can be suppressed by afatinib, but not by erlotinib or gefitinib.
Along this line, in high passage A549ΔEGFR cells which compensated the EGFR deficiency by increased ERBB family member expression and activation (Fig. S8B and S8D) afatinib was able to block compensating ERBB2 and ERBB3 activation in vitro, but not erlotinib nor gefitinib. Finally we xenografted high passage A549ΔEGFR cells into NSG mice and subjected them to treatment with afatinib or erlotinib. Confirming our hypothesis, afatinib treatment could block tumor growth of EGFR-deficient/K-RAS mutated tumor cells, whereas erlotinib did not exhibit any noticeable effect on tumor growth of these cells (Fig 6H and Fig. S8E). Altogether, these data demonstrates that both genetic and pharmacologic abrogation of EGFR mediated signaling engages a compensatory mechanism involving other ERBB family members in K-RAS mutated lung AC cells. Afatinib, as a pan-ERBB inhibitor, can efficiently suppress this compensatory machinery, therefore mediating a significant reduction of K-RAS driven tumor growth.
Discussion
Traditionally, mutated K-RAS has been considered to be locked in a constitutive active state that does not require upstream signaling. Therefore, it is commonly believed that K-RAS driven tumors are refractory to TKI therapy (31, 32). Challenging this view, recent reports using K-RASG12C irreversible inhibitors have shown that mutated K-RAS can be “hyperactivated” by upstream effectors, therefore opening the possibility of targeting receptor tyrosine kinases in K-RAS driven tumors (14, 15). Sustaining these observations, K-RAS driven pancreatic tumors show expression of EGFR and ERBB family ligands and they depend on EGFR and the ligand activating ADAM17-sheddase (33, 34). In line, we found that ERBB signaling is active in human and mouse K-RAS-driven lung AC. Of note, human mucinous lung AC (cohort I) may not be properly modeled by our murine experimental system due to the differences in NKX2-1 expression.(35) Importantly, advanced human tumors are enriched in an ERBB gene signature indicating that ERBB signaling contributes to progression of K-RAS driven AC. Supporting an EGFR active role in tumorigenesis, genetic inactivation of EGFR impaired tumor growth in different experimental models of K-RAS driven lung AC. In contrast to K-RAS driven pancreatic cancer, tumor reduction upon EGFR deletion was irrespective of the p53 status, underscoring the importance of EGFR in K-RAS driven AC. In agreement with the new proposed functional model of mutated K-RAS (14, 15), genetic deletion of EGFR downregulated activity of mutated K-RAS and downstream signaling pathways, and this may explain the observed reduction in tumorigenesis. However, over time K-RAS mutant tumors recover from EGFR deletion via increased expression and activation of remaining EGFR family members, thereby restoring downstream ERK and AKT activation levels. This further highlights the dependence on ERBB signaling for full blown tumorigenesis despite the oncogenic K-RAS mutation.
Controversially, most clinical studies using first generation TKI (erlotinib and gefitinib) showed no or little benefit in patients suffering of K-RAS driven NSCLC. In agreement with these studies, erlotinib and gefitinib failed to impair tumorigenesis in all our experimental models of K-RAS driven AC, while tumors transiently responded to genetic deletion of EGFR. This may be attributed to the inherent differences between the genetic and pharmacological approaches. Indeed, EGFR mediates kinase independent functions in cancer cell survival (36, 37), and while total EGFR knockout mice are not viable, animals with severely suppressed EGFR kinase activity display only minor epithelial defects. (38–40) However, in our study both genetic EGFR deletion and erlotinib treatment triggered a similar tumor escape mechanism relying on the activation of other non-EGFR ERBB family members, although the initiation of this mechanism was delayed upon EGFR knockout compared to TKI mediated inhibition. In fact this rapid response to TKI treatment may be responsible for the failure of the first generation TKI in K-RAS driven NSCLC. Supporting this hypothesis, afatinib (an irreversible pan-ERBB inhibitor) abrogated the activation of ERBB family members, suppressed the tumor compensatory mechanism and resulted in an efficient inhibition of K-RAS driven lung AC. In this sense, pan-ERBB inhibition using a mixture of monoclonal antibodies (pan-HER) suppresses tumorigenesis more efficiently than targeting single ERBB receptors. (41) Intriguingly, we and Kruspig et al. (co-submitted manuscript) found that irreversible TKI as afatinib or neratinib downregulated ERBB2 and ERBB3 total proteins, a similar phenomenon observed using pan-HER antibodies (41) which may contribute to the efficacy of irreversible TKI.
Altogether our data suggest that, in contrast to current opinion, resistance to first generation TKI in K-RAS driven NSCLC may not be due to constitutive activation of K-RAS but rather due to a (re)-activation of other ERBB family members. These findings suggest pan-ERBB inhibitors, such as afatinib, alone or in combination with other inhibitors, e.g. MEK (Kruspig et al., co-submitted manuscript)(42) or K-RASG12C inhibitors, as potent therapeutic agents for treatment of patients suffering from K-RAS mutated NSCLC.
Materials and Methods
Study design
The main goal of the study was to revisit the role of EGFR mediated signaling in K-RAS driven lung tumorigenesis. Hence, we used publically available datasets, biopsies of patients, in vitro model systems and mouse models for analysis, as outlined in the results section. To calculate the minimal mouse number (i.e. sample size) for Kaplan-Meier analysis we used a web-based tool (http://www.cct.cuhk.edu.hk/stat/survival/Rubinstein1981.htm) and the following parameters: α=0.0125; β=0.05; δ=0.12, Ms=5.75 month; QC=0.5; QE=0.5; T0=2 month; T-T0=8 month. For the time point analysis (10 weeks and 20 weeks post tumor initiation) we used the web-tool (http://www.quantitativeskills.com/sisa/calculations/samsize.htm) and the following parameters: Mean1 (Exp.): 10.90/21.33; Mean2 (Obs.): 6.105/6.333; SD1:2.937/12.01; SD2:3.037/4.041; Allokation Ratio=1; Power 95; Alpha: 5. 18. In the course of the experiments, calculated sample numbers were adjusted according to the availability of the respective mice. For our in vivo studies using TKI we randomly assigned mice to the different treatment groups before the start of the experiment, i.e. rAd.Cre inhalation or tumor cell injection. Also, tissue was harvested and processed in a random and blinded order. All other experiments were performed using several biological replicates (as indicated in figure legends), and all replicates were included in our data analysis. As a common guideline in our laboratory, we determine outliers and exclude them from analysis according to the following rule: An outlier in a distribution is a number that is more than 1.5 times the length of the box away from either the lower or upper quartiles. Specifically, if a number is less than Q1 - 1.5×IQR or greater than Q3 + 1.5×IQR, then it is determined as an outlier.
GEO dataset analysis
Illumina microarray data (GSE75037) (16) of K-RAS mutated lung AC and healthy adjacent parenchyma was validated by positively testing for elevated K-RAS gene signature and increased expression of genes associated with poor survival in the tumors using gene set enrichment analysis (GSEA, Fig. S1B) (17, 43). We used Illumina unique identifier ILMN_1798975 to identify EGFR, ILMN_2352131 for ERBB2, ILMN_1751346 for ERBB3, ILMN_1653728 for ERBB4, ILMN_1690733 for EGF, ILMN_1657248 for EREG, ILMN_1815313 for EPGN, ILMN_1804421 for AREG and ILMN_2083946 for TGFα. For hierarchical clustering and illustration of heatmap we used the heatmapper.ca webtool (44). Gene set enrichment analysis (GSEA) for indicated gene sets downloaded from the Molecular Signature Database (http://software.broadinstitute.org/gsea/index.jsp) was performed according to the provider’s protocol. For the comparison of stage II and higher tumors versus stage I tumors we used the ratios of mRNA expression within the tumor tissue over the expression of the adjacent non-malignant tissue of the same individuals. Gene signature dataset alveolar_KRAS_up was generated by using the top 500 significantly upregulated genes when comparing wt mouse primary alveolar type II cells versus K-RASG12D mutated mouse primary alveolar type II cells. KRAS_NSCLC_up gene signature was generated using the top 500 genes significantly upregulated in K-RAS mutated adenocarcinomas when compared to adjacent normal parenchyma tissue using the publically available GEO dataset GSE75037.
TMA
Two sets of tissue microarray (TMA) were produced. The first one included 35 cases of human mucinous lung AC harboring KRAS mutations, as well as tissue from patients with wt KRAS or mutations in EGFR, respectively (cohort I). From each patient three cores of tumor tissue and one core of non-tumor lung parenchyma were included. Antibodies for the tyrosine phosphorylation sites Y845, Y1045, Y1068, Y1148, and Y1173 on EGFR were used (all antibodies were purchased from Cell Signaling Technology). Percentage of the positive tumor cells, as well as intensity of the staining (scale 1-3) were recorded by a board-certified pathologist (HP&LB), and final results expressed as H-score (percentage of positive tumor cells x intensity). The same scoring method was performed on non-tumor lung parenchyma samples. In the second TMA (cohort II, unselected KRAS mutated lung AC) single or double cores per patient, depending on tumor heterogeneity were prepared from selected areas of formalin-fixed paraffin-embedded surgical tissue samples (TMA Master, 3DHistech). TMAs were stained with EGFR (1:100, Invitrogen, #28-0005), phospho-EGFR (Tyr1086, 1:100, Thermo, #369700), ERBB2 (1:600, Dako, #A0485), phospho-ERBB2 (Tyr1221/1222, 1:300, Cell Signaling, #2243) primary antibodies. The stained slides were digitalized with a slide scanner (Mirax Scan) and then scored by a board-certified pathologist (JM&KD) with the H-score from 0 to 300 based on both the intensity of tumor staining and the percentage of cells stained. Intensity was considered 0 for absent expression, 1+ for weak, 2+ for moderate and 3+ for strong staining. Both cohorts were stained with a NKX2-1 antibody (1:100, Agilent, M357529-2) and percentage of positive tumor cells determined by pathologists (HP&LB and JM&KD, respectively).
Animal husbandry and experiments
Animal husbandry and all experimental protocols as described below followed ethical guidelines and were approved by the Austrian Federal Ministry of Science, Research and Economy or the Competent Authority of the Comunidad de Madrid, respectively. All procedures were conducted in accordance with these protocols. For the autochthonous lung cancer GEMM we crossed mice carrying floxed EGFR alleles (21) with the K-rasLSL-G12D (K) knockin mouse (19, 20) to generate K-rasG12D: EgfrΔLep/ΔLep (ΔLep: deleted in Lung epithelial cells, KE) mice. Mice harboring floxed p53 alleles(22) were used to generate K-rasG12D:p53ΔLep/ΔLep (KP) and K-rasG12D:p53ΔLep/ΔLep:EgfrΔLep/ΔLep (KPE) mice. All mice used for the autochthonous lung tumor GEMM were genotyped using primers for the transgenes (Supplementary Table 2) and maintained on C57Bl/6 background. Lung tumors were induced in 8-10 weeks old mice through intranasal inhalation using 2.5x107 plaque forming units of an Cre-recombinase expressing adenovirus (Ad.Cre) purchased from the Viral Vector Core of the University of Iowa (30). For the syngeneic, orthotopic transplantation model we crossed 129 mice with C57Bl/6 mice and used F1 littermates for experiments as previously described (45). In this model we injected 104 368T1 cells, whereas for the orthotopic xenotransplantation model we injected 105 A549 cells through the lateral tail vein. For subcutaneous xenograft experiments and transplantation of mouse derived 368T1 cells NOD scid gamma (NSG) mice were used at 6 – 8 weeks for experiments. 2x106 cells in a 50% matrigel (Corning) suspension were injected per graft into the right and left flank of NSG mice. Tumor volume was measured using a caliper and calculated using the formula [(length x width2)x0.52] (46). For treatments with afatinib, erlotinib or gefitinib (L.C. Labs) we suspended respective inhibitors in vehicle, i.e. a physiological NaCl solution containing 0.5 (w/v) % methylcelluose 0.4 (v/v) % Tween80. Animals were treated 5 times per week via oral gavage using 5 mg inhibitor per kg body weight, unless otherwise stated. For the patient derived xenograft (PDX) model experiments female athymic nude mice (6–8 weeks old; 15–20 g), purchased from Harlan Laboratories were used. Frozen PDX tissue was thawed and cut into 3 × 3 × 3 mm pieces. Tumor pieces were incubated in Matrigel for 10 minutes, and each piece was subcutaneously inserted into the flank of each mouse. Tumor volume was measured using a caliper and calculated using the formula [(length x width2)/2]. Once the tumors reached 200mm3 animals were randomized in 4 treatment groups. The compounds used in this study were afatinib at 15mg/kg QD for 30 days and paclitaxel (Teva, Spain) at 15mg/kg once a week for 4 weeks, doses considered as MTD in this mouse strain. Animals were maintained at the Animal Facility (awarded with the AAALAC accreditation) of the Spanish National Cancer Research Centre (CNIO).
Histology
Lung tissue samples were fixed in 2% formaldehyde overnight and following dehydration in 100% ethanol embedded in paraffin. 5 μm thick sections were stained by hematoxylin and eosin (H&E). Slides were scanned and tumor area, tumor number and tumor grade were determined in a blinded manner, using TissueFaxs and HistoQuest (TissueGnostics) software, respectively. Tumor grades were classified by board certified pathologists HP, LB, KD and JM.(30) For immunohistochemistry (IHC) and immunofluorescence (IF), 5 μm sections were subjected to stainings using standard protocols and antibodies against: EGFR (1:300, BD Biosciences #610016), Ki-67 (1:400, Cell Signaling#12202), phospho-Erk1/2 (Thr202/Tyr204; 1:400, Cell Signaling#4370), Cleaved Caspase 3 (Asp175; 1:200, Cell Signaling#9661) and phospho-Akt (Ser473; 1:100, Cell Signaling#3787). For analysis, at least 5 different high power field sections per group were compared, using HistoQuest for IHC and TissueQuest (TissueGnostics) for IF, respectively, as previously described(20).
Isolation of primary pneumocytes and RNA sequencing
Isolation of primary pneumocytes was described previously(20). Briefly, lungs were minced and proteolytically digested with Dispase II (Corning) and 0.01 % DNase I (Sigma-Aldrich). After negative selection of immune cells via CD16/CD32 (BD Bioscience) coated dishes, remaining pneumocytes were cultured in F12 media (Gibco) supplemented with 2 % FBS, ITS supplements (Sigma-Aldrich), 0.8 mM CaCl2, 15 mM HEPES, 0.25 (w/v) % BSA and antibiotics. Purity of cell populations was verified by immunocytochemistry using antibodies against CC-10 and SPC (Santa Cruz). 72 hours post isolation cells were transduced with rAd.Cre at a MOI of 250. For the next three days, media was replaced daily wish fresh media, and cells were harvested 5 days following adenoviral transduction in RLT lysis buffer (Qiagen). Total RNA was isolated using the RNeasy Kit (Qiagen) and analyzed using Illumina HiSeq sequencing. To test for efficient K-rasG12D recombination and Egfr deletion, we harvested cells K and KE cells two days post transduction for DNA and protein extraction and subsequent analysis.
Cell lines
K-RAS mutated cell lines A549, A427 and SK-LU1 were obtained from ATTC and grown in RPMI (Gibco) supplemented with 10 % FBS, 2 mM glutamine (Gibco) and penicillin/streptomycin (Gibco). 368T1 cells were a kind gift of Dr. Tyler Jacks (45). Pulm24 cells were derived from the PDX model and harbor a K-RAS G12C mutation. KP and KPE cell lines were isolated in house from respective mice by enzymatic digestion of lungs harvested following survival analysis for genotyping or 6 weeks after Ad.Cre mediated tumor induction for re-transplantation and K-ras activity assay. (45) To determine the density of viable cells we used a CASY counter (Roche).
CRISPR/Cas9 based gene knockout
sgRNAs were designed and cloned into the Cas9 expressing vector pSpCas9(BB)-2A-Puro (PX459) V2.0 (Addgene) as previously described (47). Cells were transfected with respective plasmids using Lipofectamine LTX (Thermo Fisher Scientific) and selected using 2.5 μg/ml puromycin for three days, and subsequently single cells were isolated by limiting dilution. After clonal expansion, DNA was retrieved and the region spanning the targeted sequence was amplified, sequenced and analyzed using TIDE software (Netherlands Cancer institute) (48). Three different successfully targeted clones were used for experiments, and untargeted clones which underwent the same procedure were used as controls. Sequences of sgRNA used were GCGACCCTCCGGGACGGCCG for EGFR and GCTTGTAGATGGCCATGGCG for p53.
RNA extraction and real-time quantitative PCR
RNA of cells and tissue was purified using E.Z.N.A. total RNA kit (Omega Biotek) and reverse transcribed using RevertAid H Minus Reverse Transcriptase (Fermentas). Tissue was homogenized using RNA homogenizer mini columns (Omega Biotek) prior to RNA isolation. qPCR was performed using specific primers (Supplementary Table 3), iTaq Universal SYBR Green reagents (Biorad) and a Biorad CFX Connect Real Time PCR system. Relative gene expression ratios were determined according to the Pfaffl method, using ACTB as a housekeeping gene. In the A549Δp53 ortotopic xenografts, Bestkeeper values were calculated using human 28S and human TATA box binding protein (TBP), as well as mouse Actb and 28S as reference genes. (49)
Immunoblotting and RAS activity assay
Cells were harvested using a lysis buffer containing 20 mm Tris, 100 mm NaCl, 1 mm Na3VO4, 100 mm NaF, 20 mm glycerol 2-phosphate, 2.5 mm EDTA, 1 mm EGTA, 1% Nonidet P-40, and 1 mm PMSF, and Complete Protease Inhibitor Tablets (Roche). Antibody array slides were incubated with equal amounts of total protein according to the manufacturer’s protocols (Cell Signaling #12622). For Western blot analysis, cell lysates were loaded on 4–15% polyacrylamide gels (Bio-Rad), separated using an SDS-containing running buffer, and transferred to nitrocellulose membranes (GE Healthcare) by semi-dry blotting. After blocking, membranes were probed with primary antibodies against phospho-EGFR (Y1068, Cell Signaling #3777), EGFR (Santa Cruz sc1005), phospho-ERBB2 (Y1221/1222, Cell Signaling #2243), ERBB2 (Cell Signaling #4290), phospho-ERBB3 (Y1289, Cell Signaling #2842), ERBB3 (Cell Signaling #12708), p53 (Santa Cruz sc6243), phospho-ERK1/2 (T202/Y204, Cell Signaling #4376), ERK1/2 (Cell Signaling #4695), phospho-MEK1/2 (S217/221, Cell Signaling #9121), MEK1/2 (Cell Signaling #8727), phospho-AKT (S473, Cell Signaling #9271) and AKT (Cell Signaling #9272). HSC70 (Santa Cruz, sc7298) was used as a loading control.
To determine levels of activated RAS isoforms, the Active Ras Detection Kit (Cell Signaling, #8821) was used according to the manufacturer’s instructions. Subsequently, input and pulldown were subjected for Western blot analysis using the provided anti-RAS antibody and antibodies specifically against G12D mutated K-RAS (Cell Signaling #14429), K-RAS (Invitrogen, PA544339), H-RAS (Santa Cruz, sc34) and N-RAS (Santa Cruz, sc31).
Statistical Analysis
Graph Pad Prism 5.0 was used for statistical analysis. All values are given as means ± standard deviation (s.d.), as indicated in figure legends. Comparisons between two groups were made by Student’s t-test, except for Kaplan Meier analysis, were we used a log-rank test. For comparison of more than two groups we used one way analysis of variance (ANOVA) with subsequent Tukey’s multiple comparison test. We did not use any statistical method to predetermine sample size in animal studies.
Supplementary Material
One Sentence Summary.
K-RAS mutated lung adenocarcinomas depend on upstream ERBB signaling and hence pan-ERBB tyrosine kinase inhibitors, but not sole EGFR inhibitors, impair K-RAS driven lung tumorigenesis.
Acknowledgments
We thank Dr. T. Jacks for providing us the 368T1 cell line, S. Zahma for help with the preparation of tissue sections and the Core Facility Genomics of the Medical University of Vienna for RNA-seq analysis.
Funding
This project was supported by the Austrian Science Fund (FWF-P 25599-B19 to EC). HPM was supported by the Fellinger Krebsforschungsverein. RM was supported by the Austrian Science Fund (FWF), grants SFB-F4707 and SFB-F06105. BG was supported by the NVKP_16-1-2016-0037 grant of the National Research, Development and Innovation Office, Hungary. KD is the recipient of the Bolyai fellowship of the Hungarian Academy of Sciences and received support from the TÁMOP 4.2.4. A/1-11-1-2012-0001 ‘National Excellence Program’. BD acknowledges support from the Hungarian NRDI Office (K109626, K108465, KNN121510 and SNN114490).
Footnotes
Author contributions
HP Moll designed and performed experiments, performed GSEA analysis, interpreted the data and wrote the manuscript. K Pranz, B Grabner and P Aigner conducted experiments, M Musteanu and PP Lopez-Casas conducted the PDX experiment, P Stiedl and D Schramek were involved in in vivo experiments, N Hruschka, J Mohrherr, L Brcic and V Laszlo performed immunohistochemistry, L Brcic, H Popper and J Moldvay prepared the TMAs and helped with the pathology, K Dezso helped with pathology, B Győrffy gave bioinformatic support, M Hidalgo provided material, R Moriggl, R Eferl, D Stoiber, J Penninger, M Sibilia, M Barbacid, B Dome provided critical support and corrected the manuscript, E Casanova designed the study, interpreted the data and wrote the manuscript.
Competing interests
The authors declare no potential conflicts of interest.
Data and materials availability
All materials will be made available to the scientific community. RNAseq data and description of experimental design are deposited under GEO number _______.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA: a cancer journal for clinicians. 2016;66:7–30. doi: 10.3322/caac.21332. [DOI] [PubMed] [Google Scholar]
- 2.T. C. G. A. Consortium. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–550. doi: 10.1038/nature13385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Swanton C, Govindan R. Clinical Implications of Genomic Discoveries in Lung Cancer. The New England journal of medicine. 2016;374:1864–1873. doi: 10.1056/NEJMra1504688. [DOI] [PubMed] [Google Scholar]
- 4.Kobayashi S, Boggon TJ, Dayaram T, Janne PA, Kocher O, Meyerson M, Johnson BE, Eck MJ, Tenen DG, Halmos B. EGFR mutation and resistance of non-small-cell lung cancer to gefitinib. The New England journal of medicine. 2005;352:786–792. doi: 10.1056/NEJMoa044238. [DOI] [PubMed] [Google Scholar]
- 5.Pao W, Miller V, Zakowski M, Doherty J, Politi K, Sarkaria I, Singh B, Heelan R, Rusch V, Fulton L, Mardis E, et al. EGF receptor gene mutations are common in lung cancers from “never smokers” and are associated with sensitivity of tumors to gefitinib and erlotinib. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13306–13311. doi: 10.1073/pnas.0405220101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yun CH, Mengwasser KE, Toms AV, Woo MS, Greulich H, Wong KK, Meyerson M, Eck MJ. The T790M mutation in EGFR kinase causes drug resistance by increasing the affinity for ATP. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:2070–2075. doi: 10.1073/pnas.0709662105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu YL, Zhou C, Hu CP, Feng J, Lu S, Huang Y, Li W, Hou M, Shi JH, Lee KY, Xu CR, et al. Afatinib versus cisplatin plus gemcitabine for first-line treatment of Asian patients with advanced non-small-cell lung cancer harbouring EGFR mutations (LUX-Lung 6): an open-label, randomised phase 3 trial. The Lancet Oncology. 2014;15:213–222. doi: 10.1016/S1470-2045(13)70604-1. [DOI] [PubMed] [Google Scholar]
- 8.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nature reviews Drug discovery. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gysin S, Salt M, Young A, McCormick F. Therapeutic strategies for targeting ras proteins. Genes & cancer. 2011;2:359–372. doi: 10.1177/1947601911412376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linardou H, Dahabreh IJ, Kanaloupiti D, Siannis F, Bafaloukos D, Kosmidis P, Papadimitriou CA, Murray S. Assessment of somatic k-RAS mutations as a mechanism associated with resistance to EGFR-targeted agents: a systematic review and meta-analysis of studies in advanced non-small-cell lung cancer and metastatic colorectal cancer. The Lancet Oncology. 2008;9:962–972. doi: 10.1016/S1470-2045(08)70206-7. [DOI] [PubMed] [Google Scholar]
- 11.Mao C, Qiu LX, Liao RY, Du FB, Ding H, Yang WC, Li J, Chen Q. KRAS mutations and resistance to EGFR-TKIs treatment in patients with non-small cell lung cancer: a meta-analysis of 22 studies. Lung cancer (Amsterdam, Netherlands) 2010;69:272–278. doi: 10.1016/j.lungcan.2009.11.020. [DOI] [PubMed] [Google Scholar]
- 12.Rulli E, Marabese M, Torri V, Farina G, Veronese S, Bettini A, Longo F, Moscetti L, Ganzinelli M, Lauricella C, Copreni E, et al. Value of KRAS as prognostic or predictive marker in NSCLC: results from the TAILOR trial. Annals of oncology: official journal of the European Society for Medical Oncology. 2015;26:2079–2084. doi: 10.1093/annonc/mdv318. [DOI] [PubMed] [Google Scholar]
- 13.Papadimitrakopoulou V, Lee JJ, Wistuba II, Tsao AS, Fossella FV, Kalhor N, Gupta S, Byers LA, Izzo JG, Gettinger SN, Goldberg SB, et al. The BATTLE-2 Study: A Biomarker-Integrated Targeted Therapy Study in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2016 doi: 10.1200/JCO.2015.66.0084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science (New York, NY) 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, Chen Y, Kucharski JM, Feng J, Ely T, Chen JH, et al. Selective Inhibition of Oncogenic KRAS Output with Small Molecules Targeting the Inactive State. Cancer discovery. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 16.Girard L, Rodriguez-Canales J, Behrens C, Thompson DM, Botros IW, Tang H, Xie Y, Rekhtman N, Travis WD, Wistuba II, Minna JD, et al. An Expression Signature as an Aid to the Histologic Classification of Non-Small Cell Lung Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research. 2016;22:4880–4889. doi: 10.1158/1078-0432.CCR-15-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proceedings of the National Academy of Sciences of the United States of America. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han W, Zhang T, Yu H, Foulke JG, Tang CK. Hypophosphorylation of residue Y1045 leads to defective downregulation of EGFRvIII. Cancer biology & therapy. 2006;5:1361–1368. doi: 10.4161/cbt.5.10.3226. [DOI] [PubMed] [Google Scholar]
- 19.Jackson EL, Willis N, Mercer K, Bronson RT, Crowley D, Montoya R, Jacks T, Tuveson DA. Analysis of lung tumor initiation and progression using conditional expression of oncogenic K-ras. Genes & development. 2001;15:3243–3248. doi: 10.1101/gad.943001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grabner B, Schramek D, Mueller KM, Moll HP, Svinka J, Hoffmann T, Bauer E, Blaas L, Hruschka N, Zboray K, Stiedl P, et al. Disruption of STAT3 signalling promotes KRAS-induced lung tumorigenesis. Nature communications. 2015;6:6285. doi: 10.1038/ncomms7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Natarajan A, Wagner B, Sibilia M. The EGF receptor is required for efficient liver regeneration. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:17081–17086. doi: 10.1073/pnas.0704126104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jonkers J, Meuwissen R, van der Gulden H, Peterse H, van der Valk M, Berns A. Synergistic tumor suppressor activity of BRCA2 and p53 in a conditional mouse model for breast cancer. Nature genetics. 2001;29:418–425. doi: 10.1038/ng747. [DOI] [PubMed] [Google Scholar]
- 23.Jackson EL, Olive KP, Tuveson DA, Bronson R, Crowley D, Brown M, Jacks T. The differential effects of mutant p53 alleles on advanced murine lung cancer. Cancer research. 2005;65:10280–10288. doi: 10.1158/0008-5472.CAN-05-2193. [DOI] [PubMed] [Google Scholar]
- 24.Choi H, Sheng J, Gao D, Li F, Durrans A, Ryu S, Lee SB, Narula N, Rafii S, Elemento O, Altorki NK, et al. Transcriptome analysis of individual stromal cell populations identifies stroma-tumor crosstalk in mouse lung cancer model. Cell reports. 2015;10:1187–1201. doi: 10.1016/j.celrep.2015.01.040. [DOI] [PubMed] [Google Scholar]
- 25.Kamata T, Giblett S, Pritchard C. KRAS(G12D) expression in lung-resident myeloid cells promotes pulmonary LCH-like neoplasm sensitive to statin treatment. Blood. 2017;130:514–526. doi: 10.1182/blood-2017-02-770149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu X, Rock JR, Lu Y, Futtner C, Schwab B, Guinney J, Hogan BL, Onaitis MW. Evidence for type II cells as cells of origin of K-Ras-induced distal lung adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:4910–4915. doi: 10.1073/pnas.1112499109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Desai TJ, Brownfield DG, Krasnow MA. Alveolar progenitor and stem cells in lung development, renewal and cancer. Nature. 2014;507:190–194. doi: 10.1038/nature12930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mainardi S, Mijimolle N, Francoz S, Vicente-Duenas C, Sanchez-Garcia I, Barbacid M. Identification of cancer initiating cells in K-Ras driven lung adenocarcinoma. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:255–260. doi: 10.1073/pnas.1320383110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pao W, Miller VA, Politi KA, Riely GJ, Somwar R, Zakowski MF, Kris MG, Varmus H. Acquired resistance of lung adenocarcinomas to gefitinib or erlotinib is associated with a second mutation in the EGFR kinase domain. PLoS medicine. 2005;2:e73. doi: 10.1371/journal.pmed.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.DuPage M, Dooley AL, Jacks T. Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nature protocols. 2009;4:1064–1072. doi: 10.1038/nprot.2009.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pao W, Wang TY, Riely GJ, Miller VA, Pan Q, Ladanyi M, Zakowski MF, Heelan RT, Kris MG, Varmus HE. KRAS mutations and primary resistance of lung adenocarcinomas to gefitinib or erlotinib. PLoS medicine. 2005;2:e17. doi: 10.1371/journal.pmed.0020017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brugger W, Triller N, Blasinska-Morawiec M, Curescu S, Sakalauskas R, Manikhas GM, Mazieres J, Whittom R, Ward C, Mayne K, Trunzer K, et al. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo-controlled study of erlotinib maintenance therapy in advanced non-small-cell lung cancer. Journal of clinical oncology : official journal of the American Society of Clinical Oncology. 2011;29:4113–4120. doi: 10.1200/JCO.2010.31.8162. [DOI] [PubMed] [Google Scholar]
- 33.Ardito CM, Gruner BM, Takeuchi KK, Lubeseder-Martellato C, Teichmann N, Mazur PK, Delgiorno KE, Carpenter ES, Halbrook CJ, Hall JC, Pal D, Briel T, et al. EGF receptor is required for KRAS-induced pancreatic tumorigenesis. Cancer cell. 2012;22:304–317. doi: 10.1016/j.ccr.2012.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navas C, Hernandez-Porras I, Schuhmacher AJ, Sibilia M, Guerra C, Barbacid M. EGF receptor signaling is essential for k-ras oncogene-driven pancreatic ductal adenocarcinoma. Cancer cell. 2012;22:318–330. doi: 10.1016/j.ccr.2012.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kunii R, Jiang S, Hasegawa G, Yamamoto T, Umezu H, Watanabe T, Tsuchida M, Hashimoto T, Hamakubo T, Kodama T, Sasai K, et al. The predominant expression of hepatocyte nuclear factor 4alpha (HNF4 alpha) in thyroid transcription factor-1 (TTF-1)-negative pulmonary adenocarcinoma. Histopathology. 2011;58:467–476. doi: 10.1111/j.1365-2559.2011.03764.x. [DOI] [PubMed] [Google Scholar]
- 36.Ewald JA, Wilkinson JC, Guyer CA, Staros JV. Ligand- and kinase activity-independent cell survival mediated by the epidermal growth factor receptor expressed in 32D cells. Experimental cell research. 2003;282:121–131. doi: 10.1016/s0014-4827(02)00014-9. [DOI] [PubMed] [Google Scholar]
- 37.Weihua Z, Tsan R, Huang WC, Wu Q, Chiu CH, Fidler IJ, Hung MC. Survival of cancer cells is maintained by EGFR independent of its kinase activity. Cancer cell. 2008;13:385–393. doi: 10.1016/j.ccr.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- 39.Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes & development. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- 40.Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science (New York, NY) 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 41.Jacobsen HJ, Poulsen TT, Dahlman A, Kjaer I, Koefoed K, Sen JW, Weilguny D, Bjerregaard B, Andersen CR, Horak ID, Pedersen MW, et al. Pan-HER, an Antibody Mixture Simultaneously Targeting EGFR, HER2, and HER3, Effectively Overcomes Tumor Heterogeneity and Plasticity. Clinical cancer research : an official journal of the American Association for Cancer Research. 2015;21:4110–4122. doi: 10.1158/1078-0432.CCR-14-3312. [DOI] [PubMed] [Google Scholar]
- 42.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, Cottino F, Prahallad A, Grernrum W, Tzani A, Schlicker A, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. doi: 10.1016/j.celrep.2014.02.045. [DOI] [PubMed] [Google Scholar]
- 43.Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, Houstis N, et al. PGC-1 alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 44.Babicki S, Arndt D, Marcu A, Liang Y, Grant JR, Maciejewski A, Wishart DS. Heatmapper: web-enabled heat mapping for all. Nucleic acids research. 2016;44:W147–153. doi: 10.1093/nar/gkw419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Winslow MM, Dayton TL, Verhaak RG, Kim-Kiselak C, Snyder EL, Feldser DM, Hubbard DD, Du Page MJ, Whittaker CA, Hoersch S, Yoon S, Crowley D, et al. Suppression of lung adenocarcinoma progression by Nkx2-1. Nature. 2011;473:101–104. doi: 10.1038/nature09881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Euhus DM, Hudd C, La Regina MC, Johnson FE. Tumor measurement in the nude mouse. Journal of surgical oncology. 1986;31:229–234. doi: 10.1002/jso.2930310402. [DOI] [PubMed] [Google Scholar]
- 47.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nature protocols. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brinkman EK, Chen T, Amendola M, van Steensel B. Easy quantitative assessment of genome editing by sequence trace decomposition. Nucleic acids research. 2014;42:e168. doi: 10.1093/nar/gku936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pfaffl MW, Tichopad A, Prgomet C, Neuvians TP. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper--Excel-based tool using pair-wise correlations. Biotechnology letters. 2004;26:509–515. doi: 10.1023/b:bile.0000019559.84305.47. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All materials will be made available to the scientific community. RNAseq data and description of experimental design are deposited under GEO number _______.