Abstract
Antigen-inexperienced memory-like T (AIMT) cells are functionally unique T cells representing one of the two largest subsets of murine CD8+ T cells. However, differences between laboratory inbred strains, insufficient data from germ-free mice, a complete lack of data from feral mice, and unclear relationship between AIMT cells formation during aging represent major barriers for better understanding of their biology. We performed a thorough characterization of AIMT cells from mice of different genetic background, age, and hygienic status by flow cytometry and multi-omics approaches including analyses of gene expression, TCR repertoire, and microbial colonization. Our data showed that AIMT cells are steadily present in mice independently of their genetic background and hygienic status. Despite differences in their gene expression profiles, young and aged AIMT cells originate from identical clones. We identified that CD122 discriminates two major subsets of AIMT cells in a strain-independent manner. Whereas thymic CD122LOW AIMT cells (innate memory) prevail only in young animals with high thymic IL-4 production, peripheral CD122HIGH AIMT cells (virtual memory) dominate in aged mice. Co-housing with feral mice changed the bacterial colonization of laboratory strains, but had only minimal effects on the CD8+ T-cell compartment including AIMT cells.
Keywords: antigen-inexperienced memory-like T cells, AIMT, virtual memory, innate memory, T cells, aging, germ-free, feral mouse, CD8, CD122, co-housing
Introduction
AIMT cells are divided into two subsets based on the site of their origin. Innate T cells originate in the thymus, whereas virtual memory T cells are formed in the peripheral lymphoid organs [1, 2]. In BALB/c mice and several gene knock-outs on C57BL/6 background, innate memory cells are formed in the thymus via IL-4 produced by invariant natural killer T (iNKT) cells [3–5]. Wild-type C57BL/6 mice have low numbers of IL-4-producing iNKT cells in the thymus and form virtual memory T cells in a process dependent on IL-15 presentation by peripheral CD8α dendritic cells [2, 5–7]. Recently, it has been observed that mature CD8+ single positive thymic precursors of virtual memory T cells upregulate Eomes [8], a transcription factor required for their existence [7], suggesting that the differentiation process of virtual memory cells might be initiated already in the thymus and accomplished in the periphery [9]. This scenario is consistent with the observation that thymocytes developing in a neonatal thymus are more predisposed towards AIMT cell formation than thymocytes developing in an adult thymus of C57BL/6 mice [10].
In addition to these major routes, lymphopenia induces homeostatic proliferation coupled with the differentiation of peripheral naïve CD8+ T cells into AIMT cells, probably via IL-7 and/or IL-15 in C57BL/6 mice [11, 12]. Moreover, excess of IL-4, IL-7, or IL-15 induces the formation and/or expansion of AIMT cells [6, 13, 14]. Given the fact that these three cytokines signal through the common γ-chain [15], it is not surprising that all of them induce AIMT cells formation. Although it has been proposed that some genes such as Tbx21/T-bet or Klrk1/NKG2D are differentially expressed in thymic innate memory and virtual memory AIMT cells [2], a direct comparison of the gene expression between virtual memory and innate memory T cells has not been carried out.
Despite their abundance and functional uniqueness, AIMT cells are still understudied. The potential reasons for the underrepresentation of AIMT cells in the immunological research could be the confusing nomenclature [1] and unclear developmental and functional relationship between different types of AIMT cells. Moreover, only a minority of AIMT cell-related studies used germ-free mice [8, 16, 17], which might obscure the interpretation of the observations. It is unclear if the very existence of AIMT cells is physiological, because AIMT cells have been characterized exclusively in inbred mouse strains housed in clean conditions and because it has been proposed that the CD8+ T-cell compartment in laboratory mice substantially differs from feral mice [18].
Here, we show in side-by-side controlled experiments for the first time that the routes of AIMT cells formation are conserved in mice with different genetic backgrounds and hygienic conditions, including germ-free and feral mice.
Methods
Antibody
The antibodies that were used for flow cytometry were purchased from BD Pharmingen: CD4 PE-Cy7 (RM4-5) Cat# 552775, CD4 APC-Cy7 (GK1.5) Cat# 552051, CD4 Alexa Fluor 700 (RM4-5) Cat# 557956, CD4 FITC (H129.19) Cat# 553651, CD8α Pacific Blue (53-6.7) Cat# 558106, CD49d PE (R1-2) Cat# 553157, Ki67 Alexa Fluor 647 (B56) Cat# 558615, KLRG1 APC (2F1/KLRG1) Cat# 553157, Ly6c FITC (MR5-2) Cat# 553186, TCR Vα2 PE (B20.1) Cat# 553289, TCR Vα8.3 FITC (B21.14) Cat# 553376, TCR Vβ8.1/8.2 FITC (MR5-2) Cat# 553185, TCRβ APC (H57-597) Cat# 553174, TCRβ PE (H57-597) Cat# 553171, Biolegend: CD4 Brilliant Violet 650 (RM4-5) Cat# 100545, CD5 Brilliant Violet 510 (53-7-3) Cat# 100627, CD8α Brilliant Violet 421 (53-6.7) Cat# 558106, CD8α Brilliant Violet 510 (53-6.7) Cat# 558106, CD24 FITC (M1/69) Cat# 101806, CD44 Brilliant Violet 650 (IM7) Cat# 103049, CD44 PerCP-Cy5.5 (IM7) Cat# 103032, CD49d PE-Cy7 (R1-2) Cat# 103618, CD62L Alexa Fluor 700 (MEL14) Cat# 104426, CD122 FITC (TM-β1) Cat# 123207, CD127 PE-Cy7 (A7R34) Cat# 135015, CD127 PE (A7R34) Cat# L34975, CD218a Alexa Fluor 647 (BG/IL18RA) Cat# 132903, CX3CR1 PE-Cy7 (SA011F11) Cat# 149016, IFNγ Alexa Fluor 700 (XMG1.2) Cat# 505824, TCR Vα8.3 PE (B21.14) Cat# 127708, TCR γ/δ PerCP-Cy5.5 (GL3) Cat# 118117, or eBioscience: CD122 PerCP-eFluor 710 (TM-β1) Cat# 46-1222-80, EOMES Alexa Fluor 488 (Dan11mag) Cat# 53-4875-80.
Laboratory mice
Common laboratory mouse strains C57BL/6J and BALB/c and transgenic mouse strains Vβ5 [19] and C.Cg-Tg(DO11.10)10Dlo/J (henceforth DO11.10) [20] were used in this study.
Mice were bred in our facilities (SPF mice: Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic and Department of Biomedicine, University Hospital Basel, Basel, Switzerland; germ-free mice: Laboratory of Gnotobiology, Institute of Microbiology of the Czech Academy of Sciences, Novy Hradek, Czech Republic) in accordance with laws of the Czech Republic and Switzerland, respectively. Animal protocols were approved by the Cantonal Veterinary Office of Baselstadt, Switzerland, by the Czech Academy of Sciences, the Czech Republic and by the respective Committees for the Protection and Use of Experimental Animals of the Institute of Microbiology (ID 5/2019) and of the Institute of Molecular Genetics (ID 8/2017, 15/2019) of the Czech Academy of Sciences.
Mice were kept in the animal facility with 12 hours of light and dark cycle with food and water ad libitum. Males and females were used for experiments. Age- and sex-matched pairs of animals were used in the experimental groups. If possible, littermates were equally divided into the experimental groups.
Flow cytometry and cell counting
For the surface staining, cells were incubated with diluted antibodies in PBS/2% fetal calf serum on ice. LIVE/DEAD near-IR dye (Life Technologies, L34975) was used for discrimination of live and dead cells.
For the staining of feral mice, co-housed mice and control animals, cells were fixed with 2% formaldehyde at room temperature for 3 minutes prior to staining. All monoclonal antibodies used for this staining were tested for their compatibility with the fixation procedure.
For the intracellular staining, cells were fixed and permeabilized using Foxp3/Transcription Factor Staining Buffer Set (eBioscience, 00-5523-00). For some experiments, enrichment of CD8+ T cells was performed using magnetic bead separation kits EasySep (STEMCELL Technologies) or Dynabeads (Thermo Fisher Scientific) according to manufacturers’ instructions prior to the analysis or sorting by flow cytometry.
Cells were counted using Z2 Coulter Counter (Beckman) or using AccuCheck counting beads (Thermo Fisher Scientific) and a flow cytometer. Flow cytometry was carried out with a BD LSRII, BD Canto II or Cytek Aurora cytometers. Cell sorting was performed using Influx (BD Bioscience). Data were analyzed using FlowJo software (TreeStar).
Stimulation of T cells
400,000 cells/ml freshly isolated lymph node cells in IMDM/10% fetal calf serum supplemented with IL-2 were stimulated with PMA (10 ng/ml) and ionomycin (1 μM), or IL 12 (10 ng/ml) and IL 18 (10 ng/ml), or left unstimulated for eight hours. 5 μg/ml Brefeldin A (Biolegend, 420601) was added to the culture for the last four hours of the incubation. The cells were stained with LIVE/DEAD near-IR dye and fluorescently labeled antibodies to CD8α, CD44, and CD49d. After washing, the cells were fixed and permeabilized and labeled with anti-IFN-γ antibody and analyzed by flow cytometry.
Sublethal irradiation
C57BL/6J and BALB/c mice of the age 5-8 weeks were sublethally irradiated with 4 Gy on a T-200 X ray machine (Wolf-Medizintechnik). The mice were sacrificed at day 16 post irradiation and the lymphocytes from the peripheral lymph nodes and the spleen were stained for CD122, CD8α, viability, CD44, CD62L, CD49d, CD4, TCRβ, Ly6c, and CD5 and analyzed by flow cytometry. The control group consisted of non-irradiated littermates.
Adoptive transfer of cells into DO11.10 mice
Lymphocytes from peripheral lymph nodes and spleen from 8-12 weeks old BALB/c mice were harvested and stained with 5 μM Cell Trace Violet proliferation dye (Thermo Fisher, C34557) in PBS for 10 minutes at 37°C. 1×106 naïve CD8+ CD44- cells were sorted and i.v. injected into recipient DO11.10 mouse. The phenotype of the transferred cells was analyzed after 7 days.
Samples for transcriptomic analysis
10-12 weeks old BALB/c or C57BL/6J SPF mice were irradiated with 4 Gy and sacrificed 16 days post irradiation. Non-irradiated mice were SPF young (7-12 weeks) or aged (12-19 months) BALB/c or C57BL/6J mice. Lymphocytes from the peripheral lymph nodes and the spleen were stained with TCRβ, CD8, CD44, CD49d, CD122 and viability dye and sorted by Influx. Each sample consisted of a pool of lymphocytes originating from 2-5 mice.
RNA was isolated using Trizol LS (Thermo Fisher Scientific) followed by in-column DNAse digestion using RNA clean&concentrator kit (Zymo Research) according to manufacturers’ instructions.
Transcriptomic analysis
Samples were prepared with SMARTer® Stranded Total RNA-Seq – Pico Input Mammalian library preparation kit v2 (Takara). Protocol included cDNA synthesis from mRNA, ligation of sequencing adaptors and indexes, ribosomal cDNA depletion, final PCR amplification and product purification. Library size distribution was evaluated on the Agilent 2100 Bioanalyzer using the High Sensitivity DNA Kit (Agilent). Libraries were sequenced using an Illumina NextSeq 500 instrument using 76 bp single-end high-output configuration.
Read quality was assessed by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). For subsequent read processing, a bioinformatics pipeline nf-core/rnaseq version 1.1 (https://doi.org/10.5281/zenodo.1447331), was used. Individual steps included removing sequencing adaptors with Trim Galore! (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/), mapping to reference genomes GRCm38 or Balb_cJ_v1 [21] (Ensembl annotation version 94) respective to the specimen strain with HISAT2 [22] and quantifying expression on gene level with featureCounts [23]. Resulting per gene read counts for individual strains were merged together using gene orthologue map obtained from Ensembl. Merged counts served as input for differential expression analysis using DESeq2 R Bioconductor package [24]. Prior to the analysis, genes not expressed in at least two samples were discarded. Shrunken log2-fold changes using the adaptive shrinkage estimator [25] were used for differential expression analysis. Genes exhibiting minimal absolute log2-fold change value of 1 and statistical significance of adjusted p-value < 0.05 between compared groups of samples were considered as differentially expressed for subsequent interpretation and visualization.
To show sample expression relatedness with the generic effect of mouse strain erased, we employed method of removing unwanted source of variation by fitting linear model using limma R Bioconductor package [26] (function removeBatchEffect) after normalizing the data using variance stabilizing transformation.
Raw fastq files have been deposited in the ArrayExpress database at EMBL-EBI (www.ebi.ac.uk/arrayexpress) under accession number E-MTAB-9091.
Preparation of samples for TCR libraries
Lymphocytes from Vβ5 mice were sorted as viable CD8+ CD4- TCRVα2+ CD62L+ CD44- (naïve) or CD44+ (AIMT). RNA was isolated using Trizol (Thermo Fisher Scientific) followed by in-column DNAse treatment using RNA Clean&Concentrator kit (Zymo Research). cDNA synthesis was performed using a TCR specific primer (TTGGCACCCGAGAATTCCACTGHHHHHACAHHHHHACAHHHHHCACATTGATTTGGGAGTC) by using Maxima RT (Thermo scientific) according to manufacturer’s instructions. First round of PCR amplification was performed using primers CGTTCAGAGTTCTACAGTCCGACGATCHHHHACHHHHACHHHNGCAGCCCATGGTACCA GCAGTTCC and ACTGGAGTTCCTTGGCACCCGAGAATTC using HotStart Kappa HIFI ready mix (Kapa Biosystems) and the following conditions: 95°C for (5 min); 40 cycles: 98°C (20 s), 61°C (15 s), 72°C (15 s), and 72°C for (5 min). The resulting ~440 bp long PCR product was subjected to electrophoresis using an agarose gel and extracted using Gel Extraction Kit (Qiagen).
Following the first round of PCR, the TCRα copy number was quantified via qPCR (LightCycler 480 II, Roche) and compared to a dilution series of OT-I TCRα cloned in MSCV vector. 7.5×104 copies of the PCR product were then used for the second PCR amplification round using primers AATGATACGGCGACCACCGAGATCTACACGTTCAGAGTTCTACAGTCCGACGAT*C and CAAGCAGAAGACGGCATACGAGATCGTGATGTGACTGGAGTTCCTTGGCACCC*G. The polymerase was HotStart Kappa HIFI ready mix (2x) and the following conditions: 95 °C (5 min); 40 cycles: 98 °C (20 s), 62 °C for (15 s), 72°C for (15 s), and 72 °C for (5 min). The resulting ~500 bp long PCR product was subjected to electrophoresis in agarose gels and purified using Gel Extraction Kit (Qiagen). The samples were mixed in equimolar ratios to obtain 30 μl of a final concentration of 10-nM and sequenced by Illumina MiSeq (v3 kit, 300bp paired-end reads) at the Biocev, Charles University, Vestec, Czech Republic.
Analysis of TCR repertoires
FastQC was used to assess read quality. Next, MIGEC version 1.2.9 [27] was utilized for quantification of clonotypes and the resulting CDR3 sequence abundances in all samples were summarized to a clonotype abundance matrix. From this matrix, relative frequencies of CDR3 usage were computed. Clonotypes present in less than three samples were disregarded and the clonotype abundance matrix was treated similar to RNA-seq count matrix: DESeq2 was used to normalize for sequencing depth and RNA composition, and to test for differential clonotype abundance between AIMT and naïve sample groups. Normalized abundances were used for principal component analysis.
For comparison of the diversity of TCR repertoires (CDR3 clonotypes), Simpson Diversity Index was calculated using the function ‘diversity’ from package ‘vegan’ version 2.5-6 on R 4.0.2.
The sequence data has been deposited in the Sequence Read Archive and BioProject databases at NCBI (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA633838).
Feral mice
We analyzed feral mice Mus musculus musculus that were trapped in human houses or garden shelters in the region of Central Bohemia, Czech Republic. The mice used in this study were feral mice caught up to one month prior to the analysis and 8-24 months old F1 offspring of the feral mice born in captivity. These mice were housed at the Department of Zoology, Faculty of Science, Charles University in Prague. Mice were provided with food ad libitum and were kept under stable conditions in open cages (13:11 hours, D:N, 23 °C).
All animal procedures on feral mice were carried out in accordance with the law of the Czech Republic paragraph 17 no. 246/1992 and the local ethics committee of the Faculty of Science, Charles University in Prague, specifically approved this study by accreditation no. 13060/2014-MZE-17214.
Co-housing
Four SPF females were housed for eight weeks with one feral male per cage. The open co-housing cages were divided by a wire mesh with square openings (diameter was 1 cm), allowing contacts of co-housed animals. Co-housing cages were supplied with fresh bedding at the beginning of the experiment and provided with water and food ad libitum.
The age of SPF females was 4-6 weeks at the beginning of the co-housing experiment. The co-housed SPF females were matched with their littermates that remained in the SPF facility and had the same food and the same bedding as the co-housed animals. We performed two experiments with slightly modified setups. Experiment A: During the 6-week period of co-housing, the bedding was changed every 2 weeks and the feral males were transferred into another cage. Experiment B: During the 6-week period of co-housing, the bedding was not changed. Every 2 weeks, the mice were transferred to the other side of the same cage in order to be exposed to the feces of the feral mice.
Collection of the samples for the microbiome analysis
A reference sample of saliva was collected from each mouse participating in the co-housing experiment before the start of the experiment B. Throughout both A and B experiments, the saliva has been collected from co-housed animals prior to the co-housing and after every second week of co-housing by gentle flushing with a pipette using 50 μl of the 0.9% NaCl solution. At the end of the experiment, the co-housed animals as well as the non-co-housed controls have been sacrificed and the samples from duodenum, jejunum, ileum, cecum and colon were collected for high-throughput 16S rRNA sequencing of the intestinal microbiota. At the end of each experiment, two co-housed Vβ5 animals and one co-housed feral male have been sent for a health status examination: HM FELASA Annually + additional agents (SOPF). The results of this examination are provided as Appendix 1, which has been deposited on Zenodo, DOI: 10.5281/zenodo.4298697.
Microbiota analysis by 16S rRNA sequencing
Metagenomic DNA from gut and saliva samples was extracted using the PowerSoil DNA kit (MO BIO Laboratories Inc., USA). Barcoding was performed with previously published 16S rRNA primers Bact-0341-b-S-17 and S-D-Bact-0785-a-A-21 [28]. Dual-indexed Nextera sequencing libraries were prepared using a previously published two step PCR design [29]. The first PCR was performed in 10 μl and consisted of 1x KAPA HiFi Hot Start Ready Mix (Roche), each 16S rRNA primer at 0.2 μM and 3-μl DNA. PCR conditions were as follows: initial denaturation at 95°C for 3 min followed by 28 (for colon and caecum), 33 (for saliva) or 35 (ileum, duodenum, jejunum) cycles each of 95°C (30 s), 55°C (30 s), and 72°C (30 s), and a final extension at 72°C (5 min). Second PCR differed from the first PCR in the following parameters: it was performed in 20 μl volume; the concentration of each indexed primer was 1 μM; 1μl (gut samples) or 6 μl (saliva) of the first PCR product were used as a template; and the number of PCR cycles was 12. Technical duplicates were prepared to account for noise due to PCR and sequencing stochasticity. PCR products were pooled equimolarly, size-selected with Pippin Prep (Sage Science, USA) at 520 - 750 bp and sequenced using MiSeq (Illumina, USA) and v3 chemistry (i.e. 2 × 300 bp paired-end reads). Sequencing data were archived in European Nucleotide Archive under project accession PRJEB38301.
Samples were demultiplexed and primers were trimmed by skewer software [30]. Using dada2 [31], we filtered out low-quality sequences (expected number of errors per read > 1), denoised the quality-filtered fastq files and constructed abundance matrix representing reads counts for individual haplotypes (hereafter OTUs) in each sample. Using uchime [32] and gold.fna database we identified chimeric sequences and removed them from the abundance matrix. Taxonomic assignation of haplotypes was conducted by RDP classifier (80% confidence threshold, [33]) and Silva reference database (v 132, [34]).
Effect of cohousing on alpha diversity (based on Shannon diversity indices) was tested using linear mixed effect model, while considering individual identity as a random effect and the effect of treatment level (co-housed vs non-co-housed), gut section and their interactions as explanatory variables. The effect of cohousing on microbiota composition was assessed by permutation tests, where observed difference in composition (i.e. Bray-Curtis similarities) between co-housed lab vs. wild individuals and non-co-housed lab vs. wild individuals was compared with permuted average differences, where belonging to co-housed vs. non-co-housed lab group was randomized. OTUs whose relative abundances were affected by cohousing were identified by DESeq2 package [24].
Results
The frequency of AIMT cells increases in germ-free mice during aging
It has been shown that the percentage of AIMT cells increases upon aging [35–39]. However, these studies have used conventional specific pathogen free (SPF) mice and thus, the role of microbial antigens in this process cannot be excluded. Moreover, previous studies analyzed only C57BL/6 (henceforth B6) mice, but not BALB/c (henceforth Balb) mice that show substantial differences in the site of origin and cytokine dependency of AIMT cells [5, 40]. For this reason, we compared the CD8+ T-cell compartment in young and aged germ-free B6 and Balb mice. We observed that the frequency of peripheral CD44+ CD49d- AIMT cells increases in both strains upon aging (Fig. 1A-B and S1A-B), suggesting a common homeostatic mechanism of age-dependent induction of AIMT cells.
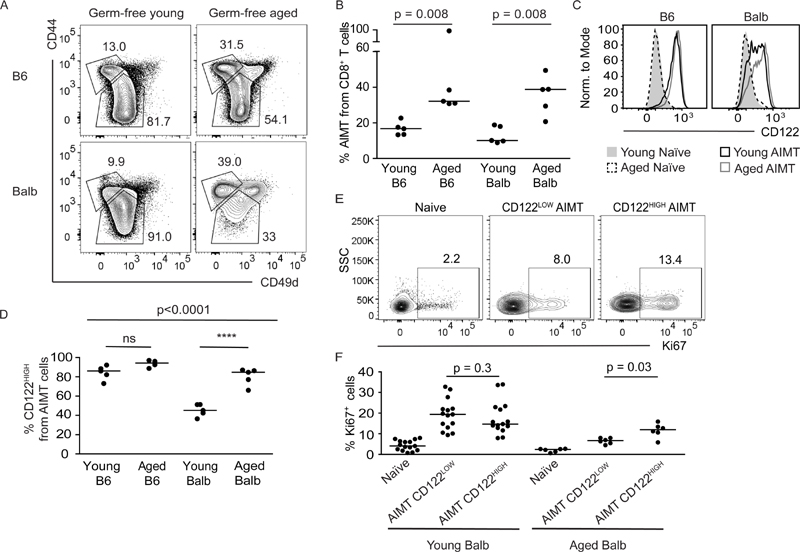
Figure 1. AIMT cells increase during aging in germ-free mice.
(A-D) T cells from spleens of germ-free young (5-12 weeks) or aged (55-75 weeks) mice were stained for viability, TCRβ, CD8α, CD44, CD49d, and CD122 and analyzed by flow cytometry.
(A) A representative staining of CD8+ T cells for CD44 and CD49d. Gates for naïve (CD44-) and AIMT (CD44+ CD49d-) cells are shown. A representative experiment out of 2 in total.
(B) Quantification of the percentage of CD44+ CD49d- AIMT cells among CD8+ T cells. n=5 mice from 2-5 independent experiments. Median is shown. The statistical significance was tested using 2-tailed Mann-Whitney test.
(C) Histograms of CD122 expression in CD44- naïve and CD44+ CD49d- AIMT CD8+ cells from indicated mice. A representative experiment out of 2 in total.
(D) Quantification of CD122HIGH cells among CD8+ AIMT cells. Median is shown. The statistical significance was tested using ANOVA (p < 0.0001) with Bonferroni’s multiple comparison posttests of indicated pairs. **** p ≤ 0.0001.
(E-F) Splenocytes from young and aged SPF Balb mice were fixed and stained for CD8α, CD44, CD49d, CD122, and Ki67.
(E) Ki67 expression in naïve, CD122HIGH and CD122LOW AIMT cells of aged mice. A representative mouse out of 6 in total.
(F) Percentage of Ki67+ cells is shown. n= 6 (aged) or 15 (naïve) mice in 6 or 3 independent experiments, respectively. The statistical significance was tested using 2-tailed Wilcoxon matched-pairs signed rank test. Median is shown.
Peripherally derived AIMT cells express high levels of CD122
AIMT cells from B6 depend on IL-15 and express high levels of CD122 (alias IL2RB), a subunit of the IL-15 receptor [5, 7, 16]. We observed that AIMT cells from young germ-free B6 mice express higher levels of CD122 than those from young Balb mice (Fig. 1C-D and S1C-D). Interestingly, a population of CD122HIGH AIMT cells appears in aged germ-free Balb mice, whereas no difference in CD122 expression was found between AIMT cells from germ-free young and aged B6 mice (Fig. 1C-D). We observed that CD122HIGH AIMT cells show slightly stronger homeostatic proliferation than CD122LOW AIMT cells in aged, but not in young Balb mice (Fig. 1E-F), which probably contributes to their accumulation in aged mice.
CD122HIGH AIMT cells in B6 are generated in the periphery via IL-15 and homeostatic TCR signaling [6, 17]. These cells are more frequent in aged mice, probably because the thymic output is low and the lymphopenic environment augments the AIMT cells formation. In contrast, CD122LOWAIMT cells are generated in the thymus of young Balb mice via IL-4 [40]. We hypothesized that during aging of Balb mice, the thymic regress impairs de novo formation of CD122LOW AIMT cells, leading to the lymphopenia-induced generation of proliferative CD122HIGH AIMT cells in the periphery. To elucidate whether CD122 expression could discriminate between thymic (innate memory) and peripheral AIMT cells (virtual memory) in Balb mice, we employed two models for lymphopenia-induced AIMT cells formation.
The first lymphopenic model was sublethal irradiation of mice. Irradiation reduces the number of T cells, which leads to homeostatic proliferation and AIMT formation of the surviving T cells in the periphery [1]. As expected, irradiation increased the frequency of AIMT cells in B6 and Balb mice (Fig. 2A-B). Irradiation-induced AIMT cells showed significantly higher CD122 expression than control AIMT cells in Balb, but not in B6, mice (Fig. 2C-D). This result is in close agreement with the hypothesis that CD122 expression is a marker of peripherally induced AIMT cells in both strains.
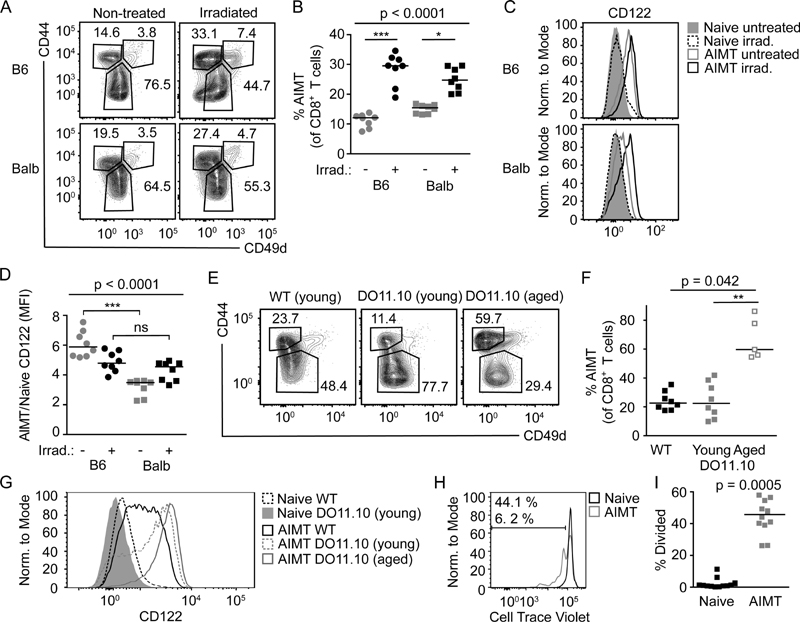
Figure 2. Lymphopenia induces CD122HIGH AIMT cells in Balb mice.
(A-D) 5-8 weeks old SPF B6 and Balb mice were sublethally irradiated or not. 16 days later, the mice were sacrificed and the splenocytes were stained for TCRβ, CD8α, CD44, CD49d, and CD122 and analyzed by flow cytometry. n=8 mice per group in 4 independent experiments.
(A) A representative experiment out of 4 in total.
(B) Quantification of the frequency of AIMT cells. Median is shown. The statistical significance was tested using Kruskall-Wallis test with Dunn’s multiple comparisons posttests. *** p < 0.001, * p < 0.05.
(C) Expression of CD122 in naïve and AIMT cells in untreated and irradiated B6 and Balb mice.
(D) Quantification of CD122 expression in AIMT cells normalized to the expression on naïve T cells. Median is shown. The statistical significance was tested using Kruskall-Wallis test with Dunn’s multiple comparisons posttests. ns p>0.05, *** p < 0.001.
(E-G) Splenocytes from young (5-14 weeks) Balb and young or aged (52-73 weeks) DO11.10 mice were stained for TCRβ, CD8α, CD44, CD49d, and CD122 and analyzed by flow cytometry. n=8 or 5 (aged DO11.10) mice in 4-5 independent experiments.
(E) Representative experiments showing CD44 and CD49d staining of CD8+ T cells.
(F) Quantification of the frequency of AIMT cells among CD8+ T cells. Median is shown. The statistical significance was tested using Kruskall-Wallis test with Dunn’s multiple comparisons posttests. ** p < 0.01
(G) Expression of CD122 in young Balb and young and aged DO11.10 naïve and AIMT CD8+ T cells.
(H-I) Cell Trace Violet-loaded naïve CD8α+ CD44- CD49dLOW T cells isolated from lymph nodes and spleen from Balb mice were adoptively transferred into DO11.10 mice. 7 days later, the lymph nodes of the recipient mice were analyzed by flow cytometry. Cell Trace Violet signal in cells that stayed naïve (CD44- CD49dLOW) and cells converted into AIMT (CD44+ CD49d-) is shown. n=12 host mice in 3 independent experiments.
(H) The Cell Trace Violet dye dilution in naïve (CD44-) and AIMT (CD44+ CD49d-) adoptively transferred cells (live TCRβ+ CD8+ TCRVβ8.1/8.2-) is shown. A representative experiment out of 3 in total.
(I) The percentage of adoptively transferred naïve or AIMT cells that underwent at least one cycle of division is shown. Median is shown. The statistical significance was tested using Wilcoxon matched-pairs signed rank test.
The second model was TCR transgenic DO11.10 Rag+ Balb mice. Most of T cells in these mice are monoclonal CD4+ T cells specific to I-Ad-OVA323-339 [41]. However, rare T cells expressing endogenous TCRα differentiate into CD8+ T cells. Thymic AIMT cells should be substantially reduced in these mice, because of the absence of IL-4-producing thymic iNKT cells that are crucial for the formation of thymic AIMT cells [40]. In addition, this mouse is lymphopenic in the CD8+ T-cell compartment, which should promote the formation of AIMT cells in the periphery.
DO11.10 mice showed comparable frequency of AIMT cells to polyclonal Balb mice. The frequency of AIMT CD8+ T cells during aging increased (Fig. 2E-F), indicating that the age-dependent increase in the AIMT-cell compartment does not depend on the presence of iNKT cells. The expression of CD122 was higher in AIMT cells from DO11.10 mice than in AIMT cells from polyclonal Balb mice (Fig. 2G), which is consistent with the hypothesis that CD122HIGH AIMT cells are formed in the periphery. When we transferred naïve polyclonal CD8+ T cells into the DO11.10 mice, we observed the formation of AIMT cells, which was coupled with their homeostatic proliferation (Fig. 2H-I).
Overall, these data indicate that CD122 expression differs between peripherally induced virtual memory AIMT cells (both in B6 and Balb mice) and thymus-derived innate memory AIMT cells of Balb mice.
The site of origin and aging fine-tune the gene expression profile of AIMT cells
The AIMT-cell compartment is heterogeneous. As mentioned above, AIMT cells are formed in different organs (thymus or peripheral lymphoid organs) and have distinct cytokine dependencies (IL-15 vs. IL-4) in different strains. Moreover, aging might change the gene expression programs of AIMT cells [36–38]. We addressed differences between particular AIMT-cell populations by high-throughput RNA sequencing using sorted naïve and AIMT cells from young B6 and Balb mice, irradiation-induced AIMT cells from B6 and Balb mice, AIMT cells from aged B6 mice, and CD122LOW and CD122HIGH AIMT cells from aged Balb mice (Fig. S1E-I).
As expected, the major source of the variability among these samples was the different genetic background of B6 and Balb mice (Fig. S2A). We normalized the gene expression data for the differences between the strains by fitting the data with a linear model using the strain identity as one of the explanatory variables (Fig. 3A). The expression of Il2rb (CD122) and Itga4 (CD49d) in the sorted populations corresponded to the sorting strategy (Fig. 3B) and the previous characterization of AIMT cells from both mouse strains (Fig. 1A-D).
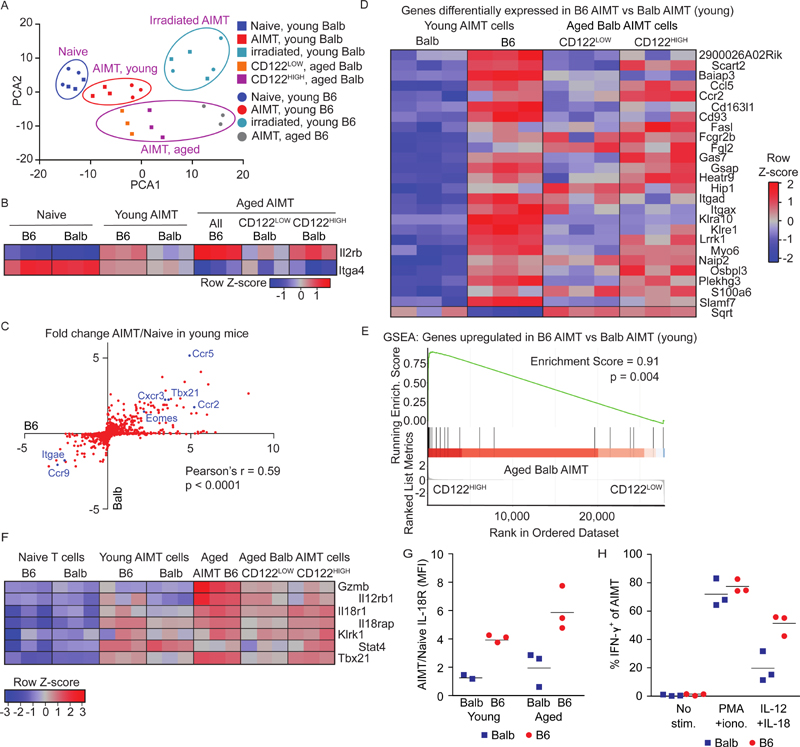
Figure 3. Similar gene expression profiles of thymic and peripheral AIMT cells.
(A-E) Naïve (CD44-) and/or AIMT (CD44+ CD49d-) CD8+ T cells were sorted from lymph nodes and spleens from young (5-12 weeks) untreated or young irradiated (4 Gy, 3 weeks) B6 and Balb or aged B6 mice (76 weeks). CD122HIGH and CD122LOW AIMT cells were sorted from lymph nodes and spleens from aged Balb mice (54-55 weeks). The transcriptome of these populations was analyzed by RNA sequencing.
(A) PCA analysis of the gene expression profiles of the individual samples normalized to avoid the strain-specific differences (top 500 variable genes).
(B) Relative expression (z-score) of Il2rb (CD122) and Itga4 (CD49d) in individual samples using normalization for strain-specific differences.
(C) Ratios of transcript levels in AIMT cells to naïve cells (young mice) were calculated for each gene and plotted as log2 for B6 and Balb strains. Selected genes are indicated. The correlation was calculated using Pearson’s correlation coefficient.
(D) Relative expression (z-score) of genes showing significantly different expression between AIMT cells from young B6 and Balb mice (after the normalization for strain-specific differences) in AIMT cells from B6 and Balb mice and in CD122HIGH and CD122LOW AIMT cells from aged Balb mice.
(E) The gene set enrichment analysis (GSEA) of genes with significantly higher expression in B6 than in Balb mice (shown in Fig. 3D) in CD122HIGH vs. CD122LOW AIMT cells from aged Balb mice.
(F) Relative expression (z-score) of selected genes involved in by-stander cytotoxicity between AIMT cells from young B6 and Balb mice (after the normalization for strain-specific differences) in AIMT cells from B6 and Balb mice and in CD122HIGH and CD122LOW AIMT cells from aged Balb mice.
(G) Expression of IL-18R on the surface of AIMT cells from the lymph nodes of young and aged Balb and B6 mice measured by flow cytometry. Geometric mean fluorescent intensity (MFI) was normalized to naïve cells in the same sample. Mean is shown. n = 3 independent experiments.
(H) Production of IFN-γ by AIMT cells (gated as CD8α+ CD44+ CD49d+) from young adult B6 or Balb mice was measured by flow cytometry. Lymph node cells were stimulated with PMA + ionomycin or with IL-12 and IL-18 or left unstimulated for 8 hours and analyzed by flow cytometry. n = 3 independent experiments.
We assumed that naïve and irradiation-induced AIMT cells represent two relatively uniform cell types present in both strains, and we used them as controls for evaluating our data normalization. The principal component analysis (PCA) of the adjusted data revealed that naïve cells and irradiation-induced AIMT cells from both strains clustered together, confirming the reliability of our adjusted data (Fig. 3A). We obtained very similar results when performing the PCA using data without strain-specific normalization, but with the exclusion of significantly differentially expressed genes between the strains (Fig. S2B).
The AIMT cells from aged mice showed moderate differences in their gene expression patterns in comparison to AIMT cells from young animals (Fig. 3A). We identified 36 genes upregulated in AIMT cells from aged mice vs. young mice in both strains (Fig. S2C). 24 of these transcripts were at least 4-fold enriched in AIMT cells from aged B6 mice in comparison to young mice in the previous study by Quinn et al. [36]. Interestingly, we did not identify a single gene significantly upregulated in AIMT cells from young mice vs. aged mice in both strains.
Young AIMT cells from both strains showed comparable global gene expression signature. The gene expression similarity between these populations was revealed as the proximity of young AIMT cells from both strains in the PCA plots (Fig. 3A, Fig. S2B) as well as a correlation of AIMT/naïve cell fold changes for individual genes plotted as B6 vs. Balb without using any correction for the strain-dependent differences (Fig. 3C). Despite the similarity of the gene expression patterns, we could identify one gene significantly upregulated, and 25 genes significantly downregulated in young Balb AIMT cells vs. B6 AIMT cells (Fig. 3D), suggesting that these genes might distinguish thymic innate memory and peripheral virtual memory subsets of AIMT cells.
We observed that the genes upregulated in AIMT cells from young B6 vs. young Balb mice are significantly enriched in aged CD122HIGH AIMT cells vs. aged CD122LOW AIMT cells from aged Balb mice (Fig. 3D-E), supporting the hypothesis that high CD122 levels predict the peripheral origin of AIMT cells irrespective of the strain.
It has been proposed that T-bet (Tbx21) and NKG2D (Klrk1) are expressed in peripheral, but not in thymic AIMT cells [2]. Moreover, a small group of genes potentially involved in by-stander cytotoxicity (Gzmb, Il12rb2, Il18r1, Il18rap, Stat4) were shown to be upregulated in peripheral AIMT cells [2, 6]. All of these genes, but Gzmb, were enriched in young AIMT cells over naïve T cells in our data (Fig. 3F). Four genes (Tbet, Klrk1, Il18r1, Il18rap) showed higher expression in AIMT cells of the proposed peripheral origin (AIMT cells from young and aged B6, and CD122HIGH AIMT cells from aged Balb mice) than in the corresponding AIMT cells of the proposed thymic origin (AIMT cells from young Balb and CD122LOW AIMT cells from aged Balb mice) (Fig. 3F).
Flow cytometry confirmed higher expression of IL-18R in AIMT cells from B6 than from Balb mice (Fig. 3G, Fig. S2D). Moreover, IL-18R was preferentially expressed in CD122HIGH AIMT cells (Fig. S2E). Accordingly, AIMT cells from B6 mice were more efficient in producing IFN-γ after stimulation with IL-12/IL-18 than AIMT cells from Balb mice, whereas the responses of these cells to PMA and ionomycine were comparable (Fig. 3H, Fig. S2F).
Overall, these data revealed a stable gene expression program in AIMT cells, which is fine-tuned by the age and genetic background. The most notable variation among AIMT cells is the differential expression of genes encoding proinflammatory receptors based on the site of their origin. Peripheral AIMT cells express higher levels of CD122 (a receptor for IL-2 and IL-15), IL-18 receptor, and activating NKG2D receptor than thymic AIMT cells. This expression pattern predisposes peripheral AIMT cells for efficient by-stander immune protection.
Young and aged AIMT cells represent identical TCR clonotypes
The frequency of murine AIMT cells changes in time. It peaks around 3 weeks of age [42] and it increases again as the mouse ages [35]. For this reason, it is unclear whether AIMT cells in young and aged animals have a different clonal origin [43]. To address this question, we analyzed TCR sequences of naïve and AIMT cells in young and aged mice. To overcome the problem with TCRα and TCRβ pairing, we analyzed TCRα chains in Vβ5 mouse that has a fixed TCRβ from the OT-I TCR.
Because central memory CD8+ CD44+ CD62L+ T cells in Vβ5 mice do not express CD49d (Fig. S3A), we gated AIMT cells in Vβ5 mice as CD44+ CD62L+ double positive. The frequency of AIMT cells in Vβ5 mice recapitulates WT mice, including the increase in the frequency of AIMT cells upon aging (Fig. S3B).
First, we quantified the usage of TRAV14 (corresponding to Vα2 epitope) and TRAV12 (corresponding to Vα8.3) by staining with specific antibodies. In line with our previous data [17], we observed a higher Vα2/Vα8.3 ratio in AIMT cells than in naïve T cells of young mice (Fig. 4A, S3C). Interestingly, the Vα2/Vα8.3 ratio in naïve cells was higher in aged mice than in young mice (Fig. S3C). The highest Vα2/Vα8.3 ratio was observed in the AIMT compartment in aged mice. Because Vα2+ T cells are on average more self-reactive than Vα8.3+ T cells in Vβ5 mice [17], our data suggest that the CD8+ T-cell compartment in aged mice is enriched for self-reactive T cells and that AIMT cells are generally more self-reactive than naïve cells in young as well as in aged mice.
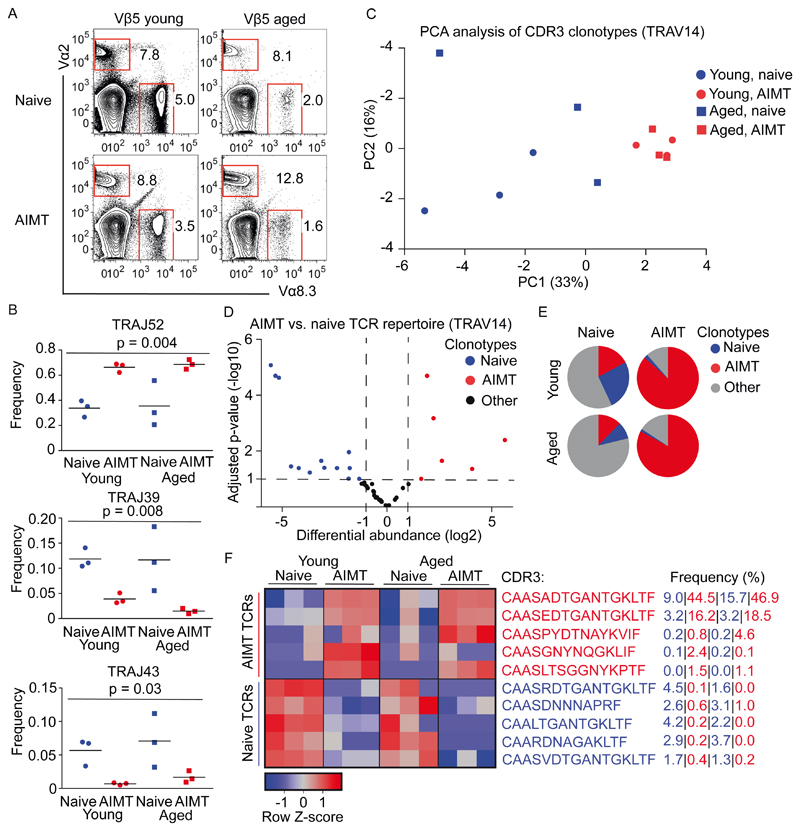
Figure 4. AIMT cells in young and aged mice represent identical clones.
(A-F) Lymph node cells from young (7-12 weeks) and aged (65-77 weeks) Vβ5 mice were enriched for CD8+ T cells by magnetic beads and stained for CD8, CD44, CD62L, CD49d, TCRVα2 and TCRVα8.3. 2-3 mice were pooled for each sample. n = 3 independent experiments.
(A) The percentage of TCRVα2+ and TCRVα8.3+ among AIMT and naïve CD8+ T cells is shown.
(B-F) AIMT (CD44+ CD62L+) and naïve (CD44- CD62L+) TCRVα2+ CD8+ T cells were FACS sorted. Library of TCR sequences was generated from isolated RNA using a two-step PCR and analyzed by deep sequencing.
(B) Frequency of TRAJ52, TRAJ39, and TRAJ43 usage among the analyzed samples. Mean is shown.The statistical significance was tested using one-way repeated measures ANOVA.
(C) PCA analysis of the TCR clonotype (based on CDR3 amino acid sequence) usage in the individual samples. Only clonotypes detected in at least 3 samples were included in the analysis.
(D) A volcano plot showing log2 differential usage between naïve and AIMT cells (both young and aged) and adjusted p-value for each individual TCR clonotype present at least in 3 samples. “AIMT”, “naïve” (2-fold difference in usage, adjusted p < 0.01) and “neutral” (non-significant) clonotypes are indicated in red, blue, and gray respectively.
(E) Pie charts showing the abundance of the “AIMT”, “naïve”, and neutral clonotypes in the T-cell repertoires of young and aged Vβ5 mice.
(F) Relative usage (z-score) of 5 most abundant “AIMT” and 5 most abundant “naïve” clonotypes in the indicated samples. The average frequency of each clone in young naïve, young AIMT, aged naïve, and aged AIMT repertoires is indicated on the right.
In the next step, we analyzed TCRs from Vα2+ T cells in greater detail using high-throughput sequencing. Naïve and AIMT cells showed differential usage of TRAJ segments, indicating that they are formed from different T-cell clonotypes (Fig. 4B, S3D), which is in line with previous results [8, 17]. Interestingly, we did not see any differences in the TRAJ usage between young and aged mice (Fig. 4B, S3D). The second step of our analysis was the comparison of individual clonotypes using CDR3 amino acid sequences. The PCA analysis separated a cluster of AIMT cells (both young and aged mice) from naïve T cells (Fig. 4C). The comparison of the usage of individual clonotypes by naïve and AIMT cells showed that some clonotypes are enriched in the naïve compartment, whereas some clonotypes are significantly enriched in AIMT cells (Fig. 4D). The AIMT cell repertoire in young and aged mice is constituted predominantly from a small number of AIMT cell-enriched clonotypes. In contrast, the naïve repertoire is more diverse and contains naïve-enriched as well as AIMT-enriched clones (Fig. 4E). The usage of the dominant clonotypes differ in naïve and AIMT subsets, but not so much between the corresponding subsets from young and aged mice (Fig. 4F). Accordingly, the diversity of the TCR repertoire is relatively high in naïve T cells and relatively low in AIMT T cells, irrespective of the age of the animals (Fig. S3E). Overall, these data showed that AIMT cells from young and aged mice have the same clonal origin, although it is possible that some minor clonotypes are lost from the AIMT repertoire during aging as indicated by the reduction of TCR Vα8.3 clonotypes.
AIMT cells in feral mice
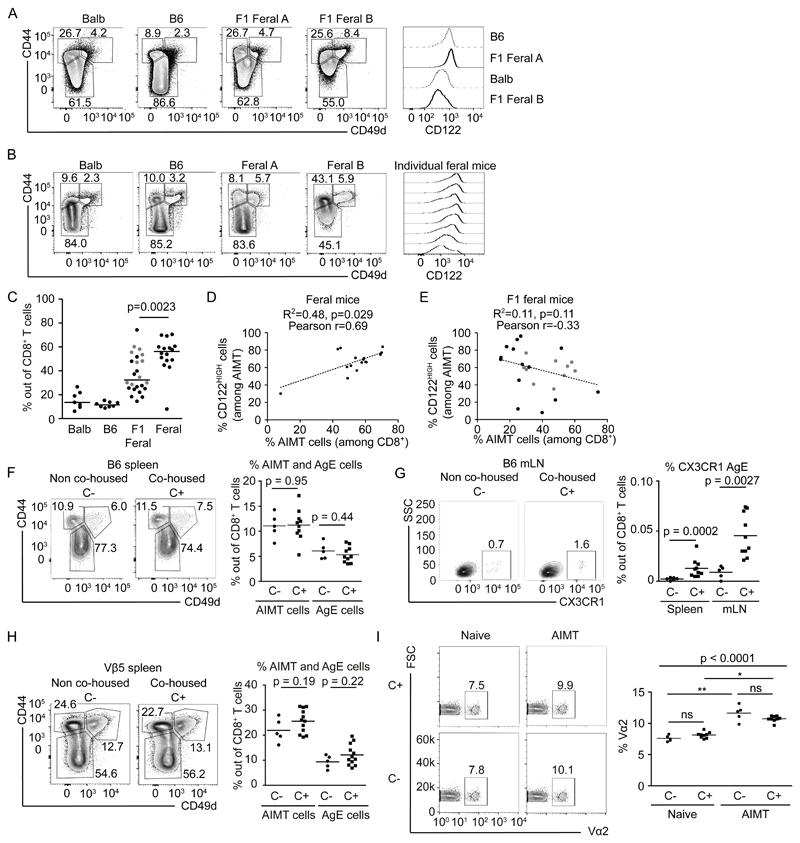
It has been suggested that the exposure to environmental antigens in feral and ‘pet shop’ mice has a substantial role in shaping the CD8+ T-cell compartment [18]. To address the possibility that the occurrence of AIMT cells might be an artifact of specific hygienic conditions in germ-free and SPF facilities, we analyzed mice with natural microbial colonization. First, bred feral mice in the animal facility and analyzed F1 offspring born in captivity. Interestingly, we observed that AIMT cells are relatively frequent in these mice (Fig. 5A, C). The expression of CD122 was variable between individual animals, suggesting variable peripheral or thymic origin of these cells likely based on the environmental or genetic factors (Fig 5A).
Figure 5. AIMT cells are present in feral mice and in laboratory mice co-housed with feral mice.
(A-C) Spleens of laboratory B6 and Balb strains and F1 offspring of feral mice (A, C) or feral mice (B, C) were isolated, fixed with formaldehyde and stained for CD8, CD44, CD49d, and CD122 and analyzed by flow cytometry. Percentage of naïve (CD44-), AIMT (CD44+ CD49d-), and antigen experienced (CD44+ CD49d+) cells among CD8+ T cells and CD122 expression in AIMT cells are shown.
(C) Percentages of AIMT cells among CD8+ T cells in B6 or Balb mice, F1 offspring of feral mice (young adult 5-8 weeks old – grey marks or aged 8-24 month old – black marks) or feral mice. Balb: n = 6 mice/6 independent experiments, B6: n = 7/7, F1 offspring of feral mice: n = 14/5, feral mice n = 16/2. Median is shown. The statistical significance was tested using two-tailed Mann-Whitney test.
(D-E) The percentages of AIMT cells among CD8+ T cells vs. CD122HIGH cells among AIMT cells were plotted for each individual mouse for feral mice (D) or for F1 offspring (young adult 5-8 weeks old – grey marks or aged 8-24 month old – black marks) of feral mice (E). The linear regression line and p value (calculated by F test) for a deviation of the regression line from a horizontal line is shown. Moreover, Pearson’s correlation coefficient is shown.
(F-I) B6 or Vβ5 mice were or were not co-housed (C+ and C-, respectively) with feral mice or F1 offspring of feral mice for 6 weeks and their splenocytes were analyzed by flow cytometry. For B6 mice, n = 5 (C-) or 10 (C+) mice in 2 independent experiments. For Vβ5 mice, n=5 (C-) or 12 (C+) mice in 2 independent experiments, if not indicated otherwise.
(F) Percentages of naïve (CD44-), AIMT (CD44+ CD49d-), and antigen experienced (CD44+ CD49d+) cells among CD8+ T cells in co-housed (C+) or non-co-housed (C-) B6 mice. A representative experiment (left) and the quantification (right) are shown. Mean is shown. Statistical significance was calculated using two-tailed Mann-Whitney test.
(G) Percentages of CX3CR1+ antigen experienced cells among CD8+ T cells from mesenteric lymph nodes (mLN) or spleens of co-housed (C+) or non-co-housed (C-) B6 mice. A representative experiment (left) and the quantification (right) are shown. Statistical significance was calculated using two-tailed Mann-Whitney test.
(H) Percentages of naïve (CD44-), AIMT (CD44+ CD49d-), and antigen experienced (CD44+ CD49d+) cells among CD8+ T cells in co-housed (C+) or non-co-housed (C-) Vβ5 mice. A representative experiment (left) and quantification (right) are shown. Mean is shown. Statistical significance was calculated using two-tailed Mann-Whitney test. Two independent experiments.
(I) Percentages of TCRVα2+ T cells among AIMT (CD44+ CD49d-) and naïve (CD44-) CD8+ T cells from co-housed (C+) or non-co-housed (C-) Vβ5 mice. A representative experiment (left) and quantification (right) are shown. n=5 (C-) or 9 (C+) mice in 2 independent experiments. Statistical significance was tested using Kruskal-Wallis test with Dunn’s multiple comparison posttests.
In the next step, we analyzed feral mice trapped in garden shelters and human houses. These mice are supposed to have even richer antigenic exposure history that the F1 feral mice born in captivity. We observed that these mice contain a large population of CD8+ CD44+ CD49d- T cells, corresponding to AIMT cells (Fig. 5B-C). The expression of CD122 in these AIMT cells largely varied among individual animals (Fig. 5B). Overall, these data suggested that the population of AIMT cells exists in feral mice. Interestingly, the percentage of AIMT cells was higher in feral mice than in F1 offspring of feral mice kept in captivity (Fig. 5C). This indicated that exposition to environmental antigens might increase the size of the AIMT-cell compartment, as was recently shown for helminth infections [44, 45]. The relative size of the AIMT-cell compartment correlated with the frequency of CD122HIGH cells among them in feral mice (Fig. 5D). This indicates that environmental factors inducing AIMT cells preferentially induce CD122HIGH peripheral virtual memory T cells. This correlation was not observed in F1 offspring of feral mice, indicating the major role of genetic background (Fig. 5E).
Co-housing of laboratory and feral mice has minimal effects on the CD8+ T-cell compartment
To observe how the AIMT-cell compartment and other CD8+ T-cell subsets change upon exposure to environmental antigens in laboratory mice, we co-housed B6 WT mice and transgenic Vβ5 mice with feral mice or F1 offspring of feral mice for 6 weeks. Surprisingly, the co-housing had no significant effect on the proportions of the major CD8+ T-cell subsets, i.e., naïve, AIMT, and antigen-experienced cells, in B6 and Vβ5 mice (Fig. 5F-H). The only observed difference between co-housed and control laboratory strains was a slightly higher percentage of CX3CR1+ CD44+ CD49d+ antigen-experienced cells in the spleen and mesenteric lymph nodes of the co-housed animals of the SPF origin (Fig. 5G).
We observed that the enrichment of the AIMT-cell compartment in Vα2+ T cells was preserved in the co-housed animals (Fig. 5I), suggesting that the co-housing did not substantially alter the composition of the repertoire of the AIMT-cell compartment. Overall, our data indicate that AIMT cells are a naturally occurring CD8+ T-cell subset in laboratory mouse strains as well as in feral mice.
Limited effects of co-housing on the microbial colonization
Because we observed a surprisingly small effect of the co-housing of laboratory strains with feral mice or F1 offspring of feral mice on their CD8+ T-cell compartment, we examined the colonization of the feral and co-housed animals with microbes and viruses. A detailed health examination of a limited number of animals detected Helicobacter, Pasteurella, MCMV, and endoparasites, but no other pathogenic agents, in the feral mice (Appendix 1, 10.5281/zenodo.4298697). Of these pathogens, only Helicobacter and endoparasites were transmitted to the co-housed animals in at least 1 case out of 4 tested co-housed mice. This was in line with the fact that none of the co-housed mice died before the end of the experiment, including Vβ5 mice, which are most likely immunocompromised because of the limited TCR repertoire.
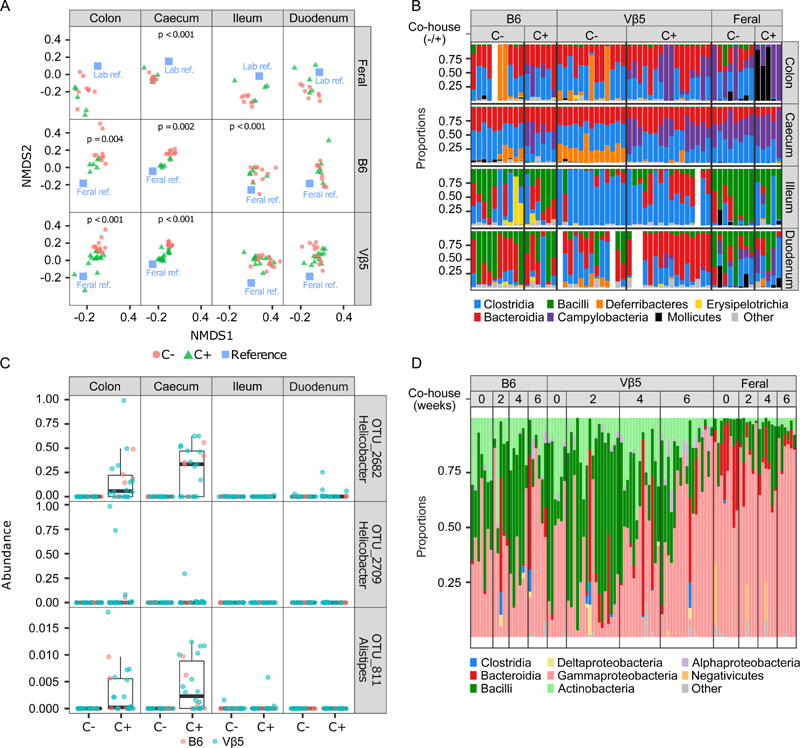
Subsequently, we addressed the effects of co-housing on bacterial colonization. We performed 16S rRNA sequence analysis of the composition of the microbial community in the gut and salivary glands of co-housed animals. The overall microbial diversity in the intestine of co-housed laboratory strains was not affected except for the ileum, where the diversity increased (Fig. S4A). Interestingly, microbial diversity decreased in the colon of feral mice upon co-housing (Fig. S4A). As expected, the microbial composition of the intestine of the laboratory strains converged to the feral mice upon co-housing (Fig. 6A, Fig. S4B). The most notable effect of the co-housing was the abundance of Campylobacteria in the colon and caecum of the co-housed mice, whereas this group of bacteria was not present in non-co-housed laboratory mice (Fig. 6B, Fig. S4C). Two different Helicobacteria taxons (Campylobacterales) and Alistipes (Bacteroidia) were detected in the colon and caecum of some of the co-housed, but not SPF, mice (Fig. 6C). In contrast, three taxonomic units from Clostridiales, Roseburia, and Ruminococcus taxons were depleted in colon, caecum and duodenum of the co-housed mice (Fig. S4D).
Figure 6. Co-housing with feral mice changes the intestinal microbial colonization of laboratory mice.
(A-C) The analysis of intestinal microbiome of co-housed and non-co-housed laboratory and feral mice (Experiment B, see Methods). Samples were taken from the colon, caecum, ileum, or duodenum. C+; co-housed mice for 6 weeks (laboratory + feral), C-; non-co-housed mice.
(A) Compositional variation in co-housed vs. non-co-housed groups assessed based on Nonmetric Multidimensional Scaling (NMDS) of Bray-Curtis dissimilarities. Centroids for a reference group (feral non-co-housed mice in the case B6 or Vβ5 and non-co-housed Vβ5 plus B6 in the case of feral mice) are indicated by blue rectangles. Permutation-based p-values indicate significant changes due to cohousing.
(B) Taxonomical composition of the intestinal microbiota of laboratory co-housed (C+) and non-co-housed (C-) B6, Vβ5, and feral mice. Color bars represent proportions of dominant bacterial classes in each sample. White space means no data.
(C) Three most abundant operational taxonomic units with higher relative abundance in co-housed than non-co-housed B6 and Vβ5 laboratory mice.
(D) Taxonomical composition of the salivary microbiota of laboratory B6 or Vβ5 mice co-housed with feral mice (Experiment B, see Methods). Color bars represent proportions of dominant bacterial classes in each sample. Samples were collected from each mouse prior to the co-housing (0 weeks) and 2, 4, and 6 weeks after the co-housing was set up.
The salivary microbiome of the laboratory strains did not converge to feral mice, but rather the microbiome of the feral mice converged to the microbiome of SPF laboratory strains during the co-housing (Fig. S4E). However, co-housing led to a dramatic increase in Bacteriodia taxon in saliva of laboratory strains (Fig. 6D, Fig. S4F). Moreover, the abundance of Clostridia and Deltaproteobacteria was detected in some individuals during, but not before, co-housing (Fig. 6D, Fig. S4F). Overall, these data document that co-housing with feral mice changed the microbial colonization of laboratory mice without a substantial impact on the CD8+ T-cell compartment.
Discussion
AIMT cells represent one of the two most numerous populations of CD8+ T cells in mice. Although it has been shown that these cells are functionally different from naïve and antigen-experienced memory T cells [6, 17, 39, 44–47], our understanding of the biology of these cells is incomplete. There are multiple potential reasons why AIMT cells are an underrepresented area of focus. First, multiple ways of AIMT cells formation were proposed involving different cytokines (IL-4, IL-7, and IL-15), different anatomical sites (thymus, secondary lymphoid organs), and different physiological states (experimentally induced lymphopenia, newborn lymphopenia, aging) [1, 2, 4]. Second, the phenotypic and functional similarities between particular AIMT-cell subsets and their differences are not well understood [1, 2, 4]. Third, only a minority of studies used germ-free mice to exclude microbial antigens [16, 17] and at the same time, no previous study addressed the presence of AIMT cells in feral mice, which were reported to have substantially different CD8+ T-cell compartment from laboratory mice [18].
In this study, we performed comprehensive phenotyping of AIMT cells to address multiple unresolved questions concerning their origin and heterogeneity. We concluded that the formation of AIMT cells represent a robust genetic differentiation program across the genetic background, aging, and hygienic status. However, these three conditions regulate AIMT-cell abundance and internal variability.
We identified CD122 as a marker discriminating CD122LOW thymic (innate memory) and peripheral CD122HIGH (virtual memory) subsets of CD8+ AIMT cells. CD122LOW AIMT cells prevail in WT young Balb mice and some feral mice. On the contrary, CD122HIGH AIMT cells are dominant in B6, in some feral mice and Balb mice with the impaired formation of CD122LOW cells because of aging, expression of a transgenic TCR disabling the formation of IL-4 producing iNKT cells, or irradiation. Collectively, our data indicate that the absence of thymic innate memory AIMT cells allows the conversion of naïve T cells into AIMT cells in the periphery, presumably via IL-4, IL-7, and/or IL-15 [1, 4, 7, 11, 12].
AIMT cells represent the largest population of CD8+ T cells in aged B6 mice [35]. Here we confirmed this observation in germ-free conditions and extended it by the analysis of aged Balb mice. The mechanism behind the high frequency of AIMT cells in aged animals was unclear. It has been proposed that the origin of these cells might differ from the AIMT cells in young animals [37]. We addressed this question by analyzing gene expression profiles and TCR profiling of young and aged AIMT cells using high-throughput sequencing. We observed that AIMT cells show a common gene expression pattern largely irrespective of their site of origin, genetic background, and the age of the host. However, our analysis revealed that the site of origin and aging fine-tune the gene expression among AIMT cells. In addition to confirming differences between AIMT cells from young and aged B6 mice as reported previously [36], we identified similarities between CD122HIGH AIMT cells from aged Balb mice and AIMT cells from B6 mice. These similarities support the concept that CD122HIGH AIMT cells are formed at the periphery in both B6 and Balb strains. Expression of key effector molecules involved in by-stander cytotoxicity as well as IL-12/IL-18 production of IFN-γ was higher in peripheral AIMT cells than in thymic AIMT cells, suggesting that these two subsets differ in their ability to elicit by-stander protective immunity as proposed previously [2]. Despite these relatively subtle differences, virtual memory T cells, mostly present in B6 mice, and innate memory T cells, mostly present in young Balb mice, exhibit very similar gene expression pattern and likely represent two flavors of the AIMT-cell lineage.
It has been proposed that aged AIMT cells are formed from T-cell clones with high level of self-reactivity [48]. Recently, we have shown that AIMT cells are formed from highly self-reactive T cells clones in young adult mice [17], which was subsequently confirmed by another study [8]. In line with these previous reports, we observed that CD8+ naïve and AIMT cells use different TCR repertoires. Clones typically present in the naïve population are virtually absent in AIMT cells, whereas typical AIMT clones can still be identified in the naïve compartment, albeit with a much lower frequency. This partial overlap is not surprising as AIMT cells are formed from naïve T cells in the periphery. However, a recent study by Miller et al. showed a clear separation of naïve and AIMT-cell repertoires [8]. This discrepancy might be caused by the difference between the employed mouse models. We analyzed TCR Vα2+ repertoire in transgenic mice expressing TCR Vβ5 adopted from H2-Kb-restricted OT-I TCR. Importantly, the Vβ5 mouse has a similar frequency of AIMT cells as WT B6 mice. Miller et al. analyzed TCRα repertoire from mice expressing TCR Vβ3 chain. This chain usage is abundant among prostate cancer-infiltrating regulatory CD4+ T cells [49] and might produce T-cell repertoire biased towards MHCII-restriction, subsequently affecting the frequency, repertoire or phenotype of CD8+ AIMT cells.
We observed that young and aged AIMT cells have comparable TCRVα2 repertoire with identical dominant T-cell clones and similar level of diversity. Moreover, we observed that AIMT cells preferentially use Vα2 over Vα8.3 TCR chains, which is more pronounced in aged mice. Thus, it is likely that some minor, relatively less self-reactive clonotypes are removed from the AIMT cell repertoire during aging, which is consistent with a previous report [50].
Overall, the probable causes of the high frequency of AIMT cells in aged B6 and Balb mice are: (i) low thymic egress of fresh naïve T cells, (ii) irreversible AIMT cells formation from highly self-reactive clones augmented by age-related lymphopenia, and (iii) the survival and proliferative advantage of AIMT cells over naïve T cells.
For the first time, we studied AIMT cells in feral mice and F1 offspring of feral mice born in captivity, suggesting that AIMT cells are not an artifact of inbred strains and/or SPF conditions. Feral mice showed an even higher frequency of AIMT cells than laboratory strains. This could be caused by the enhancement of the AIMT-cell compartment via IL-4 produced in response to helminth [44, 45] or other infections. Indeed, we observed a positive correlation between the frequency of AIMT cells and the percentage of CD122HIGH cells among AIMT cells in feral mice, but not in feral F1 offspring born in captivity. This suggests a role of environmental transient bacterial or helminth infections in feral mice and highlight the role of genetic background in housed F1 offspring.
Although it has been shown that co-housing of laboratory strains with ‘pet shop’ mice leads to dramatic changes of their CD8+ T cell compartment [18], we could detect only a minor increase of CX3CR1+ effector cells in co-housed B6 mice. Otherwise, the CD8+ T-cell compartment in the spleen, peripheral lymph nodes, and mesenteric lymph nodes of co-housed mice was unaffected. A possible explanation is that our feral mice showed much lower pathogenic burden than the ‘pet shop’ mice used by Beura et al. [18], which seems logical given the high animal density and the lack of natural predation at ‘pet shop’ farms. Although different populations of feral mice might have a different pathogenic burden, our data document that co-housing with feral mice does not always induce dramatic changes in the CD8+ T cell compartment. We observed that co-housing with feral mice substantially changes the microbial composition of the intestine and saliva of laboratory strains, including opportunistic pathogens such as Pasteurella and Helicobacter.
Altogether, our data document that AIMT cells represent a large CD8+ T-cell compartment in mice, which is present in germ-free as well as in feral conditions. The frequency of peripherally induced self-reactive CD122HIGH AIMT cells increases during aging via homeostatic mechanisms in Balb and B6 mice.
Supplementary Material
Key points.
Antigen-inexperienced memory-like T (AIMT) cells are steadily present in mice.
Aging, genetics, and hygienic conditions fine-tune AIMT cell numbers and phenotype.
Virtual memory T cells respond stronger to Th1 cytokines than innate memory T cells.
Acknowledgement
We thank Ladislav Cupak for technical assistance and genotyping of mice, Sarka Kocourkova for preparation of RNAseq libraries, Prof. Sai Reddy and Dr. Simon Friedensohn (both ETH Zürich) for their valuable help with designing the assays for TCR repertoire analysis, Dr. Dominik Filipp (Institute of Molecular Genetics, Prague) for providing us DO11.10 transgenic mice.
Abbreviations used in this article
- AgE
antigen-experienced
- AIMT
Antigen-inexperienced memory-like T
- Balb
BALB/c
- B6
C57BL/6
- DO11.10
C.Cg-Tg(DO11.10)10Dlo/J
- iNKT
invariant natural killer T
- PCA
principal component analysis
- SPF
specific pathogen free
- WT
wild-type.
Footnotes
Conflict of interest
All authors declare that they have no conflict of interest.
Footnotes
* Laboratory of Adaptive Immunity, Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic; † Laboratory of Genomics and Bioinformatics, Institute of Molecular Genetics of the Czech Academy of Sciences, Prague, Czech Republic; ‡ Department of Informatics and Chemistry, Faculty of Chemical Technology, University of Chemistry and Technology, Prague, Czech Republic; § Department of Zoology, Faculty of Science, Charles University, Prague, Czech Republic; ¶ Department of Zoology, Faculty of Science, Charles University, BIOCEV, Vestec, Czech Republic; ║ Laboratory of Immunity & Cell Communication, BIOCEV, First Faculty of Medicine, Charles University, Vestec, Czech Republic; # Research Facility Studenec, Institute of Vertebrate Biology, Czech Academy of Sciences, Brno, Czech Republic; ** Laboratory of Gnotobiology, Institute of Microbiology of the Czech Academy of Sciences, Novy Hradek, Czech Republic; †† Department of Biomedicine, University Hospital and University of Basel, Basel, Switzerland; ‡‡ Present address: Laboratory of Lymphocyte Signalling and Development, The Babraham Institute, Cambridge CB22 3AT, United Kingdom
This study was supported by the Czech Science Foundation (19-03435Y to OS, 18-17796Y to JKr, 19-02261S to HK), Swiss National Science Foundation (Promys, IZ11Z0_166538 to OS), and the European Research Council (FunDiT to OS). The Laboratory of Adaptive Immunity was supported by an EMBO Installation Grant (3259 to OS). The Laboratory of Adaptive Immunity and the Laboratory of Genomics and Bioinformatics were supported by the Institute of Molecular Genetics of the Czech Academy of Sciences core funding (RVO 68378050). OS was supported by the Purkyne Fellowship provided by the Czech Academy of Sciences. PS, RS, and TM were supported by MICOBION project funded from EU H2020 (No 810224). JN and MK were supported by “Center for Tumor Ecology - Research of the Cancer Microenvironment Supporting Cancer Growth and Spread” OP RDI CZ.02.1.01/0.0/0.0/16_019/0000785 provided by the Czech Ministry of Education, Youth and Sports and the European Regional Development Fund. LS was supported by the Charles University specific research grant (SVV 260571/2020). AM and VN were students partially supported by the Faculty of Science, Charles University, Prague. JN was a student partially supported by the University of Chemistry and Technology, Prague. The animal facility of the IMG is a part of the Czech Centre for Phenogenomics and the work there was supported in part by following grants: LM2015040, LM2018126, OP RDI CZ.1.05/2.1.00/19.0395, OP RDI BIOCEV CZ.1.05/1.1.00/02.0109 provided by the Czech Ministry of Education, Youth and Sports and the European Regional Development Fund.
AM performed most of the experiments. VN, LS, MP, AD, RS, TM, AN, KK, PS, OS performed experiments. AM, VN, and OS analyzed data and finalized figures. JKu and JM analyzed the transcriptomic data. JN analyzed the TCR profiling data. DC and JKr analyzed the S16 sequencing data. RS and PS provided feral mice. TH and HK provided germ-free mice. MK, HK, JKr, PS, and OS supervised the work. OS conceived the study. AM, VN, and OS wrote the manuscript. All the authors contributed to the manuscript and reviewed it.
Address correspondence and reprint requests to Ondrej Stepanek, Laboratory of Adaptive Immunity
Institute of Molecular Genetics of the Czech Academy of Sciences, 14220, Prague, Czech Republic. Phone: 00420241062155. E-mail address: ondrej.stepanek@img.cas.cz
References
- 1.Pribikova M, Moudra A, Stepanek O. Opinion: Virtual memory CD8 T cells and lymphopenia-induced memory CD8 T cells represent a single subset: Homeostatic memory T cells. Immunology Letters. 2018;203:57–61. doi: 10.1016/j.imlet.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 2.White JT, Cross EW, Kedl RM. Antigen-inexperienced memory CD8+ T cells: where they come from and why we need them. Nat Rev Immunol. 2017;17(6):391–400. doi: 10.1038/nri.2017.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Weinreich MA, et al. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8+ T cells. Nat Immunol. 2010;11(8):709–16. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jameson SC, Lee YJ, Hogquist KA. Innate Memory T cells. Advances in Immunology. 2015;126(126):173–213. doi: 10.1016/bs.ai.2014.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tripathi P, et al. IL-4 and IL-15 promotion of virtual memory CD8+ T cells is determined by genetic background. Eur J Immunol. 2016;46(10):2333–2339. doi: 10.1002/eji.201646404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.White JT, et al. Virtual memory T cells develop and mediate bystander protective immunity in an IL-15-dependent manner. Nat Commun. 2016;7:1–13. doi: 10.1038/ncomms11291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sosinowski T, et al. CD8alpha+ dendritic cell trans presentation of IL-15 to naive CD8+ T cells produces antigen-inexperienced T cells in the periphery with memory phenotype and function. J Immunol. 2013;190(5):1936–47. doi: 10.4049/jimmunol.1203149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Miller CH, et al. Eomes identifies thymic precursors of self-specific memory-phenotype CD8(+) T cells. Nat Immunol. 2020:567–577. doi: 10.1038/s41590-020-0653-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hussain T, Quinn KM. Similar but different: virtual memory CD8 T cells as a memory-like cell population. Immunol Cell Biol. 2019;97(7):675–684. doi: 10.1111/imcb.12277. [DOI] [PubMed] [Google Scholar]
- 10.Smith NL, et al. Developmental Origin Governs CD8(+) T Cell Fate Decisions during Infection. Cell. 2018;174(1):117–130. doi: 10.1016/j.cell.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 11.Goldrath AW, et al. Cytokine requirements for acute and Basal homeostatic proliferation of naive and memory CD8+ T cells. J Exp Med. 2002;195(12):1515–22. doi: 10.1084/jem.20020033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldrath AW, Bogatzki LY, Bevan MJ. Naive T cells transiently acquire a memory-like phenotype during homeostasis-driven proliferation. J Exp Med. 2000;192(4):557–64. doi: 10.1084/jem.192.4.557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurzweil V, LaRoche A, Oliver PM. Increased peripheral IL-4 leads to an expanded virtual memory CD8+ population. J Immunol. 2014;192(12):5643–51. doi: 10.4049/jimmunol.1301755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kieper WC, et al. Overexpression of interleukin (IL)-7 leads to IL-15-independent generation of memory phenotype CD8+ T cells. J Exp Med. 2002;195(12):1533–9. doi: 10.1084/jem.20020067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leonard WJ, Lin JX, O’Shea JJ. The gammac Family of Cytokines: Basic Biology to Therapeutic Ramifications. Immunity. 2019;50(4):832–850. doi: 10.1016/j.immuni.2019.03.028. [DOI] [PubMed] [Google Scholar]
- 16.Haluszczak C, et al. The antigen-specific CD8+ T cell repertoire in unimmunized mice includes memory phenotype cells bearing markers of homeostatic expansion. J Exp Med. 2009;206(2):435–48. doi: 10.1084/jem.20081829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Drobek A, et al. Strong homeostatic TCR signals induce formation of self-tolerant virtual memory CD8 T cells. The EMBO Journal. 2018 doi: 10.15252/embj.201798518. e98518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beura LK, et al. Normalizing the environment recapitulates adult human immune traits in laboratory mice. Nature. 2016;532(7600):512–6. doi: 10.1038/nature17655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fink PJ, et al. Both Intrathymic and Peripheral Selection Modulate the Differential Expression of V-Beta-5 among Cd4+ and Cd8+ T-Cells. Journal of Experimental Medicine. 1992;176(6):1733–1738. doi: 10.1084/jem.176.6.1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murphy KM, Heimberger AB, Loh DY. Induction by antigen of intrathymic apoptosis of CD4+CD8+TCRlo thymocytes in vivo. Science. 1990;250(4988):1720–3. doi: 10.1126/science.2125367. [DOI] [PubMed] [Google Scholar]
- 21.Zerbino DR, et al. Ensembl 2018. Nucleic Acids Research. 2018;46(D1):D754–D761. doi: 10.1093/nar/gkx1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kim D, Landmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature Methods. 2015;12(4):357–U121. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology. 2014;15(12):1–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens M. False discovery rates: a new deal. Biostatistics. 2017;18(2):275–294. doi: 10.1093/biostatistics/kxw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ritchie ME, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Research. 2015;43(7):e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shugay M, et al. Towards error-free profiling of immune repertoires. Nat Methods. 2014;11(6):653–5. doi: 10.1038/nmeth.2960. [DOI] [PubMed] [Google Scholar]
- 28.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Research. 2013;41(1):e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Glenn TC, et al. Adapterama I: universal stubs and primers for 384 unique dual-indexed or 147,456 combinatorially-indexed Illumina libraries (iTru & iNext) Peerj. 2019;7 doi: 10.7717/peerj.7755. e7755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jiang HS, et al. Skewer: a fast and accurate adapter trimmer for next-generation sequencing paired-end reads. Bmc Bioinformatics. 2014;15:1–12. doi: 10.1186/1471-2105-15-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Callahan BJ, et al. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods. 2016;13(7):581–583. doi: 10.1038/nmeth.3869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Edgar RC, et al. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics. 2011;27(16):2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang Q, et al. Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73(16):5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Quast C, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Research. 2013;41(D1):D590–D596. doi: 10.1093/nar/gks1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu BC, et al. Cutting Edge: Central Memory CD8 T Cells in Aged Mice Are Virtual Memory Cells. Journal of Immunology. 2013;191(12):5793–5796. doi: 10.4049/jimmunol.1302509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quinn KM, et al. Age-Related Decline in Primary CD8(+) T Cell Responses Is Associated with the Development of Senescence in Virtual Memory CD8(+) T Cells. Cell Rep. 2018;23(12):3512–3524. doi: 10.1016/j.celrep.2018.05.057. [DOI] [PubMed] [Google Scholar]
- 37.Renkema KR, et al. Two separate defects affecting true naive or virtual memory T cell precursors combine to reduce naive T cell responses with aging. J Immunol. 2014;192(1):151–9. doi: 10.4049/jimmunol.1301453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Decman V, et al. Defective CD8 T cell responses in aged mice are due to quantitative and qualitative changes in virus-specific precursors. J Immunol. 2012;188(4):1933–41. doi: 10.4049/jimmunol.1101098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lanzer KG, et al. Virtual memory cells make a major contribution to the response of aged influenza-naive mice to influenza virus infection. Immun Ageing. 2018;15:17. doi: 10.1186/s12979-018-0122-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weinreich MA, et al. T cells expressing the transcription factor PLZF regulate the development of memory-like CD8(+) T cells. Nature Immunology. 2010;11(8):709–U75. doi: 10.1038/ni.1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Robertson JM, Jensen PE, Evavold BD. DO11.10 and OT-II T cells recognize a C-terminal ovalbumin 323-339 epitope. Journal of Immunology. 2000;164(9):4706–4712. doi: 10.4049/jimmunol.164.9.4706. [DOI] [PubMed] [Google Scholar]
- 42.Akue AD, Lee JY, Jameson SC. Derivation and maintenance of virtual memory CD8 T cells. J Immunol. 2012;188(6):2516–23. doi: 10.4049/jimmunol.1102213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Daniels MA, Teixeiro E. Forget ‘ME’ not virtual memory T cells. Nat Immunol. 2020:499–500. doi: 10.1038/s41590-020-0668-7. [DOI] [PubMed] [Google Scholar]
- 44.Lin JS, et al. Virtual memory CD8 T cells expanded by helminth infection confer broad protection against bacterial infection. Mucosal Immunol. 2019;12(1):258–264. doi: 10.1038/s41385-018-0100-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Rolot M, et al. Helminth-induced IL-4 expands bystander memory CD8(+) T cells for early control of viral infection. Nat Commun. 2018;9(1):4516. doi: 10.1038/s41467-018-06978-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee JY, et al. Virtual memory CD8 T cells display unique functional properties. Proceedings of the National Academy of Sciences of the United States of America; 2013. pp. 13498–13503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Grau M, et al. Antigen-Induced but Not Innate Memory CD8 T Cells Express NKG2D and Are Recruited to the Lung Parenchyma upon Viral Infection. The Journal of Immunology. 2018:3635–3646. doi: 10.4049/jimmunol.1701698. [DOI] [PubMed] [Google Scholar]
- 48.Rudd BD, et al. Nonrandom attrition of the naive CD8+ T-cell pool with aging governed by T-cell receptor:pMHC interactions. Proc Natl Acad Sci U S A. 2011;108(33):13694–9. doi: 10.1073/pnas.1107594108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Malchow S, et al. Aire-dependent thymic development of tumor-associated regulatory T cells. Science. 2013;339(6124):1219–24. doi: 10.1126/science.1233913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Quinn KM, et al. Heightened self-reactivity associated with selective survival, but not expansion, of naive virus-specific CD8(+) T cells in aged mice. Proceedings of the National Academy of Sciences of the United States of America; 2016. pp. 1333–1338. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.