Abstract
Complexes containing a pair of SMC family proteins are fundamental for the 3D organization of genomes in all domains of life. The eukaryotic SMC complexes cohesin and condensin are thought to fold, respectively, interphase and mitotic chromosomes into large loop domains, although the underlying molecular mechanisms have remained unknown. We used electron cryo-microscopy to investigate the nucleotide-driven reaction cycle of condensin from the budding yeast Saccharomyces cerevisiae. Our structures of the five-subunit condensin holo complex at different functional stages suggest that ATP binding induces the transition of the SMC coiled coils from a folded-rod conformation into a more open architecture. ATP binding simultaneously triggers the exchange of the two HEAT-repeat subunits bound to the SMC ATPase head domains. We propose that these steps result in the interconversion of DNA binding sites in the catalytic core of condensin that form the basis of the DNA translocation and loop-extrusion activities.
Introduction
Multi-subunit protein complexes built around pairs of Structural Maintenance of Chromosomes (SMC) proteins are essential for the functional organization of genomes in all domains of life1,2. SMC proteins are characterized by ~50-nm-long intramolecular anti-parallel coiled coils that connect globular ‘hinge’ and ‘head’ domains at either end of the coil (Fig. 1a). The two head domains dimerize upon sandwiching two adenosine triphosphate (ATP) molecules and disengage upon nucleotide hydrolysis and release in a manner analogous to ATP-binding cassette (ABC) transporters, with whom they share their fold3. In almost all SMC complexes, a subunit of the kleisin protein family connects the heads of an SMC dimer by binding with its amino terminus to the ‘neck’ coiled-coil stem of one head and with its carboxy terminus to the distal ‘cap’ surface of the other head, respectively4. This arrangement produces a ring topology, as was first recognized for cohesin5 but is now thought to be a defining feature of probably all SMC complexes. The central regions of the kleisins bind additional subunits that are composed of either tandem winged-helix domains (WHDs), as in the case of prokaryotic Smc–ScpAB or MukBEF and eukaryotic Smc5/6 complexes, or of HEAT (Huntingtin, EF3, PP2A, TOR1) repeat motifs, as in the case of eukaryotic cohesin and condensin complexes6,7.
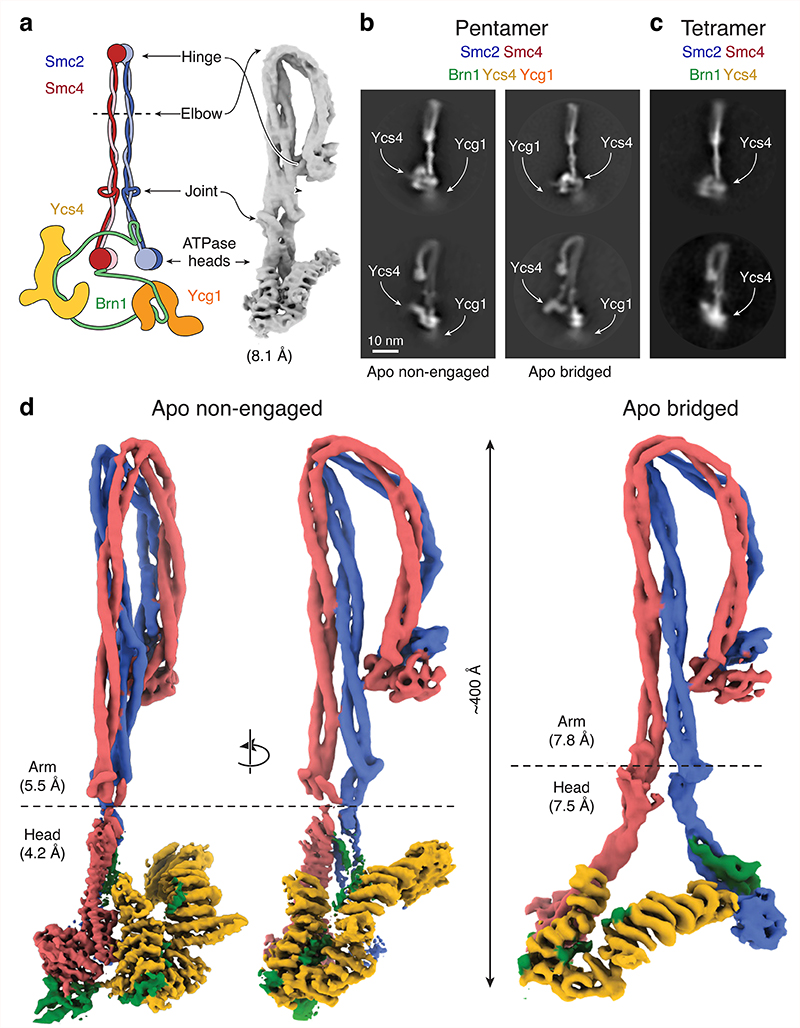
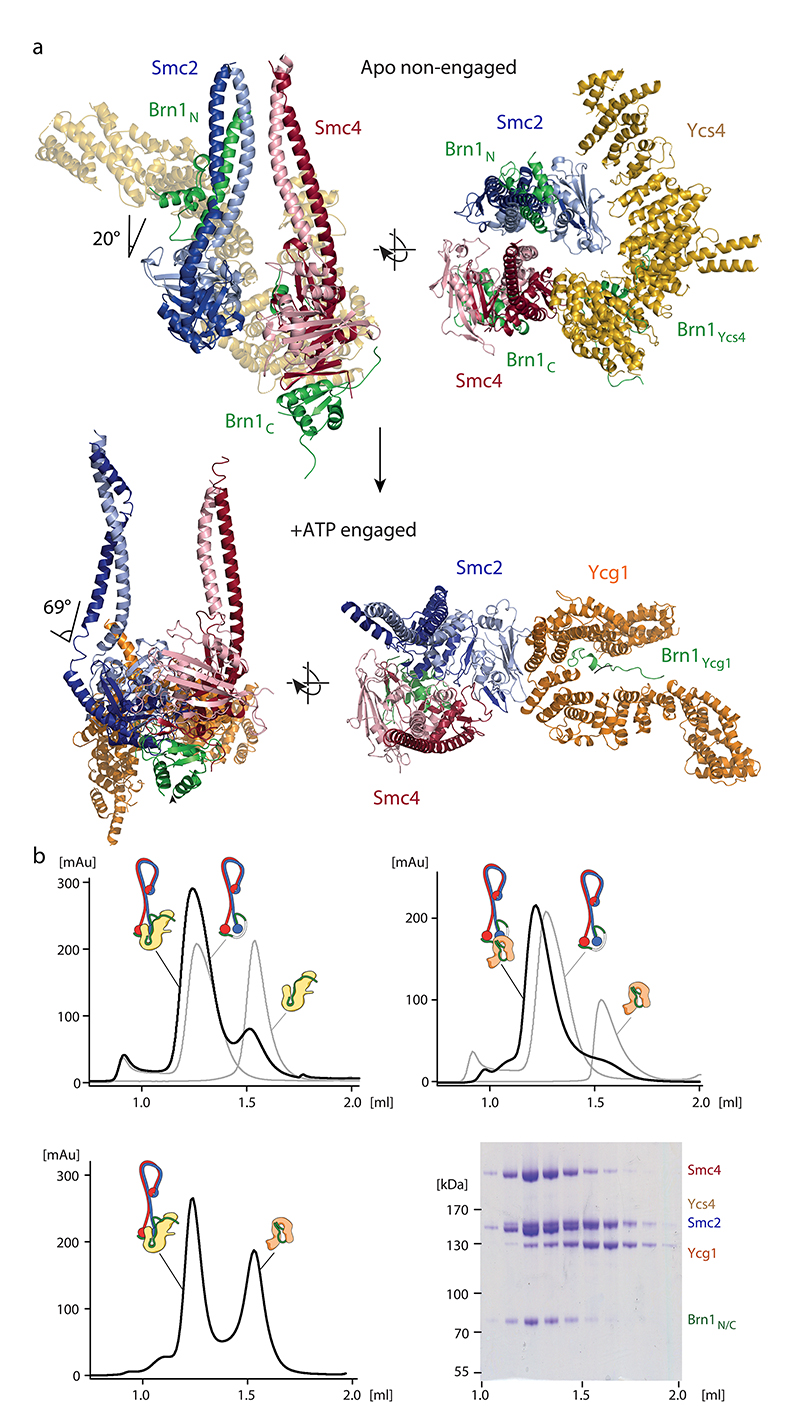
Fig. 1. Cryo-EM structures of the yeast condensin holo complex in the nucleotide-free apo form.
a, Schematic model of the S. cerevisiae condensin holo complex and 8.1 Å-resolution 3D map, showing its overall architecture. b, Representative 2D class average of the 5-subunit ‘pentamer’ holo complex in non-engaged and bridged states. c, Representative 2D class average of the 4-subunit ‘tetramer’ complex lacking Ycg1. d, Composite cryo-EM maps of the condensin complex in non-engaged and bridged apo states.
A unifying principle for the ability of SMC complexes to control a multitude of chromosomal processes is their ability to bind and processively extrude double-stranded DNA into large loops8–12. In single-molecule imaging assays, purified cohesin13,14 and condensin15,16 are able to extrude DNA loops of several kilobase pairs in length in a manner that depends on ATP hydrolysis by their SMC head domains. The consequences of DNA-loop extrusion by cohesin or condensin can explain key features of interphase17,18 or mitotic19 chromosome architecture, respectively. The molecular mechanisms that underlie DNA-loop extrusion by SMC protein complexes remain, however, to be elucidated. One reason for the lack of understanding at the molecular level is the absence of atomic structures for full-length eukaryotic SMC complexes that provide insights into their reaction cycles and concomitant conformational dynamics.
Here, we use single-particle electron cryomicroscopy (SPA cryo-EM), biochemistry and crosslinking mass spectrometry to describe and investigate three distinct states along the ATPase reaction coordinate of the budding yeast condensin complex. Our structures in the presence and absence of ATP reveal the nucleotide-dependent alternate binding of the Ycs4 and Ycg1 HEAT-repeat subunits to the Smc4 and Smc2 head domains, as well as a bridged conformation in which Ycs4 binds the head domains of both, Smc2 and Smc4. Our data is consistent with the simultaneous disengagement of the Smc2–Smc4 coiled coils from a folded rod conformation in the apo state, in which the hinge domain contacts the head-proximal region of the coils as a consequence of a sharp bent at an ‘elbow’ region, into a more open, ring-shaped architecture in the ATP state. We propose that these large-scale conformational changes form the basis for the DNA motor activities of condensin, and potentially all other SMC protein complexes.
Results
Two distinct states of apo condensin holo complexes
We first collected cryo-EM data sets for the ~633-kDa condensin (hetero-) ‘pentamer’ holo complex (consisting of S. cerevisiae subunits Smc2, Smc4, Brn1, Ycs4 and Ycg1; Supplementary Note, Supplementary Fig. 1) and of a ~515-kDa ‘tetramer’ complex that did not contain the Ycg1 subunit, in the absence of nucleotide (Table 1). These data sets revealed condensin complexes with ~40-nm-long, folded rod-shaped Smc2 and Smc4 coiled-coil ‘arms’, which were closely aligned along their entire lengths (for examples of 2D classes, see Fig. 1b and Extended Data Fig. 1a,b). Sub-classification of the pentamer dataset revealed two states with discrete orientations of the Smc2–Smc4 ATPase head domains (Extended Data Fig. 1c, d).
Table 1. Cryo-EM data collection, refinement and validation statistics of condensin apo states.
| #1 Non-engaged overall (EMD-10951, PDB 6YVU)* | #2 Non-engaged arm segment (EMD-10948) | #3 Non-engaged head segment (EMD-10947) | #4 Bridged overall (EMD-10954) | #5 Bridged arm segment (EMD-10953 | #6 Bridged head segment (EMD-10952, PDB 6YVV) | |
|---|---|---|---|---|---|---|
| Data collection and processing | ||||||
| Magnification | 130k | 105k, 130k | 105k, 130k | 130k | 130k | 130k |
| Voltage (kV) | 300 | 300 | 300 | 300 | 300 | 300 |
| Electron exposure (e–/Å2) | 46.5 | 40 ~ 55 | 40 ~ 55 | 46.5 | 46.5 | 46.5 |
| Defocus range (μm) | -0.5 ~ -0.9 | -0.5 ~ -0.9, - 1.6 ~ -3.6 |
-0.5 ~ -0.9, - 1.6 ~ -3.6 |
-0.5 ~ -0.9 | -0.5 ~ -0.9 | -0.5 ~ -0.9 |
| Pixel size (Å) | 1.065 | 1.05, 1.065, 1.085, 1.15 |
1.05, 1.065, 1.085, 1.15 |
1.065 | 1.065 | 1.065 |
| Symmetry imposed | C1 | C1 | C1 | C1 | C1 | C1 |
| Initial particle images (no.) | 4,052,794 | 2,369,713 | 1,683,081 | 1,683,081 | 1,683,081 | 1,683,081 |
| Final particle images (no.) | 100,388 | 636,446 | 576,016 | 37,639 | 24,593 | |
| Map resolution (Å) | 8.1 | 5.3 | 4.2 | 9.1 | 7.8 | 7.5 |
| FSC threshold | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 | 0.143 |
| Map resolution range (Å) | 8.1 to >10 | 5.3 to >10 | 4.2 to >10 | 9.1 to >10 | 7.8 to >10 | 7.5 to >10 |
| Refinement | ||||||
| Initial model used | Ab initio and 4RSI | Homology model derived from 6QJ2, 6QJ4, 4UX3, 6Q6E |
Homology model derived from 6QJ2, 6QJ4, 4UX3, 6Q6E |
|||
| Model resolution (Å) | 5.5 | 4.2 | 7.5 | |||
| FSC threshold | 0.143 | 0.143 | 0.143 | |||
| Model resolution range (Å) | n/a | 4.2 ~ 6.5 | n/a | |||
| Map sharpening B factor (Å2) | -276 | -200 | -334 | |||
| Model composition | ||||||
| Non-hydrogen atoms | 14,952 | |||||
| Protein residues | 1,893 | |||||
| Ligands | ||||||
| B factors (Å2) | ||||||
| Protein | 166.71 | |||||
| Ligand | ||||||
| R.m.s. deviations | 0.004 | |||||
| Bond lengths (Å) | 0.941 | |||||
| Bond angles (°) | ||||||
| Validation | ||||||
| MolProbity score | 2.46 | |||||
| Clashscore | 24.58 | |||||
| Poor rotamers (%) | 0.89 | |||||
| Ramachandran plot | ||||||
| Favored (%) | 88.77 | |||||
| Allowed (%) | 10.69 | |||||
| Disallowed (%) | 0.55 |
Pseudo-atomic sub-models of the entire complex were built and refined against the maps #2 (arm) and #3 (head segments), before being assembled into the holo-complex using the overall maps at lower resolution #1
The majority of condensin complexes (692,946 particles total, 100,388 particles refined) displayed two head domains that are in proximity, but are not in direct contact with each other as one would have expected if they had bound nucleotides (Fig. 1b, ‘non-engaged’ state). We determined the density in the vicinity of the ATPase head domains to correspond to the Ycs4Cnd1,NCAPD2 HEAT-repeat subunit, which binds predominantly to the Smc4 head domain20. We were furthermore able to assign a less well-defined density in the vicinity of the head domains to the second HEAT-repeat subunit Ycg1Cnd3,NCAPG (see below). Validating our subunit assignment, this additional density was absent in 2D classes obtained from tetrameric condensin complexes that did not contain the Ycg1 subunit (Fig. 1c, Extended Data Fig. 1a, b, Supplementary Note, Supplementary Fig. 1). A smaller fraction of complexes (136,570 particles total, 24,593 particles refined) displayed Smc2 and Smc4 heads that were separated by a considerably larger distance. In this state, the Ycs4 HEAT-repeat subunit binds to both SMC head domains and thereby serves as a spacer that bridges the two heads (Fig. 1b, ‘bridged’ state), while we did not observe clearly defined density for the Ycg1 HEAT-repeat subunit.
We used the pentamer dataset to obtain medium-resolution 3D density maps of the apo non-engaged (8.1 Å resolution; Fig. 1a, Extended Data Fig. 1c) and apo bridged (9.2 Å resolution; Extended Data Fig. 1d) states. We then performed focused refinements on the combined pentamer and tetramer data sets to obtain higher-resolution 3D maps of the Smc2–Smc4 coiled-coil arm segment (5.3 Å and 7.8 Å resolution; Extended Data Fig. 2) and of the ATPase head domain segment (4.2 Å and 7.5Å resolution; Extended Data Fig. 3) in the non-engaged and bridged states (Fig. 1d).
Conformation of the Smc2–Smc4 dimer in condensin apo complexes
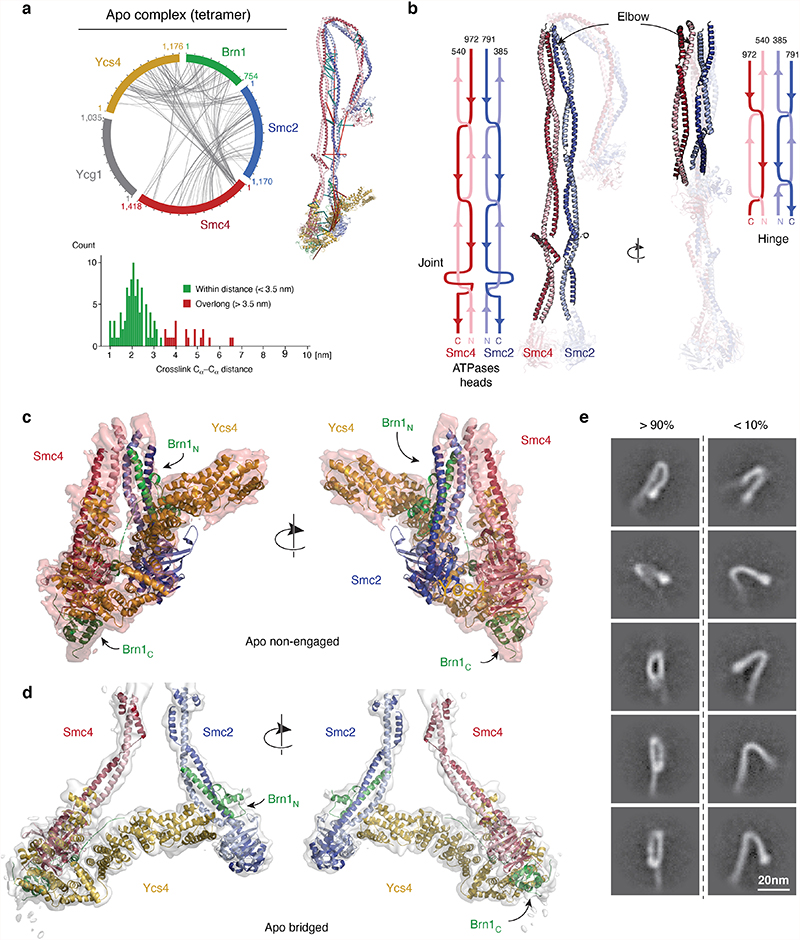
From these maps, we built complete pseudo-atomic representations of the of the nucleotide-free apo condensin complex in the non-engaged state (Fig. 2a) and in the bridged state (Fig. 2b). To obtain accurate models, we used high-resolution crystal structures of individual subcomplexes20,21 and models of the Smc2 and Smc4 coiled-coil segments based on crosslink mass spectrometry data obtained from soluble condensin pentamer complexes (Fig. 3a, Extended Data Fig. 4a–c, Supplementary Table 1). In both states, the aligned, rod-forming coiled coils of Smc2 and Smc4 bend sharply at the elbow region situated about two thirds of their lengths distant from the ATPase head domains. The elbow kink in the coils enables the Smc2–Smc4 hinge dimerization domains to fold back and contact the coils in the majority of classes (~90 %; Extended Data Fig. 4d).
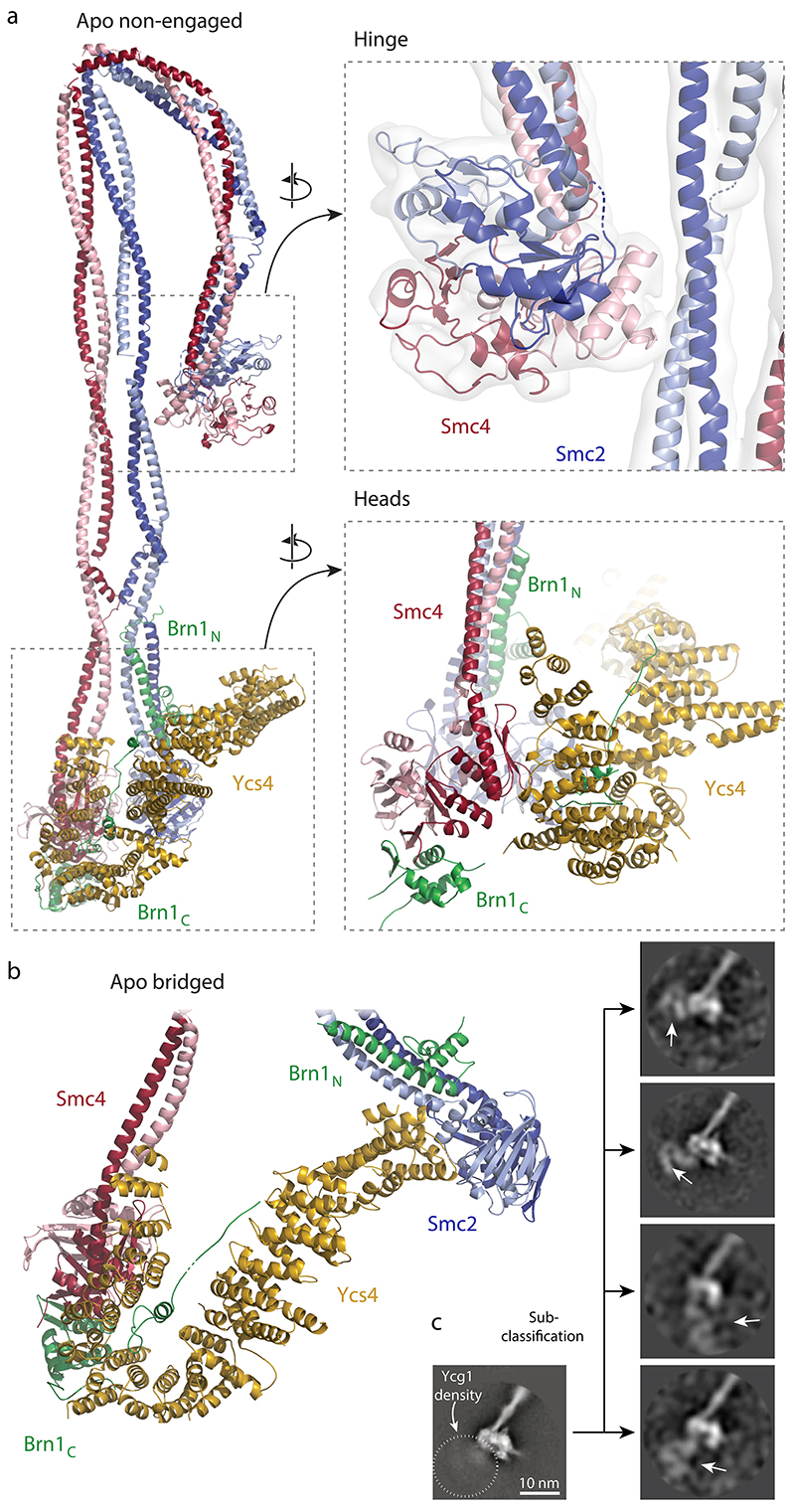
Fig. 2. Atomic models of the apo condensin holo complex.
a, Complete pseudo-atomic model of the non-engaged state. b, Pseudo-atomic model of the head segment of the bridged state c, Sub-classification of 2D class averages pinpoints positions of the Ycg1 subunits in the non-engaged state.
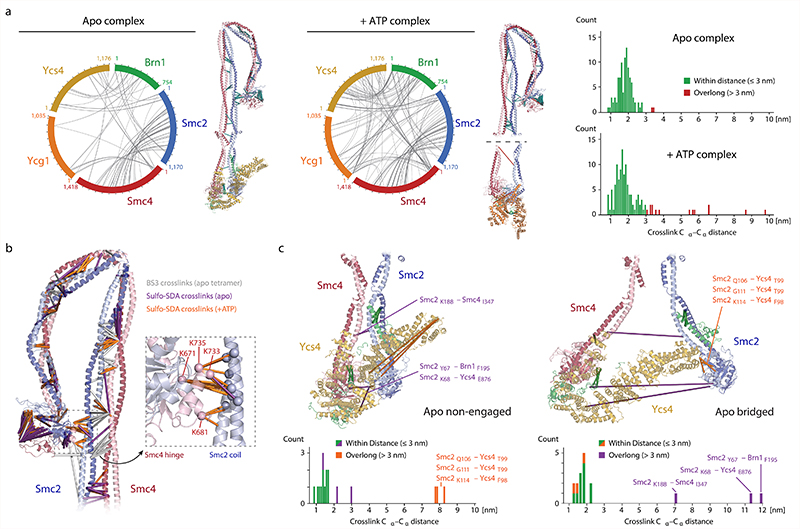
Fig. 3. Crosslink mass spectrometry.
a, Circle plots of inter-molecular sulfo-SDA crosslinks identified in condensin pentamer complexes in the absence (apo) or presence (+ATP) of nucleotide with an FDR of < 1%. Bar plots show the distance distribution of crosslinks, indicating the fraction of overlong links (>30 Å; ~5 % for apo, ~13 % for +ATP). b, Inter-molecular BS3 and sulfo-SDA crosslinks mapped on the Smc2–Smc4 coiled coil segment. c, Inter-molecular sulfo-SDA crosslinks mapped onto the non-engaged (left) and bridged (right) states on the apo head segment. Bar plots as in a, overlong links in each state are explained by the other state.
The quality of the density map allowed us to trace the Smc2 and Smc4 coiled coils through the entire elbow region and assign the two halves of the doughnut-shaped hinge dimerization domain. This revealed that only the Smc4 half of the hinge domain contacts the Smc2 coiled coil (Fig. 2a). The contact was confirmed by the previously mentioned crosslink mass spectrometry (Fig. 3b), which rules out that the folded conformation was an artifact of sample preparation for cryo-EM. The folded-elbow architecture of condensin is reminiscent of the conformations recently observed in negative-stain electron micrographs of two other SMC complexes, S. cerevisiae cohesin and Escherichia coli MukBEF22. It is furthermore consistent with coiled-coil kinking observed in low-resolution rotary shadowing electron and atomic force microscopy images of Smc2–Smc4 dimers or condensin pentamer complexes23,24.
At the apices of the coiled coils, the Smc2 and Smc4 ATPase head domains are separated by ~2 nm in the non-engaged state (Fig. 2a). Their Walker A, Walker B and ABC signature motifs face each other in roughly the proper orientation for engagement, but are too far away to sandwich two molecules of ATP between them. The lower local resolution of the Smc2 head density (~5.5 Å) suggests that, in this state, its position is not fixed rigidly with respect to the rest of the complex. This notion of flexibility is supported by the bridged state, in which the Smc2 head bends outwards by ~50° and moves away from the Smc4 head by ~10 nm (Fig. 2b, Extended Data Fig. 5a). This motion seems to be made possible by a disruption of the Smc2 coiled coil near the ‘joint’ region, which could serve as a pivot point (Supplementary Video 1). In both states, non-engaged and bridged, the Smc2 and Smc4 head domains are connected by the binding of the amino-terminal helical domain of the Brn1 kleisin subunit to the Smc2 coiled coil neck and of the Brn1 carboxy-terminal winged helix domain to the distal cap surface of the Smc4 head, respectively, as observed in previous structures of condensin sub-complexes20 and of homologous interfaces of cohesin25,26 and prokaryotic SMC complexes27.
Positions of the HEAT-repeat subunits
In the non-engaged state, the Ycs4 HEAT-repeat subunits binds to the ‘W-loop’ interface of the Smc4 head domain, in an identical orientation as observed in a recent co-crystal structure20 (Fig. 2a). In this orientation, the concave surface of Ycs4 HEAT repeats 7 and 8 forms a positively charged groove that extends between the ‘neck’ coiled coils of the disengaged Smc2 and Smc4 ATPase head domains (Extended Data Fig. 5b). Based on the findings that DNA binds to the corresponding surfaces of the Ycg1 subunit28 and the Scc3 HEAT-repeat subunit of cohesin29, as well as to the neck regions of head domains of the SMC-related Rad50–Mre11 DNA damage repair complex30, it seems justified to assume that this channel provides an interaction site for DNA within the condensin complex. A side-by-side comparison with the latter structure suggests that there is sufficient space between the neck regions of the Smc2–Smc4 coiled coils in the non-engaged state to accommodate a DNA double helix (Extended Data Fig. 5c). All SMC-associated HEAT-repeat subunits might therefore bind DNA at the concave surface of their alpha-helical solenoid. In the bridged conformation, Ycs4 maintains the contact with the Smc4 head domain via its carboxy-terminal HEAT repeats, as observed in the non-engaged state, while it simultaneously binds to the surface just above the nucleotide binding pocket of the Smc2 head domain via its first two amino-terminal HEAT repeats (Fig. 2b). The bridged state explains protein-protein crosslinks that cannot be attributed to the non-engaged state and that confirm the Smc2–Ycs4 contact (Fig. 3c).
To pinpoint the location of the Ycg1 HEAT-repeat subunit in the non-engaged state, we further sub-classified the images of the Ycs4-bound Smc2–Smc4 ATPase head domains obtained from focused refinement of the condensin pentamer data. This produced 2D class averages that clearly identified density for the Ycg1 subunit in very different positions close to the head domains (Fig. 2c, Supplementary Video 2). These data suggest that Ycg1 is, in contrast to Ycs4, only flexibly connected to the rest of the complex, most likely via its known binding to the central region of the Brn1 kleisin subunit.
Structure of the condensin complex in the presence of ATP and AMP-PNP (+ATP)
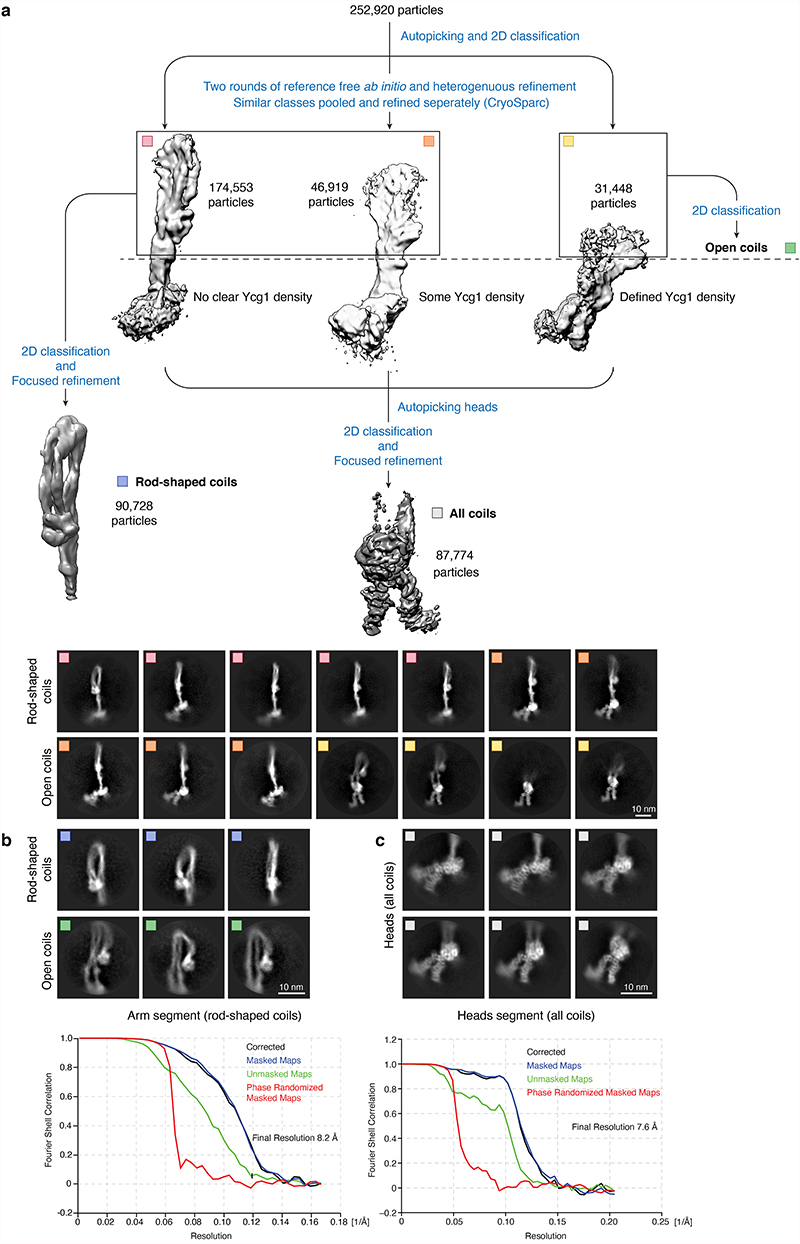
To gain insights into the conformational changes that accompany the condensin reaction cycle, we recorded cryo-EM datasets of the same S. cerevisiae condensin pentamer complex in the presence of an equimolar mixture of ATP and its non-hydrolysable analog adenylyl-imidodiphosphate (AMP-PNP; Table 2). The 2D classes generated from these images displayed a considerably higher degree of structural heterogeneity, both in the coiled-coil arm segment and in the ATPase head segment, when compared to those of the apo states (For examples of 2D classes, see Fig. 4a and Extended Data Fig. 6a). Although all classes for which we were able to trace the entire Smc2–Smc4 dimer maintained the elbow-folded coiled-coil conformation, distances between the two aligned coiled coils varied considerably, from closely aligned, rod-shaped coiled coils that matched the conformation seen in the apo structure, to coiled coils that separated by 2–3 nm in the region between the heads and the elbow. Out of 252,900 recorded particles of the ‘+ATP’ states, 174,553 particles (69 %) displayed rod-shaped coiled coil, while 78,367 particles (31 %) displayed more open coiled coils (Extended Data Fig. 6a).
Table 2. Cryo-EM data collection, refinement and validation statistics of condensin +ATP states.
| +ATP head segment (EMDB-10944) (PDB 6YVD) | +ATP rod-shaped arm segment (EMD-10964) | |
|---|---|---|
| Data collection and processing | ||
| Magnification | 81,000 | 81,000 |
| Voltage (kV) | 300 | 300 |
| Electron exposure (e–/Å2) | 45 | 45 |
| Defocus range (μm) | –1.5 ~ –2.5 | –1.5 ~ –2.5 |
| Pixel size (Å) | 1.70 | 1.70 |
| Symmetry imposed | C1 | C1 |
| Initial particle images (no.) | 425,957 | 425,957 |
| Final particle images (no.) | 87,774 | 90,728 |
| Map resolution (Å) FSC threshold | 7.6 at 0.143 FSC threshold | 8.2 at 0.143 FSC threshold |
| Map resolution range (Å) | 6.5 – 10.5 | 7.5 – 11 |
| Refinement | ||
| Initial model used (PDB code) | Homology model derived from 5OQQ, 6QJ1, 6QJ2 |
|
| Model resolution (Å) | 8.4 | |
| FSC threshold | 0.143 | |
| Model resolution range (Å) | n/a | |
| Map sharpening B factor (Å2) | -600 | |
| Model composition | ||
| Non-hydrogen atoms | 8,732 | |
| Protein residues | 1,760 | |
| Ligands | n/a | |
| B factors (Å2) | ||
| Protein | 41.20 | |
| Ligand | n/a | |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | |
| Bond angles (°) | 1.410 | |
| Validation | ||
| MolProbity score | 1.30 | |
| Clashscore | 0.80 | |
| Poor rotamers (%) | 0.00 | |
| Ramachandran plot | ||
| Favored (%) | 89.94 | |
| Allowed (%) | 8.38 | |
| Disallowed (%) | 1.68 |
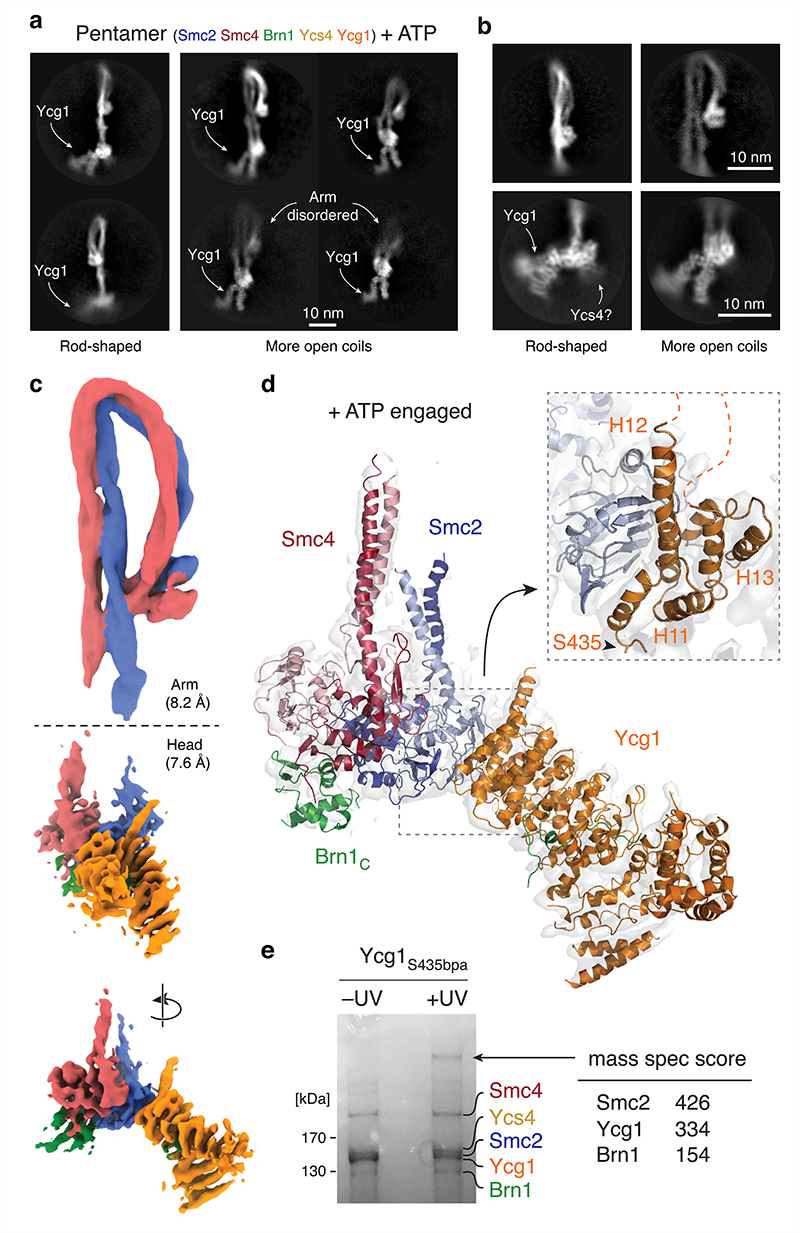
Fig. 4. Cryo-EM structure of the condensin holo complex in the presence of ATP.
Representative 2D class averages of a, pentamer complexes with rod-shaped (left) or more opened coiled coils (right) and of b, coiled-coil arm (left) or ATPase head (right) segments from a set of classes shown in Extended Data Fig. 6. c, 3D maps of arm and head segments in the +ATP state. d, Pseudo-atomic model of the +ATP head segment. e, Coomassie-stained SDS-PAGE gel of Ycg1S435bpa condensin purified from yeast before (–UV) or after (+UV) photo-crosslinking. Mass spec identification scores of proteins in the crosslinked band are listed. Uncropped images for e are available as Source Data.
In contrast to the apo structures, we no longer detected ordered density at the ATPase head segments that we could assign to the Ycs4 HEAT-repeat subunit. Instead, the additional density connected to the head domains had a different shape and position and unambiguously matched the crystal structure of the Ycg1 HEAT-repeat subunit28 (Fig. 4a). We used the same focused refinement strategy as for the apo complex to obtain 2D class averages (Fig. 4b, Extended Data Fig. 6a) and higher-resolution density maps (Fig. 4c, Extended Data Fig. 6b) of the coiled-coil arm segment in the rod-shaped conformation (8.2 Å resolution) and of the ATPase head segment (7.6 Å resolution). This selective approach revealed a conformation of the rod-shaped coiled coils that is essentially indistinguishable from the folded-elbow conformation of the coiled-coil arm segment observed in the apo states. It is important to remember, however that this state only represented 69 % of the observed particles. Crosslink mass spectrometry in the presence of ATP confirmed that the Smc2 and Smc4 coiled coils are able to align closely and that the Smc4 half of the hinge domain can contact the Smc2 coiled coil also in the +ATP state (Fig. 3a, Supplementary Table 1).
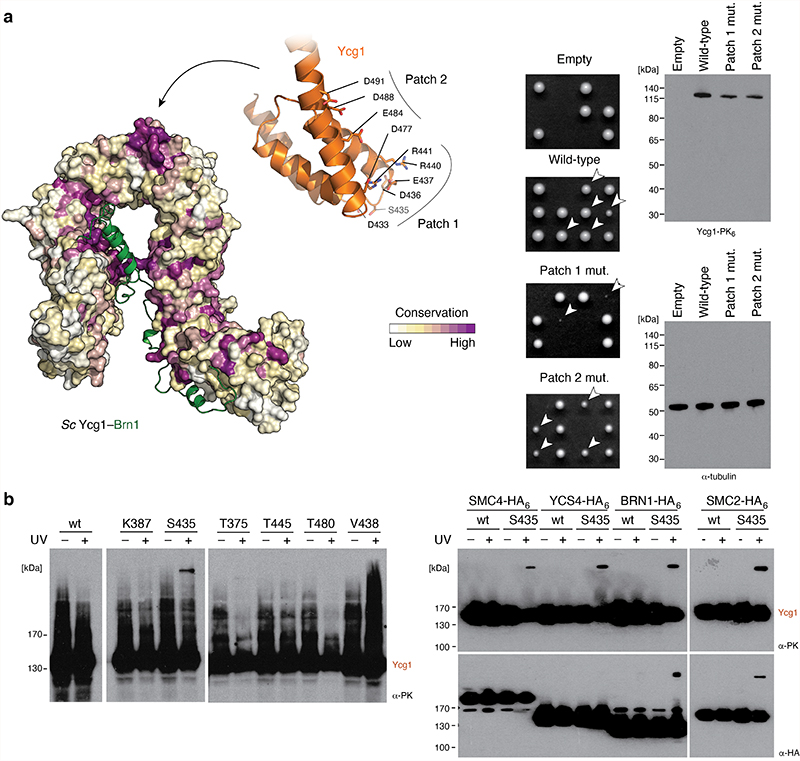
We docked individual high-resolution crystal structures of condensin subcomplexes into the 3D map of the ATPase head segment (Fig. 4d). In the resulting model, Ycg1 contacts exclusively the Smc2 head domain through HEAT repeats 11–13 of the convex side of its solenoid turn. Mutation of two patches of conserved amino acid residues at the Smc2 interface of Ycg1 reduced cell viability in budding yeast (Extended Data Fig. 7a, Supplementary Note, Supplementary Table 2), which is consistent with the notion that binding of Ycg1 to the ATP-engaged Smc2–Smc4 heads is physiologically relevant for condensin function. Mass spectrometry analysis (Fig. 4e, Supplementary Table 1) and immunoblotting (Extended Data Fig. 7b) of photo-crosslinking products between a version of Ycg1 that contains a p-benzoyl-phenylalanine (bpa) residue adjacent to the conserved patches (Supplementary Table 2) furthermore confirmed that this region binds Smc2 also in a cellular context.
Conformational changes in the presence of ATP
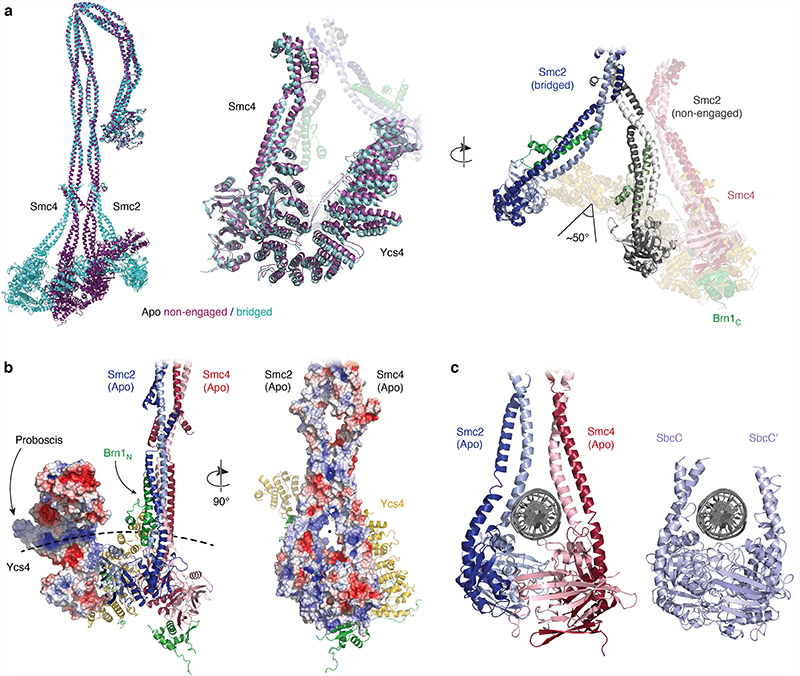
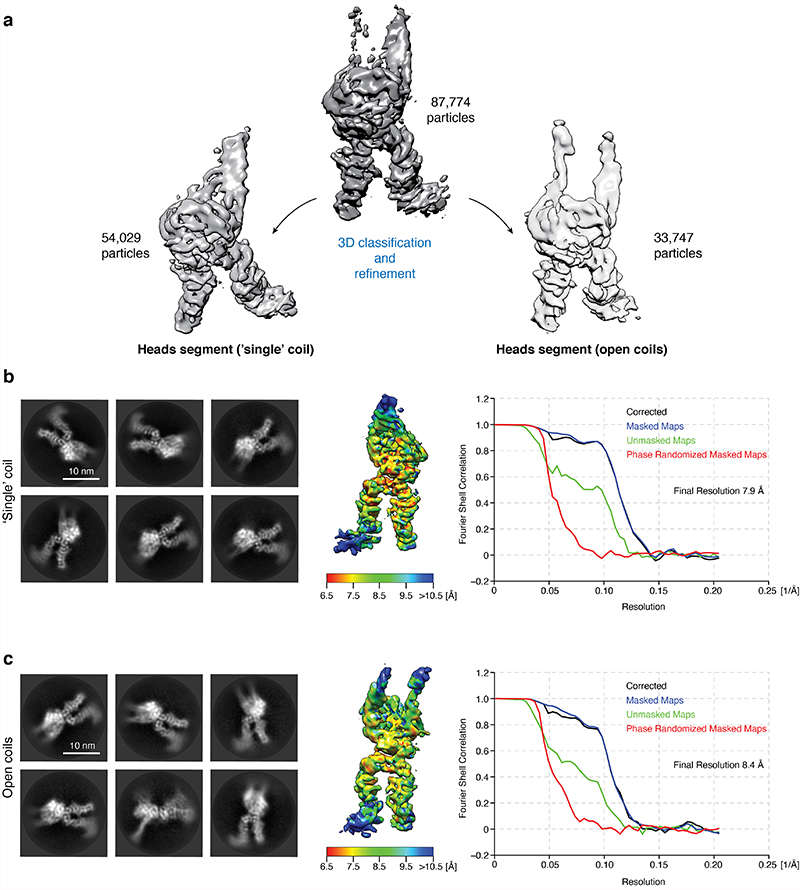
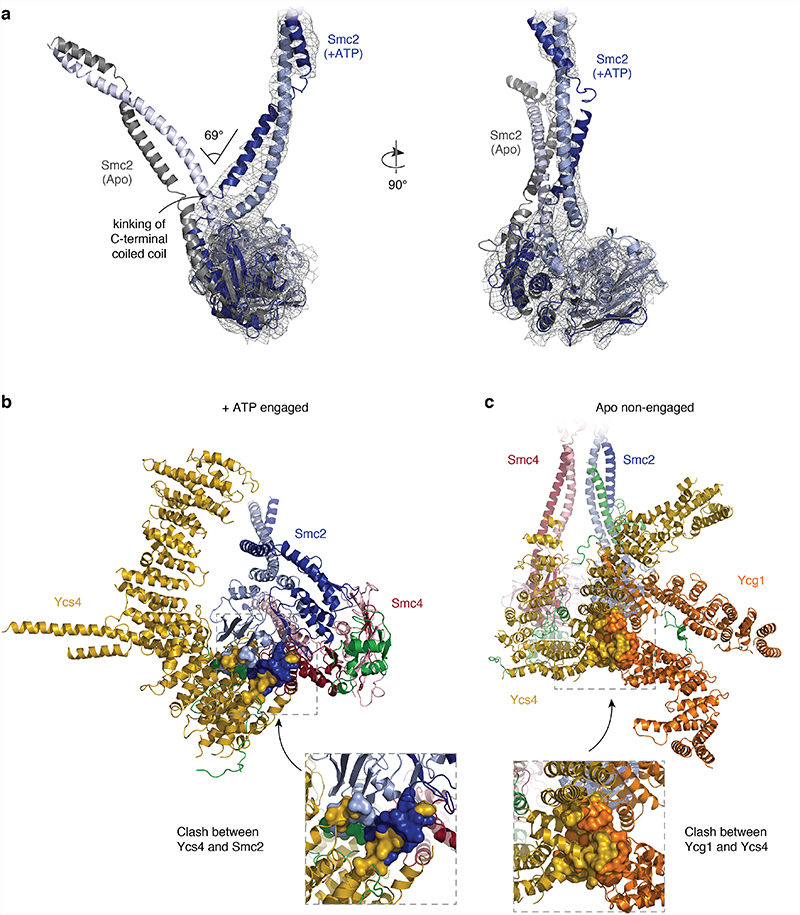
Further sub-classification of the ATPase head domain segment in the +ATP state (Extended Data Fig. 8) divided the 3D classes into one class that lacks clear density for the Smc2 coiled coil (7.9 Å resolution; 54,029 particles) and one class where both coiled-coils were clearly visible and separated further than in the non-engaged apo state (8.4 Å resolution; 33,747 particles). We generated a pseudo-atomic model for the latter class, capturing the open coiled-coil conformation (Fig. 5a, bottom). In this model, both head domains are fully engaged, as one would expect when two molecules of ATP are sandwiched between the composite nucleotide-binding pockets of Smc2 and Smc4. We therefore refer to this state as the ‘+ATP engaged’ state. Comparison to the conformation of the apo structure (Fig. 5a, top) revealed that the Smc2 coiled coil undergoes a rotational (~50 degrees) and translational (~20 Å) movement during head engagement (Supplementary Video 3). This results in the formation of a pronounced kink in the carboxy-terminal helix of the Smc2 coiled coil (Fig. 5a, Extended Data Fig. 9a). Extrapolating from earlier findings20, kinking of the Smc2 coiled coil might serve as a mechanism to temporarily eject the amino-terminal helical domain of Brn1. This notion is consistent with the absence of density for this domain in the 3D map of the engaged Smc2–Smc4 heads as revealed here.
Fig. 5. ATP-dependent exchange of the HEAT-repeat subunits at the condensin heads.
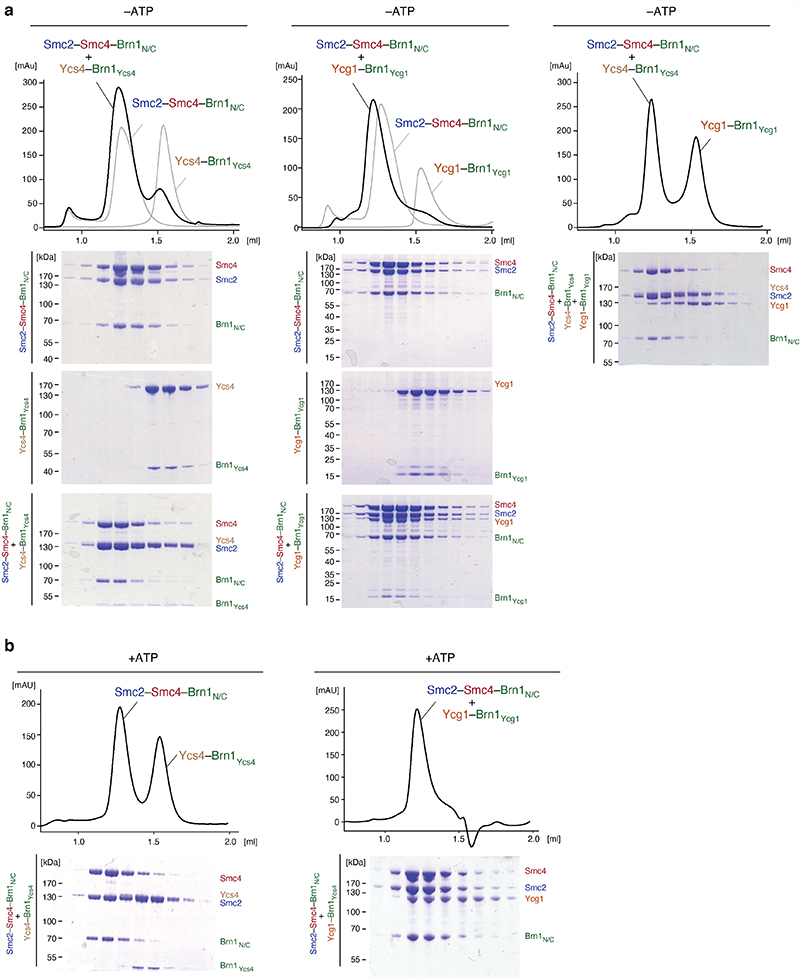
a, Comparison of the ATPase head structures in the non-engaged apo state and the +ATP engaged state. Angles indicate the kink in the carboxy-terminal Smc2 coiled-coil helix. b, Analytical size exclusion chromatography profiles of Chaetomium thermophilum Smc2–Smc4–Brn1NC with either Ycs4–Brn1Ycs4 (top left), Ycg1–Brn1Ycg1 (top right), or both (bottom left and gel bottom right; a cropped version of the same gel is also shown in Extended Data Fig. 10a).
Furthermore, we only detected density for the ‘buckle’ segment but not for the ‘latch’ segment of the Brn1Ycg1 safety belt loop within the U-shaped Ycg1 subunit, which encircles the DNA double helix bound to the composite Ycg1–Brn1Ycg1 groove in a previous co-crystal structure28 (Fig. 5a). This raises the possibility that an open safety belt conformation prevails when Ycg1 binds to the ATP-engaged Smc2–Smc4 heads. Although we were unable to find clearly defined density for the Ycs4 subunit in any of the +ATP engaged 2D classes, we observed diffuse density in the vicinity of the Smc4 head domain in some classes (Fig 4b, Extended Data Fig. 6c). This indicates that Ycs4 remains close to the position it occupies in the apo state but that the mobility of Ycs4 is greatly increased in the presence of ATP. We propose that Ycs4 maintains the connection to the rest of the complex in the +ATP state merely via its previously reported binding to the flexible central region of the Brn1 kleisin subunit31, as we found was the case for Ycg1 in the apo state.
Ycs4 and Ycg1 binding is mutually exclusive
Our structures suggest that Ycs4 binding to the Smc4 head domain not only sterically prevents Smc2–Smc4 head engagement upon nucleotide binding (Extended Data Fig. 9b), but similarly clashes with the binding of Ycg1 to the Smc2 head domains, even when the heads are not engaged (Extended Data Fig. 9c). To test this notion experimentally, we purified Smc2–Smc4 dimers bound to a minimal version of Brn1 that lacks the central binding regions for both HEAT-repeat subunits (Smc2–Smc4–Brn1NC). We then assayed whether either HEAT-repeat subunit bound to its central interaction region of Brn1 (Ycs4–Brn1Ycs4 or Ycg1–Brn1Ycg1) was able to form a complex with Smc2–Smc4–Brn1NC. Ycs4–Brn1Ycs4 or Ycg1–Brn1Ycg1 alone formed a stable complex with Smc2–Smc4–Brn1NC in the absence of nucleotide (Fig. 5b, Extended Data Fig. 10a). Addition of ATP prevented binding of Ycs4–Brn1Ycs4, as expected from previous experiments20, but not of Ycg1–Brn1Ycg1 (Extended Data Fig. 10b). When we added both, Ycs4–Brn1Ycs4 or Ycg1–Brn1Ycg1, simultaneously in the absence of nucleotide, Ycs4–Brn1Ycs4 indeed prevented binding of Ycg1–Brn1Ycg1 to the Smc2–Smc4–Brn1NC complex (Fig. 5b, Extended Data Fig. 10a). Hence, ATP-dependent release of Ycs4 from Smc4 is a prerequisite for Ycg1 binding to the engaged Smc2–Smc4 heads.
Discussion
Based on our cryo-EM structures of yeast condensin in different functional states and on biochemical data, we propose a model for the sequence of conformational changes that take place during the condensin ATPase cycle (Fig. 6). In the absence of nucleotide, the carboxy terminus of the Ycs4 HEAT-repeat subunit is tightly bound to the Smc4 ATPase head domain. The position of the Smc2 ATPase head domain presumably alternates between two states. In the non-engaged state, the Smc2 and Smc4 ATPase head domains are separated by only a couple of nanometers as a consequence of their aligned coiled coils, although the lower local resolution of the Smc2 head density suggests that it might be flexibly attached. In the bridged state, the Smc2 head binds to the amino terminus of Ycs4, thereby separating the Smc2 and Smc4 ATPase heads by a distance of ~10 nm. The transition between non-engaged and bridged states is presumably enabled by the discontinuity of the SMC coiled coils at the joint region32. Future experiments will have to address the physiological relevance of the two states, but it is tempting to speculate that a widening of the channel between the DNA-binding coiled coil segments close to the head domain in the bridged state might allow DNA to load more easily once it has entered that compartment (see below).
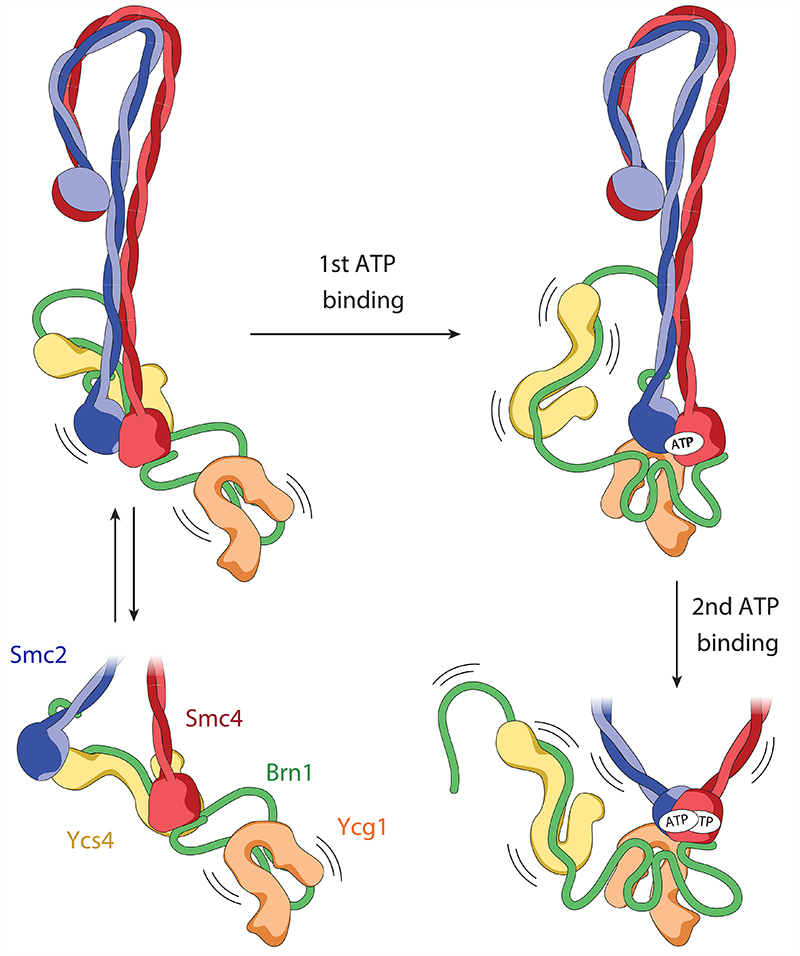
Fig. 6. Flip-flop model of the condensin reaction cycle.
In the absence of ATP, the Smc2 and Smc4 ATPase domains are either separated by ~2 nm, with Ycs4 bound to the Smc4 head (non-engaged state) or separated by ~10 nm with Ycs4 bound to both, the Smc2 and the Smc4 heads (bridged state). In both states, Ycg1 is flexibly attached to the rest of the complex only via its binding to the central region of Brn1. ATP binding to the Smc4 active site releases Ycs4, which enables Ycg1 binding to Smc2 and initiates Smc2–Smc4 head engagement. The head domains then fully engage upon ATP binding at the Smc2 active site. Kinking of the Smc2 coiled coil triggers release of Brn1N and (at least partially) opens the coiled coils from their rod-shaped conformation.
In both apo states, the rod-shaped coiled coils bend sharply at the elbow region to fold the hinge back onto the coils at a distance of ~15 nm from the ATPase heads. This distance is considerably larger than the hinge-head distance measured in the elbow-folded conformation of cohesin or MukBEF complexes22. Analysis of evolutionary conservation of amino acids in the two contact regions, especially in the Smc4 coiled coil, does not indicate increased sequence preservation (not shown). Because we could not assign the register along the Smc4 coiled coil in the contact area with absolute certainty, we were also not able to pinpoint individual residues that make the contact. It seems that an unspecific, low-affinity contact between the hinge and the coiled coil might in fact be required for the folding to be dynamic, not generating binding energy that traps the hinge domains near the coiled coils. We conclude that it might rather be the mechanical property of coiled-coil folding, rather than the point of contact between hinge and coiled coils, that is functionally important. A more detailed analysis will be required to test this notion. Although a folded-elbow conformation could not be observed in electron micrographs of Bacillus subtilis SMC complexes21, the prevalence of this feature in MukBEF, cohesin and condensin nevertheless implies that it is a conserved feature for at least some part of the SMC reaction cycle.
Consistent with previous experiments, which suggested that two molecules of ATP bind sequentially at the two active sites of the Smc2–Smc4 head domains20, our cryo-EM structures suggest that nucleotide binding to the Smc4 ATP-binding pocket unbinds Ycs4 from the Smc4 head, but not from the central region of the Brn1 kleisin. Release of Ycs4 in turn enables Ycg1 binding to the Smc2 head and the engagement of the latter with the Smc4 head upon binding of a second nucleotide at the Smc2 ATP-binding pocket (Fig. 6). Although, at the current resolution, we cannot distinguish whether AMP-PNP, ATP or ADP (as a result of nucleotide hydrolysis) is bound to each active site in the +ATP engaged state, we noticed that condensin complexes with mutations in the Smc2–Smc4 Walker B motifs that drastically reduce ATP hydrolysis rates produced very similar 2D classes in the presence of ATP as wild-type complexes in the presence of ATP and AMP-PNP (not shown). This strongly suggests that our +ATP engaged structure represents the pre-hydrolysis state, or that nucleotide hydrolysis (but not release) does not result in major conformational changes in the architecture of the condensin complex.
The exchange of the two HEAT-repeat subunits at the ATPase head domains, akin to a flip-flop mechanism, might provide the means, or the conformational flexibility, to translocate DNA molecules that are bound within the Ycg1–Brn1 safety belt28, within a groove generated by Ycs4 and the Smc2–Smc4 coiled coils immediately adjacent to the ATPase heads (Extended Data Fig. 5b,c) or both, in alternating, ATP-driven cycles. If DNA were indeed positioned between the Smc2 and Smc4 coiled coil, it is conceivable that the bridged state serves a role in allowing access or release of DNA duplexes into or out of the tight groove. The heterogeneity in the coiled-coil conformations and the opening of the coiled-coil angles in the presence of ATP suggest that the coils are then able to separate further from a rod-shaped into a larger, more open architecture21,32, similar to the ones frequently observed by atomic force and electron microscopy33. Unfortunately, the coiled-coil flexibility observed in these studies possibly makes alignments of open coiled coils in cryo-EM images difficult due to their inherent heterogeneity. To understand the architectural dynamics of condensin, it will be important to distinguish on one hand between unzipping of the rod conformation, and on the other hand a possible straightening at the elbows. Future studies will be required to capture the dynamic changes between the states of the condensin complex and its interaction with DNA. We propose that the large-scale conformational changes reported here form the basis for the ability of condensin, and likely all SMC protein complexes, to translocate along DNA and to extrude DNA loops of kilobase pairs in length.
Methods
Cryo-EM grid preparation
For the collection of condensin apo state datasets, aliquots of purified condensin tetramer or pentamer samples (see Supplementary Note) were thawed and injected onto a Superose 6 Increase 3.2/300 column (GE Healthcare) in nucleotide-free buffer (25 mM Tris-HCl, 125 mM NaCl, 1 mM TCEP, pH 7.5), prior to applying samples onto EM grids. Peak fractions were immediately used for cryo-EM grid preparations, using 3–3.5 μL of sample at a concentration of 0.15–0.25 mg/mL that was applied to freshly glow-discharged Cu/Rh 2/2 holey carbon 200 mesh grids (Quantifoil). The grids were blotted for 1.5–2 s at 4 °C and 100 % humidity, and were flash frozen using an FEI Vitrobot Mark IV (Thermo Fisher) and a liquid-ethane cryostat set to –180 °C34. For the collection of +ATP-state datasets, purified S. cerevisiae condensin pentamer complexes (see Supplementary Note) were diluted into 25 mM HEPES-NaOH pH 7.5, 125 mM NaCl, 1 mM MgCl2 and 1 mM DTT to a final concentration of 0.4 μM. After incubation with a mixture of 1 mM ATP and 1 mM AMP-PNP for 10 min, 3 μL were applied onto an UltrAufoil mesh 200 R1.2/1.3 grid (Quantifoil). Plunge freezing was carried out at 10 °C and 100 % humidity on an FEI Vitrobot Mark IV (Thermo Fisher) with a wait time of 2 min, blot force 3 and blot time 3. The blotting paper for the bottom side of the grid was replaced by a piece of Teflon to achieve one-sided blotting.
Cryo-EM data collection
Please refer to Tables 1 and 2 for data statistics. For the apo states, images were recorded on a Titan Krios electron microscope (FEI) equipped with a K2 or K3 summit direct electron detector (Gatan), mounted behind a GIF Quantum energy filter. Images were collected automatically using EPU (Thermo Fisher) or SerialEM35. A total of 13,628 micrographs of condensin tetramer grids were collected with the K2 camera in counting mode through 8 separate data collection sessions with total doses of 44–55 electrons per Å2 during exposure times of 8–12 s, dose-fractionated into 40–55 movie frames, at defocus ranges of 1.6–3.6 μm. The magnifications used were 105,000× or 130,000×, resulting in physical pixel sizes of 1.16, 1.15, 1.047, 1.055, 1.065 Å per pixel. Three of the eight tetramer datasets with the K2 camera were collected at tilts of 40, 30 and 35°, respectively. ~10,500 micrographs of condensin tetramer grids were collected using the K3 camera in super-resolution mode (at a pixel size of 1.085 Å per pixel) with a total dose of 40 electrons per Å2 during a total exposure time of 2.25 s, dose-fractionated into 40 movie frames, at a defocus range of 1.8–3.3 μm. A total of ~10,000 micrographs of condensin pentamer grids were collected with the K2 camera and a Volta phase plate (VPP)36 in counting mode (at a pixel size of 1.065 Å per pixel) with a total dose of 45 electrons per Å2 during a total exposure time of 9 s, dose-fractionated into 35–42 movie frames. A defocus range of 0.5–0.9 μm was used for VPP data collection. For the +ATP state, two datasets (4,158 micrographs and 4,894 micrographs, respectively) were acquired on a Titan Krios electron microscope (FEI) running at 300 kV, using a Gatan K2 detector in counting mode and a Gatan GIF energy filter with a slit width of 20 eV. A nominal defocus range of –1 to –2.5 μm was used with a total electron dose of 45 electrons per Å2 divided over 40 frames (0.5 s/frame) and a pixel size of 1.7 Å/pixel (81,000× magnification). Since the data showed strong orientation bias, the stage was tilted by 30° during acquisition.
Image processing and 3D reconstruction
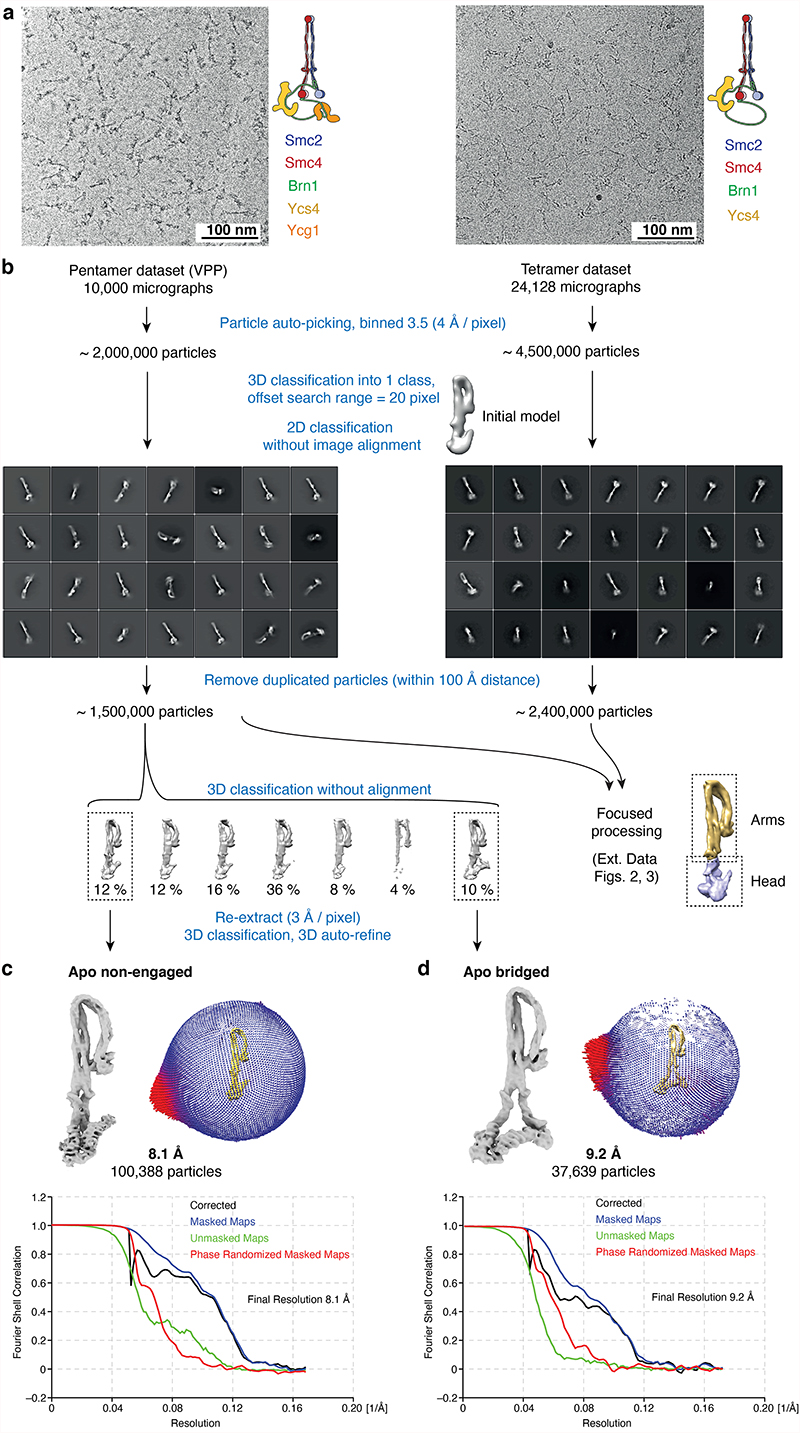
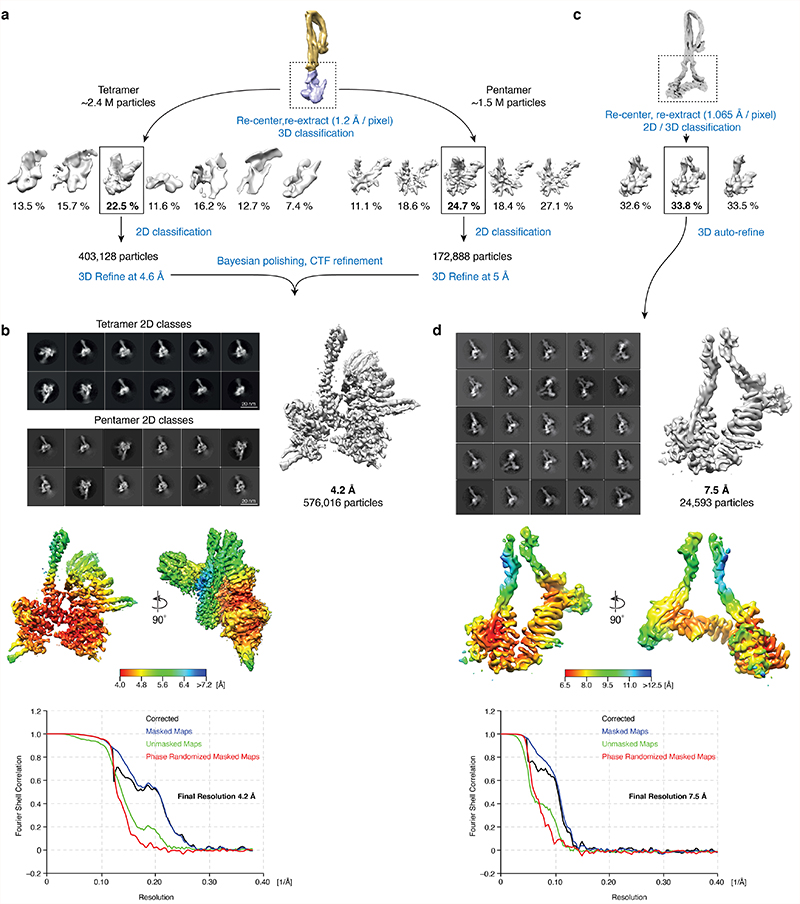
For the apo state, image processing was done in RELION 3.037. Movies were aligned using 5 × 5 patches in MotionCor2 with dose-weighting38. CTF parameters were estimated with Gctf39. For tilt datasets, local CTF estimation was performed using Gctf after initial processing. All refinements were performed using independent data half-sets (gold-standard refinement) and resolutions were determined based on the Fourier shell correlation (FSC = 0.143) criterion40. A tetramer dataset (750 images) was used for initial model calculation: Particles were picked manually from a few micrographs, and the resulting particles were binned and extracted with a 3602-pixel box size (2 Å/pixel) to generate 2D class averages. Good 2D class averages were used as a template for reference-based auto-picking. The resulting ~83 k particles were subjected to multiple rounds of 2D classifications, and a final ~19 k particles were used to calculate the initial 3D model in RELION (Extended Data Fig. 1b).
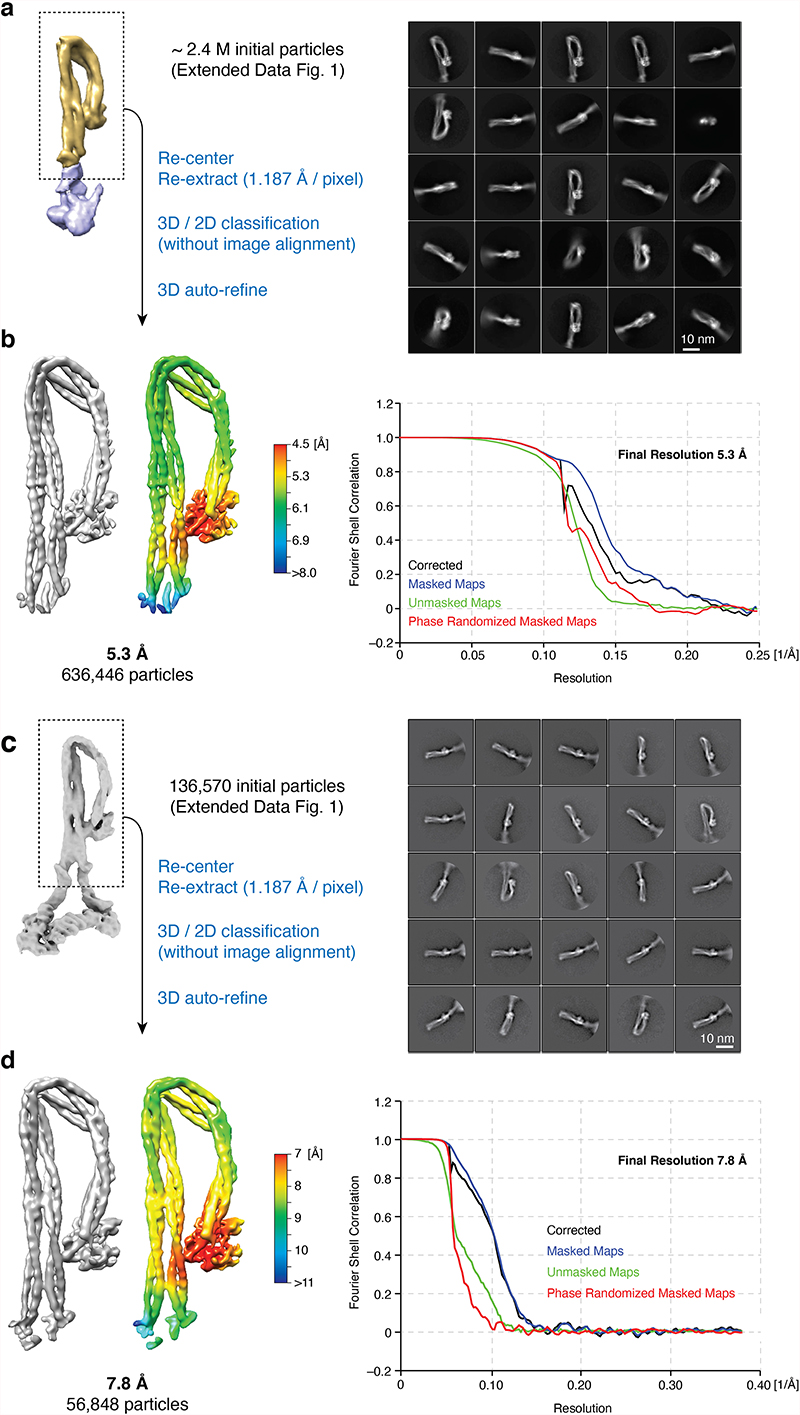
For each dataset, particle picking was performed separately using the same strategy, starting with auto-picking using initial 2D classes; the resulting particles were then downscaled to 4 Å/pixel (1802-pixel box size) for initial processing. Due to condensin’s elongated shape, most particles were not properly centered after auto-picking and rare views tended to be lost during 2D classification. To resolve this issue, particles were first aligned against the 40 Å low-pass filtered initial model by 3D classification into one class, with a large offset search range (20 pixels) and a limited E-step resolution of 10 Å. The rationale was that particle translations become consistent for all viewing angles in 3D alignment, while translations are consistent only within each class average in 2D classification. Aligned particles were then subjected to 2D classification without alignment to remove contaminations and overlapping particles. After selecting good classes, duplicated particles within 100 Å were removed. For tilted datasets, local CTF estimation was performed with the refined particle coordinates using Gctf, followed by re-extraction of particles. The resulting particles from each dataset were then merged for further processing (~ 2.4 M particle images of tetramer and ~ 1.5 M of pentamer) (Extended Data Fig. 1b). The angular assignments resulting from these initial steps were also used for focused 3D classifications for the condensin arm and head, after re-centering particles. The set of pentamer particle images obtained was subjected to further 3D classification into seven classes with no image alignment, and resulted in five similar shaped classes (apo non-engaged form) and a unique class (apo bridged form). Among the five classes of the apo non-engaged form, the best class with clear density for Smc2, Smc4, Brn1C and Ycs4 consisted of 100,388 particles. This class was selected and re-extracted in a 2402-pixel box (3 Å/pixel) and used for a final round of 3D refinement with a soft-edged mask and applying solvent-flattened FCSs. Post-processing with RELION yielded a final overall map of apo condensin at 8.1 Å resolution (Extended Data Fig. 1c). The class of the apo bridged form was re-extracted in a 2402-pixel box (3 Å/pixel) and the resulting 136,570 particles were subjected to another round of 3D classification into 3 classes. The best class consisting of 37,693 particles was selected and refined to 9.1 Å resolution (Extended Data Fig. 1d).
For the processing of the apo non-engaged condensin arm segment, which contains the coiled coil and hinge domains, the particles of the tetramer sample from the initial processing were re-centered and re-extracted with a box size of 3002 pixels (1.187 Å/pixel). An initial model for this part of condensin was produced by removing the density of the heads below the ‘joint’ from the overall apo map using UCSF Chimera41. After multiple rounds of 3D classifications and 2D classifications with no image alignments, 636,446 particle images were selected and 3D auto-refined. This resulted in a final map at 5.3 Å resolution (Extended Data Fig. 2). For focused refinement of the apo bridged arm segment, the 136,570 particles were re-centered and re-extracted in a box of 4002 pixels (1.187 Å/pixel). After further 3D and 2D classifications, the final 56,848 particles yielded a 7.8 Å resolution map (Extended Data Fig. 2c,d).
For focused refinement of the apo condensin head segment, which contains the Smc2–Smc4 ATPase domains, Brn1 and Ycs4, the particle images from the initial processing were re-centered on the heads and re-extracted in a box size of 3202 pixels (1.2 Å/pixel). Again, an initial model for this part was produced using UCSF Chimera41. The re-centered particle images of the tetramer heads (~2.4 M) were subjected to several rounds of 3D classification. The best class consisting of 403,128 particles was 3D refined to 4.6 Å resolution, followed by CTF refinement and Bayesian polishing in RELION42, resulting in a final resolution of 4.3 Å. The pentamer dataset (~1.5 M particle images) was processed with the same strategy and resulted in a ~5 Å resolution reconstruction with 172,888 particles. In the pentamer reconstruction, density for Ycg1 was not clearly visible, presumably because of its mobility with respect to the rest of the condensin complex. The final refined map of the pentamer sample was near-identical to the one obtained for the tetramer. Both datasets were hence merged at the level of refined particle images and the merged datasets yielded a combined and final 4.17 Å-resolution map of the apo non-engaged head segment (Extended Data Fig. 3b). For focused processing of the bridged head segment, the particles of the bridged class from the initial, overall processing were re-centered and re-extracted in a box size of 3382 pixels (1.065 Å/pixel). After 3D/2D classifications, the resulting 24,593 particles were selected and 3D auto-refined, resulting in a final map at 7.5 Å resolution (Extended Data Fig. 3d).
The processing strategy for the +ATP state is depicted in Extended Data Fig. 6. Pre-processing of 9,052 micrograph stacks was done using the implementations of MotionCor2 in 5×5 patches with dose-weighting38 and Gctf39 in RELION 3.037 and, if not stated otherwise, all further analyses were also carried out in RELION. Particles were auto-picked and extracted in a box of 3082 pixels (1.7 Å/pixel) and binned to a 1502-pixel box. After removing junk particles in reference-free 2D classifications, both datasets were combined and three initial maps were created de novo from 259,907 particles in cryoSPARC version 243 and then refined using heterogeneous refinement. The three refined maps were each subjected to a second round of ab initio and heterogenous refinement. Subsequently, similar classes were combined to yield the final three maps depicted in Extended Data Fig. 6a. The two classes with closed coiled coils were then combined, re-centered on the coiled-coil part, extracted in a box of 1802 pixel (3.4 Å/pixel), cleaned up by 2D and 3D classifications without image alignment and then refined to yield an 8.2 Å-resolution map (Extended Data Fig. 6b). Open coiled coils were re-centered and classified in 2D. In addition, all head parts were re-centered and extracted in a box of 1442 pixel (2.448 Å/pixel). In parallel, heads were auto-picked and combined with the re-centered head parts. After removal of duplicates within 70 Å, all head particles were cleaned up by 2D classification and refined to yield a 7.6 Å map (Extended Data Fig. 6c). A subsequent 3D classification resulted in a conformation with clear density for both coiled coils and a second form where density for the Smc2 coiled coil was largely absent (Extended Data Fig. 8a). Each class was refined to 8.4 Å and 7.9 Å, respectively (Extended Data Fig. 8b,c).
Model building
To generate a molecular model of the apo non-engaged Smc4 ATPase, the Brn1 carboxy-terminal domain and Ycs4, previous crystal structures of Smc4hd–Brn1C (PDB code 6QJ2) from C. thermophilum and Ycs4–Brn1Ycs4 from C. thermophilum (PDB code 6QJ4)20 were docked into the cryo-EM head segment map at 4.2 Å resolution using UCSF Chimera41. Manual modifications were performed in COOT44, including S. cerevisiae sequence assignment. Smc4 residues 151–365 and 1,234–1,414, Ycs4 residues 5–1,149 and Brn1 residues 643–748 were built manually based on the available density. Homology models of the Smc2 ATPase domain and the Brn1 amino-terminal domain were generated using as templates the crystal structure of Smc3hd–Scc1N from S. cerevisiae (PDB code 4UX3)25 and the NMR structure of Brn1N from C. thermophilum (PDB code 6Q6E)20. Modelling was performed using the SWISS-MODEL server45 and models were rigid-body docked into the cryo-EM map using UCSF Chimera41. Smc2 residues 2–212 and 986–1,167 and Brn1 25–108 were fit according to the map. To generate a pseudo-atomic model of the arm, elbow and hinge regions of condensin, the crystal structure of the hinge domain from S. cerevisiae (PDB code 4RSI, Smc2 residues 453–739, Smc4 residues 560–947)21 was fitted into the 5.3 Å map. Furthermore, the coiled coils were extended up to the elbow by building manually Smc2 residues 399–789 and Smc4 residues 544–969. The coiled-coil model of the region between the joint and the elbow (Smc2 residues 212–383, 792–936 and Smc4 residues 399–539, 773–1,114) was first built by placing poly-alanine chains into the density, and the sequences were tentatively assigned based on crosslink mass spec information and the map’s appearance. Finally, to obtain an overall atomic model of condensin, the above models of the arm and head regions were fitted into the overall low-resolution map at 8.1 Å resolution with UCSF Chimera41, and the missing residues linking the two parts were manually added using COOT44.
For the molecular model of the apo bridged state, Smc2hd–Brn1N (Smc2 residues 2–233, 962–1,167), Smc4hd-Brn1C (Smc4 residues 151–402, 1,159–1,414) and Ycs4–Brn1Ycs4 models were each taken from the apo non-engaged model and separately fitted by rigid-body fitting into the 7.5 Å bridged head map. Equally, the atomic model of the arm segment was taken from the non-engaged model and docked into the 7.8 Å arm segment map of the bridged state. Missing residues linking head and arm segments were added manually using COOT44.
For the molecular model of the +ATP state, the existing crystal structure of the S. cerevisiae Ycg1–Brn1Ycg1 complex (PDB code 5OQQ, chains A and C) was docked into the electron density using UCSF Chimera41. Ycg1 residues 499–507 of HEAT repeat 12 were manually added using COOT44. Brn1 residues not matching experimental electron density (residues 458–496) were deleted. For modelling the engaged Smc2–Smc4 head domains, the ‘helical’ and ‘RecA’ half-domains of C. thermophilum Smc2 (PDB code 6QJ1) and Smc4hd–Brn1C (PDB code 6QJ2) were placed individually. The Smc2 and Smc4 coiled-coil segments were adjusted in COOT44 starting from manually placed segments taken from the apo model.
Crosslink mass spectrometry
The crosslinkers BS3 (bis(sulfosuccinimidyl)suberate) and sulfo-SDA (sulfosuccinimidyl 4,4’-azipentanoate) (Thermo Scientific Pierce) were dissolved in crosslinking buffer (20 mM HEPES, 150 mM NaCl, 1 mM TCEP, and 5 % glycerol, pH 7.7) to 100 mM before use. For crosslinking with BS3, the purified condensin tetramer in crosslinking buffer (20 mM HEPES, 150 mM NaCl, 1 mM TCEP, and 5 % glycerol, pH 7.7) were incubated at 0.7 mg/mL with 2 mM BS3 for 2 h on ice and the reactions were quenched with 50 mM NH4HCO3 for 45 min at 4 °C. Reaction products were separated on a Criterion TGX 4–15 % SDS-PAGE gel (BioRad). The gel band corresponding to the crosslinked complex was excised and digested with trypsin (Thermo Scientific Pierce)46. The resulting tryptic peptides were extracted and desalted using C18 StageTips47.
For photo-crosslinking the nucleotide-free apo condensin pentamer with sulfo-SDA, 0.85 mg/mL purified complexes were incubated with sulfo-SDA using three different protein-to-crosslinker molar ratios (1:250, 1:500, 1:1,000) for 2 h at 4 °C. For photo-crosslinking of condensin in the presence of ATP, ATP (3 mM) and MgCl2 (5 mM) were added to the purified condensin pentamer at 0.85 mg/mL and crosslinking was performed using three protein-to-crosslinker molar ratios (1:300, 1:600, 1:1,200). The samples were irradiated with UV light at 365 nm for 20 min and quenched with 50 mM NH4HCO3. Subsequently, samples were denatured in 8 M urea, 100 mM NH4HCO3 derivatized with iodoacetamide and digested with LysC endoproteinase (Wako) for 4 h at 25 °C. After dilution of the sample to a urea concentration of 1.5 M, trypsin (Thermo Scientific Pierce) was added and the samples were digested for 16 h at 25 °C. The resulting tryptic peptides were extracted and desalted using C18 StageTips47.
Eluted peptides were fractionated using a Superdex Peptide 3.2/300 column (GE Healthcare) at a flow rate of 10 μL/min using 30 % (v/v) acetonitrile and 0.1 % (v/v) trifluoroacetic acid as mobile phase. 50 μl fractions were collected and dried. Samples for analysis were resuspended in 0.1 % (v/v) formic acid 1.6 % (v/v) acetonitrile. LC-MS/MS analysis was performed on an Orbitrap Fusion Lumos Tribrid mass spectrometer (Thermo Fisher) coupled on-line with an Ultimate 3000 RSLCnano system (Dionex, Thermo Fisher). Samples were separated on a 50-cm EASY-Spray column (Thermo Fisher). Mobile phase A consisted of 0.1 % (v/v) formic acid and mobile phase B of 80 % (v/v) acetonitrile with 0.1 % (v/v) formic acid. Flow rates were 0.3 μL/min using gradients optimized for each chromatographic fraction from offline fractionation, ranging from 2 % mobile phase B to 45 % mobile phase B over 90 min. Mass spec data were acquired in data-dependent mode using the top-speed setting with a three second cycle time. For every cycle, the full scan mass spectrum was recorded using the Orbitrap at a resolution of 120,000 in the range of 400 to 1,500 m/z. Ions with a precursor charge state between 3+ and 7+ were isolated and fragmented. Fragmentation by Higher-energy Collisional Dissociation (HCD) employed a decision tree logic with optimized collision energies48. The fragmentation spectra were then recorded in the Orbitrap with a resolution of 50,000. Dynamic exclusion was enabled with single repeat count and 60-seconds exclusion duration. A recalibration of the precursor m/z was conducted based on high-confidence (<1 % False Discovery Rate, FDR) linear peptide identifications. The re-calibrated peak lists were searched against the sequences and the reversed sequences (as decoys) of crosslinked peptides using the Xi software suite (v.1.6.745) for identification49. Final crosslink lists were compiled using the identified candidates filtered to 1 % FDR on link level with xiFDR v.1.4.150.
Bpa crosslinking
Yeast strains expressing Ycg1bpa constructs were generated by plasmid shuffle. A URA3-based episomal plasmid containing a wild-type YCG1 allele in a ycg1Δ background strain with a TRP1-based plasmid encoding E. coli TyrRS and tRNA CUA51 was replaced with a LEU2-based centromeric plasmid containing an ycg1bpa allele containing an amber stop codon at the indicated position. Genotypes of the resulting strains are listed in the Supplementary Note. For Western blot analysis, yeast strains were grown in 20 mL –LEU–TRP synthetic drop-out medium containing 1 mM p-benzoyl-L-phenylalanine (bpa; Bachem 4017646) at 30 °C to an OD600 of 0.6–0.8. Cells were harvested by centrifugation and resuspended in 4 mL PBS. Half of the sample was exposed in a Petri dish to a total of 10 J of 365 nm light (~50 min exposure time) on ice, whereas the other half was kept in the dark. Cells were collected by centrifugation, resuspended in 0.2 mL 100 mM NaOH and incubated for 10 min at room temperature. Cells were again collected by centrifugation and lysed in SDS loading buffer (50 mM Tris-HCl pH 6.8, 2 % (w/v) SDS, 10 % (v/v) glycerol, 0.1 % (w/v) bromophenol blue, 0.1 M DTT) by at 65 °C for 5 min prior to SDS-PAGE and Western blotting with antibodies against the PK (V5) tag (Serotec, MCA1360) or the HA tag (Abcam, ab9110).
For analysis by mass spectrometry, cells were harvested from 4 L –LEU–TRP medium containing 1 mM bpa at an OD600 of 1 and resuspended in 500 mL PBS. Half of the sample was exposed in Petri dishes to 10 J 365 nm light (~50 min exposure time), whereas the other half was kept in the dark. Cells were harvested by centrifugation, washed with 45 mL lysis buffer (50 mM Tris-HCl pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 0.25 % (v/v) Triton X-100) and resuspended in 15 mL lysis buffer containing 1 mM DTT, 1 mM PMSF and 2 × cOmplete–EDTA protease inhibitors before lysis by cryogenic grinding (SPEX Freezer/Mill 6970). Condensin complexes were immunoprecipitated using 100 μL Protein A-coupled Dynabeads that had been pre-bound to 10 μg anti-PK antibody. After elution and SDS-PAGE, gels were stained with Coomassie G250. The crosslinked band in the +UV sample and a band at the same height in the –UV control were excised for analysis by mass spectrometry as described28.
Analytical size exclusion chromatography
C. thermophilum Ycg1–Brn1Ycg1, Ycs4–Brn1Ycs4 and Smc2–Smc4–Brn1NC complexes were mixed at equimolar ratio (15 μM each) in 50 mM Tris-HCl pH 7.5, 1 mM DTT, 1 mM MgCl2 with a final concentration of 150 mM NaCl. After incubation for 15 min on ice, samples were loaded onto a Superose 6 Increase 3.2/300 column (GE Healthcare) pre-equilibrated in SEC-buffer (50 mM Tris-HCl pH 7.5, 150 mM NaCl, 1 mM DTT, 1 mM MgCl2) at a flow rate of 0.05 mL/min. 100 μL fractions were collected and the peak fractions were loaded onto a 4–12% Bis-Tris SDS-PAGE gel (Thermo Fisher) and the gel was stained with Coomassie Blue. For experiments in the presence of ATP, Smc2–Smc4–Brn1NC complexes were pre-incubated with 1 mM ATP for 15 min and the SEC-buffer contained 0.1 mM ATP.
Reporting summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Extended Data
Extended Data Fig. 1. Cryo-EM structure determination of the nucleotide-free apo condensin complex.
a, Representative micrographs of nucleotide-free (apo) condensin tetramer (Smc2–Smc4–Brn1–Ycs4, left) and pentamer (Smc2–Smc4–Brn1–Ycs4–Ycg1, right). b, Workflow of initial data processing. Representative 2D classes are shown for the tetramer and pentamer data sets. Density map, angular distribution plot and FSC curves of the overall complexes in c, the non-engaged and d, the bridged state.
Extended Data Fig. 2. Focused image processing of the arm segments of nucleotide-free apo condensin.
a, Workflow for the apo non-engaged condensin arm segment. Particles of the tetramer complex were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. b, Representative 2D classes (top), cryo-EM density map and its local resolution plot as calculated by RELION (left) and FSC curves (right) for the arm segment. c, Workflow for the apo bridged condensin arm segment. Particles of the pentamer bridged class were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. d, Representative 2D classes (top), 7.8 Å cryo-EM density map and its local resolution plot as calculated by RELION (left) and FSC curves (right) for the arm segment.
Extended Data Fig. 3. Focused image processing of the head segments of nucleotide-free apo condensin.
a, Workflow for the apo non-engaged condensin head segment. Particles of tetramer and pentamer complexes were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. b, Representative 2D classes (top), 4.2 Å cryo-EM density map and its local resolution plot as calculated by RELION (middle) and FSC curves (bottom) for the non-engaged head segment. c, Workflow for the apo bridged condensin head segment. Particles of the bridged class of the pentamer complex were initially processed and preliminary angles and translations were assigned as described in Extended Data Fig. 1. d, Representative 2D classes (top), 7.5 Å cryo-EM density map and its local resolution plot as calculated by RELION (middle) and FSC curves (bottom) for the bridged head segment.
Extended Data Fig. 4. Crosslink mass spec and pseudo-atomic model of the apo complex.
a, Circle plots of inter-molecular BS3 crosslinks identified in condensin tetramer complexes in the absence of nucleotide with an FDR of < 1%. Bar plots show the distance distribution of crosslinks (>35 Å; ~23 %). b, Model of the Smc2 and Smc4 coiled coils. c, Pseudo-atomic model of the non-engaged head segment with final electron density map. d, Pseudo-atomic model of the bridged head segment with final electron density map. e, Example images of 2D classes of conformations with the hinge folding back all the way to contact the coiled coils (left) or with lower degrees of elbow bending (right).
Extended Data Fig. 5. Conformation al changes and putative DNA binding sites in the apo condensin complex.
a, Structural alignment of pseudo-atomic models of overall apo complexes in the non-engaged and bridged states. b, Electrostatic potential maps of Ycs4 (left) and the Smc2–Smc4 heads and coiled coils (right); red: –5 keT, blue: +5 keT. The dotted line indicates a putative path for a DNA double helix that goes through the coiled coils above the ATPase heads and also includes the positively charged patch on Ycs4 near the ‘proboscis’ protrusion. c, Placement of a DNA double helix in the space between the neck regions of the Smc2 and Smc4 coiled coils in the apo non-engaged state, based on a comparison to a DNA co-crystal structure of the E. coli SbcC (Rad50) head dimer in the engaged state (PDB code 6S85).
Extended Data Fig. 6. Structure determination of the +ATP condensin complex.
a, Workflow of the initial data processing of pentameric condensin in the presence of ATP by reference-free ab initio model estimation, combined with focused refinement and representative 2D class averages of the three major classes of the overall complex. b. 2D class averages of open and rod-shaped coiled coils after re-centering and re-extraction of the arm segments (left), and of the ATPase heads after re-centering and re-extraction the ATPase head segments (right) with FSC curves of the cryo-EM density maps.
Extended Data Fig. 7. Ycg1 binds Smc2 and Brn1 via a conserved patch in vivo.
a, Surface conservation plot of S. cerevisiae Ycg1–Brn1 (left). Tetrad dissection of diploid S. cerevisiae YCG1/ycg1Δ cells expressing an ectopic PK6-tagged copy of Ycg1 harboring patch 1 or patch 2 mutations, after three days on rich media at 25 °C for 3 days (center). Expression levels of Ycg1-PK6 tested by Western blotting against the PK6 tag (right). b, Western blot analysis of photo-crosslinking products expressing Ycg1-PK6 with bpa substitutions at the indicated residues before (– UV) or after (+UV) exposure to light at 365 nm (left). Shift of endogenously HA-tagged versions Smc4, Smc2, Ycg1 or Brn1 in cells expressing unmodified wild-type (wt) or bpa-substituted Ycg1 at position S435 measured by Western blotting before (–UV) or after (+UV) light exposure (right).
Extended Data Fig. 8. Sub-classification of the +ATP condensin head domain segment.
a, 3D classification of all head particles results in two distinct maps, one with clear density for both coiled coils (open coils, right) and a second one with weak Smc2 coiled coil density (‘single’ coil, left). b, Representative 2D class averages, local resolution as plotted by ResMap52, and FSC curves of the cryo-EM density map of the ‘single’ coil state, as plotted by RELION. c, Representative 2D class averages, local resolution, and FSC curves of the cryo-EM density map of the open coil state.
Extended Data Fig. 9. Conformational changes in the head segment between the apo and +ATP states.
a, Structural alignment based on the RecA-like lobe of the Smc2 head domains of the +ATP (‘open’) form (in blue colors) and the apo (nucleotide-free) form (grey colors) highlights the extent of coiled-coil kinking in Smc2. Positioning and extent of the kinking in the carboxy-terminal Smc2 coiled coil are indicated. b, Simultaneous Smc2–Smc4 head engagement and Ycs4 binding to Smc4 would lead to steric clashes. c, Simultaneous binding of Ycs4 and Ycg1 HEAT-repeat subunits to the Smc4 and Smc2 heads, respectively, in the non-engaged state would result in major steric clashes.
Extended Data Fig. 10. Association of Ycs4 with Smc2–Smc4 prevents Ycg1 binding.
a, Size exclusion chromatography of complexes formed between a trimeric Smc2–Smc4–Brn1NC and either Ycs4–Brn1Ycs4 (left), Ycg1–Brn1Ycg1 (center) or both (right). Peak fractions were analyzed by SDS-PAGE and Coomassie Blue staining. b, as in a, but in the presence of 1 mM ATP. One chromatography run of two independent repeats shown for each experiment.
Supplementary Material
Acknowledgments
We thank D. D’Amours for sharing plasmids and yeast strains. We are grateful to L. Thärichen, S. Perović and C. Stober for help with yeast experiments and W. Hagen and F. Weis of the EMBL Cryo-EM Platform for support with cryo-EM data collection and processing (all EMBL). We thank F. Coscia, G. Cannone, A. Gonzales, J. García-Nafría, K. Zhang, S. Scheres and the MRC-LMB EM facility for assistance and advice with data collection and processing, T. Darling and J. Grimmett for computing (all MRC-LMB). We thank Alejandra F. Cid for help with purifications (MRC-LMS). We acknowledge the Diamond Light Source (eBIC), Astbury Biostructure Electron Microscopy (Leeds) and Cambridge Nano Science Centre for access and help (facilities supported by Wellcome Trust, MRC and BBSRC). TN was supported by the Japan Society for the Promotion of Science. This work was funded by the European Molecular Biology Laboratory, the European Research Council (ERC-2015-CoG 681365 to CHH), the Medical Research Council (U105184326 to JL and MC-A652-5PY00 to LA), the DFG (EXC 2008/1 - 390540038 and 329673113 to JR) and the Wellcome Trust (202754/Z/16/Z to JL and 100955/Z/13/Z to LA).
Footnotes
Author contributions
Cryo-EM of apo condensin: BGL, CC, PGE, TN, LA and JL; Cryo-EM of +ATP condensin: FM, MA, SB, MB and CHH; Biochemical and genetic assays: MK, LL, MH and CHH; crosslink mass spec: FJOR, LS and JR; Data analysis and manuscript preparation: BGL, FM, MH, JL and CHH.
Competing interests
The authors declare no competing interests.
Data and materials availability
The cryo-EM maps and the pseudo atomic model of the apo condensin structure were deposited in the EM Data Bank (EMDB) and Protein Data Bank (PDB) with accession numbers EMD-10951 (non-engaged overall), EMD-10948 (non-engaged arm segment), EMD-10947 (non-engaged head segment), EMD-10954 (bridged overall), EMD-10953 (bridged arm segment), EMD-10952 (bridged head segment), EMD-10944 (+ATP head segment), EMD-10964 (+ATP arm segment) and PDB 6YVU (non-engaged overall model), 6YVV (bridged head segment), PDB 6YVD (+ATP head segment). Crosslinking mass spectrometry data are available via ProteomeXchange with identifiers PXD019275 (BS3) and PXD019274 (sulfo-SDA). Source data are available with the paper online. All other data are available in the main text or the supplementary materials. Correspondence and requests for materials to: luis.aragon@lms.mrc.ac.uk; martin.beck@embl.de; jyl@mrc-lmb.cam.ac.uk; christian.haering@uni-wuerzburg.de.
References
- 1.Yatskevich S, Rhodes J, Nasmyth K. Organization of Chromosomal DNA by SMC Complexes. Annu Rev Genet. 2019;53:445–482. doi: 10.1146/annurev-genet-112618-043633. [DOI] [PubMed] [Google Scholar]
- 2.Uhlmann F. SMC complexes: from DNA to chromosomes. Nat Rev Mol Cell Biol. 2016;17:399–412. doi: 10.1038/nrm.2016.30. [DOI] [PubMed] [Google Scholar]
- 3.Hopfner KP, Tainer JA. Rad50/SMC proteins and ABC transporters: unifying concepts from high-resolution structures. Curr Opin Struct Biol. 2003;13:249–55. doi: 10.1016/s0959-440x(03)00037-x. [DOI] [PubMed] [Google Scholar]
- 4.Gligoris T, Lowe J. Structural Insights into Ring Formation of Cohesin and Related Smc Complexes. Trends Cell Biol. 2016;26:680–93. doi: 10.1016/j.tcb.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Haering CH, Lowe J, Hochwagen A, Nasmyth K. Molecular architecture of SMC proteins and the yeast cohesin complex. Mol Cell. 2002;9:773–88. doi: 10.1016/s1097-2765(02)00515-4. [DOI] [PubMed] [Google Scholar]
- 6.Palecek JJ, Gruber S. Kite Proteins: a Superfamily of SMC/Kleisin Partners Conserved Across Bacteria, Archaea, and Eukaryotes. Structure. 2015;23:2183–2190. doi: 10.1016/j.str.2015.10.004. [DOI] [PubMed] [Google Scholar]
- 7.Wells JN, Gligoris TG, Nasmyth KA, Marsh JA. Evolution of condensin and cohesin complexes driven by replacement of Kite by Hawk proteins. Curr Biol. 2017;27:R17–R18. doi: 10.1016/j.cub.2016.11.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goloborodko A, Imakaev MV, Marko JF, Mirny L. Compaction and segregation of sister chromatids via active loop extrusion. Elife. 2016;5 doi: 10.7554/eLife.14864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Alipour E, Marko JF. Self-organization of domain structures by DNA-loop-extruding enzymes. Nucleic Acids Res. 2012;40:11202–12. doi: 10.1093/nar/gks925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Brandao HB, Le TB, Laub MT, Rudner DZ. Bacillus subtilis SMC complexes juxtapose chromosome arms as they travel from origin to terminus. Science. 2017;355:524–527. doi: 10.1126/science.aai8982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nasmyth K. Disseminating the genome: joining, resolving, and separating sister chromatids during mitosis and meiosis. Annu Rev Genet. 2001;35:673–745. doi: 10.1146/annurev.genet.35.102401.091334. [DOI] [PubMed] [Google Scholar]
- 12.Guacci V, et al. Structure and function of chromosomes in mitosis of budding yeast. Cold Spring Harb Symp Quant Biol. 1993;58:677–85. doi: 10.1101/sqb.1993.058.01.075. [DOI] [PubMed] [Google Scholar]
- 13.Davidson IF, et al. DNA loop extrusion by human cohesin. Science. 2019;366:1338–1345. doi: 10.1126/science.aaz3418. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Shi Z, Zhang H, Finkelstein IJ, Yu H. Human cohesin compacts DNA by loop extrusion. Science. 2019;366:1345–1349. doi: 10.1126/science.aaz4475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim E, Kerssemakers J, Shaltiel IA, Haering CH, Dekker C. DNA-loop extruding condensin complexes can traverse one another. Nature. 2020;579:438–442. doi: 10.1038/s41586-020-2067-5. [DOI] [PubMed] [Google Scholar]
- 16.Ganji M, et al. Real-time imaging of DNA loop extrusion by condensin. Science. 2018;360:102–105. doi: 10.1126/science.aar7831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fudenberg G, et al. Formation of Chromosomal Domains by Loop Extrusion. Cell Rep. 2016;15:2038–49. doi: 10.1016/j.celrep.2016.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sanborn AL, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–65. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gibcus JH, et al. A pathway for mitotic chromosome formation. Science. 2018;359 doi: 10.1126/science.aao6135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hassler M, et al. Structural Basis of an Asymmetric Condensin ATPase Cycle. Mol Cell. 2019;74:1175–1188.:e9. doi: 10.1016/j.molcel.2019.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Soh YM, et al. Molecular basis for SMC rod formation and its dissolution upon DNA binding. Mol Cell. 2015;57:290–303. doi: 10.1016/j.molcel.2014.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burmann F, et al. A folded conformation of MukBEF and cohesin. Nat Struct Mol Biol. 2019;26:227–236. doi: 10.1038/s41594-019-0196-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anderson DE, Losada A, Erickson HP, Hirano T. Condensin and cohesin display different arm conformations with characteristic hinge angles. J Cell Biol. 2002;156:419–24. doi: 10.1083/jcb.200111002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshimura SH, et al. Condensin architecture and interaction with DNA: regulatory non-SMC subunits bind to the head of SMC heterodimer. Curr Biol. 2002;12:508–13. doi: 10.1016/s0960-9822(02)00719-4. [DOI] [PubMed] [Google Scholar]
- 25.Gligoris TG, et al. Closing the cohesin ring: structure and function of its Smc3-kleisin interface. Science. 2014;346:963–7. doi: 10.1126/science.1256917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haering CH, et al. Structure and stability of cohesin’s Smc1-kleisin interaction. Mol Cell. 2004;15:951–64. doi: 10.1016/j.molcel.2004.08.030. [DOI] [PubMed] [Google Scholar]
- 27.Burmann F, et al. An asymmetric SMC-kleisin bridge in prokaryotic condensin. Nat Struct Mol Biol. 2013;20:371–9. doi: 10.1038/nsmb.2488. [DOI] [PubMed] [Google Scholar]
- 28.Kschonsak M, et al. Structural Basis for a Safety-Belt Mechanism That Anchors Condensin to Chromosomes. Cell. 2017;171:588–600.:e24. doi: 10.1016/j.cell.2017.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, et al. Structural basis for Scc3-dependent cohesin recruitment to chromatin. Elife. 2018;7 doi: 10.7554/eLife.38356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kashammer L, et al. Mechanism of DNA End Sensing and Processing by the Mrell-Rad50 Complex. Mol Cell. 2019;76:382–394.:e6. doi: 10.1016/j.molcel.2019.07.035. [DOI] [PubMed] [Google Scholar]
- 31.Piazza I, et al. Association of condensin with chromosomes depends on DNA binding by its HEAT-repeat subunits. Nat Struct Mol Biol. 2014;21:560–8. doi: 10.1038/nsmb.2831. [DOI] [PubMed] [Google Scholar]
- 32.Diebold-Durand ML, et al. Structure of Full-Length SMC and Rearrangements Required for Chromosome Organization. Mol Cell. 2017;67:334–347.:e5. doi: 10.1016/j.molcel.2017.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eeftens JM, et al. Condensin Smc2-Smc4 Dimers Are Flexible and Dynamic. Cell Rep. 2016;14:1813–8. doi: 10.1016/j.celrep.2016.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Russo CJ, Scotcher S, Kyte M. A precision cryostat design for manual and semi-automated cryo-plunge instruments. Rev Sci Instrum. 2016;87:114302. doi: 10.1063/1.4967864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schorb M, Haberbosch I, Hagen WJH, Schwab Y, Mastronarde DN. Software tools for automated transmission electron microscopy. Nat Methods. 2019;16:471–477. doi: 10.1038/s41592-019-0396-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Danev R, Buijsse B, Khoshouei M, Plitzko JM, Baumeister W. Volta potential phase plate for in-focus phase contrast transmission electron microscopy. Proc Natl Acad Sci USA. 2014;111:15635–40. doi: 10.1073/pnas.1418377111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zivanov J, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. Elife. 2018;7 doi: 10.7554/eLife.42166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosenthal PB, Henderson R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol. 2003;333:721–45. doi: 10.1016/j.jmb.2003.07.013. [DOI] [PubMed] [Google Scholar]
- 41.Pettersen EF, et al. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–12. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 42.Zivanov J, Nakane T, Scheres SHW. A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. IUCrJ. 2019;6:5–17. doi: 10.1107/S205225251801463X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 44.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–32. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 45.Waterhouse A, et al. SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res. 2018;46:W296–W303. doi: 10.1093/nar/gky427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shevchenko A, Tomas H, Havlis J, Olsen JV, Mann M. In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc. 2006;1:2856–60. doi: 10.1038/nprot.2006.468. [DOI] [PubMed] [Google Scholar]
- 47.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–70. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 48.Kolbowski L, Mendes ML, Rappsilber J. Optimizing the Parameters Governing the Fragmentation of Cross-Linked Peptides in a Tribrid Mass Spectrometer. Anal Chem. 2017;89:5311–5318. doi: 10.1021/acs.analchem.6b04935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mendes ML, et al. An integrated workflow for crosslinking mass spectrometry. Mol Syst Biol. 2019;15:e8994. doi: 10.15252/msb.20198994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fischer L, Rappsilber J. Quirks of Error Estimation in Cross-Linking/Mass Spectrometry. Anal Chem. 2017;89:3829–3833. doi: 10.1021/acs.analchem.6b03745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen HT, Warfield L, Hahn S. The positions of TFIIF and TFIIE in the RNA polymerase II transcription preinitiation complex. Nat Struct Mol Biol. 2007;14:696–703. doi: 10.1038/nsmb1272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kucukelbir A, Sigworth FJ, Tagare HD. Quantifying the local resolution of cryo-EM density maps. Nat Methods. 2014;11:63–5. doi: 10.1038/nmeth.2727. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps and the pseudo atomic model of the apo condensin structure were deposited in the EM Data Bank (EMDB) and Protein Data Bank (PDB) with accession numbers EMD-10951 (non-engaged overall), EMD-10948 (non-engaged arm segment), EMD-10947 (non-engaged head segment), EMD-10954 (bridged overall), EMD-10953 (bridged arm segment), EMD-10952 (bridged head segment), EMD-10944 (+ATP head segment), EMD-10964 (+ATP arm segment) and PDB 6YVU (non-engaged overall model), 6YVV (bridged head segment), PDB 6YVD (+ATP head segment). Crosslinking mass spectrometry data are available via ProteomeXchange with identifiers PXD019275 (BS3) and PXD019274 (sulfo-SDA). Source data are available with the paper online. All other data are available in the main text or the supplementary materials. Correspondence and requests for materials to: luis.aragon@lms.mrc.ac.uk; martin.beck@embl.de; jyl@mrc-lmb.cam.ac.uk; christian.haering@uni-wuerzburg.de.