Abstract
Macrophages reside in the body cavities where they maintain serosal homeostasis and provide immune surveillance. Peritoneal macrophages are implicated in the etiology of pathologies including peritonitis, endometriosis and metastatic cancer thus understanding the factors that govern their behavior is vital. Using a combination of fate mapping techniques, we have investigated the impact of sex and age on murine peritoneal macrophage differentiation, turnover and function. We demonstrate that the sexually dimorphic replenishment of peritoneal macrophages from the bone marrow, which is high in males and very low in females, is driven by changes in the local microenvironment that arise upon sexual maturation. Population and single cell RNAseq revealed striking dimorphisms in gene expression between male and female peritoneal macrophages that was in part explained by differences in composition of these populations. By estimating the time of residency of different subsets within the cavity and assessing development of dimorphisms with age and in monocytopenic Ccr2 -/- mice, we demonstrate that key sex-dependent features of peritoneal macrophages are a function of the differential rate of replenishment from the bone marrow while others are reliant on local microenvironment signals. Importantly, we demonstrate that the dimorphic turnover of peritoneal macrophages contributes to differences in the ability to protect against pneumococcal peritonitis between the sexes. These data highlight the importance of considering both sex and age in susceptibility to inflammatory and infectious diseases.
Introduction
Macrophages are present in every tissue of the body, where they provide immune protection and orchestrate tissue repair following insult or injury. Peritoneal macrophages are arguably the most studied population of macrophages in the body, having been used extensively as a convenient source of macrophages for ex vivo analyses for decades. In spite of this body of knowledge, the heterogeneity of peritoneal macrophages and much of the biology that governs their development, differentiation, and function remains unclear. Macrophages in the peritoneal cavity are programmed for ‘silent’ clearance of apoptotic cells, maintenance of innate B1 cells through secretion of CXCL13, and for immune surveillance of the cavity and neighboring viscera (1–4). However, they are also implicated in many pathologies, including peritonitis, endometriosis, post-surgical adhesions, pancreatitis and metastatic cancer (5–15), although the exact roles they play in these processes are not fully understood.
Under physiological conditions, at least two macrophage populations are present in the murine peritoneal cavity, with those expressing high levels of F4/80, CD11b and CD102 outnumbering their F4/80loMHCII+ counterparts by approximately 10-fold. F4/80hiCD102+ macrophages (sometimes referred to as ‘large’ peritoneal macrophages (16)) rely on the transcription factors C/EBPβ and GATA6 for their differentiation and survival (17–20), with the latter under the control of retinoic acid proposed to derive, in part, from the omentum (19). The F4/80loMHCII+ compartment is heterogeneous, comprising both macrophages and conventional dendritic cells (21–24). F4/80loMHCII+ macrophages (sometimes referred to as ‘small’ peritoneal macrophages or monocyte-derived DC) rely on IRF4 for their differentiation and can be further defined by their expression of CD226 and the immunomodulatory molecule RELMα (21, 23, 24). Notably, recent studies employing lineage tracing techniques have established that F4/80loMHCII+ macrophages arise postnatally, are short-lived and replaced by Ly6Chi classical monocytes in a CCR2-dependent manner (20–23). In contrast, F4/80hiCD102+ macrophages are longer-lived cells that derive from embryonic sources, but are subsequently replaced by cells of hematopoietic stem cell (HSC) origin (21, 25). Importantly, we have recently shown that unlike resident macrophages in numerous other tissues the turnover of peritoneal F4/80hiCD102+ macrophages from the bone marrow is highly sex-dependent, with high and low rates in male and female mice, respectively (21). We have also shown that long-lived macrophages can be identified by their expression of the phagocytic receptor, Tim4, whereas most recent descendants of BM-derived cells amongst the F4/80hiCD102+ macrophage compartment are Tim4-(21). Indeed Tim4 expression has been shown to a feature of long-lived macrophages in other tissues (26–30). However, it remains unclear if further heterogeneity exists amongst these broadly-defined populations and if sexually-dimorphic turnover influences the composition and function of the F4/80hiCD102+ macrophage population in other ways.
Sex is a variable often overlooked in immunological research (31) despite strong sex biases in many pathologies including autoimmune disorders and infection susceptibility (32). Notably, sex dimorphisms in the immune system are present a diverse range of species from insects, bird, lizards, and mammals (32), suggesting that this may be an evolutionarily conserved phenomenon. It is therefore essential to understand how intrinsic factors, such as sex, control the behavior of innate immune effector cells. Specifically, sex has been proposed to affect macrophage behavior, such as influencing the differentiation of brain microglia, (33–35) and sex hormones appear able to directly regulate gene expression (36) and proliferation (37) of macrophages. Whereas previous studies have considered the effects of sex on peritoneal macrophage behavior, many of these have focused on in vitro functional assessments using macrophages elicited by injection of an irritant or inflammatory agent (38), or they have not appreciated the complexity of the peritoneal macrophage compartment (37, 39).
Here, we have used a combination of fate-mapping techniques together with population-level and single cell RNA sequencing (scRNAseq) to dissect the role of sex in the composition, environmental imprinting, and function of peritoneal macrophages. We show that the F4/80hiCD102+ macrophage population is heterogeneous, and that dimorphic turnover is associated with divergence in the heterogeneity of this compartment with age. Specifically, we demonstrate that the sexual dimorphism in replenishment from the bone marrow and phenotype arise following sexual maturation. Furthermore, we provide examples of transcriptional and functional dimorphisms that arise due to sex differences in turnover versus those arising directly from sex differences in the peritoneal microenvironment. Importantly, we identify the C-type lectin receptor CD209b (also known as Specific ICAM3-grabbing nonintegrin-related 1; SIGN-R1) as a marker whose expression is determined by replenishment that becomes increasingly dimorphic with age, and show that sex-dependent resistance to pneumococcal peritonitis arises, in part, due to dimorphic expression of CD209b.
Results
Environmental factors drive sexual dimorphism in peritoneal macrophage replenishment
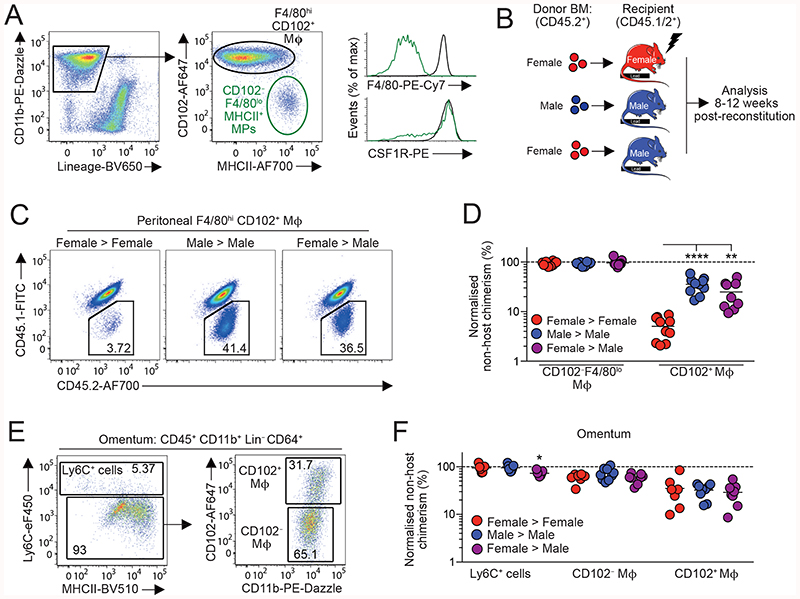
We first set out to determine if the dimorphic effects in peritoneal macrophage replenishment were due to the peritoneal environment or to cell-intrinsic differences in the ability of male and female monocytes to generate F4/80hiCD102+ macrophages at this site. To this end, we generated sex-mismatched, tissue-protected bone marrow (BM) chimeric mice to measure the turnover of peritoneal F4/80hiCD102+ macrophages from the BM (Fig. 1A & Fig. S1A) and assess the role of sex in this process. Wild type (CD45.1/.2+) mice were irradiated, with all but the head and upper torso protected with lead to prevent direct exposure to ionising radiation (Fig. 1B), before being reconstituted with sex-matched (female > female, male > male) or sex-mismatched (female > male, male > female) BM. Following at least 8 weeks reconstitution, the non-host chimerism was measured in peritoneal macrophages. Consistent with our previous work (21), only low levels of non-host chimerism could be detected amongst peritoneal F4/80hiCD102+ macrophages from female > female BM chimeric mice, whereas high levels were detected in their male > male counterparts (Fig. 1C&D, & Fig. S1B), confirming marked sex dimorphism in macrophage turnover. Importantly, this dimorphism was specific to F4/80hiCD102+ peritoneal macrophages, as all other leukocyte subsets showed identical replenishment in male and female BM chimeric mice (Fig. S1C). Strikingly, F4/80hiCD102+ peritoneal macrophages from sex mismatched (female > male) chimeras had similar levels of chimerism to male > male chimeras (Fig. 1C&D), demonstrating that female and male monocytes have equal ability to generate F4/80hiCD102+ macrophages in the male peritoneal cavity. Female recipients rejected male BM and thus chimerism in this group could not be determined.
Figure 1. Environment drives sexual dimorphism in macrophage replenishment in the peritoneal cavity.
(A) Expression of CD11b and CD3, CD19, Ly6G and SiglecF (‘Lineage’) by live CD45+ peritoneal leukocytes (left) and expression of CD102 and MHCII by CD11b+ Lin- cells (centre) from adult C57Bl/6 female mice. Histograms show expression of F4/80 and CSF1R by CD102+ and CD102-MHCII+ cells (also see Fig. S1A). (B) Scheme for the generation of sex mis-matched, tissue-protected bone marrow (BM) chimeric mice. (C-D) Representative chimerism (C) and normalized non-host chimerism (D) of peritoneal F4/80hiCD102+ macrophages from sex matched or mismatched tissue protected BM chimeric mice 8-12 weeks post-reconstitution (also see Fig. S1B). Data are normalised to the non-host chimerism of Ly6Chi monocytes. **P<0.01, ****P<0.0001 One-way ANOVA. Data represent 9 (female>male) or 10 (sex matched) mice per group pooled from two independent experiments. (E) Gating strategy to identify macrophages amongst omental isolates (also see Fig. S1D). (F) Normalized non-host chimerism of omental Ly6C+ monocytes and CD102-defined macrophages from mice in (D). Data are normalised to the non-host chimerism of Ly6Chi monocytes. Data represent 7 (female > female), 8 (female > male) or -9 (male > male) mice per group pooled from two independent experiments. *P<0.05. One-way ANOVA. Symbols represent individual animals and horizontal bars represent the mean.
The omentum has been implicated in the differentiation of F4/80hi macrophages in the peritoneal cavity, potentially acting as site of macrophage maturation (19, 40, 41). Indeed, CD102+ macrophages that co-express GATA6, can be detected amongst omental isolates (19), together with CD102-MHCII+ macrophages and a population of Ly6C+ CD11b+ cells similar to monocytes (Fig. 1E and Fig. S1D,E). To determine if the dimorphic replenishment of peritoneal F4/80hiCD102+ macrophages arises in the omentum, we assessed non-host chimerism in the macrophage populations within this site. While this showed clear differences in the turnover of CD102-defined macrophage populations from BM, with higher replenishment in the CD102- fraction, no sex dimorphism was detected in any monocyte/macrophage population within the omentum (Fig. 1F). Furthermore, the chimerism of omental and peritoneal CD102+ macrophages in male recipients was identical, rather than showing the gradation that would have been expected if omental macrophages were intermediate precursors between monocytes and cavity CD102+ cells (Fig. S1F). Thus, the sexual dimorphism in peritoneal F4/80hiCD102+ macrophage replenishment is driven by factors present in the local environment.
Sexual dimorphism in peritoneal macrophage replenishment occurs following sexual maturity
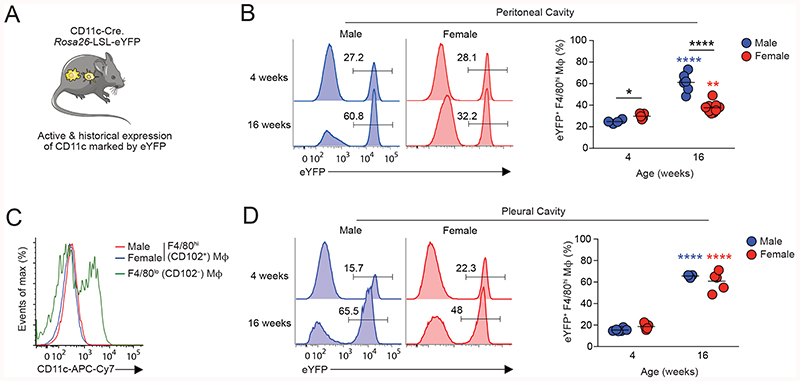
To extend these findings and to assess macrophage turnover at different stages of maturity, we next used a genetic fate mapping approach. Adoptive transfer experiments suggest F4/80loMHCII+ macrophages in the peritoneal macrophage compartment act, in part, as precursors of F4/80hiCD102+ macrophages (20) and we have recently shown that this differentiation can be mapped by exploiting their expression of CD11c (21). Thus, in CD11cCre.Rosa26 LSL-eYFP mice (Fig. 2A), in whom active or historic expression of CD11c leads to irreversible labelling with eYFP, labelled cells accumulate with age in the F4/80hiCD102+ macrophage compartment, despite these cells themselves not actively expressing CD11c (21). We therefore used CD11cCre.Rosa26 LSL-eYFP mice to compare the rate of eYFP+ cell accumulation in peritoneal F4/80hiCD102+ macrophages from male and female mice. In juvenile/prepubescent mice (4 weeks of age), the extent of eYFP labelling was relatively similar between male and female peritoneal F4/80hiCD102+ macrophages and indeed was marginally higher in female mice (Fig. 2B & Fig. S2A). By 16 weeks of age, the frequency of eYFP+ cells amongst F4/80hiCD102+ macrophages had increased in both male and female mice compared with their 4-week-old counterparts. However, although there was no difference in CD11c protein expression by male and female F4/80hiCD102+ macrophages (Fig. 2C), higher levels of eYFP labelling were detected amongst male peritoneal macrophages, consistent with more rapid accumulation of newly differentiated macrophages in male mice (Fig. 2B). Consistent with our previous findings made using tissue-protected BM chimeras (21), the sexual dimorphism in eYFP labelling was not detected in F4/80hiCD102+ macrophages from the pleural cavity (Fig. 2D), where both male and female pleural cells exhibited high levels of labelling that were equivalent to those seen in the male peritoneal cavity by 16 weeks. Collectively, these data confirm that the peritoneal environment controls macrophage turnover and suggest that dimorphisms arise in this site following sexual maturity.
Figure 2. Sexual dimorphism in peritoneal macrophage replenishment occurs following sexual maturity.
(A) Experimental scheme of CD11cCre.Rosa26 LSL-eYFP fate-mapping mice.
(B) Representative expression of eYFP by peritoneal F4/80hiCD102+ macrophages from male and female CD11cCre.Rosa26 LSL-eYFP fate-mapping mice at 4 and 16 weeks of age. Right, frequency of eYFP+ cells amongst F4/80hiCD102+ macrophages in male and female mice at the indicated ages. Symbols represent individual animals and horizontal bars represent the mean. Data represent 4 (male 4 weeks), 5 (female 4 weeks), 6 (male 16 weeks) or 9 (female 16 weeks) mice per group pooled from two independent experiments. For upstream gating strategy see Fig. S2A.
(C) Expression of CD11c by peritoneal F4/80hiCD102+ macrophages from male and female and CD102- MHCII+ cells from female mice.
(D) Representative expression of eYFP by pleural F4/80hiCD102+ macrophages from male and female CD11cCre.Rosa26 LSL-eYFP fate-mapping mice at 4 and 16 weeks of age. Right, frequency of eYFP+ cells amongst pleural F4/80hiCD102+ macrophages in male and female mice at the indicated ages. Symbols represent individual animals and horizontal bars represent the mean. Data represent 3 (male 16 weeks), 5 (female 4 & 16 weeks) or 6 (male 4 weeks) per group pooled from two independent experiments.
Ovariectomy leads to increased macrophage replenishment
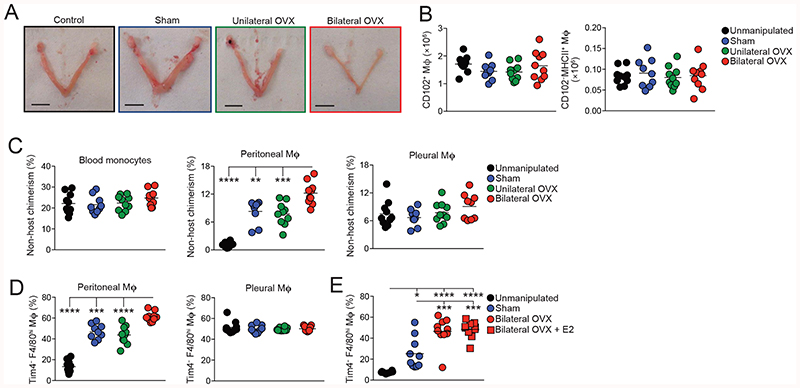
The onset of sexually dimorphic turnover of peritoneal macrophages following sexual maturation and the uniquely slow replenishment of female peritoneal macrophages suggested that factors involved in female reproductive function may drive this dimorphism. Therefore, we next assessed macrophage turnover in females after ovariectomy (OVX). Thus, female > female tissue protected BM chimeric were generated and after 8 weeks reconstitution, both ovaries were surgically removed (bilateral OVX), before measuring nonhost chimerism after another 8 weeks. To account for the potential effects of surgery on macrophage replenishment, BM chimeric mice receiving sham surgery or unilateral OVX were used as controls, together with unmanipulated BM chimeric mice. As expected, the cessation of ovarian estradiol production caused by bilateral OVX led to complete atrophy of the uterine horn; this was not seen in mice with unilateral OVX, or in other control groups (Fig. 3A). Bilateral OVX had no effect on the numbers of F4/80hiCD-102+ and CD102 MHCII’ macrophages in the peritoneal cavity when compared with the control groups (Fig. 3B). Although bilateral OVX led to increased proportions and absolute numbers of eosinophils, these differences did not attain statistical significance and the opposite pattern was found with B1 cells (Fig. S2B-I). Importantly and in striking contrast to the very low levels of chimerism (~1%) detected in unmanipulated control chimeras (Fig. 3C), sham surgery and unilateral OVX led to significant increases the level of chimerism compared with unmanipulated chimeric mice, demonstrating that minimally-invasive laparotomy itself appears to have long term effects on the dynamics of peritoneal macrophages in female mice. Nevertheless, complete removal of the ovaries further elevated macrophage turnover, with chimerism reaching approximately 12%. No difference was found between the chimerism seen after sham surgery with or without unilateral OVX, indicating that the OVX procedure itself does not exaggerate the effects of laparotomy and that it is the compete loss of ovarian function that underlies the further elevation in macrophage turnover that results from bilateral OVX. Consistent with these results, significantly more Tim4-CD102+ macrophages were present in the cavity of mice that received bilateral OVX than any other group (Fig. 3D), further supporting the idea of elevated macrophage replenishment from BM. Notably, no differences in chimerism or in Tim4-defined subsets could be detected amongst F4/80hiCD102+ macrophages from the pleural cavity, again confirming that the effect of surgery and ovariectomy on macrophage turnover are specific to the peritoneal cavity (Fig. 3B, C).
Figure 3. Ovariectomy leads to increased macrophage replenishment.
(A) Representative images of the uterine horns of tissue-protected BM chimeric mice that had received unilateral or bilateral oophorectomy (OVX), sham surgery or were completely unmanipulated (control).
(B) Absolute number of F4/80hiCD102+ macrophages and CD102-MHCII+ cells obtained from the peritoneal cavity of tissue-protected BM chimeric mice that had received surgery 8 weeks earlier. Symbols represent individual animals and horizontal bars represent the mean. Data represent 9 (sham) or 10 (control, unilateral, bilateral) mice per group pooled from two independent experiments. For upstream gating strategy see Fig. S2B.
(C) Non-host chimerism of blood Ly6Chi blood monocytes (left) and F4/80hiCD102+ macrophages obtained from the peritoneal (centre) or pleural (right) cavity of tissue-protected BM chimeric mice that had received surgery 8 weeks earlier. Symbols represent individual animals and horizontal bars represent the mean. Data represent 9 (sham) or 10 (control, unilateral, bilateral) mice per group pooled from two independent experiments. **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(D) Frequency of Tim4- cells amongst F4/80hiCD102+ macrophages obtained from the peritoneal (centre) or pleural (right) cavity of mice in (B). ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test. For representative FACS plots see Fig. S2C.
(E) Frequency of Tim4- cells amongst F4/80hiCD102+ macrophages obtained from the peritoneal cavity of unmanipulated C57Bl/6 female mice (controls) or age-matched females that received bilateral OVX or sham surgery 4 weeks earlier. One group received exogeneous estradiol (E2) thrice weekly for 3 weeks. Symbols represent individual animals and horizontal bars represent the mean. Data represent 8 (control) or 10 (sham, bilateral, bilateral + E2) mice per group pooled from two independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
Estrogens are the prototypical female sex steroid hormones, which are ablated by OVX. To assess if estrogen influences macrophage replacement, we repeated the OVX experiment with an additional group of bilateral OVX mice receiving exogenous estradiol (E2). However, while this treatment reversed OVX-mediated atrophy of the uterine horns and peritoneal eosinophilia, it had no effect on the heightened rate of replenishment of F4/80hiCD102+ peritoneal macrophages in OVX mice, suggesting that estradiol is not directly responsible for generating the sex dimorphism in peritoneal macrophage turnover (Fig. 3E). As males exhibit much greater levels of adipose tissue in the peritoneal cavity (Fig. S3), and a common feature of ovariectomy/oophorectomy in mice and humans is increased adiposity (42, 43), we combined a high fat diet (HFD) with our BM chimeric system to reveal if changes in adiposity affect replenishment of peritoneal F4/80hiCD102+ macrophage. However, replenishment was not affected by diet in either males or females, despite the expected increase in body weight and adipose tissue seen in mice on an HFD (Fig. S3). Hence, sexual maturation controls dimorphic turnover in the peritoneal cavity through a mechanism controlled at least in part by the female reproductive system, but independently of estrogen and local fat deposition.
Sex determines the transcriptional signature of peritoneal macrophages
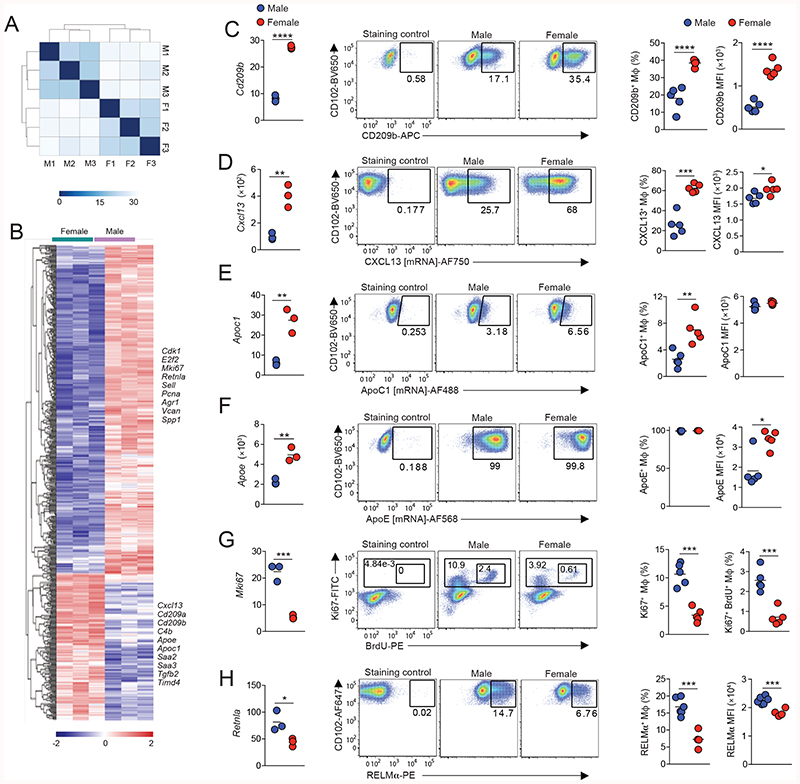
The difference in macrophage replenishment prompted us to assess the wider effects of sex on the imprinting of peritoneal macrophage identity and function. We therefore first performed population-level RNASeq on peritoneal F4/80hiCD102+ macrophages FACS-purified from unmanipulated 10-12 week old male and female mice (Fig. S4). To limit potential confounding effects of the estrous cycle, the stage of each female mouse was confirmed by vaginal cytology and samples pooled to include cells from all stages of the cycle. Furthermore, to limit the effects of circadian influence, mice in each biological replicate were euthanised at the same time each day. Unbiased clustering was then used to group the populations based on sex, with sex explaining 81% of the variance within the datasets (Fig. 4A) and differential gene expression analysis revealed that 486 mRNA transcripts were differentially expressed (>1.5fold) between female and male peritoneal CD102+ macrophages (Fig. 4B & Table S1). Analysis of the 148 mRNA transcripts more highly expressed in female peritoneal macrophages revealed that a large proportion was associated with immune function, including the C-type lectin receptors Clec4g, Cd209a and Cd209b, the complement components C4b, C1qa, and C3 the immunoregulatory cytokine Tgfb2, the B cell chemoattractant Cxcl13, and as expected the phagocytic receptor Timd4 (Fig. 4C & Table S1). Consistent with this, ‘immune response’ and ‘immune system processes’ were among the top pathways identified by gene-set enrichment analysis in genes up-regulated in female cells (Table S2). Transcripts for the apolipoproteins Apoe, Saa2, Saa3 and Apoc1 were also expressed more highly in female cells. Notably, in contrast to previous work that assessed basal gene expression by total peritoneal cells across the sexes (39), we did not detect any dimorphism in expression of toll-like receptors (TLRs), the TLR adaptor molecule MyD88 or CD14 (Fig. S5). Moreover, the dimorphic cassette of genes we identified is distinct from that recently shown to be sexually dimorphic in microglia (Fig. S5A, B), consistent with previous reports that transcriptional differences between sexes in macrophages are largely tissue-specific (44).
Figure 4. Sex determines the transcriptional signature of peritoneal macrophages.
(A) Heatmap showing distance between samples of male (M) and female (F) CD102+F4/80hi macrophages FACS-purified from the peritoneal cavity of 10-12 week old mice.
(B) Gene expression profile of the 148 differentially expressed (>1.5 fold) genes between male and female peritoneal macrophages with selected genes highlighted.
(C) Expression of Cd209b from RNAseq (FPKM; left panel), representative expression of CD209b protein (middle panels) and frequency of CD209b+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female C57BL/6 mice (right panel) and the mean fluorescence intensity (MFI) of CD209b expression by these cells (far right panel). Symbols represent individual animals and horizontal bars represent the mean. RNAseq data represent 3 mice per group and protein analysis represents 5 mice per group from one of five independent experiments. ****P<0.0001. Student’s t test. For gating strategy see Fig. S5C.
(D) Expression of Cxcl13 from RNAseq (FPKM; left panel), representative expression of CXCL13 mRNA (middle panels) and frequency of CXCL13+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female C57BL/6 mice (right panel) and the mean fluorescence intensity (MFI) of CXCL13 mRNA expression by these cells (far right panel). Symbols represent individual animals and horizontal bars represent the mean. RNAseq data represent 3 mice per group and flow cytometric analysis represents 5 mice per group from one of three independent experiments. *P<0.05, ***P<0.001. Student’s t test.
(E) Expression of Apoc1 from RNAseq (FPKM; left panel), representative expression of ApoC1 mRNA (middle panels) and frequency of ApoC1+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female C57BL/6 mice (right panel) and the mean fluorescence intensity (MFI) of ApoC1 mRNA expression by these cells (far right panel). Symbols represent individual animals and horizontal bars represent the mean. RNAseq data represent 3 mice per group and flow cytometric analysis represents 5 mice per group from one of three independent experiments. *P<0.05, ***P<0.001. Student’s t test.
(F) Expression of Apoe from RNAseq (FPKM; left panel), representative expression of ApoE mRNA (middle panels) and frequency of ApoE+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female C57BL/6 mice (right panel) and the mean fluorescence intensity (MFI) of ApoE mRNA expression by these cells (far right panel). Symbols represent individual animals and horizontal bars represent the mean. RNAseq data represent 3 mice per group and flow cytometric analysis represents 5 mice per group from one of three independent experiments. *P<0.05, ***P<0.001. Student’s t test.
(G) Expression of Mki67 from RNAseq (FPKM; left panel), representative expression of Ki67 protein and BrdU incorporation (middle panels) and the frequency of BrdU+Ki67+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female C57BL/6 mice. Symbols represent individual animals and horizontal bars represent the mean. Data represent 5 mice per group from one of two experiments. ***P<0.001. Student’s t test.
(H) Expression of Retnla from RNAseq (FPKM; left panel), representative expression of RELMa protein (middle panels) and the frequency of RELMa+ cells amongst CD102+F4/80hi peritoneal macrophages obtained from 10-12-week-old male or female Rag1-/- C57BL/6 mice. Symbols represent individual animals and horizontal bars represent the mean. *P<0.05, ***P<0.001. Student’s t test.
We used flow cytometry to confirm higher expression of Cd209b, Cxcl13, and Apoc1 by female macrophages, as these were the most differentially expressed non-X-linked genes with mapped read counts greater than 10. This analysis revealed unexpected heterogeneity within resident peritoneal macrophages. For instance, only a proportion of male and female CD102+ macrophages expressed CD209b, although the frequency of these was greater in females than in males (35% and 20% respectively). Moreover, CD209b was expressed at a higher level on a per cell basis by female CD102+ macrophages compared with their male counterparts (Fig. 4C), a finding consistent across different strains, including Rag1-/- mice, and mice from different housing environments (Fig. S6). Due to the unavailability of commercial antibodies for CXCL13, ApoE and ApoC1, we used PrimeFlow technology to measure mRNA of these genes at a single cell level using flow cytometry (Fig. S5C). Again, this revealed that a greater proportion of female CD102+ macrophages expressed mRNA for CXCL13 and ApoC1 than their male counterparts, and CXCL13 mRNA was also higher on a per cell basis in female cells (Fig. 4D, E). In contrast, PrimeFlow measurement of mRNA for ApoE, the most highly expressed of all differentially expressed genes by female cells by RNAseq, revealed that all peritoneal macrophages expressed ApoE irrespective of sex, but that expression was higher in female cells on a per cell basis. Hence, the transcriptional differences seen at population level appear to result from differential gene expression at a single cell level but also from different frequencies of gene-expressing cells amongst the CD102+ population.
The majority of genes more highly expressed by male peritoneal CD102+ macrophages were associated with cell cycle, including Cdk1, E2f2, and Mki67 (Fig. 4B & Table S1). Pathway analysis also revealed that at least 162 of the 338 genes differentially up-regulated in male CD102+ macrophages were associated with proliferation, and cell cycle-related processes predominated among the significantly enriched pathways (Table S2). Short-term BrdU pulse-chase experiments confirmed that male CD102+ macrophages have elevated levels of in situ proliferation compared with their female counterparts (Fig. 4G). These analyses also identified that Retlna, which encodes the immunomodulatory cytokine RELMa and is expressed specifically by those resident peritoneal macrophages that are most recently-derived from monocytes (21), was differentially expressed between sexes, with higher expression by male cells at both the mRNA and protein level (Fig. 4H).
Of note, a number of genes previously reported to distinguish long-lived, embryonically-derived macrophages from those of recent BM origin in the lung and liver were more highly expressed in females. These included receptors involved in phagocytosis and immunity (i.e. Timd4, Colec12, and Cd209 family members), Apoc1, as well as the bone morphogenic receptor Bmpr1a (Table S1) (26, 45, 46). Lowering the stringency of selection of differentially-expressed genes identified additional genes within the female-specific cluster that have been associated with embryonically-derived or long-lived macrophages, including Cd163 that also encodes a phagocytic receptor (26) (46). Furthermore, to discern systemic from local effects of sex, we compared gene expression by CD102+ macrophages from female peritoneal cavity to pleural CD102+ macrophages from both sexes. This analysis identified a module of 18 genes that was uniquely upregulated by female peritoneal macrophages, and that included Apoc1, Cd209b, and Colec12, as well as Saa3, C4b, and Tgfb2 (Table S3). Conversely, the 86 genes uniquely downregulated by female peritoneal macrophages compared with the other CD102+ populations were highly enriched for cell cycle related genes and pathways (Table S3&4). Thus, the more limited proliferative activity of female peritoneal macrophages and their expression of numerous immune-related genes appear either related to their slower replenishment from the bone-marrow or regulated directly by the unique signals present within the female peritoneal microenvironment.
scRNA-seq reveals equivalent cluster identities in male and female peritoneal macrophages but these exhibit dimorphisms in abundance and gene expression
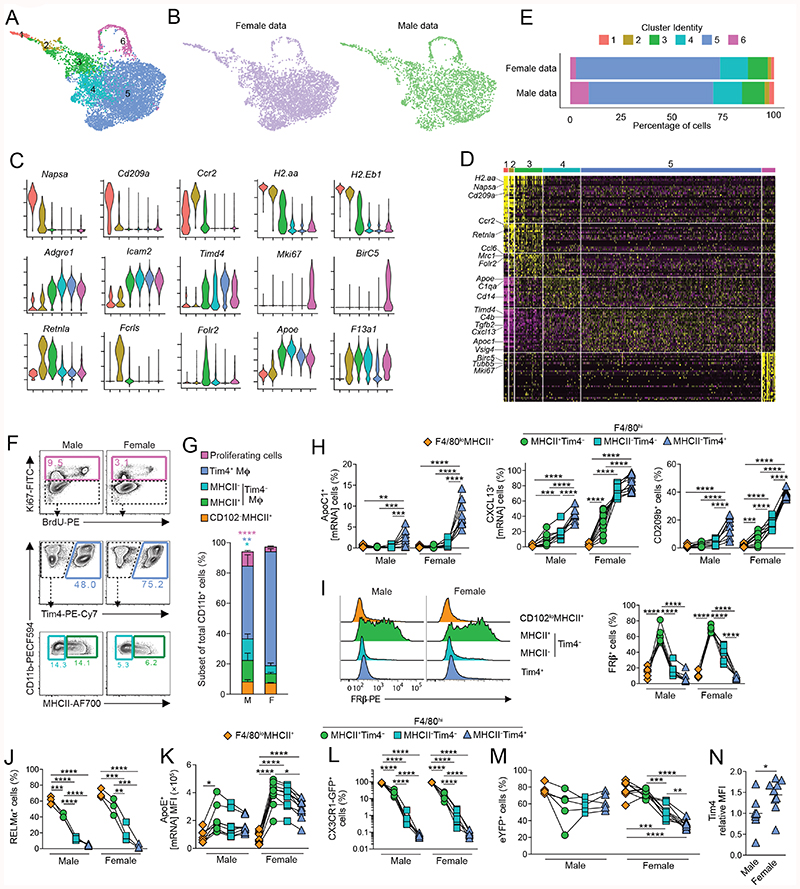
We next applied single cell RNA sequencing (scRNA-seq) to determine whether the transcriptional differences seen in our population-level data were the result of gene differences at a single cell level or if dimorphism was a reflection of differential subset composition between the sexes. A broad approach was used to capture all CD11b+ cells depleted of granulocytes and B1 B cells to allow both CD102- F4/80loMHCII+ mononuclear phagocytes and resident CD102+ cells to be examined. These cells were FACS-purified from age-matched 12-week-old male and female mice and droplet-based scRNASeq performed using the 10X Genomics platform. 10,000 sorted cells of each sex were sequenced and following quality control, analysis was performed on 4341 and 2564 cells from female and male respectively.
Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction analysis revealed 6 clusters that were present in both male and female cells (Fig. 5A,B). Given that the starting population of CD11b+ cells is known to be phenotypically heterogeneous, containing resident CD102+F4/80hi macrophages, CD102- F4/80lo MHCII+ macrophages and CD11c+MHCII+ cDC2 (20–23), we first used a panel of known markers to validate subset identity (Fig. 5B). 3 clusters of resident macrophages (3–5) could be identified on the basis of their high expression of Adgre1 (F4/80) and Icam2 (CD102). As expected, these were clearly distinct from short-lived CD102-F4/80loMHCII+ macrophages and cDC2, which were found in clusters 1 and 2 respectively, and expressed Ccr2 (Fig. 5C, D) (21). However, CD102-F4/80loMHCII+ macrophages and cDC2 could be distinguished from one another on the basis of expression of the DC markers Cd209a and Napsa (28, 47), and of Retnla and Fclrs, which we and others have shown to be signature markers of cavity CD102-F4/80loMHCII+ macrophages (21, 23, 48). Cluster 6 was defined by genes associated with cell cycle, such as Mki67 and Birc5, suggesting this cluster represents proliferating cells. In both sexes, the majority of cells was in cluster 5 (Fig. 5E), which was characterised by markers of resident F4/80hiCD102+ macrophages including Icam2, Prg4, VSig4 and Tgfb2 (Fig, 5C, D & Table S5) that form part of the core peritoneal macrophage-specific transcriptional signature (49); cluster 5 cells also expressed markers of long-lived macrophages, including Timd4 and Apoc1, confirming the findings above. Although the cells in cluster 3 expressed Icam2, they also expressed a number of genes that were highly expressed by the CD102-F4/80loMHCII+ macrophages in cluster 2, such as Retnla, H2.Aa and Ccr2, suggesting a common origin of these clusters, or a close relationship between them. This analysis also identified genes expressed more highly by cluster 3, including Folr2, which encodes the beta subunit of the folate receptor (FRβ). Although cluster 4 showed a distinct pattern of gene expression, such as high expression of Apoe, it also shared features with cluster 3 and cluster 5, suggesting it may contain differentiation intermediates. Consistent with our earlier analysis, we found that the Timd4- expressing cluster 5 was more abundant amongst female cells, whereas more male cells were found within clusters 1, 2, 3 and 6 (Fig. 5C).
Figure 5. scRNAseq analysis reveals dimorphic macrophage heterogeneity.
(A) UMAP dimensionality reduction analysis of 4341 and 2564 number of cells from the peritoneal cavity of 19 week-old male or female mice identifying 6 clusters.
(B) UMAP profile of female and male peritoneal cells.
(C) Feature plots displaying expression of individual genes by merged female/male cells.
(D) Heatmap displaying the 10 most differentially expressed genes by each cluster from A (select genes highlighted).
(E) Relative frequency of each cluster in the female and male dataset.
(F) Representative expression of Ki67 and BrdU incorporation by CD102+ macrophages (top panels), expression of Tim4 by Ki67- CD102+ macrophages (middle panels), and expression of MHCII by Tim4- CD102+ peritoneal macrophages from 10–12 week old male or female C57BL/6 mice.
(G) Relative frequency of each cluster as a proportion of all CD11b+ cells determined by flow cytometry in F.
(H) Expression of Apoc1 (mRNA), CXCL13 (mRNA) and CD209b protein by CD102loMHCII+ and Tim4/MHCII-defined CD102+ peritoneal macrophages from 10-12 week old male or female C57BL/6 mice. Data represent 8 (male) and 10 (female) mice per group pooled from three independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(I) Histograms show representative expression of FRβ by CD102loMHCII+ and Tim4-defined CD102+ peritoneal macrophages from 10-12 week old male or female C57BL/6 mice. Scatter plot show frequency of FRβ+ cells amongst CD102loMHCII+ and Tim4/MHCII-defined CD102+ peritoneal macrophages. Data represent 6 (female) or 7 (male) mice per group pooled from two independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(J) Expression of RELMa by CD102loMHCII+ and Tim4/MHCII-defined CD102+ peritoneal macrophages from 10 week old male or female C57BL/6 mice. Data represent 3 mice per group from one of at least 5 independent experiments **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(K) Frequency of ApoE+ (mRNA) cells amongst CD102loMHCII+ and Tim4/MHCII-defined CD102+ peritoneal macrophages (left) and mean fluorescence intensity of ApoE by these subsets (right) from 10-12 week old male or female C57BL/6 mice. Data represent 8 (male) and 10 (female) mice per group pooled from three independent experiments. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(L) Expression of CX3CR1-GFP by CD102loMHCII+ and Tim4/MHCII-defined CD102+ peritoneal macrophages from 15 week old male or female Cx3cr1+/gfp mice. Data represent 5 (female) or 7 (male) mice per group pooled from three independent litters. ****P<0.0001. One-way ANOVA with Tukey’s multiple comparisons test.
(M) Frequency of eYFP+ cells amongst F4/80, MHCII and Tim4-defined macrophages obtained from 16- week-old male and female CD11cCre.Rosa26 LSL-eYFP mice. Symbols represent individual animals and horizontal bars represent the mean. Data represent 5 (male) or 9 (female) mice per group pooled from two independent experiments.
(N) Relative mean fluorescence intensity of Tim4 expression by Tim4+ CD102+ peritoneal macrophages from 10-12 week old male or female C57BL/6 mice. Data represent 8 (male) or 11 (female) mice per group pooled from three independent experiments. *P<0.05. Mann Whitney test.
We next used flow cytometry to validate the additional heterogeneity uncovered by our scRNA-seq analysis. Consistent with the unbiased clustering and our previous study (21), macrophages expressing Tim4 (corresponding to cluster 5) were more abundant in female than in male mice (Fig. 5F, G). Moreover, using this strategy we validated high expression of ApoC1, V-set immunoglobulin-domain-containing 4 (VSIG4) and CXCL13 by this subset of cells (Fig. 5H and Fig. S7A). Although not identified as a cluster defining gene in our scRNAseq analysis due to low coverage, we found that CD209b displayed the same pattern of expression as CXCL13 (Fig. 5H). Again, confirming our scRNA-seq, we found the Tim4- fraction of CD102+ macrophages to be more abundant in males. Macrophages lacking Tim4 expression could be divided into MHCII+ and MHCII- subsets, which corresponded to clusters 3 and 4 in our scRNA-seq analysis (Fig. 5F,G). Consistent with this, we found that FRβ was highly and selectively expressed at protein level by Tim4-MHCII+ CD102+ macrophages (Fig. 5I), and these expressed RELMa at a higher level than all other CD102+ macrophages (Fig. 5J). While Apoe was proposed to define cluster 4 in our scRNAseq analysis, we found it was expressed by all CD102+ macrophages (Fig. 4F), although, in females, it was most highly expressed by Tim4-MHCII+ and the level decreased progressively to Tim4+MHCII– CD102+ macrophages (Fig. 5K). The phenotypic profile of cluster 3 (MHCII+Tim4 CD102’) suggested that they may represent the most derivatives from CD102-F4/80loMHCII’ macrophages. Consistent with this, they also expressed the highest level of CX3CR1, which is highly expressed by CD-102 F4/80loMHCII’ macrophages (Fig. 5L). To test this directly, we reanalysed our data from adult CD11cCre.Rosa26 LSL-eYFP mice to assess if MHCII/Tim4-defined subsets showed differential levels of replenishment. Notably, we found that MHCII-expressing Tim4-CD102+ macrophages showed equivalent labelling to CD102- F4/80loMHCII+ macrophages in female CD11cCre.Rosa26 LSL-eYFP mice, indicative of more recent derivation from CD102-F4/80loMHCII+ cells (Fig. 5M). Consistent with their intermediate transcriptional profile, Tim4-CD102+ macrophages that had lost MHCII expression showed intermediate labelling when compared with their MHCII+Tim4- and Tim4+ counterparts. No difference in eYFP labelling between Tim4-defined subsets was noted in male mice, consistent with more rapid replenishment of all subsets of macrophages in this environment. Collectively these data show that excluding proliferating cells, resident peritoneal macrophages comprise three main clusters, with Tim4- macrophages displaying an intermediate phenotype compared with F4/80loMHCII+ macrophages and Tim4+ macrophages. Furthermore, while equivalent clusters of CD102+ peritoneal macrophages are found in males and females, our analysis suggests that sexual dimorphisms in gene expression in CD102+ cells identified by population level RNA-seq arise in part due to differences in abundance of these clusters.
Finally, we performed differential gene expression analysis on our scRNA-seq datasets to distinguish dimorphisms that arise due to differential gene expression across all CD102+ cells irrespective of cluster identity from those that only occur within individual clusters, and potentially reveal sex-dependent gene expression not detected by our population-level RNAseq analysis. Excluding X and Y-link genes and mitochondrial or ribosomal related genes that can arise from stress induced during cell isolation, this identified 51 and 116 genes expressed more highly by female and male cells, respectively (Table S6). Almost 50% of these gene differences were validated by our population level RNA-seq dataset when the 1.5 log-fold cut-off limit was removed to allow detection of subtle differences (Table S6). For instance, in keeping with our PrimeFlow analysis (Fig. 5H, 5K & Table S7), Cxc113 and Apoe were found to be more highly expressed in female cells of all clusters of CD102+ macrophages (Table S6). Similarly, Saa3, C4b and Timp2 were found to be more highly expressed by female CD102+ macrophages irrespective of cluster identity, whereas Vim, Arg1, S100 family genes and SerpinB2 and Serpbin B1a were among those genes expressed more highly by male CD102+ cells irrespective of cluster. However, over 50% of differentially expressed genes were either unique to a single cluster of CD102+ macrophages or shared across only 2 of the 3 populations (Table S6). For example, in spite of being a defining marker of cluster 5, Timd4 was also identified as being differentially expressed within this cluster between the sexes. Consistent with this, female Tim4+ macrophages expressed markedly more surface Tim4 on per-cell basis (Fig. 5N). Furthermore, Folr2 and Vsig4, neither of which had been identified by population-level RNAseq analysis, were both identified by scRNA-seq (Table S6) and flow cytometry (Fig.5I & Table S7) to be expressed more highly by female cells within cluster 4 (MHCII- Tim4-). However, there were some discrepancies between datasets. For example, while Cd209b expression appeared uniquely dimorphic within cluster 5 (Table S6), our flow cytometric analysis suggested that all clusters of CD102+ macrophages exhibit a degree of differential expression of this receptor (Fig. 5H and Table S7). Furthermore, while Apoc1 was identified as differentially expressed across all clusters of female cells (Table S6), our Primeflow analysis suggests this dimorphism is restricted to cluster 5 (Fig. 5H and Table S7). Such discrepancies likely reflect detection biases between these methods and highlight the need for validation of differentially expressed genes. Nevertheless, our scRNA-seq analysis revealed that pan-cluster and cluster-specific dimorphisms in gene expression contribute to the distinct transcriptional landscape of male and female peritoneal macrophages.
Differential replenishment and environmental signals drive the dimorphic features of peritoneal macrophages
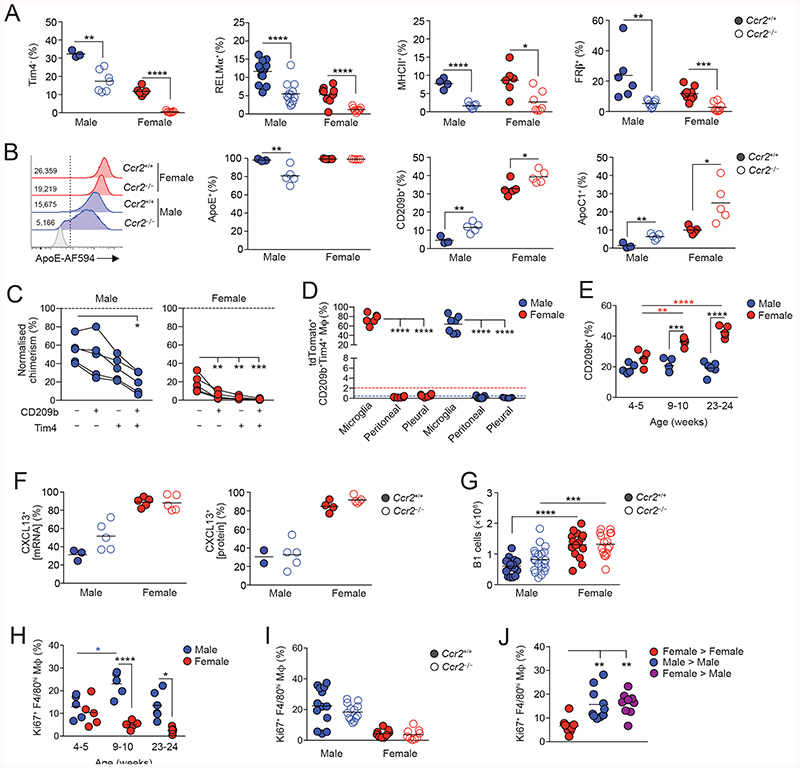
To dissect the dimorphic features of CD102+ macrophages that could be related to longevity from those more directly controlled by dimorphic environmental signals, we next assessed expression of these in Ccr2 -/- mice in whom macrophage replenishment is markedly reduced due to severe monocytopenia (50, 51). Strikingly, the frequency of Tim4- macrophages, as well as those expressing RELMa, FRβ or MHCII were markedly reduced in Ccr2 -/- mice compared with Ccr2+/+ mice irrespective of sex, confirming these cells to be recently derived from monocytes (Fig. 6A). In males, CCR2 deficiency also led to reduced expression of ApoE and emergence of an ApoE- subset of CD102+ macrophages (Fig. 6B). In contrast, a higher proportion of CD102+ macrophages in Ccr2 -/- mice expressed CD209b and ApoC1, markers that are characteristic of the Tim4+MHCII- subset, suggesting these markers may be expressed selectively by long-lived macrophages (Fig. 6B). Consistent with this, Tim4+ macrophages expressing CD209b displayed the lowest level of replacement by donor cells in BM chimeras when compared with all other CD209b/Tim4-defined macrophages, even in male mice where overall replenishment from the bone marrow is markedly higher (Fig. 6C). The low levels of replacement of peritoneal CD209b+Tim4+ macrophages does not reflect derivation from yolk sac progenitors, as, unlike microglia in the brain, these cells are not labelled in male or female Cdh5 Cre-ERT2.Rosa26 LSL-tdTomato mice (Fig. 6D), which allow tracing of cells arising from yolk sac haematopoiesis (52). Similar results were obtained with CD209b+Tim4+ macrophages in the pleural cavity. Hence, despite being long-lived, CD209b+Tim4+ macrophages derive from conventional haematopoiesis in both sexes. Importantly, temporal analysis revealed that while little difference in abundance of CD209b-expressing CD102+ macrophages was seen in pre-pubescent (4-5-week-old) male and female mice, these cells accumulated progressively in the cavity of female mice following sexual maturation. This did not occur in male mice, consistent with their higher rate of replenishment from the bone marrow and indicating that acquisition of CD209b expression appears to be associated with time-of-residency in the female cavity (Fig. 6E). Similarly, enrichment of cells expressing other markers of cluster 5, such as VSIG4, became apparent in females with age (Fig. S7B) suggesting numerous cluster 5 genes may be regulated by time-of-residency.
Figure 6. Differential replenishment and environmental signals drive the dimorphic features of peritoneal macrophages.
(A) Frequency of Tim4-, RELMα+, and MHCII+ and RFβ’ cells amongst CD102+ macrophages from the peritoneal cavity of unmanipulated age-matched Ccr2 -/- or Ccr2 -/- mice. Symbols represent individual animals and horizontal bars represent the mean. Data are pooled from two independent experiments. Tim4 data represents 3 (Ccr2+/+ males), 6 (Ccr2 +/+ females) or 7 (Ccr2 -/-) 22-28 week old mice per group. RELMa data represent with 13 male and 9 female 14-18 week old mice per group. MHCII data represent 4 (Ccr2 +/+ males), 6 (Ccr2 +/+ females) or 7 (Ccr2 +/+) 22-28 week old mice per group. FRβ data represent 6 (Ccr2 +/+ males), 8 (Ccr2 +/+ females) or 9 (Ccr2 +/+ females and Ccr2 +/+ males) 13 week old mice per group. *P<0.05, **P<0.01, ****P<0.0001. Student’s t test with Holm-Sidak correction.
(B) Representative expression of ApoE by CD102+ macrophages (histograms) and frequency of ApoE-, CD209b+ and ApoC1+ cells from the peritoneal cavity of unmanipulated age-matched Ccr2 +/+ or Ccr2 -/- mice. Symbols represent individual animals and horizontal bars represent the mean. Data are pooled from two independent experiments and represents 3 (Ccr2 +/+ males), 6 (Ccr2 +/+ females) or 7 (Ccr2 -/-) 22-28 week old mice per group. *P<0.05, **P<0.01. Student’s t test with Holm-Sidak correction.
(C) Normalized non-host chimerism of CD209/Tim4-defined subsets of CD102+ macrophages from the peritoneal cavity of sex matched tissue-protected BM chimeric mice 8 weeks post-reconstitution. Data are normalised to the non-host chimerism of Ly6Chi monocytes. Data represent 5 mice per group from one experiment. *P<0.05, ***P<0.001. Paired Student’s t test.
(D) Proportion of tdTomato+ (Ai14) cells amongst microglia, peritoneal and pleural macrophages from 15- week-old Cdh5 Cre-ERT2.Rosa26 LSL-Ai14.Cx3cr1 +/gfp mice administered 4-hydroxytamoxifen at E7.5. Data represent 6 (female) or 7 (male) mice per group from three independent litters. ****P<0.0001. One-way ANOVA followed by Tukey’s multiple comparisons test.
(E) Frequency of cells expressing CD209b amongst CD102+ macrophages obtained from the peritoneal cavity of unmanipulated C57BL/6 mice of indicated ages. Symbols represent individual animals and horizontal bars represent the mean. *P<0.05, **P<0.01, ***P<0.001, ****P<0.0001. Two-way ANOVA with Sidak’s multiple comparisons test.
(F) Frequency of cells expressing CXCL13 mRNA (left) or CXCL13 protein (right) amongst CD102+ macrophages obtained from the peritoneal cavity of unmanipulated 22-28 week old Ccr2+’+ or Ccr2 -/- mice. Symbols represent individual animals and horizontal bars represent the mean. CXCL13 mRNA data represents 3 (Ccr2 +/+ males), 5 (Ccr2 +/+ females) or 5 (Ccr2 -/-) mice per group. CXCL13 protein data represents 2 (Ccr2 +/+ males), 4 (Ccr2 +/+ females) or 5 (Ccr2 -/-) mice per group.
(G) The absolute number of B1 cells obtained from the peritoneal cavity of unmanipulated age matched 14-28 week old Ccr2+/+ or Ccr2 -/- mice. Data represent 15 (Ccr22+/+ females), 16 (Ccr2 +/+ females), 17 (Ccr2 +/+ males) or 20 (Ccr2 +/+ females) mice per group pooled from four independent experiments.
(H) Frequency of cells expressing Ki67 amongst F4/80hi macrophages obtained from the peritoneal cavity of unmanipulated C57BL/6 mice of indicated ages. Symbols represent individual animals and horizontal bars represent the mean. *P<0.05, ****P<0.0001. Two-way ANOVA with Tukey’s multiple comparisons test.
(I) Frequency of Ki67+ cells amongst peritoneal F4/80hi macrophages obtained from the peritoneal cavity of unmanipulated 14-18 week old Ccr2 +/+ or Ccr2 +/+ mice. Data represents 15 (Ccr2 females), 16 (Ccr2 -/- females), 17 (Ccr2+/+ males) or 20 (Ccr2 +/+ females) mice per group pooled from 2 experiments.
(J) Frequency of Ki67+ cells amongst peritoneal F4/80hi macrophages obtained from sex matched or mismatched tissue protected BM chimeric mice 8-12 weeks post-reconstitution. Data represent 9 (female > male) or 10 (sex matched groups) mice per group pooled from one of two independent experiments. **P<0.01. One-way ANOVA followed by Tukey’s multiple comparisons test.
Not all dimorphic features of peritoneal CD102+ macrophages were influenced by their rate of replenishment. For instance, the intrinsically higher expression of CXCL13 by female macrophages was not altered by CCR2 deficiency (Fig. 6F). In parallel, although we confirmed previous findings of a clear dimorphism in the numbers of B1 cells between adult male and female mice (15) and this developed gradually following sexual maturation (Fig. S7C), this phenomenon remained in Ccr2 -/- mice (Fig. 6G). Similarly, while the higher levels of proliferation by male CD102+ macrophages developed following sexual maturation (Fig. 6H), this was unaffected by CCR2 deficiency (Fig. 6I). This evidence that certain dimorphic features are driven by environmental factors, independent of cell replenishment was supported further by the fact that macrophages derived from female BM in the cavity of chimeric male mice showed levels of proliferation that were identical to those of male BM derived macrophages in the male cavity and were higher than those of female BM derived macrophages in female cavity (Fig. 6J). Thus, the differential proliferation of female and male macrophages is not due to cell-intrinsic differences in their proliferative activity.
Taken together these data demonstrate that both local imprinting and differential rate-of-replenishment contribute to the sexual dimorphisms seen in peritoneal macrophages.
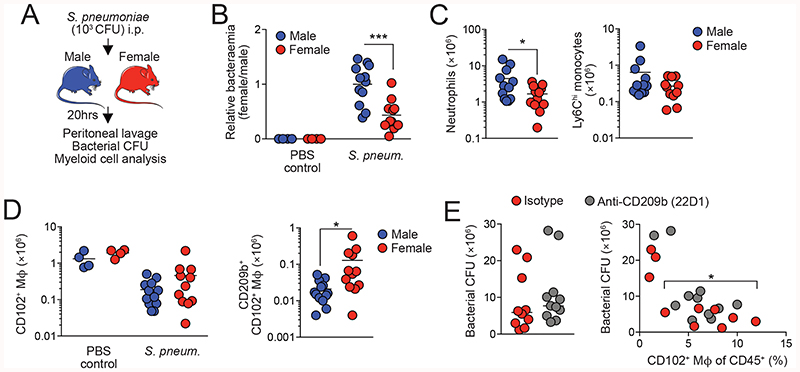
Differential CD209b expression confers an advantage on female macrophages in the setting of pneumococcal peritonitis
We postulated that differential expression of key pattern recognition receptors such as CD209b might endow female macrophages with an enhanced ability to deal with bacterial infection. To test this idea, we examined the acute peritonitis caused by infection with the gram-positive bacterium Streptococcus pneumoniae (Fig. 7A). At low doses, resident macrophages, and in particular CD209b, are indispensable for protective immunity in this model (53, 54) whereas recruitment of neutrophils is not required (53) and hence avoids any confounding effects of systemic sex-dependent effects on innate immune responses that have been reported previously (55). As CD209b is expressed exclusively by CD102+ macrophages in the peritoneal cavity (Fig. S6A), this model allowed us to directly assess the importance of differential CD209b expression by CD102+ macrophages in bacterial elimination. Strikingly, females showed enhanced capability to control S. pneumoniae infection, with lower levels of bacteria in peritoneal fluid of female mice compared with their male counterparts 20hrs after inoculation (Fig. 7B). Fewer neutrophils and Ly6Chi monocytes were present in the female cavity compared with male mice (Fig. 7C & Fig. S8), consistent with a model in which resident macrophages control infection (53). In contrast, while the well-documented macrophage ‘disappearance reaction’ (56) occurred in both male and female mice after infection (Fig. 7D), higher numbers of CD209b-expressing macrophages persisted in the female cavity (Fig. 7D). Administration of an anti-CD209b blocking antibody (22D1) (57) led to increased levels of bacteremia in female mice, although this did not attain statistical significance due to variance in bacterial counts in some mice in whom the macrophage ‘disappearance reaction’ was more pronounced (Fig. 7E).
Figure 7.
Thus, dimorphic expression of key immune receptors and molecules leads to differential ability to handle local bacterial infection.
Discussion
Understanding the extrinsic and intrinsic factors that govern tissue macrophage differentiation is a key goal in the field of macrophage biology. Here we reveal a striking effect of sex on the phenotypic and transcriptional identity of resident peritoneal macrophages and demonstrate that this contributes to the sex-dependent resistance of mice to bacterial peritonitis. Moreover, we show that this arises through a combination of dimorphic microenvironmental signals and sex-dependent differences in the rate of macrophage renewal from the bone marrow.
Using classical defining markers such as F4/80, CD11b and CD102, we found peritoneal macrophages from male and female mice to be phenotypically identical. However, mRNA sequencing revealed marked dimorphism in the transcriptional fingerprint of resident peritoneal macrophages under homeostatic conditions. Importantly, female CD102+ macrophages expressed higher levels of genes associated with lipid uptake and transport as well as immune defence/response. In contrast, the signature of male peritoneal macrophages was dominated by cell cycle associated genes consistent with their elevated levels of proliferation, a dimorphism we have reported previously (21). Although others have reported dimorphic expression of TLRs and CD14 by peritoneal macrophages (38, 39), we found no significant difference in mRNA transcripts of the adaptor protein MyD88, CD14 or any TLRs, consistent with more recent analysis of mRNA (44) and protein expression (55).
Despite a significant degree of transcriptional difference at the population level, single cell mRNA sequencing showed that male and female resident CD102+ macrophages encompassed three transcriptionally-defined clusters of cells. Of these, the cells in cluster 3 expressed CD102 together with MHCII, RELMa and CX3CR1, all of which are key markers of F4/80loMHCII+ peritoneal macrophages, suggesting that cluster 3 may be recently derived from the F4/80loMHCII+ macrophage population that is derived from blood monocytes in adult mice (21–23). As cells in cluster 4 shared features with both the MHCII-defined cluster 3 and the dominant cluster 5 population of Tim4+ macrophages, these may represent a further intermediate differentiation state. Consistent with this idea, Tim4- cells were largely ablated in monocytopenic Ccr2 -/- mice. A linear-developmental relationship that culminates at cluster 5 would be consistent with the greater abundance of cells in this cluster in females, given the slower entry of bone-marrow-derived cells into the female CD102+ macrophage pool. However, it seems unlikely that such a linear developmental relationship between clusters exists in males, as Tim4 and MHCII defined subsets were found to be replenished at similarly high rates. Hence, what dictates cluster identity in males remains unclear.
Given that the rate of replenishment from BM was markedly different between the sexes, a finding consistent with recent monocyte fate mapping using Ms4a3 Cre.Rosa26 LSL-tdTomato mice (58), this raised the possibility that transcriptional differences could reflect different ontogenies of male and female peritoneal macrophages. Indeed, a number of the genes we found to be expressed more highly by female peritoneal macrophages, including Colec12, Cd163, Bmpr1a, Cdc42bpa, Timd4, Apoc1, and members of the Cd209 family have been reported to be expressed by embryonically, but not monocyte-derived macrophages in other tissues (26, 59). However, the fact that a proportion of BM-derived peritoneal macrophages can acquire the expression of at least some of these “embryonic” signature markers (e.g. Tim4, CD209b) in the setting of tissue-protected BM chimeras, suggests that this is more likely related to their time-of-residency rather than rigid differences related to origin. Consistently, co-expression of Tim4 and CD209b identifies the longest-lived macrophages in the peritoneal cavity irrespective of sex. The concept that macrophages require prolonged residence within the tissue to acquire their characteristic features is consistent with work from the Guilliams lab showing that acquisition of Tim4 expression by monocyte-derived cells that engraft in the liver following deletion of endogenous Kupffer cells increases with time (26). Notably, human peritoneal macrophages that align with mouse F4/80hi macrophages exhibit high expression of TIMD4, CD209, COLEC12, CD163 and APOC1 (60), suggesting these may represent phylogenetically conserved markers of long-lived macrophages. Importantly, certain dimorphisms (e.g. proliferation and CXCL13 expression) appeared to be regulated independently of replenishment kinetics, consistent with previous data showing that the proliferative capacity of macrophages is determined by signals in the local microenvironment rather than their origin (21, 26, 61) While our ovariectomy studies clearly implicate the ovaries and/or their products in controlling lower levels of replenishment in females, it seems unlikely that estradiol plays a key role, as administration of exogenous estradiol failed to rescue the effects of OVX. In addition, estradiol is unlikely to be responsible for the lower proliferation of female peritoneal macrophages, as estradiol is reported to increase rather than inhibit proliferation of these cells (37). Nevertheless, although expression of receptors for progesterone and androgens did not differ between the sexes, we cannot categorically rule out a role for these steroids in generating sex dimorphisms. Thus, the exact local factor(s) driving the sex dimorphisms identified here remain to be elucidated.
The incidence and severity of sepsis and post-surgical infections are profoundly lower in women than men (67), but the mechanisms underlying these differences remain unclear. Our finding that female mice are more resistant to S. pneumoniae peritonitis is consistent with previous work on group B streptococcal peritonitis (39). However, while others attributed this to other elements of innate immune responses, such as neutrophil recruitment (55), our data suggest that the resistance of females is at least, in part, due to differences in resident peritoneal macrophages, such as elevated expression of CD209b. However, we cannot rule out other mechanisms, such as dimorphic expression of CXCL13 which plays a central role in recruiting natural IgM-producing B1 cells (2) (62, 63), or elevated expression of complement components C1q, C3, and C4b, as well as Cfb that are essential for innate resistance to against S.pneumoniae (62–64). Nonetheless, we propose that this heightened barrier function in the female peritoneum may have evolved to mitigate the risk of sexually transmitted infection disseminating from the lower female reproductive tract (65) or to protect against puerperal peritonitis.
Our studies highlight the importance of taking age and sex into account when understanding the peritoneal response to disease and implicate time-of-residency as an underlying determinant of resident macrophage function. Further work is needed to understand the molecular processes that underlie the requirement for time-of-residency on expression of these genes and to identify the local signals that govern this process. Beyond the cavity, our findings also have wider implications for the molecular mechanisms that drive dimorphic production of natural IgG by peritoneal B1 cells that provides women and infants with heightened resistance to blood-borne bacterial infections, particularly as these antibodies are lost in the absence of peritoneal macrophages (15).
Materials and methods
Study Design
We performed phenotypic, transcriptomic and functional analysis of peritoneal macrophages to identify sexual dimorphisms that may exist. We used tissue-protected bone marrow chimeric mice and genetic fate mapping to assess the replenishment kinetics under normal physiological conditions. To assess the role for sex hormones in generating/maintaining sex dimorphisms, we performed ovariectomy in C57BL/6 mice. To assess macrophage function in the context of infection, we used a model of low dose Streptococcus pneumoniae infection (51).
Experimental Animals
Wild type C57BL/6J CD45.2+, congenic CD45.1+CD45.2+ mice, Ccr2 -/-, Rag1 -/- and Balb/c mice were bred and maintained in SPF facilities at the University of Edinburgh, UK. In some experiments, C57BL/6J (Crl) mice were purchased from Charles River, UK or bred and maintained at the University of Manchester, while Ccr2-/-, Rag1-/- and control mice were bred and maintained at the University of Glasgow. ItgaxCre (66) (referred here to CD11cCre) mice were crossed with Rosa26 LSL-YFP mice (a gift from Dr. Megan Mcleod, University of Glasgow, UK) and maintained at the University of Glasgow. For Cdh5 Cre-ERT2 fate mapping, WT females aged 6-10 weeks were subjected to timed matings with Cdh5 CreERT2+/- or Cdh5 CreERT2+/+ Rosa26 tdT/tdT Cx3cr1 gfp/gfp males (experiments performed at CIML, Marseille, France). Successful mating was judged by the presence of vaginal plugs the morning after, which was considered 0.5days post conception. For induction of reporter recombination in the offspring, a single dose of 4-hydroxytamoxifen (4OHT; 1.2mg) was delivered by i.p. injections to pregnant females at E7.5. To counteract adverse effects of 4OHT on pregnancy, 4OHT was supplemented with progesterone (0.6mg). In cases when females could not give birth naturally, pups were delivered by C-section and cross-fostered with lactating CD1 females. All experimental mice were age and sex matched. To perform estrous staging, vaginal lavage was performed and cellular content examined following haematoxylin and eosin staining, as previously described (67). For high fat diet (HFD) experiments, tissue protected BM chimeric mice were placed on HFD (58 kcal% fat and sucrose, Research Diet, D1233li) for 8 weeks starting 4 weeks post reconstitution. Experiments performed at UK establishments were permitted under license by the UK Home Office and were approved by the University of Edinburgh and University of Manchester Animal Welfare and Ethical Review Body’s or the University of Glasgow Local Ethical Review Panel.
Surgery
Ovariectomy was performed on 6-week-old wild type (C57BL/6J) or tissue protected BM chimeras 8 weeks post-reconstitution (16 weeks of age). Briefly, dorsal unilateral or bilateral ovariectomy (OVX) was performed and mice allowed to recover for up to 8 weeks. Sham surgery was performed to control for the effects of surgery on the peritoneal environment. This involved identical surgery except for the excision of the ovary/ovaries. Surgery was performed under isoflurane anaesthesia followed by a postoperative analgesic, buprenorphine (0.1 mg/kg), for pain management. In some experiments following 7 days of recovery, mice received exogenous estradiol in the form of E2 valerate (E2; 0.01 mg/kg) s.c. thrice weekly for 3 weeks.
Tissue-protected BM chimeric mice
Anaesthetised 6-12 week old C57BL/6J CD45.1+CD45.2+ animals were exposed to a single dose of 9.5 Gy g-irradiation, with all but the head and upper thorax of the animals being protected by a 2 inch lead shield. Animals were subsequently given 2-5 ×106 BM cells from sex matched or mismatched congenic CD45.2+ C57BL/6J animals by i.v. injection. Unless specified, mice were left for a period of at least 8 weeks before analysis of chimerism in the tissue compartments.
BrdU injection
For labelling of proliferating cells, mice were injected s.c. with 100μl of 10mg/ml BrdU (Sigma) in Dulbecco’s PBS 2hr before culling.
Preparation of single cell suspensions
Mice were sacrificed by CO2 inhalation or by terminal anaesthesia followed by exsanguination. Peritoneal and pleural cavities were lavaged with RPMI containing 2mM EDTA and 10mM HEPES (both Invitrogen) as described previously (68). Any samples with excessive erythrocyte contamination were excluded from analysis. Omental tissue was excised, chopped finely and digested in 0.5ml pre-warmed collagenase D (1mg/ml; Roche) in RPMI 1640 media supplemented with 2% FCS for 15 minutes in a shaking incubator at 37°C. Following disaggregation with a P1000 Gilson, omental tissue was digested for a further 20mins before being placed on ice. 2.5μl of 0.5M EDTA was added to each sample to inhibit enzymatic activity. Cell suspensions were passed through an 100μm filter and centrifuged at 1700rpm for 10mins. The resulting cell suspension was subsequently passed through a 40μm strainer prior to cell counting. All cells were maintained on ice until further use. Cellular content of the preparations was assessed by cell counting using a Casey TT counter (Roche) in combination with multi-colour flow-cytometry.
Flow cytometry
Equal numbers of cells were blocked with 0.025 μg anti-CD16/32 (2.4G2; Biolegend) and 1:20 heat-inactivated mouse serum (Invitrogen), and then stained with a combination of the antibodies detailed in Supplementary Table 8. Where appropriate, cells were subsequently stained with streptavidin-conjugated fluorochromes. Dead cells were excluded using DAPI, 7-AAD or Zombie Aqua fixable viability dye (Biolegend). Fluorescence-minus-one (FMO) controls confirmed gating strategies, while discrete populations within lineage+ cells were confirmed by omission of the corresponding population-specific antibody. Erythrocytes in blood samples were lysed using 1x RBC Lysis buffer (Biolegend), as per the manufacturer’s guidelines. For intracellular staining, cells were subsequently fixed and permeabilized using FoxP3/Transcription Factor Staining Buffer Set (eBioscience), and intracellular staining performed using antibodies detailed in Supplementary Table 2. For the detection of BrdU, cells were fixed as above and incubated with 3μg DNaseI (Sigma) for 30-60mins, before being washed in PermWash (eBioscience) and then incubated with anti-BrdU antibody for 30mins at RT. Samples were acquired using a FACS LSRFortessa or AriaII using FACSDiva software (BD) and analyzed with FlowJo software (version 9 or 10; Tree Star). Analysis was performed on single live cells determined using forward scatter height (FCS-H) versus area (FSC-A) and negativity for viability dyes. For analysis of macrophage proliferation, Ki67 expression was used to determine the frequency of all CD102+/F4/80hi cells in cycle, whereas a 2h BrdU pulse before necropsy combined with Ki67 expression was used to identify cells in S phase, as described previously (69). mRNA was detected by flow cytometry using PrimeFlow technology (ThermoFisher) using probes against ApoE (probe type 10; AF568), ApoC1 (probe type 4; AF488) and CXCL13 (probe type 6; AF750) according to the manufacturer’s guidelines. For staining controls in PrimeFlow analysis, the Target Probe Hybridization step was omitted with all other steps identical to samples.
Transcriptional Analysis
Bulk RNAseq
CD102+F4/80hi cells were FACS-purified from the peritoneal and pleural cavities of unmanipulated male and female mice. For each population, 25,000 cells were sorted into 500μl RLT buffer (Qiagen) and snap frozen on dry ice. RNA was isolated using the RNeasy Plus Micro kit (Qiagen), at which point triplicates of 25,000 cells for each population were pooled. 10 ng of total RNA were amplified and converted to cDNA using Ovation RNASeq System V2 (Nugen). Sequencing was performed by Edinburgh Genomics using the Illumina HiSeq 4000 system (75PE). Raw map reads were processed with the R package DESeq2 (70) to generate the differentially expressed genes, and the normalized count reads to generate and visualize on heat maps generated by the R package pheatmap. Samples with >5% of reads mapped to ribosomal RNA were removed from analysis. DEG were determined using at least a 1.5-fold difference and adjusted p < 0.01, for each of the six pairwise comparisons. Pathway enrichment analysis was performed using the GSEA online database and the R package gskb (Gene Set data for pathway analysis in mouse) which makes predictions between each of the six pairwise comparisons, incorporating in the analysis the statistically significant differences in gene expression. The R package gskb was used to determine the chromosomal location of each of the genes and transcription factors. All R code is available upon request.
Single-cell RNAseq
10K cells for male and female sorted cells were loaded in Chromium 10x in parallel. Libraries were prepared as per manufacturer’s protocol and sequenced on Illumina Novaseq S1. Initial processing was done using Cellranger (v2.1.1) mkfastq and count (aligned to mouse assembly mm10). Preparation of analysis ready data: For each dataset (filtered data from Cell Ranger pipeline), we filtered out potentially low quality cells using dataset-specifc thresholds based on the trend of the number of genes per cell versus number of housekeeping genes per cell and number of genes per cell versus percentage of mitochondrial genes per cell curves as follows. More specifically, for the female data, we retained 4341 cells that have between 300 and 5000 genes, at least 65 housekeeping genes and percentage of mitochondrial genes over the total number of expressed genes below 2%. For the male data, we retained 2564 cells that have between 300 and 6000 genes, at least 70 housekeeping genes and percentage of mitochondrial genes over the total number of expressed genes below 2%. Finally, we filtered out genes that were expressed in less than 1% of the cells from each dataset. Clustering analysis of the data: Clustering and data merging using CCA was done using Seurat (v3.1.0). We used default parameters and 20 principal components for aligning and clustering the data. We next removed a very small cluster that lay far from all other clusters on the UMAP projection, indicating it could be either contamination or doublets and constructed a phylogenetic tree of the remaining clusters to understand the distances and relationship between them. Clusters that were closely grouped together and did not show unique markers, were merged together. The final result consists of 6890 cells grouped into 6 clusters. Identification of differentially expressed genes in CCA aligned clusters: We used MAST (v1.10) as implemented in the Seurat package and with default parameters to identify differentially upregulated genes between the identified clusters. To overcome the bias of batch effect, we found differentially upregulated genes within each dataset separately and retained the intersection of markers (conserved markers). Identification of differentially expressed genes between female and male cells: We used Student’s t-test as implemented in the Seurat package between equivalent female and male cells to identify differentially upregulated genes between male and female cells. We only retained genes with adjusted p-value based on Bonferroni correction below 0.05. For definition of cluster-specific differentially expressed genes, overlap with expression in cluster 6 was ignored given that this cluster was defined by proliferation and hence comprised cells from across other clusters.
S. pneumoniae peritonitis
S. pneumoniae were cultured overnight on blood agar plates (5% CO2, 95% air, 37 C), inoculated into Brain Heart Infusion broth, cultured for 3 h, washed, and resuspended at 104 CFU/ ml (estimated by OD595) in sterile PBS. Their concentration was verified by serial dilution and culture on blood agar plates. Groups of male and female, age-matched C57Bl/6 mice (8–14 wk of age) were inoculated intraperitoneally with 100 μl of PBS containing 103 CFU S. pneumoniae (capsular type 2 strain D39). Mice were culled 20 h later and peritoneal lavage performed using sterile PBS. 100 μl of lavage fluid was cultured for bacterial growth for 24 h. The remaining lavage fluid was centrifuged at 400g for 5 mins and the resulting cells counted and prepared for flow cytometric analysis.
Statistics
Statistics were performed using Prism 7 (GraphPad Software). The statistical test used in each experiment is detailed in the relevant figure legend.
Supplementary Material
Summary Sentence.
Peritoneal macrophages exhibit sexually dimorphic transcriptional profiles, functional capacity and replenishment kinetics.
Acknowledgements
Flow cytometry data were generated with support from the QMRI Flow Cytometry and Cell Sorting Facility, University of Edinburgh. mRNA sequencing was performed by Edinburgh Genomics, The University of Edinburgh. Edinburgh Genomics is partly supported through core grants from NERC (R8/H10/56), MRC (MR/K001744/1) and BBSRC (BB/J004243/1). Servier Medical Art was used for the generation of some of the graphics. We thank Prof’s Gerry Graham (Ccr2 -/-), Rick Maizels (Rag1 -/-) and Allan McI. Mowat (CD11cCre.Rosa26 LSL-eYFP; University of Glasgow), Prof Jeffery Pollard and Dr Erin Greaves (Ccr2 -/-; University of Edinburgh), and Dr John Grainger (C57BL/6; University of Manchester) for provision of mice. We would like to thank the ShIELD (Sheffield, Edinburgh, Newcastle,and Birmingham) consortium for access to bacterial stocks. Finally, we would like to thank J. Marshall for advice on bacteriology work.
Funding
This work was funded by the Medical Research Council UK (MR/L008076/1 to S.J.J), with additional support from the Wellcome Trust (IS3-R34 to S.J.J; PhD studentship 203909/Z/16/A to P.L.) and a Sir Henry Dale Fellowship jointly funded by the Wellcome Trust and the Royal Society (Grant Number 206234/Z/17/Z to C.C.B). SRW is funded by a Wellcome Trust Senior Clinical Fellowship (209220). PTKS received funding from the MRC (MR/N024524/1).
Footnotes
Author Contributions:
C.C.B. conceived and performed most of the experiments, analysed and interpreted the data, performed statistical analysis, wrote the manuscript and provided funding. D.A.G. designed and performed experiments and edited the manuscript. N.S. performed transcriptomic analysis (population level RNAseq) and figure generation. K.B. performed single cell RNAseq analysis and figure generation. P.L. performed experiments for generation of scRNAseq data. C.D. provided technical assistance for the design and execution of infection experiments. R.G. performed Cdh5-fate mapping experiments. M.M-P. helped with the design and execution of high fat diet experiments. T. S. performed phenotyping experiments in Manchester. C.B. helped with the design and execution of high fat diet experiments. M.B. generated and provided access to Cdh5 fate-mapper mice. S.R.W. provided reagents and assistance in the execution of infection experiments. D.D. helped with the design and execution of infection experiments. P.T.K.S. provided advice for the design and interpretation of experiments and edited the manuscript. N.B. performed the scRNAseq analysis, provided advice on the interpretation of these data, edited the manuscript and provided funding. S.J.J. conceived and performed experiments, analysed and interpreted the data, performed statistical analysis, wrote the manuscript, obtained funding and supervised the project.
Competing Interests:
The authors declare that they have no competing interests.
Data availability
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and RNA sequencing data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/). Population-level RNAseq (accession code:149014) and scRNA-seq (primary accession code GSE139999 [male: GSM4151330; female: GSM4151331]).
References
- 1.Bain CC, Jenkins SJ. The biology of serous cavity macrophages. Cell Immunol. 2018;330:126–135. doi: 10.1016/j.cellimm.2018.01.003. [DOI] [PubMed] [Google Scholar]
- 2.Ansel KM, Harris RBS, Cyster JG. CXCL13 is required for B1 cell homing, natural antibody production, and body cavity immunity. Immunity. 2002;16:67–76. doi: 10.1016/s1074-7613(01)00257-6. [DOI] [PubMed] [Google Scholar]
- 3.Roberts AW, et al. Tissue-Resident Macrophages Are Locally Programmed for Silent Clearance of Apoptotic Cells. Immunity. 2017;47:913–927.:e6. doi: 10.1016/j.immuni.2017.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang J, Kubes P. A Reservoir of Mature Cavity Macrophages that Can Rapidly Invade Visceral Organs to Affect Tissue Repair. Cell. 2016;165:668–678. doi: 10.1016/j.cell.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 5.Rana N, et al. Basal and stimulated secretion of cytokines by peritoneal macrophages in women with endometriosis. Fertil Steril. 1996;65:925–930. [PubMed] [Google Scholar]
- 6.Sikora J, Mielczarek-Palacz A, Kondera-Anasz Z. Association of the Precursor of Interleukin-1β and Peritoneal Inflammation-Role in Pathogenesis of Endometriosis. J Clin Lab Anal. 2016;30:831–837. doi: 10.1002/jcla.21944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greaves E, et al. A novel mouse model of endometriosis mimics human phenotype and reveals insights into the inflammatory contribution of shed endometrium. Am J Pathol. 2014;184:1930–1939. doi: 10.1016/j.ajpath.2014.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Greaves E, et al. Estradiol is a critical mediator of macrophage-nerve cross talk in peritoneal endometriosis. Am J Pathol. 2015;185:2286–2297. doi: 10.1016/j.ajpath.2015.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shrivastava P, Bhatia M. Essential role of monocytes and macrophages in the progression of acute pancreatitis. World J Gastroenterol. 2010;16:3995–4002. doi: 10.3748/wjg.v16.i32.3995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Burnett SH, et al. Development of peritoneal adhesions in macrophage depleted mice. J Surg Res. 2006;131:296–301. doi: 10.1016/j.jss.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 11.Burnett SH, et al. Conditional macrophage ablation in transgenic mice expressing a Fas-based suicide gene. Journal of leukocyte biology. 2004;75:612–623. doi: 10.1189/jlb.0903442. [DOI] [PubMed] [Google Scholar]
- 12.Honjo K, et al. Plasminogen activator inhibitor-1 regulates macrophage-dependent postoperative adhesion by enhancing EGF-HER1 signaling in mice. FASEB J. 2017;31:2625–2637. doi: 10.1096/fj.201600871RR. [DOI] [PubMed] [Google Scholar]
- 13.Miselis NR, Wu ZJ, van Rooijen N, Kane AB. Targeting tumor-associated macrophages in an orthotopic murine model of diffuse malignant mesothelioma. Mol Cancer Ther. 2008;7:788–799. doi: 10.1158/1535-7163.MCT-07-0579. [DOI] [PubMed] [Google Scholar]
- 14.Robinson-Smith TM, et al. Macrophages mediate inflammation-enhanced metastasis of ovarian tumors in mice. Cancer Res. 2007;67:5708–5716. doi: 10.1158/0008-5472.CAN-06-4375. [DOI] [PubMed] [Google Scholar]
- 15.Zeng Z, et al. Sex-hormone-driven innate antibodies protect females and infants against EPEC infection. Nature immunology. 2018;19:1100–1111. doi: 10.1038/s41590-018-0211-2. [DOI] [PubMed] [Google Scholar]
- 16.Ghosn EEB, et al. Two physically, functionally, and developmentally distinct peritoneal macrophage subsets. Proc Natl Acad Sci USA. 2010;107:2568–2573. doi: 10.1073/pnas.0915000107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosas M, et al. The transcription factor Gata6 links tissue macrophage phenotype and proliferative renewal. Science (New York, NY. 2014;344:645–648. doi: 10.1126/science.1251414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gautier EL, et al. Gata6 regulates aspartoacylase expression in resident peritoneal macrophages and controls their survival. The Journal of experimental medicine. 2014 doi: 10.1084/jem.20140570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Okabe Y, Medzhitov R. Tissue-specific signals control reversible program of localization and functional polarization of macrophages. Cell. 2014;157:832–844. doi: 10.1016/j.cell.2014.04.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cain DW, et al. Identification of a tissue-specific, C/EBPβ-dependent pathway of differentiation for murine peritoneal macrophages. J Immunol. 2013;191:4665–4675. doi: 10.4049/jimmunol.1300581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bain CC, et al. Long-lived self-renewing bone marrow-derived macrophages displace embryo-derived cells to inhabit adult serous cavities. Nat Commun. 2016;7:ncomms11852. doi: 10.1038/ncomms11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liao C-T, et al. IL-10 differentially controls the infiltration of inflammatory macrophages and antigen-presenting cells during inflammation. Eur J Immunol. 2016;46:2222–2232. doi: 10.1002/eji.201646528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K-W, et al. MHC II+ resident peritoneal and pleural macrophages rely on IRF4 for development from circulating monocytes. The Journal of experimental medicine. 2016;213:1951–1959. doi: 10.1084/jem.20160486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goudot C, et al. Aryl Hydrocarbon Receptor Controls Monocyte Differentiation into Dendritic Cells versus Macrophages. Immunity. 2017;47:582–596.:e6. doi: 10.1016/j.immuni.2017.08.016. [DOI] [PubMed] [Google Scholar]
- 25.Sheng J, Ruedl C, Karjalainen K. Most Tissue-Resident Macrophages Except Microglia Are Derived from Fetal Hematopoietic Stem Cells. Immunity. 2015;43:382–393. doi: 10.1016/j.immuni.2015.07.016. [DOI] [PubMed] [Google Scholar]
- 26.Scott CL, et al. Bone marrow-derived monocytes give rise to self-renewing and fully differentiated Kupffer cells. Nat Commun. 2016;7:10321. doi: 10.1038/ncomms10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Theurl I, et al. On-demand erythrocyte disposal and iron recycling requires transient macrophages in the liver. Nat Med. 2016;22:945–951. doi: 10.1038/nm.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dick SA, et al. Self-renewing resident cardiac macrophages limit adverse remodeling following myocardial infarction. Nature immunology. 2019;20:29–39. doi: 10.1038/s41590-018-0272-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw TN, et al. Tissue-resident macrophages in the intestine are long lived and defined by Tim-4 and CD4 expression. The Journal of experimental medicine. 2018;215:1507–1518. doi: 10.1084/jem.20180019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.De Schepper S, et al. Self-Maintaining Gut Macrophages Are Essential for Intestinal Homeostasis. Cell. 2018;175:400–415.:e13. doi: 10.1016/j.cell.2018.07.048. [DOI] [PubMed] [Google Scholar]
- 31.Woitowich NC, Woodruff TK. Opinion: Research community needs to better appreciate the value of sex-based research. Proc Natl Acad Sci USA. 2019;116:7154–7156. doi: 10.1073/pnas.1903586116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Klein SL, Flanagan KL. Sex differences in immune responses. Nature reviews. 2016;16:626–638. doi: 10.1038/nri.2016.90. [DOI] [PubMed] [Google Scholar]
- 33.Thion MS, et al. Microbiome Influences Prenatal and Adult Microglia in a Sex-Specific Manner. Cell. 2018;172:500–516.:e16. doi: 10.1016/j.cell.2017.11.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinhard L, et al. Sexual dimorphism of microglia and synapses during mouse postnatal development. Dev Neurobiol. 2018;78:618–626. doi: 10.1002/dneu.22568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Villa A, et al. Sex-Specific Features of Microglia from Adult Mice. Cell Rep. 2018;23:3501–3511. doi: 10.1016/j.celrep.2018.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribas V, et al. Myeloid-specific estrogen receptor alpha deficiency impairs metabolic homeostasis and accelerates atherosclerotic lesion development. Proc Natl Acad Sci USA. 2011;108:16457–16462. doi: 10.1073/pnas.1104533108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pepe G, et al. Self-renewal and phenotypic conversion are the main physiological responses of macrophages to the endogenous estrogen surge. Sci Rep. 2017;7:44270. doi: 10.1038/srep44270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marriott I, Bost KL, Huet-Hudson YM. Sexual dimorphism in expression of receptors for bacterial lipopolysaccharides in murine macrophages: a possible mechanism for gender-based differences in endotoxic shock susceptibility. J Reprod Immunol. 2006;71:12–27. doi: 10.1016/j.jri.2006.01.004. [DOI] [PubMed] [Google Scholar]
- 39.Scotland RS, Stables MJ, Madalli S, Watson P, Gilroy DW. Sex differences in resident immune cell phenotype underlie more efficient acute inflammatory responses in female mice. Blood. 2011;118:5918–5927. doi: 10.1182/blood-2011-03-340281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandache E, Moldoveanu E, Savi G. The involvement of omentum and its milky spots in the dynamics of peritoneal macrophages. Morphol Embryol (Bucur) 1985;31:137–142. [PubMed] [Google Scholar]
- 41.Wijffels JF, Hendrickx RJ, Steenbergen JJ, Eestermans IL, Beelen RH. Milky spots in the mouse omentum may play an important role in the origin of peritoneal macrophages. Res Immunol. 1992;143:401–409. doi: 10.1016/s0923-2494(05)80072-0. [DOI] [PubMed] [Google Scholar]
- 42.McCarthy AM, Menke A, Visvanathan K. Association of bilateral oophorectomy and body fatness in a representative sample of US women. Gynecol Oncol. 2013;129:559–564. doi: 10.1016/j.ygyno.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Macotela Y, Boucher J, Tran TT, Kahn CR. Sex and depot differences in adipocyte insulin sensitivity and glucose metabolism. Diabetes. 2009;58:803–812. doi: 10.2337/db08-1054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gal-Oz ST, et al. ImmGen report: sexual dimorphism in the immune system transcriptome. Nat Commun. 2019;10:4295. doi: 10.1038/s41467-019-12348-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gibbings SL, et al. Transcriptome analysis highlights the conserved difference between embryonic and postnatal-derived alveolar macrophages. Blood. 2015;126:1357–1366. doi: 10.1182/blood-2015-01-624809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beattie L, et al. Bone marrow-derived and resident liver macrophages display unique transcriptomic signatures but similar biological functions. J Hepatol. 2016;65:758–768. doi: 10.1016/j.jhep.2016.05.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Miller JC, et al. Deciphering the transcriptional network of the dendritic cell lineage. Nature immunology. 2013;13:888–899. doi: 10.1038/ni.2370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gautier EL, Ivanov S, Lesnik P, Randolph GJ. Local apoptosis mediates clearance of macrophages from resolving inflammation in mice. Blood. 2013;122:2714–2722. doi: 10.1182/blood-2013-01-478206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gautier EL, et al. Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nature immunology. 2012;13:1118–1128. doi: 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nature immunology. 2006;7:311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 51.Bain CC, et al. Constant replenishment from circulating monocytes maintains the macrophage pool in the intestine of adult mice. Nature immunology. 2014 doi: 10.1038/ni.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Gentek R, et al. Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity. 2018;48:1160–1171.:e5. doi: 10.1016/j.immuni.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 53.Preston JA, et al. Alveolar Macrophage Apoptosis-associated Bacterial Killing Helps Prevent Murine Pneumonia. Am J Respir Crit Care Med. 2019;200:84–97. doi: 10.1164/rccm.201804-0646OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lanoue A, et al. SIGN-R1 contributes to protection against lethal pneumococcal infection in mice. The Journal of experimental medicine. 2004;200:1383–1393. doi: 10.1084/jem.20040795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kay E, Gomez-Garcia L, Woodfin A, Scotland RS, Whiteford JR. Sexual dimorphisms in leukocyte trafficking in a mouse peritonitis model. Journal of leukocyte biology. 2015;98:805–817. doi: 10.1189/jlb.3A1214-601RR. [DOI] [PubMed] [Google Scholar]
- 56.Barth MW, Hendrzak JA, Melnicoff MJ, Morahan PS. Review of the macrophage disappearance reaction. Journal of leukocyte biology. 1995;57:361–367. doi: 10.1002/jlb.57.3.361. [DOI] [PubMed] [Google Scholar]
- 57.Kang Y-S, et al. The C-type lectin SIGN-R1 mediates uptake of the capsular polysaccharide of Streptococcus pneumoniae in the marginal zone of mouse spleen. Proc Natl Acad Sci USA. 2004;101:215–220. doi: 10.1073/pnas.0307124101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu Z, et al. Fate Mapping via Ms4a3-Expression History Traces Monocyte-Derived Cells. Cell. 2019;178:1509–1525.:e19. doi: 10.1016/j.cell.2019.08.009. [DOI] [PubMed] [Google Scholar]
- 59.Misharin AV, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. The Journal of experimental medicine. 2017;214:2387–2404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Irvine KM, et al. CRIg-expressing peritoneal macrophages are associated with disease severity in patients with cirrhosis and ascites. JCI Insight. 2016;1:e86914. doi: 10.1172/jci.insight.86914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science (New York, NY. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brown JS, et al. The classical pathway is the dominant complement pathway required for innate immunity to Streptococcus pneumoniae infection in mice. Proc Natl Acad Sci USA. 2002;99:16969–16974. doi: 10.1073/pnas.012669199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Boes M, Prodeus AP, Schmidt T, Carroll MC, Chen J. A critical role of natural immunoglobulin M in immediate defense against systemic bacterial infection. The Journal of experimental medicine. 1998;188:2381–2386. doi: 10.1084/jem.188.12.2381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kang Y-S, et al. A dominant complement fixation pathway for pneumococcal polysaccharides initiated by SIGN-R1 interacting with C1q. Cell. 2006;125:47–58. doi: 10.1016/j.cell.2006.01.046. [DOI] [PubMed] [Google Scholar]
- 65.Revzin MV, Mathur M, Dave HB, Macer ML, Spektor M. Pelvic Inflammatory Disease: Multimodality Imaging Approach with Clinical-Pathologic Correlation. Radiographics. 2016;36:1579–1596. doi: 10.1148/rg.2016150202. [DOI] [PubMed] [Google Scholar]
- 66.Caton ML, Smith-Raska MR, Reizis B. Notch-RBP-J signaling controls the homeostasis of CD8-dendritic cells in the spleen. The Journal of experimental medicine. 2007;204:1653–1664. doi: 10.1084/jem.20062648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Byers SL, Wiles MV, Dunn SL, Taft RA. Mouse estrous cycle identification tool and images. PLoS ONE. 2012;7:e35538. doi: 10.1371/journal.pone.0035538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bain CC, Jenkins SJ. Isolation and Identification of Murine Serous Cavity Macrophages. Methods Mol Biol. 2018;1784:51–67. doi: 10.1007/978-1-4939-7837-3_5. [DOI] [PubMed] [Google Scholar]
- 69.Jenkins SJ, et al. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. The Journal of experimental medicine. 2013;210:2477–2491. doi: 10.1084/jem.20121999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550–21. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials, and RNA sequencing data have been deposited in National Center for Biotechnology Information Gene Expression Omnibus public database (http://www.ncbi.nlm.nih.gov/geo/). Population-level RNAseq (accession code:149014) and scRNA-seq (primary accession code GSE139999 [male: GSM4151330; female: GSM4151331]).