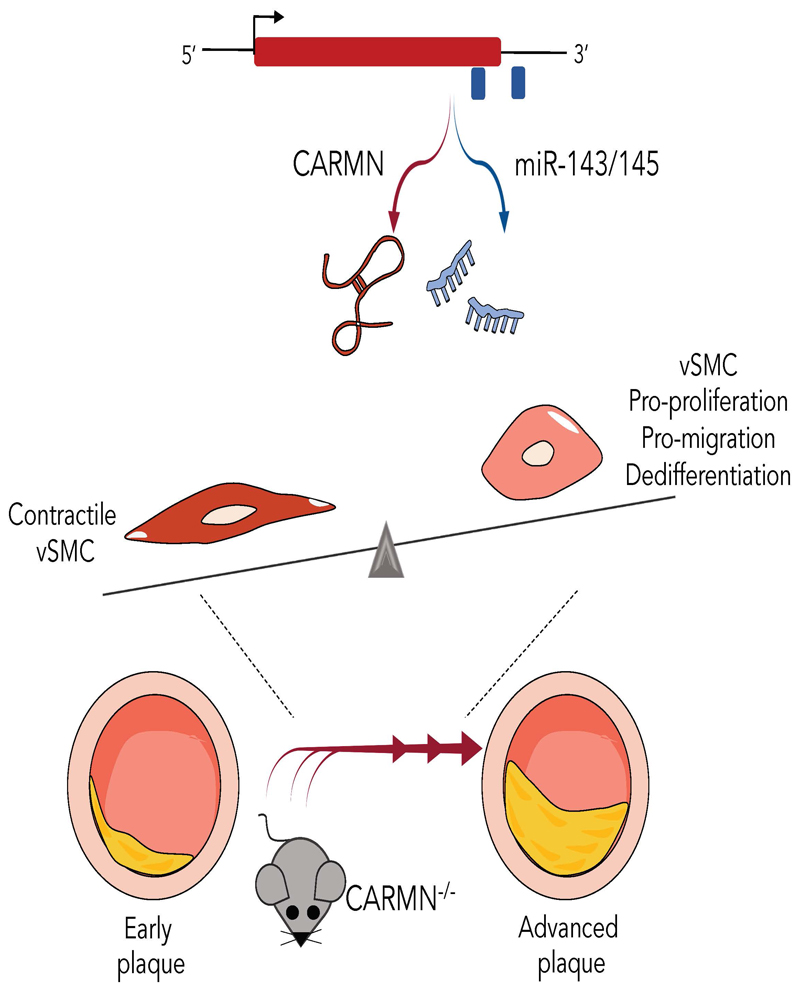
Abstract
Rationale
In the microenvironment of atherosclerotic lesions, vascular smooth muscle cells (vSMCs) switch to a dedifferentiated state but the underlying molecular mechanisms driving this switch are not fully understood. Long noncoding RNAs (lncRNAs) are dysregulated during vascular pathology, but relatively little is known about their involvement in controlling vSMCs function. CARMN is a lncRNA located immediately upstream of the microRNAs-143 and -145 (miR-143 and miR-145), both involved in vSMCs function.
Objective
We investigated the role of the lncRNA CARMN, independent from miR-143 and miR-145, as potential a regulator of vSMC phenotypes in vitro and the consequences of its loss during the development of atherosclerosis in vivo. We hypothesized that loss of CARMN is a primary event controlling the functional switch towards pro-atherogenic vSMC phenotype and accelerates the development of the plaques in vivo.
Method and Results
Expression of CARMN lncRNA was silenced using GapmeRs in human coronary arterial smooth muscle cells (hCASMCs), revealing that GapmeR-mediated loss of CARMN negatively affects miR-143 and miR-145 miRNA expression. RNA sequencing of CARMN-depleted hCASMCs revealed large transcriptomic changes, associated with vSMC proliferation, migration, inflammation, lipid metabolism and dedifferentiation. The use of miR-143 and miR-145 mimics revealed that CARMN regulates hCASMC proliferation in a miRNA-independent manner. In human and mouse, CARMN and associated miRNAs were downregulated in advanced versus early atherosclerotic lesions. Using a CRISPR (clustered regularly interspaced short palindromic repeats)- Cas9 (CRISPR-associated protein 9) knock-out approach, we explored the implications of CARMN depletion during atherosclerosis in vivo. Consistent with in vitro results, the knock-out of CARMN impaired the expression of miR-143 and miR-145 under homeostatic conditions. Importantly, when atherosclerosis was induced in these mice, CARMN knock-out increased the volume, size, pro-inflammatory LGALS3-expressing cells content and altered plaque composition, yielding an advanced phenotype.
Conclusions
We identified the early loss of CARMN lncRNA as critical event which primes vSMCs towards a pro-atherogenic phenotype in vitro and accelerates the development of atherosclerosis in vivo.
Keywords: Cholesterol, non-coding RNA, smooth muscle cells, vascular biology, atherosclerosis
Subject Terms: Animal Models of Human Disease, Coronary Circulation, Mechanisms, Smooth Muscle Proliferation and Differentiation, Vascular Biology
Introduction
Atherosclerosis is a chronic and inflammatory condition of the arterial branches and a leading cause of cardiovascular disease and cerebrovascular disease1. Vascular smooth muscle cells (vSMCs) have a critical and complex role in atherogenesis. In homeostatic conditions, adult vSMCs are largely quiescent and characterized by a specific gene expression pattern and functional properties which define their signature2. However, in response to pathological environmental stress signals that occur during atherogenesis, vSMCs lose their contractile signature and switch into a pro-atherogenic phenotype as they start to proliferate, migrate and lose their typical markers (such as Smooth Muscle Actin Alpha 2)3–5. In particular, during the formation of stable plaques, vSMC proliferation and migration toward the intimal layer are associated with the formation of a stabilizing fibrotic cap6. Whereas, during the latter stages of disease, the recruitment and activation of inflammatory cells, the decrease in collagen content and increase in metalloprotease secretion contribute to the breakdown and dynamic remodeling of the fibrotic cap and the progression towards rupture-prone plaques7. Therefore, it is critical to unravel and fully understand these complex regulatory networks to be able to target specific molecular mechanisms during the progression of atherosclerosis.
Recently, several studies have demonstrated that non-coding RNAs, namely long non-coding RNAs (lncRNAs) and miRNAs (miRNAs), are crucial regulators of many pathophysiological processes during atherosclerosis8–10. Namely, miRNAs miR-143 and miR-145 have been found to be important regulators of vSMC differentiation and disease-associated phenotypic switching in response to pro-atherogenic stimuli such as cholesterol loading (water-soluble cholesterol)11 or PDGF-BB treatment12–16. Their functions have also been highlighted in several mouse models, where the ablation of both miR-143 and miR-145 induced spontaneous neointimal lesions in the femoral artery14 17, dysregulated signaling pathways14, and regulated vSMC migration17. However, despite numerous studies uncovering the importance of miR-143 and miR-145 in diverse disease conditions14–16,18–22, their function and regulation remains conflicting in the context of atherosclerosis16,19.
A long noncoding RNA (lncRNA), named CARMN, is juxta-positioned upstream from miR-143 and miR-14523. This lncRNA was originally identified in human cardiomyocytes and named Cardiac Mesoderm Enhancer-associated Noncoding RNA (CARMN)24 25. Previous studies have demonstrated that a crosstalk may exist between the processing of lncRNA host genes, such as CARMN, and encoded miRNAs26 and that the biogenesis of these noncoding molecules might be mutually exclusive27 and produced RNAs with independent functions28. Although the functions of miR143 and miR145 have been explored, whether CARMN has an independent function is not completely elucidated in the context of atherosclerosis. Here, we demonstrated that CARMN has a function independent from miR-143 and mi-R145 in regulating vSMC proliferation, while vSMC migration or dedifferentiation processes are regulated through the modulation of miRNA expression in hCASMCs. Moreover, we uncovered a pivotal role of the CARMN/miRNA locus during the pathophysiology of atherosclerotic disease in vivo and demonstrated that in CARMN knockout mice, the development of atheroma was accelerated when compared to wild type mice, thereby inducing advanced and more vulnerable plaques.
Methods
Results
Expression and subcellular localisation of CARMN lncRNA and associated miRNAs in primary human CASMCs under basal and stimulated conditions
We first sought to understand the expression and regulation of CARMN in hCASMCs together with miRNAs miR-143 and miR-145. Publicly available data in Ensembl database 98 show several transcript variants belonging to the complex CARMN locus (Figure 1 A). Initially, we assessed the expression levels of individual isoforms in hCASMCs, using isoform-specific primers when possible (only isoform 4 was amplified in combination with isoform 9) (Figure 1 A). Expression data indicated that all the isoforms were detectable in hCASMCs (Figure 1 B). Following this, we used common primers capturing all isoforms except 205 and 211, to assess the expression levels of CARMN (Figure 1 A). Based on previous studies, the function of lncRNAs is related to their subcellular localization29,30, therefore, we next determined the localization of CARMN in hCASMCs. Subcellular fractionation as well as RNA-FISH indicated that CARMN predominantly localized in the nuclear compartment of hCASMCs (Online Figure I A, B). Of note, during the course of these experiments, there was a new release of the Ensembl annotation that includes 7 new potential isoforms (Online Figure II A), yet all remained detectable by our common primer set. However, considering that lncRNA annotation is not definitively accurate and that lncRNA are expressed in a cell-specific way, we applied a transcript discovery pipeline to publicly available RNA-sequencing data of hCASMCs obtained by Encyclopedia of DNA Elements (https://www.encodeproject.org/), which did not reveal the existence of un-annotated additional isoforms (Online Figure II A). This analysis also showed that the 3’ end of the most abundant CARMN transcripts is upstream of the miR143/miR145 loci (Online Figure II A). Moreover, we carried out 5’ Rapid Amplification of cDNA Ends and long-reads sequencing (Oxford Nanopore technology). Both techniques revealed the presence of main transcriptional start site (TSS), already described in the annotation, but also detected minor additional (TSS) (Online Figure II B, C). Moreover, the long-read sequencing revealed 4 major exonic structures all covered by the current annotation and not including miR143/145 (Online Figure II D). In addition, we found reads mapping to the miR-143/145 stem-loop which suggested the existence of fragments from the processed primary miRNA transcript (pri-miRNA) with different 5’ end, one of them shared by CARMN transcripts (Online Figure II E). Altogether, these analyses confirmed the complexity of CARMN locus.
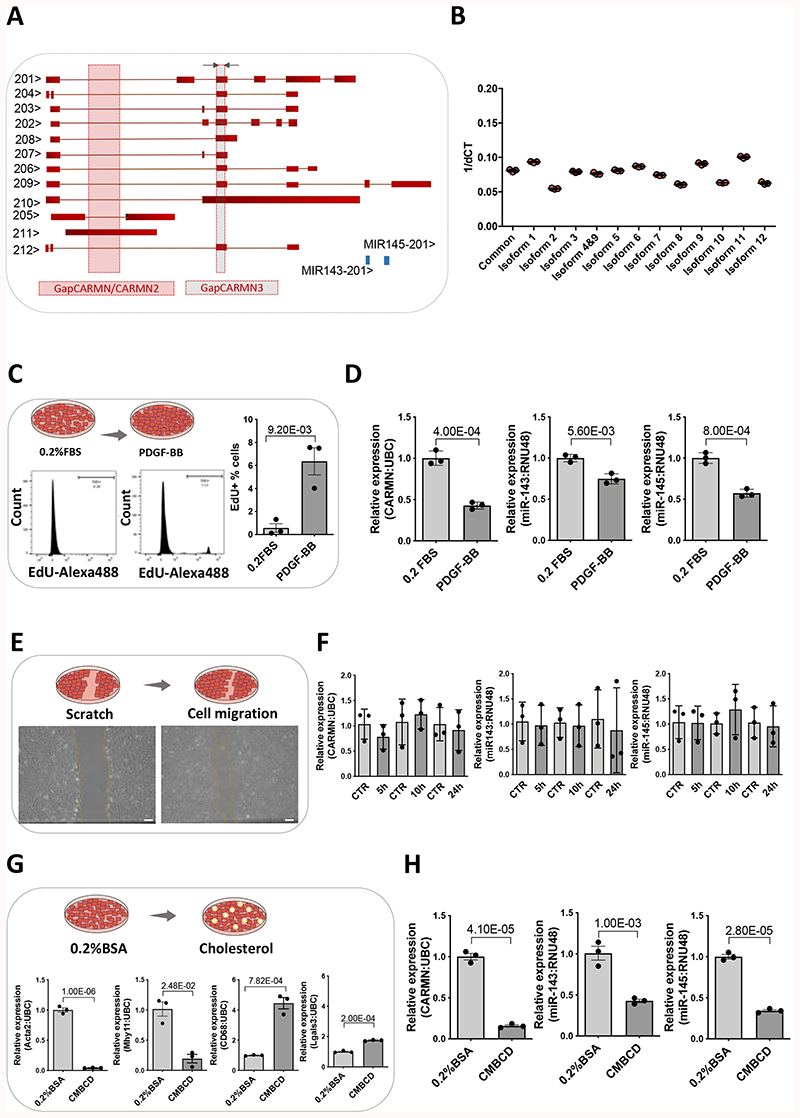
Figure 1. CARMN expression in basal and stimulated human primary CASMCs.
A) Schematic representation of human CARMN splice variants and pre-miRNAs miR-143 and miR-145 based on ENSEMBL version (GRCh38/hg38), including the position of the forward (Fw) and reverse (Rev) primers used to amplify all transcripts, grey arrows (except 4, 10 and 11) and the target region for the GapmeRs, GapCARMN, GapCARMN2 and GapCARMN3. The black arrows next to each transcript name, indicate the direction of transcription. B) Expression levels of CARMN transcripts in hCASMCs (n=3) via qRT-PCR using isoform-specific primers (Isoform 1,2,3,4+9,5,6,7,8,9,10,11, and 12) and common primers (amplifying all isoforms except 4, 10 and 11). C) Diagram of hCASMCs in basal condition (0.2% FBS) and treated for 48 hours with PDGF-BB. Histogram plots obtained by FACS showing the percentage of hCASMCs (n=3) up-tacking EdU following PDGF-BB treatment and in basal (0.2% FBS) conditions. Student’s t-test was used to assess statistical significance indicated with p values. D) QRT-PCR to show expression of CARMN, miR-143 and miR-145 in hCASMCs (n=3) treated with PDGF-BB. Student’s t-test was used to assess statistical significance indicated with p values. E) Diagram of the wound healing assay performed in hCASMCs (n=3), representative micrographs of hCASMCs scratch assay and migrating cells (Scale bar 100μm) and qRT-PCR showing the expression of CARMN and miR-143/145 G) Diagram of hCASMCs (n=3) in basal condition (0.2% BSA) and treated with cholesterol-methyl-b-cyclodextrin treated for 72 hours and expression levels of dedifferentiation markers (ACTA2, MYH11, CD68 and Lgals3), CARMN and miR-143/145 by RT-qPCR. Student’s t-test was used to assess statistical significance indicated with p values. H) qRT-PCR data showing the expression of CARMN and miR-143/145 in basal (0.2% FBS) and following CMBCD-loading treatment. Student’s t-test was used to assess statistical significance indicated with p values.
Platelet-derived growth factor (PDGF-BB) is one of the main pathogenic cytokines released at the site of injury with a potent mitogen action to vSMCs31. To gain insight into the regulation of CARMN in proliferating VSMCs, we treated serum-starved hCASMCs with PDGF-BB for 48 hours (Figure 1 C) and assessed the levels of CARMN. CARMN was significantly downregulated following PDGF-BB stimulation (Figure 1 D). Levels of expression of miR-143 and miR145 were also significantly decreased (Figure 1 D). It is well established that vSMC migration from the media to the intima is critically involved in the pathophysiology of vascular disease, although a difficult process to quantify in human lesions4. We therefore assessed whether CARMN was dynamically regulated under migratory stimuli. A scratch was made in a monolayer of serum-starved hCASMCs, and the levels of CARMN and miR-143/145 were assessed at 5, 10 and 24 hours after scratching (Figure 1 E). However, no statistical difference was observed in the lncRNA or microRNAs expression in scratched versus control cells (Figure 1 F). Additionally, we characterized the expression of the lncRNA CARMN in primary culture of hCASMCs following the treatment with water-soluble cholesterol-methyl-β-cyclodextrin (CMBCD), for 72 hours (Figure 1 G). CMBCD loading substantially decreased the expression levels of CARMN versus control cells, and similarly the expression of miR-143 and miR-145 (Figure 1 H) as previously described11. Moreover, we observed similar pattern of decrease in CARMN expression using another model of hCASMC dedifferentiation towards foamy cell-like phenotype32 by loading cells with oxidised Low-Density Lipoproteins (ox-LDL) (Online Figure III A-C). Thus, the expression levels of CARMN lncRNA and associated miRNAs are reduced by the exposure to proliferative stimuli and CMBCD loading of hCASMCs, which is a critical process during the pathophysiology of atherosclerosis.
Knockdown of the human CARMN axis drives the phenotypic transition of hCASMCs towards a pro-atherogenic phenotype
To assess the functional importance of CARMN, we utilised an locked nucleic acids antisense oligonucleotides (GapmeRs) antisense oligonucleotide (ASOs) approach. We designed two different GapmeRs targeting the introns of CARMN isoforms (named GapCARMN and GapCARMN2) (Figure 1 A). Following transfection of hCASMCs with GapCARMN and GapCARMN2, we observed a significant knockdown of CARMN versus a GapmeR Control (GapCTR) using the common primers (Figure 2 A, Online Figure III D). Importantly, we confirmed the downregulation of CARMN at the isoform level using isoform-specific primers (Online Figure III E). Notably, the levels of miR-143 and miR-145 were also significantly downregulated following GapCARMN and GapCARMN2 transfection in CASMCs (Figure 2 A and Online Figure III D). Furthermore, to support the results obtained with intronic-targeting GapmeR (GapCARMN/GapCARMN2), we used a previously described GapmeR targeting the exonic region of CARMN transcripts (named GapCARMN3 in this manuscript)25 and confirmed similar changes (Online Figure IV A).
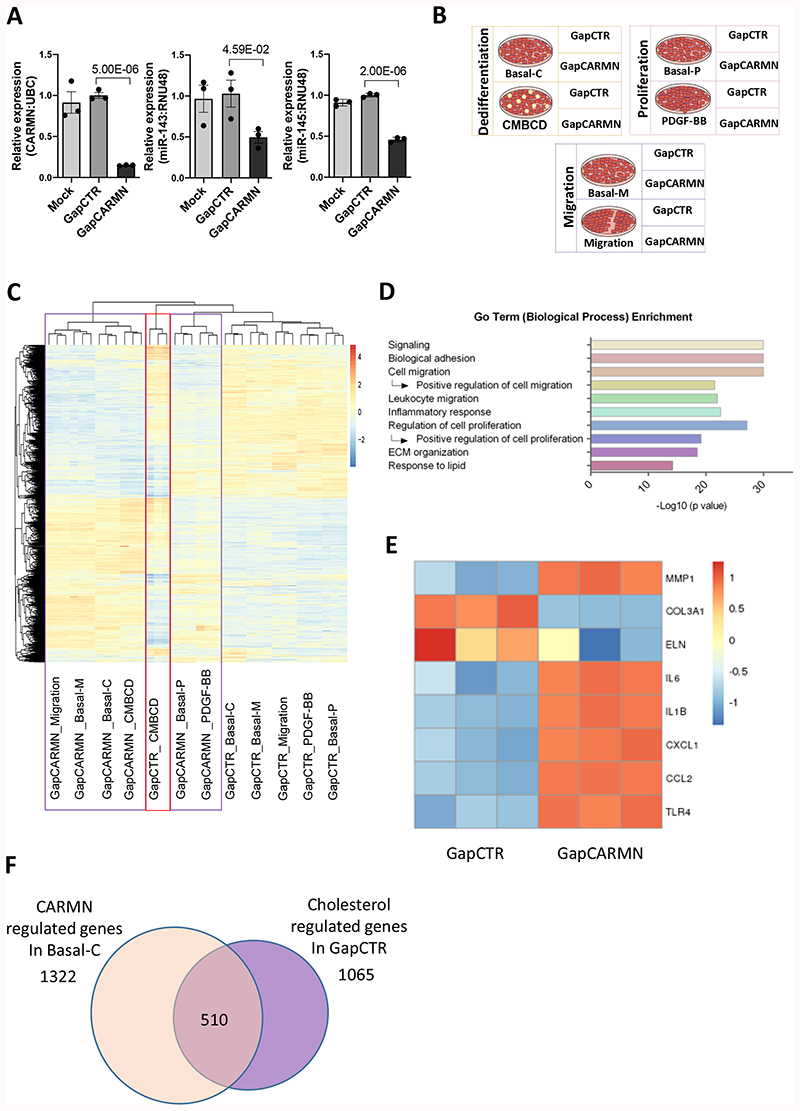
Figure 2. CARMN-regulated genes relevant to vSMC proliferation, migration and water-soluble cholesterol dependent dedifferentiation.
A) QRT-PCR results showing the expression of CARMN, miR-143 and miR145 hCASMCs (n=3) following transfection with GapmeR targeting CARMN (GapCARMN) and GapmeR control (GapCTR). Mock indicates un-transfected cells. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. B) Schematic representation of the RNA-seq study design. C) Heatmap (as z-score of log2(FPKM+1)) of differentially regulated genes by CARMN depletion in all conditions. The purple box highlights the clustering of all CARMN-knock down samples while the red box shows the CMBCD treated replicates (Cont_KD). D) Graph of selected Go Term (Biological Process). The analysis was performed on the 2315 genes regulated by CARMN depletion in any conditions. E) Heatmap (as z-score of log2(FPKM+1)) of selected differentially regulated genes by GapCARMN and GapCTR in the basal condition with the shortest time of GapmeR transfection (migration). F) Venn diagram showing the overlap between genes regulated by CARMN depletion (in Basal-C condition) and genes regulated by CMBCD treatment (in GapCTR condition) in the same direction.
To understand the role of CARMN in hCASMCs in physiological and pathological conditions, we analyzed the transcriptomic changes associated with the CARMN-axis depletion using GapCARMN in untreated hCASMCs (basal), as well as following stimulation upon proliferation, migration or CMBCD loading (Figure 2 B). The expression of CARMN was firstly assessed in RNA-seq samples to validate appropriate knockdown (Online Figure IV B). Following RNA sequencing analysis, principal component analysis (PCA) confirmed clustering of the sample replicates and showed a clear separation of the GapCARMN-treated samples from the GapCTR-treated hCASMCs in all analyzed conditions (Online Figure IV C). The PCA also confirmed an effect of the different treatments at the transcriptomic level. In particular, we noted the clear separation of the CMBCD-treated cells from their relevant control cells (Basal_C), showing the strong effect of this stimulus on the hCASMC transcriptome. We also noticed the proliferation-stimulated samples showed an intermediate phenotype which can be explained by different responses due to the diverse cell culture densities of the hCASMCs as well as the point of harvesting following GapmeR transfection. We analyzed the significant changes in gene expression in GapCARMN versus GapCTR treated cells for each condition (Online Figure IV D) and found that CARMN depletion affected a large number of genes, on average 910 genes per condition with 411 up-regulated and 499 down-regulated genes. In total, we found 2315 genes whose expression was significantly changed by CARMN depletion and analyzed their expression profiles (Figure 2 C). The heatmap revealed a largely consistent effect of CARMN-axis depletion, independent of hCASMC treatment. Gene ontology analysis of the CARMN-axis dependent genes in basal conditions already revealed the enrichment of several processes related to vSMC phenotypic transition towards de-differentiated phenotype (Figure 2 D). More specifically, we noted enrichment of terms linked to a positive effect on cell migration and proliferation that are key processes during atherogenesis. Moreover, we investigated whether the depletion of CARMN-axis affects the transcription factor regulating vSMC homeostasis, Myocardin (MYOCD)33. However, MYOCD was undetectable in both our RNA-Seq and by qRT-PCR analysis (Online Figure IV E, F). Interestingly, CARMN-dependent genes were also involved in lipid metabolism and cholesterol homeostasis. CARMN depletion also upregulated the expression of pro-inflammatory cytokine genes such as interleukins (IL1-beta, IL6) associated with atherosclerosis progression and CXCL1 and as well as CCL2 chemokines responsible of leucocyte migration and recruitment to the developing lesions (Figure 2 E)34,35. Furthermore, we noted a significant upregulation of matrix metalloproteinase 1 (MMP1) and a significant downregulation of collagen and elastase genes (COL3a1 and ELN), with expression decreasing in the context of extracellular matrix remodelling and plaque vulnerability (Figure 2 E)36,37. Genes regulated by CARMN and involved in positive regulation of proliferation, migration, regulation of inflammatory response and response to lipid are reported in Online Data set II. Related to the enrichment of ‘lipid’ terms in the gene ontology analysis, the heatmap of CARMN regulated genes revealed the clustering of CMBCD treated samples transfected with GapCTR among all GapCARMN samples, further suggesting that CARMN-dependent genes were also affected by CMBCD treatment (Figure 2 C). Therefore, we directly compared the genes regulated by CARMN in basal condition with genes changed upon CMBCD treatment. Importantly, we found that about 50% of CMBCD-regulated genes were altered in the same direction upon CARMN depletion (Figure 2 F), suggesting that CARMN depletion alone promotes a water-soluble cholesterol loaded-like phenotype in the absence of CMBCD co-incubation. These results show the impact of CARMN depletion on hCASMCs transcriptome and in particular on cholesterol-regulated genes. Collectively, these results suggest CARMN knock-down might be an important event which primes vSMCs to the phenotypical transition occurring in plaques formation and progression.
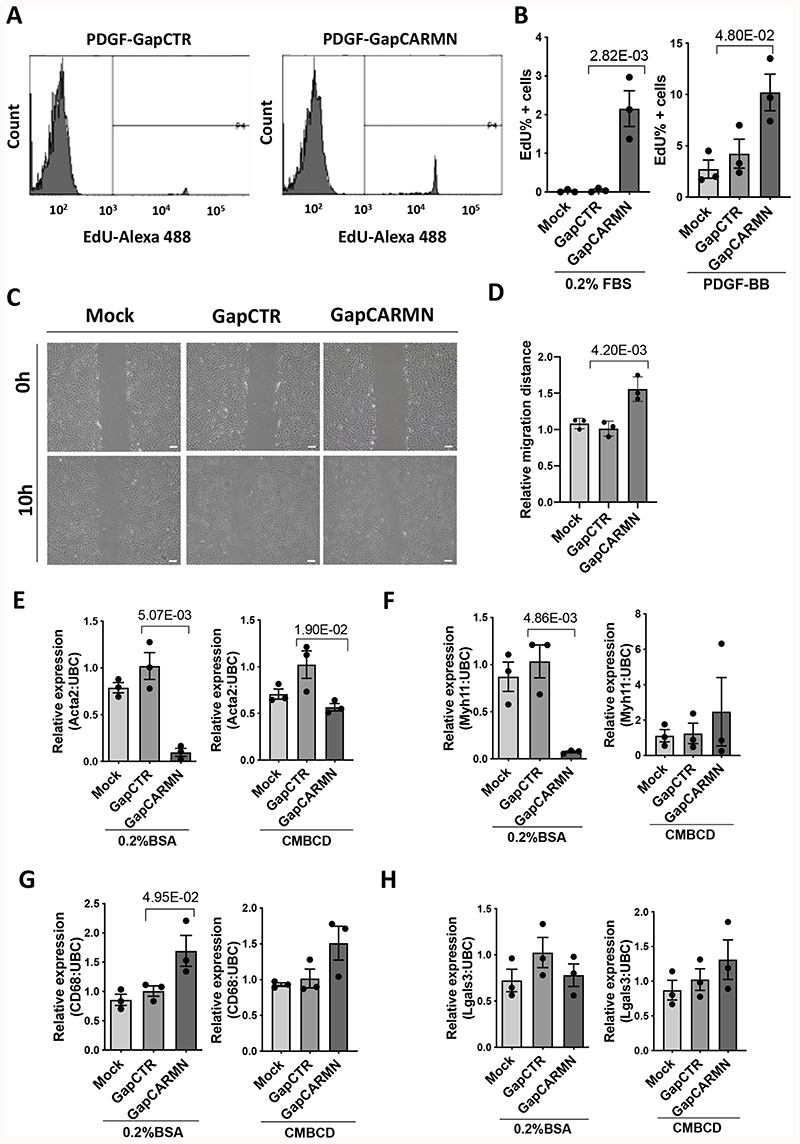
Knockdown of CARMN-axis lncRNA activates proliferation, migration and induces partial dedifferentiation of hCASMCs in vitro
Considering the results obtained from the transcriptomic analysis of CARMN silenced hCASMCs, we further deciphered the functional consequence of CARMN axis silencing on hCASMC phenotypes. Interestingly, the depletion of CARMN significantly increased the proliferation of hCASMCs in quiescent conditions (0.2%FBS) or under PDGF-BB stimulation (Figure 3 A, B). Next, we assessed the effect of CARMN depletion on hCASMC migration. Although the response to the migratory stimulus did not show significant difference in the expression levels of endogenous CARMN and miR-143/145, the downregulation of CARMN increased the migration rate of hCASMCs versus cells transfected with GapCTR (Figure 3 C, D). To further validate the involvement of CARMN in hCASMC CMBCD-induced dedifferentiation process, we analysed mRNA expression changes of the smooth muscle identity markers, Actin Alpha 2 (ACTA2) and Myosin Heavy Chain11 (MYH11), and activated macrophage-like genes, Cluster of Differentiation 68 (CD68) and Galectin 3 (Lgals3). We confirmed that CARMN depletion leads to significant decrease of Acta2 and Myh11 levels, with concomitant significant increase of CD68 expression in basal conditions (Figure 3 E, F, G). However, we did not observe significant change in LGALS3 expression in CARMN-depleted hCASMCs when compared to control group (Figure 3 H). Moreover, we assessed the expression levels of Kruppell-like factor 4 (KLF4) in CARMN-depleted hCASMCs, previously associated with vSMC dedifferentiation phenotype38. We did not observe significant change in the expression of this transcription factor in CARMN-depleted proliferative or de-differentiating hCASMCs (Online Figure V A, B), indicating likely KLF4-independent effects. Instead, we observed a significant increase in KLF4 in scratched hCASMCs with CARMN-depletion (Online Figure C). In order to robustly characterize hCASMCs phenotypic state following CARMN depletion, we also took into consideration other typical vSMCs marker genes such as Calponin1 (CNN1) and Taglin (TAGLN)39,40. However, their expression levels were not significantly changed following CARMN axis knockdown (Online Figure V D, E), therefore indicating that CARMN downregulation partially affects the vSMCs signature under unstimulated conditions. In addition, we investigated the expression of lipid-metabolism associated genes, such as ABCA1 and ABCG1 11, which were found significantly upregulated following CARMN knockdown versus control (Online Figure V F, G). We did not observe the same pattern in the case of Srebpf2, SCD and HMGS1 expression (Online Figure V H, I, J), indicating the modulation of CARMN partially primes the dedifferentiation towards a pro-inflammatory phenotype foam-cell like phenotype. These phenotypical changes observed in hCASMCs were confirmed with a second GapmeR, GapCARMN2 (Online Figure VI A-E). Collectively, these data suggest that the manipulation of the CARMN locus modulates the ability of hCASMCs to proliferate and migrate. In addition to this, the results obtained indicate that CARMN depletion affects the expression of some of the vSMC identity genes (Acta2 and Myh11) and increases the levels of some of the CMBCD-regulated genes (CD68, ABCA1, ABCG1) priming vSMC to a partial dedifferentiation process.
Figure 3. The knockdown of CARMN regulated hCASMC proliferation, migration and dedifferentiation.
A) Representative FACS plots showing the of EdU uptake in hCASMCs transfected with GapmeR targeting CARMN (GapCARMN) or GapmeR control (GapCTR). The gate (P4) indicates the EdU positive cells, the samples are named GapmeR negative control (GapCTR) and GapmeR targeting CARMN (GapCARMN). B) Percentage of EdU positive hCASMCs (n=3) following transfection with GapCARMN, GapCTR and un-transfected cells (Mock) upon PDGF-BB stimuli and in 0.2% FBS. Oneway ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. C) Representative pictures of hCASMCs at 0 and 10 hours from the scratch following transfection with GapCARMN, GapCTR and Mock in 0.2% FBS medium. Pictures were acquired at 10X magnification. Scale bar 100μm. D) Quantification of the relative migration distances of hCASMCs (n=3), obtained using ImageJ tool. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. E), F), G), H) Expression levels of dedifferentiation markers Acta2, Myh11, CD68 and Lgals3 in hCASMCs (n=3) stimulated with water-soluble cholesterol-methyl-b-cyclodextrin (CMBCD) or 0.2% BSA for 72 hours following transfection with GapCARMN, GapCTR or Mock. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values.
CARMN depletion regulates hCASMCs proliferation in a miRNAs-independent manner
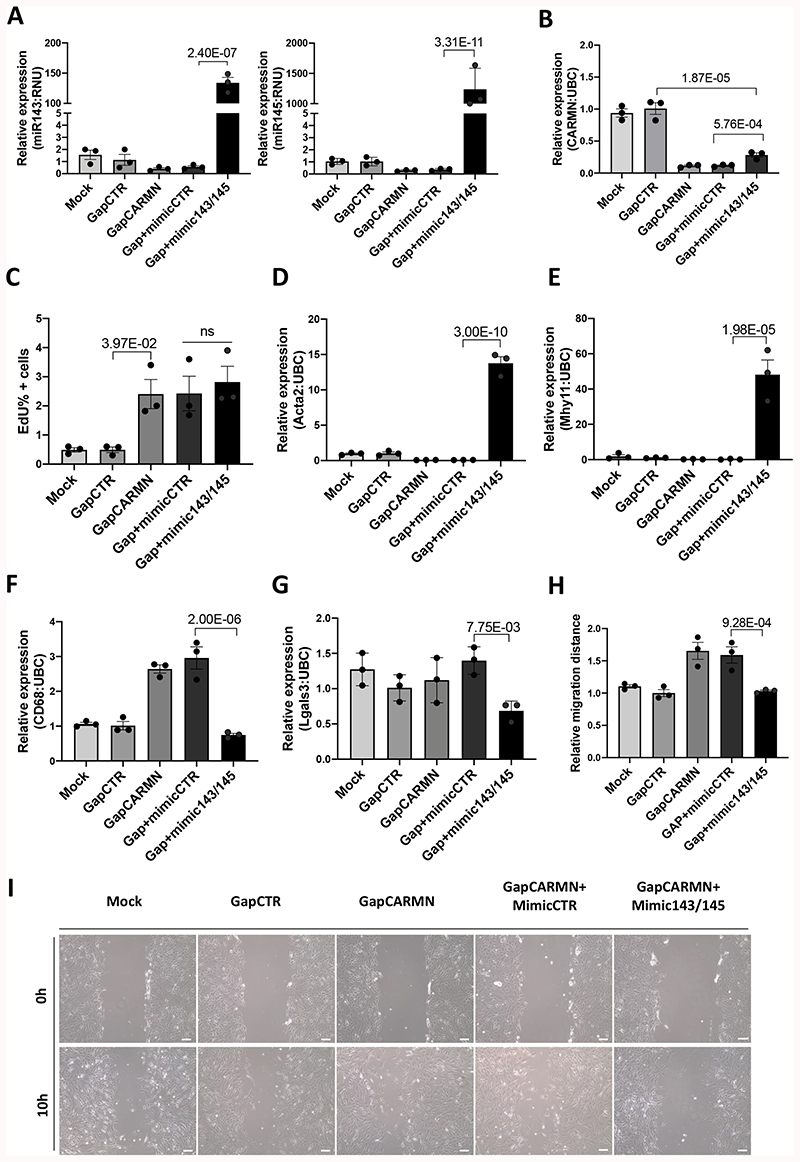
As the GapmeR strategy to downregulate CARMN also affects the expression of the miRNAs, we designed a rescue experiment to assess the contribution of both CARMN and the miRNAs to the described phenotypes. We have applied a co-transfection approach to overexpress the levels of miRNAs by treatment with miR-mimic (mimic-143 and mimic-145) in combination with GapmeR-mediated CARMN knockdown. We first assessed the efficiency of this strategy confirming a significant upregulation in the expression of both miRNAs by qRT-PCR (Figure 4 A). Moreover, to assess their functional effect, we checked the expression of two miRNAs targets, EGFR and PDGFRa41,12. These were significantly downregulated following treatment with mimic143 and mimic145 (mimic143/145) versus mimic control (mimicCTR) (Online Figure VI F, G). In addition, we observed a significant upregulation of CARMN following treatment with miR143/145 compared to mimic control. While this increase might suggest a feedback loop in the modulation of CARMN expression induced by the hosted miRNAs (Figure 4 B), this modulation did not restore CARMN expression to its basal levels. We then co-transfected hCASMCs in basal conditions with GapmeR targeting CARMN (GapCARMN) concomitantly with mimic control (mimicCTR) or a combination of mimic-143 and mimic-145. CARMN knock-down increased hCASMCs proliferation while the overexpression of miR-143 and miR-145 did not significantly change the observed increase in cell proliferation. Therefore, CARMN appears to have independent effect on the modulation of hCASMCs proliferation at basal conditions (Figure 4 C). In contrast, miR-143/145 overexpression restored the levels of vSMC identity genes modulated by CARMN depletion, ACTA2, MYH11 (Figure 4 D, E, F, G). In addition, the overexpression of miR-143/145 restored hCASMCs migration at basal levels thus rescuing the effects observed following CARMN depletion (Figure 4 H, I). Taken together, these results suggest that CARMN can regulate hCASMCs proliferation independent of miR-143 and miR-145 but that migration and dedifferentiation may be driven through CARMN-mediated regulation of the miRNAs.
Figure 4. CARMN depletion regulated hCASMCs phenotypes in miRNAs-dependent and independent mode.
A), B) QRT-PCR expression data of miR-143, miR-145 and CARMN relative to housekeeper gene (RNU and UBC respectively) in GapCARMN, control (GapCTR), a combination of GapCARMN and mimic control (Gap+mimicCTR) and a combination of GapCARMN and mimic overexpressing miR143 and miR145 (GapCARMN+mimic143/145). Mock refers to un-transfected control cells. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. C) Percentage of EdU positive hCASMCs (n=3) evaluated by FACS analysis and analyzed using FlowJo software. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. D), E), F), G) qRT-PCR expression data relative to Acta2, Myh11, CD68 and Lgals3, respectively relative to housekeeper gene UBC in the above-mentioned transfection conditions. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. H), I) Quantification of the relative migration distances of hCASMCs (n=3), obtained using ImageJ tool and representative images of hCASMCs at 0 and 10 hours from the induction of the scratch. Images acquired at 10X magnification, scale bar 100μm. One-way ANOVA with Bonferroni multiple comparison test was used to assess statistical significance indicated with p values. ns= not significant.
The expression of CARMN lncRNA and associated miRNAs are downregulated in mouse and human advanced plaques
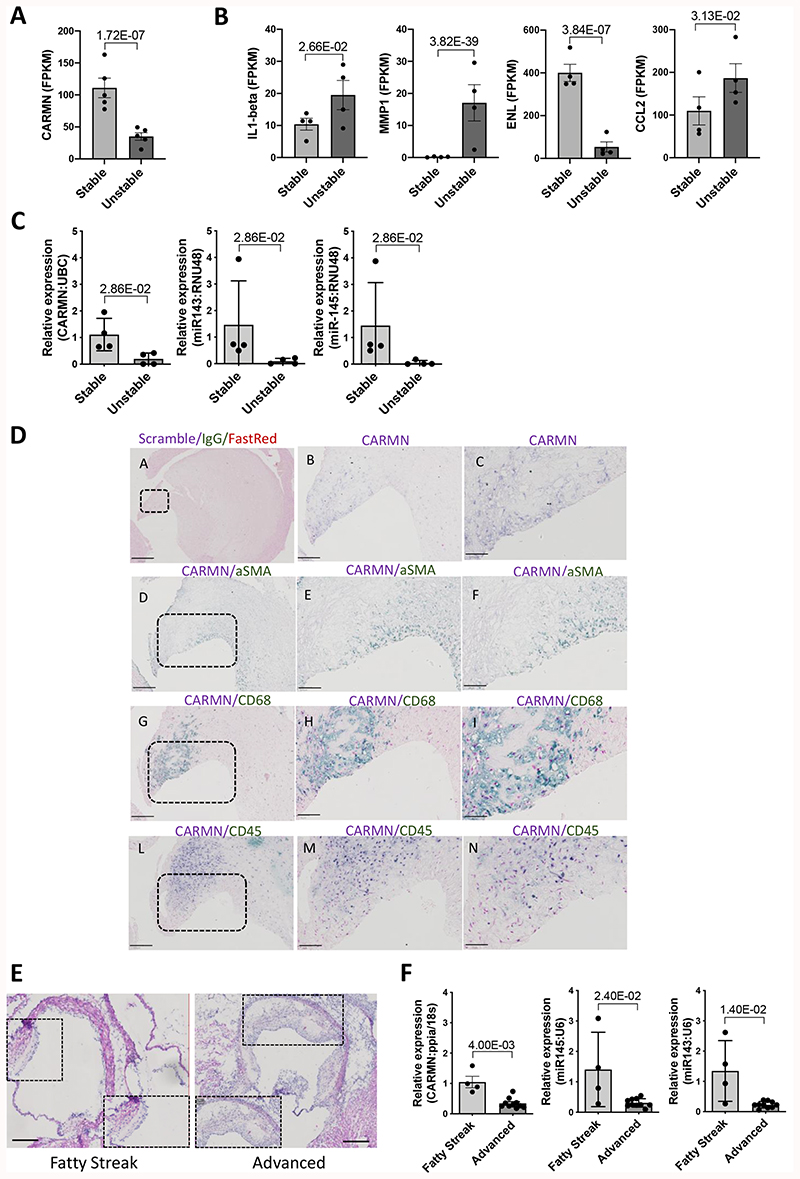
To determine the relevance of CARMN in human atherosclerosis, we assessed the expression of the lncRNA in atherosclerotic plaques isolated from symptomatic patients undergoing carotid endarterectomy. Previous RNA-seq analysis performed in stable and unstable regions of human plaques8, were interrogated and revealed a significant decrease of CARMN levels in the unstable region versus the stable plaques (Figure 5 A). Moreover, in the same RNA-Seq data, we interrogated the expression levels of previously identified CARMN-regulated genes. In agreement with the observations in CARMN knock-down samples, we found a significant upregulation of metalloproteinase (MMP1), significant downregulation of elastase (ELN) and a significant increase of pro-inflammatory genes (IL1-beta and CCL2) (Figure 5B), in unstable versus stable plaques, confirming our findings from in vitro studies through to human atherosclerosis.
Figure 5. The CARMN locus is downregulated in human and murine atherosclerosis.
A) Expression level of CARMN and B) Expression level of MMP1, ELN, IL1-beta, CCL2 as FPKM in paired, stable (n=4) and unstable (n=4) human plaque segments retrieved after endarterectomy from 4 symptomatic patients. P value obtained based on the raw read counts using DESeq2. C) Expression of CARMN and miR143/145 in human stable and unstable plaques isolated through endarterectomy (n=4) determined by qRT-PCR. Mann-Whitney was used to assess statistical significance indicated with p values. D) Representative bright field images of in-situ detection of CARMN co-localizing with α-SMA, CD68 or CD45 signal in plaques obtained from carotid artery derived from symptomatic patients at carotid endarterectomy. Pictures were acquired consecutively at 4X, 10X and 40X magnification Scale bar 50μm. E) Representative Haematoxylin & Eosin staining of aortic roots isolated from LDLR-/- mice. Pictures were acquired with 4x and 10X magnification. Scale bar 100μm. F) Expression levels of CARMN and miR143/145 in fatty streak (n=4) and advanced murine plaques (n=11) isolated by Laser Capture Microdissection. Mann-Whitney was used to assess statistical significance indicated with p values.
To validate these findings, we analysed CARMN expression levels by qRT-PCR in carotid artery stenosis plaques isolated from different patients and confirmed the downregulation of the lncRNA in advanced versus early atheroma. Additionally, we showed the downregulation of miR-143 and miR-145 in the same samples (Figure 5 C). In order to identify the cellular localization of CARMN within human plaques, In Situ Hybridization (ISH) was performed in advanced stable plaques isolated from human carotid arteries of symptomatic patients. CARMN co-localized with α-Smooth Muscle Actin (α-SMA), typical marker for vSMCs and with CD68, marker of macrophages and dedifferentiated vSMCs within the plaques (Figure 5 D). Moreover, some CARMN RNAs co-localises with the CD45 marker42 confirming the expression in immune cells. However, regions of the CD68/CARMN positive area appear negative for CD45 in parallel sections, suggesting the possibility of a non-immune origin for these cells (Figure 5D).
As CARMN and miR-143/145 are evolutionary conserved, we were able to take advantage of mouse models to evaluate the expression and characterize the function of the CARMN locus. The mouse locus harbours two annotated isoforms of CARMN, and their expression was analysed using a common primer. To further examine the role of the CARMN in the pathophysiology of disease, we assessed the expression of the lncRNA and the miRNAs at different stages of murine atherosclerosis. For this purpose, we investigated the levels of the CARMN locus in two groups of Low-Density Lipoprotein Receptor Knock Out mice (LDLR-/-): i) mice fed with a 0.25% cholesterol diet for 14 weeks to develop advanced plaques, and ii) mice fed on regular chow for 14 weeks to promote the formation of early fatty streak. We first confirmed the plaque stage though qualitative histologic analysis classifying them as fatty streak, the first visible irregular lesion consisting of atherogenic lipoproteins and macrophages occurring at the lesion site43, or advanced atheroma characterized by a large lipid-rich core, infiltrated macrophages and thin collagen-poor fibrotic cap44 (Figure 5 E). Oil Red O (ORO) staining was performed to confirm plaques stages (Online Figure VII A).
We then isolated RNA from atherosclerotic lesions from each aortic root using Laser Capture Microdissection (LCM) and assessed the expression levels of the lncRNA and miRNAs. The murine CARMN as well as miR-143 and miR-145 were found significantly downregulated in advanced lesions versus early plaques (Figure 5 F), confirming the expression pattern observed in human advanced plaques.
CARMN axis knockout accelerates atherosclerosis in mice
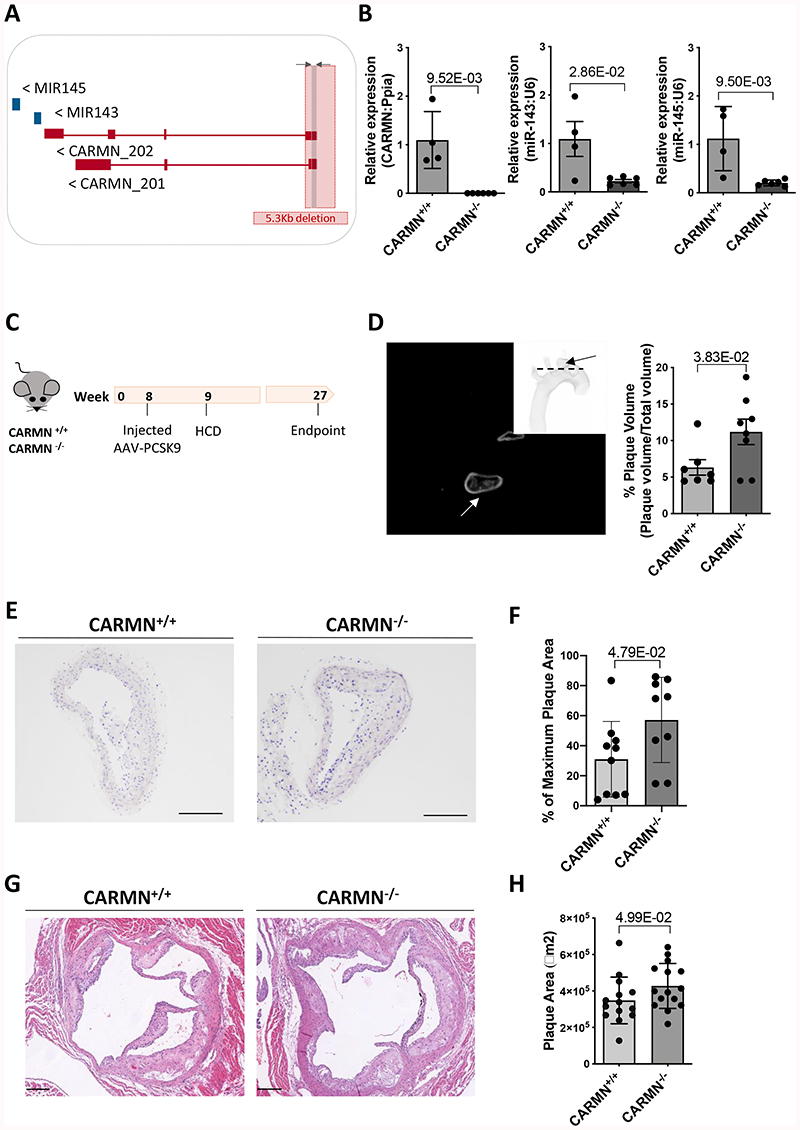
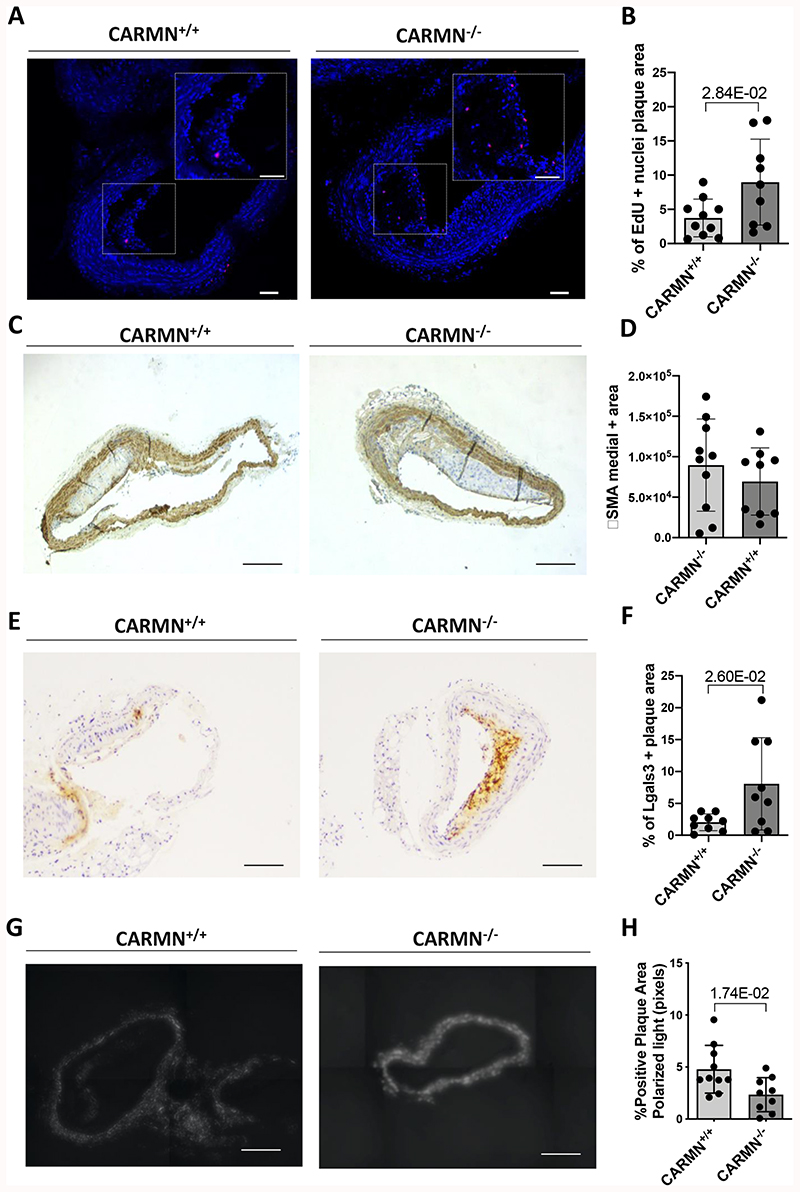
Considering the decrease of CARMN in advanced human and murine plaques and in vitro vSMC functional data, we hypothesized that the loss of murine CARMN would modulate the development of atherosclerosis in vivo. Therefore, we exploited a CRISPR/Cas9-mediated gene editing approach to constitutively knock-out (-/-) the CARMN locus. The first exon of the CARMN transcripts and a 4.8kb portion of the gene promoter were deleted, leaving exons 2/3 and the miRNA stem loops intact (Figure 6 A). Aortas of mice on normal diet were used to confirm the deletion at the RNA level by RT-qPCR using primers directed at exon 1 versus wild type controls (CARMN+/+) (Figure 6 B). Using isoform specific primers downstream of the deletion site, we confirmed the knock-down of both transcripts (Online Figure VII B, C). We also observed in the same samples the downregulation of miR-143 and miR-145. Moreover, as part of the characterization of knockout mice we examined the expression of the lncRNA Braveheart, downstream to CARMN transcript, which was previously found to be downregulated following loss-of-function of CARMN in mouse culture of CPCs8. However, qRT-PCR data indicated that Braveheart expression was not significantly changed in aortic arches isolated from CARMN-/- versus CARMN+/+ samples (Online Figure VII, D). To determine the effect of CARMN deficiency in the pathological context, we induced atherosclerosis in CARMN-/- mice and C57BL/6Ntac littermate controls using a validated mouse model of atherosclerosis [3] (Figure 6 C). To achieve long-term hypercholesterolemia, 8-weeks old CARMN+/+ and CARMN-/- mice were injected with 1012 vector genomes/mouse of the recombinant Adeno-associated Virus serotype 8 (AAV8) to induce overexpression of Proprotein convertase subtilisin/Kexin type 9 (PCSK9) in the liver. From one-week post injection, all mice received high cholesterol diet for the following 18 weeks to promote hyperlipidaemia and advanced atherosclerosis. Circulating cholesterol levels were measured at baseline (prior to virus administration), 6 and 10-weeks post injection and at sacrifice. Consistent with previous studies validating the AAV-PCSK9-induced model in wild-type animals [3][4], we found increased cholesterol levels in CARMN+/+ and CARMN-/- animals compared to baseline, demonstrating efficacy of the model (Online Figure VII E). We found no differences in body weight and plasma cholesterol in CARMN-/- versus CARMN+/+ animals (Online Figure VII F, G). Additionally, we did not observe significant differences in circulating immune cells and bone marrow progenitors in the two groups of animals, suggesting little or no effect on immune cell differentiation and proliferation (Online Figure VIII, IX, X). We next evaluated the severity of atherosclerosis by histological analysis in cross-sections in two of the main sites where atherosclerosis develops in mice, the brachiocephalic arteries and the aortic roots. To specifically evaluate the structure and the volume of the lesions in the brachiocephalic arteries, we used Optical Projection Tomography (OPT) [5]. Quantification of the resulting 3-dimensional images showed increased plaque volumes in CARMN-/- animals compared to wild type controls (Figure 6 D). Analysis of Haematoxylin/Eosin stained section confirmed an increase in plaque area in the brachiocephalic arteries of CARMN-/- versus CARMN+/+ mice (Figure 6 E-F). Furthermore, we observed a significant increase of the cross-sectional area of the lesions developed in the aortic roots of CARMN-/- versus control mice (Figure 6 G-H). We next sought to characterize the composition of the plaques in the brachiocephalic artery and aortic roots. In the pathological setting, abnormal proliferation of vSMCs4 3 and resident macrophages48 49 represent a key features of developing plaques. Cellular proliferation was analysed by EdU incorporation. A significant increase in EdU incorporation was observed in the plaques of CARMN-/- animals compared to controls (Figure 7 A, B) and thus increased proliferation rate of cells within the plaques. To further analyse the cellular composition of the plaques, cross-sections of the brachiocephalic arteries were stained for markers of vSMCs ACTA2, MYH11, and a marker for activated macrophages and de-differentiated vSMCs, LGALS3. While the overall amount of smooth muscle actin-expressing vSMCs and cells positive for Myh11 was similar among the CARMN+/+ and CARMN-/- groups (Figure 7 C-D and Online Figure X C, D), the content of LGALS3 in the plaque was found to be significantly increased in the lesions of CARMN-/- versus wild type controls (Figure 7 E-F). Moreover, the process of collagen deposition/degradation is a determinant factor in evaluating the structure and the stability of the plaques and can be used as measure of plaque vulnerability50,51. Therefore, we investigated the impact of CARMN-/- on the collagen turnover. Picrosirius red staining of the brachiocephalic arteries showed a significant decrease in collagen in CARMN-/- versus CARMN+/+ animals, in line with decreased collagen gene expression observed in vitro. (Figure 7 G, H). However, no significant change was observed in the plaque composition of the aortic roots of CARMN-/- versus wild type controls (Online Figure VII H-K). The absence of significant differences between the CARMN-/- and CARMN+/+ groups at the aortic root level could be explained by the vascular-site specific or temporal differences during plaque formation52. Overall, these data suggest that the depletion of CARMN induced the progression of atherosclerotic lesions to a more vulnerable plaque type.
Figure 6. Genetic ablation of CARMN affects the area, the volume and the composition of plaques developed in the aortic arches of CARMN-/- versus CARMN+/+ animals.
Schematic representation of CARMN locus located on mouse chromosome 18 and the CRISPR/Cas9 deletion of the first exon and 4.8 kilobases of the promoter (total 5 kilobases). The arrows next to each transcript name, indicate the direction of transcription. The grey arrows and box indicate where common primers to both isoforms were designed. B) Validation of knockout strategy by qRT-PCR using common primer set performed on the aortic arches of CARMN+/+ (n=4) and CARMN-/- (n=6) mice. Mann-Whitney was used to assess statistical significance indicated with p values. C) Schematic workflow used for the induction of mouse atherosclerosis using AAV8-mediated overexpression of PCSK9 and high cholesterol diet in CARMN+/+ and CARMN-/- animals. D) Representative tomographic reconstruction of pictures obtained by Optical Projection Tomography (black picture) and non-tomographic projection of the mouse aortic arch (white picture). The arrows indicate the brachiocephalic artery. Quantification of the plaque volumes in CARMN+/+ (n=7) and CARMN-/- (n=8) animals performed using CTan software. Student’s t-test was used to assess statistical significance indicated with p values. E) Representative Haematoxylin & Eosin staining of cross-sections from brachiocephalic artery of CARMN+/+ and CARMN-/- animals. Pictures acquired at 10X magnification. Scale bar 200μm. F) Quantification of the plaque areas at the site of maximum lesion of brachiocephalic arteries in CARMN+/+ (n=10) and CARMN-/- (n=9) mice, performed using Fiji software. Student’s t-test was used to assess statistical significance indicated with p values. G) Haematoxylin & Eosin staining of cross-sections from aortic roots of wild type and CARMN-/-animals. Pictures acquired at 5X magnification. Scale bar 200μm. H) Quantification of plaque area of aortic roots of CARMN+/+ (n=14) and CARMN-/- (n=15) animals, performed using QuPath software. Student’s t-test was used to assess statistical significance indicated with p values.
Figure 7. The genetic knock-out of CARMN increased the expression of Lgals3 in plaques developed in the aortic arch of CARMN-/- animals versus wild type controls.
A) Representative EdU staining of proliferating cells in plaques from CARMN+/+ and CARMN-/- animals. Pictures were acquired with 10X magnification. Scale bar 80μm. B) Quantification of proliferating cells in plaques of CARMN+/+ (n=10) and CARMN-/- (n=9) animals. Values are expressed as % of positive nuclei over the total cells counted in the plaque. C) and E) Representative immunostainings and D) and F) quantification of the positive area for αSMA and Lgals3 staining respectively in the plaques of CARMN+/+ (n=10) and CARMN-/- (n=9) animals. Pictures acquired at 10X magnification. Scale bar 200μm. G) Representative picrosirius red staining and H) quantification of collagen content in the plaques of CARMN+/+ (n=10) and CARMN-/-(n=9) animals. The pictures were acquired by polarized light microscopy at 10X magnification. Scale bar 200μm. Student’s t-test was used to assess statistical significance indicated with p values.
Discussion
Here, we comprehensively characterized for the first time the expression and the function of the CARMN lncRNA loci in hCASMC phenotypes in vitro and identified the functional consequence of the genetic loss of this lncRNA in the pathological setting of atherosclerosis in vivo. We show that GapmeR-mediated loss of the lncRNA primes vSMCs into a pro-pathological activated state, with concomitant reduction in the expression levels of both miR-143 and miR-145. Moreover, we functionally dissected the roles of CARMN and the miR-143/145 cluster and highlighted that in addition to its role as microRNA host gene, our data suggest that the CARMN lncRNA has miRNA-independent effects on vSMCs function. Furthermore, we show loss of CARMN in advanced human and mouse atherosclerotic lesions compared to matched tissue with stable and less advanced disease regions. Finally, we show that CRISPR/Cas9-mediated genetic targeting of CARMN in mice leads to development of more advanced lesions in cholesterol-fed mice.
We used an RNA sequencing approach to understand the functional consequence of the modulation of CARMN lncRNA expression in vitro in hCASMCs. Our results reveal that the loss of CARMN lncRNA expression significantly affects the transcriptional vSMC landscape. In particular following CARMN knockdown, hCASMCs gained a gene expression signature associated with a partially differentiated state towards a more plastic phenotype, and showed a dysregulation of fundamental biological processes such as proliferation and migration. Transcriptomic analysis of CARMN at basal condition showed a substantial overlap with CMBCD-loaded signature, particularly in metabolism and homeostasis of lipids. Accordingly, we postulated that depletion of CARMN predisposes hCASMCs to acquire early phenotypic/functional properties of foam-cell like cells, thus priming the pro-atherogenic phenotype likely contributing to the pathophysiology of CVD. However, the dedifferentiation process following CARMN and miR-143/145 depletion affected only some of the vSMC identity markers indicating a partial effect towards de-differentiating phenotype.
Recent studies have begun to unravel the complex network of interactions between those miRNAs that are embedded within so-called host genes26, whose biogenesis can be mutually exclusive27 or they can be co-expressed and exert independent functions28. In our study, we observed the expression of miR-143 and miR-145 was also affected by CARMN depletion. This effect could simply be due to the pri-miRNA precursor overlapping CARMN locus. On the other hand, it is also possible the existence of a regulatory circuit in which CARMN represents an additional level of post-transcriptional regulation and its downregulation affects the expression level of the miRNAs. We acknowledge that the presence of multiple promoters identified for the miRNAs16,20 enhances the challenge to uncover the existing lncRNA-miRNAs regulatory mechanisms. A recent study highlighted the possibility for lncRNA host genes to regulate hosted miRNAs via super-enhancer regulatory regions53 and CARMN was previously shown to be associated with a super-enhancer region and to increase its activity24. However, the demonstration of the cis-regulatory role of CARMN in vSMCs would require further studies, including chromatin marks and interaction analysis in this cell type. In addition, the literature on the regulation and function of these miRNAs in atherosclerosis is unclear.
We used a co-transfection strategy to increase the levels of miR-143/145 in CARMN-depleted vSMCs, allowing to dissect the contribution of CARMN from the miRNAs in vitro. Albeit the off-target limitation of the co-transfection strategy, our findings suggest that CARMN regulation of vSMC proliferation may possibly occur independently of the modulation of miR-143/145 expression. However, further dissociation of CARMN and miR-143 and miR-145 effect remains experimentally challenging due to the genetic complexity of the locus. Hence, further studies are required to understand the independent effects of CARMN and mir143/145 using, for example, targeted CRISPR activation, inhibition and editing approaches on the locus in primary vascular cells and mice. Our data expands the knowledge on the expression of the lncRNA at early and advanced stages of human and mouse atherosclerotic plaques. In a clinical setting, CARMN was significantly downregulated in advanced plaques versus stable lesions isolated by endarterectomy from symptomatic patients. Additionally, CARMN expression was found significantly reduced in murine advanced atheroma versus fatty streak. Therefore, we suggest that the downregulation of CARMN is a key event associated with the progression to more advanced lesions in CVD.
To determine whether the functional role of the lncRNA is conserved in vivo, and to understand the functional consequence of the loss of CARMN during the pathophysiology of atherosclerosis in vivo, we genetically modulated the expression of the lncRNA in mice exploiting the CRISPR (clustered regularly interspaced short palindromic repeats)-Cas9 (CRISPR-associated protein 9) technology. Previous studies have genetically ablated a region of the pre-miRNAs stem loop to generate miR-143/145-deficent mice14,15,17,19. We targeted the promoter region of the locus encompassing the first exon of CARMN transcripts. This resulted in the first genetic CARMN-/- model. In our in vivo atherosclerosis study using CARMN-/- mice, we found increased cellular proliferation in the atherosclerotic lesions of CARMN-/- versus CARMN+/+ animals and a more advanced plaque phenotype in the CARMN-/- mice. This result can be explained by findings from previous studies where vSMCs within advanced plaque show high proliferative index54,55,56. The evaluation of plaque composition revealed a largely unchanged content of typical vSMC markers, α-SMA and MYH11. However, considering that expression of vSMC differentiation markers, such as α-SMA and Myh11, are lost by dedifferentiated vSMCs in advanced plaques7, the interpretation of these results is limited as vSMC may no longer be recognised as such. Recent advances in vSMC-fate mapping have allowed a more reliable identification of vSMC-derived cells which contribute to intimal and atherosclerotic lesions formation in injured arteries57,58. We acknowledge the lack of lineage-tracing mouse model represents a limitation to unambiguously characterize the plaque composition in our in vivo study. The evaluation of Lgals3, marker for macrophages and dedifferentiated vSMCs revealed a significant increase in CARMN-/- versus CARMN+/+ animals. Recently, studies have provided evidence that the rapid turnover and build-up of macrophages in advanced atherosclerotic plaques is mostly due to their local proliferation48,59. Importantly, the large pool of foamy and pro-inflammatory cells expressing Lgals3, was defined among the main features of lesion progression toward the advanced state38. Accordingly, our results show that CARMN-/- increases the content of Lgals3, thus inducing the progression of the plaques toward an inflammatory and advanced stage. Collectively, these results reveal the functional role of CARMN axis to the phenotype observed in vivo and to clearly define the contribution of CARMN and microRNAs separately is an area for future research.
The decrease in collagen content within the lesions has been associated with progression to a more advanced plaque phenotype60. In this context, previous studies have reported that lower content of collagen may be due to a reduced synthesis by vSMCs and to an enhanced degradation by metalloproteases and collagenases50. In our study, the analysis of total collagen content within the plaques showed a significant decrease in CARMN-/- versus CARMN+/+ mice, confirming that CARMN-/- induced a more advanced plaque phenotype. These results are consistent with the transcriptomic analysis of hCASMCs following knock-down of CARMN, where we observed a decrease in collagen and elastase gene expression and the upregulation of genes associated with collagen breakdown (MMPs).
Collectively, our study demonstrates that loss of CARMN axis is an early event in atherosclerosis, both in mice and human. We have shown that downregulation of CARMN is detrimental for the maintenance of hCASMCs differentiated state and induces itself a phenotypic transition toward proproliferative phenotype, independently from the modulation of miR-143/145 cluster. In this research, we have characterized the effect of the dysregulation of miR-143/145 on the expression of vSMC identity genes and their ability to migrate. Finally, we have characterized the phenotypical consequence of CARMN depletion during the development of atherosclerosis in vivo and demonstrated that the knock-out of CARMN locus altered the composition of the plaques to a more advanced plaque type.
Supplementary Material
Novelty and Significance.
What Is Known?
De-differentiation of vascular smooth muscle cells (vSMCs) occurs in atherosclerotic lesions.
Non-coding RNAs, such as long non-coding RNAs (lncRNAs) and microRNAs (miRNAs), can regulate key aspects of vascular pathology.
CARMN is a lncRNA located immediately upstream of miRNAs, miR-143 and miR-145, which are involved in vSMC function.
What New Information Does This Article Contribute?
The CARMN/mir143/mir145 axis can prime vSMCs towards pro-proliferative, pro-migratory and de-differentiated phenotypes, in vitro.
CARMN lncRNA potentially acts independently of the miRNAs to regulate vSMC proliferation, whereas migration and de-differentiation phenotypes are likely modulated by mir143/145 independently.
CARMN knockout mice develop accelerated atherosclerosis and advanced plaques.
In this study, the conserved and genetically complex long non-coding RNA CARMN, and its co-located miRNAs, miR-143 and miR-145, were comprehensively characterized in human and mouse atherosclerosis. CARMN transcript down-regulation was associated with advanced plaque phenotype and CARMN knockout in mouse had increased atherosclerosis. Downregulation of the CARMN/miR-143/miR-145 axis primed vSMCs towards pro-proliferative, pro-migratory and dedifferentiated phenotypes, in vitro. Mechanistically, we provided evidence that the CARMN lncRNA acted independently of the miRNAs to regulate vSMC proliferation, whereas migration and dedifferentiation phenotypes were associated with the modulation of miRNAs expression. While both miRNAs are well known in vSMC biology, our study begins to dissect the complex genetic interplay between lncRNA host genes and their associated miRNAs in health and disease. Further, we provide evidence for the fundamental, yet complex role of lncRNAs in vSMC function.
Acknowdlegements
The authors thank Gregor Aitchison, Kathryn Newton, Michael Millar, Erwin Wijnands and George Kuriakose for their technical assistance during the study. Flow cytometry data were generated with support from the QMRI Flow Cytometry and cell sorting facility, University of Edinburgh. AAV8-PCSK9 was kindly provided by the AVU (AAV Vector Unit), I.C.G.E.B (Trieste, TS). Laser Capture Microdissection was performed in collaboration with Professor Ira Tabas at Columbia University Medical Centre (NY) and with the technical assistance of the Confocal and Specialized Microscopy Shared Resource, Columbia University Medical Centre (NY). The authors also thank Prof. Martyn Taylor, Stefan Rooke and Amanda Warr for the assistance provided during the Oxford Nanopore sequencing and data analysis.
Sources of Funding
This work is supported by the British Heart Foundation Research Excellent Award [RE/18/5/34216], BHF Chair [CH/11/2/28733] and fellowship to F.V. (FS/18/10/33413), and a BHF programme Grant to A.H.B [BHF CVR grant (RM/17/3/33381)] and the European Research Council Advanced Grant VASMIR (RE7644) to A.H.B., and Dutch heart foundation fellowship 2016T060 and a Leducq transatlantic network grant to J.C.S. and BHF project grant to A.H.B and L.D. This project has also received funding from the European Union’s Horizon 2020 Programme for Research and Innovation (825670). T.C.W is the recipient of a Wellcome Trust award [204802/Z/16/Z]. Australian National Health and Medical Research Council Early Career Fellowship [APP1072662 to M.B.C]
Nonstandard abbreviations and Acronyms
- AAV
adeno-associated virus
- ABCA1
ATP-binding Cassette Transporter 1
- ABCG1
ATP-binding Cassette Subfamily G Member 1
- Acta2
Smooth muscle actin 2
- ApoE
apolipoprotein E
- ASO
antisense oligonucleotides
- αSMA
α-Smooth Muscle Actin
- CARMN
cardiac mesoderm enhancer-associated noncoding RNA
- CARMN-/-
CARMN knock-out
- CARMN+/+
CARMN wild type
- CCL
Chemokine C-C Motif Ligand
- CD68
Cluster of Differentiation 68
- CD45
Protein Tyrosine Phosphatase Receptor Type C
- CMBCD
cholesterol-methyl-β-cyclodextrin
- CXCL1
C-X-C Motif Chemokine 1
- CNN1
Calponin 1
- COL3a1
Collagen type 3 alpha chain 1
- CRISPR
Clustered Regularly Interspaced Short Palindromic Repeats
- EdU
5-ethynyl-2’-deoxyuridine
- EGFR
Epidermal Growth Factor Receptor
- FACS
fluorescence-activated cell sorting
- FISH
Fluorescent In-Situ Hybridization
- hCASMCs
human coronary artery smooth muscle cells
- KLF4
Kruppell-like factor 4
- IL1-beta
Interleukin 1 beta
- IL6
Interleukin 6
- ISH
In-Situ Hybridization
- LCM
laser capture microdissection
- LDLR-/-
low density lipoprotein receptor knock-out
- Lgals3
Galectin 3
- lncRNA
Long Noncoding RNA
- Myh11
Myosin Heavy Chain 11
- Myocd
Myocardin
- MiRNAs
miRNAs
- MMP
Metalloproteases
- OPT
Optical Projection Tomography
- ORO
Oil Red O
- Ox-LDL
Oxidised Low-Density Lipoprotein
- PCSK9
Proprotein Convertase Subtilisin/Kexin type 9
- PDGF
Platelet-Derived Growth Factor
- PDFGRa
Platelet-Derived Growth Factor Receptor alpha
- qRT-PCR
Quantitative Real-Time Polymerase Chain Reaction
- RNA-seq
RNA sequencing
- RNU48
Small Nucleolar RNA 48
- TAGLN
Taglin
- TSS
Transcriptional Start Sites
- UBC
Ubiquitin C
- vSMCs
vascular smooth muscle cells
Footnotes
Disclosures
MBC has received support from Oxford Nanopore Technologies (ONT) to present at scientific conferences. However, ONT played no role in study design, execution, analysis or publication.
Data Availability
Expanded information about materials and methods are available in the Online Data Supplement. All supporting data and materials are available within the Online Data Supplement and available from the corresponding author upon reasonable request. All RNA-Seq data are deposited in the Gene Expression Omnibus database (Ref. GSE158972 and Ref. GSE165445).
References
- 1.Stefanadis C, Antoniou CK, Tsiachris D, Pietri P. Coronary atherosclerotic vulnerable plaque: Current perspectives. J Am Heart Assoc. 2017;6 doi: 10.1161/JAHA.117.005543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 3.Gomez D, Owens GK. Smooth muscle cell phenotypic switching in atherosclerosis. Cardiovasc Res. 2012;95:156–164. doi: 10.1093/cvr/cvs115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennett MR, Sinha S, Owens GK. Vascular Smooth Muscle Cells in Atherosclerosis. Circ Res. 2016;118:692–702. doi: 10.1161/CIRCRESAHA.115.306361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang Y, Dubland JA, Allahverdian S, Asonye E, Sahin B, Jaw JE, Sin DD, Seidman MA, Leeper NJ, Francis GA. Smooth Muscle Cells Contribute the Majority of Foam Cells in ApoE (Apolipoprotein E)-Deficient Mouse Atherosclerosis. Arterioscler Thromb Vasc Biol. 2019;39:876–887. doi: 10.1161/ATVBAHA.119.312434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Newby AC, Zaltsman AB. Fibrous cap formation or destruction - The critical importance of vascular smooth muscle cell proliferation, migration and matrix formation. Cardiovasc Res. 1999;41:345–360. [PubMed] [Google Scholar]
- 7.Allahverdian S, Chaabane C, Boukais K, Francis GA, Bochaton-Piallat ML. Smooth muscle cell fate and plasticity in atherosclerosis. Cardiovasc Res. 2018;114:540–550. doi: 10.1093/cvr/cvy022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mahmoud AD, Ballantyne MD, Miscianinov V, Pinel K, Hung J, Scanlon JP, Iyinikkel J, Kaczynski J, Tavares AS, Bradshaw AC, Mills NL, et al. The human-specific and smooth muscle cellenriched LncRNA SMILR promotes proliferation by regulating mitotic CENPF mRNA and drives cell-cycle progression which can be targeted to limit vascular remodeling. Circ Res. 2019;125:535–551. doi: 10.1161/CIRCRESAHA.119.314876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cremer S, Michalik KM, Fischer A, Pfisterer L, Jaé N, Winter C, Boon RA, Muhly-Reinholz M, John D, Uchida S, Weber C, et al. Hematopoietic deficiency of the long noncoding RNA malat1 promotes atherosclerosis and plaque inflammation. Circulation. 2019;139:1320–1334. doi: 10.1161/CIRCULATIONAHA.117.029015. [DOI] [PubMed] [Google Scholar]
- 10.Hung J, Scanlon JP, Mahmoud AD, Rodor J, Ballantyne M, Fontaine MAC, Temmerman L, Kaczynski J, Connor KL, Bhushan R, Biessen EAL, et al. Novel plaque enriched long noncoding RNA in atherosclerotic macrophage regulation (PELATON) Arterioscler Thromb Vasc Biol. 2020;40:697–713. doi: 10.1161/ATVBAHA.119.313430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vengrenyuk Y, Nishi H, Long X, Ouimet M, Savji N, Martinez FO, Cassella CP, Moore KJ, Ramsey SA, Miano JM, Fisher EA. Cholesterol loading reprograms the microRNA-143/145-Myocardin axis to convert aortic smooth muscle cells to a dysfunctional macrophage-like phenotype. Arterioscler Thromb Vasc Biol. 2015;35:535–546. doi: 10.1161/ATVBAHA.114.304029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Quintavalle M, Elia L, Condorelli G, Courtneidge SA. MicroRNA control of podosome formation in vascular smooth muscle cells in vivo and in vitro. J Cell Biol. 2010;189:13–22. doi: 10.1083/jcb.200912096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vacante F, Denby L, Sluimer JC, Baker AH. The function of miR-143, miR-145 and the MiR-143 host gene in cardiovascular development and disease. Vascul Pharmacol. 2019;112:24–30. doi: 10.1016/j.vph.2018.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boettger T, Beetz N, Kostin S, Schneider J, Krüger M, Hein L, Braun T. Acquisition of the contractile phenotype by murine arterial smooth muscle cells depends on the Mir143/145 gene cluster. J Clin Invest. 2009;119:2634–2647. doi: 10.1172/JCI38864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elia L, Quintavalle M, Zhang J, Contu R, Cossu L, Latronico MVG, Peterson KL, Indolfi C, Catalucci D, Chen J, Courtneidge SA, et al. The knockout of miR-143 and -145 alters smooth muscle cell maintenance and vascular homeostasis in mice: Correlates with human disease. Cell Death Differ. 2009;16:1590–1598. doi: 10.1038/cdd.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN, Srivastava D. MiR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460:705–710. doi: 10.1038/nature08195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xin M, Small EM, Sutherland LB, Qi X, McAnally J, Plato CF, Richardson JA, Bassel-Duby R, Olson EN. MicroRNAs miR-143 and miR-145 modulate cytoskeletal dynamics and responsiveness of smooth muscle cells to injury. Genes Dev. 2009;23:2166–2178. doi: 10.1101/gad.1842409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lovren F, Pan Y, Quan A, Singh KK, Shukla PC, Gupta N, Steer BM, Ingram AJ, Gupta M, Al-Omran M, Teoh H, et al. MicroRNA-145 targeted therapy reduces atherosclerosis. Circulation. 2012;126:S81–90. doi: 10.1161/CIRCULATIONAHA.111.084186. [DOI] [PubMed] [Google Scholar]
- 19.Cipollone F, Felicioni L, Sarzani R, Ucchino S, Spigonardo F, Mandolini C, Malatesta S, Bucci M, Mammarella C, Santovito D, De Lutiis F, et al. A unique MicroRNA signature associated with plaque instability in humans. Stroke. 2011;42:2556–2563. doi: 10.1161/STROKEAHA.110.597575. [DOI] [PubMed] [Google Scholar]
- 20.Deng L, Blanco FJ, Stevens H, Lu R, Caudrillier A, McBride M, McClure JD, Grant J, Thomas M, Frid M, Stenmark K, et al. MicroRNA-143 Activation Regulates Smooth Muscle and Endothelial Cell Crosstalk in Pulmonary Arterial Hypertension. Circ Res. 2015;117:870–883. doi: 10.1161/CIRCRESAHA.115.306806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caruso P, Dempsie Y, Stevens HC, McDonald RA, Long L, Lu R, White K, Mair KM, McClure JD, Southwood M, Upton P, et al. A role for miR-145 in pulmonary arterial hypertension: Evidence from mouse models and patient samples. Circ Res. 2012;111:290–300. doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 22.Rangrez AY, Massy ZA, Le Meuth VM, Metzinger L. MiR-143 and miR-145 molecular keys to switch the phenotype of vascular smooth muscle cells. Circ Cardiovasc Genet. 2011;4:197–205. doi: 10.1161/CIRCGENETICS.110.958702. [DOI] [PubMed] [Google Scholar]
- 23.Lagarde J, Uszczynska-Ratajczak B, Carbonell S, Pérez-Lluch S, Abad A, Davis C, Gingeras TR, Frankish A, Harrow J, Guigo R, Johnson R. High-throughput annotation of full-length long noncoding RNAs with capture long-read sequencing. Nat Genet. 2017;49:1731–1740. doi: 10.1038/ng.3988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ounzain S, Micheletti R, Arnan C, Plaisance I, Cecchi D, Schroen B, Reverter F, Alexanian M, Gonzales C, Ng SY, Bussotti G, et al. CARMEN, a human super enhancer-associated long noncoding RNA controlling cardiac specification, differentiation and homeostasis. J Mol Cell Cardiol. 2015;89:98–112. doi: 10.1016/j.yjmcc.2015.09.016. [DOI] [PubMed] [Google Scholar]
- 25.Plaisance I, Perruchoud S, Fernandez-Tenorio M, Gonzales C, Ounzain S, Ruchat P, Nemir M, Niggli E, Pedrazzini T. Cardiomyocyte Lineage Specification in Adult Human Cardiac Precursor Cells Via Modulation of Enhancer-Associated Long Noncoding RNA Expression. JACC Basic to Transl Sci. 2016;1:472–493. doi: 10.1016/j.jacbts.2016.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Profumo V, Forte B, Percio S, Rotundo F, Doldi V, Ferrari E, Fenderico N, Dugo M, Romagnoli D, Benelli M, Valdagni R, et al. LEADeR role of miR-205 host gene as long noncoding RNA in prostate basal cell differentiation. Nat Commun. 2019;10 doi: 10.1038/s41467-018-08153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Legnini I, Morlando M, Mangiavacchi A, Fatica A, Bozzoni I. A Feedforward Regulatory Loop between HuR and the Long Noncoding RNA linc-MD1 Controls Early Phases of Myogenesis. Mol Cell. 2014;53:506–514. doi: 10.1016/j.molcel.2013.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Q, Tripathi V, Yoon JH, Singh DK, Hao Q, Min KW, Davila S, Zealy RW, Li XL, Polycarpou-Schwarz M, Lehrmann E, et al. MIR100 host gene-encoded lncRNAs regulate cell cycle by modulating the interaction between HuR and its target mRNAs. Nucleic Acids Res. 2018;46:10405–10416. doi: 10.1093/nar/gky696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ulitsky I, Bartel DP. XLincRNAs: Genomics, evolution, and mechanisms. Cell. 2013;154:26. doi: 10.1016/j.cell.2013.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ponting CP, Oliver PL, Reik W. Evolution and Functions of Long Noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 31.Deaton RA, Gan Q, Owens GK. Spl-dependent activation of KLF4 is required for PDGF-BB-induced phenotypic modulation of smooth muscle. Am J Physiol - Hear Circ Physiol. 2009;296:H1027–1037. doi: 10.1152/ajpheart.01230.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chellan B, Reardon CA, Getz GS, Bowman MAH. Enzymatically modified low-density lipoprotein promotes foam cell formation in smooth muscle cells via macropinocytosis and enhances receptor-mediated uptake of oxidized low-density lipoprotein. Arterioscler Thromb Vasc Biol. 2016;36:1101–1113. doi: 10.1161/ATVBAHA.116.307306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miano JM. Myocardin in biology and disease. J Biomed Res. 2015;29:3–19. doi: 10.7555/JBR.29.20140151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ait-Oufella H, Taleb S, Mallat Z, Tedgui A. Recent advances on the role of cytokines in atherosclerosis. Arterioscler Thromb Vasc Biol. 2011;31:969–979. doi: 10.1161/ATVBAHA.110.207415. [DOI] [PubMed] [Google Scholar]
- 35.van der Vorst EPC, Döring Y, Weber C. Chemokines and their receptors in Atherosclerosis. J Mol Med. 2015;93:963–971. doi: 10.1007/s00109-015-1317-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Adiguzel E, Ahmad PJ, Franco C, Bendeck MP. Collagens in the progression and complications of atherosclerosis. Vasc Med. 2009;14:73–89. doi: 10.1177/1358863X08094801. [DOI] [PubMed] [Google Scholar]
- 37.Newby AC. Metalloproteinases and Vulnerable Atherosclerotic Plaques. Trends Cardiovasc Med. 2007;17:253–258. doi: 10.1016/j.tcm.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shankman LS, Gomez D, Cherepanova OA, Salmon M, Alencar GF, Haskins RM, Swiatlowska P, Newman AAC, Greene ES, Straub AC, Isakson B, et al. KLF4-dependent phenotypic modulation of smooth muscle cells has a key role in atherosclerotic plaque pathogenesis. Nat Med. 2015;21:628–637. doi: 10.1038/nm.3866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duband JL, Gimona M, Scatena M, Sartore S, Small JV. Calponin and SM22 as differentiation markers of smooth muscle: spatiotemporal distribution during avian embryonic development. Differentiation. 1993;55:1–11. doi: 10.1111/j.1432-0436.1993.tb00027.x. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, Jiang M, Liu Q, Han Z, Zhao Y, Ji S. miR-145-5p inhibits the proliferation and migration of bladder cancer cells by targeting TAGLN2. Oncol Lett. 2018;16:6355–6360. doi: 10.3892/ol.2018.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhu H, Dougherty U, Robinson V, Mustafi R, Pekow J, Kupfer S, Li YC, Hart J, Goss K, Fichera A, Joseph L, et al. EGFR signals downregulate tumor suppressors miR-143 and miR-145 in western diet-promoted murine colon cancer: Role of G 1 regulators. Mol Cancer Res. 2011;9:960–975. doi: 10.1158/1541-7786.MCR-10-0531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Koltsova EK, Hedrick CC, Ley K. Myeloid cells in atherosclerosis: A delicate balance of anti-inflammatory and proinflammatory mechanisms. Curr Opin Lipidol. 2013;24:371–380. doi: 10.1097/MOL.0b013e328363d298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stary HC, Chandler AB, Glagov S, Guyton JR, Insull W, Rosenfeld ME, Schaffer SA, Schwartz CJ, Wagner WD, Wissler RW. A definition of initial, fatty streak, and intermediate lesions of atherosclerosis: A report from the committee on vascular lesions of the council on arteriosclerosis, American Heart Association. Arterioscler Thromb. 1994;14:840–856. doi: 10.1161/01.atv.14.5.840. [DOI] [PubMed] [Google Scholar]
- 44.Falk E. Pathogenesis of Atherosclerosis. J Am Coll Cardiol. 2006;47:C7–12. doi: 10.1016/j.jacc.2005.09.068. [DOI] [PubMed] [Google Scholar]
- 45.Maxwell KN, Breslow JL. Adenoviral-mediated expression of Pcsk9 in mice results in a low-density lipoprotein receptor knockout phenotype. Proc Natl Acad Sci U S A. 2004;101:7100–7105. doi: 10.1073/pnas.0402133101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bjørklund MM, Hollensen AK, Hagensen MK, Dagnæs-Hansen F, Christoffersen C, Mikkelsen JG, Bentzon JF. Induction of atherosclerosis in mice and hamsters without germline genetic engineering. Circ Res. 2014;114:1684–1689. doi: 10.1161/CIRCRESAHA.114.302937. [DOI] [PubMed] [Google Scholar]
- 47.Kirkby NS, Low L, Seckl JR, Walker BR, Webb DJ, Hadoke PWF. Quantitative 3-dimensional imaging of murine neointimal and atherosclerotic lesions by optical projection tomography. PLoS One. 2011;6 doi: 10.1371/journal.pone.0016906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robbins CS, Hilgendorf I, Weber GF, Theurl I, Iwamoto Y, Figueiredo JL, Gorbatov R, Sukhova GK, Gerhardt LMS, Smyth D, Zavitz CCJ, et al. Local proliferation dominates lesional macrophage accumulation in atherosclerosis. Nat Med. 2013;19:1166–1172. doi: 10.1038/nm.3258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jenkins SJ, Ruckerl D, Thomas GD, Hewitson JP, Duncan S, Brombacher F, Maizels RM, Hume DA, Allen JE. IL-4 directly signals tissue-resident macrophages to proliferate beyond homeostatic levels controlled by CSF-1. J Exp Med. 2013;210:2477–2491. doi: 10.1084/jem.20121999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rekhter MD. Collagen synthesis in atherosclerosis: Too much and not enough. Cardiovasc Res. 1999;41:376–384. doi: 10.1016/s0008-6363(98)00321-6. [DOI] [PubMed] [Google Scholar]
- 51.Rekhter MD. How to evaluate plaque vulnerability in animal models of atherosclerosis? Cardiovasc Res. 2002;54:36–41. doi: 10.1016/s0008-6363(01)00537-5. [DOI] [PubMed] [Google Scholar]
- 52.VanderLaan PA, Reardon CA, Getz GS. Site Specificity of Atherosclerosis: Site-Selective Responses to Atherosclerotic Modulators. Arterioscler Thromb Vasc Biol. 2004;24:12–22. doi: 10.1161/01.ATV.0000105054.43931.f0. [DOI] [PubMed] [Google Scholar]
- 53.Suzuki HI, Young RA, Sharp PA. Super-Enhancer-Mediated RNA Processing Revealed by Integrative MicroRNA Network Analysis. Cell. 2017;168:1000–1014.e15. doi: 10.1016/j.cell.2017.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chappell J, Harman JL, Narasimhan VM, Yu H, Foote K, Simons BD, Bennett MR, Jørgensen HF. Extensive Proliferation of a Subset of Differentiated, yet Plastic, Medial Vascular Smooth Muscle Cells Contributes to Neointimal Formation in Mouse Injury and Atherosclerosis Models. Circ Res. 2016;119:1313–1323. doi: 10.1161/CIRCRESAHA.116.309799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Dobnikar L, Taylor AL, Chappell J, Oldach P, Harman JL, Oerton E, Dzierzak E, Bennett MR, Spivakov M, Jørgensen HF. Disease-relevant transcriptional signatures identified in individual smooth muscle cells from healthy mouse vessels. Nat Commun. 2018;9 doi: 10.1038/s41467-018-06891-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Majesky MW, Horita H, Ostriker A, Lu S, Regan JN, Bagchi A, Dong XR, Poczobutt J, Nemenoff RA, Weiser-Evans MCM. Differentiated Smooth Muscle Cells Generate a Subpopulation of Resident Vascular Progenitor Cells in the Adventitia Regulated by Klf4. Circ Res. 2017;120:296–311. doi: 10.1161/CIRCRESAHA.116.309322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Feil S, Fehrenbacher B, Lukowski R, Essmann F, Schulze-Osthoff K, Schaller M, Feil R. Transdifferentiation of vascular smooth muscle cells to macrophage-like cells during atherogenesis. Circ Res. 2014;115:662–667. doi: 10.1161/CIRCRESAHA.115.304634. [DOI] [PubMed] [Google Scholar]
- 59.Lutgens E, De Muinck ED, Kitslaar PJEHM, Tordoir JHM, Wellens HJJ, Daemen MJAP. Biphasic pattern of cell turnover characterizes the progression from fatty streaks to ruptured human atherosclerotic plaques. Cardiovasc Res. 1999;41:473–479. doi: 10.1016/s0008-6363(98)00311-3. [DOI] [PubMed] [Google Scholar]
- 60.Barnes MJ, Farndale RW. Collagens and atherosclerosis. Exp Gerontol. 1999;34:513–525. doi: 10.1016/s0531-5565(99)00038-8. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Expanded information about materials and methods are available in the Online Data Supplement. All supporting data and materials are available within the Online Data Supplement and available from the corresponding author upon reasonable request. All RNA-Seq data are deposited in the Gene Expression Omnibus database (Ref. GSE158972 and Ref. GSE165445).