Abstract
Systemic autoinflammatory diseases (SAIDs) are a growing group of disorders caused by a dysregulation of the innate immune system leading to episodes of systemic inflammation. In 1997, MEFV was the first gene identified as disease causing for Familial Mediterranean Fever, the most common hereditary SAID. In most cases, auto-inflammatory diseases have a strong genetic background with mutations in single genes. Since 1997 more than 30 new genes associated with autoinflammatory diseases have been identified, affecting different parts of the innate immune system. Nevertheless, for at least 40–60% of patients with phenotypes typical for SAIDs, a distinct diagnosis cannot be met, leading to undefined SAIDs (uSAIDs). However, SAIDs can also be of polygenic or multifactorial origin, with environmental influence modulating the phenotype. The implementation of a disease continuum model combining the adaptive and the innate immune system with autoinflammatory and autoimmune diseases shows the complexity of SAIDs and the importance of new methods to elucidate molecular changes and causative factors in SAIDs. Diagnosis is often based on clinical presentation and genetic testing. The timeline from onset to diagnosis takes up to 7.3 years, highlighting the indisputable need to identify new treatment and diagnostic targets. Recently, other factors are under investigation as additional contributors to the pathogenesis of SAIDs.
This review gives an overview of pathogenesis and etiology of SAIDs, and summarizes recent diagnosis and treatment options.
Keywords: Systemic autoinflammatory disease, Periodic fever, Inflammasomes, Innate immunity, Inflammation
1. Introduction
Systemic autoinflammatory diseases (SAIDs) are a group of disorders caused by a dysregulation of the innate immune system. As their name suggests, a shared key feature is their systemic pathobiology, which means that symptoms can affect the entire body. They can be viewed as the counterpart to autoimmune diseases, having in common that both are caused by abnormal immune responses. The main distinction is that autoimmune diseases are typically defined by a malfunction of the adaptive immune system while in SAIDs the innate immune system is affected (Table 1). The main cell types of the innate immune system are monocytes, macrophages, and neutrophils whereas the adaptive immune response is mediated by B and T cells. The initial defining criteria for SAIDs are the lack of high-titer autoantibodies or antigen-specific T cells [1] and a seemingly unprovoked systemic inflammation. Additional symptoms include fluctuating degrees of fever, as well as abdominal, articular, and cutaneous signs that may lack specificity, making a clinical diagnosis difficult [2]. The name “autoinflammatory disease” was proposed in 1999 by McDermott et al. [1]. Since its initial definition more than 30 new genes associated with autoinflammatory diseases have been identified, affecting different parts of the innate immune system [3]. This growing number is partly the result of genome wide association studies (GWAS).
Table 1. Differences between autoinflammatory and autoimmune diseases [4–10].
| Autoinflammation | Autoimmunity | |
|---|---|---|
| Immune dysregulation | Innate immune system | Adaptive immune system |
| Predominant cell types | Monocytes, macrophages, neutrophils | T cells, B cells |
| Cytokine targets used therapeutically | TNF, IFNαβ, IL-1, IL-2, IL-12, IL-23, IL-18 | IFNγ, TNFα, IL-1, IL-2, IL-4, IL-6, IL-5, IL-9, IL-10, IL-12, IL-13, IL-17, IL-22, IL-23 |
| Pathogenesis of organ damage | Neutrophil- and macrophage-mediated | Autoantibody- or autoantigen-specific T cell-mediated |
In most cases, autoinflammatory diseases have a strong genetic background with mutations in single genes. However, they can also be of polygenic or multifactorial origin, with environmental influence modulating the phenotype [3]. While there are clear differences between autoinflammatory and autoimmune diseases, they also share many similarities. In both groups, the underlying pathological processes are directed against the own body. They are systemic, involve the musculoskeletal system, and both include monogenic and polygenic diseases. The innate immune system plays a role in activating the adaptive immune system by antigen presenting cells. Thus, the innate immune system can trigger B and T cell response, and overt or longterm activation of innate immunity can result in autoimmune diseases [11–13]. Another crucial link between adaptive and innate immunity is IL-1β. It is one of the main effector molecules driving autoinflammatory processes and also acts on the effector cells of the adaptive immune system, B and T cells [14]. The similarities and connections between them led to the discussion if autoinflammatory and autoimmune diseases should be considered as one single group of diseases made up of a large spectrum of immunologic abnormalities with autoinflammatory syndromes at one end and autoimmune diseases at the other end. In this continuum model, immunological diseases are grouped along a spectrum with varying degrees of involvement of adaptive and innate immunity components. They are classified along the spectrum as autoinflammatory disease, polygenic autoinflammatory conditions, mixed pattern diseases or polygenic autoimmune conditions, and autoimmune diseases [15].
2. Inflammatory response and inflammasome activation
On a molecular basis, SAIDs result from an aberrant activation of the innate immune system. The innate immune system acts as a first line of defence, providing a large array of evolutionary conserved signalling receptors also called pattern-recognition receptors (PRR) to detect and act against pathogens. These membrane-bound and intracellular PRRs are vital for danger sensing and recognition of highly conserved structures expressed by pathogens (microbial pathogen-associated molecular patterns or PAMPs) and damaged cells (non- microbial damage-associated molecular patterns or DAMPs) [16–18].
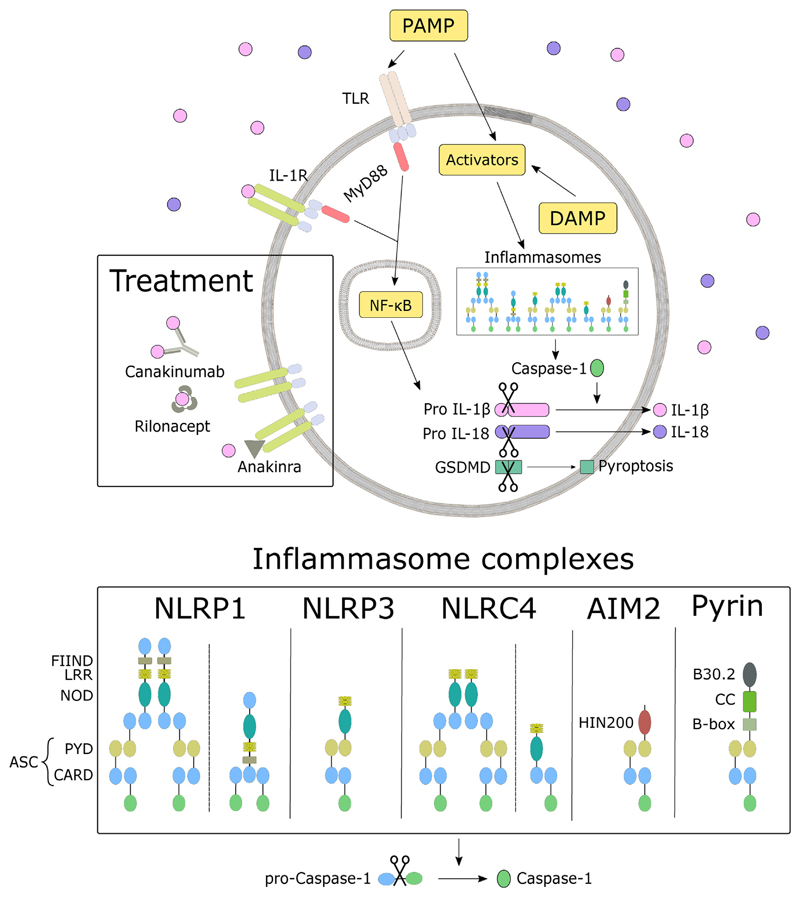
Membrane-bound PRRs include Toll-like receptors (TLRs) and C-type lectin receptors (CLRs) which are characterised by the presence of a carbohydrate-binding domain. Intracellular sensors, including Nucleotide-binding oligomerization domain (NOD)-like receptors (NLRs), Absence in melanoma 2 (AIM2)-like receptors (ALRs), and Pyrin are part of multimeric protein scaffolds called inflammasomes. Generally, inflammasomes consist of sensor molecules, adapter molecules apoptosis related speck-like protein containing caspase activation and recruitment domains (ASC), and caspase-1 as an effector molecule. The components of NLRs are a NOD domain and a C-terminal leucine-rich repeat (LRR) domain. The N-terminal domain varies between subgroups. NLRP1 has an additional C-terminal FIIND motif and a C-terminal caspase recruitment domain (CARD) domain. The AIM2 inflammasome consists of a Pyrin domain (PYD) and a HIN-200 domain mediating the binding of double-stranded DNA. Pyrin has a N-terminal PYD, a B-box domain, coiled-coiled domain (CC), and a C-terminal B30.2 domain [19]. ASC consists of PYD and CARD. It recruits caspase-1 through its CARD domain and activates it through proteolytic cleavage [20]. An exception to this mechanism is the Pyrin inflammasome that indirectly senses pathogen virulence factors modifying RhoA [21]. Even though inflammasomes differ in sensor molecules and activation signals (see Table 2), all mediate activation of caspase-1 [16].
Table 2. Inflammasome sensor proteins, their corresponding activating signals, and structure [22–24].
| Inflammasome complexes | Structure | Activating signals |
|---|---|---|
| NLRP1b | CARD - NOD - LRR - FIIND - CARD | Key virulence factors of Bacillus anthracis |
| NLRP3 | PYD - NOD - LRR | Diverse endogenous danger signals and pathogenic molecules |
| NLRC4 | CARD - NOD - LRR | Bacterial flagellin, type III secretion system components |
| AIM2 | PYD - HIN200 | Direct binding of double-stranded DNA |
| Pyrin | PYD - B-box - CC - B30.2 | Detection of bacterial modifications of Rho GTPases |
Active caspase-1 can catalyze a variety of cellular events crucial for regulating the immune response and inflammation such as the secretion of essential cytokines. The ability of caspase-1 to convert pro-IL-1β into its bioactive form IL-1β was identified in 1989 [25]. Later it was shown that caspase-1 also cleaves IL-18 [26]. Both bioactive forms are released from the cytoplasm into the bloodstream. Once released, IL-1β binds to the IL-1 receptor type I (IL-1RI) which is ubiquitously expressed. Upon binding, the myeloid differentiation primary response gene 88 (MyD88) induces the translocation of active NF- κB to the nucleus where it promotes the transcription of NF-κB-dependent genes, such as NLRP3, pro-IL-β, pro-IL-18, and IL-6, leading to induction and maintaining of the inflammatory response [22,27–29]. The inflammatory cascade is shown in Fig. 1.
Fig. 1. Inflammatory cascade.
Upon recognition of PAMP (see Table 2) and DAMP by intracellular sensors, multimeric protein scaffolds called inflammasomes are formed. As a result of this formation, pro-Caspase-1 is activated by proteolytic cleavage and further converting pro IL-1β and pro IL-18 to their bioactive form, which is released into the bloodstream. IL-1 receptors sense IL-1β and reacts by activating the NF-κB pathway. Active NF-κB is translocated to the nucleus where it promotes the transcription of NF-κB-dependent genes, such as NLRP3, pro-IL-β, pro-IL-18, and IL-6. The compositions of the inflammasome complexes are adapted from Ref. [21]. In the treatment section, different IL-1 blocking agents (Canakinumab, Rilonacept) and anti-IL-1 receptor antagonist (Anakinra) are depicted.
Furthermore, caspase-1 cleaves the membrane-pore-forming protein gasdermin-D (GSDMD), inducing pyroptosis. Pyroptosis is a rapid inflammatory form of cell death leading to a swelling of the cell and eventually membrane rupture to release alarmins in the extracellular space [19]. Contrary to pyroptosis, apoptosis is a regulated and controlled process that avoids eliciting inflammation. Mediated by a subset of caspases it leads to the formation of membrane-enclosed cellular fragments or apoptotic bodies [30].
In SAIDs many aspects of the inflammatory pathway can be dysre-gulated. A more detailed look on disease specific pathogenic mechanisms are discussed below.
3. Pathogenesis and etiology
Genetically, SAIDs can be categorized as monogenic or polygenic/ multifactorial. Monogenic SAIDs are caused by highly penetrant genetic variants in single genes and follow a clear pattern of Mendelian inheritance [31]. Polygenic or multifactorial SAIDs, like systemic juvenile idiopathic arthritis (SJIA), are more complex. They arise from permutations and combinations of common gene variants, where each variant alone confers only a small risk but together or with other extraneous influences becomes pathogenic [32,33].
Overall, polygenic and multifactorial SAIDs are more common than monogenic SAIDs, excluding the eastern Mediterranean basin where the prevalence of Familial Mediterranean Fever (FMF) is highly increased [34]. The most common monogenic SAIDs are FMF, NLRP3-associated autoinflammatory disease (NLRP3-AID; formerly known as Cryopyrin associated periodic syndromes - CAPS), and TNF receptor-associated periodic syndrome (TRAPS) [16].
3.1. Monogenic systemic autoinflammatory diseases
Generally, autoinflammatory phenotypes can be classified according to their type of mutation (see Table 3). Depending on their molecular mechanism, SAIDs can be separated into in-flammasomopathies or IL-1β-activation syndromes (FMF, NLRP3-AID, MKD, DIRA, DITRA), protein-folding disorders (TRAPS), NF-κB-activation disorders (Blau syndrome), interferonopathies (Aicardi-Goutières syndromes), and other cytokine-signalling disorders and complementopathies (i.e.: paroxysmal nocturnal hemoglobinuria, atypical hemolytic uremic syndrome) [21,35,36]. An overview of the most common monogenic SAIDs can be found in Table 4.
Table 3.
Categorization of causative mutations for autoinflammatory disorders [27,28] Abbreviations: MKD: Mevalonat kinase deficiency; DIRA: deficiency of the IL-1 receptor antagonist; DITRA: deficiency of IL-36 receptor antagonist; PLAID: PLCγ2-associated antibody deficiency and immune dysregulation; APLAID: autoin flammation and PLAID; LAID: LYN-associated autoinflammatory disease.
| Type of mutation | Effect | Disease |
|---|---|---|
| Gain-of-function mutations in genes encoding PRRs or their adaptor molecules | constitutively increased innate immune sensor function; increased or prolonged production of proinflammatory mediators | FMF, NLRP3-AID, Aicardi-Goutières syndromes, Blau Syndrome |
| Loss-of-function mutations or haploinsufficiency of molecules controlling cell homeostasis | accumulation of intracellular stressors that stimulate intracellular sensor/PRR activation and the production of proinflammatory mediators | TRAPS, MKD |
| Loss-of-function mutations of negative regulators that downregulate proinflammatory responses | loss-of-function of a cytokine receptor antagonist or anti-inflammatory cytokine; failure to terminate the release of inflammatory mediators by inflammatory cells | DIRA, DITRA |
| Mutations that alter immune receptor signaling | Hyper-responsiveness to immune signals; increased signaling through receptors controlling innate immune cell function (the resulting diseases often have more complex phenotypes with overlapping features of autoinflammation, immunodeficiencies and autoimmunity) | PLAID/APLAID, LAID, Cherubism |
Table 4. Overview of the most common hereditary monogenic SAIDs. Abbreviations: AR: autosomal recessive; AD: autosomal dominant.
| Disease | OMIM | Affected Gene | location | reported INFEVERS variants | Inheritance | Prevalence | Male/female ratio | Treatment | Mechanism | |
|---|---|---|---|---|---|---|---|---|---|---|
| FMF | #249100 | MEFV | 16pl3.3 | 365 | AR | 1:1 [75] |
|
Inflammasomopathy | ||
| NLRP3-AID |
FCAS
MWS NOMID |
#120100 #191900 #607115 |
NLRP3 | lq44 | 227 | AD | France 1:360000 [76] | 2:1 [77] 1:1 [77] 1:1 [77] |
|
Inflammasomopathy |
| MKD | #260920 | MVK | 12q24.11 | 227 | AR | Netherlands 5:1000000 [78] | 1:1 [79] |
|
Inflammasomopathy | |
| TRAPS | #142680 | TNFRS1A | 12pl3.31 | 163 | AD | 1:1000000 [68] | 3:2 [66] |
|
protein folding disorder |
Inflammasomopathies or IL-1 β-activation syndromes are characterised by an elevation of IL-1β due to inflammasome activation. Associated diseases are FMF, NLRP3-AID and Mevalonate kinase deficiency (MKD).
FMF is the most common hereditary autoinflammatory disease (MIM #249100). It is an autosomal recessive disease and mainly affects people living in the eastern Mediterranean basin, hence the name. In 1997, the International FMF consortium identified mutations in the MEFV gene on chromosome 16p as a cause for the disease [37]. MEFV encodes for Pyrin, an important component of inflammasomes that interacts with caspase-1 and other inflammasome components to regulate IL-1β production [24,38]. Even though the identification of MEFV as disease causing for FMF was in 1997, the role of Pyrin has been a subject of debate. Early studies with mice showed that Pyrin acts as an inhibitor of caspase-1 and the authors suggested an anti-inflammatory role for Pyrin [39]. Other studies demonstrated that Pyrin can assemble an inflammasome complex and act pro-inflammatory [40,41]. Later it could be shown that homozygous gain-of-function Pyrin mutations in mice result in Pyrin inflammasome activation and severe inflammatory phenotypes by generating both pyrin-deficient and knock-in mice with mutated human B30.2 domains [42]. In 2016, the mechanism of Pyrin inflammasome activation has been identified. It could be shown that the Pyrin inflammasome is regulated by RhoA-dependent phosphorylation. Phosphorylated Pyrin interacts with chaperone proteins 14-3-3, keeping Pyrin at an inactive state. Dysregulated interaction between 14-3-3 and Pyrin leads to an activated Pyrin inflammasome [43,44]. FMF is characterised by recurrent fever attacks, abdominal pain, chest pain, and arthritis [45]. The diagnosis of FMF often relies on the phenotypical Tel Hashomer [46] or Yalcinkaya-Ozen criteria [47] and can be supported by genetic analysis [48]. In 20% of patients showing a FMF phenotype, a second mutation of the MEFV gene cannot be found [49].
NLRP3-AID is a group of autosomal dominant diseases that is defined by a mutation in the NLRP3 (nucleotide-binding domain, leucine-rich repeat family, pyrin domain containing 3) gene located on chromosome 1q44 [50]. The disease group is also known as Cryopyrin associated periodic syndromes (CAPS) but it has been proposed to rename the group to NLRP3-AID, to reflect the name of the gene over its encoded protein [51]. The NLRP3-AID spectrum includes three formerly distinct diseases: familial cold autoinflammatory syndrome (FCAS; MIM #120100), Muckle-Wells syndrome (MWS; MIM #191900), and Neonatal-onset multisystem inflammatory disease (NOMID) or chronic infantile neurologic cutaneous and articular (CINCA) syndrome (MIM #607115). The three diseases differ in severity where FCAS is the milder form, NOMID/CINCA is on the severe side of the spectrum, and the MWS phenotype is moderate. Gain-of-function or single germline mutations in NLRP3 lead to a low binding affinity of cellular cyclic AMP (cAMP) [52], and a disability of CARD8 binding [53] to mutated NLRP3 protein, thus an increased IL-1β secretion. The NLRP3 inflammasome formation enables activation of caspase-1 that can cleave pro-IL-1β and pro-IL-18 to their biologically active forms. Thus, a dysregulation of the NLRP3 inflammasome leads to inflammation. The clinical manifestation of NLRP3-AID is characterised by chronic systemic and organ inflammation, and the systemic features include fatigue, headache, and influenza-like muscle aches. Organ inflammation effects the skin, muscoskeleton, eyes, ears and the central nervous system [54,55].
MKD is a rare autosomal recessive autoinflammatory disease caused by a loss-of-function mutation in the mevalonate kinase gene (MVK) on chromosome 12q24.11. The mevalonate pathway produces isoprenoids, which lead to prenylation of RhoA and further phosphorylate Pyrin to interact with 14-3-3 proteins. A dysregulated mevalonate pathway inhibits the RhoA prenylation and thus activating the Pyrin inflamma-some and increased secretion of IL-1β [44,56,57]. MKD has two associated diseases: Hyper-IgD (Immunoglobulin D) Syndrome (HIDS; MIM #260920) and Mevalonic Aciduria (MEVA; MIM #610377). The common symptoms are fever, gastrointestinal symptoms like abdominal pain and vomiting-diarrhoea, skin rashes, lymphadenopathy, hepatos-plenomegaly, arthralgia, myalgia and mucosal ulcers. As MEVA also has additional symptoms like dysmorphic features, growth retardation (pre- and postnatal), ocular, and neurological involvement, it is a more severe form of MKD than HIDS [58,59]. Contrary to the initial publication [60] and what the name implies, measurements of serum Immunoglobulin D (IgD) levels are not a reliable method to diagnose MKD [61–63].
The protein-folding disorder Tumour Necrosis Factor (TNF) Receptor - Associated periodic syndrome (TRAPS; MIM #142680) is an autosomal dominant autoinflammatory disease that was first described as Hibernian fever [64]. Missense mutations in the gene tumor necrosis factor receptor superfamily member 1A (TNFRS1A) located on chromosome 12 are associated with TRAPS affecting receptor folding and trafficking [65]. Most disease-causing mutations are located within exon 2 to 4. The missense substitutions result in a dysregulation of structurally important cysteine-cysteine disulfide bonds leading to misfolded receptors that accumulate in the cell cytoplasm. This further enhances NF-κB activation, reactive oxygen species (ROS) production, and impaired autophagy [66]. Clinical manifestations of the disease are not completely specific and can be strikingly different [67]. A survey of 158 TRAPS patients of the Eurofever/EUROTRAPS international registry (https://www.printo.it/eurofever/) reported that the most common symptoms are fever, limb pain, abdominal pain, and rash seen in more than 63% of the cohort, followed by periorbital oedema (20%) [68]. For TRAPS diagnosis relies on “suspicion” and genetic testing.
3.2. Multifactorial systemic autoinflammatory diseases
For multifactorial SAIDs, like SJIA, Behçet disease, and Periodic fever adenopathy pharyngitis (PFAPA), the pathogenesis is still unknown and requires extensive research to elucidate disease causing mechanisms. SJIA is a subgroup of juvenile idiopathic arthritis (JIA; MIM #604302) [69]. It is the most common rheumatic disease in children but the underlying etiological pathway still needs to be identified [70]. The diagnosis is based on elevated biological markers such as IL-18, S100A12, and MRP8/14 [71,72]. Clinically SJIA patients present with JIA typical arthritis and additional symptoms of systemic inflammation like periodic fever, pericarditis, peritonitis, lymphadenopathy, and organomegaly. One life-threatening complication in SJIA is macrophage activation syndrome (MAS) with a prevalence of about 10% and even higher subclinical prevalence [73].
4. Diagnosis
For defined monogenic autoinflammatory syndromes, genetic testing has become a standard and gene panel sequencing is available covering the majority of known genetic loci associated with those SAIDs [80,81]. Papa et al. designed a NGS diagnostic panel, including 41 genes related to SAIDs and additional genes reported in the INFEVERS database [82]. Touitou and Aksentijevich proposed three prerequisites for genetic testing: i) evidence for systemic inflammation (elevated C-reactive protein (CRP), erythrocyte sedimentation rate (ESR), and serum amyloid A (SAA)), ii) a likely monogenic disease, and iii) a plausible candidate causal gene. Diseases with multifactorial inheritance multiple factors, including environmental factors, have a cumulative effect on the disease. Therefore, genetic testing cannot identify multifactorial diseases [3]. The diagnosis of SAIDs often relies on suspicion, resulting in a diagnostic delay of up to 7.3 years [83].
For at least 40%–60% of patients with phenotypes typical for SAIDs, a distinct diagnosis cannot be met, leading to undefined or undifferentiated SAIDs (uSAIDs) [3,14,84]. In 2019, the Eurofever Project described the characteristics of 187 patients with uSAIDs, concluding a need for new classification criteria for monogenic SAIDs which should combine genetic and clinical variables, and the need for further research to provide insight into genotype-phenotype relation [85].
For some defined monogenic SAIDs, like FMF, a distinct diagnosis based on clinical criteria is available. The Tel Hashomer criteria are the most widely used criteria for the diagnosis of FMF in adult patients. It includes a set of ten criteria grouped into Major criteria, Minor criteria, and Supportive criteria. A diagnosis of FMF requires ≥ 1 major criterion; or ≥ 2 minor criteria, or 1 minor criterion plus ≥ 5 supportive criteria [46]. The Yalcinkaya-Ozen criteria enables a diagnosis of children with FMF. It includes the five criteria Fever, Abdominal pain, Chest pain, Arthritis, and Family history of FMF [47]. They do not necessarily require molecular confirmation. Another classification criterion set, based on the Eurofever/PRINTO projects was published by Gattorno et al., in 2019. The set combines genetic and clinical findings to classify NLRP3- AID, FMF, TRAPS, and MKD patients [86].
In many cases of multifactorial SAIDs clinical criteria are not available nor applicable. For ideal treatment and in order to prevent severe complications such as amyloidosis, destructive arthropathy, organ damage an early and accurate diagnosis is crucial. Differential diagnosis of SAIDs can be difficult due to the non-specific or overlapping symptoms and the lack of universally accepted screening protocols [3]. One characteristic feature of SAIDs are the recurrent fever attacks. Fever can also be caused by more prevalent conditions such as infectious diseases, congenital immune defects, or neoplasms. In patients with SAID, the fever is periodic, alternating between fever attacks and fever-free episodes. In contrast to chronic illnesses, SAIDs are not associated with a progressive deterioration of the patients. Therefore, it is essential to follow patients with recurrent fever attacks for at least six months before settling on the final diagnosis of autoinflammatory disease. Each individual syndrome has specific characteristics such as the type of skin rash and the areas affected by it or the duration and pattern of acute inflammatory episodes. Together they offer clues for the diagnosis and allow for differentiation [83].
5. Treatment
Treatment of patients with SAIDs is aimed at supressing the systemic inflammation. For some patients the use of corticosteroids during attacks can be an option for reducing inflammatory attacks. However, they often require higher doses as the disease progresses. Withdrawal of corticosteroids lead to frequent attack relapses or continuous symptoms [49]. Although the underlying mutations and causes for SAIDs vary widely, a common effect shared by almost all of them is the dysregulation of IL-1 signalling. Given its prominent role in innate immunity and inflammasome formation, it is a successful target for treatment. Treatments targeting IL-1 are e.g. Anakinra, Canakinumab, and Rilonacept [87]. Anakinra is a recombinant anti-IL-1 receptor antagonist that inhibits both IL-lα and IL-1β by binding to the IL-1 receptor. Canakinumab is a selective anti-IL-lβ monoclonal antibody. Rilonacept prevents the interaction of IL-lα, IL-1β, and IL-IRa with cell-surface receptors by acting as a soluble decoy receptor that binds to IL-1β [88]. Anakinra and Canakinumab are the most commonly used medications in the therapy of SAIDs. Since 1972 Colchicine has been the main therapeutic for FMF, where two-thirds of the patients respond positively, one-third are partial-responders, and 5–10% are non-responders. Colchicine exerts its anti-inflammatory effect by suppressing of pyrin oligomerization and interfering with neutrophil migration and adhesion [24]. For non-responders that do not show improvement while adapting the colchicine dose, other treatment options are added to the colchicine treatment. In 2019, El Hasbani and colleagues summarized trials and case studies using anti-IL-1 drugs (Anakinra, Canakinumab, and Rilo- nacept), anti-TNF drugs (Etanercept), anti-IL-6 drugs (Tocilizumab), and Janus kinase inhibitors (Tofacitinib). It was shown that those additional treatment options could be beneficial for colchicine non-responders [89]. TRAPS and MKD patients respond to Etanerecept, a recombinant human TNFR2-Fc fusion protein [90,91]. Treatment of SJIA is challenging, due to its heterogenous nature. IL-6 pathway inhibitor (Tocilizumab), IL-1 receptor antagonist (Anakinra), and anti-IL-1β monoclonal antibody (Canakinumab) have been reported to reduce disease activity [92,93].
For uSAIDs no specific drug is available. Currently the treatment consists of nonspecific immunosuppression with corticosteroids and other disease-modifying anti-rheumatic drugs [94].
6. Other factors driving SAIDs
The definition of immune related diseases recently departs from the strict separation between autoinflammatory and autoimmune diseases. The inflammatory spectrum also includes polygenic and multifactorial diseases, where the causes are not completely resolved. Recently, autophagy, epigenetics, microbiome dysregulations, and autoantibody signatures are under investigation as additional contributors to the pathogenesis of monogenic or multifactorial SAIDs.
The innate immune system acts as a first line of defense by sensing PAMPs and DAMPs. Autophagy is an intracellular degradation system that delivers cellular stressors like misfolded proteins, damaged organelles, or intracellular microorganisms into the lysosome. It is a crucial mechanism of the innate immune system to regulate inflammation. It could be shown that after stimulation by inducers, autophagy depleted macrophages accumulate damaged mitochondria and produce a high amount of ROS. Both ROS and mitochondrial DNA activate the NLRP3 inflammasome, leading to an excessive inflammation [95]. Pyrin, also known as TRIM20 recruits the autophagic machinery. It could be shown that FMF associated mutations in the MEFV gene alter the capacity to direct autophagy of inflammasome components, leading to increased IL-1β production [96].
Epigenetic changes that alter the expression of genes, like DNA methylation, microRNA (miRNA) expression, or Histone modifications can be efficiently analysed with microarray and NGS based approaches. In 20% of FMF patients, only one mutation in the MEFV gene is identified, leading to the suggestion of additional causes for disease development. In a study comparing methylation levels of the CpG island within the second exon of MEFV between 30 FMF patients and 21 healthy controls in peripheral leukocytes, a small but significant higher methylation level (76% versus 74%) in FMF patients was shown [97]. Another study analysed methylation levels of inflammasome related genes (IL1B, NLRC5, PYCARD, AIM2, and CASP1) in monocytes and monocyte-derived macrophages from NLRP3-AID patients compared to those of healthy controls. The methylation levels in untreated patients were lower than in healthy controls after stimulation with IL-1β. Patients treated with IL-1 drugs had levels similar to healthy controls [98].
miRNA expression is another epigenetic regulator analysed in SAIDs. miRNAs are ~21 nucleotides long, single stranded RNA molecules that regulate host gene expression by base-pair binding to messenger RNA (mRNA). It is known that they are involved in most types of inflammatory responses and contribute significantly to the magnitude of the response by impacting the development of inflammatory cell subsets and by establishing the level of immune cell function [99]. A review by Balci-Peynircioglu et al. summarizes miRNAs associated with FMF, TRAPS, NLRP3-AID, Behçet’s disease, and NOMID, with a strong emphasis on FMF specific miRNAs [100]. Studies demonstrated that during fever attacks in three FMF groups differing in MEFV mutation location (exon 10, exon 3, neither exon 3 nor exon 10), the level of 25 circulating miRNAs changed specifically for the respective group [101]. Another study showed a lower miR-204-3p expression in the serum of FMF patients during an attack inhibiting inflammatory cytokine release via the phosphoinositide 3-kinase γ pathway [102]. In FMF patients without an inflammatory attack miR-132, miR-146a, miR-15a, miR-16, miR-181a, miR-21, miR-223, miR-26a, and miR-34a were lower compared to patients during an active attack [103]. Other studies comparing the expression levels of 798 mature miRNAs in peripheral blood mononuclear cells (PBMCs) of FMF patients and healthy controls showed that miR-144-3p, miR-21-5p, miR-4454, and miR-451a were increased and miR-107, let-7d-5p, and miR-148b-3p were decreased [104]. Apart from miRNA expression analysis in FMF patients, it could be shown that the NLRP3 inflammasome activity is negatively controlled by miR-223 and the expression is associated with NLRP3-AID [105]. miR-223 is a posttranscriptional regulator of NLRP3 expression, and miR-223 deficient mice display increased immune infiltration by neutrophils and monocytes, hyperactivated NLRP3 and IL-1 β release. Upon delivery of miR-223 the inflammation, and cytokine balance could be restored [106]. Comparing TRAPS patients to controls also showed altered circulating miRNA levels, where miR-134, miR-17-5p, miR-498, miR-451a, miR-572, miR-92a-3p are downregulated, and miR-150-3p, miR-92a-3p, miR-22-3p, miR-30d-5p are upregulated [107].
The above-mentioned studies and discoveries show the importance of autophagy and epigenetic factors that could act as novel diagnostic and treatment targets.
Another important factor is the human microbiome that is acquired after birth and shaped by environmental factors and the host immune system. The composition of gut microbiome is important for health and disease and dysbiosis has a central role in the pathogenesis in patients with inflammatory bowel disease (IBD). As NLRs sense microbial activation signals (shown in Table 2), they are important in controlling the bacterial community in the intestine [108]. A study comparing the gut microbiome of FMF patients against healthy controls, showed a specific shift in composition of bacteria, where diseased patients had a reduced amount of bacterial diversity [109]. Further studies are required to investigate the influence of the microbiome in SAIDs.
Autoantibodies play a crucial role in early diagnosis and classification in autoimmune diseases [110]. SAIDs are known to lack high-titer autoantibodies [1] but as immune related diseases are placed on a spectrum between autoimmune and autoinflammatory diseases [15], like the multifactorial disease SJIA, the role of autoantibodies needs to be reconsidered. Systematic profiling using human proteome arrays presenting immobilized recombinantly expressed human proteins can be used like in cancer studies (e.g. [111,112]) to elucidate antibodyprofiles at an “immunomics”-level. Additionally, proteomics and metabolomics can be used to identify molecular-pathological layers in SAIDs in the near future by cooperative research.
7. Conclusion
Systemic autoinflammatory diseases (SAID) are a growing group of disorders caused by a dysregulated activation of the innate immune system. Most autoinflammatory diseases are related to the activation of the Interleukin-1 pathway. The pathogenesis of these conditions is often driven by mutations in genes encoding proteins that are involved in the assembly of inflammasomes [24]. Since the first discovery of mutations in MEFV as a cause for FMF in 1997, the number of identified diseases, genes, and mutations responsible for those diseases has increased, resulting from advances in technology (e.g. genome wide association studies). Even though FMF is the best described SAID, there are still several unanswered questions. Based on the NIH, 52 clinical trials for “autoinflammatory disease” are currently active (https://clinicaltrials.gov/) [113] in order to find new treatment options for these severe and often life threatening diseases.
Disease-causing mutations can be identified for only half of the patients with SAIDs and the other half is classified as undefined or undifferentiated SAIDs. Improving the clinical definitions and creating precise diagnostic criteria for autoinflammatory disorders is important in order to better understand the differences between diseases and for more efficient therapies [114]. The implementation of a disease continuum model combining the adaptive and the innate immune system with autoinflammatory and autoimmune diseases as described in McGonagle et al. [15] shows the complexity of the diseases and the importance of new methods and international collaboration to elucidate molecular changes and causative factors in SAIDs. The timeline from onset to diagnosis takes up to 7.3 years [83] highlighting the indisputable need to identify new treatment and diagnostic targets. These novel targets could be identified using methods to detect differences in autophagy, DNA methylation, microRNA expression, microbiome composition, autoantibody signatures, and others.
Due to global transition, the number of SAIDs that have a high prevalence in specific countries could increases in other countries, making clinical awareness in diagnosis crucial. This awareness can be achieved with the work of scientific societies or networks such as listed below, and by conducting active research in this area.
European Scientific Societies and Networks:
ERN RITA: European Reference Network: Immunodeficiency, Autoinflammatory and Autoimmune Diseases (http://rita.ern-net.eu/)
PRES: Paediatric Rheumatology European Association (https://www.pres.eu/)
PRINTO: Paediatric Rheumatology International Trials Organisation (https://www.printo.it/)
ESID: European Society for Immunodeficiencies (https://esid.org/)
ISSAID: International Society of Systemic Auto-Inflammatory Diseases (https://issaid.umai-montpellier.fr/)
Supplementary Material
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jaut.2020.102421.
Funding
This study is part of the INSAID project (E-rare-3 program [grant number 9003037603]) and funded by the Austrian Science Fund [I 2742 International Projects].
Footnotes
Author contribution
J.K. wrote the manuscript with the help of S.S. The manuscript was critically reviewed by A.W.
References
- [1].McDermott MF, Aksentijevich I, Galon J, McDermott EM, Ogunkolade BW, Centola M, Mansfield E, Gadina M, Karenko L, Pettersson T, McCarthy J, et al. Germline mutations in the extracellular domains of the 55 kDa TNF receptor, TNFR1, define a family of dominantly inherited autoinflammatory syndromes. Cell. 1999;97:133–144. doi: 10.1016/S0092-8674(00)80721-7. [DOI] [PubMed] [Google Scholar]
- [2].Rowczenio D, Shinar Y, Ceccherini I, Sheils K, Gijn MV, Patton SJ, Touitou I. Current practices for the genetic diagnosis of autoinflammatory diseases: results of a European Molecular Genetics Quality Network Survey. Eur J Hum Genet. 2019:1–7. doi: 10.1038/s41431-019-0439-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Touitou I, Aksentijevich I. Genetic approach to the diagnosis of autoin-flammatory diseases. In: Hashkes PJ, Laxer RM, Simon A, editors. Textb Autoinflammation. Springer International Publishing, Cham; 2019. pp. 225–237. [DOI] [Google Scholar]
- [4].Jesus AA, Goldbach-Mansky R. IL-1 blockade in autoinflammatory syndromes. Annu Rev Med. 2014;65:223–244. doi: 10.1146/annurev-med-061512-150641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Chen K, Kolls JK. Interluekin-17A (IL17A) Gene. 2017;614:8–14. doi: 10.1016/j.gene.2017.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Lai Y, Dong C. Therapeutic antibodies that target inflammatory cytokines in autoimmune diseases. Int Immunol. 2016;28:181–188. doi: 10.1093/intimm/dxv063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Dinarello CA, Simon A, van der Meer JWM. Treating inflammation by blocking interleukin-1 in a broad spectrum of diseases. Nat Rev Drug Discov. 2012;11:633–652. doi: 10.1038/nrd3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Li P, Zheng Y, Chen X. Drugs for autoimmune inflammatory diseases: from small molecule compounds to anti-TNF biologics. Front Pharmacol. 2017;8 doi: 10.3389/fphar.2017.00460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Adamichou C, Georgakis S, Bertsias G. Cytokine targets in lupus nephritis: current and future prospects. Clin Immunol. 2019;206:42–52. doi: 10.1016/j.clim.2018.08.013. [DOI] [PubMed] [Google Scholar]
- [10].Hausmann JS. Targeting cytokines to treat autoinflammatory diseases. Clin Immunol. 2019;206:23–32. doi: 10.1016/j.clim.2018.10.016. [DOI] [PubMed] [Google Scholar]
- [11].Wang L, Wang F-S, Gershwin ME. Human autoimmune diseases: a comprehensive update. J Intern Med. 2015;278:369–395. doi: 10.1111/joim.12395. [DOI] [PubMed] [Google Scholar]
- [12].Navegantes KC, de Souza Gomes R, Pereira PAT, Czaikoski PG, Azevedo CHM, Monteiro MC. Immune modulation of some autoimmune diseases: the critical role of macrophages and neutrophils in the innate and adaptive immunity. J Transl Med. 2017;15:36. doi: 10.1186/s12967-017-1141-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Waldner H. The role of innate immune responses in autoimmune disease development. Autoimmun Rev. 2009;8:400–404. doi: 10.1016/JautRev2008.12.019. [DOI] [PubMed] [Google Scholar]
- [14].Doria A, Zen M, Bettio S, Gatto M, Bassi N, Nalotto L, Ghirardello A, Iaccarino L, Punzi L. Autoinflammation and autoimmunity: bridging the divide. Autoimmun Rev. 2012;12:22–30. doi: 10.1016/JautRev2012.07.018. [DOI] [PubMed] [Google Scholar]
- [15].McGonagle D, McDermott MF. A proposed classification of the immunological diseases. PLoS Med. 2006;3:e297. doi: 10.1371/journal.pmed.0030297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Gurung P, Kanneganti T-D. Autoinflammatory skin disorders: the inflammasome in focus. Trends Mol Med. 2016;22:545–564. doi: 10.1016/J.molmed.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Saavedra V, Moghaddas F, Latz E, Masters SL. Pattern recognition receptors in autoinflammation. In: Hashkes PJ, Laxer RM, Simon A, editors. Textb Autoinflammation. Springer International Publishing, Cham; 2019. pp. 61–87. [DOI] [Google Scholar]
- [18].Schnappauf O, Chae JJ, Kastner DL, Aksentijevich I. The pyrin inflammasome in health and disease. Front Immunol. 2019;10 doi: 10.3389/fimmu.2019.01745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Awad F, Assrawi E, Louvrier C, Jumeau C, Georgin-Lavialle S, Grateau G, Amselem S, Giurgea I, Karabina S-A. Inflammasome biology, molecular pathology and therapeutic implications. Pharmacol Ther. 2018;187:133–149. doi: 10.1016/Jpharmthera.2018.02.011. [DOI] [PubMed] [Google Scholar]
- [20].Broderick L. Inflammasomes and autoinflammation. In: Hashkes PJ, Laxer RM, Simon A, editors. Textb Autoinflammation. Springer International Publishing, Cham; 2019. pp. 89–109. [DOI] [Google Scholar]
- [21].Manthiram K, Zhou Q, Aksentijevich I, Kastner DL. The monogenic autoin-flammatory diseases define new pathways in human innate immunity and inflammation. Nat Immunol. 2017;18:832–842. doi: 10.1038/ni.3777. [DOI] [PubMed] [Google Scholar]
- [22].Liu X, Lieberman J. A Mechanistic Understanding of Pyroptosis: the Fiery Death Triggered by Invasive Infection. first. Elsevier Inc; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Rathinam VAK, Fitzgerald KA. Inflammasome Complexes: Emerging Mechanisms and Effector Functions. 2016 doi: 10.1016/Jcell.2016.03.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].de Torre-Minguela C, Mesa del Castillo P, Pelegrín P. The NLRP3 and pyrin inflammasomes: implications in the pathophysiology of autoinflammatory diseases. Front Immunol. 2017;8 doi: 10.3389/fimmu.2017.00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Kostura MJ, Tocci MJ, Limjuco G, Chin J, Cameron P, Hillman AG, Chartrain NA, Schmidt JA. Identification of a monocyte specific pre-interleukin 1 beta convertase activity. Proc Natl Acad Sci Unit States Am. 1989;86:5227–5231. doi: 10.1073/pnas.86.14.5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Ghayur T, Banerjee S, Hugunin M, Butler D, Herzog L, Carter A, Quintal L, Sekut L, Talanian R, Paskind M, Wong W, et al. Caspase-1 processes IFN-γ-inducing factor and regulates LPS-induced IFN-γ production. Nature. 1997;386:619–623. doi: 10.1038/386619a0. [DOI] [PubMed] [Google Scholar]
- [27].Baroja-Mazo A, Martín-Sánchez F, Gomez AI, Martínez CM, Amores-Iniesta J, Compan V, Barberà-Cremades M, Yagüe J, Ruiz-Ortiz E, Antón J, Buján S, et al. The NLRP3 inflammasome is released as a particulate danger signal that amplifies the inflammatory response. Nat Immunol. 2014;15:738–748. doi: 10.1038/ni.2919. [DOI] [PubMed] [Google Scholar]
- [28].Dinarello CA. Interleukin-1 in the pathogenesis and treatment of inflammatory diseases. Blood. 2011;117:3720–3732. doi: 10.1182/blood-2010-07-273417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Weber A, Wasiliew P, Kracht M. Interleukin-1 (IL-1) pathway. Sci Signal. 2010;3 doi: 10.1126/scisignal.3105cm1cm1-cm1. [DOI] [PubMed] [Google Scholar]
- [30].Fink SL, Cookson BT. Apoptosis, pyroptosis, and necrosis: mechanistic description of dead and dying eukaryotic cells. Infect Immun. 2005;73:1907–1916. doi: 10.1128/IAI.73.4.1907-1916.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Gregersen PK, Olsson LM. Recent advances in the genetics of autoimmune disease. Annu Rev Immunol. 2009;27:363–391. doi: 10.1146/annuRevimmunol.021908.132653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10:241–251. doi: 10.1038/nrg2554. [DOI] [PubMed] [Google Scholar]
- [33].Lachmann HJ, Şengül B, Yavuzşen TU, Booth DR, Booth SE, Bybee A, Gallimore JR, Soytürk M, Akar S, Tunca M, Hawkins PN. Clinical and subclinical inflammation in patients with familial Mediterranean fever and in heterozygous carriers of MEFV mutations. Rheumatology. 2006;45:746–750. doi: 10.1093/rheumatology/kei279. [DOI] [PubMed] [Google Scholar]
- [34].Kastner DL, Aksentijevich I, Goldbach-Mansky R. Autoinflammatory disease reloaded: a clinical perspective. Cell. 2010;140:784–790. doi: 10.1016/Jcell.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Rodero MP, Crow YJ. Type I interferon-mediated monogenic autoinflammation: the type I interferonopathies, a conceptual overview. J Exp Med. 2016;213:2527–2538. doi: 10.1084/jem.20161596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Baines AC, Brodsky RA. Complementopathies. Blood Rev. 2017;31:213–223. doi: 10.1016/Jblre.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].The International Fmf Consortium, Ancient missense mutations in a new member of the RoRet gene family are likely to cause familial mediterranean fever. Cell. 1997;90:797–807. doi: 10.1016/S0092-8674(00)80539-5. [DOI] [PubMed] [Google Scholar]
- [38].Alghamdi M. Familial Mediterranean fever, review of the literature. Clin Rheumatol. 2017;36:1707–1713. doi: 10.1007/s10067-017-3715-5. [DOI] [PubMed] [Google Scholar]
- [39].Papin S, Cuenin S, Agostini L, Martinon F, Werner S, Beer H-D, Grütter C, Grütter M, Tschopp J. The SPRY domain of Pyrin, mutated in familial Mediterranean fever patients, interacts with inflammasome components and inhibits proIL-1 β processing. Cell Death Differ. 2007;14:1457–1466. doi: 10.1038/sJcdd.4402142. [DOI] [PubMed] [Google Scholar]
- [40].Chae JJ, Wood G, Masters SL, Richard K, Park G, Smith BJ, Kastner DL. The B30.2 domain of pyrin, the familial Mediterranean fever protein, interacts directly with caspase-1 to modulate IL-1β production. Proc Natl Acad Sci Unit States Am. 2006;103:9982–9987. doi: 10.1073/pnas.0602081103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Yu J-W, Wu J, Zhang Z, Datta P, Ibrahimi I, Taniguchi S, Sagara J, Fernandes-Alnemri T, Alnemri ES. Cryopyrin and pyrin activate caspase-1, but not NF-k B, via ASC oligomerization. Cell Death Differ. 2006;13:236–249. doi: 10.1038/sJcdd.4401734. [DOI] [PubMed] [Google Scholar]
- [42].Chae JJ, Cho Y-H, Lee G-S, Cheng J, Liu PP, Feigenbaum L, Katz SI, Kastner DL. Gain-of-Function pyrin mutations induce NLRP3 protein-independent interleukin-1β activation and severe autoinflammation in mice. Immunity. 2011;34:755–768. doi: 10.1016/Jimmuni.2011.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Gao W, Yang J, Liu W, Wang Y, Shao F. Site-specific phosphorylation and microtubule dynamics control Pyrin inflammasome activation. Proc Natl Acad Sci Unit States Am. 2016;113:E4857–E4866. doi: 10.1073/pnas.1601700113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Park YH, Wood G, Kastner DL, Chae JJ. Pyrin inflammasome activation and RhoA signaling in the autoinflammatory diseases FMF and HIDS. Nat Immunol. 2016;17:914–921. doi: 10.1038/ni.3457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Sönmezgöz E, Özer S, Gül A, Yılmaz R, Kasap T, Takcı Ş, Gümüşer R, Demir O. Clinical and demographic evaluation according to MEFV genes in patients with familial mediterranean fever. Biochem Genet. 2019;57:289–300. doi: 10.1007/s10528-018-9889-y. [DOI] [PubMed] [Google Scholar]
- [46].Livneh A, Langevitz P, Zemer D, Zaks N, Kees S, Lidar T, Migdal A, Padeh S, Pras M. Criteria for the diagnosis of familial mediterranean fever. Arthritis Rheum. 1997;40:1879–1885. doi: 10.1002/art.1780401023. [DOI] [PubMed] [Google Scholar]
- [47].Yalçınkaya F, Özen S, Özçakar ZB, Aktay N, Çakar N, Düzova A, Kasapçopur Ö, Elhan AH, Doğanay B, Ekim M, Kara N, et al. A new set of criteria for the diagnosis of familial Mediterranean fever in childhood. Rheumatology. 2009;48:395–398. doi: 10.1093/rheumatology/ken509. [DOI] [PubMed] [Google Scholar]
- [48].Berkun Y, Eisenstein EM. Diagnostic criteria of familial Mediterranean fever. Autoimmun Rev. 2014;13:388–390. doi: 10.1016/JautRev2014.01.045. [DOI] [PubMed] [Google Scholar]
- [49].Ozen S, Bilginer Y. A clinical guide to autoinflammatory diseases: familial Mediterranean fever and next-of-kin. Nat Rev Rheumatol. 2014;10:135–147. doi: 10.1038/nrrheum.2013.174. [DOI] [PubMed] [Google Scholar]
- [50].Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1β-processing inflammasome with increased activity in muckle-wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/S1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- [51].Ben-Chetrit E, Gattorno M, Gul A, Kastner DL, Lachmann HJ, Touitou I, Ruperto N. Consensus proposal for taxonomy and definition of the autoin-flammatory diseases (AIDs): a Delphi study. Ann Rheum Dis. 2018;77:1558–1565. doi: 10.1136/annrheumdis-2017-212515. [DOI] [PubMed] [Google Scholar]
- [52].Lee G-S, Subramanian N, Kim AI, Aksentijevich I, Goldbach-Mansky R, Sacks DB, Germain RN, Kastner DL, Chae JJ. The calcium-sensing receptor regulates the NLRP3 inflammasome through Ca 2+ and cAMP. Nature. 2012;492:123–127. doi: 10.1038/nature11588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ito S, Hara Y, Kubota T. CARD8 is a negative regulator for NLRP3 inflammasome, but mutant NLRP3 in cryopyrin-associated periodic syndromes escapes the restriction. Arthritis Res Ther. 2014;16:R52. doi: 10.1186/ar4483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Hoffman HM, Kuemmerle-Deschner JB, Goldbach-Mansky R. Cryopyrin-associated periodic syndromes (CAPS) In: Hashkes PJ, Laxer RM, Simon A, editors. Textb Autoinflammation. Springer International Publishing, Cham; 2019. pp. 347–365. [DOI] [Google Scholar]
- [55].Twilt M, Benseler SM. Cryopyrin-associated periodic syndromes (CAPS) In: Efthimiou P, editor. Auto-Inflamm Syndr Pathophysiol Diagn Manag. Springer International Publishing, Cham; 2019. pp. 95–109. [DOI] [Google Scholar]
- [56].Dang EV, Cyster JG. Loss of sterol metabolic homeostasis triggers inflammasomes — how and why. Curr Opin Immunol. 2019;56:1–9. doi: 10.1016/Jcoi.2018.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Akula MK, Shi M, Jiang Z, Foster CE, Miao D, Li AS, Zhang X, Gavin RM, Forde SD, Germain G, Carpenter S, et al. Control of the innate immune response by the mevalonate pathway. Nat Immunol. 2016;17:922–929. doi: 10.1038/ni.3487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].van der Burgh R, ter Haar NM, Boes ML, Frenkel J. Mevalonate kinase deficiency, a metabolic autoinflammatory disease. Clin Immunol. 2013;147:197–206. doi: 10.1016/Jclim.2012.09.011. [DOI] [PubMed] [Google Scholar]
- [59].Favier LA, Schulert GS. Mevalonate kinase deficiency: current perspectives. Appl Clin Genet. 2016 doi: 10.2147/TACG.S93933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Van Der Meer JosWM, Radl J, Meyer ChrisJLM, Vossen JaakM, Van Nieuwkoop JannyA, Lobatto S, Van Furth R. HYPERIMMUNOGLOBULINAEMIA D and periodic fever: a new syndrome. Lancet. 1984;323:1087–1090. doi: 10.1016/S0140-6736(84)92505-4. [DOI] [PubMed] [Google Scholar]
- [61].Ammouri W, Cuisset L, Rouaghe S, Rolland M-O, Delpech M, Grateau G, Ravet N. Diagnostic value of serum immunoglobulinaemia D level in patients with a clinical suspicion of hyper IgD syndrome. Rheumatology. 2007;46:1597–1600. doi: 10.1093/rheumatology/kem200. [DOI] [PubMed] [Google Scholar]
- [62].ter Haar NM, Jeyaratnam J, Lachmann HJ, Simon A, Brogan PA, Doglio M, Cattalini M, Anton J, Modesto C, Quartier P, Hoppenreijs E, et al. The phenotype and genotype of mevalonate kinase deficiency: a series of 114 cases from the eurofever registry. Arthritis Rheum. 2016;68:2795–2805. doi: 10.1002/art.39763. [DOI] [PubMed] [Google Scholar]
- [63].Simon A, Bijzet J, Voorbij HaM, Mantovani A, Meer JWMVD, Drenth JPH. Effect of inflammatory attacks in the classical type hyper-IgD syndrome on immunoglobulin D, cholesterol and parameters of the acute phase response. J Intern Med. 2004;256:247–253. doi: 10.1111/J1365-2796.2004.01359.x. [DOI] [PubMed] [Google Scholar]
- [64].Williamson LM, Hull D, Mehta R, Reeves WG, Robinson BH, Toghill PJ. Familial hibernian fever. Q J Med. 1982;51:469–480. [PubMed] [Google Scholar]
- [65].Simon A, Park H, Maddipati R, Lobito AA, Bulua AC, Jackson AJ, Chae JJ, Ettinger R, de Koning HD, Cruz AC, et al. Concerted action of wild-type and mutant TNF receptors enhances inflammation in TNF receptor 1-associated periodic fever syndrome. Proc Natl Acad Sci Unit States Am. 2010;107:9801–9806. doi: 10.1073/pnas.0914118107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Hull KM, Drewe E, Aksentijevich I, Singh HK, Wong K, Mcdermott EM, Dean J, Powell RJ, Kastner DL. The TNF receptor-associated periodic syndrome (TRAPS): emerging concepts of an autoinflammatory disorder. Medicine (Baltim) 2002;81:349. doi: 10.1097/00005792-200209000-00002. [DOI] [PubMed] [Google Scholar]
- [67].Dodé C, André M, Bienvenu T, Hausfater P, Pêcheux C, Bienvenu J, Lecron J-C, Reinert P, Cattan D, Piette J-C, Szajnert M-F, et al. The enlarging clinical, genetic, and population spectrum of tumor necrosis factor receptor-associated periodic syndrome. Arthritis Rheum. 2002;46:2181–2188. doi: 10.1002/art.10429. [DOI] [PubMed] [Google Scholar]
- [68].Lachmann HJ, Papa R, Gerhold K, Obici L, Touitou I, Cantarini L, Frenkel J, Anton J, Kone-Paut I, Cattalini M, Bader-Meunier B, et al. The phenotype of TNF receptor-associated autoinflammatory syndrome (TRAPS) at presentation: a series of 158 cases from the Eurofever/EUROTRAPS international registry. Ann Rheum Dis. 2014;73:2160–2167. doi: 10.1136/annrheumdis-2013-204184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Petty RE, Southwood TR, Manners P, Baum J, Glass DN, Goldenberg J, He X, Maldonado-Cocco J, Orozco-Alcala J, Prieur A-M, Suarez-Almazor ME, et al. I.L. of A. for Rheumatology, International League of Associations for Rheumatology classification of juvenile idiopathic arthritis: second revision, Edmonton, 2001. J Rheumatol. 2004;31:390–392. [PubMed] [Google Scholar]
- [70].Ravelli A, Martini A. Juvenile idiopathic arthritis. Lancet. 2007;369:767–778. doi: 10.1016/S0140-6736(07)60363-8. [DOI] [PubMed] [Google Scholar]
- [71].Swart JF, de Roock S, Prakken BJ. Understanding inflammation in juvenile idiopathic arthritis: how immune biomarkers guide clinical strategies in the systemic onset subtype. Eur J Immunol. 2016;46:2068–2077. doi: 10.1002/eji.201546092. [DOI] [PubMed] [Google Scholar]
- [72].Gohar F, Orak B, Kallinich T, Jeske M, Lieber M, von Bernuth H, Giese A, Weissbarth-Riedel E, Haas J-P, Dressler F, Holzinger D, et al. Correlation of secretory activity of neutrophils with genotype in patients with familial mediterranean fever. Arthritis Rheum. 2016;68:3010–3022. doi: 10.1002/art.39784. [DOI] [PubMed] [Google Scholar]
- [73].Davì S, Minoia F, Pistorio A, Horne A, Consolaro A, Rosina S, Bovis F, Cimaz R, Gamir ML, Ilowite NT, Kone-Paut I, et al. Performance of current guidelines for diagnosis of macrophage activation syndrome complicating systemic juvenile idiopathic arthritis. Arthritis Rheum. 2014;66:2871–2880. doi: 10.1002/art.38769. [DOI] [PubMed] [Google Scholar]
- [74].Tunca M, Akar S, Onen F, Ozdogan H, Kasapcopur O, Yalcinkaya F, Tutar E, Ozen S, Topaloglu R, Yilmaz E, Arici M, et al. Familial Mediterranean fever (FMF) in Turkey: results of a nationwide multicenter study. Medicine (Baltim) 2005;84:1–11. doi: 10.1097/01.md.0000152370.84628.0c. [DOI] [PubMed] [Google Scholar]
- [75].Sayarlioglu M, Cefle A, Inanc M, Kamali S, Dalkilic E, Gul A, Ocal L, Aral O, Konice M. Characteristics of patients with adult-onset familial Mediterranean fever in Turkey: analysis of 401 cases. Int J Clin Pract. 2005;59:202–205. doi: 10.1111/J1742-1241.2004.00294.x. [DOI] [PubMed] [Google Scholar]
- [76].Cuisset L, Jeru I, Dumont B, Fabre A, Cochet E, Bozec JL, Delpech M, Amselem S, Touitouthe I, FC study Group Mutations in the autoinflammatory cryopyrin-associated periodic syndrome gene: epidemiological study and lessons from eight years of genetic analysis in France. Ann Rheum Dis. 2011;70:495–499. doi: 10.1136/ard.2010.138420. [DOI] [PubMed] [Google Scholar]
- [77].Tilson H, Primatesta P, Kim D, Rauer B, Hawkins PN, Hoffman HM, Kuemmerle-Deschner J, van der Poll T, Walker UA. Methodological challenges in monitoring new treatments for rare diseases: lessons from the cryopyrin-associated periodic syndrome registry. Orphanet J Rare Dis. 2013;8:139. doi: 10.1186/1750-1172-8-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Toplak N, Dolezalovà P, Constantin T, Sedivà A, Pašić S, Čižnar P, Wolska-Kuśnierz B, Harjaček M, Stefan M, Ruperto N, Gattorno M, et al. Eastern/ central European autoinflammatory collaborating group for the paediatric Rheumatology international trials organization (PRINTO) and eurofever project, periodic fever syndromes in eastern and central European countries: results of a pediatric multinational survey. Pediatr Rheumatol. 2010;8:29. doi: 10.1186/1546-0096-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Mulders-Manders CM, Simon A. Hyper-IgD syndrome/mevalonate kinase deficiency: what is new? Semin Immunopathol. 2015;37:371–376. doi: 10.1007/s00281-015-0492-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Hügle B, Hinze C, Lainka E, Fischer N, Haas J-P. Development of positive antinuclear antibodies and rheumatoid factor in systemic juvenile idiopathic arthritis points toward an autoimmune phenotype later in the disease course. Pediatr Rheumatol. 2014;12:28. doi: 10.1186/1546-0096-12-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Gijn MEV, Ceccherini I, Shinar Y, Carbo EC, Slofstra M, Arostegui JI, Sarrabay G, Rowczenio D, Omoyımnı E, Balci-Peynircioglu B, Hoffman HM, et al. New workflow for classification of genetic variants’ pathogenicity applied to hereditary recurrent fevers by the International Study Group for Systemic Autoinflammatory Diseases (INSAID) J Med Genet. 2018;55:530–537. doi: 10.1136/jmedgenet-2017-105216. [DOI] [PubMed] [Google Scholar]
- [82].Papa R, Rusmini M, Volpi S, Caorsi R, Picco P, Grossi A, Caroli F, Bovis F, Musso V, Obici L, Castana C, et al. Next generation sequencing panel in undifferentiated autoin-flammatory diseases identifies patients with colchicine-responder recurrent fevers. Rheumatology. 2019:kez376. doi: 10.1093/rheumatology/kez376. [DOI] [PubMed] [Google Scholar]
- [83].Toplak N, Frenkel J, Ozen S, Lachmann HJ, Woo P, Koné-Paut I, Benedetti FD, Neven B, Hofer M, Dolezalova P, Kümmerle-Deschner J, et al. The paediatric Rheumatology international trials organisation (PRINTO), an international registry on autoinflammatory diseases: the eurofever experience. Ann Rheum Dis. 2012;71:1177–1182. doi: 10.1136/annrheumdis-2011-200549. [DOI] [PubMed] [Google Scholar]
- [84].Lachmann HJ. Periodic fever syndromes, best pract. Res Clin Rheumatol. 2017;31:596–609. doi: 10.1016/Jberh.2017.12.001. [DOI] [PubMed] [Google Scholar]
- [85].Haar NMT, Eijkelboom C, Cantarini L, Papa R, Brogan PA, Kone-Paut I, Modesto C, Hofer M, Iagaru N, Fingerhutová S, Insalaco A, et al. Clinical characteristics and genetic analyses of 187 patients with undefined autoinflammatory diseases. Ann Rheum Dis. 2019;78:1405–1411. doi: 10.1136/annrheumdis-2018-214472. [DOI] [PubMed] [Google Scholar]
- [86].Gattorno M, Hofer M, Federici S, Vanoni F, Bovis F, Aksentijevich I, Anton J, Arostegui JI, Barron K, Ben-Cherit E, Brogan PA, et al. Classification criteria for autoinflammatory recurrent fevers. Ann Rheum Dis. 2019;78:1025–1032. doi: 10.1136/annrheumdis-2019-215048. [DOI] [PubMed] [Google Scholar]
- [87].Havnaer A, Han G. Autoinflammatory disorders: a review and update on pathogenesis and treatment. Am J Clin Dermatol. 2019;20:539–564. doi: 10.1007/s40257-019-00440-y. [DOI] [PubMed] [Google Scholar]
- [88].Mitroulis I, Skendros P, Ritis K. Targeting IL-1β in disease; the expanding role of NLRP3 inflammasome. Eur J Intern Med. 2010;21:157–163. doi: 10.1016/Jejim.2010.03.005. [DOI] [PubMed] [Google Scholar]
- [89].El Hasbani G, Jawad A, Uthman I. Update on the management of colchicine resistant familial mediterranean fever (FMF) Orphanet J Rare Dis. 2019;14:224. doi: 10.1186/s13023-019-1201-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Bulua AC, Mogul DB, Aksentijevich I, Singh H, He DY, Muenz LR, Ward MM, Yarboro CH, Kastner DL, Siegel RM, Hull KM. Efficacy of etanercept in the tumor necrosis factor receptor-associated periodic syndrome: a prospective, open-label, dose-escalation study. Arthritis Rheum. 2012;64:908–913. doi: 10.1002/art.33416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].ter Haar N, Lachmann H, Özen S, Woo P, Uziel Y, Modesto C, Koné-Paut I, Cantarini L, Insalaco A, Neven B, Hofer M, et al. P.R.I.T.O. (PRINTO) and the E. Projects”, Treatment of autoinflammatory diseases: results from the Eurofever Registry and a literature review. Ann Rheum Dis. 2013;72:678–685. doi: 10.1136/annrheumdis-2011-201268. [DOI] [PubMed] [Google Scholar]
- [92].Kearsley-Fleet L, Beresford MW, Davies R, De Cock D, Baildam E, Foster HE, Southwood TR, Thomson W, Hyrich KL. Short-term outcomes in patients with systemic juvenile idiopathic arthritis treated with either tocilizumab or anakinra. Rheumatology. 2019;58:94–102. doi: 10.1093/rheumatology/key262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Ruperto N, Brunner HI, Quartier P, Constantin T, Wulffraat N, Horneff G, Brik R, McCann L, Kasapcopur O, Rutkowska-Sak L, Schneider R, et al. Two randomized trials of canakinumab in systemic juvenile idiopathic arthritis. N Engl J Med. 2012;367:2396–2406. doi: 10.1056/NEJMoa1205099. [DOI] [PubMed] [Google Scholar]
- [94].de Jager W, Hoppenreijs EPAH, Wulffraat NM, Wedderburn LR, Kuis W, Prakken BJ. Blood and synovial fluid cytokine signatures in patients with juvenile idiopathic arthritis: a cross-sectional study. Ann Rheum Dis. 2007;66:589–598. doi: 10.1136/ard.2006.061853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Nakahira K, Haspel JA, Rathinam VAK, Lee S-J, Dolinay T, Lam HC, Englert JA, Rabinovitch M, Cernadas M, Kim HP, Fitzgerald KA, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011;12:222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Kimura T, Jain A, Choi SW, Mandell MA, Schroder K, Johansen T, Deretic V. TRIM-mediated precision autophagy targets cytoplasmic regulators of innate immunityTRIMs direct precision autophagy. J Cell Biol. 2015;210:973–989. doi: 10.1083/jcb.201503023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Kirectepe AK, Kasapcopur O, Arisoy N, Celikyapi Erdem G, Hatemi G, Ozdogan H, Tahir Turanli E. Analysis of MEFV exon methylation and expression patterns in familial Mediterranean fever. BMC Med Genet. 2011;12:105. doi: 10.1186/1471-2350-12-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Vento-Tormo R, Álvarez-Errico D, Garcia-Gomez A, Hernández-Rodríguez J, Buján S, Basagaña M, Méndez M, Yagüe J, Juan M, Aróstegui JI, Ballestar E. DNA demethylation of inflammasome-associated genes is enhanced in patients with cryopyrin-associated periodic syndromes. J Allergy Clin Immunol. 2017;139:202–211. doi: 10.1016/Jjaci.2016.05.016e6. [DOI] [PubMed] [Google Scholar]
- [99].O’Connell RM, Rao DS, Baltimore D. microRNA regulation of inflammatory responses. Annu Rev Immunol. 2012;30:295–312. doi: 10.1146/annurev-immunol-020711-075013. [DOI] [PubMed] [Google Scholar]
- [100].Balci-Peynircioglu B, Akkaya-Ulum YZ, Akbaba TH, Tavukcuoglu Z. Potential of miRNAs to predict and treat inflammation from the perspective of Familial Mediterranean Fever. Inflamm Res. 2019 doi: 10.1007/s00011-019-01272-6. [DOI] [PubMed] [Google Scholar]
- [101].Wada T, Toma T, Matsuda Y, Yachie A, Itami S, h Taguchi Y, Murakami Y. Microarray analysis of circulating microRNAs in familial Mediterranean fever, Mod. Rheumatology. 2017;27:1040–1046. doi: 10.1080/14397595.2017.1285845. [DOI] [PubMed] [Google Scholar]
- [102].Koga T, Migita K, Sato T, Sato S, Umeda M, Nonaka F, Fukui S, Kawashiri S, Iwamoto N, Ichinose K, Tamai M, et al. MicroRNA-204-3p inhibits lipopolysaccharide-induced cytokines in familial Mediterranean fever via the phosphoinositide 3-kinase γ pathway. Rheumatology. 2018;57:718–726. doi: 10.1093/rheumatology/kex451. [DOI] [PubMed] [Google Scholar]
- [103].Hortu HO, Karaca E, Sozeri B, Gulez N, Makay B, Gunduz C, Atik T, Tekin IM, Unsal SE, Cogulu O. Evaluation of the effects of miRNAs in familial Mediterranean fever. Clin Rheumatol. 2019;38:635–643. doi: 10.1007/s10067-017-3914-0. [DOI] [PubMed] [Google Scholar]
- [104].Amarilyo G, Pillar N, Ben-Zvi I, Weissglas-Volkov D, Zalcman J, Harel L, Livneh A, Shomron N. Analysis of microRNAs in familial Mediterranean fever. PloS One. 2018;13:e0197829. doi: 10.1371/journal.pone.0197829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Bauernfeind F, Rieger A, Schildberg FA, Knolle PA, Schmid-Burgk JL, Hornung V. NLRP3 inflammasome activity is negatively controlled by miR-223. J Immunol. 2012;189:4175–4181. doi: 10.4049/jimmunol.1201516. [DOI] [PubMed] [Google Scholar]
- [106].Neudecker V, Haneklaus M, Jensen O, Khailova L, Masterson JC, Tye H, Biette K, Jedlicka P, Brodsky KS, Gerich ME, Mack M, et al. Myeloid-derived miR-223 regulates intestinal inflammation via repression of the NLRP3 inflammasomemiR-223 control of NLRP3 activation during colitis. J Exp Med. 2017;214:1737–1752. doi: 10.1084/jem.20160462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Lucherini OM, Obici L, Ferracin M, Fulci V, McDermott MF, Merlini G, Muscari I, Magnotti F, Dickie LJ, Galeazzi M, Negrini M, et al. First report of circulating MicroRNAs in Tumour necrosis factor receptor-associated periodic syndrome (TRAPS) PloS One. 2013;8:e73443. doi: 10.1371/journal.pone.0073443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Levy M, Thaiss CA, Elinav E. Metagenomic cross-talk: the regulatory interplay between immunogenomics and the microbiome. Genome Med. 2015;7:120. doi: 10.1186/s13073-015-0249-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Khachatryan ZA, Ktsoyan ZA, Manukyan GP, Kelly D, Ghazaryan KA, Aminov RI. Predominant role of host genetics in controlling the composition of gut microbiota. PloS One. 2008;3:e3064. doi: 10.1371/journal.pone.0003064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Lleo A, Invernizzi P, Gao B, Podda M, Gershwin ME. Definition of human autoimmunity — autoantibodies versus autoimmune disease. Autoimmun Rev. 2010;9:A259–A266. doi: 10.1016/JautRev2009.12.002. [DOI] [PubMed] [Google Scholar]
- [111].Luna Coronell JA, Sergelen K, Hofer P, Gyurján I, Brezina S, Hettegger P, Leeb G, Mach K, Gsur A, Weinhäusel A. The immunome of colon cancer: functional in silico analysis of antigenic proteins deduced from IgG microarray profiling. Dev Reprod Biol. 2018;16:73–84. doi: 10.1016/Jgpb.2017.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Gyurján I, Rosskopf S, Coronell JAL, Muhr D, Singer C, Weinhäusel A. IgG based immunome analyses of breast cancer patients reveal underlying signaling pathways. Oncotarget. 2019;10:3491–3505. doi: 10.18632/oncotarget.26834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Home-ClinicalTrials.gov. [Accessed date: 4 November 2019]; n.d https://clinicaltrials.gov/
- [114].Shimizu M, Yokoyama T, Yamada K, Kaneda H, Wada H, Wada T, Toma T, Ohta K, Kasahara Y, Yachie A. Distinct cytokine profiles of systemic-onset juvenile idiopathic arthritis-associated macrophage activation syndrome with particular emphasis on the role of interleukin-18 in its pathogenesis. Rheumatology. 2010;49:1645–1653. doi: 10.1093/rheumatology/keq133. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.