Abstract
The nucleolus is the largest substructure in the nucleus and forms around the nucleolar organizer regions (NORs) that comprise hundreds of ribosomal RNA (rRNA) genes. Recent evidence highlights further functions of the nucleolus that go beyond ribosome biogenesis. Data indicate that the nucleolus acts as a compartment for the location and regulation of repressive genomic domains and, together with the nuclear lamina, represents the hub for the organization of the inactive heterochromatin. In this review, we discuss recent findings that have revealed how nucleolar structure and rRNA gene chromatin states are regulated during early mammalian development and their contribution in the higher-order spatial organization of the genome.
Keywords: Nucleolus, rRNA genes, genome architecture, heterochromatin, early development, oocyte
The multitasking nucleolus: from ribosome producer to organizer of genome architecture
The nucleolus is the largest subnuclear compartment of the cell that forms around regions of chromosomes containing stretches of tandem repetitive ribosomal RNA (rRNA) genes, known as nucleolar organizer regions (NORs). The nucleolus is responsible for the production of the ribosomes, a highly regulated process that is essential for growth and development. Ribosome biogenesis is initiated in the nucleolus by RNA Polymerase I (Pol I) that together with a dedicated set of basal transcription factors, such as TIF1A, the TBP-TAFI complex SL1 and the DNA architectural Upstream Binding Factor UBF [1, 2], transcribes hundreds of rRNA genes to generate 45S/47S pre-rRNA in mammalian cells (Figure 1A). This rRNA precursor is chemically modified and processed to form 28S, 18S and 5.8S rRNAs, which are then assembled with ribosomal proteins and 5S rRNA and exported from the nucleus to give rise to active ribosomes in the cytoplasm [3].
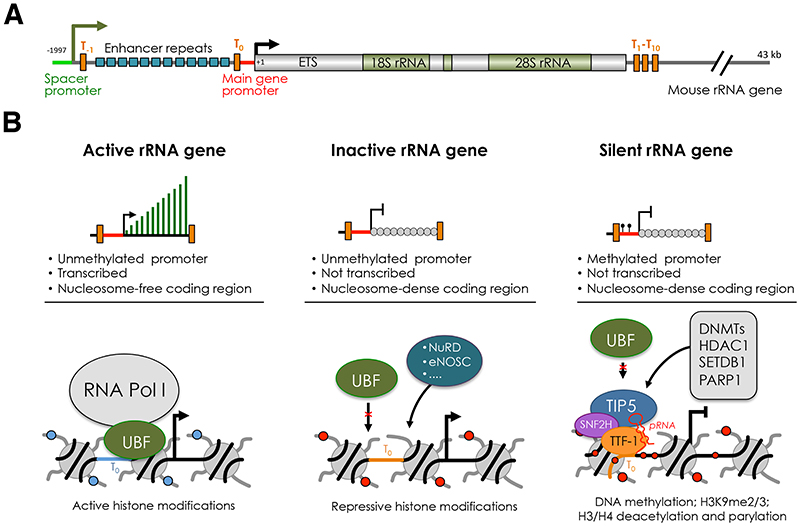
Figure 1. The three major classes of rRNA genes.
(A) Structural organization of mouse rRNA gene. The sites of transcription initiation of the 45S pre-rRNA from the main gene promoter and IGS-rRNA transcripts from the spacer promoter are indicated by arrows. Terminator elements downstream of the spacer promoter (T-1), upstream of the main gene promoter (T0) and downstream of the coding regions (T1-T10) are marked by orange bars. The repeats composing the enhancer (13 according to the sequence from Genbank accession number BK000964) are shown as blue bars. ETS, external transcribed spacer. (B) Description of active, inactive and silent rRNA genes based on transcription, chromatin and epigenetic features and factors regulating their state. The composition at the main rRNA gene promoter is described. The binding of UBF and NoRC (TIP5 and SNF2H) define active and silent rRNA genes. Inactive rRNA genes are non-transcribed repeats that lack promoter DNA methylation, are nucleosome-packed at the coding region, and are not bound by UBF or NoRC. The structure of inactive genes can be mediated by NuRD, eNOSC or other yet unknown regulators. Establishment of silent rRNA genes is described. NoRC is recruited to the promoter through the interaction with TTF-1 mediated by the lncRNA pRNA. Subsequently, NoRC recruits factors that establish epigenetic silencing, such DNA methyltransferase (DNMs) histone deacetylase (HDAC1), histone methyltransferases (SETDB1), and poly(ADP-Ribose) Polymerase 1 (PARP1).
Increasing evidence suggests that the function of nucleolus and rRNA genes go beyond ribosome biogenesis. One aspect that raised high interest in the last decade is the link of nucleolus and rRNA genes with the organization of genome architecture. Clustering of heterochromatin at nucleoli is a phenomenon known to occur in all cells. Furthermore, regions located close to nucleolus (nucleolar associated domains, NADs) have low gene densities, low transcriptional levels and repressive histone modifications [4–8]. Thus, the nucleolus, together with the nuclear lamina, can be considered the hub for the organization of the inactive chromatin in the cell [8–10].
In this review, we discuss recent findings that have revealed how nucleolar structure and rRNA gene chromatin states are regulated during gametogenesis and early mammalian development and their contribution in the higher-order spatial organization of the genome.
Nucleolus and rRNA genes in somatic cells
In somatic cells, the nucleolus is a membraneless compartment with a peculiar tripartite architecture: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC). These nucleolar subcompartments represent distinct, coexisting liquid phases [11]. Transcription of rRNA genes is thought to occur at the boundary between FC and DFC whereas rRNA processing occurs in the DFC region and pre-ribosomal subunit assembly takes place in the GC region [12, 13]. Transcription of rRNA genes in the nucleoli produces the overwhelming majority of RNAs in the cell. This large rRNA synthesis is mainly due to the high transcription rate (approx. one Pol I every 130 nt [14–16]) and the presence of multiple copies of rRNA genes (in mammalian cells about 200 per haploid genome). However, in somatic cells, even under high cell proliferative conditions, not all rRNA genes within a cell are competent for transcription [17].
rRNA genes can be subdivided in three major classes according to transcription and chromatin states: silent, inactive and active genes (Figure 1B). In mammalian cells, DNA methylation at the promoter distinguishes silent rRNA genes from the rest of the repeats [18]. Promoter methylation abrogates the formation of Pol I pre-initiation complex by impairing the binding of UBF [18]. Silent rRNA genes display constitutive heterochromatic features and associate with repressive histone marks [19, 20]. The responsible factor for the establishment of silent genes is the nucleolar remodelling complex NoRC that is composed of TIP5 (TTF-I interacting protein 5) and SNF2H, a member of the ISWI subfamily and catalytic subunit of several chromatin-remodelling complexes [19–21] (Figure 1B). NoRC is recruited to rRNA genes through its association with the long non-coding (lnc)RNA pRNA that mediates the interaction with TTF-1, which binds the terminator DNA element T0 located in the rRNA gene promoter [22–24]. NoRC establishes silent rRNA genes through the recruitment of DNA methyltransferases (DNMTs) and histone modifier enzymes, such as HDAC1, thereby establishing heterochromatic and repressive structures [19, 20, 25]. rRNA genes that lack the promoter methylation can be further classified in active and inactive repeats. Active rRNA genes associate with UBF and active histone modifications, and are nucleosome-free in the coding region (Figure 1B). In contrast, inactive genes do not interact with UBF and display nucleosome-packed chromatin as in the case of silent rRNA genes. UBF is the key factor for the establishment of active rRNA genes [26, 27]. Depletion of UBF switches genes from active to inactive states. This process is reversible as re-expression of UBF re-establishes active rRNA copies [26].
The structure and composition of nucleoli and rRNA genes observed in somatic cells undergo drastic changes in oogenesis and early development, a process thought to play important role in cell reprogramming.
Nucleolus structures and rRNA genes in gametogenesis
The process of gametogenesis, fertilization and pre-implantation developments (Box 1) are characterized by tightly coordinated molecular events that are crucial to ensure a successful embryonic development. These changes include histone and DNA modifications, higher-order chromatin organization, genome compartmentalization, and nucleolus structure.
Text box 1. Preimplantation developmental stages.
A new live multicellular organism develops from a single zygote that was created by the fusion of the two specialized and highly differentiated cells, the oocyte and the sperm, a process termed fertilization. Early mammalian embryo develops through several recognizable cellular events known as the preimplantation embryogenesis. The preimplantation development includes the period from zygote formation (1-cell embryo) until invasion of the uterine epithelium by the blastocyst [83]. After fertilization, 1-cell embryo (zygote) is formed and undergoes three rounds of cleavage division resulting in 2-cell, 4-cell, and 8-cell stage embryo, respectively, together known as blastomeres. The following formation of the morula represents the earliest point where blastomeres have differential spatial positioning. Over subsequent asymmetric cleavage divisions, embryo develops further into the blastocyst, containing two specific cell types: trophectoderm (TE) and inner cell mass (ICM) cells. At the late blastocyst stage, ICM consolidates to establish the epiblast (EPI) and the primitive endoderm (PE) lineages. At this point the epiblast cells enter the developmental “ground state,” the origin of all future embryonic lineages [84]. In the final step of preimplantation development, blastocyst is ready to hatch into to the uterus and further development takes place.
In mammals, oogenesis and spermatogenesis occur at very different times during development and achieve different endpoints. In males, meiosis is initiated at the onset of puberty [28]. In many metazoans, male germ cells undergo an extensive chromatin remodeling process in their final differentiation into sperm, during which genomic DNA becomes transcriptionally silent, highly methylated and packaged with protamines into a highly condensed configuration [29].
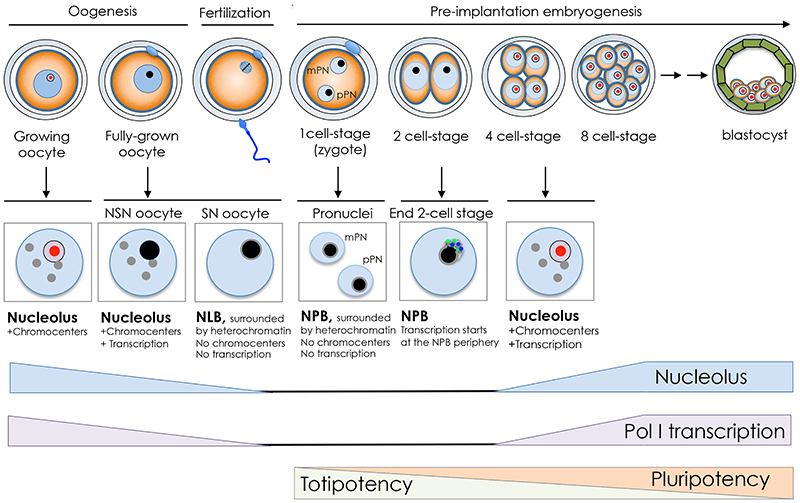
Oogenesis is initiated in the fetus well before birth, forming a finite number of gametes that are used periodically over a defined reproductive lifetime. The mammalian oocyte nucleus or germinal vesicle (GV) exhibits a unique chromatin configuration that is subject to dynamic modifications during oogenesis (Figure 2). Upon entrance into the growth phase, oocytes synthesize large amounts of material, including ribosomes, and markedly enlarge in diameter. When oocytes reach their full size, they acquire the competence to disassemble their nuclei and undergo meiotic maturation. However, not all oocytes have a full developmental potential when fertilized [30]. In the very final growth phase, oocytes acquire two main types of nuclear organization: the SN-type (Surrounded Nucleolus) and the NSN-type (Non Surrounded Nucleolus). SN type is transcriptionally inactive whereas NSN type is transcriptionally active. Although both types of oocyte are able to resume meiosis and undergo fertilization, they do not have the same developmental competence [31–33]. Following fertilization, NSN-derived embryos stop to develop at the 2-cell stage whereas SN-derived embryos pursue development until the blastocyst stage and reach full-term [31]. Remarkably, in the very final phases of the growth period, SN oocytes gradually shut down both RNA Pol I and II activities [34] and the tripartite structure of nucleoli undergoes gradual changes to transform into atypical nucleoli termed nucleolus like body (NLB) (Figure 2). At the morphological level, NLB consists of tightly packed fibers of 6-10 nm [35]. Importantly, SN oocytes display dense chromatin ring around the NLB whereas the chromatin in NSN is uncondensed. Pericentromeric heterochromatin in NSN-type oocytes is aggregated within chromocenters whereas in the SN oocyte it forms a discontinuous ring around the NLB [36]. In the following phase of oogenesis, the full-size grown oocyte undergoes meiotic maturation, with the breakdown of the nuclear envelope and consequent dispersion of NLBs into the cytoplasm [37]. Although the majority of nuclear material is retained by the eggs, at MII phase transcripts of several nucleolar proteins, such as UBF and Fibrillarin, are largely degraded and decline even further after fertilization, reaching the lowest levels at the 2-cell stage, when genome activation (EGA) occurs in the mouse [38]. The appearance of paternal transcripts at this stage indicates that the maternal transcripts are eliminated and the synthesis of these nucleolar factors is of embryo origin.
Figure 2. The nucleolar cycle in the germ line and during early development.
Illustration of mouse oogenesis and the early development timing. The nucleoli of growing oocytes in mammals are composed of fibrillar centers (red), dense fibrillar components, and granular components (purple). At the end of the growth phase, the nucleolus is transformed into nucleolus like body (NLB, black) that lacks the tripartite structure. Oocytes acquire two main types of nuclear organization: the SN-type (Surrounded Nucleolus) and the NSN-type (Non Surrounded Nucleolus). SN type is transcriptionally inactive and the NLB is surrounded by heterochromatin in a ring-like shape (grey lane). NSN type is transcriptionally active and display aggregation of heterochromatin in chromocenters (grey circles). During oocyte maturation, the nucleoli (NLBs) disappear, and upon fertilization reappear in the pronuclei of the zygote in the form of nucleolus precursor bodies (NPB, black). At the end of the 2-cell stage, transcription is initiated at the surface of NPB where UBF (green) and NoPP140 (blue) localize. From 4-cell stage, the NPB gradually transforms into somatic nucleoli that display the tripartite structure. Timing of rRNA gene transcription, nucleolus and the transition from totipotency to pluripotency are shown.
Nucleolus and rRNA genes during early embryo development
Fertilization occurs when the two highly differentiated cells, the oocyte and the sperm, fuse to form the zygote. As both parental genomes are transcriptionally silent, the initial development of the embryo exclusively depends on maternally inherited RNAs and proteins [39]. Embryonic genome activation (EGA) is established in two phases: a “minor activation”, which occurs at the 1-cell stage in most species [40], followed by a second “major activation” that in mouse takes place during the 2-cell stage [41]. rRNA gene transcription starts only at the end of the 2-cell stage and is required for further embryonic development. Embryos lacking RNA polymerase 1-2 (Rpo1-2) or UBF arrest development before morula stage [42, 43]. Furthermore, pharmacological inhibition of rRNA gene transcription using CX-5461 [44] induced developmental delay and arrest at the 4-cell stage [45].
Upon fertilization, the NLBs that disappeared during oocyte meiotic maturation reappear in both maternal and paternal pronuclei of zygotes in the form of nucleolus precursor bodies (NPBs), which persist throughout the first four cell cycles [46] (Figure 2). Later in development, during the transition from 16-cell to morula stages, NPBs gradually disappear and are replaced by somatic-type nucleoli with classic tripartite structure. In contrast to NLBs, NPBs are significantly impoverished for RNA and detectable amounts of RNA appear on the NPB surface only after resumption of rRNA gene transcription [47]. Accordingly, during the late two-cell and four-cell stages, UBF and Nopp140, two nucleolar proteins that are associated with the FC and DFC compartments of the nucleolus, localize around the periphery of the NPBs [45]. These results also suggest that active rRNA genes, which are bound by UBF [18, 19], are formed at the outer surface of the NPB. Starting at the 8-cell stage, NPBs diminish in size, disappearing by the late 16-cell stage, with the emergence of intermingled fibrillar and granular compartments and UBF positioning in the centre of the former NPB [45]. Since nucleoli and activation of rRNA genes occurs at the periphery of NPBs, it has been suggested that NPBs are the building block of the future somatic nucleoli. Data also suggest that the re-establishment of nucleoli during early embryo development is tightly linked to transcription. Inactivation of Pol I with the inhibitor CX-5461 led to a reorganization of Nopp140 and UBF, which formed nucleolar caps at both the two- and four-cell stages [45]. These caps correspond to those observed in SN oocytes and in the interphase cells treated with inhibitors of Pol I transcription. Similar structures were also observed in UBF-null embryos, where NPBs were also no longer apparent or strongly perturbed [42].
Increasing evidence suggests that the role of NPB might go beyond the simple transmission of nucleolar components from female germ cells to embryo and could participate in the remodelling of the genome during embryo development. After fertilization, the oocyte and sperm genomes undergo major structural reorganization to ensure the establishment of totipotency and the transition to pluripotency state and initial differentiations [48–51]. Genome reorganization includes the erasure of parental DNA methylation and de novo formation of heterochromatin, which are thought to be essential to ensure correct embryo development [48–50]. Remodelling is particularly evident at the paternal chromatin that is subject to active DNA demethylation and nearly genome-wide replacement of protamines by histones variant H3.3 [52]. Centromeres also undergo drastic structural changes from the 2-cell to the 4-cell stages. Centromere is organized around minor satellite repeats and flanked by major satellite repeats that constitute the pericentromeric heterochromatin, whose structure is essential for chromosome segregation [53]. In mouse interphase somatic nuclei, pericentromeric and centromeric sequences of several chromosomes cluster together, forming chromocenters. In contrast, in the zygote, centric and pericentromeric heterochromatin surrounds most of NPBs in a ring-like shape (Fig. 2). The formation of pericentromeric rings follows asymmetric parental dynamics. In the maternal pronucleus, the pericentromeres form a ring on the surface of the NPB very early, already from a few hours after pronuclear formation, whereas the positioning of the pericentromeric repeats around the male pronucleus occurs at a later time point [54]. The localization of pericentromeric repeats around NPBs occurs prior to the acquisition of their embryonic heterochromatin signature H3K27me3, suggesting that their spatial configuration is required for heterochromatic silencing [55]. Accordingly, tethering of the pericentromeric repeats from NPBs to the nuclear envelop caused defective heterochromatic silencing, impairment of chromocenters formation and developmental arrest at 2 and 8-cell stage [55]. Interestingly, rRNA genes were found positioned only around the periphery of NPBs surrounded by pericentric heterochromatin, indicating that rRNA repeats are not automatically associated with NPBs [54]. After the zygotic stage, the embryonic genome undergoes further structural and functional changes. At the beginning of the second cell cycle, major satellites still associate with the NPBs, forming thick partial rims as in zygotes [54]. At late 2-cell stage, concomitantly with the “major phase” of EGA, there is a burst in de novo major satellite transcription that rapidly decreases, becoming almost undetectable at 8-cell stage [56]. Pericentric transcripts are required for the reorganization of heterochromatin and further development [57]. Concomitant with this wave of transcription, pericentromeric heterochromatin associates with NPBs forming spherical patches, which are indicative of chromatin compaction. At the 4-cell stage, centric and pericentric heterochromatin formed structures that resemble classical chromocenters. Finally, at the blastocyst stage, the overall nuclear organization become very similar to that of somatic cell nuclei in terms of nucleolus structure and chromocenter organization [54, 58].
Further evidence for a role of NPBs in the organization of heterochromatin came from studies using enucleolation, a method that allows microsurgical removal of NLBs and NPBs from either oocytes or zygotes [59, 60]. Enucleolated oocytes are able to re-initiate meiosis and reach metaphase II at the same rates as control oocytes. They can also be fertilized and the resultant embryos form essentially normal pronuclei, which, however, do not contain NPBs [59], indicating that NLBs in the oocytes are required for the formation of NPBs in the zygote. Importantly, embryos generated with enucleolated oocytes failed to pass the first few cleavages and the replacement of nucleoli in oocytes with nucleoli from embryonic stem cells had a detrimental effect in embryo development, suggesting that NPBs of maternal origin are absolutely crucial for embryonic development. Remarkably, at 2-cell stage, embryos derived from enucleolated eggs show similar rRNA production rates and pre-rRNA processing compared to control embryos, indicating that NPBs are not necessary for pre-rRNA synthesis in embryos [38]. The presence of NPBs in embryos appears to be critical in a defined time window, approximately 8-10-h post-fertilization. The removal of NPBs 10 h after the sperm injection did not affect embryo development and new nucleoli could form after several cell divisions, indicating that the nucleoli can originate from de novo synthesized materials [61]. Data suggest that the developmental defects observed in embryos fertilized with enucleolated oocytes might probably be due to problems with chromosome segregation, mislocalization of centromeric proteins, abnormal first embryonic S-phase progression, and replication stress [38, 62]. Consistent with defects linked to genome stability, during the first embryonic cell cycle embryos lacking NPBs showed a significant reduction in major and minor satellite DNA and a decrease in satellite transcripts by more than half [38]. Interestingly, zygotes generated from enucleolated oocytes show alterations in higher chromatin organization as evident by the appearance of heterochromatin in the form of scattered dots [63]. Alterations in heterochromatin detected through DAPI staining were also reported in UBF-null embryos that lack NPBs [42]. Studies indicated an important role of nucleoplasmin 2 (NPM2) in the function of NLB and NPB linked to genome organization in early embryo development [64]. In the absence of NPM2, oocytes and embryos lack NLB and NPB and show defects in heterochromatin structures. Consistent with a role of NPM2 in nucleolus and heterochromatin, a recent study showed that the expression of NPM2 in Npm2-null oocytes sufficed to reconstitute nucleolar structures in enucleolated embryos, and rescued their first mitotic division and full-term development [62].
Taken together, these results support the idea that the nucleolus during early embryo development serves not only for the re-establishment of active somatic nucleoli and ribosome biogenesis but also it might provide a platform for the establishment of heterochromatic structures that are necessary for proper chromosome segregation and further development.
Nucleolus and rRNA genes in embryonic stem cells
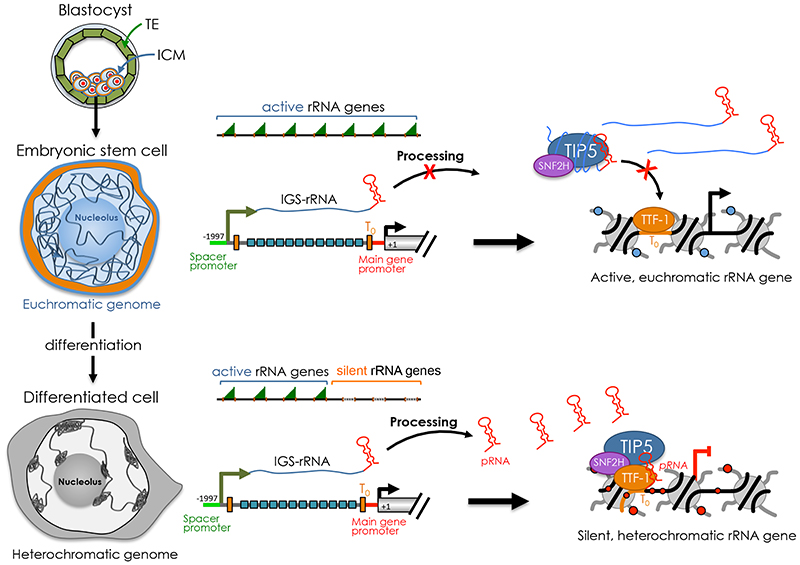
In preimplantation embryos, the parental genomes reduce DNA methylation on a global scale with the exception of parental imprints [51, 65, 66]. The loss of global DNA methylation levels continues until the blastocyst stage, where the inner cell mass (ICM) is first specified, through downregulation of the DNA methylation machinery [66–69]. The pluripotency state in ICM can be immortalized in vitro through culturing of embryonic stem cells (ESCs). Studies in ESCs revealed a chromatin state that is generally less condensed and largely devoid of compact heterochromatin blocks compared to lineage-committed cells. It is considered that this open and transcriptionally permissive state reflects the plasticity of ESC genome in order to enter any distinct transcriptional programs for lineage specification [70, 71]. The active state of ESC genome holds also true in the case of rRNA genes, which are all active in ESCs due to the lack of DNA methylation and repressive histone marks such as H3K9me2 and H3K9me3 [22, 72]. Accordingly, ESCs contain large nucleoli, indicating high ribosome biogenesis activity [73]. The formation of silent rRNA genes in ESCs is prevented through the impairment of NoRC recruitment, a process that implicates non-coding RNA-mediated mechanisms [22, 23] (Figure 3). In ESCs, DHX9-mediated processing of IGS-rRNA is impaired with consequent reduction of mature pRNA levels. The association of TIP5 with the unprocessed IGS-rRNA prevents the interaction with TTF-1 that is bound to the promoter, thereby abrogating rRNA gene silencing. Acquisition of silent rRNA genes occurs only upon differentiation and it co-occurs with the decrease of rRNA synthesis, indicating that the reduction in nucleolar transcription is an early event during the differentiation [22, 74, 75]. Accordingly, IGS-rRNA processing is reactivated only upon differentiation, thereby initiating pRNA-NoRC mediated formation of silent rRNA copies [22]. Evidence indicates that the regulation of rRNA gene transcription and chromatin state in ESCs is a critical aspect of cell fate determination. Impairment of IGS-rRNA processing and TIP5-recruitment to rRNA genes blocks the exit from pluripotency state [23]. Furthermore, constitutive expression of fibrillarin, which is indispensable for ribosome biogenesis, prolongs pluripotency of ESCs whereas inhibition of rRNA gene transcription either by fibrillarin knockdown or treatment with the Pol I inhibitor Actinomycin D induces the expression of differentiation markers and promote differentiation into neuronal lineages [75]. Similarly, differentiation of human ESCs reduces rRNA synthesis and reduction of rRNA synthesis by the Pol I inhibitor CX-5461 induces the expression of markers for all three germ layers and reduces the expression of pluripotency markers [74].
Figure 3. Establishment of silent rRNA genes during ESC differentiation.
The pluripotency state in ICM can be immortalized in vitro through culturing of embryonic stem cells (ESCs). ESCs display an open and euchromatic genome that is remodeled into a condensed and heterochromatic state, including the formation of highly compact and transcriptional repressed heterochromatic regions that cluster at the nucleolus or at the nuclear periphery. rRNA genes are all active in ESCs and establishment of silent copies occurs only upon differentiation. In ESCs, the processing of IGS-rRNA into pRNA is impaired. This process is reactivated only upon differentiation. The association of TIP5 with the unprocessed IGS-rRNA prevents the interaction with TTF-1 that is bound to the promoter, thereby abrogating rRNA gene silencing. Upon differentiation, mature pRNA is produced and promotes TIP5-TTF1 interaction that is productive for NoRC guiding to rRNA genes and formation of heterochromatin at nucleoli. The formation of silent and heterochromatic rRNA genes coincides with the remodeling of the genome from a euchromatic into a heterochromatic state that favors the exit from pluripotency.
Previous studies in somatic cells have highlighted the nucleolus as the hub for the organization of the inactive chromatin in the cell [4–8]. Recent data have also started to suggest that the nucleolus could play an active role in the remodelling of ESC genome, which during differentiation undergoes structural rearrangements and formation of highly condensed and transcriptional repressed heterochromatic regions that cluster at the nucleolus or at the nuclear periphery [22, 73, 76–78]. Addition of mature pRNA into ESCs that contain only unprocessed IGS-rRNA was not only sufficient to establish rRNA gene silencing but also induced the remodeling of the euchromatic ESC genome into a heterochromatic structure similar to differentiated cells [22]. These changes included the appearance of highly condensed heterochromatic blocks outside the nucleolus, global increase in the repressive histone mark H3K9me2, increased expression of genes involved in cell differentiation, and loss of pluripotency due the inability to form teratoma. These results indicate that the formation of heterochromatin in the nucleolus, at rRNA genes, promotes heterochromatinization of the rest of the nuclear genome and suggest that the chromatin state of rRNA genes has an impact in genome organization. A similar observation was also found in mouse embryo fibroblast cells NIH3T3, where half of rRNA genes are heterochromatic. Knockdown of TIP5 induced not only a decrease of rRNA gene silencing but also the loss of perinucleolar heterochromatic blocks and the reduction of silent histone marks at pericentric heterochromatin [79, 80]. The link between rRNA genes and genome architecture is also supported by results in Drosophila showing that deletion of rRNA repeats reduce heterochromatin content elsewhere in the genome and affect the expression of hundreds to thousands of euchromatic genes [81, 82]. Taken together these results indicate that rRNA gene chromatin states affect nucleolus structure and are implicated in genome stability, genome architecture, and cell fate decision.
Concluding Remarks and Future Perspectives
Increasing evidence indicates the nucleolus and rRNA genes as central player of genome architecture. Changes in nucleolus architecture and chromatin composition of rRNA genes, the genetic component of the nucleus, co-occur with the drastic genome re-organization during the critical phases of gametogenesis and early mammalian development. Although the exact mechanisms of this crosstalk remain to be elucidated, an attractive hypothesis is that transcription and chromatin state of rRNA genes affects the nucleolus compartment in its protein composition and structure, allowing the concentration of factors required for the establishment of repressive states. This is particularly evident during ESC differentiation where targeting of silencing in the nucleolus at rRNA genes induced heterochromatin formation in the rest of genome. Future works will be needed to improve methods to define how and which genomic regions associate with the nucleolus according to cell state and determine the molecular mechanisms of how the organization of the genome around the nucleolus is established, its functional role, and its dynamics during development and disease.
Highlights.
The nucleolus is the hub for the organization of the inactive heterochromatin
The structure of the nucleoli and chromatin states of rRNA genes undergo major changes during gametogenesis and early mammalian development
The nucleolus and the chromatin composition of rRNA genes contribute to the higher-order spatial organization of the genome
Outstanding Questions.
How are regulated the chromatin and epigenetic features of rRNA genes during gametogenesis and early embryo development?
Which mechanisms drive the formation of heterochromatin ring-like structures around NLB of SN oocytes and NPB in the pronuclei of zygotes? What is the function of these structures?
Which mechanisms regulate the crosstalk between the nucleolus and repressive chromatin domains? How does the nucleolus affect genome architecture? How do genomic regions contact the nucleolus upon ESC differentiation?
What is the temporal order of chromatin changes during ESC differentiation? Are some changes necessary for others to occur?
Is there a specific class of rRNA genes that is preferentially converted into silent copies during ESC differentiation?
Glossary.
- Centromeric heterochromatin
regions of the inner centromere that consists of minor satellite repeats in mouse cells.
- Chromocenters
structures visible in mouse interphase cells, containing pericentromeric and centromeric sequences of several chromosomes that cluster together.
- Euchromatin
chromatin regions that are less condensed, gene-rich, and more accessible to transcription. Euchromatin is typically enriched in active histone marks.
- Fertilization
Process involving the fusion of gametes to form a new organism of the same species. In animals, this process involves a sperm fusing with an oocyte, thereby forming the zygote
- Gametogenesis
the production of haploid sex cells in mammals, carrying each one-half of the genetic complement of the parents. In this process, diploid gametes undergo meiosis and differentiation. Gametes of male and female reproductive systems are known as sperms and oocytes, respectively. The corresponding processes are called as spermatogenesis and oogenesis
- Heterochromatin
chromatin regions that are highly condensed, gene-poor, and transcriptionally silent. Heterochromatin is characterized by repressive histone marks and DNA methylation in organisms showing this modification. Regions that contain a high density of repetitive DNA elements, such as clusters of satellites and transposons, are the main targets for heterochromatin formation.
- NLB
Nucleolus-like body (NLB) is an atypical nucleolus that is formed in fully grown GV oocytes. NLB structure is compact and morphologically different from nucleoli in somatic cells.
- NORs
Nucleolar Organizer Region (NOR) are chromosomal regions containing rRNA genes. In humans and apes, rRNA genes are located between the short arm and the satellite body of acrocentric chromosomes 13, 14, 15, 21, and 22. In mouse cells rRNA repeats are within the centromeric regions of chromosomes 12, 15, 16, 18 and 19.
- NPB
Nucleolus Precursor Body (NPB) is an atypical nucleolus that is formed in zygotes and persists until 8-cell stage of early development. NPBs are transcriptionally inactive and morphologically similar to the oocyte NPB, lacking the tripartite configuration of a typical somatic nucleolus.
- Nucleolus
the largest subnuclear compartment of the cell, where ribosome biogenesis takes place. The nucleolus forms around regions of chromosomes containing stretches of rRNA gene repeats, known as nucleolar organizer regions (NORs). It is a membraneless compartment. Nucleoli of somatic cells show a peculiar tripartite architecture: the fibrillar center (FC), the dense fibrillar component (DFC), and the granular component (GC).
- Pericentromeric heterochromatin
highly condensed regions of the genome that comprise major satellites, which flank the centric domain consisting of “minor” satellite repeats.
- Pluripotency
the state of a cell within the early mammalian embryo that has the capacity to generate all somatic lineages and the germline. Pluripotency is confined to the pre-implantation epiblast. In vitro pluripotency can be maintained indefinitely through derivation of embryonic stem cell lines.
- Ribosome biogenesis
a process that requires the coordinated activity of all three RNA polymerases and the orchestrated work of many (>200) transiently associated ribosome assembly factors to produce ribosomes.
- Totipotency
the state of a cell that can give rise to all the extraembryonic tissues and all tissues of the body and the germline. Establishment of totipotency initiates upon fertilization with formation of the zygote.
Acknowledgments
This work was supported by the Swiss National Science Foundation (31003A_173056) and ERC grant (ERC-AdG-787074-NucleolusChromatin).
References
- 1.Griesenbeck J, et al. Structure and Function of RNA Polymerases and the Transcription Machineries. Subcell Biochem. 2017;83:225–270. doi: 10.1007/978-3-319-46503-6_9. [DOI] [PubMed] [Google Scholar]
- 2.Moss T, et al. The chromatin landscape of the ribosomal RNA genes in mouse and human. Chromosome Res. 2019;27(1–2):31–40. doi: 10.1007/s10577-018-09603-9. [DOI] [PubMed] [Google Scholar]
- 3.Pelletier J, et al. Ribosome biogenesis in cancer: new players and therapeutic avenues. Nat Rev Cancer. 2018;18(1):51–63. doi: 10.1038/nrc.2017.104. [DOI] [PubMed] [Google Scholar]
- 4.van Koningsbruggen S, et al. High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21(21):3735–48. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nemeth A, et al. Initial genomics of the human nucleolus. PLoS Genet. 2010;6(3):e1000889. doi: 10.1371/journal.pgen.1000889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dillinger S, et al. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS One. 2017;12(6):e0178821. doi: 10.1371/journal.pone.0178821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Diesch J, et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun Biol. 2019;2:39. doi: 10.1038/s42003-019-0284-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinodoz SA, et al. Higher-Order Inter-chromosomal Hubs Shape 3D Genome Organization in the Nucleus. Cell. 2018;174(3):744–757.:e24. doi: 10.1016/j.cell.2018.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Guetg C, Santoro R. Formation of nuclear heterochromatin: The nucleolar point of view. Epigenetics. 2012;7(8):811–4. doi: 10.4161/epi.21072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kind J, et al. Single-cell dynamics of genome-nuclear lamina interactions. Cell. 2013;153(1):178–92. doi: 10.1016/j.cell.2013.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Feric M, et al. Coexisting Liquid Phases Underlie Nucleolar Subcompartments. Cell. 2016;165(7):1686–97. doi: 10.1016/j.cell.2016.04.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tiku V, Antebi A. Nucleolar Function in Lifespan Regulation. Trends Cell Biol. 2018;28(8):662–672. doi: 10.1016/j.tcb.2018.03.007. [DOI] [PubMed] [Google Scholar]
- 13.Boisvert FM, et al. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8(7):574–85. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 14.French SL, et al. In exponentially growing Saccharomyces cerevisiae cells, rRNA synthesis is determined by the summed RNA polymerase I loading rate rather than by the number of active genes. Mol Cell Biol. 2003;23(5):1558–68. doi: 10.1128/MCB.23.5.1558-1568.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harper F, Puvion-Dutilleul F. Non-nucleolar transcription complexes of rat liver as revealed by spreading isolated nuclei. J Cell Sci. 1979;40:181–92. doi: 10.1242/jcs.40.1.181. [DOI] [PubMed] [Google Scholar]
- 16.Puvion-Dutilleul F, Bachellerie JP. Ribosomal transcriptional complexes in subnuclear fractions of Chinese hamster ovary cells after short-term actinomycin D treatment. J Ultrastruct Res. 1979;66(2):190–9. doi: 10.1016/s0022-5320(79)90134-5. [DOI] [PubMed] [Google Scholar]
- 17.Conconi A, et al. Two different chromatin structures coexist in ribosomal RNA genes throughout the cell cycle. Cell. 1989;57(5):753–61. doi: 10.1016/0092-8674(89)90790-3. [DOI] [PubMed] [Google Scholar]
- 18.Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8(3):719–25. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- 19.Santoro R, et al. The nucleolar remodeling complex NoRC mediates heterochromatin formation and silencing of ribosomal gene transcription. Nat Genet. 2002;32(3):393–6. doi: 10.1038/ng1010. [DOI] [PubMed] [Google Scholar]
- 20.Zhou Y, et al. The chromatin remodeling complex NoRC targets HDAC1 to the ribosomal gene promoter and represses RNA polymerase I transcription. EMBO J. 2002;21(17):4632–40. doi: 10.1093/emboj/cdf460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Strohner R, et al. NoRC--a novel member of mammalian ISWI-containing chromatin remodeling machines. EMBO J. 2001;20(17):4892–900. doi: 10.1093/emboj/20.17.4892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savić N. lncRNA Maturation to Initiate Heterochromatin Formation in the Nucleolus Is Required for Exit from Pluripotency in ESCs. Cell Stem Cell. 2014;15(6):720–734. doi: 10.1016/j.stem.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Leone S, et al. The RNA helicase DHX9 establishes nucleolar heterochromatin, and this activity is required for embryonic stem cell differentiation. EMBO reports. 2017;18(7):1248–1262. doi: 10.15252/embr.201744330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mayer C, et al. Intergenic Transcripts Regulate the Epigenetic State of rRNA Genes. Molecular Cell. 2006;22(3):351–361. doi: 10.1016/j.molcel.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 25.Guetg C, et al. Inheritance of Silent rDNA Chromatin Is Mediated by PARP1 via Noncoding RNA. Mol Cell. 2012;45(6):790–800. doi: 10.1016/j.molcel.2012.01.024. [DOI] [PubMed] [Google Scholar]
- 26.Sanij E, et al. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183(7):1259–74. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hamdane N, et al. Conditional inactivation of Upstream Binding Factor reveals its epigenetic functions and the existence of a somatic nucleolar precursor body. PLoS Genet. 2014;10(8):e1004505. doi: 10.1371/journal.pgen.1004505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Griswold MD. Spermatogenesis: The Commitment to Meiosis. Physiol Rev. 2016;96(1):1–17. doi: 10.1152/physrev.00013.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McSwiggin HM, O’Doherty AM. Epigenetic reprogramming during spermatogenesis and male factor infertility. Reproduction. 2018;156(2):R9–R21. doi: 10.1530/REP-18-0009. [DOI] [PubMed] [Google Scholar]
- 30.Fulka H, Langerova A. Nucleoli in embryos: a central structural platform for embryonic chromatin remodeling? Chromosome Res. 2019;27(1–2):129–140. doi: 10.1007/s10577-018-9590-3. [DOI] [PubMed] [Google Scholar]
- 31.Inoue A, et al. Contribution of the oocyte nucleus and cytoplasm to the determination of meiotic and developmental competence in mice. Hum Reprod. 2008;23(6):1377–84. doi: 10.1093/humrep/den096. [DOI] [PubMed] [Google Scholar]
- 32.Monti M, et al. Developmental arrest and mouse antral not-surrounded nucleolus oocytes. Biol Reprod. 2013;88(1):2. doi: 10.1095/biolreprod.112.103887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fulka H, et al. Can Nucleoli Be Markers of Developmental Potential in Human Zygotes? Trends Mol Med. 2015;21(11):663–72. doi: 10.1016/j.molmed.2015.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Shishova KV, et al. Nucleolus-like bodies of fully-grown mouse oocytes contain key nucleolar proteins but are impoverished for rRNA. Dev Biol. 2015;397(2):267–81. doi: 10.1016/j.ydbio.2014.11.022. [DOI] [PubMed] [Google Scholar]
- 35.Kyogoku H, et al. Nucleolus precursor body (NPB): a distinct structure in mammalian oocytes and zygotes. Nucleus. 2014;5(6):493–8. doi: 10.4161/19491034.2014.990858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bonnet-Garnier A, et al. Genome organization and epigenetic marks in mouse germinal vesicle oocytes. Int J Dev Biol. 2012;56(10–12):877–87. doi: 10.1387/ijdb.120149ab. [DOI] [PubMed] [Google Scholar]
- 37.Verlhac MH, Terret ME. Oocyte Maturation and Development. F1000Res. 2016:5. doi: 10.12688/f1000research.7892.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fulka H, Langerova A. The maternal nucleolus plays a key role in centromere satellite maintenance during the oocyte to embryo transition. Development. 2014;141(8):1694–704. doi: 10.1242/dev.105940. [DOI] [PubMed] [Google Scholar]
- 39.Sirard MA. Activation of the embryonic genome. Soc Reprod Fertil Suppl. 2010;67:145–58. [PubMed] [Google Scholar]
- 40.Bouniol C, et al. Endogenous transcription occurs at the 1-cell stage in the mouse embryo. Exp Cell Res. 1995;218(1):57–62. doi: 10.1006/excr.1995.1130. [DOI] [PubMed] [Google Scholar]
- 41.Aoki F, et al. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181(2):296–307. doi: 10.1006/dbio.1996.8466. [DOI] [PubMed] [Google Scholar]
- 42.Hamdane N, et al. Disruption of the UBF gene induces aberrant somatic nucleolar bodies and disrupts embryo nucleolar precursor bodies. Gene. 2017;612:5–11. doi: 10.1016/j.gene.2016.09.013. [DOI] [PubMed] [Google Scholar]
- 43.Chen H, et al. Early pre-implantation lethality in mice carrying truncated mutation in the RNA polymerase 1-2 gene. Biochem Biophys Res Commun. 2008;365(4):636–42. doi: 10.1016/j.bbrc.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 44.Drygin D, et al. Targeting RNA polymerase I with an oral small molecule CX-5461 inhibits ribosomal RNA synthesis and solid tumor growth. Cancer Res. 2011;71(4):1418–30. doi: 10.1158/0008-5472.CAN-10-1728. [DOI] [PubMed] [Google Scholar]
- 45.Koné MC, et al. Three-Dimensional Distribution of UBF and Nopp140 in Relationship to Ribosomal DNA Transcription During Mouse Preimplantation Development. Biology of Reproduction. 2016;94(4):95. doi: 10.1095/biolreprod.115.136366. [DOI] [PubMed] [Google Scholar]
- 46.Flechon JE, Kopecny V. The nature of the ‘nucleolus precursor body’ in early preimplantation embryos: a review of fine-structure cytochemical, immunocytochemical and autoradiographic data related to nucleolar function. Zygote. 1998;6(2):183–91. doi: 10.1017/s0967199498000112. [DOI] [PubMed] [Google Scholar]
- 47.Lavrentyeva E, et al. Localisation of RNAs and proteins in nucleolar precursor bodies of early mouse embryos. Reprod Fertil Dev. 2017;29(3):509–520. doi: 10.1071/RD15200. [DOI] [PubMed] [Google Scholar]
- 48.Probst AV, Almouzni G. Heterochromatin establishment in the context of genome-wide epigenetic reprogramming. Trends Genet. 2011;27(5):177–85. doi: 10.1016/j.tig.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 49.Nestorov P, et al. H3K9/HP1 and Polycomb: two key epigenetic silencing pathways for gene regulation and embryo development. Curr Top Dev Biol. 2013;104:243–91. doi: 10.1016/B978-0-12-416027-9.00008-5. [DOI] [PubMed] [Google Scholar]
- 50.Fadloun A, et al. Mechanisms and dynamics of heterochromatin formation during mammalian development: closed paths and open questions. Curr Top Dev Biol. 2013;104:1–45. doi: 10.1016/B978-0-12-416027-9.00001-2. [DOI] [PubMed] [Google Scholar]
- 51.Seisenberger S, et al. Reprogramming DNA methylation in the mammalian life cycle: building and breaking epigenetic barriers. Philos Trans R Soc Lond B Biol Sci. 2013;368(1609):20110330. doi: 10.1098/rstb.2011.0330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lin CJ, et al. Hira-mediated H3.3 incorporation is required for DNA replication and ribosomal RNA transcription in the mouse zygote. Dev Cell. 2014;30(3):268–79. doi: 10.1016/j.devcel.2014.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Borsos M, Torres-Padilla ME. Building up the nucleus: nuclear organization in the establishment of totipotency and pluripotency during mammalian development. Genes Dev. 2016;30(6):611–21. doi: 10.1101/gad.273805.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aguirre-Lavin T, et al. 3D-FISH analysis of embryonic nuclei in mouse highlights several abrupt changes of nuclear organization during preimplantation development. BMC Dev Biol. 2012;12:30. doi: 10.1186/1471-213X-12-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jachowicz JW, et al. Heterochromatin establishment at pericentromeres depends on nuclear position. Genes Dev. 2013;27(22):2427–32. doi: 10.1101/gad.224550.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Probst AV, et al. A strand-specific burst in transcription of pericentric satellites is required for chromocenter formation and early mouse development. Dev Cell. 2010;19(4):625–38. doi: 10.1016/j.devcel.2010.09.002. [DOI] [PubMed] [Google Scholar]
- 57.Casanova M, et al. Heterochromatin reorganization during early mouse development requires a single-stranded noncoding transcript. Cell Rep. 2013;4(6):1156–67. doi: 10.1016/j.celrep.2013.08.015. [DOI] [PubMed] [Google Scholar]
- 58.Probst AV, et al. Structural differences in centromeric heterochromatin are spatially reconciled on fertilisation in the mouse zygote. Chromosoma. 2007;116(4):403–15. doi: 10.1007/s00412-007-0106-8. [DOI] [PubMed] [Google Scholar]
- 59.Ogushi S, et al. The maternal nucleolus is essential for early embryonic development in mammals. Science. 2008;319(5863):613–6. doi: 10.1126/science.1151276. [DOI] [PubMed] [Google Scholar]
- 60.Fulka J., Jr Enucleolation of porcine oocytes. Theriogenology. 2003;59(8):1879–85. doi: 10.1016/s0093-691x(02)01226-8. [DOI] [PubMed] [Google Scholar]
- 61.Kyogoku H, et al. De novo formation of nucleoli in developing mouse embryos originating from enucleolated zygotes. Development. 2014;141(11):2255–9. doi: 10.1242/dev.106948. [DOI] [PubMed] [Google Scholar]
- 62.Ogushi S, et al. Reconstitution of the oocyte nucleolus in mice through a single nucleolar protein, NPM2. J Cell Sci. 2017;130(14):2416–2429. doi: 10.1242/jcs.195875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ogushi S, Saitou M. The nucleolus in the mouse oocyte is required for the early step of both female and male pronucleus organization. J Reprod Dev. 2010;56(5):495–501. doi: 10.1262/jrd.09-184h. [DOI] [PubMed] [Google Scholar]
- 64.Burns KH, et al. Roles of NPM2 in chromatin and nucleolar organization in oocytes and embryos. Science. 2003;300(5619):633–6. doi: 10.1126/science.1081813. [DOI] [PubMed] [Google Scholar]
- 65.Guo F, et al. Active and passive demethylation of male and female pronuclear DNA in the mammalian zygote. Cell Stem Cell. 2014;15(4):447–58. doi: 10.1016/j.stem.2014.08.003. [DOI] [PubMed] [Google Scholar]
- 66.Smith ZD. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012;484:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Smith ZD, et al. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature. 2012:339–344. doi: 10.1038/nature10960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Graf U, et al. Pramel7 mediates ground-state pluripotency through proteasomal-epigenetic combined pathways. Nat Cell Biol. 2017;19(7):763–773. doi: 10.1038/ncb3554. [DOI] [PubMed] [Google Scholar]
- 69.Tang F, et al. Tracing the derivation of embryonic stem cells from the inner cell mass by single-cell RNA-Seq analysis. Cell Stem Cell. 2010:468–478. doi: 10.1016/j.stem.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gaspar-Maia A, et al. Open chromatin in pluripotency and reprogramming. Nature reviews. Molecular cell biology. 2011;12(1):36–47. doi: 10.1038/nrm3036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gorkin DU, et al. The 3D genome in transcriptional regulation and pluripotency. Cell Stem Cell. 2014;14(6):762–75. doi: 10.1016/j.stem.2014.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Schlesinger S, et al. Allelic inactivation of rDNA loci. Genes Dev. 2009;23(20):2437–47. doi: 10.1101/gad.544509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Meshorer E, Misteli T. Chromatin in pluripotent embryonic stem cells and differentiation. Nat Rev Mol Cell Biol. 2006;7(7):540–6. doi: 10.1038/nrm1938. [DOI] [PubMed] [Google Scholar]
- 74.Woolnough JL, et al. The Regulation of rRNA Gene Transcription during Directed Differentiation of Human Embryonic Stem Cells. PLoS One. 2016;11(6):e0157276. doi: 10.1371/journal.pone.0157276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Watanabe-Susaki K, et al. Biosynthesis of ribosomal RNA in nucleoli regulates pluripotency and differentiation ability of pluripotent stem cells. Stem Cells. 2014;32(12):3099–111. doi: 10.1002/stem.1825. [DOI] [PubMed] [Google Scholar]
- 76.Bhattacharya D, et al. Spatio-temporal plasticity in chromatin organization in mouse cell differentiation and during Drosophila embryogenesis. Biophys J. 2009;96(9):3832–9. doi: 10.1016/j.bpj.2008.11.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bartova E, et al. Differentiation of human embryonic stem cells induces condensation of chromosome territories and formation of heterochromatin protein 1 foci. Differentiation. 2008;76(1):24–32. doi: 10.1111/j.1432-0436.2007.00192.x. [DOI] [PubMed] [Google Scholar]
- 78.Wiblin AE, et al. Distinctive nuclear organisation of centromeres and regions involved in pluripotency in human embryonic stem cells. J Cell Sci. 2005;118(Pt 17):3861–8. doi: 10.1242/jcs.02500. [DOI] [PubMed] [Google Scholar]
- 79.Guetg C, et al. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. The EMBO Journal. 2010;29(13):2135. doi: 10.1038/emboj.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Postepska-Igielska A, et al. The chromatin remodelling complex NoRC safeguards genome stability by heterochromatin formation at telomeres and centromeres. EMBO Rep. 2013 doi: 10.1038/embor.2013.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Paredes S, Maggert KA. Ribosomal DNA contributes to global chromatin regulation. Proc Natl Acad Sci U S A. 2009;106(42):17829–34. doi: 10.1073/pnas.0906811106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Paredes S, et al. Ribosomal DNA deletions modulate genome-wide gene expression: “rDNA-sensitive” genes and natural variation. PLoS Genet. 2011;7(4):e1001376. doi: 10.1371/journal.pgen.1001376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Chazaud C, Yamanaka Y. Lineage specification in the mouse preimplantation embryo. Development. 2016;143(7):1063–74. doi: 10.1242/dev.128314. [DOI] [PubMed] [Google Scholar]
- 84.Hackett JA, Surani MA. Regulatory principles of pluripotency: from the ground state up. Cell Stem Cell. 2014;15(4):416–30. doi: 10.1016/j.stem.2014.09.015. [DOI] [PubMed] [Google Scholar]