Abstract
In plants, pathogen attack can induce an immune response known as systemic acquired resistance (SAR) that protects against a broad spectrum of pathogens. In the search for safer agrochemicals, silica nanoparticles (SiO2-NPs, food additive E551) have recently been proposed as a new tool. However, initial results are controversial, and the molecular mechanisms of SiO2-NP-induced disease resistance are unknown. Here, we show that SiO2-NPs, as well as soluble orthosilicic acid (Si(OH)4), can induce SAR in a dose-dependent manner, that involves the defence hormone salicylic acid. Nanoparticle uptake and action occurred exclusively through stomata (leaf pores facilitating gas exchange) and involved extracellular adsorption in leaf air spaces of the spongy mesophyll. In contrast to treatment with SiO2-NPs, induction of SAR by Si(OH)4 was problematic, since high concentrations caused stress. We conclude that SiO2-NPs have the potential to serve as an inexpensive, highly efficient, safe, and sustainable alternative for plant disease protection.
Nanoagrochemicals are a promising tool to improve crop yield and thus, global food security1. Silica nanoparticles (SiO2-NPs) have been proposed for the controlled nanodelivery of silicon and other active ingredients to plants but have never been systematically tested for this purpose. Silicon from orthosilicic acid (Si(OH)4, monosilicic acid), the hydrolytic degradation product of SiO2-NPs, is the only known form of silicon bioavailable for plants, and is ubiquitous in soil pore water2–4. Orthosilicic acid can promote plant growth and plant resistance against biotic and abiotic stresses3,5, thereby protecting plants against pathogen attacks or agricultural damages related to severe climate conditions3,6,7. The uptake and movement of SiO2-NPs as well as other engineered nanomaterials in plants have been studied intensively in the last decade7–10. However, it is uncertain how the nanoparticles interact with leaves at the subcellular level. Direct evidence by nm-resolution imaging for the entrance of intact nanoparticles into leaves, or intercellular movement of SiO2-NPs within leaves, is mostly missing10. It is also not known whether SiO2-NPs can induce resistance in plants, whether their performance differs from dissolved silicon species, and which molecular pathways they may induce.
To fend off potential pathogens, plants have evolved disease resistance mechanisms that share mechanistic principles with the innate immunity of animals11. An especially interesting form of plant disease resistance is the so-called induced resistance in which disease resistance of the plant can be enhanced by previous exposure to beneficial rhizosphere microorganisms, avirulent and virulent pathogens, or specific resistance-inducing chemical compounds12–14. A hallmark of induced resistance is its activity against a broad spectrum of pathogens. While the induction of plant disease resistance using chemical compounds is relatively well understood12, the benefit of using slow nano-enabled delivery systems for the same purpose has not been investigated in systematic experiments1,7.
A special form of induced resistance is systemic acquired resistance (SAR) that is characterized by the spread of locally induced disease resistance to the whole plant15,16. Systemic acquired resistance is induced in all plant parts after challenging the plant locally with a pathogen or by local application of so-called resistance-inducing compounds. Both treatments induce signal transduction pathways that lead to the production of signals that move to distant tissues14. A key signalling compound that contributes to SAR is the plant hormone salicylic acid (SA) that is responsible for the activation of pathogenesis-related (PR) genes16,17. Other factors include, e.g., nitric oxide and reactive oxygen species18,19. The fact that SAR can be activated by the application of resistance-inducing compounds12,13 makes SAR an attractive alternative strategy for controlling crop pests without the use of irreversible genetic modifications or environmentally problematic pesticides.
Systemic acquired resistance-inducing compounds such as benzothiadiazole successfully enhanced disease resistance, but also reduce crop yields20,21. Interestingly, silicon-based compounds also seem to have the capacity to induce disease resistance via a broad range of different and partially still unknown mechanisms, including the mechanical reinforcement of defensive structures of the plant architecture, most notably the cell wall3,22, but also by the activation of biochemical defenses3,23. Biochemically, root-applied silicon led, for example, to a broad-spectrum resistance against powdery mildew pathogen by increasing the activity of defence-related enzymes in leaves24. It is important to note that the protective effect of silicon seems to have, in contrast to other biostimulants such as benzothiadiazole, no negative effects on the growth and yield of plants3,25. All of this makes Si an attractive candidate to strengthen plant stress tolerance. Initial studies found that SiO2-NPs may induce stress tolerance similarly to conventional Si products, but a clear mechanistic understanding of the underlying processes is still lacking7,8,26,27.
In this work, we demonstrate the potential of SiO2-NPs in inducing local and systemic disease resistance in the widely used model plant Arabidopsis thaliana against the bacterial pathogen Pseudomonas syringae. Silicic acid was assessed in parallel to disentangle the potential differences in the mode of action of dissolved Si species compared to SiO2-NPs. We assessed the role of salicylic acid and reactive oxygen species (ROS) defence-related genes, established the therapeutic concentration range of SiO2-NPs to induce the desired beneficial effects in plants, compared the laboratory setup (infiltration of selected leaves) with the more realistic spray application, and visualized the nanoparticle-leaf interactions using transmission electron microscopy (TEM), with important implications for future strategies to apply nanoscale active ingredients for slow release on leaves.
SiO2-NPs and Subcellular Distribution within the Leaf
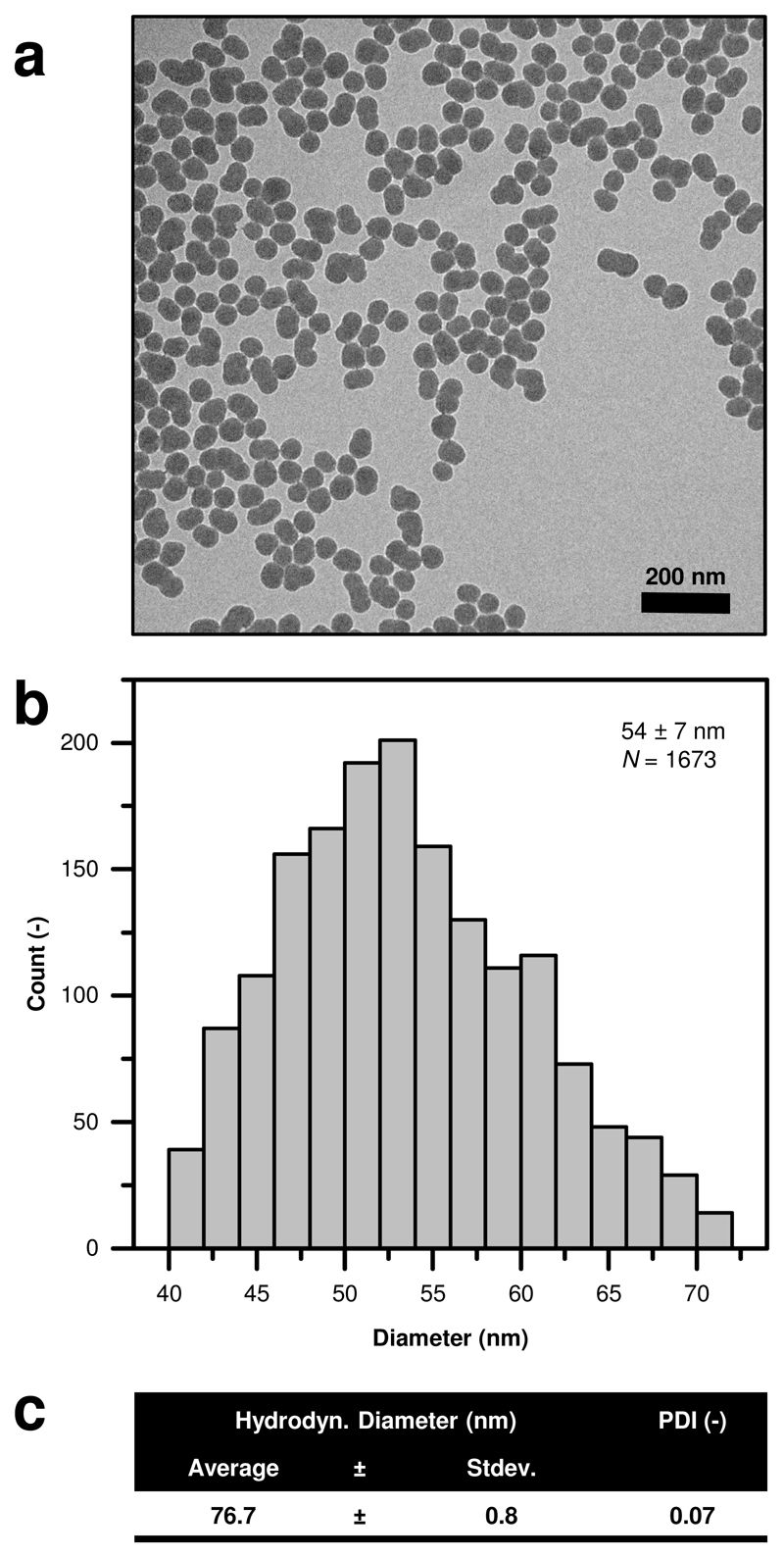
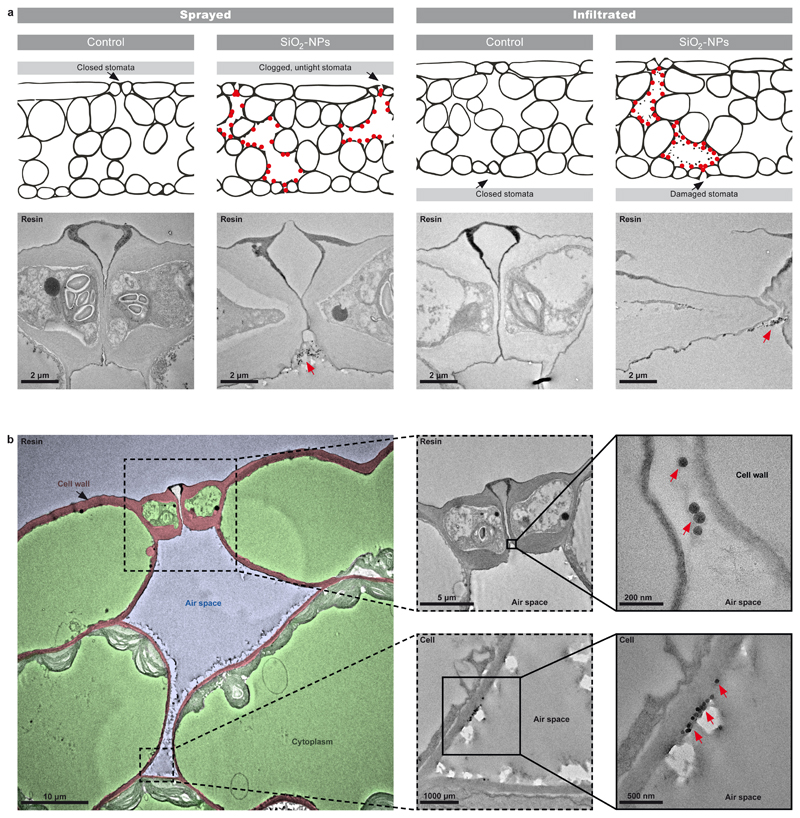
The SiO2-NP suspensions used for the dosing of the plants (Fig. 1) were well dispersed with a hydrodynamic particle size of 76.7±0.8 nm and a polydispersity index of 0.07. The primary particle size, as determined by TEM, was 54±7 nm. The interaction of the nanoparticles with the plant was assessed by TEM (Fig. 2) 2 d after application of SiO2-NPs. Preliminary experiments had shown that at this time point the SiO2-NP-exposed plants already had developed resistance. The size range of ~50-70 nm of the nanoparticles allowed them to enter the leaf exclusively through the stomata and distribute within the large extracellular air spaces of the spongy mesophyll without penetrating any cell walls (Fig. 2, Supplementary Fig. S1). The SiO2-NPs remained in the leaf air spaces during the 2 d between their application and the time point of TEM observation. At the same time, the size of the nanoparticles prevented (undesirable) nanoparticle uptake into the cytoplasm as well as cell-to-cell translocation through the plasmodesmata (Fig. 2b). This is in line with previous literature based on nm-resolution imaging of nanoparticles in plants suggesting that the cut-off for root-shoot nanoparticle translocation is at approx. <36 nm, and for cell-to-cell plasmodesmata transport <15-40 nm (basal size exclusion limits of ~3–4 nm)10. Compared to the fully closed stomata in the control plants (samples were kept in the dark for fixation), the nanoparticle-treated plants showed incompletely closed stomata due to nanoparticles stuck in between the guard cells (Fig. 2b).
Figure 1. Silica nanoparticles (SiO2-NPs) under investigation.
a Transmission electron microscopy (TEM) imaging showing the particles. b Particle size distribution based on TEM image analysis. c Dynamic light scattering (DLS) measurements of SiO2-NPs. Hydrodynamic radius consistent with the primary particle size shown in a and b. PDI: Polydispersity index. Averages ± standard deviations. For DLS, N = 10.
Figure 2. Transmission electron microscopy (TEM) of silica nanoparticle (SiO2-NP) distribution and physiological effects in Arabidopsis leaves.
Red arrows and dots: Nanoparticles. Comparison between spray application used in the field and for local defence assays, and infiltration application used in laboratory studies. Images obtained when plants already had developed resistance 2 d after exposure to SiO2-NPs. a Control leaves treated with buffer solution only. b TEM overview image and zooms into the stoma and cell-air space interface. False colours mark, in red, the cell wall (apoplast); in green, the cytoplasm (symplast); and in blue, the spaces filled with air. SiO2-NP-sprayed leaf in higher resolution shows that the stomata are not tightly closed anymore due to nanoparticle uptake and clogging. Nanoparticles entered through the stomata into the leaf air spaces, were also found adsorbed extracellularly to the outer edge of the cell walls in the air gaps of the spongy mesophyll, and were absent in the cytoplasm (intracellular space). Higher-resolution TEM in Supplementary Fig. S1.
Exogenous Application of SiO2-NPs confers SAR
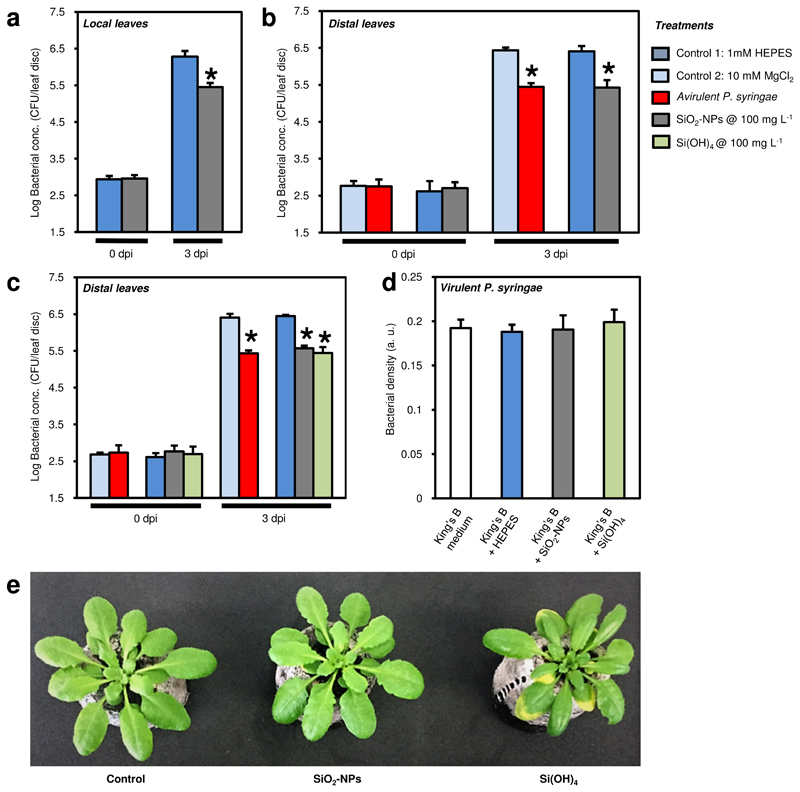
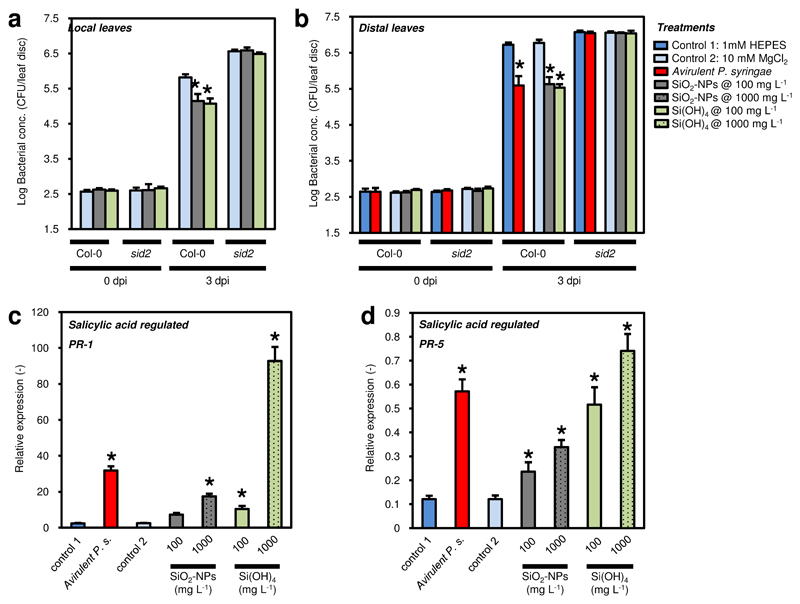
The local defence responses of Arabidopsis sprayed with SiO2-NPs or a control treatment to virulent P. syringae were quantified via the bacterial growth on leaves (Fig. 3a). Due to the lack of the avrRpt2 gene in the virulent P. syringae that is needed by the RPS2 resistance gene in Arabidopsis to induce a strong plant defence against P. syringae 28,29, a severe infection would be expected. However, a pronounced infection only occurred in the control treatment. Plants sprayed with SiO2-NPs showed an 8-fold improved basal resistance compared with 4 (2 hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer-treated control plants (Fig. 3a), demonstrating that the SiO2-NPs induced a local defence in the plant within 24 h (the nanoparticles were applied 24 h before inoculation with virulent P. syringae). The number of bacteria was reduced 8-fold in SiO2-NP-treated plants compared to the control.
Figure 3. Enhanced local and systemic disease resistance in wild type Col-0 Arabidopsis to Pseudomonas syringae induced by silica nanoparticles (SiO2-NPs) or orthosilicic acid Si(OH)4.
CFU: Colony-forming units. dpi: days post inoculation with virulent P. syringae. The bacteria in leaves were quantified 0 and 3 dpi. a Growth of virulent P. syringae in leaves. Plants were sprayed with different treatments, and virulent P. syringae was inoculated 24 h later. b Systemic acquired resistance (SAR) in distal leaves. Plants infiltrated locally with different treatments. 48 h later, virulent P. syringae was inoculated on untreated systemic leaves. c SAR in distal leaves, repetition of experimental setup in b with an additional Si(OH)4 treatment. d No effect of SiO2-NPs and Si(OH)4 on in vitro growth of virulent P. syringae bacteria in absence of plant. e Phenotype of Arabidopsis plants. Plants pre-treated with HEPES buffer (control), SiO2-NPs, or Si(OH)4 (1000 mg SiO2 L-1 each). Note the yellow leaves in the plant exposed to Si(OH)4 coinciding with the upregulated expression of the oxidative stress marker gene in Fig. 4c. In a–d, all experiments were performed twice with comparable results. Bars and whiskers are averages and standard deviations, N=3, 1-way ANOVA; post-hoc LSD, p<0.01.
The systemic responses of wild type Arabidopsis plants to SiO2-NPs and dissolved Si species are reflected in the inhibited bacterial growth in Fig. 3b-c. The positive control showed that plants previously infiltrated with the avirulent P. syringae that is known to induce systemic acquired resistance, contained as expected 10-fold less virulent P. syringae compared with MgCl2 or HEPES-preinfiltrated plants (Fig. 3b-c). Remarkably, treating local leaves with SiO2-NPs led to comparable systemic protection against virulent P. syringae as observed in the positive avirulent P. syringae control (Fig. 3a), which is equal to >90% bacterial inhibition. It is highly unlikely that a local response in distal tissue to SiO2-NPs or Si(OH)4 has caused this resistance because of the observed distribution and very slow dissolution of SiO2-NPs (Fig. 2, Supplementary Fig. S1), and the passive transport30 and high reactivity of Si(OH)4. This shows that treating Arabidopsis with SiO2-NPs induced local and systemic resistance to P. syringae.
It is well known that Si(OH)4 improves plant defences against different plant pathogens such as fungi, bacteria, and viruses5,7. We therefore also tested SAR in response to Si(OH)4 (Fig. 3c) and found that treatment with Si(OH)4 was also able to induce SAR. These results suggest that Si(OH)4 released from SiO2-NPs is at least partially responsible for the SAR-inducing ability of SiO2-NPs, and that the SiO2-NPs can act as a slow release source for Si(OH)4.
Measuring the exact amount of free Si(OH)4 directly in plantae is challenging due to the low concentrations and fragile equilibrium of dissolved Si(OH)4 and Si oligomers, and solid SiO2 species2,4,31 (refer to the Supplemental Information, section ‘Details on Si(OH)4 Analytics’). We therefore resorted to direct TEM imaging of the nanoparticles in the plants that revealed at high resolution abundant intact SiO2-NPs in stomata 2 d after the SiO2-NP treatments (Fig. 2). This demonstrates that the plants could not degrade the nanoparticles at the time point of inoculation with virulent P. syringae (in all assays nanoparticles were applied at least 24 h before inoculation). The slow nanoparticle dissolution is in line with the slow dissolution kinetics of the SiO2-NPs we measured previously in water (half-life ~66 d at pH 7)32.
To test whether SiO2-NPs and Si(OH)4 have a direct toxic effect on bacterial growth, virulent P. syringae was cultivated in vitro in the presence or absence of SiO2-NPs, or Si(OH)4 at the lowest fully effective dose of SiO2-NPs at 100 mg L-1. At these concentrations that induced strong defence in plants, neither SiO2-NPs nor Si(OH)4 harmed the growth of the virulent P. syringae bacteria alone (Fig. 3d), demonstrating that SiO2-NPs induce resistance in plants by activating defence responses in plants and not by directly inhibiting bacterial growth.
Dose Dependence of SAR Response
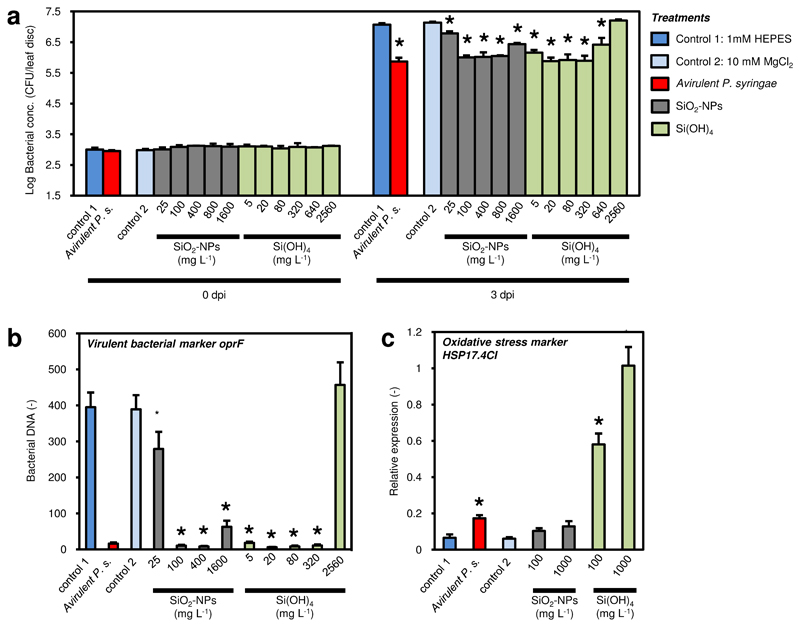
The SAR was further tested in response to different concentrations of SiO2-NPs or Si(OH)4, using for additional validation a second bacterial growth quantification method33 based on bacterial DNA (Fig. 4a-b, Supplementary Tab. S1). Treatment with SiO2-NPs at a concentration of 25 mg SiO2 L-1 already resulted in a partial reduction of 29% of bacterial growth in systemic leaves, and treatment with 100 mg L-1 resulted in maximal protection (>90%) compared to positive control plants preinfiltrated with avirulent P. syringae (Fig. 4a). As the series of concentrations in Fig. 4a shows, higher concentrations of SiO2-NPs >1600 mg SiO2 L-1 led to increased bacterial infection and were thus less effective in activating SAR. Pre-treatment with a concentration of 5 mg SiO2 L-1 of Si(OH)4 (concentration normalized to SiO2 L-1 for the sake of comparability) led to a reduction in bacterial numbers of 81% compared to the positive control. Maximal protection with a reduction similar to control plants preinfiltrated with avirulent P. syringae was achieved at concentrations between 20 to 320 mg SiO2 L-1. A higher concentration of 640 mg SiO2 L-1 was less effective, and a concentration of 2560 mg SiO2 L-1 was ineffective in inducing SAR, demonstrating a detrimental effect of high concentrations of Si(OH)4 on SAR induction.
Figure 4. Silica nanoparticles confer systemic acquired resistance (SAR) in a dose-dependent manner.
Distal leaves of wild type Col-0 Arabidopsis treated with controls or silica nanoparticles (SiO2-NPs) or orthosilicic acid Si(OH)4. CFU: Colony-forming units. dpi: days post inoculation with virulent Pseudomonas syringae. a SAR in plants infiltrated locally with different treatments. 48 h later, virulent P. syringae was inoculated on untreated systemic leaves. The bacteria in leaves were quantified 0 and 3 dpi. b qPCR transcript levels of oprF gene from virulent P. syringae using DNA templates extracted from inoculated leaves. c RT-qPCR transcript levels of oxidative stress marker gene AtHSP17.4C1 in response to different treatments. Plants were infiltrated locally with different treatments. Leaves sampled 48 h after treatments. Reference gene: At4g26410 (expG). Bars and whiskers are averages and standard deviations, N=3, 1-way ANOVA; post-hoc LSD, p<0.05. All experiments in a–c were performed twice with comparable results.
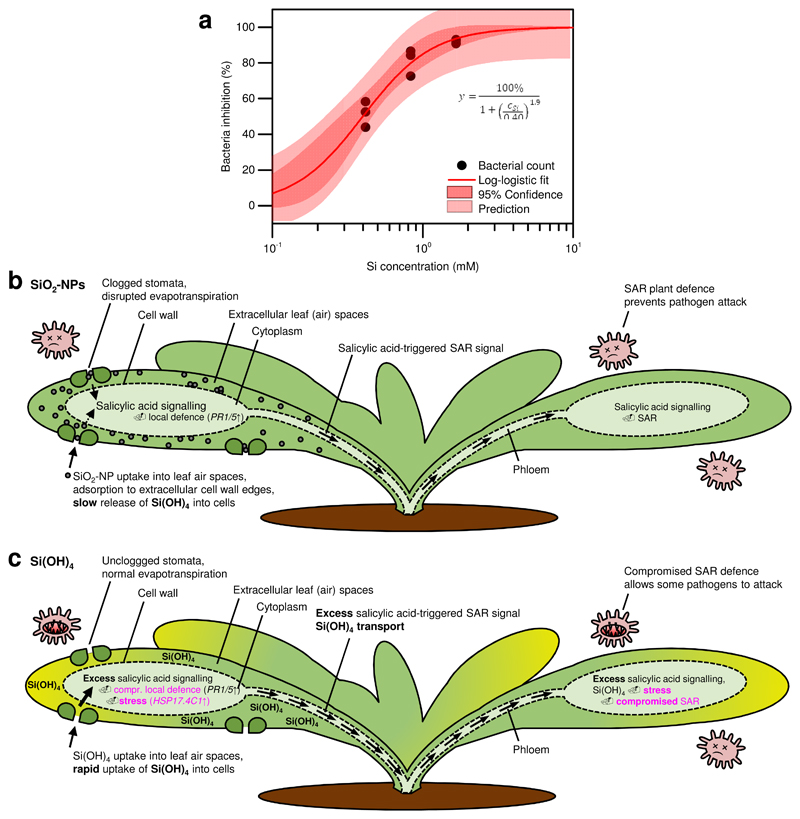
The data of Fig. 4 served to establish a dose-response relationship between SAR and SiO2-NP concentration (Fig. 5a). Using a standard log-logistic dose-response model, the dynamic range and the effective concentration at 50% bacterial inhibition (EC50) was determined at 0.4±0.04 mM Si for SiO2-NPs (i.e., 24 mg SiO2 L-1, refer to Supplementary Fig. S2 for the residual analysis and Supplementary Tab. S2 for the fitting parameters) in a range of 25 to 100 mg SiO2 L-1. For spraying, the EC50 may be similar to injected SiO2-NPs, as local (sprayed) and systemic (injected) assays at 100 mg SiO2 L-1 both resulted in disease resistance (Fig. 3, Fig. 6a-b).
Figure 5. Dynamic range for systemic acquired resistance (SAR) induced in distal leaves by silica nanoparticles (SiO2-NPs) in Arabidopsis thaliana, and model summarizing the observed plant defence-enhancing actions of SiO2-NPs and orthosilicic acid Si(OH)4.
a Data from Fig. 4a. SiO2-NP–triggered dose-dependent bacterial inhibition 3 d after infection of wild type A. thaliana with virulent Pseudomonas syringae. The effective concentration at 50% bacterial inhibition (EC50) was 0.40±0.04 mM Si for SiO2-NPs (i.e. 24 mg SiO2 L-1). Above the dynamic range, bacterial infection can increase again (Fig. 4a). Six data points at 0 mM Si are not shown due to the nature of the log axis but apparent in the detailed residual analysis in Supplementary Fig. S2 and Supplementary Tab. S2. b SiO2-NPs act by i) slowly releasing Si(OH)4 into cells, triggering SA, and thus local defence and SAR; and ii) clogging stomata, triggering SA and subsequent defences. Absence of intracellular nanoparticles confirmed by electron microscopy (Fig. 2; Supplementary Fig. S1). c Si(OH)4 instantly diffuses into cells, triggering SA and subsequent local defence and SAR. However, the instant uptake causes overdose, stress, and compromised defences. Both mechanisms are shown after treatment with the same amount of SiO2 equivalents (1000 mg SiO2 L-1). Salicylic acid (SA): plant hormone regulating SAR and PR1/5 gene expression. PR1/5: Genes encoding Pathogenesis-related protein 1 and 5. HSP17.4C1: Heat shock protein and oxidative stress marker gene.
Figure 6. Silica nanoparticles (SiO2-NPs) induce disease resistance based on salicylic acid dependent pathway.
Experiments in Arabidopsis wild type Col-0 and salicylic acid-deficient mutant sid2. CFU: Colony-forming units. dpi: days post inoculation with virulent Pseudomonas syringae. The bacteria in leaves were quantified 0 and 3 dpi. a A. thaliana wild type Col-0 and salicylic acid-deficient A. thaliana mutant sid2 were infiltrated locally with different treatments. 24 h after these treatments, virulent P. syringae was inoculated. b Systemic acquired resistance (SAR) in distal leaves of Col-0 wild type and mutant sid2. Plants were infiltrated locally with different treatments. 48 h after these treatments, virulent P. syringae was inoculated. c, d RT-qPCR analysis of gene expression of the salicylic acid-regulated genes AtPR-1 c and AtPR-5 d in response to different local treatments of wild type Arabidopsis. Leaves sampled 48 h after treatments. Reference gene: At4g26410 (expG). Bars and whiskers are averages and standard deviations, N=3, 1-way ANOVA; post-hoc LSD, p<0.02. All experiments in a-d were performed twice with comparable results.
The results based on counting bacterial colonies were confirmed by estimating the bacterial biomass based on qPCR analysis of the bacterial outer membrane protein gene oprF (Fig. 4b). The bacterial DNA levels were in good agreement with the bacterial colony counting results in Fig. 4a, in line with previous research that compared the two techniques33.
In contrast to SiO2-NPs, high concentrations of Si(OH)4 adversely affected the phenotype of treated plants (Fig. 3e). At a concentration of 1000 mg SiO2 L-1, leaves of plants treated with Si(OH)4 showed signs of chlorosis (yellowing), whereas leaves of plants treated with SiO2-NPs looked healthy (Fig. 3e). This different behaviour at higher concentrations prompted us to further investigate the negative effect of high concentrations of SiO2-NPs and Si(OH)4. The expression level of the heat shock protein AtHSP17.4C1, a molecular marker for oxidative stress34, was analysed by reverse transcription-quantitative real-time polymerase chain reaction (RT-qPCR). The HSP17.4C1 transcript levels were determined in response to avirulent P. syringae, SiO2-NPs, or Si(OH)4 (100 and 1000 mg SiO2 L-1; Fig. 4c) 2 d after the treatments. Treatment with avirulent P. syringae caused a minor increase (2.7-fold) in AtHSP17.4C1 expression compared to the control. Similarly, treatment with SiO2-NPs led to a 1.6-fold (100 mg SiO2 L-1) and 2-fold (1000 mg SiO2 L-1) increase in transcript abundance relative to control treatment that was not statistically significant. However, treatment with high concentrations of Si(OH)4 caused stress, as transcript levels of the oxidative stress marker gene HSP17.4C1 were induced 9-fold at a concentration of 100 mg SiO2 L-1 and 18-fold at 1000 mg SiO2 L-1.
SiO2-NP mediated SAR depends on Salicylic Acid
The plant hormone salicylic acid (SA) plays a core regulatory role in plant immunity35. We therefore tested the possibility that SiO2-NP-mediated SAR might function via the salicylic acid-dependent defence pathway by testing the ability of SiO2-NPs to induce local disease resistance and SAR in an Arabidopsis mutant defective in salicylic acid biosynthesis (salicylic acid deficient 2 (sid2)36). Notably, neither Si(OH)4 nor SiO2-NPs induced local disease resistance or SAR in sid2 mutant plants, while they induced basal disease resistance and SAR in wild type plants (Fig. 6a,b), demonstrating that salicylic acid-dependent defence signalling is essential for Si(OH)4- and SiO2-NP-induced disease resistance. To further support this result, we next quantified the expression of the salicylic acid-responsive marker genes Pathogenesis-related protein 1 (PR-1, gene AtPR-1) and PR-5 (gene AtPR-5) in wild type plants (Fig. 6c-d). Similar to treatment with avirulent P. syringae, treatment with Si(OH)4 and SiO2-NPs resulted in an up to 30-fold and 6-fold increase in transcript abundance of AtPR-1 (Fig. 6c) and AtPR-5 (Fig. 6d), respectively, in comparison to control treatments. Hence, both Si(OH)4 and SiO2-NPs activated salicylic acid-dependent defence reactions. Although SiO2-NPs triggered lower AtPR-1 and AtPR-5 expression levels in comparison with avirulent P. syringae infiltrated plants and Si(OH)4 treated plants, the inducing effect was sufficient to confer SAR.
Implications on the Mode of Action of Leaf-Applied SiO2-NPs
The pathosystem involving Arabidopsis and the hemibiotrophic bacterial pathogen P. syringae offers an ideal model to investigate the effect of SiO2-NPs and Si(OH)4 on plant defence. Our results summarized in the model in Fig. 5b-c show that the protective effect of SiO2-NPs and Si(OH)4 is based on the ability to induce basal resistance and SAR (Fig. 3a-c) and not on direct toxic effects as neither SiO2-NPs nor Si(OH)4 inhibited bacterial growth (Fig. 3d). Our data is in line with initial results that suggested that Si(OH)4 and sometimes SiO2-NPs can protect plants from different plant pathogens7,26,27,37, nevertheless here we show that the mechanism involved no toxic effect on the pathogen, but rather induced the defences of the plant.
Both SiO2-NPs and Si(OH)4 induce SAR in a dose-dependent manner, leading to bacterial inhibition of >90% compared to control plants treated with HEPES buffer or MgCl2 only. These results are consistent with previous results suggesting that SiO2-NPs and Si(OH)4 function in a dose-dependent manner in plants and animals26,27,38. However, instead of the previously proposed pesticidal action of SiO2-NPs, we show here that the nanoparticles caused an increase of the plant defence. Our data suggest that the SiO2-NPs used in the present study can be successfully used to slowly release Si(OH)4 to the plant from within the spongy mesophyll (Fig. 2) in close direct interaction with the diffusion layer on the plant cell walls, which is at least partially responsible for the SAR-inducing ability of SiO2-NPs. Water (vapour) secreted from the plant cell wall, or plant-induced dissolution of SiO2-NPs linked to increased secretory activity10 (exudates) may have promoted further dissolution of Si(OH)4. Based on the release rates of SiO2-NPs that were determined earlier under conditions optimized for dissolution in a continuously depleted ultrapure water system (half-life of ~66d at pH 7)32, max. ~13% of the particles could have dissolved within the 48 h of SiO2-NP exposure. Si-containing reaction byproducts of the nanoparticle synthesis were ruled out to play a significant role in the induction of defence (refer to section ‘Si Reaction Byproducts’ in the Supplemental Information). The max. released Si(OH)4 from SiO2-NPs could thus explain the bacterial inhibition; however, it cannot fully explain the lack of oxidative stress responses and higher bacterial DNA levels for SiO2-NPs in the plants (Fig. 4b). Probably, the absence of peak Si(OH)4 concentrations resulted in lower Si(OH)4 toxicity for both bacteria and plants. Other effects, such as modulated evapotranspiration due to the blockage and incomplete closure of the stomata by the nanoparticles (Fig. 2) which can cause salicylic-acid related responses similar to drought stress39, and the close interaction of the nanoparticles with cells in the spongy mesophyll may play an important role, in line with earlier research about stomata as ports of entry for pollutants and nanoparticles40,41. The exact relative contribution of each effect remains to be elucidated in follow-up studies. It is important to note that the cell walls in the mesophyll air spaces have very thin, or lack, cuticular waxes10, and therefore, in contrast to the external leaf surface, a direct interaction of the nanoparticles can take place with the cell wall and thus apoplast transport system including the xylem. Irrespective of the detailed mechanism of the nanoparticles, this is of importance for any nanoagrochemical application aiming at slow release of active ingredients, because nanoparticles in the extracellular spongy mesophyll air spaces (Fig. 2, Supplementary Fig. S1) can interact with the leaf for extended periods without being washed away by rain.
High concentrations of Si(OH)4 caused chlorotic yellowing of leaves indicative of stress (Fig. 3e). Increased expression of the oxidative stress marker gene AtHSP17.4C1 34 confirmed stress in the Si(OH)4 treatment at 100 and 1000 mg SiO2 L-1, as transcript levels of AtHSP17.4C1 were more strongly induced in comparison with avirulent P. syringae, or SiO2-NP treatments (Fig. 4c). Together, these data show that the Si(OH)4 was more toxic to plants than SiO2-NPs. Hence, impaired SAR in plants treated with high concentrations of Si(OH)4 (Fig. 4a) might be linked to enhanced oxidative stress, consistent with the fact that high levels of NO and reactive oxygen species were shown to impair the induction of SAR19,23. For SiO2-NPs, no significant increase of the oxidative stress marker gene was found (Fig. 4c). Impaired SAR for SiO2-NPs occurred only at very high concentrations in the g L-1 range (Fig. 4a), likely due to excess release of Si(OH)4 causing oxidative stress, or too intense clogging of the stomata (Fig. 2) disrupting evapotranspiration. While the low polydispersity index measured by DLS (Fig. 1c) indicates well-dispersed SiO2-NP suspensions even at high concentrations, upon contact with the leaf, heteroaggregation with mucilage in stomata, and at higher nanoparticle concentrations, probably also homoaggregation, appeared to promote the clogging of the stomata (Fig. 2a, red arrows). These results are in line with Slomberg et al.8, who found that SiO2-NP concentrations up to 1000 mg SiO2 L-1 were not phytotoxic despite the uptake of the SiO2-NPs into the root system of A. thaliana. Our results are also consistent with initial studies42,43 that found better effects of SiO2-NPs on plant growth than conventional silica fertilizers. In conclusion, the application of SiO2-NPs can reduce the risk of overdosage.
Our data demonstrate that SiO2-NPs and Si(OH)4-mediated SAR acts via the activation of the salicylic acid-dependent defence pathway, which is a key component of basal disease resistance and SAR44,45. Neither SiO2-NPs nor Si(OH)4 induced resistance in the salicylic acid-deficient mutant sid2 that has a defect in salicylic acid biosynthesis (Fig. 6a-b). The induction of resistance by SiO2-NPs was comparable to the effect of Si(OH)4 at intermediate concentrations, albeit the soluble fraction of Si(OH)4 in this treatment was far lower, as the particles dissolved only partially in the plant, if at all (Fig. 2), suggesting that SiO2-NPs can induce salicylic acid-dependent defence pathways as intact particles. Furthermore, the expression levels of two salicylic acid-responsive marker genes AtPR-1 and AtPR-5 encoding the Pathogenesis-related protein 1 (PR-1) and Pathogenesis-related protein 5 (PR-5) were induced in response to SiO2-NPs and Si(OH)4 (Fig. 6c-d). These results are in line with Fauteux et al.46, who reported that exogenous application of Si(OH)4 induced salicylic acid biosynthesis in leaves exposed to the fungal pathogen Erysiphe cichoracearum. In addition, silicon-primed tomato plants were protected against Ralstonia solanacearum via the upregulation of salicylic acid-controlled defence gene expression47. Although SiO2-NPs triggered lower AtPR-1 and AtPR-5 expression levels than plants infiltrated with avirulent P. syringae and Si(OH)4-treated plants, the achieved level of expression was sufficient to confer a full SAR response.
Conclusions
The present results show that low concentrations of SiO2-NPs efficiently protect the widely used model plant Arabidopsis from infection by the bacterial pathogen Pseudomonas, and revealed the mode of action of SiO2-NPs compared to its dissolved counterpart Si(OH)4. The protective effect of SiO2-NPs is mediated by the activation of salicylic acid-dependent plant immunity responses and is partially based on the slow release of Si(OH)4 from nanoparticles entering through the stomata and distribution within the spongy mesophyll, and likely partially by intact nanoparticle-induced salicylic acid-dependent responses.
In comparison to direct Si(OH)4 application, SiO2-NPs proved to be safer for the plant. They did not cause phytotoxicity even at concentrations 10-fold higher than the minimal dose needed for plant protection, and therefore have a broader therapeutic range than Si(OH)4. The lowest fully effective dose (100 mg SiO2 L-1) is promising because it corresponds to an extrapolated field dose of only 3 kg SiO2 kg ha-1, corresponding to >1000-fold material savings compared to solid bulk SiO2 treatments. This calculation assumes a typical 300 L ha-1 application (conventional aqueous spray volumes for pesticide application equipment48), and an uncertainty factor of 100 for the concentration. Contrary to previous assumptions about the ability of nanoparticles to penetrate the cuticle, SiO2-NP intake was clearly restricted to the stomata and extracellular spongy mesophyll, confirming our hypothesis that the leaf cuticle represents an impermeable barrier to nanoparticles10, in line with earlier fundamental research49. The spongy mesophyll is an attractive target for long-term deposition of slow-release nanoagrochemicals. Future research should extend the investigations to a broader spectrum of defence-related genes with other plant pathogens, and to biomechanic quantification of the physical effects of nanoparticles that affect leaf permeability and may trigger the salicylic acid-related responses. To further advance SiO2-NPs as nanobiostimulants and fertilizers, as this should be the case with every material or organism used in agriculture, the long-term effects of SiO2-NPs to occupationally exposed agricultural workers and non-target organisms, such as beneficial soil microorganisms or bees, must be thoroughly analysed before broad commercial application. The potential risks of nanoagrochemicals, and possible strategies for risk mitigation have been thoroughly reviewed previously1,50,51. As for amorphous SiO2-NPs, they have already been approved by the FDA as they are generally regarded as safe, and they are in use as dietary additives (E551)52 in a broad range of foodstuffs such as table salt. The daily intake of nano-scale silica from food is estimated to be 1.8 mg kg-1 53. Our own initial experiments with C. elegans nematodes used as model non-target microorganisms (Supplementary Fig. S3) have shown a ~36-fold lower ecotoxicity of SiO2-NPs compared to liquid Si(OH)4 preparations that are in use for plant nutrition since decades. Thus, compared to currently used treatments, the present SiO2-NPs alone, or in combination with other active ingredients, promise to offer a cost-effective, consumer-safe because tracelessly degradable, and sustainable alternative to protect plants against pathogens via the controlled induction of SAR, without negative effects on yield or non-target organisms that were associated with the action of previously described plant biostimulants or pesticides.
Methods
Plant Growth Conditions
Arabidopsis thaliana seeds were grown on Jiffy soil substrates (powered by Tref, Jiffy Products International B.V.). Two A. thaliana strains were grown: wild type Columbia (Col-0) plants that carry an RPS2 locus responsible for the recognition of P. syringae strains expressing the avirulence gene avrRpt2 28,29; and an A. thaliana mutant defective in salicylic acid biosynthesis (salicylic acid deficient 2, sid2 36). The seeds sown on the soil were kept at 4 °C for two days and then transferred to the growth chamber (RMC Tableaux SA). The plants were grown in a 12 h photoperiod with 60% of relative humidity, with a day temperature of 22 °C and a night temperature of 18 °C (photon flux density 100 μmmol m-2 s-1). The transplanted seedlings were covered with transparent plastic domes for 2-3 days to allow the seedlings to adapt to the new soil. Four-to five-week-old plants were used in experiments because previous experiments had shown that under the abovementioned growth conditions, this is the optimal age of the plant to induce SAR54.
Culture of Pseudomonas syringae pv. tomato
Pseudomonas syringae pv. tomato bacteria were prepared by inoculating a single colony in 10 mL of King’s B medium (1.5 g K2HPO4, 1.5 g MgSO4·7H2O, 20 g tryptone, 10 mL glycerol per 1 L of water, all from Sigma Aldrich, Switzerland, purity ≥99%) containing the appropriate antibiotics. A virulent and an avirulent strain of P. syringae were grown: P. syringae DC3000 (here termed virulent P. syringae); and P. syringae DC3000 expressing the avirulence gene avrRpt2 recognized by the A. thaliana RPS2 locus and inducing systemic acquired resistance (here termed avirulent P. syringae). The virulent P. syringae bacteria strain served to induce a strong infection with P. syringae in the plants. The avirulent P. syringae strain served as a positive control to induce systemic acquired resistance and thus an actively suppressed bacterial growth in the A. thaliana plants via recognition of the bacterial avrRpt2 gene by the plant’s RPS2 gene (refer to Chen et al. 2000 for a detailed description of the pathosystem29). The virulent P. syringae was grown with rifampicin (25 μg mL-1), and the avirulent P. syringae was grown with kanamycin (50 μg mL-1) and rifampicin (25 μg mL-1). After overnight incubation on a shaker at 28 °C in the dark (Kuhner LT-W Lab Therm Table Top Incubator Shaker, Adolf Kühner AG, Switzerland), the cells were centrifuged at 3000 rpm for 10 min, and the pellet was suspended in 10 mM MgCl2. The cell density was calculated via measurement of the light absorption of the liquid culture at the absorption wavelength 600 nm using a spectrophotometer (BioPhotometer, Eppendorf – Netheler - Hinz GmbH, Germany) and by counting the colonies plated on King’s B agar (raw data publicly available 55).
Inoculation Procedures for Local Disease Resistance
For a local disease resistance assay, three leaves per A. thaliana plant were inoculated with virulent P. syringae bacteria, and the plants were incubated at the standard A. thaliana growth conditions described above. The inoculation with the virulent P. syringae bacteria was operationally defined as 0 days post inoculation. After inoculation, leaf discs (4 mm) were harvested from the inoculated leaves at 0 and 3 days post inoculation using a cork borer (3 leaf discs from different plant leaves/sample). The leaf discs were ground and homogenized with pestles in 10 mM MgCl2 and the undiluted (0 days post inoculation) or the 103 fold diluted (3 days post inoculation) homogenates were plated on King’s B agar plates (King’s B medium as above with 15 g L-1 agar). The plates were incubated at 28°C in the dark for 48 h. Then, the bacterial colonies were counted (raw data publicly available 55).
Inoculation Procedures for Systemic Acquired Resistance Assays
For a systemic acquired resistance (SAR) assay, three leaves of 4-5 week old wild type Col-0 plants were infiltrated with 10 mM MgCl2 (negative control) or the avirulent P. syringae bacteria at 106 colony-forming unit (CFU) per millilitre in 10 mM MgCl2 (positive control). After 48 h, the distal leaves were inoculated with the virulent P. syringae bacteria (105 CFU mL-1). The inoculation with the virulent P. syringae bacteria was operationally defined as 0 days post inoculation. Leaf discs (4 mm) were harvested from the distal leaves at 0 and 3 days post inoculation using a cork borer (three times three leaf discs from different plant leaves were analysed for each treatment). The leaf discs were ground in 10 mM MgCl2, and the undiluted (0 days post inoculation) or the 103 fold diluted (3 days post inoculation) homogenates were plated on King’s B agar and incubated at 28°C for 48 h in the dark (Salvis incubator, Switzerland). Then, the bacterial colonies were counted (raw data publicly available 55). For details about this procedure, refer to Wang et al. 2014.
Plant Treatments
The SiO2-NPs (25, 100, 400 and 1600 mg SiO2 L-1, pH 7.0) and Si(OH)4 (5, 20, 80, 100, 320, 640 and 2560 mg SiO2 L-1, pH 7.0, from an aqueous potassium silicate stock solution, K2O:SiO2 1:2.60, SiO2 content 20.8 wt%, MonDroguiste, France) were prepared in in sterile distilled water in 4-(2 hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer (1-mM, pH 7.0, Sigma-Aldrich, 99.5%). The Si(OH)4 concentrations were expressed in mg SiO2 L-1 to allow for direct comparison of the effects of dissolved Si(OH)4 and solid SiO2-NPs without having to take into account the different molecular weights.
For the local disease resistance assay, the plants were sprayed with these chemicals 24 h before inoculation with virulent P. syringae. For the SAR assays, all these chemicals were injected abaxially (from the bottom of the leaf) into Arabidopsis plant leaves 2 d before inoculation using 1 mL needleless sterile disposable syringes.
SiO2-NPs and Subcellular Distribution within the Leaf
The SiO2-NPs were synthesized and characterized according to a previously established procedure31,32 adapted from earlier work56. Briefly, one equivalent of tetraethyl orthosilicate (TEOS, 10 mL, Sigma-Aldrich, >99%) was added to an equilibrated reaction mixture at 70°C containing two equivalents of ultrapure water (Milli-Q, 18.2 MΩ arium 611 DI, Sartorius Stedim Biotech, Germany), and absolute ethanol (81 mL) as a solvent under basic conditions (2.93 mL 25% NH3). The particles resulting after 3 h of hydrolysis and polycondensation of TEOS were washed by three steps of centrifugation (15 000 × g, 15 min) in ultrapure water, and by 5 or more steps of dialysis through a 14 kDa molecular weight cut-off membrane (regenerated cellulose, Carl Roth, Germany). Several batches of particles 64.8-76.7 nm in hydrodynamic diameter were prepared using the identical procedure to prevent artefacts due to suspension aging (size variability between batches 5.2 nm). Dynamic light scattering was used to quantify the hydrodynamic particle size and surface charge of diluted samples (1% v/v, Brookhaven Particle Size Analyzer Plus90, USA, scattering angle 90°, 1 min acquisition, raw data publicly available 55). Inductively coupled plasma - optical emission spectroscopy (ICP-OES) and gravimetry served to quantify the SiO2 concentration (methods described in Bossert et al.31).
For the particle characterization, and to analyse the effects of SiO2-NPs, Si(OH)4, and control treatments in the leaves, we used transmission electron microscopy (TEM). The particle size distribution was established using ImageJ (version 1.52n) analysis of TEM micrographs (raw data publicly available 55). The plants were pre-fixed in 4% glutaraldehyde solution, gently stained in the dark with 1% OsO4 solution that was centrifuged beforehand to remove potential precipitates, dehydrated using an ethanol series, and embedded in polymer resin (AGAR Low Viscosity Kit, Plano, Germany) without further staining according to a procedure described in detail in Stegemeier et al. 201557. The correct position of the stomata to cut cross-sections were identified by light microscopy examination of semi-thin resin sections before the ultramicrotoming. The TEM images were taken on an FEI Tecnai Spirit at an acceleration voltage of 120 kV (2048 × 2048 px. Resolution, Veleta CCD camera, Olympus). Besides cropping and adjustment of brightness and contrast, the micrographs were not further processed; the unprocessed raw data is publicly available55.
DNA Extraction
Plant leaf samples (five leaf discs from different inoculated plant leaves/sample) were frozen in liquid nitrogen and were homogenized using a ceramic mortar and pestle. The total DNA was extracted with a Plant DNA mini Kit (peqlab, a VWR brand, Germany). More information about the sample preparation is available in section ‘Details on DNA Extraction’ in the Supplemental Information.
RNA Extraction and complementary DNA Synthesis
Plant leaf samples (10 leaf discs taken from different infiltrated plant leaves/sample) were flash-frozen in liquid nitrogen, and the total RNA was extracted with the Spectrum Plant Total RNA Kit (Sigma Life Science, USA). One microgram of the total RNA was used for complementary DNA synthesis using the Omniscript Reverse Transcription Kit (Qiagen, Germany). More information about the sample preparation is available in section ‘Details on RNA Extraction and complementary DNA Synthesis’ in the Supplemental Information.
Quantitative real-time polymerase chain reaction (qPCR)
To validate the SAR response based on bacterial colony counts, the bacteria were also quantified via the outer membrane protein oprF gene of P. syringae in inoculated leaves (raw data publicly available 55) based on a previously established method18,33. For this bacterial DNA quantification, a reaction mixture for qPCR was prepared with 7.5 μL of 2× SensiMix™ SYBR Hi-ROX Mastermix (No. QT605-05, Bioline, Meridian Bioscience, UK), 5 μL of plant DNA, and 0.5 μL of each primer (Supplementary Tab. S1) at a concentration of 10 μM in a final volume replenished with water to 15 μL in MIC tubes (Bio Molecular Systems, Australia). Runs were performed on a MIC qPCR machine (Bio Molecular Systems, Châtel-St-Denis, Switzerland). The conditions for the qPCR were 10 min. at 95 °C initial denaturation, and then 40 cycles (95 °C for 15 s, 62 °C for 1-min, and 72 °C for 30 s). The final PCR products were analyzed by melting point analysis. The qPCR analysis software for the melting curve analysis and amplification efficiency calculation was micPCR v2.8.13 from Bio Molecular Systems. This software is designed to meet MIQE58 specifications and performs qPCR analysis automatically based on the real time runs. Five leaf discs from different plant leaves were sampled per each replicate, frozen in liquid nitrogen, and processed immediately for DNA extraction. The bacterial DNA levels of the bacterial oprF gene in Arabidopsis plants were calculated using At4g26410 (expG) as a reference gene33 and the comparative cycle threshold method (2(-ΔΔCt))59.
For oxidative stress and salicylic acid-responsive plant transcript levels, leaf discs were flash-frozen in liquid nitrogen and stored at -80°C for <24 h before being processed for RNA extraction and complementary DNA synthesis. Three independent technical replicates (ten leaf discs taken from different plant leaves) were used per treatment. The reaction mixture for RT-qPCR contained 7.5 μL of 2× SensiMix™ SYBR Hi-ROX Mastermix (No. QT605-05, Bioline, Meridian Bioscience, UK), 5 μL of complementary DNA (corresponding to 25 ng RNA), and 0.5 μL of each primer (Supplementary Tab. S1) at a concentration of 10 μM in a final volume replenished with water to 15 μL in MIC tubes (Bio Molecular Systems, Australia). Runs were performed on a MIC qPCR machine (Bio Molecular Systems, Châtel-St-Denis, Switzerland). The conditions for the qPCR and the analysis of the final PCR products by melting point analysis were analogous to the above bacterial DNA quantification. The final PCR products were analyzed by melting point analysis. The transcript levels of the oxidative stress marker (At3g46230; HSP17.4C1)34, and the salicylic acid-responsive genes AtPR-1 and AtPR-5 in Arabidopsis plants were calculated with At4g26410 (expG) as reference gene60 and the comparative cycle threshold method (2(-ΔΔCt)) as mentioned above.
The expG gene was selected because Czechowski et al.60 specifically recommends expG as one of the top five reference genes to be used in biotic stress studies due to its high stability under such conditions. This high stability was confirmed in previous work of our laboratory61 and others’ work33. In the present study, the stable expression of expG is reflected in the very small variation of the Cq for expG. In the PR1 expression experiments (Fig. 6c) for example, the average Cq ranged from 23.19-23.93 for all different tested conditions with an average relative error of only 0.63% 55. The amplification efficiencies were all very close to two and with good comparability of the reference gene to the target gene. For example, in Fig. 6c, the average amplification efficiency of expG and AtPR-1 across all the different treatment conditions (1.949±0.011 vs. 1.962±0.027) differed by only 0.7% 55. All statistical tests hereinafter were performed using IBM SPSS Statistics (version 22).
Ecotoxicity of SiO2-NPs and Si(OH)4 to C. elegans larvae
The ecotoxicity assays were conducted on larval stage one (L1) nematodes of the C. elegans wild type (ancestral; N2) genotype. Synchronized C. elegans larvae were grown according to a previously established protocol62 (raw data publicly available 55). A known number of larvae (~70) per replicate were then exposed to 0, 25, 125, 250, 500, 750, 1000, 1500, or 2000 mg SiO2 L-1 of SiO2-NPs or Si(OH)4 in 96-well plates (Corning Costar No. 3596). A 0.1% NaN3 solution served as a positive control. As a food source for the nematodes, the wells contained 10 μL of living Escherichia coli (strain OP50; final optical density at 600 nm 1 a. u., ~5×108 cells mL-1). The total volume per well was 100 μL, and the final pH installed in the phosphate-buffered saline test solutions was 7.4. After incubating the nematodes at 20 °C for 48 h in the dark, the surviving larvae were counted under a stereo microscope at 20× magnification. The resulting number of mobile nematode larvae was subtracted from the initially incubated number of larvae to calculate the percentage of immobile nematodes. The effective concentrations at 50% (EC50) were calculated using a numerically fitted standard log-logistic dose-response model (Levenberg-Marquardt iteration algorithm, Origin 2016, build 9.3.2.903, OriginLab Corporation, MA USA, Supplementary Fig. S3). The experiment comprised twelve biological replicates for each treatment and was repeated twice with comparable results.
Supplementary Material
Supplementary information is available in the online version of the paper.
Acknowledgments
M.E.-S. was supported by the Swiss State Secretariat for Education, Research, and Innovation by a Swiss Government Excellence Scholarship for Foreign Scholars. F.S. and M.M. were supported by the Swiss National Science Foundation under the Ambizione grant “Enhancing Legume Defenses” (168187) and Innosuisse (project 38515.1 IP-EE). We are grateful to Nicola F. Schäppi for his help with the graphic design and to Martine Schorderet for excellent technical assistance with microtoming. This research was also supported by the National Center of Competence in Research “Bioinspired Materials”, the Adolphe Merkle Foundation, and the University of Fribourg.
Footnotes
Author Contributions
M.E.-S., F.S., and F.M. conceived and designed the study. F.S. led the team, rationally designed the SiO2-NPs to induce optimal plant defence, performed initial germination tests to establish the dosing regimen, contributed the mechanistic understanding of silica and with plant TEM, and has drawn the artwork. M.M. synthesized and characterized the SiO2-NPs. A.M. cultured the Arabidopsis plants and conducted the C. elegans experiments. D.R. provided access to his microtome and a technician that trained F.S. in microtoming. M.E.-S. performed all the Arabidopsis experiments and their statistical evaluation and wrote the manuscript draft with contributions by F.S. (figures and text) and F.M. (text). F. M. contributed the mechanistic understanding of the gene expression results and molecular mechanisms of SAR. The manuscript was critically reviewed by A.P.-F., B.R.-R., and D.R. All co-authors read and approved the manuscript prior to submission.
Competing Interests
Any opinions, findings, conclusions, or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the Swiss NSF or the government. This work has not been subjected to SNSF review, and no official endorsement should be inferred. F.S. and M.M. have a patent pending on a SiO2-NP plant growth enhancer. F.S. was supported by Innosuisse (project No. 38515.1 IP-EE). Other than that, the authors have declared no conflict of interest and are responsible for the content and writing of the article.
Reprints and permission information is available online at http://www.nature.com/reprints.
Data Availability
The datasets that support the findings of the current study are available in the Zenodo repository with the identifier doi:10.5281/zenodo.4131137 at https://doi.org/10.5281/zenodo.4131137. Additional data related to this study are available from M. El-Shetehy and F. Schwab upon reasonable request.
References
- 1.White JC, Gardea-Torresdey J. Achieving food security through the very small. Nature Nanotechnology. 2018;13:627–629. doi: 10.1038/s41565-018-0223-y. [DOI] [PubMed] [Google Scholar]
- 2.Casey W, Kinrade S, Knight C, Rains D, Epstein E. Aqueous silicate complexes in wheat, Triticum aestivum L. Plant, Cell Environ. 2004;27:51–54. [Google Scholar]
- 3.Ma JF, Yamaji N. Silicon uptake and accumulation in higher plants. Trends Plant Sci. 2006;11:392–397. doi: 10.1016/j.tplants.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 4.Choppin GR, Pathak P, Thakur P. Polymerization and complexation behavior of silicic acid: A review. Main Group Met Chem. 2008;31:53. [Google Scholar]
- 5.Bélanger RR, Bowen PA, Ehret DL, Menzies JG. Soluble silicon - its role in crop and disease management of greenhouse crops. Plant Dis. 1995;79:329–336. [Google Scholar]
- 6.Abdel-Haliem ME, Hegazy HS, Hassan NS, Naguib DM. Effect of silica ions and nano silica on rice plants under salinity stress. Ecol Eng. 2017;99:282–289. [Google Scholar]
- 7.Luyckx M, Hausman J-F, Lutts S, Guerriero G. Silicon and plants: current knowledge and technological perspectives. Front Plant Sci. 2017;8:411. doi: 10.3389/fpls.2017.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Slomberg DL, Schoenfisch MH. Silica nanoparticle phytotoxicity to Arabidopsis thaliana. Environ Sci Technol. 2012;46:10247–10254. doi: 10.1021/es300949f. [DOI] [PubMed] [Google Scholar]
- 9.Eichert T, Kurtz A, Steiner U, Goldbach HE. Size exclusion limits and lateral heterogeneity of the stomatal foliar uptake pathway for aqueous solutes and water–suspended nanoparticles. Physiol Plant. 2008;134:151–160. doi: 10.1111/j.1399-3054.2008.01135.x. [DOI] [PubMed] [Google Scholar]
- 10.Schwab F, et al. Barriers, pathways and processes for uptake, translocation and accumulation of nanomaterials in plants–Critical review. Nanotoxicology. 2016;10:257–278. doi: 10.3109/17435390.2015.1048326. [DOI] [PubMed] [Google Scholar]
- 11.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 12.Conrath U, et al. Priming: getting ready for battle. Mol Plant-Microbe Interact. 2006;19:1062–1071. doi: 10.1094/MPMI-19-1062. [DOI] [PubMed] [Google Scholar]
- 13.Mauch-Mani B, Baccelli I, Luna E, Flors V. Defense priming: an adaptive part of induced resistance. Annu Rev Plant Biol. 2017;68:485–512. doi: 10.1146/annurev-arplant-042916-041132. [DOI] [PubMed] [Google Scholar]
- 14.Ryals JA, et al. Systemic acquired resistance. The Plant Cell. 1996;8:1809. doi: 10.1105/tpc.8.10.1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ross AF. Systemic acquired resistance induced by localized virus infections in plants. Virology. 1961;14:340–358. doi: 10.1016/0042-6822(61)90319-1. [DOI] [PubMed] [Google Scholar]
- 16.Durrant WE, Dong X. Systemic acquired resistance. Annu Rev Phytopathol. 2004;42:185–209. doi: 10.1146/annurev.phyto.42.040803.140421. [DOI] [PubMed] [Google Scholar]
- 17.Mauch F, et al. Manipulation of salicylate content in Arabidopsis thaliana by the expression of an engineered bacterial salicylate synthase. The Plant Journal. 2001;25:67–77. doi: 10.1046/j.1365-313x.2001.00940.x. [DOI] [PubMed] [Google Scholar]
- 18.Wang C, et al. Free radicals mediate systemic acquired resistance. Cell Reports. 2014;7:348–355. doi: 10.1016/j.celrep.2014.03.032. [DOI] [PubMed] [Google Scholar]
- 19.El-Shetehy M, et al. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signaling & Behavior. 2015;10:e998544. doi: 10.1080/15592324.2014.998544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Louws F, et al. Field control of bacterial spot and bacterial speck of tomato using a plant activator. Plant Dis. 2001;85:481–488. doi: 10.1094/PDIS.2001.85.5.481. [DOI] [PubMed] [Google Scholar]
- 21.Romero A, Kousik C, Ritchie D. Resistance to bacterial spot in bell pepper induced by acibenzolar-S-methyl. Plant Dis. 2001;85:189–194. doi: 10.1094/PDIS.2001.85.2.189. [DOI] [PubMed] [Google Scholar]
- 22.Kim SG, Kim KW, Park EW, Choi D. Silicon-Induced Cell Wall Fortification of Rice Leaves: A Possible Cellular Mechanism of Enhanced Host Resistance to Blast. Phytopathology. 2002;92:1095–1103. doi: 10.1094/PHYTO.2002.92.10.1095. [DOI] [PubMed] [Google Scholar]
- 23.Wang M, et al. Role of silicon on plant–pathogen interactions. Front Plant Sci. 2017;8:701. doi: 10.3389/fpls.2017.00701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang Y, Si J, Römheld V. Silicon uptake and transport is an active process in Cucumis sativus. New Phytol. 2005;167:797–804. doi: 10.1111/j.1469-8137.2005.01463.x. [DOI] [PubMed] [Google Scholar]
- 25.van Bockhaven J, et al. Silicon induces resistance to the brown spot fungus Cochliobolus miyabeanus by preventing the pathogen from hijacking the rice ethylene pathway. New Phytol. 2015;206:761–773. doi: 10.1111/nph.13270. [DOI] [PubMed] [Google Scholar]
- 26.Rouhani M, Samih M, Kalantari S. Insecticidal effect of silica and silver nanoparticles on the cowpea seed beetle, Callosobruchus maculatus F.(Col.: Bruchidae) J Entomol Res. 2013;4:297–305. [Google Scholar]
- 27.El-Helaly A, El-Bendary H, Abdel-Wahab A, El-Sheikh M, Elnagar S. The silica-nano particles treatment of squash foliage and survival and development of Spodoptera littoralis (Bosid.) larvae. J Entomol Zool. 2016;4:175–180. [Google Scholar]
- 28.Kunkel BN, Bent AF, Dahlbeck D, Innes RW, Staskawicz BJ. RPS2, an Arabidopsis disease resistance locus specifying recognition of Pseudomonas syringae strains expressing the avirulence gene avrRpt2. The Plant Cell. 1993;5:865–875. doi: 10.1105/tpc.5.8.865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen Z, Kloek AP, Boch J, Katagiri F, Kunkel BN. The Pseudomonas syringae avrRpt2 gene product promotes pathogen virulence from inside plant cells. Mol Plant-Microbe Interact. 2000;13:1312–1321. doi: 10.1094/MPMI.2000.13.12.1312. [DOI] [PubMed] [Google Scholar]
- 30.Exley C. A possible mechanism of biological silicification in plants. Front Plant Sci. 2015;6:853. doi: 10.3389/fpls.2015.00853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bossert D, et al. A hydrofluoric acid-free method to dissolve and quantify silica nanoparticles in aqueous and solid matrices. Sci Rep. 2019;9:1–12. doi: 10.1038/s41598-019-44128-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwab F, Maceroni M. A controlled release silica-based nanoparticle composition, method of production and fertilization methods. 2019 pending patent No. 62/835,030; PCT/EP2020/060765. [Google Scholar]
- 33.Ross A, Somssich IE. A DNA-based real-time PCR assay for robust growth quantification of the bacterial pathogen Pseudomonas syringae on Arabidopsis thaliana. Plant Methods. 2016;12:48. doi: 10.1186/s13007-016-0149-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sewelam N, Kazan K, Hüdig M, Maurino VG, Schenk PM. The AtHSP17. 4C1 gene expression is mediated by diverse signals that link biotic and abiotic stress factors with ROS and can be a useful molecular marker for oxidative stress. Int J Mol Sci. 2019;20:3201. doi: 10.3390/ijms20133201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.An C, Mou Z. Salicylic acid and its function in plant immunity. J Integr Plant Biol. 2011;53:412–428. doi: 10.1111/j.1744-7909.2011.01043.x. [DOI] [PubMed] [Google Scholar]
- 36.Nawrath C, Métraux J-P. Salicylic acid induction–deficient mutants of Arabidopsis express PR-2 and PR-5 and accumulate high levels of camalexin after pathogen inoculation. The Plant Cell. 1999;11:1393–1404. doi: 10.1105/tpc.11.8.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye M, et al. Priming of jasmonate-mediated antiherbivore defense responses in rice by silicon. Proc Natl Acad Sci USA. 2013;110:E3631–E3639. doi: 10.1073/pnas.1305848110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ziaee M, Ganji Z. Insecticidal efficacy of silica nanoparticles against Rhyzopertha dominica F. and Tribolium confusum Jacquelin du Val. J Plant Prot Res. 2016;56:250–256. [Google Scholar]
- 39.La VH, et al. Salicylic acid improves drought-stress tolerance by regulating the redox status and proline metabolism in Brassica rapa. Horticulture, Environment, and Biotechnology. 2019;60:31–40. [Google Scholar]
- 40.Krajíčková A, Mejstřik V. The effect of fly ash particles on the plugging of stomata. Environ Pollut A. 1984;36:83–93. [Google Scholar]
- 41.Burkhardt J, Basi S, Pariyar S, Hunsche M. Stomatal penetration by aqueous solutions—an update involving leaf surface particles. New Phytol. 2012;196:774–787. doi: 10.1111/j.1469-8137.2012.04307.x. [DOI] [PubMed] [Google Scholar]
- 42.Amrullah DS, Junaedi A. Influence of nano-silica on the growth of rice plant (Oryza sativa L.) Asian J Agric Res. 2015;9:33–37. [Google Scholar]
- 43.Karunakaran G, et al. Effect of nanosilica and silicon sources on plant growth promoting rhizobacteria, soil nutrients and maize seed germination. IET Nanobiotechnology. 2013;7:70–77. doi: 10.1049/iet-nbt.2012.0048. [DOI] [PubMed] [Google Scholar]
- 44.Cameron RK, Pavia NK, Lamb CJ, Dixon RA. Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol Mol Plant Pathol. 1999;55:121–130. [Google Scholar]
- 45.Malamy J, Carr JP, Klessig DF, Raskin I. Salicylic acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science. 1990;250:1002–1004. doi: 10.1126/science.250.4983.1002. [DOI] [PubMed] [Google Scholar]
- 46.Fauteux F, Chain F, Belzile F, Menzies JG, Bélanger RR. The protective role of silicon in the Arabidopsis–powdery mildew pathosystem. Proc Natl Acad Sci USA. 2006;103:17554–17559. doi: 10.1073/pnas.0606330103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang N, Fan X, Lin W, Wang G, Cai K. Transcriptome analysis reveals new insights into the bacterial wilt resistance mechanism mediated by silicon in tomato. Int J Mol Sci. 2019;20:761. doi: 10.3390/ijms20030761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lavers A. Guidelines on Good Practice for Ground Application of Pesticides. Food and Agriculture Organization (FAO) of the United Nations; Rome: 2001. [Google Scholar]
- 49.Schreiber L. Polar paths of diffusion across plant cuticles: New evidence for an old hypothesis. Ann Bot. 2005;95:1069–1073. doi: 10.1093/aob/mci122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kookana RS, et al. Nanopesticides: guiding principles for regulatory evaluation of environmental risks. J Agric Food Chem. 2014;62:4227–4240. doi: 10.1021/jf500232f. [DOI] [PubMed] [Google Scholar]
- 51.Kah M, Tufenkji N, White JC. Nano-enabled strategies to enhance crop nutrition and protection. Nat Nanotechnol. 2019;14:532–540. doi: 10.1038/s41565-019-0439-5. [DOI] [PubMed] [Google Scholar]
- 52.Bourquin J, et al. Biodistribution, clearance, and long-term fate of clinically relevant nanomaterials. Advanced Materials. 2018;30:e1704307. doi: 10.1002/adma.201704307. [DOI] [PubMed] [Google Scholar]
- 53.Mebert AM, Baglole CJ, Desimone MF, Maysinger D. Nanoengineered silica: Properties, applications and toxicity. Food Chem Toxicol. 2017;109:753–770. doi: 10.1016/j.fct.2017.05.054. [DOI] [PubMed] [Google Scholar]
- 55.El-Shetehy M, et al. Silica nanoparticles enhance disease resistance in Arabidopsis plants - raw data. Zenodo. 2020 doi: 10.5281/zenodo.4131137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stöber W, Fink A, Bohn E. Controlled growth of monodisperse silica spheres in the micron size range. J Colloid Interface Sci. 1968;26:62–69. [Google Scholar]
- 57.Stegemeier JP, et al. Speciation matters: Bioavailability of silver and silver sulfide nanoparticles to alfalfa (Medicago sativa) Environ Sci Technol. 2015;49:8451–8460. doi: 10.1021/acs.est.5b01147. [DOI] [PubMed] [Google Scholar]
- 58.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 59.Rao X, Huang X, Zhou Z, Lin X. An improvement of the 2ˆ (–delta delta CT) method for quantitative real-time polymerase chain reaction data analysis. Biostatistics, Bioinformatics and Biomathematics. 2013;3:71. [PMC free article] [PubMed] [Google Scholar]
- 60.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tomczynska I, Stumpe M, Mauch F. A conserved RxLR effector interacts with host RABA-type GTPases to inhibit vesicle-mediated secretion of antimicrobial proteins. The Plant Journal. 2018;95:187–203. doi: 10.1111/tpj.13928. [DOI] [PubMed] [Google Scholar]
- 62.Joller C, et al. S-methyl Methanethiosulfonate: Promising Late Blight Inhibitor or Broad Range Toxin? Pathogens. 2020;9 doi: 10.3390/pathogens9060496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets that support the findings of the current study are available in the Zenodo repository with the identifier doi:10.5281/zenodo.4131137 at https://doi.org/10.5281/zenodo.4131137. Additional data related to this study are available from M. El-Shetehy and F. Schwab upon reasonable request.